Abstract
The severity of the COVID-19 pandemic has necessitated the search for drugs against SARS-CoV-2. In this study, we explored via in silico approaches myxobacterial secondary metabolites against various receptor-binding regions of SARS-CoV-2 spike which are responsible in recognition and attachment to host cell receptors mechanisms, namely ACE2, GRP78, and NRP1. In general, cyclic depsipeptide chondramides conferred high affinities toward the spike RBD, showing strong binding to the known viral hot spots Arg403, Gln493 and Gln498 and better selectivity compared to most host cell receptors studied. Among them, chondramide C3 (1) exhibited a binding energy which remained relatively constant when docked against most of the spike variants. Chondramide C (2) on the other hand exhibited strong affinity against spike variants identified in the United Kingdom (N501Y), South Africa (N501Y, E484K, K417N) and Brazil (N501Y, E484K, K417T). Chondramide C6 (9) showed highest BE towards GRP78 RBD. Molecular dynamics simulations were also performed for chondramides 1 and 2 against SARS-CoV-2 spike RBD of the Wuhan wild-type and the South African variant, respectively, where resulting complexes demonstrated dynamic stability within a 120-ns simulation time. Protein-protein binding experiments using HADDOCK illustrated weaker binding affinity for complexed chondramide ligands in the RBD against the studied host cell receptors. The chondramide derivatives in general possessed favorable pharmacokinetic properties, highlighting their potential as prototypic anti-COVID-19 drugs limiting viral attachment and possibly minimizing viral infection.
Communicated by Ramaswamy H. Sarma
Keywords: Antiviral agents, COVID-19, molecular docking, protein-protein interactions, SARS-CoV-2 spike proteins and variants
1. Introduction
The spike (S) protein crowning SARS-CoV-2, the causative agent COVID-19, is responsible for the initial step of host cell infection (Ge et al., 2020). It is a transmembrane glycoprotein composed of two subunits: the S1 subunit essential for binding to the host cell receptor and the S2 subunit responsible for the viral fusion to the host cell membrane (Laurini et al., 2020). The receptor-binding regions of the spike protein are capable of recognizing and attaching to multiple host receptors namely ACE2, GRP78, and NRP1 (Cantuti-Castelvetri et al., 2020; Carlos et al., 2021; Shang et al., 2020a). The elucidation of its structure and function makes it an attractive target for drug discovery and vaccine development.
The angiotensin-converting enzyme 2 (ACE2) expressed at the outer surface of host cells of lung epithelia, was identified as a critical receptor for mediating SARS-CoV-2 entry (Brielle et al., 2020). The S1 subunit of the spike protein contains the receptor-binding domain (RBD) that is able to recognize ACE2. Within the RBD lies a core and a receptor binding motif with the latter mediating interactions with ACE2 (Shang et al., 2020a). It has been established that host susceptibility to SARS-CoV-2 is determined by the binding affinity of the spike protein RBD to ACE2 (Walls et al., 2020). As such, it is critical to consider biologically significant mutations occurring in the RBD that increase pathogenicity. Mutations in the RBD may lead to immunologic resistance and escape in vaccine-elicited antibodies. These impact vaccine efficacy and have implications in drug design. Hence, it is critical to screen compounds against SARS-CoV-2 spike variants with single and multiple mutations in the RBD such as those observed in the recently discovered United Kingdom, South African and Brazilian variants. Another route of SARS-CoV-2 entry is through the master chaperone protein of the unfolded protein response GRP78, which is overexpressed during cellular stress where it escapes endoplasmic reticulum retention and subsequently translocates to the cell membrane and becomes susceptible to viral recognition via the RBD of the spike protein (Ibrahim et al., 2020). In the S1-S2 junction of the spike protein also lies an important cleavage site that is proteolytically activated by furin, a known host cell protease (Shang et al., 2020b). Proteolytic cleavage by furin exposes a conserved C-terminal motif in the spike protein which can bind to neuropilin-1 (NRP1), a transmembrane protein important in angiogenesis and axon guidance (Daly et al., 2020). NRP1 has been identified as a host factor relevant for SARS-CoV-2 infection.
To accelerate anti-COVID-19 drug discovery, previously reported natural products that confer profound antiviral activity have been repurposed and screened computationally (Shanmugam et al., 2020). In this paper, we disclose the potential of known anti-Ebola virus/actin inhibitory chondramides which resulted from high throughput screening of myxobacterial metabolites in targeting the RBD of the SARS-CoV-2 S protein precluding their interaction with ACE2, GRP78, and NRP1 in silico through molecular docking screening and molecular dynamics simulations. Myxobacteria are known producers of structurally and functionally diverse metabolites with broad-spectrum antiviral activity (Mulwa & Stadler, 2018). Exploiting the diversity and evolutionary significance of metabolites produced by myxobacteria is a promising approach to discover lead structures for antiviral drug development as previously demonstrated by a range of myxobacterial secondary metabolites for example host replication factor methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) antagonist carolacton inhibiting SARS-CoV-2 Vero E6 cells at IC50 = 0.14 μM as well as sorangicin A and myxopyronin B exhibiting as computationally verified inhibitors of SARS-CoV-2 RNA-dependent RNA polymerase (Anderson et al., 2020; Elkarhat et al., 2020; Mohr, 2018). Selectivity of top metabolites towards the S protein was also determined to validate antagonistic effects on spike’s interaction with various host cell receptors. The top-binding ligands were additionally screened against biologically significant SARS-CoV-2 mutations occurring in the RBD of the S protein such as N501Y, E484K, K417N/T, A475V, I472V, L452R, V483A, F490L, S477N and N439K along with the United Kingdom, South African and Brazilian SARS-CoV-2 variants. Finally, pharmacokinetic and toxicity profiles were determined to investigate the druggability of the in silico active compounds.
2. Materials and methods
2.1. Ligand selection and preparation
A library of 72 secondary metabolites from Myxobacteria (Figures S1–S3, previously reported to exhibit activity against HIV, Ebola, Hepatitis C, CMV, and actins (Mulwa & Stadler, 2018; Herrmann et al., 2013) were screened against the receptor-binding regions of SARS-CoV-2 spike protein. The structures were converted from SMILES format to SYBYL mol2 file and optimized using Avogadro (version 1.2.0), an open-source molecular builder (Hanwell et al., 2012). The mol2 files were added to UCSF Chimera platform for docking purposes (Pettersen et al., 2004).
2.2. Target protein preparation
The RBD of the spike protein crucial for facilitating cellular entry of SARS-CoV-2 were selected as molecular target PDB ID: 6M0J and was obtained from the Protein Data Bank (https://www.rcsb.org/) and its crystal structure was added to the UCSF Chimera platform (Pettersen et al., 2004). The protein model was prepared by removing all non-standard residues and protein chains and subsequently minimized by the steepest descent method (100 steps – step size 0.02 Å) and conjugate gradient method (10 steps – step size 0.02 Å). Using the same PDB ID, the SARS-CoV-2 variants (N501Y, E484K, K417N/T, A475V, L452R, V483A, F490L, S477N, N439K) were constructed using UCSF chimera platform by editing necessary amino acids of the S protein via the swapaa command which utilizes information from a rotamer library (Shapovalov & Dunbrack, 2011). The variants were optimized following the described minimization method. The GRP78- and NRP1-binding regions were prepared using PDB ID 6VXX where missing residues and loop sequences of the protein structure (particularly residues 454–462; 486–489) were built using Modeller 9.21 package (Yang et al., 2015). The resulting structure was then minimized by steepest descent method. The ligands were similarly subjected to molecular docking against the following host cell receptors to determine selectivity: ACE2 (PDB ID 6M0J, chain A) and GRP78 (PDB ID 5E84, chain A).
2.3. Molecular docking
The three-dimensional structures of the proteins derived from RCSB PDB were subsequently used for molecular docking experiments through the UCSF Chimera platform (Pettersen et al., 2004). Docking grid coordinates are shown in Table S5. Minimization and preparation of ligand and protein structures were done by adding missing hydrogen atoms and appropriate charges to the structures employing the Gasteiger charge method computed using Amber’s Antechamber module (Wang et al., 2006). The docking procedure was done using ‘flexible ligand into flexible active site’ protocol, allowing the ligand’s translational and rotational walk within the grid box (Yang et al., 2015). Virtual screening of the prepared library was performed following the Broyden-Fletcher-Goldfarb-Shanoo (BFGS) algorithm of AutoDock Vina (version 1.5.6) with number of binding modes = 10 at maximum degree of exhaustiveness (Trott & Olson, 2010). The binding affinity of the enzyme–ligand complex conformation was determined using UCSF Chimera and was visualized and analyzed through BIOVIA Discovery Studios (version 4.1) and ChimeraX platform. Protein-protein docking was performed using HADDOCK version 4.2.
In order to improve the reliability of the docking output, we performed a molecular docking calculation using Glide program (Schrödinger LLC, NY, 2018). The energy grid for each variants and wild type protein was prepared as follows: default value of the protein atom-scaling factor (1.0 Å), with a cubic box centered on the center of the binding site considering the centroid generated by considering the following amino acids (Arg403, Tyr449, Tyr453, Leu455, Gln498, and Tyr505). After the generation of the grids, the compound was docked into the binding site. The docked poses considered for the post-docking minimization step were 1000 using Extra Precision (XP) as scoring function (the output of this calculation was provided as a supplementary file, Figure S4). The RMSD between the output obtained from AutoDock Vina and Glide software for each variant and for the wild type protein was calculated using the RMSD tool calculation available from Maestro (Schrödinger LLC, NY, 2018). Proteins and the ligand were prepared using protein preparation wizard and LigPrep, respectively, implemented in Maestro as previously reported (Brogi et al., 2017; Di Capua et al., 2016).
2.4. Molecular dynamics simulation
To understand the dynamic behavior of chondramide C3 (1)-wild-type, chondramide C (2)-wildtype, and chondramide C (2)-SA SARS-CoV-2 spike complex, molecular dynamics simulation were employed. Molecular dynamics simulation of 120 ns was performed using FF14SB for the wild-type spike protein and Generalized Amber Force Fields (GAFF) for the chondramides in AMBER16. The molecules were solvated using TIP3PBOX water model with the edge of the box at 10 Å away from the solute molecules. The solvent box size was 18.774*18.774*18.774. Throughout the simulation, each complex system is maintained at the temperature of 300 K with constant pressure. Energy minimization was done for 50,000 steps. The simulation was carried out through Particle Mesh Ewald Molecular Dynamics (PMEMD) of Amber16 (Wang et al., 2001). The heating time for the equilibration was 100 picoseconds. The thermostat used was Berendsen thermostat for temperature control (ntt = 1). The cut-off used was 8 Å and the time step was 2 femtoseconds. The trajectories were collected for every nanosecond to get insights into the interactions at the atomic level. Hydrogen bonding analysis of the protein-ligand complex was additionally performed to reveal the half-life of the interactions during the simulation. Binding free energy between the protein and the ligand is calculated by Molecular Mechanics Poisson–Boltzmann Surface Area (MM/PBSA) module of Amber Tools16 by using Eq. (1) (Sun et al., 2014):
| (1) |
2.5. Drug-likeness, ADME, and toxicity prediction
SwissADME software was used to predict absorption, distribution, metabolism and excretion of the hit compounds (Daina et al., 2017). Following Lipinski's rule-of-five, the pharmacokinetic profiles of the compounds that may influence absorption and permeation across cell membranes were evaluated. For compounds to show drug-likeness, it has to satisfy at least three of the four criteria: molecular weight < 500 Daltons (Da), calculated lipophilicity (Log P) < 5, number of hydrogen-bond acceptors < 10, and number of hydrogen-bond donors < 5. In silico toxicity was predicted using OSIRIS property explorer program (Sander et al., 2009) which considers potential mutagenicity, tumorigenicity, irritant effects, and reproductive toxicity of hit compounds (Quimque, Notarte, Letada, et al., 2021; Magpantay et al., 2021). The solubility (Log S) of the hits was also determined using OSIRIS wherein a Log S value equivalent or greater than −4 is indicative of a favorable solubility (De Leon et al., 2021; Quimque, Notarte, Fernandez, et al., 2021).
3. Results and discussion
3.1. Molecular docking to angiotensin-converting enzyme (ACE2)-receptor binding domain (RBD) of SARS-CoV-2 spike and its variants
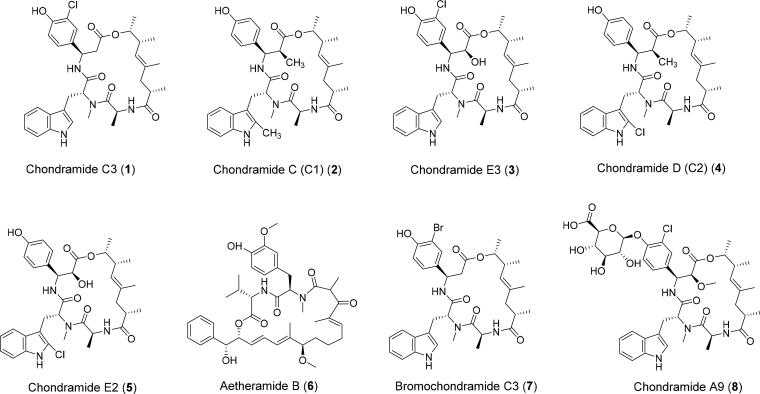
The main mechanism of entry of the virus to host cells is through binding of the S protein to ACE2. Key residue substitutions in the C-terminal domain of the S protein strengthen interaction with ACE2 leading to higher binding affinity (Wang et al., 2020). The ACE2-binding ridge in SARS-CoV-2 RBD has a more compact conformation (Shang et al., 2020a, 2020b) and a larger binding interface with more contacts against ACE2 receptor in the binding motif of its RBD compared to SARS-CoV. The stability of the protein-protein interaction between the spike RBD of SARS-CoV-2 and the ACE2 receptor involves the specific spike residues Arg403, Gln493, Gln498, Pro499 and Thr500 and were identified as viral hot spots (Laurini et al., 2020; Othman et al., 2020). Therefore, it is crucial to highlight intermolecular interactions between the ligands and the viral hot spots that can induce destabilization of spike binding to ACE2, precluding native interactions between the viral spike protein and the host cell receptor ACE2. Among the 72 potential myxobacterial compounds (Table S1–S3) able to interfere with the mentioned recognition mechanism, we identified through in silico approaches eight top-scoring chondramide derivatives (Figure 1). These cyclic depsipeptides were previously isolated and identified from Chondromyces sp. MSr9030 as a result of antiviral and actin-inhibiting compound studies (Herrmann et al., 2013). Notably, chondramide C3 (1) showed highest affinity to the RBD of the viral spike with a BE of −8.7 kcal/mol (Table 1). The phenol moiety was noted to establish pi-donor H bond with Tyr495 and conventional hydrogen bonds with Tyr505 and Gly496 (Figure 2). Additionally, the indole group strongly interacted with the following amino acid residues of spike RBD: Arg403 (pi-cation), Glu406 (H-bond), Lys417 (pi-alkyl), and Tyr453 (pi-pi stacking). Finally, two additional H-bonds were also observed with Ser494 and Tyr449, and the macrocyclic chondramide core.
Figure 1.
Top scoring chondramides 1–8 against spike receptor-binding domain.
Table 1.
Summary of the binding energies (BE, kcal/mol) and interacting residues of 1–8 against SARS-CoV-2 spike wild-type and variants.
| Cpd | Wild-type |
N501Y |
E484K |
|||
|---|---|---|---|---|---|---|
| BE | Intermolecular interactions | BE | Intermolecular interactions | BE | Intermolecular interactions | |
| 1 | −8.7 | Glu406, Tyr449, Tyr453, Ser494, Gly496, Tyr505 (H bond); Arg403 (π-cation), Lys417, Tyr505 (π-alkyl); Tyr453 (π-π stacked); Tyr495 (π-donor H bond); Gly496 (C-H bond) | −6.6 | Arg403, Ser494, Gly496 (H bond); Leu455 (π-alkyl and π-sigma); Tyr505 (π-alkyl) | −7.2 | Gly496 (H bond); Tyr505 (π-π stacked and π-π T-shaped); Arg403 (C-H bond); Tyr495 (Alkyl) |
| 2 | −8.6 | Tyr453, Tyr505 (H bond); Tyr449 (π-alkyl); Tyr505 (π-π stacked, π-alkyl, π-π T-shaped) | −9.1 | Gly496 (H bond); Tyr449 (π-alkyl); Tyr501 (π-π stacked); Tyr505 (π-π T-shaped) | −8.7 | Tyr453, Gly496, Asn501, Tyr505 (H bond); Tyr449 (π-alkyl), Tyr505 (π-alkyl, π-π stacked, π-π T-shaped) |
| 3 | −8.4 | Glu406, Tyr453, Tyr449, Ser494, Asn501 (H bond); Arg403 (π-cation); Tyr453 (π-π stacked); Gly496 (π-donor H bond) | −7.1 | Glu406, Ser494, Gly496 (H bond); Tyr453 (π-π stacked), Leu455 (π-sigma), Tyr505 (π-alkyl) | −7.5 | Gly496, Gln498 (H bond); Arg403 (π-cation); Tyr505 (π-π stacked, π-donor H bond); Gly496 (C-H bond) |
| 4 | −8.4 | Tyr453, Gly496, Asn501 (H bond); Tyr449 (π-alkyl); Tyr505 (π-π stacked, π-π T-shaped) | −8.6 | Tyr449 (π-alkyl); Tyr501 (π-π stacked); Tyr505 (π-π T-shaped) | −8.4 | Tyr453, Gly496 (H bond); Tyr449 (π-alkyl); Tyr505 (π-π stacked, π-π T-shaped) |
| 5 | −8.2 | Tyr449, Gln498, Asn501 (H bond); Tyr449 (π-alkyl); Tyr505 (π-π stacked); Gly496 (C-H bond) | −8.6 | Tyr453, Tyr505 (H bond); Tyr449 (π-alkyl); Tyr501 (π-π stacked); Tyr505 (π-π T-shaped) | −8.1 | Tyr449, Tyr453, Gln498, Asn501 (H bond); Tyr449 (π-alkyl); Tyr505 (π-π stacked); Gly496 (C-H bond) |
| 6 | −8.1 | Arg403, Asn501, Tyr505 (H bond); Tyr489 (π-π T-shaped); Gly496 (π-donor H bond); Tyr505 (π-π T-shaped); Lys417 (Alkyl), Asn501 (C-H bond) | −7.2 | Tyr449, Tyr453, Gln498 (H bond); Tyr449 (π-alkyl); Tyr495 (π-sigma); Tyr501 (π-alkyl); Tyr505 (π-alkyl); Gly446, Ser494, Tyr501 (C-H bond) | −8.1 | Tyr453, Asn501 (H bond); Tyr489 (π-π T-shaped); Gly496 (π-donor H bond); Lys417, Leu455 (Alkyl); Gly496, Asn501 (C-H bond) |
| 7 | −8.1 | Tyr453, Gly496, Asn501, Tyr505 (H bond); Tyr505 (π-π stacked, π-π T-shaped) | −7.5 | Gly496 (H bond); Tyr449, Tyr453, Tyr495 (π-alkyl), Tyr501 (π-π stacked), Tyr505 (π-π T-shaped) | −8.1 | Tyr453, Asn501, Tyr505 (H bond); Tyr505 (π-π stacked, π-π T-shaped) |
| 8 | −8 | Tyr449, Ser494 (H bond); Tyr449 (π-π stacked); Tyr505 (π-alkyl); Tyr495, Gly496 (C-H bond) | −8.6 | Arg403, Gln409, Lys417, Gly496 (H bond); Arg403 (π-cation); Tyr453 (π-π stacked); Leu455 (π-sigma); Tyr501 (C-H bond) | −7.9 | Ser349, Asn450 (H bond); Tyr449, Leu452 (π-alkyl); Asn450 (C-H bond) |
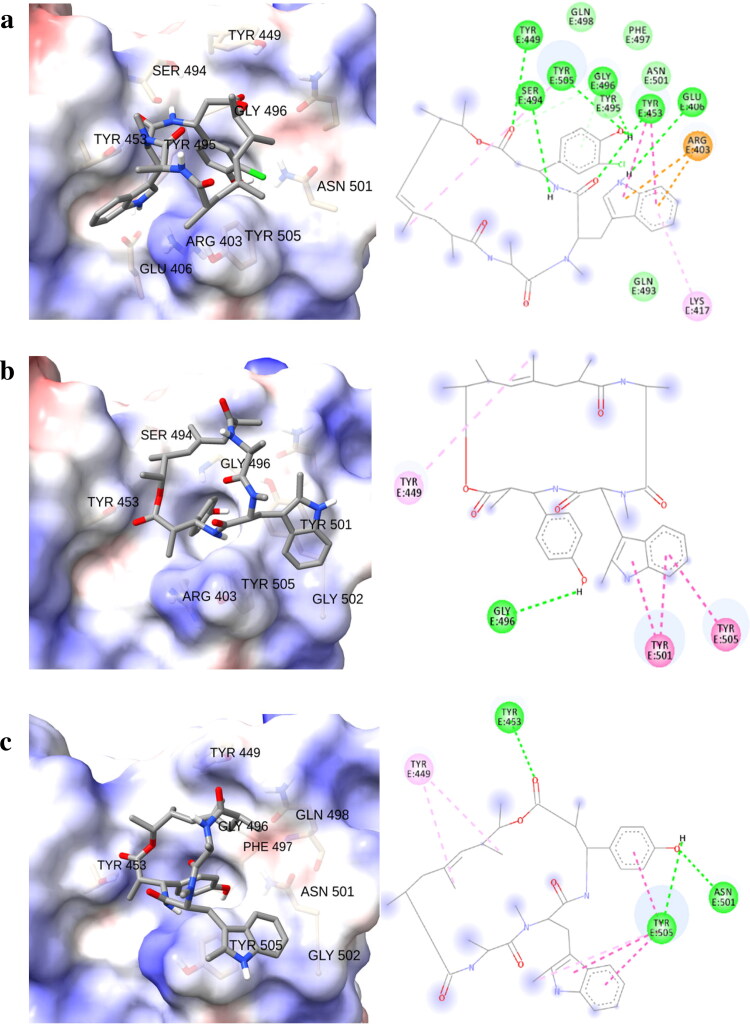
Figure 2.
Three dimensional (left) and schematic representations (right) of (a) chondramide C3 (1) against wild-type, (b) chondramide C (2) against N501Y, and (c) chondramide C (2) against E484K SARS-CoV-2 spike variants.
The various chondramides were also subjected to molecular docking analysis against the different variants of spike RBD. Li and co-workers identified single amino acid substitutions in the viral spike RBD (A475V, I472V, L452R, V483A, and F490L) which were found to affect binding of neutralizing antibodies (Li et al., 2020) and were identified to occur in higher frequencies during the time evolution of SARS-CoV-2 spike protein RBD indicating stronger transmission capacity (Chen et al., 2020). In particular, V483A is prevalent among patients with COVID-19 in the United States which has the highest mutation frequency within the receptor-binding motif of the spike indicating that the mutation makes SARS-CoV-2 more infectious (Wang et al., 2021).
Accordingly, the UK variant has been reported to be more infectious and is strongly associated with increasing rates of COVID-19 cases (Wise, 2020). Remarkably, among the mutations in the UK variant, the mutation N501Y within the RBD showed increased binding affinity to human and murine ACE2. On the other hand, the mutation E484K found in the RBD is responsible for a 10-fold decrease in neutralization by convalescent serum antibodies (Greaney et al., 2020).
This kind of mutation was also noted in both South African (B. 1.351) and Brazilian (B.1.1.28) variants (Fontanet et al., 2021). Similarly, the South African variant is characterized by two additional mutations in the RBD namely N501Y and K417N (Wibmer et al., 2021). Sharing similar E484K mutation with the South African variant, the Brazilian variant may be also associated with antibody escape. Furthermore, E484K has been linked with some cases of reinfection in Brazil (Nonaka et al., 2021).
Due to the clinical relevance of mutations, it is essential to identify compounds with high affinity to spike protein variants. To this end, we performed an extensive in silico analysis for investigating the behavior of chondramide derivatives against the current SARS-CoV-2 variants. Across all variants, the binding affinity of Chondramide C3 (1) remained constant (Tables S1–S3) except towards N501Y (−6.6 kcal/mol) and E484K (−7.2 kcal/mol) (Table 1). Based on post-docking analysis of 1 against the N501Y variant, we observed that the new residue (Tyr501) is responsible for an observable decrease of the binding affinity since it did not exhibit significant interactions with the ligand. The same was observed for depsipeptide 1 considering the E484K variant. The substituted residue Lys484 did not establish any contact with the ligand, thus pointing to a decrease in binding affinity. The point mutations found in the aforementioned variants are non-conservative which are involved in a change in the binding properties of the spike protein RBD (Figure 2). The swap to a tyrosine residue as in the case of the N501Y variant promoted hydrogen bonding with phenolic moieties and pi-pi stacked interactions with the indole feature of chondramides due to the presence of phenol moiety in tyrosine but absent in asparagine. On the other hand, the mutation to a lysine residue as observed in the E484K variant provided structural accommodation for hydrogen bonding with free hydroxyl moieties. Therefore, ligands that fail to form intermolecular interactions with these polar residues of the variants may affect binding affinity. One of the major structural features of chondramide C3 (1) is an ortho-chloro substituent on the phenol moiety. The electronegative chlorine atom may form intramolecular H-bonding with the hydroxyl group of the phenol ring deminishing its ability to form intermolecular interactions with the polar residues present in the RBD of the spike protein variants. To further explore this observed relationship between the polarity of ligands and the spike variants, we subjected the selected metabolites to molecular docking against the different SARS-CoV-2 variants. Among the derivatives, chondramide C (2) showed strong affinities against N501Y (−9.1 kcal/mol) and E484K (−8.7 kcal/mol) SARS-CoV-2 variants. Structurally, one of the major differences between depsipeptides 1 and 2 is the absence of an ortho-chloro substituent in the phenol moiety of 2. Post-docking analysis of 2 against N501Y variant revealed the presence of a conventional hydrogen bonding between the phenol residue and Gly496 and, a pi-pi stacking between the indole moiety and Tyr501 and Tyr505. Regarding the E484K variant, the outcome of the docking calculation of 2 showed that this derivative is able to form a strong H-bonds network involving the phenol moiety (against Asn501 and Tyr505) and the ester group (against 453). Other metabolites that exhibited an increase in binding affinity towards N501Y variant, compared to the wild-type are chondramides D (4), E2 (5), and A9 (8). Similar to 2, depsipeptide 4 does not have an ortho-chlorine in its phenolic functionality. Depsipeptides 5 and 8, on the other hand, are adorned with a hydroxyl/methoxy functionality alpha to the ester and a glycosylated phenol, respectively. These data support our observation that having a free (no ortho substituent) phenol and/or further oxygenation in the chondramide core may increase binding to certain RBD variants, particularly those with non-conservative mutations like N501Y and E484K where their binding receptor environment is particularly polar.
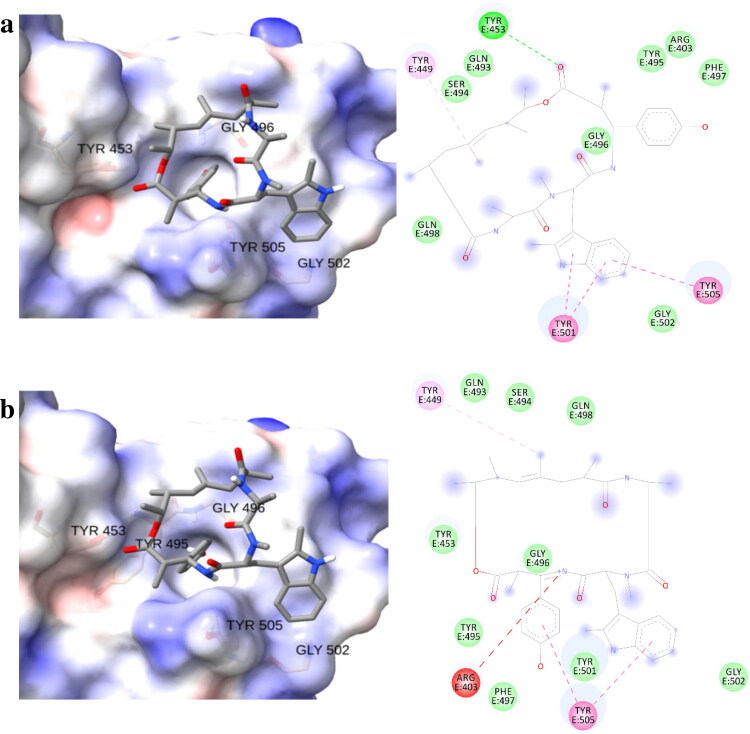
After determining the binding affinities of the chondramides towards point-mutated SARS-CoV-2 spike variants, the selected compounds were subjected to molecular docking against recently known strains with more than one amino acid substitutions in the spike RBD sequence namely, the South African variant (Wibmer et al., 2021) (N501Y-E484K-K417N) and the Brazilian variant (Nonaka et al., 2021) (N501Y-E484K-K417T). Based on computational analysis performed on chondramide C (2) against the single-substituted spike variants (N501Y and E484K), it is expected that this compound would exhibit similar strong affinities against the South African and Brazilian variants (Table 2). The mutation from lysine to either asparagine or threonine, respectively, kept the polar nature of the binding site on RBD, which probably aided in the accommodation of polar ligands (Figure 3).
Table 2.
Summary of the binding energies (BE, kcal/mol) and interacting residues of the top compounds against South African and Brazilian variants.
| Cpd | BE | South African (N501Y-E484K-K417N) | BE | Brazilian (N501Y-E484K-K417T) |
|---|---|---|---|---|
| intermolecular interactions | intermolecular interactions | |||
| 1 | −6.5 | Leu492, Ser494 (H bond); Leu452, Leu484 (π-alkyl); Phe490 (π-π stacked, π-π T-shaped) | −7.9 | Glu406, Tyr453, Ser494, Tyr501 (H bond); Arg403 (π-cation); Tyr453 (π-π stacked); Gly496 (π-donor hydrogen bond); Tyr501, Tyr505 (π-alkyl) |
| 2 | −9.3 | Tyr453 (H bond); Tyr501 (π-π stacked); Tyr505 (π-π T-shaped) | −8.4 | Tyr449 (π-alkyl); Tyr505 (π-π T-shaped) |
| 3 | −7.1 | Ser494 (H bond); Gly496 (π-donor hydrogen bond); Tyr501 (π-π stacked); Tyr505 (π-alkyl, π-π T-shaped) | −7.1 | Glu406, Ser494, Tyr501 (H bond); Tyr453 (π-π stacked); Leu455 (π-sigma); Tyr505 (π-alkyl) |
| 4 | −8.8 | Tyr453, Gly496 (H bond); Tyr449 (π-alkyl); Tyr501 (π-π stacked, π-π T-shaped) | −8.0 | Tyr453 (H bond); Tyr449 (π-alkyl); Tyr501 (π-π T-shaped); Tyr505 (π-π stacked, π-π T-shaped) |
| 5 | −8.5 | Tyr453, Tyr505 (H bond); Tyr449 (π-alkyl); Tyr501 (π-π stacked, π-π T-shaped) | −7.9 | Tyr453, Gln496, Asn505 (H bond); Tyr449 (π-alkyl); Tyr505 (π-π T-shaped) |
| 6 | −7.9 | Gln493, Gly496, Tyr501 (H bond); Leu455, Lys484, Tyr489, Tyr501 (Alkyl); Phe490 (π-π stacked) | −7.8 | Gln493, Gly496 (H bond); Leu455, Phe456, Lys484, Tyr489, Tyr495 (Alkyl); Phe490 (π-π stacked) |
| 7 | −7.6 | Tyr453, Tyr449, Tyr495 (π-alkyl), Tyr501 (π-π T-shaped), Tyr505 (π-π stacked) | −6.7 | Tyr449, Gly496, Tyr501 (H bond) |
| 8 | −7.9 | Asn450 (H bond); Tyr449, Leu452 (π-alkyl); Asn450 (C-H bond) | −8.6 | Arg403, Glu406, Thr417, Gly496 (H bond); Arg403 (π-cation); Tyr453 (π-π stacked); Leu455 (π-sigma) |
Figure 3.
Three dimensional (left) and schematic representations (right) of chondramide C (2) against (a) South African (N501Y-E484K-K417N) and (b) Brazilian (N501Y-E484K-K417T) SARS-CoV-2 variants.
3.2. Molecular docking to glucose-regulated protein 78 (GRP78) – receptor binding domain (RBD)
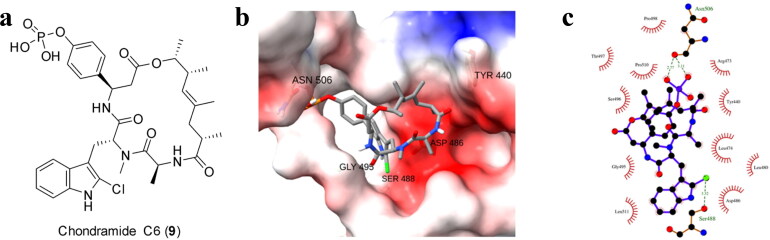
SARS-CoV-2 spike binding to GRP78 is an alternative way of viral entry to host cells. Elevated during cellular stress, GRP78 translocates from the endoplasmic reticulum to the cell membrane wherein it becomes susceptible to viral recognition (Ibrahim et al., 2020). Accordingly, it has been investigated as a potential therapeutic target (Allam et al., 2020). The binding site of SARS-CoV-2 spike protein identified for GRP78 is located in between regions III and IV wherein the latter is the main driver for the binding as predicted by in silico studies (Ibrahim et al., 2020). The 72 compounds were also investigated for assessing if they are able to disrupt the interaction between the viral spike protein and GRP78 (Table S1). Chondramide C6 (9) displayed highest affinity against the GRP78-binding region of spike with the binding energy of −8.8 kcal/mol. The presence of a phosphorylated phenol on 9 helped to stabilize its binding, to the mentioned binding region, through a hydrogen bond with Asn506 (Figure 4). Another H-bond was also observed between the adjacent amide group and Gly495. Moreover, the indole moiety is involved in pi-anion and pi-sigma interactions with Asp486 and Leu474, respectively. Both aromatic groups are able to target Pro510 via pi-alkyl interplay.
Figure 4.
(a) Structure of chondramide C6 (9), (b) Three dimensional and (c) 2 D animated representation of chondramide C6 (9) against SARS-CoV-2 S protein receptor-binding domain to GRP78.
3.3. Molecular docking to neuropilin-1 (NRP1) receptor-binding site in SARS-CoV-2 spike protein
NRP1 is a transmembrane protein involved in angiogenesis and axon guidance which has been identified as a host factor involved in SARS-CoV-2 infection (Daly et al., 2020). SARS-CoV-2 entry is facilitated by proteolytic cleavage through furin subsequently exposing a conserved C-terminal motif of the spike protein. The outcome after cleavage is a sequence conforming to a C-end peptide rule that is recognized by NRP1. Docking experiments showed low binding affinities of myxobacterial metabolites against the NRP1 binding site in the spike protein (Table S1) therefore the chondramide-NRP1 complexes were no longer subjected to further in silico experiments.
3.4. Protein–protein and selectivity docking
In addition to investigating the binding of chondramides against the different receptor-binding domains (RBD) of spike, we determined the effect of the bound ligand to the binding behavior of spike against the host receptor proteins. Protein-protein docking was performed considering spike protein, with and without the top-binding ligand, versus ACE2 and GRP78.
The spike RBD and ACE2 protein complex demonstrated a strong binding affinity (HADDOCK score) of −130.9 ± 0.2 kcal/mol. On the other hand, the interaction of chondramide C3 (1) with spike RBD resulted in a weaker affinity towards ACE2 as displayed by a decrease in HADDOCK score at −82.1 ± 6.3 kcal/mol. A similar observation was also noted between spike-GRP78 complexes where the presence of chondramides C6 (9) and C3 (1), respectively, hindered the formation of stable protein complexes. The bound chondramide 9 at the interspace between spike RBD and GRP78 resulted in a decline in a HADDOCK score of about 37.3 kcal/mol. Through protein-protein docking, we were able to further demonstrate the potential antagonistic effects of chondramides 1 and 9 against the spike protein RBD with a significant reduction of binding affinity of spike protein RBD towards ACE2 and GRP78, respectively.
Molecular docking calculations also demonstrated better binding affinity of depsipeptide 1 towards spike RBD (−8.7 kcal/mol) than with its binding to host cell ACE2 (−6.1 kcal/mol). Similarly, to represent the variants, chondramide C (2) showed selectivity towards spike protein variants (N501Y, −9.1 kcal/mol and E484K, −8.7 kcal/mol) than ACE2. Finally, chondramide C6 (9) showed selectivity towards the spike RBD (−8.8 kcal/mol) with respect to the GRP78 (−6.6 kcal/mol).
3.5. Molecular dynamics simulation
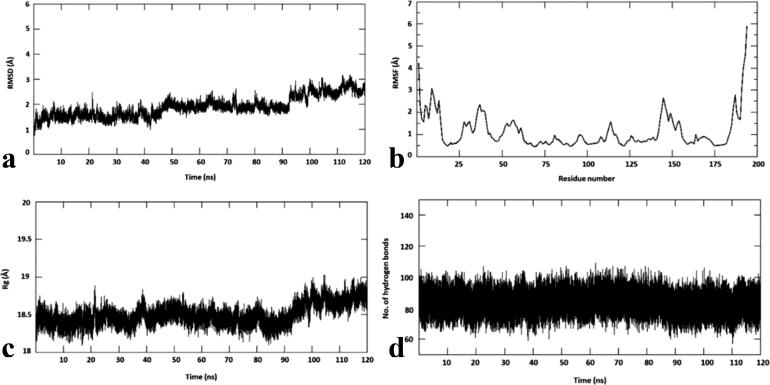
To study the conformational stability of spike RBD-chondramide C3 (1) complex, the root mean square deviation (RMSD) of the Cα atom with respect to its initial position in molecular dynamics (MD) simulations was measured and plotted against various time intervals (Figure 5a). The RMSD was measured within 1 to 3 Å for chondramide C3 (1) complex. It is shown to attain stability from 50 ns to 90 ns at 2 Å. However, there seems to be an increase from 2 Å to 3 Å from 90 ns and this is maintained until the end of the simulation. To predict the conformational flexibility of the protein-ligand complexes, the root mean square fluctuations (RMSF) of Cα atoms of each residue with respect to its initial position was measured and were plotted against the residues of the protein (Figure 5b). The RMSF plot of chondramide C3 (1) complex revealed similar fluctuation profiles with an average RMSF value of 1.8 Å. The lesser RMSF value indicates that the chondramide C3 (1) complex was stable and did not show any flexibility throughout the simulations.
Figure 5.
Molecular dynamics simulations of chondramide C3-wild-type SARS-CoV-2 spike complex. (a) root mean square deviation (RMSD), (b) root mean square fluctuation (RMSF), (c) radius of gyration (Rg) and (d) number of hydrogen bonds.
The radius of gyration (Rg), an indicator of protein structure compactness, was also measured (Figure 5c). The Rg in chondramide C3 (1) complex ranges from 18.5 to 19 Å. This shows that the complex is compact and tightly bound. Furthermore, hydrogen bonds are important non-covalent structural forces, primarily electrostatic in nature, in molecular systems. They are formed when a single hydrogen atom is effectively shared between the two heavy atoms covalently bonded to each other (the hydrogen bond donor and the hydrogen bond acceptor). Both the donor and acceptor atoms are typically quite electronegative. The average number of hydrogen bonds are tracked at each frame of the simulations and is plotted against different time intervals. The image in Figure 5d shows that the number of hydrogen bonds for the chondramide C3 (1) complex are stable throughout the simulation. The absolute value of binding free energy of the selected complex is −23.12 kcal/mol. For this complex, the binding free energy at 40 and 120 ns seem to be similar with a fluctuation at 80 ns. Notably, MM/PBSA calculation was carried out every 20 ns. Table 3 shows that the binding energies for all the intervals fall under the window of −20 kcal/mol and −30 kcal/mol, further highlighting the stability of the complex and the high affinity of the compound for spike RBD.
Table 3.
Binding free energy of chondramide C-wildtype is and chondramide C-SA variant at different intervals.
| 40 ns (kcal/mol) | 80 ns (kcal/mol) | 120 ns (kcal/mol) | |
|---|---|---|---|
| Chondramide C-wildtype | −34.24 | −30.56 | −42.76 |
| Chondramide C-SA | −32.01 | −32.97 | −40.17 |
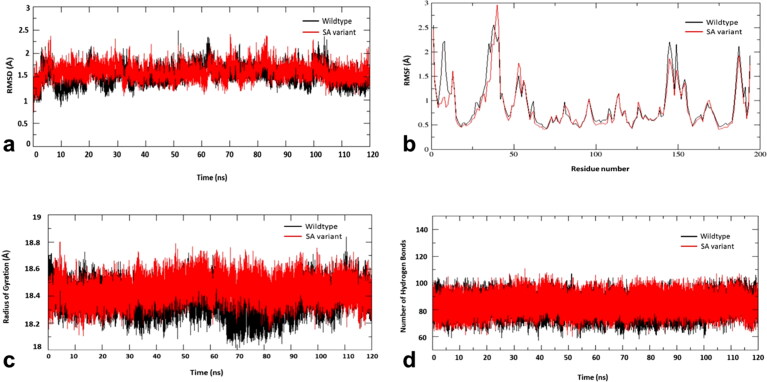
To study the conformational stability of spike RBD of the wild type and the South African (SA) variant with the compound, chondramide C (1) complexes, the root mean square deviation (RMSD) of the Cα atom with respect to their initial position in molecular dynamics (MD) simulations was measured and plotted against various time intervals (Figure 6a). The RMSD was measured within 1 to 2.5 Å for both chondramide C-wildtype and chondramide C (2)-SA variant complexes. Both the complexes are shown to have slight fluctuations at 60 ns and 100 ns and then remain stable at 1.5 Å until the end of the simulations.
Figure 6.
Molecular dynamics simulations of chondramide C (2)-wild-type SARS-CoV-2 spike complex (black) and chondramide C (2)-South African variant complex (red). (a) root mean square deviation (RMSD), (b) root mean square fluctuation (RMSF), (c) radius of gyration (Rg) and (d) number of hydrogen bonds.
To predict the conformational flexibility of the protein-ligand complexes, the root mean square fluctuations (RMSF) of Cα atoms of each residue with respect to its initial position was measured and were plotted against the residues of the protein (Figure 6b). The RMSF plot of chondramide C (2)-wildtype complex revealed similar fluctuation profiles as compared to chondramide C-SA variant complex with an RMSF value ranging between 0.5 Å to 3 Å. Both the complexes show same fluctuation profile overall. However, the initial fluctuations and the residues between 145 and 150 in the wildtype seem to be higher than the mutant variant. In the mutant, the residues 40–45 have shown to reach the highest fluctuation amongst all at 3 Å. This could be due to the mutations in SA variant.
The radius of gyration (Rg), an indicator of protein structure compactness, was also measured (Figure 6c). The Rg in chondramide C-wildtype and chondramide C (2)-SA variant complexes ranges average from 18.2 to 18.8 Å. However, Rg of chondramide C-wildtype is shown to have higher fluctuations between 70 and 90 ns reaching 18 Å and attain stability at 18.4 Å from 100 ns compared the chondramide C-SA variant. Since there are no many variations, both the complexes are shown to be tightly bound.
Furthermore, hydrogen bonds are important non-covalent structural forces, primarily electrostatic in nature, in molecular systems. They are formed when a single hydrogen atom is effectively shared between the two heavy atoms covalently bonded to each other (the hydrogen bond donor and the hydrogen bond acceptor). Both the donor and acceptor atoms are typically quite electronegative. The average number of hydrogen bonds are tracked at each frame of the simulations and is plotted against different time intervals. The image in Figure 6d shows that the number of hydrogen bonds for the chondramide C (2)-wildtype and chondramide C (2)-SA variant complexes are stable throughout the simulation.
The absolute value of binding free energy of chondramide C-wildtype and chondramide C-SA variant is −35.85 kcal/mol and −35.05 kcal/mol, respectively. For chondramide C-wildtype, the binding free energy at 80 ns seem to fluctuate lower and then at 120 ns, the energy reaches higher. For chondramide C-SA, the binding free energy at 40 ns and 80 ns seem to be similar with the energy reaching the highest at 120 ns. Notably, MM/PBSA calculation was carried out every 20 ns. Table 3 shows that the binding energies for all the intervals of both the complexes fall under the window of −30 kcal/mol and −40 kcal/mol, further highlighting the stability of the complexes and the high affinity of the compounds for the target.
3.6. In silico ADMET and drug-likeness
The pharmacokinetic properties of the top-scoring compounds were predicted using Swiss ADME (Daina et al., 2017). Based on the Lipinski’s Rule of Five, the drug-likeness of the compounds was assessed by looking into their physical properties, such as molecular weight, lipophilicity (reported as octanol-water partition coefficient), and the number of hydrogen bond donors and acceptors (Table S5). Among the top-scoring chondramides and their derivatives with high binding energy to the SARS-CoV2 spike protein, compounds 1, 2, 4, 6 and 7 were highly druggable as reflected by their favorable ADME (absorption, distribution, metabolism and excretion) profiles. Despite the prediction that the top-scoring compounds 3, 5, 8, and 9 were non-druggable according to Lipinski's rule of five, successful drug candidates could be under the extended or beyond the rule of five domain of oral availability considering the abundance of binding site hot spots that are filled by larger candidates and thereby improve affinity or selectivity (Egbert et al.,2019).
In addition, mutagenicity, tumorigenicity, irritant effect and reproductive toxicity of the top-scoring compounds (Table S6) were also evaluated using OSIRIS Property Explorer (Sander et al., 2009). Among the top-scoring chondramides, compounds 1, 2, 3, 4, 5, 7, 8 and 9 did not demonstrate any form of toxicity. Taking into consideration both drug-likeness and in silico toxicity, chondramide C3 (1) and chondramide C (2) exemplified the most promising pharmacokinetic characteristics.
4. Conclusion
In the face of this pandemic, we strive to discover treatment options for patients afflicted with COVID-19. Natural products continue to be an important source of antivirals. In this study, we explored the antagonistic prospects of myxobacterial metabolites against multiple mechanisms of viral entry highlighting their potential as drug prototypes for COVID-19. Our investigation showed that myxobacterial chondramides are selectively active in silico against target spike RBDs precluding the correct recognition of spike with its host counterparts ACE2 and GRP78. This event is an important mechanism in blocking the entry of SARS-CoV-2 to the host cell, thus, slowing viral infection. We also demonstrated that some chondramides interact with the spike protein RBD by targeting well-known viral hot spots. The relevance of a polar environment influenced by N501Y and E484K mutations in the S protein binding regions of SARS-CoV-2 variants was also highlighted as observed in the reinforcement of affinity of highly polar ligands. Molecular dynamics simulations were performed for top-ranked depsipeptides 1 and 2 against the SARS-CoV-2 spike RBD of the Wuhan wild-type within a 120-ns simulation time. To demonstrate the stability of the chondramides against the spike variants, the complex between depsipeptide 2 and the South African spike RBD variant was similarly subjected to MD. All three complexes were found to be dynamically stable. Through protein-protein docking experiments, chondramides C3 (1), C (2) and C6 (9) were shown not only to exhibit high binding scores but were also found effective in antagonizing the binding of SARS-CoV-2 S protein towards target host cell receptors. Thus, we provide a strong basis to pursue in vitro experiments for myxobacterial chondramides as inspirations for development of potential drugs against COVID-19 targeting viral entry mechanisms.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Allam, L., Ghrifi, F., Mohammed, H., El Hafidi, N., El Jaoudi, R., El Harti, J., Lmimouni, B., Belyamani, L., & Ibrahimi, A. (2020). Targeting the GRP78-dependent SARS-CoV-2 cell entry by peptides and small molecules. Bioinformatics and Biology Insights, 14, 117793222096511–117793222096550. 10.1177/1177932220965505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D. E., Cui, J., Qian, Y., Huang, B., Zu, W., Gong, J., Liu, W., Kim, S. Y., Yan, B. G., & Sigmundsson, K. (2020). Orthogonal genome-wide screenings in bat cells identify MTHFD1 as a target of broad antiviral therapy. bioRxiv. 10.1101/2020.03.29.014209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brielle, E. S., Schneidman-Duhovny, D., & Linial, M. (2020). The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. Viruses, 12, 497. 10.3390/v12050497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogi, S., Ramunno, A., Savi, L., Chemi, G., Alfano, G., Pecorelli, A., Pambianchi, E., Galatello, P., Compagnoni, G., Focher, F., Biamonti, G., Valacchi, G., Butini, S., Gemma, S., Campiani, G., & Brindisi, M. (2017). First dual AK/GSK-3β inhibitors endowed with antioxidant properties as multifunctional, potential neuroprotective agents. European Journal of Medicinal Chemistry, 138, 438–457. 10.1016/j.ejmech.2017.06.017 [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri, L., Ojha, R., Pedro, L. D., Djannatian, M., Franz, J., Kuivanen, S., van der Meer, F., Kallio, K., Kaya, T., Anastasina, M., Smura, T., Levanov, L., Szirovicza, L., Tobi, A., Kallio-Kokko, H., Österlund, P., Joensuu, M., Meunier, F. A., Butcher, S. J., … Simons, M. (2020). Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (New York, N.Y.), 370(6518), 856–860. 10.1126/science.abd2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos, A. J., Dat, P. H., Yeh, D.-W., Krieken, R. V., Tseng, C.-C., Zhang, P., Gill, P., Machida, K., & Lee, A. S. (2021). The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. The Journal of Biological Chemistry, 296, 100757–100759. 10.1016/j.jbc.2021.100759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Wang, R., Wang, M., & Wei, G. W. (2020). Mutations strengthened SARS-CoV-2 infectivity. Journal of Molecular Biology, 432(19), 5212–5226. 10.1016/j.jmb.2020.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T., Wu, D., Chen, H., Yan, W., Yang, D., Chen, G., Ma, K., Xu, D., Yu, H., Wang, H., Wang, T., Guo, W., Chen, J., Ding, C., Zhang, X., Huang, J., Han, M., Li, S., Luo, X., Zhao, J., & Ning, Q. (2020). Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ (Clinical Research ed.), 368, m1091 10.1136/bmj.m1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, J. L., Simonetti, B., Klein, K., Chen, K.-E., Williamson, M. K., Antón-Plágaro, C., Shoemark, D. K., Simón-Gracia, L., Bauer, M., Hollandi, R., Greber, U. F., Horvath, P., Sessions, R. B., Helenius, A., Hiscox, J. A., Teesalu, T., Matthews, D. A., Davidson, A. D., Collins, B. M., Cullen, P. J., & Yamauchi, Y. (2020). Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science (New York, N.Y.), 370(6518), 861–865. 10.1126/science.abd3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon, V. N. O., Manzano, J. A., Pilapil, D. Y., Fernandez, R. A., Ching, J. K., Quimque, M. T., Agbay, J. C., Notarte, K. I., & Macabeo, A. P. (2021). Anti-HIV reverse transcriptase plant polyphenolic natural products with in silico inhibitory properties on seven non-structural proteins vital in SARS-CoV-2 pathogenesis. Journal of Genetic Engineering and Biotechnology, 19(1), 104 10.1186/s43141-021-00206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Capua, A., Sticozzi, C., Brogi, S., Brindisi, M., Cappelli, A., Sautebin, L., Rossi, A., Pace, S., Ghelardini, C., Di Cesare Mannelli, L., Valacchi, G., Giorgi, G., Giordani, A., Poce, G., Biava, M., & Anzini, M. (2016). Synthesis and biological evaluation of fluorinated 1,5-diarylpyrrole-3-alkoxyethyl ether derivatives as selective COX-2 inhibitors endowed with anti-inflammatory activity. European Journal of Medicinal Chemistry, 109, 99–106. 10.1016/j.ejmech.2015.12.044 [DOI] [PubMed] [Google Scholar]
- Egbert, M., Whitty, A., Keserű, G. M., & Vajda, S. (2019). Why some targets benefit from beyond rule of five drugs. Journal of Medicinal Chemistry, 62(22), 10005–10025. 10.1021/acs.jmedchem.8b01732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkarhat, Z., Charoute, H., Elkhattabi, L., Barakat, A., & Rouba, H. (2020). Potential inhibitors of SARS-CoV-2 RNA dependent RNA polymerase protein: Molecular docking, molecular dynamics simulations and MM-PBSA analyses. Journal of Biomolecular Structure & Dynamics, 1–14. 10.1080/07391102.2020.1813628 [DOI] [PubMed] [Google Scholar]
- Fontanet, A., Autran, B., Lina, B., Kieny, M. P., Karim, S. S. A., & Sridhar, D. (2021). SARS-CoV-2 variants and ending the COVID-19 pandemic. The Lancet, 397(10278), 952–954. 10.1016/S0140-6736(21)00370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, H., Wang, X., Yuan, X., Xiao, G., Wang, C., Deng, T., Yuan, Q., & Xiao, X. (2020). The epidemiology and clinical information about COVID-19. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology, 39(6), 1011–1019. 10.1007/s10096-020-03874-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney, A. J., Loes, A. N., Crawford, K. H., Starr, T. N., Malone, K. D., Chu, H. Y., & Bloom, J. D. (2020). Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv. 10.1101/2020.12.31.425021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanwell, M. D., Curtis, D. E., Lonie, D. C., Vandermeersch, T., Zurek, E., & Hutchison, G. R. (2012). Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. Journal of Cheminformatics, 4(1), 17. 10.1186/1758-2946-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, J., Hüttel, S., & Müller, R. (2013). Discovery and biological activity of new chondramides from Chondromyces sp. Chembiochem: A European Journal of Chemical Biology, 14(13), 1573–1580. 10.1002/cbic.201300140 [DOI] [PubMed] [Google Scholar]
- Ibrahim, I. M., Abdelmalek, D. H., Elshahat, M. E., & Elfiky, A. A. (2020). COVID-19 spike-host cell receptor GRP78 binding site prediction. The Journal of Infection, 80(5), 554–562. 10.1016/j.jinf.2020.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurini, E., Marson, D., Aulic, S., Fermeglia, M., & Pricl, S. (2020). Computational alanine scanning and structural analysis of the SARS-CoV-2 spike protein/angiotensin-converting enzyme 2 complex. ACS Nano, 14(9), 11821–11830. 10.1021/acsnano.0c04674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Wu, J., Nie, J., Zhang, L., Hao, H., Liu, S., Zhao, C., Zhang, Q., Liu, H., Nie, L., Qin, H., Wang, M., Lu, Q., Li, X., Sun, Q., Liu, J., Zhang, L., Li, X., Huang, W., & Wang, Y. (2020). The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell, 182(5), 1284–1294. e9, 10.1016/j.cell.2020.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magpantay, H. D., Malaluan, I. N., Manzano, J. A. H., Quimque, M. T. J., Pueblos, K. R., Moor, N., Budde, S., Bangcaya, P. S., Lim-Valle, D., Dahse, H.-M., Khan, A., Wei, D.-Q., Alejandro, G. J. D., & Macabeo, A. P. G. (2021). Antibacterial and COX-2 inhibitory tetrahydrobisbenzylisoquinoline alkaloids from the Philippine medicinal plant Phaeanthus ophthalmicus. Plants, 10, 1–16. 10.3390/plants10030462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, K. I. (2018). Diversity of Myxobacteria-we only see the tip of the iceberg. Microorganisms, 6, 84. 10.3390/microorganisms6030084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulwa, L. S., & Stadler, M. (2018). Antiviral compounds from Myxobacteria. Microorganisms, 6, 73–88. 10.3390/microorganisms6030073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, C. K. V., Franco, M. M., Gräf, T., de Lorenzo Barcia, C. A., de Ávila Mendonça, R. N., de Sousa, K. A. F., Neiva, L. M. C., Fosenca, V., Mendes, A. V. A., de Aguiar, R. S., Giovanetti, M., & de Freitas Souza, B. S. (2021). Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerging Infectious Diseases, 27(5), 1522–1524. (online ahead of print), 10.3201/eid2705.210191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman, H., Bouslama, Z., Brandenburg, J. T., da Rocha, J., Hamdi, Y., Ghedira, K., Srairi-Abid, N., & Hazelhurst, S. (2020). Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: Similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochemical and Biophysical Research Communications, 527(3), 702–708. 10.1016/j.bbrc.2020.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Quimque, M. T. J., Notarte, K. I. R., Fernandez, R. A. T., Mendoza, M. A. O., Liman, R. A. D., Lim, J. A. K., Pilapil, L. A. E., Ong, J. K. H., Pastrana, A. M., Khan, A., Wei, D. Q., & Macabeo, A. P. G. (2021). Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms . Journal of Biomolecular Structure & Dynamics, 39(12), 4316–4318. 10.1080/07391102.2020.1776639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimque, M. T., Notarte, K. I., Letada, A., Fernandez, R. A., Pilapil, D. Y., Pueblos, K. R., Agbay, J. C., Dahse, H.-M., Wenzel-Storjohann, A., Tasdemir, D., Khan, A., Wei, D.-Q., & Gose Macabeo, A. P. and (2021). Potential cancer- and Alzheimer's disease-targeting phosphodiesterase inhibitors from Uvaria alba: Insights from in vitro and consensus virtual screening. ACS Omega, 6(12), 8403–8417. 10.1021/acsomega.1c00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, T., Freyss, J., von Korff, M., Reich, J. R., & Rufener, C. (2009). OSIRIS, an entirely in-house developed drug discovery informatics system. Journal of Chemical Information and Modeling, 49(2), 232–246. 10.1021/ci800305f [DOI] [PubMed] [Google Scholar]
- Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020b). Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America, 117(21), 11727–11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020a). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581(7807), 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam, N., Muralidharan, D., Velmurugan, D., & Gromiha, M. M. (2020). Therapeutic targets and computational approaches on drug development for COVID-19. Current Topics in Medicinal Chemistry, 20(24), 2210–2220. 10.2174/1568026620666200710105507 [DOI] [PubMed] [Google Scholar]
- Shapovalov, M. V., & Dunbrack, R. L. (2011). A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure, 19(6), 844–858. 10.1016/j.str.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Li, Y., Tian, S., Xu, L., & Hou, T. (2014). Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Physical Chemistry Chemical Physics: PCCP, 16(31), 16719–16729. 10.1039/c4cp01388c [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Chen, J., Gao, K., Hozumi, Y., Yin, C., & Wei, G.-W. (2021). Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Communications Biology, 4(1), 228 10.1038/s42003-021-01754-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., Chen, W., Zhang, Z., Deng, Y., Lian, J.-Q., Du, P., Wei, D., Zhang, Y., Sun, X.-X., Gong, L., Yang, X., He, L., Zhang, L., Yang, Z., Geng, J.-J., Chen, R., Zhang, H., Wang, B., Zhu, Y.-M., … Chen, Z.-N. (2020). CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduction and Targeted Therapy, 5(1), 283 10.1038/s41392-020-00426-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2001). Antechamber, an accessory software package for molecular mechanical calculations. Journal of the American Chemical Society, 222, U403. [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2006). Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics & Modelling, 25(2), 247–260. 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., Lu, G., Qiao, C., Hu, Y., Yuen, K.-Y., Wang, Q., Zhou, H., Yan, J., & Qi, J. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell, 181(4), 894–904.e9. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer, C. K., Ayres, F., Hermanus, T., Madzivhandila, M., Kgagudi, P., Lambson, B. E., Vermeulen, M., van den Berg, K., Rossouw, T., & Boswell, M. (2021). SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 10.1101/2021.01.18.427166 [DOI] [PubMed] [Google Scholar]
- Wise, J. (2020). Covid-19: New coronavirus variant is identified in UK. BMJ (Clinical Research ed.), 371, m4857. 10.1136/bmj.m4857 [DOI] [PubMed] [Google Scholar]
- Yang, J., Nune, M., Zong, Y., Zhou, L., & Liu, Q. (2015). Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure (London, England: 1993), 23(12), 2191–2203. 10.1016/j.str.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.