Abstract
Viruses exploit the translation machinery of an infected cell to synthesize their proteins. Therefore, viral mRNAs have to compete for ribosomes and translation factors with cellular mRNAs. To succeed, eukaryotic viruses adopt multiple strategies. One is to circumvent the need for m7G-cap through alternative instruments for ribosome recruitment. These include internal ribosome entry sites (IRESs), which make translation independent of the free 5′ end, or cap-independent translational enhancers (CITEs), which promote initiation at the uncapped 5′ end, even if located in 3′ untranslated regions (3′ UTRs). Even if a virus uses the canonical cap-dependent ribosome recruitment, it can still perturb conventional ribosomal scanning and start codon selection. The pressure for genome compression often gives rise to internal and overlapping open reading frames. Their translation is initiated through specific mechanisms, such as leaky scanning, 43S sliding, shunting, or coupled termination-reinitiation. Deviations from the canonical initiation reduce the dependence of viral mRNAs on translation initiation factors, thereby providing resistance to antiviral mechanisms and cellular stress responses. Moreover, viruses can gain advantage in a competition for the translational machinery by inactivating individual translational factors and/or replacing them with viral counterparts. Certain viruses even create specialized intracellular “translation factories”, which spatially isolate the sites of their protein synthesis from cellular antiviral systems, and increase availability of translational components. However, these virus-specific mechanisms may become the Achilles’ heel of a viral life cycle. Thus, better understanding of the unconventional mechanisms of viral mRNA translation initiation provides valuable insight for developing new approaches to antiviral therapy.
Keywords: cap-independent translation, VPg, IRES and 3′ CITE, TURBS-mediated reinitiation, circular RNAs, translation initiation factors eIF2 and eIF4F, picornaviruses PV and EMCV, flaviviruses HCV and DENV, lentiviruses HIV-1 and HIV-2, coronavirus SARS-CoV-2
DEDICATION
We wrote this review in tribute to Alexander S. Spirin, an outstanding scientist who made groundbreaking contributions to the studies of protein biosynthesis. Although the main areas of interests of Prof. Spirin were the ribosome and basic translation processes, he was also fascinated by translation of viral mRNAs. This interest can be highlighted by his studies on the structure and translational control of plant virus RNAs, on the peculiarities of translation initiation of poxviruses, usage of viral translation mechanisms to optimize cell-free protein synthesizing systems. This text is written by his colleagues, friends, students, co-authors, and collaborators. All of us highly admire him as a scientist and a person, and we dedicate our review to his memory with great respect and gratitude.
INTRODUCTION
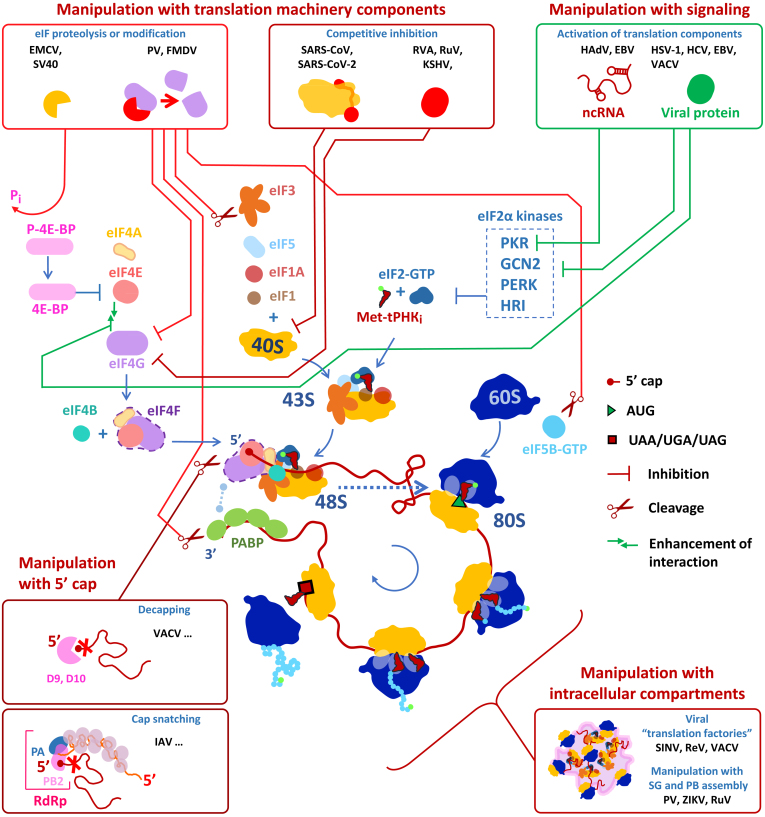
Our planet is inhabited by viruses, and many of them are pathogens of eukaryotes. Despite the fact that viral genomes can be larger in size and complexity than those of some primitive bacteria, as of now there is not a single case when they would contain a complete set of genes necessary for protein biosynthesis [1]. This makes viruses almost completely dependent on the cellular translational apparatus. Moreover, most often they do not just use what is available: many viruses are able to usurp the protein-synthesizing machinery, redirecting the lion’s share of cellular resources to the production of their own proteins. In the course of evolution, viruses have acquired the ability to manipulate different stages of the translational cycle, with translation initiation being the primary target. By hijacking or undermining translation machinery components, and using non-canonical mechanisms to recruit ribosomes, viruses gain a competitive advantage for their mRNA and halt the cellular antiviral response.
In this review, we describe some of the structural and functional features of viral mRNAs and discuss how they allow successful competition for the translational apparatus of infected cells.
CANONICAL MECHANISM OF TRANSLATION INITIATION IN EUKARYOTES
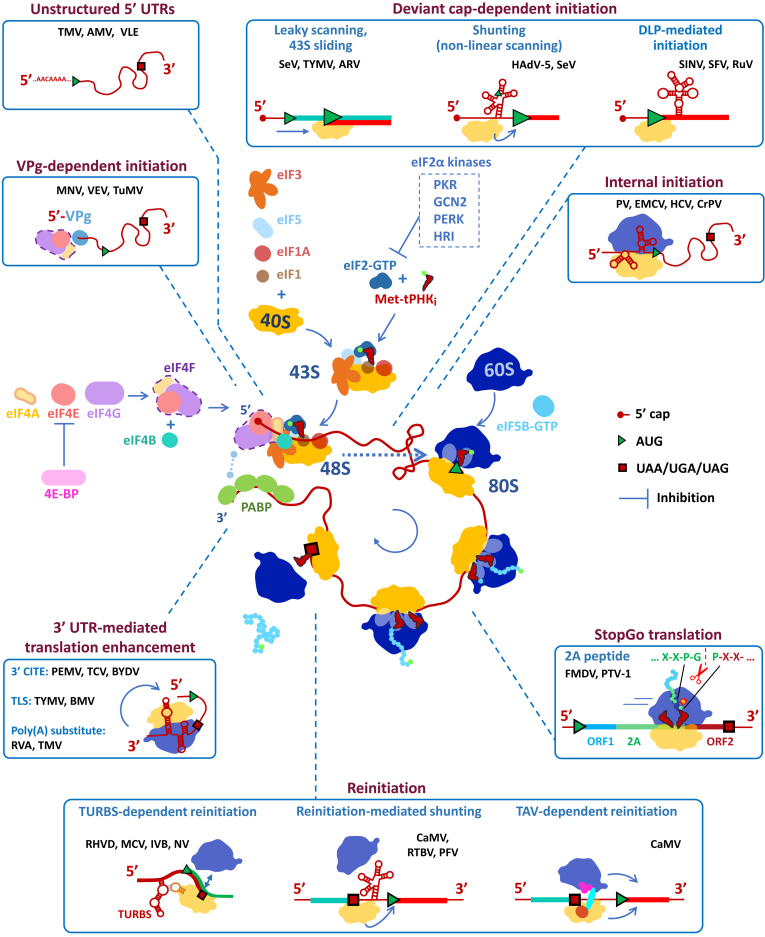
Cytoplasmic mRNAs of the eukaryotic cell have a specialized chemical structure at the 5′ end, the m7G-cap (7-methylguanosine, attached through a 5′,5′-triphosphate bridge to the first nucleotide of mRNA), and are usually equipped with a poly(A)-tail at the 3′ end. Such multifunctional “labels” are recognized in the cytoplasm by specialized proteins [2-5]. Under conditions of active translation, the 5′ cap is associated with the eIF4F, which consists of three subunits: a small cap-binding protein eIF4E, a large scaffold eIF4G, and an ATP-dependent RNA helicase eIF4A. The 3′ end of mRNA is usually associated with several molecules of the poly(A)-binding protein PABP, which, via interaction with eIF4G, shapes mRNA into a closed-loop structure (Fig. 1).
Fig. 1.
Translation cycle of the eukaryotic mRNA and major non-canonical translation initiation mechanisms used by viral mRNAs. Full names of the viruses are given in the text of the article.
Another set of translation initiation factors binds to the small subunit of the ribosome, forming the 43S pre-initiation complex [6]. The GTP-bound heterotrimer eIF2 delivers the initiator Met-tRNAi to the ribosomal P-site. Three other factors – eIF1, eIF1A, and eIF5 – bind in close proximity and control tRNA accommodation. The giant eIF3 protein, which consists of 13 subunits in mammals, wraps the 40S subunit, forming multiple contacts with almost all other initiation factors.
Due to the interaction of eIF3 and eIF4G, the 43S complex is recruited to the mRNA. Importantly, during the canonical translation initiation, the eukaryotic ribosome enters strictly at the 5′ end of the transcript accommodating it into the RNA-binding channel of the 40S subunit. The 43S complex then starts travelling towards the 3′ end, searching for an AUG triplet (“ribosomal scanning”). Recognition of the appropriate start codon (usually AUG in a suitable nucleotide context, but sometimes a near-cognate codon like CUG, ACG, GUG, etc.) is ensured by stringent monitoring of the Met-tRNAi conformation at the P-site by factors eIF1, eIF2, eIF5, and certain subunits of eIF3. During recognition of the start codon, inorganic phosphate (Pi) is released from the hydrolyzed eIF2-bound GTP due to coordinated action of the factors, which causes their sequential dissociation and irreversible arrest of the scanning ribosome. At this stage, the factor eIF5B binds to the complex and facilitates 60S joining. The resulting 80S particle is ready to accept aminoacyl-tRNA into the A-site and proceed to elongation.
This classical mechanism of translation initiation is called cap-dependent scanning and is predominant for cellular mRNAs [3, 7]. Its steps are regulated depending on the conditions the cell is exposed to. In particular, under certain types of stress, eIF2 is phosphorylated and sequestered into an inactive complex with the guanine nucleotide exchange factor eIF2B. This stops delivery of Met-tRNAi to the initiation complex and leads to translation repression. Another subject of regulation are the cap-binding factors: eIF4E–eIF4G interaction is disrupted by the 4E-BP proteins, which are activated upon dephosphorylation. Both pathways are often involved in the cellular response to viral infection.
In the course of evolution, many viruses have developed alternative modes of translation initiation, as well as various ways of manipulating its different stages and regulation. This provides viral mRNA with a competitive advantage over cellular transcripts.
“DEVIANT” CAP-DEPENDENT INITIATION EMPLOYED BY VIRAL mRNAs
In this section, we will consider the mechanisms of translation initiation of viral mRNAs that contain a functional m7G-cap structure at the 5′ end, but nevertheless use unconventional modes of ribosomal scanning or start codon selection.
Manipulations with the mechanism of start codon selection. Due to their compactness, viral genomes often contain overlapping open reading frames (ORFs). More than one protein can be synthesized from one mRNA – and they can be encoded both in different reading frames and in the same frame, starting from different start codons (in the latter case, these can be either co-terminal isoforms or individual proteins obtained by proteolytic processing). In classical cap-dependent scanning, the ribosome does not always start translation from the 5′ proximal AUG codon. The 43S scanning complex may not recognize an AUG and simply “drive” through [7]. The probability of such an event depends on the nucleotide context of the AUG, primarily on the nucleotides at positions –3 and +4. Pyrimidines in these positions (“weak” context) reduce the recognition efficiency, which leads to so-called “leaky scanning”. Purines in both positions form a “strong” context, usually referred to as Kozak’s context after M. Kozak, who first discovered this phenomenon. Recognition can also be enhanced by stable secondary structure of mRNA downstream of the considered triplet, since this slows the advance of the scanning complex (see below). However, even if the complex recognizes AUG and stops, final fixation on the selected codon requires hydrolysis of the GTP molecule bound to eIF2 and, most importantly, release of the Pi, making the hydrolysis irreversible. If this does not happen for a long time (for example, under conditions of inactive eIF5), the complex can resume scanning and reach the next suitable codon [8]. This second mechanism is called 43S sliding, and it is context-independent.
In practice, it is difficult to distinguish between leaky scanning and 43S sliding. Both have the same consequences, although the mechanism and kinetics of these processes are different. However, since 5′ proximal AUG codons are often ignored in the different contexts, both phenomena seem to be quite common.
Many viruses exploit such mechanisms for their own purposes. For example, the P/C mRNA of the murine respirovirus, better known as Sendai virus (SeV), simultaneously encodes eight products, and leaky scanning is used for initiation of the synthesis of three of them (C′, P and C in the order of initiation sites) [9]. In this case, proteins C′ and C have a common C-end and are encoded in one frame, and P – in another, which strongly overlaps with the first. For the initiation complex to reach the C protein start codon, it must skip the two previous ones. This occurs because the C′ protein start codon is ACG, and in the case of P, the AUG codon context contains pyrimidine at position –3. Similar principles allow production of several proteins from a single mRNA in the case of other viruses, often with an AUG-like triplet acting as the first of the start codons (see review in [10]). One of the most striking cases, when the coding potential of mRNA is used especially effectively by employing leaky scanning, is the subgenomic RNA (sgRNA) of some umbraviruses [11], where two large proteins are encoded in different but almost completely overlapping frames.
Some viral mRNAs violate classic leaky scanning principles. For example, in the case of the genomic RNA (gRNA) of the turnip yellow mosaic virus (TYMV), initiation frequency at the first of the two start codons depends on recognition efficiency of the second one and on the distance between them [12], which is difficult to explain from the standpoint of the classical unidirectional (5′-3′) scanning, even taking into account all the nuances [13]. An especially efficient form of leaky scanning has been shown for the S1 mRNA of avian reovirus (ARV), which allows placing the pre-initiation complex at the start codon of the ςC frame. It is possible that in these cases the primary role is played by sliding of the 43S complex or the choice of start codon changes depending on the concentration of mRNA and factors in the cell [14]. Skipping of AUG codons is possible not only in the case of cap-dependent initiation, but also in other scanning scenarios (see below). The choice of the start codon can also be influenced by specialized elements of secondary structure of the viral mRNA. Stable hairpins located at a distance of 14 nt or slightly further downstream of the initiation codon not only promote its recognition, as mentioned above, but can also somehow reduce the need of such mRNA for some initiation factors (in particular, eIF2 and eIF4F). Such structures, called DLP (downstream loop), are present in the sgRNA of some alphaviruses (for example, Sindbis virus, SINV, and Semliki forest virus, SFV), as well as in the related rubiviruses (rubella virus, RuV) [15-17]. In infected cells, phosphorylation of eIF2 by PKR kinase at the late stages of infection leads to suppression of translation of cellular mRNAs and the viral gRNA [16, 18], while sgRNA is still translated efficiently. This translation is also resistant to artificial inhibition of eIF2, and eIF4A helicase [19, 20], as well as to eIF4G cleavage [19], but all this is true only in the context of viral infection. The reason for this is not fully understood, as well as the mechanism of Met-tRNAi delivery to the initiation complex in the absence of eIF2 under these conditions [21]. During reconstruction of the SINV sgRNA translation from purified components, eIF2 can be replaced by recycling/reinitiation factors eIF2D or MCTS•DENR [22]; however, this activity is most likely a side effect and is hardly significant in vivo [23]. A conserved hairpin (cHP) in the corresponding position of the coding region, which helps in the selection of the start codon, is also present in the mRNA of some flaviviruses (for example, dengue virus, DENV) [24]; however, translation resistance to eIF2 phosphorylation has not been documented in this case. Interestingly, mRNA of DENV and related flaviviruses has reduced requirements for activity of the cap-binding apparatus (see below); however, the cHP hairpin is apparently not involved in this phenomenon [25].
Nonlinear scanning. The classical model of ribosomal scanning assumes continuous inspection of every position in the 5′ UTR by the pre-initiation complex. However, in some viral mRNAs, certain regions of the leaders seem to avoid this. In these cases, AUG codons or stable hairpins present in the 5′ UTRs, which usually prevent progression of the 43S complex, do not affect the translation levels of the main frames. This situation is termed nonlinear scanning or shunting.
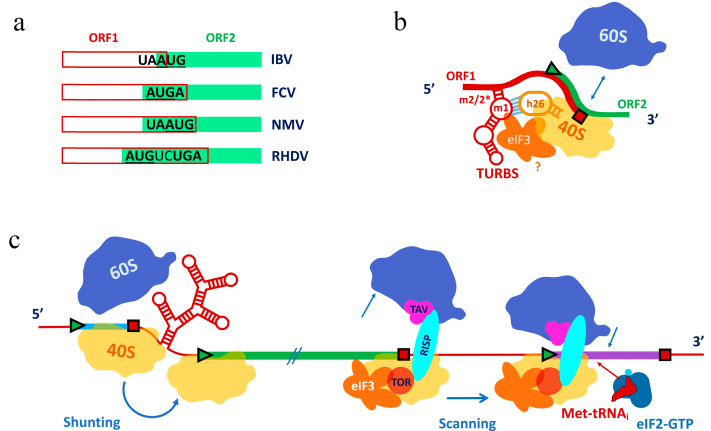
Non-linear scanning is an umbrella term. Most of the relevant reviews start with the case of 35S pre-genomic RNA (pgRNA) of the CaMV pararetrovirus (cauliflower mosaic virus), in the description of which this term was introduced [26]. However, we will break this tradition, since it is now known that what was once called “shunting” on 35S pgRNA is based not on nonlinear scanning of the 43S leader by the pre-initiation complex, but rather on a special mechanism of translation reinitiation, which is activated after reading and termination on the stop codon of the first short frame located in the 5′ UTR. Therefore, we will consider this case below in the section dealing with reinitiation.
Shunting as bona fide nonlinear scanning was first documented in 1988 when translation of the aforementioned P/C mRNA of the Sendai virus was studied [27]. While initiation of translation of the first three ORFs in this mRNA occurs by the conventional or leaky scanning mechanisms, ribosomes reach the three distal start codons (located in-frame and giving rise to the co-terminal proteins Y1, Y2, and X) bypassing the 5′ proximal region [9, 28]. This, however, requires the capped 5′ end of the P/C mRNA. The mechanism of shunting during initiation on the AUG codons Y1 and Y2 (separated by 15 nt) has been studied in great detail. After binding to the 5′ cap and scanning of the first ~50 nt of the leader, the pre-initiation complex jumps to the start codons Y1 or Y2. No discreet donor site could be delineated, and the acceptor site lies close to the AUG codons Y1 and Y2, including the 24-nt sequence necessary for shunting located immediately after the latter codon. Interestingly, in an artificial construct that directs the ribosome to the same codons by classical cap-dependent scanning, this sequence did not affect efficiency of their recognition (i.e., the need for this structure is not associated with stopping scanning, as in the case of the above-described DLP). Another unique feature is that the AUG start codons Y1 and Y2 can be replaced with other triplets without loss of shunting efficiency. Viral proteins are not required for shunting on the P/C mRNA.
The second case, also considered a classic one, is nonlinear scanning of the so-called tripartite leader (TPL) of late mRNAs of human adenovirus 5 (HAdV-5), as well as mRNA IVa2 of the same virus. The R. Schneider group showed that the 40S subunit first binds to the capped 5′ end and starts scanning, but then skips the internal highly structured part of the TPL. According to the authors, base-pairing of a certain TPL region with 18S rRNA plays an important role in this shunting variation [29]. This process can occur in uninfected cells, but it requires unidentified auxiliary protein(s) in addition to the canonical initiation factors [30]. During infection, shunting is further stimulated by adenoviral protein 100K, which simultaneously binds TPL and eIF4G [31]. The mechanism of this phenomenon is not clear, but, remarkably, the 100K protein contains an RGG motif (arginine-glycine-glycine), which is common for many cellular mRNA-binding proteins and can in fact mediate their binding to eIF4G [32]. In the case of cellular RGG proteins, however, this interaction leads to the formation of inactive ribonucleoproteins (mRNPs). The TPL-directed translation is resistant to partial inactivation of eIF4F [33], although whether this is associated with shunting is unknown.
There are other, less characterized cases of shunting: for example, on the mRNA of human papillomavirus 18 (HPV 18), which encodes the E1 protein; on the bicistronic pgRNA of duck hepatitis B virus (DHBV); on the tricistronic mRNA S1 of ARV reovirus; on the mRNA 3 of coronavirus causing transmissible gastroenteritis coronavirus (TGEV), and some others (see reviews [10, 34]). Translation initiation on all of these mRNAs requires a capped 5′ terminus, but introduction of stable hairpins and AUG codons into the region “shunted” by the ribosome does not lead to translation inhibition. Molecular mechanisms in all these cases are also not fully characterized, but they, apparently, differ from the two described above, since they do not exhibit the specific features described above.
Translation initiation on viral mRNAs with unstructured 5′ UTR. mRNAs of some viruses have entirely single-stranded leaders. This reduces their requirements for some of the initiation factors. A classic example is alfalfa mosaic virus (AMV) sgRNA 4, which contains a 36-nt long, unstructured U-rich 5′ UTR. In an in vitro system reconstituted from purified components, this mRNA can form a 48S initiation complex in the absence of ATP and eIF4 factors (eIF4A, eIF4B, eIF4F) [35]. While in the complete cell lysate translation of the AMV-4 mRNA apparently requires the complete eIF4F factor (see discussion in [36]), the mentioned structural features give this mRNA a competitive advantage over cellular templates and allow relatively efficient translation even in the absence of 5′ cap.
The A-rich omega leader of the tobacco mosaic virus (TMV) mRNA, which is capable of directing highly efficient translation in various eukaryotic cell-free systems even in the absence of the 5′ cap, apparently also has a predominantly single-stranded conformation [37]. Experiments performed at the A. Spirin lab demonstrated that an mRNA bearing this leader can form 48S initiation complexes in the absence of eIF4F and ATP in a reconstituted translation system [38]. The authors proposed a model of “diffusion wandering”, i.e., bidirectional ATP-independent scanning of this leader, although the question of whether such a process can occur in a complete cell lysate or in an intact cell, remains unanswered.
The unusual properties of single-stranded 5′ UTRs are even more pronounced in the case of transcripts with oligo(A)-leaders, which are characteristic of intermediate and late mRNAs of the vaccinia virus (VACV). According to early estimates, the length of these leaders, formed during transcription by means of non-template synthesis, is about 30-40 nt; however, later data indicate a shorter length ranging from 7-8 nt for intermediate mRNAs to 11-20 nt for late ones [39, 40], and suggest prevalence of the non-capped transcripts among the mRNAs synthesized at these stages of infection [40]. Shirokikh and Spirin [41] showed that the mRNAs with oligo(A)-leaders can operate in the 48S reconstitution system not only without eIF4F, but also without eIF3. Perhaps this property underlies the preferential translation of VACV mRNA during infection, as well as its resistance to cleavage by eIF4G and inhibition by cap analogs in vitro [42, 43]. A similar situation takes place in the case of mRNA of yeast virus-like elements (VLE) pGKL1/2, which also have oligo(A)-leaders of variable length, but usually not exceeding 12 nt [44]. As with VACV, many VLE transcripts are uncapped, and thus their translation does not require eIF4E. To effectively initiate translation in infected cells, the length of oligo(A)-leaders should not exceed 12 nt [45]. This can be explained by the fact that longer oligo(A)s are able to bind PABP [5], which would inevitably interfere with ribosome entry. In human cells infected with VACV, predominant translation of the mRNAs with oligo(A)-leaders requires phosphorylation of the ribosomal protein RACK1 by viral kinase [46], but the reason for this is unclear.
Alternative cap-binding apparatus. Viruses which encode their own cap-binding proteins that replace the eIF4F initiation factors or some of its subunits deserve mentioning. As the initiation process per se does not differ from the standard, we are just listing such cases, while interested readers can refer to the relevant papers [47, 48]. Giant protozoan viruses encode their own ortholog of the eIF4E; the cap-binding subunit PB2 of influenza virus RNA polymerase (influenza A virus, IAV) binds eIF4G and thus replaces cellular eIF4E (which is inactivated upon infection) for viral mRNA; protein N of some arenaviruses (Junin virus (JUNV), Tacaribe virus (TCRV) and Pichinde virus (PICV)) appears to have a similar activity; and the hantavirus Sin Nombre orthohantavirus (SNV), family Bunyaviridae) N protein replaces the entire eIF4F factor and has the activities of all three of its subunits.
In the next section, we will consider cases where the 5′ cap functions (all or only some) are performed by proteins covalently linked to the 5′ end of the viral mRNA.
VPg-DEPENDENT TRANSLATION INITIATION
Presence of the cap structure is not obligatory for initiation of 5′ end-dependent translation. Some viral mRNAs do not have a cap, but are able to use the same set of initiation factors, involving them in translation in the same order as during cap-dependent initiation. The VPg protein (viral protein genome-linked) is bound covalently to the 5′ end of the mRNA and can be used instead of the cap. Although the presence of VPg is a trait of many RNA viruses, where it participates in RNA replication, VPg as a cap substitution was described only for the representatives of the families Potyviridae, Caliciviridae, and Astroviridae.
Caliciviruses that infect mammals are notable examples. Their VPg can function as a substitute of 5′ cap, allowing viral mRNA binding to eIF4E or even directly to eIF4G and PABP, as described for the members of the Vesivirus and Norovirus genera [49, 50]. For example, the C-terminal VPg region of the murine norovirus (MNV) interacts with the HEAT-1 domain of the eIF4G factor, which leads to the efficient assembly of pre-initiation complexes on the viral mRNA [50]. Similar cases are known for plant viruses: VPg of potyviruses is able to compete for the cap-binding site of the eIF4E factor. Thus, on the one hand, the cap-dependent initiation of mRNA of the infected cell is suppressed, and on the other hand, translation of the viral mRNAs is promoted [51]. In addition, a synergistic effect of VPg and PABP has also been shown: PABP increases the VPg binding to eIF4F 3-4-fold, which stimulates translation of the turnip mosaic virus (TuMV) mRNA in a cell-free system from wheat germ extract (WGE). When the purified PABP was added to the WGE system depleted of eIF4F, eIFiso4F, and PABP, a 30-fold increase in translation of viral mRNA was observed [52], which was almost an order of magnitude higher than the stimulating effect of PABP on the translation of cellular mRNAs in this system. Alternative mechanisms of attracting the initiation complex are also possible: for example, VPg of the feline calicivirus (FCV) and human Norwalk virus (HNV) bind eIF3 [53].
In addition to recruiting initiation factors, VPg can perform other functions in translation: for example, the noroviral VPg interacts with G3BP1, one of the key components in the formation of stress granules, and this binding also stimulates production of viral proteins [54]. The importance of these mechanisms is highlighted by the fact that proteolytic removal of VPg (for example, in the vesicular exanthema virus (VEV), a representative of caliciviruses) results in the complete loss of infectivity of the viral mRNA [55].
All of the above indicates that the presence of VPg in potyviruses and caliciviruses is a vital necessity in the struggle for control over the cellular translational apparatus. The design of small molecule inhibitors that can specifically uncouple the interaction of VPg with its partners may be a promising direction in the treatment of diseases caused by caliciviruses in mammals and potyviruses in plants.
INTERNAL TRANSLATION INITIATION
An alternative way to initiate translation is via the use of special cis-acting RNA elements, called internal ribosome entry sites (IRESs). As a rule, IRESs are high-order RNA structures located in 5′ UTRs or in intergenic spacers of polycistronic mRNAs. Individual domains of the IRESs bind initiation factors and ribosomes, or in certain cases mimic tRNA or other translational components (Fig. 2). An IRES performs two tasks: first, it recruits the initiation complex regardless of the presence of a 5′ cap in the mRNA, and, second, it ensures remodeling of the small ribosomal subunit so that the latter can accommodate an internal region of the template into the RNA-binding channel, which is prohibited during conventional translation initiation. This second aspect of IRES activity is crucial for achieving internal initiation of translation and distinguishes IRESs from, for example, the cap-independent translation enhancers discussed below.
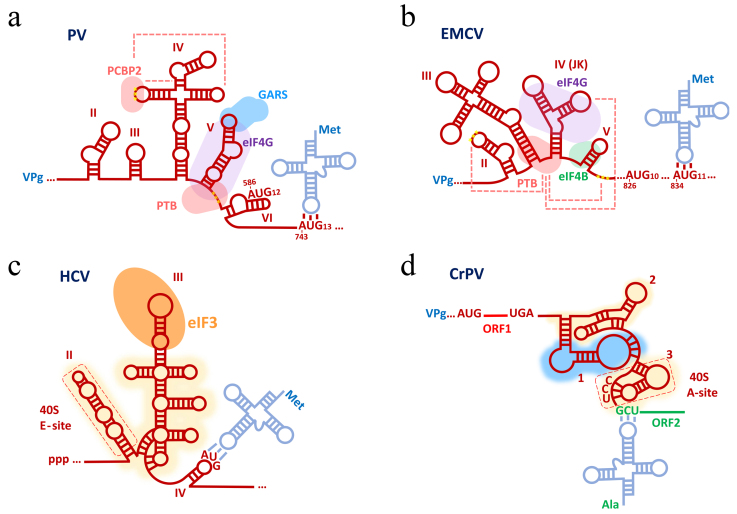
Fig. 2.
Main types of classic IRESs as exemplified by the most typical representatives (full names of viruses are given in the text of the article). Secondary structure of type I-IV elements [panels (a-d), respectively], proteins specifically binding to them, as well as areas of contact with 40S and 60S subunits (yellow and blue shading, respectively) are shown schematically. Also shown are aminoacyl-tRNAs, which ensure delivery of the N-terminal amino acid of the future protein.
Such a special route of attracting initiation complexes often allows IRES-containing mRNAs to have reduced requirements for the set of initiation factors (and in some cases to operate without any of them). This enables IRESs to function effectively under conditions when translation of cellular mRNAs is suppressed. Many viruses build their strategies of translational dominance upon this property via inactivation of individual components of the cellular translational machinery.
IRES diversity and difficulties in their classification. A wide variety of viral IRESs is known in terms of their structure and mechanism of functioning. However, not all of them have been studied thoroughly, which greatly complicates their classification. Recently discovered and/or superficially studied IRESs are sometimes assigned to new types, which further confuses the matter. In addition, a significant portion of the work on identification of IRESs (mostly of cellular origin) was performed without taking into account possible artifacts (see next section), which is why some cases may eventually turn out to be false. The situation is further complicated by the fact that viruses effectively exploit horizontal transfer between phylogenetically distant groups, which prevents reliable use of taxonomy for their classification.
In our opinion, a convenient classification of IRESs should be based on the similarity of their secondary structures, mechanism they use to attract ribosomes, and a minimal set of the required initiation factors that is predetermined by their structure. For the purposes of this review, we will highlight 4 main types of IRESs, numbered in the order of their discovery, and separately describe the groups that do not fit this classification. However, we do not insist that this classification is better than those used by our peers [47, 56-60].
Challenges in IRES research. Advances in the study of the classical viral IRESs described below have largely contributed to the opinion that many viral mRNAs use the mechanism of internal ribosome binding to initiate translation. Indeed, a number of viral mRNAs that are translated under conditions when most of the cellular mRNAs are inactive, have 5′ UTRs with a complex secondary structure and multiple uAUGs, which should greatly reduce efficiency of ribosomal scanning.
To confirm the presence of an IRES in a particular fragment of mRNA, the bicistronic assay, which was proposed in the pioneering studies on this topic [61, 62], is routinely used. It is based on the assessment of expression of two non-overlapping reporters encoded within a single mRNA. The 5′ proximal reporter is translated via the cap-dependent mechanism and serves as an internal reference. The second reporter, however, can only be efficiently translated if ribosomes are capable of binding to the intercistronic region, that is, if the latter contains an IRES. This elegant approach has become widely adopted, but there is a high risk of false positive results if it is misapplied and its limitations are overlooked [63-66].
One of the main problems with this method is the use of plasmids to deliver bicistronic reporters. When transcribed in mammalian cells, plasmid DNA, in addition to the authentic bicistronic mRNA, generates a chaotic set of aberrant transcripts – products of background promoter activity and/or uncontrolled splicing [64, 67]. Among them are monocistronic mRNAs encoding the reporter that is assayed to monitor internal initiation. Thus, in some cases, this minor product can be the exclusive source of reporter signal, despite the fact that its quantity is miniscule compared to the correct bicistronic mRNA. As an alternative approach devoid of these drawbacks, the use of bicistronic mRNAs synthesized in vitro has been proposed [63, 67] but as of now it has not become common practice. The RNA transfection method also has limitations. In particular, when cationic lipid-based reagents are used, most of the liposomes attached to the cells are not delivered into the cytoplasm; therefore, for example, it is meaningless to analyze the amount or stability of mRNA that has entered the cells with the RT-qPCR method [68].
Yet, the main flaw of the bicistronic assay emerges when one compares translation driven by the hypothetical IRESs among each other (as well as with negative control, i.e., with a bicistronic mRNA that lacks an IRES). The fundamental problem is subjectivity of the interpretation of the results of such comparison [63]. This approach is only justified if the sequence under study naturally resides in an intercistronic position (as, for example, in the case of the intergenic IRES of Dicistroviridae); however, such situations are rare. When a putative IRES originates from a 5′ UTR (and especially, if the mRNA is naturally capped), it is necessary to compare not only different bicistronic reporters, but also the bicistronic and capped monocistronic mRNAs that contain the studied 5′ UTR in either intergenic, or 5′ terminal position, respectively. Only such comparisons make it possible to evaluate the mechanism by which natural mRNA is translated. Comparable levels of translation directed by the putative IRES from 5′ UTR or internal position indeed suggest a noticeable contribution of internal initiation and, thus, represent reasonable evidence of IRES function [63]. However, this approach also does not guarantee an unambiguous conclusion, since there is a risk that incorporation of the studied RNA fragment into unnatural context may affect its functional activity.
Another source of false positive results during identification of IRESs may be the popular cell-free translation system, rabbit reticulocyte lysate (RRL) with hydrolyzed endogenous mRNA. This system does not reproduce the competitive conditions of the cell and has a depleted repertoire of RNA-binding proteins; therefore, mRNAs translated in it demonstrate a relatively weak dependence on the 5′ cap, increased sensitivity to variations in the secondary structure of the 5′ UTR, and aberrant internal initiation in the extended unstructured regions containing AUG codons (see discussion in [63, 65, 69]). In addition, some of the eIF4G molecules that make up eIF4F remain bound to the capped 5′ end fragments of the hydrolyzed mRNA and are released upon addition of the cap-dependent initiation inhibitors (m7GTP, 4E-BP1, or proteases that cut eIF4G), which leads to stimulation of translation of the uncapped or otherwise ineffectively translated capped mRNA. Aberrant internal initiation in the case of bicistronic constructs and apparent cap independence in the case of monocistronic constructs can be misinterpreted as evidence of IRES activity. These phenomena are usually not reproduced in cell-free systems prepared from cultured mammalian cells [69, 70]. However, even when working with such systems, one should remember that the results can strongly depend on the preparation conditions and concentration of the components, and be careful when correlating the data obtained in the specific cell-free system with the results in cultured cells, and even more so in vivo.
Next, we will describe the methods of IRES-mediated initiation using examples of the most studied representatives of each of the four types (Fig. 2, Table 1), and then we will touch upon those cases that are in the process of being studied and have not yet been assigned to any of the types, or require additional confirmation.
Table 1.
Classic types of internal ribosome entry sites (IRESs) and their brief characteristics
| IRES type | Representative viruses* | Family (Genus) of viruses | Location in mRNA | Required eIFs and ITAFs** | Kd, nM |
|---|---|---|---|---|---|
| I | Poliovirus (PV) | Picornaviridae (Enterovirus) | 5′ UTR | (eIF1), (eIF1A), eIF2, eIF3, eIF4A, (eIF4B), eIF4G, (PTB), PCBP2, (PCBP1, GARS, UNR, La) |
78 (eIF4G) [71] 40 (eIF4F) [71] 41 (PTB) [71] 250 (PCBP1) [72] |
| Human Coxsackievirus B3 (CVB3) | |||||
| Human rhinovirus A (HRV) | |||||
| II | Encephalomyocarditis virus (EMCV) | Picornaviridae (Cardiovirus) | 5′ UTR | (eIF1), (eIF1A), eIF2, eIF3, eIF4A, (eIF4B), eIF4G; PTB, (ITAF45) | 55 (40S) [73] |
| Foot-and-mouth disease virus (FMDV) | Picornaviridae (Aphthovirus) | ||||
| III | Hepatitis C virus (HCV) | Flaviviridae (Hepacivirus) | 5′ UTR | (eIF1A), eIF2/eIF5B, eIF3 |
1.9 (40S) [74] 35 (eIF3) [74] |
| Classical swine fever virus (CSFV) | Flaviviridae (Pestivirus) | ||||
| Seneca valley virus (SVV) | Picornaviridae (Senecavirus) | ||||
| Porcine teschovirus 1 (PTV-1) | Picornaviridae (Teschovirus) | ||||
| Porcine enterovirus 8 (PEV8) | Picornaviridae (Enterovirus) | ||||
| IV a | Cricket paralysis virus (CrPV) | Dicistroviridae (Cripavirus) | IGR | none (eEF2) |
22-26 (40S) [75] 8 (80S) [76] |
| b | Taura syndrome virus (TSV) | Dicistroviridae (Aparavirus) | none (eEF2) | 25 (80S) [76] | |
| c | Halastavi arva virus (HalV) | not assigned | none | – |
Notes. Typical representatives (the viruses from which these elements were isolated), location in the gRNA, factors and ITAFs required for translation initiation, as well as equilibrium dissociation constants (Kd) for some of them are indicated.
* The names of viruses with the most well-characterized IRESs of this type are highlighted in bold.
** Requirements for initiation factors and ITAFs are deduced primarily from experiments on reconstruction of initiation complexes from purified components, and may be specific to this particular system; names in brackets denote factors that either stimulate the 48S assembly (eIF1A, eIF4B, PTB), or are not absolutely necessary for all representatives of this type (ITAF45), or are needed for additional purposes – for example, choosing the correct start codon (eIF1), transition to elongation (eEF2) or translation in other cell-free systems (some ITAF).
Classic type I IRESs. Internal initiation of translation of eukaryotic mRNA was first demonstrated in the late 1980s using the IRES of poliovirus (poliovirus, PV, family Picornaviridae) [62, 77], which epitomizes type I IRESs. Representatives of this group were found only in the 5′ UTR of gRNAs of some picornaviruses, and PV IRES is the most studied among them (Fig. 2a).
Like in other picornaviruses, PV gRNA is not capped at its 5′ end but rather covalently linked to VPg (which, unlike the above described cases, is not involved in translation initiation). The IRES is about 650-nt long and occupies most of the ~740 nt 5′ UTR. This region contains several structural domains (II-VI) necessary for IRES activity [78, 79], followed by the weakly structured 160-nt region and the start codon AUG743, 13th from the 5′ end (Fig. 2a). At the base of domain VI is an oligopyrimidine tract (Yn) containing the conserved UUUCC sequence. Other representatives of this type of IRESs – such as those present in the gRNA of enterovirus A71 (enterovirus EV-A71), Coxsackievirus type B (Coxsackievirus B3, CVB3), and human rhinovirus A (human rhinovirus A2, HRV A2) – have a similar structure (for details, see reviews [56, 59, 80]).
A puzzling feature of this type of IRES is the presence of a “cryptic” AUG-codon (AUG586 in the case of PV) located inside domain VI, 18-20 nt downstream of the Yn motif. This codon is important for the efficient operation of the IRES [81]. It can be recognized by the initiation complex, however, due to the suboptimal nucleotide context, this AUG is not the main start codon for viral polyprotein synthesis. The authentic start codon, AUG743, lies more than 100 nt downstream of domain VI. In the case of HRV, a similar pair is formed by AUG589 and AUG626, located opposite each other near the base of the hairpin of domain VI. In different viruses, these two AUG codons can be located in the same or in different reading frames.
The results of mutagenesis experiments show that the Yn motif, the cryptic AUG codon, and the fixed-length spacer between them form a combined functional Yn-Xm-AUG module, which is important for the efficient operation of the IRES and, most likely, is the site of ribosome entry (see [81] and references therein). The Yn-AUG tandem is also typical for other types of picornavirus IRESs, but it is not always strictly necessary (see below). Particular interest in this structural element is due to the fact that it determines neurovirulence of the virus. Mutants of the highly neurovirulent mouse strain of poliovirus, in which the main initiator AUG743 was transferred to the location of the cryptic AUG of the tandem in a favorable context, showed a high degree of attenuation (decrease in pathogenicity) in experiments in mice. At the same time, these mutants largely retained the ability to multiply in cultured cells (including those of a neuronal lineage), and their RNA exhibited high translational activity in the cell-free system based on Krebs 2 ascites carcinoma cells, thereby indicating presence/absence in the cells of the central nervous system of a specific factor or factors that determine significance of the Yn-Xm-AUG module for poliovirus biology [82]. Interestingly, a recent study found [83] that in a number of enteroviruses AUG589 is not “silent”, but rather directs synthesis of the 65-aa-long peptide that affects the course of infection of intestinal epithelial cells. It is possible that some of the previously described effects of AUG586 mutations on the pathogenicity of the virus are associated with impaired synthesis of this peptide.
Another intriguing area of study is the poorly understood mechanism of ribosome relocation from the Yn-Xm-AUG region to the main start codon AUG743. Data on mutagenesis of the region between these codons, introduction of stable hairpins and additional AUGs suggest nonlinear scanning of this region, reminiscent of shunting [84, 85]. In the case of HRV, where two AUGs are located opposite each other in a stable hairpin, some ribosomes also relocate from AUG589 to AUG626 by shunting it [86].
An almost complete set of the canonical initiation factors, except eIF4E, is required for the functioning of type I IRESs (Table 2; [85]). Instead of binding eIF4E to the cap, the process begins with the interaction of eIF4G with the V domain of the IRES, and then the sequence of events is very similar to the standard for eukaryotes: eIF4G binds eIF3 and recruits the 43S pre-initiation complex, which then recognizes the AUG codon in the downstream region. There are, however, significant differences. First of all, the 40S subunit does not attach to the mRNA 5′ end but to the internal region (apparently, to the region of the cryptic AUG); it is possible that the direct affinity of IRES for the ribosome plays some role in this [73]. In addition, auxiliary proteins (IRES trans-acting factors, ITAFs), which do not take part in the canonical translation, are required for operation of these IRES.
Table 2.
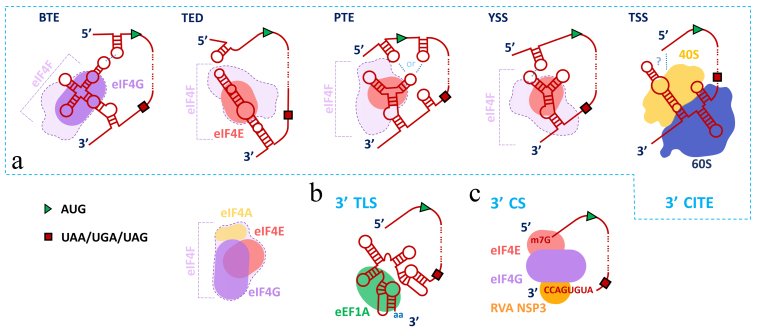
Types of 3′ end cap-independent translation enhancers (3′ CITEs) and their brief characteristics
| 3′ CITE type | Family (Genus) of viruses | Interaction with 5′ end of gRNA | Bind | Kd, nM |
|---|---|---|---|---|
|
BTE B YDV like T ranslation E nhancer |
Luteoviridae (Luteovirus); Tombusviridae (Necrovirus, Dianthovirus, Umbravirus*) | Yes (RCNMV ?)** |
eIF4G eIF4F |
177 [180] 37 [180] |
|
TED T ranslation E nhancer D omain |
Not attributed (Albetovirus); Tombusviridae (Carmovirus) | Yes (STNV ?)** |
eIF4F eIFiso4F eIF4E eIF4isoE |
17-30 [181] 33-50 [181] 660 [181] |
|
PTE P MV-like T ranslation E nhancer |
Tombusviridae (Panicovirus, Carmovirus, Aureusvirus, Umbravirus*) | Yes (PEMV-2 no) |
eIF4E eIF4F |
58 [182] 48 [182] |
|
ISS I-ShapedStructure |
Tombusviridae (Tombusvirus, Carmovirus, Aureusvirus) | Yes | eIF4F | 190 [183] |
|
YSS Y-ShapedStructure |
Tombusviridae (Tombusvirus) | Yes |
eIF4F eIFiso4F |
[184]*** [184]*** |
|
TSS T-ShapedStructure |
Tombusviridae (Carmovirus, Umbravirus*) | No |
60S 80S |
|
|
kl-TSS kissingloopT-ShapedStructure |
Tombusviridae (Umbravirus*) | Yes |
40S 60S 80S |
360 [186] 300 [186] |
Notes: * Assignment to Tombusviridae is provisional: umbraviruses contain two gRNAs and only one of them (which carries 3′ CITE) resembles other members of this family.
** In this case mutation analysis did not confirm the predicted kissing interaction with a 5′ proximal hairpin.
*** Factor binding was demonstrated by a non-equilibrium method.
As a rule, ITAFs are cellular RNA-binding proteins, which either help bind initiation factors or simply maintain the correct spatial IRES structure, thus functioning as RNA chaperones [56, 60]. Although binding to the type I IRES has been documented for dozens of different proteins, only a very small number of these interactions have clearly demonstrated functional significance. In particular, in the cell-free system reconstituted from purified components only poly(rC)-binding protein PCBP2 interacting with several sites in domain IV (see [85] and references therein) or its paralog PCBP1 were strictly required for the assembly of the 48S complex on the PV IRES out of the eight analyzed ITAFs. The efficiency of complex formation was somewhat enhanced by another ITAF, polypyrimidine-binding protein PTB/PTBP1, a classical RNA chaperone that facilitates recruitment of the eIF4G factor [87]. Other ITAFs, such as GARS [88], La/SSB [89], or UNR/CSDE1 [90], despite specific binding to regions of the IRES and stimulating its activity in other in vitro systems, did not affect assembly of the 48S complex in this experiment [85].
Nevertheless, the set of ITAFs and their interaction with individual structural regions of RNA appear to determine the tissue-specific activity of type I IRESs in vivo. This fact is important for viral pathogenesis. Thus, the effects of attenuating mutations in the internal region of the 5′ UTR of the Sabin live polio vaccine strains, which reduce affinity for the translation initiation factors [71, 91], are pronounced more in neural cells than in the cells of other lineages [92-94]. This difference, directly associated with pathogenesis of poliomyelitis, could be likely explained by the intercellular variations in the concentration or set of ITAFs or translational factors.
Another important aspect of pathogenesis is associated with the mechanisms by which viruses of this group provide translational advantage to their mRNA. In the early stages of infection, enteroviral protease 2A cleaves factor eIF4G, cutting off the eIF4E binding site from it, which entails suppression of translation of cellular mRNA (see reviews [56, 80]). PABP also undergoes degradation; however, the full-length protein disappears only by the time that the viral RNA needs to shift from a translation mode to one involving replication (picornavirus mRNAs are polyadenylated and PABP is used to stimulate translation [95]). Also, in the late stages of infection, protease 3C hydrolyses PTB and PCBP2, thereby suppressing activity of the IRES, and cleaves eIF5B (however, the proteolytic fragment thus obtained is in fact bigger than the deletion variants that are fully functional in vitro in the 80S assembly, therefore the physiological role of eIF5B proteolysis is unclear) [80].
During replication of RNA-containing viruses, which include poliovirus, a double-stranded RNA is synthesized and activates protein kinase R (PKR/EIF2AK2). This results in phosphorylation of eIF2α, but translation of the viral mRNA continues [96]. One possible explanation is that the eIF2α-specific subunit of PP1 phosphatase, CReP/PPP1R15B, is capable of retaining active eIF2α on the membrane of the endoplasmic reticulum, where translation of the viral mRNA occurs. This physically protects translational complexes from inactivating kinases [97].
Classic type II IRESs. IRESs of this type were discovered in the pioneering study by the E. Wimmer group in 1988 [61]. As classic representatives of this type, structures in the 5′ UTR of two picornaviruses: encephalomyocarditis virus (EMCV belonging to the genus Cardiovirus) and foot-and-mouth disease virus, (FMDV, a member of the genus Aphthovirus) are usually considered. These IRESs have practically the same length (about 450 nt) and very similar domain organization (domains II-V/VI or, according to another nomenclature, domains H-K/L [98], see Fig. 2b), however, they differ in the location of their start codons. In addition, some aspects of the biology of IRESs of this type have been studied in detail using the example of another cardiovirus, Theiler’s murine encephalomyelitis virus (TMEV).
Similar to the representatives of the previous group, type II IRESs contain a high-affinity eIF4G binding site located in domain IV (J-K) [99]. This way of attracting eIF4G makes the translation independent of the 5′ cap and eIF4E. However, it is important to understand that the involvement of initiating factors alone is not a sufficient condition for internal initiation. For example, when only the J-K domain is introduced into the 5′ UTR or 3′ UTR of the reporter mRNA, its translation almost completely ceases to depend on the cap, but the ribosome can still be bound exclusively to its 5′ end [100]. This example clearly shows that the picornavirus IRES has a modular structure, in which the J-K domain can be considered as a kind of CITE (see below), and other domains are required for mRNA placement into the channel.
Landing of the ribosome occurs, as in the case of PV, at the 3′ side of the IRES. In this case, the AUG of the conserved Yn-Xm-AUG module located at its border usually serves as an authentic initiation codon, and integrity of the Yn-AUG tandem is not critical for the overall activity of the IRES, yet it is important for neurovirulence (see below). Analysis of the influence of insertions and deletions introduced into this region of the TMEV IRES made it possible to formulate the concept of a “starting window” [101]. According to it, IRES places the 43S initiation complex at a specific region of mRNA, after which it can either recognize the AUG codon inside this region, or, if there is no AUG there, start scanning and choose a starting point downstream. The rules for selection of the initiation codon within the starting window do not quite correspond to those in standard scanning. On the one hand, the nucleotide context plays the same role here; on the other hand, the probability of AUG recognition greatly increases from the 5′ to the 3′ terminal boundary until it reaches a plateau [101]. This difference is clearly visible when comparing the pattern of 48S complex distribution between the AUG codons in the EMCV initiation region in two cases: when translation is directed by the IRES and when most of the IRES is removed and ribosomes scan the resulting mRNA directly from its 5′ end [8, 102]. In the first case, the complexes are predominantly formed on AUG834 (11th in a row), which is the main start codon of the EMCV polyprotein, while the upstream AUG826 is almost not recognized by the ribosomes despite its good context, because it apparently lies close to the 5′ boundary of the starting window. On the contrary, when ribosomes enter the same region by means of cap-dependent initiation, then the opposite situation is observed in complete accordance with the prediction of the scanning model. It is pertinent to note that similar rules are also characteristic of the classical 5′ end-dependent translation initiation in the case of AUG codons located near the very 5′ end – this similarity is probably due to the common features for these two cases that arise during mRNA placement into the channel of the 40S subunits. The analogy is enhanced by the fact that in the reconstituted translation system the 48S complexes on AUG826 of EMCV can be seen in the absence of the eIF1, i.e. under the same conditions in which it is possible to see the complexes on the AUG located near the 5′ end on the cap-dependent mRNA [103].
There is another AUG in the initiation area of the EMCV mRNA, the 12th in a row (AUG846), which is in the same frame with AUG834. The 43S complex described above can slide onto it under certain conditions [8], but normally it is not used as an initiation codon. In contrast, FMDV has two functional start codons (also located in the same frame), separated by an extended 84-nt spacer, that give rise to two isoforms of the leader (L) protease, with the second AUG used more frequently [80]. Apparently, the pre-initiation complex assembles on the FMDV IRES in the vicinity of the first of these AUGs, after which it is either recognized or the eIF1-dependent scanning and recognition of the second AUG takes place (see [104] and references therein).
The requirements for canonical initiation factors for the type II IRESs in the cell-free system are basically the same as for the PV-like ones [105, 106]. However, ITAFs deserve special attention here, since in some cases they determine the biology of the relevant viruses – in particular, their ability to synthesize proteins and replicate in certain cell types, as well as neurovirulence in vivo. For example, all three above-mentioned IRESs (from EMCV, TMEV, and FMDV) require PTB for the assembly of the 48S initiation complex, but FMDV also requires an additional ITAF, the Mpp1/ITAF45/PA2G4 protein [105, 106]. PA2G4 is associated with the cell cycle and is present only in proliferating cells, while it is absent in neurons, which is probably why replacement of the IRES of the neurovirulent strain GDVII of TMEV with the FMDV IRES V leads to a complete loss of their ability to multiply in neural cells [105].
There are interesting cases of the relationship between mutations in the ITAF recognition sites and loss of the viral neurovirulence without losing the ability to reproduce and synthesize proteins in other types of cells. For example, destruction of the Yn-AUG tandem in TMEV by changing critical distance between its polypyrimidine block (which probably serves as one of the PTB binding sites [107]) and AUG does not significantly affect viral reproduction in cultured BHK-21 cell and cell-free translation [101], but sharply decreases neurovirulence in mice [108]. Other similar examples are also known (see discussion in [105, 109]). The mechanism of this relationship was established by studying dependence of TMEV neurovirulence on the interaction of its IRES with various forms of PTB. The cells of the central nervous system (in particular, neurons) are deficient in the PTBP1 protein, but they produce its neuron-specific paralog – nPTB/PTBP2. Both forms of PTB bind to the same TMEV IRES regions and exhibit a comparable ability to stimulate translation. However, some mutations in PTB-binding motifs reduce the affinity of IRES to the nPTB significantly more than to the “normal” PTB. These mutations significantly reduce neurovirulence of the virus without significantly affecting its translational activity and reproduction in other cells [109].
Functional properties of the cis-elements of 5′ UTR of IRES-dependent viruses also affect the nature of clinical symptoms of the diseases they cause. The engineered TMEV mutants can cause either lethal tetraplegia or mild neurological disorders, depending on the context of the AUG codon in the starting window [110]. These examples illustrate how the peculiarities of non-canonical mechanisms of translation initiation of viral RNAs, related to the structure of the corresponding cis-elements and the variety of cellular factors interacting with them, can determine key aspects of the pathogenesis of viral diseases.
Pathogenesis of picornaviruses with type II IRESs is, of course, also associated with their mechanisms of suppression of cellular translation. For example, FMDV encodes two proteases cleaving eIF4G: 3C and L [80]. EMCV does not encode enzymes capable of cleaving this factor; however, upon infection, the cellular repressor protein 4E-BP1 is activated [111], leading to inhibition of cap-dependent translation and giving priority to the viral mRNA.
In connection with the described strategies of eIF4F repression, it is appropriate to mention one more type of picornavirus IRESs harbored in the 5′ UTR of hepatitis A virus (HAV). Despite the clear similarity with the picornavirus type II IRESs, the HAV IRES has long been placed in a separate group, since it requires the full-component eIF4F factor, including both eIF4E and intact eIF4G, for its operation [112]. In addition, its domain V, which binds eIF4G, differs in primary structure from the corresponding (J-K/IV) domains of EMCV and FMDV. However, subsequent studies have shown that the spatial structures of these domains are similar [113]. As for the dependence on eIF4E, it turned out that in the case of other picornavirus IRESs, eIF4E has a positive effect on the affinity of eIF4G and helicase activity of eIF4A [114], thus the peculiarity of HAV IRES was, rather, in the degree of the eIF4E requirement. In any case, proteolysis of eIF4G is not required for the functioning of IRESs of types I and II: viral proteases have not yet been synthesized at the early stages of infection, therefore intact eIF4F is used to attract ribosomes.
Classic type III IRESs. IRESs of this type are present in the mRNAs of several families of viruses: Flaviridae, Picornaviridae, and, possibly, individual representatives of Dicistroviridae. The characteristics of the elements of this type were best studied for the IRES from the 5′ UTR of the hepatitis C virus (HCV), a flavivirus (Fig. 2c) [115]. Translation initiation directed by this ~330-nt long 5′ UTR does not include a scanning step in contrast to the mechanisms described above. The IRES binds the 40S ribosomal subunit directly [116] with the AUG codon in the immediate vicinity of the P-site of the small ribosomal subunit. The larger domain III binds to 40S from the side facing the solution and interacts with both ribosomal proteins and rRNA, while domain II is located in the region of the E-site (see [117-120] and references therein). In addition to the 40S subunit, HCV IRES is also able to bind the eIF3 factor [121], although stability of the RNA•40S complex (Kd = 1.9 nM) is much higher than that of the RNA•eIF3 (Kd = 35 nM) [74]. Early in vitro experiments showed that only the factors eIF2 and eIF3 are fundamentally required for translation initiation on HCV mRNA [116]. eIF1A helps stabilize Met-tRNAi at the P-site [122]. Additional proteins (ITAFs) are optional, although some of them may be capable of enhancing translation directed by the HCV IRES (see review [123]). A recent study demonstrated an important role of the modified nucleotides (m6A) as well as of the m6A-binding protein YTHDC2 in the activity of the HCV IRES [124].
Later, the classical eIF2-dependent pathway was found not be the sole option for Met-tRNAi delivery. It was demonstrated that translation directed by HCV IRES could occur even when functional eIF2 was not available (phosphorylated under conditions of cellular stress or in the presence of specific inhibitors) [125, 126]. It was shown in vitro that the delivery of Met-tRNAi to the 48S complex on HCV-like IRESs is possible using eIF5B, the ortholog of bacterial IF2 [126, 127], and this was later confirmed by structural studies [128]. A functional initiation complex on HCV IRES can also be assembled from purified components using the 40S recycling and translation reinitiation factors: eIF2D or MCTS1•DENR dimer [22, 129, 130]. However, knockout of the EIF2D gene does not lead to loss of eIF2-independence of HCV IRES [122, 131], which may indicate that this factor is most likely not involved in the initiation of HCV mRNA translation in living cells (although for a more correct experiment eIF2D should be removed simultaneously with one of the components of the MCTS1•DENR dimer, since these factors are interchangeable [22, 132]). In a study by Kim et al. [133], a Met-tRNAi-delivering activity during initiation of HCV mRNA translation was also attributed to the eIF2A factor, although in the direct experiment on assembly of the initiation complex it was not active [129], and knockout of its gene did not lead to loss of resistance to inactivation of eIF2 [122, 131]. Moreover, the pre-initiation complex with Met-tRNAi on some IRESs of this type can be obtained without the participation of translational factors, as described for the IRES of simian picornavirus type 9 (SPV9) [134]. Such a complex was also obtained on the HCV IRES, but only at a nonphysiologically high concentration of Mg2+ [135], and this pathway is probably extremely ineffective [126].
Initially, it was assumed that the position of eIF3 in the initiation complex on type III IRES coincides with its position in the analogous complexes formed during the cap-dependent translation. However, structural studies have shown that the binding sites of IRES and eIF3 on the surface of the 40S subunit overlap. This suggests different orientation of eIF3 in the initiation complexes formed on HCV-like IRESs. Thus, eIF3 does not contact the 40S subunit at all in the complex of 40S, eIF2, eIF3, and DHX29 with the CSFV IRES devoid of domain II [120]. It was therefore suggested that the main purpose of the IRES binding to eIF3 is to displace the latter from the 40S subunit to gain access to the ribosome surface and reduce formation of the canonical 43S complexes, thereby favoring translation of viral mRNAs. Additional experiments should show whether this is really the case. Note, however, that the IRESs of HCV and CSFV lacking domain II are unable to form an 80S complex [127, 136], and the eIF5B-dependent mechanism of the initiator tRNA delivery requires participation of eIF3 [126, 127].
Of particular interest is the ability of this type of IRESs to capture (hijack) the translating ribosome. Single molecule spectroscopy and cryoelectron microscopy have shown that HCV IRES is able to bind a ribosome that translates another mRNA or the same mRNA in which it is located. IRES firmly binds to the platform of the 40S subunit and, presumably, remains in this state until the moment of termination. After the release of the synthesized peptide and disassembly of the ribosome, domain II of the IRES is folded into the E site, directing HCV RNA to the mRNA-binding channel [137]. This “reservation” of the ribosome probably helps viral RNA achieve translational dominance in the infected cell.
In addition to Flaviviridae, HCV-like IRESs have been also found in some groups of Picornaviridae (see review by Arhab et al. [59] for details), which confirms the existence of horizontal transfer of not only genes but also of individual regulatory elements of viral RNA.
Classic type IV IRESs. The aforementioned types of IRES require participation of at least some initiation factors to ensure internal landing of the ribosome. However, there are IRESs that do not require initiation factors, auxiliary proteins such as ITAF, or even initiator Met-tRNAi for their function [138]. Translation directed by these elements does not start with methionine [138, 139]. Such IRESs are characterized by a small length (~200 nt) and have so far been found only in representatives of the Dicistroviridae family, where they are located in the intergenic region (IGR) of gRNA [56]. Independence from initiation factors and interaction with the highly conserved intersubunit region of ribosomes allows these IRESs to initiate translation in heterologous systems – for example, cells (and their extracts) of mammals, insects, plants, protozoa, yeasts, and even bacteria [58] – which is completely uncharacteristic, for example, of type I, II and III IRESs.
At the moment, three subgroups of type IV IRESs are known. The classic representative of the subtype IVa, as well as of the whole type, is the IGR IRES of the cricket paralysis virus (CrPV), which is responsible for translation of the second cistron of its gRNA (Fig. 2d). Three domains of this IRES, containing pseudoknots (see [140] and references therein), directly bind the ribosome and functionally replace tRNA and translation factors [138, 141, 142] allowing assembly of an elongation-competent 80S ribosome, thus bypassing classical stages of initiation. The details of this mechanism were elucidated using cryo-electron microscopy: domain 3 of the CrPV IRES (and other IRESs from similar viruses) bind to the A-site of the ribosome. At the same time pseudoknot PKI, a component of the IRES, mimics tRNA in a codon-anticodon interaction with mRNA [141, 143]. During pseudotranslocation the elongation factor eEF2 promotes domain 3 movement to the P-site, freeing the A-site for eEF1A-dependent Ala-tRNAAla landing (GCU is the first coding codon of the second CrPV cistron) [75, 141, 144, 145]. Synthesis of the polypeptide chain begins after the next (second) act of translocation, immediately proceeding to elongation, with the alanine becoming the first amino acid residue in the protein [138]. It was shown by FRET that the CrPV IRES is able to bind both to the free 40S subunit and to the fully assembled 80S ribosome [75].
Subtype IVb is represented by the IGR IRESs of Taura syndrome virus (TSV), red fire ant virus (Solenopsis invicta virus 1, SINV-1), and honey bee paralysis virus (HBPV). They are distinguished from the IRESs of subtype IVa by the presence of an additional hairpin structure (SLIII) in domain 3, the role of which is not yet completely clear. Removal of this hairpin, however, does not prevent binding to the 80S ribosome or translocation activity of eEF2, but makes productive translation impossible [76, 146]. Structural studies show that SLIII is involved in mimicking tRNA and interacts with 28S rRNA [143]. In all likelihood, SLIII is necessary for the correct positioning of the IRES on the ribosome, but it may somehow affect translocation as well [76, 143].
Features of the recently characterized mechanism of translation initiation on IGR IRES of the Halastavi árva (HalV) virus isolated from the intestinal contents of freshwater carp allowed assigning it to a separate subtype IVc. Its main difference from the CrPV is the inability to bind the free 40S subunit due to the absence of the functional domain 2. The HalV IRES binds the 80S ribosome using domain 1, which interacts with the 60S subunit, while domain 3 binds to the 40S subunit immediately in P-, and not in the A-site. As a consequence, initiation of HalV on IRES does not require eEF2-dependent pseudotranslocation, which makes it the simplest of all currently known translation initiation mechanisms [147].
In addition to translating the frame coding for envelope proteins, some IGR IRES are also capable of directing translation of the alternative (+1 and +2) reading frames. The mechanism and physiological significance of this process are not yet fully understood (see discussion in [148, 149]). The second interesting feature, inherent only to some of the mentioned groups of viruses, is a very stable 14-18-bp hairpin (SLVI) at the end of the first cistron, just before the start of the IRES. Probably, this hairpin helps regulate the flow of ribosomes “en route” to the IRES, preventing its unfolding [149].
Chimeric and unclassified IRESs. Speaking about Dicistroviridae, it should be noted that the mRNAs of these viruses are modified by VPg; therefore, translation of their first cistron also cannot be cap-dependent. It is directed by IRESs, and different viruses of this family use various strategies to attract ribosomes. The first strategy is based on a 5′ UTR that contains extended single-stranded regions capable of “nonspecifically” bind the 40S subunit in the presence of the eIF3 factor [150, 151]. Such unstructured sequences are usually enriched in uridine or adenine residues, as in the case of the 5′ terminal IRESs of the bird aphid virus (Rhopalosyphum padi virus, RhPV) and HalV, respectively. Only eIF2, eIF3, and the 40S subunit are strictly required for the formation of initiation complex; however, to find the start codon by limited scanning, eIF1 is also required, while both eIF1A and factors of the eIF4 group strongly stimulate assembly [150, 151]. Due to the fact that this mechanism does not use specific binding of mRNA to the components of the translational apparatus, the 5′ terminal IRES of RhPV allows initiation of translation in any eukaryotic system, from yeast to mammals.
The 5′ terminal IRES of another member of Dicistroviridae family, the already mentioned virus CrPV, apparently has a distant functional similarity with the HCV IRES, despite differences in their structures. It specifically binds the eIF3 factor, and in this case, this interaction is strictly necessary for the 40S subunit landing [152, 153]. Like HCV IRES, the 5′ terminal CrPV IRES interacts with the “optional” ribosomal protein RACK1, which explains the previously found dependence of the translation of the first but not the second CrPV cistron on this protein [154]. Nevertheless, detailed structural and functional analysis of the reconstructed complex of this IRES with purified 40S and eIF3 revealed a number of unique features [152]. Its three domains cover the “head” of the 40S subunit interacting with proteins and rRNA, and the single-stranded region following domain III is loaded into the mRNA channel. Addition of Met-tRNAi and either eIF2 or eIF5B leads to the formation of pre-initiation complex, in which the P-site contains not the start codon AUG709, but the preceding uAUG701 codon. For complex assembly on the AUG709 start codon, additional factors are required (eIF1 and eIF1A), and in this case eIF2 can no longer be replaced by eIF5B [152]. This suggests a local scan of the initiation region, similar to that of the above-described EMCV IRES. Interestingly, uAUG701 opens a small ORF, AUGUGA; therefore, during real translation, it is impossible to exclude the ribosome reaching the AUG709 start codon also as a result of reinitiation.
Another group of IRESs that do not fall into the above classification are “chimeric” IRESs found in some picornaviruses. In terms of domain organization, they are similar to types I and II; however, some domains in their composition are more similar to the corresponding regions of the type I IRESs, while others to domains of the type II elements [59]. Among the representatives of Flaviviridae, there are also viruses carrying IRESs of the picornavirus type with poorly characterized structure [59].
Due to currently insufficient information, the IRES detected in the Triticum mosaic virus, TriMV, has not yet received an unambiguous classification. The uncapped mRNA of this virus has an unusually large 5′ UTR (739 nt) for plant viruses, containing 12 uAUGs, which excludes efficient translation initiation using a scanning mechanism. Indeed, placing a stable hairpin at the 5′ end of the viral mRNA does not suppress translation. The mechanism of operation of this IRES is poorly understood, however it has been shown to bind eIF4G [155]. This, as well as the presence of polypyrimidine sequence important for translation in front of the initiation codon, possibly makes it similar to picornavirus IRESs of types I and II [156], but conclusion is not final yet.
Cases requiring further studies. Due to the great social significance of the human immunodeficiency virus 1 (HIV-1), much research has been devoted to the study of translation of its mRNA. HIV-1 mRNA, synthesized by the cellular RNA polymerase II from proviral DNA, is capped and polyadenylated. Alternative splicing leads to formation of several transcripts encoding various viral proteins, while unspliced (aka genomic) RNA encodes gag-pol. Its 5′ UTR is 335-nt long and contains a number of secondary structure elements necessary for viral replication, and a stable hairpin called TAR, located at the very 5′ end of the gRNA. Such hairpins are believed to effectively inhibit 5′ dependent translation initiation. On the other hand, presence of the 5′ cap and absence of uAUG-codons in the 5′ UTR speak in favor of the standard mechanism of translation initiation of this mRNA. Like picornaviruses, HIV-1 encodes a protease that is capable of cleaving eIF4G (as well as PABP) [157-160]. However, it acts only on one of the eIF4G paralogs, eIF4GI/eIF4G1, without affecting the second, eIF4GII/eIF4G3 [158, 159]. The data on the effects of this cleavage are contradictory: in some studies, it resulted in the suppressed translation of only cellular mRNAs, without affecting synthesis of the gag and translation of the reporter mRNAs with picornavirus IRES; in others, it negatively affected any mRNA (including reporters with the HIV-1 leader and picornavirus IRES), except those containing IRES HCV [157-159]. On the other hand, picornavirus proteases suppress translation of the HIV-1 mRNA, which is opposite to their effect on the classical IRES-dependent initiation described in the preceding sections.
Nevertheless, attempts have been made in a number of studies to show the presence of an IRES in the genomic HIV-1 mRNA [161-163]. Unfortunately, in most of them, the authors used bicistronic plasmids and RRL, and the main approach was to compare different bicistronic constructs with each other. In the only study to date that used mRNA transfection of cells, no significant contribution of internal initiation to translation directed by the 5′ UTR of HIV-1 was found [164]. The same result was obtained in a cell-free systems based on cultured cell lysates. It was found that the hairpin structure of TAR, which strongly suppresses translation in RRL [165], has little effect in living cells [164]. Introduction of an uAUG into the 5′ UTR of HIV-1 significantly suppressed translation of the viral mRNA, which is in better agreement with cap-dependent scanning than with internal initiation [164, 166]. At the same time, it cannot be ruled out that the translation of HIV-1 mRNA, in addition to the canonical initiation factors, involves some auxiliary proteins that facilitate scanning of the structured leader such as RNA helicases RHA/DDX9 and DDX3, the viral Tat protein or a component of the nuclear cap-binding complex CBP80/NCBP1 (see discussion in [164]).
An even more extravagant hypothesis proposes existence of an IRES within the HIV-1 and HIV-2 gag coding sequences [167-169]. The idea that an IRES could be located entirely within a coding region, where its structure would be constantly disrupted by translating ribosomes, seems somewhat doubtful, and the arguments in favor of this IRES are not highly convincing. To test this hypothesis, the authors used a leaderless mRNA, that is, mRNA that does not have a 5′ UTR at all [167-169]. This excluded any influence of the natural 5′ UTR on the translation. The authors proceeded from the premise that a leaderless mRNA cannot effectively initiate translation by the 5′ end-dependent mechanism. However, this assumption is at odds with the facts: such mRNAs in eukaryotic systems can use up to four different methods of translation initiation, including the classical one [170, 171]. Another premise of the authors is that initiation at the 5′ end AUG codon of a leaderless mRNA with a hypothetical IRES in the coding part has different requirements for the concentration of initiation factors than translation from the internal AUG codons [167]. However, similar differences have been described for the common cap-dependent mRNAs containing several start codons [14, 172]. Thus, despite the abundance of studies investigating translation initiation of mRNA of the retro- and lentiviruses (see review [173]), it cannot yet be stated that it occurs by some unusual mechanism.
The non-canonical mechanism of translation initiation is undoubtedly characteristic of mRNAs of the Flaviviridae family members: Zika virus (ZIKV), West Nile virus (WNV), yellow fever virus (YFV), and the aforementioned DENV. Although these mRNAs are capped, their efficient translation continues after inactivation of eIF4E and eIF4G, and the presence of a functional cap is not necessary [25, 174, 175]. According to some data, this property is completely determined by the 5′ UTR of viral mRNA [174, 175], while according to others, it also requires interaction of 5′ and 3′ UTR [25], yet the above-described cHP hairpin in the coding part is not required for this. Although in 2006 it was demonstrated [25] that the 5′ UTR of the DENV mRNA had no IRES activity, two recent studies [174, 175] dispute this statement. Taking into account the difficulties described above in the interpretation of the results obtained using bicistronic reporters, additional studies would help resolve this issue. There is no doubt, however, that these viruses employ an unconventional translation initiation mechanism.
Another insufficiently studied case is the polypurine IRES in sgRNA of the crTMV plant virus. It directs synthesis of the CP protein encoded by the second cistron of this bicistronic mRNA. It resembles the above-described IRESs from the 5′ UTR of RhPV and HalV viruses: RNA segments rich in adenine residues seem to form extended single-stranded elements that are capable of providing internal translation initiation not only in plants, but also in yeast and mammalian cells [176]. On the other hand, like all tobamoviruses, crTMV encodes a separate monocistronic sgRNA CP, and due to this, the bicistronic mRNA contributes only about 3% of the total synthesis of CP [177]. A similar assessment of the contribution of internal initiation was made for the unstructured IRES from the turnip wrinkle virus (TCV) [178]. The physiological role of this redundancy of CP synthesis remains to be elucidated.
STRUCTURAL ELEMENTS OF VIRAL 3′ UTRs INVOLVED IN TRANSLATION INITIATION
Positioning of translation enhancers at the very 3′ end of an mRNA, i.e., at the maximum possible distance away from the initiation site, is not as strange as it may seem. First of all, translation of viral gRNAs competes with their replication, and location of a translation enhancing sequence close to the 3′ end is an elegant solution to the problem: when RNA-dependent RNA polymerase (RdRp) starts negative strand synthesis, it almost immediately “melts” the structure of the 3′ proximal elements, inhibiting translation and, thereby, ensuring unobstructed genome replication. For example, it has been shown that binding of RdRp to 3′ UTR of the TCV gRNA caused irreversible structural rearrangements in the 3′ terminal element, simultaneously initiating replication and inhibiting translation of the viral RNA, which proceed in opposite directions [179].
In addition, (+)RNA viruses often employ sgRNAs, which are usually 3′ terminal fragments of gRNAs, as templates for protein synthesis. In some cases, translation of sgRNAs is regulated by the same 3′ proximal structural elements as translation of the full-length gRNA, providing genome compaction – the strategy commonly used by viruses.
In contrast to animal viruses, the majority of plant viruses require living host cells to spread to adjacent cells via interconnecting plasmodesmata. That is why complete suppression of cellular protein synthesis by direct competition or by destroying parts of the translation machinery is uncommon for this group. Generally, gRNAs of plant viruses are not more efficient templates compared to cellular mRNAs, but binding of the initiation factors and/or ribosomal subunits to their 3′ proximal structural elements helps them maintain optimal translational activity regardless of the level of cellular protein synthesis.
3′ terminal cap-independent translation enhancers (3′ CITEs). Most gRNAs of (+)RNA plant viruses lacking a 5′-terminal cap or VPg contain in their 3′ UTR the so-called cap-independent translation enhancers (3′ CITEs), structural elements that bind translation initiation factors and/or ribosomal subunits and functionally replace the cap structure. Nearly all plant virus 3′ CITEs participate in a kissing long-distance interaction with the apical loop of a hairpin proximal to the 5′ end of gRNA, mimicking circularization of capped cellular mRNAs mediated by the interaction of eIF4F with PABP.
3′ CITEs are classified into six types according to their well-defined secondary structures, namely barley yellow dwarf virus (BYDV)-like translation enhancer (BTE), translation enhancer domain (TED), panicum mosaic virus-like translational enhancer (PTE), I-shaped structure (ISS), Y-shaped structure (YSS), and T-shaped structure (TSS) (Fig. 3a; Table 2).
Fig. 3.
Main elements located in 3′ UTRs of viral mRNAs that enhance translation. a) Five main types of 3′ CITEs and components of translation apparatus associated with them. b) Aminoacylated 3′ TLS in complex with eEF1A. c) Rotavirus 3′ CS and components closing gRNA in a ring. For abbreviations see the text and Table 2.
All 3′ CITEs, except TSS, bind the initiation factor eIF4F, but with some variations. In the case of PTE and TED, and, probably, ISS and YSS, the eIF4E subunit serves as the main binding point. Genetic, biochemical, and structural data obtained for PTE, TED, and ISS imply their interaction with the cap-binding pocket of eIF4E, with a highly mobile guanine residue mimicking the cap. It is worth mentioning that while PTE binds eIF4E with high affinity, close to that of binding the complete eIF4F, TED interacts with eIF4E much less efficiently than with the complete eIF4F (difference in Kd is more than an order of magnitude). It is likely that in the latter case, structural rearrangement in eIF4E caused by the interaction with eIF4G could increase the strength of its binding to 3′ CITE in much the same way as it occurs upon binding to the 5′ end of a capped mRNA.
BTE is the only 3′ CITE that binds eIF4G directly. At the same time, its binding to eIF4F is about 5-fold stronger, which can be the result of conformational rearrangements in eIF4G caused by its association with eIF4E. It has been shown that a minimal fragment of eIF4G sufficient for 3′ CITE binding and for enhancing translation includes the binding sites for eIF4A and eIF3, and the RNA-binding domain, while regions interacting with eIF4E and PABP are not necessary [189].
It looks plausible that the ability of 3′ CITEs to bind tightly and hence sequester the factors of the eIF4 family is used by viruses not only to stimulate synthesis of their own proteins but also to suppress the cap-dependent translation of cellular mRNAs. It has been shown that RNA fragments containing 3′ CITEs of different types served as efficient inhibitors of in vitro translation of both viral and capped reporter mRNAs [190]. In addition, translation of a capped template in a cell-free system was significantly suppressed by the parallel translation of a viral gRNA containing PTE 3′ CITE [191].
Unlike the other types of 3′ CITEs, TSS directly binds the large ribosomal subunit, as well as the 80S ribosome, and, in the case of kl-TSS from the pea enation mosaic virus 2 (PEMV-2) gRNA, also the small ribosomal subunit. Structural analysis revealed that TSS forms a tRNA-like structure, which, however, cannot be aminoacylated, since it is not located at the very 3′ end of gRNA. However, TSS can compete with aminoacyl-tRNA for ribosome binding, and preliminary cryo-EM data have confirmed that TCV TSS enters the P-site of the 80S ribosome [192].
In one of the isolates of the melon necrotic spot virus (MNSV), a short (65 nt) translation enhancer was identified, probably the result of recombination with the 3′ UTR gRNA of the Asian variant of cucurbit aphid-borne yellows virus (CABYV, Xinjiang isolate) from the Polerovirus genus, Luteoviridae family [193]. Since the secondary structure of this element, two joint short hairpins, does not resemble the structure of any known 3′ CITE, it is sometimes considered as a separate type – CXTE (CABYV-Xinjiang-like Translation Element). The mechanisms behind the CXTE-mediated translation have not yet been studied in detail. Little is known about its interaction with the 5′ proximal region of the gRNA, as well as its requirement for translation factors, except that CXTE remains active in plants with suppressed eIF4E activity.
How factors immobilized at the 3′ end of the template stimulate initiation of translation remains unclear. The majority of accumulated data indicate that in this case, ribosome landing occurs at the 5′ end of the mRNA, followed by canonical scanning in search of the correct initiation codon. The necessity of a free 5′ end was confirmed by the fact that addition of a stable hairpin to the very end of the 5′ UTR caused complete blockage or strong inhibition of viral or reporter mRNA translation, both in vivo and in vitro [194, 195]. Scanning of the 5′ UTR was confirmed by conventional tests – introduction of additional AUGs [183, 195-197] or stable hairpins between the 5′ end and the initiation codon [195, 198]. In most cases, the priority AUG codon was the closest to the 5′ end, and internal hairpins inhibited protein synthesis.
However, it is necessary to mention an unusual 3′ CITE identified in both gRNAs of the blackcurrant reversion nepovirus (BRV) that promotes efficient cap-independent translation by interacting with the 5′ UTR but also can stimulate internal initiation in protoplasts even when the 5′ UTR sequence is inserted between two reporter ORFs [199]. Hence, this 3′ CITE can be considered as a part of a composite IRES.
It is still unclear whether 3′ CITEs bind only initiation factors or whether they can assemble a 40S-containing pre-initiation complex and transfer it to the 5′ end. It has been shown that the weak binding of the 40S subunit to BYDV BTE is enhanced by the addition of eIF4F, eIF4A, eIF4B, and ATP [200], indicating formation of pre-initiation complex on this 3′ CITE. However, the prevailing view is that it is the helicase activity of initiation factors that enhances direct binding of the 40S to the BTE region complementary to 18S rRNA [189]. Recently, it was shown that the PEMV-2 gRNA with a point mutation, that prevented long-range kissing interaction and blocked transfer of the initiation components from the 3′ to the 5′ end, quantitatively bound the 40S subunit in WGE [191], while the gRNA with deleted PTE 3′ CITE did not. This is direct evidence that PTE, in addition to initiation factors, also binds the small ribosomal subunit (supposedly through a chain of interactions).
In contrast to the other types of 3′ CITEs, TSS binds the large ribosomal subunit and the whole 80S ribosome. Unlike initiation factors of the eIF4 family employed by the rest of 3′ CITEs, 60S subunits are not deficient components of the translation apparatus. Immobilization of the 60S subunits on viral gRNA to ensure its efficient translation in a competitive environment does not seem very useful. Sequestration of the 60S subunits to suppress translation of cellular mRNAs also cannot be effective, due to the relatively small amount of viral RNA in the cell. Moreover, most viral gRNAs containing TSS do not have structures facilitating interactions between the 5′ and 3′ ends, i.e., there is no mechanism for the delivery of bound subunits or ribosomes to attend translation initiation. It has been suggested based on the indirect data, that the 80S ribosome binds 5′ UTR and 3′ TSS of TCV gRNA simultaneously, closing the RNA loop [201]. However, it remains unclear how the bound ribosome can be included in the initiation process. It can be assumed, that a more or less traditional mechanism is used based on the fact that translation of the reporter RNA with 5′ and 3′ UTR of TCV gRNA was considerably suppressed in the cells deficient in the initiation factor eIF4G [202].
Apparently, precise relative positioning of 5′ UTR and 3′ CITE is not very important for efficient initiation of translation or selection of the proper start codon. Extension of the gRNA 5′ UTR [195, 196] or change in the position of the hairpin mediating the end-to-end kissing interaction [196, 197] do not dramatically affect the level of protein synthesis. In the only known case (PEMV-2 gRNA), when the 3′ terminal site of long-range kissing interaction is not a part of the main PTE 3′ CITE that binds eIF4E (eIF4F) but is a part of the separate auxiliary 3′ CITE (kl-TSS), transfer of the interaction point to PTE does not significantly change the gRNA translation level in vitro [191]. These observations suggest that neither recognition of the 5′ end nor positioning of the initiation complex on a specific AUG codon is determined by the geometry of the 5′ UTR-3′ CITE interaction. In other words, 3′ CITE only provides spatial convergence of the initiation factors (or pre-initiation complexes) with the mRNA 5′ end reducing total entropy of the process. At the same time, initiation at the second AUG is prevalent on the sgRNA of the same PEMV-2, where the 3′ CITE-dependent synthesis of two auxiliary proteins starts from closely spaced AUG codons located near the hairpin providing long-range interaction. Relative efficiency of the synthesis of these two proteins depends on the distance between the AUG codons and their position relative to the point of 5′-3′ interaction [195]. It is likely that under certain conditions interaction of the initiating 40S subunit with the RNA chain via bound 3′ CITE could determine its preferred position during scanning and thereby ensure recognition of the proper AUG codon.
It is obvious that scanning of 5′ UTR or translation of the beginning of a reading frame should destroy the RNA secondary structure and lead to the loss of long-range interactions between the 3′ CITE and the gRNA 5′ end, intermittently shutting down initiation of translation. It is assumed that such dynamic “oscillating” character of initiation is a sort of regulation, which provides the level of translation optimal for virus fitness [196, 197]. Indeed, it was shown that loading of the in vitro translated PEMV-2 gRNA with ribosomes is low compared to a capped mRNA, being limited to 1-3 ribosomes [191, 195]. This means a relatively low frequency of ribosome recruitment, which may be a consequence of the periodic blockage of translation initiation. It has been suggested [191] that such a “disperse” distribution of ribosomes on gRNA is important for the efficient synthesis of RdRp, which is provided by programmed frameshifting caused by three sequential structural elements located near the point of the ORF shift [203]. It is clear that translation of gRNA will melt these structural elements and their restoration may require significant spatial and temporal gaps between the translating ribosomes.
tRNA-like structures (TLSs). Specific elements structurally and functionally resembling tRNAs (tRNA-like structures, TLSs – Fig. 3b) were found at the 3′ end of the gRNAs of different plant viruses belonging to eight genera of three different families. All TLSs contain a pseudoknot that allows formation of an analog of the aminoacylated acceptor stem without the involvement of the RNA 5′ end. TLSs possess three key features of tRNA: they serve as a substrate for cellular CCA-nucleotidyltransferase, can be aminoacylated by specific aminoacyl-tRNA synthetases, and form a ternary complex with eEF1A•GTP.
Three main types of TLSs have been described: those aminoacylated by valine (typical for the members of the Tymoviridae family), histidine (Viraviridae), and tyrosine (Bromoviridae). Each of these types possesses characteristic primary and secondary structures that provide specificity of aminoacylation. Although the ability of TLSs to be aminoacylated and to bind elongation factor implies involvement of these structural elements in translation, so far only their role in initiation of (-)RNA strand synthesis by the viral replicase, and a telomere-like function mediated by nucleotidyltransferase have been demonstrated convincingly.
TYMV TLS is the most studied example of the tRNA-like translational enhancer. TYMV gRNA is capped, and simultaneous capping of the 5′ end and aminoacylation of the 3′ end is crucial for its efficient translation [204]. Since the ability to bind eEF1A•GTP is required for translation enhancement, and specificity of aminoacylation is not important, it was concluded that the enhancement effect is mediated by the elongation factor eEF1A. Crystal structure of the TYMV TLS confirmed similarity of its surface with the tRNA side interacting with aminoacyl-tRNA synthetase and factor eEF1A [205]. Recently, it was shown that the TYMV TLS can bind the ribosome [205] (albeit using prokaryotic 70S ribosomes only). Given the above, the question arises: do aminoacylated 3′ TLS provide the main function of aminoacyl-tRNA – inclusion of an amino acid into the synthesized protein? After the discovery of TLS aminoacylation, this function seemed obvious and efforts were made to prove it. Although several studies have shown transfer of a radioactively labeled amino acid from TYMV TLS to the synthesized protein, other studies have not confirmed these results, and later the authors themselves admitted that these results were artifacts caused by the transfer of amino acid to the corresponding tRNA (see discussion in [206]). The story repeated itself a quarter-century later, when, based on the data obtained in vitro, the Trojan horse mechanism was proposed to explain efficient initiation at the second initiation codon of TYMV gRNA, suggesting direct recognition of this codon by the ribosome associated with Val-TLS and the start of protein synthesis with N-terminal valine [207]. However, independent examination carried out with the same objects and in the same translation system did not confirm the results [208]. Evidence was provided that in this case, an unusual variant of leaky scanning takes place (see above) [12] and that the initiation of translation at the second initiation codon depends on the presence of 5′ cap, and not TLS [208]. To sum up, direct involvement of TLS in translation as a functional analog of aminoacyl-tRNA remains an interesting hypothesis that has not yet been convincingly proven.
Another example of a relatively well-studied translation enhancer is TLS of brome mosaic virus (BMV) [209]. Mutations disrupting the TLS structure led to reduced efficiency of the BMV gRNA translation in vitro [210]. In this case, aminoacylation of TLS (with tyrosine) was also important for the enhancer activity, suggesting a similar mechanism of BMV and TYMV TLS action, although the details remain unclear so far. Synergism in the action of the 5′ cap and 3′ TLS suggests that the TLS-bound eEF1A somehow interacts with the initiation factor eIF4F, substituting for the canonical chain of interactions eIF4F•PABP•poly(A). On the other hand, binding of the aminoacyl-TLS to the ribosome [205] indicates the possibility of the same translation enhancement mechanism as in the case of 3′ CITE TSS, for which the binding of ribosomes was also postulated due to mimicry of the P-site tRNA.
At this time, it is not clear whether translation enhancement is a common and necessary property of all aminoacylated TLSs. Some data raise doubt about this. For example, the TMV tRNA-like structure, similar to the TYMV TLS, has no influence on translation [211], and TLS of the peanut clump virus (PCV) does not bind eEF1A•GTP [212].
Aminoacylated 3′ TLSs have also been found in the RNA of some animal viruses, for example, members of tetraviruses (Tetraviridae) that infect insects [213, 214]. However, these structures have hardly been studied yet.
Elements functionally replacing the poly(A)-tail. (+)RNAs of all group A rotaviruses (RVA) contain the GACC consensus sequence at the 3′ end (3′ consensus sequence, 3′ CS – Fig. 3c), which specifically binds the N-terminal domain of the viral non-structural protein 3 (NSP3) [215]. The C-terminal domain of this protein interacts with eIF4G [216] competing with the poly(A)-binding protein. NSP3 is a very efficient enhancer of viral translation [217] supposedly substituting for PABP in the formation of the cyclic structure of viral RNA. In addition, NSP3 is a potent inhibitor of translation of cellular mRNAs, since displacement of PABP from the complex with eIF4G prevents circularization of polyadenylated templates [217].
A similar mechanism is realized during translation of the AMV gRNA. 3′ UTR forms a pseudoknot structure resembling TLS and is recognized by the viral replicase and tRNA-specific enzymes. However, this tRNA-like structure is destroyed upon binding of the viral coat protein (CP), causing blockage of the synthesis of (-)RNA strands and leading to increased translation [218, 219]. CP also binds eIF4G or eIFiso4G [220] mimicking the eIF4G•PABP•poly(A) interaction, providing cyclization of viral gRNA, and stimulating its translation.
A hairpin at the 3′ end of DENV gRNA [221] and a structural element containing several pseudoknots within the 3′ UTR of TMV gRNA [211] are capable of replacing 3′ poly(A) upon translation of capped reporter mRNAs. The 3′ UTR of HCV also can nonspecifically increase the rate of mRNA translation, regardless of the translation initiation mechanism [222]. Thus, these viral elements can be considered as functional equivalents of the polyadenylated mRNA 3′ end.
REINITIATION EVENTS DURING VIRAL mRNA TRANSLATION
Viral mRNAs containing several separate or partially overlapping reading frames can use the translation re-initiation mechanism to deliver ribosomes to the distal ORFs. Usually, the eukaryotic ribosome is not prone to reinitiation after reading full-length protein-coding frames, since, due to the monocistronic nature of most mRNAs, evolutionary pressure acts against aberrant reinitiation on the 3′ UTR [223]. However, there are at least two exceptions to this rule. The first is re-initiation after translation of the uORFs located in the 5′ UTR of many mRNAs. Its effectiveness can vary greatly depending on the length of the uORF, distance from the stop codon to the main frame, and presence of the cis-acting elements in the 5′ UTR. The second exception involves cases when, due to specialized mechanisms, effective reinitiation becomes possible after reading the full-length frame, and this pathway is primarily utilized by viral mRNAs.
TURBS-mediated translation reinitiation. For the synthesis of several proteins from one RNA, some viruses use a special mechanism of termination-reinitiation on partially overlapping frames [224], translation of which is coupled. In places where such frames overlap, variants of the mutual arrangement of the start and stop codons are possible, some of which are shown in Fig. 4a. Such reinitiation has been well studied for the representatives of the Caliciviridae family (rabbit hemorrhagic disease virus, RHDV, as well as the previously mentioned FCV, MNV, and WNV) and slightly less for the influenza B virus (IBV) from the Orthomyxoviridae family. Thus, one of the RHDV sgRNAs encodes the main capsid protein VP1 in the first frame and the small capsid protein VP2 in the second. The mechanism of coupled termination-reinitiation apparently allows maintaining optimal stoichiometry of the two proteins, contributing to the correct assembly of the capsid.
Fig. 4.
Viral mRNA translation mechanisms based on effective reinitiation. a) Examples of overlapping regions of translationally coupled frames in the viral mRNAs containing TURBS. b) Mechanism of operation of TURBS. c) Two mechanisms used by pgRNA CaMV: on the left – shunting mediated by reinitiation, on the right – TAV-mediated reinitiation. Start and stop codons are designated as in Fig. 2 and 3.
This mechanism is based on “forcing” the ribosome, which terminated at the stop codon of the first frame, to reinitiate translation instead of dissociating from mRNA (recycling) (Fig. 4b). It requires the presence of a special structural element termed TURBS (termination upstream ribosomal binding site), that binds the ribosome at the end of the first frame shortly before the stop codon [225-227]. TURBS contains a conserved motif 1 (m1), complementary to the loop in the h26 hairpin of the 18S rRNA, and motif 2 (m2/2 *), which forms a hairpin structure with the loop containing m1 [228, 229]. As a result of interaction of h26 with the UGGGA region common for m1 caliciviruses and IBV, some of the 40S subunits after termination and disassembly of the post-termination complex remain associated with mRNA. This allows them to reinitiate if a start codon is available in the vicinity. Thus, in a sense, TURBS provides an internal initiation of translation; however, unlike IRESs, it is able to operate only with the previously attracted ribosome, i.e., when there is no need to re-fold mRNA into the channel of the 40S subunit.
The concepts of what translational factors are required for TURBS-mediated reinitiation are controversial. In the early studies, it was shown that eIF3, like 40S, is able to bind directly to the FCV TURBS in the in vitro RRL-based system. On this basis, it was suggested that eIF3 plays a key role in this process and is strictly necessary for effective reinitiation by this mechanism [225]. This was in agreement with the important role of this factor in reinitiation on some cellular mRNAs containing uORFs [223]. However, later, it was shown in an in vitro reconstituted system that reinitiation on RHDV and human norovirus (HNV) mRNAs is possible without participation of eIF3, although its addition enhances the effect [230]. The authors also showed that reinitiation is possible in several scenarios. In one case, Met-tRNAi, eIF1, eIF1A and eIF2 are required; another variant requires Met-tRNAi and eIF2D; the third mechanism requires only Met-tRNAi for initiation and involves the ribosome not dissociated into 80S subunits [230]. Nevertheless, in other studies of TURBS found in RHDV, HNV, and IBV, reinitiation in the presence of eIF3 was efficient even on the mRNA with disrupted m1 TURBS [231, 232] leading to a conclusion that binding of eIF3 to TURBS is sufficient to maintain post-termination 40S subunit on mRNA.
Differences in the observed mechanisms of reinitiation can be explained by the fact that the process occurs slightly differently for different viruses, and concentrations of the initiating factors, magnesium and potassium ions used in the in vitro reconstituted system, may differ from the values in the complete lysate. Nevertheless, based on the above data, it can be assumed that the key role in TURBS-mediated reinitiation is played by the binding of m1 with the 40S subunit, although eIF3 can enhance, stabilize, or even replace this interaction, and also, possibly, participate in the regulation of reinitiation depending on the state of the cell or stage of the virus life cycle.
In addition to caliciviruses and IBV, other cases of effective reinitiation on overlapping frames have been described: for example, in mRNA of the HvV190S virus of the parasitic fungus Helminthosporium victoriae, human respiratory syncytial virus (HRSV), and the prototypic hypovirus (Cryphonectria hypovirus 1, CHV1-EP713) [223]. The TURBS motif was not detected in the RNA of these viruses; however, other poorly characterized regions of the nonconserved secondary structure were identified that have complementarity with 18S rRNA.
Shunting mediated by reinitiation. The 35S pgRNA of the CaMV pararetrovirus uses another unusual method of reinitiation. This mRNA has a very long 5′ UTR containing an extended internal region with complex secondary structure, which includes a stable stem at the base and branched hairpins in the distal part. This region contains several non-functional AUG codons, and a short uORF encoding a 6-residue peptide located immediately in front of the stem [26]. In a series of studies carried out by T. Hohn and his followers, it was shown that this variant of shunting requires translation of the uORF, termination at its stop codon and release of the peptide, after which the ribosome “skips” the entire region with a stable secondary structure and re-initiates on the ORF VII frame encoding the P7 protein [34]. Thus, in this case, shunting is not, in fact, nonlinear scanning (already described earlier), but acts as a special case of reinitiation (Fig. 4c, left). The authors introduced various RNA fragments into the region skipped by the ribosome including lengthy ones with containing a variable number of AUG codons, and in one of the experiments even introduced a break in the sugar-phosphate backbone – and all these modifications did not decrease the translation efficiency of ORF VII. The areas bordering this region received the figurative names of the “take-off” and “landing” sites for the ribosome, implying its detachment from the mRNA during the “jump”. Nevertheless, the present more widely accepted point of view is that the mRNA chain does not leave the channel of the small subunit during “jumping”; instead, the stable hairpin “threads” through the channel without unwinding. Such passage of an unwound hairpin through the canal can be observed in an in vitro reconstituted system in the presence of an incomplete set of initiation factors, in particular, in the absence of eIF1 [233]. Since recycling of the post-termination complex and subsequent reinitiation imply an under-characterized overlap of functions between the SUI1-domain containing factors eIF1, eIF2D, and DENR [132], the peculiarity of the 35S CaMV pgRNA may include the use of unusual combination of the factors in these post-termination processes.
CaMV is not the only virus using this unusual strategy. pgRNA of a number of other plant pararetroviruses (including most members of the Caulimoviridae family), DNA pararetroviruses (for example, rice tungro bacilliform virus, RTBV), gRNA of one of the picorna-like plant viruses (spherical rice tungrovirus spherical virus, RTSV), as well as pgRNA of spumaviruses – retroviruses with a DNA genome (in particular, human spumavirus, prototypic foamy virus, PFV), have similar structures that provide the same reinitiation with shunting of a large RNA region [34].
Mechanisms of reinitiation involving viral proteins. Plant pararetroviruses also use another method of effective reinitiation, which was characterized in a series of studies carried out under the leadership of L. Ryabova, also using CaMV mRNA as an example. This mechanism is associated with activity of the viral protein P6/TAV (transactivator viroplasmin) and is called TAV-mediated reinitiation (Fig. 4c, right).
TAV binds to the eIF3 factor in the initiation complex (probably at the moment when eIF4B factor leaves it) and retains it after joining of a large ribosomal subunit due to its affinity for several 60S proteins (for more details, see review [34]). In addition, TAV attracts TOR kinase to polysomes and promotes its activation [234]. TOR, in turn, activates kinase S6K1, which then phosphorylates the h subunit of eIF3 contributing to the retention of this factor on the ribosome during elongation and promoting reinitiation [235].
Another important player in this process is the cellular protein RISP, which acts as a TAV partner in the binding of eIF3, 40S, and 60S subunits [236]. In the recent study by the same group [237], an interesting model has been proposed, according to which an elongated RISP molecule, interacting simultaneously with the eS6/RSP6 and eL24/RPL24 proteins, acts as a clamp, holding together the large and small ribosomal subunits. In this case, binding is regulated by phosphorylation of eS6 and RISP itself by the TOR-S6K1 signaling cascade.
Apparently, under conditions of viral infection, the presence of TAV and activated RISP on the ribosome stimulates efficient reinitiation via two mechanisms acting simultaneously. First, retention of eIF3 on the elongating ribosomes allows the 40S subunit to remain bound to the mRNA after termination and 60S departure to quickly recruit initiation factors and resume scanning. Second, impaired recycling of the 60S subunit presumably promotes 80S-mediated reinitiation (which was previously shown in other eukaryotic systems [132, 230, 238, 239]). Whether these mechanisms are specific for the aforementioned plant viruses or whether they may operate in the case of other polycistronic viral mRNAs is still unknown.
STOPGO TRANSLATION
Some viruses of the families Picornaviridae, Iflaviviridae, Tetraviridae, Dicistroviridae, and Reoviridae also have another, fundamentally different way of producing an individual polypeptide encoded in an internal region of mRNA – the so-called “StopGo” (or “Stop-Carry On”) mechanism [240]. It requires a special amino acid sequence – 2A-peptide. A ribosome that synthesizes such a peptide does not form a peptide bond at a single specific position with near 100% probability, but subsequently continues normal translation. As a result, a separate protein molecule is produced, although no new act of translation initiation occurs during this process. The 2A peptide from FMDV is considered a classic example, while in practice the more active peptides E2A and T2A from the PTV-1 and TaV (Thosea asigna virus), respectively, are widely used [240, 241].
TRANSLATION OF CIRCULAR RNAs
Circular (covalently closed) protein-coding RNAs (circRNAs) are also of great interest. Such circRNAs do not have a free 5′ end and must initiate translation via internal ribosome entry; hence, they are likely to contain IRESs [242]. However, the available data on this subject are contradictory, obtained mainly by bioinformatics methods and do not always have experimental confirmation. In eukaryotic cells, circular transcripts are produced as a result of “reverse” splicing, in which a subsequent exon joins the previous one. Similar spliced RNAs were also found in viruses [243], but little is known about their translation mechanism.
One of the translated circRNAs encodes the papilloma virus E7 oncoprotein [244]. E7 induces malignant transformation of cells by modifying chromatin structure and altering transcription of protooncogenes and tumor suppressors. It has been shown that the HPV circRNA, synthesized after transfection of cervical carcinoma cells with a plasmid with the E7 minigene, contains m6A modified nucleotides and is translated in polysomes [244]. circRNAs are found in the life cycle of other viruses, for example, Epstein-Barr virus (EBV), Kaposhi sarcoma herpes virus (KSHV), HBV, etc. [245]. However, it is not certain whether they function as mRNAs.
In the world of viruses, there are also RNAs that are initially circular by design – mainly viroids and virusoids. However, very few cases are known where the viral circRNA encodes a protein. One of them is a satellite virus of rice yellow mottle virus (RYMV), whose “RNA nanogenome” is a covalently closed 220-nt long RNA. According to AbouHaidar et al. [246], this circRNA contains a ribosome binding site (AAGGA) 11 nt before the AUG codon, which provides internal initiation. The translation product is a 16-kDa protein that lacks homology with other known proteins. This polypeptide is able to bind its mRNA and is probably required for protection of the viral genome [246].
MECHANISMS ENSURING TRANSLATIONAL DOMINANCE FOR VIRAL mRNAs
Above, we described the mechanisms of translation of viral mRNAs aimed at increasing the efficiency of viral protein synthesis, as well as allowing to adapt to a wide range of stress conditions that inevitably arise in cells during infection. In this section, we briefly describe the most striking examples of mechanisms that allow viruses to preferentially translate their mRNAs. This is often achieved by inhibition, modification or destruction of the components of the cellular cap-binding apparatus or other elements of the canonical mechanism of translation initiation (Fig. 5). In addition, there are strategies based on compartmentalization of the components of the life cycle of viruses, which allow them to be spatially separated from the defense mechanisms of an infected cell.
Fig. 5.
Mechanisms used by viruses to create competitive advantage for their mRNAs. Full names of the viruses are given in the text of the article.
Manipulation of translation machinery components. Many viruses actively manipulate translational components to effectively compete for cellular resources. A striking example is the NSP1 protein of the SARS-CoV-2 coronavirus. It binds to the mRNA entry channel of the 40S subunit with its C-terminal domain acting as a stopper [247-249]. The N-terminal domain stabilizes this binding and, probably, also interacts with 40S. While interfering with cellular mRNA entering the ribosome, NSP1 still allows translation of the SARS-CoV-2 mRNA. This is due to the fact that the viral gRNA and sgRNAs have a common leader containing the hairpin structure SL1, which specifically binds to the N-terminal domain of the protein [250-252]. This interaction leads to the release of the C-terminal domain of NSP1 from the mRNA channel of the ribosome and allows translation to begin [251, 252]. Interestingly, initiation factors favor NSP1 binding to the 40S subunit, while interaction of the protein with the whole ribosome is weaker [253]. This suggests that in an infected cell, the inhibitory complex is formed specifically at the stage of translation initiation. One way or another, NSP1 is a powerful tool that helps the virus tap into the cell’s resources, but at the same time it can be a convenient target for antiviral therapy [250].
The NSP1 protein of another coronavirus, SARS-CoV, is also able to bind to the ribosome, inhibiting translation of cellular mRNAs. However, in this case, NSP1 probably recruits cellular endonucleases that selectively cleave mRNA depending on the employed mechanism of translation initiation [254].
Examples are known when viruses carry out limited proteolysis of translation initiation factors. In particular, we described viral proteases that cleave eIF4G in the sections devoted to IRESs. Such mechanisms allow viruses to increase efficiency of cap-independent translation by redirecting the liberated cellular resources [80, 255]. In some cases, initiation factors necessary for translation of the viral mRNA (subunits eIF3, eIF5B, PABP, and others) are also cleaved, which can be used to regulate the life cycle.
Other ways of influencing the initiation apparatus have also been described [255]. For example, RNA polymerase and NS1 protein of the influenza virus recruit the eIF4G factor to viral mRNAs, ensuring that they win competition for this limiting factor; the ICP6 protein of the herpes simplex virus 1 (HSV-1) enhances interaction of eIF4E with eIF4G, preventing the cell from inactivating translation via 4E-BP1 in response to infection; the EMCV virus, on the other hand, activates 4E-BP1 (see above) because it does not need eIF4E; the simian polyomavirus SV40 also activates 4E-BP1 in the late stages of infection; a number of viruses manipulate the PABP protein, modulating formation of the closed loop (rubella virus capsid protein, influenza virus NS1, KSHV virus SOX10 and K10 proteins, rotavirus NSP3 protein). More information on this topic can be found in the review [255].
Manipulations with the 5′ cap. Some viruses use another cunning strategy for winning the competition with cellular mRNAs – they disable cellular templates. Thus, the D9 and D10 proteins of the VACV virus directly bind and remove the cap of cellular mRNAs. This frees up resources for translation of non-capped viral mRNAs with the 5′ poly(A)-leader, which is necessary in the late stage of infection (see above). Interestingly, the D9 protein only inhibits translation by decapping mRNA and thereby provoking degradation, while D10 additionally stimulates translation of mRNA with poly(A)-leaders [256].
Even more devious is the mechanism of cap snatching from cellular mRNA, carried out by the (-)RNA viruses of the families Arenaviridae, Bunyaviridae, and Orthomyxoviridae. These viruses do not have their own capping enzymes, but they encode proteins that can bind the cap and cleave it, forming a 15-20-nt long capped 5′-terminal fragment of the cellular mRNA. This fragment is then used as a primer for synthesis of the (+) strand of viral RNA, which will include the capped segment of the cellular mRNA. For example, the replicase (RdRp) of the IAV influenza virus consists of three subunits: PA, PB1, and PB2, with PB2 responsible for cap binding and PA exhibiting endonuclease activity. Representatives of arenaviruses and bunyaviruses combine the functions of cap and endonuclease binding in one protein L.
Many RNA and DNA viruses replicating in the cytoplasm acquired their own capping apparatus during evolution, since the cellular machinery is confined to the nucleus. Capping enzymes are encoded in the genomes of VACV, RuV, SARS-CoV, SARS-CoV-2, Ebola virus (EBOV), vesicular stomatitis virus (VSV), protozoal giant viruses, and others. The most studied is the D1-D12 VACV heterodimer, which is widely used for capping mRNAs synthesized in vitro. More details about decapping, cap snatching, and non-canonical capping of viral RNAs can be found in the review [257].
Compartmentalized translation of viral mRNAs. Mechanisms associated with localized translation, formation of RNP granules, and generation of viral compartments within the cell deserve separate consideration [258]. In many RNA viruses, the life cycle is based on the formation of specialized “viral factories” (VFs) – intracellular structures made up of the membrane-bound organelles of the cell [259]. As a rule, VFs in (+)RNA-containing viruses serve exclusively as platforms for the synthesis of viral RNA, while in a number of viruses with dsRNA and (-)RNA genomes, they are also involved in the translation of their mRNA, protecting them from the action of cellular regulatory mechanisms. Viruses can actively recruit translational factors into such specialized compartments [258]. Association of viral mRNA translation with membranes could play a role in its resistance to the cellular response to infection even without regard to VF. This is evident from the case of poliovirus mRNA described above, which escapes the inhibitory effect of eIF2α phosphorylation due to membrane localization of the active eIF2 fraction in the infected cell [97].
The cellular response to viral infection includes formation of stress granules. Although this is not the main function of stress granules, this decreases the concentration of available translational components in the cytosol, which should interfere with the translation of viral mRNAs. However, many viruses skillfully manipulate this process, either by preventing granule formation (for example, by proteolysis of their key components), as do ZIKV, VSV, IAV, and a number of picornaviruses, or, conversely, facilitating their assembly at an advantageous time in their life cycle, like RuV [260, 261]. More details about the mechanisms associated with localized translation of viral mRNA and manipulation with the membrane and granular compartments can be found in the relevant reviews [258-261].
Manipulation of translation-related signaling pathways. Another common strategy employed by viruses to manipulate stress responses is to directly target cell signaling pathways. A classic example of this is inhibition of the eIF2 α-subunit kinases, which allows viruses to circumvent the repression of translation initiation mediated by these proteins. The proteins US1 of the HSV-1 virus, SM of EBV, E3L and K3L of VACV, NS5A of HCV bind to the PKR/EIF2AK2 kinase and inhibit its activity. The gB proteins of HSV-1, K3L of VACV, and HCV E2 act on another kinase, eIF2α, PERK/EIF2AK3. In this case, HCV E2 acts as a pseudosubstrate for both PKR and PERK. Cases are known when small noncoding viral RNAs bind to PKR, preventing its activation: for example, such activity is exhibited by the EBER RNA of EBV virus and VA RNA of adenovirus [255, 262].
Indirect effects of viral infection on the initiation of translation also deserve mentioning. For example, picornaviruses are able to affect permeability of the cell membrane and alter intracellular salt concentrations, optimizing conditions for the initiation of translation by IRESs and negatively affecting cap-dependent translation [263, 264].
Often, different viral mechanisms of selective suppression of protein biosynthesis in the cell work in a coordinated manner, simultaneously depriving cellular mRNAs of the 5′ cap, the apparatus of cap-dependent translation, and canonical delivery of Met-tRNAi, as well as of access to the compartments with active translational components. Manipulating the mechanisms of the cell stress response is an additional strategy. All this puts cells under control of the replicating virus and often deprives them of the chances not only for survival, but also for programmed death.
CONCLUSIONS
In this review, we examined the non-canonical mechanisms of translation initiation that are characteristic of viral mRNAs. During infection, these mechanisms provide them with competitive advantages, which are often empowered by the activity of specialized viral proteins. On the other hand, these features provide vulnerabilities to existing or potential drugs targeting specific components of the involved biochemical pathways [265]. Small molecules and oligonucleotides that change the complex structure of IRESs or CITEs, disrupt important RNA–RNA and RNA–protein interactions, attack ITAFs and other components critical for unconventional translation, can interfere with expression of viral mRNAs and thus block infection. Development of novel antiviral drugs based on these approaches holds great potential and will be at the forefront of future efforts to battle viruses.
Acknowledgments
The authors are grateful to Alexey Bogdanov for the invitation to contribute to the thematic collection dedicated to the memory of Alexander S. Spirin, to Alexey Agranovsky and Mikhail Schepetilnikov for the discussion of several chapters of this review, and to Alexander Alexandrov for valuable suggestions and comments. I. I. S. and S. E. D. are members of the Interdisciplinary Scientific and Educational School of Moscow University “Molecular Technologies of Living Systems and Synthetic Biology”.
Abbreviations
- CaMV
cauliflower mosaic virus
- circRNA
circular RNA
- CITE
cap-independent translation enhancer
- CrPV
cricket paralysis virus
- DENV
dengue virus
- EMCV
encephalomyocarditis virus
- FCV
feline calicivirus
- FMDV
foot-and-mouth disease virus
- gRNA
genomic RNA
- HalV
Halastavi árva virus
- HAV
hepatitis A virus
- HCV
hepatitis C virus
- HIV-1
human immunodeficiency virus type 1
- HNV
human Norwalk virus
- IBV
influenza B virus
- IGR
intergenic region
- IRES
internal ribosome entry site
- ITAF
IRES trans-acting factor
- ORF
open reading frame
- PEMV-2
pea enation mosaic virus 2
- pgRNA
pre-genomic RNA
- PTE
panicum mosaic virus-like translation enhancer
- PV
poliovirus
- RHDV
rabbit hemorrhagic disease virus
- RhPV
Rhopalosiphum padi virus
- RRL
rabbit reticulocyte lysate
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- sgRNA
subgenomic RNA
- TCV
turnip crinkle virus
- TED
translation enhancer domain
- TLS
tRNA-like structure
- TMEV
Theiler’s murine encephalomyelitis virus
- TMV
tobacco mosaic virus
- TPL
tripartite leader of late adenovirus mRNAs
- TURBS
termination upstream ribosomal binding site
- TYMV
turnip yellow mosaic virus
- UTR
untranslated region
- VACV
vaccinia virus
- VPg
viral protein genome-linked
- WGE
wheat germ extract
Funding
The work was supported by the Russian Science Foundation [project no. 19-74-20186 (K. S. V., “Structural elements of viral 3′ UTR involved in translation initiation” section) and project no. 20-14-00178 (N. O. K., all other sections)].
Ethics declarations
The authors declare no conflicts of interest in financial or any other sphere. This article does not contain any studies involving human participants or animals performed by any of the authors.
References
- 1.Rodrigues R. A. L., da Silva L. C. F., Abrahão J. S. Translating the language of giants: translation-related genes as a major contribution of giant viruses to the virosphere. Arch. Virol. 2020;165:1267–1278. doi: 10.1007/s00705-020-04626-2. [DOI] [PubMed] [Google Scholar]
- 2.Pelletier J., Sonenberg N. The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem. 2019;88:307–335. doi: 10.1146/annurev-biochem-013118-111042. [DOI] [PubMed] [Google Scholar]
- 3.Hinnebusch A. G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 4.Alekhina O. M., Vassilenko K. S. Translation initiation in eukaryotes: versatility of the scanning model. Biochemistry (Moscow) 2012;77:1465–1477. doi: 10.1134/S0006297912130056. [DOI] [PubMed] [Google Scholar]
- 5.Eliseeva I. A., Lyabin D. N., Ovchinnikov L. P. Poly(A)-binding proteins: structure, domain organization, and activity regulation. Biochemistry (Moscow) 2013;78:1377–1391. doi: 10.1134/S0006297913130014. [DOI] [PubMed] [Google Scholar]
- 6.Hinnebusch A. G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 2017;42:589–611. doi: 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/s0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terenin I. M., Akulich K. A., Andreev D. E., Polyanskaya S. A., Shatsky I. N., Dmitriev S. E. Sliding of a 43S ribosomal complex from the recognized AUG codon triggered by a delay in eIF2-bound GTP hydrolysis. Nucleic Acids Res. 2016;44:1882–1893. doi: 10.1093/nar/gkv1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Latorre P., Kolakofsky D., Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell Biol. 1998;18:5021–5031. doi: 10.1128/MCB.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firth A. E., Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taliansky M. E., Robinson D. J. Molecular biology of umbraviruses: phantom warriors. J. Gen. Virol. 2003;84:1951–1960. doi: 10.1099/vir.0.19219-0. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda D., Dreher T. W. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA. 2006;12:1338–1349. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak M. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc. Natl. Acad. Sci. USA. 1995;92:2662–2666. doi: 10.1073/pnas.92.7.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasso M. C., Milburn S. C., Hershey J. W., Jackson R. J. Selection of the 5′-proximal translation initiation site is influenced by mRNA and eIF-2 concentrations. Eur. J. Biochem. 1990;187:361–371. doi: 10.1111/j.1432-1033.1990.tb15313.x. [DOI] [PubMed] [Google Scholar]
- 15.Frolov I., Schlesinger S. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 1994;68:8111–8117. doi: 10.1128/JVI.68.12.8111-8117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ventoso I., Sanz M. A., Molina S., Berlanga J. J., Carrasco L., Esteban M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pappas C. L., Tzeng W. P., Frey T. K. Evaluation of cis-acting elements in the rubella virus subgenomic RNA that play a role in its translation. Arch. Virol. 2006;151:327–346. doi: 10.1007/s00705-005-0614-x. [DOI] [PubMed] [Google Scholar]
- 18.Gorchakov R., Frolova E., Williams B. R., Rice C. M., Frolov I. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 2004;78:8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanz M. A., Castelló A., Ventoso I., Berlanga J. J., Carrasco L. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS One. 2009;4:e4772. doi: 10.1371/journal.pone.0004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Moreno M., Sanz M. A., Pelletier J., Carrasco L. Requirements for eIF4A and eIF2 during translation of Sindbis virus subgenomic mRNA in vertebrate and invertebrate host cells. Cell Microbiol. 2013;15:823–840. doi: 10.1111/cmi.12079. [DOI] [PubMed] [Google Scholar]
- 21.Carrasco L., Sanz M. A., Gonzalez-Almela E. The regulation of translation in alphavirus-infected cells. Viruses. 2018;10:70. doi: 10.3390/v10020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skabkin M. A., Skabkina O. V., Dhote V., Komar A. A., Hellen C. U., Pestova T. V. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanz M. A., González Almela E., Carrasco L. Translation of Sindbis Subgenomic mRNA is Independent of eIF2, eIF2A and eIF2D. Sci. Rep. 2017;7:43876. doi: 10.1038/srep43876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clyde K., Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 2006;80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgil D., Polacek C., Harris E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol. 2006;80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fütterer J., Kiss-László Z., Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 27.Curran J., Kolakofsky D. Scanning independent ribosomal initiation of the Sendai virus X protein. EMBO J. 1988;7:2869–2874. doi: 10.1002/j.1460-2075.1988.tb03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Breyne S., Simonet V., Pelet T., Curran J. Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins. Nucleic Acids Res. 2003;31:608–618. doi: 10.1093/nar/gkg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yueh A., Schneider R. J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 30.Dmitriev S. E., Terenin I. M., Dunaevsky Y. E., Merrick W. C., Shatsky I. N. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′-untranslated regions. Mol. Cell. Biol. 2003;23:8925–8933. doi: 10.1128/MCB.23.24.8925-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xi Q., Cuesta R., Schneider R. J. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajyaguru P., Parker R. RGG motif proteins: modulators of mRNA functional states. Cell Cycle. 2012;11:2594–2599. doi: 10.4161/cc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolph P. J., Racaniello V., Villamarin A., Palladino F., Schneider R. J. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J. Virol. 1988;62:2059–2066. doi: 10.1128/JVI.62.6.2059-2066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pooggin M. M., Ryabova L. A. Ribosome shunting, polycistronic translation, and evasion of antiviral defenses in plant pararetroviruses and beyond. Front. Microbiol. 2018;9:644. doi: 10.3389/fmicb.2018.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pestova T. V., Kolupaeva V. G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hann L. E., Gehrke L. mRNAs containing the unstructured 5′ leader sequence of alfalfa mosaic virus RNA 4 translate inefficiently in lysates from poliovirus-infected HeLa cells. J. Virol. 1995;69:4986–4993. doi: 10.1128/JVI.69.8.4986-4993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallie D. R., Sleat D. E., Watts J. W., Turner P. C., Wilson T. M. A comparison of eukaryotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res. 1987;15:8693–8711. doi: 10.1093/nar/15.21.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agalarov S., Sakharov P. A., Fattakhova D., Sogorin E. A., Spirin A. S. Internal translation initiation and eIF4F/ATP-independent scanning of mRNA by eukaryotic ribosomal particles. Sci. Rep. 2014;4:4438. doi: 10.1038/srep04438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z., Martens C. A., Bruno D. P., Porcella S. F., Moss B. Pervasive initiation and 3′-end formation of poxvirus postreplicative RNAs. J. Biol. Chem. 2012;287:31050–31060. doi: 10.1074/jbc.M112.390054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vopálenský V., Sýkora M., Mělková Z., Mašek T., Pospíšek M. Transcripts of vaccinia virus postreplicative genes do not contain a 5′ methylguanosine cap. bioRxiv. 2020 doi: 10.1101/2020.07.15.204867. [DOI] [Google Scholar]
- 41.Shirokikh N. E., Spirin A. S. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc. Natl. Acad. Sci. USA. 2008;105:10738–10743. doi: 10.1073/pnas.0804940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulder J., Robertson M. E., Seamons R. A., Belsham G. J. Vaccinia virus protein synthesis has a low requirement for the intact translation initiation factor eIF4F, the cap-binding complex, within infected cells. J. Virol. 1998;72:8813–8819. doi: 10.1128/JVI.72.11.8813-8819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bablanian R., Goswami S. K., Esteban M., Banerjee A. K., Merrick W. C. Mechanism of selective translation of vaccinia virus mRNAs: differential role of poly(A) and initiation factors in the translation of viral and cellular mRNAs. J. Virol. 1991;65:4449–4460. doi: 10.1128/JVI.65.8.4449-4460.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vopálenský V., Sýkora M., Mašek T., Pospíšek M. Messenger RNAs of yeast virus-like elements contain non-templated 5′ Poly(A) leaders, and their expression is independent of eIF4E and Pab1. Front. Microbiol. 2019;10:2366. doi: 10.3389/fmicb.2019.02366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhungel P., Cao S., Yang Z. The 5′-poly(A) leader of poxvirus mRNA confers a translational advantage that can be achieved in cells with impaired cap-dependent translation. PLoS Pathog. 2017;13:e1006602. doi: 10.1371/journal.ppat.1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jha S., Rollins M. G., Fuchs G., Procter D. J., Hall E. A., et al. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature. 2017;546:651–655. doi: 10.1038/nature22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jan E., Mohr I., Walsh D. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 2016;3:283–307. doi: 10.1146/annurev-virology-100114-055014. [DOI] [PubMed] [Google Scholar]
- 48.Linero F., Welnowska E., Carrasco L., Scolaro L. Participation of eIF4F complex in Junin virus infection: blockage of eIF4E does not impair virus replication. Cell Microbiol. 2013;15:1766–1782. doi: 10.1111/cmi.12149. [DOI] [PubMed] [Google Scholar]
- 49.Leen E. N., Sorgeloos F., Correia S., Chaudhry Y., Cannac F., et al. A Conserved interaction between a C-terminal motif in Norovirus VPg and the HEAT-1 domain of eIF4G is essential for translation initiation. PLoS Pathog. 2016;12:e1005379. doi: 10.1371/journal.ppat.1005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung L., Bailey D., Leen E. N., Emmott E. P., Chaudhry Y., et al. Norovirus translation requires an interaction between the C terminus of the genome-linked viral protein VPg and eukaryotic translation initiation factor 4G. J. Biol. Chem. 2014;289:21738–21750. doi: 10.1074/jbc.M114.550657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coutinho de Oliveira L., Volpon L., Rahardjo A. K., Osborne M. J., Culjkovic-Kraljacic B., et al. Structural studies of the eIF4E-VPg complex reveal a direct competition for capped RNA: implications for translation. Proc. Natl. Acad. Sci. USA. 2019;116:24056–24065. doi: 10.1073/pnas.1904752116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan M. A., Goss D. J. Poly (A) binding protein enhances the binding affinity of potyvirus VPg to eukaryotic initiation factor eIF4F and activates in vitro translation. Int. J. Biol. Macromol. 2019;121:947–955. doi: 10.1016/j.ijbiomac.2018.10.135. [DOI] [PubMed] [Google Scholar]
- 53.Daughenbaugh K. F., Fraser C. S., Hershey J. W., Hardy M. E. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosmillo M., Lu J., McAllaster M. R., Eaglesham J. B., Wang X., et al. Noroviruses subvert the core stress granule component G3BP1 to promote viral VPg-dependent translation. Elife. 2019;8:e46681. doi: 10.7554/eLife.46681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burroughs J. N., Brown F. Presence of a covalently linked protein on calicivirus RNA. J. Gen. Virol. 1978;41:443–446. doi: 10.1099/0022-1317-41-2-443. [DOI] [PubMed] [Google Scholar]
- 56.Mailliot J., Martin F. Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip. Rev. RNA. 2018;9:e1458. doi: 10.1002/wrna.1458. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto H., Unbehaun A., Spahn C. M. T. Ribosomal chamber music: toward an understanding of IRES mechanisms. Trends Biochem. Sci. 2017;42:655–668. doi: 10.1016/j.tibs.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 58.Jaafar Z. A., Kieft J. S. Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 2019;17:110–123. doi: 10.1038/s41579-018-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arhab Y., Bulakhov A. G., Pestova T. V., Hellen C. U. T. Dissemination of Internal Ribosomal Entry Sites (IRES) between viruses by horizontal gene transfer. Viruses. 2020;12:612. doi: 10.3390/v12060612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwan T., Thompson S. R. Noncanonical translation initiation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2019;11:a032672. doi: 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jang S. K., Krausslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/JVI.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 63.Terenin I. M., Smirnova V. V., Andreev D. E., Dmitriev S. E., Shatsky I. N. A researcher’s guide to the galaxy of IRESs. Cell. Mol. Life Sci. 2017;74:1431–1455. doi: 10.1007/s00018-016-2409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson R. J. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 2013;5:a011569. doi: 10.1101/cshperspect.a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson S. R. So you want to know if your message has an IRES? Wiley Interdiscip. Rev. RNA. 2012;3:697–705. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andreev D. E., Terenin I. M., Dmitriev S. E., Shatsky I. N. Pros and cons of pDNA and mRNA transfection to study mRNA translation in mammalian cells. Gene. 2016;578:1–6. doi: 10.1016/j.gene.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 68.Barreau C., Dutertre S., Paillard L., Osborne H. B. Liposome-mediated RNA transfection should be used with caution. RNA. 2006;12:1790–1793. doi: 10.1261/rna.191706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dmitriev S. E., Andreev D. E., Adyanova Z. V., Terenin I. M., Shatsky I. N. Efficient cap-dependent translation of mammalian mRNAs with long and highly structured 5′-untranslated regions in vitro and in vivo. Mol. Biol. (Mosk.) 2009;43:108–113. doi: 10.1134/S0026893309010154. [DOI] [PubMed] [Google Scholar]
- 70.Dmitriev S. E., Bykova N. V., Andreev D. E., Terenin I. M. Adequate system for studying translation initiation on the human retrotransposon L1 mRNA in vitro. Mol. Biol. (Mosk.) 2006;40:20–24. doi: 10.1134/S0026893306010043. [DOI] [PubMed] [Google Scholar]
- 71.Avanzino B. C., Jue H., Miller C. M., Cheung E., Fuchs G., Fraser C. S. Molecular mechanism of poliovirus Sabin vaccine strain attenuation. J. Biol. Chem. 2018;293:15471–15482. doi: 10.1074/jbc.RA118.004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beckham S. A., Matak M. Y., Belousoff M. J., Venugopal H., Shah N., et al. Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES. Nucleic Acids Res. 2020;48:8006–8021. doi: 10.1093/nar/gkaa519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chamond N., Deforges J., Ulryck N., Sargueil B. 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of type II IRESs. Nucleic Acids Res. 2014;42:10373–10384. doi: 10.1093/nar/gku720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kieft J. S., Zhou K., Jubin R., Doudna J. A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrov A., Grosely R., Chen J., O’Leary S. E., Puglisi J. D. Multiple parallel pathways of translation initiation on the CrPV IRES. Mol. Cell. 2016;62:92–103. doi: 10.1016/j.molcel.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jang C. J., Jan E. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA. 2010;16:1182–1195. doi: 10.1261/rna.2044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jang S. K., Davies M. V., Kaufman R. J., Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J. Virol. 1989;63:1651–1660. doi: 10.1128/JVI.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pilipenko E. V., Blinov V. M., Romanova L. I., Sinyakov A. N., Maslova S. V., Agol V. I. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- 79.Skinner M. A., Racaniello V. R., Dunn G., Cooper J., Minor P. D., Almond J. W. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 1989;207:379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- 80.Martínez-Salas E., Francisco-Velilla R., Fernandez-Chamorro J., Lozano G., Diaz-Toledano R. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res. 2015;206:62–73. doi: 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 81.Pilipenko E. V., Gmyl A. P., Maslova S. V., Svitkin Y. V., Sinyakov A. N., Agol V. I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 82.Slobodskaya O. R., Gmyl A. P., Maslova S. V., Tolskaya E. A., Viktorova E. G., Agol V. I. Poliovirus neurovirulence correlates with the presence of a cryptic AUG upstream of the initiator codon. Virology. 1996;221:141–150. doi: 10.1006/viro.1996.0360. [DOI] [PubMed] [Google Scholar]
- 83.Lulla V., Dinan A. M., Hosmillo M., Chaudhry Y., Sherry L., et al. An upstream protein-coding region in enteroviruses modulates virus infection in gut epithelial cells. Nat. Microbiol. 2019;4:280–292. doi: 10.1038/s41564-018-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hellen C. U., Pestova T. V., Wimmer E. Effect of mutations downstream of the internal ribosome entry site on initiation of poliovirus protein synthesis. J. Virol. 1994;68:6312–6322. doi: 10.1128/JVI.68.10.6312-6322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sweeney T. R., Abaeva I. S., Pestova T. V., Hellen C. U. The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J. 2014;33:76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaminski A., Pöyry T. A., Skene P. J., Jackson R. J. Mechanism of initiation site selection promoted by the human rhinovirus 2 internal ribosome entry site. J. Virol. 2010;84:6578–6589. doi: 10.1128/JVI.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kafasla P., Morgner N., Robinson C. V., Jackson R. J. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andreev D. E., Hirnet J., Terenin I. M., Dmitriev S. E., Niepmann M., Shatsky I. N. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res. 2012;40:5602–5614. doi: 10.1093/nar/gks182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meerovitch K., Svitkin Y. V., Lee H. S., Lejbkowicz F., Kenan D. J., et al. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798–3807. doi: 10.1128/JVI.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hunt S. L., Hsuan J. J., Totty N., Jackson R. J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Svitkin Y. V., Pestova T. V., Maslova S. V., Agol V. I. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology. 1988;166:394–404. doi: 10.1016/0042-6822(88)90510-7. [DOI] [PubMed] [Google Scholar]
- 92.Agol V. I., Drozdov S. G., Ivannikova T. A., Kolesnikova M. S., Korolev M. B., Tolskaya E. A. Restricted growth of attenuated poliovirus strains in cultured cells of a human neuroblastoma. J. Virol. 1989;63:4034–4038. doi: 10.1128/JVI.63.9.4034-4038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.La Monica N., Racaniello V. R. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 1989;63:2357–2360. doi: 10.1128/JVI.63.5.2357-2360.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guest S., Pilipenko E., Sharma K., Chumakov K., Roos R. P. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J. Virol. 2004;78:11097–11107. doi: 10.1128/JVI.78.20.11097-11107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Svitkin Y. V., Imataka H., Khaleghpour K., Kahvejian A., Liebig H. D., Sonenberg N. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA. 2001;7:1743–1752. doi: 10.1017/S135583820100108X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.White J. P., Reineke L. C., Lloyd R. E. Poliovirus switches to an eIF2-independent mode of translation during infection. J. Virol. 2011;85:8884–8893. doi: 10.1128/JVI.00792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kastan J. P., Dobrikova E. Y., Bryant J. D., Gromeier M. CReP mediates selective translation initiation at the endoplasmic reticulum. Sci. Adv. 2020;6:eaba0745. doi: 10.1126/sciadv.aba0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pilipenko E. V., Blinov V. M., Chernov B. K., Dmitrieva T. M., Agol V. I. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kolupaeva V. G., Pestova T. V., Hellen C. U., Shatsky I. N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 100.Terenin I. M., Andreev D. E., Dmitriev S. E., Shatsky I. N. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 2013;41:1807–1816. doi: 10.1093/nar/gks1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pilipenko E. V., Gmyl A. P., Maslova S. V., Belov G. A., Sinyakov A. N., et al. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J. Mol. Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- 102.Kaminski A., Howell M. T., Jackson R. J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pestova T. V., Borukhov S. I., Hellen C. U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 104.Andreev D. E., Fernandez-Miragall O., Ramajo J., Dmitriev S. E., Terenin I. M., et al. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pilipenko E. V., Pestova T. V., Kolupaeva V. G., Khitrina E. V., Poperechnaya A. N., et al. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 106.Pestova T. V., Hellen C. U., Shatsky I. N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/MCB.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kolupaeva V. G., Hellen C. U., Shatsky I. N. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 108.Pilipenko E. V., Gmyl A. P., Maslova S. V., Khitrina E. V., Agol V. I. Attenuation of Theiler’s murine encephalomyelitis virus by modifications of the oligopyrimidine/AUG tandem, a host-dependent translational cis element. J. Virol. 1995;69:864–870. doi: 10.1128/JVI.69.2.864-870.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pilipenko E. V., Viktorova E. G., Guest S. T., Agol V. I., Roos R. P. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 2001;20:6899–6908. doi: 10.1093/emboj/20.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pilipenko E. V., Viktorova E. G., Khitrina E. V., Maslova S. V., Jarousse N., et al. Distinct attenuation phenotypes caused by mutations in the translational starting window of Theiler’s murine encephalomyelitis virus. J. Virol. 1999;73:3190–3196. doi: 10.1128/JVI.73.4.3190-3196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gingras A. C., Svitkin Y., Belsham G. J., Pause A., Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Borman A. M., Kean K. M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129–136. doi: 10.1006/viro.1997.8761. [DOI] [PubMed] [Google Scholar]
- 113.Koirala D., Shao Y., Koldobskaya Y., Fuller J. R., Watkins A. M., et al. A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites. Nat. Commun. 2019;10:3629. doi: 10.1038/s41467-019-11585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Avanzino B. C., Fuchs G., Fraser C. S. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc. Natl. Acad. Sci. USA. 2017;114:9611–9616. doi: 10.1073/pnas.1704390114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/JVI.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malygin A. A., Kossinova O. A., Shatsky I. N., Karpova G. G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013;41:8706–8714. doi: 10.1093/nar/gkt632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Quade N., Boehringer D., Leibundgut M., van den Heuvel J., Ban N. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat. Commun. 2015;6:7646. doi: 10.1038/ncomms8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yamamoto H., Collier M., Loerke J., Ismer J., Schmidt A., et al. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 2015;34:3042–3058. doi: 10.15252/embj.201592469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hashem Y., des Georges A., Dhote V., Langlois R., Liao H. Y., et al. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sizova D. V., Kolupaeva V. G., Pestova T. V., Shatsky I. N., Hellen C. U. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/JVI.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jaafar Z. A., Oguro A., Nakamura Y., Kieft J. S. Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. Elife. 2016;5:e21198. doi: 10.7554/eLife.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.King H. A., Cobbold L. C., Willis A. E. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 124.Kim G. W., Siddiqui A. N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc. Natl. Acad. Sci. USA. 2021;118:e2022024118. doi: 10.1073/pnas.2022024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Robert F., Kapp L. D., Khan S. N., Acker M. G., Kolitz S., et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2. GTP. Met-tRNA(i)(Met) ternary complex availability. Mol. Biol. Cell. 2006;17:4632–4644. doi: 10.1091/mbc.e06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Terenin I. M., Dmitriev S. E., Andreev D. E., Shatsky I. N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 127.Pestova T. V., de Breyne S., Pisarev A. V., Abaeva I. S., Hellen C. U. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yamamoto H., Unbehaun A., Loerke J., Behrmann E., Collier M., et al. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014;21:721–727. doi: 10.1038/nsmb.2859. [DOI] [PubMed] [Google Scholar]
- 129.Dmitriev S. E., Terenin I. M., Andreev D. E., Ivanov P. A., Dunaevsky J. E., et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weisser M., Schafer T., Leibundgut M., Bohringer D., Aylett C. H. S., Ban N. Structural and functional insights into human re-initiation complexes. Mol. Cell. 2017;67:447–456.e447. doi: 10.1016/j.molcel.2017.06.032. [DOI] [PubMed] [Google Scholar]
- 131.González-Almela E., Williams H., Sanz M. A., Carrasco L. The Initiation factors eIF2, eIF2A, eIF2D, eIF4A, and eIF4G are not involved in translation driven by hepatitis C virus IRES in human cells. Front. Microbiol. 2018;9:207. doi: 10.3389/fmicb.2018.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Young D. J., Makeeva D. S., Zhang F., Anisimova A. S., Stolboushkina E. A., et al. Tma64/eIF2D, Tma20/MCT-1, and Tma22/DENR recycle post-termination 40S subunits in vivo. Mol. Cell. 2018;71:761–774.e765. doi: 10.1016/j.molcel.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim J. H., Park S. M., Park J. H., Keum S. J., Jang S. K. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Breyne S., Yu Y., Pestova T. V., Hellen C. U. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA. 2008;14:367–380. doi: 10.1261/rna.696508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lancaster A. M., Jan E., Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Locker N., Easton L. E., Lukavsky P. J. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yokoyama T., Machida K., Iwasaki W., Shigeta T., Nishimoto M., et al. HCV IRES captures an actively translating 80S ribosome. Mol. Cell. 2019;74:1205–1214.e1208. doi: 10.1016/j.molcel.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 138.Wilson J. E., Pestova T. V., Hellen C. U., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 139.Sasaki J., Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pfingsten J. S., Costantino D. A., Kieft J. S. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fernández I. S., Bai X. C., Murshudov G., Scheres S. H., Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Spahn C. M., Jan E., Mulder A., Grassucci R. A., Sarnow P., Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 143.Koh C. S., Brilot A. F., Grigorieff N., Korostelev A. A. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. Proc. Natl. Acad. Sci. USA. 2014;111:9139–9144. doi: 10.1073/pnas.1406335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Murray J., Savva C. G., Shin B. S., Dever T. E., Ramakrishnan V., Fernandez I. S. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife. 2016;5:e13567. doi: 10.7554/eLife.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muhs M., Hilal T., Mielke T., Skabkin M. A., Sanbonmatsu K. Y., et al. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell. 2015;57:422–432. doi: 10.1016/j.molcel.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pfingsten J. S., Costantino D. A., Kieft J. S. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J. Mol. Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Abaeva I. S., Vicens Q., Bochler A., Soufari H., Simonetti A., et al. The Halastavi arva virus intergenic region IRES promotes translation by the simplest possible initiation mechanism. Cell Rep. 2020;33:108476. doi: 10.1016/j.celrep.2020.108476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kerr C. H., Wang Q. S., Moon K. M., Keatings K., Allan D. W., et al. IRES-dependent ribosome repositioning directs translation of a +1 overlapping ORF that enhances viral infection. Nucleic Acids Res. 2018;46:11952–11967. doi: 10.1093/nar/gky1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ren Q., Wang Q. S., Firth A. E., Chan M. M., Gouw J. W., et al. Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site. Proc. Natl. Acad. Sci. USA. 2012;109:E630–639. doi: 10.1073/pnas.1111303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Terenin I. M., Dmitriev S. E., Andreev D. E., Royall E., Belsham G. J., et al. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell Biol. 2005;25:7879–7888. doi: 10.1128/MCB.25.17.7879-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Abaeva I. S., Pestova T. V., Hellen C. U. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi arva virus IRES. Nucleic Acids Res. 2016;44:2362–2377. doi: 10.1093/nar/gkw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Neupane R., Pisareva V. P., Rodriguez C. F., Pisarev A. V., Fernandez I. S. A complex IRES at the 5′-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. Elife. 2020;9:e54575. doi: 10.7554/eLife.54575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gross L., Vicens Q., Einhorn E., Noireterre A., Schaeffer L., et al. The IRES 5′ UTR of the dicistrovirus cricket paralysis virus is a type III IRES containing an essential pseudoknot structure. Nucleic Acids Res. 2017;45:8993–9004. doi: 10.1093/nar/gkx622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Majzoub K., Hafirassou M. L., Meignin C., Goto A., Marzi S., et al. RACK1 controls IRES-mediated translation of viruses. Cell. 2014;159:1086–1095. doi: 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Roberts R., Mayberry L. K., Browning K. S., Rakotondrafara A. M. The Triticum Mosaic virus 5′ leader binds to both eIF4G and eIFiso4G for translation. PLoS One. 2017;12:e0169602. doi: 10.1371/journal.pone.0169602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jaramillo-Mesa H., Gannon M., Holshbach E., Zhang J., Roberts R., et al. The Triticum Mosaic virus internal ribosome entry site relies on a picornavirus-like YX-AUG motif to designate the preferred translation initiation site and to likely target the 18S rRNA. J. Virol. 2019;93:e01705–18. doi: 10.1128/JVI.01705-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Castelló A., Franco D., Moral-López P., Berlanga J. J., Álvarez E., et al. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Perales C., Carrasco L., Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003;533:89–94. doi: 10.1016/s0014-5793(02)03764-x. [DOI] [PubMed] [Google Scholar]
- 159.Ohlmann T., Prévôt D., Décimo D., Roux F., Garin J., et al. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002;318:9–20. doi: 10.1016/S0022-2836(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 160.Ventoso I., Blanco R., Perales C., Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl. Acad. Sci. USA. 2001;98:12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Buck C. B., Shen X., Egan M. A., Pierson T. C., Walker C. M., Siliciano R. F. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vallejos M., Carvajal F., Pino K., Navarrete C., Ferres M., et al. Functional and structural analysis of the internal ribosome entry site present in the mRNA of natural variants of the HIV-1. PLoS One. 2012;7:e35031. doi: 10.1371/journal.pone.0035031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Brasey A., Lopez-Lastra M., Ohlmann T., Beerens N., Berkhout B., et al. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/jvi.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Smirnova V. V., Terenin I. M., Khutornenko A. A., Andreev D. E., Dmitriev S. E., Shatsky I. N. Does HIV-1 mRNA 5′-untranslated region bear an internal ribosome entry site? Biochimie. 2016;121:228–237. doi: 10.1016/j.biochi.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 165.Svitkin Y. V., Pause A., Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994;68:7001–7007. doi: 10.1128/JVI.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Berkhout B., Arts K., Abbink T. E. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 2011;39:5232–5244. doi: 10.1093/nar/gkr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.de Breyne S., Chamond N., Décimo D., Trabaud M. A., André P., et al. In vitro studies reveal that different modes of initiation on HIV-1 mRNA have different levels of requirement for eukaryotic initiation factor 4F. FEBS J. 2012;279:3098–3111. doi: 10.1111/j.1742-4658.2012.08689.x. [DOI] [PubMed] [Google Scholar]
- 168.Weill L., James L., Ulryck N., Chamond N., Herbreteau C. H., et al. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010;38:1367–1381. doi: 10.1093/nar/gkp1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Herbreteau C. H., Weill L., Décimo D., Prévôt D., Darlix J. L., et al. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 170.Akulich K. A., Andreev D. E., Terenin I. M., Smirnova V. V., Anisimova A. S., et al. Four translation initiation pathways employed by the leaderless mRNA in eukaryotes. Sci. Rep. 2016;6:37905. doi: 10.1038/srep37905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Andreev D. E., Terenin I. M., Dunaevsky Y. E., Dmitriev S. E., Shatsky I. N. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol. Cell. Biol. 2006;26:3164–3169. doi: 10.1128/MCB.26.8.3164-3169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tahara S. M., Dietlin T. A., Dever T. E., Merrick W. C., Worrilow L. M. Effect of eukaryotic initiation factor 4F on AUG selection in a bicistronic mRNA. J. Biol. Chem. 1991;266:3594–3601. doi: 10.1016/S0021-9258(19)67836-1. [DOI] [PubMed] [Google Scholar]
- 173.Barrera A., Olguín V., Vera-Otarola J., López-Lastra M. Cap-independent translation initiation of the unspliced RNA of retroviruses. Biochim. Biophys. Acta Gene Regul. Mech. 2020;1863:194583. doi: 10.1016/j.bbagrm.2020.194583. [DOI] [PubMed] [Google Scholar]
- 174.Song Y., Mugavero J., Stauft C. B., Wimmer E. Dengue and Zika virus 5′ untranslated regions harbor internal ribosomal entry site functions. mBio. 2019;10:e00459–19. doi: 10.1128/mBio.00459-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fernández-García L., Angulo J., Ramos H., Barrera A., Pino K., et al. The internal ribosome entry site of the Dengue virus mRNA is active when cap-dependent translation initiation is inhibited. J. Virol. 2020 doi: 10.1128/JVI.01998-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dorokhov Y. L., Skulachev M. V., Ivanov P. A., Zvereva S. D., Tjulkina L. G., et al. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. USA. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dorokhov Y. L., Sheshukova E. V., Komarova T. V. Tobamovirus 3′-terminal Gene overlap may be a mechanism for within-host fitness improvement. Front. Microbiol. 2017;8:851. doi: 10.3389/fmicb.2017.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.May J., Johnson P., Saleem H., Simon A. E. A sequence-independent, unstructured internal ribosome entry site is responsible for internal expression of the coat protein of Turnip Crinkle virus. J. Virol. 2017;91:e02421–16. doi: 10.1128/JVI.02421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yuan X., Shi K., Meskauskas A., Simon A. E. The 3′ end of Turnipcrinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA. 2009;15:1849–1864. doi: 10.1261/rna.1708709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Treder K., Kneller E. L., Allen E. M., Wang Z., Browning K. S., Miller W. A. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gazo B. M., Murphy P., Gatchel J. R., Browning K. S. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 2004;279:13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- 182.Wang Z., Treder K., Miller W. A. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J. Biol. Chem. 2009;284:14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nicholson B. L., Wu B., Chevtchenko I., White K. A. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA. 2010;16:1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nicholson B. L., Zaslaver O., Mayberry L. K., Browning K. S., White K. A. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 2013;87:1872–1883. doi: 10.1128/JVI.02711-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Stupina V. A., Meskauskas A., McCormack J. C., Yingling Y. G., Shapiro B. A., et al. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA. 2008;14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gao F., Kasprzak W., Stupina V. A., Shapiro B. A., Simon A. E. A ribosome-binding, 3′ translational enhancer has a T-shaped structure and engages in a long-distance RNA–RNA interaction. J. Virol. 2012;86:9828–9842. doi: 10.1128/JVI.00677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zuo X., Wang J., Yu P., Eyler D., Xu H., et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of Turnip crinkle virus. Proc. Natl. Acad. Sci. USA. 2010;107:1385–1390. doi: 10.1073/pnas.0908140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gao F., Gulay S. P., Kasprzak W., Dinman J. D., Shapiro B. A., Simon A. E. The kissing-loop T-shaped structure translational enhancer of Pea enation mosaic virus can bind simultaneously to ribosomes and a 5′ proximal hairpin. J. Virol. 2013;87:11987–12002. doi: 10.1128/JVI.02005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhao P., Liu Q., Miller W. A., Goss D. J. Eukaryotic translation initiation factor 4G (eIF4G) coordinates interactions with eIF4A, eIF4B, and eIF4E in binding and translation of the barley yellow dwarf virus 3′ cap-independent translation element (BTE) J. Biol. Chem. 2017;292:5921–5931. doi: 10.1074/jbc.M116.764902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Z., Parisien M., Scheets K., Miller W. A. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure. 2011;19:868–880. doi: 10.1016/j.str.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Du Z., Alekhina O. M., Vassilenko K. S., Simon A. E. Concerted action of two 3′ cap-independent translation enhancers increases the competitive strength of translated viral genomes. Nucleic Acids Res. 2017;45:9558–9572. doi: 10.1093/nar/gkx643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sharma G., Stupina V., Pallesen J., Shapiro B., Simon A., Dinman J., Frank J. Cryo-electron microscopy (Cryo-EM) structure of a cap-independent translational enhancer of the Turnip crinkle virus (TCV) bound to the eukaryotic ribosome. Biophys. J. 2012;102:393A. doi: 10.1016/j.bpj.2011.11.2147. [DOI] [Google Scholar]
- 193.Miras M., Sempere R. N., Kraft J. J., Miller W. A., Aranda M. A., Truniger V. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014;202:233–246. doi: 10.1111/nph.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guo L., Allen E. M., Miller W. A. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- 195.Gao F., Alekhina O. M., Vassilenko K. S., Simon A. E. Unusual dicistronic expression from closely spaced initiation codons in an umbravirus subgenomic RNA. Nucleic Acids Res. 2018;46:11726–11742. doi: 10.1093/nar/gky871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rakotondrafara A. M., Polacek C., Harris E., Miller W. A. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fabian M. R., White K. A. Analysis of a 3′-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sarawaneeyaruk S., Iwakawa H. O., Mizumoto H., Murakami H., Kaido M., et al. Host-dependent roles of the viral 5′ untranslated region (UTR) in RNA stabilization and cap-independent translational enhancement mediated by the 3′ UTR of Red clover necrotic mosaic virus RNA1. Virology. 2009;391:107–118. doi: 10.1016/j.virol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- 199.Karetnikov A., Lehto K. Translation mechanisms involving long-distance base pairing interactions between the 5′ and 3′ non-translated regions and internal ribosomal entry are conserved for both genomic RNAs of Blackcurrant reversion nepovirus. Virology. 2008;371:292–308. doi: 10.1016/j.virol.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 200.Sharma S. D., Kraft J. J., Miller W. A., Goss D. J. Recruitment of the 40S ribosome subunit to the 3′-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation. J. Biol. Chem. 2015;290:11268–11281. doi: 10.1074/jbc.M115.645002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Stupina V. A., Yuan X., Meskauskas A., Dinman J. D., Simon A. E. Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′ untranslated region in cap-independent translation of turnip crinkle virus. J. Virol. 2011;85:4638–4653. doi: 10.1128/JVI.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yoshii M., Nishikiori M., Tomita K., Yoshioka N., Kozuka R., et al. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 2004;78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gao F., Simon A. E. Multiple Cis-acting elements modulate programmed -1 ribosomal frameshifting in Pea enation mosaic virus. Nucleic Acids Res. 2016;44:878–895. doi: 10.1093/nar/gkv1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Matsuda D., Dreher T. W. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 205.Colussi T. M., Costantino D. A., Hammond J. A., Ruehle G. M., Nix J. C., Kieft J. S. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature. 2014;511:366–369. doi: 10.1038/nature13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Haenni A. L., Joshi S., Chapeville F. tRNA-like structures in the genomes of RNA viruses. Prog. Nucleic Acid Res. Mol. Biol. 1982;27:85–104. doi: 10.1016/s0079-6603(08)60598-x. [DOI] [PubMed] [Google Scholar]
- 207.Barends S., Bink H. H., van den Worm S. H., Pleij C. W., Kraal B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–129. doi: 10.1016/s0092-8674(02)01256-4. [DOI] [PubMed] [Google Scholar]
- 208.Matsuda D., Dreher T. W. Cap- and initiator tRNA-dependent initiation of TYMV polyprotein synthesis by ribosomes: evaluation of the Trojan horse model for TYMV RNA translation. RNA. 2007;13:129–137. doi: 10.1261/rna.244407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gallie D. R., Kobayashi M. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene. 1994;142:159–165. doi: 10.1016/0378-1119(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 210.Barends S., Rudinger-Thirion J., Florentz C., Giegé R., Pleij C. W., Kraal B. tRNA-like structure regulates translation of Brome mosaic virus RNA. J. Virol. 2004;78:4003–4010. doi: 10.1128/jvi.78.8.4003-4010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Leathers V., Tanguay R., Kobayashi M., Gallie D. R. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell. Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331-5347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Matsuda D., Dunoyer P., Hemmer O., Fritsch C., Dreher T. W. The valine anticodon and valylatability of Peanut clump virus RNAs are not essential but provide a modest competitive advantage in plants. J. Virol. 2000;74:8720–8725. doi: 10.1128/jvi.74.18.8720-8725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gordon K. H., Johnson K. N., Hanzlik T. N. The larger genomic RNA of Helicoverpa armigera stunt tetravirus encodes the viral RNA polymerase and has a novel 3′-terminal tRNA-like structure. Virology. 1995;208:84–98. doi: 10.1006/viro.1995.1132. [DOI] [PubMed] [Google Scholar]
- 214.Sherlock M. E., Hartwick E. W., MacFadden A., Kieft J. S. Structural diversity and phylogenetic distribution of valyl tRNA-like structures in viruses. RNA. 2021;27:27–39. doi: 10.1261/rna.076968.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Poncet D., Laurent S., Cohen J. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J. 1994;13:4165–4173. doi: 10.1002/j.1460-2075.1994.tb06734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Groft C. M., Burley S. K. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell. 2002;9:1273–1283. doi: 10.1016/s1097-2765(02)00555-5. [DOI] [PubMed] [Google Scholar]
- 217.Vende P., Piron M., Castagné N., Poncet D. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 2000;74:7064–7071. doi: 10.1128/jvi.74.15.7064-7071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Olsthoorn R. C., Mertens S., Brederode F. T., Bol J. F. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen S. C., Olsthoorn R. C. In vitro and in vivo studies of the RNA conformational switch in Alfalfa mosaic virus. J. Virol. 2010;84:1423–1429. doi: 10.1128/JVI.01443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Krab I. M., Caldwell C., Gallie D. R., Bol J. F. Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J. Gen. Virol. 2005;86:1841–1849. doi: 10.1099/vir.0.80796-0. [DOI] [PubMed] [Google Scholar]
- 221.Holden K. L., Harris E. Enhancement of dengue virus translation: role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology. 2004;329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 222.Bung C., Bochkaeva Z., Terenin I., Zinovkin R., Shatsky I. N., Niepmann M. Influence of the hepatitis C virus 3′-untranslated region on IRES-dependent and cap-dependent translation initiation. FEBS Lett. 2010;584:837–842. doi: 10.1016/j.febslet.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 223.Gunišová S., Hronová V., Mohammad M. P., Hinnebusch A. G., Valášek L. S. Please do not recycle! Translation reinitiation in microbes and higher eukaryotes. FEMS Microbiol. Rev. 2018;42:165–192. doi: 10.1093/femsre/fux059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Powell M. L. Translational termination-reinitiation in RNA viruses. Biochem. Soc. Trans. 2010;38:1558–1564. doi: 10.1042/BST0381558. [DOI] [PubMed] [Google Scholar]
- 225.Pöyry T. A., Kaminski A., Connell E. J., Fraser C. S., Jackson R. J. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Meyers G. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 2007;81:9623–9632. doi: 10.1128/JVI.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Meyers G. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 2003;278:34051–34060. doi: 10.1074/jbc.M304874200. [DOI] [PubMed] [Google Scholar]
- 228.Wennesz R., Luttermann C., Kreher F., Meyers G. Structure-function relationship in the “termination upstream ribosomal binding site” of the calicivirus rabbit hemorrhagic disease virus. Nucleic Acids Res. 2019;47:1920–1934. doi: 10.1093/nar/gkz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Luttermann C., Meyers G. The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev. 2009;23:331–344. doi: 10.1101/gad.507609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zinoviev A., Hellen C. U. T., Pestova T. V. Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol. Cell. 2015;57:1059–1073. doi: 10.1016/j.molcel.2015.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Powell M. L., Leigh K. E., Poyry T. A., Jackson R. J., Brown T. D., Brierley I. Further characterisation of the translational termination-reinitiation signal of the influenza B virus segment 7 RNA. PLoS One. 2011;6:e16822. doi: 10.1371/journal.pone.0016822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Powell M. L., Napthine S., Jackson R. J., Brierley I., Brown T. D. Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA. 2008;14:2394–2406. doi: 10.1261/rna.1231008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Abaeva I. S., Marintchev A., Pisareva V. P., Hellen C. U., Pestova T. V. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Schepetilnikov M., Kobayashi K., Geldreich A., Caranta C., Robaglia C., et al. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J. 2011;30:1343–1356. doi: 10.1038/emboj.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Schepetilnikov M., Ryabova L. A. Recent discoveries on the role of TOR (Target of rapamycin) signaling in translation in plants. Plant Physiol. 2018;176:1095–1105. doi: 10.1104/pp.17.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Thiébeauld O., Schepetilnikov M., Park H. S., Geldreich A., Kobayashi K., et al. A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J. 2009;28:3171–3184. doi: 10.1038/emboj.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mancera-Martínez E., Dong Y., Makarian J., Srour O., Thiébeauld O., et al. Phosphorylation of a reinitiation supporting protein, RISP, determines its function in translation reinitiation. Nucleic Acids Res. 2021;49:6908–6924. doi: 10.1093/nar/gkab501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Skabkin M. A., Skabkina O. V., Hellen C. U., Pestova T. V. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell. 2013;51:249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Young D. J., Guydosh N. R., Zhang F., Hinnebusch A. G., Green R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′-UTRs in vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Brown, J. D., and Ryan, M. D. (2010) Ribosome “Skipping”: “Stop-Carry On” or “StopGo” Translation. in Recoding: Expansion of Decoding Rules Enriches Gene Expression (Atkins, J. F., and Gesteland, R. F., eds.) Springer New York, New York, NY, pp. 101-121.
- 241.Liu Z., Chen O., Wall J. B. J., Zheng M., Zhou Y., et al. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017;7:2193. doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21.e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tagawa T., Kopardé V. N., Ziegelbauer J. M. Identifying and characterizing virus-encoded circular RNAs. Methods. 2021 doi: 10.1016/j.ymeth.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhao J., Lee E. E., Kim J., Yang R., Chamseddin B., et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019;10:2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tan K. E., Lim Y. Y. Viruses join the circular RNA world. FEBS J. 2020 doi: 10.1111/febs.15639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.AbouHaidar M. G., Venkataraman S., Golshani A., Liu B., Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl. Acad. Sci. USA. 2014;111:14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Yuan S., Peng L., Park J. J., Hu Y., Devarkar S. C., et al. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell. 2020;80:1055–1066.e1056. doi: 10.1016/j.molcel.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Schubert K., Karousis E. D., Jomaa A., Scaiola A., Echeverria B., et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 250.Vankadari N., Jeyasankar N. N., Lopes W. J. Structure of the SARS-CoV-2 Nsp1/5′-untranslated region complex and implications for potential therapeutic targets, a vaccine, and virulence. J. Phys. Chem. Lett. 2020;11:9659–9668. doi: 10.1021/acs.jpclett.0c02818. [DOI] [PubMed] [Google Scholar]
- 251.Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., et al. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2020 doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shi M., Wang L., Fontana P., Vora S., Zhang Y., et al. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv. 2020 doi: 10.1101/2020.09.18.302901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lapointe C. P., Grosely R., Johnson A. G., Wang J., Fernandez I. S., Puglisi J. D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl. Acad. Sci. USA. 2021;118:e2017715118. doi: 10.1073/pnas.2017715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Huang C., Lokugamage K. G., Rozovics J. M., Narayanan K., Semler B. L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cantu F., Cao S., Hernandez C., Dhungel P., Spradlin J., Yang Z. Poxvirus-encoded decapping enzymes promote selective translation of viral mRNAs. PLoS Pathog. 2020;16:e1008926. doi: 10.1371/journal.ppat.1008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lashkevich K. A., Dmitriev S. E. mRNA targeting, transport and local translation in eukaryotic cells: from the classical view to a diversity of new concepts. Mol. Biol. 2021 doi: 10.1134/S0026893321030080. [DOI] [PubMed] [Google Scholar]
- 259.Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Reineke L. C., Lloyd R. E. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436:255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.McCormick C., Khaperskyy D. A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- 262.Dzananovic E., McKenna S. A., Patel T. R. Viral proteins targeting host protein kinase R to evade an innate immune response: a mini review. Biotechnol. Genet. Eng. Rev. 2018;34:33–59. doi: 10.1080/02648725.2018.1467151. [DOI] [PubMed] [Google Scholar]
- 263.Alonso M. A., Carrasco L. Reversion by hypotonic medium of the shutoff of protein synthesis induced by encephalomyocarditis virus. J. Virol. 1981;37:535–540. doi: 10.1128/JVI.37.2.535-540.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Nair C. N. Monovalent cation metabolism and cytopathic effects of poliovirus-infected HeLa cells. J. Virol. 1981;37:268–273. doi: 10.1128/JVI.37.1.268-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Dmitriev S. E., Vladimirov D. O., Lashkevich K. A. A quick guide to small-molecule inhibitors of eukaryotic protein synthesis. Biochemistry (Moscow) 2020;85:1389–1421. doi: 10.1134/S0006297920110097. [DOI] [PMC free article] [PubMed] [Google Scholar]