Abstract
The accumulation of protein aggregates and dysfunctional organelles as organisms age has led to the hypothesis that aging involves general breakdown of protein quality control. We tested this hypothesis using a proteomic and informatic approach in the fruit fly Drosophila melanogaster. Turnover of most proteins was markedly slower in old flies. However, ribosomal and proteasomal proteins maintained high turnover rates, suggesting that the observed slowdowns in protein turnover might not be due to a global failure of quality control. As protein turnover reflects the balance of protein synthesis and degradation, we investigated whether decreases in synthesis or decreases in degradation would best explain the observed slowdowns in protein turnover. We found that while many individual proteins in old flies showed slower turnover due to decreased degradation, an approximately equal number showed slower turnover due to decreased synthesis, and enrichment analyses revealed that translation machinery itself was less abundant. Mitochondrial complex I subunits and glycolytic enzymes were decreased in abundance as well, and proteins involved in glutamine-dependent anaplerosis were increased, suggesting that old flies modify energy production to limit oxidative damage. Together, our findings suggest that age-related proteostasis changes in Drosophila represent a coordinated adaptation rather than a system collapse.
Keywords: Aging, Degradation, Drosophila, Metabolism, Protein turnover, Translation
A substantial percentage of a cell’s energy is invested in maintaining proteostasis, the equilibrium in which the cell produces adequate supplies of functional proteins and efficiently degrades nonfunctional proteins (1,2). As organisms age, energy generation decreases (3), and insoluble proteins often accumulate (2). For these reasons among others, the idea that protein turnover slows down with age is intuitively appealing. However, work to date has not consistently supported this theory (4–6). Moreover, those studies reporting slower protein turnover in aging have not established an underlying mechanism. Some have attributed slower turnover to a decrease in protein synthesis capacity with age and others to failure of protein degradation systems (7,8). To distinguish between these possibilities, we measured protein turnover in the heads of old and young Drosophila melanogaster and investigated age-related changes using an informatic approach.
Method
Drosophila Strains and Culture
Fly stocks were maintained on standard cornmeal-molasses food at 25°C. The flies used for the aging study were w1118, obtained from the Bloomington Stock Center (stock 3605). The Atg7d4 and Atg7d77 alleles have been previously described (9). Atg7 null mutants were Atg7d4/Atg7d77 transheterozygotes. See our previous work (10) for a detailed description of the WT controls used for Atg7.
Life Span
Male w1118 flies (n = 238) were collected on the day of eclosion and maintained in groups of approximately 20 on standard food, changed every 2–3 days. Kaplan–Meier survival analysis was performed using GraphPad Prism software.
In Vivo Stable Isotope Labeling of Flies
Yeast labeled with [5,5,5-2H3]–leucine (D3-leucine; 99 atom % deuterium) was prepared as previously described (11). Atg7 mutants and their controls were labeled using D3-leucine yeast paste as previously described (10). In the aging study, groups of 20–30 male w1118 flies were labeled using labeled cornstarch-molasses food as previously described (11). Three biological replicates (~50 heads each) were obtained for each group and time point.
Mass Spectrometry
Frozen flies were vortexed to remove heads, and the isolated heads were homogenized in 0.1% RapiGest solution in 50 mM ammonium bicarbonate (Waters) using a 0.2-mL micro tissue grinder (Wheaton Industries). For further details of sample prep, liquid chromatography, and mass spectrometry, please see Supplementary Methods.
Calculation of Protein Turnover and Abundance
Turnover rates were calculated using Topograph software (4). For details, see Supplementary Methods.
We excluded proteins with excessive inter-replicate variability of turnover rates, defined as follows. We calculated the turnover rate separately for each biological replicate and determined the coefficient of variation across replicates. Proteins with a coefficient of variation ≥0.25 were excluded from the analysis. Proteins were analyzed only if they met inclusion criteria in both old and young (or mutant and control) flies. In total, 1438 proteins met the criteria for turnover analysis and 6805 for abundance, and 1096 had values for both turnover and abundance.
Statistical significance of fold change (FC) in half-life was calculated for groups of proteins using nested analysis of variance, and significance of turnover change for individual proteins was calculated using Student’s t tests, with multiple testing correction as discussed in Supplementary Methods. For abundance, the Wilcoxon signed-rank test was used to test for changes in groups of proteins. In both turnover and abundance studies, we performed intergroup comparisons using the Mann–Whitney U test or the Kruskal–Wallis test with Dunn post-tests, as appropriate.
We measured protein abundance from the same raw mass spectrometry data used in the turnover study, using Skyline (12) and MSstats (13). Prior to MSstats analysis, we obtained total abundance (labeled plus unlabeled) for each peptide using a custom R script. The statistical significance of intergroup differences was calculated using a linear mixed model, then adjusted for multiple comparisons by the Benjamini–Hochberg procedure with a false discovery rate of 0.05. Abundance comparisons were made at the time point when intergroup differences were most marked (120 hours for old vs young flies, 240 hours for Atg7 mutants vs controls).
Annotation and Classification of Drosophila Proteins
General
Drosophila protein localization was determined from a variety of resources as previously described (10).
Proteasome substrates
We designated a protein as a proteasome substrate (418 detected in both turnover and abundance analyses) if its mammalian ortholog was identified as a substrate in either of 2 studies (14,15). We obtained the Drosophila orthologs using the DRSC Integrative Ortholog Prediction Tool v6 (16), minimum score 5.
Endosomal microautophagy substrates
We identified microautophagy substrates by searching for targeting sequences, also called KFERQ-like motifs. These motifs were defined as sequences of 5 amino acids (AAs) that fit criteria established by Dice (17,18):
(1) The sequence begins or ends with Q.
(2) The sequence contains either 1 or 2 basic AAs (K, R), 1 or 2 bulky hydrophobic AAs (F, I, L, V), and one acidic AA (D, E).
We wrote an algorithm to search protein sequences for these motifs using Python 2.7 and applied it to the fly proteome (FASTA sequences downloaded from FlyBase). We then identified cytosolic proteins by annotation as described above and compared the effects of age on cytosolic proteins with and without KFERQ-like sequences (117 cytosolic proteins detected in both turnover and abundance analyses had KFERQ-like sequences; 92 did not).
Macroautophagy substrates
As described in the text, we considered proteins from the following organelles to be canonical autophagy substrates: mitochondria, ribosomes, endoplasmic reticulum, peroxisomes, and proteasomes.
Substrates of multiple degradation mechanisms
We considered a protein a substrate of multiple degradation mechanisms if it met the criteria for 2 or more of the 3 mechanisms listed above.
Enrichment Analysis
We performed enrichment analysis using the FlyEnrichr online tool (19), using 2 unweighted lists: (a) proteins with significantly slower turnover and significantly decreased abundance and (b) proteins with significantly slower turnover and significantly increased abundance. We chose the KEGG pathway and GO Biological Process analyses as the most informative, and the top 10 significant enrichment categories in each analysis are displayed.
Results and Discussion
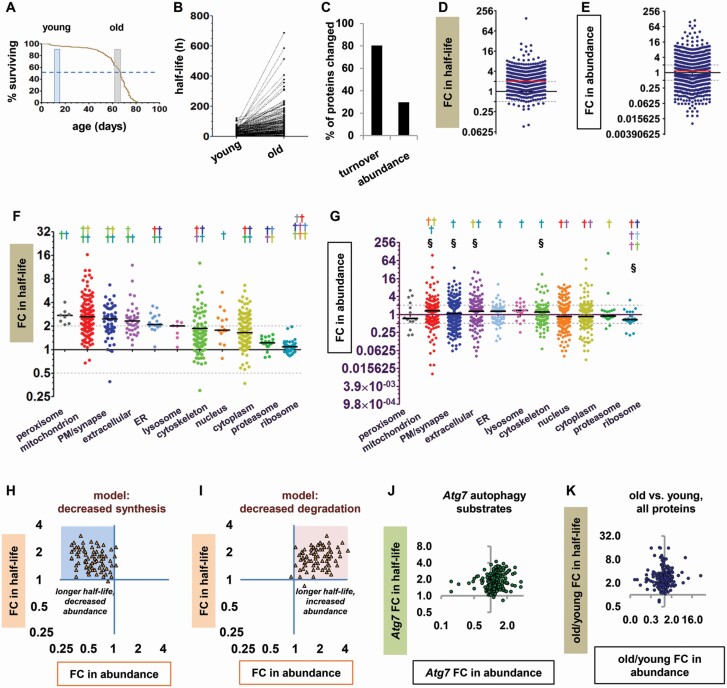
We measured the half-life and abundance of proteins from the heads of old and young flies as previously described (20). After a stable isotope labeling period of 5 or 10 days, young flies were frozen at 10–15 days of age and old flies at 62–67 days, so that the oldest flies were at the population 50% survival point (Figure 1A). FC in half-life was calculated by dividing a protein’s half-life in old flies by its half-life in young flies, such that FC > 1 indicates a protein with slower turnover in old flies and FC < 1 indicates a protein with faster turnover in old flies. We found that 80.3% of head proteins had significantly slower turnover in old flies than in young ones (Figure 1B and C; Supplementary Figure S1), with a median FC in half-life of 2.04 (Figure 1D). Many proteins also showed significant differences in abundance between old and young flies (29.7%, Figure 1C; Supplementary Figure S1); however, the median FC in abundance was only 1.17 (Figure 1E), reflecting nearly equal numbers of proteins with increased and decreased abundance in old flies relative to young ones. When we assessed the effects of age on proteins localized to different parts of the cell, we found that turnover slowed with age in all cellular components analyzed, but the magnitude of the effect varied greatly from one group of proteins to another (Figure 1F), and there was no clear relationship to group changes in abundance (Figure 1G). While peroxisomal and mitochondrial proteins had large median FCs, turnover changes in proteasomal and ribosomal proteins were minimal. As ribosomes, proteasomes, peroxisomes, and mitochondria are all degraded by autophagy, autophagy substrates thus included the protein groups with the smallest effects on turnover as well as those with the largest effects. This disparity could be explained by differential contribution of compensatory turnover mechanisms, but more fundamentally, maintaining high turnover rates for some of the cell’s most abundant proteins seemed inconsistent with a profound general failure of protein degradation.
Figure 1.
Protein turnover slows in old flies due to protein-specific changes in synthesis or degradation. (A) Life span of the male w1118 flies used in this study. Shaded windows represent the ages of the young and old flies at the time of harvest. Dashed line represents 50% survival. (B) Before–after plot showing half-lives in hours (h) of all proteins detected in young and old flies. (C) Percentages of fly head proteins significantly changed in turnover or abundance. (D) Fold change (FC) in half-life for all proteins in old versus young flies. FC > 1 indicates slower turnover in old flies than in young flies, and FC < 1 indicates faster turnover in old flies. In this and subsequent panels, solid horizontal line = median; gray dashed lines = 2-fold and 0.5-fold change. (E) FC in abundance for proteins in old versus young flies. Dot plots of abundance show only proteins significantly different between old and young flies. (F and G) FC in half-life (F) and abundance (G) for proteins from different cellular components. Each colored dagger (†) symbol indicates a significant difference from the group with the matching dot color (Kruskal–Wallis test with Dunn post-tests). In panel F, all protein groups have significantly longer half-lives in old than in young flies by nested analysis of variance (p < .05). (H and I) Theoretical models of the proteostasis effects caused by (H) decreased synthesis and (I) decreased degradation. FC in half-life is plotted against FC in abundance for individual model proteins. (H) Blue shading highlights theoretical data points with slower turnover and decreased abundance. (I) Pink shading highlights theoretical data points with slower turnover and increased abundance. (J) FC in half-life versus FC in abundance for autophagy substrate proteins in autophagy-deficient Atg7 mutants. (K) FC in half-life versus FC in abundance for all head proteins in old versus young flies. Among the proteins with significantly longer half-lives, 182 were significantly decreased in abundance and 197 were significantly increased in abundance.
As protein turnover is the net result of synthesis and degradation, we next investigated whether the observed slowdowns in protein turnover resulted from decreases in synthesis or decreases in degradation. To do this, we created theoretical models integrating changes in protein turnover with changes in abundance, similar to the approach used by Dörrbaum et al. (21). Specifically, we plotted FC in half-life against FC in abundance for each modeled protein. If synthesis decreases but degradation remains constant, proteins with slower turnover will also be decreased in abundance; in the model, this is represented by data points clustering in the upper left quadrant of the graph (Figure 1H). Conversely, if degradation decreases but synthesis remains constant, proteins with slower turnover will be increased in abundance. This is represented by data points clustering in the upper right quadrant (Figure 1I). We compared these predictions to a plot of our previously published data on autophagy substrate proteins in autophagy-deficient flies (Atg7 mutants (20)). Despite the potential for compensatory changes in Atg7 mutants, the pattern of turnover and abundance changes closely resembled the prediction for changes due to decreased degradation (Figure 1J). Finally, we plotted the full data set from old versus young flies in the same format (Figure 1K). Nearly equal numbers of data points were found in the left and right upper quadrants, in an apparent combination of the 2 predictions. This pattern suggests that the slowdown of any individual protein in the heads of old flies is as likely to arise from a decrease in synthesis as from a decrease in degradation, or from a combination of both.
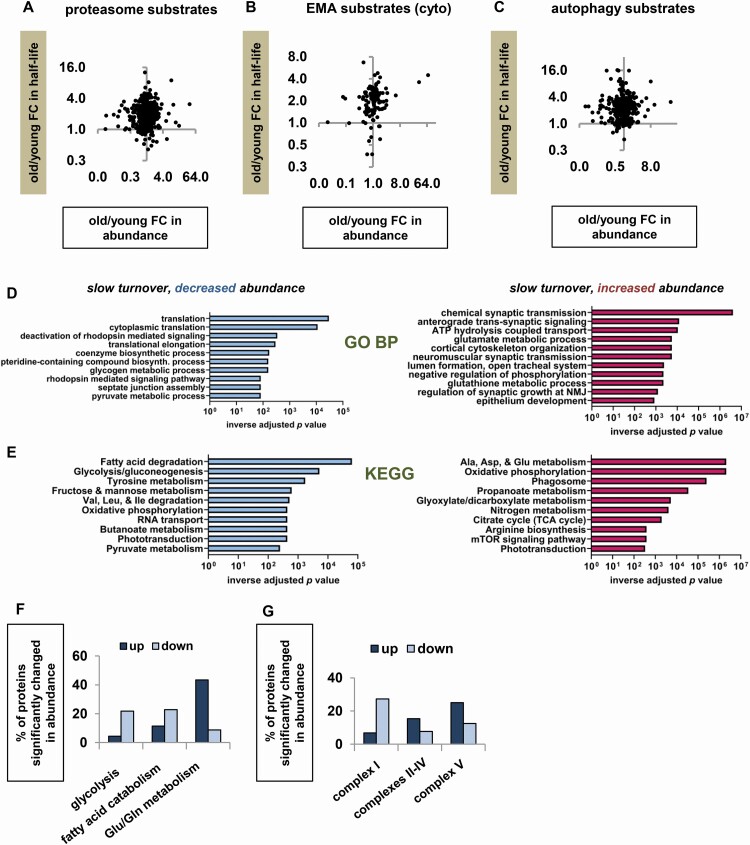
We considered the possibility that while degradation defects did not predominate among the proteins overall, they might predominate among substrates of specific degradation pathways, especially if some pathways were severely impaired and others were relatively intact. We also predicted that substrates of multiple degradation pathways would show less turnover slowdown than proteins without redundant degradation mechanisms. To test these predictions, we repeated the previous analyses (FC in turnover vs FC in abundance) for substrates of the proteasome (14,15), endosomal microautophagy (17), and autophagy (20). For each of the 3 groups of substrates, the data points were distributed across both upper quadrants (Figure 2A–C), suggesting that some of the proteins had slower turnover due to decreased degradation and some due to decreased synthesis. The same was true for substrates of more than one degradation mechanism (Supplementary Figure S2A). Finally, proteins with multiple known degradation mechanisms showed the same degree of age-related slowdown of turnover as other proteins (Supplementary Figure S2B and C). Taking all these findings together, we conclude that there is a marked slowdown of protein turnover in old flies, which appears to be caused by protein-specific decreases in synthesis and degradation.
Figure 2.
Proteostasis changes in old flies suggest a metabolic remodeling including decreased translation and changes in energy production. (A–C) FC in half-life versus FC in abundance in old versus young flies for substrates of different degradation pathways. (A) Proteasome substrates. (B) Cytosolic protein substrates of endosomal microautophagy (EMA). (C) Organellar substrates of macroautophagy. (D and E) FlyEnrichr enrichment analysis of proteins with significantly longer half-lives in old flies, comparing proteins with significantly decreased abundance (blue bars) and increased abundance (red bars). (D) Top 10 enriched categories in GO Biological Process (BP) analysis. (E) Top 10 enriched categories in KEGG pathway analysis. (F and G) Percentages of various protein categories changed in abundance in old flies, including all proteins with abundance data (not only the subset with turnover data). (F) Percentage of proteins increased and decreased in abundance in old flies for proteins involved in glycolysis, fatty acid catabolism, and metabolism of glutamine and glutamate. (G) Percentage of proteins increased and decreased in abundance in old flies for respiratory chain (RC) proteins.
We next investigated whether proteins with slower turnover due to decreased synthesis differed functionally from those with slower turnover due to decreased degradation. FlyEnrichr enrichment analysis (19) showed that the subgroup of proteins with decreased abundance included a large number of translation-related proteins, while the subgroup with increased abundance was enriched in proteins related to synaptic function, glutathione metabolism, and mTOR signaling (Figure 2D and E). We also detected evidence of wide-ranging changes in intermediary metabolism. In particular, enzymes involved in glutamine-dependent anaplerosis and the glutamine–glutamate cycle were increased in abundance in old flies (Figure 2E and F). Of note, many of these enzymes are predominantly expressed in glia rather than neurons, suggesting that metabolic changes in the fly head may vary by tissue or cell type; it would be fruitful to compare neurons and glia in future work, as well as to analyze non-head tissues with different metabolic demands (eg, thoracic muscle). Finally, we found that glycolysis proteins and complex I subunits showed decreased abundance, although subunits of other respiratory chain complexes did not (Figure 2F and G). As glycolysis and complex I–dependent respiration both generate toxic by-products (reactive oxygen species and methylglyoxal), some of the proteostasis changes in old flies could represent a shift to a less damaging way of generating energy, consistent with previous findings of energy metabolism changes in long-lived fly mutants (22).
In summary, we have found that most proteins in the fly head have slower turnover in advanced age, and that this slowdown appears to reflect a complex change in proteostatic equilibrium rather than a catastrophic failure of quality control. Reduced translation is a key component of multiple life-extending interventions (23), and our findings suggest that a moderate decrease in translation over time is also a normal and possibly adaptive part of natural aging. Although much has been written about the importance of mitochondrial quality control in healthy aging, we did not find that mitochondrial proteins were spared from the general slowdown in turnover. Instead, we found that old flies maintained turnover of ribosomal and proteasomal proteins with only slight slowing (~12% for the ribosome). The combination of preserved ribosomal protein turnover rate and decreased ribosomal protein abundance suggests that old flies have a small pool of frequently replaced ribosomes; this scenario is hypothesized to maximize translational accuracy (24), which has been linked to increased longevity (25). Taken together, our findings suggest that age-related changes in protein turnover and abundance in Drosophila may represent not proteostatic collapse but a controlled and adaptive remodeling of the aging proteome.
Supplementary Material
Acknowledgments
We thank Thomas Neufeld for providing fly stocks and Nick Shulman for writing the R script used in abundance analysis. We thank the Brewer Lab at the University of Washington for providing the BB14-3A strain and for their gracious assistance with yeast culture.
Funding
This work was funded by a grant from Samsung Corporation and by the National Institutes of Health grants R01NS94252 and R01GM104990.
Author Contributions
E.S.V. and L.J.P. conceived and designed the experiments. E.S.V. and G.E.M. performed them, with resources and knowledge provided by M.J.M. E.S.V. analyzed the data with input from R.E.T. E.S.V., R.E.T., and L.J.P. wrote the article.
Conflicts of Interest
None declared.
References
- 1.Kepp KP, Dasmeh P. A model of proteostatic energy cost and its use in analysis of proteome trends and sequence evolution. PLoS One. 2014;9:e90504. doi: 10.1371/journal.pone.0090504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne BA, Chinnery PF. Mitochondrial dysfunction in aging: much progress but many unresolved questions. Biochim Biophys Acta. 2015;1847:1347–1353. doi: 10.1016/j.bbabio.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basisty N, Meyer JG, Schilling B. Protein turnover in aging and longevity. Proteomics. 2018;18:e1700108. doi: 10.1002/pmic.201700108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popa-Wagner A, Sandu RE, Cristin C, et al. Increased degradation rates in the components of the mitochondrial oxidative phosphorylation chain in the cerebellum of old mice. Front Aging Neurosci. 2018;10:32. doi: 10.3389/fnagi.2018.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rennie MJ, Selby A, Atherton P, et al. Facts, noise and wishful thinking: muscle protein turnover in aging and human disuse atrophy. Scand J Med Sci Sports. 2010;20:5–9. doi: 10.1111/j.1600-0838.2009.00967.x [DOI] [PubMed] [Google Scholar]
- 7.Webster GC, Webster SL. Decline in synthesis of elongation factor one (EF-1) precedes the decreased synthesis of total protein in aging Drosophila melanogaster. Mech Ageing Dev. 1983;22:121–128. doi: 10.1016/0047-6374(83)90105-7 [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Cao Y, Zhao J, Fang Y, Liu N, Zhang Y. Multidimensional proteomics identifies declines in protein homeostasis and mitochondria as early signals for normal aging and age-associated disease in Drosophila. Mol Cell Proteomics. 2019;18:2078–2088. doi: 10.1074/mcp.RA119.001621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juhász G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincow ES, Thomas RE, Merrihew GE, et al. Autophagy accounts for approximately one-third of mitochondrial protein turnover and is protein selective. Autophagy. 2019;15:1592–1605. doi: 10.1080/15548627.2019.1586258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas RE, Vincow ES, Merrihew GE, MacCoss MJ, Davis MY, Pallanck LJ. Glucocerebrosidase deficiency promotes protein aggregation through dysregulation of extracellular vesicles. PLoS Genet. 2018;14:e1007694. doi: 10.1371/journal.pgen.1007694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi M, Chang CY, Clough T, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305 [DOI] [PubMed] [Google Scholar]
- 14.Wagner SA, Beli P, Weinert BT, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111.013284. doi: 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braten O, Livneh I, Ziv T, et al. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci U S A. 2016;113:E4639–E4647. doi: 10.1073/pnas.1608644113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Flockhart I, Vinayagam A, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8 [DOI] [PubMed] [Google Scholar]
- 18.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144 [DOI] [PubMed] [Google Scholar]
- 19.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincow ES, Merrihew G, Thomas RE, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorrbaum AR, Alvarez-Castelao B, Nassim-Assir B, Langer JD, Schuman EM. Proteome dynamics during homeostatic scaling in cultured neurons. eLife. 2020;9:e52939. doi: 10.7554/eLife.52939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Davis SS, Borch Jensen M, et al. JNK modifies neuronal metabolism to promote proteostasis and longevity. Aging Cell. 2019;18:e12849. doi: 10.1111/acel.12849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonskikh Y, Polacek N. Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev. 2017;168:30–36. doi: 10.1016/j.mad.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 24.Mathis AD, Naylor BC, Carson RH, et al. Mechanisms of in vivo ribosome maintenance change in response to nutrient signals. Mol Cell Proteomics. 2017;16:243–254. doi: 10.1074/mcp.M116.063255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie J, de Souza Alves V, von der Haar T, et al. Regulation of the elongation phase of protein synthesis enhances translation accuracy and modulates lifespan. Curr Biol. 2019;29:737–749.e5. doi: 10.1016/j.cub.2019.01.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.