Abstract
Purpose
BRCA1 associated protein-1 (BAP1), a nuclear deubiquitinase thought to be involved in DNA double-strand break repair is frequently mutated in mesothelioma. Because poly-(ADP-ribose) polymerase inhibitors (PARPIs) induce synthetic lethality in BRCA1/2 mutant cancers, we evaluated whether BAP1 inactivating mutations confer sensitivity to PARPIs in mesothelioma and if combination therapy with temozolomide (TMZ) is beneficial.
Methods
Ten patient-derived mesothelioma cell-lines were generated and characterized for BAP1 mutation status, protein expression, nuclear localization and sensitivity to the PARPIs olaparib and talazoparib alone or in combination with TMZ. BAP1 deubiquitinase (DUB) activity was evaluated by ubiquitin-AMC assay. BAP1-knockout (KO) mesothelioma cell-lines were generated by CRISPR/Cas9. Because Schlafen 11 (SLFN11) and O6-methylguanine-DNA methyltransferase (MGMT) also drive response to TMZ and PARPIs, we tested their expression and relationship with drug response.
Results
BAP1 mutations and/or copy-number alterations were present in all ten cell lines. However, four cell lines exhibited intact DUB activity and two had nuclear BAP1 localization. Half-maximal inhibitory concentrations of olaparib and talazoparib ranged from 4.8 μM to >50 μM and 0.039 μM to >5 μM, respectively, classifying them into sensitive (two) or resistant (seven) cells, independent of their BAP1 status. Cell lines with BAP1-KO resulted in loss of BAP1 DUB activity but did not increase sensitivity to talazoparib. Response to PARPI tended to be associated with high SLFN11 expression, and combination with temozolomide increased sensitivity of cells with low or no MGMT expression.
Conclusions
BAP1 status does not determine sensitivity to PARPIs in patient-derived mesothelioma cell-lines. Combination of PARPI with TMZ may be beneficial for patients whose tumors have high SLFN11 and low or no MGMT expression.
Keywords: Mesothelioma, Poly(ADP-ribose) polymerase inhibitors (PARPIs), BRCA1 associated protein 1 (BAP1), olaparib, talazoparib, temozolomide, synergism, Schlafen 11 (SLFN11), O6-methylguanine- DNA methyltransferases (MGMT)
INTRODUCTION
Since 2014, the US Food and Drug Administration has approved several PARP inhibitors (PARPIs) including olaparib and talazoparib for the treatment of advanced and recurrent ovarian cancer carrying BRCA1/2 mutations 1, 2. PARPIs compete with β-nicotinamide adenine dinucleotide (ß-NAD+) for binding to the catalytic pockets of PARP1 and PARP2 enzymes. As a result, PARPIs trap PARP1/2 3, 4 and block PARylation, a major step in the base excision repair (BER) pathway for DNA single-strand breaks (SSB) 2, 5, 6. Hence, cells treated with PARPIs accumulate PARP1/2-DNA complexes and unrepaired SSBs that results in replication fork stalling and formation of double-strand breaks (DSBs). DSBs and PARP trapping are lethal in homologous recombination (HR)–deficient cancer cell 3, 4, 7, 8. Therefore, cancer cells with BRCA1/2 mutations are common cellular targets for PARPIs as they encode defective key HR-mediated DNA repair proteins. Talazoparib is the most potent catalytic and PARP trapping inhibitor among the current clinical PARP inhibitors 4. Combination of DNA alkylating agent temozolomide (TMZ) further enhances the efficacy of PARPIs in several cancers 4, 9–12 by inducing base damage, causing recruitment of PARP1/2 for BER 4 and PARP trapping. In this study, we sought to determine if patient derived mesothelioma cell lines demonstrate similar synergy using talazoparib and TMZ.
Mesothelioma, an aggressive cancer of the serosal surfaces of pleura and peritoneum, is predominantly linked to asbestos exposure. Recently, many groups including ours have shown germline mutations of BAP1 (BRCA1-associated protein 1) present in mesothelioma patients. In addition, it has been found that patients with germline BAP1 mutations also had somatic “second hits” in their tumor 13–17. Other somatic studies had also uncovered loss of BAP1 nuclear staining in up to 67% of 70 malignant mesothelioma biopsies 18. Likewise, in another study, 23% of 53 with malignant pleural mesothelioma have somatic BAP1 inactivating mutation 19.
BAP1 is a nuclear deubiquitinase with N-terminal ubiquitin C-terminal hydrolase domain (UCH) and two C-terminal nuclear localization signals (NLS1, NLS2) functioning as a tumor suppressor through multiple mechanisms 20. Acting as a component of Polycomb repressive deubiquitinase complexes, BAP1 deubiquitinates histone H2A, leading to transcriptional activation of genes that regulate cell growth 21. BAP1 also regulates transcription by associating with several transcription factors and co-factors such as Ying Yang 1 (YY1), host cell factor-1 (HCF1) and E2F1 to induce transcription of genes involved in cell cycle regulation 22, 23. BAP1 plays role in DNA damage response (DDR) through various mechanisms. It modulates the E3 ligase activity of the BRCA1-BARD1 (BRCA1-associated RING domain 1) heterodimer by binding and deubiquitinating BARD1 24. It co-localizes with γ-H2AX following DNA damage 25, recruits RAD51 and BRCA1 to DSB sites 26, promotes the repair of DSBs under ATM regulation and enhances cell survival after DNA damage 25. Recently it has been shown that cytoplasmic BAP1 modulates IP3R3 mediated calcium release from mitochondria and induces apoptosis 27.
BAP1’s presumed role in HR repair led us to hypothesize that patient-derived mesothelioma cell lines harboring BAP1 mutations would be hypersensitive to PARPIs. To clarify whether BAP1 mutations/inactivation could serve as a predictive biomarker for treatment response to PARPIs in mesothelioma, we established ten patient-derived cell lines from ascites or pleural fluid. We assessed the sensitivity of these cell lines to PARPIs with respect to their BAP1 status. To further consolidate our findings, we generated isogenic BAP1-knockout and - overexpression systems from mesothelioma patient-derived cell lines and evaluated their response to PARPIs. In an effort to define patient population that would benefit from PARPI treatment, we evaluated the expression of SLFN11 in these cell lines, as recent studies have demonstrated that sensitivity to PARPIs is dependent on SLFN11 expression in cancer cell lines 10, 28. We assessed whether the toxicity of talazoparib could be enhanced by combination with the DNA alkylating agent TMZ and whether a synergistic effect would be dependent on the inactivation of O6-methylguanine-DNA methyltransferase (MGMT), which is commonly inactivated in many cancer besides glioblastomas 29.
MATERIALS AND METHODS
Patient-derived mesothelioma cell lines and drugs
Early-passage mesothelioma cell lines were established from ascites or pleural fluid obtained from mesothelioma patients treated at the NCI (Bethesda, MD) on Institutional Review Board-approved protocols. The methods for establishment of primary culture cell lines have been described previously 30. Cell line authentication was done by Genetica Cell Line Testing- a LapCorp brand (Burlington, NC) using short tandem repeat analysis (Table S1). All patient-derived cell lines were grown in RPMI (Roswell Park Memorial Institute medium) supplemented with 20% FBS (Fetal bovine serum), 1% antibiotic-antimycotic, 1% sodium pyruvate and 1% L-glutamine. Additional cell lines (HCC1937, H2052, H2452, H28, and 293T) were obtained from ATCC. Olaparib, talazoparib and temozolomide were purchased from the Selleckchem.com Inhibitor Expert (Houston, TX). They were reconstituted in dimethyl sulfoxide (DMSO) and were stored at −20°C.
Antibodies
Antibodies against BAP1 (sc-28383) and PARP1 (sc-8007) were obtained from Santa Cruz Biotechnology; Rabbit polyclonal anti-PAR polymer antibody (336-BPC-100) from Trivegin; RhoGDI (06–730) and β-Actin (A5441) from Sigma-Aldrich; anti-H2AX pS139 (REA502) from Miltenyi Biotech. Horseradish peroxidase (HRP)-conjugated antibodies to mouse (Santa Cruz, sc-516102) or rabbit (CST, 7074) were used as secondary antibodies.
Genomic characterization
Genomic DNA was extracted from early passage patient-derived cell lines using Qiagen Blood & Cell Culture DNA Mini Kit (Hilden, Germany). Next generation sequencing (NGS) was performed with a targeted multiplexed amplicon panel, the Oncomine™ Comprehensive Assay v3M (Thermo Fisher Scientific, Grand Island, NY) 31. Ion AmpliSeq™ libraries were prepared with an Ion Chef™ System (Thermo Fisher Scientific) and sequenced on an Ion S5™ XL Sequencer (Thermo Fisher Scientific). Mutation and copy number analysis were performed with Ion Reporter software and all variants were manually reviewed using Integrative Genomics Viewer (IGV).
Cancer cell line genomic and response to olaparib and talazoparib comparisons
Analyses of drug responses across cancer cell line databases were performed using the CellMinerCDB browser (http://discover.nci.nih.gov/cellminercdb) 32. Sensitivity of Olaparib and Talazoparib (BMN 673) has been tested in 860 and 920 cell lines in the NCI-60, and GDSC-MGH-Sanger databases, respectively. Correlations analyses was done from the “Univariate Analyses” tab using the “Compare Patterns” tab (https://www.youtube.com/watch?v=XljXazRGkQ8&feature=youtu.be) 32.
Western blot analysis
Whole cell lysates were prepared using RIPA buffer (ThermoScientific) supplemented with Halt Protease and Phosphatase inhibitor cocktail (ThermoScientific). Lysates were quantified using the BCA protein assay kit (ThermoScientific). Twenty micrograms of protein were separated on a TGX gel (Bio-Rad) and transferred onto PVDF membrane using Trans-Blot Turbo system (BioRad). Western blot analysis was performed according to standard procedures. RhoGDI was used as a loading control. Bound antibodies were detected using Clarity Western ECL Substrate (ThermoScientific) and blots were imaged using a LI-COR Odyssey Fc machine (Li-COR Inc.).
Immunohistochemistry
Formalin-fixed, paraffin embedded sections of each cell line were stained for BAP-1 (clone C-4, Santa Cruz Biotechnology, Santa Cruz, CA, USA) using a standard protocol on the Leica Bond Max Autostainer (Leica Biosystems, Buffalo Grove, IL). The slides were reviewed in a blinded fashion by two cytopathologists (A.F and M.R) with expertise in BAP1 IHC. Only the nuclear expression of BAP1 was considered for evaluation, despite the fact that in some cases fine granular cytoplasmic positivity was also noticed. Photographs were obtained at 200x magnification using an Olympus BX41 microscope with attached Olympus DP27 camera.
Quantitative Real-Time PCR
RNA was extracted using a Qiagen RNeasy® Mini Kit and total RNA concentration was measured using a Nanodrop Spectrophotometer (NanoDrop 200, ThermoScientific). cDNA was synthesized using SuperScript VILO master Mix (Invitrogen) and qPCR was performed in triplicate using PowerUp™ SYBR® Green (Applied Biosystem, Life Technologies). Expression of house-keeping gene Rpl7 was used as internal control. Primer sequences are listed in Table S2.
BAP1 purification by immunoprecipitation (IP) and ubiquitin-AMC assay
Whole cell lysates were prepared using NP40 lysis buffer (Invitrogen, FNN0021) following manufacturer’s instruction. BAP1 antibody was pre-coated onto protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Santa Cruz, CA) by overnight incubation at 4°C. Whole cell lysate equivalent to 250 μg of protein was incubated over night with anti-BAP1 antibody coated agarose beads in 250 μl reaction volume at 4°C. Uncoated agarose beads were used to determine non-specific protein binding. IP complex beads were washed 4× with NP40 lysis buffer. BAP1 was eluted with 50 μl NP40 lysis buffer supplemented with 2 mM DTT. To assess UCH activity, the fluorogenic DUB substrate, 7-amido-4-methylcoumarin derivatized ubiquitin (ubiquitin-AMC, Boston Biochem) was diluted to final concentration of 300 nM in assay reaction buffer (50 mM HEPES pH 7.5, 0.5 mM EDTA and 1mM DTT). 40 μl of BAP1 eluates were incubated with 10 μl of Ub-AMC substrate for 1 hour at room temperature (25 °C), and the levels of hydrolyzed AMC were measured by excitation/emission at 380/460 nm. 6.25 nM of UCH-L3 (Boston Biochem) was used as a positive control.
Cytotoxicity assays
Three thousand tumor cells were seeded in 96-well white plates (PerkinElmer Life and Analytical Sciences, Waltham, MA) in 100 μl of medium per well. Cellular sensitivity was determined by using ATPlite 1-step (PerkinElmer Life, Waltham, MA) after continuous exposure to olaparib or talazoparib as a single agent or in combination with temozolomide (SelleckChem.com) for 96 hours (single agent) and 72 hours (combination) respectively. 50 μl of ATPlite solution was added to each well and after 5 minutes, luminescence was measured by SoftMax M3 Microplate Reader (Molecular Devices). GraphPad Prism 8 was used to plot cell viability curves, sensitivity plots and calculate half maximal inhibitory concentrations (IC50). ATP concentration in untreated wells was defined as 100 % and percentage cell viability of treated cells was calculated as:
Gamma H2AX assay
One million cells were seeded in 6-well plates overnight. Next day, they were incubated with 1μM talazoparib. After 12 hours, cells were harvested and stained with anti-H2AX pS139 antibody (Miltenyi Biotech). Intranuclear staining of gamma-H2AX was carried out using eBioscience™ Foxp3/Transcription Factor Fixation/Permeabilization Concentrate and Diluent (ThermoFisher) following the instructions provided by the manufacturer. Gamma-H2AX level was measured by BD LSRII flow cytometry. Data was analyzed in FlowJo Version 10.
Generation of BAP1-knockout cells
BAP1-deleted cells were generated by using CRISPR/Cas9 technique. sgRNAs were designed using the sgRNA Scorer 2.0 tool 33 and cloned into lentiCRISPRv2 (Addgene plasmid, 52961, a gift from Feng Zhang 34). The guide RNA sequences used to target BAP1 were 5’-ACCGAAATCTTCCACGAGCAGGG-3’ and 5’-CCTCATCGCAGGTGTCAAGGGGG – 3’. A scrambled sgRNA (5’- GCACTCACATCGCTACATCA-3’) was used as a control. BAP1-targetting constructs and packaging plasmids (Applied Biological Materials, Inc.) were co-transfected into 293T cells using Lipofectamine 2000 (11668019, Invitrogen). Lentiparticles were collected 48- and 72-hours post-transfection and were used to infect HCC1937, H2052, and NCI-Meso19 cells. Cells were selected with puromycin for two weeks and single clones were generated by limiting dilution. Single clones were expanded, and knockout was confirmed by Western blot analysis and IHC.
Generation of BAP1-expressing cells
pLenti-C-mGFP-P2A-Puro (PS100093) and a plasmid with a wildtype BAP1 insert (RC200378L4) were obtained from Origene. Packaging plasmids and either the BAP1-expressing plasmid or the empty vector control plasmid were co-transfected into 293T cells using Lipofectamine 2000. Lentiparticles were collected 48- and 72-hours post-transfection and were used to infect H2452, NCI-Meso17, and NCI-Meso21 cells. Infection was followed by two weeks of puromycin selection and overexpression was confirmed by Western blot analysis and RT-qPCR.
Analysis of combination effects
Chou-Talalay method was used to determine synergism for combination effects. The combination index (CI) of each combination treatment was calculated using CompuSyn software (Combosyn. Inc.). CI 0.3–0.7, CI 0.1–0.3, and CI<0.1 were defined as synergism, strong synergism and very strong synergism, respectively 35, 36.
RESULTS
Genomic characterization of BAP1 in patient-derived mesothelioma cell lines reveal frequent BAP1 alterations
Details of cell line establishment and mutation analysis of NCI-Meso16, NCI-Meso17, NCI-Meso18, NCI-Meso19 and NCI-Meso21 were previously reported 30. In the present study, we have established five additional primary patient-derived mesothelioma cell lines. BAP1 mutational analysis was carried out on all ten cell lines using the Oncomine Comprehensive Assay v3M on the Ion S5™ XL platform. All 10 cases were found to have inactivating mutations and/or copy number (CN) abnormalities in the BAP1 gene (Table 1). Two cell lines (NCI-Meso41 and NCI-Meso63) harbored homozygous copy number loss. NCI-Meso18 had partial homozygous loss for the majority of the gene with single copy loss at the 5’-end of the gene encoding exons 1–7. Interestingly, this retained segment encoding the N-terminal UCH domain of the protein. NCI-Meso19 had a calculated CN of 1.18, consistent with loss of heterozygosity (LOH)/single copy BAP1 loss. In our previous study, NCI-Meso19 was identified as BAP1 wild- type (WT) by Sanger sequencing 30. In NCI-Meso44, an ambiguous CN of 0.61 was called possibly indicating coexisting subclones with both single-copy loss and homozygous BAP1 loss. Intra-tumoral heterogenicity and polyclonality in mesothelioma has been reported in several studies18, 37. Further analysis at the single cell level may help to understand the ambiguous result in this case. A sixth cell line, NCI-Meso21 was found to have an inactivating mutation at an allele fraction of 100% and LOH (copy number of 1). Two cell lines, NCI-Meso17 and NCI-Meso29, had BAP1 inactivating mutations at variant allele fractions of 100% and 96% with CNs of 2, most likely representing copy number neutral LOH events. Inactivating mutations were detected in NCI-Meso16 and NCI-Meso52 at variant allele fractions of 49% and 63% with copy numbers 1.7 and 2 respectively, consistent with these variants being heterozygous.
Table 1.
Genomic characterization of BAP1 in patient-derived mesothelioma cell lines using Next Generation Sequencing (NGS), Oncomine platform.
| Cell line | Genomic location | Nucleotide change# | Amino acid change | VAF^ (%) | Copy number (NGS) | LOH/zygosity |
|---|---|---|---|---|---|---|
| NCI-Meso16 | chr3:52442095 | c.256-2A>G | p.? (3’ splice site) | 49 | 1.7 | Heterozygous |
| NCI-Meso17 | chr3:52436694 | c.1985_c.1986-3del | p.? (includes 3’splice site) | 100 | 2 | Homozygous copy neutral-LOH |
| NCI-Meso18 | NA | NA | NA | NA | 0.54** | Homozygous copy number loss (partial), with retention of UCH encoding domain |
| NCI-Meso19 | NA | NA | NA | NA | 1.18 | LOH |
| NCI-Meso21 | chr3:52437839 | c.1302_1322del | p.Asp434fs | ∼100* | 1.09 | Hemizygous/LOH |
| NCI-Meso29 | chr3:52437443 | c.1717del | p.Leu573fs | 96 | 2 | Homozygous copy neutral-LOH |
| NCI-Meso41 | NA | NA | NA | NA | 0 | Homozygous copy number loss |
| NCI-Meso44 | NA | NA | NA | NA | 0.61 | LOH |
| NCI-Meso52 | chr3:52436624 | c.2050C>T | p.Gln684Ter | 63 | 2 | Heterozygous |
| NCI-Meso63 | NA | NA | NA | NA | 0 | Homozygous copy number loss |
transcript = NM_004656.3
VAF = variant allele fraction
identified by manual review of IGV
partial homozygous gene loss, fraction represents algorithmically calculated integration of retained and lost gene segments
NA: Not applicable for CN loss data, LOH: loss of heterozygosity
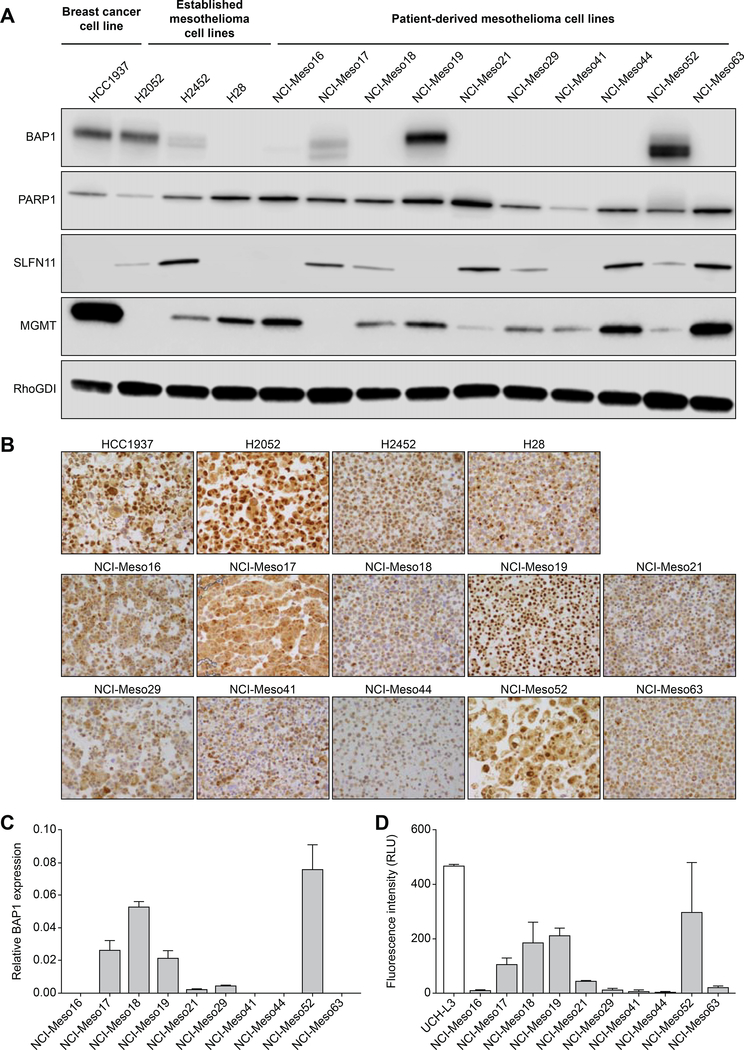
Mesothelioma cells frequently lack BAP1 protein and DUB activity
BAP1 expression was studied in patient-derived cell lines, established mesothelioma cell lines (H2052, H2452 and H28) and a breast cancer cell line (HCC1937). Since, established mesothelioma cell lines and HCC1937 cell line have varying BAP1 expression level, we included them in this study to compare the correlation of BAP1 expression and PARPIs with our patient-derived cell lines. Western blot analysis shows that two of ten patient-derived mesothelioma cell lines (NCI-Meso19 and NCI-Meso52) have high and one cell line (NCI-Meso17) has low BAP1 protein expression, despite the presence of the genomic alterations detected. While it is understandable that NCI-Meso19 with one unaffected allele would express BAP1 protein, we can only speculate the reason for protein expression in NCI-Meso52 and NCI-Meso17. Case NCI-Meso17 has a homozygous (copy number neutral LOH detected) splice site mutation at the 3’ end of the gene in exon 16 (c.1985_c.1986–3del), and similarly case NCI-Meso52 has a homozygous (copy number neutral LOH detected) nonsense mutation also located near the 3’ end of the gene at the end of exon 16 (p.Q684*), again close to the C-terminus of the protein. In both cases it is conceivable that detectable protein could be produced, as both variants are occurring relatively close to the end of the protein. In the first case the predicted effect would be a slightly smaller protein with exon 16 skipping, and for the second case the transcript may have escaped nonsense mediated decay and lead to a foreshortened protein as has been shown for other nonsense mutations located in the penultimate exon38. All other cell lines did not have detectable BAP1 expression. The established cell lines HCC1937 (breast cancer) and H2052 (mesothelioma) have high BAP1 expression, while the mesothelioma cell lines H2452 and H28 have low and undetectable expression, respectively (Figure 1A). BAP1 localization was studied in the cell lines by immunohistochemistry. Our results show nuclear localization of BAP1 in HCC1937, H2052, NCI-Meso19 and NCI-Meso52. Other cell lines did not have nuclear BAP1 localization (Figure 1B). Quantitative RT-PCR assays revealed high BAP1 relative expression in NCI-Meso 17, NCI-Meso 18, NCI-Meso 19 cells and NCI-Meso 52 cells (Figure 1C). Although NCI-Meso18 does not have detectable BAP1 protein, qRT-PCR analysis showed BAP1 gene expression. These results are consistent with the genetic evidence previously discussed indicating that NCI-Meso18 cells have retained an active UCH domain of BAP1 that is not recognized by the antibody used for Western blot analysis which recognizes a 3’ region of the protein.
Figure 1. Patient-derived mesothelioma cells either lack or have low BAP1 protein expression while most of them express PARP1 and MGMT, and a fraction of them express SLFN11 proteins.
(A) Western blot analysis of whole cell lysates demonstrating BAP1, PARP1, SLFN11 and MGMT expression in patient-derived cell lines compared to established breast and mesothelioma cell lines. RhoGDI was used as loading control. (B) Detection of BAP1 protein expression by immunohistochemistry. Nuclear BAP1 localization was noted in HCC1937, H2052, NCI-Meso19 and NCI-Meso52. Magnification 200X. (C) Quantitative RT-PCR was carried out in patient-derived mesothelioma cells to determine BAP1 gene expression level. Primer sequences are provided in Table S2. (D) BAP1 deubiquitinase activity was evaluated using ubiquitin-AMC assay. BAP1 protein was immunopurified from whole cell lysate and incubated with ubiquitin-AMC substrate. Fluorescence intensity of free AMC released due to hydrolysis was measured to determine BAP1 functionality. UCH-L3 was used as a positive control. BAP1 protein in NCI-Meso17, NCI-Meso18, NCI-Meso19 and NCI-Meso52 are functionally active and align with gene expression level measured by qRT-PCR.
Because BAP1 is a deubiquitinase (DUB) that hydrolyzes ubiquitin-linked to H2A during DNA damage repair 39, a functional assay was performed to examine DUB activity of BAP1 in the cell lines using ubiquitin-AMC (7-amino-4-methylcoumarin) as a substrate (Figure 1D). UCH-L3 was used as a positive control. DUB activity was present in the BAP1- expressing cell lines NCI-Meso17, NCI-Meso19 and NCI-Meso52, and surprisingly, in NCI-Meso18 that does not express detectable BAP1 protein expression in Western blotting. Interestingly, the latter case had partial homozygous deletions of BAP1 but retained the UCH domain on one allele as described earlier. We consider these four cell lines as functionally BAP1 positive despite the presence of genetic alterations. The remaining cell lines did not exhibit DUB activity (Figure 1D).
Though NCI-Meso19 exhibits LOH (Table 1) in NGS analysis, we have considered it as a BAP1 expressing cell line in our study because the translated BAP1 protein (Figure 1A) from the unmutated alleles (wild type) was found catalytically active (Figure 1D).
In addition to BAP1, we examined the cell lines for PARP1, SLFN11 and MGMT expression by immunoblotting (Figure 1A) as previous studies have shown that PARP1 is essential for PARP trapping 3, 4 and SFFN11 increases sensitivity to PARPI 10, 28. In addition, MGMT expression level determines sensitivity of several cancer cells to combinatorial treatment with PARPI and TMZ 9, 10. PARP1 expression was found in all the tested cell lines, independent of BAP1. By contrast, SLFN11 expression varied in both established and patient-derived cell lines. Three out of ten patient-derived cell lines (NCI-Meso16, NCI-Meso19 and NCI-Meso41) lack detectable SLFN11 expression (Figure 1A and Figure 2A), which is consistent with other cell line databases (see Figure S1A in 40), which show that SLFN11inactivation is common (~45%) in cancer cells 32, 41. Varying levels of MGMT expression was noted in all the cell lines except in H2052 and NCI-Meso17.
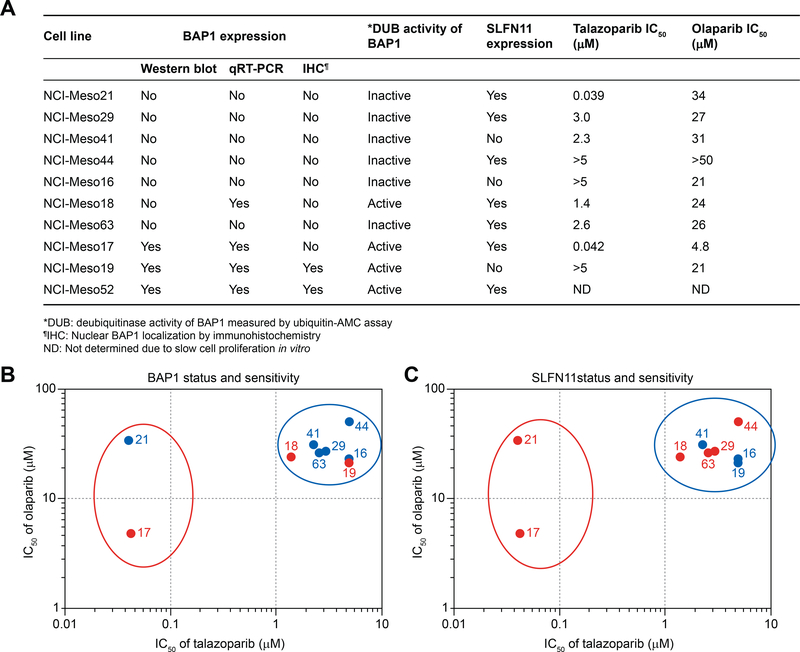
Figure 2. BAP1 DUB activity does not correlate with sensitivity to PARPIs in patient-derived mesothelioma cell lines; however, all the cell lines sensitive to PARPIs are SFN11-positive.
(A) Summary of BAP1 and SFN11 protein expression in ten patient-derived mesothelioma cell lines and their corresponding IC50 for olaparib and talazoparib. (B) Graphical scatter plot based on IC50 of olaparib vs talazoparib shows a clear separation of sensitive (red open oval) and resistant (blue open oval) cell line clusters. One (NCI-Meso17) out of three cell lines having catalytically active BAP1 (red dots) are sensitive while other two (NCI-Meso18 and NCI-Meso19) are resistant. Likewise, NCI-Meso21 (blue dot within red oval) being sensitive to PARPIs does not contain catalytically active BAP1. This data suggests that patient-derived mesothelioma cellular sensitivity to PARPIs does not depend on the activity of BAP1. (C) The sensitivity plot shows SLFN11 expression level. All the SLFN11-negative cells (blue dots) are clustered within resistant group while the SLFN11-positive cells (red dots) are present in both the clusters. Number on each dotted plot represent the cell lines having “NCI-Meso” as prefix.
Sensitivity of patient-derived mesothelioma cell lines to PARPIs does not correlate with lack of BAP1 activity
To determine if BAP1 deficiency leads to sensitivity to PARPIs, we tested two clinical PARPIs, olaparib and talazoparib, against our panel of patient derived mesothelioma cell lines. Due to the variation in half- maximal inhibitory concentration (IC50) of PARPIs against NCI-Meso52, which could be linked to the slow cell proliferation, we excluded this cell line in sensitivity analysis. IC50 of olaparib and talazoparib ranged from 4.8 μM to >50 μM and 0.039 μM to >5 μM, respectively (Figure 2A, Figure S1). Compared to talazoparib, the tested cell lines exhibited a broad range of IC50 for olaparib. NCI-Meso19 expressing BAP1 was resistant to both drugs. While NCI-Meso44 and NCI-Meso16, which lack BAP1 expression, were also highly resistant to both PARPIs. Therefore, we did not observe any negative correlation between BAP1 expression and drug sensitivity.
Based on the sensitivity of the cells to olaparib and talazoparib, a sensitivity plot was constructed, classifying them into two tightly clustered populations: sensitive (2 cell lines, demarked with red open oval) and resistant (7 cell lines, demarked with blue open oval) cells. However, both the clusters contain BAP1 catalytically active (red dots) (1 sensitive and 2 resistant) and inactive cell lines (blue dots) (1 sensitive and 5 resistant) (Figure 2B). Thus, we are unable to detect a correlation between sensitivity to PARPIs based on BAP1 activity. To reinforce these findings, we added the established mesothelioma cell lines (H2052, H2452 and H28) and the breast cancer cell line HCC1937 to the sensitivity plot. H2452 exhibiting low BAP1 protein expression, was sensitive to the drugs. H2052 and HCC1937 having moderate amount of BAP1 protein were resistant to PARPIs (Figure S2 and S3). Taken together, we conclude that there is no clear correlation between BAP1 protein expression and sensitivity to PARPIs.
To examine differential DNA damage between sensitive and resistant cell lines, NCI-Meso21 (sensitive) and NCI-Meso19 (resistant) were treated with talazoparib for 12 hours and γ-H2AX was measured by flow cytometry. We observed an increase in γ-H2AX level after talazoparib treatment in both cell lines (Figure S4). These results suggest that DNA damage is equally induced in both cell lines by talazoparib.
Neither knocking-out nor overexpressing BAP1 alters the sensitivity to talazoparib
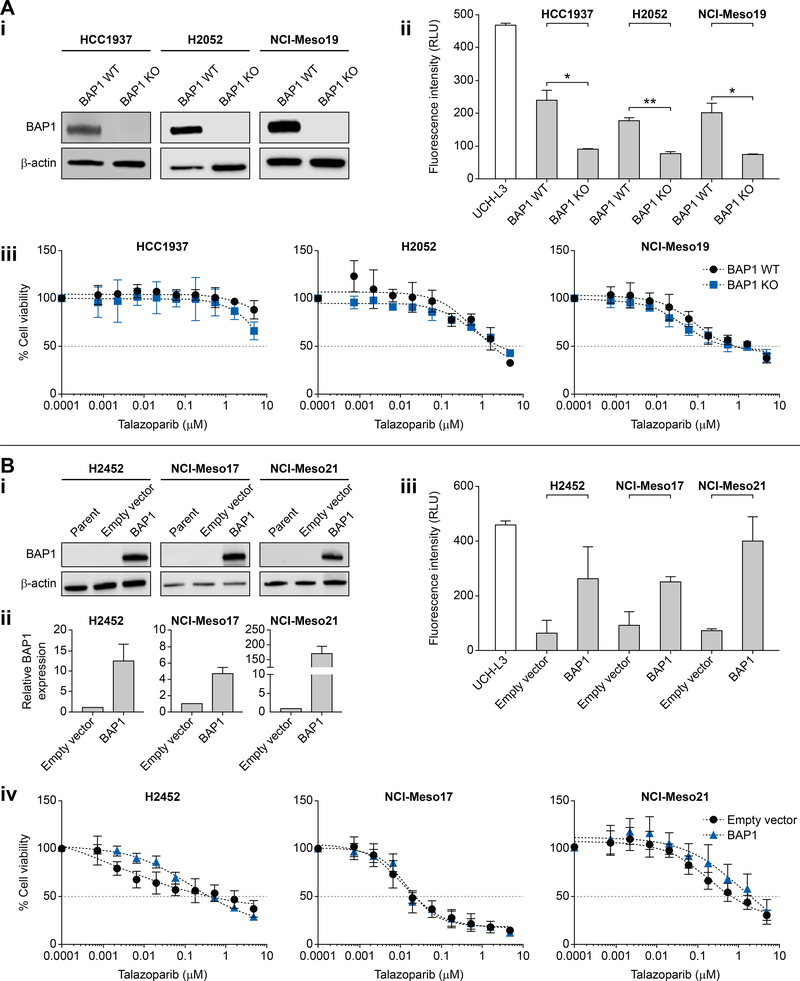
To further examine the effect of BAP1 on PARPIs sensitivity, we established both BAP1-deleted and -overexpressing cellular models using our mesothelioma patient-derived cell lines and also commercially available mesothelioma and breast cancer cell lines to compare their drug sensitivity.
Based on the published role of BAP1 in HR, we hypothesized that the lack of correlation between sensitivity to PARPIs and BAP1 status in our cell line panel could be due to the fact that the cells were non-isogenic. Hence, we tested whether deleting BAP1 would increase sensitivity to talazoparib in isogenic cells. We selected two high BAP1-expressing cell lines, H2052, a BAP1-wild-type mesothelioma cell line and NCI-Meso19, a patient-derived BAP1-wild-type cell line. As positive control, we included the breast cancer cell line, HCC1937, having strong BAP1 protein expression and resistance to PARPIs (Figure 1A and Figure S2A). Using CRISPR/Cas9, we stably knocked out BAP1 in HCC1937, H2052and NCI-Meso19 cells (Figure 3A (i)). We also evaluated H2052 and NCI-Meso19 BAP1 knockout cells for BAP1 expression by IHC. Nuclear BAP1 staining was lost in 95% of cells in H2052 KO and 99% of cells in NCI-Meso19 KO cell lines (Figure S5). DUB activity of BAP1 (Figure 3A (ii)) was assessed in both wild type and knock-out monoclonal cell populations. As expected, knocking-out BAP1 diminished the BAP1 DUB activity in all three cell line models. After isolating monoclonal cell populations by limiting dilution, 96-hour cytotoxicity assays were conducted to test sensitivity to talazoparib. All the BAP1 knockout cell lines failed to show significant alteration in IC50’s. In all cases, IC50’s remained greater than 5 μM (Figure 3A (iii)).
Figure 3: Neither knocking-out nor overexpressing BAP1 alter sensitivity of mesothelioma cells to talazoparib.
A) BAP1 knockout does not increase sensitivity to talazoparib in patient derived as well as established cancer cell lines that endogenously express BAP1. (i) Western blot analysis confirming BAP1 knockout in HCC1937 (breast cancer), H2052 (mesothelioma) and NCI-Meso19 cell lines. Whole cell lysates of cells transduced with BAP1-targeting sgRNA (BAP1 KO) or non-targeting scramble control (BAP1 WT) are included. (ii) DUB assay was performed on both the knockout and BAP1 WT cell lines to confirm the functional deactivation of BAP1, revealing diminished DUB activity in BAP1 KOs. UCH-L3 serves as a positive control of the assay. (iii) Cytotoxicity assays of BAP1 WT and BAP1 KO cell lines using talazoparib. Sensitivity did not differ significantly between the BAP1 WT and BAP1 KO cell lines.
B) Overexpressing BAP1 in H2452, NCI-Meso17 and NCI-Meso21 cell line lacking BAP1, does not decrease sensitivity to talazoparib. (i) BAP1 protein levels were determined by Western blot analysis of whole cell lysates of indicated parent cell lines, cells transduced with control plasmid (empty vector), and plasmid expressing wildtype BAP1. (ii) BAP1 mRNA levels were measured by qRT-PCR. Y-axis represents relative BAP1 expression in wild-type and empty vector -transduced cell lines shows increase in BAP1 gene expression in transduced cells. (iii) Ubiquitin-AMC assay shows increase in deubiquitinase activity in all the BAP1-deficient cell lines transfected with wild-type BAP1. (iv) Cytotoxicity assays in response to talazoparib do not demonstrate significant difference between the cell lines with differential BAP1 expression.
To verify the unaltered response to talazoparib in BAP1 WT and knockout cells, we overexpressed BAP1 by lentiviral transduction in talazoparib-sensitive cells. We hypothesized that over-expression of BAP1 would cause cells to become resistant to talazoparib. NCI-Meso 17 and NCI-Meso21 were chosen as cell lines sensitive to talazoparib with low or no BAP1 expression. The established mesothelioma cell line, H2452, which shares similar characteristics was also included in this set of experiments (Figure 1A and Figure S2B). Overexpression of BAP1 in these cell-lines was confirmed by Western blotting (Figure 3B (i)). BAP1 transcripts in H2452, NCI-Meso17 and NCI-Meso21 were increased by ~12, ~5, and ~170-fold, respectively, compared to the empty vector controls in qPCR analysis (Figure 3B (ii)). We confirmed that the exogenously expressed BAP1 was associated with enhanced DUB activity in all the engineered cells (Figure 3B (iii)). Nevertheless, the BAP1-overexpressing cells did not show any significant difference in sensitivity to talazoparib, and the IC50’s of the cells exogenously expressing BAP1 were nearly identical to the IC50’s of the cells transduced with the empty vector (Figure 3B (iv)). We conclude that BAP1 expression and catalytic activity do not determine cellular sensitivity to PARPIs in mesothelioma cells.
BAP1 status is not correlated with sensitivity to PARPIs against cell lines derived from many different human cancers
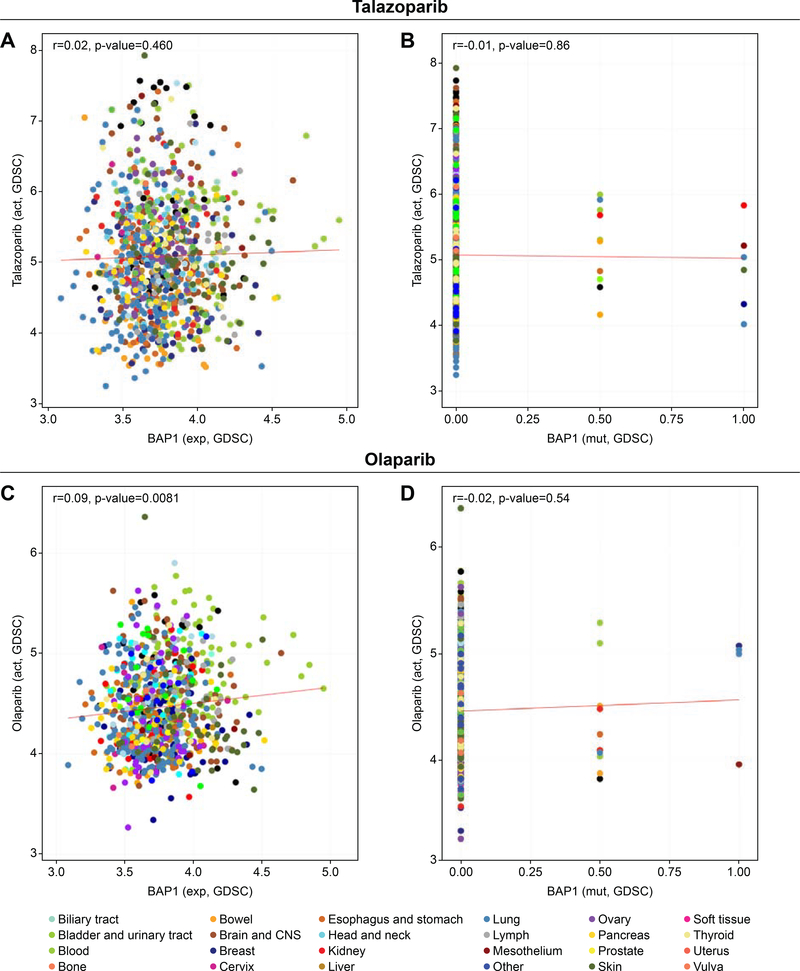
To expand our finding in mesothelioma cells to a broad type of cancer cells, we took advantage of the Genomic of Drug Sensitivity in Cancer (GDSC) database available through the web application CellMinerCBD (http://discover.nci.nih.gov/cellminercdb/) 32, 42, 43. Olaparib and talazoparib have been tested in approximately 900 cell lines originating from various cancers, and their sensitivity profiles were compared to the expression profile of BAP1 (wild-type) as well as BAP1 mutants (heterozygous mutation, homozygous mutation). As per the dataset available, BAP1 expression level as well as BAP1 mutation status poorly correlated with PARPIs sensitivity [Pearson’s correlation coefficient (r) <0.1], which is consistent with the results we obtained in the patient-derived mesothelioma cell lines (Figure 4). Hence, BAP1 expression level and BAP1 mutation status in tumor cells is not a dominant determinant of sensitivity to PARPIs.
Figure 4. BAP1 expression in different human cancer cell lines including BAP1-mutant does not correlate with sensitivity to the PARPIs talazoparib and olaparib.
The graph created using CellMiner web application (http://discover.nci.nih.gov/cellminercdb) shows representing BAP1 expression (x-axis) and drug sensitivity (y-axis) in the GDSC (Genomics of Drug Sensitivity in Cancer). Colored dot represents single cell lines from the indicated tissues of origin. Pearson’s correlation coefficient (r) and two-sided P value between BAP1 transcript and the PARPIs talazoparib (A) and olaparib (C) are indicated. BAP1-mutant tissues also fail to show any correlation between BAP1 mutations and sensitivity to talazoparib (B) or olaparib (D).
Inactivation of SLFN11 and resistance to PARPIs
SLFN11 binds to replication forks in response to DNA damage and causes replication block and cell death, with S-phase arrest 40, 44. Recent studies have revealed that SLFN11 expression is a dominant determinant of sensitivity to PARPIs 28. Studies carried out in isogenic SLFN11-expressing and –deleted cells reveal that SLFN11-positive cancer cell lines are more sensitive to talazoparib and olaparib 10, 28, 40. Examination of the NCI-60 and Sanger-MGH (GDSC) databases show a highly significant correlation between talazoparib sensitivity and SLFN11 expression (Pearson correlations and p-values of 0.6 and 5.5e-07 and 0.44 and 9.5e-45, respectively; Figure S6). Similarly, highly significant correlation was also found for olaparib (r = 0.27; p-value = 7.44e-18) in the GDSC database (http://discover.nci.nih.gov/cellminercdb/) 32.
To determine whether these findings are applicable to our patient-derived mesothelioma cell line model, we assessed SLFN11 protein expression by immunoblotting (Figure 1A). A sensitivity scatter plot of the cell lines with respect to SLFN11 expression showed that all three SLFN11-negative cells (NCI-Meso 16, NCI-Meso19 and NCI-Meso41) were resistant to PARPIs (Figure 2C). While SLFN11-positive cells were present in both the sensitive and the resistant clusters, all sensitive cells were SLFN11-positive. Although a larger sample size and detailed analyses are warranted, our study suggests that inactivation of SLFN11 may confer resistance to PARPIs in mesothelioma cells.
Temozolomide (TMZ) synergizes with talazoparib in patient-derived mesothelioma cell lines depending on their MGMT and SLFN11 status
Recently, drug combinatorial studies have been focusing on combining PARPIs with DNA damaging agents, as a new strategy to increase the efficacy of DNA damaging agents by blocking PARP-mediated DNA repair and increasing PARP trapping by damaging DNA 45. Various studies have reported the synergistic effect between temozolomide and PARPIs across multiple cancer including breast cancer, Ewing’s sarcoma, glioblastoma and small cell lung cancer 9–12, 36. To our knowledge, similar studies have not been carried out in mesothelioma.
TMZ is an alkylating agent that induces DNA damage by methylating guanine residues at their N7 and O6 positions. O6-methylguanine (O6-meG) kills cells by futile cycles of mismatch repair 29, 46. O6-methylguanine-DNA methyltransferase (MGMT), a demethylating DNA repair protein, reverses the O6-guanine methylation by sequestering the methyl group from O6-meG, thereby conferring resistance to TMZ 29, 46, 47. Promoter hypermethylation of the MGMT gene is correlated with improved tumor response and overall survival in glioblastoma patients treated with TMZ in combination with radiotherapy. Similar trend has been observed in SCLC clinical studies 48, 49. However, lack of MGMT expression is also observed in a significant fraction of cancer cells even without promoter hypermethylation 29.
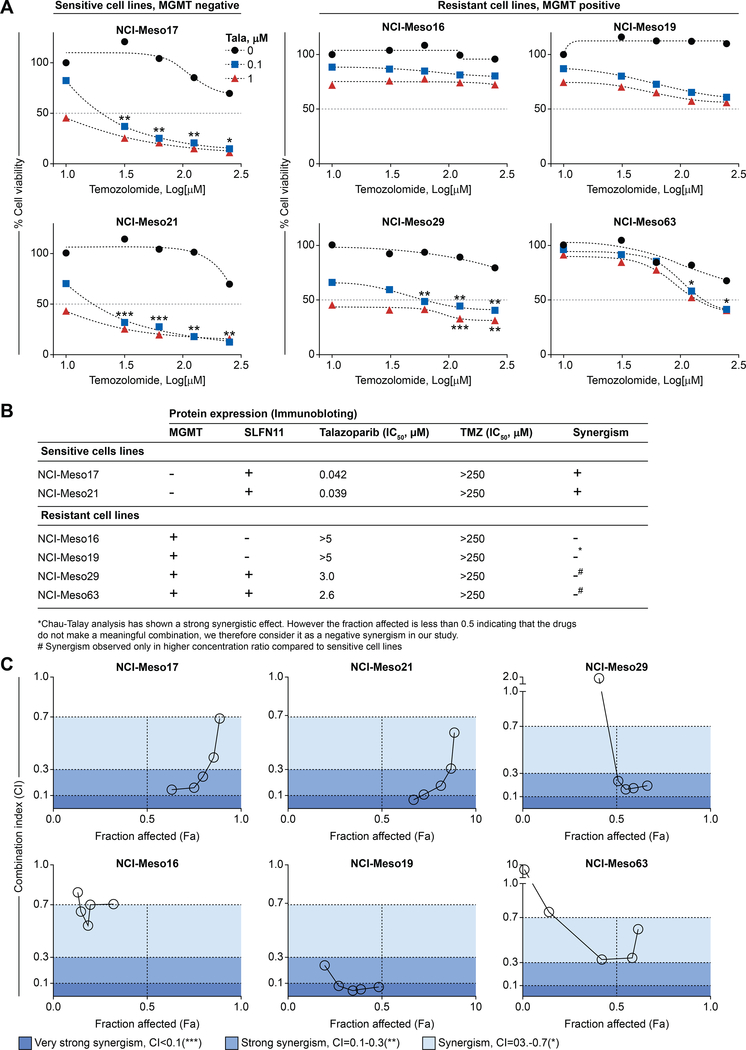
TMZ as a single agent is not effective in mesothelioma 50. However, combination of PARPIs and TMZ increases cytotoxicity by PARP trapping in cancer cell lines 36. Since our panel of patient-derived cell lines showed varying levels of MGMT and SLFN11 expression (Figure 1A), we evaluated the combination of talazoparib and TMZ. We exposed six cell lines (two sensitive and four resistant), with differential MGMT and SLFN11 protein expression levels to increasing concentrations of talazoparib and TMZ. The sensitive cell lines (NCI-Meso17 and NCI-Meso21) exhibited strong synergistic effect in a concentration-dependent manner (Figure 5A and Figure S7) while two out of the four resistant cell lines (NCI-Meso16 and NCI-Meso19) did not show cytotoxicity at any tested concentrations (Figure 5A and Figure S7). Knocking out BAP1 by CRISPER in NCI-Meso 19 expressing BAP1, did not increase sensitivity of the cell line to the combinatorial treatment (Figure S8) further ruling out the role of BAP1 in PARPI mediated sensitivity. In the case of NCI-Meso29 and NCI-Meso63, we observed reduction in cell survival at higher concentrations of temozolomide (Figure 5A and Figure S7). Notably, the sensitive cell lines (NCI-Meso17 and NCI-Meso21) exhibiting strong synergism are either deficient or have low levels of MGMT expression and have relatively higher level of SLFN11expression (Figure 1A), indicating that lack of MGMT and expression of SLFN11 increases sensitivity to TMZ. In contrast, all the resistant cells were MGMT positive. Yet, among those, NCI-Meso29 and NCI-Meso63, which demonstrated synergistic effect at higher concentrations of drugs, were SLFN11-positive as well (Figure 1A and Figure 5B). Interestingly, PARPI resistant H2052 mesothelioma cell line expressing low level of SLFN11 and undetectable level of MGMT did not show synergism (Figure S9). Resistance in this cell line might be due to other factors beyond SLFN11 known to be responsible for tolerance of cancer cells to alkylating agents in absence of MGMT 29, 51, 52. Using the Chau-Talalay synergism analysis method, we plotted fraction-affected (Fa) against combination index (CI) for all the tested cell lines (Figure 5C). We fixed the concentration of talazoparib to 0.1 μM, as this concentration was determined to be optimal for producing synergistic effect in sensitive cell lines and increased the concentrations of TMZ. The CI’s for the other tested concentrations of talazoparib are provided in Figure S7. Fa value 1.0 indicates 100 % drug response, corresponding to 100 % reduction in cell viability. Lower CI with Fa value close to 1 indicates strong synergism and meaningful combination effect. At Fa greater than 0.5, NCI-Meso17 and NCI-Meso21 showed strong synergism in a concentration-dependent manner with CI<0.3. Strong synergism observed at lower concentrations of TMZ weakened with increasing concentrations (Figure 5A and Figure 5C). For NCI-Meso29 and NCI-Meso63, strong synergism was observed at higher concentrations of TMZ. For NCI-Meso16 and NCI-Meso19, the Fa was less than 0.5, indicating lack of synergism at any concentration. Taken together, our analysis shows the potential for increased efficacy of combination therapy in MGMT negative mesothelioma cell lines. While further studies are warranted, SLFN11 status might determine sensitivity to combination therapy in MGMT positive cell lines.
Figure 5: Synergistic effect of talazoparib and TMZ observed in patient-derived mesothelioma cell lines depends on MGMT expression and/or may be influenced by SLFN11 status.
(A) Synergism is observed at lower concentrations of TMZ in MGMT-negative and SLFN11-positive sensitive cell lines (NCI-Meso17, and NCI-Meso21). All the MGMT-expressing cells are resistant and do not show synergism at lower concentrations of TMZ. However, NCI-Meso29 and NCI-Meso63 exhibit synergism at higher concentrations of TMZ. Cellular ATP concentration was used to measure cell viability after continuous drug treatment for 72 hours. The viability of untreated control cells was set as 100 %. (B) Summary reflecting combination study between talazoparib and TMZ in different cell lines having varying MGMT and SLFN11 expression status. (C) Fraction affected (Fa)-CI plot for quantitative analyses of synergistic effects in different concentration drug combination. The plot was obtained from data of Figure 5A for different concentration of TMZ ranging from 31.5 μM to 250 μM with 0.1 μM of talazoparib. Shading of color indicates the level of synergism between PARPIs and TMZ. **CI = 0.1–0.3 and ***CI<0.1, **CI = 0.1–0.3 and *CI = 0.3–0.7 reflects very strong, strong synergistic and synergic effect, respectively. Data are shown in the Figure S7.
DISCUSSION
Here we report that BAP1 is not a determinant of cellular response to PARPIs using patient-derived mesothelioma cell lines, established cancer cell lines, and isogenic BAP1-deleted and overexpressing cell lines. This finding was confirmed in external datasets of cell lines established from many different tumors. However, we observed a strong synergism between TMZ and PARPIs in MGMT-deficient and SLFN-positive patient-derived mesothelioma cell lines.
Since BRCA-mutant cells are sensitive to PARPIs, we hypothesized that the BAP1-defective cells would be sensitive to PARPIs due to synthetic lethality 53, which would open new treatment avenues for BAP1-mutated mesothelioma patients. However, our results do not show any correlation between PARPI sensitivity and BAP1 expression. To further validate the findings, we successfully created BAP1 knockout and overexpressing patient-derived mesothelioma cellular models utilizing CRISPR/Cas9 and lentiviral mediated transfection of BAP1 expressing plasmid, respectively. Because the mesothelioma cell lines in our study exhibited consistent sensitivity within narrow range of IC50 with talazoparib (0.039 μM to >5 μM) compared to olaparib (4.8 μM to >50 μM), we tested talazoparib sensitivity in the knockout and the overexpression cellular models. Both cellular models showed no significant difference in IC50 of talazoparib. Hence, regardless of knocking out or overexpressing BAP1 in these cell lines, there was no difference in sensitivity or resistance to talazoparib.
In divergence to our findings, Peña-Llopis et al., have reported that BAP1-mutant renal cell carcinoma is more sensitive to olaparib than cells reconstituted with wild-type BAP1 54. Also, Yu et al., revealed that knocking out BAP1 in DT40 chicken cells increases sensitivity to olaparib 26. However, the differences in sensitivity results could be related to the fact that DT40 cells have relatively intact genome, which make them a well-suited cellular model system to study DNA repair mechanism 3, 55. Recent study by He et al., have demonstrated that BAP1 mediated apoptosis is tissue specific. BAP1 mutation promotes cell death in mouse embryonic stem cells, fibroblasts, liver and pancreatic tissue, but not in mesothelial cells and melanocytes56. Thus, the differences observed in various studies could be attributed to different cell lines used.
Recently Bononi et al. have shown that apart from nuclear localization that is responsible for genome stability, BAP1 also localizes to endoplasmic reticulum (ER) in the cytoplasm. It deubiquitinates and stabilizes type 3 inositol-1,4,5-triphosphate receptor (IP3R3) in ER and modulates calcium release to cytoplasm and mitochondria, thereby promoting apoptosis. BAP1 mutant cells are not only deficient in HR but are severely compromised in undergoing apoptosis 27, 57. Hence, unlike BRCA1 mutant cells, BAP1 mutant cells keep proliferating despite accumulating DNA damages. Therefore, it is plausible that PARPI induced synthetic lethality might not work for BAP1 deficient cells.
Other reports support our finding that sensitivity of mesothelioma cells to PARPIs may not be related to BAP1 status 19, 58. However, our study is the first to correlate BAP1 mutation status (using NGS), BAP1 DUB functionality and PARPI’s sensitivity using patient-derived cells as well as isogenic patient-derived BAP1 knockout and overexpressing cellular model. Thus, our study provides convincing evidence that BAP1 does not play a major role to determine PARPI’s sensitivity against human mesothelioma cells.
Since the efficacy of drugs is generally enhanced by combination strategy, we explored the rationale of combining talazoparib with TMZ in our patient-derived mesothelioma cell line panel in connection with MGMT and SLFN11 expression. Our results demonstrate strong combinatorial efficacy of talazoparib and TMZ that appears dependent on low or absence of MGMT expression. Conversely, expression of SLFN11 has been shown to sensitize cells to PARPIs 10, 28. In our study, out of the four MGMT-proficient resistant cell lines, two demonstrated synergisms at higher concentrations of TMZ and were SLFN11 positive. Similar MGMT expression level dependent synergy has been observed by Lok et al., where low combination dose was found to be sufficient for synergism in MGMT- low cell lines, whereas high combination dose was required to kill MGMT-high cell lines10. We surmise that SLFN11 expression in the MGMT positive cell lines, can sensitize them to the PARPI-TMZ combination. Hence, our results suggest that TMZ-mediated synergism with talazoparib is negatively correlated with MGMT expression. In addition, SLFN11 expression might be important for synergy in MGMT-positive cells. These initial observations will need to be validated in a larger sample size. In addition, mechanistic studies evaluating the specific contribution of SLFN11 such as use of SLFN11 knockout cell lines are needed to fully understand the synergistic activity of TMZ and talazoparib.
In conclusion, though studies have shown that BAP1 helps in recruiting HR proteins to DSBs 26, its direct involvement in HR-mediated DNA repair is not well understood. Unlike BRCA1/2 mutant breast and ovarian cancers, patient-derived mesothelioma cell lines are sensitive or resistant to PARPIs regardless of BAP1 mutation status, indicating that accumulated DSBs are repaired by alternative mechanisms independent of BAP1. In depth studies are warranted to get a better insight into the role of BAP1 in HR-mediated DNA repair. In addition, patients having MGMT-negative and SLFN11-positive tumor might get benefit from TMZ and PARPI combination therapy.
Supplementary Material
ACKNOWLEDGEMENTS
FUNDING
This research is supported by the NCI Intramural Program, Center for Cancer Research (Z01-BC-006150).
DISCLOSURES
Dr. Thomas reports grants from Astra Zeneca to NCI, during the conduct of the study.
Dr. Hassan reports grants from Bayer AG, grants from Aduro BioTech, grants from Morphotek Inc., outside the submitted work.
Remaining authors have nothing to disclose.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bitler BG, Watson ZL, Wheeler LJ, et al. PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecologic Oncology 2017;147:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pommier Y, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med 2016;8:362ps317. [DOI] [PubMed] [Google Scholar]
- 3.Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res 2012;72:5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murai J, Huang SY, Renaud A, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther 2014;13:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamin RC. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem 1980;255:10493–10501. [PubMed] [Google Scholar]
- 6.Durkacz BW, Omidiji O, Gray DA, et al. (ADP-ribose)n participates in DNA excision repair. Nature 1980;283:593–596. [DOI] [PubMed] [Google Scholar]
- 7.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–921. [DOI] [PubMed] [Google Scholar]
- 8.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913–917. [DOI] [PubMed] [Google Scholar]
- 9.Gill SJ, Travers J, Pshenichnaya I, et al. Combinations of PARP Inhibitors with Temozolomide Drive PARP1 Trapping and Apoptosis in Ewing’s Sarcoma. PLoS One 2015;10:e0140988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lok BH, Gardner EE, Schneeberger VE, et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin Cancer Res 2017;23:523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith MA, Reynolds CP, Kang MH, et al. Synergistic activity of PARP inhibition by talazoparib (BMN 673) with temozolomide in pediatric cancer models in the pediatric preclinical testing program. Clin Cancer Res 2015;21:819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tentori L, Ricci-Vitiani L, Muzi A, et al. Pharmacological inhibition of poly(ADP-ribose) polymerase-1 modulates resistance of human glioblastoma stem cells to temozolomide. BMC Cancer 2014;14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan R, Morrow B, Thomas A, et al. Inherited predisposition to malignant mesothelioma and overall survival following platinum chemotherapy. Proc Natl Acad Sci U S A 2019;116:9008–9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kittaneh M, Berkelhammer C. Detecting germline BAP1 mutations in patients with peritoneal mesothelioma: benefits to patient and family members. J Transl Med 2018;16:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panou V, Gadiraju M, Wolin A, et al. Frequency of Germline Mutations in Cancer Susceptibility Genes in Malignant Mesothelioma. J Clin Oncol 2018;36:2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastorino S, Yoshikawa Y, Pass HI, et al. A Subset of Mesotheliomas With Improved Survival Occurring in Carriers of BAP1 and Other Germline Mutations. J Clin Oncol 2018:3485–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 2011;43:1022–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasu M, Emi M, Pastorino S, et al. High Incidence of Somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol 2015;10:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 2011;43:668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ventii KH, Devi NS, Friedrich KL, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res 2008;68:6953–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahtoe DD, van Dijk WJ, Ekkebus R, et al. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat Commun 2016;7:10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eletr ZM, Wilkinson KD. An emerging model for BAP1’s role in regulating cell cycle progression. Cell Biochem Biophys 2011;60:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H, Mashtalir N, Daou S, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol 2010;30:5071–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa H, Wu W, Koike A, et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res 2009;69:111–119. [DOI] [PubMed] [Google Scholar]
- 25.Ismail IH, Davidson R, Gagne JP, et al. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res 2014;74:4282–4294. [DOI] [PubMed] [Google Scholar]
- 26.Yu H, Pak H, Hammond-Martel I, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A 2014;111:285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bononi A, Giorgi C, Patergnani S, et al. BAP1 regulates IP3R3-mediated Ca(2+) flux to mitochondria suppressing cell transformation. Nature 2017;546:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murai J, Feng Y, Yu GK, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 2016;7:76534–76550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas A, Tanaka M, Trepel J, et al. Temozolomide in the Era of Precision Medicine. Cancer Res 2017;77:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalra N, Zhang J, Thomas A, et al. Mesothelioma patient derived tumor xenografts with defined BAP1 mutations that mimic the molecular characteristics of human malignant mesothelioma. BMC Cancer 2015;15:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hovelson DH, McDaniel AS, Cani AK, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 2015;17:385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajapakse VN, Luna A, Yamade M, et al. CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines. iScience 2018;10:247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chari R, Yeo NC, Chavez A, et al. sgRNA Scorer 2.0: A Species-Independent Model To Predict CRISPR/Cas9 Activity. ACS Synthetic Biology 2017;6:902–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 36.Murai J, Zhang Y, Morris J, et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther 2014;349:408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comertpay S, Pastorino S, Tanji M, et al. Evaluation of clonal origin of malignant mesothelioma. J Transl Med 2014;12:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Xin D, Wang P, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol 2009;7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carbone M, Yang H, Pass HI, et al. BAP1 and cancer. Nat Rev Cancer 2013;13:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murai J, Tang SW, Leo E, et al. SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol Cell 2018;69:371–384 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoppoli G, Regairaz M, Leo E, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A 2012;109:15030–15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinhold WC, Sunshine M, Varma S, et al. Using CellMiner 1.6 for Systems Pharmacology and Genomic Analysis of the NCI-60. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21:3841–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeeberg BR, Kohn KW, Kahn A, et al. Concordance of gene expression and functional correlation patterns across the NCI-60 cell lines and the Cancer Genome Atlas glioblastoma samples. PloS one 2012;7:e40062–e40062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murai J, Thomas A, Miettinen M, et al. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol Ther 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murai J, Pommier Y. PARP Trapping Beyond Homologous Recombination and Platinum Sensitivity in Cancers. Annual Review of Cancer Biology 2019;3:131–150. [Google Scholar]
- 46.Zhang J, Stevens MF, Bradshaw TD. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol 2012;5:102–114. [DOI] [PubMed] [Google Scholar]
- 47.Wick W, Weller M, van den Bent M, et al. MGMT testing--the challenges for biomarkerbased glioma treatment. Nat Rev Neurol 2014;10:372–385. [DOI] [PubMed] [Google Scholar]
- 48.Pietanza MC, Kadota K, Huberman K, et al. Phase II Trial of Temozolomide in Patients with Relapsed Sensitive or Refractory Small Cell Lung Cancer, with Assessment of Methylguanine-DNA Methyltransferase as a Potential Biomarker. Clin Cancer Res 2012;18:1138–1145. [DOI] [PubMed] [Google Scholar]
- 49.Hegi ME, Diserens A-C, Gorlia T, et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. 2005;352:997–1003. [DOI] [PubMed] [Google Scholar]
- 50.van Meerbeeck JP, Baas P, Debruyne C, et al. A phase II EORTC study of temozolomide in patients with malignant pleural mesothelioma. Eur J Cancer 2002;38:779–783. [DOI] [PubMed] [Google Scholar]
- 51.Karran P, Hampson R. Genomic instability and tolerance to alkylating agents. Cancer Surv 1996;28:69–85. [PubMed] [Google Scholar]
- 52.Nagel ZD, Kitange GJ, Gupta SK, et al. DNA Repair Capacity in Multiple Pathways Predicts Chemoresistance in Glioblastoma Multiforme. Cancer Res 2017;77:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature Genetics 2012;44:751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molnár J, Póti Á, Pipek O, et al. The Genome of the Chicken DT40 Bursal Lymphoma Cell Line. G3: Genes|Genomes|Genetics 2014;4:2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He M, Chaurushiya MS, Webster JD, et al. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science 2019;364:283–285. [DOI] [PubMed] [Google Scholar]
- 57.Carbone M, Adusumilli PS, Alexander HR Jr., et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J Clin 2019;69:402–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivasan G, Sidhu GS, Williamson EA, et al. Synthetic lethality in malignant pleural mesothelioma with PARP1 inhibition. Cancer Chemother Pharmacol 2017;80:861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.