ABSTRACT
Recently, several molecular subtypes with different prognosis have been found in lung adenocarcinoma (LUAD). However, the characteristics of the ferroptosis molecular subtypes and the associated tumor microenvironment (TME) cell infiltration have not been fully studied in LUAD.
Using 1160 lung adenocarcinoma samples, we explored the molecular subtypes mediated by ferroptosis-related genes, along with the associated TME cell infiltration. The ferroptosis score was constructed using the least absolute shrinkage and selection operator regression (LASSO) method to quantify the ferroptosis characteristics of a single tumor.
Three different molecular subtypes related to ferroptosis, with different prognoses, were identified in LUAD. Analysis of TME cell infiltration revealed immune heterogeneity among the three subtypes. Cluster A was characterized by immunosuppression and was associated with stromal activation. Cluster C was characterized by a large number of immune cells infiltrating the TME, promoting tumor immune response, and it was significantly enriched in immune activation-related signaling pathways. Relatively less infiltration of immune cells was a feature of cluster B. The ferroptosis score can predict tumor subtype, immunity and prognosis. A low ferroptosis score was characterized by immune activation and good prognosis, as seen in the cluster C subtype. Relative immunosuppression and poor prognosis were the characteristics of a high ferroptosis score, as seen in cluster A and B subtypes. At the same time, the anti-PD-1/L1 immunotherapy cohort demonstrated that a low ferroptosis score was associated with higher efficacy of immunotherapy.
The ferroptosis score is a promising biomarker that could be of great significance to determine the prognosis, molecular subtypes, TME cell infiltration characteristics and immunotherapy effects in patients with LUAD.
KEYWORDS: Ferroptosis, lung adenocarcinoma, tumor microenvironment, immunotherapy, molecular subtypes
Introduction
In China, malignant tumors of the lung are associated with the highest incidence and death rates.1 About 85% of lung cancer cases are non-small cell lung cancer (NSCLC),2 of which the most common is lung adenocarcinoma (LUAD).3 The occurrence of LUAD is associated with smoking, drinking and metabolic disorders, among other factors.4 Although considerable progress has been made in chemotherapy, radiotherapy, and targeted therapy for the treatment of LUAD, the survival rate of patients with this disease remains relatively low.5 In recent years, studies have shown that the traditional histological classification of LUAD has limitations for treatment, because it is a highly heterogeneous and complex cancer.6 Therefore, an increasing number of molecular subtypes are being studied to guide therapy; for example, Yang et al. divided 335 lung adenocarcinoma patients into seven subtypes according to the level of gene methylation. They found significant survival differences between the groups.7 Wang et al. divided patients with LUAD from The Cancer Genome Atlas (TCGA) into two subtypes – high-risk and low-risk subtypes – based on immune genes.8 Although these studies have provided more insight into the molecular subtypes of LUAD, the prognostic prediction of patients with LUAD is less satisfactory. Therefore, more prognostically relevant factors need to be considered. Dixon et al. found that ferroptosis plays an indispensable role in the development of cancer.9 Therefore, this process may serve as a novel target in the treatment of LUAD.
Ferroptosis is a novel cell death modality, distinct from necrosis and apoptosis.10 It is driven by the iron-dependent buildup of lipid peroxidation products.11 Ferroptosis induction holds promise as a novel therapeutic approach, especially for tumors that are resistant to traditional treatment modalities.12–14 Previous studies have identified many genes that are regulators of ferroptosis. Some genes positively regulate ferroptosis, whereas others regulate negatively.15–17 However, the relationship between ferroptosis-related genes and the prognosis of patients with LUAD is not clear. Therefore, determining the molecular characteristics of ferroptosis-related genes may help clarify the cause of heterogeneity in LUAD.
In the present study, 1160 LUAD samples were divided into three ferroptosis-related subtypes on the basis of 14 genes related to ferroptosis, and the survival and immune infiltration differences among the subtypes were explored. Additionally, a ferroptosis score was established to quantify individual ferroptosis levels. The findings showed that the ferroptosis score is a powerful prognostic marker.
Materials and methods
LUAD data sets and preprocessing
Open LUAD gene expression datasets with complete clinical information annotation were downloaded from the public databases Gene Expression Omnibus (GEO) and TCGA. Patients without survival information were excluded. A total of six available datasets, including five from GEO (GSE30219, GSE37745, GSE50081, GSE68465, GSE72094) and TCGA-LUAD dataset, were collected. The raw “cel” files of these datasets were first downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/), then use Affy and simpleaffy packages to perform background adjustment and quantitative normalization. Next, we use the R package of SVA to remove the batch effect of the merged dataset.18 Data from TCGA were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/) in June 2020. Transcriptome data (FPKM value), clinical information and mutation information of 551 lung adenocarcinoma patients were obtained from the TCGA database. The R (version 3.6.1) and R Bioconductor packages were used for all data analysis.
Unsupervised clustering for ferroptosis-related genes
Initially, 74 ferroptosis-related genes were identified from previous studies; then, 14 ferroptosis-related genes were selected via univariate Cox analysis. On the basis of the expression level of these 14 genes, 1160 LUAD cases from GEO were classified using the unsupervised clustering analysis, and a follow-up analysis was conducted. The consensus clustering algorithm was used to determine the number of clusters the samples were divided into. To verify the accuracy of the clustering, the relatively complete clinical information dataset GSE70249 was used for unsupervised clustering analysis. Finally, the ConsensusClusterPlus R package is to perform cluster analysis on the all cohorts and identify three clusters, and further to perform cluster analysis on the GSE72094 cohort and also identify three clusters. And it was used for 1000 cycles to ensure the stability of the classification.19
Functional and pathway enrichment analysis
The “GSVA” R packages were applied for enrichment analysis, to ascertain the different pathways for analyzing the differences in biological function among the different ferroptosis clusters.20 The gene sets of “c2.cp.kegg.v6.2.-symbols” were downloaded from the MSigDB database to run GSVA enrichment analysis. The R package clusterProfiler was used for functional annotation of the ferroptosis-related genes. P values less than 0.05 were considered to indicate significant differences in gene ontology.21
Evaluation of tumor microenvironment cells in patients with LUAD
The scores of tumor microenvironment (TME) cells in each LUAD sample were evaluated using the single sample gene set enrichment analysis (ssGSEA) algorithm.22 To understand the immune characteristics of the 1160 LUAD samples, CIBERSORT (https://cibersort.stanford.edu/) was used to evaluate the relative proportion of 22 immune cells. R software was used to run the CIBERSORT algorithm. Based on the 1160 sample gene expression matrix and the gene expression feature set of 22 immune cell subtypes provided by the official website, the simulation calculation was performed 1000 times, and the relative composition ratio of the 22 immune cells in each sample was finally obtained. And we used the R package of estimate to evaluate the immune score and ESTIMATE score of each patient.23
Differentially expressed genes associated with the ferroptosis subtypes
The differentially expressed genes (DEGs) among the ferroptosis clusters were identified using the empirical Bayesian approach of the limma R package.24 Genes with an adjusted P value < .001 are the DEGs of different subtypes of ferroptosis.25
Derivation of the ferroptosis prognostic signature
First, 14 ferroptosis-related genes associated with prognosis were identified by univariate Cox analysis (P value < .05). Then, the most powerful ferroptosis prognostic genes were identified using the penalized Cox regression model with least absolute shrinkage and selection operator (LASSO) penalties.26 Finally, 13 hub genes and their correlative coefficients were obtained, to construct the ferroptosis gene signature, defined as the ferroptosis score (e (each gene’s expression × correlative coefficient). And we used the surv_cutpoint function in the R package of “Survminer” to calculate the optimal cutoff value. On the basis of the best cutoff value of the score, the 1160 LUAD samples were divided into high and low ferroptosis score groups. Then, survival analysis was carried out for the two groups using the survminer R package. Finally, receiver operating characteristic (ROC) curve analysis was conducted using the survivalROC R package to obtain the area under the curve (AUC) value and evaluate the predictive power of the signature.
Identification of cohorts with immune-checkpoint blockade
Imvigor 210 (http://research-pub.gene.com/IMvigor210CoreBiologies) is a cohort of patients with urothelial carcinoma treated with PD-L1.27 It has relatively complete survival information, follow-up information and immunotherapy effect information. Samples with incomplete clinical information were removed, and 298 samples were finally obtained for follow-up analysis. The raw count data was normalized using the DEseq2 R package.
Statistics
The difference between the groups was analyzed using the Wilcoxon test. The Kruskal-Wallis and one-way ANOVA tests were used for differential analysis among the three groups.28 Correlation tests were performed using Spearman analyses. Survival curves were drawn using the log-rank and Kaplan-Meier tests. Mutations among different groups were analyzed using the maftools R package. The 95% confidence interval (CI) and hazards ratios (HR) were computed using the univariate Cox regression model. In comparisons between groups, P < .05 was considered to be statistically significant. The R 3.6.1 software was used for data processing.
Results
Identification of ferroptosis subtypes in LUAD
Ferroptosis-related patterns and the ferroptosis signature were systematically constructed (Figure S1). An LUAD meta cohort (GSE30219, GSE37745, GSE50081, GSE68465, GSE72094) including patients’ survival and clinical information was included in the study (Table S1). Univariate Cox regression identified 14 ferroptosis-related genes associated with prognosis in the 1160 patients with LUAD (Figure S2A and Table S2), and Spearman’s correlation analysis revealed the correlation of these 14 genes (Figure S2B). Next, the 14 ferroptosis genes were constructed into a network map, which enabled comprehensive analysis of the interaction and interconnection of the genes and their impact on the prognosis of patients with LUAD (Figure 1a). To explore the classification of ferroptosis in LUAD, the 1160 LUAD samples that expressed the 14 ferroptosis-related genes expression were analyzed using the unsupervised clustering analysis. From the results, three subtypes (Figure S2C-F), designated clusters A-C, respectively, were identified (Figure S2G). Cluster A included 398 cases, cluster B included 412 cases, and cluster C included 350 cases.
Figure1.
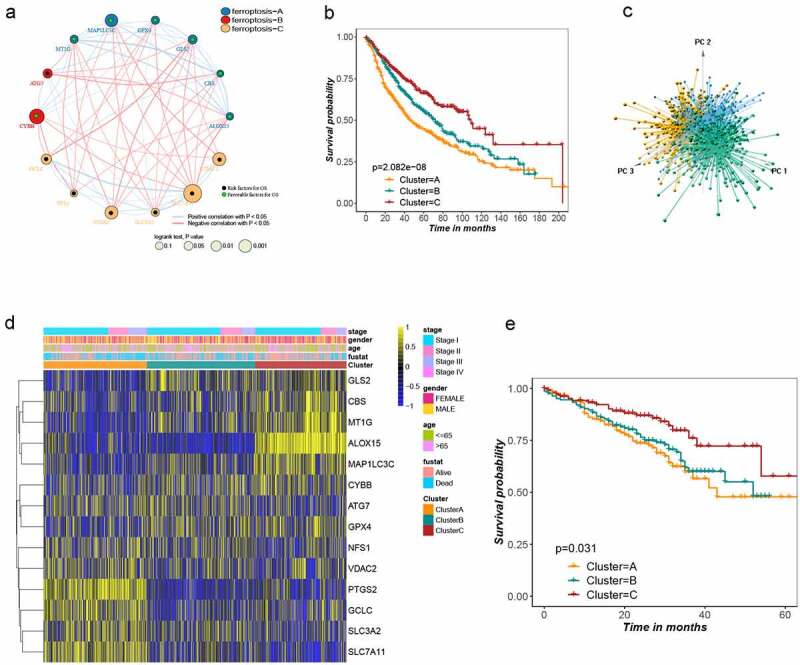
Landscape of the ferroptosis and biological characteristics of ferroptosis subtypes in lung adenocarcinoma. (a) Interaction of ferroptosis related genes in LUSD. Blue, red and yellow represent ferroptosis A, B and C, respectively. The size of the circle represents the impact of each ferroptosis gene on the prognosis, the P value calculated by log-rank test. Green dots in the circle represent protective factors and black dots represent risk factors. Links between genes represent their interactions, blue lines represent positive correlations, red lines represent negative correlations, and the thickness of the lines represents the strength of the correlation between them. (b) Survival analysis of three ferroptosis subtypes in all patients, Kaplan-Meier survival curve showed significant differences among the three subtypes (log-rank test, P < .001). (c) Principal component analysis of 14 ferroptosis genes in all lung adenocarcinoma cohorts identified three distinct subtypes. (d) Unsupervised clustering of 14 ferroptosis genes in all lung adenocarcinoma cohorts. Tumor stage, gender, age, survival status and cluster were used as patient annotations. Yellow and blue represent high and low expression of ferroptosis genes respectively. (e) Kaplan–Meier curves for overall survival (OS) of GSE72094 cohort with the ferroptosis classes (log-rank test, P = .031)
Survival analysis revealed that prognosis differed substantially among the three ferroptosis subtypes, and cluster C had considerable survival advantages (Figure 1b). There were obvious differences in the expression of the 14 ferroptosis-related genes in the three clusters (Figure S3A). Next, principal component analysis (PCA) further confirmed three remarkably different subtypes (Figure 1c), as did a thermogram. The relationship between the three subtypes and various clinical characters was studied (Figure 1d). Since the GSE72094 dataset has relatively complete clinical information and a large sample size, it was used to validate the repeatability of the clustering. Unsupervised clustering was conducted on this cohort using the “Consensus Cluster Plus” R package, and three distinct subtypes were again clearly identified (Figure S3B-E). Significant differences in survival were noted among these three subtypes (Figure 1e), further proving that there are three subtypes of LUAD.
Characteristics of TME cell infiltration and biological function in the ferroptosis subtypes
To better understand the difference of survival among the three clusters. GSVA enrichment analysis was firstly performed on the three subtypes to examine their functional and biological differences (Figure 2a,b). According to the comparison of enrichment analysis between cluster A and cluster C, cluster B and cluster C, respectively. The results showed that the cluster A was mainly enriched in some carcinogenic pathways, such as the P53, WNT, and TGF BET signaling pathways; cluster B was mainly related to carcinogenic pathways and nucleotide repair and degradation; and cluster C was mainly enriched in immune activation-related pathways, such as the B cell receptor signaling pathway, T cell receptor signaling pathway, and antigen processing and presentation. Subsequently, the TME cell infiltration of three ferroptosis subtypes was examined by Spearman analysis. As shown in Figure S4A, 28 kinds of immune cells were demonstrated significantly correlated with the 14 ferroptosis-related genes. Next, the enrichment score of the 28 kinds of immune cells was evaluated in the three subtypes using ssGSEA analysis (Figure 2c). In cluster A, the most significant immunoinfiltrating cells were activated CD4 T cells, activated dendritic cells, neutrophils and plasmacytoid dendritic cells. While the macrophages, central memory CD4T cells, effect memory CD8T cells, eosinophils, immature B cells and T-follicular helper cells showed the greatest infiltration in cluster C. The level of immune cell infiltration in cluster B was between clusters A and C. After that, the ESTIMATE score and immune score of three ferroptosis clusters were comprehensively analyzed, to find cluster C had the highest score (Figure S4B-C).
Figure 2.
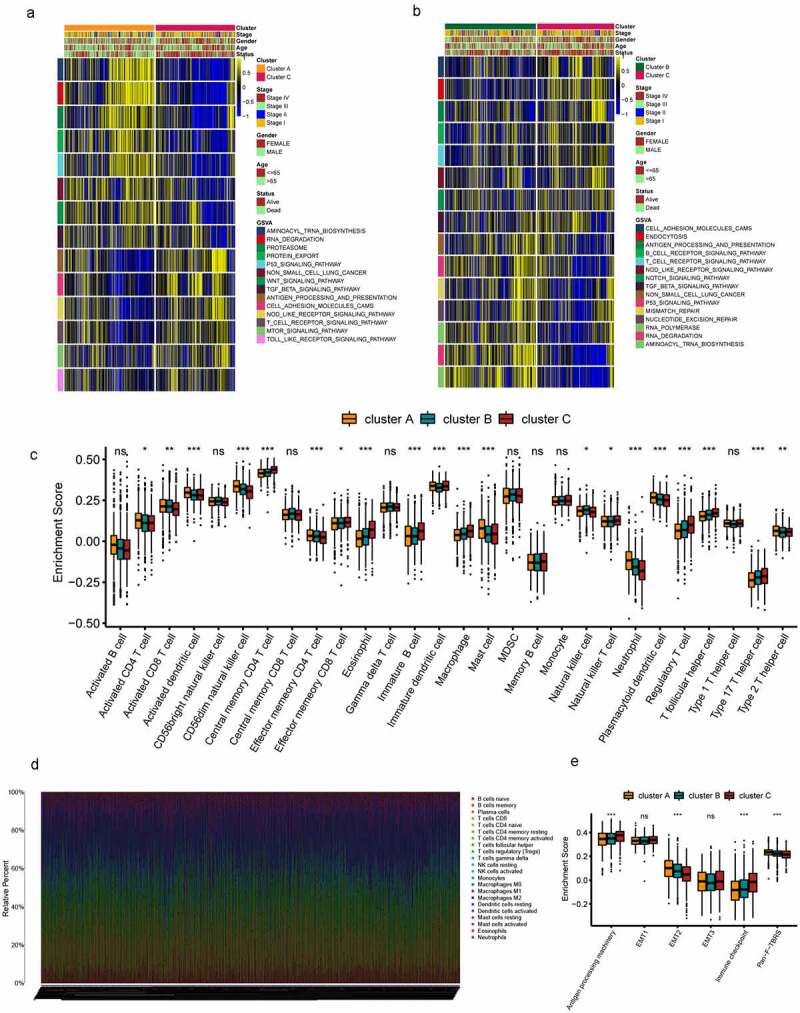
TME cell infiltration and biological characteristics in three subtypes of LUSD. (a-b) GSVA analyzed the biological pathways of three ferroptosis subtypes. Yellow represents activation of biological pathways and blue represents inhibition of biological pathways. Cluster, tumor stage, gender, age, survival state were used as sample annotations. (a) cluster A vs cluster C; (b) cluster B vs cluster C. (c) 28 TME cells infiltration abundance of three ferroptosis subtypes. The line in the box represents the median value and the asterisk represents the P value (*P < .05; **P < .01; ***P < .001). The statistical differences among the three clusters were analyzed by one-way ANOVA test. (d) The relative percentage of 22 subpopulations of immune cells in 1160 samples from all LUSD cohort. (e) Differences in interstitial activation pathways of three ferroptosis subtypes. The line in the box represents the median value and the asterisk represents the P value (*P < .05; **P < .01; ***P < .001). The statistical differences among the three clusters were analyzed by one-way ANOVA test
To further explore the differences in the composition of TME-infiltrating cells among the three clusters. The relative percentage of the 22 kinds of immune cells in each patient was calculated using the CIBERSORT algorithm (Figure 2d). Difference in TME-infiltrating cell composition was found among the three ferroptosis clusters (Figure S4D). Quantitative analysis of stroma activity in the 1160 patients with LUAD, and as expected, EMT2 and Pan−F− TBRS were significantly higher in cluster A than in the other two clusters; cluster C was found to be related to antigen-processing machinery and immune checkpoints (Figure 2e).
Comprehensive analysis of ferroptosis DEGs in LUAD
In order to further explore the potential biological function of the ferroptosis subtype in LUAD, the limma R package was used to identify the DEGs among the three clusters (log-rank test, P < .001), and 470 genes were obtained (Figure S5A). Next, gene ontology enrichment analysis showed that the DEGs that were considerably enriched were involved in positive regulation of T cell proliferation, natural killer cell activation and natural killer cell activation, all of which are part of the immune response (Figure 3a). The results showing that these DEGs are closely related to immune-related biological processes confirm that ferroptosis is significantly associated with immunity. Clustering analysis of the DEGs was carried out in the GSE72094 dataset using the “Consensus Cluster Plus” R package. The results were similar to phenotypic clustering of ferroptosis, that is, three subtypes, designated gene cluster AC, were identified (Figures 3b and S5B-E). The three subtypes could be significantly separated on the basis of the expression levels of the DEGs (Figure 3c). The survival rates of gene clusters AC were significantly different in the GSE72094 cohort (P < .001; Figure 3d). The alluvial diagram was used to better visualize the survival differences among the different gene clusters and ferroptosis clusters (Figure 3e). As expected, there were significant differences in the expression of the 14 ferroptosis-related genes among the three gene clusters (Figure 3f). Next, the relationship between the three gene clusters and 28 immune cells was explored. The results showed that gene cluster A had more infiltrated activated B cells, central memory CD4 T cells, effector memory CD8 T cells, eosinophils, macrophages and natural killer cells than the other two gene clusters (Figure S5F). This also explains why gene cluster A has the best survival advantage. Simultaneously, it proves that ferroptosis plays an important role in the TME.
Figure 3.
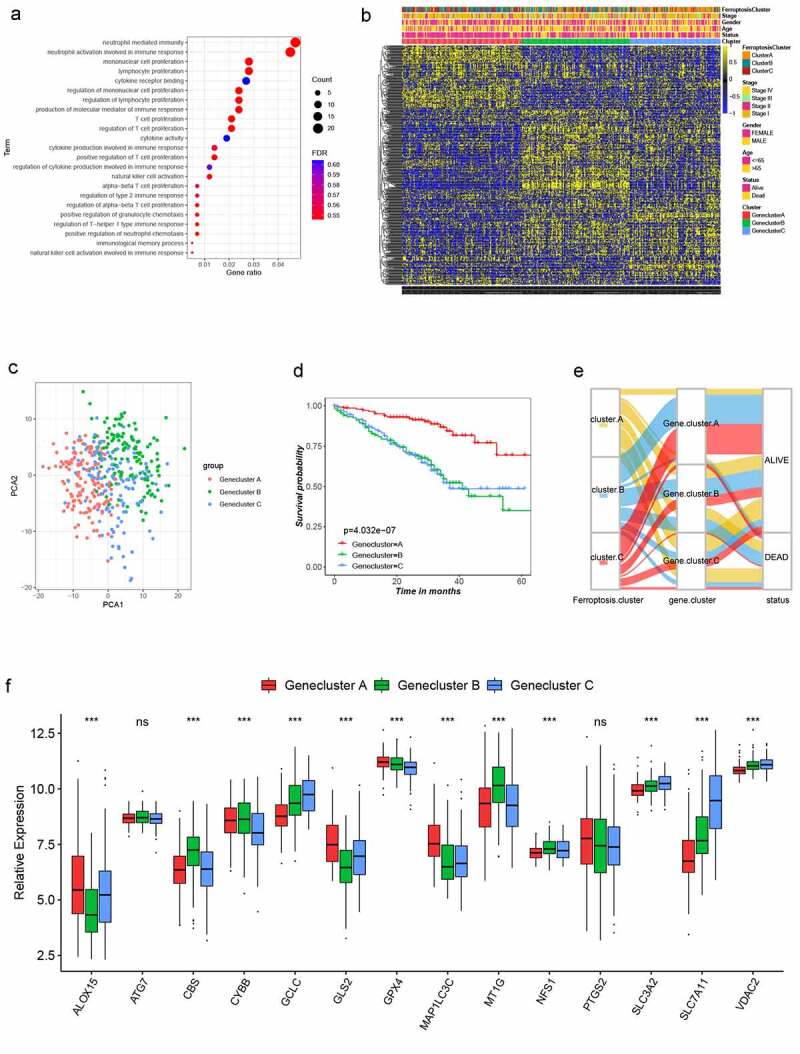
Landscape of biological characteristics of Ferroptosis gene cluster. (a) GO enrichment analysis of DEGs. The size of bubbles represents the amount of gene enrichment, and the depth of color represents the FDR value. DEGs, differentially expressed protein-coding genes; GO, Gene Ontology; FDR, False Discovery Rate. (b) Unsupervised clustering of 470 DEGs in the GSE72094 LUSD cohort. Ferroptosis Cluster, tumor stage, gender, age, survival status and cluster were used as patient annotations. Yellow and blue represent high and low expression of ferroptosis genes respectively. (c) Principal component analysis of 470 DEGs in the GSE72094 LUSD cohort identified three distinct Gene clusters. (d) Survival analysis of three Gene clusters in the GSE72094 LUSD cohort, Kaplan-Meier survival curve showed significant differences among the three gene clusters (log-rank test, P < .001). (e) Alluvial diagram showing the changes of Ferroptosis cluster, gene cluster and status. (f) Expression of 14 ferroptosis genes in three Gene clusters. The line in the box represents the median value and the asterisk represents the P value (*P < .05; **P < .01; ***P < .001). The statistical differences among the three clusters were analyzed by one-way ANOVA test
Construction of the ferroptosis signature
In order to further understand the characteristics of ferroptosis in each patient with LUAD, LASSO Cox regression was used to determine the optimal value of λ, and 13 key genes were identified (Figure S6A). A ferroptosis prognostic model was then established based on the expression of the 13 key genes, and this was defined as the ferroptosis score. The correlation coefficients are provided in Table S3. According to a cutoff value of 8.97 (Figure S6B), the 1160 patients with LUAD were divided into high ferroptosis score (n = 679) and low ferroptosis score (n = 481) groups. In all the LUAD cohorts, the survival rate was lower among those with a high ferroptosis score than among those with a low ferroptosis score, and the Kaplan-Meier survival curve showed a significant difference between the groups (Figure 4a; P < .001). In the GSE72094 cohort, obvious differences in survival were found between groups with high and low ferroptosis scores (Figure 4b). In the GSE30219, GSE50081 and GSE68465 cohorts, the ability of the model to predict patient prognosis was further verified (Figure S6C-E). However, in the GSE37745 cohort, there was no difference in survival between the high and low ferroptosis scores (Figure S6F). The ROC curve showed that the ferroptosis signature was effective in predicting the 5-year survival rate in patients with LUAD (Figure 4c). Multivariate Cox regression analysis showed that the ferroptosis score was an independent prognostic factor for LUAD (Table S4).
Figure 4.

Construction of ferroptosis signature. (a) Kaplan-Meier curves were used to analyze the survival of patients with high (679 cases) and low (481 cases) ferroptosis score in all LUSD cohorts (P < .0001, Log-rank test). (b) Kaplan-Meier curves were used to analyze the survival of patients with high (141 cases) and low (257 cases) ferroptosis score in the GSE72094 cohort (P < .0001, Log-rank test). (c) The predictive value of ferroptosis score in 5-year survival of patients with LUSD. AUC 0.75. (d) Difference of ferroptosis score among three clusters in all LUSD cohorts (P < .001, Kruskal-Wallis test). (e) Difference of ferroptosis score among three gene clusters in all LUSD cohorts (P < .001, Kruskal-Wallis test). (f) 28 TME cells infiltration abundance of high and low ferroptosis score groups. P values were showed as: *, P < .05; **, P < .01; ***, P < .001
Next, the relationship between the ferroptosis score and ferroptosis clusters A-C and ferroptosis gene clusters A-C was further explored. Cluster C was significantly correlated with a low ferroptosis score, suggesting that a low ferroptosis score may be related to immune activation. Clusters A and B were significantly correlated with the high ferroptosis score (Figure 4d). Gene cluster A was associated with a low ferroptosis score, and these patients had a better prognosis. Gene clusters B and C were significantly associated with a high ferroptosis score (Figure 4e). These results suggest that the ferroptosis score may be associated with immunity and may be helpful in predicting the ferroptosis subtype in LUAD. Next, the infiltration of 28 kinds of immune cells into the TME were further studied in the high and low ferroptosis score groups. There was a significant difference in the infiltration of immune cells between the groups. In the low ferroptosis score group, the infiltration of most immune cells involved in tumor immune activation was higher (Figure 4f). This finding confirmed the hypothesis that the low ferroptosis score is associated with immune activation, and patients with this score has a better survival rate. At the same time, we have also established a table to illustrate the characteristics of each cluster, ferroptosis score and immune cell infiltration (Table S5).
Characteristics of ferroptosis signature in the TCGA cohort
First, multivariate Cox regression was used to explore the relationship between the ferroptosis score and clinical characteristics, and the results showed that stage and ferroptosis score could be used as independent prognostic factors (HR = 1.638, 95% CI = 1.0112.653, P = .045; HR = 1.030, 95% CI = 1.0131.048, P < .001, respectively) (Figure 5a). In the TCGA cohort, the differences in survival rate between patients with high and low ferroptosis scores was statistically significant (Figure 5b; log-rank tests, P < .001). Again, this validates the use of the ferroptosis score to predict the prognosis of patients in LUAD. The predictive advantage evaluated using the ROC curve was especially reflected in older patients (Figure 5c).
Figure 5.
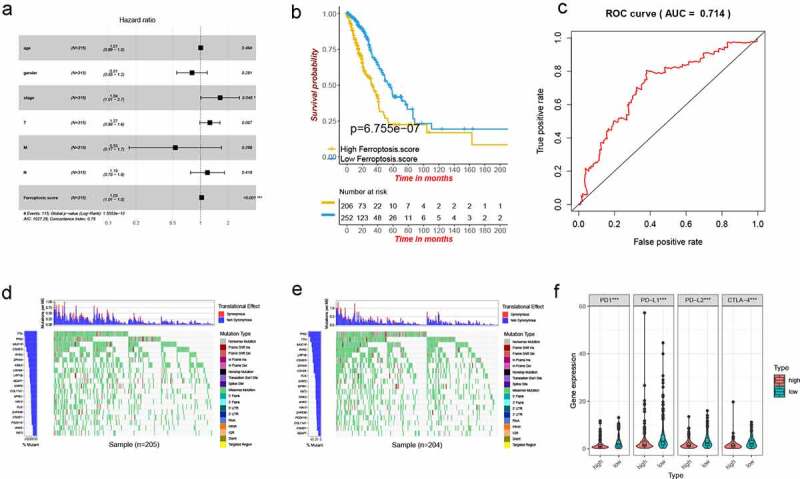
Characteristics of ferroptosis score in TCGA cohort and tumor somatic mutation. (a) Forest map of ferroptosis score and other clinicopathological parameters. (b) Kaplan–Meier plot of overall survival by ferroptosis score groups for patients in the TCGA cohort (P < .001, Log-rank test). (c) The predictive value of ferroptosis score in older patients with LUSD. AUC 0.714. (d-e) The waterfall plot showing the differences of tumor somatic mutation landscape between low and high ferroptosis score groups. (d) High ferroptosis score group. (e) Low ferroptosis score group. (f) Differential expression of immune checkpoint related molecules in high and low ferroptosis score groups. P values were showed as: ***, P < .001
Next, the relationship between the ferroptosis score and immunity was examined in the TCGA cohort. First, somatic mutations were compared in those with high and low ferroptosis scores, and the top 20 genes with the highest mutation frequency were visualized (Figure 5d,e). No significant differences in tumor mutation burden were seen between the high and low ferroptosis score groups. Then, the relationship between the groups and immune checkpoint-related molecules was examined. PD1, PD-L1, PD-L2 and CTLA-4 had significantly higher expression in the low ferroptosis score group (Figure 5f). This led us to speculate that there may be a correlation between the ferroptosis score and the effect of immunotherapy.
Relationship between the ferroptosis score and the effect of immunotherapy
At present, anti-PD1/PD-L1 therapy plays an important role in tumor immunotherapy. To further illustrate the relationship between the ferroptosis score and the efficacy of immunotherapy, an anti-PD1/PD-L1 treatment cohort (imvigor210) with relatively complete clinical data and a large sample size was investigated. Samples that had no information about the effect of immunotherapy were omitted, and 298 samples were finally included in the study. Then, using the correlation coefficient in Table S3 to construct the ferroptosis signature, the imvigor210 cohort was divided into high and low ferroptosis score groups; the prognosis of the groups differed significantly (Figure 6a). Patients with a low ferroptosis score had a better prognosis, which also provided preliminary evidence that patients with a low ferroptosis score had a better outcome with immunotherapy than those with a high ferroptosis score.
Figure 6.
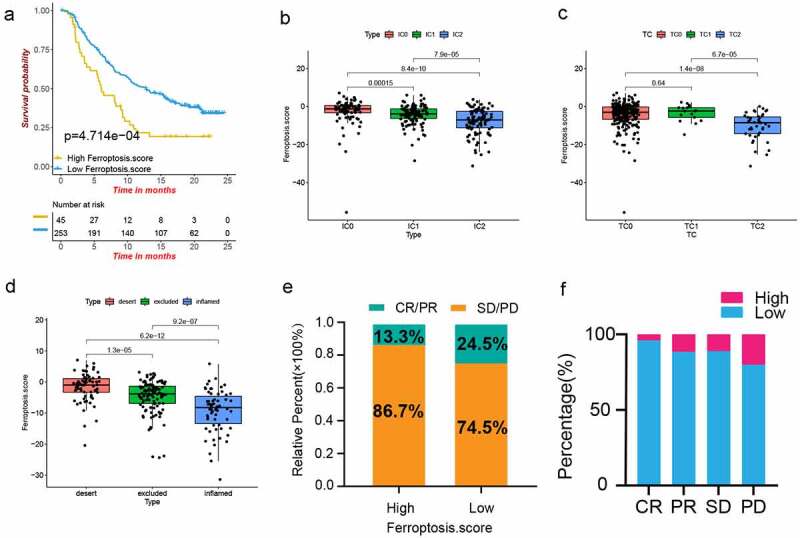
The role of ferroptosis score in anti-PD-1/L1 immunotherapy. (a) Kaplan–Meier plot of overall survival by ferroptosis score groups for patients in the IMvigor210 cohort (P < .01, Log-rank test). (b-c) The difference of ferroptosis score among PD-L1 expression of different IC (b) and TC (c) in the IMvigor210 cohort. Tumor tissue samples were scored through immunohistochemistry (IHC) for PDL1 expression on tumor-infiltrating immune cells (IC), which included macrophages, dendritic cells and lymphocytes. Specimens were scored as IHC IC0, IC1, IC2, or IC3 if <1%, ≥1% but <5%, ≥5% but <10%, or ≥10% of IC were PD-L1 positive, respectively. An exploratory analysis of PD-L1 expression on tumor cells (TC) was conducted. Specimens were scored as IHC TC0, TC1, TC2, or TC3 if <1%, ≥1% but <5%, ≥5% but <50%, or ≥50% of TC were PD-L1 positive, respectively. (d) Difference of ferroptosis score among three immune subtypes in IMvigor210 cohort (P < .001, Kruskal-Wallis test). (e) Proportions of anti-PD-L1 immunotherapy response in high and low ferroptosis score groups. PR, Partial Response, PD, Progressive Disease; SD, Stable Disease, and CR, Complete Response. (f) Proportion of patients with high and low ferroptosis score in different anti-PD-L1 immunotherapy responses
It is well known that there is a strong relationship between the expression level of PD-L1 and the efficacy of anti-PD-L1 therapy. The relationship between the ferroptosis score and the IC and TC immune types was analyzed (Figure 6b,c,d), and the ferroptosis score of IC2 was lower than that of IC0 and IC1; TC2 had a lower ferroptosis score than the other two groups; the ferroptosis score of the immune-inflamed type was lower than that of the immune-desert and immune-excluded types. These results showed that the ferroptosis score was negatively correlated with the expression level of PD-L1, and the low ferroptosis score was significantly associated with the immune-inflamed type. Further, a low ferroptosis score had a better CR/PR rate and a lower SD/PD rate (Figure 6e,f). These findings indicated that the ferroptosis score can be used to predict the efficacy of immunotherapy.
Discussion
Due to the strong heterogeneity of LUAD,29 the overall survival rate of patients with LUAD is relatively low. In recent years, progress has been made in chemotherapy for LUAD, but, when the traditional histological classification is used to guide anti-tumor treatment, tumor drug resistance is frequent.30 Therefore, accurate identification of the molecular subtypes of LUAD is vital to guide personalized therapy. Although previous studies have also identified several molecular subtypes of LUAD, there is still considerable heterogeneity among the subtypes.7,8 Therefore, more accurate classification of LUAD is urgently needed to improve patient survival. Ferroptosis, a recently proposed form of iron-dependent oxidative cell death mainly characterized by the lethal accumulation of lipid peroxides, has been strongly implicated in cancer.31 Many studies have shown that ferroptosis plays an important role in regulating cancer growth, immunotherapy and the TME.14,32–35 Extensive studies have implicated ferroptosis in silencing and downregulating genes that are initiators of tumor necroptosis.36 Further, the RAS–RAF–MEK pathway has been found to be critical to ferroptosis sensitivity.11 Cancer cell have a much higher demand for iron than normal cells, as they need it to promote their rapid proliferation. Because tumor cells are more iron dependent, this makes them more vulnerable to iron overload and reactive oxygen species accumulation, which can be useful in cancer therapies that target ferroptosis.37 Therefore, induced ferroptosis may be a new direction for cancer treatment.38 According to recent literature, ferroptosis dysfunction plays a dual role in the progression of cancer. It inhibits the growth of tumors by inducing ferroptosis, and it promotes the occurrence of cancer by altering immune suppression of tumor microenvironment.39 Ferroptosis has been investigated in a variety of cancers. For example, an LncRNA prognosis model, which was related to ferroptosis, was developed in colon cancer;40 The genes related to ferroptosis were potentially prognostic molecular markers for patients with colorectal cancer;41 Moreover, ferroptotic damage promotes tumorigenesis in pancreatic cancer patients;42 A model of liver cancer patients was constructed, and it was based on four ferroptosis-related genes. This model predicted the prognosis of patients with liver cancer;43 Studies have also been conducted to understand the correlation between ferroptosis and breast cancer patients, head and neck squamous cell carcinoma, and renal cell carcinoma.44–46 However, the relationship between LUAD and ferroptosis is not well defined. Therefore, the comprehensive role of genes associated with ferroptosis in LUAD phenotyping and TME was examined in the present study. Additionally, a signature related to ferroptosis was constructed to predict the ferroptosis subtype, prognosis and effect of immunotherapy in LUAD.
In the present research, it was found that LUAD can be divided into three subtypes on the basis of the expression of 14 ferroptosis-related genes, and the testing set proved the repeatability of the classification. Significant differences in prognosis were found among the three subtypes. In order to explore the causes of these differences, GSVA enrichment analysis was used, and it was apparent from the findings that cluster C was prominently enriched in pathways related to tumor immune activation, cluster A was more enriched in signaling pathways that promote cancer, and cluster B was mainly associated with cancer-promoting and nucleotide repair and degradation pathways. Since previous studies have shown that ferroptosis is closely related to tumor immunity.47–49 For example, cancer cells transfer tumor infiltrating immune cells that meet their demand for ferroptosis, thereby affecting the immune monitoring of tumors;50 Wang et al. explored the relationship between ferroptosis and CD8 + T cells, and they found that by activating CD8 + T cells, lipid peroxidation could be increased in tumor cells. Moreover, with an increase in ferroptosis, the anti-tumor immune effect was promoted.47 Meanwhile, cancer cells that underwent ferroptosis released a protein of HMGB1 into the TME around cancer, and they interacted with pattern- recognition receptor (PRR) that stimulated the activation of innate immune system, thereby promoting anti-tumor immunity.51,52 Therefore, we investigated the relationship between three ferroptosis subtypes and TME cell infiltration. However, the relationship between immune cells and tumors is extremely complex, and different immune cells have different roles. It is reported that CD4 T cells play a negative role in tumor immunity,53 macrophages play a complex role in tumor immunotherapy,54 and M1 macrophages exert antitumor immunity by secreting reactive oxygen species and proinflammatory cytokines. M2 macrophages promote tumor growth, invasion, and migration and inhibit tumor immunity by secreting anti-inflammatory cytokines such as interleukin-10, interleukin-13, and transforming growth factor-β.55–57 Eosinophil infiltration in the TME is generally considered to be related with better outcomes.58 Neutrophils play an important role in the occurrence and progression of many diseases. Tumor-related neutrophils are an important part of the TME and can secrete some factors to promote cancer.59–61 Plasmacytoid dendritic cells can produce a large amount of IFN-α to promote the immune response, but the damage to TLR7/9 activation in cancer leads to a decrease in IFN-α, which inhibits the tumor immune response.62 T-follicular helper cells are significantly correlated with the high expression of PD-L1, which promotes the tumor immune response.63 In the present study, activated CD4 T cells, activated dendritic cells, neutrophils and plasmacytoid dendritic cells, which can promote tumor immunosuppression, infiltrated cluster A more than they did the other two clusters. Additionally, macrophages, central memory CD4 T cells, effector memory CD8 T cells, eosinophils, image B cells and T-follicular helper cells, which promote cancer immunity, infiltrated cluster C more than they did the other two clusters. Thus, cluster A is associated with immunosuppression, and the patients with this cluster have the worst LUAD prognosis; on the other hand, cluster C is significantly associated with immune activation, and patients with this cluster have the best prognosis. In the meantime, ferroptosis cluster B showed moderate immune activity. And the latest literature shows that cancer cells with ferroptosis can be immunogenic and contribute to tumor immune activation, which means that inducing ferroptosis may lead to favorable clinical outcomes for patients.64 So, one possible mechanism for the significant differences in immune cell infiltration among the three ferroptosis clusters is that ferroptotic cancer cells have immunogenicity, which can attract immune cells to their location. This is because ferroptotic cancer cells release HMGB1, which is a key factor in cancer cell immunogenicity.52,65,66 Another possibility is that ferroptotic cancer cells can release a potential signal–amino acid oxidation products–to attract immune cells to their location to participate in immune regulation.67 However, the specific mechanism between ferroptosis and immunity is not clear. The analysis of clinical characteristics also showed that there were more advanced LUAD cases in ferroptosis cluster A, which explains the poor survival rate in this cluster. Further investigation of ferroptosis-related DEGs showed that they could also be divided into three subtypes in LUAD. Moreover, these three subtypes were found to be related to tumor immune. These findings may advance our understanding of the relationship between ferroptosis, TME cell infiltration, and LUAD.
Considering the impact of ferroptosis on the heterogeneity of LUAD and the corresponding clinical outcomes,68,69 a ferroptosis signature based on 13 key genes associated with ferroptosis was constructed to quantify ferroptosis scores. As predicted, cluster A had the highest ferroptosis score, cluster C had the lowest ferroptosis score, and cluster B had a moderate ferroptosis score. Additionally, patients with a low ferroptosis score had a better survival rate than those with a high ferroptosis score. Again, the TCGA cohort validated this outcome. This study also showed that the ferroptosis score can be an independent prognostic biomarker in patients with LUAD.
Next, it was found that the ferroptosis score was negatively correlated with immune cell infiltration and expression of immune checkpoint molecules, which indirectly indicated that the ferroptosis score may play an important role in predicting the effects of immunotherapy. Then, the relationship of the ferroptosis score and immunotherapy efficacy was further analyzed in the imvigor210 cohort of metastatic urothelial cancer treated with anti-PD-L1. The higher PD-L1 expression in IC and TC, the lower was the ferroptosis score. Comparison of the immune-inflamed subtype with the immune-desert and immune-exclusion subtypes showed that the former had the lowest ferroptosis score. These analyses indirectly illustrate that the ferroptosis score is inversely associated with the immunotherapy effect. The CR/PR rate was 11.2% higher in those with the low ferroptosis score than in the high ferroptosis score. This is a direct demonstration of a better immunotherapeutic effect in those with a low ferroptosis score.
This study has some limitations. First, a large number of LUAD samples were needed to verify the stability of the typing, and the relationship between ferroptosis and immunity requires further experimental verification.
Conclusion
In conclusion, this study found three subtypes of ferroptosis-related molecules in LUAD, all indicating a different prognosis. Ferroptosis-related genes are important contributors to the heterogeneity of the TME in LUAD. The ferroptosis score is a promising biomarker that could be of great significance to distinguish the prognosis, molecular subtypes, TME cell infiltration characteristics and immunotherapy effects of patients with LUAD.
Supplementary Material
Funding Statement
This study was funded by grants from the Key Scientific and Technological Projects in Nantong City, Jiangsu, China (No. MS22019015), Jiangsu Post-doctoral Foundation Research Project, China (No. 2019Z142), Clinical basic research youth project of Nantong University (No. 2019JQ002), Basic research project of Nantong Municipal Science (No. JCZ19103).
Disclosure statement
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–13. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7(9). doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong HX, Wang R, Jin X-Y, Zeng J, Pan J. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J Cell Physiol. 2018;233(5):4126–4136. doi: 10.1002/jcp.26215. [DOI] [PubMed] [Google Scholar]
- 4.Park CK, Cho H-J, Choi Y-D, Oh I-J, Kim Y-C. A Phase II trial of osimertinib in the second-line treatment of non-small cell lung cancer with the EGFR T790M mutation, detected from circulating tumor DNA: liquidLung-O-Cohort 2. Cancer Res Treat. 2019;51(2):777–787. doi: 10.4143/crt.2018.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.Hensing T, Chawla A, Batra R, Salgia R.. A personalized treatment for lung cancer: molecular pathways, targeted therapies, and genomic characterization. Adv Exp Med Biol. 2014;799:85–117. doi: 10.1007/978-1-4614-8778-4_5. [DOI] [PubMed] [Google Scholar]
- 7.Shi S, Xu M, Xi Y. Molecular subtypes based on DNA promoter methylation predict prognosis in lung adenocarcinoma patients. Aging (Albany NY). 2020;12(23):23917–23930. doi: 10.18632/aging.104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Li M, Yang M, Yang Y, Song F, Zhang W, Li X, Chen K. Analysis of immune-related signatures of lung adenocarcinoma identified two distinct subtypes: implications for immune checkpoint blockade therapy. Aging (Albany NY). 2020;12(4):3312–3339. doi: 10.18632/aging.102814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon SJ, Lemberg K, Lamprecht M, Skouta R, Zaitsev E, Gleason C, Patel D, Bauer A, Cantley A, Yang W, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73(11–12):2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447(7146):864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Z, Song J, Yung BC, Zhou Z, Wu A, Chen X. Emerging strategies of cancer therapy based on ferroptosis. Adv Mater. 2018;30(12):e1704007. doi: 10.1002/adma.201704007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31(51):e1904197. doi: 10.1002/adma.201904197. [DOI] [PubMed] [Google Scholar]
- 15.Jennis M, Kung C-P, Basu S, Budina-Kolomets A, Leu JIJ, Khaku S, Scott JP, Cai KQ, Campbell MR, Porter DK, et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30(8):918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai Y, Xia C, Sun Z. The inhibitory effect of 6-Gingerol on ubiquitin-specific peptidase 14 enhances autophagy-dependent ferroptosis and anti-tumor in vivo and in vitro. Front Pharmacol. 2020;11:598555. doi: 10.3389/fphar.2020.598555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gai C, Liu C, Wu X, Yu M, Zheng J, Zhang W, Lv S, Li W. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. 2020;11(9):751. doi: 10.1038/s41419-020-02939-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 19.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Kwan PW, Shi D. Sparse kernel learning with LASSO and Bayesian inference algorithm. Neural Netw. 2010;23(2):257–264. doi: 10.1016/j.neunet.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazra A, Gogtay N. Biostatistics series module 3: comparing groups: numerical variables. Indian J Dermatol. 2016;61(3):251–260. doi: 10.4103/0019-5154.182416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D, Liu Y, Bai C, Wang X, Powell CA. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett. 2020;468:82–87. doi: 10.1016/j.canlet.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 31.Yang WS, Stockwell BR. Ferroptosis: death by Lipid Peroxidation. Trends Cell Biol. 2016;26(3):165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Guo P, Xie X, Wang Y, Chen G. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J Cell Mol Med. 2017;21(4):648–657. doi: 10.1111/jcmm.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia X, Fan X, Zhao M, Zhu P. The relationship between ferroptosis and tumors: a novel landscape for therapeutic approach. Curr Gene Ther. 2019;19(2):117–124. doi: 10.2174/1566523219666190628152137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu S, Tao Y. The epigenetic regulators and metabolic changes in ferroptosis-associated cancer progression. Mol Cancer. 2020;19(1):39. doi: 10.1186/s12943-020-01157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang M, Qiao M, Zhao C, Deng J, Li X, Zhou C. Targeting ferroptosis for cancer therapy: exploring novel strategies from its mechanisms and role in cancers. Transl Lung Cancer Res. 2020;9(4):1569–1584. doi: 10.21037/tlcr-20-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Yu J, Zhang L. Necroptosis: an alternative cell death program defending against cancer. Biochim Biophys Acta. 2016;1865(2):228–236. doi: 10.1016/j.bbcan.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35(6):830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Su Y, Zhao B, Zhou L, Zhang Z, Shen Y, Lv H, AlQudsy LHH, Shang P. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020;483:127–136. doi: 10.1016/j.canlet.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021. doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- 40.Cai H-J, Zhuang Z-C, Wu Y, Zhang -Y-Y, Liu X, Zhuang J-F, Yang Y-F, Gao Y, Chen B, Guan G-X, et al. Development and validation of a ferroptosis-related lncRNAs prognosis signature in colon cancer. Bosn J Basic Med Sci. 2021. doi: 10.17305/bjbms.2020.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Guo F, Guo W, Wang Y, Song W, Fu T. Ferroptosis-related genes are potential prognostic molecular markers for patients with colorectal cancer. Clin Exp Med. 2021. doi: 10.1007/s10238-021-00697-w. [DOI] [PubMed] [Google Scholar]
- 42.Dai E, Han L, Liu J, Xie Y, Zeh HJ, Kang R, Bai L, Tang D. Ferroptotic damage promotes pancreatic tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway. Nat Commun. 2020;11(1):6339. doi: 10.1038/s41467-020-20154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang B, Zhu J, Li J, Fan K, Gao Y, Cheng S, Kong C, Zheng L, Wu F, Weng Q, et al. The ferroptosis and iron-metabolism signature robustly predicts clinical diagnosis, prognosis and immune microenvironment for hepatocellular carcinoma. Cell Commun Signal. 2020;18(1):174. doi: 10.1186/s12964-020-00663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma S, Henson ES, Chen Y, Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7(7):e2307. doi: 10.1038/cddis.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J, Jiang X, Dong Z, Hu S, Xiao M. low-concentration PTX and RSL3 inhibits tumor cell growth synergistically by inducing ferroptosis in mutant p53 hypopharyngeal squamous carcinoma. Cancer Manag Res. 2019;11:9783–9792. doi: 10.2147/CMAR.S217944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo SM, Seo S, Min K-J, Im -S-S, Nam J-O, Chang J-S, Kim S, Park J-W, Kwon T. Corosolic acid induces non-apoptotic cell death through generation of lipid reactive oxygen species production in human renal carcinoma caki cells. Int J Mol Sci. 2018;19(5). doi: 10.3390/ijms19051309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stockwell BR, Jiang X. A physiological function for ferroptosis in tumor suppression by the immune system. Cell Metab. 2019;30(1):14–15. doi: 10.1016/j.cmet.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang R, Xu J, Zhang B, Liu J, Liang C, Hua J, Meng Q, Yu X, Shi S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13(1):110. doi: 10.1186/s13045-020-00946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sacco A, Battaglia AM, Botta C, Aversa I, Mancuso S, Costanzo F, Biamonte F. Iron metabolism in the tumor microenvironment-implications for anti-cancer immune response. Cells. 2021;10(2). doi: 10.3390/cells10020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ.. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 52.Yamazaki T, Hannani D, Poirier-Colame V, Ladoire S, Locher C, Sistigu A, Prada N, Adjemian S, JP Catani, Freudenberg M, et al. Defective immunogenic cell death of HMGB1-deficient tumors: compensatory therapy with TLR4 agonists. Cell Death Differ. 2014;21(1):69–78. doi: 10.1038/cdd.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22(6):679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 54.Dehne N, Mora J, Namgaladze D, Weigert A, Brüne B. Cancer cell and macrophage cross-talk in the tumor microenvironment. Curr Opin Pharmacol. 2017;35:12–19. doi: 10.1016/j.coph.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Jeannin P, Paolini L, Adam C, Delneste Y. The roles of CSFs on the functional polarization of tumor-associated macrophages. FEBS J. 2018;285(4):680–699. doi: 10.1111/febs.14343. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26(1):78. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42(6):717–727. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 58.Davis BP, Rothenberg ME. Eosinophils and cancer. Cancer Immunol Res. 2014;2(1):1–8. doi: 10.1158/2326-6066.CIR-13-0196. [DOI] [PubMed] [Google Scholar]
- 59.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11(8):519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 60.Galdiero MR, Garlanda C, Jaillon S, Marone G, Mantovani A. Tumor associated macrophages and neutrophils in tumor progression. J Cell Physiol. 2013;228(7):1404–1412. doi: 10.1002/jcp.24260. [DOI] [PubMed] [Google Scholar]
- 61.Eruslanov EB. Phenotype and function of tumor-associated neutrophils and their subsets in early-stage human lung cancer. Cancer Immunol Immunother. 2017;66(8):997–1006. doi: 10.1007/s00262-017-1976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchell D, Chintala S, Dey M. Plasmacytoid dendritic cell in immunity and cancer. J Neuroimmunol. 2018;322:63–73. doi: 10.1016/j.jneuroim.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 63.Ma QY, Huang D-Y, Zhang H-J, Chen J, Miller W, Chen X-F. Function of follicular helper T cell is impaired and correlates with survival time in non-small cell lung cancer. Int Immunopharmacol. 2016;41:1–7. doi: 10.1016/j.intimp.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 64.Efimova I, Catanzaro E, Van der Meeren L, Turubanova VD, Hammad H, Mishchenko TA, Vedunova MV, Fimognari C, Bachert C, Coppieters F, et al. Vaccination with early ferroptotic cancer cells induces efficient antitumor immunity. J Immunother Cancer. 2020;8(2). doi: 10.1136/jitc-2020-001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wen Q, Liu J, Kang R, Zhou B, Tang D. The release and activity of HMGB1 in ferroptosis. Biochem Biophys Res Commun. 2019;510(2):278–283. doi: 10.1016/j.bbrc.2019.01.090. [DOI] [PubMed] [Google Scholar]
- 66.Yu Y, Xie Y, Cao L, Yang L, Yang M, Lotze MT, Zeh HJ, Kang R, Tang D. The ferroptosis inducer erastin enhances sensitivity of acute myeloid leukemia cells to chemotherapeutic agents. Mol Cell Oncol. 2015;2(4):e1054549. doi: 10.1080/23723556.2015.1054549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19(7):405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 68.Kim K-T, Lee HW, Lee H-O, Kim SC, Seo YJ, Chung W, Eum HH, Nam D-H, Kim J, Joo KM, et al. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol. 2015;16(1):127. doi: 10.1186/s13059-015-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hua X, Zhao W, Pesatori AC, Consonni D, Caporaso NE, Zhang T, Zhu B, Wang M, Jones K, Hicks B, et al. Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat Commun. 2020;11(1):2459. doi: 10.1038/s41467-020-16295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


