Abstract
Background
Neoantigens derived from somatic mutations correlate with therapeutic responses mediated by treatment with immune checkpoint inhibitors. Neoantigens are therefore highly attractive targets for the development of therapeutic approaches in personalized medicine, although many aspects of their quality and associated immune responses are not yet well understood. In a case study of metastatic malignant melanoma, we aimed to perform an in-depth characterization of neoantigens and respective T-cell responses in the context of immune checkpoint modulation.
Methods
Three neoantigens, which we identified either by immunopeptidomics or in silico prediction, were investigated using binding affinity analyses and structural simulations. We isolated seven T-cell receptors (TCRs) from the patient’s immune repertoire recognizing these antigens. TCRs were compared in vitro by multiparametric analyses including functional avidity, multicytokine secretion, and cross-reactivity screenings. A xenograft mouse model served to study in vivo functionality of selected TCRs. We investigated the patient’s TCR repertoire in blood and different tumor-related tissues over 3 years using TCR beta deep sequencing.
Results
Selected mutated peptide ligands with proven immunogenicity showed similar binding affinities to the human leukocyte antigen complex and comparable disparity to their wild-type counterparts in molecular dynamic simulations. Nevertheless, isolated TCRs recognizing these antigens demonstrated distinct patterns in functionality and frequency. TCRs with lower functional avidity showed at least equal antitumor immune responses in vivo. Moreover, they occurred at high frequencies and particularly demonstrated long-term persistence within tumor tissues, lymph nodes and various blood samples associated with a reduced activation pattern on primary in vitro stimulation.
Conclusions
We performed a so far unique fine characterization of neoantigen-specific T-cell responses revealing defined reactivity patterns of neoantigen-specific TCRs. Our data highlight qualitative differences of these TCRs associated with function and longevity of respective T cells. Such features need to be considered for further optimization of neoantigen targeting including adoptive T-cell therapies using TCR-transgenic T cells.
Keywords: immunotherapy, adoptive, adaptive immunity, antigens, neoplasm, CD8-positive T-lymphocytes, receptors, immunologic
Inroduction
Cancer immunotherapy has demonstrated high efficacy in the treatment of diverse malignant diseases as shown by the potency of immune checkpoint modulation.1 These therapies can unleash antitumor immune responses by conveying recognition and eradication of cancers by the patients’ own immune system, although the contribution of specific T cells and the targeted antigens are not predefined. However, not all patients benefit from this therapeutic approach, thus, understanding the nature of tumor recognition versus escape remains of fundamental importance for the development of novel immunotherapeutic approaches and the improvement of patients’ outcomes. The tumor mutational load is an important prognostic and predictive biomarker for the therapeutic success of immune checkpoint inhibitors across various disease entities, emphasizing the role of neoepitopes in tumor recognition and rejection.2 Moreover, identification and characterization of neoantigens and neoantigen-specific T-cell responses are of particular interest for the generation of new personalized immunotherapies, such as vaccination and cellular therapies.3–5 In order to fully exploit the potential of neoantigen-directed immunotherapy, more efforts are required to understand different qualities of neoepitopes and their respective T-cell receptors (TCRs) specifically recognizing them. We believe that case studies are currently a valid source to investigate the multitude of aspects involved6 and that more detailed, comprehensive data are necessary to this purpose. Recently, we have identified two neoantigens by mass spectrometry (MS) and validated these by proof of defined neoantigen-specific autologous T-cell responses in a patient with melanoma.7 We have now extended the number of immunogenic antigens in this patient using additional prediction analyses and characterized neoantigens by 3D modeling as well as experimental validation of major histocompatibility complex (MHC)–peptide stability. In addition, we identified seven TCRs recognizing these antigens. We tracked these TCRs in blood and different tissues of the patient over time and functionally characterized them in vitro as well as in vivo, providing evidence of substantial differences regarding functionality and longevity with relevance for further comprehensive immunotherapies.
Materials and methods
Primary human specimens and cell lines
An overview of the clinical course of the patient with melanoma is given in online supplemental figure 1, as well as in online supplemental material and methods in more detail. All resected samples were handled as previously described for confirmation of diagnosis and expansion of tumor-infiltrating lymphocytes (TILs) from a lung metastasis (MLung).7 Peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient centrifugation (Ficoll-Hypaque, Biochrom) immediately on receipt using EDTA-anticoagulated blood from the patient and blood or apheresis products from healthy donors followed by storage in liquid nitrogen.
jitc-2021-002754supp002.pdf (2.2MB, pdf)
jitc-2021-002754supp001.pdf (785KB, pdf)
PBMCs and T cells used for experiments were cultivated as previously described.7 CD8+ T cells used for transduction were obtained by magnetic separation from healthy donor-derived apheresis products (Dynabeads Untouched Human CD8+ T Cells Kit, Thermo Fisher; Magnet, DynaMag).
Cell lines used in this study were T2 (American Type Culture Collection - ATCC cat# CRL-1992), purchased from ATCC in 2005; lymphoblastoid cell lines (LCLs) HOM-2 (European Collection of Authenticated Cell Cultures - ECACC cat# 98092902), SWEIG007 (ECACC cat# 88052037), AMALA (Interlab Cell Line Collection - ICLC cat# HTL14002), OZB (ECACC cat# 94022545), RSH (ECACC cat# 88052021), KLO (ECACC cat# 94050324), LWAGS (ECACC cat# 88052078), and BM21 (ECACC cat# 88052043), kindly provided by Steve Marsh in 2007; human metastatic melanoma cell line A2058 (ECACC cat# 91100402) and human colon carcinoma cell line MDST8 (ECACC cat# 99011801) purchased from Sigma-Aldrich in 2018; B-cell lymphoma cell line U-698-M (DSMZ cat# ACC-4), acquired from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) in 2018; embryonal kidney cell line 293Vec-RD114 (BioVec Pharma, Québec, Canada); and NS0-IL15, kindly provided by S. R. Riddell in 2011.8 LCL (LCL8) was generated in our laboratory by infection and immortalization of healthy donor-derived B cells with Epstein-Barr virus-containing supernatant in 2011. Absence of mycoplasma infection in cell line cultures was routinely confirmed by PCR (Venor GeM mycoplasma detection kit, Minerva Biolabs). Culture conditions are described in online supplemental material and methods.
DNA extraction, exome and TCR-β sequencing
For exome and T-cell receptor beta (TCR-β) deep sequencing, genomic DNA (gDNA) was extracted from a lung biopsy (BLung), an intestinal metastasis (MInt), MLung and draining lymph nodes (LNs) after microdissections of respective regions of interest. The extraction of nucleic acids was performed on 2 µm formalin-fixed, paraffin-embedded (FFPE) tissue slides using Maxwell RSC Blood DNA Kit (Promega) following the manufacturer’s recommendations. DNA was extracted from PBMCs obtained from diverse blood withdrawals (online supplemental figure 1) and MLung-derived TILs with DNeasy Blood & Tissue Kit (Qiagen). Whole-exome sequencing and single-nucleotide variant (SNV) analyses from MInt were available from our previous publication and were afterwards obtained for MLung, as previously described.7 Next-generation sequencing of TCR-β loci was performed by Adaptive Biotechnologies with ImmunoSEQ platform at the deep level (exception made for BLung gDNA, which underwent TCR-β sequencing survey level only due to limited material).
In silico prediction of peptide ligands and human leukocyte antigen (HLA) binding affinity
Putative mutated nonamer peptide ligands originating from SNVs, identified through a stringent variant calling approach,7 were predicted by translating all sequences bearing a mutation to 23-residue long amino acid (AA) sequences (mutated AA in 12th position). Protein transcripts were downloaded from Ensembl GRCh38, release 86. In cases where the mutation was located less than 12 AA from the 3′ or 5′ terminus of the gene, the string was shorter and the mutation was not centrally located. Peptide–HLA class I binding affinity was predicted for the patient’s HLA alleles HLA-A03:01 and HLA-B27:05 using NetMHC V.4.0. Nonamer peptides were ranked by predicted affinity (cut-off <500 nM) (online supplemental tables 1 and 2). Half maximal inhibitory concentration (IC50) in vitro measurements were performed as described in online supplemental material and methods.
Immunogenicity assessment of mutated peptide ligands
For immunogenicity assessment, peptide ligands were ordered from Genscript and DgPeptides. Ligands identified by MS, including NCAPG2P333L and SYTL4S363F, were tested for recall antigen-experienced T-cell responses as described in the online supplemental material and methods. Predicted peptides were arranged in pools according to predicted affinity (online supplemental table 3) and in vitro screened for immunogenicity following protocols described in the online supplemental material and methods. Reactivity was assessed by specific Interferon-gamma (IFN-γ) release by ELISpot assay by coating ELISpot plates (MAHAS4510) with IFN-γ capture antibody (MABTECH cat# 3420-3-250) followed by application of IFN-γ-detection antibody (MABTECH cat# 3420-6-250) and visualization by streptavidin–horseradish peroxidase (MABTECH cat# 3310-9-1000).
Structural modeling of neoantigen binding to respective HLA complexes
Neoantigens and wild-type (WT) peptides were modeled on experimental structures of HLA-A03:01 (PDB-ID: 2XPG)9 and HLA-B27:05 (PDB-ID 1W0V)10 crystallized with nine AA long ligands. Peptides from the template were mutated into neoantigen/WT sequences using the IRECS program.11
Solvent-accessible surface area (SASA) values were calculated using the VMD software V.1.9.2,12 13 applying a probe radius of 1.4 Å. Further information is provided in online supplemental materials and methods. Details about heat-up parameters are provided in online supplemental table 4.
Flow cytometry-based assessment of TCR expression
Transduction efficiency was determined by staining of transgenic TCR using an antibody specifically binding to murine TCR constant beta chain (TCRmu-antibody (clone H57-597), Thermo Fisher Scientific cat# 11-5961-81) based on the isotype control (Thermo Fisher Scientific cat# 11-4888-81). For multimer staining, Strep-tagged mutated and WT peptide-Human Leucocyte Antigen (pHLA) (eg. SYTL4S363F and SYTL4WT) were multimerized on a fluorophore labeled Strep-Tactin backbone (Iba GmbH) in a 1 µg:1 µL ratio. Transduced T cells were stained with multimers, anti-CD8 (BD Biosciences cat# 557 085 and BD Biosciences cat# 555634), anti-CD3 (BD Biosciences cat# 557694) and 7-aminoactinomycin D. All flow cytometry measurements were performed with LSRII or LSRFortessa flow cytometers (BD Biosciences) and analyzed with FlowJo Software V.10.
Functional analyses of TILs and TCR-transgenic effector T cells
TCR-transduced T cells were coincubated with target cells (LCL-1) transduced with different minigene (MG) constructs or pulsed with defined peptides (1 µM). WT MGs and/or irrelevant peptides served as controls for TCR specificity. Assays for detection of cytokine secretion were performed in triplicate (effector to target (E:T)=1:1; 10,000 target and effector cells per well). Supernatants from T-cell cultures were used for assessment of a diverse cytokine panel with MACSPlex 12 Cytokines Kit (Miltenyi Biotech) following the manufacturer’s recommendations. Reactivity of TILs was assessed by IFN-γ ELISpot assay on a coculture with autologous peptide-pulsed γ-irradiated LCLs (1 µM) for 24 hours using at E:T of 1:1.
Functional avidity was assessed by incubating transgenic T cells with T2 target cells (ATCC CRL-1992) pulsed with titrated peptide concentrations (E:T=1:1; 10,000 cells/well). IFN-γ secretion was quantified by ELISA, and values obtained were fitted into a non-linear variable-slope regression curve on GraphPad Prism V.7.
Functional avidity assays were performed at least three times using at least two different transductions and donors showing comparable results.
In vitro target cell killing was assessed by impedance-based xCELLigence assays as described in online supplemental material and methods.
TCR cross-reactivity was tested by stimulation of TCR-transgenic T-cell populations with T2 cells pulsed with ala/thr substitution variants of the three neoepitopes, in direct comparison to the original mutated identified peptides. IFN-γ values from every condition were normalized on the positive control using the following formula:
Relative IFN-γ-secretion after incubation with an ala/thr variant above 50% was considered as a replaceable position.
For cross-reactivity scan of synthesized peptides based on the ScanProsite pattern, T cells were coincubated with T2 cells pulsed with 1 µM of peptide (E:T=1:1; 10,000 cells/well). Analysis was conducted for at least two different donors for each TCR construct.
For evaluation of general alloreactivity, TCR-transduced T cells were stimulated with respective neoantigens presented in the context of different HLA class I alleles as well as non-pulsed target cells. Effector cells were cultured with LCL presenting a broad variety of HLA alleles (online supplemental table 5), pulsed with mutated peptides (E:T=1:1; 10,000 cells/well). As readout, IFN-γ production was quantified by ELISA.
In vivo TCR rejection potential
NOD.CG-Prkdcscid IL2rgtm1Wjl/SzJ (NSG, The Jackson Laboratory) was maintained according to the institutional guidelines and approval of local authorities. A xenograft murine model was generated as previously described.14 15 NSG mice at the age of 6–19 weeks were subcutaneously injected with U-698-M cells (10×106 cells/flank) transduced with tandem MG constructs coding for defined neoantigens. Animal well-being was assessed daily and growth was monitored in vivo by external measurements with digital caliper. Ten days after administration of tumor cells, almost all mice developed visible and measurable tumor growth. T cells transduced with TCRs KIF2C-PBC1, KIF2C-PBC2, SYTL4-PBC1, SYTL4-TIL1, or a control TCR 2.5D6 targeting an irrelevant antigen (myeloperoxidase)14 were injected intravenously. TCR-transduced T cells (2×107) in total were administrated to eight mice per group (n=8) over 2 days, except for KIF2C-PBC1, of which 4.6×106 TCR-transduced T cells were injected in total due to lower transduction efficiency. Male and female animals as well as animals of different ages were distributed evenly across all treatment groups. Tumor growth kinetics were monitored daily for 26 days with digital caliper. Interleukin (IL)-15-producing NS0 cells (1.0–1.5×107) were administered intraperitoneally two times per week after irradiation with 80 Gy.16 On day 4, after the first T-cell injection, two mice per group were removed for flow cytometry analysis. On day 20, remaining tumors, bone marrow and spleen were retrieved and passed through a cell strainer. Blood samples were taken and anticoagulated with EDTA. Ammonium–chloride–potassium lysis (Life Technologies) was performed on blood samples. Single-cell suspensions were stained for detection of transgenic T cells by flow cytometry as explained previously.
Statistical analysis
Significance of differences within half maximal effective concentration (EC50) values of analyzed TCRs were investigated by one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test. With regard to in vivo rejection potential of the TCRs, differences in tumor growth were calculated with two-way ANOVA test (time and treatment) and Dunnett’s test for multiple comparisons. Statistical comparison of survival was performed using the Mantel-Cox test. To calculate the statistical significance of the increase in SASA, a standard independent two-sample t-test was used. Statistical analyses were performed with GraphPad Prism V.7.04 software.
Additional information on the material and methods is provided in the online supplemental file.
Results
We have previously reported MS-based identification of two immunogenic neoantigens (SYTL4S363F and NCAPG2P333L) presented by an MInt of a patient with melanoma.7 The patient was suffering from metastatic malignant melanoma and was treated sequentially with chemotherapy and ipilimumab. Two metastases were surgically removed within a period of 24 months, resulting in complete remission. Subsequently, the patient received treatment with pembrolizumab for 1 year and has remained in sustained remission since then. Based on this case, we performed multidimensional analyses using a comprehensive material collection including a variety of tissues (BLung, MLung, MInt−LN1, MInt−LN2, and MLung−LN), as well as blood specimens, collected over a period of 3 years. The disease course of this patient is explained in details in the online supplemental material and shown in online supplemental figure 1).
In silico predictions complement MS-based neoantigen identification
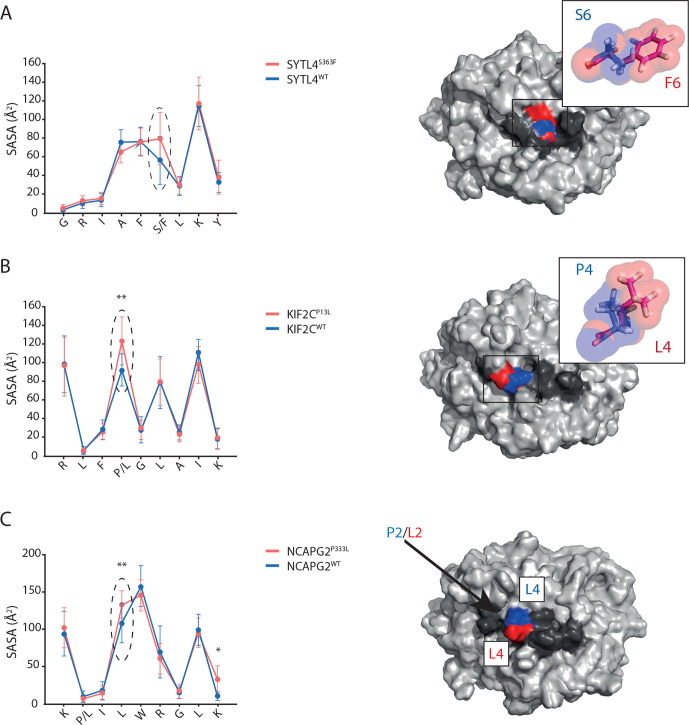
In order to investigate whether critical neoantigens may have been missed by MS, a sequence-based prediction approach was applied. A total of 1196 missense mutations were called from MInt tumor tissue, leading to prediction of ~4670 peptides (8–12 AA long) with binding affinity of <500 nM. By sorting nine AA-long putative peptides according to NetMHC predicted binding affinity, previously identified neoantigens NCAPG2P333L and SYTL4S363F ranked 24th and 6th in HLA-A03:01 and HLA-B27:05 lists, respectively (online supplemental tables 1 and 2). Most peptides demonstrated also experimentally high binding affinities; however, some binding affinities partially differed from the predicted ones (online supplemental tables 1 and 2). Investigation of immunogenicity of these top-ranked nonamers binding to HLA-A03:01 and B27:05 alleles resulted in the detection of one additional confirmed neoepitope, KIF2CP13L (online supplemental figure 2A, B and table 1), ranking position 1 of top 25 HLA-A03:01 binding peptides according to NetMHC V.4.0 (online supplemental table 2). Molecular dynamic (MD) simulations of MHC peptide binding of SYTL4S363F, KIF2CP13L, and NCAPG2P333L were subsequently performed to elucidate differences in SASAs. Clear differences between WT and mutated peptides were observed (figure 1A–C). This was even the case for the mutated P2 of NCAPG2P333L harboring the mutation inside the peptide binding cleft and not at the TCR interface (figure 1C) as the larger size of the mutated residue (Leu≥Pro) shifted the position of solvent-exposed P4 Leu, increasing its SASA significantly from 108.06 Å2 (WT) to 133.54 Å2 (mutant).
Table 1.
Characteristics of neoantigens targeted by identified TCRs
| Peptide name | Gene name Ensembl transcript ID | Peptide sequence | HLA restriction | Predicted affinity (nM), rank (%) | Experimental IC50 (nM) | MS FDR (%) | Reads exome PBMCs (Ref:Alt) | Reads exome MInt (Ref:Alt) | Reads exome MLung (Ref:Alt) |
| SYTL4S363F | SYTL4; ENST00000263033 | GRIAFFLKY | B27:05 | 18.43; 0.6 | 214.7±1.8 | 1 | 54:0 | 32:10 | 71:20 |
| SYTL4WT | GRIAFSLKY | 30.62; 0.12 | 97.32±2.1 | ||||||
| KIF2CP13L | KIF2C; ENST00000372224 | RLFLGLAIK | A03:01 | 21.6; 0.09 | 237.4±2.0 | 75:0 | 75:18 | 177:21 | |
| KIF2CWT | RLFPGLAIK | 12.10; 0.03 | 146.2±1.1 | ||||||
| NCAPG2P333L | NCAPG2; ENST00000409339 | KLILWRGLK | A03:01 | 32.6; 0.15 | 202.9±1.03 | 1 | 126:0 | 146:27 | 54:0 |
| NCAPG2WT | KPILWRGLK | 6946.48; 6.50 | 4,488.7±10.4 |
Alt, alternative read; FDR, false discovery rate; HLA, human leukocyte antigen; IC50, half maximal inhibitory concentration; MInt, intestinal metastasis; MLung, lung metastasis; PBMC, peripheral blood mononuclear cell; Ref, reference read; TCR, T-cell receptor.
Figure 1.
Mutations result in distinct antigen features as shown by structural modeling. (A–C) Residue-specific SASA values of the neoantigen (red) and its respective WT (blue) are plotted on the left side. Corresponding superimposed representative structures as extracted from MD simulations are shown on the right side for SYTL4S363F (A), KIF2CP13L (B), and NCAPG2P333L (C). HLA peptide complexes (right side) are shown in surface representation with HLA (light gray), peptide (dark gray) and central positions of increased SASA in red (neoantigen) and blue (WT). Latter positions are emphasized in the SASA plots by black arrows and dashed line ellipses. (A, B) Mutated residues are depicted in sticks and transparent spheres to illustrate physicochemical differences. (C) The arrow indicates the buried mutation site. SASA mean value and SD of each individual peptide AA were calculated out of three replicates. *P<0.05, **P<0.01. HLA, human leukocyte antigen; MD, molecular dynamic; SASA, solvent-accessible surface area; WT, wild type.
In-depth characterization of immune responses against three selected neoantigens show high antigen specificity
Immune responses against KIF2CP13L, SYTL4S363F, and NCAPG2P333L neoantigens (table 1) were further analyzed in detail, and seven TCRs with specificity to these neoantigens were isolated (online supplemental table 6). SYTL4-TIL1 and SYTL4-TIL2 TCRs were isolated from TILs expanded from lung tissue after in vitro peptide stimulation and CD137-based enrichment, whereas all other TCRs were isolated from peripheral blood after in vitro stimulation and expansion as described before7 and in the Materials and methods section. Transduction efficiency by staining of the murinized constant regions of transduced TCRs showed differences for defined TCRs, with lowest transduction rates for TCRs KIF2C-PBC1 and SYTL4-TIL2 (online supplemental figure 3). Moreover, by performing HLA class I multimer staining for all TCRs, T-cell populations binding the mutated peptide multimer could be detected only for five out of seven TCRs (online supplemental figure 4).
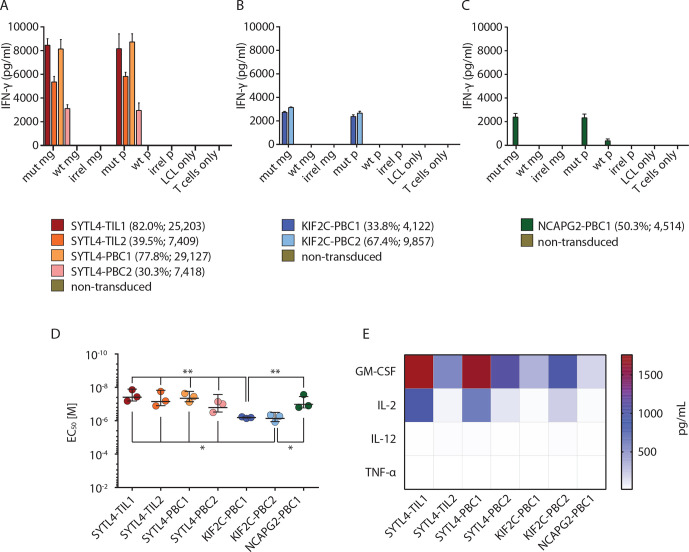
In vitro functional characterization of TCR-transgenic CD8+ T cells showed highly specific IFN-γ secretion on recognition of target cells pulsed with mutant peptides or transduced with MGs coding for neoantigens (figure 2A–C). No reactivity was observed against WT or irrelevant peptides as well as the corresponding control MGs. Of note, transduction rates, evaluated by TCRmu staining (online supplemental figure 3), did not necessarily correlate with IFN-γ secretion as both KIF2CP13L-specific TCRs showed similar reactivity to target LCL-1 cells presenting the mutant peptide despite major differences in TCR expression (figure 2B). In line with that, cytokine secretion of SYTL4S363F-specific TCRs was substantially higher in comparison to those reactive to KIF2CP13L or NCAPG2P333L, even for those TCRs with lower surface expression (figure 2A–C).
Figure 2.
High specificity and strong functional performance of identified TCRs in vitro. (A–C) Secretion of IFN-γ by CD8+ T cells transduced with TCRs specific for SYTL4S363F (A), KIF2CP13L (B), and NCAPG2P333L (C) after coculture with LCL-1-presenting neoantigens and respective WT counterparts at effector to target (E:T)=1:1 is shown. LCL-1 transduced with MGs encoding for fragments of mut sequence (mut MG), WT sequence (WT MG) or irrelevant sequence (irrel MG) were compared with peptide pulsed LCLs (mut P, WT P, irrel P; 1 µM peptide). IFN-γ secretion in supernatants was investigated by ELISA assay. Bars represent average reads from three duplicates; error bars represent SD transduction efficiencies; and Mean Fluorescent Intensity (MFI) values are indicated below the graphs. (D) Comparison of functional avidity of neoantigen-specific TCRs, calculated as EC50 of cognate mutated peptide. IFN-γ secretion was assessed on supernatants, and a non-linear curve was fit to determine the EC50 value. EC50 values deriving from three different experiments were depicted for each TCR. Bars in the graph represent the mean value, and SD significance is calculated with one-way analysis of variance and Tukey multiple comparison test (*p≤0.05, **p≤0.01). (E) Assessment of multicytokine secretion of TCR-transduced T cells on coculture with LCL-1 pulsed with mutated peptides. All experiments were performed at least with three different sets of transduced T cells derived from two different healthy donors. EC50, half maximal effective concentration; IFN, interferon; LCL, lymphoblastoid cell line; MG, minigene; mut, mutant; TCR, T-cell receptor; WT, wild type.
Functional avidity measurements using peptide-pulsed T2-A3 and T2-B27 cells showed EC50 values against respective ligands in the almost nanomolar range for all TCR-transduced T cells (figure 2D). However, TCRs specific for SYTL4S363F and NCAPG2P333L showed significantly higher avidities compared with KIF2CP13L-specific TCRs (figure 2D, online supplemental figure 5).
To further decipher functional differences between all seven TCRs, defined cytokines as granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-2, IL-12, and tumor necrosis factor alpha (TNF-α) were assessed by multiplex analysis using supernatants of TCR-transgenic CD8+ T cells stimulated with mutated or WT MG-transduced LCL-1 target cells (figure 2E). Again, T cells transduced with TCRs SYTL4-TIL1 and SYTL4-PBC1 secreted the highest concentrations of these cytokines in response to respective mutated MGs.
To analyze the dynamics of TCR-mediated cytotoxic activity in vitro, an impedance-based xCELLigence system was used to monitor growth kinetics of MG transduced target cell lines after coincubation with TCR-transduced T cells. With the exception of SYTL4-PBC2, all TCRs mediated 100% lysis of MDST8MUT and A2058MUT target cells within 12–16 hours, while WT clones were not affected (online supplemental figure 6A–C). Among SYTL4S363F-specific TCRs, SYTL4-TIL1 showed the fastest rejection dynamics (online supplemental figure 6A). With respect to KIF2CP13L-specific TCRs, rejection dynamics again appear to be similar despite different transduction rates (online supplemental figure 6B). Comparison of cytotoxic potential for all TCRs showed different dynamics dependent on the target cells but comparable final effects except for SYTL4-PBC2 displaying an inferior cytolysis pattern (online supplemental figure 6D).
TCRs show neoantigen-specific binding and low cross-reactivity patterns
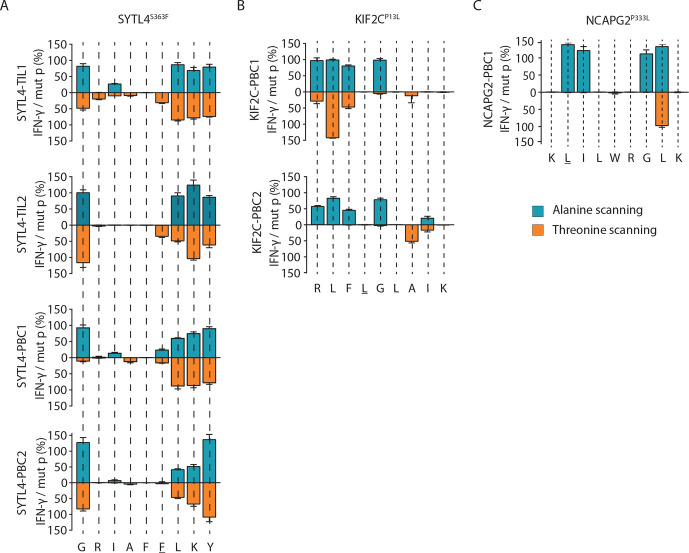
A set of altered peptide ligands containing individual alanine or threonine replacements (ala/thr scan) at every single position of the cognate neoantigen was used to investigate the cross-reactivity potential of defined TCR-transgenic T cells (figure 3). Highly similar recognition patterns of all four SYTL4S363F-specific TCRs were observed despite highly diverse TCR sequences (figure 3A and online supplemental table 7). These results suggest a similar peptide–HLA docking pattern for all TCRs with this neoantigen specificity. In contrast, recognition patterns of exchange ala/thr motifs were slightly more variable with respect to both KIF2CP13L-specific TCRs (figure 3B). This was reflected by a higher potential for cross-reactivity against naturally occurring peptides within the human proteome, as investigated by ScanProsite tool (https://prosite.expasy.org/scanprosite).17 SYTL4S363F-specific and NCAPG2P333L-specific TCRs exhibited potential cross-reactivity with four to six antigens only, while KIF2CP13L-specific TCRs might recognize >400 targets (online supplemental table 7). The potential discrepancy in cross-reactivity was tested in vitro with a selection of the ScanProsite retrieved peptides according to predicted binding affinity to HLA-A03:01 and B27:05 (online supplemental table 8). This small-scale screening did not show any reactivity of the TCRs against the selected peptides (online supplemental figure 7). Moreover, the alloreactive potential of selected TCRs tested against a panel of LCLs expressing the most frequent HLA allotypes (online supplemental table 5) with or without previous peptide pulsing showed low alloreactive potential for all neoantigen-specific TCRs (online supplemental figure 8).
Figure 3.
Cross-reactivity analyses indicate antigen-dependent TCR docking on HLA peptide complexes. (A–C) TCR cross-reactivity was tested by quantification of secreted IFN-γ on coculturing TCR-transduced T cells with T2 target cells pulsed with ala/thr scanned peptide cognates (1 µM peptide) of ligands SYTL4S363F (A), KIF2CP13L (B), and NCAPG2P333L (C). IFN-γ secretion values from single conditions were normalized against the cytokine level in response to the defined neoantigen. HLA, human leukocyte antigen; IFN, interferon; TCR, T-cell receptor.
Neoantigen-specific TCRs display in vivo antitumor potential
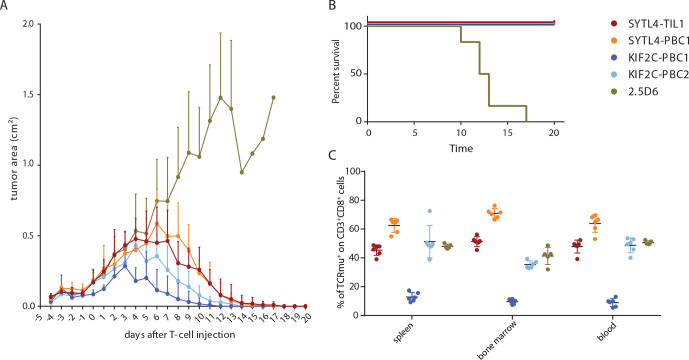
We next asked whether differences of in vitro functionality, especially with respect to SYTL4S363F-specific and KIF2CP13L-specific TCRs, may also lead to different in vivo tumor rejection patterns (figure 4). Therefore, antitumor reactivity of defined neoantigen-specific TCRs was investigated in a xenograft murine model using a MG-transduced U-698-M lymphoma model. U-698-M tumor growth in NSG mice was monitored starting from 5 days before intravenous T-cell injection. TCR-transduced T cells were normalized to approximately 62%, except for KIF2C-PBC1, for which transduction rate was about 14% (online supplemental table 9). Around day 4 post-T-cell injection, tumors of mice that received T cells expressing a control TCR kept constantly growing, while tumors in hosts receiving T cells transduced with TCRs SYTL4-TIL1, SYTL4-PBC1, KIF2C-PBC1, and KIF2C-PBC2 completely rejected the tumor (figure 4A). Of note, tumor rejection by T cells transduced with KIF2C-PBC2 and especially KIF2C-PBC1 with lower transduction rate showed very good rejection kinetics. Survival curves visualize that mice injected with control TCR 2.5D6 developed uncontrolled tumor growth with ulcerations beginning at day 10 and subsequently had to be euthanized, whereas neoantigen-specific TCRs mediated significantly prolonged survival (figure 4B). Flow cytometry data show infiltration of mutated tumors by respective TCRs at day 4 after first T-cell injection (online supplemental figure 9) as well as persistence of TCR-transgenic T cells in the spleen, bone marrow and blood at day 20 (figure 4C). Observed frequencies of TCRmu+ T cells among all CD8+ lymphocytes in these compartments reflect frequencies of transferred TCR-transgenic T cells.
Figure 4.
Performance of T cells transgenic for neoantigen-specific TCRs in vivo. (A) Growth kinetics of U-698-M tumors expressing neoantigens (area in mm2) in NSG mice. Mean values and SDs are depicted for each group of mice to monitor rejection dynamics (n=6). Animals were intravenously injected with a total of 2×107 T cells on day 0+1. Tumor growth was significantly impaired in all mice receiving neoantigen-specific TCRs in comparison to 2.5D6 at day 10 (adjusted p value of 0.0001, calculated with two-way analysis of variance (time and treatment) and multiple comparison Dunnett’s test). (B) Kaplan-Meier survival curve of tumor-bearing mice injected with different TCR-transduced T cells. Survival of animals receiving neoantigen-specific TCRs was significantly prolonged to 2.5D6 (p<0.0001, Mantel-Cox test) (C), percentage of TCRmu+ T cells among all infiltrating CD3+CD8+ T cells in spleen, bone marrow and blood. Organs of animals receiving 2.5D6-transgenic T cells were processed at the respective day of removal. One control mouse receiving 2.5D6-transgenic T cells did not exhibit a clear population of infiltrating T cells and was therefore excluded from this analysis. All other animals with transferred neoantigen-specific TCR constructs were analyzed at day 20 exhibiting complete tumor rejection. TCR, T-cell receptor.
Antigen-dependent spatial and temporal distribution of identified TCRs in blood and tissues
To investigate spatial and temporal distributions of tumor antigen-specific TCR clonotypes within the patient’s tissues and blood, TCR-β deep sequencing was performed by extracting gDNA from BLung, MInt, MLung, LNs, as well as PBMCs from different time points (online supplemental figure 1).
Overlap of TCR-β clonotypes was investigated in all three tumor tissues. All tumor samples, resected over a time span of 8 years, share 29 TCR-derived CDR3 (online supplemental figure 10A). MInt and MLung, closer in time, share 3072 TCR clonotypes (14.77% of all sequences identified in MInt and MLung). Notably, a number of 462 TCR clonotypes were shared between both resected metastases and all analyzed adjacent LNs (online supplemental figure 10B).
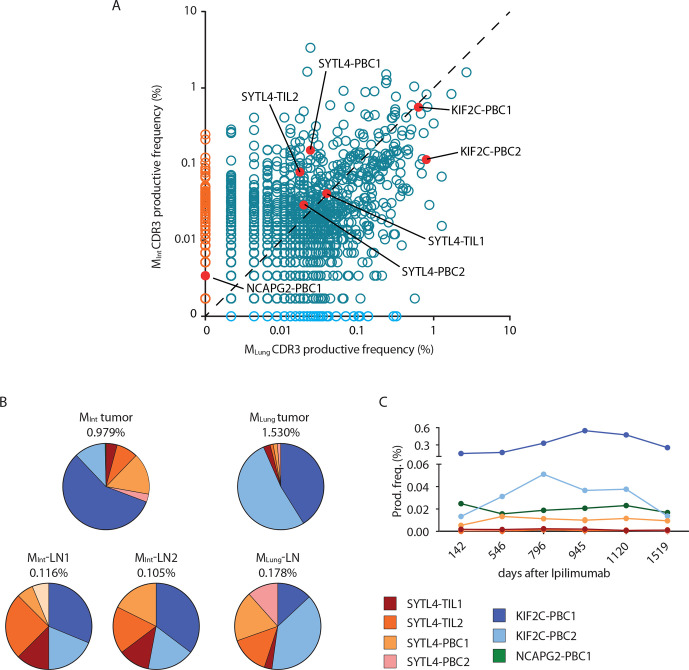
The seven characterized neoantigen-specific TCRs were tracked within metastasized tumor lesions, LNs and blood samples (figure 5 and online supplemental figure 8). Frequencies of these TCRs identified from peripheral blood specimens, LNs and tumor tissue are shown in online supplemental tables 10 and 11). KIF2CP13L-specific TCRs were the most abundant in all metastasized tumor tissues over time, and TCR KIF2C-PBC1 could be detected in BLung even prior to therapy with Ipilimumab. Overall, frequency of KIF2CP13L-specific TCRs, especially KIF2C-PBC2, was increasing in MLung compared with MInt (figure 5A and online supplemental table 10), whereas frequencies of SYTL4S363F-specific TCRs were decreasing or remained stable. Notably, TCR NCAPG2-PBC1 could not be detected in MLung, corresponding to the lack of detectability of this specific mutation in the metastasis on the exome level (figure 5A and table 1). By investigating the relative frequency of known neoantigen-specific TCRs among all identified CDR3 sequences in tumor-associated non-malignant LNs, the overall percentage of all seven cognate TCRs contributing to the analyzed TCR repertoire was calculated, ranging from 0.11% to 1.53% (figure 5B). Thereby, TCRs specific for KIF2CP13L comprised the highest relative percentage not only in tumor tissues but also in tumor-associated LNs. However, we observed relative frequency of defined TCR clonotypes being more evenly distributed in associated LNs (figure 5B).
Figure 5.
Spatial and temporal distribution of neoantigen-specific TCRs as determined by TCR-β sequencing. (A) Productive frequency of CDR3 rearrangements (amino acid) calculated as the number of sequencing templates divided by the sum of template counts for all productive rearrangements in MInt and MLung. Scatter plot dataset was generated with adaptive Biotechnologies ImmunoSEQ analysis software. (B) Distribution of neoantigen-specific TCRs in resected tumor and lymph node tissues. Percentages indicate frequency of identified neoantigen-specific TCRs in analyzed tissue repertoire as inferred from TCR-β deep sequencing data. (C) Productive frequency expressed as percentage of neoantigen-specific clonotypes in peripheral blood at different time points after first treatment with ipilimumab. TCR, T-cell receptor.
In peripheral blood, all specific TCRs could be detected in at least four out of the six sequenced samples (figure 5C). Productive frequencies of SYTL4S363F-specific TCRs ranged between 0.0008% to maximal 0.013%, whereas TCRs KIF2C-PBC1, KIF2C-PBC2, and NCAPG2-PBC1 showed considerably higher frequencies between 0.013% and 0.54% in analyzed blood samples (online supplemental table 11). Of note, TCR KIF2C-PBC1 showed again the highest frequencies over time. In contrast, frequency of TCR KIF2C-PBC2 in peripheral blood increases up to the time point of resection of the MLung and decreases thereafter corresponding to the high frequency of this TCR in MLung and associated LN (figure 5C).
Presence of identified TCR KIF2C-PBC2 within respective primary tumor tissue was verified additionally by TCR imaging using RNA hybridization BaseScope assay on tumor MLung (online supplemental figure 11). In addition, mutated ligand KIF2CP13L was also recognized by isolated TILs of the patient (online supplemental figure 12).
T cells specific for SYTL4S363F and KIF2CP13L display divergent activation profiles
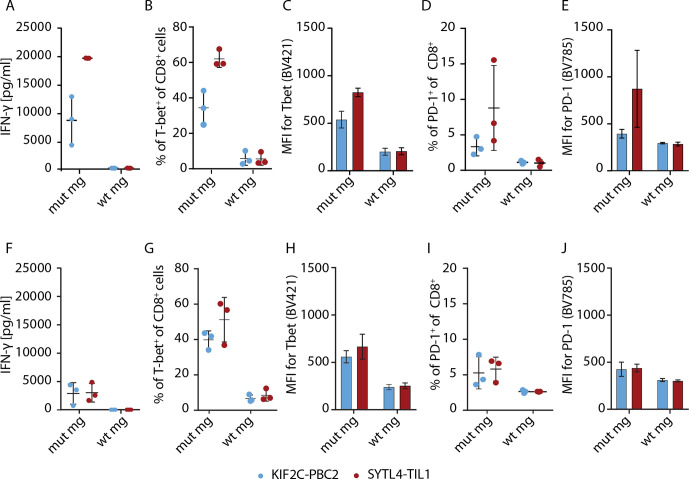
To further delineate TCR-dependent differences, we aimed to investigate activation parameters of neoantigen-specific stimulated T cells. We exemplarily explored expression of the T-box transcription factor TBX21 (T-bet) and programmed cell death protein 1 (PD-1) expression on SYLT4-TIL1, -PBC1, KIF2C-PBC1, and KIF2C–PBC2 transduced T cells after coculture with MG-transduced U-698-M. SYTL4S363F-specific TCR displayed a higher upregulation of T-bet after 24 hours compared with KIF2CP13L-specific constructs with all TCR showing a downregulation of expression to baseline after 96 hours (online supplemental figure 13A–B). To minimize the potential influence of different transduction rates, T cells transduced with TCRs SYTL4-TIL1 and KIF2C-PBC2 were enriched using near -infrared fluorescent protein (iRFP)-containing constructs. Using enriched populations, we observed marked differences of IFN-γ secretion between TCRs with defined specificities on antigen-specific stimulation (figure 6A), as shown for non-enriched transduced T cells (figure 2A, B). Investigating early T-cell activation, we observed a higher upregulation of T-bet for SYTL4-TIL1-iRFP after 24 hours compared with KIF2C-PBC2-iRFP (figure 6B, C, and online supplemental figure 13C–D). Also, upregulation of PD-1 was higher after 24 hours in SYTL4-redirected T cells in comparison to KIF2C-PBC2 with respect to frequency and MFI of PD-1+ T cells (figure 6D, E). Yet, in vitro rechallenging of neoantigen-specific T cells after 11 days contrarily revealed an almost equal IFN-γ secretion of both TCRs on stimulation with U-698-M expressing the mutated MG (figure 6F). In this regard, T-bet and PD-1 expression of KIF2C-PBC2-iRFP was at the same level as SYTL4-TIL1-iRFP (figure 6G–J).
Figure 6.
T-cell activation and dysfunction of enriched SYTL4-TIL1-iRFP and KIF2C-PBC1-iRFP TCR on neoantigen stimulation with MG-transduced U-698-M. (A) IFN-γ secretion of T cells 24 hours after coculture. (B–E) Expression of specific markers 24 hours after coculture: percentage of T-bet+ cells within CD8+ T cells (B), MFI for T-bet of CD8+ T cells (C), percentage of PD-1+ cells within CD8+ T cells (D) and MFI for PD-1 of CD8+ T cells (E). (F, J) T-cell activation and dysfunction was comparably assessed 24 hours after second stimulation of SYLT4-TIL1-iRFP and KIF2C-PBC2-iRFP transgenic T cells with MG-expressing target cells at day 11 after initial stimulation. Results of transduced T cells from three different donors are shown. MG, minigene; PD-1, programmed cell death protein 1; TCR, T-cell receptor.
Discussion
Within this study, we performed an in-depth characterization of TCR specifically recognizing neoantigens derived from somatic mutations which have been identified in malignant tissue of a patient with melanoma. We thereby reveal important insights with potential relevance for the development of advanced personalized immunotherapies targeting neoantigens.
For neoantigen identification, MS-based immunopeptidomics and in silico peptide prediction were applied using a patient-specific database of SNVs. SYTL4S363F and NCAPG2P333L neoantigens have been detected by MS as previously reported,7 whereas KIF2CP13L has been primarily identified through in silico prediction of high-affinity HLA binders. However, this peptide could be meanwhile detected by MS using novel machine learning algorithms demonstrating the power of this approach.18 Experimental affinity measurements of selected predicted peptides showed similar IC50 values for all three neoantigens, although no real correlation to predicted affinity was observed for the whole cohort of analyzed peptides. Furthermore, experimental and in silico determined affinity are not accurate immunogenicity predictors in our case. MDs confirmed previous reports pointing to the fact that antigen recognition may depend not only on a defined mutation but also on conformational changes affecting the adjacent AAs.19 Moreover, our data confirm that structural parameters as SASA can be used to improve prediction of suitable neoantigens and may be a valuable addition to comprehensive neoantigen selection algorithms.20
Interestingly, the somatic mutation coding for KIF2CP13L peptide is in close proximity to another one previously reported. The latter results in an immunogenic decamer which overlaps our neoantigen by eight AAs.21 This finding may further support the notion of existing hotspots for peptide presentation and emphasizes that genes especially activated in cancer, such KIF2C, may preferentially result in peptide ligands.22
Although several groups have published data describing neoantigen-specific TCRs,23–26 the data shown here provide a much deeper insight. We observed oligoclonal endogenous T-cell responses against two antigens within one patient. We showed recognition of endogenously processed neoantigens as well as tumor reactivity in vitro and in vivo by all identified TCRs. Of particular interest, TCRs with identical specificity showed similar characteristics with regard to temporal and spatial distribution in the body, functional avidity, functionality, and docking pattern as investigated by alanine/threonine scans, suggesting a major role of the antigen or MHC-peptide docking by the TCR driving defined immune responses. Functional avidities of all seven TCRs were in a narrow range, yet TCRs with specificity for KIF2CP13L showed significant lower avidities compared with the other receptors and a much higher frequency in both metastatic lesions, adjacent LNs as well as the peripheral blood over time. Moreover, despite inferior cytokine secretion in relation to SYTL4S363F-specific TCRs in vitro, both KIF2CP13L-specific TCRs demonstrated especially effective tumor rejection potential, with KIF2C-PBC1 showing even a markedly lower transduction rate. We suggest that highly activated TCRs may be more prone to T-cell dysfunction27 28 as previously demonstrated for CAR-T cells.29 This hypothesis is further corroborated by high PD-L1 expression on patients’ tumor cells within MLung7 potentially inducing a PD-L1-mediated dysfunctional state. In vitro data of TCR-transgenic T cells suggest a different activation signature exemplarily tested for SYTL4-TIL1 in comparison to KIF2C-PBC2, with SYTL4-TIL1 displaying a higher PD-1 and an elevated T-bet expression on neoantigen-specific stimulation. Differences in T-bet upregulation may be associated to skewing CD8+ T cells towards a short-lived effector phenotype and may contribute to a dysfunctional state.30 31 Interestingly, levels of IFN-γ secretion, T-bet and PD-1 expression substantially decreased on restimulation of highly activated SYTL4-TIL1 in contrast to KIF2C-PBC2. These data may indicate a disadvantage of SYTL4-specific T cells with respect to sustained function. Our data therefore suggest that future engineering of TCR and respective transgenic T cells may be considered as a very personalized task for the individual TCR. This may affect defined comprehensive T-cell engineering measures as PD-1 knock-down as well as affinity maturation of tumor-specific TCRs.32–34 Yet, further investigations implementing multiparametric transcriptional profiling are needed for a deeper understanding and dissection. Advantages of TCRs with lower avidity with respect to persistence have been recently reported for virus-specific T cells,35 and the observations might coincide with maintenance of such TCRs throughout presentation of similar peptides, potentially leading to cross-reactive stimulation.36 Interestingly, occurrence of side effects correlates with favorable clinical response in patients treated with immune checkpoint inhibitors, suggesting that cross-reactivity may in fact be to some extent beneficial.37 Despite this, no major side effects associated with ipilimumab were observed in the clinical history of the patient and no cross-reactivity was observed in vitro for investigated peptides. In the case of NCAPG2P333L, the coding mutation was lost during progression of disease, coinciding with low frequencies of this TCR and loss of detection of this clonotype in MLung. However, despite low frequencies, both SYTL4S363F-specific and NCAPG2P333L-specific TCRs could be still isolated from peripheral blood or TILs, indicating the presence of non-terminally exhausted memory T cells with potential for expansion. Moreover, non-malignant LNs may also represent an attractive source for isolation of such neoantigen-specific TCRs.
In retrospect, we have identified seven endogenous neoantigen-specific TCRs with high potential for transgenic modification of T cells, to be used for adoptive T-cell transfer. Our data provide evidence that TCRs display different qualities in vitro and in vivo, which seem to be TCR class dependent and need to be taken into account in the context of adoptive T-cell transfer of neoantigen-specific TCR-transgenic T cells.
Acknowledgments
The authors thank the patient for participating in the study and his continuous support. We also thank Anja Stelzl for excellent technical support as well as Jürgen Schlegel and Sandra Baur (TU München, Department of Neuropathology, Munich, Germany) for technical advice.
Footnotes
Contributors: EB, GL, and AK designed the study. EB, GL, ETA, DG, FF, HL, SA, ME, YC, KY, and CA conducted and analyzed experiments. NdAK, LW, SL, and TE performed bioinformatics. EB, GL, FF, MB, and KS validated data. FB, WW, DHB, RR, CF, IA, and AK acquired funding and provided resources. WW, DHB, RR, CF, IA, and AK supervised the project or parts of it. EB, GL, NdAK, LW, and AK wrote the manuscript. All authors reviewed and edited the manuscript.
Funding: This work was supported by grants to AMK from EIT Health (number 19638), Deutsche Krebshilfe (110281) and Wilhelm Sander-Stiftung (2015.030.1), as well as to AMK and IA from Deutsche Forschungsgemeinschaft (SFB824/C10, TRR338, SFB1035/A10 and CIPSM) and the Technical University of Munich and International Graduate School of Science and Engineering.
Competing interests: No, there are no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Informed consent of all healthy donors and the patient was obtained following requirements of the institutional review board (Ethics Commission, Faculty of Medicine, TUM) and in accordance with the principles of the Declaration of Helsinki
References
- 1.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450–61. 10.1016/j.ccell.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samstein RM, Lee C-H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547:217–21. 10.1038/nature22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547:222–6. 10.1038/nature23003 [DOI] [PubMed] [Google Scholar]
- 5.Tran E, Robbins PF, Lu Y-C, et al. T-Cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016;375:2255–62. 10.1056/NEJMoa1609279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalaora S, Wolf Y, Feferman T, et al. Combined analysis of antigen presentation and T-cell recognition reveals restricted immune responses in melanoma. Cancer Discov 2018;8:1366–75. 10.1158/2159-8290.CD-17-1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassani-Sternberg M, Bräunlein E, Klar R, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016;7:13404. 10.1038/ncomms13404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Berger C, Wong CW, et al. Engraftment of human central memory-derived effector CD8+ T cells in immunodeficient mice. Blood 2011;117:1888–98. 10.1182/blood-2010-10-310599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon RM, Friis L, Siebold C, et al. Structure of HLA-A*0301 in complex with a peptide of proteolipid protein: insights into the role of HLA-A alleles in susceptibility to multiple sclerosis. Acta Crystallogr D Biol Crystallogr 2011;67:447–54. 10.1107/S0907444911007888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hülsmeyer M, Welfle K, Pöhlmann T, et al. Thermodynamic and structural equivalence of two HLA-B27 subtypes complexed with a self-peptide. J Mol Biol 2005;346:1367–79. 10.1016/j.jmb.2004.12.047 [DOI] [PubMed] [Google Scholar]
- 11.Hartmann C, Antes I, Lengauer T. IRECS: a new algorithm for the selection of most probable ensembles of side-chain conformations in protein models. Protein Science 2007;16:1294–307. 10.1110/ps.062658307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996;14:33–8. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 13.Varshney A, Brooks JF, William J. Linearly scalable computation of smooth molecular surfaces. IEEE Comp Graph Appl 1997;14https://www.cs.umd.edu/~varshney/papers/av_cga.pdf [Google Scholar]
- 14.Klar R, Schober S, Rami M, et al. Therapeutic targeting of naturally presented myeloperoxidase-derived HLA peptide ligands on myeloid leukemia cells by TCR-transgenic T cells. Leukemia 2014;28:2355–66. 10.1038/leu.2014.131 [DOI] [PubMed] [Google Scholar]
- 15.Audehm S, Glaser M, Pecoraro M, et al. Key features relevant to select antigens and TCR from the MHC-mismatched repertoire to treat cancer. Front Immunol 2019;10:1485. 10.3389/fimmu.2019.01485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mall S, Yusufi N, Wagner R, et al. Immuno-PET imaging of engineered human T cells in tumors. Cancer Res 2016;76:4113–23. 10.1158/0008-5472.CAN-15-2784 [DOI] [PubMed] [Google Scholar]
- 17.de Castro E, Sigrist CJA, Gattiker A, et al. ScanProsite: detection of PROSITE signature matches and prorule-associated functional and structural residues in proteins. Nucleic Acids Res 2006;34:W362–5. 10.1093/nar/gkl124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilhelm M, Zolg DP, Graber M, et al. Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics. Nat Commun 2021;12:3346. 10.1038/s41467-021-23713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebrahimi-Nik H, Michaux J, Corwin WL, et al. Mass spectrometry–driven exploration reveals nuances of neoepitope-driven tumor rejection. JCI Insight 2019;4. 10.1172/jci.insight.129152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaidi N, Soban M, Chen F, et al. Role of in silico structural modeling in predicting immunogenic neoepitopes for cancer vaccine development. JCI Insight 2020;5. 10.1172/jci.insight.136991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y-C, Yao X, Crystal JS, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res 2014;20:3401–10. 10.1158/1078-0432.CCR-14-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caron E, Vincent K, Fortier Marie‐Hélène, Fortier MH, et al. The MHC I immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol Syst Biol 2011;7:533. 10.1038/msb.2011.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran E, Ahmadzadeh M, Lu Y-C, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350:1387–90. 10.1126/science.aad1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chheda ZS, Kohanbash G, Okada K, et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J Exp Med 2018;215:141–57. 10.1084/jem.20171046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuda T, Leisegang M, Park J-H, et al. Induction of neoantigen-specific cytotoxic T cells and construction of T-cell receptor–engineered T cells for ovarian cancer. Clin Cancer Res 2018;24:5357–67. 10.1158/1078-0432.CCR-18-0142 [DOI] [PubMed] [Google Scholar]
- 26.Ren L, Leisegang M, Deng B, et al. Identification of neoantigen-specific T cells and their targets: implications for immunotherapy of head and neck squamous cell carcinoma. Oncoimmunology 2019;8:e1568813. 10.1080/2162402X.2019.1568813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander-Miller MA, Leggatt GR, Sarin A, et al. Role of antigen, CD8, and cytotoxic T lymphocyte (CTL) avidity in high dose antigen induction of apoptosis of effector CTL. J Exp Med 1996;184:485–92. 10.1084/jem.184.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santa SD, Merlo A, Bobisse S, et al. Functional avidity–driven activation-induced cell death shapes CTL immunodominance. J Immunol 2014;193:4704–11. 10.4049/jimmunol.1303203 [DOI] [PubMed] [Google Scholar]
- 29.Chmielewski M, Hombach A, Heuser C, et al. T cell activation by antibody-like immunoreceptors: increase in affinity of the single-chain fragment domain above threshold does not increase T cell activation against antigen-positive target cells but decreases selectivity. J Immunol 2004;173:7647–53. 10.4049/jimmunol.173.12.7647 [DOI] [PubMed] [Google Scholar]
- 30.Intlekofer AM, Takemoto N, Kao C, et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med 2007;204:2015–21. 10.1084/jem.20070841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard GH, Kedl RM, Hunter CA. The evolving role of T-bet in resistance to infection. Nat Rev Immunol 2019;19:398–410. 10.1038/s41577-019-0145-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong S, Malecek K, Johnson LA, et al. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc Natl Acad Sci U S A 2013;110:6973–8. 10.1073/pnas.1221609110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inderberg EM, Wälchli S. Long-term surviving cancer patients as a source of therapeutic TCR. Cancer Immunol Immunother 2020;69:859–65. 10.1007/s00262-019-02468-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stadtmauer EA, Fraietta JA, Davis MM, et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020;367:eaba7365. 10.1126/science.aba7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schober K, Voit F, Grassmann S, et al. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat Immunol 2020;21:434–41. 10.1038/s41590-020-0628-2 [DOI] [PubMed] [Google Scholar]
- 36.Łuksza M, Riaz N, Makarov V, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 2017;551:517–20. 10.1038/nature24473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eggermont AMM, Kicinski M, Blank CU, et al. Association between immune-related adverse events and recurrence-free survival among patients with stage III melanoma randomized to receive pembrolizumab or placebo. JAMA Oncol 2020;6:519–27. 10.1001/jamaoncol.2019.5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-002754supp002.pdf (2.2MB, pdf)
jitc-2021-002754supp001.pdf (785KB, pdf)
Data Availability Statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.