Abstract
We have identified novel interactions between the human (h)TATA-binding protein-associated factor TAFII55 and the ligand-binding domains (LBDs) of the nuclear receptors for vitamin D3 (VDR) and thyroid hormone (TRα). Following expression in Cos cells, hTAFII55 interacts with the VDR and TRα LBDs in a ligand-independent manner whereas no interactions with the retinoid X receptors (RXRs) or with other receptors were observed. Deletion mapping indicates that hTAFII55 interacts with a 40-amino-acid region spanning α-helices H3 to H5 of the VDR and TRα LBDs but not with the equivalent highly related region of RXRγ. TAFII55 also interacts with chimeric receptors in which the H3-to-H5 region of RXRγ has been replaced with that of the VDR or TRα. Furthermore, replacement of two single amino acids of the RXRγ LBD with their VDR counterparts allows the RXRγ LBD to interact with hTAFII55 while the corresponding double substitution allows a much stronger interaction. In transfection experiments, the single mutated RXRγ LBDs activate transcription to fivefold higher levels than wild-type RXRγ while the double mutation activates transcription to a level comparable to that observed with the VDR. There is therefore a correlation between the ability of the modified RXRs to interact with hTAFII55 and transactivation. These results strongly suggest that the TAFII55 interactions with the modified RXR LBDs modulate transcriptional activation.
Transcription factor TFIID is one of the general factors required for accurate and regulated initiation by RNA polymerase II. TFIID comprises the TATA-binding protein (TBP) and TBP-associated factors (TAFIIs) (5, 9, 10, 13, 15, 17, 20, 43, 55). The cDNAs encoding many human (h)TAFIIs have been isolated, revealing a striking sequence conservation with yeast and Drosophila TAFIIs (14, 21, 22, 28–30).
The TAFII proteins are of particular interest, since they play several roles in transcriptional regulation, some of them being present not only in TFIID but also in the SAGA, PCAF, and TFTC complexes (18, 25, 35, 50). TAFIIs contribute to promoter recognition both directly by interaction of specific TAFIIs with promoter sequences (46, 47) and more generally through multiple TAFII-DNA interactions which possibly arise from the wrapping of DNA around a nucleosome-like structure formed by TAFIIs with histone fold motifs (6, 34, 35).
An increasing body of results also shows that hTAFII28, hTAFII135, and hTAFII105 can act as specific transcriptional coactivators in mammalian cells. For example, distinct domains of hTAFII135 interact specifically with Sp1, cyclic AMP response element-binding protein, and E1A and coexpression of the fragments of TAFII135 with which these activators interact has a dominant negative effect on their activity (27, 32, 41, 44). Similar experiments have shown that hTAFII105 interacts specifically with the p65 subunit of NF-κB and that TAFII105 expression strongly potentiates activation by NF-κB in mammalian cells (53). Coexpression of hTAFII28 and/or TBP also strongly potentiates activation by the viral Tax protein, and Tax interacts directly with hTAFII28 and TBP to form a ternary complex (11).
There is also evidence that TAFIIs are involved in nuclear receptor (NR) function. The activity of NR activation function 2 (AF-2) requires a ligand-induced conformational change in the ligand-binding domain (LBD) which brings the AF-2 activating domain (AD) core in α-helix H12 into the proximity of α-helix H4 of the LBD (8, 40, 48), forming a novel interaction surface and allowing the NRs to interact with putative transcriptional intermediary factors (TIFs) (4, 12, 33, 36, 39, 45, 54). Although interaction with TIFs is required for NR AF-2 function, additional direct or indirect interactions with the basal transcription apparatus may also contribute to activity. In support of this, we have shown that expression of hTAFII135 specifically potentiates activation by AF-2 of the all-trans-retinoic acid (RA) receptor (RAR), the thyroid hormone receptor (TR), and the vitamin D3 receptor (VDR) (28) while expression of hTAFII28 potentiates activation by many NRs, the most dramatic effects being seen with the receptors for the 9-cis-RA receptor (RXR), the estrogen receptor (ER), and the VDR (26).
In this report, we provide evidence that hTAFII55 is involved in the activity of some NRs. We show that hTAFII55 selectively interacts with the LBDs of the human VDR and chicken TRα following coexpression in Cos cells. Analysis with VDR deletion mutants shows that hTAFII55 interacts with a 40-amino-acid region spanning α-helices H3 to H5 and containing the NR signature. hTAFII55 interacts with the isolated H3-to-H5 region of the VDR and TRα but not with the analogous highly related region of RXRγ, thus mimicking the selective interactions observed with the corresponding LBDs. Replacement of one or two amino acids of the RXRγ H3-to-H5 region with their counterparts from the VDR resulted in interactions with hTAFII55. In transfected cells, the mutant RXRγ LBDs which interact weakly with TAFII55 activate transcription to fivefold higher levels than wild-type RXRγ while the double mutant which interacts strongly with TAFII55 activates transcription as strongly as the VDR. These results provide evidence that interaction with TAFII55 modulates the transactivation properties of the modified RXR LBDs.
MATERIALS AND METHODS
Construction of recombinant plasmids.
The hTAFII55 and NR expression vectors used were previously described (22, 26, 28, 29, 31). All of the G4-VDR, TRα, and RXR chimeras were constructed by PCR using the appropriately designed oligonucleotides with restriction sites and cloned into the vector pXJ440, encoding the DNA-binding domain (DBD) of the yeast activator GAL4 (amino acids 1 to 147; G4) (52). All plasmids were verified by automated DNA sequencing. Further details of constructions are available on request.
Transfection of Cos cells and immunoprecipitations.
Cos cells were transfected by the calcium phosphate coprecipitation technique, and immunoprecipitations were performed as previously described (22, 29). At 48 h following transfection, the cells were harvested by three freeze-thaw cycles in buffer A (50 mM Tris-HCl [pH 7.9], 20% glycerol, 1 mM dithiothreitol, 0.1% Nonidet P-40) containing 0.5 M KCl. The expression of the transfected proteins was verified on Western blots by using 10-μl cell extract samples. For immunoprecipitations, 50 μl of the cell extracts was incubated for 1 h at 4°C with 1 to 2 μg of the indicated monoclonal antibodies (MAbs), after which time 50 μl of protein G-Sepharose was added and incubation was continued for another 2 h. The protein G-Sepharose was then washed four times for 10 min each time at room temperature with buffer A containing 1.0 M KCl and once with buffer A containing 0.1 M KCl. The resin was resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiled for 5 min, and one-half of the sample was subjected to SDS-PAGE. The bound proteins were detected on Western blots with the indicated antibodies by using an ECL kit (Amersham). Where indicated, ligands were added [50 nM all-trans-RA, 9-cis-RA, and 3,5,3′-triiodo-l-thyronine, and 100 nM 1,25(OH)2D3] at the same time as the DNA-calcium phosphate coprecipitate. For chloramphenicol acetyltransferase (CAT) assays, 3 μg of the 17m5-TATA-CAT reporter plasmid was cotransfected with 2 μg of a α-galactosidase reporter as an internal control, along with the indicated concentrations of the G4-RXRγ expression vectors. After correction for transfection efficiency using β-galactosidase assays, CAT assays were performed by standard protocols and the percentage of acetylated chloramphenicol was determined by quantitative PhosporImager analysis on a Fujix BAS 2000 apparatus. In all cases, similar results (±20%) were obtained in at least three independent transfections and the results of typical experiments are shown.
Antibody preparation.
MAbs against hTAFII55 (19TA), the B10 epitope, and the G4 DBD (3GV2) were previously described (1, 22, 26, 29, 49).
RESULTS
hTAFII55 interacts selectively with the LBDs of VDR and TRα.
To look for interactions between hTAFII55 and transcriptional activators, vectors expressing chimeras comprising the NR LBDs or the full-length VDR fused to the DBD of the yeast activator G4 were cotransfected into Cos cells along with vectors expressing B10-tagged hTAFII55 (Fig. 1). Transfected-cell extracts were then prepared, and protein expression was verified on immunoblots by using MAbs directed against the ER B10 tag (1), hTAFII55 (19TA; 22), or the G4 DBD (3GV2; 49). Transfected-cell extracts were then immunoprecipitated with these MAbs, and the precipitated proteins were detected on immunoblots. Analysis of interactions with hTAFII55 is complicated by the fact that it comigrates with heavy chains of the MAbs used in the immunoprecipitations. For clarity, some of the Western blots which are presented were revealed only with MAbs against hTAFII55 or the coprecipitated G4-NR chimera and secondary antibodies against either the light or heavy chains, as indicated in the figure legends.
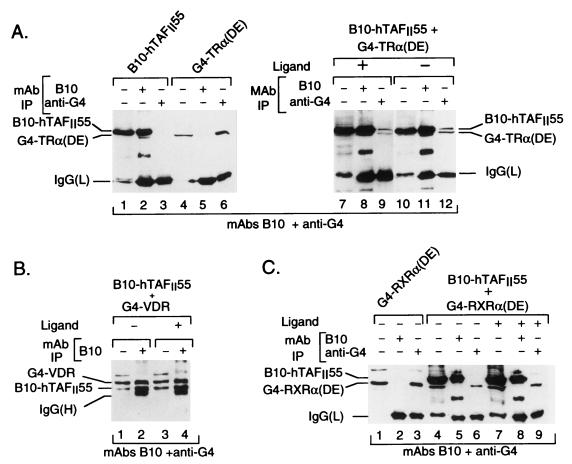
FIG. 1.
Structures of hTAFIIs and nuclear expression vectors. The pAT6 vectors contain the epitope for MAb B10 at the N terminus. The amino acid coordinates of the N- and C-terminal boundaries in each construct are shown. All of the NR expression vectors are cloned in the pXJ440 vector, where the NR sequences are fused to the G4 DBD. h, human; m, mouse; c, chicken.
When expressed alone, B10-hTAFII55 was precipitated by MAb B10 but not by the anti-G4 antibody while G4-TRα(DE) was precipitated only with the anti-G4 antibody (Fig. 2A, lanes 1 to 6). However, when coexpressed, both B10-hTAFII55 and G4-TRα(DE) were coprecipitated by each antibody, irrespective of the presence or absence of the ligand [Fig. 2A, lanes 7 to 12; while B10-hTAFII55 and the G4-TRα(DE) chimera have similar electrophoretic mobilities and are difficult to distinguish in lanes 8 and 11, both proteins can be clearly seen in lanes 9 and 12, compared with lanes 4 and 6]. Similarly, when coexpressed with B10-hTAFII55, G4-VDR could be immunoprecipitated by MAb B10 both in the presence and in the absence of the ligand (Fig. 2B, lanes 1 to 4). In contrast, no coprecipitation of hTAFII55 with the G4 chimeras of the RXR [G4-RXRα(DE); Fig. 2C, lanes 4 to 9] or RAR (data not shown) LBDs was observed. Thus, under the same stringent conditions used to detect TAF-TAF interactions (i.e., washing with a buffer containing 1.0 M KCl), hTAFII55 formed a stable, salt-resistant, immunoprecipitable complex selectively with the VDR and TR LBDs in a ligand-independent manner.
FIG. 2.
Selective interactions between hTAFII55 and the TRα and VDR LBDs. (A) The transfected expression vectors are shown at the top along with the antibodies used in the immunoprecipitations (IP) and the presence or absence of cognate ligands. Lanes 1, 4, 7, and 10 show aliquots (10 μl) of the transfected-cell extracts used for the immunoprecipitations. Due to their similar electrophoretic mobilities, coprecipitated G4-TRα(DE) is masked by the excess of B10-hTAFII55 in lanes 8 and 11 but coprecipitated B10-hTAFII55 is clearly visible in lanes 9 and 12. The antibodies used to reveal each blot are indicated at the bottom of the panel. In this panel, a peroxidase-conjugated secondary antibody was directed against the light chain. The locations of the immunoprecipitated proteins, as well as those of the immunoglobulin G MAb light chains [IgG(L)] used in the immunoprecipitations, are indicated at the sides. Note that in lanes 1, 7, and 10, when B10-hTAFII55 is transfected, there is a proteolytic breakdown product with mobility similar to that of IgG(L). In subsequent figures, either the light chain or the heavy chain [IgG(H)] of the immunoprecipitating antibody is indicated, depending on which peroxidase-conjugated secondary antibody was used. (B and C) The layout is as in panel A. IgG(H) indicates the position of the heavy chain of the MAb used in the immunoprecipitation.
A series of G4-VDR deletion mutants was then used to determine which regions of the VDR were required for interaction with hTAFII55 (see Fig. 4A). No interaction was seen with the chimera G4-VDR DE(90-195) containing the D domain and the region containing α-helices H1 and H2 of the E domain, whereas G4-VDR E(196-427), containing the remainder of the E domain, was coimmunoprecipitated with B10-hTAFII55 (data not shown). To further delineate the amino acids of the E region required for interaction with hTAFII55, a further series of G4-VDR E chimeras (see Fig. 4A) were expressed either alone or together with B10-hTAFII55 (for example, Fig. 3A). As several of these chimeric fusion proteins comigrated on SDS-PAGE with the light chains of the MAbs used in the immunoprecipitations, the transfected-cell extracts were precipitated with the anti-G4 MAb and the presence of the coprecipitated B10-hTAFII55 was revealed by using MAb B10 or vice versa.
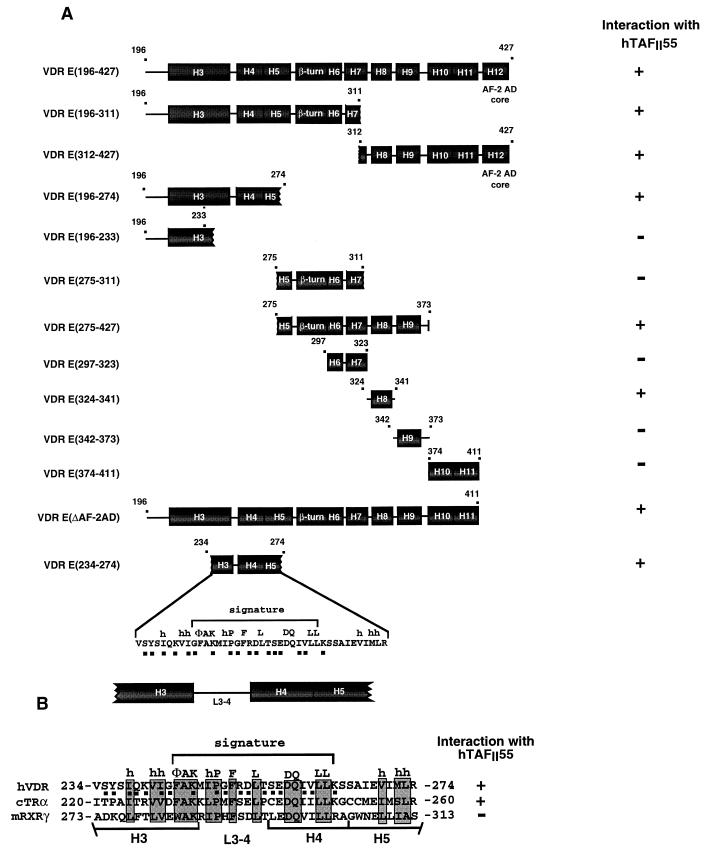
FIG. 4.
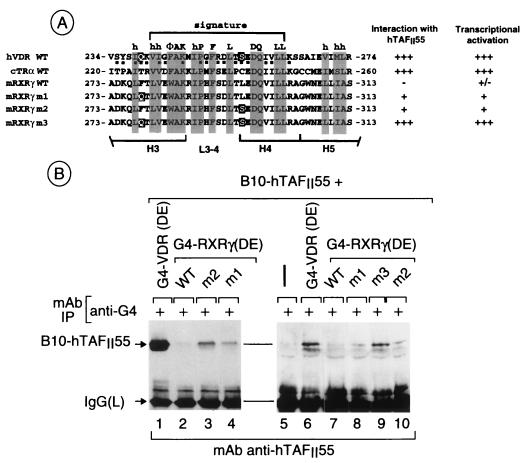
(A and B) Schematic representation of deletion mutant VDR LBDs. The positions of the predicted α-helices (H3 to H12) are indicated. The numbers above each representation show the N- and C-terminal boundaries of the constructs. The interaction (+ or −) of each region with hTAFII55 is summarized on the right. The amino acid sequence of the VDR H3-to-H5 region is shown at the bottom. Conserved amino acids which form the signature are indicated above the sequence. The filled squares below the amino acids indicate amino acids which may be exposed on the surface of the VDR LBD. (B) Alignment of the signature regions of the VDR, chicken (c) TRα, and mouse (m) RXRγ. The conserved signature amino acids are boxed, and the positions of predicted α-helices H3, H4, and H5 and the loop (L3-4) region are indicated.
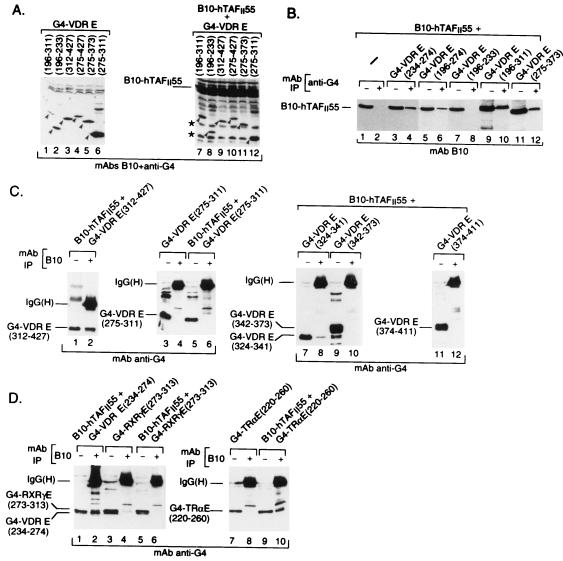
FIG. 3.
Interaction of hTAFII55 with deletion mutant forms of the VDR. Panel A shows the immunoblot of a representative set of transfected-cell extracts where G4-VDR chimeras have been expressed alone or in the presence of B10-hTAFII55. Immunoprecipitations (IP) of the transfected-cell extracts are shown in panels B and C. Interactions with the H3-to-H5 region of the VDR, TRα, and RXRγ are shown in panel D. The layout is as described in the legend to Fig. 2A, with the transfected expression vectors and MAbs used for immunoprecipitation shown at the top and the antibodies used to reveal the blot shown at the bottom. The positions of the expressed proteins and the heavy or light chains revealed by the secondary antibodies are indicated.
B10-hTAFII55 was not precipitated by the anti-G4 antibodies when expressed alone (Fig. 3B, lanes 1 and 2), whereas it was precipitated when coexpressed with the N-terminal [G4-VDR E(196-311)] moiety of the VDR E region (Fig. 3B, lanes 9 and 10). Deletions within the N-terminal half of the E region showed that B10-hTAFII55 was coimmunoprecipitated with G4-VDR E(196-274) and G4-VDR E(234-274) (lanes 3 to 6) but not with G4-VDR E(196-233) (lanes 7 and 8). In the converse immunoprecipitation, G4-VDR E(234-274) was coprecipitated by MAb B10 (Fig. 3D, lanes 1 and 2) whereas G4-VDR E(275-311) was not precipitated with B10-hTAFII55 (Fig. 3C, lanes 3 to 6). These results show that amino acids 234 to 274 of the VDR stably interact with hTAFII55 when transferred to the G4 DBD (summarized in Fig. 4A).
Although hTAFII55 interacts with amino acids 234 to 274 in the N-terminal half of the E domain, a second region of interaction in the C-terminal moiety of the VDR E domain was observed since G4-VDR E(312-427) was also coprecipitated with B10-hTAFII55 (Fig. 3C, lanes 1 and 2). Although, as indicated above, no coprecipitation of hTAFII55 and G4-VDR E(275-311) was observed, B10-hTAFII55 was coprecipitated with G4-VDR E(275-373) (Fig. 3B, lanes 11 to 12), indicating that amino acids between 311 and 373, which encompass α-helices H8 and H9, allow interaction with hTAFII55 (summarized in Fig. 4A). This second hTAFII55-interacting region was more precisely mapped. G4-VDR E(324-341), containing α-helix H8, was coprecipitated with B10-hTAFII55 (Fig. 3C, lanes 7 and 8), while no coprecipitation of G4-VDR E(342-373) or G4-VDR E(374-411), containing α-helices H9 and H10 and -11, respectively, was observed (lanes 9 to 12). In agreement with this result, interaction between VDR(DE) and hTAFII55 was not affected by deletion of the AF-2 AD core located in α-helix H12 between amino acids 411 and 427 (data not shown; Fig. 4A). Therefore, interaction between the C-terminal moiety of the VDR E region and hTAFII55 requires α-helix H8 but not α-helices H9 to H11 nor the AF-2 AD core, which is required for ligand-dependent interactions of NRs with various TIFs (summarized in Fig. 4A).
The above-described results indicate that hTAFII55 selectively interacts with two independent regions of the VDR E domain: a region spanning α-helices H3 to H5 containing the NR signature and α-helix H8.
Selective interaction of hTAFII55 with the H3-to-H5 NR signature-containing regions of the VDR and TRα.
The above-described results show that hTAFII55 interacts with the VDR H3-to-H5 region containing the NR signature. This region contains many well-conserved amino acids (boxed in Fig. 4B) involved in intramolecular interactions required to stabilize the canonical NR fold (51). The high conservation in this region of the NR LBDs (51; Fig. 4B) led us to compare the binding of hTAFII55 to the equivalent regions of chicken TRα and RXRγ. G4 chimeras containing these H3-to-H5 regions [G4-TR E(220-260) and G4-RXRγ E(273-313)] were coexpressed along with B10-hTAFII55. G4-TRα E(220-260) was specifically precipitated along with B10-TAFII55 (Fig. 3D, lanes 7 to 10). In contrast, no coprecipitation of G4-RXRγ E(273-313) was observed (Fig. 3D, lanes 3 to 6). The selective binding of hTAFII55 to the H3 to H5 region of the VDR and TRα, but not the RXRγ, LBDs therefore mimics the specificity seen when the complete LBDs of these NRs were used.
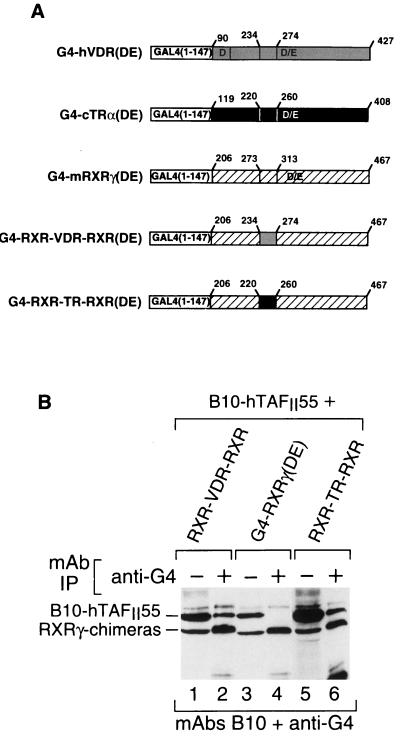
To further demonstrate that the VDR and TRα H3-to-H5 regions can promote interactions with hTAFII55, we created chimeric RXRγ(DE)s where amino acids 273 to 313 of RXRγ containing the H3-to-H5 region have been replaced with the equivalent amino acids of the VDR or TRα E domain (G4-RXR-VDR-RXR and G4-RXR-TR-RXR; Fig. 5A). These chimeras were coexpressed along with hTAFII55. As described above for G4-RXRα, no coprecipitation of hTAFII55 with G4-RXRγ(DE) was observed (Fig. 5B, lanes 3 and 4). In contrast, the RXR-VDR-RXR and RXR-TR-RXR chimeras were coprecipitated with hTAFII55 (Fig. 5B, lanes 1, 2, 5, and 6). Thus, the VDR or TR H3-to-H5 regions can mediate interactions with hTAFII55 when transferred to the G4-DBD either alone or in the context of the RXRγ DE region. In the converse experiments, deletion of the VDR and TRα H3-to-H5 regions or their replacement with that of the RXR did not abrogate TAFII interaction with these LBDs, in agreement with the observation that α-helix H8 can also interact with hTAFII55 (data not shown).
FIG. 5.
RXRγ chimeras containing the H3-to-H5 region of the VDR or TRα interact with TAFII55. (A) Schematic representation of the structures of G4-RXR chimeras. (B) Western blot of immunoprecipitated (IP) G4-RXR chimeras. The layout is as described in the legend to Fig. 2.
Single amino acid changes in the RXRγ LBD induce interactions with hTAFII55.
The selective interaction of hTAFII55 with the H3-to-H5 regions of the VDR and TRα but not RXRγ is surprising considering the high degree of sequence homology between these receptors in this region (Fig. 4B). We therefore reasoned that exchanging solvent-exposed amino acids of RXRγ with those of the VDR might recreate the TAFII55 interaction surface. Two positions on the exposed surface were chosen where the RXR amino acids are radically different from those in the VDR, i.e., F278 in H3 and L295 in H4 (Fig. 6A). At these positions, there are polar amino acids in the VDR while there are bulky hydrophobic residues in the RXR.
FIG. 6.
Interaction of hTAFII55 with mutant RXRγ LBDs. (A) Sequences of the H3-to-H5 regions of G4-NR(DE) chimeras. The amino acids mutated in G4-RXR(DE) m1, m2, and m3 are highlighted. h, human; c, chicken; m, mouse. (B) Western blot of immunoprecipitated (IP) G4-VDR and G4-RXR chimeras. The layout is as described in the legend to Fig. 2.
Several single or multiple mutations were made in G4-RXRγ(DE) (m1, F278Q; m2, L295S; m3, F278Q/L295S; Fig. 6A) and coexpressed with hTAFII55, and interactions were verified by immunoprecipitation with the anti-G4 antibody. No significant coimmunoprecipitation of B10-hTAFII55 was observed when it was expressed alone or with wild-type (WT) G4-RXR(DE) (Fig. 6B, lanes 2, 5, and 7). B10-hTAFII55 was coprecipitated with both G4-RXR(DE) m1 and m2 (lanes 3, 4, 8, and 10); however, this interaction was weaker than that observed with the VDR (lanes 1 and 6). In contrast, a strong interaction, comparable to that seen with the VDR, was observed with the double mutant m3 (compare lanes 6 and 9). Therefore, exchange of amino acids between the RXRγ and VDR LBDs can recreate a surface, allowing interactions with TAFII55. Single amino acid changes allow a weak interaction, while the double substitution allows a much stronger interaction.
Transactivation is modulated by mutations in the RXRγ H3-to-H5 region which allow interaction with hTAFII55.
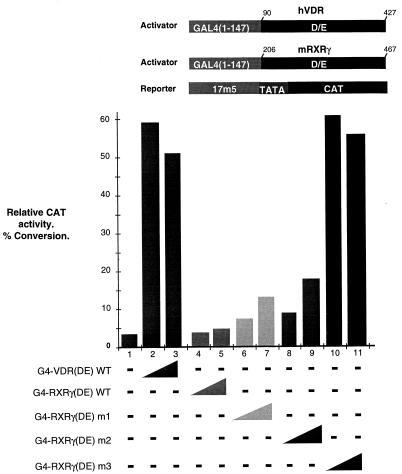
To test whether these novel interactions could affect the transcriptional activation properties of the mutated RXR LBDs, increasing quantities of vectors expressing the WT or mutated G4-RXRγ(DE) chimeras were cotransfected along with a G4-responsive CAT reporter (see Materials and Methods and references 26 and 28). As previously described (26), the WT RXRγ LBD activated transcription only weakly from this promoter (Fig. 7, lanes 4 and 5) while strong activation by the VDR LBD was seen (lanes 2 and 3). In contrast to the WT RXR, mutants m1 and m2 activated transcription up to five times more than the WT (lanes 6 to 9) whereas mutant m3 activated transcription to a level comparable to that seen with the VDR (lanes 2 and 3 and lanes 10 and 11). Immunoblot experiments showed equivalent expression levels of each of these activators (data not shown). These results reveal a correlation between the interaction of the mutated RXR LBDs with TAFII55 and their ability to activate transcription (summarized in Fig. 6A), providing evidence that this interaction is involved in transcriptional activation.
FIG. 7.
Graphic representation of CAT assay results. The structures of the activator and reporter constructs used are shown schematically above the graph. The transfected expression vectors used are shown below the graph. Transfections contained 0 (−), 0.25 (narrow end of wedge), or 0.5 (broad end of wedge) μg of the expression vectors. All transfections were performed in the presence of ligand. h, human; m, mouse.
DISCUSSION
Novel interaction surfaces in the VDR LBD.
We describe here novel interactions between the LBDs of the VDR and TRα with a component of transcription factor TFIID. Following coexpression in Cos cells, hTAFII55 could be specifically coprecipitated with the LBDs of the VDR and TRα, irrespective of the presence of the ligand. The selectivity of the interactions is shown by the observation that under the same conditions, no interactions between hTAFII55 and other NRs were observed.
hTAFII55 interacts with two separable, independent sites in the VDR LBD. The first of these is located between amino acids 234 and 274, the region spanning α-helices H3 to H5 and containing the NR signature. Fusion of this region to the G4 DBD is sufficient to mediate interactions with hTAFII55, showing that although the NR LBD is highly structured, this domain can interact with hTAFII55 even when presented in a different context. This is therefore an autonomous domain which can mediate interactions with hTAFII55. hTAFII55 interacted with the H3-to-H5 regions of the VDR and TRα, but not RXRγ, hence mimicking the selectivity seen with the corresponding full-length NR LBDs. Moreover, replacement of the RXRγ H3-to-H5 region with that of TRα or the VDR is sufficient to induce TAFII55 interactions with the RXRγ LBD.
When exposed amino acids Q278 and L295 in H3 or H4 of the RXRγ LBD are replaced with their VDR counterparts, novel interactions with TAFII55 are observed. Thus, despite the high degree of conservation of the H3-to-H5 region among the NRs, due to the presence of the signature, these regions of the VDR and TRα also contain amino acids which dictate selective NR-hTAFII interactions.
Comparison of the amino acids required for hTAFII55 interactions with those required for interaction with the LXXLL motif in several TIFs (19, 23) shows that these two sites are close to each other but not identical. VDR amino acids Q239 and S256 and their RXR equivalents are located on the surface surrounding the hydrophobic cleft created by the juxtaposition of hydrophobic amino acids of the NR signature with amino acids of H12, which is required for TIF interactions (16, 33). Interaction with TAFII55 does not require this hydrophobic cleft, as there is no requirement for the H12 helix. Moreover, mutation of the amino acids in the human TRβ LBD equivalent to Q239 and S256 (T281 and C298) had no effect on interaction with the LXXLL motif of GRIP1. The amino acids which are critical for TAFII55 interaction are not part of the canonical NR signature and are not essential for interaction with TIFs bearing LXXLL motifs. There are therefore two surfaces in this region of the VDR and TR LBDs specifying interactions with distinct cofactors.
Although mutation of amino acids F278 and L295 in RXRγ to their VDR counterparts suffices to allow interaction with TAFII55, the converse mutations in the VDR LBD do not abolish interaction with TAFII55 or transactivation (our unpublished data). It is probable that the RXRγ mutations define a minimal surface required for interaction with hTAFII55 but that in the VDR other amino acids also contribute to the interactions. In addition, it should be remembered that TAFII55 also interacts with amino acids in H8 and that this interaction is not affected by the mutations in the H3-to-H5 region.
The second VDR region interacting with hTAFIIs is located between amino acids 324 and 341, corresponding to α-helix H8. A molecular model of the VDR LBD generated from sequence alignments and comparison with the known structures of the RXR, RAR, and TR LBDs suggests that only amino acids at the N- and C-terminal extremities of H8 would be solvent exposed, the remainder being buried in the structured LBD. The C-terminal end of H8 and the L8-9 loop presents most of the exposed residues and is therefore the most likely hTAFII interaction site.
Mutant RXRγ LBDs which interact with TAFIIs have altered transactivation properties.
The results presented here provide strong evidence that the interaction between TAFII55 and the mutated RXR LBDs may promote transcriptional activation in mammalian cells. Two single amino acid substitutions in the RXRγ LBD which allow interaction with TAFII55 also enhance transcriptional activation. The double mutation which induces a much stronger interaction with the RXR LBD, comparable to that seen with the VDR, activates transcription to levels comparable to that seen with the VDR. In the case of the RXR, there is therefore a correlation between interaction with hTAFII55 and transactivation potential. The presence of multiple TAFII55 interaction sites in the VDR LBD complicates the reciprocal loss-of-function analysis which would require the simultaneous mutation of both regions. However, by analogy with what is observed with the RXR, it is possible that the VDR- or TR-TAFII55 interaction also contributes to the transcriptional activity of these activators.
The situation described here with the RXRγ LBD is somewhat analogous to that described in yeast with the GAL11 and GAL11P proteins, where a novel interaction with the G4 DBD induced by a fortuitous mutation in the holoenzyme component GAL11 (GAL11P) suffices to activate transcription (3, 38). Here, we show that mutations which induce RXR-TAFII55 interactions are also sufficient to convert the RXRγ LBD from a weak activator to a much stronger activator, providing evidence that the novel interactions with hTAFII55 potentiate transcriptional activation in mammalian cells.
Our results favor the two-step model which has been proposed for NR function. Upon ligand binding, the NR LBDs interact with the TIFs with histone acetyltransferase activities and induce chromatin remodelling (45 and references therein). Following this step, our results and those of others suggest that transactivation by NRs may involve additional interactions with TAFIIs and/or other components of the general transcription machinery, for example, TFIIB for the VDR and the TR (2, 7, 24), hTAFII30 for the ER (21), and TBP and/or drosophila TAFII110 for the RXR and the TR (37, 42). In this respect, it is important to note that, unlike hTAFII55, most TIFs interact with each of the NR LBDs with comparable affinities yet the RXR LBD is a considerably weaker activator than the VDR or TR LBD, at least with our test promoter. Therefore, although interaction with TIFs and chromatin remodelling are essential steps, additional interactions such as those described here with hTAFII55 may well contribute to activation by a given LBD.
A similar model has been proposed for the cyclic AMP response element-binding protein (CREB), where phosphorylation-induced interaction with the CREB-binding protein, also one of the NR TIFs, and a constitutive interaction with TAFII135 have been shown to be required for transactivation (32). A requirement for inducible interactions with cofactors allows activators which respond to extracellular stimuli (hormones or mitogens) to integrate these signals with basal transcription machinery interactions required for activation.
ACKNOWLEDGMENTS
We thank P. Chambon for support; L. Carré for excellent technical assistance; L. Perletti for critical comments; S. Vicaire and D. Stephane for DNA sequencing; Y. Lutz and the MAb facility, the staff of the cell culture and oligonucleotide facilities, B. Boulay, J. M. Lafontaine, R. Buchert, and C. Werlé for illustrations; and Roussel-Uclaf for providing 1,25(OH)2D3.
G.M. and A.-C.L. were supported by fellowships from the Ligue Nationale contre le Cancer and the Association pour la Recherche contre le Cancer. This work was supported by grants from the CNRS, the INSERM, the Hôpital Universitaire de Strasbourg, the Ministère de la Recherche et de la Technologie, the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, and the Human Frontier Science Programme.
A.-C.L. and G.M. contributed equally to this work.
REFERENCES
- 1.Ali S, Lutz Y, Bellocq J P, Chenard-Neu M P, Rouyer N, Metzger D. Production and characterization of monoclonal antibodies recognising defined regions of the human oestrogen receptor. Hybridoma. 1993;12:391–405. doi: 10.1089/hyb.1993.12.391. [DOI] [PubMed] [Google Scholar]
- 2.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M J, O’Malley B W. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 4.Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 5.Bell B, Tora L. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp Cell Res. 1999;246:11–19. doi: 10.1006/excr.1998.4294. [DOI] [PubMed] [Google Scholar]
- 6.Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne A C, Davidson I, Moras D. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 7.Blanco J C, Wang I M, Tsai S Y, Tsai M J, O’Malley B W, Jurutka P W, Haussler M R, Ozato K. Transcription factor TFIIB and the vitamin D receptor cooperatively activate ligand-dependent transcription. Proc Natl Acad Sci USA. 1995;92:1535–1539. doi: 10.1073/pnas.92.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 9.Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly J M, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 11.Caron C, Mengus G, Dubrowskaya V, Roisin A, Davidson I, Jalinot P. Human TAFII28 interacts with the human T cell leukemia virus type I Tax transactivator and promotes its transcriptional activity. Proc Natl Acad Sci USA. 1997;94:3662–3667. doi: 10.1073/pnas.94.8.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 13.Chiang C M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubrovskaya V, Lavigne A C, Davidson I, Acker J, Staub A, Tora L. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 15.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 16.Feng W, Ribeiro R C, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 17.Goodrich J A, Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 18.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, Workman J L. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 19.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptor. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 21.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 22.Lavigne A C, Mengus G, May M, Dubrovskaya V, Tora L, Chambon P, Davidson I. Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J Biol Chem. 1996;271:19774–19780. doi: 10.1074/jbc.271.33.19774. [DOI] [PubMed] [Google Scholar]
- 23.LeDouarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald P N, Sherman D R, Dowd D R, Jefcoat S C, Jr, DeLisle R K. The vitamin D receptor interacts with general transcription factor IIB. J Biol Chem. 1995;270:4748–4752. doi: 10.1074/jbc.270.9.4748. [DOI] [PubMed] [Google Scholar]
- 25.Martinez E, Kundu T K, Fu J, Roeder R G. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 26.May M, Mengus G, Lavigne A C, Chambon P, Davidson I. Human TAFII28 promotes transcriptional stimulation by activation function 2 of the retinoid X receptors. EMBO J. 1996;15:3093–3104. [PMC free article] [PubMed] [Google Scholar]
- 27.Mazzarelli J M, Mengus G, Davidson I, Ricciardi R P. The transactivation domain of adenovirus E1A interacts with the C terminus of human TAFII135. J Virol. 1997;71:7978–7983. doi: 10.1128/jvi.71.10.7978-7983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mengus G, May M, Carre L, Chambon P, Davidson I. Human TAFII135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 29.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 33.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 34.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 35.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 36.Perlmann T, Evans R M. Nuclear receptors in Sicily: all in the famiglia. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- 37.Petty K J, Krimkevich Y I, Thomas D. A TATA binding protein-associated factor functions as a coactivator for thyroid hormone receptors. Mol Endocrinol. 1996;10:1632–1645. doi: 10.1210/mend.10.12.8961272. [DOI] [PubMed] [Google Scholar]
- 38.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 39.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 41.Saluja D, Vassallo M F, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulman I G, Chakravarti D, Juguilon H, Romo A, Evans R M. Interactions between the retinoid X receptor and a conserved region of the TATA-binding protein mediate hormone-dependent transactivation. Proc Natl Acad Sci USA. 1995;92:8288–8292. doi: 10.1073/pnas.92.18.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanese N, Pugh B F, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 44.Tanese N, Saluja D, Vassallo M F, Chen J L, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 46.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 47.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 48.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 49.White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 51.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:206. doi: 10.1038/nsb0296-206. [DOI] [PubMed] [Google Scholar]
- 52.Xiao J H, Davidson I, Matthes H, Garnier J M, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 53.Yamit-Hezi A, Dikstein R. TAFII105 mediates activation of anti-apoptotic genes by NF-kappaB. EMBO J. 1998;17:5161–5169. doi: 10.1093/emboj/17.17.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]