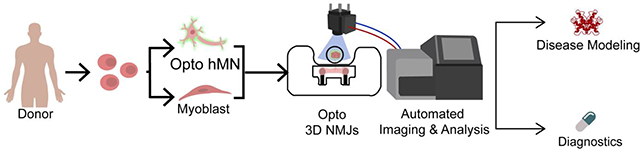
Abstract
Functional human tissues engineered from patient-specific induced pluripotent stem cells (hiPSCs) hold great promise for investigating the progression, mechanisms, and treatment of musculoskeletal diseases in a controlled and systematic manner. For example, bioengineered models of innervated human skeletal muscle could be used to identify novel therapeutic targets and treatments for patients with complex central and peripheral nervous system disorders. There is a need to develop standardized and objective quantitative methods for engineering and using these complex tissues, in order increase their robustness, reproducibility, and predictiveness across users. Here we describe a standardized method for engineering an isogenic, patient specific human neuromuscular junction (NMJ) that allows for automated quantification of NMJ function to diagnose disease using a small sample of blood serum and evaluate new therapeutic modalities. By combining tissue engineering, optogenetics, microfabrication, optoelectronics and video processing, we created a novel platform for the precise investigation of the development and degeneration of human NMJ. We demonstrate the utility of this platform for the detection and diagnosis of myasthenia gravis, an antibody-mediated autoimmune disease that disrupts the NMJ function.
Keywords: Neuromuscular junction, iPS cells, human tissue models, optogenetics, disease modeling, myasthenia gravis
Graphical Abstract
INTRODUCTION
The neuromuscular junction (NMJ) is the chemical synapse between presynaptic motor neurons and post-synaptic muscle cells that enables transmission of electrical signals from the neurons to the skeletal fibers, resulting in muscle contraction. The large size and anatomical accessibility of NMJs have made them a popular model for studying synaptic development and degeneration. Given the prevalence of neuromuscular diseases, due to either motoneuron defects (e.g., amyotrophic lateral sclerosis, spinal muscular atrophy) or directly to degeneration of the NMJ function (e.g., congenital myasthenic syndromes, myasthenia gravis), there is a critical need for bioengineered human tissue models of the NMJ to conduct mechanistic and therapeutic experimental studies. While animal models have traditionally been used to investigate the NMJ, there are significant barriers to translation. The shift towards using human in vitro models not only reduces the need for animal models, but it also provides the opportunity to directly examine relevant human pathological complexities. By utilizing human in vitro models, the molecular and cellular mechanisms of human disorders can be further elucidated to allow for better translation of results.
Human induced pluripotent stem cells (hiPSCs) [1] can be used to derive multiple human cell types, including motoneurons [2,3] and skeletal muscle cells [4,5], from the same donor. Furthermore, hiPSCs can be easily derived from patients [6,7] making them an ideal tool for disease modeling [8,9] and drug screening [10,11]. Standard monolayer co-cultures of skeletal muscle and motoneuron cells remain a mainstay for many assays, but they lack the morphology, phenotype, and organization of physiological NMJs, thus also lacking in vivo- like functional responses. In standard co-cultures, NMJs randomly form across the culture and require tedious manual evaluation of their function. The lack of consistent organization prevents repeated functional measurements on specific motor units and introduces biological and inter-evaluator variability, thus limiting the utility of these models for reproducible, systematic studies [12]. Some of the limitation of 2D systems can be overcome by advanced culture systems which allow for 3D organization and directed interactions between different cellular components. Importantly, 3D systems facilitate the application of biomechanical cues necessary for optimizing tissue structure and function, thus better mimicking in vivo physiology [13–17].
We recently demonstrated that optogenetic methods used in conjunction with tissue engineering allowed precise non-invasive control and functional evaluation of human NMJs grown in vitro [18]. Optogenetics uses genetic engineering to integrate light sensitive proteins (opsins) into the cell genome. One of the most commonly used opsins is channelrhodopsin-2 (ChR2), an ion channel that opens in response to blue light [19,20]. Introduction of opsins into the membrane of excitable cells such as neurons or myocytes allows for noncontact spatiotemporal control of cellular activity with millisecond resolution. Thus, this technology enables the functional evaluation of NMJs between optogenetic motoneurons and non-optogenetic skeletal myotubes with blue light stimulation.
Here we describe an automated method to quantify NMJ function in a human 3D tissue model and demonstrate its capability to diagnose myasthenia gravis (MG), the most common neuromuscular disorder[21]. We use advanced compartmentalized culture systems for the formation of NMJs between optogenetic motoneurons and non-optogenetic skeletal muscle tissues attached to pillars [18,22,23]. A custom optical stimulation platform integrated with microscopy is used to automatically stimulate and record the responses from a set of tissues. The video recordings are processed with custom code that allows for blinded, non-biased evaluation of NMJ function. Such a robust and reproducible methodology allows for repeated measurements of NMJ function and quantification of changes over time or in response to various inputs. Using this strategy, we were able to distinguish the differential effect of myasthenia gravis (MG) antibodies from different donors on NMJ function, with a sensitivity that was sufficient to detect changes in NMJ function in the case of double-negative MG, a disease that escapes detection by any existing diagnostic test.
MATERIALS AND METHODS
1. Generation of donor-specific engineered NMJs
To engineer isogenic NMJs, skeletal myotubes and motoneurons need to be derived from a single cell source. Here we describe two methodologies for engineering donor-specific NMJs: (1) Isolation of skeletal myoblasts from a biopsy followed by reprogramming into an hiPSC line that can then be differentiated into isogenic motoneurons and (2) direct differentiation of both cell types from a single hiPSC line by inducing expression of specific transcription factors. While the former strategy produces adult-like skeletal tissues ideal for studies of healthy NMJs, the latter allows for the generation of donor-specific NMJs from any available iPSC line, including those obtained from patients with congenital neuromuscular pathologies, allowing for personalized disease modeling.
1.1. Derivation of myotubes and motoneurons from primary myoblasts
1.1.1. Primary skeletal muscle cells and myotube differentiation.
Healthy human skeletal muscle cells were obtained from Cook Myosite (#SK-1111) and expanded in Myotonic Growth Medium (Cook Myosite #MK-4444) for a maximum of 6 passages. Myoblast fusion was induced by culturing confluent myoblasts in a series of defined media [24]. Briefly, cells were cultured in Fusion Inducing Media (FIM), components of which are listed in Table I, for 7 days, followed by 2 days in Maturation Medium Ia (MMIa), and 2 days in Maturation Media Ib (Table I). At day 11 after differentiation, media was changed to Maturation Medium II (NbActive4, BrainBits LLC #Nb4–500). Media was replaced every 2 days.
1.1.2. Generation of muscle derived-hiPSCs lines.
Primary skeletal muscle cells were reprogrammed using CytoTune-iPS 2.0 Sendai Reprogramming Kit (ThermoFisher Scientific #A16517) that contains Sendai viruses for KOS, hc-Myc and hKlf4. Briefly, 1×105 P1 skeletal muscle cells were plated in one well of a 6-well plate one day before infection. Cells were infected the next day at MOI 10:10:6 for KOS:hc-Myc:hKlf4 viruses and incubated for 24 hours. After 5 days, cells were trypsinized and plated on top of irradiated mouse embryonic fibroblasts (MEFs; Globalstem). HiPSCs colonies formed 2 weeks after transduction. Colonies were picked and expanded in MEF feeders for 5 additional passages, before switching to Matrigel-coated plates.
1.1.3. Generation of transgenic ChR2 cell lines.
A transgenic cell line was created by infection of the muscle-derived hiPSCs with the pLenti-EF1a-hChR2(H134R)-EYFP-WPRE construct (Addgene #20942). Plasmids were grown in One Shot™ Stbl3™ chemically competent E. coli (ThermoFisher Scientific #C737303) cultured in Luria Broth (ThermoFisher Scientific #10855) with 100 μg/ml ampicillin (ThermoFisher # 11593027), and isolated using E.Z.N.A.® Endo-Free Plasmid Maxi Kit (Omega Biotek #D6926–03). Human embryonic kidney cells HEK-293FT (ThermoFisher Scientific # R700–07) grown in DMEM (ThermoFisher Scientific #10566–016) supplemented with 2% v/v of fetal bovine serum (FBS) (Atlanta Biological #S11150) and 50 U/ml penicillin/streptomycin were transfected with 32.73 μg of the ChR2-YFP plasmid, 10.91 μg of viral envelope plasmid (pMD2.G Addgene #12259) and 21.82 μg of packaging construct (pCMV ΔR8.2, Addgene #12263) using polyethylenimine (Polysciences # 23966). After 60 hours, supernatant was filtered through a 0.45 mm low protein-binding Steriflip-HV, (Millpore #SE1M003M00) and the viral particles were precipitated using the Lenti-X Concentrator (Takara #631231). Viruses were added to the hiPSCs one day after passaging. YFP+ cells were selected by FACS (BD Influx™ cell sorter) and expanded.
1.1.4. Derivation of motoneurons from transgenic ChR2-expressing iPSCs.
Motoneurons were derived form ChR2-expressing transgenic hiPSC lines using a protocol adapted from Maury et al. [2]. Briefly, on day 0, 4×106 hiPSCs were transferred to petri dishes for suspension culture in 10ml of Motoneuron Suspension Culture Medium (MSCM, Table II). On day 0 MSCM was supplemented with 3 μM CHIR99021 (Tocris # 4423/10), 0.2 μM LDN193189 (Miltenyl Biotec #130–103-925), 40 μM SB431542 hydrate (Sigma Aldrich #S4317) and 5μM Y-27632 dihydrochloride. On day 2, neurospheres were isolated using 37 μm reversible strainers (STEMCELL Technologies #27215) and replated in MSCM supplemented with 3 μM CHIR99021, 0.2 μM LDN193189, 40 μM SB431542 hydrate, and 0.1 μM retinoic acid. Thereafter, media was replaced for MSCM supplemented with 0.5 μM SAG (Millpore #566660), 0.2 μM LDN193189, 40 μM SB431542, and 0.1 μM retinoic acid at day 4; 0.5 μM SAG and 0.1 μM retinoic acid at day 7; 10μM DAPT (R&D Systems # 2634/10) at day 9; and 20 ng/ml BDNF and 10 ng/ml GDNF at day 11.
1.2. Direct differentiation of myotubes and motoneurons from hiPSCs
In this second approach, both skeletal myoblasts and motoneurons were derived from wild type hiPSCs using transcription factor-mediated differentiation protocols.
1.2.1. Generation of a MyoD/OCT4 KD line.
We designed a PiggyBac plasmid for the overexpression of the transcription factor MYOD1 and knock-down of OCT4 in hiPSCs based on a recent publication by Akiyama et al [25]. We subcloned the human MYOD1sequence and OCT4-shRNA downstream of a 3rd-generation Tet-responsive (Tet-On® 3G) promoter[26]. The integrated sequence allows for the conditional activation of MYOD1 and OCT4 knock down (KD) upon addition of doxycycline. To reduce silencing of the promoter, we included a short minimal ubiquitous chromatin opening element (UCOE)[27]. To generate a stable MyoD/OCT KD line, we co-transfected the MyoD/OCT4 KD plasmid and the PiggyBac™ transposase using Lipofectamine™ Stem (ThermoFisher # STEM00003). Briefly, we seeded 1.5×106 of hiPSCs in a Matrigel-coated 6-well plate using Essential-8 medium and 10μM Y-27632. The lipofectamine mix containing the transposase and transgene plasmids was incubated for 30 minutes. HiPSCs were then transfected with 2 μg of DNA at a 1:2 ratio (transposase:DNA). After 24 hours, cells were inspected for nuclear BFP expression and selective pressure with puromycin was maintained for 14 days.
1.2.2. Derivation of skeletal myoblasts from hiPSCs (hiSkMs).
For generation of iPSC-derived skeletal myoblasts (hiSKMs), 0.5 ×106 of MyoD/OCT4 KD-iPSCs were seeded per well in a Matrigel-coated 6 well plate in Myogenic Inducing Media (MIM, Table I), supplemented with Y-27632 10 μM and 2μg/ml doxycycline [25]. The medium was replaced with fresh MIM supplemented with doxycycline the following day. On day 5, cells were passaged and plated in Myotonic Growth Medium (MGM) supplemented with 2μg/ml doxycycline. Cells were kept in this media for 2 weeks and medium was changed every other day. At day 19, myotube differentiation was induced using the same protocol as for primary myoblasts, with the addition of 10 μM SB431542 and 10μM DAPT to FIM (Table I).
1.2.3. Generation of an optogenetic hNIL line.
A cassette expressing NGN2, ISL1, and LHX3 (hNIL) under the tetracyline response element (TRE3G), CAG promoter driving constitutive expression of the reverse tetracycline transactivator (rtTA3G), and an EF-1α promoter driving constitutive expression of the neomycin resistance gene was stably integrated into the safe-harbor AAVS1 locus of the WTC11 iPSC line via TALENs [28]. Using NEBuilder® HiFi DNA Assembly Master Mix (Catalog #E2621S), the ChR2-YFP sequence from plasmid pLenti-EF1a-hChR2(H134R)-EYFP-WPRE (Addgene #20942) was cloned into plasmid pC13N-dCas9-BFP-KRAB (Addgene #127968) containing a CAG promoter and homologous regions for integration of the cassette into the CLYBL safe harbor locus which enables robust transgene expression. Plasmids were grown in One Shot™ Stbl3™ chemically competent E. coli (ThermoFisher Scientific #C737303) cultured in Luria Broth (ThermoFisher Scientific #10855) with 50 μg/mL Kanamycin (ThermoFisher # 15160054), and isolated using Plasmid Plus Midi Kit (Quiagen# 12943 and 12945). The donor plasmid containing the ChR2-YFP gene was then integrated into iPSCs using the TALEN pair pZT-C13-L1 (Addgene #62196) and pZT-C13-R1 (Addgene #62197) to cleave the genome specifically at the CLYBL locus. Briefly, 1×106 hNIL-iPSCs were nucleofected with 3 μg of each CLYBL TALEN pair (pZT-C13-L1 and pZT-C13-R1, Addgene #62196, #62197), and 6 μg of the ChR2 donor plasmid using the P3 Primary Cell 4D-Nucleofector Kit (Lonza #V4XP-3024). Starting 24 hours post-nucleofection, cells were subjected to geneticin selection (G418, ThermoFisher #10131035) in StemFit medium (Ajinomoto #AS-Bo1R) supplemented with 10 μM Y-27632. Increasing concentrations of geneticin (50 μg/mL – 150 μg/mL) were applied for 7 days until stable colonies appeared. Post-selection, single-cell colonies were manually picked and expanded. The clones were genotyped by PCR to confirm the integration of the ChR2-YFP cassette at the CLYBL safe harbor locus.
1.2.4. Derivation of motoneurons from hNIL iPSCs (opto hiMN).
To differentiate hNIL motor neurons, 4×106 hiPSCs were transferred onto suspension culture dishes in 10 ml of mTESR1 medium supplemented with 10μM of Y-27632. On day 1, culture medium was replaced with warm neural inducing medium (NIM, Table II) supplemented with 10μM Y-27632, 2μg/ml doxycycline and 0.2 μM Compound-E (Stem Cell Technologies # 73952). On day 4, media was replaced with warm NIM supplemented with Culture One supplement (ThermoFisher #A3320201), 1μg/ml of laminin, 40 ng/ml BDNF, 40 ng/ml GDNF and 20 ng/ml NT3. On day 7, half of the medium was replaced with fresh warm media. On day 10, cells were switched to neural maturation medium (NMM, Table II). Fresh media was changed every other day.
2. Engineering of photosensitive NMJs
2.1. Microfluidic platform.
Compartmentalization of the 3D culture of motoneurons and skeletal muscle was achieved using a custom-made microfluidic platform consisting of two tissue-specific culture chambers separated by a 1-mm gap for axonal sprouting and growth. The 1.5mm × 0.6m muscle compartment features a set of 2 capped pillars for muscle bundle attachment, 1mm apart, and is lined by an array of posts to ensure confinement of the gel solution upon seeding. The chamber is accessible on one end by a gel filling channel which features a necking region at the entrance to the chamber, designed to locally weaken the muscle bundle to separate the muscle in the chamber from the filling channel during gel compaction, resulting in muscle tissue anchored solely to the pillars and capable of freely contracting. The neuronal chamber, also accessible via a gel-filling channel, is delimited by 5 posts forming a 0.5 mm-diameter cup. This design allows for the seeding of an individual neurosphere aligned with the center of the muscle bundle. The gap region between the cell chambers ensures a clear physical separation between the two tissue types, which allows for axonal outgrowth and visualization. The tissue chambers are flanked by two large open reservoirs with a diameter of 6 mm, positioned to facilitate seeding and media changes with a multichannel pipette.
2.2. Fabrication of the microfluidic platform.
Polyurethane molds of the microfluidic platform were kindly provided by Professor Roger Kamm. A 10:1 base/curing agent mixture of PDMS (Ellsworth Adhesives) was degassed in a vacuum chamber for approximately 30 min until no air bubbles were left. The PDMS mix was then cast into the molds and degassed for a second cycle. The molds were then cured at 65°C for at least 4 hours and peeled off the molds once they have cooled down. Once peeled off their polyurethane molds, the individual PDMS devices were cut out and gel filling ports and medium ports/reservoirs were formed with 1- and 6-mm-diameter biopsy punches, respectively. The devices were cleaned in an ultrasonic bath (Branson 1800) using 1-hour cycles of soap, isopropranol and distilled water, before drying overnight in a 65°C oven. Next, the devices were plasma treated and bonded to a glass cover slip pretreated with 2% Pluronic® F-127 for at least 30 min. Devices were then autoclaved in water and allowed to dry in an oven 65°C at overnight.
2.3. Cell seeding in the microfluidic devices.
Primary or hiPSC-derived myoblasts were seeded at a concentration of 20 ×106 cells/mL in a 4:1 mix of 3 mg/mL collagen I (Corning #354249) and Matrigel. Briefly, collagen was diluted in PBS with Phenol Red (Sigma Aldrich #P0290) to achieve the desired concentration, and a 10% solution of NaOH was used to neutralize the gel before adding the Matrigel and resuspending the myoblasts. A 10 μl micropipette was then used to inject the cell suspension into the muscle port until the muscle chamber and channel were filled with the cell-collagen mixture (approximately 4 μl). After 20 min of polymerization at 37°C, the media reservoirs were filled with Myotonic Growth Media. Myotube differentiation was initiated the day after seeding. Two weeks after myoblast seeding, the muscle and neurosphere chambers, and the connecting channel were filled with a 4:1 mixture of collagen I (2 mg/mL) and Matrigel. hiPSC-derived neurospheres in the 200–300 μm range were selected using pluriStrainers (PluriSelect #43–50200-03 and #43–50300-03) and seeded in the motoneuron chamber using the hydrogel. Devices were incubated for 20 min at 37oC and kept in coculture medium from this point onwards.
3. Open-well bioreactor platform.
A novel open well PDMS bioreactor system was used for the compartmentalized culture of skeletal muscle tissues and motoneurons. The system consists of two separated chambers connected by an 800 μm × 250 μm × 250 μm (L×W×H) channel that allows for axonal growth. The muscle chamber contains two horizontal pillars separated by 3mm for attachment and contraction of the muscle tissue. The motoneuron chamber is approximately 800 μm in diameter. The reservoir for the seeding of the muscle chamber measures 5.5 mm × 1.5 mm × 1 mm (L×W×H). Two 1 mm × 1 mm notches on the side of this chamber allow for longer pillars while having minimal impact on symmetric seeding around the pillar heads. Each well can accommodate up to 500 μL of media. This open well system allows for easy seeding of both cell types, as well as easy access and recovery of the samples for terminal analysis.
3.1. Fabrication of the open-well bioreactor system.
Molds were machined in polyoxymethylene on a 3-axis Haas OM2 milling machine. The mold geometry was modeled in SolidWorks, and the CNC toolpaths were generated using Mastercam. To fabricate the devices, a 10:1 base/curing agent mixture of PDMS was degassed and poured into the bottom half of the mold. After a second cycle of degassing, the mold was closed by placing the top half of the mold in the right orientation (Supplementary Fig 1). Steel hexagonal rods (McMaster-Carr #91920A046) and metal c-clamps (McMaster-Carr #1705A12) were used to ensure a tight fit. Extra PDMS was then poured over the inlet holes in the top of the mold to ensure complete filling of the mold. The molds were then cured at 65°C for 4–6 hours. After curing, the molds were carefully opened to peel off the devices, that were then plasma treated (Harrik PDC-32G) and bonded to a glass cover slip that had been pretreated with 2% Pluronic® F-127 for at least 30 min at room temperature. Compressed air and scotch tape was used to remove debris from the devices before bonding. Devices were then dry autoclaved and stored until used.
3.2. Cell seeding in the open-well bioreactor.
A suspension of myoblasts in collagen I (2 ×107 myoblasts/mL) was prepared as described in section 2.3. The muscle chamber was then filled with 15 μl of the cell-collagen suspension. After 30 min of polymerization at 37°C, the media reservoirs were filled with Myotonic Growth Media. Myotube differentiation was initiated two days after seeding of the myoblasts. MN aggregates were seeded in the device at day 14. To do so, media was carefully aspirated from the neurosphere chamber and replaced by 10 μl of a 4:1 mixture of 2 mg/mL collagen I and Matrigel; this step is critical to ensure proper connectivity between the neuronal and muscle chambers. hiPSCs-derived neurospheres in the 400–500 μm range were selected using pluriStrainers (PluriSelect #43–50400-03 and #43–50500-03) and resuspended in 100 μl of the same hydrogel. A 20 μl micropipette filled with gel was used to pick up a single neurosphere and carefully place it in the bottom of the neurosphere well. After assessing the correct placement of the neurosphere under a microscope, the devices were placed in an incubator to allow polymerization for 30 min before adding 500 μL of the coculture medium, consisting of NbActiv4 supplemented with 10ng/ml GDNF and 20 ng/ml BDNF. From this point on, medium was changed every 2 days.
4. Automated stimulation and recording of NMJ function
4.1. Optical Stimulation Platform.
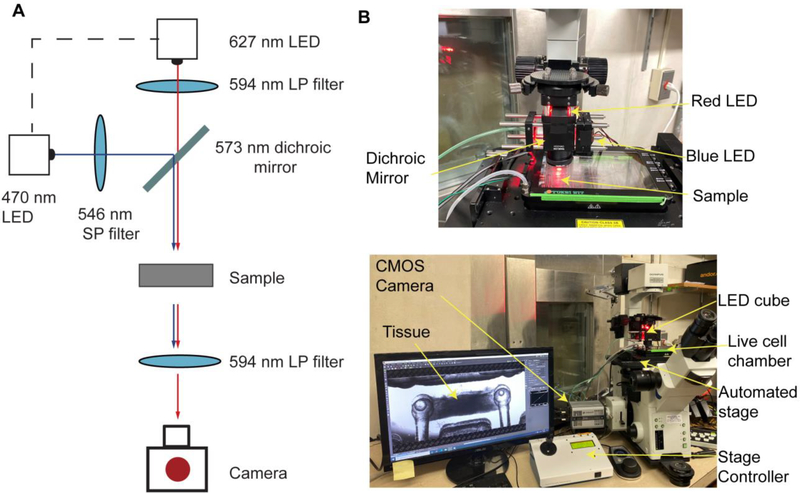
A custom-made optical stimulation platform was built using a 573 nm dichroic mirror (Semrock FF573-Di01–25×36) to couple two light sources together: red (627 nm LED (Luxeon Star LEDs SP-05-R5) filtered through a 594 nm long-pass excitation filter (Semrock BLP01–594R-25)) and blue (448 nm LED (Luxeon Star LEDs #SP-05-V4) filtered through a 546 nm short-pass excitation filter (Semrock FF01–546/SP-25)) (Fig 4A). A 30 mm cage cube system from ThorLabs (#CM1-DCH, #CP33, #ER1-P4 and #ER2-P4) was used to couple the light sources. A ring-actuated iris diaphragm (ThorLabs #SM1D12D) was attached to the bottom to control of the size of the illuminated area. The LEDs were powered with T-Cube LED drivers (ThorLabs #LEDD1B, # KPS101) controlled from an Arduino Uno Rev3 board.
Figure 4. Optical set-up for simultaneous stimulation and recording of engineered NMJs.
(A) Light path (B) Implementation of the custom optical set-up in a traditional inverted microscope provided with a live cell chamber, an automated stage and a CMOS camera.
4.2. Stimulation set-up.
For imaging, an Olympus FSX100 microscope was used in conjunction with an Andor Zyla 4.2 sCMOS camera, an automated stage (MS-2000 XYZ) connected to a multi-axis stage controller (MS-2000-WK), and a live-cell chamber with controlled temperature and CO2 (Tokai Hit STX Series Stage Top Incubator System TOKAI-HIT-STXG). The optical stimulation platform was hung directly over the sample. A 594 nm long-pass emission filter placed on top of the microscope objective, distal to the sample (Semrock BLP01–594R-25) was used to filter out the blue light used to stimulate the motoneurons (Fig 4B). The maximum intensity of the blue light illuminating the sample in this set-up was 326±8 μW/mm2.
4.3. Automated Imaging.
A custom script was loaded onto the Arduino board with the desired optical stimulation parameters prior to each imaging session (https://github.com/ofvila/NMJ-function-analysis). This script instructs the red LED to turn ON 5s after the execution of the script, and then start the blue light pulses after another 5 s. A ramped stimulation protocol with decreasing intervals was used to challenge the tissues in terms of maximum frequency of response and fatigability in one measurement. Pulse duration, pulse number, and initial and final frequencies can be adjusted to generate more or less challenging ramps, depending on the motoneurons’ responsivity to light stimulation, the level of muscle innervation and the expected changes in NMJ function. The parameters ranges used in our studies were 30–40 light pulses, with a duration of 50–100 ms, and frequency ramps from 0.2–0.5 to 2–3 Hz.
For imaging, a 594 nm long-pass emission filter (Semrock BLP01–594R-25) was placed between the sample and the imaging objective to filter out blue light pulses. The camera settings were adjusted to an exposure of 20 ms (50 fps) for high-speed imaging of the region of interest. Once the region of interest was identified, a plate (ThermoFisher #267061 and #267062) containing either 4 bioreactors (4 × 6 = 24 tissues) or 8 microfluidic platforms (8 × 4 = 48 tissues) was placed into the live-cell chamber on top of the automated stage. An NIS-Elements Software custom macro that controls stage position, the Arduino board, and the video acquisition was then executed, allowing for automated stimulation and video acquisition of every tissue in the plate without user intervention (Movie 1) (https://github.com/ofvila/NMJ-function-analysis). Medium was replaced with fresh coculture medium post-recording, and samples were returned to the incubator. Resting intervals of at least 24 hours were used between recordings to avoid tissue fatigue.
5. Automatic quantification of NMJ function
5.1. Batch movie processing.
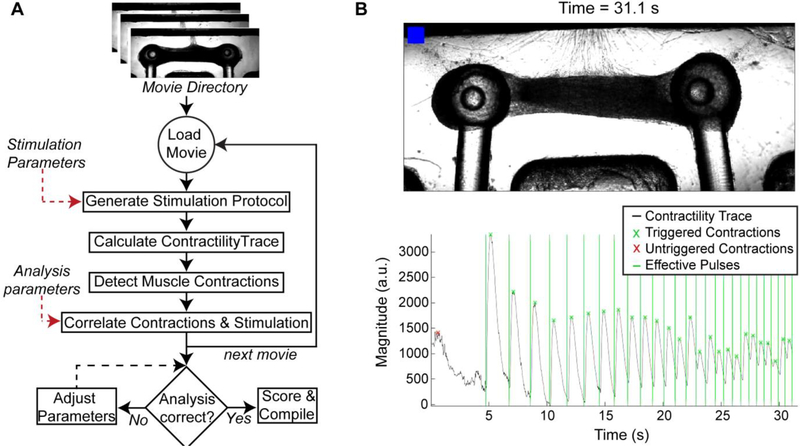
Custom MATLAB code (https://github.com/ofvila/NMJ-function-analysis) was used to perform automatic batch analysis of the acquired movies. The main functions and brief explanations are listed in Table III. Briefly, the main script recursiveOSAnalysis finds every movie file in an indicated folder and runs the function OSAnalysis using the indicated stimulation parameters and a set of initial analysis parameters. This function calculates the stimulation protocol based on the parameters (getOSProtocol), and then detects the moment that the red light turns ON and uses it to synchronize the stimulation and the recording (cutVideoToOS). It then extracts the motion from each movie by subtracting every frame from a baseline frame to output a matrix of differences.
The amount of motion at any time point is calculated as the average absolute value of the difference matrix across the frame and used to create the trace of contractile activity (getContractilityTrace). Muscle contractions are detected (getPeakHeighThreshold) and aligned against the stimulation trace (matchPulsesToOSContraction). Each stimulation pulse was determined as effective if a contraction occurred within a specific time window (matchLength).
The fraction of effective pulses (F) was calculated as the ratio of effective pulses to total light pulses. To account for the possibility that random unstimulated contractions could be correlated with the stimulation pulses by happenstance, an expected fraction of effective pulses (E) was calculated as the expected fraction of pulses to be labeled as effective if the contractions were randomly distributed throughout the time course (total number of contractions × matchLength/total time). The final tissue score was then calculated as (F-E)/(1-E), with a range from 0 to 1.
At the end of this first analysis, recursiveOSAnalysis displays graphs showing the results of the analysis for each tissue to the user (OSPostAnalysis). The user at this point has the option to either approve to the analysis or change some of the analysis parameters to refine the detection of the contractions. Finally, once every analysis has been reviewed and approved, an excel table that compiles the scores of each individual tissue is generated (compileData).
5.2. Determining Analysis Parameters.
Our code utilizes a series of default parameters to evaluate NMJ function during the first round of analysis, but these can be adjusted in individual movies during the analysis if required (i.e. peaks and minima are misidentified, Supplementary Fig 2). At the end of the analysis, MATLAB displays a graph with the result of the analysis for each tissue and ask the user if any parameter requires readjustment. To help with this assessment, the graph will show the contractility trace, the light pulses (vertical lines) the detected peaks (asps) and the minima for each peak (blue circles).
The following parameters can be adjusted to optimize the analysis:
Baseline time (baselineTime): This is the frame that the code uses as reference for tissue at rest. The default value is set at the very beginning of the recording, before the NMJ is stimulated (0.01). However, spontaneous activity of the skeletal tissue may cause the tissue to be contracted at this point and result in inaccurate analysis which presents incorrect peak shape (Supplementary Fig 2A). In that case, the option to change baseline time (option 1) should be selected when prompted at the end of the analysis and a new baseline time should be picked.
Peak threshold (peakThreshold): This parameter is a value that allows the code to count as contractions only peaks over a certain magnitude level. The default value of this parameter is automatically set at 25% of the highest peak, which we found to be a good threshold for most cases. If this threshold is too high, the code might inaccurately omit peaks (Supplementary Fig 2B). In that case, the option to change the peak threshold (option 2) should be selected when prompted at the end of the analysis and a new threshold should be chosen.
Minimum minima height and width (minMinProminence and minMinWidth): These two parameters determine the sensitivity when detecting the start of each peak. The time between a light pulse and the start of a peak determines if the peak is considered a triggered or untriggered contraction. The code starts searching minima starting from the peak maximum towards the beginning of the movie, therefore low values of minMinProminence and/or minMinWidth result in high sensitivity in the detection of minima, which can lead the code to tag some point in the middle of the peak as the beginning (Supplementary Fig 2C). On the other hand, high minMinProminence and/or minMinWidth values can result in minima being detected before the start of the contraction (Supplementary Fig 2D). Note that in this case, unlike in the high peakThreshold example, the peaks are detected correctly (red crosses) but the minima are not being detected corrected.
6. Generate video files.
The script recursiveOSMovie can be run on any folder that has been previously analyzed to generate movies that visualize the tissue contractions, the optical stimulation (indicated by a blue blinker in the top-left) and the contractility analysis.
Notes: Code is compatible with nd2 and czi formats. Code requires parallel processing activated in MATLAB, as well as the bioformats package.
RESULTS
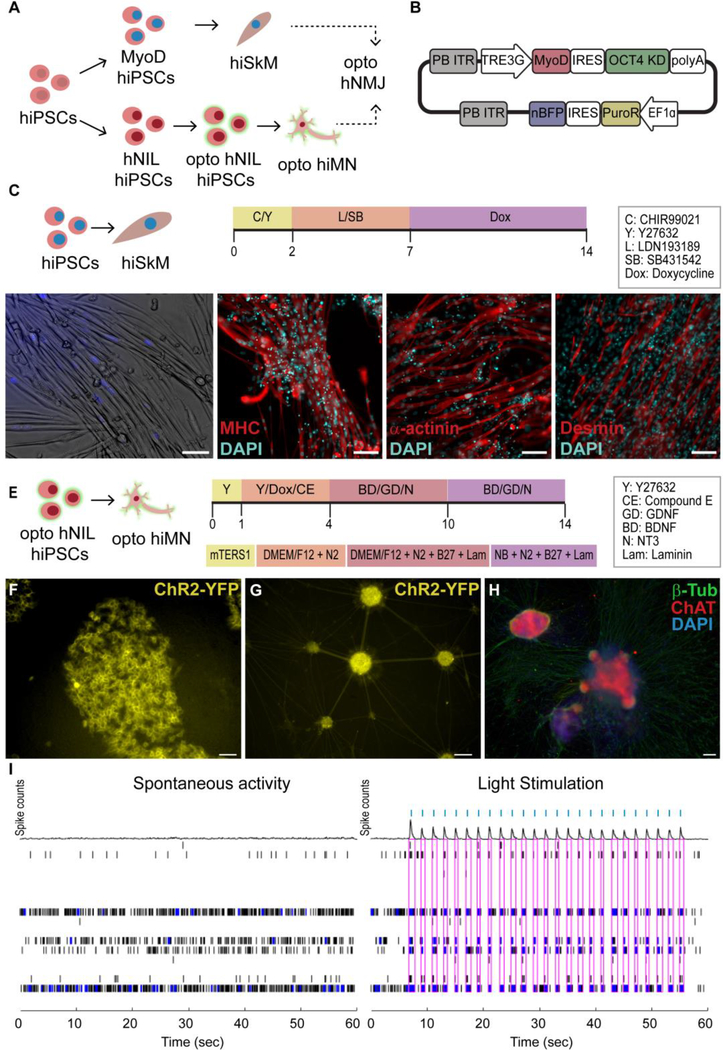
The first overall strategy to produce NMJs from a specific donor utilized human primary skeletal myoblasts as the starting cell source and reprogrammed them into hiPSCs using Sendai viruses to create isogenic motoneurons (Fig 1A). The human primary skeletal myoblasts were differentiated into myotubes utilizing a 14-day protocol (Fig 1B), resulting in elongated, multinucleated cells that expressed the skeletal muscle markers myosin heavy chain (MHC), α-actinin and desmin (Fig 1C). In parallel, the myoblast-derived hiPSCs were infected with lentivirus carrying the channelrhodopsin-2 tagged with a yellow fluorescent protein (ChR2-YPF) and differentiated into hMNs using a dual SMAD inhibitor protocol [2] (Fig 1D). Both the optogenetic iPSCs and motoneurons showed membrane localization of the ChR2-YFP construct (Fig 1E, F). Successful differentiation of optogenetic hiPSCs into hMNs with persistent expression of ChR2-YFP was confirmed by fluorescent microscopy showing co-expression of ChR2-YFP and choline acetyltransferase (ChAT) (Fig 1G). The ability to induce electrical depolarization of the optogenetic hMNs using light was confirmed with MEA recordings (Fig 1H) that showed the generation of electrical spikes in response to blue light stimulation.
Figure 1. Generation of myotubes and motoneurons (MN) from human primary skeletal myoblasts (hSkM).
(A) Overall strategy (B) Myotube differentiation protocol (MM = Maturation Media) (C) Immunofluorescence (IF) images of skeletal markers in hSkM-derived myotubes. (D) human induced pluripotent stem cells (hiPSC) MN differentiation protocol. (E) Membrane expression of YFP-Channelrhodopsin-2 (ChR2) in optogenetic iPSCs and (F) MNs. (G) Co-expression of ChR2 (green) and choline acetyltransferase (ChAT, red) in opto-hMNs. (H) Spontaneous electrical depolarization vs light-induced electrical depolarization in opto-hMNs as confirmed by multi-electrode array (MEA) recordings, in which each row shows the electrical activity of one electrode, blue dots represent spikes and pink boxes indicate the presence of burst (synchronous activity in the electrodes), while blue ticks signal the light stimulation pulses. Scale bars 100 um.
Alternatively, a second method of producing isogenic hNMJs consisted of using a single source of hiPSCs which were then differentiated into its respective components, skeletal muscle cells and motoneurons independently (Fig 2A). For this strategy, we utilized transcription factor overexpression, which circumvents some of the challenges associated with small molecule pathway inhibitor-mediated differentiation protocols, such as low efficiency and variable sensitivity of different hiPSC lines or clones to small molecules.
Figure 2. Transcription-factor mediated differentiation of myotubes and motoneurons from iPSCs.
(A)Overall strategy. (B) PiggyBac plasmid for the expression of MYOD1 and knock down of OCT4 in hiPSCs. (C) hiPSC-derived skeletal myoblast (hiSkM) differentiation protocol (D) Bright field and (E) skeletal muscle marker IF images (F) hNIL- MN differentiation protocol. (G) Membrane expression of ChR2-YFP in optogenetic hNIL iPSCs and (H) MNs. (I) IF images for β-tubulin (green) and ChAT (red) (J) MEA recording showing spontaneous and light-stimulated electrical activity in optogenetic hNIL-MNs. Each row shows the electrical activity of one electrode, blue dots represent spikes,pink boxes indicate the presence of burst (synchronous activity in the electrodes), and blue ticks indicate light stimulation pulses. Scale bars 100 μm.
Overexpression of the master myogenic regulator MYOD1 has been widely used to induce direct myogenic transdifferentiation of fibroblasts [29] as well as to differentiate PSC -derived mesoderm cells into myoblasts [30]. However, direct MYOD1 conversion in pluripotent cells without the generation of a mesodermal intermediate is very inefficient [31]. Coupling MYOD1 overexpression with OCT4 knockdown can overcome this limitation, allowing for rapid and robust differentiation of myoblast from iPSCs [25]. Thus, we created a PiggyBac™ plasmid that allowed for inducible and simultaneous MYOD1 overexpression and OCT4 knockdown (Fig 2B) and used it to generate a MyoD/OCT4 KD cell line in the WTC11 background. These hiPSCs were then differentiated into hiPSC-derived skeletal myoblasts (hiSkM) that showed nuclear expression of the blue fluorescence protein (nBFP), indicative of successful transfection (Fig 2 C,D). Further maturation and fusion into myotubes was achieved using a combination of serum-free media and small molecules [32] (Fig 2D).
Similarly to MYOD1, overexpression of the master neuronal transcriptional regulator neurogenin-2 (NGN2) results in rapid, one-step differentiation of hiPSCs to functionally mature glutamatergic neurons [33,34]. Motoneurons can be generated in a similar manner by overexpressing the transcription factors Islet-1 (ISL1) and LIM Homeobox 3 (LHX3) along with NGN2 [28]. As described in the methods, we created a stable hiPSC line that allowed for the inducible expression of the human NGN2, ISL1, and LHX3 cassette (hNIL) (Fig 2E). This hNIL-iPSC line was then further edited for constitutive expression of the ChR2-YFP gene to generate optogenetic hNIL-iPSCs (Fig 2F). These opto-hNIL-iPSCs were then differentiated into optogenetic hiMNs through inducible transcription factor overexpression (Fig 2E). Phenotype and function were validated by membrane expression of ChR2-YFP (Fig 2G), IF stains for ChAT (Fig 2H) and MEA recordings demonstrating optical control of neuronal depolarization (Fig 2I).
To form distinct, controllable photosensitive hNMJs, we used compartmentalized culture systems with physically separated chambers for the culture of skeletal muscle microtissues and motoneuron aggregates that allow for axonal sprouting from the neuronal aggregates towards the skeletal tissues. Two culture systems of different scales were used to demonstrate that the same methodology can be implemented in any optically accessible culture systems. The first culture system used is a microfluidic device that allows the muscle tissue to anchor itself between two capped pillars connected to a well for motoneuron aggregate seeding by a channel for axonal growth and sprouting (Fig 3A). The resulting tissues are 1 mm long and contain approximately 20,000 myoblasts. The second is a larger open-well system that creates 4 mm-long tissues containing approximately 300,000 myoblasts (Fig 3B).The larger size of these tissues, combined with their accessibility, allows for the integration of functional analysis with molecular and histological assays, unlike the microfluidic system, which is more suitable for high-throughput functional studies than the open well system, since it requires lower cell numbers. The timeline for the generation of 3D NMJs was the same for both platforms (Fig 3C). Briefly, primary or iPSC-derived skeletal muscle were expanded for 2–3 weeks before the start of the experiment. On day 0, myoblasts were seeded into the muscle chamber using a collagen gel, and myotube differentiation was started. At the same time, 3D differentiation of ChR2-iPSCs into motoneurons was started in suspension dishes separate to the reactor. On day 14, the motoneuron neurosphere was seeded into the reactor and axonal innervation of the skeletal tissue occurred about 7 to 12 days later (Fig 3D, E).
Figure 3. Advanced culture systems for the formation of 3D NMJs.
A) Microfluidic platform. Right panel depicts the microfluidic chambers inside the platform, located between each pair of media reservoirs. B) Open-well system C) Timeline showing differentiation and seeding strategies. D)Innervated primary myoblast-derived skeletal muscle tissue formed in the microfluidic platform. E) Innervated iPSC-derived skeletal muscle tissue cultured in the open-well bioreactor. Scale bar 0.5 mm.
, A custom optoelectronic platform was developed to image and stimulate hNMJs. A 627 nm (red) LED to provide brightfield illumination and a 470 nm (blue) LED for MN stimulation were coupled together with a dichroic mirror and filtered with a combination of long and short pass filters (Fig 4A). The stimulation apparatus is placed above the NMJ tissue samples on the automated stage (Fig 4B, Movie 1). A live-cell chamber was used to keep the tissues under physiologic parameters for an extended period of time while the automated stage and custom script captured data for batches of tissues.
To evaluate and measure hNMJ function, movies are batch-processed by a custom MATLAB algorithm (Fig 5A). The output of the automated analysis including triggered/untriggered contractions and effective stimulation pulses is visually synced with the original recording for user review (Fig 5B). The code allows for user input to optimize the analysis parameters until satisfactory results are obtained and a final score of NMJ function is calculated, with values between 0 to 1. This automated process enables repeated, unbiased quantification of hNMJ function of a large number of samples and can be used to analyze contractility of tissues derived from different cell sources (Movie 2: primary skeletal myoblasts; Movie 3: hiPSC-derived myoblasts) in both of the culture systems presented in this work (Movie 2: Open well bioreactor, Movie 4: microfluidic platform). Analysis of the evolution of NMJ function over time demonstrated that while the limiting factor for NMJ function for primary skeletal myoblast-derived tissues was motoneuron to muscle innervation, skeletal tissue maturity was the limiting factor for iPSC-derived myoblast muscle. This is evident in the reduced NMJ function that improves long after complete innervation of iPSC-derived myoblast muscle is achieved (Supplementary Fig 3). Similarly, there are some differences on the evolution of NMJ tissues in the open-well bioreactors vs the microfluidic system, with function peaking later in the larger tissues (day 23 in the bioreactor vs day 14 in the smaller microfluidic devices, Supplementary Fig 4).
Figure 5. Automated analysis of NMJ function.
(A) Video-processing algorithm for the batch analysis of movies taken during optical stimulation of NMJ cultures (B) Representative frame of the output generated by the analysis software showing the video frame and time stamp on the top, and the contractility trace graph in the bottom. The blue blinker indicates when the blue stimulation is happening, shown in the graph by the vertical lines. Light-pulses followed by a contraction (x) are counted as triggered and marked in green.
To assess the translational utility of our system, we recapitulated pathological changes in NMJ function caused by MG, the most common NMJ disease.MG is an autoimmune disorder caused by autoantibodies against the nicotinic acetylcholine receptors (nAChR), leading to muscular weakness due to a decrease in NMJ function. MG diagnosis is routinely performed based on symptomatology, blood tests for specific antibodies, and electrodiagnostic tests. However, the antibody titers correlate poorly with disease severity [35]. Furthermore, not all antibodies contributing to MG have been identified, leaving seronegative patients with MG symptoms undiagnosed and untreated [36]. Electrodiagnosis, in the other hand, is an invasive and painful technique [37] and it cannot be relied on as a standalone diagnosis tool because the results can be confused with other pathologies such as Lambert-Eaton myasthenic syndrome, botulism, or motoneuron disease.
To recapitulate the myasthenic phenotype in our system, we first incorporated pooled sera from 5 patients carrying MG autoantibodies. Steinbeck et al., and others, previously reported the ability to model MG in co-cultures of optogenetic, pluripotent stem cell - derived motoneurons and skeletal myotubes by manually illuminating randomly selected fields in the coculture and measuring myotube contraction. [38,39].
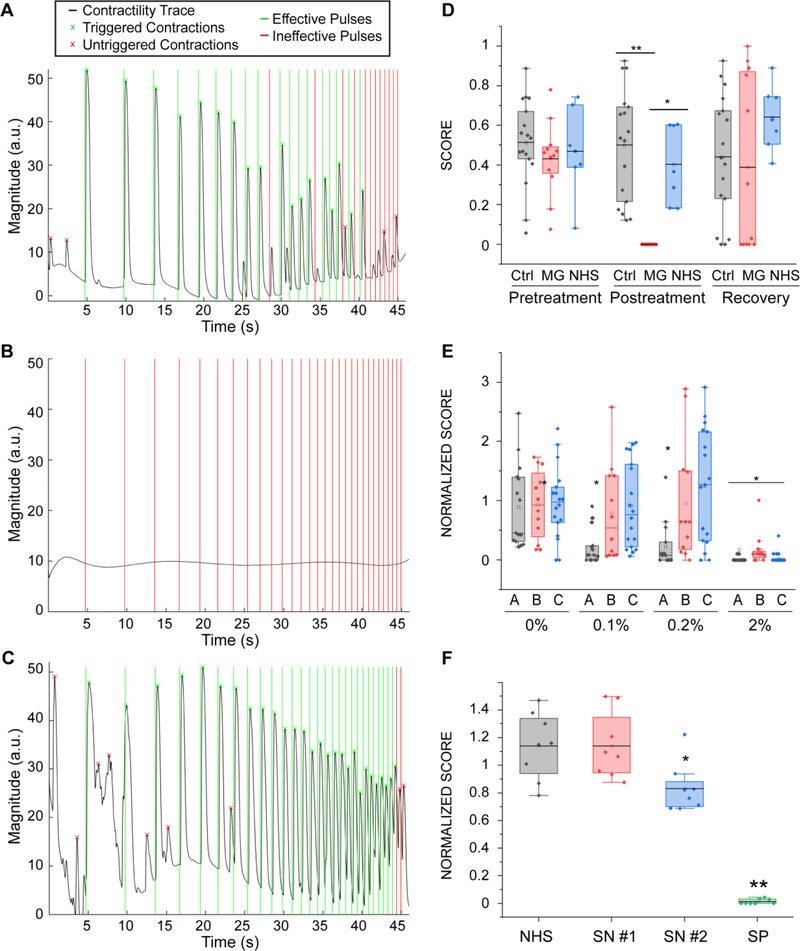
Our system allowed us to perform repeated precise measurements of the same tissues before exposure (Fig 6A, Movie 4) and after incubation with MG serum (Fig 6B, Movie 5). After washout of the antibodies (Fig 6C, Movie 6) NMJ function was recovered. For this set of experiments, we used the microfluidic platform, since it allowed us to use smaller amounts of patient sera, a limited resource. Similarly, we used myotubes and motoneurons derived from human primary myoblasts (as depicted in Fig 1), since we didn’t require a specific genetic background. Quantification of the results for tissues treated with 20% of MG serum for 48 hours showed drastically impaired function (before treatment score mean = 0.43 ± 0.05; after treatment score mean = 10−5 ± 0.00) compared to controls treated with serum from healthy donors (before treatment score mean=0.47±0.07; after treatment score = 0.40 ± 0.07, p=0.0015) or non-treated controls (before treatment score mean=0.51±0.05; after treatment score = 0.47 ± 0.07, p=3.5·10−6). Evaluation of tissue function 48 hours after serum removal showed total functional recovery (recovery score mean = 0.49 ± 0.11) (Fig 6D). These results demonstrate the capability of our system to detect and quantify changes in NMJ function, and to model human diseases in vitro.
Figure 6. Myasthenia Gravis effect on NMJ function.
(A-C) Contractility traces for tissue engineered NMJs using myotubes and MNs generated from hSkM in a microfluidic platform (as depicted in Fig 1) (A) before and (B) after 48 hours of treatment with 20% of MG sera and (C) 48 hours after its removal. (D) Quantification of NMJ function of treated and control groups before and after treatment and post-recovery, (n = 12; after treatment post-hoc ANOVA F = 0.0002; *indicates p = 0.0015 **indicates p = 3.5·10−6) (MG = Myasthenia Gravis; NHS = normal human serum). (E) Effect of sera from different patients (A, B, C) on tissue function (n = 15; after treatment post-hoc ANOVA F = 2·10−5; * indicates p< 0.01 vs control group). Treatment was started on day 14 with sequential dose increments at days 16, 18 and 20 (F) Effect of IgG isolated from seronegative patients on tissue function (SN = seronegative, SP = seropositive), (n=8, *indicates p = 0.02). Boxes= 25–75 percentiles; brackets = 1.5 standard deviations. n indicates the number of biological replicates.
To demonstrate the potential of our system as a diagnosis and evaluation tool for MG and other neuromuscular diseases, we tested individual serum from 3 different patients at increasing doses. Our results show that the system is capable of detecting changes in NMJ function at doses as low as 0.1% for one of the patients, whereas it was necessary to increase the serum concentration by 20 times to elicit an observable effect for the other two patients (Fig 6E). These results prove that our system can detect differential effects from different patients and suggest that determining the lowest dilution at which the serum has an effect on the NMJ function could be a quantifiable strategy to evaluate the severity of the disease.
Finally, we assessed the potential of our strategy as a diagnostic tool for double-seronegative MG patients, a subpopulation of MG patients that present with typical symptoms of the disease without detectable antibodies to the nAChR or to muscle-specific tyrosine kinase (MuSK). Through the Columbia Neurological Institute, we obtained sera from two double-seronegative MG donors. We didn’t observe a significant decrease in NMJ function upon addition of 20% seronegative sera to the media, presumably because the concentration of antibodies present was not high enough, so we proceeded to concentrate the immunoglobulin (IgG) fraction of the sera samples. Treatment with concentrated IgGs equivalent to adding 40% serum to the media resulted in a 27% decrease of function in tissues treated with sera from one of the patients (SN#2, score normalized to day 0 for control = 1.14 ± 0.08, seronegative patient 2 = 0.83 ± 0.06, p = 0.017), whereas tissues treated with sera from the other patient (SN#1) showed no difference with the non-treated control (seronegative patient 1 = 0.83±0.06), (Fig 6F). The IgG fraction isolated from a seropositive patient was used as a positive control and resulted in complete disruption of the NMJ function (seropositive patient = 0.006 ± 0.06, p = 2×10−6).
DISCUSSION
We report on a platform for high-throughput, automated analysis of NMJ function in an all-human patient-specific model. Previously, quantification of NMJ function proved to be labor-intensive, time-consuming, and inconsistent. The combination of scientific and technological advancements in cell line derivation, differentiation, bioreactor design, tissue engineering, optoelectronic hardware, and automated analysis, enabled our improved NMJ model to yield reproducible assays of many samples in parallel for practical applications in disease modeling and drug testing.
We describe two strategies for the derivation of cells from a single donor in order to generate patient-specific NMJs. The first strategy relies on reprogramming primary skeletal myoblasts into hiPSCs, which can then be subsequently differentiated into photosensitive motoneurons. This method results in robust and fully functional tissues because it uses primary skeletal myoblasts, which are ideal for the study of healthy NMJs. However, it requires an of skeletal muscle biopsy (an invasive procedure), and its limited by the growth potential of primary myoblasts ex vivo. The second strategy is to directly differentiate motoneurons and myotubes from hiPSCs. While we have incorporated transcription-factor differentiation techniques into our platform, it still warrants further optimization for long-term success. The transcription factor approach is inherently more scalable given the existence of large hiPSC biobanks and effectively indefinite self-renewal potential of stem cells. Furthermore, generation of de novo iPSC lines can be achieved with less invasive sampling of skin fibroblasts or blood samples, instead of skeletal muscle biopsies. For the MG experiments presented in this paper, we used with primary skeletal myoblasts from healthy patients since the phatophysiology of MG suggests it is an acquired disease with normal muscle at baseline.
In both the microfluidic and open-well platforms described, separate chambers are connected by a channel for axonal growth and sprouting, to mimic the native human NMJ physiology. Compartmentalization allows for defined microenvironments and independent manipulations of each chamber while the three-dimensional nature of the model allows for improved muscle architecture and increased contractile function compared to two-dimensional systems [40]. Compared to the open-well design, the microfluidic platform requires fewer cells per sample and smaller amounts of incubation media, thus being more amenable to higher throughput and more cost-efficient experiments. However, the open-well format is much easier to retrieve samples for terminal analysis and provides greater tissue mass per sample for analysis. The two systems are highly complementary and can be selected based on the chosen application. For example, in the case of the MG assays, we used the microfluidic platform as the patient sera was a limiting resource for our experiments.
Although previous NMJ models have used compartmentalization to selectively stimulate motoneurons or skeletal muscle [41], optogenetics allows for greater versatility by easily targeting specific cell types in culture when compared to traditional electrical or chemical stimulation strategies. In particular, the genotype-specific spatiotemporal control offered by optogenetics is easily incorporated into hiPSC-based models, and simultaneously simplifies bioreactor design given the lack of electrodes. We have further utilized the potential of these models to combine optogenetic motoneurons with automated microscopy, standardized stimulation protocols, and computer vision. In comparison to previous optogenetic models [38,22,23,39], our system is much more amenable to the parallel workflows necessary for the high-throughput assays typically required for disease modeling and drug development. Furthermore, while our software is programmed to measure motion, it can be easily adapted to utilize strain [42] or other optically measured parameters.
To demonstrate the capabilities of our system, we were able to quantify dose-dependent and patient-specific changes in individual NMJs before and after exposure to MG serum with a high degree of sensitivity, using clinically relevant MG sera concentrations. Furthermore, we were able to quantify a decrease in NMJ function for double-seronegative MG, which has been undetectable through existing diagnostics due to a lack of specific biomarkers, a problem which has hindered the diagnosis and clinical management of these patients [43,44]. By interrogating NMJ function rather than serum biomarkers, our platform provides a promising approach for the diagnosis of this specific subtype of MG, as well as for other diseases that cannot be diagnosed through existing methods.
One avenue for future development of our system would be the incorporation of additional cell types. Incorporating glial cells known to be crucial in the development and health of the NMJ (e.g., Schwann cells), or other neuronal circuit components (e.g., interneurons and sensory neurons), will increase the physiologic fidelity of our system and allow for more complex modeling (e.g. neuronal synaptogenesis, reflex arcs). Incorporating non-neuronal cell types can also be used to study the autonomic nervous system (e.g. parasympathetic innervation of cardiac nodal cells).
In parallel with increased cellular diversity, our system can be improved through the use of multicolor optogenetic actuation [45] or the incorporation of optogenetic sensors for voltage/calcium imaging [46], or probes for muscle metabolism such as the ATP-FRET sensor [47]. This would provide the ability to precisely stimulate and record various physiologic activities from multiple cell types in the circuit. Likewise, advanced optical tools, such as lasers and micro-mirror arrays could be used to stimulate specific cells or regions with different wavelengths using more complex patterns [48,49]. One example would be a NMJ chip where both the motoneurons and skeletal muscle are optogenetic, and selective activation of one or the other can be achieved by spatial control of light stimulation. Such platform would expand the capabilities of the current system by allowing us to distinguish between muscle weakness caused due to NMJ disruption or direct muscle damage.
A combination of these additions would allow for specific and targeted perturbations of individual cell populations contributing to the complex neuronal circuit, as well as for multi-modal downstream analysis of the effects of these perturbations, ultimately providing a platform to dissect the functional contributions of each synaptic partner in a well-defined and systematic manner.
In summary, this platform provides a reproducible and versatile method for studying the human NMJ and can be used to study both healthy and diseased NMJs in a patient-specific manner. It is sensitive enough to detect dose-dependent effects of patient serum in MG and can be similarly applied to drug development. Our system has the advantages of simplicity and scalability, and lays the groundwork for more complex systems to address future applications in personalized medicine.
Supplementary Material
Movie 1. Automated stimulation and video acquisition. The custom script moves the automated stage moves to the specified coordinates for each tissues, and then starts the video acquisition and the Arduino program that controls the optical set up. Once the stimulation is done, the script moves to the next tissue until the whole plate has been recorded.
Movie 2. Automated stimulation and evaluation of NMJ function. Movie and contractility analysis of functional neuromuscular tissue comprising iPSC-derived motoneurons and a skeletal tissue derived from primary myoblasts seeded in an open-well bioreactor system. Blue blinker indicates blue light pulses (50 ms). Graph correlates light pulses (vertical lines) with tissue contractions (black trace). The ratio of effective pulses vs total number of pulses is used to score the tissues, in this case score = 1. Movie was taken at day 13 of the coculture. Movie correspond to traces shown in Figure 5B.
Movie 3. Automated stimulation and evaluation of NMJ function in a iPSC-derived skeletal tissue. Movie and contractility analysis of functional neuromuscular tissue comprising motoneurons and a skeletal myoblasts derived from the same iPSC line using transcription-factor mediated differentiations in an open-well bioreactor system. Movie was taken at day 37 of the coculture. The resulting movie score was 0.84.
Movie 4. Automated analysis of NMJ function before MG treatment. Movie and analysis of a representative tissue before treatment with 20% sera from myasthenia gravis patients (day 15 of coculture). The neuromuscular tissue comprises iPSC-derived motoneurons and a skeletal tissue derived from primary myoblasts seeded in a microfluidic platform. Score = 0.78. Movie correspond to traces shown in Figure 6A.
Movie 5. Automated analysis of NMJ function after MG treatment. Movie and analysis of the tissue in Movie 4 after 48h treatment with 20% sera from myasthenia gravis patients shows impaired NMJ function (score = 0.00) (day 17 of coculture). Movie correspond to traces shown in Figure 6B.
Movie 6. Automated analysis of NMJ function after recovery from MG treatment. Movie and analysis of the tissue in Movies 4 & 5 taken 48h after wash-out of MG sera (day 19 of coculture) shows recovery of function (score = 1). Movie correspond to traces shown in Figure 6C.
ACKNOWLEDGMENTS
We gratefully acknowledge funding support by the NIH [grant numbers EB025765, EB027062 and U01ES032673], DOD [award number W81XWH-18-1-0095], and the Roddenberry Foundation and the UCSF Health Innovation via Engineering (HIVE Fellowship). We gratefully acknowledge the Columbia Stem Cell Core for their help and guidance with cell reprogramming and Trevor Nash for his help with the microscope scripts.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
GVN is a co-founder of Tara Biosystems, a Columbia University start-up that is commercializing “heart on a chip” platform for drug discovery. She holds equity, serves on the Board of Directors, is an inventor on licensed by Tara, and has received financial compensation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY
The authors confirm that all data obtained in this study are included in the manuscript and supplemental information.
REFERENCES
- [1].Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S, Induction of pluripotent stem cells from adult human fibroblasts by defined factors., Cell. 131 (2007) 861–872. 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- [2].Maury Y, Côme J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, Peschanski M, Martinat C, Nedelec S, Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes., Nat Biotechnol. 33 (2015) 89–96. 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- [3].Bianchi F, Malboubi M, Li Y, George JH, Jerusalem A, Szele F, Thompson MS, Ye H, Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling., Stem Cell Res. 32 (2018) 126–134. 10.1016/j.scr.2018.09.006. [DOI] [PubMed] [Google Scholar]
- [4].Rao L, Qian Y, Khodabukus A, Ribar T, Bursac N, Engineering human pluripotent stem cells into a functional skeletal muscle tissue, Nature Communications. 9 (2018) 1–12. 10.1038/s41467-017-02636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N, Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs, ELife. 4 (2015) e04885. 10.7554/eLife.04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors., Cell. 126 (2006) 663–676. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- [7].Kim JS, Choi HW, Choi S, Do JT, Reprogrammed pluripotent stem cells from somatic cells, Int J Stem Cells. 4 (2011) 1–8. 10.15283/ijsc.2011.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Turan S, Farruggio AP, Srifa W, Day JW, Calos MP, Precise Correction of Disease Mutations in Induced Pluripotent Stem Cells Derived From Patients With Limb Girdle Muscular Dystrophy, Molecular Therapy. 24 (2016) 685–696. 10.1038/mt.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lodi R, Schapira AH, Manners D, Styles P, Wood NW, Taylor DJ, Warner TT, Abnormal in vivo skeletal muscle energy metabolism in Huntington’s disease and dentatorubropallidoluysian atrophy., Ann Neurol. 48 (2000) 72–76. [PubMed] [Google Scholar]
- [10].Ebert AD, Liang P, Wu JC, Induced pluripotent stem cells as a disease modeling and drug screening platform, J Cardiovasc Pharmacol. 60 (2012) 408–416. 10.1097/FJC.0b013e318247f642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lin C-Y, Yoshida M, Li L-T, Ikenaka A, Oshima S, Nakagawa K, Sakurai H, Matsui E, Nakahata T, Saito MK, iPSC-derived functional human neuromuscular junctions model the pathophysiology of neuromuscular diseases, JCI Insight. 4 (2019) e124299. 10.1172/jci.insight.124299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Centeno EGZ, Cimarosti H, Bithell A, 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling, Molecular Neurodegeneration. 13 (2018) 27. 10.1186/s13024-018-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N, Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs, ELife Sciences. 4 (2015) e04885. 10.7554/eLife.04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Okano T, Matsuda T, Tissue Engineered Skeletal Muscle: Preparation of Highly Dense, Highly Oriented Hybrid Muscular Tissues, Cell Transplantation. 7 (1998) 71–82. 10.1016/S0963-6897(97)00067-5. [DOI] [PubMed] [Google Scholar]
- [15].Powell CA, Smiley BL, Mills J, Vandenburgh HH, Mechanical stimulation improves tissue-engineered human skeletal muscle, American Journal of Physiology-Cell Physiology. 283 (2002) C1557–C1565. 10.1152/ajpcell.00595.2001. [DOI] [PubMed] [Google Scholar]
- [16].Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G, Advanced maturation of human cardiac tissue grown from pluripotent stem cells, Nature. 556 (2018) 239–243. 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thavandiran N, Dubois N, Mikryukov A, Massé S, Beca B, Simmons CA, Deshpande VS, McGarry JP, Chen CS, Nanthakumar K, Keller GM, Radisic M, Zandstra PW, Design and formulation of functional pluripotent stem cell-derived cardiac microtissues, PNAS. 110 (2013) E4698–E4707. 10.1073/pnas.1311120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vila OF, Uzel SGM, Ma SP, Williams D, Pak J, Kamm RD, Vunjak-Novakovic G, Quantification of human neuromuscular function through optogenetics, Theranostics. 9 (2019) 1232–1246. 10.7150/thno.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K, Millisecond-timescale, genetically targeted optical control of neural activity, Nat Neurosci. 8 (2005) 1263–1268. 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- [20].Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E, Channelrhodopsin-2, a directly light-gated cation-selective membrane channel, Proc Natl Acad Sci USA. 100 (2003) 13940. 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Phillips LH, The epidemiology of myasthenia gravis, Ann. N. Y. Acad. Sci. 998 (2003) 407–412. [DOI] [PubMed] [Google Scholar]
- [22].Uzel SGM, Platt RJ, Subramanian V, Pearl TM, Rowlands CJ, Chan V, Boyer LA, So PTC, Kamm RD, Microfluidic device for the formation of optically excitable, three-dimensional, compartmentalized motor units, Sci Adv. 2 (2016). 10.1126/sciadv.1501429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Osaki T, Uzel SGM, Kamm RD, Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons, Sci Adv. 4 (2018) eaat5847. 10.1126/sciadv.aat5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guo X, Greene K, Akanda N, Smith A, Stancescu M, Lambert S, Vandenburgh H, Hickman J, In vitro Differentiation of Functional Human Skeletal Myotubes in a Defined System, Biomater Sci. 2 (2014) 131–138. 10.1039/C3BM60166H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Akiyama T, Sato S, Chikazawa-Nohtomi N, Soma A, Kimura H, Wakabayashi S, Ko SBH, Ko MSH, Efficient differentiation of human pluripotent stem cells into skeletal muscle cells by combining RNA-based MYOD1-expression and POU5F1-silencing, Sci Rep. 8 (2018). 10.1038/s41598-017-19114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu W, Hill SE, Nathan WJ, Paiano J, Callen E, Wang D, Shinoda K, van Wietmarschen N, Colón-Mercado JM, Zong D, De Pace R, Shih H-Y, Coon S, Parsadanian M, Pavani R, Hanzlikova H, Park S, Jung SK, McHugh PJ, Canela A, Chen C, Casellas R, Caldecott KW, Ward ME, Nussenzweig A, Neuronal enhancers are hotspots for DNA single-strand break repair, Nature. 593 (2021) 440–444. 10.1038/s41586-021-03468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Skipper KA, Hollensen AK, Antoniou MN, Mikkelsen JG, Sustained transgene expression from sleeping beauty DNA transposons containing a core fragment of the HNRPA2B1-CBX3 ubiquitous chromatin opening element (UCOE), BMC Biotechnol. 19 (2019) 75. 10.1186/s12896-019-0570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fernandopulle MS, Prestil R, Grunseich C, Wang C, Gan L, Ward ME, Transcription Factor–Mediated Differentiation of Human iPSCs into Neurons, Current Protocols in Cell Biology. 79 (2018) e51. 10.1002/cpcb.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davis RL, Weintraub H, Lassar AB, Expression of a single transfected cDNA converts fibroblasts to myoblasts, Cell. 51 (1987) 987–1000. 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- [30].Tanaka A, Woltjen K, Miyake K, Hotta A, Ikeya M, Yamamoto T, Nishino T, Shoji E, Sehara-Fujisawa A, Manabe Y, Fujii N, Hanaoka K, Era T, Yamashita S, Isobe K, Kimura E, Sakurai H, Efficient and Reproducible Myogenic Differentiation from Human iPS Cells: Prospects for Modeling Miyoshi Myopathy In Vitro, PLoS ONE. 8 (2013) e61540. 10.1371/journal.pone.0061540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Albini S, Coutinho P, Malecova B, Giordani L, Savchenko A, Forcales SV, Puri PL, Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres, Cell Rep. 3 (2013) 661–670. 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Selvaraj S, Mondragon-Gonzalez R, Xu B, Magli A, Kim H, Lainé J, Kiley J, Mckee H, Rinaldi F, Aho J, Tabti N, Shen W, Perlingeiro RC, Screening identifies small molecules that enhance the maturation of human pluripotent stem cell-derived myotubes, ELife. 8 (2019) e47970. 10.7554/eLife.47970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Südhof TC, Rapid single-step induction of functional neurons from human pluripotent stem cells, Neuron. 78 (2013) 785–798. 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Busskamp V, Lewis NE, Guye P, Ng AHM, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, Stadler M, Weiss R, Church GM, Rapid neurogenesis through transcriptional activation in human stem cells, Mol Syst Biol. 10 (2014) 760. 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sanders DB, Burns TM, Cutter GR, Massey JM, Juel VC, Hobson-Webb L, Muscle Study Group, Does change in acetylcholine receptor antibody level correlate with clinical change in myasthenia gravis?, Muscle Nerve. 49 (2014) 483–486. 10.1002/mus.23944. [DOI] [PubMed] [Google Scholar]
- [36].Vernino S, Unraveling the Enigma of Seronegative Myasthenia Gravis, JAMA Neurol. 72 (2015) 630–631. 10.1001/jamaneurol.2015.0205. [DOI] [PubMed] [Google Scholar]
- [37].Tim RW, Sanders DB, Repetitive nerve stimulation studies in the Lambert-Eaton myasthenic syndrome, Muscle Nerve. 17 (1994) 995–1001. 10.1002/mus.880170906. [DOI] [PubMed] [Google Scholar]
- [38].Steinbeck JA, Jaiswal MK, Calder EL, Kishinevsky S, Weishaupt A, Toyka KV, Goldstein PA, Studer L, Functional Connectivity under Optogenetic Control Allows Modeling of Human Neuromuscular Disease, Cell Stem Cell. 18 (2016) 134–143. 10.1016/j.stem.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Afshar Bakooshli M, Lippmann ES, Mulcahy B, Iyer N, Nguyen CT, Tung K, Stewart BA, van den Dorpel H, Fuehrmann T, Shoichet M, Bigot A, Pegoraro E, Ahn H, Ginsberg H, Zhen M, Ashton RS, Gilbert PM, A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction, Elife. 8 (2019). 10.7554/eLife.44530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Machado CB, Pluchon P, Harley P, Rigby M, Sabater VG, Stevenson DC, Hynes S, Lowe A, Burrone J, Viasnoff V, Lieberam I, In Vitro Modeling of Nerve–Muscle Connectivity in a Compartmentalized Tissue Culture Device, Advanced Biosystems. 3 (2019) 1800307. 10.1002/adbi.201800307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Santhanam N, Kumanchik L, Guo X, Sommerhage F, Cai Y, Jackson M, Martin C, Saad G, McAleer CW, Wang Y, Lavado A, Long CJ, Hickman JJ, Stem cell derived phenotypic human neuromuscular junction model for dose response evaluation of therapeutics, Biomaterials. 166 (2018) 64–78. 10.1016/j.biomaterials.2018.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Czirok A, Isai DG, Kosa E, Rajasingh S, Kinsey W, Neufeld Z, Rajasingh J, Optical-flow based non-invasive analysis of cardiomyocyte contractility, Sci Rep. 7 (2017). 10.1038/s41598-017-10094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zisimopoulou P, Brenner T, Trakas N, Tzartos SJ, Serological diagnostics in myasthenia gravis based on novel assays and recently identified antigens, Autoimmunity Reviews. 12 (2013) 924–930. 10.1016/j.autrev.2013.03.002. [DOI] [PubMed] [Google Scholar]
- [44].Berrih-Aknin S, Frenkian-Cuvelier M, Eymard B, Diagnostic and clinical classification of autoimmune myasthenia gravis, Journal of Autoimmunity. 48–49 (2014) 143–148. 10.1016/j.jaut.2014.01.003. [DOI] [PubMed] [Google Scholar]
- [45].Bugaj LJ, Lim WA, High-throughput multicolor optogenetics in microwell plates, Nature Protocols. 14 (2019) 2205–2228. 10.1038/s41596-019-0178-y. [DOI] [PubMed] [Google Scholar]
- [46].Nguyen C, Upadhyay H, Murphy M, Borja G, Rozsahegyi EJ, Barnett A, Brookings T, McManus OB, Werley CA, Simultaneous voltage and calcium imaging and optogenetic stimulation with high sensitivity and a wide field of view, Biomed Opt Express. 10 (2019) 789–806. 10.1364/BOE.10.000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Depaoli MR, Karsten F, Madreiter-Sokolowski CT, Klec C, Gottschalk B, Bischof H, Eroglu E, Waldeck-Weiermair M, Simmen T, Graier WF, Malli R, Real-Time Imaging of Mitochondrial ATP Dynamics Reveals the Metabolic Setting of Single Cells, Cell Reports. 25 (2018) 501–512.e3. 10.1016/j.celrep.2018.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tsuda S, Kee MZL, Cunha C, Kim J, Yan P, Loew LM, Augustine GJ, Probing the function of neuronal populations: Combining micromirror-based optogenetic photostimulation with voltage-sensitive dye imaging, Neuroscience Research. 75 (2013) 76–81. 10.1016/j.neures.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Arrenberg AB, Stainier DYR, Baier H, Huisken J, Optogenetic Control of Cardiac Function, Science. 330 (2010) 971–974. 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Automated stimulation and video acquisition. The custom script moves the automated stage moves to the specified coordinates for each tissues, and then starts the video acquisition and the Arduino program that controls the optical set up. Once the stimulation is done, the script moves to the next tissue until the whole plate has been recorded.
Movie 2. Automated stimulation and evaluation of NMJ function. Movie and contractility analysis of functional neuromuscular tissue comprising iPSC-derived motoneurons and a skeletal tissue derived from primary myoblasts seeded in an open-well bioreactor system. Blue blinker indicates blue light pulses (50 ms). Graph correlates light pulses (vertical lines) with tissue contractions (black trace). The ratio of effective pulses vs total number of pulses is used to score the tissues, in this case score = 1. Movie was taken at day 13 of the coculture. Movie correspond to traces shown in Figure 5B.
Movie 3. Automated stimulation and evaluation of NMJ function in a iPSC-derived skeletal tissue. Movie and contractility analysis of functional neuromuscular tissue comprising motoneurons and a skeletal myoblasts derived from the same iPSC line using transcription-factor mediated differentiations in an open-well bioreactor system. Movie was taken at day 37 of the coculture. The resulting movie score was 0.84.
Movie 4. Automated analysis of NMJ function before MG treatment. Movie and analysis of a representative tissue before treatment with 20% sera from myasthenia gravis patients (day 15 of coculture). The neuromuscular tissue comprises iPSC-derived motoneurons and a skeletal tissue derived from primary myoblasts seeded in a microfluidic platform. Score = 0.78. Movie correspond to traces shown in Figure 6A.
Movie 5. Automated analysis of NMJ function after MG treatment. Movie and analysis of the tissue in Movie 4 after 48h treatment with 20% sera from myasthenia gravis patients shows impaired NMJ function (score = 0.00) (day 17 of coculture). Movie correspond to traces shown in Figure 6B.
Movie 6. Automated analysis of NMJ function after recovery from MG treatment. Movie and analysis of the tissue in Movies 4 & 5 taken 48h after wash-out of MG sera (day 19 of coculture) shows recovery of function (score = 1). Movie correspond to traces shown in Figure 6C.
Data Availability Statement
The authors confirm that all data obtained in this study are included in the manuscript and supplemental information.