Summary
Protein-tyrosine phosphatases (PTPs) are pleomorphic regulators of eukaryotic cellular responses to extracellular signals that function by modulating the phosphotyrosine of specific proteins. A handful of PTPs have been implicated in germline and somatic human disease. Using exome sequencing, we identified missense and truncating variants in PTPN4 in six unrelated individuals with varying degrees of intellectual disability or developmental delay. The variants occurred de novo in all five subjects in whom segregation analysis was possible. Recurring features include postnatal growth deficiency or excess, seizures, and, less commonly, structural CNS, heart, or skeletal anomalies. PTPN4 is a widely expressed protein tyrosine phosphatase that regulates neuronal cell homeostasis by protecting neurons against apoptosis. We suggest that pathogenic variants in PTPN4 confer risk for growth and cognitive abnormalities in humans.
Keywords: Neurodevelopment, Developmental Delay, Intellectual Disability, Macrocephaly, Somatic growth anomaly, PTPN4, Protein tyrosine phosphatase
Using exome and targeted sequencing, we identified six primarily de novo PTPN4 variants and report an associated phenotype that converges on neurodevelopment and growth. Conservation at variant loci combined with analysis of variant effect on protein trafficking and catalytic activity for selected variants establish a distinct PTPN4-related developmental disorder.
Introduction
Protein-tyrosine phosphatases (PTPs) represent the largest family of human phosphatases.1 PTPs are signaling molecules that regulate a variety of cellular processes, including proliferation, differentiation, and oncogenic transformation.2 Protein-tyrosine phosphatase, non-receptor-type, 4 (PTPN4 [MIM: 176878]) encodes a non-receptor tyrosine phosphatase that in neurons interacts with glutamate receptor delta 2 and epsilon subunits and mediates signaling downstream of the glutamate receptors through tyrosine dephosphorylation.3 PTPN4 is broadly expressed, with highest expression in human neurons and fetal astrocytes, as well as thyroid.3, 4, 5, 6, 7, 8 Genome Aggregation Database (gnomAD) data indicate that PTPN4 has a high probability of loss-of-function (LoF) intolerance with a loss-of-function observed/expected upper bound fraction (LOEUF) of 0.28, indicating that no more than 28% of the expected loss-of-function variants were observed and therefore PTPN4 is likely under selection against loss-of-function variants.9
Here, we describe six individuals with neurodevelopmental phenotypes and growth or morphological anomalies who carry rare, putatively deleterious sequence variants in PTPN4. Clinical data were obtained through written informed consent. In all cases, the procedures followed were in accordance with the ethical standards of the respective institution’s committee on human research and were in keeping with national standards. PTPN4 sequence variants were identified in all of these individuals by clinically based exome sequencing (except subject 2 who had a targeted gene panel) performed in CLIA- or ISO15189-certified laboratories and confirmed by Sanger sequencing. The clinical phenotypes of these individuals and a description of their PTPN4 sequence changes—including in silico prediction of effects on protein function via PolyPhen-2,9 SIFT,10 and MutationTaster11 and allele frequencies reported in the gnomAD, ClinVar, and Human Gene Mutation Database (HGMD)—are summarized in Table S1.
Material and methods
Conservation analysis
The protein sequences for 300 unique PTPN4 orthologs (Vertebrata) were downloaded from the NCBI Ortholog database entry for PTPN4. The sequences were aligned in Geneious Prime 2020.2.2 (Biomatters, Auckland, New Zealand) using the Clustal Omega function to calculate the amino acid conservation at each position (% identity based on frequency).
Biochemical analysis
Clone preparation
DNA sequence corresponding to amino acids (aa) 611–926 of PTPN4 was amplified via PCR from cDNA clone ID BC010674. PCR product was purified using NucleoSpin PCR clean-up kit (Clontech) and cloned into pET24-based vector using InFusion HD cloning kit (Clontech). Transformations and plasmid isolations were performed using standard protocols. Resulting PTPN4 expression construct contained N-terminal 6× his tag followed by TEV protease cleavage site.
PTPN4 mutagenesis
p.Arg808His variant was introduced in the wild-type (WT) construct via PCR using InFusion HD kit. PCR primers encoded mutated DNA sequences. Variants were verified by DNA sequencing.
PTPN4 WT and p.Arg808His purification
Rosetta2 cells were transformed with WT or p.Arg808His encoding construct of PTPN4. Overnight starter cultures were prepared by inoculating 100 mL of Luria-Bertani (LB) media (Thermo Fisher Scientific) with several colonies and grown overnight at 37°C. Large-scale cultures were grown in 2× TY media at 37°C until optical density (OD) reached ∼0.6 and then induced with 0.25 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown at 18°C overnight. The cells were harvested by centrifugation and lysed in buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 10% glycerol, 0.25 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP,Sigma-Aldrich), 5 mM MgCl2, and protease inhibitor cocktail. PTPN4s were purified using Roche cOmplete Ni-NTA resin using manufacturer’s instructions. Proteins were further purified using Superdex 75 (GE Healthcare) size exclusion column in the final formulation buffer (50 mM HEPES [pH 7.5], 10% glycerol, 500 mM NaCl, 0.25 mM TCEP).
PTPN4 p.Arg808His purification
Arctic Express cells were transformed with p.Arg808His encoding construct of PTPN4. Overnight starter culture was prepared by inoculating 100 mL of LB media with several colonies and grown O/N at 37°C. Large-scale culture was grown in Terrific Broth (TB) media (Sigma-Aldrich) at 37°C until OD reached ∼1, then induced with 0.25 mM IPTG and grown at 12°C for 24 h. The cells were harvested by centrifugation and lysed in buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 10% glycerol, 0.25 mM TCEP, 5 mM MgCl2, and benzamidine protease inhibitor cocktail. PTPN4 was purified using Roche cOmplete NiNTA resin, employing a 2 M urea wash to remove excess chaperonin protein. Protein was further purified using Superdex 75 (GE Healthcare) size exclusion column in the final formulation buffer (50 mM HEPES [pH 7.5], 10% glycerol, 500 mM NaCl, 0.25 mM TCEP).
Kinetics experiments
Kinetics experiments were carried out on a BMG Pherastar FS (BMG Labtech). Absorbance values were measured at 405 nm at 25°C for each time point. Experiments were performed by mixing increasing concentrations of each of the PTPN4 variants, ranging from 100 nM top concentration with two-fold serial dilution versus a fixed concentration of pNPP (5 mM) in a 50 μL reaction volume, with a 10 μL 1 N NaOH stop reaction. The buffer was phosphate-buffered saline (PBS), 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl. Initial linear reaction rates were calculated during a 20–60 min reaction. The enzyme kinetics were deduced from fitting the Michaelis-Menten equation with the GraphPad Prism software.
Circular dichroism
70 μg of WT and R808H PTPN4 were buffer exchanged into 20 mM sodium phosphate (pH 7.5), 150 mM NaCl and concentrated to ≤400 μL using Amicon centrifugal filters (Millipore). Note that TCEP was left out of the buffer to avoid any spectral artifacts due to oxidation of TCEP, which is unstable in phosphate buffers. The buffer-exchanged samples were spun at 10,000 rpm for 5 min at 4°C to pellet any aggregates. No pellet was observed. Circular dichroism measurements were taken using a Jasco J-815 spectrophotometer with an MPTC-490 6-cell 52 Peltier thermostatted cell holder. Spectra were recorded at 20°C from 190 to 250 nm at 0.1 nm intervals using a 1 mm pathlength quartz cuvette. The samples were scanned at 50 nm/min with a 1 nm bandwidth and a 4 s data integration time (DIT) four times and plotted as an average of all four reads. A spectrum of buffer alone was subtracted from the data. Thermal denaturation curves were collected by measuring the change in CD signal at 209 nm and 222 nm as a function of temperature from 20°C to 88°C with a temperature ramp rate of 1°C/min and a DIT of 4 s. WT data were collected every 1°C from 20°C–40°C and 65°C–88°C, and every 0.5°C from 40°C–65°C. R808H data were collected every 1°C from 20°C–35°C and 60°C–88°C, and every 0.5°C from 35°C–60°C. WT was measured at 0.175 mg/mL, and R808H was measured at 0.167 mg/mL. The CD signal in mdeg was buffer subtracted and converted to mean molar ellipticity per residue (MRE) using the following equation:
with molecular weight (M) in g/dmol, concentration (c) in g/mL, pathlength (l) in cm, number of residues (nr), and ellipticity (θdeg) in degrees. For the thermal denaturation curves, the MRE was converted to fraction denatured as follows. The pre-transition points (characteristic of the native state) were fitted to a linear curve and MRE values extrapolated from this and denoted yN. The post-transition points (characteristic of the denatured state) were fitted to a linear curve and MRE values extrapolated from this and denoted yD. MRE values (y) were converted to fraction denatured (fD) using the following equation: fD = (y − yN)/(yD − yN). The data were fit to a Boltzmann sigmoid using Prism to extract the temperature of unfolding (Tm).
Neuronal studies
PTPN4, PTPN4 p.Gly239Arg, and PTPN4 p.Arg808His cloning into the expression vectors
The synthetic human gene PTPN4 was assembled from synthetic oligonucleotides and/or PCR products by the GeneArt Gene Synthesis (Thermo Fisher Scientific) as reported previously.12 The single-nucleotide substitution variants PTPN4mutG715A (amino acid sequence PTPN4 p.Gly239Arg) and PTPN4mutG2423A (amino acid sequence PTPN4 p.Arg808His) were ordered from the GeneArt Gene Synthesis (Thermo Fisher Scientific) as the variants of PTPN4 sequence. The mutated genes were inserted into pMA cloning vectors. The final constructs were verified by sequencing. The PTPN4, PTPN4 p.Gly239Arg, and PTPN4 p.Arg808His sequences were next cloned separately into the pSyn-EGFP-N1 expression vector (with restriction enzymes SacI and AgeI flanking the desired sequences) for PTPN4-EGFP or PTPN4 p.Gly239Arg-EGFP or PTPN4 p.Arg808His-EGFP fusion protein expression in neurons. The ligation of 50 ng pSyn-EGFP-N1 vector with either 100 ng PTPN4 or 100 ng PTPN4 p.Gly239Arg or 100 ng PTPN4 p.Arg808His insert sequences was performed overnight at room temperature (RT) using 5 U ExpressLink T4 DNA ligase (Invitrogen). Next, 2 μL of ligation reaction was transformed into chemically competent Escherichia coli TOP10 strain, and the bacteria were plated onto LB agar plates containing 50 μg/mL kanamycin. The plasmid was isolated from 3 mL single-colony bacterial cultures using GenElute Plasmid Miniprep kit (Sigma-Aldrich) according to manufacturer’s protocol. The correct clones were selected by NheI and HindIII restriction enzyme digestion, and the sequence was verified by sequencing with the primers designed in Primer3Plus. Finally, QIAfilter Plasmid Midi Kit (QIAGEN) was used for purification and isolation of the larger amount of plasmid DNA.
Primary neuronal cultures
The culture of primary hippocampal neurons was set from postnatal day 0 Wistar rats of either sex. The hippocampi were dissected in ice-cold Hank’s balanced salt solution (HBSS, Sigma-Aldrich), washed 3 × 10 mL of ice-cold HBSS, and treated with 10 μL per one hippocampus of 2.5% trypsin enzyme (Gibco) in 4 mL HBSS for 20 min at 37°C water bath with occasional flicking. After 3 × 10 mL warm HBSS wash and dissociation, the neurons were plated at a density of 120,000 cells per 18 mm diameter coverslip (Assistent) coated with 50 μg/mL poly-D-lysine (Sigma-Aldrich) and 2.5 μg/mL laminin (Roche) in Neurobasal A medium (Gibco) supplemented with 2% B-27 (Gibco), 1% penicillin-streptomycin (Sigma-Aldrich), 2 mM GlutaMAX (Gibco), and 12.5 μM glutamate (Sigma-Aldrich). The cultures were developed in humidified 5% CO2 incubator at 37°C.
Transfection
The rat primary hippocampal neurons (DIV16) were transfected with pSyn-PTPN4-EGFP or pSyn-PTPN4, p.Gly239Arg-EGFP, or pSyn-PTPN4 p.Arg808His-EGFP expression vectors. Additionally, pSyn-EGFP plasmid was used as a control. Briefly, 1 μL Lipofectamine 3000 (Thermo Fisher Scientific) or 0.5 μg plasmid and 1 μL P300 Reagent were incubated separately with 150 μL Neurobasal A each for 5 min at RT. Next, the plasmid and transfecting reagent were mixed and incubated 20 min at RT. The transfecting mix was incubated with cells for 1 h and replaced with the conditioned medium. Following 48 h after transfection, the neurons were fixed with 4% paraformaldehyde and 4% sucrose in PBS for 7 min, washed 3 × 10 min with PBS, and subjected to immunostaining. The transfections were repeated on three independent neuronal cultures.
Immunofluorescence staining
The neurons were permeabilized using 0.1% Triton X-100 in PBS (PBST) for 10 min and blocked with 5% bovine serum albumin, 5% normal goat serum in PBST for 1 h at RT. Subsequently, the cells were incubated with primary antibodies in blocking solution overnight at 4°C. The antibodies used were chicken anti-GFP (1:500, ab13970, Abcam) to enhance the EGFP signal, mouse anti-PSD95 (1:500, MAB1598, Merck Millipore) to visualize glutamatergic synapses, and rabbit anti-MAP2 (1:500, #4542, Cell Signaling) to visualize dendrites. After washing 3 × 10 min with PBST, the cells were incubated with secondary antibodies (Alexa Fluor anti-chicken 488, anti-mouse 647, anti-rabbit 555, 1:1,000, Life Technologies) for 1 h at RT, washed 3 × 10 min with PBST, and mounted with DAPI Fluoromount-G (SouthernBiotech).
Imaging and quantification
All images were acquired using confocal microscope LSM 700, Axio Imager Z2 (Zeiss), using EC Plan-Neofluar 40×/1.30 Oil DIC M27 and Plan-Apochromat 63×/1.40 Oil DIC M27 objectives and Zen software (Zeiss). The images were captured by snap as well as in a stack of nine 0.3-μm-thick z sections (2.7 μm total range) by frame scan mode. For the whole neuron visualization, the 40× snap images are presented with the 0.8× zoom factor on frame size 1,024 × 1,024 (pixel size is 0.20 μm). For dendrite images, the maximum intensity projection pictures of 63× flattened z stacks are presented with 0.7× zoom factor on frame size 2,048 × 2,048 (pixel size equals 0.07 μm). The images were analyzed using Zen Blue software (Zeiss). For quantification, the number of EGFP-expressing dendritic spines was counted on 50 μm length of three secondary dendrites from 14 neurons (n = 42 dendrites per variant). The statistical analysis was performed using unpaired Student’s t test in GraphPad Prism version 7.03.
Results
Case summaries
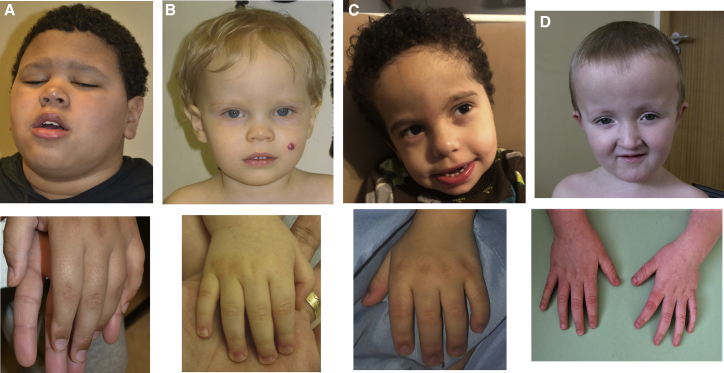
Subject 1 (Figure 1A) is an 11-year-old male of mixed European and African American descent with a de novo c.715G>A change in PTPN4 that results in a p.Gly239Arg amino acid change. This variant was predicted to be benign by PolyPhen-2, tolerated by SIFT, disease-causing by MutationTaster, and classified as a variant of uncertain significance (VUS) using Varsome10 following American College of Medical Genetics (ACMG) guidelines.11 His clinical features include severe developmental delay (DD), autism, limited speech, congenital dermal melanocytosis, several cafe-au-lait macules, and generalized overgrowth-obesity and macrocephaly (>99th percentile for height, weight and head circumference). From the age of 7 years, when consistent measurements were obtained, he was 88th percentile or greater for weight and height, with a body mass index (BMI) of 75th percentile or greater. He stood with his knees flexed and was unable actively to extend them fully and ambulated with a crouch gait with symmetrical internal rotation of the hips and bilateral hamstring tightness. Spine X-rays showed no notable bony abnormalities. A brain magnetic resonance imaging (MRI) scan obtained at 10 years of age was normal. Family history is significant for a maternal half-brother diagnosed with autism at age 14.
Figure 1.
Individuals with PTPN4 variants
(A) Subject 1 at 11 years of age. Macrocephaly (upper panel) and tapering fingers (lower panel). (B) Subject 2 at approximately 2 years showing macrocephaly with prominent forehead and facial hemangioma (upper panel) and tapering fingers with broad distal phalanges (lower panel). (C) Subject 3 at 4 years showing macrocephaly with tall, broad forehead (upper panel) and broad distal phalanges (lower panel). (D) Subject 4 at 4.5 years showing macrocephaly also with prominent forehead (upper panel) and tapering fingers (lower panel).
Subject 2 (Figure 1B) is a 23-month-old European male who carries a de novo c.191T>G (p.Leu64Trp) substitution in PTPN4 predicted to be probably damaging by PolyPhen-2, disease-causing by SIFT, damaging by MutationTaster, and classified as a VUS. His clinical features included speech and motor developmental delays, with axial and appendicular hypotonia with a wide-based gait. He had minor dysmorphic features as follows: prominent forehead, deep-set eyes with mildly upslanting palpebral fissures, a flat nasal bridge and wide nose, a wide philtrum, and thin upper lip with normal palate and dentition. In addition, he had a facial hemangioma, a single abdominal cafe au lait spot, and post-natal growth deficiency with relative macrocephaly (weight: 5th percentile, height: 2nd percentile, head circumference, 79th percentile). Brain MRI at 23 months of age demonstrated a small multiloculated cystic area in his right frontal lobe likely representing large perivascular spaces or a benign neuroepithelial or neuroglial cyst. He has no confirmed seizures and a negative electroencephalogram (EEG), although he has had some staring episodes. This subject also harbored a de novo c.655-2A>G:IVS5-2A>G variant in intron 5 of the Cullin 3 gene (CUL3 [MIM: 603136]). This variant is not found in gnomAD, HGMD, or ClinVar. Loss-of-function variants in CUL3 have been associated with autism13, 14, 15 and (in the case of exon 9 splicing defects) with pseudohypoaldosteronism, type IIE (MIM: 614496).16 The CUL3 variant in this subject was interpreted as a pathogenic variant by ACMG criteria. Neurobehavioral issues aside, however, CUL3 is not generally associated with growth anomalies in humans.
Subject 3 (Figure 1C) is a 3-year-old European/African American male who carries a de novo c.393_396del (p.Gln132ThrfsTer17) frameshift change in PTPN4. This change is predicted to severely truncate the 926-amino-acids-long protein causing haploinsufficiency and classified as a pathogenic variant by ACMG criteria. Pregnancy was the result of natural conception and there was no evidence of maternal diabetes mellitus or other suspected teratogen exposure. Chromosomal microarray performed on amniocytes suggested normal male pattern. He was evaluated at 20 months of age for multiple congenital anomalies including congenital heart defect (pulmonary valve stenosis, aortic valve insufficiency, large ventricular septal defect, and aberrant right subclavian artery), unilateral right-sided radial deficiency with oligodactyly (two digits present; Figure S1), and unilateral right talipes equinovarus as well as vertebral anomalies (thoracic region block vertebrae with rib fusions and an isolated left lumbosacral hemivertebra). Other features include developmental delay, post-natal growth deficiency (height < 1st percentile, weight 2nd percentile), and progressive macrocephaly (increase from 20th percentile at birth to >99th percentile at 16 months). Brain MRI obtained at 20 months revealed mildly diminished cerebral white matter volume with prominence of the ventricles and supratentorial extra-axial spaces. Renal ultrasound was normal, and there was no tracheoesophageal fistula or anal anomalies. A differential diagnosis of VACTERL association (vertebra-anal-cardiac-tracheoesophageal fistula-renal-limb; MIM: 192350) was considered. CHARGE syndrome (MIM: 214800) was also considered; however, ophthalmologic examination was normal and Chromodomain helicase DNA-binding protein 7 (CHD7 [MIM: 608892]) gene analysis was normal. Split hand/foot malformation (SHFM) with long bone deficiency (SHFLD) (MIM: 119100) was previously noted in association with 2q14.2 chromosomal breakpoints without localization of the responsible gene(s).17,18 Subject 3’s limb defects (Figure S1), taken in isolation, could lie within the heterogeneous SFHM spectrum.
Subject 4 (Figure 1D) is a 19-year-old male of European descent who carries a de novo heterozygous c.2171T>C (p.Ile724Thr) substitution in PTPN4 that was found to be possibly damaging by PolyPhen-2, damaging by SIFT, disease causing by MutationTaster, and classified as a VUS by ACMG criteria. Birth weight was measured at the 65th percentile, with a head circumference at the 99th percentile (+2.5 SD). Childhood height generally tracked the mid-percentile range (71st percentile at 14 years), and head circumference remained at the top of the range, measuring at 99th percentile (+2.6 SD) at age 14 years. His clinical features included moderate intellectual disability (ID), attention deficit disorder (ADHD), autism spectrum disorder (ASD), astigmatism, hyperacusis, joint laxity, and general unsteadiness, but no seizures. Primary dentition was extracted at age 3 years due to decay. A brain MRI scan at 3.5 years of age revealed a large cavum septum pellucidum and vergae, a slight reduction in white matter around the posterior horns, and associated thinning of this region of the corpus callosum. He had an early working diagnosis of Sotos syndrome I (MIM: 117550). Nuclear-receptor-binding set domain protein 1 (NSD1 [MIM: 606681]) testing at age 4.5 years identified neither a pathogenic variant nor copy-number variant (CNV) at this locus.
Subject 5 is a 15-year-old female of Asian descent who carries a c.2512 C>T (p.Arg838Ter) change in PTPN4. This variant was present in gnomAD in 1 person (1/251,432) and dbSNP (rs1259252500; frequency T = 0.000004, derived from gnomAD data above), but was absent from ClinVar. Parental DNA samples could not be obtained for testing. The variant was predicted damaging by SIFT and disease causing by MutationTaster. Symptoms included borderline intellectual functioning with a full-scale IQ of 71, motor tics, ADHD, and recurrent absence seizures. EEG showed generalized spike wave discharges. A brain MRI at age 8 years was normal; however, spinal MRI showed a diastematomyelia associated with a lipoma from L3–L4 extending into her sacrum, and L5 butterfly vertebrae. Her height was 30th percentile, weight 63rd percentile, head circumference 7th percentile (consistent with relative microcephaly). Besides hypermetropia and astigmatism, she had eye movement abnormalities.
Subject 6, a 17-year-old male of European descent, carries a de novo c.1738G>T (p.Asp580Tyr) variant in PTPN4 that was found to be probably damaging by PolyPhen-2, damaging by SIFT, disease causing by MutationTaster, and classified as a VUS by ACMG criteria. His clinical features included mild intellectual disability, ADHD, and growth abnormalities (weight > 99th percentile, height 2nd percentile, occipitofrontal circumference [OFC] 98th percentile, consistent with relative macrocephaly). Seizures started at 17 months of age. EEG showed numerous paroxysmal generalized bursts of spikes-waves. The epilepsy was treatment resistant until the age of 10 years, when treatment with zonisamide was initiated. There was a family history of paternal childhood seizures and a 14-year-old male sibling with ASD and rare fever-induced seizures between 1 month to 7 years of age.
In addition to these six primary subjects, other subjects with PTPN4 variants either (1) of unclear significance, or (2) culled from the literature are included in Table S1 for comparison as follows (subjects A–D, in red text):
Subject A, a female of European descent, was born at 39 weeks of gestation with symmetric intrauterine growth retardation and deceased at 7 days of life. Her birth weight was 2.42 kg (2nd percentile), birth length was 44 cm (∼0.8 percentile), and head circumference was 28 cm (<1 percentile). She was found to harbor a maternally inherited c.2423G>A (p.Arg808His) change in PTPN4, which was present in her mother in 9/96 reads for 9.3% mosaicism (95% confidence interval, 4.3%–17.1%). This PTPN4 change is predicted to be probably damaging by PolyPhen-2, tolerated by SIFT, disease causing by MutationTaster, and classified as a VUS by Varsome. The proband was also found to harbor a maternally inherited c.2533+1G>A (p.IleValSer25+1Gly>Ala) change in Stromal Antigen 2 (STAG2 [MIM: 300826]), which is predicted to destroy the canonical splice donor site in intron 25, leading to abnormal gene splicing. This type of change is not amenable to evaluation by PolyPhen-2, SIFT, or MutationTaster but was classified as a pathogenic variant by Varsome. The individual had severe microcephaly, semilobar holoprosencephaly with a formed but thin splenium of the corpus callosum, and hypoplastic left heart syndrome. STAG2 encodes a member of the kinesin family of proteins that acts as a plus-end-directed microtubule motor, participating in transport of cellular cargo in neurons as well as in other cell types. STAG2 variants have been implicated in X-linked recessive Mullegama-Klein-Martinez syndrome (MIM: 301022). De novo STAG2 variants have additionally been described in holoprosencephaly,19,20 and subject A was previously reported in Corona-Rivera et al.18 with STAG2 discussed as a candidate for her phenotype. Because the X-linked STAG2 variant was inherited from a healthy, non-mosaic mother, its pathogenicity and the exact relative contributions of STAG2 and PTPN4 variants to this subject’s phenotype are challenging to disentangle.
Szczałuba et al.12 reported a 5-year-old European individual with a mosaic de novo c.215 T>C (p.Leu72Ser) PTPN4 variant, predicted to be probably damaging by PolyPhen-2, damaging by SIFT, and disease causing by MutationTaster. His clinical features include psychomotor and speech delay, developmental delay, ritualized behavior and sensory deficits, various upper limb stereotypies, and autistic-like features but without seizures or growth or brain MRI abnormalities. Here, we refer to him as subject B.
Christodolou et al.21 reported a de novo deletion of PTPN4 in monozygotic twins with delayed motor and cognitive development, limited speech, seizures, and substantial postnatal deceleration in growth (height, weight, OFC). These subjects are referred to as subjects C and D.
For analysis within the phenotypic table (Table S1), we excluded subjects A–D. The coexistence of a STAG2 variant in subject A obscures interpretation of the phenotype in that individual, which was more severe than what was observed in the other individuals. Subjects B–D have previously been described. Subject B has a mosaic variant, and subjects 9 and 10 had genetic evaluation that was limited to CNV analysis. Because of these limitations, we include them here only for comparison.
Subjects 1–6 have a range of cognitive findings, from borderline intellectual function to intellectual disability or developmental delay, and 4/6 had speech delays. Our cohort also demonstrates a variety of dysregulated growth patterns: 4/6 had longitudinal growth anomalies (height and/or weight), 4/6 had absolute or relative macrocephaly and 1/6 had absolute or relative microcephaly; 3/6 had gait or ambulatory anomalies; 3/6 had brain MRI anomalies, particularly cystic or ventricular anomalies; 2/6 had seizures or abnormal EEGs; 4/6 had ADHD, two of those individuals with comorbid autism; 2/6 had skin anomalies; and 2/6 had spinal anomalies, with both having butterfly vertebrae and one having a split cord malformation (diastematomyelia). Congenital heart defects were seen in 1/6.
Characterization and conservation of variant substitutions
PTPN4 is a large modular 926-amino-acid protein localized in the cytoplasm and at the plasma membrane. It contains an N-terminal FERM (Band 4.1, Ezrin, Radixin, and Moesin) domain, a PDZ (PSD-95/Dlg/ZO-1) domain, and a C-terminal catalytic tyrosine phosphatase domain, within which lies a catalytic motif (amino acids HCSAGIGR, residues 851–858).2,3,22,23 After the suppression of the FERM domain, which is responsible for protein anchoring to the plasma membrane,22,24 the phosphatase is exclusively cytoplasmic.25 PTPN4 is proteolyzed in the cell by calpain in response to physiological stimuli, leading to enzyme activation.22 The active form of PTPN4 consists of the PDZ protein-protein interaction domain23,24 and the PTP domain, which is responsible for substrate dephosphorylation.26
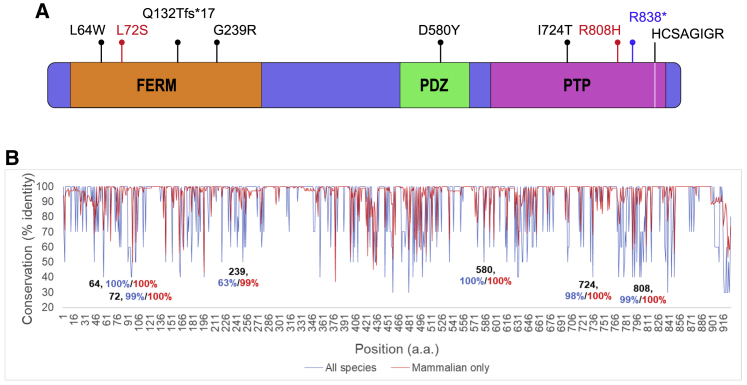
The variants in this study comprise two truncations and four missense variants. Two missense variants lie within the FERM domain, one missense variant lies within the PDZ domain, while two truncations and one missense variant lie within the phosphatase domain but not within the HCSAGIGR catalytic motif (Figure 2A). Analysis of nucleotide sequence conservation across 300 vertebrate orthologs (blue; 140 mammalian orthologs, red) is shown in Figure 2B. Of the six missense amino acid changes in Table S1, five variants are located at highly conserved residues (98%–100% conservation across all orthologs). Residue 239 was located at a locus with 60%–70% conservation across all orthologs but showed mammalian conservation at the level of 99%. For residue 239, one sequence (Mus caroli) has a substitution of glycine with alanine, which in some contexts can be neutral. However, due to glycine’s unique conformational flexibility attributable to a hydrogen-containing side chain, even apparently neutral variants can be forbidden in certain contexts. In aggregate, there is a high degree of conservation at the loci of variant substitutions in our subjects.
Figure 2.
Domain structure, conservation, and locations of observed variation in PTPN4
(A) Variation observed in PTPN4 is shown for the 926-amino-acid protein, GenBank: NP_002821.1. Protein domains include Band 4.1, Ezrin, Radixin, and Moesin (FERM), PSD-95/Dlg/ZO-1 (PDZ), and tyrosine phosphatase (PTP) domains. Protein-altering variants are shown above the schematic representation of PTPN4. De novo variants in subjects 1–6 are shown in black text, and those of unknown inheritance are shown in green. Variants in subjects A and B are shown in red. (B) Conservation at sites of missense substitutions: for sequence comparisons across all 300 orthologs, the conservation scores were binned over every 10% to facilitate visualization (e.g., a score of 100% indicates a range of 91%–100% sequence identity). For sequence comparisons of the 140 mammalian orthologs, the scores were not binned and are shown as is (i.e., a score of 100% indicates that the amino acid at that position is the same across all orthologs analyzed). Conservation across all orthologs is marked in blue, while mammalian ortholog conservation is marked in red.
Subcellular localization of p.Gly239Arg within neurons
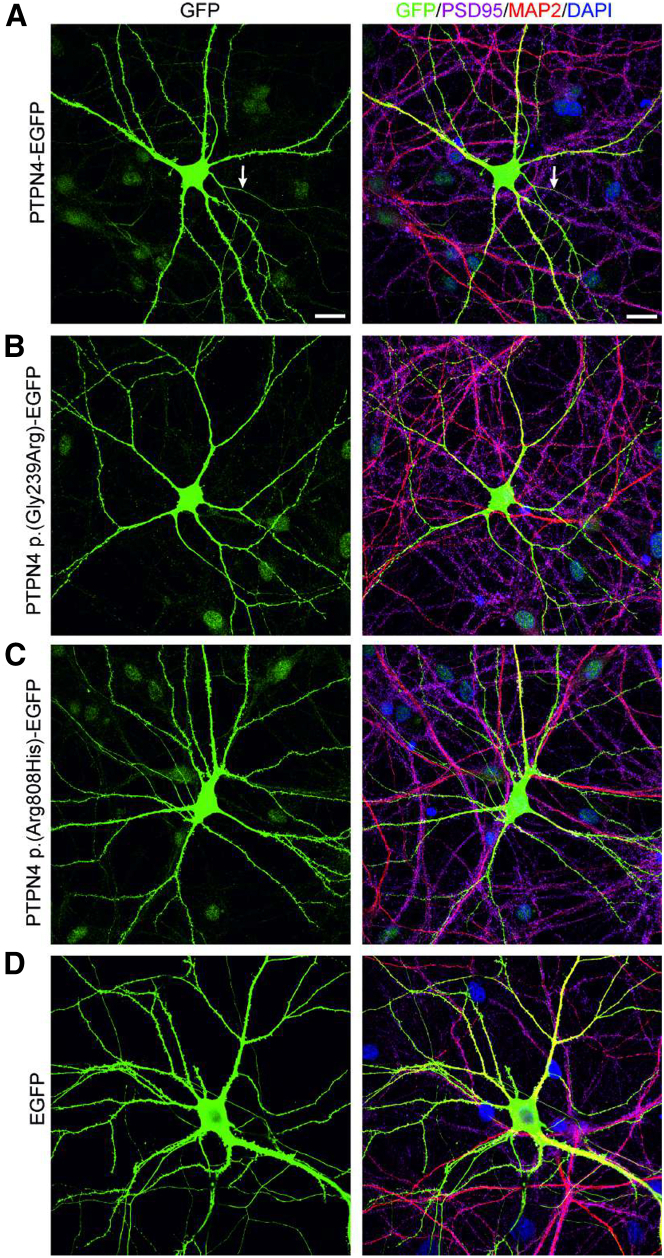
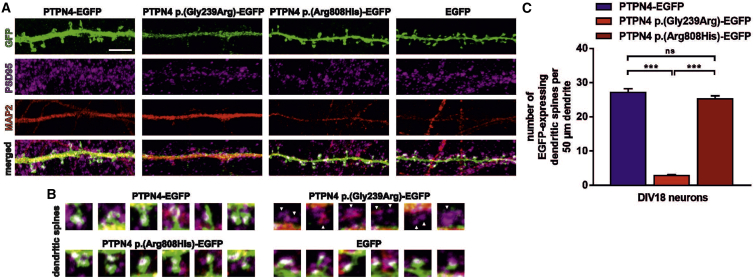
The PTPN4 p.Gly239Arg variant (subject 1) is located in the non-catalytic FERM domain that anchors the protein to the plasma membrane and promotes PTPN4 interaction with its target membrane receptors.22,24 To assess whether the p.Gly239Arg and p.Arg8080His variants affect proper subcellular distribution of the protein, we transfected mature rat hippocampal neurons (DIV13) with human PTPN4-EGFP-, PTPN4 p.Gly239Arg-EGFP-, or PTPN4 p.Arg808His-EGFP-expressing vectors to produce PTPN4 variants fused with the EGFP protein. Additionally, the fluorescence was enhanced by immunostaining with anti-GFP antibody to determine the localization of these constructs in neurons (Figures 3A–3D). All three constructs were efficiently expressed in the cells, and the fluorescent fusion protein was present in the cytoplasm and nucleus of hippocampal neurons, as well as in neuronal processes, axons, and dendrites (Figure 3). Compared to PTPN4-EGFP and PTPN4 p.Arg808His EGFP, PTPN4 p.Gly239Arg-EGFP exhibited a weaker and more granular pattern of staining within dendrites. Imaging of transfected dendrites by confocal microscopy revealed that the PTPN4 p.Gly239Arg-EGFP protein variant failed to localize to dendritic spines. To specifically determine dendrites, we used MAP2 antibody, and for synapses PSD-95 staining was used (Figures 4A and 4B). For the statistical analysis, the number of EGFP-expressing dendritic spines was counted per 50 μm length of secondary dendrite transfected with PTPN4-EGFP, PTPN4 p.Gly239Arg-EGFP, and PTPN4 p.Arg808His-EGFP (Figure 4C). The results indicate that the simple amino acid change in the FERM domain of PTPN4 can prevent its delivery to synapses. In contrast, the PTPN4 p.Arg808His-EGFP phosphatase domain variant does not impede protein trafficking to its synaptic destination.
Figure 3.
Expression of human PTPN4, PTPN4 p.Gly239Arg, and PTPN4 p.Arg808His proteins in rat primary hippocampal neurons
The neurons were transfected with PTPN4-EGFP, PTPN4 p.Gly239Arg-EGFP, PTPN4 p.Arg808His-EGFP, or control EGFP expression vectors and immunostained with antibodies recognizing: GFP (to delineate PTPN4 localization), PSD95 (to show glutamatergic synapses), MAP2 (to visualize dendrites), and DAPI (for nuclear staining). (A–C) An overview of DIV18 rat hippocampal neurons showing PTPN4 and PTPN4 p.Gly239Arg and PTPN4 p.Arg808His protein ubiquitous localization within the cytoplasm and nucleus of neuronal cell body. (A) The PTPN4 protein is also present in axons (designated as white arrow), dendrites, and dendritic spines. (B) The PTPN4 p.Gly239Arg protein is present in dendrites, but it does not localize to dendritic spines. (C) PTPN4 p.Arg808His protein is expressed in both dendrites and dendritic spines. (D) The visualization of neuron transfected with EGFP as a control. Scale bars, 20 μm.
Figure 4.
PTPN4 p.Gly239Arg protein is not present in dendritic spines, while PTPN4 p.Arg808His protein localization is not impaired
(A) The rat hippocampal neurons (fixed at DIV18) transfected with PTPN4-EGFP, PTPN4 p.Gly239Arg-EGFP, PTPN4 p.Arg808His-EGFP, or control EGFP expression vectors, respectively, were immunostained with antibodies against: GFP (to designate PTPN4 localization), PSD95 (to show glutamatergic synapses), and MAP2 (to visualize dendrites). The PTPN4 and PTPN4 p.Arg808His protein is expressed in dendritic spines, while no PTPN4 p.Gly239Arg protein was detected there. Bottom panel presents the merged image. Scale bar, 5 μm. (B) The dendritic spines of neurons transfected with PTPN4-EGFP, PTPN4 p.Gly239Arg-EGFP, PTPN4 p.Arg808His-EGFP, or control EGFP expression vectors. The white arrows point at the absence of PTPN4 p.Gly239Arg protein in dendritic spines of PTPN4 p.Gly239Arg-EGFP-transfected neurons. (C) Quantification. The graph shows the number of EGFP-expressing dendritic spines counted per 50 μm length of secondary dendrite from PTPN4-EGFP (mean, 25.95 ± 1.05 SEM), PTPN4 p.Gly239Arg-EGFP (mean, 2.38 ± 0.58 SEM), and PTPN4 p.Arg808His-EGFP (mean, 21.81 ± 1.73 SEM) transfected neurons (∗∗∗p < 0.0001 by unpaired t test, n = 42 dendrites from 14 neurons per variant; ns, not significant).
PTPN4 dephosphorylates the NMDA receptor subunit GluRϵ1 (also known as GRIN2A).3 Another important function of PTPN4 in neurons is the direct dephosphorylation of GluA2 AMPA receptors, necessary for proper long-term depression (LTD) expression in cerebellar neurons27 and requiring the anchoring of PTPN4 to the post-synaptic density (PSD) via its direct interaction with GluRδ2 (also known as GRID2).3,27 Non-catalytic domains of PTPs control protein subcellular distribution and thereby indirectly regulate PTP activity by limiting access to specific substrates.28 For example, FERM domain suppression of PTPN4 leads to cytoplasmic retention in T cells,29 and the FERM-containing Preso protein subsequently fails to target dendritic spines after FERM domain deletion.30 Our analysis shows that the PTPN4 p.Gly239Arg protein aggregates in dendrites and does not enter dendritic spines, in contrast to WT PTPN4. This mislocalization of PTPN4 p.Gly239Arg protein within neuronal cells is likely to impair its function. The same mislocalization of variant PTPN4 was previously described for the p.Leu72Ser variant harbored in mosaic form by subject 8.12 While it is unclear how PTPN4 p.Gly239Arg mislocalization in neurons leads to a downstream phenotype, it is possible that the lack of PTPN4 p.Gly239Arg at synapses leads to excessive phosphorylation of PTPN4 phosphatase substrates (e.g., GluRδ2, GluRε1) and impacts signaling at these receptors.
Biochemical analysis of p.Arg808His substitution
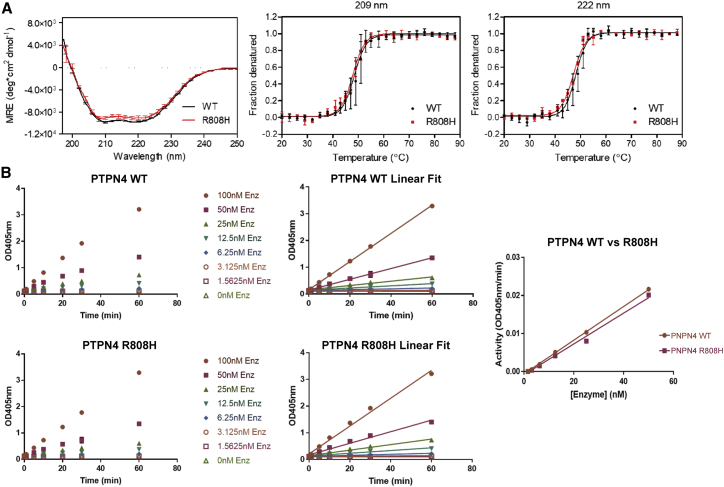
An individual with holoprosencephaly and ectrodactyly was previously described in association with a t(2;4)(q14.2;q35) translocation.18 Since holoprosencephaly was a phenotypic outlier in our cohort, we wished to clarify the extreme outlier genotype-phenotype correlation observed in subject A to determine if the p.Arg808His variant was a contributor to the holoprosencephaly phenotype. This variant was a (likely post-zygotic) mosaic variant present in only a portion of the healthy parent’s cells and transmitted to the affected offspring. Identification of such parental-mosaic transmissions have been shown to be an important complement to the established approach of screening for de novo substitutions.31, 32, 33 As the CUPSAT34 and DUET35 software programs both suggest the p.Arg808His substitution would be destabilizing to the protein, we used circular dichroism spectroscopy to evaluate the overall conformation of p.Arg808His constructs in comparison to wild-type PTPN4. Using Escherichia coli-expressed constructs, spectra were recorded from 250 to 190 nm (Figure 5). PTPN4 constructs display a double-negative double peak at 208 and 222 nm and positive ellipticity below 200 nm, consistent with the presence of substantial α-helical structure. CD spectra of both are superimposable (Figure 5A, left panel). Thermal denaturation of PTPN4 was monitored by CD at 209 and 222 nm (Figure 5A, middle and right panels). The R808H variant does not significantly affect the unfolding temperature (209 nm: p = 0.55; 222 nm: p = 0.30) or the cooperativity of unfolding (209 nm: p = 0.17; 222 nm: p = 0.19) of PTPN4. At 209 nm, WT denatures at 49.0°C ± 1.4°C, and R808H denatures at 48.1°C ± 0.5°C. At 222 nm, WT denatures at 48.4°C ± 1.0°C, and R808H denatures at 47.2°C ± 0.1°C. Aggregation of both PTPN4 constructs was observed when the cuvette was removed from the sample cell, indicating that PTPN4 does not unfold and refold reversibly when thermally denatured. In summary, the R808H substitution does not alter the secondary structure composition of PTPN4 compared to WT, nor does the variant affect PTPN4’s resistance to thermal denaturation or the cooperativity of its unfolding by thermal denaturation.
Figure 5.
Biochemical characteristics and phosphatase activity of variant p.Arg808His PTPN4
(A) Left panel: far-UV CD spectra of PTPN4 in 20 mM sodium phosphate (pH 7.5), 150 mM NaCl. The three replicates are plotted as an average. Error bars represent SD. The spectra of wild-type (WT) and R808H are essentially identical, which indicates that they have very similar secondary structure compositions. Middle and right panels: thermal denaturation of PTPN4 in 20 mM sodium phosphate (pH 7.5), 150 mM NaCl monitored by mean molar ellipticity per residue (MRE) at 209 and 222 nm. Error bars represent the SD of three replicates. For visualization purposes, not every data point is shown. The data were fit to a Boltzmann sigmoid using Prism. The R808H variant does not significantly affect the unfolding temperature (209 nm: p = 0.55; 222 nm: p = 0.30) or the cooperativity of unfolding (209 nm: p = 0.17; 222 nm: p = 0.19) of PTPN4. (B) Saturation kinetics of wild-type and variant PTPN4. His-tagged versions of the PTPN4 protein segments comprising the two catalytic domains containing WT and p.Arg808His variant sequences were expressed in bacteria and purified using nickel affinity chromatography. Equal amounts of WT and variant proteins were used to evaluate enzyme kinetics. The rate of hydrolysis of substrate (dNPP) is plotted against increasing substrate concentration. Data were fitted to the Michaelis-Menten equation.
To further investigate the properties of the p.Arg808His missense substitution and to delineate the respective contributions of inherited PTPN4 and STAG2 variants in subject A, we examined the effect of p.Arg808His on PTPN4 phosphatase function. We constructed the PTPN4 p.Arg808His vector for expression in Escherichia coli. The purified (His)6-tagged PTPN4 linker-phosphatase construct was tested for the ability to dephosphorylate the commonly used p-nitrophenyl phosphate (p-NPP) substrate in vitro.36,37 Within the limits of experimental error, the Michaelis-Menten kinetic properties of p.Arg808His and WT are similar (Figure 5B).
Thus, WT and p.Arg808His appear indistinguishable in terms of their in vitro catalytic, structural, and chemical properties. Overall, we label this PTPN4 variant as being of undetermined pathogenicity. The STAG2 gene has been reported to be disrupted in holoprosencephaly with hypoplastic left heart syndrome in males and some females.19,20 While a contribution from PTPN4 is suggested by the inheritance of the X-linked STAG2 variant from a healthy mother who also harbors the autosomal PTPN4 variant only in a low-mosaic form, the phenotype mirrors the known STAG2 phenotype more closely than phenotypes we have observed here in subjects 1–6.
Discussion
In summary, we describe a cohort of individuals with predominantly de novo variants in PTPN4 manifesting various degrees of developmental and growth anomalies. Silencing of PTPN4 markedly increases neuronal apoptosis.26,38 PTPN4 interacts with glutamate receptor subunits, which are required for neural development and function.3 Consistent with a neural role, Ptpn4-deficient mice on a 129SvJ background have defective motor learning and cerebellar long-term depression.5 Furthermore, in Drosophila, loss of expression of PTPMEG (the single homolog of PTPN3 and PTPN4 in this species) results in defective axon growth associated with impaired development of mushroom bodies.39 While the closest human PTPN4 homolog, PTPN3, dephosphorylates the growth hormone receptor (GHR) and male Ptpn3-deficient mice display increased weight gain on a mixed 129 SvEv × C57BL/6 genetic background,40,41 no change in body mass of Ptpn4-deficient or Ptpn3/Ptpn4 double-deficient mice was observed in another study with mice on a 129P2/Ola Hsd background or a C57BL/6 background.7
Subjects with FERM domain variants were noted to have absolute or relative macrocephaly but tended to lack seizures or EEG anomalies. The only subject in the cohort with a PDZ domain variant also has relative macrocephaly, while one individual with a phosphatase domain variant had macrocephaly (possibly familial, taking parental measurements into account). Butterfly vertebrae and bony abnormalities were found only in individuals with truncations (2/2). While cognitive or developmental delay was frequent, one person with a truncation (subject 5) had normal intelligence, albeit with borderline intellectual functioning (tested full-scale IQ of 71). It remains possible that cognitive function within the range of normal might ultimately manifest in the younger subjects with developmental delay in our cohort.42 Subject 2 had a likely pathogenic missense variant in CUL3, a gene for whom loss-of-function variants have been associated with autism.13, 14, 15 At age 23 months, subject 2 does not carry an autism diagnosis.
The downstream pathogenic mechanisms by which FERM-domain mislocalization substitutions lead to phenotypic manifestations may not wholly overlap with mechanisms attributable to phosphatase-domain, PDZ-domain, or truncation variants. Subject 5’s PTPN4 variant is present in gnomAD, and this variant, as well as another truncation, has been reported in individuals with autism43 as a parent-transmitted protein-truncating variant (PTV). Both PTVs, c.16C>T (p.Arg6Ter) and c.2512C>T (p.Arg838Ter), were described in an analysis of a large set of de novo unique PTVs in autism/developmental delay, which were compared to de novo ASD/developmental delay PTVs that are also observed as standing variation in the population. As a class, singleton PTVs with one allele in ExAC showed no over-transmission. However, removal of PTVs present in ExAC or in a loss-of-function-tolerant gene (pLI < 0.9) led to a modest excess of transmitted PTVs in ASD individuals. Overall ∼1/3 of de novo variants identified in neurodevelopmental disease cohorts were also present as standing variation in ExAC, were enriched for more mutable CpG sites, and conferred no detectable risk to intellectual disability/developmental disorders and ASDs, consistent with benign variation. However, the class of inherited singleton PTVs with one variant in ExaC and a pLI > 0.9 (which includes PTPN4) includes a number of high-penetrance neurodevelopmental genes that are associated with autosomal dominant loss-of-function intellectual disability or autism phenotypes, including ARID2, CADPS2, CHD8, CUX1, KMT2C, KMT2D, MBD5, NIPBL, and PTEN. Thus, there are many genes within this class for which loss-of-function variation is clearly deleterious. Interestingly, subject 5 does not have intellectual disability or autism.
Abnormalities of head growth in our subjects are notable. The phosphatase activity of PTPN4 has been implicated in the regulation of cytoskeletal events.44 Overexpression of PTPN4 in COS-7 cells decreased colony formation, inhibited cell growth, and decreased saturation density of these cells.45 Furthermore, PTPN4 shows human-specific brain expression patterns. Primary human versus macaque cell type comparisons, as well as human-chimpanzee organoid comparisons, show that PTPN4 undergoes concerted differential gene expression as well as differential expression in neuronal cell types, including intermediate progenitor cells and radial glia cells, which are particularly likely to influence the evolutionary expansion of the brain.46,47 Analyses of organoid models, which preserve gene regulatory networks and developmental processes and are suitable for analysis of recently evolved gene expression patterns (within six million years), implicate PTPN4 as one of 261 overlapping genes that are likely to have derived human-specific gene expression patterns during cortical development.48 A second phosphatase identified within this gene set is the ubiquitously expressed Phosphatase and tensin homolog (PTEN [MIM: 601728]). Interestingly, heterozygous PTEN deleterious human variants show substantial variability in penetrance and expressivity, manifesting as a range of syndromes generally with macrocephaly and/or overgrowth but also in some cases with normal cognitive development.49, 50, 51, 52 Loss of mouse Pten results in progressive macrocephaly, enlarged neuronal soma size, and increased proliferation and self-renewal of neural stem cells without a major disturbance of cell fate commitments.53, 54, 55, 56
The range of phenotypic outcomes observed in association with heterozygous PTPN4 variants in the subjects described in this paper suggests that incomplete penetrance and variable expressivity are likely to influence the outcome of deleterious variants in PTPN4, and genetic background likely plays a role in phenotypic expression (including normal cognition) similar to what is described with other genes or CNVs associated with disorders of development.57, 58, 59
In conclusion, we have identified six individuals harboring rare, heterozygous PTPN4 variants that are likely to be deleterious, in conjunction with neurodevelopmental deficits, growth anomalies, and variable seizures, structural CNS anomalies, gait abnormalities, congenital heart defects, dermatological manifestations, and spinal and vertebral defects. We believe these observations strongly support the conclusion that damaging variation in PTPN4 is causally related to a neurodevelopmental disorder (NDD). The key points of evidence are as follows: (1) excluding one individual where both parents were unavailable, none of the variants described in subjects 1–6 were found to be inherited, and 5 of 6 observed variants are absent from gnomAD, HGMD, and ClinVar; the variant present in gnomAD is found in a subject with normal IQ in the borderline intellectual functioning range; (2) PTPN4 is relatively intolerant to protein-altering variation in the population; (3) all variants in probands are either predicted to result in loss of function or are missense variants at highly conserved residues in mammals; (4) all variants are computationally predicted to be deleterious with one or more software prediction programs; (5) model organism evidence suggests a role for PTPN4 in neurodevelopment; and (6) functional analysis implicates impaired subcellular targeting of FERM-variant protein in neurons. In summary, these data collectively implicate PTPN4 as an NDD-related gene, adding to the growing list of human protein tyrosine-phosphatase related disease conditions60, 61, 62, 63, 64, 65 and confirming the importance of these pleiotropic proteins in diverse aspects of human development.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We would like to thank the families who kindly consented to participate in our genetics study. We would also like to acknowledge Erin Torti of GeneDx for assistance in subject accrual and data retrieval. This work was supported by a NCATS/NIH/Case Western Reserve Clinical and Translational Science Collaborative (CTSC) Award (A.A.), the US National Institutes of Health (R01MH113106, A.W.B.), and NCN grants (2014/14/E/NZ3/00375, M.D.; 2017/27/N/NZ1/01381, J.J.C.). Subject 4 was ascertained through the Deciphering Developmental Disorders (DDD) study. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant no. HICF-1009-003). This study makes use of DECIPHER (http://www.deciphergenomics.org), which is funded by Wellcome. See Nature PMID: 25533962 or https://www.ddduk.org/access.html for full acknowledgement.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100033.
Data and code availability
Variant information for all variants described in this manuscript is publicly available through ClinVar under the following accession numbers: p.Gly239Arg: ClinVar: SUB8550358; p.Leu64Trp: ClinVar: SUB8550428; p.Gln132ThrfsTer17: ClinVar: SUB8550246; p.Arg838Ter: ClinVar: SUB8561208; p.Asp580Tyr: ClinVar: SUB9192275; p.Arg808His: ClinVar: SUB8550877. Variant information for p.Ile724Thr is available through the DECIPHER database (ID: 262775) and 100,000 Genomes Project (ID: 119000445).
Web resources
Database of Genomic Variants, http://dgv.tcag.ca/dgv/app/home
DECIPHER, https://www.deciphergenomics.org/
Genomics England, https://www.genomicsengland.co.uk/
Human Gene Mutation Database, http://www.hgmd.cf.ac.uk/ac/index.php
International Genome Sample Resource, https://www.internationalgenome.org/
National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/gene/5775/ortholog/?scope=89593#genes-tab
NHLBI Exome Sequencing Project, https://evs.gs.washington.edu/EVS/
OMIM, https://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental information
References
- 1.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Alonso A., Pulido R. The extended human PTPome: a growing tyrosine phosphatase family. FEBS J. 2016;283:2197–2201. doi: 10.1111/febs.13748. [DOI] [PubMed] [Google Scholar]
- 3.Hironaka K., Umemori H., Tezuka T., Mishina M., Yamamoto T. The protein-tyrosine phosphatase PTPMEG interacts with glutamate receptor delta 2 and epsilon subunits. J. Biol. Chem. 2000;275:16167–16173. doi: 10.1074/jbc.M909302199. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kina S., Tezuka T., Kusakawa S., Kishimoto Y., Kakizawa S., Hashimoto K., Ohsugi M., Kiyama Y., Horai R., Sudo K., et al. Involvement of protein-tyrosine phosphatase PTPMEG in motor learning and cerebellar long-term depression. Eur. J. Neurosci. 2007;26:2269–2278. doi: 10.1111/j.1460-9568.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- 6.Carithers L.J., Moore H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank. 2015;13:307–308. doi: 10.1089/bio.2015.29031.hmm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauler T.J., Hendriks W.J., King P.D. The FERM and PDZ domain-containing protein tyrosine phosphatases, PTPN4 and PTPN3, are both dispensable for T cell receptor signal transduction. PLoS ONE. 2008;3:e4014. doi: 10.1371/journal.pone.0004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G., et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopanos C., Tsiolkas V., Kouris A., Chapple C.E., Albarca Aguilera M., Meyer R., Massouras A. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szczałuba K., Chmielewska J.J., Sokolowska O., Rydzanicz M., Szymańska K., Feleszko W., Włodarski P., Biernacka A., Murcia Pienkowski V., Walczak A., et al. Neurodevelopmental phenotype caused by a de novo PTPN4 single nucleotide variant disrupting protein localization in neuronal dendritic spines. Clin. Genet. 2018;94:581–585. doi: 10.1111/cge.13450. [DOI] [PubMed] [Google Scholar]
- 13.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., et al. DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D., et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A., et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyden L.M., Choi M., Choate K.A., Nelson-Williams C.J., Farhi A., Toka H.R., Tikhonova I.R., Bjornson R., Mane S.M., Colussi G., et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babbs C., Heller R., Everman D.B., Crocker M., Twigg S.R., Schwartz C.E., Giele H., Wilkie A.O. A new locus for split hand/foot malformation with long bone deficiency (SHFLD) at 2q14.2 identified from a chromosome translocation. Hum. Genet. 2007;122:191–199. doi: 10.1007/s00439-007-0390-7. [DOI] [PubMed] [Google Scholar]
- 18.Corona-Rivera A., Corona-Rivera J.R., Bobadilla-Morales L., García-Cobian T.A., Corona-Rivera E. Holoprosencephaly, hypertelorism, and ectrodactyly in a boy with an apparently balanced de novo t(2;4) (q14.2;q35) Am. J. Med. Genet. 2000;90:423–426. doi: 10.1002/(sici)1096-8628(20000228)90:5<423::aid-ajmg12>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 19.Aoi H., Lei M., Mizuguchi T., Nishioka N., Goto T., Miyama S., Suzuki T., Iwama K., Uchiyama Y., Mitsuhashi S., et al. Retraction note to: Nonsense variants in STAG2 result in distinct sex-dependent phenotypes. J. Hum. Genet. 2020;65:811. doi: 10.1038/s10038-020-0782-2. [DOI] [PubMed] [Google Scholar]
- 20.Kruszka P., Berger S.I., Casa V., Dekker M.R., Gaesser J., Weiss K., Martinez A.F., Murdock D.R., Louie R.J., Prijoles E.J., et al. Cohesin complex-associated holoprosencephaly. Brain. 2019;142:2631–2643. doi: 10.1093/brain/awz210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson S.L., Ellaway C.J., Peters G.B., Pelka G.J., Tam P.P., Christodoulou J. Deletion of protein tyrosine phosphatase, non-receptor type 4 (PTPN4) in twins with a Rett syndrome-like phenotype. Eur. J. Hum. Genet. 2015;23:1171–1175. doi: 10.1038/ejhg.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu M., Majerus P.W. The properties of the protein tyrosine phosphatase PTPMEG. J. Biol. Chem. 1996;271:27751–27759. doi: 10.1074/jbc.271.44.27751. [DOI] [PubMed] [Google Scholar]
- 23.Maisonneuve P., Caillet-Saguy C., Raynal B., Gilquin B., Chaffotte A., Pérez J., Zinn-Justin S., Delepierre M., Buc H., Cordier F., Wolff N. Regulation of the catalytic activity of the human phosphatase PTPN4 by its PDZ domain. FEBS J. 2014;281:4852–4865. doi: 10.1111/febs.13024. [DOI] [PubMed] [Google Scholar]
- 24.Caillet-Saguy C., Toto A., Guerois R., Maisonneuve P., di Silvio E., Sawyer K., Gianni S., Wolff N. Regulation of the Human Phosphatase PTPN4 by the inter-domain linker connecting the PDZ and the phosphatase domains. Sci. Rep. 2017;7:7875. doi: 10.1038/s41598-017-08193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maisonneuve P., Caillet-Saguy C., Vaney M.C., Bibi-Zainab E., Sawyer K., Raynal B., Haouz A., Delepierre M., Lafon M., Cordier F., Wolff N. Molecular Basis of the Interaction of the Human Protein Tyrosine Phosphatase Non-receptor Type 4 (PTPN4) with the Mitogen-activated Protein Kinase p38γ. J. Biol. Chem. 2016;291:16699–16708. doi: 10.1074/jbc.M115.707208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babault N., Cordier F., Lafage M., Cockburn J., Haouz A., Prehaud C., Rey F.A., Delepierre M., Buc H., Lafon M., Wolff N. Peptides targeting the PDZ domain of PTPN4 are efficient inducers of glioblastoma cell death. Structure. 2011;19:1518–1524. doi: 10.1016/j.str.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Kohda K., Kakegawa W., Matsuda S., Yamamoto T., Hirano H., Yuzaki M. The δ2 glutamate receptor gates long-term depression by coordinating interactions between two AMPA receptor phosphorylation sites. Proc. Natl. Acad. Sci. USA. 2013;110:E948–E957. doi: 10.1073/pnas.1218380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonks N.K. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gjörloff-Wingren A., Saxena M., Han S., Wang X., Alonso A., Renedo M., Oh P., Williams S., Schnitzer J., Mustelin T. Subcellular localization of intracellular protein tyrosine phosphatases in T cells. Eur. J. Immunol. 2000;30:2412–2421. doi: 10.1002/1521-4141(2000)30:8<2412::AID-IMMU2412>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 30.Lee H.W., Choi J., Shin H., Kim K., Yang J., Na M., Choi S.Y., Kang G.B., Eom S.H., Kim H., Kim E. Preso, a novel PSD-95-interacting FERM and PDZ domain protein that regulates dendritic spine morphogenesis. J. Neurosci. 2008;28:14546–14556. doi: 10.1523/JNEUROSCI.3112-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halvorsen M., Petrovski S., Shellhaas R., Tang Y., Crandall L., Goldstein D., Devinsky O. Mosaic mutations in early-onset genetic diseases. Genet. Med. 2016;18:746–749. doi: 10.1038/gim.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright C.F., Prigmore E., Rajan D., Handsaker J., McRae J., Kaplanis J., Fitzgerald T.W., FitzPatrick D.R., Firth H.V., Hurles M.E. Clinically-relevant postzygotic mosaicism in parents and children with developmental disorders in trio exome sequencing data. Nat. Commun. 2019;10:2985. doi: 10.1038/s41467-019-11059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X., Yang X., Wu Q., Liu A., Yang X., Ye A.Y., Huang A.Y., Li J., Wang M., Yu Z., et al. Amplicon Resequencing Identified Parental Mosaicism for Approximately 10% of “de novo” SCN1A Mutations in Children with Dravet Syndrome. Hum. Mutat. 2015;36:861–872. doi: 10.1002/humu.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parthiban V., Gromiha M.M., Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pires D.E., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai A., Mieskes G. Inhibitory effect of okadaic acid on the p-nitrophenyl phosphate phosphatase activity of protein phosphatases. Biochem. J. 1991;275:233–239. doi: 10.1042/bj2750233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pot D.A., Woodford T.A., Remboutsika E., Haun R.S., Dixon J.E. Cloning, bacterial expression, purification, and characterization of the cytoplasmic domain of rat LAR, a receptor-like protein tyrosine phosphatase. J. Biol. Chem. 1991;266:19688–19696. [PubMed] [Google Scholar]
- 38.Préhaud C., Wolff N., Terrien E., Lafage M., Mégret F., Babault N., Cordier F., Tan G.S., Maitrepierre E., Ménager P., et al. Attenuation of rabies virulence: takeover by the cytoplasmic domain of its envelope protein. Sci. Signal. 2010;3:ra5. doi: 10.1126/scisignal.2000510. [DOI] [PubMed] [Google Scholar]
- 39.Whited J.L., Robichaux M.B., Yang J.C., Garrity P.A. Ptpmeg is required for the proper establishment and maintenance of axon projections in the central brain of Drosophila. Development. 2007;134:43–53. doi: 10.1242/dev.02718. [DOI] [PubMed] [Google Scholar]
- 40.Pasquali C., Curchod M.L., Wälchli S., Espanel X., Guerrier M., Arigoni F., Strous G., Hooft van Huijsduijnen R. Identification of protein tyrosine phosphatases with specificity for the ligand-activated growth hormone receptor. Mol. Endocrinol. 2003;17:2228–2239. doi: 10.1210/me.2003-0011. [DOI] [PubMed] [Google Scholar]
- 41.Pilecka I., Patrignani C., Pescini R., Curchod M.L., Perrin D., Xue Y., Yasenchak J., Clark A., Magnone M.C., Zaratin P., et al. Protein-tyrosine phosphatase H1 controls growth hormone receptor signaling and systemic growth. J. Biol. Chem. 2007;282:35405–35415. doi: 10.1074/jbc.M705814200. [DOI] [PubMed] [Google Scholar]
- 42.Field M., Fox N., Radcliffe J. Predicting IQ change in preschoolers with developmental delays. J. Dev. Behav. Pediatr. 1990;11:184–189. [PubMed] [Google Scholar]
- 43.Kosmicki J.A., Samocha K.E., Howrigan D.P., Sanders S.J., Slowikowski K., Lek M., Karczewski K.J., Cutler D.J., Devlin B., Roeder K., et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu M.X., York J.D., Warshawsky I., Majerus P.W. Identification, cloning, and expression of a cytosolic megakaryocyte protein-tyrosine-phosphatase with sequence homology to cytoskeletal protein 4.1. Proc. Natl. Acad. Sci. USA. 1991;88:5867–5871. doi: 10.1073/pnas.88.13.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu M., Meng K., Majerus P.W. The effect of overexpression of the protein tyrosine phosphatase PTPMEG on cell growth and on colony formation in soft agar in COS-7 cells. Proc. Natl. Acad. Sci. USA. 1996;93:12980–12985. doi: 10.1073/pnas.93.23.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noctor S.C., Flint A.C., Weissman T.A., Dammerman R.S., Kriegstein A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 47.Rakic P. Developmental and evolutionary adaptations of cortical radial glia. Cereb. Cortex. 2003;13:541–549. doi: 10.1093/cercor/13.6.541. [DOI] [PubMed] [Google Scholar]
- 48.Pollen A.A., Bhaduri A., Andrews M.G., Nowakowski T.J., Meyerson O.S., Mostajo-Radji M.A., Di Lullo E., Alvarado B., Bedolli M., Dougherty M.L., et al. Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell. 2019;176:743–756.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orloff M.S., Eng C. Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene. 2008;27:5387–5397. doi: 10.1038/onc.2008.237. [DOI] [PubMed] [Google Scholar]
- 50.Eng C. PTEN: one gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 51.Busch R.M., Chapin J.S., Mester J., Ferguson L., Haut J.S., Frazier T.W., Eng C. Cognitive characteristics of PTEN hamartoma tumor syndromes. Genet. Med. 2013;15:548–553. doi: 10.1038/gim.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frazier T.W., Embacher R., Tilot A.K., Koenig K., Mester J., Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol. Psychiatry. 2015;20:1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwon C.H., Zhu X., Zhang J., Knoop L.L., Tharp R., Smeyne R.J., Eberhart C.G., Burger P.C., Baker S.J. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat. Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 54.Clipperton-Allen A.E., Page D.T. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum. Mol. Genet. 2014;23:3490–3505. doi: 10.1093/hmg/ddu057. [DOI] [PubMed] [Google Scholar]
- 55.Page D.T., Kuti O.J., Prestia C., Sur M. Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proc. Natl. Acad. Sci. USA. 2009;106:1989–1994. doi: 10.1073/pnas.0804428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groszer M., Erickson R., Scripture-Adams D.D., Lesche R., Trumpp A., Zack J.A., Kornblum H.I., Liu X., Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 57.Girirajan S., Rosenfeld J.A., Coe B.P., Parikh S., Friedman N., Goldstein A., Filipink R.A., McConnell J.S., Angle B., Meschino W.S., et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bijlsma E.K., Gijsbers A.C., Schuurs-Hoeijmakers J.H., van Haeringen A., Fransen van de Putte D.E., Anderlid B.M., Lundin J., Lapunzina P., Pérez Jurado L.A., Delle Chiaie B., et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur. J. Med. Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 59.Todarello G., Feng N., Kolachana B.S., Li C., Vakkalanka R., Bertolino A., Weinberger D.R., Straub R.E. Incomplete penetrance of NRXN1 deletions in families with schizophrenia. Schizophr. Res. 2014;155:1–7. doi: 10.1016/j.schres.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 60.Au A.C., Hernandez P.A., Lieber E., Nadroo A.M., Shen Y.M., Kelley K.A., Gelb B.D., Diaz G.A. Protein tyrosine phosphatase PTPN14 is a regulator of lymphatic function and choanal development in humans. Am. J. Hum. Genet. 2010;87:436–444. doi: 10.1016/j.ajhg.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wayne S., Robertson N.G., DeClau F., Chen N., Verhoeven K., Prasad S., Tranebjärg L., Morton C.C., Ryan A.F., Van Camp G., Smith R.J. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 62.Kontaridis M.I., Swanson K.D., David F.S., Barford D., Neel B.G. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 63.Marsh D.J., Dahia P.L., Zheng Z., Liaw D., Parsons R., Gorlin R.J., Eng C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 64.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M., et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 65.Hendriks W.J., Pulido R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim. Biophys. Acta. 2013;1832:1673–1696. doi: 10.1016/j.bbadis.2013.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Variant information for all variants described in this manuscript is publicly available through ClinVar under the following accession numbers: p.Gly239Arg: ClinVar: SUB8550358; p.Leu64Trp: ClinVar: SUB8550428; p.Gln132ThrfsTer17: ClinVar: SUB8550246; p.Arg838Ter: ClinVar: SUB8561208; p.Asp580Tyr: ClinVar: SUB9192275; p.Arg808His: ClinVar: SUB8550877. Variant information for p.Ile724Thr is available through the DECIPHER database (ID: 262775) and 100,000 Genomes Project (ID: 119000445).