Abstract
Saccharomyces cerevisiae CDC7 encodes a serine/threonine kinase required for G1/S transition, and its related kinases are present in fission yeast as well as in higher eukaryotes, including humans. Kinase activity of Cdc7 protein depends on the regulatory subunit, Dbf4, which also interacts with replication origins. We have identified him1+ from two-hybrid screening with Hsk1, a fission yeast homologue of Cdc7 kinase, and showed that it encodes a regulatory subunit of Hsk1. Him1, identical to Dfp1, previously identified as an associated molecule of Hsk1, binds to Hsk1 and stimulates its kinase activity, which phosphorylates both catalytic and regulatory subunits as well as recombinant MCM2 protein in vitro. him1+ is essential for DNA replication in fission yeast cells, and its transcription is cell cycle regulated, increasing at middle M to late G1. The protein level is low at START in G1, increases at the G1/S boundary, and is maintained at a high level throughout S phase. Him1 protein is hyperphosphorylated at G1/S through S during the cell cycle as well as in response to early S-phase arrest induced by nucleotide deprivation. Deletion of one of the motifs conserved in regulatory subunits for Cdc7-related kinases as well as alanine substitution of three serine and threonine residues present in the same motif resulted in a defect in checkpoint regulation normally induced by hydroxyurea treatment. The alanine mutant also showed growth retardation after UV irradiation and the addition of methylmethane sulfonate. In keeping with this result, a database search indicates that him1+ is identical to rad35+. Our results reveal a novel function of the Cdc7/Dbf4-related kinase complex in S-phase checkpoint control as well as in growth recovery from DNA damage in addition to its predicted essential function in S-phase initiation.
Eukaryotic chromosomal replication is tightly regulated so that the ordered activation of replication origins occurs only during specific times of the cell cycle (9, 20, 62). It is also flexible, in that programs of origin activation vary depending on the stage of development and on the specific cell type (1, 29, 39) and in that genetic manipulation which inactivates subsets of active origins can be tolerated (13, 16).
Genetic and biochemical studies of DNA replication in yeast as well as in Xenopus eggs led to the discovery of cell cycle-regulated alteration of protein-DNA complexes assembled at the replication origins (11, 12, 14, 40). Prereplicative complexes (preRC) generated at late M to early G1 phase of cell cycle are prerequisite for origin activation at S phase, and rapidly turn into postreplicative complexes (postRC) after the firing of the origins (54). Origin activation and DNA chain elongation are under the control of external stimuli such as growth factors and DNA-damaging agents. PreRC need to be triggered in order for S phase to be initiated. This triggering process involves serine/threonine kinases whose activities are under cell cycle control.
Among them, Cdc7 kinase of Saccharomyces cerevisiae has been known to be required at the onset of S phase (27, 28). More recently, it was reported that function of Cdc7 is required throughout S phase to activate each individual origin (6, 15). Cdc7 requires a regulatory subunit, Dbf4, for its kinase activity (31, 38, 47, 61). Dbf4 not only activates Cdc7 kinase but is also tethered at the origins, presumably through association with components of preRC (18). MCM components may be among physiologically important targets of Cdc7-mediated phosphorylation, although the precise mechanisms of origin activation by Cdc7 kinase are not known (7, 42, 60). Kinases related to Cdc7 have been identified in fission yeast, Xenopus, mice, and humans, suggesting conserved functions of Cdc7-related kinases in S-phase initiation (32, 35, 46, 60).
Fission yeast hsk1+ was identified on the basis of its structural similarity (46). hsk1+ is essential for viability and a significant fraction of a null mutant of hsk1 undergoes premature mitosis in the absence of DNA replication (replication checkpoint defect). In order to search for a putative regulatory subunit for Hsk1 kinase, we searched for Hsk1-interacting molecules. Among the clones isolated, we report here that the him1+ (for Hsk1-interacting molecule 1) gene product is able to bind and stimulate Hsk1 kinase activity. him1+ is identical to dfp1+, which was recently identified in the EMBL database and was shown to associate with the Hsk1 protein (7). It is essential for G1/S transition in fission yeast, and its expression is cell cycle regulated. The level of Him1 protein is low in G1 and increases at late G1 through S. Him1 protein, recovered in Triton-insoluble fractions, becomes hyperphosphorylated at late G1 to S as well as upon block of replication fork progression by nucleotide deprivation. Furthermore, we have identified a mutant of Him1 protein which retains mitotic function, but is sensitive to hydroxyurea (HU), UV, and methylmethane sulfonate (MMS). We further showed that HU sensitivity is caused by a defect in S-phase checkpoint control. After a search of the database, we discovered identity of him1+ with rad35+, recently isolated as a radiation-sensitive mutant gene. Our results revealed a novel function of Cdc7-related kinase complex in cells’ responses to replication fork blocks by HU or those to DNA damage in addition to its predicted essential function in the initiation and progression of S phase.
MATERIALS AND METHODS
Yeast strains, media, and genetics.
Schizosaccharomyces pombe strains used in this study are listed in Table 1 and were grown in rich (YE5S) or minimal (EMM) medium containing the required supplements. Crosses and sporulation were performed on SPA and MEA (25). General genetic methods (25) and transformation (56) were performed as described previously. To induce expression from the nmt1 or modified nmt1 promoter (48), cells were grown to midexponential phase in EMM containing 10 μg of thiamine/ml, spun down and washed three times with EMM, before being resuspended in fresh medium lacking thiamine at a density calculated to produce 107 cells/ml at the time of peak expression from the nmt1 promoter. To disrupt him1+, the 0.4-kb HindIII fragment located in the middle of 1.9-kb him1+ cDNA (amino acids 223 to 364) was replaced with the 1.8-kb ura4+ gene in vitro. The fragment containing the disrupted him1 gene was used for gene disruption as previously described (57). Cell survival analysis for DNA replication block or DNA damage was performed as described previously (2).
TABLE 1.
Description of strains used in this study
| Strain | Genotype |
|---|---|
| NT145 | h− leu1-32 |
| NI289 | h− cdc25-22 |
| NI291 | h+ leu1-32 cdc19-P1 |
| NI293 | h+ leu1-32 ura4-D18 ade6-M216 cdc22 |
| JY765 | h+/h− leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M216/ade6-M210 |
| NI297 | h+/h− leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M216/ade6-M210 him1::ura4+/him1+ |
| NI298 | h+/h− leu1-32/leu1-32 ura4-D18/ura4+ ade6-M216/ade6-M210 |
| NI320 | h− orp1HA ura4-D18 |
| NI453 | h− leu132 ura4-D18 cds1::ura4+ |
| NI454 | h− leu1-32 cdc10-V50 |
| NI460 | h− leu1-32 ura4-D18 cds1::ura4+ chk1::ura4+ |
Plasmid DNA.
The coding frame of pGAD181 clone expressing Him1 protein (missing the first 13 amino acids) was amplified by PCR and subcloned at SalI-BamHI sites of pREP2-HA or pREP41-HA to generate pREP2-HA-him1 and pREP41-HA-him1, respectively. pREP1-myc was constructed by inserting two hybridizing oligonucleotides (5′-ATATGGAGCAAAAGCTGATTTCTGAGGAGGATCTGGCGGCCGCCGTCGACTCTAGAGGTACCG-3′ and 5′-GAT CCGGTACCTCTAGAGTCGACGCGGCCGCCAGATCCTCCTCAGAAAT CAGCTTTTGCTCCAT-3′) at NdeI-BamHI sites of pREP1 DNA. The SalI-BamHI fragment containing him1+, described above, was inserted at the SalI-BamHI sites of pREP1-myc, resulting in the expression of myc epitope-tagged Him1 protein.
Expression of Hsk1-Him1 kinase complex in insect cells.
Hsk1, Him1, and their derivatives were expressed on pVL1392 and pVL1393 plasmids in insect cells Sf9 (Invitrogen, Inc.). hsk1+ cDNA was inserted at EcoRI-BamHI sites of pVL1392 (pVL1392-Hsk1). KK mutation, in which two consecutive lysine residues at 129 and 130 were replaced with arginine and serine, was transferred to pVL1392-Hsk1 by replacing an EcoRV-BamHI fragment containing the C-terminal two-thirds of the coding frame with the same fragment containing a mutation. pVL1393-HA-him1 was constructed by inserting the NdeI (filled-in)-BamHI fragment of pREP2-HA-him1 at SmaI-BglII sites of pVL1393. Propagation of Sf9 cells, transfection of DNA, and infection of virus solutions were conducted according to the supplier’s instructions.
Antibodies.
GST-Hsk1C containing the C-terminal 84 amino acids of Hsk1 protein and GST-Him1(ΔSpeI) containing the N-terminal 367 amino acids of Him1 protein were purified as previously described (30) and were used as antigens to produce anti-Hsk1 and anti-Him1 protein antibodies in rabbit. Anti-Him1(ΔSpeI) antibody was further affinity purified.
Overexpression and purification of GST-SpMCM2N protein.
The cDNA encoding amino acids 1 to 220 of the fission yeast MCM2 was amplified by reverse transcription-PCR with a set of primers possessing NotI or SalI site at each end. The amplified fragment was digested with NotI plus SalI and was subcloned at the NotI-SalI sites of pGEX-5X-3 (Pharmacia). The resulting plasmid expressed a 60-kDa GST-SpMCM2N fusion protein in C600lon− cells, which was purified as previously described (30).
Immunoprecipitation and kinase assays.
One microgram of affinity-purified antibody or 10 μg of protein A-affinity column-purified antibody was added to the 200-μl extract (1 mg of protein), and protein-antibody complexes were collected onto protein A-Sepharose beads. After several washings of the beads with immunoprecipitation (IP) buffer (40 mM HEPES-KOH [pH 7.6], 100 mM potassium glutamate, 1 mM EGTA, 2 mM dithiothreitol, and protease inhibitors [100 μg of tolylsulfonyl phenylalanyl chloromethyl ketone {TPCK}/ml, 0.1 μg of aprotinin/ml, 0.5 μg of leupeptin/ml, 0.1 μg of chymostatin/ml, 1 μg of pepstatin A/ml, and 10 μg/ml bacitracin]), they were run on sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGs) or used for kinase assays.
Phosphatase treatment.
Immunoprecipitates, washed extensively with IP buffer, were resuspended in λ phosphatase buffer containing 2 mM MnCl2 and divided into two equal aliquots. To one tube, 400 U of λ phosphatase and 10 U of calf intestine alkaline phosphatase were added, and both tubes were incubated for 1 h at 30°C.
Chromatin purification.
The procedure for chromatin purification is based on the method described previously (17). Spheroplasts were prepared by treating S. pombe cells growing in a vegetative state with 0.1 mg of Zymolyase-100T/ml (0.1 mg/ml; Seikagakukogyo Co., Ltd.) and glusulase (0.5% [vol/vol]; Dupont Company). Spheroplasts were lysed in 10 mM PIPES-KOH (pH 6.8), 2 mM magnesium acetate, 100 mM potassium glutamate, protease inhibitors, and 1% Triton X-100. After incubation on ice for 20 min, supernatant and pellets (chromatin enriched) were separated by centrifugation. Pellets were further treated with IP buffer containing NaCl at the concentration indicated in the figure legend, on ice, for 20 min. Alternatively, they were digested with micrococcal nuclease (MNase) (8 μg/ml) in 10 mM Tris-Cl (pH 8.0) and 2 mM CaCl2 or with DNase I (160 μg/ml) in 10 mM Tris-Cl (pH 8.0), 2 mM CaCl2 and 5 mM MgCl2 for 30 min at 30°C. After centrifugation, the supernatant and pellet were separated.
Preparation of extracts from fission yeast cells and insect cells.
The whole-cell extracts of fission yeast cells were prepared as follows (53a). The harvested cells (from 5 ml of culture) were resuspended in 250 μl of water, boiled at 90°C for 5 min, and 300 μl of 2×-concentrated Laemmli’s SDS sample buffer containing 8 M urea and 500 μl of acid-washed glass beads were added. After vigorous vortexing for 5 min, the samples were heated at 90°C for 5 min and sonicated for 1 min, and the supernatant were recovered by centrifugation. Concentrated extracts of fission yeast cells were prepared as previously described (46). Cell lysates were also prepared by vortexing the cells with glass beads in buffer containing 40 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 0.5 M NaCl, 8 M urea, 0.1% sodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride. Extracts were also prepared by lysing the spheroplasts in buffer containing Triton X-100 and 0.5 M NaCl. Insect cells were disrupted in 0.5 M NaCl, 40 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 10% glycerol, 2 mM dithiothreitol, 0.1% Nonidet P-40, and protease inhibitors by homogenization in a glass hand homogenizer. After centrifugation, the supernatant was recovered.
Mutagenesis.
PCR-mediated mutagenesis was conducted to introduce amino acid substitutions and deletions into him1+ and hsk1+ coding frames. The presence of mutations and the absence of undesired mutations were verified by direct sequencing of the amplified region.
Flow cytometry, DAPI staining, and indirect immunofluorescence.
Cells (5 × 106 to 1 × 107) were spun down, washed once with water, and then fixed in 70% ethanol. The fixed cells were treated with RNase A (0.1 mg/ml), stained with propidium iodide (2 μg/ml), and processed for flow cytometry, as described previously (51). Becton-Dickinson FACScan was used for flow cytometry. Indirect immunofluorescence was conducted basically as previously described (36). Briefly, cells fixed with 3.7% formaldehyde in 0.1 M KH2PO4 at pH 6.5 for 90 min at room temperature were treated with Zymolyase and glusulase. Anti-HA (12CA5) antibody and rhodamine-conjugated anti-mouse (Jackson ImmunoResearch Laboratory, Inc.) were used for immunofluorescence labeling. The labeled cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml) (65) in phosphate-buffered saline (PBS) for 5 min and washed with PBS. Slides for fluorescence microscopy were prepared in p-phenylendiamine–90% glycerol as described previously (33).
Northern blotting analysis.
Total RNA was prepared from synchronized cdc25 cells, and RNA blotting analysis was performed as described by Thomas (64).
RESULTS
Identification of a protein which binds and activates Hsk1, the fission yeast homologue of Cdc7 kinase.
We searched for Hsk1-interacting molecules by two-hybrid screening and obtained several clones which indicated interaction with Hsk1 in auxotroph selection on histidine-deficient plates. Among the cDNA clones, clones 32, 72, 76, and 181 gave a high level of lacZ activity after retransformation and hybridization analyses of insert DNA indicated they are derived from the same gene.
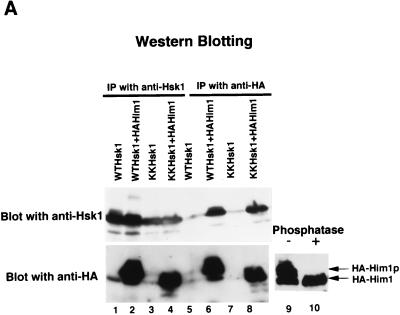
The open reading frame encoded by clone 181 (pGAD181), containing the entire coding frame of him1+ except for the first 13 amino acids, was expressed as an HA epitope-tagged polypeptide (HA-Him1) in insect cells. HA-Him1 polypeptide was coexpressed with Hsk1 protein in insect cells, and extracts were prepared. Both wild-type and kinase-attenuated Hsk1 protein (KK) was coimmunoprecipitated by anti-HA antibody (Fig. 1A, lanes 6 and 8), and reciprocally, anti-Hsk1 antibody immunoprecipitated both Hsk1 and HA-Him1 protein (Fig. 1A, lanes 2 and 4). In the presence of the wild-type Hsk1 protein, mobility of Him1 protein on SDS-PAGs was shifted upward (Fig. 1A, lower panel, lanes 2 and 6). This mobility shift was caused by phosphorylation, since it was erased by phosphatase treatment (Fig. 1A, lanes 9 and 10). This shift was not observed with the KK mutant, in which conserved lysine residues at positions 129 and 130 were replaced with arginine and serine, respectively (Fig. 1A, lower panel, lanes 4 and 8), indicating that Hsk1 phosphorylated Him1 in insect cells. A similar mobility shift was observed when hsk1+ and him1+ were overexpressed in yeast cells (data not shown).
FIG. 1.
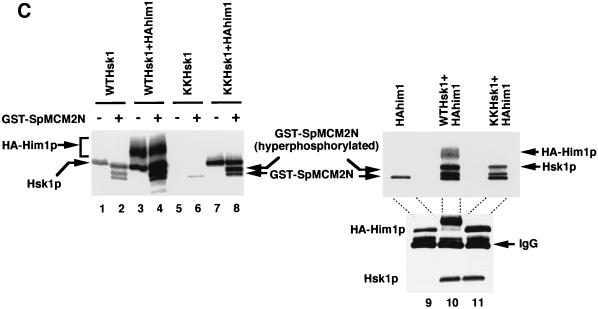
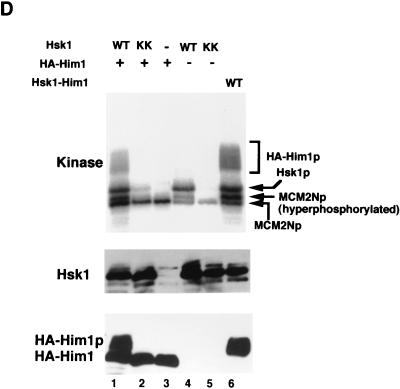
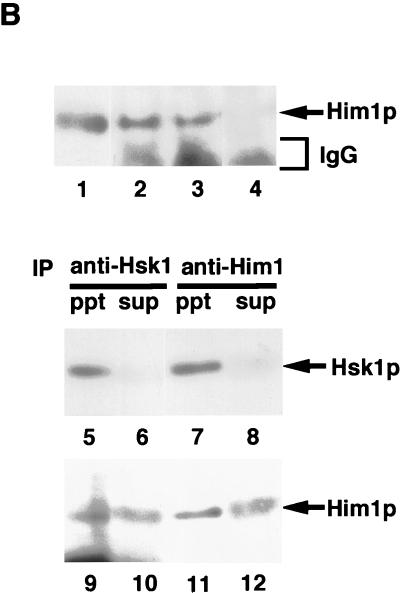
Him1 and Hsk1 proteins form an active kinase complex. (A) Extracts were prepared from insect cells expressing Hsk1 (WT, wild-type; KK, KK129, 130RS mutant) with or without HA-tagged Him1 protein as indicated in the figure, and immunoprecipitates made with anti-Hsk1 antibody or anti-HA antibody were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis through Western blotting with the indicated antibody. Immunoprecipitate from the extract expressing both the wild-type Hsk1 and HA-tagged Him1 with anti-Hsk1 antibody was incubated with (lane 10) or without (lane 9) the mixture of λ phosphatase and calf intestine alkaline phosphatase for 60 min at 30°C before being applied to SDS-PAGs. (B) Concentrated extracts were prepared from the wild-type fission yeast cells as previously described (46) and then immunoprecipitated with anti-Him1 antibody (lane 2), anti-Hsk1 antibody (lane 3), or a control antibody (lane 4), followed by Western blotting with the anti-Him1 antibody. Lane 1, an insect cell extract expressing HA-tagged Him1 protein lacking the N-terminal 13 amino acids. The immunoprecipitates (ppt) (lanes 5, 7, 9, and 11) or their supernatants (sup) (lanes 6, 8, 10, and 12) prepared from the same extract were analyzed by Western blotting with anti-Hsk1 (lanes 5 to 8) or with anti-Him1 (lanes 9 to 12) antibody. (C) The anti-HSK immunoprecipitates used in panel A were assayed in kinase reactions in the presence of GST-SpMCM2N protein as a substrate as described in Materials and Methods (lanes 1 to 8). Immunoprecipitates were prepared with anti-HA antibody from extracts expressing HA-tagged Him1 with or without Hsk1 and were used for kinase assays in the presence of GST-SpMCM2N protein (lanes 9 to 11, upper panel). The presence of Him1 or Hsk1 protein was examined by Western blotting (lanes 9 to 11, middle and lower panels). (D) Hsk1 (wild-type or KK) and HA-tagged Him1 protein were separately expressed and immunoprecipitated with anti-Hsk1 and anti-HA antibody, respectively. IPs were mixed in vitro as indicated in the figure (lanes 1 to 5). − and +, the absence and the presence of the IP indicated to the left in the reaction mixtures. HA-Him1p represent hyperphosphorylated HA-tagged Him1 protein. Lane 6, IP from extract coexpressing Hsk1 and HA-Him1 proteins. All the reaction mixtures contained GST-SpMCM2N protein as a substrate.
Endogenous Him1 protein, the size of which was identical with that of HA-Him1 protein in which the first 13 amino acids are replaced with an HA tag of the same length, could be immunoprecipitated by anti-Him1 antibody but not by a control antibody (Fig. 1B, lanes 1 to 4). Immunoprecipitation from the concentrated extract prepared from the wild-type fission yeast cells indicated that endogenous Hsk1 and Him1 proteins were coimmunoprecipitated by antibody against either protein (Fig. 1B). More than 80% of endogenous Hsk1 protein was immunoprecipitated by anti-Him1 antibody, suggesting that Hsk1 functions through interaction with Him1 protein. These results establish that Hsk1 and Him1 form a complex.
Him1 stimulates kinase activity of Hsk1 in vitro.
In vitro phosphorylation assays with the immunoprecipitates indicated that Hsk1 protein autophosphorylated in the absence of Him1 protein (Fig. 1C, lane 1). This phosphorylation was significantly diminished with the KK mutant Hsk1 (Fig. 1C, lane 5), confirming that kinase activity of Hsk1 is responsible for this phosphorylation. In the presence of Him1 protein, both Hsk1 and Him1 proteins were phosphorylated (Fig. 1C, lane 3). Although the level of autophosphorylation of the wild-type Hsk1 protein was only slightly stimulated in the presence of Him1 protein, autophosphorylation of the mutant Hsk1 protein was stimulated to a level close to that of the wild type under the same conditions (Fig. 1C, lane 7), suggesting that Him1 could stimulate the kinase activity of this attenuated Hsk1 mutant. However, the mutant Hsk1 did not stimulate the level of phosphorylation of Him1. Stimulation of the wild-type Hsk1 kinase activity by Him1 protein was more clearly shown with an exogenously added substrate, a recombinant protein containing N-terminal 220 amino acids of S. pombe MCM2 protein. This recombinant MCM2 was phosphorylated by the wild-type Hsk1 alone, and the level of this phosphorylation and the extent of its mobility shift were significantly enhanced by the presence of Him1 protein (Fig. 1C, compare lanes 2 and 4 and lanes 6 and 8). A similar result was obtained with immunoprecipitates prepared with anti-HA antibody. Phosphorylation of MCM2, which was observed with Him1 IP alone to a small extent, presumably due to a coprecipitated unknown kinase, was significantly stimulated when Hsk1, either wild type or KK mutant, was coexpressed (Fig. 1C, lanes 9 to 11). Furthermore, Him1 protein, expressed separately and mixed with Hsk1 protein in vitro, could stimulate kinase activity of the latter protein, as shown by induction of mobility shift of Him1 protein and MCM2 protein (Fig. 1D, compare lanes 1 and 4). Slight stimulation of Hsk1 autophosphorylation and MCM2 phosphorylation was also observed with the KK mutant (Fig. 1D, lane 2). These results demonstrate that Him1 is a regulatory subunit of Hsk1 kinase and stimulates the intrinsic kinase activity of Hsk1.
Structure of him1+: two conserved motifs.
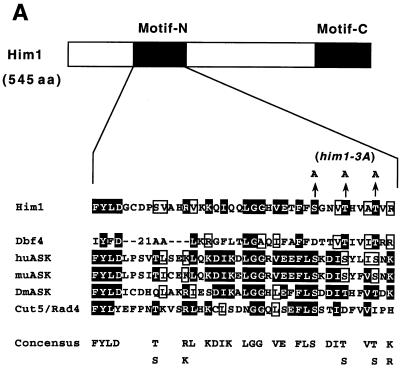
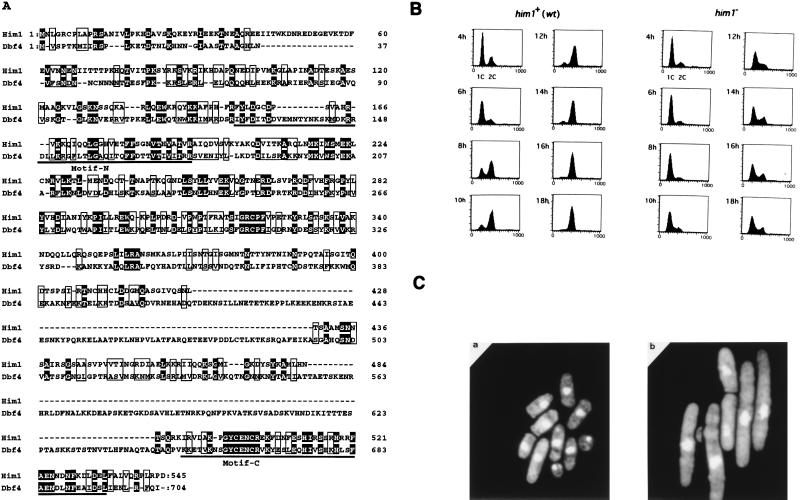
Sequencing of cDNA clone pGAD181 revealed the presence of an open reading frame encoding a protein possessing detectable similarity to S. cerevisiae Dbf4. Screening of a genomic library with the cDNA clone 181 as a probe led to the isolation of a clone containing a 5.0-kb BamHI-BglII genomic DNA fragment. Comparison of the genomic sequence with cDNA indicated the absence of introns in the him1+ gene. The open reading frame carried by pGAD181 lacked the first 13 amino acids but apparently is functional, since the HA-tagged construct can restore the growth of the him1+ null strain, as shown in Fig. 4A and 6B. The endogenous Him1 protein was detected by Western analysis, and migrated on SDS-PAGs at the position identical with that expressed from a plasmid containing the putative full-length him1+, confirming that the proposed coding frame is correct (data not shown; also Fig. 1B). him1+ is identical to dfp1+, which was identified on a database and was reported to be a subunit of Hsk1 kinase (7). Overall identity between him1+ and DBF4 is about 25% and similarity is 46%, although there are at least two large gaps of 83 and 91 amino acids on Him1 (Fig. 2A). The highest degree of identity (48%; 24 of 50 residues) was detected in the C-terminal regions of both proteins, which are also conserved in regulatory subunits for Cdc7-related kinases from higher eukaryotes and were designated motif C (underlined region in Fig. 2A) (41). Among the clones isolated from two-hybrid screening, clone no. 72 contained only the C-terminal 50 amino acids of Him1, suggesting that motif C was sufficient for binding to Hsk1 in two-hybrid assays. A segment of S. cerevisiae Dbf4 containing the corresponding motif C was previously reported to be sufficient for interaction with Cdc7 (26). Another high degree of homology was detected in the center of the protein (from residues 167 through 359; 32% identity and 57% similarity). Overlapping with this homologous segment, another stretch of amino acids, motif N, was discovered, which was also conserved in Cdc7 regulatory subunits of higher eukaryotes. Motif N is weakly conserved in Dbf4, although there appears to be an insertion within this motif of Dbf4 (Fig. 2A, see also Fig. 6A). A database search revealed 100% identity of the him1+ with the rad35+ gene (isolated by Struck and Schmidt; GenBank accession no. SPY17146). Functions of Him1 protein in DNA damage responses are discussed below.
FIG. 4.
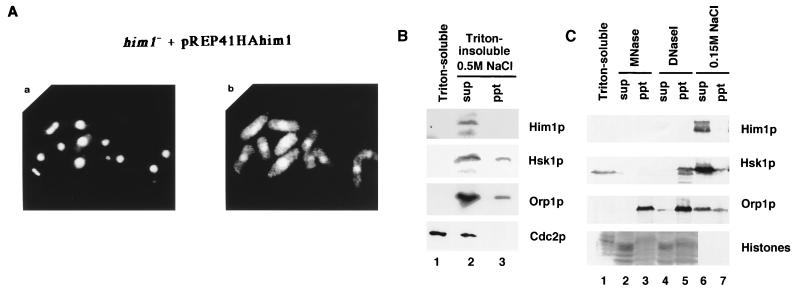
Him1 protein is localized in the nucleus. (A) Indirect immunofluorescence analysis of Him1 protein in him1− cells complemented with HA-tagged Him1 protein. (a) DAPI staining of DNA; (b) immunofluorescence image using anti-HA antibody. (B) Orp1HA-tagged cells (24) were harvested and treated with Zymolyase. Spheroplasts were lysed with 1% Triton X-100 in the buffer containing 100 mM potassium glutamate (Materials and Methods). After centrifugation, supernatants (lane 1, Triton soluble) were saved and pellets (Triton insoluble) were further treated with the buffer containing 0.5 M NaCl, and supernatants (lane 2) and pellets (lane 3) were separated by centrifugation. Each fraction from equivalent cell numbers were separately run on 8% SDS-PAGs. Proteins were blotted with anti-Him1, anti-Hsk1, anti-HA, or anti-PSTAIRE antibody (from the top). (C) Spheroplasts of Orp1HA-tagged cells were lysed as described above with 1% Triton X-100, and after centrifugation the supernatant (lane 1) and pellet was separated. More than 80% of MCM5 was present in the Triton-soluble fraction (data not shown). The pellet was further treated with MNase (lanes 2 and 3), DNase I (lanes 4 and 5) or buffer containing 0.15 M NaCl (lanes 6 and 7). After centrifugation, supernatant (sup) (lanes 2, 4, and 6) and pellet (ppt) (lanes 3, 5, and 7) were separated. Each fraction from equivalent cell numbers was run separately on 8% SDS-PAGs. Proteins were blotted with specific antibodies to detect proteins indicated. Histones were detected by Coomassie blue staining of the gel.
FIG. 6.
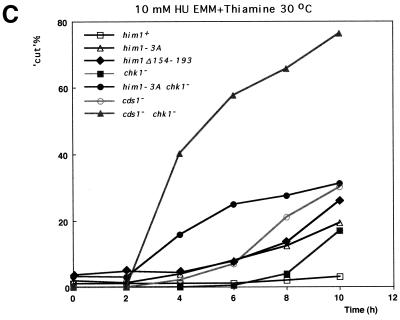
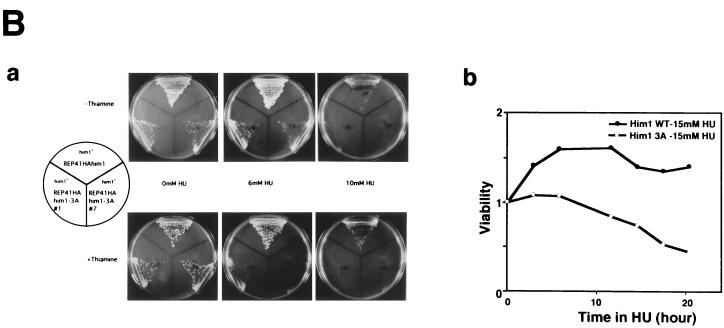
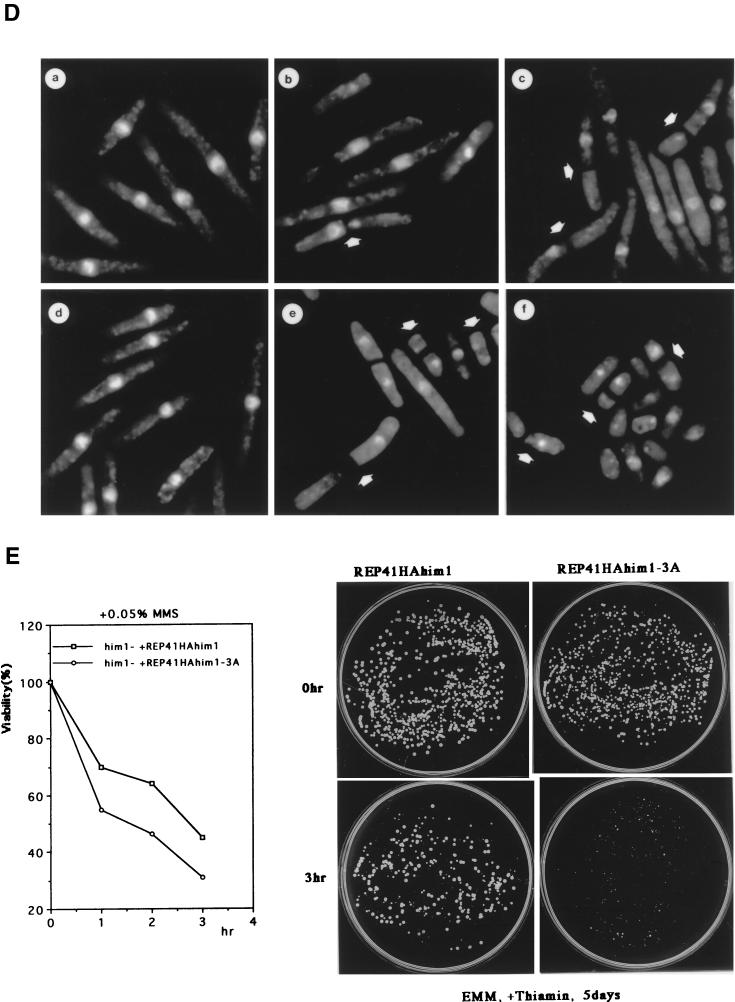
An alanine substitution mutant of Him1 protein exhibits sensitivity to HU and DNA-damaging agents. (A) Alignment of motif N conserved in Him1 and regulatory subunits (ASK) for Cdc7-related kinases from higher eukaryotes (41) as well as in fission yeast Cut5 protein (59). Three conserved serine/threonine residues mutated to alanine are indicated. Residues conserved in more than three members or those not identical but with similar functionality are shown as white or boxed letters, respectively. (B) Effect of HU on growth of him1− cells carrying a plasmid expressing wild-type or 3A mutant him1 (two independent clones); growth on plates (a) or survival rate in liquid culture (b). Plates shown are with (lower panel) or without (upper panel) thiamine. (C) Appearance of cut-like cells in HU-treated him1Δ154-193 and him1-3A mutant cells. Various vegetatively growing transformants or mutant strains (5 × 106 cells/ml) were grown in the presence of 10 mM HU and thiamine. Aliquots were sampled every 2 h to count the number of cut cells under the fluorescence microscope. (D) Cut-like morphology of him1− transformants or deletion strains. Cells arrested with HU for 8 h described in panel C were stained with DAPI and were examined by fluorescence microscopy. (a) him1− cells plus pREP41HAhim1; (b) him1− cells plus pREP41HAhim1-3A; (c) cds1− cells; (d) him1− chk1− pREP41HAhim1; (e) him1− chk1− cells plus REP41HAhim1-3A; (f) cds1− chk1−. White arrows indicate cut-like cells. (E) Growth recovery from DNA damage in him1− cells expressing Him1-3A. Vegetatively growing cells (5 × 106 cells/ml) were treated with MMS in the presence of thiamine, and aliquots were sampled every hour. One thousand cells were plated on EMM to examine survival of DNA damage. Survival curve (left panel) and colony formation assay (right panel) are shown.
FIG. 2.
him1+ is essential for G1/S transition in fission yeast cells and is related to S. cerevisiae DBF4. (A) Primary structures of Him1 protein as deduced from cDNA and genomic sequences in comparison with S. cerevisiae Dbf4 protein. Identical residues are shown in white letters. Motif N and motif C, conserved in regulatory subunits for Cdc7-related kinases, are underlined. (B) DNA content of germinating him1− cells. NI298 (him1+/him1+ ura4-D18/ura4+) or NI297 (him1+/him1::ura4+) was sporulated in EMM(-N) supplemented with leucine. The resulting spores were inoculated into EMM containing leucine and adenine at the concentration of 1 × 107 cells/ml and incubated at 30°C. Aliquots were sampled every 2 h, stained with propidium iodide, and then analyzed by fluorescence-activated cell sorting, as previously described (46). (C) Terminal morphology of him1− cells. Germinating cells described in (B) were stained with DAPI and examined by fluorescence microscopy. (a) wild type; (b) him1−.
him1+ is essential for viability, and a null mutant arrests with 1C DNA content.
To address the in vivo functions of him1+, a heterozygous diploid, in which one of the him1+ alleles was disrupted by replacement of the 0.4-kb HindIII fragment located in the middle of the coding frame (amino acids 223 to 364) of chromosomal him1+ with ura4+ gene by homologous recombination, was constructed. After sporulation, the diploid strain produced two nonviable, presumably ura+, segregants and two viable ura− segregants, indicating that him1+ is essential for cell growth (data not shown). Flow cytometry was used to analyze the DNA content of the germinating spores carrying him1−. The majority (over 80%) of the him1− cells had 1C DNA content even at 18 h after germination, whereas him1+ cells contained 2C DNA content at the same time point (Fig. 2B), consistent with predicted essential functions for G1/S transition in S. pombe. Most of the nonviable germinating spores carrying him1− arrested as an elongated shape with abnormally deformed nuclei (Fig. 2C, panel b). Approximately 10% of the cells displayed a cut-like morphology (data not shown) which was also observed in hsk1− germinating cells, albeit with a higher frequency (46). The lethal phenotype of the him1 null strain could be completely rescued by the plasmid carrying the HA-tagged cDNA described above (Fig. 4A and 6B).
Expression of him1+ during the mitotic cell cycle.
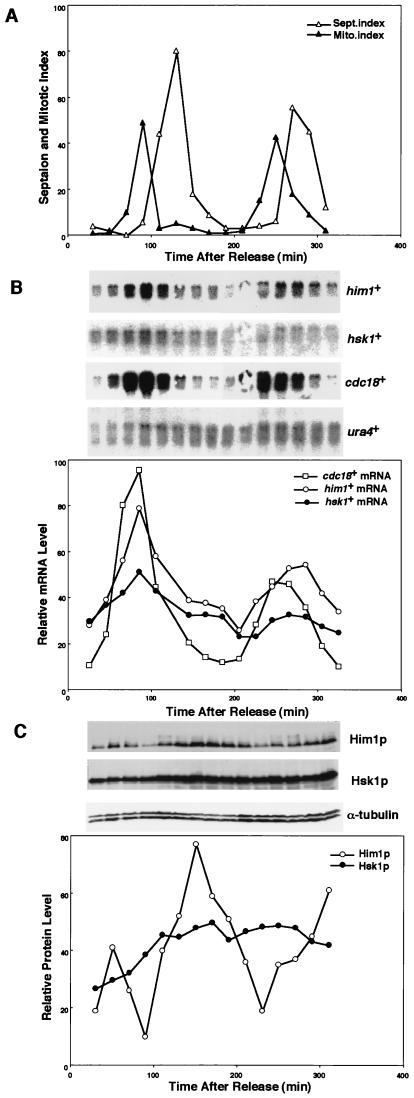
Transcription of him1+ was examined in the synchronized cell populations obtained from G2-arrested cells of a cdc25 mutant (Fig. 3A). As a control, the cdc18+ transcript increased at late M and decreased at late G1 through S phase (3). Similarly, the him1+ transcription increased at late M to G1 and then decreased during S phase. Similar to DBF4 of S. cerevisiae, a low but significant level of the him1+ transcription was observed even during G2/M (Fig. 3B). In contrast, transcription of hsk1+ is relatively constant during the mitotic cell cycle, although there appears to be a slight increase of the hsk1+ transcript level at late G1 (Fig. 3B). The protein level was also examined in the same synchronized cell cultures. It was low during M to G1, increased at late G1 to S, and then decreased again by the next G1 (Fig. 3C). In contrast, the Hsk1 protein was present at a relatively constant level during the cell cycle. These results established that the expression of him1+ is cell cycle regulated and is highest during S phase at the time of its function.
FIG. 3.
Expression of Hsk1 and Him1 transcripts and proteins during the cell cycle. (A) cdc25 cells were arrested at the restrictive temperature for 2.5 h and then released into growth at the permissive condition (25°C). Cells were harvested every 20 min, and mitotic and septation index were measured at each time point. (B) RNA was prepared from each culture, and Northern blot analysis was conducted with the probe indicated in the figure. (C) Whole-cell extracts were prepared from cells at each time point and analyzed by Western blotting to detect Him1, Hsk1, and α-tubulin proteins.
Association of Hsk1-Him1 kinase complex with insoluble nuclear fractions.
Indirect immunofluorescence analysis of HA-tagged Him1 protein expressed from a plasmid indicated that it is exclusively localized in nuclei most likely throughout the mitotic cell cycle (Fig. 4A). We then examined the subcellular localization of Him1 and Hsk1 proteins. Spheroplasts were lysed with Triton X-100 in the presence of 100 mM potassium glutamate. The supernatant was saved, and the insoluble proteins were extracted from the pellet in a solution containing 0.5 M NaCl. Him1 protein was not detected in the Triton-soluble fraction under low-salt conditions, while more than 50% of Cdc2 protein was present in this fraction. Nearly all Him1 protein and more than 80% of Hsk1 protein was recovered in the 0.5 M NaCl soluble fraction, as was the case for the major part of Orp1 protein, which is a component of the six-protein complex, ORC, and is presumably located at or close to replication origins (53) (Fig. 4B). Hsk1 and Him1 proteins were recovered in supernatant under conditions above 150 mM NaCl (Fig. 4C). Although histones in the insoluble fraction were released into the soluble fraction by treatment with MNase or DNase I (Fig. 4C), Hsk1 protein after treatment with DNase I stayed in the pellet as for Orp1 after treatment with either nuclease. Him1 protein appears to be degraded during digestion and was not detected in either fraction except for smaller presumably degraded polypeptides in the pellet fraction (Fig. 4C and data not shown). These results indicate that Hsk1 protein and, very likely, Him1 protein as well, like Orp1 (24, 53), are associated with a nuclear structure or nuclear matrix.
Hyperphosphorylation of Him1 protein during S phase and in response to early S-phase arrest induced by nucleotide deprivation.
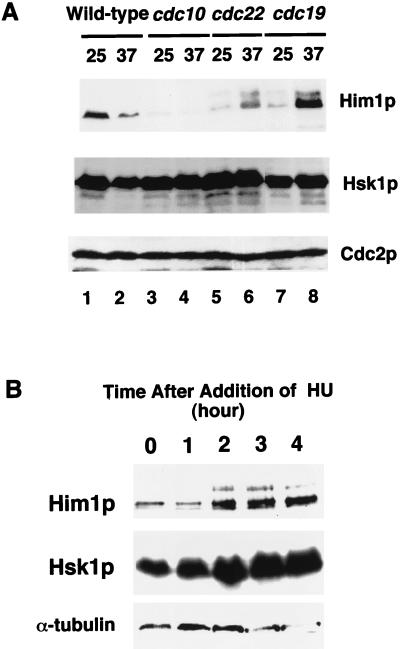
Extracts were made from various cdc(Ts) mutants which had been incubated at a nonpermissive temperature for 3 h and thus had been arrested at different cell cycle stages. Western blotting with Him1 and Hsk1 antibodies indicated that Hsk1 protein is present at a relatively constant level at different cell cycle stages, consistent with the result of synchronized cells (Fig. 5A). The level of Him1 protein was generally low in cdc(Ts) strains at a permissive temperature. At a nonpermissive temperature, it significantly increased in cdc22 and cdc19 mutants, which are arrested at early S phase and during S phase, respectively (21) (Fig. 5A), whereas it decreased in a cdc10 mutant arrested at START in G1. These results are in agreement with those of synchronized cultures (Fig. 3C) and support our conclusion that Him1 protein level is low in G1 and increases during S phase.
FIG. 5.
Him1 protein is hyperphosphorylated in response to cell cycle arrest at S phase. (A) Whole-cell extracts were prepared from the wild type and various cdc mutants grown at a permissive temperature or arrested by incubation at a nonpermissive temperature for 3 h and were run on 8% SDS-PAGE, followed by Western blotting to detect the proteins indicated. (B) Concentrated extracts were prepared from the wild-type fission yeast cells at various times after the addition of HU (10 mM), and analyzed by Western blotting by using an antibody raised against the protein indicated to the left of each panel.
Interestingly, in the cdc22 mutant, which is defective in ribonucleotide reductase (19), an additional slow-migrating band appeared by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 5A). This band, which was detected albeit at a reduced level even at a nonpermissive temperature in this mutant, disappeared after treatment with phosphatase (data not shown), indicating that the band shift is caused by phosphorylation. Mobility shift of Him1 protein was observed in a cdc19 mutant as well as in synchronized cell populations (Fig. 3C and 5A). In the latter case, the shifted band appeared at late G1 to S phase, coincident with the increase of Him1 protein level. The extent of mobility shift in cdc22 arrested cells was slightly greater than that observed in cdc19, indicating that early S-phase arrest in cdc22 may lead to phosphorylation distinct from or added to that normally observed in S phase. A similar mobility shift of Him1 protein was observed when the wild-type cells were treated with HU (Fig. 5B). A slow-migrating band, which was eliminated by phosphatase treatment (data not shown), appeared at 2 h after the addition of HU addition and persisted at least until 4 h, with concomitant increase of the amount of the total protein. No significant change was observed in the amount and mobility of Hsk1 protein under the same conditions (Fig. 5B). These results indicate that S-phase arrest by nucleotide deprivation induces hyperphosphorylation of Him1 protein.
Identification of Him1 mutants which render cells sensitive to DNA replication block and DNA damage.
In an attempt to dissect the structure of Him1 protein in relation to its functions, we constructed a mutant him1, which lacks either motif C or motif N. A C-terminally truncated Him1 lacking motif C could not complement the growth of the him1 null mutant, indicating that motif C is essential for mitotic function (data not shown). On the other hand, a him1 mutant with an internal deletion (him1Δ154-193) of motif N was able to support growth of the him1 disruptant and did not appreciably affect its mitotic function. However, we noticed that the growth of this strain was sensitive to HU (data not shown). Furthermore, about 25% of the population exhibited a cut phenotype at 10 h after addition of HU, indicative of a defect in checkpoint control (Fig. 6C). We noticed the presence of three conserved serine/threonine residues in the motif N and have mutated these three residues to alanine and examined the function of this him1-3A mutant protein (Fig. 6A). The him1 disruptant carrying pREP41HAhim1-3A was able to grow, albeit at a reduced rate compared to that of the wild type. However, this transformant did not grow on a minimal plate containing thiamine and 6 mM HU, whereas the wild type formed colonies on a similar plate containing 10 mM HU. On the plate lacking thiamine, the 3A mutant grew on 6 mM HU but did not grow on 10 mM HU (Fig. 6B). Thus, HU sensitivity in him1-3A appears to be partly compromised by the elevated level of expression of the mutant protein. In liquid culture, the cells started to lose viability at 6 h after HU addition, and the number of viable cells continued to decline thereafter (Fig. 6B).
We examined the effect of the him1-3A mutation on the morphology of the yeast cells in the presence of HU. In the following experiments, cells were incubated with 10 mM HU at 30°C in the presence of thiamine. Fluorescence-activated cell sorting analysis indicated that him1− cells carrying him1+ arrested with 1C DNA content at around 2 h after the addition of HU restarted DNA replication after another 2 h (data not shown) and exhibited elongated morphology with one nucleus (Fig. 6C and D). The majority of him1-3A cells elongated in a manner similar to that of the wild-type cells, and cut-like cells, in which the aberrant mitosis occurred in elongated cells without DNA replication, appeared with a frequency and kinetics similar to those seen in cds1− background (Fig. 6C and D). The cds1+ gene, originally isolated as a high-copy suppressor of swi7H4, one of DNA polymerase α temperature-sensitive alleles, is required for S-phase checkpoint control (43, 52). The results indicate that him1-3A as well as him1Δ154-193 are weakly defective in S-phase checkpoint control. chk1+ also encodes a protein which plays an essential role in both DNA replication and DNA damage checkpoint controls (22, 23, 66, 67). The cds1− chk1− double mutant is totally devoid of replication checkpoint control, and very small cut cells appear at as early as 2 h after the addition of HU (4, 68) (Fig. 6C and D). In him1-3A chk1− cells, similar small cut cells started to appear at 2 h after HU addition albeit with a lower frequency than that of the cds1− chk1− double mutant, followed by appearance of elongated cut cells at later time points (Fig. 6C and D). Thus, him1-3A mutation can synergistically enhance replication checkpoint defect of chk1− cells, although the effect is not as great as cds1− mutation combined with chk1−. We have concluded that S-phase checkpoint control is partly impaired in the him1-3A mutant.
The him1-3A mutant cells were sensitive to UV and MMS as well (Fig. 6E). Although the survival rate of the him1-3A cells after treatment with 0.05% MMS was only slightly lower than that observed in the wild-type him1+ (Fig. 6E, left panel), the rate of growth recovery was much slower in him1-3A cells than that in the wild type (Fig. 6E, right panel). Similar growth retardation of him1-3A cells was also observed after treatment with UV (data not shown). The mutant cells were arrested at G2 after DNA damage, most of the cells were elongated with one nucleus, and few cut cells were observed, suggesting normal DNA damage checkpoint functions in him1-3A cells. Thus, the Hsk1-Him1 complex may function in some aspect of growth recovery from DNA damage-induced arrest.
DISCUSSION
Cdc7 kinase of S. cerevisiae plays a key role in initiation of DNA replication (27, 61). Its kinase activity is regulated by association with the regulatory subunit, Dbf4, whose expression is cell cycle-regulated and increases at the G1/S boundary (10, 31, 37). Genetic and biochemical evidence suggests that MCM may be an important target of Cdc7 kinase (32, 33, 60). Although the conserved presence of the Cdc7 catalytic subunits has been reported in organisms ranging from yeasts to humans (32, 35, 46, 60), it has not been known whether activities of Cdc7-related kinases are regulated by a regulatory subunit related to Dbf4 protein. Only recently, Brown and Kelly reported association of Dfp1 with Hsk1 and proposed that it is a fission yeast homologue of Dbf4 (7). Furthermore, we have isolated a putative regulatory subunit (ASK, originally called H37) for human Cdc7-related kinase (huCdc7) (41).
him1+ encoding a Hsk1 binding protein activates Hsk1 kinase in vitro.
Here we report a fission yeast gene, him1+, which was isolated by two-hybrid screening for Hsk1-interacting molecules. It encodes a 545-amino-acid protein which possesses regional homology to DBF4 and was identical to dfp1+. Him1 protein forms a complex with Hsk1 protein. Unlike budding yeast Cdc7 and huCdc7, which are totally inactive as a kinase on its own (31, 41, 47), Hsk1 protein exhibited intrinsic kinase activity when expressed singly in insect cells or overexpressed in yeast cells (Fig. 1C and data not shown). Coexpression of Him1 protein stimulated kinase activity of Hsk1, as measured by the level of autophosphorylation as well as by that of MCM2 phosphorylation. Him1 was able to activate a severely kinase-attenuated mutant Hsk1 protein. Furthermore, Him1 protein, expressed separately and mixed with Hsk1 protein in vitro, could stimulate kinase activity of the latter protein. It was reported that Dfp1 activates the phosphorylation of exogenous substrates but not the level of autophosphorylation, and it was suggested that the role of Dfp1 is to alter the substrate specificity of Hsk1 kinase (7). Our results for recombinant wild-type Hsk1-Him1 kinase complex are in agreement with this report, and the presence of Him1 significantly stimulated only the phosphorylation of exogenously added GST-SpMCM2N. However, Him1 protein dramatically stimulated kinase activity (including autophosphorylation) of an Hsk1 KK mutant, which was inactive by itself. The basal intrinsic kinase activity of the wild-type Hsk1 appears to be sufficient for autophosphorylation as well as for low-level phosphorylation of exogenous substrates, although further activation of its kinase activity by association with Him1/Dfp1 is required for full-level phosphorylation of exogenous substrates. Our results are consistent with the general notion that Dbf4-like molecules are activators of protein kinase activity of their cognate Cdc7-like catalytic subunits.
K129A mutant was reported be kinase negative, but its overexpression did not repress DNA replication in a dominant negative manner (7). We also observed that overexpression of our KK mutant as well as that of K129D and K129N mutants did not cause dominant inhibition of growth (data not shown). This is probably due to the presence of a residual kinase activity in these mutants rather than to reduced affinity of these mutant Hsk1 to Him1/Dfp1, as suggested previously. In consistent with this prediction, the KK mutant could complement the growth of hsk1-89 temperature-sensitive mutant upon overproduction (62a). We observed no significant difference in efficiency of the complex formation or stability of the complexes between the wild type and the mutants (data not shown).
him1+ is essential for DNA synthesis.
him1+ is essential for viability of fission yeast cells, and him1 null cells are defective in chromosomal replication, in consistent with predicted essential function of the Hsk1-Him1 kinase complex in G1/S transition. Germinating cells of the him1 disruptant exhibited various aberrant nuclear morphology with somewhat elongated cells (Fig. 2C). The percentage of cut-like cells was about 10% compared to 20 to 25% in hsk1− disruptant cells (46), although 80% of the him1− disruptant cells arrested with 1C DNA content. It was previously reported that a large fraction of cells lacking those genes essential for initiation of DNA replication, such as cdc18+ (34) and cut5+ (59), underwent aberrant mitosis without entering S phase, resulting in cut-like morphology (lack of preinitiation checkpoint control). hsk1+ appears to belong to this gene category. him1+ was expected to behave similarly, judged from its essential function for DNA replication as a regulatory subunit of Hsk1 kinase. Why does a him1− disruptant exhibit less striking preinitiation checkpoint defect than hsk1− cells in spite of more stringent 1C arrest? We speculate that basal kinase activity of Hsk1 can convert preRC to postRC to some extent in the absence of Him1, thus activating the postinitiation checkpoint control. Our in vitro data indicate that Hsk1 kinase can phosphorylate MCM2 on its own, albeit at a reduced rate compared to Hsk1-Him1 complex (Fig. 1C). Nevertheless, him1− cells cannot undergo any measurable DNA synthesis (Fig. 2B). This may be due to the requirement of fully activated Hsk1 kinase for phosphorylation of key substrate(s) for DNA synthesis or to the requirement of Him1 protein for further activation of replication complexes, such as recruitment of key replication component(s) at the origins, or to both.
Two motifs conserved in Cdc7 regulatory subunits.
Alignment of the amino acid sequences of Him1 with those of Dbf4 indicated the presence of considerable homology between the two proteins. Higher homology was detected generally in the central part of the Him1 protein, from amino acids 167 to 359, and the more C terminal the sequence, the more it diverged, except for the very C-terminal 60 or so amino acids which show the highest identity between the two. Comparison of Him1 protein with Cdc7 regulatory subunits from higher eukaryotes indicated the presence of two conserved stretches of amino acids (motif N and motif C). Motif N has limited homology with BRCT motifs found in many repair and replication proteins (5). Functional characterization of deletion derivatives of Him1 protein indicated that the motif C is essential for mitotic function of Him1 protein, while motif N is dispensable. In two-hybrid assays, the C-terminal 50 amino acids containing motif C is sufficient for interaction with Hsk1, and Him1 lacking the motif C cannot activate Hsk1 in vitro to the full extent (data not shown). Motif C may play a critical role in binding and activation of Hsk1 kinase activity. The possible function of motif N will be discussed below.
Expression of Him1 protein is cell cycle regulated on both transcription and protein levels.
Transcription of him1+ is cell cycle regulated and reaches maximum at middle to late G1 and decreases at G2/M. The transcription starts to increase with a timing similar to that of cdc18+. A low but significant level of the him1+ transcription was detected even at G2/M phase, whereas the cdc18+ transcription was almost nondetectable at G2/M as reported previously (34). Furthermore, unlike cdc18+, expression of him1+ may not be regulated by Cdc10-Res1-Res2 transcription factor (8, 44, 45, 50, 63, 69), since the him1+ transcript was detected in a cdc10(Ts) mutant at both permissive and nonpermissive temperatures (data not shown). It should be of interest what factors regulate transcription of him1+ during the cell cycle.
Him1 protein levels also oscillate during the cell cycle. It is low during early to middle G1 and increases at late G1 through S phase. Consistent with this, it is low in a cdc10 mutant arrested at START in G1. It is noteworthy that the Him1 protein level is lowest when the transcript level is highest. We are now investigating whether Him1 protein is actively degraded during G1 phase. A mobility-shifted band, presumably a hyperphosphorylated form of Him1, appears by sodium dodecyl sulfate-polyacrylamide gel electrophoresis at the G1/S boundary. This may be caused by Hsk1 and may reflect increase of its kinase activity during S phase.
The majority of Hsk1-Him1 protein was present in an insoluble fraction, when spheroplasts were lysed under low-salt conditions. Both proteins were solubilized with a buffer containing salt (NaCl) at concentrations higher than 150 mM, at which a bulk of MCM and ORC proteins also dissociated from chromatin (data not shown). Nuclease treatment of the insoluble fractions resulted in release of histones in the soluble fraction, but not the Orp1 or Hsk1 protein, indicating that Hsk1 kinase complex, similar to ORC, may be associated with nuclear structures. We detected physical interaction between Hsk1-Him1 with Orp1 and Orp2 as well as with MCM2 protein (data not shown). The Hsk1-Him1 complex may be recruited at the replication origins through interaction with these prereplicative components. S. cerevisiae Dbf4 protein was shown to interact with replication origins in vivo (18). In view of the lack of apparent DNA binding motifs in Dbf4 or Him1 protein, the next important issue is to identify the chromatin components to which Hsk1/Dbf4 binds and to understand how this binding is regulated in a cell cycle-dependent manner.
him1+ is identical to rad35+ and is involved in the cells’ response to replication fork arrest induced by HU or in growth recovery from DNA-damaging agents.
Unexpectedly, him1+ was found to be identical to rad35+. Consistent with this finding, we identified mutant Him1 proteins which rendered the yeast cells sensitive to HU and various DNA-damaging agents. We first discovered that deletion of motif N resulted in HU sensitivity with appearance of cut cells after treatment with HU, although the mutant could support normal mitotic growth in the him1 null cells. We then made a him1 mutant by replacing the three conserved serine/threonine residues in the motif N with alanine. The resulting 3A mutant could support mitotic growth of fission yeast cells, albeit at a reduced efficiency. The growing population of him1-3A cells contained small fractions of 1C DNA cells, which are not observed in the wild-type cells. Growth of him1-3A cells was more sensitive to HU than the wild type. In the presence of HU, the mutant cells were arrested with 1C DNA content and became elongated, and the cut cells started to increase at 4 h after addition of HU, reaching 20% of the total population by 10 h. This is similar to S phase checkpoint defect observed in cds1− mutant, although this delayed appearance of elongated cut cells in him1-3A may be caused simply by inefficient S-phase progression during recovery from HU-induced arrest.
However, the early appearance of small cut cells in him1-3A chk1− double mutant, with kinetics similar to that of cds1− chk1− double mutant, indicates that Him1 may be more directly involved in S-phase checkpoint control in collaboration with Chk1. The Him1-3A protein can bind and activate Hsk1 with efficiency similar to that of the wild-type Him1 (data not shown). Thus, the him1-3A mutant, although not lethal and capable of assembling an active kinase complex, may be specifically defective in interaction with other components involved in signal transduction during the S-phase checkpoint control. Him1 protein may play a critical role in transmitting the checkpoint signals induced by alteration of replication structures to cell cycle machinery through Chk1 and Cds1. Our results are consistent with the notion that Him1 protein plays an important role in S-phase checkpoint control induced by replication fork blocks after nucleotide deprivation.
The 3A mutant was sensitive to UV and MMS as well. Although the viability of him1-3A cells after MMS treatment was similar to that of the wild type, the rate of growth recovery was significantly slower in the mutant strain (Fig. 6E). DNA damage checkpoint control is apparently intact in the 3A mutant, since the cells sustain the cell division after exposure to DNA damages. Pulsed-field gel electrophoresis analyses of chromosomal DNA after MMS treatment indicated no detectable difference in formation and repair of double-strand DNA breaks between the mutant and the wild type (data not shown), indicating that the Hsk1-Him1 kinase complex may function during the growth recovery from DNA damage.
Hyperphosphorylation of Him1 protein during G1/S and in response to S-phase arrest.
We discovered that Him1 is hyperphosphorylated in response to early S-phase arrest induced by HU treatment or by a cdc22 mutation or in cdc19 mutant at a nonpermissive temperature. The hyperphosphorylated form of Him1 also started to appear at 2 h after HU addition and persisted for several hours. In the proliferating cell cycle, a mobility-shifted form of Him1 protein was detected at late G1 through S phase. This coincides with the increase of Him1 protein, and it is likely to reflect activation of Hsk1 kinase during this period of the cell cycle. It remains to be investigated whether the phosphorylation of Him1 during late G1 through S plays any role in the initiation and progression of S phase.
The function of motif N is still not clear. Derivatives of Him1 lacking the entire motif N (him1Δ154-193) can support mitotic growth but display sensitivity to HU as seen in the 3A mutant (Fig. 6C) (55). Thus, motif N is dispensable for the mitotic function of Him1 but is required for cells’ responses to replication fork block by HU. It is interesting that motif N is also present in Cut5/Rad4 protein, which is essential for S phase and for DNA damage and replication checkpoint control (59) (Fig. 6A). It is an intriguing possibility that a common protein may interact with motif N in cells’ checkpoint response to nucleotide deprivation and/or in recovery from DNA-damaging agents. It is of interest whether HU-induced hyperphosphorylation of Him1 protein is involved in checkpoint control and whether the three serine/threonine residues in motif N are the targets of this phosphorylation. We have observed that the patterns of mobility-shifted bands on sodium dodecyl sulfate-polyacrylamide gel electrophoresis differed between the wild-type and 3A mutant after HU treatment, suggesting that the targets of HU-induced phosphorylation may be present among the mutated three serine/threonine residues (data not shown). However, we do see mobility shift of the 3A mutant after HU treatment, and there may be other residues of Him1 protein phosphorylated in response to HU-mediated growth arrest. We are currently mapping the residues of Him1 protein which are phosphorylated during S phase as well as in response to HU.
In summary, we report a fission yeast gene, him1+, encoding a regulatory subunit for Hsk1, a fission yeast homologue of budding yeast Cdc7 kinase. In addition to expected essential function of Him1 in initiation of S phase, we present evidence for a novel function of Him1, and presumably of the Hsk1-Him1 kinase complex, in cells’ checkpoint control after replication fork blocks and growth recovery from DNA damage. Identity of him1+ with rad35+ further supports our conclusions. Similar dual functions were previously reported for fission yeast Cut5/Rad4 protein (49, 58, 59).
ACKNOWLEDGMENTS
We are grateful to Takahisa Hachiya and Katsuyuki Tamai (MBL) for help in generation of antibodies against Him1 protein. We thank Masashi Uchiyama for help in synchronization experiments. We are also grateful to Koichi Tanaka and Hiroto Okayama for the gift of cdc10-V50 and cds1− strains and to Paul Nurse for the gift of Orp1HA-tagged strain. We thank Hiromi Iiyama for excellent technical assistance and Keiji Tanaka, Asako Sawano, and Masafumi Shibuya for advice on handling insect cells and for permitting us access to their laboratory facility. We also thank Noriko Sato for critical reading of the manuscript and our colleagues in the laboratory for valuable discussions and comments.
REFERENCES
- 1.Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 2.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J F, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoints and feedback control in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum B, Nishitani H, Yanow S, Nurse P. Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J. 1998;17:5689–5698. doi: 10.1093/emboj/17.19.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinase Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 6.Bousset K, Diffley J F. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown G W, Kelly T J. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 8.Caligiuri M, Beach D. Sct1 functions in partnership with cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell. 1993;72:607–619. doi: 10.1016/0092-8674(93)90079-6. [DOI] [PubMed] [Google Scholar]
- 9.Campbell J L, Newlon C S. Chromosomal DNA replication. In: Broach J R, Pringe J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 41–146. [Google Scholar]
- 10.Chapman J W, Johnston L H. The yeast gene, DBF4, essential for entry into S phase is cell cycle regulated. Exp Cell Res. 1989;180:419–428. doi: 10.1016/0014-4827(89)90068-2. [DOI] [PubMed] [Google Scholar]
- 11.Chong J P, Thommes P, Blow J J. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- 12.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffeley J F X. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 13.Dershowitz A, Newlon C S. The effect on chromosome stability of deleting replication origins. Mol Cell Biol. 1993;13:391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diffley J F X, Cocker J H, Dowell S J, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson A D, Fangman W L, Brewer B J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;15:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donovan S, Diffley J F X. Replication origins in eukaryotes. Curr Opin Genet Dev. 1996;6:203–207. doi: 10.1016/s0959-437x(96)80051-7. [DOI] [PubMed] [Google Scholar]
- 17.Donovan S, Harwood J, Drury L S, Diffley J F X. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowell S J, Romanowski P, Diffley J F X. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez S M-J, McInerny C, Harris P, Gordon C, Fantes P. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet. 1993;238:241–251. doi: 10.1007/BF00279553. [DOI] [PubMed] [Google Scholar]
- 20.Forsburg S L, Nurse P. Cell cycle regulation in yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annu Rev Cell Biol. 1991;7:227–256. doi: 10.1146/annurev.cb.07.110191.001303. [DOI] [PubMed] [Google Scholar]
- 21.Forsburg S L, Nurse P. The fission yeast cdc19+ gene encodes a member of the MCM family of replication proteins. J Cell Sci. 1994;107:2779–2788. doi: 10.1242/jcs.107.10.2779. [DOI] [PubMed] [Google Scholar]
- 22.Francesconi S, Grenon M, Bouvier D, Baldacci G. p56chk1 protein kinase is required for the DNA replication checkpoint at 37°C in fission yeast. EMBO J. 1997;16:1332–1341. doi: 10.1093/emboj/16.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 24.Grallert B, Nurse P. The ORC1 homologue orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 25.Gutz H, Heslot H, Leupold U, Lopreno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics 1. New York, NY: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 26.Hardy C F J, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–6782. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartwell L H. Genetic control of the cell cycle in yeast. II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 28.Hartwell L H. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1973;115:966–974. doi: 10.1128/jb.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda M, Arai K-I, Masai H. A fusion protein library: an improved method for rapid screening and characterization of DNA binding or interacting proteins. Gene. 1996;181:167–174. doi: 10.1016/s0378-1119(96)00497-0. [DOI] [PubMed] [Google Scholar]
- 31.Jackson A L, Pahl P M B, Harrison K, Rosamond J, Sclafani R A. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci USA. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson G D, de C. Nogueira Araujo G M. A simple method of reducing the fading of immunofluorescence during microscopy. J Immunol Methods. 1981;43:349–350. doi: 10.1016/0022-1759(81)90183-6. [DOI] [PubMed] [Google Scholar]
- 34.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 35.Kim J M, Sato N, Yamada M, Arai K-I, Masai H. Growth regulation of the expression of mouse cDNA and gene encoding a serine/threonine kinase related to Saccharomyces cerevisiae CDC7 essential for G1/S transition. J Biol Chem. 1998;273:23248–23257. doi: 10.1074/jbc.273.36.23248. [DOI] [PubMed] [Google Scholar]
- 36.Kirmatin J V, Adams A E M. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitada K, Johnson A L, Johnston L H, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitada K, Johnston L H, Sugino T, Sugino A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S transition. Genetics. 1992;131:21–29. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human beta-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 40.Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumagai H, Sato N, Yamada M, Mahony D, Seghezzi W, Lees E, Arai K-I, Masai H. A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol Cell Biol. 1999;19:5083–5095. doi: 10.1128/mcb.19.7.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindsay H D, Griffiths D J F, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowndes N F, Johnson A L, Johnston J H. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated trans-factor. Nature. 1991;350:247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- 45.Lowndes N F, McInerny A L, Johnson A L, Fantes P A, Johnston L H. Control of DNA synthesis genes in fission yeast by the cell cycle gene cdc10+ Nature. 1992;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- 46.Masai H, Miyake T, Arai K-I. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masai, H., and K.-I. Arai. Unpublished data.
- 48.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 49.McFarlane R J, Carr A M, Price C. Characterisation of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol Gen Genet. 1997;255:332–340. doi: 10.1007/s004380050504. [DOI] [PubMed] [Google Scholar]
- 50.Miyamoto M, Tanaka K, Okayama H. res2+, a new member of cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 1994;13:1873–1880. doi: 10.1002/j.1460-2075.1994.tb06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno S, Clar A, Nurse P. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 52.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 53.Muzi-Falconi M, Kelly T J. Orp1, a member of the Cdc18/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Nakafuku, M. Personal communication.
- 54.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 55.Ogino, K., T. Takeda, K.-I. Arai, and H. Masai. Unpublished results.
- 56.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rothstein R. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 58.Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:363–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 60.Sato N, Arai K-I, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sclafani R A, Jackson A L. Cdc7 protein kinase for DNA metabolism comes of age. Mol Microbiol. 1994;11:805–810. doi: 10.1111/j.1365-2958.1994.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 62.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 62a.Takeda, T., et al. Unpublished data.
- 63.Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugyiama A, Nojima H, Okayama H. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+ and SWI4 gene products. EMBO J. 1992;11:4923–4932. doi: 10.1002/j.1460-2075.1992.tb05599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas P. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toda T, Yamamoto M, Yanagida M. Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombe: video fluorescence microscopy of synchronously growing wild-type and cold-sensitive cdc mutants by using a DNA-binding fluorescent probe. J Cell Sci. 1981;52:271–287. doi: 10.1242/jcs.52.1.271. [DOI] [PubMed] [Google Scholar]
- 66.Wolworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 67.Wolworth N C, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 68.Zeng Y, Forbes K C, Wu Z, Moereno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 69.Zhu Y, Takeda T, Nasmyth K, Jones N. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8:885–898. doi: 10.1101/gad.8.8.885. [DOI] [PubMed] [Google Scholar]