Abstract
Engineered probiotic bacteria represent an innovative approach for treating and detecting a wide range of diseases including those caused by infectious agents. Antimicrobial peptides (AMPs) are promising alternatives to conventional antibiotics for combating antibiotic-resistant infections. These molecules can be delivered orally to the gut by using engineered probiotics, which confer protection against AMP degradation, thus enabling numerous applications including treating drug-resistant enteric pathogens and remodeling the microbiota in real time. Here, we provide an update on the current state of the art on AMP-producing probiotics, discuss methods to enhance gut colonization, and end by outlining future perspectives.
Keywords: Probiotics, synthetic biology, bioengineering, genetic circuits, antimicrobial peptides, living medicines
Introduction
The human microbiota is comprised of a wide variety of microorganisms that colonize surfaces on and inside our bodies, establishing a symbiotic relationship with the host, which has been termed the holobiont (1). Bacteria are one of the main constituents of the microbial flora, outnumbering human cells in the body (2). Disruptions in these bacterial communities (i.e., dysbiosis) have been shown to influence human diseases such as obesity (3), autoimmune disease (4,5), and even neurological disorders (6,7). Metagenomic analyses and large-scale sequencing carried out in the Human Microbiome Project have led to groundbreaking discoveries on microbiome composition and function (8).
Antimicrobial peptides (AMPs), also referred to as host defense peptides (HDPs), are produced by a wide range of organisms to confer protection against pathogenic bacteria and reduce niche competition (9,10). They are short, usually positively charged, and are produced by either mRNA ribosomal translation or nonribosomal protein synthesis (11,12). Ribosomally produced AMPs have been investigated for their therapeutic potential (10,13,14), making them good candidates for recombinant production and rational design (15–17). Producing AMPs by using biological factories is also cheaper than using standard chemical synthesis methodologies (18–20). Furthermore, AMPs primarily act by targeting the bacterial membrane thus reducing the likelihood of bacterial resistance development (21–23). In contrast, conventional antibiotics are restricted by their limited promiscuity, which readily selects for bacterial resistance (24). Moreover, AMPs can be highly selective compared to antibiotics and may be tuned by peptide engineering to favor specificity towards a particular target bacterial strain (25,26).
In the context of the gut environment, AMPs are produced by commensal bacteria and intestinal epithelial cells (27). These molecules are thus crucial in maintaining intestinal homeostasis by modulating microbiome composition and avoiding colonization by enteric pathogens (27,28). The intestinal microbiota has been reported as a reservoir for drug-resistant bacteria due to colonization by exogenous bacteria, or via horizontal gene transfer (29). In hospital settings, drug-resistant enteric bacteria constitute a severe health threat because they can colonize the microbiota of patients after extended exposure to antibiotics (29,30). The Center for Disease Control and Prevention (CDC) has identified enteric pathogens such as Clostridioides Difficile, vancomycin-resistant Enterococci, and drug-resistant Salmonella as urgent and serious health threats in the US (31). AMPs arise as an alternative for treating enteric infections because of their potency, lower probability of resistance, and potential to be tuned to target select pathogens (25,26,32), which may be used to prevent dysbiosis. AMPs have been administered dermally to treat skin pathogens, and intra-nasally or intra-tracheally for the successful treatment of lung infections (25,33). However, oral delivery of AMPs to the gut is often problematic because these molecules are susceptible to degradation (e.g., due to proteolytic cleavage and pH gradients) prior to reaching the site of infection in an adequate quantity, which leads to treatment failure (30,34).
Probiotics are living microorganisms that when administered in correct amounts confer health benefits to the host (35). A successful strategy to achieve drug delivery to the gut involves employing engineered probiotics for localized production and delivery of therapeutics, also referred to as living therapeutics (36). The goal of these therapies is to detect an environmental or external signal (e.g. rhamnose, arabinogalactan, anhydrotetracycline), to then, as a consequence, produce and deliver therapeutics in situ (36). Living therapies have been previously designed to deliver AMPs to the gut (30,37,38) using different strains of probiotic bacteria as chassis (Fig. 1). This approach holds great potential because it 1) allows for effective delivery of AMPs to the gut while avoiding degradation; 2) mitigates potential off-target effects (36,39); and 3) significantly reduces AMP production costs as self-replicating probiotics are less expensive than peptides generated through chemical synthesis (36,38). Here, we review current state-of-the-art strategies for AMP production using probiotic bacteria. Such approaches have already been used successfully in animal models and in veterinary practice, and promise to have applications for human health.
Figure 1. Mechanism of action of an ideal AMP-producing living therapeutic.
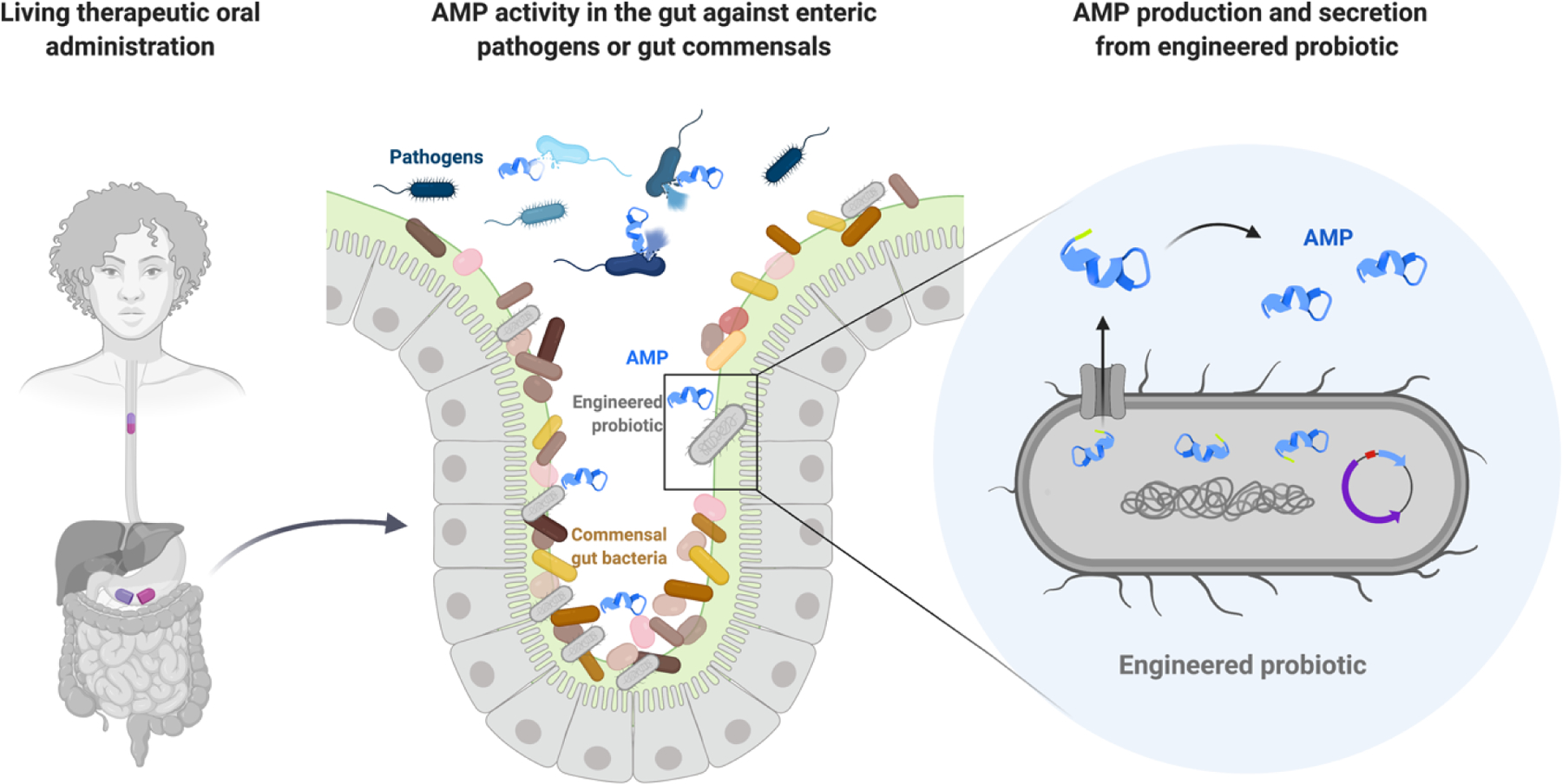
The engineered probiotic living therapeutic is administered orally (left panel). Upon induction or constitutively, the probiotic will produce and deliver the desired AMP (light blue) to the gut lumen (right panel). The AMP should have specific activity against a bacterial strain in the gut for pathogen (blue bacteria) eradication or microbiota composition remodeling in situ (middle panel).
Strategies for antimicrobial peptide production using probiotics
Recombinant protein technologies have historically enabled the production of inexpensive therapeutic proteins at large scale, which have reached clinical settings (40). One example of the application of recombinant technology for the production of proteins includes the various approaches utilized to heterologously express human insulin using E. coli (20). In order to recombinantly produce AMPs in bacteria, requirements common to any basic expression system are needed along with certain specific considerations pertinent to the nature of these peptides (Fig. 2). For example, these peptides may be lethal to the host strain (i.e., chassis), unstable, and prone to degradation by proteases (41). To address these issues, AMPs have been produced as fusion proteins using fusion partners (42), or by encoding the entire natural secretion machinery of the specific AMP within the bacterial chassis, including immunity genes that prevent host death upon peptide expression (37,38,43). Delivery of AMPs to the gut has been achieved by expressing and secreting such agents from probiotics through the use of fusion proteins coupled with secretion sequences (38,44), and by inducing lysis of the probiotic chassis to enable release of the peptides from within the bacteria (39,45). Another important consideration when engineering probiotics is the need to devise biocontainment strategies. Since AMP-producing probiotics have only been used in animals to date, no biocontainment mechanisms have been programmed within their genetic circuits. However, moving forward to applications in humans and more sophisticated genetic circuit designs, biocontainment is expected to be more widely incorporated into living therapeutics.
Figure 2. Strategies for AMP production using probiotics.
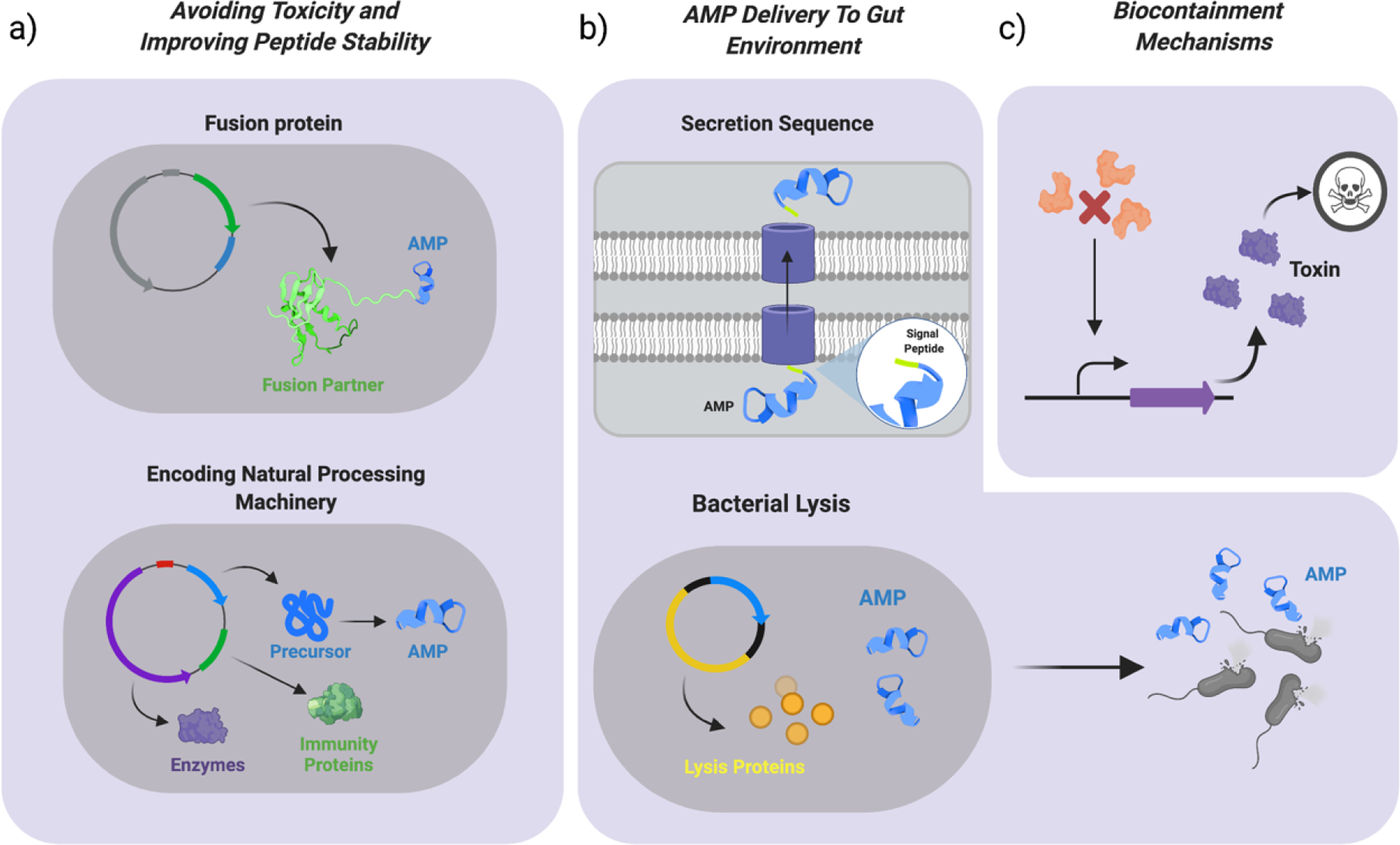
a) To improve stability of the AMP and avoid toxicity to the chassis, strategies such as producing AMPs as a fusion protein and encoding the natural AMP processing machinery in the chassis, incuding the immunity genes, have been used. b) To achieve delivery of AMPs to the gut environment from the probiotic chassis, strategies such as expressing the AMP fused to a strain-specific signal peptide for secretion and implementing a bacterial lysis mechanism have been used. In the latter, lysis proteins will be produced by the bacteria until a threshold is reached causing bacterial lysis and AMP release in the gut. c) Next-generation AMP-probiotics should ideally have a biocontainment mechanism that prevents dissemination into the environment. Some of these include bacterial killing mechanisms, which consist in the production of a toxic gene in the bacteria (purple), in this case triggered by the absence of certain externally supplemented molecule (orange).
Biocontainment strategies
An AMP-producing probiotic that disseminates into the environment could cause various deleterious issues such as contributing to antibiotic resistance or eventually leading to dysbiosis in other organisms. Therefore, biocontainment strategies must be built-in when engineering genetically modified organisms (GMO), as shown previously in the case of probiotics (46). Such strategies can be divided into two main categories; passive and active mechanisms (47). Common passive mechanisms are often the most simple to implement and involve deletion of genes encoding for essential amino acids or metabolites, which are later supplemented exogenously to enable bacterial growth (47,48). These are not optimal approaches because the auxotrophy can be reverted by cross-feeding from other bacteria in the environment. Hence, a new generation of passive mechanisms have been developed by creating strains addicted to non-canonical or synthetic amino acids, which cannot be supplemented from the surrounding environment (46,49,50). On the other hand, active mechanisms use more complex genetic circuits to sense the presence or absence of environmental signals in order to activate a circuit or enable a killing mechanism (36) (Fig. 2c). For example, two families of thermal bioswitches were designed for use in microbial therapeutics that worked orthogonally and were activated for gene transcription within a biologically relevant temperature range of 32° - 46° C (51). In another study, a temperature sensitive killing mechanism named “cryodeath” was constructed, which relied on the toxin-antitoxin mechanism CcdB/CcdA. Toxin overproduction occurred at temperatures 22°C or lower, leading to bacterial cell death. However, this mechanism had an escape frequency of 1 in 105 genetically modified organisms (52) , which surpasses the limits imposed by the NIH (National Institute of Health) of 1 in 108 (53). Therefore, more efficient killing mechanisms for microbial therapeutics should be designed. In addition, developing strains with a lower mutation rate and/or unable to horizontally transfer genetic material constitute additional levels of containment to be considered when engineering bacteria (54).
Antimicrobial peptide production strategies using probiotics
E. coli Nissle
E. coli Nissle 1917 (EcN) is a non-pathogenic strain of E. coli that has been widely employed as a probiotic given its ability to combat enteropathogens and its immunomodulatory effects (55). Multiple synthetic biology approaches have been used to engineer EcN to function as a living therapeutic, and a vast array of genetic tools have been developed. For example, EcN has been successfully engineered to treat metabolic disorders like phenylketonuria by expressing enzymes necessary for the conversion of phenylalanine to phenylpyruvate (56). EcN can also be genetically modified to diagnose diseases. For example, an EcN platform named PROP-Z was developed for diagnosing metastatic liver tumors in mice (57) .
The programmability of EcN makes it a convenient chassis for in vivo AMP production (Table 1), as reported in the literature (37–39,42). EcN has been modified to produce the AMP human β – defensin 2 (HBD2) in order to treat Crohn’s disease. The AMP was secreted by means of a fusion protein using the fusion partner YebF and its expression was under the control of the strong and inducible T7 promoter. YebF carries peptides linked to its C-terminus outside of the cell and into the medium in E. coli laboratory strains and EcN (58). The fusion protein was successfully secreted and HBD2 showed antimicrobial activity (47). Furthermore, EcN has been engineered to produce a class of small AMPs known as microcins, which target enteric pathogens. A probiotic based therapeutic against foodborne nontyphoidal Salmonella in poultry was designed by constructing an expression system that efficiently secreted microcin J25 under the control of the ProTeOn (59) promoter, which is induced by addition of anhydrotetracycline. After carrying out a trial using 300 turkeys with the genetically modified probiotic strain, a significant 97% reduction in the Salmonella carriage was observed in the treated group (37). Another study using EcN described an expression system utilizing the ttrBCA promoter from Salmonella, which is activated by tetrathionate during gut inflammation. This system enabled production and secretion of microcin H47, inhibiting Salmonella growth under anaerobic conditions (43,60). Both of the previously mentioned studies rely on encoding the natural microcin production machinery, immunity, and secretion proteins in their respective expression vectors, which protects the engineered bacteria from toxicity and spares the need to use unnatural signal peptides.
Table 1.
Toolbox of plasmids for engineering probiotic bacteria to produce AMPs, as discussed throughout the text.
| Bacteria | Plasmid name | Characteristics | References |
|---|---|---|---|
| Escherichia coli Nissle | pEAS106 | Contains yebF and peptide MHBD2 Encodes for YebFMHBD2 fusion protein Promoter: T7 |
(47) |
| pBF25 | Contains the Microcin J25 expression machinery (mcjABCD) Promoter: ProTeOn |
(36) | |
| pttrMcH47 | Tetrathionate-induced production of Microcin H47 Promoter: ttrBCA |
(41,59) | |
| pMPES | Modular peptide expression system Includes MccV secretion machinery Promoter: ProTeOn+ |
(37) | |
| pMPES2 | Reduced version of pMPES2 with a modular cloning site for peptide insertion | (30) | |
| pSED | Contains “sense-kill circuit” in E. coli Nissle Constitutive expression of LasR Induced expression of S5 pyocin, E7 lysis protein and Dispersin B Promoter: LuxR |
(44) | |
| Lactic Acid Bacteria | pMSP3545 | Optimized for controlled gene expression Promoter: PnisA Signal peptide: Usp45 |
(42,68) |
| pBK2idTZ | Tuned for accurate cCF10 pheromone detection and tight promoter regulation Promoter: Pq Signal peptide: Usp45 |
(70) | |
| pAMJ165 | Promoter: P170 Signal peptide: Sp30mut2 |
(71) |
A modular peptide expression system called pMPES allowed for production and secretion of a variety of AMPs in EcN (38). The vector encodes for the Microcin V (MccV) expression system and relies on a strong synthetic promoter, ProTeOn+, which was modified from ProTeOn (59). Seven AMPs from the MccV secretion machinery were produced and secreted independently from each construct utilizing the MccV signal peptide, signal peptide Vsp (SPVsp) or the AMP’s native signal peptide and all of them showed antimicrobial activity. In those constructs that showed activity against the producer strain (E. coli Mc1061 F’), the MccV immunity genes were included to prevent AMP-mediated toxicity to the chassis bacteria. Simultaneous production of two different AMPs from the same construct was also achieved using the pMPES system (38). In another study, Enterocin A, Enterocin B, and Hiracin JM79 were produced to target and kill Enterococcus. In this case, the plasmid pMPES was modified to a reduced version (pMPES2) with a different origin of replication and an added modular multiple cloning site for rapid peptide insertion. EcN harboring the pMPES2 vector encoding for the three AMPs and using SPVsp as the signal peptide significantly reduced intestinal colonization by vancomycin-resistant Enterococcus in a mouse model (30).
Modules for sensing and production of therapeutic molecules in E. coli may be compartmentalized using the “sense-kill” mechanism. This approach is based on a sensing module that constitutively produces the transcription factor LasR, which binds to the Pseudomonas aeruginosa quorum-sensing molecule homoserine lactone (HSL) (61), activating the therapeutic modules under the control of the PluxR promoter (activated by the complex LasR-3OC12HSL). The first study using this mechanism consisted on the HSL-LasR dependent transcription of the AMP pyocin S5 and the lysis E7 protein. In the presence of HSL, production of both (pyocin S5 and E7 protein) took place upon reaching the E7 threshold concentration that causes the chassis to lyse, leading to AMP release to the medium and Pseudomonas aeruginosa targeting (45). This mechanism was successfully reused in an EcN chassis, whose antimicrobial activity was successfully tested in Caenorhabditis elegans and mice (39).
Lactic Acid Bacteria
Lactic acid bacteria (LAB) are commonly used in the food industry due to their sugar fermentation properties (62) and because they are considered to be safe for human consumption or “food-grade”(63). They are also probiotic strains that constitute promising candidates for recombinant protein production (62) and therapeutic development because of their ability to colonize the human gut, survive passage through the GI tract, and in some cases efficiently bind to mucus (63). Furthermore, they are natural producers of a type of AMPs known as bacteriocins (64). In addition, LAB have been shown to have immunomodulatory effects (65). Lactobacillus and Lactococci are the most widely studied LAB and, hence, this section will be focused on the relevant advances and tools available for AMP production pertinent to these genera.
Engineered LAB have been used successfully as living medicines utilizing recombinant protein technologies to express an array of different molecules. Lactococcus lactis, for example, has been thoroughly studied for the development of vaccines that elicit mucosal immunization (65). This bacterium was used to develop a vaccine against human papillomavirus type 16 (HPV-16) leading to a high immune response in mice by inducible expression of the E7 protein, co-expressed alongside a signal peptide for cell wall anchoring (66). Furthermore, LAB can be leveraged as potential therapeutics for metabolic disorders, such as hyperoxaluria. Lactobacillus plantarum was used as a chassis to constitutively produce and secrete oxalate decarboxylase while colonizing the gut to prevent the formation of kidney stones in rats (67). LAB have also been developed into diagnostics. For example, synthetic gene circuits were installed into L. lactis in order to detect quorum-sensing molecules from V. cholerae and produce β-lactamase as a detectable output that was visualized by means of a chromogenic substrate in mouse feces (68).
To express AMPs in LAB, similar approaches to those used for EcN engineering have been adopted (Table 1). In a study, peptides were screened against E. coli and Salmonella, from which AMPs A3APO and Alysteserin were chosen based on their effectiveness against the target pathogens and their low activity against the bacterial chassis L. lactis. Plasmid pMSP3545, optimized for controlled gene expression (69), was used to drive the production of the selected AMPs under the control of a PnisA inducible promoter and fused to the signal peptide Usp45 (SPUsp45). The engineered strains achieved up to 20-fold inhibition of E. coli and Salmonella growth (44). Furthermore, based on the previous design, a LAB system was created to both inhibit and detect Enterococcus faecalis (70). The pheromone activated pCF10 system bearing promoter Pq from E. faecalis was utilized. Plasmid pBK2idTZ was tuned for accurate cCF10 pheromone detection from E. faecalis and tight promoter regulation. Next, genes encoding for three bacteriocins (i.e., enterocin A, hiracin JM79, and enterocin P) active against E. faecalis were cloned separately downstream of the promoter along with SPUsp45 and with their corresponding immunity genes. A plasmid bearing the three bacteriocins was also constructed. The engineered LAB inhibited the growth of E. faecalis and E. faecium in vitro, and increased antimicrobial activity was observed when the three AMPs were expressed from the same construct simultaneously (71). In a recent study, the pAMJ165 plasmid was used to drive heterologous expression of a novel chimeric AMP named cLFchimera (lactoferrampin + lactoferricin) in L. lactis (72). Protein expression in this plasmid was controlled by lactic-acid inducible p170 promoter (73) and up-regulated by low pH. The signal peptide sp310 mut2 was used to drive peptide secretion (74). This signal peptide is naturally found in L. lactis but was genetically modified to improve secretion efficiency. Using this approach, peptide secretion was successful and the peptide displayed antibacterial, antibiofilm, and antioxidant activities against an array of food pathogens in vitro (72).
Improving gut colonization by engineered probiotics
Gut commensal bacteria have co-evolved with the human host acquiring an ability to colonize select niches within the GI tract (75). Probiotic species vary in terms of their ability to effectively colonize the human gut, which is clearly mirrored in the structure of the gut microbiome (76). Colonization is mediated by adhesion of bacteria to the mucosal layer that coats intestinal epithelial cells, through the interaction between host mucin glycoproteins and cell surface mucus binding proteins (MUBs) present on the colonizing bacteria (77). MUBs are strain-specific adhesins and account for the base of competitive exclusion taking place in the microbiota environment, as different resident bacteria compete for binding to the same mucosal domains (77–79). Hence, establishing persistent colonization of living probiotic therapeutics is often a major obstacle when developing successful in situ treatments, since engineered microbes are often less competitive colonizers than natural ones due to the burden imposed by genetic modification (80). This will most certainly be reflected when AMP production is attempted in humans because some level of toxicity or burden is expected, even if not lethal to the host bacteria, thus compromising probiotic survival in the GI tract. Achieving effective epithelial adhesion and consequent colonization is key for increasing the transit time of the probiotic throughout the GI tract, a key step for ensuring therapeutic success. AMPs have been produced in this context at sufficient levels to ensure killing of their target bacteria (78). Enhancing gut colonization would reduce the time needed for the probiotic to exert its therapeutic effect, which is usually high as demonstrated for AMP-producing probiotics and other living therapies in previous studies (e.g. daily doses of 5 ×108 CFU/mL of engineered bacteria over a period of more than 10 days of treatment) (30,56). Another challenge to ensure stable gut colonization is that this is a highly strain and patient-specific phenomenon (80–82). Gut microbiota heterogeneity can be confounded by numerous factors, including dietary habits (83). It has been demonstrated that supplemented probiotics, natural or engineered, rarely perdure for more than a week in the human gut, and typically only achieve brief or no colonization (80,81,84). Thus, modulating the survival and endurance of engineered AMP-probiotics is critical. Prolonged AMP production is not expected to cause dysbiosis in the gut microbiome as long as “selectivity” is taken into account in the genetic design. Thus, the peptide sequence could be engineered to specifically target a certain pathogen, as has been shown in previous studies (25,26,85), while not affecting commensal strains. The probiotic may also be carefully designed and engineered to produce and deliver an AMP only upon encountering an ongoing infection, in which case it would kill the disease-causing pathogen while avoiding dysbiosis due to prolonged production (39,45). Furthermore, including a biocontainment mechanism in the genetic circuit should be considered to eliminate engineered bacteria if and when needed (Fig. 2c). Different genetic engineering approaches have been developed to overcome deficient gut colonization by probiotics, as we will outline next.
Mucus binding proteins
Recently, a new method was developed to quantify mucus adhesion in probiotics based on covalently functionalizing 96-well plates with mucus to generate mucus-binding curves specific to each bacterial strain tested (80). This technique helps circumvent the confounding effects encountered with common methods where bacteria have been shown to bind to the polystyrene as strongly as to the mucus coating (86). Using this technique, researchers found that commonly engineered probiotics, such as EcN 1917 and L. lactis MG1363, had poor mucus binding capacities. Engineered L. lactis MG1363 expressing MUB CmbA on its surface drastically improved mucus binding ability (80). This approach, which overexpresses MBUs on the bacterial surface, has been used successfully with other bacterial genera to improve colonization, thus further underscoring the importance of identifying strain-specific MBUs and genes involved in their expression. Using these principles, Bifidobacteria strains were modified to express the lipoprotein BopA from Bifidobacterium bifidum, improving adhesion to intestinal epithelial cells (87) (Fig. 3a).
Figure 3. Strategies to improve gut colonization of engineered probiotics.
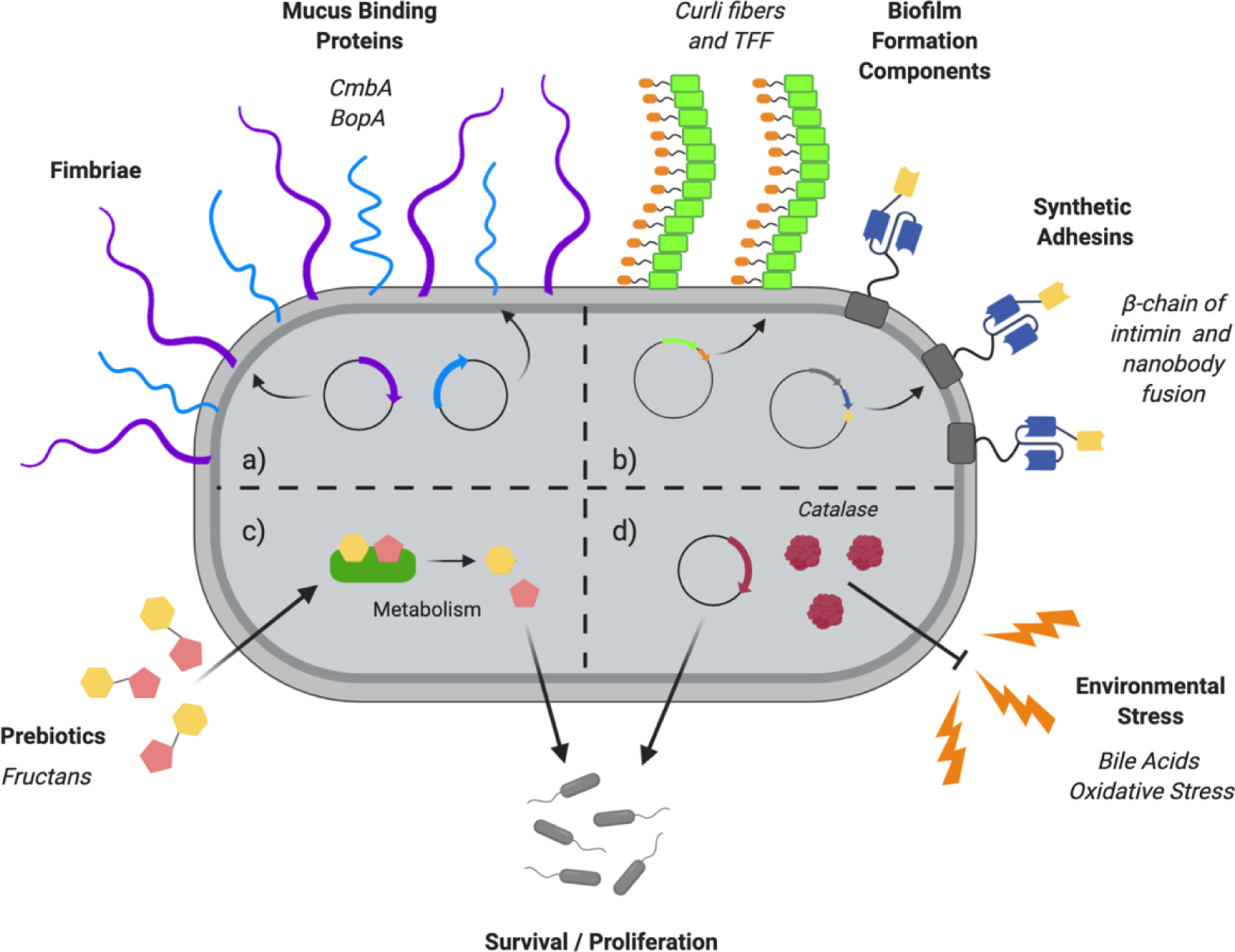
a) Overexpression of mucus binding proteins (light blue), such as CmbA (79) or BopA (85), and other bacterial surface components such as fimbriae, pilli or flagella (purple) (86). b) Expression of engineered biofilm formation (green) components or synthetic adhesins (dark blue and yellow) on the bacterial surface (88–91). c) Use of specific diets rich in prebiotics, such as fructans, that favor growth of certain bacterial strains (98,99). d) Overproduction of genes that encode for enzymes that counteract the stress conditions that the probiotic encounters through their passage along the GI tract, enhancing their survival and proliferation potential. For example, overexpression of catalase (red) to withstand oxidative stress (94,95,96).
Flagella, fimbriae and pili
Other components of the bacterial cell surface, such as flagella, fimbriae, and pili, mediate mucus binding (80). These can also be overexpressed to improve probiotic colonization, depending on the properties of the bacteria and specific strains considered. For example, EcN was engineered to express F4 and F18 fimbriae from enterotoxigenic E. coli (ETEC) on its surface, leading to improved adhesion to porcine intestinal epithelial cells compared to regular EcN (88). However, this also elicited an anticipated immune response against the synthetic fimbriae since the objective was to confer immunity to ETEC. These results raise concerns over the immunogenicity of engineered strains due to their augmented superficial epitopes, a variable that needs to be considered when choosing a mucus-binding molecule for overexpression (Fig. 3a).
Biofilm formation components
Despite the vast genetic toolbox available for E. coli engineering, its colonization capacity in humans is noticeably poor (79,84). Mucus binding molecules expressed by this strain are not well characterized and the theory that flagella mediate mucus binding (89) has been challenged (80). Expression of foreign MBUs from other probiotic bacteria on the surface of EcN or exploring alternatives such as biofilm formation, could make EcN a robust and convenient candidate for living therapeutic development. In fact, overexpression in EcN of CsgA, the main E. coli biofilm determinant and curli fiber gene, was shown to trigger biofilm formation (90–92). The versatility of curli fibers was also explored by fusing them with trefoil factors (TFF) for external anchored expression. This approach conferred E. coli with increased mucus binding capacity in vitro (91), but could not be replicated in vivo with EcN, likely because expression of the trefoil factor hampered bacterial survival (90) (Fig. 3b).
Synthetic Adhesins
Furthermore, synthetic modular adhesins have been constructed for surface expression in E. coli. They are composed of the β-chain of intimin and a single-domain antibody, also known as nanobodies, providing bacteria with high target affinity to specific cell types. Administration of a low inoculum of engineered E. coli colonized the target tumor tissue in vivo (93). Adapting a similar approach for targeting nanobodies toward intestinal epithelial cells (94), improved adherence of EcN and other probiotics to the GI tract and enhanced AMP action and biodistribution (Fig. 3b). Nanobody-displaying probiotic bacteria could also be engineered to adhere to each other creating a synthetic consortia (95), which could further boost gut colonization and, ultimately, AMP efficacy.
Persistence enhancing factors
Persistence of bacteria in the GI tract can also be tuned by genetic modifications to enhance colonization under stringent environmental conditions. For example, heterogenous expression of catalase and superoxide dismutase in B. longum improved its viability upon encountering oxidative stress (96). In addition, expression of the Listeria monocytogenes bile resistance mechanism in Bifidobacterium and Lactococcus boosted their survival in the murine GI tract (97). Also, a urease cluster has been described in Lactobacillus reuteri whose main function is withstanding the presence of high levels of gastric acid through hydrolysis of urea in gastric acid. This cluster may be produced in other bacteria or overexpressed, for example, in L. reuteri to improve persistence in the GI tract (98) (Fig. 3d).
Prebiotics
Finally, the gut microbiota is greatly influenced by diet since microbial species can utilize different nutrients from ingested food to promote their growth (99). Hence, the colonization and propagation of a particular strain or species can be modulated by certain types of diets. One example is the manipulation of communities in mouse models formed by Bacteroides, a major genera in the human microbiota, by the implementation of fructan selective diets. Providing mice with a B2–1 fructan diet shifted the Bacteroides population towards B. caccae, whereas a diet rich in B2–6 fructans skewed the community towards B. thetaiotamicron (100) (Fig. 3c). The intrinsic ability of Bacteroides strains to grow in the presence of certain carbohydrates is facilitated by gene clusters, known as polysaccharide utilization loci (PULS), that vary between strains (101). Exploring similar pathways in probiotic bacteria could help enhance gut colonization by specific prebiotic supplementation (102).
Future perspectives
The expansion of synthetic biology tools to probiotic bacteria has enabled their genetic manipulation and triggered the development of living medicines. Engineered probiotic therapies should be able to produce a therapeutic molecule on demand and achieve GI tract colonization and integration into the resident microbial community. Producing AMPs in situ in the gut represents an exciting avenue for treating infections and potentially other diseases in animals and, ultimately, in humans. The model probiotic bacteria EcN and LAB have been used as chassis for AMP production in several studies and thus the toolkit of expression vectors and characterized biological parts for these bacteria are extensive. However, other commensals for which tools do not yet exist, such as those belonging to the genera Clostridium and Bacteroides, present more desirable therapeutic properties such as GI tract persistence and colonization capacity. In addition, developing tools for engineering commensal bacteria that colonize other organs, such as the lungs or skin, would open new avenues for peptide delivery to target pathogens that cause respiratory or skin infections.
Consequently, it is crucial to expand the synthetic biology toolset to include these non-model bacteria. Other approaches towards peptide production and secretion are yet to be explored, ranging from using bacterial nanocompartments to self-cleaving intein fusion partners. For instance, cell wall-anchored AMPs could be produced as long as they do not act against the chassis bacteria and while ensuring the peptide remains active upon being tethered to the membrane. Production of membrane-anchored and active AMPs was achieved in E. coli and other Gram-negative bacteria through the use of engineered peptides with tethers that were sufficiently short to prevent interactions with other bacteria (103). Engineering a longer tether could enable activity against other bacteria by allowing interaction of the tethered AMP with the membrane of the targeted bacteria. Furthermore, advances in the production of non-standard amino acids (49) could pave the way towards engineering probiotic pharmacies that generate AMPs containing D-amino acids with improved stability and potency (104). AMP-producing living medicines have the potential to treat enteric pathogens, and may be promising tools for microbiome engineering if peptide sequences targeting specific commensal bacteria are developed. Thus far, AMP-producing probiotic technology has only been used in animal models and veterinary practice. However, we expect this area of study to acquire great relevance in the coming years as living therapeutics are developed for the treatment of enteric infections or to engineer the microbiome. Applications in humans would require additional safeguards such as biocontainment mechanisms and the establishment of regulations to prevent misuse (e.g. specific Food and Drug Administration guidelines) as established for other probiotics (105) and AMPs in medical settings (106).
Acknowledgements
Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the Nemirovsky Prize, the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1–21–1–0014).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Simon JC, Marchesi JR, Mougel C, Selosse MA. Host-microbiota interactions: From holobiont theory to analysis. Microbiome [Internet]. 2019January11 [cited 2020 Jun 29];7(1):5. Available from: 10.1186/s40168-019-0619-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol [Internet]. 2016August19 [cited 2020 May 25];14(8):e1002533. Available from: 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005August2;102(31):11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: An integrative view. Vol. 148, Cell. NIH Public Access; 2012. p. 1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011January;5(1):82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De La Fuente-Nunez C, Meneguetti BT, Franco OL, Lu TK. Neuromicrobiology: How Microbes Influence the Brain. Vol. 9, ACS Chemical Neuroscience. American Chemical Society; 2018. p. 141–50. [DOI] [PubMed] [Google Scholar]
- 7.Scheperjans F, Aho V, Pereira PAB, Koskinen K, Paulin L, Pekkonen E, et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord. 2015March1;30(3):350–8. [DOI] [PubMed] [Google Scholar]
- 8.Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, et al. A framework for human microbiome research. Nature. 2012June14;486(7402):215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock REW, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Vol. 24, Nature Biotechnology. Nature Publishing Group; 2006. p. 1551–7. [DOI] [PubMed] [Google Scholar]
- 10.Hancock REW. Cationic antimicrobial peptides: Towards clinical applications. Vol. 9, Expert Opinion on Investigational Drugs. Ashley Publications Ltd; 2000. 1723–9. [DOI] [PubMed] [Google Scholar]
- 11.Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: An emerging category of therapeutic agents. Vol. 6, Frontiers in Cellular and Infection Microbiology. Frontiers Media S.A.; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinkauf H, Von Döhren H. Peptide antibiotics. In: Biotechnology, Second, Completely Revised Edition, Volume 7: Products of Secondary Metabolism. wiley; 2008. p. 277–322. [Google Scholar]
- 13.De La Fuente-Núñez C, Cardoso MH, De Souza Cândido E, Franco OL, Hancock REW. Synthetic antibiofilm peptides. Biochim Biophys Acta -Biomembr [Internet]. 2016May1 [cited 2020 Aug 25];1858(5):1061–9. Available from: https://pubmed.ncbi.nlm.nih.gov/26724202/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De La Fuente-Núñez C, Korolik V, Bains M, Nguyen U, Breidenstein EBM, Horsman S, et al. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob Agents Chemother [Internet]. 2012May [cited 2020 Aug 25];56(5):2696–704. Available from: https://pubmed.ncbi.nlm.nih.gov/22354291/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Der Torossian Torres M, De La Fuente-Nunez C. Reprogramming biological peptides to combat infectious diseases. Chem Commun [Internet]. 2019December12 [cited 2020 Aug 25];55(100):15020–32. Available from: https://pubs.rsc.org/en/content/articlehtml/2019/cc/c9cc07898c [DOI] [PubMed] [Google Scholar]
- 16.de la Fuente-Nunez C Toward Autonomous Antibiotic Discovery. mSystems [Internet]. 2019June11 [cited 2020 Aug 25];4(3). Available from: http://msystems.asm.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porto WF, Irazazabal L, Alves ESF, Ribeiro SM, Matos CO, Pires ÁS, et al. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat Commun [Internet]. 2018December1 [cited 2020 Aug 25];9(1):1–12. Available from: www.nature.com/naturecommunications [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J, de la Fuente-Nunez C, Wen Ou R, Der Torossian Torres M, Pande SG, Sinskey AJ, et al. Yeast-Based Synthetic Biology Platform for Antimicrobial Peptide Production. 2020. [cited 2020 Aug 25]; Available from: https://pubs.acs.org/sharingguidelines [DOI] [PubMed]
- 19.Cao J, Perez-Pinera P, Lowenhaupt K, Wu MR, Purcell O, De La Fuente-Nunez C, et al. Versatile and on-demand biologics co-production in yeast. Nat Commun [Internet]. 2018December1 [cited 2020 Aug 25];9(1):1–13. Available from: www.nature.com/naturecommunications [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baeshen NA, Baeshen MN, Sheikh A, Bora RS, Morsi M, Ahmed M, et al. Cell factories for insulin production [Internet]. 2014. [cited 2020 Jun 29]. Available from: http://www.microbialcellfactories.com/content/13/1/141 [DOI] [PMC free article] [PubMed]
- 21.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Vol. 4, Nature Reviews Microbiology. Nat Rev Microbiol; 2006. p. 529–36. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich CL, Rozek A, Patrzykat A, Hancock REW. Structure and Mechanism of Action of an Indolicidin Peptide Derivative with Improved Activity against Gram-positive Bacteria. J Biol Chem. 2001June29;276(26):24015–22. [DOI] [PubMed] [Google Scholar]
- 23.Magana M, Pushpanathan M, Santos AL, Leanse L, Fernandez M, Ioannidis A, et al. The value of antimicrobial peptides in the age of resistance [Internet]. The Lancet Infectious Diseases. Lancet Publishing Group; 2020. [cited 2020 Aug 25]. Available from: https://pubmed.ncbi.nlm.nih.gov/32653070/ [Google Scholar]
- 24.Smith PA, Romesberg FE. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Vol. 3, Nature Chemical Biology. Nature Publishing Group; 2007. p. 549–56. [DOI] [PubMed] [Google Scholar]
- 25.Torres MDT, Pedron CN, Higashikuni Y, Kramer RM, Cardoso MH, Oshiro KGN, et al. Structure-function-guided exploration of the antimicrobial peptide polybia-CP identifies activity determinants and generates synthetic therapeutic candidates. Commun Biol [Internet]. 2018December1 [cited 2020 Sep 12];1(1):221. Available from: https://www.nature.com/articles/s42003-018-0224-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres MDT, Cao J, Franco OL, Lu TK, De La Fuente-Nunez C. Synthetic Biology and Computer-Based Frameworks for Antimicrobial Peptide Discovery [Internet]. ACS Nano. American Chemical Society; 2021. [cited 2021 Apr 6]. Available from: 10.1021/acsnano.0c09509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muniz LR, Knosp C, Yeretssian G, Moser B, Proost P, Leuven KU, et al. Intestinal antimicrobial peptides during homeostasis, infection, and disease. 2012. [cited 2021 Mar 25]; Available from: www.frontiersin.org [DOI] [PMC free article] [PubMed]
- 28.Zong X, Fu J, Xu B, Wang Y, Jin M. Interplay between gut microbiota and antimicrobial peptides. Vol. 6, Animal Nutrition. KeAi Communications Co.; 2020. p. 389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casals-Pascual C, Vergara A, Vila J. Intestinal microbiota and antibiotic resistance: Perspectives and solutions. Vol. 9, Human Microbiome Journal. Elsevier Ltd; 2018. p. 11–5. [Google Scholar]
- 30.Geldart KG, Kommineni S, Forbes M, Hayward M, Dunny GM, Salzman NH, et al. Engineered E. coli Nissle 1917 for the reduction of vancomycin-resistant Enterococcus in the intestinal tract . Bioeng Transl Med. 2018September;3(3):197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Centers for Disease Control U. Antibiotic Resistance Threats in the United States, 2019. [cited2020 Aug 25]; Available from: 10.15620/cdc:82532. [DOI]
- 32.de la Fuente-Nunez C, Torres MD, Mojica FJ, Lu TK. Next-generation precision antimicrobials: towards personalized treatment of infectious diseases [Internet]. Vol. 37, Current Opinion in Microbiology. Elsevier Ltd; 2017. [cited 2021 Jun 6]. 95–102. Available from: /pmc/articles/PMC5669808/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Mangoni ML, Di YP. In vivo therapeutic efficacy of frog skin-derived peptides against Pseudomonas aeruginosa-induced pulmonary infection. Sci Rep [Internet]. 2017December1 [cited 2021 Apr 6];7(1):1–13. Available from: www.nature.com/scientificreports/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardiner GE, Rea MC, O’Riordan B, O’Connor P, Morgan SM, Lawlor PG, et al. Fate of the two-component lantibiotic lacticin 3147 in the gastrointestinal tract. Appl Environ Microbiol [Internet]. 2007November [cited 2021 Mar 25];73(21):7103–9. Available from: https://pubmed.ncbi.nlm.nih.gov/17766459/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7(9):503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedrolli DB, Ribeiro NV, Squizato PN, de Jesus VN, Cozetto DA, Tuma RB, et al. Engineering Microbial Living Therapeutics: The Synthetic Biology Toolbox [Internet]. Vol. 37, Trends in Biotechnology. Elsevier Ltd; 2019. [cited 2020 Jun 28]. p. 100–15. Available from: http://www.cell.com/article/S0167779918302580/fulltext [DOI] [PubMed] [Google Scholar]
- 37.Forkus B, Ritter S, Vlysidis M, Geldart K, Kaznessis YN. Antimicrobial Probiotics Reduce Salmonella enterica in Turkey Gastrointestinal Tracts. Sci Rep. 2017January17;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geldart K, Forkus B, McChesney E, McCue M, Kaznessis YN. pMPES: A modular peptide expression system for the delivery of antimicrobial peptides to the site of gastrointestinal infections using probiotics. Pharmaceuticals. 2016December1;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun. 2017April11;8(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen R Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol Adv. 2012September1;30(5):1102–7. [DOI] [PubMed] [Google Scholar]
- 41.Parachin NS, Mulder KC, Viana AAB, Dias SC, Franco OL. Expression systems for heterologous production of antimicrobial peptides. Vol. 38, Peptides. Elsevier; 2012. p. 446–56. [DOI] [PubMed] [Google Scholar]
- 42.Seo EJ, Weibel S, Wehkamp J, Oelschlaeger TA. Construction of recombinant E. coli Nissle 1917 (EcN) strains for the expression and secretion of defensins. Int J Med Microbiol. 2012November1;302(6):276–87. [DOI] [PubMed] [Google Scholar]
- 43.Palmer JD, Piattelli E, McCormick BA, Silby MW, Brigham CJ, Bucci V. Engineered Probiotic for the Inhibition of Salmonella via Tetrathionate-Induced Production of Microcin H47. ACS Infect Dis. 2018January12;4(1):39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volzing K, Borrero J, Sadowsky MJ, Kaznessis YN. Antimicrobial Peptides Targeting Gram-negative Pathogens, Produced and Delivered by Lactic Acid Bacteria. 2013. [cited 2020 Jun 5]; Available from: https://pubs.acs.org/sharingguidelines [DOI] [PMC free article] [PubMed]
- 45.Saeidi N, Wong CK, Lo T, Nguyen HX, Ling H, Leong SSJ, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol [Internet]. 2011January16 [cited 2020 Jun 3];7(1):521. Available from: 10.1038/msb.2011.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JW, Chan CTY, Slomovic S, Collins JJ. Next-generation biocontainment systems for engineered organisms. Nat Chem Biol [Internet]. 2018June1 [cited 2020 Jun 28];14(6):530–7. Available from: https://www.nature.com/articles/s41589-018-0056-x [DOI] [PubMed] [Google Scholar]
- 47.Lee P Biocontainment strategies for live lactic acid bacteria vaccine vectors. Bioeng Bugs [Internet]. 2010. [cited 2020 Jun 28];1(1):75–7. Available from: 10.4161/bbug.1.1.10594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol [Internet]. 2003July1 [cited 2020 Jun 28];21(7):785–9. Available from: https://www.nature.com/articles/nbt840 [DOI] [PubMed] [Google Scholar]
- 49.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature [Internet]. 2015February5 [cited 2020 Jun 28];518(7537):55–60. Available from: https://www.nature.com/articles/nature14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature [Internet]. 2015February5 [cited 2020 Jun 28];518(7537):89–93. Available from: https://www.nature.com/articles/nature14095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piraner DI, Abedi MH, Moser BA, Lee-Gosselin A, Shapiro MG. Tunable thermal bioswitches for in vivo control of microbial therapeutics. In: Food, Pharmaceutical and Bioengineering Division 2017 - Core Programming Area at the 2017 AIChE Annual Meeting [Internet]. AIChE; 2017. [cited 2020 Jun 28]. p. 695–702. Available from: https://www.nature.com/articles/nchembio.2233 [Google Scholar]
- 52.Stirling F, Bitzan L, O’Keefe S, Redfield E, Oliver JWK, Way J, et al. Rational Design of Evolutionarily Stable Microbial Kill Switches. Mol Cell. 2017November16;68(4):686–697.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson DJ. NIH guidelines for research involving recombinant DNA molecules. Account Res [Internet]. 1993. [cited 2020 Jun 28];3(2–3):177–85. Available from: 10.1080/08989629308573848 [DOI] [PubMed] [Google Scholar]
- 54.Torres B, Jaenecke S, Timmis KN, García JL, Díaz E. A dual lethal system to enhance containment of recombinant micro-organisms. Microbiology [Internet]. 2003. [cited 2020 Jun 28];149(12):3595–601. Available from: https://pubmed.ncbi.nlm.nih.gov/14663091/ [DOI] [PubMed] [Google Scholar]
- 55.Sonnenborn U, Schulze J. The non-pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microb Ecol Health Dis [Internet]. 2009January26 [cited 2020 May 30];21(3–4):122–58. Available from: 10.3109/08910600903444267 [DOI] [Google Scholar]
- 56.Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol. 2018October1;36(9):857–67. [DOI] [PubMed] [Google Scholar]
- 57.Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, et al. Programmable probiotics for detection of cancer in urine. Sci Transl Med. 2015May27;7(289):289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang G, Brokx S, Weiner JH. Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli. Nat Biotechnol. 2006January20;24(1):100–4. [DOI] [PubMed] [Google Scholar]
- 59.Volzing K, Biliouris K, Kaznessis YN. ProTeOn and proTeOff, new protein devices that inducibly activate bacterial gene expression. ACS Chem Biol. 2011October21;6(10):1108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010September23;467(7314):426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gray KM, Passador L, Iglewski BH, Greenberg EP. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. Vol. 176, Journal of Bacteriology. American Society for Microbiology; 1994. p. 3076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.García-Fruitós E Lactic acid bacteria: A promising alternative for recombinant protein production. Vol. 11, Microbial Cell Factories. BioMed Central; 2012. p. 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berlec A, Strukelj B. Novel Applications of Recombinant Lactic Acid Bacteria in Therapy and in Metabolic Engineering. Recent Pat Biotechnol. 2009July3;3(2):77–87. [DOI] [PubMed] [Google Scholar]
- 64.Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol [Internet]. 2016April10 [cited 2020 Jun 5];100(7):2939–51. Available from: 10.1007/s00253-016-7343-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bahey-El-Din M Lactococcus lactis-based vaccines from laboratory bench to human use: An overview. Vol. 30, Vaccine. Elsevier; 2012. p. 685–90. [DOI] [PubMed] [Google Scholar]
- 66.Bermúdez-Humarán LG, Cortes-Perez NG, Le Loir Y Le, Alcocer-González JM, Tamez-Guerra RS, Montes De Oca-Luna R, et al. An inducible surface presentation system improves cellular immunity against human papillomavirus type 16 E7 antigen in mice after nasal administration with recombinant lactococci. J Med Microbiol. 2004May1;53(5):427–33. [DOI] [PubMed] [Google Scholar]
- 67.Paul E, Albert A, Ponnusamy S, Mishra SR, Vignesh AG, Sivakumar SM, et al. Designer probiotic Lactobacillus plantarum expressing oxalate decarboxylase developed using group II intron degrades intestinal oxalate in hyperoxaluric rats. Microbiol Res. 2018October1;215:65–75. [DOI] [PubMed] [Google Scholar]
- 68.Mao N, Cubillos-Ruiz A, Cameron DE, Collins JJ. Probiotic strains detect and suppress cholera in mice. Sci Transl Med. 2018June13;10(445). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bryan EM, Bae T, Kleerebezem M, Dunny GM. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000September1;44(2):183–90. [DOI] [PubMed] [Google Scholar]
- 70.Dunny GM. Enterococcal Sex Pheromones: Signaling, Social Behavior, and Evolution. Annu Rev Genet. 2013November23;47(1):457–82. [DOI] [PubMed] [Google Scholar]
- 71.Borrero J, Chen Y, Dunny GM, Kaznessis YN. Modified Lactic Acid Bacteria Detect and Inhibit Multiresistant Enterococci. 2020. [cited 2020 May 27];16:36. Available from: https://pubs.acs.org/sharingguidelines [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanhaeian A, Mirzaii M, Pirkhezranian Z, Sekhavati MH. Generation of an engineered food-grade Lactococcus lactis strain for production of an antimicrobial peptide: In vitro and in silico evaluation. BMC Biotechnol [Internet]. 2020March30 [cited 2020 Jun 6];20(1):19. Available from: 10.1186/s12896-020-00612-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jørgensen CM, Vrang A, Madsen SM. Recombinant protein expression in Lactococcus lactis using the P170 expression system. [cited 2020 Jun 6]; Available from: http://www.jurag.dk [DOI] [PubMed]
- 74.Ravn P, Arnau J, Madsen SM, Vrang A, Israelsen H. Optimization of signal peptide SP310 for heterologous protein production in Lactococcus lactis. Microbiology. 2003August1;149(8):2193–201. [DOI] [PubMed] [Google Scholar]
- 75.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature [Internet]. 2013August18 [cited 2020 Jun 25];501(7467):426–9. Available from: https://www.nature.com/articles/nature12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Structure, Function and Diversity of the Healthy Human Microbiome The Human Microbiome Project Consortium HHS Public Access. 2013. [cited 2020 May 25]; Available from: http://www.nature.com/authors/editorial_policies/license.html#termswww.nature.com/nature.
- 77.Juge N Microbial adhesins to gastrointestinal mucus. Vol. 20, Trends in Microbiology. Elsevier Current Trends; 2012. p. 30–9. [DOI] [PubMed] [Google Scholar]
- 78.Monteagudo-Mera A, Rastall RA, Gibson GR, Charalampopoulos D, Chatzifragkou A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health [Internet]. Vol. 103, Applied Microbiology and Biotechnology. Springer Verlag; 2019. [cited 2020 Jun 25]. p. 6463–72. Available from: 10.1007/s00253-019-09978-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Etzold S, Kober OI, Mackenzie DA, Tailford LE, Gunning AP, Walshaw J, et al. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ Microbiol [Internet]. 2014March1 [cited 2020 Jun 26];16(3):888–903. Available from: 10.1111/1462-2920.12377 [DOI] [PubMed] [Google Scholar]
- 80.Mays ZJS, Chappell TC, Nair NU. Quantifying and Engineering Mucus Adhesion of Probiotics. ACS Synth Biol [Internet]. 2020February21 [cited 2020 Jun 25];9(2):356–67. Available from: 10.1021/acssynbio.9b00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Derrien M, van Hylckama Vlieg JET. Fate, activity, and impact of ingested bacteria within the human gut microbiota [Internet]. Vol. 23, Trends in Microbiology. Elsevier Ltd; 2015. [cited 2020 Jun 25]. p. 354–66. Available from: http://www.cell.com/article/S0966842X15000566/fulltext [DOI] [PubMed] [Google Scholar]
- 82.MacKenzie DA, Jeffers F, Parker ML, Vibert-Vallet A, Bongaerts RJ, Roos S, et al. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology [Internet]. 2010November [cited 2020 Jun 26];156(11):3368–78. Available from: https://pubmed.ncbi.nlm.nih.gov/20847011/ [DOI] [PubMed] [Google Scholar]
- 83.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012June14;486(7402):207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ou B, Yang Y, Tham WL, Chen L, Guo J, Zhu G. Genetic engineering of probiotic Escherichia coli Nissle 1917 for clinical application. Vol. 100, Applied Microbiology and Biotechnology. Springer Verlag; 2016. p. 8693–9. [DOI] [PubMed] [Google Scholar]
- 85.Silva ON, Torres MDT, Cao J, Alves ESF, Rodrigues LV., Resende JM, et al. Repurposing a peptide toxin from wasp venom into antiinfectives with dual antimicrobial and immunomodulatory properties. Proc Natl Acad Sci U S A [Internet]. 2020October27 [cited 2021 Jun 6];117(43):26936–45. Available from: www.pnas.org/cgi/doi/10.1073/pnas.2025351118PNAS2021Vol.118No.3e2025351118www.pnas.org/cgi/doi/10.1073/pnas.2025351118PNAS2021Vol.118No.3e2025351118https://doi.org/10.1073/pnas.2025351118%7Chttps://doi.org/10.1073/pnas.2025351118%7C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laparra JM, Sanz Y. Comparison of in vitro models to study bacterial adhesion to the intestinal epithelium. Lett Appl Microbiol [Internet]. 2009December1 [cited 2020 Jun 26];49(6):695–701. Available from: 10.1111/j.1472-765X.2009.02729.x [DOI] [PubMed] [Google Scholar]
- 87.Gleinser M, Grimm V, Zhurina D, Yuan J, Riedel CU. Improved adhesive properties of recombinant bifidobacteria expressing the Bifidobacterium bifidum-specific lipoprotein BopA. Microb Cell Fact [Internet]. 2012June13 [cited 2020 Jun 26];11. Available from: https://pubmed.ncbi.nlm.nih.gov/22694891/ [DOI] [PMC free article] [PubMed]
- 88.Ou B, Jiang B, Jin D, Yang Y, Zhang M, Zhang D, et al. Engineered recombinant escherichia coli probiotic strains integrated with f4 and f18 fimbriae cluster genes in the chromosome and their assessment of immunogenic efficacy in vivo. ACS Synth Biol [Internet]. 2020February21 [cited 2020 Jun 26];9(2):412–26. Available from: https://pubmed.ncbi.nlm.nih.gov/31944664/ [DOI] [PubMed] [Google Scholar]
- 89.Troge A, Scheppach W, Schroeder BO, Rund SA, Heuner K, Wehkamp J, et al. More than a marine propeller - the flagellum of the probiotic Escherichia coli strain Nissle 1917 is the major adhesin mediating binding to human mucus. Int J Med Microbiol. 2012December1;302(7–8):304–14. [DOI] [PubMed] [Google Scholar]
- 90.Praveschotinunt P, Duraj-Thatte AM, Gelfat I, Bahl F, Chou DB, Joshi NS. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun. 2019December1;10(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Duraj-Thatte AM, Praveschotinunt P, Nash TR, Ward FR, Joshi NS. Modulating bacterial and gut mucosal interactions with engineered biofilm matrix proteins. Sci Rep [Internet]. 2018December1 [cited 2020 Jun 26];8(1). Available from: https://pubmed.ncbi.nlm.nih.gov/29472619/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nguyen PQ, Botyanszki Z, Tay PKR, Joshi NS. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun [Internet]. 2014September17 [cited 2020 Jun 26];5(1):1–10. Available from: https://www.nature.com/articles/ncomms5945 [DOI] [PubMed] [Google Scholar]
- 93.Piñero-Lambea C, Bodelón G, Fernández-Periáñez R, Cuesta AM, Álvarez-Vallina L, Fernández LÁ. Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth Biol [Internet]. 2015April17 [cited 2020 Jun 29];4(4):463–73. Available from: 10.1021/sb500252a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bakshi S, Sanz Garcia R, Van der Weken H, Tharad A, Pandey S, Juarez P, et al. Evaluating single-domain antibodies as carriers for targeted vaccine delivery to the small intestinal epithelium. J Control Release. 2020May10;321:416–29. [DOI] [PubMed] [Google Scholar]
- 95.Timmis K, Timmis JK, Brüssow H, Fernández LÁ. Synthetic consortia of nanobody-coupled and formatted bacteria for prophylaxis and therapy interventions targeting microbiome dysbiosis-associated diseases and comorbidities. Microb Biotechnol [Internet]. 2019January1 [cited 2020 Jun 29];12(1):58–65. Available from: 10.1111/1751-7915.13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zuo F, Yu R, Feng X, Khaskheli GB, Chen L, Ma H, et al. Combination of heterogeneous catalase and superoxide dismutase protects Bifidobacterium longum strain NCC2705 from oxidative stress. Appl Microbiol Biotechnol [Internet]. 2014June6 [cited 2020 Jun 26];98(17):7523–34. Available from: 10.1007/s00253-014-5851-z [DOI] [PubMed] [Google Scholar]
- 97.Watson D, Sleator RD, Hill C, Gahan CGM. Enhancing bile tolerance improves survival and persistence of Bifidobacterium and Lactococcus in the murine gastrointestinal tract. BMC Microbiol [Internet]. 2008October9 [cited 2020 Jun 26];8(1):176. Available from: 10.1186/1471-2180-8-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krumbeck JA, Marsteller NL, Frese SA, Peterson DA, Ramer-Tait AE, Hutkins RW, et al. Characterization of the ecological role of genes mediating acid resistance in L actobacillus reuteri during colonization of the gastrointestinal tract. Environ Microbiol [Internet]. 2016July1 [cited 2020 Jun 26];18(7):2172–84. Available from: 10.1111/1462-2920.13108 [DOI] [PubMed] [Google Scholar]
- 99.Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota [Internet]. Vol. 16, Nature Reviews Gastroenterology and Hepatology. Nature Publishing Group; 2019. [cited 2020 Jun 26]. p. 35–56. Available from: https://www.nature.com/articles/s41575-018-0061-2 [DOI] [PubMed] [Google Scholar]
- 100.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010June25;141(7):1241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe. 2008November13;4(5):447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell [Internet]. 2018September6 [cited 2021 Mar 29];174(6):1388–1405.e21. Available from: https://pubmed.ncbi.nlm.nih.gov/30193112/ [DOI] [PubMed] [Google Scholar]
- 103.Tucker AT, Leonard SP, DuBois CD, Knauf GA, Cunningham AL, Wilke CO, et al. Discovery of Next-Generation Antimicrobials through Bacterial Self-Screening of Surface-Displayed Peptide Libraries. Cell. 2018January25;172(3):618–628.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao Y, Zhang M, Qiu S, Wang J, Peng J, Zhao P, et al. Antimicrobial activity and stability of the d-amino acid substituted derivatives of antimicrobial peptide polybia-MPI. AMB Express [Internet]. 2016December1 [cited 2020 Aug 17];6(1):122. Available from: /pmc/articles/PMC5128008/?report=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoffmann DE, Fraser CM, Palumbo F, Ravel J, Rowthorn V, Schwartz J. Probiotics: achieving a better regulatory fit. Food Drug Law J [Internet]. 2014. [cited 2021 Apr 6];69(2):237. Available from: www.nutraceuticalsworld.com/contents/view_breaking-news/2013-06-10/functional-foods-lead-the-probiotic-markets-continued- [PMC free article] [PubMed] [Google Scholar]
- 106.Kazemzadeh-Narbat M, Cheng H, Chabok R, Alvarez MM, de la Fuente-Nunez C, Phillips KS, et al. Strategies for antimicrobial peptide coatings on medical devices: a review and regulatory science perspective [Internet]. Vol. 41, Critical Reviews in Biotechnology. Taylor and Francis Ltd.; 2021. [cited 2021 Apr 6]. p. 94–120. Available from: 10.1080/07388551.2020.1828810 [DOI] [PubMed] [Google Scholar]


