Abstract
Skeletal muscle wasting drives negative clinical outcomes and is associated with a spectrum of pathologies including cancer. Cancer cachexia is a multi-factorial syndrome that encompasses skeletal muscle wasting and remains understudied, despite being a frequent and serious co-morbidity. Deviation from the homeostatic balance between breakdown and regeneration leads to muscle wasting disorders, such as cancer cachexia. Muscle stem cells (MuSCs) are the cellular compartment responsible for muscle regeneration, which makes MuSCs an intriguing target in the context of wasting muscle. Molecular studies investigating MuSCs and skeletal muscle wasting largely focus on transcriptional changes, but our group and others propose that metabolic changes are another layer of cellular regulation underlying MuSC dysfunction in CC. In the present study, we combined gene expression and non-targeted metabolomic profiling of myoblasts exposed to wasting conditions (cancer cell conditioned media) to derive a more complete picture of the myoblast response to wasting factors. After mapping these features to annotated pathways, we found that more than half of the mapped pathways were amino acid-related, linking global amino acid metabolic disruption to conditioned media-induced myoblast defects. Notably, arginine metabolism was a highly enriched pathway in combined metabolomic and transcriptomic data. Arginine catabolism generates nitric oxide (NO), an important signaling molecule known to have negative effects on mature muscle. We hypothesize that tumor-derived disruptions in Nitric Oxide Synthase (NOS)2-regulated arginine catabolism impair differentiation of MuSCs. The work presented here further investigates the effect of NOS2 overactivity on myoblast proliferation and differentiation. We show that NOS2 inhibition is sufficient to rescue wasting phenotypes associated with inflammatory cytokines. Ultimately, this work provides new insights into MuSC biology and opens up potential therapeutic avenues for addressing disrupted MuSC dynamics in cancer cachexia.
Keywords: NOS2, nitric oxide, myoblast, cancer cachexia, TNFα, IFNγ
INTRODUCTION
Adult skeletal muscle possesses a remarkable capacity to regenerate damaged tissue after acute injury. Despite this ability, disorders of chronic muscle loss (or muscle wasting) are a persistent clinical problem affecting diverse patient populations1,2. Cancer cachexia is a muscle wasting disorder affecting approximately 60% of end-stage cancer patients1,3. Cancer cachexia is primarily characterized by a loss in lean mass, which contributes to decreased quality of life and increased mortality in this patient population4. Established mechanisms of cancer cachexia include proteolysis and autophagy, while impaired cellular/tissue regeneration is gaining increased attention as an important contributor to this complex syndrome5-7. Clinically targeting catabolic processes in cancer cachexia has been largely unsuccessful at slowing or reversing muscle loss8-10. We propose that a deeper understanding of the role of impaired anabolic processes in cancer cachexia may open doors to alternative and innovative therapeutic approaches.
Muscle stem cells (MuSCs) are an adult stem cell population that resides on the periphery of muscle myofibers, existing in a quiescent state until an injury stimulus causes these cells to activate, differentiate, and eventually fuse into myofibers. Myogenic regulatory factors (MRFs) control this process, which is known as myogenesis11. Previous studies have largely focused on how tumor-associated/inflammatory cytokines perturb MRFs and thus promote cancer cachexia6,7,12,13. One example is the NFkB signaling ligand tumor necrosis factor alpha (TNFα), which has been implicated in cancer cachexia development but remains controversial regarding its mechanism of action14. TNFα is particularly interesting as a tumor-derived factor because it has been detected in the serum of lung cancer patients as well as in lung cancer-associated cachexia mouse models15-19. Tumor-secreted factors likely play an important role in cancer cachexia development, but the limited efficacy of therapeutics targeting individual tumor-secreted factors suggests that the wasting environment is far more complex than we currently understand20.
In the present study, we were interested in evaluating changes in myoblasts after exposure to a milieu of tumor-derived factors by exploring combined metabolomic and transcriptomic changes to myoblasts after exposure to conditioned media from lung adenocarcinoma cells. We previously established that these media contain factors capable of disrupting myogenesis12. While previous work in cancer cachexia has focused on transcriptional changes, we and others propose that metabolic changes are another layer of cellular regulation underlying organ dysfunction in cancer cachexia21,22. Indeed, our data implicate nitric oxide synthase (NOS) 2-driven arginine catabolism as a central perturbed pathway after conditioned media treatment. NOS enzymes catabolize arginine and produce nitric oxide (NO) as a byproduct. NO is an important signaling molecule in several cell types and can exert its effects via reaction with superoxide to form the unstable radical peroxynitrite or via nitrosylation of cysteine residues in proteins23,24. NOS2 is of particular interest because in other cell types, such as macrophages, NOS2 expression is induced by cytokine stimulation24,25. Cytokines implicated in NOS2 stimulation, such as TNFα and interferon gamma (IFNγ), are also involved in cancer cachexia14,26. While anti-TNFα therapies have been largely unsuccessful in clinical trials, an understanding of the downstream metabolic and signaling changes associated with known cachectic factors could open doors to more targeted therapeutics14,27.
Current understanding of the NOS2 catabolic pathway in the context of cancer cachexia is convoluted28-32. For example, NOS2 knock out mice experience impaired muscle regeneration after injury, and NOS2 elevation in myotubes impairs MRF expression, but elevated NOS2 transcript, and citrulline, was found to benefit atrophic myotubes in vitro28,31,33. A recent study showed that NOS2 inhibition is sufficient to protect tumor-bearing mice from cancer-associated muscle loss34. While this study seems to solidify the potential of NOS2 inhibitors as anti-cachectic therapeutics, a gap in this and other related studies is a lack of attention to MuSC/myoblast dynamics. Indeed, the aforementioned study highlights the need to better understand the full impact of NOS2 dysregulation in different muscle cell populations before advancing NOS2-directed therapies.
Studies in myotubes have shown that NO can regulate MRF expression, in particular, MyoD and myogenin, yet dynamic expression of these MRFs typically occurs prior to myotube formation26,31,35. Therefore, we assessed the effects of NOS2 upregulation on myoblasts during the differentiation process. Specifically, we studied NOS2 overactivity in the context of myoblast proliferation and differentiation, two critical myogenic processes. Although our initial observations of NOS2 dysregulation were made using lung adenocarcinoma conditioned media, we utilized TNFα and IFNγ as a simplified and precise model of elevated NOS2 expression26,34,36-39 in order to establish a proof-of-concept link between NOS2 and myogenesis. The intersection of tumor-derived inflammatory factors with known mechanisms of NO/arginine metabolism, and the recent attention to muscle regeneration in cancer cachexia, make NO/arginine metabolism a strong candidate to investigate further in models of cancer cachexia. In the work presented here, we contribute to understanding the role of NO/arginine metabolism in cachexia by conducting a detailed assessment of the role of NOS2 with respect to in vitro myogenesis.
METHODS
Cell culture
KrasLA1/+;p53R172H/Δg/+ lung adenocarcinoma cell lines (307P, 393P, 531LN2, 344SQ) were generated as previously described, and maintained by growing on standard tissue culture treated dishes in growth media consisting of high glucose DMEM supplemented with 10% fetal bovine serum, and 1% penicillin/streptomycin40. Conditioned media from lung adenocarcinoma cells was collected by culturing cells (~70-90% confluence) in serum free (DMEM) media for 8 hours, followed by centrifugation of cell debris and adding serum as needed (10% FBS for growth experiments, 2% horse serum for differentiation experiments). All conditioned media was used at 100% strength (not diluted with regular media). For conditioned media experiments, the control treatment was fresh DMEM with added serum (noted above). C2C12 cells (ATCC CRL-1772) were grown on tissue culture treated dishes in growth media consisting of high glucose DMEM media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Differentiation media contains 2% horse serum in place of 10% fetal bovine serum. Differentiating cultures were maintained by alternating fresh media additions and complete media changes on a daily basis for 5 days (see Fig. 5A for cytokine/drug pulse-chase method). The following final concentrations were used (all diluted in aqueous solution): TNFα 40 ng/mL, INFγ 40 ng/mL, 1400W 100 μM. Live cell analyses: Cell proliferation and cell death were measured by live cell analysis (Incucyte Zoom Live-Cell Imaging System, Essen Bioscience). Apoptotic cells were tracked in control or treated cultured C2C12 cells with a fluorogenic substrate for activated caspase 3/7 (CellEvent Caspase 3/7 green, Invitrogen) according to the manufacturer, at a final concentration of 5 μM. Apoptotic cells were tracked for 48 hours. Incucyte Zoom software was used to measure stained myotube eccentricity.
MTT assay
C2C12 cells were plated at a density of 5000 cells/well in a 96-well plate, and allowed to proliferate for 48 hours prior to testing MTT reactivity. 5 mg/mL MTT reagent (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) diluted in water was diluted directly into the culture media to a final concentration of 10 μg/mL as previously described41. Plates were incubated with MTT reagent for 45-75 minutes, until purple granules were visible with the naked eye in control wells. Media was carefully aspirated off without disturbing the cell monolayer or MTT granules. 150 uL dimethyl sulfoxide (DMSO) was added to each well, then plate was placed on rocker for 10 minutes. Absorbance was read at 570 nm on a plate reader.
Immunostaining
Once fixed, cells were permeabilized with 0.5% Triton-X100 in PBS followed by blocking with 3% BSA, 0.2% Triton-X and 0.2% Tween-20 in PBS. Primary antibody incubations occurred at 4°C overnight, followed by incubation with secondary antibody at RT for 45 minutes in buffer described above. MF-20 (Developmental Hybridoma Bank) was used to stain myosin heavy chain I, DAPI and nuclear ID red (Enzo) were used for nuclear stain. Secondary antibodies were all Alexa fluorescent conjugates (488, 555, or 647) from Invitrogen or Jackson ImmunoResearch.
Flow cytometry
C2C12 cells were treated with TI for 24 hours, then washed once with DPBS, and trypsin was used to lift cells from the plate. Cells were washed and pelleted in DPBS. All steps were done according to the respective manufacturer’s protocol. Live cells were blocked with FCR Block (Miltenyi), then stained with Vioblue (Miltenyi) for live/dead discrimination. After viability staining, cells were fixed using eBioscience Intracellular Fixation and Permeabilization Buffer Set (ThermoFisher). NOS2 staining was performed using iNOS-PE conjugated antibody (Invitrogen #12-5920-80). After staining, cells were immediately analyzed by flow cytometry on a MACSquant 10 analyzer (Miltenyi Biotec). Data were analyzed using MACSquantify software (Miltenyi Biotec).
Nitrite Quantification
Measure-IT High-Sensitivity Nitrite Assay Kit (Invitrogen) was used to assess extracellular nitrite levels. 10 μL directly from the culture media was used in the assay. The assay was conducted following the manufacturer’s protocol and fluorescent results were read at excitation 365 nm, emission 450 nm on a plate reader.
Quantitative metabolite analyses
Sample preparation:
Cells were treated for 48 hours with cancer cell conditioned media (CC-CM), washed 3 times with PBS, and stored at −80C until metabolite extraction.
Amino Acids:
Amino acids and their metabolites were measured by LCMS as previously described42,43. Briefly, 50μl of either cell media or 1xPBS was added to the cell pellets followed by addition of internal standard solution. The sample mixture was sonicated and deproteinized with cold methanol followed by centrifugation at 18,000g for 15 minutes. The supernatant was dried down and then derivatized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate according to Waters’ MassTrak kit. A 11-point calibration standard curve underwent similar derivatization procedure after the addition of internal standards. Both derivatized standards and samples were analyzed on on the Thermo TSQ Quantum Ultra mass spectrometer (West Palm Beach, FL) coupled with a Waters Acquity UPLC system (Milford, MA). Data acquisition was done using select ion monitor (SRM). Concentrations of 42 analytes of each unknown were calculated against each perspective calibration curve.
Untargeted metabolomics analyses
Three biological replicates of control and each CC-CM treated C2C12 cells were prepared for untargeted metabolomics analyses as follows: cells were grown to 60% confluency and then treated with CC-CM for 48 hours, at which time plates were washed with PBS and cells were scraped and pelleted, followed by storage at −80° C. Samples were submitted to the Metabolomics Core at Mayo Clinic (Rochester, MN, USA) for metabolomics profiling by liquid chromatography-mass spectrometry (LC/MS) using 6550 iFunnel Quadrupole Time of Flight (Q-TOF) mass spectrometer (Agilent). Metabolites were then identified using METLIN database with Mass Profiler Professional software (Agilent). Annotated metabolites with a p value less than 0.05 and a fold change greater than 2 were further analyzed using MetaboAnalyst network explorer44 (metabolite-metabolite interactions) and enrichment analysis (pathway associated metabolite sets) tools. KEGG and HMDB identifiers were used in MetaboAnalyst.
RNA sequencing analysis
Sample/library preparation:
Three biological replicates of control and each CC-CM treated C2C12 cells were prepared for RNA sequencing analyses as follows: cells were grown to 60% confluency and then treated with CC-CM for 48 hours, at which time plates were washed with PBS and cells were scraped and pelleted, followed by storage at −80° C. RNA extraction was performed using Qiagen RNeasy columns, and frozen samples were submitted to the Mayo Clinic Medical Genome Facility where RNA quality was determined using the Fragment Analyzer from AATI. All samples had RQN values >9 and proceeded to library prep. cDNA libraries were prepared using 200 ng of good quality total RNA according to the manufacturer’s instructions for the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA), employing poly-A mRNA enrichment using oligo dT magnetic beads. The final adapter-modified cDNA fragments were enriched by 12 cycles of PCR using Illumina TruSeq PCR primers. The concentration and size distribution of the completed libraries were determined using a Fragment Analyzer (AATI, Ankeny, IA) and Qubit fluorometry (Invitrogen, Carlsbad, CA). Libraries were sequenced following Illumina’s standard protocol using the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit, yielding approximately 46-67 million total reads per sample. The flow cells were sequenced as 50 X 2 paired end reads on an Illumina HiSeq 4000 using HiSeq 3000/4000 sequencing kit and HD v3.4.0.38 collection software. Base-calling was performed using Illumina’s RTA version 2.7.7. Bioinformatics/Data Processing: All bioinformatics analyses were done through the Mayo Clinic Bioinformatics Core. The raw RNA sequencing paired-end reads for the samples were processed through the Mayo RNA-Seq bioinformatics pipeline, MAP-RSeq version 3.0.045. Briefly, MAP-RSeq employs the splice-aware aligner, STAR46, to align reads to the reference mouse genome build mm10. Gene and exon expression quantification were performed using the Subread47 package to obtain both raw and normalized (RPKM – Reads Per Kilobase per Million mapped reads) reads.
Statistics
Data are represented as the mean ± SD using GraphPad Prism unless noted otherwise in the figure legends. For all analyses, a p<0.05 was considered significant (denoted with *) unless noted otherwise in the figure legend. Specific statistical tests are noted in figure legends.
Data availability
The RNAseq dataset generated and analyzed during the current study will be available in the Sequence Read Archive (SRA) (National Center for Biotechnology Information, NCBI). All other datasets generated during the current study are available from the corresponding author upon request.
RESULTS
Lung adenocarcinoma conditioned media disrupts the myoblast transcriptome and metabolome.
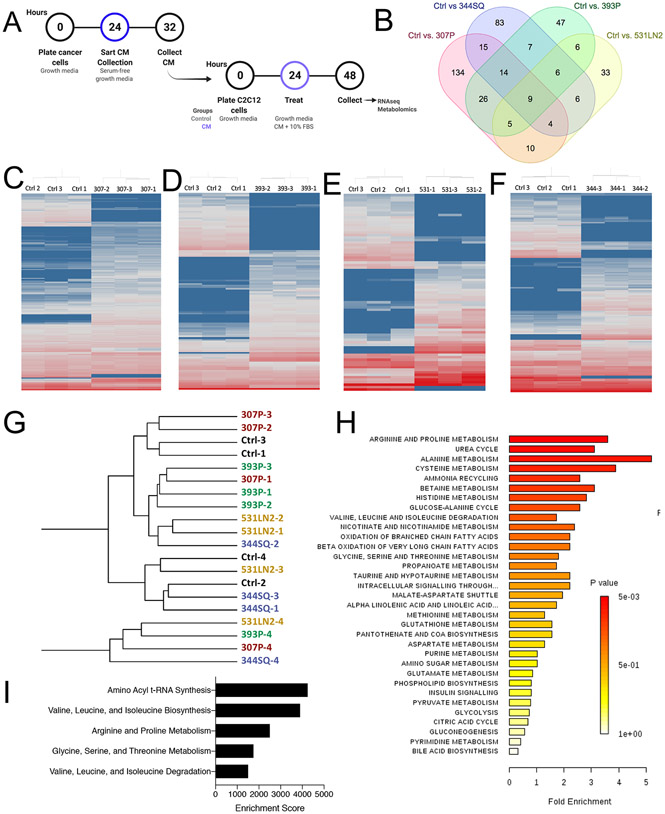
Many prior studies have evaluated the effects of individual tumor-derived factors on myoblast function, but in this study, we sought a unique approach to assess the downstream transcriptional and metabolic changes associated with exposure to a milieu of tumor cell-derived factors12,14,21. Our goal in this study was to identify pathways altered in myoblasts that may contribute to disrupted muscle homeostasis in individuals with cancer. We previously showed that tumor-derived factors contained in lung adenocarcinoma conditioned media (cancer cell conditioned media, CC-CM) are capable of disrupting myoblast differentiation12. In this study, we leveraged the same CC-CM protocol to treat and study proliferating myoblasts: CC-CM was collected under serum-free conditions for 8 hours, then serum was supplemented back in prior to treating C2C12 proliferating myoblasts (Fig. 1A). Myoblasts were exposed to CC-CM for 24 hours, then collected for transcriptomics or non-targeted metabolomics analysis (Fig. S1A,B). In total we assessed the effects of CC-CM from four distinct lung adenocarcinoma cell lines, all derived from the same murine model system: 307P, 393P, 344SQ, and 531LN248. Principal component analysis of the total detected transcriptome showed little separation of different CC-CM treatments, indicating overall similarity between groups (Fig. S2A,B). Pairwise comparisons between each CC-CM treatment and control identified sets of unique, differentially expressed genes (DEGs) for each condition (307P=134, 344SQ=83, 393P=47, 531LN2=33) (Fig. 1B, S2C-F). Hierarchical clustering based on significant DEGs (fold change cut off = 2, pval =0.05) resulted in clear separation of CC-CM treatments from control, in each treatment group (Fig. 1C-F).
Figure 1:
Lung adenocarcinoma conditioned media disrupts the transcriptome and metabolome of myoblasts. A: Schematic depicting the treatment paradigm for conditioned media (CM) collection and myoblast treatment. B: Venn diagram depicting the differentially expressed genes unique to each pairwise comparison, and those intersecting each grouping of comparisons. Created with molbiotools.com C-F: Heatmaps depicting hierarchical clustering of differentially expressed genes from pairwise comparisons of control vs. 307P (C), 393P (D), 531LN2 (E), and 344SQ (F) (cut offs of pval<0.05, FC>2) from log2 normalized FPKM values. Red = high expression, blue = low expression. G: Unsupervised hierarchical clustering of non-targeted metabolomics data. H: Pathway enrichment analysis generated in Metaboanalyst, based on ANOVA analysis of non-targeted metabolomics on control- and CM-treated myoblasts. I: Top 5 pathways from enrichment analysis (metaboanalyst) based on significant features from metabolomics and transcriptomics data sets.
In total, 3,370 LC-MS/MS peaks were processed in the metabolomics analysis. Hierarchical clustering based on total detected features did not show clear separation of CC-CM treatments from each other, mirroring the variance observed in the transcriptomics data (Fig. 1G). Due to the large number of detected metabolites, it is often easier to interpret non-targeted metabolomics data after conducting pathway enrichment analysis. Our pathway enrichment analysis, performed on significant features identified by ANOVA, revealed significant enrichment several amino acid related pathways, including the top enriching pathway “Arginine and Proline Metabolism” (Fig. 1H). To incorporate the transcriptional changes as well as metabolic changes into a comprehensive myoblast response model, we performed a combined enrichment analysis using both the differentially abundant metabolites and transcripts. The top 5 enriched pathways in this analysis were all amino acid related pathways: amino acyl t-RNA synthesis; valine, leucine, and isoleucine biosynthesis; arginine and proline metabolism; glycine, serine, and threonine metabolism; valine, leucine, and isoleucine degradation (Fig. 1I). While disrupted amino acid metabolism has long been associated with whole muscle wasting, perturbations in these pathways have been minimally characterized in the myoblasts49,50.
Disrupted NOS2 dynamics are observed in myoblasts treated with inflammatory factors.
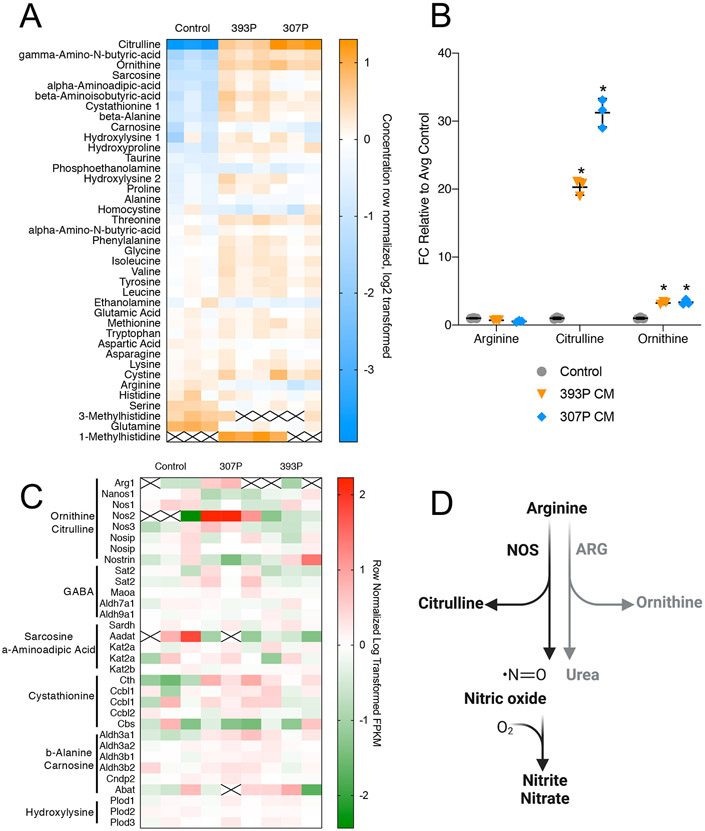
Based on the widespread enrichment of amino acid metabolism pathways by non-targeted metabolomics studies, we next performed quantitative, targeted metabolomics analysis of common amino acid metabolites, using two differed CC-CM treatments (307P and 393P). Among the highest differentially abundant metabolites were several amino acids that are intermediates or degradation products (citrulline, ornithine, sarcosine, alpha-aminoadipic-acid, beta-arminoisobutryic-acid, and cystathionine) (Fig. 2A). We focused on the abundance of metabolites and transcripts involved in “Arginine and Proline Metabolism,” in part due to the strong enrichment of this pathway in both separate analyses, as well as the combined analysis (Fig. 1H,I). After either 307P or 393P CC-CM treatment, we saw that there was a significant increase in intracellular citrulline (Fig. 2B), paired with up-regulation of NOS2 (Fig. 2C). NOS proteins are responsible for one branch of arginine catabolism, which produces citrulline and nitric oxide (NO) (Fig. 2D). Expression patterns of NOS1 and NOS3 were unchanged, indicating that the up-regulation of NOS2 is likely the driver of increased intracellular citrulline. We also observed a small but significant increase in intracellular ornithine, the product of arginine catabolism by the enzyme arginase (ARG), however, the expression change of ARG1 was less robust than that of NOS2 (Fig. 2B,C). Based on these observations, we chose to further probe NOS2-driven arginine catabolism as a disruptor of myogenesis.
Figure 2:
Amino acid metabolism is disrupted in myoblasts exposed to CC-CM. A: Heatmap depicting row-normalized, log2 transformed concentrations of amino acids detected via targeted metabolomics analysis. Orange = high abundance, blue = low abundance. White box with “x” = not detected. B: Fold change concentration of metabolites in the arginine catabolism pathway (arginine, citrulline, ornithine). Two-way ANOVA with multiple testing correction * = pval<0.05. C: Heatmap representing selected genes associated with the top 10 differentially abundant metabolites from targeted analysis (2A). 4 types of CM were used: 307P, 344SQ, 393P, and 531LN2. N = 3 for each treatment group. D: Schematic depicting arginine catabolism via NOS or ARG.
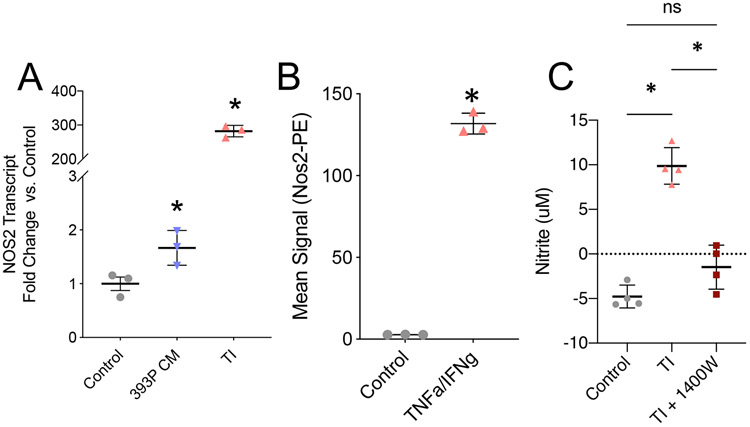
NOS2 has been critically studied in a number of cellular systems, including mature/differentiated myotubes and whole muscle25,29,31,51. NOS2 is unique among other NOS isoforms because it is expressed after cellular exposure to inflammatory cytokines such as TNFα and interferon gamma IFNγ. This is well established in macrophage populations, where the induction of NOS2 is critical for macrophage differentiation25. The transcriptional response to increased NOS2 is not clearly defined in proliferative myoblasts. We therefore tested the ability of both CC-CM and TNFα/IFNγ cocktail (TI) to induce NOS2 in C2C12 myoblasts. We observed induction of Nos2 transcript (qPCR) as well as NOS2 protein (antibody-based flow cytometry) (Fig. 3A,B, S3A-C). Lastly, we assessed the abundance of extracellular nitrites. NO is an unstable free radical that is quickly oxidized into nitrite and nitrate, which are more stable in cell culture medium (Fig. 2D). We used nitrite abundance as a proxy for NO levels and observed that TI treatment for 24 hours significantly increased nitrite abundance. To clearly demonstrate that the nitrite accumulation was due to changes in NOS2 activity, we utilized the NOS2-specific inhibitor 1400W and observed a complete restoration of nitrite abundance back to control levels (Fig. 3C). These novel findings indicate that myoblasts regulate arginine catabolism via NOS2 activity after exposure to inflammatory stimuli. Based on these observations we hypothesized that the induction of NOS2 and production of NO contributes to impaired myoblast dynamics. Importantly, we also established that TI treatment, which has been used as an atrophy stimulus in mature myotubes, is a controlled method for elevating NOS2 expression in proliferative myoblasts.
Figure 3:
NOS2 expression in myoblasts is induced by inflammatory stimuli. A: Nos2 transcript level, assessed by qPCR, in myoblasts treated with control media, 393P CM, orTNFα/IFNγ (TI). N=3, data points are representative of individual experiments B: NOS2 total protein, detected by antibody-based flow cytometry, in control or TI treated myoblasts. N=3, data points are representative of individual experiments C: Nitrite concentration in control, TI, and TI + 1400W treated myoblasts at 24 hours of treatment. Dashed line indicates lower detection limit of assay. N = 4 independent replicate experiments. For all panels, student’s t-test * = pval <0.05
Impaired myoblast proliferation after induction of NOS2 activity.
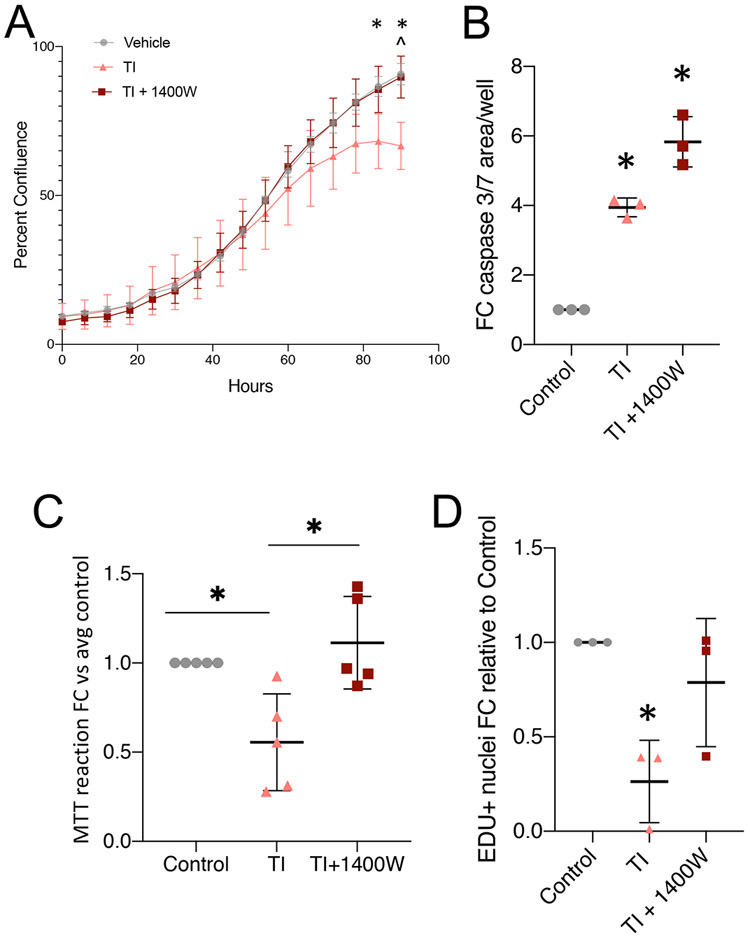
In vivo muscle regeneration begins with activation and proliferation of the MuSCs, a critical step in maintaining cell numbers for both differentiation and self-renewal. Therefore, we first assessed the effects of induced NOS2 activity in proliferating myoblasts. We hypothesized that disrupted amino acid metabolism would lead to impaired proliferation. We again utilized the NOS2 inhibitor 1400W to determine the extent to which any observed proliferation deficits were due to the induction of NOS2 activity. First, we tracked proliferation of myoblasts grown in TI or TI + 1400W conditions for 4 days. Relative to control myoblast proliferation, we observed that TI caused a significant drop in myoblast confluence/expansion after 3 days of exposure. Co-treatment with 1400W rescued proliferation back to control levels for the entire time course (Fig. 4A). We noted that the drop in expansion rate was delayed relative to the induction of NOS2 (after just 24 hours), so we hypothesized that TI treatment may be stimulating apoptosis. We tested this hypothesis by measuring caspase 3/7 activity using CellEvent™ Caspase-3/7 Green Detection Reagent. While TI treatment did cause a significant increase in caspase 3/7 activity after 48 hours, co-treatment with 1400W did not rescue this effect (Fig. 4B). This indicates that the rescue in proliferation is due to cellular proliferation that outweighs any increases in apoptosis. To confirm this hypothesis, we queried proliferation more directly using two methods: an MTT assay (Fig. 4C) and an EdU assay (Fig. 4D). Both of these methods showed significant reductions in proliferation after TI treatment for 24 hours, and complete rescue after co-treatment with 1400W. These data show that inflammatory induction of NOS2 is sufficient to disrupt the proliferation of myoblasts, a stage that is critical for eventual differentiation and in vivo muscle repair.
Figure 4:
NOS2 elevation impairs myoblast proliferation A: Proliferation curve over 4 days for control-, TI-, and TI + 1400W-treated myoblasts. Multiple t-test analysis * = pval <0.05 Ctrl vs TI, ^ = pval <0.05 TI+1400W vs TI. Representative of 3 independent replicates. Error bars represent error within 4 well replicates. B: Caspase activity in myoblasts treated for 48 hours. N = 3 independent replicates. C: MTT reaction product detected in myoblasts treated for 24 hours. N = 5 independent replicates. D: EDU incorporation assay after 24-hour treatment with TI or TI+1400W. (B-D) * = pval <0.05 by one-way ANOVA.
Acute inflammatory stimuli induce NOS2 activity and impair myoblast differentiation.
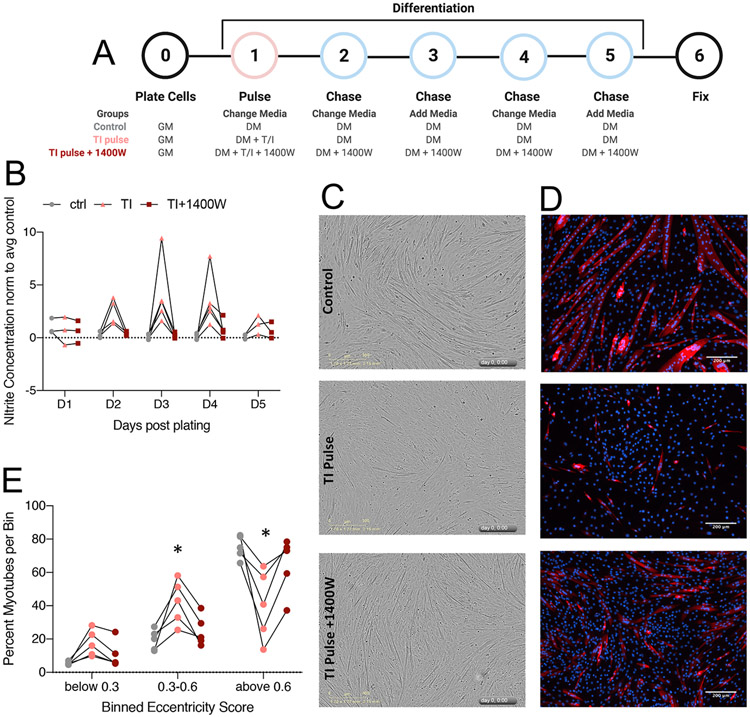
Based on our myoblast proliferation studies, we were next interested in how TI impacted differentiation of myoblasts into myotubes. We began by developing a treatment paradigm that would isolate the effects of inflammatory stimuli on differentiation, minimizing the effects we already observed on proliferation. To do this, we plated C2C12 cells at a high density and allowed them to adhere for 24 hours prior to changing the media. After 24 hours, we switched cells to differentiation media and concomitantly began a pulse-chase treatment with TI or TI + 1400W. We pulsed cells with TI-containing media for 24 hours, followed by a full change to media without TI. For the following 3 days, we continued our normal differentiation protocol, which includes full changes every other day, and fresh media supplementation in between (Fig. 5A). We began by asking if there were natural changes to nitrite (NO) abundance over the course of differentiation; we did not observe significant increases or decreases in nitrite during control differentiation over 5 days (Fig. 5B, grey dots). Pulse treatment with TI caused a sustained increase in nitrite levels through day 4 of differentiation (Fig. 5B, pink dots). As expected, the addition of 1400W prevented the accumulation of nitrites in the media (Fig. 5B, dark red dots). We first assessed differentiation at experimental endpoint by analyzing phase-contrast images. We observed that cells treated with TI pulse conditions had disrupted morphology compared to control and TI pulse + 1400W conditions (Fig. 5C). This was more evident after staining cultures for myosin heavy chain one (MyHC1), which is a marker of terminal differentiation. In TI pulse conditions we saw fewer stained myotubes, and those that were present were less elongated than control. Co-treatment with 1400W resulted in a higher density of MyHC1+ myotubes (Fig. 5D). High eccentricity (above 0.6) indicates that stained objects are approaching a linear (myotube) shape, as compared to low eccentricity (below 0.6) where objects are round in shape (myoblasts). Although the myotubes in the TI pulse + 1400W treatment appeared smaller than the control myotubes, myotube eccentricity analysis shows that there was a significant rescue in high eccentricity scores after co-treatment with 1400W (Fig. 5E). Together, these results demonstrate that TI-induced expression of NOS2 is sufficient to impair myogenic differentiation.
Figure 5:
NOS2 inhibition rescues differentiation defects associated with inflammatory markers of cachexia. A: Schematic depicting the pulse-chase set up for treatments during myoblast differentiation. GM = growth media (10% FBS), DM = differentiation media (2% HS). “Change media” indicates a full media change, “add media” indicates spiking in 50% fresh media. B: Nitrite concentrations for days 1-5 of differentiation, following the treatment outline in (A). All points were not significant (pval >0.05 by multiple t-test). C: Phase-contrast images of differentiated myotubes at day 5. Images were acquired at 10x, scale bar represents 300μm. D: Fluorescence images of differentiated myotubes stained for myosin heavy chain I (red), and DAPI (blue) Images were acquired at 20x, scale bar represents 200μm. E: Eccentricity scores for myosin heavy chain-positive objects, binned into 3 sizes. Eccentricity of 0 = round, 1 = linear. * = pval <0.05 by ANOVA.
DISCUSSION
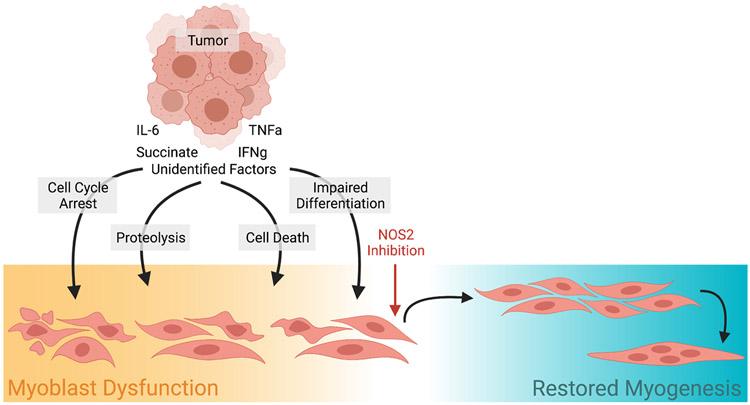
Despite several studies assessing the effects of NO on mature muscle, the field has lacked a definitive study assessing the effects of excessive NOS2 activity during early stages of myoblast differentiation30. In the present study we show that myoblasts respond to inflammatory stimuli by upregulating NOS2 expression, which results in elevated NOS2 activity. This study is one of a very small group of studies that specifically assess early-stage myoblasts, as most others examine mature myotubes14,30,31. We go on to demonstrate that upregulation of NOS2 impairs myoblast proliferation and differentiation. TI treatment is known to exert complex signaling cascades, but we show that, in myoblasts, functional impairments are specific to NOS2 upregulation as the NOS2-specific inhibitor, 1400W, was capable of rescuing both impaired proliferation and differentiation (Fig. 6). These data point toward NOS inhibition as a promising therapeutic for future testing in pre-clinical models of cachexia, especially given the efficacy of certain pan-NOS inhibitors in preventing lean mass loss in TNFα-overexpressing tumor mice32.
Figure 6:
Tumor-derived factors elicit a multi-factorial response in myoblasts including disrupted proliferation, differentiation, cell death, and protein homeostasis. These processes lead to dysfunctional myogenesis, which may contribute to skeletal muscle wasting in cancer cachexia. Our study showed that TNFα and IFNγ have complex effects on myoblasts, but a single inhibitor for NOS2, 1400W, is able to reverse these effects and restore myogenesis. Created with BioRender.com.
The study presented here was designed specifically to identify pathways in myoblasts that were modulated by a milieu of tumor-derived factors. Numerous studies have focused on single tumor-derived factors as drivers of cachexia, most with little success clinically6,7,12,13. We hypothesized that the complex inflammatory signature of CC-CM may cause a central cluster of transcriptional and metabolic changes in myoblasts. We successfully identified disrupted amino acid metabolism as a metabolic cluster that was disrupted after myoblast exposure to CC-CM. Within this cluster, we found that NOS2-driven arginine catabolism is upregulated in response to inflammatory signaling. We utilized inflammatory cytokine stimuli (TI treatment) to model NOS2 upregulation in vitro and were able to identify a role for NOS2 in controlling myogenesis. We believe that a similar strategy could be used to further elucidate the role of other amino acid catabolic pathways in disrupted myogenesis and muscle regeneration in cancer cachexia.
Although our in vitro studies show near complete rescue of TI-induced impairments in myoblast proliferation and differentiation, it is important to consider relevant differences between in vitro models, pre-clinical animal models, and the patient population. Strengths of our in vitro system include the ability to profile complex transcriptomic and metabolomic changes in an isolated system, and the ability to track myoblast dynamics longitudinally. In particular, we were able to assess metabolic changes in the absence of confounding variables associated with systemic inflammatory or metabolic processes. Muscle stem cells are notoriously difficult to extract from animals or patients for downstream profiling/omics analyses, making C2C12 myoblasts a more pragmatic model in these instances. The most notable weakness of both in vitro and in vivo studies is that even the most promising of pre-clinical models have failed to result in efficacious therapies. The current study provides impetus to assess NOS2 inhibitors in mouse models of cancer cachexia, and future work will focus on assessing NOS2 regulation in mice and patient populations.
Finally, we are able to reach several thought-provoking conclusions about myoblast biology from the current study. Despite complex NFκB and STAT3 signaling cascades known to be initiated by TNFα/IFNγ treatment, we were able to ameliorate negative consequences of this treatment using a single inhibitor, targeting a single downstream signaling cascade. Previous work has shown the importance of this pathway using in vitro models of cancer cachexia, but has not identified the sufficiency of NOS2 overactivity alone to disrupt myogenesis14. Outside the context of cancer cachexia, the present study underscores the importance of amino acid homeostasis in myogenesis and raises a number of interesting questions including: How does NOS2 regulate myogenesis in vivo? Are amino acid imbalances associated with NOS2 overactivity driving defects in myogenesis? How might NO production by myoblasts impact other cells in the muscle microenvironment? These and other muscle stem cell biology questions will be important to address as muscle wasting research moves forward. Ultimately, our study confirms that, in addition to benefitting whole muscle retention in tumor-bearing mouse models, NOS2 inhibitors could positively affect myogenesis. This makes NOS2 an interesting target in future pre-clinical cachexia therapeutic studies, and highlights NOS2-mediated catabolism as an important myogenic regulator.
Supplementary Material
HIGHLIGHTS.
Amino acid metabolism is impacted in myoblasts treated with cancer cell conditioned media
Inflammatory cytokines induce nitric oxide synthase 2 in myoblasts
Elevated nitric oxide synthase 2 activity impairs myoblast proliferation and differentiation
ACKNOWLEDGEMENTS
The authors wish to thank members of the Doles lab for helpful discussions and manuscript suggestions (Rebecca Schmitt, PhD, Aneesha Dasgupta, PhD, and Alexandra Ducharme). J.D. was supported by the National Institutes of Health (award #s: R00AR66696, R35GM128594), Mayo Clinic start-up funds, Career Development Awards from the Mayo Clinic SPORE in Pancreatic Cancer (NIH/National Cancer Institute (NCI) CA102701) and the American Association for Cancer Research/Pancreatic Cancer Action Network, and the Glenn Foundation for Medical Research. P.C.A-W was supported by the Mayo Clinic Regenerative Sciences Training Program (RSTP). Metabolomics studies were made possible by the Mayo Clinic Metabolomics Resource Core through NIH/National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) U24DK10049 originating from the NIH Director’s Common Fund. The Mayo Clinic Genome Analysis Core provided experimental support for the RNA sequencing study. Lung adenocarcinoma cell lines were a generous gift from Yanan Yang, PhD.
ABBREVIATIONS:
- ANOVA
Analysis of Variance
- ARG1
Arginase 1
- CC-CM
Cancer Cell-Conditioned Media
- DAPI
4′,6-diamidino-2-phenylindole
- DEG
Differentially Expressed Gene
- DMEM
Dulbecco’s Modified Essential Medium
- IFNγ
Interferon Gamma
- MRF
Myogenic Regulatory Factor
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MuSC
Muscle Stem Cell
- MyHC1
Myosin Heavy Chain 1
- NFkB
Nuclear Factor Kappa-light-chain-enhancer of activated B cells
- NO
Nitric Oxide
- NOS
Nitric Oxide Synthase
- PBS
Phosphate Buffered Saline
- PCR
Polymerase Chain Reaction
- qPCR
Quantitative Polymerase Chain Reaction
- TI
TNFα/IFNγ co-treatment
- TNFα
Tumor Necrosis Factor alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS STATEMENT
The authors have no competing interests to declare.
REFERENCES
- 1.Suzuki H, Asakawa A, Amitani H, Nakamura N & Inui A Cancer cachexia--pathophysiology and management. J Gastroenterol 48, 574–594, doi: 10.1007/s00535-013-0787-0 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan CR et al. Pancreatic cancer cachexia: a review of mechanisms and therapeutics. Front Physiol 5, 88, doi: 10.3389/fphys.2014.00088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Haehling S & Anker SD Cachexia as major underestimated unmet medical need: facts and numbers. International journal of cardiology 161, 121–123 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Fearon K et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol 12, 489–495, doi: 10.1016/S1470-2045(10)70218-7 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Baracos VE, DeVivo C, Hoyle DH & Goldberg AL Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Am J Physiol 268, E996–1006 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Pettersen K et al. Cancer cachexia associates with a systemic autophagy-inducing activity mimicked by cancer cell-derived IL-6 trans-signaling. Sci Rep 7, 2046, doi: 10.1038/s41598-017-02088-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He WA et al. NF-κB-mediated Pax7 dysregulation in the muscle microenvironment promotes cancer cachexia. J Clin Invest 123, 4821–4835, doi: 10.1172/JCI68523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaughan VC, Martin P & Lewandowski PA Cancer cachexia: impact, mechanisms and emerging treatments. Journal of cachexia, sarcopenia and muscle 4, 95–109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madeddu C et al. Randomized phase III clinical trial of a combined treatment with carnitine+ celecoxib±megestrol acetate for patients with cancer-related anorexia/cachexia syndrome. Clinical nutrition 31, 176–182 (2012). [DOI] [PubMed] [Google Scholar]
- 10.McMillan D et al. A prospective randomized study of megestrol acetate and ibuprofen in gastrointestinal cancer patients with weight loss. British journal of cancer 79, 495–500 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pownall ME, Gustafsson MK & Emerson CP Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol 18, 747–783, doi: 10.1146/annurev.cellbio.18.012502.105758 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Hogan KA et al. Tumor-derived cytokines impair myogenesis and alter the skeletal muscle immune microenvironment. Cytokine, doi: 10.1016/j.cyto.2017.11.006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearon KC, Glass DJ & Guttridge DC Cancer cachexia: mediators, signaling, and metabolic pathways. Cell metabolism 16, 153–166 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Ma JF et al. STAT3 promotes IFNγ/TNFα-induced muscle wasting in an NF-κB-dependent and IL-6-independent manner. EMBO Mol Med 9, 622–637, doi: 10.15252/emmm.201607052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arneson-Wissink PC, Ducharme AM & Doles JD A novel transplantable model of lung cancer-associated tissue loss and disrupted muscle regeneration. Skeletal Muscle 10, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiappalupi S et al. Targeting RAGE prevents muscle wasting and prolongs survival in cancer cachexia. Journal of cachexia, sarcopenia and muscle 11, 929–946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tas F et al. Serum levels of leptin and proinflammatory cytokines in advanced-stage non-small cell lung cancer. Medical oncology 22, 353–358 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Derin D et al. Serum levels of apoptosis biomarkers, survivin and TNF-alpha in nonsmall cell lung cancer. Lung cancer 59, 240–245 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Shang G-S, Liu L & Qin Y-W IL-6 and TNF-α promote metastasis of lung cancer by inducing epithelial-mesenchymal transition. Oncology letters 13, 4657–4660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fearon K, Arends J & Baracos V Understanding the mechanisms and treatment options in cancer cachexia. Nat Rev Clin Oncol 10, 90–99, doi: 10.1038/nrclinonc.2012.209 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Arneson-Wissink PC et al. The wasting-associated metabolite succinate disrupts myogenesis and impairs skeletal muscle regeneration. JCSM Rapid Communications 3, 56–69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koopman R, Ly CH & Ryall JG A metabolic link to skeletal muscle wasting and regeneration. Front Physiol 5, 32, doi: 10.3389/fphys.2014.00032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess DT, Matsumoto A, Kim S-O, Marshall HE & Stamler JS Protein S-nitrosylation: purview and parameters. Nature reviews Molecular cell biology 6, 150–166 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hall DT, Ma JF, Marco SD & Gallouzi I-E Inducible nitric oxide synthase (iNOS) in muscle wasting syndrome, sarcopenia, and cachexia. Aging (Albany NY) 3, 702–715, doi: 10.18632/aging.100358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rath M, Müller I, Kropf P, Closs EI & Munder M Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front Immunol 5, 532, doi: 10.3389/fimmu.2014.00532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttridge DC, Mayo MW, Madrid LV, Wang C-Y & Albert SSB NF-κB-Induced Loss of MyoD Messenger RNA: Possible Role in Muscle Decay and Cachexia. Science 289, 2363–2366, doi: 10.1126/science.289.5488.2363 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Jatoi A et al. A placebo-controlled, double-blind trial of infliximab for cancer-associated weight loss in elderly and/or poor performance nonsmall cell lung cancer patients (N01C9). Lung Cancer 68, 234–239, doi: 10.1016/j.lungcan.2009.06.020 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ham DJ et al. L-Citrulline Protects Skeletal Muscle Cells from Cachectic Stimuli through an iNOS-Dependent Mechanism. PLoS One 10, e0141572, doi: 10.1371/journal.pone.0141572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramamoorthy S, Donohue M & Buck M Decreased Jun-D and myogenin expression in muscle wasting of human cachexia. Am J Physiol Endocrinol Metab 297, E392–401, doi: 10.1152/ajpendo.90529.2008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams G, Brown T, Becker L, Prager M & Giroir BP Cytokine-induced expression of nitric oxide synthase in C2C12 skeletal muscle myocytes. Am J Physiol 267, R1020–1025, doi: 10.1152/ajpregu.1994.267.4.R1020 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Di Marco S et al. NF-kappa B-mediated MyoD decay during muscle wasting requires nitric oxide synthase mRNA stabilization, HuR protein, and nitric oxide release. Mol Cell Biol 25, 6533–6545, doi: 10.1128/MCB.25.15.6533-6545.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buck M & Chojkier M Muscle wasting and dedifferentiation induced by oxidative stress in a murine model of cachexia is prevented by inhibitors of nitric oxide synthesis and antioxidants. EMBO J 15, 1753–1765 (1996). [PMC free article] [PubMed] [Google Scholar]
- 33.Rigamonti E et al. Requirement of inducible nitric oxide synthase for skeletal muscle regeneration after acute damage. The Journal of Immunology 190, 1767–1777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadek J et al. Pharmacological or genetic inhibition of iNOS prevents cachexia-mediated muscle wasting and its associated metabolism defects. EMBO Molecular Medicine, e13591 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martiínez-Moreno M et al. Nitric oxide down-regulates caveolin-3 levels through the interaction with myogenin, its transcription factor. Journal of Biological Chemistry 282, 23044–23054 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Eley HL, Russell ST & Tisdale MJ Attenuation of depression of muscle protein synthesis induced by lipopolysaccharide, tumor necrosis factor, and angiotensin II by β-hydroxy-β-methylbutyrate. American Journal of Physiology-Endocrinology and Metabolism 295, E1409–E1416 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Hirasaka K et al. Isoflavones derived from soy beans prevent MuRF1-mediated muscle atrophy in C2C12 myotubes through SIRT1 activation. Journal of nutritional science and vitaminology 59, 317–324 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Wang D-T et al. Resveratrol prevents TNF-α-induced muscle atrophy via regulation of Akt/mTOR/FoxO1 signaling in C2C12 myotubes. International Immunopharmacology 19, 206–213 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Yuan L et al. Muscle-specific E3 ubiquitin ligases are involved in muscle atrophy of cancer cachexia: an in vitro and in vivo study. Oncol Rep 33, 2261–2268, doi: 10.3892/or.2015.3845 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Gibbons DL et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 23, 2140–2151, doi: 10.1101/gad.1820209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dasgupta A et al. SIRT1-NOX4 signaling axis regulates cancer cachexia. Journal of Experimental Medicine 217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanza IR et al. Quantitative metabolomics by 1 H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PloS one 5, e10538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkins J, Sakrikar D, Petterson X-M, Lanza IR &Trushina EA comprehensive protocol for multiplatform metabolomics analysis in patient-derived skin fibroblasts. Metabolomics 15, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia J, Sinelnikov IV, Han B &Wishart DS MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res 43, W251–257, doi: 10.1093/nar/gkv380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalari KR et al. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC bioinformatics 15, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y, Smyth GK & Shi W The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research 41, e108–e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbons DL et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PloS one 4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Q-J et al. Serum and urine metabolomics study reveals a distinct diagnostic model for cancer cachexia. J Cachexia Sarcopenia Muscle, doi: 10.1002/jcsm.12246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunz HE et al. Methylarginine metabolites are associated with attenuated muscle protein synthesis in cancer-associated muscle wasting. Journal of Biological Chemistry 295, 17441–17459 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buono R et al. Nitric oxide sustains long-term skeletal muscle regeneration by regulating fate of satellite cells via signaling pathways requiring Vangl2 and cyclic GMP. Stem Cells 30, 197–209, doi: 10.1002/stem.783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq dataset generated and analyzed during the current study will be available in the Sequence Read Archive (SRA) (National Center for Biotechnology Information, NCBI). All other datasets generated during the current study are available from the corresponding author upon request.