Abstract
Activation of the mitogen-activated protein kinase (MAPK) pathway enhances long-range transactivation by the β-globin locus control region (LCR) (W. K. Versaw, V. Blank, N. M. Andrews, and E. H. Bresnick, Proc. Natl. Acad. Sci. USA 95:8756–8760, 1998). The enhancement requires tandem recognition sites for the hematopoietic transcription factor NF-E2 within the hypersensitive site 2 (HS2) subregion of the LCR. To distinguish between mechanisms of induction involving the activation of silent promoters or the increased efficacy of active promoters, we analyzed basal and MAPK-stimulated HS2 enhancer activity in single, living cells. K562 erythroleukemia cells stably transfected with constructs containing the human Aγ-globin promoter linked to an enhanced green fluorescent protein (EGFP) reporter, with or without HS2, were analyzed for EGFP expression by flow cytometry. When most cells in a population expressed EGFP, MAPK augmented the activity of active promoters. However, under conditions of silencing, in which cells reverted to a state with no measurable EGFP expression, MAPK activated silent promoters. Furthermore, studies of populations of EGFP-expressing and non-EGFP-expressing cells isolated by flow cytometry showed that MAPK activation converted nonexpressing cells into expressing cells and increased expression in expressing cells. These results support a model in which MAPK elicits both graded and stochastic responses to increase HS2-mediated transactivation from single chromatin templates.
Transcription of the β-globin genes is regulated by four erythroid-cell-specific DNaseI-hypersensitive sites (HSs) at the 5′ end of the β-globin locus (12, 46), termed the locus control region (LCR) (16). Although the HSs function together to confer long-range transactivation in transfection assays (5) and transgenic mice (7), HS2 and HS3, individually, have strong erythroid-cell-specific enhancer activity when positioned near a promoter (9, 18, 20, 27, 29, 38, 41, 44). Recently, we reported that LCR-mediated transactivation in stable transfection assays increased upon activation of the mitogen-activated protein kinase (MAPK) signaling pathway (48). The MAPK-dependent stimulation required HS2 and was independent of the distance between HS2 and the promoter. Tandem recognition sites within HS2 for the erythroid-cell- and megakaryocyte-specific transcription factor NF-E2 (1, 33, 37), which are critical for strong enhancer activity, were necessary for induction. Besides the requirement of NF-E2 sites, expression of tethered NF-E2, with covalently linked p45 and p18 subunits, conferred induction in CB3 erythroleukemia cells (30) that lack NF-E2. We postulated that phosphorylation of NF-E2 facilitates the recruitment of a coactivator that partially overcomes a limitation to long-range activation (48). Enhancement of NF-E2-mediated transactivation by MAPK with synthetic promoter constructs in transient expression assays has also been described (36).
Two simple mechanisms could explain the MAPK-dependent stimulation of HS2 enhancer activity. First, MAPK may enhance the formation of transcription complexes on inactive promoters. Alternatively, MAPK may augment the activity of preassembled complexes. To distinguish between these possibilities, we established a system for assaying gene expression in single, living cells using cell lines containing integrated constructs with the enhanced green fluorescent protein (EGFP) reporter gene.
Gene expression can be quantitatively assayed in single cells with β-galactosidase or GFP reporter genes and flow cytometric analysis. Studies on the mechanism of enhancer action in single cells have found that enhancers act via a stochastic or binary mechanism to increase the probability of gene expression from a given template (11, 15, 24, 32, 40, 43, 50, 53). Accordingly, the enhancer determines whether the promoter will be active or inactive rather than augmenting the efficacy of an active promoter. Thus, there are three possible activity states in a diploid cell, each characterized by either two inactive alleles, one active allele, or two active alleles. Stochastic behavior can be explained by the enhancer-dependent, all-or-none recruitment of the RNA polymerase II holoenzyme (45) to an inactive promoter. As the positioning of a nucleosome on a promoter can occlude cis-acting elements (26, 49), the chromatin could prevent initiation complex assembly and thus maintain the silent state. A signaling pathway that targets an enhancer binding factor could regulate its ability to engage in protein-protein interactions necessary for the assembly of the enhancer complex. Only when the complete complex is assembled, which can be an all-or-none event (4), would the chromatin be disrupted, allowing for holoenzyme recruitment.
The alternative graded mechanism (23), involving the augmented efficacy of active promoters, would result from a factor modification that modulates the activity of the enhancer complex rather than the all-or-none complex assembly. Based on the importance of coactivators in transactivation (19), the modification may increase the efficiency of coactivator recruitment and thus increase promoter activity. Although activation may involve exclusively stochastic or graded responses, certain mechanisms may elicit both responses. For example, coactivators that modify chromatin structure to control DNA accessibility could influence both initiation complex assembly and the activity of preassembled complexes. Here, we describe the influence of MAPK on HS2 enhancer activity in single, living cells. Increased enhancer activity is manifested as both graded and stochastic responses, and the implications of this are discussed with respect to a model for HS2-mediated long-range transactivation.
MATERIALS AND METHODS
Plasmid construction.
The KpnI-HindIII multiple cloning site of plasmid pGFPemd-Basic (Packard) was replaced with the 390-bp KpnI-HindIII fragment of pGL3RIγluc (5), which contains the multiple cloning site and the human Aγ-globin promoter (−299 to +35), to yield pγEGFP. The 1,460-bp KpnI fragment of pHS2γluc (5) containing human HS2 was subcloned into pγEGFP to yield pHS2γEGFP. To physically separate HS2 from the promoter, phage λ DNA fragments (either 2.2 kb or one each of 2.4 and 5.1 kb) were subcloned into the MluI site of pHS2γEGFP to yield pHS2(2.2)γEGFP and pHS2(7.5)γEGFP. The 1,950-bp KpnI-EcoRI fragment of pHS3γluc (5) containing human HS3 was subcloned into pγEGFP to yield pHS3γEGFP. The expression vector encoding constitutively active human MEK1/R4F (cMEK1) was described previously (54).
Cell culture.
The human erythroleukemia cell line K562 was propagated in Iscove’s modified Eagle’s medium (IMEM; Biofluids) containing gentamicin (25 μg/ml) and 10% fetal calf serum (Gibco-BRL). Cell lines were grown in a humidified incubator at 37°C in the presence of 5% carbon dioxide. The cell density was maintained between approximately 1 × 105/ml and 8 × 105/ml. Stably transfected clones of K562 cells were selected and maintained in the presence of hygromycin B (0.2 and 0.1 mg/ml; Sigma) as described previously (5, 25).
Flow cytometric analysis.
Cells (2 × 105) were isolated by centrifugation at 240 × g for 5 min at 4°C, washed by resuspension in 1 ml of phosphate-buffered saline, and recentrifuged. Washed cells were resuspended in phosphate-buffered saline and 1% bovine serum albumin and then analyzed for EGFP fluorescence by using a FACScan flow cytometer and CellQuest software (Becton Dickinson). Propidium iodide (0.3 μg/ml) was added to restrict the analysis to live cells. Untransfected K562 cells were used to define nonexpressing cells. The EGFP fluorescence of uninduced and tetradecanoyl phorbol acetate (TPA)-induced samples of a given clone (see Tables 1 and 2) were measured at the same time with identical instrument settings, and they are directly comparable. In contrast, mean fluorescence values for different clones are not directly comparable, due to variations in gating between measurements made on different days and the different instrument settings required for optimal peak resolution.
TABLE 1.
Influence of TPA treatment on EGFP expression in stably transfected K562 clonal lines
| Clone | Copy no. | % EGFP-positive cells
|
Mean fluorescence
|
Induced/uninduced ratio | ||
|---|---|---|---|---|---|---|
| With TPA | Without TPA | With TPA | Without TPA | |||
| γEGFP-1 | 1 | 71.1 | 73.5 | 7.7 | 8.3 | 0.9 |
| γEGFP-4 | 8 | 99.9 | 99.8 | 492 | 531 | 0.9 |
| γEGFP-5 | 1 | 2.4 | 2.3 | 1.5 | 4.2 | 0.4 |
| HS2γEGFP-1 | 14 | 99.7 | 99.8 | 778 | 344 | 2.3 |
| HS2γEGFP-3 | 3 | 99.9 | 99.9 | 34 | 27 | 1.2 |
| HS2γEGFP-4 | 5 | 99.9 | 99.9 | 125 | 106 | 1.2 |
| HS2γEGFP-7 | 11 | 99.9 | 99.9 | 506 | 270 | 1.9 |
| HS2γEGFP-8 | 3 | 99.9 | 99.8 | 66 | 52 | 1.3 |
| HS2γEGFP-9 | 16 | 99.9 | 99.8 | 371 | 256 | 1.4 |
| HS2γEGFP-10 | 14 | 99.8 | 99.7 | 390 | 335 | 1.2 |
| HS3γEGFP-2 | 3 | 98.9 | 98.9 | 329 | 319 | 1.0 |
| HS3γEGFP-3 | 3 | 99.9 | 99.9 | 561 | 663 | 0.8 |
| HS3γEGFP-5 | 4 | 99.8 | 99.9 | 1,973 | 2,306 | 0.9 |
| HS3γEGFP-6 | 2 | 99.8 | 99.9 | 464 | 512 | 0.9 |
| HS3γEGFP-7 | 2 | 99.8 | 99.9 | 903 | 925 | 1.0 |
| HS3γEGFP-8 | 2 | 97.4 | 97.8 | 80 | 88 | 0.9 |
TABLE 2.
Influence of HS2-promoter distance and TPA treatment on EGFP expression in stably transfected K562 clonal lines
| Clone | Copy no. | % EGFP-positive cells
|
Mean fluorescence
|
Induced/ uninduced ratio | ||
|---|---|---|---|---|---|---|
| With TPA | Without TPA | With TPA | Without TPA | |||
| HS2(2.2)γEGFP-2 | 5 | 99.9 | 99.9 | 427 | 166 | 2.6 |
| HS2(2.2)γEGFP-3 | 3 | 99.3 | 96.2 | 1,383 | 262 | 5.3 |
| HS2(2.2)γEGFP-4 | 2 | 89.2 | 85.1 | 260 | 129 | 2.0 |
| HS2(2.2)γEGFP-5 | 10 | 98.3 | 99.1 | 925 | 119 | 7.8 |
| HS2(2.2)γEGFP-6 | 1 | 99.7 | 99.6 | 81 | 36 | 2.3 |
| HS2(2.2)γEGFP-7 | 4 | 67.4 | 58.1 | 58 | 33 | 1.8 |
| HS2(2.2)γEGFP-8 | 1 | 97.5 | 91.1 | 22 | 9.7 | 2.2 |
| HS2(7.5)γEGFP-1 | 1 | 99.7 | 99.9 | 25 | 15 | 1.7 |
| HS2(7.5)γEGFP-2 | 21 | 35.5 | 7.6 | 188 | 73 | 2.6 |
| HS2(7.5)γEGFP-3 | 1 | 50.4 | 43.0 | 41 | 35 | 1.2 |
| HS2(7.5)γEGFP-4 | 1 | 92.3 | 85.4 | 1,220 | 613 | 2.0 |
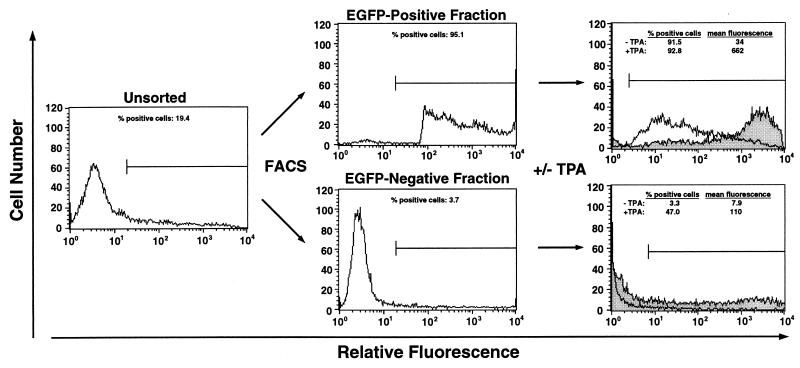
In the experiment shown in Fig. 6, HS2γEGFP-1 was grown without hygromycin for 133 and 135 days and then sorted into EGFP-expressing and non-EGFP-expressing cells with a FACStar-plus instrument (Becton Dickinson). Replicate aliquots (4 × 105) of sorted cells were treated with 5 nM TPA or vehicle for 20 h, and then EGFP fluorescence was measured by flow cytometric analysis.
FIG. 6.
Physical separation of EGFP-expressing and non-EGFP-expressing cells revealed both stochastic and graded responses to TPA treatment. Clone HS2γEGFP-1 was grown without hygromycin for 133 or 135 days and then sorted by FACS into EGFP-expressing and non-EGFP-expressing pools. Pools were treated with 5 nM TPA (shaded) or vehicle (unshaded) for 20 h, and then the percentage of EGFP-positive cells and the mean fluorescence of viable cells were measured by flow cytometric analysis. Representative flow cytometric profiles are shown. The horizontal line represents the EGFP-expressing cells. The values represent the means of two independent experiments in which cells were grown for either 133 or 135 days without hygromycin.
Reporter assays.
For the detection of EGFP in cell lysates, 2 × 105 cells were harvested by centrifugation at 240 × g for 5 min at 4°C. Cells were lysed by adding 100 μl of lysis buffer (Promega), incubating for 15 min at room temperature, and then centrifuging at 18,700 × g for 2 min at 4°C. Aliquots of cell lysate (5 to 20 μl) were added to 1 ml of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA, and the relative fluorescence was measured with a SLM 8000C spectrofluorometer. The excitation and emission settings were 484 nm and 508 nm, respectively, and the band-pass was set to 4 nm. The amount of EGFP in each sample was determined with a standard curve of recombinant EGFP (Clontech). EGFP values were normalized by protein concentration and, in certain cases, by the copy number of the integrated construct as determined by Southern blot analysis. The protein concentration was estimated by using the Bradford assay with γ-globulin as a standard. As the EGFP levels in cell lysates were determined with a standard curve of purified EGFP, values for lysates from different clonal lines are directly comparable. The TPA induction was consistently lower when calculated from measurements of EGFP in lysates than when calculated by flow cytometric analysis. This is likely related to the fact that the flow cytometric assay measured EGFP expression in only living cells.
Transient transfections.
Clonal lines containing stably integrated EGFP reporter genes were transiently transfected with a constitutive expression vector encoding enhanced blue fluorescent protein (EBFP) (Clontech) and either a plasmid encoding cMEK1 or the empty vector pcDNA3 (Invitrogen). EBFP was used as a marker for transfection, allowing us to test the influence of transiently expressed cMEK1 on the activity of the stably integrated EGFP reporter gene. A 3:1 ratio of cMEK1 or pcDNA3 to EBFP was used to increase the probability that EBFP-positive cells contained cMEK1 or pcDNA3. Cells (5 × 105) were collected by centrifugation at 240 × g for 5 min at 4°C, resuspended in 0.5 ml of IMEM containing 10% fetal calf serum and gentamicin (25 μg/ml), and mixed with 3.5 ml of identical medium in each well of a six-well plate. Plasmid DNA (6 μg in 150 μl of IMEM) was incubated with 24 μl of Superfect transfection reagent (Qiagen) for 15 min at room temperature and then added to cells. Cells were incubated for 48 h, harvested, and then subjected to flow cytometric analysis with a FACStar-plus instrument as described above.
RESULTS
TPA treatment stimulates HS2 enhancer activity by increasing the efficacy of active promoters.
To determine whether the stimulation of HS2 enhancer activity by TPA occurs via the facilitated assembly of transcription complexes on inactive promoters or the augmented efficacy of active promoters, we generated stably transfected clonal cell lines containing EGFP reporter constructs. The constructs consisted of a human Aγ-globin promoter linked to EGFP, with or without HS2 or HS3. We measured EGFP expression in several clonal lines containing each construct by flow cytometric and fluorometric analyses.
Clonal cell lines containing the Aγ-globin promoter alone showed considerable variability in EGFP expression between clones and between cells of the same clone. The percentage of EGFP-positive cells averaged 58.5% ± 35.7% (mean ± standard error, n = 3) (Fig. 1 and Table 1). The variability is consistent with the expected strong influence of chromosomal position on the promoter. The activation of MAPK in K562 cells is routinely done by treatment with the protein kinase C (PKC) activator TPA (39, 54). Paradoxically, MAPK activation can result in either the erythroid or megakaryocytic differentiation of K562 cells depending on the TPA concentration and the length of incubation. As reported previously (48), treatment with 5 nM TPA for 20 h did not influence the morphology of our K562 clonal lines. TPA did not increase EGFP expression from the promoter-only constructs, regardless of whether the basal activity was high or low (ratio of induced cells to uninduced cells [herein referred to as “induced/uninduced ratio”] = 0.74 ± 0.19, n = 3). (Fig. 1 and Table 1).
FIG. 1.
TPA induction of HS2 enhancer activity analyzed in single cells. Cultures were treated with 5 nM TPA (shaded) or vehicle (unshaded) for 20 h before each analysis. Histograms show representative flow cytometric analyses of stably transfected K562 clonal cell lines containing γEGFP, HS2γEGFP, or HS3γEGFP constructs. The number of integrated copies of the reporter plasmid is shown. Nonexpressing cells had a relative fluorescence of no more than 1 on this scale.
In contrast to the promoter-only constructs, EGFP expression by all constructs containing HS2 was induced by TPA (Fig. 1 and Table 1). Cell lines containing HS2 near the promoter (20 bp) showed very little variability in the percentage of EGFP-positive cells (99.8 ± 0.03%, n = 7) (Fig. 1 and Table 1). However, the level of expression per copy was quite variable, consistent with data showing that HS2 inefficiently overcomes position effects (5). In all cases, TPA treatment increased EGFP expression. Considering that nearly all cells expressed EGFP in the basal state, it can be inferred that this increase resulted from augmented expression in expressing cells. The TPA induction (induced/uninduced ratio = 1.49 ± 0.16, n = 7) averaged less than the two- to sixfold reported previously for clones containing constructs with luciferase reporter genes. This is likely related to the fact that EGFP has a half-life greater than 24 h (55); preexisting EGFP would have diluted the pool of newly synthesized EGFP, resulting in a lower apparent induction. Nevertheless, the induction was reproducible and observed in all cell lines containing constructs with integrated HS2.
Previously, we reported that the stimulation of LCR enhancer activity by TPA in stable transfection assays requires HS2 (48). As HS3 also has erythroid-cell-specific enhancer activity and could not mediate the response, this finding was inconsistent with a general facilitation of enhancer function. To determine whether the HS2 specificity was also evident at the single-cell level, we analyzed EGFP expression in cell lines containing constructs with HS3 (Fig. 1 and Table 1). The percentage of EGFP-positive cells was very high (99.4% ± 0.39%, n = 6). In contrast to constructs containing HS2, TPA treatment did not influence EGFP expression (induced/uninduced ratio = 0.92 ± 0.03, n = 6), consistent with previous studies of luciferase reporter constructs (48).
The tandem NF-E2 binding sites within HS2 are necessary for the stimulation of HS2 enhancer activity by TPA in stably transfected pools of K562 cells (48). To assess the importance of the NF-E2 sites at the single-cell level, we generated K562 clonal cell lines containing an NF-E2 site deletion mutant of HS2 and tested these cells for inducibility by TPA. The percentage of EGFP-positive cells in these clonal lines was high (93.3% ± 2.9%, n = 9). Five clones failed to respond to TPA treatment, whereas induction was measured in four clones by flow cytometric analysis (induced/uninduced ratio = 1.2 ± 0.05, n = 9). Thus, in agreement with our previous studies with luciferase-based constructs in stably transfected pools of K562 cells, the NF-E2 sites are important to ensure induction at any integration site.
Although HS2 is a strong enhancer when positioned near (2 to 3 kb) an Aγ-globin promoter in stable transfection assays, the enhancer activity is strongly distance dependent (5). However, the stimulation of HS2 enhancer activity by TPA is evident when HS2 was positioned either 2.2 or 7.3 kb from an Aγ-globin promoter, suggesting that the induction is distance independent (48). To assess the impact of HS2-promoter distance and the consequences of TPA treatment at the single-cell level, we generated clonal lines containing HS2 positioned 2.2 or 7.5 kb upstream of an Aγ-globin promoter linked to an EGFP reporter gene. Similar to lines containing HS2 positioned 20 bp from the promoter, the percentage of EGFP-positive cells was very high (89.9% ± 13.1%, n = 7) when HS2 was 2.2 kb from the promoter (Fig. 2 and Table 2). TPA treatment enhanced EGFP expression in all lines (induced/uninduced ratio = 3.43 ± 0.86, n = 7) (Fig. 2 and Table 2). Cell lines containing HS2 positioned 7.5 kb upstream of the promoter showed greater variability in EGFP expression between different clones and between cells of the same clone (59.0% ± 24.2%, n = 4) (Fig. 2 and Table 2). However, TPA treatment increased EGFP expression in all lines (induced/uninduced ratio = 1.86 ± 0.29, n = 4), similar to previous results with luciferase constructs (48). Importantly, several lines [HS2(2.2)γEGFP-6 and -8 and HS2(7.5)γEGFP-1, -3, and -4] contained a single copy of the integrated construct, and these lines did not obviously differ in expression or inducibility from lines containing multiple copies.
FIG. 2.
Distance independence of TPA-induced HS2 enhancer activity. Cultures were treated with 5 nM TPA (shaded) or vehicle (unshaded) for 20 h before each analysis. Histograms show representative flow cytometric analyses of stably transfected K562 clonal cell lines containing HS2(2.2)γEGFP or HS2(7.5)γEGFP constructs. The number of integrated copies of the reporter plasmid is shown. Nonexpressing cells had a relative fluorescence of no more than 1 on this scale.
In the experiments described above, TPA treatment increased EGFP expression in active cells (Fig. 1 and 2 and Tables 1 and 2), and in certain cases [e.g., HS2(7.5)γEGFP-2] (Fig. 2 and Table 2) it increased the percentage of expressing cells. Based on previous observations that stably transfected lines grown without a selecting antibiotic silence integrated constructs (50, 51), we examined the influence of silencing on TPA-induced HS2 enhancer activity.
Graded and stochastic changes in gene expression induced by MAPK.
We routinely generate stably transfected clonal K562 cell lines by cotransfecting a linearized luciferase or EGFP reporter construct with a selection construct containing a thymidine kinase promoter controlling a hygromycin resistance gene. These cell lines are grown in the presence of hygromycin to reduce the probability of gene silencing. In related systems, the expression of integrated genes is silenced, in a time-dependent fashion, upon withdrawal of the selecting antibiotic (50, 51). We have asked whether growing cells without hygromycin results in the silencing of EGFP-expressing clones and whether silencing precludes the TPA induction of HS2 enhancer activity.
The HS2γEGFP-1 cell line was grown for up to 78 days without hygromycin. At various times, EGFP expression was measured by flow cytometric analysis and fluorometry. In the presence of hygromycin, nearly all cells expressed EGFP and the percentage of EGFP-positive cells remained relatively constant with time (Fig. 3A and B). As was observed previously in Fig. 1 and 2, TPA treatment increased EGFP expression in expressing cells. In contrast, the growth of cells without hygromycin resulted in a time-dependent decrease in the percentage of EGFP-positive cells (Fig. 3B) and in EGFP levels in cell lysates (Fig. 3C). Under these conditions, TPA treatment resulted in a higher percentage of EGFP-positive cells (Fig. 3B) and increased EGFP levels in cell lysates (Fig. 3C).
FIG. 3.
Silencing of EGFP expression reveals stochastic and graded responses to TPA treatment. Clone HS2γEGFP-1 was grown for the indicated times with or without hygromycin (hygro) and then treated with 5 nM TPA (shaded) or vehicle (unshaded) for 20 h before each analysis. (A) Flow cytometric analysis was used to measure the percentage of EGFP-positive cells in the viable cell population. (B) The percentage of EGFP-positive cells determined in panel A is expressed as a function of culture time. (C) EGFP was quantitated in cell lysates by fluorometry and normalized by protein concentration and copy number of the integrated reporter construct.
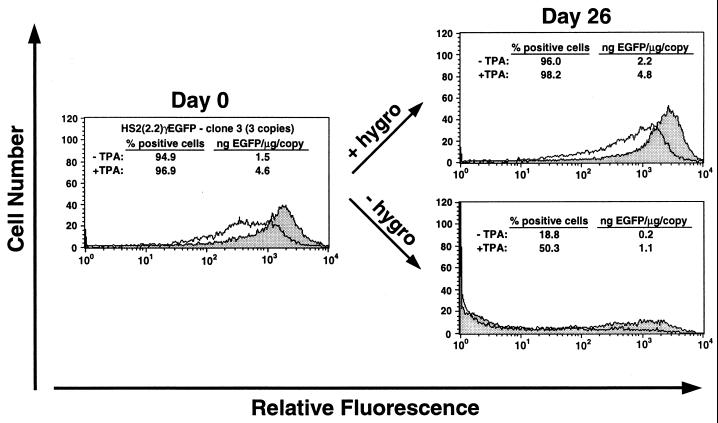
To assess whether the results with clone HS2γEGFP-1 were representative of any HS2-containing clonal line, we also measured EGFP expression in clone HS2(2.2)γEGFP-3 (Fig. 4) the single-copy clones HS2(2.2)γEGFP-6 and -8 (data not shown) and in clone HS2(7.5)γEGFP-4 (Fig. 5) after growth with or without hygromycin for up to 54 days. Nearly all HS2(2.2)γEGFP-3 cells (96.0%) expressed EGFP when grown in media containing hygromycin (Fig. 4); in contrast, the percentage of EGFP-positive cells of this clone decreased to 18.8% after growth for 26 days without hygromycin. TPA treatment increased the percentage of expressing cells from 18.8% to 50.3%. Importantly, cell lines containing single copies of the reporter gene behaved similarly. In the presence of hygromycin, the percentage of expressing cells remained high, and TPA treatment increased the level of expression from active templates. Withdrawal of hygromycin resulted in the silencing of EGFP expression, and TPA treatment increased the percentage of expressing cells up to 2.5-fold (Fig. 5). Thus, the conditions of gene silencing reveal a stochastic component of the mechanism of TPA-induced transactivation besides the graded-response characteristic of cell populations in which most cells express EGFP in the basal state.
FIG. 4.
Silencing of EGFP expression in clone HS2(2.2)γEGFP-3. The clone was grown for 26 days with or without hygromycin (hygro) and then treated with 5 nM TPA (shaded) or vehicle (unshaded) for 20 h before each analysis. The percentage of EGFP-positive cells in the viable cell population was measured by flow cytometric analysis. EGFP was quantitated in cell lysates by fluorometry and normalized by protein concentration and the copy number of the integrated reporter construct.
FIG. 5.
Silencing of EGFP expression in single-copy clone HS2(7.5)γEGFP-4. The clone was grown for 26 or 36 days with or without hygromycin (hygro) and then treated with 5 nM TPA (shaded) or vehicle (unshaded) for 20 h before each analysis. The percentage of EGFP-positive cells in the viable cell population was measured by flow cytometric analysis. EGFP was quantitated in cell lysates by fluorometry and normalized by protein concentration and the copy number of the integrated reporter construct.
To further confirm that TPA treatment can exert both graded and stochastic effects on gene expression, we physically separated EGFP-expressing and non-EGFP-expressing cells (HS2γEGFP-1) by fluorescence-activated cell sorting (FACS) (Fig. 6). We used cells grown for 133 and 135 days without hygromycin so that adequate numbers of expressing and nonexpressing cells could be isolated. Sorted pools of expressing and nonexpressing cells were treated with TPA, and then EGFP expression was analyzed by flow cytometry. TPA treatment resulted in the conversion of nonexpressing cells to expressing cells (15.5-fold increase) and in increased expression by expressing cells (19.5-fold increase). These results support the conclusion from the silencing experiments that TPA treatment elicits both graded and stochastic changes in gene expression.
Direct activation of MAPK with cMEK1 elicits graded and stochastic changes in gene expression.
We routinely activate MAPK indirectly with the PKC agonist TPA. Previously, we showed that the MEK1 inhibitor PD98059 prevented TPA induction of luciferase mRNA from a stably integrated HS2γ-luciferase construct (48), consistent with a role for MAPK in the response. However, PKC activation can have multiple effects on cells, which may be MAPK dependent or independent. Thus, we asked whether direct activation of MAPK via expression of cMEK1 mimics the effect of TPA on EGFP expression.
cMEK1 was transiently expressed in clones γEGFP-1 and -4 and HS2(2.2)γEGFP-3 grown with hygromycin and in HS2γEGFP-1 grown with and without hygromycin (at least 125 days). An expression vector encoding EBFP was cotransfected with the cMEK1 vector so that transfected cells could be identified by flow cytometric analysis. Figure 7 shows the flow cytometric detection of blue and green fluorescence after the transfection of control K562 cells and HS2γEGFP-1 with EBFP or EGFP expression vectors. Expression of the integrated EGFP reporter gene was analyzed in EBFP-positive cells. cMEK1 expression in clones γEGFP-1 and -4 did not influence EGFP expression (Table 3), consistent with the nonresponsiveness of these clones to TPA. In contrast, expression of cMEK1 in the clones containing constructs with HS2 stimulated EGFP expression, similar to expression after TPA treatment. Importantly, cMEK1 induced changes in both the percentage of expressing cells and in the level of expression per copy (Table 3).
FIG. 7.
Expression of cMEK1 elicits graded and stochastic effects on gene expression. K562 cells were transiently transfected with either constitutive EGFP or EBFP expression vectors to establish conditions for discriminating between green and blue fluorescence. Clone HS2γEGFP-1, grown in media containing hygromycin, was transiently transfected with either EBFP and cMEK1 expression vectors or EBFP and pcDNA3. EGFP expression was analyzed in EBFP-positive cells (Table 3). Regions containing green, blue, and green plus blue cells are delineated. The high level of EGFP expression in clone HS2γEGFP-1 resulted in an upward extension of the green signal. Note that the flow cytometric data for cells transfected with cMEK1 and EBFP shows more green and blue cells than data for cells transfected with pcDNA3 and EBFP; this was not seen in all experiments and did not influence the quantitation of green fluorescence in the cell population.
TABLE 3.
Expression of cMEK1 stimulates graded and stochastic effects on gene expression
| Clone | % EGFP-positive cells
|
Mean fluorescence
|
Induced/ uninduced ratio | ||
|---|---|---|---|---|---|
| With cMEK1 | Without cMEK1 | With cMEK1 | Without cMEK1 | ||
| γEGFP-1 | 99.7 | 99.2 | 1,078 | 1,071 | 1.0 |
| γEGFP-4 | 98.0 | 99.2 | 1,374 | 1,620 | 0.8 |
| HS2γEGFP-1 (with hygromycin) | 98.6 | 98.8 | 931 | 651 | 1.4 |
| 99.6 | 99.4 | 1,625 | 1,091 | 1.5 | |
| 99.7 | 91.7 | 1,480 | 832 | 1.8 | |
| HS2γEGFP-1 (without hygromycin) | 38.6 | 29.1 | 70 | 19 | 3.6 |
| 22.2 | 6.8 | 38 | 15 | 2.5 | |
| 22.4 | 8.9 | 33 | 13 | 2.6 | |
| HS2(2.2)γEGFP-3 | 94.1 | 85.4 | 538 | 224 | 2.4 |
DISCUSSION
Regulation of LCR-mediated transactivation by MAPK.
We have described herein the influence of the MAPK signaling pathway on enhancer-mediated gene expression in single, living cells. Rather than finding that MAPK influences gene expression in an exclusively stochastic or graded manner, we have measured both stochastic and graded responses, depending on the expression state of the integrated constructs. The presence of hygromycin in the media maintained gene expression for 78 days by ensuring that most cells in the population expressed EGFP. Without hygromycin, silencing analogous to that reported in related but distinct systems, which are discussed below, occurred. TPA treatment or expression of cMEK1 could partially reverse silencing through a stochastic mechanism. However, in populations of predominantly expressing cells, TPA treatment or cMEK1 expression induced a graded response. As this is the first description of the influence of a signaling pathway involving a kinase cascade on enhancer-mediated gene expression at the single-cell level, it is unclear whether this represents a typical or novel way of controlling gene expression.
Several studies have evaluated the mechanism of enhancer function in single cells (11, 15, 24, 32, 40, 43, 50, 53). Multiple studies have shown that enhancers elicit stochastic effects on gene expression. Weintraub reported that the simian virus 40 enhancer facilitates the assembly of transcription complexes on inactive promoters without stimulating active promoters (53). This stochastic mechanism was also apparent in stably transfected cells containing the glucocorticoid-responsive mouse mammary tumor virus promoter linked to a lacZ reporter gene (24). The dose-dependent glucocorticoid activation of the mouse mammary tumor virus promoter caused the conversion of nonexpressing cells to expressing cells without altering the activity of expressing cells. Evidence for a stochastic mechanism of enhancer function has also come from studies of gene silencing.
A powerful assay to study enhancer function takes advantage of the fact that expression from integrated constructs containing a promoter linked to a drug resistance gene is silenced when the cells are grown without the selecting antibiotic. Martin and colleagues have used this system to analyze the function of HS2 (50, 51), the αHS-40 enhancer from the α-globin locus (43), and the metal-inducible metallothionein promoter (32) in single cells. The enhancers were linked to a β-geo reporter gene which confers β-galactosidase activity and hygromycin resistance. Withdrawal of hygromycin from the medium resulted in the silencing of stably transfected constructs, and enhancers decreased the rate of silencing. Both the silencing and the enhancer activity suppressing silencing were stochastic processes. However, metal induction of the metallothionein promoter suppressed silencing and enhanced transcription in expressing cells, suggesting that the two functions were linked. A stochastic mechanism of HS2 enhancer function was also described by Graubert et al. (15). An extensive analysis of transgenic mice containing human HS2 linked to a fused β-globin promoter/lacZ reporter gene showed that variable transgene expression resulted from different numbers of expressing cells. In contrast, a related study with the human β-globin LCR linked to a β-globin promoter/lacZ transgene found that variegation of transgene expression resulted from a graded mechanism (17). As there were technical differences between the studies, the basis for opposite conclusions is unclear. Nevertheless, it is clear that transactivation mechanisms in multiple systems can involve a stochastic component. Lastly, Bouhassira et al. showed that single HSs of the LCR elicit stochastic effects on promoter activity, whereas multiple HSs increased promoter activity through a graded mechanism (3). The studies described above have assessed basal enhancer function and are therefore fundamentally different from our work in evaluating the influences of signals that modulate enhancer activity.
Molecular basis of graded and stochastic transcriptional responses.
An obvious question is whether distinct molecular mechanisms confer exclusively stochastic or graded responses or combined stochastic and graded responses. The activation of MAPK itself can be stochastic, which can lead to an all-or-none biological response, the maturation of Xenopus oocytes (10). Another important parameter relevant to a stochastic mechanism is the assembly of the enhancer complex, as the formation of a functional enhancer can be an all-or-none process (4). Does MAPK-mediated stimulation of HS2 enhancer activity result from the facilitated assembly of the HS2 complex? If MAPK activation stimulates the assembly of HS2 complexes, only a fraction of the templates would be active in the basal state if promoter activity is absolutely dependent on HS2. When HS2 is 20 bp or 2.2 kb from the promoter, most of the cells in the population are active before induction. Under these conditions, the MAPK response involves the increased efficacy of preexisting HS2 complexes. A simple model to explain this graded response involves the phosphorylation of a component which increases the affinity of a coactivator for an LCR binding protein(s). The importance of the NF-E2 sites of HS2 to ensure a response at all integration sites may relate to the ability of NF-E2 to interact with the coactivator cyclic AMP response element binding protein (CREB) binding protein (CBP and its homolog p300) (8, 13). This may be analogous to the PKA-mediated phosphorylation of CREB, which increases the affinity of CREB for CBP/p300 (14, 35). CBP/p300 is believed to mediate transactivation through the acetylation of histone and nonhistone components.
The acetylation of amino-terminal tails of core histones is an important chromatin modification involved in transcriptional regulation (19). Histone acetylation increases the accessibility of DNA in chromatin to transacting factors (26, 49) and thus would be expected to influence multiple steps of transcription. Such a chromatin modification could influence initiation complex assembly, promoter clearance, and elongation; these would be manifested as both stochastic and graded effects on gene expression. Histone acetyltransferases are recruited to genes through specific protein-protein interactions with transcription factors (6, 47). Considering that CBP/p300 has been implicated as being critical for strong HS2 enhancer activity (13) and that CBP/p300 is phosphorylated directly by MAPKs, which increases its transactivation properties when fused to GAL4 (21, 28), the utilization of CBP/p300 by HS2 may be modulated by MAPK activation. The importance of the NF-E2 sites for MAPK response is consistent with the requirement of these sites for sensitivity to the CBP/p300 inhibitor E1A (13).
Importance of MAPK signaling in erythropoiesis.
The MAPK signaling pathway is one of several signaling pathways activated by erythropoietin (2, 22, 34, 42), the polypeptide hormone that drives erythropoiesis (52). As MAPK is an important mediator of erythropoietin-stimulated erythroid cell proliferation and differentiation in vivo (22), and based on the influence of MAPK on LCR-mediated transactivation, the LCR represents a potential MAPK target. Consistent with this, Nagai et al. (36) used transient-transfection assays to provide evidence for a role of MAPK in regulating NF-E2-mediated transactivation from synthetic promoter constructs. Dimethyl sulfoxide induction of MEL cells resulted in increased NF-E2 transactivation activity, and this response could be mimicked by activation of MAPK. However, it is unclear whether this response resulted from the phosphorylation of NF-E2 or another component by MAPK.
In contrast to the well-established erythropoietin-dependent hemoglobinization of erythroid cells involving MAPK activation (52), the endogenous γ-globin gene in K562 cells is paradoxically repressed by TPA treatment (31). In addition, γ-globin mRNA has a 3′ element that can mediate RNA destabilization upon the treatment of K562 cells with TPA (31). Our previous Northern blot analysis of endogenous γ-globin mRNA in K562 cells did not detect such a destabilization upon TPA treatment (48), which may reflect our use of a fourfold-lower concentration of TPA than in the study by Lumelsky and Forget (31). Based on the erythropoietin-dependent MAPK activation associated with erythropoiesis, it is unclear whether the destabilization of γ-globin mRNA induced by TPA is unique to certain cell lines or stages of erythropoiesis.
How do our results on the regulation of HS2-mediated transactivation by MAPK relate to the intact LCR? MAPK activation also enhances transactivation mediated by the mini-LCR, which contains the four HSs, showing that the signaling effect is not unique for HS2 alone (48). MAPK-induced mini-LCR activity was apparent when the mini-LCR was positioned 5.1 or 7.3 kb from an Aγ promoter but not when it was placed 20 bp from the promoter, in which it had a very high basal activity. This suggests that MAPK partially overcomes a limitation to long-range activation. The identification of the target of MAPK phosphorylation and the coactivator(s) that mediates the response will be crucial for understanding the molecular basis of how MAPK elicits graded and stochastic effects on gene expression from single chromatin templates, which should be relevant to multiple genes regulated by MAPK during cell growth and development.
ACKNOWLEDGMENTS
We thank Moshe Sadofsky for a critical review of the manuscript.
We acknowledge support from the Milwaukee Foundation, the Leukemia Society of America, the Hemophilia Association of New York (grant 133BK04), and the National Institutes of Health (grant DK50107). E.H.B. is a Leukemia Society of America Scholar and a Shaw Scientist.
REFERENCES
- 1.Andrews N C, Erdjument-Bromage H, Davidson M B, Tempst P, Orkin S H. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 2.Bittorf T, Jaster R, Brock J. Rapid activation of the MAP kinase pathway in hematopoietic cells by erythropoietin, granulocyte-macrophage colony-stimulating factor and interleukin-3. Cell Signal. 1994;6:305–311. doi: 10.1016/0898-6568(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 3.Bouhassira E E, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 4.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnick E H, Tze L. Synergism between hypersensitive sites confers long-range gene activation by the human beta-globin locus control region. Proc Natl Acad Sci USA. 1997;94:4566–4571. doi: 10.1073/pnas.94.9.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 7.Bungert J, Dave U, Lim K C, Lieuw K H, Shavit J A, Liu Q, Engel J D. Synergistic regulation of human beta-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 8.Cheng X, Reginato M J, Andrews N C, Lazar M A. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enver T, Li Q, Gale K B, Hu M, May G E, Karlinsey J E, Jimenez G, Papayannopoulou T, Costantini F. Analysis of the developmental and transcriptional potentiation functions of 5′HS2 of the murine beta-globin locus control region in transgenic mice. Dev Biol. 1994;165:574–584. doi: 10.1006/dbio.1994.1277. [DOI] [PubMed] [Google Scholar]
- 10.Ferrell J E, Machleder E M. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 11.Fiering S, Northrop J P, Nolan G P, Mattila P S, Crabtree G R, Herzenberg L A. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 12.Forrester W C, Thompson C, Elder J T, Groudine M. A developmentally stable chromatin structure in the human β-globin locus. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg, E. C., K. Johnson, T. N. Zaboikina, E. A. Mosser, and E. H. Bresnick. Requirement of an E1A-sensitive factor for long-range transactivation by the β-globin locus control region. J. Biol. Chem., in press. [DOI] [PubMed]
- 14.Goldman P S, Tran V K, Goodman R H. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1997;52:103–119. [PubMed] [Google Scholar]
- 15.Graubert T A, Hug B A, Wesselschmidt R, Hsieh C L, Ryan T M, Townes T M, Ley T J. Stochastic, stage-specific mechanisms account for the variegation of a human globin transgene. Nucleic Acids Res. 1998;26:2849–2858. doi: 10.1093/nar/26.12.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 17.Guy L G, Kothary R, Wall L. Position effects in mice carrying a lacZ transgene in cis with the beta-globin LCR can be explained by a graded model. Nucleic Acids Res. 1997;25:4400–4407. doi: 10.1093/nar/25.21.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardison R, Xu J, Jackson J, Mansberger J, Selifonova O, Grotch B, Biesecker J, Petrykowska H, Miller W. Comparative analysis of the locus control region of the rabbit beta-like gene cluster: HS3 increases transient expression of an embryonic epsilon-globin gene. Nucleic Acids Res. 1993;21:1265–1272. doi: 10.1093/nar/21.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imhof A, Wolffe A P. Transcription: gene control by targeted histone acetylation. Curr Biol. 1998;8:R422–R424. doi: 10.1016/s0960-9822(98)70268-4. [DOI] [PubMed] [Google Scholar]
- 20.Jane S M, Ney P A, Vanin E F, Gumucio D L, Nienhuis A W. Identification of a stage selector element in the human gamma-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the beta-promoter. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janknecht R, Nordheim A. MAP kinase-dependent coactivation by Elk-1 and its cofactor CBP. Biochem Biophys Res Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 22.Klingmuller U, Wu H, Hsiao J G, Toker A, Duckworth B C, Cantley L C, Lodish H F. Identification of a novel pathway important for proliferation and differentiation of primary erythroid progenitors. Proc Natl Acad Sci USA. 1997;94:3016–3021. doi: 10.1073/pnas.94.7.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko M S. Induction mechanism of a single gene molecule: stochastic or deterministic. Bioessays. 1992;14:341–346. doi: 10.1002/bies.950140510. [DOI] [PubMed] [Google Scholar]
- 24.Ko M S, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam L T, Bresnick E H. A novel DNA-binding protein HS2NF5 interacts with a functionally important sequence of the human β-globin locus control region. J Biol Chem. 1996;271:32421–32429. doi: 10.1074/jbc.271.50.32421. [DOI] [PubMed] [Google Scholar]
- 26.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, Chang J C, Moi P, Liu W, Kan Y W, Curtin P T. Dissection of the enhancer activity of beta-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc Natl Acad Sci USA. 1992;89:3899–3903. doi: 10.1073/pnas.89.9.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y Z, Chrivia J C, Latchman D S. Nerve growth factor up-regulates the transcription ability of CBP through activation of the p42/p44 (MAPK) cascade. J Biol Chem. 1998;273:32400–32407. doi: 10.1074/jbc.273.49.32400. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd J A, Krakowsky J M, Crable S C, Lingrel J B. Human γ- to β-globin gene switching using a mini construct in transgenic mice. Mol Cell Biol. 1992;12:1561–1567. doi: 10.1128/mcb.12.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu S J, Rowan S, Bani M R, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci USA. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumelsky N L, Forget B G. Negative regulation of globin gene expression during megakaryocytic differentiation of a human erythroleukemic cell line. Mol Cell Biol. 1991;11:3528–3536. doi: 10.1128/mcb.11.7.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magis W, Fiering S, Groudine M, Martin D I. An upstream activator of transcription coordinately increases the level and epigenetic stability of gene expression. Proc Natl Acad Sci USA. 1996;93:13914–13918. doi: 10.1073/pnas.93.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mignotte V, Eleouet J F, Raich N, Romeo P H. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci USA. 1989;86:6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura Y, Miura O, Ihle J N, Aoki N. Activation of the mitogen-activated protein kinase pathway by the erythropoietin receptor. J Biol Chem. 1994;269:29962–29969. [PubMed] [Google Scholar]
- 35.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 36.Nagai T, Igarashi K, Akasaka J, Furuyama K, Fujita H, Hayashi N, Yamamoto M, Sassa S. Regulation of NF-E2 activity in erythroleukemia cell differentiation. J Biol Chem. 1998;273:5358–5365. doi: 10.1074/jbc.273.9.5358. [DOI] [PubMed] [Google Scholar]
- 37.Ney P A, Andrews N C, Jane S M, Safer B, Purucker M E, Weremowicz S, Morton C C, Goff S C, Orkin S H, Nienhuis A W. Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol Cell Biol. 1993;13:5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philipsen S, Talbot D, Fraser P, Grosveld F. The beta-globin dominant control region: hypersensitive site 2. EMBO J. 1990;9:2159–2167. doi: 10.1002/j.1460-2075.1990.tb07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Racke F K, Lewandowska K, Goueli S, Goldfarb A N. Sustained activation of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway is required for megakaryocytic differentiation of K562 cells. J Biol Chem. 1997;272:23366–23370. doi: 10.1074/jbc.272.37.23366. [DOI] [PubMed] [Google Scholar]
- 40.Robertson G, Garrick D, Wilson M, Martin D I, Whitelaw E. Age-dependent silencing of globin transgenes in the mouse. Nucleic Acids Res. 1996;24:1465–1471. doi: 10.1093/nar/24.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan T M, Behringer R R, Martin N C, Townes T M, Palmiter R D, Brinster R L. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human beta-globin gene expression in transgenic mice. Genes Dev. 1989;3:314–323. doi: 10.1101/gad.3.3.314. [DOI] [PubMed] [Google Scholar]
- 42.Sui X, Krantz S B, You M, Zhao Z. Synergistic activation of MAP kinase (ERK1/2) by erythropoietin and stem cell factor is essential for expanded erythropoiesis. Blood. 1998;92:1142–1149. [PubMed] [Google Scholar]
- 43.Sutherland H G E, Martin D I K, Whitelaw E. A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol Cell Biol. 1997;17:1607–1614. doi: 10.1128/mcb.17.3.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbot D, Philipsen S, Fraser P, Grosveld F. Detailed analysis of the site 3 region of the human beta-globin dominant control region. EMBO J. 1990;9:2169–2177. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson C M, Koleske A J, Chao D M, Young R A. The multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 46.Tuan D, London I M. Mapping of DNase I-hypersensitive sites in the upstream DNA of human embryonic epsilon-globin gene in K562 leukemia cells. Proc Natl Acad Sci USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Utley R K, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 48.Versaw W K, Blank V, Andrews N C, Bresnick E H. Mitogen activated protein kinases enhance long-range activation by the beta-globin locus control region. Proc Natl Acad Sci USA. 1998;95:8756–8760. doi: 10.1073/pnas.95.15.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 50.Walters M C, Fiering S, Eidemiller J, Magis W, Groudine M, Martin D I. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 52.Watowich S S, Wu H, Socolovsky M, Klingmuller U, Constantinescu S N, Lodish H F. Cytokine receptor signal transduction and the control of hematopoietic differentiation. Annu Rev Cell Dev Biol. 1996;12:91–128. doi: 10.1146/annurev.cellbio.12.1.91. [DOI] [PubMed] [Google Scholar]
- 53.Weintraub H. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc Natl Acad Sci USA. 1988;85:5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whalen A M, Galasinski S C, Shapiro P S, Nahreini T S, Ahn N G. Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol Cell Biol. 1997;17:1947–1958. doi: 10.1128/mcb.17.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang T T, Sinai P, Green G, Kitts P A, Chen Y T, Lybarger L, Chervenak R, Patterson G H, Piston D W, Kain S R. Improved fluorescence and dual color detection with enhanced blue and green variants of the green fluorescent protein. J Biol Chem. 1998;273:8212–8216. doi: 10.1074/jbc.273.14.8212. [DOI] [PubMed] [Google Scholar]