Abstract
Cancer greatly affects the quality of life of humans worldwide and the number of patients suffering from it is continuously increasing. Over the last century, numerous treatments have been developed to improve the survival of cancer patients but substantial progress still needs to be made before cancer can be truly cured. In recent years, antitumor immunity has become the most debated topic in cancer research and the booming development of immunotherapy has led to a new epoch in cancer therapy. In this review, we describe the relationships between tumors and the immune system, and the rise of immunotherapy. Then, we summarize the characteristics of tumor‐associated immunity and immunotherapeutic strategies with various molecular mechanisms by showing the typical immune molecules whose antibodies are broadly used in the clinic and those that are still under investigation. We also discuss important elements from individual cells to the whole human body, including cellular mutations and modulation, metabolic reprogramming, the microbiome, and the immune contexture. In addition, we also present new observations and technical advancements of both diagnostic and therapeutic methods aimed at cancer immunotherapy. Lastly, we discuss the controversies and challenges that negatively impact patient outcomes.
Keywords: adverse effects, cancer, hyperprogressive disease, immune checkpoints, immunity, immunotherapy, metabolic reprogramming, microbiome, mutation
In this review, we present a clear view of the major factors and regulators associated with cancer immunotherapy and to provide our point of view on the latest available technologies and treatment methods for resolving clinical problems.
Abbreviations
- ACTs
adoptive cell therapies
- AMP
adenosine monophosphate
- APCs
antigen‐presenting cells
- Arg1
arginase‐1
- ASCO
American Society of Clinical Oncology
- ATP
adenosine triphosphate
- BAs
bile acids
- bTMB
tumor mutational burden in blood
- CAR‐Ms
chimeric antigen receptor macrophages
- CAR‐T
chimeric antigen receptor T cells
- Cebpb
CCAAT enhancer binding protein beta
- cfDNA
circulating cell‐free DNA
- cGAS
cyclic guanosine monophosphate‐adenosine monophosphate synthase
- CRC
colorectal cancer
- CTCs
circulating tumor cells
- ctDNA
circulating tumor DNA
- CTLA‐4
cytotoxic T lymphocyte‐associated antigen 4
- CTLs
cytotoxic T lymphocytes
- CYT
cytolytic activity
- DAMPs
damage‐associated molecular patterns
- DCs
dendritic cells
- dMMR
mismatch repair‐deficient
- eATP
adenosine triphosphate in the extracellular space
- EOMES
eomesodermin
- ESMO
European Society for Medical Oncology
- FAO
fatty acid oxidation
- FDA
the Food and Drug Administration
- FMT
fecal microbiota transplantation
- GBM
glioblastoma multiforme
- GEP
gene expression profile
- GM‐CSF
granulocyte‐macrophage colony stimulating factor
- HLA
human leukocyte antigen
- HMAs
hypomethylating agents
- HMGB1
high mobility group box 1
- HNSCC
head and neck squamous cell carcinoma
- HPD
hyperprogressive disease
- HSV‐1
type 1 herpes simplex virus
- HTAN
The Human Tumor Atlas Network
- ICB
immune checkpoint blockade
- ICIs
immune checkpoint inhibitors
- IDO
indoleamine‐pyrrole 2,3‐dioxygenase
- IGF1R
insulin like growth factor 1 receptor
- IFNs
interferons
- IL
interleukin
- irAEs
immune‐related adverse events
- ITIM
immunoreceptor tyrosine‐based inhibitory motif
- JAK
janus kinase
- LAG‐3
lymphocyte activation gene‐3
- LUSC
lung squamous cell carcinoma
- mAb
monoclonal antibody
- MDSCs
myeloid‐derived suppressor cells
- MerTK
c‐mer proto‐oncogene tyrosine kinase
- mIHC
multiplex immunohistochemistry
- MRR
mismatch repair
- MSI
microsatellite instability
- MSI‐H
high microsatellite instability
- MSI‐L
low microsatellite instability
- MSS
microsatellite stability
- mTOR
mechanistic target of rapamycin kinase
- NGS
next‐generation sequencing
- NICHE
Nivolumab, Ipilimumab and COX2‐inhibition in Early Stage Colon Cancer: an Unbiased Approach for Signals of Sensitivity
- NK cells
natural killer cells,
- NKT cells
natural killer T cells
- NSCLC
non‐small cell lung cancer
- nsSNVs
Nonsynonymous single‐nucleotide variants
- OS
overall survival
- OVs
oncolytic viruses
- OXPHOS
oxidative phosphorylation
- p21
cyclin‐dependent kinase inhibitor 1
- PAMPs
pathogen‐associated molecular patterns
- PD‐1
programmed death 1
- PFS
progression‐free survival
- PKA
protein kinase ANF‐kB: nuclear factor‐κB
- PI3K
phosphoinositide 3‐kinase
- pMMR
mismatch repair ‐ proficient
- PTMs
posttranslational modifications
- SH2
src homology 2
- SHP‐1
src homology 2 domain‐containing protein tyrosine phosphatase 1
- SHP‐2
src homology 2 domain‐containing protein tyrosine phosphatase 2
- SIGLEC‐10
sialic acid binding Ig‐like lectin 10
- SIGLEC‐15
sialic acid binding Ig‐like lectin 15TCAA: T cell activity array
- SIRPα
signal regulatory protein α
- ST2
suppression of tumorigenicity 2
- STAT
signal transducer and activator of transcription
- 3D
three‐dimensional
- TAAs
tumor‐associated antigens
- TAMs
tumor‐associated macrophages
- TCGA
The Cancer Genome Atlas
- TCR
T cell receptor
- TCR‐T
TCR‐engineered T cells
- Th cells
CD4+ helper T cells
- Tfh cells
follicular helper T cells
- TIM‐3
T cell immunoglobulin and mucin domain‐containing protein 3
- TIGIT
T cell immunoglobulin and ITIM domain
- TMB
tumor mutational burden
- TMB‐H
high tumor mutational burden
- TILs
tumor‐infiltrating lymphocytes
- TME
tumor microenvironment
- Tregs
regulatory T cells
- T‐VEC
talimogene laherparepvec
- VEGF
vascular endothelial growth factorTLSs: tertiary lymphoid structures
- VISTA
V‐domain immunoglobulin suppressor of T cell activation
1. BACKGROUND
Cancer is one of the biggest medical problems limiting the life span of humans and is also one of the leading challenges to overcome for patients and doctors worldwide. During the last century, scientists have developed diversified treatments for cancer, including surgery, radiotherapy, and chemotherapy. Although great progresses have been achieved via these conventional strategies, many cancer patients are still facing the following two major issues: i) late diagnosis leading to advanced‐stage disease due to nonspecific clinical symptoms or inadequate screening facilities, and ii) low efficacy of conventional treatments due to the rapid spread of cancer and/or drug resistance.
In recent years, immunotherapy has become a major focus among tumor treatments, as it provides insights for further prolonging the overall survival (OS) of patients while improving the patients’ quality of life [1, 2]. The history of immunotherapy can be traced back to 1893 when the American surgeon William Coley found that live or inactivated bacteria could cause remission in sarcomas [3]. However, it was not until the 1980s and 1990s that scientists discovered the interaction between immune cells and melanoma, and conceptualized the idea of cancer immunotherapy[4, 5]. Based on the notable benefits resulting from the clinical utilization of new treatments such as cancer vaccines and immune checkpoint inhibitors (ICIs) [6, 7, 8], Science chose ‘cancer immunotherapy’ as the ‘breakthrough of the year’ in 2013 [9]. Furthermore, the discovery of programmed death 1 (PD‐1) and the targeting of cytotoxic T lymphocyte‐associated antigen 4 (CTLA‐4) in cancer led to the Nobel Prize for Physiology or Medicine honoring the scientists Tasuku Honjo and James Allison in 2018 [10].
Tumors possess immunogenicity characteristics similar to those of other pathogenic agents while also reserving many specific biological reactions. The process of antitumor immunity requires the participation of various immune cells. In most cases, the first step in antitumor immunity is the exposure of tumor‐associated antigens (TAAs) to antigen‐presenting cells (APCs), particularly dendritic cells (DCs) and macrophages [11]. In complex with human leukocyte antigen (HLA) class I and II molecules, TAAs are presented by DCs to CD8+ T cells (cytotoxic T lymphocytes, CTLs) and CD4+ helper T (Th) cells, respectively [12, 13, 14]. After activation, Th1‐ and Th2‐subtype cells are also able to further activate CTLs by secreting cytokines such as interferons (IFNs) and interleukin (IL)‐2 [15]. Then, CTLs and innate immune cells, such as natural killer (NK) cells, natural killer T (NKT) cells and γδ T cells, are recruited to tumor sites to exert antitumor effects [16]. Recently, the significant roles of B cells and follicular helper T (Tfh) cells in this process were reported [17, 18]. One report on CD4+ T cells described antitumor cytotoxicity mediated via cytokines in human bladder cancer [19]. However, the majority of tumor cells exploit immune tolerance instead of being eliminated by immune surveillance [15]. Usually, the condition of the tumor microenvironment (TME) and the infiltration of immune cells determine the survival of malignant cells in tissues and organs [20, 21, 22, 23, 24]. Surprisingly, a large number of immune cells do not play a positive role in the TME, but instead, actively participate in cancer immune evasion, resulting in an extremely complicated relationship between cancer and immune cells [25, 26, 27, 28, 29]. In addition, the heterogeneity of individual bodies or cells, such as the tumor mutational burden (TMB), metabolic status, microbiome and other specific characteristics, also exert crucial influences on the TME and outcomes of immunotherapy.
Based on current literature, in this review, we discuss the various intracellular and extracellular factors, and regulators associated with cancer and immunity. The latest available technologies and treatment methods for resolving clinical problems in cancer immunotherapy are also discussed, including the controversies and limitations in this field.
2. TYPICAL MOLECULES INVOLVED IN ANTITUMOR IMMUNITY AND THEIR CLINICAL APPLICATION
2.1. Immune checkpoints
2.1.1. Known immune checkpoints
CTLA‐4 was the first negative regulator identified to be expressed on T cells. After T cell receptor (TCR) engagement, the expression of CTLA‐4 on the T cell surface is upregulated, CTLA‐4 is trafficked to the immunologic synapse, and expression finally peaks 2 to 3 days after T cell activation [30, 31]. With a better affinity than the T cell costimulatory molecule CD28, CTLA‐4 suppresses T cell function by competitively binding to its ligands CD80 (B7.1) and CD86 (B7.2), which are also the main ligands for CD28 [32, 33]. Therefore, the primary mechanism of CTLA‐4 blockade is the release of CD28‐mediated positive costimulatory signals such as the phosphoinositide 3‐kinase (PI3K) and AKT signaling pathways [34]. In 2011, the Food and Drug Administration (FDA) first approved ipilimumab, a monoclonal antibody (mAb) drug targeting the immune checkpoint molecule CTLA‐4, which signaled the beginning of immune checkpoint blockade (ICB) immunotherapy. However, scientists observed that patients with heterozygous germline mutations in CTLA‐4 (in human) and Ctla‐4 (in animals) knockout mice always exhibited severe immune dysregulation [35, 36], and many oncologists also observed that anti‐CTLA‐4 monoclonal antibodies (mAbs) frequently induced autoimmune reactions in patients [37]. Further study revealed that this could be attributed to the high expression of CTLA‐4 on regulatory T cells (Tregs) [38]. Therefore, there is still a need to investigate how the utility of anti‐CTLA‐4 therapy could be optimized to maximize its efficacy and minimize associated adverse reactions. Many clinical trials evaluating the therapeutic effect of CTLA‐4 blockade alone or in combination with other therapies in many types of cancers, such as advanced renal cell carcinoma, non‐small cell lung cancer (NSCLC), and metastatic melanoma, have shown promising results [39, 40, 41, 42, 43].
PD‐1 is mainly expressed on the surface of activated T cells, and its primary biological function is to form a negative feedback loop to suppress local T cell responses and minimize excessive damage to self‐tissues [44, 45]. The main ligands binding to PD‐1, programmed death‐ligand 1 (PD‐L1; CD274) and PD‐L2 (CD273), are widely expressed on the surface of nonlymphoid cells such as tumor cells and DCs [46]. When engaged by PD‐L1 or PD‐L2, PD‐1 transduces a negative costimulatory signal to attenuate T cell activation through the tyrosine phosphatase src homology 2 (SH2) domain‐containing protein tyrosine phosphatase 2 (SHP‐2), which then dephosphorylates CD28 [47]. One study showed that PD‐1 engagement could suppress glycolysis and promote fatty acid oxidation and lipid catabolism, which are crucial for T cell functions [48]. Hence, when a T cell is activated, the expression of some inhibitory receptors, such as PD‐1, is upregulated to suppress the proliferative capacity and cytotoxic potential, which is defined as T cell exhaustion. Therefore, the primary mechanism of PD‐1 signaling axis blockade is to reverse the exhausting function of CTLs [49]. Numerous clinical trials have shown that anti‐PD‐1 and anti‐PD‐L1 mAbs are associated with better OS and tolerance than conventional therapies in several types of cancers [50, 51, 52, 53, 54, 55]. In 2020, the Nivolumab, Ipilimumab and COX2‐inhibition in Early Stage Colon Cancer: an Unbiased Approach for Signals of Sensitivity (NICHE) study showed that 20/20 (100%) patients with microsatellite‐instability (MSI) tumors and 4/15 (27%) patients with microsatellite‐stable (MSS) tumors had impressive pathological response after receiving ipilimumab combined with nivolumab as neoadjuvant immunotherapy [56]. This was consistent with the results of neoadjuvant ICI treatments in many other cancers [57, 58, 59]. Based on findings of these results, ICIs not only yielded promising clinical outcomes in cancer patients with refractory tumors when used as adjuvant therapy but also showed the potential to be used as a new standard therapy in early‐stage cancers.
Compared with PD‐L2, PD‐L1 is expressed more widely in normal and tumor cells, so there are many studies exploring how to disrupt the PD‐L1‐PD‐1 interaction for cancer therapy [60]. The FDA has already approved some mAbs against PD‐L1, such as atezolizumab, durvalumab and avelumab, for the treatment of many types of cancer. Furthermore, apart from neutralizing PD‐L1 mainly expressed on the tumor cell membrane, many researchers are exploring approaches to reduce the wide expression of PD‐L1. Therefore, it is of great necessity to fully understand the upstream regulation of PD‐L1, which may aid in the development of new strategies to manipulate the expression of PD‐L1 to enhance the response rates of ICI immunotherapy. There are studies that described the PD‐L1 regulation, including genomic alterations, epigenetic regulation, posttranscriptional and posttranslational modifications (PTMs), noncoding RNA‐based regulation and exosomal transport, and offered novel insights on how to reverse ICI resistance [46, 61, 62]. Moreover, other checkpoints may also be regulated by some unique pathways and molecules, which may be future targets to enhance the antitumor response.
2.1.2. Promising immune checkpoints
It is well known that due to metabolic or hypoxic stress, tumor cells release a large amount of adenosine triphosphate (ATP) into the extracellular space (eATP), and the metabolic product adenosine is able to drive tumor progression by suppressing antitumor immune responses while enhancing the proliferation of immunosuppressive cells [63, 64]. Extracellular adenosine activates G protein‐coupled receptors, particularly A2a and A2b receptors, which are mainly expressed on immune cells such as phagocytes, NK cells, and T cells [65], and ultimately suppresses immune responses through the cAMP‐protein kinase A (PKA)‐mediated inhibition of nuclear factor‐κB (NF‐kB) and Janus kinase‐signal transducer and activator of transcription (JAK‐STAT) signaling pathways [66]. Therefore, in recent years, researchers have focused on metabolic pathways of adenosine that can be targeted to suppress tumor development. Once ATP is released into the extracellular space, CD39 hydrolyzes ATP into adenosine monophosphate (AMP), which is further converted into adenosine by CD73 [67]. There are many types of cancers expressing CD73 and CD39, which are both correlated with poor prognosis [68, 69, 70, 71]. In 2013, Allard et al. [72] discovered that CD73 blockade had a synergistic effect with anti‐PD‐1 and anti‐CTLA‐4 mAbs in mouse models. Additionally, in 2019, Li et al. [64] found that CD39 blockade resulted in accumulation of eATP and increased intratumoral T cell numbers through the eATP‐P2×7‐ASC‐NALP3‐inflammasome‐IL‐18 pathway which could overcome anti‐PD1 therapy resistance. The impressive results seen in preclinical experiments have given rise to many clinical trials.
Furthermore, it is also widely known that tumor cells express many “don't eat me” signals to evade clearance by the innate immune system. One typical molecule mediating this phenomenon is CD47, whose expression is upregulated in many types of cancers [73, 74, 75, 76]. Signal regulatory protein α (SIRPα) is a specific ligand that binds to CD47 and is mainly expressed on myeloid cells [77, 78]. Upon engagement with CD47, SIRPα recruits inhibitory molecules such as src homology 2 domain‐containing protein tyrosine phosphatase 1 (SHP‐1) and SHP‐2 through its intracellular immunoreceptor tyrosine‐based inhibitory motif (ITIM) domain and subsequently suppresses phagocytosis by macrophages and prevents macrophages from clearing tumor cells [79, 80]. Furthermore, CD47 blockade seems to produce a synergistic effect with different antitumor treatments in animal models [80, 81, 82]. Additionally, in 2019, Weissman et al. [83] found a completely new “don't eat me” signal, CD24, which could promote immune evasion by interacting with sialic acid‐binding Ig‐like lectin 10 (SIGLEC‐10) and ultimately suppress phagocytosis by macrophages. In 2018, a phase 1b clinical trial showed that the CD47 inhibitor 5F9 combined with rituximab had promising activity in patients with relapsed or refractory lymphoma [84]. In addition, the favorable tolerance of the therapeutic strategy targeting this “don't eat me” signal is being investigated in ongoing clinical trials.
In 2019, Chen et al. [85] found a completely new immune checkpoint, SIGLEC‐15, through a genome‐scale T cell activity array (TCAA). SIGLEC‐15 is mainly expressed on myeloid cells and tumor cells and directly binds to T cells. SIGLEC‐15 inhibits antigen‐specific T cell responses mainly by regulating cell growth and promoting immune evasion but the specific mechanism underlying these effects is still unclear. More recently, in 2020, Yan et al. [86] discovered that selective inhibition of c‐mer proto‐oncogene tyrosine kinase (MerTK) on macrophages could also suppress efferocytosis by macrophages to increase the release of ATP and tumor DNA, which activated the cyclic guanosine monophosphate‐adenosine monophosphate synthase (cGAS) signaling pathway and enhanced type I IFN‐dependent antitumor immune responses. Lu et al. [87] discovered that the depletion of interleukin‐1 receptor‐like 1 (also known as suppression of tumorigenicity 2, ST2), a receptor of IL‐33 that is particularly expressed on tumor‐associated macrophages (TAMs), had a synergistic effect with anti‐PD‐1 treatment in colorectal cancer. Moreover, tumor‐secreted granulocyte‐macrophage colony‐stimulating factor (GM‐CSF) further activated c‐Rel through the NF‐κB signaling pathway in myeloid precursor cells which initiated myeloid‐derived suppressor cell (MDSC) differentiation by enhancing the expression of MDSC signature genes such as CCAAT enhancer‐binding protein beta (Cebpb) and arginase‐1 (Arg1), and eventually inhibited T cell‐mediated antitumor responses and promoted tumor progression [88].
Although clinical success has been demonstrated with anti‐CTLA‐4, anti‐PD‐1, and anti‐PD‐L1 mAbs, a large number of patients still do not derive benefit from them. One reason could be that there are many other immune checkpoints, such as lymphocyte activation gene‐3 (LAG‐3), expressed on the T cell surface, T cell immunoglobulin and mucin domain‐containing protein‐3 (TIM‐3), T cell immunoglobulin and ITIM domain (TIGIT) and V‐domain immunoglobulin suppressor of T cell activation (VISTA) [89, 90, 91, 92]. The variable interactions among these checkpoints are shown in Figure 1. Although there are many preclinical experiments confirming the antitumor responses induced by targeting these novel immune checkpoint molecules and an increasing number of immune checkpoints are being discovered [93, 94, 95], there is still much work that needs to be done to identify additional novel immune checkpoints and develop new therapeutic strategies to target these molecules alone or in combination with other agents to maximize clinical benefits.
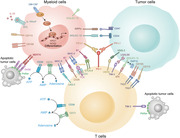
FIGURE 1.
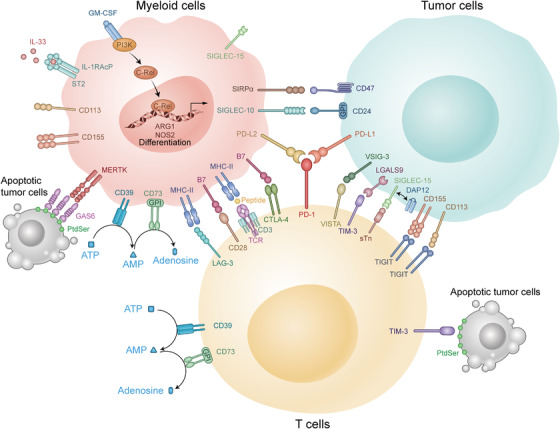
Variable interactions among immune checkpoints in the TME. There are many immune checkpoints in the TME. Some of them are expressed mainly on T cells including PD‐1, CTLA‐4, and LAG‐3, which could suppress the function of CTLs. The others are mainly expressed on myeloid cells, such as c‐Rel and MerTK, which could enhance the inhibitory function of MDSCs to tumor cells. Abbreviations: TME: tumor microenvironment; CTLs: cytotoxic T lymphocytes; IL: interleukin; CD: cluster of differentiation; MHC: major histocompatibility complex; PD‐1: programmed cell death‐1; PD‐L1: programmed cell death‐Ligand 1; PD‐L2: programmed cell death‐Ligand 2; CTLA‐4: cytotoxic T lymphocyte‐associated antigen 4; VISTA: V‐domain immunoglobulin suppressor of T cell activation; TIGIT: T cell immunoglobulin and ITIM domain; TIM‐3: T cell immunoglobulin and mucin domain‐containing protein 3; LAG‐3: lymphocyte activation gene‐3; ATP: adenosine triphosphate; AMP: adenosine monophosphate; GPI: glycosylphosphatidylinositol; SIRPα: signal regulatory protein α; SIGLEC‐15: sialic acid binding Ig‐like lectin 15; GM‐CSF: granulocyte‐macrophage colony stimulating factor; IL‐RAcP: interleukin‐1 receptor accessory protein; ST2: suppression of tumorigenicity 2; MERTK: c‐mer proto‐oncogene tyrosine kinase; GAS6: growth arrest specific 6; PtdSer: phosphatidylserine; TCR: T cell receptor; SIGLEC‐10: sialic acid binding Ig‐like lectin 10; VSIG‐3: V‐set and immunoglobulin domain‐containing protein 3; LGALS9: galectin‐9; DAP12: DNAX‐activation protein 12; sTn: sialyl‐Tn; PI3K: phosphoinositide 3‐kinase; ARG1: arginase‐1; NOS2: nitric oxide synthase 2
2.2. Soluble cytokines
More than 30 years ago, scientists purified the protein IL‐2, and found that it plays an important role in activating T cell growth [96]. It is mainly produced by activated T cells. IL‐2 was approved for the treatment of metastatic renal cell carcinoma by the FDA and has shown promising results as a single agent [97]. However, in addition to stimulating T cell proliferation and memory T cell differentiation, IL‐2 also promotes the generation and function of Tregs [98, 99, 100] which has been associated with many side effects and has limited its further use [97]. Therefore, the most sensible next step was to establishing clinical trials to evaluate the effect of IL‐2 in combination with other drugs, including IFN‐α, cisplatin, dacarbazine, and adoptive cell therapies (ACTs) [101, 102, 103, 104]. However, a phase I/II study showed that the objective response rate of IL‐2 in combination with CTLA‐4 blockade was not better than that of IL‐2 alone [105]. Therefore, whether IL‐2 can enhance the efficacy of immunotherapies may require more experimental and clinical investigations.
In addition to IL‐2, IFN‐γ plays a pivotal role in the development of tumors. The biological functions of IFN‐γ are mediated mainly through the JAK‐STAT signaling pathway which regulates the transcription of many genes [106]. However, the functions of IFN‐γ are contradictory because this cytokine can also suppress tumors in multiple ways. For example, IFN‐γ suppresses tumor cell proliferation through cyclin‐dependent kinase inhibitor 1 (p21) [107] and promotes tumor cell apoptosis through the upregulation of caspase, Fas and Fas ligand expression [108, 109]. IFN‐γ also promotes the recruitment of CTLs through a variety of chemokines such as CXCL9, CXCL10, and CXCL11 [110]. IFN‐γ was approved for the treatment of severe malignant osteopetrosis in 2000 [111]. An IFN‐related gene signature can be used to predict the efficacy of different therapies, including immunotherapy, in many types of malignancies [112, 113, 114]. There are many studies showing that ICI treatment can improve the effects of targeted therapy and radiation in an IFN‐γ‐dependent manner [115, 116]. In addition, tumors produce high levels of IFN‐γ after ICI treatment [117, 118]. On the other hand, IFN‐γ is capable of supporting tumorigenesis. IFN‐γ can induce tumor cells and TAMs to express many inhibitory receptors such as PD‐L1 and PD‐L2 [119]. IFN‐γ can also upregulate the expression of indoleamine‐pyrrole 2,3‐dioxygenase (IDO) in melanoma cells, which recruits Tregs to avoid immune attack [120]. Therefore, IFN‐γ has not shown significant clinical benefit in many types of cancers such as metastatic renal cell carcinoma, breast cancer, and colon cancer [121, 122, 123]. There are some reasons that partially account for this result; for instance, IFN‐γ is rapidly cleared after intravenous administration [124], so it cannot be delivered locally at a sufficient concentration to achieve a therapeutic effect [111]. As such, the functions of IFN‐γ are quite complex, and the efficacy and tolerance of IFN‐γ‐based therapies require further investigation.
There have been studies confirming that some other T cell growth factors, such as IL‐15, IL‐7, IL‐12, and IL‐21, also have antitumor potential through modulation of T cell expansion, survival and functions [125, 126, 127, 128], and may have synergistic effects with ICIs or other immunotherapies [129]. However, many soluble cytokines are negatively correlated with the clinical benefit derived from ICI immunotherapy. For example, many researchers observed that elevated serum IL‐8 levels were associated with increased intratumoral neutrophil levels and a relatively poor prognosis for ICI immunotherapy [130, 131]. TGF‐β also suppresses antitumor immune responses in the process of cancer progression [132]. Thus, the application of these soluble cytokines seems promising, and more in‐depth research must be conducted.
Taken together, among the molecules mentioned above, the applications of PD‐1, PD‐L1, PD‐L2, and CTLA‐4 are the most widespread. Drugs targeting other molecules are still under development. Whether they could be applied in the clinic remains unclear. Therefore, further research should be performed to utilize these targets for immunotherapy. We expect to obtain more effective drugs for patients through different clinical trials.
3. FUNDAMENTAL MODULATORS OF ANTITUMOR IMMUNITY
3.1. Genome, epigenetic regulators, and posttranscriptional modulation
There are a variety of intracellular and extracellular factors that influence the efficiency of cancer immunotherapy. The genome and mutations in the chromatin of tumor cells should be mentioned first because genes determine the phenotype of a single cell and the basic immune response. For example, Asian patients with NSCLC may obtain greater benefit from atezolizumab in terms of OS than Caucasian patients [133], and male patients with melanoma show higher TMB and PD‐L1 expression than female patients, while female patients with lung squamous cell carcinoma (LUSC) have a higher T cell‐inflamed gene expression profile (GEP) and cytolytic activity (CYT) than males [134].
Similar to normal cells, tumor cells contain HLA genes that encode MHC protein expression on the cell surface. Then, antigens specific to tumor cells can be recognized by immune cells mediating an immune response. Thorsson et al. [135] summarized 6 immune subtypes of cancer‐based on over 10,000 samples from 33 different tumor types in The Cancer Genome Atlas (TCGA): TGF‐β Dominant, Wound Healing, Lymphocyte Depleted, IFN‐γ Dominant, Immunologically Quiet, and Inflammatory. In addition, they listed some specific driver mutations associated with leukocyte levels, including mutations in TP53, BRAF, CASP8, NRAS, CTNNB1, and IDH1. The TMB has already been recognized as a favorable prognostic marker for immunotherapy in different kinds of tumors [136, 137, 138]. Nonsynonymous single‐nucleotide variants (nsSNVs) are the main sources of the TMB and serve as immunogenic peptides for immune responses [139]. In 2020, the FDA identified high TMB (TMB‐H) as an indication for PD‐1 inhibitor Keytruda use, regardless of the tumor type. Furthermore, deficiency in mismatch repair (MRR) genes with high microsatellite instability (MSI‐H) is a reason for an increased TMB, especially in colorectal cancer (CRC). In 2017, two ICIs (pembrolizumab and nivolumab) were first approved by the FDA for the treatment of patients with MMR‐deficient (dMMR) CRC [140]. Unfortunately, almost 85% of CRC cases are classified as MMR proficient (pMMR), representing a status of MSS or low microsatellite instability (MSI‐L) [141]. For patients with pMMR CRC, many immunotherapy clinical trials have failed to demonstrate a benefit, probably due to low infiltration of immune cells [142, 143, 144]. Emerging neoantigens and mutations that promote oncogenic outgrowth could be targets of the immune system and limit the spread of malignancies [145]. In contrast, tumor cell death following immune attack or other intrinsic stresses may also result in the release of damage‐associated molecular patterns (DAMPs), such as extracellular ATP, high mobility group box 1 (HMGB1) and adenosine, causing escape from immune surveillance [146, 147].
During the transcription and translation of antigenic proteins, epigenetic and posttranscriptional modulators also exert a significant influence on antigenic protein expression. DNA or histone methylation is the most common mechanism of epigenetic regulation in cells. For instance, excessive methylation of the MMR gene MLH1 represses its expression and leads to the accumulation of cellular mutations in CRC [148, 149]. In addition to cancer cells, immune cells also exhibit a reprogrammed epigenome which is a common factor associated with prognostic value [150]. Chronic viral infections have been reported to induce T cell exhaustion via de novo methylation [151]. For posttranscriptional modulation, ubiquitination is a representative and useful process to manipulate immune responses. Diverse E3 ubiquitin ligases mediate the degradation of some key proteins in various cancer cells and immune cells, affecting their interaction, such as destabilizing Foxp3 to attenuate Treg functions [152, 153]. In addition, other regulatory networks, such as histone acetylation, noncoding RNAs and autophagic degradation, have shown unique roles with therapeutic potential in antitumor immunity [154, 155, 156, 157]. Currently, many epigenetic therapeutics targeting DNA, including methyltransferase inhibitors and histone deacetylase inhibitors, have been widely used in clinical trials, and some of them are being tested in combination with ICIs [158]. Notably, the application of such inhibitors, such as hypomethylating agents (HMAs), may also induce immunosuppression [159] and which warrants caution.
3.2. Metabolism and diet
Metabolism comprises a very large system that maintains the stability of cells and promotes their adaptation to changes of the external environment. Tumor and immune cells have many metabolic similarities and differences. Most cells tend to generate ATP through oxidative phosphorylation (OXPHOS) instead of aerobic glycolysis when oxygen resources are abundant. In contrast, activated glycolysis is the preferred glucose metabolism pathway in malignant cells [160, 161]. For T cells, there is a shift in dominance between glycolysis and OXPHOS during cellular differentiation [162, 163]. Naïve T cells and memory T cells depend more on OXPHOS than glycolysis, while effector T cells metabolize more glucose through glycolysis [164, 165]. Lipid accumulation is another crucial profile in the metabolic reprogramming of the TME [166, 167]. Long‐chain fatty acids accumulate in T cells, restricting their mitochondrial function [168]. As an important process for tumor survival, fatty acid oxidation (FAO) is usually suppressed in effector T cells but is increased in naïve T cells and memory T cells [165, 169]. On the other hand, both OXPHOS and FAO are critical for the function of Tregs [170]. In addition, T cells take up plentiful glutamine, one of the most abundant amino acids in the body, which is similar to the ‘glutamine traps’ in tumors [163]. Therefore, the metabolic process overlaps between cancer cells and immune cells causing a ‘nutrition battle’ between them based on the basic metabolic status of the patient (Figure 2). For example, accelerated glycolysis in tumor cells may cause glucose deprivation in T cells, attenuating their activity [171, 172]. Moreover, there are also competitions for nutrients, such as arginine, and alternative energy sources, such as inosine, among immune cells, making the interplay in the TME much more complicated [173, 174]. These metabolic overlaps and competitions challenge the traditional view that we can kill cancer cells in vivo by editing their metabolic processes, as this approach may also impair the effects of antitumor immunity. Thus, appropriate metabolic regulation of the TME is needed to enhance immunotherapy. Selective inhibition of some metabolically redundant pathways may be a good option for treatment [175].
FIGURE 2.
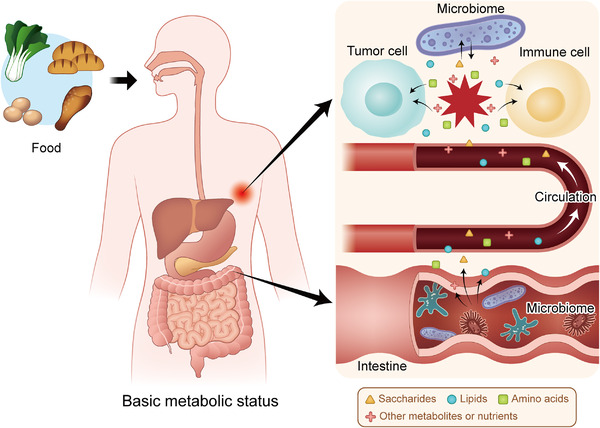
Metabolic interactions in the TME are based on the basic status of the patient. Food is digested and decomposed into metabolites and nutrients based on the basic metabolic status of the human body. The intestinal microbiome participates in the metabolism of these small molecules and influences their levels in the blood. Then, the metabolites and nutrients are sent to the tumor site, forming a competition for nutrients between tumor cells and immune cells in the TME, which is also affected by the local microbiome. Abbreviations: TME: tumor microenvironment
Among different metabolic diseases and disorders, obesity is the most striking global health problem and is correlated with many types of cancers [176]. In addition, obesity not only affects the anabolism or catabolism of fat but also disturbs the transition processes associated with other energy elements. For example, obesity‐related insulin resistance increases blood glucose levels, disturbing macrophage phagocytosis [165, 177, 178]. Obesity also causes reduced glycolysis and increased FAO in effector CD8+ T cells, which negatively impacts their functions [179]. However, there are also some contradictory findings associated with obesity [180]. For instance, obesity leads to elevated levels of pro‐inflammatory adipokine leptin, which enhances the activity of both CD8+ T cells and CD4+ T cells [181, 182, 183]. However, leptin also causes increased PD‐1 expression and CD8+ T cell functional exhaustion, attenuating antitumor immunity [180, 184]. Currently, important questions regarding the association between obesity and immunity remain unanswered, and what is already discovered may only be the beginning.
Dietary adjustment may be a good solution for people with obesity or metabolic syndrome. Previous studies have confirmed that dieting improves the effects of chemotherapy [185, 186, 187]. Moreover, fasting is also linked to elevated adiponectin levels and reduced glucose, insulin, and leptin levels following the downregulation of cAMP‐ PKA and insulin‐like growth factor 1 receptor (IGF1R)‐AKT‐mechanistic target of rapamycin kinase (mTOR)‐S6K signaling [188]. Fasting‐mimicking diets can reduce HO1 levels, leading to an increase in CD8+ tumor‐infiltrating lymphocytes (TILs) and a reduction in Tregs [189]. The interaction between the diet and immune system is extremely complicated as patients may need a larger energy supply during treatment. Therefore, much work is probably needed before doctors can create a proper food management system for patients with cancer.
3.3. Microbiome
Apart from creating direct metabolic alterations, changes to our diet may also influence the microscopic organisms that live within us [190] (Figure 2). The gut microbiome is composed of bacteria, viruses, fungi and some special microbiomes, such as archaea [191]. The influences of these commensals on carcinogenesis, metastasis and the effects of conventional therapeutics are broadly recognized [192, 193, 194, 195], and many studies have also emphasized their impacts on immunotherapy [196, 197, 198]. In 2015, two teams of researchers identified positive correlations between the flora Bacteroides and Bifidobacterium species in the intestinal tract of mice and the effects of ICIs [199, 200]. Subsequently, several intestinal microbe families have been discovered to have specific influences on immunotherapy. The intestinal microbiota is not only indispensable for maintaining the structure and functions of the mucous layer but also interacts with some immune cells, such as Th17 cells, Tregs and DCs. Recently, Zhang et al. [201] found that MDSCs were selectively recruited by Fusobacterium nucleatum, impairing the immune response of the host against CRC. In addition, some gut commensals are deeply involved in the metabolism of certain nutrients or act as facilitators of nutrients such as bile acids (BAs) to modulate inflammatory signaling in immune cells and the proportions of some T cell subtypes [202, 203]. Therefore, dysbiosis is deeply associated with the affected response and toxicity of antitumor immunotherapy [192]. As a consequence, there have been reports of antibiotics negatively impacting the outcomes of patients undergoing immunotherapy [198, 204]. In contrast, fecal microbiota transplantation (FMT) and administration of prebiotics or probiotics have been shown to have great therapeutic potential in animal experiments and clinical trials [200, 205, 206].
Unlike the well‐known intestinal commensals, the microbiome within tumor tissues has not attracted enough attention even though many studies have indicated that some of the bacteria existing inside tumor cells have the potential to affect therapeutic effects [207, 208, 209, 210]. To obtain a clear understanding of intracellular bacteria, Nejman et al. [211] comprehensively analyzed 7 types of tumors (melanoma, bone, brain, lung, breast, ovarian, and pancreatic tumors) compared with adjacent normal tissues. They found that the content of bacteria in these tumors varied from 14.3% for melanoma to 62.7% for breast tumors. Even some solid tumors without direct contact with the external environment, such as glioblastoma multiforme (GBM), possess live microbes in both cancer and immune cells (Figure 2). Additionally, different kinds of cancer exhibited distinct colonies of bacteria associated with specific metabolic signaling pathways. For example, some bacteria with the capacity to degrade cigarette smoke metabolites were more likely to be enriched in the NSCLC cells of smokers than in those of never‐smokers. Moreover, melanoma with a relatively large proportion of Clostridium showed higher sensitivity to immunotherapy, while Gardnerella vaginalis was associated with a poor response to ICIs in patients with melanoma. Thus, the intracellular microbiota plays critical roles in tumor development and the immune response, and more investigations on its biological process are expected to be performed.
3.4. Immune contexture
The immune contexture is a concept similar to the TME that emphasizes the density, distribution and functional interactions among different cells, including but not limited to tumor cells (especially in solid malignancies), immune cells, stromal cells and vessel cells. This concept can be traced back to 2007, when Jérôme Galon, Wolf‐Herman Fridman and Franck Pagès elucidated the adaptive immune microenvironment in CRC [212]. Their team summarized three major types of immune contextures, ‘hot’, ‘altered’ and ‘cold’, based on immunoscores evaluating the infiltration of CD3+ and CD8+ lymphocytes [213]. The ‘altered’ phenotype with an intermediate immunoscore was further divided into two categories (altered‐excluded and altered‐immunosuppressed) to form a more comprehensive classification together with hot and cold phenotypes [213, 214]. Hot or inflamed tumors are infiltrated by abundant TILs, so they show considerable response to ICIs, while cold or noninflamed tumors exhibit the opposite condition with a low TMB and the absence of antigen presentation. Altered‐excluded tumors contain TILs mainly at the margin with a reprogrammed TME while altered‐immunosuppressed tumors allow only a few TILs to infiltrate without further expansion or recruitment but possess a relatively high proportion of immunosuppressive cells and molecules. Moreover, the formation of the immune contexture depends on some important cytokines or chemokines, such as IFN‐γ, perforin and IL‐15 [215, 216], key enzymes or factors, such as vascular endothelial growth factor (VEGF) and IDO1 [217, 218], and diverse pathways, including the NF‐κB, STAT3 and WNT‐β‐catenin pathways [219, 220, 221]. In 2016, Fridman et al. [222] proposed another classification system based on the infiltration and tertiary lymphoid structures (TLSs) of immune cells as well as vascularization, namely, ‘immunogenic’ (with good prognosis), ‘inflammatory’ (with poor prognosis) and ‘immune neglected’ (with intermediate prognosis). Regardless of how the categories were partitioned, the identification of these profiles was meant to predict the baseline antitumor immunity from a complex TME. Therefore, turning cold or altered tumors into hot tumors with a better immune response could be one of the most pivotal treatment strategies in the future.
Cancer and immune cells interact with each other in a dynamic manner. Thus, we should consider the immune contexture as an adaptive and adjustable system. One noteworthy phenomenon is T cell exhaustion, which usually arises from chronic infection or continuous exposure of T cells to antigens [223, 224]. Viral or bacterial infection and local inflammation can lead to ongoing TCR activation along with sustained expression of inhibitory receptors, such as PD‐1, LAG‐3 and TIM‐3, limiting chronic stimulation [225, 226, 227, 228]. This process affects both CD4+ and CD8+ T cells by dramatically reducing their abilities to proliferate and generate cytokines [229, 230]. In contrast, immunosuppressive cells such as Tregs and MDSCs may be increased and aggravate the dysfunction of effector and memory CD8+ T cells by releasing immunosuppressive cytokines [224, 231, 232]. Furthermore, exhausted T cells may express some exclusive molecules, including T‐bet and eomesodermin (EOMES) [233], so identifying exhausted T cells with these markers can help facilitate a more precise understanding of the immune contexture.
There are also other aspects of the immune contexture that should be emphasized. For example, sufficient infiltration of some immune cells, such as TAMs and MDSCs, is tightly linked with immune tolerance and cancer progression [234, 235, 236]. Another important factor is the effect of tissue‐specific immunity during immunotherapy treatment, since different organs exhibit distinct immunological characteristics, the TME of the primary lesion is different from that in metastases and thus influences the efficiency of immunotherapy in tumors in different organs [237]. In summary, we must assess the immune contexture from a dynamic perspective to guide and adjust the immunotherapy regimen.
Take together, the immune response of tumors is a dynamic process that is not only based on the genome, epigenetic regulators and posttranscriptional modulation but also changes with the evolving metabolism and immune contexture. Adjustment of diet and the microbiome in the body could exert a great influence on the TME and the outcomes of cancer immunotherapy.
4. ADVANCED CLINICAL ASSESSMENT AND TREATMENT TECHNOLOGY
4.1. Predicting the clinical outcome of immunotherapy
As mentioned above, the emergence of ICIs has revolutionized the treatment of many types of tumors. However, although clinical benefit has been observed in numerous patients, there are still many patients who do not respond to currently available immunotherapies. Therefore, it is of great importance to distinguish patients who will respond to immunotherapy to maximize the cost‐benefit ratio.
4.1.1. Surgical or tissue biopsy specimens
First, multiplex immunohistochemistry (mIHC) is commonly used to assess tissue pathology [238]. As described above, the infiltration of immune cells into the TME can directly influence antitumor effects. Therefore, TILs have been regarded as a significant index correlated with the outcomes of immunotherapy [239, 240, 241]. Moreover, the most direct approach to distinguish whether a patient can respond to ICI immunotherapy is to detect whether the targeted immune checkpoint molecule is expressed in tumor specimens. Compared with patients with lower PD‐L1 expression levels, those whose tumors were ≥50% PD‐L1+ have been shown to have a higher response rate to anti‐PD‐1 mAbs and better survival [242]. Based on this result, the assessment of PD‐L1 expression in tissue specimens by mIHC was approved by the FDA as a companion diagnostic test for pembrolizumab treatment in advanced NSCLC in October 2015 [243]. The immunohistochemical detection of other immune checkpoint molecules, including CD73, LAG‐3 and TIM‐3, in tissue sections has also been widely investigated [69, 244].
In addition, as sequencing technology gradually matures, there is an increasing number of next‐generation sequencing (NGS)‐based assays to identify critical somatic mutations that can predict response to immunotherapy. For example, in 2017, one meta‐analysis including 27 tumor types or subtypes revealed that the TMB had a significantly positive correlation with the objective response rate in patients treated with anti‐PD‐1 or anti‐PD‐L1 therapy [245]. In 2019, a combined analysis including 47,721 patients found POLE/POLD1 mutations to be independent predictive biomarkers for a positive clinical benefit in many types of cancer [246]. These results suggest that genome sequencing data for tumor specimens can play a critical role in guiding the selection of patients who will respond to immunotherapy.
Finally, although great progress has been made in other traditional sequencing technologies, including DNA sequencing, RNA sequencing and protein sequencing, regarding precision and throughput of diagnosis, these technologies are based on bulk cell populations and provide information that is a steady‐state average of thousands of cells. However, the heterogeneity of a cell population, which is mainly governed by genetic heterogeneity, epigenetic and developmental programs, and extrinsic and spatial factors, may determine many essential complicated biological behaviors such as tumor growth, metastasis, and treatment resistance [247], which traditional sequencing technology can overlook. Single‐cell sequencing can overcome these shortcomings as it examines the sequence information of individual cells and provides a better understanding of the heterogeneity and evolutionary relationships of various cells in a microenvironment. Therefore, it is widely utilized in tumor research [248, 249]. In 2019, Suvà and Tirosh [247] reviewed the common themes, opportunities and challenges, and emerging single‐cell genomic methods of single‐cell sequencing, especially single‐cell RNA sequencing in cancer, and then provided new methods and ideas for the investigation of cancer biology. In 2020, Regev et al. [250] introduced the concept of a dynamic three‐dimensional (3D) atlas of cancer transitions for a diverse set of precancerous lesions and established tumors at single‐cell resolution, called the Human Tumor Atlas Network (HTAN), and is planned to be finished within the next five years. Once completed, this work could greatly improve the understanding, diagnosis, monitoring, drug development, and biomarker discovery of tumors, which could eventually boost developments in precision medicine.
4.1.2. Liquid biopsy
In clinical diagnosis, tumor tissue biopsy is regarded as the standard of choice to investigate whether a patient will respond to immunotherapy [251]. Liquid biopsy is a newly developed technique that can detect the genomic profile of patients with cancer utilizing bodily fluids such as blood, urine, saliva, pleural effusion, and cerebrospinal fluid. Circulating cell‐free DNA (cfDNA), circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), proteins, exosomes and other circulating components can all be analyzed through liquid biopsy [252]. Compared with traditional tissue biopsy, liquid biopsy is less invasive and more cost‐effective, accessible, and replicable [251]. Subsequently, there are also several studies exploiting the application of liquid biopsy in cancer management [253, 254, 255, 256].
For immunotherapy, liquid biopsy also exhibits unique potential to predict the prognosis of patients with cancer. In 2015, Mazel et al. [257] first detected PD‐L1 expression on CTCs in patients with breast tumors and created a new method to monitor PD‐L1 expression in solid tumors. Later, in 2018, Chen et al. [258] discovered that PD‐L1 expression on circulating exosomes before or during anti‐PD‐1 treatment reflected the different states of antitumor immunity. In addition, a proof‐of‐principle study revealed that the detection of ctDNA 8 weeks after anti‐PD‐1 treatment was negatively correlated with survival in patients with NSCLC, uveal melanoma, or MSI CRC [259]. Gandara et al. [260] demonstrated that the TMB in the blood (bTMB) could be a promising biomarker to predict clinical benefit from ICI immunotherapy. The inconsistency in results from previous studies may be partly due to sample instability, immature quantification and isolation methods, low sensitivity, and high cost [252, 261]. liquid biopsy has the potential to exhibit a correlation with the clinical outcomes of immunotherapy and improve clinical diagnosis.
4.2. Advanced treatment technology
With the rapid development of oncoimmunology, immunotherapy has been approved for the treatment of multiple cancers and has achieved surprising clinical outcomes in recent decades. Nevertheless, we still face great challenges in translating experimental observations into clinical practice. First, as mentioned above, not all patients respond to ICIs. Second, the main approach to delivering immunotherapy is through systematic administration, which may result in many immune‐related adverse events (irAEs) because of off‐target effects and the fact that many drugs cannot reach solid tumors at sufficient levels, especially when facing many delivery barriers [262, 263]. Therefore, developing other advanced treatment technologies to improve the safety and efficacy of immunotherapy is urgently needed. Some of these novel technologies are described below.
4.2.1. Cell‐based immunotherapies
T cells are the major type of cells that directly kill tumor cells in the TME. Cell‐based immunotherapy is mainly referred to as chimeric antigen receptor (CAR)‐T cell immunotherapy, in which doctors collect T cells from the patient (autologous) or a healthy person (allogenic), genetically engineer the cells to express an artificial CAR specifically targeting an antigen presented on tumor cells, and then administer the cells in patient to eradicate tumor cells [264]. The first experimental target was CD19, which is mainly expressed on B cell leukemia and lymphomas. The first patient treated with anti‐CD19 CAR‐T cells achieved promising outcome, and this success encouraged a series of studies [265]. The FDA approved anti‐CD19 CAR‐T cells for the treatment of refractory pre‐B cell acute lymphoblastic leukemia and diffuse large B cell lymphoma in 2017, which revolutionized cancer immunotherapy [266]. Moreover, the application of CAR‐T cells is not limited to only CD19 and it has been extended to CD22 and BCMA [267, 268].
However, there are still many challenges for this therapy. First, cytokine release syndrome is frequently observed during treatment [269]. Second, although CAR‐T cell therapy has achieved excellent results in hematological tumors, it has not been as effective in solid tumors [270, 271]. To overcome this limitation, one study tried to use bioengineered polymer scaffolds, which when implanted near or at the resection sites of tumors were able to stimulate and expand tumor‐reactive T cells, producing a curative effect in mouse models of solid tumors [272]. In addition, scientists are also investigating several novel methods such as synthetic APCs and nanoparticle‐functionalized T cells [273, 274]. Nevertheless, more evidence is required to show whether these methods can be further developed and introduced into clinical trials.
Although CAR‐T cell immunotherapy has exhibited impressive results in hematological malignancies, it usually does not provide enough value in solid tumors [275]. In contrast, TCR‐engineered T (TCR‐T) cells may show greater promise in solid tumors because engineered TCRs consist of a glycoprotein alpha‐beta chain heterodimer that can bind to peptides presented by MHC molecules [276]. As a result, TCR‐T cells can recognize both surface proteins and intracellular proteins, targeting many more antigens and penetrating tumors better than CAR‐T cells [277]. Most of the clinical trials on TCR‐T cells are still in phase 1 or phase 2, and the cost and time‐consuming process of TCR cloning limit the broad application of TCR‐T cell therapy [276]. Furthermore, several researchers believe that a combination of CAR‐T and TCR‐T cell immunotherapies will have therapeutic effects on solid tumors in the future because their mechanisms of action and resistance are completely different from those of traditional CAR‐T cell immunotherapy.
Similarly, to overcome the challenge of solid tumors not satisfactorily responding to CAR‐T cell immunotherapy, Gill et al. [278] focused on macrophages, which are very abundant in the TME of many types of solid tumors. The authors produced chimeric antigen receptor macrophages (CAR‐Ms) that secreted proinflammatory cytokines and upregulated antigen presentation, ultimately enhancing their antitumor ability. In humanized mouse models, CAR‐M therapy significantly diminished the TMB and prolonged OS [278]. However, there are still many limitations in CAR‐M therapy. For example, CAR‐Ms could not proliferate in vitro or in vivo, and the biodistribution of CAR‐Ms after systemic administration also largely influenced the response rate. Maybe we could combine CAR‐Ms with some other treatments such as cytokine therapy or nanoparticle‐based approach to overcome these shortcomings.
DCs are the most powerful APCs in the human body and have garnered considerable attention in the development of novel therapeutic cancer vaccines. The manufacture of DC vaccines generally starts with the isolation of autologous DCs, followed by exposing them to an appropriate source of tumor‐associated antigens (TAAs) ex vivo and then, refusing them back into the patient. A series of clinical trials have already been undertaken in patients representing many types of cancers, which demonstrated the safety and therapeutic profile of DC vaccines [279]. However, DC vaccination may have limitations as monotherapy because of the immunosuppressive microenvironment, so the combination of DC vaccination with other therapies, such as ICB, chemotherapy, and radiation therapy, may enhance antigen‐specific antitumor immunity [280].
4.2.2. Oncolytic virus immunotherapies
Scientists have invented various vaccines against viruses to prevent oncogenesis [281], but they are also currently using viruses as immunotherapy. As described above, ‘cold tumors’ usually do not respond well to immunotherapy. To overcome resistance to ICIs, there are many combination therapies under investigation, including approaches turning ‘cold tumors’ into ‘hot tumors’ [282]. Due to their abilities to infect tumor cells and propagate within these cells, oncolytic viruses (OVs) can selectively kill cancer cells, resulting in the release of TAAs, additional DAMPs, viral pathogen‐associated molecular patterns (PAMPs), and other molecules, such as cytokines, to induce an antitumor immune response [283]. The first approved OV therapy was talimogene laherparepvec (T‐VEC), which is based on a type 1 herpes simplex virus (HSV‐1). With the deletion of ICP34.5 and ICP47, T‐VEC can specifically replicate in and lyse tumor cells, ultimately inducing local and global antitumor immunity [284]. A phase III clinical trial investigating the efficacy and tolerance of T‐VEC successfully treated advanced‐stage melanoma [285]. However, this current OV therapy works only in a few select types of tumors. Although there are many novel methods of drug delivery under investigation, the dose‐effect relationship of OVs cannot be predicted easily due to their self‐replication [284]. Moreover, the application of OV therapy in combination with other immunotherapies remains in the experimental stage.
4.2.3. Neoantigen vaccines
In addition to OV administration, increasing the expression of neoantigens is another way to turn ‘cold tumors’ into ‘hot tumors’ which could induce specific antitumor immune responses. In 2017, two successful cases of personalized neoantigen‐based tumor vaccines for the treatment of advanced melanoma were published in Nature at the same time [129, 286]. One report was about an RNA‐based polypeptide vaccine that produced sustained progression‐free survival (PFS) in over 60% (8/13) of patients [286]. The other was about a vaccine that targets up to 20 predicted personal tumor neoantigens which led to no recurrence in nearly 70% (4/6) of patients for 25 months [129]. Although the sample numbers were relatively limited, the results are extremely inspiring. Later, in 2020, one team generated personalized neoantigen vaccines based on NGS with their in‐house pipeline iNeo‐Suite. This was the first pan‐cancer clinical study concentrating on personalized neoantigen vaccine monotherapy that showed promising feasibility, safety, and efficacy [287].
4.2.4. Nanoparticle‐based approaches
To date, a diversity of immunotherapies, including ICIs, ACTs, tumor vaccines, OVs and cytokine therapies, have been established. The effects of these therapies rely largely on their interactions with targeted molecules or cells, so an efficient delivery technology could strikingly improve the effect and safety of these therapies [262]. A typical example of nanoparticle‐based approaches is nanoparticle‐programmed CAR‐T cells [288]. Nanoparticles that encapsulate tumor CAR‐encoding DNA recognize circulating T cells through CD3 molecules in the blood, releasing the DNA into the T cells to achieve sufficient cellular CAR expression to eradicate tumor cells. This method has achieved great success in mouse models of B cell lymphoblastic leukemia, which provided a novel idea for making CAR‐T cell therapy possible in hospitals without the need to engineer T cells ex vivo in a special laboratory. Moreover, another study used nanoparticles to deliver tumoral mRNA in vivo [289]. Researchers have used lipid‐based nanoparticles to package mRNA transcripts encoding tumor neoantigens. Systemic administration into multiple mouse models of established tumors then led to the expression of TAAs by local APCs and subsequently induced durable type I IFN‐dependent antigen‐specific immunity. In summary, nanotechnology can improve the safety and efficacy of immunotherapy by better controlling the dose, location, release, and penetration of immunotherapeutic drugs or cells as well as by optimizing the treatment process. As a consequence, nanotechnology could make tumor immunotherapy more comprehensive and more effective. Despite these achievements, more research and clinical trials on nanoparticles are needed to further confirm their therapeutic effects.
Currently, the application of these new technologies is not as universal as ICIs, and some of them are still under preclinical study. Nevertheless, they could enhance the immunological function of our body through different mechanisms, and they are powerful complements for existing immunotherapies. We believe that if we could utilize them more reasonably on basis of thorough exploration, a new chapter on immunotherapy could be started in the near future.
5. CONTROVERSIES AND LIMITATIONS OF CANCER IMMUNOTHERAPY
5.1. Adverse effects of cancer immunotherapy
For over a century, until the development of anti‐PD therapy (anti‐PD‐1 and anti‐PD‐L1 antibodies), cancer immunotherapy was not popular. Despite great efforts being made in the investigation of cancer immunotherapy, low efficacy and frequent irAEs have limited its clinical application.
Before anti‐PD therapy, only five types of cancer immunotherapy were approved for clinical use, i.e., IFN (for hairy cell leukemia, kidney cancer, and melanoma) [290, 291, 292], IL‐2 (for kidney cancer and melanoma) [293, 294], sipuleucel‐T (a cancer vaccine for prostate cancer) [295], CAR‐T cells (for B cell lymphoma/leukemia) [296], and anti‐CTLA‐4 mAbs (for melanoma) [297]. Most of the above therapies resulted in frequent irAEs and low objective response rates, which dramatically limited their use to only several cancer types. For example, although only a small proportion of melanoma patients respond to anti‐CTLA‐4 mAbs (14% on average), grade 3‐4 AEs are common (27%) [38]. Similarly, IL‐2 can induce a general capillary leak syndrome that severely affects multiple organs [298], resulting in frequent grade 3‐4 AEs (>43%) [38].
In contrast, newly developed anti‐PD therapies have a higher objective response rate with many fewer irAEs (Figure 3). For example, CheckMate 067, a phase III clinical trial comparing nivolumab (an anti‐PD‐1 antibody) and ipilimumab (an anti‐CTLA‐4 antibody) in previously untreated patients with metastatic melanoma, showed that nivolumab produced more than two times the number of antitumor responses than ipilimumab (43.7% versus 19.0%), while the number of grade 3‐4 treatment‐related AEs was lower with nivolumab than with ipilimumab (16.3% versus 27.3%) [299]. A systematic review involving 20,128 patients showed that the severe toxicity (grades 3‐5) rate of anti‐PD therapy, including anti‐PD‐1 and anti‐PD‐L1 antibodies, was only 14.0%. The most common all‐grade AEs were fatigue (18.26%), pruritus (10.61%), and diarrhea (9.47%). The most common grade 3 or higher AEs were fatigue (0.89%), anemia (0.78%), and an increased aspartate aminotransferase level (0.75%). Hypothyroidism (6.07%) and hyperthyroidism (2.82%) were the most frequent all‐grade endocrine irAEs [300]. The safety profile of anti‐PD therapy is also better than that of conventional chemotherapy. A meta‐analysis involving 22 randomized immunotherapy trials, 21 of which were anti‐PD therapy trials, versus standard‐of‐care chemotherapy trials showed that immunotherapy was associated with a significant reduction in the likelihood of developing severe AEs (16.56% versus 41.09%) and AEs of any grade (65.82% versus 85.19%) [301].
FIGURE 3.
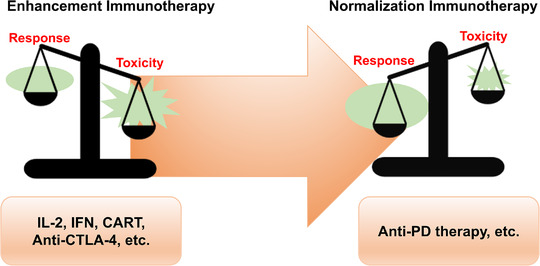
The balance between response and toxicity in anti‐PD therapy compared with other immunotherapy methods. Toxicity outweighs the response in immunotherapy methods such as IL‐2, IFNs, CAR‐T cells, and anti‐CTLA‐4 antibodies, resulting in quite limited indications. For anti‐PD therapy, the response outweighs toxicity considerably, which leads to a rather broad application. Abbreviations: CAR‐T: chimeric antigen receptor T cells; CTLA‐4: cytotoxic T lymphocyte‐associated antigen 4; IFN: interferon; IL‐2: interleukin‐2; PD: programmed death
Despite the improved safety profile, greater attention should be given to the special irAEs of anti‐PD therapy, which can be fatal. IrAEs most commonly affect the gastrointestinal tract, endocrine glands, skin, and liver [302], but severe pneumonitis, myocarditis, colitis, hepatitis and neurological toxicities can also occur and lead to a dismal prognosis [303, 304, 305, 306, 307]. For instance, fulminant ICI‐related myocarditis has been reported to increase in occurrence with the increased use of ICIs [304]. A retrospective analysis of 101 severe myocarditis cases following the administration of ICIs, where the majority of the patients were treated with anti‐PD monotherapy or combination therapy, showed that death occurred in 46% of these patients [308]. Therefore, the precise administration of anti‐PD therapy and early recognition of severe irAEs are of great clinical significance.
Hitherto, most irAEs can be observed and reported only in clinical trials and real‐world studies, and the study of irAEs in preclinical and translational investigations is still limited [309]. Early data suggest that ICIs can cause malfunctions in the self‐tolerance system: the diversification and subcompartmental expansion of lymphocytes activated by ICIs probably gives rise to autoreactive T cells and B cells, causing autoimmune responses [310, 311, 312]. In addition, after the activation of immune cells, normal cells distributed in or near the TME may also be damaged, which can result in the release of self‐antigens and a subsequent autoimmune response [313]. Moreover, the inflammatory factors secreted by activated immune cells may lead to immune‐mediated damage in organs predisposed to autoimmune diseases, as the levels of peripheral cytokines are often elevated posttreatment in patients suffering from irAEs [314, 315]. On the other hand, off‐target effects may explain some types of irAEs. For example, CTLA‐4 is also expressed on hypothalamic and pituitary tissues; thus, the administration of anti‐CTLA‐4 therapy would probably elicit hypophysitis [316]. Therefore, although corticosteroids remain the first‐line management approach for irAEs, the underlying mechanisms may still vary among organs and specific conditions; thus, precise immunohistopathological results are necessary to guide the prescription of appropriate immunomodulatory biological agents, especially when corticosteroids are ineffective [309].
5.2. Cancer immunotherapy strategies: enhancement versus normalization
Among the different types of cancer immunotherapy methods, anti‐PD therapy has achieved the most extensive success. Consequently, researchers have begun to reconsider the strategies underlying the development of cancer immunotherapy.
Based on the understanding of the cancer‐immunity cycle [317], various types of immunotherapies were developed before anti‐PD therapy. As mentioned above, those therapies generally activate the immune system and enhance the antitumor response. However, these general approaches do not aim to correct any deficiencies in the normal antitumor immune response. They simply activate the total immune response and push the immune system to supraphysiological levels to constrain tumors [38]. Therefore, the objective tumor response rates of these strategies are usually low, while the risk of accompanying irAEs is rather high. These general approaches were later categorized together as ‘‘enhancement cancer immunotherapy’’ by the team of Professor Lieping Chen [38].
In contrast, based on the understanding that tumors can develop various ways to escape elimination by the immune system [318], whereby tumor cells inhibit immune cell activity in the TME, which results in a local but not systemic immunosuppression status [319], new approaches, such as anti‐PD therapy, have been developed. These new approaches were categorized and termed “normalization cancer immunotherapy” because they aimed to restore a lost antitumor immune response [38]. These new strategies target a tumor‐induced immune escape mechanism, selectively modulating and resetting immunity in the TME. A clear example is anti‐PD therapy, which blocks the PD‐1/PD‐L1 pathway, a tumor‐induced immune escape mechanism, to restore antitumor immunity in the TME without leading to general activation of the immune system. This approach has been proved to be effective across multiple cancer types and to be associated with a relatively low risk of accompanying irAEs. Therefore, Sanmamed et al. [38] proposed a strategic shift from enhancement to normalization in cancer immunotherapy development and emphasized the importance of identifying the critical immune escape mechanism in each patient.
Targeting SIGLEC‐15, another TME‐specific immune checkpoint target mentioned above, was inspired by this conceptual shift [85]. A monoclonal antibody that blocks SIGLEC‐15‐mediated immune suppression showed encouraging single‐agent antitumor activity in a phase I clinical trial [320]. Therefore, we believe that in addition to PD‐1/PD‐L1, SIGLEC‐15 may be the first in a series of novel targets for normalization cancer immunotherapy, which would progress cancer treatment to another level.
5.3. Hyperprogressive disease: the dark side of anti‐PD therapy
Although anti‐PD therapy can substantially improve the survival of patients with advanced cancers, there is a paradoxical phenomenon whereby some patients exhibit accelerated disease progression when treated with anti‐PD‐1/PD‐L1 antibodies and have subsequent dramatically reduced survival durations, namely, hyperprogressive disease (HPD) [321].
HPD after anti‐PD therapy was first reported at the 2016 European Society for Medical Oncology (ESMO) meetings by Lahmar et al. [322] who observed eight HPD cases (9%) in a cohort of 89 NSCLC patients receiving anti‐PD therapy. Later, Champiat et al. [323] demonstrated that HPD could also occur in other examined cancer types, including melanoma, urothelial carcinoma, colorectal carcinoma, lymphoma, ovarian carcinoma, cholangiocarcinoma, and uveal melanoma. Additionally, in clinical trials comparing anti‐PD therapy with chemotherapy, a crossover of the two survival curves was observed in the initial three to six months, which was considered to be due to disease progression and death occurring at a higher proportion in the anti‐PD therapy arm within the early period of the trials [41, 324–327].
Researchers have expended great efforts to explore the biological mechanism of HPD, which is not just the manifestation of primary resistance to anti‐PD therapy. It is accelerated tumor growth triggered by the administration of anti‐PD‐1/PD‐L1 antibodies through undefined mechanisms. Several hypotheses regarding the HPD mechanism have been proposed, including: i) the expansion of PD‐1‐expressing Tregs due to the loss of contrasuppression, thus, leading to enhanced immunosuppression and tumor boosting; ii) the exhaustion of compensatory T cells due to the compensatory upregulation of the expression of other immune checkpoint molecules; iii) the modulation of tumor‐promoting cells and secretion of immunosuppressive cytokines; iv) aberrant inflammation mediated by Th1 and Th17 cells; and v) the activation of an oncogenic signaling pathway [321]. However, evidence supporting these possible biological mechanisms is mostly preliminary and requires further investigation, which would certainly provide a better understanding of HPD and an approach for the management of this condition.
On the other hand, predictive biomarkers that identify people at high risk of HPD are also urgently needed. Patients with a local recurrence of head and neck squamous cell carcinoma (HNSCC) in the field of irradiation [328], NSCLC patients with two or more metastatic sites [329], and older patients are reported to be at a higher risk of HPD [323]. With regard to genomic biomarkers, MDM2 amplification was the first genomic biomarker reported to be associated with an increased risk of HPD [330]; in addition, EGFR alteration and 11q13 amplification were also reported to be associated with HPD [330, 331]. In summary, additional predictive biomarkers are needed, and the comprehensive integration of these biomarkers may lead to the identification of patients susceptible to HPD.
In addition to the passive clinical management of HPD such as informing patients, paradigm changes in tumor assessment and the choice of subsequent therapy [321], splicing in a limited course of chemotherapy to immunotherapy could potentially provide rapid disease control, making it a practical method to tackle HPD (Figure 4). In CheckMate 9LA, a phase 3 clinical trial evaluating nivolumab plus ipilimumab plus a limited course of chemotherapy versus chemotherapy for first‐line treatment of stage IV or recurrent NSCLC, the crossover of the survival curves was eliminated by adding the limited course of chemotherapy compared with the results of CheckMate 227 [41]. Although Reck et al. [332] did not report HPD data for CheckMate 9LA at the 2020 American Society of Clinical Oncology (ASCO) meetings, the proportion of patients with disease progression during the first evaluation in the experimental arm was shown to be lower than that in the control arm (9% versus 13%). Therefore, the use of a limited course of chemotherapy to address HPD is worthy of further investigation.
FIGURE 4.
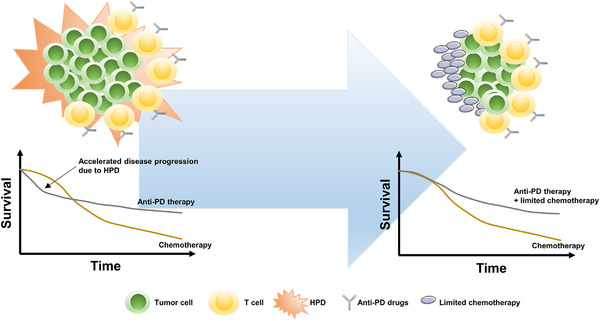
Adding a limited course of chemotherapy to immunotherapy to address HPD. Accelerated disease progression due to HPD significantly compromised the total survival benefit of anti‐PD therapy over chemotherapy, which could be rescued by adding a limited course of chemotherapy to anti‐PD therapy. Abbreviations: HPD: hyperprogressive disease; PD: programmed death
In summary, preferred cancer immunotherapy must have prominent response but less toxicity, such as anti‐PD therapy. More efficient and novel targets will be developed in accordance with the strategy of normalization cancer immunotherapy. Nevertheless, special features such as fatal irAEs and HPD should be carefully managed for better application of cancer immunotherapy.
6. CONCLUSION
The current oncotherapy has being quickly evolved with the rise of antitumor immunotherapy. We are in an epoch with many opportunities for lengthening the lifespan of patients suffering from cancer. With the development of precision medicine, more appropriate individual treatments based on the comprehensive analysis of both tumors and the TME can be offered to patients . However, opportunities are always accompanied by challenges. In 2020, Hegde et al [333] listed the 10 largest challenges in cancer immunotherapy, covering issues ranging from preclinical experiments to therapeutic endpoints, that indicated the existence of insufficient recognition in cancer‐related immunity. Despite all the setbacks in this field, we still hold full confidence in the potential of immunotherapy which can be realized with the use of more advanced medical devices and newly developed experimental methods. Immunity is considered either the weapon or the armor with which humans are born, so the mobilization and usage of this system should be an ideal option for disease control. When treating cancer with new therapeutic methods, we should not only pay attention to the dynamic alterations in the TME but also consider the relationship between local lesions and the basic status of patients. As scientists gradually expand the knowledge of immunotherapy, medical research and cancer treatments will likely advance significantly in the next decade.
DECLARATIONS
AUTHORS’ CONTRIBUTIONS
Conception: all authors. Manuscript writing: Y Wang, M Wang and HXW. Manuscript revision: RHX. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
We thank Professor Feng Wang, Hai‐Yan Zhang and Xia Zhao, Doctor Jia‐Bo Zheng, Zi‐Xian Wang and Jia‐Huan Lu for their valuable scientific advice and discussion. This work was supported by grants from the National Natural Science Foundation of China (81930065).
Wang Y, Wang M, Wu H‐X, Xu R‐H. Advancing to the era of cancer immunotherapy. Cancer Commun. 2021;41:803–829. 10.1002/cac2.12178
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruger S, Ilmer M, Kobold S, Cadilha BL, Endres S, Ormanns S, et al. Advances in cancer immunotherapy 2019 ‐ latest trends. J Exp Clin Cancer Res. 2019;38(1):268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coley WB. The treatment of inoperable sarcoma by bacterial toxins (the mixed toxins of the streptococcus erysipelas and the bacillus prodigiosus). Proc R Soc Med. 1910;3(Surg Sect):1–48. [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SA, Yannelli JR, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, et al. Treatment of patients with metastatic melanoma with autologous tumor‐infiltrating lymphocytes and interleukin 2. J Natl Cancer Inst. 1994;86(15):1159–66. [DOI] [PubMed] [Google Scholar]
- 5.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–7. [DOI] [PubMed] [Google Scholar]
- 6.Yarchoan M, Johnson BA, 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin U, Tureci O. Personalized vaccines for cancer immunotherapy. Science. 2018;359(6382):1355–60. [DOI] [PubMed] [Google Scholar]
- 8.Abril‐Rodriguez G, Ribas A. SnapShot: immune checkpoint inhibitors. Cancer Cell. 2017;31(6):848–e1. [DOI] [PubMed] [Google Scholar]
- 9.Couzin‐Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342(6165):1432–3. [DOI] [PubMed] [Google Scholar]
- 10.Altmann DM. A Nobel Prize‐worthy pursuit: cancer immunology and harnessing immunity to tumour neoantigens. Immunology. 2018;155(3):283–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markiewicz MA, Gajewski TF. The immune system as anti‐tumor sentinel: molecular requirements for an anti‐tumor immune response. Crit Rev Oncog. 1999;10(3):247–60. [PubMed] [Google Scholar]
- 12.Alatrash G, Jakher H, Stafford PD, Mittendorf EA. Cancer immunotherapies, their safety and toxicity. Expert Opin Drug Saf. 2013;12(5):631–45. [DOI] [PubMed] [Google Scholar]
- 13.Parham P, Benjamin RJ, Chen BP, Clayberger C, Ennis PD, Krensky AM, et al. Diversity of class I HLA molecules: functional and evolutionary interactions with T cells. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):529–43. [DOI] [PubMed] [Google Scholar]
- 14.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14(2):135–46. [DOI] [PubMed] [Google Scholar]
- 15.Sambi M, Bagheri L, Szewczuk MR. Current challenges in cancer immunotherapy: multimodal approaches to improve efficacy and patient response rates. J Oncol. 2019;2019:4508794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol. 2015;33:445–74. [DOI] [PubMed] [Google Scholar]
- 17.Hollern DP, Xu N, Thennavan A, Glodowski C, Garcia‐Recio S, Mott KR, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179(5):1191–206 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol. 2016;34:335–68. [DOI] [PubMed] [Google Scholar]
- 19.Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, et al. Intratumoral CD4(+) T cells mediate anti‐tumor cytotoxicity in human bladder cancer. Cell. 2020;181(7):1612–25 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SS, Cheah YK. The interplay between MicroRNAs and cellular components of tumour microenvironment (TME) on non‐small‐cell lung cancer (NSCLC) progression. J Immunol Res. 2019;2019:3046379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drake CG, Stein MN. The immunobiology of kidney cancer. J Clin Oncol. 2018:JCO2018792648. [DOI] [PubMed] [Google Scholar]
- 23.Gajewski TF, Corrales L, Williams J, Horton B, Sivan A, Spranger S. Cancer immunotherapy targets based on understanding the T cell‐inflamed versus non‐T cell‐inflamed tumor microenvironment. Adv Exp Med Biol. 2017;1036:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan S, Zhan Y, Chen X, Wu B, Liu B. Bladder cancer exhibiting high immune infiltration shows the lowest response rate to immune checkpoint inhibitors. Front Oncol. 2019;9:1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015;125(9):3335–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour‐associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP‐dependent MDSC infiltration impairs tumor progression. Cancer Discov. 2016;6(1):80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno Ayala MA, Li Z, DuPage M. Treg programming and therapeutic reprogramming in cancer. Immunology. 2019;157(3):198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatta W, Gotoda T, Oyama T, Kawata N, Takahashi A, Yoshifuku Y, et al. A scoring system to stratify curability after endoscopic submucosal dissection for early gastric cancer: “eCura system”. Am J Gastroenterol. 2017;112(6):874–81. [DOI] [PubMed] [Google Scholar]
- 30.Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, et al. CTLA‐4 can function as a negative regulator of T cell activation. Immunity. 1994;1(5):405–13. [DOI] [PubMed] [Google Scholar]
- 31.Egen JG, Allison JP. Cytotoxic T lymphocyte antigen‐4 accumulation in the immunological synapse is regulated by TCR signal strength. Immunity. 2002;16(1):23–35. [DOI] [PubMed] [Google Scholar]
- 32.van der Merwe PA, Bodian DL, Daenke S, Linsley P, Davis SJ. CD80 (B7‐1) binds both CD28 and CTLA‐4 with a low affinity and very fast kinetics. J Exp Med. 1997;185(3):393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linsley PS, Greene JL, Brady W, Bajorath J, Ledbetter JA, Peach R. Human B7‐1 (CD80) and B7‐2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA‐4 receptors. Immunity. 1994;1(9):793–801. [DOI] [PubMed] [Google Scholar]
- 34.Fong L, Small EJ. Anti‐cytotoxic T‐lymphocyte antigen‐4 antibody: the first in an emerging class of immunomodulatory antibodies for cancer treatment. J Clin Oncol. 2008;26(32):5275–83. [DOI] [PubMed] [Google Scholar]
- 35.Kuehn HS, Ouyang W, Lo B, Deenick EK, Niemela JE, Avery DT, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014;345(6204):1623–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla‐4. Science. 1995;270(5238):985–8. [DOI] [PubMed] [Google Scholar]
- 37.Weber J, Mandala M, Del Vecchio M, Gogas HJ, Arance AM, Cowey CL, et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N Engl J Med. 2017;377(19):1824–35. [DOI] [PubMed] [Google Scholar]
- 38.Sanmamed MF, Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell. 2018;175(2):313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motzer RJ, Tannir NM, McDermott DF, Aren Frontera O, Melichar B, Choueiri TK, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal‐cell carcinoma. N Engl J Med. 2018;378(14):1277–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motzer RJ, Rini BI, McDermott DF, Aren Frontera O, Hammers HJ, Carducci MA, et al. Nivolumab plus ipilimumab versus sunitinib in first‐line treatment for advanced renal cell carcinoma: extended follow‐up of efficacy and safety results from a randomised, controlled, phase 3 trial. Lancet Oncol. 2019;20(10):1370–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier‐Valette C, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tawbi HA, Forsyth PA, Algazi A, Hamid O, Hodi FS, Moschos SJ, et al. Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N Engl J Med. 2018;379(8):722–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmer L, Livingstone E, Hassel JC, Fluck M, Eigentler T, Loquai C, et al. Adjuvant nivolumab plus ipilimumab or nivolumab monotherapy versus placebo in patients with resected stage IV melanoma with no evidence of disease (IMMUNED): a randomised, double‐blind, placebo‐controlled, phase 2 trial. Lancet. 2020;395(10236):1558–68. [DOI] [PubMed] [Google Scholar]
- 44.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD‐1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong H, Zhu G, Tamada K, Chen L. B7‐H1, a third member of the B7 family, co‐stimulates T‐cell proliferation and interleukin‐10 secretion. Nat Med. 1999;5(12):1365–9. [DOI] [PubMed] [Google Scholar]
- 46.Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms controlling PD‐L1 expression in cancer. Mol Cell. 2019;76(3):359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokosuka T, Takamatsu M, Kobayashi‐Imanishi W, Hashimoto‐Tane A, Azuma M, Saito T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J Exp Med. 2012;209(6):1201–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, et al. PD‐1 alters T‐cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun. 2015;6:6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20(3):326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antonia SJ, Borghaei H, Ramalingam SS, Horn L, De Castro Carpeno J, Pluzanski A, et al. Four‐year survival with nivolumab in patients with previously treated advanced non‐small‐cell lung cancer: a pooled analysis. Lancet Oncol. 2019;20(10):1395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vokes EE, Ready N, Felip E, Horn L, Burgio MA, Antonia SJ, et al. Nivolumab versus docetaxel in previously treated advanced non‐small‐cell lung cancer (CheckMate 017 and CheckMate 057): 3‐year update and outcomes in patients with liver metastases. Ann Oncol. 2018;29(4):959–65. [DOI] [PubMed] [Google Scholar]
- 52.Wan X, Zhang Y, Tan C, Zeng X, Peng L. First‐line nivolumab plus ipilimumab vs sunitinib for metastatic renal cell carcinoma: a cost‐effectiveness analysis. JAMA Oncol. 2019;5(4):491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eng C, Kim TW, Bendell J, Argiles G, Tebbutt NC, Di Bartolomeo M, et al. Atezolizumab with or without cobimetinib versus regorafenib in previously treated metastatic colorectal cancer (IMblaze370): a multicentre, open‐label, phase 3, randomised, controlled trial. Lancet Oncol. 2019;20(6):849–61. [DOI] [PubMed] [Google Scholar]
- 54.Mansfield AS, Kazarnowicz A, Karaseva N, Sanchez A, De Boer R, Andric Z, et al. Safety and patient‐reported outcomes of atezolizumab, carboplatin, and etoposide in extensive‐stage small‐cell lung cancer (IMpower133): a randomized phase I/III trial. Ann Oncol. 2020;31(2):310–7. [DOI] [PubMed] [Google Scholar]
- 55.Cohen EEW, Soulieres D, Le Tourneau C, Dinis J, Licitra L, Ahn MJ, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head‐and‐neck squamous cell carcinoma (KEYNOTE‐040): a randomised, open‐label, phase 3 study. Lancet. 2019;393(10167):156–67. [DOI] [PubMed] [Google Scholar]
- 56.Chalabi M, Fanchi LF, Dijkstra KK, Van den Berg JG, Aalbers AG, Sikorska K, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR‐proficient and MMR‐deficient early‐stage colon cancers. Nat Med. 2020;26(4):566–76. [DOI] [PubMed] [Google Scholar]
- 57.Amaria RN, Reddy SM, Tawbi HA, Davies MA, Ross MI, Glitza IC, et al. Neoadjuvant immune checkpoint blockade in high‐risk resectable melanoma. Nat Med. 2018;24(11):1649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forde PM, Chaft JE, Smith KN, Anagnostou V, Cottrell TR, Hellmann MD, et al. Neoadjuvant PD‐1 blockade in resectable lung cancer. N Engl J Med. 2018;378(21):1976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blank CU, Rozeman EA, Fanchi LF, Sikorska K, van de Wiel B, Kvistborg P, et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat Med. 2018;24(11):1655–61. [DOI] [PubMed] [Google Scholar]
- 60.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD‐1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Dang F, Ren J, Wei W. Biochemical aspects of PD‐L1 regulation in cancer immunotherapy. Trends Biochem Sci. 2018;43(12):1014–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD‐L1 checkpoint. Immunity. 2018;48(3):434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young A, Mittal D, Stagg J, Smyth MJ. Targeting cancer‐derived adenosine: new therapeutic approaches. Cancer Discov. 2014;4(8):879–88. [DOI] [PubMed] [Google Scholar]
- 64.Li XY, Moesta AK, Xiao C, Nakamura K, Casey M, Zhang H, et al. Targeting CD39 in cancer reveals an extracellular ATP‐ and inflammasome‐driven tumor immunity. Cancer Discov. 2019;9(12):1754–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatfield SM, Sitkovsky M. A2A adenosine receptor antagonists to weaken the hypoxia‐HIF‐1alpha driven immunosuppression and improve immunotherapies of cancer. Curr Opin Pharmacol. 2016;29:90–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palmer TM, Trevethick MA. Suppression of inflammatory and immune responses by the A(2A) adenosine receptor: an introduction. Br J Pharmacol. 2008;153(Suppl 1):S27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yegutkin GG. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit Rev Biochem Mol Biol. 2014;49(6):473–97. [DOI] [PubMed] [Google Scholar]
- 68.Ma XL, Shen MN, Hu B, Wang BL, Yang WJ, Lv LH, et al. CD73 promotes hepatocellular carcinoma progression and metastasis via activating PI3K/AKT signaling by inducing Rap1‐mediated membrane localization of P110beta and predicts poor prognosis. J Hematol Oncol. 2019;12(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buisseret L, Pommey S, Allard B, Garaud S, Bergeron M, Cousineau I, et al. Clinical significance of CD73 in triple‐negative breast cancer: multiplex analysis of a phase III clinical trial. Ann Oncol. 2018;29(4):1056–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van de Veen W, Globinska A, Jansen K, Straumann A, Kubo T, Verschoor D, et al. A novel proangiogenic B cell subset is increased in cancer and chronic inflammation. Sci Adv. 2020;6(20):eaaz3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagate Y, Ezoe S, Fujita J, Okuzakis D, Motooka D, Ishibashi T, et al. Ectonucleotidase CD39 is highly expressed on ATLL cells and is responsible for their immunosuppressive function. Leukemia. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Allard B, Pommey S, Smyth MJ, Stagg J. Targeting CD73 enhances the antitumor activity of anti‐PD‐1 and anti‐CTLA‐4 mAbs. Clin Cancer Res. 2013;19(20):5626–35. [DOI] [PubMed] [Google Scholar]
- 73.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, GibbsKD, Jr., et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weiskopf K, Jahchan NS, Schnorr PJ, Cristea S, Ring AM, Maute RL, et al. CD47‐blocking immunotherapies stimulate macrophage‐mediated destruction of small‐cell lung cancer. J Clin Invest. 2016;126(7):2610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Betancur PA, Abraham BJ, Yiu YY, Willingham SB, Khameneh F, Zarnegar M, et al. A CD47‐associated super‐enhancer links pro‐inflammatory signalling to CD47 upregulation in breast cancer. Nat Commun. 2017;8:14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138(2):271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown EJ, Frazier WA. Integrin‐associated protein (CD47) and its ligands. Trends Cell Biol. 2001;11(3):130–5. [DOI] [PubMed] [Google Scholar]
- 78.Adams S, van der Laan LJ, Vernon‐Wilson E, Renardel de Lavalette C, Dopp EA, Dijkstra CD, et al. Signal‐regulatory protein is selectively expressed by myeloid and neuronal cells. J Immunol. 1998;161(4):1853–9. [PubMed] [Google Scholar]
- 79.Matozaki T, Murata Y, Okazawa H, Ohnishi H. Functions and molecular mechanisms of the CD47‐SIRPalpha signalling pathway. Trends Cell Biol. 2009;19(2):72–80. [DOI] [PubMed] [Google Scholar]
- 80.Weiskopf K, Ring AM, Ho CC, Volkmer JP, Levin AM, Volkmer AK, et al. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science. 2013;341(6141):88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao XW, van Beek EM, Schornagel K, Van der Maaden H, Van Houdt M, Otten MA, et al. CD47‐signal regulatory protein‐alpha (SIRPalpha) interactions form a barrier for antibody‐mediated tumor cell destruction. Proc Natl Acad Sci U S A. 2011;108(45):18342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chao MP, Alizadeh AA, Tang C, Myklebust JH, Varghese B, Gill S, et al. Anti‐CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non‐Hodgkin lymphoma. Cell. 2010;142(5):699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 signalling through macrophage Siglec‐10 is a target for cancer immunotherapy. Nature. 2019;572(7769):392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Advani R, Flinn I, Popplewell L, Forero A, Bartlett NL, Ghosh N, et al. CD47 blockade by Hu5F9‐G4 and rituximab in non‐Hodgkin's lymphoma. N Engl J Med. 2018;379(18):1711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M, et al. Siglec‐15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat Med. 2019;25(4):656–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou Y, Fei M, Zhang G, Liang WC, Lin W, Wu Y, et al. Blockade of the phagocytic receptor MerTK on tumor‐associated macrophages enhances P2X7R‐dependent STING activation by tumor‐derived cGAMP. Immunity. 2020;52(2):357–73 e9. [DOI] [PubMed] [Google Scholar]
- 87.Van der Jeught K, Sun Y, Fang Y, Zhou Z, Jiang H, Yu T, et al. ST2 as checkpoint target for colorectal cancer immunotherapy. JCI Insight. 2020;5(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li T, Li X, Zamani A, Wang W, Lee C‐N, Li M, et al. c‐Rel is a myeloid checkpoint for cancer immunotherapy. Nature Cancer. 2020;1(5):507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Triebel F, Jitsukawa S, Baixeras E, Roman‐Roman S, Genevee C, Viegas‐Pequignot E, et al. LAG‐3, a novel lymphocyte activation gene closely related to CD4. J Exp Med. 1990;171(5):1393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, et al. Th1‐specific cell surface protein Tim‐3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415(6871):536–41. [DOI] [PubMed] [Google Scholar]
- 91.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10(1):48–57. [DOI] [PubMed] [Google Scholar]
- 92.Wang L, Rubinstein R, Lines JL, Wasiuk A, Ahonen C, Guo Y, et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208(3):577–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kraman M, Faroudi M, Allen NL, Kmiecik K, Gliddon D, Seal C, et al. FS118, a Bispecific antibody targeting LAG‐3 and PD‐L1, enhances T‐cell activation resulting in potent antitumor activity. Clin Cancer Res. 2020;26(13):3333–44. [DOI] [PubMed] [Google Scholar]
- 94.He X, Feng Z, Ma J, Ling S, Cao Y, Gurung B, et al. Bispecific and split CAR T cells targeting CD13 and TIM3 eradicate acute myeloid leukemia. Blood. 2020;135(10):713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chiu DK, Yuen VW, Cheu JW, Wei LL, Ting V, Fehlings M, et al. Hepatocellular carcinoma cells up‐regulate PVRL1, stabilizing PVR and inhibiting the cytotoxic T‐cell response via TIGIT to mediate tumor resistance to PD1 inhibitors in mice. Gastroenterology. 2020;159(2):609–23. [DOI] [PubMed] [Google Scholar]
- 96.Taniguchi T, Matsui H, Fujita T, Takaoka C, Kashima N, Yoshimoto R, et al. Structure and expression of a cloned cDNA for human interleukin‐2. Nature. 1983;302(5906):305–10. [DOI] [PubMed] [Google Scholar]
- 97.Jiang T, Zhou C, Ren S. Role of IL‐2 in cancer immunotherapy. Oncoimmunology. 2016;5(6):e1163462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self‐tolerance maintained by activated T cells expressing IL‐2 receptor alpha‐chains (CD25). Breakdown of a single mechanism of self‐tolerance causes various autoimmune diseases. J Immunol. 1995;155(3):1151–64. [PubMed] [Google Scholar]
- 99.Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, et al. An essential role for the IL‐2 receptor in Treg cell function. Nat Immunol. 2016;17(11):1322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL‐2Rbeta‐deficient mice. Implications for the nonredundant function of IL‐2. Immunity. 2002;17(2):167–78. [DOI] [PubMed] [Google Scholar]
- 101.Atkins MB, Sparano J, Fisher RI, Weiss GR, Margolin KA, Fink KI, et al. Randomized phase II trial of high‐dose interleukin‐2 either alone or in combination with interferon alfa‐2b in advanced renal cell carcinoma. J Clin Oncol. 1993;11(4):661–70. [DOI] [PubMed] [Google Scholar]
- 102.Dutcher JP, Fisher RI, Weiss G, Aronson F, Margolin K, Louie A, et al. Outpatient subcutaneous interleukin‐2 and interferon‐alpha for metastatic renal cell cancer: five‐year follow‐up of the Cytokine Working Group Study. Cancer J Sci Am. 1997;3(3):157–62. [PubMed] [Google Scholar]
- 103.Atkins MB. Cytokine‐based therapy and biochemotherapy for advanced melanoma. Clin Cancer Res. 2006;12(7 Pt 2):2353s–8s. [DOI] [PubMed] [Google Scholar]
- 104.Sockolosky JT, Trotta E, Parisi G, Picton L, Su LL, Le AC, et al. Selective targeting of engineered T cells using orthogonal IL‐2 cytokine‐receptor complexes. Science. 2018;359(6379):1037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maker AV, Phan GQ, Attia P, Yang JC, Sherry RM, Topalian SL, et al. Tumor regression and autoimmunity in patients treated with cytotoxic T lymphocyte‐associated antigen 4 blockade and interleukin 2: a phase I/II study. Ann Surg Oncol. 2005;12(12):1005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Majoros A, Platanitis E, Kernbauer‐Holzl E, Rosebrock F, Muller M, Decker T. Canonical and non‐canonical aspects of JAK‐STAT signaling: lessons from interferons for cytokine responses. Front Immunol. 2017;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin‐dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272(5262):719–22. [DOI] [PubMed] [Google Scholar]
- 108.Fulda S, Debatin KM. IFNgamma sensitizes for apoptosis by upregulating caspase‐8 expression through the Stat1 pathway. Oncogene. 2002;21(15):2295–308. [DOI] [PubMed] [Google Scholar]
- 109.Xu X, Fu XY, Plate J, Chong AS. IFN‐gamma induces cell growth inhibition by Fas‐mediated apoptosis: requirement of STAT1 protein for up‐regulation of Fas and FasL expression. Cancer Res. 1998;58(13):2832–7. [PubMed] [Google Scholar]
- 110.Melero I, Rouzaut A, Motz GT, Coukos G. T‐cell and NK‐cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4(5):522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ. Interferon‐gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol. 2018;9:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, et al. An interferon‐related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105(47):18490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN‐gamma‐related mRNA profile predicts clinical response to PD‐1 blockade. J Clin Invest. 2017;127(8):2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mo X, Zhang H, Preston S, Martin K, Zhou B, Vadalia N, et al. Interferon‐gamma signaling in melanocytes and melanoma cells regulates expression of CTLA‐4. Cancer Res. 2018;78(2):436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stagg J, Loi S, Divisekera U, Ngiow SF, Duret H, Yagita H, et al. Anti‐ErbB‐2 mAb therapy requires type I and II interferons and synergizes with anti‐PD‐1 or anti‐CD137 mAb therapy. Proc Natl Acad Sci U S A. 2011;108(17):7142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Twyman‐Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, et al. Radiation and dual checkpoint blockade activate non‐redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liakou CI, Kamat A, Tang DN, Chen H, Sun J, Troncoso P, et al. CTLA‐4 blockade increases IFNgamma‐producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Natl Acad Sci U S A. 2008;105(39):14987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shi LZ, Fu T, Guan B, Chen J, Blando JM, Allison JP, et al. Interdependent IL‐7 and IFN‐gamma signalling in T‐cell controls tumour eradication by combined alpha‐CTLA‐4+alpha‐PD‐1 therapy. Nat Commun. 2016;7:12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garcia‐Diaz A, Shin DS, Moreno BH, Saco J, Escuin‐Ordinas H, Rodriguez GA, et al. Interferon receptor signaling pathways regulating PD‐L1 and PD‐L2 expression. Cell Rep. 2017;19(6):1189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brody JR, Costantino CL, Berger AC, Sato T, Lisanti MP, Yeo CJ, et al. Expression of indoleamine 2,3‐dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle. 2009;8(12):1930–4. [DOI] [PubMed] [Google Scholar]
- 121.Gleave ME, Elhilali M, Fradet Y, Davis I, Venner P, Saad F, et al. Interferon gamma‐1b compared with placebo in metastatic renal‐cell carcinoma. Canadian Urologic Oncology Group. N Engl J Med. 1998;338(18):1265–71. [DOI] [PubMed] [Google Scholar]
- 122.Vahdat LT, Cohen DJ, Zipin D, Lo KS, Donovan D, Savage D, et al. Randomized trial of low‐dose interleukin‐2 vs cyclosporine A and interferon‐gamma after high‐dose chemotherapy with peripheral blood progenitor support in women with high‐risk primary breast cancer. Bone Marrow Transplant. 2007;40(3):267–72. [DOI] [PubMed] [Google Scholar]
- 123.Wiesenfeld M, O'Connell MJ, Wieand HS, Gonchoroff NJ, Donohue JH, FitzgibbonsRJ, Jr., et al. Controlled clinical trial of interferon‐gamma as postoperative surgical adjuvant therapy for colon cancer. J Clin Oncol. 1995;13(9):2324–9. [DOI] [PubMed] [Google Scholar]
- 124.Kurzrock R, Rosenblum MG, Sherwin SA, Rios A, Talpaz M, Quesada JR, et al. Pharmacokinetics, single‐dose tolerance, and biological activity of recombinant gamma‐interferon in cancer patients. Cancer Res. 1985;45(6):2866–72. [PubMed] [Google Scholar]
- 125.Zhang M, Wen B, Anton OM, Yao Z, Dubois S, Ju W, et al. IL‐15 enhanced antibody‐dependent cellular cytotoxicity mediated by NK cells and macrophages. Proc Natl Acad Sci U S A. 2018;115(46):E10915–E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu Y, Zhang M, Ramos CA, Durett A, Liu E, Dakhova O, et al. Closely related T‐memory stem cells correlate with in vivo expansion of CAR.CD19‐T cells and are preserved by IL‐7 and IL‐15. Blood. 2014;123(24):3750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, et al. Successful anti‐PD‐1 cancer immunotherapy requires T cell‐dendritic cell crosstalk involving the cytokines IFN‐gamma and IL‐12. Immunity. 2018;49(6):1148–61 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Al‐Chami E, Tormo A, Khodayarian F, Rafei M. Therapeutic utility of the newly discovered properties of interleukin‐21. Cytokine. 2016;82:33–7. [DOI] [PubMed] [Google Scholar]
- 129.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yuen KC, Liu LF, Gupta V, Madireddi S, Keerthivasan S, Li C, et al. High systemic and tumor‐associated IL‐8 correlates with reduced clinical benefit of PD‐L1 blockade. Nat Med. 2020;26(5):693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schalper KA, Carleton M, Zhou M, Chen T, Feng Y, Huang SP, et al. Elevated serum interleukin‐8 is associated with enhanced intratumor neutrophils and reduced clinical benefit of immune‐checkpoint inhibitors. Nat Med. 2020;26(5):688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tauriello DVF. From poor prognosis to promising treatment. Science. 2019;363(6431):1051. [DOI] [PubMed] [Google Scholar]
- 133.Qian J, Nie W, Lu J, Zhang L, Zhang Y, Zhang B, et al. Racial differences in characteristics and prognoses between Asian and white patients with nonsmall cell lung cancer receiving atezolizumab: An ancillary analysis of the POPLAR and OAK studies. Int J Cancer. 2020;146(11):3124–33. [DOI] [PubMed] [Google Scholar]
- 134.Ye Y, Jing Y, Li L, Mills GB, Diao L, Liu H, et al. Sex‐associated molecular differences for cancer immunotherapy. Nat Commun. 2020;11(1):1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–30 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA‐4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–57. [PubMed] [Google Scholar]
- 142.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N Engl J Med. 2015;372(26):2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair‐deficient/microsatellite instability‐high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–9. [DOI] [PubMed] [Google Scholar]
- 144.Liu S, Kong P, Wang X, Yang L, Jiang C, He W, et al. Tumor microenvironment classification based on T‐cell infiltration and PD‐L1 in patients with mismatch repair‐proficient and ‐deficient colorectal cancer. Oncol Lett. 2019;17(2):2335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. [DOI] [PubMed] [Google Scholar]
- 146.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. 2017;17(2):97–111. [DOI] [PubMed] [Google Scholar]
- 147.Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity. 2018;48(3):417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.de Vogel S, Weijenberg MP, Herman JG, Wouters KA, de Goeij AF, van den Brandt PA, et al. MGMT and MLH1 promoter methylation versus APC, KRAS and BRAF gene mutations in colorectal cancer: indications for distinct pathways and sequence of events. Ann Oncol. 2009;20(7):1216–22. [DOI] [PubMed] [Google Scholar]
- 149.Coppede F, Migheli F, Lopomo A, Failli A, Legitimo A, Consolini R, et al. Gene promoter methylation in colorectal cancer and healthy adjacent mucosa specimens: correlation with physiological and pathological characteristics, and with biomarkers of one‐carbon metabolism. Epigenetics. 2014;9(4):621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Spranger S. Mechanisms of tumor escape in the context of the T‐cell‐inflamed and the non‐T‐cell‐inflamed tumor microenvironment. Int Immunol. 2016;28(8):383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, et al. De novo epigenetic programs inhibit PD‐1 blockade‐mediated T cell rejuvenation. Cell. 2017;170(1):142–57 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fujita Y, Tinoco R, Li Y, Senft D, Ronai ZA. Ubiquitin ligases in cancer immunotherapy ‐ balancing antitumor and autoimmunity. Trends Mol Med. 2019;25(5):428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhao Y, Guo H, Qiao G, Zucker M, Langdon WY, Zhang J. E3 ubiquitin ligase Cbl‐b regulates thymic‐derived CD4+CD25+ regulatory T cell development by targeting foxp3 for ubiquitination. J Immunol. 2015;194(4):1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Woods DM, Woan KV, Cheng F, Sodre AL, Wang D, Wu Y, et al. T cells lacking HDAC11 have increased effector functions and mediate enhanced alloreactivity in a murine model. Blood. 2017;130(2):146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu CJ, Cho S, Huang HY, Lu CH, Russ J, Cruz LO, et al. MiR‐23∼27∼24‐mediated control of humoral immunity reveals a TOX‐driven regulatory circuit in follicular helper T cell differentiation. Sci Adv. 2019;5(12):eaaw1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xu J, Shao T, Song M, Xie Y, Zhou J, Yin J, et al. MIR22HG acts as a tumor suppressor via TGFbeta/SMAD signaling and facilitates immunotherapy in colorectal cancer. Mol Cancer. 2020;19(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC‐I. Nature. 2020;581(7806):100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Loo Yau H, Ettayebi I, De Carvalho DD. The cancer epigenome: exploiting its vulnerabilities for immunotherapy. Trends Cell Biol. 2019;29(1):31–43. [DOI] [PubMed] [Google Scholar]
- 159.Ehx G, Fransolet G, de Leval L, D'Hondt S, Lucas S, Hannon M, et al. Azacytidine prevents experimental xenogeneic graft‐versus‐host disease without abrogating graft‐versus‐leukemia effects. Oncoimmunology. 2017;6(5):e1314425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ju HQ, Ying H, Tian T, Ling J, Fu J, Lu Y, et al. Mutant Kras‐ and p16‐regulated NOX4 activation overcomes metabolic checkpoints in development of pancreatic ductal adenocarcinoma. Nat Commun. 2017;8:14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Renner K, Singer K, Koehl GE, Geissler EK, Peter K, Siska PJ, et al. Metabolic hallmarks of tumor and immune cells in the tumor microenvironment. Front Immunol. 2017;8:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Turbitt WJ, Buchta Rosean C, Weber KS, Norian LA. Obesity and CD8 T cell metabolism: implications for anti‐tumor immunity and cancer immunotherapy outcomes. Immunol Rev. 2020;295(1):203–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Al‐Khami AA, Zheng L, Del Valle L, Hossain F, Wyczechowska D, Zabaleta J, et al. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor‐associated myeloid‐derived suppressor cells. Oncoimmunology. 2017;6(10):e1344804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li X, Wenes M, Romero P, Huang SC, Fendt SM, Ho PC. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat Rev Clin Oncol. 2019;16(7):425–41. [DOI] [PubMed] [Google Scholar]
- 168.Manzo T, Prentice BM, Anderson KG, Raman A, Schalck A, Codreanu GS, et al. Accumulation of long‐chain fatty acids in the tumor microenvironment drives dysfunction in intrapancreatic CD8+ T cells. J Exp Med. 2020;217(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang Y, Lu JH, Wang F, Wang YN, He MM, Wu QN, et al. Inhibition of fatty acid catabolism augments the efficacy of oxaliplatin‐based chemotherapy in gastrointestinal cancers. Cancer Lett. 2020;473:74–89. [DOI] [PubMed] [Google Scholar]
- 170.Beier UH, Angelin A, Akimova T, Wang L, Liu Y, Xiao H, et al. Essential role of mitochondrial energy metabolism in Foxp3(+) T‐regulatory cell function and allograft survival. FASEB J. 2015;29(6):2315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162(6):1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gupta S, Roy A, Dwarakanath BS. Metabolic cooperation and competition in the tumor microenvironment: implications for therapy. Front Oncol. 2017;7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bronte V, Kasic T, Gri G, Gallana K, Borsellino G, Marigo I, et al. Boosting antitumor responses of T lymphocytes infiltrating human prostate cancers. J Exp Med. 2005;201(8):1257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang T, Gnanaprakasam JNR, Chen X, Kang S, Xu X, Sun H, et al. Inosine is an alternative carbon source for CD8(+)‐T‐cell function under glucose restriction. Nat Metab. 2020;2(7):635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu L, Hollinshead KER, Hao Y, Au C, Kroehling L, Ng C, et al. Niche‐selective inhibition of pathogenic Th17 cells by targeting metabolic redundancy. Cell. 2020;182(3):641–54 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lauby‐Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, et al. Body fatness and cancer–viewpoint of the IARC working group. N Engl J Med. 2016;375(8):794–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest. 2017;127(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ingels C, Vanhorebeek I, Van den Berghe G. Glucose homeostasis, nutrition and infections during critical illness. Clin Microbiol Infect. 2018;24(1):10–5. [DOI] [PubMed] [Google Scholar]
- 179.Zhang C, Yue C, Herrmann A, Song J, Egelston C, Wang T, et al. STAT3 activation‐induced fatty acid oxidation in CD8(+) T effector cells is critical for obesity‐promoted breast tumor growth. Cell Metab. 2020;31(1):148–61 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang Z, Aguilar EG, Luna JI, Dunai C, Khuat LT, Le CT, et al. Paradoxical effects of obesity on T cell function during tumor progression and PD‐1 checkpoint blockade. Nat Med. 2019;25(1):141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hursting SD, Berger NA. Energy balance, host‐related factors, and cancer progression. J Clin Oncol. 2010;28(26):4058–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Saucillo DC, Gerriets VA, Sheng J, Rathmell JC, Maciver NJ. Leptin metabolically licenses T cells for activation to link nutrition and immunity. J Immunol. 2014;192(1):136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Batra A, Okur B, Glauben R, Erben U, Ihbe J, Stroh T, et al. Leptin: a critical regulator of CD4+ T‐cell polarization in vitro and in vivo. Endocrinology. 2010;151(1):56–62. [DOI] [PubMed] [Google Scholar]
- 184.Wang X, Ping FF, Bakht S, Ling J, Hassan W. Immunometabolism features of metabolic deregulation and cancer. J Cell Mol Med. 2019;23(2):694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee C, Raffaghello L, Brandhorst S, Safdie FM, Bianchi G, Martin‐Montalvo A, et al. Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci Transl Med. 2012;4(124):124ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dorff TB, Groshen S, Garcia A, Shah M, Tsao‐Wei D, Pham H, et al. Safety and feasibility of fasting in combination with platinum‐based chemotherapy. BMC Cancer. 2016;16:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bauersfeld SP, Kessler CS, Wischnewsky M, Jaensch A, Steckhan N, Stange R, et al. The effects of short‐term fasting on quality of life and tolerance to chemotherapy in patients with breast and ovarian cancer: a randomized cross‐over pilot study. BMC Cancer. 2018;18(1):476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nencioni A, Caffa I, Cortellino S, Longo VD. Fasting and cancer: molecular mechanisms and clinical application. Nat Rev Cancer. 2018;18(11):707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Di Biase S, Lee C, Brandhorst S, Manes B, Buono R, Cheng CW, et al. Fasting‐mimicking diet reduces HO‐1 to promote T cell‐mediated tumor cytotoxicity. Cancer Cell. 2016;30(1):136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Soldati L, Di Renzo L, Jirillo E, Ascierto PA, Marincola FM, De Lorenzo A. The influence of diet on anti‐cancer immune responsiveness. J Transl Med. 2018;16(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. 2019;68(3):385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330(6005):831–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rutkowski MR, Stephen TL, Svoronos N, Allegrezza MJ, Tesone AJ, Perales‐Puchalt A, et al. Microbially driven TLR5‐dependent signaling governs distal malignant progression through tumor‐promoting inflammation. Cancer Cell. 2015;27(1):27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ke W, Saba JA, Yao CH, Hilzendeger MA, Drangowska‐Way A, Joshi C, et al. Dietary serine‐microbiota interaction enhances chemotherapeutic toxicity without altering drug conversion. Nat Commun. 2020;11(1):2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–79. [DOI] [PubMed] [Google Scholar]
- 197.Song W, Tiruthani K, Wang Y, Shen L, Hu M, Dorosheva O, et al. Trapping of lipopolysaccharide to promote immunotherapy against colorectal cancer and attenuate liver metastasis. Adv Mater. 2018;30(52):e1805007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli E, et al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol. 2020;78(2):195–206. [DOI] [PubMed] [Google Scholar]
- 199.Sivan A, Corrales L, Hubert N, Williams JB, Aquino‐Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science. 2015;350(6264):1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Dong X, Pan P, Zheng DW, Bao P, Zeng X, Zhang XZ. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor‐immune microenvironment against colorectal cancer. Sci Adv. 2020;6(20):eaba1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sipe LM, Chaib M, Pingili AK, Pierre JF, Makowski L. Microbiome, bile acids, and obesity: how microbially modified metabolites shape anti‐tumor immunity. Immunol Rev. 2020;295(1):220–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581(7809):475–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mohiuddin JJ, Chu B, Facciabene A, Poirier K, Wang X, Doucette A, et al. Association of antibiotic exposure with survival and toxicity in patients with melanoma receiving immunotherapy. J Natl Cancer Inst. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–7. [DOI] [PubMed] [Google Scholar]
- 206.Elkrief A, Derosa L, Zitvogel L, Kroemer G, Routy B. The intimate relationship between gut microbiota and cancer immunotherapy. Gut Microbes. 2019;10(3):424–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Geller LT, Barzily‐Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. 2016;82(16):5039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type‐specific intracellular bacteria. Science. 2020;368(6494):973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Galon J, Fridman WH, Pages F. The adaptive immunologic microenvironment in colorectal cancer: a novel perspective. Cancer Res. 2007;67(5):1883–6. [DOI] [PubMed] [Google Scholar]
- 213.Camus M, Tosolini M, Mlecnik B, Pages F, Kirilovsky A, Berger A, et al. Coordination of intratumoral immune reaction and human colorectal cancer recurrence. Cancer Res. 2009;69(6):2685–93. [DOI] [PubMed] [Google Scholar]
- 214.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18(3):197–218. [DOI] [PubMed] [Google Scholar]
- 215.Galon J, Costes A, Sanchez‐Cabo F, Kirilovsky A, Mlecnik B, Lagorce‐Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–4. [DOI] [PubMed] [Google Scholar]
- 216.Mlecnik B, Bindea G, Angell HK, Sasso MS, Obenauf AC, Fredriksen T, et al. Functional network pipeline reveals genetic determinants associated with in situ lymphocyte proliferation and survival of cancer patients. Sci Transl Med. 2014;6(228):228ra37. [DOI] [PubMed] [Google Scholar]
- 217.Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019;36(4):418–30 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Triplett TA, Garrison KC, Marshall N, Donkor M, Blazeck J, Lamb C, et al. Reversal of indoleamine 2,3‐dioxygenase‐mediated cancer immune suppression by systemic kynurenine depletion with a therapeutic enzyme. Nat Biotechnol. 2018;36(8):758–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nishio H, Yaguchi T, Sugiyama J, Sumimoto H, Umezawa K, Iwata T, et al. Immunosuppression through constitutively activated NF‐kappaB signalling in human ovarian cancer and its reversal by an NF‐kappaB inhibitor. Br J Cancer. 2014;110(12):2965–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Han JH, Yoon JS, Chang DY, Cho KG, Lim J, Kim SS, et al. CXCR4‐STAT3 axis plays a role in tumor cell infiltration in an orthotopic mouse glioblastoma model. Mol Cells. 2020;43(6):539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang H, Jiang R, Zhou J, Wang J, Xu Y, Zhang H, et al. CTL attenuation regulated by PS1 in cancer‐associated fibroblast. Front Immunol. 2020;11:999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Becht E, Giraldo NA, Dieu‐Nosjean MC, Sautes‐Fridman C, Fridman WH. Cancer immune contexture and immunotherapy. Curr Opin Immunol. 2016;39:7–13. [DOI] [PubMed] [Google Scholar]
- 223.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman‐Saint Victor C, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–54 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Honda T, Egen JG, Lammermann T, Kastenmuller W, Torabi‐Parizi P, Germain RN. Tuning of antigen sensitivity by T cell receptor‐dependent negative feedback controls T cell effector function in inflamed tissues. Immunity. 2014;40(2):235–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Grosso JF, Goldberg MV, Getnet D, Bruno TC, HR Yen, Pyle KJ, et al. Functionally distinct LAG‐3 and PD‐1 subsets on activated and chronically stimulated CD8 T cells. J Immunol. 2009;182(11):6659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim‐3 and PD‐1 pathways to reverse T cell exhaustion and restore anti‐tumor immunity. J Exp Med. 2010;207(10):2187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single‐cell sequencing. Cell. 2017;169(7):1342–56 e16. [DOI] [PubMed] [Google Scholar]
- 230.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–9. [DOI] [PubMed] [Google Scholar]
- 231.Veiga‐Parga T, Sehrawat S, Rouse BT. Role of regulatory T cells during virus infection. Immunol Rev. 2013;255(1):182–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Goh C, Narayanan S, Hahn YS. Myeloid‐derived suppressor cells: the dark knight or the joker in viral infections? Immunol Rev. 2013;255(1):210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338(6111):1220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhou W, Ke SQ, Huang Z, Flavahan W, Fang X, Paul J, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour‐associated macrophages and promotes malignant growth. Nat Cell Biol. 2015;17(2):170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sumitomo R, Hirai T, Fujita M, Murakami H, Otake Y, Huang CL. M2 tumor‐associated macrophages promote tumor progression in non‐small‐cell lung cancer. Exp Ther Med. 2019;18(6):4490–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Marvel D, Gabrilovich DI. Myeloid‐derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. 2015;125(9):3356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pao W, Ooi CH, Birzele F, Ruefli‐Brasse A, Cannarile MA, Reis B, et al. Tissue‐specific immunoregulation: a call for better understanding of the “immunostat” in the context of cancer. Cancer Discov. 2018;8(4):395–402. [DOI] [PubMed] [Google Scholar]
- 238.Tan WCC, Nerurkar SN, Cai HY, Ng HHM, Wu D, Wee YTF, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun (Lond). 2020;40(4):135–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hamid O, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med. 2011;9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD‐1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6(8):827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non‐small‐cell lung cancer. N Engl J Med. 2015;372(21):2018–28. [DOI] [PubMed] [Google Scholar]
- 243.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism‐driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Datar I, Sanmamed MF, Wang J, Henick BS, Choi J, Badri T, et al. Expression analysis and significance of PD‐1, LAG‐3, and TIM‐3 in human non‐small cell lung cancer using spatially resolved and multiparametric single‐cell analysis. Clin Cancer Res. 2019;25(15):4663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD‐1 inhibition. N Engl J Med. 2017;377(25):2500–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang F, Zhao Q, Wang YN, Jin Y, He MM, Liu ZX, et al. Evaluation of POLE and POLD1 mutations as biomarkers for immunotherapy outcomes across multiple cancer types. JAMA Oncol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Suva ML, Tirosh I. Single‐cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell. 2019;75(1):7–12. [DOI] [PubMed] [Google Scholar]
- 248.Wu T, Wu X, Wang HY, Chen L. Immune contexture defined by single cell technology for prognosis prediction and immunotherapy guidance in cancer. Cancer Commun (Lond). 2019;39(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Tang X, Huang Y, Lei J, Luo H, Zhu X. The single‐cell sequencing: new developments and medical applications. Cell Biosci. 2019;9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rozenblatt‐Rosen O, Regev A, Oberdoerffer P, Nawy T, Hupalowska A, Rood JE, et al. The human tumor atlas network: charting tumor transitions across space and time at single‐cell resolution. Cell. 2020;181(2):236–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rijavec E, Coco S, Genova C, Rossi G, Longo L, Grossi F. Liquid biopsy in non‐small cell lung cancer: highlights and challenges. Cancers (Basel). 2019;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–48. [DOI] [PubMed] [Google Scholar]
- 253.Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic landscape of cell‐free DNA in patients with colorectal cancer. Cancer Discov. 2018;8(2):164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Best MG, Sol N, In ’t Veld S, Vancura A, Muller M, Niemeijer AN, et al. Swarm intelligence‐enhanced detection of non‐small‐cell lung cancer using tumor‐educated platelets. Cancer Cell. 2017;32(2):238–52 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8(4):444–57. [DOI] [PubMed] [Google Scholar]
- 256.Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–61. [DOI] [PubMed] [Google Scholar]
- 257.Mazel M, Jacot W, Pantel K, Bartkowiak K, Topart D, Cayrefourcq L, et al. Frequent expression of PD‐L1 on circulating breast cancer cells. Mol Oncol. 2015;9(9):1773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD‐L1 contributes to immunosuppression and is associated with anti‐PD‐1 response. Nature. 2018;560(7718):382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, et al. Circulating tumor DNA changes for early monitoring of anti‐PD1 immunotherapy: a proof‐of‐concept study. Ann Oncol. 2017;28(8):1996–2001. [DOI] [PubMed] [Google Scholar]
- 260.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood‐based tumor mutational burden as a predictor of clinical benefit in non‐small‐cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–8. [DOI] [PubMed] [Google Scholar]
- 261.Hofman P, Heeke S, Alix‐Panabieres C, Pantel K. Liquid biopsy in the era of immuno‐oncology: is it ready for prime‐time use for cancer patients? Ann Oncol. 2019;30(9):1448–59. [DOI] [PubMed] [Google Scholar]
- 262.Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18(3):175–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pollack MH, Betof A, Dearden H, Rapazzo K, Valentine I, Brohl AS, et al. Safety of resuming anti‐PD‐1 in patients with immune‐related adverse events (irAEs) during combined anti‐CTLA‐4 and anti‐PD1 in metastatic melanoma. Ann Oncol. 2018;29(1):250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hartmann J, Schussler‐Lenz M, Bondanza A, Buchholz CJ. Clinical development of CAR T cells‐challenges and opportunities in translating innovative treatment concepts. EMBO Mol Med. 2017;9(9):1183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler‐Stevenson M, Feldman SA, et al. Eradication of B‐lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116(20):4099–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol. 2019;16(6):372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Fry TJ, Shah NN, Orentas RJ, Stetler‐Stevenson M, Yuan CM, Ramakrishna S, et al. CD22‐targeted CAR T cells induce remission in B‐ALL that is naive or resistant to CD19‐targeted CAR immunotherapy. Nat Med. 2018;24(1):20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Borrello I, Imus PH. BCMA CAR T cells: the winding path to success. J Clin Invest. 2019;129(6):2175–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Fitzgerald JC, Weiss SL, Maude SL, Barrett DM, Lacey SF, Melenhorst JJ, et al. Cytokine release syndrome after chimeric antigen receptor T cell therapy for acute lymphoblastic leukemia. Crit Care Med. 2017;45(2):e124‐e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Migliorini D, Dietrich PY, Stupp R, Linette GP, PoseyAD, Jr., June CH. CAR T‐cell therapies in glioblastoma: a first look. Clin Cancer Res. 2018;24(3):535–40. [DOI] [PubMed] [Google Scholar]
- 271.Beckermann KE, Jolly PC, Kim JY, Bordeaux J, Puzanov I, Rathmell WK, et al. Clinical and immunologic correlates of response to PD‐1 blockade in a patient with metastatic renal medullary carcinoma. J Immunother Cancer. 2017;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Stephan SB, Taber AM, Jileaeva I, Pegues EP, Sentman CL, Stephan MT. Biopolymer implants enhance the efficacy of adoptive T‐cell therapy. Nat Biotechnol. 2015;33(1):97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Stephan MT, Moon JJ, Um SH, Bershteyn A, Irvine DJ. Therapeutic cell engineering with surface‐conjugated synthetic nanoparticles. Nat Med. 2010;16(9):1035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Meyer RA, Sunshine JC, Perica K, Kosmides AK, Aje K, Schneck JP, et al. Biodegradable nanoellipsoidal artificial antigen presenting cells for antigen specific T‐cell activation. Small. 2015;11(13):1519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kato D, Yaguchi T, Iwata T, Morii K, Nakagawa T, Nishimura R, et al. Prospects for personalized combination immunotherapy for solid tumors based on adoptive cell therapies and immune checkpoint blockade therapies. Nihon Rinsho Meneki Gakkai Kaishi. 2017;40(1):68–77. [DOI] [PubMed] [Google Scholar]
- 276.Zhang J, Wang L. The emerging world of TCR‐T cell trials against cancer: a systematic review. Technol Cancer Res Treat. 2019;18:1533033819831068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Garber K. Driving T‐cell immunotherapy to solid tumors. Nat Biotechnol. 2018;36(3):215–9. [DOI] [PubMed] [Google Scholar]
- 278.Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol. 2020;38(8):947–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sprooten J, Ceusters J, Coosemans A, Agostinis P, De Vleeschouwer S, Zitvogel L, et al. Trial watch: dendritic cell vaccination for cancer immunotherapy. Oncoimmunology. 2019;8(11):e1638212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. 2020;20(1):7–24. [DOI] [PubMed] [Google Scholar]
- 281.Melief CJ, van Hall T, Arens R, Ossendorp F, van der Burg SH. Therapeutic cancer vaccines. J Clin Invest. 2015;125(9):3401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sharma P, Hu‐Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14(9):642–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Harrington K, Freeman DJ, Kelly B, Harper J, Soria JC. Optimizing oncolytic virotherapy in cancer treatment. Nat Rev Drug Discov. 2019;18(9):689–706. [DOI] [PubMed] [Google Scholar]
- 285.Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33(25):2780–8. [DOI] [PubMed] [Google Scholar]
- 286.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, et al. Personalized RNA mutanome vaccines mobilize poly‐specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–6. [DOI] [PubMed] [Google Scholar]
- 287.Fang Y, Mo F, Shou J, Wang H, Luo K, Zhang S, et al. A pan‐cancer clinical study of personalized neoantigen vaccine monotherapy in treating patients with various types of advanced solid tumors. Clin Cancer Res. 2020. [DOI] [PubMed] [Google Scholar]
- 288.Smith TT, Stephan SB, Moffett HF, McKnight LE, Ji W, Reiman D, et al. In situ programming of leukaemia‐specific T cells using synthetic DNA nanocarriers. Nat Nanotechnol. 2017;12(8):813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534(7607):396–401. [DOI] [PubMed] [Google Scholar]
- 290.Hagberg H. Alpha interferon treatment in hairy cell leukemia. Nord Med. 1992;107(3):74–5. [PubMed] [Google Scholar]
- 291.Motzer RJ, Murphy BA, Bacik J, Schwartz LH, Nanus DM, Mariani T, et al. Phase III trial of interferon alfa‐2a with or without 13‐cis‐retinoic acid for patients with advanced renal cell carcinoma. J Clin Oncol. 2000;18(16):2972–80. [DOI] [PubMed] [Google Scholar]
- 292.Sanlorenzo M, Vujic I, Carnevale‐Schianca F, Quaglino P, Gammaitoni L, Fierro MT, et al. Role of interferon in melanoma: old hopes and new perspectives. Expert Opin Biol Ther. 2017;17(4):475–83. [DOI] [PubMed] [Google Scholar]
- 293.Rosenberg SA, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, Parkinson DR, et al. Treatment of 283 consecutive patients with metastatic melanoma or renal cell cancer using high‐dose bolus interleukin 2. JAMA. 1994;271(12):907–13. [PubMed] [Google Scholar]
- 294.Rosenberg SA. IL‐2: the first effective immunotherapy for human cancer. J Immunol. 2014;192(12):5451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel‐T immunotherapy for castration‐resistant prostate cancer. N Engl J Med. 2010;363(5):411–22. [DOI] [PubMed] [Google Scholar]
- 296.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor‐modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, et al. High‐dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999;17(7):2105–16. [DOI] [PubMed] [Google Scholar]
- 299.Larkin J, Chiarion‐Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wang Y, Zhou S, Yang F, Qi X, Wang X, Guan X, et al. Treatment‐related adverse events of PD‐1 and PD‐L1 inhibitors in clinical trials: a systematic review and meta‐analysis. JAMA Oncol. 2019;5(7):1008–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Magee DE, Hird AE, Klaassen Z, Sridhar SS, Nam RK, Wallis CJD, et al. Adverse event profile for immunotherapy agents compared with chemotherapy in solid organ tumors: a systematic review and meta‐analysis of randomized clinical trials. Ann Oncol. 2020;31(1):50–60. [DOI] [PubMed] [Google Scholar]
- 302.Weber JS, Hodi FS, Wolchok JD, Topalian SL, Schadendorf D, Larkin J, et al. Safety profile of nivolumab monotherapy: a pooled analysis of patients with advanced melanoma. J Clin Oncol. 2017;35(7):785–92. [DOI] [PubMed] [Google Scholar]
- 303.Naidoo J, Wang X, Woo KM, Iyriboz T, Halpenny D, Cunningham J, et al. Pneumonitis in patients treated with anti‐programmed death‐1/programmed death ligand 1 therapy. J Clin Oncol. 2017;35(7):709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Johnson DB, Balko JM, Compton ML, Chalkias S, Gorham J, Xu Y, et al. Fulminant myocarditis with combination immune checkpoint blockade. N Engl J Med. 2016;375(18):1749–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Yanai S, Nakamura S, Matsumoto T. Nivolumab‐induced colitis treated by infliximab. Clin Gastroenterol Hepatol. 2017;15(4):e80‐e1. [DOI] [PubMed] [Google Scholar]
- 306.Tian Y, Zhang Z, Yang X, Li D, Zhang L, Li Z, et al. The risk ratio of immune‐related colitis, hepatitis, and pancreatitis in patients with solid tumors caused by PD‐1/PD‐L1 inhibitors: a systematic review and meta‐analysis. Front Oncol. 2020;10:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Mirabile A, Brioschi E, Ducceschi M, Piva S, Lazzari C, Bulotta A, et al. PD‐1 inhibitors‐related neurological toxicities in patients with non‐small‐cell lung cancer: a literature review. Cancers (Basel). 2019;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Moslehi JJ, Salem JE, Sosman JA, Lebrun‐Vignes B, DB Johnson. Increased reporting of fatal immune checkpoint inhibitor‐associated myocarditis. Lancet. 2018;391(10124):933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Esfahani K, Elkrief A, Calabrese C, Lapointe R, Hudson M, Routy B, et al. Moving towards personalized treatments of immune‐related adverse events. Nat Rev Clin Oncol. 2020;17(8):504–15. [DOI] [PubMed] [Google Scholar]
- 310.Oh DY, Cham J, Zhang L, Fong G, Kwek SS, Klinger M, et al. Immune toxicities elicted by CTLA‐4 blockade in cancer patients are associated with early diversification of the T‐cell repertoire. Cancer Res. 2017;77(6):1322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Das R, Bar N, Ferreira M, Newman AM, Zhang L, Bailur JK, et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J Clin Invest. 2018;128(2):715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Subudhi SK, Aparicio A, Gao J, Zurita AJ, Araujo JC, Logothetis CJ, et al. Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab‐induced toxicities. Proc Natl Acad Sci U S A. 2016;113(42):11919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.June CH, Warshauer JT, Bluestone JA. Corrigendum: Is autoimmunity the Achilles' heel of cancer immunotherapy? Nat Med. 2017;23(8):1004. [DOI] [PubMed] [Google Scholar]
- 314.Lim SY, Lee JH, Gide TN, Menzies AM, Guminski A, Carlino MS, et al. Circulating cytokines predict immune‐related toxicity in melanoma patients receiving anti‐PD‐1‐based immunotherapy. Clin Cancer Res. 2019;25(5):1557–63. [DOI] [PubMed] [Google Scholar]
- 315.Khan S, Khan SA, Luo X, Fattah FJ, Saltarski J, Gloria‐McCutchen Y, et al. Immune dysregulation in cancer patients developing immune‐related adverse events. Br J Cancer. 2019;120(1):63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Iwama S, De Remigis A, Callahan MK, Slovin SF, Wolchok JD, Caturegli P. Pituitary expression of CTLA‐4 mediates hypophysitis secondary to administration of CTLA‐4 blocking antibody. Sci Transl Med. 2014;6(230):230ra45. [DOI] [PubMed] [Google Scholar]
- 317.Chen DS, Mellman I. Oncology meets immunology: the cancer‐immunity cycle. Immunity. 2013;39(1):1–10. [DOI] [PubMed] [Google Scholar]
- 318.Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35(Suppl):S185–S98. [DOI] [PubMed] [Google Scholar]
- 319.Chen L, Han X. Anti‐PD‐1/PD‐L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125(9):3384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Gutierrez M, Hamid O, Shum E, Wise DR, Balar AV, Weber JS, et al. Trial in progress: a phase I/II, open‐label, dose‐escalation, safety and tolerability study of NC318 in subjects with advanced or metastatic solid tumors. Journal of Clinical Oncology. 2020;38(15_suppl):TPS3166‐TPS. [Google Scholar]
- 321.Champiat S, Ferrara R, Massard C, Besse B, Marabelle A, Soria JC, et al. Hyperprogressive disease: recognizing a novel pattern to improve patient management. Nat Rev Clin Oncol. 2018;15(12):748–62. [DOI] [PubMed] [Google Scholar]
- 322.Lahmar J, Mezquita L, Koscielny S, Facchinetti F, Bluthgen MV, Adam J, et al. Immune checkpoint inhibitors (IC) induce paradoxical progression in a subset of non‐small cell lung cancer (NSCLC). Annals of Oncology. 2016;27. [Google Scholar]
- 323.Champiat S, Dercle L, Ammari S, Massard C, Hollebecque A, Postel‐Vinay S, et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti‐PD‐1/PD‐L1. Clin Cancer Res. 2017;23(8):1920–8. [DOI] [PubMed] [Google Scholar]
- 324.Ferris RL, BlumenscheinG, Jr., Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous‐cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, et al. Pembrolizumab as second‐line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376(11):1015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Powles T, Duran I, van der Heijden MS, Loriot Y, Vogelzang NJ, De Giorgi U, et al. Atezolizumab versus chemotherapy in patients with platinum‐treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open‐label, phase 3 randomised controlled trial. Lancet. 2018;391(10122):748–57. [DOI] [PubMed] [Google Scholar]
- 327.Borghaei H, Paz‐Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non‐small‐cell lung cancer. N Engl J Med. 2015;373(17):1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Saada‐Bouzid E, Defaucheux C, Karabajakian A, Coloma VP, Servois V, Paoletti X, et al. Hyperprogression during anti‐PD‐1/PD‐L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2017;28(7):1605–11. [DOI] [PubMed] [Google Scholar]
- 329.Ferrara R, Mezquita L, Texier M, Lahmar J, Audigier‐Valette C, Tessonnier L, et al. Hyperprogressive disease in patients with advanced non‐small cell lung cancer treated with PD‐1/PD‐L1 inhibitors or with single‐agent chemotherapy. JAMA Oncol. 2018;4(11):1543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after Immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res. 2017;23(15):4242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Singavi AK, Menon S, Kilari D, Alqwasmi A, Ritch PS, Thomas JP, et al. Predictive biomarkers for hyper‐progression (HP) in response to immune checkpoint inhibitors (ICI) – analysis of somatic alterations (SAs). Annals of Oncology. 2017;28. [Google Scholar]
- 332.Reck M, Ciuleanu T‐E, Dols MC, Schenker M, Zurawski B, Menezes J, et al. Nivolumab (NIVO) + ipilimumab (IPI) + 2 cycles of platinum‐doublet chemotherapy (chemo) vs 4 cycles chemo as first‐line (1L) treatment (tx) for stage IV/recurrent non‐small cell lung cancer (NSCLC): CheckMate 9LA. Journal of Clinical Oncology. 2020;38(15_suppl):9501. [Google Scholar]
- 333.Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52(1):17–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


