Abstract
Background
Resistance to platinum-based chemotherapy is a major cause of therapeutic failure during the treatment of epithelial ovarian cancer (EOC) patients. Our study aims to elucidate the molecular mechanisms by which ZNF711 down regulation promotes CISPLATIN resistance in EOC.
Methods
ZNF711 expression in 150 EOC specimens was examined using immunohistochemistry. ZNF711 expression and the survival of EOC patients were assessed with a Kaplan-Meier analysis. The effects of ZNF711 expression on CDDP resistance were studied by IC50, Annexin V, and colony formation in vitro, and in an in vivo intra-peritoneal tumor model. The molecular mechanism was determined using a luciferase reporter assay, ChIP assay, CAPTURE approach, and co-IP assay.
Findings
ZNF711 down-regulation exerts a great impact on CDDP resistance for EOC patients by suppressing SLC31A1 and inhibiting CDDP influx. ZNF711 down-regulation promoted, while ZNF711 overexpression drastically inhibited CDDP resistance, both in vivo and in vitro. Mechanistically, the histone demethylase JHDM2A was recruited to the SLC31A1 promoter by ZNF711 and decreased the H3K9me2 level, resulting in the activation of SLC31A1 transcription and enhancement of CDDP uptake. Importantly, co-treatment with the histone methylation inhibitor, BIX-01294, increased the therapeutic efficacy of CDDP treatment in ZNF711-suppressed EOC cells.
Interpretation
These findings both verified the clinical importance of ZNF711 in CDDP resistance and provide novel therapeutic regimens for EOC treatment.
Funding
This work was supported by the Natural Science Foundation of China; Guangzhou Science and Technology Plan Projects; Natural Science Foundation of Guangdong Province; The Fundamental Research Funds for the Central Universities; and China Postdoctoral Science Foundation.
Keywords: Epithelial ovarian cancer, Chemo resistance, ZNF711, Histone methylation, BIX-01294
Research in context.
Evidence before this study
The negative relationship between the level of SLC31A1 expression and platinum resistance has been demonstrated in ovarian cancer and triple-negative breast cancer. However, the inhibitory modulation of SLC31A1 in EOC remains largely unknown.
Added value of this study
Here-in, we found that ZNF711 was down regulated in EOC patients with CDDP resistance, which was negatively correlated with the overall and relapse-free survival of EOC patients following CDDP treatment. ZNF711 down regulation promoted, while up-regulation of ZNF711 weakened CDDP resistance in EOC patients, both in vivo and in vitro. Mechanistically, histone demethylase JHDM2A was recruited to the SLC31A1 promoter via ZNF711 and decreased the level of H3K9me2, resulting in activation of SLC31A1 transcription and enhanced CDDP influx. Importantly, co- treatment with the histone methylation inhibitor, BIX-01294, greatly augmented CDDP sensitivity in ZNF711-downregulated EOC cells.
Implications of all the available evidence
Implications of all available evidence these findings demonstrate that ZNF711 may serve as a pivotal bio-marker and therapeutic target of CDDP resistance in EOC patients.
Alt-text: Unlabelled box
1. Introduction
Epithelial ovarian cancer (EOC) represents one of the most common gynecological malignancies and fifth leading cause of cancer-associated death in women throughout the world [1]. Globally, it is estimated that the number of new cases of ovarian cancer will reach 300,000 cases per year, which causes approximately 180,000 deaths [[1], [2], [3]]. At present, the gold standard therapy for EOC patients is cyto-reductive surgery followed by platinum-based chemotherapy [4,5]. Although most patients with ovarian cancer are sensitive to the initial chemotherapy drugs, about 80% EOC patients suffer a relapse and become resistant to chemotherapy, leading to a poor five-year survival rate of EOC patients of only 30% [6,7]. Thus, the identification of the molecular mechanism and crucial targets of platinum resistance in EOC is of great clinical value.
Platinum resistance is attributed to a variety of factors, including changes to the tumor microenvironment, epigenetic abnormalities, and drug accumulation disorders, among which the inhibition of drug uptake is closely related to CISPLATIN (CDDP) resistance in EOC [[8], [9], [10]]. Abundant evidence demonstrates that solute carrier family 31 member 1 (SLC31A1; also known as copper transporter 1, CTR1) contributes to the substantial role of CDDP influx [11,12]. SLC31A1 is a member of the copper transporter family that regulates copper homeostasis, impacting the intracellular concentration of platinum drugs by an influx of 60% − 70% cisplatin and 30% − 40% carboplatin into the cells [13,14]. Deletion of both SLC31A1 alleles drastically dampens the uptake of CDDP in both yeast and mammalian cells, resulting in the occurrence of drug resistance [15,16]. Additionally, previous studies have shown that the inhibition of SLC31A1 degradation sensitizes patients to CDDP therapy in osteosarcoma [17]. The negative relationship between the level of SLC31A1 expression and platinum resistance has also been demonstrated in ovarian cancer and triple-negative breast cancer [18,19]; however, the inhibition of SLC31A1 expression in EOC remains largely obscure.
Histone modifications modulate gene regulation through altering the chromatin structure, exerting a vital role in the development of cancer, including carcinogenesis, cell proliferation, and chemo resistance [[20], [21], [22]]. Histone methylation is currently recognized to be a vital histone modification linked to both transcriptional activation and repression, depending on which lysine or arginine residues of the methyl groups has been added [23,24]. Numerous histone methylation modifiers have been reported in cancer proliferation, migration, and chemo-resistance, including lysine methyl transferases (KMTs), protein arginine methyl transferases (PRMTs), and lysine demethylases (KDMs) [23,25]. For instance, methyl transferases zeste homolog 2 (EZH2) overexpression has been found to lead to increased H3K27me3 and the suppression of tumor suppressor genes (e.g., RUNX3 and CDH1), which promotes the development of digestive cancer [26]. Moreover, JmjC-domain-containing histone demethylase 2A (JHDM2A, also known as JMJD1A) promotes spermatogenesis by removing the repressive histone modification, H3K9me1/2 from the promoter of transition nuclear protein 1 (Tnp1) and protamine 1 (Prm1) gene, which are required for the packaging and condensation of sperm chromatin [27]. These studies demonstrated that aberrant histone modification contributes to development and progression of cancer. Additionally, it has been suggested that histone modification plays a crucial role in chemo-resistance in hepatocellular carcinoma and ovarian cancer [28,29]. Thus, to investigate the regulatory role of histone modification in CDDP resistance might be beneficial for improving the prognosis of EOC patients.
Zinc-finger proteins are transcription factors characterized by the presence of a “fingerlike” domain, and are widely involved in the tumorigenesis and progression of multiple cancer types, which have been validated to function as co-factors and recruiters of histone modifiers [[30], [31], [32]]. A newly discovered zinc-finger protein, ZNF711, has been shown to be decreased in breast cancer and serves as a predictor of patient prognosis, whereas the associated biological function and molecular mechanism remains largely unknown [33,34]. Herein, we found that ZNF711 was down regulated in EOC patients with CDDP resistance, which was negatively correlated with the overall and relapse-free survival of EOC patients following CDDP treatment. Moreover, ZNF711 down regulation has been shown to promote, whereas up-regulation of ZNF711 weakens CDDP resistance in EOC patients, both in vivo and in vitro. Mechanistically, histone demethylase JHDM2A was recruited to the SLC31A1 promoter by ZNF711 and decreased the level of H3K9me2, resulting in the activation of SLC31A1 transcription and enhancement of the CDDP influx.
Importantly, co-treatment with the histone methylation inhibitor, BIX-01294, substantially augmented CDDP sensitivity in ZNF711-suppressed EOC cells. The findings of this study demonstrate that ZNF711 serves as a pivotal biomarker and therapeutic target of CDDP resistance in EOC patients.
2. Methods
2.1. Ethics
Prior donor consent was obtained from all patients. Approval from the Sun Yat- sen University Cancer Center Institutional Research Ethics Committee was obtained for this study. The animal experiment in this study strictly adhered to principles to minimize the use, pain and suffering of the experimental animals (approval no. 2020- 062).
2.2. EOC patients, cancer tissue samples, and cells
A total of 150 paraffin-embedded, archived EOC specimens and 10 freshly collected EOC tissues were collected from Sun Yet-sen University Cancer Center (Guangdong, China), which have been histo-pathologically and clinically diagnosed as EOC between 2006-2016. The clinical pathological characteristics of EOC samples used in this study have been shown in Supplementary Table S1-2. All EOC patients received standardized platinum-based chemotherapy. We defined chemo-resistance or chemo-sensitivity as a relapse/progression within six months or after six months from the ending day of last platinum-based chemotherapy, respectively. Human Ovarian Surface Epithelial Cell (HOSEpiC) and normal ovarian epithelial IOSE80 cells were purchased from ScienCell Research Laboratories and cultured in ovarian epithelial cell medium according to the manufacturer's instructions. The ovarian cancer cells lines, including COV362.4, SKOV3, HEY, A2780, TOV-112D, OV90, EFO-21 and COV644 were grown in the DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT). All of the cell lines used in the current study were tested for mycoplasma contamination and authenticated by short tandem repeat (STR) fingerprinting at the Medicine Lab of Forensic Medicine Department of Sun Yat-Sen University (Guangzhou, China).
2.3. Immunoblotting (IB) assays
Immunoblotting assays were performed in accordance with standard procedures. Briefly, cell lysates were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore). The membranes were probed with antibodies against ZNF711 (1:100, LifeSpan BioSciences, LS‑C109806), SLC31A1 (1:200, Abcam, ab129067), JHDM2A (1:100, Abcam, ab191389) and EHMT2 (1:1000, Abcam, ab185050), overnight at 4°C, and then incubated with indicated horseradish peroxidase–conjugated secondary antibodies (PIERCE, 31430) for 1 h at room temperature. The blotting membranes were stripped and re-probed with an anti-α-Tubulin antibody (1:1000, Abcam, ab7291) or anti-GAPDH antibody (1:1000, Abcam, ab8245).
2.4. Vectors, retroviral infection and transfection
The retroviral vector pMSCV-puro was used to generate pMSCV-puro/ZNF711 recombinant plasmid by sub-cloning the PCR-amplified human ZNF711 coding sequence into pMSCV vector. To silence endogenous ZNF711, two shRNA oligonucleotides were cloned into the pSuper-retro-puro vector to generate pSuper- retro-ZNF711-sh#1 and pSuper-retro-ZNF711-sh#2, respectively. All the detailed sequences of siRNA oligonucleotides are shown in Supplementary Table S3. Stable cell lines were selected for 10 days with puromycin (0.5μg/mL) 48 hrs after infection. The ZNF711 protein level was detected by SDS–PAGE gel after ten-day selection to confirm stable expression of ZNF711 in indicated cell lines. The reporter plasmids containing the promoter region of human SLC31A1 were subcloned into the pGL3- Control vector luciferase reporter (E1751, Promega), respectively. The plasmid phRL- tk was used as the internal control for transfection efficiency and cytotoxicity of test chemicals (E6241, Promega). Recombinant plasmids or siRNA transfections in this study were performed using Lipofectamine 3000 (Life Technologies) in accordance with the manufacturer's instructions.
2.5. Reverse transcription (RT) and real-time PCR
In this study, total RNA from cultured cells or EOC tissue samples was extracted using TRIzol (Life Technologies) according to the manufacturer's instructions.
Reverse transcription (RT) of total mRNA was performed using a PrimeScript RT Reagent kit (TaKaRa). cDNAs were amplified and quantified in Bio-Rad CFX qRT- PCR detection system (Applied Biosystems Inc., Foster City, CA, USA) via using FastStart Universal SYBR Green Master kit (ROX; Roche, Toronto, ON, Canada) and quantified by using the ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA). The expression data were normalized to housekeeping gene GAPDH to control the variability in expression levels and calculated as 2−[(C t of gene) – (C t of GAPDH)], where Ct represents the threshold cycle for each transcript. The primer sequences were obtained from the Genome database was shown in Primers and Oligonucleotides table in Supplementary Table S3.
2.6. Immunohistochemistry (IHC) assay
IHC assay was performed to study altered protein expression in 150 human paraffin-embedded EOC samples with anti-ZNF711 (1:200, Abcam, ab254776), anti- SLC31A1 (1:200, Abcam, ab133385) and anti-PCNA (1:500, Abcam, ab92552). The degree of immuno-staining of formalin-fixed, paraffin-embedded sections were reviewed and scored separately by two independent pathologists uninformed of the histo-pathological features and patient data of the samples. The immunohistochemistry images were captured by Axio Imager.Z2 system (Carl Zeiss Co. Ltd., Jena, Germany). Degrees of immuno-staining were reviewed and scored independently by two pathologists who were uninformed of the clinical information of the EOC samples. The IHC scores were given based on both the proportion of positively stained tumor cells and the intensity of staining. The proportion of positive-stained tumor cells were graded according to the following standard: 0 (no positive tumor cells); 1 (<10% positive tumor cells); 2 (10%–35% positive tumor cells); 3 (35%–75% positive tumor cells); 4 (>75% positive tumor cells). Staining intensity was scored as follows: 1 (no staining); 2 (weak staining, light yellow); 3 (moderate staining, yellow brown); 4 (strong staining, brown). The staining index (SI) was calculated as follows: = staining intensity × proportion of positively stained tumor cells (with possible scores of 0, 2, 3, 4, 6, 8, 9, 12, and 16). Samples with a SI > 8 were determined as high expression and samples with a SI ≤ 8 were determined as low expression. Cutoff values were determined on the basis of a measure of heterogeneity using the log-rank test with respect to overall and relapse-free survival.
2.7. Colony formation Assay
The colony formation Assay in this study was carried out in accordance with the previously published paper [35]. Briefly, indicated EOC Cells with different expression level of ZNF711 were cultured in 6-well plates (2 × 103 cells per plate) for 10 days with or without CDDP treatment (5 μM). The colonies were then fixed with 10% formaldehyde for 5 min and subsequently stained with 1% crystal violet for 30 min. The number of colonies (defined as cell clusters consisting of more than 50 cells) was quantified by Analysis software (Olympus Biosystems).
2.8. Annexin V Assay
For the evaluation of apoptosis, Annexin-V assay was performed by using the PE Annexin V Apoptosis Detection Kit I (KeyGEN, KGA108). According to the manufacturer's instructions, indicated treated cells were collected and washed with PBS and suspended in the Annexin-V binding solution, and 150 μL Annexin-V antibody,1.5 μL of PI at 1 mg/mL in binding buffer was added and incubated for 15 min at room temperature in the dark. After washing with the Annexin-V Binding Buffer, positive Annexin-V staining was visualized under a fluorescence microscope equipped with a filter for fluorescein isothiocyanate (excitation: 490 nm, emission: 525 nm), and PI staining was assessed with the filter for Texas red (excitation: 570 nm, emission: 610 nm).
2.9. MTT cell viability assay
Indicated HEY or SKOV3 cells (2 × 103 cells per plate) in 48-well plates were transfected appropriate siRNAs. After 48 h, cells were further treated for 24 h with CDDP (5 μM) or BIX-01294 (5 μM), then stained with 100 μL sterile 3-(4, 5- Dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) dye (0.5 mg/mL, Sigma) for 4 h at 37 °C. The culture medium was removed and 150 μL of dimethyl sulphoxide (DMSO, Sigma-Aldrich, D8418) was added. The absorbance was measured at 570 nm, with 655 nm as the reference wavelength. Dose-response curves were plotted on a semilog scale as the percentage of the control cell number, which was obtained from the sample with no drug exposure. The IC50 values were calculated using the GraphPad Prism® 5 (Version 5.01, GraphPad Software, Inc., USA).
2.10. Determination of intercellular, DNA-bound and tumor platinum
The assays used to determine the intercellular platinum was conducted in accordance with our previous published paper [35]. Briefly, indicated cells were at 37°C in the presence of CISPLATIN (5 μM) for 4 h. Cells were centrifuged and then digested overnight at 60°C with 1 M benzethonium hydroxide (Sigma-Aldrich, 498- 77-1). The platinum amount was determined by flame atomic absorption spectroscopy (Varian) and protein concentrations were determined using a BCA Protein Assay Reagent Kit (Pierce Biotechnology, #23225).
For DNA-bound and tumor platinum, high-molecular-weight DNA of indicated cells incubated with CISPLATIN (5 μM) was isolated from cell pellets according to standard procedures. Briefly, the pellets were lysed and treated with proteinase K (1 mg/mI) at 37°C for 1 h and then at 50°C for 2 h. The DNA content was extracted and were assessed by spectrophotometry at 260 and 280 nm, and the amount of platinum in the sample was determined by flame atomic absorption spectroscopy.
For tumor platinum, tumors were harvested and digested and the amount of platinum in the sample was determined directly by flame atomic absorption spectroscopy.
2.11. Xenografted tumor models
Female BALB/c nude mice were purchased from Vitalriver (Beijing, China) and housed under SPF level conditions. In the intra-peritoneal tumor model, the indicated cells stably expressing luciferase (1 × 106 cells) were intra-peritoneally injected into female nude mice. When the luminescence signal monitored by living image software reached 2 × 107 p/sec/cm2/sr, the mice were intra-peritoneally treated with either a vehicle (control), CDDP (5 mg/kg), or combination of CDDP (5 mg/kg) and BIX- 01294 (5 mg/kg) twice times per week (as per cycle) for up to six weeks. Six weeks after the drug administration, mice were sacrificed and xenograft tumors were harvest when moribund as determined by an observer who was blinded to the treatment. The tumors were excised and embedded in paraffin for subsequent experiments. All the in vivo experiments in this study were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. Serial 4.0 μm sections were cut and subjected to immunohistochemical (IHC) and hemotoxylin and eosin (H&E) staining. After deparaffinization, sections were H&E-stained with Mayer's hematoxylin solution, or IHC-stained using antibodies of ZNF711 (1:200; abcam, ab254776), PCNA (1:500, BD, 610665), or stained with TUNEL (KeyGEN, KGA700) according to manufacturer's protocols. The images were captured by the AxioVision Rel.4.6 computerized image analysis system (Carl Zeiss). All of the animal procedures were approved by the Sun Yat-sen University Animal Care Committee.
2.12. Chromatin immunoprecipitation (ChIP) assay
In accordance with the manufacturer's instructions, ChIP assay was performed by using a ChIP assay kit (Upstate/Millipore, Billerica, MA). Briefly, the indicated HEY cells were grown to 70%–80% confluence on a 100 mm culture dish and treated with 1% formaldehyde for 10 min, which were collected and sonicated to shear the DNA into small, uniform fragments. Equal aliquots of chromatin supernatants were immuno-precipitated overnight at 4°C using ZNF711 (ab254776, Abcam), or anti-JHDM2A (ab191389, Abcam), anti-Flag (F3165, Sigma), anti-Pol II (Millipore, 07-1802), anti-EHMT2 (ab185050, Abcam), anti-H3K9me2 (ab1220, Abcam), anti- H3K9me1 (ab176880, Abcam), anti-H3K27me3 (ab6002, Abcam), anti-H4R17me1 (PTM-697, PTM biolabs, Chicago, IL, USA), antibodies, or an anti-anti-IgG antibody (a negative control, Millipore, Billerica, MA) respectively, with protein G magnetic beads (10003D, Invitrogen). A total of 2 μg of each antibody was used per 107 cells for each ChIP assay. The cross-linked protein/DNA complexes were collected by magnetic pull down, and subsequently eluted from beads using elution buffer. After reverse cross-linking the protein/DNA complexes to free DNA, the enriched DNA fragments was conducted by PCR assays by specific primers. All ChIP primers used in this study are listed in Supplementary Table S3.
2.13. Streptavidin affinity purification of dCas9-captured DNA and proteins
Streptavidin affinity purification of dCas9-Captured DNA and proteins was performed following the published protocol [36]. Briefly, ZNF711-overexpressing and vector control HEY cells (5 × 107) were transfected with an FB-dCas9 plasmid and SLC31A1 promoter sgRNA was harvested following treatment with 1% formaldehyde to cross-link the proteins to DNA. The cells were centrifuged to isolate the nuclei. Nuclei cell lysates were lysed and sonicated to shear the chromatin fragments. Streptavidin T1 Dyna-beads (Thermo-Fisher Scientific) was used for incubating the cell lysates. The ChIP DNA was purified by QIA quick Spin columns (QIAGEN) and proteins were separated by SDS-PAGE gel by immunoblotting assays, which were subsequently analyzed by IP-MS analysis (Shanghai Applied Protein Technology Co.Ltd.).
2.14. Luciferase reporter assay
Briefly, the reporter plasmids containing the promoter region of human SLC31A1 were sub-cloned into the pGL3-Control vector luciferase reporter (Promega, USA), respectively. Twenty-four hours after EOC cells being seeded in 48-well plates (3 × 103 cells/well), Luciferase reporter plasmids (100 ng) with 1 ng of pRL-TK Renilla plasmid (E6241, Promega) were transfected into EOC cells using Lipofectamine 3000 (L3000001, Life Technologies) in accordance with the manufacturer's instructions.
The luciferase and Renilla signals in indicated cells were determined 48 h after transfection using a Dual Luciferase Reporter Assay Kit (E1910, Promega).
2.15. Methylation-Specific PCR (MSP)
The Methylation-Specific PCR (MSP) has been conducted according to the according to the previous published papers [37,38]. Briefly, DNA was extracted using the Genomic DNA Kit (51304, Qiagen) and then bisulphite-modified by EZ DNA Methylation Kit (D5001, Zymo Research) according to manufacturer's protocol. PCR products were generated from separate PCR reactions using three sets of primers, and each set contains two pairs of primers specific for methylated (M) or unmethylated (U) DNA template, respectively. The PCR products were then electrophoresed on agarose gel stained with ethidium bromide. All primer sequences were listed in Supplementary Table S3.
2.16. 5-aza-20-deoxycytidine treatment
DNA methyltransferase inhibitor 5-aza-20-deoxycytidine (5AZA; A3656, Sigma) was firstly dissolved in 50% acetic acid at 100 mmol/L. Cells were treated with 5- AZA (5 mmol/L) or vehicle control (50% acetic acid) for 72 h to achieve demethylation. RNA was extracted and subsequently subjected to expression analysis of SLC31A1.
2.17. Statistics
All the data analysis was conducted by using SPSS 21.0 statistical software (IBM, Armonk, NY, USA). Statistical tests for data analysis included a Fisher's exact test, log-rank test, Chi-square test, Student's two-tailed t-test, one-way ANOVA, Mann- Whitney test and Kruskal-Wallis test. The data with normal distribution were compared and analyzed using unpaired t-test or one-way ANOVA followed by Bonferroni's multiple comparisons test. The data with skewed distributions were tested using Mann-Whitney or Kruskal-Wallis tests. Pairwise comparisons among groups were done with Bonferroni's correction. Bivariate correlations between the study variables were calculated by Spearman's rank correlation coefficients. Survival curves of EOC patients or tumor-bearing mice were plotted according to the Kaplan- Meier method and compared using a log-rank test. The significance of each of the variables on the survival time was analyzed using uni-variate and multivariate Cox regression analyses. Data were presented as the mean ± SD. P values of 0.05 or less were considered to be statistically significant.
2.18. Role of funding source
The Funders didn't have any role in study design, data collection, data analyses, interpretation, or writing of report.
3. Results
3.1. ZNF711 is negatively correlated with CDDP resistance in EOC
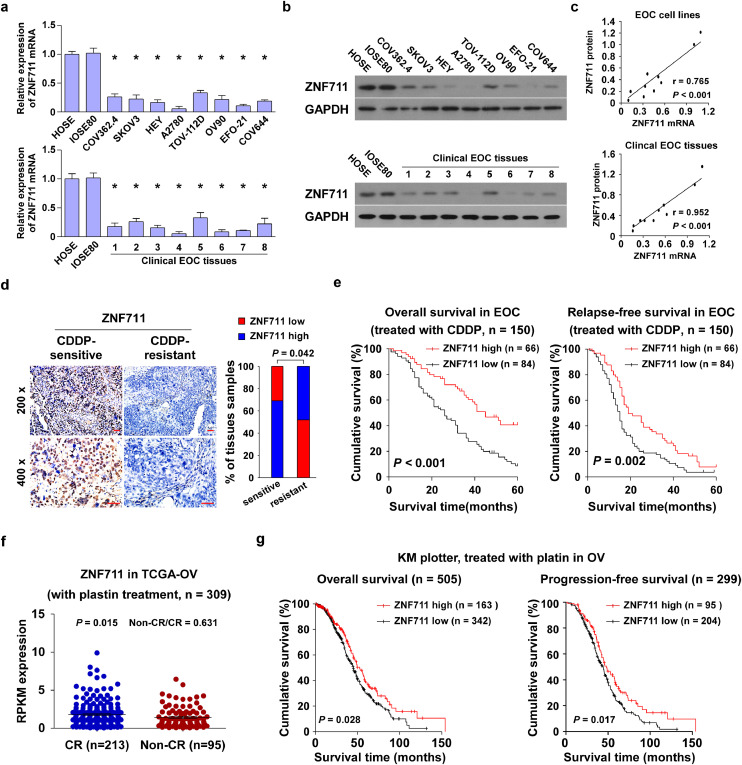
Zinc-finger proteins are vital transcription factors characterized as co-factors and recruiters of histone modifiers, which exerts great impact in the tumorigenesis and progression of multiple cancer types. [[30], [31], [32]]. To identify the essential biological role of ZNF711, a newly discovered zinc-finger protein, in the development of EOC progression, real-time PCR and immuno-blotting assays indicated that both the level of ZNF711 mRNA and protein expression in ovarian cancer cell lines and clinical tissues were lower than those in human ovarian surface epithelial (HOSE) cells and normal ovarian (IOSE80) cells (Fig. 1a and b). Additionally, the expression level of ZNF711 mRNA and protein was positively correlated in EOC cell lines and clinical EOC tissues (r = 0.765, p < 0.001; r = 0.952, p < 0.001; Pearson correlation analysis) (Fig. 1c). Analysis of the Gene Expression Profile Interactive Analysis (GEPIA) database further indicated that the level of ZNF711 expression was significantly down-regulated in ovarian cancer tissues compared to adjacent normal ovarian tissues (Supplemental Fig.1a; n = 514, p < 0.001, unpaired t-test). More importantly, the expression of ZNF711 was correlated with a shorter overall survival of ovarian cancer patients from GEPIA and the Cancer Genome Atlas (TCGA) database (Supplemental Fig.1b and c; n = 423, p = 0.023; n = 557, p = 0.034; Kaplan–Meier analysis).
Fig. 1.
ZNF711 is negatively correlated with CDDP resistance in EOC. (a) Real- time PCR analysis revealed that the level of ZNF711 mRNA was down regulated in EOC cells or tissues compared with the normal ovarian cells; Each bar shown in the figure represents the mean ± SD of three independent experiments. *p < 0.05 (one- way ANOVA followed by Bonferroni's multiple comparisons test). (b) An immuno blotting (IB) assay showed that ZNF711 protein expression was down regulated in EOC cells or tissues compared with the normal ovarian cells. (c) The correlation analysis of the mRNA and protein expression of ZNF711 in EOC cell lines and clinical EOC tissues. (d) An immunohistochemistry (IHC) assay was performed in CDDP-sensitive and CDDP-resistant epithelial ovarian cancer (EOC) tissues using an anti-ZNF711 antibody (200 × , upper; 400 × , lower, Scale bars, 50 μm.). (e) Kaplan-Meier survival analysis of the association of ZNF711 protein expression with overall and relapse-free in EOC patients treated with CDDP. (f) The level of ZNF711 mRNA expression was down regulated in patients with non-CR (partial response, progression and stable) versus CR (complete response) upon CDDP treatment. (g) Kaplan-Meier analysis of the overall and relapse-free survival of the indicated ovarian cancer patients, the cut-off used to define low and high tumors was auto selection of best cut-off.
Furthermore, the Gene Set Enrichment Analysis (GSEA) showed that the level of ZNF711 expression was inversely correlated with CDDP resistance (Supplemental Fig.1d, p < 0.05; permutation test). Furthermore, the cell survival rates of EOC cell lines in response to CDDP have been studied and we found that the expression level of ZNF711 was positively related with CISPLATIN sensitivity in EOC cell lines (Supplemental Fig.1e; r = -490.748, p < 0.001; Pearson correlation analysis). We also performed an immunohistochemistry (IHC) assay in 150 paraffin-embedded, archived EOC tissues treated with CDDP (Fig. 1d and e, Supplemental Table S1). As shown in Fig. 1d and e, the average staining intensity of ZNF711 in CDDP-resistant EOC tissues was noticeably weaker than that in the CDDP-sensitive EOC tissues, which was also positively related with the overall and relapse-free survival time of EOC patients following CDDP treatment (Fig. 1d and e, p = 0.042; p < 0.001; p = 0.002; Kaplan–Meier analysis). Importantly, the uni-variate and multivariate analyses verified that ZNF711 may serve as a prognostic factor in ovarian cancer, which was positively related with the chemo-resistance and survival of EOC patients (Supplemental Table S1). Consistently, in the cohort of EOC patients treated with CDDP in the TCGA dataset, the level of ZNF711 mRNA expression was significantly higher in patients who exhibited a complete response (CR) compared to those with a Non-CR (partial response, stable disease, and progression) after the first course of platinum-based treatment [39] (Fig. 1f, p = 0.015; unpaired t-test). Moreover, EOC patients with lower ZNF711 expression exerts a shorter overall and progression-free survival compared to those with a higher ZNF711 expression (Fig. 1g, p = 0.028; p = 0.017; Kaplan–Meier analysis). Taken together, these results indicate that ZNF711 is down regulated in EOC and negatively correlated with CDDP resistance.
3.2. ZNF711 modulates the CDDP uptake and CDDP resistance in EOC
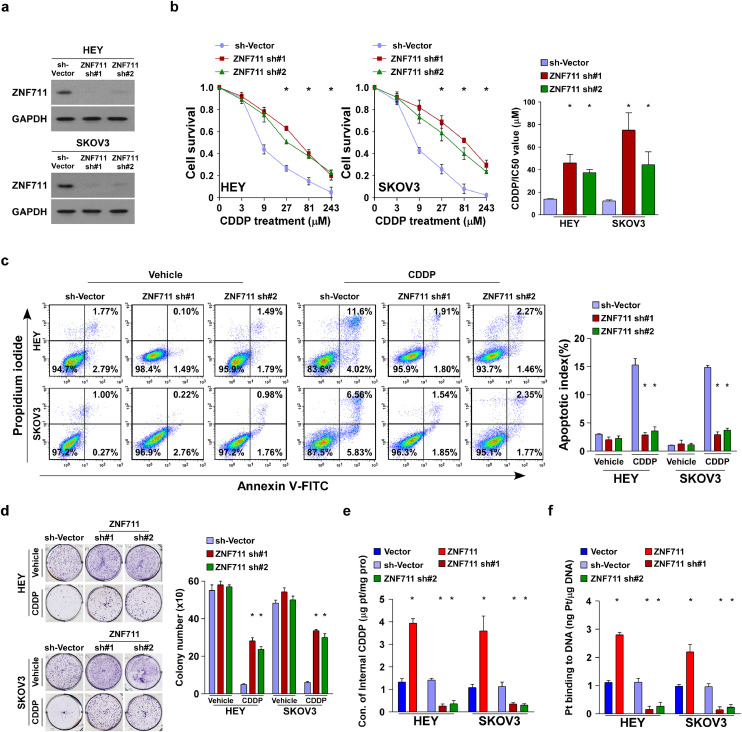
To validate the effect on ZNF711-associated chemo-resistance in EOC, both the HEY and SKOV3 ovarian cancer cell lines stably expressing ZNF711 or ZNF711 shRNAs were established, respectively (Fig. 2a and Supplemental Fig.2a). We found that knockdown of ZNF711 significantly promoted, while overexpression of ZNF711 inhibited, the IC50 value of CDDP in ovarian cancer cells (Fig. 2b and Supplemental Fig.2c). Additionally, ZNF711 down regulation drastically decreased the apoptotic percentage of EOC cells and increased the colony numbers in EOC cells. (Fig. 2c and d). In the ZNF711-silenced EOC cell lines, much higher doses of CDDP (10 μM and 50 μM) were tried and we found that the cell survival percentage of EOC cell lines were much higher in ZNF711-silenced cells than that in vector control cells (Supplemental Fig.2b). In accordance with this finding, ZNF711 up-regulation increased the CDDP-induced apoptotic death and decreased colony formation in ovarian cancer cells (Supplemental Fig.2d-e). Moreover, two primary cell lines OV-1 and OV-2 have been isolated from clinical EOC patient tissues according to our previous published paper [35]. Consistent with the results obtained from EOC cell lines, overexpression of ZNF711 decreased, while inhibition of ZNF711 increased the IC50 value of CISPLATIN (CDDP) both in OV-1 and OV-2 cells (Supplemental Fig.2f- g). These results confirm that the knockdown of ZNF711 promoted, whereas ZNF711 overexpression weakened, the CDDP resistance of ovarian cancer cells to CDDP in vitro.
Fig. 2.
ZNF711 down regulation inhibits CDDP uptake and leads to CDDP resistance in EOC in vitro. (a) Immuno-blotting (IB) assays were performed to verify ZNF711 expression in the indicated stable HEY and SKOV3 cell lines. (b) An MTT cell viability assay (left) and half-maximal inhibitory concentration (IC50) value of CDDP (right) in HEY and SKOV3 cells stably expressing ZNF711 sh#1 or ZNF711 sh#2 compared to the cells expressing SH-vector control. (c) FACS analysis of Annexin V staining (left) and quantification (right) of indicated HEY and SKOV3 stable cell lines treated with vehicle or CDDP (5 μM, 24 h). (d) Representative images (left) and quantification (right) of colony number of the indicated cells treated with vehicle or CDDP (5 μM). (e) Content of intercellular CDDP in the indicated HEY and SKOV3 cells. (f) The quantification of DNA-bound CDDP (Pt) in the indicated HEY and SKOV3 cells. Each bar shown in the figure represents the mean ± SD of three independent experiments. *p < 0.05 (Kruskal-Wallis test with Bonferroni's correction).
Previous reports have demonstrated that decreased influx velocity represents a major cause of CDDP resistance in cancer treatment [9]. Of note, ZNF711-silenced ovarian cancer cells exerted much lower concentrations of CDDP while ZNF711- up-regulated cells accumulated higher concentrations of CDDP compared with the vector control cells/tumors, respectively (Fig. 2e and Supplemental Fig.2h). The content of genomic DNA-bound CDDP was decreased in ZNF711-silenced tumors/cells, and increased in ZNF711-overpressing tumors/cells (Fig. 2f and Supplemental Fig.2i). Therefore, ZNF711 down regulation impairs CDDP influx and confers CDDP chemo resistance in EOC.
3.3. Silencing ZNF711 augments the CDDP resistance of EOC cells in vivo
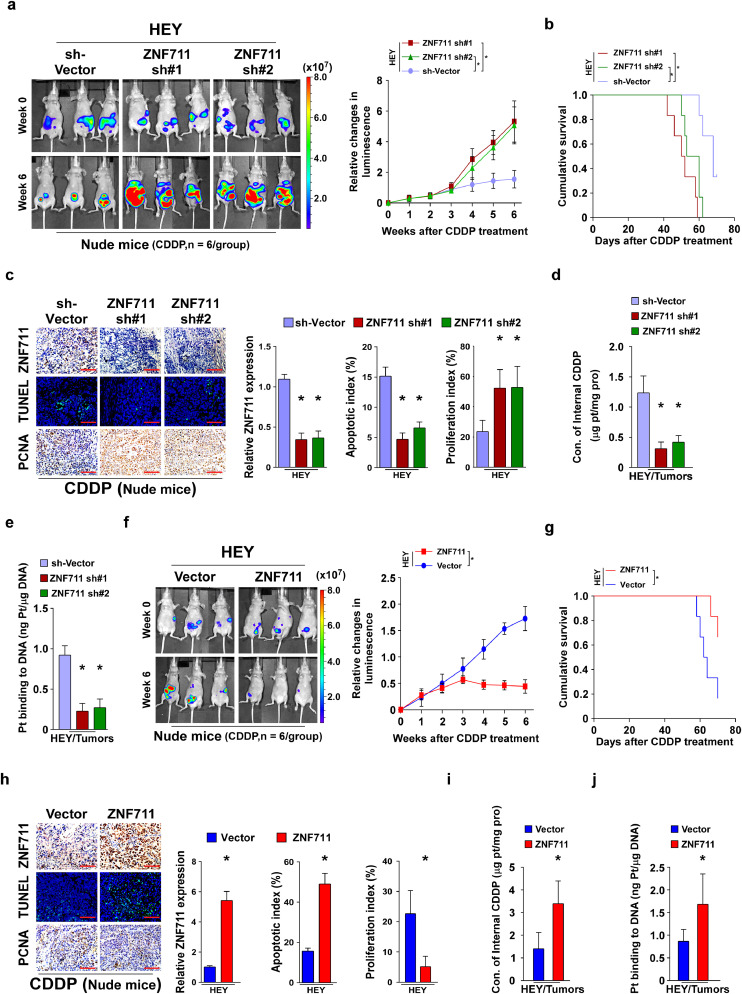
Consistent with the in vitro results, the HEY/sh-vector, /ZNF711 sh#1, and /ZNF711 sh#2 cells were intra-peritoneally injected into female nude mice and an intra-peritoneal tumor model has been established. When the luminescence signal of the xenograft tumors reached 2 × 107 p/sec/cm2/sr, each group of tumor-bearing mice were separated into two groups, which were intra-peritoneally injected with PBS or CDDP (5 mg/kg, twice a week for six weeks), respectively. After six weeks of treatment, PBS-treated xenograft tumors formed by HEY/ZNF711 sh#1, /ZNF711 sh#2 showed similar tumor growth kinetic compared with SH-vector cells, which suggests that the expression of ZNF711 did not affect tumor growth (Supplemental Fig.3a). However, CDDP treatment significantly promoted the tumor growth curve of tumors formed by HEY/ZNF711 sh#1 and HEY/ZNF711 sh#2 cells compared with that formed by HEY/sh-vector cells (Fig. 3a). More importantly, ZNF711 suppression resulted in a shorter overall survival of tumor-bearing mice treated with six weeks/cycles of CDDP therapy (Fig. 3b). Consistently, tumors formed by the ZNF711-silenced HEY cells exhibited stronger PCNA signals and fewer terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells compared with the HEY/sh-vector cells (Fig. 3c). Moreover, the content of internal CDDP and genomic DNA-bound CDDP was decreased in ZNF711-silenced tumors (Fig. 3d and e). Taken together, the ZNF711 knockdown inhibited the CDDP uptake and augmented the CDDP resistance in EOC cancer cells.
Fig. 3.
ZNF711 modulates CDDP resistance in EOC in vivo. (a) A intra-peritoneal tumor model was established in nude mice by inoculating the indicated stable HEY cell lines, and representative images of tumor-bearing nude mice were presented at the initial time of CDDP treatment (upper) and six weeks after CDDP treatment (lower) (left panel, n = 6/group). Relative changes in the bioluminescence signal of intra-peritoneal tumors in nude mice upon CDDP chemotherapy at the indicated time points (right panel). (b) Kaplan-Meier survival of tumor-bearing mice after CDDP treatment. (c) IHC staining of ZNF711 and PCNA and TUNNEL staining (left) and quantification (right) of ZNF711 expression, apoptotic rate, and proliferation index in the indicated xenograft tumors. Scale bars, 50 μm. (d) Content of intercellular CDDP in the indicated xenograft tumors. (e) The quantification of DNA-bound CDDP (Pt) in the indicated xenograft tumors. (f) Representative images of tumor-bearing nude mice formed by HEY/vector and HEY/ZNF711 cells (left) and relative changes in the bioluminescence signal upon CDDP chemotherapy at the indicated time points (right). (g) Overall survival of tumor-bearing mice after CDDP treatment. (h) IHC staining of ZNF711 and PCNA and TUNNEL staining (left) and quantification (right) of ZNF711 expression, apoptotic rate, and proliferation index in the indicated xenograft tumors. Scale bars, 50 μm. (i-j) The content of internal CDDP (i) and DNA-binding CDDP (j) in indicated tumors. Each bar shown in the figure represents the mean ± SD of three independent experiments. n = 6, *p < 0.05 (Fig. 3a-e using one-way ANOVA with Bonferroni's correction; Fig. 3f-j using Unpaired t test).
The effect of ZNF711 overexpression on CDDP resistance was further investigated in HEY cells (Fig. 3f). We found that ZNF711 restoration drastically inhibited tumor growth and prolonged the survival time of tumor-bearing mice following CDDP treatment, while PBS-treated xenograft tumors growth changed weakly (Fig. 3g and Supplemental Fig.3b). Moreover, ZNF711 up-regulation substantially enhanced the killing effect of CDDP in HEY cells in mice, as indicated by decreased PCNA signals and increased TUNEL-positive cells (Fig. 3h). Further, ZNF711 up-regulation augmented the CDDP uptake in xenograft tumors (Fig. 3i and j). Thus, ZNF711 overexpression could promote CDDP uptake restore the CDDP sensitivity in EOC cells in vivo.
3.4. Clinical relevance of ZNF711 and CDDP resistance in EOC
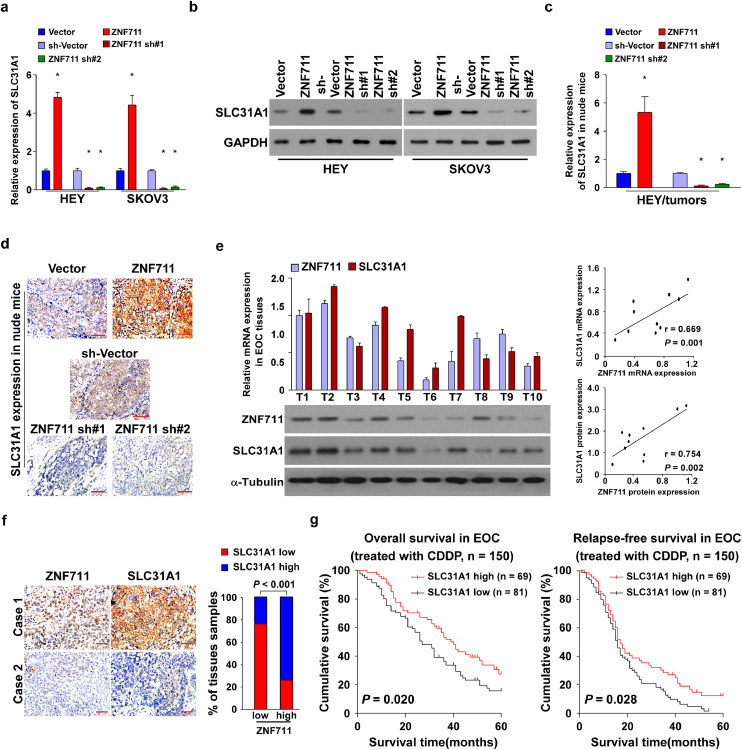
Ample evidence supports the biological and clinical involvement of the drug importer, SLC31A1, in CDDP resistance of cancer cells [15, 16]. We tested the protein expression of SLC31A1 in EOC cell lines, which showed positive correlation with the expression of ZNF711 (Supplemental Fig. 4a; r = 0.750, p = 0.001). We further examined the correlation of the mRNA expression levels of the ZNF711 and SLC3A1 in the GSEA and TCGA data base. Statistical analysis revealed that the mRNA expression levels of ZNF711 and SLC31A1 is positively correlated in the GSE28739 (n = 50; r = 0.413, p = 0.003; Pearson correlation analysis), GSE23553 (n = 56; r = 0.438, p < 0.001; Pearson correlation analysis) and TCGA data base (n = 419; r = 0.22, p < 0.001; Pearson correlation analysis) (Supplemental Fig.4b).
Moreover, both the mRNA and protein expression of SLC31A1 were elevated in ZNF711-upregulated EOC cell lines and primary tumor cells, and decreased in the ZNF711-downregulated cells compared with the corresponding control cells (Fig. 4a and b, Supplemental Fig.4c and d). In addition, the mRNA and protein expression of SLC31A1 were elevated in ZNF711-overexpressing tumors, while decreased in ZNF711-silenced tumors (Fig. 4c and d). Importantly, SLC31A1 down regulation could reverse the inhibitory effect of CDDP resistance in ZNF711-upregulated cells, as indicated by the decreased IC50 value and increased CDDP concentration (Supplemental Fig.4e and f). These results suggested that ZNF711 modulate CDDP resistance by regulating SLC31A1 expression in EOC cells.
Fig. 4.
The clinical relevance of ZNF711 and SLC31A1 in EOC tissues. (a-b) The mRNA (a) and protein (b) expression of SLC31A1 in indicated cells with different expression levels of ZNF711. *p < 0.05 (one-way ANOVA with Bonferroni's correction). (c-d) SLC31A1 mRNA and protein expression in the indicated xenograft tumors. *p < 0.05 (one-way ANOVA with Bonferroni's correction). (e) Analysis of expression (left) and correlation (right) of mRNA and protein expression level of ZNF711 and SLC31A1 in 10 freshly collected EOC samples. (f) Representative image of ZNF711 and SLC31A1 expression in EOC tissues. Scale bars, 50 μm. (g) KM-plotter analysis was conducted to confirm the negative relation between SLC31A1 expression and survival of EOC patients treated with platin. Each bar shown in the figure represents the mean ± SD of three independent experiments.
To determine whether the ZNF711/SLC31A1 axis was clinically relevant in the EOC specimens, the mRNA and protein expressions of both ZNF711 and SLC31A1 were evaluated. The mRNA and protein expression of ZNF711 and SLC31A1 was positively correlated in 10 freshly collected EOC tissues (Fig. 4e; r = 0.669, p = 0.001; r = 0.754, p = 0.002; Pearson correlation analysis). Furthermore, the GSEA assays revealed that SLC31A1 expression was negatively correlated with CISPLATIN resistance (Supplemental Fig.4g). According to the KM-plotter analysis, ovarian cancer patients with lower SLC31A1 expression were associated with a shorter overall and relapse- free survival time compared to those exhibiting higher SLC31A1 expression (Supplemental Fig.4h and i; p = 0.005, p < 0.001; p = 0.013, p < 0.001; Kaplan– Meier analysis). Consistent with these findings, the IHC analysis revealed that the level of ZNF711 expression was positively correlated with SLC31A1 in 150 clinical EOC tissues (Fig. 4f; p < 0.001; chi-square test). In addition, the level of SLC31A1 expression was positively correlated with the overall and relapse-free survival of EOC patients (Fig. 4g, p = 0.020, p = 0.028; Kaplan–Meier analysis). These results strongly suggest that knockdown of ZNF711downregulates the expression of SLC31A1, which promotes CDDP resistance and ultimately leads to poor survival of patients with ovarian cancer.
3.5. ZNF711 up-regulates SLC31A1 promoter activity in EOC
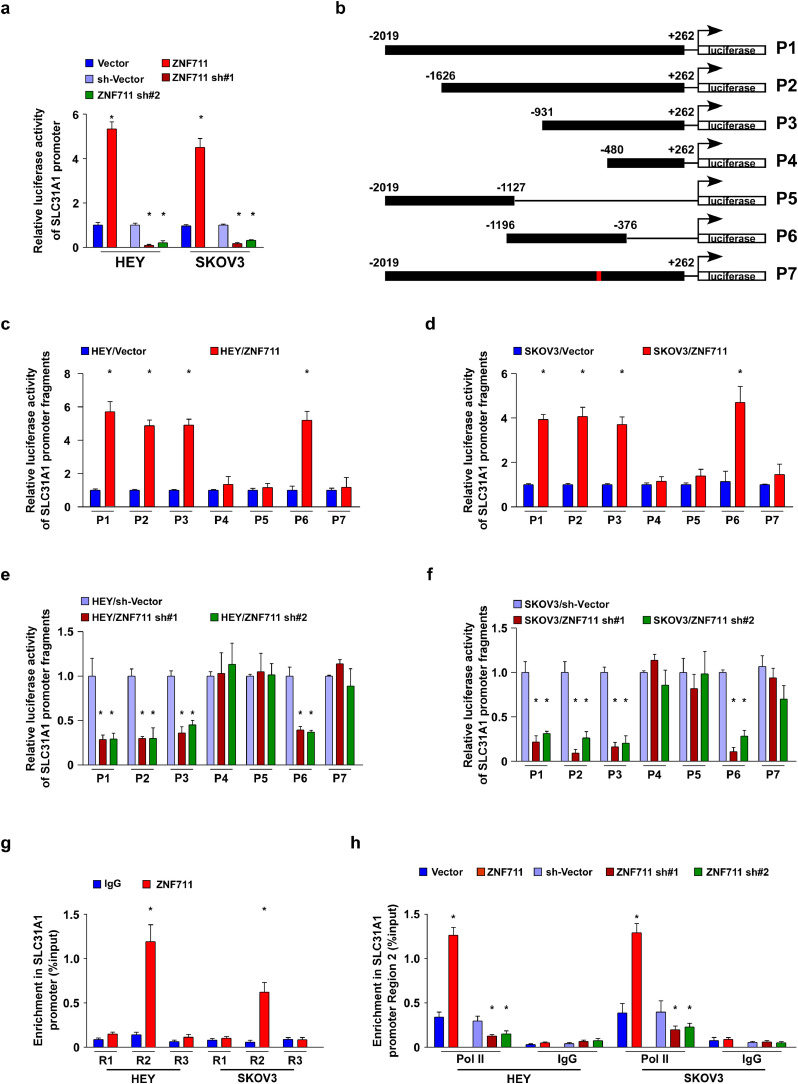
Interestingly, according to the JASPAR and ConSite database, three ZNF711 binding sites were predicted in the promoter region of SLC31A1 (Supplemental Fig.5a). To elucidate the regulatory mechanism of ZNF711, luciferase reporter assays were performed and we found that up-regulation of ZNF711 enhanced, whereas down-regulation of ZNF711 inhibited the luciferase activity of the SLC31A1 promoter in both HEY and SKOV3 cells, as well as in OV-1 and OV-2 primary tumor cells (Fig. 5a and Supplemental Fig.5b). Furthermore, six fragments of the SLC31A1 promoter were constructed and inserted into the luciferase reporter pGL3 vector, respectively, including P1(nucleotides -2019 to +262), P2 (nucleotides -1626 to +262), P3 (nucleotides -931 to +262), P4 (nucleotides -480 to +262), P5 (nucleotides - 2019 to -1127) and P6 (nucleotides -1196 to -376) (Fig. 5b). As shown in Fig. 5c-f, the luciferase activity of fragments P1, P2, P3, and P6 were significantly increased in ZNF711-upregulated ovarian cancer cells but reduced in the ZNF711-silenced cells.
Fig. 5.
ZNF711 regulates SLC31A1 promoter activity in EOC cells. (a) The SLC31A1 promoter luciferase reporter plasmids and Renilla pRL-TK plasmids were transfected into the indicated empty vector-transduced, ZNF711-transduced, control shRNA-transduced, and the ZNF711-shRNA transduced HEY and SKOV3 cells. After 48 h, the luciferase signal was examined and presented. (b) Schematic illustration of the P1-P7 cloned fragments of the human SLC31A1 promoter. (c and d) Luciferase activity of the SLC31A1 promoter fragments in control vector cells and ZNF711-overexpressing EOC cells. (e and f). Luciferase activity of SLC31A1 promoter fragments in shRNA control vector cells and ZNF711 knockdown EOC cells. (g) ChIP assays were performed using an anti-Flag antibody to screen three predicted ZNF711-binding sites on the SLC31A1 promoter. IgG was used as a negative control. (h) ChIP assay showing the enrichment of Pol II on region 2 in the SLC31A1 promoter in the indicated HEY and SKOV3 cells. Each bar shown in the figure represents the mean ± SD of three independent experiments. *p < 0.05 (unpaired t-test or one-way ANOVA with Bonferroni's correction).
However, the activity of the luciferase reporter inserted with fragments P4 or P5 remained largely unaffected (Fig. 5c-f). Moreover, the mutation of nucleotides -533 to -530 abrogated the regulatory effect of ZNF711 on the SCL31A1 promoter, indicating a critical role of nucleotides -533 to -530 in ZNF711-mediated modulation (Fig. 5c-f). Next, a chromatin immuno-precipitation (ChIP) assay was conducted, and showed that ZNF711 bound to region 2 within the SLC31A1 promoter sequence (Fig. 5g). We also found that RNA polymerase II (Pol II) was recruited to the SLC31A1 promoter in ZNF711-upregulated ovarian cancer cells (Fig. 5h). The above-mentioned results indicate that ZNF711 transcriptionally up-regulates SLC31A1 expression via binding to the SLC31A1 promoter.
3.6. Histone demethylase JHDM2A is recruited to the SLC31A1 promoter by ZNF711
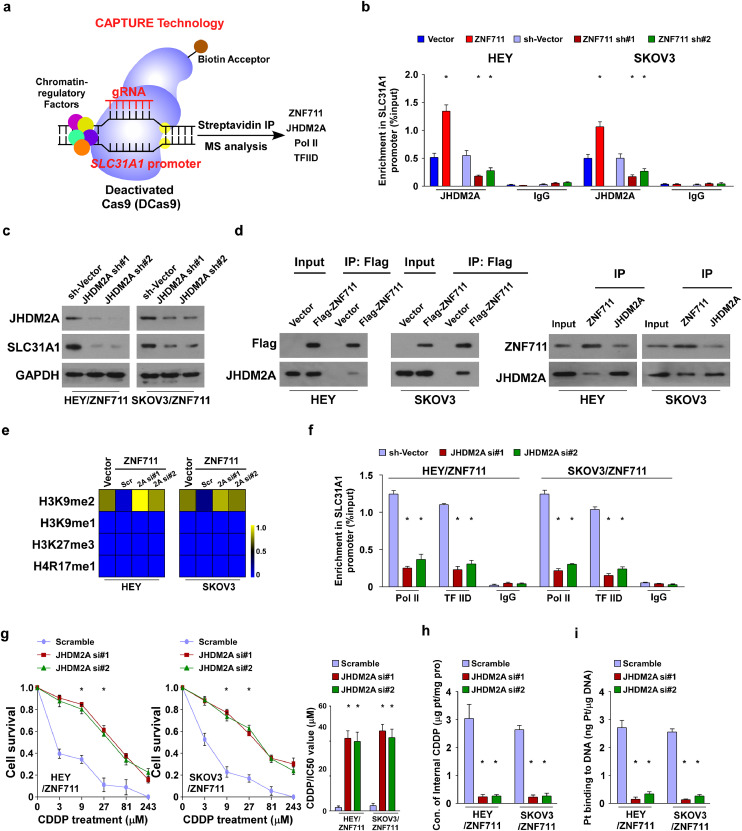
To further investigate the molecular regulation of SLC31A1, a CRISPR affinity purification in situ of regulatory elements (CAPTURE) approach was used and subsequent IP-mass data has been conducted. As shown in Supplemental Table S4, total 15 proteins with Log2 (fold change) > 1were identified by CAPTURE/quantitative MS approach (Supplemental Table S4). Further, eight proteins with more than five peptides identified by MS were selected for further examination. We found that along with the essential transcriptional activator POLR2A and TBP, only silencing ZNF711 or JHDM2A resulted in significant reduction of SLC31A1 expression in HEY cells (Supplemental Fig.6a). This result suggested that JHDM2A might be a transcriptional co-factor that contributes to the association of ZNF711 with promoters of SLC31A1 in EOC cells, and showed that ZNF711 might form a complex with histone demethylase JHDM2A on the SLC31A1 promoter in EOC cells, which also interacted with Pol II and TFIID (Fig. 6a). Consistent with this finding, JHDM2A recruitment in the SLC31A1 promoter was significantly enhanced in ZNF711-upregulated cells but was substantially decreased in ZNF711-silenced cells, both in EOC cell lines and primary tumor cells (Fig. 6b and Supplemental Fig.6b). Furthermore, a knockdown of JHDM2A by specific shRNA reversed the promoting effect of SLC31A1 transcription induced by ZNF711 overexpression in ovarian cancer cells (Supplemental Fig.6c-h and Fig.6c). Moreover, a co- immuno-precipitation (co-IP) assay was performed, which confirmed the interaction between ZNF711 and JHDM2A in HEY and SKOV3 cells, both endogenously and exogenously (Fig. 6d). To further unveil how ZNF711 modulates the transcriptional activity of SCL31A1, a systematic chromatin immuno-precipitation (ChIP) assay was performed to identify the post-translational modifications of histone proteins. We found that a knockdown of JHDM2A abrogated H3K9me2 enrichment on the SLC31A1 promoter in ZNF711-transduced cells, whereas the enrichment of H3K9me1, H3K27me3, and H4R17me1 were rarely altered (Fig. 6e), H3K27me3 and H4R17me1 have been reported as inhibitory marker and active marker of histone modulation, respectively [40, 41], which have been chosen for negative control for enrichment assays. Moreover, the enrichment of Pol II and TFIID on the SLC31A1 promoter was significantly decreased in ZNF711-overexpressing EOC cells transfected with JHDM2A siRNA (Fig. 6f). As shown in Supplemental Fig.6i, methylation-specific PCR (MSP) for the SLC31A1 promoter region and electrophoresis of PCR products have been performed. The genomic DNA of OV-1 cells treatment with SssI Methyltransferase was used as positive control and water (H2O) was used as a negative control for PCR. As shown in Supplemental Fig.6i, the methylation level of SLC31A1 promoter was strongly detected in positive control, while we did not see the evidence of methylation of SLC31A1 promoter in normal ovarian epithelial cells, cancer cell lines and primary ovarian cancer cells.
Fig. 6.
ZNF711 recruits demethylase JHDM2A and activates the transcription of SLC31A1. (a) Schematic of the CRISPR CAPTURE approach following quantitative mass spectrometry to identify the trans-regulatory factors targeting the SLC31A1 promoter in EOC cells. (b) Enrichment in the SLC31A1 promoter of JHDM2A in the indicated HEY and SKOV3 cells. (c) IB assays were performed to clarify the level of JHDM2A and SLC31A1 expression in JHDM2A knockdown EOC cells compared with the control cells. (d) Co-IP assays were performed in the indicated cells using anti-Flag antibody (left), anti-ZNF711, or anti-JHDM2A antibodies in the indicated cells. (e) A ChIP assay of the enrichment of H3K9me2, H3K9me1, H3K27me3, and H4R17me1 on the SLC31A1 promoter. The range 0-1 was defined by the fold change (Log2) of the enrichment level in SLC31A1 promoter in ZNF711-overexpressing EOC cells transfected with JHDM2A siRNAs or scramble controls. (f) ChIP assay revealed that JHDM2A downregulation reduced the binding of Pol II and TFIID on the SLC31A1 promoter in ZNF711-overexpressing cells. (g) The IC50 value of CDDP in ZNF711-overexpressing EOC cells transfected with JHDM2A siRNAs or scramble controls. (h-i) The concentration of intracellular CDDP (h) and DNA-binding CDDP (i) in the indicated cells. Each bar shown in the figure represents the mean ± SD of three independent experiments. *p < 0.05 (one-way ANOVA with Bonferroni's correction).
Furthermore, treatment with the DNA methyltransferase inhibitor 5-aza-dC (5-AZA) barely restored the expression of SLC31A1 in EOC cell lines and primary ovarian cancer cells compared with normal ovarian cells (Supplemental Fig.6j). Thus, this result suggests that the DNA methylation of SLC31A1 promoter might not be the decisive factor which induced the down regulation of SLC31A1 in EOC cells.
Importantly, the CDDP sensitivity mediated by ZNF711-overexpression was abrogated by suppressing the expression of JHDM2A in EOC cells, as indicated by the increased IC50 value of CDDP (Fig. 6g). Moreover, the concentration of intercellular CDDP and DNA-binding Pt was reduced in JHDM2A-downregulated cells (Fig. 6h and i). These results suggest that demethylase JHDM2A is required for the ZNF711-induced CDDP response in EOC cells.
3.7. BIX-01294 restores CDDP sensitivity in ZNF711-silenced EOC cells
Histone methyltransferase euchromatic histone lysine methyltransferase (EHMT2) has been identified as a pivotal histone methyltransferase and correlated with cancer development [42, 43]. Interestingly, using a ChIP assay, we found that the enrichment of EHMT2 was enhanced in the SLC31A1 promoter in ZNF711-downregulated EOC cells while inhibited in ZNF711-upregulated EOC cells (Supplemental Fig.7a). Consistent with this finding, EHMT2 knockdown abrogated the suppression of SLC31A1 mediated by ZNF711 down regulation (Supplemental Fig.7b-d).
Moreover, silencing EHMT2 decreased the IC50 and increased the CDDP uptake in ZNF711-downregulating EOC cells (Supplemental Fig.7e and f).
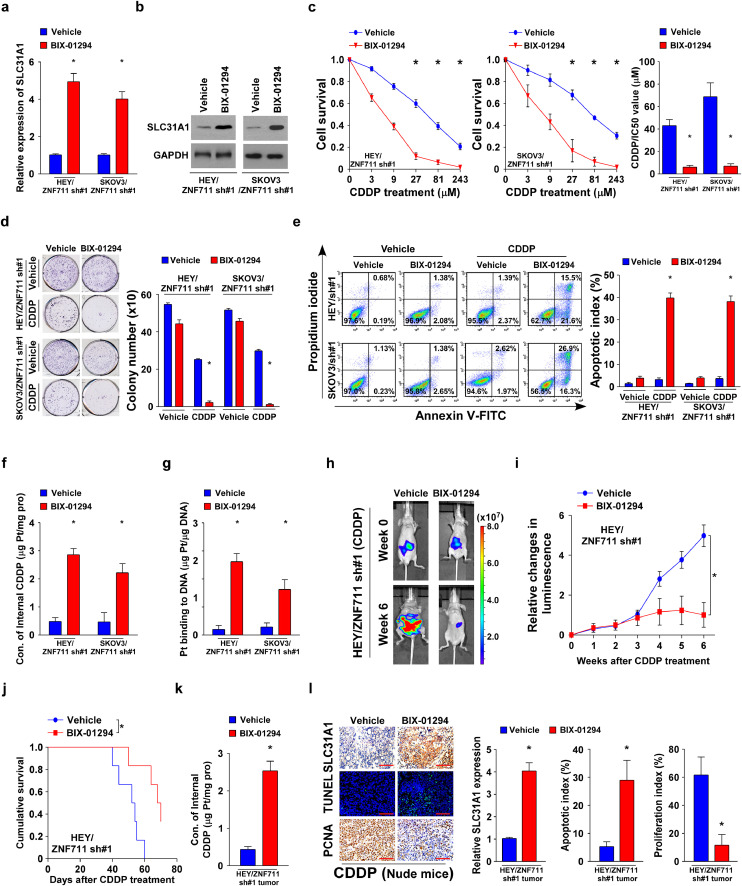
BIX-01294 is a diazepin-quinazolinamine derivative that has been recognized as a histone methyltransferase inhibitor that specifically reduces the level of H3K9me2 at the chromatin regions of the target genes [43, 44]. BIX-01294 significantly up regulated the transcriptional expression of SLC31A1 in the EOC cell lines and primary tumor cells (Fig. 7a-b and Supplemental Fig.7g-h). In line with the above- mentioned results, BIX-01294 decreased the IC50 value both in EOC cell lines and primary tumor cells (Fig. 7c and Supplemental Fig.7i). In addition, co-treatment BIX- 01294 with CDDP impaired the capability of colony formation and promoted the percentage of apoptosis in ZNF711-downregulated EOC cells (Fig. 7d and e).
Fig. 7.
Combined BIX-01294 sensitizes ZNF711-downregulated EOC cells to CDDP therapy. (a-b) SLC31A1 mRNA (a) and protein expression (b) in ZNF711- down regulated EOC cells treated with BIX-01294 or vehicle control. (c) MTT analysis (left) and IC50 value (right) of CDDP in the indicated cells. (d) Representative images (left) and quantification (right) of the number of colonies in the indicated cells treated with either the vehicle or BIX-01294. (e) FACS analysis of Annexin V staining (left) and quantification (right) of indicated stable HEY and SKOV3 cell lines treated with a vehicle or BIX-01294. (f) Intercellular CDDP content in the indicated cells. (g) The quantification of DNA-bound CDDP (Pt) in the indicated cells. (h) Representative images of the indicated nude mice treated with a vehicle or BIX-01294. (i) Relative change in the luminescence signals of the indicated xenograft tumors at the indicated time points. (j) Kaplan-Meier survival analysis of the indicated tumor-bearing mice. (k) Concentration of intercellular CDDP in indicated cells. (l) IHC staining of SLC31A1, PCNA, and TUNEL staining (left) and quantification (right) of SLC31A1 expression, apoptotic rate, and proliferation index in the indicated xenograft tumors. Scale bars, 50 μm. Each bar shown in the figure represents the mean ± SD of three independent experiments. *p < 0.05 (one-way ANOVA with Bonferroni's correction).
Furthermore, BIX01294 also promoted the CDDP uptake in ZNF711-downregulated EOC cells following CDDP treatment (Fig. 7f-g and Supplemental Fig.7j). Two EOC cell lines TOV-112D and A2780 with high or low level of ZNF711 respectively have been chosen to test the therapeutic efficacy of BIX-01294 and CDDP co- treatment. As shown in Supplemental Fig.7k-l, the expression of SLC31A1 and apoptotic index mediated by CDDP treatment was significantly augmented by BIX- 01294 co-therapy in A2780 cells, which exhibited low expression level of ZNF711. In contrast, BIX-01294 treatment only barely increased the expression level of SLC31A1 and the apoptotic percentage of CDDP-treated TOV-112D cells, which exhibited high expression level of ZNF711. Importantly, the in vivo model further supported the finding that BIX-01294 significantly reduced tumor growth and SLC31A1 expression following CDDP treatment in ZNF711-silenced cells, which prolonged the survival time of tumor-bearing mice (Fig. 7h-j). The inhibitory effect of SLC31A1 mediated by BIX-01294 in the ZNF711-downregulated tumors was further confirmed, along with a decreased proliferation rate, increasing apoptotic index, and CDDP uptake (Fig. 7k and l). These results reveal that BIX-01294 could restore the CDDP sensitivity of ZNF711-downregulated EOC cells, both in vivo and in vitro.
4. Discussion
Chemo-resistance is a major cause of therapeutic failure when treating EOC patients with CDDP [45,46]. The suppression of CDDP uptake serves as an important approach to generate CDDP resistance in multiple cancer types [[12], [13], [14]]. Unveiling the molecular mechanism underlying CDDP uptake in EOC may shed insight into the development of improved regimens for EOC treatment. In this study, we confirmed that ZNF711 down regulation exerted a substantial impact on CDDP resistance for EOC patients by suppressing SLC31A1 and inhibiting CDDP uptake. Using in vivo and in vitro assays, we further noticed that suppression of ZNF711 promoted the CDDP resistance, while overexpression of ZNF711 drastically augmented CDDP sensitivity in EOC cell lines, primary tumor cells and clinical EOC tissues. More importantly, co-treatment with BIX-01294, a histone methylation inhibitor, restored the CDDP sensitivity of ZNF711-silenced EOC cells, both in vivo and in vitro. Thus, our study uncovered the modulatory effect of CDDP uptake in EOC and might provide a potential target for improving the efficiency of CDDP treatment.
Meanwhile, because SLC31A1 assisted the influx of both CDDP and carboplatin [15], the latter is currently used as the gold-standard chemotherapeutic regimen for EOC patients. The resistance effect of ZNF711 down regulation on carboplatin therapy is currently under investigation in our laboratory.
SLC31A1 is a trans membrane protein maintaining copper homeostasis that has been widely recognized to promote the internalization of a significant fraction of CISPLATIN and its analogs in tumor cells [13,15,16]. Moreover, a negative correlation between the level of SLC31A1 expression and DDP resistance in cancer cells has been confirmed in various studies [[37], [38], [39], [40]]. In Hela cells, SLC31A1 overexpression resulted in a 2.2-fold increase in CDDP accumulation compared with the vector control cells [47]. Ishida et al. also demonstrated that the level of SLC31A1 mRNA was correlated with the quantification of DNA-CISPLATIN adducts in a cervical cancer model [14]; however, the detailed regulatory mechanism of SLC31A1 has rarely been studied. In this study, we found that the level of SLC31A1 expression in EOC patients was primarily modulated by ZNF711 binding at nucleotides -533 to -530 of the SLC31A1 promoter, which recruited histone demethylase JHDM2A and led to transcriptional activation by removal of the repressive transcription marker, H3K9me2. Furthermore, we also found that ZNF711 down regulation played an important role in repression of SLC31A1 expression in CDDP resistant EOC patients. Thus, our findings illustrate the epigenetic regulation of SLC31A1 in EOC and provide a new regulatory layer for adjusting the level of SLC31A1 in cancer treatment.
ZNF711, a newly discovered zinc-finger protein, is located at Xq21.1-q21.2 and has been associated with cognitive disability [48]. Moreover, there are 12 Zn-C2H2 domains at 383 to 755 aa of ZNF711, which may be associated with sequence-specific DNA binding and transcriptional activation [33,48]. ZNF711 has been reported to be predominantly expressed in the brain and testis and serves as a vital co-factor in the development of X-linked mental retardation (XLMR); however, its biological function and modulatory mechanisms remain largely unknown [49,50]. In the current study, the negative association between the level of ZNF711 expression and CDDP resistance in EOC patients was confirmed both in vivo and in vitro. Moreover, ZNF711 was found to be significantly correlated with the overall and relapse-free survival in EOC patients, and may serve as an independent prognostic indicator for EOC patients. Previous study has demonstrated that ZNF711 binds with PHF8 and demethylates the H3K9me2 level of target genes [50]. By using CAPTURE approach following the quantitative mass spectrometry, we identify that demethylase JHDM2A was recruited to the promoter of SLC31A1 by ZNF711, which abrogates the H3K9me2 modification level and activates the transcription of SLC31A1. The correlation between ZNF711 and SLC31A1 has also been verified in clinical tissues and intraperitoneal tumor models. Therefore, our study not only identified ZNF711 as a novel bio-marker of CDDP resistance for EOC patients, but also explicitly unraveled the regulatory mechanism of ZNF711 and might provide a new perspective for discovering molecular target for enhancing the curing effect of CDDP treatment. Interestingly, since SLC31A1 is widely known to be the main importer of copper in mammalian cells, whether the ectopic expression of ZNF711 can disturb copper homeostasis and lead to corresponding pathologies is worthy of further investigation.
BIX-01294, diazepin-quinazolinamine derivative, has been known as a histone methyltransferase inhibitor that specifically reduces the generation of H3K9me2 at the promoter of the target genes [25,26]. Previous studies have proved that BIX-01294 might inhibit tumor growth and serve as a novel anti-tumor agent in various cancer types, including breast cancer, colon cancer and lung cancer [25]. However, the appliance of BIX-01294 in ovarian cancer and the effect of BIX-01294 in CDDP response remain largely unknown. Our study indicated that BIX-01294 could increase the sensitivity of CDDP treatment in EOC cells, both in vivo and in vitro. Further, we clarified that BIX-01294 modulates chemo resistance via elevating the expression level of SLC31A1 and adjusting the CDDP uptake in EOC cells. Thus, this study not only proved that co-treatment with BIX-01294 induced the sensitization of CDDP-resistant EOC cells, but also elucidated the molecular mechanism of BIX- 01294 in regulating CDDP resistance.
In summary, the findings of our study reveal that ZNF711 down regulation enhanced CDDP resistance in EOC, which may serve as a prognostic factor for EOC patients. ZNF711 affects the level of SLC31A1 expression by binding to its promoter, recruiting the demethylase JHDM2A, and transcriptionally activating SLC31A1. This subsequently regulates the sensitivity of EOC cells to CDDP by modulating CISPLATIN uptake. Co-treatment with BIX-01294 induced the sensitization of ZNF711-silenced EOC to CDDP therapy. Thus, our evidence provides a novel perspective for the treatment of EOC chemo resistance.
Contributors
G.W., L.S. and Z.L. designed the experiments. G.W., M.T. and M.Y. performed the xenograft tumor experiments. G.W., Y.H. and Z.L. performed in vitro cell line studies. H.P. performed immuno-histochemical and pathological analysis. G.W., J.W. M.T. and M.Y. performed the co-IP and ChIP experiment. G.W., L.S. and J.L. wrote the paper. G.W., L.S. and Z.L. verified the underlying data.
Data sharing
All data are available upon request to the corresponding authors.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by Natural Science Foundation of China (No. 81830082, 82030078,91740119, 82072609, 81621004, 91740118, 82003128, 81773106 and 81530082); Guangzhou Science and Technology Plan Projects (201803010098); Natural Science Foundation of Guangdong Province (2018B030311009, 2016A030308002, 2018B030311060, 2019A1515110118); The Fundamental Research Funds for the Central Universities [No. 19ykpy177]; China Postdoctoral Science Foundation (2019M653220).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103558.
Contributor Information
Zheng Li, Email: lengyueds@outlook.com.
Libing Song, Email: songlb@sysucc.org.cn.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD. Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Primers. 2016;2:16061. doi: 10.1038/nrdp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med. 2017;14(1):9–32. doi: 10.20892/j.issn.2095-3941.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Della Pepa C, Tonini G, Pisano C, Di Napoli M, Cecere SC, Tambaro R. Ovarian cancer standard of care: are there real alternatives? Chin J Cancer. 2015;34(1):17–27. doi: 10.5732/cjc.014.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goff BA. Advanced ovarian cancer: what should be the standard of care? J Gynecol Oncol. 2013;24(1):83–91. doi: 10.3802/jgo.2013.24.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le T, Faught W, Hopkins L, Fung Kee Fung M. Primary chemotherapy and adjuvant tumor debulking in the management of advanced-stage epithelial ovarian cancer. Int J Gynecol Cancer. 2005;15(5):770–775. doi: 10.1111/j.1525-1438.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- 7.da Costa A, Baiocchi G. Genomic profiling of platinum-resistant ovarian cancer: The road into druggable targets. Semin Cancer Biol. 2020 doi: 10.1016/j.semcancer.2020.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Samimi G, Safaei R, Katano K, Holzer AK, Rochdi M, Tomioka M. Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin Cancer Res. 2004;10(14):4661–4669. doi: 10.1158/1078-0432.CCR-04-0137. [DOI] [PubMed] [Google Scholar]
- 9.Rottenberg S, Disler C, Perego P. The rediscovery of platinum-based cancer therapy. Nat Rev Cancer. 2021;21(1):37–50. doi: 10.1038/s41568-020-00308-y. [DOI] [PubMed] [Google Scholar]
- 10.Reyes ME, de La Fuente M, Hermoso M, Ili CG, Brebi P. Role of CC Chemokines Subfamily in the Platinum Drugs Resistance Promotion in Cancer. Front Immunol. 2020;11:901. doi: 10.3389/fimmu.2020.00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safaei R, Howell SB. Copper transporters regulate the cellular pharmacology and sensitivity to Pt drugs. Crit Rev Oncol Hematol. 2005;53(1):13–23. doi: 10.1016/j.critrevonc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Kuo MT, Chen HH, Song IS, Savaraj N, Ishikawa T. The roles of copper transporters in cisplatin resistance. Cancer Metastasis Rev. 2007;26(1):71–83. doi: 10.1007/s10555-007-9045-3. [DOI] [PubMed] [Google Scholar]
- 13.Wee NK, Weinstein DC, Fraser ST, Assinder SJ. The mammalian copper transporters CTR1 and CTR2 and their roles in development and disease. Int J Biochem Cell Biol. 2013;45(5):960–963. doi: 10.1016/j.biocel.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 14.Ishida S, McCormick F, Smith-McCune K, Hanahan D. Enhancing tumor-specific uptake of the anticancer drug cisplatin with a copper chelator. Cancer Cell. 2010;17(6):574–583. doi: 10.1016/j.ccr.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson CA, Blair BG, Safaei R, Howell SB. The role of the mammalian copper transporter 1 in the cellular accumulation of platinum-based drugs. Mol Pharmacol. 2009;75(2):324–330. doi: 10.1124/mol.108.052381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holzer AK, Manorek GH, Howell SB. Contribution of the major copper influx transporter CTR1 to the cellular accumulation of cisplatin, carboplatin, and oxaliplatin. Mol Pharmacol. 2006;70(4):1390–1394. doi: 10.1124/mol.106.022624. [DOI] [PubMed] [Google Scholar]
- 17.Cheng C, Ding Q, Zhang Z, Wang S, Zhong B, Huang X. PTBP1 modulates osteosarcoma chemoresistance to cisplatin by regulating the expression of the copper transporter SLC31A1. J Cell Mol Med. 2020;24(9):5274–5289. doi: 10.1111/jcmm.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombo PE, Fabbro M, Theillet C, Bibeau F, Rouanet P, Ray-Coquard I. Sensitivity and resistance to treatment in the primary management of epithelial ovarian cancer. Crit Rev Oncol Hematol. 2014;89(2):207–216. doi: 10.1016/j.critrevonc.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Lv X, Song J, Xue K, Li Z, Li M, Zahid D. Core fucosylation of copper transporter 1 plays a crucial role in cisplatin-resistance of epithelial ovarian cancer by regulating drug uptake. Mol Carcinog. 2019;58(5):794–807. doi: 10.1002/mc.22971. [DOI] [PubMed] [Google Scholar]
- 20.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 21.Chung D. Histone modification: the 'next wave' in cancer therapeutics. Trends Mol Med. 2002;8(4 Suppl):S10–S11. doi: 10.1016/s1471-4914(02)02303-1. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 23.Michalak EM, Burr ML, Bannister AJ, Dawson MA. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat Rev Mol Cell Biol. 2019;20(10):573–589. doi: 10.1038/s41580-019-0143-1. [DOI] [PubMed] [Google Scholar]
- 24.Tran TQ, Lowman XH, Kong M. Molecular pathways: metabolic control of histone methylation and gene expression in Cancer. Clin Cancer Res. 2017;23(15):4004–4009. doi: 10.1158/1078-0432.CCR-16-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam H, Gu B, Lee MG. Histone methylation modifiers in cellular signaling pathways. cellular and molecular life sciences. CMLS. 2015;72(23):4577–4592. doi: 10.1007/s00018-015-2023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei JS. EZH2 Mediates epigenetic silencing of neuro-blastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res. 2012;72(1):315–324. doi: 10.1158/0008-5472.CAN-11-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450(7166):119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Wang Z, Wang F, Wang C, Zhang H. IL-6 and IL-8 are involved in JMJD2A- regulated malignancy of ovarian cancer cells. Arch Biochem Biophys. 2020;684 doi: 10.1016/j.abb.2020.108334. [DOI] [PubMed] [Google Scholar]
- 29.Shi L, Chen ZG, Wu LL, Zheng JJ, Yang JR, Chen XF. miR-340 reverses CISPLATIN resistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Asian Pac J Cancer Prev. 2014;15(23):10439–10444. doi: 10.7314/apjcp.2014.15.23.10439. [DOI] [PubMed] [Google Scholar]
- 30.Ye Q, Liu J, Xie K. Zinc finger proteins and regulation of the hallmarks of cancer. Histol Histopathol. 2019;34(10):1097–1109. doi: 10.14670/HH-18-121. [DOI] [PubMed] [Google Scholar]
- 31.Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23(1):53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang M, Chen Y, Han D, Lei Z, Chu X. Role of the zinc finger and SCAN domain- containing transcription factors in cancer. Am J Cancer Resh. 2019;9(5):816–836. [PMC free article] [PubMed] [Google Scholar]
- 33.Rhie SK, Yao L, Luo Z, Witt H, Schreiner S, Guo Y. ZFX acts as a transcriptional activator in multiple types of human tumors by binding downstream of transcription start sites at the majority of CpG island promoters. Genome Res. 2018 doi: 10.1101/gr.228809.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Tian L, Zhang L, Xu B, Zhang Y, Li Q. Clinical Significance of ZNF711 in Human Breast Cancer. Onco Targets and Ther. 2020;13:6593–6601. doi: 10.2147/OTT.S251702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G, Cao L, Zhu J, Tan Z, Tang M, Li Z. American Association for Cancer Research; 2018. Loss of RBMS3 Confers Platinum Resistance in Epithelial Ovarian Cancer via Activation of miR-126-5p/beta-catenin/CBP signaling. Clinical cancer research: an official journal of the. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K. Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell. 2017;170(5):1028–1043. doi: 10.1016/j.cell.2017.08.003. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28(7):1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazarini M, Machado-Neto JA, Duarte AD, Pericole FV, Vieira KP, Niemann FS. BNIP3L in myelodysplastic syndromes and acute myeloid leukemia: impact on disease outcome and cellular response to decitabine. Haematologica. 2016;101(11):e445. doi: 10.3324/haematol.2016.142521. -e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD. An integrated TCGA Pan-Cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416. doi: 10.1016/j.cell.2018.02.052. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li N, Zhao Z, Liu P, Zheng Y, Cai S, Sun Y. Up-regulation of deubiquitinase USP7 by transcription factor FOXO6 promotes EC progression via targeting the JMJD3/CLU axis. Mol Ther Oncolytics. 2021;20:583–595. doi: 10.1016/j.omto.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain K, Jin CY, Clarke SG. Epigenetic control via allosteric regulation of mammalian protein arginine methyl transferases. PNAS. 2017;114(38):10101–10106. doi: 10.1073/pnas.1706978114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8(2):188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 43.Jan S, Dar MI, Wani R, Sandey J, Mushtaq I, Lateef S. Targeting EHMT2/G9a for cancer therapy: Progress and perspective. Eur J Pharmacol. 2021;893 doi: 10.1016/j.ejphar.2020.173827. [DOI] [PubMed] [Google Scholar]
- 44.Chae YC, Kim JY, Park JW, Kim KB, Oh H, Lee KH. FOXO1 degradation via G9a- mediated methylation promotes cell proliferation in colon cancer. Nucleic Acids Res. 2019;47(4):1692–1705. doi: 10.1093/nar/gky1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tendulkar S, Dodamani S. Chemo-resistance in Ovarian Cancer: prospects for new drugs. anti-cancer agents in medicinal chemistry. 2020. [DOI] [PubMed]
- 46.Savant SS, Sriramkumar S, O'Hagan HM. The role of inflammation and inflammatory mediators in the development, progression, metastasis, and chemo resistance of Epithelial Ovarian Cancer. Cancers (Basel) 2018;10(8) doi: 10.3390/cancers10080251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du X, Wang X, Li H, Sun H. Comparison between copper and CISPLATIN transport mediated by human copper transporter 1 (hCTR1) Metallomics. 2012;4(7):679–685. doi: 10.1039/c2mt20021j. [DOI] [PubMed] [Google Scholar]
- 48.van der Werf IM, Van Dijck A, Reyniers E, Helsmoortel C, Kumar AA, Kalscheuer VM. Mutations in two large pedigrees highlight the role of ZNF711 in X-linked intellectual disability. Gene. 2017;605:92–98. doi: 10.1016/j.gene.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 49.Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C. A systematic, large- scale re-sequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41(5):535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38(2):165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.