Abstract
In the ciliate Tetrahymena thermophila, thousands of DNA segments of variable size are eliminated from the developing somatic macronucleus by specific DNA rearrangements. It is unclear whether rearrangement of the many different DNA elements occurs via a single mechanism or via multiple rearrangement systems. In this study, we characterized in vivo cis-acting sequences required for the rearrangement of the 1.1-kbp R deletion element. We found that rearrangement requires specific sequences flanking each side of the deletion element. The required sequences on the left side appear to span roughly a 70-bp region that is located at least 30 bp from the rearrangement boundary. When we moved the location of the left cis-acting sequences closer to the eliminated region, we observed a rightward shift of the rearrangement boundary such that the newly formed deletion junction retained its original distance from this flanking region. Likewise, when we moved the flanking region as much as 500 bp away from the deletion element, the rearrangement boundary shifted to remain in relative juxtaposition. Clusters of base substitutions made throughout this critical flanking region did not affect rearrangement efficiency or accuracy, which suggests a complex nature for this regulatory sequence. We also found that the right flanking region effectively replaced the essential sequences identified on the left side, and thus, the two flanking regions contain sequences of analogous function despite the lack of obvious sequence identity. These data taken together indicate that the R-element flanking regions contain sequences that position the rearrangement boundaries from a short distance away. Previously, a 10-bp polypurine tract flanking the M-deletion element was demonstrated to act from a distance to determine its rearrangement boundaries. No apparent sequence similarity exists between the M and R elements. The functional similarity between these different cis-acting sequences of the two elements is firm support for a common mechanism controlling Tetrahymena rearrangement.
Developmentally programmed DNA rearrangement is an integral part of the life cycle of many organisms. One of the best-known examples is the rearrangement of immunoglobulin genes that occurs during lymphocyte development, giving rise to the vast diversity of the vertebrate immune system (reviewed in reference 36). Such DNA rearrangement events must be precisely controlled to avoid deleterious effects of aberrant reorganization. For example, chromosomal translocations involving the immunoglobulin locus are frequently associated with lymphoid malignancies (reviewed in references 27 and 39). The deleterious potential of failed rearrangement underlies the importance of understanding the molecular mechanisms guiding these events.
The most dramatic examples of DNA rearrangement have been termed chromatin diminution, which refers to the developmentally programmed elimination of large portions of genetic material from all somatic progenitor cells. This phenomenon was first described a century ago by Boveri for Ascaris (7) and has since been observed in many organisms (6, 28, 30; reviewed in references 14 and 32). Chromatin diminution is ubiquitous among the ciliated protozoa studied (32). Most ciliates exhibit a nuclear duality, maintaining distinct sets of genetic material for germ line and somatic functions. The DNA of the germ line micronucleus is the full genetic complement, whereas the DNA of the somatic macronucleus is a highly rearranged subset of the germ line DNA. The form and extent of the DNA rearrangements observed in this diverse group of organisms vary greatly. For example, the sizes of eliminated regions range from tens of base pairs to tens of kilobase pairs, and the quantities of eliminated DNA range from ∼10% to as much as 95% of the germ line genome. The relationships between the rearrangements that occur in different ciliate species or even within the same species are not well understood.
Among organisms that undergo large-scale DNA rearrangement, the ciliate Tetrahymena thermophila is particularly amenable to molecular genetic analysis. In Tetrahymena, conjugation initiates a developmental program that results in the formation of new germ line micronuclei and somatic macronuclei, as well as the destruction of old macronuclei. The genome of a developing macronucleus undergoes extensive reorganization. Chromosome fragmentation occurs at 50 to 200 sites (1, 13) defined by the chromosome breakage sequence (50), and specific DNA rearrangements remove 10 to 15% of the germ line genome from roughly 6,000 internal chromosomal sites (46, 47). The segments of micronucleus-limited sequences are referred to both as deletion elements and internal eliminated sequences. They consist of unique and/or moderately repetitive DNA sequences and range in size from several hundred base pairs to greater than 10 kbp. Most Tetrahymena deletion elements have been found outside of coding sequences, although one has been found within an intron (22). The eight deletion elements examined by sequence analysis share few obvious similarities other than a strong A+T nucleotide bias and the presence of short direct repeats of 1 to 8 bp at the rearrangement boundaries (4, 5, 12, 22, 25, 40).
The R and M elements were the first Tetrahymena deletion elements to be sequenced (4, 5) and remain the most extensively characterized. The R element is eliminated during macronuclear development by a 1.1-kbp deletion event (2). The M element is eliminated from the macronucleus by two alternative deletion events of 0.6 and 0.9 kbp (2). These two eliminated forms share a common right boundary but utilize different left boundaries that are 0.3 kbp apart (5). Alternate rearrangement boundaries may be used by as many as 25% of deletion elements (12). Different rearrangement events between the same boundaries of a given element usually produce the same junction sequence; even so, rearrangement of most elements exhibits some heterogeneity, producing variant junction sequences that differ by a few base pairs (3, 29, 31).
Deletion elements placed on Tetrahymena rDNA-based transformation vectors rearrange accurately when introduced into conjugating cells (20). By using this transformation assay to study M-element rearrangement, an essential cis-acting regulatory sequence, 5′-AAAAAGGGGG-3′ (A5G5), was identified, providing the first mechanistic insight into these site-specific deletion events. This sequence is located ∼45 bp outside each end of the micronucleus-limited region in a specific orientation (A5G5 on the left; C5T2AT2 on the right) and functions to position the rearrangement boundaries a short distance away (20). Moving the location of this sequence repositions the rearrangement boundary to within 41 to 54 bp of the new location (19). This A5G5 sequence is not found near any of the other sequenced deletion elements, and it is the only cis-regulatory sequence that has been clearly defined.
The distance-dependent action of the M-element A5G5 sequence, as well as its position outside the deletion element, is unique among known rearrangement systems. The lack of any common, identifiable cis-acting sequence among the other known deletion elements, together with their size and sequence diversity, has challenged our understanding of these rearrangement events. It is still not known whether elimination of the estimated 6,000 deletion elements occurs via a common mechanism or involves several distinct rearrangement pathways. To better understand the relationship between the rearrangement of different elements in Tetrahymena, we have characterized cis-acting sequences involved in the rearrangement of the R deletion element. In this study, we have found that sequences outside the micronucleus-limited region are required for deletion. We show that these cis-acting sequences serve to position the rearrangement boundary a short distance away. The function of these flanking regulatory sequences is very similar to that determined for the M-element A5G5 sequence (19, 20). The finding that different flanking regulatory sequences of these two elements perform the same function provides strong evidence for a common mechanism controlling DNA rearrangement in Tetrahymena.
MATERIALS AND METHODS
Strains.
T. thermophila inbred B strains CU427 [Chx/Chx (VI, cy-s)] and CU428 [Mpr/Mpr (VII, mp-s)] (obtained from Peter Bruns, Cornell University) were used for all transformation experiments described below. Maintenance and growth of these strains were carried out under standard conditions as previously described (21).
Plasmid constructions.
Recombinant DNA techniques were executed essentially as described by Sambrook et al. (33). For transformation analyses, all modified R elements were inserted into the polylinker sequence located downstream of the transcribed region of the rDNA in the Tetrahymena vector pD5H8 (20). In some cases, the polylinker of this vector had been previously modified to introduce additional cloning sites by inserting the 31-bp NSXBK/NKBXS linker sequence given in Table 1 into the unique NotI site to create pD5H8N1.
TABLE 1.
Oligonucleotides used
| Name | Sequence (5′ to 3′) |
|---|---|
| NSXBK | GGCCGCCCGGGCTCGAGGTTACCGGTACCGC |
| NKBXS | GGCCGCGGTACCGGTAACCTCGAGCCCGGGC |
| 5R266 | AACAGTGTAAAACCCAAAAAGC |
| 3R1483RC | GATTTACTGTAAGATAGTTCTAG |
| 5R297RCA | TGGGCCCTTTTTAATTAGCTTTTTGGG |
| 5R265RCA | TGGGCCCTAAAATTATAATTATATTAA |
| R328A | TGGGCCCAGTGATTCAAAAAAATGGTG |
| R353A | TGGGCCCTTTTGTATTTTTTGGTTAAA |
| 5R211RCA | TGGGCCCATTTCATTTTATTTATTTT |
| 5R264A | ATTGGGCCCAGTGTAAAACCCAAAAAGC |
| 5R228RC | ATATTAATTTCTATTCTAACTTAAG |
| 3R1430A | AAGGGCCCACAATTTGAATGAAAAA |
| 3R1548RC | GGAATTCTGCAGTTTAATATTCTAAGCA |
| 3R1747RC | TTGAATTCTGCAGTATGCTTAAACCATT |
| 5R001 | GTTAGAGTTTGATAATATTACACC |
| Kalldown | CGTAAATCTTTTGTAGACGA |
| HSS6-2 | GGCCGCGGATCCGGGCAACG |
| 5R228 | CTTAAGTTAGAATAGAAATTAATAT |
| 5R243 | AAATTAATATAATTATAATTTTAA |
| 5R1438RC | AAAATAATTTTTTCATTCA |
| 3R1499RC | CTAAATATTTAAATAAGATTTACTG |
| RPM285K | AAAGGTACCTAGTATCAAAATCTATAAATC |
| RPM297RK | TTAGGTACCTTTTGGGTTTTACA |
| RPM273K | TGTGGATCCGATGAACCTAATTAAAAATC |
| RPM286RK | TTTGGGTACCACACTGTTTAAAATTA |
| RPM262K | ATTGGTACCACAGTAAAACCCAAAAAGC |
| RPM275RK | ACTGGATCCAATTATAATTATATTA |
| RPM249K | TAAGGTACCTAGTATTTTAAACAGTGTAA |
| RPM260RK | ATAGGTACCTTAATTTCTATTCTAAC |
| RPM232K | TAAGGTACCATCTAAATTAATATAATTATA |
| RPM245RK | TATGGTACCTTAAGATTTCATTTTA |
| J1110R | GATTTATAAACAACAATTTGAATG |
The construction of pDLCR6, which contains R-element sequences from −312L to ∼−900r, was previously described by Chalker et al. (11). The construction of R-element subclones containing various lengths of flanking sequence is described below. DNA fragments containing R-element sequences −203L/−391r and −100L/−391r were generated by digestion with restriction endonucleases, AccI-NsiI and AflII-NsiI, respectively. The ends of these DNA fragments were made blunt and inserted into the SmaI site of pUC19 (41). The −203L/−116r construct was created by exonuclease III digestion of right flanking sequence between −391r and −117r present in the −203L/−391r construct. These three R-element subclones were excised from pUC19 by digestion with endonucleases EcoRI and SphI and inserted as blunt-end fragments into the SmaI site of pD5H8N1. The −63L/−70r construct was created by inserting R-element sequence generated by PCR amplification with oligonucleotides 5R266 and 3R1483RC (Table 1) into the SmaI site of pD5H8N1.
Small internal deletions of left flanking sequence were created by inverse PCR of plasmid pSR3 (20), a pHSS6-based plasmid (37) containing R-element sequences from ∼−1400L to −900r, using the following oligonucleotide pairs: the Δ−31L:+3 construct, oligonucleotides 5R297RCA and R328A; the Δ−31L:+24 construct, oligonucleotides 5R297RCA and R353A; the Δ−63L:−2L construct, oligonucleotides 5R265RCA and R328A; and the Δ−63L:+24 construct, oligonucleotides 5R265RCA and R353A. Oligonucleotide sequences are listed in Table 1. Each oligonucleotide contains an ApaI endonuclease recognition site near its 5′ end. After amplification, PCR products were digested with endonuclease ApaI. The digested DNA fragments were ligated under dilute DNA concentrations to favor intramolecular ligation. The resulting ligations were transformed into Escherichia coli to recover the circularized plasmids. The Δ−76L:+24 construct was similarly created by inverse PCR of plasmid pDLCR4 (11) with oligonucleotides 5R228RC and R353A, followed by blunt-end ligation to circularize the PCR fragment. A modified version of the Δ−31L:+3 construct, pDLCR4Ed, that contains only 312 bp of left flanking sequence was created by substituting an AccI/HindIII restriction fragment from the deletion construct with the same region of plasmid pDLCR4. Subsequently, the Δ−101L:+3 construct was made by inserting an ApaI/EcoRI-digested fragment that had been generated by PCR amplification of pDLCR4 with oligonucleotides 5R211RCA and HSS6-2 into ApaI/EcoRI-digested pDLCR4Ed. The Δ−101L:−61L construct was then created by inserting an ApaI/HindIII-digested fragment generated by PCR amplification of pDLCR4 with oligonucleotides 5R264A and 3R1499RC into ApaI/HindIII-digested Δ−101L:+3 plasmid. After isolation of each of the above constructs, the DNA sequence of the left flanking region was verified. The expected right deletion endpoint for the Δ−31L:+3 construct was −2L; however, a cloning artifact resulted in the +3 endpoint. Tetrahymena transformation vectors containing these modified R elements were created by inserting NotI-digested DNA fragments of these plasmids into the NotI site of the pD5H8 polylinker.
To insert DNA into the left flanking region of the R element, the Δ−31L:+3 R-element construct in pD5H8 was digested with ApaI. The resulting 4-bp 3′ extensions were digested with T4 DNA polymerase. Short, blunt-ended DNA fragments were generated by HaeIII digestion of pUC19 and pHSS6 and ligated into the blunt-ended Δ−31L:+3 R element vector. The resulting plasmids were recovered by E. coli transformation. The approximate size of each insert was determined by restriction endonuclease digestion followed by agarose gel electrophoresis analysis, and the sequence of each insert was subsequently verified.
Clusters of point mutations in the left macronucleus-destined region were generated by inverse PCR of plasmid pDLCR4, using overlapping oligonucleotides (Table 1) containing sequence altered at five or six positions relative to the wild-type sequence. Each set of base changes creates a KpnI site. After the inverse PCRs, the resulting amplification products were digested with KpnI, and the fragments were circularized by ligation under dilute DNA concentrations. The resulting R-element plasmids containing point mutations, RPM1 to RPM5, were recovered by E. coli transformation, and their structures were verified. NotI fragments of these modified R elements were inserted into the NotI site of the pD5H8.
Three R elements that replaced the left flanking sequences with the corresponding sequences from the right side were constructed. For two constructs, all left flanking sequences were replaced with sequences from the right. To construct these two plasmids, DNA fragments containing right flanking sequence from −1r to −135r or from −1r to −334r were generated by PCR using oligonucleotides 3R1430A and 3R1548RC or 3R1430A and 3R1747RC, respectively. Each fragment was digested with ApaI and PstI (at sites introduced as part of the oligonucleotide primers) and inserted into ApaI/PstI-digested Δ−31L:+3 construct in vector pHSS6 to replace the excised left side. For the third construct, only the first ∼100 bp of left flanking sequence were replaced with sequences from the right side. These 100 bp were removed by digestion of the −312L/−900r construct in pHSS6 with AflII (the 5′ overhang was made blunt by fill-in using T4 DNA polymerase) and HindIII. The −1r to −135r region was removed from the first construct above by digestion with PstI (the 3′ overhang was made blunt by digestion with T4 DNA polymerase) and HindIII and was inserted in place of +3 to −101L sequences. Each of the three constructs above was digested with NotI, and the DNA fragments containing the modified R elements were inserted into pD5H8 for transformation of Tetrahymena.
Tetrahymena transformations.
Transformation of Tetrahymena with R-element-containing rDNA vectors was performed by microinjection or electroporation. Logarithmically growing cells were prepared for transformation by starvation for several hours in 10 mM Tris-Cl (pH 7.4) prior to mixing strains to initiate conjugation (20). Microinjection of mating pairs was performed as described previously (10, 38, 49). Electroporation of mating cells was performed as described by Gaertig and Gorovsky (18). Transformants generated by microinjection were used only to determine the rearrangement boundaries of some modified R elements. Transformants obtained by electroporation were used to determine rearrangement activity and boundary sites.
DNA isolation and analysis.
Whole-cell DNA was isolated from transformants as previously described (4). For Southern blot analysis, DNA was digested with restriction enzymes under the conditions recommended by the suppliers. These samples were fractionated by electrophoresis in 0.8 to 1.2% agarose gels. Lambda DNA digested with either HindIII or PstI was used as a size standard. DNA was then transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, N.H.) by pressure using a PosiBlot apparatus (Stratagene, La Jolla, Calif.) and then cross-linked to the membranes by UV light. Immobilized DNA was hybridized to an R-element-specific probe in 6× SSC (20× SSC is 3 M sodium chloride plus 0.3 M sodium citrate [pH 7.0]), 0.1 M Tris-HCl (pH 7.5), 0.5% sodium dodecyl sulfate, and 2× Denhardt’s reagent (50× Denhardt’s reagent is 1% Ficoll, 1% polyvinylpyrrolidone, and 1% bovine serum albumin) at 65°C overnight (12 to 20 h). This probe was an EcoRI/PstI restriction fragment from pDLCR5 (11) that corresponds to the rearranged form of the R element with 0.3 kbp of right and 0.9 kbp left of the rearrangement junction that was radiolabeled with [32P]dATP (16, 17). Hybridized membranes were washed three to four times in 1× SSC–0.5%SDS at 65°C for 20 to 30 min and then exposed to X-ray film. The amounts of rearranged and unrearranged R element were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). To detect DNA fragments resulting from accurate rearrangement, membranes were hybridized and washed under similar conditions at 37°C to an end-labeled oligonucleotide, J1110R (Table 1), that is specific for the predominant chromosomal deletion junction (4). Autoradiograms were captured as digital images with a flatbed scanner (Epson America, Torrance, Calif.) and Photoshop version 4.0 LE (Adobe Systems) and displayed by using Canvas version 3.5.5 (Deneba Systems).
The products of R-element rearrangement were recovered from transformant DNA preparations by PCR amplification with different combinations of oligonucleotide primers 5R001, 3R1548RC, 3R1747RC, 3R1499RC, Kalldown, HSS6-2, 5R228, and 5R243 (Table 1). To determine the DNA sequence spanning the rearrangement junctions, the resulting DNA fragments were sequenced directly with oligonucleotide 5R228, 5R264A, or 5R1438RC. In some cases, the amplified rearrangement products were cloned into plasmid pUC18 or pUC19 prior to sequence analysis. Sequence reactions were performed by using either [35S]dATP with a Sequenase version 2.0 sequencing kit (United States Biochemical, Cleveland, Ohio) or 32P-end-labeled primers with a double-stranded DNA cycle sequencing system (Life Technologies, Gaithersburg, Md.).
RESULTS
The R deletion element (Fig. 1A) is a 1,084-bp micronucleus-limited sequence (4) that is located ∼2.7 kbp right of the M element (2) on micronuclear chromosome 4 (9). To aid in its description, we have divided the sequence of the element into three parts as described in the legend to Fig. 1A.
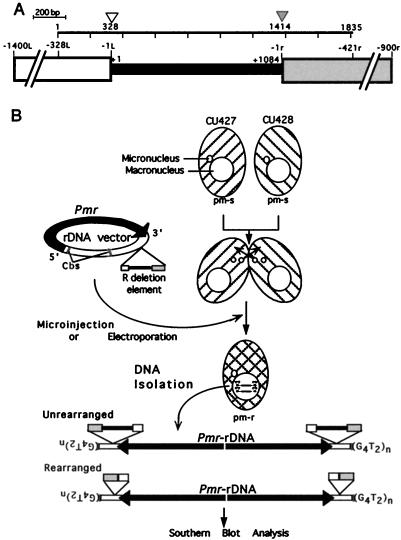
FIG. 1.
The R element and the rearrangement assay. (A) Schematic diagram of the R element. The bar above the diagram indicates the region of the element originally sequenced by Austerberry and Yao (4) and is divided into 200-bp increments. We have separated the element into three parts and assigned a numbering system to each. The micronucleus-limited region that is eliminated during rearrangement is shown as a narrow, solid box and is numbered left to right from +1 to +1084. Positions +1 and +1084 correspond to nucleotides 329 and 1413 as assigned in the original published sequence. The macronucleus-destined region on the left is represented as the wide, open box and numbered right to left from −1L to −328L. The right macronucleus-destined region is shown as the wide, shaded box and is numbered left to right from −1r to −421r. For positions beyond −328L and −421r, the distances are approximate. Positions of the predominant left and right rearrangement boundaries formed by elimination of the endogenous R element are designated by the open and shaded arrowheads, respectively. The nucleotide positions, 328 and 1414, joined by rearrangement are given under these arrowheads and correspond to the first nucleotides of the left (−1L) and right (−1r) flanking regions. (B) The rearrangement activity of vector-borne deletion elements are tested by transformation of conjugating Tetrahymena cells. Conjugation is initiated by mixing prestarved strains CU427 and CU428. Approximately 8 to 9 h after mixing, the transformation vectors are introduced into the developing macronuclei by microinjection or electroporation. The transformants are identified by their growth in the presence of the antibiotic paromomycin. DNA is isolated from the transformants, digested with restriction endonucleases to liberate the DNA fragment containing the deletion element from the transforming rDNA molecules, and analyzed by gel electrophoresis and Southern blot hybridization.
The rearrangement assay.
To identify sequences that are required in cis to control R-element rearrangement, we used a transformation assay (Fig. 1B) that was developed previously to investigate the control of M-element rearrangement (20). In this assay, the Tetrahymena rDNA-based vector, pD5H8, that contains a modified R element is transformed into conjugating T. thermophila CU427 and CU428. Upon transformation, the rDNA including the R element inserted into the 3′ nontranscribed region is cleaved from the circular plasmid at the 5′ and 3′ chromosomal breakage sequences. Telomeres are added near the 3′ breakage site, and the entire molecule is converted to a palindromic minichromosome and amplified to ∼9,000 copies per cell (48; reviewed in reference 42). The vector-carried R element undergoes rearrangement during processing of the transforming rDNA. Transformants are selected by their resistance to the antibiotic paromomycin (Pmr phenotype), which is conferred by rRNA synthesized from the vector-borne, Pmr rDNA allele (8).
After selection of transformants, DNA is isolated from the Pmr cells, digested with restriction enzymes that liberate the R element construct from the rDNA vector, and analyzed by Southern blot hybridization (as shown in Fig. 2, 3, and 5 to 7). Each lane contains a pool of DNA taken from at least five independent transformants, which gives a better estimate of the rearrangement efficiency than DNA from a single transformant. The hybridization was quantified with a PhosphorImager. The proportion of the rearranged species relative to the unrearranged element is a qualitative measure of the rearrangement activity of a given construct. Our largest intact R-element construct, −312L/−900r (Fig. 2), is our standard of rearrangement activity and always showed at least 50% rearrangement. Therefore, in the analyses below, we will consider any construct that shows ≥50% rearrangement as displaying normal activity, any construct for which rearrangement is easily detectable but has <50% rearrangement as displaying reduced activity, any R element for which rearrangement is barely detectable (≤10%) as showing greatly reduced activity, and any construct that shows no detectable rearrangement (<2%) as inactive.
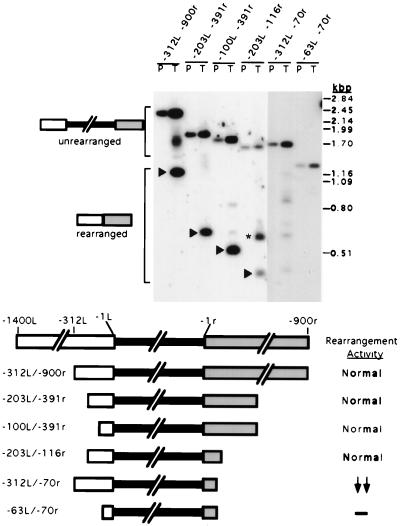
FIG. 2.
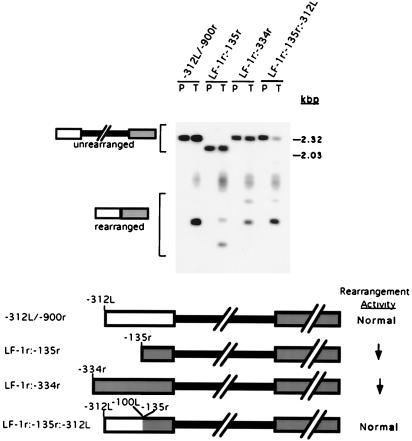
Analysis of external deletion of sequences flanking the micronucleus-limited region. Plasmid constructs containing progressively larger deletions of sequences flanking the R element were assayed for the ability to undergo precise deletion upon transformation. Plasmid DNA (P) and DNA isolated from transformants (T) were digested with NotI prior to electrophoresis and transfer to nitrocellulose membranes. Southern blot hybridization analysis with a probe specific to the macronuclear DNA from −312L to ∼−900r of the R element is shown at the top. A longer exposure of the right-hand lanes is shown to allow visualization of less abundant fragments. Positions of the unrearranged and rearranged elements are indicated to the left; positions of PstI-digested lambda DNA size standards are shown to the right. PhosphorImager analysis was used to quantify hybridization. To determine whether rearrangement was accurate, filters were hybridized separately with an end-labeled oligonucleotide, J1110R, that detects specifically the predominant rearrangement junction of chromosomal R elements. These hybridizing fragments are indicated by the arrowheads. The major rearranged product for the −203L/−116r is ∼200 bp larger than the accurately rearranged species and is denoted by the asterisk. The amount of DNA flanking the R element in each construct is indicated above each set of lanes. A diagram of the constructs is given at the bottom. The solid box represents the micronucleus-limited sequences; the wider open and shaded boxes represent the left and right macronucleus-destined regions, respectively. The rearrangement activity (ratio of hybridization to rearranged forms and the unrearranged construct) relative to an intact R element is shown on the bottom right: Normal, normal activity (>50%); ↓, reduced activity (11 to 49%); ↓ ↓, greatly reduced activity (<10%); and −, no detectable rearrangement.
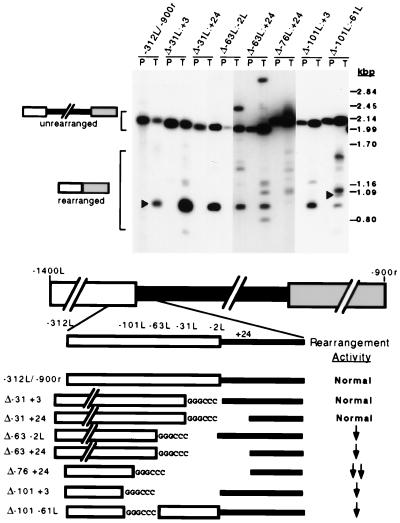
FIG. 3.
Analysis of small sequence deletions at the left flank of the R element. R-element constructs containing small <105-bp deletions at the left boundary of the micronucleus-limited region were transformed into conjugating Tetrahymena cells. Southern blot hybridization analysis used to assess the rearrangement of various R-element constructs is shown at the top. Plasmid DNA (P) and DNA isolated from transformants (T) were digested with AccI and NotI (−312L/−900r, Δ−31L:+3, Δ−31L::+24, Δ−62L:−1L, and Δ−62L:+24) or NotI alone (Δ−76L::+24, Δ−101L:+3, and Δ−101L:−61L) prior to electrophoresis and transfer to nitrocellulose membranes. The probes were the same as used for Fig. 2. A longer exposure for some lanes is given to allow visualization of DNA fragments in low abundance. Positions of PstI-digested lambda DNA size standards are shown to the right. Arrowheads indicate the fragments that hybridized to the oligonucleotide probe that detects the major chromosomal junction sequence. The region deleted from each construct is indicated above each set of lanes. A diagram showing an enlargement of the left flanking region of each construct is given at the bottom. Nucleotide positions of the deletion endpoints are indicated above the enlargement. The 6-bp ApaI site, GGGCCC, was inserted in place of the sequences removed. The efficiency of rearrangement relative to the activity of an intact R element is given to the right of each diagram: Normal, normal activity; ↓, reduced activity; and ↓ ↓, greatly reduced activity.
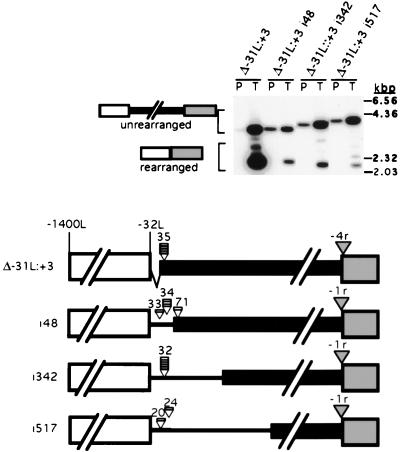
FIG. 5.
The rearrangement boundary and the flanking region remain in relative juxtaposition in constructs containing sequence insertions at their left ends. DNA fragments of 48, 342, and 517 bp were inserted into the ApaI site of deletion construct Δ−31L:+3, and the rearrangement of each construct was determined by Southern blot hybridization of NotI-digested plasmid (P) or transformant (T) DNA. The probes were the same as used for Fig. 2. The positions of HindIII-digested lambda DNA size standards are shown to the right. A diagram of the constructs is given at the bottom. The solid boxes represent the micronucleus-limited sequence; the wider open and shaded boxes represent the macronucleus-destined flanking regions; the wide lines represent the inserted sequence. Positions of the rearrangement boundaries on the right are indicated by the shaded arrowheads and were found to be at the same position, −1r or −4r, for each junction analyzed for a given construct. Each shaded arrowhead indicates the position of the left rearrangement boundary; each bar represents an independently rearranged element with its left boundary observed to be at the position indicated by the associated arrowhead. The nucleotide positions given above each arrowhead denote the observed distance of each left boundary from the −32L position. Eleven of twelve left side boundaries are within a 14-bp region right of this position.
FIG. 7.
The right flanking sequence effectively substitutes for the essential left flanking region. R-element constructs containing substitutions of the sequence immediately flanking the left end of the micronucleus-limited region with sequence flanking the right end were transformed into conjugating Tetrahymena cells. Southern blot hybridization analysis used to assess the rearrangement of each construct is shown at the top. Plasmid DNA (P) and DNA isolated from transformants (T) was digested with BamHI prior to electrophoresis and transfer to the nitrocellulose membrane. The positions of HindIII-digested lambda DNA size standards are shown to the right. The sequences flanking the left end of the eliminated region are indicated above each set of lanes. A diagram of the flank substitution constructs is shown at the bottom. In each schematic, the narrow black box represents the micronucleus-limited sequences. The wide white and shaded boxes represent the sequences flanking the eliminated region to the left and right, respectively.
A diffuse band that migrated between the unrearranged and rearranged species was seen sporadically throughout this study (e.g., see lane 312L/−900r T in Fig. 2). This band is indicative of linkage of the telomeric 3′ end of the rDNA with the R-element-containing fragment and presumably is due to some aberrant rDNA processing that is inherent to the transformation system (45). This aberrant processing obscured the rearrangement of the linked R element; therefore, we did not factor this hybridizing species into the quantification of rearrangement activity.
DNA sequences flanking the micronucleus-limited region are necessary for efficient rearrangement.
We used the above transformation assay to determine whether regions flanking the micronucleus-limited R element are required for efficient rearrangement. To this end, we created a series of deletion constructs that lacked progressively more of the left and right flanking regions and introduced vectors containing these modified R elements into conjugating Tetrahymena cells. Southern blot hybridization of DNA isolated from the resulting transformants by using a probe specific to the R-element flanking regions is shown in Fig. 2. For each construct, the plasmid used for transformation (lane P) is shown adjacent to the transformant DNA (lane T). The plasmid served as a marker for the size of the unrearranged R element. Both unrearranged and rearranged forms of the constructs were observed in the transformant DNA preparations. Each of the R-element constructs, −312L/−900r, −203L/−116r, and −100L/−391r, that contained at least 100 bp of left and 116 bp of right flanking sequences exhibited normal (i.e., >50%) rearrangement activity. In contrast, the construct that contained only 70 bp right of the micronucleus-limited region, −312L/−70r, displayed greatly reduced rearrangement activity. The construct that contained only 63 bp left and 70 bp right of the micronucleus-limited DNA, −63L/−70r, was inactive. These data indicate that the micronucleus-limited region alone is insufficient and that the R element minimally requires ∼100 bp of macronucleus-destined sequences on each side to efficiently undergo DNA rearrangement. We cannot formally exclude the alternative possibility that constructs with <100 bp of flanking DNA rearrange poorly due to the inhibitory action of vector sequences brought to within interfering distance of the boundary; however, we find this explanation unlikely and inconsistent with further analysis of the left flanking region presented below.
To determine whether the R-element rearrangement was accurate, Southern blots were hybridized separately to an oligonucleotide probe that is specific for the predominant chromosomal deletion junction (4). In the case of most constructs that showed normal rearrangement activity, the major rearranged species hybridized with this junction sequence probe, as indicated by the arrowheads in Fig. 2. The one exception was the −203L/−116r construct, for which a species ∼200 bp larger was about twice as abundant as the species containing the native junction. The nature of this aberrant rearrangement is not clear. Although the total rearrangement of this construct is >50%, the appearance of this larger species may indicate that sequences to the right of −116r increase the accuracy of rearrangement.
To further investigate the requirement for the flanking regions, we constructed several R elements with small (34- to 104-bp) internal sequence deletions at the left end of the micronucleus-limited DNA. In each construct, the sequence removed was replaced with the 6-bp ApaI restriction endonuclease recognition site, 5′-GGGCCC-3′. These modified R-element constructs were used to transform conjugating Tetrahymena cells. Southern blot hybridization analysis of their rearrangement activities is shown in Fig. 3. The Δ−31L:+3 construct, lacking the 31 bp immediately outside the micronucleus-limited region, rearranged with normal efficiency. Likewise, the Δ−31L:+24 construct, missing an additional 21 bp of the micronucleus-limited DNA, displayed normal activity. These two constructs rearrange efficiently even though both lack the left-end, 6-bp direct repeat sequence found bordering the micronucleus-limited DNA. Similarly, the direct repeat at the right end of the M element has been shown to be dispensable for its rearrangement (19). Although direct repeats border almost all known deletion elements, they do not contribute significantly to efficient rearrangement.
R elements missing the first 31 bp of the left flanking region displayed normal rearrangement efficiency. In contrast, constructs lacking an additional 31 bp of flanking DNA (Δ−63L:−2L and Δ−63L:+24) showed reduced rearrangement activity (Fig. 3). Because our external deletion analysis showed that ∼100 bp was sufficient for normal rearrangement efficiency, we expected that removal of all of these sequences would abolish rearrangement activity. To our surprise, constructs lacking most (Δ−76L:+24) or all (Δ−101L:+3) of the first 100 bp on the left still rearranged (Fig. 3). In fact, the rearrangement activity of the Δ−101L:+3 construct was not dramatically different from that of the constructs lacking only the first 63 bp (Δ−63L:−2L and Δ−63L:+24). These data lead us to suggest that although the flanking sequences beyond −100L are not required for the rearrangement (Fig. 2), they are able to partially substitute for the cis-acting sequences found within the first 100 bp of the left side of the R element.
The above data showed that sequences between −31L and −63L were required for fully efficient rearrangement activity; however, they did not provide sufficient evidence to show that the sequences beyond −63L contributed to rearrangement efficiency. We thus removed sequences between −61L and −101L (Δ−101L:−61L). This construct displayed reduced activity that is comparable to that of the Δ−63L:−2L and Δ−101L:+3 constructs. Thus, the −61L to −101L region also contains sequences that are important for efficient rearrangement.
Removing sequences beyond −31L not only resulted in reduced rearrangement activity but also introduced significant heterogeneity in the size of the rearranged elements (Fig. 3). Rearrangement of some constructs produced several different rearranged species. For example, the Δ−101L:−61L construct produced at least three other species of abundance equal to or greater than that of the accurately rearranged species containing the major junction (Fig. 3). Based on the observed heterogeneity, it appears that sequences beyond −31L are necessary for both efficient and accurate rearrangement.
Sequences in the left macronucleus-destined region control the position of the left deletion boundary.
The M-element A5G5 polypurine tract has been shown to specify the deletion boundaries at a distance 41 to 54 bp 3′ of the proximal G nucleotide (19). If the region flanking the R element also serves to determine the rearrangement boundary, then the small deletions (Fig. 3) effectively move this cis-acting control region closer to the micronucleus-limited region and therefore should produce a rightward shift of the junction formed by rearrangement. Alternatively, if the boundary is specified by sequences present within the micronucleus-limited region, removal of flanking sequences should not change the position of the rearrangement junction. To examine this issue, we characterized junctions created by rearrangement of four of these modified R elements (Fig. 4). We PCR amplified the rearranged elements from two to four independent transformants derived from each construct, cloned the amplified products, and determined the sequence spanning the junction. For each construct, the right side of the junction was formed at one of two positions, −1r or −4r (Fig. 4). These two positions are the same right-end boundaries that constitute the major and minor junction sites, respectively, resulting from the rearrangement of chromosomal R elements (4).
FIG. 4.
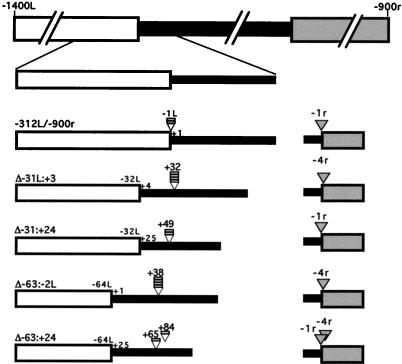
The left rearrangement boundary shifts rightward into the micronucleus-limited region a distance corresponding to the length of sequence removed from the left flanking DNA. The R element including an enlargement of the left end is shown schematically at the top. The narrow and wide boxes represent the micronucleus-limited and macronucleus-destined flanking regions, respectively, and are shaded as in prior figures. The name of each construct is indicated at the left edge of each schematic. Shaded arrowheads indicate positions of the right rearrangement boundaries; open arrowheads indicate positions of the left rearrangement boundaries. Each bar represents an independently rearranged element with its left boundary observed to be at the position indicated by the associated arrowhead. The number above each arrowhead denotes the position of the boundary relative to the start of the micronucleus-limited region as described in Fig. 1.
For each of these deletion constructs, the left side of the junction was shifted rightward into what is normally micronucleus-limited sequence. The left side of the rearrangement junction of the Δ−31L:+3 construct was found to be at position +32 (Fig. 4). Thus, replacing these 34 bp with the ApaI site (a net 28-bp deletion) resulted in a rightward shift of the left deletion boundary by 31 bp. Similarly, removal of 49 bp caused the junction of the Δ−31L:+24 construct to shift rightward by 49 bp to position the left boundary at +50, and removal of a net 56 bp produced a 38-bp rightward shift of the Δ−63L:−2L construct’s left boundary to position +39 (Fig. 4). Two different left junction sites were observed for the 81-bp net deletion of the Δ−63L:+24 construct. One was located at position +66, a rightward shift of 65 bp, and the other was located at +84, 83 bp into the micronucleus-limited DNA (Fig. 4). Thus, for all four constructs, the rightward shift of the left deletion boundary roughly corresponded to the amount of sequence removed from the construct. The shift of the deletion boundary observed for the two Δ−31L constructs more precisely matched the amount of sequence removed than the shift observed for the two Δ−63 constructs. This apparent loss of some accuracy of deletion is consistent with our finding that removal of sequences between −32L and −63L reduced rearrangement efficiency. Nonetheless, the overall conservation of distance between the flanking region and the left boundary indicates that the left side cis-acting sequences determine the position of the rearrangement boundary.
We further examined the ability of the macronucleus-destined sequences to control the deletion boundary by increasing the distance between the micronucleus-limited region and these putative regulatory sequences. These constructs were created by ligating blunt-ended DNA fragments from E. coli plasmid pUC19 or pHSS6 into the unique ApaI restriction site of deletion construct Δ−31L:+3 (after removal of the 3′ 4-bp extensions left after ApaI digestion). The rearrangement of constructs with 48-, 342-, and 517-bp insertions is shown in Fig. 5. Each construct produced a significant portion of rearranged products upon transformation, although the overall efficiency of rearrangement was lower than for the original Δ−31L:+3 construct. Three other constructs, with inserts of 240, 434, and 550 bp, rearranged rather poorly (data not shown). We suspect that particular DNA sequences can block the ability of the flanking regulatory region to act on the neighboring eliminated region when they are placed between the two regions.
The size of the rearranged R element in the transformants was consistent with the elimination of both the micronucleus-limited region and most of the inserted sequence. We PCR amplified the rearranged products from individual transformants of each of the three insertion constructs and determined the sequence spanning the junction. For each construct, the right side of the deletion junction was located at position −1r, the major right boundary of chromosomal R element rearrangement (Fig. 5). On the other hand, the left boundaries of the junctions for each construct were found in novel locations such that they retained relative juxtaposition with the left flanking region. To describe these boundary sites, we will use position −32L as a reference point. The major chromosomal rearrangement junction is located 31 bp to the right at position −1L, and the junction observed for the Δ−31L:+3 construct was found to be located 35 bp to the right at +32. For the 48-bp insertion, examination of five transformants identified three left-end junction sites. The left boundary was either 33 (one example), 34 (three examples), or 71 (one example) bp right of position −32L (Fig. 5). Five examples of a single left-end junction were observed for the 342-bp insertion, all located 32 bp to the right. Two different left-end boundary sites were observed for the 517-bp insertion construct, one 20 bp and the other 24 bp right of position −32L. Therefore, 11 of 12 of the observed junction sequences of these three constructs had the left boundary within a 14-bp range, 20 to 34 bp right of position −32L. These data again demonstrate that the left flanking region positions the deletion boundary a short distance to the right, even when this region is more than 500 bp away from the micronucleus-limited region.
The flanking regulatory region appears to span an extended sequence.
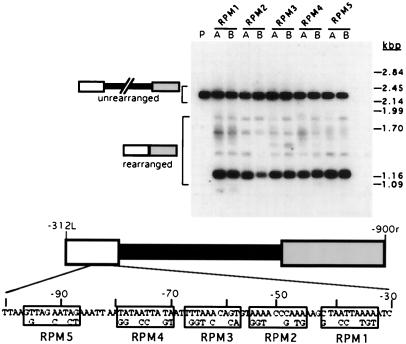
The R-element flanking region appears to control the rearrangement boundary much like the M-element A5G5; nevertheless, the actual sequence must be quite different. An A5G5-like motif does not exist in this region, nor does inspection of the sequence reveal any obvious candidate for a simple cis-acting sequence motif. Our deletion analysis above indicated that cis-acting sequences were present within the 70-bp region between −32L and −101L, but it could not determine whether the important sequences consisted of one or more simple motifs or, alternatively, a single, rather extended sequence. For instance, we could account for our data by invoking the presence of a somewhat simple motif that spans the −63L position since this region is altered or removed by all deletions that show reduced rearrangement activity. To determine whether any particular sequence within the flanking regulatory region is critical to its function, we introduced clusters of point mutations in 10-bp blocks throughout the region between −32L and −95L. The rearrangement activities of five such RPM constructs were examined. The introduced sequence changes (Fig. 6) are as follows: construct RPM1, six nucleotide changes between −33L and −42L; RPM2, six changes between −46L and −55L; RPM3, six changes between −58L and −67L; RPM4, six nucleotide changes between −70L and −79L; and RPM5, five changes between −86L and −95L. Southern blot analysis of their rearrangement activities is also shown in Fig. 6. We expected that mutation of a simple cis-acting sequence would reduce rearrangement activity similarly as was observed upon removal of these sequences (Fig. 3). However, we found that all five of these altered R elements displayed normal rearrangement activity. Hybridization with an oligonucleotide probe that is specific for the major chromosomal deletion junction showed that the rearrangement of each construct was accurate (data not shown). These results argue against the existence of a simple sequence in this region regulating rearrangement. In particular, normal rearrangement of the RPM3 construct, which contains six changes between −58L and −67L, argues against the existence of a simple regulating sequence spanning the −63L position. Because we changed only about half (29 of 63) of the nucleotides between −32L and −95L, it is possible that we failed to mutate key nucleotide positions. However, the sequences that we did not alter represent mostly blocks of A and T nucleotides that are abundant throughout this region and thus unlikely to contain such a specific signal. We believe that these data indicate that the nature of the cis-acting sequence is somewhat complex and tolerant of mutation.
FIG. 6.
Clustered mutations in the left flanking region have little effect on rearrangement efficiency or accuracy. Each R-element construct contains five or six changes within 10-bp blocks of the left flanking region between −32L and −95L. Southern blot hybridization analysis of two pools, A and B, each containing DNA from >10 transformants was used to assess the ability of each construct to undergo accurate rearrangement. The plasmids, RPM1 to RPM5, used for transformation are indicated above the pair of lanes. DNA was digested with BamHI prior to electrophoresis and transfer to a nitrocellulose membrane. Lane P is plasmid RPM1 digested with BamHI. The positions of PstI-digested lambda DNA size standards are shown to the right. A diagram of the R element and an enlargement of the left flanking sequence between −30L and −100L are shown at the bottom. The clusters of mutations that create each construct, boxed and labeled with their corresponding construct names, are shown in bold beneath this sequence.
The right-side flanking control region can substitute for the sequences flanking the left end of the micronucleus-limited region.
All data collected thus far indicate that sequences in the left-end R-element flanking region control the rearrangement boundary from a distance like the M-element A5G5. An additional property of the A5G5 sequence is that two copies, one outside each boundary, are required for efficient rearrangement. Even the boundary-controlling sequence flanking the R element cannot effectively substitute for one of the A5G5 sequences that is located outside the M element (20). The two R-element flanking regions share no obvious sequence identity. Nevertheless, if the R-element flanking regulatory sequences are truly analogous to the A5G5 sequence, then the same functional sequence found flanking the left side should also be contained outside the right end. To address this issue, we created three chimeric R-element constructs each containing sequences originally from the right flanking region immediately flanking both right and left sides of the R element. Diagrams of these constructs are shown in Fig. 7. The first two constructs replaced the entire left side with either the first 135 bp or first 334 bp from the right side of the R element (LF−1r:−135r and LF−1r:−334r, respectively). The third construct, LF−1r:−135r:−312L, contained the first 135 bp from the right side in place of the first 100 bp of the left side but retained the left side flanking sequences between −101L and −312L.
Southern blot hybridization analysis of DNA from these transformants is shown in Fig. 7. The R element in the first construct, LF−1r:−135r, rearranged rather well, albeit with somewhat reduced activity. Increasing the amount of the duplicated right flanking sequence on the left side increased rearrangement efficiency almost to the level of normal activity. Rearrangement occurred very efficiently in the LF−1r:−135r:−312L construct that also retained some of the left flanking region. These data indicate that sequences from the right flanking DNA are able to functionally replace the sequences that specify the left deletion boundary.
The rearrangement supported by these constructs appears to be accurate. The predominant hybridizing species has the size expected for complete deletion of the micronucleus-limited region. In addition to this major rearranged species, we detected a minor rearranged species that was ∼200 bp larger. We were unable to determine the exact sequence at the junction of the major rearranged products since the products of precise deletion contain palindromic sequences that could not be amplified or cloned effectively. However, we were able to identify a junction that joined the repeated −1r position on the left with position +880 on the right for both the LF−1r:−135r construct and the LF−1r:−135r:−312L construct. Identification of this junction sequence demonstrates that the duplicated right flanking sequence can specify the same boundary point even when placed out of normal context on the left side of the R element.
A wide variety of sequences at the boundaries can participate in rearrangement.
In the study above, we collected a large number of novel deletion junction sequences (Table 2). As most of our constructs were altered on their left sides, most of these junctions retained the normal right rearrangement boundary but utilized a unique left boundary. We aligned these unique boundary sites and examined the sequences for nucleotide preferences at the eight positions on each side of the junction. There appears to be no sequence bias at these 16 positions. Previously, Saveliev and Cox (34, 35) found that DNA breaks that occur during conjugation at the ends of chromosomal M and R elements have a common structure. These breaks all had four-nucleotide 5′ extensions, and 11 of 12 breaks terminated with a 3′ adenosine residue on the recessed strand. Based on this observation, the 3′ adenosine was proposed to be an important sequence feature at the rearrangement boundary. Many of the novel boundaries identified in this study have an appropriately positioned adenosine to fit this proposed consensus, but at least one-third do not. Therefore, a 3′ adenosine appears not to be required at each boundary, although it may be a preferred structure that has been selected for at chromosomal boundaries. We must note that we can only infer breakage sites from our junction sequences since the mechanism of breakage and joining is still largely unknown. Nevertheless, we find that the rearrangement boundary is highly permissive of sequence variation.
TABLE 2.
Unique junction sequences of modified R constructs
| Construct | Boundarya
|
No. observed | |
|---|---|---|---|
| Left | Right | ||
| Chromosomal | TTTATAAACA−1L gtgattca | cttccttaa−1r ACAATTTGAATG | Majorb |
| R element | TAAACAGTGA+3 ttcaaaaa | cttaaacaa−4r TTTGAATGAAAA | Minor |
| TAAACAGTGA+3 ttcaaaaa | cttccttaa−1r ACAATTTGAATG | Minor | |
| −203L/−391r | TAAACAGTGA−3 ttcaaaa | gctttaatt+416 GAAAAAAAACGT | 1 |
| Δ−31L:+3 | ATTTTTGTAT+32 tttttggtt | cttaaacaa−1r TTTGAATGAAAA | 4 |
| Δ−31:+24 | TAAAATAAAC+50 ttttttatg | cttccttaa−1r ACAATTTGAATG | 2 |
| Δ−63L:−2L | TATTTTTTGG+39 ttaaaataa | cttaaacaa−4r TTTGAATGAAAA | 4 |
| Δ−63L/+24 | ATGAAAATAA+66 aattttatt | cttaaacaa−4r TTTGAATGAAAA | 1 |
| TGTTGCCAAA+84 tattatttt | cttccttaa−1r ACAATTTGAATG | 1 | |
| Δ−31L/+3 i48c | AAAGGCCAGCi34 aaaaggcc | cttccttaa−1r ACAATTTGAATG | 3 |
| AAAAGGCCAGi33 caaaaggcc | cttccttaa−1r ACAATTTGAATG | 1 | |
| AAAATGGTGG+20 gaatttttg | cttccttaa−1r ACAATTTGAATG | 1 | |
| Δ−31L/+3 i342c | CTATCAGGACi32 atagcgttg | cttccttaa−1r ACAATTTGAATG | 5 |
| Δ−31L/+3 i517c | GCGAAGAACTi24 ccagcatga | cttccttaa−1r ACAATTTGAATG | 1 |
| TGGGCGAAGi20 aactccagc | cttccttaa−1r ACAATTTGAATG | 1 | |
| −1r:−135r | TTCATTCAAALF−1r ttgtttaag | taatagtaa+880 TTAATTGAGTAT | 1 |
| −312L:−1r:−135r | TTCATTCAAALF−1r ttgtttaag | taatagtaa+880 TTAATTGAGTAT | 1 |
| TTCTTTATAA−146L tctgactc | cttccttaa−1r ACAATTTGAATG | 1 | |
Macronucleus-retained sequences are capitalized, and eliminated sequences are in lowercase. Positions of the left and right nucleotides joined at the junction are as defined in Fig. 1. Underlined nucleotides indicate repetition of the bases on each end of the deleted segment which makes the exact boundaries ambiguous. In these cases, the boundaries were assigned by using previously identified junctions as a guide.
The predominant junction observed from the rearrangement of endogenous R elements.
When the junctions were observed in the non-R-element sequences inserted into the construct, the position is given as the distance from position −32L.
DISCUSSION
The A5G5 polypurine tracts, which are located ∼45 bp outside each end of the M element, were shown previously to specify the positions of the rearrangement boundaries (19, 20). Prior to this study, A5G5 was the only characterized cis-acting sequence known to control deletion element rearrangement. No identical or similar sequence had been found flanking the seven other characterized deletion elements, which raised the possibility that the M element utilizes a unique mechanism of rearrangement. Yet, it seems unlikely that the thousands of deletion elements are eliminated by different rearrangement systems. In this study, we have shown that the sequences located outside the micronucleus-limited region of the R element are required for accurate and efficient rearrangement. Upon altering the spacing between these sequences and the micronucleus-limited region, we found that the rearrangement boundary always shifted to remain in relative juxtaposition with the flanking region. This was true even when the flanking region was several hundred base pairs removed from the micronucleus-limited region. Furthermore, sequences from the right side flanking region effectively substituted for essential sequences on the left side, indicating that the two flanking regions of the R element are interchangeable despite lacking obvious sequence identity. These data demonstrate that the R-element flanking regions contain sequences that function similarly to the M-element A5G5 tracts in that they serve to position the deletion boundaries at a specific distance. We believe that this study suggests that many, and perhaps most, Tetrahymena deletion elements use flanking regulatory sequences to specify the location of rearrangement boundaries.
Although the R-element flanking sequences perform the same function as the A5G5 tract, the nature of the sequence recognition, at least superficially, seems different. Mutations within the G5 portion of the M element are sufficient to block rearrangement (20). In contrast, we were able to localize the essential regulatory sequences on the left side of the R element only to within a 70-bp region (−32L to −101L). Clusters of point mutations introduced throughout this region did not reduce rearrangement activity (Fig. 6). It has been proposed that the M and R elements both belong to a class of deletion elements that have a common rearrangement mechanism, but that they are members of different families that are distinguished by their use of particular flanking regulatory sequences (19). Our finding that the M- and R-element flanking regions contain different yet functionally equivalent sequences provides the first direct support for this view. The apparent complexity of the R-element flanking regulatory region helps us to reconcile with the difficulty of identifying common controlling sequences among the different known elements.
Aside from the A5G5 tract, the flanking regulatory regions of the M element may be quite similar to those of other elements. Like our modified R elements, M-element constructs that have less than 100 bp of flanking sequence displayed reduced rearrangement activity (20). In fact, the A5G5-containing minimal-flanks construct (which has 65 bp flanking the left boundary and 70 bp flanking the right) rearranges rather poorly (20). The rearrangement efficiency of this construct is probably not much different from that of the R element −312L/−70r construct (Fig. 2). Even though the A5G5 tract is the primary boundary determinant, additional flanking sequences appear to greatly facilitate the use of this signal. In light of the fact that the Δ−101L:−61L construct produces a wild-type junction (Fig. 3), the R element may also contain a sufficient boundary determinant within the first 60 bp, a location very similar to that of the M-element A5G5. However, this accurate junction is a minor product of rearrangement for this construct, indicating that these sequences function poorly without the flanking DNA between −60L and −100L. A third deletion element, mse 2.9, also requires >66 bp of flanking DNA in order to accurately rearrange (29). These observations may imply that the flanking regulatory sequences are bipartite, containing some sequences that primarily determine accuracy and others that enhance efficiency of rearrangement.
We found that the R-element construct with only 100 bp of the left flanking sequence displays normal rearrangement efficiency. Following this observation, we were quite intrigued that a construct (Δ−101L:−2L) completely lacking these sequences also displays appreciable rearrangement activity. It appears that flanking sequences beyond the first 100 bp contain some functional redundancy with the flanking region proximal to the micronucleus-limited region. Since we did not identify an obligatory role for these distal sequences in normal rearrangement, it is unclear as to whether these sequences normally contribute to efficient rearrangement activity or, alternatively, that removal of the primary regulatory sequences revealed a cryptic controlling sequence.
In this study, we did not address whether sequences within the micronucleus-limited region are required for R-element rearrangement. An M element lacking roughly half of its micronucleus-limited region displays reduced rearrangement (20). It appears that both M and R elements contain similar cis-acting sequences within their micronucleus-limited regions that function independently of orientation and position (44). The role of these internal sequences is otherwise unknown. It is clear from this study of the R element and previous studies of the M element (19, 20) that the flanking regulatory sequences are sufficient to position the boundaries. We have proposed that the internal sequences serve to target an element for elimination and rely on the flanking sequences to limit the extent of deletion (14, 43). The interaction between the functions of these two different types of cis-acting sequences remains to be determined.
We believe that using flanking regulatory sequences to position the boundary offers distinct advantages for accurate rearrangement. The pairwise use of identical, orientation-dependent regulatory sequences is likely to increase specificity and limit aberrant rearrangement. Pairing of recombination signals appears to occur prior to V(D)J recombination (23). It is compelling to speculate that Tetrahymena rearrangement requires pairing of flanking regulatory sequences, and such pairing is a common strategy used to ensure specificity of a variety of DNA deletion events. Based on the analysis of M-R-element chimeras, a boundary determined by an A5G5 tract does not effectively join to a boundary specified by an R-element flanking regulatory sequence (20). The use of different sequences to control the rearrangement of adjacent elements would greatly minimize aberrant recombination between elements. In addition, the use of external sequences to limit deletion appears to be an efficient way to eliminate elements of widely varying size. Increasing the size of the R element by 50% (the Δ−31L:+3 i517 construct) did not affect the accuracy of rearrangement (Fig. 5). This feature of the mechanism may in part account for the large variability in size and sequence of the Tetrahymena deletion elements.
The finding that DNA breaks at the ends of chromosomal M and R elements have a common structure (34, 35) provides further evidence that these elements share a rearrangement mechanism. The breaks identified are consistent with a mechanism that is similar to transposition processes. At least some internal eliminated sequences of hypotrichous ciliates appear to be related to transposable elements (15, 24, 26). In contrast, the Tetrahymena deletion elements lack most of the structural characteristics normally associated with transposable elements (reviewed in reference 14). The use of sequences exclusively outside the element to position the sites of excision is not known to occur for any transposon-like process. Still, it is logical to think that developmentally programmed DNA rearrangements found in different ciliate species have a common origin, although we lack the knowledge to clearly recognize the connections. Our identification of a probable common mechanism controlling deletion element rearrangement is an important step toward understanding the regulation and origin of developmentally programmed genome reorganization.
ACKNOWLEDGMENTS
We thank C. Randolph, M. DuBois, and D. Frank for critical reading of the manuscript.
This work was supported by National Research Service Award GM16315 to D.L.C. and Public Health Service grant GM26210 to M.-C.Y., both from the National Institutes of Health.
REFERENCES
- 1.Altschuler M I, Yao M C. Macronuclear DNA of Tetrahymena thermophila exists as defined subchromosomal-sized molecules. Nucleic Acids Res. 1985;13:5817–5831. doi: 10.1093/nar/13.16.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austerberry C F, Allis C D, Yao M C. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci USA. 1984;81:7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austerberry C F, Snyder R O, Yao M C. Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic Acids Res. 1989;17:7263–7272. doi: 10.1093/nar/17.18.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austerberry C F, Yao M C. Nucleotide sequence structure and consistency of a developmentally regulated DNA deletion in Tetrahymena thermophila. Mol Cell Biol. 1987;7:435–443. doi: 10.1128/mcb.7.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austerberry C F, Yao M C. Sequence structures of two developmentally regulated, alternative DNA deletion junctions in Tetrahymena thermophila. Mol Cell Biol. 1988;8:3947–3950. doi: 10.1128/mcb.8.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerman S. The diminution of heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda) Chromosoma. 1977;60:297–344. doi: 10.1007/BF00292858. [DOI] [PubMed] [Google Scholar]
- 7.Boveri T. Uber Differenzierung der Zellkerne wahrend der Furchung des Eies von Ascaris megalocephala. Anat Anz. 1887;2:688–693. [Google Scholar]
- 8.Bruns P J, Katzen A L, Martin L, Blackburn E H. A drug resistant mutation maps in the ribosomal DNA of Tetrahymena. Proc Natl Acad Sci USA. 1985;82:2844–2846. doi: 10.1073/pnas.82.9.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassidy-Hanley D, Yao M C, Bruns P J. A method for mapping germ line sequences in Tetrahymena thermophila using the polymerase chain reaction. Genetics. 1994;137:95–106. doi: 10.1093/genetics/137.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalker D L, Ward J G, Randolph C, Yao M-C. Microinjection of Tetrahymena thermophila. Methods Cell Biol. 1999;62:469–484. doi: 10.1016/s0091-679x(08)61551-4. [DOI] [PubMed] [Google Scholar]
- 11.Chalker D L, Yao M-C. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol Cell Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chau M-F, Orias E. Developmentally programmed DNA rearrangement in Tetrahymena thermophila: isolation and sequence characterization of three new alternative deletion systems. Biol Cell. 1996;86:111–120. doi: 10.1016/0248-4900(96)84773-3. [DOI] [PubMed] [Google Scholar]
- 13.Conover R K, Brunk C F. Macronuclear DNA molecules of Tetrahymena thermophila. Mol Cell Biol. 1986;6:900–905. doi: 10.1128/mcb.6.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne R S, Chalker D L, Yao M-C. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- 15.Doak T G, Doerder F P, Jahn C L, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common D35E motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg A P, Vogelstein B. Addendum—a technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 18.Gaertig J, Gorovsky M A. Efficient mass transformation of Tetrahymena thermophila by electroporation of conjugants. Proc Natl Acad Sci USA. 1992;89:9196–9200. doi: 10.1073/pnas.89.19.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godiska R, James C, Yao M C. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev. 1993;7:2357–2365. doi: 10.1101/gad.7.12a.2357. [DOI] [PubMed] [Google Scholar]
- 20.Godiska R, Yao M C. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell. 1990;61:1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- 21.Gorovsky M A, Yao M C, Keevert J B, Pleger G L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 22.Heinonen T Y, Pearlman R E. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J Biol Chem. 1994;269:17428–17433. [PubMed] [Google Scholar]
- 23.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 24.Jahn C L, Doktor S Z, Frels J S, Jaraczewski J W, Krikau M F. Structures of the Euplotes crassus Tec1 and Tec2 elements: identification of putative transposase coding regions. Gene. 1993;133:71–78. doi: 10.1016/0378-1119(93)90226-s. [DOI] [PubMed] [Google Scholar]
- 25.Katoh M, Hirono M, Takemasa T, Kimura M, Watanabe Y. A micronucleus-specific sequence exists in the 5′-upstream region of calmodulin gene in Tetrahymena thermophila. Nucleic Acids Res. 1993;21:2409–2414. doi: 10.1093/nar/21.10.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klobutcher L A, Herrick G. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog Nucleic Acid Res Mol Biol. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- 27.Korsmeyer S J. Chromosomal translocations in lymphoid malignancies reveal novel proto-oncogenes. Annu Rev Immunol. 1992;10:785–807. doi: 10.1146/annurev.iy.10.040192.004033. [DOI] [PubMed] [Google Scholar]
- 28.Kubota S, Kuro-O M, Mizuno S, Kohno S. Germ-line restricted, highly repeated DNA sequences and their chromosomal localization in a Japanese hagfish (Eptatretus okinoseanus) Chromosoma. 1993;102:163–173. doi: 10.1007/BF00387731. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Pearlman R E. Programmed DNA rearrangement from an intron during nuclear development in Tetrahymena thermophila: molecular analysis and identification of potential cis-acting sequences. Nucleic Acids Res. 1996;24:1943–1949. doi: 10.1093/nar/24.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakai Y, Kubota S, Kohno S. Chromatin diminution and chromosome elimination in four Japanese hagfish species. Cytogenet Cell Genet. 1991;65:196–198. doi: 10.1159/000133087. [DOI] [PubMed] [Google Scholar]
- 31.Patil N S, Hempen P M, Udani R A, Karrer K M. Alternate junctions and microheterogeneity of Tlr1, a developmentally regulated DNA rearrangement in Tetrahymena thermophila. J Eukaryot Microbiol. 1997;44:518–522. doi: 10.1111/j.1550-7408.1997.tb05733.x. [DOI] [PubMed] [Google Scholar]
- 32.Prescott D M. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E T, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Saveliev S V, Cox M M. Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J. 1996;15:2858–2869. [PMC free article] [PubMed] [Google Scholar]
- 35.Saveliev S V, Cox M M. Transient DNA breaks associated with programmed genomic deletion events in conjugating cells of Tetrahymena thermophila. Genes Dev. 1995;9:248–255. doi: 10.1101/gad.9.2.248. [DOI] [PubMed] [Google Scholar]
- 36.Schatz D G, Oettinger M A, Schlissel M S. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 37.Seifert H S, Chen E Y, So M, Hefron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tondravi M M, Yao M C. Transformation of Tetrahymena thermophila by microinjection of ribosomal RNA genes. Proc Natl Acad Sci USA. 1986;83:4369–4373. doi: 10.1073/pnas.83.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tycko B, Sklar J. Chromosomal translocations in lymphoid neoplasia: a reappraisal of the recombinase model. Cancer Cells. 1990;2:1–8. [PubMed] [Google Scholar]
- 40.Wells J M, Ellingson J L, Catt D M, Berger P J, Karrer K M. A small family of elements with long inverted repeats is located near sites of developmentally regulated DNA rearrangement in Tetrahymena thermophila. Mol Cell Biol. 1994;14:5939–5949. doi: 10.1128/mcb.14.9.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 42.Yao M-C. Amplification of ribosomal RNA gene in Tetrahymena. In: Busch H, Rothblum L, editors. The cell nucleus. Vol. 12. New York, N.Y: Academic Press, Inc.; 1982. pp. 127–153. [Google Scholar]
- 43.Yao M-C. Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet. 1996;12:26–30. doi: 10.1016/0168-9525(96)81385-0. [DOI] [PubMed] [Google Scholar]
- 44.Yao, M.-C., R. Godiska, R. C. Callahan, and C.-H. Yao. Unpublished data.
- 45.Yao M-C, Yao C-H, Monks B. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- 46.Yao M C, Choi J, Yokoyama S, Austerberry C F, Yao C H. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell. 1984;36:433–440. doi: 10.1016/0092-8674(84)90236-8. [DOI] [PubMed] [Google Scholar]
- 47.Yao M C, Gorovsky M A. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48:1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- 48.Yao M C, Kimmel A R, Gorovsky M A. A small number of cistrons for ribosomal RNA in the germinal nucleus of a eukaryote, Tetrahymena pyriformis. Proc Natl Acad Sci USA. 1974;71:3082–3086. doi: 10.1073/pnas.71.8.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao M C, Yao C H. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol Cell Biol. 1989;9:1092–1099. doi: 10.1128/mcb.9.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao M C, Zheng K, Yao C H. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell. 1987;48:779–788. doi: 10.1016/0092-8674(87)90075-4. [DOI] [PubMed] [Google Scholar]