Abstract
A site in the Epstein-Barr virus (EBV) transforming protein LMP1 that constitutively associates with the tumor necrosis factor receptor 1 (TNFR1)-associated death domain protein TRADD to mediate NF-κB and c-Jun N-terminal kinase activation is critical for long-term lymphoblastoid cell proliferation. We now find that LMP1 signaling through TRADD differs from TNFR1 signaling through TRADD. LMP1 needs only 11 amino acids to activate NF-κB or synergize with TRADD in NF-κB activation, while TNFR1 requires ∼70 residues. Further, LMP1 does not require TRADD residues 294 to 312 for NF-κB activation, while TNFR1 requires TRADD residues 296 to 302. LMP1 is partially blocked for NF-κB activation by a TRADD mutant consisting of residues 122 to 293. Unlike TNFR1, LMP1 can interact directly with receptor-interacting protein (RIP) and stably associates with RIP in EBV-transformed lymphoblastoid cell lines. Surprisingly, LMP1 does not require RIP for NF-κB activation. Despite constitutive association with TRADD or RIP, LMP1 does not induce apoptosis in EBV-negative Burkitt lymphoma or human embryonic kidney 293 cells. These results add a different perspective to the molecular interactions through which LMP1, TRADD, and RIP participate in B-lymphocyte activation and growth.
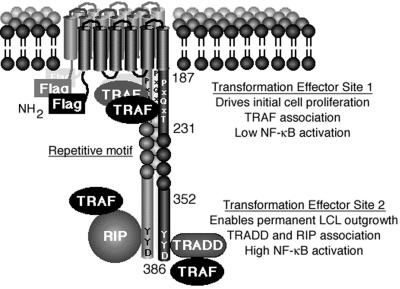
Epstein-Barr virus (EBV) latent infection membrane protein 1 (LMP1) is essential for EBV-mediated growth transformation of resting primary human B lymphocytes into indefinitely proliferating lymphoblastoid cell lines (LCLs) (30). Genetic and biochemical evidence indicates that the signal transduction pathway through which LMP1 mediates B-lymphocyte growth transformation resembles those of activated tumor necrosis factor receptors (TNFR) (26, 28, 31, 43). LMP1 consists of a 24-amino-acid N-terminal cytoplasmic domain, six transmembrane domains, and a 200-amino-acid C-terminal cytoplasmic tail (Fig. 1). Although no specific sequence of the N-terminal cytoplasmic domain is essential for growth transformation, the N terminus fulfills a critical structural role by tethering the first transmembrane domain to the cytoplasm (25, 30). The six transmembrane domains collectively enable LMP1 to self-aggregate in the plasma membrane similarly to a capped receptor. Aggregation of LMP1 in the plasma membrane causes two specific sites within the C-terminal cytoplasmic tail to constitutively mediate essential transforming signals through association with proteins that ordinarily mediate ligand-induced signals from activated TNFR family members (7, 12, 15, 16, 19, 22, 25, 31, 50, 56). These two transformation effector sites (TES1 and TES2) are within C-terminal NF-κB activating regions CTAR1 and CTAR2 (24, 42).
FIG. 1.
Diagram of LMP1. The Flag epitope was introduced at the amino terminus (NH2). LMP1 residues 187, 231, 352, and 386 are marked. LMP1 constitutively aggregates in the plasma membrane and associates with TRAFs, TRADD, and RIP. TES1 aggregates TRAFs to mediate low-level NF-κB activation and initial B-lymphocyte growth transformation. TES2 aggregates RIP or TRADD, both of which associate with TRAFs to mediate high-level NF-κB activation and enable permanent LCL outgrowth.
TES1/CTAR1 (residues 187 to 231 [Fig. 1]) interacts with TNFR-associated factors (TRAFs) and is sufficient for mediating initial B-lymphocyte growth transformation (31, 43). TRAF1, TRAF2, and TRAF5 mediate NF-κB activation from TES1/CTAR1 (4, 10, 43, 49). Deletion of the TRAF interaction site results in EBV recombinants that are unable to growth transform primary B lymphocytes (26). The TES1/CTAR1 amino acid sequence is similar to sites in CD40 and CD30 that are critical for NF-κB activation (9, 40). CD40 is the receptor for a T-cell surface ligand that activates B-lymphocyte growth and differentiation (2, 8, 32, 44), and CD30 is the receptor for a T-cell surface ligand that can negatively affect heavy-chain class switching (5, 36). In retrospect, LMP1 mimicry of an activated TNFR is not surprising given the similar effects of LMP1 and CD40 on B-lymphocyte growth and gene activation (2, 8, 9, 23, 29, 32, 44, 47).
TES2/CTAR2 is at the LMP1 C terminus and activates NF-κB through the TNFR1-associated death domain protein TRADD (28). The adverse consequences of mutation of the three C-terminal LMP1 codons from YYD to ID on TRADD interaction in the yeast Saccharomyces cerevisiae, on TRADD association in mammalian cells, on NF-κB activation, on TRADD synergy with LMP1 in NF-κB activation, and on B-lymphocyte growth transformation link TRADD to these TES2/CTAR2 effects (28). The tyrosines are not specifically required since substitution with codons encoding phenylalanine is indistinguishable from wild type in TRADD association in lymphocytes, in NF-κB activation, and in B-lymphocyte growth transformation (28). Thus, at this level of genetic analysis, TES2 appears to coincide with CTAR2 (15), and TRADD is a key signal-transducing intermediate. The constitutive association of LMP1 with TRADD in LCLs (28) contrasts with TNFR1, which recruits TRADD in response to ligand binding and receptor aggregation (22, 21).
The objective of the experiments reported here was to investigate further the biochemistry of LMP1 TES2/CTAR2 interaction with TRADD and the expected proapoptotic effects. Since receptor-interacting protein (RIP) is implicated in TNFR1 and TRADD-mediated activation of NF-κB and apoptosis (20, 33, 51, 54), we also investigated the role of RIP in TES2/CTAR2-mediated signaling.
MATERIALS AND METHODS
Viruses, cells, and DNA clones.
SVT35 Jurkat T-lymphoma cells and RIP-deficient 35.3.13 cells (54) were cultivated in RPMI 1640 (Life Technologies) in 10% fetal bovine serum (Gemini) supplemented with penicillin-streptomycin and glutamine (Life Technologies). RIP vector pRK-F-RIP, a gift from David Goeddel (Tularik) (20), was transferred to pcDNA3 in order to Myc tag the protein. Lymphoblastoid and human embryonic kidney 293 (HEK293) cell lines are described elsewhere (25, 26). N-terminally Flag-tagged LMP-1 (F-LMP1) expression vectors were derived by endonuclease deletion or codon insertions as described before (26).
Yeast two-hybrid assay.
The methods used for cell culture, yeast transformation, and β-galactosidase detection in S. cerevisiae Y190 are described elsewhere (13). Gal4 DNA binding domain (DBD)-LMP1 fusions are derived from F-LMP1 clones (see above) or from a prior study (28). The Gal4-activation domain (AD)-RIP fusion was derived from pRK-F-RIP subcloned into pACTII.
Coimmunoprecipitation analyses.
Analyses were done with an M2 affinity gel (Kodak) as before (9, 26) except that immunoblots were analyzed with RIP antibody (Pharmingen).
NF-κB activation.
3x-κB-L luciferase reporter and mut-κB-L negative control were gifts from Tom Mitchell and Bill Sugden (University of Wisconsin, Madison) (42). Methods for electroporation and analysis are described elsewhere (9, 26, 54).
Apoptosis assays.
BJAB cells were electroporated as before (10) with 30 μg of LMP1 vector DNA and 10 μg of green fluorescent protein (GFP) vector pEGFP (Clontech). Dead cells were removed by Ficoll-Hypaque gradient centrifugation. Viable cells were cultured for 10 h and then treated for 8 h with cycloheximide or left untreated. Next, cells were fixed in 1% paraformaldehyde, permeabilized with 70% ethanol, and stained for DNA with 30 μg of propidium iodide (Molecular Probes) per ml in Dulbecco’s phosphate-buffered saline PBS (Gibco) with 1% fetal bovine serum and 0.2 mg of RNase A (Sigma) per ml.
The DNA content of GFP-positive cells was quantitated by fluorescence-activated cell sorting (FACS) analysis (Becton Dickinson FACS Calibur). HEK293 cells (5 × 105) were cotransfected by using Superfect (Qiagen) with 2 μg of pcDNA3 vector (Invitrogen), pcDNA3 LMP1 vector DNA, or pcDNA3 F-LMPΔ188-352 (41) and with 0.35 μg of 3x-κB-L NF-κB, 0.35 μg of pGK-β-galactosidase, and where indicated 0.25 μg of nondegradable I-κBα vector pCMV4 I-κBα S32AS36A (a gift from Dean Ballard, Vanderbilt University) (6). After 24 h, cells were fixed with 1% paraformaldehyde, stained for β-galactosidase, and photographed. A duplicate was tested for activation of the NF-κB reporter and then analyzed by Western immunoblotting for LMP1 with S12 and M5 antibodies (Kodak) as described before (9, 26).
RESULTS
LMP1 residues 376 to 386 engage TRADD to mediate NF-κB activation.
TRADD interaction with LMP1 was originally identified as the result of a yeast two-hybrid screen with LMP1 amino acids 355 to 386 (28). A yeast two-hybrid assay was used to delineate more precisely the TES2/CTAR2 residues (355 to 386 [Fig. 1]) required for TRADD interaction. LMP1 amino acids 182 to 386 comprising TES1/CTAR1 and TES2/CTAR2 (Fig. 1) were fused to the Gal4 DBD so that interaction of TES1/CTAR1 with a Gal4 AD-TRAF3 fusion could serve as a positive control for bait expression. TRADD interacted with the LMP1 bait at a substantially lower level than TRAF3 (Table 1). Deletion of amino acids 356 to 365, 366 to 371, or 372 to 375 (in F-LMP1Δ356-365, F-LMP1Δ366-371, or F-LMP1Δ372-375) did not substantially weaken TRADD interaction, whereas deletion of amino acids 374 to 386 abrogated TRADD interaction, similar to the effect of the Y384YD386-to-ID mutation (in F-LMP1ΔID) (Table 1). None of these deletions affected TRAF3 interaction. Since LMP1 amino acids 355 to 386 are sufficient for TRADD interaction and residues 356 to 365, 366 to 371, or 372 to 375 are not critical, the critical residues are likely to be from positions 376 to 386.
TABLE 1.
LMP1 interaction with TRADD and TRAF3 in a yeast two-hybrid assaya
| Gal4 DBD-LMP1 (codons of LMP1) | Interaction with:
|
|
|---|---|---|
| Gal4 AD-TRADD | Gal4 AD-TRAF3 | |
| 182–386 | + | ++ |
| 182–386Δ356–365 | + | ++ |
| 182–386Δ366–371 | + | ++ |
| 182–386Δ372–375 | + | ++ |
| 182–373 | − | ++ |
| 182–383ID | − | ++ |
S. cerevisiae Y190 was transformed with pAS1 vectors expressing the Gal4 DBD fused to the indicated codons of LMP1 and pACT vectors expressing the Gal4 AD fused to TRADD codons 195 to 312 or TRAF3 codons 312 to 568. Transformed yeast cells were selected on medium lacking tryptophan and leucine. Colonies were transferred to filters and lysed by freeze-thawing, and β-galactosidase was detected with X-Gal. Blue colonies after 1 h at 37°C were scored ++, and light blue colonies were scored +. Yeast cells transformed with these Gal4 DBD-LMP1 DNAs alone scored negative for β-galactosidase.
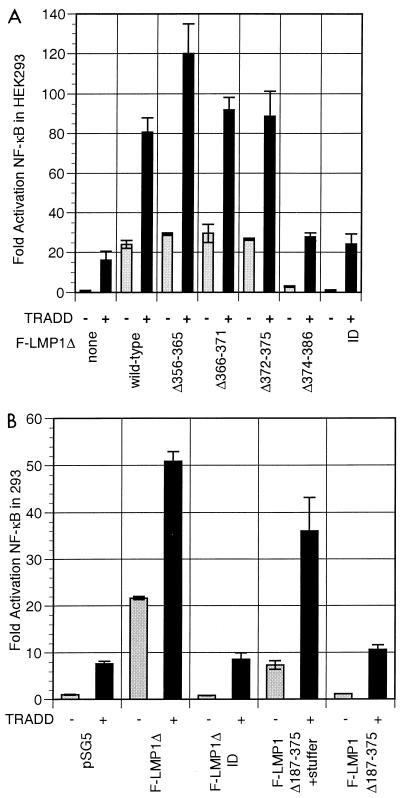
To correlate the TRADD-LMP1 interaction in yeast cells with NF-κB activation in mammalian cells, an F-LMP1 TES2/CTAR2 expression vector, F-LMP1Δ, was constructed by fusing codons 352 to 386 in frame with and 3′ to codons encoding the LMP1 amino terminus and six transmembrane domains (Fig. 1). F-LMP1Δ and TRADD are potent activators of NF-κB in HEK293 cells (Fig. 2A). Consistent with the yeast data, deletion of residues 356 to 365, 366 to 372, or 373 to 375 had minimal effect on F-LMP1Δ-mediated NF-κB activation, whereas deletion of residues 374 to 386 or mutation of the terminal Y384YD386 residues to ID did not activate NF-κB (Fig. 2A). TRADD overexpression activated NF-κB and synergistically activated NF-κB with F-LMP1Δ356-365, F-LMP1Δ366-372, or F-LMP1Δ373-375 vectors. We speculate that TRADD overexpression increases TRADD concentration and TRADD association with LMP1, resulting in synergistic NF-κB activation. No synergy was found with F-LMP1Δ374-386 or for F-LMP1ΔID. These results confirm that LMP1 amino acids 374 to 386 are critical for TES2/CTAR2-mediated NF-κB activation and for synergy with TRADD in NF-κB activation.
FIG. 2.
(A) Characterization of the LMP1 residues critical for engaging TRADD to synergistically activate NF-κB. Five million HEK293 cells were electroporated with 30 μg of pSG5 or TES2/CTAR2 vector F-LMP1Δ, 5 μg of TRADD vector where indicated, 2.5 μg of 3x-κB-L luciferase reporter, which has three copies of a major histocompatibility complex class I κB element and minimal fos promoter, and 2.5 μg of glucokinase promoter/β-galactosidase reporter to monitor transfection efficiency; 18 h later, lysates were analyzed for luciferase (Promega) and β-galactosidase (Tropix) according to the manufacturers’ directions on an Opticomp I luminometer. The base wild-type F-LMP1Δ vector is deleted for residues 187 to 351. Further deletions are indicated. ID indicates mutation of the terminal residues Y384YD386 to ID. Standard error of means are reported. In data not shown, protein levels of TRADD or LMP1 mutants were equivalent in separate transfections as analyzed by Western immunoblotting. (B) The 11 terminal residues of LMP1 are sufficient to engage TRADD to synergistically activate NF-κB. F-LMP1 vectors deleted for residues 187 to 375 or with a short stuffer consisting of residues HGHLGASLQY inserted between residues 186 and 376 were electroporated into HEK293 cells and analyzed as described for panel A.
To test whether LMP1 residues 376 to 386 are sufficient for TRADD interaction and NF-κB activation, codons for these residues were cloned in frame and 3′ to codons for the LMP1 N terminus and six membrane-spanning domains to create F-LMP1Δ187-375. When transfected into HEK293 cells, F-LMP1Δ187-375 did not activate NF-κB or synergize with TRADD (Fig. 2B). Since the inactivity of the terminal 11 amino acids could be due to their proximity to the plasma membrane, we tested another construct that has a 10-amino-acid stuffer (HGHLGASLQY) inserted between the last transmembrane domain and amino acids 376 to 386. This construct, F-LMP1Δ187-375+stuffer, induced almost 50% of the NF-κB activity of F-LMP1Δ (Fig. 2B). Further, F-LMP1Δ187-375+stuffer synergized with TRADD in NF-κB activation and conveyed 70% of the synergistic effect of F-LMP1Δ (Fig. 2B). These results indicate that the C-terminal 11 amino acids are sufficient for TRADD interaction and NF-κB activation.
LMP1 can synergize with TRADD mutants that are unable to engage TRAFs or efficiently interact with the TNFR1 death domain.
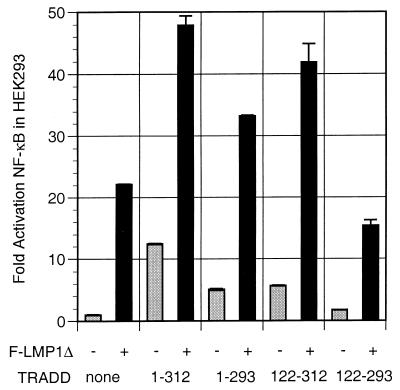
The TRADD cDNA clone that interacted in the yeast two-hybrid screen with LMP1 residues 355 to 386 consists of amino acids 195 to 312 and comprises the entire TRADD death domain (22, 28). To determine whether a smaller TRADD sequence could interact with LMP1, TRADD deletion mutations were tested for the ability to synergize with LMP1 in NF-κB activation (Fig. 3). TRADD 1-293 is competent for TRAF interactions and for some death domain interactions but is markedly diminished in TNFR1 death domain interaction and self-aggregation-mediated NF-κB activation (22, 45). TRADD 122-312 is unable to interact with TRAFs but has a complete death domain that can activate NF-κB when overexpressed. TRADD 122-293 is unable to interact with TRAFs and has an incomplete death domain that cripples TNFR1 interaction or self-aggregation-mediated NF-κB activation (22, 45). While it was, as expected, more than 50% deficient in NF-κB activation, TRADD lacking residues 294 to 312 (TRADD 1-293) retained 70% of wild-type TRADD synergy with LMP1 TES2/CTAR2 (Fig. 3). LMP1 TES2/CTAR2 synergized even better with TRADD 122-312 despite the inability of TRADD 122-312 to directly recruit TRAFs. Since a dominant negative TRAF2 can block TES2/CTAR2 induction of TES2/CTAR2 and TRADD synergy in NF-κB activation (28), these data indicate that TRADD 122-312 can aggregate with wild-type TRADD, RIP, or other proteins that can recruit TRAFs (20, 33, 51, 54). Overexpression of TRADD 122-293, which lacks a complete death domain and cannot interact with TRAFs, failed to activate NF-κB and reduced LMP1 TES2/CTAR2-mediated NF-κB activation by about 30%. This dominant negative effect is consistent with the notion that TRADD signaling from TES2/CTAR2 requires the ability of TRADD to recruit TRAFs or other proteins that can recruit TRAFs. Deletion of TRADD residues 294 to 312 probably compromises recruitment of death domain proteins that can recruit TRAF2, while the deletion of residues 1 to 121 abrogates direct TRAF recruitment.
FIG. 3.
LMP1 synergistically activates NF-κB with TRADD mutants that are unable to engage TRAFs or efficiently interact with the TNFR1 death domain. Five micrograms of full-length TRADD 1-312 vector DNA, 30 μg of TRADD 1-293 DNA (crippled for death domain association), 15 μg of TRADD 122-312 DNA (crippled for TRAF2 aggregation), or 15 μg of TRADD 122-293 DNA was cotransfected with 30 μg of pSG5 or TES2/CTAR2 vector F-LMP1Δ into HEK293 cells along with NF-κB and β-galactosidase reporter DNAs; analysis and reporting of data are as described for Fig. 2A. LMP1 and TRADD protein levels monitored by Western immunoblotting were equivalent between the individual transfections.
RIP interacts with LMP1.
The potential interaction of RIP with LMP1 was evaluated in a yeast two-hybrid assay in comparison with TRAF3 and TRADD (Table 2). RIP interacted directly with LMP1 residues 182 to 386, and mutation of the LMP1 C-terminal Y384YD386 to ID weakened RIP interaction. Overall, the interaction of LMP1 with RIP was somewhat weaker than that with TRADD. As expected, deletion of LMP1 residues 185 to 211, which includes the core TRAF binding site, abrogated TRAF3 interaction but did not affect RIP interaction. Moreover, RIP did not interact with residues 187 to 231 whereas TRAF3 bound efficiently to residues 187 to 231. Surprisingly, RIP did not interact with LMP1 amino acids 355 to 386 although this sequence is sufficient for TRADD interaction (28). These data indicate that RIP interacts with LMP1, probably through TES2/CTAR2, as evidenced by the adverse effect of the YYD-to-ID mutation, but with a broader or different sequence requirement than TRADD.
TABLE 2.
LMP1 interaction with RIP and TRAF3 in a yeast two-hybrid assaya
| Gal4 DBD-LMP1 (codons of LMP1) | Interaction with:
|
|
|---|---|---|
| Gal4 AD-RIP | Gal4 AD-TRAF3 | |
| 182–386 | + | ++ |
| 182–383ID | +/− | ++ |
| 182–386Δ185–211 | + | − |
| 182–386Δ212–240 | + | Not done |
| 187–231 | − | ++ |
| 355–386 | − | − |
S. cerevisiae Y190 was transformed with pAS1 vectors expressing the Gal4 DBD fused to the indicated codons of LMP1 and pACT vectors expressing the GAL4 AD fused to RIP codons 1 to 671 or TRAF3 codons 312 to 568. Transformed yeast cells were selected on medium lacking tryptophan and leucine. Colonies were transferred to filters and lysed by freeze thawing, and β-galactosidase was detected with X-Gal. Blue colonies after 1 h at 37°C were scored ++, and light blue colonies were scored +. Yeast cells transformed with these Gal4-DBD LMP1 DNAs alone scored negative for β-galactosidase.
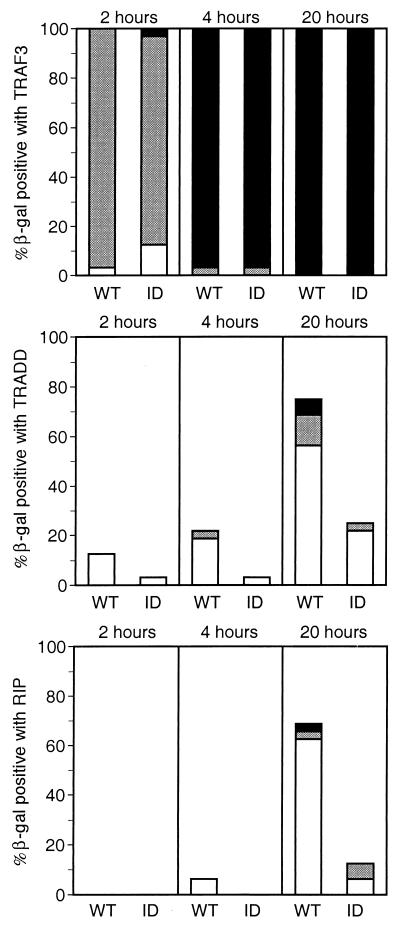
We further investigated RIP, TRADD, and TRAF3 binding to LMP1 by doubly transforming yeast with Gal4 DBD-LMP1 C-terminal tail vectors as bait and Gal4 AD-RIP, -TRADD, and -TRAF3 fusion protein expression vectors as prey. The extent of interaction was then assayed by in situ 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) color conversion using doubly selected, actively growing individual yeast colonies. This assay assesses the extent of interaction among different yeast clones where individual clones have different numbers of copies of the two expression vectors. β-Galactosidase production was monitored by assessing the chronology and intensity of color development over 20 h (Fig. 4). TRAF3 interacted strongly with wild-type LMP1 amino acids 182 to 386 or with ID-mutated LMP1. Most doubly transfected yeast clones were blue by 2 h and dark blue by 4 h. TRADD and RIP interacted with LMP1 at substantially lower levels, with blue to dark blue coloration developing in only 19% of the TRADD clones and 7% of the RIP clones by 20 h. Dark blue coloration was unusual even at 20 h. A similar hierarchy of slightly stronger TRADD than RIP interaction was evident in light blue coloration at 2 h, with 12% for TRADD and none for RIP. Both TRADD and RIP interacted better with wild-type LMP1 than with the ID mutant. In summary, TES1/CTAR1 interaction with TRAF3 is considerably stronger than TES2/CTAR2 interaction with TRADD, and interaction with TRADD is stronger than interaction with RIP.
FIG. 4.
Yeast two-hybrid assay of LMP1 interaction with TRAF3, TRADD, or RIP monitored by β-galactosidase conversion of X-Gal. S. cerevisiae Y190 was transformed with pAS1 vectors expressing the Gal4 DBD fused to LMP1 amino acids 187 to 386 (wild type [WT]) or fused to LMP1 amino acids 187 to 386 with a mutation of Y384YD386 to ID (ID) and with pACT2 vectors expressing the Gal4 AD fused to TRAF3 (residues 312 to 568), TRADD (residues 1 to 312), or RIP (residues 1 to 671). Cotransformed yeast cells were selected on medium deficient in tryptophan and leucine; 32 colonies were individually transferred to filters, frozen, and thawed twice, incubated at 37°C with 1 mg of X-Gal per ml in buffer (100 mM sodium phosphate [pH 7.0], 10 mM KCl, 0.13 mM 2-mercaptoethanol), and monitored for blue-colored product. Intensity was scored as dark blue (■), blue (░⃞), or light blue (□).
RIP is associated with LMP1 in an EBV-transformed LCL.
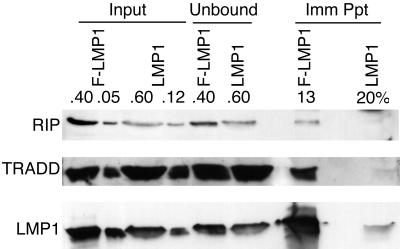
RIP association with LMP1 was evaluated in an LCL transformed by an EBV recombinant that expresses F-LMP1. F-LMP1 was precipitated with antibody M2, and coimmunoprecipitated proteins were detected by Western immunoblotting (Fig. 5). Another LCL transformed in parallel by a wild-type EBV that has an LMP1 gene without a Flag epitope served as a specificity control for the immunoprecipitation. RIP, TRADD, and LMP1 levels were similar in the input lysates from the two LCLs. F-LMP1 was highly enriched in the Flag antibody immunoprecipitate, whereas only a trace of LMP1 was nonspecifically immunoprecipitated by the Flag antibody. After correction for the efficiency of F-LMP1 precipitation, approximately 8% of TRADD and 4% of RIP were stably associated with F-LMP1. Neither TRADD nor RIP was detected in the Flag antibody control immunoprecipitate from the cell line transformed with wild-type EBV.
FIG. 5.
Coimmunoprecipitation of RIP or TRADD with F-LMP1. Proteins from LCLs (2.0 × 108 cells) infected with an EBV recombinant expressing F-LMP1 or wild-type LMP1 were solubilized by Dounce disruption in 0.5% Brij 58–100 mM NaCl–50 mM Tris (pH 7.2) and immunoprecipitated with a Flag-specific M2 affinity gel (Kodak). Precipitated proteins were Western blotted with antisera to RIP (Pharmingen), TRADD (Santa Cruz Biotechnology) or S12 monoclonal antibody to LMP1. Input lanes represent unfractionated cell proteins, unbound lanes represent proteins not precipitated with M2 affinity gel, and Imm Ppt lanes represent immunoprecipitated proteins. Percentages indicate fractions of total samples analyzed.
RIP overexpression has an additive effect on LMP1-mediated NF-κB activation.
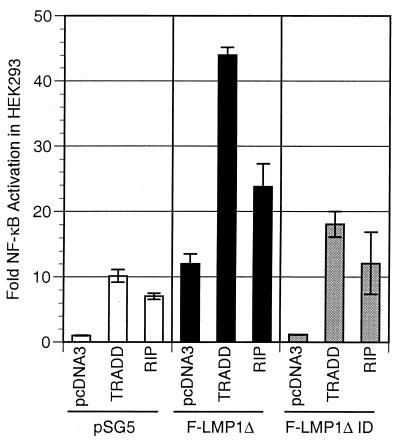
HEK293 cells were cotransfected with an LMP1 TES2/CTAR2 vector (wild-type F-LMP1Δ or mutated F-LMP1ΔID), a TRADD or RIP vector, and a luciferase reporter with three NF-κB sites. Consistent with the lower level of interaction with LMP1 in yeast and the lower level of association in LCLs, RIP did not synergize with LMP1 TES2/CTAR2 in NF-κB activation. While LMP1 TES2/CTAR2 activated NF-κB about 12-fold and RIP overexpression resulted in 7-fold NF-κB activation, cotransfection of LMP1 TES2/CTAR2 with RIP resulted in 24-fold activation, marginally better than the 19-fold expected from a simple additive effect (Fig. 6). By comparison, TRADD activated 10-fold and LMP1 TES2/CTAR2 cotransfected with TRADD activated 44-fold, substantially more than the 22-fold expected from an additive effect (Fig. 6). ID-mutated LMP1 TES2/CTAR2 activated NF-κB similarly to vector alone, and cotransfection of ID-mutated LMP1 TES2/CTAR2 with RIP activated NF-κB 12-fold, marginally better than the eightfold expected from an additive effect. Similarly, ID-mutated LMP1 TES2/CTAR2 and TRADD cotransfection activated NF-κB 18-fold, marginally better than the 11-fold expected from an additive effect. These data indicate that while TRADD synergizes with LMP1 TES/CTAR2 in NF-κB activation, RIP has only slightly more than an additive effect on LMP1 TES2/CTAR2-mediated NF-κB activation.
FIG. 6.
TRADD but not RIP synergistically activates NF-κB with LMP1. Five million HEK293 cells were electroporated with 3 μg of pSG5, TES2/CTAR2 vector F-LMP1Δ, or 6 μg of F-LMP1ΔID vector 3 μg of TRADD vector or 0.35 μg of RIP vector, and the NF-κB and β-galactosidase reporters used for Fig. 2A. Results were analyzed as described for Fig. 2A.
RIP is critical for TNFR1 but less critical for TES2/CTAR2-mediated NF-κB activation.
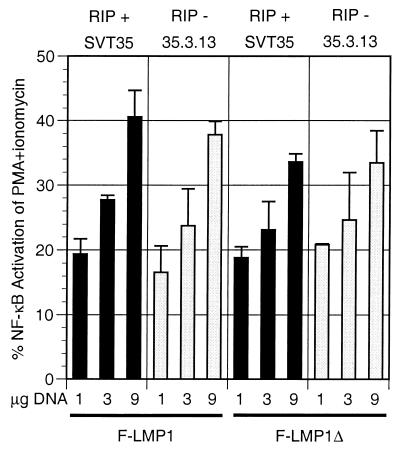
Selection of mutagenized Jurkat (SVT35) T-lymphoma cells for mutants deficient in TNFR1-mediated NF-κB activation yielded a clone (35.3.13) that lacks RIP expression and in which TNFR1-mediated NF-κB activation can be restored by transfection with a RIP expression vector (54). These results point to a critical RIP function in TNFR1-mediated NF-κB activation. Since TRADD is the proximal mediator of NF-κB activation from both TNFR1 and TES2/CTAR2, RIP might be expected to be an effector of LMP1 TES2/CTAR2-mediated NF-κB. Surprisingly, transfection of increasing amounts of TES2/CTAR2 vector F-LMP1Δ into SVT35 or 35.3.13 cells resulted in similar levels and parallel increases in NF-κB activation in both cell types (Fig. 7). F-LMP1 (both TES1/CTAR1 and TES2/CTAR2) was slightly more active overall than F-LMP1Δ but also was not different in NF-κB activation in SVT35 or 35.3.13 cells. The 35.3.13 cells were confirmed to lack RIP and for TNF-α to be unable to activate NF-κB, whereas SVT35 cells were confirmed to express RIP and for TNF-α to activate NF-κB (data not shown). Thus, RIP is not an obligate intermediary for LMP1 TES2/CTAR-mediated NF-κB activation through TRADD.
FIG. 7.
LMP1 or LMP1 TES2/CTAR2 deleted for residues 187 to 351 activates NF-κB in RIP-positive SVT35 Jurkat cells and RIP-deficient 35.3.13 Jurkat cells. Fifteen million SVT35 or 35.3.13 cells were electroporated with 9 μg of pSG5 or the indicated amounts of F-LMP1 vector or TES2/CTAR2 vector F-LMP1Δ and with 1 μg of 3x-κB-L, an NF-κB-responsive luciferase reporter. After 18 h, cultures were divided in two. One-half was untreated, while the other half was stimulated with 200 nM phorbol myristate acetate (PMA) and 10 nM ionomycin for 4 h, at which time both samples were tested and compared for luciferase activity. The results are the means of relative light unit (RLU) emission from cells transfected with LMP1 vector divided by the RLU emission of cells further stimulated with PMA and ionomycin minus baseline values for pSG5 vector-transfected cultures. In SVT35 (RIP-positive) cells, unstimulated and maximally activated luciferase activities were 2,800 and 109,000 RLU, respectively (baseline 2.6% of maximum), whereas in 35.3.13 (RIP-negative) cells, unstimulated and maximally activated luciferase activities were 7,800 and 111,000 RLU, respectively (baseline 7.1% of maximum). Levels of F-LMP1 and F-LMP1Δ were monitored by Western immunoblotting and were equivalent at the same DNA dosage for both cell lines. Further, RIP expression in SVT35 and deficiency in 35.3.13 was confirmed by Western immunoblotting (data not shown).
LMP1 TES2/CTAR2 does not induce apoptosis in BJAB B-lymphoma or HEK293 cell lines, while TRADD induces apoptosis in both cell types.
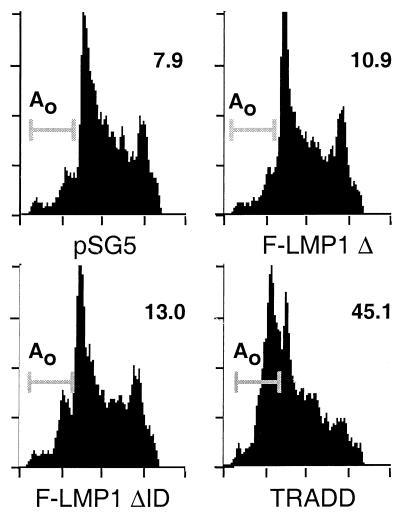
TNF-α induces TNFR1 to recruit TRADD and then FADD, which mediates caspase 8 recruitment and initiation of apoptosis signaling (reviewed in reference 1). LMP1 overexpression has been reported to induce apoptosis in RHEK cells (37) and to be toxic in BALB/c 3T3, HEp-2, 143/EBNA1, and B-lymphoblast cells (18), based on a lower yield of stable cell lines with LMP1 expression vector versus controls. To investigate whether these effects are due to TRADD-mediated apoptosis, BJAB B-lymphoma cells were transiently cotransfected with a GFP expression vector and either TRADD, LMP1 TES2/CTAR2, ID-mutated LMP1 TES2/CTAR2, or pSG5 vector. One hour after transfection, live cells were recovered by centrifugation on a Ficoll-Hypaque step gradient and cultured for 10 h. Each culture was then divided in two; one-half was treated with cycloheximide to enhance apoptosis, and the other half was untreated. Eight hours later, transfected cells were fixed, DNA was stained with propidium iodide, and the frequency of hypodiploid transfected cells was assessed by FACS analysis. Approximately 50% of the cells were GFP positive, and LMP1 TES2/CTAR2 expression did not affect GFP expression (data not shown). LMP1 TES2/CTAR2 and ID-mutated TES2/CTAR2 were expressed at similar levels as assessed by Western blotting (data not shown). The percentages of hypodiploid cells were between 8 and 13% in LMP1 TES2/CTAR2, ID-mutated LMP1 TES2/CTAR2, or pSG5 vector-transfected cells, while TRADD induced apoptosis in 45% of the transfected cells (Fig. 8). Cycloheximide treatment very slightly increased apoptosis in LMP1 TES2/CTAR2, ID-mutated LMP1 TES2/CTAR2, or pSG5 vector-transfected cells, while TRADD-mediated apoptosis was substantially enhanced (data not shown). These results indicate that TES2/CTAR2 does not activate apoptosis in BJAB B-lymphoma cells despite the ability to engage and constitutively associate with TRADD and RIP.
FIG. 8.
F-LMP1Δ and F-LMP1ΔID do not activate programmed cell death, whereas TRADD induces apoptosis. BJAB B-lymphoma cells were cotransfected with GFP vector pEGFP (Clontech) and with pSG5, TES2/CTAR2 vector F-LMP1Δ, F-LMP1ΔID vector, or TRADD vector. After transfection, dead cells were removed by gradient centrifugation. Viable cells were cultured for 10 h and then treated for 8 h with cycloheximide or left untreated. Next, cells were fixed in paraformaldehyde, permeabilized with ethanol, and stained for DNA with propidium iodide. DNA content in GFP-positive cells was quantitated by fluorescence-activated cytometry and plotted as propidium iodide fluorescence intensity (x axis) versus cell frequency (y axis). Apoptotic cells have hypodiploid DNA content (Ao) and are indicated by a line at the left and percentage at the right. Results shown are for cells that were not cycloheximide treated.
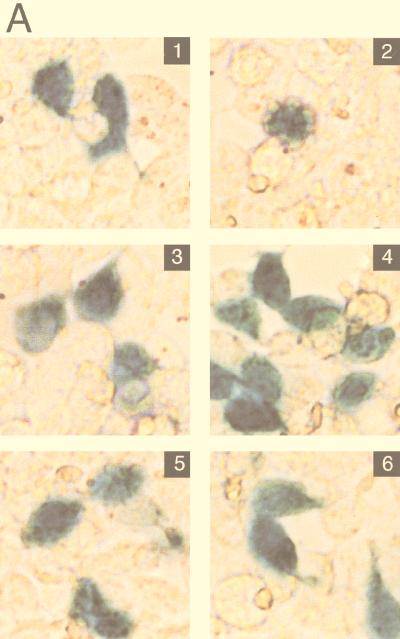
Since BJAB cells have a high constitutive level of activated NF-κB and NF-κB can have antiapoptotic effects, we examined whether LMP1 could cause apoptosis in cells that have low basal NF-κB activation. We chose HEK293 cells since they are used to demonstrate TRADD-mediated apoptosis (22) and switched to pcDNA3 as the vector for expression of LMP1 or LMP1 TES2/CTAR2 because of the increased activity of the cytomegalovirus promoter in HEK293 cells. The constitutively expressed reporter pGK-β-galactosidase was used as a marker for transfected cells. In this type of assay, apoptotic cells exhibit characteristic changes in cell morphology (rounding up) or detach from the substratum. Transfection with LMP1 (Fig. 9A, panel 3) or LMP1 CTAR2/TES2 (panel 5) expression vectors did not markedly increase the percentage of β-galactosidase-positive cells scored as apoptotic compared with vector-transfected cells (panel 1). TRADD vector transfection, in contrast, substantially increased the percentage of β-galactosidase-positive, apoptotic cells to 57% (panel 2).
FIG. 9.
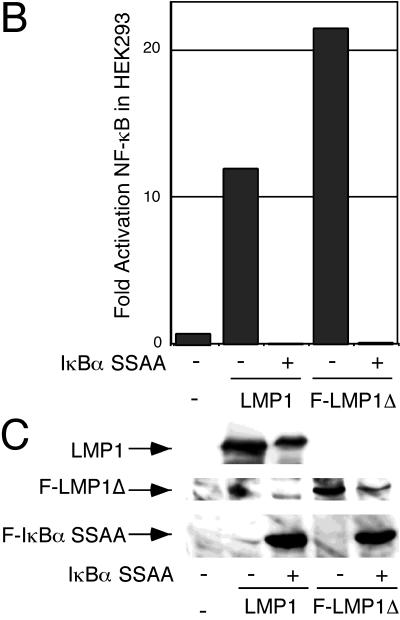
LMP1 and F-LMP1Δ do not activate apoptosis when NF-κB activation is blocked by the presence of a nondegradable IκBα (IκBα SSAA). HEK293 cells were cotransfected with pcDNA3 (panel 1), TRADD (panel 2), pcDNA3 LMP1 (panel 3), pcDNA3 LMP1 and IκBα SSAA (panel 4), TES2/CTAR2 vector pcDNA3 F-LMP1Δ (panel 5), or pcDNA3 F-LMP1Δ and IκBα SSAA (panel 6) and with β-galactosidase and NF-κB reporter DNAs. (A) Cells were fixed and then incubated in X-Gal prior to photography. The percentage of apoptotic, β-galactosidase-positive cells was 4% for pcDNA3 (panel 1), 57% for TRADD (panel 2), 8% for pcDNA3 LMP1 (panel 3), 3% for pcDNA3 LMP1 and IκBα SSAA (panel 4), 2% for pcDNA3 F-LMP1Δ (panel 5), and 3% for pcDNA3 F-LMP1Δ and IκBα SSAA (panel 6). (B) NF-κB activation as described for Fig. 2A. (C) Western immunoblot assay for LMP1 and IκBα SSAA. Equivalent amounts of protein were size separated in denaturing polyacrylamide gels, blotted to nitrocellulose, and probed with antibody M2 (Kodak) to F-LMP1Δ or Flag-tagged IκBα SSAA or antibody S12 to F-LMP1.
To eliminate the possibility that LMP1-induced NF-κB activation was protecting the cells from LMP1 TES2/CTAR2-TRADD-mediated apoptosis, a nondegradable form of IκBα with alanine substitutions for serines 32 and 36 (IκBα SSAA [6]) was coexpressed with an LMP1 or LMP1 TES2/CTAR2 vector. Cells were also cotransfected with an NF-κB-responsive luciferase reporter to monitor the effect of IκBα SSAA on NF-κB activation. NF-κB activation by LMP1 TES2/CTAR2 was completely inhibited by the coexpression of IκBα SSAA (Fig. 9B). However, apoptosis was not observed in either LMP1- or LMP1 TES2/CTAR2-expressing cells even with IκBα SSAA expression (Fig. 9A, panels 4 and 6). IκBα SSAA expression had no effect on LMP1 or LMP1 TES2/CTAR2 expression (Fig. 9C). These experiments demonstrate that LMP1 TES2/CTAR2 is substantially different from TNFR1 in inducing apoptosis.
DISCUSSION
These experiments indicate that TES2/CTAR2 signaling is similar but not identical to TNFR1 death domain signaling. TES2/CTAR2 and TNFR1 directly interact with the TRADD death domain and stably associate with TRADD when their cytoplasmic tails are aggregated. The six hydrophobic transmembrane domains of each LMP1 molecule mediate constitutive oligomeric aggregation in the plasma membrane (Fig. 1), whereas trimeric TNF-α ligand mediates trimerization of TNFR1 in the plasma membrane (20–22, 28). Both LMP1 and trimerized TNFR1 associate with TRADD, recruit TRAF2, activate NIK and IκB kinases, phosphorylate IκB, and activate NF-κB (1, 11, 28, 38, 39, 46, 52, 58, 59). LMP1 TES2/CTAR2 and TNFR1 also share the ability to activate c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) (14, 34, 35).
The studies reported here also delineate differences between LMP1 TES2/CTAR2 and TNFR1 that advance our understanding of how LMP1 and TNFR1 alter cell growth and survival. Eleven amino acids of LMP1 are sufficient for engaging the TRADD death domain to activate NF-κB, whereas the TNFR1 death domain that engages the TRADD death domain is about 70 residues (53). Further, while TRADD residues 296 to 299 or 300 to 302 are critical for TRADD interaction with itself or with TNFR1 as well as for NF-κB activation or for apoptosis (22, 45), these TRADD residues are not required for synergy with TES2/CTAR2 in NF-κB activation. A mutant TRADD that is deleted for the C-terminal residues 294 to 312 and for most of the N-terminal TRAF interaction domain even had a partial dominant negative effect on TES2/CTAR2 mediated NF-κB activation. Since the C-terminal residues of the TRADD are not required for LMP1 to synergize with TRADD in NF-κB activation, this TRADD mutant likely binds to TES2/CTAR2 and fails to signal because of the absence of both a wild-type death domain and a wild-type TRAF recruitment domain. The importance of TRADD residues 296 to 299 or 300 to 302 for TNFR1 interaction and for downstream effects and their lack of importance for TES2/CTAR2 signaling are consistent with a model in which the smaller TES2/CTAR2 domain interacts with part of the TRADD death domain and thereby propagates only a subset of the TNFR1 inducible TRADD effects.
While TNFR1 signaling through TRADD results in death domain-mediated recruitment of FADD and FADD-initiated apoptosis, LMP1 TES2/CTAR2 signaling through TRADD is deficient in induction of apoptosis. Preliminary results from yeast two-hybrid analyses reveal a low-level LMP1 TES2/CTAR2 association with FADD. Further, F-LMP1 immunoprecipitation from LCLs results in little or no specific coprecipitation of FADD (27). TNFR1-induced apoptosis is accentuated by blocking de novo protein synthesis with cycloheximide, by expression of a nondegradable IκBα, or by other interventions that inhibit NF-κB or JNK/SAPK pathways (3, 48, 55, 57). Neither cycloheximide treatment nor inhibition of NF-κB by expression of a nondegradable IκBα enabled LMP1 TES2/CTAR2 to cause apoptosis. Thus, either LMP1 TES2/CTAR2 is intrinsically unable to transmit a proapoptotic signal through TRADD or TES2/CTAR2 activates an antiapoptotic pathway that is independent of NF-κB or new protein synthesis. LMP1 thus appears able to specifically signal through TRADD and separate NF-κB and JNK/SAPK activation from the induction of apoptosis. Separation of TRADD-mediated effects has been previously described for a TRADD mutant that induces apoptosis but not NF-κB activation (45).
Another difference between LMP1 TES2/CTAR2 and TNFR1 is the ability of TES2/CTAR2 to directly interact with another death domain containing protein the Fas receptor-interacting protein RIP (17, 51, 54). RIP interaction was diminished by the Y384YD386-to-ID TES2/CTAR2 mutation that substantially diminishes TRADD interaction, NF-κB activation, and LCL outgrowth (28). Although TES2/CTAR2 associates with RIP at a lower level than TRADD and does not synergize with RIP in NF-κB activation, the RIP interaction is substantial and formally opens the possibility that RIP or another death domain protein is, in addition to TRADD, important in signaling from TES2/CTAR2. RIP is critical for TNFR1-mediated NF-κB activation in Jurkat cells and mouse fibroblasts (33, 54), and other roles are likely. The RIP kinase domain has no known function, and RIP-deficient mice exhibit runting, neonatal lethality, and lymphoid defects that are not fully explained by defects in TNF-α signaling (33). In sum, the interaction of RIP with TES2/CTAR2, the association of RIP with LMP1 in LCLs, the additive effect of RIP with LMP1 TES2/CTAR2 in NF-κB activation, and the strong negative effect of the YYD-to-ID mutation on these activities as well as on growth transformation are consistent with the possibility that RIP has a role in LMP1 signaling that mediates growth transformation.
A quite surprising aspect of the experiments reported here was the finding that LMP1 differs strikingly from TNFR1 in not requiring RIP for NF-κB activation in Jurkat cells (33, 54). LMP1 TES2/CTAR2 activates NF-κB as well in a RIP-deficient Jurkat cell line as in a wild-type Jurkat cell line. The simplest model to explain this discrepancy is that RIP is a critical part of the TNFR1-TRADD signaling complex that activates NF-κB but is not an essential mediator of NF-κB activation downstream of TRADD. In RIP-deficient cells, the TES2/CTAR2-TRADD complex activates NF-κB by recruitment of TRAFs to the TRADD N terminus, as evidenced by the dominant negative effect of a N-terminal TRAF2 deletion mutant on TES2/CTAR2-mediated NF-κB activation (28).
These and previous experiments indicate that LMP1 uses TNFR signaling molecules to accomplish an EBV-specific task (9, 10, 26, 29, 31). The LMP1 amino terminus and six transmembrane domains constitutively aggregate the C-terminal cytoplasmic domains independently of TNF-α ligand (Fig. 1). TES1/CTAR1 associates with TRAF1, -2, -3, and -5, activates NF-κB, and contributes to initial resting B-lymphocyte growth transformation, while TES2/CTAR2 associates with TRADD and RIP, activates NF-κB and JNK/SAPK, and enables long-term lymphoblastoid cell outgrowth.
ACKNOWLEDGMENTS
This research was supported by PHS grant CA47006 from the National Cancer Institute, National Institutes of Health.
Danielle Rizzo provided excellent technical assistance.
REFERENCES
- 1.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi J P, van Kooten C, Liu Y J, Rousset F, Saeland S. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 3.Beg A A, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 4.Brodeur S R, Cheng G, Baltimore D, Thorley-Lawson D. Localization of the major NF-kappaB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J Biol Chem. 1997;272:19777–19784. doi: 10.1074/jbc.272.32.19777. [DOI] [PubMed] [Google Scholar]
- 5.Cerutti A, Schaffer A, Shah S, Zan H, Liou H-C, Goodwin R G, Casali P. CD30 is a CD40-inducible molecule that negatively regulates CD40-mediated immunoglobulin class switching in non-antigen-selected human B cells. Immunity. 1998;9:247–256. doi: 10.1016/s1074-7613(00)80607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 7.Cheng G, Baltimore D. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-kappaB activation. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- 8.Cheng G, Cleary A M, Ye Z S, Hong D I, Lederman S, Baltimore D. Involvement of CRAF1, a relative of TRAF, in CD40 signaling. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 9.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devergne O, McFarland E C, Mosialos G, Izumi K M, Ware C F, Kieff E. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 12.Duckett C S, Thompson C B. CD30-dependent degradation of TRAF2: implications for negative regulation of TRAF signaling and the control of cell survival. Genes Dev. 1997;11:2810–2821. doi: 10.1101/gad.11.21.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durfee T, Becherer K, Chen P-L, Yeh S, Yang Y, Kilburn A E, Lee W, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 14.Eliopoulos A G, Young L S. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 15.Floettmann J E, Rowe M. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C-terminus and requires oligomerisation for NF-kappaB activation. Oncogene. 1997;15:1851–1858. doi: 10.1038/sj.onc.1201359. [DOI] [PubMed] [Google Scholar]
- 16.Gires O, Zimber S U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm S, Stanger B Z, Leder P. RIP and FADD: two “death domain”-containing proteins can induce apoptosis by convergent, but dissociable, pathways. Proc Natl Acad Sci USA. 1996;93:10923–10927. doi: 10.1073/pnas.93.20.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt W, Sugden B, Baichwal V R. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J Virol. 1989;63:2469–2475. doi: 10.1128/jvi.63.6.2469-2475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatzivassiliou E, Miller W E, Raab-Traub N, Kieff E, Mosialos G. A fusion of the EBV latent membrane protein 1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidemal growth factor receptor expression, nuclear factor-kappaB and stress-activated protein kinase. J Immunol. 1998;160:1116–1121. [PubMed] [Google Scholar]
- 20.Hsu H, Huang J, Shu H B, Baichwal V, Goeddel D V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 21.Hsu H, Shu H B, Pan M G, Goeddel D V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 22.Hsu H, Xiong J, Goeddel D V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 23.Hu H M, O’Rourke K, Boguski M S, Dixit V M. A novel RING finger protein interacts with the cytoplasmic domain of CD40. J Biol Chem. 1994;269:30069–30072. [PubMed] [Google Scholar]
- 24.Huen D S, Henderson S A, Croom C D, Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 25.Izumi K M, Kaye K M, Kieff E D. Epstein-Barr virus recombinant molecular genetic analysis of the LMP1 amino-terminal cytoplasmic domain reveals a probable structural role, with no component essential for primary B-lymphocyte growth transformation. J Virol. 1994;68:4369–4376. doi: 10.1128/jvi.68.7.4369-4376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumi K M, Kaye K M, Kieff E D. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1152. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi, K. M., and E. D. Kieff. Unpublished data.
- 28.Izumi K M, Kieff E D. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaye K M, Devergne O, Harada J N, Izumi K M, Yalamanchili R, Kieff E, Mosialos G. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-κB activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaye K M, Izumi K M, Mosialos G, Kieff E. The Epstein-Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J Virol. 1995;69:675–683. doi: 10.1128/jvi.69.2.675-683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehry M R. CD40-mediated signaling in B cells. Balancing cell survival, growth, and death. J Immunol. 1996;156:2345–2348. [PubMed] [Google Scholar]
- 33.Kelliher M A, Grimm S, Ishida Y, Kuo F, Stanger B Z, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 34.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilger E, Kieser A, Baumann M, Hammerschmidt W. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S Y, Lee S Y, Kandala G, Liou M L, Liou H C, Choi Y. CD30/TNF receptor-associated factor interaction: NF-kappa B activation and binding specificity. Proc Natl Acad Sci USA. 1996;93:9699–9703. doi: 10.1073/pnas.93.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J J-Y, Chen J-Y, Hsu T-Y, Yu W C Y, Su I-J, Yang C-S. Induction of apoptosis in epithelial cells by Epstein-Barr virus latent membrane protein 1. J Gen Virol. 1996;77:1883–1892. doi: 10.1099/0022-1317-77-8-1883. [DOI] [PubMed] [Google Scholar]
- 38.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 39.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 40.Miller W E, Cheshire J L, Raab-Traub N. Interaction of tumor necrosis factor receptor-associated factor signaling proteins with the latent membrane protein 1 PXQXT motif is essential for induction of epidermal growth factor receptor expression. Mol Cell Biol. 1998;18:2835–2844. doi: 10.1128/mcb.18.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller W E, Mosialos G, Kieff E, Raab T N. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 44.Noelle R J. CD40 and its ligand in host defense. Immunity. 1996;4:415–419. doi: 10.1016/s1074-7613(00)80408-2. [DOI] [PubMed] [Google Scholar]
- 45.Park A, Baichwal V J. Systematic mutational analysis of the death domain of the tumor necrosis factor receptor 1-associated protein TRADD. J Biol Chem. 1996;271:9858–9862. doi: 10.1074/jbc.271.16.9858. [DOI] [PubMed] [Google Scholar]
- 46.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 47.Rothe M, Sarma V, Dixit V M, Goeddel D V. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 48.Roulston A, Reinhard C, Amiri P, Williams L T. Early activation of c-Jun N-terminal kinase and p38 kinase regulate cell survival in response to tumor necrosis factor alpha. J Biol Chem. 1998;273:10232–10239. doi: 10.1074/jbc.273.17.10232. [DOI] [PubMed] [Google Scholar]
- 49.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP-1’s association with TRAF1, TRAF2, and TRAF3. J Virol. 1997;71:4649–4656. doi: 10.1128/jvi.71.6.4649-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shu H B, Takeuchi M, Goeddel D V. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c- IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci USA. 1996;93:13973–13978. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanger B Z, Leder P, Lee T H, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 52.Sylla B S, Hung S C, Davidson D M, Hatzivassiliou E, Malinin N L, Wallach D, Gilmore T D, Kieff E, Mosialos G. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc Natl Acad Sci USA. 1998;95:10106–10111. doi: 10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tartaglia L A, Ayres T M, Wong G H, Goeddel D V. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 54.Ting A T, Pimentel-Muinos F, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 55.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 56.VanArsdale T L, VanArsdale S L, Force W R, Walter B N, Mosialos G, Kieff E, Reed J C, Ware C F. Lymphotoxin-beta receptor signaling complex: role of tumor necrosis factor receptor-associated factor 3 recruitment in cell death and activation of nuclear factor kappaB. Proc Natl Acad Sci USA. 1997;94:2460–2465. doi: 10.1073/pnas.94.6.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C Y, Mayo M W, Baldwin A J. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 58.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 59.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]