INTS11, the catalytic subunit of the Integrator complex, regulates hematopoietic stem cell function through PRC2 proteins.
Abstract
INTS11, the catalytic subunit of the Integrator (INT) complex, is crucial for the biogenesis of small nuclear RNAs and enhancer RNAs. However, the role of INTS11 in hematopoietic stem and progenitor cell (HSPC) biology is unknown. Here, we report that INTS11 is required for normal hematopoiesis and hematopoietic-specific genetic deletion of Ints11 leads to cell cycle arrest and impairment of fetal and adult HSPCs. We identified a novel INTS11-interacting protein complex, Polycomb repressive complex 2 (PRC2), that maintains HSPC functions. Loss of INTS11 destabilizes the PRC2 complex, decreases the level of histone H3 lysine 27 trimethylation (H3K27me3), and derepresses PRC2 target genes. Reexpression of INTS11 or PRC2 proteins in Ints11-deficient HSPCs restores the levels of PRC2 and H3K27me3 as well as HSPC functions. Collectively, our data demonstrate that INTS11 is an essential regulator of HSPC homeostasis through the INTS11-PRC2 axis.
INTRODUCTION
Hematopoietic stem and progenitor cells (HSPCs) are a rare population of hematopoietic cells, having capabilities of both self-renewal and differentiation to all lineages of hematopoietic cells, such as myeloid, erythroid, T, and B cells (1). A tight balance between self-renewal and differentiation is crucial to maintain homeostasis of the hematopoietic system. The ability of hematopoietic stem cells (HSCs) to repopulate the hematopoietic compartment is regulated by both intrinsic and extrinsic mechanisms (2). The cooperation between the stochastic and epigenetic changes fine-tunes the transcriptome profiles required for normal hematopoiesis (3, 4).
Integrator (INT) is an evolutionarily conserved complex and contains at least 14 subunits (5). INT was discovered by Baillat and colleagues in 2005 and was characterized originally as an important factor for U-rich small nuclear RNA (snRNA) 3′-end processing (6). Multiple studies demonstrate additional roles for INT in RNA polymerase II (RNAPII) transcription initiation, pause-release, elongation, and termination at protein-coding genes (7–9). In addition, INT is crucial for the biogenesis of enhancer RNA (eRNA) (10). More recently, INT has been shown to trigger premature transcription termination and attenuate gene expression (11–13). All these studies point to the essential role of INT family members in RNA biogenesis.
INT components ints3 and ints9 were shown to regulate snRNA processing and the differential fate of adult stem cells in Schmidtea mediterranea (14). Tao and colleagues (15) reported that ints5 knockdown in zebrafish embryos leads to blockage of red blood cell differentiation because of perturbation of smad1/5 splicing. Knockdown of INTS13 with small hairpin RNAs in human myeloid HL-60 cells revealed that INTS13 is required to activate specific enhancers during myeloid differentiation (16). In a recent study, Yoshimi and colleagues (17) demonstrated that aberrant splicing of INTS3 is associated with leukemogenesis, and overexpression of INTS3 slows down leukemia progression in vivo. INTS11, the only subunit of the INT protein family with endonuclease activity (18), has been shown to play critical roles in mouse adipose differentiation and ciliogenesis in cultured human cells (19, 20). Unexpectedly, Drosophila IntS11 is not essential for neural development (21). These studies imply a tissue-specific function of INTS11. Up to date, the role of INT in adult stem cell and specifically HSC regulation remains unknown.
In the current study, we investigated the role of INTS11 in HSPCs both in vitro and in vivo using our newly generated hematopoietic-specific Ints11 knockout mouse model system. We show that INTS11 is essential for normal hematopoiesis, as Ints11 deletion led to G1-S cell cycle arrest and resulted in HSC impairment. Intriguingly, we identify a previously unknown mechanistic function for INTS11 in cooperation with the Polycomb repressive complex 2 (PRC2) to maintain histone H3 lysine 27 trimethylation (H3K27me3) at genes critical for cell cycle progression and HSPC functions. Loss of INTS11 destabilizes the PRC2 complex, decreases the H3K27me3 level, and derepresses PRC2 target genes. Reexpression of INTS11 or PRC2 proteins in Ints11-deficient HSPCs restores the levels of PRC2 and H3K27me3 as well as HSPC functions. Our study demonstrates an indispensable role of INTS11 in normal hematopoiesis regulating the expression of genes critical for HSPC functions through the newly discovered INTS11/PRC2 axis.
RESULTS
INTS11 is required for hematopoiesis
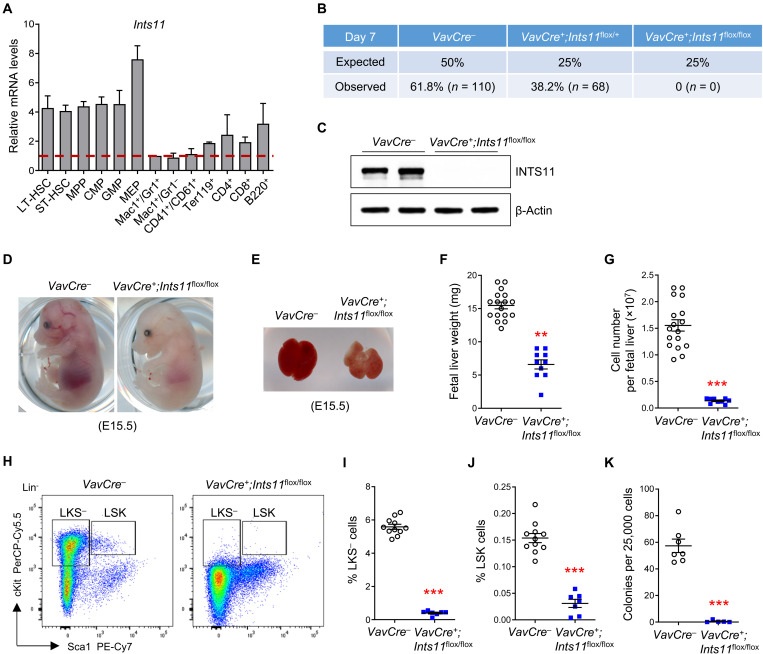
To determine the expression levels of Ints11 in wild-type (WT) hematopoietic lineages, different hematopoietic cell subpopulations were sorted and subjected to quantitative polymerase chain reaction (qPCR). Ints11 mRNA was ubiquitously expressed in all the hematopoietic lineages examined, with a higher level in the HSPCs (Fig. 1A). To investigate the function of INTS11 in hematopoiesis, we generated Ints11flox/flox mice (fig. S1A) and bred them with the Vav1-Cre mice (22). However, VavCre+;Ints11flox/flox animals were never observed, suggesting embryonic lethality (Fig. 1B). Subsequent kinetic dissection of embryos from embryonic day (E) 13.5 to 18.5 confirmed the lethality upon deletion of Ints11 at the embryonic stage (fig. S1B). VavCre+;Ints11flox/flox embryos were observed at a normal rate of Mendelian ratios up to E17.5, but a much lower frequency at E18.5 than VavCre− embryos. The successful deletion of Ints11 was confirmed in the tail and CD45+ fetal liver by PCR and qPCR, respectively (fig. S1, C and D). Western blotting demonstrated complete deletion of INTS11 protein in fetal liver cells at E13.5 (Fig. 1C). While there was no difference in the sizes between the two genotypes of the embryos, VavCre+;Ints11flox/flox embryos were pale (Fig. 1D and fig. S1E). The liver of VavCre+;Ints11flox/flox embryos was also pale, which was significantly smaller and less cellular than that of the control embryos (Fig. 1, E to G). Flow cytometric analysis of HSPC populations in VavCre+;Ints11flox/flox fetal livers revealed a severe reduction in the cell populations of Lin−cKit+Sca1− (LKS−), Lin−Sca1+cKit+ (LSK), long-term (LT-) HSCs (Lin−Sca1+cKit+CD34−CD135−), short-term (ST-) HSCs (Lin−Sca1+cKit+CD34+CD135−), and multipotent progenitors (MPPs; Lin−Sca1+cKit+CD34+CD135+) (Fig. 1, H to J, and fig. S1, F and G), indicative of impaired development of HSPCs in Ints11-deficient embryos. Consistently, colony-forming unit cell (CFU-C) assays demonstrated that fetal liver cells of VavCre+;Ints11flox/flox embryos were unable to form colonies (Fig. 1K and fig. S1H). Further phenotypic analysis showed a significant decrease in erythroid precursors in Ints11-deficient fetal livers (fig. S1, I and J). These data indicate that INTS11 is essential for sustaining fetal hematopoiesis.
Fig. 1. INTS11 is required for fetal hematopoiesis.
(A) Relative mRNA levels of Ints11 in different hematopoietic cell populations from the BM of WT mice. n = 2 independent experiments; the hematopoietic populations were sorted from five WT mice in each experiment. Long-term HSC (LT-HSC), Lin−Sca1+cKit+CD34−CD135−; short-term HSC (ST-HSC), Lin−Sca1+cKit+CD34+CD135−; multipotent progenitor (MPP), Lin−Sca1+cKit+CD34+CD135+; common myeloid progenitor (CMP), LKS−CD34+CD16/32intermediate; granulocyte-macrophage progenitor (GMP), LKS−CD34+CD16/32high; megakaryocyte-erythroid progenitor (MEP), LKS−CD34−CD16/32low. (B) Genotype distribution in the offspring of the intercross Vav1Cre+;Ints11flox/+ × Vav1Cre−;Ints11flox/flox mice on day 7 after birth. (C) Vav1Cre-mediated INTS11 depletion was verified in fetal liver CD45+ cells from E13.5 Vav1Cre+;Ints11flox/flox embryos. (D and E) Representative embryos (D) and livers (E) from Vav1Cre+;Ints11flox/flox and control littermates at E15.5. Photo credit: Peng Zhang, University of Texas Health Science Center at San Antonio. (F and G) Weight (F) and cellularity (G) of fetal livers from Vav1Cre+;Ints11flox/flox (n = 10) and control littermates (n = 17) at E13.5. (H) Flow cytometric analysis of HSPCs (LSK and LKS−) from control and Vav1Cre+;Ints11flox/flox fetal livers at E13.5. Lineage-negative cells are shown. (I and J) Quantification of the percentages of LKS− (I) and LSK cells (J) from control (n = 11) and Vav1Cre+;Ints11flox/flox (n = 7) fetal livers at E13.5. (K) CFU-C assay using E13.5 fetal liver cells from control (n = 7) and Vav1Cre+;Ints11flox/flox embryos (n = 5) was assessed in semisolid medium in the presence of SCF, IL-3, TPO, GM-CSF, EPO, and IL-6. Data are means ± SEM. Unpaired Student’s t test: **P < 0.01 and ***P < 0.001.
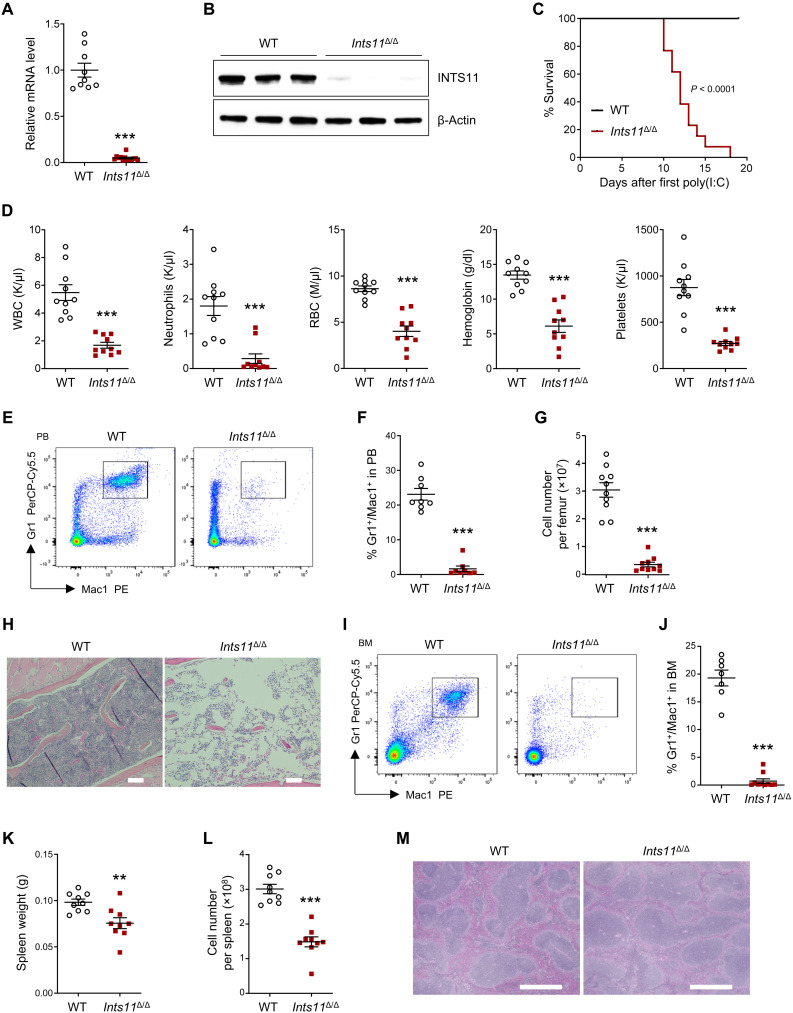
To determine whether INTS11 is required for adult hematopoiesis, we crossed Ints11flox/flox mice with Mx1-Cre mice and induced Ints11 deletion (Ints11Δ/Δ) by three daily intraperitoneal injections of polyinosine-polycytidine [poly(I:C)]. Recombination of the Ints11 floxed allele occurred 72 hours after the first poly(I:C) injection (fig. S2A). There was a >90% reduction of Ints11 mRNA and the protein in the bone marrow (BM) Lin−cKit+ (LK) cells of Ints11Δ/Δ mice compared with Mx1Cre+ (WT) controls (Fig. 2, A and B). Adult mice lacking Ints11 expression died within 3 weeks of poly(I:C) injection (Fig. 2C). All the mice developed pancytopenia (Fig. 2D). Flow cytometric analysis of peripheral blood (PB) showed that Ints11Δ/Δ mice had a marked reduction of myeloid cells (Gr1+/Mac1+) but relatively increased percentages of T and B cells (Fig. 2, E and F, and fig. S2, B and C). BM cellularity was markedly reduced (Fig. 2G). Analysis of bone histological sections revealed a substantial reduction of BM cellularity of Ints11Δ/Δ mice (Fig. 2H). In addition, flow cytometric analysis demonstrated that the percentages of myeloid (Gr1+/Mac1+), erythroid progenitor cells (CD71+/Ter119+), and lymphoid (T and B220+ cells) cells were significantly reduced in the BM of Ints11Δ/Δ mice compared with those of WT control mice (Fig. 2, I and J, and fig. S2, D to G). Despite the comparable body weights between WT and Ints11Δ/Δ mice (fig. S2H), Ints11Δ/Δ mice exhibited lower spleen weight and significantly decreased cellularity compared with WT controls (Fig. 2, K and L, and fig. S2I). The histological analysis of Ints11Δ/Δ spleen sections showed a decreased proportion of myeloid cells (Fig. 2M). Flow cytometric analysis of the spleen showed that the percentage of myeloid cells was significantly reduced, whereas the percentages of T and B cells were relatively higher in the spleens of Ints11Δ/Δ than in WT controls (fig. S2, J to M).
Fig. 2. INTS11 deletion leads to hematopoietic failure.
(A) Ints11 mRNA levels in BM Lin−cKit+ (LK) cells from WT (n = 9) and Ints11Δ/Δ mice (n = 13) 72 hours after first poly(I:C) injection. (B) INTS11 protein levels in LK cells from WT and Ints11Δ/Δ mice. (C) Kaplan-Meier survival curve representing percent survival of Ints11Δ/Δ mice following poly(I:C) injection. Log-rank (Mantel-Cox) test; WT, n = 10; Ints11Δ/Δ, n = 13. (D) PB counts of WT and Ints11Δ/Δ mice (n = 10 per genotype). WBC, white blood cells; RBC, red blood cells. (E) Flow cytometric analysis of myeloid cells in PB from WT and Ints11Δ/Δ mice 12 days after first poly(I:C) injection. (F) Frequencies of Gr1+/Mac1+ cells in PB from WT and Ints11Δ/Δ mice (n = 8 per genotype). (G) BM cellularity of WT and Ints11Δ/Δ mice (n = 10 per genotype) 12 days after poly(I:C) injection. (H) Representative hematoxylin and eosin (H&E) staining of bone sections. Scale bar, 100 μm. (I) Flow cytometric analysis of myeloid cells in BM from WT and Ints11Δ/Δ mice 12 days after poly(I:C) injection. (J) Frequency of Gr1+/Mac1+ cells in BM from WT (n = 7) and Ints11Δ/Δ mice (n = 10). (K and L) Weight (K) and cellularity (L) of the spleen from WT and Ints11Δ/Δ mice (n = 9 per genotype). (M) Representative H&E staining of spleen sections. Scale bar, 10 μm. Data are means ± SEM. Unpaired Student’s t test: **P < 0.01 and ***P < 0.001.
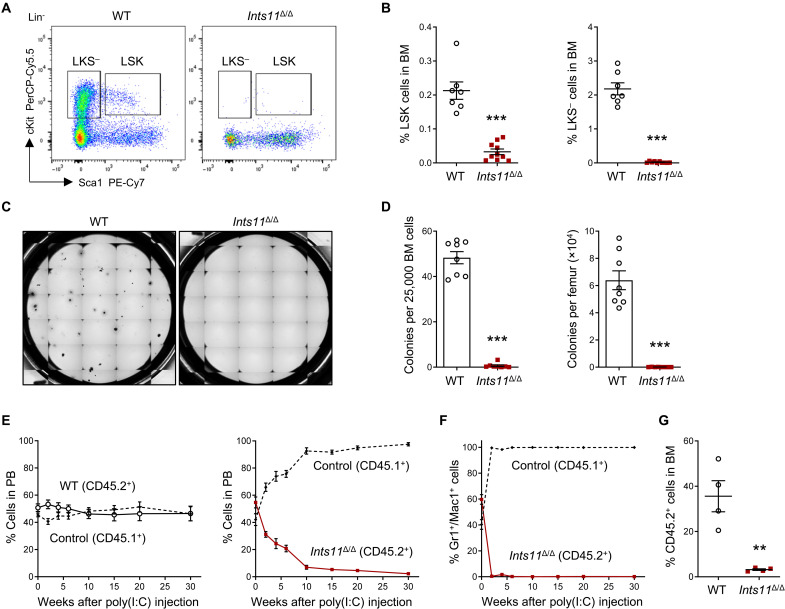
We next examined the effects of Ints11 deletion on BM HSPC populations 72 hours after poly(I:C) injection. We observed no difference in the cellularity of BM between Ints11Δ/Δ and their littermate controls (fig. S3A). Flow cytometric analysis revealed that the numbers of LSK cells and ST-HSCs were significantly decreased in the BM of Ints11Δ/Δ mice compared with WT controls (fig. S3, B and C), while the numbers of LKS− cells, LT-HSCs, MPPs, as well as cKit+ cells and LK cells were comparable (fig. S3, B to E). By day 12 after poly(I:C) injection, Ints11 deletion led to a marked reduction of HSPCs in the BM of Ints11Δ/Δ mice (Fig. 3, A and B, and fig. S3, F to H). CFU-C assays confirmed that Ints11-deficient BM cells formed minimum number of colonies (Fig. 3, C and D). Together, these results indicate that INTS11 is required for the maintenance of the HSPC pool.
Fig. 3. Loss of Ints11 impairs HSPC development.
(A) Flow cytometric analysis of HSPCs in BM cells from WT and Ints11Δ/Δ mice 12 days after first poly(I:C) injection. (B) Quantification of the percentages of LSK and LKS− cells from WT (n = 7) and Ints11Δ/Δ (n = 10) mice. (C) Representative images of colony formation for Ints11-deficient BM cells and WT controls. The images were taken on the seventh day of the assay. (D) CFU-C assay using BM cells from WT and Ints11Δ/Δ mice (n = 8 per genotype) 12 days after poly(I:C) injection. (E) Percentages of donor-derived (WT or Ints11Δ/Δ CD45.2+) versus CD45.1+ cells in the PB of recipient animals at indicated time points (n = 4 per genotype). (F) Percentages of Ints11Δ/Δ-derived (CD45.2+) versus CD45.1+ cells in the myeloid population of PB of recipient animals at indicated time points (n = 4). (G) Quantification of CD45.2+ cells in BM of recipient animals at 30 weeks after poly(I:C) injection (n = 4 per genotype). Data are means ± SEM. Unpaired Student’s t test: **P < 0.01 and ***P < 0.001.
INTS11 function in hematopoiesis is cell autonomous
To verify the cell-intrinsic effects of Ints11 deletion in HSC functions, we performed competitive BM transplantation assays by injecting equal numbers of BM cells from BoyJ mice (CD45.1+) and Mx1Cre+;Ints11flox/flox mice (CD45.2+) or Mx1Cre+ littermates (CD45.2+) into lethally irradiated BoyJ recipient mice (CD45.1+) (fig. S3I). Ints11 deletion was induced by poly(I:C) injection upon confirmation of comparable engraftment rates of CD45.1+ versus CD45.2+ cells in the PB of the recipient mice. Ints11-deficient CD45.2+ cells were rapidly outcompeted by cotransplanted CD45.1+ WT cells (Fig. 3E). Myeloid cells were the first diminished cell population in the PB of the recipient mice (Fig. 3F) followed by a persistent reduction of T and B cell populations over 30 weeks after poly(I:C) injection (fig. S3, J and K). The CD45.2+ Ints11Δ/Δ cells were almost eliminated in the BM 30 weeks after the poly(I:C) injection (Fig. 3G). These data indicate a cell-autonomous requirement for INTS11 in fetal and adult hematopoiesis.
Up-regulated genes in Ints11-deficient LK cells are enriched for PRC2 targets
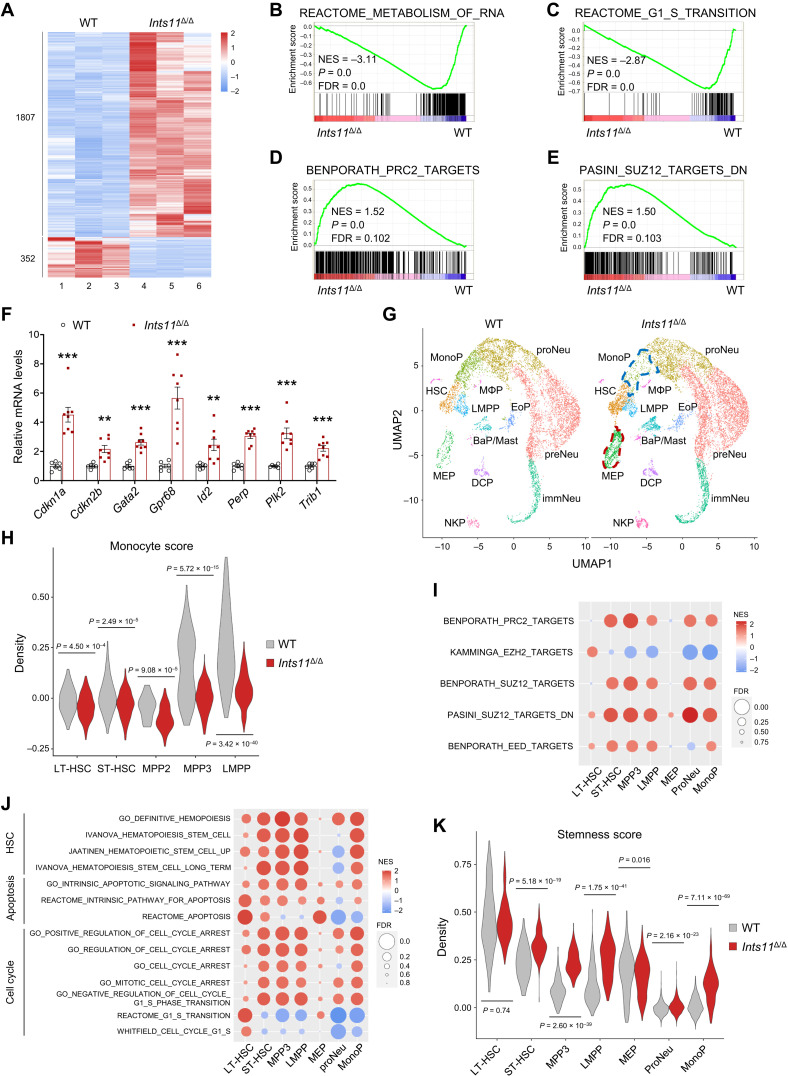
To identify the molecular pathways regulated by INTS11 in HSPCs, we performed RNA sequencing (RNA-seq) using Ints11Δ/Δ BM LK cells. It is known that snRNAs are relatively stable, as their levels only marginally decrease over 72 hours (11, 12). We thus examined genome-wide expression profiles in WT and Ints11Δ/Δ LK cells 72 hours after poly(I:C) injection. Compared with WT cells, the deletion of Ints11 resulted in an aberrant gene expression signature in LK cells consisting of 1807 up-regulated and 352 down-regulated genes [Fig. 4A and fig. S4A; fold change > 1.8 and false discovery rate (FDR) < 0.05]. Ints11 deletion decreased the Ints7 mRNA level in LK cells compared with WT cells, while the expression of other INT subunits remained at the same levels in Ints11Δ/Δ BM LK cells as in WT cells (fig. S4, B and C). Gene set enrichment analysis (GSEA) showed that a subset of the down-regulated genes in Ints11-deficient cells was enriched for the metabolism of RNA (Fig. 4B), supporting the previous finding that INTS11 is required for RNA processing (6, 10). GSEA further revealed that genes associated with the regulation of G1-S cell cycle transition and apoptosis were dysregulated in Ints11Δ/Δ LK cells (Fig. 4C and fig. S4, D and E). Unexpectedly, PRC2 target genes were up-regulated in Ints11Δ/Δ LK cells (Fig. 4, D and E, and fig. S4F). qPCR confirmed the changes on selected PRC2 target genes, including genes associated with cell cycle, apoptosis, and HSC functions (Fig. 4F and fig. S4G).
Fig. 4. Loss of Ints11 derepresses PRC2 target genes in HSPCs.
(A) Heatmap depicting significantly dysregulated (1807 up-regulated and 352 down-regulated) genes in Ints11Δ/Δ LK cells compared with WT controls (FDR < 0.05 and |fold change| > 1.8). (B to E) GSEA shows that genes involved in the regulation of RNA metabolism (B) and cell cycle transition (C) are down-regulated, and PRC2 targets (D and E) are up-regulated, in Ints11Δ/Δ LK cells. The normalized enrichment score (NES), P value, and FDR are shown. (F) Relative mRNA expression of genes associated with cell cycle, apoptosis, and PRC2 targets in LK cells of WT (n = 6) and Ints11Δ/Δ mice (n = 8) 72 hours after first poly(I:C) injection. Data are means ± SEM. Unpaired Student’s t test: **P < 0.01 and ***P < 0.001. (G) Uniform manifold approximation and projection (UMAP) visualization of hematopoietic cell clusters identified from cKit+ cells of WT and Ints11Δ/Δ mice. Each dot represents one cell, and cluster identity is color-coded (Seurat). LMPP, lymphoid-primed MPP; NeuP, neutrophil/granulocyte progenitor (proNeu, early specifying; preNeu, late committed; and immNeu, immature neutrophil); BaP/Mast, basophil progenitor and mast cells; EoP, eosinophil progenitor; DCP, dendritic cell–committed progenitor; NKP, natural killer cell–committed progenitor; MΦP, macrophage-restricted progenitor. (H) Violin plot of monocyte transcription signature in indicated populations. The score was calculated on the basis of the expression values for genes in a given gene set. Mann-Whitney U test. (I and J) GSEA with NES and FDR values for gene sets of PRC2 (I) and HSC, apoptosis, and cell cycle (J) associated with HSPC populations in Ints11Δ/Δ cells. The colors reflect scaled NES, representing the degree of expression change. The size of the circle represents the FDR value. (K) Violin plot of stemness transcription signature in indicated populations. Mann-Whitney U test.
Enrichment of cell cycle–related gene signatures led us to investigate whether deletion of Ints11 changes the cell cycle profile of HSPCs. In line with the gene expression changes, the cell cycle analysis demonstrated that the proportion of lineage-negative cells in the BM in G0-G1 was significantly increased and the S phase was significantly reduced upon Ints11 deletion, suggesting a failure of entering S phase from G0-G1 in Ints11Δ/Δ cells (fig. S4, H and I). Cyclin-dependent kinase inhibitor p21Cip1 (Cdkn1a for gene name) plays a predominant role in maintaining HSC quiescence (23, 24). Consistently, Western blot analysis revealed an up-regulation of p21Cip1, which may contribute to cell cycle arrest and G1-to-S phase transition in Ints11Δ/Δ HSPCs (fig. S4J). In addition, there was a mild increase in apoptotic cells [7-amino-actinomycin D–negative (7-AAD−)/annexin V+] in Ints11Δ/Δ lineage–negative BM cells compared with WT controls (fig. S4K).
Recently, single-cell RNA-seq (scRNA-seq) has emerged as a powerful tool to dissect hematopoietic progenitors and other cell populations (25). Given the heterogeneity of HSPCs, we performed a scRNA-seq analysis of 21,495 BM cKit+ cells from WT and Ints11Δ/Δ mice to capture the impact of Ints11 deletion on transcriptomic changes in HSPC populations. Fifteen major clusters (subpopulations) were identified by the unsupervised clustering method after integrating WT and Ints11Δ/Δ cells (Fig. 4G and fig. S5A). Loss of Ints11 skewed the cluster distribution with increased megakaryocyte-erythroid progenitors (MEPs; 7.43 versus 2.87%) and decreased monocyte progenitors (MonoPs; 2.53 versus 9.64%) (Fig. 4G and fig. S5B). We observed that there was a slight increase of LT-HSCs and ST-HSCs in Ints11Δ/Δ cells compared with WT controls (fig. S5, A and B). MPP subsets emerge from HSCs, and myeloid-biased MPP2 and MPP3 work together with lymphoid-primed MPP (LMPP) to control blood production (26). After scoring individual cells with several module scores based on the expression levels of population-specific gene signatures, we found that a higher erythroid/megakaryocyte score and a lower monocyte score were significantly enriched in all Ints11-deleted HSCs and MPPs (Fig. 4H and fig. S5C). These data support the observation of increased MEP and reduced MonoP populations.
We next performed GSEA analysis to assess the transcription signatures associated with specific HSPC populations. Consistent with the findings observed in bulk RNA-seq, Ints11Δ/Δ cells showed a significant increase in the expression of PRC2 targets as well as the genes controlling cell cycle arrest and apoptosis in almost all HSC and MPP populations (Fig. 4, I and J). Unexpectedly, genes up-regulated in HSC signatures were enriched in all the HSCs and MPPs of Ints11Δ/Δ mice (Fig. 4J). Moreover, we observed higher scores of stemness and apoptosis, but modest changes of proliferation score in Ints11Δ/Δ HSCs and MPPs compared with WT cells (Fig. 4K and fig. S5, D and E). Consistently, further examination of the cell cycle stage of individual cells in each population revealed a higher proportion of cells at the G1 phase and a low proportion of cells in the S phase in almost all Ints11Δ/Δ HSPC subsets compared with WT cells (fig. S5F). The up-regulated cell cycle–, PRC2-, and HSC-associated genes, such as Gata2 and Cdkn1a, which were identified at higher expression levels in bulk RNA-seq of Ints11Δ/Δ LK cells, were also up-regulated in almost all HSPC subpopulations following the Ints11 deletion (fig. S5, G and H).
INTS11 interacts with the PRC2 complex
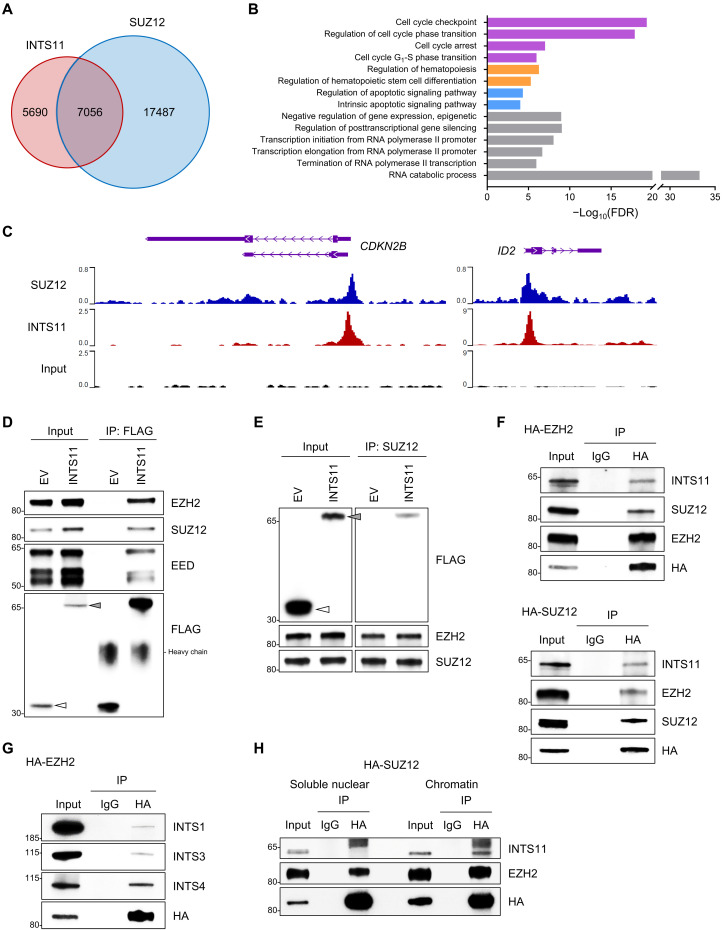
PRC2 consists of four core components: enhancer of zeste homolog 1/2 (EZH1/2), suppressor of zeste 12 homolog (SUZ12), embryonic ectoderm development (EED), and retinoblastoma-binding protein 4 and 7 (RBBP4/7) (27, 28). To determine whether INTS11 colocalizes with PRC2 genome-wide in the hematopoietic cells, we first investigated the chromatin targets of INTS11 in hematopoietic cells by analyzing the published INTS11 chromatin immunoprecipitation sequencing (ChIP-seq) data by Barbieri and colleagues (16). Genomic distribution analysis of the INTS11 peaks showed that 77.91% of INTS11-associated regions were localized at promoter regions (≤1 kb of transcription start sites; fig. S6A). Further comparison with the SUZ12 ChIP-seq dataset for hematopoietic cells (29) revealed that the peaks of INTS11 highly overlapped with the peaks of SUZ12 (55.4%; Fig. 5A). Gene ontology analysis demonstrated that the overlapping target genes among INTS11 and SUZ12 were enriched in RNA processing, cell cycle, apoptosis, hematopoiesis, and transcription regulation (Fig. 5, B and C, and fig. S6B).
Fig. 5. INTS11 forms a complex with PRC2.
(A) Venn diagram showing the overlap among binding sites (peaks) of INTS11 (GSE106359) and SUZ12 (GSE59090) in hematopoietic cells. (B) Gene ontology analysis of the genes that were co-occupied by INTS11 and SUZ12. (C) Representative ChIP-seq tracks show that INTS11 and SUZ12 co-occupy PRC2 target gene promoters in hematopoietic cells. (D) Nuclear extracts of 293T cells transfected with FLAG-tagged INTS11 or empty vector control (EV) were immunoprecipitated (IP) with anti-FLAG and probed for PRC2 proteins. Arrowhead indicates the FLAG-fusion protein. (E) Reciprocal SUZ12 IP from nuclear extracts of 293T cells transfected with FLAG-tagged INTS11 or EV, and representative immunoblot analysis. (F) Nuclear extracts of 293T cells transfected with HA-tagged EZH2 (top) or SUZ12 (bottom) were IP with anti-HA or mouse IgG, and Western blotting was performed. (G) Western blot analysis of the samples above (F) using antibodies against INT subunits INTS1, INTS3, and INTS4. (H) Soluble nuclear and chromatin-bound fractions from 293T cells transfected with HA-tagged SUZ12 were subjected to IP with anti-HA antibody followed by Western blot analysis with indicated antibodies.
The largely overlapping targets among INTS11 and SUZ12 prompted us to probe for interaction between INTS11 and PRC2 complex proteins. To test this hypothesis, we generated a lentiviral vector expressing FLAG-INTS11 (fig. S6C) and performed IP assays. All IPs were performed in the presence of benzonase to ensure that the protein-protein interactions were DNA independent. We used an anti-FLAG antibody to pull down INTS11 in the nuclear extracts from 293T cells expressing FLAG-INTS11, followed by Western blot using antibodies against EZH2, SUZ12, and EED, the core subunits of the PRC2 complex. We found that INTS11 interacts with endogenous EZH2, SUZ12, and EED (Fig. 5D). Reciprocal IP of either EZH2 or SUZ12 led to the co-IP of INTS11 (Fig. 5E and fig. S6D). To determine whether PRC2 could interact with endogenous INTS11 in vivo, we transfected plasmids expressing hemagglutinin (HA)–EZH2 or HA-SUZ12 individually into 293T cells. IP analysis of nuclear extracts using the anti-HA antibody recognizing endogenous INTS11 confirmed that INTS11 forms a complex with PRC2 (Fig. 5F). INTS1, INTS3, and INTS4 were also present (Fig. 5G), which suggests that multiple INT subunits are associated with this complex. We further performed HA-IP in the soluble nuclear fraction and chromatin-bound fraction prepared from 293T cells expressing HA-SUZ12. INTS11 was observed in the chromatin samples but not in the soluble nuclear (Fig. 5H), suggesting that INTS11 interacts with PRC2 on chromatin.
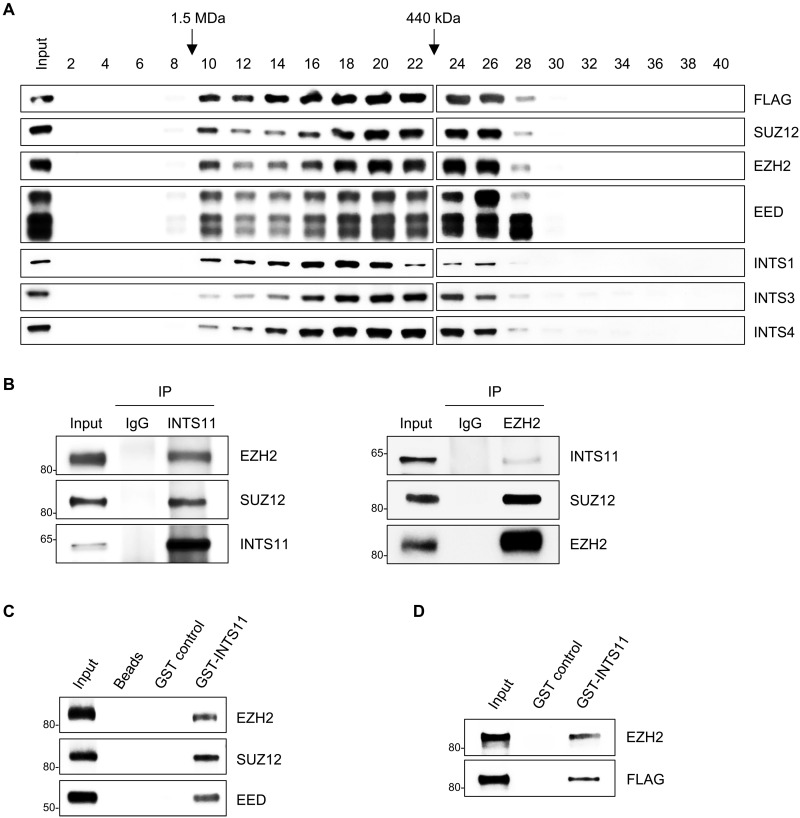
To provide additional evidence that INTS11 and PRC2 proteins exist in one complex, we subjected nuclear extracts of 293T cells expressing FLAG-INTS11 to fractionation on a size exclusion column (Superose 6). Western blot analysis showed that INTS11 coeluted with EZH2, SUZ12, and EED as well as other INT subunits in high–molecular weight fractions (Fig. 6A), suggesting that INTS11 is part of a large multiprotein complex that includes PRC2 proteins. We then assessed whether endogenous INTS11 formed a complex with PRC2 members in hematopoietic cells. We performed IPs for INTS11 or EZH2 and analyzed for INTS11, EZH2, and SUZ12 in nuclear extracts of human K562 cells. Both EZH2 and SUZ12 coimmunoprecipitated with INTS11, whereas INTS11 and SUZ12 coimmunoprecipitated with EZH2 (Fig. 6B). These results demonstrate that INTS11 interacts with PRC2 in hematopoietic cells.
Fig. 6. INTS11 physically interacts with EZH2.
(A) Western blot analysis of even fraction numbers collected from a Superose 6 size exclusion chromatography run using nuclear extracts from 293T cells expressing FLAG-INTS11. (B) INTS11 interacts with PRC2 in human K562 hematopoietic cells. Western blot analysis for the indicated proteins after IP of INTS11 (left), EZH2 (right), or IgG. (C and D) Bacterially expressed GST-INTS11 fusion protein was immobilized on glutathione-agarose beads and incubated with recombinant PRC2 protein complex (C) or FLAG-tagged EZH2 (D). Bound proteins were analyzed by Western blotting. Immobilized GST protein or glutathione-agarose beads were used as negative controls.
To assay whether INTS11 directly interacts with PRC2, we next perform a glutathione S-transferase (GST) pull-down assay. GST-INTS11 fusion proteins were captured with glutathione beads (fig. S6E) and were incubated with recombinant PRC2 protein complex (EZH2/SUZ12/EED). The results showed that GST-INTS11, but not GST or empty glutathione beads, precipitated the PRC2 core subunits EZH2, SUZ12, and EED (Fig. 6C). To further determine which subunit of PRC2 interacts with INTS11, we incubated GST-INTS11 fusion protein with recombinant protein of FLAG-tagged EZH2, SUZ12, or EED, respectively. Compared with GST alone control, the GST-INTS11 fusion protein was able to pull down EZH2, but not SUZ12 or EED (Fig. 6D and fig. S6F). These findings indicate that INTS11 physically interacts with PRC2.
Decreased H3K27me3 enrichment upon Ints11 deletion contributes to the up-regulation of PRC2 targets
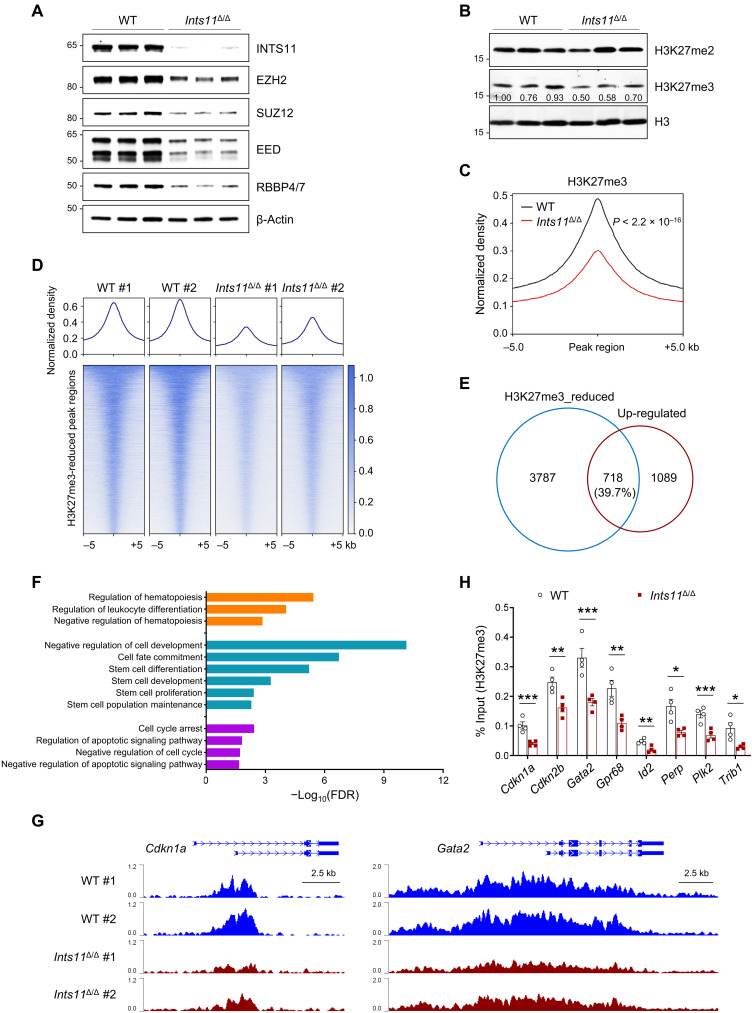
To determine whether the loss of Ints11 affects PRC2 activities and functions, we evaluated the mRNA and protein levels of PRC2 subunits in BM LK cells of WT and Ints11Δ/Δ mice 72 hours after poly(I:C) injection. The protein levels of EZH2, SUZ12, EED, and RBBP4/7 were greatly reduced in Ints11Δ/Δ cells compared with WT controls (Fig. 7A), whereas the levels of CTCF and HDAC1 were comparable between Ints11Δ/Δ and WT cells (fig. S7A). Unexpectedly, there was no change in mRNA levels of Ezh2, Suz12, Eed, and Rbbp4/7 in Ints11Δ/Δ cells (fig. S7B). The PRC2 protein complex is known to catalyze methylation on lysine 27 of histone H3 (H3K27) (27). We next performed Western blot analysis to compare the levels of H3K27me3 between WT and Ints11Δ/Δ cells. The results showed that Ints11Δ/Δ LK cells exhibited a decreased level of global H3K27me3 compared with WT LK cells, while the level of H3K27me2 was identical between Ints11Δ/Δ and WT cells (Fig. 7B). Further analysis using fetal liver cells pooled from VavCre+;Ints11flox/flox or VavCre− embryos revealed that the levels of H3K27me3, EZH2, SUZ12, and EED were also decreased in VavCre+;Ints11flox/flox compared with those in VavCre− controls (fig. S7C). These data suggest that loss of INTS11 destabilizes the PRC2 complex proteins, thus decreasing the enzymatic activity of the PRC2 complex on H3K27 trimethylation.
Fig. 7. Decreased H3K27me3 enrichment contributes to the up-regulation of gene expression in Ints11-deficient LK cells.
(A) Western blot analysis of indicated members of the PRC2 complex in LK cells from WT and Ints11Δ/Δ mice 72 hours after poly(I:C) injection. (B) Western blot showing the levels of H3K27me2 and H3K27me3 in Ints11Δ/Δ LK cells. H3 was used as a loading control. (C) Global levels of H3K27me3 at peaks and 5-kb regions surrounding the peak midpoint. The coverages were normalized by the sequencing depth and averaged in two biological replicates. P value was calculated by paired Student’s t test. (D) Heatmaps of normalized H3K27me3 ChIP-seq read densities centered on the midpoints of 5985 H3K27me3-decreased regions. Each row represents a single region. (E) Venn diagram showing the overlap between genes with reduced H3K27me3 and up-regulated genes in Ints11Δ/Δ LK cells. The number of genes in each section of the diagram is shown. (F) Functional enrichment analysis for 718 overlapping genes in (E). Representative significantly enriched pathways were displayed (FDR < 0.05). (G) Normalized H3K27me3 signals on the Cdkn1a and Gata2 gene loci. (H) ChIP-qPCR verified the reduction of H3K27me3 occupancies at the promoter regions of the genes in Fig. 4F (n = 4 per genotype). Data are means ± SEM. Unpaired Student’s t test: *P < 0.05, **P < 0.01, and ***P < 0.001.
To evaluate whether the dysregulated gene expression following Ints11 deletion was associated with the changes of histone modifications, we performed ChIP-seq for H3K27me3 and H3K4me3 using WT and Ints11Δ/Δ LK cells. Normalized global read density and locus-level enrichment revealed a significant reduction in genome-wide H3K27me3 occupancy in Ints11Δ/Δ cells compared with WT cells (P < 2.2 × 10−16) (Fig. 7, C and D, and fig. S7D). However, there were no significant changes of H3K4me3 peaks globally between WT and Ints11Δ/Δ LK cells (fig. S7E), suggesting a limited impact of Ints11 on H3K4me3 in HSPCs.
To determine the impact of decreased H3K27me3 on gene expression in Ints11Δ/Δ LK cells, we integrated the RNA-seq and ChIP-seq data. We found that 718 (39.7%) up-regulated genes were associated with reduced H3K27me3 peaks (Fig. 7E). Gene ontology analysis revealed that the up-regulated genes were enriched in hematopoiesis, stem cell development, cell cycle, and apoptosis (Fig. 7F). We also examined the occupancies of H3K27me3 on genes associated with cell cycle, apoptosis, and HSC function, respectively. Up-regulated genes, including Cdkn1a, Gata2, Perp, and Trib1, in Ints11Δ/Δ LK cells, were correlated with a lower intensity of H3K27me3 (Fig. 7G and fig. S7F). ChIP-qPCR confirmed significant decreases in H3K27me3 at the promoters of selected genes that were activated in Ints11Δ/Δ LK cells, in contrast to minimal changes in H3K4me3 occupancies (Fig. 7H and fig. S7G). These data suggest that the key up-regulated genes are directly targeted by PRC2 in Ints11-deficient HSPCs. The association of PRC2 with INTS11 is required for transcriptional repression in HSPCs.
Reexpression of INTS11 increases the PRC2 activity and rescues the Ints11-deficient phenotypes
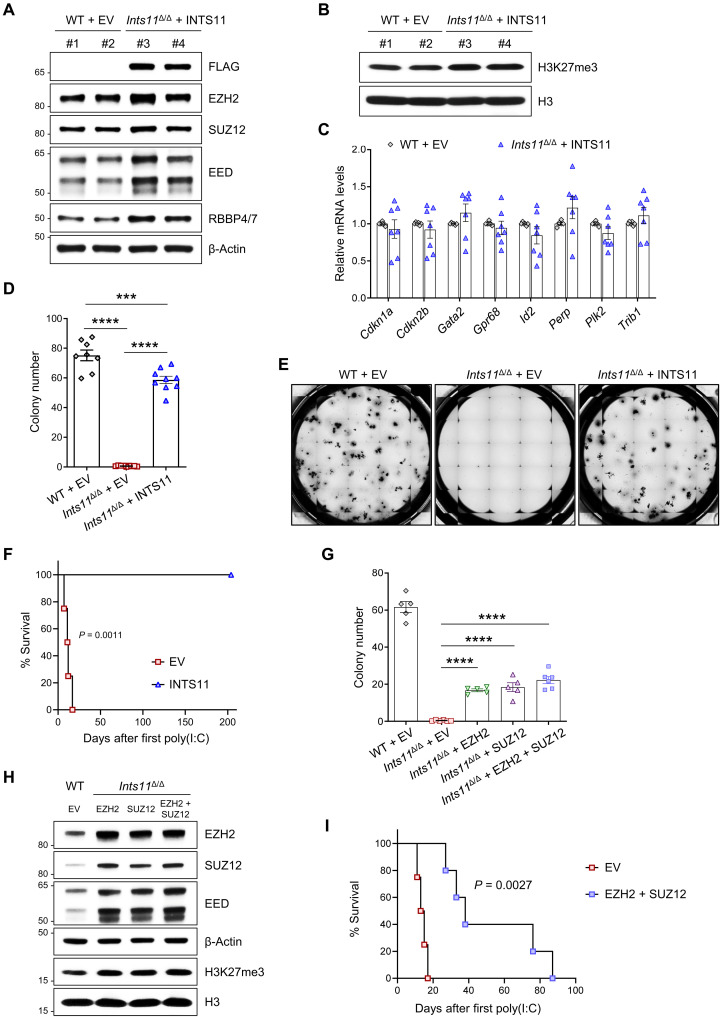
To determine whether altered PRC2 activity is responsible for the defective Ints11Δ/Δ HSPCs, we harvested BM cKit+ cells from Ints11Δ/Δ mice 72 hours after poly(I:C) injection and then transduced FLAG-INTS11 into cells and performed CFU-C assays. Reexpression of INTS11 increased the protein levels of PRC2 subunits and H3K27me3 (Fig. 8, A and B, and fig. S8A) and reduced the expression of PRC2 targets, including Cdkn1a and Perp (Fig. 8C). Further analysis by ChIP-qPCR revealed normalized H3K27me3 occupancies on these PRC2 target genes (fig. S8B). Accordingly, reexpression of INTS11 significantly increased the colony-forming capacity of Ints11Δ/Δ cells (Fig. 8, D and E). To assess the effect of reexpression of INTS11 in Ints11Δ/Δ cells on HSPC functions in vivo, we performed phenotypic analysis following the transplantation of Mx1Cre+;Ints11flox/flox BM cells (CD45.2+) expressing INTS11 or empty vector (EV) control into lethally irradiated BoyJ (CD45.1+) recipient mice (fig. S8C). Ints11 deletion was induced upon confirmation of engraftment. Similar to the primary Ints11Δ/Δ mice, the recipients receiving the EV cells died within 3 weeks of poly(I:C) injection (Fig. 8F). In contrast, the reexpression of INTS11 rescued the survival of the recipient mice transplanted with Ints11Δ/Δ BM cells expressing INTS11 (Fig. 8F). Flow cytometric analysis revealed that reexpression of INTS11 markedly increased the frequencies of LKS− and LSK cell populations in the BM (fig. S8, D and E).
Fig. 8. Reexpression of INTS11 increases the PRC2 activity and rescues the Ints11-deficient phenotypes.
(A and B) Western blot showing the levels of PRC2 subunits (A) and H3K27me3 (B) in Ints11Δ/Δ cells transduced with FLAG-INTS11. (C) qPCR showing the expression levels of up-regulated genes (Fig. 4F) in Ints11Δ/Δ cells transduced with FLAG-INTS11. Each dot represents an individual mouse (n = 6 to 7 per group). (D) Reexpression of INTS11 increased the colony-forming capacity of Ints11Δ/Δ cKit+ cells. Each dot represents an individual mouse (n = 8 to 9 per group). (E) Representative images of colony formation for Ints11-deficient cKit+ cells transduced with FLAG-INTS11 and controls. The images were taken on the seventh day of the assay. (F) Kaplan-Meier survival analysis of recipients receiving Ints11Δ/Δ cells expressing FLAG-INTS11. Log-rank (Mantel-Cox) test; FLAG-INTS11, n = 6; EV controls, n = 4. (G) Enhanced expression of PRC2 partially restored the colony-forming capacity of Ints11Δ/Δ cKit+ cells. Each dot represents an individual mouse (n = 5 to 6 per group). (H) Western blot showing the levels of PRC2 subunits and H3K27me3 in Ints11Δ/Δ cells transduced with EZH2 and/or SUZ12. (I) Kaplan-Meier survival analysis of recipients. Log-rank (Mantel-Cox) test; EZH2 + SUZ12, n = 5; EV controls, n = 4. Data are means ± SEM. One-way analysis of variance with Tukey’s multiple comparisons test: ***P < 0.001 and ****P < 0.0001.
Deletion of Ints11 reduced PRC2 protein levels along with up-regulation of PRC2 target genes in LK cells. We then asked whether enhanced expression of EZH2 or SUZ12 in Ints11Δ/Δ cKit+ cells could restore the colony-forming activity of Ints11Δ/Δ cells. Ints11Δ/Δ cKit+ cells transduced with EZH2 and/or SUZ12 significantly increased the frequencies of CFU-C compared with the cells transduced with EV control (Fig. 8G and fig. S8F). Accordingly, enhanced expression of EZH2 and/or SUZ12 increased the H3K27me3 levels and repressed the PRC2 target gene in Ints11Δ/Δ cells (Fig. 8H and fig. S8, G to I). Enhanced expression of EZH2 and SUZ12 in Ints11Δ/Δ cells extended the survival of the recipients (Fig. 8I). In summary, these data demonstrate that alteration of PRC2 function is at least partially responsible for the effects of INTS11 on normal hematopoiesis.
DISCUSSION
In the current study, we identify an essential role for INTS11 as a key regulator of HSC homeostasis in vivo. We show that deletion of Ints11 in the hematopoietic system results in cell cycle arrest of G1-to-S phase transition, leading to HSC dysfunction. We characterize a previously unidentified function of INTS11 in gene regulation and demonstrate that INTS11 interacts with PRC2 to regulate gene expression through the histone mark H3K27me3. We found that loss of Ints11 destabilized the PRC2 complex and decreased the levels of H3K27me3 in HSPCs. The up-regulated genes in Ints11Δ/Δ cells, which are key for cell cycle, HSC function, and apoptosis, were associated with the reduced H3K27me3 levels. Reexpression of INTS11 or EZH2/SUZ12 in Ints11Δ/Δ HSPCs increased the levels of PRC2 and H3K27me3 along with the restoration of the HSC function.
The mechanisms that maintain the balance between self-renewal and differentiation in HSCs are critical for preventing exhaustion of the stem cell pool or the development of hematopoietic malignancies (30). Posttranslational modifications (PTMs) on histone tails can promote either transcriptional silencing or activation. PRC2 is responsible for the methylation of lysine-27 on histone H3. The H3K27me3 signature is highly correlated with silent loci and is important for regulating developmental and oncogenic genes (31). Recent studies showed that transcript cleavage by INTS11 may not be the main event to trigger transcription termination after pausing (32). Our work demonstrates that INTS11 interacts with the PRC2 complex directly and regulates the expression of genes through H3K27me3. Previous reports using loss-of-function approaches targeting individual PRC2 subunits have suggested that PRC2 proteins are essential regulators for normal hematopoiesis (33–36). The catalytic subunit EZH2 is essential for fetal hematopoiesis, but is dispensable for adult BM HSC function, due to compensation by EZH1 (33). In contrast, the core subunits of PRC2, SUZ12, and EED are required for both fetal and adult hematopoiesis. The hematopoietic specific knockout of either Suz12 or Eed in mice results in HSC exhaustion at the fetal or adult stage (34–36). Our data indicate that INTS11 and PRC2 share the molecular mechanisms in the regulation of HSPC function. However, we cannot exclude that the observed interactions could result from nonspecific hydrophobic or electrostatic interactions because of spatial proximity. Future studies are warranted to determine the specific domain of INTS11 mediating the interaction with PRC2 and evaluate the function of INTS11 upon modulation of PRC2 proteins in HSPCs.
A growing body of evidence suggests that individual INT subunits might have a function beyond their roles in the 14-subunit complex (37). For example, INTS3 and INTS6 have been shown to stably interact with human single-stranded DNA binding protein 1 and play crucial roles in the DNA damage response and homologous recombination–dependent repair of DNA double-strand breaks (38–42). In addition, Barbieri and colleagues (16) showed that INTS13 functions as an independent submodule to mediate activation of monocytic enhancers through EGR1 and NAB2. Other INT subunits, including INTS11, INTS1, and INTS6, were absent at the INTS13-specific sites (16). Our results suggest that INTS11 functions through the PRC2 complex and several INT subunits may be involved in the process. Increasing evidence suggests that PTMs, including methylation, phosphorylation, and acetylation on the PRC2 components, can affect PRC2 stability, assembly, and catalytic activity (43). However, the specific enzymes catalyzing most of these PTMs remain to be identified, and their biological significance is largely unknown. Further studies are needed to investigate how INTS11 stabilizes the PRC2 and whether other subunits’ loss influences the function of PRC2 in hematopoietic cells.
Our study shows the complex consequences of INTS11 loss for hematopoiesis and demonstrates its essential roles in HSC self-renewal and differentiation. Our findings reveal a novel noncanonical INTS11 function via a PRC2 complex in gene regulation in HSPCs.
MATERIALS AND METHODS
Experimental design
The objective of the study was to investigate the functions of INTS11 in HSPCs. A novel Ints11 conditional knockout mouse model was generated and used to determine the effects of Ints11 deletion in the hematopoietic system.
Ints11 conditional knockout mouse model
This mouse model was provided by R. Shiekhattar (University of Miami). Targeted Ints11 knockout mice were generated by replacing exon 2 sequences of the Ints11 gene with LoxP, neomycin, and FRT-containing cassette (fig. S1A). Ints11-targeted embryonic stem cell clones were expanded and injected into blastocysts. The generated mice were then bred to Flp recombinase transgenic mice to create Ints11flox/+ mice. These mice were back-crossed to WT C57BL/6 mice for six rounds of breeding. Mx1-Cre, Vav1-Cre, and BoyJ (CD45.1+) mice were purchased from The Jackson Laboratory. Mx1Cre-induced gene deletion was done by intraperitoneal injection of poly(I:C) (5 mg/kg; InvivoGen) three times daily for three consecutive days. The genotyping PCR primers are listed in table S1. All animal studies were performed with approval from the University of Texas Health Science Center at San Antonio Institutional Animal Care and Use Committee and conducted following institutional and national regulatory standards.
Phenotypic analyses of mice
PB was collected by tail vein bleeding and subjected to an automated blood count (Hemavet System 950FS, Drew Scientific). For histopathology analyses, sternums were fixed for >24 hours in 10% neutral buffered formalin at room temperature and demineralized in 12% EDTA for 2 weeks. The specimens were dehydrated using ethanol and cleared in xylenes. The specimens were then embedded in melted paraffin and allowed to harden. Thin sections (4 to 5 μm) were cut and floated onto microscope slides. The slides were stained with hematoxylin and eosin (H&E) and visualized under a Keyence BZ-X810 microscope.
Flow cytometry, cell sorting, and colony assays
Single-cell suspensions from PB, BM, spleen, or fetal livers were stained with panels of fluorochrome-conjugated antibodies (table S2). Flow cytometric analysis of HSPCs was performed as previously described (44). The analyses were performed using a BD FACSCelesta or LSRFortessa flow cytometer (BD Biosciences). Apoptosis was assessed using the PE Annexin V Apoptosis Detection Kit (BD Biosciences). Briefly, freshly isolated BM cells were stained with lineage antibodies and phycoerythrin (PE)–annexin V/7-AAD followed by fluorescence-activated cell sorting (FACS) analysis within an hour of staining. For cell cycle analysis, BM cells were labeled with 5-bromo-2′-deoxyuridine (BrdU) for an hour in vitro, and BrdU incorporation was detected using fluorescein isothiocyanate (FITC)–conjugated anti-BrdU antibody followed by 7-AAD staining per the manufacturer’s protocol (BD Biosciences). All data were analyzed with FlowJo v10 software (Ashland, OR). For LK cell selection, magnetic-activated cell sorting was applied. BM cells were first isolated using the Direct Lineage Cell Depletion Kit (Miltenyi Biotec), and then the lineage-negative cells were sorted with cKit (CD117) MicroBeads. The purity of selected cells was routinely over 95%. For CFU-C assays, BM or fetal liver cells were plated in triplicate in methylcellulose medium (MethoCult M3134, STEMCELL Technologies) supplemented with mouse stem cell factor (mSCF; 100 ng/ml), interleukin-3 (mIL-3; 10 ng/ml), thrombopoietin (mTPO; 50 ng/ml), granulocyte-macrophage colony-stimulating factor (mGM-CSF; 10 ng/ml), human erythropoietin (hEPO; 4 U/ml), and interleukin-6 (hIL-6; 50 ng/ml, PeproTech). The colonies were then scored on day 7 of the cultures in an incubator at 37°C and 5% CO2.
Transplantation assay
Competitive repopulation assay was performed by transplanting a mixture of 1 × 106 4-week-old BoyJ (CD45.1+) BM competitor cells and 1 × 106 of either 4-week-old Mx1Cre+ or Mx1Cre+;Ints11flox/flox (CD45.2+) BM cells into lethally irradiated (9.5 Gy) BoyJ recipients through the tail vein injection. One month after the transplantation, poly(I:C) (10 mg/kg) was injected in the recipient mice to induce Ints11 deletion. Flow cytometric analyses were performed to monitor the chimerism by examining the percentages of CD45.1+ and CD45.2+ over 30 weeks after the poly(I:C) injection.
For in vivo rescue assay, BM cells were collected from 5-fluorouracil (5-FU; Selleckchem)–treated Mx1Cre+;Ints11flox/flox (CD45.2+) mice. Mice were given a single dose of 5-FU (150 mg/kg) by intraperitoneal injections and were sacrificed 48 hours after injection. Mononuclear cells were transduced with the packaged viruses in the culture medium supplemented with mSCF (100 ng/ml) and hIL-6 (50 ng/ml) and then transplanted into lethally irradiated BoyJ recipient mice (CD45.1+) through the tail vein injection. Four weeks after the transplantation, recipients were injected with a total of five doses of poly(I:C) (10 mg/kg, every other day) to induce Ints11 deletion.
Quantitative PCR analysis
Total RNA was extracted with TRIzol reagent (Invitrogen), and complementary DNA (cDNA) was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. qPCR was performed in triplicate using an Applied Biosystems QuantStudio 3 system with the Fast SYBR Green Master Mix (Applied Biosystems). The expression of gene of interest was calculated using the 2−ΔΔCt method by normalization to the housekeeping gene β-actin. All qPCR primers used are listed in table S1.
Plasmid constructs, cell culture, and lentiviral transduction
The full-length cDNAs of mouse Ints11, Ezh2, or Suz12 were tagged with 3×FLAG or HA and independently cloned into a lentiviral vector, as previously described (45). The human hematopoietic cell line K562 was obtained from the American Type Culture Collection (ATCC) and cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. 293TN cells (System Biosciences) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and transfected with the plasmid of Ints11 full-length (FLAG-INTS11), an EV (FLAG-EV), HA-EZH2, or HA-SUZ12 using Lipofectamine 3000 (Thermo Fisher Scientific). The viral supernatant was harvested 48 hours after transfection. BM cKit+ cells purified from Ints11Δ/Δ mice 72 hours after poly(I:C) injection were transduced with the packaged viruses, and the positive cells were then sorted for further experiments using a BD FACSAria machine.
Western blot, IP, and GST pull-down assays
Lysates from BM LK cells were prepared and then resolved on NuPAGE 4 to 12% bis-tris gels (Invitrogen). Immunoblotting was performed with the following antibodies (table S2): rabbit polyclonal anti-INTS11 (1:1000), rabbit polyclonal anti-H3K27me3 (1:1000), rabbit polyclonal anti-mouse H3K27me2 (1:1000), rabbit polyclonal anti-H3 (1:2000), rabbit monoclonal anti-EZH2 (1:1000), rabbit monoclonal anti-SUZ12 (1:1000), rabbit monoclonal anti-EED (1:1000), rabbit polyclonal anti-RBBP4/7 (1:800), rabbit monoclonal anti-p21Cip1 (1:1000), rabbit polyclonal anti-mouse CTCF (1:1000), rabbit polyclonal anti-mouse HDAC1 (1:1000), mouse monoclonal anti–β-actin (1:4000), mouse monoclonal anti-mouse FLAG (1:1000), and mouse monoclonal anti-mouse HA (1:1000). After incubation with anti-rabbit immunoglobulin G (IgG) or anti-mouse IgG antibodies conjugated with horseradish peroxidase (GE Healthcare), signals were detected using the Prometheus ProSignal ECL reagents (Genesee Scientific). Images were taken on a KwikQuant Imager system (Kindle Biosciences).
For the IP assays, nuclear extracts were prepared as previously described with the presence of benzonase (46). Lysates were incubated overnight with the indicated antibodies. After washing of the beads four times with IP buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA (pH 8.0), 1% Triton X-100, and protease inhibitor cocktail], the associated proteins were collected and subjected to Western blot analysis. The chromatin-bound fraction and the soluble nuclear fraction were prepared using the Subcellular Protein Fractionation Kit (Thermo Fisher Scientific).
The full-length human INTS11 was cloned into the pET expression vector downstream of the GST tag. Escherichia coli strain BL21 (DE3) was transformed with pET-hINTS11 or the EV expressing GST alone. GST fusion proteins were purified on Pierce Glutathione Agarose beads (Thermo Fisher Scientific). An equal amount of GST protein was incubated with PRC2 recombinant proteins (table S2) in binding buffer [50 mM tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 0.1% NP-40, 10 mM MgCl2, 1 mM dithiothreitol, and protease inhibitors]. After washing the beads, the bounded proteins were analyzed by Western blotting.
Size exclusion chromatography
Chromatography was performed using 0.5 ml of nuclear extracts obtained from 293T cells expressing FLAG-INTS11 by fractionation on a Superose 6 Increase 10/300 GL column (Cytiva) in a buffer composed of 50 mM tris-HCl (pH 7.4) and 150 mM NaCl with a flow rate of 0.5 ml/min. Fractions (0.5 ml) were collected across the entire protein elution profile and analyzed by Western blotting.
Bulk RNA-seq and data analysis
BM LK cells from Ints11Δ/Δ and WT littermates were purified for 72 hours after first poly(I:C) injection (same time point applied in scRNA-seq and ChIP-seq). Total RNA from individual mice was isolated with TRIzol reagent (Invitrogen). The RNA libraries were prepared using the Illumina TruSeq strand-specific mRNA sample preparation system followed by sequencing with a read length of 2 × 75 base pairs (bp) using Illumina NextSeq 500.
After being trimmed by Trimmomatic (v0.38) (47), more than 135 million reads per sample were aligned to the mouse genome (GRCm38/mm10) using STAR (v2.7.0e) (48). The raw read counts of each gene were calculated by HTSeq (v0.11.2) (49). Then, the count matrix was used to identify differentially expressed genes by DESeq2 (50) with a cutoff of FDR < 0.05 and |fold change| > 1.8. The fragments per kilobase of transcript per million mapped (FPKM) matrix transformed by count matrix was used for GSEA (51).
scRNA-seq and data analysis
For scRNA-seq, the cKit+ cells were pooled from two mice of each genotype. The libraries for each single-cell sample were prepared using the Chromium Single Cell 3′ Reagent Kit (10X Genomics) with the Chromium Controller. The libraries were sequenced on the Illumina NovaSeq 6000 sequencing system. Sequencing results were demultiplexed and converted to FASTQ format by CellRanger (v3.0.2; www.10xgenomics.com) with a parameter of mkfastq. The sequencing reads were aligned to the mouse reference genome (mm10) and quantified using CellRanger with default parameters. The filtered results (count matrix, features, and barcodes) were imported into Seurat (v3.1.5) (52) to create Seurat objects for each sample. To exclude low-quality cells and potential doublets, the cells that meet the following criteria were removed: (i) with less than 500 or more than 6000 detected features; (ii) with mitochondrial transcripts that accounted for more than 10% of the total detected transcripts; and (iii) predicted to be doublets by Scrublet (v0.2.1) (53). Moreover, the genes that were detected in less than three cells for each sample were also excluded in the following analysis. After filtering, 7872 WT and 10,657 Ints11Δ/Δ cells were eventually retained for further analysis. To remove the influence of technical characteristics among different cells and different samples, we normalized unique molecular identifier counts and regressed out the variable of mitochondrial transcripts percent for each sample separately with sctransform (v0.2.1) (54). The normalized data from the two samples were integrated with IntegrateData.
To reveal the overall and population-specific transcriptomic changes caused by Ints11 loss, we first performed principal components analysis (PCA) on the scaled expression value of the top 3000 highly variable genes with RunPCA for dimensionality reduction. The top 16 principal components were selected for uniform manifold approximation and projection (UMAP) visualization (RunUMAP) and find neighbors for clustering according to the elbow of PC SD. All cells are classified into 26 clusters after running the Louvain clustering algorithm with FindClusters (resolution = 1.2). According to the expression levels of known population-specific markers (25, 55–57), we annotated each cluster and then merged the clusters with the same annotations. Moreover, to deeply explore the impact of Ints11 loss on HSC, we performed dimensionality reduction and clustering again (top 10 principal components, resolution = 0.5) after extracting the cells in cluster 2. Last, four subclusters were eventually formed and annotated (56).
For each above annotated population, differentially expressed genes and differentially expressed pathways were identified with FindMarkers and GSEA analysis, respectively. Cell cycle scores and stages were calculated for each cell using the CellCycleScoring function with cell cycle gene sets in Seurat (converting these gene symbols to mouse homologs). Using the same method of scoring the cell cycle, we calculated the erythroid/megakaryocyte score (58–61), monocyte score, stemness score, proliferation score (25), and apoptosis score (gene ontology database), respectively.
ChIP-seq and data analysis
ChIP assays were performed using LK cells, as previously reported (44). Genomic DNA regions of interest were isolated using antibodies against H3K27me3 (MilliporeSigma, 07-449) and H3K4me3 (Diagenode, C15410003). The ChIP libraries were prepared using the MicroPlex Library Preparation Kit (Diagenode) and sequenced with a read length of 75 bp on an Illumina NextSeq 500 system.
Raw data of each sample were filtered and trimmed by TrimGalore (v0.5.0; https://github.com/FelixKrueger/TrimGalore). Clean reads were aligned to the mouse reference genome (mm10) using bowtie2 (v2.3.4.3) (62, 63). The raw SAM files generated by bowtie2 were sorted using SAMtools (v1.9) (64). PCR duplications were removed by Sambamba (v0.6.8) (65). Peak calling was performed with MACS2 (v2.1.2) (66) with the following options: -g mm -B -q 0.05. For histone modifications, peaks with high confidence (treatment/input > 5) were retained. The BAM files were normalized using deepTools (v3.1.3) (67) to generate a BigWig file for visualization with the following options: --normalize Using CPM --binSize 50. Differential peaks were detected by DiffBind (68) using the embedded method DESeq2 with the threshold of FDR < 0.05. ChIPpeakAnno (69) and ChIPseeker (70) were used to annotate the total or differential peaks. All figures were visualized in R and GraphPad Prism software.
Statistical analysis
Differences between experimental groups were determined by the log-rank test, unpaired two-tailed Student’s t test, or one-way analysis of variance followed by an appropriate post hoc correction. P values of less than 0.05 were considered significant. The statistical methods used for comparisons are indicated in the relevant figure legends.
Acknowledgments
We thank R. Shiekhattar (University of Miami, Miami, FL) for providing the Ints11flox mouse model. We thank the members of the Flow Cytometry Shared Resource Facility at the UT Health San Antonio. Funding: This work was supported by grants from the NIH (HL149318 to F.-C.Y., CA172408 to F.-C.Y. and M.X., and CA240139 to S.D.N.), Evan’s Foundation (to F.-C.Y.), and the Leukemia and Lymphoma Society Specialized Center of Research grant (to F.-C.Y., M.X., and S.D.N.). C.M.R. is supported by a T32 (AG 021890) through the UT Health San Antonio Biology of Aging program. P. Sung is a CPRIT Scholar of Cancer Research and the Robert A. Welch Distinguished Chair in Chemistry (AQ-0012). Author contributions: P.Z. and F.-C.Y. conceived the project and designed the study. P.Z., S.C., Y.G., Y.L., G.G., G.Z., H.Y., and C.M.R. performed the experiments. P. Sui and P.Z. performed the bulk RNA-seq, scRNA-seq, and ChIP-seq analyses. P.Z., P. Sui, P. Sung, S.D.N., M.X., and F.-C.Y. discussed and analyzed the data. P.Z., P. Sui, and F.-C.Y. wrote the manuscript. All authors reviewed, edited, and approved the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All the sequencing datasets generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database (GSE159405).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
REFERENCES AND NOTES
- 1.Orkin S. H., Zon L. I., Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zon L. I., Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature 453, 306–313 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Shaknovich R., De S., Michor F., Epigenetic diversity in hematopoietic neoplasms. Biochim. Biophys. Acta 1846, 477–484 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kato H., Igarashi K., To be red or white: Lineage commitment and maintenance of the hematopoietic system by the “inner myeloid”. Haematologica 104, 1919–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baillat D., Wagner E. J., Integrator: Surprisingly diverse functions in gene expression. Trends Biochem. Sci. 40, 257–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baillat D., Hakimi M. A., Naar A. M., Shilatifard A., Cooch N., Shiekhattar R., Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Gardini A., Baillat D., Cesaroni M., Hu D., Marinis J. M., Wagner E. J., Lazar M. A., Shilatifard A., Shiekhattar R., Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell 56, 128–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadelmayer B., Micas G., Gamot A., Martin P., Malirat N., Koval S., Raffel R., Sobhian B., Severac D., Rialle S., Parrinello H., Cuvier O., Benkirane M., Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat. Commun. 5, 5531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skaar J. R., Ferris A. L., Wu X., Saraf A., Khanna K. K., Florens L., Washburn M. P., Hughes S. H., Pagano M., The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 25, 288–305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai F., Gardini A., Zhang A., Shiekhattar R., Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elrod N. D., Henriques T., Huang K. L., Tatomer D. C., Wilusz J. E., Wagner E. J., Adelman K., The integrator complex attenuates promoter-proximal transcription at protein-coding genes. Mol. Cell 76, 738–752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatomer D. C., Elrod N. D., Liang D., Xiao M. S., Jiang J. Z., Jonathan M., Huang K. L., Wagner E. J., Cherry S., Wilusz J. E., The integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev. 33, 1525–1538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang K. L., Jee D., Stein C. B., Elrod N. D., Henriques T., Mascibroda L. G., Baillat D., Russell W. K., Adelman K., Wagner E. J., Integrator recruits protein phosphatase 2A to prevent pause release and facilitate transcription termination. Mol. Cell 80, 345–358.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt D., Reuter H., Huttner K., Ruhe L., Rabert F., Seebeck F., Irimia M., Solana J., Bartscherer K., The Integrator complex regulates differential snRNA processing and fate of adult stem cells in the highly regenerative planarian Schmidtea mediterranea. PLOS Genet. 14, e1007828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao S., Cai Y., Sampath K., The Integrator subunits function in hematopoiesis by modulating Smad/BMP signaling. Development 136, 2757–2765 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Barbieri E., Trizzino M., Welsh S. A., Owens T. A., Calabretta B., Carroll M., Sarma K., Gardini A., Targeted enhancer activation by a subunit of the integrator complex. Mol. Cell 71, 103–116.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimi A., Lin K. T., Wiseman D. H., Rahman M. A., Pastore A., Wang B., Lee S. C., Micol J. B., Zhang X. J., de Botton S., Penard-Lacronique V., Stein E. M., Cho H., Miles R. E., Inoue D., Albrecht T. R., Somervaille T. C. P., Batta K., Amaral F., Simeoni F., Wilks D. P., Cargo C., Intlekofer A. M., Levine R. L., Dvinge H., Bradley R. K., Wagner E. J., Krainer A. R., Abdel-Wahab O., Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y., Albrecht T. R., Baillat D., Wagner E. J., Tong L., Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance. Proc. Natl. Acad. Sci. U.S.A. 114, 4394–4399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otani Y., Nakatsu Y., Sakoda H., Fukushima T., Fujishiro M., Kushiyama A., Okubo H., Tsuchiya Y., Ohno H., Takahashi S., Nishimura F., Kamata H., Katagiri H., Asano T., Integrator complex plays an essential role in adipose differentiation. Biochem. Biophys. Res. Commun. 434, 197–202 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Jodoin J. N., Shboul M., Albrecht T. R., Lee E., Wagner E. J., Reversade B., Lee L. A., The snRNA-processing complex, Integrator, is required for ciliogenesis and dynein recruitment to the nuclear envelope via distinct mechanisms. Biol. Open 2, 1390–1396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Koe C. T., Tan Y. S., Ho J., Tan P., Yu F., Sung W. K., Wang H., The integrator complex prevents dedifferentiation of intermediate neural progenitors back into neural stem cells. Cell Rep. 27, 987–996.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Georgiades P., Ogilvy S., Duval H., Licence D. R., Charnock-Jones D. S., Smith S. K., Print C. G., VavCre transgenic mice: A tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 34, 251–256 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Steinman R. A., Cell cycle regulators and hematopoiesis. Oncogene 21, 3403–3413 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Hao S., Chen C., Cheng T., Cell cycle regulation of hematopoietic stem or progenitor cells. Int. J. Hematol. 103, 487–497 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Giladi A., Paul F., Herzog Y., Lubling Y., Weiner A., Yofe I., Jaitin D., Cabezas-Wallscheid N., Dress R., Ginhoux F., Trumpp A., Tanay A., Amit I., Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20, 836–846 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Pietras E. M., Reynaud D., Kang Y. A., Carlin D., Calero-Nieto F. J., Leavitt A. D., Stuart J. M., Gottgens B., Passegue E., Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margueron R., Reinberg D., The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang P., Xu M., Yang F.-C., The role of ASXL1/2 and their associated proteins in malignant hematopoiesis. Curr. Stem Cell Rep. 6, 6–15 (2020). [Google Scholar]
- 29.Xu J., Shao Z., Li D., Xie H., Kim W., Huang J., Taylor J. E., Pinello L., Glass K., Jaffe J. D., Yuan G. C., Orkin S. H., Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell 57, 304–316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tesio M., Tang Y., Mudder K., Saini M., von Paleske L., Macintyre E., Pasparakis M., Waisman A., Trumpp A., Hematopoietic stem cell quiescence and function are controlled by the CYLD-TRAF2-p38MAPK pathway. J. Exp. Med. 212, 525–538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youmans D. T., Schmidt J. C., Cech T. R., Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev. 32, 794–805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabath K., Staubli M. L., Marti S., Leitner A., Moes M., Jonas S., INTS10-INTS13-INTS14 form a functional module of Integrator that binds nucleic acids and the cleavage module. Nat. Commun. 11, 3422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mochizuki-Kashio M., Mishima Y., Miyagi S., Negishi M., Saraya A., Konuma T., Shinga J., Koseki H., Iwama A., Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 118, 6553–6561 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Lee S. C. W., Miller S., Hyland C., Kauppi M., Lebois M., Di Rago L., Metcalf D., Kinkel S. A., Josefsson E. C., Blewitt M. E., Majewski I. J., Alexander W. S., Polycomb repressive complex 2 component Suz12 is required for hematopoietic stem cell function and lymphopoiesis. Blood 126, 167–175 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Xie H., Xu J., Hsu J. H., Nguyen M., Fujiwara Y., Peng C., Orkin S. H., Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 14, 68–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu W., Zhang F., Wang S., Fu Y., Chen J., Liang X., Le H., Pu W. T., Zhang B., Depletion of polycomb repressive complex 2 core component EED impairs fetal hematopoiesis. Cell Death Dis. 8, e2744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendoza-Figueroa M. S., Tatomer D. C., Wilusz J. E., The integrator complex in transcription and development. Trends Biochem. Sci. 45, 923–934 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J., Gong Z., Ghosal G., Chen J., SOSS complexes participate in the maintenance of genomic stability. Mol. Cell 35, 384–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Bolderson E., Kumar R., Muniandy P. A., Xue Y., Richard D. J., Seidman M., Pandita T. K., Khanna K. K., Wang W., HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J. Biol. Chem. 284, 23525–23531 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F., Wu J., Yu X., Integrator3, a partner of single-stranded DNA-binding protein 1, participates in the DNA damage response. J. Biol. Chem. 284, 30408–30415 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skaar J. R., Richard D. J., Saraf A., Toschi A., Bolderson E., Florens L., Washburn M. P., Khanna K. K., Pagano M., INTS3 controls the hSSB1-mediated DNA damage response. J. Cell Biol. 187, 25–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F., Ma T., Yu X., A core hSSB1-INTS complex participates in the DNA damage response. J. Cell Sci. 126, 4850–4855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y., Li G., Post-translational modifications of PRC2: Signals directing its activity. Epigenetics Chromatin 13, 47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P., He F., Bai J., Yamamoto S., Chen S., Zhang L., Sheng M., Zhang L., Guo Y., Man N., Yang H., Wang S., Cheng T., Nimer S. D., Zhou Y., Xu M., Wang Q. F., Yang F. C., Chromatin regulator Asxl1 loss and Nf1 haploinsufficiency cooperate to accelerate myeloid malignancy. J. Clin. Invest. 128, 5383–5398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi H., Yamamoto S., Sheng M., Bai J., Zhang P., Chen R., Chen S., Shi L., Abdel-Wahab O., Xu M., Zhou Y., Yang F. C., ASXL1 plays an important role in erythropoiesis. Sci. Rep. 6, 28789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu C., Li L., Zhang Z., Bi M., Wang H., Su W., Hernandez K., Liu P., Chen J., Chen M., Huang T. H., Chen L., Liu Z., A non-canonical role of YAP/TEAD Is required for activation of estrogen-regulated enhancers in breast cancer. Mol. Cell 75, 791–806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolock S. L., Lopez R., Klein A. M., Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hafemeister C., Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahlin J. S., Hamey F. K., Pijuan-Sala B., Shepherd M., Lau W. W. Y., Nestorowa S., Weinreb C., Wolock S., Hannah R., Diamanti E., Kent D. G., Gottgens B., Wilson N. K., A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood 131, e1–e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tikhonova A. N., Dolgalev I., Hu H., Sivaraj K. K., Hoxha E., Cuesta-Dominguez A., Pinho S., Akhmetzyanova I., Gao J., Witkowski M., Guillamot M., Gutkin M. C., Zhang Y., Marier C., Diefenbach C., Kousteni S., Heguy A., Zhong H., Fooksman D. R., Butler J. M., Economides A., Frenette P. S., Adams R. H., Satija R., Tsirigos A., Aifantis I., The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muench D. E., Olsson A., Ferchen K., Pham G., Serafin R. A., Chutipongtanate S., Dwivedi P., Song B., Hay S., Chetal K., Trump-Durbin L. R., Mookerjee-Basu J., Zhang K., Yu J. C., Lutzko C., Myers K. C., Nazor K. L., Greis K. D., Kappes D. J., Way S. S., Salomonis N., Grimes H. L., Mouse models of neutropenia reveal progenitor-stage-specific defects. Nature 582, 109–114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tusi B. K., Wolock S. L., Weinreb C., Hwang Y., Hidalgo D., Zilionis R., Waisman A., Huh J. R., Klein A. M., Socolovsky M., Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noetzli L. J., French S. L., Machlus K. R., New insights into the differentiation of megakaryocytes from hematopoietic progenitors. Arterioscler. Thromb. Vasc. Biol. 39, 1288–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Psaila B., Barkas N., Iskander D., Roy A., Anderson S., Ashley N., Caputo V. S., Lichtenberg J., Loaiza S., Bodine D. M., Karadimitris A., Mead A. J., Roberts I., Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol. 17, 83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pessoa Rodrigues C., Herman J. S., Herquel B., Valsecchi C. I. K., Stehle T., Grun D., Akhtar A., Temporal expression of MOF acetyltransferase primes transcription factor networks for erythroid fate. Sci. Adv. 6, eaaz4815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B., Wilks C., Antonescu V., Charles R., Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 35, 421–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng J., Liu T., Qin B., Zhang Y., Liu X. S., Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramirez F., Dundar F., Diehl S., Gruning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ross-Innes C. S., Stark R., Teschendorff A. E., Holmes K. A., Ali H. R., Dunning M. J., Brown G. D., Gojis O., Ellis I. O., Green A. R., Ali S., Chin S. F., Palmieri C., Caldas C., Carroll J. S., Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu L. J., Gazin C., Lawson N. D., Pages H., Lin S. M., Lapointe D. S., Green M. R., ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2