STAT1 serves as a gatekeeper for naïve CD8+ T cell homeostasis by restraining type I IFN-STAT4-mTOR signaling axis.
Abstract
Naïve CD8+ T cell quiescence is maintained at a steady state. Although this state of quiescence involves various cell-intrinsic and cell-extrinsic regulators, the mechanisms underlying this regulation remain incompletely understood. Here, we found that signal transducer and activator of transcription 1 (STAT1), a key transcription factor downstream of interferon receptor (IFNR) signaling, plays a cell-intrinsic role in maintaining naïve CD8+ T cell quiescence. STAT1-deficient mice showed enhanced proliferation of peripheral naïve CD8+ T cells, which resulted in an abnormal increase in the number of CD44hi memory/activated phenotype cells and an enlargement of secondary lymphoid tissues. This phenomenon was not observed in IFNR-deficient mice but was paradoxically dependent on type I interferon and its alternative signaling pathway via the STAT4–RagD–lysosomal mTORC1 axis. Collectively, these findings underline the importance of STAT1 in regulating the homeostasis of peripheral naïve CD8+ T cells by suppressing their responsiveness to homeostatic cues at a steady state.
INTRODUCTION
Naïve CD8+ T cells in the periphery are constantly exposed to various extrinsic factors, even under steady-state conditions, which are either vital or dispensable for their life (1). Nevertheless, these cells are stably maintained at resting, i.e., the G0 interphase, without any overt activation (2). This state of functional quiescence is not passively inherited as a default but rather acquired actively through a mechanism that is tightly regulated, which allows the avoidance of abnormal activation and proliferation before cognate antigenic stimulation (3). Therefore, the existence of an active mechanism for controlling the responsiveness of quiescent naïve CD8+ T cells to various extrinsic cues could be assumed. However, the nature of such cues and the precise mechanism of their regulation remain incompletely understood.
Interleukin-7 (IL-7) is one such extrinsic cue, and its responsiveness to naïve CD8+ T cells was recently found to be tightly controlled via the FOXP1-mediated regulation of IL-7 receptor (IL-7R) expression (4). FOXP1-deficient CD8+ T cells exhibited enhanced IL-7R expression, leading to abnormal proliferation to even physiological levels of IL-7 in vivo. In addition to FOXP1 deficiency, perturbations to steady-state T cell quiescence have been observed in response to deficiencies of several other transcription factors (TFs), including KLF2 (5), ELF4 (6), FOXO1 (7, 8), TOB (9), and RUNX1 (10, 11), as well as non-TF proteins that exert various cellular functions, such as Schlafen2 (12), TSC1 (13), KDEL1 (14), PELI (15), GIMAP5 (16), PTPN2 (17), and PTPN22 (18). These studies suggest the requirement of many different cell-intrinsic regulators for the maintenance of a quiescent state of naïve CD8+ T cells and imply that this quiescence must be regulated at multiple different layers depending on the nature and type of certain homeostatic cues.
Type I interferon (T1IFN) consists of a group of cytokines that exert pleiotropic effects on the innate and adaptive immune system and has antiviral, antitumor, and immunoregulatory functions (19). Despite the well-known proinflammatory activity of T1IFN upon pathogen infection, its role in maintaining naïve T cell homeostasis at a steady state is less well understood. Here, we demonstrated the previously unidentified role of T1IFN in peripheral naïve CD8+ T cells using mice lacking signal transducer and activator of transcription 1 (STAT1), which is a crucial component downstream of T1IFN receptor signaling. Under STAT1-deficient conditions, T1IFN failed to induce a STAT1-dependent canonical response involving the activation of IFN-stimulated genes (ISGs) but did induce STAT4-dependent alternative signaling to activate the RagD-mechanistic target of rapamycin (mTOR) pathway. Consequently, a Stat1−/− naïve CD8+ T cell population exhibited abnormal proliferation, comprised an increased number of CD44hi memory/activated phenotype (MP) cells, and caused severe forms of inflammatory diseases in an experimental setting of inflammatory bowel disease (IBD). Therefore, this study establishes the previously unidentified finding that STAT1 serves as a crucial cell-intrinsic regulator for actively maintaining naïve CD8+ T cell quiescence under continuous exposure to the atypical homeostatic cue T1IFN.
RESULTS
Stat1−/− mice show an enhanced number of CD44hi MP CD8+ T cells
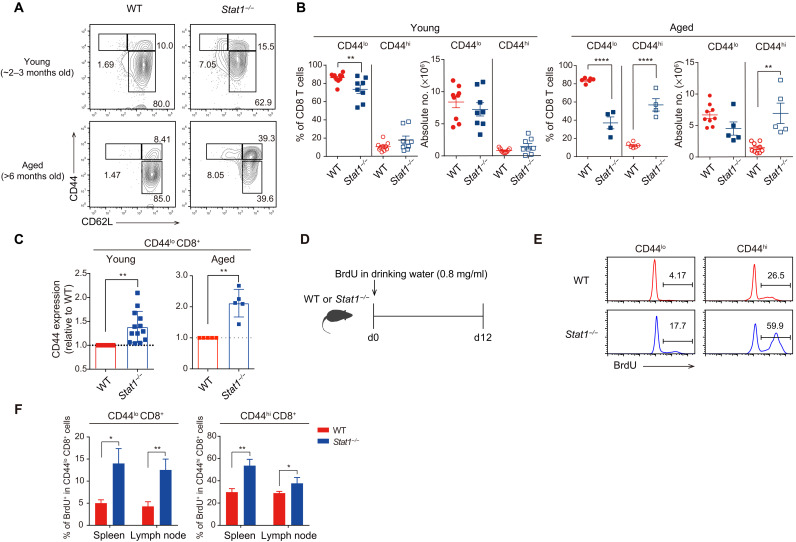
To investigate the role of STAT1 in regulating the homeostasis of CD8+ T cell populations at a steady state, we monitored the phenotype and proportion of these populations in Stat1−/− mice. Stat1−/− and wild-type (WT) mice aged ~2 to 3 months showed similar percentages and numbers of CD44hi MP and CD44lo naïve CD8+ T cells in the spleen (Fig. 1, A and B). Stat1−/− mice aged >6 months showed an approximately fivefold higher percentage and number of MP cells than age-matched WT mice (Fig. 1, A and B). Furthermore, naïve CD8+ T cells from both aged and young Stat1−/− mice presented substantially enhanced CD44 expression levels compared with those of their WT counterparts, as evidenced by the mean CD44 fluorescence intensity on a per-cell basis (Fig. 1C). This alteration in Stat1−/− mice was not observed in the thymus, as demonstrated by the lack of a difference in the CD44 expression levels in CD4+CD8+ double-positive and CD4−CD8+ single-positive thymocytes (fig. S1, A and B), which implies post-thymic alterations.
Fig. 1. Stat1−/− mice exhibit an enhanced number of CD44hi MP CD8+ T cells.
(A) Flow cytometry for CD44 and CD62L expression in splenic CD8+ T cells from young versus aged WT and Stat1−/− mice. (B) Percentage and number of CD44lo and CD44hi CD8+ T cells from WT and Stat1−/− mice. (C) Relative levels of CD44 expression in WT versus Stat1−/− CD44lo CD8+ T cells. (D) Experimental scheme of in vivo BrdU uptake. (E and F) BrdU incorporation of CD44lo and CD44hi CD8+ T cells from WT and Stat1−/− mice. The results are presented as means ± SEM. Data are representative of three to four independent experiments. *P < 0.05, **P < 0.01, and ****P < 0.0001.
Because STAT1 is required for type I and II IFN receptor signaling (20), the abovementioned data might result from a failure to respond to these cytokines. However, the enhanced MP cell numbers and CD44 expression levels (in CD44lo naïve cells) observed in Stat1−/− mice were not detected in mice lacking the T1IFN receptor (Ifnar−/−) or both type I and II receptors (Ifnar−/−.Ifngr−/−) (fig. S1, C and D). These data suggest that the effect of STAT1 deficiency might not simply reflect the failure of IFN signaling. To further understand the mechanisms underlying the enhanced generation of CD44hi cells in Stat1−/− mice, WT and Stat1−/− mice were administered 5-bromo-2′-deoxyuridine (BrdU) for 12 days, and its uptake in peripheral CD8+ T cells was recorded (Fig. 1D). Both CD44hi MP cells and CD44lo naïve cells from Stat1−/− mice showed approximately two- to three-fold higher BrdU uptake than their WT counterparts (Fig. 1, E and F). Similarly, Stat1−/− CD8+ T cells also showed an enhanced proportion of Ki67+ cells compared with WT CD8+ T cells (fig. S1E). Collectively, these results strongly suggest that STAT1 deficiency alters peripheral CD8+ T cell homeostasis, leading to enhanced proliferation and conversion of CD44lo cells into CD44hi cells.
Stat1−/− naïve CD8+ T cells show enhanced in vivo proliferation in a cell-intrinsic manner
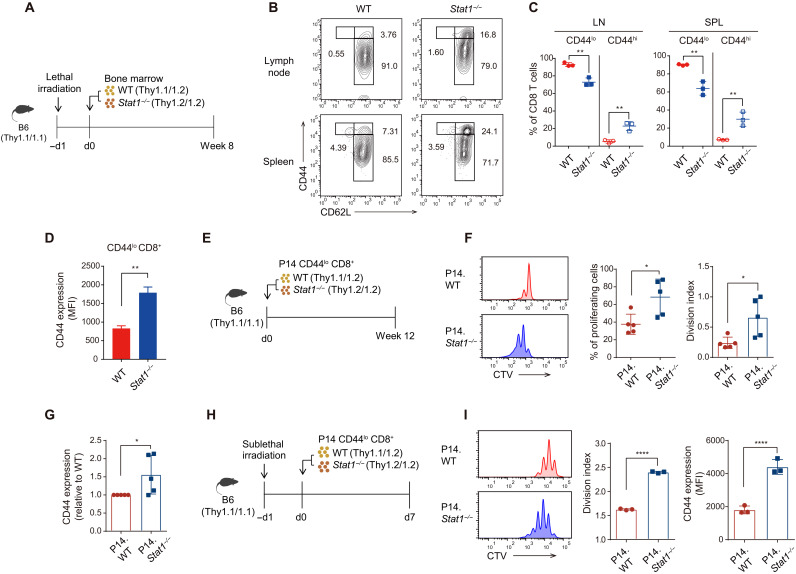
The enhanced proliferation of Stat1−/− CD8+ T cells might result from antigenic stimulation. However, this possibility was excluded by the results obtained from Stat1−/− mice with a P14 T cell receptor (TCR) transgenic background (P14.Stat1−/−), which also showed increased CD44 and Ki67 expression (fig. S1F), indicating an antigen-independent alteration. To further investigate whether the perturbed CD8+ T cell homeostasis observed in Stat1−/− mice was due to a cell-intrinsic autonomous effect, bone marrow (BM) chimeric mice were generated by injecting equal numbers of WT and Stat1−/− BM cells into lethally irradiated C57BL/6 (B6) mice (Fig. 2A). Eight weeks after BM reconstitution, Stat1−/− BM-derived CD8+ T cells showed a significant enhancement of CD44hi cells and a reduction of CD44lo cells in terms of both percentage and number (Fig. 2, B and C). In addition, an augmented CD44 expression was observed in CD44lo naïve cells on a per-cell basis, but this enhancement was not observed in their WT BM-derived counterparts (Fig. 2D). This phenomenon was apparent only in the periphery but not in the thymus, as demonstrated by similar levels of CD44 expression in WT and Stat1−/− BM-derived CD4−CD8+ single-positive thymocytes (fig. S1G). Therefore, these data suggest that alterations in CD8+ T cell homeostasis occur post-thymically through a cell-intrinsic effect of STAT1 deficiency.
Fig. 2. Stat1−/− naïve CD8+ T cells show enhanced proliferation in a cell-intrinsic manner.
(A) Experimental scheme of generating BM chimera for (B) to (D). (B) Flow cytometry for CD44 and CD62L expression in CD8+ T cells derived from WT and Stat1−/− BM. (C) Percentage of CD44lo and CD44hi CD8+ T cells derived from WT and Stat1−/− BM. (D) CD44 expression levels in WT and Stat1−/− BM-derived CD44lo CD8+ T cells. MFI, mean fluorescence intensity. (E) Experimental scheme of adoptive transfer into B6 recipients for (F) and (G). (F) In vivo proliferation of WT and Stat1−/− P14 donor cells at 12 weeks after transfer. (G) Levels of CD44 expression in WT and Stat1−/− P14 donor cells of pre- and post-transfer. (H) Experimental scheme of adoptive transfer into irradiated (500 cGy) B6 recipients for (I). (I) In vivo proliferation and CD44 level of WT and Stat1−/− P14 donor cells at 7 days after transfer. The results are presented as means ± SEM. Data are representative of three to four independent experiments. *P < 0.05, **P < 0.01, and ****P < 0.0001.
To further confirm the role of STAT1 in a normal steady-state environment, CellTrace Violet (CTV)–labeled WT and Stat1−/− P14 naïve CD8+ T cells were cotransferred at a 1:1 ratio into B6 hosts, and their proliferation and CD44 expression were analyzed 12 weeks later (Fig. 2E). Stat1−/− P14 donor cells underwent approximately two- to three-fold more division and showed significantly increased CD44 expression compared with their WT P14 counterparts during this period (Fig. 2, F and G). Similar data were also obtained in an experiment performed under lymphopenia conditions, in which WT and Stat1−/− P14 naïve CD8+ T cells were cotransferred into sublethally irradiated B6 hosts to induce homeostatic proliferation (HP; Fig. 2H). Once again, Stat1−/− P14 donor cells exhibited higher HP and CD44 expression than their WT P14 counterparts (Fig. 2I). Together, these data further strengthen the notion that STAT1 plays a cell-intrinsic role in regulating peripheral CD8+ T cell homeostasis at a steady state.
Stat1−/− naïve CD8+ T cells can respond to T1IFN and IL-7 in vitro
Because lymphopenia-induced HP markedly depends on two tonic signals, IL-7 and TCR contacts with self-ligands (21–23), the aforementioned enhancement of the HP of Stat1−/− CD8+ T cells might reflect their greater responsiveness to these cues. However, in our in vitro culture experiment, Stat1−/− naïve CD8+ T cells showed equivalent (or even slightly decreased) rather than enhanced responses to IL-7 and/or TCR (anti-CD3) stimulation compared with their WT counterparts, as evidenced by the induction of proliferation and STAT5 phosphorylation by IL-7 (fig. S2, A and B), the induction of proliferation and activation of ZAP70, phospholipase Cγ (PLCγ), and extracellular signal–regulated kinase (ERK) in response to anti-CD3 (fig. S2, C and D), and the induction of proliferation by both anti-CD3 and IL-7 (fig. S2E).
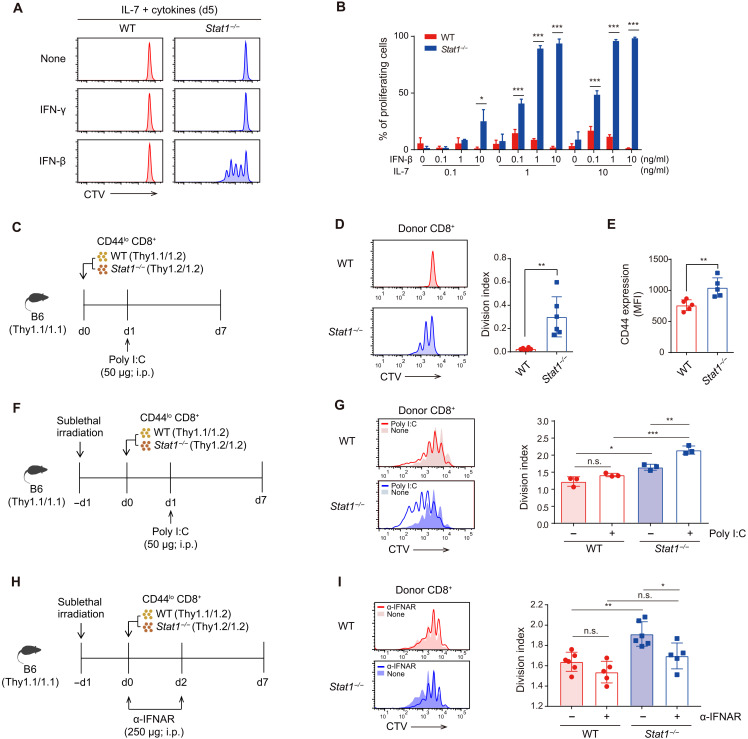
To further determine the relevant factors causing the enhancement in the in vivo HP of Stat1−/− naïve CD8+ T cells, the role of tonic self-TCR signals was considered unlikely because these cells still showed a better proliferative response than the WT cells, even in irradiated Tap1−/− hosts deprived of self-ligands (fig. S2, F and G). Therefore, we focused on examining soluble cell-extrinsic factors, such as cytokines, as a major inducer of strong HP. Of the various cytokines tested in vitro, IFN-β but not other cytokines, including IFN-γ, strongly induced Stat1−/− naïve CD8+ T cell proliferation (Fig. 3A), which was unexpected because both IFNs depend on STAT1 for mediating their biological functions (20). Moreover, the effect of IFN-β observed in Stat1−/− naïve CD8+ T cells was dependent on IL-7; thus, it was pronounced only in the culture with IL-7 but not in the cultures with other gamma-chain (γc) cytokines, such as IL-2, IL-9, IL-15, and IL-21 (Fig. 3B and fig. S2H).
Fig. 3. Stat1−/− naïve CD8+ T cells proliferate in response to T1IFN and IL-7 in vitro and in vivo.
(A) In vitro proliferation of WT and Stat1−/− naïve CD8+ T cells in culture with indicated cytokines. (B) Percentage of proliferating WT and Stat1−/− naïve CD8+ T cells in culture with IL-7 and IFN-β. (C) Experimental scheme for (D) and (E). i.p., intraperitoneally. (D) In vivo proliferation and (E) CD44 expression levels of WT and Stat1−/− CD8+ donor cells from B6 recipients. (F) Experimental scheme for (G). (G) In vivo proliferation of WT and Stat1−/− CD8+ donor cells from irradiated (500 cGy) B6 recipients. (H) Experimental scheme of in vivo T1IFN blockade for (I). (I) In vivo proliferation of WT and Stat1−/− CD8+ donor cells from untreated or αIFNAR-treated irradiated B6 recipients. The results are presented as means ± SEM. Data are representative of three to four independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001. n.s., not significant.
Stat1−/− naïve CD8+ T cells show enhanced in vivo proliferation in a T1IFN-dependent manner
The main implication from the above in vitro data is that the enhanced HP of Stat1−/− naïve CD8+ T cells observed in vivo might depend on T1IFN. To test this hypothesis, CTV-labeled WT and Stat1−/− naïve CD8+ T cells were cotransferred into B6 hosts and then injected with the Toll-like receptor 3 agonist polyinosinic:polycytidylic acid (poly I:C), which is known as a potent T1IFN inducer (Fig. 3C) (24). Following this process, WT donor cells failed to undergo cell division 7 days after transfer, whereas Stat1−/− donor cells showed an enhanced proliferative response and increased CD44 expression (Fig. 3, D and E). Similar data were also obtained from irradiated B6 hosts cotransferred with WT and Stat1−/− naïve CD8+ T cells (Fig. 3, F and G); Poly I:C treatment markedly improved the lymphopenia-induced HP of Stat1−/− but not WT donor cells.
Given the effect of poly I:C (T1IFN-rich environment), we subsequently tested opposite settings, i.e., T1IFN-deprived conditions, to determine whether the enhanced HP observed in Stat1−/− donor cells was dependent on T1IFN. To this end, the same adoptive transfer experiments shown in Fig. 3F (without the poly I:C injection) were performed with anti-IFNAR antibody (Ab) treatment to block the T1IFN receptor signal in vivo (Fig. 3H). The enhanced HP of Stat1−/− donor cells was significantly reduced by T1IFN blockade, whereas the response of WT donor cells was unchanged (Fig. 3I). Collectively, these data strongly suggest that STAT1 controls peripheral CD8+ T cell responses to tonic T1IFN by maintaining their quiescence in the presence of sufficient levels of STAT1 but inducing abnormal proliferation under STAT1-deficient conditions.
Stat1−/− naïve CD8+ T cells activate the STAT4, mTOR, and cMYC pathways in response to T1IFN and IL-7
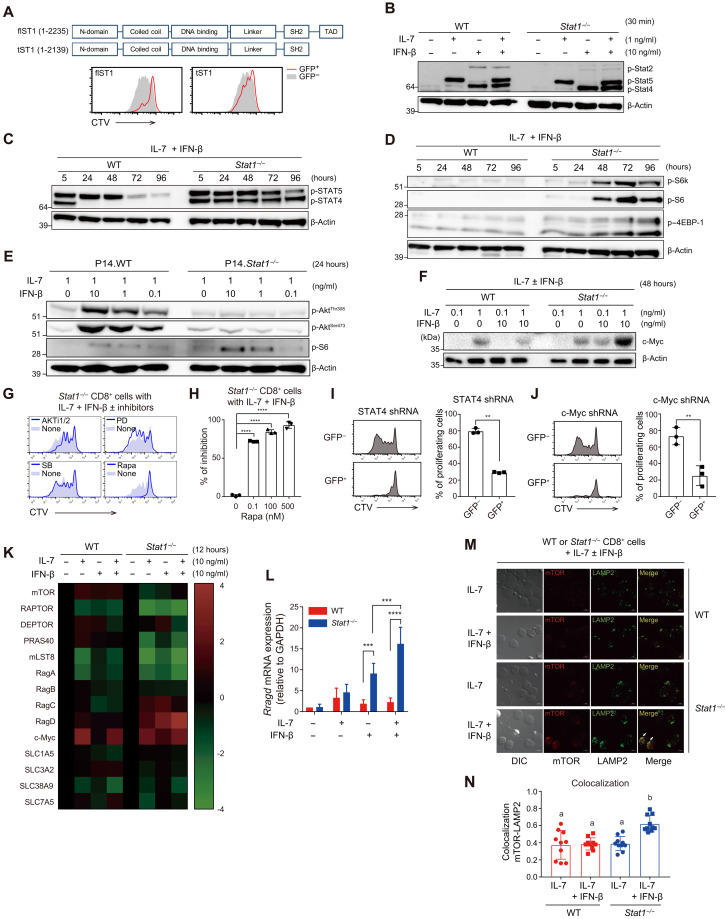
Because T1IFN induces the activation of STAT1 and its transcriptional activity to express various ISGs (19), we aimed to elucidate the precise mechanisms through which Stat1−/− cells undergo cell activation and division, which are otherwise quiescent in response to T1IFN.Therefore, we first investigated whether STAT1 and its transcriptional activity are directly involved in the maintenance of CD8+ T cell quiescence. For this purpose, Stat1−/− CD8+ T cells were transduced with retroviral vectors encoding either the full-length Stat1 gene (flST1) or a truncated Stat1 gene lacking the transcription activation domain (TAD; tST1) (Fig. 4A, top). The in vitro proliferative responses to T1IFN and IL-7 were substantially reduced in flST1-transduced, but not tST1-transduced, Stat1−/− cells (Fig. 4A, bottom). Similar results were also observed in vivo when the retrovirally transduced Stat1−/− cells were transferred into B6 hosts treated with poly I:C (fig. S2, I and J): Reduced proliferation was apparent in flST1-transduced, but not tST1-transduced, Stat1−/− donor cells. These data indicate that the transcriptional activity of STAT1 is necessary for suppressing cell activation and proliferation upon T1IFN and IL-7 exposure.
Fig. 4. Stat1−/− naïve CD8+ T cells respond to T1IFN and IL-7 via activating STAT4-RagD-mTOR and cMYC signaling pathways.
(A) In vitro proliferation of Stat1−/− CD8+ T cells transduced with retroviral vectors encoding either full-length (fl) or truncated (t) construct of Stat1 gene in response to IL-7 and IFN-β. (B) Phosphorylation of STAT2, STAT4, and STAT5 in WT and Stat1−/− naïve CD8+ T cells at 30 min after culture with IL-7 and IFN-β. (C) Phosphorylation of STAT4 and STAT5 and of (D) S6K, S6, and 4EBP-1 in WT and Stat1−/− naïve CD8+ T cells at various time points after culture with IL-7 and IFN-β. (E) Phosphorylation of AKT in WT and Stat1−/− naïve CD8+ T cells at 24 hours after culture with various concentrations of IL-7 and IFN-β. (F) Expression of c-Myc in WT and Stat1−/− naïve CD8+ T cells at 48 hours after culture with IL-7 and/or IFN-β. (G and H) In vitro proliferation of Stat1−/− CD8+ T cells by IL-7 and IFN-β in the presence or absence of various indicated inhibitors. (I and J) In vitro proliferation of Stat1−/− CD8+ T cells transduced with retroviral vectors encoding (I) STAT4 shRNA or (J) c-Myc shRNA in response to IL-7 and IFN-β. (K) Heatmap for the relative expression of gene transcripts related to mTOR signaling pathways in WT and Stat1−/− naïve CD8+ T cells at 12 hours after culture with IL-7 and IFN-β. (L) Expression levels of mRNA encoding RagD as shown in (K). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (M and N) Colocalization of mTOR and LAMP2 in WT and Stat1−/− naïve CD8+ T cells at 72 hours after culture with IL-7 and IFN-β. The results are presented as means ± SEM. Data are representative of three to four independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001 by unpaired Student’s t test. The letters “a” and “b” (P < 0.05) in (N) are calculated by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test.
To further understand how STAT1 exerts the above-described regulatory function, WT and Stat1−/− CD8+ T cells were cultured with IFN-β and/or IL-7, and the activation of various signaling proteins was then analyzed at different time points. The following three observations were made: First, Stat1−/− cells showed robust, long-term activation of STAT4 in an IFN-β–dependent manner, and its phosphorylation was detectable as early as within 30 min and stable for up to 96 hours during in vitro culture (Fig. 4, B and C, and fig. S3A). The activation of STAT5, unlike STAT4, was strongly dependent on IL-7, and the level of activation in WT cells was comparable to that in Stat1−/− cells (with the exception of poor STAT2 activation in Stat1−/− cells, as expected) (Fig. 4, B and C, and fig. S3A). Second, Stat1−/− cells showed strong activation of mTOR pathways, as evidenced by the phosphorylation of S6 kinase (S6K), S6, and eukaryotic translation initiation factor 4E-binding protein 1 (4EBP-1), and this effect was pronounced only in the culture with both IFN-β and IL-7 (Fig. 4D and fig. S3, B and C). In contrast, Stat1−/− cells treated with IFN-β and IL-7 exhibited a substantial reduction in AKT activation (Fig. 4E and fig. S3D), although it serves as a positive regulator of mTOR activation (25). P38 and ERK activation was similar or slightly increased in Stat1−/− cells compared with WT cells (fig. S3D). Third, Stat1−/− cells (relative to WT cells) showed similar levels of IL-7–induced cMYC expression, which depended on the activation of STAT5 (26), but the levels were even further promoted by coculture with IFN-β (Fig. 4F and fig. S3E), which was in line with its ability to enhance mTOR activity (27). Thus, these data suggest a role of STAT1 in negatively regulating STAT4, mTOR, and cMYC activation in response to T1IFN and IL-7.
The induction of Stat1−/− naïve CD8+ T cell proliferation by T1IFN and IL-7 is mediated through activation of the STAT4–lysosomal mTOR–cMYC signaling axis
To gain insight into the functional relevance of the aforementioned activated signaling pathways, CTV-labeled WT and Stat1−/− naïve CD8+ T cells were treated with various pharmacochemical inhibitors and cultured with IFN-β and IL-7. Notably, the mTOR inhibitor rapamycin markedly reduced the proliferative responses of Stat1−/− cells, whereas ERK, P38, and AKT inhibitors (PD98059, SB203580, and AKTi, respectively) did not have a similar effect (Fig. 4, G and H). To determine the role of enhanced STAT4 and cMYC activation, Stat1−/− cells were transduced with retroviral vectors encoding STAT4 or cMYC short hairpin RNA (shRNA). Both virally transduced Stat1−/− cells exhibited substantially reduced proliferation in response to IFN-β and IL-7 (Fig. 4, I and J).
To elucidate possible cross-talk among the STAT4, mTOR, and cMYC pathways, rapamycin-treated Stat1−/− cells exhibited decreased cMYC expression without affecting STAT4 activation (fig. S3F), and this finding suggested that STAT4 is an upstream regulator of mTOR activation, which, in turn, promotes cMYC expression. Therefore, we subsequently addressed how STAT4 activates mTOR signaling. For this purpose, the expression of a panel of genes related to mTOR activation in WT and Stat1−/− naïve CD8+ T cells cultured with IFN-β and/or IL-7 was analyzed. Of the various genes tested, the expression of the small guanosine triphosphatase (GTPase) RagD (and, to a lesser extent, RagC) was markedly enhanced in Stat1−/−, but not WT, cells after culture with IFN-β alone or with both IFN-β and IL-7 (Fig. 4, K and L). Similar data were observed for RagD protein expression (fig. S3G). IL-7 alone, unlike IFN-β, failed to induce RagD (Fig. 4, K and L, and fig. S3G), which indicated a role of T1IFN-dependent (and not IL-7–dependent) STAT4 transcriptional activity in RagD expression. In support of this finding, promoter regions of the rragd gene contained a putative binding site for STAT4 but not STAT5, as evidenced by strong STAT4 interaction with a chromatin positioned −222 kb of the rragd transcription start site (fig. S3H). Furthermore, STAT4-induced RagD was closely associated with mTOR activation (i.e., S6 phosphorylation) via a mechanism independent of AKT activation (fig. S3I).
Given that T1IFN-induced STAT4 signaling induced mTOR-dependent proliferation of Stat1−/− CD8+ T cells, we also examined whether a similar response would occur even in WT CD8+ T cells by IL-12, as the IL-12/IL-12R primarily activates STAT4 activation. In vitro culture with IL-12 (along with IL-7) led to the proliferation of CD44hi MP and, to a lesser extent, CD44lo naïve CD8+ T cells, and the proliferation was also mTOR dependent, as evidenced by no proliferative response with rapamycin treatment (fig. S3J). In line with these findings, MP cells freshly isolated from normal B6 mice showed a relatively high level of phospho-STAT4 (p-STAT4), RagD, and p-S6 compared to naïve counterparts (fig. S3K). These data suggest that STAT4 may serve as a bridge linking various cytokine receptors (e.g., IFNAR and IL-12R) with mTOR signaling pathway, presumably by inducing RagD expression.
Because RagD is a crucial component for mTOR activation, particularly on the membrane of subcellular lysosomes (28, 29), we subsequently examined whether the STAT4-RagD signaling axis converges on lysosomal mTOR activation. To this end, WT and Stat1−/− naïve CD8+ T cells were cultured with IFN-β and/or IL-7, and the colocalization of mTOR and lysosomal membrane–associated protein 2 (LAMP2) was analyzed. Stat1−/− cells showed markedly enhanced mTOR colocalization with LAMP2 in culture with both IFN-β and IL-7 (not IL-7 alone), but WT cells did not exhibit a similar colocalization (Fig. 4, M and N). To further confirm the requirements of lysosomal activity for mTOR activation, WT and Stat1−/− naïve CD8+ T cells were cultured with IFN-β and IL-7 in the presence of MG132 (N-carbobenzyloxy-l-leucyl-l-leucyl-l-leucinal), NH4Cl, and leupeptin, which are inhibitors that can block lysosomal functions (30, 31). Stat1−/− cells showed substantial reduction of mTOR activation (i.e., reduced S6 phosphorylation) after treatment with these inhibitors, and the STAT4-RagD pathways remained intact (fig. S3L). Furthermore, consistent with the reduction in mTOR activation, these inhibitors suppressed the proliferative responses of Stat1−/− cells cultured with IFN-β and IL-7 (fig. S3M). Therefore, these data strongly support the notion that STAT1 negatively regulates both the STAT4–RagD–lysosomal mTOR and cMYC signaling pathways in response to T1IFN and IL-7.
Stat1−/− naïve CD8+ T cells induce severe forms of colonic inflammation in an IBD setting
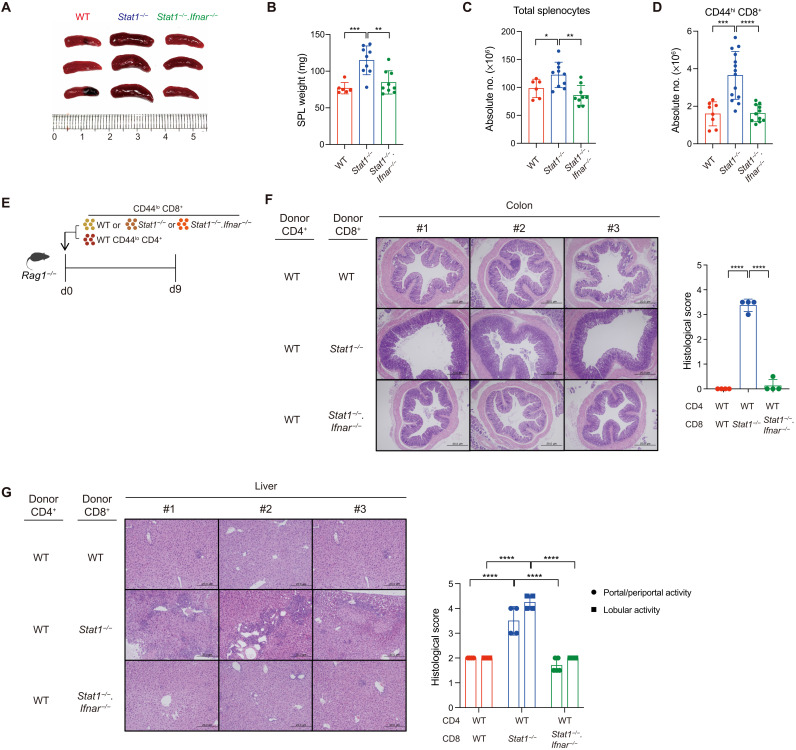
Given the uncontrolled responsiveness of Stat1−/− naïve CD8+ T cells to T1IFN and IL-7, we examined the pathophysiological impact of alterations in peripheral CD8+ T cell homeostasis in Stat1−/− mice. Consistent with the accumulation of CD44hi MP cells during aging, Stat1−/− mice aged 4 months showed clear symptoms of splenomegaly accompanied by increases in the spleen weight and total splenocyte counts, particularly that of CD44hi CD8+ T cells (Fig. 5, A to D). The numbers of B, natural killer (NK), and T regulatory (Treg) cell populations were unchanged (fig. S4A). The phenomena observed in Stat1−/− mice were completely restored in mice showing deficiency in both STAT1 and IFNAR (Stat1−/−.Ifnar−/−; Fig. 5, A to D), which supported the crucial requirement of in vivo T1IFN signaling in perturbing the quiescence of Stat1−/− naïve CD8+ T cells at a steady state.
Fig. 5. Stat1−/− naïve CD8+ T cells induce severe forms of colonic inflammation in an IBD setting.
Size (A) and weight (B) of the spleens of 20-week-old WT, Stat1−/−, and Stat1−/−.Ifnar−/− mice. (C and D) Absolute cell number of total splenocytes and splenic CD44hi CD8+ T cells from mice in (B). (E) Experimental scheme of inducing IBD for (F) and (G). (F) Representative histochemistry [left hematoxylin and eosin (H&E) staining] and histopathological score (right bar graph) of the distal colon sections of Rag1−/− recipient mice. (G) Representative histochemistry (left) and histopathological score (right) of the liver sections of Rag1−/− recipient mice. The results are presented as means ± SD. Data are representative of three to four independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Despite the enlarged spleens, the aged Stat1−/− mice did not develop any signs of severe inflammation or autoimmune diseases, presumably because of unaltered Treg cell counts (fig. S4A). To further confirm a possible pathological impact of unrestrained Stat1−/− naïve CD8+ T cells, we used a T cell transfer model to induce IBD in Rag1−/− hosts (32). To this end, Rag1−/− hosts were adoptively cotransferred with a mixture of WT naïve CD4+ T cells and either WT or Stat1−/− naïve CD8+ T cells, and 9 days later, the degree of colonic inflammation was analyzed in these hosts (Fig. 5E). At this time point, the colons from Rag1−/− hosts transferred with WT CD8+ cells were relatively normal, as demonstrated by nearly intact luminal shapes, whereas those with Stat1−/− CD8+ cells showed enhanced histological signatures of severe inflammation (Fig. 5F, top two rows). Moreover, consistent with this phenomenon, a significant increase in the number of Stat1−/− CD8+ donor cells that could produce IL-17A and IFN-γ was observed (fig. S4, B and C); a significant increase in the number of WT CD4+ donor cells was also observed only when cotransferred with Stat1−/− CD8+ cells. Note, however, that, despite the substantial increase in absolute cell number, the relative proportion of cytokine-producing donor T cells was unchanged (fig. S4D), suggesting that the severe colitis reflects a quantitative rather than a qualitative change. The aggressive colonic inflammation caused by Stat1−/− CD8+ cells was heavily dependent on T1IFN because this pathological effect was completely restored by cotransfer with Stat1−/−.Ifnar−/− naïve CD8+ T cells (Fig. 5F, bottom row). Similar pathological effects with Stat1−/−, but not Stat1−/−.Ifnar−/−, donor cells were also detectable in the liver (Fig. 5G). Collectively, these data indicate that STAT1 plays a crucial role in maintaining the homeostasis of peripheral naïve CD8+ T cells by suppressing the responsiveness to T1IFN.
DISCUSSION
STAT1 transmits a T1IFN signal by forming transcriptional complexes with STAT2 and IRF9 (interferon regulatory factor 9), which leads to various immunological functions, particularly in an inflammatory context of pathogen infection (20). In this situation, the role of STAT1 is mostly attributed to its ability to induce a panel of ISGs, and accordingly, abnormal outcomes resulting from STAT1 deficiency largely phenocopy those resulting from mice lacking the T1IFN signals (33–35). In the present study, we addressed a novel regulatory function of STAT1 per se, uncoupling its role from the abovementioned canonical T1IFN signaling pathway and highlighting a noncanonical pathway of activating the STAT4–RagD–lysosomal mTORC1 axis under steady-state conditions without any infectious episodes. As a result of STAT1 deficiency, T1IFN (along with IL-7) can transmit a stimulatory signal by recruiting and stably activating STAT4, which, in turn, triggers RagD expression and induces lysosomal mTOR activation, and these effects release naïve CD8+ T cells from the quiescent state, which results in abnormal proliferation and increased CD44hi cell generation.
A precise mechanism underlying STAT1-mediated negative regulation for the T1IFN-induced STAT4-mTOR activation remains to be further elucidated. In this respect, we demonstrated the following two observations: (i) no inhibition with TAD domain–lacking STAT1 transduced in Stat1−/− CD8+ T cells (Fig. 4A) and (ii) equally robust level of both STAT1 and STAT4 phosphorylation at early time point (0.5 to 5 hours) after IFN-β treatment in WT CD8+ T cells (Fig. 4C). Therefore, we suggest that the inhibitory role of STAT1 is primarily dependent on its transcriptional activity but not its simple physical interference for STAT4 binding to the receptor (IFNAR).
Because the T1IFN-STAT1 signaling is known to induce a large variety of genes (i.e., ISGs) that mediate various functional responses (20, 36), we assume that either single or a group of certain ISG proteins induced by STAT1 may act as a negative regulator for STAT4/mTOR pathway to primarily target the most proximal stage of IFNAR signaling, namely, Janus kinase (JAK)/STAT. In this regard, SOCS family proteins may be the most relevant candidate, as they have been shown to negatively regulate various cytokine receptor signaling, including type I and II IFN, through inhibiting enzymatic activity of JAK kinases or physical interactions with its substrates STATs (20, 37–39). Besides SOCS proteins, USP18 is also another possible candidate known as a negative feedback loop for T1IFN/IFNAR signaling by inhibiting JAK/STAT pathway (40). Hence, one possible explanation is that these regulatory proteins that are induced at a later time point by T1IFN in a STAT1-dependent manner may dampen T1IFN-induced JAK1/TYK2 activation, presumably ceasing STAT4 activation and its further downstream RagD–lysosomal mTORC1 activation. Future studies are needed to clarify this possibility.
Although the information on the physiological role of T1IFN at a steady state is currently limited, the findings obtained in our present study suggest the existence and importance of T1IFN as an atypical homeostatic cue for the maintenance of peripheral CD8+ T cell quiescence, particularly under conditions in which STAT1 is functionally intact. This notion is fully supported by the fact that the abnormal phenomena observed in Stat1−/− naïve CD8+ T cells completely return to normal by abrogating the T1IFN response in vivo. Because many cytokine receptors share STAT1 for their signaling (41–43), further determination of the possible outcomes of quiescent naïve CD8+ T cell responses to other homeostatic cues in the absence of STAT1 would be beneficial to generalizing its regulatory role. In addition to the STAT4-associated phenomena described in the current study, whether other STAT proteins are alternatively activated depending on the relevant cytokines remains to be elucidated.
Given the strong connection between prolonged T1IFN/IFNAR-induced STAT4 activation and lysosomal mTORC1 activation only in Stat1−/− CD8+ T cells, it is tempting to speculate that the similar phenomena may also occur in WT CD8+ T cells by IL-12/IL-12R engagement, as this cytokine induces STAT4, but not STAT1, activation. A study by Rao et al. (44) demonstrated that IL-12–induced STAT4 is crucial for the proliferation of effector CD8+ T cells in an mTOR-dependent manner. Likewise, we found that prolonged in vitro exposure of IL-12 (along with IL-7) can induce significant proliferation of both memory phenotype and, to a lesser extent, naïve CD8+ T cells, and notably, the proliferation was completely inhibited by adding rapamycin, implying an mTOR-dependent response. In line with these findings, we also observed a relatively high level of p-STAT4, RagD, and p-S6 in freshly isolated memory phenotype CD8+ T cells compared to naïve counterparts. Therefore, it is reasonable to assume that STAT4 has a key role in triggering mTOR pathway in general (45), although the degree of such mTOR activation may vary depending on the type of cytokine receptors involved (i.e., IFNAR or IL-12R).
Given the many reported Stat1 gene mutations in humans, whether the phenomenon observed in Stat1−/− mice is similarly reproduced in clinical patients lacking functional STAT1 might be an interesting topic to investigate. The fact that some clinical cases of inflammatory diseases have been associated with Stat1 gene polymorphism (46–48) supports this possibility, although the compounding effect of defective STAT1 cannot be easily excluded because of increased susceptibility to pathogen infection. For instance, one recent study by Zhang et al. (49) demonstrated that some patients with STAT1 gain-of-function mutation exhibit lymphopenia with a smaller number of T cells in the blood, whereas one-fourth of patients with STAT1 loss-of-function mutation show lymphadenopathy, which are in close agreement with our findings in Stat1−/− mice (Fig. 5A). As for Stat1 gene mutation, Stat4 gene mutation has also been reported in human with several autoimmune diseases such as systemic lupus erythematosus (50). These studies suggest that the uncontrolled function of STAT1 and STAT4 in T cells may cause autoimmune diseases, and therefore, the adequate balance between STAT1 and STAT4 activation is perhaps broadly relevant in both mice and humans to maintain normal T cell homeostasis.
In summary, our results further extend our understanding that STAT1 serves as an important gatekeeper to maintain the quiescence of peripheral naïve CD8+ T cells by suppressing their responsiveness to the atypical homeostatic cue T1IFN. The mechanisms underlying the IFNAR–STAT4–RagD–lysosomal mTOR axis elucidated in the present study might aid the development of optimal therapeutic strategies for modulating CD8+ T cell immunity.
MATERIALS AND METHODS
Mice
C57BL/6 (B6), B6.SJL (Ly5.1), or B6.PL (Thy1.1) mice were purchased from The Jackson Laboratory. Stat1−/−, Ifnar−/−, Ifnar.Ifngr−/−, Tap1−/−, and Rag1−/− mice (all on a B6 background) were obtained from Pohang University of Science and Technology (POSTECH). P14 TCR Tg (Thy1.1) mice were obtained from POSTECH and crossed with Stat1−/− to generate P14.Stat1−/− mice. Stat1−/− mice were crossed with Ifnar−/− mice to generate Stat1−/−.Ifnar−/− double knockout mice. Mice were maintained under specific pathogen–free conditions. Unless described otherwise, all mice were used sex-matched at 8 to 12 weeks of age, according to protocols approved by the Institutional Animal Care and Use Committee at POSTECH and Chonnam National University (CNU).
Reagents
Recombinant murine IL-2, IL-7, IL-9, IL-15, IL-21, and IFN-γ were purchased from PeproTech. Mouse IFN-β was purchased from PBL Biomedical Laboratories. PD98059, SB203580, AKTi, MG132, leupeptin, NH4Cl, and rapamycin inhibitors were all purchased from Sigma-Aldrich.
Abs and flow cytometry
Cell suspensions were prepared and stained for fluorescence-activated cell sorting (FACS) analysis of cell surface markers using phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) and 0.05% sodium azide with the following fluorochrome-conjugated Abs to (purchased from BioLegend, eBioscience, and BD Biosciences unless otherwise described): anti-CD4 (RM4-5), anti-CD8α (53-6.7), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-CD24 (1M/69), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD90 (5E10), anti-CD90.1 (HIS51), anti-CD90.2 (53-2.1), and anti-CD5 (53-7.3). Flow cytometry samples were run using LSR II or FACSCanto II (BD Biosciences) and analyzed with FlowJo software (Tree Star).
BrdU incorporation assay
To analyze proliferation ability, indicated mice were intraperitoneally injected with BrdU (1 mg per mouse; Sigma-Aldrich) once and then treated with BrdU (0.8 mg/ml) in their drinking water for 7 days before sacrifice. BrdU staining was performed with BrdU Staining Kit for Flow Cytometry FITC (eBioscience) according to the manufacturer’s protocol. Briefly, splenocytes were stained with surface markers and then fixed and treated with deoxyribonuclease I. Cells were stained with fluorochrome-conjugated anti-BrdU and analyzed with flow cytometry. For flow cytometry analysis of intracellular Ki67, splenocytes were stained for cell surface markers, fixed and permeabilized using Foxp3/Transcription Factor Staining Buffer Set (eBioscience), and then stained with fluorochrome-conjugated anti-Ki67 (SolA15, BD Biosciences).
T cell purification and in vitro proliferation assay
Single-cell suspensions from pooled lymph nodes or spleen were stained with fluorochrome-conjugated Abs to CD8α, CD5, or CD44 and then sorted to obtain CD44lo naïve CD8+ T cells using MoFlo Astrios or MoFlo XDP (Beckman Coulter). Purity was routinely tested after cell sorting and was >99%. For in vitro proliferation assay, FACS-sorted CD44lo CD8+ T cells were labeled with CTV (Thermo Fisher Scientific), plated to 96-well cell culture plates, cultured with various indicated stimuli, and analyzed for CTV dilution by flow cytometry.
Generation of mixed BM chimera
BM cells were obtained from femur bones of WT congenic mice (Thy1.1/1.2) and Stat1−/− (Thy1.2/1.2) mice. T and B cells were removed by MACS (magnetic-activated cell sorting) magnetic purification using anti-CD3 or anti-B220 Abs (eBioscience) conjugated with biotin. A mixture of WT and Stat1−/− (at a 1:1 ratio) was transferred intravenously into lethally irradiated (960 cGy) WT congenic mice (Thy1.1/1.1). After 8 weeks, the mixed BM chimera mice were sacrificed for analysis.
In vivo T cell proliferation
For normal homeostatic turnover or lymphopenia-induced HP, FACS-sorted naïve CD8+ T cells from P14.Stat1+/+ (Thy1.1/1.2) or P14.Stat1−/− (Thy1.2/1.2) mice were labeled with CTV and transferred intravenously to either non-irradiated or sublethally irradiated (500 cGy) B6 (Thy1.1/1.1) mice. Recipient mice were sacrificed at the indicated time points, and donor cells were analyzed by flow cytometry. Division index was calculated according to the FlowJo protocols.
In vitro T cell culture with inhibitors
For analyzing effects of mitogen-activated protein kinase (MAPK) and mTOR pathway on proliferation, FACS-sorted naïve CD8+ T cells from Stat1−/− mice were preincubated for 30 min with the indicated inhibitors PD98059 (1 μM), SB203580 (1 μM), AKTi (1 μM), and rapamycin (0.1 to 500 nM, as indicated) and cultured for 2 days with IL-7 and IFN-β (10 ng/ml), followed by washing and culturing for additional 2 days. For analyzing a role of lysosomal activity, the aforementioned experiment was performed in the presence of inhibitors MG132 (1 μM), NH4Cl (4 mM), and leupeptin (10 μM).
Western blot
Purified naïve CD8+ T cells cultured under the conditions indicated were harvested, washed with ice-cold PBS, and lysed on ice for 15 to 30 min in a lysis buffer [20 mM tris (pH7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, aprotinin (1 μg/ml), and leupeptin]. Cell lysates were resolved by 4 to 12% bis-tris SDS–polyacrylamide gel electrophoresis gel (Invitrogen), transferred onto nitrocellulose membrane (Invitrogen), blocked with 5% dry nonfat milk in tris-buffered saline (pH 7.4) containing 0.1% Tween 20, and probed with the following Abs to (Abs were used at 1:1000 and purchased from Cell Signaling Technology unless otherwise described): p-ERK1/2 (Thr202/Tyr204; D13.14.4E), p-PLCγ (Tyr783; rabbit polyclonal), p-ZAP70 (Tyr319; 65E4), p-AKT (Thr308; D25E6), p-AKT (Ser473; D9E), p-p38 (Thr180/Tyr182; D3F9), p-S6K (Thr389; rabbit polyclonal), p-S6 (Ser235/236; D57.2.2E), p-4EBP-1 (Thr37/46; 236B4), c-Myc (E5Q6W), RagD (polyclonal; Novus Biotechnology), p-STAT2 (Tyr690; D3P2P), p-STAT3 (Tyr705; D3A7), p-STAT4 (Try693; D2E4), p-STAT5A/B (Tyr694/699; A11W; Millipore), and β-actin (AC-15, AC-74; Sigma-Aldrich). Immunoreactivity was detected by enhanced chemiluminescence detection system according to the manufacturer’s instructions (GE Healthcare).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was done according to the manufacturer’s instructions. Briefly, naïve CD8+ T cells were purified from Stat1+/+ or Stat1−/− mice and cultured with IL-7 (1 ng/ml) and IFN-β (10 ng/ml) for 2 days. Cells were fixed with 37% of formaldehyde, resuspended in the supplied ChIP buffer, and lysed by sonication. Abs for STAT4 (C46B10; Cell Signaling Technology) and STAT5 (D2O6Y; Cell Signaling Technology) were added to the ChIP sample, followed by incubation of the samples overnight at 4°C with rotation. ChIP-grade protein G magnetic beads (Cell Signaling Technology) were added to the samples, and the tubes were placed in the Magnetic Separation Rack (Invitrogen) to isolate the Ab-bound chromatins. Chromatins were eluted from the Ab/magnetic beads by elution buffer at 65°C with vortexing and purified in the spin column. Real-time polymerase chain reaction (PCR) was done with purified DNA and primers for the predicted RagD promoter or enhancer sites, which can be bound by STAT TFs by JASPAR2020 (51): rragd -864 primer: 5′-GAAAGCAGCCACCCAGTTAG-3′ (forward) and 5′-TAGGGGTCCTCGGAAAAATC-3′ (reverse); rragd -337 primer: 5′-CCGAGGGCGTTATACACTTT-3′ (forward) and 5′-GACATGTTTCCGGAGAAGTCA-3′ (reverse); and rragd -222 primer: 5′-TCACGAGACATGTGACTTCTCC-3′ (forward) and 5′-AGCAGCTGCCTCCTAAAGTG-3′ (reverse).
Confocal staining
Purified naïve CD8+ T cells from Stat1+/+ or Stat1−/− mice were cultured with IL-7 (1 ng/ml) and IFN-β (10 ng/ml) for 2 to 3 days. After the culture, 2 × 105 cells were washed twice with ice-cold PBS and placed on a poly-l-lysine–coated glass slide (Sigma-Aldrich) for 10 min and allowed to adhere to the slide for 5 min at room temperature. The cells were fixed for 20 min with cold 4% paraformaldehyde in PBS, permeabilized for 5 min with 0.1% Triton X-100 in PBS, and then blocked for 15 min with 5% normal goat serum in PBS containing 1% bovine serum albumin. Cells were stained with rabbit anti-mouse mTOR (7C10; Cell Signaling Technology), rabbit anti-mouse LC3B (E7X4S; Cell Signaling Technology), rat anti-mouse LAMP2 (GL2A7; Abcam) for 45 min, washed twice, blocked, and then reincubated with anti-rabbit Alexa or anti-rat Alexa for 30 min. The final slides were washed with PBS and mounted in ProLong Gold Antifade Reagent (Invitrogen) and analyzed using a Zeiss LSM 700 laser scanning confocal microscope (Carl Zeiss) for acquiring fluorescence images.
Real-time reverse transcription PCR
Purified naïve CD8+ T cells from Stat1+/+ or Stat1−/− mice were cultured with IL-7 and IFN-β for indicated time points. RNA was isolated with NucleoZOL (Macherey-Nagel), and complementary DNA (cDNA) was synthesized with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo dT (Bioneer). Real-time reverse transcription PCR was performed with the SYBR Green PCR Master Mix using the StepOnePlus Real-Time PCR System (Applied Biosystems). The following primers (Bioneer) were used: Mtor: 5′-CCAGCGCTATGATGTGCTTA-3′ (forward) and 5′-ATGGGTCCTGTCTCAACTGG-3′ (reverse); Raptor: 5′-GGTACAAGCAGAGCCTCGAC-3′ (forward) and 5′-GACCCAGACCTCTCCATTCA-3′ (reverse); Deptor: 5′-GATTGTTGGTGACGCAGTTG-3′ (forward) and 5′-GCCATTGACAGAGACGACAA-3′ (reverse); Pras40: 5′-TGCTCCTAGTCCACCACCTC-3′ (forward) and 5′-GTGGCATCCTCATCCATCAT-3′ (reverse); Mlst8: 5′-CCAACCAGGCAGAACTCATT-3′ (forward) and 5′-TAGCATCTGGGTCGATGTGA-3′ (reverse); Rraga: 5′-AATCGTGTCTGGAAGCCATC-3′ (forward) and 5′-GACAAACGCCTCAGGTCTTC-3′ (reverse); Rragb: 5′-ACCTGGTTTTGAACCTGTGG-3′ (forward) and 5′-AGTTCACGGCTCTCCACATC-3′ (reverse); Rragc: 5′-GGGAAGAGAGCTTTGAACGA-3′ (forward) and 5′-GGCTTTCAGACTTGGAGCAC-3′ (reverse); Rragd: 5′-AGCTCCTTCGTCAACTTCCA-3′ (forward) and 5′-GCCAGCGCTTCCATATAGTC-3′ (reverse); Myc: 5′-AGTGCTGCATGAGGAGACAC-3′ (forward) and 5′-CTCGGGATGAAGATGAGCCC-3′ (reverse); Slc1a5: 5′-GGTCCAGCTTCTCTGTGAGG-3′ (forward) and 5′-GGTGGCATCATTGAAGGAGT-3′ (reverse); Slc3a2: 5′-GAGGACAGGCTTTTGATTGC-3′ (forward) and 5′-ATTCAGTACGCTCCCCAGTG-3′ (reverse); Slc38a9: 5′-CCATTGGGCTCTGCCTATAA-3′ (forward) and 5′-CTGTTTTATGCCCCAAGGAA-3′ (reverse); and Slc7a5: 5′-CTGGCCATCATCATCTCCTT-3′ (forward) and 5′-CAGGACATGACACCCAAGTG-3′ (reverse).
Retroviral vectors for overexpression and short hairpin (shRNA) knockdown
For overexpression, full-length Stat1 and truncated Stat1 (1-2139) cDNA from mouse splenocyte were cloned into the Bgl II and Xho I sites of the MIGR1 plasmid (Addgene) with primers for cloning: 5′-GGGCCCAGATCTATGTCACAGTGGTTCGAG-3′ (forward) and 5′-GGGCCCCTCGAGTTATACTGTGCTCATCCT-3′ (reverse) (for flST1) and 5′-GGGCCCCTCGAGTTAAATCAACTCAGTCTT-3′ (reverse) (for tST1). Retroviral shRNA LMP vector was purchased from Open Biosystems. The sequences of the shRNAs targeting Myc are “TGCTGTTGACAGTGAGCGCCTGGTGCATAAACTGACCTAATAGTGAAGCCACAGATGTATTAGGTCAGTTTATGCACCAGATGCCTACTGCCTCGGA” and
“TGCTGTTGACAGTGAGCGCACGACGAGAACAGTTGAAACATAGTGAAGCCACAGATGTATGTTTCAACTGTTCTCGTCGTTTGCCTACTGCCTCGGA” and targeting Stat4 are “TGCTGTTGACAGTGAGCGCTCCTGCGAGACTACAAGGTTATAGTGAAGCCACAGATGTATAACCTTGTAGTCTCGCAGGATTGCCTACTGCCTCGGA” and
“TGCTGTTGACAGTGAGCGCCACAGTTCAGTCTAACTACAATAGTGAAGCCACAGATGTATTGTAGTTAGACTGAACTGTGATGCCTACTGCCTCGGA.”
Plasmids were transfected to the Platinum E cell line with FuGENE HD (Promega), and supernatants containing viral particles were collected at 48 hours. Purified naïve CD8+ T cells were culture with plate-bound anti-CD3 (5 μg/ml) and anti-CD28 (2 μg/ml) for 20 hours and transduced with retroviral supernatants with spin infection (2500 rpm, 2 hours, 37°C) with polybrene (8 μg/ml; Merck). Cells were then washed and rested for 2 days with IL-7 (5 ng/ml), labeled with CTV, and further cultured with IL-7 (1 ng/ml) and IFN-β (10 ng/ml) for additional 4 days for flow cytometry.
Induction of experimental colitis
FACS-sorted naïve (CD44lo CD62Lhi) CD8+ T cells from Stat1+/+, Stat1−/−, or Stat1−/−. Ifnar−/− mice (5 × 105 cells per mouse) were cotransferred intravenously with B6 naïve (CD44lo CD62Lhi CD25−) CD4+ T cells (1 × 105 cells per mouse) into Rag1−/− recipient mice. In this experiment, to avoid NK cell–mediated cytotoxicity to Stat1−/− donor cells, Rag1−/− mice were intraperitoneally injected with NK cell depletion Ab (PK136; BioXCell) three times in 2-day intervals before adoptive transfer. At 9 days after adoptive transfer, mice were sacrificed for flow cytometry [mesenteric lymph node (mLN) and colon] and histology (colon and liver).
Histology
Colon and liver collected from mice were treated with 4% paraformaldehyde (Tech & Innovation; BPP-9004) for 24 hours and embedded into paraffin block for hematoxylin and eosin (H&E) staining. Sections (5 μm) of the colon and liver tissues were stained with H&E for histological analysis. Clinical scoring of colitis was graded on a scale of 0 to 4; average scores of two different parameters are as follows: epithelial damage score (0, none; 1, minimal loss of goblet cells; 2, extensive loss of goblet cells; 3, minimal loss of crypts and extensive loss of goblet cells; and 4, extensive loss of crypts) and infiltration score (0, none; 1, infiltrate around crypt bases; 2, infiltrate in muscularis mucosa; 3, extensive infiltrate in muscularis mucosa and edema; and 4, infiltration of submucosa). The histopathological alterations of liver were graded as described previously (52). Histological evaluation was conducted in a blinded fashion.
Intracellular cytokine staining
Freshly isolated cells from large intestine and mLN were cultured with cell stimulation cocktail plus protein transport inhibitors (Invitrogen) for 4 hours. For intracellular cytokine staining, cells were stained for cell surface markers, fixed and permeabilized using Cytofix/Cytoperm buffer (BD Biosciences), and then stained with fluorochrome-conjugated Abs to anti–IFN-γ (XMG1.2; BD Biosciences) and anti–IL-17A (ebio17B7; eBioscience) using Perm/Wash buffer (BD Biosciences).
Statistical analysis
Using Prism 7 (GraphPad), statistical differences were determined using t test or one-way analysis of variance (ANOVA) with Tukey’s test. Differences were considered significant at P < 0.05.
Acknowledgments
We thank K. S. Kim, S. H. Lim, and S. W. Lee (POSTECH) for providing various transgenic and knockout mice; J. H. Lee (CNU) for helpful discussion and comments; K. H. Lee (CNU) for scoring tissue histology data; M. J. Ryu and S. M. An (CNU) for administrative assistance; POSTECH and CNU flow cytometric core facilities for assistance with cell sorting; and G. S. Lee (POSTECH) and M. S. Kim (CNU) for mice breeding and care. Funding: This work was supported by the Cooperative Research Program for Agriculture Science and Technology Development (project no. PJ01336401 for C.-H.Y.) and supported by National Research Foundation (NRF) funded by the Korean Ministry of Science and ICT (2018R1A2B2006793 for C.-H.Y. and 2020R1A5A2031185 and 2020M3A9G3080281 for J.-H.C.). Author contributions: Y.-C.K., C.-H.Y., and J.-H.C. initiated and designed the main idea of this study. Y.-C.K. performed all major experiments. G.-W.L. performed IBD experiments. S.-W.L., Y.-J.J., and H.-O.K. performed some experiments for extended data. Y.-C.K., C.-H.Y., and J.-H.C. analyzed and interpreted data and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.Surh C. D., Sprent J., Homeostasis of naive and memory T cells. Immunity 29, 848–862 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Takada K., Jameson S. C., Naive T cell homeostasis: From awareness of space to a sense of place. Nat. Rev. Immunol. 9, 823–832 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Hamilton S. E., Jameson S. C., CD8 T cell quiescence revisited. Trends Immunol. 33, 224–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng X., Wang H., Takata H., Day T. J., Willen J., Hu H., Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 12, 544–550 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinreich M. A., Takada K., Skon C., Reiner S. L., Jameson S. C., Hogquist K. A., KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity 31, 122–130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada T., Park C. S., Mamonkin M., Lacorazza H. D., Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nat. Immunol. 10, 618–626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerdiles Y. M., Beisner D. R., Tinoco R., Dejean A. S., Castrillon D. H., DePinho R. A., Hedrick S. M., Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10, 176–184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang W., Beckett O., Flavell R. A., Li M. O., An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzachanis D., Freeman G. J., Hirano N., van Puijenbroek A. A. F. L., Delfs M. W., Berezovskaya A., Nadler L. M., Boussiotis V. A., Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2, 1174–1182 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Egawa T., Tillman R. E., Naoe Y., Taniuchi I., Littman D. R., The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204, 1945–1957 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu F. C., Shapiro M. J., Dash B., Chen C.-C., Constans M. M., Chung J. Y., Arocha S. R. R., Belmonte P. J., Chen M. W., McWilliams D. C., Shapiro V. S., An essential role for the transcription factor Runx1 in T cell maturation. Sci. Rep. 6, 23533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger M., Krebs P., Crozat K., Li X., Croker B. A., Siggs O. M., Popkin D., du X., Lawson B. R., Theofilopoulos A. N., Xia Y., Khovananth K., Moresco E. M. Y., Satoh T., Takeuchi O., Akira S., Beutler B., An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat. Immunol. 11, 335–343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K., Neale G., Green D. R., He W., Chi H., The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat. Immunol. 12, 888–897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamimura D., Katsunuma K., Arima Y., Atsumi T., Jiang J. J., Bando H., Meng J., Sabharwal L., Stofkova A., Nishikawa N., Suzuki H., Ogura H., Ueda N., Tsuruoka M., Harada M., Kobayashi J., Hasegawa T., Yoshida H., Koseki H., Miura I., Wakana S., Nishida K., Kitamura H., Fukada T., Hirano T., Murakami M., KDEL receptor 1 regulates T-cell homeostasis via PP1 that is a key phosphatase for ISR. Nat. Commun. 6, 7474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang M., Jin W., Chang J. H., Xiao Y., Brittain G. C., Yu J., Zhou X., Wang Y. H., Cheng X., Li P., Rabinovich B. A., Hwu P., Sun S. C., The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat. Immunol. 12, 1002–1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta P., Webb L. M., Avdo I., Pascall J., Butcher G. W., Survival of mature T cells in the periphery is intrinsically dependent on GIMAP1 in mice. Eur. J. Immunol. 47, 84–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiede F., La Gruta N. L., Tiganis T., PTPN2 attenuates T-cell lymphopenia-induced proliferation. Nat. Commun. 5, 3073 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Salmond R. J., Brownlie R. J., Morrison V. L., Zamoyska R., The tyrosine phosphatase PTPN22 discriminates weak self peptides from strong agonist TCR signals. Nat. Immunol. 15, 875–883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivashkiv L. B., Donlin L. T., Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platanias L. C., Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. Y., Pobezinsky L. A., Guinter T. I., Thomas J., Adams A., Park J. H., Tai X., Singer A., IL-7 signaling must be intermittent, not continuous, during CD8+ T cell homeostasis to promote cell survival instead of cell death. Nat. Immunol. 14, 143–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan J. T., Dudl E., LeRoy E., Murray R., Sprent J., Weinberg K. I., Surh C. D., IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc. Natl. Acad. Sci. U.S.A. 98, 8732–8737 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J. T., Ernst B., Kieper W. C., LeRoy E., Sprent J., Surh C. D., Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. J. Exp. Med. 195, 1523–1532 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urban S. L., Berg L. J., Welsh R. M., Type 1 interferon licenses naïve CD8 T cells to mediate anti-viral cytotoxicity. Virology 493, 52–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laplante M., Sabatini D. M., mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinz S., Unser S., Rascle A., Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer. BMC Mol. Biol. 17, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R., Dillon C. P., Shi L. Z., Milasta S., Carter R., Finkelstein D., McCormick L., Fitzgerald P., Chi H., Munger J., Green D. R., The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M., Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M., The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C., Wang X., Li X., Qiu K., Jiao F., Liu Y., Kong Q., Liu Y., Wu Y., Proteasome inhibition activates autophagy-lysosome pathway associated with TFEB dephosphorylation and nuclear translocation. Front. Cell Dev. Biol. 7, 170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard V. M., Valdor R., Patel B., Singh R., Cuervo A. M., Macian F., Macroautophagy regulates energy metabolism during effector T cell activation. J. Immunol. 185, 7349–7357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostanin D. V., Bao J., Koboziev I., Gray L., Robinson-Jackson S. A., Kosloski-Davidson M., Price V. H., Grisham M. B., T cell transfer model of chronic colitis: Concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G135–G146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xing Y., Wang X., Jameson S. C., Hogquist K. A., Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 17, 565–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crouse J., Bedenikovic G., Wiesel M., Ibberson M., Xenarios I., von Laer D., Kalinke U., Vivier E., Jonjic S., Oxenius A., Type I interferons protect T cells against NK cell attack mediated by the activating receptor NCR1. Immunity 40, 961–973 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Jennings R. N., Grayson J. M., Barton E. S., Type I interferon signaling enhances CD8+ T cell effector function and differentiation during murine gammaherpesvirus 68 infection. J. Virol. 88, 14040–14049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David M., Signal transduction by type I interferons. Biotechniques 2002, 58–65 (2002). [PubMed] [Google Scholar]
- 37.Piganis R. A., De Weerd N. A., Gould J. A., Schindler C. W., Mansell A., Nicholson S. E., Hertzog P. J., Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon alpha receptor (IFNAR1)-associated tyrosine kinase Tyk2. J. Biol. Chem. 286, 33811–33818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hertzog P. J., Williams B. R., Fine tuning type I interferon responses. Cytokine Growth Factor Rev. 24, 217–225 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Chmiest D., Sharma N., Zanin N., Viaris de Lesegno C., Shafaq-Zadah M., Sibut V., Dingli F., Hupé P., Wilmes S., Piehler J., Loew D., Johannes L., Schreiber G., Lamaze C., Spatiotemporal control of interferon-induced JAK/STAT signalling and gene transcription by the retromer complex. Nat. Commun. 7, 13476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honke N., Shaabani N., Zhang D. E., Hardt C., Lang K. S., Multiple functions of USP18. Cell Death Dis. 7, e2444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Saout C., Luckey M. A., Villarino A. V., Smith M., Hasley R. B., Myers T. G., Imamichi H., Park J.-H., O’Shea J. J., Lane H. C., Catalfamo M., IL-7–dependent STAT1 activation limits homeostatic CD4+ T cell expansion. JCI Insight 2, e96228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas S., Ghilardi N., Li J., de Sauvage F. J., IL-27 regulates IL-12 responsiveness of naive CD4+ T cells through Stat1-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. U.S.A. 100, 15047–15052 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters A., Fowler K. D., Chalmin F., Merkler D., Kuchroo V. K., Pot C., IL-27 induces Th17 differentiation in the absence of STAT1 signaling. J. Immunol. 195, 4144–4153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao R. R., Li Q., Odunsi K., Shrikant P. A., The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity 32, 67–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saleiro D., Platanias L. C., Intersection of mTOR and STAT signaling in immunity. Trends Immunol. 36, 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chapgier A., Kong X. F., Boisson-Dupuis S., Jouanguy E., Averbuch D., Feinberg J., Zhang S. Y., Bustamante J., Vogt G., Lejeune J., Mayola E., de Beaucoudrey L., Abel L., Engelhard D., Casanova J. L., A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 119, 1502–1514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Brown Z. K., Van Nostrand E. L., Higgins J. P., Kim S. K., The inflammatory transcription factors NFκB, STAT1 and STAT3 drive age-associated transcriptional changes in the human kidney. PLOS Genet. 11, e1005734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saraiva A. M., de Fátima Correia Silva J., Alves e Silva M. R. M., da Costa J. E., Gollob K. J., Moreira P. R., Dutra W. O., Transcription factor STAT1 gene polymorphism is associated with the development of severe forms of periodontal disease. Inflamm. Res. 62, 551–554 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Zhang W., Chen X., Gao G., Xing S., Zhou L., Tang X., Zhao X., An Y., Clinical relevance of gain- and loss-of-function germline mutations in STAT1: A systematic review. Front. Immunol. 12, 654406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagberg N., Rönnblom L., Interferon-α enhances the IL-12-induced STAT4 activation selectively in carriers of the STAT4 SLE risk allele rs7574865[T]. Ann. Rheum. Dis. 78, 429–431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fornes O., Castro-Mondragon J. A., Khan A., van der Lee R., Zhang X., Richmond P. A., Modi B. P., Correard S., Gheorghe M., Baranašić D., Santana-Garcia W., Tan G., Chèneby J., Ballester B., Parcy F., Sandelin A., Lenhard B., Wasserman W. W., Mathelier A., JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodman Z. D., Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J. Hepatol. 47, 598–607 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4