Abstract
DNA supercoiling controls a variety of cellular processes, including transcription, recombination, chromosome replication, and segregation, across all domains of life. As a physical property, DNA supercoiling alters the double helix structure by under‐ or over‐winding it. Intriguingly, the evolution of DNA supercoiling reveals both similarities and differences in its properties and regulation across the three domains of life. Whereas all organisms exhibit local, constrained DNA supercoiling, only bacteria and archaea exhibit unconstrained global supercoiling. DNA supercoiling emerges naturally from certain cellular processes and can also be changed by enzymes called topoisomerases. While structurally and mechanistically distinct, topoisomerases that dissipate excessive supercoils exist in all domains of life. By contrast, topoisomerases that introduce positive or negative supercoils exist only in bacteria and archaea. The abundance of topoisomerases is also transcriptionally and post‐transcriptionally regulated in domain‐specific ways. Nucleoid‐associated proteins, metabolites, and physicochemical factors influence DNA supercoiling by acting on the DNA itself or by impacting the activity of topoisomerases. Overall, the unique strategies that organisms have evolved to regulate DNA supercoiling hold significant therapeutic potential, such as bactericidal agents that target bacteria‐specific processes or anticancer drugs that hinder abnormal DNA replication by acting on eukaryotic topoisomerases specialized in this process. The investigation of DNA supercoiling therefore reveals general principles, conserved mechanisms, and kingdom‐specific variations relevant to a wide range of biological questions.
Keywords: DNA gyrase, DNA replication, histones, nucleoid‐associated proteins, topoisomerases, transcription
1. INTRODUCTION
Deoxyribonucleic acid (DNA) is the genetic material of living cells. Its linear succession of nucleotides defines an organism by specifying the products that are made and the circumstances in which they are made. However, some genetic information is encoded outside of the DNA sequence, in the structure of the DNA itself, in what is called DNA supercoiling. In all domains of life, DNA supercoiling plays a critical role in key cellular processes such as transcription, DNA replication and repair, and recombination. This article examines the regulation of DNA supercoiling across the domains of life.
2. DNA SUPERCOILING IS A PHYSICAL PROPERTY OF DNA THAT EMERGES FROM THE NATURE OF THE DOUBLE HELIX
The DNA molecule is typically organized as a double helix. DNA can present in several forms (see Reference 1 for a review on the topic). This review focuses on the more common B‐form DNA. In the absence of stress, the B‐form double helix is right‐handed, with a periodicity of h 0 = 10.6 bp.2 One can define a linking number at rest Lk0 for such molecule as how often the two strands of DNA cross each other. It therefore follows that for a DNA molecule of length N:
DNA supercoiling defines the phenomenon whereby the actual linking number Lk differs from Lk0. The handedness of supercoils is positive when they are in the same direction as the double helix (right‐handed), that is, DNA is over‐wound (Lk >Lk0), and negative when DNA is under‐wound (Lk <Lk0). Supercoils can take different physical forms, most commonly twist (when the two strands cross each other) and writhe (when the double helix crosses itself; Figure 1a). These two forms are spontaneously interconvertible.3 A common measurement for DNA supercoiling is the supercoiling density σ, defined as follows:
For example, an exponentially growing Escherichia coli cell will have a supercoil density of −0.06, meaning that for every 100 turns the double helix should have in the rest state, 6 are missing.
FIGURE 1.
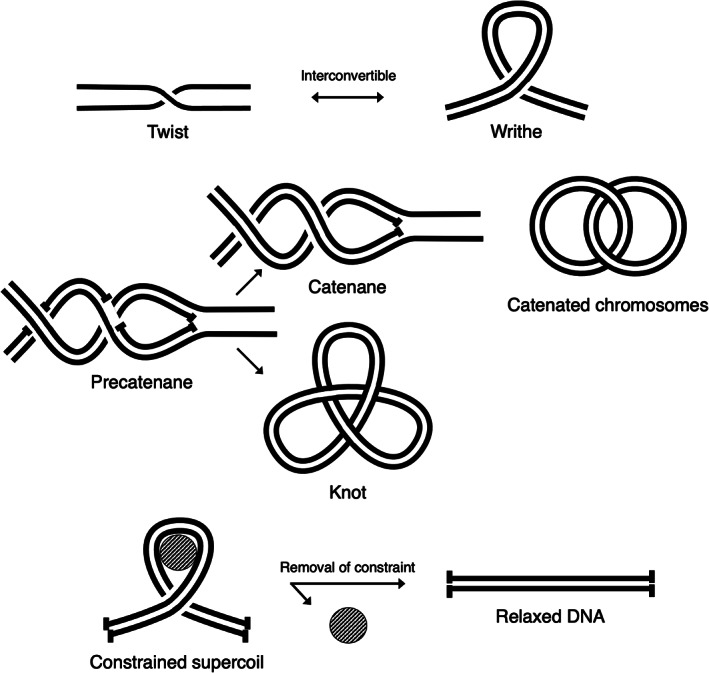
Forms of DNA supercoiling and processes that generate supercoiling. (a) DNA supercoiling is present in two interconvertible forms, namely twist (each strand crossing itself) and writhe (the two strands crossing each other). (b) Precatenanes are formed during replication and can become catenanes (intermolecular entanglements) or knots (same‐molecule entanglements). (c) Supercoils can be constrained by DNA‐binding proteins (represented as a circle) but are lost upon removal of the constraint
One key property naturally emerges from the double helix structure: DNA supercoiling is invariant as long as the two strands of DNA are intact. The corollary is that a linear, unconstrained DNA molecule cannot have DNA supercoiling: any attempt to introduce DNA supercoiling would be lost in a freely rotating end. Likewise, an unconstrained single‐ or double‐strand break dissipates all DNA supercoiling. The various kingdoms of life have evolved both divergent and shared ways to maintain DNA supercoiling, from circular chromosomes (bacteria and archaea) to proteins constraining DNA supercoiling (archaeal and eukaryotic histones and, to a lesser extent, bacterial nucleoid‐associated proteins [NAPs]) and barriers to supercoil diffusion, thereby allowing the maintenance of local, unconstrained DNA supercoiling, as detailed in the rest of this review.
3. UNIVERSAL, ESSENTIAL CELLULAR PROCESSES NATURALLY CREATE UNCONSTRAINED SUPERCOILS AS A BYPRODUCT
DNA supercoiling emerges naturally from certain cellular processes. Namely, transcription and DNA replication generate supercoils in eukaryotes and bacteria. Transcription is carried out by enzymes called RNA polymerases that melt the DNA double helix to separate the two DNA strands and form an open complex, and then transcribe RNA from the template DNA. During transcription, bacterial RNA polymerase applies torque to the DNA,4 which causes rotation of the DNA.5 However, as described in Section 4, DNA is constrained, meaning that this rotation is hindered and therefore converted to DNA supercoiling.6 Torque causes negative supercoiling behind RNA polymerase, favoring melted DNA,4 and positive supercoiling ahead of RNA polymerase without melting DNA, as positively supercoiled DNA is more resistant to melting.7 Though not directly demonstrated, torque generation by eukaryotic RNA polymerases is postulated to exist due to their similarity with the transcription elongation mechanisms carried out by bacterial RNA polymerases.8 This model of opposite supercoil generation by transcription is called “twin‐domain.”6
During transcription, it is estimated that one positive supercoil and one negative supercoil are generated for every 10 transcribed base pairs.9 Accordingly, in bacteria, a template transcribed in the presence of bacterial topoisomerase I, which only relaxes negative supercoils, becomes positively supercoiled,10 and a template transcribed in the presence of DNA gyrase, which has a strong preference toward positive supercoils, becomes negatively supercoiled.11 Both phenomena were observed in a mutant yeast deprived of endogenous topoisomerases and instead expressing bacterial topoisomerases,12 consistent with the existence of twin‐domain supercoil generation in eukaryotes. Genome‐wide transcription data in bacteria13 and eukaryotes14 are consistent with gene expression creating DNA supercoiling and impacting genes neighboring the one being transcribed (Table 1).
TABLE 1.
Distribution and properties of topoisomerases in the various kingdoms of life
| Type | Common name | Cofactors | Main role |
|---|---|---|---|
| Eukaryotes | |||
| 1B | TOP1 | Mg2+ a, polyaminesa | Remove both positive and negative supercoiling |
| IIA | TOP2b | Mg2+, ATP | Decatenation |
| 1A | TOP3b | Mg2+ | Decatenation? |
| Bacteria | |||
| IA | Topo I | Mg2+ | Remove negative supercoiling |
| IIA | Gyrase | Mg2+, ATP, polyaminesa | Introduce negative supercoiling |
| IA | Topo III | Mg2+ | Decatenation? |
| IIA | Topo IV | Mg2+, ATP | Remove positive supercoiling and decatenation |
| Archaea | |||
| IA | Topo III | Mg2+ | Decatenation? |
| IA | Reverse gyraseb | Mg2+, ATP | Introduce positive supercoiling |
| IIB | Topo VI | Mg2+, ATP | Remove both positive and negative supercoiling and decatenation. |
Significantly enhances enzymatic activity but not an absolute requirement.
Duplications exist in certain species.
DNA replication also generates DNA supercoiling. Unlike transcription, DNA replication involves full separation of the two DNA strands and use of each strand as a template to synthesize a new strand.15 In bacteria, DNA replication generates melted DNA behind the fork, that is, a strong negative DNA supercoiling.16 Because there is no DNA strand break, total supercoiling must be conserved, leading to the formation of positive supercoils ahead of the fork,17 which can be visualized in vitro by electron microscopy.18 However, positive supercoils hinder DNA replication, and a fraction are dissipated in the form of precatenanes behind the fork18 (Figure 1b). Precatenanes result from the intertwining of the two replicated DNA strand pairs and are made possible by the single‐strand breaks originating from Okazaki fragments.17 If not dissipated, precatenanes become catenanes (two chromosomes linked together and incapable of segregation) or knots (entanglements within the same chromosome) until resolved (Figure 1b). Dissipation of positive supercoils, precatenanes, catenanes, and knots is performed by topoisomerases (discussed in Section 6).
Because the fundamental mechanisms of DNA replication are the same in bacteria and eukaryotes, positive supercoils ahead of the replication fork and precatenanes behind the fork are also formed and resolved in the latter.19 Generation of supercoils by transcription and DNA replication has not been formally demonstrated in archaea. Nevertheless, their RNA polymerase is related to the eukaryotic Pol II,20 and their DNA replication resembles that of eukaryotes.21 Both processes generate supercoils as described above. It is therefore probable that archaea likewise generate supercoils during DNA replication and transcription.
DNA damage and repair also contribute to DNA supercoiling. Unlike transcription and replication, in which no unconstrained strand breaks occur, DNA damage and repair can cause single‐ and double‐ strand breaks, which result in DNA relaxation and are commonly believed to be the reason why DNA supercoiling changes during these processes. For example, X‐ray‐mediated DNA damage causes transient DNA relaxation in mitochondria.22 Oxidative23 and radiation24 stress, as well as double‐strand breaks caused by restriction enzymes,25 do likewise in the bacterium E. coli. These changes may be the direct effects of DNA damage, repair, and/or signaling pathways sensitive to DNA damage. Moreover, they raise the questions of whether and how repair machineries affect DNA supercoiling.
The eukaryotic minor mismatch repair complex Mlh1‐Mlh3 actively causes nicks in supercoiled DNA, resulting in relaxation.26 Many other repair mechanisms involve the degradation of damaged DNA, resulting in a transient single‐stranded DNA,27 which is expected to relax DNA. However, an in vitro system evaluating eukaryotic DNA supercoiling by whole cell extracts found an increase in DNA supercoiling in repaired DNA compared to nonrepaired damaged DNA.28 Likewise, an in vivo system evaluating the repair of nicks on the F‐plasmid DNA in bacteria found that nicked and repaired DNA molecules had a nonzero supercoiling,24 indicating the existence of a mechanism to preserve preexisting supercoils. Given that the F‐plasmid is 100 kb long, one would expect ~10 independent supercoiling domains (see Section 5), and lack of supercoil diffusion may be the reason why supercoiling is partially conserved. Because repair machineries and pathways differ among bacteria, archaea, and eukaryotes, establishing whether DNA repair machineries actively contribute to the maintenance of DNA supercoiling may open new therapeutic avenues aimed at perturbing their role in maintaining DNA supercoiling.
4. KINGDOM‐SPECIFIC DNA‐BINDING PROTEINS GENERATE CONSTRAINED SUPERCOILS
All domains of life use constrained supercoiling, which differs from all the unconstrained supercoils discussed above. Constrained supercoils are linked to the binding and wrapping of DNA around a protein and lost when the protein is removed (Figure 1c).
In eukaryotes and archaea, constrained supercoils are mediated by histones, a family of four proteins that assemble in heterooctamers (for a detailed review on histone assembly and structure, see Reference 29), typically organized as (H2A‐H2B)(H3‐H4)2(H2A‐H2B). In eukaryotes, one histone octamer normally constrains ~1.5 negative superhelical turns over a length of 146 bp.30 This number can vary based on the acetylation status of histones: a highly acetylated nucleosome constrains ~20% fewer supercoils than a nonacetylated one.31 In addition, the H3‐H4 heterotetramer is capable of constraining positive supercoiling in vitro,32 although in vivo constraining of positive supercoils rarely occurs33 because the H2A‐H2B dimers favor the constraining of negative supercoils.34
Most Archaea have histones. Though the monomeric forms of archaeal and eukaryotic histones are structurally similar, archaeal histones exist as dimers or tetramers.35 Dimers are capable of bending DNA without wrapping,36 whereas tetramers are capable of wrapping DNA like a eukaryotic octamer, capturing ~130 bp and 1.5 supercoils.37 Depending on physiochemical conditions, archaeal histones can trap both positive and negative supercoils.38
All Bacteria and some Archaea use a distinct class of molecules to constrain supercoils: NAPs. Despite being structurally unrelated to histones, NAPs are often referred to as “histone‐like” due to the similar function they perform. NAPs are defined as abundant DNA‐binding proteins capable of altering chromosomal structure. Bacterial proteins universally recognized as NAPs are HU (Heat‐Unstable nucleoid structuring protein), H‐NS (Heat‐Stable Nucleoid Structuring Protein, also referred to Histone‐Like Nucleoid Structuring Protein), FIS (Factor for Inversion Stimulation), Dps (DNA protection in starved cells), and IHF (Integration Host Factor). ~10–12 HU dimers bound in 9 bp intervals can constrain one negative supercoil.39 Similarly, ~6 H‐NS dimers bound in 50 bp intervals can constrain one negative supercoil.40 FIS is also capable of constraining negative supercoils, though less efficiently than HU or H‐NS: ~40 FIS dimers in 40 bp intervals can constrain one negative supercoil.41 Computer simulations suggest that any bacterial NAP capable of bending DNA (like Dps or IHF) can affect supercoiling.42 If confirmed experimentally, this would imply that constraining of DNA supercoiling in organisms that depend on NAPs is achieved modularly, by the cooperative action of many different proteins that also perform other functions, rather than by a single, specialized group of molecules like histones in eukaryotes. It may be hard to develop an antibacterial strategy based on the disruption of constrained supercoils because multiple NAPs would need to be simultaneously inhibited.
5. TRANSCRIPTION CREATES BARRIERS TO SUPERCOILING DIFFUSION THAT PRESERVE SUPERCOILING DESPITE DNA DAMAGE
Single‐ and double‐strand DNA breaks dissipate all supercoiling. The frequency with which DNA breaks occur suggests that it should be extremely difficult for organisms to maintain any DNA supercoiling. However, living organisms divide their DNA into supercoiling domains that are topologically insulated from each other, so that a break in one domain does not impact supercoiling of a neighboring domain. The existence of such domains was demonstrated in E. coli 25 and Caulobacter crescentus 43 and is hypothesized to exist in all bacteria. These domains likely correspond to the side loops on isolated nucleoids observed by electron microscopy.44 The size of these domains ranges from 2 to 65 kb, with an average size of ~10 kb.25, 44 These domains are dynamic and defined, in part, by regions of high transcription,43 which presumably reflects the drag imposed by the heavy transcription‐translation machinery in bacteria in which transcription and translation are generally coupled. Recent technical breakthroughs, such as the development of a fluorescent method to examine DNA supercoiling in living bacteria,45 will allow easy measurement of local DNA supercoiling and therefore lead to advances in our understanding of topological domains and their boundaries.
Eukaryotes also maintain highly structured, topologically insulated domains with a median size of 100 kb.46 The nature of the domain boundaries remains unclear. They have a weak correlation with boundaries between GC‐rich and AT‐rich regions, suggesting a contribution from the transcription factor CTCF, which binds to such boundaries in isolated domains.46 A correlation was also found between RNA polymerase occupancy and the negative supercoiling state of a domain, leading to the hypothesis that transcription also shapes domains in eukaryotes.46 This correlation may not represent causation since negative supercoiling facilitates gene transcription.
The existence of stable, unconstrained positive supercoiling in many Archaea strongly suggests the existence of barriers to supercoiling diffusion. Archaeal barriers to supercoiling diffusion have not been demonstrated yet but likely follow the same principles as in eukaryotes and bacteria, where a combination of regions of high transcription and specific insulating proteins would prevent supercoil diffusion.
6. TOPOISOMERASES COMPRISE A WIDE VARIETY OF KINGDOM‐SPECIFIC PROTEINS THAT DIRECTLY CHANGE DNA SUPERCOILING
Transcription and DNA replication are supercoiling‐sensitive processes that generate DNA supercoiling.47, 48 How, then, do cells manage the supercoils generated by these cellular processes? And how do bacteria and archaea manage their unconstrained DNA supercoiling? These functions are carried out by a class of enzymes designated topoisomerases that introduce breaks in DNA to change supercoiling. Topoisomerases are divided into two major types based on the nature of the DNA break: type I topoisomerases (Figure 2a,b) make single‐strand breaks, and type II topoisomerases (Figure 2c,d) make double‐strand breaks. Further subclasses are summarized in Table 1 and detailed in the text below.
FIGURE 2.
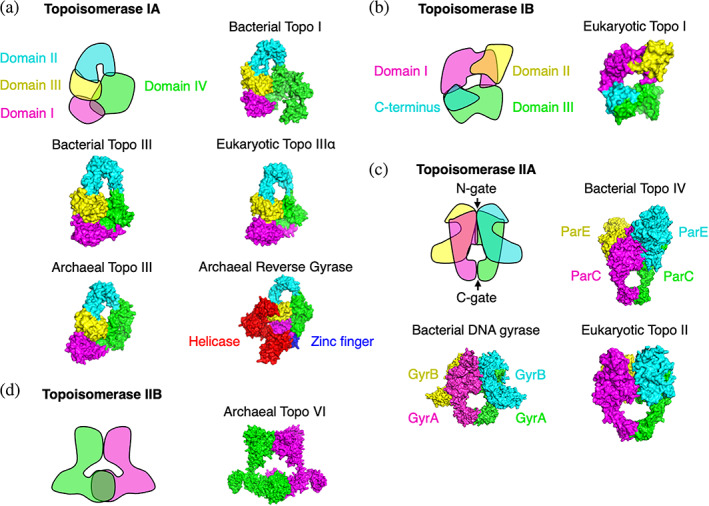
Comparison of the structures of the main topoisomerases. For each type, a general schematic is given, followed by representative experimental X‐ray structures for each kingdom. (a) Type IA topoisomerases. (b) Type IB topoisomerases. (c) Type IIA topoisomerases. Note that DNA gyrase and Topo IV are heterotetramers, unlike the eukaryotic Topo II, which is a homodimer, with the GyrA‐like and GyrB‐like domains fused together in each monomer. (d) Type IIB topoisomerases. PDB structures used in this figure: 1EJ9, 2O5C, 2Q2E, 4CGT, 4DDX, 4I3H, 4RUL, 5GWK, 6K8O, and 6RKV
All kingdoms have at least one topoisomerase dedicated to dissipating the excessive negative supercoils caused by transcription. In bacteria and eukaryotes, this task is performed by a type I enzyme (IA for bacteria and IB for eukaryotes). Types IA and IB are not structurally, mechanistically, or evolutionarily related but share the ability to operate in an adenosine triphosphate (ATP)‐independent manner. The type IA bacterial enzyme Topo I (Figure 2a) relaxes negative supercoils but not positive supercoils, unless there is an exposed region of single‐stranded DNA.49 Topo I cuts and binds to a 5′ phosphate group on DNA and performs strand passage, resulting in the removal of one supercoil.50 In contrast to bacterial Topo I, the type IB eukaryotic Topo I (Figure 2b) relaxes both types of supercoils, binds to a 3′ phosphate group on DNA, and performs strand rotation, resulting in the removal of one supercoil.51 DNA topoisomerases are generally Mg2+ dependent.52 Bacterial Topo I follows this general rule,53 but a notable particularity of the eukaryotic Topo I is that, while 16‐fold activated by Mg2+, it does not strictly require it.54 Furthermore, Mg2+ can be substituted by Mn2+, Ba2+, or Ca2+ without loss of activity.54 The Topo I enzymes also differ in that the eukaryotic Topo I is stimulated by the polyamines spermidine and spermine,54 whereas bacterial Topo I is insensitive to spermidine and its precursor putrescine.55
Archaea generally lack an equivalent type I topoisomerase. The dissipation of excessive supercoils is instead dependent on a type IIB enzyme—called Topo VI (Figure 2d)—that differs greatly from its bacterial and eukaryotic counterparts. Topo VI is an ATP‐dependent enzyme capable of relaxing both positive and negative supercoils.56 In addition, Topo VI exhibits significant decatenase activity,56 and its activity is Mg2+ dependent,56 like that of other type II enzymes. A notable exception is the Archaea Methanopyrus kandleri, which has a type I topoisomerase called Topo V that is fully Mg2+‐ and ATP‐independent and relaxes both positive and negative supercoils.57 Because this enzyme has yet to be found in other species, its evolutionary origin remains unknown.
Mobile genetic elements in bacteria and archaea sometimes encode Topo VIII, a distant relative of Topo VI that has weak relaxation and decatenation activities.58 How does a mobile genetic element benefit from encoding Topo VIII when the host genome species specifies one or more topoisomerases? One possibility is that the Topo VIII is uniquely suited to operate with the DNA replication proteins, such as transposases, specified on the mobile genetic element.
Bacteria and eukaryotes use a dedicated enzyme for decatenation. Thus, they differ from Archaea, which use Topo VI for both DNA relaxation and decatenation. In bacteria, decatenation and unknotting are primarily performed by Topo IV, a type IIA enzyme (Figure 2c). Beyond its primary role in decatenation and unknotting,59 Topo IV shares with Topo I the ability to relax excessive supercoils, exhibiting 3‐60 to 20‐fold61 higher in vitro activity on positive supercoils compared to negative supercoils. In eukaryotes, decatenation and unknotting are performed by the TOP2A and TOP2B proteins, which are homologs of topoisomerase IV and often collectively referred to as Topo II (Figure 2(C)). Topo II has significant DNA relaxation activity,62 is required for the chromosome condensation taking place before cell division,63 and is involved in a variety of cell division‐related activities alongside the proteins cohesin and condensin (for an in‐depth recent review, see Reference 64).
Bacteria also have a unique type IIA topoisomerase called DNA gyrase65 (Figure 2c). The particularity of this enzyme among other type II topoisomerases is that it is capable of introducing negative supercoils into DNA at the expense of ATP,66 which results in bacteria maintaining a negatively supercoiled DNA at all times. DNA gyrase can also relax positive60 and negative67 supercoils and perform decatenation and unknotting,68 albeit with a low efficiency compared to its paralog Topo IV.59, 69 DNA gyrase is also specified in certain eukaryotic genomes, such as in the plant A. thaliana 70 and the apicomplexan parasite Plasmodium falciparum,71 as well as in the euryarchaea Thermoplasma acidophilum 72 and Archaeblobus profundus.73 The sporadic distribution of DNA gyrase‐specifying genes suggests acquisition of these genes via horizontal gene transfer.74 Notably, the DNA gyrases of A. thaliana and P. falciparum are targeted to organelles70, 71 rather than acting on nuclear DNA.
Archaea have a unique type IA enzyme called reverse gyrase (Figure 2a) that relaxes negative (but not positive) supercoils and introduces positive supercoils in a relaxed template at the expense of ATP.75 The enzyme is Mg2+ dependent75 and generally possesses two zinc finger domains76 that contribute to its enzymatic activity.77 The N‐terminus of reverse gyrase is unique in that it resembles a eukaryotic‐like helicase78 (Figure 2a). While helicase activity could not be measured for reverse gyrase, the helicase‐like domain contributes to reverse gyrase function.79 Reverse gyrase is found in hyperthermophilic bacteria such as Thermotoga maritima,80 which likely acquire it by horizontal gene transfer.74
Topo III is a type IA topoisomerase that exists in all kingdoms of life (Figure 2a). It is a Mg2+‐dependent yet ATP‐independent enzyme. It displays decatenase activity in both bacteria68, 81 and eukaryotes.82 Though it is also capable of relaxing negatively supercoiled DNA,49, 83 the bacterial Topo III operates at a slower rate than Topo I.49, 84 Topo III is incapable of relaxing positive supercoils.49 Unlike Topo I, Topo IV, and gyrase, Topo III is not essential in bacteria.85 A Topo III null mutant has increased frameshift mutation rates,86 suggesting a role related to DNA replication. Yeasts have a single TOP3, but higher eukaryotes have two paralogs of the protein—TOP3α and TOP3β—only one of which (TOP3α) is essential.87 The widespread presence of Topo III across all kingdoms suggests an early appearance in the evolution of living cells, indicating that it likely performed a key function in early cellular life. That Topo III is generally nonessential in current organisms raises intriguing evolutionary questions: was Topo III an early, generic topoisomerase that performed DNA relaxation and decatenation but was later supplanted by specific, more efficient topoisomerases? Or did it perform a different function yet to be identified that other topoisomerases cannot, explaining its conservation to this day?
7. EXPRESSION OF CERTAIN TOPOISOMERASES IS REGULATED ACCORDING TO THEIR FUNCTION
The essentiality of many topoisomerases implies that they must be present at least at a basal level all of the time. However, organisms tune the expression of some topoisomerases in response to specific signals (summarized in Table 2 and detailed in the text below). As discussed in the previous section, the transcriptional regulation of eukaryotic topoisomerases reflects their restricted roles in managing supercoils created as a result of transcription and DNA replication.
TABLE 2.
Summary of known regulation for the main topoisomerases in bacteria and eukaryotes
| Name | Gene expression | Transcript | Enzyme | |
|---|---|---|---|---|
| DNA relaxation | ||||
| Bacteria | Topo I | Induced by supercoiled DNA | N/D | Constitutive, possibly inhibited by anaerobiosis |
| Eukaryote | Topo I | Constitutive | N/D | Activated by polyamines |
| DNA compaction | ||||
| Bacteria | DNA gyrase | Induced by relaxed DNA | N/D | Activity correlates with [ATP]/[ADP] ratio, activated by polyamines. Putative chaperone |
| Decatenases | ||||
| Bacteria | Topo IV | Cell cycle‐regulated in Caulobacter | N/D | N/D |
| Eukaryote | TOP2A | Peaks in G2/M phase, regulated by many cell cycle‐ and tissue‐dependent transcription factors | Stabilized in G2/M phases | Activity and stability regulated by ubiquitinylation, SUMOylation, and phosphorylation |
| TOP2B | Mostly constitutive, some tissue dependence | N/D | Activity regulated by SUMOylation and phosphorylation |
Abbreviation: N/D, no data available.
TOP1 encodes the eukaryotic type I topoisomerase. The TOP1 regulatory region bears binding sites for the ubiquitous transcription factors SP1 and OCT‐1.88 SP1 is expressed at a basal level and activated by phosphorylation in response to DNA damage,89 whereas OCT‐1 is fully constitutive.90 SP1‐mediated induction is generally considered minor, and therefore TOP1 is overall constitutively expressed, in agreement with its role in the housekeeping process of preventing supercoil accumulation.
TOP2 encodes the single yeast Topo II. Unlike Topo I, Topo II is both more specialized in supporting DNA replication, thanks to its decatenation and unknotting activity, and subjected to more regulation. TOP2 ensures proper chromosome segregation during mitosis91 and meiosis.92 The TOP2A and TOP2B genes encode two distinct Topo II in higher eukaryotes. TOP2A appears to play critical roles in embryonic development and cell proliferation. By contrast, TOP2B behaves more as a housekeeping enzyme. Both TOP2A and TOP2B display tissue‐, developmental‐, and cell cycle‐dependent expression and are highly expressed in the thymus of both rats and mice.93, 94 TOP2A also shows strong expression in the spleen and sporadic expression in other tissues, while TOP2B is more broadly expressed.93, 94 TOP2A is expressed in rapidly proliferating cell types and tissues.93, 95 In addition, TOP2A (but not TOP2B) is cell cycle regulated, with a peak of expression in the G2 and M phases.96 This peak is ascribed to a combination of transcriptional and post‐transcriptional regulation. The transcription factors SP1,97 NF‐Y,97, 98 ICBP90,99 MYB,100 P53,101 ATF,102 and others not yet identified103 regulate TOP2A. Some of these factors, notably ICBP90, display the same tissue‐dependent expression as TOP2A,99 which may account, in part, for its expression behavior. Though the TOP2A regulator P53 exhibits cell cycle‐dependent activity and abundance, the cell cycle control of TOP2A expression remains unclear because P53 acts on a minimal TOP2A promoter deprived of any regulatory element,101 which argues that its regulatory effect is nonspecific. An alternative hypothesis has been proposed whereby NF‐Y, despite being constitutively expressed and binding to the TOP2A promoter region, is responsible for the cell cycle‐dependent regulation by recruiting histone acetyltransferases to indirectly activate TOP2A expression in the G2/M phases.98 Post‐transcriptional regulation is also hypothesized to contribute to the cell cycle‐dependent expression of TOP2A because transcript stability depends on the cell cycle.104
As far as the eukaryotic Topo III is concerned, the TOP3A and TOP3B genes are expressed constitutively across all investigated tissues. Though TOP3A is subject to alternative splicing,105 the regulatory consequences of splicing are unknown. Understanding how TOP3A and TOP3B are regulated, especially in comparison with their conserved archaeal and bacterial counterparts, could shed light on the function of Topo III.
Bacterial topoisomerases have a key function not performed by their eukaryotic counterparts: maintaining a certain level of global negative DNA supercoiling. This key function accounts for the regulation of the bacterial Topo I and DNA gyrase differing from that of other topoisomerases. Expression of the Topo I‐encoding topA gene increases when bacteria experience high negative DNA supercoiling, a condition that promotes repression of the DNA gyrase‐specifying gyrA and gyrB genes. It makes physiological sense for transcription of the enzyme promoting DNA relaxation to increase when global DNA supercoiling increases. Mechanistically, topA transcription follows the general rule that the more supercoiled a DNA is, the easier it is to melt and thus transcribe.106
By contrast, the supercoiling sensitivity of gyrA and gyrB transcription is less clear because the −10 element of the gyrA and gyrB promoters is necessary and sufficient to confer supercoiling sensitivity107, 108 despite sharing many features with promoters that are not supercoiling sensitive. In addition to sequence features of these two promoters, the relative positioning of the −10 and −35 elements appears to play a role in their activity because mutations that reduce promoter flexibility (i.e., the physical capacity of the regulatory region to bend) also reduce their DNA supercoiling sensitivity.109 Solving the mechanism responsible for the DNA supercoiling sensitivity of a promoter would represent a foundational advance in understanding regulatory networks in bacteria, allowing the prediction and modeling of expression behaviors that depend on DNA supercoiling, thereby greatly enhancing our knowledge of the contribution of DNA supercoiling to cellular physiology.
DNA supercoiling induction of Topo I transcription by high negative supercoiling is also displayed by Mycobacterium tuberculosis, and M. smegmatis, 110 and Streptococcus pneumoniae.111 Furthermore, the opposite sensitivity of Topo I and DNA gyrase transcription to DNA supercoiling was reported in Streptomyces coelicolor.112 Conservation of the transcriptional regulation of topo I and DNA gyrase across bacterial evolution suggests an ancestral, conserved mechanism by which bacteria homeostatically maintain DNA supercoiling.
Topo III is encoded by the topB gene, which forms an operon with the upstream selD gene. Though selD is generally considered constitutively expressed in E. coli,113 expression of topB and selD decreases during late stationary phase.114 The operon organization of the selD and topB genes is not conserved beyond the Escherichia, Shigella, and Salmonella genera, and therefore regulation of topB beyond these genera is unknown.
The Topo IV‐encoding parC and parE genes are cell cycle regulated in C. crescentus.115 The promoters of both genes are activated right before DNA replication,115 consistent with the role of Topo IV in decatenating replicated chromosomes.59 It would be interesting to learn whether this regulation exists in bacterial species in which, unlike C. crescentus, DNA replication and cell division happen simultaneously.
The investigation of archaeal topoisomerases has revealed expression responses to environmental changes. In the extreme salinity‐inhabiting Halobacterium salinarum, the Topo VI‐encoding top6A and top6B genes are mildly induced in response to UV light,116 suggesting a role for Topo VI in DNA repair. In the extreme thermophilic bacterium Sulfolobus solfarticus, both the transcript117 and protein118 amounts of the reverse gyrase TopR1 are anticorrelated with temperature. Because TopR1‐specific activity increases with temperature,118 changes in TopR1 amounts may be a homeostatic mechanism to maintain constant total activity.
8. POST‐TRANSLATIONAL FACTORS ACT ON TOPOISOMERASES TO ALTER DNA SUPERCOILING
Most topoisomerases are Mg2+‐dependent enzymes,52 suggesting that changes in the intracellular Mg2+ concentration should affect DNA supercoiling. However, the difficulty of measuring bioavailable Mg2+ has prevented a direct test of this hypothesis. Nevertheless, starvation for extracellular Mg2+ impacts DNA supercoiling in a variety of organisms, presumably by altering the Mg2+ concentration in the cytosol or specific organelles. For example, the formation of DNA complexes by the human Topo II decreases in cells experiencing Mg2+ starvation.119
In enteric bacteria, the activities of both Topo I53 and DNA gyrase120 require Mg2+. That these enzymes exert opposite effects on DNA supercoiling precludes a straightforward relationship between DNA supercoiling and Mg2+ concentration. In S. Typhimurium, Mg2+ starvation causes DNA relaxation,55, 121 which may reflect that DNA gyrase is more sensitive to Mg2+ availability than Topo I. Curiously, excess Mg2+ also causes relaxation in S. Typhimurium.55 The latter effect results from a decrease in the concentration of polyamines,55 which are required for DNA gyrase activity.55, 120
The in vitro activities of bacterial DNA gyrase55 and eukaryotic Topo I54 are polyamine dependent. In vivo, bacterial DNA supercoiling is directly correlated with the concentration of the polyamines putrescine and spermidine.55 Thus, DNA supercoiling is intimately connected with polyamine metabolism, import, and export. The identity of the specific polyamine controlling DNA gyrase in vivo often differs across closely related bacterial species. For example, spermidine and putrescine are the main polyamines controlling DNA supercoiling in E. coli and S. Typhimurium, respectively.55 Putrescine is converted into spermidine by the enzyme SpeE.122 Thus, inactivation of the speE gene has opposite effects on E. coli and S. Typhimurium: it increases global DNA supercoiling in the former but decreases it in the latter.55
Bacteria are unique in that the enzymes that govern global DNA supercoiling exhibit a different dependence on ATP: the DNA‐compacting DNA gyrase is ATP dependent, whereas the DNA‐relaxing Topo I is not. In addition, DNA gyrase activity is inhibited by ADP.123 Therefore the [ATP]/[ADP] ratio is a key driver of DNA supercoiling in bacteria,124 as it shifts the equilibrium between the activities of DNA gyrase versus Topo I. A similar dependence has not been reported in other kingdoms, presumably because the archaeal reverse gyrase and topo VI are both ATP dependent, whereas eukaryotes do not maintain global supercoiling.
In eukaryotes, Topo II activity and abundance are controlled post‐translationally. The vertebrate TOP2A can be directed for degradation by ubiquitinylation.125 The opposite action of the anaphase‐promoting complex that promotes ubiquitinylation126 versus the deubiquitylase USP15127 controls TOP2A ubiquitinylation and therefore degradation in a cell cycle‐dependent manner, with TOP2A stability being maximal in G2/M phases. By contrast, neither TOP2B128 nor the unique TOP2 found in nonvertebrates129 appears to have a similar regulation. Topo II activity is also dependent on other post‐translational modifications, namely SUMOylation (a polypeptidic modification) and phosphorylation, which both contribute to the cell cycle dependence of Topo II activity (for a recent review on the topic, see Reference 64).
9. BACTERIA AND ARCHEAE CHANGE DNA SUPERCOILING IN RESPONSE TO EXTRACELLULAR STRESSES
Bacterial DNA supercoiling is sensitive to a wide variety of extracellular stresses. For example, an increase in osmolarity generally increases DNA supercoiling. However, the molecular mechanism(s) responsible for this phenomenon is unclear because NaCl shock causes a transient increase in DNA supercoiling in E. coli that closely correlates with the [ATP]/[ADP] ratio,130 whereas performing the shock with KCl causes the same transient increase in DNA supercoiling independently of the [ATP]/[ADP] ratio.131 Similar responses have been reported in S. Typhimurium,132 B. subtilis,133 and S. aureus,134 indicating widespread conservation of the phenomenon, but its link with the [ATP]/[ADP] ratio remains unknown.
Oxidative stress decreases bacterial DNA supercoiling transiently. E. coli experiences a decrease in DNA supercoiling upon treatment with H2O2 that is followed by a return to pretreatment levels within 30 min.23 In the phytopathogenic enterobacterium Dickeya dadantii, the same effect is dependent on the NAPs FIS and H‐NS.135 The dependence on FIS and H‐NS suggests the participation of specific regulatory networks in the changes in DNA supercoiling resulting from oxidative stress. Conversely, oxygen limitation and anaerobiosis increase DNA supercoiling in E. coli.136, 137 In the latter case, however, DNA supercoiling is not correlated with the [ATP]/[ADP] ratio.136 Increased supercoiling appears to result from a decrease in Topo I activity, which was lower in crude extracts from cells subjected to anaerobiosis than in control bacteria.138 The concurrent decrease in Topo I activity and increase in DNA supercoiling triggered by anaerobiosis have also been observed in S. Typhimurium,139 but the mechanism(s) by which they occur remains unknown.
A mildly acidic pH durably relaxes DNA in S. Typhimurium 140 and D. dadantii.135 In the latter, this relaxation is H‐NS dependent.135 In vitro, DNA gyrase activity is inhibited at pH <5.140 It has been proposed that a decrease in external pH causes a drop in cytoplasmic pH, explaining the observed decrease in DNA supercoiling.140 This model, however, suffers from major inconsistencies. First, mesophilic bacteria such as E. coli and S. Typhimurium maintain an intracellular pH no lower than 6.7 even when the extracellular pH drops to 5.141, 142 Second, an acidic pH favors entry of the drug commonly used to decrease DNA supercoiling in vivo—novobiocin—into the bacterial cell.143 Therefore, the reported synergy in bacterial growth inhibition resulting from novobiocin and acidic pH may result from increased permeability to novobiocin rather than from direct inhibition of DNA gyrase by a decrease in pH.140 Surprisingly, E. coli did not significantly change its DNA supercoiling in conditions where S. Typhimurium's did,144 indicating species‐specific changes in the underlying regulatory network. A supercoiling response to alkaline pH has not been reported.
An increase in temperature from 17 to 47°C results in increased negative DNA supercoiling in E. coli.145, 146 This increase is attributed to changes in the [ATP]/[ADP] ratio, which correlates with DNA supercoiling during an upshift from 30 to 47°C.145 Oddly, DNA gyrase is 90% inactivated within 20 min of treatment at 46°C in vitro,147 implying that a factor such as a protein chaperone maintains DNA gyrase activity in organisms experiencing high temperatures. DNA gyrase is essential in all investigated bacteria, and therefore factors that protect DNA gyrase may be an attractive target for antibacterial agents.
As discussed above, many archaea and notably hyperthermophiles maintain a positively supercoiled DNA due to the high resistance of positively supercoiled DNA to melting.7 However, positively supercoiled DNA is detrimental to transcription and DNA replication, so archaea must find a balance between DNA stability and capacity to carry out these essential processes. The archaeal topoisomerase VI, which conducts most of the DNA relaxation, is inhibited by high temperatures,56, 148 while reverse gyrase activity is stimulated.148 Therefore, opposing variations in enzymatic activities likely cause increased DNA supercoiling at higher temperatures. This is reminiscent of the transcriptional homeostatic control of the balance between topoisomerase I and DNA gyrase in bacteria to maintain negative supercoiling, although the archaeal equivalent for positive supercoils appears due to enzymatic activity rather than transcription.
Extracellular factors influencing DNA supercoiling in vivo in eukaryotes have yet to be reported. This likely stems from the fact that eukaryotic DNA supercoiling is only local, and suitable methods to measure it are laborious.149 Studying this control especially in pathogenic fungi may lead to benefits, notably in the agricultural sector, where fungal infections are common.
10. CONCLUDING REMARKS: EXPLOITING SIMILARITIES AND DIFFERENCES IN DNA SUPERCOILING REGULATION FOR THERAPEUTIC PURPOSES
DNA supercoiling is a fundamental property of DNA that exists in some form across all domains of life. It likely emerged alongside the DNA molecule itself and the machineries that transcribe and replicate it, and which still create and depend on supercoiling to this day. Importantly, each kingdom has evolved distinct strategies for regulating supercoiling, some even co‐opting it for gene regulation: from the opposition of the ATP‐dependent bacterial DNA gyrase to the ATP‐independent topoisomerase I, to the eukaryotic topo II associated with cell division proteins, and the polyamine dependence of only certain specific topoisomerases. Taken together, the essentiality and the kingdom‐specific (sometimes species‐specific) regulation of DNA supercoiling present extraordinary therapeutic potential. It may be possible to target the regulatory adaptations that have evolved only in certain branches, as has been done with fluoroquinolone antibiotics targeting the bacteria‐specific DNA gyrase. Similarly, the requirement of eukaryotic topoisomerases for fast proliferation has opened avenues for anticancer drugs like anthracyclines. Ultimately, deeper knowledge of the regulatory networks that underlie the maintenance of DNA supercoiling in all kingdoms has the potential to lead to new, transformative medicines.
AUTHOR CONTRIBUTIONS
Alexandre Duprey: Conceptualization (lead); funding acquisition (supporting); visualization (lead); writing—original draft (lead); writing—review and editing (equal). Eduardo A. Groisman: Funding acquisition (lead); writing—review and editing (equal).
Duprey A, Groisman EA. The regulation of DNA supercoiling across evolution. Protein Science. 2021;30:2042–2056. 10.1002/pro.4171
Funding information National Institute of Allergy and Infectious Diseases, Grant/Award Number: AI120558
Contributor Information
Alexandre Duprey, Email: alexandre.duprey@yale.edu.
Eduardo A. Groisman, Email: eduardo.groisman@yale.edu.
REFERENCES
- 1.Ghosh A, Bansal M. A glossary of DNA structures from A to Z. Acta Cryst D. 2003;59:620–626. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes D, Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980;286:573–578. [DOI] [PubMed] [Google Scholar]
- 3.Saenger W. Higher organization of DNA. In: Saenger W, editor. Principles of nucleic acid structure. New York, NY: Springer, 1984; p. 432–458. [Google Scholar]
- 4.Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, Kinosita K. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–115. [DOI] [PubMed] [Google Scholar]
- 6.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng H, Bosman J, van der Heijden T, van Noort J. Coexistence of twisted, plectonemic, and melted DNA in small topological domains. Biophys J. 2014;106:1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J, Wang MD. DNA supercoiling during transcription. Biophys Rev. 2016;8:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert N, Allan J. Supercoiling in DNA and chromatin. Curr Opin Genet Dev. 2014;25:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockshon D, Morris DR. Positively supercoiled plasmid DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucleic Acids Res. 1983;11:2999–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruss GJ, Drlica K. Topoisomerase I mutants: The gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc Natl Acad Sci US A. 1986;83:8952–8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. [DOI] [PubMed] [Google Scholar]
- 13.Sobetzko P. Transcription‐coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016;44:1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer S, Beslon G. Torsion‐mediated interaction between adjacent genes. PLoS Comput Biol. 2014;10:e1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberts B. DNA replication and recombination. Nature. 2003;421:431–435. [DOI] [PubMed] [Google Scholar]
- 16.Higgins NP. Chromosome structure. Encyclopedia of Life Sciences. Macmillan Publishers Ltd, Nature Publishing Group, 2007. 10.1002/9780470015902.a0001486.pub2 [DOI] [Google Scholar]
- 17.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: Conformations at the fork. Proc Natl Acad Sci U S A. 2001;98:8219–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. [DOI] [PubMed] [Google Scholar]
- 19.Baxter J, Sen N, Martínez VL, et al. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331:1328–1332. [DOI] [PubMed] [Google Scholar]
- 20.Hirata A, Murakami KS. Archaeal RNA polymerase. Curr Opin Struct Biol. 2009;19:724–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Z, Liu J, Yang H, Xiang H. DNA replication origins in archaea. Front Microbiol. 2014;5:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Li N, Wang Y, Wang Y, Zhang X, Zhang H. Effects of X‐irradiation on mitochondrial DNA damage and its supercoiling formation change. Mitochondrion. 2011;11:886–892. [DOI] [PubMed] [Google Scholar]
- 23.Weinstein‐Fischer D, Elgrably‐Weiss M, Altuvia S. Escherichia coli response to hydrogen peroxide: A role for DNA supercoiling, topoisomerase I and Fis. Mol Microbiol. 2000;35:1413–1420. [DOI] [PubMed] [Google Scholar]
- 24.Pettijohn DE, Pfenninger O. Supercoils in prokaryotic DNA restrained in vivo. Proc Natl Acad Sci U S A. 1980;77:1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. Mlh1‐Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2‐Msh3‐stimulated endonuclease. J Biol Chem. 2014;289:5664–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatterjee N, Walker GC. Mechanisms of DNA damage, repair and mutagenesis. Environ Mol Mutagen. 2017;58:235–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins AR, Fleming IM, Gedik CM. In vitro repair of oxidative and ultraviolet‐induced DNA damage in supercoiled nucleoid DNA by human cell extract. Biochim Biophys Acta BBA Gene Struct Expr. 1994;1219:724–727. [DOI] [PubMed] [Google Scholar]
- 29.Mariño‐Ramírez L, Kann MG, Shoemaker BA, Landsman D. Histone structure and nucleosome stability. Expert Rev Proteomics. 2005;2:719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. [DOI] [PubMed] [Google Scholar]
- 31.Norton VG, Imai BS, Yau P, Bradbury EM. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989;57:449–457. [DOI] [PubMed] [Google Scholar]
- 32.Hamiche A, Carot V, Alilat M, et al. Interaction of the histone (H3‐H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: Potential flipping of the protein from a left‐ to a right‐handed superhelical form. Proc Natl Acad Sci U S A. 1996;93:7588–7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive supercoils. Cell. 2009;138:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alilat M, Sivolob A, Révet B, Prunell A. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3‐H4)2 tetramer‐DNA particle1. J Mol Biol. 1999;291:815–841. [DOI] [PubMed] [Google Scholar]
- 35.Pereira SL, Grayling RA, Lurz R, Reeve JN. Archaeal nucleosomes. Proc Natl Acad Sci U S A. 1997;94:12633–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandman K, Krzycki JA, Dobrinski B, Lurz R, Reeve JN. HMf, a DNA‐binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc Natl Acad Sci U S A. 1990;87:5788–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musgrave DR, Sandman KM, Reeve JN. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc Natl Acad Sci U S A. 1991;88:10397–10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musgrave D, Forterre P, Slesarev A. Negative constrained DNA supercoiling in archaeal nucleosomes. Mol Microbiol. 2000;35:341–349. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka H, Yasuzawa K, Kohno K, et al. Role of HU proteins in forming and constraining supercoils of chromosomal DNA in Escherichia coli . Mol Gen Genet. 1995;248:518–526. [DOI] [PubMed] [Google Scholar]
- 40.Tupper AE, Owen‐Hughes TA, Ussery DW, et al. The chromatin‐associated protein H‐NS alters DNA topology in vitro. EMBO J. 1994;13:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider R, Lurz R, Lüder G, Tolksdorf C, Travers A, Muskhelishvili G. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 2001;29:5107–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dahlke K, Sing CE. Influence of nucleoid‐associated proteins on DNA supercoiling. J Phys Chem B. 2019;123:10152–10162. [DOI] [PubMed] [Google Scholar]
- 43.Le TB, Laub MT. Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. EMBO J. 2016;35:1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kavenoff R, Bowen BC. Electron microscopy of membrane‐free folded chromosomes from Escherichia coli . Chromasoma. 1976;59:89–101. [DOI] [PubMed] [Google Scholar]
- 45.Duprey A, Groisman EA. FEDS: A novel fluorescence‐based high‐throughput method for measuring DNA supercoiling in vivo. mBio. 2020;11:e01053‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naughton C, Avlonitis N, Corless S, et al. Transcription forms and remodels supercoiling domains unfolding large‐scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorman CJ. DNA supercoiling and transcription in bacteria: A two‐way street. BMC Mol Cell Biol. 2019;20:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schvartzman JB, Martínez‐Robles M‐L, Hernández P, Krimer DB. The benefit of DNA supercoiling during replication. Biochem Soc Trans. 2013;41:646–651. [DOI] [PubMed] [Google Scholar]
- 49.Terekhova K, Gunn KH, Marko JF, Mondragón A. Bacterial topoisomerase I and topoisomerase III relax supercoiled DNA via distinct pathways. Nucleic Acids Res. 2012;40:10432–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mills M, Tse‐Dinh Y‐C, Neuman KC. Direct observation of topoisomerase IA gate dynamics. Nat Struct Mol Biol. 2018;25:1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. [DOI] [PubMed] [Google Scholar]
- 52.Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37:702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domanico PL, Tse‐Dinh YC. Mechanistic studies on E. coli DNA topoisomerase I: Divalent ion effects. J Inorg Biochem. 1991;42:87–96. [DOI] [PubMed] [Google Scholar]
- 54.Stewart L, Ireton GC, Parker LH, Madden KR, Champoux JJ. Biochemical and biophysical analyses of recombinant forms of human topoisomerase I. J Biol Chem. 1996;271:7593–7601. [DOI] [PubMed] [Google Scholar]
- 55.Duprey A, Groisman EA. DNA supercoiling differences in bacteria result from disparate DNA gyrase activation by polyamines. PLoS Genet. 2020;16:e1009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergerat A, Gadelle D, Forterre P. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J Biol Chem. 1994;269:27663–27669. [PubMed] [Google Scholar]
- 57.Slesarev AI, Stetter KO, Lake JA, Gellert M, Krah R, Kozyavkin SA. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. [DOI] [PubMed] [Google Scholar]
- 58.Gadelle D, Krupovic M, Raymann K, Mayer C, Forterre P. DNA topoisomerase VIII: A novel subfamily of type IIB topoisomerases encoded by free or integrated plasmids in archaea and bacteria. Nucleic Acids Res. 2014;42:8578–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site‐specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashley RE, Dittmore A, McPherson SA, Turnbough CL, Neuman KC, Osheroff N. Activities of gyrase and topoisomerase IV on positively supercoiled DNA. Nucleic Acids Res. 2017;45:9611–9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single‐molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salceda J, Fernández X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JH, Berger JM. Cell cycle‐dependent control and roles of DNA topoisomerase II. Genes. 2019;10:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tretter EM, Lerman JC, Berger JM. A naturally chimeric type IIA topoisomerase in Aquifex aeolicus highlights an evolutionary path for the emergence of functional paralogs. Proc Natl Acad Sci U S A. 2010;107:22055–22059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugino A, Higgins NP, Brown PO, Peebles CL, Cozzarelli NR. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978;75:4838–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc Natl Acad Sci U S A. 1996;93:14416–14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hiasa H, DiGate RJ, Marians KJ. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J Biol Chem. 1994;269:2093–2099. [PubMed] [Google Scholar]
- 69.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli . J Biol Chem. 1996;271:31549–31555. [DOI] [PubMed] [Google Scholar]
- 70.Wall MK, Mitchenall LA, Maxwell A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc Natl Acad Sci U S A. 2004;101:7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dar MA, Sharma A, Mondal N, Dhar SK. Molecular cloning of apicoplast‐targeted plasmodium falciparum DNA gyrase genes: Unique intrinsic ATPase activity and ATP‐independent dimerization of PfGyrB subunit. Eukaryot Cell. 2007;6:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamashiro K, Yamagishi A. Characterization of the DNA gyrase from the thermoacidophilic archaeon Thermoplasma acidophilum . J Bacteriol. 2005;187:8531–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.López‐García P, Forterre P, van der Oost J, Erauso G. Plasmid pGS5 from the hyperthermophilic archaeon Archaeoglobus profundus is negatively supercoiled. J Bacteriol. 2000;182:4998–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duguet M. Reverse Gyrase. In: Eckstein F, Lilley DMJ, editors. Nucleic acids and molecular biology. Berlin, Heidelberg: Springer, 1995; p. 84–114. [Google Scholar]
- 76.Rodríguez AC, Stock D. Crystal structure of reverse gyrase: Insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rudolph MG, del Toro DY, Jungblut SP, Ganguly A, Klostermeier D. Crystal structures of Thermotoga maritima reverse gyrase: Inferences for the mechanism of positive DNA supercoiling. Nucleic Acids Res. 2013;41:1058–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M. Reverse gyrase: A helicase‐like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci U S A. 1993;90:4753–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Déclais AC, Marsault J, Confalonieri F, de La Tour CB, Duguet M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J Biol Chem. 2000;275:19498–19504. [DOI] [PubMed] [Google Scholar]
- 80.Guipaud O, Marguet E, Noll KM, de la Tour CB, Forterre P. Both DNA gyrase and reverse gyrase are present in the hyperthermophilic bacterium Thermotoga maritima. Proc Natl Acad Sci U S A. 1997;94:10606–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nurse P, Levine C, Hassing H, Marians KJ. Topoisomerase III can serve as the cellular decatenase in Escherichia coli . J Biol Chem. 2003;278:8653–8660. [DOI] [PubMed] [Google Scholar]
- 82.Cejka P, Plank JL, Dombrowski CC, Kowalczykowski SC. Decatenation of DNA by the S. cerevisiae Sgs1–Top3–Rmi1 and RPA complex: A mechanism for disentangling chromosomes. Mol Cell. 2012;47:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hanai R, Caron PR, Wang JC. Human TOP3: A single‐copy gene encoding DNA topoisomerase III. Proc Natl Acad Sci U S A. 1996;93:3653–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli . J Biol Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- 85.Perez‐Cheeks BA, Lee C, Hayama R, Marians KJ. A role for topoisomerase III in Escherichia coli chromosome segregation. Mol Microbiol. 2012;86:1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uematsu N, Eda S, Yamamoto K. An Escherichia coli topB mutant increases deletion and frameshift mutations in the supF target gene. Mutat Res. 1997;383:223–230. [DOI] [PubMed] [Google Scholar]
- 87.Li W, Wang JC. Mammalian DNA topoisomerase IIIα is essential in early embryogenesis. Proc Natl Acad Sci U S A. 1998;95:1010–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kunze N, Yang GC, Dölberg M, Sundarp R, Knippers R, Richter A. Structure of the human type I DNA topoisomerase gene. J Biol Chem. 1991;266:9610–9616. [PubMed] [Google Scholar]
- 89.Loshchenova PS, Sergeeva SV, Fletcher SC, Dianov GL. The role of Sp1 in the detection and elimination of cells with persistent DNA strand breaks. NAR Cancer. 2020;2:zcaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pance A. Oct‐1, to go or not to go? That is the PolII question. Biochim Biophys Acta Gene Regul Mech. 2016;1859:820–824. [DOI] [PubMed] [Google Scholar]
- 91.Jensen S, Redwood CS, Jenkins JR, Andersen AH, Hickson ID. Human DNA topoisomerases IIα and IIβ can functionally substitute for yeastTOP2 in chromosome segregation and recombination. Mol Gen Genet. 1996;252:79–86. [PubMed] [Google Scholar]
- 92.Hartsuiker E, Bähler J, Kohli J. The role of topoisomerase II in meiotic chromosome condensation and segregation in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsutsui K, Tsutsui K, Okada S, et al. Molecular cloning of partial cDNAs for rat DNA topoisomerase II isoforms and their differential expression in brain development. J Biol Chem. 1993;268:19076–19083. [PubMed] [Google Scholar]
- 94.Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim Biophys Acta Gene Struct Expr. 1992;1132:43–48. [DOI] [PubMed] [Google Scholar]
- 95.Zandvliet DWJ, Hanby AM, Austin CA, et al. Analysis of foetal expression sites of human type II DNA topoisomerase α and β mRNAs by in situ hybridisation. Biochim Biophys Acta Gene Struct Expr. 1996;1307:239–247. [DOI] [PubMed] [Google Scholar]
- 96.Austin CA, Lee KC, Swan RL, et al. TOP2B: The first thirty years. Int J Mol Sci. 2018;19:2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Magan N, Szremska AP, Isaacs RJ, Stowell KM. Modulation of DNA topoisomerase IIα promoter activity by members of the Sp (specificity protein) and NF‐Y (nuclear factor Y) families of transcription factors. Biochem J. 2003;374:723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Adachi N, Nomoto M, Kohno K, Koyama H. Cell‐cycle regulation of the DNA topoisomerase IIα promoter is mediated by proximal CCAAT boxes: Possible involvement of acetylation. Gene. 2000;245:49–57. [DOI] [PubMed] [Google Scholar]
- 99.Hopfner R, Mousli M, Jeltsch JM, et al. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase II alpha expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 100.Brandt TL, Fraser DJ, Leal S, Halandras PM, Kroll AR, Kroll DJ. C‐Myb trans‐activates the human DNA topoisomerase IIα gene promoter. J Biol Chem. 1997;272:6278–6284. [DOI] [PubMed] [Google Scholar]
- 101.Sandri MI, Isaacs RJ, Ongkeko WM, et al. p53 regulates the minimal promoter of the human topoisomerase IIα gene. Nucleic Acids Res. 1996;24:4464–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lim K, Lee JI, Yun KA, et al. Reduced level of ATF is correlated with transcriptional repression of DNA topoisomerase II alpha gene during TPA‐induced differentiation of HL‐60 cells. Biochem Mol Biol Int. 1998;46:35–42. [DOI] [PubMed] [Google Scholar]
- 103.Herzog CE, Zwelling LA. Evaluation of a potential regulatory role for inverted CCAAT boxes in the human topoisomerase IIα promoter. Biochem Biophys Res Commun. 1997;232:608–612. [DOI] [PubMed] [Google Scholar]
- 104.Goswami PC, Roti Roti JL, Hunt CR. The cell cycle‐coupled expression of topoisomerase II alpha during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol Cell Biol. 1996;16:1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Glatt SJ, Chandler SD, Bousman CA, et al. Alternatively spliced genes as biomarkers for schizophrenia, bipolar disorder and psychosis: A blood‐based spliceome‐profiling exploratory study. Curr Pharmacogenomics Pers Med. 2009;7:164–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tse‐Dinh Y‐C, Beran RK. Multiple promoters for transcription of the Escherichia coli DNA topoisomerase I gene and their regulation by DNA supercoiling. J Mol Biol. 1988;202:735–742. [DOI] [PubMed] [Google Scholar]
- 107.Menzel R, Gellert M. Modulation of transcription by DNA supercoiling: A deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci U S A. 1987;84:4185–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Straney R, Krah R, Menzel R. Mutations in the −10 TATAAT sequence of the gyrA promoter affect both promoter strength and sensitivity to DNA supercoiling. J Bacteriol. 1994;176:5999–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Unniraman S, Nagaraja V. Axial distortion as a sensor of supercoil changes: A molecular model for the homeostatic regulation of DNA gyrase. J Genet. 2001;80:119–124. [DOI] [PubMed] [Google Scholar]
- 110.Ahmed W, Menon S, Karthik PVDNB, Nagaraja V. Autoregulation of topoisomerase I expression by supercoiling sensitive transcription. Nucleic Acids Res. 2016;44:1541–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferrándiz M‐J, Martín‐Galiano AJ, Arnanz C, Camacho‐Soguero I, Tirado‐Vélez J‐M, de la Campa AG. An increase in negative supercoiling in bacteria reveals topology‐reacting gene clusters and a homeostatic response mediated by the DNA topoisomerase I gene. Nucleic Acids Res. 2016;44:7292–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Szafran MJ, Gongerowska M, Gutkowski P, Zakrzewska‐Czerwińska J, Jakimowicz D. The coordinated positive regulation of topoisomerase genes maintains topological homeostasis in Streptomyces coelicolor . J Bacteriol. 2016;198:3016–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sawers G, Heider J, Zehelein E, Böck A. Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium‐containing formate dehydrogenase isoenzyme. J Bacteriol. 1991;173:4983–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Srikumar S, Kröger C, Hébrard M, et al. RNA‐seq brings new insights to the intra‐macrophage transcriptome of Salmonella typhimurium . PLoS Pathog. 2015;11:e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ward DV, Newton A. Cell cycle expression and transcriptional regulation of DNA topoisomerase IV genes in caulobacter. J Bacteriol. 1999;181:3321–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boubriak I, Ng WL, DasSarma P, DasSarma S, Crowley DJ, McCready SJ. Transcriptional responses to biologically relevant doses of UV‐B radiation in the model archaeon, Halobacterium sp. NRC‐1. Saline Syst. 2008;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Garnier F, Nadal M. Transcriptional analysis of the two reverse gyrase encoding genes of Sulfolobus solfataricus P2 in relation to the growth phases and temperature conditions. Extremophiles. 2008;12:799–809. [DOI] [PubMed] [Google Scholar]
- 118.Couturier M, Gadelle D, Forterre P, Nadal M, Garnier F. The reverse gyrase TopR1 is responsible for the homeostatic control of DNA supercoiling in the hyperthermophilic archaeon Sulfolobus solfataricus . Mol Microbiol. 2020;113:356–368. [DOI] [PubMed] [Google Scholar]
- 119.Covacci V, Bruzzese N, Sgambato A, Ganapathi R, Cittadini A, Wolf FI. Effect of extracellular magnesium on topoisomerase II activity and expression in human leukemia HL‐60 cells. J Cell Biochem. 2000;78:325–333. [DOI] [PubMed] [Google Scholar]
- 120.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: An enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976;73:3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bogue MM, Mogre A, Beckett MC, Thomson NR, Dorman CJ. Network rewiring: Physiological consequences of reciprocally exchanging the physical locations and growth‐phase‐dependent expression patterns of the Salmonella fis and dps genes. mBio. 2020;11:e02128‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tabor CW, Tabor H, Xie QW. Spermidine synthase of Escherichia coli: Localization of the speE gene. Proc Natl Acad Sci U S A. 1986;83:6040–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li T‐K, Liu LF. Modulation of gyrase‐mediated DNA cleavage and cell killing by ATP. Antimicrob Agents Chemother. 1998;42:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Workum M, Dooren SJ, Oldenburg N, et al. DNA supercoiling depends on the phosphorylation potential in Escherichia coli . Mol Microbiol. 1996;20:351–360. [DOI] [PubMed] [Google Scholar]
- 125.Salmena L, Lam V, Peter McPherson J, Goldenberg GJ. Role of proteasomal degradation in the cell cycle‐dependent regulation of DNA topoisomerase IIα expression. Biochem Pharmacol. 2001;61:795–802. [DOI] [PubMed] [Google Scholar]
- 126.Eguren M, Álvarez‐Fernández M, García F, et al. A synthetic lethal interaction between APC/C and topoisomerase poisons uncovered by proteomic screens. Cell Rep. 2014;6:670–683. [DOI] [PubMed] [Google Scholar]
- 127.Fielding AB, Concannon M, Darling S, et al. The deubiquitylase USP15 regulates topoisomerase II alpha to maintain genome integrity. Oncogene. 2018;37:2326–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation‐ and cell cycle‐dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH‐3T3 cells. Cell Growth Differ. 1991;2:209. [PubMed] [Google Scholar]
- 129.Whalen AM, McConnell M, Fisher PA. Developmental regulation of drosophila DNA topoisomerase II. J Cell Biol. 1991;112:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hsieh LS, Rouviere‐Yaniv J, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: Changes associated with salt shock. J Bacteriol. 1991;173:3914–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meury J, Kohiyama M. Potassium ions and changes in bacterial DNA supercoiling under osmotic stress. FEMS Microbiol Lett. 1992;99:159–164. [DOI] [PubMed] [Google Scholar]
- 132.Higgins CF, Dorman CJ, Stirling DA, et al. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli . Cell. 1988;52:569–584. [DOI] [PubMed] [Google Scholar]
- 133.Alice AF, Sanchez‐Rivas C. DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr Microbiol. 1997;35:309–315. [DOI] [PubMed] [Google Scholar]
- 134.Sheehan BJ, Foster TJ, Dorman CJ, Park S, Stewart GS. Osmotic and growth‐phase dependent regulation of the eta gene of Staphylococcus aureus: A role for DNA supercoiling. Mol Gen Genet. 1992;232:49–57. [DOI] [PubMed] [Google Scholar]
- 135.Ouafa Z‐A, Reverchon S, Lautier T, Muskhelishvili G, Nasser W. The nucleoid‐associated proteins H‐NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii . Nucleic Acids Res. 2012;40:4306–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hsieh L‐S, Burger RM, Drlica K. Bacterial DNA supercoiling and [ATP][ADP]: Changes associated with a transition to anaerobic growth. J Mol Biol. 1991;219:443–450. [DOI] [PubMed] [Google Scholar]
- 137.Dorman CJ, Barr GC, Bhriain NN, Higgins CF. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol. 1988;170:2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cortassa S, Aon MA. Altered topoisomerase activities may be involved in the regulation of DNA supercoiling in aerobic‐anaerobic transitions in Escherichia coli . Mol Cell Biochem. 1993;126:115–124. [DOI] [PubMed] [Google Scholar]
- 139.Yamamoto N, Droffner ML. Mechanisms determining aerobic or anaerobic growth in the facultative anaerobe Salmonella typhimurium . Proc Natl Acad Sci U S A. 1985;82:2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Colgan AM, Quinn HJ, Kary SC, et al. Negative supercoiling of DNA by gyrase is inhibited in Salmonella enterica serovar typhimurium during adaptation to acid stress. Mol Microbiol. 2018;107:734–746. [DOI] [PubMed] [Google Scholar]
- 141.Hickeyt EW, Hirshfield IN. Low‐pH‐induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium . Appl Env Microbiol. 1990;56:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Olsen KN, Budde BB, Siegumfeldt H, Rechinger KB, Jakobsen M, Ingmer H. Noninvasive measurement of bacterial intracellular pH on a single‐cell level with green fluorescent protein and fluorescence ratio imaging microscopy. Appl Environ Microbiol. 2002;68:4145–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pomares MF, Corbalán NS, Adler C, et al. Macrophage environment turns otherwise MccJ25‐resistant salmonella into sensitive. BMC Microbiol. 2013;13:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Quinn HJ, Cameron ADS, Dorman CJ. Bacterial regulon evolution: Distinct responses and roles for the identical OmpR proteins of Salmonella typhimurium and Escherichia coli in the acid stress response. PLoS Genet. 2014;10:e1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Camacho‐Carranza R, Membrillo‐Hernández J, Ramírez‐Santos J, Castro‐Dorantes J, Chagoya de Sánchez V, Gómez‐Eichelmann MC. Topoisomerase activity during the heat shock response in Escherichia coli K‐12. J Bacteriol. 1995;177:3619–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goldstein E, Drlica K. Regulation of bacterial DNA supercoiling: Plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci U S A. 1984;81:4046–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Peebles CL, Higgins NP, Kreuzer KN, et al. Structure and activities of Escherichia coli DNA gyrase. Cold Spring Harb Symp Quant Biol. 1979;43:41–52. [DOI] [PubMed] [Google Scholar]
- 148.Lopez‐Garcia P, Forterre P. Control of DNA topology during thermal stress in hyperthermophilic archaea: DNA topoisomerase levels, activities and induced thermotolerance during heat and cold shock in Sulfolobus. Mol Microbiol. 1999;33:766–777. [DOI] [PubMed] [Google Scholar]
- 149.Bermúdez I, García‐Martínez J, Pérez‐Ortín JE, Roca J. A method for genome‐wide analysis of DNA helical tension by means of psoralen‐DNA photobinding. Nucleic Acids Res. 2010;38:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]


