Abstract
We investigated the protein associations and enzymatic requirements for the Xenopus histone deacetylase catalytic subunit RPD3 to direct transcriptional repression in Xenopus oocytes. Endogenous Xenopus RPD3 is present in nuclear and cytoplasmic pools, whereas RbAp48 and SIN3 are predominantly nuclear. We cloned Xenopus RbAp48 and SIN3 and show that expression of RPD3, but not RbAp48 or SIN3, leads to an increase in nuclear and cytoplasmic histone deacetylase activity and transcriptional repression of the TRβA promoter. This repression requires deacetylase activity and nuclear import of RPD3 mediated by a carboxy-terminal nuclear localization signal. Exogenous RPD3 is not incorporated into previously described oocyte deacetylase and ATPase complexes but cofractionates with a component of the endogenous RbAp48 in the oocyte nucleus. We show that RPD3 associates with RbAp48 through N- and C-terminal contacts and that RbAp48 also interacts with SIN3. Xenopus RbAp48 selectively binds to the segment of the N-terminal tail immediately proximal to the histone fold domain of histone H4 in vivo. Exogenous RPD3 may be targeted to histones through interaction with endogenous RbAp48 to direct transcriptional repression of the Xenopus TRβA promoter in the oocyte nucleus. However, the exogenous RPD3 deacetylase functions to repress transcription in the absence of a requirement for association with SIN3 or other targeted corepressors.
Transcriptional coactivators and corepressors are large multifunctional molecular machines that have the potential to regulate promoter activity both through modification of the chromatin environment and through interactions with the basal transcriptional machinery (20, 43, 63). Several coactivators are histone acetyltransferases (10, 47, 60, 78). These proteins share limited homology in the acetyltransferase domain (46) and have little overall similarity in the remainder of their sequences (43). Coactivators interact with transcriptional activation domains (for examples, references 13, 15, 18, 28, and 48) and can associate with each other to assemble large complexes that function even more effectively than the individual proteins to augment transcription (11, 60).
In contrast to the rich diversity of known histone acetyltransferases, only two families of histone deacetylases have been defined (40, 61). The plant nucleolar histone deacetylases so far form only a relatively minor group, whereas the RPD3-like family of deacetylases represents a large group of highly conserved polypeptides found in organisms as diverse as yeast and humans (36). In yeast, two distinct histone deacetylases of the RPD3-like family (RPD3 and HDA1) have been identified, both of which are present in different multiprotein complexes (30, 55). HDA1 homologs have recently also been identified in the mouse (65). In metazoans, RPD3 homologs have been found associated with diverse multiprotein complexes, many of which contain known transcriptional corepressors or repressors (1, 21–23, 25, 33, 34, 41, 44, 70, 79). Thus, unlike the structural diversity of the histone acetyltransferases, the histone deacetylase-containing corepressor complexes that have been defined to date all contain a member of the conserved family of RPD3 deacetylases.
The conservation of the RPD3 deacetylase both in terms of polypeptide sequence and as a component of different corepressor complexes raises the important question of what polypeptide sequences within the protein are important for enzymatic activity and for protein-protein interactions. Both enzymatic activity and protein interactions might be anticipated to influence the capacity of RPD3 to direct transcriptional silencing. Two recent studies have examined the requirements for enzymatic activity in transcriptional repression (22, 26). The human RPD3 homolog HDAC1 deacetylates histones in the absence of other cofactors (22). A point mutation, H141A, of HDAC1 lowers deacetylase activity sixfold and repressive activity sevenfold when activity is assayed with HDAC1 fused to a heterologous DNA binding domain. This reduction in activity occurs without influencing association with SIN3 or RbAp48 (22) (see below). In yeast, a point mutation at the same site in RPD3 eliminates enzymatic activity, but reduces repressive activity only twofold when fused to a heterologous DNA binding domain, again without influencing association with SIN3 (26). Other mutants which were unable to interact with SIN3 in human cells were indistinguishable from wild-type (wt) RPD3 in coimmunoprecipitation experiments with SIN3 in yeast (22, 26). These studies suggest that deacetylase activity alone has a role in transcriptional repression but that other mechanisms may also contribute to establishing the repressed state in yeast and human cells.
The vertebrate RPD3 homologs have been found in association with two proteins, retinoblastoma (Rb)-associated p48 (RbAp48) (61, 70, 79) and SIN3 (1, 21–23, 25, 33, 34, 45, 58). The SIN3 gene was originally defined in yeast as a negative regulator of HO that contained four paired amphipathic helix motifs (72, 74). Targeting of SIN3 by fusion to a DNA binding domain will direct transcriptional repression (73). Genetic studies indicated a close functional relationship between SIN3 and the RPD3 deacetylase (68, 69). Subsequent experiments established that SIN3 and the RPD3 deacetylase coexisted in large regulatory complexes (29, 30). The vertebrate connection between SIN3 and transcriptional regulation came from studies on the Mad1 DNA-binding protein (6, 7, 12, 24, 31). The Mad-Max heterodimer represses transcription and suppresses cell transformation (8, 12, 24, 54). The Mad-Max heterodimer mediates these functions in large part through the recruitment of SIN3 and the mammalian RPD3 homologs HDAC1 and 2 (1, 6, 21, 31). SIN3 has subsequently been shown to recruit the RPD3 deacetylase to regulatory sites targeted for transcriptional silencing by association with a wide variety of other DNA-binding proteins and corepressors (1, 21–23, 25, 44, 45, 58). However, deacetylase complexes exist that are deficient in SIN3 (70) and direct interactions of transcriptional repressors YY1 and Rb have been suggested to occur with the deacetylase (9, 41, 77).
RbAp48 was originally characterized as an Rb-binding protein (51, 52) that cofractionates with HDAC1 (61). Subsequent work has established that RbAp48 and the related protein RbAp46 interact with core histones H2A and H4 (67). The RbAps are present in diverse protein complexes involved in histone deacetylation (61, 79), histone acetylation (49, 67), nucleosome disruption (42, 70), and nucleosome assembly (32, 64, 66). Many studies of deacetylase and transcriptional repression rely on expression of exogenous RPD3 in tissue culture (for example, references 9, 41, 58, and 77). It is often assumed that this exogenous RPD3 is incorporated into large multiprotein complexes with endogenous proteins. Although some of these components can be coimmunoprecipitated, the functional significance of the interaction of exogenous RPD3 with endogenous proteins such as SIN3 and RbAp48 has not been determined for any of these complexes.
Here we examine the requirements for the repression of transcription by exogenous Xenopus RPD3 synthesized from microinjected mRNA in vivo (75). We cloned Xenopus SIN3 (xSIN3) and RbAp48 and show that these proteins are exclusively nuclear. In contrast, endogenous and expressed exogenous RPD3 accumulates in both the oocyte nucleus and the cytoplasm. Expression of exogenous RPD3, but not SIN3 and RbAp48, leads to an increase in nuclear deacetylase activity and represses transcription of the TRβA promoter. Mutagenesis of RPD3 shows that repression is dependent on deacetylase activity and nuclear localization of RPD3 through a C-terminal nuclear localization signal (NLS). Exogenous RPD3 is not incorporated into the endogenous RPD3 deacetylase complexes that we have previously described (25, 70) but cofractionates with a component of RbAp48 in the nucleus. Coimmunoprecipitation experiments show that RbAp48 forms N- and C-terminal contacts with RPD3 and that RbAp48 is also coimmunoprecipitated with SIN3. In addition, Xenopus RbAp48 shows a direct interaction with histone H4 in vivo. Our results establish that exogenous Xenopus RPD3 is competent to deacetylate histones and silence transcription in Xenopus oocyte nuclei without being incorporated into the large endogenous histone deacetylase complexes. We suggest that RPD3 may be targeted to histones by interaction with endogenous RbAp48.
MATERIALS AND METHODS
Cloning.
Subcloning and DNA manipulation were carried out by standard methods (56). RNA was isolated from Xenopus oocytes, embryos, or tissues by using RNAzol B as recommended by the manufacturer (Tel-Test, Inc.). Northern blot analyses were carried out by standard methods (56). A portion of the Xenopus laevis RbAp48 cDNA (corresponding to amino acids [aa] 295 to 387 of human RbAp48) was cloned from total Xenopus liver RNA by standard reverse transcription-PCR techniques, using the following degenerate oligonucleotides: 5′GACAAGACTGTTGC(A/C)(C/T)T(G/T/C)TGGGA(T/C)3′ (sense) and 5′AGGTTCATTGGG(A/G)TTCCA(G/A)G(A/T)(G/A)AA3′ (antisense).
Full-length cDNAs were isolated from a Xenopus oocyte cDNA library in λZAP. More than 30 cDNAs containing the entire protein coding sequence were obtained following phage screening. Several clones were sequenced in their entirety on both strands. For the cloning of xSIN3A, degenerate oligonucleotides were used to generate a reverse transcription-PCR product for library screening. First-strand cDNA was generated by using primer SIN3-3 (5′-CTGGTATGTRTGCARRATYTCCARRAA) and SuperScript II reverse transcriptase (Life Technologies, Inc.). PCR was performed with Taq DNA polymerase (Promega, Inc.) between primers SIN5-1 (5′GCCCTGTCCTAYCTRGAYCAGGTRAAR) and SIN3-3 to generate a 632-bp product. This fragment was radiolabeled by the random priming method (Amersham Redi-Prime kit) and used to screen a unidirectional λ phage oocyte cDNA library created with a Uni-Zap kit (Stratagene). Positive plaques were rescreened through tertiary platings, and single positive clones were inserted into pBluescript as instructed by the manufacturer. The first xSIN3 clone, xSIN3-1 (aa 28 to 407), was used to rescreen the library, with two new clones isolated, xSIN3-2 (98-bp untranslated region [aa 327]) and xSIN3-3 (aa 358 to 936). xSIN3-3 was used to rescreen the library, and xSIN3-4 (aa 847 [323-bp 3′ untranslated region]) was isolated. The four overlapping clones were sequenced and ligated together, using unique restriction enzyme sites (NdeI, BclI, and NsiI). The resulting clone, encoding the entire xSIN3A coding sequence, was cloned into pT7TS for in vitro transcription.
RPD3 and RbAp48 constructs for in vitro transcription were generated in plasmid pMSII, which had been obtained from pSP64 (Promega) by insertion of a linker in the unique EcoRI site downstream of the poly(A) stretch. This modification provided additional sites for linearization prior to in vitro transcription (modified vector kindly donated by Melissa Stolow). Constructs expressing deletion mutants of RPD3 were generated from the full-length Xenopus cDNA sequence in pMSII (75) by PCR using Vent DNA polymerase (New England Biolabs). PCR primers contained restriction sites (BamHI for 5′ primers and SacI for 3′ primers) for subcloning back into the RNA production vector pMSII. Primers used for generating other constructs were as follows: for pRPD3(134-347), D.R10SP (5′-GCGCG GATCC AAAGA TGTGG TCTGG TGGCC TTCAT CATGC A-3′) and D.R18S (5′-GGGGG AGCTC TCATG GACTG ATGTG AAGTT TGAAG TC-3′); for pRPD3(1-135), D.R5 (5′-GCGCG GATCC AAAGA TGGCG CTGAG TCAAG GA-3′) and D.R19S (5′-GGGGG AGCTC TCACC AGTTC ACTGA AATGT CCGTC TG-3′); for pRPD3(1-347), D.R5 and D.R18S; and for pRPD3(134-480), D.R10SP and D.R6 (5′-GGGGG AGCTC TCAGA CTGAT TTGGT CTCTT CTTTT ACCCG TTTGC T-3′). A construct to produce full-length, N-terminally FLAG-tagged RPD3 protein [pFNRPD3(wt)] was generated from pRPD3(wt) by using primers D.R24 (5′-TCTAG AGGAT CCAAA GATGG ACTAC AAGGA CGACG ATGAC AAGGC GCTGA GTCAA GGAAC AAAGA AG-3′) and D.R6. A full-length, C-terminally FLAG-tagged RPD3 construct [pFCRPD3(wt)] was obtained from pRPD3(wt) by using primers D.R5 and D.R28 (5′-TTGGG AGCTC TCACT TGTCA TCGTC GTCCT TGTAG TCGAC TGATT TGGTC TCTTC TTTTA CCCG-3′). An RPD3 point mutant was obtained by using a Promega Gene Editor in vitro site-directed mutagenesis kit with primer D.M1 (5′-GGTGG CCTTC ATGCT GCCAA GAAAT C-3′) to mutagenize His 141 to Ala [construct pRPD3(H141A)]. RbAp48 constructs in pMSII were generated from the Xenopus RbAp48 cDNA after PCR amplification with primers containing HindIII (5′) or BamHI (3′) restriction sites. Full-length RbAp48-encoding construct pRbAp48(wt) was obtained by using primers D.P1 (5′-GGCCA AGCTT AAAGA TGGCT GATAA AGAAG CTGCG TTC-3′) and D.P2 (5′-CGCGG GATCC TTAGG AACCT TGACC CTCTG GATC-3′); N- and C-terminally FLAG-tagged RbAp48-expressing constructs pFNRbAp48 and pFCRbAp48 were obtained by using primer pairs D.P7 (5′-GAATA CAAGC TTAAA GATGG ACTAC AAGGA CGACG ATGAC AAGGC TGATA AAGAA GCTGC GTTCG AT-3′)-D.P2 and D.P1-D.P8 (5′-CTCGC CCGGG GATCC TTACT TGTCA TCGTC GTCCT TGTAG TCGGA ACCTT GACCC TCTGG ATCAA C-3′), respectively; and N- and C-terminally deleted RbAp48 constructs pRbAp48(53-425) and pRbAp48(1-405) were generated by using primer pairs D.P2-D.P5 (5′-GGGGA AGCTT AAAGA TGGTT ACCAG ACCCG ATGGG AAAGA T-3′) and D.P1-D.P6 (5′-GGGGG GATCC TTACGC CATTT GCCAG ACCTG CATGA T-3′), respectively. Constructs for the TRβA promoter and for producing mRNA encoding N-terminally FLAG-tagged core histones and H4 deletions have been described elsewhere (16, 76).
Microinjection and processing of oocytes.
Oocytes were prepared and maintained as described elsewhere (2, 3). 5′-capped and 3′-polyadenylated mRNA was produced with an Ambion message machine kit for SP6 polymerase after linearization of pMSII DNA constructs with an appropriate unique restriction enzyme. mRNA dissolved in water was microinjected into the oocyte cytoplasm (2, 3). The amount of mRNA injected varied from less than 1 ng to 2 ng (as specified in figure legends). At least 10 but usually 25 oocytes or fertilized eggs were microinjected for each condition. Oocytes were maintained for the required incubation periods in the presence or absence of [35S]methionine (10−2 mCi/ml) to allow mRNA translation, resulting in [35S]methionine-labeled or unlabeled proteins. For transcription assays, unlabeled oocytes were microinjected with 1 ng or less of double-stranded or single-stranded plasmid containing the TRβA promoter and maintained overnight before collection. Primer extension was carried out as described elsewhere (75). Primer I (5′-ATCCT TATAA ACGGT GAGTA GTGAT GTCAT-3′) yields a 107-nucleotide extension product from the TRβA promoter (76). An H4 primer (5′-GAGGC CGGAG ATGCG CTTGA C-3′) annealed to the endogenous H4 transcript gives a 182-nucleotide extension product which serves as an internal control for RNA recovery and loading. For protein analysis, whole oocytes, oocyte cytoplasm, or nuclei were collected and immediately homogenized in buffer A(150) (20 mM HEPES [pH 7.5], 0.15 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 5 mM β-glycerophosphate, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 2 μg of pepstatin A per ml) before freezing. For collection of nuclei, oocytes were transferred into 10 mM Tris-HCl (pH 8)–10 mM MgCl2, dissected manually, immediately homogenized in buffer A(150), and frozen. Soluble oocyte homogenates were prepared from whole oocytes and cytoplasm of nuclei homogenized in buffer A(150) by collecting the clear, middle layer and leaving the yolk and pigment granules behind after centrifugation at 14,000 rpm in Eppendorf tubes for 10 min at 4°C. These extracts were used for immunoprecipitation, histone deacetylation assays, or protein fractionation or were diluted with sodium dodecyl sulfate (SDS) loading dye and electrophoresed directly on SDS-polyacrylamide gels. [35S]methionine-labeled proteins were visualized and quantitated with a PhosphorImager using the ImageQuaNT software (Molecular Dynamics).
Protein fractionation.
Extracts from 150 to 200 whole oocytes, oocyte cytoplasm, or nuclei in a total volume of no more than 300 μl were loaded onto sucrose gradients [5 to 20% in buffer A(150); formed with a Biocomp Gradient Master] and centrifuged at 41,000 rpm for 28 h (Sorvall SW41) at 4°C. Gradients were fractionated into 24 fractions of approximately 500 μl each and stored at −70°C. Fractions were assayed for deacetylase activity or precipitated with 2 volumes of cold acetone and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Immunoprecipitation.
Protein homogenates from 30 or more [35S]methionine-labeled oocytes were adjusted to 250 μl in buffer A(150) containing 0.02% Nonidet P-40. Homogenates were rotated at 4°C for 1 h with 20 μl of FLAG M2 beads (Kodak) and centrifuged briefly, and the supernatant was removed. Beads were washed three times with 500 μl of cold buffer A(150) containing 0.02% Nonidet P-40, boiled in 15 to 20 μl of SDS loading buffer, and electrophoresed on SDS–10% polyacrylamide gels.
Histone deacetylation assays.
Histone deacetylase activity was measured by incubating protein fractions with free core histones (1 μg, 12,000 cpm) which had been acetylated in vitro with recombinant Hat1p and [3H]acetyl coenzyme A and then purified on a Biorex 70 column (25, 70). The final volume of the incubation mixture was 200 μl. After 1 h at 30°C, the reaction was quenched with 50 μl of 0.1 M HCl–0.1 M acetic acid and extracted with 600 μl of ethyl acetate; 450 μl of the sample was counted by liquid scintillation. Deacetylation activity is reported as counts per minute of tritium (released acetate).
Western blots.
SDS-polyacrylamide gels were incubated for 10 min in Transfer buffer (0.039 M glycine, 0.048 M Tris-HCl [pH 8], 15% methanol, 0.037% SDS) and transferred to nitrocellulose membranes (Hybond ECL; Amersham) in a semidry blotting apparatus (Semi-Phor; Hoefer Scientific Instruments) for 90 min at 10 mA/cm2. Transfer was verified by staining the blot with Ponceau S solution (Sigma) before blocking for 1 h at room temperature or overnight at 4°C in 10% nonfat dry milk–0.3% Tween–1× PBS (phosphate-buffered saline [56]). The blot was rinsed with water and incubated for 1 h at room temperature or overnight at 4°C in primary antibody solution. Primary antibodies were diluted to the required, empirically determined working concentration in 1× PBS–0.2% Tween–10% fetal calf serum. Anti-RPD3, SIN3, and RbAp48 antibodies were raised against Xenopus proteins expressed in bacteria (25, 70). Anti-FLAG tag monoclonal antibody M2 was purchased from Kodak. The blot was rinsed with water, washed three times for 10 min in washing solution (1× PBS, 0.3% Triton X-100, 0.5 M NaCl), incubated in secondary antibody solution (7 μl of anti-rabbit or anti-mouse immunoglobulin, horseradish peroxidase-linked whole antibody [Amersham] per 25 ml of 1% milk–1× PBS–0.02% Tween) for 40 min to 1 h at room temperature, and washed three more times as before. Visualization was through chemiluminescence using an equal volume of luminol enhancer and peroxide solution (Supersignal chemiluminescence reagents; Pierce). Western blots were stripped by two washes of 10 min each in 0.05% milk–1× PBS–0.2% Tween at room temperature followed by a 30-min wash in 2% SDS–100 mM β-mercaptoethanol–62.5 mM Tris-HCl (pH 6) at 70°C.
Nucleotide sequence accession numbers.
The xSIN3A and Xenopus RbAp48 cDNAs have been assigned GenBank accession no. AF154112 and AF073787, respectively.
RESULTS
Xenopus RPD3, SIN3, and RbAp48 differ in distribution between the oocyte nucleus and cytoplasm.
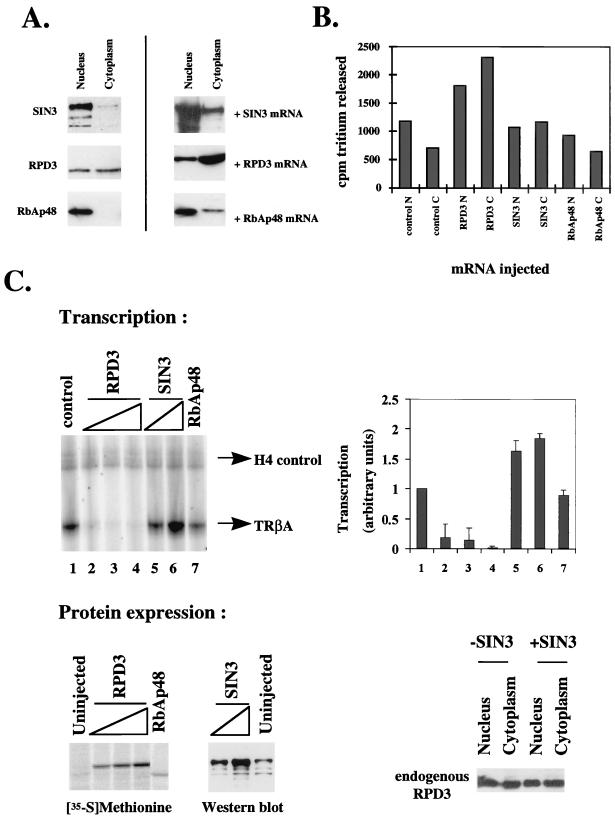
In earlier work, we isolated a complex which includes RPD3, RbAp46/48, and stoichiometric quantities of either SIN3 and MeCP2 (25) or the Mi-2 ATPase from Xenopus egg extracts (70). MeCP2 interactions with SIN3 and deacetylase have been identified in mammalian cells (45). The abundant Mi-2 ATPase complex which possesses both nucleosome remodeling and deacetylase activity (62, 70) has also been characterized in mammalian cell extracts (62, 80). To better define the properties and activities of Xenopus RPD3, RbAp48, and SIN3, we made use of antibodies (25, 70) to examine the distribution of the endogenous proteins in nuclear and cytoplasmic homogenates prepared by manual dissection of Xenopus oocytes. We find that xSIN3 and Xenopus RbAp48 are almost entirely nuclear in the Xenopus oocyte, whereas RPD3 is equally distributed between nucleus and cytoplasm (Fig. 1A). We expressed exogenous RPD3 in oocytes following the injection of mRNA encoding the protein and found that the levels of both nuclear and cytoplasmic RPD3 were elevated, with cytoplasmic levels exceeding those in the nucleus (Fig. 1A, +RPD3 mRNA). Exogenous RPD3 accumulates preferentially in the cytoplasm at low levels of expression (data not shown). Expression of SIN3 or RbAp48 also leads to elevated nuclear as well as cytoplasmic protein levels (Fig. 1A, +SIN3 mRNA and +RbAp48 mRNA). We conclude that endogenous Xenopus RPD3 is present in nuclear and cytoplasmic pools whereas RbAp48 and SIN3 are predominantly nuclear (Fig. 1A). The levels of RPD3, RbAp48, and SIN3 are significantly increased in the oocyte nucleus and cytoplasm following microinjection of the respective mRNAs into the cytoplasm (Fig. 1A).
FIG. 1.
Nuclear distribution, transcriptional repression, and deacetylase activity of RPD3, SIN3, and RbAp48 in Xenopus oocytes. (A) Nuclear and cytoplasmic distribution of SIN3, RbAp48, and RPD3. Uninjected oocytes or oocytes which had been microinjected with SIN3, RPD3, or RbAp48 mRNA (+mRNA) were manually separated into nuclei and cytoplasm. Soluble material of five oocyte nuclei or cytoplasm equivalents was analyzed by Western blotting after SDS-PAGE. Antibodies were against SIN3, RbAp48, and RPD3. Endogenous proteins were all detected on the same Western blot. (B) Deacetylase activity of oocytes in the presence or absence of exogenous RPD3, SIN3, or RbAp48. Oocytes were uninjected (control) or microinjected with mRNA encoding RPD3, SIN3, or RbAp48, resulting in similar levels of expression of exogenous protein. Oocytes were manually dissected into nuclei (N) and cytoplasm (C), and equivalent oocyte homogenates were assayed for deacetylase activity. The error in the deacetylase assays is ±5%. (C) Exogenous RPD3 but not SIN3 or RbAp48 represses transcription from the TRβA promoter in oocytes. Oocytes were microinjected with 0.5, 1, and 1.5 ng of mRNA for RPD3 (lanes 2, 3, and 4), 1 and 1.5 ng of mRNA for SIN3 (lanes 5 and 6), or 1.5 ng of mRNA for RbAp48 (lane 7). Oocytes were maintained for 6 h to allow translation and then microinjected with 0.5 ng of double-stranded DNA for TRβA promoter. After overnight incubation, the transcript levels were analyzed by primer extension (top left). H4 serves as an internal control. The primer extension results were quantitated with a PhosphorImager and are represented graphically (top right). Protein expression was verified by labeling with [35S]methionine (RPD3 or RbAp48) or by Western blotting (SIN3) (bottom left). The distribution of endogenous RPD3 between the nucleus and cytoplasm in the presence (+SIN3) or absence (−SIN3) of exogenous SIN3 was determined by detection of RPD3 by Western blotting (bottom right).
Expression of exogenous Xenopus RPD3, but neither SIN3 nor RbAp48, leads to elevated histone deacetylase activity in the Xenopus oocyte nucleus and cytoplasm.
Next we wished to determine whether expression of Xenopus RPD3, SIN3, or RbAp48 leads to a change in deacetylase activity in the oocyte nucleus or cytoplasm. In the deacetylase assay, we used histones which had been enzymatically acetylated by recombinant yeast Hat1p (25, 70). We find that both nuclear and cytoplasmic deacetylase levels increase in the presence of exogenous Xenopus RPD3 (Figure 1B; compare lane 1 with lane 3 and lane 2 with lane 4), whereas deacetylase activity remains unchanged in the presence of exogenous RbAp48 (lanes 7 and 8). Exogenous SIN3 leads to a slight increase in cytoplasmic deacetylase activity (compare lanes 2 and 6). We conclude that exogenous Xenopus RPD3, but neither SIN3 nor RbAp48, is the limiting component determining deacetylase activity in Xenopus oocytes.
Exogenous Xenopus RPD3, but neither SIN3 nor RbAp48, directs transcriptional repression in the oocyte nucleus.
We have previously shown that Xenopus RPD3 synthesized from mRNA microinjected into the cytoplasm of Xenopus oocytes represses transcription of the TRβA promoter microinjected into Xenopus oocyte nuclei (75). Under these conditions, transcriptional repression is established in the absence of unliganded thyroid hormone receptor and represents the repression of basal transcription dependent only on chromatin assembly. In contrast to the 12-fold transcriptional repression of the TRβA promoter established by exogenous Xenopus RPD3 with this protocol (Fig. 1C; upper panel, lanes 1 to 4) (75), we observe a twofold increase in transcription by exogenous SIN3 and no change in transcription in the presence of exogenous RbAp48 (compare lanes 1, 4, 6, and 7). RPD3 and RbAp48 synthesis was assayed by radiolabeling, and SIN3 levels were analyzed by immunoblotting (Fig. 1C, lower panels). We conclude that exogenous RPD3, but neither SIN3 nor RbAp48, directs repression of transcription in the oocyte. The repression of transcription by exogenous RPD3 (Fig. 1C) is coincident with the increase of RPD3 protein in the nucleus (Fig. 1A). In Fig. 1B, we noted an apparent increase in the cytoplasmic deacetylase level in the presence of exogenous SIN3 (lane 6). It was possible that the increase in transcription in the presence of exogenous SIN3 could be explained by titration of endogenous RPD3 into the cytoplasm. However, the distribution of endogenous RPD3 between the nucleus and the cytoplasm remains the same in the presence or absence of exogenous SIN3 (Fig. 1C, lower panels). It remains possible that SIN3 titrates a deacetylase other than RPD3 into the cytoplasm. Since SIN3 appears to be a constituent of several activating and repressive complexes within the oocyte nucleus (25), the modest increase in transcription in the presence of exogenous SIN3 is probably due to an interaction with an activator or titration of a repressor other than endogenous RPD3.
Cloning and sequence comparison of xSIN3A and RbAp48.
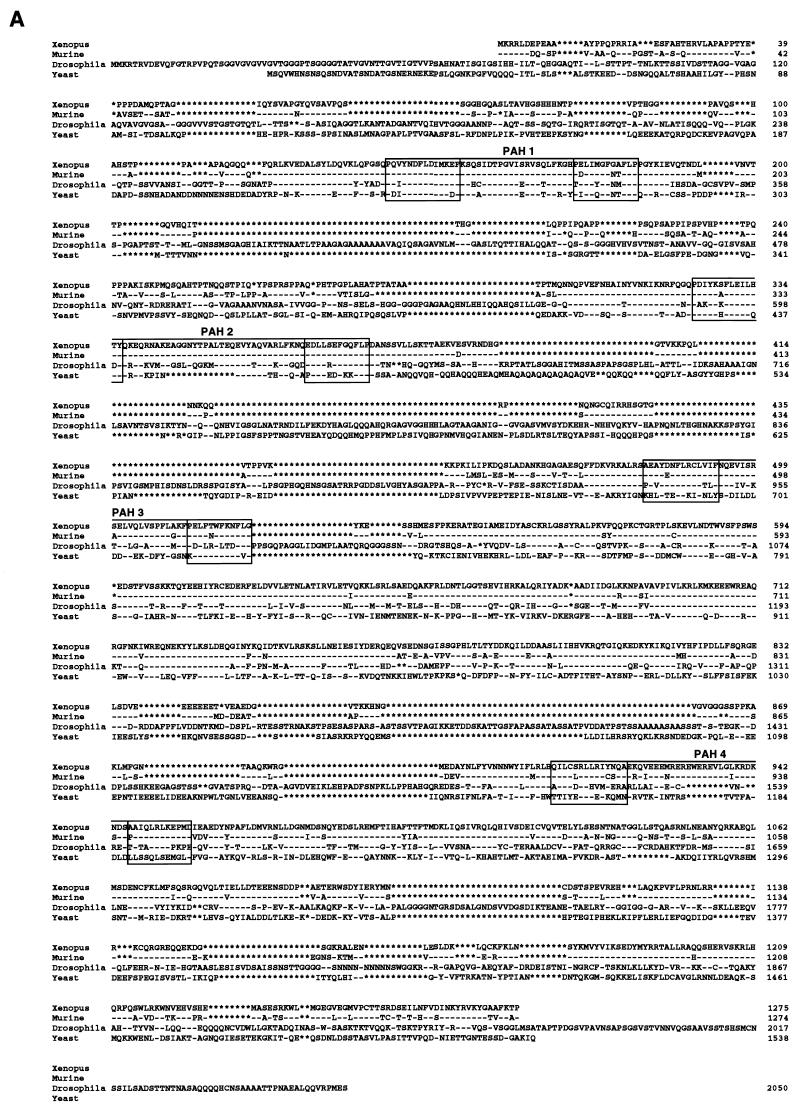
To further characterize the components of a functional deacetylase complex in Xenopus oocytes, we cloned the xSIN3A and RbAp48 cDNAs from an oocyte library. A full-length cDNA for the xSIN3A protein was cloned from an X. laevis oocyte cDNA library. This cDNA was pieced together from four overlapping cDNA fragments and consists of an open reading frame with the capacity to encode a 1,276-aa protein with a molecular mass of 150 kDa. The predicted protein sequence for xSIN3A shows very high homology with the murine SIN3A (mSIN3A) protein (Fig. 2A) (6, 53) (75% amino acid identity, 81% similarity) throughout the protein, with the four paired amphipathic helices (PAH1 to PAH4) exhibiting extremely high conservation (94, 98, 91, and 83% identity, respectively) (Fig. 2C). Across species, xSIN3A, mSIN3A, Drosophila melanogaster SIN3 (dSIN3), and Saccharomyces cerevisiae SIN3 show an overall high conservation in PAH1 (68% identity; 81% similarity), PAH2 (50% identity; 63% similarity), and PAH3 (23% identity; 55% similarity). However, the hydrophobicity of residues at the critical positions 3, 7, 8, 10, and 14 of helix A and positions 3, 7, and 10 of helix B for each pair is totally conserved across species with one exception (in dSIN3 PAH2, position A3 is lysine). This high level of conservation suggests that the xSIN3A protein may have many of the functional properties of the other described Sin3 proteins.
FIG. 2.
Sequence comparison of xSIN3A and Xenopus RbAp48/p46 with family members from other organisms. (A) Comparison of the deduced sequence of xSIN3A with sequences of mSIN3A dSIN, and yeast SIN3A (6, 50, 74). PAH1 to PAH4 are boxed. Asterisks represent gaps and dashes identities in the sequence comparisons. (B) Manual alignment of the deduced sequence of the Drosophila and human RbAp proteins with the X. laevis sequence. WD repeats were deduced from the consensus derived from mammalian β-transducin (59, 71).
The deduced amino acid sequence of the Xenopus RbAp48 cDNA is 97% identical to the human ortholog (51). As previously noted for yeast, Drosophila, and mammalian RbAp48/46 homologs (42, 49, 51, 52, 64), X. laevis RbAp48 contains seven copies of the WD repeat sequence motif and is predicted to form a β-propeller structure similar to that of mammalian β-transducin (59, 71). Interestingly, the RbAp48/46 family differs slightly from that of β-transducin by the number of intervening amino acids between individual WD repeats (Fig. 2B; note in particular WD repeats 2 and 7). These regions are predicted to be solvent exposed, and we speculate that they may be important in protein-protein interactions. Several attempts to isolate an oocyte cDNA corresponding to Xenopus RbAp46 were unsuccessful despite the recovery of more than 30 full-length RbAp48 clones (data not shown).
Nuclear localization, transcriptional repression, and histone deacetylase properties of Xenopus RPD3.
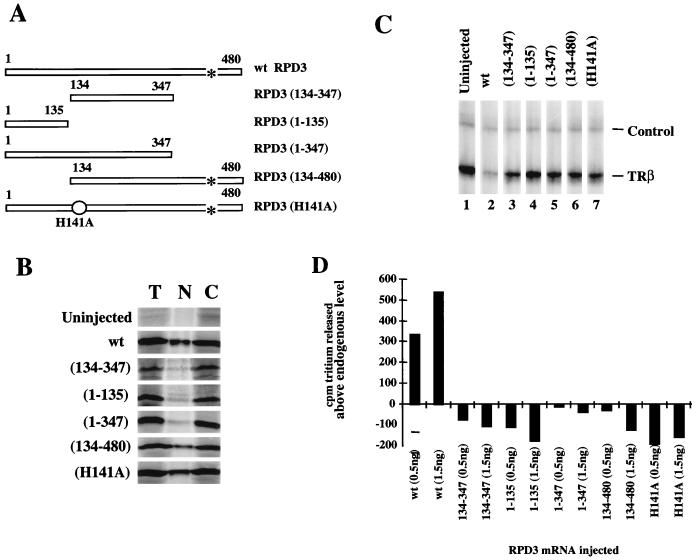
Earlier work using inhibitors and point mutations of histone deacetylase had demonstrated the importance of enzymatic activity for transcriptional repression (21, 22, 26, 75). We have shown that injection of mRNA encoding Xenopus RPD3 leads to an increase in total RPD3 levels and deacetylase activity in the nucleus and cytoplasm, correlating with increased transcriptional repression (Fig. 1 and reference 75). Since the capacity of RPD3 to gain access to the nucleus may contribute to variation in deacetylase activity and transcriptional repression, we investigated the nuclear localization of RPD3 mutants in Xenopus oocytes. The RPD3 mutants used in our experiments (Fig. 3A) are all efficiently translated in Xenopus oocytes following injection of mRNA into the cytoplasm (Fig. 3B). Nuclear uptake varies widely, with the H141A point mutation and N-terminal deletion mutant [RPD3(134-480)] being taken up into the nucleus with near-wt efficiency (Fig. 3B). The other mutants in which the C terminus is deleted remain mainly in the cytoplasm. We conclude that nuclear localization of RPD3 depends on a C-terminal NLS. Inspection of the amino acid sequence reveals a candidate NLS (KKVKRVK [75]).
FIG. 3.
Comparison of wt and mutant RPD3. (A) Schematic representation of RPD3 mutants. wt RPD3 is the full-length 480-aa protein (75). Numbers in parentheses indicate amino acids present in the deletion mutants. A point mutation of His to Ala at position 141 is indicated by a circle. A putative NLS (aa 438 to 444) is indicated by an asterisk. (B) Expression and nuclear localization of RPD3 constructs in oocytes. Oocytes were microinjected with 1.5 ng of mRNA encoding wt or mutant RPD3. Two oocyte equivalents of [35S]methionine-labeled protein from total oocytes (T), nuclei (N), or cytoplasm (C) were analyzed by SDS-PAGE. (C) Transcriptional repression of the TRβA promoter by wt and mutant RPD3. Oocytes were microinjected with 0.5 or 1.5 ng of mRNA, maintained for 6 h to allow translation, and then microinjected with 0.5 ng of double-stranded DNA for the TRβA promoter. After overnight incubation, the transcript levels were analyzed by primer extension. H4 serves as an internal control. (D) Histone deacetylase activity of wt and mutant RPD3 in oocyte nuclear homogenates. Nuclear oocyte extract (10 μl [one oocyte equivalent]) was assayed for histone deacetylation activity (released acetate measured as counts per minute of tritium). The level of released tritium in the absence of injected mRNA (1,100 cpm) was subtracted to yield the values shown. The error of the deacetylase assays is ±5%.
None of the mutant RPD3 proteins have the capacity to silence transcription from the TRβA promoter (Fig. 3C, lanes 3 to 7), nor do they enhance nuclear histone deacetylase activity (Fig. 3D). In contrast, expression of wt RPD3 both silences the TRβA promoter (Fig. 3C; compare lanes 1 and 2) and enhances nuclear histone deacetylase activity (Fig. 3D). Point mutations of the histone deacetylase HDAC1 have been shown to influence association with RbAp48 and SIN3 (22). The point mutant H141A, which retains the ability to associate with RbAp48 and SIN3 (22), neither silences transcription nor augments nuclear deacetylase in Xenopus oocytes (Fig. 3B to D). Our results substantiate the conclusion that the catalytic activity of histone deacetylase is a key component in establishing transcriptional repression (22, 26).
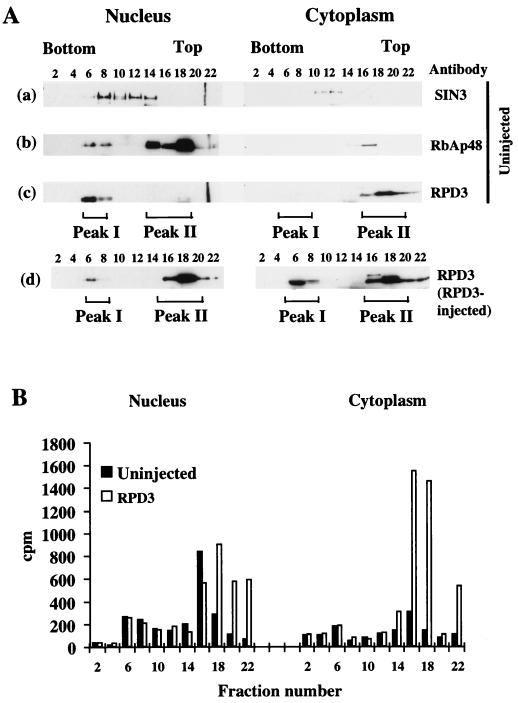
Exogenous RPD3 is not incorporated into the endogenous nuclear RPD3 complex but cofractionates with a component of the endogenous RbAp48.
Exogenous RPD3 is usually assumed to be incorporated into large complexes containing SIN3, RbAp48, or other proteins which target the deacetylase to specific promoters which are subsequently transcriptionally repressed. We therefore wished to determine whether the exogenous RPD3 which represses transcription in Xenopus oocytes functions within the large nuclear histone deacetylase complexes which we have previously purified (70). We first examined the distribution of endogenous SIN3, RbAp48, and RPD3 within nuclear and cytoplasmic homogenates that were fractionated on sucrose gradients. We find that the endogenous nuclear RPD3 and RbAp48 cofractionate with a component of the endogenous nuclear SIN3 in peak I (Fig. 4A, fractions 6 to 8), toward the bottom of the sucrose gradient. The large Mi-2 deacetylase complex that we have previously isolated from egg extracts migrates at an identical position on sucrose gradients, with a molecular mass of approximately 1 to 1.5 MDa (70). The MeCP2-SIN3 deacetylase complex also migrates as a very large complex of approximately 700 kDa (25). Very little nuclear SIN3 cofractionates with endogenous RbAp48 in peak II toward the top of the nuclear gradients (Fig. 4A, fractions 16 to 18). In contrast, the endogenous cytoplasmic RPD3 is almost entirely in fractions toward the top of the sucrose gradient with an apparent molecular size of 50 to 200 kDa. Assays for histone deacetylase activity (Fig. 4B; note that activity derives both from RPD3 homologs and distinct deacetylases [unpublished observations]) indicate that most of the endogenous deacetylase activity is nuclear and is at the top of the sucrose gradient (fractions 16 to 18). We find that exogenous RPD3 is not incorporated into the large peak I complex in the nucleus but instead accumulates predominantly at the top of the sucrose gradients both in the cytoplasm and in the nucleus (Fig. 4A). In the cytoplasm, the exogenous RPD3 accumulates in both peak I and peak II with an even distribution. The reason for the accumulation of exogenous RPD3 in the cytoplasmic peak I is unknown. The absence of endogenous RPD3 in peak I and the absence of any increase in enzymatic activity in peak I indicates that this accumulation is nonphysiological and perhaps due to aggregation. Note that RPD3 was untagged and detected with an anti-RPD3 antibody, and so the signal is the sum of endogenous and expressed proteins. The presence of exogenous RPD3 in the peak II fractions is consistent with the distributions of elevated histone deacetylase activity (Fig. 4B). We conclude that exogenous RPD3 is not incorporated into either the endogenous nuclear Mi-2 complex or MeCP2-SIN3 complexes. Expression of RPD3 leads to the enhancement of histone deacetylase activity in nuclear and cytoplasmic compartments (Fig. 4). However, it is only in the nucleus that interactions of RPD3 with SIN3 and RbAp48 may occur (Fig. 1). Although some of the endogenous nuclear RbAp48 present in peak II is known to be involved in interactions with other proteins (24a), the cofractionation of exogenous RPD3 and RbAp48 suggests that these proteins may be associated in vivo in the nucleus. The expression of exogenous RPD3 did not alter the distribution of RbAp48 between peak I and peak II fractions in the oocyte nucleus (65a). We interpret this observation to indicate that RbAp48 is stably sequestered into the large macromolecular complexes present in peak I, such as the Mi-2 deacetylase (70).
FIG. 4.
Distribution of nuclear and cytoplasmic RPD3, RbAp48, SIN3, and histone deacetylation activity on sucrose gradients. (A) Fractionation of nuclear and cytoplasmic endogenous RPD3, SIN3, and RbAp48 and of exogenous RPD3 on sucrose gradients. Extracts from nuclei and cytoplasm of uninjected oocytes or oocytes which had been microinjected with RPD3 mRNA were fractionated on sucrose gradients (200 oocyte equivalents per gradient). One-third of every second fraction was analyzed by Western blotting with the indicated antibodies. SIN3, RPD3, and RbAp48 were detected on the same blot for uninjected oocytes. Size markers indicated that fractions 8, 12, and 16 corresponded to 669, 443, and 150 kDa, respectively (25). (B) Expressed RPD3 cofractionates with increased histone deacetylase activity in nuclei and cytoplasm. Histone deacetylation activity of 7 μl of every second fraction was determined in uninjected and RPD3-injected nuclear and cytoplasmic fractions. Released acetate as counts per minute of tritium is indicated by numbers on the y axis. The error of the deacetylase assays is ±5%.
Interactions between Xenopus RPD3, RbAp48, and SIN3.
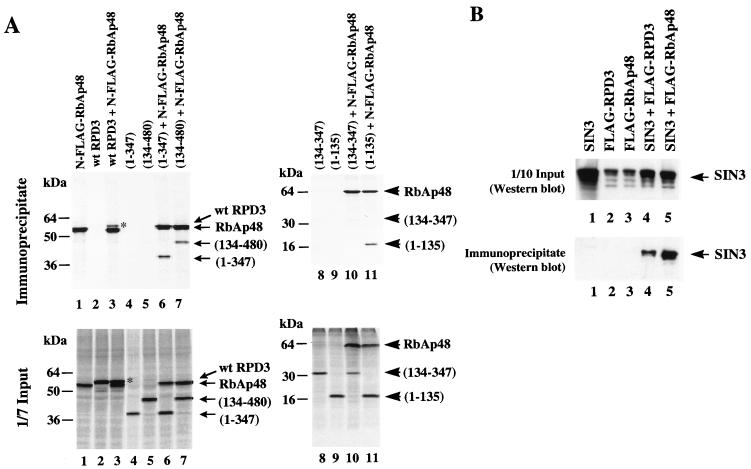
Hassig and colleagues have coexpressed SIN3, RbAp48, and HDAC1 in mammalian cells and have established that these proteins coimmunoprecipitate (22). However, point mutations of HDAC1 that inhibit association with RbAp48 also inhibit association with SIN3. Thus, it remains possible that SIN3 interacts with both RbAp48 and HDAC1 and is necessary to hold these proteins in a common complex. RbAp48 and HDAC1b can be coimmunoprecipitated from cell extracts expressing the proteins simultaneously, but not if cell extracts in which the proteins were separately expressed were mixed (81). Our data so far suggest that exogenous Xenopus RPD3 and endogenous RbAp48 potentially interact in vivo (Fig. 4A). We next examined the requirements for interaction between RPD3 and RbAp48 directly. mRNAs encoding RPD3 and N-terminally FLAG-tagged RbAp48 were translated individually or in combination following microinjection into Xenopus oocyte cytoplasm (Fig. 5A, lanes 1 to 5). These proteins accumulate in the nuclear peak II fractions as previously described (65a) (Fig. 4A). We find that both N- and C-terminally FLAG-tagged RbAp48 interact with RPD3 (Fig. 5A and data not shown). Repeated attempts to immunoprecipitate RbAp48 with either an N- or a C-terminally FLAG-tagged RPD3 protein were unsuccessful, probably due to interference of the FLAG tag with RbAp48 binding (81).
FIG. 5.
Interactions between RPD3, RbAp48, and SIN3. (A) RPD3 is coimmunoprecipitated with FLAG-tagged RbAp48. mRNAs encoding N-terminally FLAG-tagged RbAp48 and wt RPD3 or RPD3 deletion mutants were microinjected in the indicated combinations, and immunoprecipitation was carried out with anti-FLAG antibodies. RPD3 deletion mutants are indicated by amino acid numbers present in the resultant truncated proteins (Fig. 3A). One-seventh of the input extracts and all of the immunoprecipitated [35S]methionine-labeled proteins were analyzed by SDS-PAGE. Full-length RPD3 protein is marked on the upper right-hand corner with an asterisk. (B) SIN3 coimmunoprecipitates with RPD3 and with RbAp48. mRNAs encoding SIN3 and FLAG-tagged RPD3 or RbAp48 were injected as indicated above each lane. One-tenth of the input extracts and anti-FLAG-immunoprecipitated proteins were analyzed by Western blotting with anti-SIN3 antibodies.
We next used the large deletions of RPD3 removing either the C- or N-terminal sequences (Fig. 3A) in immunoprecipitation experiments. We found that both RPD3 deletion mutants retained the capacity to bind to RbAp48 in vivo (Fig. 5A, lanes 6 and 7). The N-terminal 135 aa of RPD3 will stably associate with RbAp48 (Fig. 5A, lane 11); however, removal of both N and C termini from RPD3 abolishes binding (lane 10). Our results indicate that there are multiple contacts between RPD3 and RbAp48, because both N and C termini in RPD3 appear to be important for association with RbAp48.
We examined whether RPD3 and RbAp48 bind to SIN3. As previously reported (21, 33), RPD3 binds to SIN3 (Fig. 5B, lane 4). The FLAG-tagged RPD3 can therefore interact with SIN3 in the absence of interactions with RbAp48, since the FLAG-tagged RPD3 will not interact with RbAp48. We have found that the FLAG-tagged RPD3 is compromised in repression of the TRβA promoter and for enzymatic activity (65a). RbAp48 also coimmunoprecipitates with SIN3 (Fig. 5B, lane 5). In this assay, the sensitivity of antibody detection precludes detection of interactions of endogenous SIN3 with FLAG-RPD3 and FLAG-RbAp48 (Fig. 5B, lanes 2 and 3). We conclude that RPD3 interacts with RbAp48 through N- and C-terminal contacts which may be mediated by endogenous SIN3 in the oocyte.
Specific recognition of core histone H4 by Xenopus RbAp48.
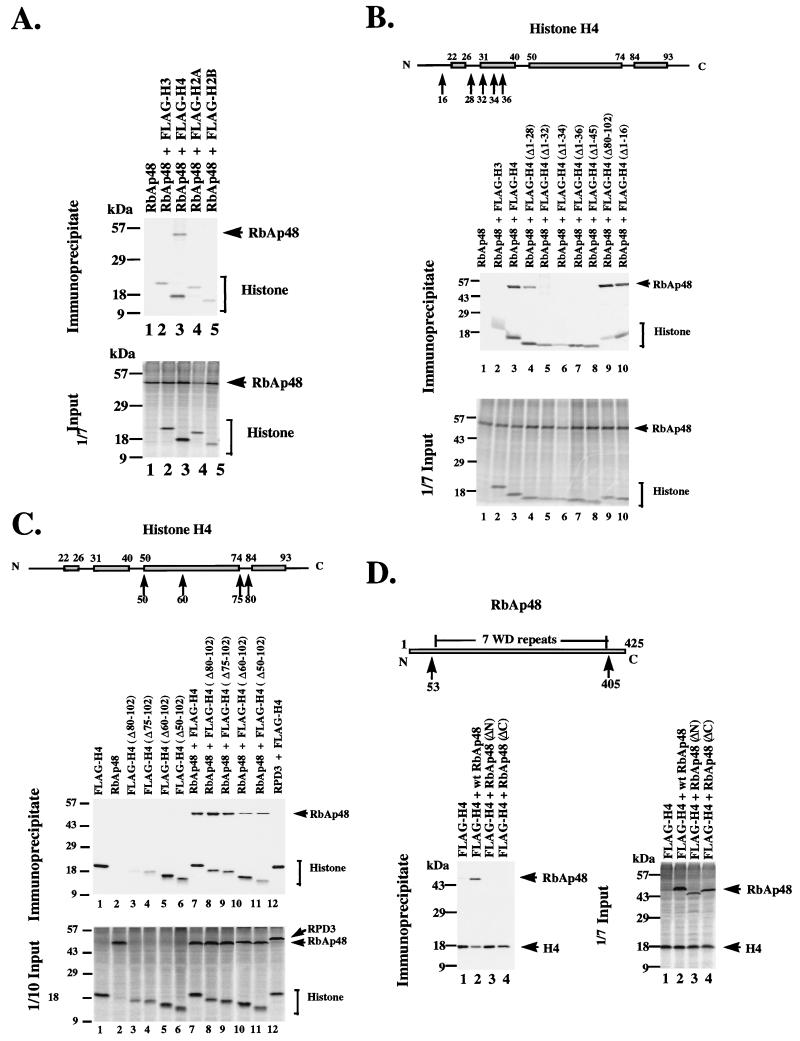
Verreault et al. (67) used in vitro glutathione S-transferase pull-down assays to demonstrate that human RbAp48 interacts with both H2A and H4 but not with histones H2B and H3. These investigators also found similar results with RbAp46 and demonstrated for RbAp46 that key contacts with histone H4 were made between aa 21 and 41 within the N terminus of histone H4. RbAp48 did not recognize the C terminus of histone H4 from aa 35 to 102 or the N terminus in isolation from aa 1 to 34. It was concluded that the α helix in the N terminus of H4 between aa 31 and 41 is recognized by RbAp46 (67). This same α-helical domain is essential for the assembly of histone H4 into chromatin in vivo (16). However, RbAp46 binds to histone acetyltransferase 1 whereas RbAp48 does not in humans (67). Thus, RbAp46 and RbAp48 may differ in their recognition of histone domains. We therefore tested the selectivity of histone association and the sequence requirements for histone recognition in vivo.
We microinjected mRNA encoding Xenopus RbAp48 together with mRNA encoding individual FLAG-tagged core histones into Xenopus oocyte cytoplasm. Immunoprecipitation with antibody against the FLAG epitope shows strong association of RbAp48 with histone H4 (Fig. 6A, lane 3). No significant binding to RbAp48 is detected for H3, H2A, and H2B (lanes 2, 4, and 5). We next examined N-terminal deletions of H4 for association with RbAp48. We find that deletions of the N terminus of H4 to aa 32 severely inhibit association with RbAp48, whereas deletions of the N terminus to aa 28 still allow strong association (Fig. 6B). This result suggests that the segment of the N-terminal tail immediately proximal to the amino-terminal helix of the histone fold domain is important for association with RbAp48. The C-terminal domain of histone H4 has a modest influence on association with RbAp48 (Fig. 6C). Even deletions as extreme as H4 (Δ50-102), in which only the amino-terminal 50 amino acids of H4 remain, bind to RbAp48 (Fig. 6C, lane 11). As a control for the specificity of the association of histone H4 with RbAp48, we examined the capacity of histone H4 to associate with RPD3. Histone H4 does not interact with RPD3 (Fig. 6C, lane 12). Finally, we examined the capacity of histone H4 to associate with N- and C-terminal deletion mutants of RbAp48. We find that an N-terminal deletion of 52 aa or a C-terminal deletion of 20 aa completely prevents association of H4 with RbAp48 (Fig. 6D).
FIG. 6.
Interaction of RbAp48 with core histones. (A) RbAp48 interacts specifically with core histone H4. mRNAs encoding FLAG-tagged core histones were coinjected with RbAp48 mRNA as indicated, and immunoprecipitation was carried out with anti-FLAG antibodies. One-seventh of the input extracts and immunoprecipitated [35S]methionine-labeled proteins were analyzed by SDS-PAGE. (B) RbAp48 interacts with a region in the N-terminal helix of the histone fold motif of H4. Details are as for panel A except that mRNAs encoding FLAG-tagged full-length and N-terminally deleted H4 were coinjected with RbAp48 mRNA as indicated. Deleted amino acids are preceded by Δ. (C) The C-terminal region of H4 up to aa 50 is dispensable for the interaction with RbAp48. Details are as for panel A except that mRNAs encoding FLAG-tagged full-length and C-terminally deleted H4 were coinjected with RbAp48 mRNA as indicated. (D) Removal of either the N- or C-terminal region of RbAp48 abolishes interaction with H4. Details are as for panel A except that mRNA encoding FLAG-tagged full-length H4 was coinjected with full-length and N- or C-terminally deleted RbAp48 mRNA [RbAp48(ΔN) or RbAp48(ΔC)] as indicated. For comparison, the first WD repeat is from aa 55 to 94 and the seventh and last WD repeat is from aa 363 to 403.
In the crystal structure of the heterotrimeric Gαβγ protein, the WD repeats of the β-transducin subunit can be likened to the blades of a propeller (59, 71). The Gα and Gγ subunits bind to two distinct interfaces of Gβ. This propeller structure might be conserved in other WD repeat proteins, allowing them to act as scaffolds for the assembly of multiprotein complexes. If the structure of RbAp48 is conserved with that of the β-transducin WD protein, then the N- and C-terminal domains will exit from the propeller structure assumed by the WD repeats in close proximity (59, 71). In the β-transducin structure, the N-terminal tail forms an α helix which interacts with other subunits. In the case of RbAp48, the interaction region with H4 might be formed by the N- and C-terminal tails of RbAp48, leaving the long loop regions located between WD1 and WD2, WD2 and WD3, and WD6 and WD7 available for interaction with other proteins such as RPD3 (Fig. 2). An alternative explanation could be that removal of the tails disrupts the folding of the protein, but this seems unlikely from comparison with the β-transducin structure. We conclude that the interaction between RbAp48 and the N-terminal α helix of histone H4 is mediated by the N- and C-terminal tails of RbAp48.
DISCUSSION
The SIN3-RbAp48-RPD3 complex.
Our results extend existing information on the function of the RPD3 family of histone deacetylases, SIN3, and RbAp48 in transcriptional repression. We show that deacetylase activity is essential for transcriptional repression by RPD3 in Xenopus oocytes and depends on the nuclear import of exogenous protein through a C-terminal NLS. We find that the network of protein-protein interactions in which SIN3 binds RPD3 (21, 33) and where RbAp48 interacts with histone H4 (67) can be extended by our direct demonstration that RbAp48 interacts with both RPD3 and SIN3 (Fig. 5). Our controls demonstrate that RPD3 does not bind directly to H4 (Fig. 6C). Thus, there is a network of protein-protein interactions, SIN3-RPD3-RbAp48-H4, that might contribute to targeted transcriptional silencing either by the recruitment of SIN3 in association with corepressors such as silencing mediator of retinoid and thyroid receptors (SMRT) and nuclear hormone corepressor (NCoR) (1, 23, 33, 44) or by association with DNA-binding repressors such as Mad-Max and MeCP2 (6, 7, 23, 33, 45). It is also possible that direct interactions of transcriptional repressors with RPD3 recruit deacetylase for silencing (77), or RPD3-RbAp48 might be recruited as an distinct entity, for example, by Rb itself (9, 41).
The functional form of the exogenously expressed histone deacetylase catalytic subunit RPD3.
We find that endogenous pools of RPD3 are equally distributed between nucleus and cytoplasm (Fig. 1). In contrast, endogenous SIN3 and RbAp48 are almost entirely nuclear (Fig. 1). Previously characterized deacetylase complexes present in the oocyte that contain SIN3 and/or RbAp48 are nuclear and migrate in sucrose gradient fractions at sizes greater than 600 kDa (25, 70). This would correspond to fractions at the bottom of the sucrose gradient (i.e., fractions 6 to 10) (Fig. 4). The majority of endogenous nuclear SIN3 and all of the endogenous RPD3 appear to exist in such large complexes in oocyte nuclei (Fig. 4A); however, the endogenous nuclear RbAp48 is distributed into large complexes as well as in much smaller forms at the top of the sucrose gradient. Endogenous cytoplasmic RPD3 is also found in these low-molecular-weight fractions (Fig. 4A). Expression of exogenous RPD3 leads to an increase in histone deacetylase activity within the low-molecular-weight fractions in both nucleus and cytoplasm but not in the larger complexes that migrate at the bottom of the sucrose gradients (Fig. 4A and B). We conclude that exogenous nuclear RPD3 is functionally competent to direct both histone deacetylation and transcriptional repression during chromatin assembly, without incorporation into the large nuclear deacetylase complexes that contain the endogenous protein.
In numerous cotransfection experiments, repression has never been observed in the absence of targeting (for example, reference 77). It is therefore highly unlikely that exogenous RPD3 is capable of deacetylating histones in chromatin on its own. Although free, recombinant deacetylases have been shown to deacetylate histone H4 in vitro (21), exogenous Xenopus RPD3 does not bind to coexpressed H4 in vivo (Fig. 6C), nor is it incorporated into the high-molecular-weight endogenous Xenopus RPD3-containing complexes that have been characterized (Fig. 4; references 25 and 70). Exogenous Xenopus RbAp48 does, however, bind H4 and also binds exogenous RPD3 (Fig. 5 and 6). Furthermore, endogenous RbAp48 is detected in overlapping nuclear fractions with expressed RPD3 (Fig. 4). These data are consistent with a model where exogenous RPD3 is targeted to histones by RbAp48 in vivo.
Earlier work had shown that the mixing of Hat1p with RbAp46 greatly stimulates acetyltransferase activity (67). Taken together, these results strongly suggest that the histone targeting function of the RbAps (Fig. 6) plays an important role in sequestering the core histone substrate for modification. In yeast, the Hat2 protein component of the Hat1 acetyltransferase is a structural homolog of the vertebrate RbAps (49). Hat2p is structurally related to Cac3p, which is found in yeast chromatin assembly factor 1 (CAF-1) (32). Disruption of CAC3 leads to sensitivity to UV light and derepression of telomeric silencing (32). This is consistent with the observations that CAF-1 has a role in chromosomal repair after DNA damage and is important for chromatin assembly (17, 57). In contrast, disruption of HAT2 has essentially no phenotype. This suggests that specialized functions may exist for the vertebrate homologs of Cac3p and Hat2p. Thus, distinct activities might be anticipated for the vertebrate RbAp48 and RbAp46, respectively. Any role of Cac3p and/or Hat2p in deacetylase complexes in yeast has not yet been determined. In humans, RbAp48 is found associated with CAF-1 (67). However CAF-1 lacking RbAp48 will still associate with histones, whereas Hat1p lacking RbAp46 does not show such a stable association. It has therefore been proposed that the sole function of the RbAp48 in CAF-1 is to attract the RPD3 histone deacetylase to sites of newly synthesized histone deposition (67).
RbAp48 interacts with the N-terminal tail immediately proximal to the histone fold domain of H4.
RbAp48 fails to bind to histone H4 if the N-terminal tail is deleted past aa 28 (Fig. 6); similarly, the assembly of histone H4 into embryonic chromatin is severely reduced if N-terminal deletions extend past aa 32 (16). We find that RbAp48 requires aa 28 through 32 in the context of an intact C terminus for association with H4 (Fig. 6). Helix 1 of histone H4, comprising aa 31 to 41 relative to the N terminus, contributes to heterodimerization with H3 (5) and makes contact with nucleosomal DNA (38, 39). Removal of this helix through aa 36 prevents the assembly of H4 into chromatin entirely (16). Thus, it appears that the determinants of chromatin assembly are very similar to those for association with RbAp48.
RbAp48 does not appear capable of gaining access to its interaction site within H4 when assembled into nucleosomes (38, 39, 67). Other endogenous proteins or chromatin remodeling events may be required to allow access of RPD3-RbAp48 to chromatin in vivo. The ability of RbAp48 to bind to H4 in the absence of H3 (Fig. 6A; reference 16) indicates that an RPD3-RbAp48 complex may recognize and deacetylate histones during their assembly into chromatin in vivo. An attractive model is that RPD3-RbAp48 may modify histones during nucleosome assembly onto DNA templates. Successive replacement of the histone acetyltransferase Hat1p by CAF-1 followed by the histone deacetylase, all of which contain RbAp46, RbAp48, or homologs and interact with core histone H4, may facilitate this process (70a). Histone H4 is stored in Xenopus oocytes in the diacetylated state modified at lysines 5 and 12 and as a heterodimer with H3 (3, 14). Association with N1 and N2 might prevent deacetylation prior to incorporation into chromatin during replication through CAF-1-dependent pathways (3, 4).
Deposition-related deacetylation by RPD3-RbAp48 would account for the untargeted repression of all promoters and for the requirement for a physiological density of nucleosomes to be assembled to exhibit repression due to the maturation of the deacetylated, assembled chromatin (75). Implicit in this model is the requirement for maintenance of some level of acetylation on assembled plasmids in the absence of exogenous RPD3. This may be the result of less efficient deacetylation of diacetylated H4 during assembly in the absence of exogenous RPD3 or of some level of histone acetylation by histone acetyltransferase activity on templates assembled in the absence of RPD3. This type of nontargeted histone modification might be necessary to restore basal levels of gene activity on the removal of transcriptional activation functions, for example, on removal of hormone from the glucocorticoid receptor (35) or on addition of a histone deacetylase inhibitor to an unliganded nuclear receptor (75).
One problem with the suggested model for H4 deacetylation during chromatin assembly through the successive replacement of Hat1p, CAF-1, and RPD3 on H4 as mediated by RbAp46 and RbAp48 homologs is the timing of H4 deacetylation with respect to nucleosome assembly. Nucleosomes are assembled on newly replicated DNA within a few minutes of DNA synthesis (2, 4, 57, 66). In contrast, the complete deacetylation of histone H4 takes 30–60 minutes or more (4a, 56a, 64a). If RbAp48 cannot gain access to chromatin when H4 is assembled into a bona fide nucleosome, then the RbAp48-RPD3 complex may cease any association with newly assembled nucleosomes within minutes of H4 deposition long before the deacetylation of newly synthesized H4 is completed. Under these circumstances, the deacetylation of newly assembled chromatin would require the action of other endogenous proteins or remodeling events or an RbAp48-independent mechanism. This possibility provides an interesting area for future investigation.
ACKNOWLEDGMENTS
P. L. Jones was supported by the PRAT Fellowship program, NIGMS/NIH. D. Vermaak was supported by the Foundation for Advanced Education in the Sciences in the joint Johns Hopkins University–NIH Ph.D. program.
We are grateful to Thuy Vo for manuscript preparation.
The first three authors made equal contributions to this work.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, De Pinho R A. Role of NCoR and histone deacetylase in Sin3-mediated transcriptional and oncogenic repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Almouzni G, Wolffe A P. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- 3.Almouzni G, Wolffe A P. Nuclear assembly, structure, and function: the use of Xenopus in vitro systems. Exp Cell Res. 1993;205:1–15. doi: 10.1006/excr.1993.1051. [DOI] [PubMed] [Google Scholar]
- 4.Almouzni G, Mechali M. Assembly of spaced chromatin by DNA synthesis in extracts from Xenopus eggs. EMBO J. 1988;7:664–672. doi: 10.1002/j.1460-2075.1988.tb02861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Annunziato A T. Histone acetylation during chromatin replication and nucleosome assembly. Nucleus. 1995;1:31–58. [Google Scholar]
- 5.Arents G, Burlingame R W, Wang B W, Love W E, Moudrianakis E N. The nucleosomal core histone octamer at 3.1Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 7.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayer D E, Eisenman R N. A switch from Myc-Max to Mad-Max accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 9.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:601–605. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 10.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Willingham T, Margraf L R, Schreiber-Agus N, De Pinho R A, Nisen P D. Effects of the MYC oncogene antagonist, MAD on proliferation, cell cycling and the malignant phenotype of human brain tumor cells. Nat Med. 1995;1:638–643. doi: 10.1038/nm0795-638. [DOI] [PubMed] [Google Scholar]
- 13.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 14.Dimitrov S, Wolffe A P. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histone H1 and H1° from chromatin and the acquisition of transcriptional competence. EMBO J. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- 15.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livington D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-KD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;7:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 16.Freeman L, Kurumizaka H, Wolffe A P. Functional assays for assembly of histones H3 and H4 into the chromatin of Xenopus embryos. Proc Natl Acad Sci USA. 1996;93:12780–12785. doi: 10.1073/pnas.93.23.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaillard P-H L, Martini E M-D, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor 1. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 18.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgel P T, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory P D, Horz W. Life with nucleosomes: chromatin remodelling in gene regulation. Curr Opin Cell Biol. 1998;10:339–345. doi: 10.1016/s0955-0674(98)80009-4. [DOI] [PubMed] [Google Scholar]
- 21.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin 3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 22.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinzel T, Laviusky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J T, Yang W-M, Brard C, Ngo S G, Davie J R, Seto E, Eisenman R M, Rose D W, Glass C K, Rosenfeld M G. N-CoR, mSIN3, and histone deacetylase in a complex required for repression by nuclear receptors and Mad. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 24.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1996;15:2030–2040. [PMC free article] [PubMed] [Google Scholar]
- 24a.Imhof, A. Unpublished data.
- 25.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamakaka R T, Bulger M, Kaufman P D, Stillman B, Kadonaga J T. Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor I. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokama R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 29.Kasten M M, Stillman D J. Identification of the Saccharomyces cerevisiae STB1-STB5 encoding Sin3p binding proteins. Mol Gen Genet. 1997;256:376–386. doi: 10.1007/s004380050581. [DOI] [PubMed] [Google Scholar]
- 30.Kasten M M, Dorland S, Stillman D J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasten M M, Ayer D E, Stillman D J. SIN3 dependent transcriptional repression by interaction with the Mad1 DNA binding protein. Mol Cell Biol. 1996;16:4215–4221. doi: 10.1128/mcb.16.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-1. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 33.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R M. Histone deacetylase associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 34.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J M, Mullen T M, Davie J R, Rose D W, Glass C K, Rosenfeld M G, Ayer D E, Eisenman R N. SAP30, a component of the mSin3 corepressor complex involved in N-CoR mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 35.Lee H H, Archer T K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vitro. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leipe D D, Landsman D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 1997;25:3693–3697. doi: 10.1093/nar/25.18.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzyko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y presents chromatin to potentiate p300 and acetylation responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luger K, Rechsteiner T J, Flaus A J, Wayne M M Y, Richmond T J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 39.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. X-ray structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–259. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 40.Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 41.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Balbas M A, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 45.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 46.Neuwald A F, Landsman D. GCN5-related histone N-acetyltransferases belong to a superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 47.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 48.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 49.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin assembly and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 50.Pennetta G, Pauli D. The Drosophila SIN3 gene encodes a widely distributed transcription factor essential for embryonic viability. Dev Genes Evol. 1998;208:531–536. doi: 10.1007/s004270050212. [DOI] [PubMed] [Google Scholar]
- 51.Qian Y W, Wang Y-C J, Hollingsworth R E J, Jones D, Ling N, Lee E Y-H P. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 52.Qian Y W, Lee E Y-H P. Dual retinoblastoma-binding proteins with properties related to a negative regulator of Ras in yeast. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 53.Rao G, Alland L, Guida P, Schreiber-Agus N, Chin L, Chen K, Rochelle J M, Seldin M F, Skoultchi A I, De Pinho R A. Mouse SIN3A interacts with and can functionally substitute for the amino-terminal repression of the Myc antagonist Mxi1. Oncogene. 1995;12:1165–1172. [PubMed] [Google Scholar]
- 54.Roussel M F, Ashmun R A, Sherr C J, Eisenman R N, Ayer D E. Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol Cell Biol. 1996;16:2796–2801. doi: 10.1128/mcb.16.6.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56a.Shimamura A, Worcel A. The assembly of regularly spaced nucleosomes in the Xenopus oocyte S150 extract is accompanied by deacetylation of histone H4. J Biol Chem. 1989;264:14524–14530. [PubMed] [Google Scholar]
- 57.Smith S, Stillman B W. Purification and characterization of CAF1, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 58.Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Siser C, Loidl P, Luscher B. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- 59.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Crystal structure of a G-protein β γ dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. . (Erratum, 379:847.) [DOI] [PubMed] [Google Scholar]
- 60.Spencer T E, Jenster G, Bercin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Steroid receptor coactivator one is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 61.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to a yeast transcriptional regulator Rpd3. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 62.Tong J K, Hassig C A, Schnitzer G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 63.Torchia J, Glass C, Rosenfeld M G. Coactivators and corepressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 64.Tyler J K, Bulger M, Kamakaka R T, Kobayashi R, Kadonaga J T. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64a.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription nucleosome mobility and positioning and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 65a.Vermaak D. Functional studies of linker histone H1 domains and histone acetylation in Xenopus development. Dissertation. Baltimore, Md: The Johns Hopkins University; 1999. [Google Scholar]
- 66.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 67.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone binding subunit of the human hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 68.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vidal M, Strich R, Esposito R E, Gaber R F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wade P A, Jones P L, Vermaak D, Wolffe A P. The multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 70a.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 71.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. The structure of the G protein heterotrimer Gi α1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 72.Wang H, Stillman D J. In vitro regulation of a SIN3-dependent DNA-binding activity by stimulatory and inhibitory factors. Proc Natl Acad Sci USA. 1990;87:9761–9765. doi: 10.1073/pnas.87.24.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, Stillman D J. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H, Clark I, Nicholson P R, Herskowitz I, Stillman D J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990;10:5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong J, Patterton D, Imhof A, Shi Y-B, Wolffe A P. Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J. 1998;17:520–534. doi: 10.1093/emboj/17.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong J, Shi Y B. Coordinated regulation of and transcriptional activation by Xenopus thyroid hormone and retinoid X receptors. J Biol Chem. 1995;270:18479–18483. doi: 10.1074/jbc.270.31.18479. [DOI] [PubMed] [Google Scholar]
- 77.Yang W M, Inouye C, Zeng Y, Bearss D, Soto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral E1A oncoprotein. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]