Abstract
A dynamic continuum exists from free-living environmental microbes to strict host-associated symbionts that are vertically inherited. However, knowledge of the forces that drive transitions in symbiotic lifestyle and transmission mode is lacking. Arsenophonus is a diverse clade of bacterial symbionts, comprising reproductive parasites to coevolving obligate mutualists, in which the predominant mode of transmission is vertical. We describe a symbiosis between a member of the genus Arsenophonus and the Western honey bee. The symbiont shares common genomic and predicted metabolic properties with the male-killing symbiont Arsenophonus nasoniae, however we present multiple lines of evidence that the bee Arsenophonus deviates from a heritable model of transmission. Field sampling uncovered spatial and seasonal dynamics in symbiont prevalence, and rapid infection loss events were observed in field colonies and laboratory individuals. Fluorescent in situ hybridisation showed Arsenophonus localised in the gut, and detection was rare in screens of early honey bee life stages. We directly show horizontal transmission of Arsenophonus between bees under varying social conditions. We conclude that honey bees acquire Arsenophonus through a combination of environmental exposure and social contacts. These findings uncover a key link in the Arsenophonus clades trajectory from free-living ancestral life to obligate mutualism, and provide a foundation for studying transitions in symbiotic lifestyle.
Subject terms: Microbial ecology, Molecular evolution, Bacterial evolution, Bacterial genetics, Phylogenetics
Introduction
Microbes that associate with hosts span a continuum from mutualism to parasitism and employ disparate transmission strategies [1]. Heritable bacterial symbionts, such as Wolbachia and Arsenophonus, transmit vertically (VT) from parent to offspring [2]. Other microbial symbioses form via horizontal transmission (HT) during host development, by acquisition from environmental reservoirs or via infected conspecifics or other host taxa [3, 4]. In some cases, a combination of these routes operate, creating a complex transmission landscape [5, 6]. Across this transmission axis lies the impact of the symbiosis on each partner—whether the host and microbe benefit from the interaction, and whether one party requires the other to complete their life cycle. Symbioses vary from being facultative (where one partner does not require the other) to obligate (where there is dependence). Obligacy may be a component of symbiont or host biology, or in some cases both parties are mutually dependent [2, 7, 8].
Clades of heritable symbionts commonly include strains that are both facultative and obligate from the host perspective. In contrast, for the microbe, life is commonly obligately symbiotic, generally lacking replicative or dormant phases outside of host organisms [7]. Despite this dependence, many of these symbionts can also transmit horizontally (inter and intra-specifically) over evolutionary timescales [9–11]. However, it is generally accepted that VT predominantly drives population dynamics [12], but see [13]. In contrast, exclusively horizontally acquired symbionts must be transmitted to new hosts via infected con-/hetero-specifics or environmental reservoirs [14, 15]. As a result, symbiotic life for these microbes is commonly facultative [3]. The absence of VT can promote higher rates of partner switching [2, 16] and a weaker association between host and symbiont fitness. Consequently, selection for higher virulence and trajectories toward parasitism are commonly assumed to be favoured more readily in HT symbionts [1, 17, 18], but see [19, 20].
Symbioses thus exist on an evolutionary landscape in which transitions between transmission mode and lifestyles occur. However, study of the drivers and impact of transitions in transmission mode is inhibited by a lack of clades in which different lifestyles co-occur among members. Clades encapsulating diverse transmission biology enable understanding of both the ecological and evolutionary mechanisms driving transitions, and the consequences for processes such as genome evolution [21–24]. For instance, the emergence of a horizontally transmitted vertebrate pathogen (Coxiella burnetti) from a clade of maternally inherited tick endosymbionts [22] provides insight into how infectious transmission can emerge from a heritable, obligate clade. Likewise, the presence of an opportunistic human infective Sodalis provides an important comparator for understanding the evolution of symbiosis, through comparison to insect-associated mutualistic lineages of Sodalis [21].
As a monophyletic clade of heritable Enterobacteriaceae, Arsenophonus provides a valuable base for exploring the evolution of a heritable lifestyle [25], due to its wide host distribution (est. 5% of arthropod species) [26] and diversity in symbiotic lifestyle [27]. Members of the clade include reproductive parasites [28], facultative mutualists [29], and highly coevolved obligate endosymbionts undergoing genomic decay [30–33]. Despite this diversity, all strains characterised to date are VT transmitted. While frequent HT over ecological timescales has been shown for a number of strains, including the male-killer Arsenophonus nasoniae [13, 34, 35] and two species of insect-vectored phytopathogens (Arsenophonus phytopathogenicus and Phlomobacter fragariae) [36–38], this route occurs alongside vertical transmission. As a result, Arsenophonus is commonly reported from arthropod screening efforts and presumed heritable without further characterisation [26].
An economically important eusocial host, the Western honey bee (Apis mellifera), has previously been associated with Arsenophonus [39–43] and infection has been linked to poor health outcomes [44] including colony collapse disorder [45]. Whilst this interaction has attracted interest from the community, basic information on the epidemiology and transmission of Arsenophonus in honey bee populations is lacking. Symbionts in eusocial hosts are exposed to markedly different selection pressures from those in solitary species. This difference arises from a higher density of hosts, greater host relatedness and a homoeostatic nest environment, as well as reproductive division of labour, overlapping generations and cooperative brood care [46, 47]. Specialised social behaviours [48] additionally foster the transmission of microbes by direct contact, such as via proctodeal (anus—mouth feeding) and stomodeal trophallaxis (mouth—mouth feeding) [49]. Thus, host sociality is an important driver of symbiont phenotypes [47, 49, 50], and indeed interesting heterogeneities in symbiont infections are emerging based on caste, sex [51–55] and degree of sociality [50]. Despite this, the potential effects of host sociality on symbiont ecology and evolution, including important phenotypes such as transmission, remain largely unexplored.
This study focuses on characterising the ecology and transmission of Arsenophonus from a eusocial host. We use a phylogenomic approach to show robust placement of the strain within the Arsenophonus clade and genomic analysis to report on its predicted metabolic properties and genome features. We then present data tracking the prevalence of the symbiont in honey bee colonies over space and time, to search for indicators of stable maintenance (implying VT) or changes in prevalence (implying infectious transmission epidemics). These data are complemented by fluorescence in situ hybridisation (FISH) analysis highlighting tissue interactions with honey bees. Finally, the capacity for vertical transmission is assessed directly by screening for Arsenophonus across host life history, and the ability to horizontally transmit intra-specifically is investigated under differing social conditions. These data demonstrate the presence of an infectiously transmitting Arsenophonus without vertical transmission, highlighting transitions in life history within this important genus of insect-associated microbes.
Methods
Phylogenomic position of Arsenophonus from honey bees and comparative genomic analysis
To assess the relatedness of the Arsenophonus strain associated with honey bees to other Arsenophonus, a phylogenomic approach was adopted. To this end, a draft genome was assembled (bioproject accession: PRJEB39047) using Illumina paired end shotgun library reads obtained in a previous study derived from haemolymph template purified through a Nycodenz gradient (see Gauthier et al. [56]). The assembled contigs were annotated using prokka v1.14.0 [57] and completeness was assessed using BUSCO v4.1.4 based on the presence of 124 universal bacterial marker genes [58]. The phylogenetic position of the Arsenophonus strain was estimated in relation to other Arsenophonus, with the free-living species Proteus mirabilis and Providencia stuartii used as outgroups, using first an alignment of 53 ribosomal proteins and then a concatenated set of 155 single copy core orthologus proteins. In addition, we compared the predicted metabolic potential of Arsenophonus from bees to other strains, and in greater detail examined functional differences and the degree of synteny to A. nasoniae (see Supplementary information for details).
Spatial and seasonal dynamics of Arsenophonus in honey bee colonies
To monitor Arsenophonus prevalence over time and space, adult workers were collected from the outer frames of 159 colonies stemming from 45 apiaries in ten counties across England. Colonies were sampled from April to November during 2014–2018, and repeated screening of some colonies resulted in a total of 230 sampling events. Apiaries are defined here as a collection of colonies that are colocalised within a small area. Bees were preserved in 70% EtOH at −20 °C until DNA extraction. For each colony, posterior legs were pulled from workers (n = 12), pooled in groups of four and exposed to UV light for 10 min to cross link DNA from surface microbes. Legs were used as this tissue is a reliable marker of Arsenophonus association in bees and did not inhibit downstream PCR assays when DNA was extracted using a high throughput Chelex protocol [59]. Extraction quality was verified by amplification of host DNA (EF1-α) [60] and the presence of Arsenophonus spp. established by PCR assays targeting fbaA (adapted from Duron et al. [34], see Supplementary information) and Sanger sequencing of the product to determine broad identity of the Arsenophonus strains. Sensitivity of PCR assays was established through serial dilution, and was robust over two orders of magnitude (see Supplementary information).
To determine if Arsenophonus is lost from honey bee colonies during overwintering, the status of 25 colonies (from seven apiaries) was tracked from autumn to spring at a greater depth than described above. In the autumn, 15 worker bees were screened from each colony (three individuals pooled per extraction) to determine Arsenophonus prevalence. If 80% of extractions (i.e. 4 of 5) were positive for Arsenophonus the colony was included in the infected cohort (group A, n = 19). The uninfected cohort (group B, n = 6) comprised of colonies where all extractions returned negative. Colonies were left to overwinter, and the sampling process was repeated in the spring for colonies that overwintered successfully. To determine infection status in spring a total of 24 bees were screened per colony (eight pools of three bees). DNA was extracted from whole bees (heads removed prior to extraction to minimise PCR inhibition [61]) by Promega Wizard purification, with PCR detection of Arsenophonus (see Supplementary information).
Localisation of Arsenophonus within the gut
To visualise Arsenophonus using FISH, whole guts were dissected from live worker bees and placed into Carnoy’s fixative for 24–48 h, washed with 100% EtOH (×3) and incubated with hybridisation buffer at room temperature in the dark (~15 h, see Supplementary information for detail). The symbiont was targeted using an Arsenophonus specific probe (TCATGACCACAACCTCCA) [62] with a 5′ Alexa Fluor 647 fluorochrome. Tissues were washed with pre-heated buffer (∼48 °C, ×3 for 10 min) and mounted on glass slides with DAPI counter-staining. Tissues cured for 24 h before visualisation by confocal microscopy (ZEISS LSM 88, ×40 objective lens), after which multiple optical sections were assembled into Z-stacks under maximum intensity settings using ImageJ [63]. Gut tissue of bees from colonies not associated with Arsenophonus underwent the same process to function as negative controls (see Supplementary information). In addition to gut imaging, faecal samples were collected from workers (n = 22) isolated in sterile petri dishes and extracted by Promega Wizard purification (see Supplementary information). Material was then tested for Arsenophonus DNA through PCR assay.
Assessing the heritability of Arsenophonus and acquisition from infected conspecifics
To determine heritability of Arsenophonus in honey bees, infection was assessed across life history. Frames of worker brood were removed from managed field colonies (N = 8, A+ = 6, A− = 2) where Arsenophonus status had previously been determined with high confidence. Eggs (n = 29), larvae (n = 67), pupae (n = 49), newly emerged workers (NEWs) (n = 36), adult workers (n = 45) and where possible drones (n = 22) were collected concurrently. To asses if Arsenophonus association is heritable but only becomes detectable later in the life history, a further 64 NEWs were removed from colonies and left to develop to forager age (25 ± 2 days post-emergence) in the company of other NEWs only. DNA was extracted individually from eggs and early stage larvae using a Qiagen DNeasy Blood & Tissue Kit, for all other life stages Promega Wizard purification was used. Molecular detection was completed as previously.
To assess if infections were maintained in the absence of the colony and foraging environment, infected individuals were detained in the laboratory and their Arsenophonus status tracked over 15 days. Adult workers were collected from two colonies (total N = 76, Colony A = 36, Colony B = 40) with a high prevalence of Arsenophonus in 15 workers (>90%) or one control colony where none of 15 workers tested positive (N = 32) (based on Arsenophonus detection of individual bees, see Supplementary information). On entering the laboratory (day 1) the Arsenophonus status of all individuals was established by removal of posterior tarsus tissue (‘leg snip’) and PCR assays for symbiont presence (see Supplementary information). Additional bees (‘no leg snip control’, N = 40) from the infected source colonies were not subjected to leg snips to establish if this method biased results. Individuals were marked and maintained in groups of four. To track the maintenance of Arsenophonus over time, individuals were randomly selected from each group and culled at days 4, 8 and 15. Individual Arsenophonus status was determined using the same methodology as day 1, but using opposing tarsus tissue.
To assess the capacity for horizontal acquisition of the symbiont, adult workers (donors) were taken from colonies (N = 6) with a high prevalence of Arsenophonus in 15 adult bees (>85% infected, see Supplementary information) and mixed in 340 ml pots with uninfected NEWs (recipients). Each pot contained ten donors and five recipients. Two transmission treatments were established, each with ten replicates. In the ‘general contact’ treatment, bees were allowed to freely contact one another within the pot and food (50% sucrose solution) was openly available. In the ‘trophallaxis’ treatment, recipients with no direct access to food were separated from donors (with food) by fine mesh, forcing infected bees to feed uninfected individuals by trophallaxis, but preventing other social contact. Contacts were allowed for 5 days, after which the fate (dead or alive) of all individuals was recorded, i.e. those that had died during the course of the experiment (prior to final cull) were labelled dead and analysed separately. In each case, experiments were compared to controls in which uninfected bees were mixed with recipient bees. DNA was extracted individually from whole bees (test; n = 300, control; n = 165) by Promega Wizard purification.
Statistical methods
For statistical analyses, generalised linear models (GLM) and mixed models (GLMM) were fitted in R version 3.3.1 [64] with binomial error distributions and logit link function, using the packages glm and lme4 [65]. Minimum adequate models were selected by Akaike information criteria [66, 67] and likelihood ratio tests (LRT), with the latter being used to assess the significance of fixed and random effects [68]. Overdispersion was assessed using the Blemco package [64]. See Supplementary information tables detailing selection of statistical models.
Results
Phylogenomic position of the honey bee Arsenophonus in the wider clade and metabolic competence
Phylogenomic analysis based on 53 ribosomal protein sequences unambiguously placed the honey bee associated strain within the Arsenophonus clade (Fig. 1) alongside known heritable symbionts. The strain was established as a sister strain, with strong support, to Arsenophonus strains infecting other hymenopteran hosts (parasitoid wasps), including the male-killing reproductive parasite A. nasoniae. These results were mirrored in analysis of a wider set of core genes, in which the Arsenophonus from honey bees again clustered as sister to A. nasoniae (Supplementary information Fig. 1).
Fig. 1. Transmission mode and lifestyle diversity in the Arsenophonus clade.
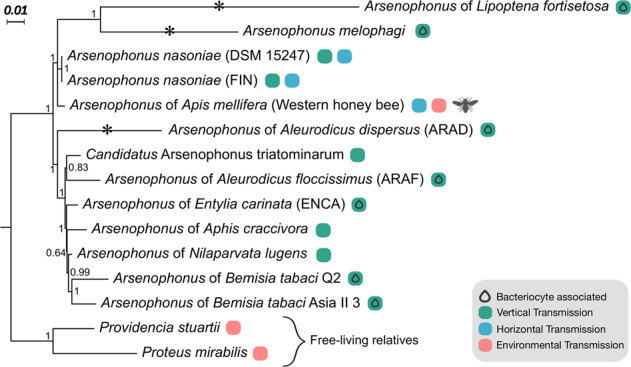
Phylogenomic position of the Arsenophonus associated with honey bees (this study) among Arsenophonus spp. with available genomes. Bacterial species names are given, if formally recognised, else the associated insect host is given. Symbols indicate known bacteriocyte associations and transmission modes, vertical transmission (green), horizontal transmission (blue), if an environmental transmission route is additionally inferred this is indicated in pink. Analysis based on 53 ribosomal proteins support for Bayesian inference (posterior probabilities) is shown at nodes (Color figure online).
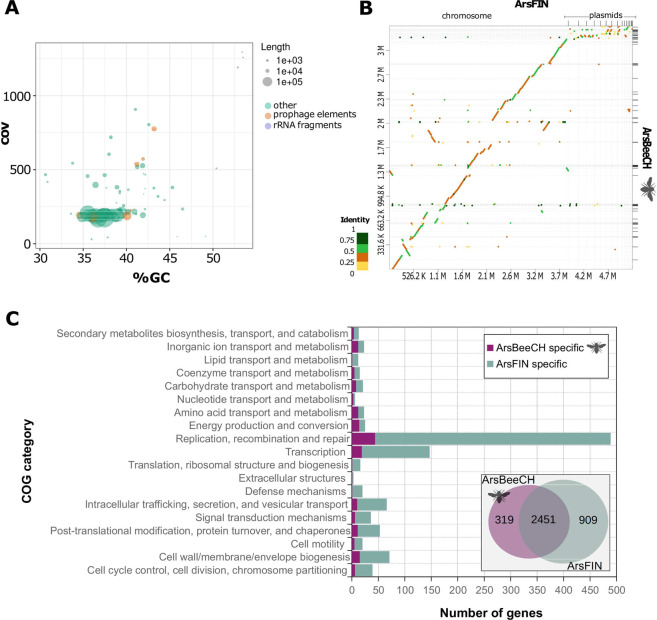
We assembled a draft genome for Arsenophonus from bees and examined synteny and compared gene content to A. nasoniae (Fig. 2), which is a closed completed genome [69]. At 3.3 Mb, the draft genome was larger than most other sequenced Arsenophonus with the exception of A. nasoniae and Candidatus Arsenophonus triatominarum (Supplementary information Table 1). BUSCO completeness score for the bee Arsenophonus strain mirrored that of A. nasoniae (C:99.2% [S:99.2%, D:0.0%], F:0.8%, M:0.0%, n:124) (Supplementary information Fig. 2) suggesting a near complete genome. Synteny analysis supported the close relationship of the two strains, with evidence of some structural differences including at least one inversion event and several indels (Fig. 2). As expected, accessory elements (plasmids) were less well conserved between the two genomes. Overall, 2451 orthologous groups of proteins were shared between the two symbionts, with 319 found solely in Arsenophonus from bees, and 909 solely in A. nasoniae. This higher number of unique orthologous genes in A. nasoniae may reflect the difference between draft and polished genomes, particularly in accessory genes carried on plasmids and prophage elements.
Fig. 2. Overview of the genome assembly of Arsenophonus from honey bees (ArsBeeCH) and comparison with close relative Arsenophonus nasoniae (ArsFIN).
A GC% vs coverage plot of the ArsBeeCH draft assembly. Contigs with putative phage origin were identified using PHASTER web server based on sequence similarities searches. B Syntenic comparison between ArsBeeCH and ArsFIN genomes as determined by D-Geneis and minimap2. C COG functional annotation of strain-specific genes. The inset Venn diagram shows the number of shared and unique ortholog groups between the two Arsenophous strains.
Arsenophonus from bees also clustered with A. nasoniae in analysis of predicted metabolic potential (Supplementary information Figs. 3 and 4). These strains shared higher predicted competency for the glyoxylate cycle, polyamine biosynthesis compared to other sequenced Arsenophonus, and reduced capacity for cysteine biosynthesis. Predicted differences in metabolic capacity between Arsenophonus from bees and A. nasoniae were modest, making similar metabolic potential and culture requirements likely.
Spatial and seasonal dynamics of Arsenophonus in honey bee colonies
Arsenophonus was found associated with 38.9% (95% CI: 32.5–45.6%, N = 159 colonies) of honey bee colonies across the UK. Infections were spatially widespread and identifiable in all ten county regions tested (Fig. 3A). County region was not a significant predictor of Arsenophonus infection (GLMM, LRT: X2 = 0, df = 1, p = 1), but spatial patterns were evident at a local scale, with apiary emerging as an important covariate, with colonies within an apiary having correlated infection status (GLMM, LRT: X2 = 13.8, df = 1, p = 0.002**). All Arsenophonus detected had identical sequence at the fbaA locus (N = 159).
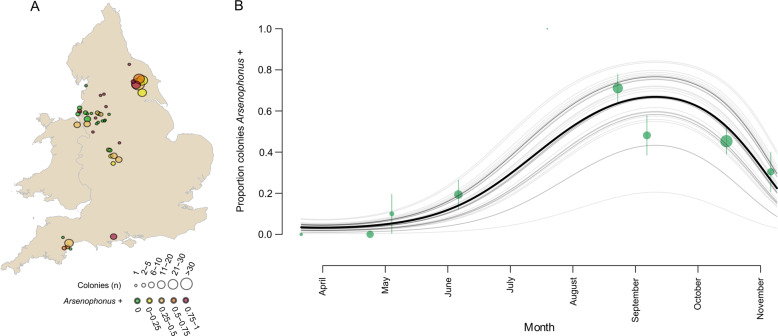
Fig. 3. Spatial and seasonal dynamics in the Arsenophonus–honey bee association.
A Circles show the approximate locations of 45 honey bee apiaries (colonies colocalised within a small area) sampled for Arsenophonus across England (2014–2018). Circle size reflects the number of colonies sampled within an apiary location and colour indicates the proportion of colonies associated with Arsenophonus. B The proportion of colonies testing positive for Arsenophonus from spring to autumn was modelled using 229 binomial observations of Arsenophonus status from 159 colonies (2014–2018). An overall prediction for all apiaries (black) and individual predictions for each apiary (grey, n = 45) are shown. Green circles represent the mean prevalence of Arsenophonus by month at the mean monthly sampling date, size is proportional to the square root of the number of colony samplings. Error bars represent binomial CI (Color figure online).
Strong seasonal dynamics in Arsenophonus infections (Fig. 3B) were evident, with prevalence of the bacterium changing in a non-linear fashion during the main foraging period of the host (March to October). Arsenophonus prevalence is lowest during the spring (2.63% of colonies, 95% CI: 0.0670–13.8%, N = 38) with the first infected colony detected in May. Prevalence continues to rise, reaching a peak in late summer (August: 71% of colonies, 95% CI: 55.7–83.6, N = 45), before dropping into Autumn (43.5% of colonies, 95% CI: 34.3–53.0, N = 115). Significant temporal variation by year was observed for Arsenophonus prevalence (GLMM, LRT: X2 = 3.87, df = 1, p = 0.049*). See Supplementary information Table 2 for a full summary.
In temperate environments, overwintering conditions represent a distinct state for honey bees, with foraging ceasing temporarily and survival dependent on stored colony resources. The Arsenophonus infection status of 25 honey bee colonies was tracked from autumn to spring (Fig. 4). Of the 23 colonies that survived winter, all of those infected with Arsenophonus in the autumn (Group A, n = 17) had lost the bacterium by spring. Of the colonies where Arsenophonus was not detected in the autumn (Group B, n = 6), five remained uninfected in spring, and one case gained Arsenophonus. Confidence in this newly positive colony is high, as all pools tested (N = 8 pools of three bees) were positive for Arsenophonus. Notably, this colony was sampled latest in the spring season.
Fig. 4. Overwintering loss of Arsenophonus at a colony level.
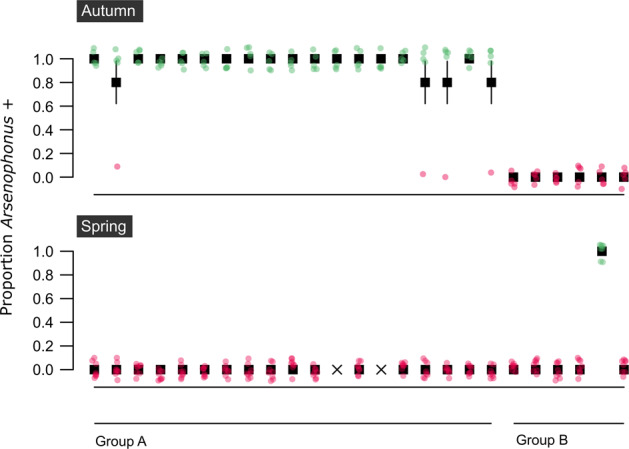
Group A colonies (n = 19) were infected with Arsenophonus in autumn but the bacterium was undetectable by the following spring. Group B colonies (n = 6) were uninfected with Arsenophonus in autumn, one colony gained Arsenophonus association by spring. Colonies that did not survive the winter (n = 2) are denoted by an X. The proportion of bees testing positive for Arsenophonus in each colony is shown. Note, fewer bees were sampled in Autumn (n = 15 bees per colony, pooled in groups of 3) than spring (n = 25 bees per colony, pooled in groups of 3). Pink dots (0 = Arsenophonus−) and green dots (1 = Arsenophonus+) show the raw binomial data jittered. Error bars indicate binomial SE (Color figure online).
Localisation of Arsenophonus within the gut
Previous work using metagenomic analysis has reported Arsenophonus in the gut [40, 41], on the cuticular surface [39] and in haemolymph [56]. To obtain confirmation of symbiosis, targeted imaging of gut tissue from infected honey bees was conducted. This analysis showed aggregations of Arsenophonus (coloured red) in the midgut (Fig. 5) sufficiently large to imply colonisation and replication. More sporadic infections were apparent in the crop, and also the rectum (consistent with low representation of Arsenophonus in honey bee gut microbiome studies that focus on the hindgut). Control images of uninfected bee tissue suggested neither autofluorescence nor inadequate probe removal contributed to artefactual Arsenophonus visualisation. The absence of a strong signal in the rectum is consistent with specific binding, as there was no widespread probe binding in the rectum of Arsenophonus positive individuals (see Supplementary information Fig. 5). Arsenophonus was also detected by PCR assay in 59.1% of faeces from infected bees (95% CI: 36.4–79.3) suggesting the bacterium may be shed from the gut, however viability of the symbiont was not confirmed.
Fig. 5. Localisation of Arsenophonus bacteria in the midgut of a worker honey bee.
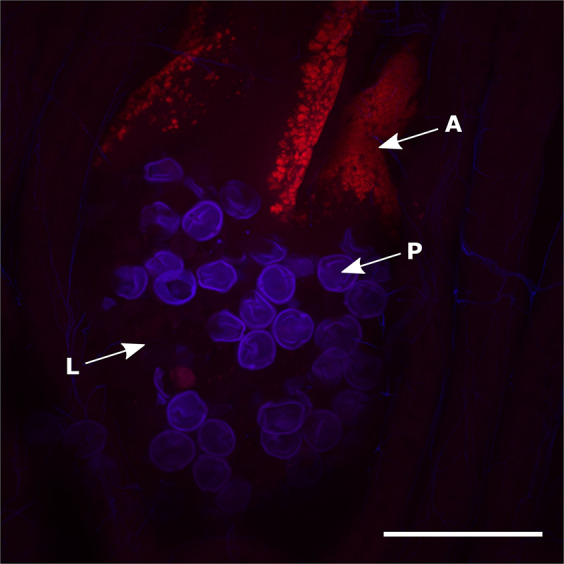
Confocal microscopic image of a whole honey bee gut mount with an Arsenophonus specific Alexa Fluor® 647 labelled probe (red fluorescence) and DAPI counter-staining (blue fluorescence). Hybridisation of an Arsenophonus specific probe (A) is visible within the lumen (L) of the midgut (ventriculus) alongside pollen grains (P). The image is a composite Z-stack comprised of 32 optical slices (ZEISS LSM 880) assembled in ImageJ under maximum intensity, scale bar represents 100 μm. Amplicon based approaches also report Arsenophonus from haemolymph [56]. For confocal images of whole gut mounts from honey bees uninfected with Arsenophonus see Supplementary information Fig. 5 (Color figure online).
Assessing the heritability of Arsenophonus and acquisition from infected conspecifics
To date, all characterised Arsenophonus strains show vertical transmission in their arthropod hosts. To assess if the honey bee strain conforms to this transmission strategy, the presence of Arsenophonus was assessed across host life history (Fig. 6). There was no evidence of transovarial transmission of Arsenophonus with eggs consistently testing negative, corroborating previous findings in honey bees [43]. In other early life stages Arsenophonus was detected only at low frequencies, with 5.97% of larvae (95% CI: 1.65–14.6, p < 0.001***), 4.081% of pupae (95% CI: 0.498–14.0, p < 0.001***) and 2.78% of NEWs (95% CI: 0.07–14.5, p < 0.001***) testing positive for Arsenophonus. In contrast, adult life stages showed high incidences of Arsenophonus, with the bacterium detectable in 54.5% of drones (95% CI: 32.2–75.6, p = 0.0198*) and 82.2% of workers (95% CI: 67.9–91.0). Workers isolated from the colony since emergence also did not develop Arsenophonus association on reaching forager age (95% CI: 0.00–0.06, p < 0.001***).
Fig. 6. Vertical transmission is largely absent in the Arsenophonus–honey bee association.
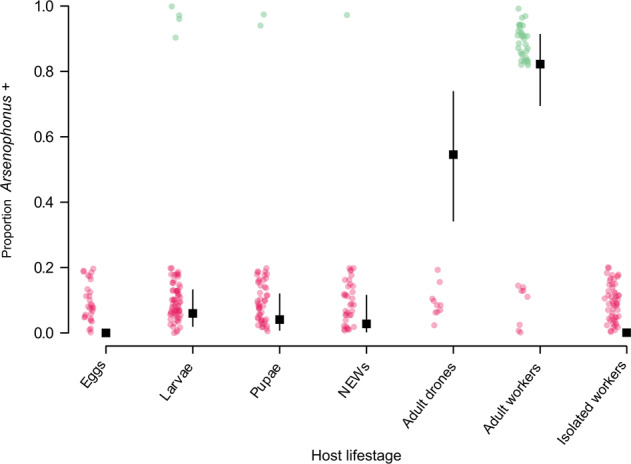
Samples from across the host life history were taken from colonies (n = 6) identified as Arsenophonus positive (based on workers) and screened for Arsenophonus. Eggs (n = 29), larvae (n = 67), pupae (n = 49) and newly emerged workers (NEWs) (n = 36) were all of worker caste. Isolated workers (n = 45) were removed from colonies as NEWs and allowed access only to other NEWs before screening for Arsenophonus at forager age (25 days ± 2). Binomial GLM predictions for the proportion of each life-stage infected are shown with 95% CI. Pink dots (0 = Arsenophonus−) and green dots (1 = Arsenophonus+) show the raw binomial data jittered (Color figure online).
The maintenance of Arsenophonus in worker bees in the absence of sources of infection was then tested (Fig. 7). Bees were removed from two infected colonies (colony A and colony B) and maintained in the laboratory for 15 days with access only to sterile food. At day 0 all individuals in the leg snip group were confirmed positive for Arsenophonus. Individuals in the ‘no snip control’ group were presumed, from identical colony origin, to be infected at a similar prevalence. There was no significant effect of treatment (leg snip or no snip) on the proportion of bees infected with Arsenophonus at each time point (LRT: X2 = 0.253, df = 1, p = 0.615), suggesting the leg snip did not impact our results. Overall, the proportion of bees infected with Arsenophonus decreased significantly over time (p < 0.001***) indicating loss of the bacterium. However, variation was evident at a colony level, and a significant effect of colony was observed (LRT: X2 = 20.2, df = 1, p < 0.001***). Colony B lost Arsenophonus at a significantly faster rate (p < 0.001***), with fewer than 20% of individuals infected by day 15. Control bees taken from uninfected colonies maintained 0% infection throughout the study. See Supplementary information Table 3 for a full summary.
Fig. 7. Loss of Arsenophonus in individuals removed from the colony and foraging environment.
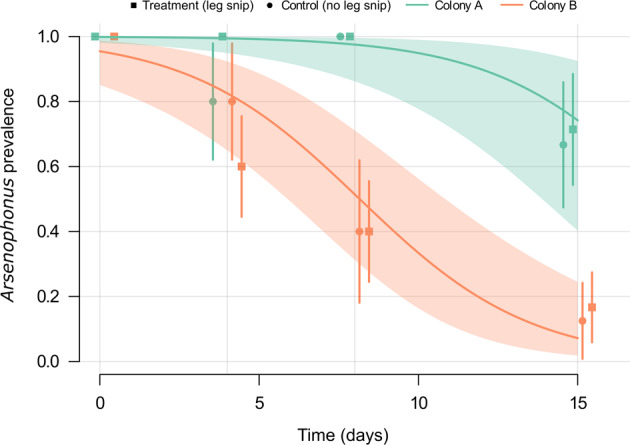
Binomial GLMM predictions (with 95% CI) show the proportion of individual honey bees that remain infected with Arsenophonus over time when removed from colonies and detained under lab conditions. The initial Arsenophonus status of each bee (n = 76) was determined by a leg snip (treatment group). An additional group of bees (n = 40) from the same colonies did not undergo leg snip (control group), and thus Arsenophonus starting prevalence was unknown. Overall predictions are shown for colony A (green) and colony B (orange), and the proportion of honey bees infected at days 0, 4, 8, and 15 is plotted independently for each colony and treatment (Color figure online).
Horizontal acquisition of Arsenophonus was tested in a separate experiment, where uninfected recipient bees were exposed to infected donor bees, either through trophallaxis or general contact. Acquisition of Arsenophonus among recipient bees was observed under both of these social conditions (Fig. 8A). The rate of Arsenophonus gain for recipients was higher under general contact exposure (recipients, 40.0%, 95% CI: 26.4–54.8) compared to trophallaxis (recipients, 22.0%, 95% CI: 11.5–36.0) and social context had a significant effect in the model (GLMM, LRT: X2 = 5.22, df = 1, p = 0.0223*).
Fig. 8. Horizontal transmission and maintenance of Arsenophonus in honey bees under two social conditions.
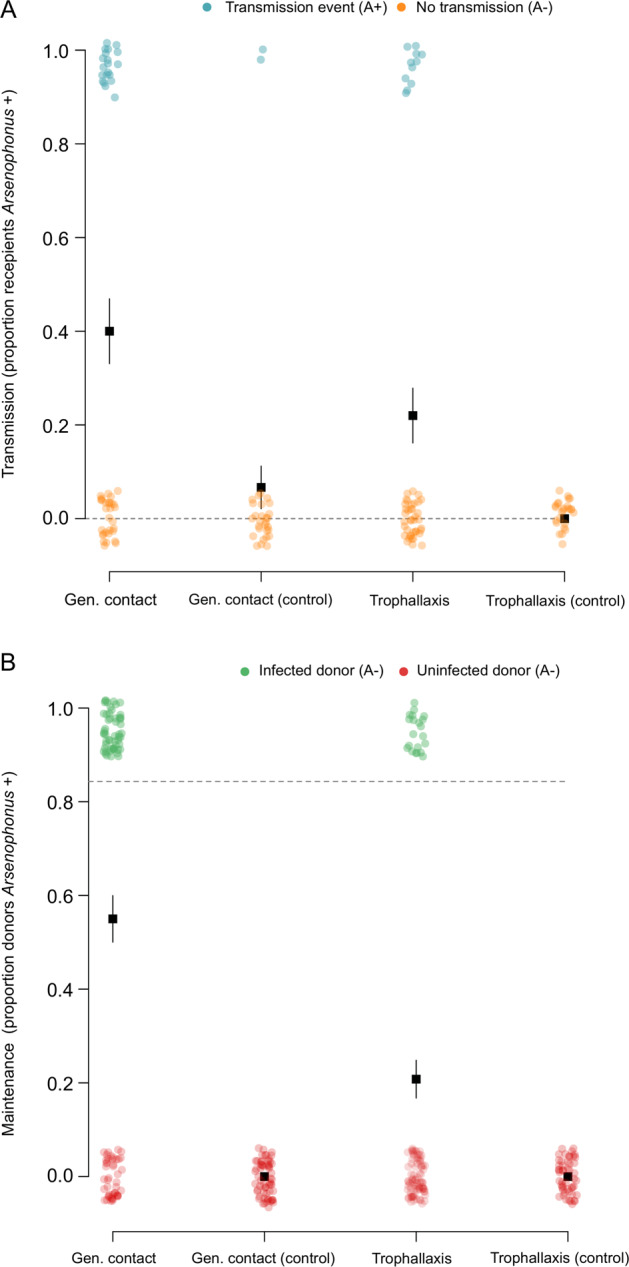
A Uninfected (recipient) bees were mixed with (B) infected (donor) bees and allowed either general contact or contact via trophallaxis only. For control groups, recipients were mixed with uninfected bees. Dotted lines indicate the prevalence of Arsenophonus in recipients and donors at the start of the transmission period. Note, all control bees started uninfected (Arsenophous prevalence = 0%). After 5 days of social interaction the Arsenophonus status of recipients (A) and donors (B) is shown. Transmission to recipients occurred under general contact and trophallaxis, but at varying levels. Recipients mixed with uninfected (control) bees did not acquire Arsenophonus, with the exception of two individuals in the general contact treatment. Coloured dots show raw binomial data jittered (0 = Arsenophonus−, 1 = Arsenophonus+) and error bars indicate binomial SE. Each group was replicated (n = 10), with five donor bees and ten recipient bees per replicate.
Within this experiment, a subset of donors lost the infection from the 85% prevalence starting threshold (Fig. 8B), paralleling the previous observation that infection was unstable under laboratory isolation. Horizontal acquisition between donor and recipient bees contrasted to control pots where the same contact was with uninfected ‘donor’ bees. Here, recipient controls under trophallaxis remained uninfected (control recipients, 0%, 95% CI: 0–13.7), however two control recipients under general contact tested positive for Arsenophonus (control recipients, 6.66%, 95% CI: 0.818–22.1). The fate of individuals (live/dead at end of transmission period) emerged as an important predictor of infection status (GLMM, LRT: X2 = 7.64, df = 1, p = 0.00571**), with individuals that had died during the study more commonly associated with Arsenophonus infection (see Supplementary information Fig. 6 and Supplementary information Table 4 for a full summary).
Discussion
The diversity of existing symbiotic interactions represents the result of historical evolutionary transitions between lifestyles. Here, we investigate the symbiotic lifestyle of Arsenophonus associated with honey bees. A significant contingent of the Arsenophonus clade is in the latter stages of an evolutionary trajectory towards obligate intracellular life [30–32], and until now all characterised members of the genus demonstrate a heritable lifestyle, at least to some degree [26, 27]. We here show that Arsenophonus associated with honey bees deviates significantly from this heritable model, instead demonstrating strong seasonal patterns and a dynamic association that appears to be driven by social transmission with an additional, unidentified, environmental reservoir. These results shed new light on evolutionary transitions and symbiotic diversity within a clade of heritable arthropod symbionts.
We have identified the Arsenophonus–honey bee strain as the first within the clade to show little evidence of heritability, with evidence for this coming from multiple sources. First, the absence of the bacterium in eggs of infected colonies corroborates previous findings that Arsenophonus is not transovarially transmitted in honey bees [43], while very low prevalence in early host life stages provides new evidence that no alternative routes of VT are operating in the association. Secondly, the pronounced seasonal dynamics we observed for Arsenophonus are not consistent with the epidemiology of heritable symbionts, whose infection prevalence generally remains relatively static within a host generation though may vary dynamically between generations [70]. Within these dynamics, colonies appear to lose Arsenophonus over the winter, or the association declines to a level undetectable by our survey. Finally, experiments show first that workers bees with Arsenophonus infection lose it under laboratory conditions under social isolation, and also that social contact can result in acquisition of infection. Overall, Arsenophonus presents as a horizontally acquired infection of honey bees rather than a persistent VT transmitted one.
This acquisition-loss cycle occurs for a bacterium that is otherwise nested into a clade of heritable microbes, with phylogenomic analysis indicating Arsenophonus from bees lies in the same subclade as the male-killing microbe, Arsenophonus nasoniae. Whilst predominantly associated with the parasitic Nasonia wasp, A. nasoniae is an extracellular symbiont that also has phases outside of the wasp host [71], is culturable [28], and is also maintained by a combination of vertical and infectious transmission [13]. Arsenophonus nasoniae and the bee Arsenophonus also share the capacity to establish through the gut [71], indicating this pathway is preserved in the clade. The genome of Arsenophonus from the honey bee is, like A. nasoniae, relatively large in size and not subject to the degradation process that occurs in the obligate symbionts in the genus. The two strains have markedly similar predicted metabolic competence profiles, which further supports the likely culturability of the Arsenophonus from Apis, and its capacity to survive and replicate outside of the host environment. Taken together, the A. nasoniae/honey bee Arsenophonus clade is most parsimoniously regarded as a retained free-living, insect-associated microbe in a clade that has evolved into endosymbiosis, and indeed obligate endosymbiosis, in other cases. The alternate model—reacquisition of free-living capacity—is less probable as this would require the re-establishment of multiple genetic systems allowing growth outside of a host environment. This scenario is not, however, impossible, as previous work on Coxiella indicated the striking emergence of an infectious vertebrate pathogen from a clade of heritable, and sometimes obligate, endosymbionts of arthropods [22].
Our data thus indicate that the clade Arsenophonus contains more transmission diversity than previously considered. Knowledge of the selective forces that drive the emergence of symbiotic diversity is imperative for understanding important transitions such as the emergence of pathogenic agents from commensal partners, and vice versa [21, 22, 72]. Enterobacteriaceae genera appear to be well adapted for transitioning between ecological and symbiotic niches [73, 74]; for example, Pantoea, Sodalis and Serratia include notable symbionts of insects [75–77] but are largely comprised of representatives from soil, plants and clinical settings [21, 74, 77]. Likewise, the genus Photorhabdus includes both defensive symbionts and environmentally acquired pathogens of invertebrates [78, 79], but also a primary pathogen of humans with an unidentified transmission route [80, 81]. Among Enterobacteriaceae, the Arsenophonus genus differs, in that all strains to date are believed to be arthropod host-restricted and previously environmental acquisition has not been shown to occur routinely [27].
Given that vertical transmission is not operating in the honey bee–Arsenophonus association, the questions arises: what are the main drivers of HT? Do honey bees acquire infection predominantly from infected conspecifics, or are infections acquired from the wider environment (e.g. additional host species or reservoirs)? While the symbiotic phenotype of the honey bee–Arsenophonus remains unknown, we identified horizontal (infectious) transmission when general contact between conspecifics was allowed, and when contact was via trophallaxis exclusively. For a heritable symbiont, maintenance or reversal to the ancestral (horizontal) transmission route has a clear adaptive benefit in a eusocial host, typified by high host density and low genetic diversity [82], and social acquisition can prevent workers being the evolutionary dead ends they are often considered for heritable symbionts [83]. Localisation in the gut and detection in faeces is consistent with a faecal—oral route, which could allow effective social transmission within a colony and to other hosts via shared floral resources [84–86]. Indeed, Arsenophonus has previously been detected on flowers [87], but until now direct evidence for the potential capacity to transmit via this route was lacking.
The marked seasonal incidence of Arsenophonus, peaking in autumn, has a number of potential drivers. Shedding and accumulation of the bacterium in the foraging environment (e.g. on flowers) may generate increased infection risk as the foraging season proceeds, as observed for some parasite species [88]. However, this would be contingent on the survival of Arsenophonus in the environment, a trait conventionally considered absent from this class of insect symbionts, despite relatively large symbiont genomes and cell free cultivability found within the clade [89, 90]. Alternatively, but not mutually exclusively, Arsenophonus dynamics may reflect the activity period of an environmental reservoir or additional host species that is responsible for transmission to honey bees. Comparable seasonality is observed for Spiroplasma in honey bees, with peak prevalence of S. melliferum aligning with peak flowering periods [91, 92]. This latter hypothesis may explain our observation that apiary is a significant predictor of a colonies Arsenophonus status, while over broad spatial scales infection prevalence does not vary notably. Here, localised spatial pattern reflects shared exposure to reservoirs of infection, with colonies originating from the same apiary often overlapping in foraging area [93]. Relevant here are reports of Arsenophonus associated with pollen and nectaring sites [42, 87], providing further evidence for environmental transmission [87]. Other processes, such as drifting of infected bees within apiaries, could drive inter-colonial transmission of Arsenophonus and feedback to drive the localised infection prevalence we observed.
The position of the honey bee Arsenophonus on the parasitism-mutualism continuum remains speculative. Our observations of HT, systemic infections and a higher prevalence among dead hosts strengthens interpretations of previous work correlating Arsenophonus with poor health outcomes in bees [44, 45, 94]. HT is considered to increase the scope for the evolution of virulence [17, 18], although this trait alone is insufficient to draw conclusions regarding symbiont phenotype, as many avirulent or beneficial microbes are transmitted horizontally [15, 95]. The association with dead hosts may implicate Arsenophonus directly or represent opportunistic proliferation in a compromised host. Alternatively, saprophytic growth on cadavers may be occurring, a capacity demonstrated by A. nasoniae in fly puparia [96]. Further work is needed to determine if health outcomes are causal in honey bees and to characterise the transmission phenotype of Arsenophonus reported from other bee species [94, 97–99].
To conclude, the honey bee–Arsenophonus contrasts in its phenotype from the rest of the heritable clade, as vertical transmission is not the predominant transmission mode. Instead, HT occurs via social interactions, colonisation can occur in the gut and environmental reservoirs appear to be necessary for maintenance of the association. Our data establish the honey bee–Arsenophonus as a key link in a symbiont clades trajectory from free-living ancestral life to obligate mutualism threatened by mutational decay. This will provide new avenues for research into the emergence of symbiotic diversity.
Supplementary information
Acknowledgements
We thank Dr. Katherine Roberts, Dr. Laurent Gauthier and Dr. Kirsty Stainton for assistance on the project, Dr. Andri Manser for statistical input, and three anonymous reviewers for improving the manuscript. Jack Wilford, and many other bee keepers, were pivotal in sampling efforts. This work was financially supported by a BBSRC iCASE studentship to GCD (BB/L016133/1) and Bee Disease Insurance Ltd. Microscopy was conducted at the Centre for Cell Imaging (CCI), using equipment under grant BB/M012441/1.
Author contributions
The project was conceived initially by GEB, CLF and GDDH. GCD designed and completed survey, experimental and statistical analysis, with input from GDDH and GEB. PN, OY and SS completed the sequencing and phylogenomic analysis elements. GCD wrote the paper with GDDH and SS, which was edited by OY, PN and GEB with the final draft approved by all authors.
Data availability
All data associated with this study can be publicly accessed at 10.6084/m9.figshare.c.5073479.v1.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00977-z.
References
- 1.Ewald PW. Transmission modes and evolution of the parasitism-mutualism continuum. Ann NY Acad Sci. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- 2.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–90. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 3.Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;51:505–10. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salem H, Florez L, Gerardo N, Kaltenpoth M. An out-of-body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proc R Soc B. 2015;282:1804. doi: 10.1098/rspb.2014.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebert D. The epidemiology and evolution of symbionts with mixed-mode transmission. Annu Rev Ecol Evol Syst. 2013;44:623–43. doi: 10.1146/annurev-ecolsys-032513-100555. [DOI] [Google Scholar]
- 6.Webster JP, Borlase A, Rudge JW. Who acquires infection from whom and how? Disentangling multi-host and multimode transmission dynamics in the ‘elimination’ era. Philos Trans R Soc B Biol Sci. 2017;372:20160091. [DOI] [PMC free article] [PubMed]
- 7.Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–76. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Law R, Dieckmann U. Symbiosis through exploitation and the merger of lineages in evolution. Proc R Soc B. 1998;265:1245–53. doi: 10.1098/rspb.1998.0426. [DOI] [Google Scholar]
- 9.Cordaux R, Michel-Salzat A, Bouchon D. Wolbachia infection in crustaceans: novel hosts and potential routes for horizontal transmission. J Evol Biol. 2001;14:237–43. doi: 10.1046/j.1420-9101.2001.00279.x. [DOI] [Google Scholar]
- 10.Russell JA, Latorre A, Sabater-Muñoz B, Moya A, Moran NA. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol. 2003;12:1061–75. doi: 10.1046/j.1365-294X.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- 11.Zug R, Koehncke A, Hammerstein P. Epidemiology in evolutionary time: the case of Wolbachia horizontal transmission between arthropod host species. J Evol Biol. 2012;25:2149–60. doi: 10.1111/j.1420-9101.2012.02601.x. [DOI] [PubMed] [Google Scholar]
- 12.Werren JH, O’Neill SL. The evolution of heritable symbionts. In: O’Neill SL, Hoffmann AA, Werren JH (eds). Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. 1997. Oxford University Press, Oxford, pp 1–41.
- 13.Parratt SR, Frost CL, Schenkel MA, Rice A, Hurst GDD, King KC. Superparasitism drives heritable symbiont epidemiology and host sex ratio in a wasp. PLoS Pathog. 2016;12:1–22. doi: 10.1371/journal.ppat.1005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon ERL, McFrederick QS, Weirauch C. Comparative phylogenetic analysis of bacterial associates in Pyrrhocoroidea and evidence for ancient and persistent environmental symbiont reacquisition in Largidae (Hemiptera: Heteroptera) Appl Environ Microbiol. 2016;82:064022. doi: 10.1128/AEM.02114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol. 2007;73:4308 LP–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchner P. Endosymbiosis of animals with plant microorganisms. Z Für Allg Mikrobiol. 1967;7:168. doi: 10.1002/jobm.19670070220. [DOI] [Google Scholar]
- 17.Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:211–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- 18.Frank SA. Host-symbiont conflict over the mixing of symbiotic lineages. Proc Biol Sci. 1996;263:339–44. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- 19.Sachs JL, Essenberg CJ, Turcotte MM. New paradigms for the evolution of beneficial infections. Trends Ecol Evol. 2011;26:202–9. doi: 10.1016/j.tree.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro JW, Turner PE. The impact of transmission mode on the evolution of benefits provided by microbial symbionts. Ecol Evol. 2014;4:3350–61. doi: 10.1002/ece3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clayton AL, Oakeson KF, Gutin M, Pontes A, Dunn DM, Von AC, et al. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect—bacterial symbioses. Plos Genet. 2012;8:11. doi: 10.1371/journal.pgen.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duron O, Noël V, Mccoy KD, Bonazzi M, Sidi K, Morel O, et al. The recent evolution of a maternally- inherited endosymbiont of ticks led to the emergence of the Q fever pathogen Coxiella burnetii. Plos Pathog. 2015;11:1–23. doi: 10.1371/journal.ppat.1004892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo WS, Huang YY, Kuo CH. Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiol Rev. 2016;40:855–74. doi: 10.1093/femsre/fuw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toft C, Andersson SGE. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 2010;11:465–75. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 25.Wilkes TE, Duron O, Darby AC, Hypša V, Nováková E, Hurst GDD. The Genus Arsenophonus. In: Bourtzis K, Zchori-Fein E, editors. Manipulative tenants: bacteria associated with arthropods. Boca Raton: CRC Press; 2011. p. 225–44.
- 26.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstädter J, et al. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008;6:1–12. doi: 10.1186/1741-7007-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nováková E, Hypša V, Moran NA. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 2009;9:1–14. doi: 10.1186/1471-2180-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gherna RL, Werren JH, Weisburg W, Cote R, Woese CR, Mandelco L, et al. Notes: Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int J Syst Bacteriol. 1991;41:563–5. doi: 10.1099/00207713-41-4-563. [DOI] [Google Scholar]
- 29.Qu LY, Lou YH, Fan HW, Ye YX, Huang HJ, Hu MQ, et al. Two endosymbiotic bacteria, Wolbachia and Arsenophonus, in the brown planthopper Nilaparvata lugens. Symbiosis. 2013;61:47–53. doi: 10.1007/s13199-013-0256-9. [DOI] [Google Scholar]
- 30.Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci USA. 2010;107:12168–73. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nováková E, Hypša V, Nguyen P, Husník F, Darby AC. Genome sequence of Candidatus Arsenophonus lipopteni, the exclusive symbiont of a blood sucking fly Lipoptena cervi (Diptera: Hippoboscidae) Stand Genom Sci. 2016;11:72. doi: 10.1186/s40793-016-0195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perotti MA, Allen JM, Reed DL, Braig HR. Host-symbiont interactions of the primary endosymbiont of human head and body lice. FASEB J. 2007;21:1058–66. doi: 10.1096/fj.06-6808com. [DOI] [PubMed] [Google Scholar]
- 33.Nováková E, Husník F, Šochová E, Hypša V. Arsenophonus and Sodalis symbionts in louse flies: An analogy to the Wigglesworthia and Sodalis system in tsetse flies. Appl Environ Microbiol. 2015;81:6189–99. doi: 10.1128/AEM.01487-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duron O, Wilkes TE, Hurst GDD. Interspecific transmission of a male-killing bacterium on an ecological timescale. Ecol Lett. 2010;13:1139–48. doi: 10.1111/j.1461-0248.2010.01502.x. [DOI] [PubMed] [Google Scholar]
- 35.Huger AM, Skinner SW, Werren JH. Bacterial infections associated with the son-killer trait in the parasitoid wasp Nasonia (= Mormoniella) vitripennis (Hymenoptera: Pteromalidae) J Invertebr Pathol. 1985;46:272–80. doi: 10.1016/0022-2011(85)90069-2. [DOI] [PubMed] [Google Scholar]
- 36.Bressan A. Emergence and evolution of Arsenophonus bacteria as insect-vectored plant pathogens. Infect Genet Evol. 2014;22:81–90. doi: 10.1016/j.meegid.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Bressan A, Terlizzi F, Credi R. Independent origins of vectored plant pathogenic bacteria from arthropod-associated Arsenophonus endosymbionts. Micro Ecol. 2012;63:628–38. doi: 10.1007/s00248-011-9933-5. [DOI] [PubMed] [Google Scholar]
- 38.Bressan A, Sémétey O, Arneodo J, Lherminier J, Boudon-Padieu E. Vector transmission of a plant-pathogenic bacterium in the Arsenophonus clade sharing ecological traits with facultative insect endosymbionts. Phytopathology. 2009;99:1289–96. doi: 10.1094/PHYTO-99-11-1289. [DOI] [PubMed] [Google Scholar]
- 39.Aizenberg-Gershtein Y, Izhaki I, Halpern M. Do honeybees shape the bacterial community composition in floral nectar? PLoS ONE. 2013;8:e67556. doi: 10.1371/journal.pone.0067556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babendreier D, Joller D, Romeis J, Bigler F, Widmer F. Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS Microbiol Ecol. 2007;59:600–10. doi: 10.1111/j.1574-6941.2006.00249.x. [DOI] [PubMed] [Google Scholar]
- 41.Corby-Harris V, Maes P, Anderson KE. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE. 2014;9:e95056. doi: 10.1371/journal.pone.0095056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donkersley P, Rhodes G, Pickup RW, Jones KC, Wilson K. Bacterial communities associated with honeybee food stores are correlated with land use. Ecol Evol. 2018;8:4743–56. doi: 10.1002/ece3.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yañez O, Gauthier L, Chantawannakul P, Neumann P. Endosymbiotic bacteria in honey bees: Arsenophonus spp. are not transmitted transovarially. FEMS Microbiol Lett. 2016;363:1–7. doi: 10.1093/femsle/fnw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budge GE, Adams I, Thwaites R, Pietravalle S, Drew GC, Hurst GDD, et al. Identifying bacterial predictors of honey bee health. J Invertebr Pathol. 2016;141:41–4. doi: 10.1016/j.jip.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Cornman RS, Tarpy DR, Chen Y, Jeffreys L, Lopez D, Pettis JS, et al. Pathogen webs in collapsing honey bee colonies. PLoS ONE. 2012;7:e43562. [DOI] [PMC free article] [PubMed]
- 46.Hughes DP, Pierce NE, Boomsma JJ. Social insect symbionts: evolution in homeostatic fortresses. Trends Ecol Evol. 2008;23:672–7. doi: 10.1016/j.tree.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Schmid-Hempel P. Parasites and their social hosts. Trends Parasitol. 2017;33:453–62. doi: 10.1016/j.pt.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Wilson EO. The insect societies. Harvard University Press: Cambridge, MA, 1971.
- 49.Onchuru TO, Javier Martinez A, Ingham CS, Kaltenpoth M. Transmission of mutualistic bacteria in social and gregarious insects. Curr Opin Insect Sci. 2018;28:50–58. doi: 10.1016/j.cois.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Rubin BER, Sanders JG, Turner KM, Pierce NE, Kocher SD. Social behaviour in bees influences the abundance of Sodalis (Enterobacteriaceae) symbionts. R Soc Open Sci. 2018;5:180369. [DOI] [PMC free article] [PubMed]
- 51.Anderson KE, Russell JA, Moreau CS, Kautz S, Sullam KE, Hu Y, et al. Highly similar microbial communities are shared among related and trophically similar ant species. Mol Ecol. 2012;21:2282–96. doi: 10.1111/j.1365-294X.2011.05464.x. [DOI] [PubMed] [Google Scholar]
- 52.Frost CL, FernÁndez-MarÍn H, Smith JE, Hughes WOH. Multiple gains and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol Ecol. 2010;19:4077–85. doi: 10.1111/j.1365-294X.2010.04764.x. [DOI] [PubMed] [Google Scholar]
- 53.Keller L, Liautard C, Reuter MAX, Brown WD, Chapuisat M, Sundstro L. Sex ratio and Wolbachia infection in the ant Formica exsecta. Heredity. 2001;87:227–33. doi: 10.1046/j.1365-2540.2001.00918.x. [DOI] [PubMed] [Google Scholar]
- 54.Van Borm S, Wenseleers T, Billen J, Boomsma JJ. Wolbachia in leafcutter ants: a widespread symbiont that may induce male killing or incompatible matings. J Evol Biol. 2001;14:805–14. doi: 10.1046/j.1420-9101.2001.00321.x. [DOI] [Google Scholar]
- 55.Wenseleers T, Sundström L, Billen J. Deleterious Wolbachia in the ant Formica truncorum. Proc R Soc B Biol Sci. 2002;269:623–9. doi: 10.1098/rspb.2001.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gauthier L, Cornman S, Hartmann U, Cousserans F, Evans JD, De Miranda JR, et al. The Apis mellifera filamentous virus genome. Viruses. 2015;7:3798–815. doi: 10.3390/v7072798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 59.Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–13. [PubMed] [Google Scholar]
- 60.Lourenço AP, Mackert A, Cristino A, dos S, Simões ZLP. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie. 2008;39:372–85. doi: 10.1051/apido:2008015. [DOI] [Google Scholar]
- 61.Boncristiani H, Li J, Evans JD, Pettis J, Chen Y. Scientific note on PCR inhibitors in the compound eyes of honey bees, Apis mellifera. Apidologie. 2011;42:457–60. doi: 10.1007/s13592-011-0009-9. [DOI] [Google Scholar]
- 62.Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, et al. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 2008;22:2591–9. doi: 10.1096/fj.07-101162. [DOI] [PubMed] [Google Scholar]
- 63.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R Core Team. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. http://www.R-project.org/.
- 65.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models Using lme4. J Stat Softw. 2015;1:1–48.
- 66.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–23. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 67.Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. Springer Science & Business Media: New York, NY, 2003.
- 68.Zuur AF, Ieno EN, Elphick CS. A protocol for data exploration to avoid common statistical problems. Methods Ecol Evol. 2009;1:3–14. doi: 10.1111/j.2041-210X.2009.00001.x. [DOI] [Google Scholar]
- 69.Frost CL, Siozios S, Nadal-Jimenez P, Brockhurst MA, King KC, Darby AC, et al. The hypercomplex genome of an insect reproductive parasite highlights the importance of lateral gene transfer in symbiont biology. mBio. 2020;11:e02590–19. doi: 10.1128/mBio.02590-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith AH, Łukasik P, O’Connor MP, Lee A, Mayo G, Drott MT, et al. Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol Ecol. 2015;24:1135–49. doi: 10.1111/mec.13095. [DOI] [PubMed] [Google Scholar]
- 71.Nadal-Jimenez P, Griffin JS, Davies L, Frost CL, Marcello M, Hurst GDD. Genetic manipulation allows in vivo tracking of the life cycle of the son-killer symbiont, Arsenophonus nasoniae, and reveals patterns of host invasion, tropism and pathology. Environ Microbiol. 2019;21:3172–82. doi: 10.1111/1462-2920.14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc Biol Sci. 2006;273:2097–106. doi: 10.1098/rspb.2006.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108:10800–7. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walterson AM, Stavrinides J. Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev. 2015;39:968–84. doi: 10.1093/femsre/fuv027. [DOI] [PubMed] [Google Scholar]
- 75.Chrudimský T, Husník F, Nováková E, Hypša V. Candidatus Sodalis melophagi sp. nov.: phylogenetically independent comparative model to the tsetse fly symbiont Sodalis glossinidius. PLoS ONE. 2012;7:e40354. doi: 10.1371/journal.pone.0040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dale C, Maudlin I. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol. 1999;1:267–75. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 77.Kenyon LJ, Meulia T, Sabree ZL. Habitat visualization and genomic analysis of ‘Candidatus Pantoea carbekii,’ the primary symbiont of the brown marmorated stink bug. Genome Biol Evol. 2015;7:620–35. doi: 10.1093/gbe/evv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fischer-Le Saux M, Viallard V, Brunel B, Normand P, Boemare NE. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov. Int J Syst Evol Microbiol. 1999;49:1645–56. doi: 10.1099/00207713-49-4-1645. [DOI] [PubMed] [Google Scholar]
- 79.Forst S, Dowds B, Boemare N, Stackebrandt E. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 80.Costa SCP, Girard PA, Brehélin M, Zumbihl R. The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect Immun. 2009;77:1022–30. doi: 10.1128/IAI.01064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gerrard J, Waterfield N, Vohra R, ffrench-Constant R. Human infection with Photorhabdus asymbiotica: an emerging bacterial pathogen. Microbes Infect. 2004;6:229–37. doi: 10.1016/j.micinf.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 82.Schmid-Hempel P. Parasites in social insects. Princeton University Press: Princeton, NJ, 1998.
- 83.Frost CL, Pollock SW, Smith JE, Hughes WOH. Wolbachia in the flesh: symbiont intensities in germ-line and somatic tissues challenge the conventional view of Wolbachia transmission routes. PLoS ONE. 2014;9:e95122. [DOI] [PMC free article] [PubMed]
- 84.Graystock P, Goulson D, Hughes WOH. Parasites in bloom: Flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc R Soc B Biol Sci. 2015;282:1471–2954. doi: 10.1098/rspb.2015.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Graystock P, Goulson D, Hughes WOH. The relationship between managed bees and the prevalence of parasites in bumblebees. PeerJ. 2014;2:e522. doi: 10.7717/peerj.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koch H, Abrol DP, Li J, Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol. 2013;22:2028–44. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- 87.McFrederick QS, Thomas JM, Neff JL, Vuong HQ, Russell KA, Hale AR, et al. Flowers and wild megachilid bees share microbes. Micro Ecol. 2017;73:188–200. doi: 10.1007/s00248-016-0838-1. [DOI] [PubMed] [Google Scholar]
- 88.Satterfield DA, Altizer S, Williams MK, Hall RJ. Environmental persistence influences infection dynamics for a butterfly pathogen. PLoS ONE. 2017;12:1–16. doi: 10.1371/journal.pone.0169982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Darby AC, Choi JH, Wilkes T, Hughes MA, Werren JH, Hurst GDD, et al. Characteristics of the genome of Arsenophonus nasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol Biol. 2010;19:75–89. doi: 10.1111/j.1365-2583.2009.00950.x. [DOI] [PubMed] [Google Scholar]
- 90.Dale C, Beeton M, Harbison C, Jones T, Pontes M. Isolation, pure culture, and characterization of ‘Candidatus Arsenophonus arthropodicus,’ an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl Environ Microbiol. 2006;72:2997–3004. doi: 10.1128/AEM.72.4.2997-3004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clark T. Honeybee spiroplasmosis, a new problem for beekeepers. Am Bee J. 1978;118:18–19. [Google Scholar]
- 92.Schwarz RS, Teixeira ÉW, Tauber JP, Birke JM, Martins MF, Fonseca I, et al. Honey bee colonies act as reservoirs for two Spiroplasma facultative symbionts and incur complex, multiyear infection dynamics. MicrobiologyOpen. 2014;3:341–55. doi: 10.1002/mbo3.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levin MD. Interactions among foraging honey bees from different apiaries in the same field. Insectes Sociaux. 1961;8:195–201. doi: 10.1007/BF02224008. [DOI] [Google Scholar]
- 94.Parmentier A, Billiet A, Smagghe G, Vandamme P, Deforce D, Van Nieuwerburgh F, et al. A prokaryotic–eukaryotic relation in the fat body of Bombus terrestris. Environ Microbiol Rep. 2018;10:644–50. doi: 10.1111/1758-2229.12673. [DOI] [PubMed] [Google Scholar]
- 95.Nussbaumer AD, Fisher CR, Bright M. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature. 2006;441:345–8. doi: 10.1038/nature04793. [DOI] [PubMed] [Google Scholar]
- 96.Werren JH, Skinner SW, Huger AM. Male-killing bacteria in a parasitic wasp. Science. 1986;231:990–2. doi: 10.1126/science.3945814. [DOI] [PubMed] [Google Scholar]
- 97.Gerth M, Saeed A, White JA, Bleidorn C. Extensive screen for bacterial endosymbionts reveals taxon-specific distribution patterns among bees (Hymenoptera, Anthophila) FEMS Microbiol Ecol. 2015;91:1–12. doi: 10.1093/femsec/fiv047. [DOI] [PubMed] [Google Scholar]
- 98.McFrederick QS, Mueller UG, James RR. Interactions between fungi and bacteria influence microbial community structure in the Megachile rotundata larval gut. Proc R Soc B Biol Sci. 2014;281:1471–2954. doi: 10.1098/rspb.2013.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saeed A, White JA. Surveys for maternally-inherited endosymbionts reveal novel and variable infections within solitary bee species. J Invertebr Pathol. 2015;132:111–4. doi: 10.1016/j.jip.2015.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study can be publicly accessed at 10.6084/m9.figshare.c.5073479.v1.