This study shows that elevated apolipoprotein E in PDAC mediates immune suppression and high serum apolipoprotein E levels correlate with poor patient survival.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy with few effective therapeutic options. PDAC is characterized by an extensive fibroinflammatory stroma that includes abundant infiltrating immune cells. Tumor-associated macrophages (TAM) are prevalent within the stroma and are key drivers of immunosuppression. TAMs in human and murine PDAC are characterized by elevated expression of apolipoprotein E (ApoE), an apolipoprotein that mediates cholesterol metabolism and has known roles in cardiovascular and Alzheimer's disease but no known role in PDAC. We report here that ApoE is also elevated in peripheral blood monocytes in PDAC patients, and plasma ApoE protein levels stratify patient survival. Orthotopic implantation of mouse PDAC cells into syngeneic wild-type or in ApoE−/− mice showed reduced tumor growth in ApoE−/− mice. Histologic and mass cytometric (CyTOF) analysis of these tumors showed an increase in CD8+ T cells in tumors in ApoE−/− mice. Mechanistically, ApoE induced pancreatic tumor cell expression of Cxcl1 and Cxcl5, known immunosuppressive factors, through LDL receptor and NF-κB signaling. Taken together, this study reveals a novel immunosuppressive role of ApoE in the PDAC microenvironment.
Significance:
This study shows that elevated apolipoprotein E in PDAC mediates immune suppression and high serum apolipoprotein E levels correlate with poor patient survival.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy, with a 5-year survival rate of ∼10% (1). PDAC is characterized by a fibroinflammatory stroma comprised in large part by cancer-associated fibroblasts (CAF) and tumor-associated macrophages (TAM; ref. 2). Complex cellular cross-talk between these populations and cancer cells drives the formation of a highly immunosuppressive tumor microenvironment (TME; ref. 3), which serves to dampen the effectiveness of otherwise-promising immune-checkpoint therapies (4, 5).
Apolipoprotein E (ApoE) is secreted at high levels by hepatocytes and macrophages to mediate lipid metabolism and is involved in diseases such as Alzheimer's disease and atherosclerosis (6). Increased ApoE levels are observed in several tumor types (7), including non–small cell lung (8), gastric (9), ovarian (10), and bladder cancers (11). APOE's roles in cancer are context dependent: in acute myeloid leukemia (AML), it mediates T-cell suppression (12, 13), whereas in melanoma, it promotes cytotoxic T-cell responses (14, 15).
We found that APOE is expressed predominately by TAMs and CAFs in mouse and human PDAC. Tumors in ApoE knockout (ApoE−/−) mice have lower tumor burden, less fibrosis, reduced innate immune response, and increased adaptive immune cell infiltration. ApoE−/− mouse tumors had fewer immature myeloid cells and regulatory T cells (Treg), suggesting reduced immunosuppression, consistent with increased tumor-infiltrating CD8+ T cells. RNA sequencing (RNA-seq) of PDAC cells treated with recombinant ApoE showed upregulation of the chemokines Cxcl1 and Cxcl5, which are known to impair T-cell infiltration in PDAC (16). Gene set enrichment analysis (GSEA) revealed that treatment of tumor cells with recombinant ApoE upregulates NF-κB signaling, which in turn upregulates Cxcl1 and Cxcl5 expression. We propose that ApoE actively promotes an immunosuppressive TME in PDAC.
Materials and Methods
Study approvals
Animal experiments were performed at the University of Michigan in compliance with, and approved by, the Institutional Animal Care and Use Committee. Human research was performed in accordance with the Declaration of Helsinki and the ethical standards and guidelines approved by the University of Michigan Institutional Review Board. All patients provided written informed consent before procedures were performed. Peripheral blood was collected from patients who received diagnostic endoscopic ultrasound for pancreatic mass under IRB HUM00041280 or surgical resection under IRB HUM00025339.
APOE expression analysis and human PDAC stratification
RNA-seq data from laser capture microdissected tumor epithelium and matched stroma (n = 65) were downloaded from NCBI Gene-Expression Omnibus (GEO; accession number GSE93326). For stratification of patient tumors by APOE expression, we used The Cancer Genome Atlas (TCGA) PDAC RNA-seq data (150 samples) downloaded from cBioPortal (https://www.cbioportal.org/). Tumor samples were split into two groups, APOE top and bottom quartile. Genes differentially expressed between the two groups were identified using limma package in R software (v 3.5.2) with an adjusted P < 0.05.
Single-cell RNA-seq
Human single-cell RNA-seq (scRNA-seq) data were previously published in (17). The processed data are available at the NIH GEO database (accession #GSE155698). Both raw and processed data are available at the NIH dbGaP database (accession #phs002071.v1.p1; ref. 17), with full clinical annotation. Raw and processed scRNA-seq data sets for the orthotopic KPC tumor and normal mouse pancreas are available at GEO (accession #GSE158356). Downstream analysis was performed using Seurat V3.2.2 in R Studio V1.3.1093. Code is publicly available on GitHub.com (https://github.com/sam-kemp/Apoe_ImmuneSuppression_scRNAseq).
Human peripheral blood mononuclear cells and plasma isolation
Human peripheral blood mononuclear cells (PBMC) and plasma were prepared from whole blood as previously described (17).
Enzyme-linked immunosorbent assay
APOE in human plasma was measured using the Human Apolipoprotein E Human ELISA Kit (Abcam, ab108813). ApoE in mouse macrophage media was measured with Mouse Apolipoprotein E SimpleStep ELISA Kit (Abcam, ab215086). Human CXCL1 was measured with Human CXCL1/GRO alpha Quantikine ELISA Kit (R&D Systems, DGR00B). Mouse bone marrow cells were plated in 1:1 tumor cell conditioned media (CM):DMEM with 10% FBS for seven days before media were removed. For APOE detection, human plasma and macrophage CM were diluted 1:1,000 and 1:100, respectively, and measured in duplicate. APOE or CXCL1 levels in PDAC patients were stratified by APOE or CXCL1 levels (top and bottom quartile). Survival analysis was performed using log-rank test.
Mice
Wild-type C57BL/6J mice and ApoE−/− mice on the C57BL/6 background were purchased from the Jackson Laboratory, (#000664; #002052) and housed at the University of Michigan Rogel Cancer Center.
Orthotopic transplantation
For orthotopic allografts, 5 × 104 7940b KPC (C57BL/6) cells were injected into the pancreas as previously described (18).
Doxycycline treatment
iKras* mice (19) were fed doxycycline chow (BioServ, F3949) to induce KrasG12D expression for 72 hours. Pancreatitis was induced by two days of 8 intraperitoneal injections of caerulein (Sigma, 75 μg/kg) with continuous administration of doxycycline as previously described (19). Doxycycline chow was administered for 3 weeks for Kras-ON time points and replaced with regular chow for Kras-OFF time points. Littermate control mice lacking the full set of alleles received caerulein and doxycycline at indicated time points.
Histopathology
Tissues were fixed overnight in 10% neutral-buffered formalin, dehydrated, and paraffin-embedded. Hematoxylin and eosin and Gomori Trichrome (Thermo Fisher, #87021) staining was performed according to the manufacturer's guidelines. IHC was performed using a Ventana Discovery Ultra XT autostainer. For immunofluorescence staining, deparaffinized slides were blocked with 1% bovine serum albumin in PBS for 1 hour at RT. Primary antibody was diluted in blocking buffer and incubated overnight at 4°C, followed by secondary antibody (Alexa Fluor secondaries, 1:300) for 45 minutes at RT. Nuclei were counterstained with Prolong Diamond Antifade Mountant with DAPI (Invitrogen). Antibodies are found in Supplementary Table S1. Inverted fluorescence images were taken on an Olympus BX53F microscope and confocal images on either Leica SP5 or SP8 microscopes. ImageJ, Fiji V2.0.0-rc-69/1.52p was used for quantitation using at least three 20× magnification fields across three or more biological replicates.
Macrophage polarization
Bone marrow cell isolation from C57BL/6J mouse femurs and macrophage polarization was performed as previously described (20, 21).
qRT-PCR
Tumor tissues were flash-frozen in liquid nitrogen and stored in RNAlater-ICE (Thermo Fisher, AM7030) overnight at −20°C. Tissues and cell lines were homogenized in Buffer RLT (Qiagen, 79216). RNA was extracted according to RNeasy Plus instructions (Qiagen, 74134). cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, 4368814). For qRT-PCR, samples were prepared with either Fast SYBR Green PCR Master Mix (Applied Biosystems, 4385612) or TaqMan Universal Master Mix (Applied Biosystems, 4364340). Cyclophilin A/Ppia was used for normalization. Primers and TaqMan probes are in Supplementary Table S2.
Flow cytometry
Mouse splenocytes were prepared by mincing spleens and filtering through 40-μm mesh. Flow cytometry was performed as previously described (22). Cyan ADP analyzer (Beckman Coulter) was used for data analysis. Downstream analysis was performed using FlowJo v10 software. Antibodies used can be found in Supplementary Table S1.
Mass cytometry
Mouse tumors were disrupted mechanically and enzymatically (1 mg/mL Collagenase P:DMEM) for 30 minutes at 37°C. Samples were washed with DMEM with 10% FBS. Single-cell suspensions were created by filtering through 40-μm mesh. Samples were prepared for CyTOF staining according to the manufacturer's guidelines (Fluidigm, PN 400276 A4). Samples were washed with Maxpar PBS (Fluidigm, 201058). Up to 1 × 106 cells were stained with Cell-ID Cisplatin (Fluidigm, #201064) for 5 minutes at RT, to label dead cells. Cells were washed with PBS and stained with a panel of 16 surface mouse antibodies (Supplementary Table S1) for 30 minutes at room temperature. Samples were washed with PBS twice and left pelleted in 1 mL Cell-ID Intercalator-IR (Fluidigm, 201192A) and run on the CyTOF2 at the University of Rochester Flow Cytometry Core. Files were normalized to internal bead controls. Downstream analysis was performed using Premium CytoBank Software (cytobank.org). Live singlets were gated using the DNA Intercalator Ir191, event length, and Cisplatin Pt195. tSNE and SPADE visualizations were performed in CytoBank on representative samples.
T-cell depletion
C57BL/6 and ApoE−/− mice were administered anti-CD8 antibody (Bio X Cell; #BP0060) and anti-CD4 antibody (Bio X Cell; # BE0003-1) via intraperitoneal injection at 200 μg/mouse three times weekly. Control mice were administered IgG2b isotype control (Bio X Cell; #BE0090).
Cell culture
KPC cell lines: 7940b (gift from Dr. Gregory Beatty, University of Pennsylvania), mT3, mT4, and mT5 (gift from Dr. David Tuveson, Cold Spring Harbor Laboratory). BLK6318 was generated from normal mouse (C57BL/6) pancreas. FB1 CAF line was generated from an iKras* p53* mouse (23) by fluorescence-activated cell sorting of PDGFRα+;EpCAM− cells. DMEM with 10% FBS and 1% penicillin/streptomycin (Thermo Fisher, 15140163) was used for all cell lines. Murine recombinant ApoE (Abcam, ab226314) was used at 0.3 μg/mL for all experiments. 50,000–100,000 cells were plated into a 6-well dish and allowed to adhere for 24 hours. ApoE was spiked into the culture media for 1 to 48 hours, depending on the experiment. For NF-κB inhibition, cells were pretreated with 5 or 10 μmol/L BAY 11–7082 (Abcam, ab141228) for 2 hours, before ApoE addition.
RNA-seq
Cells were lysed in Buffer RLT (Qiagen, 79216). RNA was extracted using RNeasy Plus Mini Kits (Qiagen, 74134). RNA quality was determined using both NanoDrop and an RNA Integrity Number >9, and then reverse transcribed. The University of Michigan Advanced Genomics Core prepared libraries, which underwent paired-end sequencing on the NovaSeq6000 (Illumina). Sample reads were aligned with HISAT2 v2.2.0 using prebuilt index of Mus musculus UCSC reference genome mm10 obtained from http://daehwankimlab.github.io/hisat2/download/. Gene assembly and quantification was done with Stringtie 2.1.1, counts of duplicated genes were averaged, and differential gene expression was determined using DESeq2 package (v 1.22.2) in R software after filtering out genes with low counts (total raw counts <20). Pathway enrichment and gene ontology analyses were performed with GSEA v4.0.3 (pre-ranked) using differentially expressed genes and with DAVID v6.8 (https://david.ncifcrf.gov/summary.jsp). Bulk RNA-seq data are available at GEO (accession #GSE160592).
siRNA transfection
7940b KPC cells were seeded at 60% confluency in a 6-well plate. The next day, two separate low-density lipoprotein receptor (LDLR) siRNA (Thermo, #s69153, #s69154) and scrambled negative control (Thermo, 4390843) were diluted in Opti-MEM (Gibco, 31985062). Lipofectamine RNAiMAX transfection reagent (Thermo, 13778075) was diluted in Opti-MEM. Diluted siRNA was mixed with diluted lipofectamine RNAiMAX at 1:1 ratio and incubated for 5 minutes at RT. siRNA and lipofectamine complexes were added to adherent cells at a final concentration of 25 pmol/L. Cells were incubated for 48 hours at 37°C.
Immunoblotting
Cells were lysed in RIPA buffer with protease (Sigma) and phosphatase (Roche) inhibitors. Protein samples were electrophoresed on 4%–20% precast polyacrylamide gels (BioRad, 4561094) and transferred to PVDF membrane (BioRad, 1620177). Membranes were blocked with 5% milk for one hour at RT, incubated with primary antibodies overnight at 4°C followed by secondary antibody for 2 hours at RT. Membranes were washed, incubated in Western Lightning Plus-ECL (PerkinElmer, 509049323) and imaged using BioRad Chemidoc. ImageJ was used for quantitation.
Statistical analysis
GraphPad Prism was used for statistical analyses. Data are presented as means ± standard deviation (SD). Two-tailed Student t test and one-way ANOVA with Tukey test were performed for comparison between groups. P < 0.05 was considered significant. Log-rank test was used for Kaplan–Meier analysis.
Results
APOE is highly expressed in the PDAC stroma and plasma levels correlate with patient survival
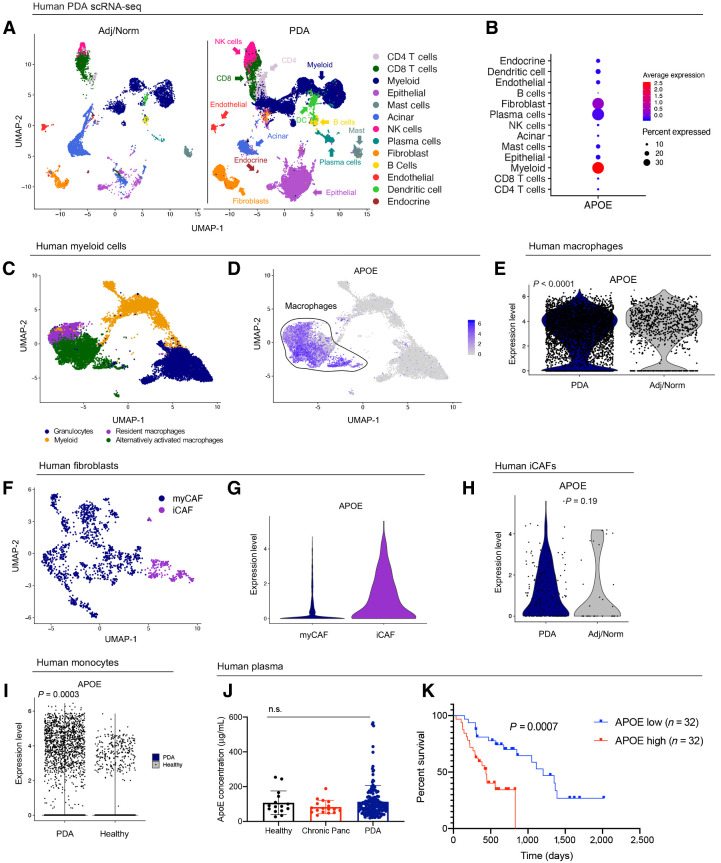
APOE is highly expressed in PDAC-associated myeloid cells and fibroblasts (24, 17), but has no known function. Because both myeloid cells and fibroblasts represent heterogeneous cell populations, we sought to describe APOE expression in further detail by interrogating a scRNA-seq data set (17), including 16 PDAC patient tumor and 3 adjacent benign pancreas samples (Fig. 1A). As expected, APOE expression was highest in myeloid cells and fibroblasts (Fig. 1B; Supplementary Fig. S1A). By extracting total myeloid cells from the data, we identified and classified individual subpopulations based on marker expression (Fig. 1C; Supplementary Fig. S1B). APOE was highly expressed in resident macrophages and alternatively activated macrophages, while expression in other myeloid populations was low (Fig. 1D). Comparing expression levels, APOE expression was higher in tumor macrophages compared with macrophages in adjacent tissue (Fig. 1E). Using a similar approach, we identified populations of myofibroblastic-CAFs (myCAF) and inflammatory-CAFs (iCAF; Fig. 1F; Supplementary Fig. S1C; ref. 25) and found APOE expression was enriched in human iCAFs compared with myCAFs (Fig. 1G). However, unlike in macrophages, APOE expression in fibroblasts did not differ between benign and cancer samples (Fig. 1H).
Figure 1.
Human APOE levels are elevated in PDAC and correlate to patient survival. A, UMAP analysis of the 13 identified cell populations in human adjacent/normal pancreas (n = 3) and PDAC tumors (n = 16). B, Dot plot of APOE in all identified cell populations in human single-cell data set. Color represents average expression. Size of the dot represents expression frequency. C, UMAP visualization of four identified myeloid cell subpopulations in the human PDAC tissue. D, Feature plot of APOE expression in all identified myeloid cell populations in human PDAC. Gray, low expression; blue, high expression. Black outline denotes APOE-positive macrophages. E, Violin plot of normalized gene expression of APOE in PDAC and adjacent normal pancreas macrophages in human PDAC. Statistical significance was determined using nonparametric Wilcoxon rank sum test. F, UMAP visualization of human PDAC fibroblast subpopulations. G, Violin plot of normalized gene expression of APOE in human myCAF and iCAF populations. H, Violin plot of normalized gene expression of APOE in PDAC and adjacent normal pancreas iCAFs in human PDAC. Statistical significance was determined using nonparametric Wilcoxon rank sum test. I, Violin plot of normalized expression of APOE in human monocytes. Statistical significance was determined using nonparametric Wilcoxon rank sum test. J, Human APOE concentration (μg/mL) in plasma from healthy donors (n = 15), chronic pancreatitis patients (n = 17), and PDAC patients (n = 155). Statistical significance was determined using one-way ANOVA with Tukey test for multiple comparisons. n.s., not significant. K, Survival analysis of PDAC patients stratified by plasma APOE levels. APOE low, n = 32; APOE high, n = 32. Statistical significance was determined using log-rank (Mantel–Cox) test.
We then examined APOE expression in human PBMC, using scRNA-seq data from 16 PDAC patients and 4 healthy donors (17). Among four distinct monocyte populations identified, APOE expression was prevalent in only one (Supplementary Fig. S1D and S1E). Finally, APOE expression was higher in peripheral monocytes of PDAC patients compared with healthy individuals, suggesting that elevated monocyte APOE expression represents a systemic response to the tumor (Fig. 1I). We then measured overall APOE protein levels in plasma from 155 PDAC patients but found no significant difference when compared with plasma from 15 healthy donors and 17 chronic pancreatitis patients (Fig. 1J). However, noting outliers with very high APOE levels, we stratified patients based on APOE plasma levels and performed survival analysis. We found PDAC patients with high plasma levels of APOE (n = 32) had shorter survival, compared with patients with low levels (n = 32; Fig. 1K). Thus, APOE gene expression is elevated in monocytes and macrophages of PDAC patients, and protein levels correlate with poor survival, suggesting a functional role in the disease.
Finally, we explored APOE expression in other data sets. Using published laser capture microdissection data from matched epithelial and stromal samples from human PDAC (26), we confirmed APOE was upregulated in the stroma compared with the epithelium (Supplementary Fig. S1F). Using TCGA PDAC data set that include 150 PDAC samples, we stratified patients into APOE low and APOE high. We observed a positive correlation between APOE expression and expression of MARCO and TREM2, markers of alternatively activated macrophages (Supplementary Fig. S1G; refs. 27, 28), supporting the notion that APOE-expressing macrophages are enriched in PDAC.
APOE protein is highly expressed by tumor-associated macrophages
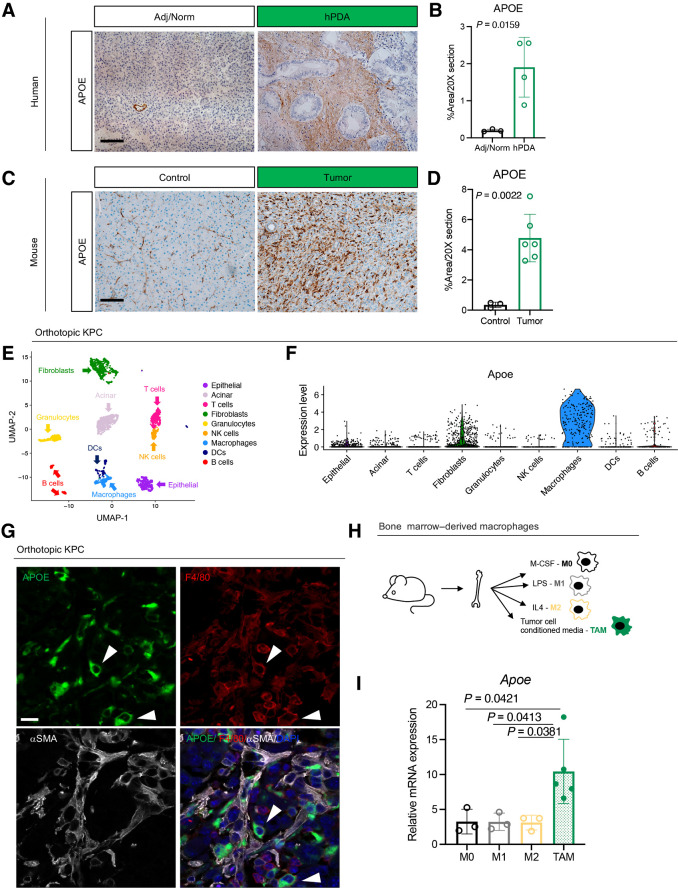
IHC revealed elevated APOE expression in the stroma of human PDAC compared with adjacent tissue (Fig. 2A and B). To evaluate mouse PDAC, we used a syngeneic orthotopic murine model, using 7940b cells derived from the KrasLSL-G12D/+; Trp53LSL-R172H/+; Pdx1-Cre (KPC) mouse model of PDAC (29). Immunostaining showed elevated ApoE expression in mouse tumors compared with healthy pancreas (Fig. 2C and D). To determine the source of ApoE, we performed scRNA-seq (Fig. 2E; Supplementary Fig. S2A). Consistent with human tumors, the highest levels of ApoE expression were in macrophages, followed by fibroblasts (Fig. 2F). Coimmunofluorescence staining for ApoE, macrophages (F4/80), and fibroblasts (alpha-smooth muscle actin, αSMA) confirmed ApoE expression most commonly in F4/80+ macrophages (Fig. 2G). APOE+ macrophages were more abundant (Supplementary Fig. S2B) and ApoE expression levels in macrophages were higher in orthotopic KPC tissue compared with normal pancreas (Supplementary Fig. S2C).
Figure 2.
APOE is highly expressed by TAM. A, IHC analysis of APOE in human benign pancreas and PDAC samples. Scale bars, 100 μm. B, Quantitation of positive APOE staining as a percentage of area in a 20× field of view. At least three fields of view were averaged per sample. Adjacent/normal pancreas, n = 3; hPDA, n = 4. Statistical significance was determined using two-tailed t test. C, IHC analysis of APOE in normal mouse pancreas and orthotopic KPC tumor. Scale bars, 100 μm. D, Quantitation of positive APOE staining as a percentage of area in a 20× field of view. Five fields of view were averaged per mouse. Control, n = 3; orthotopic tumor, n = 6. Statistical significance was determined using two-tailed t test. E, UMAP visualization of 9 identified populations in orthotopic KPC tumors (n = 2). F, Violin plot of normalized expression of Apoe in identified cell populations in orthotopic KPC tumors (n = 2). G, Coimmunofluorescence of orthotopic KPC tumor with single channels of APOE (green), F4/80 (red), αSMA (white), and merge to show APOE and F4/80 colocalization. Two examples of APOE and F4/80 colocalization are denoted by white arrowheads. Scale bars, 25 μm. H, Experimental design for bone marrow–derived macrophage polarization assay. I, qRT-PCR analysis of Apoe mRNA levels relative to Cyclophilin A in four macrophage conditions (M0, M1, M2, and TAM). Statistical significance was determined using one-way ANOVA with Tukey test for multiple correction.
Macrophages are plastic cells that exist on a polarization spectrum from proinflammatory (M1) to immunosuppressive (M2; ref. 30). TAMs are distinct from both M1 and M2 macrophages, expressing markers of both (31). To determine which macrophage population produce ApoE, we performed an in vitro macrophage polarization assay. We treated bone marrow–derived macrophage cultures with either M-CSF, LPS, IL4, or PDAC cell CM to polarize to M0, M1, M2, and TAM, respectively (Fig. 2H), and assessed ApoE expression. Strikingly, ApoE was specifically upregulated in TAMs compared with other macrophage populations (Fig. 2I).
To assess if ApoE expression was induced in macrophages specifically by tumor cells, we cultured bone marrow cells with CM from normal pancreas duct cells (HPNE) or tumor cells (7940b). Unlike tumor cell CM, HPNE CM was unable to sustain macrophage differentiation from bone marrow progenitors, making it impossible to assess ApoE expression (Supplementary Fig. S2D). As an alternative, we differentiated macrophages from bone marrow progenitors by adding M-CSF to the medium. Once differentiated to macrophages, we treated cultures with either HPNE or 7940b CM (Supplementary Fig. S2E). HPNE CM induced ApoE expression, but induction was higher with tumor cell CM (Supplementary Fig. S2F). Macrophages treated with tumor cell CM had the lowest expression of the inflammatory macrophage marker Tnfα (Supplementary Fig. S2G), suggesting that ApoE-expressing macrophages were associated primarily with the immunosuppressive phenotype.
To determine if oncogenic Kras in the epithelium regulates ApoE expression in vivo, we utilized the iKras* mouse model of PDAC (19), where oncogenic Kras (Kras*) expression is inducible and reversible. We induced Kras* expression, followed by induction of acute pancreatitis, and harvested pancreata after 3 weeks of continuous Kras* expression, or from mice where Kras* was expressed for three weeks and then inactivated for 3 or 7 days (Supplementary Fig. S2H). In this model, 3 weeks of continuous Kras* expression results in replacement of the majority of the parenchyma with pancreatic intraepithelial neoplasia (PanIN) and extensive fibroinflammatory stroma. Inactivation of Kras* led to lesion regression, redifferentiation of acinar cells, and remodeling of the stroma. ApoE+ cells were more abundant in iKras* pancreata compared with control (Supplementary Fig. S2I and S2J). The number of ApoE+ cells decreased after extinguishing Kras* expression (Supplementary Fig. S2I and S2J). Thus, ApoE is upregulated in TAMs during early carcinogenesis and depends on epithelial expression of oncogenic Kras.
APOE ablation reduces tumor burden and reprograms the TME
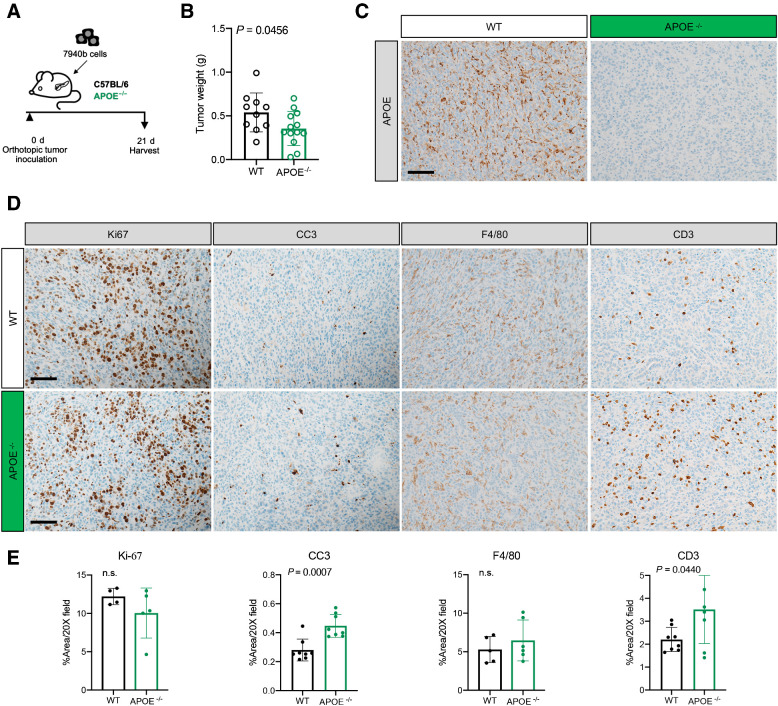
To evaluate the function of ApoE, we utilized germline ApoE knockout mice (ApoE−/−), to eliminate both myeloid and fibroblast ApoE production. We implanted 7940b KPC cells (C57/BL6J) into the pancreata of ApoE−/− or wild-type (WT) C57BL6/J mice (n = 10–13/cohort; Fig. 3A). ApoE−/− mice had moderately, but significantly smaller, tumors at endpoint compared with WT controls (Fig. 3B). IHC revealed complete absence of ApoE within the tumor compared with WT mice, as expected (Fig. 3C) with no other differences noted (Supplementary Fig. S3A). Ki-67 staining revealed no difference in proliferation (Fig. 3D and E). However, levels of apoptosis, measured by cleaved caspase-3 (CC3) staining, were increased in tumors implanted in ApoE−/− mice compared with controls (Fig. 3D and E). We observed fewer αSMA+ fibroblasts along with reduced collagen deposition visualized by Trichrome stain (Supplementary Fig. S3A). We next evaluated infiltration of macrophages by F4/80 staining and found no difference in tumors grown in ApoE−/− or WT mice (Fig. 3D and E). However, we noted an increased infiltration of CD3+ T cells in the tumors of ApoE−/− mice (Fig. 3D and E), suggesting changes in the immune microenvironment. The greater T-cell infiltration in ApoE–/− mice was not attributable to them having generally more T cells systemically, as flow cytometry of spleens from ApoE–/− mice showed a slight reduction in total, CD4+ and CD8+ T cells compared with controls (Supplementary Fig. S3B).
Figure 3.
Loss of APOE results in reduced tumor burden and fibrosis. A, Experimental scheme for orthotopic transplantation of 7940b, KPC tumor cells. B, Final tumor weight (g) in WT (n = 10) and ApoE–/− (n = 13) mice. Statistical significance was determined using two-tailed t test, with a P < 0.05 considered statistically significant. C, Representative IHC for APOE in WT and ApoE–/− mice. Scale bar, 100 μm. D, Representative IHC staining for Ki-67, cleaved caspase-3 (CC3), F4/80, and CD3 in WT and ApoE–/− mice. Scale bars, 100 μm. E, Quantitation of IHC stain as a percentage area per 20× field in WT (n = 4–8) and ApoE–/− mice (n = 5–8). Statistical significance was determined by two-tailed t tests. n.s., not significant.
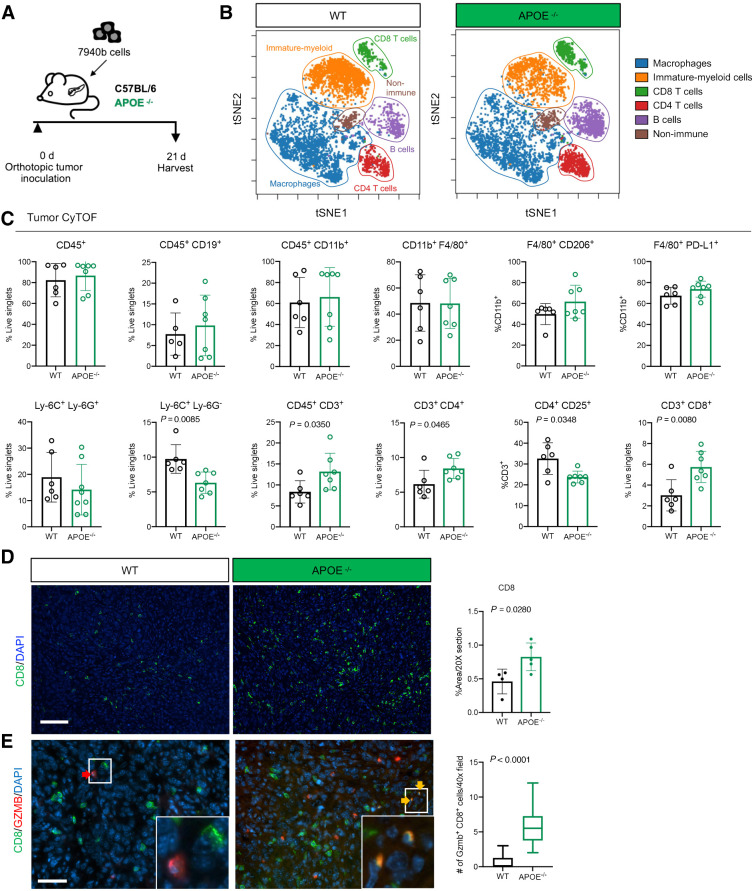
To expand immune cell characterization, we performed mass cytometry (CyTOF) on tumors from WT and ApoE−/− mice (Fig. 4A). Unbiased clustering visualized through t-distributed stochastic neighbor embedding (t-SNE) revealed populations of macrophages, immature myeloid cells, CD8 T cells, CD4 T cells, and B cells along with a small population of nonimmune cells (Fig. 4B; Supplementary Fig. S4A). Manual gating of identified populations revealed no significant difference in total immune cells (CD45+), B cells (CD45+ CD19+), total myeloid cells (CD45+ CD11b+), macrophages (CD11b+ F4/80+), or changes in macrophage subpopulations (F4/80+ CD206+; F4/80+ PD-L1+; Fig. 4C). Macrophage polarization, determined by expression of programmed death-ligand 1 (Cd274), arginase-1 (Arg1), mannose receptor C-type 1 (Mrc1), and inducible nitric oxide synthase (Nos2), did not differ between tumors of WT and ApoE−/− mice (Supplementary Fig. S4B). Finally, we isolated bone marrow–derived monocytes and polarized to TAMs in both WT and ApoE−/− mice and saw more Arg1, and less Cd274 and Tnf-α in ApoE−/− TAMs, suggesting loss of ApoE has minor effects on macrophage polarization in vitro (Supplementary Fig. S4C).
Figure 4.
ApoE−/− mice have fewer monocytic-MDSCs and increased CD8+ T-cell infiltration. A, Experimental scheme for orthotopic transplantation of 7940b, KPC tumor cells. B, tSNE visualization of the 6 cell populations identified using CyTOF in WT and ApoE–/− tumors. Populations identified include macrophages (blue), immature myeloid cells (orange), CD8 T cells (green), CD4 T cells (red), B cells (purple), and nonimmune (brown). C, Manual gating quantitation of cell populations in WT (n = 5–6) and ApoE–/− (n = 7) tumors. Populations include total immune (CD45+), B cells (CD45+ CD19+), total myeloid (CD45+ CD11b+), macrophages (CD11b+ F4/80+), TAMs (F4/80+ CD206+; F4/80+ PD-L1+), granulocytic-MDSCs (Ly-6C+ Ly6G+), monocytic-MDSCs (Ly-6C+ Ly-6G−), total T cells (CD45+ CD3+), CD4 T cells (CD3+ CD4+), Tregs (CD4+ CD25+), and CD8 T cells (CD3+ CD8+). D, Representative immunofluorescence staining of CD8 (green) and DAPI (blue) in WT and ApoE−/− tumors. Scale bars, 100 μm. Right, quantitation of percent CD8-positive area in a 20× field in WT (n = 4) and ApoE–/− mice (n = 5). Statistical significance was determined by two-tailed t test. E, Representative coimmunofluorescence staining of CD8 (green), GZMB (red), and DAPI (blue) in WT and ApoE–/− tumors. Scale bars, 50 μm. Right, quantitation of the number of Gzmb+ CD8+ double-positive cells in at least three, 40× fields in WT (n = 4) and ApoE–/− mice (n = 4). Statistical significance was determined by two-tailed t test.
Another prominent population identified in the tumors were immature myeloid cells, often described as myeloid-derived suppressor cells (MDSC), distinguished based on their Ly-6G expression. Granulocytic-MDSCs (Ly-6C+ Ly-6G+) did not differ (Fig. 4C), while the monocytic-MDSC population (Ly-6C+ Ly-6G−) was smaller in ApoE−/− mice compared with controls, suggesting a less immunosuppressive TME in the former (Fig. 4C). To explore this possibility further, we evaluated T-cell populations and found an increase in total T cells (CD45+ CD3+), CD4 T cells (CD3+ CD4+), and CD8 T cells (CD3+ CD8+; Fig. 4C). There was also a decrease in potential Tregs (CD4+ CD25+; Fig. 4C). We confirmed a reduction of Tregs in ApoE−/− mice by immunostaining for the Treg marker FOXP3 (Supplementary Fig. S4D). Coimmunofluorescence staining confirmed the increase in CD8+ T cells in tumors in ApoE−/− mice (Fig. 4D). We performed coimmunofluorescence for the cytotoxic marker granzyme B (Gzmb) and CD8 to assess the functional status of infiltrating CD8+ T cells. Tumors in ApoE−/− mice had an increase in Gzmb+ CD8+ T cells compared with tumors in WT mice, suggesting enhanced cytotoxic activity (Fig. 4E). Expression of immune checkpoints Pdcd1 and Ctla4 trended lower in tumors in ApoE−/− mice, while expression of the exhaustion marker Eomes was significantly decreased (Supplementary Fig. S4E). Taken together, loss of monocytic-MDSCs and Tregs, along with the increase in cytotoxic T-cell infiltration in tumors implanted in ApoE−/− mice, supports the notion that ApoE promotes immune suppression in the pancreatic TME.
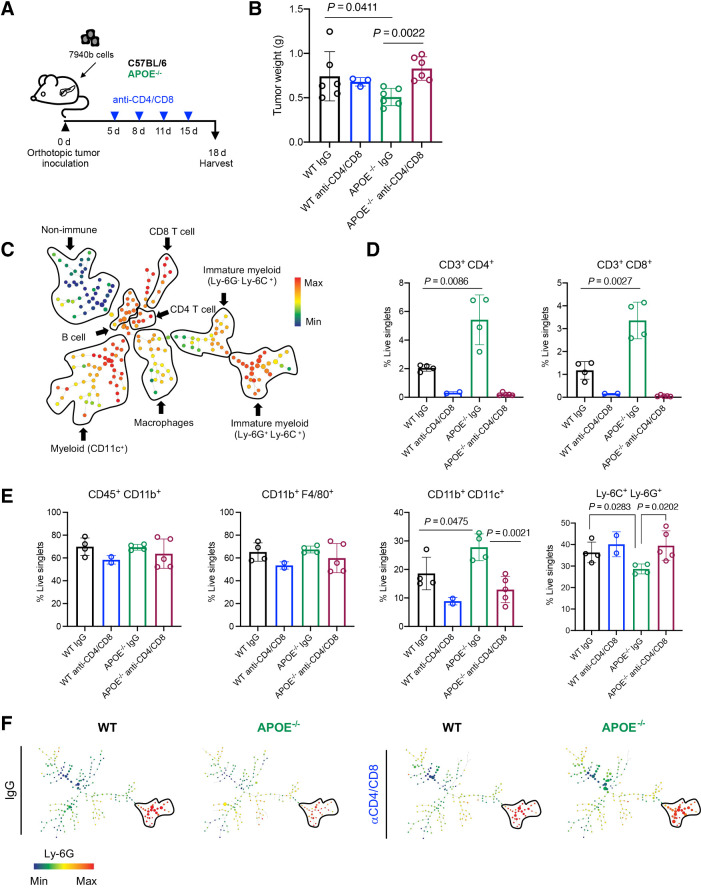
To test if reduced tumor growth in ApoE−/− mice was due to T-cell–driven immune responses, we implanted pancreatic tumors in WT or ApoE−/− mice and randomized mice (n = 3–6/cohort) for treatment with isotype control IgG or anti-CD4 and anti-CD8 depletion antibodies (Fig. 5A). Depletion of T cells indeed rescued tumor growth (Fig. 5B). We performed CyTOF to evaluate changes in immune infiltration and analyzed the data using Spanning-tree Progression Analysis of Density-normalized Events (SPADE; Fig. 5C; Supplementary Fig. S5A). Compared with controls, tumors in ApoE−/− mice had more abundant CD4+ and CD8+ T cells (Fig. 5D), and treatment with anti-CD4 and CD8 antibodies efficiently depleted T cells (Fig. 5D). There was no difference in total myeloid cells (CD45+ CD11b+) or total macrophages (CD11b+ F4/80+; Fig. 5E). However, CD11b+ CD11c+ myeloid cells were more abundant, while immature myeloid cells (Ly-6C+ Ly-6G+) were decreased in tumors implanted in ApoE−/− mice (Fig. 5E and F), and rescued upon T-cell depletion (Fig. 5E and F). The fibroblast population was decreased in ApoE−/− mice, consistent with immunostaining data (Supplementary Fig. S5B). Taken together, these data suggest the antitumor phenotype in ApoE−/− mice is T-cell dependent.
Figure 5.
Antitumor phenotype in ApoE–/− mice is rescued upon T-cell depletion. A, Experimental design schematic for T-cell depletion in WT and ApoE–/− mice. B, Final tumor weight (g) from WT (n = 6), WT anti-CD4/CD8 (n = 3), ApoE–/− (n = 6), and ApoE–/− anti-CD4/CD8 (n = 6). Statistical significance was determined with a nonparametric Mann–Whitney test. C, Representative SPADE analysis of cellular infiltrate in WT tumor. Identified populations include nonimmune cells, CD8 T cells, CD4 T cells, B cells, immature myeloid cells, macrophages, and CD11c+ myeloid cells. The SPADE plot is colored to indicate CD45 expression. Red, high expression; blue, low expression. D, Manual gating quantitation of cell populations in WT (n = 4), WT anti-CD4/CD8 (n = 2), ApoE–/− (n = 4), and ApoE–/− anti-CD4/CD8 (n = 5) tumors. Populations include CD4 T cells (CD3+ CD4+) and CD8 T cells (CD3+ CD8+) E, total myeloid cells (CD45+ CD11b+), macrophages (CD11b+ F4/80+), CD11c+ myeloid cells (CD11b+ CD11c+), and immature myeloid cells (Ly-6C+ Ly-6G+). Statistical significance was determined by two-tailed t tests between groups. F, Representative SPADE analysis colored by Ly-6G expression in WT, WT anti-CD4/CD8, ApoE−/−, and ApoE–/− anti-CD4/CD8 tumors. Red, high expression; blue, low expression.
APOE regulates Cxcl1 expression in tumor cells and fibroblasts
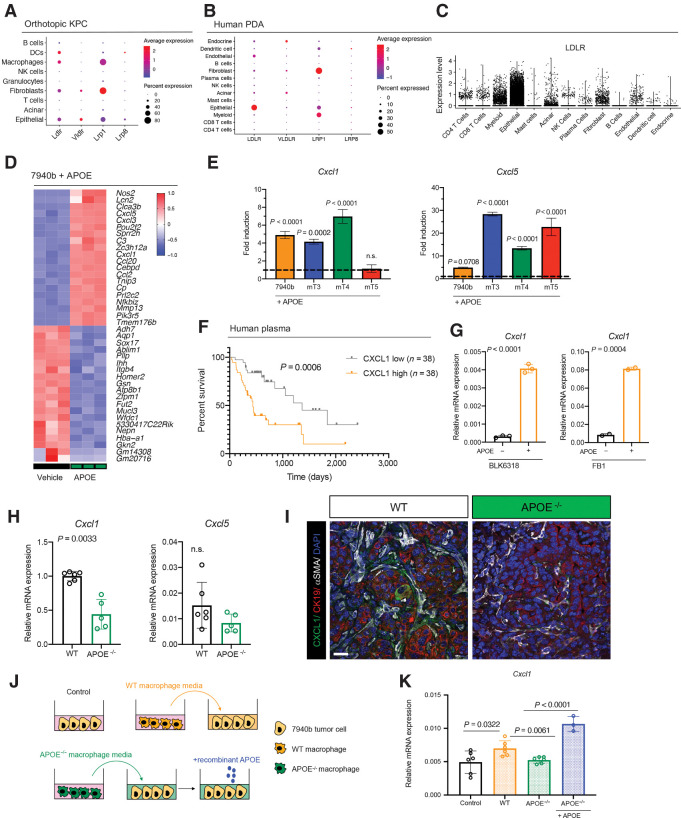
Although ApoE can act directly on T cells (13, 32, 33), we noted in our scRNA-seq data that KPC tumor cells express 4 of the major ApoE receptors: LDLR, VLDLR, LRP1, and LRP8; Fig. 6A; ref. 34). In human PDAC epithelia, the most highly expressed of these was LDLR; thus, we focused on this receptor for functional work (Fig. 6B and C). We treated 7940b tumor cells with recombinant murine ApoE (rApoE) and performed bulk RNA-seq compared with vehicle-treated controls (Fig. 6D) to assess direct effects of ApoE on epithelial cells. GSEA revealed upregulation of inflammation-associated transcriptional signatures in rApoE-treated tumor cells (Supplementary Fig. S6A). Differential expression analysis revealed tumor cells treated with rApoE showed increased expression of the chemokines Ccl2, Cxcl1, and Cxcl5, all chemoattractants for myeloid cells (Fig. 6D; refs. 35, 36). Cxcl1 and Cxcl5 bind the Cxcr2 receptor expressed on immature myeloid cells, which, in turn, inhibit T-cell infiltration in PDAC (16, 37), consistent with our phenotype. We also found a significant increase in Cxcl1 and Cxcl5 in 3 additional murine KPC tumor cell lines when treated with rApoE (Fig. 6E). To explore whether CXCL1 is important in human PDAC, we measured CXCL1 levels in patient plasma. By stratifying patient CXCL1 into high and low groups, we discovered that high CXCL1 plasma levels (n = 38) were associated with reduced overall survival compared with patients with low CXCL1 plasma levels (n = 38; Fig. 6F).
Figure 6.
APOE regulates Cxcl1 expression in tumor cells and fibroblasts. A, Dot plot of Ldlr, Vldlr, Lrp1, and Lrp8 in orthotopic KPC samples. Color represents average expression, while size of the dot represents expression frequency. B, Dot plot of LDLR, VLDLR, LRP1, and LRP8 in human PDAC. Color represents average expression, while size of the dot represents expression frequency. C, Violin plot of normalized LDLR expression in human PDAC. D, Heat map of differentially expressed genes in in vitro 7940b KPC cells treated with vehicle (n = 3) compared with 7940b KPC cells treated with 0.3 μg/mL murine recombinant APOE (n = 3) for 48 hours. Red, high expression; blue, low expression. E, qRT-PCR analysis of Cxcl1 and Cxcl5 mRNA levels relative to Cyclophilin A in four KPC cell lines (7940b, mT3, mT4, mT5). Dotted line represents fold induction compared with vehicle-treated cells normalized to 1. Statistical significance was determined using one-way ANOVA with Tukey test for multiple correction. F, Survival analysis of PDAC patients stratified by plasma CXCL1 levels. CXCL1 low, n = 38; CXCL1 high, n = 38. Statistical significance was determined using log-rank (Mantel–Cox) test. G, qRT-PCR analysis for Cxcl1 mRNA levels relative to Cyclophilin A in WT fibroblasts (BLK6318) and CAFs (FB1) treated with vehicle (n = 2–3) or 0.3 μg/mL recombinant ApoE (n = 2–3) for 48 hours. Statistical significance was determined by two-tailed t tests. H, qRT-PCR analysis of Cxcl1 and Cxcl5 mRNA levels relative to Cyclophilin A in WT (n = 6) and ApoE–/− (n = 5) tumors. Statistical significance was determined using two-tailed t test. n.s., not significant. I, Coimmunofluorescence staining of CXCL1 (green), CK19 (red), αSMA (white), and DAPI (blue) in WT and ApoE–/− orthotopic KPC tumors. J, Experimental design schematic. K, qRT-PCR analysis of Cxcl1 mRNA levels relative to Cyclophilin A in 7940b tumor cells alone control (n = 6), 7940b cells cultured with WT macrophage CM (n = 6), 7940b cells cultured with ApoE–/− macrophage CM (n = 6), and 7940b cells cultured with ApoE–/− macrophage CM with 0.3 μg/mL recombinant ApoE (n = 3). Statistical significance was determined by two-tailed t tests between groups.
We used scRNA-seq data from an orthotopic KPC tumor sample to assess the cellular source of Cxcl1, Cxcl2, and Cxcl5 (Supplementary Fig. S6B). Cxcl2 was highly expressed by granulocytes, while Cxcl1 and Cxcl5 were expressed by epithelial cells and fibroblasts (Supplementary Fig. S6B). Cxcl1 was elevated in tumor epithelium compared with normal pancreas (Supplementary Fig. S6C). To determine if fibroblast Cxcl1 expression was also upregulated by ApoE, we treated WT mouse pancreatic fibroblasts (BLK6318) and cancer-associated mouse fibroblasts (FB1) with rApoE; in both lines, Cxcl1 expression was induced (Fig. 6G). We then investigated regulation of Cxcl1 in vivo. We detected significantly reduced Cxcl1 and a trend toward lower Cxcl5 in tumors implanted in ApoE−/− mice (Fig. 6H). Accordingly, we observed reduced Cxcl1 protein in both tumor cells and fibroblasts in tumors from ApoE−/− mice (Supplementary Fig. S6D and S6I).
ApoE is a secreted protein, and macrophages are one of the main sources (Supplementary Fig. S6E). To test whether macrophage-secreted ApoE upregulates Cxcl1 expression in cancer cells, bone marrow–derived monocytes were isolated from WT mice and ApoE−/− mice and polarized to TAMs by culturing them in tumor cell CM (20). We then cultured tumor cells in WT or ApoE−/− macrophage CM for 48 hours (Fig. 6J) and Cxcl1 levels were measured by qRT-PCR (Fig. 6K). Tumor cells cultured in WT, but not ApoE−/−, macrophage CM induced Cxcl1 expression in tumor cells, suggesting that ApoE is required for this induction. To confirm that ApoE was the primary factor missing from ApoE−/− CM, we added rApoE to Apoe−/− macrophage CM, which rescued induction of Cxcl1 expression (Fig. 6K). We conclude that macrophage-secreted ApoE induces tumor cell Cxcl1 expression.
APOE induces tumor cell Cxcl1 expression through LDLR and NF-κB signaling
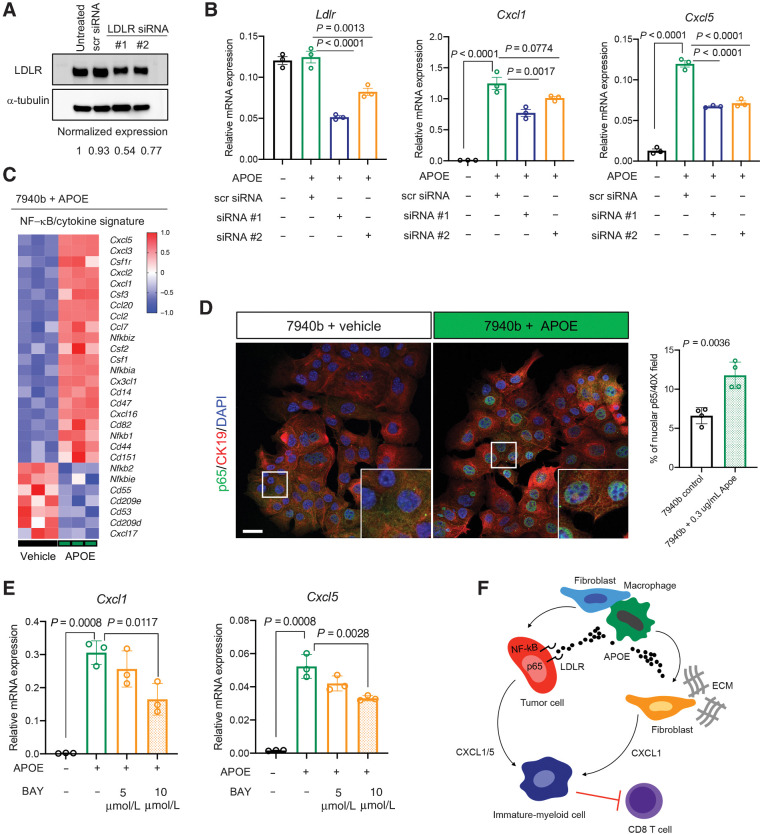
We next explored which ApoE receptor mediated the induction of Cxcl1 and Cxcl5 expression response in pancreatic tumor cells (Fig. 6). Because LDLR was highly expressed in human and mouse PDAC cells (Fig. 6A–C), we used small interfering RNA (siRNA) to knockdown LDLR in 7940b cells (Fig. 7A). Optimized LDLR targeting by siRNA resulted in a maximum of ∼50% knockdown. However, even this partial knockdown was sufficient to reduce induction of Cxcl1 and Cxcl5 expression by rApoE (Fig. 7B). These data suggest ApoE signals, in part, through LDLR to induce Cxcl1 and Cxcl5 expression.
Figure 7.
APOE regulates tumor cell Cxcl1 production via NF-κB signaling. A, Representative Western blot analysis of 7940b KPC tumor cells that were either untreated, treated with scrambled siRNA-negative control, or with LDLR siRNA for 24 hours. α-Tubulin was used as a loading control. Normalized protein expression is denoted under each lane. B, qRT-PCR analysis of Ldlr, Cxcl1, and Cxcl5 mRNA levels relative to Cyclophilin A in 7940b KPC cells that underwent LDLR knockdown for 48 hours and were treated with 0.3 μg/mL recombinant ApoE (n = 3) for 1 hour. Statistical significance was determined using one-way ANOVA with Tukey test for multiple correction. C, Heat map of NF-κB/cytokine signatures in 7940b KPC cells treated with 0.3 μg/mL recombinant ApoE (n = 3) compared with vehicle (n = 3) for 48 hours. Red, high expression; blue, low expression. D, Representative coimmunofluorescence staining of p65 (green), CK19 (red), and DAPI (blue) in 7940b tumor cells in vitro treated with vehicle or 0.3 μg/mL recombinant ApoE for 48 hours. Scale bars, 25 μm. Quantitation of percent nuclear p65 in a 40× field in 7940b cells (n = 4) and 7940b cells treated with 0.3 μg/mL recombinant ApoE (n = 4) for 48 hours. White box represents higher magnification. Statistical significance was determined using two-tailed t tests. E, qRT-PCR analysis of Cxcl1, and Cxcl5 mRNA levels relative to Cyclophilin A in 7940b cells (n = 3), 7940b cells treated with 0.3 μg/mL recombinant ApoE for 2 hours (n = 3), 7940b cells pretreated with 5 μmol/L BAY11-7082 for 1 hour and treated with 0.3 μg/mL recombinant ApoE for 2 hours (n = 3), and 7940b cells pretreated with 10 μmol/L BAY11-7082 for 1 hour and treated with 0.3 μg/mL recombinant ApoE for 2 hours (n = 3). Statistical significance was determined using one-way ANOVA with Tukey test for multiple comparisons. F, Working model. PDAC tumors with active ApoE secretion regulate CXCL1 production from tumor cells and fibroblasts, which in turn recruits immature myeloid cells, resulting in suppression of CD8+ T-cell infiltration.
To determine the mechanism of Cxcl1 induction by ApoE, we noted that Cxcl1 expression is activated by NF-κB signaling (38) and that ApoE activates NF-κB in AML (12). To examine if APOE upregulates CXCL1 through activation of NF-κB in PDAC, we turned to GSEA of TCGA PDAC data, revealing a positive correlation of APOE expression with NF-κB signaling components (Supplementary Fig. S7A). In concordance, GSEA of RNA-seq data from 7940b cells treated with rApoE revealed an upregulation of components of NF-κB signaling (Supplementary Fig. S7B) and many NF-κB/cytokine signature genes (Fig. 7C). We then performed immunofluorescence for the p65 subunit of NF-κB, which undergoes nuclear translocation upon activation of the NFκB pathway, in 7940b cells treated with rApoE or vehicle (Fig. 7D). rApoE treatment clearly increased nuclear translocation of p65 (Fig. 7D). To confirm that NF-κB activity is required for ApoE induction of Cxcl1/5 expression, we treated cells with BAY11-7082 (39), which inhibits IκB kinase (IKK), preventing IκB phosphorylation and p65 nuclear translocation. As before, rApoE induced Cxcl1 and Cxcl5 expression in tumor cells, while treatment with BAY 11-7082 blunted this induction (Fig. 7E). Finally, using LDLR-targeting siRNA, we found that LDLR knockdown greatly reduced nuclear p65 levels (Supplementary Fig. S7C). Thus, ApoE regulates tumor cell derived Cxcl1 and Cxcl5 expression, at least in part, through binding to LDLR and activation of NF-κB signaling (Fig. 7F).
Discussion
We show that ApoE is expressed in mouse and human PDAC and mediates cross-talk between cancer cells and the immune system. Our data suggest that macrophage-derived ApoE signals directly to tumor cells to enhance Cxcl1 and Cxcl5 expression. Cxcl1 and Cxcl5 act as chemoattractants for immature myeloid cells. These cells in turn prevent T-cell infiltration in PDAC (16). We further show that ApoE induction of Cxcl1 and Cxcl5 expression is at least partially dependent on it binding to LDL receptor on tumor cells, activating the NF-κB pathway. While we cannot eliminate a functional role of other ApoE receptors, especially VLDLR, which is highly expressed in mouse but not human PDAC epithelia (Fig. 6A–C), the proportional response of Cxcl1/5 expression and NF-κB activation to a partial knockdown of LDLR suggests it plays a prominent role.
Interestingly, inactivation of cholesterol signaling in mouse PDAC delays carcinogenesis and prolongs survival. When tumors do develop, they have more aggressive, basal-like features, possibly indicating that classical tumors are uniquely dependent on cholesterol (40). An interesting consideration, as it relates to our study, is that one of the physiologic consequences of ablating ApoE is higher plasma cholesterol, which may contribute to the suppression of tumor growth that we observe in vivo.
In the current study, we focused on ApoE produced by macrophages because they are critical in establishing the immunosuppressive TME (41). Directly targeting macrophage infiltration into pancreas tumors shows promise in combination with chemotherapy (42) and immunotherapy (43). However, ApoE is also highly expressed by CAFs and ApoE receptors are expressed on many other cell types that may play a role in tumor progression, including immune cells (Fig. 6C; ref. 44). ApoE can directly inhibit T-cell function in vitro (45). In vivo, the role of ApoE in different malignancies is context dependent. In some solid tumors (including ovarian cancer and melanoma), ApoE promoted antitumor immunity by binding the LRP8 receptor on MDSCs (15). Conversely, in AML, ApoE directly bound the LILRB4 receptor on tumor cells and promotes immune escape (12). In PDAC, it remains to be determined which cell types, in addition to epithelial cells, are targets of ApoE and what the functional role is in each compartment.
We observed that higher serum APOE levels correlate with worse overall survival in PDAC patients, suggesting that it could be a useful prognostic marker. Unlike mice, humans have three different variants of APOE: APOE2, APOE3, and APOE4, with distinct functions (46). In Alzheimer's disease APOE4 is a risk factor for disease (47), while APOE2 plays a protective role (48). APOE variants have not been extensively studied in cancer, but in one melanoma study, APOE4 plays a favorable role in disease, while APOE2 promotes worse outcome (49). This finding correlates with APOE4's higher affinity for LDL receptor (50). With ApoE's contrary effect in PDAC, we would predict that APOE4 promotes immune suppression more effectively and is associated with worse patient outcome.
Authors' Disclosures
A. Pacheco reports grants from NCI during the conduct of the study. C.A. Lyssiotis reports personal fees from Astellas, Inc. outside the submitted work; in addition, C.A. Lyssiotis has a patent for “Methods for Diagnosing and Treating Oncogenic Kras-Associated Cancer” (015126580-A1) issued. A. Rao reports grants from NIH, ACS, and institutional funds from University of Michigan Ann Arbor during the conduct of the study; grants from NIH, and ACS, institutional funds from University of Michigan Ann Arbor, other support from CHI, Genophyll, LLC, and Voxel analytics, LLC outside the submitted work; and A. Rao is a member of Voxel analytics, LLC. H.C. Crawford reports grants from NIH during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Table S1
Table S2
Acknowledgments
This study was funded by the NIH U01CA224145 to H.C. Crawford and M. Pasca di Magliano and the University of Michigan Cancer Center Support Grant (P30CA046592), including an Administrative Supplement to H.C. Crawford and M. Pasca di Magliano. This project was additionally supported by NIH/NCI grants R01CA151588, R01CA198074, and the American Cancer Society to M. Pasca di Magliano. S.B. Kemp was supported by NIH T32-GM113900 and NCI F31-CA247076. Z.C. Nwosu was supported by the Michigan Postdoctoral Pioneer Program, University of Michigan Medical School. E.S. Carpenter was supported by the American College of Gastroenterology Clinical Research Award and by T32-DK094775. N.G. Steele was funded by American Cancer Society Postdoctoral Award PF-19-096-01 and the Michigan Institute for Clinical and Healthy Research (MICHR) Postdoctoral Translational Scholar Program fellowship award. N.G. Steele and K.L. Donahue were supported by T32-CA009676. C.A. Lyssiotis was supported by NIH grant R37CA237421. A. Rao and S. The were funded by institutional startup funds from the University of Michigan, a gift from Agilent Technologies, NCI grant R37CA214955, and a Research Scholar Grant from the American Cancer Society (RSG-16-005-01). The funders had no role in the planning or execution of this study or the preparation of this manuscript. The authors thank the Advanced Genomics core at the University of Michigan. They thank the Flow Cytometry Core at the University of Rochester, specifically Matthew Cochran and Terry Wightman, for help with CyTOF panel design and analysis. The authors thank Kevin Brown and Vinicius Motta from Fluidigm and Dr. Gregory Beatty at the University of Pennsylvania for his generous gift of the 7940b KPC cell line. They thank Dr. David Tuveson, at Cold Spring Harbor Laboratory for the generous gift of the mT3, mT4, and mT5 KPC cell lines. They thank Michael Scales for generation of the BLK6318 mouse WT fibroblast cell line.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
S.B. Kemp: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, writing–original draft, writing–review and editing. E.S. Carpenter: Resources, data curation, formal analysis, investigation, writing–review and editing. N.G. Steele: Data curation, formal analysis, investigation, writing–review and editing. K.L. Donahue: Formal analysis, investigation. Z.C. Nwosu: Data curation, formal analysis, writing–review and editing. A. Pacheco: Investigation. A. Velez-Delgado: Investigation. R.E. Menjivar: Investigation. F. Lima: Investigation. S. The: Resources and software. C.E. Espinoza: Investigation. K. Brown: Investigation. D. Long: Investigation. C.A. Lyssiotis: Resources, writing–review and editing. A. Rao: Resources and software. Y. Zhang: Formal analysis, investigation, writing–review and editing. M. Pasca di Magliano: Conceptualization, resources, supervision, funding acquisition, visualization, writing–original draft, project administration, writing–review and editing. H.C. Crawford: Conceptualization, resources, supervision, funding acquisition, visualization, writing–original draft, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Zhang Y, Crawford HC, Pasca di Magliano M. Epithelial-stromal interactions in pancreatic cancer. Annu Rev Physiol 2019;81:211–33. [DOI] [PubMed] [Google Scholar]
- 3. Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res 2007;67:9518–27. [DOI] [PubMed] [Google Scholar]
- 4. Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother 2010;33:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med 2016;94:739–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren L, Yi J, Li W, Zheng X, Liu J, Wang J, et al. Apolipoproteins and cancer. Cancer Med 2019;8:7032–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trost Z, Marc J, Sok M, Cerne D. Increased apolipoprotein E gene expression and protein concentration in lung cancer tissue do not contribute to the clinical assessment of non-small cell lung cancer patients. Arch Med Res 2008;39:663–7. [DOI] [PubMed] [Google Scholar]
- 9. Sakashita K, Tanaka F, Zhang X, Mimori K, Kamohara Y, Inoue H, et al. Clinical significance of ApoE expression in human gastric cancer. Oncol Rep 2008;20:1313–9. [PubMed] [Google Scholar]
- 10. Chen YC, Pohl G, Wang TL, Morin PJ, Risberg B, Kristensen GB, et al. Apolipoprotein E is required for cell proliferation and survival in ovarian cancer. Cancer Res 2005;65:331–7. [PubMed] [Google Scholar]
- 11. Urquidi V, Goodison S, Ross S, Chang M, Dai Y, Rosser CJ. Diagnostic potential of urinary alpha1-antitrypsin and apolipoprotein E in the detection of bladder cancer. J Urol 2012;188:2377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng M, Gui X, Kim J, Xie L, Chen W, Li Z, et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 2018;562:605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gui X, Deng M, Song H, Chen Y, Xie J, Li Z, et al. Disrupting LILRB4/APOE interaction by an efficacious humanized antibody reverses T-cell suppression and blocks AML development. Cancer Immunol Res 2019;7:1244–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell 2012;151:1068–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tavazoie MF, Pollack I, Tanqueco R, Ostendorf BN, Reis BS, Gonsalves FC, et al. LXR/ApoE activation restricts innate immune suppression in cancer. Cell 2018;172:825–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity 2018;49:178–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Steele NG, Carpenter ES, Kemp SB, Sirihorachai VR, The S, Delrosario L, et al. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat Cancer 2020;1:1097–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aiello NM, Rhim AD, Stanger BZ. Orthotopic injection of pancreatic cancer cells. Cold Spring Harb Protoc 2016;2016:pdb prot078360. [DOI] [PubMed] [Google Scholar]
- 19. Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 2012;122:639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Yan W, Mathew E, Kane KT, Brannon A 3rd, Adoumie M, et al. Epithelial-myeloid cell crosstalk regulates acinar cell plasticity and pancreatic remodeling in mice. Elife 2017;6:e27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Halbrook CJ, Pontious C, Kovalenko I, Lapienyte L, Dreyer S, Lee HJ, et al. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab 2019;29:1390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Yan W, Mathew E, Bednar F, Wan S, Collins MA, et al. CD4+ T lymphocyte ablation prevents pancreatic carcinogenesis in mice. Cancer Immunol Res 2014;2:423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collins MA, Brisset JC, Zhang Y, Bednar F, Pierre J, Heist KA, et al. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One 2012;7:e49707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov 2019;9:1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017;214:579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maurer C, Holmstrom SR, He J, Laise P, Su T, Ahmed A, et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 2019;68:1034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol 2006;177:3520–4. [DOI] [PubMed] [Google Scholar]
- 28. van der Laan LJ, Dopp EA, Haworth R, Pikkarainen T, Kangas M, Elomaa O, et al. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J Immunol 1999;162:939–47. [PubMed] [Google Scholar]
- 29. Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7:469–83. [DOI] [PubMed] [Google Scholar]
- 30. Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS−) vs. alternatively activated macrophages. Front Immunol 2019;10:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci 2019;26:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bonacina F, Coe D, Wang G, Longhi MP, Baragetti A, Moregola A, et al. Myeloid apolipoprotein E controls dendritic cell antigen presentation and T cell activation. Nat Commun 2018;9:3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tenger C, Zhou X. Apolipoprotein E modulates immune activation by acting on the antigen-presenting cell. Immunology 2003;109:392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lane-Donovan C, Herz J. ApoE, ApoE receptors, and the synapse in Alzheimer's disease. Trends Endocrinol Metab 2017;28:273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 2009;29:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol 2016;37:208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steele CW, Karim SA, Leach JDG, Bailey P, Upstill-Goddard R, Rishi L, et al. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell 2016;29:832–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burke SJ, Lu D, Sparer TE, Masi T, Goff MR, Karlstad MD, et al. NF-kappaB and STAT1 control CXCL1 and CXCL2 gene transcription. Am J Physiol Endocrinol Metab 2014;306:E131–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee J, Rhee MH, Kim E, Cho JY. BAY 11-7082 is a broad-spectrum inhibitor with anti-inflammatory activity against multiple targets. Mediators Inflamm 2012;2012:416036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gabitova-Cornell L, Surumbayeva A, Peri S, Franco-Barraza J, Restifo D, Weitz N, et al. Cholesterol pathway inhibition induces TGF-beta signaling to promote basal differentiation in pancreatic cancer. Cancer Cell 2020;38:567–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res 2014;74:5057–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nywening TM, Wang-Gillam A, Sanford DE, Belt BA, Panni RZ, Cusworth BM, et al. Targeting tumour-associated macrophages with CCR2 inhibition in combination with FOLFIRINOX in patients with borderline resectable and locally advanced pancreatic cancer: a single-centre, open-label, dose-finding, non-randomised, phase 1b trial. Lancet Oncol 2016;17:651–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med 2012;2:a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Macy M, Okano Y, Cardin AD, Avila EM, Harmony JA. Suppression of lymphocyte activation by plasma lipoproteins. Cancer Res 1983;43:2496s–502s. [PubMed] [Google Scholar]
- 46. Mahley RW, Rall SC Jr. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet 2000;1:507–37. [DOI] [PubMed] [Google Scholar]
- 47. Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A 1993;90:1977–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr., et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 1994;7:180–4. [DOI] [PubMed] [Google Scholar]
- 49. Ostendorf BN, Bilanovic J, Adaku N, Tafreshian KN, Tavora B, Vaughan RD, et al. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat Med 2020;26:1048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamamoto T, Choi HW, Ryan RO. Apolipoprotein E isoform-specific binding to the low-density lipoprotein receptor. Anal Biochem 2008;372:222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Table S1
Table S2