Abstract
Specialized mechanisms ensure proper expression of critically important genes such as those specifying cell identity or conferring protection from environmental stress. Investigations of the heat shock response have been critical in elucidating basic concepts of transcriptional control. Recent studies demonstrate that in response to thermal stress, heat shock-responsive genes associate with high levels of transcriptional activators and co-activators and those in yeast intensely interact across and between chromosomes, coalescing into condensates. In mammalian cells, cell identity genes that are regulated by super-enhancers (SEs) are also densely occupied by transcriptional machinery that form phase-separated condensates. We suggest that the stress-remodeled yeast nucleome bears functional and structural resemblance to mammalian SEs, and will reveal fundamental mechanisms of gene control by transcriptional condensates.
Keywords: 3D genome topology, Heat Shock Factor 1, heat shock response, inter-chromosomal interactions, super-enhancers, transcriptional condensates, phase separation
3D genome structure and transcriptional condensates
The eukaryotic genome is hierarchically organized into discrete structural loops and higher order domains. Regulatory signals are typically relayed via chromatin loops that facilitate the process of transcription by reducing the effective distance between enhancers and their cognate promoters [1, 2]. Additional loops also form, including those between gene promoters and terminators [3–6], gene regulatory and coding regions [7–10] and loci bound by architectural proteins [11, 12]. These units afford remarkable spatial contraction without which physical distances between genomic regulatory elements would be very large.
Regulatory loops assemble within higher-order structural units termed topologically associating domains (TADs). Such domains are delimited by strong boundaries occupied by insulator proteins that suppress contacts between neighboring TADs [1, 2]. Loops permit chromatin interactions, while TADs increase the frequency and specificity of such contacts. Together, loops and domains modulate gene activity by either facilitating or restricting regulatory element interactions. In principle, these multi-layered genomic topologies reduce search time and space, and thereby coordinate a transcriptional response that is both more rapid and more robust. At the molecular level, this is achieved when multiple DNA elements, including enhancers and promoters, and their regulatory factors engage in a network of interactions within common sites of transcription. While enhancer-promoter (E-P) loops are thought to be a general feature of eukaryotic transcription, large clusters of regulatory elements, termed “super-enhancers” (SEs), have been observed at key genes involved in development and disease [13]. To cooperate, two regulatory elements must be located within a distance of 100 to 300 nm, but need not physically interact [14]. Multiple factors occupying these sites bridge the interactions, and liquid-liquid phase transitions have been suggested to underpin the process, at least at SEs [15–17]. Many transcriptional regulators, including gene-specific and general transcription factors (TFs), the Mediator complex and RNA Pol II itself, are multivalent and/or contain intrinsically disordered regions (IDRs) [15, 18, 19]. The residues within the IDRs are further subject to reversible chemical modifications that can potentially modulate their interaction with histones, nucleic acids and other transcriptional and post-transcriptional regulators and contribute to the formation of large multi-molecular assemblies [20]. These assemblies, or “condensates”, can spontaneously separate from the surrounding solution phase and thereby function as membraneless compartments [21, 22].
Condensates are likely to contribute to coordinated transcriptional control. Cell type-specific genes – including immunoglobulin, erythroid-specific and cytokine-responsive genes – have been observed to associate within “transcription factories” [23–25]. In addition, the multi-enhancer olfactory receptor (OR) hub that drives OR gene expression in mice serves as an extreme example of coordinate regulation [26]. These examples suggest that intergenic clustering enables coordinate regulation of transcriptional programs during growth and differentiation.
Three-dimensional genome topology is likely to play an important role in other physiological processes, such as cellular stress responses. Following temperature upshift, oxidant injury, nutrient deprivation or exposure to heavy metals, cells in all kingdoms of life activate a common transcriptional program known as the heat shock response (HSR), characterized by the induction of molecular chaperones [27]. In eukaryotes, the HSR is under the control of the master transcription factor, Heat Shock Factor 1 (HSF1), and this transcriptional response has proven to be a powerful model system for understanding mechanisms of gene regulation. In this perspective, we consider regulation of the HSR and discuss global changes in 3D genome organization that occur in response to stress. We contend that the dynamic chromatin restructuring that occurs during the HSR in the budding yeast S. cerevisiae is structurally, functionally and mechanistically related to super-enhancers and multi-chromosomal enhancer hubs that control cell identity in mammals.
Deep conservation of the function and regulation of the heat shock response
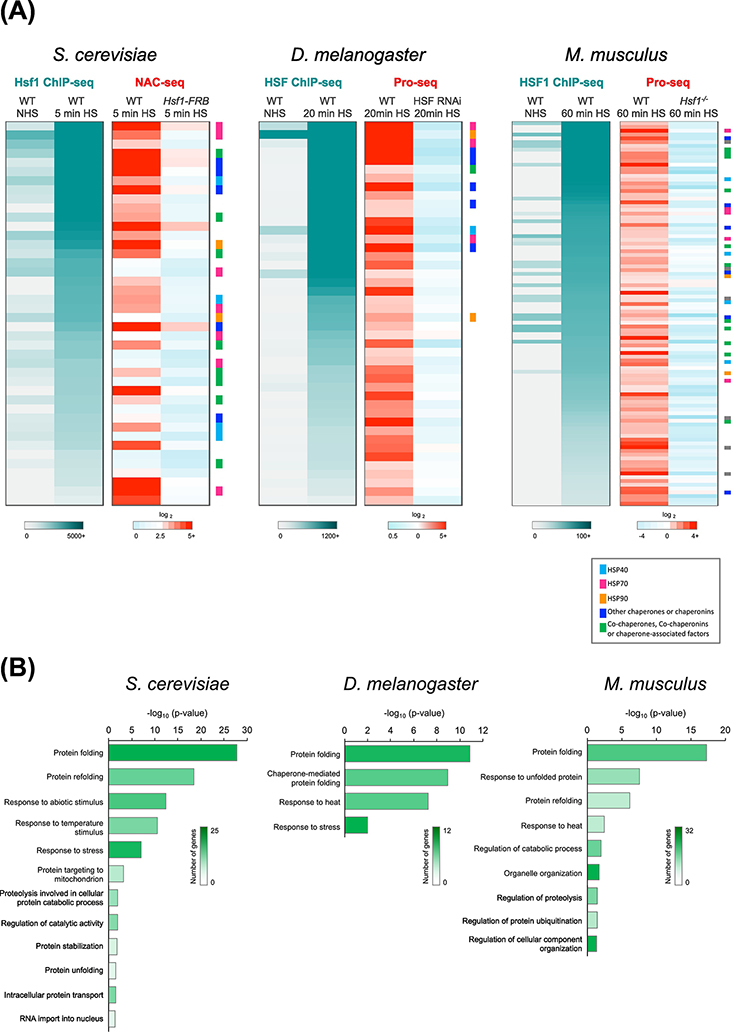
The HSR is among the most ancient and conserved gene expression programs. The interaction between the transcription factor HSF1 and its cognate DNA binding site, the heat shock element (HSE), has remained intact since the last eukaryotic common ancestor more than a billion years ago. While it has taken on species-specific roles and elaborations, the core function of the HSR – to regulate the expression level of molecular chaperones according to the fluctuating demands of the cell – has proven to be indispensable. It is therefore unsurprising that HSR has been linked to numerous human diseases such as cancer and neurodegeneration [28, 29]. HSF1 responds to a variety of environmental and genetic stressors through the formation of a DNA-binding homotrimer that specifically recognizes the HSE [30]. A large subset of HSF1 target genes across organisms encode chaperones and other heat shock proteins (HSPs) (Figure 1A) that function in a myriad of cellular processes (Figure 1B). Thus, maintaining protein homeostasis (“proteostasis”) appears to be the responsibility of HSF1 across eukaryotes.
Figure 1. Conservation of the Heat Shock Response in Eukaryotes.
A) Heat Shock Factor 1 (HSF1) binding as determined by ChIP-seq (left, gray to teal) and fold-change in nascent RNA levels as determined by NAC-seq or Pro-seq (right, blue to red) at genes bound and induced by HSF1 in S. cerevisiae (n=42), D. melanogaster S2 cells (n=44) and mouse embryonic fibroblasts (MEFs; n=102). HSF1 occupancy was determined under both non-heat shock (NHS) and heat shock (HS) conditions. Fold-change in NAC-seq or Pro-seq reads (HS / NHS) was determined for WT and HSF1-depleted cells. Genes are sorted in descending order of HSF1 ChIP-seq counts (HS condition). Genes encoding molecular chaperones are color coded as indicated by the key. Data were obtained from YeastMine [73], FlyMine [74], Mouse Genome Informatics (http://www.informatics.jax.org) and UniProt [75]. Yeast NAC-seq and Hsf1 ChIP-seq data were obtained from a previous study [37]; Drosophila HSF ChIP-seq and Pro-seq data were provided by F.M. Duarte and M.J. Guertin [42, 44] and MEF HSF1 ChIP-seq and Pro-seq data were provided by D.B. Mahat [43].
B) Gene Ontology (GO) comparison of HSF1-dependent genes in yeast, fly and mouse, respectively, using the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system [76, 77]. Heatmap indicates the number of genes in each GO category. The length of each bar corresponds to the P-value corrected for multiple comparison testing using the Bonferroni method. Test was for enrichment of genes from panel A mapping to the given GO category in comparison to expected distribution of genes in the same category in the reference genome.
A key feature of HSF1-driven transcriptional activation is that it’s remarkably rapid and robust, yet evanescent. In budding yeast, local chromatin changes at HSP gene promoters and coding regions are detected within 60 seconds of temperature upshift (30° to 39°C). Accompanying the rapid, and nearly quantitative, displacement of nucleosomes at yeast HSP genes are large increases in the occupancy of Hsf1, RNA Pol II, and transcriptional coactivators along with >100-fold increase in the transcription of several HSP genes [9, 31–37] (Figure 1A, left; Figure 2A, red bar). These phenomena are transient, as both factor occupancy and mRNA levels typically return to pre-heat shock levels within 30 – 60 minutes even while the temperature remains elevated.
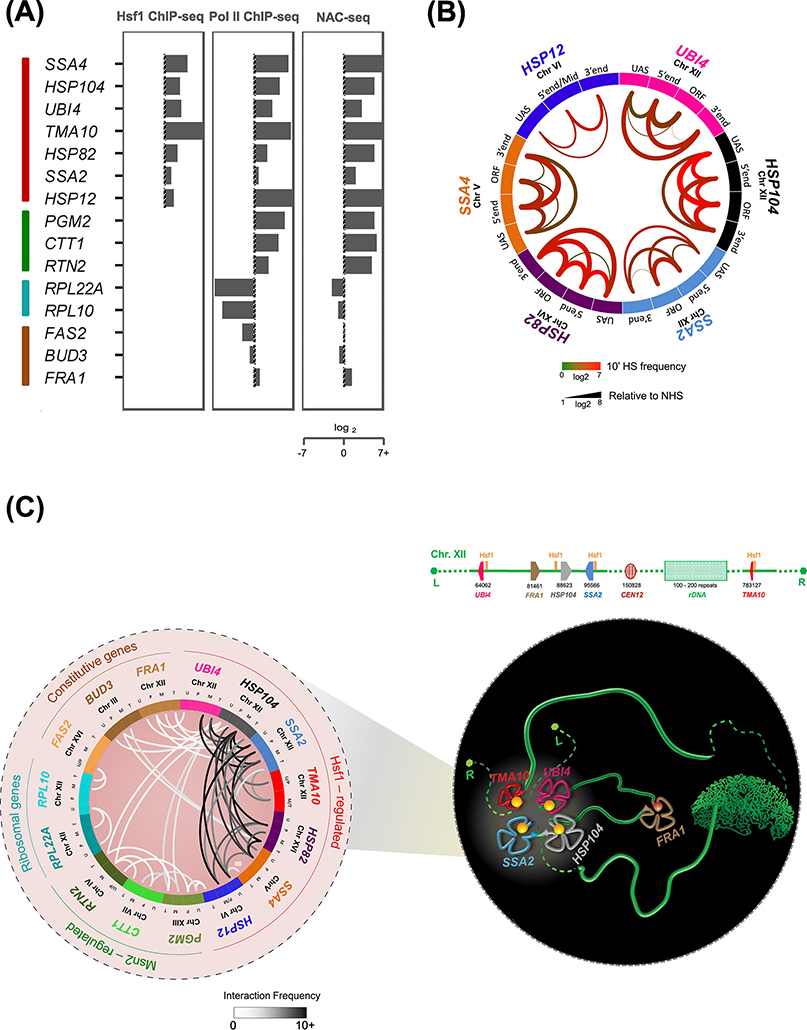
Figure 2. Hsf1-regulated Genes in S. cerevisiae Loop, Crumple and Coalesce upon Transcriptional Activation.
A) Fold-change nascent RNA, Pol II and Hsf1 densities at representative genes (heat shock-inducible: Hsf1- and Msn2/4-regulated (red and green); heat shock-repressed: ribosomal protein coding (RPL; blue); and constitutively expressed (brown) in budding yeast. Log2 fold-change levels are calculated at 5 min HS relative to NHS. NAC-seq and Hsf1 ChIP-seq data were derived from [37] and Rpb1 ChIP-seq data from [61].
B) Circos plot depicting intragenic interactions within indicated Hsf1-target genes as determined by Taq I-3C. Data were obtained from NHS and 10 min HS-induced cells [7, 9]. Arcs depict chromatin interactions between indicated gene regions. Color of each arc reflects the interaction frequency detected under 10 min HS conditions; thickness represents fold-change of HS relative to NHS.
C)(Left) Circos plot depicting intergenic interactions detected between the indicated genes in acutely heat-shocked cells by Taq I-3C [7, 9]. Arcs depict chromatin interactions between indicated genic regions; shade is proportional to the frequency of interaction. For each gene pair, top two interaction frequencies are shown. U, UAS; P, promoter; M, mid-ORF region; T, terminator. (Right) Schematic model of Chromosome XII (green ribbon-like structure), depicting heat shock-driven interactions between Hsf1-target genes. UBI4, HSP104 and SSA2 on the left arm physically interact with TMA10 located on the right arm, circumventing the physical barrier imposed by the nucleolus (green crescent shaped structure). In contrast, FRA1, a constitutively active gene interposed between UBI4 and HSP104, is excluded from coalescence. L, left telomere; R, right telomere. Physical map is shown above.
Transcriptional activation of Drosophila and mammalian HSP genes is likewise associated with substantial coactivator recruitment [38, 39] and rapid, robust and transient induction. In Drosophila, elongating RNA Pol II has been detected within 75 seconds of temperature upshift (22° to 37°C) [40, 41], and ~400-fold induction has been reported for several HSP genes (Figure 1A, center) [42]. Studies in mammalian cells have demonstrated similar levels of HSP gene induction (≥300-fold) in response to thermal upshift (37° to 42°C) (Figure 1A, right) where changes in Pol II density are evident within 2.5 minutes of heat shock [43]. Together, these studies illustrate that near-instantaneous induction is a conserved feature of the HSR.
While activation of HSF1 gene targets leads to prolific levels of transcription, the number of genes actually dependent on HSF1 is comparatively small. In yeast, Hsf1 binds to 74 loci and induces the expression of 52 genes [37] (Figure 1A, left, NAC-seq). In Drosophila, HSF binds to 400 loci [44], yet regulates only 44 genes [42] (Figure 1A, center, Pro-seq). In mouse cells, HSF1 binds to 602 loci yet only 102 genes are induced in an HSF1-dependent fashion [43] (Figure 1A, right, Pro-seq). The discord between HSF1 binding and activation is also mirrored in non-canonical HSF1 targets in human cancer cells, where only a fraction of bound genes requires HSF1 for activation [45]. Thus, rather than controlling a broad transcriptional program, HSF1 regulates a highly conserved and focused set of targets comprising <1% of the genes in diverse organisms.
Notably, HSF1 targets include only a small fraction of the genes that change expression upon heat shock. In yeast, other stress-activated TFs such as Msn2, Msn4, Yap1 and Skn7 induce more genes during heat shock than Hsf1[46–49]. In mammalian cells, serum response factor (SRF) appears to be involved in heat shock-dependent induction of the large set of HSF1-independent genes [43]. Moreover, several hundred genes in yeast [37, 46], more than a thousand in Drosophila [42, 50] and up to 4000 in mammals [43] are repressed in response to heat shock. This repression is HSF1-independent and has been termed “stress-induced transcriptional attenuation” (SITA) [51]. Among these heat shock-regulated transcriptional processes, the HSR is unique in its functional and regulatory conservation. The rapid dynamics, robust induction and conserved components of the HSR make it an ideal system to study transcription-associated changes in genome architecture.
3D genome remodeling in response to heat shock
Recent studies in budding yeast using TaqI-3C, a highly sensitive chromosome conformation capture (3C) assay [52], have revealed that changes in 3D genome topology accompany the HSR. Heat shock induces both intragenic rearrangements within HSP genes and intergenic interactions among them. These alterations include intragenic chromatin contacts between [i] the upstream activation sequences (UASs) and promoters of Hsf1-dependent HSP genes; [ii] their 5’ and 3’ ends; and [iii] their regulatory and coding regions (Figure 2B). Such intragenic interactions are remarkably dynamic and strongly correlate with transcriptional activity [7].
In addition, chromosomally linked as well as unlinked HSP genes coalesce upon heat shock and physically interact (Figure 2C). These intergenic interactions – undetectable under control (non-heat shock [NHS]) conditions – are Hsf1-dependent and display an intensity and specificity comparable to intragenic interactions. Moreover, interactions between HSP genes extend beyond their regulatory regions, as all combinations of UAS, promoter, coding region and terminator contacts are found [7, 9]. Similar intra- and inter-chromosomal interactions have been observed in cells exposed to chemical stress (8.5% ethanol) (personal communication, L.S. Rubio), demonstrating that Hsf1 activation, and not thermal stress per se, underlies HSP gene coalescence.
Importantly, other transcriptionally active genes, while adopting similar looped and folded states as stress-induced HSP genes, show little or no evidence of intergenic interaction. In fact, genes interposed between HSP genes, despite their proximity to one member of the interacting pair, fail to physically contact the HSP loci (e.g., FRA1, interposed between UBI4 and HSP104 [see Figure 2C, right and Figure 3A]) [9]. Genes transcriptionally activated in response to heat stress by Msn2/Msn4 also fail to form detectable intergenic interactions among themselves or with HSP genes [9] (Figure 2C, white lines). Intergenic interactions among yeast HSP genes are therefore unprecedented in their specificity and dynamics. Why genes under exclusive Msn2/4 regulation fail to detectably coalesce in heat shock-induced cells is unknown, although it is notable that Msn2 has been shown to activate its targets in a linear, non-cooperative manner [53] while Hsf1 acts cooperatively [54].
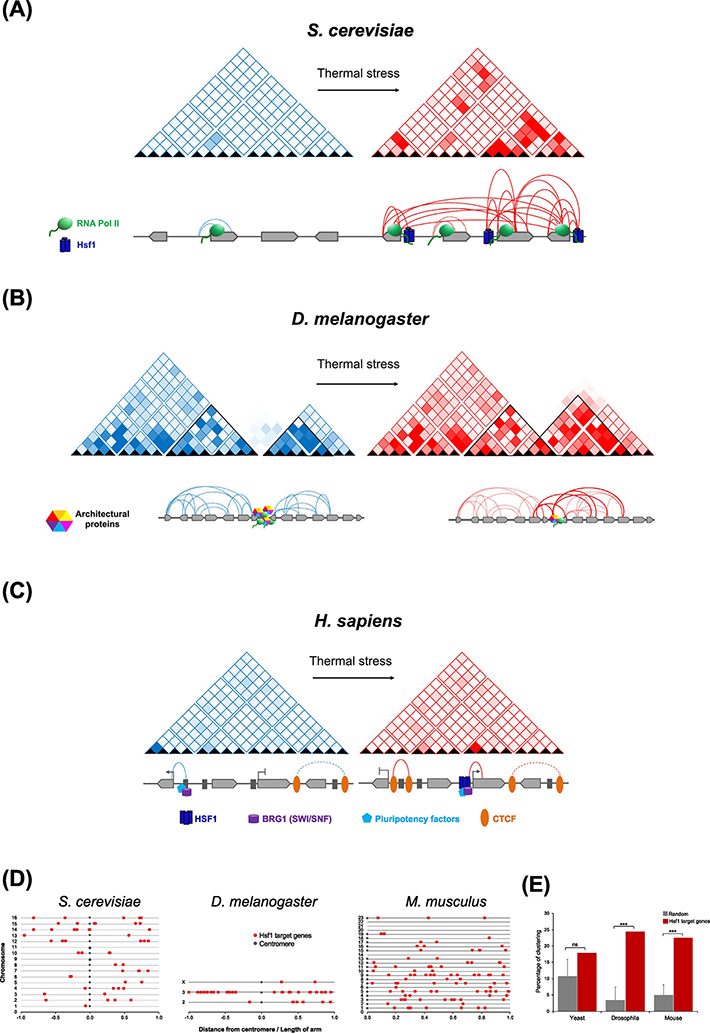
Figure 3. HSF1-target Gene Topology and Chromosomal Distribution in Yeast, Fly and Mammals.
A) In S. cerevisiae under NHS conditions (blue matrix), Hsf1-target genes UBI4, HSP104 and SSA2, located on the left arm of Chr. XII, exhibit minimal transcriptional activity and remain unstructured, while the constitutively transcribed FRA1 gene undergoes intragenic looping (dotted arcs). Acute thermal stress results in dramatic increase in HSP gene transcription and intra- and intergenic interactions (red matrix and solid arcs) that are dependent on Hsf1 (navy blue) and RNA Pol II (green). The non-Hsf1-regulated FRA1 gene does not participate in intergenic interactions despite its proximity to the other three [9]. Genes are depicted as light grey block arrows.
B) In Drosophila Kc167 cells, heat shock-dependent inter-TAD interactions involving genomic loci associated with Polycomb complex, along with redistribution of architectural proteins, was reported [55]. Under NHS conditions, intra-TAD interactions (blue matrices and dotted arcs) are more frequent. TAD borders are enriched in architectural proteins (multicolored hexagons) that enforce insulation between neighboring TADs. Upon heat shock, architectural proteins are relocated from TAD borders to their interior, and result in weakened borders and higher inter-TAD interactions (solid red arcs).
C) In hESCs, heat shock-induced increase in HSF1 binding, as well as that of pluipotency factors and chromatin remodeling complexes, takes place; this is accompanied by de novo formation of E-P contacts (solid red arc) [57]. Decommissioning is marked by gain in CTCF (orange) loops. Genes are depicted as light grey block arrows; enhancers, dark grey rectangles.
D) Illustrated is the location of Hsf1-target genes on the chromosomes of yeast, Drosophila and mouse, depicted as the distance from the respective telomere divided by the length of each chromosomal arm.
E) A two-sided Kolmogorov–Smirnov test, using Cluster Locater [78], was done to compare the distribution of Hsf1 target genes along each chromosome to 1000 randomly generated gene lists. While such an analysis revealed that Hsf1 target genes are not significantly clustered in S. cerevisiae (P=0.08), a significant fraction is clustered in groups of 2–4 genes in both Drosophila (P=8.3 × 10−8) and mouse (P=1.2 × 10−8).
Heat shock-dependent 3D genome restructuring has also been observed in Drosophila Kc167 cells, in which global alterations in transcriptional patterns were accompanied by restructuring of TADs [55]. Under control conditions, chromatin contacts were confined within a TAD; following exposure to temperature stress, architectural proteins from TAD borders were redistributed to the interior. This caused a decline in TAD border strength and a consequential increase in heat shock-specific inter-TAD interactions involving genomic loci associated with Polycomb complex [55] (Figure 3B). In contrast, a more recent study observed no significant changes in TAD structure upon HS of Drosophila S2 cells [56]. This latter study reported that HSF1 target genes adopt a pre-set chromatin architecture, including enhancer-promoter looping, and these contacts do not measurably change upon heat shock.
In human embryonic stem cells (hESCs), heat shock-induced de novo enhancer-promoter (E-P) contacts correlate with enhancer activation, and a corresponding loss of E-P contacts correlates with enhancer deactivation [57]. Activation involved increased binding of HSF1 and other transcription factors, as well as the architectural proteins CTCF and cohesin and the chromatin remodeler BRG1. Inhibition of HSF1 led to a decrease in HS-induced cohesin loops (E-P contacts), suggesting fewer changes in 3D genome architecture in the absence of functional HSF1 [57] (Figure 3C). However, in erythroleukemia cells, chromatin interactions between HSF1-occupied enhancers and their cognate promoters were mostly pre-established, and large changes in transcription and local chromatin structure occurring upon heat shock were not accompanied by changes in TADs, TAD boundaries or compartments [56]. Nonetheless, the latter study observed that a subset of E-P interactions showed small changes in contact frequency upon heat shock irrespective of HSF1 binding.
Metazoan studies therefore argue that changes in the 3D architecture of heat shock-responsive genes are subtler than in yeast. It is probable that the substantial increase in DNA polymer length in metazoans provides constraints in diffusion and/or mobility not present in the compact chromosomes of S. cerevisiae. In addition, the absence of long-range chromatin interactions in metazoans may reflect a practical constraint of CTCF-mediated insulation of TADs. It is notable, however, that in both the Drosophila and mouse genomes, HSF1 target genes are distributed in the genome in a non-uniform manner and cluster together in groups of 2–4 genes within small chromosomal segments with few other genes interspersed. By contrast, in yeast, Hsf1-target genes are not significantly more clustered than a random set of genes (Figures 3D, 3E). While it is tempting to speculate that these clustered HSF1 target genes in metazoan genomes – particularly those that fall within a single TAD – engage in intergenic interactions upon heat shock, testing this will require the application of higher-resolution 3C approaches than have been used to date.
In addition to being linked to 3D genome remodeling, HSF1 coalesces into prominent intranuclear foci in human cells in response to heat shock as well as during oncogenesis [58, 59]. Such foci arise from HSF1 binding to non-coding satellite III repetitive DNA sequences and pericentromeric regions [60] and may reflect formation of phase-separated condensates [59]. These foci likely represent inactive HSF1, as they are most prominent in cells with low levels of chaperone expression. Thus, such HSF1-mediated structures are unlikely to be either functionally or structurally related to the heat-shock-induced changes in chromatin organization associated with transcriptional activation. However, the presence of these large and stable HSF1 foci at satellite III repeats does not preclude the existence of smaller and more transient HSF1-containing condensates at actively transcribed target genes.
HSP gene clusters in yeast: primordial super-enhancers?
The structures formed by heat shock-triggered coalescence of HSP genes share three key attributes with mammalian super-enhancers (SEs) (schematically summarized in Figure 4A). First, there are relatively few SEs in any given cell type, yet this small number of genomic loci concentrates a large fraction of the total cellular transcriptional machinery. Similarly, Hsf1-dependent genes are relatively few in number yet concentrate a disproportionate amount of the transcriptional machinery during heat shock: we estimate that 13% of all Pol II transcription in acutely stressed cells is Hsf1-dependent, while <1% of all protein-encoding genes are Hsf1 targets [37]. Rank-order analyses of Mediator occupancy at mouse ESC enhancers [13] and of Pol II occupancy at heat shock-induced yeast promoters [61] underscore the similarity of SEs and Hsf1 target genes in yeast (Figure 4B). As is evident, both SEs and Hsf1 targets lie above the inflection point of their respective curves, readily distinguishable from typical enhancers and housekeeping gene promoters.
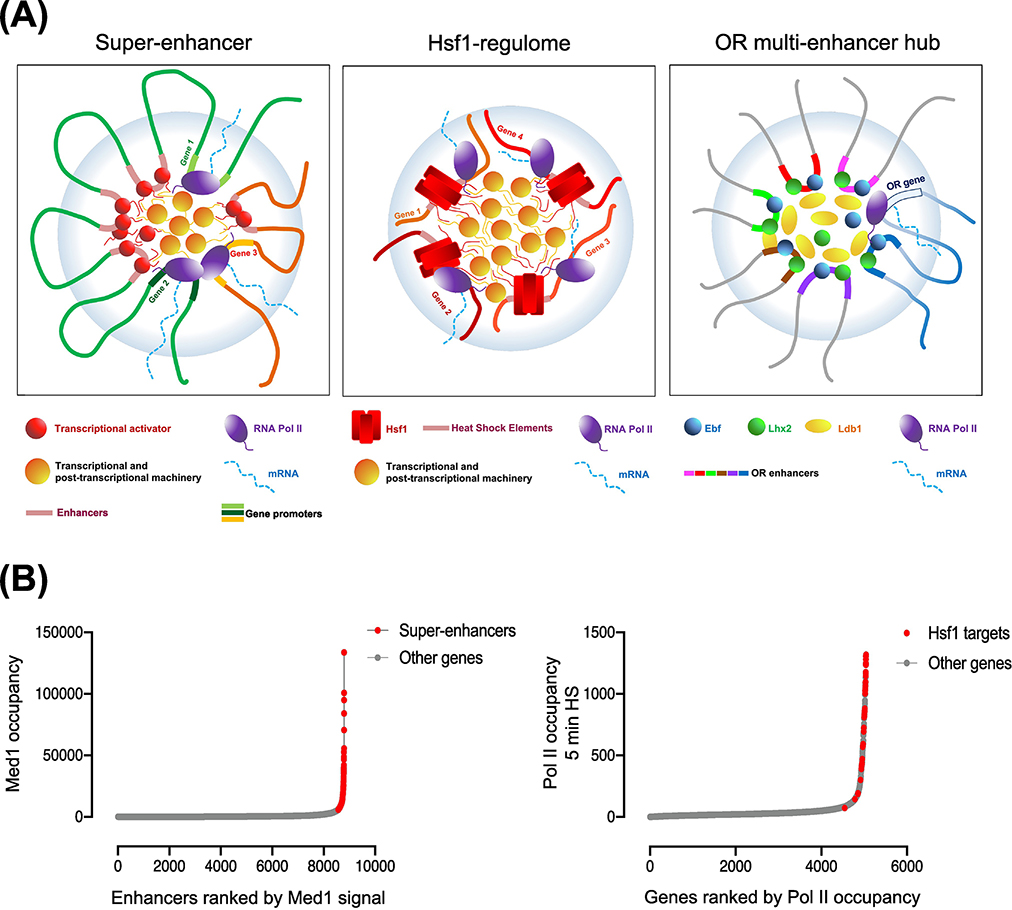
Figure 4. HSP Gene Coalescence Shares Key Attributes with Super-Enhancers and Olfactory Receptor Multi-Enhancer Hubs.
A) (Key Figure)(Left) Super-enhancers (SEs) comprise clusters of enhancers (red spheres) that engage in physical interactions with promoters of associated genes (genes 1–3). SEs are densely occupied by transcriptional activators (red spheres) and other transcriptional and post-transcriptional apparatus (orange ovals), and a network of multivalent cooperative interactions exist between them. The increase in density and affinity of interactions between these components contribute towards SE assembly within a phase-separated condensate (light blue bubble). (Center) Yeast HSP genes (orange) are occupied by unusually high densities of activator (Hsf1, red rectangles), RNA Pol II (purple ovals) and other transcriptional and post-transcriptional factors (orange ovals), and exhibit strong inter-chromosomal interactions upon their induction. (Right) Inter-chromosomal clustering of multiple olfactory receptor (OR) enhancers lead to the formation of an OR enhancer hub in mouse olfactory sensory neurons. The OR enhancers, co-bound by transcription factors (Ebf and Lhx2) and coactivators (Ldb1), stochastically converge on a single olfactory receptor gene to activate its transcription.
B)(Left) Mediator (MED1) occupancy across 8794 enhancers in mouse ESC cells arranged according to their rank order [13]. (Right) Pol II occupancy across 5047 genes in S. cerevisiae in the 5 min heat shock condition, arranged according to their rank order (data from [61]). Both SEs and Hsf1-regulated genes fall above inflection points of their respective curves, suggesting exceptionally high occupancy levels of transcriptional machinery at these genes.
Second, formation of SEs involves the 3D clustering of multiple enhancer modules and their associated gene promoters [62, 63]. This can result in the simultaneous activation of multiple promoters [64] as well as in multi-way contacts between SEs [10]. Likewise, HSP genes engage in a network of chromatin contacts; as described above, coalescence involves intergenic contacts between UASs and promoters of Hsf1-regulated genes as well as between their coding regions [7, 9]. While the inter-chromosomal interactions observed upon HSP activation may not be a typical characteristic of mammalian SEs, the high local concentration (multivalency) of transcription factor binding sites – provided in cis in SEs and in trans in the case of HSP gene coalescence – is a core feature of both SEs and HSP gene clusters.
Finally, SE-associated TFs such as OCT4 form cooperative assemblies with Mediator that underpin “demixing” of super-enhancers into phase-separated condensates [15, 16] (Figure 4A, left). Similarly, Hsf1 forms discrete intra-nuclear puncta in yeast in response to heat shock [9]; strikingly, such puncta overlap those formed by Mediator and Pol II and exhibit properties suggestive of phase-separated condensates (unpublished) (Figure 4A, center). Together, these characteristics of heat shock-induced HSP gene clusters in yeast – their disproportionate concentration of the transcriptional machinery, their complex 3D interaction landscape and their organization in phase-separated condensates – suggest that these yeast structures capture the salient features of mammalian SEs.
HSP gene clusters recapitulate features of multi-enhancer OR hubs
As alluded above, a regulatory system with a strong parallel to stress-activated yeast HSP genes is the multi-chromosomal, multi-enhancer olfactory receptor (OR) hub that drives OR gene expression in mice. Within each murine olfactory sensory neuron (OSN), only one of ~1400 OR genes is activated during OSN differentiation, determining the type of odor that the mature neuronal cell will perceive [65]. Monogenic OR expression is achieved within a multi-enhancer hub, where multiple enhancers cooperatively bound by the TFs Ebf and Lhx2 converge with the selected OR allele and engage in extensive interactions with each other as well as with the selected locus, thereby activating it (Figure 4A, right) [26].
HSP gene coalescence shares key attributes with multi-enhancer OR hubs. First, both systems involve inter-chromosomal interactions between participating DNA sequences that occur under specific contexts. In OSN cells, OR enhancers engage in physical and/or functional interactions with the promoter of the OR allele selected for expression [66, 67]. Similarly, in yeast, inter-chromosomal contacts occur only within heat shock-activated cells. An important difference between the two is the timeframe within which the reconfiguration of chromatin topology takes place. In differentiating mouse olfactory sensory neurons, global remodeling takes place over a period of days to weeks, whereas comparable changes in the nuclear architecture of yeast takes place over a period of seconds to minutes. As discussed above, the small size of S. cerevisiae chromosomes and the relatively uniform distribution of HSP genes might facilitate their rapid restructuring; a limiting attribute of the metazoan genome in this regard is the diffusion constant of its large chromosomes.
Second, inter-chromosomal contacts in the OR hubs are highly specific, as unrelated neighboring genes are excluded from such interactions [26]. Likewise, HSP gene coalescence appears specific to genes under the control of Hsf1. Third, OR enhancers, like SEs, are occupied by high levels of transcriptional activators and coactivators that drive OR gene activation [67]. Similarly, HSP genes are associated with prolific quantities of Hsf1, Mediator and other components of the transcriptional machinery. And fourth, deletion of an OR enhancer results in loss of interaction of its locus with the rest of the multi-enhancer hub, whereas interactions between remaining enhancers are unaffected [67]. Likewise, locus-specific deletion of the Hsf1 enhancer upstream of HSP12, while having only a minimal effect on HSP12 transcription, obviates the interaction of HSP12 with other HSP genes while sparing all other intergenic interactions tested [9]. Thus, as in the case of SEs, HSP gene clusters recapitulate key features of multi-enhancer OR hubs.
Concluding Remarks
We have argued that the yeast heat shock response shares specialized mechanisms with mammalian cell identity programs that enforce robust expression of critically important genes. In budding yeast, whose compact genome is parceled into 16 chromosomes, transcriptional induction that occurs in response to heat shock is accompanied by 3D reorganization of HSP genes that culminates in their physical coalescence. Such organization, which includes novel cis- and trans-chromatin contacts, bears similarity to mammalian super-enhancers and multi-enhancer olfactory receptor hubs. The observation that homologous 3D structures in the nucleus – with similar organization, components and biophysical properties – underlie these diverse processes in evolutionarily distant organisms suggests that transcriptional condensates are ancient and flexible structures that predated the divergence of yeast and metazoans. Practically speaking, this means that fundamental principles of organization and regulation of transcriptional condensates are now subject to mechanistic dissection in yeast.
There are several avenues of future exploration. First, it will be important to understand what contributes to the specificity of transcriptional condensate formation. Hsf1-target genes form detectable condensates commensurate with their transcriptional activation, whereas Msn2-target genes do not. The availability of powerful genetic tools in yeast may help shed light on this. Second, what is the role of nascent RNA production in regulating the intra- and inter-chromosomal clustering seen in SEs, OR hubs and HSP gene clusters? This question is of particular interest in light of recent evidence implicating nascent RNAs in stimulating condensate formation and shaping local nuclear space [68, 69]. Third, might yeast HSP gene and mammalian SE condensates contribute to their subnuclear localization – in particular, to their physical association with nuclear pore complexes [70, 71]?
Although it remains to be fully explored, we speculate that intergenic coalescence of co-regulated genes may prove to be the rule rather than the exception for inducible transcriptional programs in yeast. Evidence that this may be the case comes from molecular and imaging studies suggestive of intergenic coalescence for genes under the regulation of the Met4 and Ino2/Ino4 TFs [70, 72]. Beyond fungi, it may be that intergenic transcriptional condensates commonly form in lineages with relatively compact genomes. Moreover, as we have described, inter-chromosomal interactions can occur in metazoan cells under specific circumstances that have likely yet to be fully enumerated. The common features yet distinct functions of HSP gene clusters, SEs and OR hubs underscore the conservation and plasticity of transcriptional condensates.
Outstanding Questions.
What contributes to the specificity of transcriptional condensate formation? Does the Hsf1 transcription factor use a distinctive activation mechanism to drive the cis- and trans-intergenic interactions of its target genes in yeast?
How does 3D chromatin organization of HSF1-target genes influence target gene selection and regulation under distinct conditions, such as during oncogenesis?
Does HSP gene coalescence impact the transcriptional output of individual genes or their kinetics of activation in response to a stimulus?
What is the mechanistic contribution of individual factors found within Hsf1 transcriptional condensates?
Do high local concentrations of nascent RNA contribute to the formation of SEs, OR hubs and HSP gene clusters?
Do other eukaryotes with compact genomes exhibit inter-chromosomal clustering or condensate formation of select gene regulons?
Are there other examples of multi-chromosomal enhancer control in mammals?
How do transcriptional condensates promote survival and developmental fitness?
Highlights.
The rapid dynamics, robust induction and conserved components of the HSF1-driven heat shock response make it an ideal system to study eukaryotic gene regulation.
Heat shock (acute thermal stress) induces both short- and long-range changes in the chromosomal topology of budding yeast Heat Shock Protein (HSP) genes, culminating in their physical coalescence.
HSP gene coalescence shares several key attributes with mammalian super-enhancers and multi-enhancer olfactory receptor hubs, including exceptional concentration of transcription factors and coactivators, extensive DNA looping and clustering, and cooperative assembly into phase-separated condensates.
Elucidating the molecular basis for HSP gene coalescence could reveal fundamental mechanisms of gene control during critical cellular processes such as differentiation and development.
Acknowledgements
We thank F.M. Duarte, M.J. Guertin and D.B. Mahat for providing ChIP-seq and Pro-seq data. Work on this project was supported by startup funds from the Biological Sciences Division at The University of Chicago to D.P. and by grants from the NSF (MCB-1518345) and NIH (GM128065 and GM138988) to D.S.G.
Glossary
- Chromatin
Complex of DNA, proteins and RNA that constitutes the physiological state of the genome
- Nucleosome
A bead-like structure consisting of nearly two superhelical turns of DNA wrapped around an octamer of histones — two H2A–H2B dimers and an H3–H4 tetramer. Fundamental repeating subunit of chromatin
- Enhancer / Upstream Activating Sequence (UAS)
Regulatory DNA sequence that serves as a binding site for transcriptional activators, with a mean size (in mammalian cells) of 0.5 – 1 kb
- Promoter
Regulatory DNA sequence that serves as a binding site for RNA polymerase and associated general transcription factors (GTFs). Comprised of the transcription start site (TSS) and typically other motifs, including a TATA box, to which GTFs bind
- Transcriptional activators (gene-specific transcription factors, GSTFs)
Sequence-specific DNA-binding proteins that bind to enhancer elements and coordinate promoter opening and transcriptional activation of associated genes
- Transcriptional coactivator
A protein that transiently interacts with a DNA-bound gene-specific transcription factor to facilitate transcriptional activation of a gene or set of genes
- Mediator
Conserved multisubunit transcriptional coactivator of RNA Pol II (Pol II) transcribed genes. Serves as a physical and functional bridge between GSTFs and Pol II
- Super-enhancers (SEs)
Clusters of enhancers that control transcription of cell-identity and tissue-specific genes in mammalian cells; can span >10 kb. SEs exhibit an unusually strong enrichment of GSTFs, RNA Pol II and transcriptional coactivators
- Transcription factories
Discrete subnuclear sites of nascent RNA production. Typically contain clusters of genes whose expression is coordinated by locally enriched RNA polymerases, gene-specific and general transcription factors, coactivators and other components of transcriptional and post-transcriptional machinery
- Biomolecular condensates
Subcellular compartments that form through liquid-liquid phase separation, driven by interactions of multivalent molecules. Exhibit a higher concentration of certain biological molecules than the surrounding milieu while continuously exchanging with it. Typically lack a surrounding membrane
- Liquid-liquid demixing (phase separation)
A physical process that involves spontaneous separation of a super-saturated solution of components into two co-existing liquid-like phases, a dense phase and a dilute phase
- Multivalent interactions
Nexus of interactions where a molecule engages with other molecules via multiple binding sites
- IDR (Intrinsically Disordered Regions)
Regions of proteins that do not have a defined three-dimensional structure and exist as ensemble of dynamic conformations
- Nucleome
Molecular, genetic and physical state of nuclear components of the cell
- Chromosome Conformation Capture (3C)
A molecular technique that allows for the determination of DNA contact frequencies in chromatin and insight into chromosome topology
Footnotes
Declaration of Interests
The authors declare no competing interests.
Websites:
D. Pincus https://mgcb.uchicago.edu/faculty/david-pincus-phd
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dekker J (2014) Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin 7 (1), 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wendt KS and Grosveld FG (2014) Transcription in the context of the 3D nucleus. Curr. Opin. Genet. Dev 25, 62–67. [DOI] [PubMed] [Google Scholar]
- 3.Ansari A and Hampsey M (2005) A role for the CPF 3’-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19 (24), 2969–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan-Wong SM et al. (2008) Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A 105 (13), 5160–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Sullivan JM et al. (2004) Gene loops juxtapose promoters and terminators in yeast. Nat. Genet 36 (9), 1014–1018. [DOI] [PubMed] [Google Scholar]
- 6.Rowley MJ et al. (2019) Condensin II counteracts cohesin and RNA Polymerase II in the establishment of 3D chromatin organization. Cell Rep 26 (11), 2890–2903 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhary S et al. (2017) Heat shock protein genes undergo dynamic alteration in their three-dimensional structure and genome organization in response to thermal stress. Mol Cell Biol 37: e00292–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K et al. (2015) Dynamic enhancer-gene body contacts during transcription elongation. Genes Dev. 29 (19), 1992–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhary S et al. (2019) Heat shock factor 1 drives intergenic association of its target gene loci upon heat shock. Cell Rep 26 (1), 18–28 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beagrie RA et al. (2017) Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543 (7646), 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuin J et al. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 111 (3), 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weintraub AS et al. (2017) YY1 is a structural regulator of enhancer-promoter loops. Cell 171 (7), 1573–1588 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whyte WA et al. (2013) Master transcription factors and Mediator establish super-enhancers at key cell identity genes. Cell 153 (2), 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furlong EEM and Levine M (2018) Developmental enhancers and chromosome topology. Science 361 (6409), 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boija A et al. (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175 (7), 1842–1855 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho WK et al. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361 (6400), 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehning M et al. (2018) RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol 25 (9), 833–840. [DOI] [PubMed] [Google Scholar]
- 19.Chong S et al. (2018) Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361: eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D et al. (2017) A phase separation model for transcriptional control. Cell 169 (1), 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banani SF et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18 (5), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. [DOI] [PubMed] [Google Scholar]
- 23.Papantonis A et al. (2012) TNFα signals through specialized factories where responsive coding and miRNA genes are transcribed. EMBO J. 31 (23), 4404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SK et al. (2014) Pronounced cohabitation of active immunoglobulin genes from three different chromosomes in transcription factories during maximal antibody synthesis. Genes Dev. 28 (11), 1159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoenfelder S et al. (2010) Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet 42 (1), 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bashkirova E and Lomvardas S (2019) Olfactory receptor genes make the case for inter-chromosomal interactions. Curr Opin Genet Dev 55, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12, 3788–3796. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Pastor R et al. (2018) Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 19 (1), 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santagata S et al. (2013) Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 341 (6143), 1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pincus D (2020) Regulation of Hsf1 and the heat shock response. Adv Exp Med Biol 1243, 41–50. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J et al. (2005) Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol Cell Biol 25 (20), 8985–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shivaswamy S and Iyer VR (2008) Stress-dependent dynamics of global chromatin remodeling in yeast: A dual role for SWI/SNF in the heat shock stress response. Mol. Cell. Biol 28, 2221–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kremer SB and Gross DS (2009) SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression. J Biol Chem 284 (47), 32914–32931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S and Gross DS (2013) Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and mediator tail subunits Med15 and Med16. J Biol Chem 288 (17), 12197–12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anandhakumar J et al. (2016) Evidence for multiple mediator complexes in yeast independently recruited by activated heat shock factor. Mol Cell Biol 36 (14), 1943–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinayachandran V et al. (2018) Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res.28: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pincus D et al. (2018) Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome. Mol Biol Cell 29 (26), 3168–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JM et al. (2001) Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8, 9–19. [DOI] [PubMed] [Google Scholar]
- 39.Takii R et al. (2019) The pericentromeric protein shugoshin 2 cooperates with HSF1 in heat shock response and RNA Pol II recruitment. EMBO J 38 (24), e102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehm AK et al. (2003) Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol 23 (21), 7628–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Brien T and Lis JT (1993) Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 13, 3456–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duarte FM et al. (2016) Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev 30 (15), 1731–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahat DB et al. (2016) Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell 62 (1), 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guertin MJ and Lis JT (2010) Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet 6 (9), e1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendillo ML et al. (2012) HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150 (3), 549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasch AP et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morano KA et al. (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190 (4), 1157–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raitt DC et al. (2000) The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol Biol Cell 11 (7), 2335–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solis EJ et al. (2016) Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol. Cell 63, 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teves SS and Henikoff S (2011) Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev 25 (22), 2387–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aprile-Garcia F et al. (2019) Nascent-protein ubiquitination is required for heat shock-induced gene downregulation in human cells. Nat Struct Mol Biol 26 (2), 137–146. [DOI] [PubMed] [Google Scholar]
- 52.Chowdhary S et al. (2020) Chromosome conformation capture that detects novel cis- and trans-interactions in budding yeast. Methods 170, 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart-Ornstein J et al. (2013) Msn2 coordinates a stoichiometric gene expression program. Curr Biol 23 (23), 2336–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erkine AM et al. (1999) Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol Cell Biol 19 (3), 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L et al. (2015) Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 58 (2), 216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray J et al. (2019) Chromatin conformation remains stable upon extensive transcriptional changes driven by heat shock. Proc Natl Acad Sci U S A 116 (39), 19431–19439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyu X et al. (2018) Architectural proteins and pluripotency factors cooperate to orchestrate the transcriptional response of hESCs to temperature stress. Mol Cell 71 (6), 940–955 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolly C et al. (1997) HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J Cell Sci 110 ( Pt 23), 2935–2941. [DOI] [PubMed] [Google Scholar]
- 59.Gaglia G et al. (2020) HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat Cell Biol 22 (2), 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eymery A et al. (2010) Heat shock factor 1 binds to and transcribes satellite II and III sequences at several pericentromeric regions in heat-shocked cells. Exp Cell Res 316 (11), 1845–1855. [DOI] [PubMed] [Google Scholar]
- 61.Albert B et al. (2019) A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. Elife 8: e45002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dowen JM et al. (2014) Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159 (2), 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang J et al. (2018) Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun 9 (1), 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukaya T et al. (2016) Enhancer control of transcriptional bursting. Cell 166 (2), 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monahan K and Lomvardas S (2015) Monoallelic expression of olfactory receptors. Annu Rev Cell Dev Biol 31, 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Markenscoff-Papadimitriou E et al. (2014) Enhancer interaction networks as a means for singular olfactory receptor expression. Cell 159 (3), 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monahan K et al. (2019) LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565 (7740), 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinodoz SA et al. (2020) RNA promotes the formation of spatial compartments in the nucleus. bioRxiv, 2020.08.25.267435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henninger JE et al. (2021) RNA-mediated feedback control of transcriptional condensates. Cell 184 (1), 207–225 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brickner DG et al. (2012) Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell 22 (6), 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ibarra A et al. (2016) Nucleoporin-mediated regulation of cell identity genes. Genes Dev 30 (20), 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du M et al. (2017) Three distinct mechanisms of long-distance modulation of gene expression in yeast. PLoS Genet 13 (4), e1006736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balakrishnan R et al. (2012) YeastMine – an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012, bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lyne R et al. (2007) FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol 8 (7), R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.UniProt C (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47 (D1), D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ashburner M et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25 (1), 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.The Gene Ontology C (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47 (D1), D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pazos Obregon F et al. (2018) Cluster Locator, online analysis and visualization of gene clustering. Bioinformatics 34 (19), 3377–3379. [DOI] [PubMed] [Google Scholar]