Summary
A conceptual framework for understanding abnormal endometrial decidualization, with considerable significance for the diagnosis and treatment of abnormal decidualization-related changes in non-receptive endometrium in implantation failure during early pregnancy is very important. Here, we found the expression levels of miR-29a in endometrial tissues were associated with the menstrual phases and pregnancy outcome. Inhibition of miR-29a led to decreased decidualization of endometrial stromal cells (ESCs) in vitro, whereas Tet methylcytosine dioxygenase 3 (TET3) and its potential demethylation target, the collagen type I alpha 1 chain (Col1A1), were restored. The binding capacity of TET3 to the Col1A1 promoter could be enhanced by the inhibition of miR-29a. Finally, deletion of TET3 rescued the inhibitory effect of the miR-29a antagomir on the proliferation of decidualized ESCs in vitro and embryo implantation in vivo. Thus, loss of miR-29a causes implantation failure because of the limitation of ESCs decidualization-related changes in non-receptive endometrium during early pregnancy.
Subject areas: Molecular biology, Cell biology, Developmental biology
Graphical abstract

Highlights
-
•
Loss of miR-29a inhibits decidualization of ESCs
-
•
TET3 demethylates the Col1A1 promoter
-
•
Loss of miR-29a enhances the binding capacity of TET3 to the Col1A1 promoter
-
•
Loss of miR-29a suppresses embryo implantation during early pregnancy in mice
Molecular biology; Cell biology; Developmental biology
Introduction
Maternal-foetal communication is the basis for maintaining a successful and healthy pregnancy (Vento-Tormo et al., 2018). Endometrial stromal cells (ESCs) proliferate and differentiate significantly to achieve the states of decidualization during early pregnancy (Okada et al., 2018; Percharde et al., 2018). Dysregulated decidualization of ESCs impairs the invasion of the endometrium by trophoblast cells, which may lead to the development of a variety of pregnancy-associated issues, including infertility, early pregnancy loss (EPL), intrauterine growth retardation (IUGR), and preeclampsia (Ning et al., 2016; Conrad et al., 2017; Kliman and Frankfurter, 2019). Previous studies have shown that homeobox A10 (HOXA10) or interleukin 6 (IL-6) regulate cell proliferation and decidualization of ESCs (Li et al., 2016; Piltonen et al., 2015). Steroid hormones and adhesion molecules are also reportedly involved in the regulation of ESC proliferation and decidualisation, which further affect embryo implantation and pregnancy (Salsano et al., 2019; Nallasamy et al., 2019). However, the underlying molecular mechanism of the decidualization defect remains unclear.
MicroRNAs (miRNAs) are a class of small, noncoding RNAs that negatively regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of target mRNA (Saadatpour et al., 2016). Recent studies have found that miRNAs are differentially expressed in the endometrium during the process of decidualization and the window period of embryo implantation (Liang et al., 2017; Ferlita et al., 2018). It has been demonstrated that downregulation of miR-98 promotes proliferation of ESCs and inhibits apoptosis of ESCs by targeting Bcl-xl in the rat uterus during the receptive phase (Xia et al., 2014b). A recent study has found that miR-24 is downregulated in the decidual tissue from patients with recurrent miscarriage, thus affecting the pregnancy process by regulating the proliferation and apoptosis of ESCs (Li and Li, 2016). However, there are numerous unknown miRNAs that control the decidualization process of ESCs. MiR-29a reportedly induces trophoblast cell apoptosis and regulates the function of trophoblast cells in recurrent miscarriage and placenta accreta (Gu et al., 2016; Yang et al., 2018). It has been revealed that miR-29a promotes granulosa cell proliferation by reducing aromatase expression and oestradiol biosynthesis in polycystic ovary syndrome (Li et al., 2019). Furthermore, it has been demonstrated that miR-29a-5p inhibits endometrial cancer-derived cell proliferation by targeting TPX2 (Jiang et al., 2018). Notably, another previous study has revealed that miR-29a inhibits apoptosis of ESCs by targeting apoptotic factors during embryo implantation (Xia et al., 2014a). However, the effects of miR-29a on the proliferation and decidualization of ESCs remain to be elucidated.
Here, we found aberrant expression of miR-29a in endometrial tissues from the secretory phase, proliferative phase, normal early pregnancy, and EPL. The inhibition of miR-29a by miR-29a antagomir treatment led to decreased decidualization and cell viability of ESCs in vitro. Previous studies have proved that there are many aberrant DNA methylation events during decidualization (Hong et al., 2020; Gao et al., 2012). TET family members, involved in DNA demethylation, reportedly play an important role in embryo implantation and development, and were reported to be potentially targeted by miR-29 (Kremer et al., 2018). Interestingly, TET3 is regulated by multiple miRNAs in the progress of epigenetic regulation (Dong et al., 2020). Therefore, we hypothesized the correlation between miR-29 and TET family molecules during decidualization. Here, we found TET3 negatively associated with miR-29a in decidual tissues from patients with EPL and decidualized ESCs in vitro. Using bioinformatics analysis, TET3 was predicted to be a potential direct downstream target of miR-29a, and this was further confirmed by luciferase reporter assay.
Col1A1, a component of type I collagen, is involved in the regulation of decidualization in ESCs. Collagen type I alpha 1 chain (Col1A1), an effective component of collagen fibers and bone marrow, participates in cell proliferation, infiltration, metastasis, and angiogenesis (Kim et al., 2019a, 2019b). The aberrant transcriptional activation of Col1A1 causes a defect of decidualization in mice and in humans (Kim et al., 2019a, 2019b). Col1A1 expression can be affected by DNA methylation events and promoter demethylation of COL1A1 is related to its expression and functional properties of human embryonic stem cells (Hewitt et al., 2011; Kim et al., 2019a, 2019b). In addition, previous study showed that TET3 promotes multiple TGF-β pathway gene expression by demethylation function (Xu et al., 2020). Col1A1 was a key molecule of the TGF-β pathway and we found loss of miR-29a up-regulated the expression of Col1A1. However, whether the signaling axis miR-29 and Col1A1 participates in decidualization of ESCs via affecting DNA methylation events remains unclear. In this study, miR-29a could also inhibit the recruitment of TET3 to the Col1A1 promoter and could decrease the demethylation function of TET3. Deletion of TET3 was prevented by the inhibitory effect of the miR-29a antagomir on the proliferation of decidualized ESCs in vitro and embryo implantation in vivo. These results demonstrated that loss of miR-29a suppressed decidualization of ESCs through the regulation of the TET3-Col1A1 axis during embryo implantation.
Results
Loss of miR-29a suppresses proliferation and decidualization of human ESCs
MiR-29a was reported to be activated in the process of embryo implantation and suppressed the apoptosis of ESCs by targeting the pro-apoptotic factors Bak1 and Bmf (Kim et al., 2019a). To investigate the potential role of miR-29a during endometrial tissue development, endometrial tissues from the proliferative phase or the secretory phase, as well as decidual tissues from patients with EPL or normal early pregnancy, were collected to determine the expression status of miR-29a by qRT-PCR. MiR-29a was preferentially expressed in the endometrial tissues from the secretory phase relative to those from the proliferative phase, whereas miR-29a was downregulated in the decidual tissues from EPL patients compared to those from normal early pregnancy donors (Figure 1A). To study the significance of the abnormal expression levels of miR-29a, ESCs were isolated from human endometrial tissues and confirmed by IF staining of vimentin (Figure 1B) and morphology analysis (Figure 1C). To understand the mechanism by which loss of miR-29a contributed to decidualization and decreased viability of ECSs, oestradiol, progesterone, and cAMP were introduced to modulate the decidualization of human ESCs. The normal ESCs that did not undergo decidualization were used as control. As a result, miR-29a levels were significantly elevated in the decidualized ESCs compared with the control and inhibition of miR-29a significantly decreased the miR-29a levels in the decidualized ESCs compared with control antagomir, whereas miR-29a levels in miR-29a antagomir-treated ECSs were higher than those in the control group (Figure 1D). Cell viability was also increased in the decidualized ESCs compared to that observed in the initial state (Figure 1E), whereas inhibition of miR-29a significantly attenuated the enhanced viability of decidualized ESCs (Figure 1E). Interestingly, we found that the cell viability in miR-29a antagomir-treated ECSs was lower than that in the control group, which was not consistent with the miR-29a levels, suggesting that inhibition of miR-29a significantly impaired the viability of the decidualized ESCs, whereas upregulation of miR-29a led to a slight increase in cell viability of the decidualized ESCs. Additionally, levels of the decidualized biomarkers PRL and IGFBP1 were both elevated after decidualization but decreased when miR-29a expression was inhibited (Figures 1F and 1G).
Figure 1.
Inhibition of miR-29a attenuated proliferation and decidualization of the human ESCs
(A) The expression status of miR-29a in different stages of endometrial tissues and decidual tissues. qRT-PCR analysis of miR-29a on endometrial tissues and decidual tissues. All results were relative to endometrial tissues of proliferative phase, and U6 served as an internal control.
(B) Immunofluorescence for Vimentin in human ESCs showing cellular localization. Scale bar: 50 μm.
(C) Phase contrast images of human ESCs. Scale bar: 100 μm.
(D) qRT-PCR analysis of miR-29a levels in decidualized ESCs transfected with Control/miR-29a antagomir relative to control. U6 served as an internal control.
(E–G) (E) CCK-8 assays were performed to evaluate decidualized ESCs viability transfected with Control/miR-29a antagomirfor 48h. Western blot analysis (F) and quantification (G) of PRL and IGFBP1 in decidualized ESCs transfected with Control/miR-29a antagomir (n = 3). Data are represented as mean ± SEM. An unpaired two-tailed Student's t-test was used for conducting direct comparison between two groups. Student's t test: n.s. = not significant, ∗p < 0.05, ∗∗p < 0.01.
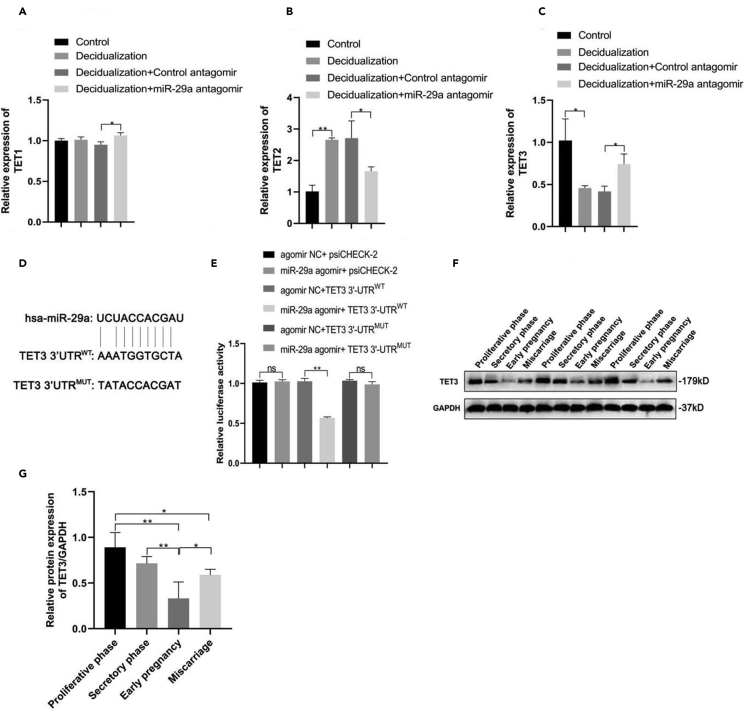
TET3 is a target of miR-29a
TET3, involved in DNA demethylation, reportedly plays an important role in embryo implantation and development (Han et al., 2019). To define the association between miR-29a and the TET family, we examined the levels of TET1, TET2, and TET3 mRNA following decidualization of ESCs and miR-29a inhibition. Interestingly, ESC decidualization had no effect on TET1 levels but significantly increased TET2 levels, whereas it downregulated TET3 expression levels (Figures 2A–2C, respectively). However, loss of miR-29a in decidualized ESCs led to a significant decrease in TET2 levels, whereas TET1 and TET3 levels were both elevated (Figures 2A–2C, respectively). Using the miRNA target prediction program analysis, miR-29a was predicted to target the 3′UTR of TET3 (Figure 2D). Moreover, luciferase activity of the TET3 3′UTRWT luciferase construct was increased significantly in the absence of miR-29a, whereas it remained unchanged in the control or TET3 3′UTRMUT constructs, indicating that miR-29a might target the 3′-UTR of TET3 in ESCs (Figure 2E). Furthermore, we found that TET3 protein levels were significantly downregulated in the decidual tissues from early pregnancy compared with the endometrial tissues from patients in either proliferative phase or secretory phase, whereas TET3 expression was remarkably upregulated in the decidual tissues from patients with EPL compared with the decidual tissues from early pregnancy (Figures 2F and 2G), suggesting that the expression of TET3 was negatively associated with miR-29a expression in different stages of endometrial and decidual tissues.
Figure 2.
TET3 is a target gene of miR-29a
(A–C) qRT-PCR analysis of TET1 (A), TET2 (B) and TET3 (C) mRNA levels in decidualized ESCs transfected with Control/miR-29a antagomir.
(D) Prediction of wild-type (WT) and mutant-type (MUT) sequences of TET3 3′UTR with binding site to miR-29a.
(E–G) (E) Luciferase activity in ESCs co-transfected with the NC/miR-29a agomir and WT/MUT TET3 luciferase reporter constructs.Western blot analysis (F) and quantification (G) of TET3 in different stages of endometrial tissues and the decidual tissues (n = 3). Data are represented as mean ± SEM. An unpaired two-tailed Student's t-test was used for conducting direct comparison between two groups. Student's t test: n.s. = not significant, ∗p < 0.05, ∗∗p < 0.01.
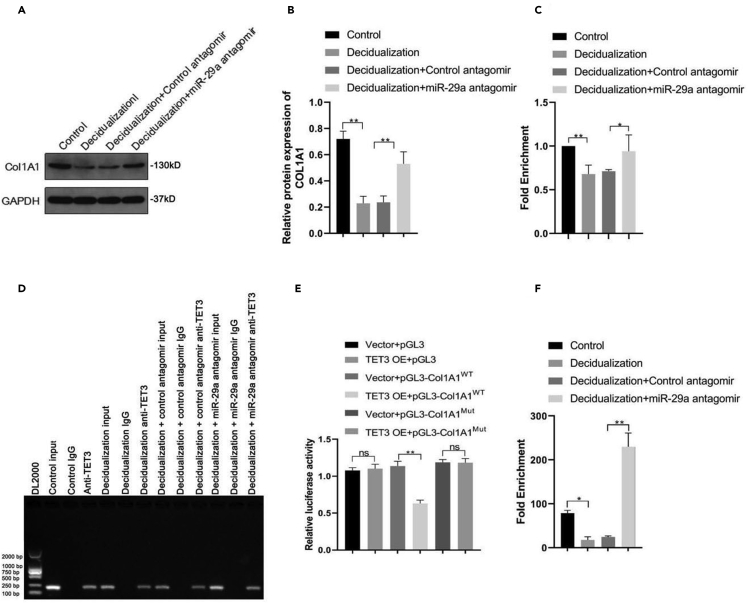
Loss of miR-29a induces interaction of TET3 and Col1A1 promoter in human ESCs
Col1A1, a component of type I collagen, is involved in the regulation of decidualization in ESCs (Kim et al., 2019b). In contrast, promoter demethylation of Col1A1 is related to its expression and functional properties of human embryonic stem cells (Hewitt et al., 2011). Based on the role of TET3 in DNA demethylation during embryo implantation and development, we hypothesized that Col1A1 was a downstream factor of TET3. First, we found that inhibition of miR-29a significantly upregulated Col1A1 expression in the decidualized ESCs (Figures 3A and 3B), which might be the consequence of demethylation of TET3. To confirm this hypothesis, ChIP analysis was performed to determine the binding capacity of TET3 with the promoter region of Col1A1 in decidualized ESCs. The fold enrichment of TET3 occupancy on the Col1A1 promoter region was attenuated in the decidualized ESCs, whereas it was restored following transfection with miR-29a antagomir, suggesting that the loss of miR-29a might induce the recruitment of TET3 to the Col1A1 promoter in the human ESCs (Figures 3C and 3D). To further prove this hypothesis, Col1A1 promoter-luciferase reporter plasmids were constructed. Compared with the control reporter, TET3 overexpression did not affect the relative luciferase activity of the pGL3 empty vector or MUT Col1A1 promoter-luciferase reporter, while significantly inhibiting the WT Col1A1 promoter-luciferase activity, indicating that TET3 could specifically bind to the Col1A1 promoter region in ESCs (Figure 3E). Subsequently, hMeDIP-qPCR was performed to determine whether inhibition of miR-29a could affect the enrichment of 5-hmC at the Col1A1 promoter. The results showed that the 5-hmC level of the Col1A1 promoter decreased significantly in the decidualized ESCs; however, it was restored by addition of miR-29a antagomir (Figure 3F), suggesting that the inhibition of miR-29 promoted the demethylation function of TET3 to suppress the promoter methylation of Col1A1, and leading to enhanced Col1A1 expression. Taken together, it can be inferred miR-29a may play a role in targeting TET3 and inhibiting TET3 from binding to the Col1A1 promoter region, thus methylating the promoter and suppressing the transcription of Col1A1.
Figure 3.
The binding capacity of TET3 to Col1A1 promoter was enhanced in the absence of miR-29a in human ESCs
(A and B) Western blot analysis (A) and quantification (B) of Col1A1 in decidualized ESCs transfected with Control/miR-29a antagomir (n = 3).
(C) ChIP analysis of TET3 occupancy in the promoter regions of Col1A1 in indicated ESCs relative to Control. The fold enrichment represents the binding amount normalized to respective inputs.
(D) Control ESCs, decidualised ESCs, control/miR-29a antagomir-challenged decidualised ESCs were collected to perform CHIP assay using antibodies against IgG and TET3. Agarose gel electrophoresis showed PCR results from each ChIP assay. The band indicated the Col1A1 promoter was binded to TET3.
(E) Luciferase reporter assays were performed to detect the luciferase activity in ESCs after co-transfected with TET3 vectors and WT or Mut of pGL3-Col1A1 luciferase reporters.
(F) hMeDIP-qPCR analysis of 5-hmC enrichment in the Col1A1 promoter in indicated ESCs. Data are represented as mean ± SEM. An unpaired two-tailed Student's t-test was used for conducting direct comparison between two groups. Student's t test: n.s. = not significant, ∗p < 0.05, ∗∗p < 0.01.
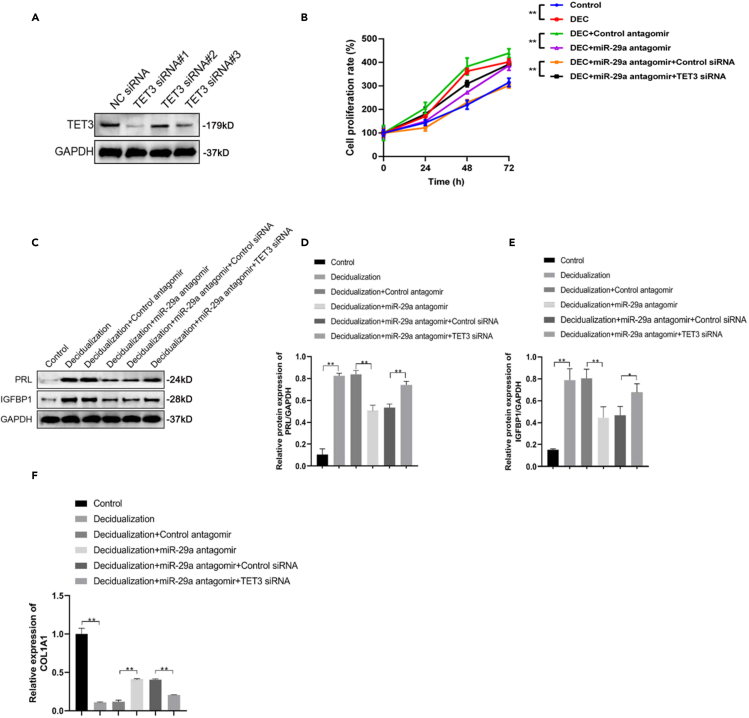
Loss of miR-29a suppresses proliferation and decidualization by activation of TET3/Col1A1 signaling
To determine whether miR-29a regulated ESC proliferation and decidualization through TET3, we introduced three small, interfering RNAs targeting TET3, and the knockdown efficiency was confirmed by western blotting (Figure 4A). The most effective RNA was selected for the subsequent experiments. Knockdown of TET3 reversed the inhibitory effect of miR-29a antagomir on the proliferation of decidualized ESCs (Figure 4B). In addition, PRL and IGFBP1 protein levels were also partially restored in the absence of TET3 (Figures 4C–4E). The elevated mRNA level of Col1A1 in the presence of the miR-29a antagomir in decidualized ESCs was abrogated by TET3 knockdown, indicating that TET3 was a key effector in miR-29a-associated proliferation and decidualization of human ESCs (Figure 4F).
Figure 4.
Loss of miR-29a suppressed proliferation and decidualization via TET3
(A) Western blot analysis of TET3 in ESCs transfected with siRNAs.
(B–E) (B) CCK-8 assays were performed to evaluate human ESCs viability post decidualization and transfected with miR-29a antagomir and TET3 siRNA. Western blot analysis (C) and quantification of PRL (D) and IGFBP1 (E) in indicated ESCs (n = 3).
(F) qRT-PCR analysis of COL1A1 mRNA levels in indicated ESCs.
Data are represented as mean ± SEM. An unpaired two-tailed Student's t-test was used for conducting direct comparison between two groups. Student's t test: ∗p < 0.05, ∗∗p < 0.01.
Loss of miR-29a impairs embryo implantation in vivo
Intravenous injections with either miR-29a inhibitor (antagomir) or adenovirus containing TET3 shRNA were administered to 6- to 8-week-old C57BL/6 female mice, resulting in different embryo implantation on gestation day 8. Implantation sites were visualised by using the Chicago Sky Blue 6B reaction and were represented as blue spots on gestation day 8. As shown in Figure 5A, embryos were observed in the uterine horns of the pregnant mice. Compared with the pregnant mice treated with inhibitor NC adenovirus vectors, the number of embryos implanted in the uterus and the weight of the uterus were significantly reduced in the pregnant mice treated with the miR-29a inhibitor-containing adenovirus vectors (Figures 5B and 5C). Moreover, the number of uterine embryo implantation events and the weight of the uterus remained unchanged in the pregnant mice treated with control adenovirus vectors (scramble) compared with the pregnant mice treated with miR-29a inhibitor-containing adenovirus vectors (Figures 5B and 5C). In addition, the number of uterine embryo implantation events and uterine weight recovered in the absence of TET3 (Figures 5B and 5C), thereby demonstrating that TET3 acted as a downstream factor of miR-29a in the embryo implantation process. The expression levels of miR-29a in the endometrial tissue were significantly increased in the normal pregnancy mice compared with the non-pregnant control group, whereas they were downregulated in the endometrial tissue of the pregnant mice injected with miR-29a inhibitor-containing adenovirus vectors (Figure 5D). Furthermore, downregulation of TET3 did not significantly change the expression levels of miR-29a in the endometrium of pregnant mice compared with miR-29a inhibitor-containing adenovirus vector-treated pregnant mice (Figure 5D). These results showed that miR-29a inhibitor-containing adenovirus vectors could significantly inhibit the increase in miR-29a content, whereas downregulation of TET3 demonstrated a less remarkable effect on the content of miR-29a in the endometrial tissue of pregnant mice.
Figure 5.
Loss of miR-29a impaired embryo implantation in vivo
On gestation D1 (Day 1), 100 μL adenovirus containing control inhibitor, miR-29a inhibitor, miR-29a inhibitor and scramble, or miR-29a inhibitor and TET3 shRNA were intravenously (i.v.) injected into female C57BL/6 mice for two consecutive days. Animals were sacrificed on day 8, the uterus was photographed, and the number of embryonic implants and the weight of the uterus were determined.
(A and B) On gestation day 8, uterus samples were acquired and photographed. Subsequently, the implantation sites and fetal number were determined in the uterine following treatment with miR-29a inhibitor and TET3 shRNA adenovirus vectors on gestation day 8 (n = 5 per group).
(C) The uterine weight (g) was measured following treatment with miR-29a inhibitor and TET3 shRNA adenovirus vectors on gestation day 8 (n = 5 per group).
(D) The expression levels of miR-29a in the uterine following various treatments as analyzed by qRT-PCR. Data are represented as mean ± SEM. An unpaired two-tailed Student's t-test was used for conducting direct comparison between two groups. Student's t test: n.s. = not significant, ∗∗p < 0.01.
Loss of miR-29a suppresses pinopodes of endometrium during embryo implantation
To determine the effect of miR-29a inhibitor on the histological morphology of the endometrium during embryo implantation, the decidua tissues of the uterus were collected from mice subjected to different treatments. The nucleus was round or oval with larger nucleoli and darker staining of cytoplasm was observed in samples obtained from the pregnant mice injected with inhibitor NC-containing adenovirus vectors compared to that observed in samples in the non-pregnant control group (Figures 6A and 6B). Moreover, the nucleus was smaller and cytoplasmic staining was less intense in miR-29a inhibitor-containing adenovirus vector-treated pregnant mice than that observed in the inhibitor NC-containing adenovirus vector-treated mice (Figures 6A and 6B). Furthermore, the morphology of decidual tissues showed vacuolization, and the cell arrangement was disordered in the pregnant mice injected with miR-29a inhibitor-containing adenovirus vectors compared with the inhibitor NC-containing adenovirus vector-treated mice (Figures 6A and 6B). However, we found that downregulation of TET3 improved the miR-29a inhibitor-associated detrimental effects on histological morphology of decidual tissue in the pregnant mice (Figures 6A and 6B). The number of pinocytes in the endometrium obtained from different groups was observed by scanning electron microscopy. As shown in Figure 6C, the pinopodes were in the development stage, and few had short microvilli in their endometrial surface of the non-pregnant control group. The number of pinocytes in the normal pregnancy group was significantly higher compared to that in the non-pregnant control group, whereas the miR-29a inhibitor treatment significantly decreased the number of pinocytes in the endometrium compared to the normal pregnancy group (Figure 6C). Interestingly, the downregulation of TET3 expression partially rescued the miR-29a inhibitor-associated effects and decreased the number of pinocytes on the endometrial surface (Figure 6C).
Figure 6.
Loss of miR-29a impaired histological morphology of endometrium during embryo implantation
On gestation day 1, adenovirus containing inhibitor, miR-29a inhibitor, miR-29a inhibitor and scramble, or miR-29a inhibitor and TET3 shRNA were intravenously (i.v.) injected into female C57BL/6 mice for two consecutive days. Uterus samples in each group were acquired on gestation day 8. After embedding with paraffin, 5-μm sections were used to perform HE staining assay.
(A and B) (A) H&E (10X, scale bar: 100 μm) and (B) H&E (40X, scale bar: 20 μm) staining of decidual tissue collected from different treatment groups.
(C) Scanning electron microscopy photomicrograph from the endometrial surface. The endometrium tissues were acquired from C57BL/6 mice in different treatment groups, scale bar: 2μm. Representative SEM photomicrograph showed the pinopodes of endometrial surface in different groups (n = 5 per group). Red arrow: pinocytes.
Discussion
MiRNAs can participate in the post-transcriptional regulation of multiple physiological and pathological processes, including gene expression, modification, transcription, and translation (Bhaskaran and Mohan, 2014). MiR-29a is a key regulator of cell differentiation in diverse cell types, including bone marrow cells, T cells, hematopoietic stem cells, and myoblasts (Hu et al., 2015; Galimov et al., 2016; Xuan et al., 2017). A previous study has shown that highly expressed miR-29a is necessary for establishing early pregnancy in the rat uterus during embryo implantation (Xia et al., 2014a). Our study demonstrated that the expression of miR-29a in decidual tissues from early pregnant women was higher than that in non-pregnant women, whereas miR-29a expression was remarkably downregulated in the decidual tissues from patients with EPL compared to those from normal controls, suggesting that miR-29a might be involved in the decidualization of the endometrium during early pregnancy. The important prerequisite for embryo implantation is decidualization of the endometrium when the uterus enters a receptive state (Lessey and Kim, 2017). The number of embryos implanted in the uterus was reduced in the miR-29a inhibitor treated pregnant mice, which indicated that miR-29a exerted an important role in embryo implantation in mice in vivo. Moreover, we showed inhibition of miR-29a impeded the pinopodes of endometrium in pregnant mice, which suggested that loss of miR-29a might be associated with impaired endometrial decidualization as well as miR-29a was critical to the receptive endometrium in pregnant mice.
Decidualization of ESCs is a complicated biological mechanism involving crosstalk between multiple genes involved in a myriad cellular processes, including ESC proliferation and differentiation, which affects embryo implantation, maintenance of pregnancy, placental development, and fetal growth (Okada et al., 2014, 2018). Although miR-29a is highly expressed in the rat uterus during early pregnancy and targets the pro-apoptotic factor genes Bak1 and Bmf in ESCs (Xia et al., 2014a), whether miR-29a can regulate proliferation and decidualization of ESCs and the underlying molecular mechanism remain unknown. In this study, we found that the expression of miR-29a was significantly elevated in decidualized ESCs and that inhibition of miR-29a significantly attenuated the proliferation of ESCs and the decidualized biomarkers PRL and IGFBP1, which suggested that miR-29a expression was required for the proliferation and decidualization of ESCs.
DNA methylation and demethylation are critical for the regulation of pregnancy as a pathway of epigenetic modification (Guo et al., 2014). A study has revealed that DNA methylation of Chromobox homolog 4 initiates stromal cell decidualization during early pregnancy (Gao et al., 2012). DNA demethylation is a chemical process which results in the removal of a methyl group (CH3) from a methylated variant of cytosine, 5-hydroxymethylcytosine (5-hmC), to generate 5-methylcytosine (5-mC). It has been demonstrated that demethylation modifications of H3K27 regulate the states of decidualization in uterine stromal cells during early and late pregnancy stages (Nancy et al., 2018). Previous studies have shown that TET-mediated demethylation is essential for normal embryo implantation and development (Khoueiry et al., 2017; Blaschke et al., 2013). In addition, another study has demonstrated that TET levels are implicated in embryo development during pregnancy (Vasconcelos et al., 2019). In this study, we found that the expression of TET3, but not TET1 and TET2, was negatively regulated by miR-29a in decidualized ESCs. The luciferase activity of the 3′-UTR of wild-type TET3 was significantly inhibited by the treatment with miR-29a agomir, suggesting that TET3 was a potential target of miR-29a in ESCs. Moreover, we showed that TET3 was significantly upregulated in the decidual tissues of patients with EPL, in contrast to the expression status of miR-29a. A previous study has shown that miR-29a may regulate the methylation level of mouse embryos by targeting Dnmt3a and Dnmt3b (Movahed et al., 2019). Our results revealed that downregulation of miR-29a increased the 5-hmC level of Col1A1 in the decidualized ESCs, indicating that miR-29a might inhibit the demethylation capacity of TET3 in decidualized ESCs. A recent study has shown that TET3-mediated demethylation promotes embryo development by regulating the pluripotency gene levels (Cheng et al., 2019). Interestingly, we found that downregulation of miR-29a decreased PRL and IGFBP1 protein expression levels, whereas it reversed the loss of TET3, suggesting that miR-29a-mediated negative regulation of TET3 demethylation might be associated with the process of ESC decidualization, which contributed to the pathophysiology of impaired decidualization in women with EPL.
Col1A1, a component of type I collagen, is reportedly targeted by histone deacetylase 3 (HDAC3), which regulates decidualization of ESCs (Kim et al., 2019b). Here, we demonstrated that Col1A1 expression was significantly upregulated following downregulation of miR-29a expression in the decidualized ESCs, suggesting that downregulation of miR-29a enhanced the demethylation function of TET3 to suppress the promoter methylation of Col1A1, leading to an enhanced Col1A1 expression. Loss of HDAC3 causes a defect in decidualization through interaction with Col1A1 (Kim et al., 2019b), indicating that Col1A1 affects decidualization and may be involved in the interaction with HDAC3. However, miR-29a could not directly target the 3′-UTR of Col1A1 (Figures S1A and S1B). Interestingly, we found that loss of miR-29a enhanced the interaction of TET3 and Col1A1 and reversed the inhibitory effect of the miR-29a antagomir on the proliferative activity and the expression of decidualized biomarkers in the decidualized ESCs. In addition, inhibition of the TET3 downregulated miR-29a antagomir enhanced the expression of Col1A1 in the decidualized ESCs, which was involved in the loss of TET3 demethylation function. Thus, the binding of TET3 to Col1A1 functioned as a critical switch during miR-29a-mediated decidualization. Promoter demethylation of COL1A1 is observed during the differentiation of human embryonic stem cells (Hewitt et al., 2011). As described previously, TET3 demethylation might be associated with the process of ESC decidualization. Thus, a miR-29a downregulation-mediated increased expression of Col1A1 might depend on the function of TET3 demethylation. In fact, the hMeDIP-qPCR assay confirmed that TET3 demethylated the Col1A1 promoter in decidualized ESCs, which showed a more pronounced effect in the absence of miR-29a. These results suggested that the loss of miR-29a suppressed proliferation and decidualization of ESCs by activation of TET3/Col1A1 signaling. Embryo implantation in vivo revealed that inhibition of miR-29a significantly reduced the number of uterine embryo implantation and uterine weight compared with the NC group, whereas knockdown of TET3 restored the number of uterine embryo implantation and uterine weight, suggesting that TET3 was a downstream factor of miR-29a and downregulation of TET3 could reverse miR-29a inhibitor impaired embryo implantation in mice. Interestingly, loss of miR-29a impaired the histological morphology and decreased the number of pinocytes, whereas downregulation of TET3 could restore the effect of miR-29a inhibitor on the histological morphology and pinopodes in the endometrium during embryo implantation in mice, which suggested that downregulation of TET3 could also reverse miR-29a inhibitor impaired endometrial decidualization and receptive state in pregnant mice. These results indicated that the miR-29a/TET3 signaling was a key regulator in endometrial decidualization and embryo implantation in mice.
In conclusion, our study found that loss of miR-29a could suppress proliferation and decidualization of ESCs by enhancing the function of TET3 demethylation and the interaction of TET3 and Col1A1 in vitro, and that it impaired embryo implantation in vivo, which elucidated the important role of miR-29a/TET3/Col1A1 in regulating the process of decidualization and maintenance of decidual function during early pregnancy. These results suggest that the loss of miR-29a causes implantation failure because of the limitation of ESC decidualization-related changes in non-receptive endometrium during early pregnancy. Thus, our results provide a conceptual framework for understanding abnormal endometrial decidualization, with considerable significance for the diagnosis and treatment of abnormal decidualization-related changes in non-receptive endometrium in implantation failure during early pregnancy.
Limitations of the study
Despite miR-29a/TET3 signaling was a key regulator in endometrial decidualization and embryo implantation in mice, the changes and role of Col1A1 in uterine embryo implantation in miR-29a- or TET3-shRNA challenged mice remains to be explored by more research. Second, the role of miR-29a/TET3/Col1A1 in regulating the process of decidualization and maintenance of decidual function during early pregnancy will be further validated in the clinic.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Vimentin (1:100) | Proteintech | Cat#10366-1-AP; RRID: AB_2273020 |
| Rabbit monoclonal anti-IGFBP1 (1:1000) | Cell Signaling Technology | Cat#31025; RRID: AB_2798998 |
| Rabbit Polyclonal anti-PRL (1:1000) | Affinity Biosciences | Cat#DF6506; RRID: AB_2838468 |
| Rabbit Polyclonal anti-TET3 (1:1000) | Merck Millipore | Cat#ABS463 |
| Rabbit Polyclonal anti-Col1A1 (1:1000) | Abcam | Cat#ab34710; RRID: AB_731684 |
| Rabbit Polyclonal anti-GAPDH (1:3000) | Abcam | Cat#ab9485; RRID: AB_307275 |
| HRP-conjugated secondary antibody (1:5000) | Abcam | Cat#ab6721; RRID: AB_955447 |
| Cy3-tagged secondary antibody (1:500) | Boster | Cat#BA1032; RRID: AB_2716305 |
| Rabbit monoclonal anti-5hmC | Abcam | Cat#ab214728; RRID: AB_2797407 |
| Rabbit Polyclonal anti-TET3 (ChIP) | Sigma-Aldrich | Cat#ABE290 |
| Bacterial and virus strains | ||
| BJ5183 | Unibio | Cat#ST1039 |
| XL1-Blue | Unibio | Cat#ST1054 |
| Biological samples | ||
| Human endometrial tissues of proliferative-phase women and secretory-phase women | Department of Reproductive Endocrinology, Women's Hospital, School of Medicine, Zhejiang University | N/A |
| Human decidual tissues of EPL patients and normal early pregnancy women | Department of Reproductive Endocrinology, Women's Hospital, School of Medicine, Zhejiang University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol | Invitrogen | Cat#15596-026 |
| DMEM/F12 | Procell | Cat#PM150312 |
| Fetal bovine serum (FBS) | Procell | Cat#164210-500 |
| Trypsin | Procell | Cat#PB180225 |
| Penicillin-Streptomycin Solution | Procell | Cat#PB180120 |
| PBS | Procell | Cat#PB180327 |
| Collagenase I | Sigma | Cat#C0130-100MG |
| TritonX-100 | Beyotime | Cat#ST795 |
| PFA | Beyotime | Cat#P0099 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat#L3000-015 |
| RIPA lysis buffer | Beyotime | Cat#P0013K |
| PMSF | Beyotime | Cat#ST505 |
| DAPI | Beyotime | Cat#C1002 |
| Oestradiol | Targetmol | Cat#T1048 |
| Progesterone | Targetmol | Cat#T5040 |
| cAMP | Targetmol | Cat#TC02745 |
| DL2000 DNA Marker | TIANGEN | Cat#MD114-02 |
| Critical commercial assays | ||
| SYBR Green PCR kit | VAZYME | Cat#Q111-02 |
| Plasmid Midi kit | Qiagen | Cat#12143 |
| CCK-8 assay kit | Beyotime | Cat#C0037 |
| BCA Protein Assay Kit | Beyotime | Cat#P0010S |
| Dual Luciferase Reporter Kit | Beyotime | Cat#RG027 |
| hMeDIP Kit | Abcam | Cat#ab117134 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Vital River Animal Center, Beijing, China |
Cat#213 |
| Oligonucleotides | ||
| Primers for miR-29a-3p: 5’-TGCGCTAGCACCATCTGAAATC-3’ and 5’-CCAGTGCAGGGTC CGAGGTATT-3’ | This paper | N/A |
| Primers for U6: 5’-CGCTTCGGCAGCACATATAC-3’ and 5’-AAATATGGAA CGCTTCACGA-3’ | This paper | N/A |
| Primers for TET1: 5’-CCTGCTCACGGCTCGGTTTTGATTG-3’ and 5’-G GGAGAGGCGGGTTGGATGATTACG-3’ | This paper | N/A |
| Primers for TET2: 5’-CCATTCCTGATACCATCAC CTCCCA-3’ and 5’-GGCTTACCCCGAAGTTACGTCTTTC-3’ | This paper | N/A |
| Primers for TET3: 5’-CCAAC GAGGAAATAGCGATTGACTG-3’ and 5’-CCCATTGTAGAGGTTATGCTGGTCC -3’ | This paper | N/A |
| Primers for Col1A1: 5’-GTTCCTCCCTGCTCTCCATC-3’ and 5’-GAGGTCCACAAAG CTGAACA-3’ | This paper | N/A |
| Primers for GAPDH: 5’-TCAAGAAGGTGGTGAAGCAGG-3’ and 5’-TCAA AGGTGGAGGAGTGGGT-3’ | This paper | N/A |
| MiR-29a agomir and antagomir | GenePharma | N/A |
| Control agomir and antagomir | GenePharma | N/A |
| Recombinant DNA | ||
| pAd5 vector | Addgene | Cat#122555 |
| pGL3 reporter vector | promega | Cat#E1751 |
| pRL-TK reporter | promega | Cat#E2241 |
| Software and algorithms | ||
| SPSS 17.0 software | SPSS Inc. | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Aixia Liu (liuaixia@zju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Sample collection
Human endometrial tissues of proliferative-phase women (10 cases) and secretory-phase women (15 cases), as well as human decidual tissues of EPL patients (23 cases) and normal early pregnancy women (22 cases) with the age of 24 to 38 years old, were collected from December 2018 to December 2019 at the Department of Reproductive Endocrinology, Women's Hospital, School of Medicine, Zhejiang University, Hangzhou, China. The EPL diagnosis was conducted based on medical history, physical examination, and transvaginal ultrasound scanning (detection of embryo sac without foetal heart activity in the uterine cavity). Inclusion criteria included presence of embryo at 8–10 gestational weeks at the time of diagnosis, with no maternal history of recurrent spontaneous abortion, chromosomal abnormalities, endocrine disease, anatomical abnormalities of the genital tract, immunological disease, infection, or trauma. All samples were washed in normal saline to remove excess blood. Samples were immediately snap-frozen in liquid nitrogen and stored at −80°C. Informed consent was obtained from all participants prior to sample collection. This study was performed in accordance with the ethical standards of the Declaration of Helsinki. This study protocol was approved by the Ethical Committee of the Women’s Hospital, School of Medicine, Zhejiang University, Hangzhou, China (file no. 20180192).
Mice
C57BL/6 mice (female=30, male=15; age: 6- to 8-week-old) were bred at a room temperature of approximately 22°C and a 12L:12D photoperiod. The handling of all animals complied with the relevant regulations for experimental animals. Animal care followed the guidelines recommended by the Animal Care and Use Committee (ACUC) of the School of Medicine, Zhejiang University.
Method details
In vivo mice embryo implantation
Female and male mice were placed in cages at a ratio of 2:1, and the day of observation of the first vaginal plug was recorded as the first day of pregnancy (Day 1). Mice were randomly divided into five animals per group. Mice were injected with control, miR-29a inhibitor (antagomir), and TET3 shRNA adenovirus vectors (1×109 pfu per mouse in a 100 μL volume) through the tail vein for two consecutive days from Day 1. Animals were sacrificed on Day 8, and uterus samples were acquired for subsequent treatment. On Day 8, the uterus was photographed, and the number of embryonic implants and the weight of the uterus were determined.
RNA isolation and RT-PCR
RNA was isolated using TRIzol following the manufacturer’s instructions. In total, 20 μL template, 175 ng cDNA, 400 nM primers, and 10 μL of the RT-PCR Master Mix (2X) were gently mixed together. RT-qPCR was performed according to the manufacturer's instructions using the SYBR Green PCR kit. Primers used in the present study are listed in Supplementary tables. U6 and GAPDH served as internal miRNAs and mRNA controls, respectively. All relative expressions were calculated using the 2-ΔΔCt method and reported as fold change relative to the internal control and control groups.
Isolation, culture, and identification of ESCs
ESCs were isolated from the normal endometrium. Briefly, endometrial tissue was rinsed twice with sterile phosphate-buffered saline (PBS) and cut into 0.5–1-mm3 fragments with ophthalmic scissors, and fragments were then digested in collagenase solutions for 1 h at 37°C. The digestion reactions were terminated using DMEM/F12 containing 10% fetal bovine serum (FBS), and the samples were filtered through wire sieves with various pore sizes to remove cell impurities and epithelial cells. After performing centrifugation at 800 g for 5 min, ESCs were resuspended in DMEM/F12 containing 10% FBS and incubated in a 5% CO2 incubator at 37°C. Immunofluorescence (IF) staining was performed to determine the purity of isolated ESCs using monoclonal anti-Vimentin. To induce decidualisation, ESCs were cultured with DMEM/F12 containing 2% FBS in the presence of 10 nM oestradiol, 1 μM progesterone, and 0.5 mM cAMP for 5 consecutive days, as per methods previously reported (Le et al., 2017). decidualization was confirmed by the induction of IGFBP-1 and PRL protein expression, which are specific markers of decidualization (Grasso et al., 2018).
MiRNA, adenoviral vectors, and transfection
Cells were seeded in 6-well plates at a density of 2 × 105 cells per well. At 24 h after seeding, miR-29a agomir and antagomir or control agomir and antagomir were transfected into the cells using Lipofectamine 3000 according to the manufacturer's instructions. Cells were harvested for subsequent assays 48 h after transfection. The shuttle vectors were linearised and subjected to homologous recombination with the linearised Ad5 backbone in the E. coli BJ5183 strain. The pAd5 plasmids were amplified in the E. coli strain XL1-Blue and prepared using the Plasmid Midi kit according to the manufacturer's instructions. The pAd5 plasmids were then linearised and transfected into HEK-293 cells to produce the respective adenovirus vectors. The cell lysates and culture media were harvested 48 h post-infection.
Cell proliferation assay
Cell proliferation was examined using the CCK-8 assay kit (He et al., 2019). Briefly, cells were transfected with miR-29a-antagomir or control-antagomir using the Lipofectamine 3000 reagent. After 5 days of transfection, 10 μL of the CCK-8 reagent was added and the mixture was incubated at 37°C for 2 h. The absorbance at 450 nm was measured with a Multiskan spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Western blot analysis
Treated ESCs were lysed in RIPA lysis buffer containing 1% 100mM phenylmethanesulfonyl fluoride (PMSF). Protein concentrations were quantified for all samples using the BCA Protein Assay Kit. The samples were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels followed by electrophoretic transfer onto polyvinylidene fluoride (PVDF) membranes (Meck Millipore, Burlington, MA, USA). After blocking with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBST), the membranes were incubated overnight at 4°C with primary antibodies as follows: anti-TET3 (1:1000), PRL (1:1000), IGFBP1 (1:1000), Col1A1(1:1000), or GAPDH (1:3000). The membranes were then washed, incubated with HRP-conjugated secondary antibodies for 1 h at room temperature (RT), and subjected to chemiluminescent detection. The western blot bands of the same size in the same gel were first treated with stripping buffer and re-probed with other antibodies.
Bioinformatics analysis
The putative targets of miR29 were determined using the TargetScan, PITA, miRnada, PicTar, and miRDB databases. Subsequently, the candidate MREs of miR29 were determined according to the AgoExpNum value, which was analysed based on the results of Ago CLIP-Seq.
Dual luciferase assays
The fragments containing the wild-type (WT) and the mutant type (MUT) 3’ UTR of TET-3 were cloned into their respective luciferase reporter target expression vector. Cells were co-transfected with miR-29a agomir and WT or MUT 3ʹUTR-TET-3 luciferase reporter. For TET-3 and Col1A1 promoter detection, the fragments containing the of Col1A1 promoter WT and MUT, respectively, were cloned into the pGL3 reporter vectors. Cells were co-transfected withTET-3 overexpression vectors, WT or MUT pGL3 Col1A1 luciferase reporter vectors, and pRL-TK reporter vectors. The luciferase activity of the cells was detected using a multifunctional microplate reader (Molecular Devices). Renilla activity was used to normalise the luciferase reporter activity.
Chromatin immunoprecipitation (ChIP) analysis
Cells were treated with 1% formaldehyde in PBS and chromatin was crosslinked for 15 min at 37°C. Crosslinking reactions were terminated by the addition of 125 mM glycine for 5 min at room temperature. Cells were washed with PBS, lysed, and the chromatin was solubilised to the desired length by sonication. The immunoprecipitation of crosslinked proteins was observed by addition of anti-TET3 (2 μg). The IgG antibody (1 μg) served as ChIP assay control. Immunoprecipitated DNA was analysed by performing PCR using specific primers for the Col1A1 promoter flanking the TET3-binding sites.
Methylated and hydroxymethylated DNA immunoprecipitation (hMeDIP)
Immunoprecipitation of 5-hmC was performed using the hMeDIP Kit. DNA in the samples was sonicated into short fragments for 4 min with on and off cycles of 15 s and was heat-denatured at 95°C for 5 min. An amount of 0.5 μg of the sonicated DNA was immunoprecipitated with 2 μL of anti-5-hydroxymethylcytosine (anti-5hmC). After incubation at 4°C overnight, mouse anti-IgG magnetic beads were added to the DNA-antibody mixture and incubated at 4°C for 2 h. Immunoprecipitated DNA was purified and subjected to RT-PCR analysis. The percentage of enrichment was calculated relative to the amount of DNA used in the IP reaction.
Immunofluorescence staining
The ESCs were fixed and permeabilised with 4% PFA containing 0.5% Triton X-100 for 20 min. Fixed cells were washed and blocked for 1 h in blocking buffer (1% BSA in PBS) and probed with anti-Vimentin (1:100). The cells were washed with PBS and probed with Cy3-tagged secondary antibodies. The nuclei were counterstained with DAPI. Images were captured under a fluorescence microscope (Olympus, Tokyo, Japan).
Hematoxylin and eosin (HE) staining
The decidua tissue at the embryo implantation site and the endometrial tissue of the control group were prepared as paraffin-embedded sections, and histological morphology was observed by HE staining on Day 8. The tissues were fixed in 10% formalin at room temperature for 24 h. The formalin-fixed tissues that were embedded in paraffin were cut into 4 μm-thick sections. The sections were then stained with HE and observed under a light microscope.
Scanning electron microscopy
The mice were euthanised, and the uteri were dissected and immersed in 2% glutaraldehyde. Thereafter, the endometrium tissues were fixed in 1% osmium tetroxide solution and were dehydrated in ethanol at increasing concentrations. The specimens were dried, mounted, and coated with gold, and they were observed by scanning electron microscopy.
Quantification and statistical analysis
Statistical analysis was conducted using the SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). All experiments were performed in at least three independent biological replicates and reported as mean ± SEM. An unpaired two-tailed Student’s t-test was used for conducting direct comparison between two groups. Differences were considered statistically significant when P values were < 0.05.
Acknowledgements
This work was supported by the National Key R&D Program of China (no. 2018YFC1004400).
Author contributions
AL and DW conceived and designed the experiments; YZ and MT collected the clinical samples; DW, MJ (Mengmeng Jin) and LX performed the experiments and acquired the data; MJ (Mengyu Jing) and YZ analyzed the data; DW drafted the manuscript and AL revised the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure that the study questionnaires were prepared in an inclusive way. We worked to ensure diversity in experimental samples through the selection of the cell lines. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103065.
Contributor Information
Aixia Liu, Email: liuaixia@zju.edu.cn.
Dimin Wang, Email: deane14@163.com.
Supplemental information
Data and code availability
All relevant data and analyses are included in the present manuscript and its supplemental information. This study did not generate original code. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- Bhaskaran M., Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014;51:759–774. doi: 10.1177/0300985813502820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Zhang J., Zhang S., Zhai Y.H., Jiang Y., An X.L., Ma X., Zhang X., Li Z., Tang B. Tet3 is required for normal in vitro fertilization preimplantation embryos development of bovine. Mol. Reprod. Dev. 2019;86:298–307. doi: 10.1002/mrd.23105. [DOI] [PubMed] [Google Scholar]
- Conrad K.P., Rabaglino M.B., Uiterweer E.D.P. Emerging role for dysregulated decidualization in the genesis of preeclampsia. Placenta. 2017;60:119–129. doi: 10.1016/j.placenta.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Xiangbo W., Yanfen Z., Weina W., Zhenshan W. The microRNA/TET3/REST axis is required for olfactory globose basal cell proliferation and male behavior. EMBO Rep. 2020;21:e49431. doi: 10.15252/embr.201949431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlita A., Battaglia R., Andronico F., Caruso S., Cianci A., Purrello M., Pietro C.D. Non-coding RNAs in endometrial physiopathology. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19072120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimov A., Merry T.L., Luca E., Rushing E.J., Mizbani A., Turcekova K., Hartung A., Croce C.M., Ristow M., Krützfeldt J. MicroRNA-29a in adult muscle stem cells controls skeletal muscle regeneration during injury and exercise downstream of fibroblast growth factor-2. Stem Cells. 2016;34:768–780. doi: 10.1002/stem.2281. [DOI] [PubMed] [Google Scholar]
- Gao F., Ma X.H., Rusie A., Hemingway J., Ostmann A.B., Chung D., Das S.K. Epigenetic changes through DNA methylation contribute to uterine stromal cell decidualization. Endocrinology. 2012;153:6078–6090. doi: 10.1210/en.2012-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso E., Gori S., Paparini D., Soczewski E., Fernandez L., Gallino L., Salamone G., Martinez G., Irigoyen M., Ruhlmann C. VIP induces the decidualization program and conditions the immunoregulation of the implantation process. Mol. Cell Endocrinol. 2018;460:63–72. doi: 10.1016/j.mce.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Gu Y.Z., Bian Y.H., Xu X.F., Wang X.T., Zuo C.T., Meng J.L., Li H., Zhao S., Ning Y., Cao Y. Downregulation of miR-29a/b/c in placenta accreta inhibits apoptosis of implantation site intermediate trophoblast cells by targeting MCL1. Placenta. 2016;48:13–19. doi: 10.1016/j.placenta.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Guo H.S., Zhu P., Yan L.Y., Li R., Hu B.Q., Lian Y., Yan J., Ren X., Lin S., Li J. The DNA methylation landscape of human early embryos. Nature. 2014;511:606. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Han C.Q., Cui C.C., Xing X.P., Lu Z.Z., Zhang J.C., Liu J., Zhang Y. Functions of intrinsic disorder in proteins involved in DNA demethylation during pre-implantation embryonic development. Int. J. Biol. Macromolecules. 2019;136:962–979. doi: 10.1016/j.ijbiomac.2019.06.143. [DOI] [PubMed] [Google Scholar]
- He W.H., Jin M.M., Liu A.P., Zhou Y., Hu X.L., Zhu Y.M., Liu A.X. Estradiol promotes trophoblast viability and invasion by activating SGK1. Biomed. Pharmacother. 2019;117 doi: 10.1016/j.biopha.2019.109092. [DOI] [PubMed] [Google Scholar]
- Hewitt K.J., Shamis Y., Hayman R.B., Margvelashvili M., Dong S.M., Carlson M.W., Garlick J.A. Epigenetic and phenotypic profile of fibroblasts derived from induced pluripotent stem cells. Plos One. 2011;6 doi: 10.1371/journal.pone.0017128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Xiaobo H., Gil M., Liao. A. Epigenetic modifications working in the decidualization and endometrial receptivity. Cell Mol. Life Sci. 2020;77:2091–2101. doi: 10.1007/s00018-019-03395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.H., Dooley J., Chung S.S., Chandramohan D., Cimmino L., Mukherjee S., Mason C.E., de Strooper B., Liston A., Park C.Y. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 2015;125:2206–2216. doi: 10.1182/blood-2014-06-585273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T.C., Sui D.M., You D., Yao S.M., Zhang L.R., Wang Y.J., Zhao J., Zhang Y. MiR-29a-5p inhibits proliferation and invasion and induces apoptosis in endometrial carcinoma via targeting TPX2. Cell Cycle. 2018;17:1268–1278. doi: 10.1080/15384101.2018.1475829. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Khoueiry R., Sohni A., Thienpont B., Luo X.L., Velde J.V., Bartoccetti M., Boeckx B., Zwijsen A., Rao A., Lambrechts D. Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat. Genet. 2017;49:1061. doi: 10.1038/ng.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.Y., Ko J.M., Park M.H., Koo S.K. Generation of a patient-specific induced pluripotent stem cell line, KSCBi006-A, for osteogenesis imperfecta type I with the COL1A1, c.3162delT mutation. Stem Cell Res. 2019;41 doi: 10.1016/j.scr.2019.101622. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Yoo J.Y., Choi K.C., Shin J.H., Leach R.E., Fazleabas A.T., Young S.L., Lessey B.A., Yoon H.G., Jeong J.W. Loss of HDAC3 results in nonreceptive endometrium and female infertility. Sci. Transl Med. 2019;11 doi: 10.1126/scitranslmed.aaf7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman H.J., Frankfurter D. Clinical approach to recurrent implantation failure: evidence-based evaluation of the endometrium. Fertil. Sterility. 2019;111:618–628. doi: 10.1016/j.fertnstert.2019.02.011. [DOI] [PubMed] [Google Scholar]
- Kremer E.A., Gaur N., Lee M.A., Engmann O., Bohacek J., Mansuy I.M. Interplay between TETs and microRNAs in the adult brain for memory formation. Sci. Rep. 2018;8:1678. doi: 10.1038/s41598-018-19806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A., Wang Z.H., Dai X.Y., Xiao T.H., Zhuo R., Zhang B., Xiao Z., Fan X. Icaritin inhibits decidualisation of endometrial stromal cells. Exp. Ther. Med. 2017;14:5949–5955. doi: 10.3892/etm.2017.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessey B.A., Kim J.J. Endometrial receptivity in the eutopic endometrium of women with endometriosis: it is affected, and let me show you why. Fertil. Steril. 2017;108:19–27. doi: 10.1016/j.fertnstert.2017.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.D., Li J. Association of miR-34a-3p/5p, miR-141-3p/5p, and miR-24 in decidual natural killer cells with unexplained recurrent spontaneous abortion. Med. Sci. Monit. 2016;22:922–929. doi: 10.12659/Msm.895459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu Y.D., Zhou X.Y., Chen S.L., Chen X., Zhe J., Zhang J., Zhang Q.Y., Chen Y.X. MiR-29a regulates the proliferation, aromatase expression, and estradiol biosynthesis of human granulosa cells in polycystic ovary syndrome. Mol. Cell Endocrinol. 2019;498 doi: 10.1016/j.mce.2019.110540. [DOI] [PubMed] [Google Scholar]
- Li D.D., Zhao S.Y., Yang Z.Q., Duan C.C., Guo C.H., Zhang H.L., Geng S., Yue Z.P., Guo B. Hmgn5 functions downstream of Hoxa10 to regulate uterine decidualisation in mice. Cell Cycle. 2016;15:2792–2805. doi: 10.1080/15384101.2016.1220459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.J., Wang S.Y., Wang Z.G. Role of microRNAs in embryo implantation. Reprod. Biol. Endocrinol. 2017;15 doi: 10.1186/s12958-017-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahed E., Soleimani M., Hosseini S., AkbariSene A., Salehi M. Aberrant expression of miR-29a/29b and methylation level of mouse embryos after in vitro fertilization and vitrification at two-cell stage. J. Cell. Physiol. 2019;234:18942–18950. doi: 10.1002/jcp.28534. [DOI] [PubMed] [Google Scholar]
- Nallasamy S., Kaya Okur H.S., Bhurke A., Davila J., Li Q., Young S.L., Taylor R.N., Bagchi M.K., Bagchi I.C. MsxHomeobox genes act downstream of BMP2 to regulate endometrial decidualisation in mice and in humans. Endocrinology. 2019;160:1631–1644. doi: 10.1210/en.2019-00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nancy P., Siewiera J., Rizzuto G., Tagliani E., Osokine I., Manandhar P., Dolgalev I., Clementi C., Tsirigos A., Erlebacher A. H3K27me3 dynamics dictate evolving uterine states in pregnancy and parturition. J. Clin. Invest. 2018;128:233–247. doi: 10.1172/JCI95937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning F., Liu H.S., Lash G.E. The role of decidual macrophages during normal and pathological pregnancy. Am. J. Reprod. Immunol. 2016;75:298–309. doi: 10.1111/aji.12477. [DOI] [PubMed] [Google Scholar]
- Okada H., Tsuzuki T., Murata H. Decidualization of the human endometrium. Reprod. Med. Biol. 2018;17:220–227. doi: 10.1002/rmb2.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Tsuzuki T., Shindoh H., Nishigaki A., Yasuda K., Kanzaki H. Regulation of decidualization and angiogenesis in the human endometrium: mini review. J. Obstet. Gynaecol. Res. 2014;40:1180–1187. doi: 10.1111/jog.12392. [DOI] [PubMed] [Google Scholar]
- Percharde M., Lin C.J., Yin Y.F., Guan J., Peixoto G.A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X., Ramalho-Santos M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell. 2018;174:391–+. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piltonen T.T., Chen J.C., Khatun M., Kangasniemi M., Liakka A., Spitzer T., Tran N., Huddleston H., Irwin J.C., Giudice L.C. Endometrial stromal fibroblasts from women with polycystic ovary syndrome have impaired progesterone-mediated decidualisation, aberrant cytokine profiles and promote enhanced immune cell migration in vitro. Hum. Reprod. 2015;30:1203–1215. doi: 10.1093/humrep/dev055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadatpour L., Fadaee E., Fadaei S., Mansour R.N., Mohammadi M., Mousavi S.M., Goodarzi M., Verdi J., Mirzaei H. Glioblastoma: exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther. 2016;23:415–418. doi: 10.1038/cgt.2016.48. [DOI] [PubMed] [Google Scholar]
- Salsano S., Perez-Deben S., Quinonero A., Gonzalez-Martin R., Dominguez F. Phytoestrogen exposure alters endometrial stromal cells and interferes with decidualisation signaling. Fertil. sterility. 2019;112:947–958.e3. doi: 10.1016/j.fertnstert.2019.06.014. [DOI] [PubMed] [Google Scholar]
- Vasconcelos S., Ramalho C., Marques C.J., Doria S. Altered expression of epigenetic regulators and imprinted genes in human placenta and fetal tissues from second trimester spontaneous pregnancy losses. Epigenetics. 2019;14:1234–1244. doi: 10.1080/15592294.2019.1634988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polański K., Goncalves A. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H.F., Jin X.H., Cao Z.F., Hu Y., Ma X. MicroRNA expression and regulation in the uterus during embryo implantation in rat. FEBS J. 2014;281:1872–1891. doi: 10.1111/febs.12751. [DOI] [PubMed] [Google Scholar]
- Xia H.F., Jin X.H., Cao Z.F., Shi T., Ma X. MiR-98 is involved in rat embryo implantation by targeting Bcl-xl. FEBS Lett. 2014;588:574–583. doi: 10.1016/j.febslet.2013.12.026. [DOI] [PubMed] [Google Scholar]
- Xu Y., Sun X., Zhang R., Cao T., Cai S.Y., Boyer J.L., Zhang X., Li D., Huang Y. A positive feedback loop of TET3 and TGF-beta1 promotes liver fibrosis. Cell Rep. 2020;30:1310–1318.e5. doi: 10.1016/j.celrep.2019.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan J., Guo S.L., Huang A., Xu H.B., Shao M., Yang Y., Wen W. MiR-29a and miR-652 attenuate liver fibrosis by inhibiting the differentiation of CD4+T cells. Cell Struct. Funct. 2017;42:95–103. doi: 10.1247/csf.17005. [DOI] [PubMed] [Google Scholar]
- Yang Q., Gu W.W., Gu Y., Yan N.N., Mao Y.Y., Zhen X.X., Wang J.M., Yang J., Shi H.J., Zhang X. Association of the peripheral blood levels of circulating microRNAs with both recurrent miscarriage and the outcomes of embryo transfer in an in vitro fertilization process. J. Transl. Med. 2018;16 doi: 10.1186/s12967-018-1556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data and analyses are included in the present manuscript and its supplemental information. This study did not generate original code. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.