Abstract
Many natural polysaccharides have significant anticancer activity with low toxicity, but the complex chemical structures make in-depth studies of the involved mechanisms extremely difficult. The purpose of this study was to investigate the effect of the marine bacterial exopolysaccharide (exopolysaccharide 11 [EPS11]) on liver cancer metastasis to explore the underlying target protein and molecular mechanism. We found that EPS11 significantly suppressed cell adhesion, migration, and invasion in liver cancer cells. Proteomic analysis showed that EPS11 induced downregulation of proteins related to the extracellular matrix–receptor interaction signaling pathway. In addition, the direct pharmacological target of EPS11 was identified as collagen I using cellular thermal shift assays. Surface plasmon resonance and pull-down assays further confirmed the specific binding of EPS11 to collagen I. Moreover, EPS11 was shown to inhibit tumor metastasis by directly modulating collagen I activity via the β1-integrin–mediated signaling pathway. Collectively, our study demonstrated for the first time that collagen I could be a direct pharmacological target of polysaccharide drugs. Moreover, directly targeting collagen I may be a promising strategy for finding novel carbohydrate-based drugs.
Keywords: exopolysaccharide, collagen I, tumor metastasis, β1-integrin, target
Abbreviations: cDNA, complementary DNA; CETSA, cellular thermal shift assay; Dil, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbo-cyanine perchlorate; DMEM, Dulbecco's modified Eagle's medium; ECM, extracellular matrix; EMT, epithelial–mesenchymal transition; EPS, exopolysaccharide; EPS11, exopolysaccharide 11; FBS, fetal bovine serum; NR, normalization; qRT-PCR, quantitative RT-PCR; SPR, surface plasmon resonance
Liver cancer is a major health problem, with an incidence of more than 850,000 new cases worldwide each year (1). It is currently the second leading cause of cancer-related mortality in the world (approximately 800,000 per year), a figure that is on the rise (2). Over the past decades, there has been an improvement in the treatment of the disease. Five treatments can extend the life expectancy of patients with liver cancer: early stage tumors that are amenable to curative therapies—resection, liver transplantation, or local ablation; at more-developed stages, only chemoembolization (for intermediate tumors) and sorafenib (for advanced tumors) have shown survival benefits (3). There are still major unmet needs in the management of liver cancer that might be addressed through discovery of new therapies.
The intensive communication between tumor microenvironment and tumor cells regulates growth, differentiation, and migration of tumor cells. An increasing amount of recent research has concentrated on the function of the tumor microenvironment in favoring cancer progression (4). The extracellular matrix (ECM), a three-dimensional structure with distinct biochemical and biomechanical properties, is an important component of tumor microenvironment (5). ECM is increasingly recognized as an important contributor to tumor behaviors, including tumor growth, angiogenesis, and metastatic progression. The major receptors mediating ECM–cell interactions are integrins, a family of heterodimeric transmembrane proteins composed of noncovalently associated α and β subunits. β1-integrin is the key transmembrane protein that conducts extracellular biochemical and biomechanical signals into the cell, regulating cell proliferation, migration, invasion, and survival (6). The β1-integrin subfamily includes a variety of receptors, such as collagen, fibronectin, and laminin for ECM proteins. The expression level of β1-integrin was higher in cancer tissues than that in adjacent nontumor tissues (5). And researchers have explored the possibility of developing integrin-targeted therapy to manage cancer (7). Previous study has demonstrated that the activation of β1-integrin by collagen I promoted the epithelial–mesenchymal transition (EMT) in pancreatic cancer (8). In breast cancer, inhibition of the β1-integrin activity by monoclonal antibody AIIB2 dramatically enhances radiotherapy efficacy and increases sensitivity to human epidermal growth factor receptor 2–targeting agents (9).
Collagen, as a large family of triple helical proteins, is the major constituent of the tumor stroma environment. The collagen superfamily now includes more than 20 collagen types. Collagen I, as a major component of ECM, plays vital roles on the disruption of cellular adhesion and EMT (10, 11, 12), which influence tissue integrity and allow tumor cells to disseminate from the primary tumor, subsequently results in invasion and metastasis (13), which are largely responsible for poor prognosis of liver cancer. Recently, the expression of collagen I was investigated in liver cancer tissues. It has been reported that the expression of collagen I was significantly upregulated in 83.7% of human liver cancer specimens compared with their adjacent nontumor tissues (5). A better understanding of the role of ECM in the progression of liver cancer will allow the development of new diagnostic and therapeutic strategies in cancer treatment. Disruption of tumor ECM integrity has shown promising results in halting tumor metastasis in preclinical studies (14). Methylumbelliferone, a hyaluronan synthesis inhibitor, was effective in preventing bone metastasis of lung cancer in vivo (15). Pirfenidone, a novel antifibrotic and anti-inflammatory agent, was reported to partially inhibit EMT through the direct inhibition of collagen I expression (16). Therefore, pharmacological perturbation of collagen I can exert effective regulation of ECM to delay progressive tumors.
The oceans harbor a great diversity of organisms, ranging from unicellular bacteria to large multicellular mammals, and have been recognized as an important source of new compounds with nutritional and therapeutic potential (17). Among these compounds, carbohydrate-based compounds are of particular interest because they exhibit numerous important pharmacological activities, such as antitumor (18), antiviral (19), anticoagulants (20), antioxidants (21), and anti-inflammatory (22). Up to now, among the 17 marine drugs approved for marketing in the world, there are six marine carbohydrate-based drugs, including cytarabine, sodium alginate, mannan oligosaccharide diacid (23), and so on. Marine microbial exopolysaccharides (EPSs) produced by EPS-producing bacteria, are widely present in marine ecosystems and can be isolated from the water column, sediments, animals, etc. In recent years, marine EPSs show the extraordinary efficacy as anti-cancer, anti-oxidant and immune-stimulatory activities that made EPSs gained increasing attention as a promising source of potential new drugs for cancer (24).
In our previous studies, we obtained a novel marine bacterial EPS (exopolysaccharide 11 [EPS11]) from marine bacterium Bacillus sp. 11. It has been reported that EPS11 not only significantly inhibits cancer growth in vitro and in vivo (25) but also suppresses tumor metastasis in vitro and in vivo (26), indicating that EPS11 has dual antitumor effects of inhibiting cancer growth and metastasis. Tumor metastasis accounts for the vast majority of cancer deaths in patients, and EPS11 possesses significant antimetastasis activity with low toxicity and high safety. Therefore, the inhibitory effects of EPS11 on the metastasis in human hepatocarcinoma cells were explored deeply in this work. Moreover, we observed that EPS11 directly bonds to collagen I and displayed antimetastasis activity via β1-integrin signaling pathway, indicating that collagen I protein is a crucial pharmacological target for the effect of EPS11 on tumor metastasis.
Results
EPS11 inhibits migration, invasion, and colony formation of liver cancer cells
In our previous study, we found that EPS11 inhibited the growth and migration of Huh7.5 liver cancer cells via blocking cell adhesion and attenuating filiform structure formation (26). In order to determine whether EPS11 has the same inhibition on other liver cancer cells, further investigations were thus performed in this study.
Similarly, both HepG2 and Bel-7402 liver cancer cells in the untreated group displayed regular growth and attachment with long and multiple filiform structures, whereas the EPS11-treated cells had the destroyed filiform structures with abnormal cellular morphology of becoming round shape (Fig. 1A), suggesting the loss of the adhesive capacity to the ECM in the two kinds of hepatocellular carcinoma cells. Filiform structure is a key factor determining cell adhesion and migration in cancer cells (27). Hence, we next sought to disclose the effects of EPS11 on the migration and invasion of liver cancer cells using transwell migration and invasion assays, respectively. Compared with the untreated cells, the numbers of migratory and invasive cells (Huh7.5, HepG2, and Bel-7402 liver cancer cells) treated with EPS11 were dramatically decreased in a concentration- and time-dependent manner (Fig. 1, B and C). We next examined the inhibitory effects of EPS11 on the colony formation of liver cancer cells. And the results of colony formation assay showed that Huh7.5, HepG2, and Bel-7402 liver cancer cells in the control group formed markedly large-sized colonies, whereas smaller and even almost no colony formation was observed in the EPS11-treated cells (Fig. 1D). These in vitro results suggest that EPS11 could significantly block the migratory and invasive biological functions of hepatocarcinoma cells.
Figure 1.
EPS11 inhibits migration, invasion, and colony formation of liver cancer cells (Huh7.5, Bel-7402, and HepG2 cell lines).A, observation of the filiform structures in Bel-7402 and HepG2 cells after treatment of EPS11 via scanning electron microscopy. Liver cancer cells were treated with indicated concentrations of EPS11 (0, 0.1, and 0.2 mg/ml) for 6 and 12 h, respectively. Transwell assay showed that EPS11 inhibited migratory (B) and invasive (C) ability of liver cancer cells (Huh7.5, Bel-7402, and HepG2 cells) after treatment with EPS11 (0, 0.1, and 0.2 mg/ml) for 6 and 12 h, respectively. D, EPS11 could obviously inhibit colony formation of liver cancer cells (Huh7.5, Bel-7402, and HepG2 cells) after incubation with EPS11 (0, 0.1, and 0.2 mg/ml) for 5 and 10 days, respectively. The experiments were performed in triplicate. EPS11, exopolysaccharide 11.
EPS11-FITC is located on or outside the cell membrane of Huh7.5 liver cancer cells
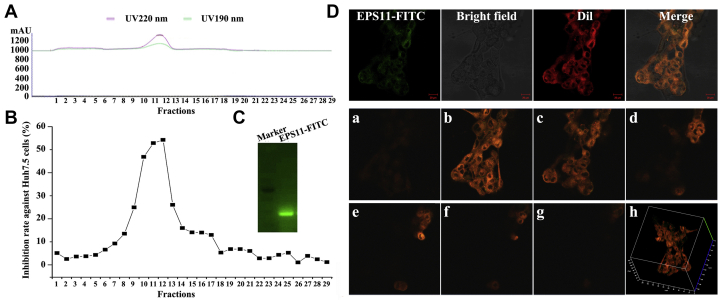
To find out the subcellular localization of EPS11 in the hepatocarcinoma cells, we labeled EPS11 with FITC. After dialysis, anion exchange, and gel filtration, EPS11-FITC was finally eluted as a single peak on the gel filtration chromatogram, indicating that EPS11-FITC was a relatively pure compound (Fig. 2A). Meanwhile, the cytotoxic effects of each eluted fraction after gel filtration chromatography of EPS11-FITC on Huh7.5 cells were evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. And the data showed that the position of the active fractions coincided with the elution peak location (fractions 10–12) of EPS11-FITC on the gel chromatography (Fig. 2B), and we then collected the corresponding active components for later use. EPS11-FITC was further analyzed with native-PAGE electrophoresis, and it was displayed as a single band with green fluorescence in the gel under the 488-nm excitation light (Fig. 2C), suggesting the successful synthesis of EPS11-FITC. To observe colocalization of EPS11 with membranes more clearly, Huh7.5 liver cancer cells were first labeled with membrane-specific dye, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbo-cyanine perchlorate (Dil) and then treated with EPS11-FITC for 3 h. The observations of confocal laser scanning microscopy showed that EPS11-FITC induced evident cell aggregation (Fig. 2D) similar to that of EPS11 (Fig. 3B), indicating that the conjugation of FITC does not change the activity and structure of EPS11. The green fluorescence emitted by EPS11-FITC and the red fluorescence emitted by Dil overlapped completely on the membranes of Huh7.5 cells (Fig. 2D), clearly suggesting that EPS11 is associating with membranes. Besides, EPS11/membrane floatation assay was also performed in our work, and the result showed that the green fluorescence signals of EPS11-FITC were only detected in fractions of crude cell lysates and cell membranes, but not in fraction of cellular mitochondria, further indicating the colocation between EPS11 and membranes (Fig. S1).
Figure 2.
Subcellular localization analysis of EPS11 in Huh7.5 liver cancer cells.A and B, purification and activity assay of EPS11-FITC. The fractions of EPS11-FITC in the gel filtration (55–65 ml, fractions 10–12) were collected and monitored for the cell proliferation determined at an absorbance at 570 nm using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. C, the purified EPS11-FITC on the native-PAGE gel exhibited green fluorescence signals under the 488-nm excitation light. D, laser confocal microscopic observations showed that EPS11-FITC and Dil were colocated on the cell membrane of Huh7.5 liver cancer cells. Images of stacks of confocal Z series of EPS11-FITC and Dil in the surface of Huh7.5 cells are shown (a–g). In addition, peripheral visualization of merge of Z-stack images is shown (h), demonstrating the surface location of EPS11. Dil, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbo-cyanine perchlorate; EPS11, exopolysaccharide 11.
Figure 3.
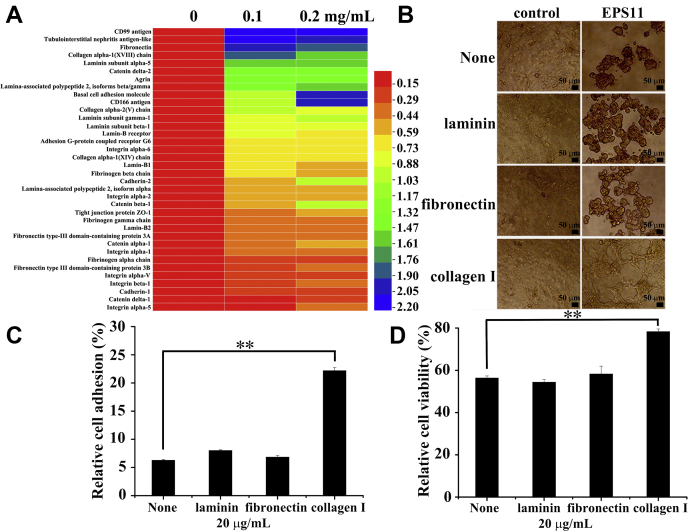
Interaction analysis of ECM and EPS11 in Huh7.5 liver cancer cells.A, proteomic analysis showed that EPS11 downregulated the ECM–receptor interaction signaling pathway. Huh7.5 cells were treated with different concentrations of EPS11 (0, 0.1, and 0.2 mg/ml) for 24 h, and cell lysates were separated and identified using liquid chromatography electrospray ionization tandem MS analysis. The relative abundances of differentially expressed proteins (fold change ≤ 0.67 and p ≤ 0.05) were imported for clustering analysis using HemI software. B–D, coating with collagen I on the surface of the plates rescued the abnormal cell morphology (B), cell adhesion reduction (C), and cell viability inhibition (D) induced by EPS11 in Huh7.5 cells, whereas coating with laminin and fibronectin had no effects. The data were presented as means ± SD of three experiments. ∗p ≤ 0.05, ∗∗p ≤ 0.01. ECM, extracellular matrix; EPS11, exopolysaccharide 11.
Interaction analysis of ECM and EPS11 in Huh7.5 liver cancer cells
To better understand the inhibitory effects of EPS11 on the liver cancer cells, we performed a proteomic analysis to identify differentially expressed proteins after EPS11 treatment. Information for all protein identifications are provided as Supporting information. For the single-peptide identifications of proteins, mass-labeled MS/MS spectra were also provided (Supporting figure).
Compared with that in the untreated cells, there were 1140 and 1127 proteins differentially expressed (1.5-fold change cutoff and p value less than 0.05) in 0.1 and 0.2 mg/ml EPS11-treated groups, respectively (Table 1). Kyoto Encyclopedia of Genes and Genomes pathway–based enrichment analysis indicated that the most significantly impacted pathway in the EPS11-treated versus EPS11-untreated group comparison was the ECM–receptor interaction signaling pathway. About 36 proteins were significantly downregulated in this pathway after Huh7.5 liver cancer cells were treated with EPS11 (0.1 and 0.2 mg/ml) for 24 h (Fig. 3A), suggesting that ECM is likely to be involved in the anticancer activity of EPS11.
Table 1.
Summary of differentially expressed proteins analyzed by proteomic analysis
| Label | Upregulated | Downregulated | All regulated |
|---|---|---|---|
| 0.1 mg/ml EPS11-treated versus untreated group | 778 | 362 | 1140 |
| 0.2 mg/ml EPS11-treated versus untreated group | 765 | 362 | 1127 |
1.5-fold change cutoff and p value less than 0.05.
Collagen I, laminin, and fibronectin are the main ECM proteins that can participate in cell–ECM interaction and play a key role in cell migration. After coating culture wells with collagen I (20 μg/ml), the aggregation of Huh7.5 cells induced by EPS11 was evidently reversed (Fig. 3B). In contrast, laminin (20 μg/ml) and fibronectin (20 μg/ml) showed no obvious reversal effects on the cell aggregation (Fig. 3B). The effects of ECM proteins on cancer cell adhesion were assessed by crystal violet staining. Compared with the noncoated condition, collagen I coating significantly improved the cell adhesion in Huh7.5 cells (Fig. 3C). However, laminin showed no further stimulation, and neither did fibronectin (Fig. 3C). In addition, the inhibitory effect of EPS11 on the viability of Huh7.5 cells was clearly rescued by collagen I, whereas stimulation of cell viability was not observed in the presence of laminin and fibronectin (Fig. 3D). These data demonstrated that collagen I, but not laminin and fibronectin, appeared to be mainly involved in EPS11-induced tumor inhibition, and it could specifically and effectively facilitate cell adhesion and viability in Huh7.5 cells.
Collagen I is the potential target of EPS11 in antimetastasis of liver cancer
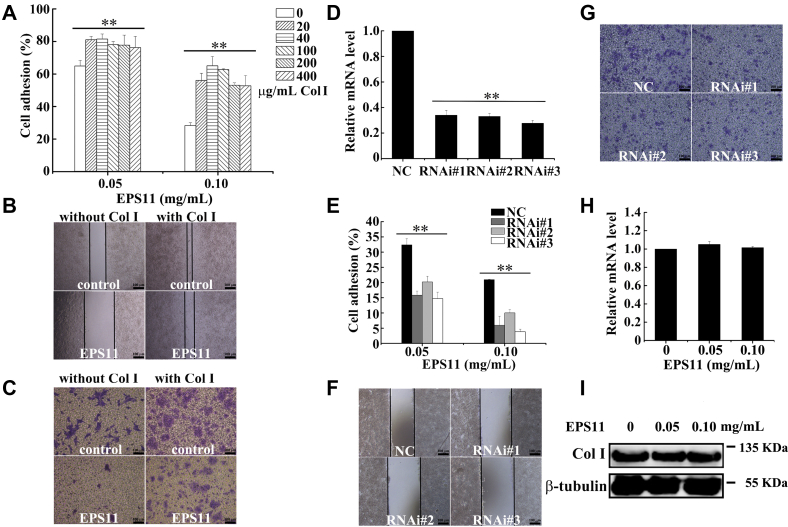
To further evaluate the efficiency of collagen I on antimetastasis of liver cancer induced by EPS11, culture wells were first coated with different doses of collagen I (0, 20, 40, 100, 200, and 400 μg/ml), and Huh7.5 cells were then seeded with EPS11 treatment (0, 0.05, and 0.1 mg/ml). Figure 4A showed that compared with the uncoated group, coating with collagen I significantly attenuated the suppression of cell adhesion induced by EPS11, and 40 μg/ml collagen I had the most obviously stimulatory effect. Thus, 40 μg/ml collagen I was used in the following experiments. The effect of collagen I on cell migration was assessed by the wound-healing assay. As shown in Figure 4B, compared with the initial wounds when time is zero (Fig. S2), all the wounds became narrower because of cell migration. Collagen I (40 μg/ml) coating significantly restored the migratory ability of Huh7.5 cancer cells inhibited by EPS11, when compared with the noncoated condition (Fig. 4B). Boyden chamber inserts coated with or without collagen I on both sides were applied to evaluate cell invasion. The result showed that EPS11 suppressed cell invasion in Huh7.5 cells without collagen I coating, whereas cell invasion in Huh7.5 cells with collagen I coating (40 μg/ml) was almost unaffected by EPS11 (Fig. 4C). Knockdown of endogenous collagen I was conducted by RNAi, and the data of quantitative RT-PCR (qRT-PCR) analysis showed that the mRNA expression levels of collagen I decreased after Huh7.5 cells were transfected with three siRNAs of collagen I for 48 h (Fig. 4D). The inhibition on cell adhesion, migration, and invasion in Huh7.5 cells caused by EPS11 was further strengthened with the depressed mRNA levels of collagen I (Fig. 4, E–G). In conclusion, collagen I appeared to be the most promising mediator in antimetastasis of liver cancer induced by EPS11.
Figure 4.
Collagen I is the potential target of EPS11 in anti-metastasis of liver cancer. Collagen I reduced the inhibitory effects of EPS11 on cell adhesion (A), migration (B), and invasion (C) in the Huh7.5 cells. D, the expression levels of collagen I were reduced after Huh7.5 cells were transfected with the siRNAs (RNAi #1, RNAi #2, and RNAi #3) of collagen I for 48 h. Knockdown of collagen I expression increased the inhibitory activities of EPS11 to cell adhesion (E), migration (F), and invasion (G) in the Huh7.5 cells. H and I, EPS11 had no effects on the expressions of collagen I at the mRNA and protein levels in the Huh7.5 cells. Col I stands for collagen I. The data were presented as means ± SD of three experiments. ∗p ≤ 0.05, ∗∗p ≤ 0.01. EPS11, exopolysaccharide 11.
Moreover, qRT-PCR and Western blotting showed that the mRNA and protein expression levels of collagen I were not affected by EPS11 (Fig. 4, H and I), suggesting that EPS11 had no effect on the gene transcription and expression of collagen I. Therefore, we speculated that collagen I was a key target for EPS11 exerting antimetastasis activity.
EPS11 directly targets collagen I
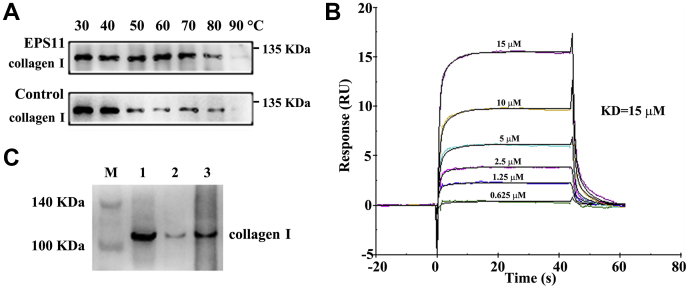
To test the interaction of EPS11 with collagen I, we first performed target identification by cellular thermal shift assay (CETSA). CETSA experiment revealed that EPS11 treatment efficiently protected collagen I protein from temperature-dependent degeneration (Fig. 5A). Immunofluorescence staining showed that collagen I was widely distributed inside and outside Huh7.5 cells, whereas EPS11 labeled with FITC was distributed more on or outside the Huh7.5 cells (Fig. S3). The distribution of EPS11 and collagen I overlapped, and they were colocalized on or outside the cells (Fig. S3). To verify the direct interaction between EPS11 with collagen I, we next performed surface plasmon resonance (SPR) assay. SPR analysis revealed that target affinity (equilibrium dissociation constant value) of EPS11 binding to collagen I was 15 μM (Fig. 5B), showing a specific binding of EPS11 with collagen I. In addition, FITC pull-down assay was further performed to confirm the binding of EPS11 with collagen I in cells. FITC magnetic beads were first incubated with cell lysates, and after a series of washing steps, the complexes of FITC beads and target molecules were finally obtained for Western blotting analysis. As shown in Figure 5C, collagen I was detected in all three fractions, including total cell lysates, flow through (the unbound proteins), and FITC pull down (FITC-beads bound proteins), further confirming the previously proposed interaction between EPS11 and collagen I.
Figure 5.
EPS11 directly targets collagen I.A, EPS11 protected target protein (collagen I) against temperature-dependent degeneration in Huh7.5 cells. B, the target affinity (equilibrium dissociation constant [KD] value) of EPS11 binding to collagen I was detected by SPR. C, collagen I was pulled down using FITC beads. FITC beads were incubated with cell lysates of Huh7.5 cells, and after washing steps, the complexes were finally obtained for Western blotting analysis. M, marker; 1, total cell lysates; 2, flow through (the bead-unbound proteins); 3, FITC pull-down (the bead-bound proteins). EPS11, exopolysaccharide 11; SPR, surface plasmon resonance.
EPS11 displays antimetastasis activity through collagen I/β1-integrin signaling
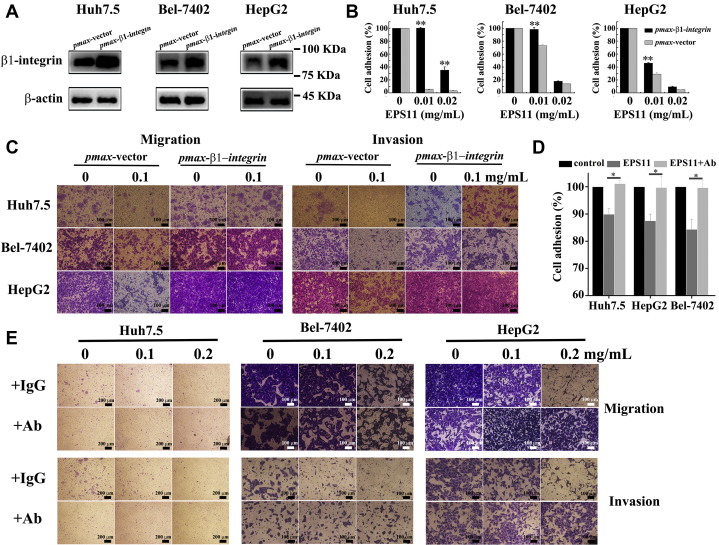
β1-Integrin is a transmembrane receptor, which plays a vital role in cell–ECM interactions. It is reported that β1-integrin is positively correlated with the level of collagen I in liver cancer tissue. To further figure out whether β1-integrin is involved in the antimetastasis of liver cancer induced by EPS11, β1-integrin is cloned and overexpressed in three liver cancer cell lines (Huh7.5, Bel-7402, and HepG2) through transfection. As shown in Figure 6A, the expression levels of β1-integrin in these three liver cancer cells were dramatically increased after transfection for 48 h. Indeed, the decreased cell adhesion caused by EPS11 in these three liver cancer cells was all restored by overexpressions of β1-integrin (Fig. 6B). Besides, the overexpressions of β1-integrin also alleviated the inhibiting effects of EPS11 on cell migration (Fig. 6C, left panels) and invasion (Fig. 6C, right panels) in these three liver cancer cells. Blocking antibody against β1-integrin was used to perturb the signaling pathways related with β1-integrin in our study. As shown in Figure 6E, blocking antibody against β1-integrin could decrease the migratory and invasive ability of untreated liver cancer cells (Huh7.5, Bel-7402, and HepG2), suggesting that β1-integrin is involved in cell migration and invasion. EPS11 caused the reduction of cell adhesion in all three liver cancer cell lines but had no inhibitory effects on cell adhesion when cells were treated with blocking antibody against β1-integrin (Fig. 6D). Similarly, the inhibition on cell migration (Fig. 6E, upper panels) and invasion (Fig. 6E, lower panels) of three cell lines induced by EPS11 were completely reversed by addition of blocking antibody against β1-integrin. These data further support that EPS11 exerts the antimetastasis of liver cancer via β1-integrin.
Figure 6.
β1-integrin plays vital roles in the inhibition of EPS11 on adhesion, migration, and invasion of liver cancer cells (Huh7.5, Bel-7402, and HepG2).A, the expression levels of β1-integrin were detected by Western blotting after Huh7.5, Bel-7402, and HepG2 cells were transfected with pmax-β1-integrin for 48 h, respectively. The overexpression of β1-integrin rescued the EPS11-induced reduction of cell adhesion (B), migration (C, left panels), and invasion (C, right panels) in three liver cancer cell lines (Huh7.5, Bel-7402, and HepG2). EPS11 had no inhibitory effects on cell adhesion (D, 0.01 mg/ml EPS11), migration (E, upper panels), and invasion (E, lower panels) in three liver cancer cell lines (Huh7.5, Bel-7402, and HepG2) with blocking antibody against β1-integrin. Ab stands for the blocking antibody against β1-integrin. Data were presented as means ± SD of three independent experiments (n = 3). ∗p < 0.05, ∗∗p < 0.01. EPS11, exopolysaccharide 11.
In conclusion, the obtained data demonstrate that the antimetastasis of liver cancer cells induced by EPS11 is directly based on collagen I via β1-integrin signaling pathway.
Discussion
As the culprit behind most cancer-related deaths, metastasis is the ultimate challenge in our effort to fight cancer as a life-threatening disease (28). Thus, development of effective therapeutic agents and strategies for the prevention of metastatic cancer will be a promising strategy for cancer. In the current study, it has been demonstrated that natural marine carbohydrate-based compound EPS11 could effectively inhibit metastasis of liver cancer by directly targeting collagen I through β1-integrin signaling pathway, indicating a novel and key drug candidate for treating metastatic liver cancer. Previous studies mainly focus on the discovery of new agents directly targeting cancer cells; however, tumor microenvironment, especially ECM proteins, has been paid less attention. In this study, our data revealed that EPS11 could specially target extracellular collagen I to suppress metastasis of liver cancer. To our knowledge, these findings have not aroused extensive attentions before.
During tumor development, tumor cells constantly communicate with the surrounding microenvironment through both biochemical and biophysical cues (14). And in particular, the tumor microenvironment can facilitate local invasion and distant metastasis by instructing cancer cells to undergo a morphogenesis program termed EMT. EMT refers to a global cellular and molecular transition during tumor metastasis, by which polarized epithelial cells gain mesenchymal properties to migrate (14, 29). In the tumor microenvironment, ECM plays an important role in tumorigenesis and tumor development (30). The critical role of ECM in promoting EMT was already reported in the original experiments conducted by Greenburg and Hay. They showed that epithelial cells from embryonic and adult anterior lens cultured in three-dimensional collagen gels can elongate and migrate as individual cells (31). Recently, Zhang et al. (32) unraveled that ECM structural protein collagen I could promote tumor cell invasion by activating downstream SRC/extracellular signal–regulated kinase 2 to stabilize SNAIL1 in breast tumor cells. Another ECM structural component, fibronectin, was reported to stimulate EMT by inducing SNAIL1 expression in tumor cells (33). Thus, the ECM-targeted therapy may be a potential strategy for the treatment of tumor metastasis.
In our study, proteomic analysis revealed that EPS11 had strong inhibition effect on the ECM–receptor interaction signaling pathway of Huh7.5 liver cancer cells (Fig. 3A), indicating that there was a strong link between the antimetastasis of EPS11 and ECM. To determine which component of ECM is responsible for the metastasis inhibition, we treated Huh7.5 cells with EPS11 on plates coated with collagen I, laminin, or fibronectin. This experiment showed that Huh7.5 cells cultured on collagen I had a higher proliferation and adhesion rates than those on laminin and fibronectin (Fig. 3, B–D). Further study showed that collagen I was an important potential target of EPS11 (Fig. 4). CETSA, immunofluorescence, SPR, and pull-down studies finally proved that there was a direct interaction between EPS11 and collagen I (Fig. 5 and Fig. S3).
Integrin, a typical adhesion molecule in cancer cells, often mediates cancer cell behavior, especially when combined with collagen (4). Different types of collagen bind to various integrins in cancer cells, mediating a broad spectrum of cellular functions, such as differentiation, proliferation, survival, and progression (34). The expression level of β1-integrin was often upregulated in human liver cancer specimens, positively correlated with the level of collagen I. Besides, the upregulated expressions of β1-integrin and collagen I in liver cancer cells were closely associated with cancer proliferation and metastasis (5). In our study, β1-integrin is identified as the metastasis-inhibitory receptor for EPS11 via binding to collagen I (Fig. 6). Integrin-targeted therapy has revealed promising results in both preclinical and clinical studies in breast cancer, melanoma, glioblastoma, and other solid tumors (35). It has been reported that tumor cell migration and invasion was strongly inhibited by anti-β1-integrin monoclonal antibodies in the pancreatic carcinoma (36). Knockdown of β1-integrin significantly reduces primary tumor growth and inhibits pancreatic cancer metastasis (37). Consistently, our data provide additional insights to the inhibition of β1-integrin as a potential intervention for cancer metastasis.
In recent years, studies have shown that natural polysaccharides have significant anticancer activity, and the toxic side effects of the human body are relatively small, making the antitumor activity of polysaccharides become a hot topic of domestic and foreign experts and scholars in the pharmaceutical fields (38). Bacterial EPSs with low cytotoxicity and side effects are considered to serve as a good substitute to the synthetic antitumor agents (39) and have drawn a great deal of attention in the fields of tumor therapy. At present, many anticancer EPSs have been found, including EPSs isolated from Rhizopus nigricans (40), Lactobacillus delbrueckii subsp bulgaricus (41), and marine Bacillus amyloliquefaciens 3MS 2017 (42). The mechanism of antitumor action of EPSs consists of the stimulation of the immune system, induction of cell apoptosis, and activation of autophagy, but so far, there are no reports about the exact targets of antitumor effects of EPSs. EPSs are usually long-chain high molecular-mass polymers with an enormous structural diversity, composed of the sugar units mainly glucose, galactose and rhamnose, mannose, fructose, arabinose, and xylose in different ratios or some sugar derivatives such as N-acetylgalactosamine and N-acetylglucosamine (39), thus making the studies of in-depth mechanisms extremely difficult. Our study has demonstrated for the first time that EPS can directly target collagen I to display its antimetastasis activity, which will contribute to the research and development of new polysaccharides in medicine.
In summary, we demonstrate here that marine bacterial EPS, EPS11, can directly interact with collagen I. And then the blockade of collagen I by EPS11, in large part, inhibits the metastasis of liver cancer cells via β1-integrin signaling pathway. Considering the general research situation of polysaccharides in medicine, our data present a novel mechanism underlying the antitumor metastasis of polysaccharides in medicine and provide a theoretical basis for developing more carbohydrate-based drugs.
Experimental procedures
Materials
RPMI1640 and high-glucose Dulbecco's modified Eagle's medium (DMEM) were purchased from Thermo Fisher Scientific. Fetal bovine serum (FBS) was bought from PAN Biotech. Transwell plates were obtained from Corning. Collagen I, laminin, and fibronectin were ordered from Beyotime Institute of Biotechnology. siRNA and transfection reagent siRNA-Mate were supplied by GenePharma. FITC was provided by Solarbio. Antibodies against collagen I, β1-integrin, β-actin, and β-tubulin were the products of Bioss. Blocking monoclonal anti-β1-integrin (clone P5D2) was purchased from R&D Systems. FITC magnetic beads were obtained from Aladdin. Carboxymethylated 5 sensor chip was purchased from GE Healthcare. All other reagents used in the experiment were of analytical grade or higher.
Cell culture
Huh7.5 liver cancer cells were cultured in RPMI1640 medium with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Both HepG2 liver cancer cells and Bel-7402 liver cancer cells were cultured in high-glucose DMEM with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. All experiments were carried out with the same batch of cell line between passages 2 and 8.
Scanning electron microscopy observation
HepG2 and Bel-7402 cells in the logarithmic phase were seeded onto the glass coverslips and treated with different doses of EPS11 (0, 0.1, and 0.2 mg/ml) for 6 or 12 h. Cells were then fixed by 5% glutaric dialdehyde, washed by PBS three times, and gradually dehydrated in ethanol (30%, 50%, 70%, 90%, and 100% for 10 min at each step). Finally, scanning electon microscopy was performed to observe the ultrastructural changes of cells on the coverslips.
Cell adhesion assay
Hepatocellular carcinoma cells (Huh7.5, Bel-7402, and HepG2) were seeded in 96-well plates, which were filled with RPMI1640 or DMEM containing EPS11. After incubation for 12 h, nonadherent cells were removed by washing with PBS twice, whereas adhered cells were fixed with 95% ethanol for 30 min at room temperature and stained with 0.1% crystal violet for 20 min. After redundant crystal violet was removed, 100 μl acetic acid was added into each well with gentle shaking for 10 min. The absorbance at 590 nm was then measured by a multidetection microplate reader (Infinite M1000 Pro; TECAN).
Cell migration assay
For wound-healing assays, three human hepatocarcinoma cell lines (Huh7.5, Bel-7402, and HepG2 cells) were seeded in 96-well plates. After 24 h, a scratch was made on the bottom of each well using sterile pipette tips. Washed twice with PBS, the cells were exposed to medium supplemented with or without EPS11. After incubation for 6 or 12 h, three fields of each wound were selected and photographed with an inverted microscope (NIKON TS100) equipped with a digital camera.
For transwell-migration assays, first, the lower compartment of transwell plates contained 0.6 ml of medium supplemented with 20% FBS. Hepatocellular carcinoma cells (Huh7.5, Bel-7402, and HepG2) were then resuspended in 100-μl medium containing 5% FBS with or without EPS11 and seeded into the upper compartment of each well. Cells were further incubated for 6 or 12 h to allow cell migration through the filter membrane to the lower side of the insert. After washed with PBS three times, cells were fixed with 95% ethanol and stained with 0.1% crystal violet. Then, the nonmigrated cells on the upper side of the insert were gently removed using cotton swabs, and the migrated cells on the lower side of the insert were observed and photographed by an inverted microscope (NIKON TS100).
Cell invasion assay
The invasive potential of cells was also assessed in the transwell chamber, thereby allowing the cells to pass through a polycarbonate membrane (8-μm pore size) coated with Matrigel. Briefly, 2 mg/ml of Matrigel was coated on the chambers for 6 h at 37 °C. Cells (Huh7.5, Bel-7402, and HepG2) were then resuspended in RPMI1640 or DMEM (100 μl) containing EPS11 and were added into the upper chamber of each well. The lower chambers were also filled with 500 μl of RPMI1640 or DMEM containing 20% FBS. After incubation for 6 or 12 h, cells on the lower surface were fixed with 95% ethanol for 30 min, followed by 0.1% crystal violet staining for 20 min at room temperature. Afterward, the cells on the upper surface were removed via gentle scraping with cotton swabs. Images of the invading cells on the lower surface of the chambers were captured using an inverted microscope (NIKON TS100).
Colony formation assay
The colony formation assay was carried out in 6-well plates. The plates containing 3000 cells (Huh7.5, Bel-7402, and HepG2) were incubated at 37 °C for 5 to 10 days, after which the colonies were stained with 0.1% crystal violet staining for 20 min at room temperature, and images of colonies were captured by an inverted microscope (NIKON TS100).
Preparation and purification of EPS11-FITC
EPS11 was prepared according to the procedures described before (25). The reaction of fluorescent labeling was performed with a molar ratio of 10:1 FITC to EPS11. EPS11 was diluted with 100 mM sodium carbonate (pH 9.3) to 0.5 mg/ml and further dialyzed against this buffer for 24 h. Dilute FITC solution with 100 mM sodium carbonate (pH 9.3) to 0.05 mg/ml. About 0.5 mg/ml EPS11 solution was mixed with a freshly made 0.05 mg/ml FITC solution (in reaction buffer) in the 50-ml tube. The tube was wrapped in aluminum foil for light protection and was agitated for 30 min at 900 rpm. After fluorescent labeling, the reaction mix was loaded onto Hiload 16/600 Sephadex 200 column (GE Healthcare) to purify EPS11-FITC.
Confocal laser scanning microscopy observation
Huh7.5 cells were seeded in glass bottom dishes (35-mm dish with 14-mm bottom well) for live cell microscopy measurement. After incubation at 37 °C for 24 h, cells were first labeled with membrane-specific dye Dil (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) and then treated with EPS11-FITC (0.1 mg/ml) for 3 h. After being washed twice with PBS, cells were observed by the laser scanning confocal microscope LSM 710 (Carl Zeiss) for colocalization analysis.
Proteomic analysis
Proteomic analysis of Huh7.5 cells was performed by PTM-Bio labs Co, Ltd. Briefly, Huh7.5 cells seeded in dishes (100 mm) were, respectively, treated with EPS11 (0, 0.1, and 0.2 mg/ml) for 24 h, and cells were then collected and lysed to obtain total cellular protein. Protein samples were digested, and the tryptic peptides were dissolved in solvent A (0.1% formic acid and 2% acetonitrile in water), directly loaded onto a reversed-phase analytical column (25-cm length, 100 μm i.d.). Peptides were separated with a gradient from 7% to 23% solvent B (0.1% formic acid in 90% acetonitrile) over 26 min, 23% to 35% in 8 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nl/min on an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific).
The separated peptides were analyzed in Q Exactive (Thermo Fisher Scientific) with a nanoelectrospray ion source. The electrospray voltage applied was 2.1 kV. The full MS scan resolution was set to 70,000 for a scan range of 350 to 1800 m/z. Up to 20 most abundant precursors were then selected for further MS/MS analyses with 30-s dynamic exclusion. The higher energy collisional dissociation fragmentation was performed at normalized collision energy of 28% and 31%. The fragments were detected in the Orbitrap at a resolution of 17,500. Fixed first mass was set as 100 m/z. Automatic gain control target was set at 5E4, with an intensity threshold of 1E4 and a maximum injection time of 200 ms.
The resulting MS/MS data were processed using MaxQuant search engine (version 1.5.2.8, https://www.maxquant.org/). Tandem mass spectra were searched against the human SwissProt database (9606_SP_Human_201701.fasta, 20,130 entries) concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to two missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in first search and 5 ppm in main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as fixed modification. Acetylation on N-terminal protein and oxidation on Met were specified as variable modifications. Tandem mass tag-6plex quantification was performed. False discovery rate was adjusted to <1%, and minimum score for peptides was set as >40.
The database search and analysis results of the mass spectrum data give the signal intensity value of each peptide in different samples. According to this information, the relative quantification of the protein is calculated by the following steps:
-
(1)
The signal intensity value (I) of the peptide in different samples is changed by centralization, and then the relative quantification value (R) of the peptide in different samples is obtained. The calculation formula is as follows: where i represents the sample and j represents the peptide.
-
(2)
In order to eliminate the systematic error of the sample amount of different samples in the MS detection, the relative quantitative value of the peptide needs to be corrected by the median normalization (NR) method.
-
(3)
The relative quantitative value of the protein is expressed as the median value of the relative quantitative value of the specific peptide corresponding to the protein. The calculation formula is as follows: where k represents protein. j represents the specific peptide to which the protein belongs.
Preparation of coated surfaces
Preparation of coated surfaces with 20 μg/ml extracellular matrices (collagen I, laminin, and fibronectin) was performed as mentioned previously (43). To coat culture plates, laminin (20 μg/ml), fibronectin (20 μg/ml), or collagen I (20 μg/ml) diluted with PBS were added into each well and incubated for 1 h at 37 °C. Afterward, the coated surfaces were thoroughly rinsed with PBS and water twice, respectively, and were air dried before adding medium or cells.
Cell proliferation assay
Huh7.5 cells (7 × 103/well) in exponential phase of growth were seeded in the 96-well plate at 37 °C for 12 h and then were treated with EPS11. After treatment for 24 h, 20 μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide solution (5 mg/ml) was added into each well and incubated for another 4 h. About 100 μl of “triplex solution” (10% SDS, 5% isobutanol, and 12 mM HCl) was added to each well for 12 h to dissolve purple crystals of formazan. Absorbance at 570 nm was measured by a multidetection microplate reader (Infinite M1000 Pro; TECAN).
Gene silencing by siRNA
The RNAi sequences (RNAi#1, 5′-GCUGUCUUAUGGC UAUGAUTT-3′), (RNAi#2, 5′-CUGGAAAGAAUGGAGAUGATT-3′), and (RNAi#3, 5′-GCAAGACAGUGAUUGAAUATT-3′) against human collagen I were synthesized by GenePharma. Huh7.5 cells growing in the RPMI1640 medium at 40% to 50% confluence were transfected with siRNA against human collagen I using siRNA-Mate (GenePharma) according to the manufacturer's instructions. Three collagen I siRNAs were applied separately, and a nonsilencing siRNA was used as a negative control for nonspecific silencing events. The final concentration of siRNA was 100 nM. After 48 h, the transfected cells were further maintained in fresh medium before experimentation, and the expression levels of collagen I in transfected cells were detected by Western blotting analysis.
qRT-PCR
RNA was isolated according to the manufacturer's protocol of TRIpure reagent (Aidlab). RNA was reversely transcribed into complementary DNA (cDNA) using a reverse transcription kit (Toyobo) with the DNase I-treated total RNA as the template. GAPDH was used as internal control. Real-time qRT-PCR was performed in triplicates with ABI 7900 Real-Time PCR System (Applied Biosystems), and data were calculated by 2−ΔΔCT method.
Western blotting analysis
Huh7.5 cells were collected by trypsinization and centrifugation, washed with ice-cold PBS three times, and lysed with radioimmunoprecipitation buffer. Afterward, protein samples were resolved on 10% SDS-PAGE gels, electrotransferred to nitrocellulose membranes, and incubated with primary antibodies and secondary antibodies, and finally detected by enhanced chemiluminescence kit using Tanon 5200SF Imaging Analysis System (Tanon).
Target identification by CETSA
To find out the possible target responsible for EPS11-mediated anticancer activity, CETSA was performed as described previously (44). In brief, Huh7.5 cells were incubated with EPS11 (0.1 mg/ml) for 12 h followed by heating for 3 min at the desired temperatures (30, 40, 50, 60, 70, 80, and 90 °C) and placed on ice until the range of heat treatments was completed. Then the lysates were prepared using radioimmunoprecipitation buffer and further analyzed by Western blotting analysis.
SPR analysis
The whole flow path was first primed by running buffer (PBS with 150 mM NaCl, 0.05% surfactant P20, and pH 7.5) for three times. In SPR measurements, reaction temperature was set at 25 °C. Collagen I protein (0.5 mg/ml) in 10 mM sodium acetate buffer (pH 4.5) was immobilized on a carboxymethylated 5 sensor chip. Gradient concentrations of EPS11 (0–15 μM) were injected to Biacore T200 system at a flow rate of 30 μl/min. Results were analyzed with Biacore evaluation software (GE, T200, version 1.0) and fitted to kinetic model. Equilibrium dissociation constant was derived by fitting to a 1:1 Langmuir binding model.
FITC pull-down assay
After incubated with EPS11-FITC for 2 h at 4 °C, FITC magnetic beads were washed three times by PBS to remove the unbound EPS11-FITC. Huh7.5 cell lysates were then incubated with EPS11-bound beads overnight at 4 °C. Wash away the unbound proteins, and the bead-bound proteins were separated by SDS-PAGE and examined by Western blotting analysis.
DNA constructs, transfection, and overexpression
Extraction of total RNA and synthesis of cDNA were performed as mentioned previously (45). The full-length cDNA sequence of β1-integrin was obtained by PCR. The PCR products were gel purified, digested with restriction enzymes, and ligated into the pmax-vector (Biovector). The constructed plasmid (pmax-β1-integrin) was transformed into Escherichia coli DH5α competent cells, and the cloned gene was confirmed by sequencing. The correct plasmid pmax-β1-integrin was transfected into liver cancer cells (Huh7.5, Bel-7402, and HepG2) using the Lipofectamine 2000 (Thermo Scientific) according to the manufacturer's protocol. The transfected cells were collected at 48 h post-transfection, and the overexpression levels of β1-integrin in three liver cancer cell lines were further confirmed by Western blotting analysis.
β1-integrin–blocking assay
For blocking β1-integrin signaling, Huh7.5 cells were pretreated for 45 min with 20 μg/ml anti-β1-integrin blocking antibody at 37 °C and 5% CO2. After incubation, cell adhesion, migration, and invasion were detected as described previously.
Statistical analysis
All data are expressed as means ± SD. Statistical significances are analyzed by two-tailed Student's t test using SPSS 17.0 (IBM). Differences of p ≤ 0.05 were considered statistically significant (∗p ≤ 0.05, ∗∗p ≤ 0.01).
Data availability
All MS data/data of proteomic analysis are deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier: PXD025155).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work is funded by the National Natural Science Foundation of China (42006081), China Ocean Mineral Resources R&D Association Grant (grant no.: DY135-B2-14), National Key R and D Program of China (grant no.: 2018YFC0310800), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no.: XDA22050301), the Taishan Young Scholar Program of Shandong Province (grant no.: tsqn20161051), Natural Science Foundation of Shandong Province (grant no.: ZR2019BH024), and the Basic Applied Research program of Qingdao (grant no.: 19-6-2-35-cg).
Author contributions
G. L. and C. S. conceptualization; R. L. and Y. S. methodology; G. L. investigation; G. L. data curation; G. L. writing–original draft; R. L., Y. S., and C. S. writing–review and editing; C. S. supervision; C. S. funding acquisition.
Edited by Gerald Hart
Supporting information
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J., Han K.H., Gores G., Llovet J.M., Mazzaferro V. Liver cancer: Approaching a personalized care. J. Hepatol. 2015;62:S144–S156. doi: 10.1016/j.jhep.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forner A., Reig M., Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 4.Xu S., Xu H., Wang W., Li S., Li H., Li T., Zhang W., Yu X., Liu L. The role of collagen in cancer: From bench to bedside. J. Transl. Med. 2019;17:309. doi: 10.1186/s12967-019-2058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng X., Liu W., Xiang J., Liu P., Ke M., Wang B., Wu R., Lv Y. Collagen I promotes hepatocellular carcinoma cell proliferation by regulating integrin β1/FAK signaling pathway in nonalcoholic fatty liver. Oncotarget. 2017;8:95586–95595. doi: 10.18632/oncotarget.21525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casar B., Rimann I., Kato H., Shattil S.J., Quigley J.P., Deryugina E.I. In vivo cleaved CDCP1 promotes early tumor dissemination via complexing with activated β1 integrin and induction of FAK/PI3K/Akt motility signaling. Oncogene. 2014;33:255–268. doi: 10.1038/onc.2012.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keely P.J. Mechanisms by which the extracellular matrix and integrin signaling act to regulate the switch between tumor suppression and tumor promotion. J. Mammary Gland Biol. Neoplasia. 2011;16:205–219. doi: 10.1007/s10911-011-9226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan W., Ma J., Ma Q., Xu Q., Lei J., Han L., Li X., Wang Z., Wu Z., Lv S., Ma Z., Liu M., Wang F., Wu E. The activation of β1-integrin by type I collagen coupling with the Hedgehog pathway promotes the epithelial-mesenchymal transition in pancreatic cancer. Curr. Cancer Drug Targets. 2014;14:446–457. doi: 10.2174/1568009614666140402105101. [DOI] [PubMed] [Google Scholar]
- 9.Wu T., Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 10.Koenig A., Mueller C., Hasel C., Adler G., Menke A. Collagen type I induces disruption of E-cadherin-mediated cell-cell contacts and promotes proliferation of pancreatic carcinoma cells. Cancer Res. 2006;66:4662–4671. doi: 10.1158/0008-5472.CAN-05-2804. [DOI] [PubMed] [Google Scholar]
- 11.Shintani Y., Maeda M., Chaika N., Johnson K.R., Wheelock M.J. Collagen I promotes epithelial-to-mesenchymal transition in lung cancer cells via transforming growth factor-beta signaling. Am. J. Respir. Cell Mol. Biol. 2008;38:95–104. doi: 10.1165/rcmb.2007-0071OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkland S.C. Type I collagen inhibits differentiation and promotes a stem cell-like phenotype in human colorectal carcinoma cells. Br. J. Cancer. 2009;101:320–326. doi: 10.1038/sj.bjc.6605143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A., Zhou T., Guo L., Si J. Collagen type I regulates beta-catenin tyrosine phosphorylation and nuclear translocation to promote migration and proliferation of gastric carcinoma cells. Oncol. Rep. 2010;23:1247–1255. doi: 10.3892/or_00000757. [DOI] [PubMed] [Google Scholar]
- 14.Jung H.Y., Fattet L., Yang J. Molecular pathways: Linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin. Cancer Res. 2015;21:962–968. doi: 10.1158/1078-0432.CCR-13-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futamura N., Urakawa H., Arai E., Kozawa E., Ishiguro N., Nishida Y. Hyaluronan synthesis inhibitor supplements the inhibitory effects of zoledronic acid on bone metastasis of lung cancer. Clin. Exp. Metastasis. 2013;30:595–606. doi: 10.1007/s10585-012-9563-4. [DOI] [PubMed] [Google Scholar]
- 16.Hisatomi K., Mukae H., Sakamoto N., Ishimatsu Y., Kakugawa T., Hara S., Fujita H., Nakamichi S., Oku H., Urata Y., Kubota H., Nagata K., Kohno S. Pirfenidone inhibits TGF-β1-induced over-expression of collagen type I and heat shock protein 47 in A549 cells. BMC Pulm. Med. 2012;12:24. doi: 10.1186/1471-2466-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasconcelos A.A., Pomin V.H. Marine carbohydrate-based compounds with medicinal properties. Mar. Drugs. 2018;16:233. doi: 10.3390/md16070233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y., Wang Z.J., Xie D., Sun X., Yang W., Zhao X., Xu N. Characterization and potential antitumor activity of polysaccharide from Gracilariopsis lemaneiformis. Mar. Drugs. 2017;15:100. doi: 10.3390/md15040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talarico L.B., Damonte E.B. Interference in dengue virus adsorption and uncoating by carrageenans. Virology. 2007;363:473–485. doi: 10.1016/j.virol.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 20.Pomin V.H. Structure-function relationship of anticoagulant and antithrombotic well-defined sulfated polysaccharides from marine invertebrates. Adv. Food Nutr. Res. 2012;65:195–209. doi: 10.1016/B978-0-12-416003-3.00012-3. [DOI] [PubMed] [Google Scholar]
- 21.Jarmila V., Vavríková E. Chitosan derivatives with antimicrobial, antitumour and antioxidant activities--a review. Curr. Pharm. Des. 2011;17:3596–3607. doi: 10.2174/138161211798194468. [DOI] [PubMed] [Google Scholar]
- 22.Lee S.H., Ko C.I., Ahn G., You S., Kim J.S., Heu M.S., Kim J., Jee Y., Jeon Y.J. Molecular characteristics and anti-inflammatory activity of the fucoidan extracted from Ecklonia cava. Carbohydr. Polym. 2012;89:599–606. doi: 10.1016/j.carbpol.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 23.Gao Y., Zhang L., Jiao W. Marine glycan-derived therapeutics in China. Prog. Mol. Biol. Transl. Sci. 2019;163:113–134. doi: 10.1016/bs.pmbts.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Abdelnasser S.M., SM M.Y., Mohamed W.F., Asker M.M., Abu Shady H.M., Mahmoud M.G., Gadallah M.A. Antitumor exopolysaccharides derived from novel marine bacillus: Isolation, characterization aspect and biological activity. Asian Pac. J. Cancer Prev. 2017;18:1847–1854. doi: 10.22034/APJCP.2017.18.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao R., Jin W., Shan Y., Wang J., Liu G., Kuang S., Sun C. Marine bacterial polysaccharide EPS11 inhibits cancer cell growth via blocking cell adhesion and stimulating anoikis. Mar. Drugs. 2018;16:85. doi: 10.3390/md16030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Liu G., Ma W., Lu Z., Sun C. Marine bacterial polysaccharide EPS11 inhibits cancer cell growth and metastasis via blocking cell adhesion and attenuating filiform structure formation. Mar. Drugs. 2019;17:50. doi: 10.3390/md17010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang F.K., Han S., Xing B., Huang J., Liu B., Bordeleau F., Reinhart-King C.A., Zhang J.J., Huang X.Y. Targeted inhibition of fascin function blocks tumour invasion and metastatic colonization. Nat. Commun. 2015;6:7465. doi: 10.1038/ncomms8465. [DOI] [PubMed] [Google Scholar]
- 28.Wan L., Pantel K., Kang Y. Tumor metastasis: Moving new biological insights into the clinic. Nat. Med. 2013;19:1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- 29.De Craene B., Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 30.Fang S., Dai Y., Mei Y., Yang M., Hu L., Yang H., Guan X., Li J. Clinical significance and biological role of cancer-derived type I collagen in lung and esophageal cancers. Thorac. Cancer. 2019;10:277–288. doi: 10.1111/1759-7714.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenburg G., Hay E.D. Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J. Cell Biol. 1982;95:333–339. doi: 10.1083/jcb.95.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K., Corsa C.A., Ponik S.M., Prior J.L., Piwnica-Worms D., Eliceiri K.W., Keely P.J., Longmore G.D. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat. Cell Biol. 2013;15:677–687. doi: 10.1038/ncb2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J., Schwarzbauer J.E. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene. 2014;33:1649–1657. doi: 10.1038/onc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arneth B. Tumor microenvironment. Medicina (Kaunas) 2019;56:15. doi: 10.3390/medicina56010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J., Zhou S., Siech M., Habisch H., Seufferlein T., Bachem M.G. Pancreatic stellate cells promote hapto-migration of cancer cells through collagen I-mediated signalling pathway. Br. J. Cancer. 2014;110:409–420. doi: 10.1038/bjc.2013.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryschich E., Khamidjanov A., Kerkadze V., Büchler M.W., Zöller M., Schmidt J. Promotion of tumor cell migration by extracellular matrix proteins in human pancreatic cancer. Pancreas. 2009;38:804–810. doi: 10.1097/MPA.0b013e3181b9dfda. [DOI] [PubMed] [Google Scholar]
- 37.Grzesiak J.J., Tran Cao H.S., Burton D.W., Kaushal S., Vargas F., Clopton P., Snyder C.S., Deftos L.J., Hoffman R.M., Bouvet M. Knockdown of the β(1) integrin subunit reduces primary tumor growth and inhibits pancreatic cancer metastasis. Int. J. Cancer. 2011;129:2905–2915. doi: 10.1002/ijc.25942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y., Shen M., Song Q., Xie J. Biological activities and pharmaceutical applications of polysaccharide from natural resources: A review. Carbohydr. Polym. 2018;183:91–101. doi: 10.1016/j.carbpol.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Rahbar Saadat Y., Yari Khosroushahi A., Pourghassem Gargari B. A comprehensive review of anticancer, immunomodulatory and health beneficial effects of the lactic acid bacteria exopolysaccharides. Carbohydr. Polym. 2019;217:79–89. doi: 10.1016/j.carbpol.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Yu W., Chen G., Zhang P., Chen K. Purification, partial characterization and antitumor effect of an exopolysaccharide from Rhizopus nigricans. Int. J. Biol. Macromol. 2016;82:299–307. doi: 10.1016/j.ijbiomac.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Adebayo-Tayo B., Fashogbon R. In vitro antioxidant, antibacterial, in vivo immunomodulatory, antitumor and hematological potential of exopolysaccharide produced by wild type and mutant Lactobacillus delbureckii subsp. bulgaricus. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibrahim A.Y., Youness E.R., Mahmoud M.G., Asker M.S., El-Newary S.A. Acidic exopolysaccharide produced from marine Bacillus amyloliquefaciens 3MS 2017 for the protection and treatment of breast cancer. Breast Cancer (Auckl.) 2020;14 doi: 10.1177/1178223420902075. 1178223420902075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaschke F., Stawowy P., Goetze S., Hintz O., Gräfe M., Kintscher U., Fleck E., Graf K. Hypoxia activates beta(1)-integrin via ERK 1/2 and p38 MAP kinase in human vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2002;296:890–896. doi: 10.1016/s0006-291x(02)02033-8. [DOI] [PubMed] [Google Scholar]
- 44.Luo J., Zheng M., Jiang B., Li C., Guo S., Wang L., Li X., Yu R., Shi D. Antidiabetic activity in vitro and in vivo of BDB, a selective inhibitor of protein tyrosine phosphatase 1B, from Rhodomela confervoides. Br. J. Pharmacol. 2020;177:4464–4480. doi: 10.1111/bph.15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Liu G., Liu R., Wei M., Zhang J., Sun C. EPS364, a novel deep-sea bacterial exopolysaccharide, inhibits liver cancer cell growth and adhesion. Mar. Drugs. 2021;19:171. doi: 10.3390/md19030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MS data/data of proteomic analysis are deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier: PXD025155).