Summary
The specific interaction between rhizobia and legume roots leads to the development of a highly regulated process called nodulation, by which the atmospheric nitrogen is converted into an assimilable plant nutrient. This capacity is the basis for the use of bacterial inoculants for field crop cultivation. Legume plants have acquired tools that allow the entry of compatible bacteria. Likewise, plants can impose sanctions against the maintenance of nodules occupied by rhizobia with low nitrogen‐fixing capacity. At the same time, bacteria must overcome different obstacles posed first by the environment and then by the legume. The present review describes the mechanisms involved in the regulation of the entire legume–rhizobium symbiotic process and the strategies and tools of bacteria for reaching the nitrogen‐fixing state inside the nodule. Also, we revised different approaches to improve the nodulation process for a better crop yield.
The present review describes the mechanisms involved in the regulation of the entire legume‐rhizobium symbiotic process and the strategies and tools of bacteria for reaching the nitrogen‐fixing state inside the nodule. Also, we revised different approaches to improve the nodulation process for a better crop yield.
Introduction
Legumes (soybeans, clover, lotus, pea, bean and chickpea bean) are a fundamental source for human food and livestock feed. The selection of legume varieties with better growth relies on the application of traditional agronomic techniques. In the last years, transgenic legume plants with a desirable trait have been obtained using molecular biotechnology tools. Such is the case of glyphosate‐resistant transgenic soybean, an herbicide used to eliminate weed growth. In the 2018–2019 season, 364 million tons of soybean have been commercialized (FAO, 2019), giving an idea of the importance of this crop in the world economy.
Nitrogen is one of the dominant rate‐limiting nutrients in natural systems (Ferguson et al., 2010; Guignard et al., 2017). Legumes have developed a particular ability to establish a symbiotic relationship with certain bacteria from the soil, whereby they can utilize the atmospheric nitrogen (Sprent, 2008). Thus, the application of nitrogen fertilizers can be avoided by means of this symbiosis and the consequent biological nitrogen fixation (BNF) (Ferguson et al., 2010). This is auspicious because chemical fertilizers generate a negative effect on the ecosystem and involve the use of non‐renewable fossil energies (Ferguson et al., 2010; Galloway et al., 2013). The term rhizobia refers to bacterial species that can interact with the roots of legumes and induce the formation of structures called nodules, where gaseous di‐nitrogen is transformed into ammonium (BNF) and can thus be assimilated by the plant (Lindström and Mousavi, 2019). Most of the rhizobial species belong to families of the alpha‐proteobacteria class, including Rhizobiaceae (Rhizobium, Sinorhizobium, Allorhizobium, Pararhizobium, Neorhizobium and Shinella), Phyllobacteriaceae (Mesorhizobium, Aminobacter, Phyllobacterium), Brucellaceae (Ochrobactrum), Methylobacteriaceae (Methylobacterium, Microvirga), Bradyrhizobiaceae (Bradyrhizobium), Xanthobacteraceae (Azorhizobium) and Hyphomicrobiaceae (Devosia) (Lindström and Mousavi, 2019). Some species of the Burkholderiaceae family (Paraburkholderia, Cupriavidus) of the beta‐proteobacteria class can also induce an active nodulation process on legumes (Lindström and Mousavi, 2019). The nodulation process ranges from the interaction of bacteria with the root hairs to the formation of root nodules, where bacteria inside organelles called symbiosomes differentiate into nitrogen‐fixing bacteroids. While the plant benefits from the nitrogen supply, bacteria get carbon compounds provided by the plant. This is a specific process by which certain species of rhizobia induce the formation of nodules on particular legumes. However, some legumes such as Glycine max and Phaseolus vulgaris may be nodulated by more than one species of rhizobia (Ji et al., 2017; Andrews and Andrews, 2017) and some rhizobia like Rhizobium sp. NGR234 can nodulate different legume genera (Pueppke and Broughton, 1999). Using in vitro culture methods and more recently by applying metagenomic approaches, it has been found that symbiotic rhizobia cohabit in the nodules with other non‐nodulating rhizobia and non‐rhizobial species (Busby et al., 2016; Lu et al., 2017), which may affect process performance (Peix et al., 2015; Gano‐Cohen et al., 2016; Lu et al., 2017).
The nodulation process is controlled primarily by the plant (Sachs et al., 2018). Legumes benefit from and have evolved to allow this symbiosis, but such energy‐demanding process also needs to be regulated (Sachs et al., 2018). The plant has developed mechanisms to enable and prevent the entry of compatible and incompatible bacteria respectively (Clúa et al., 2018; Wang et al., 2018). Additionally, legumes can display sanctions against the maintenance of nodules occupied by rhizobia with low nitrogen‐fixing capacity (Kiers et al., 2003). Bacteria must overcome different obstacles posed first by the environment and then by the legume. In this context, rhizobia often adopt tools to survive under certain adverse soil conditions and have also developed mechanisms and strategies to facilitate the nodulation of a given legume (Soto et al., 2006; Clúa et al., 2018; Syska et al., 2019). Below we will discuss the different pathways and mechanisms behind the limitations of the nodulation process and the tools by which bacteria could adapt to and/or overcome them.
Mechanisms behind the limitations of the nodulation process and bacterial tools to overcome them
Bacterial survival in the rhizosphere
The diversity of soil bacterial communities decreases in close proximity to the root. Root exudation modulates the microbial community composition in the rhizosphere, that is, the soil in contact to plant roots (Lundberg et al., 2012; Bulgarelli et al., 2013; Philippot et al., 2013; Eng et al., 2020; Vives‐Peris et al., 2020). The maintenance of a bacterial population in the rhizosphere community partly depends on its chemotaxis towards certain root‐secreted components and its ability to use them as substrates (Bais et al., 2006; Badri et al., 2009; Chaparro et al., 2013). Flagellar function and bacterial motility are required to reach the root and can also influence the effectiveness of nodulation (López García et al., 2009). Competitive and cooperative microbe–microbe interactions also contribute to shape the overall community structure in the rhizosphere (Hassani et al., 2018; Han et al., 2020). In microbe–microbe interactions, the competitive effect depends on the growth rate, the ability to capture limiting nutrients such as iron or the presence of toxic compounds (Hassani et al., 2018; Eng et al., 2020). The example of iron limitation is very illustrative. Bacteria secrete siderophores to capture ferric iron, exhibiting membrane proteins for the uptake of the ferric‐siderophore complexes. Several rhizobial species secrete siderophores and may present receptors for both their own siderophores and heterologous ones (Geetha and Joshi, 2013). Competition for iron will depend on the quality and quantity of siderophores and receptors displayed by each strain (Geetha and Joshi, 2013; Eng et al., 2020).
Numerous bacterial species have information to produce toxins, which are used to restrict the growth of competitors. The structures, the mechanisms of action and the genomic organization of toxin‐encoding loci are highly diverse (Zhang et al., 2012; Jamet and Nassif, 2015; Scholl, 2017). Polymorphic toxins are multidomain proteins that can be secreted by different protein secretion systems (Zhang et al., 2012). Some bacteria translocate polymorphic toxins directly into target bacteria through the Type VI secretion system (T6SS) (Zhang et al., 2012; Ho et al., 2014). Bacteria possessing this toxic activity, which may have the function of DNase, RNase, phospholipase, amidase or muramidase, also code for the corresponding immunity protein (antitoxin) (Russell et al., 2011; Russell et al., 2013; Ma et al., 2014; Whitney et al., 2014). Interbacterial competition activity mediated by T6SS‐secreted toxins has been described for some soil bacteria as Agrobacterium tumefaciens or Pseudomonas putida (Bondage et al., 2016; Bernal et al., 2017). Despite the available information on T6SS and related toxin‐immunity protein pairs in different rhizobial strains (Bondage et al., 2016; Bernal et al., 2018; Salinero‐Lanzarote et al., 2019; Zalguizuri et al., 2019), a role for rhizobial putative T6SS toxins in interbacterial competition has not been demonstrated thus far (Lin et al., 2018). Another type of interbacterial toxins is bacteriocins (Holtsmark et al., 2008). The secretion of trifolitoxin, a bacteriocin‐type peptide with antibiotic activity, has been described in Rhizobium leguminosarum bv. trifolii strain T25 (Triplett and Barta, 1987). This peptide inhibits the growth of most species in the alpha‐proteobacteria class.
The rhizosphere community structure is influenced by cooperative effects in microbe–microbe interactions, probably due to nutritional interdependences, biofilm formation and molecular communication through microbial compounds (Hassani et al., 2018). The arbuscular mycorrhizal mycelium can release energy‐rich organic compounds, modifying the composition of plant exudates and the ensuing growth of other soil microbes in the rhizosphere (Barea et al., 2005).
The environment may offer adverse conditions such as watering, dryness and salinity, or present harmful components. Bacterial multiplication in these soils will rely on the capacity to tolerate the stress conditions and the presence of specific enzymes to eliminate the harmful components. For instance, the ability of rhizobia to accumulate trehalose promotes their growth and nodulation capacity in saline or dry soils (Reina‐Bueno et al., 2012; Moussaid et al., 2015), whereas glyphosate tolerance is a good example of tolerance to harmful compounds. Glyphosate inhibits the activity of 5‐enolpyruvylshikimic acid‐3‐phosphate synthase (EPSPS). This enzyme is found in plants, bacteria and fungi and is required for the biosynthesis of aromatic amino acids, which are fundamental for growth (Jaworski, 1972). Some microorganisms have a glyphosate‐resistant EPSPS enzyme (Barry et al., 1992). While soybean EPSPS is naturally sensitive to glyphosate, transgenic soybeans carry the EPSPS gene from Agrobacterium sp. CP4, which codes for a resistant enzyme (Padgette et al., 1995). Thus, glyphosate can be used as an herbicide in transgenic soybean crops. The presence of a glyphosate‐resistant enzyme in rhizobia would favour their survival in soils treated with this compound.
Factors affecting bacterial survival in the rhizosphere are schematized in Fig. 1.
Fig. 1.
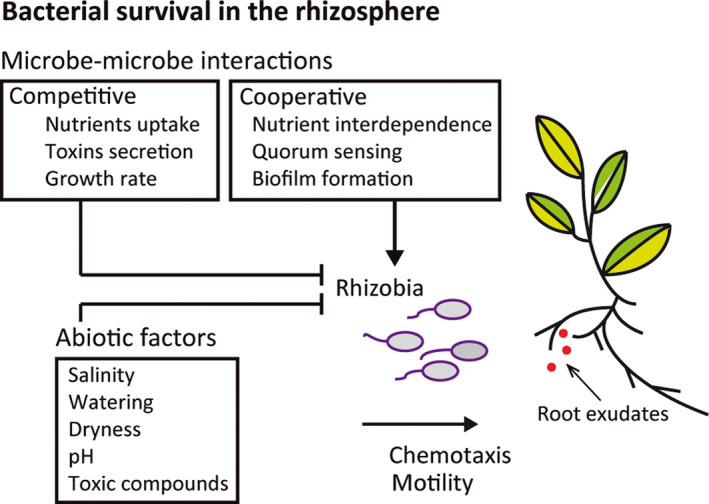
Factors affecting bacterial survival in the rhizosphere. Competitive and cooperative microbe–microbe interactions, along with the environmental conditions and root‐secreted compounds, contribute to shape the overall microbial community structure in the rhizosphere.
Specific recognition for initiation of the nodulation process
Flavonoids and isoflavonoids are components of the plant root exudates. Rhizobia express a transcriptional factor called NodD, which is activated by the specific flavonoid secreted by the compatible legume (Broughton et al., 2000; Chen et al., 2005; Peck et al., 2006). Different rhizobial species respond to different flavonoids. Often, rhizobia contain multiple nodD copies that encode different NodD isoforms (Ferguson et al., 2020). These isoforms may perform divergent roles, may be involved in distinct stages during symbiotic infection, or may be required under different environmental conditions, and have been described to enhance nodulation competitiveness or to extend symbiotic host range through perception of different plant signalling molecules (Demont et al., 1994; del Cerro et al., 2015, 2017, 2020; Kelly et al., 2018; Acosta‐Jurado et al., 2019; Ferguson et al., 2020). Once activated, NodD induces the expression of bacterial genes involved in the synthesis of the so‐called Nod factors (Broughton et al., 2000). Nod factors are lipochito‐oligosaccharides with different substituents that constitute a characteristic molecule which will be recognized by specific receptors (LysM receptor‐like kinases) on the root of the compatible legume (D’Haeze and Holsters, 2002; Madsen et al., 2003; Radutoiu et al., 2003; Smit et al., 2007). Once the legume recognizes the specific Nod factors, a cascade of signals is initiated in the plant tissue, promoting the physiological, morphological and molecular changes that characterize the nodulation process. The molecular signal cascade induced in the plant by the specific Nod factor recognition has been extensively revised (Ferguson et al., 2010; Oldroyd et al., 2011; Liu et al., 2018; Tang and Capela, 2020).
Morphological changes start with curling of the root hair tips, cell multiplication and differentiation in the root cortex, leading to the formation of the nodule primordium (Gage, 2004). Simultaneously, bacteria are entrapped in the curled root hair, proliferate and infect it through a tube‐like structure made of the plant cell wall, known as the infection thread (Gage, 2004). Infection threads grow towards the base of the root hair where rhizobia are released into the nodule primordium by endocytosis of the plasma membrane (Patriarca et al., 2002). Once inside the nodule cell, bacteria are surrounded by a membrane of plant origin called peribacteroid membrane, and differentiate into nitrogen‐fixing bacteroids (Brewin, 1998; Geurts and Bisseling, 2002; Gage, 2004). Depending on the nodulated legume, two types of nodules have been described, determinate and indeterminate. Determinate nodules have no persistent meristem, originate by multiplication of cells in the outer cortex and are round‐shaped (Crespi and Gálvez, 2000; Patriarca et al., 2002). Within them, bacteria differentiate reversibly to a bacteroid state. Determinate nodules are observed in soybean, lotus and bean. Indeterminate nodules derive from the multiplication of cells of the inner cortex; they are elongated, present persistent apical meristem and different areas corresponding to different states of development. In these nodules, bacterial differentiation to the bacteroid state is irreversible. Such nodules are formed in alfalfa, pea and clover, among others. Albeit to a lesser extent, another nodulation mechanism occurs through a crack‐entry infection process of plant tissues (Oldroyd and Downie, 2008; Markmann et al., 2012).
The molecular consequence of the plant signalling cascade induced by the Nod factors is a transcriptional and translational reprogramming that involves the activity of different transcriptional factors, micro RNAs, enzymes and phytohormones (Libault et al., 2010; Breakspear et al., 2014; Dalla Via et al., 2015; Boivin et al., 2016; Liu et al., 2018; Huang et al., 2019; Traubenik et al., 2020). More recently, another bacterial component involved in the specific recognition process, the exopolysaccharide (EPS), has been described (Kawaharada et al., 2015). Different bacterial species and even different strains from the same species differ in the structure of their EPS (Skorupska et al., 2006). The role of EPS in the infection process has been extensively studied. Mutants affected in EPS biosynthesis show defects in their ability to initiate or elongate infection threads (Breedveld et al., 1993; Cheng and Walker, 1998). Interestingly, a specific Mesorhizobium japonicum R7A EPS receptor called EPR3 (LysM receptor‐like kinase) was described in Lotus japonicus Gifu. This receptor distinguishes between EPS variants, acting positively towards compatible EPS and negatively towards incompatible EPS by restricting the entry of incompatible strains into epidermal cells of the root (Kawaharada et al., 2015; Kawaharada et al., 2017).
Recently, rhizobial transfer RNA (tRNA)‐derived small RNA fragments (tRFs) were described as positive regulators of nodulation through silencing of legume host genes engaged in a negative regulation of this process (Ren et al., 2019). Host gene repression is achieved through mRNA base‐pairing at the so‐called target sites with the rhizobial tRFs and their subsequent cleavage (Ren et al., 2019). Silencing of tRFs or overexpressing their target host genes inhibits nodule formation, suggesting a new mechanism by which bacteria can regulate nodulation through tRFs (Ren et al., 2019). This mechanism was described for B. japonicum–Glycine max interaction, but tRFs were also identified in other rhizobia (Ren et al., 2019). Further research is needed to determine the relevance of this host gene regulation in other rhizobium–legume interactions.
Plant defence response
In response to pathogen attack, plants trigger a first defence response through the recognition of general elicitors known as pathogen‐/microbe‐associated molecular patterns (PAMPs/MAMPs) (Wanke et al., 2021). Flagella, elongation factor thermo unstable (EF‐Tu), different polysaccharides as well as different bacterial surface proteins can be recognized as PAMPs by the plant (Chisholm et al., 2006; Wanke et al., 2021). The same as pathogens, rhizobia that present MAMPs will induce a defence response in the plant. Unlike pathogens, rhizobial flagella are not recognized as MAMPs (López‐Gómez et al., 2011). The recognition of PAMPs occurs through plant‐specific receptors (LysM receptor‐like kinases); this first defence response is called PAMP‐triggered immunity (PTI). Bacteria have acquired type three secretion system (T3SS) through either horizontal gene transfer or adaptation of the flagellar apparatus, by which they translocate effector proteins into the plant cells and thus suppress the PTI defence (Mudgett, 2005; Chisholm et al., 2006). In turn, the plant encodes for resistance proteins that recognize these effectors (or molecules modified by them) and induce a hypersensitivity response (HR) called effector‐triggered immunity (ETI) (Mudgett, 2005; Chisholm et al., 2006). It has been reported that bacterial T3SS secretes additional effectors that would suppress HR (Chisholm et al., 2006). Several rhizobia also code for a T3SS (Viprey et al., 1998; Krause et al., 2002; Krishnan et al., 2003; Hubber et al., 2004; Vinardell et al., 2004; Sánchez et al., 2009). In addition to the induction of nod genes expression, flavonoid‐activated NodD also upregulates the expression of TtsI transcriptional factor, which in turn induces transcription of components and effectors of the T3SS (Krause et al., 2002; Marie et al., 2004). A role in suppressing the defence response has been described for some rhizobial T3SS effectors (Bartsev et al., 2004; Xin et al., 2012). Accordingly, mutant strains deficient in the synthesis or secretion of T3SS effectors have shown reduced symbiotic properties on specific host plants (Skorpil et al., 2005; Kambara et al., 2009; Sánchez et al., 2012). On the other hand, direct or indirect recognition of T3SS effectors by plant cells expressing specific resistance (R) proteins can result in effector‐triggered defence responses that negatively affect rhizobial infection (Staehelin and Krishnan, 2015; Sugawara et al., 2018). Thus, T3SS participates in the specific recognition that determines which strain can nodulate a given legume. The determination of compatibility by T3SS effectors could occur at the infection and post‐infection stages (Kusakabe et al., 2020). The presence of specific R genes in the plant implies a restriction towards rhizobia displaying specific effectors (Yasuda et al., 2016; Fan et al., 2017) which may create a host‐range limitation for a given rhizobium strain as a result of T3SS functionality (Hubber et al., 2004; Jiménez‐Guerrero et al., 2020). For example, M. japonicum MAFF303099 does not form effective nodules in Leucaena leucocephala unless its T3SS or effectors are mutated (Hubber et al., 2004). Examples of altered compatibility have also been reported for other legume–rhizobium partnerships (Jiménez‐Guerrero et al., 2021). Rhizobial strains carrying a mutation in the T3SS or in a specific T3SS effector exhibited an extension of their host range to other legumes (Jiménez‐Guerrero et al., 2021). Thus, both positive and negative effects on symbiotic performance have been attributed to T3SS effectors. The combination of these effects will determine whether the T3SS acts positively, negatively, or has no effect on nodule formation in a particular legume (Skorpil et al., 2005; Kambara et al., 2009; Sánchez et al., 2012).
In addition to the T3SS, other bacterial protein secretion systems have been described as having a role in the symbiotic process: type IV and type VI secretion system (T4SS and T6SS respectively) (Nelson and Sadowsky, 2015; Lin et al., 2018; Salinero‐Lanzarote et al., 2019). In M. japonicum R7A, T4SS has an analogous role to that attributed to T3SS in M. japonicum MAFF303099 (Hubber et al., 2004).
Bacterial polysaccharides also modulate the plant defence response (Battisti et al., 1992; Aslam et al., 2008; Jones et al., 2008; Wanke et al., 2021). It was hypothesized that the EPS receptor 3 (EPR3)‐compatible EPS recognition step in M. japonicum R7A–Lotus symbiosis could be the signal for suppressing the plant defence response and the subsequent sustained infection (Kawaharada et al., 2015). Another polysaccharide involved in the nodulation process is cyclic glucan (Geremía et al., 1987; D´Antuono et al., 2005). It has been determined that cyclic glucan had a role in the suppression of the plant defence response in the plant pathogen Xanthomonas (Rigano et al., 2007). In Nicotiana benthamiana, inoculation of a Xanthomonas mutant strain affected in cyclic glucan biosynthesis induced an observable defence response by the increase in callose deposition, while prior infiltration of the leaves with purified cyclic glucan suppressed such defence response (Rigano et al., 2007). Cyclic glucan was shown to be essential for the onset of infection in rhizobia–legume interactions; however, the pleiotropic effect of its mutation, affecting membrane stability, must be considered (D'Antuono et al., 2005). This would explain why cyclic glucan mutants cannot penetrate infection threads. Empty pseudo‐nodules formed after inoculation with cyclic glucan mutants had a higher level of phenolic compounds, indicating an increase in the defence response (D’Antuono et al., 2005). However, the increased defence response may be due to the absence of bacteria and not to the specific absence of glucan. It has been reported that bacterial cyclic glucan competes with a hepta‐beta‐glucose for its binding to a solubilized membrane fraction of soybean roots, suggesting the presence of glucan receptors in the plant (Bhagwat et al., 1999). So far, there is no clear evidence of its role as a defence suppressor in rhizobia–legume symbiosis.
Lipopolysaccharide (LPS) structure was relevant for bacterial symbiotic capacity in some rhizobia–legume interactions (Noel et al., 2000; D´Antuono et al., 2005; Di Lorenzo et al., 2020). LPSs have been attributed a role in protecting against (D´Antuono et al., 2005) and suppressing (Perotto et al., 1994; Scheidle et al., 2005; Tellström et al., 2007) the plant defence response. It has been recently demonstrated that Nod factors possess the ability to suppress the PAMP‐induced plant defence in both legumes and non‐legumes (Liang et al., 2013). Plant transcriptome analysis revealed many induced and repressed defence genes throughout the nodulation process (Kouchi et al., 2004; Lohar et al., 2006). Comparative analyses of the transcriptional profile of legume genes upon inoculation with wild‐type and mutant strains affected in different bacterial components allowed to explore their involvement in plant defence response regulation (Kouchi et al., 2004; D´Antuono et al., 2008; Dalla Via et al., 2015). Defence response induction results in callose deposition, production of phenolic compounds and phytoalexins, increased levels of reactive oxygen species (ROS) and induction of phytohormones such as salicylic acid and ethylene (Soto et al., 2009; Katagiri and Tsuda, 2010; Liu et al., 2018; Syska et al., 2019). Besides the mechanisms for suppression of the plant defence response, bacteria have also developed tools to tolerate and eliminate ROS through the activity of ROS‐scavenging enzymes such as catalase, superoxide dismutase and peroxidase, or by accumulating antioxidant compounds such as glutathione (Ma et al., 2002; Soto et al., 2006; Becana et al., 2010; Syska et al., 2019). Some bacteria produce rhizobitoxine, an ethylene synthesis inhibitor, to decrease the plant ethylene levels locally generated, while others possess the enzyme ACC deaminase which degrades 1‐aminocyclopropane‐1‐carboxylate (ACC), the substrate for ethylene biosynthesis (Ma et al., 2002).
Nodule‐cysteine‐rich (NCR) peptides
Host–rhizobium compatibility is defined also at the late stage of the nodulation process, that is, during bacterial differentiation into bacteroids and in the determination of bacteroids viability. A diversity of peptides is expressed in the nodules formed in inverted repeat‐lacking clade (IRLC) legumes such as Medicago spp. (alfalfa), that is, indeterminate nodules where bacteria are irreversibly differentiated into bacteroids. These peptides are known as NCR peptides due to the presence of various cysteine residues (Mergaert et al., 2003; Alunni et al., 2007; Lima et al., 2020). Several of these peptides present in vitro antimicrobial activity (Van de Velde et al., 2010) that may affect membrane permeability and cause cell elongation, DNA duplication and, eventually, bacterial death. The NCR peptides have evolved from defensins, which form part of the innate immunity of plants (Sathoff and Samac, 2018). The activity of NCR peptides in the nodule does not necessarily lead to the death of bacteria, but rather to irreversible terminal bacteroid differentiation (the nitrogen fixation state), characterized by increased membrane permeability, polyploidy, larger size and alteration of the cell shape (Kondorosi et al., 2013). The NCR peptides enter the bacteria through membrane protein BacA (Wehmeier et al., 2010; Guefrachi et al., 2015). The fact that lethal effects were not observed inside the nodule has been attributed to different factors, such as the presence of sublethal concentrations of NCR peptides or the transport function of BacA, which prevents the accumulation of NCR peptides in the membrane and therefore their negative effect on it (Van de Velde et al., 2010; Haag et al., 2011). Some strains of S. meliloti have plasmids that code for a metalloprotease that degrades NCRs (Hrrp protease). This protease, while promoting bacterial viability, has a negative effect on nitrogen fixation (Price et al., 2015). The outcome of the action of Hrrp protease depends on both the host variety and the bacterial strain (Price et al., 2015). Besides, NCR peptides also affect bacteroid viability. Their involvement in the recognition and subsequent elimination of incompatible bacteria has been described (Yang et al., 2017). In M. truncatula, NCRs encoded by NSF1 and NSF2 genes recognize and degrade bacteroids of the Sinorhizobium meliloti Rm R41 strain, giving the Fix– phenotype (formation of non‐fixing nodules) and advancing senescence. On the other hand, they do not affect bacteroids of the A145 strain (Yang et al., 2017). In conclusion, the dialogue between NCRs in nodules, the extent of NCR antibacterial activity, the sensitivity of the rhizobial strain and the mechanisms it has developed to tolerate or eliminate these NCR peptides will collectively determine the nitrogen fixation phenotype of the formed nodule (Price et al., 2015; Lindström and Mousavi, 2019; Syska et al., 2019). Given the multiple bacterial targets of NCRs, the development of resistance against them is unlikely (Lima et al., 2020). In non‐ICRL legumes such as soybean, lotus and bean, determinate nodules are induced and bacteria are reversibly differentiated into bacteroids. These legumes do not code for NCR peptides (Mergaert et al., 2003). While the existence of incompatibility manifested at the nitrogen fixation step has been described for certain combinations of non‐ICRL legumes–bacterial strains (Parniske et al., 1994), the underlying mechanism has not yet been elucidated (Wang et al., 2018).
Autoregulation of nodule number
Legumes have a systemic mechanism for nodule number regulation that prevents from unnecessary waste of energy (Caetano‐Anollés and Gresshoff, 1991). Autoregulation of nodulation (AON) is a negative feedback system mediated by long‐distance signalling between shoots and roots. Small peptides (12–13 aminoacids) that constitute the signal generated when the nodulation process begins at roots are involved in AON (Mortier et al., 2012a). The CLAVATA3/embryo surrounding region‐related (CLE) small peptides named LjCLE‐RS1 and LjCLE‐RS2 are nodule‐specific and were first described in L. japonicus (Okamoto et al., 2009). Orthologues of these peptides have also been described in other legumes (Reid et al., 2011; Mortier et al., 2012a; Hastwell et al., 2017). The negative effect of these CLE peptides on nodulation involves specific kinase receptors in the shoot (Liu et al., 2018). It was suggested that CLE peptides travel through the xylem from the roots to the shoot where they bind to their specific receptor (Okamoto et al., 2013). The CLE‐specific receptor in L. japonicus is LjHAR1 (Okamoto et al., 2009), and orthologues of this receptor have been reported in other leguminous species (Krussell et al., 2002; Searle et al., 2003; Schnabel et al., 2005). The perception of shoot CLE peptides induces the biosynthesis of a shoot‐derived inhibitor (SDI) for further nodule development (Sasaki et al., 2014). In Lotus japonicus, it was proposed that cytokinin hormone could act as a SDI (Sasaki et al., 2014). The signal transduction cascade induced by Nod factors that will give rise to nodule formation will also induce the AON mechanism (Mortier et al., 2012b). Besides, it has been described that Nod factor signalling induces the expression of other factors which will negatively regulate the AON mechanism (Lei et al., 2019). This suggests that nodule number regulation is the result of a delicate balance involving different signal pathways induced by rhizobia (Lei et al., 2019). Specific CLE peptides also participate in the mechanism of inhibition of nodule formation in soils with high nitrate levels (Reid et al., 2011).
Sanctions for poor nitrogen fixation
Nodules containing nitrogen‐fixing rhizobia have been shown to develop normally, while ineffective nodules tend to remain small (Sachs et al., 2010; Oono et al., 2011; Regus et al., 2015). The capacity of legume hosts to target ineffective and less‐effective rhizobia and reduce their fitness relative to beneficial genotypes is termed sanctions (Kiers et al., 2003; Oono et al., 2011). This has been observed in both determinate and indeterminate nodules. The plant can sanction ineffective bacteria in the nodule by inducing a cellular senescence process (Regus et al., 2017). Nodule senescence is the nodule ageing process, also defined as the nodule organ break down (Thomas, 2013). Nodules containing both effective and ineffective rhizobia may include different sectors with different bacterial genotypes. The senescence characteristics are located only in cells occupied by ineffective rhizobia (Regus et al., 2017). The mechanism by which the lack of nitrogen fixation results in accelerated senescence is not yet known. Despite the plant ability to eliminate uncooperative strains, nodules occupied by less‐effective rhizobia are observed in nature, probably because of the existence of a variation in the degree of control exerted by different host genotypic variants on these rhizobia (Wendlandt et al., 2019).
Figure 2 presents a scheme of the different mechanisms developed by legumes to regulate or limit the nodulation process, together with the bacterial tools or strategies to overcome them.
Fig. 2.
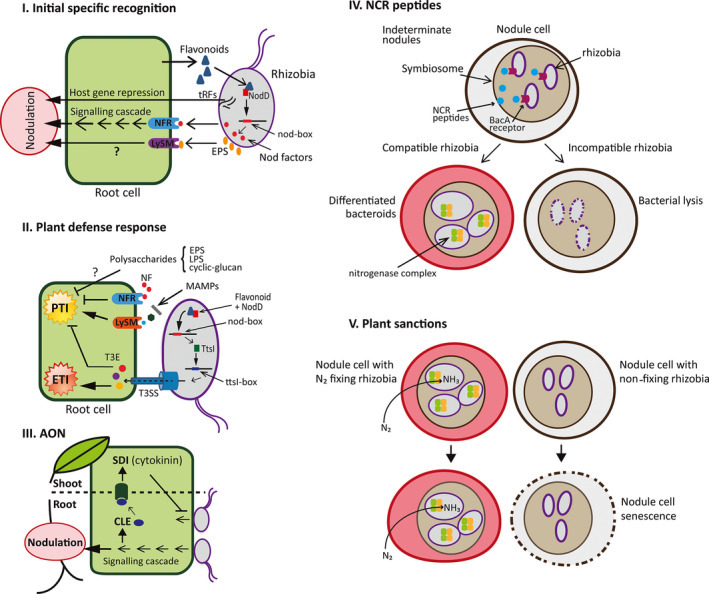
Mechanisms behind the limitations of nodulation and bacterial tools for overcoming them. (I) Flavonoids exuded by the legume induce, through the activation of the transcriptional factor NodD, the production of Nod factors in compatible bacteria. Nod factors are recognized by specific legume receptors and induce a signal cascade in the plant tissue responsible for the first physiological, biochemical and transcriptional changes that lead to the initiation of the nodulation process. EPS is also recognized by root‐specific receptors, being involved in the initial specific recognition. Rhizobial transfer RNA (tRNA)‐derived small RNA fragments (tRFs) are also involved in the regulation of the nodulation process through silencing of legume host genes. (II) Following MAMPs recognition, a first defence response is induced in the plant (PTI). Bacterial T3SS and its effectors, induced through NodD activation by plant flavonoids, participate in the suppression of the plant defence response. Resistance proteins recognize T3SS effectors (or molecules modified by them) and induce a plant defence response (ETI). T3SS effectors would also participate in the suppression of ETI (not drawn). A role for polysaccharides and Nod factors (NF) in the suppression of the plant defence response has also been proposed. (III) Autoregulation mechanism of nodule number. The initial signalling cascade induced by Nod factors leads to the induction of CLE peptide biosynthesis on roots. These peptides are specifically recognized by shoot cell receptors, inducing the production of a shoot‐derived inhibitor (SDI) of further nodule development. It was proposed that cytokinin hormone would act as an SDI. (IV) In indeterminate nodules, NCR peptides are involved in the differentiation of compatible bacteria to bacteroids. In addition, a role for NCR peptides in the recognition of incompatible bacteria and their subsequent elimination has been described. Transport of NCR through BacA avoids NCR accumulation at bacteroid membranes. (V) The plant can sanction ineffective bacteria present in the nodule by inducing a senescence process in the cells containing them.
Improve the nodulation process to improve legume crop yield
Evolutionarily, the legume–rhizobium interaction has been going in the direction of mutual benefits. Thus, the plant obtains assimilable nitrogen and bacteria obtain the necessary nutrients for their development and a niche without competition to multiply. Taking advantage of this beneficial interaction, bacterial inoculants can be used on legume crops to overcome the problem of nitrogen limitation in soils. However, it is often observed that the inoculated strain does not predominantly occupy the nodules of the cultivated plant (Rodriguez Blanco et al., 2010). The symbiotic process depends on many variables that will determine whether a strain can compete efficiently and will lead to an effective nitrogen fixation process. The aim of the inoculant industry is to use those bacteria that allow the best crop yield. In search of this higher performance, it looks for.
Improved competitiveness
Inoculated bacteria must compete with native soil rhizobia (van Dillewijn et al., 2001; Rodriguez Blanco et al., 2010; Geetha and Joshi, 2013; Irisarri et al., 2019). Nodulation competitiveness refers to the ability of the inoculated bacteria to induce more nodules on the roots of the compatible legume than the native bacteria from the rhizosphere (Brewin et al., 1983). Nodulation competitiveness could be the consequence of a better performance of the inoculated bacteria at some or several steps of the process. At first, a higher competitive capacity could arrive from a better soil survival capacity. Therefore, the relevance of enzymatic functions and activities for survival in a particular soil condition or within the microbial community or favouring a chemotactic response or a higher multiplication in the rhizosphere or plant surface by an increased capacity to utilize plant exudates should be considered. For instance, proline dehydrogenase is required to metabolize the proline found in roots exudates. Its mutation in rhizobia negatively affects colonization and therefore nodulation competitiveness of S. meliloti on alfalfa roots (Jiménez‐Zurdo et al., 1995).
If we focus on the competitive capacity for the nodulation process itself, a better performance in either of the subsequent stages (initiation of infection, multiplication in the infection threads, symbiosome formation and transformation to bacteroids) will contribute to a better nodulation competitiveness. In this respect, the alteration of bacterial components involved in some of these steps may affect competitiveness. For instance, modifications in the Nod factors (Lamrabet et al., 1999; Madinabeitia et al., 2002) or the EPS (Kawaharada et al., 2015), involved in the initial recognition step, negatively affected nodulation competitiveness. Similar results were described for a modification in the LPS structure that provokes bacterial accumulation in infection threads (D´Antuono et al., 2005) and for a mutation in a bacterial peptidase that protects against NCR peptides (Arnold et al., 2017). There are also examples in which an altered bacterial function results in improved competitiveness. In M. japonicum MAFF303099, mutants in T3SS effectors showed higher competitiveness on Lotus tenuis cv. Esmeralda than the wild‐type strain (Sánchez et al., 2012). Disruption of S. medicae nolR, a negatively acting transcriptional regulator of Nod factor biosynthesis, led to increased competitiveness on Medicago truncatula (Sugawara and Sadowsky, 2014). Rhizobial strains overexpressing its own ACC deaminase (Conforte et al., 2010) or expressing an exogenous one (Ma et al., 2004) were more competitive in nodulation.
Higher number of fixing nodules per root
Several mechanisms regulate nodule number. Local regulation is provided by the level of different phytohormones (Mortier et al., 2012b; Ferguson and Mathesius, 2014). Increased ethylene levels generated after Nod factors perception (Reid et al., 2018) negatively regulate nodule number (Nukui et al., 2000; Nascimento et al., 2018). Rhizobia can degrade the substrate for ethylene biosynthesis through ACC deaminase activity, resulting in decreased ethylene local levels and increased number of nodules (Ma et al., 2004; Nascimento et al., 2018). As previously mentioned, the number of fixing nodules per root is also determined by the plant systemic AON mechanism, which relies on the induction of CLE peptides in the plant (Liu et al., 2018). While this induction is already observed in the nodule primordium, the strength and range of the signal will depend on the number, activity and state of development of the nodules (Mortier et al., 2012b). Mature nitrogen‐fixing nodules display greater inhibition for further nodulation than less effective ones (Li et al., 2009). This could be the reason why an increase in nodule number is usually observed in mutants that induce non‐fixing nodules, as is the case with mutants in cyclic glucan (D'Antuono et al., 2005) or the Rieske iron/sulphur protein (a component of the respiratory electron transport chain required for nitrogen fixation inside the nodule) (Pickering et al., 2012; Basile et al., 2018).
Improved nitrogen fixation efficiency
Efficiency during the nitrogen fixation step is concerned with bacterial nitrogenase activity. The nitrogenase complex, coded by the nifHDK genes, is functional under microoxic conditions. These conditions are provided by the barrier that constitutes the external wall of the nodule and by plant leghaemoglobin (Ott et al., 2005). The presence of more efficient nitrogenases could increase the amount of fixed nitrogen and, therefore, crop yield. Likewise, a greater nitrogenase expression would also contribute to an improvement in performance. Regulation of the nitrogen fixation process is under nifA, fixLJ and fixK genes (Lindström and Mousavi, 2019). Besides the microoxic conditions required, nitrogenase expression can be controlled through the action of several inducers and suppressors (Soberón et al., 2001; Mesa et al., 2008; Torres‐Quesada et al., 2010; Quelas et al., 2016). For example, it has been suggested that the mutation on PhaR in B. diazoefficiens could have a positive effect on the amount of nitrogen fixed, since it has been described as a negative regulator of the FixK2 transcriptional factor (Quelas et al., 2016).
The release of hydrogen in the nitrogen fixation process implies loss of energy (Schubert and Evans, 1976). It was proposed that the hydrogenase enzyme involved in H2 recycling and present in some rhizobial strains (Baginsky et al., 2002) would help improve fixation efficiency (Schubert et al., 1978; Albrecht et al., 1979).
In the nodule, nitric oxide (NO) levels increase (Baudouin et al., 2006) due to the utilization of nitrite instead of O2 as the final electron acceptor in the mitochondrial electron transport chain under microoxic conditions (Igamberdiev and Hill, 2009). Nodule NO levels have also been observed to increase under stress conditions (Fancy et al., 2017; Syska et al., 2019). Nitric oxide not only participates in signal transduction in the plant–symbiont interaction (Ferrarini et al., 2008; Boscari et al., 2013), but also has a toxic effect. It has been described that NO inhibits nitrogenase activity (Trinchant and Rigaud, 1982; Kato et al., 2010) as well as nitrogen‐fixing activity through repression of the nitrogenase genes (Sánchez et al., 2010). The plant has mechanisms for NO detoxification (Berger et al., 2019) and bacteria also possess NO‐degrading enzymes (Cam et al., 2012; Berger et al., 2019; Syska et al., 2019). Both symbiotic partners would contribute to balance different signalling, metabolic and toxic effects, and to maintain symbiotic N2 fixation (Berger et al., 2019).
An exhaustive revision about the different processes involved in the effectiveness of nitrogen fixation in rhizobia can be found in Lindström and Mousavi (2019).
Increased duration of the nitrogen fixation stage
A longer nitrogen fixation stage implies maintaining bacteroid viability and/or delaying senescence. Senescence is part of the normal nodule development; it depends on plant regulation and is the least studied stage of the nodulation process. When the plant needs to direct the carbon source to seed development, it no longer supports the costly process of nitrogen fixation in the nodule. Senescence is followed by the release of rhizobia (differentiated bacteroids from indeterminate nodules and undifferentiated cells from determinate nodules) to the soil (Denison and Kiers, 2004). Besides, senescence can be induced early when the plant is under stress or by ineffective nitrogen fixation (Regus et al., 2017). A role for NCR peptides in early nodule senescence has been described in Medicago truncatula (Yang et al., 2017). High levels of NO, such as those induced by mutants in NO‐scavenging enzymes, also induced premature senescence in nodules (Cam et al., 2012). In contrast, overexpression of these enzymes led to a delay in nodule senescence (Cam et al., 2012). The activity of ROS‐scavenging enzymes has also been shown to delay senescence (Redondo et al., 2009).
The toxin–antitoxin (TA) system has been described to participate in some bacterial symbiotic processes. This system is involved in the regulation of bacterial metabolism in response to environmental stress (Goeders and Van Melderen, 2014; Lobato‐Márquez et al., 2016; Syska et al., 2019). While toxins can affect protein synthesis, DNA replication, cell wall synthesis and DNA or RNA integrity, among other functions (Lobato‐Márquez et al., 2016), antitoxins inhibit this activity. There are different types of TA systems based on the nature (RNA or protein) and mode of action of the antitoxin (Page and Peti, 2016). In type II TA system, the antitoxin is a protein which counteracts the effect of the toxin by forming a complex with it (Yamaguchi and Inouye, 2011). Since the antitoxin is more unstable than the toxin, it is eliminated more quickly by protease degradation under a stress situation (Gerdes et al., 2005; Lobato‐Márquez et al., 2016). Once the toxin is free of its antitoxin, it acts on bacterial metabolism, resulting in the arrest of bacterial growth and persistence or cell death (Hayes and Kędzierska, 2014). In S. meliloti, several TA modules have been described. One of them is vapBC‐5, corresponding to the type II TA system (Lipuma et al., 2014). Deletion of the complete TA vapBC‐5 module had no effect on the nodulation and nitrogen fixation phenotype of M. sativa. The antitoxin mutation produced less efficient nodules for nitrogen fixation and lower plant yields. In contrast, the toxin mutation produced higher nitrogen fixation efficiency and plant performance compared with the wild‐type strain (Lipuma et al., 2014). Besides, analysis of the expression of markers of active nitrogen‐fixing zones showed that the mutation in the toxin delayed the senescence phenotype (Lipuma et al., 2014). The role of other TA modules in S. meliloti has also been studied (Oláh et al., 2001). Inside the nodule, bacteria must adapt to stress conditions given by acidic pH, microoxia, ROS and antimicrobial peptides. It was proposed that the activity of different bacterial TA modules would play a role in the adaptation to these stresses by limiting the symbiotic interaction and contributing to the appearance of the senescence stage (Syska et al., 2019). It was also suggested that the inactivation of toxin genes in S. meliloti could constitute a strategy for improving alfalfa production (Lipuma et al., 2014).
Strategies to improve inoculants
In addition to the leguminous specie to be nodulated, soil conditions should be considered when choosing a suitable inoculant. The carrier used and the way the inoculation is applied are among the variables that determine the inoculant efficiency. We will not discuss these topics here; they can be revised in (Hungria et al., 2005; Albareda et al., 2008; Ruiz‐Valdiviezo et al., 2015; O’Callaghan, 2016; Sanches Santos et al., 2019). The inoculant potential will depend mainly on the microbial strain (or strains) used. The strategy is to develop an inoculant based on the use of rhizobial strains that can survive in the soil conditions that are competitive over the native strains that properly complete the whole process and have an optimal nitrogen fixation capacity. The inoculant may also contain non‐nodulating strains that promote the process, such as plant growth‐promoting rhizobacteria (PGPR) or mycorrhizas. The different strategies that could be used for the development of an appropriate inoculant are discussed below.
Selection of native strains that are favoured by the mentioned characteristics
Numerous studies have tried to find highly competitive and highly efficient nitrogen‐fixing native soil strains. One strategy consists of isolating the different strains found in the nodules, selecting highly efficient nitrogen‐fixing strains and, finally, selecting more competitive strains than at least the commercial strain through competitiveness assays performed under laboratory and field conditions (Melchiorre et al., 2011; Irisarri et al., 2019). Competitiveness between different strains over a given legume may vary depending on soil conditions (Ji et al., 2017). Therefore, the optimal strain for each crop in different types of soil may not be the same. The selection of native strains ensures the best adaptation to a particular soil. For instance, in Ethiopia, a local strain isolated from acidic soils, highly adapted to those conditions, produced enhanced soybean yields (Muleta et al., 2017). New strains of Mesorhizobium have been identified in nodules of Lotus tenuis grown in flooded soils (Estrella et al., 2020) and saline–alkaline soils (Sannazzaro et al., 2018) in Argentina. A possible strategy would be the isolation of native glyphosate‐tolerant Bradyrhizobium strains from glyphosate‐treated soils to be used as inoculant (Kuykendall, 2012). In soils with a history of legume crops, native strains lacking the ability to nodulate and/or fix nitrogen can acquire it from inoculated strains through horizontal gene transfer. This can give rise to new strains with greater adaptability to the soil, together with the ability to nodulate. Searching for these strains may be a good source of inoculants with a better capacity to inhabit the nodules (Melchiorre et al., 2011).
Co‐inoculation with PGPR and mycorrhizas
Both PGPR and mycorrhizas improve plant growth. The former are soil bacteria with the ability to either be part of the rhizosphere or colonize the root; some can even penetrate and grow between root cells (endophytes). The name of these bacteria is derived from their role in favouring plant growth by producing phytohormones as indole acetic acid (IAA), by fixing N2 in free life, and by producing siderophores (Bhattacharyya and Jha, 2012; Sanches Santos et al., 2019). They can also prevent the multiplication of plant pathogens through spatial or nutrient competition or via the secretion of toxic compounds or the induction of plant defence responses (Vacheron et al., 2013; Peix et al., 2015). Arbuscular mycorrhiza (root‐associated fungi) can solubilize the insoluble phosphate present in soil (Baslam et al., 2014) and increase surface absorption through their association with roots (Barea et al., 2017). The co‐inoculation of PGPR and mycorrhizas has a synergic effect on both root colonization by mycorrhizas and bacterial growth, which results in the stimulation of plant growth (Artursson et al., 2006). In some systems, rhizobial co‐inoculation with PGPR, mycorrhizas or both elicited a positive effect on nodulation process efficacy and plant performance (Hungria et al., 2015; Korir et al., 2017; Nascimento et al., 2018; Raklami et al., 2019; Alemneh et al., 2020; Swarnalakshmi et al., 2020). For instance, rhizobial co‐inoculation with endophytes having ACC deaminase activity increased the nodulation abilities of both alpha‐ and beta‐rhizobia, resulting in increased leguminous plant growth (Nascimento et al., 2019). The dual inoculation of Rhizobium with either Pseudomonas putida, P. fluorescens or Bacillus cereus on pigeon pea significantly increased plant growth and nodulation (Tilak et al., 2006). Co‐inoculation of Rhizobium tropici and Azospirillum brasilense strains on common bean enhanced crop yields (Souza and Ferreira, 2017). Rhizobium etli nodulation efficiency on Phaseolus vulgaris was improved by co‐inoculation with Rhizobium fabae, showing that the quorum‐sensing signals from the commensal bacteria were involved in this better performance (Miao et al., 2018). Dual inoculation of R. leguminosarum and a mixture of arbuscular mycorrhizal fungi on fava bean legumes grown on alkaline soils significantly increased nodule mass and number and dry weight of roots and shoots compared with individual inoculation (Abd‐Alla et al., 2014). Currently, numerous inoculants are designed as rhizobial combinations with PGPR and/or mycorrhiza (Bashan et al., 2014; Vejan et al., 2016; Sanches Santos et al., 2019).
Genetic engineering in bacteria
Knowledge of the molecular basis of the rhizobium–legume interaction enables the design of new bacterial strains with increased or added capacities that could improve some steps of the nodulation process, leading to successful nodulation. This is usually intended to modify a single function in strains used as inoculants. Several studies have reported the construction of such strains as well as improvements in nodulation performance and plant yields in controlled laboratory experiments (Robleto et al., 1997; Geetha and Joshi, 2013). Yet, examples of field experiments are scarce (Scupham et al., 1996; Dillewijn et al., 2001; Hirsch, 2005). The modification of a strain with the traits sought could be an appropriate strategy, despite restrictions to accept genetically engineered strains as inoculants (Geetha and Joshi, 2013). Modifications for nodulation improvement might consist in the heterologous expression of a new function (Robleto et al., 1997), the expression of an endogenous function at an increased level (Conforte et al., 2010) or the deletion of a specific function (Hubber et al., 2004). The incorporation of a new or an increased function should be achieved via chromosomal integration instead of being carried on a plasmid to avoid its transfer to other soil bacteria or loss in the absence of selection pressure (Conforte et al., 2010). In case, the modification involves genes present in the symbiotic island, the fact that these regions may also be transferred to another bacterium in the rhizosphere should be considered (Bamba et al., 2019). The use of molecular methodologies that enable the desired modification without the need to incorporate a permanent selection marker such as an antibiotic resistance gene is required. For instance, point mutagenesis (Quelas et al., 2021) is a useful tool for achieving modifications on rhizobial endogenous information.
The properties and strategies that can potentially improve inoculant performance are summarized in Table 1.
Table 1.
Properties and strategies that can potentially improve inoculant performance.
| Inoculants: desired properties |
|
Improved competitiveness Soil survival, Initial recognition steps, Multiplication in infection threads, Symbiosome formation, Transformation to bacteroid |
|
Higher number of fixing nodules per root Inhibition of phytohormones production |
|
Improved nitrogen fixation efficiency Regulation of nitrogenase activity, Hydrogenase and NO‐degrading enzymes |
|
Increased duration of the nitrogen fixation stage NO and ROS‐scavenging enzymes, TA modules |
| Strategies to improve inoculant performance |
|
Native strain isolation Strain selection based on competitive and nitrogen efficiency assays |
| Carrier and application mode |
| Co‐inoculation with PGPR and mycorrhizas |
|
Genetic modification of bacterial functions Chromosomal integration, avoiding of antibiotic selection marker |
We will give below examples of genetically engineered strains with positive effects on the nodulation process with reference to competitiveness, nitrogen fixation and plant yield. A very complete list of rhizobial genetic modifications and the corresponding effects on nodulation was recently published (Goyal et al., 2021). As mentioned above, competitiveness may be given by increased survival or proliferation and niche occupation in the rhizosphere at the expense of other competing bacteria. So, a good strategy could be the release of toxic compounds by rhizobia. Such is the case of R. etli production of the antibiotic trifolitoxin. Isogenic strains that differed only in their antibiotic‐producing capacity were generated and co‐inoculated on Phaseolus vulgaris under agricultural conditions (Robleto et al., 1997). It was found that the antibiotic‐producing strain showed increased nodule occupation (Robleto et al., 1997). The ability to metabolize the proline present in root exudates was also used to obtain more competitive strains. An engineered strain that constitutively expressed proline dehydrogenase (encoded by the putA gene) presented greater competitiveness for the nodulation process (van Dillewijn et al., 2001). This is one of the few examples in which experiments were conducted in microcosms with non‐sterile soil and controlled field conditions. A higher nitrogen fixation has been achieved through the introduction of a chimeric copy of the nitrogenase operon (nifHDK) under the activity of a strong promoter into R. etli (Peralta et al., 2004). This construction was added to the two endogenous copies that are naturally encoded under the regulation of low‐activity promoters, resulting in increased nitrogen activity and plant weight (Peralta et al., 2004). Recently, heterologous hydrogenase was expressed in rhizobial strains lacking endogenous hydrogenase activity and used as commercial inoculants of Phaseolus vulgaris L. The resulting strains presented increased nodule efficiency and seed nitrogen content as compared with the corresponding wild‐type strains (Ribeiro Torres et al., 2020).
The effect of ethylene on the initiation of nodule formation and the ability of some bacterial species to decrease ethylene levels is highly studied. As stated above, this is achieved through the activity of ACC deaminase, which degrades the substrate for ethylene biosynthesis (Nascimento et al., 2018). The heterologous ACC deaminase expression of R. leguminosarum in S. meliloti, which does not naturally possess this activity, produced more competitive strains (Ma et al., 2004). In M. japonicum MAFF303099, ACC deaminase expression was induced under microoxic conditions (Uchiumi et al., 2004; Nukui et al., 2006). Its constitutive expression led to an increase in competitiveness for nodulation (Conforte et al., 2010).
Overexpression of bacterial adhesin RapI in R. leguminosarum promoted its nodulation on red clover roots (Mongiardini et al., 2009). The hydroxamate‐type siderophore receptor of E. coli (Fhu) was expressed in two Rhizobium spp. strains which do not naturally code for this receptor. The heterologous expression led to greater nodule number and fresh weight of Pigeon pea plants (Geetha et al., 2009). The engineered strains presented higher nodule occupation than the original ones, even in non‐sterile soils (Geetha et al., 2009). In Sinorhizobium meliloti, overexpression of flavodoxin, a protein involved in the oxidative stress response, delayed nodule senescence (Redondo et al., 2009) and provided enhanced tolerance to salinity stress in the nodules (Redondo et al., 2012). With regards to glyphosate tolerance, Bradyrhizobium diazoefficiens USDA 110 is naturally sensitive to this herbicide (Zablotowicz and Reddy, 2004). A glyphosate‐resistant engineered Bradyrhizobium strain expressing the EPSPS from A. tumefaciens strain CP4 has already been patented (King and Purcell, 2005). The strain thus engineered displayed tolerance to the compound in in vitro assays. However, it would be unstable for the information incorporated considering that it carries two copies of the EPSPS gene (endogenous and heterologous). Recently, our research group obtained a glyphosate‐tolerant strain by changing only two nucleotides in the active site of the EPSPS enzyme of B. diazoefficiens USDA110 (Quelas et al., 2021). This strain does not contain any exogenous information and proved to be more competitive than the wild‐type strain in laboratory co‐inoculation trials performed in the presence of glyphosate (Quelas et al., 2021).
Conclusions
Based on the hypothesis that the rhizobia–legume symbiotic process has evolved towards the selection of strains that best interact and induce efficient nodules for nitrogen fixation, is there a way to improve it? Despite inoculants composed of highly efficient nitrogen‐fixing rhizobia were used, the populations found in plant nodules were often diverse, with a high proportion of native strains. Native strains are certainly better adapted to the environment than those making up the inoculant. Their competitiveness for nodulation may be greater, equal, or less than that of the inoculant and may be therefore very, little or not efficient at all in nitrogen fixation. At the beginning of the nodulation process, the plant cannot discriminate rhizobia with respect to their nitrogen fixation efficiency. Once inside the nodule, some plants have the capacity to sanction non‐fixing bacteria, without preventing them from still being in the rhizosphere and competing for nodulation. In relation to the question raised, we believe that the symbiotic process between legumes and rhizobia can still be improved by seeking strains able to adapt to the environment, more competitive in the nodulation process than native strains and, at the same time, having a better capacity to fix nitrogen. This can be achieved by selecting strains with good nitrogen fixation efficiency from the most competitive ones, and then finding the appropriate inoculation conditions to increase their presence in the proximity of the root. Also, by accompanying efficient nitrogen‐fixing strains with other PGPR strains or mycorrhizas that favour proliferation in the rhizosphere and root colonization, or by modifying some functions through genetic engineering of rhizobia. The genetic modification may be targeted to improving bacterial adaptation to a particular soil condition, improving one of the multiple stages involved in the nodulation process itself or increasing the nitrogen fixation capacity. However, overexpression or mutation of a function may have pleiotropic effects that should be avoided. The resulting mechanism of action of different bacterial components may be dual and depend on the variety of the nodulated legume. As shown in various field campaigns, different varieties of the same legume species may be grown and the genetic modifications on these bacterial components may not be desirable. Accordingly, a thorough knowledge of the molecular mechanisms involved in the different stages of the nodulation process is required for the selection of strains by genetic engineering.
Conflict of interest
None declared.
Microb. Biotechnol. (2021) 14(5), 1897–1917
Funding information
No funding information provided.
References
- Abd‐Alla, M.H., El‐Enany, A.W.E., Nafady, N.A., Khalaf, D.M., and Morsy, F.M. (2014) Synergistic interaction of Rhizobium leguminosarum bv. viciae and arbuscular mycorrhizal fungi as a plant growth promoting biofertilizers for faba bean (Vicia faba L.) in alkaline soil. Microbiol Res 169: 49–58. [DOI] [PubMed] [Google Scholar]
- Acosta‐Jurado, S., Rodríguez‐Navarro, D.N., Kawaharada, Y., Rodríguez‐Carvajal, M.A., Gil‐Serrano, A., Soria‐Díaz, M.E., et al. (2019) Sinorhizobium fredii HH103 nolR and nodD2 mutants gain capacity for infection thread invasion of Lotus japonicus Gifu and Lotus burttii . Environ Microbiol 21: 1718–1739. [DOI] [PubMed] [Google Scholar]
- Albareda, M., Rodríguez‐Navarro, D.N., Camacho, M., and Temprano, F.J. (2008) Alternatives to peat as a carrier for rhizobia inoculants: solid and liquid formulations. Soil Biol Biochem 40: 2771–2779. [Google Scholar]
- Albrecht, S.L., Maier, R.J., Hanus, F.J., Russell, S.A., Emerich, D.W., and Evans, H.J. (1979) Hydrogenase in Rhizobium japonicum increases nitrogen fixation by nodulated soybeans. Science 203: 1255–1257. [DOI] [PubMed] [Google Scholar]
- Alemneh, A.A., Zhou, Y., Ryder, M.H., and Denton, M.D. (2020) Mechanisms in plant growth‐promoting rhizobacteria that enhance legume‐rhizobial symbioses. J Appl Microbiol 129: 1133–1156. [DOI] [PubMed] [Google Scholar]
- Alunni, B., Kevei, Z., Redondo‐Nieto, M., Kondorosi, A., Mergaert, P., and Kondorosi, E. (2007) Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule‐specific gene families in Medicago truncatula . Mol Plant Microbe Interact 20: 1138–1148. [DOI] [PubMed] [Google Scholar]
- Andrews, M., and Andrews, M.E. (2017) Specificity in legume‐Rhizobia symbioses. Int J. Mol Sciences 18: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, M.F., Shabab, M., Penterman, J., Boehme, K.L., Griffitts, J.S., and Walker, G.C. (2017) Genome‐wide sensitivity analysis of the microsymbiont Sinorhizobium meliloti to symbiotically important, defensin‐like host peptides. <span>mBio</span> 8:e01060‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artursson, V., Finaly, R.D., and Jansson, J.K. (2006) Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ Microbiol 8: 1–10. [DOI] [PubMed] [Google Scholar]
- Aslam, S.N., Newman, M.‐A., Erbs, G., Morrissey, K.L., Chinchilla, D., Boller, T., et al. (2008) Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Badri, D.V., Weir, T.L., van der Leile, D., and Vivanco, J.M. (2009) Rhizosphere chemical dialogues: plant‐microbe interactions. Curr Opin Biotechnol 20: 642–650. [DOI] [PubMed] [Google Scholar]
- Baginsky, C., Brito, B., Imperial, J., Palacios, J.M., and Ruiz‐ Argüeso, T. (2002) Diversity and evolution of hydrogenase systems in Rhizobia. Appl Environ Microbiol 68: 4915–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais, H.P., Weir, T.L., Perry, L.G., Gilroy, S., and Vivanco, J.M. (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266. [DOI] [PubMed] [Google Scholar]
- Bamba, M., Aoki, S., Kajita, T., Setoguchi, H., Watano, Y., Sato, S., et al. (2019) Exploring genetic diversity and signatures of horizontal gen transfer in nodule bacteria associated with Lotus japonicus in natural environments. Mol Plant Microbe Interactions 32: 1110–1120. [DOI] [PubMed] [Google Scholar]
- Barea, J.M., Azcón, R., and Azcón‐Aguilar, C. (2017) Mycorrhizosphere interactions to improve a sustainable production of legumes. In Microbes for Legume Improvement, Zaidi, A., Khan, M., and Musarrat, J. (eds). Cham: Springer, pp. 199–225. [Google Scholar]
- Barea, J.M., Pozo, M.J., Azcón, R., and Azcón‐Aguilar, C. (2005) Microbial co‐operation in the rhizosphere. J Exp Botany 56: 1761–1778. [DOI] [PubMed] [Google Scholar]
- Barry, G.F., Kishmore, G.M., and Padgette, S.R. (1992) A herbicidal composition comprising glyphosate in the form of a mixture of the potassium and ammonium salts. International Patent 92: 04449. [Google Scholar]
- Bartsev, A.V., Deakin, W.J., Boukli, N.M., Mcalvin, C.B., Stacey, G., Brouthton, W.J., et al. (2004) NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol 134: 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashan, Y., de‐Bashan, L.E., Prabhu, S.R., and Hernandez, J.‐P. (2014) Advances in plant growth‐promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant Soil 378: 1–33. [Google Scholar]
- Basile, L.A., Zalguizuri, A., Briones, G., and Lepek, V.C. (2018) Two Rieske Fe/S proteins and TAT system in Mesorhizobium loti MAFF303099: differential regulation and roles on nodulation. Front Plant Sci 9: 1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslam, M., Qaddoury, A., and Goicoechea, N. (2014) Role of native and exotic mycorrhizal symbiosis to develop morphological, physiological, and biochemical responses coping with water drought of date palm, Phoenix dactylifera . Trees 28: 161–172. [Google Scholar]
- Battisti, L., Lara, J.C., and Leigh, J.A. (1992) Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc Natl Acad Sci USA 89: 5625–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin, E., Pieuchot, L., Engler, G., Pauly, N., and Puppo, A. (2006) Nitric oxide is formed in Medicago truncatula‐Sinorhizobium meliloti functional nodules. Mol Plant Microbe Interact 19: 970–975. [DOI] [PubMed] [Google Scholar]
- Becana, M., Matamoros, M.A., Udvardi, M., and Dalton, D.A. (2010) Recent insights into antioxidant defenses of legume root nodules. New Phytol 188: 960–976. [DOI] [PubMed] [Google Scholar]
- Berger, A., Boscari, A., Frendo, P., and Brouquisse, R. (2019) Nitric oxide signaling, metabolism and toxicity in nitrogen‐fixing symbiosis. J Exp Bot 70: 4505–4520. [DOI] [PubMed] [Google Scholar]
- Bernal, P., Allsopp, L.P., Filloux, A., and Llamas, M.A. (2017) The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J 11: 972–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal, P., Llamas, M.A., and Filloux, A. (2018) Type VI secretion systems in plant‐associated bacteria. Environ Microbiol 20: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat, A.A., Mithöfer, A., Pfeffer, P.E., Kraus, C., Spickers, N., Hotchkiss, A., et al. (1999) Further studies of the role of cyclic beta‐glucans in symbiosis. An NdvC mutant of Bradyrhizobium japonicum synthesizes cyclodecakis‐(1–3)‐beta‐glucosyl. Plant Physiol 119: 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, P.N., and Jha, D.K. (2012) Plant growth‐promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28: 1327–1350. [DOI] [PubMed] [Google Scholar]
- Boivin, S., Fonouni‐Farde, C., and Frugier, F. (2016) How auxin and cytokinin phytohormones modulate root microbe interactions. Frontiers Plant Science 7: 1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondage, D.D., Lin, J.S., Ma, L.S., Kuo, C.H., and Lai, E.M. (2016) VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor‐effector complex. Proc Natl Acad Sci USA 113: E3931–E3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscari, A., Del Giudice, J., Ferrarini, A., Venturini, L., Zaffini, A.‐L., Delledonne, M., et al. (2013) Expression dynamics of the Medicago truncatula transcriptome during the symbiotic interaction with Sinorhizobium meliloti: which role for nitric oxide? Plant Physiol 161: 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., et al. (2014) The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedveld, M.w., Cremers, H.c., Batley, M., Posthumus, M.a., Zevenhuizen, L.p., Wijffelman, C.a., and Zehnder, A.j. (1993) Polysaccharide synthesis in relation to nodulation behavior of Rhizobium leguminosarum . J Bacteriol 175: 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin, N.J. (1998) Tissue and cell invasion by Rhizobium: The structure and development of infection threads and symbiosomes. In The Rhizobiaceae. Spaink, H.P., Kondorosi, A., and Hooykaas, P.J.J. (eds). Dordrecht, The Netherlands: Kluwer Academic Publishers, pp. 417–429. [Google Scholar]
- Brewin, N.J., Wood, E.A., and Young, J.P.W. (1983) Contribution of the symbiotic plasmid to the competitiveness of Rhizobium leguminosarum . Microbiol 129: 2973–2977. [Google Scholar]
- Broughton, W.J., Jabbouri, S., and Perret, X. (2000) Keys to symbiotic harmony. J Bacteriol 182: 5641–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli, D., Schlaeppi, K., Spaepen, S., Loren, V., van Themaat, E., and Schulze‐Lefert, P. (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64: 807–838. [DOI] [PubMed] [Google Scholar]
- Busby, R.R., Rodriguez, G., Gebhart, D.L., and Yannarell, A.C. (2016) Native Lespedeza species harbor greater non‐rhizobial bacterial diversity in root nodules compared to the coexisting invader, L. cuneata . Plant Soil 401: 427–436. [Google Scholar]
- Caetano‐Anollés, G., and Gresshoff, P.M. (1991) Plant genetic control of nodulation. Annu Rev Microbiol 45: 345–382. [DOI] [PubMed] [Google Scholar]
- Cam, Y., Pierre, O., Boncompagni, E., Jérouart, D., Meilhoc, E., and Bruand, C. (2012) Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytol 196: 548–560. [DOI] [PubMed] [Google Scholar]
- del Cerro, P., Ayala‐García, P., Buzón, P., Castells‐Graells, R., López‐Baena, F.J., Ollero, F.J., and Pérez‐Montaño, F. (2020) OnfD, an AraC‐type transcriptional regulator encoded by Rhizobium tropici CIAT 899 and involved in Nod factor synthesis and symbiosis. Appl Environ Microbiol 86: e01297‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Cerro, P., Pérez‐Montaño, F., Gil‐Serrano, A., López‐Baena, F.J., Megías, M., Hungria, M., and Ollero, F.J. (2017) The Rhizobium tropici CIAT 899 NodD2 protein regulates the production of Nod factors under salt stress in a flavonoid‐independent manner. Sci Rep 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Cerro, P., Rolla‐Santos, A.A.P., Gomes, D.F., Marks, B.B., Pérez‐Montaño, F., Rodríguez‐Carvajal, M.Á., et al. (2015) Regulatory nodD1 and nodD2 genes of Rhizobium tropici strain CIAT 899 and their roles in the early stages of molecular signaling and host‐legume nodulation. BMC Genom 16: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, J.M., Badri, D.V., Bakker, M.G., Sugiyama, A., Manter, D.K., and Vivanco, J.M. (2013) Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8: e55731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.C., Feng, J., Hou, B.H., Li, F.Q., and Hong, G.F. (2005) Modulating DNA bending affects NodD‐mediated transcriptional control in Rhizobium leguminosarum . Nucleic Acids Res 33: 2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H.P., and Walker, G.C. (1998) Succinoglycan is required for initiation and elongation of its during nodulation of alfalfa by Rhizobium meliloti . J Bacteriol 180: 5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006) Host‐microbe interaction: shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Clúa, J., Roda, C., Zanetti, M.E., and Blanco, F.A. (2018) Compatibility between legumes and rhizobia for the establishment of a successful nitrogen‐fixing symbiosis. Genes 9: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforte, V.P., Echeverría, M., Sánchez, C., Ugalde, R.A., Menéndez, A.B., and Lepek, V.C. (2010) Engineered ACC deaminase‐expressing free‐living cells of Mesorhizobium loti show increased nodulation efficiency and competitiveness on Lotus spp. J Gen Appl Microbiol 56: 3331–3338. [DOI] [PubMed] [Google Scholar]
- Crespi, M., and Gálvez, S. (2000) Molecular mechanisms in root nodule development. J Plant Growth Regul 19: 155–166. [DOI] [PubMed] [Google Scholar]
- D’Antuono, A.L., Casabuono, A., Couto, A., Ugalde, R.A., and Lepek, V.C. (2005) Nodule development induced by Mesorhizobium loti mutant strains affected in polysaccharide synthesis. Mol Plant Microbe Interact 18: 446–457. [DOI] [PubMed] [Google Scholar]
- Dalla Via, V., Narduzzi, C., Aguilar, O.M., Zanetti, M.E., and Blanco, F.A. (2015) Changes in the common bean (Phaseolus vulgaris) transcriptome in response to secreted and surface signal molecules of Rhizobium etli . Plant Physiol 169: 1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antuono, A.L., Ott, T., Krusell, L., Voroshilova, V., Ugalde, R.A., Udvardi, M., and Lepek, V.C. (2008) Defects in rhizobial cyclic glucan and lipopolysaccharide synthesis alter legume gene expression during nodule development. Mol Plant Microbe Interact 21: 50–60. [DOI] [PubMed] [Google Scholar]
- Demont, N., Ardourel, M., Maillet, F., Prome, D., Ferro, M., Promé, J.C., and Denarié, J. (1994) The Rhizobium meliloti regulatory nodD3 and syrM genes control the synthesis of a particular class of nodulation factors N‐acylated by (omega‐1)‐hydroxylated fatty acids. EMBO J 13: 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison, R.F., and Kiers, E.T. (2004) Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Lett 237: 187–193. [DOI] [PubMed] [Google Scholar]
- D'Haeze, W., and Holsters, M. (2002) Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12: 79R–105R. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo, F., Speciale, I., Silipo, A., Alías‐Villegas, C., Acosta‐Jurado, S., Rodríguez‐Carvajal, M.‐Á., et al. (2020) Structure of the unusual Sinorhizobium fredii HH103 lipopolysaccharide and its role in symbiosis. J Biol Chem 295: 10969–10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, T., Herbert, R.A., Martinez, U., Wang, B., Chen, J.C., Brown, J.B., et al. (2020) Iron supplementation eliminates antagonistic interactions between root‐associated bacteria. Front Microbiol 11: 1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella, M.J., Fontana, M.F., Cumpa Velásquez, L.M., Torres Tejerizo, G.A., Diambra, L., Hansen, L.H., et al. (2020) Mesorhizobium intechi sp. nov. isolated from nodules of Lotus tenuis in soils of the Flooding Pampa, Argentina. Syst Appl Microbiol 43: 126044. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Liu, J., Lyu, S., Wang, Q.i., Yang, S., and Zhu, H. (2017) The soybean Rfg1 gene restricts nodulation by Sinorhizobium fredii USDA193. Front Plant Sci 8: 1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy, N.N., Bahlmann, A.K., and Loake, G.J. (2017) Nitric oxide function in plant abiotic stress. Plant Cell Environ 40: 462–472. [DOI] [PubMed] [Google Scholar]
- FAO (2019) Food Outlook – Biannual Report on Global Food Markets. Rome, Italy. Licence: CC BY‐NC‐SA 3.0 IGO. ISBN 978‐92‐5‐131448‐7. [Google Scholar]
- Ferguson, B.J., Indrasumunar, A., Hayashi, S., Lin, M.‐H., Lin, Y.‐H., Reid, D.E., et al. (2010) Molecular analysis of legume nodule development autoregulation. J Integrative Plant Biol 53: 61–76. [DOI] [PubMed] [Google Scholar]
- Ferguson, B.J., and Mathesius, U. (2014) Phytohormone regulation of legume‐rhizobia Interactions. J Chem Ecol 40: 770–790. [DOI] [PubMed] [Google Scholar]
- Ferguson, S., Major, A.S., Sullivan, J.T., Bourke, S.D., Kelly, S.J., Perry, B.J., and Ronson, C.W. (2020) Rhizobium leguminosarum bv. trifolii NodD2 enhances competitive nodule colonization in the clover‐rhizobium symbiosis. Appl Environ Microbiol 86: e01268‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini, A., De Stefano, M., Baudouin, E., Pucciariello, C., Polverari, A., Puppo, A., et al. (2008) Expression of Medicago truncatula genes responsive to nitric oxide in pathogenic and symbiotic conditions. Mol Plant Microbe Interact 21: 781–790. [DOI] [PubMed] [Google Scholar]
- Gage, D.J. (2004) Infection and invasion of roots by symbiotic, nitrogen‐fixing rhizobia during nodulation of temperate legumes. Microbial Mol Biol Rev 68: 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway, J.N., Leach, A.M., Bleeker, A., and Erisman, J.W. (2013) A chronology of human understanding of the nitrogen cycle. Phil Trans R Soc B 368: 20130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gano‐Cohen, K.A., Stokes, P.J., Blanton, M.A., Wendlandt, C.E., Hollowell, A.C., Regus, J.U., et al. (2016) Nonnodulating Bradyrhizobium spp. modulate the benefits of legume‐Rhizobium mutualism. Appl Environ Microbiol 82: 5259–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha, R., Desai, A.J., and Archana, G. (2009) Effect of the expression of Escherichia coli fhuA gene in Rhizobium sp. IC3123 and ST1 in planta: its role in increased nodule occupancy and function in pigeon pea . Appl Soil Ecol 43: 185–190. [Google Scholar]
- Geetha, S.J., and Joshi, S.J. (2013) Engineering rhizobial bioinoculants: a strategy to improve iron nutrition. Scientific World J 2013: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, K., Christensen, S.K., and Lobner‐Olesen, A. (2005) Prokaryotic toxin‐antitoxin stress response loci. Nat Rev Microbiol 3: 371–382. [DOI] [PubMed] [Google Scholar]
- Geremía, R., Cavaignac, S., Zorreguieta, A., Toro, N., Olivares, J., and Ugalde, R. (1987) A Rhizobium meliloti mutant that forms ineffective pseudonodules in alfalfa produces exopolysaccharides but fails to form β‐(1‐2) glucan. J Bacteriol 169: 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts, R., and Bisseling, T. (2002) Rhizobium Nod factor perception and signaling. Plant Cell 14: S239–S249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders, N., and Van Melderen, L. (2014) Toxin‐antitoxin systems as multilevel interaction systems. Toxins (Basel) 6: 304–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal, R.K., Schmidt, M.A., and Hynes, M.F. (2021) Molecular biology in the improvement of biological nitrogen fixation by rhizobia and extending the scope to cereals. Microorganism 9: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guefrachi, I., Pierre, O., Timchenko, T., Alunni, B., Barriere, Q., Czernic, P., et al. (2015) Bradyrhizobium BclA is a peptide transporter required for bacterial differentiation in symbiosis with Aeschynomene legumes. Mol Plant Microbe Interact 28: 1155–1166. [DOI] [PubMed] [Google Scholar]
- Guignard, M.S., Leitch, A.R., Acquisti, C., Eizaguirre, C., Elser, J.J., Hessen, D.O., et al. (2017) Impacts of nitrogen and phosphorus: from genomes to natural ecosystems and agriculture. Front Ecol Evol 5: 70. [Google Scholar]
- Haag, A.F., Baloban, M., Sani, M., Kerscher, B., Pierre, O., Farkas, A., et al. (2011) Protection of Sinorhizobium against host cysteine‐rich antimicrobial peptides is critical for symbiosis. PLoS Biol 9: e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Q., Ma, Q., Chen, Y., Tian, B., Xu, L., Bai, Y., et al. (2020) Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J 14: 1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani, M.A., Durán, P., and Hacquard, S. (2018) Microbial interactions within the plant holobiont. Microbiome 6: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastwell, A.H., de Bang, T.C., Gresshoff, P.M., and Ferguson, B.J. (2017) CLE peptide‐encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis . Sci Rep 7: 9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, F., and Kędzierska, B. (2014) Regulating toxin‐antitoxin expression: controlled detonation of intracellular molecular timebombs. Toxins (Basel) 6: 337–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, P.R. (2005) Release of transgenic bacterial inoculants‐rhizobia as a case study. Plant Soil 266: 1–10. [Google Scholar]
- Ho, B.T., Dong, T.G., and Mekalanos, J.J. (2014) A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtsmark, I., Eijsink, V.G.H., and Brurberg, M.B. (2008) Bacteriocins from plant pathogenic bacteria. FEMS Microbiol Lett 280: 1–7. [DOI] [PubMed] [Google Scholar]
- Huang, C.‐Y., Wang, H., Hu, P., Hamby, R., and Jin, H. (2019) Small RNAs‐big players in plant‐microbe interactions. Cell Host Microbe 26: 173–182. [DOI] [PubMed] [Google Scholar]
- Hubber, A., Vergunst, A.C., Sullivan, J.T., Hooykaas, P.J.J., and Ronson, C.W. (2004) Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol Microbiol 54: 561–574. [DOI] [PubMed] [Google Scholar]
- Hungria, M., Loureiro, M.F., Mendes, J.C., Campo, R.J., and Graham, P.H. (2005) Inoculant preparation, production and application. In Nitrogen Fixation in Agriculture, Forestry, Ecology and the Environment. Werner, D., and Newton, W.E. (eds). Dordrecht, The Netherlands: Springer, pp. 223–253. [Google Scholar]
- Hungria, M., Nogueira, M.A., and Araujo, R.S. (2015) Soybean seed co‐inoculation with Bradyrhizobium spp. and Azospirillum brasilense: a new biotechnological tool to improve yield and sustainability. Am J Plant Sci 6: 811–817. [Google Scholar]
- Igamberdiev, A.U., and Hill, R.D. (2009) Plant mitochondrial function during anaerobiosis. Annals Botany 103: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irisarri, P., Cardozo, G., Tartaglia, C., Reyno, R., Gutiérrez, P., Lattanzi, F.A., et al. (2019) Selection of competitive and efficient rhizobia strains for white clover. Frontiers Microbiol 10: 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet, A., and Nassif, X. (2015) New players in the toxin field: polymorphic toxin systems in bacteria. mBio 6: e00285‐15. 10.1128/mBio.00285-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski, E.G. (1972) Mode of action of N‐phosphonomethyl‐glycine inhibition of aromatic amino acid biosynthesis. J Agric Food Chem 20: 1195–1198. [Google Scholar]
- Ji, Z.J., Yan, H., Cui, Q.G., Wang, E.T., Chen, W.F., and Chen, W.X. (2017) Competition between rhizobia under different environmental conditions affects the nodulation of a legume. Systemic Appl Microbiol 40: 114–119. [DOI] [PubMed] [Google Scholar]
- Jiménez‐Guerrero, I., Acosta‐Jurado, S., Medina, C., Ollero, F.J., Alias‐Villegas, C., Vinardell, J.M., et al. (2020) The Sinorhizobium fredii HH103 type III secretion system effector NopC blocks nodulation with Lotus japonicus Gifu. J Exp Botany 7: 6043–6056. [DOI] [PubMed] [Google Scholar]
- Jiménez‐Guerrero, I., Moreno‐De Castro, N., and Pérez‐Montaño, F. (2021) One door closes, another opens: when nodulation impairment with natural hosts extends rhizobial host‐range. Environ Microbiol 23: 1837–1841. [DOI] [PubMed] [Google Scholar]
- Jiménez‐Zurdo, J.I., van Dillewijn, P., Soto, M.J., de Felipe, M.R., Olivares, J., and Toro, N. (1995) Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol Plant Microbe Interact 8: 492–498. [DOI] [PubMed] [Google Scholar]
- Jones, K.M., Sharopova, N., Lohar, D.P., Zhang, J.Q., VandenBosch, K.A., and Walker, G.C. (2008) Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide‐deficient mutant. Proc Natl Acad Sci USA 105: 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara, K., Ardissone, S., Kobayashi, H., Saad, M.M., Schumpp, O., Broughton, W.J., and Deakin, W.J. (2009) Rhizobia utilize pathogen‐like effector proteins during symbiosis. Mol Microbiol 71: 92–106. [DOI] [PubMed] [Google Scholar]
- Katagiri, F., and Tsuda, K. (2010) Understanding the plant immune system. Mol Plant Microbe Interact 23: 1531–1536. [DOI] [PubMed] [Google Scholar]
- Kato, K., Kanahama, K., and Kanayama, Y. (2010) Involvement of nitric oxide in the inhibition of nitrogenase activity by nitrate in Lotus root nodules. J Plant Physiol 167: 238–241. [DOI] [PubMed] [Google Scholar]
- Kawaharada, Y., Kelly, S., Nielsen, M.W., Hjuler, C.t., Gysel, K., Muszyński, A., et al. (2015) Receptor‐mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312. [DOI] [PubMed] [Google Scholar]
- Kawaharada, Y., Nielsen, M.W., Kelly, S., James, E.K., Andersen, K.R., Rasmussen, S.R., et al. (2017) Differential regulation of the Epr3 receptor coordinates membrane‐restricted rhizobial colonization of root nodule primordia. Nature commun 8: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, S., Sullivan, J.T., Kawaharada, Y., Radutoiu, S., Ronson, C.W., and Stougaard, J. (2018) Regulation of Nod factor biosynthesis by alternative NodD proteins at distinct stages of symbiosis provides additional compatibility scrutiny. Environ Microbiol 20: 97–110. [DOI] [PubMed] [Google Scholar]
- Kiers, E.T., Rousseau, R.A., West, S.A., and Denison, R.F. (2003) Host sanctions and the legume‐rhizobium mutualism. Nature 425: 78–81. [DOI] [PubMed] [Google Scholar]
- King, C.A., and Purcell, L.C. (2005) Herbicide resistant dinitrogen fixing bacteria and method of use. US Patent 6,872,562.
- Kondorosi, E., Mergaert, P., and Kereszt, A. (2013) A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol 67: 611–628. [DOI] [PubMed] [Google Scholar]
- Korir, H., Mungai, N.W., Thuita, M., Hamba, Y., and Masso, C. (2017) Co‐inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front Plant Sci 8: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi, H., Shimomura, K., Hata, S., Hirota, A., Wu, G.J., Kumagai, H., et al. (2004) Large‐scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus . DNA Res 11: 263–274. [DOI] [PubMed] [Google Scholar]
- Krause, A., Doerferl, A., and Göttfert, M. (2002) Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum . Mol Plant Microbe Interact 15: 1228–1235. [DOI] [PubMed] [Google Scholar]
- Krishnan, H.B., Lorio, L., Kim, W.S., Jiang, G., Kim, K.Y., DeBoer, M., et al. (2003) Extracellular proteins involved in soybean cultivar‐specific nodulation are associated with pilus‐like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol Plant Microbe Interact 16: 617–625. [DOI] [PubMed] [Google Scholar]
- Krusell, L., Madsen, L.H., Sato, S., Aubert, G., Genua, A., Szczyglowski, K., et al. (2002) Shoot control of root development and nodulation is mediated by a receptor‐like kinase. Nature 420: 422–426. [DOI] [PubMed] [Google Scholar]
- Kusakabe, S., Higasitani, N., Kaneko, T., Yasuda, M., Miwa, H., Okazaki, S., et al. (2020) Lotus accessions possess multiple checkpoints triggered by different type III secretion system effectors of the wide‐host‐range symbiont Bradyrhizobium elkanii USDA61. Microbes Environ 35: ME19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuykendall, L.D. (2012) Herbicide‐resistant inoculant strains, US Patent 20120266332.
- Lamrabet, Y., Bellogín, R.A., Cubo, T., Espuny, R., Gil, A., Krishnan, H.B., et al. (1999) Mutation in GDP‐fucose synthesis genes of Sinorhizobium fredii alters Nod factors and significantly decreases competitiveness to nodulate soybeans. Mol Plant Microbe Interact 12: 207–217. [DOI] [PubMed] [Google Scholar]
- Lei, Y., Su, S., He, L., Hu, X., and Luo, D. (2019) A member of the ALOG gene family has a novel role in regulating nodulation in Lotus japonicus . J. Integ Plant Biol 61: 463–477. [DOI] [PubMed] [Google Scholar]
- Li, D., Kinkema, M., and Gresshoff, P.M. (2009) Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signaling events associated with both nodule primordial development and nitrogen fixation. J Plant Physiol 166: 955–967. [DOI] [PubMed] [Google Scholar]
- Liang, Y., Cao, Y., Tanaka, K., Thibivilliers, S., Wan, J., Choi, J., et al. (2013) Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341: 1384. [DOI] [PubMed] [Google Scholar]
- Libault, M., Farmer, A., Brechenmacher, L., Drnevich, J., Langley, R.J., Bilgin, D.D., et al. (2010) Complete transcriptome of the soybean root hair cell, a single‐cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol 152: 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, R.M., Kylarová, S., Mergaert, P., and Kondorosi, E. (2020) Unexplored arsenals of legume peptides with potential for their applications in medicine and agriculture. Front Microbiol 11: 1307. 10.3389/fmicb.2020.01307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H.H., Huang, H.M., Yu, M., Lai, E.M., Chien, H.L., and Liu, C.T. (2018) Functional exploration of the bacterial type VI secretion system in mutualism: Azorhizobium caulinodans ORS571‐Sesbania rostrata as a research model. Mol Plant Microbe Interact 31: 856–867. [DOI] [PubMed] [Google Scholar]
- Lindström, K., and Mousavi, S.A. (2019) Effectiveness of nitrogen fixation in rhizobia. Microb Biotechnol 13: 1314–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma, J., Cineg, G., Bodogai, M., Oláh, B., Kjiers, A., Endre, G., et al. (2014) A vapBC‐type toxin‐antitoxin module of Sinorhizobium meliloti influences symbiotic efficiency and nodule senescence of Medicago sativa . Environ Microbiol 16: 3714–3729. [DOI] [PubMed] [Google Scholar]
- Liu, H., Zhang, C., Yang, J., Yu, N., and Wang, E. (2018) Hormone modulation of legume‐rhizobial symbiosis. J Integ Plant Biol 60: 632–648. [DOI] [PubMed] [Google Scholar]
- Lobato‐Márquez, D., Díaz‐Orejas, R., and García‐del Portillo, F. (2016) Toxin‐antitoxins and bacterial virulence. FEMS Microbiol Rev 40: 592–609. [DOI] [PubMed] [Google Scholar]
- Lohar, D.P., Sharopova, N., Endre, G., Penuela, S., Samac, D., Town, C., et al. (2006) Transcript analysis of early nodulation events in Medicago truncatula . Plant Physiol 140: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López García, S.L., Perticari, A., Piccinetti, C., Ventimiglia, L., Arias, N., De Battista, J.J., et al. (2009) In‐furrow inoculation and selection for higher motility enhances the efficacy of Bradyrhizobium japonicum nodulation. Agron J 101: 357–363. [Google Scholar]
- López‐Gómez, M., Sandal, N., Stougaard, J., and Boller, T. (2011) Interplay of flg22‐induced defense responses and nodulation in Lotus japonicus . J Exp Bot 63: 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., Yang, F., Wang, S., Ma, H., Liang, J., and Chen, Y. (2017) Co‐existence of rhizobia and diverse non‐rhizobial bacteria in the rhizosphere and nodules of Dalbergia odorifera seedlings inoculated with Bradyrhizobium elkanii, Rhizobium multihospirium‐like and Burkholderia pyrrocinia‐like strains. Front Microbiol 8: 2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, D.S., Lebeis, S.L., Paredes, S.H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.S., Hachani, A., Lin, J.S., Filloux, A., and Lai, E.M. (2014) Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W., Charles, T.C., and Glick, B.R. (2004) Expression of an exogenous 1‐aminocyclopropane‐1‐carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl Environ Microbiol 70: 5891–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W., Penrose, D.M., and Glick, B.R. (2002) Strategies used by rhizobia to lower plant ethylene levels and increase nodulation. Can J Microbiol 48: 947–954. [DOI] [PubMed] [Google Scholar]
- Madinabeitia, N., Bellogín, R.A., Buendía‐Clavería, A.M., Camacho, M., Cubo, T., Espuny, M.R., et al. (2002) Sinorhizobium fredii HH103 has a truncated nolO gene due to a‐1 frameshift mutation that is conserved among other geographically distant S. fredii strains. Mol Plant Microbe Interact 15: 150–159. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Marie, C., Deakin, W.J., Ojanen‐Reuhs, T., Diallo, E., Reuhs, B., Broughton, W.J., and Perret, X. (2004) TtsI, a key regulator of Rhizobium species NGR234 is required for type III‐dependent protein secretion and synthesis of rhamnose‐rich polysaccharides. Mol Plant Microbe Interact 17: 958–966. [DOI] [PubMed] [Google Scholar]
- Markmann, K., Radutoiu, S., and Stougaard, J. (2012) Infection of Lotus japonicus roots by Mesorhizobium loti . In Perotto, S. & Baluška, F. (eds). Signaling and Communication in Plant Symbiosis. Berlin, Heidelberg, Germany: Springer, pp. 31–50. [Google Scholar]
- Melchiorre, M., de Luca, M.J., Gonzalez Anta, G., Suarez, P., Lopez, C., Lascano, R., and Racca, R.W. (2011) Evaluation of bradyrhizobia strains isolated from field‐grown soybean plants in Argentina as improved inoculants. Biol Fertil Soils 47: 81–89. [Google Scholar]
- Mergaert, P., Nikovics, K., Kelemen, Z., Maunoury, N., Vaubert, D., Kondorosi, A., et al. (2003) A novel family in Medicago trucatula consisting of more than 300 nodule‐specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa, S., Hauser, F., Friberg, M., Malaguti, E., Fischer, H.‐M., and Hennecke, H. (2008) Comprehensive assessment of the regulons controlled by the FixLJ‐FixK2‐FixK1 cascade in Bradyrhizobium japonicum . J Bacteriol 190: 6568–6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, J., Zhang, N., Liu, H., Wang, H., Zhong, Z., and Zhu, J. (2018) Soil commensal rhizobia promote Rhizobium etli nodulation efficiency through CinR‐mediated quorum sensing. Arch Microbiol 200: 685–694. [DOI] [PubMed] [Google Scholar]
- Mongiardini, E.J., Pérez‐Giménez, J., Athabegoiti, M.J., Covelli, J., Quelas, J.I., López‐García, S.L., et al. (2009) Overproduction of tha rhizobial adhesion RapA1 increases competitiveness for nodulation. Soil Biol Biochem 41: 2017–2020. [Google Scholar]
- Mortier, V., De Wever, E., Vuylsteke, M., Holsters, M., and Goormachtig, S. (2012b) Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J 70: 367–376. [DOI] [PubMed] [Google Scholar]
- Mortier, V., Holsters, M., and Goormachtig, S. (2012a) Never too many? How legumes control nodule numbers. Plant Cell Environ 35: 245–258. [DOI] [PubMed] [Google Scholar]
- Moussaid, S., Domínguez‐Ferreras, A., Muñoz, S., Aurag, J., Berraho, E.B., and Sanjuán, J. (2015) Increased trehalose biosynthesis improves Mesorhizobium ciceri growth and symbiosis establishment in saline conditions. Symbiosis 67: 103–111. [Google Scholar]
- Mudgett, M.B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56: 509–531. [DOI] [PubMed] [Google Scholar]
- Muleta, D., Ryder, M.H., and Denton, M.D. (2017) The potential for rhizobial inoculation to increase soybean grain yields on acid soils in Ethiopia. Soil Sci Plant Nutr 63: 1–11. [Google Scholar]
- Nascimento, F.X., Rossi, M.J., and Glick, B.R. (2018) Ethylene and 1‐Aminocyclopropane‐1‐carboxylate (ACC) in plant‐bacterial interactions. Front Plant Sci 9: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento, F.X., Tavares, M.J., Franck, J., Ali, S., Glick, B.R., and Rossi, M.J. (2019) ACC deaminase plays a major role in Pseudomonas fluorescens YsS6 ability to promote the nodulation of Alpha‐ and Betaproteobacteria rhizobial strains. Arch Microbiol 201: 817–822. [DOI] [PubMed] [Google Scholar]
- Nelson, M.S., and Sadowsky, M.J. (2015) Secretion systems and signal exchange between nitrogen‐fixing rhizobia and legumes. Front Plant Sci 6: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel, K.D., Forsberg, L.S., and Carlson, R.W. (2000) Varying the abundance of O antigen in Rhizobium etli and its effect on symbiosis with Phaseolus vulgaris . J Bacteriol 182: 5317–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukui, N., Ezura, H., Yuhashi, K., Yasuta, T., and Minamisawa, K. (2000) Effect of ethylene precursor and inhibitors for ethylene biosynthesis and perception on nodulation in Lotus japonicus and Macroptilium atropurpureum . Plant Cell Physiol 41: 893–897. [DOI] [PubMed] [Google Scholar]
- Nukui, N., Minamisawa, K., Ayabe, S., and Aoki, T. (2006) Expression of the 1‐aminocyclopropane‐1‐carboxyloc acid deaminase gene requires symbiotic nitrogen‐fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl Environ Microbiol 72: 4964–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan, M. (2016) Microbial inoculation of seed for improved crop performance: issues and opportunities. Appl Microbiol Biotechnol 100: 5729–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, S., Ohnishi, E., Sato, S., Takahashi, H., Nakazono, M., Tabata, S., and Kawaguchi, M. (2009) Nod factor/nitrate‐induced CLE genes that drive HAR1‐mediated systemic regulation of nodulation. Plant Cell Phyiol 50: 67–77. [DOI] [PubMed] [Google Scholar]
- Okamoto, S., Shinohara, H., Mori, T., Matsubayashi, Y., and Kawaguchi, M. (2013) Root‐derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun 4: 2191. [DOI] [PubMed] [Google Scholar]
- Oláh, B., Kiss, E., Györgypál, Z., Borzi, J., Cinege, G., Csanádi, G., et al. (2001) Mutation in the ntrR gene, a member of the vap gene family, increases the symbiotic efficiency of Sinorhizobium meliloti . Mol Plant Microbe Interact 14: 887–894. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Downie, J.A. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., Murray, J.M., Poole, P.S., and Downie, J.A. (2011) The rules of engagement in the legume‐rhizobial symbiosis. Annu Rev Genet 45: 119–145. [DOI] [PubMed] [Google Scholar]
- Oono, R., Anderson, C.G., and Denison, R.F. (2011) Failure to fix nitrogen by non‐reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc R Soc B 278: 2698–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, T., van Dongen, J.T., Günther, C., Krusell, L., Desbrosses, G., Vigeolas, H., et al. (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531–535. [DOI] [PubMed] [Google Scholar]
- Padgette, S.R., Kolacz, K.H., Delannay, X., Re, D.B., LaVallee, B.J., Tinius, C.N., et al. (1995) Development, identification, and a characterization of a glyphosate‐tolerant soybean line. Crop Sci 35: 1451–2146. [Google Scholar]
- Page, R., and Peti, W. (2016) Toxin‐antitoxin systems in bacterial growth arrest and persistence. Nature Chem Biol 12: 2018–2214. [DOI] [PubMed] [Google Scholar]
- Parniske, M., Schmidt, P., Kosch, K., and Müller, P. (1994) Plant defense responses of host plants with determinate nodules induced by EPS‐defective exob mutants of Bradyrhizobium japonicum . Mol Plant Microbe Interact 7: 631–638. [Google Scholar]
- Patriarca, E.J., Tatè, R., and Iaccarino, M. (2002) Key role of bacterial NH4+ metabolism in rhizobium‐plant symbiosis. Microbiol Mol Biol Rev 66: 203–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, M.C., Fisher, R.F., and Long, S.R. (2006) Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti . J Bacteriol 188: 5417–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peix, A., Ramírez‐Bahena, M.H., Velázquez, E., and Bedmar, E.J. (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34: 17–42. [Google Scholar]
- Peralta, H., Mora, Y., Salazar, E., Encarnación, S., Palacios, R., and Mora, J. (2004) Engineering the nifH promoter region and abolishing poly‐β‐hydroxybutyrate accumulation in Rhizobium etli enhance nitrogen fixation in symbiosis with Phaseolus vulgaris . Appl Environ Microbiol 70: 3272–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotto, S., Brewin, N., and Kannenberg, E. (1994) Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide‐defective mutants of Rhizobium leguminosarum strain 3841. Mol Plant Microbe Interact 7: 99–112. [Google Scholar]
- Philippot, L., Raaijmakers, J.M., Lemanceau, P., and van der Putten, W.H. (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nature Rev Microbiol 11: 789–799. [DOI] [PubMed] [Google Scholar]
- Pickering, B.S., Yudistira, H., and Oresnik, I.J. (2012) Characterization of the twin‐arginine transport secretome in Sinorhizobium meliloti and evidence for host‐dependent phenotypes. Appl Environ Microbiol 78: 7141–7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, P.A., Tanner, H.R., Dillon, B.A., Shabab, M., Walker, G.C., and Griffitts, J.S. (2015) Rhizobial peptidase HrrP cleaves host‐encoded signaling peptides and mediates symbiotic compatibility. Proc Natl Acad Sci USA 112: 15244–15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueppke, S.G., and Broughton, W.J. (1999) Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host range. Mol Plant Microbe Interact 12: 293–318. [DOI] [PubMed] [Google Scholar]
- Quelas, J.I., Lastra, R.A., Lorenze, C., Escobar, M., and Lepek, V.C. (2021) Site‐directed mutagenesis of Bradyrhizobium diazoefficiens USDA 110 aroA improves bacterial growth and competitiveness for soybean nodulation in the presence of glyphosate. Environ Microbiol Rep., 13 464–469. [DOI] [PubMed] [Google Scholar]
- Quelas, J.I., Mesa, S., Mongiardini, E.J., Jendrossek, D., and Lodeiro, A.R. (2016) Regulation of poyhydroxybutyrate synthesis in the soil bacterium Bradyrhizobium diazoefficiens . Appl Environ Microbiol 82: 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Grønlund, M., et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor‐like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Raklami, A., Bechtaou, N., Tahiri, A.‐I., Anil, M., Meddich, A., and Oufdou, K. (2019) Use of rhizobacteria and mycorrhizae consortium in the open field as a strategy for improving crop nutrition productivity and soil fertility. Front Microbiol 10: 1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo, F.J., Coba de la Peña, T., Morcillo, C.N., Lucas, M.M., and Pueyo, J.J. (2009) Overexpression of Flavodoxin in bacteroids induces changes in antioxidant metabolism leading to delayed senescence and starch accumulation in alfalfa root nodules. Plant Physiol 149: 1166–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo, F.J., Coba de la Peña, T., Lucas, M.M., and Pueyo, J.J. (2012) Alfalfa nodules elicited by a flavodoxin‐overexpressing Ensifer meliloti strain display nitrogen‐fixing activity with enhanced tolerance to salinity stress. Planta 236: 1687–1700. [DOI] [PubMed] [Google Scholar]
- Regus, J.U., Gano, K.A., Hollowell, A.C., Sofish, V., and Sachs, J.L. (2015) Lotus hosts delimit the mutualism‐parasitism continuum of Bradyrhizobium . J Evol Biol 28: 447–456. [DOI] [PubMed] [Google Scholar]
- Regus, J.U., Quides, K.W., O'Neill, M.R., Suzuki, R., Savory, E.A., Chang, J.H., and Sachs, J.L. (2017) Cell autonomous sanctions in legumes target ineffective rhizobia in nodules with mixed infections. Am J Bot 104: 1299–1312. [DOI] [PubMed] [Google Scholar]
- Reid, D.E., Ferguson, B.J., and Gresshoff, P.M. (2011) Inoculation‐ and nitrate‐induced CLE peptides of soybean control NARK‐dependent nodule formation. Mol Plant Microb Interact 24: 606–618. [DOI] [PubMed] [Google Scholar]
- Reid, D., Liu, H., Kelly, S., Kawaharada, Y., Mun, T., Andersen, S.U., et al. (2018) Dynamics of ethylene production in response to compatible Nod factor. Plant Physiol 176: 1764–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina‐Bueno, M., Argandoña, M., Nieto, J.J., Hidalgo‐García, A., Iglesias‐Guerra, F., Delgado, M.J., and Vargas, C. (2012) Role of trehalose in heat and desiccation tolerance in the soil bacterium Rhizobium etli . BMC Microbiol 12: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, B., Wang, X., Duan, J., and Ma, J. (2019) Rhizobia tRNA‐derived small RNAs are signal molecules regulating plant nodulation. Science 365: 919–922. [DOI] [PubMed] [Google Scholar]
- Rigano, L.A., Payette, C., Brouillard, G., Marano, M.R., Abramowicz, L., Torres, P.S., et al. (2007) Bacterial cyclic β‐(1–2)‐glucan acts in systemic suppression of plant immune responses. Plant Cell 19: 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robleto, E.A., Scupham, A.J., and Triplett, E.W. (1997) Trifolitoxin production in Rhizobium etli strain CE3 increases competitiveness for rhizosphere colonization and root nodulation of Phaseolus vulgaris in soil. Mol Plant Microbe Interact 10: 228–233. [Google Scholar]
- Rodriguez Blanco, A., Sicardi, M., and Frioni, L. (2010) Competition for nodule occupancy between introduced and native strains of Rhizobium leguminosarum biovar trifolii . Biol Fertil Soils 46: 419–425. [Google Scholar]
- Ruiz‐Valdiviezo, V.M., Ventura Canseco, L.M.C., Castillo Suárez, L.A., Gutiérrez‐Miceli, F.A., Dendooven, L., and Rincón‐Rosales, R. (2015) Symbiotic potential and survival of native rhizobia kept on different carriers. Brazilian J Microbiol 46: 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.B., Hood, R.D., Bui, N.K., LeRoux, M., Vollmer, W., and Mougous, J.D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475: 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.B., LeRoux, M., Hathazi, K., Agnello, D.M., Ishikawa, T., Wiggins, P.A., et al. (2013) Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 25: 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, J.L., Quides, K.W., and Wendlandt, C.E. (2018) Legumes versus rhizoid: a model for ongoing conflict in symbiosis. New Phytol 218: 1199–1206. [DOI] [PubMed] [Google Scholar]
- Sachs, J.L., Russell, J.E., Lii, Y.E., Black, K.C., Lopez, G., and Patil, A.S. (2010) Host control over infection and proliferation of a cheater symbiont. J Evol Biol 23: 1919–1927. [DOI] [PubMed] [Google Scholar]
- Salinero‐Lanzarote, A., Pacheco‐Moreno, A., Domingo‐Serrano, L., Durán, D., Ormeño‐Orrillo, E., Martínez‐Romero, E., et al. (2019) The Type VI secretion system of Rhizobium etli Mim1 has a positive effect in symbiosis. FEMS Microbiol Ecol 95: fiz054. [DOI] [PubMed] [Google Scholar]
- Sanches Santos, M., Nogueira, M.A., and Hungria, M. (2019) Microbial inoculants: reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express 9: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez, C., Gates, A.J., Meakin, G.E., Uchiumi, T., Girard, L., Richardson, D.J., et al. (2010) Production of nitric oxide and nitrosylleghemoglobin complexes in soybean nodules in response to flooding. Mol Plant Microbe Interact 23: 702–711. [DOI] [PubMed] [Google Scholar]
- Sánchez, C., Iannino, F., Deakin, W.J., Ugalde, R.A., and Lepek, V.C. (2009) Characterization of the Mesorhizobium loti MAFF303099 type‐three protein secretion system. Mol Plant Microbe Interact 22: 519–528. [DOI] [PubMed] [Google Scholar]
- Sánchez, C., Mercante, V., Babuin, M.F., and Lepek, V.C. (2012) Dual effect of Mesorhizobium loti T3SS functionality on the symbiotic process. FEMS Microbiol Lett 330: 148–156. [DOI] [PubMed] [Google Scholar]
- Sannazzaro, A.I., Torres Tejerizo, G., Fontana, M.F., Cumpa Velásquez, L.M., Hansen, L.H., Pistorio, M., and Estrella, M.J. (2018) Mesorhizobium sanjuanii sp. nov., isolated from nodules of Lotus tenuis in the saline‐alkaline lowlands of Flooding Pampa. Argentina. Int J Syst Evol Microbiol 68: 2936–2942. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Suzaki, T., Soyano, T., Kojima, M., Sakakibara, H., and Kawaguchi, M. (2014) Shoot‐derived cytokinins systemically regulate root nodulation. Nat Commun 5: 1–9. [DOI] [PubMed] [Google Scholar]
- Sathoff, A.E., and Samac, D.A. (2018) Antibacterial activity of plant defensins. Mol Plant Microbe Interact 32: 507–514. [DOI] [PubMed] [Google Scholar]
- Scheidle, H., Groß, A., and Niehaus, K. (2005) The lipid a substructure of the Sinorhizobium meliloti lipopolysaccharides is sufficient to suppress the oxidative burst in host plants. New Phytol 165: 559–566. [DOI] [PubMed] [Google Scholar]
- Schnabel, E., Journet, E.‐P., de Carvalho‐Nierbel, F., Duc, G., and Frugoli, J. (2005) The Medicago truncatula SUNN gene encodes a CLV1‐like leucine‐rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822. [DOI] [PubMed] [Google Scholar]
- Scholl, D. (2017) Phage tail‐like bacteriocins. Annu Rev Virol 4: 453–467. [DOI] [PubMed] [Google Scholar]
- Schubert, K.R., and Evans, H.J. (1976) Hydrogen evolution: a major factor affecting the efficiency of nitrogen fixation in nodulated symbionts. Proc Natl Acad Sci USA 73: 1207–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, K.R., Jennings, N.T., and Evans, H.J. (1978) Hydrogen reactions of nodulated leguminous plants: II. Effects on dry matter accumulation and nitrogen fixation. Plant Physiol 61: 398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scupham, A.J., Bosworth, A.H., Ellis, W.R., Wacek, T.J., Albrecht, K.A., and Triplett, E.W. (1996) Inoculation with Sinorhizobium meliloti RMBPC‐2 increases alfalfa yield compared with inoculation with a nonengineered wild‐type strain. Appl Environm Microbiol 62: 4260–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, I.R., Men, A.E., Buzas, D.M., Iturbe‐Ormaetxe, I., Carroll, B.J., and Gresshoff, P.M. (2003) Long‐distance signaling in nodulation directed by a CLAVATA1‐like receptor kinase. Science 299: 109–112. [DOI] [PubMed] [Google Scholar]
- Skorpil, P., Saad, M.M., Boukli, N.M., Kobayashi, H., Ares‐Orpel, F., Broughton, W.J., et al. (2005) NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii . Mol Microbiol 57: 1304–1317. [DOI] [PubMed] [Google Scholar]
- Skorupska, A., Janczared, M., Marczak, M., Mazur, A., and Drol, J. (2006) Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb Cell Fact 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, P., Limpens, E., Geurts, R., Fedorova, E., Dolgikh, E., Gough, C., and Bisseling, T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón, M., Morera, C., Kondorosi, A., Lopez, O., and Miranda, J. (2001) A Purine‐related metabolite negatively regulates fixNOQP expression in Sinorhizobium meliloti by modulation of fixK expression. Mol Plant Microbe Interact 14: 572–576. [DOI] [PubMed] [Google Scholar]
- Soto, M.J., Domínguez‐Ferreras, A., Pérez‐Mendoza, D., Sanjuán, J., and Olivares, J. (2009) Mutualism versus pathogenesis: the give‐and‐take in plant‐bacteria interactions. Cell Microbiol 11: 381–388. [DOI] [PubMed] [Google Scholar]
- Soto, M.J., Sanjuán, J., and Olivares, J. (2006) Rhizobia and plant‐pathogenic bacteria: common infection weapons. Microbiol 152: 3167–3174. [DOI] [PubMed] [Google Scholar]
- Souza, J.E.B., and Ferreira, E.P.B. (2017) Improving sustainability of common bean production systems by co‐inoculating rhizobia and azospirilla. Agric Ecosyst Environ 237: 250–257. [Google Scholar]
- Sprent, J.I. (2008) 60Ma of legume nodulation. What’s new? What’s changing? J Exp Bot 59: 1081–1084. [DOI] [PubMed] [Google Scholar]
- Staehelin, C., and Krishnan, H.B. (2015) Nodulation outer proteins: double‐edged swords of symbiotic rhizobia. Biochem J 470: 263–274. [DOI] [PubMed] [Google Scholar]
- Sugawara, M., and Sadowsky, M.J. (2014) Enhanced nodulation and nodule development by nolR mutants of Sinorhizobium medicae on specific Medicago host genotypes. Mol Plant Microbe Interact 27: 328–335. [DOI] [PubMed] [Google Scholar]
- Sugawara, M., Takahashi, S., Umehara, Y., Iwano, H., Tsurumaru, H., Odake, H., et al. (2018) Variation in bradyrhizobial NopP effector determines symbiotic incompatibility with Rj2‐soybeans via effector‐triggered immunity. Nat Commun 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarnalakshmi, K., Yadav, V., Tyagi, D., Dhar, D.W., Kannepalli, A., and Kumar, S. (2020) Significance of plant growth promoting rhizobacteria in grain legumes: growth promotion and crop production. Plants 9: 1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syska, C., Brouquisse, R., Alloing, G., Pauly, N., Frendo, P., Bosseno, M., et al. (2019) Molecular weapons contribute to intracellular rhizobia accommodation within legume host cell. Front Plant Sci 10: 1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, M., and Capela, D. (2020) Rhizobium diversity in the light of evolution. Adv Bot Res 94: 251–288. [Google Scholar]
- Tellström, V., Usadel, B., Thimm, O., Stitt, M., Küster, H., and Niehaus, K. (2007) The lipopolysaccharide of Sinorhizobium meliloti suppresses defense‐associated gene expression in cell cultures of the host plant Medicago truncatula . Plant Physiol 143: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, H. (2013) Senescence, ageing and death of the whole plant. New Phytol 197: 696–711. [DOI] [PubMed] [Google Scholar]
- Tilak, K.V.B.R., Ranganayaki, N., and Manoharachari, C. (2006) Synergistic effects of plant‐growth promoting rhizobacteria and Rhizobium on nodulation and nitrogen fixation by pigeonpea (Cajanus cajan). Eur J Soil Sci 57: 67–71. [Google Scholar]
- Torres, A.R., Brito, B., Imperial, J., Palacios, J.M., Ciampitti, I.A., Ruiz‐Argüeso, T., and Hungria, M. (2020) Hydrogen‐uptake genes improve symbiotic efficiency in common beans (Phaseolus vulgaris L.). Antonie Van Leeuwenhoek 113: 687–696. [DOI] [PubMed] [Google Scholar]
- Torres‐Quesada, O., Oruezabal, R.I., Peregrina, A., Jofré, E., Lloret, J., Rivilla, R., et al. (2010) The Sinorhizobium meliloti RNA chaperon Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol 10: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traubenik, S., Reynoso, M.A., Hobecker, K., Lancia, M., Hummel, M., Rosen, B., et al. (2020) Reprogramming of root cells during nitrogen‐fixing symbiosis involves dynamic polysome association of coding and non‐coding RNAs. Plant Cell 32: 352–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchant, J.C., and Rigaud, J. (1982) Nitrite and nitric oxide as inhibitors of nitrogenase from soybean bacteroids. Appl Environ Microbiol 44: 1385–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett, E.W., and Barta, T.M. (1987) Trifolitoxin production and nodulation are necessary for the expression of superior nodulation competitiveness by Rhizobium leguminosarum bv. trifolii strain T24 on clover. Plant Physiol 85: 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi, T., Ohwada, T., Itakura, M., Mitsui, H., Nukui, N., Dawadi, P., et al. (2004) Expression islands clustered on the symbiosis island of the Mesorhizobium loti genoma. J Bacteriol 186: 2439–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron, J., Desbrosses, G., Bouffaud, M.‐L., Touraine, B., Moënne‐Loccoz, Y., Muller, D., et al. (2013) Plant growth‐promoting rhizobacteria and root system functioning. Front Plant Sci 4: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde, W., Zehirov, G., Szatmari, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126. [DOI] [PubMed] [Google Scholar]
- Van Dillewijn, P., Soto, M.J., Villadas, P.J., and Toro, N. (2001) Construction and environmental release of a Sinorhizobium meliloti strain genetically modified to be more competitive for alfalfa nodulation. Appl Environ Microbiol 67: 3860–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., and Boyce, A.N. (2016) Role of plant growth promoting rhizobacteria in agricultural sustainability – a review. Molecules 21: 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinardell, J.M., Ollero, F.J., Hidalgo, A., López‐Baena, F.J., Medina, C., Ivanov‐Vangelov, K., et al. (2004) NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol Plant Microbe Interact 17: 676–685. [DOI] [PubMed] [Google Scholar]
- Viprey, V., Del Greco, A., Golinowski, W., Broughton, W.J., and Perret, X. (1998) Symbiotic implications of type III protein secretion machinery in Rhizobium . Mol Microbiol 28: 1381–1389. [DOI] [PubMed] [Google Scholar]
- Vives‐Peris, V., de Ollas, C., Gómez‐Cadenas, A., and Pérez‐Clemente, R.M. (2020) Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep 39: 3–17. [DOI] [PubMed] [Google Scholar]
- Wang, Q.I., Liu, J., and Zhu, H. (2018) Genetic and molecular mechanisms underlying symbiotic specificity in legume‐rhizobium interactions. Front Plant Sci 9: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke, A., Malisic, M., Wawra, S., and Zuccaro, A. (2021) Unraveling the sugar code: the role of microbial extracellular glycans in plant‐microbe interactions. J Exp Bot 72: 15–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeier, S., Arnold, M.F., Marlow, V.L., Aouida, M., Myka, K., Fletcher, V., et al. (2010) Internalization of a thiazole‐modified peptide in Sinorhizobium meliloti occus by BacA‐dependent and independent mechanisms. Microbiology 156: 2702–2713. [DOI] [PubMed] [Google Scholar]
- Wendlandt, C.E., Regus, J.U., Gano‐Cohen, K.A., Hollowell, A.C., Quides, K.W., Lyu, J.Y., et al. (2019) Host investment into symbiosis varies among genotypes of the legume Acmispon strigosus, but host sanctions are uniform. New Phytol 221: 446–458. [DOI] [PubMed] [Google Scholar]
- Whitney, J.C., Beck, C.M., Goo, Y.A., Russell, A.B., Harding, B.N., De Leon, J.A., et al. (2014) Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol 92: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, D.‐W., Liao, S., Xie, Z.‐P., Hann, D.R., Steinle, L., Boller, T., and Staehelin, C. (2012) Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog 8: e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y., and Inouye, M. (2011) Regulation of growth and death in Escherichia coli by toxin‐antitoxin systems. Nat Rev Microbiol 9: 779–790. [DOI] [PubMed] [Google Scholar]
- Yang, S., Wang, Q., Fedorova, E., Liu, J., Qin, Q., Zheng, Q., et al. (2017) Microsymbiont discrimination mediated by a host‐secreted peptide in Medicago truncatula . Proc Natl Acad Sci USA 114: 6848–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, M., Miwa, H., Masuda, S., Takebayashi, Y., Sakakibara, H., and Okazaki, S. (2016) Effector‐triggered immunity determines host genotype‐specific incompatibility in legume‐Rhizobium symbiosis. Plant Cell Physiol 57: 1791–1800. [DOI] [PubMed] [Google Scholar]
- Zablotowicz, R.M., and Reddy, K.N. (2004) Impact of glyphosate on the Bradyrhizobium japonicum symbiosis with glyphosate‐resistant transgenic soybean: a minireview. J Environ Qual 33: 825–831. [DOI] [PubMed] [Google Scholar]
- Zalguizuri, A., Caetano‐Anollés, G., and Lepek, V.L. (2019) Phylogenetic profiling, an untapped resource for the prediction of secreted proteins and its complementation with sequence‐based classifiers in bacterial type III, IV and VI secretion systems. Briefings Bioinformatics 20: 1395–1402. [DOI] [PubMed] [Google Scholar]
- Zhang, D., de Souza, R.F., Anantharaman, V., Iyer, L.M., and Aravind, L. (2012) Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]


