Abstract
Since Darwin, evolutionary biologists have sought to understand the drivers and mechanisms of natural trait diversity. The field advances toward this goal with the discovery of phenotypes that vary in the wild, their relationship to ecology, and their underlying genes. Here, we established resistance to extreme low temperature in the free-living nematode Caenorhabditis briggsae as an ecological and evolutionary model system. We found that C. briggsae strains of temperate origin were strikingly more cold-resistant than those isolated from tropical localities. Transcriptional profiling revealed expression patterns unique to the resistant temperate ecotype, including dozens of genes expressed at high levels even after multiple days of cold-induced physiological slowdown. Mutational analysis validated a role in cold resistance for seven such genes. These findings highlight a candidate case of robust, genetically complex adaptation in an emerging model nematode, and shed light on the mechanisms at play.
Keywords: Caenorhabditis briggsae, cold tolerance, temperate and tropical clusters, transcriptome, cold-evoked expression
Significance
A main goal of evolutionary genetics is to understand how organisms acquire new traits that allow them to thrive in their environments. To meet this challenge, we need to study populations of a species of interest that have adapted to different niches and are genetically tractable—and such systems remain at a premium in the field. Here, we discover a qualitative difference in cold resistance between wild nematode worm populations, and we demonstrate the power of molecular tools ported from lab raised worms to study the molecular mechanisms of the trait.
Introduction
Understanding how and why organisms differ in the wild is a key goal of evolutionary biology. Some traits are evolutionary accidents, and others arise under local adaptation, fixing in a population when they promote fitness in a new niche. Dissecting these processes requires case studies where we can establish the underlying ecology, the phenotypes that have evolved, and, ultimately, the molecular mechanisms. Invertebrates can provide exceptional power toward this end (Sanford and Kelly 2011; Savolainen et al. 2013; Adrion et al. 2015; Kraemer and Boynton 2017), but even in lower eukaryotes, genetically tractable ecological study systems remain at a premium in the field. Of particular interest are cases in which tools from the lab can be brought to bear to study evolution in natural settings.
Caenorhabditis briggsae, like its relative the model organism Caenorhabditis elegans, is a free-living nematode that has been isolated all over the world (Cutter et al. 2006; Prasad et al. 2011; Baird and Stonesifer 2012; Stegeman et al. 2013; Okahata et al. 2016). Phylogenetic analyses have established several well-defined C. briggsae populations (Thomas et al. 2015), with a particularly well-characterized distinction between strains isolated from temperate and tropical localities (Graustein et al. 2002; Cutter et al. 2006; Thomas et al. 2015; Ferrari et al. 2017). The contrast between these populations serves as a useful framework for the study of ecological diversification. Under current models, ancestral C. briggsae occupied the tropical niche, with recent colonization of and expansion into temperate latitudes, possibly associated with human activity (Cutter et al. 2006). Elegant reports have characterized differences between temperate and tropical C. briggsae in temperature-dependent fecundity (Prasad et al. 2011) and behavior (Cutter et al. 2006; Baird and Stonesifer 2012; Stegeman et al. 2013, 2019), with a focus on chronic response to warm and hot conditions (14–30 °C). Resistance to acute temperature shock, as it has evolved among C. briggsae populations, remains less well understood. The response to extreme low temperature is a compelling potential character under ecological pressures in the C. briggsae system, as it is likely to manifest in the winter season of temperate latitudes and not in tropical regions (Lacher and Goldstein 1997).
In this study, we investigated the response to cold stress across C. briggsae strains isolated from different niches. We found that temperate strains survived cold conditions in which most animals of tropical origin died. Using two strains, AF16 and HK104, as representative of tropical and temperate ecotypes respectively, we profiled cold-responsive transcriptomes to achieve molecular insight into the divergence in the cold resistance trait. Against a backdrop of thousands of genes with cold-evoked expression patterns shared between the strains, we found >100 genes with high expression unique to cold-treated HK104. In mutational tests, we confirmed the role of seven such factors in cold resistance.
Results
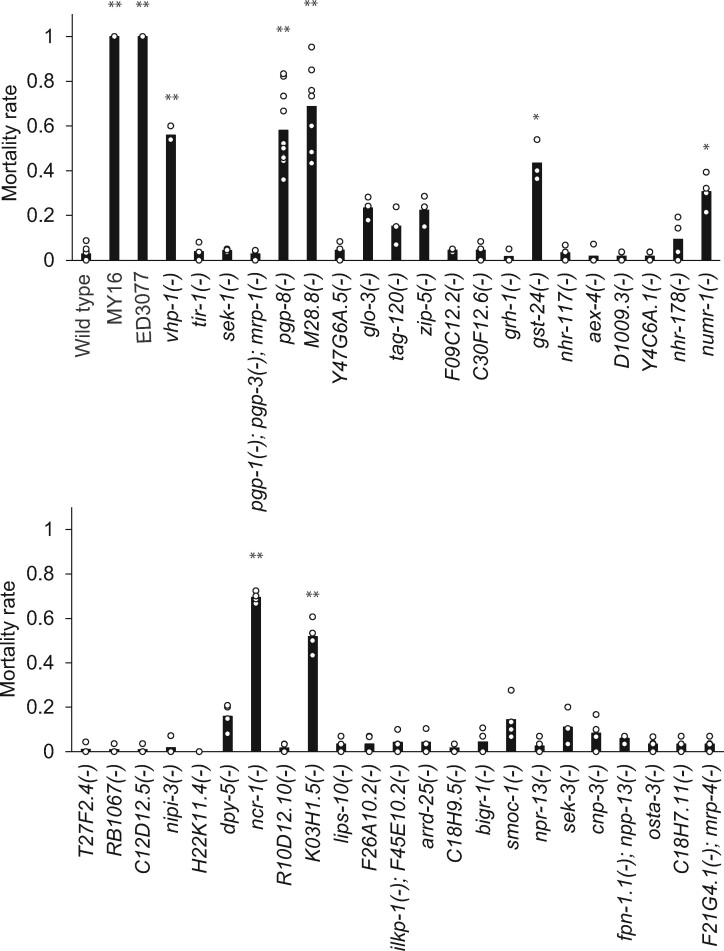
Wild C. briggsae Isolated from Temperate but Not Tropical Regions Survive Hypothermia Independent of Rearing Temperature
Since temperature is one of the major factors that distinguish temperate and tropical climates (Lacher and Goldstein 1997; Prasad et al. 2011), we hypothesized that C. briggsae strains from these two niches would respond differently to hypothermia. To test this, we first focused on C. briggsae strains that, in previous phylogenetic analysis (Cutter et al. 2006), had been assigned to a population referred to as Tropical (AF16, VT847, ED3083, JU726, and QX1410), and strains from a population with the name Temperate (HK104, PB826, JU439, EG4181, and VX34) (table 1). We cultivated each strain at 20 °C until the animals reached adulthood, and then switched them to 4 °C. All worms were immobile after 60 h of cold treatment, but upon recovery, Temperate strains survived at ∼100%, whereas Tropical strains died at high rates (fig. 1a). We next turned to additional C. briggsae strains (table 1) that had been isolated in localities qualifying as temperate and tropical, but whose genomes did not affiliate them with the named Temperate or Tropical populations (Cutter and Choi 2010; Thomas et al. 2015). Phenotyping revealed cold resistance in C. briggsae from Quebec, a sister clade to the Temperate population (Thomas et al. 2015), but not in strains from tropical sites, including those from the early-diverging Kerala population (fig. 1b). These data implicate cold resistance as a derived character in C. briggsae, with a likely origin in the ancestor of the Quebec and Temperate populations—the only ones in our survey from cooler localities.
Table 1.
Collection Localities of Wild Caenorhabditis briggsae Strains
| Strain | Locality of Origin | Phylogeographic Group | Latitude | Approximate Elevation (m) | Mean Winter Temperature (°C min, max) | Mean Summer Temperature (°C min, max) |
|---|---|---|---|---|---|---|
| AF16 | Ahmedabad, India | Tropical | 23°01′N | 50 | 14.4, 29.4 | 26.1, 33.9 |
| VT847 | HI, USA | Tropical | 20°57′N | 930 | 18.9, 26.1 | 22.8, 29.4 |
| ED3083 | Johannesburg, S. Africa | Tropical | 26°10′N | 1,750 | 2.8, 18.9 | 15.6, 27.2 |
| JU726 | Chengyang Village, China | Tropical | 44°28′N | 11 | 3.9, 12.8 | 24.4, 34.4 |
| QX1410 | St. Lucia | Tropical | 13°22′N | 14 | 23.0, 27.0 | 25.0, 31.0 |
| HK104 | Okayama, Japan | Temperate | 34°40′N | 30 | 0.0, 8.3 | 22.8, 30.6 |
| PB826 | OH, USA | Temperate | 39°34′N | 270 | −6.1, 2.8 | 17.8, 30.0 |
| JU439 | Reykjavik, Iceland | Temperate | 64°08′N | 16 | −2.2, 2.2 | 8.9, 14.4 |
| EG4181 | Salt Lake City, UT, USA | Temperate | 40°42′N | 1,290 | −7.8, 2.8 | 15.6, 34.4 |
| VX34 | Guangshui, Hubei, China | Temperate | 30°58′N | 80 | −6.1, 0.0 | 13.8, 23.9 |
| ED3101, ED3032, ED3034 | Nairobi, Kenya | Nairobi | 1°17′S | 1,798 | 11.0, 23.0 | 14.0, 28.0 |
| JU1341, JU1345, JU1348 | Kerala, India | Kerala | 10°51′N | 900 | 23.0, 32.0 | 25.5, 36.0 |
| QR24, QR25 | Quebec, Canada | Quebec | 46°49′N | 98 | −15.0, −7.0 | 15.0, 25.0 |
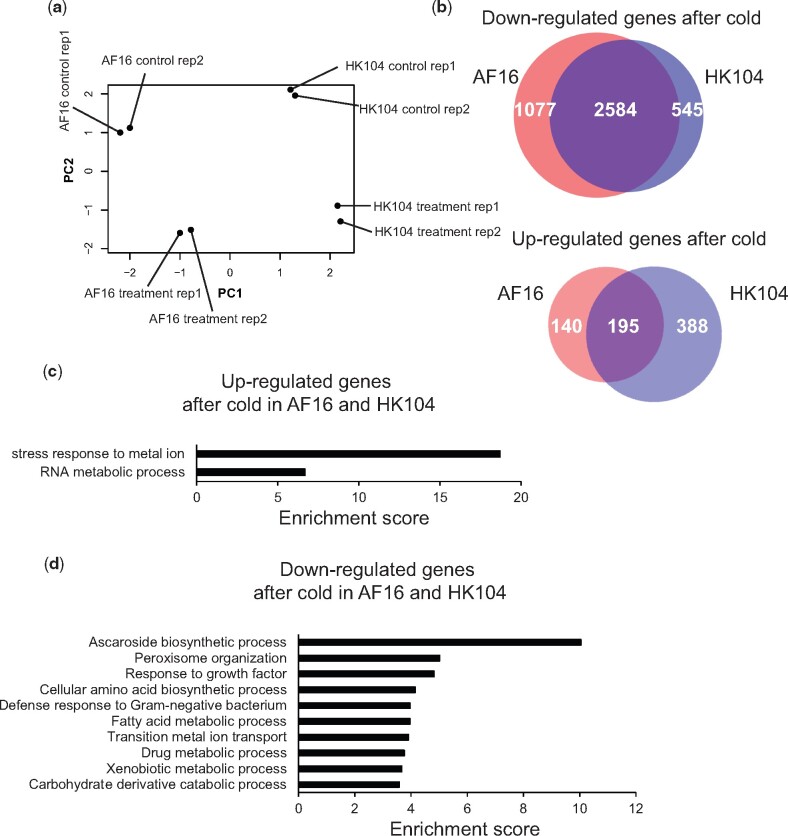
Fig. 1.
Cold survival differentiates tropical and temperate Caenorhabditis briggsae. Each bar reports the mean proportion of animals of the indicated C. briggsae strain surviving after development at 20 °C followed by 60 h of incubation at 4 °C. Each point reports results from one biological replicate. See table 1 for strain information.
Cultivation temperature during development can affect the cold tolerance of adult worms (Ohta et al. 2014; Okahata et al. 2016). To examine this effect in the C. briggsae system, we used AF16, isolated from tropical India, and HK104, from a temperate niche in Japan, as representatives of the respective ecotypes. As expected (Ohta et al. 2014), cold tolerance decreased in both AF16 and HK104 animals that had gone through development at higher temperatures (fig. 2) and in worms treated with longer cold shock (supplementary fig. 1, Supplementary Material online). However, at all rearing temperatures, HK104 was far more likely to survive cold treatment than was AF16 (fig. 2). Tests in F1 hybrids from the mating of AF16 and HK104 indicated a partly dominant basis for the phenotype, suggesting appreciable genetic complexity (supplementary fig. 2, Supplementary Material online).
Fig. 2.
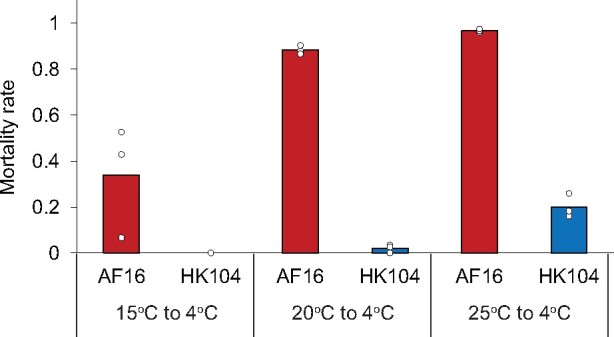
The temperate Caenorhabditis briggsae strain HK104 is much more cold-tolerant than tropical strain AF16 regardless of rearing temperature. Symbols are as in figure 1, except that rearing temperature was 15 °C (left), 20 °C (middle), or 25 °C (right) before cold shock.
Transcriptional Responses to Cold Stress Unique to and Shared between C. briggsae Strains
To gain molecular insight into differences in cold stress response between tropical and temperate C. briggsae, we took a transcriptional approach, again making use of the AF16 and HK104 isolates as a model comparison. Transcriptome profiles of these strains before and after cold treatment revealed robust clustering by temperature and strain background, and tight agreement between replicates (fig. 3a and supplementary table 1, Supplementary Material online).
Fig. 3.
Cold-evoked expression programs shared between AF16 and HK104. (a) Shown are the results of principal component analysis of transcriptomes of Caenorhabditis briggsae temperate HK104 and tropical AF16 before and after cold treatment (60 h of incubation at 4 °C after development at 20 °C). Each point reports values of the first (x axis) and second (y axis) principal component from one replicate of the indicated strain and condition. (b) Top, each circle reports the number of genes in the indicated strain with lower expression upon cold treatment than in control conditions. White values report the number of such genes detected only in AF16 (left), only in HK104 (right), or in both strains (center). Bottom, data are as at top except that genes with higher expression in cold conditions were analyzed. (c) Each bar reports enrichment (as a ratio of the number observed to the number expected under a genomic null) of genes with the indicated function among those detected in AF16 and HK104 as downregulated under cold treatment. (d) Data are as in (c) except that genes with higher expression in cold conditions were analyzed.
We first analyzed AF16 and HK104 separately with respect to cold-evoked changes in gene expression, and inspected commonalities between the strains. In these data, a pattern of declining RNA levels predominated in both AF16 and HK104, with 2584 genes expressed at lower levels upon cold treatment in both strains (at a false discovery rate of 0.15; fig. 3b). Functional-genomic analyses of the latter revealed enrichment for a variety of gene groups involved in growth and cellular metabolism (fig. 3d), as expected if cold-treated animals of both strains slow or arrest many biological processes (Robinson and Powell 2016; Jiang et al. 2018). By contrast, relatively few genes in each strain were expressed at higher levels in the cold relative to an untreated control (fig. 3b). Only 195 genes exhibited high RNA levels in cold conditions in both AF16 and HK104, with enrichment for functions in RNA processing and metal ion stress (fig. 3c). More salient from these data was the apparent bias for a program unique to temperate HK104, in which we detected an excess of cold-evoked genes that were not called in the analogous tests on AF16 transcriptomes (fig. 3b).
We thus turned our attention to a more rigorous search for strain-by-temperature expression effects (see Materials and Methods). The results revealed 191 genes for which the expression response to cold shock was distinct between AF16 and HK104 (at a false discovery rate of 0.15; supplementary table 1, Supplementary Material online). These cases of expression divergence were largely specific to cold treatment, with few striking difference between the strains at 20 °C (supplementary fig. 3, Supplementary Material online). Most (164 genes) followed a pattern of high cold-evoked expression in the temperate strain, HK104, and dropped in expression in the cold in AF16 (fig. 4 and supplementary fig. 3, Supplementary Material online).
Fig. 4.
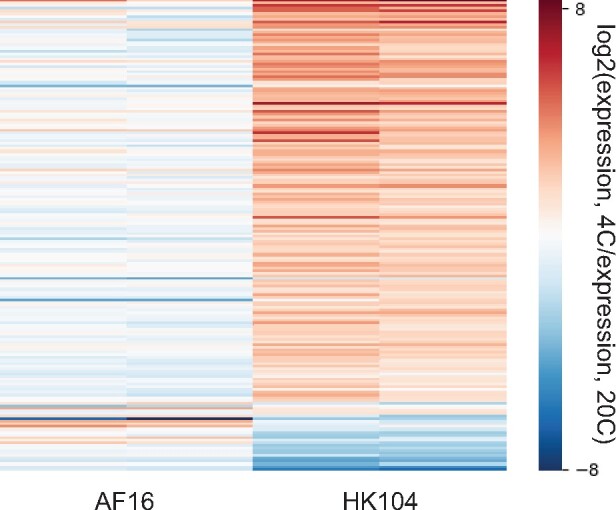
Cold-evoked expression programs that differ between AF16 and HK104. Shown are genes with significant strain-by-temperature effects on expression, in analyses of transcriptomes from figure 3. Each row reports expression measurements for one gene; each column reports one replicate comparison of the indicated temperatures for the indicated strain.
We thus formulated a model in which HK104 expressed a unique program of cold-protective factors, whose components could underlie cold resistance at the organismal level, and we earmarked this program for mechanistic follow-up. In transcriptional profiles of AF16 x HK104 hybrid animals, we saw little evidence for cis-regulatory divergence at these focal genes (supplementary table 1, Supplementary Material online), implicating trans-acting variants as the underlying cause of the expression patterns of interest.
Natural Variation and Genetics of Cold Tolerance in C. elegans
We set out to use gene ablation to test the phenotypic role of our candidate genes, those expressed at high levels in HK104 during survival of hypothermia. We reasoned that an expedient paradigm for this purpose could use C. elegans. Biologically, this model species is much better characterized than C. briggsae (Hillier et al. 2007), and genetic mutants are widely available, as opposed to the few mutants generated to date in C. briggsae (Hillier et al. 2007); many developmental, behavioral, and physiological phenotypes are conserved between the species (Hillier et al. 2007; Yin et al. 2018; Culp et al. 2020).
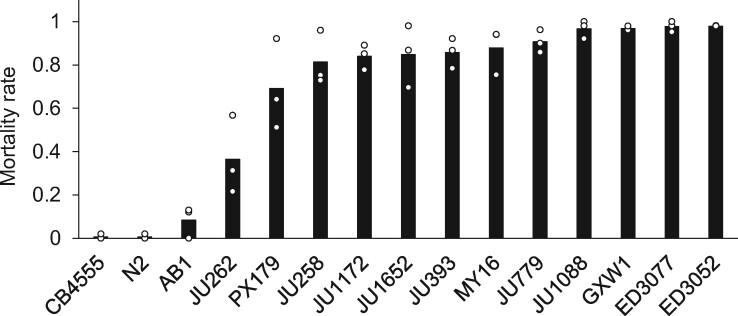
To explore the utility of C. elegans as a model for cold resistance, we first assayed cold tolerance in 14 wild C. elegans strains from temperate locales, and one from a tropical region (supplementary table 2, Supplementary Material online). We used a regimen of rearing at 20 °C and cold shock at 4 °C for 60 h, in which the C. elegans laboratory strain N2 exhibits robust resistance (Ohta et al. 2014). Our results revealed complete lethality in response to cold shock in most other C. elegans isolates (fig. 5). Beside N2, originally isolated from England, and CB4555, an N2 descendant (Sterken et al. 2015), only AB1, an Australian isolate previously shown to acclimate rapidly to cold (Okahata et al. 2016), exhibited cold resistance on par with that of temperate-clade C. briggsae (fig. 5). These data indicated that, although rare among our sampled C. elegans isolates, the cold tolerance manifesting in the laboratory strain could serve as a useful model for that of temperate C. briggsae, with the potential for insights from genetic analyses.
Fig. 5.
Cold tolerance variation across wild Caenorhabditis elegans isolates. Symbols are as in figure 1 except that wild C. elegans strains were analyzed; see supplementary table 2, Supplementary Material online, for strain information.
To this end, we carried out a mutant survey in C. elegans of cold-induced genes in HK104. To cast the widest possible net for genes of interest, we selected them from our expression data with more lenient cutoffs than we had used for initial genomic analyses (see Materials and Methods), amounting to 43 total genes for the survey. For each, we acquired a strain of the N2 background harboring a mutation in the respective gene, in some cases in a background also including other lesions. Assays for cold resistance revealed seven genes as necessary for the trait in C. elegans N2 (fig. 6): vhp-1 (encoding a MAPK phosphatase), pgp-8 (an ABC transporter), ncr-1 (involved in cholesterol trafficking), gst-24 (glutathione-S-transferase), numr-1 (a hypothesized splicing factor), and the uncharacterized genes M28.8 and K03H1.5. By virtue of their role in cold tolerance in C. elegans, and their unique cold-evoked induction profile in C. briggsae HK104, this set of genes represented our top candidates for determinants of cold resistance in the latter.
Fig. 6.
Caenorhabditis elegans mutant survey reveals seven genes that impair cold tolerance. Each bar reports cold tolerance in a C. briggsae wild strain (MY16 and ED3077; see table 1); the C. elegans laboratory strain N2 (wild-type); or a strain isogenic to N2 harboring a mutation (-) in the indicated gene(s). Symbols are as in figure 1 except that cold treatment was 120 h. Asterisks report results from a one-sided t-test comparing the indicated strain to wild-type N2; *P < 0.05; **P < 0.01.
Genetic Determinants of Cold Resistance in Temperate C. briggsae
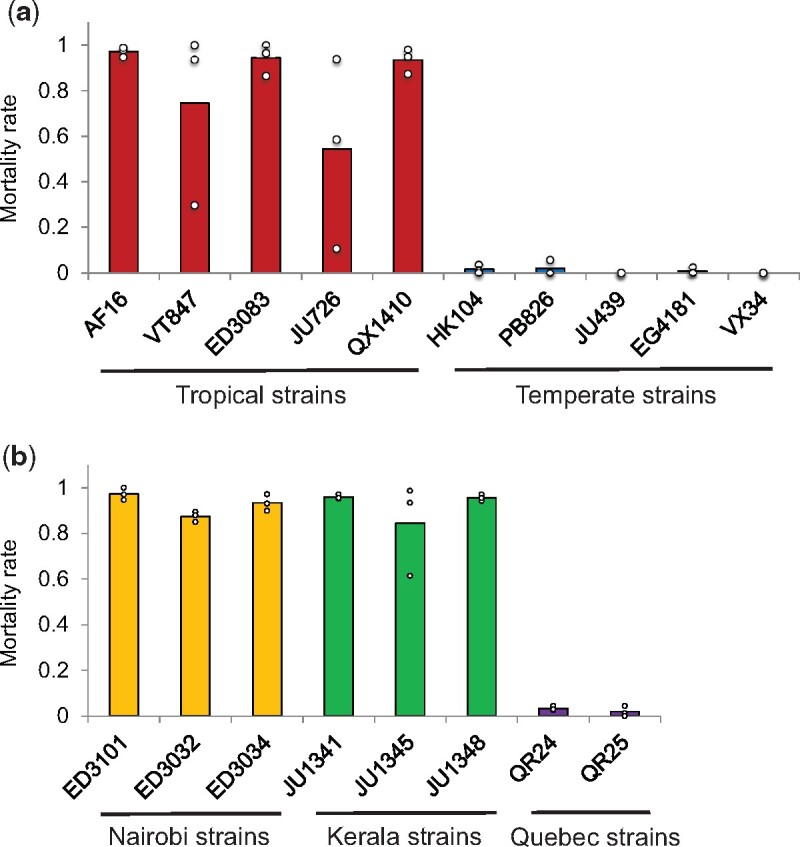
To explore the phenotypic role of our candidate genes in C. briggsae, we made use of a CRISPR-Cas9 system for targeted mutations, for which we chose to focus on five genes with the largest effect size in our C. elegans cold resistance mutant survey (vhp-1, pgp-8, M28.8, ncr-1, and K03H1.5). Of these, we were unable to develop homozygous mutants for ncr-1 in the HK104 temperate C. briggsae strain, suggesting an essential function for this gene. For each of the remaining genes in turn (vhp-1, pgp-8, M28.8, and K03H1.5), we established an HK104 line and, separately, a line of tropical C. briggsae AF16 harboring a premature stop codon in the coding region (supplementary table 3, Supplementary Material online). We then investigated the cold survival phenotype of each such mutant strain. For this purpose, given the extreme cold resistance of HK104 (figs. 1 and 2), we subjected mutants in this background to five days of cold treatment alongside a wild-type control. Results revealed a robust and significant increase in cold shock lethality in pgp-8, M28.8, and K03H1.5 mutants, confirming the importance of these genes in the HK104 phenotype (fig. 7a). In the AF16 background, which is radically cold-sensitive (figs. 1 and 2), we were required to use a shorter-duration cold-shock assay design. Here, we observed no effect of vhp-1, pgp-8, M28.8, or K03H1.5 mutation at any timepoint (fig. 7b). These data establish that several of our top genes contribute uniquely to the cold resistance phenotype in HK104, validating our inference from this strain’s expression profiles (fig. 4) as a resource for mechanistic insights in this system.
Fig. 7.
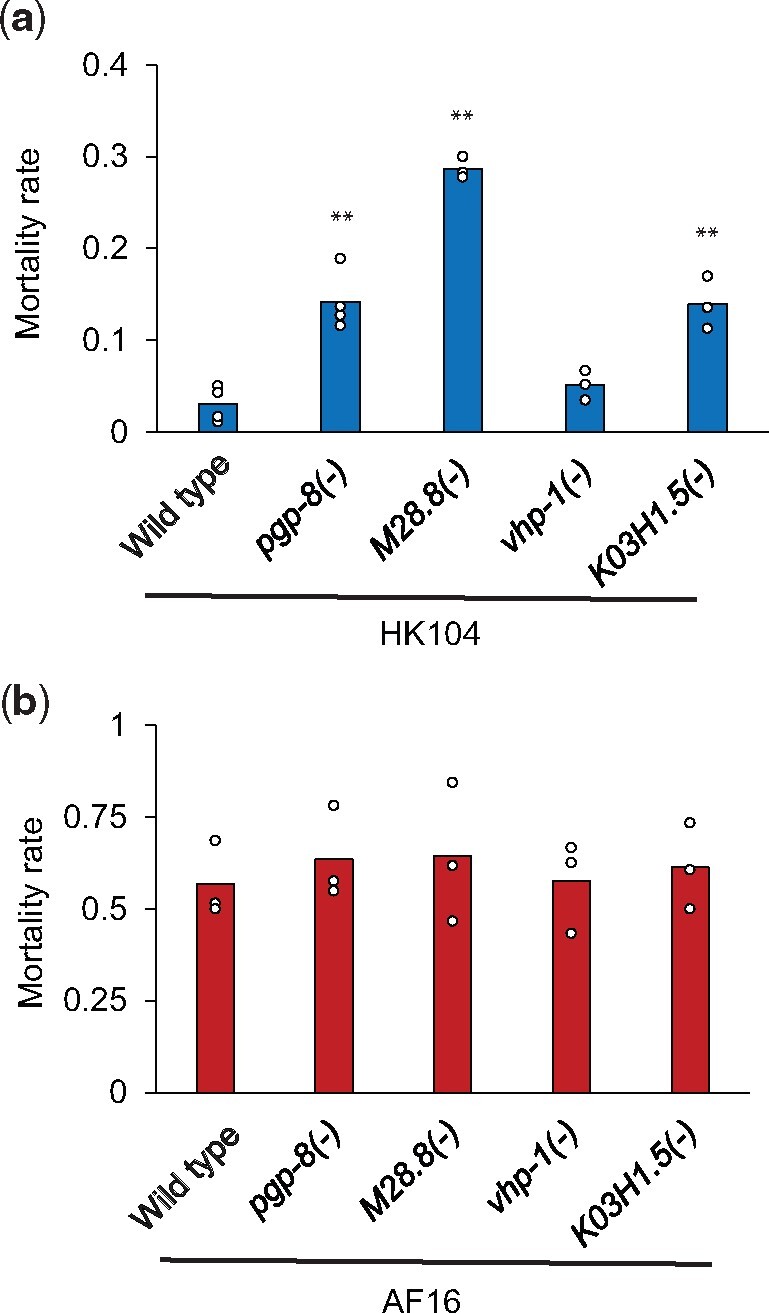
Identification of three genes important for HK104 cold tolerance. (a) Each bar reports cold tolerance in the temperate Caenorhabditis briggsae HK104 strain (wild-type) or an isogenic strain harboring a mutation (-) in the indicated gene. Symbols are as in figure 1 except that cold exposure was for five days. (b) Symbols are as in (a) except that the tropical C. briggsae AF16 strain was analyzed, and cold exposure was for 48 h. Asterisks report results from a one-sided t-test comparing the indicated strain to the wild-type of its respective background; *P < 0.05; **P < 0.01. See supplementary table 3, Supplementary Material online, for strain information.
Discussion
Understanding diversity in the natural world requires the discovery of traits that have changed in response to ecological factors, and the use of molecular tools to understand their mechanisms. Heat and cold responses are particularly ripe for evolutionary study in ectotherms, whose body temperature depends on that of their environment (Flouris and Piantoni 2015). In this work, we have established resistance to cold shock as a character distinguishing the Temperate and Quebec clades of the nematode C. briggsae from cold-sensitive tropical isolates. Our findings dovetail with the known fecundity disadvantage of Temperate C. briggsae at high temperature relative to Tropical strains (Prasad et al. 2011), as well as behavioral (Cutter et al. 2006; Stegeman et al. 2013, 2019) and transcriptional (Mark et al. 2019) differences between the clades under hot conditions.
Our focus on cold was motivated by the cooler extreme winter temperatures in the collection localities of Temperate C. briggsae (Stegeman et al. 2013; Adrion et al. 2015). By contrast, the average summer temperature is comparable in most such collection sites, regardless of whether the region is temperate or tropical (Prasad et al. 2011). Given these climatic factors, and the striking phenotype we report here, it is tempting to speculate that selection for cold survival in the winter months has been a key driver of local adaptation in C. briggsae as in other nematodes (McGaughran and Sommer 2014). Assuming that the trait arose in the ancestor of the Temperate-Quebec clades of C. briggsae, these evolutionary events would have happened during the tens of thousands of generations that separate this branch from its closest relatives (Thomas et al. 2015).
Evolutionary pressures on the trait in C. elegans remain less clear, since we and others (Okahata et al. 2016) have found strains of this species from temperate localities to run the gamut from cold-resistant to cold-sensitive. This may be the product of migration and admixture in the worldwide C. elegans population, preventing local adaptation in hot or cold environments, or in any one niche (although important exceptions have been reported: Petersen et al. 2015; Crombie et al. 2019; Lee et al. 2021). As C. elegans and C. briggsae diverged ∼20 Ma (Cutter 2008), cold tolerance in the two species likely arose independently. The vast divergence in their genomes (Stein et al. 2003) would represent differences in the chassis on which the trait was built, reflected in the fact that some genes we tested affected cold shock survival in resistant C. elegans but not C. briggsae.
Our evolutionary analysis of wild worms complements an extensive literature on the basic biology of hypothermia response in laboratory C. elegans. An important thread of this prior work has highlighted the dieoff of animals reintroduced into warm temperatures after cold shock, and its physiological and transcriptomic correlates (Robinson and Powell 2016; Jiang et al. 2018). For our expression profiling, instead of the rewarming recovery phase, we focused on the end of the cold exposure regimen, and the genes with high RNA levels at this timepoint. Our validation of a causal role in cold tolerance for seven such genes (in C. elegans, temperate C. briggsae, or both) suggests that cold resistance hinges in part on physiology during the treatment itself. Under one compelling model, protective factors expressed during cold stress could mitigate tissue damage before recovery even starts. Additionally, proteins expressed in the cold could set up a poised state for rapid signaling and repair during recovery. Ultimately, as a complete picture of the biology of cold resistance becomes clear, it will likely integrate mechanisms operating in the two phases; and the failures in tropical C. briggsae could prove to manifest in either one. The transcription factor gene ZIP-10, known to promote death during recovery after cold treatment (Jiang et al. 2018), was expressed less in tropical AF16 than in temperate HK104 in our profiles (supplementary table 1, Supplementary Material online), suggesting that the trait divergence between the strains hinges on a mechanism distinct from the ZIP-10 pathway.
That said, among the many other known molecular mechanisms of cold resistance in C. elegans (Murray et al. 2007; Ma et al. 2015; Robinson and Powell 2016; Sonoda et al. 2016; Jiang et al. 2018; Ujisawa et al. 2018; Okahata et al. 2019; Takagaki et al. 2020), some are echoed in the genes we have validated in the trait. For instance, regulating membrane lipid composition and fluidity is well-characterized as a strategy for cold tolerance in the worm (Murray et al. 2007; Ma et al. 2015); the cholesterol processing gene ncr-1, which we found to be required for cold tolerance in C. elegans, is likely to act through this mechanism. Likewise, detailed studies have established the role of sensory neurons in cold resistance (Ohta et al. 2014; Sonoda et al. 2016; Ujisawa et al. 2018; Okahata et al. 2019; Takagaki et al. 2020). This system could well involve M28.8, which we identified as required for the trait in C. elegans and temperate C. briggsae, is expressed in dopaminergic neurons, and is the ortholog of a Drosophila photoreceptor gene (Nie et al. 2012).
Furthermore, in our larger set of cold-evoked expression changes, several emergent patterns parallel those seen in the broader literature. In previous analysis of C. elegans, recovery after cold shock was associated with changes in lipid and amino acid metabolism (Jiang et al. 2018), groups from which many genes exhibited low expression during cold shock itself in our C. briggsae study. Likewise, RNA binding and stabilization factors are activated by cold in mammalian cells (Sonna et al. 2002) and in yeast and bacteria (Aguilera et al. 2007; Keto-Timonen et al. 2016), just as we have seen in C. briggsae. Metal ion stress response, another cold-induced gene set in mammalian cells (Sonna et al. 2002), also featured among upregulated genes in our C. briggsae data. Additional stress response genes are induced in cold-shock studies of other organisms, including heat shock factors (Sonna et al. 2002; Aguilera et al. 2007; Rinehart et al. 2007; Shore et al. 2013; Keto-Timonen et al. 2016), whose relevance in the worm remains to be elucidated.
In summary, we have established cold resistance in C. briggsae as a rapidly evolved, and likely adaptive, product of ecological diversification, and we have traced the attendant expression changes in genes required for cold tolerance. These findings underscore the power of ecological-genetic studies in wild strains of a tractable model organism, which hold promise for continued progress in the discovery of how nature builds new traits.
Materials and Methods
Worm Strains
Wild-type strains of C. briggsae (AF16, VT847, ED3083, JU726, QX1410, HK104, PB826, JU439, EG4181 VX34, ED3101-4, JU1341, JU1345, JU1348, and QR24-25) and C. elegans (CB4555, N2, AB1, JU262, PX179, JU258, JU1172, JU1652, JU393, MY16, JU779, JU1088, GXW1, ED3077, and ED3052) are described in table 1 and supplementary table 2, Supplementary Material online, respectively. To generate and transcriptionally profile AF16 x HK104 F1 hybrids, we first made a marked strain of AF16, CP161 (Cbr-unc-119(nm67) III; nmIs7 [Cni-mss-1(+) + Cni-mss-2(+) + Cbr-myo-2::GFP + unc-119(+)]). We then crossed HK104 hermaphrodites with CP161 males, picked labeled hermaphrodite F1 progeny at the L4 stage, and used them as input into cold treatment and expression profiling procedures as detailed below.
Caenorhabditis elegans mutant strains profiled in figure 5 are as follows: JT366 vhp-1(sa366) II, IG685 tir-1(tm3036) III, KU4 sek-1(km4) X, NL152 pgp-1(pk17) IV; pgp-3(pk18) X; mrp-1(pk89) X, RB1916 pgp-8(ok2489) X, RB1840 M28.8(ok2380) II, VC2677 Y47G6A.5(gk1098) I, GH403 glo-3(kx94) X, VC422 tag-120(gk221) V, VC1392 zip-5(gk646) V, RB792 F09C12.2(ok582) II, RB1284 C30F12.6(ok1381) I, VC2072 grh-1(gk960) I, RB2200 gst-24(ok2980) II, VC1499 nhr-117(gk707) V, JT5244 aex-4(sa22) X, RB1267 D1009.3(ok1349) X, RB2171 Y4C6A.1(ok2938) IV, VC2214 nhr-178(gk1005) V, RB1749 numr-1(ok2239) III, RB2499 T27F2.4(ok3462) V, RB1067 his-24(ok1024) X, VC1544 C12D12.5(gk700) X, IG544 nipi-3(fr4) X, RB1362 H22K11.4(ok1529) X, BC15170 dpy-5(e907) I, COP677 ncr-1(knu4) X, swt-6(tm5930), K03H1.5(tm10908), lips-10(tm7601), F26A10.2(tm549), F45E10.2(tm5965), arrd-25(tm12435), C18H9.5; C18H9.1(tm12783), bigr-1(tm6317), smoc-1(tm7000), npr-13(tm1504), sek-3(tm1344), cnp-3(tm2950), fpn-1.1; npp-13(tm6914), osta-3(tm5747), C18H7.11(tm12727), F21G4.1; mrp-4(tm10068).
Cold Tolerance Assays
Cold tolerance assays were performed as described (Jiang et al. 2018). For a given biological replicate of a given strain or treatment condition, 20–30 worms were spread on each of two to three plates and raised at 20 °C until 24 h after they reached the L4 stage. Plates were then spread with equal distance in a box and transferred to a constant 4 °C cold room for the duration indicated below and in figure captions. Plates were then moved to room temperature for 2 h before survival scoring, in which worms that failed to respond to a gentle prodding with a platinum wire were scored as dead. For each strain at least three independent experiments were performed.
RNA Isolation and Library Preparation for RNA-Seq
RNA isolation was performed essentially as described (Wang et al. 2018). For a given biological replicate of a given strain, 100–200 synchronized mid/late L4 stage worms were picked and incubated at 20 °C for 24 h, after which one cohort of worms was harvested immediately, representing the untreated control, and another cohort was subjected to 4 °C treatment for 60 h as above, followed by harvest. Two replicates for each strain and each condition were performed. Collected worms were homogenized in 1 ml Tri-reagent (ThermoFisher) for 30 min at room temperature. About 0.1 ml of bromochloropropane (BCP, Sigma) was added to the sample and mixed well. The sample was then spun at 12,000×g for 15 min at 4 °C and the aqueous phase was transferred to a new tube. About 0.5 ml isopropanol was added to the sample, followed by incubation at room temperature for 10 min and centrifugation at 12,000×g for 10 min. The RNA pellet was washed twice with 75% EtOH and dissolved in water. The RNA sample was then purified to remove DNA using the RNeasy Mini Kit (Qiagen), and used as input into RNA-seq library preparation using the KAPA RNA hyper prep kit (Roche).
Amended Reference Genome Construction
We established a reference genome for C. briggsae strain HK104 as follows. Raw genome sequencing reads for HK104 were downloaded from the NCBI (project accession PRJNA509247). These reads were aligned to genome assembly CB4 of the reference sequence of the C. briggsae AF16 strain (GenBank Assembly accession number: GCA_000004555.3) using bowtie2 with default parameters. Samtools, bcftools, and bgzip were used to call SNPs, retaining those with a quality score of >20 and combined depth of >5 and <71. We then generated a pseudogenome by replacing the reference AF16 allele with that of HK104 at each SNP using bcftools, totaling 441,227 SNPs. For allele-specific mapping of RNA-seq reads from the HK104 x AF16 F1 hybrid (see below), this new amended HK104 genome was then concatenated to the reference AF16 genome to form a master AF16-HK104 genome.
RNA-Seq Data Analysis
RNA-seq data analysis was essentially as described (Wang et al. 2018). For each library, ∼20M 150-bp-length paired-end reads were generated. Low quality reads were removed using the FASTX Toolkit. Illumina primer sequences (adaptors) were removed from read sequences using cutadapt.
For transcriptomes of purebred AF16 and HK104, the resulting trimmed reads were aligned to the respective reference genome using the HISAT2 2.2.0 alignment program with default parameters. Mapped reads were then input into HTSeq 0.11.1 to calculate normalized counts for each gene. Genes with fewer than 20 reads mapped to them in all samples were removed and the remaining genes were used as input to test for differential expression using the generalized linear model framework in edgeR (in the module glmQLFTest) as follows. For figure 3, each strain’s transcriptomes were used as input to a test for genes whose expression changed between cold and untreated samples. For figure 4, the complete set of transcriptomes from both strains and both conditions was used as input to a test for genes at which the impact of cold on expression was different between the strains.
For transcriptomes of the AF16 x HK104 F1 hybrid, trimmed reads were aligned to a concatenation of the two strains’ reference genomes, and only reads that mapped to a single location in this concatenated reference were retained for analysis, reflecting allele-specific expression at the respective strain’s allele of the respective gene. Normalized read counts and significance testing were as for figure 4 above, reporting cases of temperature effects that differed between the two strains’ alleles.
All raw transcriptome data are available at GEO (accession number: GSE171725) and SRA (accession number: SRP314054).
Principal component analysis was performed using the built-in function in edgeR. Gene Ontology term enrichment analysis was performed using the web tool at geneontology.org.
Genes for the mutant survey of figure 5 in C. elegans were chosen as those that exhibited higher expression under cold treatment than in control condition in HK104 when analyzed separately, at nominal P < 0.01; exhibited stronger cold-evoked expression change in HK104 than in AF16 in the strain-by-temperature analysis, at nominal P < 0.01; and were annotated with one-to-one orthology between C. elegans and C. briggsae.
CRISPR-Mediated Genome Editing
CRISPR-mediated genome editing for C. briggsae was performed using an established protocol (Culp et al. 2020) modified from the analogous protocol for C. elegans (Friedland et al. 2013). For generating mutations in WBGene00025434, WBGene00037162, WBGene00025987, WBGene00034955, and WBGene00031437, the C. briggsae orthologs of vhe-1, pgp-8, M28.8, ncr-1, and K03H1.5 genes respectively, day 1 adult animals of AF16 and HK104 were injected with pDD162 (Peft-3::Cas9::tbb-2 3′-UTR), pCFJ90 (pmyo-2::mCherry), and 2–4 different pU6::sgRNAs for the gene of interest. Surviving worms were separated, and F1 mCherry-positive animals were collected; their progeny, representing the F2 generation, were genotyped for the respective gene. Mutant genotypes are reported in supplementary table 3, Supplementary Material online.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by NIH GM120430-A1 to J.L.G. and R.B.B., NIH R35GM119828 to J.L.G., and NIH 1S10OD021686 for the COPAS large particle sorter housed in the Buck Institute Morphology and Imaging Core; the American Federation for Aging Research; and the Glenn Foundation for Medical Research. QX1410 and VX34 were kind gifts from Dr Erik Andersen (Northwestern University). All other strains were provided by the Caenhorhabditis Genome Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), and the National BioResource Project of Japan, supported by the Japanese Ministry of Education, Culture, Science, Sports and Technology. We thank members of the Brem laboratory (UC Berkeley) and Garrison laboratory (Buck Institute for Research on Aging) for insightful discussions.
Data Availability
Raw sequence data for the RNA-sequencing data set used in this study are available at the Gene Expression Omnibus (GSE171725) and the Short Read Archive (SRP314054).
Contributor Information
Wenke Wang, Buck Institute for Research on Aging, Novato, California, USA; Department of Plant and Microbial Biology, UC Berkeley, USA.
Anna G Flury, Buck Institute for Research on Aging, Novato, California, USA; Department of Plant and Microbial Biology, UC Berkeley, USA.
Jennifer L Garrison, Buck Institute for Research on Aging, Novato, California, USA; Leonard Davis School of Gerontology, University of Southern California, USA; Department of Cellular and Molecular Pharmacology, UC San Francisco, USA; Global Consortium for Reproductive Longevity & Equality, Novato, California, USA.
Rachel B Brem, Buck Institute for Research on Aging, Novato, California, USA; Department of Plant and Microbial Biology, UC Berkeley, USA; Leonard Davis School of Gerontology, University of Southern California, USA.
Literature Cited
- Adrion JR, Hahn MW, Cooper BS. 2015. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 31(8):434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera J, Randez-Gil F, Prieto JA. 2007. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol Rev. 31(3):327–341. [DOI] [PubMed] [Google Scholar]
- Baird SE, Stonesifer R. 2012. Reproductive isolation in Caenorhabditis briggsae: dysgenic interactions between maternal- and zygotic-effect loci result in a delayed development phenotype. Worm 1(4):189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie TA, et al. 2019. Deep sampling of Hawaiian Caenorhabditis elegans reveals high genetic diversity and admixture with global populations. Elife 8:e50465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp E, et al. 2020. Genome editing in the nematode Caenorhabditis briggsae using the CRISPR/Cas9 system. Biol Methods Protoc. 5(1):bpaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 25(4):778–786. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Choi JY. 2010. Natural selection shapes nucleotide polymorphism across the genome of the nematode Caenorhabditis briggsae. Genome Res. 20(8):1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Félix MA, Barrière A, Charlesworth D. 2006. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173(4):2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari C, et al. 2017. Ephemeral-habitat colonization and neotropical species richness of Caenorhabditis nematodes. BMC Ecol. 17(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouris AD, Piantoni C. 2015. Links between thermoregulation and aging in endotherms and ectotherms. Temperature 2(1):73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, et al. 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 10(8):741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graustein A, Caspar JM, Walters JR, Palopoli MF. 2002. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 161(1):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LDW, et al. 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5(7):e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, et al. 2018. A genetic program mediates cold-warming response and promotes stress-induced phenoptosis in C. elegans. Elife 7:e35037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keto-Timonen R, et al. 2016. Cold shock proteins: a minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front Microbiol. 7: 1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SA, Boynton PJ. 2017. Evidence for microbial local adaptation in nature. Mol Ecol. 26(7):1860–1876. [DOI] [PubMed] [Google Scholar]
- Lacher TE, Goldstein MI. 1997. Tropical ecotoxicology: status and needs. Environ Toxicol Chem. 16(1):100–111. [Google Scholar]
- Lee D, et al. 2021. Balancing selection maintains hyper-divergent haplotypes in Caenorhabditis elegans. Nat Ecol Evol. 5(6):794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, et al. 2015. Acyl-CoA dehydrogenase drives heat adaptation by sequestering fatty acids. Cell 161(5):1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark S, et al. 2019. Genome structure predicts modular transcriptome responses to genetic and environmental conditions. Mol Ecol. 28(16):3681–3697. [DOI] [PubMed] [Google Scholar]
- McGaughran A, Sommer RJ. 2014. Natural variation in cold tolerance in the nematode Pristionchus pacificus: the role of genotype and environment. Biol Open. 3(9):832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P, Hayward SAL, Govan GG, Gracey AY, Cossins AR. 2007. An explicit test of the phospholipid saturation hypothesis of acquired cold tolerance in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 104(13):5489–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, et al. 2012. Cross species analysis of Prominin reveals a conserved cellular role in invertebrate and vertebrate photoreceptor cells. Dev Biol. 371(2):312–320. [DOI] [PubMed] [Google Scholar]
- Ohta A, Ujisawa T, Sonoda S, Kuhara A. 2014. Light and pheromone-sensing neurons regulates cold habituation through insulin signalling in Caenorhabditis elegans. Nat Commun. 5:4412–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahata M, et al. 2016. Natural variations of cold tolerance and temperature acclimation in Caenorhabditis elegans. J Comp Physiol B. 186(8):985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahata M, Wei AD, Ohta A, Kuhara A. 2019. Cold acclimation via the KQT-2 potassium channel is modulated by oxygen in Caenorhabditis elegans. Sci Adv. 5(2):eaav3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Dirksen P, Schulenburg H. 2015. Why we need more ecology for genetic models such as C. elegans. Trends Genet. 31(3):120–127. [DOI] [PubMed] [Google Scholar]
- Prasad A, Croydon-Sugarman MJF, Murray RL, Cutter AD. 2011. Temperature-dependent fecundity associates with latitude in Caenorhabditis briggsae. Evolution 65(1):52–63. [DOI] [PubMed] [Google Scholar]
- Rinehart JP, et al. 2007. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc Natl Acad Sci U S A. 104(27):11130–11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JD, Powell JR. 2016. Long-term recovery from acute cold shock in Caenorhabditis elegans. BMC Cell Biol. 17:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford E, Kelly MW. 2011. Local adaptation in marine invertebrates. Ann Rev Mar Sci. 3:509–535. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. 2013. Ecological genomics of local adaptation. Nat Rev Genet. 14(11):807–820. [DOI] [PubMed] [Google Scholar]
- Shore AM, et al. 2013. Cold-induced changes in gene expression in brown adipose tissue, white adipose tissue and liver. PLoS One 8(7):e68933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonna LA, Fujita J, Gaffin SL, Lilly CM. 2002. Invited review: effects of heat and cold stress on mammalian gene expression. J. Appl. Physiol. 92(4):1725–1742. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Ohta A, Maruo A, Ujisawa T, Kuhara A. 2016. Sperm affects head sensory neuron in temperature tolerance of Caenorhabditis elegans. Cell Rep. 16(1):56–65. [DOI] [PubMed] [Google Scholar]
- Stegeman GW, Baird SE, Ryu WS, Cutter AD. 2019. Genetically distinct behavioral modules underlie natural variation in thermal performance curves. G3 (Bethesda) 9(7):2135–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman GW, De Mesquita MB, Ryu WS, Cutter AD. 2013. Temperature-dependent behaviours are genetically variable in the nematode Caenorhabditis briggsae. J Exp Biol. 216(Pt 5):850–858. [DOI] [PubMed] [Google Scholar]
- Stein LD, et al. 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1(2):E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, Snoek LB, Kammenga JE, Andersen EC. 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 31(5):224–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki N, et al. 2020. The mechanoreceptor DEG‐1 regulates cold tolerance in Caenorhabditis elegans. EMBO Rep. 21(3):e48671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, et al. 2015. Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis. Genome Res. 25(5):667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujisawa T, et al. 2018. Endoribonuclease ENDU-2 regulates multiple traits including cold tolerance via cell autonomous and nonautonomous controls in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 115(35):8823–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, et al. 2018. SET-9 and SET-26 are H3K4me3 readers and play critical roles in germline development and longevity. Elife 7:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D, et al. 2018. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 359(6371):55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data for the RNA-sequencing data set used in this study are available at the Gene Expression Omnibus (GSE171725) and the Short Read Archive (SRP314054).