Abstract
Neural datasets are increasing rapidly in both resolution and volume. In neuroanatomy, this trend has been accelerated by innovations in imaging technology. As full datasets are impractical and unnecessary for many applications, it is important to identify abstractions that distill useful features of neural structure, organization, and anatomy. In this review article, we discuss several such abstractions and highlight recent algorithmic advances in working with these models. In particular, we discuss the use of generative models in neuroanatomy; such models may be considered “meta-abstractions” that capture distributions over other abstractions.
Introduction
As the scale of neuroanatomy data has grown, algorithms and abstractions have been developed to distill high-dimensional data into usable forms. Such approaches have allowed us to address questions such as: What is the density of synapses in a specific region of the brain? What is the connectivity between an area of interest and the rest of the brain? What is the best way to divide a brain area into subregions? As the number of data points grows yet further, however, it is possible to ask a different kind of question about variation across different samples or different individuals. These questions can be thought of as “how” instead of “what”: How does neuroprotective treatment alter the density of synapses? How does learning affect the sparseness of connections in a network? How does the modularity of brain networks vary across subjects?
The goal of this article is to discuss how generative approaches in machine learning can be used to address such questions in large-scale neuroanatomy. A generative model captures the variability between samples in a dataset, or between entire datasets, by generating artificial examples with similar statistics to the real data. For example, a generative modeling approach can be used to sketch artificial neurons that are structurally similar to genuine ones, or to simulate a connectome for which the network properties match those observed from microscope data. Generally, the model itself incorporates randomness in order to simulate the true probability distribution over data. A perfect generative model would parameterize the underlying data distribution exactly, allowing the entire dataset to be recreated algorithmically.
We start by describing three main classes of abstractions widely used in neuroanatomy: counts or densities to model the spatial distribution of discrete objects like cells or synapses, connectomes to model the connectivity between either cells or brain areas, and modular or hierarchical models that describe how data are organized into groups. We then describe generative models that are matched to these various abstractions. For example, Poisson models can generate count data of objects such as cells or synapses [1], stochastic block models can be used to build graphs [2], and hidden Markov models can be used to generate the dendritic trees of neurons [3]. In each case, we describe both the algorithmic approach and the conclusions that can be drawn from these abstractions.
After providing an overview of generative models that are built on top of these popular abstractions, we outline generative models that are not built upon any lower-level abstraction. Instead, models such as generative adversarial networks (GANs) [4], [5] and variational autoencoders (VAEs) [6] can generate very high-dimensional data, including entire images. Such models can be used to analyze the sources of variability in observed images [7], to augment observed data, or to interpolate between different imaging modalities [8].
Abstractions
In this section, we highlight key classes of abstractions used in neuroanatomy and describe approaches to estimate these models from high-dimensional and complex brain data (see Figure 1). Each of these abstractions can be considered in terms of (i) what data sources it is commonly derived from, (ii) what questions it can be used to answer, (iii) what information it retains and what it discards from the full-dimensional data, and (iv) algorithms used to derive the abstraction.
Figure 1: Abstractions and Generative Models for Neuroanatomy.
(1) COUNTS & DISTRIBUTIONS: From left to right, we show a (left) retinal dataset before and after cell detection [9], (middle) a depiction of how count data can be represented as a density function, and (right) a Poisson model for generating new count data. (2) CONNECTOMES: From left to right, we show (left) an electron microscope image of a thin slice of cortical brain tissue before and after dense segmentation to build a connectome, (middle) a depiction of a connectome as a graph, and (right) an example of the random overlapping communities model for sparse graphs with three communities displayed as different colors [10]. (3) Modular and Hierarchical Representations: From left to right, we show (left) a light microscope image with a biocytin filled neuron in two views before and after tracing (from the Allen Institute for Brain Science’s Cell Types Atlas [11]), (middle) a hierarchical representation of a dendrite, and (right) example neuronal morphologies generated after an iterative sampling procedure [3], where the iteration number is displayed over each generated morphology.
Counts and Densities.
In neuroanatomy, quantification of brain structure often starts by counting cells, synapses, spines, or other objects in the brain. Counts, or the number of discrete objects in a interval/bin of fixed size, provide the data necessary to compute density estimates from many samples. A large body of work in neuroanatomy involves modeling changes in densities across multiple samples or conditions.
Example data sources: Cellular densities can be resolved in Nissl- or DAPI-stained brain images [12] and retinal datasets (Figure 1a) [13], as well as X-ray microCT [14]. Synapses can be resolved in electron microscopy (EM) [15], [16] and array tomography [17] datasets. Individual mRNAs can now be resolved in brain tissue with spatial transcriptomics (mFISH) [18], multiplexed error-correcting FISH (merFISH) [19], and expansion microscopy-based FISH [20].
Type of conclusion drawn: The spatial distribution and patterns of discrete objects like cells or synapses. Micro and macroscale architecture can also be detected by analyzing spatial patterns in the data. Counts can also be used to track changes to the nervous system in development [21], disease [22], or aging [23].
Information included: The spatial position of objects is included but the connectivity between these objects is not modeled. In some cases, each count can also be associated with additional metadata or ‘marks’ like the object’s size.
Algorithms used to create abstraction: Segmentation and density-based methods have been developed to quantify the spatial organization and distributions of cells [14], [24] (Figure 1c), synapses [25]–[28], neuronal arbors [29], organelles [30], and spines [31].
Connectomes.
Graphs are some of the most widely used abstractions for neuroanatomical data. They are typically used to convey observed physical connectivity between individual neurons or neuronal assemblages. Such graphs are commonly referred to as “structural” connectomes (in contradistinction to “functional” connectomes, which capture correlations between observed activity of neurons). At the microscale, cellular connectomes have nodes for neurons and (weighted) edges for synapses. In meso- or macro-scale connectomes, nodes represent local or global brain areas, while edges represent projections between the areas. Such graphs are also referred to as “projectomes”.
Example data sources: At the microscale, connectomes can be extracted from EM [16] (Figure 1) and expansion microscopy (ExM) [32] datasets. Projectome mapping methods have made use of viral tracing methods and whole-brain serial two-photon microscopy (STP) and MOST [33]–[36] to reveal long-range connections. Projectome data has also been obtained from humans using magnetic resonance imaging (MRI) [37]–[40], mainly through the use of diffusion tensor imaging.
Type of conclusion drawn: Connectomes and projectomes can be used to understand learning and plasticity, as well as constrain models of neural information processing.
Information included: The connectivity between neurons or brain areas is included in these models. In some cases, the strength of connections can also be estimated and included to produce a weighted graph. The spatial position of each node is often excluded in a graphical representation of the data.
Algorithms used to create abstraction: There has been extensive recent work on automatic labeling of EM and ExM images to segment neurons [41]–[49] and synapses [26], [50]. On the computational side, Majka et al. [51] demonstrate tools for coregistering projectomes to create a common map of primate (marmoset) cortex, while other algorithms have been developed to infer higher resolution completions of partial connectivity data [52], [53].
Modular and Hierarchical Models.
Finally, we consider modular and hierarchical abstractions which divide data into groups based upon which examples/segments have similar characteristics. One example is representing a large brain volume as a collection of brain regions, modules, or spatially-defined regions of interest [54], [55]. This principle can be iterated by expressing data examples in terms of a hierarchical model, where discrete groups are divided into subgroups at many scales. For example, the morphology of a neuron can be described with a hierarchical format, with a coarse division into soma, axon, and dendrite which is further broken down into individual branches.
Example data sources: Serial two-photon and fMOST for whole-brain imaging have been used to obtain parcellations of the brain [34]. Morphological reconstructions for modeling the components of neurons can be extracted from light microscopy datasets [56].
Type of conclusion drawn: The high-level organization of the data and which parts of the signal are similar and thus belong to the same group. A hierarchical format for data can be advantageous in representing similarities in data across multiple spatial or evolutionary divisions/scales.
Information included: Modular representations group the structure of many nearby segments of a neuron (parts) or nearby parts of a brain region into one bulk class. The membership of data to a class is preserved and perhaps the average (centroid) of the class is also maintained. Hierarchical models further provide information about the distance between different groups as relative to their multi-scale dependencies.
Algorithms used to create abstraction: To obtain an informative parcellation and simplification of the data, clustering algorithms [57] such as k-means and spectral clustering methods [58] can be used to group spatial loci that have similar statistics in terms of their measured anatomical signal. Semi-automated approaches have recently been shown to provide new insights into structurally and functionally distinct areas in whole human brains with multi-modal measurements [59].
Generative Models for Abstractions
In this section, we describe different generative models that are built on top of the previously discussed abstractions. Each generative model represents a design choice about what features of the true data are most important to capture, based upon the questions under consideration.
Generative models for count-valued data
A generative model for count-valued data (i.e., how many objects are in a region of interest) creates a synthetic dataset where objects are placed across space according to the underlying statistics of real data. Which statistics are important represent a design choice. For example, a model might be designed so that the density functions of real and synthetic data match or so as to preserve nearest-neighbor properties of the counts (e.g., the Ripley k-function [60]).
The simplest generative model used for count-valued data is a Poisson process. Here, we assume that the number of objects observed in a bin/interval is a Poisson-distributed random variable with mean given by an intensity (density) function, and where the numbers of objects in different bins are conditionally independent. Thus, given the potentially spatially-varying intensity of the process, samples can be generated to create a simulated dataset. To extend the independence assumption of Poisson models to ensure that objects are separated by a minimum distance, random sequential adsorption (RSA) processes have been used to model synapses throughout all cortical layers [27]. Point process models can also be constrained to generate counts along a graph structure, for instance in the modeling of spines along a neurite [61].
See [1] for a review of spatial point process models and their applications in neuroanatomy.
To model more complex spatial properties of the data, the underlying intensity function can be approximated by a sparse combination of simpler functions. LaGrow et al. [9], [62] show that by using a basis that can capture change points in the density, this enables the efficient estimation of mesoscopic properties of the density, like the layering structure in the cortex.
Generative models for connectomes
To create a realistic generative model for graphs, we need to first specify the property of the graph we wish to capture. One such property is the average degree of all nodes. Random graph theory provides a wealth of resources for generating graphs that have certain edge and high-level properties [63]. A more complex generative model could ask that graph metrics like clustering and modularity match between real and synthesized data. Such generative models can be introduced by building on random graph models like the widely used Erdős-Renyi random graph model [64], in which each pair of nodes is assigned an edge with some fixed probability p. The random overlapping communities (ROC) model is a good example of a generative model that can generate overlapping communities as observed in neural circuits, and has provable convergence in terms of its desired properties [10]. In this model, many subsets of the overall graph are chosen at random, and dense Erdős-Renyi random graphs are constructed on these (possibly overlapping) subsets. Additionally, stochastic block models (SBMs) are a class of generative models for synthesizing graphs [65], which have been used to model hierarchical modules within a connectome. In an SBM, the nodes of the graph are divided into several blocks, and the probability of connection between two nodes depends only on the blocks in which they lie. Jonas and Kording [66] introduce a variant of SBMs to model connectivity between neurons, where the blocks of the model correspond to cell types, and where distances also affect the probability of connection. They use Markov Chain Monte Carlo (MCMC) methods to fit the parameters of the model, thereby automatically inferring cell types from connectomics data.
Hidden Markov model (HMMs) have also been applied successfully to the graph structures representing the branching of individual neurons. HMMs model the growth of a graph or other data structure over time using a Markov chain that depends on hidden variables that can be statistically inferred but are not observed directly. For example, the hidden state of a neuron as it grows might include biochemical factors that are not directly observable, even though they lead to observable data such as the morphology of the neuron. In Farhoodi et al. [67], the branching patterns of different types of neurons are learned and incorporated into a generative model by analyzing single-neuron morphological data compiled by neuromorpho.org [68]. The HMM inferred by Farhoodi et al. suggests that the probability of branching within a neuron depends on the distance to the soma, whether whether the branching occurs in a main branch or a side branch, and what the type of neuron is. The model thus yields both insights into the underlying factors that may be at play in neural branching and also a procedure for generative artificial neuronal morphologies. See also Farhoodi and Kording [3] for a generative approach to neuron morphologies based on Markov chain Monte Carlo (MCMC) sampling.
Modular and hierarchical generative models
Generative models built on top of hierarchical abstractions, typically will generate a sequence of items wherein the probabilistic model depends upon what was generated at previous generate samples. To ensure that our generative model matches the distribution of data, the sequence of steps must generate an output that matches the same sequence generation of real data.
SBMs (defined in the preceding section) are well-suited to dissecting graphical data into hierarchically organized modules. Lyzinski et al. [2] combine SBMs with clustering algorithms to decompose a partial Drosophila connectome into blocks, which are then clustered into similar subnetworks (motifs). The process is then repeated to generate a hierarchy of motifs. Priebe et al. [69] apply another generalization of SBMs to the Drosophila connectome to explain variation in cells that is fit poorly by simple clusters.
Generative Models for Image Data
Whereas the generative models highlighted in the previous section require (often intensive) pre-processing steps to first build an abstraction from image data, modern machine learning methods make it possible to learn a generative model from images directly. Learning models from images directly could potentially allow us to by-pass the initial steps of building an abstraction (e.g., segmenting or finding objects in images). Rather than specifying what to look for in an image, the generative model would be able to automatically pull out features from the image data that are important for building realistic representations of neural structure and capturing variability across examples. In this section, we highlight matrix factorization and deep learning approaches for learning generative models from collections of images, and discuss their applications in neuroanatomy.
Latent variable models
Learning to model the distribution of high-dimensional image data is extremely challenging. A first step is often to form a low-dimensional representation that is easier to model. A simple and widely used linear approach for learning latent factors from data is principal component analysis (PCA); PCA fits a k-dimensional linear approximation to a dataset with many examples, such as a collection of many brain images. Other dimensionality reduction techniques such as non-negative matrix factorization [70], probabilistic PCA [71], and sparse PCA [72] can all be used to form a low-dimensional representation of collection of data (see [73] for a comprehensive review of dimensionality reduction techniques and their applications in analyzing measurements of neural activity).
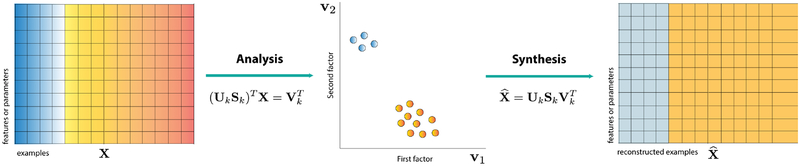
After distilling data into a low-dimensional space, image data can be reconstructed by inverting the low-dimensional model learned in the analysis step (Figure 2, left). This synthesis operation is visualized in Fig. 2 for a linear system learned in PCA. In this case, a new image is created by either: reconstructing an input (pass in a noisy signal and the output is a clean version) or generating a new sample in the low-dimensional space and then using the decoder to synthesize a new image as output. This interpretation of linear matrix factorization (PCA) as a generative model provides a simple strategy for creating high-dimensional images when the data lie near a linear subspace.
Figure 2: Linear matrix factorization methods like PCA and their interpretation as a generative modeling procedure.
From left to right, we show the decomposition of a data matrix consisting of examples along its columns, into a low-dimensional format. In the middle, we depict the low-d representation of the data in two dimensions. On the right, we show a reconstructed or synthesized data matrix that uses the inverse mapping to expand the low-d data back into a high-dimensional space again.
Autoencoders
Generative models that rely on PCA and other matrix factorization approaches use linear transformations of data. It is also possible to find nonlinear low-dimensional representations of data. Autoencoders are now routinely used for this task [74]. Autoencoders can be constructed through different neural network architectures, encompassing models such as stacked convolutional autoencoders [75] and variational autoencoders [6]. Essentially, an autoencoder functions by passing high-dimensional input through a sequence of layers, including a low-dimensional “bottleneck” layer, then reconstructing the full-dimensional input again in the output layer (see Figure 3, left). The bottleneck layer thus learns a low-dimensional latent representation of the data. The first part (up to the bottleneck) is the encoder and the remainder (reconstructing the input) is the decoder. Thus, the encoder compresses the data to the latent representation, and the decoder is a generative model that recreates data from this latent representation. Thus autoencoders provide an analogous architecture for generative modeling for the nonlinear case as that depicted for the linear case in Figure 2.
Figure 3: Generative models for synthesizing structural brain images.
On the left, we depict an autoencoder consisting of an input layer, a low-dimensional hidden layer (latent space), and output layer. In the training phase, a low-dimensional model for data is learned and in the synthesis phase, a sample from this model is generated and used to generate a new image. This architecture is applied to auto-fluorescence images of 1,700 different brains (25 micron resolution) to synthesize new images: on the right, a synthetically generated image (top), example of a real image used to train the network (bottom), and a denoised (reconstructed) version of the image displayed on the bottom.
Generative adversarial networks
Within deep learning, generative adversarial networks (GANs) have recently been developed to learn from an unlabeled training dataset to generate artificial data resembling examples from the dataset. Like autoencoders, GANs learn a nonlinear generative process for data via an artificial neural network. However, unlike autoencoders, which learn both an “encoding” step and a “decoding” (generative) step, GANs learn by pitting two network algorithms against each other, with one (the generator) attempting to generate plausible examples from a dataset, while the other (the discriminator) tries to tell the difference between real and fake examples, thus forcing the generator to improve. While extensive applications to neuroanatomy have yet to be developed, GANs have already been used to simulate neuron morphologies [76] and spike trains [77]. A similar approach (using deep learning methods distinct from GANs) uses the output of one imaging modality to simulate the result of another imaging modality [78].
It is tempting to consider using the output of a GAN to augment real data in fitting additional algorithms. However, there is so far no magical algorithm that replaces the power of large real datasets. For example, while a GAN might be used to learn from a thousand images and then create a million more similar-seeming images, the artificial images would likely either fail in subtle ways to be truly realistic or would fail to capture the full diversity of real-world data. We therefore believe the function of generative algorithms in neuroanatomy should be, for the moment, more in modeling than in augmenting data for training.
Conclusions
As neuroanatomy datasets become more numerous and higher-dimensional, there is increasing need for generative models that capture variability across data samples and subjects. Where traditional abstractions such as connectomes compress data, generative “meta-abstractions” compress distributions over data or over abstractions. We believe that an understanding of the breadth of available abstractions and concomitant generative models, each suited to different questions and data modalities, is essential to present-day neuroanatomy.
Many of the generative models we have described make strong assumptions about the structure of the data-for example, that it is well-approximated by a density function or succinctly described by a Markov chain. By contrast, generative algorithms from deep learning typically have no such prior assumptions, and the models they learn are often “black boxes” that are hard to interpret. Interpretation becomes increasingly difficult as we move to full images and raw data because it is not always clear what properties of the data are being modeled and how. New approaches for ‘disentangling representations’ [79] aim to mitigate these issues and build architectures that reveal more interpretable factors. An important line of research is to build deep learning architectures that are interpretable and can be used to draw inferences about disease, inter-subject variability, and other changes to neural structure.
Traditionally, neuroscience has provided views of the structure of the nervous system that resolve or model one aspect of the anatomy at a time. Neuroscience methods are, however, increasingly moving towards resolving multiple types of structures simultaneously to provide multi-modal and multi-scale structural information for large volumes, in some cases up to whole brains [12], [34]. With increasing access to multi-modal information, it is critical to develop abstractions and generative models that distill the data into a usable simplification that leverages the multi-modal data provided. Because traditional methods for modeling neuroanatomy have focused on modeling a single attribute of the data (a graph, or a density), in some cases it is not clear how best to integrate data formats and models across different modalities of information. It is exceedingly likely that different aspects of anatomy (change in density of synapses or cells, or strengthening of connections in a specific region of the brain) co-vary in complex and nonlinear ways and multi-modal datasets will be necessary to reveal these relationships.
Generative models are now being used to learn increasingly complex attributes of a wide range of datasets. We believe that they will be a useful tool moving forward for modeling variability in large-scale neuroanatomy datasets.
Highlights.
Modern neuroscience has now entered the age of big data. Low-dimensional models and abstractions are needed to distill high-dimensional neuroanatomical data into a simpler format.
Generative models provide a powerful strategy for modeling large-scale neuroscience datasets. Such approaches aim to synthesize new data, rather than analyzing data.
Deep learning-based generative modeling frameworks like generative adversarial networks and autoencoders are still difficult to interpret; however, they have the potential to reveal hidden factors that control variability across complex large-scale neuroanatomy datasets.
Acknowledgements
We would like to thank Konrad Körding and Chethan Pandarinath for helpful feedback on the manuscript. We would also like to acknowledge Samantha Petti and Santosh Vempala for providing us an example of a ROC (generative graph model), Yaron Meirovitch for segmented EM images for connectomics, and Roozbeh Farhoodi for providing us with the dendrogram and generative model figures for hierarchical models, all of which were used to create Figure 1. ED was supported by the National Institute of Mental Health of the National Institutes of Health under Award Number R24MH114799. DR was supported by the National Science Foundation under Award Number 1803547.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors report no conflicts of interest.
References
- [1].Budd JM, Cuntz H, Eglen SJ, and Krieger P, “Quantitative analysis of neuroanatomy,”Frontiers in neuroanatomy, vol. 9, p. 143, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lyzinski V, Tang M, Athreya A, Park Y, and Priebe CE, “Community detection and classification in hierarchical stochastic blockmodels,” IEEE Transactions on Network Science and Engineering, vol. 4, no. 1, pp. 13–26, 2017. [Google Scholar]
- [3].Farhoodi R and Körding KP, “Sampling neuron morphologies,” BioRxiv, p. 248 385,2018. [Google Scholar]
- [4].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, “Generative adversarial nets,” in Advances in neural information processing systems, 2014, pp. 2672–2680. [Google Scholar]
- [5].Radford A, Metz L, and Chintala S, “Unsupervised representation learning with deep convolutional generative adversarial networks,” ArXiv preprint arXiv:1511.06434, 2015. [Google Scholar]
- [6].Kingma DP and Welling M, “Auto-encoding variational bayes,” ArXiv preprint arXiv:1312.6114,2013. [Google Scholar]
- [7].Subakan C, Kojeyo O, and Smaragdis P, “Learning the base distribution in implicit generative models,” ArXiv preprint arXiv:1803.04357, 2018. [Google Scholar]
- [8].Iglesias JE, Konukoglu E, Zikic D, Glocker B, Van Leemput K, and Fischl B, “Is synthesizing MRI contrast useful for inter-modality analysis?” In International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2013, pp. 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].LaGrow TJ, Moore M, Prasad JA, Davenport MA, and Dyer EL, “Approximating cellular densities with sparse recovery techniques,” BioArxiv, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Petti S and Vempala SS, “Approximating sparse graphs: The random overlapping communities model,” ArXiv preprint arXiv:1802.03652, 2018. [Google Scholar]
- [11].Allen institute cell types atlas, 2018. [Online]. Available: http://celltypes.brain-map.org/experiment/morphology/607124114.
- [12].Amunts K, Lepage C, Borgeat L, Mohlberg H, Dickscheid T, Rousseau M-É, Bludau S,Bazin P-L, Lewis LB, Oros-Peusquens A-M, et al. , “BigBrain: An ultrahigh-resolution 3D human brain model,” Science, vol. 340, no. 6139, pp. 1472–1475, 2013. [DOI] [PubMed] [Google Scholar]
- [13].Chang B, Hawes N, Pardue M, German A, Hurd R, Davisson M, Nusinowitz S, Rengarajan K, Boyd A, Sidney S, and Phillips M, “Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cgmp phosphodiesterase gene,” Vision Research, vol. 47, no. 5, pp. 624–633, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dyer EL, Roncal WG, Prasad JA, Fernandes HL, Gürsoy D, De Andrade V, Fezzaa K, Xiao X, Vogelstein JT, Jacobsen C, et al. , “Quantifying mesoscale neuroanatomy using x-ray microtomography,” ENeuro, vol. 4, no. 5, ENEURO–0195, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Merchán-Pérez A, Rodríguez J-R, González S, Robles V, DeFelipe J, Larrañaga P, and Bielza C, “Three-dimensional spatial distribution of synapses in the neocortex: A dual-beam electron microscopy study,” Cerebral Cortex, vol. 24, no. 6, pp. 1579–1588, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, KnowlesBarley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, et al. , “Saturated reconstruction of a volume of neocortex,” Cell, vol. 162, no. 3, pp. 648–661, 2015. [DOI] [PubMed] [Google Scholar]
- [17].Micheva KD and Smith SJ, “Array tomography: A new tool for imaging the molecular architecture and ultrastructure of neural circuits,” Neuron, vol. 55, no. 1, pp. 25–36, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shah S, Lubeck E, Zhou W, and Cai L, “In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus,” Neuron, vol. 92, no. 2, pp. 342–357, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen KH, Boettiger AN, Moffitt JR, Wang S, and Zhuang X, “Spatially resolved, highly multiplexed rna profiling in single cells,” Science, vol. 348, no. 6233, aaa6090, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang J-B, Marblestone A, Church GM, et al. , “Nanoscale imaging of RNA with expansion microscopy,” Nature methods, vol. 13, no. 8, p. 679, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bandeira F, Lent R, and Herculano-Houzel S, “Changing numbers of neuronal and non-neuronal cells underlie postnatal brain growth in the rat,” Proceedings of the National Academy of Sciences, vol. 106, no. 33, pp. 14 108–14 113, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kreczmanski P, Heinsen H, Mantua V, Woltersdorf F, Masson T, Ulfig N, Schmidt-Kastner R, Korr H, Steinbusch HW, Hof PR, et al. , “Volume, neuron density and total neuron number in five subcortical regions in schizophrenia,” Brain, vol. 130, no. 3, pp. 678–692, 2007. [DOI] [PubMed] [Google Scholar]
- [23].Andrade-Moraes CH, Oliveira-Pinto AV, Castro-Fonseca E, da Silva CG, Guimaraes DM, Szczupak D, Parente-Bruno DR, Carvalho LR, Polichiso L, Gomes BV, et al. , “Cell number changes in Alzheimer’s disease relate to dementia, not to plaques and tangles,” Brain, vol. 136, no. 12, pp. 3738–3752, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang C, Yan C, Ren M, Li A, Quan T, Gong H, and Yuan J, “A platform for stereological quantitative analysis of the brain-wide distribution of type-specific neurons,” Scientific Reports, vol. 7, no. 1, p. 14 334, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li L, Tasic B, Micheva KD, Ivanov VM, Spletter ML, Smith SJ, and Luo L, “Visualizing the distribution of synapses from individual neurons in the mouse brain,” PloS one, vol. 5, no. 7, e11503, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Roncal WG, Pekala M, Kaynig-Fittkau V, Kleissas DM, Vogelstein JT, Pfister H, Burns R, Vogelstein RJ, Chevillet MA, and Hager GD, “Vesicle: Volumetric evaluation of synaptic interfaces using computer vision at large scale,” ArXiv preprint arXiv:1403.3724, 2014. [Google Scholar]
- [27].Anton-Sanchez L, Bielza C, Merchán-Pérez A, Rodríguez J-R, DeFelipe J, and Larrañaga P, “Three-dimensional distribution of cortical synapses: A replicated point pattern-based analysis,” Frontiers in neuroanatomy, vol. 8, p. 85, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Domínguez-Álvaro M, Montero-Crespo M, Blazquez-Llorca L, Insausti R, DeFelipe J, and Alonso-Nanclares L, “Three-dimensional analysis of synapses in the transentorhinal cortex of Alzheimer’s disease patients,” Acta neuropathologica communications, vol. 6, no. 1, p. 20, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sümbül U, Zlateski A, Vishwanathan A, Masland RH, and Seung HS, “Automated computation of arbor densities: A step toward identifying neuronal cell types,” Frontiers in neuroanatomy, vol. 8, p. 139, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Perez AJ, Seyedhosseini M, Deerinck TJ, Bushong EA, Panda S, Tasdizen T, and Ellisman MH, “A workflow for the automatic segmentation of organelles in electron microscopy image stacks,” Frontiers in neuroanatomy, vol. 8, p. 126, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Anton-Sanchez L, Larrañaga P, Benavides-Piccione R, Fernaud-Espinosa I, DeFelipe J, and Bielza C, “Three-dimensional spatial modeling of spines along dendritic networks in human cortical pyramidal neurons,” PloS one, vol. 12, no. 6, e0180400, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen F, Tillberg PW, and Boyden ES, “Expansion microscopy,” Science, p. 1 260 088,2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, and Chandrashekar J, “A platform for brain-wide imaging and reconstruction of individual neurons,” Elife, vol. 5, e10566, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L,Henry AM, et al. , “A mesoscale connectome of the mouse brain,” Nature, vol. 508, no. 7495, p. 207, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, and Osten P, “Serial two-photon tomography for automated ex vivo mouse brain imaging,” Nature methods, vol. 9, no. 3, p. 255, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li A, Gong H, Zhang B, Wang Q, Yan C, Wu J, Liu Q, Zeng S, and Luo Q, “Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain,” Science, vol. 330, no. 6009, pp. 1404–1408, 2010. [DOI] [PubMed] [Google Scholar]
- [37].Gray WR, Bogovic JA, Vogelstein JT, Landman BA, Prince JL, and Vogelstein RJ, “Magnetic resonance connectome automated pipeline: An overview,” IEEE pulse, vol. 3, no. 2, pp. 42–48, 2012. [DOI] [PubMed] [Google Scholar]
- [38].Craddock RC, Jbabdi S, Yan C-G, Vogelstein JT, Castellanos FX, Di Martino A,Kelly C, Heberlein K, Colcombe S, and Milham MP, “Imaging human connectomes at the macroscale,” Nature methods, vol. 10, no. 6, p. 524, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, and Sporns O, “Mapping the structural core of human cerebral cortex,” PLoS biology, vol. 6, no. 7, e159, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].He Y, Chen ZJ, and Evans AC, “Small-world anatomical networks in the human brain revealed by cortical thickness from mri,” Cerebral cortex, vol. 17, no. 10, pp. 2407–2419, 2007. [DOI] [PubMed] [Google Scholar]
- [41].Lee K, Zlateski A, Ashwin V, and Seung HS, “Recursive training of 2D-3D convolutional networks for neuronal boundary prediction,” in Advances in Neural Information Processing Systems, 2015, pp. 3573–3581. [Google Scholar]
- [42].Meirovitch Y, Matveev A, Saribekyan H, Budden D, Rolnick D, Odor G, Jones SK-BTR, Pfister H, Lichtman JW, and Shavit N, “A multi-pass approach to large-scale connectomics,” ArXiv preprint arXiv:1612.02120, 2016. [Google Scholar]
- [43].Maitin-Shepard JB, Jain V, Januszewski M, Li P, and Abbeel P, “Combinatorial energy learning for image segmentation,” in Advances in Neural Information Processing Systems, 2016, pp. 1966–1974. [Google Scholar]
- [44].Januszewski M, Maitin-Shepard J, Li P, Kornfeld J, Denk W, and Jain V, “Flood-filling networks,” ArXiv preprint arXiv:1611.00421, 2016. [DOI] [PubMed] [Google Scholar]
- [45].Januszewski M, Kornfeld J, Li PH, Pope A, Blakely T, Lindsey L, Maitin-Shepard JB,Tyka M, Denk W, and Jain V, “High-precision automated reconstruction of neurons with flood-filling networks,” BioRxiv, p. 200 675, 2017. [DOI] [PubMed] [Google Scholar]
- [46].Matveev A, Meirovitch Y, Saribekyan H, Jakubiuk W, Kaler T, Odor G, Budden D, Zlateski A, and Shavit N, “A multicore path to connectomics-on-demand,” in Proceedings of the ACM SIGPLAN Symposium on Principles and Practice of Parallel Programming, ACM, 2017, pp. 267–281. [Google Scholar]
- [47].Rolnick D, Meirovitch Y, Parag T, Pfister H, Jain V, Lichtman JW, Boyden ES, and Shavit N, “Morphological error detection in 3D segmentations,” ArXiv preprint arXiv:1705.10882, 2017. [Google Scholar]
- [48].Yoon Y-G, Dai P, Wohlwend J, Chang J-B, Marblestone AH, and Boyden ES, “Feasibility of 3D reconstruction of neural morphology using expansion microscopy and barcode-guided agglomeration,” Frontiers in computational neuroscience, vol. 11, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zung J, Tartavull I, Lee K, and Seung HS, “An error detection and correction framework for connectomics,” in Advances in Neural Information Processing Systems, 2017, pp. 6821–6832. [Google Scholar]
- [50].Santurkar S, Budden D, Matveev A, Berlin H, Saribekyan H, Meirovitch Y, and Shavit N, “Toward streaming synapse detection with compositional convnets,” ArXiv preprint arXiv:1702.07386, 2017. [Google Scholar]
- [51].Majka P, Chaplin TA, Yu H-H, Tolpygo A, Mitra PP, Wójcik DK, and Rosa MG, “Towards a comprehensive atlas of cortical connections in a primate brain: Mapping tracer injection studies of the common marmoset into a reference digital template,” Journal of Comparative Neurology, vol. 524, no. 11, pp. 2161–2181, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Harris KD, Mihalas S, and Shea-Brown E, “High resolution neural connectivity from incomplete tracing data using nonnegative spline regression,” in Advances in Neural Information Processing Systems, 2016, pp. 3099–3107. [Google Scholar]
- [53].Knox JE, Harris KD, Graddis N, Whitesell JD, Zeng H, Harris JA, Shea-Brown E, and Mihalas S, “High resolution data-driven model of the mouse connectome,” BioRxiv,p. 293 019, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Van Essen DC, “Windows on the brain: The emerging role of atlases and databases in neuroscience,” Current opinion in neurobiology, vol. 12, no. 5, pp. 574–579, 2002. [DOI] [PubMed] [Google Scholar]
- [55].Amunts K and Zilles K, “Architectonic mapping of the human brain beyond brodmann,”Neuron, vol. 88, no. 6, pp. 1086–1107, 2015. [DOI] [PubMed] [Google Scholar]
- [56].Armañanzas R and Ascoli GA, “Towards the automatic classification of neurons,”Trends in neurosciences, vol. 38, no. 5, pp. 307–318, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jain AK, “Data clustering: 50 years beyond k-means,” Pattern recognition letters, vol.31, no. 8, pp. 651–666, 2010. [Google Scholar]
- [58].Von Luxburg U, “A tutorial on spectral clustering,” Statistics and computing, vol. 17, no.4, pp. 395–416, 2007. [Google Scholar]
- [59].Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. , “A multi-modal parcellation of human cerebral cortex,” Nature, vol. 536, no. 7615, pp. 171–178, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ripley BD, “Modelling spatial patterns,” Journal of the Royal Statistical Society. SeriesB (Methodological), pp. 172–212, 1977. [Google Scholar]
- [61].Baddeley A, Jammalamadaka A, and Nair G, “Multitype point process analysis of spines on the dendrite network of a neuron,” Journal of the Royal Statistical Society: Series C (Applied Statistics), vol. 63, no. 5, pp. 673–694, 2014. [Google Scholar]
- [62].LaGrow TJ, Moore M, Prasad JA, Davenport MA, and Dyer EL, “Approximating cellular densities from high-resolution neuroanatomical imaging data,” in Proceedings of the IEEE Engineering in Medicine and Biology Society Conference, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sporns O, Tononi G, and Edelman GM, “Theoretical neuroanatomy: Relating anatomical and functional connectivity in graphs and cortical connection matrices,” Cerebral cortex, vol. 10, no. 2, pp. 127–141, 2000. [DOI] [PubMed] [Google Scholar]
- [64].Erdős P and Rényi A, “On the evolution of random graphs,” Publ. Math. Inst. Hung. Acad. Sci, vol. 5, no. 1, pp. 17–60, 1960. [Google Scholar]
- [65].Holland PW, Laskey KB, and Leinhardt S, “Stochastic blockmodels: First steps,” Social networks, vol. 5, no. 2, pp. 109–137, 1983. [Google Scholar]
- [66].Jonas E and Körding K, “Automatic discovery of cell types and microcircuitry from neural connectomics,” Elife, vol. 4, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Farhoodi R, Rolnick D, and Körding K, “Neuron dendrograms uncover asymmetrical motifs,” in Computational Systems Neuroscience (COSYNE) Annual Meeting, 2018. [Google Scholar]
- [68].Parekh R and Ascoli GA, “Neuronal morphology goes digital: A research hub for cellular and system neuroscience,” Neuron, vol. 77, no. 6, pp. 1017–1038, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Priebe CE, Park Y, Tang M, Athreya A, Lyzinski V, Vogelstein JT, Qin Y, Co-canougher B, Eichler K, Zlatic M, et al. , “Semiparametric spectral modeling of the Drosophila connectome,” ArXiv preprint arXiv:1705.03297, 2017. [Google Scholar]
- [70].Lee DD and Seung HS, “Learning the parts of objects by non-negative matrix factorization,” Nature, vol. 401, no. 6755, p. 788, 1999. [DOI] [PubMed] [Google Scholar]
- [71].Tipping ME and Bishop CM, “Probabilistic principal component analysis,” Journal of the Royal Statistical Society: Series B (Statistical Methodology), vol. 61, no. 3, pp. 611–622, 1999. [Google Scholar]
- [72].Zou H, Hastie T, and Tibshirani R, “Sparse principal component analysis,” Journal of computational and graphical statistics, vol. 15, no. 2, pp. 265–286, 2006. [Google Scholar]
- [73].Cunningham JP and Byron MY, “Dimensionality reduction for large-scale neural recordings,” Nature neuroscience, vol. 17, no. 11, p. 1500, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Vincent P, Larochelle H, Bengio Y, and Manzagol P-A, “Extracting and composing robust features with denoising autoencoders,” in Proceedings of the 25th International Conference on Machine Learning, ACM, 2008, pp. 1096–1103. [Google Scholar]
- [75].Masci J, Meier U, Cireşan D, and Schmidhuber J, “Stacked convolutional auto-encoders for hierarchical feature extraction,” in International Conference on Artificial Neural Networks, Springer, 2011, pp. 52–59. [Google Scholar]
- [76].Farhoodi R, Ramkumar P, and Körding K, “Deep learning approach towards generating neuronal morphology,” Computational and Systems Neuroscience, 2018. [Google Scholar]
- [77].Molano-Mazon M, Onken A, Piasini E, and Panzeri S, “Synthesizing realistic neural population activity patterns using generative adversarial networks,” in Proceedings of the International Conference on Learning Representations, 2018. [Google Scholar]
- [78].Christiansen EM, Yang SJ, Ando DM, Javaherian A, Skibinski G, Lipnick S, Mount E, O’Neil A, Shah K, Lee AK, et al. , “In silico labeling: Predicting fluorescent labels in unlabeled images,” Cell, vol. 173, no. 3, pp. 792–803, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chen X, Duan Y, Houthooft R, Schulman J, Sutskever I, and Abbeel P, “Infogan: Interpretable representation learning by information maximizing generative adversarial nets,” in Advances in neural information processing systems, 2016, pp. 2172–2180. [Google Scholar]