Abstract
Studies over the past four decades have elucidated the role of Wnt/β-catenin mediated regulation in cell proliferation, differentiation and migration. These processes are fundamental to embryonic development, regeneration potential of tissues, as well as cancer initiation and progression. In this review, we focus on the epigenetic players which influence the Wnt/β-catenin pathway via modulation of its components and coordinated regulation of the Wnt target genes. The role played by crosstalk with other signaling pathways mediating tumorigenesis is also elaborated. The Hippo/YAP pathway is particularly emphasized due to its extensive crosstalk via the Wnt destruction complex. Further, we highlight the recent advances in developing potential therapeutic interventions targeting the epigenetic machinery based on the characterization of these regulatory networks for effective treatment of various cancers and also for regenerative therapies.
Keywords: Wnt signaling, β-catenin, YAP, epigenetics, cancer, therapeutics
Introduction
The Wnt signaling pathway remains one of the most extensively studied signaling pathways to date and yet there is a long way to fully understand its functionality. The Wnt pathway was discovered in 1982 with the identification of the int1 (wnt1) gene responsible for tumor growth (Nusse and Varmus, 1982). The hallmark Wnt pathway gene wnt1 (int1 at the time of identification) was discovered as the first proto-oncogene in mice mammary tumors using the pro-viral tagging screening method in 1982 (Nusse and Varmus, 1982). The Wnt pathway is associated with the widest array of biological processes, including cell proliferation, differentiation, organogenesis, regeneration and diseases such as neurodevelopmental diseases and cancer (Nusse and Clevers, 2017). Although the mammalian wnt1 gene was discovered as a part of screening for proto-oncogenes, its Drosophila homolog wingless was already known to play a role in segmentation during development (Nüsslein-Volhard and Wieschaus, 1980). Other genes in this pathway (including, armadillo, arrow, disheveled, zeste-white 3, porcupine) were also identified due to segmentation polarity defects during Drosophila development (Peifer et al., 1991; Siegfried et al., 1992; Noordermeer et al., 1994; Kadowaki et al., 1996; Bafico et al., 2001). Overexpression studies of Wnt1 and GSK3β in Xenopus showed axis duplication (McCrea et al., 1993; Dominguez et al., 1995). Studies in planaria and Hydra have established the role of the Wnt pathway in regeneration as well (Hobmayer et al., 2000; Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008; Reddy et al., 2020). Due to the important roles of the Wnt pathway in the processes of development and regeneration, the interest in investigating its role in human cancers climbed steadily. APC was found to be the most frequently mutated gene in inherited forms of human cancer, causing multiple polyps in intestines (Familial Adenomatous Polyposis or FAP) (Su et al., 1992). Sporadic cases of cancers, on the other hand, didn’t show alterations in the APC gene. Rather, other components of the Wnt pathway, such as CTNNB1 and Axin showed mutations. Apart from genetic alterations in the Wnt pathway components, there are several epigenetic changes associated with tumor initiation and progression in Wnt driven cancers. Additionally, there are epigenetic changes which are a result of Wnt activation. There are three modes of mechanism for the epigenetic machinery to work. It can change the status of DNA methylation, histone modification profiles and can also work through a plethora of non-coding RNAs (Dai et al., 2020). Even though these do not change the DNA sequence but these are heritable changes and thus can contribute extensively toward tumorigenesis. Very recently, drugs targeting epigenetic machinery have been adopted into the chemotherapeutic regime for cancers reflecting the importance of epigenetic modulation in cancers (Topper et al., 2020).
This review is specifically aimed at providing an overview of all well characterized epigenetic mechanisms which modulate the expression and function of the canonical Wnt pathway components and therefore affect Wnt target genes in various cancer types. In addition to this, the review will also cover advances made in the therapeutic interventions targeting the Wnt signaling pathway, especially through the epigenetic players.
Wnt Signaling Network – The Recent Most Snapshot
Wnt signaling is broadly classified into canonical and non-canonical Wnt signaling based on β-catenin -dependent and independent responses, respectively (Semenov et al., 2007). Canonical Wnt signaling is an intricate pathway involving 19 Wnt ligands, 10 Frizzled (FZD) receptors and 3 Disheveled (DVL) proteins. The selective combination of Wnt-FZD generates a wealth of information, modulating the signaling outcome crucial for normal development, multitude of cellular processes and development of various diseases (Semenov et al., 2007; Dijksterhuis et al., 2015; Voloshanenko et al., 2017). In unstimulated cells, the transcriptional co-activator β-catenin is engaged by a large cytoplasmic destruction complex composed of Adenomatous polyposis coli (APC), Axis inhibition protein (AXIN), Glycogen synthase kinase 3 (GSK3) and Casein kinase 1 (CK1), inducing sequential phosphorylation of β-catenin at Serine 33, Serine 37, Serine 45, and Threonine 41 by CK1 and GSK3β. The phosphorylated β-catenin is then ubiquitinated by βTrCP, an E3 ubiquitin ligase, followed by its degradation, preventing its nuclear transport (MacDonald et al., 2009). The stability of the destruction complex is very critical for continuous degradation of β-catenin. In canonical signaling, Wnt ligands bind to Frizzled and low-density lipoprotein receptor related proteins (LRP5/6) to stimulate Wnt signaling by recruiting the DVL proteins to the plasma membrane. Binding of DVLs to the membrane and sequential phosphorylation on the cytoplasmic domain of LRP5/6 promotes its interaction with AXIN, thereby destabilizing the destruction complex and inducing dis-engagement of β-catenin from the destruction complex. Moreover, the inactivation of ubiquitination and proteasomal degradation of β-catenin in the intact destruction complex have been shown to saturate the destruction complex, preventing further engagement and thereby degradation of β-catenin (Li et al., 2012). Disengaged β-catenin evades phosphorylation due to the multivesicular sequestration of GSK3β which prevents recognition by βTrCP E3 ubiquitin ligase and hence the degradation by the proteasome pathway (Taelman et al., 2010). Dephosphorylated β-catenin stabilizes and accumulates in the cytoplasm with subsequent translocation into the nucleus (MacDonald et al., 2009). Nuclear β-catenin interacts with the transcription factors of the TCF/LEF (T cell factor/lymphoid enhancer-binding factor) family by replacing corepressor, Groucho, to induce expression of Wnt/β-catenin target genes (Daniels and Weis, 2005).
Multiple factors have been shown to regulate the precise stimulation and inhibition of Wnt signaling. For instance, the R-spondin/LGR/RNF43 module is the key signaling paradigm that potentiates Wnt signaling. R-spondin family proteins regulate Wnt signaling via a common mechanism involving interaction with the Leucine-rich repeat-containing G-protein coupled (LGR) family of receptors (Kim et al., 2008). In the absence of R-spondins, two homologous ubiquitin E3 ligases, ZNRF3/RNF43 bind to the FZD receptors and target it for degradation. However, in the presence of R-spondins, ZNRF3/RNF43 interact with LGR4-6, leading to the inhibition of ZNRF3/RNF43 and dickkopf glycoproteins family member, DKK1 activity allowing subsequent accumulation of FZD receptors, thereby potentiating Wnt signaling (Kim et al., 2008; Wang et al., 2013; Xie et al., 2013; Figure 1). A recent study suggests that LGR5 potentiates Wnt signaling without sequestering ZNRF3/RNF43 by directly enhancing the Wnt signalosome mediated LRP6 phosphorylation (Park et al., 2020). This finding establishes that LGR4 and LGR5 have non-equivalent functions adopting different routes to stringently potentiate Wnt signaling.
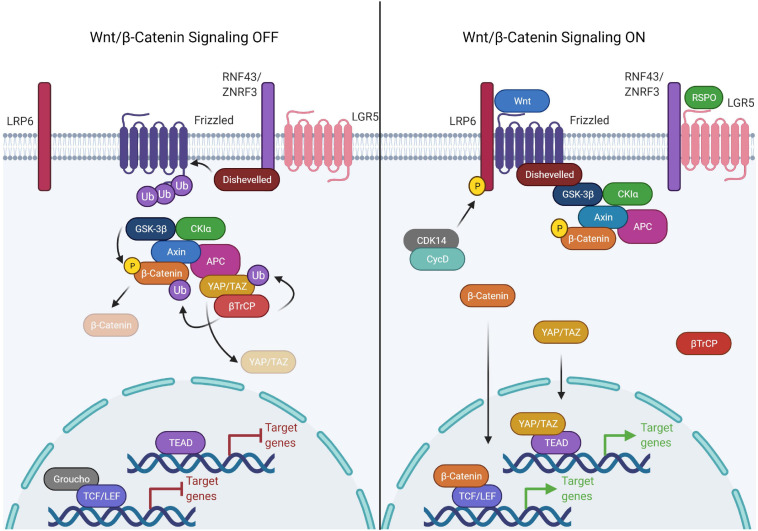
FIGURE 1.
Components of the canonical Wnt signaling pathway. The canonical Wnt pathway is primarily dependent on the transcriptional activity of β-catenin. In the Wnt OFF state, the Wnt destruction complex is actively engaged in the phosphorylation and subsequent ubiquitination mediated degradation of β-catenin. This is achieved by sequestering β-catenin to the Wnt destruction complex, consisting of APC, AXIN (scaffold proteins), GSK3β, and CK1α (kinases). Upon phosphorylation by the kinases, β-catenin is ubiquitinated by βTrCP which is recruited to the destruction complex by YAP/TAZ. The DNA binding partner of β-catenin, TCF/LEF, is bound to the DNA in a repressive complex with Groucho/TLE. Wnt signaling is activated upon binding of Wnt ligand causing complex formation between LRP6 and frizzled which allows Disheveled to bind to LRP. This activated form of Disheveled is able to bind and recruit the Wnt destruction complex thereby disrupting the binding of βTrCP. The receptor bound Wnt destruction complex is ubiquitination incompetent, hence β-catenin can’t be targeted for proteasomal degradation, therefore leading to saturation of the destruction complex with phospho-β-catenin. The new pool of β-catenin is free to translocate into the nucleus and activate the transcription of Wnt responsive genes by binding the TCF/LEF family of transcription factors. In the same process, YAP/TAZ is unable to recruit β-TrCP to the destruction complex and escape degradation. YAP/TAZ translocates into the nucleus as well and binds to the TEAD family of transcription factors.
Non-canonical Wnt signaling, consisting of Planar cell polarity (PCP) and Ca+ signaling pathways, has been shown to converge with multiple pathways and regulate a diverse number of cellular processes required for development and organogenesis. The combination of receptor/co-receptor at the plasma membrane binding to Wnts acts as a regulatory switch to activate either β-catenin-dependent or β-catenin-independent pathways. In the PCP pathway, Wnt ligands, Wnt5a and Wnt11 bind to receptors FZD3 or FZD6 along with tyrosine kinase co-receptors ROR1, ROR2 or RYK to orient toward Wnt/PCP signaling (Oishi et al., 2003; Martinez et al., 2015). Subsequently, this leads to recruitment of DVLs, leading to the activation of small GTPases of the Rho family [Cdc42, Rac1, RhoA and DVL-associated activator of morphogenesis1 (DAAM1)] (Habas et al., 2001; Liu et al., 2008). This cascade leads to the activation of downstream c-Jun kinase and Rho associated kinase required for actin cytoskeletal remodeling and cell contractility (Sebbagh and Borg, 2014; Martinez et al., 2015). The Wnt/PCP pathway is also activated independent of DVLs and the cascade of signaling is transduced to activate Nemo like kinase (NLK) which is required for cell fate determination and cell movement (Thorpe and Moon, 2004). In the Wnt/Ca+ pathway, Wnt ligands bind to FZD with concomitant recruitment of DVLs leading to the production of intracellular signaling molecules, inositol 1,4,5-triphosphate (IP3), 1,2 diacylglycerol (DAG), and Ca2+ from the membrane-bound phospholipid phosphatidylinositol 4,5-bisphosphate via the action of membrane-bound enzyme phospholipase C (De, 2011). The network of this signaling paradigm leads to the activation of downstream signaling proteins such as protein kinase C, calcineurin and Ca2+/calmodulin-dependent protein kinase II (CaMKII), thereby regulating cell adhesion and cell migration (Malbon et al., 2000; Figure 2).
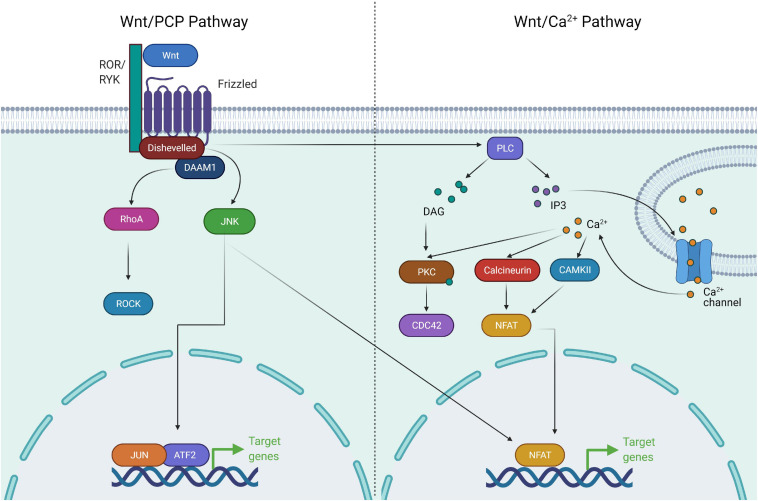
FIGURE 2.
Components of non-canonical Wnt signaling pathway. The non-canonical Wnt pathway is independent of β-catenin. Wnt activation leads to complex formation between ROR and Frizzled which allows activation of Disheveled. Disheveled is then able to activate DAAM1 and RHOA GTPase. RHOA in turn activates ROCK. Activation of ROCK and JNK leads to cytoskeletal remodeling. Wnt/Ca+ signaling involves activation of Disheveled upon Wnt activation, followed by activation of PLC. PLC activates the downstream effectors PKC, Calcineurin, and CAMKII via release of Ca+. These factors drive NFAT mediated transcriptional program and cytoskeletal changes.
Wnt Signaling in Cancers
Tumorigenesis involves progressive accumulation of genetic, epigenetic and molecular changes altering the cellular phenotype. Wnt signaling is a complex network of proteins and regulates molecular processes in a regulatory manner. Any imbalance in its regulation induces untoward cellular changes responsible for the development of various diseases and tumorigenesis. Dysregulation of Wnt signaling has been described in multiple cancers and plays a dramatic role in the progression of cancers (Bugter et al., 2021). Colorectal cancer is most commonly caused due to dysregulation of the Wnt pathway. It is sporadic although 30% of them are genetic in nature (Byrne and Tsikitis, 2018). Despite the role of the genetic mutations linked to colorectal cancer being well studied, the precise mechanism of the disease is not clearly understood. Mutations in the critical regulatory switches of the Wnt signaling pathway have been implicated in multiple cancers and loss of APC is a hallmark of colorectal cancer initiation and progression (Zhang and Shay, 2017). In most colorectal cancers, both the alleles of APC gene are mutated, especially in the region required for interaction with the armadillo (arm) repeats of β-catenin (Rubinfeld et al., 1997). Such mutations prevent the degradation of β-catenin and induce cellular changes critical for colorectal cancer initiation. Mutations in the APC gene are responsible for approximately 1% of all colorectal cancer cases (Bienz and Clevers, 2000; Half et al., 2009). The main feature of Wnt signaling driven cancers is the constitutive nuclear localization of β-catenin. Numerous studies have identified mutations in the serine and threonine residues of β-catenin causing abrogation of phosphorylation by GSK3β and CK1 kinases, ultimately leading to the hyperactivation of Wnt signaling (Sparks et al., 1998). Mutation in the serine/threonine regulatory domain of β-catenin has been found in 48% of colorectal cancers lacking APC mutations (Sparks et al., 1998). High throughput cancer genomics studies have cataloged mutations in the genes encoding multiple components of the Wnt signaling pathway, including the ligands, receptors and intracellular components. Besides APC and CTNNB1, mutations have been found in the regions of AXIN1 gene coding for domains which are critical for its interaction with APC, GSK3β and β-catenin, resulting in destabilization of the destruction complex with concomitant increase in cytoplasmic as well as nuclear levels of β-catenin (Jin et al., 2003; Salahshor and Woodgett, 2005). Similarly, a mutation in AXIN2 identified in colorectal cancers promotes GSK3β inhibition and stabilization of Snail, a positive regulator of Wnt signaling (Mazzoni et al., 2015). The increased expression of LRPs have also been linked with hyperactivation of Wnt signaling and development of colorectal cancers (Rismani et al., 2017; Boulagnon-Rombi et al., 2018; Yao et al., 2020). The R-spondin/LGR5/RNF43 module has also been implicated in Wnt signaling driven cancers. A deleterious mutation in RNF43 has been reported in 19% of colorectal cancers lacking APC mutations (Giannakis et al., 2014). Further, a high number of R-spondin mutations and fusion proteins causing hyperactivation of Wnt signaling has been described in 10% of colon cancers lacking the APC mutations (Seshagiri et al., 2012; Giannakis et al., 2014).
Among gastric cancers, 30% exhibit nuclear β-catenin as a prominent acquired change (Clements et al., 2002). Around 18% of gastric cancers have mutations in APC and RNF43 as the primary cause of the disease (Clements et al., 2002; Fang et al., 2002; Wang K. et al., 2014; Flanagan et al., 2019). In addition to the genetic mutations, epigenetic changes (such as promoter methylation) have also been identified in Wnt antagonists such as the secreted frizzled-related proteins (sFRPs) and Wnt negative-modulators, the dickkopf family of glycoproteins (DKK1–3), leading to activation of Wnt signaling (Nojima et al., 2007; Yu et al., 2009; Flanagan et al., 2017). These will be elaborated in the following sections.
In cancers, heterogenous levels of activation determine the adversity and fate of different cancers. For example, 37% of APC-mutant gastric cancers have mutation in RNF43, suggesting a compound activation of Wnt signaling in the same tumor. However, only 5.5% of colon cancers have mutations both in APC and RNF43, suggesting a differential mechanism of optimal activation of Wnt signaling in gastric and colon cancers for tumor growth and progression (Albuquerque et al., 2002). The role of Wnt signaling in hematopoietic stem cells and leukemia progression has also been described wherein β-catenin appears to be essential for leukemia initiating cells and their self-renewal (Albuquerque et al., 2002; Luis et al., 2012; Lento et al., 2013). Chronic lymphocytic leukemia shows higher expression of multiple Wnt proteins with enhanced β-catenin-dependent Wnt signaling (Lu et al., 2004). Further, epigenetic silencing of Wnt signaling inhibitors DKK1/2 and somatic mutations in FZD5 is associated with the development of chronic lymphocytic leukemia in 14% of the cases (Moskalev et al., 2012; Wang L. et al., 2014). Table 1 summarizes the mutations in different Wnt components reported in various cancers. Recently, the synergistic interplay of the canonical and non-canonical Wnt signaling playing a major role in the development of cancers has also been established (Katoh, 2017; Flores-Hernández et al., 2020).
TABLE 1.
Mutations in Wnt signaling components in cancers.
| Gene | Alteration | Cancer type | Outcome | References |
| APC | Exon 15 | CRC | Loss of interaction with β-catenin | Rubinfeld et al., 1997; Bienz and Clevers, 2000; Half et al., 2009 |
| APC | Exon 15 | GC | Loss of interaction with β-catenin | Fang et al., 2002; Flanagan et al., 2019 |
| β-catenin | Codons 33, 37, 41, and 45 in exon 3 | CRC | Loss of phosphorylation by GSK3β and Ck1α | Sparks et al., 1998 |
| AXIN1 | Exon 1 and 5 | CRC | Loss of interaction with APC, GSK3β and β-catenin | Jin et al., 2003; Salahshor and Woodgett, 2005 |
| AXIN2 | Stop codon at codon 663 in exon 7 | CRC | Inhibition of GSK3β and stabilization of Snail | Mazzoni et al., 2015 |
| RNF43 | Truncation mutations | CRC | Inactivation of RNF43 and increased cell surface abundance of FZD5 | Giannakis et al., 2014 |
| RNF43 | Truncation mutations | GC | Inactivation of RNF43 and increased cell surface abundance of FZD5 | Wang K. et al., 2014 |
| R-spondins | Gene fusions | CRC | Gene fusion of R-spondins (RSPO2 and RSPO3) with PTPRK and/or EIF3E causing elevated expression of R-spondins | Seshagiri et al., 2012 |
Convergence of Wnt Pathway With Other Signaling Networks in Cancer
Signaling pathways do not operate linearly nor exclusively. Rather, there are multiple points of crosstalk between all the pathways which ultimately fine tune target gene expression. The Wnt pathway is known to work in conjunction with other pathways to regulate the fundamental cellular processes critical for normal development and cancer.
Wnt and Hippo Signaling Pathways: Partners in Action
Like the Wnt pathway, the Hippo signaling pathway was also identified in Drosophila. The pathway gets its name owing to the phenotype it induces in flies. Flies with mutations in the Hippo pathway components have abnormal growth due to loss of organ size control (Justice et al., 1995; Xu et al., 1995; Jia et al., 2003; Udan et al., 2003). Studying crosstalk between the Wnt and Hippo pathways has been of particular interest to researchers. It is due to the fact that the effector proteins of these pathways, β-catenin and YAP/TAZ, directly interact with each other and their stability is dependent on the Wnt destruction complex (Azzolin et al., 2012, 2014). The Hippo pathway does not consist of a defined set of receptors and has diverse upstream regulators (Pan, 2010; Totaro et al., 2018). Unlike other pathways, the activation of the Hippo pathway leads to inactivation of its effector molecules, YAP and TAZ. These regulators can either be components of other signaling pathways or, often, are part of the cell’s mechanosensory machinery (Zhao et al., 2007; Lei et al., 2008). The mechanosensory proteins are the ones that are involved in the maintenance of cell-cell adhesion, apico-basal polarity and cell junctions, for example, E-cadherin, KIBRA, NF2, α-catenin and the Crumbs complex (Genevet et al., 2010; Varelas et al., 2010b; Zhang et al., 2010; Silvis et al., 2011; Benham-Pyle et al., 2015). Proteins, such as NF2, act as a scaffold for the assembly of Hippo kinases (Lallemand et al., 2003). Activation of Hippo signaling leads to the activation of a cascade of kinases, namely, Macrophage-stimulating protein (MST1/2), Salvador (SAV1), Large tumor suppressor (LATS1/2), and Mps one binder kinase (MOB1). MST1/2 is the upstream kinase. It gets activated upon phosphorylation and forms an active complex with SAV1. This complex further activates LATS1/2 via phosphorylation. LATS1/2, along with MOB1 leads to the phosphorylation of YAP/TAZ (Chan et al., 2005; Callus et al., 2006; Zhao et al., 2007; Praskova et al., 2008; Varelas et al., 2010b). The phosphorylated form of YAP/TAZ is sequestered in the cytoplasm and subsequently degraded due to ubiquitination (Zhao et al., 2007, 2010; Lei et al., 2008). Absence of Hippo signaling allows nuclear accumulation of YAP/TAZ which then bind to the TEAD (transcriptional enhanced associate domain) family of DNA binding proteins, leading to transcription of its downstream targets (Vassilev et al., 2001; Mahoney et al., 2005). YAP/TAZ can have other DNA binding partners as well, such as p75, RUNXs and SMADs (Yagi et al., 1999; Strano et al., 2001; Varelas et al., 2008). Due to its involvement in regulating organ size control and cell adhesion properties, its role in multiple cancer types is not surprising. It has been reported to be involved in cases of breast cancer, lung cancer, liver cancer, prostate cancer, etc. (Chan et al., 2008; Zhou et al., 2011; Chen Q. et al., 2014; Ma et al., 2014; Chen H.-Y. et al., 2015; Seo et al., 2017). Similar to the Wnt signaling pathway, the Hippo pathway is involved in the regulation of intestinal regeneration and is therefore frequently dysregulated in colorectal cancers as well (Cai et al., 2010; Gregorieff et al., 2015). Interestingly, even though the Hippo pathway is frequently perturbed in various cancer types, mutations in its components are uncommon (Harvey et al., 2013).
The first link between the Wnt and Hippo pathways was established in a study which showed TAZ as the negative regulator of the Wnt pathway. It was shown that TAZ inhibits interaction of DVL with CK1δ/ε thereby preventing its phosphorylation upon Wnt activation leading to inhibition of Wnt signaling (Varelas et al., 2010a). Recently, LATS1/2 have been shown to maintain the intestinal stem cell niche by sustaining Wnt signaling (Li Q. et al., 2020). The study shows that when the Hippo pathway is inactive, nuclear YAP/TAZ interacts with Groucho and prevents the Wnt mediated transcriptional program (Figure 3). Interestingly, LATS1/2 depletion was able to silence the expression of the Wnt target gene, MYC, even in the APC mutant intestinal cells, making it a good candidate for intestinal tumor therapy. Other evidence of positive regulation by the Hippo pathway showed that S127D mutant YAP (phosphomimetic form and hence constitutively cytoplasmic) inhibited the nuclear accumulation of β-catenin (Imajo et al., 2015). In contrast, few reports have suggested a negative role of the Hippo pathway in the regulation of Wnt signaling. A study showed that Taz knockout in mice kidney led to a minimal increase in nuclear β-catenin (Makita et al., 2008). An increased expression of β-catenin was observed in the case of cardiomyocytes from Salvador (Sav1) knockout mice (Heallen et al., 2011). Additionally, it was shown that YAP/TEAD and β-catenin/TCF complexes bind cooperatively to a few of their target genes, for e.g., snail2 and sox2 (Heallen et al., 2011). Most importantly, all the three effectors, β-catenin, YAP and TAZ, are involved in regulating βTrCP mediated ubiquitination and subsequent degradation of each other. YAP and TAZ themselves are part of the Wnt destruction complex (Azzolin et al., 2012). In Wnt off conditions, YAP/TAZ binds to the destruction complex and recruits βTrCP, thereby promoting degradation of β-catenin as well as YAP/TAZ (Azzolin et al., 2014). Similarly, β-catenin phosphorylated by GSK3β can bind to TAZ and initiate its ubiquitination (Azzolin et al., 2012). A recent report showed that the balance between β-catenin and YAP is crucial for the maintenance of the intestinal stem cell niche. YAP overexpression or LATS depletion resulted in the inhibition of Lgr5+ intestinal stem cell growth by re-programming these cells into a state of low Wnt and high Klf6, driving the cells toward a ‘wound-healing state’. The same study showed a similar effect in cases of organoids, patient derived xenografts as well as colorectal cancer cell lines (Cheung et al., 2020).
FIGURE 3.
Interplay of the Wnt pathway with other pathways. Summary of the interaction of the Wnt pathway components (depicted in green) with other signaling pathways, including the Hippo pathway (depicted in blue), TGF-β pathway (depicted in magenta), Hedgehog pathway (depicted in purple), and Notch pathway (depicted in red). The Hippo pathway helps in the stabilization of β-catenin by inhibiting YAP’s interaction with the Wnt destruction complex, thereby preventing the recruitment of βTrCP. TGF-β signaling has synergistic effects with Wnt signaling. SMAD7 can help in preventing the GSK-3β mediated β-catenin degradation. The Hedgehog effector Gli1/2 can inhibit Wnt target gene expression by directly binding to the β-catenin/TCF complex. It can also drive the expression of the Wnt inhibitor sFRP1. The Notch signaling transcription factor, NICD, interacts with β-catenin. NICD can promote Wnt signaling by promoting the expression of the Wnt ligands. However, the Wnt inhibitor DKK2 is also a target of NICD. NICD itself is a substrate of GSK-β.
TGF-β Signaling Pathway
The interplay between TGF-β and Wnt pathways is context-dependent and can result in different outcomes. For example, Wnt3a stimulation increases TGF-β expression as well as SMAD2 phosphorylation and promotes differentiation in the case of myofibroblasts (Carthy et al., 2011). Wnt and TGF-β activation have synergistic effects during EMT in alveolar epithelial cells (Zhou et al., 2012). TGF-β and Wnt activation promotes expression of mesendodermal (ME) related genes in human embryonic stem cells (hESCs). YAP recruits Negative elongation factor (NELF) and inhibits the expression of ME related genes (Estarás et al., 2015). Inhibitory SMAD7 promotes β-catenin binding to plasma membrane through E-cadherin and prevents its degradation in cancer epithelial cells and helps in regulation of cell–cell adhesion. Complex formation between SMAD7 and AXIN prevents interaction of β-catenin with GSK3β and E3 ligase Smurf2 via AXIN. This prevents degradation of β-catenin but increases its localization on the membrane instead of nucleus (Tang et al., 2008). On the other hand, an earlier study showed that SMAD7 can promote the nuclear translocation of β-catenin in prostate cancer cells (Edlund et al., 2005). AXIN proteins have been shown to play a contrasting role in the regulation of TGF-β signaling. Earlier studies showed that AXIN acts as a positive regulator of TGF-β signaling by promoting phosphorylation of SMAD3 upon TGF-β activation (Furuhashi et al., 2001; Dao et al., 2007). However, another report showed that AXIN is able to mediate binding of SMAD3 (not SMAD2) to GSK3β and promotes SMAD3 phosphorylation and degradation in the absence of TGF-β signaling (Figure 3). Depletion of AXIN increased the transcriptional activity of SMAD3 even in the absence of TGF-β activation (Guo et al., 2008). As mentioned earlier, SMADs are known DNA-binding partners of YAP/TAZ, apart from the TEAD proteins. YAP and TAZ interact with activated SMAD2/4 and SMAD3/4 complexes and increase their nuclear localization (Hiemer et al., 2014). TGF-β activation also leads to an increase in TAZ mRNA and not YAP, in a SMAD3-independent manner (Miranda et al., 2017). TAZ has been shown to enhance the metastatic properties of breast cancer cells via expression of BMP4 and subsequent activation of BMP signaling (Lai and Yang, 2013). However, when the Hippo pathway is activated and YAP/TAZ accumulates in the cytoplasm, the nuclear translocation of SMADs is blocked (Barrios-Rodiles, 2005).
Hedgehog Signaling
The crosstalk between Hedgehog and Wnt signaling is stage-specific. The inhibitory feedback between the two pathways is responsible for driving developmental processes. Canonical Hedgehog signaling is an inhibitor of Wnt signaling especially in the context of intestinal homeostasis. Hedgehog signaling suppresses Wnt signaling by regulating the expression of sFRP1 via its effector proteins, Gli1 and Gli2 (He et al., 2006; Figure 3). Activation of Hedgehog signaling prevents the nuclear accumulation of β-catenin in liver cancers as well (He et al., 2006). However, the non-canonical Hedgehog signaling, which is Patched1 (PTCH1)-dependent and Smoothened (SMO) and Gli-independent, has been shown to positively regulate Wnt signaling in the intestinal tumors by promoting the undifferentiated state of cancer stem cells (Regan et al., 2017). Interestingly, one study showed that the advanced stages of colon cancer have a higher activation level of Hedgehog and not Wnt signaling. Activation of Hedgehog signaling also led to the expression of embryonic genes such as, POU5F1 and NANOG as well as inhibition of β-catenin/TCF transcriptional activity via Gli1. Activation of embryonic genes therefore might be responsible for increasing cancer stemness and metastatic properties in Hedgehog driven cases (Varnat et al., 2010). In contrast, Gli2 was shown to promote nuclear localization of β-catenin and together these factors drive the proliferation of osteosarcoma cells (Ma J. et al., 2019; Xu et al., 2019). Sonic hedgehog has been reported to positively regulate YAP in medulloblastoma. It not only drives the expression of YAP but also promotes its nuclear accumulation (Fernandez-L et al., 2012). However, in case of the intestinal mesenchyme, YAP/TAZ promote proliferation and self-renewal whereas Hedgehog signaling drives differentiation into smooth muscle (Cotton et al., 2017).
FGF Signaling
The Wnt-FGF gradient is a paradigm which has been very well established in the context of development. It plays an extensive role during axis formation in development (Dyer et al., 2014; Oginuma et al., 2017). Due to similar reasons, the interplay between the two pathways is important in cancer as well. Wnt1 and Fgf3 expression is upregulated in cases of breast cancer. Breast cancer cell lines overexpressing both these ligands and therefore with high Wnt and Fgf activity, exhibited higher mitochondrial biogenesis which correlated with enhanced mammosphere forming capacity and stemness (Lamb et al., 2015). An extensive axis between Wnt and FGF regulates the stem cell niche in the lungs. In a study by Volckaert et al. (2017), it was shown that YAP expression promotes the basal stem cell niche in the lung epithelium. Activation of the Hippo pathway keeps the differentiated epithelium in a quiescent state. However, injury leads to the activation of YAP caused due to the degradation of Merlin, an upstream regulator of the Hippo pathway. Yap in turn causes induction of Wnt7b which in turn drives the expression of FGF10 which is required for the maintenance of basal stem cells (Volckaert et al., 2017).
Notch Signaling
Notch activity is Wnt-dependent and it cooperates with Wnt in development as well as cancer. Cancer stem cells isolated from clear cell renal cell cancer specifically showed upregulation of the Wnt and Notch related genes including TCF7L2, TCF3, LGR4, AXIN2, EP300RBPJ, NOTCH3, HES1, and JAG1 (Fendler et al., 2020). At a mechanistic level, it has been shown that the Notch intracellular domain (NICD) is a substrate for GSK3 kinases (Espinosa et al., 2003). The activity of GSK3-β is inturn dependent on the Wnt activation status. In Wnt inactive state, GSK3-β-dependent phosphorylation primes Notch for proteasomal degradation, thereby preventing its transcriptional activity (Figure 3; Espinosa et al., 2003). β-catenin can directly bind to NICD and enhance its transactivation. However, overexpression of LEF1 hinders this process by competing with NICD to interact with β-catenin (Jin et al., 2009). In another study, it was shown that Notch signaling is required for maintaining the stemness of mammary associated stem cells. Notch activation through stem cells in macrophages eventually led to expression of multiple Wnt ligands- Wnt3, 10, and 16 which ultimately promoted self-renewal in stem cells (Chakrabarti et al., 2018). Wnt signaling has been shown to prevent differentiation of hESCs into Medial Ganglionic Eminence (MGE)-like cells and promoted expansion of progenitors by driving the expression of the Notch ligand, JAG1 (Ma L. et al., 2019). However, it has also been shown that DKK2 is a target of Notch signaling in the intestinal stem cells, indicating a negative feedback for Wnt by Notch signaling (Katoh and Katoh, 2007).
Transcription Factors
Wnt signaling pathway partners with various transcriptional factors and chromatin remodelers, mainly via interaction with β-catenin, presumably assisting the expression of target genes for these transcription factors in a Wnt-dependent manner. A few instances have been reported in which these transcription factors can act as negative regulators of Wnt signaling as well. Some of these include SOX17, CBP, FOXO, MLL1, TBP, and HIF1a. A number of these factors are reviewed in Valenta et al. (2012) and Söderholm and Cantù (2020). In this review, we have focused on the two protein families that have been recently identified to work alongside the Wnt pathway.
The Homeodomain (HOX) group of proteins are involved in embryogenesis. In adults, these are required for maintenance and homeostasis. HOXA5 has been shown to coordinate with the Wnt pathway in driving stem cell differentiation. HOXA5 acts as a tumor suppressor and promotes intestinal cell differentiation by inhibiting the Wnt/β-catenin pathway. Wnt pathway, on the other hand, inhibits HOXA5 via MYC which is a direct target of the Wnt signaling (Ordóñez-Morán et al., 2015). Another study using cervical cancer cells showed that HOXA5 can inhibit Wnt pathway genes by transactivating TP53. TP53 regulates the expression of the miR-200 family of microRNAs which target multiple Wnt pathway components, as mentioned in the proceeding sections (Ma et al., 2020). In contrast, HOXA13 exerts an opposite effect. It binds to β-catenin and helps in the nuclear translocation and subsequent activation of β-catenin/TCF/LEF mediated transcription (Gu et al., 2020). HOXB4 is another HOX gene family member which appears to have both tumor suppressive and oncogenic potential depending on the cancer type. In a recent study, it was shown to inhibit cancer cell proliferation and further tumorigenesis by inhibiting β-catenin at the transcriptional level by binding to its promoter in cervical cancer cells. Interestingly, several other Wnt related genes were found to be downregulated in cells overexpressing HOXB4, including MYC, whereas genes such as FZD8, DKK1, and AXIN2 were upregulated (Lei et al., 2021).
The Specificity Protein (SP) family of zinc finger proteins have been shown to regulate Wnt signaling as well. SP1, one of the earliest transcription factors to be identified, has been shown to regulate the stability of β-catenin by modulating its interaction with the Wnt destruction complex. Interestingly, SP1 depletion led to an increased phosphorylation and ubiquitination of β-catenin in colorectal cancer cell lines (Mir et al., 2018). SP5, on the other hand, acts as a feedback inhibitor of Wnt signaling. Its expression is induced upon Wnt activation, especially in cells with stemness, including embryonic carcinoma cells (Huggins et al., 2017). Further, Huggins et al. (2017) hypothesized that SP5 might compete with SP1 for binding to a limited set of loci upon Wnt activation in case of human pluripotent stem cells. These loci are mainly Wnt target genes which are downregulated by SP5 (Huggins et al., 2017). Interestingly, SP5 is also strongly expressed in Lgr5+ intestinal stem cells (Barker et al., 2010). Another study showed that SP5 and SP8 both assist the binding of β-catenin to LEF1 upon Wnt activation. This study also pointed out that SP5 and SP8 are responsible for regulating only a subset of Wnt target genes (Kennedy et al., 2016).
Wnt Signaling and Epigenetic Regulation
Modes of Epigenetic Regulation
Cells in multicellular organisms are homogenous at the genetic level but their phenotypic heterogeneity is a key hallmark at the structural and functional level. This heterogeneity is considered crucial in the regulation of gene expression and such heritable alterations that do not affect the DNA sequence are referred to as epigenetic changes. The epigenetic pathways respond to various signaling cues such as DNA methylation, histone variants, histone modifications, chromatin structure, nucleosome remodeling, and epigenetic interactions (Allis and Jenuwein, 2016). DNA methylation is one of the major epigenetic modifications regulating gene expression by altering DNA conformation, chromosome structure and by recruiting other epigenetic regulators (Denis et al., 2011; Hervouet et al., 2018; Laisné et al., 2018). DNA methylation is catalyzed by three methyltransferases DNMT1, DNMT3a and DNMT3b. DNMT3a and DNMT3b catalyzes de novo methylation while DNMT1 is required for maintenance and is critical for the inheritance of DNA methylation (Moore et al., 2013; Greenberg and Bourc’his, 2019). Around 1.5% of the human genome is methylated and the majority of methylation occurs at CpG sites on gene promoters. Aberrant hypermethylation at CpG sites is associated with diverse cancers (Robertson, 2001).
The selective incorporation of histone modifications at gene promoters and enhancers acts as a regulatory switch to determine the fate of gene expression. Trimethylation of histone H3 at lysine 4 (H3K4me3) is enriched at the active promoters whereas trimethylation of histone H3 lysine 27 (H3K27me3) and histone H3 lysine 9 (H3K9me3) are typically seen at inactive promoters. The enhancers are enriched with histone H3 lysine 4 monomethylated (H3K4me) and acetylated histone H3 lysine 27 (H3K27Ac) modifications. The histone methyltransferases (KMTs) and histone demethylases (KDMs) add and remove methyl groups to/from histones respectively. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) add and remove acetyl groups to/from histones, respectively (Hon et al., 2009; Hawkins et al., 2011; Albini et al., 2019). The incorporation of these modifications involves dynamic regulation and any deviation from the regulation is associated with developmental defects and diseases (Hon et al., 2009; Hawkins et al., 2011).
MicroRNAs (miRs) are small non-coding RNAs typically consisting of 20–21 nucleotides and are essential for various developmental processes and diseases. They regulate gene expression post-transcriptionally either by inhibiting translation or inducing mRNA degradation (Filipowicz et al., 2008). Another group of non-coding RNAs called the long non-coding RNAs (lncRNAs) with less protein coding potential (also referred as transcription noise) are 200 nucleotide long and have been shown to be essential for regulating transcription, translation, splicing, chromatin modification and structure (Jarroux et al., 2017; Kopp and Mendell, 2018).
Epigenetic Regulation of Wnt Signaling Components
Wnt signaling is a complex pathway modulated by various cellular and environmental cues to precisely regulate the myriad of cellular processes during development and disease. Wnt driven cancers have either hyperactivation of positive regulators or hypoactivation of negative regulators. In this section, we provide the details on the epigenetic modulations undergone by the components of the Wnt pathway. Table 2 summarizes the epigenetic regulation of Wnt components discussed in the following section.
TABLE 2.
Epigenetic mechanisms regulating the Wnt components in different types of cancer.
| Wnt component | Epigenetic alteration | Cancer type | Outcome | References |
| Wnt2 | ‘Loss of H3K27me3 | CRC | Upregulation | Jung et al., 2015 |
| miR-548b downregulation | CRC | Upregulation | Xu et al., 2020 | |
| Wnt5a | Promoter hypermethylation | CRC | Downregulation | Ying et al., 2008 |
| Wnt9a | Promoter hypermethylation | CRC | Downregulation | Galamb et al., 2016 |
| Wnt10b | Promoter hypermethylation | CRC | Downregulation | Yoshikawa et al., 2007 |
| DKK1 | Promoter hypermethylation | CRC, CLL, BC | Downregulation | Nojima et al., 2007; Yu et al., 2009; Felipe de Sousa et al., 2011; Moskalev et al., 2012; Flanagan et al., 2017 |
| loss of H3Ac | Glioma, BC | Downregulation | Kim H.-Y. et al., 2015 | |
| loss of H3K4me3 | BC | Downregulation | Huang et al., 2013 | |
| miR-375 upregulation | BC | Downregulation | Taube et al., 2013 | |
| miR-376 upregulation | BC | Downregulation | Cui et al., 2016 | |
| DKK3 | Promoter hypermethylation | CRC, BC | Downregulation | Xiang et al., 2013 |
| sFRP1 | Promoter hypermethylation | CRC | Downregulation | Suzuki et al., 2002; Fujikane et al., 2010; Li et al., 2018 |
| miR-139-5p and miR-9 upregulation | BC | Downregulation | Yang et al., 2015 | |
| sFRP2 | Promoter hypermethylation | CRC, HCC, BC | Downregulation | Suzuki et al., 2002; Veeck et al., 2008; Yu et al., 2019; Xiang et al., 2021 |
| sFRP4 | Promoter hypermethylation | CRC | Downregulation | Suzuki et al., 2002 |
| sFRP5 | Promoter hypermethylation | CRC | Downregulation | Suzuki et al., 2002 |
| WIF1 | Promoter hypermethylation | CRC, BC, OC | Downregulation | Ai et al., 2006; Paluszczak et al., 2015 |
| miR-603 downregulation | Glioma | Downregulation | Hu et al., 2018 | |
| Guo et al., 2014 | ||||
| Fzd4 | miR-505 downregulation | CC | Upregulation | Ma et al., 2017 |
| loss of H3K27me3 | GC | Upregulation | Li Z.-T. et al., 2020 | |
| Fzd6 | miR-199a-5p downregulation | CRC | Upregulation | Kim B.-K. et al., 2015 |
| miR-130-30p downregulation | BC | Upregulation | Poodineh et al., 2020 | |
| Fzd7 | miR-613 downregulation | PC | Upregulation | Song et al., 2017 |
| Fzd8 | miR-100 downregulation | BC | Upregulation | Jiang et al., 2016 |
| Fzd10 | Gain of H3K9Ac | BC | Upregulation | Gong et al., 2014 |
| LRP6 | miR-34 downregulation | CRC, BC | Upregulation | Kim et al., 2011 |
| miR-130-30p downregulation | BC | Upregulation | Poodineh et al., 2020 | |
| miR-487b downregulation | CRC | Upregulation | Hata et al., 2017 | |
| miR-181 upregulation | OC | Upregulation | Ruan et al., 2020 | |
| LGR5 | Promoter hypermethylation | CRC | Downregulation | Su et al., 2015 |
| miR-363 downregulation | CRC | Upregulation | Tsuji et al., 2014 | |
| miR-34 downregulation | CRC | Upregulation | Hahn et al., 2013 | |
| APC | Promoter hypermethylation | CRC | Downregulation | Su et al., 1992, 1993; Arnold et al., 2004 |
| miR-135 upregulation | CRC | Downregulation | Segditsas et al., 2008; Liang et al., 2017 | |
| miR-142 upregulation | BC | Downregulation | Nagel et al., 2008; Isobe et al., 2014 | |
| AXIN2 | Promoter hypermethylation | CRC | Downregulation | Koinuma et al., 2006 |
| miR-34 upregulation | CRC | Downregulation | Kim et al., 2013 | |
| miR-103/107 upregulation | CRC | Downregulation | Kim et al., 2013; Chen H.-Y. et al., 2019 | |
| GSK3-β | miR-224 upregulation | CRC | Downregulation | Li T. et al., 2016 |
| miR-1229 upregulation | BC | Downregulation | Tan et al., 2016 | |
| βTrCP | miR-182 upregulation | CRC | Downregulation | Wang S. et al., 2016 |
| miR-135b upregulation | OS | Downregulation | Jin et al., 2017 | |
| CK1α | miR-155 upregulation | CRC, BC | Downregulation | Zhang et al., 2012 |
| miR 135b upregulation | OS | Downregulation | Jin et al., 2017 | |
| YAP1 | miR-506 downregulation | BC | Upregulation | Hua et al., 2015 |
| miR-506 upregulation | CRC | Downregulation | Krawczyk et al., 2017 | |
| miR-1224/CREB promoter | HCC | Downregulation | Yang et al., 2021 | |
| hypermethylation | BC | Downregulation | Real et al., 2018 | |
| TAZ1 | LINC00174 upregulation | CRC | Upregulation | Shen et al., 2018 |
| miR-125a downregulation | CRC | Upregulation | Yang M. et al., 2019 | |
| LATS1/2 | Promoter hypermethylation | CRC, HNSCC, BC, RCC | Downregulation | Okami et al., 2005; Steinmann et al., 2009; Chen et al., 2013; Wierzbicki et al., 2013; Chen K.-H. et al., 2014 |
| MST1/2 | Promoter hypermethylation | HNSCC, Mesothelioma | Downregulation | Steinmann et al., 2009; Maille et al., 2019 |
| β-catenin | miR-200a downregulation | CRC | Upregulation | Tian et al., 2014 |
| miR-141 downregulation | BC | Upregulation | Abedi et al., 2015 | |
| miR-340 downregulation | BC | Upregulation | Abedi et al., 2015 | |
| miR-520f-3p downregulation | GC | Upregulation | Chen et al., 2020 | |
| TCFL1/2 | Promoter hypomethylation | CRC | Upregulation | Guo et al., 2015 |
| TCF7 | miR-29 downregulation | CRC | Upregulation | Subramanian et al., 2014 |
| LEF1 (transcript from P2) | miR-34 | BC | Upregulation | Kim et al., 2011 |
| gain of H3K9me3 | BC | Downregulation | Yokoyama et al., 2010 | |
| TCF7 | LOC728196 upregulation | Glioma | Upregulation | Wang O. et al., 2018 |
Ligands
Several tumor types are dependent on the hyperactivation of Wnt ligands because it provides an advantage to the tumor cells. Wnt secretion not only activates autocrine signaling but can also act on neighboring cells via paracrine signaling (Carbone et al., 2018; Eyre et al., 2019). Cancer cells seem to utilize multiple mechanisms to upregulate the expression of Wnt ligands. For example, Wnt2 expression can be upregulated in colorectal cancer via loss of repressive histone mark H3K27me3 by EZH2 (Jung et al., 2015). Wnt2 expression is also enhanced by downregulation of its regulatory miRNAs. In case of colorectal cancer, miR-548b overexpression can suppress cell proliferation by targeting Wnt2 (Xu et al., 2020). Several upstream regulators can also be targeted to achieve higher expression of the Wnt ligands. miR-600 targets SCD1 (stearoyl desaturase 1), an enzyme required for the lipid modification of the Wnts. Downregulation of miR-600 has been shown to promote the self-renewal of breast cancer stem cells whereas overexpression promotes their differentiation (El Helou et al., 2017). However, contrasting reports of promoter hypermethylation in case of multiple Wnt genes, including WNT5A, WNT9A, and WNT10B, suggest a potential tumor suppressive role (Yoshikawa et al., 2007; Ying et al., 2008; Galamb et al., 2016). The role of Wnt ligands in colorectal cancer has been reviewed in Nie et al. (2020). Promoter hypermethylation is also involved in silencing the Wnt negative-modulators, the DKK family, leading to hyperactivation of Wnt signaling in cancers (Nojima et al., 2007; Yu et al., 2009; Moskalev et al., 2012; Flanagan et al., 2017). Interestingly, DKK1 promoter methylation has been reported in the later stages of cancer (Felipe de Sousa et al., 2011). Therefore, it might be assumed that it plays an important role in cancer progression instead of initiation. Promoter hypermethylation of DKK3 has been reported in both colorectal cancer and breast cancer. However, its effect is not via suppression of LRP5/6 activity. Rather, it promotes membrane localization of β-catenin from the nucleus (Xiang et al., 2013). Promoter hypermethylation of DKK2 has been reported but no correlation was observed with its expression (Silva et al., 2014). Apart from promoter methylation, DKK1 promoter is also targeted by removal of activatory histone marks, such as H3Ac and H3K4me3 due to loss of p300 and recruitment of HDACs to its promoter, leading to reduced expression (Kim H.-Y. et al., 2015; Bao et al., 2017; Huang et al., 2013). DKK1 expression is also inhibited due to the upregulation of certain miRNAs, such as miR-375 and miR-376 (Taube et al., 2013; Cui et al., 2016). Silencing the Wnt antagonistic protein sFRPs by promoter hypermethylation resulting in the aberrant activation of Wnt signaling has been identified in diverse cancers with increased tumorigenic risk (Yu et al., 2019). Promoter hypermethylation of SFRP1, 2, 4, and 5 has been reported which correlates with the expression and cancer stage in case of breast cancer and colorectal cancer (Suzuki et al., 2002; Fujikane et al., 2010; Li et al., 2018). However, SFRP3 does not harbor a CpG island in the promoter region (Suzuki et al., 2002). sFRP1 is also downregulated via multiple miRNAs in breast cancer (Yang et al., 2015). Downregulation of sFRP1 by hypermethylation has been shown to be essential for self-renewal of colorectal cancer stem cells (Li et al., 2018). SFRP2 promoter hypermethylation occurs in HBV-associated hepatocellular carcinoma and breast cancer (Veeck et al., 2008; Yu et al., 2019; Xiang et al., 2021). Downregulation of WIF1 occurs by promoter hypermethylation in colorectal cancer, breast cancer, ovarian cancer and several other cancer types (Ai et al., 2006; Paluszczak et al., 2015; Hu et al., 2018). miR-603 is known to target WIF1 in glioma (Guo et al., 2014).
Receptors
Almost all members of the Wnt receptor family, such as FZD4, 6, 7, 8, and 10 have been reported to be regulated by different miRNAs in multiple cancer types including colorectal cancer, prostate cancer, breast cancer (Gong et al., 2014; Kim B.-K. et al., 2015; Jiang et al., 2016; Song et al., 2017; Li Z.-T. et al., 2020). In a few cases, upregulation of miRNAs regulating the negative regulators of FZD proteins can also contribute toward the severity of cancer. For example, miR-106b is upregulated in breast cancer which suppresses BRMS1L. BRMS1L is responsible for regulating the expression of FZD10. It recruits HDAC1 to FZD10 which leads to promoter H3K9Ac (Gong et al., 2014). miR-130-30p, which is downregulated in triple negative breast cancer, acts as a Wnt antagonist by targeting multiple Wnt components, including FZD6 and LRP6 (Poodineh et al., 2020). Another lncRNA, GATA4-AS1 inhibits Wnt signaling in gastric cancer by reducing FZD4 expression. It recruits EZH2 and increases H3K27me3 occupancy at the promoter region (Ma et al., 2017; Li Z.-T. et al., 2020). LRP6 is also under the regulation of multiple miRNAs which are downregulated in colorectal cancer and breast cancer (Kim et al., 2011; Hata et al., 2017; Poodineh et al., 2020). MEST (mesoderm specific transcript) is a known regulator of LRP6 (Ruan et al., 2020). It blocks LRP6 maturation via glycosylation. Inhibition of MEST, therefore, enhances Wnt signaling (Ruan et al., 2020). MEST itself is targeted by miR-181 in ovarian cancer cells thereby promoting Wnt signaling. However, in another study, MEST was shown to be a positive regulator of the Wnt pathway. It was found to be under the regulation of zinc finger protein ZFP57, a transcription factor involved in maintaining DNA methylation in ESCs via its interaction with DNMTs. Promoter hypermethylation at MEST locus by ZFP57 was shown to be lost in breast cancer which led to enhanced Wnt signaling (Chen L. et al., 2019). Upregulation of the upstream positive regulators for LGR5 have been reported to serve toward cancer progression. For instance, loss of miR-363 promotes GATA6 expression and loss of miR-34 promotes ZNF281 expression, both of which are required for the expression of LGR5 in colorectal cancer (Hahn et al., 2013; Tsuji et al., 2014). However, promoter hypermethylation in case of LGR5 suggests a tumor suppressive role instead. Interestingly, the degree of methylation correlated with the grade of tumor (Schuebel et al., 2005; Felipe de Sousa et al., 2011; Su et al., 2015).
Destruction Complex Components
Adenomatous polyposis coli is the most frequently mutated gene in Wnt driven cancer cases, especially in the case of familial adenomatous polyposis. APC is also downregulated due to promoter hypermethylation, which can occur with or without mutations in the gene itself (Su et al., 1992, 1993; Arnold et al., 2004; Segditsas et al., 2008; Liang et al., 2017). It is also regulated by multiple miR-RNAs, such as miR-135 in colorectal cancer and miR-142 in breast cancer (Nagel et al., 2008; Isobe et al., 2014). miR-135b suppresses APC and is itself part of a positive feedback loop with TAZ (Shen S. et al., 2017). AXIN2 is often induced after Wnt activation and is a part of a negative feedback loop. It limits the duration of Wnt signaling. It is primarily found to be expressed in colon cells (Lustig et al., 2002). AXIN2 repression helps in prolonging the duration of Wnt signaling by blocking the negative feedback loop as well as increasing the duration of Wnt target genes expression (Lustig et al., 2002). Therefore, AXIN2 is frequently targeted in colorectal cancer by promoter hypermethylation (Koinuma et al., 2006). Interestingly, AXIN2 is also downregulated by miR-34 in colorectal cancer which should inhibit Wnt signaling but miR-34 targets a number of Wnt activators as well, thus limiting the action of AXIN2 (Kim et al., 2013). Recently, miR-103/107 were also shown to promote colorectal cancer stemness by targeting AXIN2. miR-103/107 also promotes colorectal cancer by targeting LATS2. AXIN2 overexpression also led to nuclear accumulation of GSK3-β which would ultimately promote Wnt signaling (Kim et al., 2013; Chen H.-Y. et al., 2019). As expected, GSK3-β, βTrCP and CK1α, are downregulated in Wnt driven cancers, thereby promoting β-catenin stabilization. Table 2 summarizes the miRNAs reported to be responsible for negative regulation of these proteins in multiple cancers (Zhang et al., 2012; Li T. et al., 2016; Tan et al., 2016; Wang S. et al., 2016; Jin et al., 2017). DVL1, 2, and 3 are under the positive regulation by SIRT1 (a member of histone deacetylase family) in case of breast cancer (Holloway et al., 2010; Simmons et al., 2014). DACT (Disheveled Binding Antagonist of β-catenin) family of proteins are known negative regulators of Wnt signaling. DACT3 undergoes transcriptional repression due to the deposition of bivalent histone modifications H3K27me3 (repressive) and H3K4me3 (activatory) (Jiang et al., 2008).
Due to the dual role of YAP/TAZ in regulating the Wnt signaling pathway, there have been contrasting reports showing both tumor suppressive as well as oncogenic roles of YAP and TAZ. For example, miR-506, which targets YAP, is downregulated in colorectal cancer but upregulated in breast cancer (Hua et al., 2015; Krawczyk et al., 2017). The YAP promoter also shows aberrant hypermethylation in breast cancer (Real et al., 2018). However, the majority of studies point toward an oncogenic role of YAP and TAZ. Some of these have been listed in Table 2 (Shen et al., 2018; Yang M. et al., 2019; Yang et al., 2021). Hippo kinases and their upstream regulators are not a part of the Wnt signaling but it is important to mention them since these regulate the nuclear levels of YAP/TAZ and as a result, β-catenin. LATS has been reported as a tumor suppressor in colorectal cancer, head and neck squamous cell carcinoma, breast cancer and renal cell carcinoma. The promoter of LATS1/2 undergoes hypermethylation (Okami et al., 2005; Steinmann et al., 2009; Chen et al., 2013; Chen K.-H. et al., 2014; Wierzbicki et al., 2013). Several miRNAs have also been reported to control the levels of LATS1/2 (Lee et al., 2009). Promoter hypermethylation has also been reported for MST1/2 (Steinmann et al., 2009; Maille et al., 2019). MORC2 (microrchidia) acts as a negative regulator of Hippo signaling. MORC proteins are epigenetic readers as well as chromatin remodelers which help in DNA methylation (Pastor et al., 2014; Li S. et al., 2016). It forms a complex with DNMT3A and causes hypermethylation at the promoter regions of NF2 and KIBRA which are upstream regulators of the Hippo pathway. Depletion of NF2 and KIBRA in turn increases the nuclear levels of YAP/TAZ and promotes stemness and oncogenicity in hepatocellular carcinoma (Wang T. et al., 2018).
Transcriptional Regulators
Promoter hypermethylation at CTNNB1 gene is not observed frequently in colorectal cancer (Galamb et al., 2016). miR-34, which itself is a target of p53, regulates β-catenin. Loss of p53 in colorectal cancer and breast cancer in turn leads to upregulation of β-catenin (Kim et al., 2011). Some miRNAs responsible for regulating β-catenin levels are mentioned in Table 2 (Tian et al., 2014; Abedi et al., 2015). Interestingly, miR-520f-3p attenuates nuclear localization of β-catenin by targeting degradation of SOX9 mRNA in gastric cancer (Chen et al., 2020). JMJD1A, a histone demethylase, is responsible for upregulation of β-catenin expression in colorectal cancer. JMJD1A can also directly interact with β-catenin leading to its transactivation and upregulation of expression of multiple Wnt target genes, such as MYC, CCND1 and MMP9 (Peng et al., 2018). TCF7L1 and TCF7L2 promoters have hypomethylation in colorectal cancer (Guo et al., 2015). Several miRNAs regulating various members of the TCF/LEF family are downregulated in cancers (Table 2; Kim et al., 2011; Subramanian et al., 2014; Wang O. et al., 2018). Interestingly, the LEF1 gene has two promoters, P1 and P2. P1 is under the regulation of Wnt signaling, therefore upregulation of Wnt leads to increased expression of LEF1. Transcription from P2, on the other hand, produces a dominant negative form of LEF1. Transcription from P2 promoter is lost in case of breast cancer due to the repressive mark H3K9me3 mediated by YY1 (Yokoyama et al., 2010). Chromatin modification-induced alterations in Groucho expression are not frequent. However, downregulation of Groucho has been reported in colorectal cancer via the action of HDAC3 (Godman et al., 2008).
Epigenetic Regulation Downstream of Wnt Signaling
Recent advances in cancer biology have identified the role of epigenetic regulation in cancers and numerous studies have identified mutations in the genes encoding both, the components and the regulators of Wnt signaling to aberrantly activate the Wnt signaling pathway in various cancers. In this section, we will elaborate upon the epigenetic modulations introduced by the Wnt pathway that promote cancer initiation as well as progression.
DNA Methylation
Multiple pathways and molecular players influence the outcome of the Wnt pathway. A study by Song et al. (2015) has shown that β-catenin interacts with DNMT1 and their interaction is required for mutual stabilization. Further, β-catenin complex with DNMT1 is a key requirement for DNMT1 dependent promoter methylation and Wnt/β-catenin signaling dependent target gene expression (Song et al., 2015). In another study, tumor suppressor- Na+/H+ exchanger regulatory factor 1 (NHERF1/EBP50), an adaptor molecule known to suppress Wnt signaling has been observed to be downregulated in colon cancer cells and is associated with decreased survival and increased intestinal tumor burden (Georgescu et al., 2016). Promoter methylation by DNMT1 has been implicated in reducing the expression of NHERF1/EBP50. Furthermore, Wnt signaling dependent upregulation of DNMT1 has been shown to be required to trigger hypermethylation of NHERF1 promoter in colon cancer (Guo et al., 2018). Mechanistically, whether β-catenin recruits DNMT1 differentially on tumor suppressor genes and oncogenes, and what other molecular partners may be required to decide DNA methylation dependent and independent roles has not been fully explored. A recent study adds another layer of complexity by the Wnt signaling via crosstalk with various chromatin modifiers. The study shows that EZH2 dependent protein stability of LSD1, HDAC1, DNMT1, β-catenin, or SMAD2/4, via recruitment of deubiquitinase USP7, is key in suppressing neuronal genes and sustaining the expression of Wnt and TGFβ target genes in cancer cells (Lei et al., 2019). Understanding the role of β-catenin in DNMT1 regulation and recruitment would be crucial in developing future therapeutics.
Histone Modifications
The Wnt pathway employs diverse factors to regulate target gene expression and converges with epigenetic signaling at Wnt response elements through recruitment of epigenetic modulators. Upon Wnt stimulation, β-catenin translocates to the nucleus and interacts with the TCF family proteins by replacing co-repressors with subsequent recruitment of CBP/p300 HATs, thereby promoting H3 and H4 lysine acetylation. This reverses the HDAC-dependent chromatin compaction and induces molecular events to promote Wnt target gene expression (Parker et al., 2008). Although this study has emphasized on the role of β-catenin as an transcriptional activator, dependent on the recruitment of CBP/p300 HATs, it has been demonstrated that CBP/p300 can act as a bimodal regulator, regulating TCF/β-catenin interaction and also facilitating transactivation of β-catenin (Li et al., 2007). The ability of β-catenin to recruit multiple factors on promoters and the regulatory role of Wnt/β-catenin signaling in determining heterogeneity at the cellular level suggest that β-catenin could facilitate a poised chromatin state during developmental programs. Furthermore, information on how β-catenin regulated gene loci are selected as poised chromatin loci during the unfolding of developmental programs is still incomplete. Blythe et al. (2010) found that in Xenopus, prior to the midblastula transition during embryogenesis, Wnt/β-catenin signaling uncouples the activation of dorsal specific genes by establishing a poised chromatin state through recruitment of arginine methyltransferase PRMT2 at target gene loci. The PRMT2 catalyzes histone H3 arginine 8 (H3R8me2) asymmetric methylation. The advantage of such a poised nature of chromatin is to enable synchronous and rapid activation of tissue specific genes (Blythe et al., 2010). Another study showed that the C-terminal transactivation of β-catenin domain interacts with TRIPP/TIP60 and mixed-lineage leukemia (MLLI/MLL2) SET1-chromatin modifying complexes and recruits H3K4me3 at the promoters of target genes (Sierra et al., 2006; Zhu et al., 2019). These studies provide a mechanistic clue that during embryogenesis, pattern and combinations of these modifications could serve as critical molecular cues deciding tissue specific gene expression. Also, PRMT2-dependent poised state during early developmental stages could subsequently be followed by MLL1/MML2 driven gene activation in later stages. A recent study has shown that the dual pattern of histone modifications dependent on Spindlin1, a multivalent epigenetic reader, which potentiates Wnt/β-catenin signaling by recognizing the dual histone modification pattern ‘H3K4me3-H3R8me2’ (Su et al., 2014). Recently, Spindlin1 has been shown to drive the growth of colorectal carcinoma which have high levels of β-catenin. Spindlin1 is expressed in Lgr5+ intestinal stem cells and is required for self-renewal. It also promotes cancer stemness in a β-catenin-dependent manner (Su et al., 2014; Grinat et al., 2020). Previously, a report had shown that the MLL1/CBP/β-catenin complex promotes tumor initiation and metastasis in the head and neck squamous cell carcinoma by increasing the activation associated mark H3K4me3 at the promoters of target genes (Qiang et al., 2016). Interestingly, SETD7, a histone lysine methyltransferase, has been shown to be the part of the destruction complex itself. SETD7 mediated methylation of YAP aids in Wnt induced nuclear localization of β-catenin and thus is essential for intestinal regeneration and also plays a role in promoting tumorigenesis (Oudhoff et al., 2016). The studies demonstrating crosstalk of β-catenin transactivation with the epigenetic code provides new avenues to be tested in future for therapeutic intervention.
Wnt signaling and burst of chromatin reorganization
The genetic information on DNA is organized into the structural and regulatory framework of chromatin thereby allowing different machineries and signaling networks to switch off or switch on the gene expression. The major component of eukaryotic chromatin is the nucleosome comprising of 146 bp DNA wrapped around an octamer of core histones H2A, H2B, H3, and H4. The different machineries of transcription gain access to this complex structural framework by posttranslational modification of histones (Shilatifard, 2006; Berger, 2007; Fu et al., 2016). Depending upon the type of modifications and recruitment of modifiers, mediators and suppressors, gene activation or silencing is promoted (Kouzarides, 2007).
Wnt signaling drives molecular changes through the nuclear function of a prominent member of the ARM repeats containing protein, β-catenin. The central ARM repeats bound to TCF are critical for its function and their deletion completely abrogates β-catenin function (Xing et al., 2008). In the absence of nuclear β-catenin, TCF is bound to the co-repressor complex to stimulate compression of chromatin and suppress Wnt target gene expression (Cavallo et al., 1998; Brantjes et al., 2001; Courey and Jia, 2001). The N-terminal and C-terminal regions of β-catenin are transactivation domains known to interact with multiple cofactors to regulate Wnt target gene expression. The C-terminal domain (CTD) of β-catenin interacts with histone acetyltransferases (CBP or p300) resulting in chromatin modifications promoting the activation of Wnt target genes (Hecht, 2000; Takemaru and Moon, 2000; Courey and Jia, 2001). Upon Wnt stimulation, in CBP-dependent manner, nucleosomes are acetylated up to 30 kilobases with enhanced rate and saturated within 5.5 h, inducing a widespread wave of chromatin acetylation and re-organization (Figure 4A; Parker et al., 2008). The widespread use of acetylation and rapid activation of target genes is suggested to be dependent on the nature of TCF binding. TCF belongs to the high mobility group of proteins which are known to induce strong DNA bending upon binding. TCF induced bending facilitates the architectural framework for large chromatin regions to be organized together to stimulate a burst of chromatin reorganization dependent on β-catenin bound HAT (CBP) activity (Heintzman et al., 2007; Parker et al., 2008). This architectural framework of chromatin by the TCF/β-catenin/HAT complex provides a robust mechanism for rapid activation of Wnt target genes. The β-catenin CTD acts as a scaffold to recruit multiple diverse factors, such as, the SWI/SNF factors BRG1 and ISWI, HMTs, the Mediator component MED12, and the polymerase-associated factor 1 PAF1 (Waltzer et al., 2001; Kim et al., 2006; Mosimann et al., 2006; Sierra et al., 2006). These factors rearrange histone positions, modify the histones post-translationally and orient RNA Pol II, thereby providing the sequential exchange platform for remodeling chromatin architecture to induce rapid gene expression (Figure 4C; Mosimann et al., 2009). β-catenin has been shown to pre-pattern chromatin signatures during differentiation in an OCT4-dependent manner (Ying et al., 2015). Wnt signaling driven β-catenin acts as a molecular switch for the chromatin-associated high mobility group protein (HMG-17) to de-repress the inhibitory complex HMG-17/PITX2. The inhibitory complex HMG-17/PITX2 binds to specific chromatin regions primed for transcriptional activation and β-catenin forms a ternary complex with HMG-17/PITX2, thereby switching the inhibitory complex, modulating chromatin structure and inducing spatiotemporal expression of genes during embryogenesis (Amen et al., 2008; Ying et al., 2015). The histone methyltransferase and demethylase work in concert to stringently regulate gene expression. Protein lysine demethylases (KDMs) modify chromatin by demethylation of histones, especially repressive mark histone H3 lysine 9 trimethylation (Shi, 2007). The transactivation potential of β-catenin is very crucial for its nuclear function to target gene expression. Several studies have shown that β-catenin interacts with KDMs to epigenetically modulate target gene expression. For instance, β-catenin has been shown to interact with KDM3 on Wnt target genes inducing histone H3 lysine 9 (H3K9me2) demethylations. The β-catenin/KDM3 complex further drives MLL1-dependent H3K4 methylation to promote recruitment of BCL9 and Pygo2 to chromatin, thus promoting Wnt/β-catenin-dependent target gene expression (Li et al., 2019). In another study, KDM4 has been shown to physically interact with β-catenin and demethylated H3K9me3 on the promoters of Wnt target genes to promote target gene expression (Figure 4B; Peng et al., 2019). The complex nature of the cellular context requires multiple factors to work in concert for precise activation of target genes. For example, β-catenin recruits BRG1-dependent KDM4 isoforms on the promoters of Disintegrin and Metalloproteinase (ADAM) to regulate their expression in colorectal cancer cells (Sun et al., 2020). Thus, these findings establish that β-catenin recruits KDM to remove repressor marks and facilitate concomitant recruitment of activator complexes. However, the context-dependent and temporal nature of these β-catenin driven modulations require further investigation. The Wnt signaling-dependent activation of β-catenin and nuclear interaction with several transcription factors and chromatin acts as a molecular switch to regulate global gene activation. For example, β-catenin interacts with the chromatin organizer special AT-rich binding protein 1 (SATB1) and mediates the expression of specific set of genes during T helper-cell differentiation (Notani et al., 2010) and colorectal cancer progression (Mir et al., 2016), presumably via alterations in chromatin domain organization (Galande et al., 2007). The precise spatiotemporal activation of gene expression during development is a critical event dependent on chromatin organization and Wnt signaling directs the chromatin modifying machinery to establish a poised state at promoters (Shahbazian and Grunstein, 2007; Blythe et al., 2010). Wnt signaling has been shown to regulate differential developmental outcomes; however, it is not clear which mechanism governs this. Recently, Esmaeili et al. (2020) established that loss of chromatin architecture at Wnt target gene promoters is responsible for differential outcome of β-catenin bound chromatin without changing the inductive signal. The studies so far have established that Wnt signaling driven β-catenin modulates multiple aspects of gene regulation by acting as a signaling switch to regulate diverse cellular processes during development and tissue homeostasis.
FIGURE 4.
Epigenetic machinery downstream to the Wnt signaling pathway. (A) β-catenin can form complexes with different epigenetic modulators and activate gene expression upon Wnt activation. CBP is a well-known interaction partner of β-catenin which helps in opening up of the chromatin by catalyzing H3K9 acetylation. (B) Histone demethylase KDM4 is recruited via BRG1, an interactor of β-catenin which stimulates the expression of the ADAM group of genes. (C) β-catenin can also form an elaborate complex with the chromatin remodelers composed of the MLL complex, Pygo and Bcl-9 leading to H3K4me3 modification. This activity requires prior ubiquitination of H2BK120.
Non-coding RNAs
Wnt signaling and microRNAs play essential roles in embryonic development and tissue homeostasis. MicroRNAs are involved in tumorigenic pathways. They play differential roles either regulating oncogenes or tumor suppressors to promote cancer progression (Xu and Mo, 2012). A recent study by Roman et al., has shown in cell-based experiments that microRNAs can modulate Wnt signaling by either regulating positive regulators or negative regulators of the Wnt pathway (Anton et al., 2011). Aberrant activation of Wnt signaling in various cancers regulate a wide range of gene expression at transcript level including microRNAs (Table 3). Oncogenic miRs-miR-182, miR30e, miR122, and miR21 are directly regulated by Wnt signaling in tumorigenesis (Liao and Lönnerdal, 2010; Lan et al., 2012; Chiang et al., 2013). In another study, miR215 and miR137 are repressed and miR708, miR135b, and miR3 are upregulated in APC mutant tumors (Necela et al., 2011). In gastric cancers, the expression of miR4739, miR210, miR135-5P, and miR12334-3P are significantly increased and the expression of miR20a-3p, miR-23b-5p, miR335-3p, miR423-5p, and miR455-3p is significantly reduced upon knockdown of β-catenin (Wang et al., 2009; Dong et al., 2015). In hepatocellular carcinoma, Wnt/β-catenin signaling upregulates miR770, miR183, miR96, and miR182 (Leung et al., 2015; Wu et al., 2016) and downregulates miR375 (Ladeiro et al., 2008). In breast cancer, miR182 and miR125b are upregulated by Wnt signaling and miR Let-7 is downregulated (Cai et al., 2013; Chiang et al., 2013; Liu et al., 2013). Aberrant activation of Wnt/β-catenin and microRNAs also involve autoregulatory feedback loop in cancers, for example, miR218 expression is induced by Wnt signaling and in turn, it augments the Wnt signaling pathway by targeting Wnt signaling inhibitors (SOST, DKK2, and sFRP2) (Hassan et al., 2012). Similarly, miR146a expression is induced by Wnt signaling in CRC, and in turn, it hyperactivates Wnt signaling by reducing the expression of Numb (Hwang et al., 2014). In summary, crosstalk between Wnt/β-catenin and miRs is crucial for the development of cancers and further understanding of this intricate signaling paradigm will provide insights for future therapeutics.
TABLE 3.
miRNAs and lncRNAs regulated by Wnt pathway in various cancer types.
| miRNAs | |||
| Cancer type | Upregulated | Downregulated | References |
| CRC | miR-708, miR-31, miR-135b, miR-21, miR-182, miR574-3p, miR-30e | miR-215, miR-137 | Liao and Lönnerdal, 2010; Lan et al., 2012; Chiang et al., 2013; Necela et al., 2011 |
| GC | miR-20a-3p, miR-23b-5p, miR-335-3p, miR-423-5p, miR-455-3p | miR-1234-3p, miR-135b-5p, miR-210, miR4739, miR-122a | Wang et al., 2009; Dong et al., 2015 |
| HCC | miR-770, miR-183, miR-96, miR-182 | miR-375 | Ladeiro et al., 2008; Leung et al., 2015; Wu et al., 2016 |
| BC | miR-182, miR-125b | Let-7 | Cai et al., 2013; Chiang et al., 2013; Liu et al., 2013 |
| IncRNAs | |||
| Cancer Type | IncRNAs | Outcome | References |
| CRC | KIAA0125 | Downregulated | Yang Y. et al., 2019 |
| CC | CALML3-AS1 | Upregulated | Liu et al., 2019 |
| BC | LUCAT1 | Upregulated | Zheng et al., 2019 |
| LC | LINC00673-v4 | Upregulated | Lin et al., 2020 |
| LC | AK126698 | Downregulated | Fu et al., 2016 |
| Pituitary adenoma | CLRN1-AS1 | Downregulated | Wang et al., 2019 |
| Glioma | SNHG17, MIR22HG, HOXC13-AS, LSINCT5, H19, BLACAT1, SNHG5, LINC01503, AGAP2-AS1, OIP5-AS1, DANCR, SNHG7, NEAT1, AB073614, MIR155HG, CCND2-AS1, CCAT2 and MALAT1 | Upregulated | Han et al., 2020 |
| Glioma | CASC7, Linc00320, TUNAR, MEG3, CASC2 and PTCSC3 | Downregulated | Han et al., 2020 |
| Colon cancer | CCAT2 | Upregulated | Ling et al., 2013 |
| LC | LINC00673-v4 | Upregulated | Guan et al., 2019 |
| Liver cancer | TCF7 | Upregulated | Wang et al., 2015 |
CRC, colorectal cancer; CLL, chronic lymphocytic leukemia; BC, breast cancer; HCC, hepatocellular carcinoma; OC, ovarian cancer; CC, cervical cancer; GC, gastric cancer; HNSCC, head and neck squamous cell carcinoma; RCC, renal cell carcinoma; LC, lung cancer; PC, pancreatic cancer; OS, osteosarcoma.
Numerous studies have shed light on the Wnt signaling-dependent role of lncRNAs in diverse cancers (Table 3) such as colorectal cancer (Yang Y. et al., 2019), cervical cancer (Liu et al., 2019; Yang Y. et al., 2019), breast cancer (Zheng et al., 2019), non-small cell lung cancer (Lin et al., 2020), and brain (Wang et al., 2019). Multiple mechanisms are operational via long non-coding RNAs to differentially regulate Wnt signaling. For instance, a gene desert located upstream of the MYC gene is known to express Wnt related lncRNAs such as CCAT1-L, CCAT1-S, CCAT2, and CASC11 (Shen P. et al., 2017). CCAT2 (Colon cancer associated transcript 2) is known to enhance Wnt/β-catenin signaling activity by binding to TCF and increasing its transcriptional activity subsequently activating the cascade of downstream gene expression crucial for cancer progression and invasion (Ling et al., 2013; Shen P. et al., 2017). lncRNA LINC00673-v4 enhances TCF/LEF activity and increases the binding of DEAD-box helicase 3 X-linked (DDX3) and casein kinase 1 (CK1), thereby potentiating Wnt signaling (Guan et al., 2019). Apart from acting as transcription factors, lncRNAs are also shown to interact with chromatin modifying enzymes to promote spatiotemporal expression of target genes (Marchese and Huarte, 2014; Guan et al., 2019). For instance, the lncRNA 34a regulates miR-34a by recruiting repressor epigenetic machinery DNMT3a/PHB2 and HDAC1 at the promoter of miR-34a, thereby inducing DNA methylation and histone deacetylation to repress its expression (Wang L. et al., 2016). miR-34a is a key component in the Wnt signaling pathway and it regulates TCF7 expression (Chen W.-Y. et al., 2015).
In summary, lncRNAs act as regulatory switches to modulate the signaling outcome by targeting various crucial components of the pathway. The lncRNA-Wnt signaling module employs several mechanisms to regulate the expression of Wnt target genes. LncRNAs provide signaling scaffolds and act as transcription factors, chromatin co-modifiers and mediators. The significant involvement of lncRNAs in Wnt signaling driven cancers makes them attractive targets for drug discovery and cancer therapy.
Therapeutics
Epigenetic regulation plays a dynamic role in Wnt signaling driven cancers. Epigenetic alterations are reversible by using pharmacological inhibitors. Multiple approaches have been employed for epigenetic therapy in cancers. These include DNA methylating inhibitors, histone methyltransferase inhibitors and HDAC inhibitors. To reverse aberrant DNA methylation acquired in cancers, DNMT inhibitors azacitidine and decitabine were developed and approved for acute myeloid leukemia (AML) (Derissen et al., 2013). Aberrant methylation of histone tails is linked with gene silencing in cancers. Several methyltransferases such as EZH2, SETD2 and Dot1L have been implicated in cancers (Albert and Helin, 2010; Hamamoto and Nakamura, 2016). Currently, tazemetostat is the most advanced therapeutic drug against EZH2 whereas valemetostat, CPI-1205, and CPI-0209 are in clinical phase 2 trials (Gulati et al., 2018). Inhibition of EZH2 reverses the silencing of genes induced by EZH2 hyperactivation. Another tested mode of epigenetic therapy is the reversal of histone deacetylation promoted by HDACs. Four HDAC inhibitors- vorinostat, romidepsin, belinostat, and panobinostat have been approved by the FDA (Bates, 2020). Inhibition of HDACs causes concomitant increase in HAT activity resulting in global increase in acetylation, subsequently relaxing chromatin and inducing transcription of genes responsible for promoting cell death (Zhao et al., 2020). The mechanism of HDAC inhibitors inducing cell death is not clearly understood and needs to be addressed in future to have a deeper understanding of their mechanism of action for therapeutic use. The epigenetic therapies have been shown to reduce cell proliferation, induce cell death in hematological malignancies (Altucci and Minucci, 2009), but they have not been seen to be effective in solid tumors (Graham et al., 2009). The HDAC inhibitors are mostly non-isoform selective and with recent advances in drug discovery, isoform specific and dual capability HDAC inhibitors have been extensively developed for cancer treatment (Peng et al., 2020). Current knowledge and complexity of how HDAC inhibitors work in the cellular context and combination therapies targeting the epigenome could be useful for designing future therapeutic interventions.
Targeting the Wnt pathway is an attractive model for therapeutics and cancer therapy. Aberrant activation of Wnt signaling is associated with poor cancer outcomes and recent strategies involve targeting the pathway in diverse cancers (Figure 5). Inhibiting the stimulation of the Wnt pathway by using monoclonal antibodies against Wnt1 has been tested in many cancer cells and has resulted in reduction of downstream components and apoptosis (He et al., 2004; Esmaeili et al., 2020). Similarly, an anti-FZD antibody (Vantictumab) has been used to target the frizzled receptors and shown to reduce the tumor burden. This antibody was in clinical trials with patients of breast cancer and pancreatic cancer but eventually discontinued because of bone toxicity observed in these patients (Gurney et al., 2012; Smith et al., 2013). Ipafricept, a recombinant fusion protein consisting of FZD8 extracellular domain and human IgG1 FC fragment blocks Wnt ligands and xenograft assays have shown efficacy of this drug (Wall and Arend, 2020). It is in the first phase of clinical trials and is administered after chemotherapeutic agents (Fischer et al., 2017). Porcupine inhibitors, such as WNT974, inhibit Wnt secretion and are effective in reducing tumor growth. Porcupine catalyzes palmitoylation on Wnts required for their secretion. Currently, WNT974 is in phase 1 clinical trial (Koo et al., 2015; Agarwal et al., 2017). Another group of inhibitors such as IWR-1 and XAV939 target the stability of the β-catenin destruction complex, including inhibitors of tankyrase 1 and tankyrase 2; these induce AXIN protein stability. These inhibitors reduce Wnt signaling, EMT and angiogenesis (Arqués et al., 2016; Shetti et al., 2019).
FIGURE 5.
Chemotherapeutic interventions targeting the components of the Wnt signaling pathway. Drugs leading to the activation of the Wnt response inhibitors and repression of the Wnt response activators are being used as therapeutic agents for cancer therapy (or are under clinical trials). Wnt1 monoclonal antibody (MAb), Vantictumab and Ipafricept target the upstream regulators of the Wnt pathway. WNT974 blocks PORCN which prevents processing of Wnt ligands. IWR-1 and XAV939 act by activating AXIN, thereby stabilizing the Wnt destruction complex. PRI-724 acts by inhibiting the interaction of β-catenin with CBP/p300. PKF115-584, CGPO49090, PKF222-815, iCRT3, iCRT5, and iCRT14 act by inhibiting interaction of β-catenin with TCF/LEF.
The downstream signaling network of Wnt signaling pathway involves β-catenin nuclear signaling through interaction with various nuclear factors to drive target gene expression and tumorigenic phenotypic changes. Several attempts have been made to target this β-catenin and TCF interface to modulate target gene expression and induce regression of tumors. For instance, PKF115-584, CGPO49090, and PKF222-815 are fungal derived drugs and inhibit β-catenin’s interaction with TCF. These are currently in phase I and phase II of clinical trials (Lepourcelet et al., 2004). High throughput screening has identified three inhibitors, β-catenin responsive transcription inhibitor 3 (iCRT3), iCRT5, and iCRT14 specifically known to inhibit β-catenin and TCF interaction (Lepourcelet et al., 2004; Gonsalves et al., 2011). But clinical trials for these are not yet complete and potential side effects remain unknown.
The interaction of β-catenin with various co-factors is crucial for potentiating the Wnt pathway driven gene expression and associated cellular consequences, especially when the pathway is hyperactivated, has also been exploited to design drugs for cancer treatment. For instance, PRI-724 disrupts the interaction between β-catenin and its co-transcriptional activator CBP (Ko et al., 2016). Several natural compounds have also been used to treat Wnt signaling driven cancers such as resveratrol, curcumin, and Genistein (Cheng et al., 2019). The combinatorial use of epigenome modulating drugs along with conventional chemotherapeutic drugs will be required to treat epigenomic alteration driven cancers.
Discussion
Summary
In this review, we have provided an overview of the regulation of the Wnt signaling pathway in various cancer types, with a special emphasis on the crosstalk with the epigenetic machinery. Apart from undergoing alterations at the genetic level, the Wnt signaling pathway components are dysregulated due to various ncRNAs and chromatin modifiers. In turn, the Wnt effector β-catenin is also involved in various epigenetic processes due to its interaction with a multitude of epigenetic players. We have also provided an overview of the signaling pathways that work in conjunction or restrict the activity of the Wnt signaling pathway.
Therapeutic Potential
Aberrant Wnt pathway activation seems to be the root cause for the development and progression of multiple cancers. However, even four decades after the discovery of the Wnt signaling, therapeutic intervention of this key pathway remains far from achieving its potential. Several factors seem to be responsible such as: the pathway is intricate and regulated by multiple combinations of Wnt ligands and their receptors, crosstalk of canonical and non-canonical pathways, and crosstalk with other signaling pathways. Further, multiple mutations of the Wnt signaling components cause constitutive activation of β-catenin, making it difficult to target the upstream players. Additionally, the physiological roles of the Wnt/β-catenin signaling are important for normal cellular functions. It is the combination of various signaling pathways that results in an imbalance in the rate of activation. Hence, future therapeutics will be aimed at optimizing the rate of activation and directing it toward maintaining the cellular homeostasis.
Using certain synthetic biochemical tags that could regulate levels of crucial components of the Wnt signaling pathway will be useful to develop future therapeutic intervention. Proteolysis-targeting chimeras (PROTACs) have emerged as an efficient technology to degrade proteins of interest in vivo (Sakamoto et al., 2001). One such strategy was employed recently using PROTAC chemistry to degrade β-catenin in Wnt signaling driven colorectal cancers. Membrane permeable PROTACs were designed to tether β-catenin with the ubiquitin system to induce controlled degradation of β-catenin to regress the tumor burden in Apc mutant mice (Liao et al., 2020). Similarly, PROTACs using biochemical inhibitors were designed to tag SGK3 kinase with VHL ubiquitin ligases to facilitate selective and efficient degradation of SGK3 in breast cancers compared to conventional inhibitors because of their sub-stoichiometric catalytic mode of action (Tovell et al., 2019). Recently characterized roles of the lncRNAs and chromatin in Wnt signaling regulation make them an attractive target for therapeutics in the future. Using a combination of PROTAC chemistry for crucial components of the Wnt pathway and chromatin, and nanoparticle based targeting of non-coding RNAs could enhance the chances of better therapies in the future. The precise and balanced stimulation of Wnt/β-catenin signaling governs cellular outcomes, tissue homeostasis and developmental paradigms and disruption of this balance may lead to a multitude of processes either in their favor or disfavor. Therefore, future therapeutics will depend on understanding this complexity employing multiple ways to curb the signaling imbalance.
Future Perspectives: Open Questions
Wnt signaling crosstalks with several signaling and regulatory networks including epigenetic networks, mechano-sensing and environmental cues to modulate the expression of target genes crucial for developmental paradigms and tumorigenesis. Hence, holistic understanding of the molecular changes acquired during the initiation of Wnt signaling driven cancers is critical for disease etiology and future therapeutics. Identifying the pioneering molecular cues will be useful for developing better cancer therapies. The role of nuclear factors driving the Wnt/β-catenin signaling response raises hope of using drugs targeting these effectors. Obtaining a better picture of protein-protein interaction complexes regulating Wnt/β-catenin signaling will be helpful in identifying future therapeutic targets. Understanding the comprehensive picture of nuclear regulators and/or missing partner(s) crucial for convergence with other pathways will be essential for the treatment of Wnt signaling related diseases.
Author Contributions
All the authors contributed to design the concept and structure of the review article, and approved the submitted version. AS and RM collected and analyzed the literature, and wrote bulk of the manuscript. AS prepared the figures. RM and SG provided critical inputs as the corresponding authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Sudiksha Mishra for help with the bibliography.
Footnotes
Funding. SG is a recipient of the J. C. Bose National Fellowship (JCB/2019/000013) from the Scientific and Engineering Research Board, Government of India. RM is a recipient of the Ramalingaswami Fellowship (Grant No. BT/RLF/RE-ENTRY/29/2018) from the Department of Biotechnology, Government of India. AS is supported by a Senior Research Fellowship from the University Grants Commission, Government of India.
References
- Abedi N., Mohammadi-Yeganeh S., Koochaki A., Karami F., Paryan M. (2015). miR-141 as potential suppressor of β-catenin in breast cancer. Tumour Biol. 36 9895–9901. 10.1007/s13277-015-3738-y [DOI] [PubMed] [Google Scholar]
- Agarwal P., Zhang B., Ho Y., Cook A., Li L., Mikhail F. M., et al. (2017). Enhanced targeting of CML stem and progenitor cells by inhibition of porcupine acyltransferase in combination with TKI. Blood 129 1008–1020. 10.1182/blood-2016-05-714089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai L., Tao Q., Zhong S., Robert Fields C., Kim W.-J., Lee M. W., et al. (2006). Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis 27 1341–1348. 10.1093/carcin/bgi379 [DOI] [PubMed] [Google Scholar]
- Albert M., Helin K. (2010). Histone methyltransferases in cancer. Semin. Cell Dev. Biol. 21 209–220. [DOI] [PubMed] [Google Scholar]
- Albini S., Zakharova V., Ait-Si-Ali S. (2019). “Chapter 3 – histone modifications,” in Epigenetics and Regeneration, ed. Palacios D. (Cambridge, MA: Academic Press; ), 47–72. [Google Scholar]
- Albuquerque C., Breukel C., van der Luijt R., Fidalgo P., Lage P., Slors F. J. M., et al. (2002). The “just-right” signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum. Mol. Genet. 11 1549–1560. 10.1093/hmg/11.13.1549 [DOI] [PubMed] [Google Scholar]
- Allis C. D., Jenuwein T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Altucci L., Minucci S. (2009). Epigenetic therapies in haematological malignancies: searching for true targets. Eur. J. Cancer 45 1137–1145. 10.1016/j.ejca.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Amen M., Espinoza H. M., Cox C., Liang X., Wang J., Link T. M. E., et al. (2008). Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/beta-catenin signaling. Nucleic Acids Res. 36 462–476. 10.1093/nar/gkm1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton R., Chatterjee S. S., Simundza J., Cowin P., Dasgupta R. (2011). A systematic screen for micro-RNAs regulating the canonical Wnt pathway. PLoS One 6:e26257. 10.1371/journal.pone.0026257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C. N., Goel A., Niedzwiecki D., Dowell J. M., Wasserman L., Compton C., et al. (2004). APC promoter hypermethylation contributes to the loss of APC expression in colorectal cancers with allelic loss on 5q1. Cancer Biol. Ther. 3 960–964. 10.4161/cbt.3.10.1113 [DOI] [PubMed] [Google Scholar]
- Arqués O., Chicote I., Puig I., Tenbaum S. P., Argilés G., Dienstmann R., et al. (2016). Tankyrase inhibition blocks Wnt/β-Catenin pathway and reverts resistance to PI3K and AKT inhibitors in the treatment of colorectal cancer. Clin. Cancer Res. 22 644–656. 10.1158/1078-0432.ccr-14-3081 [DOI] [PubMed] [Google Scholar]
- Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., et al. (2014). YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158 157–170. 10.1016/j.cell.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Azzolin L., Zanconato F., Bresolin S., Forcato M., Basso G., Bicciato S., et al. (2012). Role of TAZ as mediator of Wnt signaling. Cell 151 1443–1456. 10.1016/j.cell.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Bafico A., Liu G., Yaniv A., Gazit A., Aaronson S. A. (2001). Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol. 3 683–686. 10.1038/35083081 [DOI] [PubMed] [Google Scholar]
- Bao Z., Xu X., Liu Y., Chao H., Lin C., Li Z., et al. (2017). CBX7 negatively regulates migration and invasion in glioma via Wnt/β-catenin pathway inactivation. Oncotarget 8 39048–39063. 10.18632/oncotarget.16587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H. J., van Es J. H., et al. (2010). Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6 25–36. 10.1016/j.stem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M. (2005). High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307 1621–1625. 10.1126/science.1105776 [DOI] [PubMed] [Google Scholar]
- Bates S. E. (2020). Epigenetic therapies for cancer. N. Engl. J. Med. 383 650–663. [DOI] [PubMed] [Google Scholar]
- Benham-Pyle B. W., Pruitt B. L., Nelson W. J. (2015). Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348 1024–1027. 10.1126/science.aaa4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447 407–412. 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- Bienz M., Clevers H. (2000). Linking colorectal cancer to Wnt signaling. Cell 103 311–320. 10.1016/s0092-8674(00)00122-7 [DOI] [PubMed] [Google Scholar]
- Blythe S. A., Cha S.-W., Tadjuidje E., Heasman J., Klein P. S. (2010). beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev. Cell 19 220–231. 10.1016/j.devcel.2010.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulagnon-Rombi C., Schneider C., Leandri C., Jeanne A., Grybek V., Bressenot A. M., et al. (2018). LRP1 expression in colon cancer predicts clinical outcome. Oncotarget 9 8849–8869. 10.18632/oncotarget.24225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantjes H., Roose J., van De Wetering M., Clevers H. (2001). All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29 1410–1419. 10.1093/nar/29.7.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugter J. M., Fenderico N., Maurice M. M. (2021). Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat. Rev. Cancer 21 5–21. 10.1038/s41568-020-00307-z [DOI] [PubMed] [Google Scholar]
- Byrne R. M., Tsikitis V. L. (2018). Colorectal polyposis and inherited colorectal cancer syndromes. Ann. Gastroenterol. Hepatol. 31 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Zhang N., Zheng Y., de Wilde R. F., Maitra A., Pan D. (2010). The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24 2383–2388. 10.1101/gad.1978810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.-Y., Wei T.-Z., Luo Q.-C., Wu Q.-W., Liu Q.-F., Yang M., et al. (2013). The Wnt-β-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J. Cell Sci. 126 2877–2889. [DOI] [PubMed] [Google Scholar]
- Callus B. A., Verhagen A. M., Vaux D. L. (2006). Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 273 4264–4276. 10.1111/j.1742-4658.2006.05427.x [DOI] [PubMed] [Google Scholar]
- Carbone C., Piro G., Gaianigo N., Ligorio F., Santoro R., Merz V., et al. (2018). Adipocytes sustain pancreatic cancer progression through a non-canonical WNT paracrine network inducing ROR2 nuclear shuttling. Int. J. Obes. 42 334–343. 10.1038/ijo.2017.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthy J. M., Garmaroudi F. S., Luo Z., McManus B. M. (2011). Wnt3a induces myofibroblast differentiation by upregulating TGF-β signaling through SMAD2 in a β-catenin-dependent manner. PLoS One 6:e19809. 10.1371/journal.pone.0019809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., et al. (1998). Drosophila Tcf and Groucho interact to repress wingless signalling activity. Nature 395 604–608. 10.1038/26982 [DOI] [PubMed] [Google Scholar]
- Chakrabarti R., Celià-Terrassa T., Kumar S., Hang X., Wei Y., Choudhury A., et al. (2018). Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science 360:eaan4153. 10.1126/science.aan4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. H. Y., Nousiainen M., Chalamalasetty R. B., Schäfer A., Nigg E. A., Silljé H. H. W. (2005). The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24 2076–2086. 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- Chan S. W., Lim C. J., Guo K., Ng C. P., Lee I., Hunziker W., et al. (2008). A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 68 2592–2598. 10.1158/0008-5472.can-07-2696 [DOI] [PubMed] [Google Scholar]
- Chen C., Wang L., Liao Q., Huang Y., Ye H., Chen F., et al. (2013). Hypermethylation of EDNRB promoter contributes to the risk of colorectal cancer. Diagn. Pathol. 8:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-Y., Lang Y.-D., Lin H.-N., Liu Y.-R., Liao C.-C., Nana A. W., et al. (2019). miR-103/107 prolong Wnt/β-catenin signaling and colorectal cancer stemness by targeting Axin2. Sci. Rep. 9:9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-Y., Yu S.-L., Ho B.-C., Su K.-Y., Hsu Y.-C., Chang C.-S., et al. (2015). R331W missense mutation of oncogene YAP1 is a germline risk allele for lung adenocarcinoma with medical actionability. J. Clin. Oncol. 33 2303–2310. 10.1200/jco.2014.59.3590 [DOI] [PubMed] [Google Scholar]
- Chen J.-Q., Huang Z.-P., Li H.-F., Ou Y.-L., Huo F., Hu L.-K. (2020). MicroRNA-520f-3p inhibits proliferation of gastric cancer cells via targeting SOX9 and thereby inactivating Wnt signaling. Sci. Rep. 10:6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.-H., He J., Wang D.-L., Cao J.-J., Li M.-C., Zhao X.-M., et al. (2014). Methylation-associated inactivation of LATS1 and its effect on demethylation or overexpression on YAP and cell biological function in human renal cell carcinoma. Int. J. Oncol. 45 2511–2521. 10.3892/ijo.2014.2687 [DOI] [PubMed] [Google Scholar]
- Chen L., Wu X., Xie H., Yao N., Xia Y., Ma G., et al. (2019). ZFP57 suppress proliferation of breast cancer cells through down-regulation of MEST-mediated Wnt/β-catenin signalling pathway. Cell Death Dis. 10:169. 10.1038/s41419-019-1335-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang N., Gray R. S., Li H., Ewald A. J., Zahnow C. A., et al. (2014). A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev. 28 432–437. 10.1101/gad.233676.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-Y., Liu S.-Y., Chang Y.-S., Yin J. J., Yeh H.-L., Mouhieddine T. H., et al. (2015). MicroRNA-34a regulates WNT/TCF7 signaling and inhibits bone metastasis in Ras-activated prostate cancer. Oncotarget 6 441–457. 10.18632/oncotarget.2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Xu X., Chen D., Zhao F., Wang W. (2019). Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed. Pharmacother. 110 473–481. 10.1016/j.biopha.2018.11.082 [DOI] [PubMed] [Google Scholar]
- Cheung P., Xiol J., Dill M. T., Yuan W.-C., Panero R., Roper J., et al. (2020). Regenerative reprogramming of the intestinal stem cell state via hippo signaling suppresses metastatic colorectal cancer. Cell Stem Cell 27 590–604.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C.-H., Hou M.-F., Hung W.-C. (2013). Up-regulation of miR-182 by β-catenin in breast cancer increases tumorigenicity and invasiveness by targeting the matrix metalloproteinase inhibitor RECK. Biochim. Biophys. Acta 1830 3067–3076. 10.1016/j.bbagen.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Clements W. M., Wang J., Sarnaik A., Kim O. J., MacDonald J., Fenoglio-Preiser C., et al. (2002). beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 62 3503–3506. [PubMed] [Google Scholar]
- Cotton J. L., Li Q., Ma L., Park J.-S., Wang J., Ou J., et al. (2017). YAP/TAZ and hedgehog coordinate growth and patterning in gastrointestinal mesenchyme. Dev. Cell 43 35–47.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Jia S. (2001). Transcriptional repression: the long and the short of it. Genes Dev. 15 2786–2796. [DOI] [PubMed] [Google Scholar]
- Cui Y., Ma W., Lei F., Li Q., Su Y., Lin X., et al. (2016). Prostate tumour overexpressed-1 promotes tumourigenicity in human breast cancer via activation of Wnt/β-catenin signalling. J. Pathol. 239 297–308. 10.1002/path.4725 [DOI] [PubMed] [Google Scholar]
- Dai Z., Ramesh V., Locasale J. W. (2020). The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 21 737–753. 10.1038/s41576-020-0270-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Weis W. I. (2005). Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12 364–371. 10.1038/nsmb912 [DOI] [PubMed] [Google Scholar]
- Dao D. Y., Yang X., Chen D., Zuscik M., O’Keefe R. J. (2007). Axin1 and Axin2 are regulated by TGF- and mediate cross-talk between TGF- and Wnt signaling pathways. Ann. N. Y. Acad. Sci. 1116 82–99. 10.1196/annals.1402.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A. (2011). Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim. Biophys. Sin. 43 745–756. 10.1093/abbs/gmr079 [DOI] [PubMed] [Google Scholar]
- Denis H., Ndlovu M. N., Fuks F. (2011). Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 12 647–656. 10.1038/embor.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derissen E. J. B., Beijnen J. H., Schellens J. H. M. (2013). Concise drug review: azacitidine and decitabine. Oncologist 18 619–624. 10.1634/theoncologist.2012-0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijksterhuis J. P., Baljinnyam B., Stanger K., Sercan H. O., Ji Y., Andres O., et al. (2015). Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J. Biol. Chem. 290 6789–6798. 10.1074/jbc.m114.612648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez I., Itoh K., Sokol S. Y. (1995). Role of glycogen synthase kinase 3 beta as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc. Natl. Acad. Sci. U. S. A. 92 8498–8502. 10.1073/pnas.92.18.8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Deng J., Sun Z.-M., Pan A.-P., Xiang X.-J., Zhang L., et al. (2015). Interference with the β-catenin gene in gastric cancer induces changes to the miRNA expression profile. Tumour Biol. 36 6973–6983. 10.1007/s13277-015-3415-1 [DOI] [PubMed] [Google Scholar]
- Dyer C., Blanc E., Hanisch A., Roehl H., Otto G. W., Yu T., et al. (2014). A bi-modal function of Wnt signalling directs an FGF activity gradient to spatially regulate neuronal differentiation in the midbrain. Development 141 63–72. 10.1242/dev.099507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund S., Lee S. Y., Grimsby S., Zhang S., Aspenström P., Heldin C.-H., et al. (2005). Interaction between Smad7 and β-Catenin: importance for transforming growth factor β-induced apoptosis. Mol. Cell. Biol. 25 1475–1488. 10.1128/mcb.25.4.1475-1488.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Helou R., Pinna G., Cabaud O., Wicinski J., Bhajun R., Guyon L., et al. (2017). miR-600 acts as a bimodal switch that regulates breast cancer stem cell fate through WNT signaling. Cell Rep. 18 2256–2268. 10.1016/j.celrep.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Esmaeili M., Blythe S. A., Tobias J. W., Zhang K., Yang J., Klein P. S. (2020). Chromatin accessibility and histone acetylation in the regulation of competence in early development. Dev. Biol. 462 20–35. 10.1016/j.ydbio.2020.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa L., Inglés-Esteve J., Aguilera C., Bigas A. (2003). Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278 32227–32235. 10.1074/jbc.m304001200 [DOI] [PubMed] [Google Scholar]
- Estarás C., Benner C., Jones K. A. (2015). SMADs and YAP compete to control elongation of β-Catenin:LEF-1-Recruited RNAPII during hESC differentiation. Mol. Cell 58 780–793. 10.1016/j.molcel.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre R., Alférez D. G., Santiago-Gómez A., Spence K., McConnell J. C., Hart C., et al. (2019). Microenvironmental IL1β promotes breast cancer metastatic colonisation in the bone via activation of Wnt signalling. Nat. Commun. 10:5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D. C., Luo Y. H., Yang S. M., Li X. A., Ling X. L., Fang L. (2002). Mutation analysis of APC gene in gastric cancer with microsatellite instability. World J. Gastroenterol. 8 787–791. 10.3748/wjg.v8.i5.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe de Sousa E. M., Colak S., Buikhuisen J., Koster J., Cameron K., et al. (2011). Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9 476–485. 10.1016/j.stem.2011.10.008 [DOI] [PubMed] [Google Scholar]
- Fendler A., Bauer D., Busch J., Jung K., Wulf-Goldenberg A., Kunz S., et al. (2020). Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 11:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-L A., Squatrito M., Northcott P., Awan A., Holland E. C., Taylor M. D., et al. (2012). Oncogenic YAP promotes radioresistance and genomic instability in medulloblastoma through IGF2-mediated Akt activation. Oncogene 31 1923–1937. 10.1038/onc.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9 102–114. 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- Fischer M. M., Cancilla B., Yeung V. P., Cattaruzza F., Chartier C., Murriel C. L., et al. (2017). WNT antagonists exhibit unique combinatorial antitumor activity with taxanes by potentiating mitotic cell death. Sci. Adv. 3:e1700090. 10.1126/sciadv.1700090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan D. J., Barker N., Di Costanzo N. S., Mason E. A., Gurney A., Meniel V. S., et al. (2019). Frizzled-7 is required for wnt signaling in gastric tumors with and without Apc mutations. Cancer Res. 79 970–981. 10.1158/0008-5472.can-18-2095 [DOI] [PubMed] [Google Scholar]
- Flanagan D. J., Vincan E., Phesse T. J. (2017). Winding back Wnt signalling: potential therapeutic targets for treating gastric cancers. Br. J. Pharmacol. 174 4666–4683. 10.1111/bph.13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Hernández E., Velázquez D. M., Castañeda-Patlán M. C., Fuentes-García G., Fonseca-Camarillo G., Yamamoto-Furusho J. K., et al. (2020). Canonical and non-canonical Wnt signaling are simultaneously activated by Wnts in colon cancer cells. Cell. Signal. 72:109636. 10.1016/j.cellsig.2020.109636 [DOI] [PubMed] [Google Scholar]
- Fu X., Li H., Liu C., Hu B., Li T., Wang Y. (2016). Long noncoding RNA AK126698 inhibits proliferation and migration of non-small cell lung cancer cells by targeting Frizzled-8 and suppressing Wnt/β-catenin signaling pathway. Onco. Targets Ther. 9 3815–3827. 10.2147/ott.s100633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikane T., Nishikawa N., Toyota M., Suzuki H., Nojima M., Maruyama R., et al. (2010). Genomic screening for genes upregulated by demethylation revealed novel targets of epigenetic silencing in breast cancer. Breast Cancer Res. Treat. 122 699–710. 10.1007/s10549-009-0600-1 [DOI] [PubMed] [Google Scholar]
- Furuhashi M., Yagi K., Yamamoto H., Furukawa Y., Shimada S., Nakamura Y., et al. (2001). Axin facilitates Smad3 activation in the transforming growth factor beta signaling pathway. Mol. Cell. Biol. 21 5132–5141. 10.1128/mcb.21.15.5132-5141.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galamb O., Kalmár A., Péterfia B., Csabai I., Bodor A., Ribli D., et al. (2016). Aberrant DNA methylation of WNT pathway genes in the development and progression of CIMP-negative colorectal cancer. Epigenetics 11 588–602. 10.1080/15592294.2016.1190894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande S., Purbey P. K., Notani D., Kumar P. P. (2007). The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 17 408–414. 10.1016/j.gde.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Genevet A., Wehr M. C., Brain R., Thompson B. J., Tapon N. (2010). Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18 300–308. 10.1016/j.devcel.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu M.-M., Gagea M., Cote G. (2016). NHERF1/EBP50 suppresses Wnt-β-Catenin pathway-driven intestinal neoplasia. Neoplasia 18 512–523. 10.1016/j.neo.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis M., Hodis E., Jasmine, Mu X., Yamauchi M., Rosenbluh J., et al. (2014). RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 46 1264–1266. 10.1038/ng.3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godman C. A., Joshi R., Tierney B. R., Greenspan E., Rasmussen T. P., Wang H.-W., et al. (2008). HDAC3 impacts multiple oncogenic pathways in colon cancer cells with effects on Wnt and vitamin D signaling. Cancer Biol. Ther. 7 1570–1580. 10.4161/cbt.7.10.6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Qu S., Lv X.-B., Liu B., Tan W., Nie Y., et al. (2014). BRMS1L suppresses breast cancer metastasis by inducing epigenetic silence of FZD10. Nat. Commun. 5:5406. [DOI] [PubMed] [Google Scholar]
- Gonsalves F. C., Klein K., Carson B. B., Katz S., Ekas L. A., Evans S., et al. (2011). An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 108 5954–5963. 10.1073/pnas.1017496108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J. S., Kaye S. B., Brown R. (2009). The promises and pitfalls of epigenetic therapies in solid tumours. Eur. J. Cancer 45 1129–1136. 10.1016/j.ejca.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Greenberg M. V. C., Bourc’his D. (2019). The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20 590–607. 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- Gregorieff A., Liu Y., Inanlou M. R., Khomchuk Y., Wrana J. L. (2015). Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526 715–718. 10.1038/nature15382 [DOI] [PubMed] [Google Scholar]
- Grinat J., Heuberger J., Vidal R. O., Goveas N., Kosel F., Berenguer-Llergo A., et al. (2020). The epigenetic regulator Mll1 is required for Wnt-driven intestinal tumorigenesis and cancer stemness. Nat. Commun. 11:6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Gu J., Shen K., Zhou H., Hao J., Li F., et al. (2020). HOXA13 promotes colon cancer progression through β-catenin-dependent WNT pathway. Exp. Cell Res. 395:112238. 10.1016/j.yexcr.2020.112238 [DOI] [PubMed] [Google Scholar]
- Guan H., Zhu T., Wu S., Liu S., Liu B., Wu J., et al. (2019). Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/β-catenin signaling. Proc. Natl. Acad. Sci. U. S. A. 116 14019–14028. 10.1073/pnas.1900997116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati N., Béguelin W., Giulino-Roth L. (2018). Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk. Lymphoma 59 1574–1585. 10.1080/10428194.2018.1430795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Wang G., Khadarian K., Zheng Y., Ha A. D., Shen J. (2014). Abstract 4354: miR-603 targets WIF1 to promote glioma tumorigenesis via Wnt/β-catenin pathway. Mol. Cell. Biol. 74:Abstract nr 4354. 10.1158/1538-7445.am2014-4354 [DOI] [Google Scholar]
- Guo X., Ramirez A., Waddell D. S., Li Z., Liu X., Wang X.-F. (2008). Axin and GSK3- control Smad3 protein stability and modulate TGF- signaling. Genes Dev. 22 106–120. 10.1101/gad.1590908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Lee J. H., Shu L., Huang Y., Li W., Zhang C., et al. (2015). Association of aberrant DNA methylation in Apc(min/+) mice with the epithelial-mesenchymal transition and Wnt/β-catenin pathways: genome-wide analysis using MeDIP-seq. Cell Biosci. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Wang M., Jia X., Zhu H., Zhi Y., Yuan L. (2018). Wnt signaling pathway upregulates DNMT1 to trigger NHERF1 promoter hypermethylation in colon cancer. Oncol. Rep. 40 1165–1173. [DOI] [PubMed] [Google Scholar]
- Gurley K. A., Rink J. C., Sánchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319 323–327. 10.1126/science.1150029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney A., Axelrod F., Bond C. J., Cain J., Chartier C., Donigan L., et al. (2012). Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc. Natl. Acad. Sci. U. S. A. 109 11717–11722. 10.1073/pnas.1120068109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas R., Kato Y., He X. (2001). Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107 843–854. 10.1016/s0092-8674(01)00614-6 [DOI] [PubMed] [Google Scholar]
- Hahn S., Jackstadt R., Siemens H., Hünten S., Hermeking H. (2013). SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 32 3079–3095. 10.1038/emboj.2013.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Half E., Bercovich D., Rozen P. (2009). Familial adenomatous polyposis. Orphanet J. Rare Dis. 4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto R., Nakamura Y. (2016). Dysregulation of protein methyltransferases in human cancer: an emerging target class for anticancer therapy. Cancer Sci. 107 377–384. 10.1111/cas.12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Wang S., Fritah S., Wang X., Zhou W., Yang N., et al. (2020). Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain 143 512–530. 10.1093/brain/awz406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K. F., Zhang X., Thomas D. M. (2013). The Hippo pathway and human cancer. Nat. Rev. Cancer 13 246–257. [DOI] [PubMed] [Google Scholar]
- Hassan M. Q., Maeda Y., Taipaleenmaki H., Zhang W., Jafferji M., Gordon J. A. R., et al. (2012). miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J. Biol. Chem. 287 42084–42092. 10.1074/jbc.m112.377515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata T., Mokutani Y., Takahashi H., Inoue A., Munakata K., Nagata K., et al. (2017). Identification of microRNA-487b as a negative regulator of liver metastasis by regulation of KRAS in colorectal cancer. Int. J. Oncol. 50 487–496. 10.3892/ijo.2016.3813 [DOI] [PubMed] [Google Scholar]
- Hawkins R. D., Hon G. C., Yang C., Antosiewicz-Bourget J. E., Lee L. K., Ngo Q.-M., et al. (2011). Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 21 1393–1409. 10.1038/cr.2011.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., You L., Uematsu K., Xu Z., Lee A. Y., Matsangou M., et al. (2004). A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia 6 7–14. 10.1016/s1476-5586(04)80048-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Sheng T., Stelter A. A., Li C., Zhang X., Sinha M., et al. (2006). Suppressing Wnt signaling by the hedgehog pathway through sFRP-1. J. Biol. Chem. 281 35598–35602. 10.1074/jbc.c600200200 [DOI] [PubMed] [Google Scholar]
- Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R. L., et al. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332 458–461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A. (2000). The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19 1839–1850. 10.1093/emboj/19.8.1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39 311–318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Hervouet E., Peixoto P., Delage-Mourroux R., Boyer-Guittaut M., Cartron P.-F. (2018). Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin. Epigenetics 10:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemer S. E., Szymaniak A. D., Varelas X. (2014). The transcriptional regulators TAZ and YAP direct transforming growth factor β-induced tumorigenic phenotypes in breast cancer cells. J. Biol. Chem. 289 13461–13474. 10.1074/jbc.m113.529115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobmayer B., Rentzsch F., Kuhn K., Happel C. M., von Laue C. C., Snyder P., et al. (2000). WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407 186–189. 10.1038/35025063 [DOI] [PubMed] [Google Scholar]
- Holloway K. R., Calhoun T. N., Saxena M., Metoyer C. F., Kandler E. F., Rivera C. A., et al. (2010). SIRT1 regulates Dishevelled proteins and promotes transient and constitutive Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 107 9216–9221. 10.1073/pnas.0911325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G. C., Hawkins R. D., Ren B. (2009). Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 18 R195–R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Li B., Zhou C., Ying X., Chen M., Huang T., et al. (2018). Diagnostic value of WIF1 methylation for colorectal cancer: a meta-analysis. Oncotarget 9 5378–5386. 10.18632/oncotarget.23870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K., Yang W., Song H., Song J., Wei C., Li D., et al. (2015). Up-regulation of miR-506 inhibits cell growth and disrupt the cell cycle by targeting YAP in breast cancer cells. Int. J. Clin. Exp. Med. 8 12018–12027. [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Li S., Song W., Li X., Li Q., Zhang Z., et al. (2013). Correction: lysine-specific demethylase 1 (LSD1/KDM1A) contributes to colorectal tumorigenesis via activation of the Wnt/B -Catenin pathway by Down-Regulating Dickkopf-1 (DKK1). PLoS One 8:e70077. 10.1371/annotation/d090733e-1f34-43c5-a06a-255456946303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins I. J., Bos T., Gaylord O., Jessen C., Lonquich B., Puranen A., et al. (2017). The WNT target SP5 negatively regulates WNT transcriptional programs in human pluripotent stem cells. Nat. Commun. 8:1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.-L., Jiang J.-K., Yang S.-H., Huang T.-S., Lan H.-Y., Teng H.-W., et al. (2014). MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 16 268–280. 10.1038/ncb2910 [DOI] [PubMed] [Google Scholar]
- Iglesias M., Gomez-Skarmeta J. L., Saló E., Adell T. (2008). Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development 135 1215–1221. 10.1242/dev.020289 [DOI] [PubMed] [Google Scholar]
- Imajo M., Ebisuya M., Nishida E. (2015). Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat. Cell Biol. 17 7–19. 10.1038/ncb3084 [DOI] [PubMed] [Google Scholar]
- Isobe T., Hisamori S., Hogan D. J., Zabala M., Hendrickson D. G., Dalerba P., et al. (2014). miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. Elife 3:e01977. 10.7554/eLife.01977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarroux J., Morillon A., Pinskaya M. (2017). History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 1008 1–46. 10.1007/978-981-10-5203-3_1 [DOI] [PubMed] [Google Scholar]
- Jia J., Zhang W., Wang B., Trinko R., Jiang J. (2003). The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17 2514–2519. 10.1101/gad.1134003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., He M., Guan S., Ma M., Wu H., Yu Z., et al. (2016). MicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/β-catenin signaling pathway. Tumor Biol. 37 5001–5011. 10.1007/s13277-015-4342-x [DOI] [PubMed] [Google Scholar]
- Jiang X., Tan J., Li J., Kivimäe S., Yang X., Zhuang L., et al. (2008). DACT3 is an epigenetic regulator of Wnt/β-Catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 13 529–541. 10.1016/j.ccr.2008.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Luo S., Wang Y., Liu C., Piao Z., Xu M., et al. (2017). miR-135b stimulates osteosarcoma recurrence and lung metastasis via notch and Wnt/β-Catenin signaling. Mol. Ther. Nucleic Acids 8 111–122. 10.1016/j.omtn.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L.-H., Shao Q.-J., Luo W., Ye Z.-Y., Li Q., Lin S.-C. (2003). Detection of point mutations of the Axin1 gene in colorectal cancers. Int. J. Cancer 107 696–699. 10.1002/ijc.11435 [DOI] [PubMed] [Google Scholar]
- Jin Y. H., Kim H., Ki H., Yang I., Yang N., Lee K. Y., et al. (2009). Beta-catenin modulates the level and transcriptional activity of Notch1/NICD through its direct interaction. Biochim. Biophys. Acta 1793 290–299. 10.1016/j.bbamcr.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Jung Y.-S., Jun S., Lee S. H., Sharma A., Park J.-I. (2015). Wnt2 complements Wnt/β-catenin signaling in colorectal cancer. Oncotarget 6 37257–37268. 10.18632/oncotarget.6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. (1995). The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9 534–546. 10.1101/gad.9.5.534 [DOI] [PubMed] [Google Scholar]
- Kadowaki T., Wilder E., Klingensmith J., Zachary K., Perrimon N. (1996). The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev. 10 3116–3128. 10.1101/gad.10.24.3116 [DOI] [PubMed] [Google Scholar]
- Katoh M. (2017). Canonical and non-canonical WNT signaling in cancer stem cells and their niches: cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review). Int. J. Oncol. 51 1357–1369. 10.3892/ijo.2017.4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M., Katoh M. (2007). WNT antagonist, DKK2, is a Notch signaling target in intestinal stem cells: augmentation of a negative regulation system for canonical WNT signaling pathway by the Notch-DKK2 signaling loop in primates. Int. J. Mol. Med. 19 197–201. 10.3892/ijmm.19.1.197 [DOI] [PubMed] [Google Scholar]
- Kennedy M. W., Chalamalasetty R. B., Thomas S., Garriock R. J., Jailwala P., Yamaguchi T. P. (2016). Sp5 and Sp8 recruit β-catenin and Tcf1-Lef1 to select enhancers to activate Wnt target gene transcription. Proc. Natl. Acad. Sci. U. S. A. 113 3545–3550. 10.1073/pnas.1519994113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-K., Yoo H.-I., Kim I., Park J., Yoon S. K. (2015). FZD6 expression is negatively regulated by miR-199a-5p in human colorectal cancer. BMB Rep. 48 360–366. 10.5483/bmbrep.2015.48.6.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-Y., Park J.-H., Won H.-Y., Lee J.-Y., Kong G. (2015). CBX7 inhibits breast tumorigenicity through DKK-1-mediated suppression of the Wnt/β-catenin pathway. FASEB J. 29 300–313. 10.1096/fj.14-253997 [DOI] [PubMed] [Google Scholar]
- Kim K.-A., Wagle M., Tran K., Zhan X., Dixon M. A., Liu S., et al. (2008). R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol. Biol. Cell 19 2588–2596. 10.1091/mbc.e08-02-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. H., Cha Y. H., Kang S. E., Lee Y., Lee I., Cha S. Y., et al. (2013). p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle 12 1578–1587. 10.4161/cc.24739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. H., Kim H. S., Kim N.-G., Lee I., Choi H.-S., Li X.-Y., et al. (2011). p53 and MicroRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 4:ra71. 10.1126/scisignal.2001744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Xu X., Hecht A., Boyer T. G. (2006). Mediator is a transducer of Wnt/beta-catenin signaling. J. Biol. Chem. 281 14066–14075. 10.1074/jbc.m602696200 [DOI] [PubMed] [Google Scholar]
- Ko A. H., Gabriela Chiorean E., Kwak E. L., Lenz H.-J., Nadler P. I., Wood D. L., et al. (2016). Final results of a phase Ib dose-escalation study of PRI-724, a CBP/beta-catenin modulator, plus gemcitabine (GEM) in patients with advanced pancreatic adenocarcinoma (APC) as second-line therapy after FOLFIRINOX or FOLFOX. J. Clin. Oncol. 34:e15721. 10.1200/jco.2016.34.15_suppl.e15721 [DOI] [Google Scholar]
- Koinuma K., Yamashita Y., Liu W., Hatanaka H., Kurashina K., Wada T., et al. (2006). Epigenetic silencing of AXIN2 in colorectal carcinoma with microsatellite instability. Oncogene 25 139–146. 10.1038/sj.onc.1209009 [DOI] [PubMed] [Google Scholar]
- Koo B.-K., van Es J. H., van den Born M., Clevers H. (2015). Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc. Natl. Acad. Sci. U. S. A. 112 7548–7550. 10.1073/pnas.1508113112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F., Mendell J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172 393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Krawczyk P., Powrózek T., Olesiński T., Dmitruk A., Dziwota J., Kowalski D., et al. (2017). Evaluation of miR-506 and miR-4316 expression in early and non-invasive diagnosis of colorectal cancer. Int. J. Colorectal Dis. 32 1057–1060. 10.1007/s00384-017-2814-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeiro Y., Couchy G., Balabaud C., Bioulac-Sage P., Pelletier L., Rebouissou S., et al. (2008). MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology 47 1955–1963. 10.1002/hep.22256 [DOI] [PubMed] [Google Scholar]
- Lai D., Yang X. (2013). BMP4 is a novel transcriptional target and mediator of mammary cell migration downstream of the Hippo pathway component TAZ. Cell. Signal. 25 1720–1728. 10.1016/j.cellsig.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Laisné M., Gupta N., Kirsh O., Pradhan S., Defossez P.-A. (2018). Mechanisms of DNA methyltransferase recruitment in mammals. Genes 9:617. 10.3390/genes9120617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand D., Curto M., Saotome I., Giovannini M., McClatchey A. I. (2003). NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 17 1090–1100. 10.1101/gad.1054603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R., Bonuccelli G., Ozsvári B., Peiris-Pagès M., Fiorillo M., Smith D. L., et al. (2015). Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: understanding WNT/FGF-driven anabolic signaling. Oncotarget 6 30453–30471. 10.18632/oncotarget.5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Yue X., Han L., Shi Z., Yang Y., Pu P., et al. (2012). Genome-wide identification of TCF7L2/TCF4 target miRNAs reveals a role for miR-21 in Wnt-driven epithelial cancer. Int. J. Oncol. 40 519–526. [DOI] [PubMed] [Google Scholar]
- Lee K.-H., Goan Y.-G., Hsiao M., Lee C.-H., Jian S.-H., Lin J.-T., et al. (2009). MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp. Cell Res. 315 2529–2538. 10.1016/j.yexcr.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Lei A., Chen L., Zhang M., Yang X., Xu L., Cao N., et al. (2019). EZH2 regulates protein stability via recruiting USP7 to mediate neuronal gene expression in cancer cells. Front. Genet. 10:422. 10.3389/fgene.2019.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei D., Yang W.-T., Zheng P.-S. (2021). HOXB4 inhibits the proliferation and tumorigenesis of cervical cancer cells by downregulating the activity of Wnt/β-catenin signaling pathway. Cell Death Dis. 12:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q.-Y., Zhang H., Zhao B., Zha Z.-Y., Bai F., Pei X.-H., et al. (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28 2426–2436. 10.1128/mcb.01874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lento W., Congdon K., Voermans C., Kritzik M., Reya T. (2013). Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb. Perspect. Biol. 5:a008011. 10.1101/cshperspect.a008011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepourcelet M., Chen Y.-N. P., France D. S., Wang H., Crews P., Petersen F., et al. (2004). Small-molecule antagonists of the oncogenic Tcf/β-catenin protein complex. Cancer Cell 5 91–102. 10.1016/s1535-6108(03)00334-9 [DOI] [PubMed] [Google Scholar]
- Leung W. K. C., He M., Chan A. W. H., Law P. T. Y., Wong N. (2015). Wnt/β-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 362 97–105. 10.1016/j.canlet.2015.03.023 [DOI] [PubMed] [Google Scholar]
- Li J., Sutter C., Parker D. S., Blauwkamp T., Fang M., Cadigan K. M. (2007). CBP/p300 are bimodal regulators of Wnt signaling. EMBO J. 26 2284–2294. 10.1038/sj.emboj.7601667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yu B., Deng P., Cheng Y., Yu Y., Kevork K., et al. (2019). Author Correction: KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/β-catenin signalling. Nat. Commun. 10:5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Sun Y., Jarugumilli G. K., Liu S., Dang K., Cotton J. L., et al. (2020). Lats1/2 sustain intestinal stem cells and Wnt activation through TEAD-dependent and independent transcription. Cell Stem Cell 26 675–692.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Han Z., Zhao N., Zhu B., Zhang Q., Yang X., et al. (2018). Inhibition of DNMT suppresses the stemness of colorectal cancer cells through down-regulating Wnt signaling pathway. Cell. Signal. 47 79–87. 10.1016/j.cellsig.2018.03.014 [DOI] [PubMed] [Google Scholar]
- Li S., Yen L., Pastor W. A., Johnston J. B., Du J., Shew C. J., et al. (2016). Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc. Natl. Acad. Sci. U.S.A 113 E5108–E5116. 10.1073/pnas.1609709113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Lai Q., Wang S., Cai J., Xiao Z., Deng D., et al. (2016). MicroRNA-224 sustains Wnt/β-catenin signaling and promotes aggressive phenotype of colorectal cancer. J. Exp. Clin. Cancer Res. 35:21. 10.1186/s13046-016-0287-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li V. S. W., Ng S. S., Boersema P. J., Low T. Y., Karthaus W. R., Gerlach J. P., et al. (2012). Wnt signaling through inhibition of β-Catenin degradation in an intact Axin1 complex. Cell 149 1245–1256. 10.1016/j.cell.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Li Z.-T., Zhang X., Wang D.-W., Xu J., Kou K.-J., Wang Z.-W., et al. (2020). Overexpressed lncRNA GATA6-AS1 inhibits LNM and EMT via FZD4 through the Wnt/β-Catenin signaling pathway in GC. Mol. Ther. Nucleic Acids 19 827–840. 10.1016/j.omtn.2019.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liang T.-J., Wang H.-X., Zheng Y.-Y., Cao Y.-Q., Wu X., Zhou X., et al. (2017). APC hypermethylation for early diagnosis of colorectal cancer: a meta-analysis and literature review. Oncotarget 8 46468–46479. 10.18632/oncotarget.17576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Li X., Zhao L., Wang Y., Wang X., Wu Y., et al. (2020). A PROTAC peptide induces durable β-catenin degradation and suppresses Wnt-dependent intestinal cancer. Cell Discov. 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Lönnerdal B. (2010). Beta-catenin/TCF4 transactivates miR-30e during intestinal cell differentiation. Cell. Mol. Life Sci. 67 2969–2978. 10.1007/s00018-010-0366-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Zhen Y., Guan Y., Yi H. (2020). Roles of Wnt/β-Catenin signaling pathway regulatory long non-coding RNAs in the pathogenesis of non-small cell lung cancer. Cancer Manag. Res. 12 4181–4191. 10.2147/cmar.s241519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H., Spizzo R., Atlasi Y., Nicoloso M., Shimizu M., Redis R. S., et al. (2013). CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23 1446–1461. 10.1101/gr.152942.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.-N., Zhang H.-Y., Liu C.-L., Wang C.-C. (2019). Upregulation of lncRNA CALML3-AS1 promotes cell proliferation and metastasis in cervical cancer via activation of the Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 23 5611–5620. [DOI] [PubMed] [Google Scholar]
- Liu W., Sato A., Khadka D., Bharti R., Diaz H., Runnels L. W., et al. (2008). Mechanism of activation of the formin protein Daam1. Proc. Natl. Acad. Sci. U. S. A. 105 210–215. 10.1073/pnas.0707277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Liu H., Desai S., Schmitt D. C., Zhou M., Khong H. T., et al. (2013). miR-125b functions as a key mediator for snail-induced stem cell propagation and chemoresistance. J. Biol. Chem. 288 4334–4345. 10.1074/jbc.m112.419168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Zhao Y., Tawatao R., Cottam H. B., Sen M., Leoni L. M., et al. (2004). Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 101 3118–3123. 10.1073/pnas.0308648100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis T. C., Ichii M., Brugman M. H., Kincade P., Staal F. J. T. (2012). Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia 26 414–421. 10.1038/leu.2011.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Karsten U., et al. (2002). Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 22 1184–1193. 10.1128/mcb.22.4.1184-1193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Xu B., Husaiyin S., Wang L., Wusainahong K., Ma J., et al. (2017). MicroRNA-505 predicts prognosis and acts as tumor inhibitor in cervical carcinoma with inverse association with FZD4. Biomed. Pharmacother. 92 586–594. 10.1016/j.biopha.2017.04.028 [DOI] [PubMed] [Google Scholar]
- Ma H.-M., Cui N., Zheng P.-S. (2020). HOXA5 inhibits the proliferation and neoplasia of cervical cancer cells via downregulating the activity of the Wnt/β-catenin pathway and transactivating TP53. Cell Death Dis. 11:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Huang X., Li Z., Shen Y., Lai J., Su Q., et al. (2019). FOXE1 supports the tumor promotion of Gli2 on papillary thyroid carcinoma by the Wnt/β-catenin pathway. J. Cell. Physiol. 234 17739–17748. 10.1002/jcp.28399 [DOI] [PubMed] [Google Scholar]
- Ma L., Wang J., Lin J., Pan Q., Yu Y., Sun F. (2014). Cluster of differentiation 166 (CD166) regulated by phosphatidylinositide 3-Kinase (PI3K)/AKT signaling to exert its anti-apoptotic role via yes-associated protein (YAP) in liver cancer. J. Biol. Chem. 289 6921–6933. 10.1074/jbc.m113.524819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Wang Y., Hui Y., Du Y., Chen Z., Feng H., et al. (2019). WNT/NOTCH pathway is essential for the maintenance and expansion of human MGE progenitors. Stem Cell Rep. 12 934–949. 10.1016/j.stemcr.2019.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B. T., Tamai K., He X. (2009). Wnt/β-Catenin signaling: components, mechanisms, and diseases. Dev. Cell 17 9–26. 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney W. M., Jr., Hong J.-H., Yaffe M. B., Farrance I. K. G. (2005). The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem. J. 388 217–225. 10.1042/bj20041434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maille E., Brosseau S., Hanoux V., Creveuil C., Danel C., Bergot E., et al. (2019). MST1/Hippo promoter gene methylation predicts poor survival in patients with malignant pleural mesothelioma in the IFCT-GFPC-0701 MAPS Phase 3 trial. Br. J. Cancer 120 387–397. 10.1038/s41416-019-0379-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita R., Uchijima Y., Nishiyama K., Amano T., Chen Q., Takeuchi T., et al. (2008). Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am. J. Physiol. Renal Physiol. 294 F542–F553. [DOI] [PubMed] [Google Scholar]
- Malbon C. C., Kühl M., Sheldahl L. C., Moon R. T. (2000). Ca2/Calmodulin-dependent protein kinase II is stimulated by Wnt and frizzled homologs and promotes ventral cell fates in xenopus. J. Biol. Chem. 275 12701–12711. 10.1074/jbc.275.17.12701 [DOI] [PubMed] [Google Scholar]
- Marchese F. P., Huarte M. (2014). Long non-coding RNAs and chromatin modifiers: their place in the epigenetic code. Epigenetics 9 21–26. 10.4161/epi.27472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez S., Scerbo P., Giordano M., Daulat A. M., Lhoumeau A.-C., Thomé V., et al. (2015). The PTK7 and ROR2 protein receptors interact in the vertebrate WNT/Planar Cell Polarity (PCP) pathway. J. Biol. Chem. 290 30562–30572. 10.1074/jbc.m115.697615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni S. M., Petty E. M., Stoffel E. M., Fearon E. R. (2015). An AXIN2 mutant allele associated with predisposition to colorectal neoplasia has context-dependent effects on AXIN2 protein function. Neoplasia 17 463–472. 10.1016/j.neo.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea P. D., Brieher W. M., Gumbiner B. M. (1993). Induction of a secondary body axis in Xenopus by antibodies to beta-catenin. J. Cell Biol. 123 477–484. 10.1083/jcb.123.2.477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir R., Pradhan S. J., Patil P., Mulherkar R., Galande S. (2016). Wnt/β-catenin signaling regulated SATB1 promotes colorectal cancer tumorigenesis and progression. Oncogene 35 1679–1691. 10.1038/onc.2015.232 [DOI] [PubMed] [Google Scholar]
- Mir R., Sharma A., Pradhan S. J., Galande S. (2018). Regulation of transcription factor SP1 by the β-Catenin destruction complex modulates wnt response. Mol. Cell. Biol. 38 10.1128/MCB.00188-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M. Z., Bialik J. F., Speight P., Dan Q., Yeung T., Szászi K., et al. (2017). TGF-β1 regulates the expression and transcriptional activity of TAZ protein via a Smad3-independent, myocardin-related transcription factor-mediated mechanism. J. Biol. Chem. 292 14902–14920. 10.1074/jbc.m117.780502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. D., Le T., Fan G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38 23–38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C., Hausmann G., Basler K. (2006). Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125 327–341. 10.1016/j.cell.2006.01.053 [DOI] [PubMed] [Google Scholar]
- Mosimann C., Hausmann G., Basler K. (2009). Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 10 276–286. 10.1038/nrm2654 [DOI] [PubMed] [Google Scholar]
- Moskalev E. A., Luckert K., Vorobjev I. A., Mastitsky S. E., Gladkikh A. A., Stephan A., et al. (2012). Concurrent epigenetic silencing of wnt/β-catenin pathway inhibitor genes in B cell chronic lymphocytic leukaemia. BMC Cancer 12:213. 10.1186/1471-2407-12-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R., le Sage C., Diosdado B., van der Waal M., Oude Vrielink J. A. F., Bolijn A., et al. (2008). Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 68 5795–5802. 10.1158/0008-5472.can-08-0951 [DOI] [PubMed] [Google Scholar]
- Necela B. M., Carr J. M., Asmann Y. W., Thompson E. A. (2011). Differential expression of microRNAs in tumors from chronically inflamed or genetic (APC(Min/+)) models of colon cancer. PLoS One 6:e18501. 10.1371/journal.pone.0018501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X., Liu H., Liu L., Wang Y.-D., Chen W.-D. (2020). Emerging roles of Wnt ligands in human colorectal cancer. Front. Oncol. 10:1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima M., Suzuki H., Toyota M., Watanabe Y., Maruyama R., Sasaki S., et al. (2007). Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene 26 4699–4713. 10.1038/sj.onc.1210259 [DOI] [PubMed] [Google Scholar]
- Noordermeer J., Klingensmith J., Perrimon N., Nusse R. (1994). dishevelled and armadillo act in the wingless signalling pathway in Drosophila. Nature 367 80–83. 10.1038/367080a0 [DOI] [PubMed] [Google Scholar]
- Notani D., Gottimukkala K. P., Jayani R. S., Limaye A. S., Damle M. V., Mehta S., et al. (2010). Global regulator SATB1 recruits β-Catenin and regulates TH2 differentiation in Wnt-dependent manner. PLoS Biol. 8:e1000296. 10.1371/journal.pbio.1000296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Clevers H. (2017). Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169 985–999. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. (1982). Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31 99–109. 10.1016/0092-8674(82)90409-3 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Oginuma M., Moncuquet P., Xiong F., Karoly E., Chal J., Guevorkian K., et al. (2017). A gradient of glycolytic activity coordinates FGF and Wnt signaling during elongation of the body axis in amniote embryos. Dev. Cell 40 342–353.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I., Suzuki H., Onishi N., Takada R., Kani S., Ohkawara B., et al. (2003). The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells 8 645–654. 10.1046/j.1365-2443.2003.00662.x [DOI] [PubMed] [Google Scholar]
- Okami K., Sakai A., Onuki J., Hamano T., Iida M., Takahashi M. (2005). [Promoter hypermethylation of tumor-associated genes in head and neck cancer]. Nihon Jibiinkoka Gakkai Kaiho 108 207–213. 10.3950/jibiinkoka.108.207 [DOI] [PubMed] [Google Scholar]
- Ordóñez-Morán P., Dafflon C., Imajo M., Nishida E., Huelsken J. (2015). HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell 28 815–829. 10.1016/j.ccell.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Oudhoff M. J., Braam M. J. S., Freeman S. A., Wong D., Rattray D. G., Wang J., et al. (2016). SETD7 controls intestinal regeneration and tumorigenesis by regulating Wnt/β-Catenin and hippo/YAP signaling. Dev. Cell 37 47–57. 10.1016/j.devcel.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Paluszczak J., Sarbak J., Kostrzewska-Poczekaj M., Kiwerska K., Jarmuż-Szymczak M., Grenman R., et al. (2015). The negative regulators of Wnt pathway—DACH1, DKK1, and WIF1 are methylated in oral and oropharyngeal cancer and WIF1 methylation predicts shorter survival. Tumor Biol. 36 2855–2861. 10.1007/s13277-014-2913-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. (2010). The hippo signaling pathway in development and cancer. Dev. Cell 19 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Wu L., Tu J., Yu W., Toh Y., Carmon K. S., et al. (2020). Unlike LGR4, LGR5 potentiates Wnt-β-catenin signaling without sequestering E3 ligases. Sci. Signal. 13:eaaz4051. 10.1126/scisignal.aaz4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. S., Ni Y. Y., Chang J. L., Li J., Cadigan K. M. (2008). Wingless signaling induces widespread chromatin remodeling of target loci. Mol. Cell. Biol. 28 1815–1828. 10.1128/mcb.01230-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W. A., Stroud H., Nee K., Liu W., Pezic D., Manakov S., et al. (2014). MORC1 represses transposable elements in the mouse male germline. Nat. Commun. 5:5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Rauskolb C., Williams M., Riggleman B., Wieschaus E. (1991). The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation. Development 111 1029–1043. 10.1242/dev.111.4.1029 [DOI] [PubMed] [Google Scholar]
- Peng K., Kou L., Yu L., Bai C., Li M., Mo P., et al. (2019). Histone demethylase JMJD2D interacts with β-Catenin to induce transcription and activate colorectal cancer cell proliferation and tumor growth in mice. Gastroenterology 156 1112–1126. 10.1053/j.gastro.2018.11.036 [DOI] [PubMed] [Google Scholar]
- Peng K., Su G., Ji J., Yang X., Miao M., Mo P., et al. (2018). Histone demethylase JMJD1A promotes colorectal cancer growth and metastasis by enhancing Wnt/β-catenin signaling. J. Biol. Chem. 293 10606–10619. 10.1074/jbc.ra118.001730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Sun Z., Kuang P., Chen J. (2020). Recent progress on HDAC inhibitors with dual targeting capabilities for cancer treatment. Eur. J. Med. Chem. 208:112831. 10.1016/j.ejmech.2020.112831 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2008). Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319 327–330. 10.1126/science.1149943 [DOI] [PubMed] [Google Scholar]
- Poodineh J., Sirati-Sabet M., Rajabibazl M., Mohammadi-Yeganeh S. (2020). MiR-130a-3p blocks Wnt signaling cascade in the triple-negative breast cancer by targeting the key players at multiple points. Heliyon 6:e05434. 10.1016/j.heliyon.2020.e05434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M., Xia F., Avruch J. (2008). MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18 311–321. 10.1016/j.cub.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang R., Cai N., Wang X., Wang L., Cui K., Wang X., et al. (2016). MLL1 promotes cervical carcinoma cell tumorigenesis and metastasis through interaction with β-catenin. Onco Targets Ther. 9 6631–6640. 10.2147/ott.s114370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real S. A. S., Parveen F., Rehman A. U., Khan M. A., Deo S. V. S., Shukla N. K., et al. (2018). Aberrant promoter methylation of YAP gene and its subsequent downregulation in indian breast cancer patients. BMC Cancer 18:711. 10.1186/s12885-018-4627-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P. C., Gungi A., Ubhe S., Galande S. (2020). Epigenomic landscape of enhancer elements during Hydra head organizer formation. Epigenetics Chromatin 13:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan J. L., Schumacher D., Staudte S., Steffen A., Haybaeck J., Keilholz U., et al. (2017). Non-canonical hedgehog signaling is a positive regulator of the WNT pathway and is required for the survival of colon cancer stem cells. Cell Rep. 21 2813–2828. 10.1016/j.celrep.2017.11.025 [DOI] [PubMed] [Google Scholar]
- Rismani E., Fazeli M. S., Mahmoodzadeh H., Movassagh A., Azami S., Karimipoor M., et al. (2017). Pattern of LRP6 gene expression in tumoral tissues of colorectal cancer. Cancer Biomark. 19 151–159. 10.3233/cbm-160175 [DOI] [PubMed] [Google Scholar]
- Robertson K. D. (2001). DNA methylation, methyltransferases, and cancer. Oncogene 20 3139–3155. 10.1038/sj.onc.1204341 [DOI] [PubMed] [Google Scholar]
- Ruan Z., Lu L., Zhang L., Dong M. (2020). Bone marrow stromal cells-derived microRNA-181-containing extracellular vesicles inhibit ovarian cancer cell chemoresistance by downregulating MEST via the Wnt/β-catenin signaling pathway. Cancer Gene Ther. 10.1038/s41417-020-0195-6 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Albert I., Porfiri E., Munemitsu S., Polakis P. (1997). Loss of beta-catenin regulation by the APC tumor suppressor protein correlates with loss of structure due to common somatic mutations of the gene. Cancer Res. 57 4624–4630. [PubMed] [Google Scholar]
- Sakamoto K. M., Kim K. B., Kumagai A., Mercurio F., Crews C. M., Deshaies R. J. (2001). Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U. S. A. 98 8554–8559. 10.1073/pnas.141230798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahshor S., Woodgett J. R. (2005). The links between axin and carcinogenesis. J. Clin. Pathol. 58 225–236. 10.1136/jcp.2003.009506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel K. E., Chen W., Cope L., Glockner S. C., Suzuki H., Yi J. M., et al. (2005). Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 3:1709–1723. 10.1371/journal.pgen.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbagh M., Borg J.-P. (2014). Insight into planar cell polarity. Exp. Cell Res. 328 284–295. 10.1016/j.yexcr.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Segditsas S., Sieber O. M., Rowan A., Setien F., Neale K., Phillips R. K. S., et al. (2008). Promoter hypermethylation leads to decreased APC mRNA expression in familial polyposis and sporadic colorectal tumours, but does not substitute for truncating mutations. Exp. Mol. Pathol. 85 201–206. 10.1016/j.yexmp.2008.09.006 [DOI] [PubMed] [Google Scholar]
- Semenov M. V., Habas R., Macdonald B. T., He X. (2007). SnapShot: noncanonical Wnt signaling pathways. Cell 131:1378. [DOI] [PubMed] [Google Scholar]
- Seo W. I., Park S., Gwak J., Ju B. G., Chung J. I., Kang P. M., et al. (2017). Wnt signaling promotes androgen-independent prostate cancer cell proliferation through up-regulation of the hippo pathway effector YAP. Biochem. Biophys. Res. Commun. 486 1034–1039. 10.1016/j.bbrc.2017.03.158 [DOI] [PubMed] [Google Scholar]
- Seshagiri S., Stawiski E. W., Durinck S., Modrusan Z., Storm E. E., Conboy C. B., et al. (2012). Recurrent R-spondin fusions in colon cancer. Nature 488 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M. D., Grunstein M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76 75–100. 10.1146/annurev.biochem.76.052705.162114 [DOI] [PubMed] [Google Scholar]
- Shen P., Pichler M., Chen M., Calin G., Ling H. (2017). To Wnt or lose: the missing non-coding linc in colorectal cancer. Int. J. Mol. Sci. 18:2003. 10.3390/ijms18092003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Huang K., Wu Y., Ma Y., Wang J., Qin F., et al. (2017). A miR-135b-TAZ positive feedback loop promotes epithelial–mesenchymal transition (EMT) and tumorigenesis in osteosarcoma. Cancer Lett. 407 32–44. 10.1016/j.canlet.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Shen Y., Gao X., Tan W., Xu T. (2018). STAT1-mediated upregulation of lncRNA LINC00174 functions a ceRNA for miR-1910-3p to facilitate colorectal carcinoma progression through regulation of TAZ. Gene 666 64–71. 10.1016/j.gene.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Shetti D., Zhang B., Fan C., Mo C., Lee B. H., Wei K. (2019). Low dose of paclitaxel combined with XAV939 attenuates metastasis, angiogenesis and growth in breast cancer by suppressing Wnt signaling. Cells 8:892. 10.3390/cells8080892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2007). Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 8 829–833. 10.1038/nrg2218 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (2006). Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75 243–269. 10.1146/annurev.biochem.75.103004.142422 [DOI] [PubMed] [Google Scholar]
- Siegfried E., Chou T. B., Perrimon N. (1992). wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell 71 1167–1179. 10.1016/s0092-8674(05)80065-0 [DOI] [PubMed] [Google Scholar]
- Sierra J., Yoshida T., Joazeiro C. A., Jones K. A. (2006). The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20 586–600. 10.1101/gad.1385806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.-L., Dawson S. N., Arends M. J., Guttula K., Hall N., Cameron E. A., et al. (2014). Boosting Wnt activity during colorectal cancer progression through selective hypermethylation of Wnt signaling antagonists. BMC Cancer 14:891. 10.1186/1471-2407-14-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvis M. R., Kreger B. T., Lien W.-H., Klezovitch O., Rudakova G. M., Camargo F. D., et al. (2011). α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal. 4:ra33. 10.1126/scisignal.2001823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G. E., Pandey S., Nedeljkovic-Kurepa A., Saxena M., Wang A., Pruitt K. (2014). Frizzled 7 expression is positively regulated by SIRT1 and β-Catenin in breast cancer cells. PLoS One 9:e98861. 10.1371/journal.pone.0098861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. C., Rosen L. S., Chugh R., Goldman J. W., Xu L., Kapoun A., et al. (2013). First-in-human evaluation of the human monoclonal antibody vantictumab (OMP-18R5; anti-Frizzled) targeting the WNT pathway in a phase I study for patients with advanced solid tumors. J. Clin. Oncol. 31:2540. 10.1200/jco.2013.31.15_suppl.2540 [DOI] [Google Scholar]
- Söderholm S., Cantù C. (2020). The WNT/β-catenin dependent transcription: a tissue-specific business. Wiley Interdiscip. Rev. Syst. Biol. Med. 13:e1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Nan Y., Wang X., Zhang G., Zong S., Kong X. (2017). MicroRNA-613 inhibits proliferation and invasion of renal cell carcinoma cells through targeting FZD7. Mol. Med. Rep. 16 4279–4286. 10.3892/mmr.2017.7076 [DOI] [PubMed] [Google Scholar]
- Song J., Du Z., Ravasz M., Dong B., Wang Z., Ewing R. M. (2015). A Protein Interaction between β-Catenin and Dnmt1 regulates Wnt signaling and DNA methylation in colorectal cancer cells. Mol. Cancer Res. 13 969–981. 10.1158/1541-7786.mcr-13-0644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks A. B., Morin P. J., Vogelstein B., Kinzler K. W. (1998). Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 58 1130–1134. [PubMed] [Google Scholar]
- Steinmann K., Sandner A., Schagdarsurengin U., Dammann R. H. (2009). Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol. Rep. 22 1519–1526. [DOI] [PubMed] [Google Scholar]
- Strano S., Munarriz E., Rossi M., Castagnoli L., Shaul Y., Sacchi A., et al. (2001). Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 276 15164–15173. 10.1074/jbc.m010484200 [DOI] [PubMed] [Google Scholar]
- Su L. K., Kinzler K. W., Vogelstein B., Preisinger A. C., Moser A. R., Luongo C., et al. (1992). Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256 668–670. 10.1126/science.1350108 [DOI] [PubMed] [Google Scholar]
- Su L. K., Vogelstein B., Kinzler K. W. (1993). Association of the APC tumor suppressor protein with catenins. Science 262 1734–1737. 10.1126/science.8259519 [DOI] [PubMed] [Google Scholar]
- Su S., Hong F., Liang Y., Zhou J., Liang Y., Chen K., et al. (2015). Lgr5 Methylation in cancer stem cell differentiation and prognosis-prediction in colorectal cancer. PLoS One 10:e0143513. 10.1371/journal.pone.0143513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Zhu G., Ding X., Lee S. Y., Dou Y., Zhu B., et al. (2014). Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev. 28 622–636. 10.1101/gad.233239.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M., Rao S. R., Thacker P., Chatterjee S., Karunagaran D. (2014). MiR-29b downregulates canonical Wnt signaling by targeting BCL9L and other coactivators of β-catenin in human colorectal cancer cells. J. Cell. Biochem. 115 1974–1984. 10.1002/jcb.24869 [DOI] [PubMed] [Google Scholar]
- Sun L., Chen B., Wu J., Jiang C., Fan Z., Feng Y., et al. (2020). Epigenetic regulation of a disintegrin and metalloproteinase (ADAM) transcription in colorectal cancer cells: involvement of β-Catenin, BRG1, and KDM4. Front. Cell Dev. Biol. 8:581692. 10.3389/fcell.2020.581692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Gabrielson E., Chen W., Anbazhagan R., van Engeland M., Weijenberg M. P., et al. (2002). A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 31 141–149. 10.1038/ng892 [DOI] [PubMed] [Google Scholar]
- Taelman V. F., Dobrowolski R., Plouhinec J.-L., Fuentealba L. C., Vorwald P. P., Gumper I., et al. (2010). Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143 1136–1148. 10.1016/j.cell.2010.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K. I., Moon R. T. (2000). The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149 249–254. 10.1083/jcb.149.2.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Zheng H., Liu X., Zhang W., Zhu J., Wu G., et al. (2016). MicroRNA-1229 overexpression promotes cell proliferation and tumorigenicity and activates Wnt/β-catenin signaling in breast cancer. Oncotarget 7 24076–24087. 10.18632/oncotarget.8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Liu Z., Zhao L., Clemens T. L., Cao X. (2008). Smad7 stabilizes beta-catenin binding to E-cadherin complex and promotes cell-cell adhesion. J. Biol. Chem. 283 23956–23963. 10.1074/jbc.m800351200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube J. H., Malouf G. G., Lu E., Sphyris N., Vijay V., Ramachandran P. P., et al. (2013). Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Sci. Rep. 3:2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe C. J., Moon R. T. (2004). nemo-like kinase is an essential co-activator of Wnt signaling during early zebrafish development. Development 131 2899–2909. 10.1242/dev.01171 [DOI] [PubMed] [Google Scholar]
- Tian Y., Pan Q., Shang Y., Zhu R., Ye J., Liu Y., et al. (2014). MicroRNA-200 (miR-200) cluster regulation by achaete scute-like 2 (Ascl2): impact on the epithelial-mesenchymal transition in colon cancer cells. J. Biol. Chem. 289 36101–36115. 10.1074/jbc.m114.598383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper M. J., Vaz M., Marrone K. A., Brahmer J. R., Baylin S. B. (2020). The emerging role of epigenetic therapeutics in immuno-oncology. Nat. Rev. Clin. Oncol. 17 75–90. 10.1038/s41571-019-0266-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A., Panciera T., Piccolo S. (2018). YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20 888–899. 10.1038/s41556-018-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovell H., Testa A., Zhou H., Shpiro N., Crafter C., Ciulli A., et al. (2019). Design and characterization of SGK3-PROTAC1, an isoform specific SGK3 Kinase PROTAC degrader. ACS Chem. Biol. 14 2024–2034. 10.1021/acschembio.9b00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji S., Kawasaki Y., Furukawa S., Taniue K., Hayashi T., Okuno M., et al. (2014). The miR-363-GATA6-Lgr5 pathway is critical for colorectal tumourigenesis. Nat. Commun. 5:3150. 10.1038/ncomms4150 [DOI] [PubMed] [Google Scholar]
- Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G. (2003). Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5 914–920. 10.1038/ncb1050 [DOI] [PubMed] [Google Scholar]
- Valenta T., Hausmann G., Basler K. (2012). The many faces and functions of β-catenin. EMBO J. 31 2714–2736. 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X., Miller B. W., Sopko R., Song S., Gregorieff A., Fellouse F. A., et al. (2010a). The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell 18 579–591. [DOI] [PubMed] [Google Scholar]
- Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B. M., Dembowy J., et al. (2008). TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 10 837–848. 10.1038/ncb1748 [DOI] [PubMed] [Google Scholar]
- Varelas X., Samavarchi-Tehrani P., Narimatsu M., Weiss A., Cockburn K., Larsen B. G., et al. (2010b). The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev. Cell 19 831–844. 10.1016/j.devcel.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Varnat F., Zacchetti G., Altaba A. R. I. (2010). Hedgehog pathway activity is required for the lethality and intestinal phenotypes of mice with hyperactive Wnt signaling. Mech. Dev. 127 73–81. 10.1016/j.mod.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Vassilev A., Kaneko K. J., Shu H., Zhao Y., DePamphilis M. L. (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15 1229–1241. 10.1101/gad.888601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeck J., Noetzel E., Bektas N., Jost E., Hartmann A., Knüchel R., et al. (2008). Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumor-specific epigenetic marker in human breast cancer. Mol. Cancer 7:83. 10.1186/1476-4598-7-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T., Yuan T., Chao C.-M., Bell H., Sitaula A., Szimmtenings L., et al. (2017). Fgf10-hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43 48–59.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshanenko O., Gmach P., Winter J., Kranz D., Boutros M. (2017). Mapping of Wnt-Frizzled interactions by multiplex CRISPR targeting of receptor gene families. FASEB J. 31 4832–4844. 10.1096/fj.201700144r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. A., Arend R. C. (2020). Harnessing Wnt signaling as a targetable therapy in epithelial ovarian cancer. Ann. Transl. Med. 8:837. 10.21037/atm.2020.03.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L., Vandel L., Bienz M. (2001). Teashirt is required for transcriptional repression mediated by high wingless levels. EMBO J. 20 137–145. 10.1093/emboj/20.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Tan C., Wen Y., Zhang D., Li G., Chang L., et al. (2019). FOXP1-induced lncRNA CLRN1-AS1 acts as a tumor suppressor in pituitary prolactinoma by repressing the autophagy via inactivating Wnt/β-catenin signaling pathway. Cell Death Dis. 10:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Huang B., Zhang S., Yu X., Wu W., Wang X. (2013). Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 27 1339–1344. 10.1101/gad.219360.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Yuen S. T., Xu J., Lee S. P., Yan H. H. N., Shi S. T., et al. (2014). Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 46 573–582. 10.1038/ng.2983 [DOI] [PubMed] [Google Scholar]
- Wang L., Bu P., Ai Y., Srinivasan T., Chen H. J., Xiang K., et al. (2016). A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife 5:e14620. 10.7554/eLife.14620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shalek A. K., Lawrence M., Ding R., Gaublomme J. T., Pochet N., et al. (2014). Somatic mutation as a mechanism of Wnt/β-catenin pathway activation in CLL. Blood 124 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang O., Huang Y., Wu H., Zheng B., Lin J., Jin P. (2018). LncRNA LOC728196/miR-513c axis facilitates glioma carcinogenesis by targeting TCF7. Gene 679 119–125. 10.1016/j.gene.2018.08.081 [DOI] [PubMed] [Google Scholar]
- Wang S., Ji J., Song J., Li X., Han S., Lian W., et al. (2016). MicroRNA-182 promotes pancreatic cancer cell proliferation and migration by targeting β-TrCP2. Acta Biochim. Biophys. Sin. 48 1085–1093. 10.1093/abbs/gmw105 [DOI] [PubMed] [Google Scholar]
- Wang T., Qin Z.-Y., Wen L.-Z., Guo Y., Liu Q., Lei Z.-J., et al. (2018). Epigenetic restriction of Hippo signaling by MORC2 underlies stemness of hepatocellular carcinoma cells. Cell Death Differ. 25 2086–2100. 10.1038/s41418-018-0095-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lam E. K. Y., Zhang J., Jin H., Sung J. J. Y. (2009). MicroRNA-122a functions as a novel tumor suppressor downstream of adenomatous polyposis coli in gastrointestinal cancers. Biochem. Biophys. Res. Commun. 387 376–380. 10.1016/j.bbrc.2009.07.034 [DOI] [PubMed] [Google Scholar]
- Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., et al. (2015). The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16 413–425. 10.1016/j.stem.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Wierzbicki P. M., Adrych K., Kartanowicz D., Stanislawowski M., Kowalczyk A., Godlewski J., et al. (2013). Underexpression of LATS1 TSG in colorectal cancer is associated with promoter hypermethylation. World J. Gastroenterol. 19 4363–4373. 10.3748/wjg.v19.i27.4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.-J., Shi J., Hu G., Yu X., Lu H., Yang M.-L., et al. (2016). Wnt/β-catenin signaling inhibits FBXW7 expression by upregulation of microRNA-770 in hepatocellular carcinoma. Tumour Biol. 37 6045–6051. 10.1007/s13277-015-4452-5 [DOI] [PubMed] [Google Scholar]
- Xiang L., Chen L.-M., Zhai Y.-J., Sun W.-J., Yang J.-R., Fan Y.-C., et al. (2021). Hypermethylation of secreted frizzled related protein 2 gene promoter serves as a noninvasive biomarker for HBV-associated hepatocellular carcinoma. Life Sci. 270:119061. 10.1016/j.lfs.2021.119061 [DOI] [PubMed] [Google Scholar]
- Xiang T., Li L., Yin X., Zhong L., Peng W., Qiu Z., et al. (2013). Epigenetic silencing of the WNT antagonist Dickkopf 3 disrupts normal Wnt/β−catenin signalling and apoptosis regulation in breast cancer cells. J. Cell. Mol. Med. 17 1236–1246. 10.1111/jcmm.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zamponi R., Charlat O., Ramones M., Swalley S., Jiang X., et al. (2013). Interaction with both ZNRF3 and LGR4 is required for the signalling activity of R-spondin. EMBO Rep. 14 1120–1126. 10.1038/embor.2013.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Takemaru K.-I., Liu J., Berndt J. D., Zheng J. J., Moon R. T., et al. (2008). Crystal structure of a full-length beta-catenin. Structure 16 478–487. 10.1016/j.str.2007.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Mo Y. Y. (2012). The Akt-associated microRNAs. Cell. Mol. Life Sci. 69 3601–3612. 10.1007/s00018-012-1129-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Zhang H., Chong Y., Guan B., Guo P. (2019). YAP promotes VEGFA expression and tumor angiogenesis though Gli2 in human renal cell carcinoma. Arch. Med. Res. 50 225–233. 10.1016/j.arcmed.2019.08.010 [DOI] [PubMed] [Google Scholar]
- Xu T., Wang W., Zhang S., Stewart R. A., Yu W. (1995). Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121 1053–1063. 10.1242/dev.121.4.1053 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhong Y.-D., Zhao X.-X. (2020). MiR-548b suppresses proliferative capacity of colorectal cancer by binding WNT2. Eur. Rev. Med. Pharmacol. Sci. 24 10535–10541. [DOI] [PubMed] [Google Scholar]
- Yagi R., Chen L. F., Shigesada K., Murakami Y., Ito Y. (1999). A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18 2551–2562. 10.1093/emboj/18.9.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Tang X., Wang Z., Wu X., Tang D., Wang D. (2019). miR-125 inhibits colorectal cancer proliferation and invasion by targeting TAZ. Biosci. Rep. 39 10.1042/BSR20190193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Jiang W., Yang W., Yang C., Yang X., Chen K., et al. (2021). Epigenetically modulated miR-1224 suppresses the proliferation of HCC through CREB-mediated activation of YAP signaling pathway. Mol. Ther. Nucleic Acids 23 944–958. 10.1016/j.omtn.2021.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xing Y., Liang C., Hu L., Xu F., et al. (2015). Crucial microRNAs and genes of human primary breast cancer explored by microRNA-mRNA integrated analysis. Tumor Biol. 36 5571–5579. 10.1007/s13277-015-3227-3 [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhao Y., Hu N., Zhao J., Bai Y. (2019). lncRNA KIAA0125 functions as a tumor suppressor modulating growth and metastasis of colorectal cancer via Wnt/β-catenin pathway. Cell Biol. Int. 43 1463–1470. 10.1002/cbin.11196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q., An Y., Hou W., Cao Y.-N., Yao M.-F., Ma N.-N., et al. (2020). Correction: LRP6 promotes invasion and metastasis of colorectal cancer through cytoskeleton dynamics. Oncotarget 11:3102. 10.18632/oncotarget.27710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying J., Li H., Yu J., Ng K. M., Poon F. F., Wong S. C. C., et al. (2008). WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/ -Catenin signaling, and is frequently methylated in colorectal cancer. Clin. Cancer Res. 14 55–61. 10.1158/1078-0432.ccr-07-1644 [DOI] [PubMed] [Google Scholar]
- Ying L., Mills J. A., French D. L., Gadue P. (2015). OCT4 coordinates with WNT signaling to pre-pattern chromatin at the SOX17 locus during human ES cell differentiation into definitive endoderm. Stem Cell Rep. 5 490–498. 10.1016/j.stemcr.2015.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N. N., Pate K. T., Sprowl S., Waterman M. L. (2010). A role for YY1 in repression of dominant negative LEF-1 expression in colon cancer. Nucleic Acids Res. 38 6375–6388. 10.1093/nar/gkq492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H., Matsubara K., Zhou X., Okamura S., Kubo T., Murase Y., et al. (2007). WNT10B functional dualism: β-Catenin/Tcf-dependent growth promotion or independent suppression with deregulated expression in cancer. Mol. Biol. Cell 18 4292–4303. 10.1091/mbc.e06-10-0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tao Q., Cheng Y. Y., Lee K. Y., Ng S. S. M., Cheung K. F., et al. (2009). Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer 115 49–60. 10.1002/cncr.23989 [DOI] [PubMed] [Google Scholar]
- Yu J., Xie Y., Li M., Zhou F., Zhong Z., Liu Y., et al. (2019). Association between SFRP promoter hypermethylation and different types of cancer: a systematic review and meta-analysis. Oncol. Lett. 18 3481–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shay J. W. (2017). Multiple roles of APC and its therapeutic implications in colorectal cancer. J. Natl. Cancer Inst. 109:djw332. 10.1093/jnci/djw332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bai H., David K. K., Dong J., Zheng Y., Cai J., et al. (2010). The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19 27–38. 10.1016/j.devcel.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Bill K., Liu J., Young E., Peng T., Bolshakov S., et al. (2012). MiR-155 is a liposarcoma oncogene that targets casein kinase-1α and enhances β-catenin signaling. Cancer Res. 72 1751–1762. 10.1158/0008-5472.can-11-3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C.-Y., Guan K.-L. (2010). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24 72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Dong H., Xu Q., Zhang Y. (2020). Histone deacetylase (HDAC) inhibitors in cancer: a patent review (2017-present). Expert Opin. Ther. Pat. 30 263–274. 10.1080/13543776.2020.1725470 [DOI] [PubMed] [Google Scholar]
- Zheng A., Song X., Zhang L., Zhao L., Mao X., Wei M., et al. (2019). Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J. Exp. Clin. Cancer Res. 38:305. 10.1186/s13046-019-1315-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Liu Y., Kahn M., Ann D. K., Han A., Wang H., et al. (2012). Interactions between β-catenin and transforming growth factor-β signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP). J. Biol. Chem. 287 7026–7038. 10.1074/jbc.m111.276311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Hao Y., Liu N., Raptis L., Tsao M.-S., Yang X. (2011). TAZ is a novel oncogene in non-small cell lung cancer. Oncogene 30 2181–2186. 10.1038/onc.2010.606 [DOI] [PubMed] [Google Scholar]
- Zhu Q., Fang L., Heuberger J., Kranz A., Schipper J., Scheckenbach K., et al. (2019). The Wnt-driven Mll1 epigenome regulates salivary gland and head and neck cancer. Cell Rep. 26 415–428.e5. [DOI] [PubMed] [Google Scholar]