Abstract
The severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) infection displays a wide array of clinical manifestations. Although some risk factors for coronavirus disease 2019 (COVID-19) severity and outcomes have been identified the underlying biologic mechanisms are still not well understood. The surface SARS-CoV-2 proteins are heavily glycosylated enabling host cell interaction and viral entry. Angiotensin-converting enzyme 2 (ACE2) has been identified to be the main host cell receptor enabling SARS-CoV-2 cell entry after interaction with its S glycoprotein. However, recent studies report SARS-CoV-2 S glycoprotein interaction with other cell receptors, mainly C-type lectins which recognize specific glycan epitopes facilitating SARS-CoV-2 entry to susceptible cells. Here, we are summarizing the main findings on SARS-CoV-2 interactions with ACE2 and other cell membrane surface receptors and soluble lectins involved in the viral cell entry modulating its infectivity and potentially playing a role in subsequent clinical manifestations of COVID-19.
Graphical abstract
Keywords: COVID-19, Glycans, Lectins, SARS-CoV-2, S glycoprotein
Introduction
The severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) has caused a pandemic in 2019 and became one of the major threats to human health around the world [1, 2]. SARS-CoV-2 is an enveloped virus, and its genome consists of a positive-sense single-stranded RNA, which encodes four structural proteins, the spike (S) glycoprotein, the membrane (M) protein, the envelope (E) protein, and the nucleocapsid (N) protein [3]. SARS-CoV-2 is the seventh coronavirus (CoV) known to infect humans [4].
CoVs interact with the host cell membrane through a homotrimeric S glycoprotein extending from their viral envelope. Proteolytic activation of SARS-CoV-2 S glycoprotein by furin cleavage at S1/S2 site results in the generation of S1 subunit, containing receptor-binding domain (RBD), and S2 subunit, containing cell fusion machinery of SARS-CoV-2. Moreover, S1 stabilizes the S2 subunit in its pre-fusion conformation [5]. Interaction of RBD with SARS-CoV-2 cell entry receptor and proteolytic cleavage at S2’ site results in additional conformational changes in S2 subunit allowing viral and host membrane fusion [6].
Each protomer of the homotrimeric S glycoprotein contains 22 N-glycosylation sites and three O-glycosylation sites [4, 7, 8]. Such heavy glycosylation is important for host cell recognition, binding, and viral entry [8]. Several glycoproteomic studies characterized S glycoprotein glycosylation, but with some contradictory results. Watanabe et al. showed that all 22 N-glycosylation sites are occupied most of the time, predominantly with high-mannose or complex N-glycans with a low abundance of hybrid glycans [7]. On the other hand, Shajahan et al. observed partial N-glycan occupancy on 17 out of 22 N-glycosylation sites exclusively with high-mannose and complex-type glycans and found no hybrid type N-glycans [9]. Recent in-depth N-glycan and O-glycopeptide analysis of S glycoprotein mainly detected oligomannose and complex-type glycans with relative abundances of individual glycans differing between full-length recombinant S glycoprotein, S1 and S2 domains. Oligomanose N-glycans were mainly detected in the full-length S glycoprotein, while most complex glycans were core fucosylated and a large portion was neutral with N-acetylglucosamine (GlcNAc) or N-acetyllactosamine (LacNAc) as terminal monomers [10]. Differences between these data indicate the complexity of glycoproteomic analysis and more importantly shed light on the importance of the origin of S glycoprotein (e.g. expression system, culture conditions) on its final glycosylation status and consequently structure and function.
S glycoprotein is glycosylated by hijacking the host glycosylation mechanisms after SARS-CoV-2 replication resulting in a glycosylation pattern that has a signature of the host cells. By using host glycosylation machinery, viruses incorporate „self “glycans in their structure enabling them to evade the host immune response and potentially influence binding to other cell receptors to increase infectivity. On the other hand, clusters of glycans, e.g. high-mannose glycans on HIV-1 gp120, present glycoepitopes serving as immune system targets [11, 12].
Moreover, the binding of SARS-CoV-2 to host cell receptor angiotensin-converting enzyme 2 (ACE2) is facilitated by the interaction of glycosaminoglycan heparan sulfate of cellular glycocalyx with the RBD of SARS-CoV-2 S glycoprotein, in a way to shift the spike structure to an open conformation [13]. Also, Awasthi et al. found that S glycoprotein from SARS-CoV-2 has a sialic acid-binding pocket at its N-terminus, indicating that sialic acid-containing carbohydrates play a role in SARS-CoV-2 host cell binding [14].
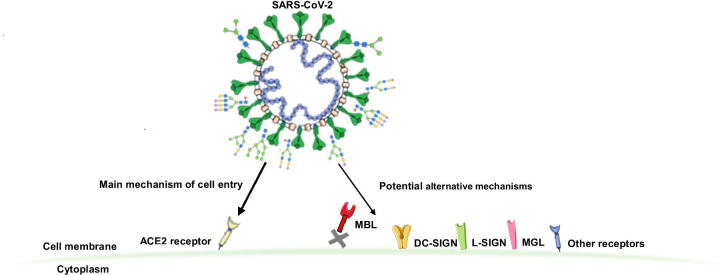
Although ACE2 is broadly expressed in different mammalian tissues [15, 16] and has been recognized as the main host cell receptor for SARS-CoV-2 cell entry [17–19], some studies have reported low expression of ACE2 in the respiratory system [15, 20, 21] indicating that S glycoprotein could interact with other receptors to enter the host cells causing systemic infection [10, 22]. Recent NMR studies showed that N-glycans of the RBD at glycosylation sites N331 and N343 bind to macrophage galactose lectin (MGL), human lectins galectin-3, -7 and -8, sialic acid-binding immunoglobulin-type lectin (Siglec)-10 and dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) [23]. These lectins recognize specific epitopes in complex-type glycans, indicating that additional mechanisms could be involved in tropism and SARS-CoV-2 binding to host cells analogous to SARS-CoV-1 [24].
In this review, we will summarize the main findings on SARS-CoV-2 interactions with ACE2 and other cell receptors, primarily lectins which bind to specific glycan epitopes on SARS-CoV-2 potentially enabling its cell entry and subsequent infection.
Angiotensin-converting enzyme 2
ACE2 is a zinc-containing carboxypeptidase in the renin-angiotensin system that converts angiotensin I to angiotensin II regulating heart function, mainly by maintaining fluid and electrolyte homeostasis and blood pressure [19]. Several research studies have shown that SARS-CoV-2 entries a host cell via binding of RBD of its S glycoprotein to a human receptor ACE2 (hACE2) [6] by electrostatic interactions [25]. The affinity of SARS-CoV-2 to hACE2 was shown to be 10–20 times higher compared to SARS-CoV-1 affinity to the same receptor [26], while other studies have shown small differences in affinity for hACE2 between these two viruses [27] and shown different transmission rates [28]. Several studies have reported structures of SARS-CoV-2 RBD and ACE2 complex revealing their detailed interactions [29–32]. It was shown that 17 amino acids in the extended loop of RBD, referred to as the receptor-binding motif (RBM), are in a network of hydrophilic interactions with 20 amino acids in ACE2. Additionally, Lys417 (Val404 in SARS-CoV-1) in the core subdomain of SARS-CoV-2 RBD, outside the RBM, forms strong ionic interactions with Asp30 of ACE2 [31, 33, 34].
Glycans of both S glycoprotein and hACE2 affect their interactions and this binding occurs by S glycoprotein transiently sampling the hACE2 functional conformation. It was demonstrated by molecular dynamic simulations that S glycoprotein N-glycans at sites N165 and N234 directly modulate the interaction with ACE2 by stabilization of RBD conformation [8, 35]. After binding of N-terminal part of S1 domain to ACE2, receptor transmembrane protease serine 2 (TMPSRR2) cleaves SARS-CoV-2 S glycoprotein between domains S1 and S2. Conformational changes of the S2 domain after the S1 detachment enable fusion of viral and host cell membrane [19].
It was suggested that one of the mechanisms for SARS-CoV-2 high transmission rates is its binding to receptors other than hACE2, mainly human lectins. Previous studies have shown that eight glycosylation sites of SARS-CoV-1 interact with receptors DC-SIGN and liver/lymph node-specific intercellular adhesion molecule-3-grabbing non-integrin (L-SIGN), of which six of them are conserved in SARS-CoV-2 implicating they might be involved in similar interactions [10, 24, 36]. Moreover, hACE2 expression in lung and upper airway, thymus, spleen, pancreas, liver, ileum, and colon was shown to be generally low and is mainly restricted to epithelial cells of severe COVID-19 cases. On the other hand, mannose receptor (MR) and MGL were expressed at a higher level, especially in lung and upper airway cells, and mostly restricted to macrophages and dendritic cells (DCs), which also produce DC-SIGN but to a lower extent. L-SIGN is mostly expressed in epithelial alveolar cells, lymphatic tissues in the ileum, endothelial cells in the pancreas and liver, and at lower levels in immune cells [10, 22]. Expression levels of MGL and DC-SIGN were increased in severe COVID-19 patients with an elevated amount of proinflammatory monocyte-derived macrophages and inflammatory cytokines and chemokines (including IL-6, IL-1B, TNF, CXCL10, CXCL8, CCL2, and CCL3) [10]. Moreover, genetic variants in mannose-binding lectin (MBL) were associated with COVID-19 severity [37]. Low expression of ACE2 in bronchoalveolar and other innate immune cells, and higher expression of C-type lectins (CTLs) indicate that MR, MGL, and DC-SIGN might act as alternative receptors and entry routes for SARS-CoV-2 [10].
In addition to CTLs, other receptors like toll-like receptors (TLR), neuropilin-1 (NRP1), and the non-immune receptor glucose-regulated protein 78 (GRP78) have been reported to interact with SARS-CoV-2 [38].
C-type lectin receptors
C-type lectin receptors (CLRs) are found on antigen-presenting cells (APCs), mainly DCs and macrophages, which are widely present in lung tissue and respiratory mucosa. CLRs are involved in the detection of carbohydrate molecular patterns found on pathogens, and consequently in immune response [39, 40]. The expression of a wide variety of CLRs on different types of innate immune cells enables fine-tuning of immune response after carbohydrate ligand binding [41]. Under inflammatory conditions, monocytes that usually circulate in the blood, bone marrow, and spleen infiltrate mucosal or inflamed tissues where they differentiate into DCs or macrophages. While DCs are localized at mucous and epithelial interfaces (including nasopharyngeal and pulmonary mucosae), macrophages are found in the lung alveoli. It was postulated that CLR and APCs play a role in SARS-CoV-2 cell attachment and immune system activation [41]. Approximately one-third of SARS-CoV-2 S glycoprotein N-glycans are of oligomannose type [7] making them suitable ligands for DC-SIGN, L-SIGN, and Langerin.
Many enveloped viruses use CLRs and other molecules at the host cell surface as means to an end, e.g. for transfer to their cell entry receptors that will enable viral and host membranes fusion. For example, HIV uses DC-SIGN for DCs infection (cis-infection) and T cells infection (trans-infection) [42]. Enhancement of SARS-CoV-1 infection has been associated with DC-SIGN and L-SIGN involvement [43–45], while L-SIGN has also been implicated to have a protective role in SARS-CoV-1 infection [46]. Earlier research has demonstrated the binding of Fc-CLR constructs based on MGL and DC-SIGN to a commercial SARS-CoV-2 RBD domain [47]. Of cautionary note that Fc-CRD constructs might have different avidity properties compared to the entire CLR ectodomain, and RBD on the other hand contains 2 N-glycosylation sites, while 66 N-glycosylation sites are found on the whole S glycoprotein, potentially leading to results that would otherwise not have been found in the case of full-structure proteins [41].
Recent research has confirmed that CTLs DC-SIGN, L-SIGN, MGL, and Langerin bind glycans of the SARS-CoV-2 S-glycoprotein via multiple interactions [41]. Interactions of S glycoprotein with DC-SIGN, L-SIGN, MGL, and additionally MR have also been reported by another study [10]. It was shown that ACE2, DC-SIGN, L-SIGN, MR and MGL all bind SARS-CoV-2 S glycoprotein in a dose-dependent manner, with ACE2 showing the highest affinity; DC-SIGN, MR and MGL strong affinity, and L-SIGN moderate affinity [10]. The interaction of DC-SIGN, L-SIGN, MGL, and MR with SARS-CoV-2 was shown to be Ca2+-dependent and S glycoprotein glycosylation-dependent [10]. Moreover, it was shown that DC-SIGN and L-SIGN promote SARS-CoV-2 transfer to permissive ACE2+ Vero E6, as well as Calu-3 cells [41]. Both expressions of DC-SIGN and L-SIGN are induced by proinflammatory cytokines, e.g. IL-4, IL-6, IL-10, and IL-13, that are overexpressed in severe COVID-19 patients and SARS [41, 48]. It can be speculated that DC-SIGN binding to SARS-CoV-2 and increased IL-10 levels in severe COVID-19 patients serve as an immune-inhibitory mechanism stimulated by the rapid accumulation of inflammatory cytokines, via a negative feedback loop; while on the other hand, a significant increase of IL-10 stimulates the production of other cytokine storm mediators potentially leading to hyperactivation of immune system exacerbating COVID-19 severity [49]. Also, glycan binding by MGL generally suppresses the immune response which is characterized by increased IL-10 production and induction of regulatory and effector T cells potentially promoting SARS-CoV-2 infection [50, 51].
Langerin is a CLR of Langerhans’ cells, a type of DCs present in all stratified squamous epithelium of mammals [52], specific for high-mannose and sulfated glycans [53]. Although it binds SARS-CoV-2 S glycoprotein glycans (with at least one order of magnitude lower affinity than DC-SIGN, L-SIGN, and MGL) it doesn’t mediate trans-infection of SARS-CoV-2 pseudovirions in a T-lymphocyte cell line [41]. Its role in SARS-CoV-2 infectivity remains inconclusive.
It was suggested by Gadanec et al. [38] that another CLR, C-type lectin-like receptor 2 (CLEC2), could bind SARS-CoV-2 via its O-glycans facilitating SARS-CoV-2 capture and transfer between platelets since it was previously shown that it binds sialylated O-glycans [54]. However, no study has confirmed this yet.
Although recent studies didn’t find any evidence of SARS-CoV-2 binding to other CTL receptors like Dectin-1 [47] and Dectin-2 [10], their potential interaction with SARS-CoV-2 S glycoprotein and role in COVID-12 remains an open question. It is known that Dectin-1 can associate with some TLRs resulting in receptor synergism and amplification of inflammatory response induced by TLRs, and TLR2 has been reported to sense SARS-CoV-2 E protein leading to murine lung inflammation [55].
Mannose-binding lectin
The MBL binds to carbohydrates on the surface of viruses, bacteria, fungi, and protozoa and activates the complement [56, 57]. MBL gene polymorphisms have been reported to associate with susceptibility to SARS-CoV-1 infection [58] and SARS-CoV-2 infection severity [37]. A single N-glycosylation site on SARS-CoV-1 S glycoprotein is critical for interaction with MBL facilitating inhibition of SARS-CoV-1 cell entry [59]. Moreover, MBL can interfere with RBD/ACE2 interaction blocking SARS-CoV-1 attachment to the receptor [60].
In the case of SARS-CoV-2, MBL was shown to bind to the recombinant S glycoprotein trimer from different sources, but not to a non-covalent trimer of S glycoprotein or individual S1 and S2 subunits [37]. It was indicated that this interaction is dependent on SARS-CoV-2 S glycoprotein glycosylation since GlcNAc and D-mannose, MBL ligands, inhibited MBL binding to the S glycoprotein. Molecular modeling results suggested that MBL interacted with glycans on the sites N603, N709 and N1074 (with N709 on a different chain) or N603, N801 and N1074 bridging the S1 and S2 subunits, possibly preventing the S1 detachment and inhibiting SARS-CoV-2 cell entry [37]. Moreover, MBL recognized SARS-CoV-2 variants of concern, showed antiviral activity as well as activated the complement lectin pathway [37].
The SARS-CoV-2 N protein binds to mannose-binding protein-associated serine protease 2 (MASP-2), the key serine protease in the MBL pathway of complement activation. This binding of N protein and MASP-2 results in aberrant complement activation and aggravated inflammatory lung injury [61].
These results suggest that in the early stage of SARS-CoV-2 infection MBL may block the virus entry to the cells, while in the later stages of COVID-19 it may contribute to uncontrolled inflammation via complement activation [37]. Despite these studies, detailed mechanisms remain to be elucidated.
DC-SIGN
DC-SIGN was characterized as a receptor for SARS-CoV-1 S glycoprotein [44] increasing viral cell entry by DC transfer to ACE2+ pneumocytes [45] and recently identified as a mediator of genetic risk in COVID-19 through ABO locus [62]. It recognizes high-mannose glycans as well as ligands containing fucose (some Lewis-type glycans).
Recent research has suggested that both DC-SIGN and L-SIGN bind to S glycoprotein via oligomannose and complex N-glycans [10]. This binding was specific to the full-length recombinant S glycoprotein and S1 domain usually involved in hACE2 recognition, while neither of these two lectins bound to the recombinant S2 domain involved in cell fusion [10]. In the same study, it was shown by N-glycan microarray analysis that core fucose significantly attenuates DC-SIGN and L-SIGN binding of GlcNAc-terminating S glycoprotein N-glycans, making them unlikely ligands for DC-SIGN and L-SIGN [10]. Moreover, Lewis A/X epitope was quantified to be relatively highly abundant in the full-length S glycoprotein and S1 domain potentially serving as a ligand of DC-SIGN and L-SIGN, while N,N’-diacetyllactosamine (LacdiNAc) epitope was most abundant in both S1 and S2 domain potentially serving as a ligand of MGL [10].
In kinetic studies, it was shown that the formation of the complex between DC-SIGN and SARS-CoV-2 S glycoprotein is fast with several areas interacting with DC-SIGN, while the complex of L-SIGN and SARS-CoV-2 S glycoprotein might be more stable [41]. After strong binding of recombinant SARS-CoV-2 S glycoprotein by DC-SIGN+ cells virus was internalized by the cells. The same result was obtained with L-SIGN + cells but to a lesser extent [10]. Binding of DC-SIGN, L-SIGN, and MR was confirmed for native SARS-CoV-2 S glycoprotein expressed by SARS-CoV-2-infected Vero E6 cells, while MGL didn’t bind native S glycoprotein from Vero E6 cells contrary to its binding to recombinant S glycoprotein expressed in HEK293 cells [10]. This finding supports the fact that glycosylation of SARS-CoV-2 S glycoprotein largely depends on the expression system like it was shown for previous studies reporting different results on SARS-CoV-2 S glycoprotein glycosylation pattern [7–9, 27]. Additionally, differential glycosylation of SARS-CoV-2 S glycoprotein can modulate its binding to different cell surface receptors, primarily CTLs, potentially influencing infectivity.
Moreover, DC-SIGN promoted efficient SARS-CoV-2 trans-infection from monocyte-derived dendritic cells (MDDCs) to susceptible Vero E6 cells, the reference ACE2 + cell line for SARS-CoV-2 cell culture, as well as human lung cell line Calu-3 [41]. This SARS-CoV-2 trans-infection of Vero E6 and Calu-3 cells mediated by DC-SIGN was blocked by a DC-SIGN glycomimetic antagonist—PM26, which inhibits the interaction of the lectin receptor and S glycoprotein [41]. Both DC-SIGN and L-SIGN were shown to mediate trans-infection of SARS-CoV-2 pseudovirions in a T-lymphocyte cell line. However, this was not the case for Langerin [41]. Raji B cell line transfected with DC-SIGN didn’t capture SARS-CoV-2 effectively, while transfection with Siglec-1 enabled SARS-CoV-2 capture [63].
L-SIGN
L-SIGN is expressed in endothelial cells and type II alveolar cells in human lungs. Like DC-SIGN, it was characterized as a receptor for SARS-CoV-1 S glycoprotein [43] and it is highly specific for high-mannose glycans.
The role of L-SIGN has been investigated in the SARS-CoV-1 and it was shown that L-SIGN mediates trans- but not cis-infection with SARS-CoV-1 [46]. This was confirmed by Jeffers et al. who have also shown that L-SIGN is a less efficient SARS-CoV-1 receptor than ACE2 [43]. Therefore, in addition to ACE2 and DC-SIGN, L-SIGN may be a receptor facilitating the cellular entry of SARS-CoV-2.
Currently, there are two under review manuscripts that studied the role of L-SIGN as receptors for SARS-CoV-2 cell entry. Gao et al. showed that L-SIGN can recognize SARS-CoV-2 leading to its internalization stimulating infectivity. In the same study, it was determined that ACE2 had the highest binding affinity for SARS-CoV-2, followed by DC-SIGN, while L-SIGN had a lower affinity for SARS-CoV-2 [10]. Amraei et al. showed that L-SIGN binds to the RBD of SARS-CoV-2 to mediate cell entry as well as interacts with ACE2, when both are present, suggesting that heterodimerization of L-SIGN and ACE2 has a role in SARS-CoV-2 cell entry and infection [22]. Furthermore, soluble L-SIGN-Fc neutralized viral entry by 48%, while overexpression of L-SIGN in HEK293 cells increased SARS-CoV-2 viral replication [22].
A study published by Thépaut et al. [41] demonstrated that L-SIGN promoted efficient trans-infection of Vero E6 and Calu-3 cells which was significantly inhibited by anti-DC/L-SIGN antibody. These results suggested a role of L-SIGN in the pathogenesis and cellular entry of SARS-CoV-2.
Macrophage galactose type C-type lectin (MGL)
Second macrophage membrane C-type lectin, MGL has been shown to play a role in viral pathology [64] and it has been suggested as a potential receptor for SARS-CoV-2 cell entry. MGL is expressed in DCs and macrophages in human lungs and upper airway tissue [64]. It recognizes glycans bearing terminal galactose or GlcNAc residues and it was shown to interact with SARS-CoV-2 S glycoprotein [41], indicating that complex N-glycans can anchor SARS-CoV-2 S protein to the cell surface via different receptors, including MGL [64].
It was demonstrated that MGL binding to SARS-CoV-2 S glycoprotein is attenuated by sialic acid since neuraminidase treatment slightly increases MGL binding to S glycoprotein [10]. Chiodo et al. suggest that O-glycans located at Thr323 or Ser325 on the S glycoprotein are involved in MGL binding to SARS-CoV-2 [47] while Gao et al. have identified Thr323 as the only O-glycosylation site containing Tn antigen (N-acetylgalactosamine, GalNAc) which is a ligand for MGL [10]. These findings indicate that both N-glycans and O-glycans of SARS-CoV-2 are involved in MGL binding.
Mannose receptor
The MR is a glycoprotein CLR expressed on DCs and macrophages [65]. Recent research has suggested that MR binds to the highly mannosylated regions of the SARS-CoV-2 S glycoprotein with a strong affinity in HEK293 cells [10]. Also, it was shown that during fungal infection, human monocytes cooperate between TLR2, TLR4, and MR, indicating that receptor synergism between MR and TLR2 and TLR4 may account for the severe inflammation [66]. Altogether, this indicates that the level of COVID-19 severity may at least in part be a consequence of the heavily glycosylated mannose regions of the S glycoprotein binding to MR and synergism with TLRs.
Lectin-like oxidized low-density lipoprotein receptor-1
Lectin-like oxidized low-density lipoprotein receptor-1 (LOX-1) is expressed on DCs [67], and so far, there are no studies that show LOX-1 as a direct receptor for the SARS-CoV-2 S glycoprotein. It was only shown that LOX-1 enhances the expression of DCs, as well as DC-SIGN and L-SIGN, to aid in SARS-CoV-2 cellular entry [38]. Also, LOX-1 can trigger intracellular signaling and increase cellular processes linked to an increased risk of cardiovascular disease [68]. Combadiere et al. showed that COVID-19 patients with severe symptoms have increased expression of LOX-1 compared to those with mild symptoms, indicating that LOX-1 may be a plausible characteristic to identify [69].
Although there are no studies up to this date that investigate the role of LOX-1 as a receptor mediating SARS-CoV-2 cell entry, it is possible that LOX-1 may help other receptors involved in SARS-CoV-2 entry.
Liver and lymph node sinusoidal endothelial cell C-type lectin
Liver and lymph node sinusoidal endothelial cell C-type lectin (LSECtin) is a glycan-binding receptor expressed in hepatic and lymph node sinusoidal endothelial cells, DCs, macrophages and Kupffer cells [65]. LSECtin identifies carbohydrate motifs and glycoprotein structures that contain (N-acetyl-)glucosamine, fucose, and mannose [65]. It has been shown to play a role during a viral invasion, recognition of self- and pathogenic-glycoprotein and carbohydrate signatures present on pathogens [70], clearance of apoptotic cells by macrophages [71], antigen capture and pathogen binding [72] as well as inhibition of T cell activation [73]. Also, it has been shown that LSECtin directly associates with glycans present on the surface of SARS-CoV-1 S glycoprotein, by acting as a co-receptor in conjunction with ACE2 to promote viral entry into host cells [74].
Although the role of LSECtin in human SARS-CoV-2 infection is poorly investigated, studies involving ferrets, intranasally inoculated with SARS-CoV-2, suggest that LSECtin may be potentially involved in viral infection and may utilize additional receptors for infectivity [75]. Therefore, LSECtin has been proposed as a possible receptor employed by SARS-CoV-2 to infect cells through recognition of mannosylated N-glycan and O-glycan moieties present on SARS-CoV-2 S glycoprotein [9, 35]. However, further studies are needed.
Toll-like receptors (TLRs)
TLRs are transmembrane type I glycoproteins expressed in innate immune cells consisting of three structural components [76]. They provide host surveillance by detecting foreign- and self-molecular signatures and identify pathogen-associated molecular patterns (PAMPs) through inflammatory response to eliminate invading pathogens [38, 77]. TLRs are expressed in the nasal cavity (TLR1-7, TLR9) [78], macrophages (TLR1-9) [79], natural killer cells (TLR1, TLR2, TLR4, TLR5, TLR6) [80], neutrophils (TLR1, TLR2, TLR4, TLR5, TLR9) [81], airway epithelial cells (TLR1-10) [82] and in other cell populations [82–85]. It was suggested that TLRs located on the membranes of intracellular organelles that recognize pathogenic nucleic acids could facilitate viral clearance of SARS-CoV-2 [38]. Also, a recent study reported increased expression of TLR2 with COVID-19 severity and significantly increased expression of TLR1, TLR4, TLR5, TLR8 and TLR9 in severe and critical COVID-19 patients [55].
Although the role of TLR1 and TLR6 together with TLR2 is known in fungi [86], gram-positive bacteria [87], and mycobacteria [88], their role during COVID-19 infection still begins to unravel. Choudhury et al., demonstrated by in silico studies that SARS-CoV-2 S glycoprotein could bind to TLR1, TLR4, and TLR6 through hydrogen bonding and hydrophobic interactions, of which, TLR4 would have the highest affinity for the S glycoprotein S1 domain as determined by the molecular docking approach [89]. Also, the association between TLR1 and TLR6 could be facilitated by SARS-CoV-2 S glycoprotein oligomannose-type glycans [38]. Moreover, it was recently reported that TLR2 senses the SARS-CoV-2 E protein leading to induction of TLR2-dependent signaling and production of inflammatory cytokines, including TNF-α and INF-γ. On the other hand, blocking TLR2 signaling in vivo protected against SARS-CoV-2-induced lethality [55].
TLR4 endocytosis of microbial products has been reported during bacterial infection and although it hasn’t been investigated in viral infection it has been proposed by Gadanec et al. that after interaction of surface glycans on the S1 domain of SARS-CoV-2 with TLR4 the virus gets internalized [38]. This pathway would be another alternative route for SARS-CoV-2 cell entry independently of ACE2 expression.
Neuropilin-1
NRP1 is a transmembrane polypeptide [90] mostly expressed in the epithelial surface layer of the human respiratory and olfactory epithelium [91], which acts as a co-receptor for different growth factors [92], and facilitates the regulation of biological processes, including angiogenesis, ganglion genesis [93], and vascular permeability [94]. Recent studies report that NRP1, as a host receptor, mediates cellular entry and infectivity of SARS-CoV-2 [90]. The SARS-CoV-2 S glycoprotein contains a polybasic cleavage site (RRARS) between the S1 and S2 subunits enabling cleavage by furin and other proteases [91, 95]. NRP1 located on the cell surface, on the other hand, is known to bind and is activated by furin-cleaved substrates [91]. Cleavage at the S1/S2 junction has been suggested as an important role for viral infectivity and SARS-CoV-2 pathogenicity [91].
Cantuti-Castelvetri et al. showed that NRP1 alone did not increase HEK293 cell susceptibility to viral infectivity, compared to ACE2. However, NRP1 in co-expression with ACE2, or ACE2 and transmembrane protease serine 2 (TMPRSS2), significantly increased wild type SARS-CoV-2 (containing furin-cleavage site) infectivity [91] suggesting that NRP1 acts in concordance with other host factors, such as ACE2 and TMPRSS2.
Glucose-regulated protein 78 (GRP78)
GRP78 is a non-immune receptor and essential endoplasmic reticulum chaperone protein that controls the unfolded protein response initiated by the accumulation of incorrectly folded or unfolded proteins [38]. Upon accumulation of misfolded and unfolded proteins, GRP78 releases from its receptors and translocates to the plasma membrane where it can interact with viruses via its substrate-binding domain and mediate their cell entry [38].
It was shown that COVID-19 patients have increased gene expression and serum concentrations of GRP78 suggesting its release by damaged airway epithelial cells in response to pulmonary trauma and injury during SARS-CoV-2 infection [96]. Recent predictive in silico study has investigated GRP78 affinity to SARS-CoV-2 S glycoprotein and demonstrated preferred potential binding of RBD regions III and IV of S glycoprotein to substrate-binding domain-beta of GRP78 [97]. However, glycosylation involvement in this modeled interaction hasn’t been studied and remains to be investigated.
Siglecs
Siglecs are a family of carbohydrate receptors expressed on almost all immune cells that bind sialylated glycans and play critical roles in immune cell signaling [47, 98]. Siglec-1 has been reported to participate in HIV-1 trans-infection by mature DCs [99] and is shown to be upregulated on APCs in SARS-CoV-2 infected individuals [100]. It is highly expressed on pulmonary macrophages and activated myeloid cells and was recently shown to recognize different SARS-CoV-2 variants of concern via interaction with ganglioside GM1 containing sialyllactose anchored on the viral membrane [63]. Moreover, Siglec-1-mediated transfer of SARS-CoV-2 to susceptible cells was shown to be more effective than DC-SIGN-mediated trans-infection and was successfully blocked by anti-Siglec-1 monoclonal antibodies [63]. While Siglec-1 expressed on DCs facilitated SARS-CoV-2 trans-infection of target cells, Siglec-1 on macrophages resulted in the higher release of cytokines compared to DCs after viral capture triggering proinflammatory responses and potentially participating in a cytokine-storm associated with severe COVID-19 [63].
Another study has also explored the binding of Siglec-3, -5, -7, -9 and -10 to SARS-CoV-2 S glycoprotein and shown a specific binding of SARS-CoV-2 S glycoprotein to Siglec-3, Siglec-9 and Siglec-10 expressed on myeloid immune cells [47]. Siglec-3 is highly expressed on monocytes and macrophages and predominantly interacts with α2,6 linked sialic acids. Siglec-9 is highly expressed on monocytes, neutrophils, DCs, NK and T cells and predominantly binds to α2,3 linked sialic acids on fucosylated or sulfated oligosaccharides, while Siglec-10 is highly expressed on monocytes, B cells and eosinophils and predominantly recognizes α2,6 linked sialic acids. These results suggest a role of Siglecs in the modulation of monocytes, macrophages, neutrophils, eosinophils and B cells function in COVID-19 hypothetically leading to immune suppression [47].
Galectins
Galectin-3 is a β-galactoside-binding protein expressed on epithelial cells and various inflammatory cells, including DCs, macrophages and Kupffer cells. It’s involved in cell–cell and cell-extracellular matrix interactions and activates various types of cells, e.g. recruits APCs and inflammatory cells to affected sites modulating biological response [101]. Galectin-3 promotes the release of several cytokines (e.g. IL-6, TNF-α) driving a proinflammatory response contributing to airway inflammation. Recently, it was recognized as a potential prognostic biomarker of severe COVID-19, with improved classification performance when used in combination with CRP and albumin, regardless of initial disease severity [102]. Galectin-3 also interacts with neutrophils, whose mobilization is essential in the innate immune response, further driving a pro-inflammatory response. Inhibitors of galectin-3 have been suggested as a potential therapy to attenuate the hyperinflammation in COVID-19 infection [103].
Another galectin, galectin-9 has been identified as a potential biomarker of COVID-19 since it can differentiate SARS-CoV-2 infected and healthy individuals with high specificity/sensitivity [104]. It is secreted by different immune and nonimmune cells and mediates various biological processes, e.g. induces chemotaxis, activates eosinophils, promotes DC maturation, enhances the function of DCs and NK cells, etc. Plasma galectin-9 in COVID-19 patients was positively correlated with a wide range of proinflammatory biomarkers (e.g. IL-6, TNF-α) whose expression and production by immune cells of COVID-19 patients was enhanced by galectin-9 treatment in vitro. Moreover, galectin-9 was downregulated in neutrophils in COVID-19 [104].
Conclusions
Recent research has demonstrated the capabilities of other receptors, in addition to hACE2, to bind to SARS-CoV-2 S glycoprotein enabling trans-infection of susceptible cells potentially facilitating viral transmission. Interaction of SARS-CoV-2 S glycoprotein with additional receptors could emphasize severe inflammation and cytokine release (leading to cytokine storm). While Amraie et al. [22] suggested CLRs could serve as alternative receptors to hACE2 in non-susceptible cells, this hasn’t been confirmed by Thépaut et al. [41]. Detailed trans-infection mechanism and SARS-CoV-2 fate after interaction with CLRs remain an open question.
These late results on glycosylation-mediated CTL interactions with SARS-CoV-2 indicate that SARS-CoV-2 glycosylation possibly determines the immune receptors with which SARS-CoV-2 interact potentially leading to SARS-CoV-2 clearance or resulting in its spread to other organs, and/or other hosts. Moreover, interactions of SARS-CoV-2 with CTLs and subsequent internalization suggest a possible mechanism for asymptomatic COVID-19 clinical manifestation as a result of decreasing the available titer of free SARS-CoV-2 via endocytosis by resident innate immune cells in the lung [10].
In addition to CTLs, other receptors like TLRs, NRP1, GRP78, LOX-1 etc., have been implicated to interact with SARS-CoV-2 and/or potentially facilitate its cell binding and infectivity, although their exact role still has to be investigated.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- APC
Antigen-presenting cell
- CoV
Coronavirus
- CLEC2
C-type lectin-like receptor 2
- CLR
C-type lectin receptor
- COVID-19
Coronavirus disease 2019
- CTL
C-type lectin
- DC
Dendritic cells
- DC-SIGN
Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- GalNAc
N-Acetylgalactosamine
- GlcNAc
N-Acetylglucosamine
- GRP78
Glucose-regulated protein 78
- L-SIGN
Liver/lymph node-specific intercellular adhesion molecule-3-grabbing non-integrin
- LacdiNAc
N,N’-Diacetyllactosamine
- LacNAc
N-Acetyllactosamine
- LSECtin
Liver and lymph node sinusoidal endothelial cell C-type lectin
- LOX-1
Lectin-like oxidized low-density lipoprotein receptor-1
- MASP-2
Mannose-binding protein-associated serine protease 2
- MDDC
Monocyte-derived dendritic cell
- MGL
Macrophage galactose lectin
- MR
Mannose receptor
- NRP1
Neuropilin-1
- PAMPs
Pathogen-associated molecular patterns
- RBD
Receptor-binding domain
- RBM
Receptor-binding motif
- SARS-CoV-1
Severe acute respiratory syndrome-related coronavirus-1
- SARS-CoV-2
Severe acute respiratory syndrome-related coronavirus-2
- Siglec
Sialic acid-binding immunoglobulin-type lectin
- TLR
Toll-like receptors
- TMPRSS2
Transmembrane protease serine 2
Authors' contributions
GL had the idea for the article, IT-A and TP performed the literature search and wrote the first and final version of the manuscript. GL critically revised the work. All authors approved the final version to be published.
Funding
This work has been supported in part by Croatian Science Foundation under the project IP-CORONA-2020–04-2052.
Data Availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Code availability
(software application or custom code).
Code sharing not applicable to this article as no codes were generated during the current study.
Compliance with ethical standards
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of interest
GL is the founder and owner of Genos Ltd, a private research organization that specializes in high-throughput glycomic analysis and has several patents in this field. IT-A and TP are employees of Genos Ltd.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou, P., Yang, X. Lou, Wang, X.G., Hu, B., Zhang, L., Zhang, W., Si, H.R., Zhu, Y., Li, B., Huang, C.L., Chen, H.D., Chen, J., Luo, Y., Guo, H., Jiang, R. Di, Liu, M.Q., Chen, Y., Shen, X.R., Wang, X., Zheng, X.S., Zhao, K., Chen, Q.J., Deng, F., Liu, L.L., Yan, B., Zhan, F.X., Wang, Y.Y., Xiao, G.F., Shi, Z.L.: A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273 (2020). 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed]
- 3.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, Y., Zhang, J., Xiao, T., Peng, H., Sterling, S.M., Walsh, R.M., Rawson, S., Rits-Volloch, S., Chen, B.: Distinct conformational states of SARS-CoV-2 spike protein. Science (80). 369, 1586–1592 (2020). 10.1126/science.abd4251 [DOI] [PMC free article] [PubMed]
- 6.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe, Y., Allen, J.D., Wrapp, D., McLellan, J.S., Crispin, M.: Site-specific glycan analysis of the SARS-CoV-2 spike. Science (80). 369, 330–333 (2020). 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed]
- 8.Zhao P, Praissman J, Grant O, Cai Y, Xiao T, Rosenbalm K, Aoki K, Kellman B, Bridger R, Barouch D, Brindley M, Lewis N, Tiemeyer M, Chen B, Woods R, Wells L. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe. 2020;28:586–601. doi: 10.1101/2020.06.25.172403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shajahan, A., Supekar, N.T., Gleinich, A.S., Azadi, P.: Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 1–20 (2020). 10.1093/glycob/cwaa042 [DOI] [PMC free article] [PubMed]
- 10.Gao, C., Zeng, J., Jia, N., Stavenhagen, K., Matsumoto, Y., Zhang, H., Li, J., Hume, A.J., Mühlberger, E., van Die, I., Kwan, J., Tantisira, K., Emili, A., Cummings, R.D.: SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv Prepr. Serv. Biol. (2020). 10.1101/2020.07.29.227462
- 11.Miller NL, Clark T, Raman R, Sasisekharan R. Glycans in Virus-Host Interactions: A Structural Perspective. Front. Mol. Biosci. 2021;8:1–10. doi: 10.3389/fmolb.2021.666756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritchard, L.K., Spencer, D.I.R., Royle, L., Bonomelli, C., Seabright, G.E., Behrens, A.J., Kulp, D.W., Menis, S., Krumm, S.A., Dunlop, D.C., Crispin, D.J., Bowden, T.A., Scanlan, C.N., Ward, A.B., Schief, W.R., Doores, K.J., Crispin, M.: Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat. Commun. 6, (2015). 10.1038/ncomms8479 [DOI] [PMC free article] [PubMed]
- 13.Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, Thacker BE, Glass CA, Yang Z, Torres JL, Golden GJ, Bartels PL, Porell RN, Garretson AF, Laubach L, Feldman J, Yin X, Pu Y, Hauser BM, Caradonna TM, Kellman BP, Martino C, Gordts PLSM, Chanda SK, Schmidt AG, Godula K, Leibel SL, Jose J, Corbett KD, Ward AB, Carlin AF, Esko JD. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awasthi M, Gulati S, Sarkar DP, Tiwari S, Kateriya S, Ranjan P, Verma SK. The sialoside-binding pocket of SARS-CoV-2 spike glycoprotein structurally resembles MERS-CoV. Viruses. 2020;12:1–10. doi: 10.3390/v12090909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hikmet, F., Méar, L., Edvinsson, Å., Micke, P., Uhlén, M., Lindskog, C.: The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 16, 1–16 (2020). 10.15252/msb.20209610 [DOI] [PMC free article] [PubMed]
- 16.Sun, K., Gu, L., Ma, L., Duan, Y.: Atlas of ACE2 gene expression reveals novel insights into transmission of SARS-CoV-2. Heliyon. 7, (2021). 10.1016/j.heliyon.2020.e05850 [DOI] [PMC free article] [PubMed]
- 17.Xiantian X, Ping C, Jingfang W, Jiannan F, Hui Z, Xuan L, Wu Z, Pei H. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. CHINA Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunyavanich S, Do A, Vicencio A. Nasal Gene Expression of Angiotensin-Converting Enzyme 2 in Children and Adults. JAMA - J. Am. Med. Assoc. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganier, C., Du-Harpur, X., Harun, N., Wan, B., Arthurs, C., Luscombe, N.M., Watt, F.M., Lynch, M.D., MD1, 3: CD147 (BSG) but not ACE2 expression is detectable in vascular endothelial cells within single cell RNA sequencing datasets derived from multiple tissues in healthy individuals. bioRxiv Prepr. Serv. Biol. (2020)
- 22.Amraei R, Yin W, Napoleon MA, Suder EL, Berrigan J, Zhao Q, Olejnik J, Chandler KB, Xia C, Feldman J, Hauser BM, Caradonna TM, Schmidt AG, Gummuluru S, Mühlberger E, Chitalia V, Costello CE, Rahimi N, Muhlberger E, Chitalia V. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. ACS Cent. Sci. 2021;7:1156–1165. doi: 10.1101/2020.06.22.165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenza MP, Oyenarte I, Diercks T, Quintana JI, Gimeno A, Coelho H, Diniz A, Peccati F, Delgado S, Bosch A, Valle M, Millet O, Abrescia NGA, Palazón A, Marcelo F, Jiménez-Osés G, Jiménez-Barbero J, Ardá A, Ereño-Orbea J. Structural Characterization of N-Linked Glycans in the Receptor Binding Domain of the SARS-CoV-2 Spike Protein and their Interactions with Human Lectins. Angew. Chemie - Int. Ed. 2020;59:23763–23771. doi: 10.1002/anie.202011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han DP, Lohani M, Cho MW. Specific Asparagine-Linked Glycosylation Sites Are Critical for DC-SIGN- and L-SIGN-Mediated Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/jvi.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen HL, Lan PD, Thai NQ, Nissley DA, O’Brien EP, Li MS. Does SARS-CoV-2 bind to human ACE2 more strongly than does SARS-CoV? J. Phys. Chem. B. 2020;124:7336–7347. doi: 10.1021/acs.jpcb.0c04511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrapp, D., De Vlieger, D., Corbett, K.S., Torres, G.M., Wang, N., Van Breedam, W., Roose, K., van Schie, L., Team, V.-C.C.-19 R., Hoffmann, M., Po ¨hlmann, S., Graham, B.S., Callewaert, N., Schepens, B., Saelens, X., McLella, J.S.: Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell. 181, 1004–1015 (2020) [DOI] [PMC free article] [PubMed]
- 27.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen E, Koopmans M, Go U, Hamer DH, Petrosillo N, Castelli F, Storgaard M, Al Khalili S, Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 32.Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., Zhou, Q.: Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (80). 367, 1444–8 (2020) [DOI] [PMC free article] [PubMed]
- 33.Watanabe C, Okiyama Y, Tanaka S, Fukuzawa K, Honma T. Molecular recognition of SARS-CoV-2 spike glycoprotein: quantum chemical hot spot and epitope analyses. Chem. Sci. 2021;12:4722–4739. doi: 10.1039/d0sc06528e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rangel, H.R., Ortega, J.T., Maksoud, S., Pujol, F.H., Serrano, M.L.: SARS-CoV-2 host tropism: An in silico analysis of the main cellular factors. Virus Res. 289, 198154 (2020) [DOI] [PMC free article] [PubMed]
- 35.Casalino L, Gaieb Z, Goldsmith JA, Hjorth CK, Dommer AC, Harbison AM, Fogarty CA, Barros EP, Taylor BC, Mclellan JS, Fadda E, Amaro RE. Beyond shielding: The roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih Y-P, Chen C-Y, Liu S-J, Chen K-H, Lee Y-M, Chao Y-C, Chen Y-MA. Identifying Epitopes Responsible for Neutralizing Antibody and DC-SIGN Binding on the Spike Glycoprotein of the Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2006;80:10315–10324. doi: 10.1128/jvi.01138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stravalaci, M., Pagani, I., Paraboschi, E.M., Pedotti, M., Doni, A., Scavello, F., Mapelli, S.N., Sironi, M., Varani, L., Matkovic, M., Cavalli, A., Cesana, D., Gallina, P., Pedemonte, N., Capurro, V., Clementi, N., Mancini, N., Invernizzi, P., Rappuoli, R., Duga, S., Bottazzi, B., Uguccioni, M., Asselta, R., Vicenzi, E., Mantovani, A., Garlanda, C.: Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. medRxiv. 2021.06.07.21258350 (2021). 10.1101/2021.06.07.21258350
- 38.Gadanec LK, McSweeney KR, Qaradakhi T, Ali B, Zulli A, Apostolopoulos V. Can SARS-CoV-2 virus use multiple receptors to enter host cells? Int. J. Mol. Sci. 2021;22:1–35. doi: 10.3390/ijms22030992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geijtenbeek TBH, Gringhuis SI. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi O, Akira S. Pattern Recognition Receptors and Inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Thépaut M, Luczkowiak J, Vivès C, Labiod N, Bally I, Lasala F, Grimoire Y, Fenel D, Sattin S, Thielens N, Schoehn G, Bernardi A, Delgado R, Fieschi F. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLOS Pathog. 2021;17:e1009576. doi: 10.1371/journal.ppat.1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Witte L, Nabatov A, Geijtenbeek TBH. Distinct roles for DC-SIGN+-dendritic cells and Langerhans cells in HIV-1 transmission. Trends Mol. Med. 2008;14:12–19. doi: 10.1016/j.molmed.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD, Thackray LB, Young MD, Mason RJ, Ambrosino DM, Wentworth DE, DeMartini JC, Holmes KV. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marzi A, Gramberg T, Simmons G, Möller P, Rennekamp AJ, Krumbiegel M, Geier M, Eisemann J, Turza N, Saunier B, Steinkasserer A, Becker S, Bates P, Hofmann H, Pöhlmann S. DC-SIGN and DC-SIGNR Interact with the Glycoprotein of Marburg Virus and the S Protein of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/jvi.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z-Y, Huang Y, Ganesh L, Leung K, Kong W-P, Schwartz O, Subbarao K, Nabel GJ. pH-Dependent Entry of Severe Acute Respiratory Syndrome Coronavirus Is Mediated by the Spike Glycoprotein and Enhanced by Dendritic Cell Transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/jvi.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan VSF, Chan KYK, Chen Y, Poon LLM, Cheung ANY, Zheng B, Chan KH, Mak W, Ngan HYS, Xu X, Screaton G, Tam PKH, Austyn JM, Chan LC, Yip SP, Peiris M, Khoo US, Lin CLS. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 2006;38:38–46. doi: 10.1038/ng1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiodo F, Bruijns S, Rodriguez E, Li RJE, Molinaro A, Silipo A, Di Lorenzo F, Garcia-Rivera D, Valdes-Balbin Y, Verez-Bencomo V, van Kooyk Y. Novel ACE2-Independent Carbohydrate-Binding of SARS-CoV-2 Spike Protein to Host Lectins and Lung Microbiota. Biorxiv. 2020 doi: 10.1101/2020.05.13.092478. [DOI] [Google Scholar]
- 48.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Obaid A, Lu-Culligan A, Nelson A, Brito A, Nunez A, Martin A, Watkins A, Geng B, Kalinich C, Harden C, Todeasa C, Jensen C, Kim D, McDonald D, Shepard D, Courchaine E, White EB, Song E, Silva E, Kudo E, DeIuliis G, Rahming H, Park HJ, Matos I, Nouws J, Valdez J, Fauver J, Lim J, Rose KA, Anastasio K, Brower K, Glick L, Sharma L, Sewanan L, Knaggs L, Minasyan M, Batsu M, Petrone M, Kuang M, Nakahata M, Campbell M, Linehan M, Askenase MH, Simonov M, Smolgovsky M, Sonnert N, Naushad N, Vijayakumar P, Martinello R, Datta R, Handoko R, Bermejo S, Prophet S, Bickerton S, Velazquez S, Alpert T, Rice T, Khoury-Hanold W, Peng X, Yang Y, Cao Y, Strong Y, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu L, Zhang H, Dauphars DJ, He Y-W. A Potential Role of Interleukin 10 in COVID-19 Pathogenesis. Trends Immunol. 2021;42:3–5. doi: 10.1016/j.it.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Vliet SJ, Gringhuis SI, Geijtenbeek TBH, van Kooyk Y. Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 2006;7:1200–1208. doi: 10.1038/ni1390. [DOI] [PubMed] [Google Scholar]
- 51.van Vliet SJ, Bay S, Vuist IM, Kalay H, García-Vallejo JJ, Leclerc C, van Kooyk Y. MGL signaling augments TLR2-mediated responses for enhanced IL-10 and TNF-α secretion. J. Leukoc. Biol. 2013;94:315–323. doi: 10.1189/jlb.1012520. [DOI] [PubMed] [Google Scholar]
- 52.Jaitley S, Saraswathi TR. Pathophysiology of Langerhans cells. J. Oral Maxillofac. Pathol. 2012;16:239–244. doi: 10.4103/0973-029X.99077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tateno H, Ohnishi K, Yabe R, Hayatsu N, Sato T, Takeya M, Narimatsu H, Hirabayashi J. Dual specificity of langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J. Biol. Chem. 2010;285:6390–6400. doi: 10.1074/jbc.M109.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cummings, R.D., McEver, R.P.: C-Type Lectins. In: Varki, A., Cummings, R., and Esko, J. (eds.) Essentials of Glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press (2017)
- 55.Zheng M, Karki R, Williams EP, Yang D, Fitzpatrick E, Vogel P, Jonsson CB, Kanneganti TD. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22:829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dommett, R.M., Klein, N., Turner, M.W.: Dommett et al (2006) - Mannose‐binding lectin in innate immunity past, present and future. Tissue Antigens. 63, 193–209 (2006) [DOI] [PMC free article] [PubMed]
- 57.Van Asbeck EC, Hoepelman AIM, Scharringa J, Herpers BL, Verhoef J. Mannose binding lectin plays a crucial role in innate immunity against yeast by enhanced complement activation and enhanced uptake of polymorphonuclear cells. BMC Microbiol. 2008;8:1–10. doi: 10.1186/1471-2180-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ip, W.K.E., Hung Chan, K., Law, H.K.W., Tso, G.H.W., Kong, E.K.P., Wong, W.H.S., Fai To, Y., Yung, R.W.H., Chow, E.Y., Leung Au, K., Chan, E.Y.T., Lim, W., Jensenius, J.C., Turner, M.W., Peiris, J.S.M., Lau, Y.L.: Mannose-Binding Lectin in Severe Acute Respiratory Syndrome Coronavirus Infection. J. Infect. Dis. 191, 1697–1704 (2005). 10.1111/j.1442-200x.2005.02092.x [DOI] [PMC free article] [PubMed]
- 59.Zhou Y, Lu K, Pfefferle S, Bertram S, Glowacka I, Drosten C, Pöhlmann S, Simmons G. A Single Asparagine-Linked Glycosylation Site of the Severe Acute Respiratory Syndrome Coronavirus Spike Glycoprotein Facilitates Inhibition by Mannose-Binding Lectin through Multiple Mechanisms. J. Virol. 2010;84:8753–8764. doi: 10.1128/jvi.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keyaerts E, Vijgen L, Pannecouque C, Van Damme E, Peumans W, Egberink H, Balzarini J, Van Ranst M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 2007;75:179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao, T., Hu, M., Zhang, X., Li, H., Zhu, L., Liu, H., Dong, Q., Zhang, Z., Wang, Z., Hu, Y., Fu, Y., Jin, Y., Li, K., Zhao, S., Xiao, Y., Luo, S., Li, L., Zhao, L., Liu, J., Zhao, H., Liu, Y., Yang, W., Peng, J., Chen, X., Li, P., Liu, Y., Xie, Y., Song, J., Zhang, L., Ma, Q., Bian, X., Chen, W., Liu, X., Mao, Q., Cao, C.: Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv. (2020). 10.1101/2020.03.29.20041962
- 62.Katz, D., Tahir, U., Ngo, D., Benson, M., Bick, A., Pampana, A., Gao, Y., Keyes, M., Correa, A., Sinha, S., Shen, D., Yang, Q., Robbins, J., Chen, Z.-Z., Cruz, D., Peterson, B., Natarajan, P., Vasan, R., Smith, J.G., Wang, T., Wilson, J., Gerszten, R.: Proteomic Profiling in Biracial Cohorts Implicates DC-SIGN as a Mediator of Genetic Risk in COVID-19. medRxiv Prepr. Serv. Heal. Sci. (2020). 10.1101/2020.06.09.20125690
- 63.Perez-Zsolt, A.D., Muñoz-Basagoiti, J., Rodon, J., Paiardini, M., Chojnacki, J., Erkizia, I., Muñiz, X., Tarrés-Freixas, F., Blanco, I., Guallar, V., Carrillo, J., Perez-Zsolt, D., Muñoz-Basagoiti, J., Rodon, J., Elousa, M., Raïch-Regué, D., Risco, C., Sachse, M., Pino, M., Gumber, S., Paiardini, M., Chojnacki, J., Erkizia, I., Muñiz, X., Ballana, E., Riveira-Muñoz, E., Noguera, M., Paredes, R., Trinité, B., Tarrés-Freixas, F., Blanco, I., Guallar, V., Carrillo, J., Blanco, J., Telenti, A., Heyn, H., Segalés, J., Clotet, B., Martinez-Picado, J., Vergara-Alert, J., Izquierdo-Useros, N.: Siglec-1 on dendritic cells mediates SARS-CoV-2 trans-infection of target cells while on macrophages triggers proinflammatory responses. bioRxiv. 2021.05.11.443572 (2021). 10.1101/2021.05.11.443572
- 64.Upham JP, Pickett D, Irimura T, Anders EM, Reading PC. Macrophage Receptors for Influenza A Virus: Role of the Macrophage Galactose-Type Lectin and Mannose Receptor in Viral Entry. J. Virol. 2010;84:3730–3737. doi: 10.1128/jvi.02148-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Apostolopoulos V, Thalhammer T, Tzakos AG, Stojanovska L. Targeting Antigens to Dendritic Cell Receptors for Vaccine Development. J. Drug Deliv. 2013;2013:1–22. doi: 10.1155/2013/869718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakaira-Takahagi E, Golim MA, Bannwart CF, Puccia R, Peraçoli MTS. Interactions between TLR2, TLR4, and mannose receptors with gp43 from Paracoccidioides brasiliensis induce cytokine production by human monocytes. Med. Mycol. 2011;49:694–703. doi: 10.3109/13693786.2011.565485. [DOI] [PubMed] [Google Scholar]
- 67.Joo H, Li D, Dullaers M, Kim T, Duluc D, Upchurch K, Xue Y, Zurawski S, Grand RL, Liu Y, Kuroda M, Zurawski G, Oh S. C-Type Lectin-Like Receptor LOX-1 Promotes Dendritic Cell-Mediated Class-Switched B Cell Responses. Immunity. 2014;41:592–604. doi: 10.1016/j.immuni.2014.09.009.C-Type. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sorokin AV, Karathanasis SK, Yang ZH, Freeman L, Kotani K, Remaley AT. COVID-19—Associated dyslipidemia: Implications for mechanism of impaired resolution and novel therapeutic approaches. FASEB J. 2020;34:9843–9853. doi: 10.1096/fj.202001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Combadiere, B., Adam, L., Quentric, P., Rosenbaum, P., Dorgham, K., Bonduelle, O., Parizot, C., Sauce, D., Mayaux, J., Luyt, C.-E., Boissonnas, A., Amoura, Z., Pourcher, V., Miyara, M., Gorochov, G., Guihot, A., Combadière, C.: LOX-1 + immature neutrophils predict severe COVID-19 patients at risk of thrombotic complications. bioRxiv. 2020.09.15.293100 (2020)
- 70.Zhao D, Han X, Zheng X, Wang H, Yang Z, Liu D, Han K, Liu J, Wang X, Yang W, Dong Q, Yang S, Xia X, Tang L, He F. The Myeloid LSECtin Is a DAP12-Coupled Receptor That Is Crucial for Inflammatory Response Induced by Ebola Virus Glycoprotein. PLoS Pathog. 2016;12:1–20. doi: 10.1371/journal.ppat.1005487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Z, Li Q, Wang X, Jiang X, Zhao D, Lin X, He F, Tang L. C-type lectin receptor LSECtin-mediated apoptotic cell clearance by macrophages directs intestinal repair in experimental colitis. Proc. Natl. Acad. Sci. U. S. A. 2018;115:11054–11059. doi: 10.1073/pnas.1804094115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Domínguez-Soto, Á., Aragoneses-Fenoll, L., Gómez-Aguado, F., Corcuera, M.T., Clária, J., García-Monzón, C., Bustos, M., Corbí, A.L.: The pathogen receptor liver and lymph node sinusoidal endotelial cell C-type lectin is expressed in human Kupffer cells and regulated by PU.1. Hepatology. 49, 287–296 (2009). 10.1002/hep.22678 [DOI] [PMC free article] [PubMed]
- 73.Wang, J., Manick, B., Renelt, M., Gerassenkov, T., Bi, M., Kalabokis, V., Person, A., Wu, G.: LSECtin interacts with BTN3A1 to inhibit T cell activation. J. Immunol. 204, 78.6 (2020)
- 74.Gramberg T, Hofmann H, Möller P, Lalor PF, Marzi A, Geier M, Krumbiegel M, Winkler T, Kirchhoff F, Adams DH, Becker S, Münch J, Pöhlmann S. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340:224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli KP, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Godfroy JI, Roostan M, Moroz YS, Korendovych IV, Yin H. Isolated Toll-like Receptor Transmembrane Domains Are Capable of Oligomerization. PLoS ONE. 2012;7:1–14. doi: 10.1371/journal.pone.0048875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeda K, Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 78.Van Tongeren, J., Röschmann, K.I.L., Reinartz, S.M., Luiten, S., Fokkens, W.J., De Jong, E.C., Van Drunen, C.M.: Expression profiling and functional analysis of Toll-like refile:///D:/Genos - Znanstveni radovi/COVID-19 and glycosylation/Koller et al (2008) - TLR Expression on Neutrophils at the Pulmonary Site of Infection.pdf file:///D:/Genos - Znanstveni radovi/COV. Clin. Transl. Allergy. 5, 1–9 (2015). 10.1186/s13601-015-0086-3 [DOI] [PMC free article] [PubMed]
- 79.Hoppstädter J, Diesel B, Zarbock R, Breinig T, Monz D, Koch M, Meyerhans A, Gortner L, Lehr CM, Huwer H, Kiemer AK. Differential cell reaction upon Toll-like receptor 4 and 9 activation in human alveolar and lung interstitial macrophages. Respir. Res. 2010;11:1–15. doi: 10.1186/1465-9921-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Freeman CM, Martinez FJ, Han MLK, Washko GR, McCubbrey AL, Chensue SW, Arenberg DA, Meldrum CA, McCloskey L, Curtis JL. Lung CD8+ T cells in COPD have increased expression of bacterial TLRs. Respir. Res. 2013;14:1. doi: 10.1186/1465-9921-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koller B, Kappler M, Latzin P, Gaggar A, Schreiner M, Takyar S, Kormann M, Kabesch M, Roos D, Griese M, Hartl D. TLR Expression on Neutrophils at the Pulmonary Site of Infection: TLR1/TLR2-Mediated Up-Regulation of TLR5 Expression in Cystic Fibrosis Lung Disease. J. Immunol. 2008;181:2753–2763. doi: 10.4049/jimmunol.181.4.2753. [DOI] [PubMed] [Google Scholar]
- 82.Mayer AK, Muehmer M, Mages J, Gueinzius K, Hess C, Heeg K, Bals R, Lang R, Dalpke AH. Differential Recognition of TLR-Dependent Microbial Ligands in Human Bronchial Epithelial Cells. J. Immunol. 2007;178:3134–3142. doi: 10.4049/jimmunol.178.5.3134. [DOI] [PubMed] [Google Scholar]
- 83.Esnault S, Bernau K, Torr EE, Bochkov YA, Jarjour NN, Sandbo N. RNA-sequencing analysis of lung primary fibroblast response to eosinophil-degranulation products predicts downstream effects on inflammation, tissue remodeling and lipid metabolism. Respir. Res. 2017;18:1–16. doi: 10.1186/s12931-017-0669-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kulka M, Metcalfe DD. TLR3 activation inhibits human mast cell attachment to fibronectin and vitronectin. Mol. Immunol. 2006;43:1579–1586. doi: 10.1016/j.molimm.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 85.Thorley, A.J., Grandolfo, D., Lim, E., Goldstraw, P., Young, A., Tetley, T.D.: Innate immune responses to bacterial ligands in the peripheral human Lung - Role of alveolar epithelial TLR expression and signalling. PLoS One. 6, (2011). 10.1371/journal.pone.0021827 [DOI] [PMC free article] [PubMed]
- 86.Fuchs, K., Cardona Gloria, Y., Wolz, O., Herster, F., Sharma, L., Dillen, C.A., Täumer, C., Dickhöfer, S., Bittner, Z., Dang, T., Singh, A., Haischer, D., Schlöffel, M.A., Koymans, K.J., Sanmuganantham, T., Krach, M., Roger, T., Le Roy, D., Schilling, N.A., Frauhammer, F., Miller, L.S., Nürnberger, T., LeibundGut‐Landmann, S., Gust, A.A., Macek, B., Frank, M., Gouttefangeas, C., Dela Cruz, C.S., Hartl, D., Weber, A.N.: The fungal ligand chitin directly binds TLR 2 and triggers inflammation dependent on oligomer size. EMBO Rep. 19, 1–14 (2018). 10.15252/embr.201846065
- 87.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting Edge: Role of Toll-Like Receptor 1 in Mediating Immune Response to Microbial Lipoproteins. J. Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 88.Misch, E.A., Macdonald, M., Ranjit, C., Sapkota, B.R., Wells, R.D., Siddiqui, M.R., Kaplan, G., Hawn, T.R.: Human TLR1 deficiency is associated with impaired mycobacterial signaling and protection from leprosy reversal reaction. PLoS Negl. Trop. Dis. 2, (2008). 10.1371/journal.pntd.0000231 [DOI] [PMC free article] [PubMed]
- 89.Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daly, J.L., Simonetti, B., Klein, K., Chen, K.E., Williamson, M.K., Antón-Plágaro, C., Shoemark, D.K., Simón-Gracia, L., Bauer, M., Hollandi, R., Greber, U.F., Horvath, P., Sessions, R.B., Helenius, A., Hiscox, J.A., Teesalu, T., Matthews, D.A., Davidson, A.D., Collins, B.M., Cullen, P.J., Yamauchi, Y.: Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science (80 ). 370, 861–865 (2020). 10.1126/science.abd3072 [DOI] [PMC free article] [PubMed]
- 91.Cantuti-Castelvetri, L., Ojha, R., Pedro, L.D., Djannatian, M., Franz, J., Kuivanen, S., van der Meer, F., Kallio, K., Kaya, T., Anastasina, M., Smura, T., Levanov, L., Szirovicza, L., Tobi, A., Kallio-Kokko, H., Österlund, P., Joensuu, M., Meunier, F.A., Butcher, S.J., Winkler, M.S., Mollenhauer, B., Helenius, A., Gokce, O., Teesalu, T., Hepojoki, J., Vapalahti, O., Stadelmann, C., Balistreri, G., Simons, M.: Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (80). 370, 856–860 (2020). 10.1126/science.abd2985 [DOI] [PMC free article] [PubMed]
- 92.Huang, X., Ye, Q., Chen, M., Li, A., Mi, W., Fang, Y., Zaytseva, Y.Y., O’Connor, K.L., Vander Kooi, C.W., Liu, S., She, Q.B.: N-glycosylation-defective splice variants of neuropilin-1 promote metastasis by activating endosomal signals. Nat. Commun. 10, (2019). 10.1038/s41467-019-11580-4 [DOI] [PMC free article] [PubMed]
- 93.Maden CH, Gomes J, Schwarz Q, Davidson K, Tinker A, Ruhrberg C. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev. Biol. 2012;369:277–285. doi: 10.1016/j.ydbio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roth L, Prahst C, Ruckdeschel T, Savant S, Weström S, Fantin A, Riedel M, Héroult M, Ruhrberg C, Augustin HG. Neuropilin-1 mediates vascular permeability independently of vascular endothelial growth factor receptor-2 activation. Sci. Signal. 2016;9:1–11. doi: 10.1126/scisignal.aad3812. [DOI] [PubMed] [Google Scholar]
- 95.Örd M, Faustova I, Loog M. The sequence at Spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV. Sci. Rep. 2020;10:1–10. doi: 10.1038/s41598-020-74101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Palmeira A, Sousa E, Köseler A, Sabirli R, Gören T, Türkçüer İ, Kurt Ö, Pinto MM, Vasconcelos MH. Preliminary virtual screening studies to identify grp78 inhibitors which may interfere with sars-cov-2 infection. Pharmaceuticals. 2020;13:1–13. doi: 10.3390/ph13060132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duan S, Paulson JC. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 2020;38:365–395. doi: 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- 99.Izquierdo-Useros, N., Lorizate, M., Puertas, M.C., Rodriguez-Plata, M.T., Zangger, N., Erikson, E., Pino, M., Erkizia, I., Glass, B., Clotet, B., Keppler, O.T., Telenti, A., Kräusslich, H.G., Martinez-Picado, J.: Siglec-1 Is a Novel Dendritic Cell Receptor That Mediates HIV-1 Trans-Infection Through Recognition of Viral Membrane Gangliosides. PLoS Biol. 10, (2012). 10.1371/journal.pbio.1001448 [DOI] [PMC free article] [PubMed]
- 100.Bedin AS, Makinson A, Picot MC, Mennechet F, Malergue F, Pisoni A, Nyiramigisha E, Montagnier L, Bollore K, Debiesse S, Morquin D, Veyrenche N, Renault C, Foulongne V, Bret C, Bourdin A, Le Moing V, Van De Perre P, Tuaillon E. Monocyte CD169 Expression as a Biomarker in the Early Diagnosis of Coronavirus Disease 2019. J. Infect. Dis. 2021;223:562–567. doi: 10.1093/infdis/jiaa724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sano H, Hsu DK, Yu L, Apgar JR, Kuwabara I, Yamanaka T, Hirashima M, Liu F-T. Human Galectin-3 Is a Novel Chemoattractant for Monocytes and Macrophages. J. Immunol. 2000;165:2156–2164. doi: 10.4049/jimmunol.165.4.2156. [DOI] [PubMed] [Google Scholar]
- 102.Cervantes-Alvarez, E., Rosa, N.L. la, Salgado-de la Mora, M., Valdez-Sandoval, P., Palacios-Jiménez, M., Rodriguez-Alvarez, F., Vera-Maldonado, B.I., Aguirre-Aguilar, E., Escobar-Valderrama, J.M., Alanis-Mendizabal, J., Méndez-Guerrero, O., Tejeda-Dominguez, F., Torres-Ruíz, J., Gómez-Martín, D., Colborn, K.L., Kershenobich, D., Huang, C.A., Navarro-Alvarez, N.: Galectin-3 as a potential prognostic biomarker of severe COVID-19 in SARS-CoV-2 infected patients. medRxiv. 2021.02.07.21251281 (2021). 10.1101/2021.02.07.21251281
- 103.Caniglia JL, Guda MR, Asuthkar S, Tsung AJ, Velpula KK. A potential role for Galectin-3 inhibitors in the treatment of COVID-19. PeerJ. 2020;8:e9392. doi: 10.7717/peerj.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bozorgmehr, N., Mashhouri, S., Rosero, E.P., Xu, L., Shahbaz, S., Sligl, W., Osman, M., Kutsogiannis, D.J., Macintyre, E., O’neil, C.R., Elahi, S.: Galectin-9, a player in cytokine release syndrome and a surrogate diagnostic biomarker in SARS-CoV-2 infection. MBio. 12, (2021). 10.1128/mBio.00384-21 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
(software application or custom code).
Code sharing not applicable to this article as no codes were generated during the current study.