Abstract
Species are shifting their distributions in response to climate change. This geographic reshuffling may result in novel co-occurrences among species, which could lead to unseen biotic interactions, including the exchange of parasites between previously isolated hosts. Identifying potential new host–parasite interactions would improve forecasting of disease emergence and inform proactive disease surveillance. However, accurate predictions of future cross-species disease transmission have been hampered by the lack of a generalized approach and data availability. Here, we propose a framework to predict novel host–parasite interactions based on a combination of niche modelling of future host distributions and parasite sharing models. Using the North American ungulates as a proof of concept, we show this approach has high cross-validation accuracy in over 85% of modelled parasites and find that more than 34% of the host–parasite associations forecasted by our models have already been recorded in the literature. We discuss potential sources of uncertainty and bias that may affect our results and similar forecasting approaches, and propose pathways to generate increasingly accurate predictions. Our results indicate that forecasting parasite sharing in response to shifts in host geographic distributions allow for the identification of regions and taxa most susceptible to emergent pathogens under climate change.
This article is part of the theme issue ‘Infectious disease macroecology: parasite diversity and dynamics across the globe’.
Keywords: parasite sharing, climate change, host–parasite interactions, North American ungulates, niche modelling
1. Introduction
The need to understand and predict future risk of cross-species infections is underscored by the COVID-19 pandemic. Over the past few decades, zoonotic diseases have increased in frequency, prevalence, severity, host range and geographic distribution [1,2]. Global trade, transport and the introduction of exotic species have likely been important in facilitating the emergence of zoonoses [3]. These trends may worsen under climate change [4,5]. For example, many animal host species are already shifting their geographic range as a response to recent environmental global change (e.g. [6,7]), generating new species assemblages [8] and promoting opportunities for parasite exchange. When previously allopatric host species come into sympatry, novel host–parasite interactions may emerge if parasites are able to successfully infect newly exposed hosts. Understanding and predicting how interaction networks will reorganize under climate change, identifying the host pairs most likely to exchange parasites, and the parasites most likely to shift to infect novel hosts presents a major challenge in global change biology and parasite macroecology [9–12].
The role of climate change in restructuring host–parasite interactions through shifts in host ranges is poorly understood outside the crop pest literature [13,14]. To generate predictions of future host–parasite interactions, we need data on host distributions and on host–parasite interactions, as well as predictive models to project future host ranges under climate change, and link-prediction models for host–parasite interactions. Here, we present a framework for predicting future parasite sharing networks that incorporates shifts in host distributions due to climate change, host evolutionary history, and ecological traits. We first review recent progress in modelling host–parasite interactions and their potential links to climate change as our theoretical foundation. We then outline a generalized analytical framework to predict future host–parasite interactions and use North American mammals as a case study to demonstrate the value of our approach for predicting the risk of novel disease emergence events. Finally, we discuss remaining challenges faced by our approach and suggest ways to move the field forward.
(a) . Spread of disease due to climate change
A growing body of literature has focused on the geographic spread and transmission of infectious diseases under various climate change scenarios. Pathogens that have been modelled under future climate change include Ebola virus [15–17], Aedes-borne arboviruses such as dengue and Zika [18,19], and West Nile virus [20–22]. Most of these examples are characterized by strong relationships between environmental or climatic indicators and the spatial-temporal distribution of the disease, its vector or host [19]. The emerging consensus is that effects of climate change on the transmission of vector-borne diseases tends to increase disease spread, risk and burden, if mitigation measurements are not adopted [18,23]. These examples are understandably skewed to identifying potential hotspots of disease transmission to humans, using predictions on projected shifts in vector geographic distribution and, if available, on vector population density [19,21]. While these approaches can provide highly accurate predictions on the geographic areas with the highest probability of occurrence of the intermediate host, they usually ignore the definitive host and are not directly extendable to non-vector-borne diseases.
Research on climate change and human diseases has expanded rapidly, whereas the likely impact of climate change on diseases of wildlife is much less understood [24]. Climate is an important factor in diseases of both wildlife [25] and domesticated species [26], but whether ongoing changes in climate will necessarily lead to an increase in disease occurrence has been challenged [27]. There remains important debate on whether disease distributions will expand or shift geographically under projected climate change, and how climate factors might interact with non-climate factors to drive disease distribution and prevalence [28,29]. There are, for example, contrasting expectations regarding the effects of climate change on disease burden within a given host [24,30]. However, beyond a few well-studied species [31–33], few wildlife species have high-quality disease data for long time series that includes both climatic and non-climatic drivers [28] necessary for making robust predictions. In a recent multi-host study, Cohen et al. [24] generated forecasts of parasite prevalence under climate change using thermal performance data and reported markedly different responses across taxa and regions. Examples such as this could be extended to include less well-studied species given the appropriate analytical framework, but also illustrate the challenges of generalizing trends across different taxa and biogeographic regions.
(b) . Multi-host pathogens and the biogeography of parasite sharing
The patterns of co-occurrence of parasites and their hosts can shape the fundamental properties of ecological communities, impacting host persistence, coexistence and geographic range extents [34]. Recent efforts have focused on bridging gaps in our knowledge of parasite distributions relative to their host distributions [35–37], host–parasite interactions [12] and parasite biogeography [38,39]. Some general trends have emerged. The richness of parasites seems to track the richness of hosts, at least for zoonotic pathogens ([40], but see [41]), and while many parasites infect only one or a few host species, others have a wide host breadth, sometimes encompassing multiple orders [42,43]. The latter—multi-host parasites—are responsible for a large proportion of the emerging infectious diseases most threatening to humans and wildlife [44–46].
Parasite sharing networks characterize the distribution of interactions among parasites, forming the basis for predictions of undocumented and future cross-species parasite sharing [47–50]. However, our ability to make accurate predictions is challenged by the sparseness of most existing host–parasite networks. The detection of a host–parasite interaction requires extensive sampling of the host and screening for the parasite species of interest, which is resource intensive and rather time consuming. Thus, it is common to find sparsely connected host–parasite networks, with a large portion of interactions unsampled [51]. Recent research has focused on inferring biotic interactions using proxies—a powerful approach when documenting all interactions within an ecological network is not feasible [52]—and computational algorithms for ‘filling in’ host–parasite interaction networks by inferring missing links [53]. However, such approaches have largely overlooked how shifting host distributions with climate change has the potential to reshape these networks.
Geography, often presented as a condition for exposure, has been shown to be a key predictor in the probability of parasite sharing among hosts [49,54] and is thus critical in structuring parasite sharing networks. Further, in mammalian parasite sharing networks, habitat is a good predictor for network modularity [47], and host geographic ranges together with population density can explain host centrality in the network [48]. A viral sharing network of mammals illustrates the heterogeneity in parasite sharing across both geography and taxonomy of hosts, with more sharing of viruses in the tropics among rodents and bats [55]. While research on forecasting shifts in parasite sharing networks with shifts in host geographic distributions is still incipient [13], such approaches would allow the identification of regions and taxa most susceptible to emergent pathogens with the geographic reshuffling of host distributions under climate change. Here, we develop a general probabilistic framework to explore how climate change might alter current host–parasite networks and, as a case study, we apply our framework to the parasite sharing network of North American ungulates.
2. Heuristic framework to infer shifts in parasite sharing networks
We propose here a simple three-step approach to forecasting parasite sharing networks under future climate change scenarios (figure 1) that:
-
(1)
models current and future geographic distribution of hosts to identify which currently non-co-occurring pairs of hosts would co-occur under different climate change scenarios (figure 1a);
-
(2)
models the probability of a given parasite being shared between each pair of hosts (figure 1b);
-
(3)
combines steps 1 and 2 to build a probabilistic parasite sharing network allowing us to identify which host species may be most vulnerable to novel pathogens and which parasite species will be more likely transmitted to novel hosts (figure 1c).
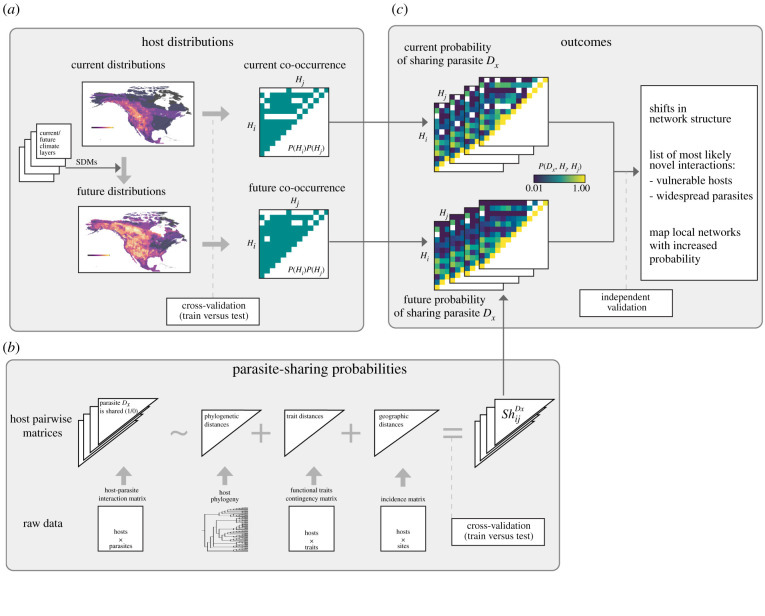
Figure 1.
Diagram summarizing the three major steps describing our framework for inferring parasite sharing networks under current and forecasted patterns of host co-occurrence. The first step (a) uses species distribution modelling techniques to forecast host distributions under future climate. These distributions inform co-occurrence among hosts. The second step (b) models ShDxij, or the probability of each pair of hosts i and j sharing a given parasite Dx. For each parasite, models fit the relationships of observed pairwise parasite sharing against phylogenetic, trait and geographic proxies. The third step (c) combines host co-occurrences (thresholded or probabilistic) with host parasite sharing probabilities to generate both current and future predictions of probabilistic parasite sharing networks mediated by host co-occurrence. Comparing future versus current networks can inform about potential impacts of climate change on network structure, on the emergence of novel interactions, or locations where local networks may experience an increase in parasites. (Online version in colour.)
(a) . Step 1: modelling host geographic shifts
The first step in our framework (figure 1a) addresses the challenge of modelling and forecasting host distributions. We used species distribution models (SDMs hereafter)—i.e. statistical models fitting the geographic distribution of a species as a function of a set of environmental predictors [56–58]—following usual recommendations in the field [59]. We optimized model fit using cross-validation, whereby for a pre-set number of iterations, data are divided into training and testing subsets, models are calibrated using training data and their average accuracy is checked against the testing subset of data. The SDM literature on model thresholds, evaluation and validation is abundant [59–62]. We chose commonly used metrics and approaches (AUC, Kappa, D2, the threshold maximizing sensitivity and specificity) to assess model fit, but note that alternative approaches could be similarly applied. Once models were fitted, predicted distributions of hosts under both current and future climate were transformed into host co-occurrence networks based on pairwise host geographical overlap measured either as a binary or as a probabilistic value.
(b) . Step 2: probabilistic conceptualization for parasite sharing interactions
Based on the previous work by Gravel et al. [63], our framework employs models of the probability that any pair of hosts, Hi and Hj, share a link with any parasite or disease Dx known to infect at least two or more of the hosts in the analysed host–parasite data, P(Dx,Hi,Hj). We focus our framework here on host–parasite interactions but it could be equally applied to other types of interaction networks such as herbivory, mutualism or predation with little transformation. Generally, for two host species to share a previously unshared parasite, the parasite must be capable of infecting both hosts, and hosts must co-occur in space. We can formulate this as follows:
| 2.1 |
where the leftmost term of the right-hand side of equation 2.1, P(Dx|Hi,Hj), represents the probability that parasite Dx infects a given host Hi, while another host Hj is present. The term on the right, P(Hi,Hj), is the host co-occurrence probability calculated from Step 1. Decomposing equation (2.1), we define the probability of a given parasite Dx infecting a given host Hi, P(Hi,Dx) as follows:
| 2.2 |
where P(Hi|Dx) is the probability that parasite Dx is found in host Hi, and P(Dx) is the probability that the parasite is actually present in a local network. While we know the distribution of Hi, we typically do not know the distribution of Dx, because parasites tend to be spatially aggregated [64] and may not be present throughout the entire ranges of their hosts [65,66]. As a simple first approximation, we use the distribution of hosts that are potential reservoirs for the parasite, for example, any other host Hj that may or may not contain the parasite and may or may not be present in the local network. We thus specify the distribution of the parasite as follows:
| 2.3 |
Since the parasite cannot be found where there is no host, we assume that P(Dx|Hj = 0) = 0, and thus equation (2.3) simplifies to
| 2.4 |
plugging equation (2.4) into equation (2.2) we get:
| 2.5 |
We make two extra assumptions to deal with unknown information in equation (2.5). First, we assume that the parasite is always present when the host is present, such that P(Dx|Hj = 1) = 1. Thus, a parasite is assumed to occur across the entire host range, which is known to be unrealistic [39,66] but simplifies calculations for the sake of our example, though data on parasite distributions within host ranges may be included if available. Further, we assume that the distribution of the host is independent of the distribution of the parasite, such that P(Hi|Dx) = P(Hi). These simplifying assumptions allow us to describe the co-distribution of the host and the parasite as a function of the co-distribution of the two host species:
| 2.6 |
(c) . Step 3: combining probabilities of host co-occurrences and host parasite sharing probability
Three proxies are commonly used to infer biotic interactions [52]—geography, phylogeny and traits—and each has been proven to be informative in modelling the probability of an interaction between a host and parasite species [48,53,55,67,68]. We therefore model the sharing of a given parasite—coded as a binary pairwise interaction matrix—as a function of their summed effects:
| 2.7 |
where ShDxij is the observed sharing of parasite Dx by hosts Hi and Hj as a function of: Cij, a measure of host co-occurrence, that can be simplified as a binary variable coding for sympatry between host pairs; Phyij, the phylogenetic distance between hosts; and Tij, a measure of trait distance between each pair of hosts Hi and Hj (figure 1).
Two hosts may share a given parasite even if they do not currently co-occur, P(Hi)P(Hj) = 0, as a legacy from past sympatry or geographic bridging by extinct hosts (see [69] this issue, among others). Nevertheless, for simplicity, we assume here that host i and j must have a non-zero probability of co-occurrence, P(Hi)P(Hj) > 0, such that the probability that they share a parasite is given by
| 2.8 |
Importantly, P(Hi)P(Hj) may vary depending on whether current or future host distributions are considered. Our approach therefore allows us to model effects of climate change-induced host range shifts on the probability of parasite sharing.
(d) . A case study for North American ungulates
The orders Artiodactyla and Perissodactyla (i.e. ungulates or hoofed mammals) comprise a diverse set of hosts, for which high-quality data on geographic distributions, biology and host–parasite associations [49] is available. Several ungulate species are directly used by humans through either domestication or game and are thus of particular interest as potential reservoirs of diseases that impact human and livestock health [70]. We restricted our analyses here to ungulate species within North America because: (i) the relatively low species richness of ungulates in this region (13 species) provides a highly tractable dataset; (ii) species distributions within the Nearctic are well-resolved; (iii) ungulates have a high number of reported parasitic interactions relative to the number of species, resulting in one of the most information-dense host–parasite systems in North American; and (iv) climate change is projected to have a large impact across this biogeographic region. Here, we combine data on host–parasite associations [12], present-day host species distributions [36], climatic variables for both current and future climate [71], and pairwise host phylogenetic and trait distances [49] (see electronic supplementary material, Appendix S1 for details on data and methods). In the following three sections, we apply each step of our framework to North American ungulates as a case study, report main results and discuss associated challenges.
3. Host distributions and shifts in parasite richness
Climate change may have profound impacts on the geographic distribution of both hosts and their parasites. Species distribution models for host current distributions showed high overall accuracy, with AUC values above 0.928 for all species (median = 0.993; see electronic supplementary material, table S1). These strong distribution–climate relationships suggest that climate change will likely result in tight tracking of ungulate ranges with climate change. While the inclusion of additional abiotic predictors such as elevation and vegetation, or human-related predictors such as urbanization or land use would have likely contributed to increasing model accuracy, i.e. highest ungulate diversity concentrates in mountainous regions (electronic supplementary material, figure S1), some of those predictors such as elevation would be invariant with climate change. Assuming parasites faithfully track the distribution of their ungulate hosts, we can see large differences between current (figure 2a) and forecasted (figure 2b) parasite richnesses even in the absence of novel host–parasite interactions. The current distribution of parasites of North American ungulates highlights three distinct regions of parasite diversity: the Rockies and US east coast, where the maximum richness of parasites is estimated above 300 species; Northern Canada and Alaska, with the lowest parasite diversity (less than 50 species); and, Southern Canada, the Great Plains, the US east coast and Mexico with intermediate values. Assuming that ungulate hosts shift their distributions to track changing climate, and there are host shifts, the peak of parasite richness would expand northwards and eastwards outside the Rockies (figure 2b). While parasite diversity is projected to increase across the majority of the continent, with the highest relative increases forecasted for northernmost Canada and Alaska, in some areas, parasite diversity is projected to decrease as host ranges contract (figure 2c). Because we assume that parasites occupy the entire range of their hosts, parasite richness maps closely match host richness maps (see electronic supplementary material, figure S1), though the correlation is not perfect (rCURRENT = 0.815; rFUTURE = 0.824).
Figure 2.
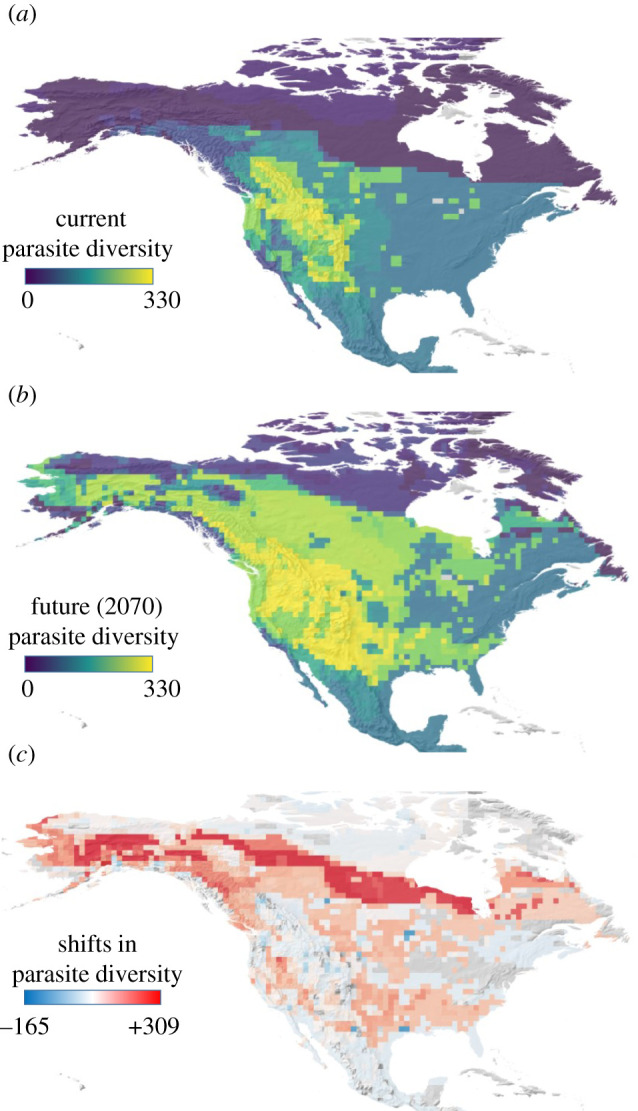
Maps of species richness and shifts in species richness of parasites infecting ungulate hosts in North America. Species richness of parasites is calculated by summing the number of unique parasites across all co-occurring ungulate hosts using the information on contemporary (a) and future projected distributions of hosts (b) with climate change. Mapping assumes parasites are evenly distributed across the entire range of their hosts, which is likely unrealistic [64,65], but we use this simplifying assumption here for illustration. Forecasted shifts in species richness of parasites due to climate change (c) are computed as the difference between contemporary (data compiled in the Global Parasite Mammal Database up to 2010) parasite richness and parasite richness after host range shifts (distributions for the year 2070 under Representative Concentration Pathway 8.5).
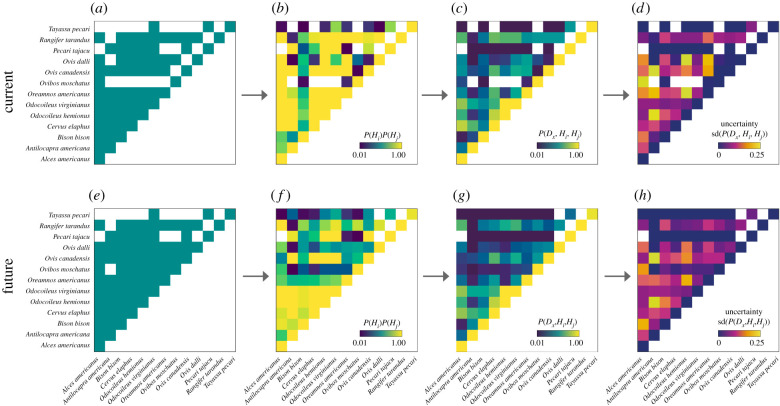
Host distributional patterns and shifts translate into co-occurrence meta-networks (figure 3). Our SDMs generally predict host range expansion and thus greater co-distribution of hosts, i.e. host pairs not co-occurring currently but predicted to do so in the future, and we do not observe any instances where host pairs co-occurring currently become allopatric in the future (figure 3e). However, we note that these projections do not take into account current or future range restrictions due to human settlements or anthropogenic land use change. According to thresholded predictions, novel host co-occurrences include those between Ovis dalli (Dall's sheep), Ovis canadensis (bighorn sheep) or Ovibos moschatus (musk ox) (figure 3a,e). If, instead of binary predictions, we use probabilistic predictions instead (see Methods section in electronic supplementary material, Appendix S1 for details on probability calculation), then additional novel co-occurrences emerge (figure 3b,f). For example, the white-lipped peccary, Tayassu pecari, would co-occur with eight hosts other than Pecari tajacu (collared peccary) and Odocoileus virginianus (white-tailed deer), though with low probabilities (figure 3b,f).
Figure 3.
Sequential calculation of pairwise parasite sharing among North American ungulate hosts. The framework starts by fitting models for co-occurrence among pairs of ungulate hosts according to their contemporary (a,b) and forecasted (e,f) future distributions for the year 2070 under RCP 8.5; (a,e) show the networks computed based on binary distributions of hosts (co-occurring hosts are marked in green). The next column to the right (b,f) shows the joint probability with which each pair of hosts co-occurs. Joint probabilities shown here are summarized across the study area as the upper quartile of sites where each host from a host pair has a non-zero probability of occurrence. The framework finishes by fitting models of parasite sharing for current (c) and forecasted (g) co-occurrence among ungulate hosts and each parasite in the dataset, here illustrated with an Orbivirus (blue tongue disease). Example of modelled predictions corresponds to averaged probabilities across 50 iterations of the parasite sharing model. Uncertainty around predicted probabilities is also reported for current (d) and future (h) host co-occurrence. Note that this same figure can be reproduced for all other parasites in the dataset. (Online version in colour.)
A major assumption likely influencing our results is that we assume unconstrained dispersal, which has shown to impact forecasts of host–parasite interactions in viruses [13]. For example, our projections indicate that T. pecari could dramatically expand its range with climate change, but it is currently classified as vulnerable to extinction (IUCN Red List) and may be more likely to experience population declines and range reduction as current extinction drivers, including deforestation destruction and hunting, intensify. The practical utility and limitations of SDMs have been extensively reviewed, documented and discussed elsewhere [72,73], we therefore refrain from repeating this discussion here so we are able to focus on the more conceptual advances of our modelling approach. We note, however, that any future shifts in the distributions of ungulate hosts will likely be more strongly influenced by humans than by climate.
4. Parasite sharing among host pairs
Step 2 of our framework (figure 1b) models parasite sharing among co-occurring hosts as a function of host pairwise phylogenetic distance, geographic sympatry and ecological or trait distance. In our case study, we use logistic Generalised Linear Models (GLMs; see equation (2.7)) rather than more sophisticated algorithms which tend to overfit (e.g. Random Forest, Generalized Boosted Models; [74]). The higher uncertainty around GLM predictions may be more suitable for the multiple sources of uncertainty conditioning future interactions. Nevertheless, any modelling approach for fitting statistical relationships with a binary response variable—i.e. presence or absence of a host interaction with a given parasite—could be used. In principle, a similar cross-validation approach as applied to SDMs could be used to tune model fitting—i.e. splitting data randomly, calibrating models with a training subset (e.g. two-thirds of the data) and testing it against a testing subset (e.g. the remaining third of the data) (see electronic supplementary material, Appendix S1 for details).
Our parasite sharing models fitted for each multi-host parasite species in the North American ungulate dataset were accurate on average (AUC > 0.7 for 87.9% of 58 parasite species with models subjected to cross-validation; median AUC = 0.846; see electronic supplementary material, table S2 and Appendix S1 for details). These models yield the probability with which each host pair will share (and thus exchange) a given parasite currently and with future climate change. Generalist parasites infecting a high number of hosts are more often predicted to be shared by co-occurring hosts, compared to more specialized parasites (electronic supplementary material, table S2). We found a high correlation between the number of parasites a host infects and its average probability of being shared between any host pair (r = 0.909; p < 0.001). There was a high level of variability in interaction probabilities across parasite species (see electronic supplementary material, table S2; average P(Dx, Hi, Hj) ranging between 0.012 and 0.367, median = 0.072). Protozoans had the highest probabilities of being shared (n = 2; median P(Dx, Hi, Hj) = 0.193), followed by viruses (n = 21; median P(Dx, Hi, Hj) = 0.111), arthropods (n = 13; median P(Dx, Hi, Hj) = 0.079), helminths (n = 4; median P(Dx, Hi, Hj) = 0.076) and bacteria (n = 17; median P(Dx, Hi, Hj) = 0.076). The taxa level median probability values are summarized across all host pairs, many of which had predicted probabilities of zero. To explore whether fitting models with a larger dataset would increase model accuracy, we repeated this step using all ungulate species in the Global Mammal Parasite Database (GMPD; [49]). Overall, parasite sharing models with data restricted to North American ungulates performed better than models for all ungulates globally (see electronic supplementary material, figure S3), possibly due to increased data quality for North American species. Expanding the taxonomic or regional extent of the data does not necessarily improve model accuracy and performance.
Our approach to modelling parasite sharing limits the scope of our inference in two major ways. First, we only model parasites that are documented to infect more than one host, because the model can only be fit for multi-host parasites. By definition, a single-host parasite cannot be shared by two or more hosts, and thus the parasite sharing response variable built for a single-host parasite would result in a vector of zeros. This issue could be partially addressed by employing link-prediction approaches designed to deal with unobserved interactions among different taxa (e.g. occupancy models [75] or trait-matching approaches; [76]). However, given our current framework, we may be able to increase the number of parasites that can be modelled by expanding the taxonomic or geographic extent of the data used to fit the models. A second, related shortcoming has to do with cross-validation approaches being only applicable to multi-host parasite species infecting more than two hosts—otherwise either train and test data subsets may frequently yield all-zero associations. Similarly, cross-validation would only be applicable to parasites infecting less than n – 2, n being the number of hosts in the dataset. These conditions effectively reduced the amount of cross-validated parasite species by 84%—i.e. 261 single-host parasites and 41 parasites infecting two hosts were not evaluable. The challenge of modelling single-host parasites is not limited to our approach, but applies more widely to bipartite link-prediction algorithms [53].
5. Forecasts of shifts in parasite sharing due to climate change
In Step 3 (figure 1c), our framework builds probabilistic parasite sharing networks for each parasite by combining host pairwise information on projected host co-occurrences (Step 1) and predictions from the parasite sharing model (Step 2). Since co-occurrence networks—either binary or probabilistic—are easily produced using host contemporary distributions and projected distributions under climate change, probabilistic parasite sharing networks for current and future host distributions can be simply derived from equation (2.8). Consequently, we are able to compare the parasite sharing networks inferred for current and future points in time, and evaluate potential impacts of climate change on host–parasite interactions (figure 1c). With this approach, it is possible to quantify shifts in network properties and structure, and to identify and rank novel host–parasite associations (i.e. parasites with a high average probability of becoming shared by hosts). We can also construct geographic projections of the networks to identify areas with the largest increases in parasite sharing probabilities. In this way, we can predict potentially vulnerable areas where shifts in host distributions may lead to increased numbers of spillover events. Using our approach, the change in parasite sharing across periods is solely a product of shifts in host distributions. This simplification makes strong assumptions, for example, that pairwise parasite sharing probabilities do not change with time and are constant across each host range. Nonetheless, we believe predictions from our approach are informative as they tend to reflect the extreme of possible scenarios because they maximize the geographic spread of the parasites.
For North American ungulates, we find climate change is likely to have quantifiable effects on the structure of the parasite sharing network (figure 3c,d,g,h), by increasing the connectivity, amount of links, centrality and closeness of each host pair in the network (Δdegree = 9.0%, Δbetweenness = 21.0%, Δcentrality = 15.1%, Δcloseness = 60.1%). Further, by comparing current and future parasite sharing networks, we identified parasites likely to increase in their probability of being shared (electronic supplementary material, table S3). From these, we identified the parasites that could be shared by newly co-occurring pairs of hosts (i.e. hosts that were previously not recorded as a realized interaction in the GMPD v. 2.0). Examples of novel parasite sharing included Francisella tularensis infecting Bison bison (bison) and Cervus elaphus (elk), Nematodirus odocoilei infecting Ovibos moschatus (mule deer), or Protostrongylus stilesi, and Orf virus infecting B. bison, C. elaphus and Odocoileus hemionus (see electronic supplementary material, table S1 for a complete list).
While we likely need to wait to validate predictions from this type of framework, if model predictions are accurate, it is probable that some predicted interactions already exist in nature but are simply not included in the database used to train the link-prediction model. To validate predictions from Step 3, we evaluated the list of potential novel host–parasite interactions (table 1) against independent data sources for evidence of any record of a positive association between host and parasite, for example, in experimental cross-infections or the veterinary literature. While direct evidence for predicted interactions was found for 34.15% of the cases, indirect evidence of infection—evidence of the parasite infecting a congeneric host in a different region or a congeneric parasite infecting the target host—increased the proportion of links with evidence to 60.9% (table 1). These numbers suggest that our approach is able to make realistic predictions of undocumented host–parasite associations. This also emphasizes that large-scale host–parasite datasets (e.g. [12]) are likely to be incomplete [51], due to biased sampling across taxa, omitting interactions only reported in the ‘grey‘ literature and lack of taxonomic standardization, and become quickly outdated as our knowledge of host–parasite association expands [68,104]. In addition, as digitization efforts have been increasing in recent years, older literature is resurfacing, which may identify rare interactions that may have been missed during the compilation of literature-based interaction databases.
Table 1.
Independent validation of model predictions. This table provides a summary of already realized host–parasite interactions predicted to emerge under climate change by our models. To be included, a predicted host–parasite association had to meet two conditions: an increase in the probability in the parasite sharing model, and the predicted host–parasite interaction not being recorded in the source data (GMPD v. 2.0; [12]). We assessed if independent evidence for the interaction was found after a literature search and recorded the type of interaction. Note that most of the supporting evidence for the ‘novel’ host–parasite interactions suggest gaps in the GMPD database and only a few were recent papers published after the GMPD data compilation. See electronic supplementary material, table S4 in Appendix S2 for more details on direct and indirect interactions and on host–parasite interactions for which we did not find records in the literature, and for a complete list of references. Abbreviations in the region column: Eur, Europe; NAm, North America; cosm, cosmopolite; capt, captivity; exp, experimental setting.
| host species (common name) | parasite species | region | references |
|---|---|---|---|
| direct evidence | |||
| Bison bison (American bison) | Arcanobacterium pyogenes | NAm | Zamke & Schlater [77]; Rzewuska et al. [78] |
| Bison bison (American bison) | Nematodirus helvetianus | NAm | Wade et al. [79] |
| Bison bison (American bison) | Teladorsagia circumcincta | NAm | Knapp et al. [80]; Samuel et al. [81] |
| Bison bison (American bison) | Toxoplasma gondii | NAm | Dubey [82]; Moskwa et al. [83] |
| Odocoileus hemionus (mule deer) | Marshallagia marshalli | NAm | Walker & Becklund [84] |
| Odocoileus hemionus (mule deer) | Nematodirus helvetianus | NAm | Russell [85] |
| Odocoileus hemionus (mule deer) | Ostertagia trifurcata | NAm | Walker & Becklund [84] |
| Odocoileus hemionus (mule deer) | Parapoxvirus | NAm | Tryland et al. [86] |
| Oreamnos americanus (mountain goat) | Marshallagia marshalli | NAm | Kerr & Holmes [87]; Aleuy et al. [88] |
| Oreamnos americanus (mountain goat) | Nematodirus helvetianus | NAm | Samuel et al. [81]; Aleuy et al. [88] |
| Oreamnos americanus (mountain goat) | Bluetongue virus | NAm | Williams & Barker [89] |
| Oreamnos americanus (mountain goat) | Protostrongylus stilesi | NAm | Thorne & Honess [90]; Jenkins et al. [91] |
| Oreamnos americanus (mountain goat) | Teladorsagia circumcincta | NAm | Kerr & Holmes [87] |
| Ovis canadensis (bighorn sheep) | Brucella abortus | exp | Kreeger et al. [92] |
| indirect evidence (congeneric hosts) | |||
| Bison bison (American bison) | Francisella tularensis | Eur | Krzysiak et al. [93] |
| Bison bison (American bison) | Ostertagia trifurcata | Eur | Karbowiak et al. [94] |
| Cervus elaphus (red deer) | Parapoxvirus | Eur | Scagliarini et al. [95] |
| Oreamnos americanus (mountain goat) | Arcanobacterium pyogenes | NAm | Tell et al. [96] |
| Oreamnos americanus (mountain goat) | Bovine viral diarrhea virus 1 | capt | Doyle & Heuschele [97]; Williams & Barker [89] |
| Oreamnos americanus (mountain goat) | Human parainfluenza virus 3 | Eur | Frölich [98]; Ataseven et al. [99] |
| Oreamnos americanus (mountain goat) | Toxoplasma gondii | cosm | Pavone et al. [100] |
| Oreamnos americanus (mountain goat) | Bovine herpesvirus 1 | cosm | Williams and Barker [89] |
| indirect evidence (congeneric parasites) | |||
| Bison bison (American bison) | Marshallagia marshalli | Eur | Kuzmina et al. [101] |
| Bison bison (American bison) | Parapoxvirus Orf virus | NAm | Robin et al. [102] |
| Odocoileus virginianus (white-tailed deer) | Alcelaphine herpesvirus 1 | NAm | Li et al. [103] |
Our results demonstrate how forecasts of biotic interactions, such as those between hosts and parasites, can be used to describe a landscape of potential future interactions. However, they also highlight the Eltonian shortfall in biodiversity science by showing that many predicted interactions might already be realized, but are not yet captured in current databases [52].
6. Challenges and limitations
Our case study demonstrates how it is possible to predict the effects of climate change on parasite sharing networks, but also raises important questions. Most of these derive from the strong (but operationally useful) assumptions we make in our analyses. Important questions include: (i) how are parasites typically distributed across the ranges of their hosts?; (ii) is the probability of parasite sharing between hosts proportional to the degree of host geographic overlap?; (iii) how closely will host distributions track climate change?; (iv) how well will hosts track changes in climate in the face of dispersal restrictions?; and (v) how do climatic conditions influence arthropod vectors or parasite survival outside of their hosts? While many assumptions we make may be unrealistic [13,39,65,66], we suggest they allow us to make predictions as to the upper limits of the geographic spread of hosts and their parasites. As such, these results can be used to target areas and species at higher risk, and thus higher priority for monitoring.
(a) . Uncertainty propagation
An important consideration across all steps of our framework is how uncertainty is propagated through the different modelling stages. Both Step 1 and Step 2 generate multiple predictions, which contain uncertainty (see electronic supplementary material, figure S2; figure 3b,f, respectively). For illustrative purposes, we have averaged over this uncertainty before moving onto the next step, but understanding the true extent of variability in our predictions will require that uncertainties from earlier steps be propagated through and accounted for when their associated predictions are combined in Step 3. Although uncertainty in estimated parameters and predictions can become rather large when combining uncertainty from different sub-models (see e.g. [105]), appropriately propagating uncertainty is a requirement of serious risk assessment approaches. To date, uncertainty propagation has been underexplored in the literature aimed at inferring biotic interactions (but see [106]). Operationally, it is unclear how to best propagate uncertainty in our framework—e.g. how to weight uncertainties from different sub-models, whether to present alternative scenarios resulting from sets of assumptions at each stage, or if future work would benefit from the development of a fully joint model that estimates all parameters simultaneously. We suggest that future work incorporating Steps 1–3 into a single model would be a fruitful research avenue to explore, as it would allow the propagation of uncertainty and the inclusion of other processes such as observation error directly into the model. Such efforts would be particularly worthwhile when making host–parasite forecasts aimed at informing policy or management [107–109].
(b) . Evaluation and independent validation
Model evaluation and validation for SDMs (figure 1a) has been extensively discussed elsewhere [59–62]. However, there is less literature contrasting methods for validating predictions of biotic interactions, and thus, there is currently no consensus on best practices for predicting networks, or splitting data into training and testing sets for prediction of ecological interactions (but see [110,111]). In the case of parasite sharing networks, previous work has examined the frequencies with which different hosts share viruses [13,55]; this validation scheme differs from ours in that its focus is on viral sharing, not on predicting individual host–parasite associations. The cross-validation approach we employ allows us to consider individual network links, but has the inconvenience of only being operative for multi-host parasites, which decreases the number of modelled parasites (see §4). Expanding the number of hosts to include species from other regions or even other taxa could help predictions as this will tend to re-classify single-host parasites in our database as multi-host parasites, but could also inflate the sparseness of the matrix, or increase the number of single-host parasites if the additional hosts and parasites are less well-studied. One way to reduce this bias would be to explicitly apply thresholds to predicted probabilities (see e.g. [53]), similarly to approaches in the SDM literature. This would be another interesting avenue to explore further.
(c) . Applying models to single-host parasites (specialists versus generalists)
The focus of our methods on documented multi-host parasite species is both a strength and a limitation. Focusing on multi-host parasites could be considered conservative. If a parasite has demonstrated the ability to transfer between two or more hosts, then that parasite could be predicted to be more likely to gain an additional host than a historically single-host parasite [112]. However, specialist parasite species also have the potential to gain the ability to infect multiple hosts [113] or may already be generalists with undersampled host ranges. One approach that could allow us to include predictions for single-host pathogens would be to consider their true host range as unknown, and model their expected host breadth, for example, using phylogenetic imputations [114]. In addition to phylogenetic imputation, information on ecological proximity, when available, could also be incorporated to better inform projections of host–parasite associations [48]. However, such approaches would require information not only on the host phylogeny, but also on the phylogeny of parasites, which is often less well known.
(d) . Types of transmission and intermediate hosts
Our framework may be considered ‘host-centric’ because even though it is applied individually to each parasite species, the sharing of that parasite depends on host properties and may shift with host distributions. Our predicted parasite sharing networks assume that geographic range overlap provides a useful proxy for potential parasite transmission. However, our model, as presented, does not consider the different modes of transmission that characterize different parasites, and this may have a large impact on predictions and on the need for additional layers of information to be incorporated into the models. For example, ectoparasites (e.g. arthropods) and parasites with free-living developmental stages (e.g. eggs and larvae in helminths) could have additional climatic constraints on their geographic distributions [41,65]. Similarly, the distribution of non-ungulate hosts should also be modelled for parasites with intermediate hosts or vector-borne parasites. For example, in the North American dataset, the bacteria Ehrlichia chaffeensis is predicted with moderate accuracy by our models (AUC = 0.88; see electronic supplementary material, table S2), but it uses the lone star tick, Amblyomma americanum, as a vector [115], and thus the tick's distribution should be also accounted for when forecasting responses to climate change. Adding in these additional steps would be methodologically straightforward, for example, including an extra probability Hk to the framework either in Step 2, or thresholding co-occurrence networks in Step 1 (figure 1). However, the availability of the high-quality data needed to accurately model the distribution of intermediate hosts or vectors presents an additional constraint on model fitting. We suggest a useful first step would be to explore model accuracy for parasites with different transmission modes. An additional step would incorporate the complex interactions among climatic factors that may determine parasite distributions [116], especially for ectoparasites and parasite species with free-living stages.
(e) . Mechanistic versus phenomenological models
Common approaches to inferring host–parasite interaction networks have relied on phenomenological, correlative models, particularly so when targeted networks encompass multiple hosts and parasites, or broad geographic extents (e.g. [13,53,68]). These approaches are constrained by data availability and trade off with how realistic model outcomes are. Mechanistic, process-based models incorporating demography or epidemiology in a spatially explicit context can lead to substantially more realistic predictions of host–parasite interactions (e.g. [117]). However, implementing mechanistic models at macro-scales may only be feasible for a few, intensely studied diseases [118] and to date are more often used in the crop literature [119]. Developing ‘full joint models’ may be an alternative in the middle ground between simplistic approaches and more sophisticated ones. Doing so would help incorporate uncertainty across different steps of the model (rather than picking single alternative scenarios at different points in the modelling process).
7. Concluding remarks and future directions
Our approach to forecast parasite sharing models to future scenarios of shifted co-occurrence among hosts enlarges the toolbox aimed at predicting disease threats resulting from climate change. Such frameworks can help prioritize regions and species that may become increasingly vulnerable to emerging diseases in the future [12,25]. We present a novel framework to address this challenge and demonstrate our approach with a case study of host–parasite interactions for North American ungulates. Our results are promising as both cross-validations of parasite sharing models and independent validation suggest high accuracy in our predictions of novel interactions (table 1; electronic supplementary material, table S2). We also highlight key limitations of our approach and suggest some critical research to improve predictions. We believe our framework provides a useful first step and allows us to present a possible scenario of maximal disease spread under future climate change scenarios. Here, we outline how predictions could be further refined and suggest some potentially useful ways forward:
-
(1)
Targeted parasite surveillance—our framework makes predictions on likely hosts and regions of parasite sharing; we suggest these predictions could help in the targeting of parasite surveillance programmes. This exercise could also help fill gaps in parasite databases and thus feedback to improve the information available for predictive models.
-
(2)
Forecasting zoonotic diseases—making accurate forecasts of host–parasite interactions is important for both wildlife and people. Many human diseases have zoonotic origins [120,121]. Expanding forecasts to encompass humans and other potential wildlife reservoirs will help identify areas and taxa with a high risk of disease spillover (see e.g. figure 2c), and allow for proactive measures to reduce disease risk.
-
(3)
Increased modelling realism—all models are a simplification, and we make several simplifying assumptions in our framework. By incorporating additional layers of information, it may be possible to further refine our future prediction. For example, spatial information on current and future land use change and vegetation distribution would aid in refining forecasts of host geographic distributions and subsequent parasite sharing potential [122]. Information on human population density and connectivity could additionally help in identifying areas with high risk of zoonotic disease emergence [123,124].
-
(4)
Parasite distributions within host ranges—potential variation in the distribution of parasites within their host geographic extents is a major limitation of data feeding our models. One approach to address this imitation would be to fit SDMs for the parasite species directly; while we often lack geolocation data on parasite occurrences, such approaches will be increasingly possible as more parasite occurrences become available (see [39]). Parasite SDMs could then be combined with host SDMs to project geographical landscapes of host–parasite interactions, or host distributions could be included as predictors within parasite SDMs.
-
(5)
Transcending phenomenological models—SDMs are a powerful tool for projecting species distributions, and when applied to host extents, they provide us with the foundations on which spatial parasite sharing networks can be constructed. Nevertheless, mechanistic models [117] may help avoid simplifying assumptions which are no longer required if demographic, epidemiologic or other relevant processes could be modelled directly. Clearly, process-based modelling approaches trade off with more simplistic ones in their data needs, and thus, which model to apply will ultimately depend on what data is available for which geographic and taxonomic scope.
The framework presented here represents a step forward to forecasting host–parasite associations as climate continues changing and host redistributions lead to novel interactions. In the scenario of maximum parasite spread depicted by our models, climate change can have profound consequences on the disease risk landscape for North American ungulates by increasing host sympatry and facilitating parasite transmission. As data accumulate and deeper methodological insights are gained, we should be able to move towards increasingly process-based models, allowing precise predictions and high-resolution disease risk maps that will be able to inform policy and better prepare for the future disease risk landscape.
Acknowledgements
We thank RCN participants for their constructive feedback and P. Stephens for sharing readily formatted pairwise data on ungulate species.
Data accessibility
Source data for analyses are available and accessible through previous publications (see electronic supplementary materials for details). Derived data and custom computer R code to perform analyses are freely available at GitHub, https://github.com/MoralesCastilla/HostParasiteForecasting/.
Authors' contributions
I.M.C. conceived of the paper, analysed the data and wrote initial drafts; D.G. contributed analytical expertise and helped design the analytical framework; T.J.D. and M.F. provided critical feedback since early stages; P.P. and A.A.A. extracted and analysed independent validation data. All authors provided contextual expertise and helped write the paper.
Competing interests
We declare we have no competing interests
Funding
We thank the Macroecology of Infectious Disease Research Coordination Network, funded by NSF (grant no. DEB 1316223), for facilitating discussion among the authors. I.M.C. acknowledges funding from the Spanish Ministry for Science and Innovation (grant no. PID2019-109711RJ-I00 to I.M.C., grant CGL2017-86926-P to M.Á.R.) as well as from Comunidad de Madrid and University of Alcalá (funders of I.F.W. through grant CM/BG/2021-003 to I.M.C.). S.H. was supported by the German Science Foundation (grant no. DFG, HU 2748/1-1).
References
- 1.Smith KF, Goldberg M, Rosenthal S, Carlson L, Chen J, Chen C, Ramachandran S. 2014Global rise in human infectious disease outbreaks. J. R. Soc. Interface 11, 20140950. ( 10.1098/rsif.2014.0950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008Global trends in emerging infectious diseases. Nature 451, 990-993. ( 10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith KF, Guégan JF. 2010Changing geographic distributions of human pathogens. Annu. Rev. Ecol. Evol. Syst. 41, 231-250. ( 10.1146/annurev-ecolsys-102209-144634) [DOI] [Google Scholar]
- 4.Gould EA, Higgs S. 2009Impact of climate change and other factors on emerging arbovirus diseases. Trans. R. Soc. Trop. Med. Hyg. 103, 109-121. ( 10.1016/j.trstmh.2008.07.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindgren E, Andersson Y, Suk JE, Sudre B, Semenza JC. 2012Monitoring EU emerging infectious disease risk due to climate change. Science 336, 418-419. ( 10.1126/science.1215735) [DOI] [PubMed] [Google Scholar]
- 6.Parmesan C, Yohe G. 2003A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37-42. ( 10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 7.Chen I-C, Hill JK, Ohlemüller R, Roy DB, Thomas CD. 2011Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024-1026. ( 10.1126/science.1206432) [DOI] [PubMed] [Google Scholar]
- 8.Williams JW, Jackson ST. 2007Novel climates, no-analog communities, and ecological surprises. Front. Ecol. Environ. 5, 475-482. ( 10.1890/070037) [DOI] [Google Scholar]
- 9.Kutz S, Hoberg EP, Polley L, Jenkins E. 2005Global warming is changing the dynamics of Arctic host–parasite systems. Proc. R. Soc. B 272, 2571-2576. ( 10.1098/rspb.2005.3285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks DR, Hoberg EP. 2007How will global climate change affect parasite–host assemblages? Trends Parasitol. 23, 571-574. ( 10.1016/j.pt.2007.08.016) [DOI] [PubMed] [Google Scholar]
- 11.Woodward G, et al. 2010Ecological networks in a changing climate. Adv. Ecol. Res. 42, 71-138. ( 10.1016/B978-0-12-381363-3.00002-2) [DOI] [Google Scholar]
- 12.Stephens PR, et al. 2017Global mammal parasite database version 2.0. Ecology 98, 1476. ( 10.1002/ecy.1799) [DOI] [PubMed] [Google Scholar]
- 13.Carlson CJ, Albery GF, Merow C, Trisos CH, Zipfel CM, Eskew EA, Olival KJ, Ross N, Bansal S. 2020Climate change will drive novel cross-species viral transmission. BioRxiv918755. ( 10.1101/2020.01.24.918755) [DOI] [PubMed]
- 14.Pickles RS, Thornton D, Feldman R, Marques A, Murray DL. 2013Predicting shifts in parasite distribution with climate change: a multitrophic level approach. Glob. Change Biol. 19, 2645-2654. ( 10.1111/gcb.12255) [DOI] [PubMed] [Google Scholar]
- 15.Schmidt JP, Park AW, Kramer AM, Han BA, Alexander LW, Drake JM. 2017Spatiotemporal fluctuations and triggers of Ebola virus spillover. Emerg. Infect. Dis. 23, 415-422. ( 10.3201/eid2303.160101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olivero J, et al. 2017Recent loss of closed forests is associated with Ebola virus disease outbreaks. Sci. Rep. 7, 1-9. ( 10.1038/s41598-017-14727-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redding DW, Atkinson PM, Cunningham AA, Iacono GL, Moses LM, Wood JL, Jones KE. 2019Impacts of environmental and socio-economic factors on emergence and epidemic potential of Ebola in Africa. Nat. Commun. 10, 1-11. ( 10.1038/s41467-018-07882-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan SJ, Carlson CJ, Mordecai EA, Johnson LR. 2019Global expansion and redistribution of Aedes-borne virus transmission risk with climate change. PLoS Negl. Trop. Dis. 13, e0007213. ( 10.1371/journal.pntd.0007213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero D, Olivero J, Real R, Guerrero JC. 2019Applying fuzzy logic to assess the biogeographical risk of dengue in South America. Parasit. Vectors 12, 1-13. ( 10.1186/s13071-019-3691-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrigan RJ, Thomassen HA, Buermann W, Smith TB. 2014A continental risk assessment of West Nile virus under climate change. Glob. Change Biol. 20, 2417-2425. ( 10.1111/gcb.12534) [DOI] [PubMed] [Google Scholar]
- 21.Morin CW, Comrie AC. 2013Regional and seasonal response of a West Nile virus vector to climate change. Proc. Natl Acad. Sci. USA 110, 15 620-15 625. ( 10.1073/pnas.1307135110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paz S. 2015Climate change impacts on West Nile virus transmission in a global context. Phil. Trans. R. Soc. B 370, 20130561. ( 10.1098/rstb.2013.0561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caminade C, McIntyre KM, Jones AE. 2019Impact of recent and future climate change on vector-borne diseases. Ann. N. Y. Acad. Sci. 1436, 157. ( 10.1111/nyas.13950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen JM, Sauer EL, Santiago O, Spencer S, Rohr JR. 2020Divergent impacts of warming weather on wildlife disease risk across climates. Science 370, eabb1702. ( 10.1126/science.abb1702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, Samuel MD. 2002Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158-2162. ( 10.1126/science.1063699) [DOI] [PubMed] [Google Scholar]
- 26.Gubbins S, Carpenter S, Baylis M, Wood JL, Mellor PS. 2008Assessing the risk of bluetongue to UK livestock: uncertainty and sensitivity analyses of a temperature-dependent model for the basic reproduction number. J. R. Soc. Interface 5, 363-371. ( 10.1098/rsif.2007.1110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafferty KD. 2009Calling for an ecological approach to studying climate change and infectious diseases. Ecology 90, 932-933. ( 10.1890/08-1767.1) [DOI] [PubMed] [Google Scholar]
- 28.Ostfeld RS. 2009Biodiversity loss and the rise of zoonotic pathogens. Clin. Microbiol. Infect. 15, 40-43. ( 10.1111/j.1469-0691.2008.02691.x) [DOI] [PubMed] [Google Scholar]
- 29.Gething PW, Smith DL, Patil AP, Tatem AJ, Snow RW, Hay SI. 2010Climate change and the global malaria recession. Nature 465, 342-345. ( 10.1038/nature09098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atkinson CT, Dusek RJ, Woods KL, Iko WM. 2000Pathogenicity of avian malaria in experimentally-infected Hawaii Amakihi. J. Wildl. Dis. 36, 197-201. ( 10.7589/0090-3558-36.2.197) [DOI] [PubMed] [Google Scholar]
- 31.Cook R, White M, Trainer D, Glazener W. 1971Mortality of young white-tailed deer fawns in south Texas. J. Wildl. Manage. 35, 47-56. ( 10.2307/3799870) [DOI] [Google Scholar]
- 32.Packer C, Altizer S, Appel M, Brown E, Martenson J, O'Brien SJ, Roelke-Parker M, Hofmann-Lehmann R, Lutz H. 1999Viruses of the Serengeti: patterns of infection and mortality in African lions. J. Anim. Ecol. 68, 1161-1178. ( 10.1046/j.1365-2656.1999.00360.x) [DOI] [Google Scholar]
- 33.Mathieu-Bégné E, Loot G, Mazé-Guilmo E, Mullet V, Genthon C, Blanchet S. 2021Combining species distribution models and population genomics underlines the determinants of range limitation in an emerging parasite. Ecography 44, 307-319. ( 10.1111/ecog.05301) [DOI] [Google Scholar]
- 34.Ricklefs RE. 2011A biogeographical perspective on ecological systems: some personal reflections. J. Biogeogr. 38, 2045-2056. ( 10.1111/j.1365-2699.2011.02520.x) [DOI] [Google Scholar]
- 35.Chowdhury N, Aguirre AA (eds). 2001. Helminths of wildlife. Enfield, NH: Science Publishers, Inc. [Google Scholar]
- 36.Schipper J, et al. 2008The status of the World's land and marine mammals: diversity, threat, and knowledge. Science 322, 225-230. ( 10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 37.Faurby S, Davis M, Pedersen RØ, Schowanek SD, Antonelli A, Svenning JC. 2018PHYLACINE 1.2: the phylogenetic atlas of mammal macroecology. Ecology 99, 2626. ( 10.1002/ecy.2443) [DOI] [PubMed] [Google Scholar]
- 38.Morand S, Krasnov BR. 2010The biogeography of host-parasite interactions. Oxford, UK: Oxford University Press. [Google Scholar]
- 39.Pappalardo P, Morales-Castilla I, Park AW, Huang S, Schmidt JP, Stephens PR. 2020Comparing methods for mapping global parasite diversity. Glob. Ecol. Biogeogr. 29, 182-193. ( 10.1111/geb.13008) [DOI] [Google Scholar]
- 40.Dunn RR, Davies TJ, Harris NC, Gavin MC. 2010Global drivers of human pathogen richness and prevalence. Proc. R. Soc. B. 277, 2587-2595. ( 10.1098/rspb.2010.0340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dallas T, Gehman AM, Aguirre AA, Budischak SA, Drake JM, Farrell MJ, Ghai R, Huang S, Morales-Castilla I. 2019Contrasting latitudinal gradients of body size in helminth parasites and their hosts. Glob. Ecol. Biogeogr. 28, 804-813. ( 10.1111/geb.12894) [DOI] [Google Scholar]
- 42.Pedersen AB, Altizer S, Poss M, Cunningham AA, Nunn CL. 2005Patterns of host specificity and transmission among parasites of wild primates. Int. J. Parasitol. 35, 647-657. ( 10.1016/j.ijpara.2005.01.005) [DOI] [PubMed] [Google Scholar]
- 43.Park A, et al. 2018Characterizing the phylogenetic specialism–generalism spectrum of mammal parasites. Proc. R. Soc. B 285, 20172613. ( 10.1098/rspb.2017.2613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daszak P, Cunningham AA, Hyatt AD. 2000Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287, 443-449. ( 10.1126/science.287.5452.443) [DOI] [PubMed] [Google Scholar]
- 45.Cleaveland S, Laurenson M, Taylor L. 2001Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil. Trans. R. Soc. Lond. B 356, 991-999. ( 10.1098/rstb.2001.0889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. 2002Biological and biomedical implications of the coevolution of pathogens and their hosts. Nat. Genet. 32, 569-577. ( 10.1038/ng1202-569) [DOI] [PubMed] [Google Scholar]
- 47.Pilosof S, Morand S, Krasnov BR, Nunn CL. 2015Potential parasite transmission in multi-host networks based on parasite sharing. PLoS ONE 10, e0117909. ( 10.1371/journal.pone.0117909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dallas TA, Han BA, Nunn CL, Park AW, Stephens PR, Drake JM. 2019Host traits associated with species roles in parasite sharing networks. Oikos 128, 23-32. ( 10.1111/oik.05602) [DOI] [Google Scholar]
- 49.Stephens PR, Altizer S, Ezenwa VO, Gittleman JL, Moan E, Han B, Huang S, Pappalardo P. 2019Parasite sharing in wild ungulates and their predators: effects of phylogeny, range overlap, and trophic links. J. Anim. Ecol. 88, 1017-1028. ( 10.1111/1365-2656.12987) [DOI] [PubMed] [Google Scholar]
- 50.Albery GF, Becker DJ, Kenyon F, Nussey DH, Pemberton JM. 2019The fine-scale landscape of immunity and parasitism in a wild ungulate population. Integr. Comp. Biol. 59, 1165-1175. ( 10.1093/icb/icz016) [DOI] [PubMed] [Google Scholar]
- 51.Dallas T, Park AW, Drake JM. 2017Predicting cryptic links in host-parasite networks. PLoS Comput. Biol. 13, e1005557. ( 10.1371/journal.pcbi.1005557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morales-Castilla I, Matias MG, Gravel D, Araújo MB. 2015Inferring biotic interactions from proxies. Trends Ecol. Evol. 30, 347-356. ( 10.1016/j.tree.2015.03.014) [DOI] [PubMed] [Google Scholar]
- 53.Elmasri M, Farrell MJ, Davies TJ, Stephens DA. 2020A hierarchical Bayesian model for predicting ecological interactions using scaled evolutionary relationships. Ann. Appl. Stat. 14, 221-240. ( 10.1214/19-AOAS1296) [DOI] [Google Scholar]
- 54.Davies TJ, Pedersen AB. 2008Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. R. Soc. B 275, 1695-1701. ( 10.1098/rspb.2008.0284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albery GF, Kirkpatrick L, Firth JA, Bansal S. 2020Unifying spatial and social network analysis in disease ecology. J. Anim. Ecol. 90, 45-61. ( 10.1111/1365-2656.13356) [DOI] [PubMed] [Google Scholar]
- 56.Araújo MB, Pearson RG, Thuiller W, Erhard M. 2005Validation of species–climate impact models under climate change. Glob. Change Biol. 11, 1504-1513. ( 10.1111/j.1365-2486.2005.01000.x) [DOI] [Google Scholar]
- 57.Araújo MB, Alagador D, Cabeza M, Nogúes-Bravo D, Thuiller W. 2011Climate change threatens European conservation areas. Ecol. Lett. 14, 484-492. ( 10.1111/j.1461-0248.2011.01610.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morales-Castilla I, Davies TJ, Pearse WD, Peres-Neto P. 2017Combining phylogeny and co-occurrence to improve single species distribution models. Glob. Ecol. Biogeogr. 26, 740-752. ( 10.1111/geb.12580) [DOI] [Google Scholar]
- 59.Araújo MB, et al. 2019Standards for distribution models in biodiversity assessments. Sci. Adv. 5, eaat4858. ( 10.1126/sciadv.aat4858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lobo JM, Jiménez-Valverde A, Real R. 2008AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17, 145-151. ( 10.1111/j.1466-8238.2007.00358.x) [DOI] [Google Scholar]
- 61.Barbet-Massin M, Jiguet F, Albert CH, Thuiller W. 2012Selecting pseudo-absences for species distribution models: how, where and how many? Methods Ecol. Evol. 3, 327-338. ( 10.1111/j.2041-210X.2011.00172.x) [DOI] [Google Scholar]
- 62.Liu C, Newell G, White M. 2016On the selection of thresholds for predicting species occurrence with presence-only data. Ecol. Evol. 6, 337-348. ( 10.1002/ece3.1878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gravel Det al. 2019Bringing Elton and Grinnell together: a quantitative framework to represent the biogeography of ecological interaction networks. Ecography 42, 401-415. ( 10.1111/ecog.04006) [DOI] [Google Scholar]
- 64.Poulin R. 2013Explaining variability in parasite aggregation levels among host samples. Parasitology 140, 541. ( 10.1017/S0031182012002053) [DOI] [PubMed] [Google Scholar]
- 65.Cumming G. 1999Host distributions do not limit the species ranges of most African ticks (Acari: Ixodida). Bull. Entomol. Res. 89, 303-327. ( 10.1017/S0007485399000450) [DOI] [Google Scholar]
- 66.Shenbrot G, Krasnov B, Lu L. 2007Geographical range size and host specificity in ectoparasites: a case study with Amphipsylla fleas and rodent hosts. J. Biogeogr. 34, 1679-1690. ( 10.1111/j.1365-2699.2007.01736.x) [DOI] [Google Scholar]
- 67.Pedersen AB, Davies TJ. 2009Cross-species pathogen transmission and disease emergence in primates. EcoHealth 6, 496-508. ( 10.1007/s10393-010-0284-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farrell MJ, Elmasri M, Stephens DA, Davies TJ. 2020Predicting missing links in global host-parasite networks. bioRxiv 965046. ( 10.1101/2020.02.25.965046) [DOI] [Google Scholar]
- 69.Farrell M, Park A, Cressler C, Dallas T, Huang S, Mideo N, Morales-Castilla I, Davies J, Stephens P. 2021The ghost of hosts past: impacts of host extinction on parasite specificity. Phil. Trans. R. Soc. B 376, 20200350. ( 10.1098/rstb.2020.0351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farrell MJ, Davies TJ. 2019Disease mortality in domesticated animals is predicted by host evolutionary relationships. Proc. Natl Acad. Sci. USA 116, 7911-7915. ( 10.1073/pnas.1817323116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fick SE, Hijmans RJ. 2017WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302-4315. ( 10.1002/joc.5086) [DOI] [Google Scholar]
- 72.Elith J, Leathwick JR. 2009Species distribution models: ecological explanation and prediction across space and time. Ann. Rev. Ecol. Evol. Syst. 40, 677-697. ( 10.1146/annurev.ecolsys.110308.120159) [DOI] [Google Scholar]
- 73.Araújo MB, Peterson AT. 2012Uses and misuses of bioclimatic envelope modeling. Ecology 93, 1527-1539. ( 10.1890/11-1930.1) [DOI] [PubMed] [Google Scholar]
- 74.Hastie T, Tibshirani R, Friedman J. 2009The elements of statistical learning, pp. 587-604. Berlin, Germany: Springer. [Google Scholar]
- 75.Walker JG, Plein M, Morgan ER, Vesk PA. 2017Uncertain links in host–parasite networks: lessons for parasite transmission in a multi-host system. Phil. Trans. R. Soc. B 372, 20160095. ( 10.1098/rstb.2016.0095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bartomeus I, Gravel D, Tylianakis JM, Aizen MA, Dickie IA, Bernard-Verdier M. 2016A common framework for identifying linkage rules across different types of interactions. Funct. Ecol. 30, 1894-1903. ( 10.1111/1365-2435.12666) [DOI] [Google Scholar]
- 77.Zamke RL, Schlater LK. 1984Abscesses in a free-ranging bison in Alaska. J. Wildl. Dis. 20, 151-152. ( 10.7589/0090-3558-20.2.151) [DOI] [PubMed] [Google Scholar]
- 78.Rzewuska M, Stefańska I, Osińska B, Kizerwetter-Świda M, Chrobak D, Kaba J, Bielecki W. 2012Phenotypic characteristics and virulence genotypes of Trueperella (Arcanobacterium) pyogenes strains isolated from European bison (Bison bonasus). Vet. Microbiol. 160, 69-76. ( 10.1016/j.vetmic.2012.05.004) [DOI] [PubMed] [Google Scholar]
- 79.Wade S, Haschek W, Georgi J. 1979Ostertagiosis in captive bison in New York State: report of nine cases. Cornell Vet. 69, 198-205. [PubMed] [Google Scholar]
- 80.Knapp S, Marley S, Button S, Rognlie M. 1993Bibliography and Abstracts: Parasites of the American bison, Bison bison Linnaeus. (Miscellaneous Publication 363). Bozeman, MT: Montana State University. [Google Scholar]
- 81.Samuel WM, Pybus MJ, Kocan AA. 2001Parasitic diseases of wild mammals, 2nd edn. Ames, IA: Iowa State University Press. [Google Scholar]
- 82.Dubey JP. 1983Experimental infection of bison with Toxoplasma gondii oocysts. J. Wild Dis. 19, 148-149. ( 10.7589/0090-3558-19.2.148) [DOI] [PubMed] [Google Scholar]
- 83.Moskwa B, Kornacka A, Cybulska A, Cabaj W, Reiterova K, Bogdaszewski M, Steiner-Bogdaszewska Z, Bien J. 2018Seroprevalence of Toxoplasma gondii and Neospora caninum infection in sheep, goats, and fallow deer farmed on the same area. Sci. J. Anim. Sci. 96, 2468-2473. ( 10.1093/jas/sky122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walker ML, Becklund WW. 1970Checklist of the internal and external parasites of deer, Odocoileus hemionus and O. virginianus, in the United States and Canada. Washington, DC: US Department of Agriculture. [Google Scholar]
- 85.Russell LJ. 1967Parasites of the whitetail deer (Odocoileus virginianus ochrourus). PhD thesis, University of British Columbia. [Google Scholar]
- 86.Tryland M, Beckmen KB, Burek-Huntington KA, Breines EM, Klein J. 2018Orf virus infection in Alaskan mountain goats, Dall's sheep, muskoxen, caribou and Sitka black-tailed deer. Acta Vet. Scand. 60, 12. ( 10.1186/s13028-018-0366-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kerr GR, Holmes JC. 1966Parasites of mountain goats in west central Alberta. J. Wildl. Manag. 30, 786-790. ( 10.2307/3798286) [DOI] [Google Scholar]
- 88.Aleuy OA, Ruckstuhl K, Hoberg EP, Veitch A, Simmons N, Kutz SJ. 2018Diversity of gastrointestinal helminths in Dall's sheep and the negative association of the abomasal nematode, Marshallagia marshalli, with fitness indicators. PLoS ONE 13, e0192825. ( 10.1371/journal.pone.0192825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Williams ES, Barker IK (eds). 2001. Infectious diseases of wild mammals. Ames, IA: Iowa State University Press. [Google Scholar]
- 90.Thorne ET, Honess RE. 1982Diseases of wildlife in Wyoming. Cheyenne, WY: Wyoming Game and Fish Department. [Google Scholar]
- 91.Jenkins EJ, Veitch AM, Kutz SJ, Hoberg EP, Polley L. 2006Climate change and the epidemiology of protostrongylid nematodes in northern ecosystems: Parelaphostrongylus odocoilei and Protostrongylus stilesi in Dall's sheep (Ovis d. dalli). Parasitology 132, 387-401. ( 10.1017/S0031182005009145) [DOI] [PubMed] [Google Scholar]
- 92.Kreeger TJ, Cook WE, Edwards WH, Cornish T. 2004Brucellosis in captive Rocky Mountain bighorn sheep (Ovis canadensis) caused by Brucella abortus biovar 4. J. Wildl. Dis. 40, 311-315. ( 10.7589/0090-3558-40.2.311) [DOI] [PubMed] [Google Scholar]
- 93.Krzysiak MK, Iwaniak W, Kęsik-Maliszewska J, Olech W, Larska M. 2017Serological study of exposure to selected arthropod-borne pathogens in European bison (Bison bonasus) in Poland. Transbound Emerg. Dis. 64, 1411-1423. ( 10.1111/tbed.12524) [DOI] [PubMed] [Google Scholar]
- 94.Karbowiak G, Demiaszkiewicz A, Pyziel A, Wita I, Moskwa B, Werszko J, Bień J, Goździk K, Lachowicz J, Cabaj W. 2014The parasitic fauna of the European bison (Bison bonasus) (Linnaeus, 1758) and their impact on the conservation. Part 1. The summarising list of parasites noted. Acta Parasitol. 59, 363-371. ( 10.2478/s11686-014-0252-0) [DOI] [PubMed] [Google Scholar]
- 95.Scagliarini A, Vaccari F, Turrini F, Bianchi A, Cordioli P, Lavazza A. 2011Parapoxvirus infections of red deer, Italy. Emerg. Infect. Dis. 17, 684-687. ( 10.3201/eid1704.101454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tell LA, Brooks JW, Lintner V, Matthews T, Kariyawasam S. 2011Antimicrobial susceptibility of Arcanobacterium pyogenes isolated from the lungs of white-tailed deer (Odocoileus virginianus) with pneumonia. J. Vet. Diagn. Invest. 23, 1009-1013. ( 10.1177/1040638711416618) [DOI] [PubMed] [Google Scholar]
- 97.Doyle L, Heuschele W. 1983Bovine viral diarrhea virus infection in captive exotic ruminants. J. Am. Vet. Med. Assoc. 183, 1257-1259. [PubMed] [Google Scholar]
- 98.Frölich K. 2000Viral diseases of northern ungulates. Ran 20, 83. ( 10.7557/2.20.2-3.1505) [DOI] [Google Scholar]
- 99.Ataseven V, Başaran Z, Yılmaz V, Dağalp S. 2010Seroprevalence of parainfluenza virus-3 (PIV-3) and bovine herpesvirus type 1 (BHV-1) infections in goats of Van region. Yüzüncü yıl Üniversitesi Veteriner Fakültesi Dergisi 21, 7-9. [Google Scholar]
- 100.Pavone S, Crotti S, Cruciani D, D'Avino N, Zema J, Morelli S, Gobbi M, Madeo L. 2020Fatal systemic toxoplasmosis in a 3-month-old young Tibetan goat (Capra hircus). BMC Vet. Res. 16, 423. ( 10.1186/s12917-020-02641-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuzmina T, Kharchenko V, Malega A. 2010Helminth fauna of roe deer (Capreolus capreolus) in Ukraine: biodiversity and parasite community. Vestnik Zool. 44, e-12. [Google Scholar]
- 102.Robin C, Abdel Wahab S, Donnell R, Kania S. 2013Detection of antibodies to ovine herpesvirus-2 interleukin-10 homologue in sheep-associated malignant catarrhal fever. Suez Canal Vet. Med. J. 18, 159-166. ( 10.21608/SCVMJ.2013.78289) [DOI] [Google Scholar]
- 103.Li H, Dyer N, Keller J, Crawford TB. 2000Newly recognized herpesvirus causing malignant catarrhal fever in white-tailed deer (Odocoileus virginianus). J. Clin. Microbiol. 38, 1313-1318. ( 10.1128/JCM.38.4.1313-1318.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gibb Ret al.2021Data proliferation, reconciliation, and synthesis in viral ecology. bioRxiv 426572. ( 10.1101/2021.01.14.426572) [DOI] [Google Scholar]
- 105.García‐Valdés R, Zavala MA, Araujo MB, Purves DW. 2013Chasing a moving target: projecting climate change‐induced shifts in non‐equilibrial tree species distributions. J. Ecol. 101, 441-453. ( 10.1111/1365-2745.12049) [DOI] [Google Scholar]
- 106.Cirtwill AR, Eklöf A, Roslin T, Wootton K, Gravel D. 2019A quantitative framework for investigating the reliability of empirical network construction. Methods Ecol. Evol. 10, 902-911. ( 10.1111/2041-210X.13180) [DOI] [Google Scholar]
- 107.Becker DJ, et al. 2020Predicting wildlife hosts of betacoronaviruses for SARS-CoV-2 sampling prioritization: a modeling study. bioRxiv 111344. ( 10.1101/2020.05.22.111344) [DOI]
- 108.Carlson CJ. 2020From PREDICT to prevention, one pandemic later. The Lancet Microbe 1, e6-e7. ( 10.1016/S2666-5247(20)30002-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han BA, et al. 2019Confronting data sparsity to identify potential sources of Zika virus spillover infection among primates. Epidemics 27, 59-65. ( 10.1016/j.epidem.2019.01.005) [DOI] [PubMed] [Google Scholar]
- 110.Calatayud J, et al. 2020Positive associations among rare species and their persistence in ecological assemblages. Nat. Ecol. Evol. 4, 40-45. ( 10.1038/s41559-019-1053-5) [DOI] [PubMed] [Google Scholar]
- 111.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. 2017Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646-650. ( 10.1038/nature22975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Budria A, Candolin U. 2014How does human-induced environmental change influence host-parasite interactions? Parasitology 141, 462. ( 10.1017/S0031182013001881) [DOI] [PubMed] [Google Scholar]
- 113.Agosta SJ, Janz N, Brooks DR. 2010How specialists can be generalists: resolving the ’parasite paradox’ and implications for emerging infectious disease. Zoologia (Curitiba) 27, 151-162. ( 10.1590/S1984-46702010000200001) [DOI] [Google Scholar]
- 114.Bruggeman J, Heringa J, Brandt BW. 2009PhyloPars: estimation of missing parameter values using phylogeny. Nucleic Acids Res. 37(Suppl. 2), W179-W184. ( 10.1093/nar/gkp370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Allan BF, Goessling LS, Storch GA, Thach RE. 2010Blood meal analysis to identify reservoir hosts for Amblyomma americanum ticks. Emerg. Infect. Dis. 16, 433-440. ( 10.3201/eid1603.090911) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McIntyre KM, Setzkorn C, Hepworth PJ, Morand S, Morse AP, Baylis M. 2017Systematic assessment of the climate sensitivity of important human and domestic animals pathogens in Europe. Sci. Rep. 7, 7134. ( 10.1038/s41598-017-06948-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tomkins M, Kliot A, Marée AF, Hogenhout SA. 2018A multi-layered mechanistic modelling approach to understand how effector genes extend beyond phytoplasma to modulate plant hosts, insect vectors and the environment. Curr. Opin. Plant Biol. 44, 39-48. ( 10.1016/j.pbi.2018.02.002) [DOI] [PubMed] [Google Scholar]
- 118.Redding DW, Moses LM, Cunningham AA, Wood J, Jones KE. 2016Environmental-mechanistic modelling of the impact of global change on human zoonotic disease emergence: a case study of Lassa fever. Methods Ecol. Evol. 7, 646-655. ( 10.1111/2041-210X.12549) [DOI] [Google Scholar]
- 119.Castex V, de Cortázar-Atauri IG, Calanca P, Beniston M, Moreau J. 2020Assembling and testing a generic phenological model to predict Lobesia botrana voltinism for impact studies. Ecol. Modell. 420, 108946. ( 10.1016/j.ecolmodel.2020.108946) [DOI] [Google Scholar]
- 120.Taylor LH, Latham SM, Woolhouse ME. 2001Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B 356, 983-989. ( 10.1098/rstb.2001.0888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Han BAet al.2019Confronting data sparsity to identify potential sources of Zika virus spillover infection among primates. Epidemics 27, 59-65. ( 10.1016/j.epidem.2019.01.005) [DOI] [PubMed] [Google Scholar]
- 122.Carlson CJ, Albery GF, Merow C, Trisos CH, Zipfel CM, Eskew EA, Olival KJ, Ross N, Bansal S. 2020Climate change will drive novel cross-species viral transmission. bioRxiv 918755. ( 10.1101/2020.01.24.918755) [DOI] [PubMed] [Google Scholar]
- 123.Allen T, Murray KA, Zambrana-Torrelio C, Morse SS, Rondinini C, Di Marco M, Breit N, Olival KJ, Daszak P. 2017Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 8, 1124. ( 10.1038/s41467-017-00923-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hassell JM, Begon M, Ward MJ, F'evre EM. 2017Urbanization and disease emergence: dynamics at the wildlife–livestock–human interface. Trends Ecol. Evol. 32, 55-67. ( 10.1016/j.tree.2016.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source data for analyses are available and accessible through previous publications (see electronic supplementary materials for details). Derived data and custom computer R code to perform analyses are freely available at GitHub, https://github.com/MoralesCastilla/HostParasiteForecasting/.