Abstract
Animals and fungi produce cholesterol and ergosterol, respectively, while plants produce the phytosterols stigmasterol, campesterol, and β‐sitosterol in various combinations. The recent sequencing of many algal genomes allows the detailed reconstruction of the sterol metabolic pathways. Here, we characterized sterol synthesis in two sequenced Chlorella spp., the free‐living C. sorokiniana, and symbiotic C. variabilis NC64A. Chlamydomonas reinhardtii was included as an internal control and Coccomyxa subellipsoidea as a plant‐like outlier. We found that ergosterol was the major sterol produced by Chlorella spp. and C. reinhardtii, while C. subellipsoidea produced the three phytosterols found in plants. In silico analysis of the C. variabilis NC64A, C. sorokiniana, and C. subellipsoidea genomes identified 22 homologs of sterol biosynthetic genes from Arabidopsis thaliana, Saccharomyces cerevisiae, and C. reinhardtii. The presence of CAS1, CPI1, and HYD1 in the four algal genomes suggests the higher plant cycloartenol branch for sterol biosynthesis, confirming that algae and fungi use different pathways for ergosterol synthesis. Phylogenetic analysis for 40 oxidosqualene cyclases (OSCs) showed that the nine algal OSCs clustered with the cycloartenol cyclases, rather than the lanosterol cyclases, with the OSC for C. subellipsoidea positioned in between the higher plants and the eight other algae. With regard to why C. subellipsoidea produced phytosterols instead of ergosterol, we identified 22 differentially conserved positions where C. subellipsoidea CAS and A. thaliana CAS1 have one amino acid while the three ergosterol producing algae have another. Together, these results emphasize the position of the unicellular algae as an evolutionary transition point for sterols.
Keywords: algal sterol composition, Chlorella sorokiniana , Chlorellavariabilis NC64A, Clotrimazole, Coccomyxa subellipsoidea , Ketoconazole, oxidosqualene cyclase, terbinafine
Abbreviations
- BBM
Bold's basal medium
- CAS
cycloartenol synthase
- ER
endoplasmic reticulum
- %GC
percent guanine cytosine
- GC/MS
gas chromatography mass spectroscopy
- HMG CoA
3‐hydroxy‐3‐methylglutaryl coenzyme A
- IPP
isopentenyl pyrophosphate
- LAS
lanosterol synthase
- MBBM
modified Bold's basal medium
- MEP
methyl‐D‐erythritol 4‐phosphate
- MVA
mevalonate
- OSC
2,3‐oxidosqualene cyclase
Sterols are a type of lipid found within the membrane of animals, fungi, and plants and contribute to membrane stability and other important cellular functions (He et al. 2003). Sterols are found in many forms: free sterols, sterol esters, sterol alkyl ethers, sterol sulfates, or linked to a glycoside moiety (Benveniste 2004). As integral components of the cell membrane, free sterols are crucial for the integrity, fluidity, and permeability of the lipid bilayer (Benveniste 2004). In addition to their importance for cell membrane stability, they affect membrane‐bound protein composition and influence the functionality of enzymes, receptors, and channels (Porsbring et al. 2009) and play an important role in host defense during viral infection (Blanc et al. 2011).
Interestingly, despite their consistent presence, sterols are found in distinctly different compositions between the kingdoms. Whereas cholesterol is the primary zoosterol, ergosterol is the principal mycosterol, and separately, a handful of phytosterols such as stigmasterol, campesterol, and β‐sitosterol dominate the plant membrane. In all cases, sterol biosynthesis occurs in the endoplasmic reticulum (ER) through a functional complex of enzymes that display specific protein‐protein interactions. In fungi, this complex of enzymes has been termed the “ergosome” (Mo and Bard 2005). In comparison, the sterol biosynthetic pathway present in plants differs from fungi and animals because plants produce a wide variety of phytosterols and intermediates in the phytosterol pathway. While the sterol composition of most algae is well established, the recent availability of full genomes allows a detailed reconstruction of the sterol metabolic pathways.
Our aim was to study sterol biosynthesis in four unicellular, freshwater green algae. Our approach took advantage of well‐characterized inhibitors which block sterol production in fungi, leading either to an accumulation of sterol precursors or to a different suite of sterols. Here, we report the sterol composition of four green algae: Chlorella variabilis NC64A, Chlorella sorokiniana, Chlamydomonas reinhardtii, and Coccomyxa subellipsoidea. A major difference among these algae was that ergosterol was the major sterol in both the Chlorella spp. and C. reinhardtii, whereas C. subellipsoidea did not have ergosterol but contained three phytosterols instead. Using the sterol biosynthetic pathways identified in Arabidopsis thaliana, (Benveniste 2004), Saccharomyces cerevisiae (Parks and Casey 1995, Mo and Bard 2005), and C. reinhardtii (Brumfield et al. 2017) as plant, fungal, and algal models, respectively, we describe an ergosterol biosynthetic pathway present in both Chlorella spp. This pathway fits with the modified sterols and sterol precursors observed following treatment with the antibiotics ketoconazole and clotrimazole. Finally, we tackled the plant‐like sterol composition of C. subellipsoidea by multiple sequence alignment of relevant oxidosqualene cyclase proteins. We identified 22 residues where the 3 ergosterol‐containing algae used one amino acid while C. subellipsoidea and the A. thaliana CAS used another.
MATERIALS AND METHODS
Cell cultures and growth conditions
All species were grown under normal growth conditions as follows. Chlorella variabilis NC64A and Coccomyxa subellipsoidea cultures were grown in modified Bolds basal medium (MBBM; Van Etten et al. 1983) shaken at 100 RPM, 22°C, and a light intensity of 30 µE. Chlorella sorokiniana UTEX 1230 was obtained from the University of Texas Culture Collection and grown in liquid Bolds Basal Medium (BBM; Nichols and Bold 1965), shaken at 115 RPM, 25°C, and a light intensity of 58 µE. Chlamydomonas reinhardtii CC124, obtained from Dr. Donald Weeks, was grown in Tris‐Acetate‐Phosphate medium (Gorman and Levine 1965), shaken at 100 RPM, 22°C, and a light intensity of 30 µE. Cell abundance was determined using a Coulter Multisizer II instrument (Beckman Coulter, Fullerton, CA, USA).
Sterol standards and inhibitors
A plant sterols kit (cat. #1123, Matreya, Pleasant Gap, PA, USA) was used for standards. Cholestanol (1 mg · mL‐1) was used as a sterol internal standard for GC‐MS analysis. Obtusifoliol was a gift from Prof. David Nes, Texas Tech University. Antifungal inhibitors Atorvastatin (A7658), Terbinafine (T1672), Clotrimazole (C4657), Ketoconazole (K1676), and Fluconazole (F4682) were purchased from LKT Laboratories (St. Paul, MN, USA). Thiolutin was obtained from Tocris Bioscience (Bristol, UK).
Sterol extraction and analysis by gas chromatography‐mass spectrometry (GC/MS)
Sterols were extracted in triplicate by chloroform‐methanol, purified by silica‐solid phase extraction, and analyzed by GC/MS. One mg/ml of cholestanol standard (Matreya, Pleasant Gap, PA, USA) was added to freeze‐dried algal pellets followed by extraction three times with chloroform: MeOH (1:1, v:v). Samples were dried in a stream of nitrogen, dissolved in chloroform, and loaded onto silica SPE columns. A purified sterol fraction was eluted from the column with 30% 2‐propanol in hexane. Purified sterol extracts were dried under nitrogen and then converted to trimethylsilyl ether (TMS‐ether) derivatives using bis (trimethylsilyl) trifluoracetamide (BSFTA‐TMCS 99:1; Sigma, St. Louis, MO, USA). Dried sterol samples were suspended in 100 µl hexane for GC/MS analysis. Initial gas chromatographic analysis was carried out using the Agilent 6890 Series Gas Chromatograph System equipped with a DB‐5ms capillary column (30.0 m x 250.00 µm, 0.25 µm, J&W 122‐5532, J&W Scientific, Inc., Folsom, CA, USA). Helium was the carrier gas at a linear velocity of 48 cm/sec and constant flow of 1.5 mL · min‐1. A dual ramp temperature program was used with the oven heated from 250 to 270°C for 30 min and then from 270 – 280°C for 3.5 min. The detector temperature was 270°C. Sterols were initially identified using the NIST98 library (Scientific Instrument Services, Inc., Ringoes, NJ, USA) followed by comparisons to published spectra based on their mass fragmentation patterns and retention times (see Table 1). Peak areas of identified sterols were quantified relative to the cholestanol standard. Values reported are the average of triplicate experiments.
Table 1.
Retention times of sterol standards.
| Sterol standard | Rfa | m/z (TMS‐ether) | PubChem CID | NIST |
|---|---|---|---|---|
| Ergostatetraenol | 1.14 | 466 | 23724485 | 17866 |
| Ergosterol | 1.15 | 468 | 444679 | 23726 |
| Campesterol | 1.17 | 472 | 173183 | 331835 |
| 14α‐Methylergosta‐8,24(28)‐dienolb | 1.18 | 484 | ||
| 14α‐Methylergost‐8‐enolb | 1.21 | 486 | 129703316 | |
| Stigmasterol | 1.22 | 484 | 5280794 | 331824 |
| Ergosta‐5,7,24(28)‐trienolc | 1.23 | 468 | 42608412 | 23726d |
| Ergosta‐5,7‐dienol | 1.27 | 470 | 5326970 | |
| Ergost‐7‐enol | 1.32 | 472 | 5283646 | 17938 |
| 7‐Dehydroporiferasterol | 1.33 | 482 | 20843308 | 17146e |
| 4α,14α‐Dimethylergosta‐8,22‐dienol | 1.33 | 498 | 101614321 | |
| Obtusifoliol | 1.37 | 498 | 65252 | 16573 |
| β‐sitosterol | 1.37 | 486 | 222284 | 331677 |
| 24(28)‐Dihydroobtusifoliolf | 1.39 | 500 | 23258269 | |
| Cycloartenol | 1.40 | 500 | 313075 | |
| 24(28)‐Dihydrocycloeucalenol | 1.56 | 500 | 6427296g | |
| 24‐Methylenecycloartenol | 1.57 | 512 | 94204 | |
| 24‐Methylcycloartenol | 1.62 | 514 | 13784482 | |
| 9,19‐Cyclolanost‐25‐en‐3β‐ol, 24‐methyl | 1.66 | 512 | ||
| Cyclolaudenol | 1.75 | 512 | 101729 |
Growth curves and MIC values
The inhibitory concentrations of terbinafine and the azole drugs were determined using methanol (MeOH) solutions of the drug ranging from 0‐50 mM. A culture of Chlorella sorokiniana was diluted with fresh BBM to a final cell density of 1x106 cells · mL‐1. The culture was partitioned into 5 mL volumes and treated with 5 µL of a given stock solution, which resulted in a 1:1000 dilution of the stock drug solution. Other samples were treated with 5 µL of MeOH as solvent controls. Trials were run in duplicate. After six days, the absorbance at 750 nm was measured via a Biotek Synergy H1 Hybrid reader (Winooski, VT, USA). Inhibitory concentrations were determined by graphing absorbance vs. drug concentration and identifying the lowest concentrations at which growth was inhibited. Growth curves were similar in design except that absorbance was read every day. To determine if a given drug was algicidal (killing the algal cultures) or algistatic (merely arresting cell growth), cultures that had been inhibited for 2‐3 d were centrifuged at 4200 RPM for three min and then transferred into 5 mL of fresh BBM media to allow growth.
Sterol biosynthesis inhibition
Algal cultures were grown to mid‐log phase (ca. 1x106 cells · mL‐1). The sterol inhibitors were added to each culture at final concentrations of 100 µM atorvastatin, 4 µM terbinafine, 30 µM ketoconazole, and 5 µM clotrimazole. These concentrations were chosen based on the MIC values and growth inhibition we observed for C. sorokiniana. For terbinafine, ketoconazole, and clotrimazole, the concentrations were slightly higher than those needed for 50% inhibition of cell growth while for the noninhibitory atorvastatin, 100 µM was the highest concentration tested. Methanol (0.1%) was added to control cultures. Algal cultures were incubated for an additional 48h before cells were harvested by centrifugation. Pelleted algal cells were freeze‐dried and stored at −80°C for further analysis of their sterol composition. Peaks were identified based on their retention times and MS fragmentation patterns (Table 1). Values reported are the average of triplicate experiments.
Sequence retrieval
The complete Chlorella variabilis genome assembly (http://genome.jgi.psf.org/ChlNC64A_1/ChlNC64A_1.home.html;v1.0; Blanc et al. 2010) was used to search and identify sterol biosynthetic pathway genes as well as the recently sequenced and annotated Chlorella sorokiniana UTEX 1230 genome (H.D. Cerutti, unpub. data). The genomes for Arabidopsis thaliana (NCBI taxon ID: 3702), Chlamydomonas reinhardtii (NCBI taxon ID: 3055), and Coccomyxa subellipsoidea (NCBI taxon ID: 574566) were accessed through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Saccharomyces cerevisiae (NCBI taxon ID: 4932) ergosterol (ERG) genes were used to identify homologues and elucidate the ergosterol pathway in C. variabilis. The number of transmembrane domains for each protein in each species was predicted using version 2.0 of TMHMM (Sonnhammer et al. 1998).
Phylogenetic analysis
The 2,3‐oxidosqualene cyclase (OSC) sequences used for the phylogenetic analyses were taken from the results of a BLASTp search (version 2.2.30) against the nonredundant protein database using default settings (Altschul et al. 1990). Sequences from lanosterol‐producing vertebrates and fungi as well as cycloartenol‐producing plants, diatoms, and algae were chosen to represent a broad representation of each lineage. The squalene‐hopane cyclase from Acetobacter tropicalis was included as an out‐group to derive the root of the tree, but not shown in the final phylogeny to maintain readability due to its divergence from the eukaryotic OSC genes. The protein sequences were globally aligned using version 7.402 of MAFFT (Katoh et al. 2019). The maximum likelihood phylogenic tree was produced with version 20120412 of PhyML with 1000 bootstrap pseudoreplicates (Guindon et al. 2010).
OSC structure prediction
The tertiary structure of the Coccomyxa subellipsoidea CAS protein was predicted using the template‐based structure prediction from the RaptorX webserver (Källberg et al. 2012). The predictions were based primarily on the human OSC in complex with lanosterol (PDB 1W6K). The key structures were confirmed using the template‐free contact prediction from the RaptorX webserver (Wang et al. 2017). The predicted structure was visualized and annotated using version 2.3.2 of PyMOL (Schrödinger 2019).
RESULTS
We took a 4‐prong approach to study sterol biosynthesis in four unicellular algae. We determined [A] which sterols were synthesized under standard growth conditions, [B] which of six known antifungal inhibitors were also inhibitory toward the microalgae, [C] how these inhibitors influenced the sterol compositions of the respective algae, and [D] which homologs of known sterol biosynthetic genes from Arabidopsis thaliana, Saccharomyces cerevisiae, and Chlamydomonas reinhardtii were present in C. subellipsoidea, C. variabilis and C. sorokiniana.
Sterol composition of four microalgae
The major sterols were identified by GC/MS for four unicellular, freshwater green algae Chlorella variabilis, C. sorokiniana, Chlamydomonas reinhardtii, and Coccomyxa subellipsoidea. Four different patterns were observed (Table 2). Chlamydomonas reinhardtii and the two Chorella sp. contained ergosterol as the dominant sterol. Chlamydomonas reinhardtii contained ergosterol (63%), ergost‐7‐enol (10%), and 7‐dehydroporiferasterol (27%; Table 2). Note that ergosterol (C24ß‐methyl) and 7‐dehydroporiferasterol (C24ß‐ethyl) differ by only a single carbon. Our results for C. reinhardtii serve as an internal control in that they confirm prior reports of ergosterol and 7‐dehydroporiferasterol as the major sterols (Miller et al. 2012, Brumfield et al. 2017). The distinction between C. variabilis having both ergosterol and ergostatetraenol and C. sorokiniana having ergosterol only (Table 2) reflects in part the absence of the final enzyme C‐24 (28) sterol reductase in C. variabilis (Table 3) with the concomitant necessity of recruiting a less efficient reductase to make ergosterol. Ergostatetraenol is the precursor for ergosterol (Fabris et al. 2014, Berkow et al. 2015, Brumfield et al. 2017; Fig. 1) and the two molecules have very similar retention times by GC/MS (Table 1). Thus, a small difference in the % completion of the last step could determine whether ergostatetraenol appeared as a separate peak (C. variabilis) or as a shoulder on the ergosterol peak (C. sorokiniana). In contrast, C. subellipsoidea resembled the vascular plants because it only contained the phytosterols campesterol (48%), β‐sitosterol (43%), and stigmasterol (9%; Table 2).
Table 2.
Percentage of major sterols in four microalgae.
| Name | Chlorella variabilis | Chlorella sorokiniana | Coccomyxa subellipsoidea |
Chlamydomonas reinhardtii |
|---|---|---|---|---|
| Ergostatetraenol | 42% | |||
| Ergosterol | 53% | 99% | 63% | |
| Campesterol | 48% | |||
| Stigmasterol | 9% | |||
| Ergosta‐5,7,24(28)‐trienol | 2% | |||
| Ergosta‐5,7‐dienol | 3% | |||
| Ergost‐7‐enol | 1% | 10% | ||
| 7‐dehydroporiferasterol | 27% | |||
| beta‐sitosterol | 43% | |||
| Total Sterols (nmol · mg dw‐1) | 2.5 | 3.2 | 3.6 | 3.4 |
Table 3.
Sterol biosynthetic pathway genes identified in algae. (NF = not found, number in parentheses is protein length in aa).
| Enzyme | Chlamydomonas reinhardtii | Chlorella sorokiniana | Chlorella variabilis | Coccomyxa subellipsoidea |
|---|---|---|---|---|
| Stage I plastid ‐ MEP Pathway | ||||
| 1‐deoxy‐D‐xylulose 5‐phosphate synthase | Cre07.g356350 (736) | sca185.g101550 (712) | 59788 (721) | 16525 (733) |
| 1‐deoxy‐D‐xylulose 5‐phosphate reductoisomerase | Cre12.g546050 (456) | sca079.g101000 (469) | 29723 (456) | 47418 (476) |
| 2‐C‐methyl‐D‐erythritol 4‐phospate cytidyltransferase | Cre16.g679669 (320) | sca011.g102250 (304) | 141910 (168) | 35670 (260) |
| 4‐diphosphocytidyl‐2‐C‐methyl‐D‐erythritol kinase | Cre02.g145050 (348) | sca085.g104500 (366) | 33454 (354) | 35952 (301) |
| 2‐C‐methyl‐D‐erythritol 2,4‐cyclo‐diphosphate synthase | Cre12.g503550 (208) | sca055.g102250 (140) | 12213 (167) | 18339 (208) |
| 1‐hydroxy‐2‐methyl‐2E‐butenyl‐4‐diphosphate synthase | Cre12.g490350 (682) | sca127.g100200 (1157) | 144676 (699) | 27377 (749) |
| 4‐hydroxy‐3‐methyl‐2E‐butenyl‐4‐diphosphate reductase | Cre08.g372950 (466) | sca012.g106300 (402) | 59658 (506) | 53820 (439) |
| Isopentenyl pyrophosphate isomerase | Cre11.g467544 (254) | sca136.g102550 (306) | 53578 (244) | 36851 (230) |
| Stage I cytosol ‐ MVA pathway | ||||
| Acetoacetyl‐CoA thiolase | Cre02.g146050 (491) | sca243.g102350 (533) | 27161 (401) | 37141 (404) |
| 3‐Hydroxy‐3‐methylglutaryl‐CoA synthase | Cre16.g678850 (718) | sca110.g104250 (476) | 138158 (513) | 27385 (520) |
| 3‐Hydroxy‐3‐methylglutaryl‐CoA reductase | N.F. | N.F. | N.F. | N.F. |
| Mevalonate kinase | N.F. | N.F. | N.F. | N.F. |
| Phosphomevalonate kinase | N.F. | N.F. | N.F. | N.F. |
| Mevalonate diphosphate decarboxylase | N.F. | N.F. | N.F. | N.F. |
| Stage II ER | ||||
| Farnesyl diphosphate synthetase | Cre03.g207700 (361) | sca140.g100250 (358) | 33543 (347) | 22862 (340) |
| Farnesyl diphosphate farnesyl transferase | Cre03.g175250 (462) | sca085.g101650 (437) | 11287 (187) | 35284 (348) |
| Monooxygenase/hydrolase (squalene epoxidase) | Cre17.g734644 (524) | sca094.g100900 (545) | 137251 (525) | 53930 (509) |
| Stage III ER | ||||
| Cycloartenol synthase | Cre01.g011100 (763) | sca081.g101700 (695) | 22200 (762) | 54267 (754) |
| Lanosterol synthase | N.F. | N.F. | N.F. | N.F. |
| Sterol C‐24 methyltransferase | Cre12.g500500 (388) | sca033.g100850 (412) | 26131 (333) | 27964 (389) |
| Cycloeucanol cycloisomerase | Cre16.g657300 (276) | sca047.g103200 (298) | 11379 (282) | 65481 (298) |
| Sterol C‐14 demethylase | Cre02.g092350 (496) | sca139.g104650 (498) | 142513 (613) | 54016 (492) |
| C(8,7) sterol isomerase | Cre12.g557900 (219) | sca034.g101250 (297) | 34496 (224) | 13584 (235) |
| C‐4 sterol methyl oxidase | Cre06.g261200 (308) | sca222.g101000 (518) | 57760 (290) | 11014 (308) |
| C‐3 sterol dehydrogenase | Cre12.g518650 (402) | sca089.g108000 (277) | 49861 (361) | 54657 (357) |
| 3‐keto sterol reductase | N.F. | N.F. | N.F. | N.F. |
| Endoplasmic reticulum protein | Cre13.g567901 (74) | sca039.g105150 (149) | 59539 (149) | 14819 (124) |
| C‐5 sterol desaturase | Cre16.g663950 (346) | sca134.g102400 (337) | 37407 (286) | 15746 (275) |
| C‐22 sterol desaturase | Cre11.g467527 (516) | sca146.g107050 (528) | 30281 (422) | 29437 (539) |
| C‐24 (28) sterol reductase | Cre02.g076800 (426) | sca232.g100350 (444) | N.F. | 26214 (436) |
Fig. 1.
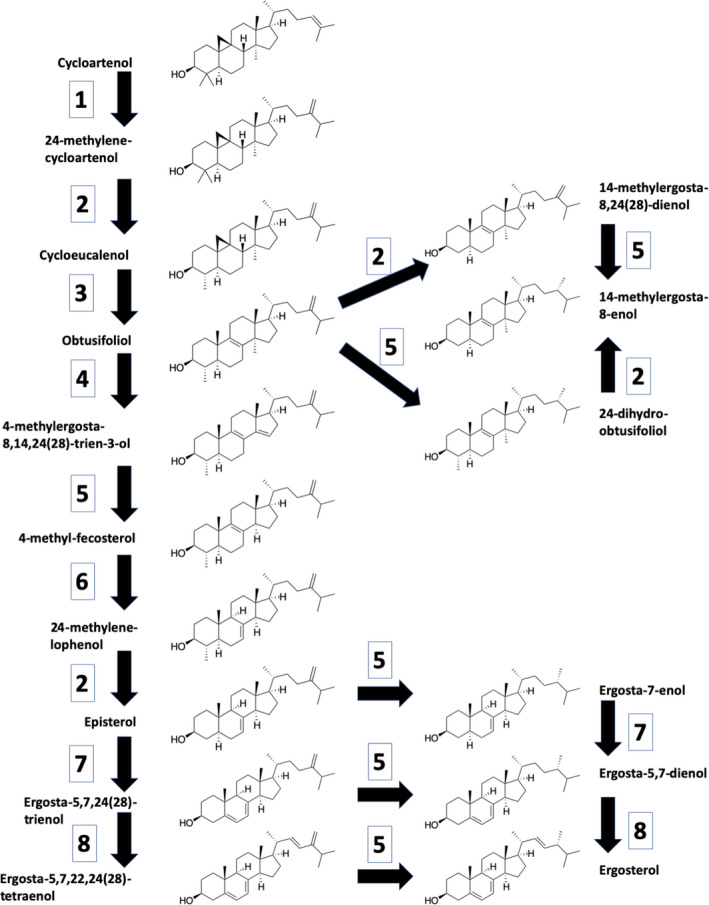
Putative sterol pathway in Chlorella variabilis and Chlorella sorokiniana showing structures of the biosynthetic intermediates and the enzymes which catalyze each step, starting for convenience with cycloartenol, the product of Cas1p. 1: SMT1‐sterol C‐24 methyltransferase; 2: SMO2‐C‐4 sterol methyloxidase; 3: CPI1‐cycloeucanol cycloisomerase; 4: CYP51‐sterol C‐14 demethylase; 5: ERG4/24‐C‐24(28) sterol reductase; 6: HYD1‐C (8, 7) sterol isomerase; 7: STE1‐C‐5 sterol desatirase; 8: CYP710‐C‐22 sterol desaturase. The biosynthetic pathway follows that suggested by Brumfield et al (2017) for C. reinhardtii, overlaid with our data showing the sterol intermediates which accumulate following treatment with ketoconazole or clotrimazole, drugs thought to inhibit the two cytochrome P450 enzymes, CYP51 and CYP710, here labeled 4 and 8, respectively.
Sensitivity to known antimycotic compounds
Chlorella sorokiniana cultures (1x106 cells · mL‐1) were tested for their sensitivity to six antifungal antibiotics, five of which (terbinafine, clotrimazole, ketoconazole, fluconazole, and atorvastatin) block sterol biosynthesis in fungi (Macreadie et al. 2006), whereas thiolutin inhibits RNA polymerases. Two of the antibiotics, fluconazole and atorvastatin, were not inhibitory at any concentrations tested up to 50 and 100 µM, respectively. This finding is not surprising for atorvastatin, a well‐known inhibitor of HMG CoA reductase used to lower cholesterol levels in humans, because neither Chlorella sp. has the gene for HMG CoA reductase (Table 3). For another inhibitor, terbinafine, the cultures did not have differing sterol profiles, but, compared with the controls, their total ergosterol levels decreased by ca. 40%, from 4.3 nmol · mg‐1 to 2.5 nmol · mg‐1 in C. variabilis and 3.2 nmol · mg‐1 to 2.0 nmol · mg‐1 dry weight for C. sorokiniana (Table 4). These results are close to those of Lu et al. (2014) who reported a 20% decrease in total sterol content for terbinafine‐treated Nannochloropsis oceanica. Together these observations are consistent with a mode of action for terbinafine in algae identical with that established in fungi, that is, inhibition of squalene epoxidase. This mode of action would reduce the entry of carbon into stage III of the sterol pathway but not alter the remaining sterol profiles.
Table 4.
Antifungal inhibition of Chlorella spp. sterol biosynthetic pathway and resulting sterol compositions.
| Chlorella sorokiniana | Chlorella variabilis | |||||
|---|---|---|---|---|---|---|
| Sterol | Control | Clotrimazole | Ketoconazole | Control | Clotrimazole | Ketoconazole |
| Ergostatetraenol | 41.7% | 24.1% | 28.2% | |||
| Ergosterol | 99% | 57% | 47% | 53% | 38.8% | 44.3% |
| 14α‐Methylergosta‐8,24(28)‐dienol | 1% | 18% | 17.8% | 7.3% | ||
| 14α‐Methylergost‐8‐enol | 24% | |||||
| Ergosta‐5,7,24(28)‐trienol | 2.3% | 3.6% | ||||
| Ergosta‐5,7‐dienol | 13% | 2.9% | 6.9% | 5.9% | ||
| Ergost‐7‐enol | 1% | 6% | 0.4% | |||
| Obtusifoliol | 22% | 10% | 8.8% | 13.8% | ||
| Cyclolaudenol | 2% | |||||
| Total Sterols (nmol · mg dw‐1) | 3.2 | 2.6 | 2.6 | 4.3 | 3.5 | 4.0 |
Terbinafine, clotrimazole, thiolutin, and ketoconazole, blocked growth of Chlorella sorokiniana at minimal inhibitory concentrations of 2, 2, 10, and 20 µM respectively. Each of these four active drugs was algistatic rather than algicidal, in agreement with the fungistatic nature of their activity toward fungi (Odds et al. 1985). Thus, if their modes of action in algae parallel those in fungi, terbinafine, clotrimazole, and ketoconazole should be effective inhibitors of algal sterol synthesis while thiolutin, as a transcription inhibitor, would allow the determination of algal mRNA half‐lives (Kebaara et al. 2006). The two inhibitory azoles, clotrimazole and ketoconazole, are both imidazoles whereas fluconazole is a triazole, and this structural difference may contribute to their sterol inhibition, or lack thereof.
Sterol biosynthesis inhibition
The azole antifungals inhibit key enzymes in sterol biosynthesis, leading to the accumulation of diagnostic intermediates that allow us to study the intermediates, and in doing so better understand their sterol biosynthetic pathways. Thus, Chlorella sorokiniana, C. variabilis, Chlamydomonas reinhardtii, and Coccomyxa subellipsoidea were treated with clotrimazole (5 µM) and ketoconazole (30 µM). These concentrations were chosen to be slightly higher than those needed for 50% inhibition of cell growth for C. sorokiniana in liquid culture. Methanol‐only controls were used in each case. Although the clotrimazole and ketoconazole concentrations were chosen based on their activity toward C. sorokiniana, their general suitability for the other green algae is shown by their dramatically altered sterol profiles in all cases (Tables 4 and 5, Table S1 in the Supporting Information).
Table 5.
Antifungal inhibition of Coccomyxa subellipsoidea sterol biosynthetic pathway and resulting sterol compositions.
| Sterol | Control | Clotrimazole | Ketoconazole |
|---|---|---|---|
| Campesterol | 48.1% | 40.1% | 31.7% |
| 14α‐Methylergost‐8‐enol | 4.2% | ||
| Stigmasterol | 9.0% | 6.1% | 5.2% |
| 4α,14α‐Dimethylergosta‐8,22‐dienol | 4.7% | ||
| β‐Sitosterol | 42.9% | 42.7% | 31.1% |
| Obtusifoliol | 4.3% | 2.6% | |
| 24,25‐dihydrolanosterol | 2.5% | ||
| 24‐Methylenecycloartenol | 0.9% | ||
| 9,19‐Cyclolanost‐22‐en‐3β‐ol, 24‐methyl | 9.2% | ||
| Cyclolaudenol | 14.3% | ||
| Total Sterols (nmol · mg dw‐1) | 3.6 | 4.6 | 3.7 |
In Chlorella sorokiniana, the decreased ergosterol in the ketoconazole treated cells was accompanied by the appearance of 10% obtusifoliol, 18% 14α‐methylergosta‐8,24(28)‐dienol (also known as 4‐desmethyl obtusifoliol), and 24% 14α‐methylergost‐8‐enol (Table 4). This observation is consistent with ketoconazole inhibiting the P450 CYP51 C‐14 demethylase whose normal substrate is obtusifoliol (step 4 in Fig. 1). CYP51 is an ortholog of ERG 11, the primary target of the azole drugs in fungi. Thus, inhibition of CYP51 leads to the accumulation of obtusifoliol and subsequent conversion to 14‐methylergost‐8‐enol by the combined action of Erg 4/24 and the SM02/BSD1/Erg28 complex, acting in either order. Next, the sterols detected in clotrimazole treated cells (Table 4) suggest that clotrimazole inhibits CYP710, the P450 C‐22 sterol desaturase, as well as CYP51 because clotrimazole treated cells also accumulated ergost‐7‐enol and ergost‐5,7‐dienol (Table 4). Inhibition of CYP710 (step 8 in Fig. 1) would lead to the accumulation of episterol and ergosta‐5, 7, 24 (28) trienol which can both act as substrates for Erg 4/24, being converted to ergost‐7‐enol and ergosta‐5, 7‐dienol, respectively (Fig. 1).
The sterol profiles for Chlorella variabilis following azole treatment are very similar to those for C. sorokiniana (Table 4) but in this case ketoconazole also appears to inhibit CYP710 as well as CYP51. Previously, Doyle et al. (1971) noted that triparanol‐treated Chlorella emersonii accumulated both 14‐methylergost‐8‐enol and 14‐methylergosta‐8, 24 (28)‐dienol while Chan et al. (1974) observed that triparanol‐treated C. sorokiniana accumulated large amounts of ergost‐7‐enol and ergost‐8‐enol in addition to their normal ergosterol and ergosta‐5, 7‐dienol.
The sterol inhibition patterns for the other two algae were more complicated. For Chlamydomonas reinhardtii, ketoconazole and clotrimazole dramatically reduced the levels of both ergosterol and ergost‐7‐enol with the concomitant appearance of several intermediates from the cycloartenol side of the oxidosqualene cyclase pathway (Table S1). However, the levels of 7‐dehydroporiferasterol remained relatively unchanged. At present we have no explanation for the different responses of the two major sterols in C. reinhardtii. Finally, in the presence of the azoles, Coccomyxa subellipsoidea, which normally synthesizes the phytosterols campesterol, stigmasterol, and β‐sitosterol (Table 5), produced seven new intermediates from the cycloartenol pathway including obtusfoliol and cyclolaudenol. We will revisit why C. subellipsoidea synthesizes a different suite of sterols a little later when we analyze the amino acid sequence of the Coccomyxa CAS1.
Sequence retrieval and curation
Protein sequences from known Arabidopsis thaliana sterol biosynthetic genes were used in BLASTp searches against the Chlorella sorokiniana genome to identify their orthologs. The C. sorokiniana protein sequences identified were then used in BLASTp searches against the remaining algal sequences to identify their orthologs in each species. All of the identified orthologs were used in reciprocal BLASTp searches of the NCBI database to confirm the identities of these genes. Genes for sterol biosynthesis in the three model systems, Arabidopsis thaliana, Saccharomyces cerevisiae, and Chlamydomonas reinhardtii, are shown in Tables S2 and S3 in the Supporting Information while the biosynthetic genes for the four unicellular algae are in Table 3. Complete BLASTp statistics along with values for gene length in amino acids, % GC content, % identity, and numbers of exons and transmembrane domains are shown in Tables [Link], [Link], [Link], [Link] in the Supporting Information, for C. sorokiniana (Table S4), C. variabilis (Table S5), C. reinhardtii (Table S6), and Coccomyxa subellipsoidea (Table S7). Similar data for the model systems A. thaliana and S. cerevisiae are in Tables S3 and S8 in the Supporting Information, respectively. The gene lengths and % GC values are consistent with the respective genes being orthologs to one another, with no indication of their arrival by horizontal gene transfer.
The sterol biosynthetic genes were compared in three groups: Stage 1, forming isopentenyl pyrophosphate (IPP); Stage 2, IPP to 2,3‐oxidosqualene; and Stage 3, oxidosqualene to completion. For stage 1, Arabidopsis thaliana has both the plastid‐localized MEP pathway and the cytosolic MVA pathway, while the yeast Saccharomyces cerevisiae has only the cytosolic MVA pathway (Table S2). In contrast, all of the algae lack the last four enzymes (HMGR, MK, PMK, and MVD) of the MVA pathway including HMG CoA reductase; instead, they only contain the plastidal MEP pathway for IPP synthesis (Table 3). The absence of HMG CoA reductase was supported by the negligible effect of atorvastatin on growth and sterol production (as previously discussed), suggesting that Chlorella spp. only use the plastidic MEP pathway to produce IPP (Table 3). This feature was also demonstrated for Chlamydomonas reinhardtii by Schwender et al. (1997) and for the diatom Haslea ostrearia by Massé et al. (2004); however, Fabris et al. (2014) showed that the diatom Phaeodactylum tricornutum retained and used the MVA pathway to make IPP.
Stage 2 of sterol biosynthesis – IPP to 2, 3‐oxidosqualene – has three enzymatic steps and homologous genes were present in the three model systems (Table S2) and all four of the algae for each of these steps (Table 3). Thus, our data agree with the conclusion from Benveniste (2004) that the biosynthetic pathway from IPP to 2, 3‐oxidosqualene is the same in all eukaryotes.
For stage 3, from 2, 3‐oxidosqualene to completion, there are at least six points of interest: 1/ Arabidopsis thaliana has two oxidosqualene cyclase (OSC) genes, designated CAS1 for cycloartenol synthase and LAS1 for lanosterol synthase (Tables S2 and S3), while the other organisms have only one. The yeast Saccharomyces cerevisiae has ERG7, a lanosterol synthase (Tables S2 and S8), while the four algae have only a single cycloartenol synthase (Table 3). The assignment of these algal genes as CAS rather than LAS is based primarily on their functionality and the chemical identity of their downstream products and is supported by the bioinformatic analyses shown later. 2/ The presence of the three genes CAS1, CPI1, and HYD1 in the genomes for Chlorella sorokiniana, C. variabilis, Coccomyxa subellipsoidea, and Chlamydomonas reinhardtii strongly suggests that these algae use the higher plant cycloartenol branch of sterol biosynthesis, which is likely a general feature of the green algae. 3/ In yeasts, the C‐4 demethylation step is catalyzed by a complex encoded by ERG 25, 26, 27, and 28; however, C. reinhardtii (Brumfield et al. 2017) and the other algae have orthologs to ERG 25, 26, and 28, but not to ERG 27 (Table 3). 4/ The ergosterol biosynthetic pathway is composed of membrane‐associated enzymes assembled as a multi‐enzyme complex. In yeasts, the noncatalytic protein ERG28 functions as a scaffold protein, anchoring enzymes and creating a “hub” for enzymatic interactions with substrates (Mo and Bard 2005, Winkel 2009). We have identified an ERG28 homolog in C. variabilis (59539), C. sorokiniana (039.g105150.t1), and C. subellipsoidea (141819) as well as in C. reinhardtii and other algae (Brumfield et al. 2017). 5/ For C. sorokiniana, the 11 genes for stages 1 and 2 are all high % GC: they range from 63.6 to 70.8% GC, averaging 67.1% GC. Similarly, the 11 predicted genes for Stage 3 are also high % GC, ranging from 61.8 to 67.1% GC, averaging 65.1% GC (Table S4). There are no apparent differences in % GC for the genes encoding plastid‐localized components versus ER‐localized components (Table S4). The only notable difference based on localization is the number of transmembrane domains (Table S4). No further conclusions can be drawn from these high % GC values because the genomic GC value for C. sorokiniana is 63% GC. 6/ RNA‐Seq experiments with C. sorokiniana showed that all 22 of the predicted sterol biosynthetic genes were expressed as full‐length transcripts during growth (W. Riekhof, unpub. data). Thus, the direct gene comparison reinforces the premise that the green algae as a group produce sterols using the cycloartenol pathway of higher plants and algae rather than the lanosterol pathway of fungi and animals.
Comparison of Chlorella sp., Coccomyxa subellipsoidea, and Chlamydomonas reinhardtii
Sterol biosynthesis in the two Chlorella spp. and Coccomyxa subellipsoidea (Table 3) are very similar to Chlamydomonas reinhardtii both biochemically and genetically (Table S2; Miller et al. 2012, Brumfield et al. 2017). All four algae make IPP via the plastid MEP pathway and are missing the cytosolic MVA pathway (Table 3). Likewise, all four contain the same suite of 14 stage II and III genes (Table 3). These 14 genes (Tables [Link], [Link], [Link], [Link]) are roughly equivalent in their percent identity, length of coding sequence, and number of introns present. One notable exception is SMO2 (C‐4 sterol methyl oxidase), which stands out by being only 29% identical between C. sorokiniana (Table S4) and C. reinhardtii (Table S6). Additionally, C. sorokiniana is notable for having several more exons (up to twice as many) than the other species for nearly all sterol‐related genes, consistent with the increased number of exons across the rest of its genome (Table S4).
Phylogenetic analysis of OSC
Saccharomyces cerevisiae has a single oxidosqualene cyclase OSC (ERG 7) which cyclizes 2, 3‐oxidosqualene to make lanosterol, while Arabidopsis thaliana has 13 OSCs, one of which (LAS1, At3g45130) makes lanosterol and another (CAS1, At2g07050) makes cycloartenol (Tables S2 and S3; Xue et al. 2012). To determine the number of OSC genes in the four algae, we performed BLASTp searches against all 13 A. thaliana OSCs (Table S9 in the Supporting Information). For all four species, the only significant hits to any of the A. thaliana OSCs is the single OSC reported. Additionally, in every case, the alignment for the algal OSC protein is 5‐6% higher with CAS1 than LAS1.
To investigate the algal OSCs further, we constructed a phylogenetic tree for forty OSCs (Fig. 2). The sequences separated into four clades or clusters. The first included OSCs that preferentially produce the sterol precursor cycloartenol, forming the branch for higher plants and algae. The second clade included Metazoa using lanosterol, while the third clade included the fungi using lanosterol. A fourth clade included two OSCs from the diatoms Thalassiosira pseudonana and Fistulifera solaris (Fig. 2). All nine of the microalgal OSCs were part of the cycloartenol‐forming plant OSCs (Fig. 2). The Chlorella spp. have ergosterol as their major sterol (Table 2) and it is likely synthesized via CAS and cycloartenol because only cycloartenol/obtusifoliol intermediates were formed after inhibition by ketoconazole and clotrimazole (Tables 4 and 5). It is well known that single‐site mutations in the Arabidopsis thaliana CAS gene can produce lanosterol instead of cycloartenol (Segura et al. 2002, Benveniste 2004). However, as pointed out by Brumfield et al. (2017), multiple sequence alignment shows that the four algal OSCs all have the highly conserved amino acids expected for cycloartenol synthase (CAS) rather than for lanosterol synthase (LAS; Fig. 3).
Fig. 2.
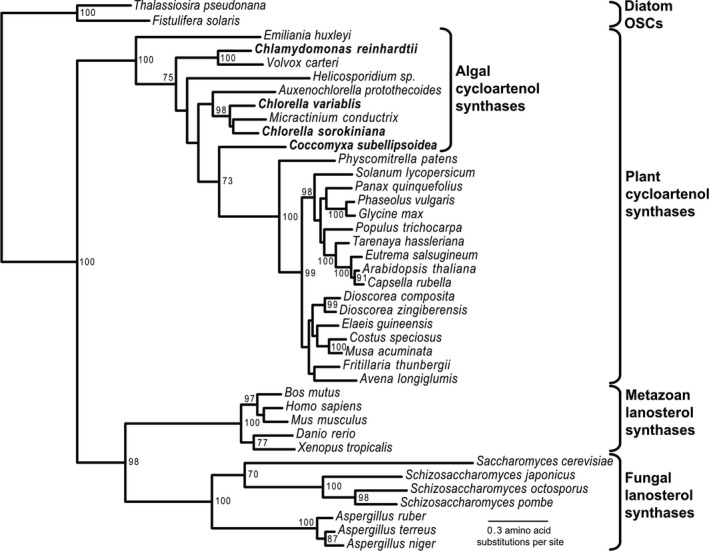
Phylogenetic tree for 40 oxidosqualene cyclase (OSC) proteins showing the position of genes from 9 algal species relative to their plant, animal, fungal, and diatom equivalents. Acetobacter tropicallis (not shown) was used as an out‐group to derive the root.
Fig. 3.

Multiple sequence alignment of OSC proteins presented in Tables 3 and S2. The three residues identified by Brumfield et al. (2017) as conserved in CAS proteins are colored in orange. The catalytic aspartic acid (D483 for Arabidopsis thaliana) is colored in blue. Residues that are conserved across Chlamydomonas reinhardtii, Chlorella sorokiniana, and C. variabilis but are differently conserved in A. thaliana CAS1 and Coccomyxa subellipsoidea are highlighted in yellow.
The Coccomyxa question
Coccomyxa subellipsoidea appears to be intermediary between the other algae and higher plants, both in its OSC phylogeny (Fig. 2) and in its dramatically different sterol composition (Tables 2 and 6). Additionally, its % GC composition ranges from 51 to 62% for the sterol biosynthetic genes in the MEP pathway and from 49 to 63% for the genes for stages 2 and 3 (Table S7) whereas the genes for the two Chlorella sp. (Tables S4 and S5) were more tightly clustered around their average values of 66% GC. To investigate possible causes for this transition, we conducted a multiple sequence alignment of the OSC proteins from Arabidopsis thaliana (both CAS1 and LAS1), Saccharomyces cerevisiae, and the four algae we have studied (Fig. 3). The three residues used by Brumfield et al. (2017) as characteristic of cycloartenol directed synthesis (marked in orange) and the catalytic aspartic acid (marked in blue) are identical for the four algae and the A. thaliana CAS (Fig. 3), and thus they cannot explain the synthesis of the three phytosterols by C. subellipsoidea. Instead, we have identified 22 amino acid positions in the CAS protein that are differentially conserved. That is, the C. subellipsoidea CAS and A. thaliana CAS1 have one amino acid, but the three ergosterol‐containing algae have a different amino acid. These 22 amino acids are shown in yellow on the multiple sequence alignment (Fig. 3). Most of these 22 amino acids are located either around the catalytic site or on the outer surface of the protein (Fig. 4).
Fig. 4.
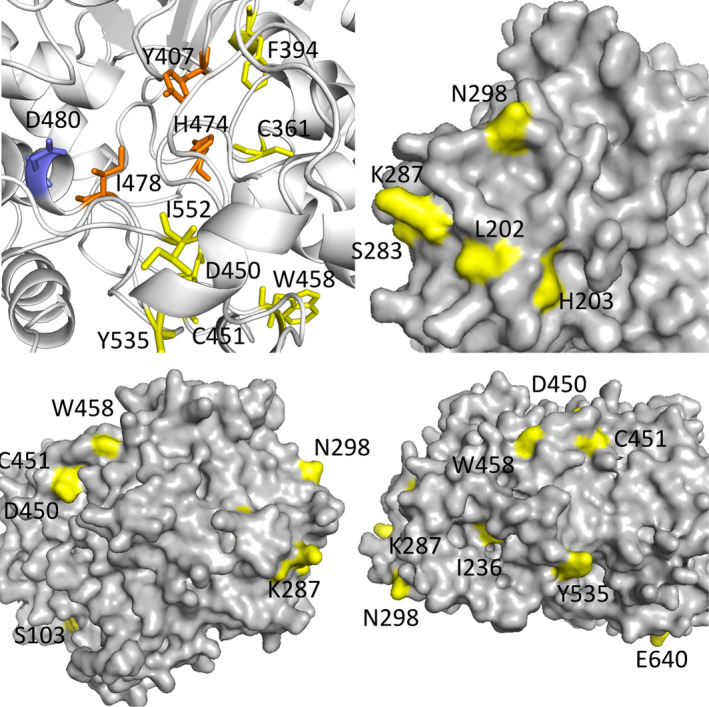
Four views of the tertiary structure prediction of Coccomyxa subellipsoidea CAS. Positions are given for the residues on the C. subellipsoidea sequence (Fig. 3). Yellow residues are conserved across Chlamydomonas reinhardtii, Chlorella sorokiniana, and C. variabilis, but are differentially conserved in Arabidopsis thaliana CAS1 and C. subellipsoidea. The catalytic D480 residue is colored in blue. Residues identified by Brumfield et al. (2017) as conserved in CAS proteins are colored in orange. 15 of the 22 differently conserved residues are shown. These residues cluster on the outer surface of the protein and around the catalytic region and could alter the protein‐protein binding of the CAS proteins and/or reaction dynamics to explain the shift from ergosterol to phytosterol production in C. subellipsoidea.
DISCUSSION
Unicellular algae as an evolutionary transition point for sterols
The biosynthesis of sterols in animals, fungi, and plants differs in terms of precursors, enzymatic steps, and final sterol composition. It is widely accepted that the major sterols present in animals and fungi are cholesterol and ergosterol, respectively, and that both employ the cytosol localized MVA pathway leading to the 5‐carbon precursor IPP. Subsequent steps are shared in animals and fungi up to lanosterol, after which a bifurcated pathway produces their respective distinct sterols. Plants differ in that they use the plastid‐localized MEP pathway to make IPP and then use the precursor cycloartenol (instead of lanosterol) to produce a wide array of phytosterols. Currently, algae have been grouped with plants as producing cycloartenol and utilizing the phytosterol biosynthetic pathway. We studied ergosterol biosynthesis in Chlorella variabilis NC64A and C. sorokiniana because of their recently sequenced and annotated genomes (Blanc et al. 2010; H.D. Cerutti, unpub. data). Coccomyxa subellipsoidea was included because its sterol lipid profile appears to be transitionary between algae and plants, which is supported by the OSC phylogenetic analysis (Table 2). We combined these bioinformatic data (Table 3) with the antibiotic inhibition of key steps in sterol biosynthesis (Table 4) to create a putative ergosterol biosynthetic pathway for C. variabilis and C. sorokiniana (Fig. 1).
In terms of their sterol contents, unicellular algae exhibit greater diversity than either multicellular algae or plants. Our results emphasize the unique position of the unicellular algae as an evolutionary transition point for sterols and suggest that the unicellular algae provide the evolutionary precursor for the cycloartenol based pathways in higher plants (Fig. 2; Haubrich et al. 2014, Vriet et al. 2015).
Sterol diversity
In addition to the four patterns we observed in Table 2, previous studies on multicellular algae reported the presence of cholesterol in the marine red algae and fucosterol in the marine brown algae (Fieser and Fieser 1959) while Lu et al. (2014) reported that the oleaginous microalga Nannochloropsis oceanica had both cholesterol and fucosterol/isofucosterol as the major and minor sterols, respectively. These general classifications were extended by Duperon et al. (1983) to identify and quantify the free sterols, sterol esters, steryl glycosides, and acetylated steryl glycosides present in marine algae. More recently, this evolutionary diversity was illustrated by the studies of Rampen et al. (2010) who examined 106 diatom cultures looking for an unambiguous diatom biomarker. They detected 44 sterols, of which 11 were major sterols in that they comprised ≥ 10% of the total sterols in one or more diatom. However, none of the major sterols qualified as a diatom‐specific biomarker because all of them were common sterols in other algae (Rampen et al. 2010).
In algae, the intricate roles and functions of sterols are not completely understood. Our putative pathway (Fig. 1) helps explain the diversity in sterols in unicellular algae, which may contribute to our understanding their specific functions. Factors influencing sterol differences among algal species could be morphological, environmental, or evolutionary. From a morphological viewpoint, having an alternating life cycle or multiple distinct cell types may require distinct sterols, possibly using both cycloartenol and lanosterol precursors. Different membrane structures might also be related to whether the algae had evolved as the photosynthetic partner in a symbiosis, with the added complication that both primary and secondary plastid endosymbiotic events may have occurred. From an environmental perspective, water composition, temperature, salinity, nutrient, and ion composition are highly variable between freshwater and marine ecosystems (Porsbring et al. 2009). For the algae we have studied, Chlorella sorokiniana can grow at warmer temperatures than C. variabilis while Coccomyxa subellipsoidea is unusual in that it can still grow at extreme, near‐polar temperatures. These temperature adaptations likely require different sterol compositions (Starr and Parks 1962).
Why ergosterol?
Changing environmental conditions, such as light intensity and day length, have been shown to change the ratio of sterols present in higher plants (Rahier and Taton 1997). Algae within the class Chlorophyceae have many species that contain ergosterol and/or other Δ5, 7‐ sterols not found in higher orders of algae and land plants (Patterson et al. 1991). One idea is based on the absorption spectrum of sterols containing two conjugated double bonds. The evolution away from cell membranes containing Δ5, 7‐ sterols to those with either Δ5 ‐ or Δ7‐sterols took place as the earth’s ozone layer developed, because Δ5,7‐ sterols are more efficient in absorbing UV radiation (Patterson et al. 1991). Indeed, ergosterol is commonly assayed by its absorption at 280 nm. Hence, once the ozone layer was established, the selective advantage of the Δ5,7‐ sterols may have disappeared. Higher plants contain a cocktail of three Δ5 phytosterols – campesterol (24‐methyl), stigmasterol (24‐ethyl), and β‐sitosterol (24‐ethyl) (Mercer 1993, Holmberg et al. 2002) and this phytosterol composition also occurs in the unicellular green alga Coccomyxa (Table 2). The size and direction of the 24‐alkyl group inserted by sterol methyltransferase are indicative of either a primitive (24 β‐methyl) or advanced (24 α‐ethyl) organism (Zhou et al. 2007).
Another more recent idea is that the sterol composition may be constrained by the other aspects of an interlocking membrane composition (i.e., the need for sterols to partner with particular sphingolipids) providing lipid rafts and other distinctive capabilities for the algal membranes (Guan et al. 2009, Gulati et al. 2010, Hannich et al. 2011). An attractive feature of sterol‐sphingolipid pairing is that it provides a rationale why organisms as distinct, ecologically, evolutionarily, and metabolically as algae and Saccharomyces utilize the same sterol, ergosterol.
Authors Contributions
SLR, JLVE, AV, and KWN designed the research; SLR, NTMC, MK, and AV performed the experiments; all authors analyzed data; and AV and KWN wrote the paper.
Supporting information
Table S1. Antifungal inhibition of Chlamydomonas reinhardtii sterol biosynthetic pathway and resulting sterol compositions.
Table S2. Sterol biosynthetic pathway genes in model organisms.
Table S3. Stage I, II, and III genes in Arabidopsis thaliana.
Table S4. BLASTp results for protein identification in Chlorella sorokiniana.
Table S5. BLASTp results for protein identification in Chlorella variabilis.
Table S6. BLASTp results for protein identification in Chlamydomonas reinhardtii.
Table S7. BLASTp results for protein identification in Coccomyxa subellipsoidea.
Table S8. BLASTp results for protein identification in Saccharomyces cerevisiae.
Table S9. BLASTp results for all 13 Arabidopsis thaliana OSC proteins in algae.
ACKNOWLEDGEMENT
NTMC was supported by the Integrated Development of Bioenergy Systems (IDBS) Summer Research Program at the University of Nebraska‐Lincoln.Funding for this work was provided by the NSF‐EPSCoR grant EPS‐1004094 (to JVE and WRR), the COBRE program of the National Center for Research Resources Grant P20‐RR15535 (JVE), and Ann L Kelsall and the Farnesol and Candida albicans Research Fund, University of Nebraska Foundation (KWN). We thank Heriberto Cerutti for allowing us to use the C. sorokiniana UTEX‐1230 genome sequence data prior to publication.
Contributor Information
Adam Voshall, Email: adam.voshall@childrens.harvard.edu.
Kenneth W. Nickerson, Email: knickerson1@unl.edu.
References
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–10. [DOI] [PubMed] [Google Scholar]
- Benveniste, P.2004. Biosynthesis and accumulation of sterols. Ann. Rev. Plant Biol. 55:429–57. [DOI] [PubMed] [Google Scholar]
- Berkow, E. L., Manigaba, K., Parker, J. E., Barker, K. S., Kelly, S. L. & Rogers, P. D. 2015. Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis . Antimicrob. Agents Chemother. 59:5942–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., Duncan, G., Agarkova, I., Borodovsky, M., Gurnon, J., Kuo, A., Lindquist, E. et al. 2010. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22:2943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, M., Hsieh, W. Y., Robertson, K. A., Watterson, S., Shui, G., Lacaze, P., Khondoker, M. et al. 2011. Host defense against viral infection involves interferon mediated down‐regulation of sterol biosynthesis. PLoS Biol. 9:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden, B. N. & Palmer, M. A. 1975. 24‐Dihydro obtusifoliol from sorghum‐vulgare caryopses. Phytochemistry 14:1140–1. [Google Scholar]
- Brumfield, K. M., Laborde, S. M. & Moroney, J. V. 2017. A model for the ergosterol biosynthetic pathway in Chlamydomonas reinhardtii . Eur. J. Phycol. 52:64–74. [Google Scholar]
- Chan, J. T., Patterson, G. W., Dutky, S. R. & Cohen, C. F. 1974. Inhibition of sterol biosynthesis in Chlorella sorokiniana by triparanol. Plant Physiol. 53:244–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, P. J., Patterson, G. W., Dutky, S. R. & Cohen, C. F. 1971. 14alpha‐methyl‐5alpha‐ergost‐8‐en‐3beta‐ol and 14alpha‐methyl‐5alpha‐ergosta‐8:24(28)‐dien‐3beta‐ol from triparanol‐treated Chlorella emersonii . Phytochem. 10:2093–8. [Google Scholar]
- Duperon, R., Thiersault, M. & Duperon, P. 1983. Occurrence of steryl glycosides and acylated steryl glycosides in some marine algae. Phytochem. 22:535–8. [Google Scholar]
- Fabris, M., Matthijs, M., Carbonelle, S., Moses, T., Pollier, J., Dasseville, R., Baart, G. J. E., Vyverman, W. & Goossens, A. 2014. Tracking the sterol biosynthesis pathway of the diatom Phaeodactylum tricornutum . New Phytol. 204:521–35. [DOI] [PubMed] [Google Scholar]
- Fieser, L. F. & Fieser, M. 1959. Steroids. Reinhold Pub. Corp, New York, NY, USA, 964 pp. [Google Scholar]
- Gorman, D. S. & Levine, R. P. 1965. Cytochrome F and plastocyanin ‐ their sequence in photosynthetic electron transport chain of Chlamydomonas reinhardtii . Proc. Natl. Acad. Sci. USA 54:1665–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, X. L., Souza, C. M., Pichler, H., Dewhurst, G., Schaad, O., Kajiwara, K., Wakabayashi, H. et al. 2009. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol. Biol. Cell 20:2083–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. 2010. New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biol. 59:307–21. [DOI] [PubMed] [Google Scholar]
- Gulati, S., Liu, Y., Munkacsi, A. B., Wilcox, L. & Sturley, S. L. 2010. Sterols and sphingolipids: Dynamic duo or partners in crime? Prog. Lipid Res. 49:353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich, J. T., Umebayashi, K. & Riezman, H. 2011. Distribution and functions of sterols and sphingolipids. Cold Spring Harb. Perspect Biol. 3:a004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubrich, B. A., Collins, E. K., Howard, A. L., Wang, Q., Snell, W. J., Miller, M. B., Thomas, C. D., Pleasant, S. K. & Nes, W. D. 2014. Characterization, mutagenesis and mechanistic analysis of an ancient algal sterol C24‐methyltransferase: Implications for understanding sterol evolution in the green lineage. Phytochem. 113:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J. X., Fujioka, S., Li, T. C., Kang, S. G., Seto, H., Takatsuto, S., Yoshida, S. & Jang, J. C. 2003. Sterols regulate development and gene expression in Arabidopsis . Plant Physiol. 131:1258–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg, N., Harker, M., Gibbard, C. L., Wallace, A. D., Clayton, J. C., Rawlins, S., Hellyer, A. & Safford, R. 2002. Sterol C‐24 methyltransferase type 1 controls the flux of carbon into sterol biosynthesis in tobacco seed. Plant Physiol. 130:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källberg, M., Wang, H. P., Wang, S., Peng, J., Wang, Z. Y., Lu, H. & Xu, J. B. 2012. Template‐based protein structure modeling using the RaptorX web server. Nat. Protoc. 7:1511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., Rozewicki, J. & Yamada, K. D. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20:1160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebaara, B. W., Nielsen, L. E., Nickerson, K. W. & Atkin, A. L. 2006. Determination of mRNA half‐lives in Candida albicans using thiolutin as a transcription inhibitor. Genome 49:894–9. [DOI] [PubMed] [Google Scholar]
- Lu, Y., Zhou, W., Wei, L., Li, J., Jia, J., Li, F., Smith, S. M. & Xu, J. 2014. Regulation of the cholesterol biosynthetic pathway and its integration with fatty acid biosynthesis in the oleaginous microalga Nannochloropsis oceanica . Biotechnol. Biofuels 7:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macreadie, I. G., Johnson, G., Schlosser, T. & Macreadie, P. I. 2006. Growth inhibition of Candida species and Aspergillus fumigatus by statins. FEMS Microbiol. Lett. 262:9–13. [DOI] [PubMed] [Google Scholar]
- Massé, G., Belt, S. T., Rowland, S. J. & Rohmer, M. 2004. Isoprenoid biosynthesis in the diatoms Rhizosolenia setigera (Brightwell) and Haslea ostrearia (Simonsen). Proc. Natl. Acad. Sci. USA 101:4413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer, E. I.1993. Inhibitors of sterol biosynthesis and their applications. Prog. Lipid Res. 32:357–416. [DOI] [PubMed] [Google Scholar]
- Miller, M. B., Haubrich, B. A., Wang, Q., Snell, W. J. & Nes, W. D. 2012. Evolutionarily conserved Delta(25(27)‐olefin ergosterol biosynthesis pathway in the alga Chlamydomonas reinhardtii . J. Lipid Res. 53:1636–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, C. Q. & Bard, M. 2005. A systematic study of yeast sterol biosynthetic protein‐protein interactions using the split‐ubiquitin system. Biochim. Biophys. Acta. 1737:152–60. [DOI] [PubMed] [Google Scholar]
- Nichols, H. W. & Bold, H. C. 1965. Trichosarcina polymorpha gen. et. sp. nov. J. Phycol. 1:34–8. [Google Scholar]
- Odds, F. C., Cockayne, A., Hayward, J. & Abbott, A. B. 1985. Effects of imidazole‐ and triazole‐derivative antifungal compounds on the growth and morphological development of Candida albicans hyphae. J. Gen. Microbiol. 131:2581–9. [DOI] [PubMed] [Google Scholar]
- Parks, L. W. & Casey, W. M. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49:95–116. [DOI] [PubMed] [Google Scholar]
- Patterson, G.W., Nes, W.D., & American Oil Chemists' Society . 1991. Physiology and biochemistry of sterols. American Oil Chemists' Society, Champaign, IL, USA, pp. 141–50. [Google Scholar]
- Porsbring, T., Blanck, H., Tjellstrom, H. & Backhaus, T. 2009. Toxicity of the pharmaceutical clotrimazole to marine microalgal communities. Aquat Toxicol. 91:203–11. [DOI] [PubMed] [Google Scholar]
- Rahier, A. & Taton, M. 1997. Fungicides as tools in studying postsqualene sterol synthesis in plants. Pestic Biochem Phys 57:1–27. [Google Scholar]
- Rampen, S. W., Abbas, B. A., Schouten, S. & Damste, J. S. S. 2010. A comprehensive study of sterols in marine diatoms (Bacillariophyta): Implications for their use as tracers for diatom productivity. Limnol Oceanogr 55:91–105. [Google Scholar]
- Schrödinger, L. L. C.2019. The PyMOL Molecular Graphics System. Version 2.3.2.
- Schwender, J., Zeidler, J., Groner, R., Muller, C., Focke, M., Braun, S., Lichtenthaler, F. W. & Lichtenthaler, H. K. 1997. Incorporation of 1‐deoxy‐D‐xylulose into isoprene and phytol by higher plants and algae. FEBS Lett. 414:129–34. [DOI] [PubMed] [Google Scholar]
- Segura, M. J. R., Lodeiro, S., Meyer, M. M., Patel, A. J. & Matsuda, S. P. T. 2002. Directed evolution experiments reveal mutations at cycloartenol synthase residue His477 that dramatically alter catalysis. Org. Lett 4:4459–62. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E. L., von Heijne, G. & Krogh, A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–82. [PubMed] [Google Scholar]
- Starr, P. R. & Parks, L. W. 1962. Effect of temperature on sterol metabolism in yeast. J. Cell Comp. Physiol. 59:107–10. [DOI] [PubMed] [Google Scholar]
- Van Etten, J. L., Burbank, D. E., Xia, Y. & Meints, R. H. 1983. Growth‐cycle of a virus, PBCV‐1, that infects Chlorella‐like algae. Virology 126:117–25. [DOI] [PubMed] [Google Scholar]
- Vriet, C., Lemmens, K., Vandepoele, K., Reuzeau, C. & Russinova, E. 2015. Evolutionary trials of plant steroid genes. Trends Plant Sci. 20:301–8. [DOI] [PubMed] [Google Scholar]
- Wang, S., Sun, S., Li, Z., Zhang, R. & Xu, J. 2017. Accurate de novo prediction of protein contact map by ultra‐deep learning model. PLoS Comp. Biology 13:e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel, B. S.2009. Metabolite Channeling and Multi‐enzyme Complexes. InOsbourn, A. & Lanzotti, V. [Eds.] Plant‐derived Natural Products. Springer, New York, NY, pp. 195–208. [Google Scholar]
- Xue, Z., Duan, L., Liu, D., Guo, J., Ge, S., Dicks, J., ÓMáille, P., Osbourn, A. & Qi, X. 2012. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 193:1022–38. [DOI] [PubMed] [Google Scholar]
- Zhou, W. X., Cross, G. A. M. & Nes, W. D. 2007. Cholesterol import fails to prevent catalyst‐based inhibition of ergosterol synthesis and cell proliferation of Trypanosoma brucei . J. Lipid Res. 48:665–73. [DOI] [PubMed] [Google Scholar]
- Zhou, W. X., Lepesheva, G. I., Waterman, M. R. & Nes, W. D. 2006. Mechanistic analysis of a multiple product sterol methyltransferase implicated in ergosterol biosynthesis in Trypanosoma brucei . J. Biol. Chem. 281:6290–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Antifungal inhibition of Chlamydomonas reinhardtii sterol biosynthetic pathway and resulting sterol compositions.
Table S2. Sterol biosynthetic pathway genes in model organisms.
Table S3. Stage I, II, and III genes in Arabidopsis thaliana.
Table S4. BLASTp results for protein identification in Chlorella sorokiniana.
Table S5. BLASTp results for protein identification in Chlorella variabilis.
Table S6. BLASTp results for protein identification in Chlamydomonas reinhardtii.
Table S7. BLASTp results for protein identification in Coccomyxa subellipsoidea.
Table S8. BLASTp results for protein identification in Saccharomyces cerevisiae.
Table S9. BLASTp results for all 13 Arabidopsis thaliana OSC proteins in algae.


