Abstract
Understanding the origin and maintenance of adaptive phenotypic novelty is a central goal of evolutionary biology. However, both hybridization and incomplete lineage sorting can lead to genealogical discordance between the regions of the genome underlying adaptive traits and the remainder of the genome, decoupling inferences about character evolution from population history. Here, to disentangle these effects, we investigated the evolutionary origins and maintenance of Batesian mimicry between North American admiral butterflies (Limenitis arthemis) and their chemically defended model (Battus philenor) using a combination of de novo genome sequencing, whole-genome resequencing, and statistical introgression mapping. Our results suggest that balancing selection, arising from geographic variation in the presence or absence of the unpalatable model, has maintained two deeply divergent color patterning haplotypes that have been repeatedly sieved among distinct mimetic and nonmimetic lineages of Limenitis via introgressive hybridization.
Keywords: hybridization, mimicry, introgression, ancestral polymorphism, incomplete lineage sorting
Introduction
Hybridization can promote or constrain adaptive divergence depending on the balance of gene flow, recombination, and selection (Felsenstein 1981; Barton and Hewitt 1985; Harrison 1993; Abbott et al. 2013). Unfortunately, hybridization also greatly complicates efforts to disentangle the history of adaptive trait evolution from population demography because recombination decouples patterns of variation at neutral and selected sites. Understanding the evolutionary history of alleles underlying adaptive traits among hybridizing lineages is further confounded by the fact that shared genetic variation between populations may arise due to either incomplete lineage sorting (ILS) of ancestral polymorphism or introgression following secondary contact (Maddison 1997; Degnan and Rosenberg 2009; Zhou et al. 2017). In spite of these challenges, distinguishing between genealogical patterns produced by the neutral demographic history of populations and those that reflect the history of character evolution for traits experiencing selection is essential to our understanding of how adaptive phenotypic novelty arises, the timing and mode of phenotypic evolution, and the role that selection on such traits plays in the speciation process (Stinchcombe and Hoekstra 2008; Nadeau and Jiggins 2010; Stapley et al. 2010).
Few systems offer better opportunities to investigate how selection and gene flow shape the history of adaptive trait evolution than examples of wing pattern diversity in butterflies. Butterflies display extraordinarily diverse color patterns that function in many aspects of thermoregulation, predator avoidance, and mate recognition (Kingsolver 1985; Beldade and Brakefield 2002; McMillan et al. 2002; Boggs et al. 2003). Much of this diversity stems from strong natural selection for Batesian and Müllerian mimicry (Joron and Mallet 1998; Mallet and Joron 1999; Kapan 2001; Gilbert 2003; Kunte 2009), and the last decade has witnessed rapid progress in identifying the genes responsible for the adaptive diversity in these mimetic color pattern traits (Joron et al. 2011; Reed et al. 2011; Martin et al. 2012; Gallant et al. 2014; Kunte et al. 2014; Nishikawa et al. 2015; Nadeau et al. 2016; Mazo-Vargas et al. 2017; Van Belleghem et al. 2017; Westerman et al. 2018).
Here, we investigate the evolutionary origin and spread of Batesian mimicry among hybridizing lineages of North American admiral butterflies in the genus Limenitis (fig. 1). Mimetic (Limenitis arthemis astyanax) and nonmimetic (Limenitis arthemis arthemis) populations of the polytypic Limenitis arthemis species complex meet and hybridize across a broad secondary contact zone in the eastern United States (Platt and Brower 1968; Mullen et al. 2008; Frentiu et al. 2015; fig. 1B). Predator-mediated natural selection for Batesian mimicry between Limenitis and its unpalatable model, Battus philenor, appears to partially limit gene flow between these two subspecies and maintains the position of the phenotypic hybrid zone (Mullen et al. 2008; Ries and Mullen 2008).
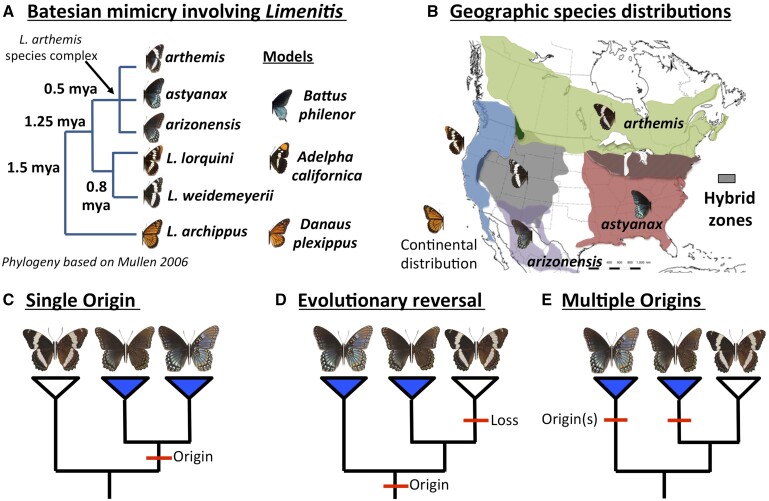
Fig. 1.
(Panel A) The evolutionary relationships among North American Limenitis species (Mullen 2006) and known examples of mimetic convergence. (Panel B) Parapatric distributions of Limenitis species and subspecies showing known areas of hybridization (diagonal lines). (Panels C–E) Competing hypotheses for the evolutionary origins of mimicry assuming it: (C) evolved once in the common ancestor of the two mimetic subspecies (Limenitis arthemis astyanax + Limenitis arthemis arizonensis), (D) evolved once and was lost due to an evolutionary reversal in nonmimetic, white-banded populations of Limenitis arthemis arthemis, or (E) evolved twice independently.
Recently, using a combination of positional cloning, population genomic resequencing, and functional assays, Gallant et al. (2014) demonstrated that mimetic variation in the distribution of melanin on the fore- and hind-wings of L . arthemis is associated with a large (30 kb) segregating haplotype, comprised 173 fixed SNPs in perfect linkage disequilibrium (LD), located ∼23 kb upstream of the 5′ coding region of the signaling ligand gene WntA. However, a second, geographically isolated subspecies of L . arthemis occurs in the southwestern United States (Limenitis arthemis arizonensis), which may have independently evolved mimicry with Battus, and, despite repeated attempts to reconstruct the evolutionary history of this complex (Mullen 2006; Prudic and Oliver 2008; Savage and Mullen 2009), the origin of the mimetic haplotype remains unclear.
To address this issue, we leveraged the recent identification of the genomic region responsible for adaptive mimetic variation in Limenitis (Gallant et al. 2014) to gain insights about the evolutionary origin and history of mimetic character evolution in this system. Specifically, we used a combination of de novo reference genome assembly, whole-genome population resequencing, and sliding window analyses to test competing phylogenetic hypotheses that mimicry 1) evolved once (fig. 1C) in the common ancestor of the two mimetic subspecies (L. a. astyanax + L. a. arizonensis), 2) evolved once and was lost due to an evolutionary reversal (fig. 1D) in nonmimetic, white-banded populations of L. a. arthemis, or 3) evolved twice independently (fig. 1E). Our results indicate that the two mimetic subspecies share a nearly identical haplotype upstream of WntA, rejecting the evolutionary reversal hypothesis, and suggest that mimicry spread via adaptive introgression from L. a. arizonensis into ancestrally white-banded populations of L. a. astyanax. In addition, comparisons of polymorphism and divergence across the color-patterning interval suggest that maintenance of the extensive LD observed within the mimetic haplotype is most likely due to multi-locus epistatic selection rather than a consequence of undetected structural variation (SV). Surprisingly, our results also provide evidence for introgression among white-banded lineages of Limenitis, implying that color pattern evolution in this group reflects a complex history of balancing selection on deeply divergent color patterning alleles, and introgression leading to the repeated sieving of ancestral polymorphism dependent on the selective environment provided by the presence or absence of the model, B. philenor.
Results
Reference Genome Assembly and Annotation
To facilitate genome-wide analyses necessary to disentangle the evolution of mimicry and the L. arthemis species complex, we sequenced and assembled the L. arthemis genome de novo using bacterial artificial chromosome (BACs; Gallant et al. 2014), Illumina, and PacBio sequencing data from several wild-caught individuals, collected from the phenotypic hybrid zone, and/or inbred, lab-reared, progeny (supplementary table S1 and methods; supplementary information, Supplementary Material online). Limenitis scaffolds were then mapped and reordered relative to other lepidopteran genomes by synteny comparison (supplementary table S2 and methods, Supplementary Material online). The resulting Limenitis draft genome assembly comprises 306.3 megabases (Mb) in 4,786 scaffolds (N 50 2.16 Mb), with the longest scaffold approaching 32 Mb. We identified Z-linked scaffolds using our mapping information and male versus female read depth and excluded these scaffolds from downstream analyses; the smaller effective population size of Z relative to autosomes can bias summary statistics (Materials and Methods).
We assessed the completeness of the full L. arthemis genome using three methods. First, we used BUSCO to assess single copy ortholog statuses and found levels comparable to the other published nymphalid genomes (table 1; International Silkworm Genome Consortium 2008; Zhan et al. 2011; Dasmahapatra et al. 2012; You et al. 2013; Ahola et al. 2014; Tang et al. 2014; Cong et al. 2015; Challis et al. 2016; Davey et al. 2016). Second, we annotated the L. arthemis genome using previously collected RNA-seq data from imaginal wing discs and the MAKER pipeline (Materials and Methods; Campbell et al. 2014; Gallant et al. 2014). The final annotation comprised 13,138 gene protein-coding gene models, slightly fewer than other nymphalids like Danaus plexippus (15,130; Zhan et al. 2011), Heliconius erato (14,613; Lewis et al. 2016), and Melitaea cinxia (16,571; Ahola et al. 2014). We also manually inspected and annotated a suite of melanin (n = 19) and ommochrome (n = 31) synthesis pathway genes and found that, consistent with the results of our automated pipeline, 72% of the annotated genes were complete (i.e., contained a start and stop codon, were without missing or duplicated exons; supplementary table S3 and methods, Supplementary Material online). Finally, we aligned the physical BAC sequence (134 kb) housing the color pattern gene WntA to the draft assembly (supplementary fig. S2, Supplementary Material online), and found that the complete sequence was contained within a large, 650 kb scaffold. Thus, our L. arthemis assembly provides a robust if slightly incomplete reference for genome-wide analyses comparable to other nymphalids.
Table 1.
Comparison of Limenitis BUSCO Results with Other Lepidopteran Genomes.
| Lepidopteran Genomes | Complete (Dup.) | Partial | Missing |
|---|---|---|---|
| Bicyclus anynana 1.2 | 89.6% (0.7%) | 3.9% | 6.5% |
| Bombyx mori | 89.6% (0.4%) | 4.4% | 6.0% |
| Danaus plexippus v3 | 96.6% (1.9%) | 2.3% | 1.1% |
| Heliconius melpomene v2.5 | 86.2% (0.4%) | 4.1% | 9.7% |
| Limenitis arthemis | 81.7% (1.4%) | 1.9% | 16.4% |
| Melitaea cinxia | 57.1% (0.2%) | 11.8% | 31.1% |
| Papilio xuthus | 93.4% (0.2%) | 2.3% | 4.3% |
Quality statistics for the de novo Limenitis genome assembly (italics) are noted in underline.
Species-Tree Inference and Demographic History
To infer the species-level phylogeny required for our downstream introgression analyses, we generated whole-genome resequencing data from 64 Limenitis samples (supplementary table S4, Supplementary Material online), representing each of the North American species (Limenitis lorquini, n = 10; Limenitis weidemeyerii, n = 10; and Limenitis archippus, n = 9) and the three focal subspecies of L. arthemis (L. a. arthemis, n = 13; L. a. astyanax, n = 11; and L. a. arizone n sis, n = 11). These data were then aligned to the Limenitis reference genome using Bowtie2 (Langmead and Salzberg 2012) and SNPs were called using GATK (McKenna et al. 2010; DePristo et al. 2011; Van der Auwera et al. 2013). We inferred the relationships between the six groups using maximum likelihood and concatenated genome-wide SNPs for 12 representative samples with the best sequencing depth (supplementary fig. S1, Supplementary Material online; RAxML v.8.0, Stamatakis 2014). Our result supports a topology similar to previous estimates of the relationships among these species obtained using gene genealogies (fig. 1A; Mullen 2006; Ries and Mullen 2008; Mullen et al. 2011) and genome-wide RADseq data (Ebel et al. 2015). Specifically, we recovered a species tree indicating that 1) L. archippus is the sister lineage to the rest of the North American radiation, 2) the two western taxa, L. lorquini and L. weidemeyerii are closely related sister species and the closest relatives of the poltypic L. arthemis complex, and 3) within the L. arthemis complex, L. a. arizonensis (mimetic) is sister to a monophyletic clade containing L. a. arthemis (nonmimetic) and L. a. astyanax (mimetic). These results are consistent with previous efforts to infer phylogeny and likely represent the true consensus species tree (Mullen 2006; Prudic and Oliver 2008; Savage and Mullen 2009). However, the relationships depicted by the species-tree are potentially confounded by hybridization and, therefore, may not accurately reflect the history of mimetic trait evolution in this system.
To address this concern, we applied a Bayesian coalescent-based model, implemented using the Generalized Phylogenetic Coalescent Sampler (G-PhoCS; Gronau et al. 2011; supplementary table S5–8 and methods, Supplementary Material online), to infer the neutral demographic history of divergence and gene flow among different lineages of Limenitis. Preliminary results, allowing no migration, suggested that L. a. arthemis and L. a. astyanax diverged recently relative to the split with L. a. arizonensis, and produced estimates of ancestral population size (supplementary table S6, Supplementary Material online) that were significantly lower than expected given prior inferences (Ries and Mullen 2008). However, the results of the full G-PhoCS model (supplementary fig. S3, Supplementary Material online), allowing migration (supplementary tables S7 and S8, Supplementary Material online), were more consistent with previous attempts to understand the demographic history of these populations and indicate that the common ancestor of the L. arthemis complex split from the western two species ∼250 ka before diverging into the three subspecific lineages soon afterward. The full model also recovered extensive evidence of historical gene flow between 1) the two eastern subspecies of L. arthemis (L. a. arthemis and L. a. astyanax, and 2) the two western taxa (L. lorquini and L. weidemeyerii), supporting the hypothesis that population history and character evolution in this system may be decoupled by hybridization.
Mimetic Trait Evolution
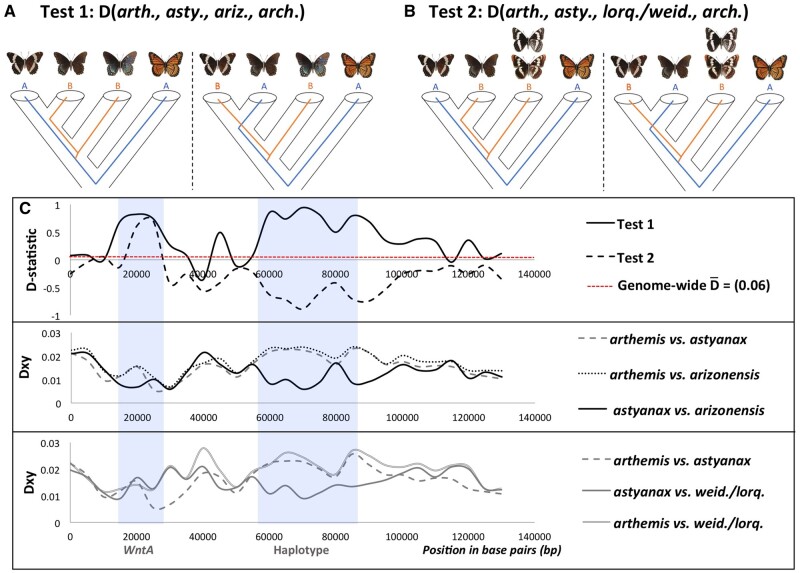
Given the potential for hybridization and recombination to confound efforts to reconstruct character evolution, we next calculated Patterson’s D-statistic (ABBA/BABA; Patterson et al. 2012) to test for evidence of allele sharing across the WntA color patterning scaffold. We found strong evidence for hybridization between astyanax and arizonensis by analyzing D-statistics in 50-kb windows for all putative autosomal scaffolds (supplementary fig. S4, Supplementary Material online; genome-wide average D for (((arthemis, astyanax), arizonensis), archippus) = 0.0684 ± 0.0022; P < 0.001, based on a two-tailed z-test that D is significantly different from zero). We then performed two additional tests for allele sharing across the WntA scaffold using smaller 5 kb windows. Test 1 assessed evidence of allele sharing between the allopatric mimetic subspecies (L. a. arizonensis) and the two eastern subspecies (((arthemis, astyanax), arizonensis), archippus). Test 2 assessed evidence of allele sharing between the two eastern subspecies and the western, white-banded species (((arthemis, astyanax), lorquini+weidemyerii), archippus). Test 1 identified two peaks with significantly positive D-values, corresponding to the WntA coding region and the upstream haplotype identified by Gallant et al. (2014). Test 2 recovered a peak with significantly negative D-values that also corresponded the region of the associated haplotype, suggesting a history of allele sharing between white-banded arthemis and the two white-banded western species (fig. 2).
Fig. 2.
Evidence of allele sharing between Limenitis arthemis astyanax and Limenitis arthemis arizonensis (test 1, panels A + C) or between Limenitis arthemis arthemis and the two nonmimetic western species (test 2, panels B + C) of Limenitis (Limenitis lorquini/Limenitis weidemeyerii). Panel C (top row) displays Patterson’s D-statistic (calculated based on ABBA-BABA patterns; 5-kb windows) across the WntA scaffold for each test of allele sharing. The middle and bottom rows (Panel C) indicate estimates of absolute divergence between Limenitis populations compared for each test. The position of the WntA protein coding gene and associated haplotype (identified by Gallant et al. 2014) are indicated with light blue shaded columns.
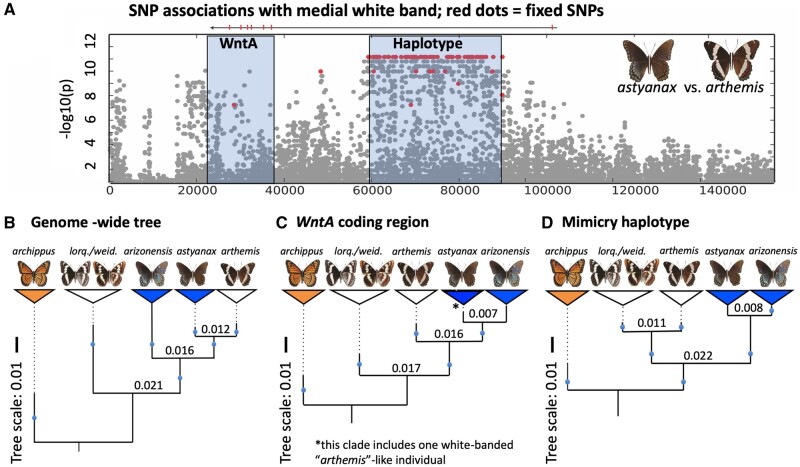
We then took several approaches to determine if these patterns of allele sharing were due to incomplete lineage sorting or introgression. First, we calculated dXY within the color patterning interval, with the expectation that introgressed regions would display lower dXY than genome-wide estimates (Smith and Kronforst 2013; table 2, supplementary table S9, Supplementary Material online). We found that, as predicted, the two mimetic subspecies of L. arthemis display reduced divergence (dXY; table 2) within the regions corresponding to positive peaks of introgression identified in Test 1 (fig. 2, Panel C, middle row). Interestingly, dXY was also reduced between white-banded L. a. arthemis and the two western, white-banded species of Limenitis for the peak centered on the nonmimetic haplotype upstream of WntA (fig. 2, Panel C, bottom row). Maximum likelihood trees for these two putatively introgressed regions, corresponding to the position of both WntA protein coding region and haplotype associated with mimetic variation, recovered strongly supported topologies that are highly discordant from the inferred species tree (fig. 3; supplementary figs. S5 and S6, Supplementary Material online). To assess how common such discordant patterns were in the genome as a whole, we employed Martin and Van Belleghem’s (2017) topology weighting method by iterative sampling subtrees (“Twisst”) using 50-kb sliding-windows. We found that 60,490 subtrees support the genome-wide species tree topology, 18,741 subtrees support monophyly of mimetic L. a. arthemis, and 17,889 support a sister relationship between mimetic L. a. arizonensis and nonmimetic L. a. arthemis.
Table 2.
Mean Absolute Divergence, dXY, between Mimetic and Nonmimetic Forms of Limenitis arthemis.
| Population 1 | Population 2 | Genome-Wide dXY | WntA Gene | Associated Haplotype |
|---|---|---|---|---|
| L. a. arthemis | L. a. astyanax | 0.012 | 0.015 | 0.022 |
| L. a. arthemis | L. a. arizonensis | 0.016 | 0.016 | 0.023 |
| L. a. astyanax | L. a. arizonensis | 0.015 | 0.007 | 0.008 |
| L. a. arthemis | L. lorquini | 0.021 | 0.017 | 0.011 |
Fig. 3.
Comparison of genealogical patterns among sampled Limenitis species. (Panel A) Physical map of the WntA scaffold displaying the relative position of the WntA protein coding gene and casual haplotype. Grey dots represent polymorphic SNPs between Limenitis arthemis arthemis and Limenitis arthemis astyanax. Red dots are fixed SNPs between these two taxa. (Panel B–D) Maximum-likelihood trees with branch lengths extended (dashed lines) to align the terminal taxon labels. (Panel B) Species tree estimated from genome-wide SNPs with branch lengths (see also supplementary fig. S1, Supplementary Material online). (Panel C) Gene tree for the WntA protein coding region; *note that in panel C one nonmimetic individual (L. a. arthemis) grouped with all mimetic individuals of L. a. astyanax (see also supplementary fig. S4, Supplementary Material online). (Panel D) Gene tree for the associated haplotype showing monophyly of all mimetic samples (L. a. astyanax + Limenitis arthemis arizonensis). Blue circles indicate branches with >95% bootstrap support for all tree figures; trees were simplified by collapsing individuals into monophyletic groups represented by colored triangles. Numbers above internal nodes in Panels B–D represent absolute divergence (dXY) between taxa.
Although the reciprocal monophyly of mimetic and nonmimetic individuals we recovered for the associated haplotype region implies a single origin of the causative variant responsible for mimetic convergence between Battus and Limenitis, this discordant genealogical pattern could be explained by either introgression or differential sorting of ancestral polymorphism at this locus. To assess this latter possibility (termed “hemiplasy”; Guerrero and Hahn 2018), we quantified the hemiplasy risk factor (HRF) along branches of the Limenitis phylogeny using 700 randomly chosen 100-kb windows (supplementary fig. S7, Supplementary Material online). Our results suggest that hemiplasy is unlikely to explain discordance between the gene tree for the WntA haplotype and the species tree. Next, we conducted a PhyloNet-HMM analysis using phylogenetic networks (Than et al. 2008) combined with hidden Markov models (HMMs; Liu et al. 2014; Liu et al. 2015) to simultaneously assess the potential reticulate history while also accounting for ILS and local recombination dependencies within the genome. This statistical introgression mapping approach follows methods developed by Yu et al. (2012) to convert each phylogenetic network into a multi-labeled (MUL) tree, allowing coalescent-based calculations of gene tree probabilities in cases where hybridization may have occurred. Each MUL-tree, encoded by a species network, can then be represented as a “row” of HMM states with gene tree topologies corresponding to locally evolving coalescent histories represented as distinct states within the “row.” Introgression is indicated by HMM switching from one “row” of states to another, whereas switching between states within the same “row” is indicative of ILS and/or recombination (see Wuyun et al. 2019 for further details). Therefore, the introgression probability indicates the probability that the genealogical pattern observed for a particular genomic region was generated by introgression rather than ILS.
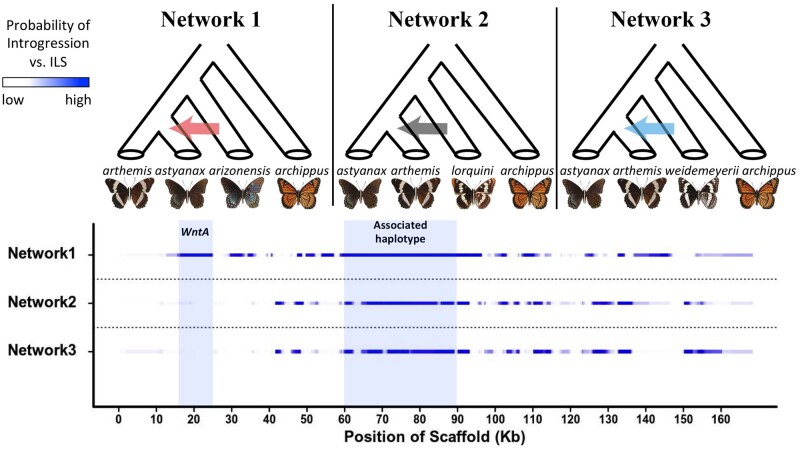
We used PhyloNet –HMM to test three distinct phylogenetic networks (fig. 4) that assessed the probability of introgression between the two mimetic taxa or between nonmimetic L. a. arthemis and either L. lorquini or L. weidemeyerii. Specifically, we calculated the posterior probability that a site’s coalescent history involved introgression versus strict tree-like descent, while also accounting for potential ILS of ancestral polymorphism within each of these fixed species tree topologies. We focused our analyses on the largest 30 scaffolds in the Limenitis reference assembly, due to the long computational runtimes associated with the HMM, and then assessed patterns of allele sharing due to introgression and/or ILS across the WntA scaffold. Our results indicate more evidence for genome-wide introgression between the two mimetic lineages as tested in network 1 relative to the other two networks investigated (supplementary figs. S8–S10, Supplementary Material online). More importantly, we found that the strongest evidence for introgression (probability >95%, fig. 4) between the two mimetic lineages was centered over the discordant haplotype region identified by Gallant et al. (2014) and that this introgression tract was longer (37 kb) than 99.2% of all tracts identified genome-wide (supplementary figs. S11 and S12, Supplementary Material online). Signatures of introgression between nonmimetic species (L. arthemis vs. L. lorquini or L. weidemeyerii) were also detected across the WntA interval (fig. 4) but these introgression blocks were smaller and more fragmented (presumably by recombination; fig. 4) than between the two mimetic lineages. This strongly implies that gene flow between the mimetic forms occurred more recently.
Fig. 4.
PhyloNet-HMM analysis of Limenitis WntA scaffold. Each row corresponds to one of three distinct species network hypotheses. Per-site introgression probabilities inferred using PhyloNet-HMM are shown, where probabilities between 0 and 1 are colored using a continuous gradient from white to blue, respectively.
Origin and Maintenance of the Mimetic Haplotype
To gain further insights into the relative timing of gene flow among mimetic and nonmimetic lineages of Limenitis, we then calculated LD across the WntA scaffold and compared it to genome-wide estimates of LD. We found that LD (mean r 2) between SNPs within the color patterning region was 1) elevated among mimetic individuals (L. a. astyanax +L. a. arizonensis) relative to genome-wide estimates, and 2) decayed more slowly as a function of physical distance (in base pairs) than among nonmimetic samples (L. a. arthemis, L. lorquini, L. weidemeyerii; supplementary fig. S13, Supplementary Material online). In contrast, we observed similar levels of LD and rates of decay among nonmimetic individuals both at the scale of the whole genome and within the genomic region housing WntA. We also found that absolute divergence (dXY) between the two mimetic forms of L. arthemis (L. a. astyanax and L. a. arizonensis) was significantly lower within the WntA scaffold, as noted above (fig. 2), than between L. a. arthemis and either of the two western species of Limenitis (dXY(arth, weid) vs. dXY(asty, ariz) ‖ 0.0180 vs. 0.0137, W = 770, P = 0.01815; dXY(arth, lorq) vs. dXY(asty, ariz) ‖ 0.0179 vs. 0.0137, W = 763, P = 0.02296). These results indicate that gene flow between the mimetic subspecies of L. a. arthemis occurred more recently than gene exchange between L. a. arthemis and either L. lorquini or L. weidemeyerii, and are consistent with the inferred demographic history (supplementary fig. S2, Supplementary Material online).
Comparisons of additional population genomic summary statistics (e.g., F ST, r 2, and π) among the subspecies of the L. arthemis complex further support the hypothesis that mimicry spread via recent introgression between L. a. arizonensis and L. a. astyanax. For example, we found that measurements of F ST (supplementary fig. S14, Supplementary Material online) and mean r 2 between all SNPs within 500-bp sliding windows (50-bp steps; supplementary fig. S15, Supplementary Material online) were sharply reduced between the two mimetic forms in the region corresponding to the divergent haplotype upstream of WntA relative to comparisons involving mimetic and nonmimetic forms. In addition, we found that the mimetic subspecies have lower overall levels of nucleotide diversity (π) within the color patterning interval than L. a. arthemis or any of the other species of Limenitis that we sampled (supplementary fig. S16, Supplementary Material online). These results, which corroborate the findings of Gallant et al. (2014), are consistent with the hypothesis of a selective sweep of the mimetic haplotype in L. a. astyanax.
To identify potential mechanisms maintaining the extended LD observed between the two hybridizing color pattern forms in the eastern United states (L. a. arthemis and L. a. astyanax) we examined patterns of SV across the color patterning interval using delly (Rausch et al. 2012) and pindel (Ye et al. 2009). Both programs identified a high-quality, precise deletion relative to the reference between positions 78760 and 84272 bp, corresponding roughly to the position of the long interspersed nuclear element (LINE) reported by Gallant et al. (2014). However, the deletion breakpoints 1) fall outside the region corresponding to the LINE itself, as defined by RepeatMasker (Smit et al. 2015), 2) were not influenced by different mapping quality filters/thresholds, and 3) were supported in each case by at least one read spanning the breakpoint (supplementary fig. S17, Supplementary Material online). The absence of the LINE in all of our samples suggests that it is not associated with the color pattern phenotype, and its presence in the previous study appears to have been an artifact of aligning short-read data to the physical BAC reference (∼134 kb) rather than to the entire genome (∼306 Mb). We also failed to detect any other structural variants (e.g., -insertions, deletions, inversions, or duplications) that partitioned the two color pattern phenotypes or that were associated with the region of upstream of WntA, suggesting that SV itself is unlikely to be responsible for the maintenance of LD across the 30-kb region corresponding to the associated haplotype (supplementary fig. S17, Supplementary Material online).
Although the lack of recombination seen in this system within the extended genomic region associated with color pattern phenotypes in this system prevents further fine mapping of potentially causative mutations, comparisons of patterns of polymorphism and divergence within the color patterning interval revealed 32 SNPs that were fixed in both L. a. astyanax and L. a. arizonensis, and at a frequency of <0.3 in L. a. arthemis (supplementary table S10, Supplementary Material online). In comparison, application of these same filters to an additional 3.2 Mb of genomic scaffolds, representing ∼100× more sequence data than the 30-kb WntA haplotype, revealed just four SNPs that fit this pattern. Half of these putative “mimetic” alleles cluster near the beginning of the haplotype region (60,315–74,060 bp) upstream of WntA and are not found in L. arthemis. A second cluster of such fixed SNPs also occurs at the other end of the haplotype (85,455–86,759 bp).
Discussion
Here, we have investigated the evolutionary origins and maintenance of Batesian mimicry among hybridizing lineages of mimetic and nonmimetic populations of the polymorphic L . arthemis species complex. Assembly of the reference genome allowed us to 1) determine the species tree for North American Limenitis using genome-wide SNPs, 2) infer the demographic history of this butterfly radiation, 3) investigate patterns of introgression and phylogenetic discordance across the WntA color patterning scaffold to reconstruct the history of mimetic character evolution, and 4) explore patterns of SV, divergence (F ST, dXY), and diversity (π) among the three L. arthemis subspecies to test competing hypotheses about the evolutionary mechanism that maintains extensive LD across the mimicry haplotype upstream of WntA.
Bayesian inference of the demographic history for North American Limenitis, based on putatively neutral loci sampled across the genome, indicated that divergence among lineages occurred primarily during the Pleistocene (supplementary fig. S2, Supplementary Material online; ∼250–200 ka). G-PhoCS also recovered evidence for high-levels of historical gene flow between the hybridizing mimetic and nonmimetic subspecies of the L. arthemis from the eastern United Sates and between the two western species, L. lorquini and L. weidemeyerii, which are also known to hybridize extensively. We interpret these results to mean that divergence with gene flow has characterized the history of speciation within this radiation, supporting the hypothesis that recombination resulting from gene flow among lineages has decoupled patterns of character evolution from the history of populations.
This view is further substantiated by the results of our genome-wide versus locus-specific inferences of the evolutionary relationships among species. Specifically, we recovered gene trees from the WntA protein-coding region and the upstream haplotype associated with mimetic polymorphism that were well supported and highly discordant from the species tree inferred from genome-wide SNP data (fig. 3). Surprisingly, the genealogical history of the genomic region responsible for mimetic color pattern variation is also incongruent with all of our a priori hypotheses about character evolution (fig. 1). Although the gene tree recovered for this region is consistent with a single origin of the mimetic haplotype (fig. 1C), the results of our hemiplays risk factor analysis (Guerrero and Hahn 2018), which calculates the potential contribution of individual branches to incongruent trait patterns arising via hemiplasy due to ILS, indicate that the observed discordance between the genome-wide species tree (fig. 3A) and gene tree recovered for the causal haplotype (fig. 3D) is unlikely to have arisen as a consequence of ILS. Instead, the low HRF values, in combination with evidence of genome-wide and allele-specific introgression between the two mimetic taxa, strongly support a single, ancient origin of the mimetic haplotype followed by postdivergence gene flow and introgression between L. a. arizonensis and L. a. astyanax.
This is interesting because adaptive introgression of color-pattern mimicry appears to be widespread among many species of Neotropical Heliconius butterflies (Dasmahapatra et al. 2012; Pardo-Diaz et al. 2012; Kronforst and Papa 2015; Zhang et al. 2016) and was recently shown to have been important in the origin of “supergene” mimicry in this group (Jay et al. 2018). Intriguingly, we also found evidence for gene flow among nonmimetic, white-banded species of Limenitis in the region of the associated haplotype that appears to predate gene flow among the mimetic lineages based on patterns of absolute divergence (dXY), the strength and extent (i.e., rate of decay) of LD, and the distribution of introgression tract lengths. Therefore, we conclude that both haplotypes associated with mimetic polymorphism in this system have been impacted by introgressive hybridization at different points during the history of lineage divergence.
Taken together with genealogical evidence that the two haplotypes at the mimicry locus are deeply divergent, our results suggest that the causal variant(s) responsible for mimetic convergence between Limenitis and Battus originated prior to the divergence events that gave rise to the current geographically distinct subspecies of L. arthemis. Furthermore, they are consistent with the hypothesis that the mimetic haplotype originated in allopatric populations of L. a. arizonensis and spread via adaptive introgression into L. a. astyanax, coincident with historical expansion of the model’s range throughout the southeastern United States. Previous work (Mullen et al. 2008; Ries and Mullen 2008) suggests that the broad phenotypic hybrid zone observed between mimetic (L. a. astyanax) and nonmimetic (L. a. arthemis) populations of this complex in the eastern United states arose by secondary contact and that its geographical position is maintained by strong natural selection for Batesian mimicry. This implies that the maintenance of the two deeply divergent haplotypes segregating upstream of WntA reflects a long history of balancing selection acting at this locus related to the presence or absence of the model, B . phileno r.
Although these findings clearly implicate introgression as the source of mimetic variation in this system, they do not explain how LD is maintained across the haplotype associated with mimetic variation in the face of extensive and ongoing hybridization that occurs between mimetic and nonmimetic populations of L. arthemis in eastern North America. Structural variation has been demonstrated to play an important role in maintaining adaptive phenotypic diversity in other mimetic butterflies (Joron et al. 2006, 2011; Kunte et al. 2014; Iijima et al. 2018) but our analyses revealed no evidence for inversions, indels, or copy-number variants associated with phenotype. One potential caveat to this conclusion is that our ability to detect SVs is dependent on the quality of our genome assembly and, therefore, more work will be necessary to definitively rule this possibility out. Alternatively, LD might simply be elevated due to the recent timing of introgression. However, theory predicts (Chakraborty 1986; Racimo et al. 2015) that a selective sweep should generate a region of reduced nucleotide diversity flanked by regions of higher diversity outside the region of the sweep, which we observe (see supplementary fig. S16, Supplementary Material online). This suggests that the elevated LD across the mimetic haplotype reflects a history of strong selection and adaptive introgression. One final possibility is that there are multiple sites within the haplotype that influence color pattern and that epistatic selection on these sites maintains LD (sensu Schumer and Brandvain 2016). Two observations support this hypothesis. First, comparisons of polymorphism and divergence between mimetic (L. a. astyanax + L. a. arizonensis) and nonmimetic (L. a. arthemis) populations revealed two clusters of fixed SNPs near the start and end of the region of high LD, suggesting that the multiple sites may be the targets of selection. Second, phenotypic variation in the hybrid zone between L. a. arthemis and L. a. astyanax, as well as evidence from genetic crosses (Platt and Brower 1968; Platt 1975; Gallant et al. 2014), indicates that several modifying loci impact the penetrance of the white-band in the heterozygous condition, suggesting that there may be different combinations of SNPs within the haplotype that produce similar phenotypic outcomes (e.g., Linnen et al. 2013). However, future work employing CRISPR/Cas9 will be necessary to functionally dissect this locus and identify the causal SNP(s).
In conclusion, our results highlight the importance of distinguishing between introgression and ILS to fully understand the evolutionary origins and maintenance of shared adaptive variation among hybridizing lineages. Furthermore, they illustrate the power of coalescent-based introgression mapping approaches to disentangle the complex genealogical patterns produced by the combined effects of demography, selection, and gene flow during phenotypic divergence and/or speciation.
Materials and Methods
To generate the reference genome, genomic DNA was isolated from wild-caught L. a. astyanax (n = 5) and used to construct short-read and mate-pair libraries for sequencing. These data were assembled using Platanus (Kajitani et al. 2014) and combined with trimmed, quality filtered PacBio long reads and BAC sequences using Redundans (Pryszcz and Gabaldón 2016). Z-linked and autosomal scaffolds were identified and ordered relative to other lepidopteran genome assemblies using a custom BLAT pipeline. The resulting Limenitis assembly was annotated with MAKER (v3.01.02; Campbell et al. 2014) using RNA-seq data generated by Gallant et al. (2014) and available protein sequences from the UniProt/SwissProt protein database and other lepidopteran genomes. We also manually annotated a set of pigmentation genes to validate the MAKER annotations and performed a BUSCO analysis using BUSCO v3 (Waterhouse et al. 2018) to test the overall quality of the reference genome. We then generated genome resequencing data for 65 butterflies (supplementary table S4, Supplementary Material online) to infer the demographic history of this radiation using G-PhoCS (Gronau et al. 2011), conducted sliding window phylogenetic inferences using RAxML (Stamatakis 2014), quantified the risk of hemiplasy across branches (Guerrero and Hahn 2018), and calculated genome-wide estimates of allele sharing between populations using Patterson’s D-statistic. Finally, we used PhyloNet-HMM (Liu et al. 2014) to distinguish between genealogical patterns produced by introgression versus ILS. For additional information, please see supplementary methods, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Associate Editor and two anonymous reviewers for their comments on an earlier version of the manuscript. This study was funded by awards to S.P.M. (National Science Foundation DEB-1342712), M.R.K. (National Science Foundation DEB-1342790, National Science Foundation IOS-1452648, National Instituties of Health RO1 GM108626), K.J.L. (National Science Foundation CCF-1714417, CCF-1565719), R.I.H. (National Science Foundation DEB-1342706), and A.B.D. (National Science Foundation IOS-1656260 and National Science Foundation DEB-1342759).
Author Contributions
S.P.M., M.R.K., and A.B.D. conceived the study. S.P.M. and M.R.K. supervised the genome assembly, genome annotation, and population genomic analyses. S.P.M. authored the first draft of the manuscript. S.N. led the de novo genome assembly. A.B.D. annotated the pigmentation genes. N.W.V. performed genome quality assessment and annotation, structural variant detection, and population genomic analyses. W.Z. performed variant calling on the population genomic resequencing data, performed the demographic analysis, and contributed to summary statistic calculations. Q.W. and K.L. led the statistical introgression mapping analyses. R.I.H. provided butterfly specimens. E.K. assisted with data generation and figure production. All authors contributed to revisions of the manuscript.
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJ, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, et al. 2013. Hybridization and speciation. J Evol Biol. 26(2):229–246. [DOI] [PubMed] [Google Scholar]
- Ahola V, Lehtonen R, Somervuo P, Salmela L, Koskinen P, Rastas P, Välimäki N, Paulin L, Kvist J, Wahlberg N, et al. 2014. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun. 5(1):4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. 1985. Analysis of hybrid zones. Annu Rev Ecol Syst. 16(1):113–148. [Google Scholar]
- Beldade P, Brakefield PM. 2002. The genetics and evo–devo of butterfly wing patterns. Nat Rev Genet. 3(6):442–452. [DOI] [PubMed] [Google Scholar]
- Boggs CL, Watt WB, Ehrlich PR. 2003. Butterflies: ecology and evolution taking flight. Chicago and London: University of Chicago Press. [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 48(1):4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis RJ, Kumar S, Dasmahapatra KKK, Jiggins CD, Blaxter M. 2016. Lepbase: the Lepidopteran genome database. BioRxiv doi: 10.1101/056994. [DOI] [Google Scholar]
- Chakraborty R. 1986. Gene admixture in human populations: models and predictions. Am J Phys Anthropol. 29(S7):1–43. [Google Scholar]
- Cong Q, Borek D, Otwinowski Z, Grishin NV. 2015. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 10(6):910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra KK, Walters JR, Briscoe AD, Davey JW, Whibley A, Nadeau NJ, Zimin AV, Hughes DS, Ferguson LC, Martin SH, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Chouteau M, Barker SL, Maroja L, Baxter SW, Simpson F, Merrill RM, Joron M, Mallet J, Dasmahapatra KK, et al. 2016. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 (Bethesda) 6:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol (Amst) 24(6):332–340. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel ER, DaCosta JM, Sorenson MD, Hill RI, Briscoe AD, Willmott KR, Mullen SP. 2015. Rapid diversification associated with ecological specialization in Neotropical Adelpha butterflies. Mol Ecol. 24(10):2392–2405. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1981. Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution 35(1):124–138. [DOI] [PubMed] [Google Scholar]
- Frentiu FD, Yuan F, Savage WK, Bernard GD, Mullen SP, Briscoe AD. 2015. Opsin clines in butterflies suggest novel roles for insect photopigments. Mol Biol Evol. 32(2):368–379. [DOI] [PubMed] [Google Scholar]
- Gallant JR, Imhoff VE, Martin A, Savage WK, Chamberlain NL, Pote BL, Peterson C, Smith GE, Evans B, Reed RD, et al. 2014. Ancient homology underlies adaptive mimetic diversity across butterflies. Nat Commun. 5(1):4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. 2003. Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic “tool box” from synthetic hybrid zones and a theory of diversification. In: Ecology and evolution taking flight: butterflies as model systems. Chicago and London p. 281–318. [Google Scholar]
- Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. 2011. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 43(10):1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Hahn MW. 2017. Speciation as a sieve for ancestral polymorphism. Mol Ecol. 26(20):5362–5368. [DOI] [PubMed] [Google Scholar]
- Guerrero RF, Hahn MW. 2018. Quantifying the risk of hemiplasy in phylogenetic inference. Proc Natl Acad Sci USA. 115(50):12787–12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG. 1993. Hybrids and hybrid zones: historical perspective. In: Hybrid zones and the evolutionary process. Oxford: Oxford University Press. p. 3–12.
- Iijima T, Kajitani R, Komata S, Lin C-P, Sota T, Itoh T, Fujiwara H. 2018. Parallel evolution of Batesian mimicry supergene in two Papilio butterflies, P. polytes and P. memnon. Sci Adv. 4(4):eaao5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 38(12):1036–1045. [DOI] [PubMed] [Google Scholar]
- Jay P, Whibley A, Frézal L, de Cara MÁR, Nowell RW, Mallet J, Dasmahapatra KK, Joron M. 2018. Supergene evolution triggered by the introgression of a chromosomal inversion. Curr Biol. 28(11):1839–1845. [DOI] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR, Whibley A, Becuwe M, Baxter SW, Ferguson L, et al. 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477(7363):203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Mallet JL. 1998. Diversity in mimicry: paradox or paradigm? Trends Ecol Evol. 13:461–466. [DOI] [PubMed] [Google Scholar]
- Joron M, Papa R, Beltrán M, Chamberlain N, Mavárez J, Baxter S, Abanto M, Bermingham E, Humphray SJ, Rogers J, et al. 2006. A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 4(10):e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapan DD. 2001. Three-butterfly system provides a field test of Müllerian mimicry. Nature 409(6818):338–340. [DOI] [PubMed] [Google Scholar]
- Kingsolver JG. 1985. Thermal ecology of Pieris butterflies (Lepidoptera: Pieridae): a new mechanism of behavioral thermoregulation. Oecologia 66(4):540–545. [DOI] [PubMed] [Google Scholar]
- Kronforst MR, Papa R. 2015. The functional basis of wing patterning in Heliconius butterflies: the molecules behind mimicry. Genetics 200(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte K. 2009. The diversity and evolution of Batesian mimicry in Papilio swallowtail butterflies. Evolution 63(10):2707–2716. [DOI] [PubMed] [Google Scholar]
- Kunte K, Zhang W, Tenger-Trolander A, Palmer D, Martin A, Reed R, Mullen S, Kronforst M. 2014. Doublesex is a mimicry supergene. Nature 507(7491):229–232. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JJ, van der Burg KR, Mazo-Vargas A, Reed RD. 2016. ChIP-Seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in Lepidoptera. Cell Rep. 16(11):2855–2863. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Poh Y-P, Peterson BK, Barrett RD, Larson JG, Jensen JD, Hoekstra HE. 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339(6125):1312–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KJ, Dai J, Truong K, Song Y, Kohn MH, Nakhleh L. 2014. An HMM-based comparative genomic framework for detecting introgression in eukaryotes. PLoS Comput Biol. 10(6):e1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KJ, Steinberg E, Yozzo A, Song Y, Kohn MH, Nakhleh L. 2015. Interspecific introgressive origin of genomic diversity in the house mouse. Proc Natl Acad Sci USA. 112(1):196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP. 1997. Gene trees in species trees. Syst Biol. 46(3):523–536. [Google Scholar]
- Mallet J, Joron M. 1999. Evolution of diversity in warning color and mimicry: polymorphisms, shifting balance, and speciation. Annu Rev Ecol Syst. 30(1):201–233. [Google Scholar]
- Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G, Jiggins CD, Kronforst MR, Long AD, McMillan WO, et al. 2012. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci USA. 109(31):12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Van Belleghem SM. 2017. Exploring evolutionary relationships across the genome using topology weighting. Genetics 206(1):429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo-Vargas A, Concha C, Livraghi L, Massardo D, Wallbank RW, Zhang L, Papador JD, Martinez-Najera D, Jiggins CD, Kronforst MR, et al. 2017. Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc Natl Acad Sci USA. 114(40):10701–10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan WO, Monteiro A, Kapan DD. 2002. Development and evolution on the wing. Trends Ecol Evol. 17(3):125–133. [Google Scholar]
- Mullen SP. 2006. Wing pattern evolution and the origins of mimicry among North American admiral butterflies (Nymphalidae: Limenitis). Mol Phylogenet Evol. 39(3):747–758. [DOI] [PubMed] [Google Scholar]
- Mullen SP, Dopman EB, Harrison RG. 2008. Hybrid zone origins, species boundaries, and the evolution of wing-pattern diversity in a polytypic species complex of North American admiral butterflies (Nymphalidae: Limenitis). Evolution 62(6):1400–1417. [DOI] [PubMed] [Google Scholar]
- Mullen SP, Savage WK, Wahlberg N, Willmott KR. 2011. Rapid diversification and not clade age explains high diversity in neotropical Adelpha butterflies. Proc R Soc B. 278(1713):1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Jiggins CD. 2010. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 26(11):484–492. [DOI] [PubMed] [Google Scholar]
- Nadeau NJ, Pardo-Diaz C, Whibley A, Supple MA, Saenko SV, Wallbank RW, Wu GC, Maroja L, Ferguson L, Hanly JJ, et al. 2016. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534(7605):106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, Sugano S, Fujiyama A, Kosugi S, Hirakawa H, et al. 2015. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet. 47(4):405–409. [DOI] [PubMed] [Google Scholar]
- Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, McMillan WO, Jiggins CD. 2012. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet. 8(6):e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt AP. 1975. Monomorphic mimicry in Nearctic Limenitis butterflies: experimental hybridization of the L. arthemis–astyanax complex with L. archippus. Evolution 29(1):120–141. [DOI] [PubMed] [Google Scholar]
- Platt AP, Brower LP. 1968. Mimetic versus disruptive coloration in intergrading populations of Limenitis arthemis and astyanax butterflies. Evolution 22(4):699–718. [DOI] [PubMed] [Google Scholar]
- Prudic KL, Oliver JC. 2008. Once a Batesian mimic, not always a Batesian mimic: mimic reverts back to ancestral phenotype when the model is absent. Proc R Soc B. 275(1639):1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz LP, Gabaldón T. 2016. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44(12):e113–e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E. 2015. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 16(6):359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28(18):i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, et al. 2011. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333(6046):1137–1141. [DOI] [PubMed] [Google Scholar]
- Ries L, Mullen SP. 2008. A rare model limits the distribution of its more common mimic: a twist on frequency-dependent Batesian mimicry. Evolution 62(7):1798–1803. [DOI] [PubMed] [Google Scholar]
- Savage WK, Mullen SP. 2009. A single origin of Batesian mimicry among hybridizing populations of admiral butterflies (Limenitis arthemis) rejects an evolutionary reversion to the ancestral phenotype. Proc R Soc B. 276(1667):2557–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Brandvain Y. 2016. Determining epistatic selection in admixed populations. Mol Ecol. 25(11):2577–2591. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2015. RepeatMasker Open-4.0. 2013–2015. [Google Scholar]
- Smith J, Kronforst MR. 2013. Do Heliconius butterfly species exchange mimicry alleles. Biol Lett. 9(4):20130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Reger J, Feulner PG, Smadja C, Galindo J, Ekblom R, Bennison C, Ball AD, Beckerman AP, Slate J. 2010. Adaptation genomics: the next generation. Trends Ecol Evol. 25:705–712. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. 2008. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 100(2):158–170. [DOI] [PubMed] [Google Scholar]
- Tang W, Yu L, He W, Yang G, Ke F, Baxter SW, You S, Douglas CJ, You M. 2014. DBM-DB: the diamondback moth genome database. Database 2014:bat087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than C, Ruths D, Nakhleh L. 2008. PhyloNet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinformatics. 9(1):322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belleghem SM, Rastas P, Papanicolaou A, Martin SH, Arias CF, Supple MA, Hanly JJ, Mallet J, Lewis JJ, Hines HM, et al. 2017. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 1:0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 43:11–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman EL, VanKuren NW, Massardo D, Tenger-Trolander A, Zhang W, Hill RI, Perry M, Bayala E, Barr K, Chamberlain N, et al. 2018. Aristaless controls butterfly wing color variation used in mimicry and mate choice. Curr Biol. 28(21):3469–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyun Q, VanKuren N, Kronforst M, Mullen SP, Liu KJ. 2019. Scalable statistical introgression mapping using approximate coalescent-based inference. Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. Niagra Falls, NY, USA: ACM. p. 504–513.
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25(21):2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Yue Z, He W, Yang X, Yang G, Xie M, Zhan D, Baxter SW, Vasseur L, Gurr GM, et al. 2013. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 45(2):220–225. [DOI] [PubMed] [Google Scholar]
- Yu Y, Degnan JH, Nakhleh L. 2012. The probability of a gene tree topology within a phylogenetic network with applications to hybridization detection. PLoS Genet. 8(4):e1002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. 2011. The monarch butterfly genome yields insights into long-distance migration. Cell 147(5):1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Dasmahapatra KK, Mallet J, Moreira GR, Kronforst MR. 2016. Genome-wide introgression among distantly related Heliconius butterfly species. Genome Biol. 17(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Duvaux L, Ren G, Zhang L, Savolainen O, Liu J. 2017. Importance of incomplete lineage sorting and introgression in the origin of shared genetic variation between two closely related pines with overlapping distributions. Heredity 118(3):211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.