Abstract
MCMs are a family of proteins related to ATP-dependent helicases that bind to origin recognition complexes and are required for initiation of DNA replication. We report that antibodies against MCM2(BM28) specifically inhibited transcription by RNA polymerase II (Pol II) in microinjected Xenopus oocytes. Consistent with this observation, MCM2 and other MCMs copurified with Pol II and general transcription factors (GTFs) in high-molecular-weight holoenzyme complexes isolated from Xenopus oocytes and HeLa cells. Pol II and GTFs also copurified with MCMs isolated by anti-MCM3 immunoaffinity chromatography. MCMs were specifically displaced from the holoenzyme complex by antibody against the C-terminal domain (CTD) of Pol II. In addition, MCMs bound to a CTD affinity column, suggesting that their association with holoenzyme depends in part on this domain of Pol II. These results suggest a new function for MCM proteins as components of the Pol II transcriptional apparatus.
Transcription by RNA polymerase II (Pol II) is carried out with the aid of many accessory proteins, including the general transcription factors (GTFs) TFIIA, -B, -D, -E, -F, and -H (45). Large pol II holoenzyme complexes, which contain GTFs, have been isolated from both yeast and mammalian cells (28, 38, 46, 47). In addition to GTFs, the holoenzyme contains many other components, some of which make contacts with the C-terminal domain (CTD) of the pol II large subunit. Antibodies against the CTD disrupt the yeast holoenzyme into core Pol II and a mediator subcomplex, which contains the Srbs and other proteins (20, 27, 42). Temperature-sensitive alleles of the SRB4 and SRB6 genes showed that these mediator subunits are essential for expression of most mRNAs in budding yeast (56). Other holoenzyme components, such as Srb2, -5, and -7 to -11, and SWI/SNF proteins, Sin4, Rgr1, Med2, Med9/Cse2, Med10/Nut2, Med11, Gal11 and Pgd1 (18, 20, 34, 43, 63, 18), are not essential for transcription of most genes but do contribute to the response to transactivators and repressors (reviewed in references 6 and 17). In addition to its role in the response to transcriptional regulators, the holoenzyme may also integrate transcription with RNA processing, DNA repair, and replication. In support of this idea, the DNA repair factors DNA Pol ɛ, XPC, XPF, XPG, Ku, and RAD51 (38); BRCA1 (52); RNA helicase A (1); the replication factors RP-A and RP-C (38); and the cleavage/polyadenylation factors CPSF and CstF (40) have been identified in Pol II holoenzyme preparations. Holoenzyme purified by different procedures differs in its composition, indicating that there are multiple forms of this complex in vivo (7). It has been estimated that HeLa cells contain approximately 8,000 copies of a 2- to 4-MDa Pol II holoenzyme complex, which corresponds to 10% of the total Pol II and 0.5% of soluble protein in cell extracts (47). The complexity of the mammalian Pol II holoenzyme suggests that many of its components remain to be identified.
Replication of genomic DNA is limited to a single round per cell cycle by a licensing factor, which binds to origins of replication in M phase and is released after the origins have fired in S phase (4). One component of licensing factor is a complex of six MCM proteins which bind to the origin recognition complex (ORC) (reviewed in references 25 and 44). The MCM genes were originally identified in budding yeast, where they are required for minichromosome maintenance (37). As predicted by the licensing model, most MCMs are released from chromatin during S phase and reassociate at the end of mitosis (2, 8, 35, 53, 58). In addition to promoting replication, MCMs may also aid replication fork movement (2). The precise biochemical function of MCMs remains unclear; however, they have a conserved DNA-dependent ATPase domain shared with DNA helicases (29), and they copurify with helicase activity (23). They also bind with high affinity to core histone H3-H4 dimers (24), indicating a possible chromatin-remodeling function (2). In both yeast and mammalian cells, MCMs are far more abundant than replication origins (10, 67). Mammalian cells have at least 106 copies of the MCMs per nucleus, which is at least an order of magnitude greater than the number of replication origins (5, 58). The excess of MCMs over origins suggests that these proteins may have functions in addition to replication licensing. Indeed, a role in transcriptional activation is suggested by the recent report that MCM5 interacts with the activation domain of Stat1α and that overexpression of MCM5 stimulates transcription (68).
In this paper, we demonstrate a functional and physical interaction between MCM proteins and the general Pol II transcription machinery. Antibodies against MCM2, originally termed BM28 (59), specifically inhibited Pol II transcription in injected Xenopus oocytes. Furthermore, MCM proteins copurified with holoenzyme complexes containing Pol II and general transcription factors.
MATERIALS AND METHODS
Oocyte injection and RNase protection.
The mouse c-myc (pSX943) and the adenovirus VA1 (pSPVA), pGal5-P2mycCAT (65), and pHIV2-LTR-CAT-556/+156) (11) plasmids have been described previously. Template DNAs were injected at 0.46 ng/oocyte, and Gal4-AH was injected at 4.6 ng/oocyte in 46 nl. Seven to sixty nanograms of antigen affinity-purified immunoglobulin (Ig) was injected per oocyte. These amounts of antibody are expected to saturate most of the endogenous antigen pools. Total protein in injection samples was made up to 1 mg/ml with bovine serum albumin (BSA). The injected antibodies were concentrated, if necessary, and dialyzed against 10 mM HEPES (pH 7.5)–70 mM NaCl–0.2 mM EDTA–0.1 mM ZnCl2.
RNase protection of pSX943, pGal5-P2mycCAT, and pHIV2-LTR-CAT-556/+156 transcripts has been described previously (65). One oocyte equivalent of total RNA (∼5 μg) was hybridized to 50,000 cpm of each antisense probe (specific activity, 80 Ci/mmol of [32P]UTP). Hybridization was in 0.4 M NaCl, 0.5 mM EDTA, 20 mM PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.4), 80% formamide at 50°C. RNase digestion was in 0.3 M NaCl, 10 mM Tris (pH 7.5), 5 mM EDTA, 5 μg of RNase T1/ml, 1 μg of RNase A/ml for 30 min at 37°C. Scanned autoradiographs were quantified with NIH Image version 1.61.
Antibodies.
The following rabbit antibodies were used: anti-BM28-N, anti-BM28-C, and anti-BM28-P against amino acids 1 to 591, 592 to 892, and 1 to 412 of human MCM2(BM28), respectively (59); anti-Xenopus MCM3 (36); anti-human MCM5 (50); anti-Xenopus MCM7 (50); and anti-Xenopus ORC1 and ORC2 (49). Anti-GST, -TFIIB, -p34(TFIIE), and -rap74(TFIIF) were raised against recombinant proteins. Anti-CstFp77 and -CPSFp160 were raised against the peptides VPPVHDIYRARQQKRIR and TPDIILDDLLETDRVTAHF. Anti-Pol Iβ, anti-Pol III RPC62 and RPC82, and anti-CDK8 antibodies (61) were provided by L. Rothblum, R. Roeder, and E. Lees. Anti-TBP was from Upstate Biotechnology. All polyclonal antibodies except the anti-Pol I and anti-Pol III antibodies were affinity purified.
The following monoclonal antibodies were used as purified IgG: anti-Pol II CTD (8WG16) (57); anti-c-myc (9E10) (12); anti-TFIIH p62 (3C9) (13); anti-CDK7 (2F8) (51); and anti-RP-A p34 (34-A) and anti-RP-A p70 (70-C) (26).
The three antibodies against MCM2(BM28) and the antibodies against MCM3, MCM5, MCM7, ORC1, ORC2, TFIIB, Pol II CTD, and p70(RP-A) reacted with both the human and Xenopus homologous peptides, and all recognized a single major band in Xenopus extracts as determined by Western blotting (data not shown) (36, 49, 66).
Western blotting.
Proteins were transferred to Immobilon P membrane by semidry electroblotting. Blots were developed by ECL (Amersham) with horseradish peroxidase coupled to protein A (Sigma) or to secondary antibody (Dako).
Recombinant proteins.
For blocking of injected anti-MCM2(BM28) antibodies (Fig. 1A and B), soluble MCM2(BM28) was expressed with recombinant baculovirus and purified from Sf9 cells by Q-Sepharose and Phenyl Sepharose chromatography. In experiments not shown, antibodies were blocked by incubation with renatured bacterially expressed, His6-MCM2 coupled to Affigel-10 (Bio-Rad) at 1 to 2 mg/ml.
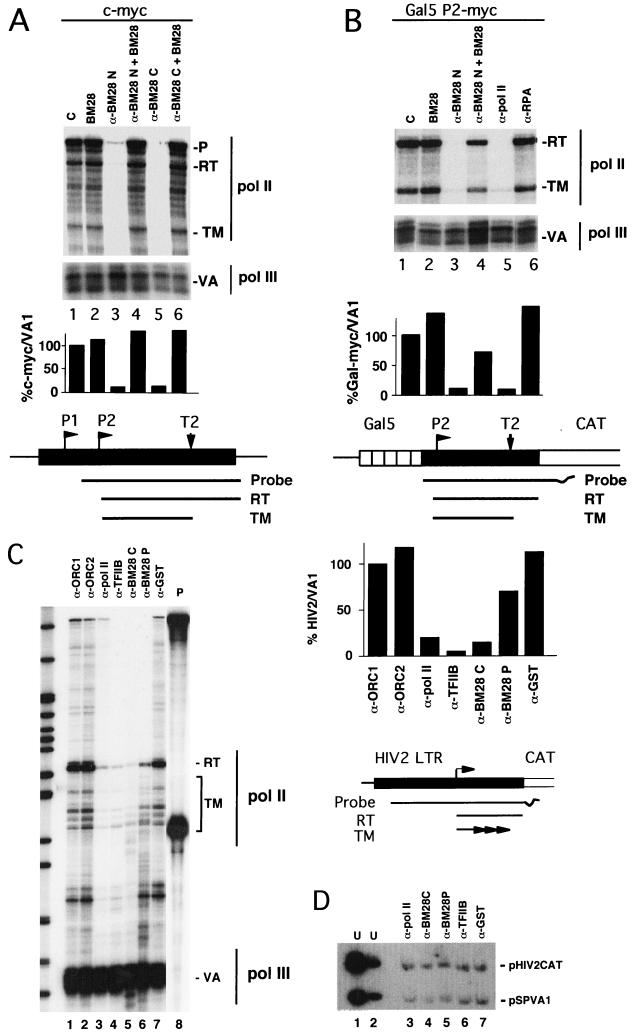
FIG. 1.
Anti-MCM2(BM28) antibodies inhibit Pol II transcription in Xenopus oocytes. (A) RNase protection analysis of c-myc and VA1 transcripts from injected oocytes. Anti-BM28-N (0.15 mg/ml; lanes 3 and 4) and anti-BM28-C antibodies (0.22 mg/ml; lanes 5 and 6) were coinjected with mouse c-myc exon I plasmid pSX943 and adenovirus VA1 plasmid pSPVA. Full-length recombinant MCM2(BM28) was coinjected at 0.55 mg/ml (lanes 2, 4, and 6). C, control oocytes injected with 1 mg of BSA/ml. Readthrough (RT) and terminated (TM) transcripts and VA1 Pol III transcripts are indicated. Transcripts from the P1 promoter and those which read around the plasmid protect the full-length probe (P). The RNase protection strategy is diagrammed with P1 and P2 promoters and the T2 terminator. RNase protection signals were quantified from scanned images, and the ratios of myc to VA1 are shown in the histogram, with the control normalized to 100%. (B) RNase protection analysis of pGal5-P2mycCAT and VA1 transcripts. Transcription was activated by injection of recombinant Gal4-AH. Anti-BM28-N (0.15 mg/ml; lanes 3 and 4), anti-Pol II CTD (0.15 mg/ml; lane 5), and anti-RP-A p70 (0.15 mg/ml; lane 6) were coinjected as indicated. Recombinant MCM2(BM28) was coinjected at 0.55 mg/ml (lanes 2 and 4). C, control oocytes injected with 1 mg of BSA/ml. The results were quantified as in panel A. (C) RNase protection analysis of HIV-2 CAT and VA1 transcripts. Anti-ORC1 (1 mg/ml), anti-ORC2 (1.3 mg/ml), anti-Pol II CTD (0.15 mg/ml), anti-TFIIB (0.5 mg/ml), anti-BM28-C (0.15 mg/ml), anti-BM28-P (0.25 mg/ml), and anti-GST (1 mg/ml) antibodies were coinjected with the pHIV2-LTR-CAT-556/+156 and pSPVA1 plasmids as indicated. Undigested HIV and VA probes marked P (10% of total) are shown in lane 8. Readthrough (RT) and terminated (TM) transcripts and VA1 Pol III transcripts are indicated. The results were quantified as in panel A. Size markers in the left-hand lane are MspI-digested pBR322, from 404 to 67 bases. (D) Southern blot of pSPVA1 and pHIV2-LTR plasmids recovered from injected oocytes. One oocyte equivalent of the samples, analyzed in panel C, lanes 3 to 7, was RNase treated, electrophoresed on an agarose gel, blotted, and hybridized to RNA probes complementary to pSPVA1 and pHIV2-LTR-CAT-556/+156. Lanes 1 and 2 were loaded with a mixture of uninjected supercoiled plasmids (U).
Glutathione S-transferase (GST), GST-VP16(410-490), GST-TFIIS (residues 1 to 301 of mouse TFIIS), and GST-mutant CTD were expressed in Escherichia coli with derivatives of the pGEX2T vector (Pharmacia). The GST-mutant CTD fusion protein contains 15 consensus CTD repeats with a Ser-to-Ala substitution at position 5 (62). The GST fusion to full-length mouse CTD was cloned into pET21a. Purification of Gal4-AH has been described previously (65).
HeLa cell whole-cell extract.
HeLa cell whole-cell extract was prepared by lysis in hypotonic buffer (20 mM HEPES 7.9, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 15 mM 2-glycerophosphate, 1 mM Na3O4, 2 mM NaF, 2 mM benzamidine, 0.4 mM dithiothreitol [DTT], 0.2% Nonidet P-40 (NP-40), 1 μM microcystin, 1 μg of pepstatin/ml, 1 μg of leupeptin/ml, 2 μg of aprotinin/ml, 50 μg of phenylmethylsulfonyl fluoride PMSF, 0.2% NP-40) and subsequent extraction with 0.41 M (NH4)2SO4. The extract was buffer exchanged with PD10 columns (Bio-Rad) against chromatography buffer (CB) (10 mM HEPES 7.9, 0.2 mM EDTA, 0.2 mM EGTA, 5 mM 2-glycerophosphate, 1 mM Na3VO4, 1 mM NaF, 1 mM benzamidine, 1 mM DTT, 50 μM ZnCl2, 1 μM microcystin, 1 μg of pepstatin/ml, 1 μg of leupeptin/ml, 2 μg of aprotinin/ml 12% glycerol, 0.05% NP-40) plus 50 mM NaCl and clarified by centrifugation (40 min at 50,000 × g) prior to the chromatography experiments presented in Fig. 3, 6, and 7. For the preparation of holoenzyme in the experiment shown in Fig. 5, whole-cell extract was prepared by extraction with 0.41 M (NH4)2SO4 and dialysis against 20 mM Tris acetate (pH 7.9), 0.1 M K acetate, 1 mM EDTA, 5% glycerol, 2 mM DTT (52).
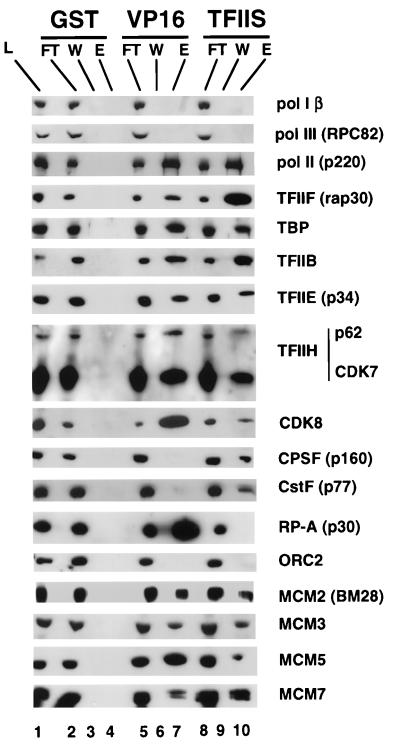
FIG. 3.
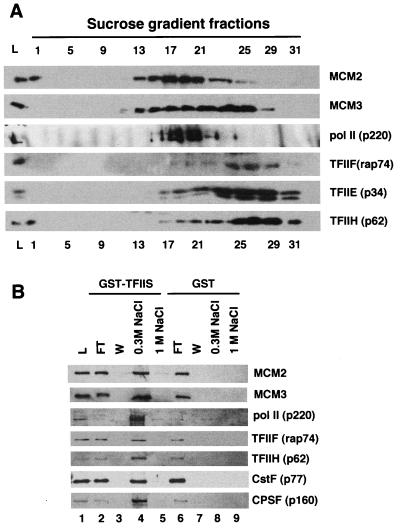
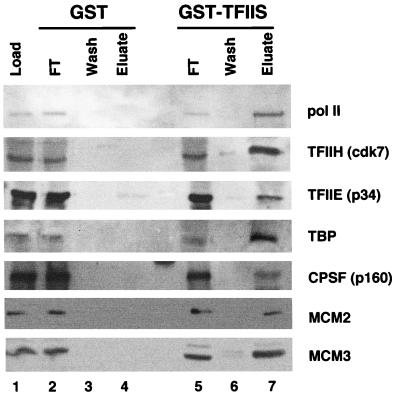
HeLa MCMs and Pol II holoenzyme components bind to VP16 and TFIIS affinity columns. Affinity columns (1 ml) containing GST (10 mg/ml), GST-VP16 (6 mg/ml), or GST-TFIIS (10 mg/ml) were loaded in parallel with 240 mg of HeLa whole-cell extract, washed five times with 3 ml of CB plus 50 mM NaCl, and eluted with 5 ml of CB plus 0.325 mM NaCl. A total of 0.025% of the load (L) and the flowthrough (FT) fractions and 0.5% of the final wash (W) and eluate (E) fractions were analyzed by Western blotting with the indicated antibodies. The data are representative of five independent experiments.
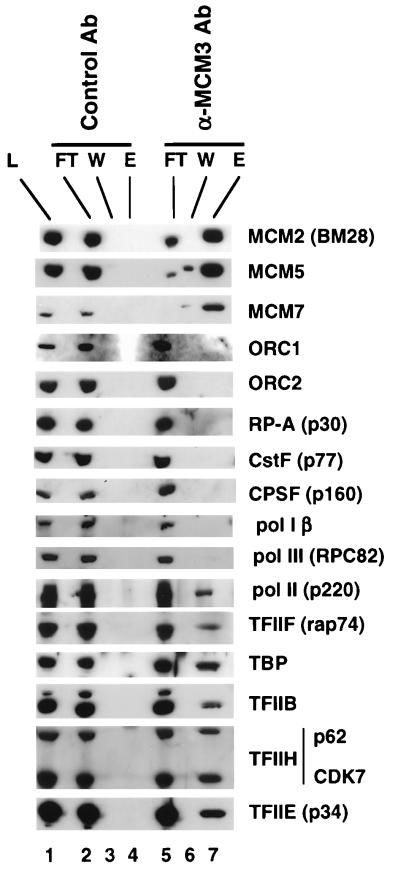
FIG. 6.
Coimmunopurification of Pol II holoenzyme and MCMs with anti-MCM3 antibody. Anti-MCM3 and control rabbit IgG immunoaffinity columns were loaded with HeLa whole-cell extract, washed, and eluted with 1 M NaCl. A total of 0.25% of the load (L) and flowthrough (FT) and 10% of the final wash (W) and eluate (E) fractions were analyzed by Western blotting with the indicated antibodies (Ab).
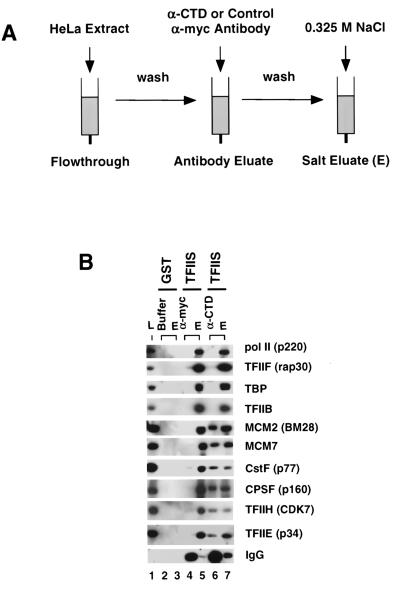
FIG. 7.
Anti-CTD antibody disrupts association of MCMs with the holoenzyme. (A) Diagram of the antibody disruption experiment. GST-TFIIS and GST control columns (100 μl) were loaded with HeLa extract and washed with CB plus 50 mM NaCl. Control (anti-myc; 9E10) or anti-CTD (8WG16) antibody (6 μg in 50 μl of CB plus 50 mM NaCl) was added to the GST-TFIIS resins. The GST column was eluted with buffer only. After 1 h, the antibody eluates were collected and the columns were washed and eluted with CB plus 0.325 M NaCl. (B) Western blots of holoenzyme components displaced by anti-CTD antibody. A total of 0.25% of the load (L), 16% of the control (α-myc) and anti-CTD antibody eluates, and 20% of the high-salt eluates (E) were analyzed with the indicated antibodies. The buffer eluate of the control GST column is shown in lane 2.
FIG. 5.
Copurification of Pol II holoenzyme and MCMs by cation-exchange chromatography, sucrose gradient sedimentation, and TFIIS affinity chromatography. (A) HeLa whole-cell extract was fractionated on Biorex 70, and the 0.3 to 0.6 M K acetate fraction (L) was separated on a 10 to 60% sucrose gradient (see Materials and Methods). The Load (L) and alternate fractions from the gradient were analyzed by Western blotting with the indicated antibodies. Note the comigration of Pol II with MCM2 and -3. (B) Pol II-containing sucrose gradient fractions (15 to 20) were chromatographed on GST and GST-TFIIS affinity columns (125 μl) in the presence of ethidium bromide. The columns were washed five times with 0.5 ml CB plus 50 mM NaCl and eluted with CB plus 0.325 M NaCl and CB plus 1 M NaCl. A total of 2.5% of the load (L) and the flowthrough (FT) fractions, 12% of the final wash (W), and 10% of the 0.325 and 1 M NaCl eluates were analyzed by Western blotting with the indicated antibodies.
Xenopus laevis oocyte extract.
Oocytes were defolliculated with 1 mg of collagenase/ml in MBS buffer (10 mM HEPES [pH 7.6], 88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.8 mM MgSO4, 0.7 mM CaCl2, 50 μg of gentamicin/ml), washed with XL extraction buffer (30 mM Tris HCl [pH 8], 100 mM KCl, 10 mM 2-glycerophosphate, 2 mM EGTA, 1 mM DTT, 2 mM benzamidine), and snap frozen. Frozen oocytes were combined with an equal volume of XL extraction buffer plus 2 μg of pepstatin/ml, 2 μg of leupeptin/ml, 2 μg of aprotinin/ml, and 2 mM DTT and broken by two strokes of a loose Dounce homogenizer. The homogenate was overlaid with one-fifth volume of mineral oil and centrifuged for 15 min at 25,000 × g. The translucent midphase was recentrifuged for 15 min at 25,000 × g, and the supernatant was cleared by centrifugation for 90 min at 225,000 × g. Glycerol was added to 15%, and aliquots were snap frozen.
Purification of Pol II holoenzyme.
Affinity columns containing 5 to 10 mg of immobilized GST, GST-VP16(410-490), or GST-TFIIS per ml of resin were loaded in parallel with HeLa or Xenopus whole-cell extract (100 to 200 mg/ml of resin), washed extensively with CB plus 50 mM NaCl, and eluted with CB plus 0.325 mM NaCl (40, 47). Some experiments were performed in the presence of 0.4 mg of ethidium bromide/ml (31). GST-TFIIS 0.325 M NaCl eluate (1.5 to 2 ml) was chromatographed on a Sepharose CL-2B column (60 by 1.6 cm; 0.4 ml/min) equilibrated in CB with 8% glycerol and 50 mM NaCl. Four-milliliter fractions were collected and concentrated by trichloroacetic acid TCA precipitation. The column was calibrated with dextran blue 2000 (2-MDa) and thyroglobulin (660-kDa) markers before each run.
In the experiment shown in Fig. 5, HeLa Pol II holoenzyme was purified by chromatography of whole-cell extract (150 mg) on Biorex 70 (5 ml) as described previously (52), and the 0.3 to 0.6 M K acetate fraction (6 ml) was loaded on a 30-ml 10 to 60% sucrose gradient and centrifuged for 16 h at 25,000 rpm (Beckman SW28 rotor), and 1-ml fractions were collected. The pooled peak of fractions containing Pol II (15 to 20) was chromatographed on GST and GST-TFIIS columns in the presence of 0.4 mg of ethidium bromide/ml as described above.
Immunoaffinity chromatography.
Affinity-purified rabbit anti-MCM3 and control rabbit IgG were coupled to 100 μl of protein A-Sepharose 4B (Pharmacia) at 2.9 mg of antibody/ml of resin. The columns were loaded with 14 mg of HeLa cell extract, washed seven times with 1 ml of CB plus 50 mM NaCl, and eluted with 0.9 ml of CB plus 1 M NaCl. The eluates were concentrated by TCA precipitation.
GST-CTD affinity chromatography.
Glutathione-Sepharose 4B resins contained GST (17 mg/ml of resin), GST-mutant CTD (3 mg/ml), or GST–wild-type CTD (3 mg/ml). HeLa nuclear extract (16 mg) in 20 mM HEPES (pH 7.9), 0.1 M NaCl, 0.1 mM EDTA, 2 mM DTT, 20% glycerol, 0.1% NP-40, 0.5 μM microcystin, 1 mM 2-glycerophosphate, 0.4 mg of ethidium bromide/ml was chromatographed on 250-μl columns as described previously (40). Bound proteins were eluted in buffer containing 1 M NaCl.
RESULTS
Anti-MCM2(BM28) antibodies inhibit Pol II transcription in Xenopus oocytes.
We analyzed transcription from three different promoters on plasmids injected into X. laevis oocytes: the human immunodeficiency virus type 2 (HIV-2) long terminal repeat (LTR), the mouse c-myc P1+P2 promoters, and a minimal c-myc P2 promoter with five upstream binding sites for Gal4 (pGal5-P2mycCAT). The HIV-2 LTR and c-myc promoters were activated by endogenous oocyte factors, whereas pGal5-P2mycCAT was activated by coinjected recombinant Gal4-AH (14). Under the conditions used (0.92 ng of DNA/oocyte), the plasmid DNA is all assembled into chromatin (16) and the promoters are transcribed exclusively by Pol II (3). The adenovirus VA1 gene, which is transcribed by Pol III, was included as a control for injection efficiency and RNA recovery. Affinity-purified antibodies against MCM2(BM28), Pol II general transcription factors, and replication factors were coinjected with the DNA templates. Any effect of the antibodies is independent of replication, since oocytes do not replicate double-stranded plasmid DNA.
Figure 1A demonstrates the effects of two antibodies against different regions of MCM2(BM28) on expression of a murine c-myc reporter gene. Correctly initiated transcripts from the P2 promoter were detected by RNase protection (Fig. 1). Both anti-MCM2(BM28) antibodies virtually eliminated both readthrough and terminated transcripts from this promoter (Fig. 1A, compare lanes 1, 3, and 5). The antibodies also abolished transcripts starting at the P1 promoter and transcripts that read around the plasmid (Fig. 1A). Note that this plasmid lacks a poly(A) site. The reduction in c-myc transcripts was completely reversed by blocking the antibodies with recombinant MCM2(BM28) (Fig. 1A, lanes 4 and 6), verifying that the effect is indeed due to reactivity with the antigen. Injected recombinant MCM2(BM28) alone had no effect on c-myc transcripts (Fig. 1A, lane 2). In contrast to c-myc, VA1 transcripts made by Pol III were not significantly reduced by the anti-MCM2(BM28) antibodies (Fig. 1A, lanes 3 and 5). The relative levels of c-myc and VA1 transcripts are quantified in the bar chart in Fig. 1A.
The effect of anti-MCM2(BM28) antibody on transcription from the c-myc P2 basal promoter activated by Gal4-AH is shown in Fig. 1B. Anti-BM28-N severely inhibited accumulation of transcripts from this template, and the effect was reversed by blocking the antibody with recombinant MCM2(BM28) (Fig. 1B, lanes 3 and 4). The magnitude of the effect of anti-BM28-N was comparable to that of antibody against Pol II itself (Fig. 1B, lane 5). In contrast, the antibody against replication protein A (RP-A; p70) did not affect Pol II transcription relative to the BSA control (Fig. 1B, lanes 1 and 6). The antibodies had little or no effect on the amount of VA1 RNA made by Pol III (Fig. 1B).
To address whether the effect of anti-MCM2(BM28) antibodies was peculiar to c-myc promoters, we also tested the HIV-2 LTR fused to chloramphenicol acetyltransferase (CAT) (pHIV2-LTR-CAT-556/+156) (11). This plasmid was coinjected into oocytes with antibodies against ORC1, ORC2, Pol II CTD, TFIIB, MCM2(BM28), or GST (Fig. 1C, lanes 1 to 7). Two anti-MCM2(BM28) antibodies both inhibited HIV-2 LTR transcription relative to the anti-GST control (Fig. 1C, compare lanes 5 to 7). In this experiment, inhibition of Pol II transcription was less complete than that shown in Fig. 1A and B, but the effects of the anti-MCM2(BM28-C) antibody was still comparable to those of anti-Pol II and anti-TFIIB (Fig. 1C, compare lanes 3 to 5). In contrast, anti-ORC1 and -ORC2 antibodies had little effect (Fig. 1C, lanes 1 and 2). None of the antibodies significantly affected Pol III transcription of the VA1 gene. As we observed for c-myc, anti-MCM2(BM28) antibodies reduced both readthrough and prematurely terminated transcripts from the HIV-2 template.
To evaluate the state of the DNA templates in antibody-injected oocytes, the same samples used for RNase protection (Fig. 1C) were also analyzed by Southern blotting (Fig. 1D). The recovered VA1 and HIV-2 CAT plasmids (Fig. 1D, lanes 3 to 7) comigrated with uninjected supercoiled marker plasmids (Fig. 1D, lanes 1 and 2) as expected for chromatinized plasmids (64). Furthermore, the amounts of recovered plasmid were not affected by the coinjected antibodies. Southern blots of plasmids recovered from the injected oocytes in Fig. 1A showed similar results (data not shown). The anti-MCM2(BM28) antibodies therefore did not reduce the accumulation of Pol II transcripts by destabilizing the microinjected template DNAs. The simplest explanation for these results is that the anti-MCM2(BM28) antibodies inhibited transcription by Pol II but not Pol III.
MCM proteins copurify with Xenopus and HeLa Pol II holoenzyme.
Inhibition of transcription by antibodies against MCM2(BM28) indicates that this protein may interact with the Pol II transcription apparatus. We therefore investigated whether Xenopus MCMs copurified with Pol II holoenzyme complexes prepared by GST-TFIIS affinity chromatography (47). Oocyte extract was loaded on GST and GST-TFIIS columns, and the columns were extensively washed with low-salt buffer and eluted with 0.325 M NaCl. Western blots of the load, flowthrough, final wash, and high-salt eluate fractions are shown in Fig. 2. The high-salt eluates from the GST-TFIIS but not the GST control column contained Pol II, TFIIH, TFIIE, the TATA binding protein TBP, and the cleavage/poly(A) factor CPSF (Fig. 2, lane 7) as previously observed for HeLa Pol II holoenzyme (40, 47). Significantly, the holoenzyme fraction also contained MCM2 and MCM3 (Fig. 2, lane 7). Furthermore, the efficiency of retention of oocyte MCM2 and -3 on the TFIIS column (approximately 1 to 5%) was comparable with that of TFIIE and TFIIH (Fig. 2, lanes 1 and 7). Because oocytes contain very small amounts of DNA relative to total protein, it is unlikely that MCMs artifactually copurify with holoenzyme by binding to DNA. As a further precaution against this possibility, the TFIIS affinity chromatography was performed in the presence of ethidium bromide, which efficiently disrupts protein-DNA interactions (31). In summary, these results suggest that MCMs associate with Pol II holoenzyme in oocytes and that anti-MCM2 antibodies could therefore be inhibiting transcription (Fig. 1) by binding to this protein complex.
FIG. 2.
MCM2 and MCM3 copurify with Xenopus oocyte Pol II holoenzyme. Affinity columns (250 μl) containing GST or GST-TFIIS at 10 mg/ml were loaded in parallel with 50 mg (2 ml) of X. laevis oocyte extract in the presence of ethidium bromide, washed five times with 1 ml of CB plus 50 mM NaCl, and eluted with 1.2 ml of CB plus 0.325 mM NaCl. A total of 0.25% of the load and the flowthrough (FT), 8% of the final wash, and 10% of the eluate fractions were analyzed by Western blotting with the indicated antibodies.
We addressed whether association with MCMs is a general property of Pol II holoenzyme by asking whether they also copurify with Pol II from HeLa cells. HeLa Pol II holoenzyme was enriched by three different procedures: (i) GST-VP16 affinity chromatography (20), (ii) GST-TFIIS affinity chromatography (47) (Fig. 3), and (iii) a combination of Biorex 70 cation-exchange chromatography and sucrose gradient sedimentation (52) followed by GST-TFIIS chromatography (see Fig. 5). The VP16 activation domain and the elongation factor TFIIS are unrelated proteins which probably bind to different surfaces of the holoenzyme. Whereas VP16 is highly acidic (pI, 3.30), TFIIS is slightly basic (pI, 8.40). HeLa whole-cell extract was chromatographed in parallel on GST, GST-TFIIS, and GST-VP16 affinity columns. The resins were eluted with 0.325 M NaCl, which was previously shown to elute the holoenzyme but not core Pol II from TFIIS (47). Western blots of the peptides in the load, flowthrough, final wash, and 0.325 M NaCl eluate fractions are shown in Fig. 3. As expected, the TFIIS and VP16 columns, but not the GST control, retained Pol II, TFIIB, -E, -F, and -H, TBP, and CDK8, but not Pol I or Pol III (Fig. 3, lanes 7 and 10). In agreement with previous reports, the experiment shown in Fig. 3 shows that the cleavage/poly(A) factors CPSF and CstF bound to GST-TFIIS (41) but not to GST-VP16. Conversely, CDK8 bound better to GST-VP16 than to GST-TFIIS (15, 47) (Fig. 3, lanes 7 and 10) and RP-A bound efficiently to VP16 (19, 32) but not to TFIIS. Importantly, MCM2(BM28), MCM3, MCM5, and MCM7 were retained on both the VP16 and TFIIS affinity resins but not on the GST control (Fig. 3, lanes 7 and 10). MCM2(BM28) also did not bind to a mutant of the VP16 activation domain in which four Phe residues were replaced with Ala (data not shown). We do not know if these fractions also contain MCM4 and -6. The possibility of artifactual binding of MCMs to the columns via association with contaminating chromatin is unlikely because binding was resistant to 0.4 mg of ethidium bromide/ml (data not shown) and because the holoenzyme fractions did not contain ORC2 (Fig. 3, lanes 7 and 10), a subunit of the protein complex that tethers MCMs to DNA at replication origins.
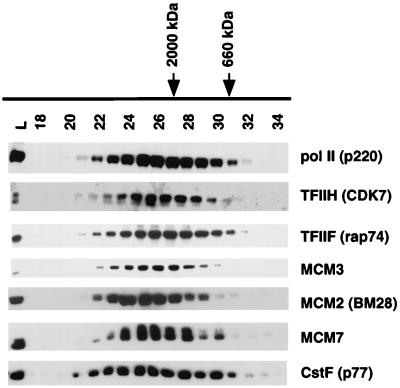
MCMs could bind to VP16 and TFIIS as a complex with Pol II holoenzyme, or alternatively, MCMs could bind these proteins independently of holoenzyme. Pol II holoenzyme complexes have apparent molecular masses of 2 to 4 MDa (38, 47), whereas previously characterized MCM complexes have much lower molecular masses. The MCM2-MCM7 complex RLF-M is about 400 to 600 kDa, and other complexes of MCM3-MCM4-MCM5 and MCM2-MCM4-MCM6-MCM7 are presumably even smaller (30, 48, 54). If MCMs and Pol II holoenzyme bind independently to the TFIIS column, they should be easily separated by gel filtration. Fractionation of the GST-TFIIS 0.325 M NaCl eluate on a Sepharose CL-2B column demonstrated precise copurification of Pol II, TFIIF, TFIIH, CstF, MCM2(BM28), MCM3, and MCM7 in a single peak with an apparent molecular mass greater than 2 MDa (Fig. 4). Although a relatively small fraction of total MCMs was present in the GST-TFIIS eluate (Fig. 3, lanes 8 and 10), essentially all the MCMs in this fraction copurified with Pol II during gel filtration (Fig. 4). In contrast, other proteins in the TFIIS eluate, including the SR family of splicing factors, migrated at significantly lower apparent molecular mass (data not shown). The migration of MCM proteins in the gel filtration column rules out the possibility that previously characterized MCM complexes bind to TFIIS independently of Pol II holoenzyme. Instead, the results are consistent with the model in which Pol II holoenzyme and MCMs associate with one another in a single complex, which binds to GST-TFIIS. These results do not eliminate the possibility, however, that a previously undiscovered high-molecular-weight form of MCM fortuitously binds to TFIIS and comigrates with Pol II holoenzyme on a gel filtration column.
FIG. 4.
TFIIS-bound MCMs and Pol II holoenzyme comigrate on a gel filtration column. The holoenzyme fraction from a GST-TFIIS affinity column (0.325 M NaCl eluate) was fractionated on a Sepharose CL-2B column. A total of 0.5% of the load (L) and 5% of each fraction (fraction numbers are at the top of each lane) were analyzed by Western blotting with the indicated antibodies. The migrations of dextran blue 2000 (2-MDa) and thyroglobulin (660-kDa) mass markers are indicated. This experiment is representative of four independent fractionations.
To reduce the possibility of such coincidental copurification, we purified Pol II holoenzyme by an independent procedure involving ion-exchange chromatography and sucrose gradient sedimentation (52). Fractionation on Biorex 70 separates Pol II holoenzyme (0.3 to 0.6 M K acetate eluate) from core Pol II (0.6 to 1.5 M K acetate eluate) (52). In our experiments about 30% of Pol II was present in the 0.3 to 0.6 M K acetate fraction, with the remainder in the 0.6 to 1.5 M K acetate fraction (not shown). Sucrose gradient sedimentation of the 0.3 to 0.6 M K acetate fraction demonstrated that a minor portion of TFIIE, TFIIF, TFIIH, CPSF, and CstF and a substantial fraction of MCM2 and MCM3 cosedimented with Pol II (Fig. 5A). The Pol II-containing sucrose gradient fractions (15 to 20) were pooled and chromatographed on parallel GST and GST-TFIIS affinity columns as described in the legend to Fig. 2. Western blots of the load, flowthrough, final wash, and 0.325 and 1 M NaCl eluates from the columns are shown in Fig. 5B. Consistent with the properties of holoenzyme (47), most of the Pol II in sucrose gradient fractions 15 to 20 was retained by the GST-TFIIS but not by the GST resin and eluted at 0.325 M NaCl (Fig. 5B, lanes 4 and 5). As expected for a fraction that is enriched for holoenzyme, significant amounts (more than 25%) of CstF, CPSF, TFIIF, and TFIIH (Fig. 5B) bound to GST-TFIIS and eluted together with Pol II. Importantly, more than 25% of MCM2 and MCM3 in the sucrose gradient-purified holoenzyme preparation also bound to GST-TFIIS and coeluted with Pol II (Fig. 5B, lanes 1 and 4). In contrast, only 2 to 4% of MCM2 and -3 in whole-cell extract bound to GST-TFIIS (Fig. 3, lanes 8 and 10). This experiment indicates that MCMs do not bind GST-TFIIS independently of Pol II and GTFs but together in the context of a holoenzyme complex.
Coimmunopurification of Pol II and GTFs with MCMs.
If MCMs are tightly associated with the holoenzyme as predicted by the results shown in Fig. 2 to 5, then Pol II and GTFs would be expected to immunoprecipitate with anti-MCM antibodies. To test this idea, HeLa cell extract was passed through immunoaffinity columns containing control rabbit IgG or rabbit anti-MCM3 antibodies. The columns were washed extensively with low-salt buffer and eluted with high salt. Load, flowthrough, final wash, and high-salt eluates from the columns were analyzed by Western blotting. MCM2, -5, and -7 bound to the anti-MCM3 affinity column as expected, since MCMs associate with one another (Fig. 6, lane 7). In contrast ORC1, ORC2, RP-A, Pol I, and Pol III were not retained on the anti-MCM3 column (Fig. 6, lane 7). Remarkably, Pol II and the general transcription factors TFIIB, -E, -F, and -H and TBP were all specifically retained on the anti-MCM3 column but not on the control IgG column (Fig. 6, lanes 4 and 7). The apparent efficiency of Pol II and GTF retention on the anti-MCM3 column was two- to fourfold lower than that of MCM retention on VP16 or TFIIS columns (Fig. 2 and 3). This discrepancy could be due to destabilization of the holoenzyme by anti-MCM3 or to incomplete elution from the antibody column. We did not detect CPSF or CstF binding to anti-MCM3, possibly because MCMs and polyadenylation factors occur in distinct holoenzyme complexes. The presence of Pol II and GTFs in anti-MCM3 immunoprecipitates provides further independent evidence that MCMs are components of a form of Pol II holoenzyme.
CTD-dependent binding of MCMs to holoenzyme.
The Pol II CTD plays a central role in maintaining the integrity of the Pol II holoenzyme. We assayed whether the interaction of MCMs with mammalian Pol II holoenzyme was dependent on the CTD. The experimental strategy is shown in Fig. 7A. HeLa extract was chromatographed on GST-TFIIS columns to purify holoenzyme. The complex immobilized on the affinity resin was then challenged with monoclonal anti-CTD (8WG16) or control anti-myc (9E10) antibody. Proteins, which associate with the holoenzyme in a CTD-dependent manner, are expected to be displaced specifically by the anti-CTD antibody (27). Proteins remaining bound to the resin after antibody treatment were eluted with high salt. Anti-myc antibody did not displace any of the analyzed holoenzyme components (Fig. 7B, lane 4). Instead, Pol II, GTFs, polydenylation factors, and MCMs were present exclusively in the high-salt eluate (Fig. 7B, lane 5). In contrast, the anti-CTD monoclonal antibody displaced significant amounts of TFIIE, TFIIH, CstF, CPSF, MCM2(BM28), and MCM7 (Fig. 7B, lane 6). MCM3 and MCM5 were also eluted by the anti-CTD antibody (data not shown). The anti-CTD antibody did not displace TFIIB, TFIIF, TBP, or Pol II itself (Fig. 7B, lane 6). A significant amount of anti-CTD antibody was retained on the affinity column and eluted by high salt (Fig. 7B, lane 7), consistent with binding to the immobilized Pol II. These data indicate that the association of MCM proteins, as well as CPSF, CstF, TFIIE, and TFIIH, with the Pol II holoenzyme is at least partly mediated by direct or indirect contacts with the CTD. A significant portion of MCMs, CPSF, CstF, TFIIE, and TFIIH were not displaced by anti-CTD antibody (Fig. 7), either because of incomplete antibody binding or because there are additional CTD-independent contacts between these factors and the holoenzyme. The displacement of MCMs by anti-CTD antibody argues that these proteins are not contaminants that coincidentally copurify with Pol II holoenzyme but rather are genuine subunits of the complex.
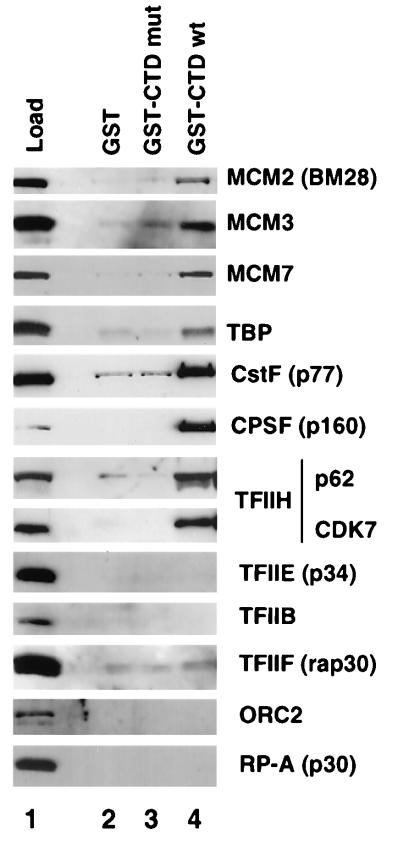
The displacement of MCMs from Pol II holoenzyme by anti-CTD antibodies suggests a direct or indirect physical interaction with the CTD. We investigated this possibility by chromatography of HeLa nuclear extract on mutant and wild-type GST-CTD affinity columns. The high-salt eluates from the columns were analyzed by Western blotting (Fig. 8). In agreement with previous reports (40, 60), we observed specific binding of TBP, CstF, and CPSF to the wild-type CTD resin (Fig. 8, lane 4). Notably, we also observed specific binding of MCM2(BM28), MCM3, and MCM7 to the wild-type CTD (Fig. 6, lane 4) above background binding by the GST and mutant CTD control columns (Fig. 8, lanes 2 and 3). TFIIH also bound to the wild-type CTD resin; however, under these conditions, we did not observe binding of RP-A, ORC2, TFIIB, or TFIIF (Fig. 8, lane 4). For reasons we do not understand, TFIIE binding to GST-CTD was not detected (Fig. 8), although this factor was displaced from the holoenzyme by anti-CTD antibody (Fig. 7). These results show that MCMs bind directly or indirectly to recombinant CTD. We suggest that this interaction contributes to the association of MCMs with Pol II holoenzyme.
FIG. 8.
MCM proteins bind to recombinant CTD. HeLa nuclear extract was chromatographed on GST (lane 2), GST-mutant CTD (mut; lane 3), and GST–wild-type CTD (wt; lane 4) affinity resins. Western blots with the indicated antibodies of 0.05% of the load and 1% of the eluates are shown.
DISCUSSION
Anti-MCM2 antibodies inhibit Pol II transcription.
We report functional and biochemical evidence for a role of MCM proteins in the Pol II transcriptional apparatus. The functional data demonstrates a specific inhibition of Pol II transcript accumulation in vivo by three antibodies against different regions of the MCM2 protein (Fig. 1). In injected Xenopus oocytes, anti-MCM2 antibodies were approximately as effective as antibodies against Pol II and TFIIB in reducing transcript levels from three different promoter constructs. The effect of anti-MCM2 antibodies was specific to Pol II, as there was no significant decrease in Pol III transcripts from a coinjected reporter gene. The effect of anti-MCM2 on accumulation of Pol II transcripts was reversed by incubating the antibodies with purified recombinant MCM2, proving that it was caused by reactivity with this antigen rather than a contaminant in the antibody preparations. The anti-MCM2 antibodies did not destabilize the template DNA or inhibit its supercoiling due to chromatin assembly (Fig. 1D). The simplest explanation of these results is that anti-MCM2 antibodies specifically inhibit transcription by Pol II.
We performed biochemical experiments to seek an explanation for the unexpected effect of anti-MCM2 antibodies on Pol II transcription. The results of these studies show that MCM2 and other MCMs are in fact components of high-molecular-weight Pol II holoenzyme complexes isolated from Xenopus oocytes and HeLa cells by several different procedures. It is therefore plausible that anti-MCM2 inhibits transcription in injected oocytes by interfering with Pol II holoenzyme. Although MCM3, MCM5, and MCM7 are found in Pol II holoenzyme complexes (Fig. 2 to 4), antibodies against these peptides did not affect transcription in oocytes (data not shown). This negative result may mean that in contrast to MCM2, the association of MCM3, MCM5, and MCM7 with the holoenzyme is not important for Pol II transcription. Alternatively, it could simply reflect poor antibody accessibility in vivo.
Pol II holoenzyme contains MCM proteins.
MCM proteins copurified with Pol II and GTFs on two unrelated affinity columns with the immobilized activation domain of herpes simplex transcription factor VP16 or the Pol II-associated elongation factor TFIIS (Fig. 2 and 3). The holoenzyme fraction of Pol II is specifically eluted from both affinity resins at 0.325 M NaCl (47), along with about 2 to 4% of the MCM2, -3, -5, and -7 present in extracts from both HeLa cells and Xenopus oocytes (Fig. 2 and 3). Note that oocytes contain far less DNA relative to protein than HeLa cells, yet MCM proteins from both sources bound equally well to the TFIIS affinity resin. It is therefore highly unlikely that the binding is an artifact of chromatin contamination. Essentially 100% of the detectable MCM proteins in the eluate from a TFIIS column precisely comigrated with Pol II holoenzyme in a single peak with an apparent molecular mass of 2 to 4 MDa on a Sepharose CL-2B gel filtration column (Fig. 4). The apparent molecular masses of the MCMs in this fraction are much greater than that of previously reported MCM complexes (30, 48, 54). This experiment shows that a novel protein complex of MCMs binds to TFIIS and comigrates with Pol II holoenzyme by gel filtration. Because MCMs are about 100-fold more abundant than Pol II holoenzyme (5, 47), it is not surprising that the small fraction which copurifies with Pol II in the 2 to 4 MDa range has not been detected previously. The most likely explanation of these results is that MCMs are in fact associated with holoenzyme and bind to TFIIS as a complex with Pol II and GTFs. Alternatively, Pol II holoenzyme and a novel minor form of MCM could have coincident molecular masses and bind independently to TFIIS.
We performed three experiments to eliminate the latter possibility: (i) additional purification of holoenzyme by cation-exchange chromatography and sucrose gradient sedimentation, (ii) immunoprecipitation with anti-MCM3 antibody, and (iii) displacement of holoenzyme-associated factors by antibody against the CTD of the Pol II large subunit.
Holoenzyme can be significantly enriched by chromatography of whole-cell extract on Biorex 70 followed by sucrose gradient sedimentation (52). We observed that a fraction of MCMs cofractionated with Pol II and GTFs through these two purification steps and that holoenzyme isolated by this procedure bound specifically to TFIIS (Fig. 5). It is significant that the percentage of MCMs which bound to the TFIIS resin was much higher for the sucrose gradient-purified holoenzyme fraction than it was for whole-cell extract (Fig. 5B). This observation is not consistent with independent binding of MCMs and holoenzyme to the TFIIS affinity column. Rather, it supports the hypothesis that a single complex containing MCMs, Pol II, and GTFs binds to TFIIS.
The copurification of MCMs with holoenzyme was observed not only with the two affinity resins that bind components of the Pol II transcriptional apparatus (VP16 and TFIIS) but also with a resin that binds directly to MCMs. An anti-MCM3 immunoaffinity column specifically retained not only MCMs but also Pol II, TBP, and TFIIB, -E, -F, and -H (Fig. 6). The observation that affinity resins designed to bind either MCMs or basal transcription components retain a common set of proteins provides independent support for the idea that MCMs and Pol II holoenzyme are parts of the same complex.
The exact composition and stoichiometry of MCMs associated with holoenzyme is not clear, however, we have detected MCM2, -3, -5, and -7 in these complexes (Fig. 3). There is heterogeneity among yeast and mammalian holoenzymes isolated in different ways (7), and MCMs may not be present in all forms of holoenzyme. Such heterogeneity is suggested by the fact that holoenzyme complexes purified by Biorex 70 chromatography, sucrose gradient sedimentation, and TFIIS binding procedure contain both MCMs and polyadenylation factors (Fig. 5), whereas the complexes purified by anti-MCM3 chromatography lack polyadenylation factors (Fig. 6).
The CTD and association of MCMs with Pol II holoenzyme.
The specificity of the protein-protein contacts that tether MCMs to holoenzyme was investigated by asking if the interaction required the CTD of the Pol II large subunit. The importance of the CTD for the integrity of holoenzyme complexes was demonstrated by the fact that anti-CTD antibody specifically displaces the mediator complex from yeast Pol II holoenzyme (27). We applied the same strategy to probe mammalian holoenzyme and observed that a subset of associated factors, including TFIIE, TFIIH, CstF, and CPSF, was specifically displaced. Notably, MCM proteins were among those proteins displaced from holoenzyme by anti-CTD antibody (Fig. 7B). The reversal of binding between MCMs and Pol II holoenzyme by anti-CTD antibody eliminates the possibility that a nonspecific interaction is responsible for this association. Furthermore, this experiment strongly suggests that a protein contact with the CTD is required for association of MCMs with holoenzyme. Consistent with this conclusion, we observed that MCMs bind specifically to a CTD affinity column (Fig. 8). These experiments do not distinguish whether MCM binding to the CTD is direct or indirect, however.
What is the function of MCM proteins in Pol II holoenzyme?
MCM proteins were previously identified as components of the replication licensing factor, and their exact function in the holoenzyme complex is not clear. The presence of MCMs could reflect some function of the Pol II holoenzyme in replication. In agreement with this idea, proteins which function primarily in DNA repair and replication have been found previously in mammalian holoenzyme complexes (38, 52). In addition, transcriptional activation domains, which presumably recruit Pol II holoenzyme, can stimulate DNA replication when tethered to viral or cellular origins of replication (9, 39). A link between Pol II transcription and DNA replication is also suggested by the tight correlation between the potency of transactivators in enhancing transcription and replication (21, 33). The mechanism underlying this phenomenon is poorly understood, but there is evidence that transactivators recruit chromatin-remodeling factors (21) to origins, probably by a mechanism similar to the way that they recruit holoenzyme to promoters. Our experiments suggest that transactivators could recruit MCM proteins to origins of replication via contacts with Pol II holoenzyme and thereby stimulate DNA replication. The possibility that the MCMs in the Pol II holoenzyme function in DNA replication is not inconsistent with inhibition of transcription by anti-MCM2 antibodies (Fig. 1). Antibody binding anywhere on its surface could sequester holoenzyme or prevent its assembly, thereby inhibiting transcription even if the epitope recognized is in a subunit that does not directly participate in the transcription reaction.
Although a role of Pol II holoenzyme in control of replication remains possible, the most straightforward interpretation of our data is that MCMs have a previously unsuspected role in transcription. This hypothesis is in agreement with recent observations of a correlation between transcriptional activation by Stat1α and its ability to bind MCM5 (68). Our results imply that the interaction between Stat1α and MCM5 may serve to recruit Pol II holoenzyme to promoters, which are targeted by Stat1α. A transcriptional function of MCMs is also indicated by the fact that mcm5 mutants in budding yeast show genetic interactions with mutants in the RPB1 gene encoding the Pol II large subunit (7a). MCMs bind to histones and have sequence homology with ATP-dependent DNA helicases (22, 23, 29). On the basis of their apparent movement with DNA polymerase at replication forks, it has been suggested that MCMs facilitate fork movement by remodeling chromatin (2). Such a remodeling activity associated with Pol II holoenzyme could facilitate transcription of chromatin templates. More needs to be learned about the interaction of MCM proteins with chromatin in order to address this question.
ACKNOWLEDGMENTS
We thank N. Fong, M. Pandes, E. Lees, L. Rothblum, R. Roeder, N. Thompson, G. Evan, J.-M. Egly, and R. Wood for gifts of antibodies and J. Douglas, Dept. of Comparative Medicine, University of Toronto, and the ICRF animal unit for supplying Xenopus oocytes. We are also grateful to J. Greenblatt, J. Diffley, S. Mason, B. McNeil, J. Parvin, E. Rosonina, A. Wildeman, D. Evans, and A. Hilliker for valuable discussions and T. Boudreau for secretarial help.
This work was supported by a start-up grant to K.Y. from the University of Guelph and by grants to D.L.B. from the Medical Research Council of Canada and NIH GM-58613-01.
REFERENCES
- 1.Anderson S F, Schlegel B P, Nakajima T, Wolpin E S, Parvin J D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio O, Weinstein O, Bell S. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and cdc45 during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Bentley D L, Brown W L, Groudine M. Accurate, TATA box-dependent polymerase III transcription from promoters of the c-myc gene in injected Xenopus oocytes. Genes Dev. 1989;3:1179–1189. doi: 10.1101/gad.3.8.1179. [DOI] [PubMed] [Google Scholar]
- 4.Blow J J, Laskey R A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 5.Burkhart R, Schulte D, Hu D, Musahl C, Goehring F, Knippers R. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur J Biochem. 1995;228:431–438. [PubMed] [Google Scholar]
- 6.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Chang M, Jaehning J J. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Cilli, K., and D. Bentley. Unpublished data.
- 8.Coue M, Kearsey S, Mechali M. Chromatin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 1996;15:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 9.DePamphilis M L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988;52:635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- 10.Donovan S, Harwood J, Drury L S, Diffley J F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerman M, Guyader M, Montagnier L, Baltimore D, Muesing M A. The specificity of the human immunodeficiency virus type 2 transactivator is different from that of human immunodeficiency virus type 1. EMBO J. 1987;6:3755–3760. doi: 10.1002/j.1460-2075.1987.tb02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evan G, Lewis G, Ramsay G, Bishop J. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer L, Gerard M, Chalut C, Lutz Y, Humbert S, Kanno M, Chambon P, Egly J M. Cloning of the 62-kilodalton component of basic transcription factor btf2. Science. 1992;257:1392–1395. doi: 10.1126/science.1529339. [DOI] [PubMed] [Google Scholar]
- 14.Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 15.Gold M O, Tassan J P, Nigg E A, Rice A P, Herrmann C H. Viral transactivators e1a and vp16 interact with a large complex that is associated with ctd kinase-activity and contains cdk8. Nucleic Acids Res. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurdon J B, Melton D A. Gene transfer in amphibian eggs and oocytes. Annu Rev Genet. 1981;15:189–218. doi: 10.1146/annurev.ge.15.120181.001201. [DOI] [PubMed] [Google Scholar]
- 17.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han S J, Lee Y C, Gim B S, Ryu G-H, Park S J, Lane W S, Kim Y-J. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol Cell Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Z G, Brinton B T, Greenblatt J, Hassell J A, Ingles C J. The transactivator protein VP16 and protein Gal4 bind replication factor A. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 20.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y F, Hao Z L, Li R. Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev. 1999;13:637–642. doi: 10.1101/gad.13.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishimi Y, Komamura Y, You Z, Kimura H. Biochemical function of mouse minichromosome maintenance 2 protein. J Biol Chem. 1988;273:8369–8375. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- 23.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6 and -7 complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 24.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. MCM protein complex is associated with Histone H3. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 25.Kearsey S E, Labib K. MCM proteins: evolution, properties, and role in DNA replication. Biochim Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 26.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 27.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 28.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 29.Koonin E V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R, Botchan M R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 33.Li R, Yu D S, Tanaka M, Zheng L, Berger S L, Stillman B. Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol Cell Biol. 1998;18:1296–1302. doi: 10.1128/mcb.18.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Bjorklund S, Jiang Y W, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madine M A, Khoo C Y, Mills A D, Laskey R A. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 37.Maine G, Sinha P, Tye B-K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C, Linn S, Reinberg D. A human RNA polymerase II associated complex with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 39.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 40.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G H, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples messenger RNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 41.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myers L C, Gustafson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers L C, Gustafson C M, Hayashibara K C, Brown P O, Kornberg R D. Mediator protein mutations that selectively abolish activated transcription. Proc Natl Acad Sci USA. 1999;96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 45.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 46.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 47.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 48.Richter A, Knippers R. High molecular mass complexes of human minichromosome maintenance proteins in mitotic cells. Eur J Biochem. 1997;247:136–141. doi: 10.1111/j.1432-1033.1997.00136.x. [DOI] [PubMed] [Google Scholar]
- 49.Romanowski P, Madine M A, Laskey R A. XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication. Proc Natl Acad Sci USA. 1996;93:10189–10194. doi: 10.1073/pnas.93.19.10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanowski P, Madine M A, Rowles A, Blow J J, Laskey R A. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- 51.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J, Egly J M. The MO15 cell-cycle kinase is associated with the TFIIH transcription DNA-repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 52.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 54.Thoemmes P, Kubota Y, Takisawa H, Blow J J. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 56.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson N, Aronson D, Burgess R. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. J Biol Chem. 1990;265:7069–7077. [PubMed] [Google Scholar]
- 58.Todorov I T, Attaran A, Kearsey S E. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Todorov I T, Pepperkok R, Philipova R N, Kearsey S E, Ansorge W, Werner D. A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J Cell Sci. 1994;107:253–265. doi: 10.1242/jcs.107.1.253. [DOI] [PubMed] [Google Scholar]
- 60.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Luo T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West M, Corden J. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Rna polymerase II holoenzyme contains Swi/Snf regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 64.Wyllie A H, Laskey R, Finch J, Gurdon J B. Selective DNA conservation and chromatin assembly after injection of SV40 DNA into Xenopus oocytes. Dev Biol. 1978;64:178–188. doi: 10.1016/0012-1606(78)90069-6. [DOI] [PubMed] [Google Scholar]
- 65.Yankulov K, Blau J, Purton T, Roberts S, Bentley D. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 66.Yankulov K Y, Pandes M, McCracken S, Bouchard D, Bentley D L. TFIIH functions in regulating transcriptional elongation by RNA polymerase II in Xenopus oocytes. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young M A, Tye B K. MCM2 and MCM3 are constitutive nuclear proteins that exhibit distinct isoforms and bind chromatin during specific cell cycle stages of Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:1587–1601. doi: 10.1091/mbc.8.8.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J J, Zhao Y, Chait B T, Lathem W W, Ritzi M, Knippers R, Darnell J E. Ser727-dependent recruitment of MCM5 by stat1alpha in IFN-gamma-induced transcriptional activation. EMBO J. 1998;17:6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]