Abstract
Members of the 160-kDa nuclear receptor coactivator family (p160 coactivators) bind to the conserved AF-2 activation function found in the hormone binding domains of nuclear receptors (NR) and are potent transcriptional coactivators for NRs. Here we report that the C-terminal region of p160 coactivators glucocorticoid receptor interacting protein 1 (GRIP1), steroid receptor coactivator 1 (SRC-1a), and SRC-1e binds the N-terminal AF-1 activation function of the androgen receptor (AR), and p160 coactivators can thereby enhance transcriptional activation by AR. While they all interact efficiently with AR AF-1, these same coactivators have vastly different binding strengths with and coactivator effects on AR AF-2. p160 activation domain AD1, which binds secondary coactivators CREB binding protein (CBP) and p300, was previously implicated as the principal domain for transmitting the activating signal to the transcription machinery. We identified a new highly conserved motif in the AD1 region which is important for CBP/p300 binding. Deletion of AD1 only partially reduced p160 coactivator function, due to signaling through AD2, another activation domain located at the C-terminal end of p160 coactivators. C-terminal coactivator fragments lacking AD1 but containing AD2 and the AR AF-1 binding site served as efficient coactivators for full-length AR and AR AF-1. The two signal input domains (one that binds NR AF-2 domains and one that binds AF-1 domains of some but not all NRs) and the two signal output domains (AD1 and AD2) of p160 coactivators played different relative roles for two different NRs: AR and thyroid hormone receptor.
Transcriptional activator proteins modulate gene transcription by binding to specific enhancer elements associated with the promoters of their target genes. The subsequent transcriptional activation of the gene involves local modifications in chromatin structure and recruitment of a transcription initiation complex containing RNA polymerase II to the promoter (3, 52). Recent studies have shown that DNA-bound transcriptional activator proteins accomplish these two tasks with the assistance of a class of proteins called transcriptional coactivators (19, 47, 48, 51). Coactivators are generally not DNA binding proteins but rather are recruited to the promoter through protein-protein contacts with the transcriptional activators. They may be thought of as adaptors or components in a signaling pathway that transmit transcriptional activation signals from DNA-bound activator proteins to the chromatin and transcription machinery.
The nuclear receptors (NR) comprise a large superfamily of more than 100 structurally related proteins, many of which bind to and serve as transcriptional regulators for specific genes (2, 13, 14, 38, 51, 52). The most well-characterized group of NRs include receptors for steroid and thyroid hormones, retinoic acid, and vitamin D. Binding of the appropriate hormones to the steroid hormone receptors causes a conformational change that allows the receptors to bind (usually as homodimers) to enhancer elements in the promoters of target genes, recruit transcriptional coactivators, and activate transcription. The receptors for thyroid hormones, retinoic acid, and vitamin D bind to their enhancer elements (generally as heterodimers with retinoid X receptors) even in the absence of hormone and repress transcription; hormone binding triggers a conformational change like that in the steroid receptors which allows recruitment of coactivators and thus causes transcriptional activation. The hallmark of all NRs is a highly conserved DNA-binding domain (DBD) located in the central part of the polypeptide chain. The hormone binding domain (HBD), which is also conserved but somewhat less than the DBD, is a large C-terminal domain; in addition to binding hormone, it also contains an important highly conserved activation function, AF-2, which is one of two domains primarily responsible for activation of transcription by the hormone-activated, DNA-bound NR (12, 14, 22, 32, 34). The other activation function, AF-1, found in the N-terminal domains of most NRs, is not conserved in length or sequence. The relative importance of AF-1 and AF-2 varies among different NRs (20, 29, 34) and also can be influenced by ligand, cell type, and target gene promoter (39, 53).
Very little is known about the mechanism of action of AF-1 domains of NRs. In contrast, AF-2 domains of essentially all NRs that function as transcriptional activators are highly conserved in sequence and three-dimensional structure (8, 9, 15, 58). A substantial number of candidate coactivator proteins have been identified, by virtue of their ability to bind HBDs (AF-2 domains) of hormone-activated NRs (26, 51). While a number of these proteins have been shown to enhance NR function, the most well characterized among these is a family of three structurally related but genetically distinct 160-kDa proteins called the NR coactivators or p160 coactivators. The three family members are steroid receptor coactivator 1 (SRC-1), glucocorticoid receptor interacting protein 1 (GRIP1; also called TIF2), and pCIP (also named RAC3, ACTR, AIB1, and TRAM1) (51). There is approximately 40% amino acid sequence identity between any two different family members (1, 24); functionally distinct splicing isoforms of these proteins have been reported (27, 30). Transient expression of any one of these proteins enhances transcriptional activation by hormone binding NRs and some orphan NRs (51). Antibody microinjection experiments in mammalian cells (50) and functional assays for NRs in yeast cells (24, 56) indicate that these coactivators are required for efficient NR function.
The current study is focused on defining the signal input domains of the p160 coactivators, which interact with NRs, and the signal output domains which transmit the transcriptional activation signal to the chromatin and transcription machinery. Previous studies suggested that there was only one primary type of signal input domain and one primary signal output domain. The conserved AF-2 domain of any hormone-activated NR binds to the conserved NR interaction domain (NID) found in the central part of the polypeptide chains of the three p160 coactivators (see Fig. 1A) (9, 10, 21, 50, 55). The NID contains three conserved sequences called NR boxes, which contain the motif LXXLL, where L is leucine and X is any amino acid. Binding of the NID to the NR AF-2 domain is sufficient to recruit the coactivator to enhance NR function. Recently, it was shown that the AF-1 domains of the progesterone receptor (PR) and the estrogen receptor (ER) also interact with p160 coactivators, although it is not known whether PR and ER AF-1 bind to the same or different sites on the coactivators (43, 57). Whether AF-1 regions of other NRs also function in this manner remains to be determined and is addressed here for the androgen receptor (AR). The lack of homology in the AF-1 regions of NRs suggests that they may function by a variety of different mechanisms.
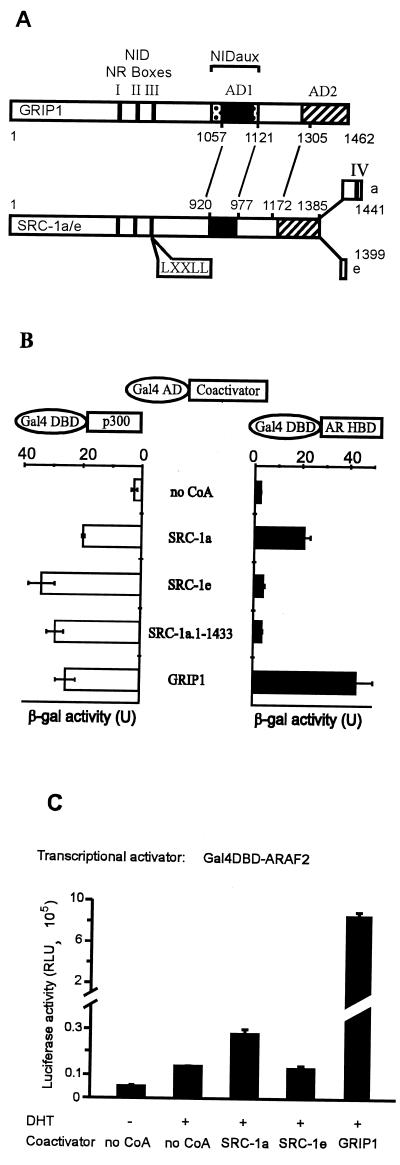
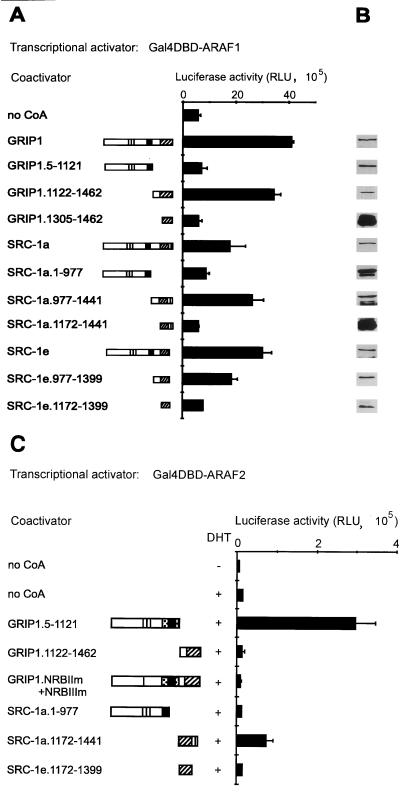
FIG. 1.
Binding of GRIP1, SRC-1a, and SRC-1e to AR HBD correlates with coactivator effect on AR AF-2. (A) Alignment and functional domains of p160 coactivators. NID, NR HBD interaction domain; vertical solid bars, NR boxes I, II, and III (LXXLL motifs); NIDaux (spotted box), auxiliary domain in GRIP1 required in addition to NR boxes for efficient binding of AR HBD but not TR HBD; AD1 (solid box) and AD2 (striped box), two autonomous activation domains; numbers, amino acids of GRIP1 and SRC-1. The lines between GRIP1 and SRC-1 amino acid numbers indicate alignment of homologous regions. NIDaux and AD1 overlap within GRIP1 amino acids 1011 and 1121; SRC-1 lacks NIDaux and SRC-1a contains a fourth NR box (IV). (B) Interaction of p160 coactivators with AR HBD in yeast two-hybrid assays. Gal4 AD, fused to the indicated coactivator, was expressed in yeast strain SFY526 along with a Gal4DBD-ARHBD or Gal4DBD-p300 fusion protein; when AR HBD was present, yeast cells were grown in 100 nM DHT. Activation of the integrated β-Gal reporter gene controlled by Gal4 binding sites was determined by measuring β-Gal activity in cell extracts. (C) Enhancement of AR AF-2 function by p160 coactivators. CV-1 cells were transfected with 0.5 μg of pM vector encoding Gal4DBD-ARAF2, 0.5 μg of GK1 reporter gene, and 1.0 μg of pSG5.HA (no CoA) or the same vector encoding the indicated coactivator. Luciferase activity, shown in relative light units (RLU), observed with or without DHT treatment is shown.
A single domain in p160 coactivators that contributes to signal output has also been defined. This conserved domain, called activation domain 1 (AD1) is located approximately between amino acids 1040 and 1120 of GRIP1, which has 1,462 amino acids (see Fig. 1A) (7, 25, 55). The signaling function of AD1 is due to AD1’s ability to bind CREB binding protein (CBP) or p300, two related proteins that serve as secondary coactivators for NRs; i.e., CBP or p300 is recruited to the promoter by its interaction with the primary p160 coactivators. While a full mechanistic understanding of CBP and p300 function awaits further study, their action is at least partly due to their ability to acetylate histones, transcriptional activators, and/or components of the transcription initiation complex such as basal transcription factors (18, 28, 31, 42). Acetylation of the N-terminal tails of histones alters nucleosome and chromatin structure by interfering with the association between the basic histone tails and the acidic sugar-phosphate backbone of the DNA helices wrapped around the nucleosomes (35, 36, 47). Mutations in the AD1 region greatly reduced or eliminated the ability of the p160 proteins to bind CBP or p300 and to serve as coactivators for NRs (7, 55), suggesting that AD1 was the principle coactivator domain responsible for downstream signaling.
Another putative AD, AD2, located in the highly conserved C-terminal region of the p160 proteins, was initially identified by its ability to activate transcription when fused to Gal4 DBD but was not a binding site for CBP or p300 (7, 55). To date, there has been no evidence of AD2 function in assays that test the ability of p160 proteins to act as coactivators, i.e., in true coactivator assays. In the current study we tested AD2’s ability to serve as a signal output domain for p160 proteins in true coactivator assays. This effort was facilitated partly by identification of a new AD1 deletion that eliminated CBP/p300 binding without compromising other coactivator functional domains. The relative importance of the AF-1 and AF-2 binding sites as signal input domains and the relative importance of AD1 and AD2 as signal output domains of p160 proteins were studied by testing the ability of p160 proteins and various p160 protein fragments and mutants to serve as coactivators for AR and the thyroid hormone receptor (TR).
MATERIALS AND METHODS
Plasmids.
Mammalian expression vector pSG5.HA, which we used to express proteins fused with an N-terminal hemagglutinin (HA) tag in mammalian cells from simian virus 40 promoter and in vitro from a T7 promoter, was described previously, as were pSG5.HA-GRIP1 (full length), pSG5.HA-GRIP11122–1462, and pSG5.HA-SRC-1a (full length) (6). pSG5.HA-GRIP1.NRBIIm+IIIm, encoding full-length GRIP1 containing mutations in NR boxes II and III (leucines 693, 694, 748, and 749 all changed to alanines), was constructed by inserting the EcoRI insert from the corresponding pGAD424 vector (10) into pSG5.HA. Deletion of GRIP1 codons 1057 to 1109 (Δ1057–1109) and 1095 to 1106 (Δ1095–1106) was performed with the Promega Gene Editor kit. PCR-amplified DNA fragments encoding parts of mouse GRIP1 or human SRC-1 were inserted into pSG5.HA by using the indicated restriction endonuclease sites: GRIP15–1121 and GRIP11305–1462 were EcoRI-XhoI fragments inserted into the corresponding sites; SRC-1e1–1399 (full length), SRC-1a1–977, SRC-1a977–1441, SRC-1a1172–1441, SRC-1e977–1399, and SRC-1e1172–1399 were SmaI-SalI fragments inserted into EcoRI (blunted with Klenow polymerase) and XhoI sites.
Mammalian expression vectors for human NRs, pSVAR0 (4), pCMV.AR0 (5), and pCMX.hTRβ1 (15) were described previously. Plasmids encoding Gal4DBD-ARAF1 and Gal4DBD-ARAF2 were constructed by inserting, respectively, an EcoRI-XhoI PCR fragment encoding hAR1–555 and a BamHI-PstI fragment encoding hAR644–919 into pM (Clontech). Luciferase reporter plasmids for mammalian cells included MMTV-LUC, containing the native mouse mammary tumor virus promoter, and MMTV(TRE)-LUC, in which native glucocorticoid response elements (GRE) were replaced by a single palindromic thyroid receptor response element (TRE) (54); and GK1, controlled by a minimal adenovirus E1b promoter and five tandem Gal4 response elements (57). Chloramphenicol acetyltransferase (CAT) reporter gene rPB-CAT was constructed by inserting a PCR-amplified SalI-XbaI genomic DNA fragment representing nucleotides −376 to +41 of the androgen responsive rat probasin promoter (44) into pCAT-basic (Promega).
Yeast expression vectors encoding Gal4 DBD fused with AR HBD (23), TRβ1 HBD (10), or the p300 C terminus (25) or encoding Gal4 AD fused with full-length GRIP1 (wild type or with mutations in NR boxes II and III) or full-length SRC-1a (10) were described previously. The yeast expression vectors for the fusion proteins Gal4AD–SRC-1e and Gal4AD–SRC-1a1–1433 were constructed by inserting SmaI/SalI PCR fragments into pGAD424 (Clontech). The expression vectors coding for Gal4AD-GRIP1Δ1057–1109 and Gal4AD-GRIP1Δ1095–1106 fusion proteins were constructed by moving an EcoRI fragment encoding full-length GRIP1 containing each deletion from the pSG5.HA vector described above into pGAD424.
Bacterial expression vectors encoding glutathione S-transferase (GST) fusion proteins were constructed by inserting PCR-amplified cDNA fragments into vectors as follows: a BamHI/EcoRI fragment encoding GRIP1563–1121 and a BamHI-XhoI fragment encoding AR AF-1 (hAR amino acids 1–555) were inserted into pGEX-2TK (Pharmacia); an EcoRI-SalI fragment encoding GRIP11122–1462 and an EcoRI-XhoI fragment encoding CBP2041–2240 were inserted into pGEX-4T1 (Pharmacia).
Cell culture and transfections.
CV-1 cells (16) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Approximately 20 h before transfection, 105 cells were seeded into each well (3.3 cm in diameter) of six-well dishes. Cells in each well were transfected with SuperFect Transfection Reagent (Qiagen) according to manufacturer’s protocol with a total of 2.1 μg DNA, including 0.1 μg of β-galactosidase (β-Gal) expression vector pCMV–β-Gal (37) as an internal control to monitor transfection efficiency. After transfection, cells were grown in medium supplemented with 5% charcoal-stripped fetal bovine serum for 40 h before harvest; where indicated, medium was supplemented with 20 nM dihydrotestosterone (DHT) for AR or 20 nM 3,5,5′-triiodo-l-thyronine (T3) for TR during the last 30 h of growth. Luciferase and β-Gal assays were performed by using the Promega Luciferase Assay Kit and Promega β-Galactosidase Assay Kit according to manufacturer’s protocols. CAT assays were described previously (60). Luciferase and CAT activities are shown as the mean and standard deviation of three transfected wells and are representative of at least three independent experiments.
Immunoblots.
COS-7 cells (16) were grown and transfected as described above except that 60-mm dishes were seeded with 2 × 105 cells and transfected with 6 μg of pSG5.HA plasmid encoding coactivators or their fragments. At 40 h after transfections, cells were harvested in 150 μl of radioimmunoprecipitation assay buffer (45), and 25 μl of each cell extract was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoblotting was performed as described previously (10) with rat monoclonal antibody 3F10 against the HA epitope (Boehringer Mannheim) at 100 ng/ml as the primary antibody and horseradish peroxidase-conjugated anti-rat immunoglobulin G (sc-2006; Santa Cruz Biotechnology) at 160 ng/ml (1:2,500 dilution) as the secondary antibody.
Protein-protein interaction assays.
Yeast two-hybrid assays were performed as previously described (10). Where indicated, yeast cultures were incubated for approximately 15 h before harvest with 100 nM DHT for AR and 10 μM T3 for TR. Data presented are the mean and standard deviation from three yeast transformants and are representative of two or more independent experiments.
GST pulldown assays were performed as described previously (23) except that 35S-labeled proteins were produced with the TNT T7-coupled reticulocyte lysate system (Promega), and GST fusion proteins were produced in Escherichia coli BL21. To prepare inactive and hormonally activated AR, separate translations of AR were performed in the absence or presence of 1 μM DHT.
RESULTS
Correlation of physical and functional interactions between AR AF-2 and p160 coactivators.
Interactions between NR HBDs (i.e., the AF-2 activation functions) and the NR boxes of p160 coactivators allow the coactivators to mediate reporter gene activation by NRs (10, 21, 50). While these interactions are essentially universal for NRs that act as transcriptional activators, the efficiency of the interaction is influenced by a variety of coactivator sequences adjacent to and away from the NR boxes; the specific additional coactivator sequences required vary for each NR HBD (9, 10, 25, 40). For example, the HBDs (i.e., the AF-2 regions) of most NRs tested (e.g., TR) can interact efficiently with a small coactivator protein fragment representing the central NID of any of the three p160 coactivators. However, efficient binding of the AR HBD and the glucocorticoid receptor HBD to GRIP1 requires the NID plus an auxiliary region, NIDaux, located near the CBP binding domain (Fig. 1A) (9, 25). SRC-1 lacks the NIDaux function, and so the central NID of SRC-1 binds poorly to AR HBD. However, one splicing variant of SRC-1, SRC-1a, has a unique fourth NR box motif at its extreme C terminus, and this motif binds AR HBD (10). Another SRC-1 splicing variant, SRC-1e, lacks the fourth NR box motif (Fig. 1A), and therefore we predicted that it would not bind the AR HBD (10).
We therefore tested whether GRIP1, SRC-1a, and SRC-1e differ in their abilities to bind AR HBD and thus to serve as coactivators for AR (10). Protein-protein interactions were examined in yeast two-hybrid assays; AR HBD fused to Gal4 DBD and p160 coactivators fused with Gal4 AD were expressed in a yeast strain containing a chromosomally integrated β-Gal reporter gene controlled by Gal4 binding sites, and β-Gal activity was used as an indication of the protein-protein interaction. As we showed previously, GRIP1 interacted with AR HBD somewhat more efficiently than did SRC-1a, and both interactions were androgen dependent (Fig. 1B) (10). In contrast, SRC-1e or a truncated SRC-1a with a deleted C-terminal NR box IV failed to interact with AR HBD (Fig. 1B). All four of these coactivator proteins interacted well with a C-terminal fragment of p300, indicating that the coactivator fusion proteins were expressed and capable of interacting in the yeast two-hybrid assays.
Given their different AR AF-2 binding efficiencies, we tested the abilities of GRIP1, SRC-1a, and SRC-1e to enhance transcriptional activation by AR AF-2. The AR HBD, containing the AF-2 domain, was fused with Gal4 DBD and expressed transiently in CV-1 cells with various p160 coactivators and a reporter gene controlled by Gal4 response elements. The hormone-dependent activity of the AR AF-2 fusion protein was dramatically enhanced by GRIP1 and only modestly enhanced by SRC-1a; SRC-1e had no effect on AR AF-2 activity (Fig. 1C). Thus, the ability of the coactivators to enhance the activity of AR AF-2 correlated with binding of the coactivators to AR AF-2 (compare Fig. 1B and C).
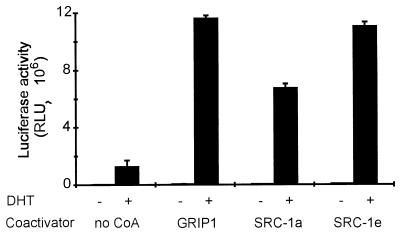
GRIP1, SRC-1a, and SRC-1e have approximately equal coactivator function for full-length AR.
In spite of their differential binding to and different coactivator function levels for AR AF-2 (Fig. 1B and C), GRIP1, SRC-1a, and SRC-1e were similar in their abilities to serve as coactivators for full-length AR in transiently transfected mammalian CV-1 cells (Fig. 2). The ability of AR to activate expression of a luciferase reporter gene (MMTV-LUC) with an androgen responsive mouse mammary tumor virus promoter was enhanced six- to ninefold by each of these coactivators, compared with the activity of AR alone. Since AR HBD bound SRC-1a weakly and did not bind SRC-1e, these results suggest the existence of an interaction between p160 coactivators and full-length AR in addition to the already known AF-2/NR box interaction.
FIG. 2.
p160 coactivator function with full-length AR does not correlate with binding to AR AF-2. CV-1 cells were transiently transfected with 0.25 μg of pSVAR0, 0.5 μg of MMTV-LUC reporter gene, and 1.25 μg of pSG5.HA (no CoA) or the indicated pSG5.HA-coactivator expression vector. Luciferase activity was measured after cells were grown with or without 20 nM DHT.
Physical and functional interactions between AR AF-1 and p160 coactivators.
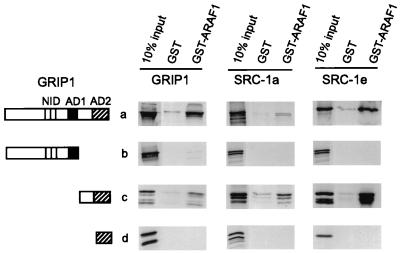
The fact that SRC-1e is an efficient coactivator for full-length AR but completely lacks the ability to bind to or enhance the activity of AR AF-2, suggests that there may be another coactivator-AR interface that is different from the classical NR box–AF-2 interaction defined in previous studies. We therefore used GST-dependent protein-protein interaction assays, i.e., GST pulldown assays, to test for a physical interaction between AR AF-1 and p160 coactivators or coactivator fragments. [35S]GRIP1, SRC-1a, or SRC-1e, or else N-terminal or C-terminal fragments of these coactivators, were synthesized in vitro and tested for binding to a fusion protein of GST and AR AF-1. Full-length GRIP1, SRC-1a, and SRC-1e, as well as C-terminal regions of these coactivators downstream from AD1, bound specifically to GST-ARAF1, compared with GST alone (Fig. 3). In contrast, the N-terminal coactivator fragments containing the central NID and AD1, as well as the shorter C-terminal fragments containing AD2 but not the region between AD1 and AD2, failed to bind to AR AF-1 in vitro (Fig. 3). Thus, AR AF-1 bound to a C-terminal region of p160 coactivators located downstream from AD1, and the region between AD1 and AD2 was essential for this binding. We designated this new NID as NIDAF-1.
FIG. 3.
Interaction of AR AF-1 with C-terminal regions of p160 coactivators in vitro. Glutathione-Sepharose-bound GST or GST-ARAF1 was incubated with 35S-labeled full-length GRIP1, SRC-1a, SRC-1e, or their fragments translated in vitro from pSG5.HA vectors. Bound proteins were eluted and analyzed by SDS-PAGE and autoradiography; shown for comparison is 10% of the total labeled protein incubated in each binding reaction (10% input). Functional domains in diagrams of the coactivator fragments are represented as in Fig. 1A. Coactivator fragments translated in vitro include the following amino acids: GRIP1 5 to 1462 (full length), 5 to 1121, 1122 to 1462, and 1305 to 1462; SRC-1a 1 to 1441 (full length), 1 to 977, 977 to 1441, and 1172 to 1441; SRC-1e 1 to 1399 (full length), 1 to 977, 977 to 1399, and 1172 to 1399.
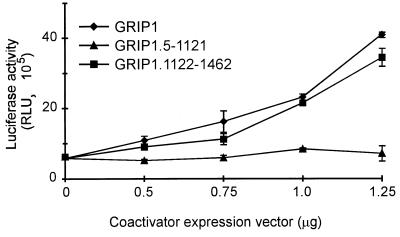
To determine whether this physical interaction could allow the p160 proteins to serve as coactivators for AR AF-1, we tested the ability of the p160 proteins to enhance reporter gene activation by AR AF-1 fused to Gal4 DBD in transiently transfected CV-1 cells. As expected, AR AF-1 was a hormone-independent AD. Full-length GRIP1 enhanced AR AF-1 activity up to sevenfold, and the degree of enhancement increased with the amount of coactivator expression vector transfected (Fig. 4). Thus, GRIP1 can bind to and serve as a potent coactivator for AR AF-1.
FIG. 4.
NR box-independent and AD1-independent enhancement of AR AF-1 activity by GRIP1 and its C-terminal fragment. CV-1 cells were cotransfected with 0.25 μg of pM vector encoding Gal4DBD-ARAF1, 0.5 μg of GK1 reporter gene, and the indicated amount of pSG5.HA vector encoding GRIP1 or GRIP1 fragments. Luciferase activity of the cell extracts is shown.
Both AD1 and AD2 can function as downstream signaling domains for p160 coactivators.
To explore the domains of p160 coactivators required to enhance AR AF-1 and AF-2 functions, the same fragments of GRIP1 and SRC-1 used for in vitro binding studies (Fig. 3) were tested for their ability to serve as coactivators for Gal4DBD-ARAF1 and Gal4DBD-ARAF2 fusion proteins in CV-1 cells. The N-terminal coactivator fragments contained the central NID, with its three NR boxes, and the AD1 signaling domain (Fig. 1A). The C-terminal p160 protein fragments contained NIDAF-1 and AD2; the SRC-1a C-terminal fragment also contained NR box IV, but GRIP1 and SRC-1e lack this extra NR box. The N-terminal fragment of GRIP1 failed to enhance AR AF-1 activity at all concentrations tested (Fig. 4). In contrast, the C-terminal GRIP1 fragment was almost as good as full-length GRIP1 as a coactivator for AR AF-1. The coactivator activity of the GRIP1 C-terminal fragment confirmed the functional relevance of the physical interaction between AR AF-1 and the GRIP1 NIDAF-1 region. Furthermore, this result indicated that GRIP1 AD2 can function as a downstream signaling domain. To our knowledge this is the first demonstration that AD2 can function as a downstream signaling domain in a true coactivator assay.
SRC-1a and SRC-1e also enhanced AR AF-1 activity with efficacies similar to that of GRIP1 (Fig. 5A). C-terminal fragments of SRC-1a (amino acids 977 to 1441) and SRC-1e (amino acids 977 to 1399) corresponding approximately (by sequence alignment) to GRIP11122–1462 also functioned as efficient coactivators for AR AF-1. Shorter C-terminal fragments, GRIP11305–1462, SRC-1a1172–1441, and SRC-1e1172–1399, failed to enhance AR AF-1 activity (Fig. 5A), a result consistent with their failure to bind AR AF-1 (Fig. 3). The full-length p160 coactivators and their various fragments were all expressed at similar levels in transiently transfected COS-7 cells, except that the shortest C-terminal fragments of GRIP1 and SRC-1a were expressed at higher levels (Fig. 5B). Thus, the failure of some fragments to serve as coactivators for AR AF-1 was not due to lack of expression or stability.
FIG. 5.
Activity of two signal input and two signal output domains in p160 coactivator fragments. (A) Enhancement of AR AF-1 function by C-terminal p160 coactivator fragments containing NIDAF-1 and AD2. CV-1 cells were transiently transfected with 0.25 μg of pM vector encoding Gal4DBD-ARAF1, 0.5 μg of GK1 reporter gene, and 1.25 μg of pSG5.HA vector (no CoA) or the same vector encoding the indicated coactivator or coactivator fragment; luciferase activity was measured. Functional domains in diagrams of coactivator fragments are represented as in Fig. 1A. The activity of the Gal4 DBD alone was 0.1 × 105 RLU (data not shown). (B) Expression levels of p160 coactivators and their fragments in transfected cells. COS-7 cells were transfected with pSG5.HA vectors encoding the indicated coactivator or fragment. Cell extracts were subjected to immunoblot analysis with antibodies against the HA epitope. (C) AD1-dependent or AD2-dependent enhancement of AR AF-2 function by fragments of p160 coactivators. CV-1 cells were transfected with 0.5 μg of pM vector encoding Gal4DBD-ARAF2, 0.5 μg of GK1 reporter gene, and 1.0 μg of pSG5 vector encoding no coactivator (no CoA) or the indicated coactivator fragment or mutant, with or without DHT treatment as indicated. Functional domains in diagrams of coactivator fragments are represented as in Fig. 1A. GRIP1.NRBIIm+IIIm is full-length GRIP1 with LL-to-AA mutations in NR boxes II and III.
The same p160 protein fragments were tested as coactivators for Gal4DBD-ARAF2. The N-terminal fragment of GRIP1 was a strong coactivator for AR AF-2 (Fig. 5C), presumably by using the combined NID and NIDaux (25) to interact with AR AF-2 and by using AD1 (7, 55) to transmit the signal downstream to the transcription machinery. The C-terminal fragment of GRIP1 was inactive as a coactivator for AR AF-2, which was consistent with our previous finding that this region of GRIP1 cannot interact with AF-2 domains of any NRs (10). Note that the same two GRIP1 fragments yielded the opposite results as coactivators for AR AF-1 (Fig. 5A). Full-length GRIP1 with mutations in NR boxes II and III (GRIP1.NRBIIm+IIIm) cannot bind to NR HBDs (10) and also did not enhance AR AF-2 function.
In contrast to the results with GRIP1, the N-terminal domain of SRC-1 was unable to serve as a coactivator for AR AF-2 (Fig. 5C); this reflected our previous findings that this region of SRC-1 does not interact with AR AF-2 (10) due to the lack of an NIDaux function in SRC-1 (25). However, the C-terminal region of SRC-1a exhibited a moderate coactivator activity for AR AF-2 (Fig. 5C). In this coactivator fragment NR box IV provided a binding site for AR AF-2 (10); since AD1 was absent from this fragment, downstream signaling must have been due to AD2. The C-terminal fragment of SRC-1e did not enhance AR AF-2 activity (Fig. 5C), which was consistent with its inability to bind AR AF-2 (Fig. 1B). Thus, AD2 can serve as a downstream signaling domain when a small C-terminal fragment of SRC-1a binds AR AF-2 (Fig. 5C) or when a larger C-terminal fragment of GRIP1, SRC-1a, or SRC-1e binds to AR AF-1 (Fig. 5A).
Independent function of p160 activation domains AD1 and AD2 with full-length AR.
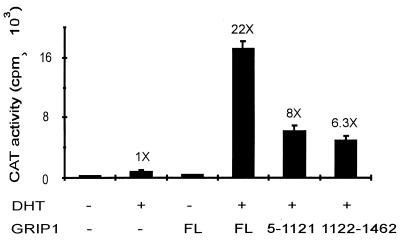
In the foregoing experiments reporter gene activation by individual activation functions of AR, fused to Gal4 DBD, was enhanced by fragments of the p160 coactivators. By using a more physiologically relevant test, we found that each of these coactivator fragments also substantially enhanced the activity of full-length AR, in this case with the rat probasin promoter; probasin is a prostate-specific gene (17, 44). The GRIP1 N-terminal and C-terminal fragments each substantially enhanced the activity of full-length AR but had about one-third the coactivator activity of full-length GRIP1 (Fig. 6). Thus, both p160 protein fragments were capable of functioning independently as coactivators for full-length AR, and their actions are at least additive in the context of full-length GRIP1.
FIG. 6.
GRIP1 N-terminal and C-terminal fragments independently enhance activity of full-length AR. CV-1 cells were transiently transfected with 0.25 μg of pSVAR0 encoding full-length AR, 0.5 μg of rPB-CAT reporter gene (CAT gene driven by rat probasin promoter), and 1.25 μg of pSG5.HA vector encoding no coactivator (−), full-length (FL) GRIP1, or the indicated GRIP1 fragment. CAT activity was determined from extracts of cells grown with or without DHT as indicated. Numbers above data bars indicate the activity relative to that with AR in the presence of DHT and absence of GRIP1.
Mapping of the p300/CBP binding site.
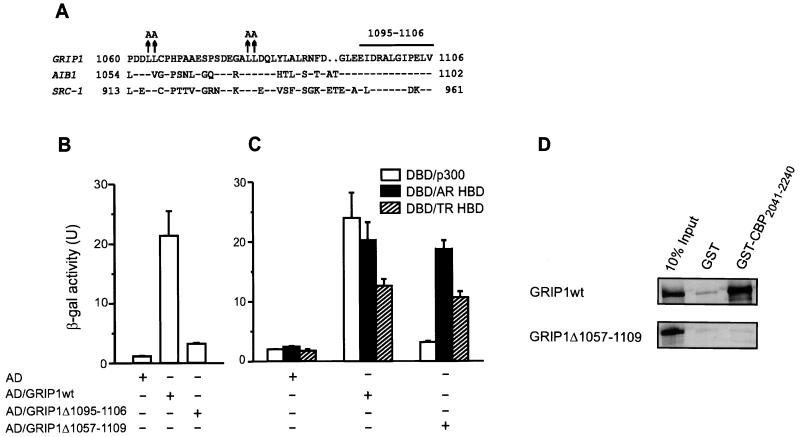
In order to test the contributions of AD1 and AD2 in the context of full length GRIP1, we wanted to identify mutations that would eliminate AD1 activity without affecting functions of other domains in GRIP1, especially NIDaux which is essential for efficient binding of AR HBD (25). GRIP1 amino acids 1011 to 1121 are sufficient for binding p300 (25). Several sequence motifs in this region are highly conserved among p160 coactivators (Fig. 7A). Substitution of alanines for conserved leucines 1079 and 1080 in GRIP1 reduced p300 binding by about 70%, whereas alanine substitutions for leucines 1063 and 1064 had no effect (25, 55). To map further the sequences important for p300 binding, we deleted another highly conserved motif in this region, amino acids 1095 to 1106 (Fig. 7A). In yeast two-hybrid assays this caused a loss of approximately 90% of the p300 binding activity (Fig. 7B), demonstrating that this conserved motif is an important part of the p300/CBP binding site. A deletion of all the previously mentioned conserved sequence motifs (amino acids 1057 to 1109) resulted in an essentially complete loss of p300 binding, but had no effect on binding of AR or TR HBDs (Fig. 7C). Since efficient binding of AR HBD requires the NIDaux function located near the p300/CBP binding site, in addition to the central NID (25), this result indicates that NIDaux is located within amino acids 1011 to 1121 but outside of amino acids 1057 to 1109. In vitro, the GRIP1Δ1057–1109 mutation also caused loss of GRIP1 binding to a C-terminal CBP fragment fused to GST (Fig. 7D), in agreement with the results obtained with p300 in the yeast two-hybrid system (Fig. 7C).
FIG. 7.
Mapping the p300/CBP binding site (AD1) of GRIP1. (A) Sequence of the core p300/CBP binding region of p160 coactivators. Amino acids identical to those in GRIP1 are indicated (−), as are gaps introduced for optimal sequence alignment (. .). Previously characterized mutations (↑) (25, 55) and a new deletion (horizontal bar) described here are shown above the GRIP1 sequence. (B and C) Binding of wild-type and mutant GRIP1 to p300 and NR HBDs. Yeast two-hybrid assays were conducted as in Fig. 1B. AD, Gal4 AD; DBD, Gal4 DBD. (D) Binding of wild-type or mutant GRIP1 to a C-terminal fragment of CBP in vitro. Wild-type or mutant GRIP1 was translated in vitro from a pSG5.HA vector, and binding to GST-CBP2041–2240 was measured as in Fig. 3.
Differential use of GRIP1’s signal input and signal output domains by AR and TR.
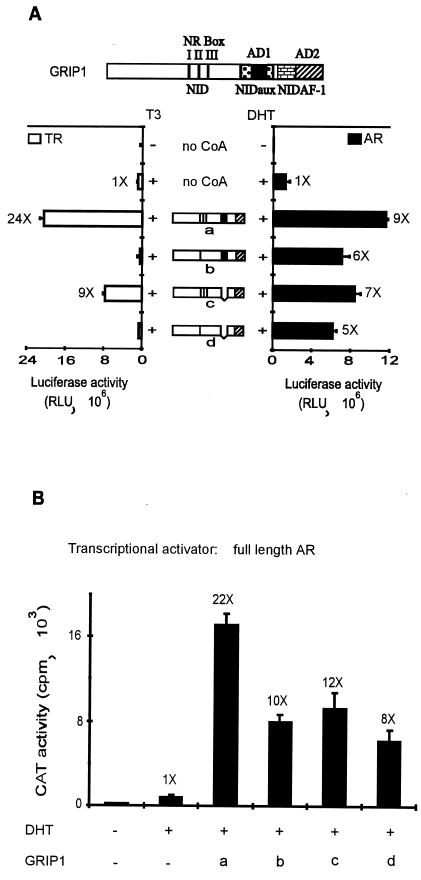
The experiment in Fig. 6 demonstrated that in the context of GRIP1 fragments the central NID (which binds AR AF-2) and NIDAF-1 (which binds AR AF-1) can each serve as a functional NR-coactivator interface and AD1 and AD2 can each serve as a downstream signaling domain. To examine the roles of these two signal input and signal output domains in the context of full-length GRIP1, we expressed full-length GRIP1 with two types of mutations. First, NR boxes II and III were inactivated with leucine-to-alanine substitutions that changed the LXXLL motifs to LXXAA; this mutant GRIP1 cannot bind to NR HBDs but binds p300 as strongly as wild-type GRIP1 (10). Second, AD1 was deleted; this mutation eliminated p300 binding but did not affect binding to NR HBDs (Fig. 7C). These GRIP1 mutants were compared for their ability to serve as coactivators for AR and TRβ1. Both of these NRs have conserved AF-2 functions that are enhanced by p160 coactivators (7, 27). Both AF-1 functions are important for robust enhancement of target gene transcription (20, 29) but differ dramatically in length and sequence.
Wild-type GRIP1 was a potent coactivator for full-length AR and full-length TR in transiently transfected CV-1 cells (Fig. 8A, construction a). The reporter genes were controlled by a mouse mammary tumor virus promoter containing either the native GREs (for AR) or a TRE substituted for the native GREs (for TR). The NR box mutations completely eliminated GRIP1’s coactivator function for TR but reduced coactivator function for AR by only ca. 40% (mutant b). This result reflects the ability of AR to interact with GRIP1 through either the AR AF-1 or AF-2 domain, and it demonstrates that the AF-1 interaction is sufficient to recruit full-length GRIP1 as a functional coactivator. In contrast, TR can apparently only interact with GRIP1 through the AF-2 interaction with GRIP1’s NR boxes.
FIG. 8.
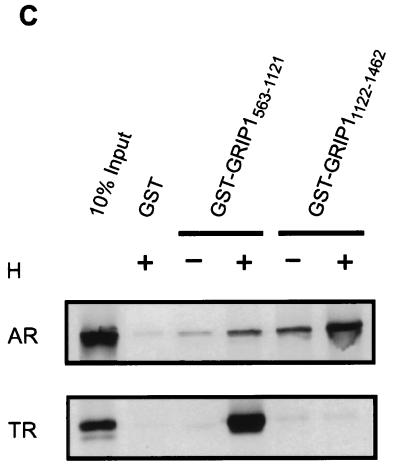
Differential use of NR-binding regions and ADs of GRIP1 by AR and TR. (A) Coactivator function of GRIP1 mutants with AR and TR. CV-1 cells were transiently transfected with 0.25 μg of pSVAR0 or pCMX.hTRβ1, 0.5 μg of reporter gene MMTV-LUC (for AR) or MTV(TRE)-LUC (for TR), and 1.25 μg of pSG5.HA vector encoding no coactivator (no CoA) or full-length GRIP1 with a wild-type sequence (a) or with mutations in the NR boxes II and III (b), deletion of AD1 (c), or both mutations (d). Luciferase activity was determined from extracts of cells grown without or with 20 nM DHT (for AR) or 20 nM T3 (for TR) as indicated. Functional domains in diagrams are as in Fig. 1A; in addition, the brick pattern represents NIDAF-1. Numbers beside data bars indicate the activity relative to that with AR or TR in the presence of hormone and absence of GRIP1. (B) Coactivator function of GRIP1 mutants with AR tested on a rat probasin promoter. CV-1 cells were transiently transfected with 0.25 μg of pSVAR0, 0.5 μg of rPB-CAT reporter gene, and pSG5.HA vectors encoding wild-type or mutant GRIP1; letters a to d represent GRIP1 protein species as in panel A. (C) Binding of GRIP1 N- or C-terminal fragments to AR and TR in vitro. 35S-labeled full-length AR or TR was synthesized in vitro and then incubated with glutathione-Sepharose-bound GST or GST-GRIP1 fusion proteins in the absence or presence of the appropriate hormone (H, 1 μM DHT for AR or 1 μM T3 for TR) as indicated. Bound proteins were eluted and analyzed by SDS-PAGE and autoradiography.
When AD1 was deleted, GRIP1 lost only about 30% of its coactivator function for AR and about 70% of its coactivator function for TR (mutant c). This result indicates that the GRIP1 AD2 is an effective downstream signaling domain in the context of full-length GRIP1 and suggests that the relative contributions of AD1 and AD2 to downstream signaling may vary somewhat, depending on which NR interacts with GRIP1. When the NR box and AD1 mutations were combined, the mutant GRIP1 was inactive with TR but still retained about 50% of wild-type GRIP1 function for AR (mutant d). In this last experiment, GRIP1 presumably interacted with AR through AR AF-1 and signaled to the transcription machinery through AD2, thus demonstrating the ability of each of these domains to contribute to coactivator function in the context of full-length GRIP1. As in the above experiments with the mouse mammary tumor virus promoter (Fig. 8A), the same GRIP1 mutants retained the ability to substantially enhance AR activity with the rat probasin promoter (Fig. 8B), demonstrating that NIDAF-1 and the AD2 signaling domain of GRIP1 can function with different types of promoters to which AR is bound.
The fact that GRIP1’s ability to serve as a coactivator for TR was completely eliminated by mutations in NR boxes II and III suggests that TR may interact with GRIP1 only through the TR AF-2 domain, while AR interacts through both AF-1 and AF-2 domains. In vitro GST-dependent protein-protein interaction studies were used to compare interactions of TR and AR with the central NID of GRIP1 (amino acids 563 to 1121), containing the three NR boxes and NIDaux, and the C-terminal GRIP1 fragment (amino acids 1122 to 1462) containing NIDAF-1. Full-length AR and TR were synthesized in vitro and tested for binding to the GRIP1 fragments in the presence or absence of the appropriate hormone, DHT or T3, respectively. Consistent with previous findings, the 35S-labeled AR and TR bound in a hormone-stimulated manner to GST-GRIP1563–1121, which has been shown previously to bind the HBDs of a wide range of hormone-activated NRs (10). In contrast, GST-GRIP11122–1462 bound AR but not TR (Fig. 8C). The lack of an interaction between TR and the GRIP1 C-terminal region is consistent with our finding that TR was completely dependent on its AF-2 domain to interact functionally with GRIP1 (Fig. 8A) and indicates that TR AF-1 does not interact with the GRIP1 C-terminal region. The interaction between AR and the GRIP1 C-terminal fragment was stimulated by DHT, suggesting that the hormonally induced conformational change of AR is necessary for efficient interaction of GRIP1 with both the AF-2 and AF-1 domains of AR.
DISCUSSION
Multiple physical and functional interactions between p160 coactivators and AR.
It appears that all NRs that function as transcriptional activators have conserved AF-2 domains that can bind to NR boxes (LXXLL motifs) of p160 coactivator proteins (9, 10, 21, 50, 55). However, within the apparent universality of this interaction between the NR and p160 coactivator protein families, there are many elements of NR binding specificity. Each NR box can efficiently bind a broad but not universal set of NR AF-2 domains (10, 50), and this specificity is determined by the amino acid sequences within and surrounding the LXXLL motif (9, 40). Coactivator sequences separate from the NR boxes (e.g., NIDaux in GRIP1) contribute to efficient binding of some NR AF-2 domains (e.g., AR and glucocorticoid receptor HBDs) (25). In addition, efficient binding of another subset of NRs (e.g., TR, retinoic acid receptors, and peroxisome proliferator-activated receptors) apparently requires simultaneous interaction of two adjacent NR boxes with the two AF-2 domains of the NR dimer (40, 41).
Among NRs AR AF-2 has a particularly complex interaction with p160 coactivators, and the efficiency and mechanism of this interaction vary dramatically among different coactivator family members and splicing isoforms. AR AF-2 binds efficiently to GRIP1 through cooperative effects of NR box III and NIDaux (10, 25), with moderate efficiency to SRC-1a through NR box IV (10), and poorly or not at all with SRC-1e (Fig. 1B). These differences in AR AF-2 binding efficiencies correspond to the relative coactivator activities of these p160 proteins for the AR AF-2 domain in the absence of AR AF-1 (Fig. 1C). In contrast, all three p160 proteins function as efficient coactivators for full-length AR (Fig. 2) and for AR AF-1 (Fig. 5A), due to the binding interaction reported here between AR AF-1 and the NIDAF-1 site in the C-terminal region of the p160 coactivators (Fig. 3).
Thus far the minimum p160 protein fragment that can bind AR AF-1 is GRIP11122–1462 or SRC-1a977–1441, which contains AD2 as well. The results of coactivator assays (Fig. 5) and protein-protein interaction studies (Fig. 3) for the SRC-1a C-terminal fragments provide additional information on the relative locations of NIDAF-1 and AD2 within this C-terminal fragment. SRC-1a1172–1441 worked as a coactivator for AR AF-2 (Fig. 5C) and therefore must contain an intact AD2 for downstream signaling, as well as an AF-2 interaction site (the C-terminal NR box IV). This same SRC-1a fragment was not a coactivator for AR AF-1, while the longer SRC-1a977–1441 was (Fig. 5A). Since the ability to serve as a coactivator for AR AF-1 correlated with the AR AF-1 binding activity of the two fragments (Fig. 3), we conclude that the binding site for AR AF-1 involves the region between AD1 and AD2, e.g., SRC-1a amino acids 977 to 1172, but may also include more C-terminal residues.
The conserved nature of this AR AF-1 binding function among the three p160 family members suggests that it resides within the several conserved sequence motifs found in this region of the p160 protein family (1, 24). Whereas essentially all NR AF-2 domains are structurally conserved (58) and interact with NR boxes of p160 coactivators (10), the AF-1 domains are not conserved in sequence, and only a subset of NR AF-1 domains interact with NIDAF-1. AR AF-1 interacted physically and functionally with NIDAF-1, but TR AF-1 did not (Fig. 8). To date, the AF-1 domains of two other steroid hormone receptors have been shown to interact with p160 coactivators. The AF-1 region of ERα (but not ERβ) also bound to GRIP11122–1462, and as reported here for AR this interaction was sufficient to allow the coactivator to enhance transcriptional activation by the steroid receptor (57). PR AF-1 was shown to bind a large SRC-1 fragment composed of amino acids 361 to 1139 (43). Since this SRC-1 fragment partially overlaps with the C-terminal fragments of SRC-1 and GRIP1 which we have shown to bind AR AF-1 (Fig. 3), it is possible that the AF-1 regions of AR, ERα, and PR all bind to the same site in the C-terminal regions of the p160 coactivators.
While recent studies with coactivators and corepressors have shed considerable new light on the mechanism of action of NR AF-2 domains (51), very little is known about the mechanisms of NR AF-1 function. Our finding of a direct interaction between AR AF-1 and p160 coactivators provides a new insight into the mechanism of AR AF-1 action and furthermore suggests that the AF-1 domains of at least some steroid hormone receptors surprisingly function through a common mechanism. Thus, in spite of their lack of primary sequence homology, it is possible that parts of the AF-1 domains of PR, ER, and AR have a similar three-dimensional structure that binds a common site on the C-terminal domain of p160 coactivators. Previous studies with AR have demonstrated that AR AF-1 and AR AF-2 can interact directly, and this interaction also contributes to synergistic transcriptional activation by these two domains (11, 33). A recent study found that SRC-1 enhanced binding between AR AF-1 and AF-2, thus providing indirect evidence that p160 coactivators may in some way mediate cooperative interactions between AF-1 and AF-2 (27). Combined with the above evidence, our finding that AR AF-1 binds directly to p160 coactivators suggests that three pairs of binding interactions between AR AF-1, AR AF-2, and p160 coactivators may be involved in the AR transcriptional activation mechanism, i.e., AF-1 plus AF-2, AF-2 plus p160 NID, and AF-1 plus p160 NIDAF-1.
Role for the AD2 domain of p160 coactivators in signaling to the transcription machinery.
After interacting with the DNA bound NRs, p160 coactivators must use one or more ADs to transmit the activating signal from the NRs to the transcription machinery. Two potential ADs were identified by testing fragments of the p160 proteins for their ability to activate transcription when fused to the DBD of a heterologous protein (7, 55). Such assays can only identify potential activation domains, since they test protein fragments outside of their natural context. Specific coactivator assays are required to ascertain the true roles of these domains in the coactivator protein. Studies of the effects of deletion mutations on the ability of p160 proteins to serve as coactivators for NRs established a clear and important role for AD1 in downstream signaling and suggested in fact that AD1 might be the primary or only major signaling domain for p160 coactivators (7, 50, 55). However, here we used a newly defined AD1 deletion mutation and C-terminal p160 fragments lacking AD1 to show that AD2 can also participate in the downstream signaling by p160 coactivators and that the contribution of AD2 is comparable in magnitude to that of AD1. With full-length AR, the effects of AD1 and AD2 were approximately additive or modestly synergistic (Fig. 6). AD2 contributed to coactivator signaling in the context of either the full-length p160 coactivator (Fig. 6 and 8) or a C-terminal p160 protein fragment that lacks AD1 (Fig. 4 to 6). AD2 signaling was triggered by binding of the coactivator to an NR AF-1 domain or AF-2 domain. The former was seen when AR was tested with a C-terminal coactivator fragment (Fig. 6) or with full-length GRIP1 containing NR box mutations and an AD1 deletion (Fig. 8A and B). The latter is best illustrated by experiments where the activity of full-length TR was enhanced by a GRIP1 mutant with a deleted AD1 region (Fig. 8A) and where a C-terminal SRC-1a fragment that lacked AD1 enhanced the function of AR AF-2 (Fig. 5C).
AD1 functions by binding to CBP or p300 (7, 50), which have acetyltransferase activities and may also directly contact elements of the basal transcription machinery (31, 42, 49, 59). The mechanism of AD2 signaling remains to be established. A histone acetyltransferase activity was reported in the C-terminal fragment of SRC-1a (46) and ACTR (7), but whether this activity contributes to p160 coactivator function has not been established. Furthermore, the relationship of this activity to AD2 remains to be determined; preliminary deletion mapping studies suggest that regions responsible for the transcriptional activation and acetyltransferase activities do not overlap (7, 46, 55). Recent studies in our lab may be relevant to the mechanism of AD2 function. We have identified a novel protein, coactivator associated arginine methyltransferase 1 (CARM1), which binds to the C-terminal region of the p160 coactivators and stimulates the activity of this p160 protein fragment fused to Gal4 DBD; when coexpressed with p160 coactivators, CARM1 also enhances the activity of NRs above that observed with the p160 coactivators alone. We propose that CARM1 serves as a secondary coactivator for NRs and represents a downstream target of the AD2 domain of p160 coactivators (6).
Differential use of the multiple signal input and output domains of p160 coactivators by different NRs.
The different results obtained for TR and AR with the various mutants of GRIP1 (Fig. 8A) suggest that, while the p160 proteins are universally used as coactivators by all NRs that are transcriptional activators, different NRs use different combinations of coactivator signal input and output domains. With regard to p160 signal input domains, AR, ER, and PR bind to p160 coactivators through two interfaces: NR AF-2 binds p160 NR boxes, with an assist from GRIP1 NIDaux when GRIP1 binds to AR AF-2 (10, 21, 25, 50, 55), and NR AF-1 binds NIDAF-1 (43, 57) (Fig. 3). In contrast, TR uses the AF-2 interface but not the AF-1 interface (Fig. 8A and C). With regard to signal output domains, the differences in utilization of AD1 and AD2 were less dramatic; but AD2 made a stronger relative contribution to AR function and a weaker contribution to TR function (Fig. 8A). We are currently investigating the specific usage patterns of the p160 coactivator input and output domains by other NRs. It is also possible that the relative contributions of AD1 and AD2 could be influenced by promoter or cellular context, e.g., different cell types may express different types or amounts of the downstream targets for AD1 and AD2, and the ADs and their target proteins could be regulated by posttranslational modifications or interactions with other proteins.
ACKNOWLEDGMENTS
We thank M.-J. Tsai and B. W. O’Malley (Baylor College of Medicine, Houston, Tex.) for cDNA encoding hSRC-1a, C. K. Glass (University of California at San Diego, La Jolla) for cDNA encoding hSRC-1e, and A. O. Brinkmann (Erasmus University, Rotterdam, The Netherlands) for the plasmids encoding hAR.
This work was supported by U.S. Public Health Service grants DK43093 (to M.R.S.), CA/OD72821 (to G.A.C.), and DK51083 (to P.J.K.) from the National Institutes of Health. H.M. was supported in part by a predoctoral traineeship from the University of California Breast Cancer Research Program, and R.A.I. was supported by a predoctoral traineeship from the National Institutes of Health.
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 3.Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann A O, Faber P W, van Rooij H C J, Kuiper G G J M, Ris C, Klaassen P, van der Korput J A G M, Voorhorst M M, van Laar J H, Mulder E, Trapman J. The human androgen receptor: domain structure, genomic organization and regulation of expression. J Steroid Biochem Mol Biol. 1989;34:307–310. doi: 10.1016/0022-4731(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain N L, Driver E D, Miesfeld R L. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. 1994;22:3181–3186. doi: 10.1093/nar/22.15.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D, Ma H, Hong H, Koh S S, Huang S-M, Schurter B T, Aswad D W, Stallcup M R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 8.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 11.Doesburg P, Kuil C W, Berrevoets C A, Steketee K, Faber P W, Mulder E, Brinkmann A O, Trapman J. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36:1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 12.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis-retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enmark E, Gustafsson J A. Orphan nuclear receptors—the first eight years. Mol Endocrinol. 1996;10:1293–1307. doi: 10.1210/mend.10.11.8923456. [DOI] [PubMed] [Google Scholar]
- 14.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng W, Ribeiro R C J, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 16.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg N M, DeMayo F J, Sheppard P C, Barrios R, Lebovitz R, Finegold M, Angelopoulou R, Dodd J G, Duckworth M L, Rosen J M, Matusik R J. The rat probasin gene promoter directs hormonally and developmentally regulated expression of a heterologous gene specifically to the prostate in transgenic mice. Mol Endocrinol. 1994;8:230–239. doi: 10.1210/mend.8.2.8170479. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 19.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 20.Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka B M, Samuels H H. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor alpha is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 22.Hollenberg S M, Evans R M. Multiple and cooperative transactivation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 23.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional co-activator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong H, Darimont B D, Ma H, Yang L, Yamamoto K R, Stallcup M R. An additional region of coactivator GRIP1 required for interaction with the hormone-binding domains of a subset of nuclear receptors. J Biol Chem. 1999;274:3496–3502. doi: 10.1074/jbc.274.6.3496. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 27.Ikonen T, Palvimo J J, Jänne O A. Interaction between the amino- and carboxy-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 28.Imhof A, Yang X-J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 29.Jenster G, van der Korput H A G M, van Vroonhoven C, van der Kwast T H, Trapman J, Brinkmann A O. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 30.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 32.Kumar V, Green S, Stack G, Berry M, Jin J-R, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 33.Langley E, Zhou Z X, Wilson E M. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 34.Lees J A, Fawell S E, Parker M G. Identification of two transactivation domains in the mouse oestrogen receptor. Nucleic Acids Res. 1989;17:5477–5489. doi: 10.1093/nar/17.14.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 36.Luger K, Richmond T J. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 37.Ma W-J, Holz R W, Uhler M D. Expression of a cDNA for a neuronal calcium channel α1 subunit enhances secretion from adrenal chromaffin cells. J Biol Chem. 1992;267:22728–22732. [PubMed] [Google Scholar]
- 38.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 39.McDonnell D P, Clemm D L, Hermann T, Goldman M E, Pike J W. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- 40.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 42.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 43.Oñate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M-J, Edwards D P, O’Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 44.Rennie P S, Bruchovsky N, Leco K J, Sheppard P C, McQueen S A, Cheng H, Snoek R, Hamel A, Bock M E, MacDonald B S, Nickel B E, Chang C, Liao S, Cattini P A, Matusik R J. Characterization of two cis-acting DNA elements involved in the androgen regulation of the probasin gene. Mol Endocrinol. 1993;7:23–36. doi: 10.1210/mend.7.1.8446105. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 18–38. [Google Scholar]
- 46.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 47.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 48.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 49.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 50.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 51.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 52.Tsai M-J, O’Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 53.Tzukerman M T, Esty A, Santiso-Mere D, Danielian P, Parker M G, Stein R B, Pike J W, McDonnell D P. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- 54.Umesono K, Evans R M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 55.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walfish P G, Yoganathan T, Yang Y-F, Hong H, Butt T R, Stallcup M R. Yeast hormone response element assays detect and characterize GRIP1 coactivator-dependent activation of transcription by thyroid and retinoid nuclear receptors. Proc Natl Acad Sci USA. 1997;94:3697–3702. doi: 10.1073/pnas.94.8.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen M P, Chen D, Huang S-M, Subramanian S, McKinerney E, Katzenellenbogen B S, Stallcup M R, Kushner P J. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 58.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 59.Yao T-P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Danielsen M. Selective effects of 8-Br-cAMP on agonists and antagonists of the glucocorticoid receptor. Endocrine. 1995;3:5–12. doi: 10.1007/BF02917442. [DOI] [PubMed] [Google Scholar]