Abstract
COVID-19 is an infectious and contagious virus. As of this writing, more than 160 million people have been infected since its emergence, including more than 125,000 in Algeria. In this work, We first collected a dataset of 4986 COVID and non-COVID images confirmed by RT-PCR tests at Tlemcen hospital in Algeria. Then we performed a transfer learning on deep learning models that got the best results on the ImageNet dataset, such as DenseNet121, DenseNet201, VGG16, VGG19, Inception Resnet-V2, and Xception, in order to conduct a comparative study. Therefore, We have proposed an explainable model based on the DenseNet201 architecture and the GradCam explanation algorithm to detect COVID-19 in chest CT images and explain the output decision. Experiments have shown promising results and proven that the introduced model can be beneficial for diagnosing and following up patients with COVID-19.
Keywords: Xception, Densenet-121, Densenet-201, COVID-19, Imagenet
MSC: Deep learning; Transfer learning; VGG16; VGG19,
1. Introduction
The Severe Acute Respiratory Syndrome COronaVirus 2 (SARS-CoV-2), also called COVID-19, is a contagious virus and one of the most severe problems facing our lives. This pandemic was identified in Wuhan, China, in December 2019 before spreading to the rest of the world [8], [28].
Several researchers and laboratories have focused on the search for a vaccine. However, despite the existence of few vaccines, the virus continues to spread with great speed, especially after the new strain has emerged.
The most effective screening technique is Reverse Transcription Polymerase Chain Reaction (RT-PCR). Nevertheless, RT-PCR has low sensitivity, and the world is currently experiencing a shortage of these testing kits [26]. In this circumstance, numerous sick persons are not confined, which accelerates the spread of COVID-19 and obliges the health structures to use other diagnostic techniques to control better the pandemic [17], [31]. They recommend the use of medical imaging (Computed Tomography (CT) and X-Ray) as a complement to RT-PCR [5]. Chest CT-scan is a quick and easy exam to be obtained and it has shown a much higher sensitivity than RT-PCR for examining COVID-19 [23].
Unfortunately, given the number of COVID-19 cases, which to date has exceeded 160 million worldwide, the number of radiologists remains very low. Therefore, the use of Deep Learning (DL) techniques is helpful for the automatic detection of COVID-19. These techniques allow the process of large image datasets with a high degree of precision to reassure health experts about the reliability of these models.
In our previous study [13], we have addressed the problem of the classification of pneumonia using Chest X-ray images, and we have concluded that in some databases, it is not mandatory to use complex architecture or transfer learning to classify images. After a discussion with several doctors at the hospital of Tlemcen in Algeria, they asked us to provide them with a decision support system for the COVID-19. They also confirmed that they rely on CT-scan images to diagnose COVID-19 since this contains more information than X-ray images. For this reason, and to offer a model that will be useful for physicians, we have proposed an Explainable DL architecture for the classification of chest CT-scan images.
To train these DL models, we need large labeled datasets. There are public databases on the internet, but they do not include information on the acquisition and confirmation of infection by RT-PCR.
Due to the limitation of existing datasets, we proposed, in this work, collecting a new COVID-19 chest CT dataset from infected patients admitted to the hospital of Tlemcen in Algeria. A collection of new COVID-19 CT-scans datasets from confirmed and unconfirmed patients with RT-PCR test. Accordingly, the highlights of the manuscript are as follows:
-
•
Collecting of a new COVID-19 CT-scan dataset from patients confirmed positive or negative with RT-PCR test;
-
•
The development of a customized architecture using the transfer learning based on the Densenet201 model and the GradCam algorithm.
2. Related works
In recent years, DL has realized great success in several fields of medical imaging; it has improved the capability to detect and classify features in images using deep models. Some works focused on the use of DL techniques for the automatic detection of several lung diseases such as pneumonia [13],[11], and tuberculosis [14],[10], [19].
In today’s world, with the rapid spread of the Coronavirus, a significant amount of research has been focused to diagnose COVID-19 based on CT scans and chest X-Ray images using DL approaches. In this area, [3]used a database composed of 1428 chest x-ray images (224 patients positive for COVID-19, 700 cases of pneumonia, and the rest are healthy patients) in order to evaluate the effectiveness of the advancing Convolutional Neural Network (CNN) architectures such as transfer learning in COVID-19 classification. The researchers proposed a pre-trained VGG-19 model composed of nineteen deep convolutional layers[7]. detected the COVID-19 virus by applying COVIDX-Net based on DL using chest x-ray images[29]. proposed a CNN model in order to distinguish between COVID-19, influenza A, and viral pneumonia; the proposed architecture offered an accuracy of 86.7%[25]. have developed a prediction model based on DL approaches using the modified technique of Inception based on transfer learning. The obtained accuracy by this method is 89.5% which makes it better compared to the results of [29] [16]. applied another more complex pre-trained model of ImageNet with transfer learning for the prediction of COVID-19 in chest x-ray images. The researchers used the Inception-ResNetV2, InveptionV3, and ResNet50 models. ResNet50 model reached the best accuracy with an accuracy of 98%, which is better and more significant than [29] and [25].
On the other hand, recent research has shown that chest CT provides a lower false-positive rate than chest X-Ray images [1]. In this context, some works have been proposed using CT scan images[15]. developed a model named COVNet to extract visual characteristics from chest CT for the detection of the COVID-19 virus.
[22] proposed a DL technique called VB-net using chest-CT for automatic segmentation of all lungs and infected regions of the dataset. In another related work, [25], applied COVID-19 detection techniques based on Inception architecture modified by transfer learning on CT images to screen COVID-19 patients. The results obtained are 89.5%, 88%, and 87% for accuracy, specificity, and sensitivity, respectively.
[18] performed a comprehensive study for 16 ImageNet models based on transfer learning. In addition, he confirmed the robustness of DenseNet201 [9] for this task. Pharm also concluded that data augmentation has a negative effect on transfer learning.
In some works proposed to detect COVID-19 cases, new architectures of deep neural networks have been explored [21]. proposed CTnet-10, a 10-layer CNN. The results are promising and show that this model is more efficient than the existing architecture [30]. proposed DeCovNet, which can operate on a full 3D CT volume.
In the work of [24], a model called Details Relation Extraction neural network (DRE-Net) was used to extract the deep characteristics of CT images. In this study, the authors used CT images of 275 patients, including 88 patients with COVID-19, 101 patients awaiting bacterial pneumonia, and 86 healthy people. They reported an accuracy of 94%.
In some research, explainable methods have been exploited to visualize network predictions [12]. and [4] examined importance variations in chest CT scan images using Gradient-weighted Class Activation Mapping (Grad-CAM) ([20]).
To control the spread of COVID-19 quickly and effectively, DL techniques are necessary for proper isolation and treatment. In the literature, several methods have been proposed based on DL techniques for COVID-19 diagnosis. Some works have been based on chest x-ray images using advanced CNN architectures. Other more recent studies used CT images to detect the most important virus characteristics and classify the disease correctly. In this paper, we proposed an explainable DL method that allowed us to extract the characteristics of COVID-19 from the CT images to reduce false negatives, give an early clinical analysis, and prides visual explanation of the decision.
3. Materials and methods
3.1. Materials
Our experiments were based on chest CT image data collected at the Hospital of Tlemcen in Algeria. All executions were performed on a local machine with an i7-8750H processor, an Nvidia GeForce GTX 1070 GPU card, and 16GB of RAM. This paper used transfer learning techniques on CNN models widely used in applications where the data set is not significant. Transfer learning is to retrain a pre-trained model on large datasets such as ImageNet. This technique reduces the overall training time of the model and allows a comparatively smaller dataset to be used with relatively complex architecture.
3.1.1. Collection of dataset
The dataset was collected locally between June 2020 and October 2020 at the University Hospital of Tlemcen, Algeria. It consists of 177 patients (69 infected patients and 108 non-infected ones). This dataset is composed of 4986 CT images of suspected cases, including 1868 for patients infected with COVID-19 and confirmed with RT-PCR and 3118 images for patients not infected with COVID-19 but with other lung diseases. All these images have undergone pre-processing to remove all unnecessary scanners. Indeed, CT is a set of images. This image set contains black slices at the beginning of each volume and other images that do not contain any part of the lung. We then performed a pre-processing, which consists of removing these images to not distort the diagnosis. Figure1 shows a sample of the data collected in this study.
Fig. 1.
Samples from our collected dataset.
3.2. Methods
This study aims to find the best architecture to classify patients suspected of COVID-19 (positive or negative). For this purpose, we have selected several CNN architectures that have achieved the best results on the ImageNet dataset. We have used the data augmentation method to automatically increase the dataset and prevent the overfitting [27], which significantly affects the size and quality of the dataset and the model’s capacity during training. In this case, the data augmentation tool firstly rescaled the images (reduction or enlargement during the augmentation process). Then, a random rotation was performed during the training. After that, we did a height and width shift to move the images horizontally and vertically. The zoom range is fixed at 30% random. Finally, the images of the dataset were inverted horizontally. Table 1 shows the parameters used in the data augmentation method. The dataset is randomly divided in the experiment, where 80% is adopted for the training and the rest for the test. Afterward, we experimented with several CNN architectures for all the deep CNN models in the classification phase, modifying only the fully connected layers to adapt the model with the appropriate outputs, keeping the convolution and pooling layers. These two types of layers are used in the feature extraction step. In our study, we have two outputs, and the Softmax function is used to classify those two classes, one for healthy cases, i.e., negative, and the other for infected cases, i.e., positive.
Table 1.
Settings for the image augmentation.
| methods | setting |
|---|---|
| Rotation range | 30 |
| Width shift | 0.2 |
| Height shift | 0.2 |
| Rescale | 1/255 |
| Shear range | 0.2 |
| Zoom range | 0.2 |
| Horizontal flip | true |
| Vertical flip | true |
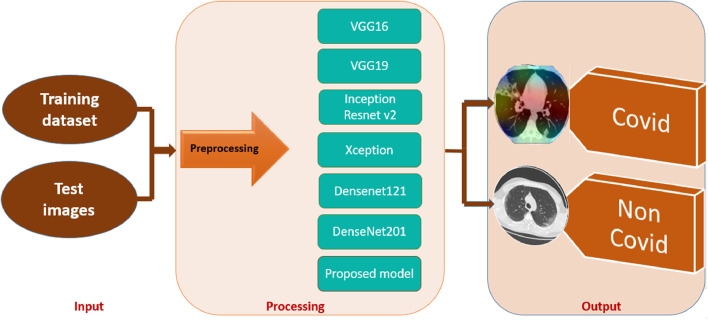
The block diagram of this work is presented in Fig. 2 . For all the proposed models, images are resized to 128 x 128 x 3, batch size and epoch were fixed at 64 and 200, respectively, using Stochastic Gradient Descent (SGD) as a specific optimizer. For better visualization of the results, the GradCam algorithm was used to explain the model’s decision.
Fig. 2.
Block diagram of this work.
3.2.1. Densenet201
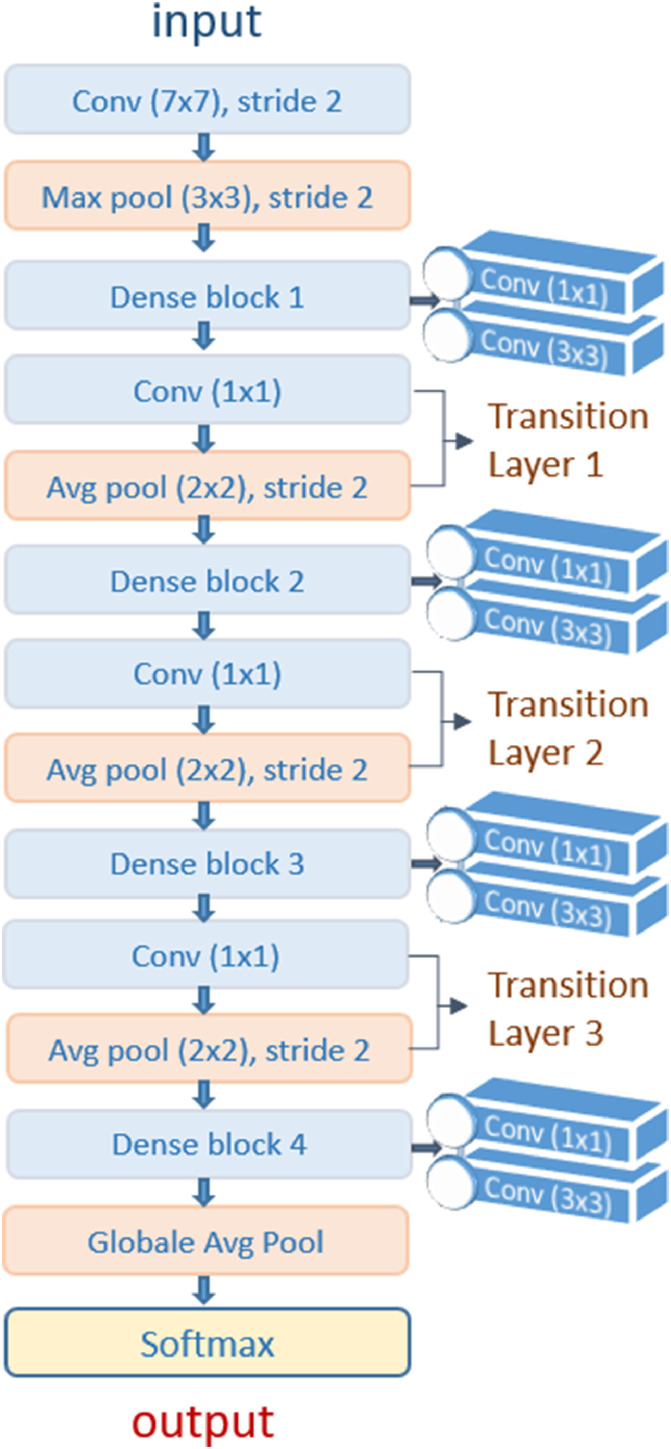
Transfer learning has shown its robustness in limited data classification problems. Results can be improved by using hyper-tuning of Deep Transfer Learning (DTL). In our contribution, the DenseNet201 model by transfer learning was proposed to extract features automatically and use their weights learned on the ImageNet dataset that reduces the effort of the calculation. DenseNet proved its effectiveness on different datasets such as CIFAR-100 [6] and ImageNet. Besides, this architecture provides simple and easy-to-build models. Also, it is possible to re-use features by different layers, which makes the parameters of this architecture highly efficient and allows to increase the variation in the following layers and improve the performance.
The features of the last deep layers can be projected by those of all the previous set of layers (this means that the deep network layers can re-use all the features produced by the previous layers) under the form of :
| (1) |
Where is a composite function of three operations, including batch normalization (BN). Followed by a ReLu function and a convolution layer (3,3). In classical CNNs architectures, the convolution layers are usually followed by down-sampling layers to reduce the size of the feature maps to half. Therefore, concatenation of feature maps around the down-sampling layers can cause a multiplicity of sizes. To solve this issue, dense blocks were designed before the down-sampling layers, while the dense block layers are tightly connected as shown in Fig. 3 , which makes the size of the feature maps constant in all dense blocks and reduced to half after down-sampling. Thus, for X layers of a dense block, the overall number of links between the layers is / 2, unlike a traditional convolutional network which equals X. However, the computation will be huge if the layers are deep because the number of concatenated feature maps entered in the layers is high. Thus, the control of the number of feature maps newly created by a growth rate K is necessary. The total number of feature maps in the last layer of a dense block is in the form of: . Where is the number of channels in the first input layer. To solve the computational effort, 1 x 1 bottleneck layers were applied before each 3 x 3 convolution layer followed by transition layers that significantly improve the network compactness by controlling the number of output feature maps. Furthermore, it is usually found after the dense block at a certain depth, which explains why there is no transition after the first dense block, as shown in Fig. 3.
Fig. 3.
Architecture of transferred DenseNet201.
3.2.2. Proposed method
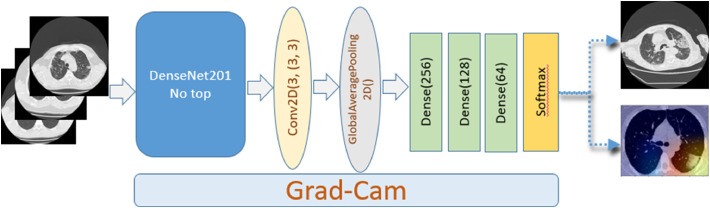
In this work, we proposed a customized DenseNet201-based model for the screening of COVID-19. We used the pre-trained DenseNet201 by keeping only the feature extraction layers. Then, we added a convolution layer followed by the GlobalAveragePooling layer and added three fully connected layers with 256, 128 and 64 neurons. The last classification layer contains two neurons and uses the Softmax function, where all the neurons of the same layer are connected to the next layer. After and to avoid overfitting our model, we considered the Dropout layers defined at 20%, 30%, and 30%, respectively.
To provide visual evidence and improve the explainability of our architecture and highlight the relevant areas that drive our model decision, we used the Grad-CAM algorithm. Without an additional manual annotation, this heat map is produced entirely by the DL model. The network predictions are interpreted by generating heat maps to visualize the most representative areas of the image using gradient-weighted class activation maps. The methodology followed in this work has been summarized in the flowchart shown in Fig. 4 .
Fig. 4.
Flowchart of the proposed study.
4. Results and discussion
In this work, we have collected a new dataset from the Hospital of Tlemcen in Algeria. The labeling of each image (positive or negative) is done by a Radiologist and confirmed by RT-PCR test.
The dataset is then pre-processed. We have readjusted the dataset so that all images will be 128x128 in size. For a better generalization, we used a data augmentation techniques by applying random transformations on the input images. To ensure the diversity of the data, several transformations have been performed, such as rotation and zoom with a probability of 0 to 1.
This study compared our proposed model with five other architectures: VGG16, VGG19, Xception, Inception_V2_Resnet, and DenseNet121. We have trained all models using batches of size 32 for 200 epochs using the Stochastic Gradient Descent (SGD) method to reduce the loss function. Results have been compared taking into account different metrics such as accuracy (Eq. 2), precision (Eq. 3), Recall (Eq. 4), and F1-score (Eq. 6).
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Where:
P: total number of COVID-19 patients
N: total number of Non-COVID-19 patients
True positive (TP): prediction is COVID, and the image is COVID.
True negative (TN): prediction is Non-COVID, and the patient is healthy.
False positive (FP): prediction is COVID, and the patient is healthy.
False negative (FN): prediction is Non-COVID, and the image is COVID.
Table 3 shows the obtained performances of different models. We notice that our proposed Densenet201 model gives better results compared to the other architectures on the accuracy, precision, and F1-score.
Table 3.
Comparisons of the results between the different architectures applied to our dataset.
| Model | Accuracy | Precision | Sensitivity | Specificity | F1-score | AUC |
|---|---|---|---|---|---|---|
| VGG16 | 94.69% | 96.21% | 95.12% | 94.00% | 95.67% | 94.56% |
| VGG19 | 94.38% | 93.47% | 97.72% | 89.03% | 95.55% | 93.38% |
| Xception | 96.59% | 97.08% | 97.4% | 95.3% | 97.24% | 96.35% |
| Inception_ | ||||||
| V2_Resnet | 96.49% | 97.54% | 96.75% | 96.08% | 97.14% | 96.42% |
| DenseNet121 | 96.99% | 98.99% | 96.09% | 98.43% | 97.52% | 97.27% |
| DenseNet201 | 98.1% | 99.01% | 97.89% | 98.43% | 98.45% | 98.16% |
| Our Work | 98.8% | 99.50% | 98.54% | 99.22% | 99.02% | 98.88% |
Additionally, Densenet201 gives the best recall (also known as sensitivity), which is good since, in medical diagnostic aid in general and decision support for COVID-19, this metric is crucial. Indeed, high sensitivity is equivalent to a shallow false-negative rate. False negatives are patients who have been diagnosed as Non-COVID by the system but are positive. Such an error can cause the patient’s death and does not have the same impact as a false positive error that can be quickly corrected with additional tests. Also, a person classified as healthy while infected with COVID-19 can transmit the virus and causes more losses. Our model also offers good results in terms of false positives as shown in Table 2. Such an error can cause panic and emotional distress to the patient. We can therefore deduce that our model is the best in terms of performance.
Table 2.
TP, TN, FP and FN ratio obtained from the testing dataset.
| Model | TP | FN | FP | TN |
|---|---|---|---|---|
| VGG16 | 585 | 30 | 23 | 360 |
| VGG19 | 601 | 14 | 42 | 341 |
| Xception | 599 | 16 | 18 | 365 |
| Inception_ | ||||
| V2_Resnet | 595 | 20 | 15 | 368 |
| DenseNet121 | 593 | 24 | 6 | 375 |
| DenseNet201 | 602 | 13 | 6 | 377 |
| Our Work | 606 | 9 | 3 | 380 |
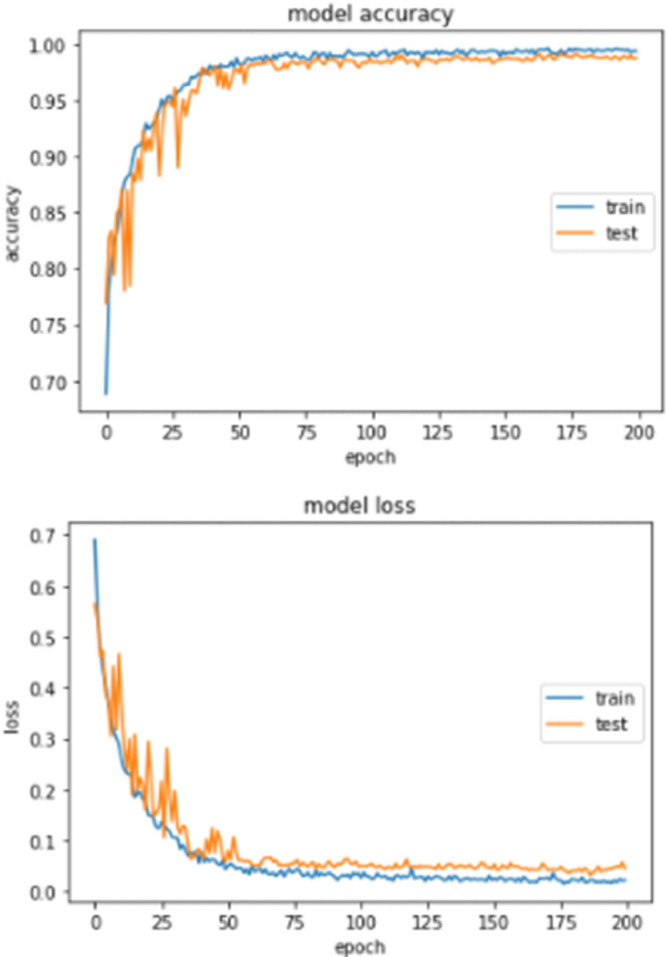
Fig. 5 shows the training and validation accuracy curves of the proposed work. The first remark is that our proposed method Densenet201 exceeded 90% just after the 15th epoch and gains more accuracy. This clearly shows that the proposed model can be used effectively for the early diagnosis of COVID-19. We can also notice that most models have learned well from the dataset and suffer neither from over- nor under-fitting. This can be explained by the fact that SGD contains the error, stays stable and prevents overfitting even without regularization.
Fig. 5.
Training and validation analysis over 200 epochs: Training and Testing accuracy analysis and Training and Testing loss analysis.
In order to prove the robustness of our proposed model, we used another public database ([2]). Table 4 shows the performance results. Here, the sensitivity obtained is 98.20%, and the specificity is 98.17% for COVID-19 positive cases. From the obtained results, we can conclude that the proposed model can correctly detect true positives (i.e., COVID-19 infected patients) with an ACCURACY of 98.18%. In addition, the proposed model showed good performance in terms of accuracy and F-1 score.
Table 4.
Comparisons of the results between the different architectures applied to the dataset of [2].
| Model | Accuracy | Precision | Sensitivity | Specificity | F1-score | AUC |
|---|---|---|---|---|---|---|
| VGG16 | 94.37% | 91.49% | 96.41% | 92.70% | 93.89% | 94.96% |
| VGG19 | 96.37% | 95.96% | 95.96% | 96.72% | 95.96% | 96.33% |
| Xception | 91.95% | 91.40% | 90.58% | 93.07% | 91.00% | 91.82% |
| Inception_ | ||||||
| V2_Resnet | 94.57% | 93.36% | 94.62% | 94.52% | 93.98% | 91.82% |
| DenseNet121 | 95.77% | 91.73% | 99.55% | 92.73% | 95.48% | 96.12% |
| DenseNet201 | 97.38% | 95.26% | 99.10% | 95.98% | 97.14% | 97.54% |
| Our Work | 98.18% | 97.76% | 98.20% | 98.17% | 97.98% | 98.82% |
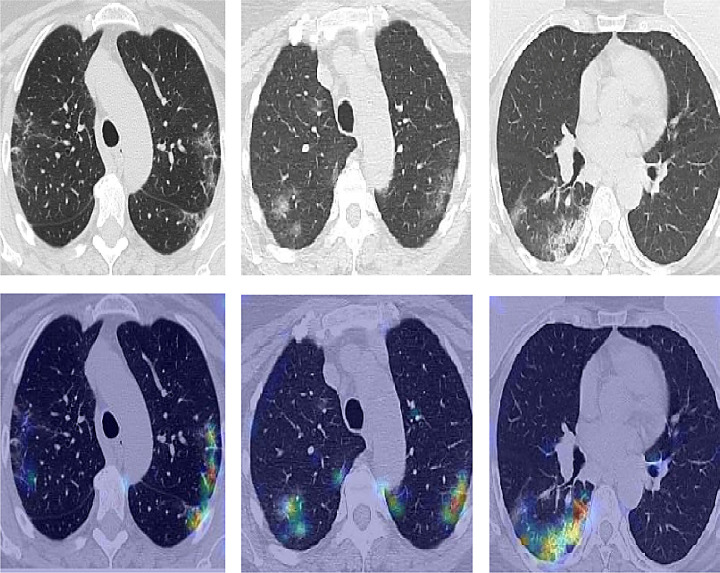
To convince physicians of the accuracy and usefulness of the proposed tool and to visualize the areas that motivate the model’s decision, we have applied the Grad-CAM algorithm.
Fig. 6 shows the heatmaps on the suspect areas, which prove that our algorithm focuses on the infected areas while neglecting the other normal regions. The network predictions are interpreted based on heatmaps to visualize the most significant areas on an image by applying gradient-weighted class activation maps.
Fig. 6.
Explanation with heatmap on COVID-19 images using Grad-CAM.
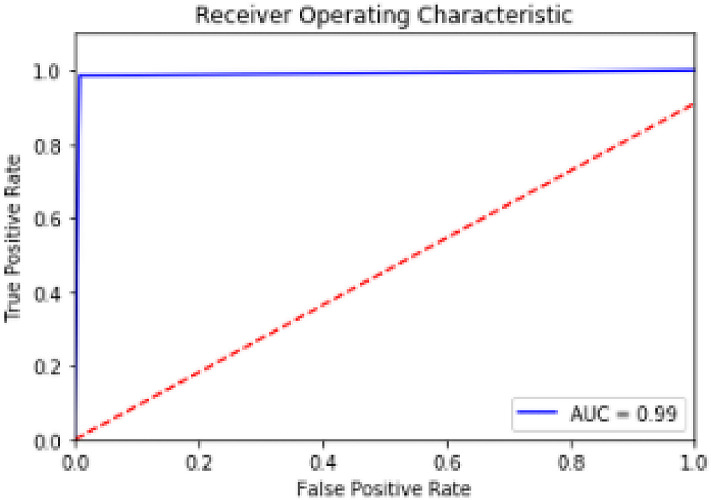
To better evaluate the performance of our model, we plotted the ROC curve (Receiver Operating Characteristic) and calculated the AUC (Area Under the Curve). The ROC curve is a graph that allows asserting the classification efficiency of a model according to the true positives and false positives. As shown in Fig. 7 , the model proposed by DTL with Densenet201 offers an AUC of 98.88%. We remark that this classification model delivers better results compared to the existing classification models as seen in Table4.
Fig. 7.
Area Under Curve for proposed model.
5. Conclusion
This research introduced a benchmarking analysis and study on different models to classify CT-scan images of COVID-19. This tool is necessary for managing the COVID-19 crisis, given the worldwide shortage of RT-PCR screening kits. For that, we have collected a new CT-scan dataset from the hospital of Tlemcen in Algeria. We then applied a transfer learning on six well-known DL architectures (VGG16, VGG19, Xception, Inception V2 Resnet, DenseNet121, Densenet201), then proposed a model based on DenseNet201 and the GradCam algorithm. According to the carried out experiments, the proposed architecture has shown its reliability with 98.8% of accuracy. It also provides a visual explanation, which proves that the proposed model can be considered as an alternative for screening COVID-19 and the follow-up of patients. Thus, the DL methods can help in the fight against the spread of the virus. We intend to add more classes containing other lung diseases to help health experts and general practitioners.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Edited by Maria De Marsico
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.patrec.2021.08.035
Appendix A. Supplementary materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
References
- 1.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in china: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelov P., Almeida Soares E. Sars-cov-2 ct-scan dataset: a large dataset of real patients ct scans for sars-cov-2 identification. medRxiv. 2020 [Google Scholar]
- 3.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai H.X., Wang R., Xiong Z., Hsieh B., Chang K., Halsey K., Tran T.M.L., Choi J.W., Wang D.-C., Shi L.-B., et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296(3):E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basavegowda H.S., Dagnew G. Deep learning approach for microarray cancer data classification. CAAI Trans. Intell. Technol. 2020;5(1):22–33. [Google Scholar]
- 6.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE conference on computer vision and pattern recognition. Ieee; 2009. Imagenet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 7.E.E.-D. Hemdan, M.A. Shouman, M.E. Karar, Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images, arXiv preprint: 2003.11055(2020).
- 8.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, china. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 10.Hwang S., Kim H.-E., Jeong J., Kim H.-J. Medical imaging 2016: computer-aided diagnosis. volume 9785. International Society for Optics and Photonics; 2016. A novel approach for tuberculosis screening based on deep convolutional neural networks; p. 97852W. [Google Scholar]
- 11.Jaiswal A.K., Tiwari P., Kumar S., Gupta D., Khanna A., Rodrigues J.J. Identifying pneumonia in chest x-rays: a deep learning approach. Measurement. 2019;145:511–518. [Google Scholar]
- 12.Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X., Deng L., Zheng C., Zhou J., Shi H., et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 2020;11(1):1–14. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahsaini I., El Habib Daho M., Chikh M.A. Proceedings of the 1st International Conference on Intelligent Systems and Pattern Recognition. 2020. Convolutional neural network for chest x-ray pneumonia detection; pp. 55–59. [Google Scholar]
- 14.Lakhani P., Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 15.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2) doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Narin, C. Kaya, Z. Pamuk, Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks, arXiv preprint: 2003.10849(2020). [DOI] [PMC free article] [PubMed]
- 17.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. Deep transfer learning based classification model for COVID-19 disease. IRBM. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham T.D. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci. Rep. 2020;10(1):1–8. doi: 10.1038/s41598-020-74164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi G., Wang H., Haner M., Weng C., Chen S., Zhu Z. Convolutional neural network based detection and judgement of environmental obstacle in vehicle operation. CAAI Trans. Intell. Technol. 2019;4(2):80–91. [Google Scholar]
- 20.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE international conference on computer vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 21.Shah V., Keniya R., Shridharani A., Punjabi M., Shah J., Mehendale N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg. Radiol. 2021:1–9. doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.F. Shan, Y. Gao, J. Wang, W. Shi, N. Shi, M. Han, Z. Xue, Y. Shi, Lung infection quantification of covid-19 in ct images with deep learning, arXiv preprint: 2003.04655(2020).
- 23.Singh D., Kumar V., Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Diseas. 2020:1–11. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Zhao H., Jie Y., Wang R., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. 2020 doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19) MedRxiv. 2020 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-cov-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 28.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in china. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv. 2020 [Google Scholar]
- 31.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from china. Radiology. 2020:200490. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/