Abstract
mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin 3 (IL-3) signaling pathways and plays an important role in the viability response of these cytokines. In this study, we demonstrated that cytokine stimulation of mcl-1 mRNA and protein expression were attenuated by pretreatment of cells with phosphatidylinositol 3-kinase (PI3-K) inhibitors. Reporter gene assays further showed that the PI3-K/Akt signaling pathway was involved in IL-3 activation of mcl-1 gene transcription. Analysis of the mcl-1 promoter revealed that both promoter elements, SIE at position −87 and CRE-2 at −70, contribute to IL-3 stimulation of mcl-1 gene expression. Although either the SIE site or the CRE-2 site alone was sufficient to confer IL-3 inducibility on a heterologous promoter, only IL-3 activation of the CRE-2 reporter was mediated via the PI3-K/Akt pathway. The SIE binding activity was constitutively high in cells deprived of or stimulated by IL-3. In contrast, the CRE-2 binding activity was low in cytokine-starved cells and was strongly induced within 1 h following cytokine treatment of cells. In addition, cytokine induction of the CRE-2 but not of the SIE binding activity was dependent on activation of the PI3-K/Akt signaling pathway. Lastly, we showed that CREB was one component of the CRE-2 binding complex and played a role in IL-3 activation of the mcl-1 reporter gene. Taken together, our results suggest that both PI3-K/Akt-dependent and -independent pathways contribute to the IL-3 activation of mcl-1 gene expression. Activation of mcl-1 by the PI3-K/Akt-dependent pathway is through a transcription factor complex containing CREB.
Granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin 3 (IL-3) are two members of a family of cytokine growth factors that play an important role in regulating the viability, differentiation, proliferation, and function of multipotential hematopoietic progenitors as well as various other hematopoietic cells (4). They function by binding to their cognate receptors and triggering a cascade of signaling events leading to various biological responses. The receptors for GM-CSF and IL-3 are composed of two subunits, the cytokine-specific α chain and the common β chain, which is also possessed by the receptor for another cytokine, IL-5. Ligand binding to these cytokine receptors induces rapid tyrosine phosphorylation of several cellular proteins, including the receptor β chain itself, the Jak2 kinase, Shc, vav, fps, STAT5A, and STAT5B (5, 17, 22, 42, 44, 53). Cytokine-activated receptors also lead to the activation of the phosphatidylinositol 3-kinase (PI3-K) and the Ras/Raf/mitogen-activated protein (MAP) kinase pathway (10, 20, 29, 50, 51) and transcriptional activation of some immediate-early genes like c-jun, c-fos, c-myc, cis, and mcl-1 (7, 9, 60). By deletion analysis, the cytoplasmic domains of the common β chain responsible for activation of some of these cellular targets have been defined. The membrane-proximal domain is important for the induction of c-myc and cis and for the activation of the Jak2 kinase and the STAT5 proteins (44, 46, 50, 60); the membrane distal domain is crucial for the induction of c-jun, c-fos, and mcl-1 and for the activation of the PI3-K and the Ras/Raf/MAP kinase cascade (7, 50). The activation of the latter two kinase pathways has been shown to be important for the antiapoptotic activity of GM-CSF and IL-3 (29, 55).
The mcl-1 gene was originally identified as an early gene induced during differentiation of ML-1 myeloid leukemia cells (33). Its encoded gene product contains some structural motifs that characterize it as a member of the Bcl-2 family protein. Unlike other members of this protein family, Mcl-1 has an extended N-terminal domain which is rich in PEST sequences (33). The PEST sequence is probably responsible for the short half-life of this protein (7, 57). Overexpression of Mcl-1 delays apoptosis induced by various inducers such as c-Myc overexpression, growth factor withdrawal, and other cytotoxic agents (7, 48, 61). We recently demonstrated that mcl-1 is another immediate-early gene activated by the GM-CSF and IL-3 signaling pathways and that the mcl-1 gene product is one component of the viability response of these cytokines (7). Cytokine activation of the mcl-1 gene is regulated at the transcriptional level and requires the membrane-distal region between amino acids 573 and 755 of the common β chain (7). Using transient transfection assays with luciferase reporters driven by various regions of the mcl-1 promoter, we further demonstrated that the upstream sequence between −197 and −69 is responsible for cytokine activation of the mcl-1 gene (7).
The signaling pathway from PI3-K to the serine/threonine protein kinase Akt/protein kinase B (PKB) is involved in some cellular responses induced by growth factors such as insulin, platelet-derived growth factor (PDGF), epidermal growth factor (EGF), insulin-like growth factor I (IGF-I), basic fibroblast growth factor, IL-2, and IL-3 (1, 6, 11, 19, 32, 34, 41, 55). Overexpression of the constitutively active PI3-K or Akt/PKB kinase protects apoptosis induced by various stimuli, including cellular differentiation and serum withdrawal in conjunction with induced myc activity (18, 26, 27). With the use of specific chemical inhibitors and/or the dominant negative mutants, the survival effects generated by some of the above-mentioned growth factors and some transforming oncogenes were shown to be mediated through activation of the PI3-K and Akt/PKB signaling pathway (1, 16, 28, 34, 41, 54, 55). However, the downstream effectors of this kinase pathway that are directly responsible for the antiapoptotic activity of various survival factors are largely unknown.
In this study, we dissected the signaling pathway responsible for survival factor activation of Mcl-1 expression. We demonstrated that cytokine stimulation of mcl-1 gene transcription was mediated through both the PI3-K/Akt-dependent and -independent pathways. We further showed that CREB was one component of the transcription factor complex activated by the PI3-K/Akt-dependent pathway and played a role in IL-3 stimulation of mcl-1 gene expression.
MATERIALS AND METHODS
Cell culture.
TF-1 cells (30) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 50 μM β-mercaptoethanol, 2 mM l-glutamine, 100 U of penicillin G/ml, 100 μg of streptomycin/ml, and 1 ng of GM-CSF/ml. Human GM-CSF was kindly provided by Schering-Plough Ltd., Taipei, Taiwan. For GM-CSF depletion experiments, TF-1 cells were washed three times in medium without cytokine and seeded in RPMI 1640 supplemented with 0.5% FBS, 2 mM l-glutamine and 50 μM β-mercaptoethanol. During restimulation experiments, only GM-CSF (10 ng/ml) was added back to the cells that had been previously cultivated in low-serum medium containing no cytokine. Murine IL-3-dependent pro-B (Ba/F3) cells were maintained in RPMI 1640 supplemented with 10% FBS and 1% conditioned medium from WEHI 3B as a source of IL-3. Ba/F3Akt*-A1 and Ba/F3Akt*-D2 are two representatives clones of Ba/F3 derivatives which stably overexpress the constitutively active form of the Akt protein (M-Akt [34]). Ba/F3DNAkt-9H and Ba/F3DNAkt-19M are two other representative clones of Ba/F3 cells which stably overexpress the dominant negative mutant of the Akt protein (AktK179M [34]). Ba/F3hMcl-1 cells are Ba/F3 derivatives stably overexpressing the human Mcl-1 protein. Two representative clones (hMcl/17 and hMcl/19) were analyzed. Ba/F3 cells transfected with the empty expression vectors (Ba/F3Neo) were used as controls. All Ba/F3 derivatives were generated by electroporation with respective expression vectors and selected in growth medium supplemented with 500 μg of G418/ml. For cytokine depletion and restimulation experiments, cells were treated essentially as described above for TF-1 cells but stimulated with 100 U of recombinant IL-3 (R & D Systems, Minneapolis, Minn.)/ml. For experiments with chemical inhibitors, unless specified, the following concentrations and compounds were used: wortmannin (0.1 μM), LY294002 (50 μM), PD98059 (50 μM), rapamycin (50 nM), and cycloheximide (10 mg/ml). All inhibitors were purchased from Calbiochem.
Immunoblotting.
Cells treated under various conditions were lysed in radioimmunoprecipitation assay buffer and analyzed as previously described (7). Briefly, 50 μg of cell lysates were resolved on a sodium dodecyl sulfate (SDS)-containing 12% polyacrylamide gel, transferred to polyvinylidene difluoride nylon membranes (Millipore, Bedford, Mass.), and probed with antibodies specific to Bcl-2, Mcl-1 (Santa Cruz Biotechnology, Santa Cruz, Calif.), HA tag (Boehringer, Mannheim, Germany), CREB (a gift of Ming-Zong Lai, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan) (see reference 24), phosphorylated (S-133) CREB (Upstate Biotechnology, Inc., Lake Placid, N.Y.) or α-tubulin (Amersham, Buckinghamshire, England). The membrane was then probed with either horseradish peroxidase-conjugated goat anti-mouse or goat-anti-rabbit antibody. The specific bands were visualized by an ECL (enhanced chemiluminescence) Western blot system (Amersham, Buckinghamshire, England).
Reporter plasmids and luciferase assay.
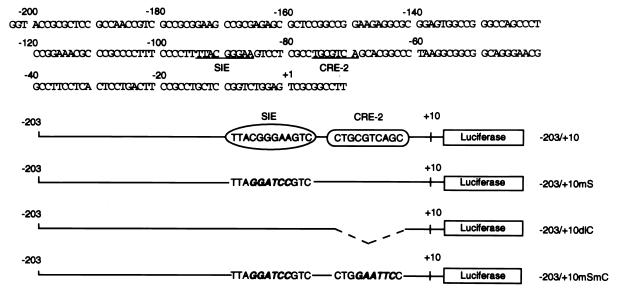
The luciferase reporter plasmids containing various regions of the mcl-1 promoter [p(−1288/+10)mcl-luc, p(−203/+10)mcl-luc, and pB-dl(−197/−69)] have been previously described (7). The numbers in parentheses indicate the nucleotide position with respect to the transcriptional initiation site (7). Other reporter plasmids shown in Fig. 6 were derived from p(−203/+10)mcl-luc by site-directed mutagenesis of each individual region as indicated. During the construction of these reporter plasmids, some DNA fragments to be cloned were PCR amplified by using appropriate primers and subcloned into pGL-2-basic vector (Promega). All plasmids constructed in this way were sequenced to confirm their primary structure. Plasmids pGL2-1XCRE-2, pGL2-1XSIE, and pGL2-1XSC were derived by inserting one copy of the DNA fragment containing CRE-2, SIE, or both sites (see below for individual sequences) into the SmaI site of the pGL2-promoter vector (Promega). To analyze the promoter activity of these reporter genes, Ba/F3 cells were transiently transfected with these plasmids by electroporation by using the Bio-Rad gene pulser II RF module system as previously described (7). Electroporated cells were seeded in growth medium with or without murine IL-3 (mIL-3). Twelve hours after reseeding, cells were harvested and assayed for luciferase activity. A cytomegalovirus-driven chloramphenicol acetyltransferase (CAT) reporter gene was cotransfected to correct for variations in transfection efficiency. For analyzing the effects of various dominant negative mutants in the reporter gene assays, electroporated cells were recovered in mIL-3-containing medium for 12 h and then deprived of mIL-3 for 8 h before mIL-3 restimulation was initiated. Three hours after mIL-3 restimulation, cell lysates were prepared and assayed for luciferase activity. This modified procedure was found to be necessary for obtaining maximal effect of the dominant negative mutants. The sense strand sequences of the double-stranded oligonucleotides used in the plasmid construction and the gel-shift assay (see below) are as follows: SIE, 5′CTTTTACGGGAAGTCC3′ (wild type) and 5′CTTTTAGGATCCGTCC3′ (mt); CRE-2, 5′TCGCCTGCGTCAGCACG3′ (wt) and 5′TCGCCTGGAATTCCACG3′ (mt); and SC fragment, 5′GTACCCCTTTTACGGGAAGTCCTCGCCTGCGTCAGCACGGC3′.
FIG. 6.
Schematic representation of constructs used in this study. The nucleotide sequence encompassing the murine mcl-1 gene promoter from −203 to +10 (7) is shown at the top. The core sequences of the SIE and CRE-2 sites are underlined. The mutated sequence in each individual construct is shown in italics. In construct p(−203/+10)dlC, the CRE-2 core sequence was deleted and is shown as a dashed line.
Transient transfection and detection of protein expression by flow cytometry.
TF-1 cells were transiently transfected with various expression plasmids by using the liposome-mediated gene transfer method as previously described (7). The expression vector driving the synthesis of green fluorescence protein (GFP) was cotransfected at a one-sixth molar amount of the construct of interest to assist the identification of the transfected cells by flow cytometry. Twenty-four hours after transfection, cells were analyzed for the expression of GFP and various proteins of interest by flow cytometry as previously described (7).
Apoptosis detection by annexin-V staining.
Cells transiently transfected with constructs of interest plus an expression vector encoding a GFP protein were fixed in 2% paraformaldehyde at room temperature for 15 min. After being rinsed with the binding buffer (10 mM HEPES-NaOH [pH 7.4], 140 mM NaCl, and 5 mM CaCl2), fixed cells were incubated in the same buffer containing biotin-conjugated annexin-V (Boehringer, Mannheim, Germany) for 20 min at room temperature. After several washes, annexin-V-bound cells were revealed by binding to phycoerythrin-conjugated streptavidin, and the percentages of double positive cells were analyzed by flow cytometry.
Preparation of nuclear extracts and gel-shift assays.
Cells treated under various conditions were lysed, and nuclear extracts were prepared according to the method described by Dignam et al. (14). The gel-shift assay was carried out essentially as previously described (58). Briefly, the double-stranded oligonucleotide containing SIE or the CRE-2 site (same oligonucleotides as described above for reporter gene construction) was 32P labeled by a kinase or a fill-in reaction. The labeled probe (∼0.2 ng) was incubated with extracts for 20 min at room temperature in a buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, and 10% glycerol. The specific protein complexes were resolved in a 4% native polyacrylamide gel (acrylamide/bisacrylamide ratio, 80:1) at 4°C. The gel was dried, and signals were visualized by autoradiography. For antibody-supershifting experiments, 1 μg of the CREB or control antibody was included in the binding reaction.
RESULTS
The PI3-K/Akt kinase pathway is involved in the viability response of IL-3 and GM-CSF in Ba/F3 and TF-1 cells, respectively.
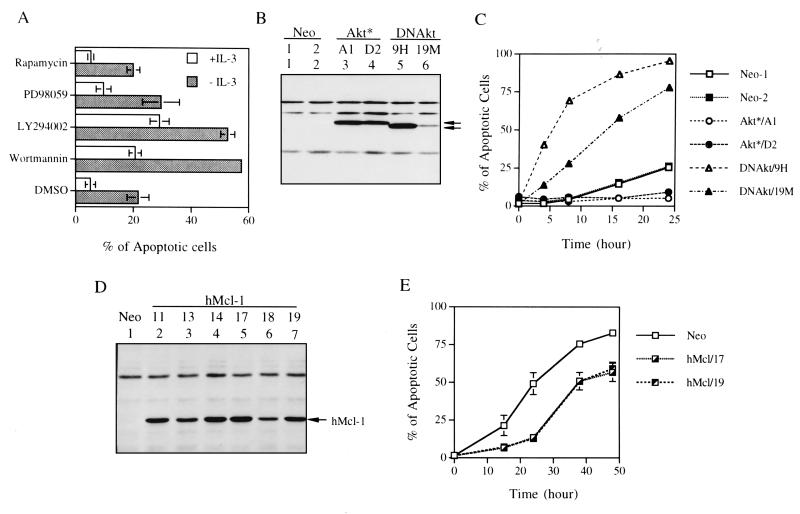
Activation of the PI3-K/Akt kinase pathway mediates the survival activity of IL-3 in 32D cells (55). We demonstrated that this was also the case in another IL-3-dependent pro-B cell line (Ba/F3) (Fig. 1). In this experiment, addition of chemical inhibitors of PI3-K (wortmannin or LY294002) to Ba/F3 cells accelerated apoptosis of these cells cultivated either in the presence or absence of IL-3 (Fig. 1A). This cell death acceleration effect was not present in cells treated with rapamycin, a p70S6 kinase inhibitor, and was observed only marginally in cells treated with the MEK inhibitor PD98059. The involvement of Akt kinase in IL-3 viability response of Ba/F3 cells was evident from studies with Ba/F3 cells stably overexpressing the constitutively active (Ba/F3Akt*) or the dominant negative mutant (Ba/F3DNakt) of the Akt protein (see Materials and Methods) (Fig. 1B). As shown in Fig. 1C, when IL-3 was removed from the growth medium, Ba/F3Akt* cells (two representative clones A1 and D2) maintained their viability much longer than cells transfected with the vector control (Ba/F3Neo). In contrast, Ba/F3DNAkt cells (clones 9H and 19M) underwent apoptosis much faster than the Ba/F3Neo cells. The rate of apoptosis of the latter two clones correlated well with the expression levels of the DNAkt protein (compare Fig. 1B and C) and was consistent with their requirement of higher doses of IL-3 for survival (ED50 for Ba/F3Neo and clones 19M and 9H were 0.01, 0.03, and 0.14 ng of IL-3/ml, respectively, data not shown).
FIG. 1.
The PI3-K/Akt kinase pathway is involved in the viability response of IL-3 in Ba/F3 cells. (A) Ba/F3 cells cultivated in growth medium with or without IL-3 were treated with various chemical inhibitors as indicated. Fifteen hours after each treatment, cells were harvested and the percentage of apoptotic cells under each condition was quantified by flow cytometric analysis of cells with a sub-G1 DNA content. Results shown are means ± standard deviations from three independent experiments. (B) Protein expression of Ba/F3 cells overexpressing M-Akt (Ba/F3Akt*) or AktK179M (Ba/F3DNAkt). One hundred micrograms of protein lysates from Ba/F3Akt* (clones A1 and D2) or from Ba/F3DNAkt (clones 9H and 19M) were analyzed by immunoblotting with anti-HA antibody (both proteins are HA tagged). Specific bands are indicated by arrows. (C) Ba/F3 derivatives as indicated were deprived of IL-3, and the percentage of apoptotic cells at various time points was measured as described in panel A. During IL-3 deprivation, these cells were kept in medium containing 10% instead of 0.5% FBS (see Materials and Methods) to slow the death rate of Ba/F3DNAkt cells. Results shown here are the averages of two independent experiments. (D) Same as in panel B except that the cells analyzed were Ba/F3 cells stably overexpressing human Mcl-1 protein (Ba/F3hMcl-1) and the immunoblot probed with anti-human Mcl-1 antibody, which did not cross-react with the murine homologue. Several individual clones, as indicated by numbers, were analyzed. (E) Ba/F3hMcl-1 cells were deprived of IL-3 by the standard procedure as described in Materials and Methods, and the percentage of cells undergoing apoptosis at various time points was analyzed as described in panel A. Only results from two representative clones (hMcl/17 and hMcl/19) are shown. Neo represents control cells that are stably transfected by the empty expression vector and is a mixture of two stable lines. Results shown are means ± standard deviations from two independent experiments.
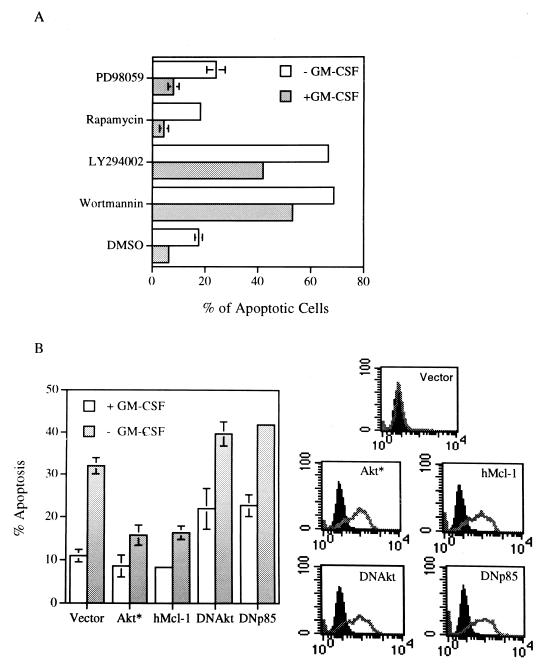
Due to possession of the common β subunit of the receptor, GM-CSF elicits many biological responses that overlap with those induced by IL-3. We next examined whether GM-CSF also mediated its survival activity via the PI3-K/Akt signaling pathway. To address this issue, similar experiments as described above for the Ba/F3 cells were performed in the TF-1 myeloid progenitor cell line, whose growth is dependent on human GM-CSF (hGM-CSF) or IL-3 (30). As shown in Fig. 2A, TF-1 cells underwent apoptosis upon deprivation of their dependent cytokine as previously reported (59). Furthermore, as in the case with Ba/F3 cells, addition of wortmannin or LY294002 but not rapamycin or PD98059 to growth medium significantly accelerated apoptosis of these cells cultivated either in the presence or absence of GM-CSF. The involvement of the PI3-K/Akt kinase pathway in GM-CSF viability response was further supported by the results of the transient-transfection experiment as shown in Fig. 2B. In this experiment, GM-CSF withdrawal-induced apoptosis was significantly inhibited by transient overexpression of the constitutively active form of Akt, whereas transient overproduction of the dominant negative mutant of PI3-K(Δp85 [23]) or of Akt enhanced apoptosis of TF-1 cells cultivated in the presence or absence of GM-CSF (Fig. 2B). These results suggest that in TF-1 cells the PI3-K/Akt kinase pathway is indeed involved in the survival activity of GM-CSF.
FIG. 2.
The PI3-K/Akt kinase pathway is involved in the viability response of GM-CSF in TF-1 cells. (A) TF-1 cells cultivated in growth medium with or without GM-CSF were treated with various inhibitors for 15 h and analyzed as described in the legend to Fig. 1A. The percentage of apoptotic cells under each condition was expressed as the mean ± standard deviation from two independent experiments. (B) TF-1 cells were transiently transfected with the constructs of interest plus GFP expression vectors. Twenty-four hours after transfection, cells were placed in medium with or without GM-CSF for another 18 h before they were fixed, stained, and analyzed as described in Materials and Methods. The GFP-positive and annexin-V bound (apoptotic) cells were quantified by flow cytometry, and the results are plotted in the left panel. The right-hand panel displays the flow cytometric results showing expression of each individual protein in the transfected (GFP positive, gray lines), but not in the nontransfected (GFP negative, solid peaks), fractions. All proteins were HA tagged and detected by HA-specific antibody. Vector, Akt*, hMcl-1, DNAkt, and DNp85 denote cells transfected with an empty expression vector or vectors expressing M-Akt, human Mcl-1 protein, AktK179M, and the dominant negative mutant of PI3-K (Δp85), respectively.
The PI3-K/Akt kinase pathway is involved in cytokine activation of Mcl-1 expression.
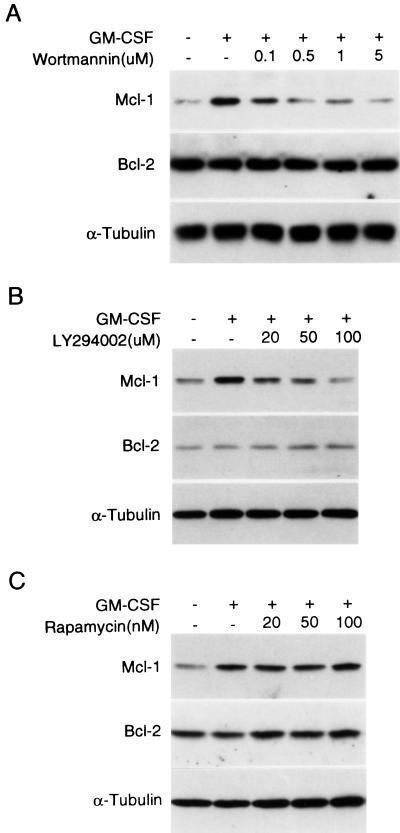
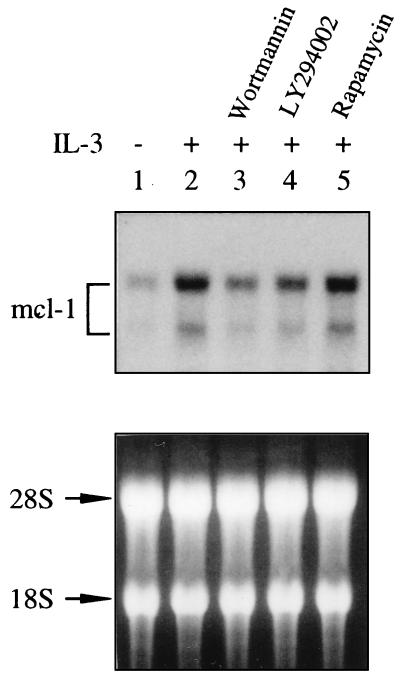
We recently demonstrated that mcl-1 is an immediate-early gene activated by the GM-CSF and IL-3 signaling pathways and is one component of the viability response of these cytokines (7). These findings together with the fact that both GM-CSF and IL-3 activate the PI3-K/Akt pathway in many factor-dependent cells, including TF-1 and Ba/F3 (55, 56a) and the fact that Mcl-1 and the activated Akt kinase both protect apoptosis ([7] Fig. 1C and E, and Fig. 2B) prompted us to examine the possibility that the PI3-K/Akt kinase pathway is involved in the cytokine activation of the mcl-1 gene. We first examined if GM-CSF stimulation of Mcl-1 expression was affected by wortmannin or LY294002. As illustrated in Fig. 3, when TF-1 cells were pretreated with either of these two inhibitors, the GM-CSF induction of Mcl-1 protein expression was inhibited in a dose-dependent manner (panels A and B), whereas the p70S6 kinase inhibitor, rapamycin, did not have any effect on Mcl-1 induction under the same conditions (panel C). As a control, the Bcl-2 protein level, previously shown not to be affected by GM-CSF treatment (7) was not inhibited by any of these three inhibitors (Fig. 3). We next determined whether a similar inhibitory effect could be observed in IL-3 stimulation of Mcl-1 expression in Ba/F3 cells. Due to lack of a good antibody to detect the murine Mcl-1 protein, Northern blotting was employed. The results shown in Fig. 4 indicated that both PI3-K inhibitors, but not rapamycin, inhibited IL-3 stimulation of mcl-1 mRNA expression in Ba/F3 cells.
FIG. 3.
GM-CSF induction of Mcl-1 expression is attenuated by PI3-K inhibitors. TF-1 cells deprived of cytokine were pretreated with various doses of wortmannin (A), Ly294002 (B), or rapamycin (C) for 30 min prior to stimulation with GM-CSF for 1 h. After stimulation, cells were lysed and 100 μg of protein lysates were resolved by SDS-polyacrylamide gel electrophoresis, blotted to membranes, and analyzed by immunoblotting with antibodies specific to Mcl-1, Bcl-2, and α-tubulin, respectively.
FIG. 4.
(Top panel) IL-3 induction of mcl-1 expression is attenuated by PI3-K inhibitors. Ba/F3 cells deprived of cytokine were pretreated with various inhibitors as indicated for 30 min prior to stimulation with IL-3 for 1 h. After stimulation, total RNA was prepared from these cells and analyzed by Northern blotting by using a 32P-labeled probe specific for detection of the murine mcl-1 mRNA. (Bottom panel) The same RNA samples stained with ethidium bromide before probing.
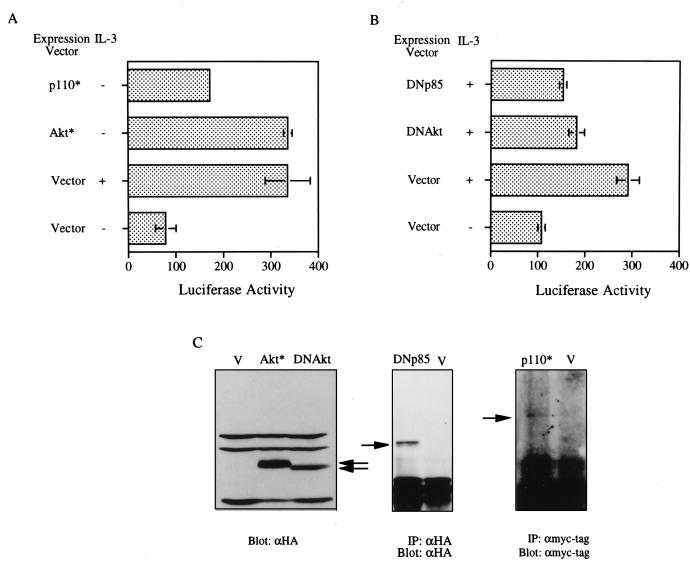
As the inhibitory effect on Mcl-1 expression by wortmannin and LY294002 was also observed at the mRNA levels in TF-1 cells (data not shown), we next tried to determine if the PI3-K/Akt pathway was involved in cytokine activation of the mcl-1 gene transcription. To address this issue, the mcl-1 luciferase reporter gene, p(−1288/+10)mcl-luc, was transiently cotransfected into Ba/F3 cells, with expression plasmids driving the synthesis of the constitutively active PI3-K (P110* [31]), M-Akt, the dominant negative mutant of PI3-K, or the dominant negative mutant of Akt. As shown in Fig. 5A, both P110* and M-Akt activated the mcl-1 reporter gene in the absence of IL-3. The weaker transactivation effect of p110* was probably due to inefficient expression (Fig. 5C) or partial activation of this protein (31) compared to that of M-Akt. On the other hand, overexpression of the dominant negative mutant of PI3-K or of Akt efficiently inhibited IL-3’s ability to stimulate mcl-1 promoter activity (panel B). A similar inhibitory effect was also observed for GM-CSF induction of the same reporter gene in Ba/F3 cells stably overexpressing the hGM-CSF receptor (data not shown). Taken together, these results suggest that the PI3-K/Akt pathway is indeed involved in IL-3 and GM-CSF activation of mcl-1 gene expression.
FIG. 5.
The PI3-K/Akt signaling pathway is involved in IL-3 stimulation of mcl-1 reporter gene expression. Ba/F3 cells were transfected with the mcl-1 reporter gene [p(−1288/+10)mcl-luc] and various expression vectors as indicated by a procedure detailed in Materials and Methods. Twelve (A) or 3 (B) hours after stimulation with IL-3, cell lysates were prepared and analyzed by luciferase assays. Data shown here are representative results from three independent experiments performed in duplicate. Luciferase activities are plotted in arbitrary units. (C) Immunoblots confirming expression of various proteins encoded by individual expression vectors (see legend to Fig. 2 for identification of these vectors). Specific bands are indicated by arrows.
The mcl-1 gene promoter between −197 and −69 is essential for PI3-K/Akt activation.
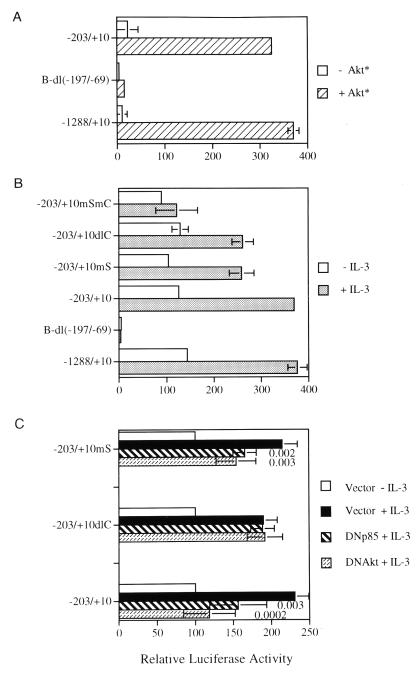
We recently reported that the mcl-1 promoter sequence between −197 and −69 is required for IL-3 and GM-CSF stimulation of this gene (7). We next sought to determine if the same region is necessary for PI3-K/Akt activation of the promoter. By transient cotransfection of Ba/F3 cells with the expression vector encoding the constitutively active Akt and various previously characterized mcl-1 luciferase reporters (see Fig. 6 in reference 7), we demonstrated that mutants with 5′ deletion of the promoter sequence down to −203 (Fig. 6) still retained full inducibility by M-Akt [only data for reporters p(−1288/+10)mcl-luc and p(−203/+10)mcl-luc are shown in Fig. 7A], a result similar to the same reporter activated by IL-3 (compare Fig. 7A and B). However, deletion of the internal region between −197 and −69 from the p(−1288/+10)mcl-luc reporter [i.e., the pB-dl(−197/−69) construct] completely blocked Akt activation of this reporter gene (Fig. 7A). This result suggests that the PI3-K/Akt signaling pathway mediates some stimulatory effects of IL-3 on mcl-1 gene transcription by activation of some transcription factor(s) that directly or indirectly recognize the promoter region between −197 and −69.
FIG. 7.
Mapping of promoter elements required for PI3-K/Akt and IL-3 activation of the mcl-1 reporter gene. (A) Ba/F3 cells cotransfected with the indicated reporter gene and an empty expression vector (−Akt*) or vector encoding M-Akt (+Akt*) were left in medium without IL-3 for 12 h before cell lysates were analyzed for luciferase activity. (B) Same as panel A except that cells were transfected with the indicated reporter construct alone and were left untreated (−IL-3) or stimulated with IL-3 (+IL-3) for 12 h. (C) Ba/F3 cells transfected with the reporter plasmids as indicated plus an expression vector encoding nothing (Vector), DNAkt, or DNp85 were starved and restimulated with IL-3 for 3 h before cell lysates were prepared and analyzed for luciferase activity. The results shown are averages from five independent transfection assays and are plotted as relative activities to cells transfected by the same reporter plasmid and deprived of IL-3. The latter activity was considered to be 100. Numbers inside the bar graph are P values (calculated by the Student t test) for various conditions as indicated.
The CRE-2 and SIE binding motifs contribute to IL-3 stimulation of mcl-1 gene expression via two distinct pathways.
Within the mcl-1 promoter region between −197 and −69, there are DNA elements similar to the SIE and CRE-2 binding motifs (25, 56) (underlined sequences in Fig. 6). We wanted to determine whether these two sequence motifs were involved in IL-3 activation of the mcl-1 gene promoter. By using reporter genes with mutation of either one or both motifs (Fig. 6), we found that mutation of the CRE-2 or SIE site alone (−203/+10dlC and −203/+10mS, respectively) diminished IL-3 induction of the promoter by ∼40% and that mutation of both sites (−203/+10mSmC) resulted in a reporter that lost nearly all IL-3 inducibility (Fig. 7B). These results suggest that both the CRE-2 site and the SIE site do indeed play an important role in the IL-3 stimulation of mcl-1 gene transcription. Since these two sites are located in the promoter region which is essential to PI3-K/Akt activation of the mcl-1 gene (Fig. 7A), we next wanted to determine if mutation of the CRE-2 or the SIE-2 site would affect IL-3 activation of mcl-1 expression through the PI3-K/Akt pathway. For this experiment, the dominant negative mutants of PI3-K or Akt were cotransfected into Ba/F3 cells with the parental reporter (−203/+10) or the same reporter without the SIE (−203/+10mS) or the CRE-2 site (−203/+10dlC) to check if they could block IL-3 stimulation of these reporters. As shown in Fig. 7C, the dominant negative mutant of PI3-K or of Akt inhibited IL-3 activation of both −203/+10 and −203/+10mS reporters. In contrast, IL-3 activation of the mcl-1 reporter without the CRE-2 site (−203/+10dlC) was not affected by either mutant. These results suggest that although CRE-2 and SIE sites both contribute to IL-3 stimulation of mcl-1 gene expression, they apparently play two distinct roles in this process.
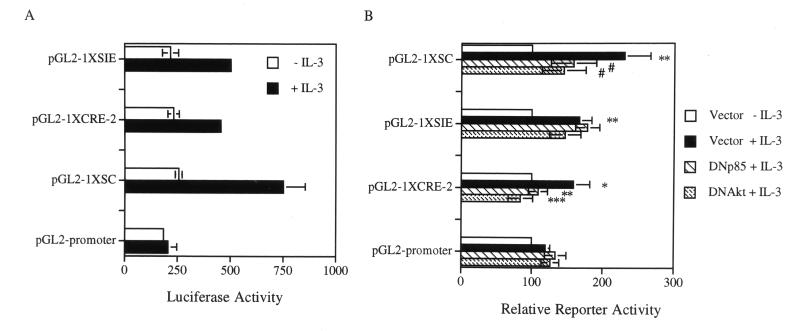
To differentiate further the roles of these two elements in IL-3 signaling, a heterologous promoter placed downstream of a short DNA fragment containing the SIE, CRE-2, or both sites (the SC fragment) was constructed (see Materials and Methods) and analyzed. For this experiment, the simian virus 40 (SV40) promoter in the pGL2 promoter vector (Promega) was selected as the heterologous system, because this promoter had a minimal activity and was not significantly affected by IL-3 in the Ba/F3 cell line used in this assay (Fig. 8). Insertion of a CRE-2 (pGL2-1XCRE-2) or SIE (pGL2-1XSIE) site alone was sufficient to confer IL-3 inducibility on the SV40 promoter (P values calculated by the Student t test for pGL2-1XCRE-2 and pGL2-1XSIE are between 0.01 and 0.02 and between 0.001 and 0.01, respectively), albeit the induction was somewhat weak (≤twofold, compare luciferase activities with or without IL-3 stimulation in Fig. 8A and B). When both sites (the SC fragment) were inserted into this reporter system (pGL2-1XSC), the IL-3 inducibility of the SV40 promoter was additively increased (Fig. 8). We next examined whether IL-3 activation of these reporter genes was mediated through the PI3-K/Akt pathway. To examine this possibility, the dominant negative mutants of PI3-K or of Akt were cotransfected in the same reporter gene assays to determine whether they could block IL-3 stimulation of these reporters. While expression of the dominant negative mutant of PI3-K or of Akt did not have any significant effect on the IL-3 stimulation of the SIE reporter (pGL2-1xSIE), the expression of either dominant negative mutant completely inhibited IL-3 stimulation of the CRE-2 reporter (pGL2-1xCRE, P < 0.001 and 0.001 < P < 0.01 for the dominant negative mutant of Akt and that of PI3-K, respectively) (Fig. 8B). Interestingly, the IL-3 inducibility of the SC reporter (pGL2-1XSC) was only partially inhibited (0.02 < P < 0.05 for both mutants). Taken together, these results clearly indicate that IL-3 stimulation of the mcl-1 promoter is mediated through modulation of at least two transcription factors, the CRE-2 and SIE binding proteins, via activation of the PI3-K/Akt-dependent and -independent pathways, respectively.
FIG. 8.
(A) CRE-2 and SIE binding motifs can individually confer IL-3 inducibility on a heterologous promoter (SV40 minimal promoter) and can work additively in IL-3 stimulation of the same reporter system. Ba/F3 cells transfected with various reporter plasmids as indicated were left untreated (−IL-3) or stimulated with IL-3 (+IL-3) before cell lysates were prepared and analyzed for luciferase activity. Data shown here are representative of three independent experiments performed in triplicate. (B) IL-3 stimulation of the CRE-2 but not the SIE reporter is mediated through the PI3-K/Akt kinase pathway. Ba/F3 cells transfected with the reporter plasmids as indicated plus an expression vector encoding nothing (Vector), DNAkt, or DNp85 were processed and analyzed for the luciferase activities as described in the legend to Fig. 7C. The data shown here are averages from five to six independent experiments, and the reporter activities of cells transfected with each reporter but treated under various conditions are plotted relative to cells transfected by the same reporter plasmid and deprived of IL-3. The latter activity was considered to be 100. The IL-3 induction of each reporter was statistically significant compared to that of the parental reporter (pGL2 promoter), as was the dominant negative effect of DNp85 or DNAkt on the CRE-2 or the SC reporter. The P values are as follows: ∗, 0.01 < P < 0.02; ∗∗, 0.001 < P < 0.01; ∗∗∗, P < 0.001; #, 0.02 < P < 0.05.
CRE-2 but not SIE binding activity is stimulated by IL-3 via the PI3-K/Akt pathway.
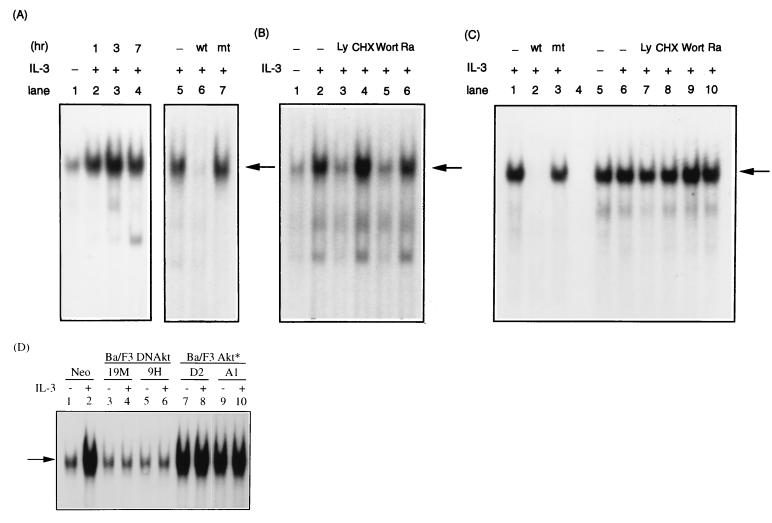
To further examine how the IL-3 signaling pathway modulates the activities of CRE-2 and SIE binding proteins, gel-shift assays with nuclear extracts prepared from cells after various treatments were carried out. As shown in Fig. 9A, judging from the band (indicated by an arrow) that was specifically inhibited by the wild-type but not by the mutant oligonucleotide (compare lanes 6 and 7), CRE-2 binding activity was very low in cells deprived of cytokine (Fig. 9A, lane 1) and was rapidly induced within 1 h after stimulation of cells with IL-3 (Fig. 9A, lanes 2 to 4). The fact that cytokine stimulation of mcl-1 gene expression does not require any new protein synthesis (7) prompted us to examine the possibility that the CRE-2 binding protein(s) preexisted in cells prior to IL-3 stimulation. The result shown in Fig. 9B indicates that this was indeed the case, since pretreatment of cells with the protein synthesis inhibitor, cycloheximide, did not interfere with IL-3 stimulation of CRE-2 binding activity (lane 4). Furthermore, since IL-3 stimulation of CRE-2 reporter activity was mediated via the PI3-K/Akt pathway, we next examined whether CRE-2 binding activity was also affected by this signaling pathway. The result shown in Fig. 9B indicates that CRE-2 binding activity was greatly reduced in extracts prepared from cells pretreated with wortmannin or LY294002 prior to IL-3 stimulation (compare lanes 3 and 5 to lane 2). Interestingly, rapamycin, which did not exert any inhibitory effect on IL-3 induction of mcl-1 expression (Fig. 4), did not have any significant effect on the IL-3 stimulation of CRE-2 binding activity (Fig. 9B, lane 6). As to the SIE binding activity, a rather different pattern was observed. SIE binding activity was quite abundant in cytokine-deprived cells and was not further induced by IL-3 treatment (compare lanes 5 and 6 in Fig. 9C). Furthermore, this binding activity was not affected by any of the above-mentioned inhibitors (Fig. 9C, compare lanes 7 to 10 to lane 6).
FIG. 9.
CRE-2 but not SIE binding activity is activated by IL-3 through the PI3-K/Akt-dependent pathway. (A) The gel-shift assay with probe CRE-2 and nuclear extracts from Ba/F3 cells under various treatments was carried out as described in Materials and Methods. Specific DNA-protein complexes (pointed by an arrow) were resolved on a nondenaturing gel and visualized by autoradiography. Lane 1, extracts from cells deprived of IL-3; lanes 2 to 4, extracts from cells deprived of cytokine and restimulated with IL-3 for 1, 3, or 7 h, respectively; lane 5, same as lane 2; lanes 6 and 7, same as lane 5 except that 100 molar excess of nonlabeled oligonucleotide containing the wild type (lane 6) or mutant (lane 7) CRE-2 site was included in the binding reaction. (B) Same experiment as that shown in panel A except that extracts were made from cells deprived of cytokine (lane 1), from cells restimulated with IL-3 for 1 h (lane 2), or from cells that were preincubated with Ly294002 (lane 3), cycloheximide (lane 4), wortmannin (lane 5), or rapamycin (lane 6), respectively, for 30 min before stimulation with IL-3. (C) Same experiment as carried out in panels A and B except that the labeled SIE probe was used in the gel-shift assay. Lanes 2 and 3 show the results of competition experiments with a 100 molar excess of wild-type or mutant SIE oligonucleotide probe, respectively; lane 4, a lane without loading any sample; lanes 5 to 10, same treatments as those shown in lanes 1 to 6 of panel B. (D) Results for the same experiment as that whose results are shown in lanes 1 and 2 of panel B, except that extracts were made from various cells, as indicated at the top. Odd-numbered lanes, extracts made from cells deprived of cytokine; even-numbered lanes, extracts from cells restimulated with IL-3 for 1 h.
Next, we examined if activation of the Akt kinase was required for IL-3 activation of CRE-2 binding activity. For this experiment, similar gel-shift assays were performed with extracts made from Ba/F3 cells stably overexpressing the constitutively active (Ba/F3Akt*) or the dominant negative mutant (Ba/F3DNAkt) of the Akt kinase. As shown in Fig. 9D, CRE-2 binding activity was no longer induced by IL-3 in either clone (9H and 19M) of Ba/F3DNAkt cells (lanes 3 to 6), whereas this binding activity was constitutively active (irrespective of the presence or absence of IL-3) in both clones (A1 and D2) of Ba/F3Akt* cells. These results, together with that shown in Fig. 9B, strongly indicated that CRE-2 binding activity was activated by IL-3 via the PI3-K/Akt pathway. Taken together, these results indicate that although both CRE-2 and SIE motifs contribute to IL-3 stimulation of mcl-1 gene expression, proteins binding to these two sites are differentially modulated by the IL-3 signaling pathway.
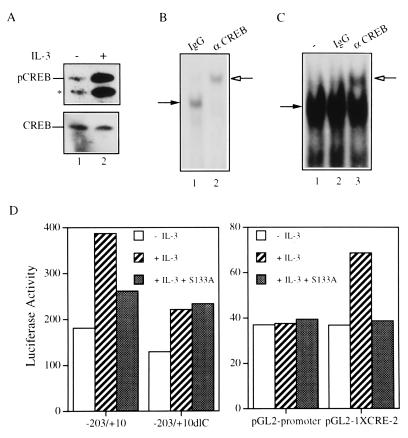
CREB exists in the CRE-2 binding complex and plays a role in IL-3 regulation of mcl-1 gene expression.
IL-3 was reported to induce phosphorylation of CREB on serine 133 in TF-1 cells (35). This was also the case in Ba/F3 cells, as revealed by an increased amount of S-133 phosphorylated CREB in cells stimulated with IL-3 (compare lanes 1 and 2 in Fig. 10A). The CRE-2 site was demonstrated to bind CREB in rat liver nuclear extracts (45). We therefore examined whether CREB exists in the CRE-2 binding complex characterized in Ba/F3 cells. Using gel-shift assays, we observed that CREB synthesized in vitro with the TNT coupled reticulocyte lysate system (Promega) bound specifically to the CRE-2 probe at an affinity lower than that with which it bound to an oligonucleotide containing the CRE site (45, 56a). The addition of the CREB-specific antibody (Fig. 10B, lane 2), but not that of the control rabbit immunoglobulin G (IgG) (Fig. 10B, lane 1), supershifted the specific protein complex. Figure 10C shows that the same CREB antibody also supershifted the protein complex formed on the CRE-2 site in extracts made from IL-3-stimulated cells (compare lanes 2 and 3). This result indicates that a CREB-like protein is one component of the CRE-2 binding complex and suggests that CREB or a closely related member plays a role in the IL-3 regulation of mcl-1 gene expression. To examine this latter possibility, we tested whether blocking CREB activity by its dominant negative mutant (CREBS133A) interferes with IL-3 stimulation of mcl-1 reporter activity. From three independent transfection experiments, we consistently observed that IL-3 stimulation of luciferase activities in both mcl-1 and CRE-2 reporters [p(−203/+10)mcl-luc and pGL2-1XCRE-2] was significantly reduced in cells cotransfected with an expression vector driving synthesis of CREBS133A. However, IL-3 induction of the mcl-1 reporter lacking the CRE-2 site (−203/+10dlC) was not affected by the coexpression of the CREBS133A protein (Fig. 10D).
FIG. 10.
CREB is one component of the CRE-2 binding complex and plays a role in the IL-3 stimulation of the mcl-1 and CRE-2 reporters. (A) Ba/F3 cells deprived of IL-3 overnight (lane 1) or restimulated with IL-3 for 5 min (lane 2) were lysed, and equal amounts of cell lysates were analyzed by immunoblotting by using antibodies specific to S133-phosphorylated CREB (upper panel) or to all forms of CREB (lower panel). The asterisk denotes a cross-reactive protein (possibly phosphorylated ATF-1) recognized by the anti-pCREB antibody. (B) The CREB protein synthesized in vitro with the TNT-coupled reticulocyte lysate system was allowed to bind to the CRE-2 probe in the presence of a control rabbit IgG (lane 1) or purified rabbit anti-CREB antibody (lane 2) prior to being resolved on the nondenaturing gel. (C) Results for the same experiment as that whose results are shown in panel B, except that IL-3-stimulated (1 h) Ba/F3 cell extracts were used in the gel-shift assay. Lane 1, no antibody was present in the binding reaction; lanes 2 and 3, binding assays with control or CREB-specific antibody, respectively. The solid arrow indicates the specific DNA-protein complex, and the open arrow indicates the band supershifted by the antibody. (D) Ba/F3 cells transiently transfected with a reporter gene, as indicated, plus a control or CREB S133A expression vector were deprived of IL-3 or restimulated with IL-3 for 3 h before cell lysates were prepared and analyzed for luciferase activity. Very similar results were obtained from three independent transfection assays, and data from one representative experiment are shown.
DISCUSSION
The PI3-K/Akt signaling pathway is involved in the survival effect of many growth factors and some transforming oncogenes (1, 16, 28, 34, 41, 54, 55). Recent findings demonstrate that the proapoptotic protein Bad is one cellular target that PKB may phosphorylate to protect cells from apoptosis (12, 13). However, Bad has a restricted tissue distribution, and not every survival signal that activates Akt stimulates Bad phosphorylation (52), suggesting that PKB may exert its antiapoptotic effect via the activation or inactivation of other cellular targets. In the present study, we provided evidence that Mcl-1, the survival factor activated by GM-CSF and IL-3, is another cellular target of the PI3-K/Akt signaling pathway. Unlike the effect on Bad, the PI3-K/Akt pathway upregulates Mcl-1 expression at the transcriptional level. As activation of the PI3-K/Akt pathway and Mcl-1 expression are both essential to the survival effect of GM-CSF and IL-3 (7, 55, and this report), our results strongly suggest that transcriptional activation of mcl-1 gene expression by the PI3-K/Akt pathway is one important mechanism by which GM-CSF and IL-3 exert their survival activity. Our previous results (7) and the one shown in Fig. 1E indicate that overexpression of Mcl-1 alone delays but does not completely abolish cytokine withdrawal-induced apoptosis, suggesting that other cytokine-activated events are required for full survival activity of GM-CSF and IL-3. It is possible that PI3-K/Akt pathway-mediated phosphorylation of Bad and transcriptional activation of mcl-1 gene expression plus other yet-to-be-identified events together contribute to the survival effect of GM-CSF and IL-3. The antiapoptotic protein A1 is another Bcl-2 family member that is transcriptionally activated by the GM-CSF signaling pathway (38). It would be interesting to examine if the PI3-K and Akt pathway is also involved in the GM-CSF activation of the A1 gene.
Activation of Mcl-1 expression by the IL-3 and GM-CSF signaling pathways occurs at the transcriptional level (7). In the present study, deletion and site-directed mutagenesis analysis indicated that both SIE and CRE-2 DNA elements located in the mcl-1 gene promoter contribute to cytokine activation of mcl-1 gene transcription. Deletion of either element diminished IL-3 response by ∼40%. Either element alone was sufficient to confer IL-3 inducibility on a heterologous promoter, suggesting that both SIE and CRE-2 elements can function independently of each other but can also work additively to mediate IL-3 stimulation of mcl-1 gene expression. Although both SIE and CRE-2 binding factors may be regulated by the same IL-3-activated signaling pathway, results from this study do not support this hypothesis and, to the contrary, provide strong evidence that the SIE and CRE-2 elements are regulated by different IL-3-activated signaling pathways. The CRE-2 element is activated via the PI3-K/Akt pathway, whereas activation of SIE is mediated through another pathway yet to be identified.
mcl-1 is an immediate-early gene activated by IL-3 and GM-CSF, and cytokine activation of this gene does not require new protein synthesis (7). Consistent with this result, in the present study, we demonstrated that the protein factor(s) binding to the CRE-2 element is induced by IL-3 with kinetics similar to those for the IL-3 induction of mcl-1 mRNA expression, and the induction persists even in the presence of the protein synthesis inhibitor. This finding suggests that the CRE-2 binding protein(s) plays a major role in the cytokine regulation of mcl-1 gene transcription. The CRE-2 site, TGCGTCA, has sequence homology to the AP-1/TRE (TGACTCA) (3, 37) and the ATF/CRE sequence motifs (TGACGTCA) (36, 43) and is an essential element for transcriptional activation of the proenkephalin gene in response to cyclic AMP (cAMP) and phorbol ester stimulation (8). Many members from the Fos/Jun and the ATF/CREB protein family have been reported to bind to the CRE-2 element in vitro (21, 45). However, the protein complexes formed on this site in vivo appear to vary in a gene- and cell type-specific manner (39). Although the full identity of the CRE-2 binding complex formed in IL-3-stimulated Ba/F3 cells remains to be characterized, our results strongly suggest that CREB is one component of this protein complex. Furthermore, based on the following observations, CREB is likely to be the nuclear target that mediates PI3-K/Akt activation of mcl-1 gene expression. First, IL-3-activated CRE-2 binding and transcriptional activities are both dependent on the PI3-K/Akt kinase pathway. Second, IL-3 induction of the mcl-1 reporter containing the CRE-2 site, but not that of the same reporter without the CRE-2 site, is dependent on activation of the CREB protein (Fig. 10). Third, CREB has recently been shown to be a regulatory target for the Akt kinase (15). The fact that Akt translocates to the nucleus upon growth factor stimulation (2, 40) suggests that upon IL-3 stimulation, Akt translocates to the nucleus, where it phosphorylates CREB directly or indirectly and activates transcription of the mcl-1 gene. This potential scenario warrants further investigation.
As mentioned earlier, another DNA element located at position −87 (the SIE site) also contributes to IL-3 activation of the mcl-1 gene. Like the CRE-2 element, the SIE site alone is sufficient to confer IL-3 inducibility on a heterologous promoter. However, IL-3 regulation of this DNA element appears to be quite different from that of the CRE-2 DNA motif. First, IL-3 activates the SIE site through a PI3-K/Akt-independent pathway. Second, the SIE binding activity is constitutively high in Ba/F3 cells and is not enhanced upon IL-3 stimulation. This result suggests that the SIE binding protein preassociates with the mcl-1 gene promoter on its binding site and that its transcriptional activity is activated by IL-3 through a pathway yet to be identified. The core sequence (shown in uppercase letters below) of the SIE site located in the mcl-1 gene promoter (gacttCCCGTAaatc) differs from that of the SIE in the c-fos gene promoter (cagttCCCGTCaatc) by one nucleotide (underlined base) and is identical to that of the synthetic, high-affinity SIE site (m67, catttCCCGTAaatc) as previously reported (56). In addition, five of nine nucleotides in the flanking sequence (lowercase letters) differ between the SIE in the mcl-1 gene and that in the c-fos promoter or in the m67 oligonucleotide. The SIE site contributes to the GM-CSF activation of the c-fos promoter, and the protein binding to the high-affinity SIE site (m67 oligonucleotide probe) is very low in cells deprived of growth factors and is highly inducible by GM-CSF in TF-1 (47) or by EGF in A431 cells (49). In contrast, the protein(s) binding to the SIE site in the mcl-1 gene promoter is quite abundant, and its binding activity is not affected by IL-3, suggesting that different protein complexes form on different SIE sites and that the binding components also vary among the cell types studied. The STAT family of proteins bind to the SIE site in the c-fos promoter (47). Whether any STAT protein contributes to IL-3 activation of the SIE element in the mcl-1 gene promoter remains to be determined. Further experiments are required to reveal the identity of proteins formed on the SIE site in the mcl-1 gene promoter and to explore how this protein complex may work additively with the CRE-2 binding protein in the IL-3 regulation of mcl-1 gene expression.
ACKNOWLEDGMENTS
We thank R.-H. Chen for plasmid p110*, Anke Klippel for plasmids expressing M-Akt and AktK179M, Masato Kasuga for Δp85 expression vector, and M.-Z. Lai for anti-CREB antibody and critical reading of the manuscript.
This work was supported in part by an intramural fund from Academia Sinica and by grant NSC-87-2311-B-001-096 from the National Science Council of Taiwan to H.-F. Yang-Yen.
REFERENCES
- 1.Ahmed N N, Grimes H L, Bellacosa A, Chan T O, Tsichlis P N. Transduction of interleukin-2 antiapoptotic and proliferative signals via Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:3627–3632. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andjelkovic M, Alessi D R, Meier R, Fernanadez A, Lamb N J, Frech M, Cron P, Cohen P, Lucocq J M, Hemmings B A. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 3.Angel P, Imagawa M, Chiu R, Stein B, Imbra R J, Rahmsdorf H J, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 4.Arai K, Lee F, Miyajima A, Arai N, Yokota T. Cytokines: coordinators of immune and inflammatory responses. Annu Rev Biochem. 1990;59:783–836. doi: 10.1146/annurev.bi.59.070190.004031. [DOI] [PubMed] [Google Scholar]
- 5.Azam M, Erdjument-Bromage H, Kreider B L, Xia M, Quelle F, Basu R, Saris C, Tempst P, Ihle J N, Schindler C. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgering B M T, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 7.Chao J-R, Wang J-M, Lee S-F, Peng H-W, Lin Y-H, Chou C-H, Li J-C, Huang H-M, Chou C-K, Kuo M-L, Yen J J-Y, Yang-Yen H-F. mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is one component of the GM-CSF viability response. Mol Cell Biol. 1998;18:4883–4898. doi: 10.1128/mcb.18.8.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comb M, Mermod N, Hyman S E, Pearlberg J, Ross M E, Goodman H M. Proteins bound at adjacent DNA elements act synergistically to regulate human proenkephalin cAMP inducible transcription. EMBO J. 1988;7:3793–3805. doi: 10.1002/j.1460-2075.1988.tb03264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conscience J F, Verrier B, Martin G. Interleukin-3-dependent expression of the c-myc and c-fos proto-oncogenes in hemopoietic cell lines. EMBO J. 1986;5:317–323. doi: 10.1002/j.1460-2075.1986.tb04215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corey S, Eguinoa A, Puyana-Theall K, Bolen J B, Cantley L, Mollinedo F, Jackson T R, Hawkins P T, Stephens L R. Granulocyte macrophage-colony stimulating factor stimulates both association and activation of phosphoinositide 3-OH-kinase and src-related tyrosine kinase(s) in human myeloid derived cells. EMBO J. 1993;12:2681–2690. doi: 10.1002/j.1460-2075.1993.tb05929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 12.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 13.Del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 14.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 16.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 17.Duronio V, Clark-Lewis I, Federsppiel B, Wieler J S, Schrader J W. Tyrosine phosphorylation of receptor beta subunits and common substrates in response to interleukin-3 and granulocyte-macrophage colony-stimulating factor. J Biol Chem. 1992;267:21856–21863. [PubMed] [Google Scholar]
- 18.Eves E M, Xiong W, Bellacosa A, Kennedy S G, Tsichlis P N, Rosner M R, Hay N. Akt, a target of phosphatidylinositol 3-kinase, inhibits apoptosis in a differentiating neuronal cell line. Mol Cell Biol. 1998;18:2143–2152. doi: 10.1128/mcb.18.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 20.Gold M R, Duronio V, Saxena S P, Schrader J W, Aebersold R. Multiple cytokines activate phosphatidylinositol 3-kinase in hemopoietic cells. Association of the enzyme with various tyrosine-phosphorylated proteins. J Biol Chem. 1994;269:5403–5412. [PubMed] [Google Scholar]
- 21.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanazono Y, Chiba S, Sasaki K, Mano H, Miyajima A, Arai K, Yazaki Y, Hirai H. c-fps/fes protein-tyrosine kinase is implicated in a signaling pathway triggered by granulocyte-macrophage colony-stimulating factor and interleukin-3. EMBO J. 1993;12:1641–1646. doi: 10.1002/j.1460-2075.1993.tb05809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson T R, Hawkins P T, Dhand R, Clark A E, Holman G D, Waterfield M D, Kasuga M. Phosphatidylinositol 3-kinase activity is required for insulin-stimulated glucose transport but not for RAS activation in CHO cells. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsueh Y P, Lai M-Z. Overexpression of activation transcriptional factor 1 in lymphomas and in activated lymphocytes. J Immunol. 1995;154:5675–5683. [PubMed] [Google Scholar]
- 25.Hyman S E, Comb M, Lin Y-S, Pearlberg J, Green M R, Goodman H M. A common trans-acting factor is involved in transcriptional regulation of neurotransmitter genes by cyclic AMP. Mol Cell Biol. 1988;8:4225–4233. doi: 10.1128/mcb.8.10.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 27.Kennedy S G, Wagner A J, Conzen S D, Jordan J, Bellacosa A, Tsichlis P N, Hay N. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 28.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinoshita T, Yokota T, Arai K, Miyajima A. Suppression of apoptotic death in hematopoietic cells by signalling through the IL-3/GM-CSF receptors. EMBO J. 1995;14:266–275. doi: 10.1002/j.1460-2075.1995.tb07000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao Y F, Miyazono K, Urabe A, Takaku F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 31.Klippel A, Reinhard C, Kavanaugh W M, Apell G, Escobedo M-A, Williams L T. Membrane localization of phosphatidylinositol 3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol Cell Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohn A D, Kovacina K S, Roth R A. Insulin stimulates the kinase activity of RAC-PK, a pleckstrin homology domain containing ser/thr kinase. EMBO J. 1995;14:4288–4295. doi: 10.1002/j.1460-2075.1995.tb00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozopas K M, Yang T, Buchan H L, Zhou P, Craig R W. Mcl-1, a gene expressed in programmed myeloid cell differentiation has sequence similarity to Bcl2. Proc Natl Acad Sci USA. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulik G, Klippel A, Weber M J. Antiapoptotic signalling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H-J J, Mignacca R C, Sakamoto K M. Transcriptional activation of egr-1 by granulocyte-macrophage colony-stimulating factor but not interleukin 3 requires phosphorylation of cAMP response element-binding protein (CREB) on serine 133. J Biol Chem. 1995;270:15979–15983. doi: 10.1074/jbc.270.27.15979. [DOI] [PubMed] [Google Scholar]
- 36.Lee K A, Hai T Y, SivaRaman L, Thimmappaya B, Hurst H C, Jones N C, Green M R. A cellular protein, activating transcription factor, activates transcription of multiple ElA-inducible adenovirus early promoters. Proc Natl Acad Sci USA. 1987;84:8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 38.Lin E Y, Orlofsky A, Berger M S, Prystowsky M B. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J Immunol. 1993;151:1979–1988. [PubMed] [Google Scholar]
- 39.MacArthur L. AP-1-related proteins bind to the enkephalin CRE-2 element in adrenal chromaffin cells. J Neurochem. 1996;67:2256–2264. doi: 10.1046/j.1471-4159.1996.67062256.x. [DOI] [PubMed] [Google Scholar]
- 40.Meier R, Alessi D R, Cron P, Andjelkovic M, Hemmings B A. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase B beta. J Biol Chem. 1997;272:30491–30497. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- 41.Minshall C, Arkins S, Freund G G, Kelley K W. Requirement for phosphatidylinositol 3′-kinase to protect hemopoietic progenitors against apoptosis depends upon the extracellular survival factor. J Immunol. 1996;156:939–947. [PubMed] [Google Scholar]
- 42.Miyajima A, Mui A L-F, Ogorochi T, Sakamaki K. Receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3 and interleukin-5. Blood. 1993;82:1960–1974. [PubMed] [Google Scholar]
- 43.Montminy M R, Sevarino K A, Wagner J A, Mandel G, Goodman R H. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mui A L-F, Wakao H, O’Farrell A M, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony-stimulating factor and interleukin-5 transduce signals through two STAT5 homologues. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nichols M, Weih A, Schmid W, DeVack C, Kowenz-Leutz E, Luckow B, Boshart M, Schutz G. Phosphorylation of CREB affects its binding to high and low affinity sites: implications for cAMP induced gene. EMBO J. 1992;11:3337–3346. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quelle F W, Sato N, Witthuhn B A, Inhorn R C, Eder M, Miyajima A, Griffin J D, Ihle J N. Jak2 associates with the βc chain of the receptor for granulocyte-macrophage colony-stimulating factor, and its activation requires the membrane-proximal region. Mol Cell Biol. 1994;14:4335–4341. doi: 10.1128/mcb.14.7.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajotte D, Sadowski H B, Haman A, Gopalbhai K, Meloche S, Liu L, Krystal G, Hoang T. Contribution of both STAT and SRF/TCF to c-fos promoter activation by granulocyte-macrophage colony-stimulating factor. Blood. 1996;88:2906–2916. [PubMed] [Google Scholar]
- 48.Reynolds J E, Yang T, Qian L, Jenkinson J D, Zhou P, Eastman A, Craig R W. Mcl-1, a member of the Bcl-2 family, delays apoptosis induced by c-Myc overexpression in Chinese hamster ovary cells. Cancer Res. 1994;54:6348–6352. [PubMed] [Google Scholar]
- 49.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 50.Sato N, Sakamaki K, Terada N, Arai K, Miyajima A. Signal transduction by the high-affinity GM-CSF receptor: two distinct cytoplasmic regions of the common beta subunit responsible for different signaling. EMBO J. 1993;12:4181–4189. doi: 10.1002/j.1460-2075.1993.tb06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheid M P, Lauener R W, Duronio V. Role of phosphatidylinositol 3-OH-kinase activity in the inhibition of apoptosis in haemopoietic cells: phosphatidylinositol 3-OH-kinase inhibitors reveal a difference in signalling between interleukin-3 and granulocyte-macrophage colony stimulating factor. Biochem J. 1995;312:159–162. doi: 10.1042/bj3120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheid M P, Duronio V. Dissociation of cytokine-induced phosphorylation of Bad and activation of PKB/akt: involvement of MEK upstream of Bad phosphorylation. Proc Natl Acad Sci USA. 1998;95:7439–7444. doi: 10.1073/pnas.95.13.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silvennoinen O, Witthuhn B, Quelle F W, Cleveland J L, Yi T, Ihle J N. Structure of the Jak2 protein tyrosine kinase and its role in IL-3 signal transduction. Proc Natl Acad Sci USA. 1993;90:8429–8433. doi: 10.1073/pnas.90.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi J K, Trotta R, Wlodarski P, Perrotti D, Chan T O, Wasik M A, Tsichlis P N, Calabretta B. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Interleukin 3-dependent survival by the Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Wang et al. Unpublished results.
- 57.Yang T, Kozopas K M, Craig R W. The intracellular distribution and pattern of expression of Mcl-1 overlap with, but are not identical to, those of Bcl-2. J Cell Biol. 1995;128:1173–1184. doi: 10.1083/jcb.128.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang-Yen H-F, Chambard J-C, Sun Y-L, Smeal T, Schmidt T J, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 59.Yen J J-Y, Hsieh Y C, Yen C L, Chang C C, Lin S, Yang-Yen H F. Restoring the apoptosis suppression response to IL-5 confers on erythroleukemic cells a phenotype of IL-5-dependent growth. J Immunol. 1995;154:2144–2152. [PubMed] [Google Scholar]
- 60.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou P, Qian L, Kozopas K M, Craig R W. Mcl-1, a bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood. 1997;89:630–643. [PubMed] [Google Scholar]