Summary
Thoracic aortic aneurysm (TAA) is characterized by dilation of the aortic root or ascending/descending aorta. TAA is a heritable disease that can be potentially life threatening. While 10%–20% of TAA cases are caused by rare, pathogenic variants in single genes, the origin of the majority of TAA cases remains unknown. A previous study implicated common variants in FBN1 with TAA disease risk. Here, we report a genome-wide scan of 1,351 TAA-affected individuals and 18,295 control individuals from the Cardiovascular Health Improvement Project and Michigan Genomics Initiative at the University of Michigan. We identified a genome-wide significant association with TAA for variants within the third intron of TCF7L2 following replication with meta-analysis of four additional independent cohorts. Common variants in this locus are the strongest known genetic risk factor for type 2 diabetes. Although evidence indicates the presence of different causal variants for TAA and type 2 diabetes at this locus, we observed an opposite direction of effect. The genetic association for TAA colocalizes with an aortic eQTL of TCF7L2, suggesting a functional relationship. These analyses predict an association of higher expression of TCF7L2 with TAA disease risk. In vitro, we show that upregulation of TCF7L2 is associated with BCL2 repression promoting vascular smooth muscle cell apoptosis, a key driver of TAA disease.
Keywords: thoracic aortic aneurysm, GWAS, regulatory variant, TCF7L2, VSMC, apoptosis
Introduction
Thoracic aortic aneurysm (TAA) is characterized by dilation of the aortic root, ascending aorta, the aortic arch, and descending aorta. TAA often develops asymptomatically over a period of time, and thus, early diagnosis of TAA is quite challenging. Aneurysms in thoracic aorta are frequently followed by rupture or dissection of aortic tissue, which can be fatal. Early diagnosis through selective screening of individuals with the highest genetic risk of TAA can potentially reduce the risk of death through early surgical intervention.1
Approximately 10%–20% of TAA cases are monogenic, i.e., due to rare, pathogenic variants in single genes.2,3 Variants in at least 11 such genes (ACTA2, COL3A1, FBN1, MYH11, SMAD3, TGFB2, TGFBR1, TGFBR2, MYLK, LOX, and PRKG1) are associated with strong risk of TAA4 and were primarily identified through family studies. Monogenic TAAs are often present with syndromic heritable thoracic aortic disorders, including Marfan, Loeys-Dietz, Ehlers-Danlos, and smooth muscle dysfunction syndromes.5,6 Moreover, approximately 50% of individuals with congenital heart malformations, such as bicuspid aortic valve (BAV), later develop TAA.7 Yet the origin of the majority of TAA cases remains unknown. It is likely that these cases have more complex origins, involving common genetic variants affecting many genes. Genome-wide association studies (GWASs) have been particularly successful in identifying common genetic variants that increase disease risk. Previously, common variants in FBN1 were found to be associated with TAA.8 Another study implicated LRP1 and ULK4 in acute thoracic aortic dissection.9
In an attempt to identify genetic associations with TAA, we performed a GWAS of 1,351 TAA-affected individuals and 18,295 control individuals and identified a previously unpublished genome-wide significant locus in the third intron of TCF7L2. We replicated this signal with meta-analysis of four additional independent cohorts. At this locus, an inverse association between TAA and type 2 diabetes (T2D) was observed. The association signal colocalizes with an eQTL of TCF7L2 in aorta, providing functional insight into the genetic association. In vitro, we show a potential role of TCF7L2 in vascular smooth muscle cell (VSMC) apoptosis, a key TAA disease mechanism.
Material and methods
Description of the discovery cohorts
The Cardiovascular Health Improvement Project (CHIP) is a cohort with genotype data linked to electronic health records, family history, and aortic tissue from individuals seen at Michigan Medicine. The Michigan Genomics Initiative (MGI) is a hospital-based cohort with genotype data linked to electronic health records from individuals recruited during pre-surgical encounters at Michigan Medicine. Both studies were approved by the Institutional Review Board of the University of Michigan Medical School (IRBMED) (HUM00052866 and HUM00071298) and informed consent was obtained from study participants.
Genotype, QC, and imputation
CHIP and MGI samples were genotyped with two array versions of customized Illumina Infinium CoreExome-24 bead arrays: UM_HUNT_Biobank_11788091_A1 and UM_HUNT_Biobank_v1-1_20006200_A. A detailed description of these two arrays can be found in Fritsche et al.10 Genotype calling was performed with Illumina GenomeStudio. Extensive quality control (QC) was performed prior to imputation. For CHIP samples, we first excluded samples with (1) rotated/swapped plates or DNA extraction batches, (2) duplicate/redundant samples with lower call rate, (3) identical ID and discordant genotypes or different ID but identical genotypes (both samples excluded), (4) gender discordance (unusual XY composition/no gender information available; no gender check possible/reported gender different from inferred sex), (5) large copy number variant, (6) estimated contamination > 2.5% (BAF Regress11), and (7) sample call rate < 99%. In all samples, we excluded variants with (1) call rate < 90%, (2) GenTrain score < 0.15, and (3) cluster separation < 0.3. After this, individual batches in each array version were merged. We also obtained MGI samples with similar processing as described in Fritsche et al.10 Next, from three sets (two CHIP array versions and MGI), we excluded samples with (1) outlier heterozygosity and (2) non-European ancestry as identified by laser12 and variants with (1) call rate < 99% or (2) deviation from Hardy-Weinberg equilibrium (p value < 10−4). Further analysis was restricted to variants present in both the MGI and two CHIP array versions among unrelated (>3rd degree) individuals identified via KING.13 Variants with allele frequency differences between two array versions in CHIP (Chi-square; p < 0.001) were removed. Monomorphic variants were also excluded at this stage, generating 457,758 polymorphic variants from 42,119 samples after QC. As a preparation for imputation with the Haplotype Reference Consortium (HRC) panel,14 we further excluded 71,942 variants from this set by using McCarthy group tools. The 385,816 variants that passed QC were used as input in the Michigan Imputation server. Imputation was performed with the HRC reference panel via Minimac415 with Rsq filter 0.3.
Case-control selection and association analysis
TAA-affected individuals were identified from both CHIP and MGI while control individuals were identified from MGI. Affected individuals in CHIP (n = 956) were defined as having an aneurysm in the thoracic aorta following diagnosis by cardiologists. Individuals with a known diagnosis of BAV were excluded because this is a strong risk factor of TAA, presumably because of altered haemodynamic blood flow and aortopathy instead of shared genetic architecture or molecular mechanisms.16,17 Affected individuals in MGI (n = 395) were identified via International Classification of Diseases (ICD) codes (ICD9, 441.1/441.2; ICD10, I71.1/I71.2) excluding samples with a diagnosis of BAV (ICD9, 746.4; ICD10, Q23.1). After removing samples with any related diseases from the potential control individuals (phecodes 440–449.99), we used a case-control matching strategy to identify 18,295 control individuals from MGI (Table S1). We used R package MatchIt and applied nearest neighbor matching for birth year, PC1–4 (using Mahalanobis-metric matching), and exact matching for sex and array version. Association analysis was performed via SAIGE18 with sex, birth year, array version, and PC1–4 as covariates. We reported results for all variants with minimum minor allele frequency (MAF) of 0.01 and minor allele count of 5.
Replication data and meta-analysis
The replication study from the University of Texas Health Science Center at Houston was approved by the institutional review board (IRB) (HSC-MS-01-251) and all subjects signed consent forms at recruitment to the study. The phenotyping and case-control association analysis is described in LeMaire et al.8 Briefly, this data comprises 765 individuals of European ancestry with sporadic ascending aortic aneurysms or classic aortic dissection of the ascending or descending thoracic aorta (Stanford types A and B, respectively) and 875 control individuals. A logistic regression model was used for testing of variant association with the phenotype.
The VA Million Veteran Program (MVP) is comprised of veterans aged 18 to >100 years recruited from VA healthcare centers across the United States with genetic data linked to electronic health records. The study was approved by the VA central IRB and all participants provided informed consent. In MVP, 6,554 unrelated TAA-affected individuals of European ancestry were identified on the basis of the occurrence of a TAA ICD code (ICD9, 441.1/ 441.2; ICD10, I71.1/I71.2) on two distinct dates (individuals with BAV were excluded; ICD9, 746.4; ICD10, Q23.1) and compared to 329,971 unrelated control individuals who were free of any codes listed in Table S2. Genotyping, imputation, and genomic quality control was performed as described elsewhere.19 DNA variants were tested for their association with TAA via logistic regression in PLINK2 and controlled for age at time of analysis, age, gender, HARE ancestry assignment,20 and the first five genetic principal components of ancestry (PCAs). Sensitivity analyses were then performed with participants who had undergone any form of thoracic aortic repair and subgroups of ascending and arch repair as cases based on Current Procedural Terminology (CPT) codes provided in Table S2. Covariates and controls were included in the same manner as the primary analysis.
The Trøndelag Health Study (the HUNT Study)21 is a longitudinal population-based health survey conducted in Norway. Participation in the HUNT Study is based on informed consent, and the study has been approved by the Norwegian Data Protection Authority and the Regional Ethics Committee for Medical Research in Norway. Genotyping, quality control and imputation is described elsewhere.22 We used ICD9 441.1/441.2 and ICD10 I71.1/I71.2 to identify 380 affected individuals and 69,255 control individuals from HUNT. Association analysis was performed via SAIGE18 v.43.3 with sex, birth year, batch, and four PCs as covariates.
The UK Biobank23 is a population-based cohort collected from multiple sites in the United Kingdom. We used ICD10 I71.1/I71.2 (excluding Q23.1) to identify 220 affected individuals among those of white British ancestry. After excluding samples with ICD10 I71.0–I71.9, 3,253 control individuals were selected by case-control matching as described in the discovery cohort. Association analysis was performed via SAIGE18 with sex, birth year, array version, and four PCs as covariates.
Meta-analysis of three variants in the discovery and replication cohorts was performed by METAL24 with the standard error approach.
Additional datasets
We downloaded the T2D GWAS in Europeans as “Mahajan.NatGenet2018b.T2D.European.gz” from the DIAGRAM consortium. The eQTL data in aorta as “Artery_Aorta.v8.signif_variant_gene_pairs.txt.gz” and “GTEx_v8_finemapping_CAVIAR.tar” were downloaded from the GTEx portal. DNase I hypersensitive site and H3K27ac of thoracic/ascending aorta (Table S3) were obtained from ENCODE. Promoter capture Hi-C data, as described in Jung et al.,25 were obtained as “AO.po.txt.zip.”
Conditional analysis
Approximate conditional analyses were performed with GCTA.26 We used the –cojo-cond option to run conditional analyses with specific variants. We used the full summary statistics but limited the analysis to ± 500 kb of the conditional variant by using the –extract-region-snp parameter. For conditional analysis, imputed variants from CHIP+MGI (European ancestry) were used as the linkage disequilibrium (LD) reference. We reported the p value of rs7903146 in T2D GWAS conditional on rs4073288 and the p value of rs4073288 in TAA GWAS conditional on rs7903146 and other T2D variants (Table S4).
Colocalization
Colocalization analyses were performed with the R package coloc.27 Coloc performs an approximate Bayes factor analysis with association statistics. We used the function coloc.abf()to calculate the posterior probabilities for the following: (H0) no association with either trait, (H1/H2) association with one of the two tested traits, (H3) association for both traits but different causal variants, and (H4) association for both traits with the same causal variant. A high posterior probability for H4 (PP4) indicates colocalization of the two trait associations. Variants in the ± 500 kb region surrounding rs4073288 were used for colocalization. The default prior probability for colocalization was used for both T2D and eQTL analyses.
Transcriptome-wide association study
To evaluate the evidence of colocalization between the TAA GWAS and eQTL association, we used the paradigm of transcriptome-wide association study (TWAS). TWAS methods perform gene-based association tests. We used these methods to test the association between gene expression predicted by cis-eQTLs and phenotype. We used the MetaXcan package28 to run TWAS with TAA summary statistics. Briefly, we used GWAS tools from the MetaXcan package for summary statistics harmonization and imputation. The imputation step imputes missing GWAS variants via present GWAS variants and the GTEx genotypes. Next, we ran SPrediXcan with the imputed variants and the MASHR expression model (eQTL) of aorta from GTEx v.8.29 We reported the Z scores and p values from this analysis. We did not observe any inflation of test statistics (λ = 0.99). No other genes except TCF7L2 had significant association after multiple testing correction for 13,951 genes present in the aortic expression model (p value significance threshold; 0.05/13,951 = 3.58 × 10−6).
Fine-mapping
We employed FINEMAP,30 a Bayesian fine-mapping method that implements a stochastic shotgun search algorithm incorporating the LD structure and associations (Z scores) in a locus, to identify potential causal variants. Variants with MAF > 5% in the ± 500 kb surrounding rs4073288 were used for this analysis. The LD structure in this locus was determined via PLINK31 from imputed variants in CHIP/MGI. The fine-mapping was carried out with the assumption of a single causal variant. We constructed the 95% credible set by including the variants with highest posterior probabilities and until the sum of posterior probabilities was ≥0.95. We used RegulomeDB32 to annotate variants. RegulomeDB uses public databases to aggregate information about eQTL, transcription factor binding/motif, DNase peak/footprint, etc. for each tested variant. The method assigns a rank to each variant that represents the likelihood of a variant’s being a regulatory variant. We prioritized two variants by using this approach (Table S5). Because RegulomeDB operates in a tissue-agnostic manner, we checked the overlap of fine-mapped variants with the DNase I hypersensitive site and H3K27ac of thoracic/ascending aorta from ENCODE.33 We configured custom tracks in UCSC genome browser to visualize the overlap of variant and chromatin marks. We obtained the promoter capture Hi-C in aorta from Jung et al.25 The p values of chromatin interaction between promoter (chr10: 114,703,220–114,715,198) and variant (respective genomic bin) were reported.
LD score regression (LDSC)
We used “munge_sumstats.py” in LDSC34 to format summary statistics. For further analyses, only variants with MAF > 0.01 and imputation Rsq > 0.9 and that were from Hapmap3 were incorporated. Heritability calculation was performed via the –h2 parameter (with –samp-prev and –pop-prev) in LDSC. Genetic correlations between TAA and other traits were calculated in LD Hub.35 Sources of summary statistics of these traits are listed in Table S6.
Adenoviruses
The Myc-tagged human full-length TCF7L2 pcDNA3 plasmid was previously reported36 (Addgene plasmid # 32738; RRID: Addgene_32738). The coding region of Myc-TCF7L2 from that plasmid was cloned into PCR8/GW/TOPO TA (entry) vector (K250020, Thermo Fisher Scientific) and recombined from the entry vector to the pAd/CMV/V5-DEST Vector (V49320, Thermo Fisher Scientific). The adenovirus carrying LacZ was described before.37 The adenoviruses were packaged in HEK293A cells and purified by CsCl2 density gradient ultracentrifugation. Adenovirus titration was determined by the Adeno-XTM quantitative PCR titration kit (Clontech, CA, USA).
Cell line growth and TCF7L2 gain and loss of function in vitro
Primary human aortic smooth muscle cells (HASMCs) (Lonza, CC2571, lot # 000335663, Walkersville, MD, USA) were grown in smooth muscle cell growth medium 2 containing 5% fetal bovine serum (PromoCell, Germany) and 1% penicillin/streptomycin solution (GIBCO, #15140122) at 37°C with 5% CO2 in a humidified incubator and subjected to quiescence in Opti-MEM for 48 h when so required. HASMCs were used from passage 5 to 8. For overexpression of TCF7L2, adenoviruses containing myc-tagged TCF7L2 or LacZ (control) under the control of the cytomegalovirus (CMV) promoter were used for infection at 30 MOI. For knockdown, cells were transfected with siRNA against TCF7L2 (Horizon D-003816-01 sequence: GAUGGAAGCUUACUAGAUU) or siRNA control (Horizon D-001206-14) at a concentration of 30 nM with Lipofectamine RNAiMAX for 72 h in serum-free medium (Opti-MEM) before use for apoptosis studies.
Apoptosis studies
HASMCs with gain or loss of TCF7L2 as above were treated with 100 ng/mL Fas ligand (FasL) (Enzo Life Science: ALX-522-020) for the indicated times in Opti-MEM, as we previously reported37 (48 h after adenoviral infection or 72 h after siRNA treatment). For annexin V studies, cells were treated for 4 h with FasL in Opti-MEM medium (31985062, GIBCO). Cells were dissociated with 0.25% trypsin (25200056, GIBCO) and stained with the FITC Annexin V Apoptosis Detection Kit I (556548, BD Biosciences), as we previously reported.37 In brief, the cells were washed with cold PBS twice, suspended in 1× binding buffer and stained with FITC annexin V and propidium iodide (PI). The flow cytometry analysis was performed by the flow cytometry core at the University of Michigan with a MoFlo Astrios Cell Sorter (Beckman Coulter, Brea, CA). Apoptotic cells are defined as FITC annexin V positive and PI negative. For immunoblot, cells were treated for 6 h with FasL and subjected to immunoblot with standard protocols as described below.
Quantitative real-time PCR and immunoblot
HASMCs were grown and treated as indicated in each case. For quantitative real-time PCR, we isolated total RNA from HASMCs with the RNeasy Mini Kit (QIAGEN, #74106) followed by the SuperScript III First-Strand Synthesis System (Invitrogen, #18080051) by using random hexamers to generate cDNA, as suggested by the respective manufacturers. Gene expression was quantified in triplicates by quantitative real-time PCR with IQ SYBR Green Supermix (Bio-Rad, #1708882). Relative levels were determined via the ΔΔCt method with expression of the ACTB (Actin Beta) gene as the internal control. The qPCR primer sequences are listed in Table S7. For immunoblot, cell extracts were prepared with RIPA lysis buffer supplemented with complete EDTA-free protease inhibitor cocktail (Roche, #11836170001, Penzberg, Germany) and PhosSTOP phosphatase inhibitor (Roche, #4906845001). The protein concentration was determinedwith the Bradford assay (Bio-Rad, #5000002) and a GloMax Explorer Multimode Microplate Reader. A total of 40 μg of total protein/lane was resolved in SDS-PAGE gels (Bio-Rad 4561094) and transferred to nitrocellulose membranes (Bio-Rad, #1620115, Hercules, CA) with standard techniques. Antibodies were used as follows: the BCL2 antibody (ab182858, 1.2 μg/mL) from Abcam (UK); β-actin (#3700, 0.65 μg/mL), PARP (#9542, 0.1 μg/mL), Caspase 3 (#9662, 0.05 μg/mL), cleaved Caspase 3 (#9664, 0.04 μg/mL), BAX (#5023, 0.14 μg/mL), and myc (#2276, 0.3 μg/mL) from Cell Signaling Technology (CST, Danvers, MA); and the secondary antibodies, IRDye 800CW Donkey anti-Rabbit IgG and IRDye 680RD Donkey anti-Mouse IgG at 0.2 μg/mL, from LI-COR Bioscience (Lincoln, NE). We collected data by using the Odyssey CLx Imaging System (LI-COR Bioscience) and quantified data with the LI-COR Image Studio Software to determine the protein abundance relative to β-actin.
Results
We performed genetic discovery association analysis of 1,351 affected individuals and 18,295 control individuals (European ancestry) from the Cardiovascular Health Improvement Project (CHIP) and Michigan Genomics Initiative (MGI) at the University of Michigan, Michigan Medicine. Imputation with the HRC panel in the Michigan Imputation Server enabled testing of 22.9 million variants. We did not observe any evidence of inflation (λGC = 1.04) of the association test statistics (Figure S1A). Using LD score regression with sample- and population-level prevalence (6% and 0.34%,38 respectively), we estimated SNP heritability of TAA as 0.11 (SE = 0.04) in liability scale. Next, genetic correlation between TAA and known risk factors was examined with GWAS summary statistics (Table S6). We observed a positive genetic correlation between TAA and height (rg = 0.32, SE = 0.11, p = 0.003). This correlation remained significant after excluding variants within 500 kb from rs591519 (FBN1, a gene associated with Marfan syndrome and related disorders) from TAA summary statistics. We did not observe a significant genetic correlation between TAA and smoking or lipid traits, two factors strongly correlated with abdominal aortic aneurysm (AAA).39 On the contrary, diastolic blood pressure is positively correlated with TAA (rg = 0.27, SE = 0.09, p = 0.003) but systolic blood pressure is not (rg = −0.006, SE = 0.08, p = 0.93).
Association with TAA at the TCF7L2 locus
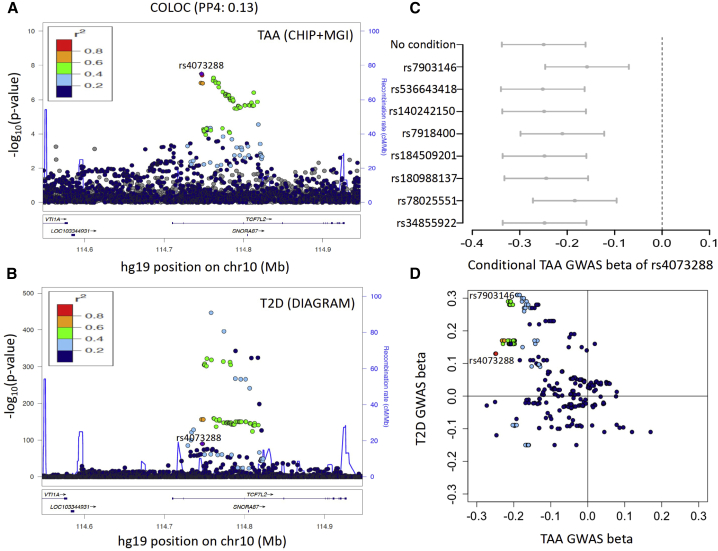
57 variants from two loci reached genome-wide significance (p < 5 × 10−8) in our analysis (Figure S1B). First, we replicated a significant association at the previously known FBN1 locus (index variant rs591519, A/T, effect allele: T, β = 0.42, SE = 0.06, OR 95% CI = 1.35–1.71, p = 1.1 × 10−9). Second, we identified a previously unpublished TAA association in the intronic region of TCF7L2 in chromosome 10 (Figure 1A). The index variant rs4073288 (Table 1) is a common variant with an MAF of 0.32 in cases and 0.37 in controls. We observed 49 common variants (MAF range: 0.37–0.47) with p < 5 × 10−6 in this locus (three genotyped; 46 imputed, Rsq: 0.88 to 0.94). A conditional analysis on rs4073288 did not identify any secondary independent variants in this locus. At both the TCF7L2 and FBN1 loci, we observed a reduced effect in MGI compared to CHIP cases (Table S8). Among 956 CHIP cases, approximately 80% and 8% were classified as ascending and descending TAA, respectively. Interestingly, at the TCF7L2 locus, we observed stronger effect in descending TAA, while the FBN1 effects were stronger in ascending TAA (Table S8). We observed lower effects in the same direction for association with AAA39 at TCF7L2 (rs4073288, effect allele: A, β = −0.05, SE = 0.02, OR 95% CI = 0.91–0.99, p = 3.1 × 10−3) as well as FBN1 (rs591519, effect allele: T, β = 0.06, SE = 0.03, OR 95% CI = 1–1.13, p = 0.03). These variants were not significant (p > 0.05) for association with intracranial aneurysm.40
Figure 1.
TCF7L2 locus in TAA and T2D
(A) LocusZoom plot of TCF7L2 locus in TAA GWAS from CHIP+MGI.
(B) LocusZoom plot of TCF7L2 locus in T2D GWAS from DIAGRAM. LD colors are with respect to the TAA index variant rs4073288.
(C) Conditional effect sizes (beta, in SD units) of TAA index variant rs4073288 in TAA GWAS. Eight independent T2D variants were used for conditioning. Error bars represent 95% confidence intervals.
(D) Comparison of effect sizes (beta) in TAA and T2D GWAS. Variants in this locus have opposite direction in effect sizes in TAA versus T2D GWAS.
Table 1.
Discovery and replication of TCF7L2 variants in TAA
| Variant | EA | OA |
Discovery |
Replication |
Discovery + replication |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE |
OR 95% CI |
p | Beta | SE |
OR 95% CI |
p | Beta | SE |
OR 95% CI |
p | |||
| rs4073288 (index) | A | G | −0.25 | 0.04 | 0.72–0.84 | 3.1 × 10−8 | −0.07 | 0.02 | 0.90–0.97 | 4.2 × 10−5 | −0.10 | 0.02 | 0.87–0.94 | 5.2 × 10−9 |
| rs7904519 (genotyped) | G | A | −0.21 | 0.04 | 0.75–0.88 | 3.6 × 10−7 | −0.07 | 0.02 | 0.90–0.97 | 1.7 × 10−5 | −0.09 | 0.01 | 0.90–0.93 | 3.3 × 10−9 |
| rs4074718 (fine-mapped) | A | G | −0.23 | 0.04 | 0.73–0.86 | 1.1 × 10−7 | −0.07 | 0.02 | 0.90–0.97 | 4.0 × 10−5 | −0.09 | 0.02 | 0.88–0.95 | 5.8 × 10−9 |
EA, effect allele; OA, other allele.
Next, we sought replication of the TCF7L2 locus in a meta-analysis of four independent datasets of European ancestry from (1) University of Texas Health Science Center at Houston (UT, 765 affected individuals), (2) The Trøndelag Health Study (the HUNT Study, 380 affected individuals), (3) UK Biobank (220 affected individuals), and (4) the Million Veteran Program (MVP, 6,554 affected individuals). We observed consistent direction of effect sizes in all replication cohorts (Figure S2). At both the TCF7L2 and FBN1 loci, substantial differences in effect estimates among cohorts were observed (Figure S2). TCF7L2 effect estimates were similar between the Michigan discovery and UT datasets, two cohorts that were primarily ascertained from aortic clinics, whereas the other datasets used ICD-based phenotypes from large biobanks. In MVP, we further identified a smaller subset of TAA-affected individuals in whom surgical repair was performed (730 affected individuals). We observed stronger effect estimates in these affected individuals compared to ICD-based phenotypes (three TCF7L2 variants, OR = 0.86–0.89 for surgical repair TAA versus OR = 0.94 for all ICD-derived TAA-affected individuals; Table S9). This observation highlights the limitation of replication with cohorts with different phenotype definitions and/or case-ascertainment strategies. Nonetheless, after combining all replication datasets, we observed an association p value < 5 × 10−5 for all three tested variants at TCF7L2 (Table 1).
Comparison of the TCF7L2 association with TAA and type 2 diabetes
TCF7L2 is known to be associated with T2D.41 A common variant in the 3rd intron of TCF7L2 is associated with T2D and most likely causes a defect in insulin secretion in the pancreas.42 The TAA index variant rs4073288 is in relatively low LD with the T2D index variant rs7903146 (R2 = 0.28 in Europeans, ∼11 kb apart, 3rd intron). We further examined GWAS summary statistics of T2D from Mahajan et al.43 (rs7903146, PT2D = 1.5 × 10−447; rs4073288, PT2D = 3.8 × 10−91) (Figure 1B). Although the same region within TCF7L2 (3rd intron) is associated with both TAA and T2D risk, the posterior probabilities from colocalization analysis indicated independent signals for TAA and T2D (COLOC, PP3 = 0.85, PP4 = 0.13). Also, a reciprocal conditional analysis did not diminish either signal completely (TAA, Pcond:rs7903146 rs4073288 = 4.5 × 10−4; T2D, Pcond:rs4073288 rs7903146 = 3.4 × 10−262). Mahajan et al. reported seven other secondary independent variants associated with T2D in this locus, which may be functional in different tissues.43 All of these secondary T2D variants have very low correlation with the TAA index variant rs4073288 (maximum R2 = 0.24; Table S4). The association between rs4073288 and TAA remained significant when we conditioned on each of these eight T2D variants (Figure 1C). These observations suggest the presence of different causal variants for TAA and T2D. Finally, we noted an opposite direction of effect (Figure 1D) for TAA and T2D at variants in this region (rs4073288, effect allele: A, βTAA = −0.24 [0.04], βT2D = 0.13 [0.006]; rs7903146, effect allele: T, βTAA = −0.18 [0.04], βT2D = 0.31 [0.006]). The opposite direction of effect for T2D and TAA was consistent in all variants with PTAA < 5 × 10−6 in this locus. We did not observe a significant genetic correlation between TAA and T2D (rg = 0.006, SE = 0.15, p = 0.96) via GWAS summary statistics.
Higher expression of TCF7L2 in aorta is associated with TAA risk
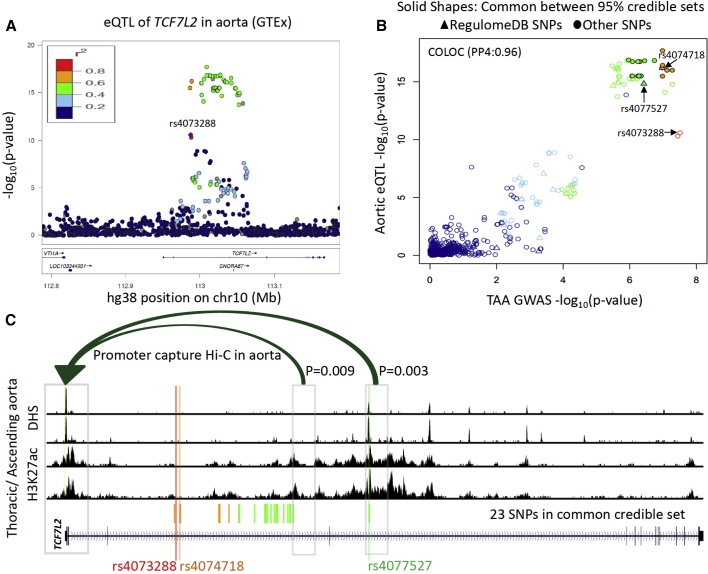
To investigate a potential effect of TAA GWAS variants on expression of TCF7L2, we looked for tissue-specific cis-eQTLs from GTEx v.8.29 The TAA index variant rs4073288 is an eQTL of TCF7L2 in aorta (effect allele: A, normalized effect size = −0.27, p = 2.6 × 10−11, N = 387). The TAA risk allele (G in rs4073288) is associated with higher expression of TCF7L2 in aorta (Figure S3A). This eQTL association was not observed in any other tissue (Figure S3B). Further analysis revealed colocalization (Figure 2B) between the GWAS and eQTL signal (COLOC, PP3 = 0.03, PP4 = 0.96). Next, we performed a TWAS analysis by using SPrediXcan28 with the MASHR aortic expression model from GTEx v.8. This resulted in a significant association between predicted expression levels of TCF7L2 and TAA (Z score = 5.35; p = 8.38 × 10−8; Bonferroni corrected threshold: 0.05/13,951 = 3.58 × 10−6; Figure S3C). A positive Z score in TWAS indicated that higher expression of TCF7L2 is associated with TAA disease. This observation suggests a probable disease mechanism of TAA-associated variants via an increase of TCF7L2 expression in aorta.
Figure 2.
Functional characterization of TCF7L2 locus
(A) LocusZoom plot of aortic eQTL of TCF7L2. LD colors are with respect to the TAA index variant rs4073288.
(B) Comparison of p values in TAA GWAS and aortic eQTL from GTEx. These two associations colocalize with posterior probability 0.96 by COLOC. The solid shapes represent 23 variants that were in both GWAS and eQTL 95% credible sets. Two variants were prioritized by RegulomeDB (rs4074718 and rs4077527).
(C) Regulatory landscape of TCF7L2 gene body in thoracic aorta/ascending aorta from ENCODE. The fine-mapped variant rs4077527 intersects with DNase I hypersensitive site (DHS) and H3K27ac in this tissue. Using promoter capture Hi-C in aorta, we highlighted only genomic bins that significantly interact (p < 0.01) with TCF7L2 promoter. Promoter capture Hi-C p values for 23 variants are given in Figure S4B.
Investigation of the causal variant in the TCF7L2 locus
We hypothesize that the TAA causal variant affects the expression of TCF7L2 via alteration of some regulatory element in this region. In an attempt to identify the causal variant, we applied a Bayesian fine-mapping approach, FINEMAP.30 The 95% credible set for the GWAS consisted of 32 variants from this locus. We also obtained the fine-mapped variants in eQTL by CAVIAR44 from GTEx. Out of 32 variants in the GWAS credible set, 23 were also present in the 95% causal set of eQTL (Figure S4A). Next, we used RegulomeDB32 to annotate variants with known or predicted regulatory elements. We could further prioritize (Table S5) two variants in the common credible set (Figure 2B). Both these variants (rs4074718 and rs4077527) have larger effect sizes for the TCF7L2 eQTL than the index variant rs4073288. While both variants are from regions annotated as enhancers in multiple tissues, rs4077527 intersects with accessible chromatin (DNase I hypersensitive site) in adult thoracic/ascending aorta in ENCODE33 data (Figure 2C, Table S3). We also observed strong evidence of enhancer activity (H3K27ac) in these samples overlapping rs4077527. Because both these variants are quite far from the TCF7L2 promoter (rs4074718, ∼40 kb; rs4077527, ∼10 0kb), we examined promoter capture Hi-C data from aorta.25 A significant long-distance chromatin interaction (p < 0.01, as per methods in Jung et al.25) between rs4077527 and the TCF7L2 promoter region (hg19; chr10: 114,703,220–114,715,198) was observed (Figure 2C, Figure S4B). Together, these observations predict rs4077527 as a strong candidate for the causal TAA variant in this locus, most likely interacting with the promoter to impact TCF7L2 expression in the aorta.
TCF7L2 enhances human vascular smooth muscle cell apoptosis
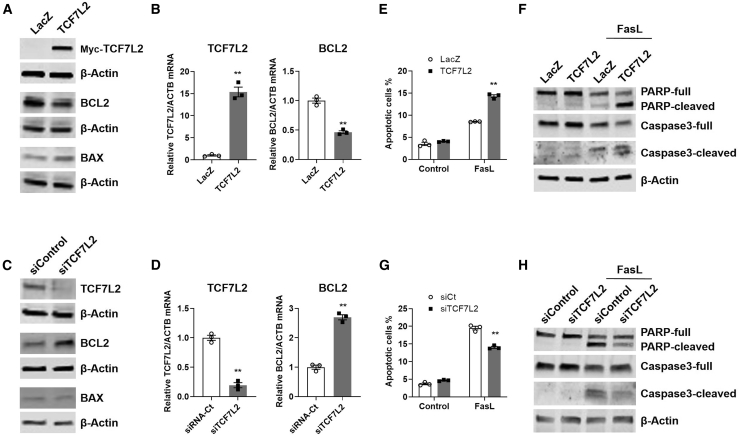
TAA is characterized by vascular smooth muscle cell (VSMC) apoptosis in association with loss of their contractile phenotype, inflammatory status, and increased extracellular matrix degradation as drivers of adverse outcomes.45 Loss of VSMC in TAA is recognized to be the result of increased apoptosis through the endogenous pathway regulated via BCL2.45,46 As a distal effector of the Wnt/β-catenin/TCF-LEF pathway, TCF7L2 is known to regulate homeostasis of VSMC, the media layer of blood vessels, including phenotypic modulation and proliferation.47, 48, 49 TCF7L2 may exert pro-50,51 or anti-apoptotic52,53 effects in different contexts. Yet, contribution of TCF7L2 to VSMC apoptosis in aneurysm remains unaddressed. We experimentally assessed whether gain and loss of function of TCF7L2 contributes to VSMC apoptosis via differential regulation of BCL2 expression in human aortic smooth muscle cells (HASMCs) in vitro. Adenoviral-mediated overexpression of TCF7L2 resulted in reduced BCL2 expression of both mRNA (54% reduction, p = 0.0004) and protein (24% reduction, p = 0.0011) (Figures 3A and 3B, Figure S5A) compared with the adenoviral-mediated expression of LacZ used as control. Conversely, siRNA-mediated downregulation of TCF7L2, which resulted in a significant reduction in endogenous TCF7L2 mRNA (80% reduction, p = 0.0002 versus siRNA control) and protein (77% reduction, p = 0.0145 versus siRNA control), demonstrated a concomitant increase in BCL2 protein (1.6-fold increase, p = 0.0154 versus shRNA control) and mRNA abundance (2.7-fold increase, p = 0.0001) (Figures 3C and 3D, Figure S5B). Changes in TCF7L2 showed no significant effects on BAX protein or mRNA expression (Figures 3A–3D, Figures S5A–S5D). Consequently, we determined a significant BAX/BCL2 protein ratio between TCF7L2 overexpression and knockdown (1.3222 ± 0.1024 versus 0.6426 ± 0.1176, p = 0.0168, respectively) in basal growth conditions (Figures 3A and 3B, Figure S5A), suggesting a potential pro-apoptotic effect of TCF7L2 upregulation in HASMCs.
Figure 3.
TCF7L2 expression is directly associated with vascular smooth muscle cell apoptosis in vitro
HASMCs were infected with adenovirus (30 MOI)-encoding LacZ (control) or myc-tagged TCF7L2, and after 48 h, (A) the abundance of the indicated proteins in the cell extracts was determined by immunoblot (representative of three independent experiments) and (B) the mRNA expression of the indicated genes, relative to ACTB, by quantitative real-time PCR. A LacZ control was set at 1 as reference (n = 3; representative of three independent experiments). Data are mean ± SEM. ∗∗p ≤ 0.0004. HASMCs were transfected with siRNA control or siRNA against TCF7L2 via Lipofectamine RNAiMax, and after 72 h, (C) the abundance of the indicated proteins and (D) the mRNA expression of the indicated genes were determined as described above with an siRNA control set at 1 as reference. ∗∗p ≤ 0.0002. HASMCs were infected with adenovirus as in (A). After 48 h in serum-free medium, they were treated with Fas ligand (FasL, 100 ng/mL) in serum-free medium and apoptosis was assessed by (E) annexin V via FACS analysis after treatment for 4 h (n = 3; representative of three independent experiments) or (F) the abundance of the indicated proteins in the cell extracts after 6 h as determined by immunoblot (representative of three independent experiments). HASMCs were transfected with the indicated siRNA as in (C). After 72 h in serum-free medium, they were treated with FasL (100 ng/mL) for (G) annexin V assay after 4 h as in (E) or (H) immunoblot after 6 h as in (F) and subjected to immunoblot as in (E). Densitometry for (A), (B), (E), (F), and mRNA expression of BAX are provided in Figure S5.
We further examined the expectation that increased TCF7L2 expression enhances apoptosis of aortic smooth muscle cells through treatment with FasL, a potent inducer of VSMC apoptosis produced by inflammatory cells in the aneurysmal tissues.45,54 In these experimental conditions, FasL treatment resulted in significant increase in annexin V positive cells (1.7-fold, p ≤ 0.0001), indicative of enhanced early apoptosis when TCF7L2 was overexpressed (Figure 3E, Figure S5E), while siRNA-mediated TCF7L2 knockdown significantly reduced the apoptotic response by 28% (p ≤ 0.0001) (Figure 3G, Figure S5F), as determined by fluorescence-activated cell sorting (FACS). These data were in agreement with progression into apoptosis upon TCF7L2 overexpression, as indicated by significantly increased cleavage of PARP and caspase-3 compared to the corresponding full-length proteins (Figure 3F, Figure S5G). Conversely, siRNA-mediated knockdown of TCF7L2 significantly reduced cleavage of both apoptosis markers (Figure 3H, Figure S5H). Overall, these data indicate that increased expression of TCF7L2 may result in enhanced apoptosis of vascular smooth muscle cells—a likely mechanism of action of TAA-associated variants in TCF7L2.
Discussion
In this study, we identified a locus in the intronic region of TCF7L2 that is associated with TAA, to our knowledge, the first locus identified for TAA via genome-wide approaches. Using eQTL from aorta, we provide evidence that TAA-associated variants are associated with altered expression of TCF7L2 and we highlight one variant in the locus that overlaps with an enhancer active in aorta. Common variants (rs7903146, index variant) in this locus are the strongest known genetic risk factor for T2D.42 Vinuela et al.55 demonstrated that the T2D risk allele of rs7903146 decreases pancreatic islet expression of the last (3′) exon of TCF7L2. Although there is evidence of different causal variants for TAA and T2D, we observed an opposite direction in effect sizes. Interestingly, epidemiological studies have historically suggested an inverse relationship between aortic aneurysm and T2D.56, 57, 58 Our genetic results suggest that, at least for the TCF7L2 locus, shared but independent etiology (gene-level horizontal pleiotropy) may be partially responsible for this association. These results most likely indicate that largely independent sets of causal variants lead to increased TCF7L2 expression in the human aorta and decreased TCF7L2 expression in pancreatic islets.
The T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors (including TCF7L2) are signal integrators and effectors of the Wnt/β-catenin pathway with contextual tissue and cell-type-specific functions59 and can exhibit differential effects because of epigenetic modifications of the target genes. Wnt/β-catenin is an evolutionarily conserved pathway that regulates various aspects of development, including cell proliferation, migration, polarity, and apoptosis.60 Dysregulation of the Wnt/β-catenin signaling is involved in a wide range of disorders, including Alzheimer disease, cancer, and metabolic diseases.61 Current research supports a role of TCF7L2 in vascular development.62, 63, 64 In cardiovascular diseases, the Wnt/β-catenin pathway was found to be dysregulated in restenosis,65 and more recently, TCF7L2 was demonstrated to regulate VSMC plasticity in both GATA6-dependent and -independent fashions.47 Regarding aneurysm, Krishna et al. reported activation of the Wnt/β-catenin pathway in AAA via epigenetic silencing of the Wnt inhibitor sclerostin.66 Recently, dysregulated Notch/BMP/WNT pathways were found in aortic endothelial cells isolated from individuals with TAA compared to healthy donors,67 including increased TCF7L2 mRNA expression and activation of the Wnt/β-catenin pathway in the endothelium. Our analyses indicate that variants associated with higher expression of TCF7L2 are also associated with TAA. TAA is a disease characterized by VSMC death as a driver of adverse outcomes.45 Therefore, we specifically addressed the potential effects of TCF7L2 on VSMC apoptosis in vitro. We show that TCF7L2 expression is directly associated with VSMC apoptosis and that this effect could be mediated, at least in part, through TCF7L2 repression of BCL2 expression, a known target of the pathway. Our data from VSMC, together with the report from endothelial cells,67 clearly indicate that upregulation of TCF7L2 may play a fundamental role in TAA pathology.
Mendelian, monogenic inheritance of TAA is primarily driven by pathogenic variants in genes from the TGF-β signaling pathway (FBN1, SMAD3, TGFB2, TGFBR1, and TGFBR2) and VSMC contraction pathway (ACTA2, MYH11, MYLK, and PRKG1). The TGF-β/SMAD3 pathway interacts with β-catenin in the canonical Wnt/β-catenin signaling pathway toward activation of TCF/LEF transcription factors (including TCF7L2) (Figure S6). Future studies should focus on understanding whether the effects observed in our experiments are dependent on its interaction with β-catenin68 or other co-repressors or co-activators69, 70, 71 or whether they may reflect the ability of TCF7L2 to integrate signaling from other pathways, including TGF-β via SMAD3.72
Current treatment of aortic aneurysms involves surgical repair to reduce the risk of rupture, yet only a fraction of patients are eligible and those who are eligible are at high risk for post-surgery complications. Therapies for stabilization of aortic aneurysms have yet to prove efficacy in controlled clinical trials. Because of the involvement of this pathway in various diseases, present efforts in the drug development field are aimed at identifying inhibitors of the β-catenin-TCF7L2 interactions73 and the design of specific approaches targeting TCF7L2 stability.74 Our findings here highlight a potential path for TCF7L2 as a target for intervention in individuals with TAA or for possible prevention in individuals at risk of TAA.
Acknowledgments
We thank all that contributed to the Cardiovascular Health Improvement Project and Michigan Genomics Initiative, including the financial support of the Frankel Cardiovascular Center (FCVC), Michigan Medicine, the Aikens Aortic Discovery Program of the FCVC, and the University of Michigan Medical School Central Biorepository for providing biospecimen management and distribution services. The Center for Statistical Genetics in the Department of Biostatistics at the School of Public Health provided genotype data curation, imputation, and management in support of this research (for MGI cohort). C.J.W. is supported by NIH grants R35-HL135824, R01-HL142023, and R01-HL109946. Y.E.C. is supported by NIH grants HL134569, HL109946, HL147527, and HL137214. J.Z. is supported by NIH grant HL138139. L.C. is supported by NIH grant HL122664. D.M. is supported by R01-HL109942-09, John Ritter Foundation, and Remembrin’ Benjamin. S.M.D. is supported by IK2-CX001780. P.S.T. is supported by I01-BX003362. B.Y. is supported by K08-HL130614, R01-HL141891, and R01-HL151776. K.A.E. is supported by WL Gore and Marfan Foundation. B.N.W. is supported by NSF Graduate Research Fellowship (DGE 1256260). W.Z. is supported by NIH NHGRI under award number T32HG010464. HUNT-MI study, which comprises the genetic investigations of the HUNT Study, is a collaboration between investigators from the HUNT study and University of Michigan Medical School and the University of Michigan School of Public Health. The K.G. Jebsen Center for Genetic Epidemiology is financed by Stiftelsen Kristian Gerhard Jebsen; Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology (NTNU), and Central Norway Regional Health Authority.
Declaration of interests
The spouse of C.J.W. is an employee of Regeneron. All other authors declare no competing interests.
Published: July 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.016.
Contributor Information
Minerva T. Garcia-Barrio, Email: minerva@med.umich.edu.
Cristen J. Willer, Email: cristen@umich.edu.
Data and code availability
TAA GWAS summary statistics from CHIP+MGI are available here: http://csg.sph.umich.edu/willer/public/TAA2021/. Analyses were performed by open-source software and cited in the material and methods.
Web resources
DIAGRAM, https://diagram-consortium.org
ENCODE, https://www.encodeproject.org/
GTEx Portal, http://www.gtexportal.org/home/index.html
Promoter capture Hi-C, http://www.3div.kr/capture_hic
Supplemental information
References
- 1.Hiratzka L.F., Bakris G.L., Beckman J.A., Bersin R.M., Carr V.F., Casey D.E., Jr., Eagle K.A., Hermann L.K., Isselbacher E.M., Kazerooni E.A. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with Thoracic Aortic Disease: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. Circulation. 2010;121:e266–e369. doi: 10.1161/CIR.0b013e3181d4739e. [DOI] [PubMed] [Google Scholar]
- 2.Pinard A., Jones G.T., Milewicz D.M. Genetics of Thoracic and Abdominal Aortic Diseases. Circ. Res. 2019;124:588–606. doi: 10.1161/CIRCRESAHA.118.312436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolford B.N., Hornsby W.E., Guo D., Zhou W., Lin M., Farhat L., McNamara J., Driscoll A., Wu X., Schmidt E.M. Clinical Implications of Identifying Pathogenic Variants in Individuals With Thoracic Aortic Dissection. Circ Genom Precis Med. 2019;12:e002476. doi: 10.1161/CIRCGEN.118.002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renard M., Francis C., Ghosh R., Scott A.F., Witmer P.D., Adès L.C., Andelfinger G.U., Arnaud P., Boileau C., Callewaert B.L. Clinical Validity of Genes for Heritable Thoracic Aortic Aneurysm and Dissection. J. Am. Coll. Cardiol. 2018;72:605–615. doi: 10.1016/j.jacc.2018.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietz H.C., Cutting G.R., Pyeritz R.E., Maslen C.L., Sakai L.Y., Corson G.M., Puffenberger E.G., Hamosh A., Nanthakumar E.J., Curristin S.M. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 6.Loeys B.L., Schwarze U., Holm T., Callewaert B.L., Thomas G.H., Pannu H., De Backer J.F., Oswald G.L., Symoens S., Manouvrier S. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N. Engl. J. Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 7.Rooprai J., Boodhwani M., Beauchesne L., Chan K.L., Dennie C., Nagpal S., Messika-Zeitoun D., Coutinho T. Thoracic Aortic Aneurysm Growth in Bicuspid Aortic Valve Patients: Role of Aortic Stiffness and Pulsatile Hemodynamics. J. Am. Heart Assoc. 2019;8:e010885. doi: 10.1161/JAHA.118.010885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeMaire S.A., McDonald M.L., Guo D.C., Russell L., Miller C.C., 3rd, Johnson R.J., Bekheirnia M.R., Franco L.M., Nguyen M., Pyeritz R.E. Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning FBN1 at 15q21.1. Nat. Genet. 2011;43:996–1000. doi: 10.1038/ng.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo D.C., Grove M.L., Prakash S.K., Eriksson P., Hostetler E.M., LeMaire S.A., Body S.C., Shalhub S., Estrera A.L., Safi H.J. Genetic Variants in LRP1 and ULK4 Are Associated with Acute Aortic Dissections. Am. J. Hum. Genet. 2016;99:762–769. doi: 10.1016/j.ajhg.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritsche L.G., Gruber S.B., Wu Z., Schmidt E.M., Zawistowski M., Moser S.E., Blanc V.M., Brummett C.M., Kheterpal S., Abecasis G.R., Mukherjee B. Association of Polygenic Risk Scores for Multiple Cancers in a Phenome-wide Study: Results from The Michigan Genomics Initiative. Am. J. Hum. Genet. 2018;102:1048–1061. doi: 10.1016/j.ajhg.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jun G., Flickinger M., Hetrick K.N., Romm J.M., Doheny K.F., Abecasis G.R., Boehnke M., Kang H.M. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taliun D., Chothani S.P., Schönherr S., Forer L., Boehnke M., Abecasis G.R., Wang C. LASER server: ancestry tracing with genotypes or sequence reads. Bioinformatics. 2017;33:2056–2058. doi: 10.1093/bioinformatics/btx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang B., Zhou W., Jiao J., Nielsen J.B., Mathis M.R., Heydarpour M., Lettre G., Folkersen L., Prakash S., Schurmann C. Protein-altering and regulatory genetic variants near GATA4 implicated in bicuspid aortic valve. Nat. Commun. 2017;8:15481. doi: 10.1038/ncomms15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasta S., Rinaudo A., Luca A., Pilato M., Scardulla C., Gleason T.G., Vorp D.A. Difference in hemodynamic and wall stress of ascending thoracic aortic aneurysms with bicuspid and tricuspid aortic valve. J. Biomech. 2013;46:1729–1738. doi: 10.1016/j.jbiomech.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter-Zinck H., Shi Y., Li M., Gorman B.R., Ji S.G., Sun N., Webster T., Liem A., Hsieh P., Devineni P. Genotyping Array Design and Data Quality Control in the Million Veteran Program. Am. J. Hum. Genet. 2020;106:535–548. doi: 10.1016/j.ajhg.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang H., Hui Q., Lynch J., Honerlaw J., Assimes T.L., Huang J., Vujkovic M., Damrauer S.M., Pyarajan S., Gaziano J.M. Harmonizing Genetic Ancestry and Self-identified Race/Ethnicity in Genome-wide Association Studies. Am. J. Hum. Genet. 2019;105:763–772. doi: 10.1016/j.ajhg.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krokstad S., Langhammer A., Hveem K., Holmen T.L., Midthjell K., Stene T.R., Bratberg G., Heggland J., Holmen J. Cohort Profile: the HUNT Study, Norway. Int. J. Epidemiol. 2013;42:968–977. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., Zhou W., Skov M.W., Graham S.E., Herron T.J., McCarthy S., Schmidt E.M., Sveinbjornsson G. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung I., Schmitt A., Diao Y., Lee A.J., Liu T., Yang D., Tan C., Eom J., Chan M., Chee S. A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet. 2019;51:1442–1449. doi: 10.1038/s41588-019-0494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benner C., Spencer C.C., Havulinna A.S., Salomaa V., Ripatti S., Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder M.P., Gingeras T.R., Moore J.E., Weng Z., Gerstein M.B., Ren B., Hardison R.C., Stamatoyannopoulos J.A., Graveley B.R., Feingold E.A. Perspectives on ENCODE. Nature. 2020;583:693–698. doi: 10.1038/s41586-020-2449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Pourcain B.S. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tetsu O., McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 37.Lu H., Sun J., Liang W., Chang Z., Rom O., Zhao Y., Zhao G., Xiong W., Wang H., Zhu T. Cyclodextrin Prevents Abdominal Aortic Aneurysm via Activation of Vascular Smooth Muscle Cell Transcription Factor EB. Circulation. 2020;142:483–498. doi: 10.1161/CIRCULATIONAHA.119.044803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kälsch H., Lehmann N., Möhlenkamp S., Becker A., Moebus S., Schmermund A., Stang A., Mahabadi A.A., Mann K., Jöckel K.H. Body-surface adjusted aortic reference diameters for improved identification of patients with thoracic aortic aneurysms: results from the population-based Heinz Nixdorf Recall study. Int. J. Cardiol. 2013;163:72–78. doi: 10.1016/j.ijcard.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 39.Klarin D., Verma S.S., Judy R., Dikilitas O., Wolford B.N., Paranjpe I., Levin M.G., Pan C., Tcheandjieu C., Spin J.M. Genetic Architecture of Abdominal Aortic Aneurysm in the Million Veteran Program. Circulation. 2020;142:1633–1646. doi: 10.1161/CIRCULATIONAHA.120.047544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakker M.K., van der Spek R.A.A., van Rheenen W., Morel S., Bourcier R., Hostettler I.C., Alg V.S., van Eijk K.R., Koido M., Akiyama M. Genome-wide association study of intracranial aneurysms identifies 17 risk loci and genetic overlap with clinical risk factors. Nat. Genet. 2020;52:1303–1313. doi: 10.1038/s41588-020-00725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 42.Gloyn A.L., Braun M., Rorsman P. Type 2 diabetes susceptibility gene TCF7L2 and its role in beta-cell function. Diabetes. 2009;58:800–802. doi: 10.2337/db09-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hormozdiari F., Kostem E., Kang E.Y., Pasaniuc B., Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He R., Guo D.C., Estrera A.L., Safi H.J., Huynh T.T., Yin Z., Cao S.N., Lin J., Kurian T., Buja L.M. Characterization of the inflammatory and apoptotic cells in the aortas of patients with ascending thoracic aortic aneurysms and dissections. J. Thorac. Cardiovasc. Surg. 2006;131:671–678. doi: 10.1016/j.jtcvs.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Durdu S., Deniz G.C., Balci D., Zaim C., Dogan A., Can A., Akcali K.C., Akar A.R. Apoptotic vascular smooth muscle cell depletion via BCL2 family of proteins in human ascending aortic aneurysm and dissection. Cardiovasc. Ther. 2012;30:308–316. doi: 10.1111/1755-5922.12007. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava R., Rolyan H., Xie Y., Li N., Bhat N., Hong L., Esteghamat F., Adeniran A., Geirsson A., Zhang J. TCF7L2 (Transcription Factor 7-Like 2) Regulation of GATA6 (GATA-Binding Protein 6)-Dependent and -Independent Vascular Smooth Muscle Cell Plasticity and Intimal Hyperplasia. Arterioscler. Thromb. Vasc. Biol. 2019;39:250–262. doi: 10.1161/ATVBAHA.118.311830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivastava R., Zhang J., Go G.W., Narayanan A., Nottoli T.P., Mani A. Impaired LRP6-TCF7L2 Activity Enhances Smooth Muscle Cell Plasticity and Causes Coronary Artery Disease. Cell Rep. 2015;13:746–759. doi: 10.1016/j.celrep.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Adhikari N., Li Q., Hall J.L. LDL receptor-related protein LRP6 regulates proliferation and survival through the Wnt cascade in vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2004;287:H2376–H2383. doi: 10.1152/ajpheart.01173.2003. [DOI] [PubMed] [Google Scholar]
- 50.Liu X., Huang Y., Zhang Y., Li X., Liu C., Huang S., Xu D., Wu Y., Liu X. T-cell factor (TCF/LEF1) binding elements (TBEs) of FasL (Fas ligand or CD95 ligand) bind and cluster Fas (CD95) and form complexes with the TCF-4 and b-catenin transcription factors in vitro and in vivo which result in triggering cell death and/or cell activation. Cell. Mol. Neurobiol. 2016;36:1001–1013. doi: 10.1007/s10571-015-0290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma B., Zhong L., van Blitterswijk C.A., Post J.N., Karperien M. T cell factor 4 is a pro-catabolic and apoptotic factor in human articular chondrocytes by potentiating nuclear factor κB signaling. J. Biol. Chem. 2013;288:17552–17558. doi: 10.1074/jbc.M113.453985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., Ma Y., Chen B., Shi J. miR-182 enhances acute kidney injury by promoting apoptosis involving the targeting and regulation of TCF7L2/Wnt/β-catenins pathway. Eur. J. Pharmacol. 2018;831:20–27. doi: 10.1016/j.ejphar.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y., Zhang E., Berggreen C., Jing X., Osmark P., Lang S., Cilio C.M., Göransson O., Groop L., Renström E., Hansson O. Survival of pancreatic beta cells is partly controlled by a TCF7L2-p53-p53INP1-dependent pathway. Hum. Mol. Genet. 2012;21:196–207. doi: 10.1093/hmg/ddr454. [DOI] [PubMed] [Google Scholar]
- 54.Henderson E.L., Geng Y.J., Sukhova G.K., Whittemore A.D., Knox J., Libby P. Death of smooth muscle cells and expression of mediators of apoptosis by T lymphocytes in human abdominal aortic aneurysms. Circulation. 1999;99:96–104. doi: 10.1161/01.cir.99.1.96. [DOI] [PubMed] [Google Scholar]
- 55.Viñuela A., Varshney A., van de Bunt M., Prasad R.B., Asplund O., Bennett A., Boehnke M., Brown A.A., Erdos M.R., Fadista J. Genetic variant effects on gene expression in human pancreatic islets and their implications for T2D. Nat. Commun. 2020;11:4912. doi: 10.1038/s41467-020-18581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel K., Zafar M.A., Ziganshin B.A., Elefteriades J.A. Diabetes Mellitus: Is It Protective against Aneurysm? A Narrative Review. Cardiology. 2018;141:107–122. doi: 10.1159/000490373. [DOI] [PubMed] [Google Scholar]
- 57.Takagi H., Umemoto T., ALICE (All-Literature Investigation of Cardiovascular Evidence) Group Negative Association of Diabetes With Thoracic Aortic Dissection and Aneurysm. Angiology. 2017;68:216–224. doi: 10.1177/0003319716647626. [DOI] [PubMed] [Google Scholar]
- 58.Raffort J., Lareyre F., Clément M., Hassen-Khodja R., Chinetti G., Mallat Z. Diabetes and aortic aneurysm: current state of the art. Cardiovasc. Res. 2018;114:1702–1713. doi: 10.1093/cvr/cvy174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi F., Brubaker P.L., Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J. Biol. Chem. 2005;280:1457–1464. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- 60.Komiya Y., Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng L.F., Kaur P., Bunnag N., Suresh J., Sung I.C.H., Tan Q.H., Gruber J., Tolwinski N.S. WNT Signaling in Disease. Cells. 2019;8:E826. doi: 10.3390/cells8080826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Facchinello N., Tarifeño-Saldivia E., Grisan E., Schiavone M., Peron M., Mongera A., Ek O., Schmitner N., Meyer D., Peers B. Tcf7l2 plays pleiotropic roles in the control of glucose homeostasis, pancreas morphology, vascularization and regeneration. Sci. Rep. 2017;7:9605. doi: 10.1038/s41598-017-09867-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang S., Huang H., Xiang H., Gu B., Li W., Chen L., Zhang M. Wnt Signaling Modulates Routes of Retinoic Acid-Induced Differentiation of Embryonic Stem Cells. Stem Cells Dev. 2019;28:1334–1345. doi: 10.1089/scd.2019.0065. [DOI] [PubMed] [Google Scholar]
- 64.Corada M., Nyqvist D., Orsenigo F., Caprini A., Giampietro C., Taketo M.M., Iruela-Arispe M.L., Adams R.H., Dejana E. The Wnt/beta-catenin pathway modulates vascular remodeling and specification by upregulating Dll4/Notch signaling. Dev. Cell. 2010;18:938–949. doi: 10.1016/j.devcel.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X., Xiao Y., Mou Y., Zhao Y., Blankesteijn W.M., Hall J.L. A role for the beta-catenin/T-cell factor signaling cascade in vascular remodeling. Circ. Res. 2002;90:340–347. doi: 10.1161/hh0302.104466. [DOI] [PubMed] [Google Scholar]
- 66.Krishna S.M., Seto S.W., Jose R.J., Li J., Morton S.K., Biros E., Wang Y., Nsengiyumva V., Lindeman J.H., Loots G.G. Wnt Signaling Pathway Inhibitor Sclerostin Inhibits Angiotensin II-Induced Aortic Aneurysm and Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2017;37:553–566. doi: 10.1161/ATVBAHA.116.308723. [DOI] [PubMed] [Google Scholar]
- 67.Kostina A., Bjork H., Ignatieva E., Irtyuga O., Uspensky V., Semenova D., Maleki S., Tomilin A., Moiseeva O., Franco-Cereceda A. Notch, BMP and WNT/β-catenin network is impaired in endothelial cells of the patients with thoracic aortic aneurysm. Atheroscler. Suppl. 2018;35:e6–e13. doi: 10.1016/j.atherosclerosissup.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 68.Graham T.A., Ferkey D.M., Mao F., Kimelman D., Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat. Struct. Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- 69.Valenta T., Lukas J., Korinek V. HMG box transcription factor TCF-4's interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res. 2003;31:2369–2380. doi: 10.1093/nar/gkg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brantjes H., Roose J., van De Wetering M., Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hecht A., Stemmler M.P. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 2003;278:3776–3785. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- 72.Labbé E., Letamendia A., Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc. Natl. Acad. Sci. USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian W., Han X., Yan M., Xu Y., Duggineni S., Lin N., Luo G., Li Y.M., Han X., Huang Z., An J. Structure-based discovery of a novel inhibitor targeting the β-catenin/Tcf4 interaction. Biochemistry. 2012;51:724–731. doi: 10.1021/bi201428h. [DOI] [PubMed] [Google Scholar]
- 74.Zhang H., Rong X., Wang C., Liu Y., Lu L., Li Y., Zhao C., Zhou J. VBP1 modulates Wnt/β-catenin signaling by mediating the stability of the transcription factors TCF/LEFs. J. Biol. Chem. 2020;295:16826–16839. doi: 10.1074/jbc.RA120.015282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TAA GWAS summary statistics from CHIP+MGI are available here: http://csg.sph.umich.edu/willer/public/TAA2021/. Analyses were performed by open-source software and cited in the material and methods.