Summary
Interpretation of the function of non-coding risk loci for neuropsychiatric disorders and brain-relevant traits via gene expression and alternative splicing quantitative trait locus (e/sQTL) analyses is generally performed in bulk post-mortem adult tissue. However, genetic risk loci are enriched in regulatory elements active during neocortical differentiation, and regulatory effects of risk variants may be masked by heterogeneity in bulk tissue. Here, we map e/sQTLs, and allele-specific expression in cultured cells representing two major developmental stages, primary human neural progenitors (n = 85) and their sorted neuronal progeny (n = 74), identifying numerous loci not detected in either bulk developing cortical wall or adult cortex. Using colocalization and genetic imputation via transcriptome-wide association, we uncover cell-type-specific regulatory mechanisms underlying risk for brain-relevant traits that are active during neocortical differentiation. Specifically, we identified a progenitor-specific eQTL for CENPW co-localized with common variant associations for cortical surface area and educational attainment.
Keywords: cell-type specificity, common genetic variants, expression/splicing quantitative loci, neuropsychiatric disorders, neurogenesis, genome-wide association study, transcriptome-wide association study
Introduction
Genome wide association studies (GWASs) have identified many common non-coding variants associated with risk for neurodevelopmental disorders, or inter-individual variability in brain structure and other brain-related traits.1, 2, 3, 4, 5, 6, 7 However, it is challenging to determine the mechanism of these non-coding variants because, in general, (1) the genes impacted by non-coding risk variants are unknown, (2) the cell type(s) and developmental period(s) where the variants have an effect are not known, and (3) there may be limited availability of cells or tissue representing the causal developmental stage and cell type.
One potential mechanism by which non-coding genetic variation can influence brain traits is through alterations in gene expression or expression quantitative trait loci (eQTLs). Genetic variation also impacts transcript splicing,8, 9, 10 and several studies have implicated genetically mediated alterations in splicing as important risk factors for neuropsychiatric disorders.11, 12, 13
Most current efforts to explain the function of these risk loci rely on mapping local expression and splicing quantitative trait loci (e/sQTLs) in bulk adult brain tissue, which has been a fruitful approach.14,15 However, neuropsychiatric disorder genetic risk loci are enriched in cell types relevant for neocortical differentiation that are not present in the adult brain.16,17 e/sQTL studies performed on human fetal brain bulk cortical tissue have demonstrated the importance of developmental stage and cell composition, by identifying thousands of fetal brain-specific e/sQTLs.18, 19, 20 However, these studies necessarily focus on one developmental time point for each individual and heterogeneity in bulk tissue may mask cell-type-specific allelic effects.21, 22, 23, 24
Utilizing a cell-type-specific in vitro model system including neural progenitors (ndonor = 85) and their virally labeled and sorted neuronal progeny (ndonor = 74) derived from a multi-ancestry population, here we investigated how common genetic variants impact brain-related traits through gene expression and splicing during human neurogenesis. We discovered 2,079/872 eQTLs in progenitors and neurons and 5,900/4,396 sQTLs in progenitors and neurons, respectively. Importantly, 66.1%/47% of eQTLs and 79.3%/73.4% of sQTLs in progenitor/neuron were unique and not found in fetal bulk brain e/sQTLs from a largely overlapping sample19 or in adult bulk e/sQTL data from GTEx.25 We showed both eQTLs and sQTLs colocalized with known GWAS loci for neuropsychiatric disorders and other brain-relevant traits in a cell-type-specific manner. By integrating the dataset generated here with cell-type-specific chromatin accessibility from the same cell lines17 and brain structure GWAS,4 we propose a regulatory mechanism whereby genetic variation influences educational attainment, a proxy for human intelligence, across multiple levels of biology. Furthermore, we genetically imputed cell-type-specific and temporal specific gene expression and alternative splicing associated with brain-relevant traits and neuropsychiatric disorder risk using transcriptome-wide association studies (TWASs).
Material and methods
Cell culture
Generation of human neural progenitor cells was previously described.17,26 Briefly, human fetal brain tissue was acquired from the UCLA Gene and Cell Therapy Core following IRB regulations from approximately 14–21 gestation weeks (inferred to be 12–19 post conception weeks). The tissue was derived from voluntary terminations of pregnancy. We excluded known trisomy 21 cases. We were not aware of any fetal anomalies in any body system. For a small subset of intact samples, cortical tissue was dissected to generate primary human neural progenitor cells (phNPCs). For most samples that were non-intact, flat and sheet-like pieces of brain tissue that were presumed to originate from the cortex were selected to generate phNPCs. The tissue was then dissociated and cultured as neurospheres as previously described.26 Neurospheres were plated on laminin/fibronectin and polyornithine-coated plates for an average of 2.5 ± 1.8 SD passages, and cryopreserved.
Cryopreserved phNPCs were transferred to UNC Chapel Hill, after material transfer agreement, where all downstream culture and analyses were completed. Donors processed for ATAC-seq (described previously17) and RNA-seq (described here) were cultured simultaneously. The overall design of the experiment and media used for culture was previously described.17 Briefly, we cultured 89 unique donors for subsequent RNA-seq library preparation. We first randomly assigned the approximately 8–9 donors into 12 rounds for a feasible cell culture workload. We thawed one round every 3 weeks. To reduce batch effects, we processed each round on the same day of the week and designated the same person to do each task as much as possible. Cells were isolated at two time points: progenitor and their differentiated and virally labeled neuronal progeny. Progenitors were cultured in proliferation media including growth factors for 3 weeks (see Liang et al.17), and we lifted them with trypsin to prepare RNA-seq libraries. Differentiation in the absence of growth factors was performed for 5 weeks, after which the culture was transduced with AAV2-hSyn1-eGFP virus, that specifically expresses a reporter gene in neurons without integrating into the genome, at 20,000 multiplicity of infection (MOI) and then differentiated for another 3 weeks. FACS sorting (using either BD FACS Aria II or Sony SH800S) at 56 days (8 weeks) post-differentiation was used to isolate EGFP-labeled neurons (Figure S1A). After cells were isolated as either progenitors or neurons, we added Qiazol and stored the mixture at −80°C for randomized RNA isolation to reduce batch effects.
Immunofluorescence labeling and imaging
At the progenitor stage or after 8 weeks of differentiation, we fixed the cells by incubating them in 4% PFA and performed permeabilization with 0.4% Triton in PBST. We used 10% goat serum dissolved in PBST for blocking. We incubated blocked samples with primary antibodies dissolved in PBST solution with 3% goat serum at 4°C overnight followed by washing 3 times with PBST. Samples were subject to incubation in fluorophore-conjugated secondary antibodies, for 1 h at room temperature, then they were stained with DNA-binding dye DAPI with 10 min incubation. We used antibodies with concentrations listed as follows: SOX2 (1:400, rabbit, Millipore #AB5603), Ki67 (1:1,000, rat, Invitrogen #14-5698-82), HOPX (1:1,000, Sigma-Aldrich, Catalog#:HPA030180, Lot#: C105752), TUJ1 (1:2,000, mouse, Biolegend #801202), GFP (1:500, Millipore, Catalog#: AB16901, Lot#:2712295), Alexa Fluor 568 (1:1,000, goat anti-rabbit, Invitrogen #A11036), Alexa Fluor 647 (1:1,000, goat anti-rat, Invitrogen #A21247), Alexa Fluor 488 (1:1,000, goat anti-mouse, Invitrogen #A11001).
RNA-seq library preparation
We isolated RNA from progenitors and neurons using the QIAGEN miRNeasy Minelute kit, quantified RNA concentration with a Qubit 2.0 fluorometer, and assessed RNA integrity via eRIN scores using the Agilent Tapestation. We prepared libraries for sequencing using Kapa Biosystems KAPA Stranded RNA-seq with Riboerase (HMR) kit by loading 50 ng of total RNA into the initial reaction. We followed the manufacturer’s instructions for fragmentation and PCR steps. To obtain ∼350 bp average insert size, we fragmented cDNA at 85°C for 6 min. Final library concentrations were determined using Qubit 2.0 fluorometer and pooled to a normalized input library. Pools were sequenced on a NovaSeq S2 flowcell using 150 bp PE reads with an average read depth of 99.8M ± 29.8 SD read pairs per sample.
RNA-sequencing data processing
We merged fastq files from the same library when sequenced on multiple flow cells and trimmed the adapters using sequences provided by Illumina with Cutadapt/1.15.27 Quality control of each library was performed with FastQC. For alignment, we first integrated the sequence of AAV2-hSyn1-eGFP plasmid used for labeling neurons into GRCh38 release92 reference genome. Then, we aligned the fastq files to this combined reference genome by implementing STAR/2.6.0a aligner.28
We processed aligned data further with different steps based on downstream analyses. To estimate gene expression levels, we quantified reads with the union exon based approach using featureCounts, where for each gene, all overlapping exons were merged to form union exons, and the reads mapped to those union exons with the same strandedness were counted.29 Gene models were identified using the GTF file Homo_sapiens.GRCh38.92 merged with AAV2-hSyn1-eGFP plasmid.
For allele-specific expression and splicing quantification, we remapped the aligned data with WASP software (v2018-07)30 to reduce reference mapping bias. First, we identified reads overlapping with bi-allelic SNPs within our acquired genotype data. Following this, the genotype of any reads overlapping with a SNP was swapped with the other allele, and re-mapped. WASP discarded re-mapped reads that did not map to the same genomic position. As a final step, we implemented the rmdup.py script provided in the WASP software which removes duplicate reads randomly, regardless of their mapping score.
Mycoplasma contamination test
Adaptor trimmed reads (see above) were mapped using STAR to a combined reference including the GRCh38 release 92 human reference genome, AAV2-hSyn1-eGFP plasmid, and more than 1,400 mycoplasma genomes. Alignment parameters allowed for simultaneous mapping of reads to one or more human and mycoplasma genomes. No sample exceeded 0.11% of total reads mapping to any mycoplasma genome, indicating none of our cultures were contaminated with mycoplasma. This mapping strategy was only used for mycoplasma contamination analysis and not for subsequent analyses.
Genotype processing
We performed genotyping using Illumina HumanOmni2.5 or HumanOmni2.5Exome platform and exported SNP genotypes to PLINK format following the procedure previously described.17 Briefly, we converted SNP marker names from Illumina KGP IDs to rsIDs using the conversion file provided by Illumina. We performed quality control with PLINK v.1.90b3 software31 as follows. We filtered out SNPs with the following criteria: variant missing genotype rate >5% (--geno 0.05), deviations from Hardy-Weinberg equilibrium at p < 1 × 10−6 (--hwe 10−6), minor allele frequency <1% (--maf 0.01). We also filtered out individuals with missing genotype rate >10% (--mind 0.10). We obtained 1,760,704 directly genotyped variants surviving our QC procedure. Lastly, we called sex from genotype data using PLINK v.1.90b3 software based on heterozygosity on the X chromosome. When there was an ambiguity for sex assessment based on genotype data, we checked XIST expression. We estimated the population structure of our study cohort by implementing multidimensional scaling (MDS) for genotype data of our samples and genotype data from HapMap3, following the protocol from the ENIGMA consortium. By plotting MDS1 versus MDS2, we visually show each donor’s ancestry relative to known populations (Figure S2B).
Imputation
After filtering genotype data, we pre-phased the data with SHAPEIT v.2.837.32 For our imputation reference panel, we used 1000 Genomes Project Phase 3 that contains a total of 37.9 million SNPs in 2,504 individuals with multiple ancestries, including those from West Africa, East Asia, and Europe.33 Imputation was implemented using Minimac4 software34 (v.1.0.0). On the X chromosome, we separately performed pre-phasing and imputation steps for the pseudoautosomal region and non-pseudoautosomal regions. Following imputation, we retained any variants with missing genotype rate lower than 0.05, Hardy-Weinberg equilibrium p value greater than 1 × 10−6, and minor allele frequency (MAF) bigger than 1%. We retained SNPs with sufficient imputation quality (R2 > 0.3), and obtained approximately 13.6 million SNPs in total.
Sample quality control
One library with missing eRIN score and one library with missing final cDNA concentration from neurons were removed. In order to detect sample swaps or mixing between samples, we evaluated consistency of genotypes called from the RNA-seq and genotyping array via VerifyBamID v.1.1.3.35 We removed the RNA-seq libraries file with [FREEMIX] > 0.04 or [CHIPMIX] > 0.04 (nlibrary = 14). Also, we corrected samples where we detected swaps (nlibrary = 8). After quality control, we retained 85 unique donors for progenitors, and 74 unique donors for neurons for subsequent analyses.
Replicate correlation and determination of technical factors correlating with gene expression
Quantified RNA-seq reads with featureCounts were imported to generate a gene count matrix in DESeqDataSet format from DESeq2 R package.36 We filtered out the lowly expressed genes (those where fewer than 10 read counts of a gene were observed in fewer than 5% of samples), and normalized the data via variance stabilizing transformation (vst()) function from DESeq2 R package.36 We included genes on the X and Y chromosomes and genes transcribed from mitochondrial DNA meeting the expression criteria. We subset the normalized gene expression matrix into progenitor- and neuron-specific samples. To identify major axes of variation in gene expression across samples, we computed principal components of gene expression with prcomp() function from stats R package for each cell type separately and reported the proportion of variance explained by each component.
We recorded biological and technical variables for each sample which may potentially impact gene expression: cell type, postconception week, sex, tissue acquisition date, researcher extracting RNA and preparing libraries, RNA input amount, index number and bases, final cDNA concentration, BioAnalyzer run date, average fragment size of BioAnalyzer cDNA, sequencing pool, cell input, Qiazol lot number and addition date, eRIN, RNA extraction date, RNA tapestation date, QIAGEN extraction kit lot number, FACS sorting date and time, total live cells during sorting, FACS machines used, researcher performing FACS sorting, papain lot number and addition date, differentiation rank (a qualitative assessment of cell health evaluated under the microscope), well location in the 6-well plate, date to plate for differentiation, researcher washing and differentiating cells and date, virus addition date, researcher adding virus, PBS lot number used for cell proliferation and differentiation, laminin, polyornithine lot numbers used for proliferation and differentiation, donor ID, round, media lot numbers used for proliferation, passage number, split dates, researcher performing each split, rank for proliferation (qualitative assessment of cell health), trypsin lot number used for splitting cells, and fibronectin lot number. To identify technical covariates impacting expression levels, we assessed whether any recorded biological or technical variables were significantly correlating with the first 10 expression PCs separately for each cell type. We observed that different FACS machines (Sony SH800S with ndonor = 8; FACS Aria II with ndonor = 66) used to isolate GFP-labeled neurons had a strong impact on global gene expression in neurons (PC1: r = 0.59, p value = 1.782e−08; PC2: r = 0.58, p value = 3.972e−08) (Figure S2D). To remove the impact of sorter on global neuron expression profiles prior to differential expression analysis, we implemented limma::removeBatchEffect function.37 Then, we combined the gene expression matrix from batch-corrected neurons with progenitors gene expression data.
We cultured 20 donors multiple times during the course of the experiment in order to quantify cell culture-induced noise. We calculated Pearson’s correlation of gene expression between libraries from the same donors (nlibrary-library pairs = 15 in progenitors and nlibrary-library pairs = 12 in neurons), and between each library across donors in a pairwise manner (nlibrary-library pairs = 11,556 for progenitors; nlibrary-library pairs = 9,312 for neurons). For neurons, we used gene expression values after batch correction with the limma R package for the sorter type, as described above. We performed an unpaired two-sided t test for statistical assessment of mean difference between these two categories after fisher’s z transformation of correlation r values (Figure S1C).
Differential gene expression analysis
We identified differentially expressed genes between progenitors and neurons by using vst normalized expression values corrected for sorter with limma R package.37 We retained the genes if at least 10 counts of the gene were present in more than 5% of the samples from either one of the cell types. To perform a paired differential gene expression analysis, which inherently controls for donor-related differences, we established the following design matrix: model.matrix(∼CellType + as.factor(DonorID) + RIN, data). Following this, we adjusted p values for each gene via multiple test correction with the Benjamini-Hochberg procedure38 and defined significant differentially expressed genes as adjusted p value < 0.05.
Gene Ontology analysis
We performed gene ontology enrichment analysis by using the gprofiler2 package as the R interface to the g:Profiler tools by using GO:BP database.39 For differentially expressed genes, after performing DGE analysis, we categorized the genes into two groups as upregulated in progenitors (logFC < −1.5 and adjusted p value < 0.05) and upregulated in neurons (logFC > 1.5 and adjusted p value < 0.05) (Figure 1D). For each enrichment analysis, we applied multiple test correction and considered only pathway enrichments with adjusted p value lower than 5% false discovery rate as statistically significant.
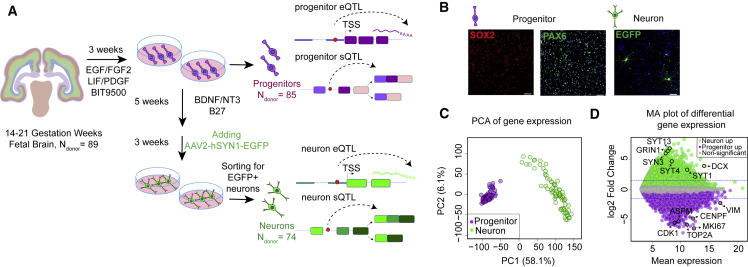
Figure 1.
Study design and cell-type-specific expression
(A) Study design illustrating the fetal brain tissue derived cell-type-specific system used to perform eQTL and sQTL analysis.
(B) Immunofluorescence of the cells showed that undifferentiated progenitors were SOX2 (in red) and PAX6 (in green) positive, and 8-week differentiated neurons labeled with AAV2-hSyn1-EGFP were positive for EGFP (in green) (scale bar is 100 m, DAPI in blue).
(C) Principal component analysis of progenitor (purple) and neuron (green) transcriptomes from each donor indicates cell-type-specific clustering.
(D) MA plot showing differentially expressed genes in progenitor versus neurons. log2FC > 0 and adjusted p value < 0.05 indicates genes upregulated in neurons shown in green (neuron up), log2FC < 0 and adjusted p value < 0.05 indicates genes upregulated in progenitors shown in purple (progenitor up) and genes not significantly differentially expressed between two cell types are shown in gray. Blue lines indicate |log2FC| > 1.5.
Transition mapping (TMAP)
To evaluate the transcriptomic similarity between our in vitro culture system and the in vivo brain, we performed transition mapping analysis as described in previous work.26,40 To evaluate transcriptomic similarity to cortical laminae in the developing brain, we used previously published laminar expression data from laser capture microdissections of prenatal human brain41 (H376.IIIB.02. female, 16 pcw, brainspan.org). In our comparison, genes were retained which showed expression in either cell type and were present on the array in which the in vivo data were acquired. We used gene symbols to find ensemblIDs and used ensemblIDs to match with in vitro data. When multiple probes were present for a given gene on the array, the probe with the highest expression per gene was used. We quantile normalized the gene expression and we performed in vivo differential gene expression via limma between every two laminae. Similarly, we performed differential expression analysis in our in vitro cultures as described above. We applied transition mapping via RRHO2 R package with “stratified approach” to avoid misinterpretation of the discordant overlaps.42 In this algorithm, first genes were ranked based on their degree of differential expression (DDE) (i.e., −log10(p value) × signed effect size) separately for in vivo and in vitro data. Following ranking, a hypergeometric test was applied to assess enrichment for each overlap between two datasets for a series of arbitrary step sizes. By employing a stratified algorithm, we computed the degree of overlap. Finally, we visualized the hypergeometric test −log10(p values) as a heatmap (Figure S1G).
Cell-type-specific local eQTL mapping
To perform local eQTL analysis, we conducted an association test between gene expression (retaining genes if at least 10 counts of the gene were present in more than 5% of the samples of that cell type, resulting in 24,778 and 27,649 genes for progenitors and neurons, respectively) with genetic variants within ±1 Mb window of gene TSS for both autosomal chromosomes and X chromosome, for progenitors and neurons separately. Each gene TSS was defined as the transcription start site of the gene isoform with the most upstream exon based on GTF file Homo_sapiens.GRCh38.92.
We removed variants of low allele frequency in order to prevent one donor from strongly influencing association results. For variant selection, PLINK v.1.90b3 software function was implemented to obtain donor counts per genotype group for each variant. We included only variants with at least two heterozygous donors and no homozygous minor allele donors, or at least two minor allele homozygous donors for autosomal chromosomes, and for X chromosome we retained the variants with at least two haploid allele counts in addition to this criteria.
For eQTL mapping, we used a linear mixed effects regression model to control for population stratification and cryptic relatedness with EMMAX software.43 To compute the kinship matrix, we implemented emmax-kin -v -h -d algorithm creating the identity by state (IBS) kinship matrix by excluding all genetic variants located on the same chromosome as the tested variant from non-imputed genotype data for each single variant association test (MLMe method; see Yang et al.44). We used additional ancestry control by including the first ten MDS components from the genotype data as covariates.45 In order to control for unmeasured technical variables impacting gene expression, we computed global gene expression PCs. To optimize eQTL discovery, we sequentially added gene expression PCs and re-ran the genetic associations via EMMAX. For neurons, we included a covariate for FACS sorter for each run given its strong impact on gene expression.
The full association model for neurons was:
expression ∼SNP + 10 MDS of global genotype + kinship matrix + FACS sorter + PCs of global gene expression
The full model for progenitors was:
expression ∼SNP + 10 MDS of global genotype + kinship matrix + PCs of global gene expression
For each run, we adjusted nominal values of all gene variant associations, and defined significant associations with nominal p value lower than 5% false discovery rate (FDR).38 We found that 10 PCs and 12 PCs of gene expression resulted in a maximum number of eGenes discovery in progenitors and neurons, respectively (Figure S2E). Our final eQTL model was:
Neuron:
expression ∼SNP + 10 MDS of global genotype + kinship matrix + FACS sorter + 12 PCs of global gene expression
Progenitors:
expression ∼SNP + 10 MDS of global genotype + kinship matrix + 10 PCs of global gene expression
In order to stringently control our association results for both number of variants and genes tested, we further implemented a hierarchical correction procedure called eigenMT-FDR46 for the models optimized above. Using this method, as step 1, we adjusted the nominal p values of the all cis SNPs separately for each gene to compute locally adjusted p values with the eigenMT method that resulted in the estimation of effective number of independent tests from the genotype correlation matrix including cis SNPs.47 In step 2, locally adjusted minimum p values for all genes were then subjected to FDR procedure to obtain globally adjusted p values. In step 3, we defined eGenes as genes with globally adjusted p value lower than 0.05. Then, to find other independent SNPs for those eGenes, we set the significance threshold as the maximum nominal p value from step 1 that had corresponding globally adjusted p value lower than 0.05.
We performed conditional analysis by using this threshold p value gathered from the eigenMT-FDR multiple correction method to identify independent significant eQTLs. To identify conditionally independent eQTLs, for each eGene (a gene significantly associated with at least one variant), we iteratively included the hard call genotype of the variant with the strongest association with eGene as a covariate and re-ran the regression model specified above (Figure S3A). We defined a variant as “conditionally independent” from the variant conditioned on, if the association of the variant with the eGene was still significant based on the initial threshold p value. Then, we conditioned on those variants that met threshold p value condition at the first round plus the primary variant and identified third conditionally independent eQTLs. We applied this procedure iteratively until no additional significant eQTLs remained.48,49
Bulk fetal brain eQTL mapping
We utilized bulk fetal cortical wall eQTL data described previously.19 We re-analyzed data in this study with the following modifications to harmonize with the eQTL approach implemented in this study: (1) we controlled for population stratification using a linear mixed effects model as described above and (2) we included 23 additional donors which were genotyped after the publication of the previous manuscript. We used rRNA-depleted RNA-seq data from flash frozen human fetal brain cortical wall tissues derived from 240 donors at 14–21 gestation weeks (inferred to be 12–19 post conception weeks). We excluded 4 donors for sample swap and contamination based on verifyBAMID analysis, and 1 donor with sex ambiguity, resulting in 235 unique donors for eQTL analysis (35 of unique donors shared with cell-type-specific data). Gene-based annotations of the genome were derived from Homo sapiens gene ensembl v.92 (GRCh38) for eQTLs. We included only genes with at least 10 counts in 5% of donors. We normalized the data with the VST method to be used as phenotype in eQTL analysis. We also extracted genomic DNA from the same donors and performed genotyping on a dense array (Illumina Omni 2.5+Exome) and imputation to a common reference panel (1000 Genomes Phase 3; described above). Variants were retained in the analysis if there were at least two heterozygous donors and no homozygous minor allele donors, or if there were at least two minor allele homozygous donors as for cell-type-specific eQTLs, as described above.
We performed local eQTL analysis to test the association between each gene’s expression and variants within the ±1 Mb window of the transcription start site of each gene. We applied linear mixed model association software EMMAX43 to control for population stratification and cryptic relatedness (as described above for cell-type-specific eQTL analysis). We used the linear mixed effects regression model testing association between expression of each gene and nearby genetic variants, controlling for ten MDS genotype components, ten PCs of gene expression, and a kinship matrix as random effect excluding the chromosome genotypes testing with the MLMe approach.44 After association, nominal p values were corrected for hierarchical multiple testing using the eigenMT-FDR method as described above, and we obtained independent eQTLs performing conditional analysis as described for cell-type-specific eQTLs above.
Enrichment of eQTLs within functional genomic annotations
To identify enrichment of eQTLs and sQTLs within functionally annotated genomic regions, we implemented GARFIELD software to control for the distance to TSS, LD, and minor allele frequency (MAF) of QTLs.50 We used functional genomic annotations from 25 chromatin states given in the ChromHMM BED files of Roadmap Epigenomics project from human male fetal brain51,52 lifted over from hg19 to hg38. For all eQTLs, we extracted the p value from the strongest association for each variant (with minimum p value) in the case that one variant was associated with multiple genes. To create annotation files, we considered a variant overlapping with a functional element if the variant itself or any of the variants in high LD within 500 kb (r2 > 0.8) overlapped with each of the annotation categories. LD pruning50 was performed at r2 > 0.01 within GARFIELD software. Following this, a logistic regression model controlling for the distance to TSS of the gene with the strongest association to the tested SNP, LD proxies, and MAF binned for five quantiles was performed with GARFIELD software for enrichment at eigenMT-FDR p value thresholds defined in eQTL analysis. The effective number of annotations were estimated and multiple testing adjusted p values were computed by the software to identify enrichment of eQTLs within defined annotations.
Enrichment of eGenes within likely in vitro artifacts
To determine whether eQTL discovery was driven by in vitro artifacts, we performed an enrichment analysis via fgsea software53 to test whether discordant genes between in vivo laminar expression data41 and our cell-type-specific in vitro data were enriched among cell-type-specific eGenes. To define discordant genes, we used two lists of differentially expressed genes from in vivo oSVZ versus SP (selecting these regions as the most overlapping with our cell types in Figure S1G) and separately from in vitro progenitor versus neurons as described for TMAP analysis. We defined the discordant genes as genes with adjusted p value lower than 0.01 and opposing sign of log fold change. Then, for each cell type, we tested for enrichment of discordant genes among all eGenes ranked by their ascending m-value (from low values for cell-type-specific to high values for shared effect size).
Allele-specific expression analysis pipeline
To identify sites with allele specific expression (ASE), we initially extracted uniquely mapped reads from the RNA-seq data remapped with WASP to reduce mapping bias and to discard duplicate reads; then, we applied the ASEReadCounter algorithm from GATK tools.54 For each donor, we counted allele-specific reads overlapping with bi-allelic variants identified in the genotypeVCF files. We retained only variants with at least five heterozygous donors and at least ten counts from either allele (at least two counts supporting each allele). ASE can be falsely called when genotyping errors are present in the dataset. We used two approaches to identify and remove potential genotyping errors. (1) We detected wrongly called variant genotypes by assessing concordance between genotypes called by DNA versus RNA.55 We removed variants that were called homozygous based on the genotype data when at least ten counts of the alternate allele were present in the RNA-seq data, and (2) we discarded variants where at least seven heterozygous donors based on genotype data have zero counts for one of the alleles, which may indicate a donor falsely called as heterozygote when in truth the donor is a homozygote (given that (1/2)7 = 0.008, meaning that probability of having all donors receiving an imprinted allele from either mother or father is low). Because ASEReadCount does not disambiguate the strandedness of reads, it is not possible to confidently assign reads overlapping with multiple gene annotations to a specific gene.54 Therefore, if a variant overlapped with more than one gene annotation, we removed the variant by implementing findOverlaps function from IRanges R package56 for genes based on their genomic coordinates defined GTF file Homo_sapiens.GRCh38.92.
To evaluate allelic imbalance, we used DESeq2 with the design: design = ∼0 + RNAid + Allele. Excluding homozygous donors, we computed the log2 fold change of non-reference allele counts over reference allele counts and used a Wald test to detect allelic imbalance by setting fitType = “mean” after visual inspection of dispersion. Multiple test correction was performed with the Benjamini and Hochberg method, and we defined significant ASE sites as those with adjusted p values lower than 0.05.
To compare eQTLs with ASE sites (Figures 2B, S4F, and S4G), we extracted eQTLs associations with the variants tested for ASE analysis (at least 5 heterozygous donors and overlapping with at least 10 RNA-seq reads). We also extracted eGenes (defined based on significant eigenMT-FDR global p value) with at least 10 counts per donor. To calculate allelic fold change (aFC) for the eQTLs in this list, we applied aFC software57 using VST normalized genes and controlling for the same fixed effect covariates used for eQTL analysis.
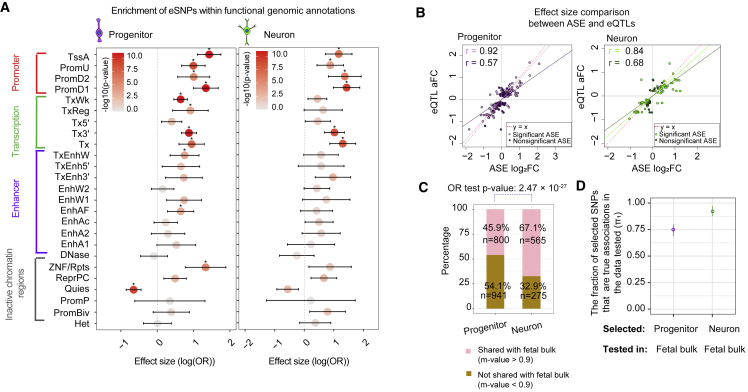
Figure 2.
Cell-type-specific eQTL analysis
(A) Enrichment of progenitor eSNPs (left) and neuron eSNPs (right) within chromatin states in the fetal brain from chromHMM listed on the y axis. The x axis shows the effect size of enrichment with 95% upper and lower confidence interval and the plot is color-coded based on −log10(p value) value from enrichment analysis. Significant enrichments are shown with an asterisk. Enrichment was tested using eQTLs thresholded at the eigenMT-FDR p value.
(B) Comparison of the effects of shared ASE sites and eQTLs in progenitors (left in purple) and neurons (right in green). Nonsignificant ASE sites are shown as darker colors for both cell types, and significant ASE sites are shown as lighter colors. Correlation coefficient (r) values are indicated in colors for each category and the red dashed line indicates y = x.
(C) Overlap percentage of cell-type-specific eSNP-eGene pairs shared with fetal bulk eQTLs in progenitors and neurons at m-value > 0.9. Odds ratio (OR) test p values are shown.
(D) The fraction of progenitor/neuron primary eGene-eSNP pairs that are true associations (π1) in fetal bulk eQTLs. 95% upper and lower confidence interval are shown.
Quantification of intron excisions
To identify alternatively excised introns, separately for each cell type, we extracted exon-exon junctions from uniquely mapped reads from WASP-mapped RNA-seq data in BAM format via regtools function where reads map to a minimum of 6 nt of each exon.58 Next, we processed those junctions that are called intron excisions or exon-exon junctions with the pipeline provided by LeafCutter software.59 First, intron excisions with shared splice junctions were clustered together applying an iterative procedure until each cluster has at least 50 reads across donors and introns with maximum 50 kb length, separately for progenitors and neurons. For differential splicing analysis, we performed clustering by combining exon-exon junctions files from each cell type. For each cluster, intron excisions supported by at least one count in more than five donors were retained (within each set of donors contributing to the three different sQTL analyses for that cell type [progenitor, neuron] or tissue class [fetal brain bulk]; or for differential splicing analysis across donors from both cell types used [progenitor + neuron]). We further calculated intron excision ratios and filtered out introns represented in less than 40% of donors (within each set of donors contributing to the three different sQTL analyses for that cell type [progenitor, neuron] or tissue class [fetal brain bulk]; or for differential splicing analysis across donors from both cell types used [progenitor + neuron]) with prepare_phenotype_table.py. We referred to each intron excision ratio as percent spliced in (PSI) that corresponds to the usage of each intron compared to other introns in the same cluster. Standardized and quantile normalized intron excision ratios, and global alternative splicing PCs computed with those ratios were used for downstream analysis.
Differential splicing analysis
To perform differential splicing analysis, we used quantile normalized PSI values as input to the limma package.37 Identical to differential expression analysis, neuron splice ratios were corrected for batch including FACS machine used for sorting with limma::removeBatchEffect function. Batch corrected neuron splice ratios were combined with progenitor data. We implemented a paired differential splicing analysis inherently controlling donor-related differences with the design matrix: model.matrix(∼CellType + as.factor(DonorID) + RIN, data). We defined intron junctions with adjusted p values via multiple test correction with Benjamini-Hochberg procedure38 lower than 0.05 as significant differentially spliced introns.
Splicing QTL mapping
We performed cell-type-specific splicing QTL analysis by testing the association of PSI with the genetic variants located within the ±200 kb window from starting and end points of the splice junctions for autosomal chromosomes and the X chromosome. Identical to local eQTL analysis, we used only genetic variants that met the following criteria: if there were at least two heterozygous donors and no homozygous minor allele donors, or if there were at least two minor allele homozygous donors.
We used standardized and normalized intron excision ratios (percent spliced in) calculated by LeafCutter as the phenotype for sQTL mapping. EMMAX43 was used to test for association between SNPs within a cis-region of ±200 kb of the intron cluster and intron ratios within cluster. We controlled for population stratification and cryptic relatedness as described above for eQTL mapping. Also, we controlled for unmeasured technical variables impacting alternative splicing via computed global splicing PCs. Similar to eQTL analysis, we optimized sQTL discovery by sequentially adding global splicing PCs to the genetic associations via EMMAX. Again for neurons, we additionally controlled for FACS sorter for each run given its strong impact on splicing.
The full model for neurons was:
PSI ∼SNP + 10 MDS of global genotype + kinship matrix + FACS sorter + PCs of global splicing
The full model for progenitors was:
PSI ∼SNP + 10 MDS of global genotype + kinship matrix + PCs of global splicing
For every run, we adjusted nominal values of all PSI variant associations and defined significant associations with lower than at 5% false discovery rate (FDR).38 We found that 1 PC and 1 PC across the PSI matrix resulted in a maximum number of intron excisions with at least one significant association in progenitors and neurons, respectively (Figure S6B). Our final sQTL model was:
Neuron:
PSI ∼SNP + 10 MDS of global genotype + kinship matrix + FACS sorter + 1 PCs of global splicing
Progenitors:
PSI ∼SNP + 10 MDS of global genotype + kinship matrix + 1 PCs of global splicing
Implementing the same hierarchical correction procedure as for eQTLs (eigenMT-FDR46) first, we adjusted the p values of the all cis SNPs strongest association separately for each intron excision to compute locally adjusted p values with the eigenMT method,47 and then locally adjusted minimum p values for all intron excisions were subjected to the BH procedure giving globally adjusted p values. Intron excision with corresponding global p value lower than 0.05 were considered as significant alternative splicing events. In order to find other independent significant sQTLs in addition to the ones associated with lowest p values, we applied conditional analysis at eigenMT-FDR p value threshold as described for eQTL analysis.
For bulk fetal cortical tissue sQTL mapping, we applied the same strategy used for cell-type-specific sQTLs and found the following model maximized significant intron junctions discovery:
PSI ∼SNP + 10 MDS of global genotype + kinship matrix + 6 PCs of global splicing
After calculating eigenMT-FDR threshold p value, we performed conditional analysis to define independent significant sQTLs.
To find genes overlapping with intron excision, we annotated intron junctions by using LeafCutter based on genomic coordinates and gene model provided in GTF file Homo_sapiens.GRCh38.104. Intron junctions assigned as cryptic 5', cryptic 3', or novel annotated pair were considered as novel splicing events for the genes overlapped with junctions including unannotated splice sites for the ARL14EP.
RNA binding protein motif analysis
We performed enrichment of sSNPs in RNA binding protein binding sites via GARFIELD as described above with the only difference being controlling for the distance to the intron with the strongest association to the tested SNP. In this analysis, we used BED files including RNA binding protein sites from a CLIP-seq database as annotation files60 and assessed significant enrichment of cell-type-specific sQTLs for binding sites of each RBP.
Comparison of QTL association methods
To determine the impact and reproducibility of a linear mixed effects model as compared to a standard linear regression on QTL results, we applied the FastQTL61 method in nominal pass mode for different models to run eQTL analysis on autosomal chromosomes. To test for impacts on population stratification, we performed FastQTL (1) without controlling either for population structure or technical confounders, (2) controlling for only technical confounders, (3) controlling for 10 MDS of global genotype and global gene expression PCs. Following this analysis, we compared genomic inflation factors (λGC) across those three groups to our data where we controlled for 10 MDS of global genotype and global gene expression, as well as the cryptic relatedness with kinship matrix.
We also compared autosomal eGenes/significant introns and primary eGene-eSNP/intron-sSNP pairs detected via either EMMAX or FastQTL. For EMMAX analysis, eGenes/significant introns and primary eGene-eSNP/intron-sSNP pairs were defined using the eigenMT-FDR approach with 5% FDR. For FastQTL, eGenes/significant introns and primary eGene-eSNP/intron-sSNP pairs were defined by fitting nominal p values of the most highly associated pairs extrapolated from a beta distribution to adaptive permutations with the setting –permute 1000 10000 as previously described.62 Then, Storey’s q value method63 was applied on permutation p values derived from beta approximation across genes/introns for multiple correction with 5% FDR.
QTL sharing
We estimated m-values to assess cell type specificity of SNP-gene or SNP-intron excision pairs with Metasoft.64 Prior to software implementation, we extracted e/sQTLs from the neuron data corresponding to primary progenitor eSNP-eGene/sSNP-introns junction pairs to determine overlap of sharing significant progenitor e/sQTLs with neuron eQTLs. Similarly, we extracted e/sQTLs from the progenitor data corresponding to neuron primary eSNP-eGene/sSNP-introns junction pairs to determine overlap of sharing significant neuron eQTLs with progenitor e/sQTLs. We estimated standard errors by dividing beta estimates from EMMAX by t-statistics for each association p value. We defined associations shared across different QTLs as m-value > 0.9. Similarly, in order to find significant progenitor/neuron e/sQTLs shared with fetal bulk e/sQTLs, we extracted e/sQTLs from the fetal bulk data corresponding to progenitor/neuron primary eSNP-eGene/sSNP-introns junction pairs and defined shared QTLs at m-value > 0.9.
We also applied the π1 statistic63 to quantify QTL sharing for progenitor versus neuron and progenitor/neuron versus fetal bulk primary eSNP-eGene pairs/sSNP-intron junction pairs using the R qvalue package.65 To find the fraction of progenitor/neuron primary eSNP-eGene pairs that are true associations in neuron/progenitor eQTLs (π1), we extracted nominal p values from neuron/progenitor eQTLs for corresponding progenitor/neuron primary eSNP-eGene pairs. Using the qvalue() function by setting lambda seq (0.2,0.8,0.1), we computed the π0 value and defined the π1 as 1 − π0. The previously described π1 statistic requires the gene to be detectable in both cell types, which may underestimate cell type specificity. To account for this, in a separate analysis, when a SNP-gene pair was not tested in a cell type, we assigned a random p value (sampled from a uniform distribution). We applied the same strategy to find the fraction of progenitor/neuron primary sSNP-intron junction pairs that are true associations in neuron/progenitor sQTLs. Similarly, in order to find the fraction of progenitor/neuron primary eSNP-eGene or sSNP-intron junction pairs that are true associations in fetal bulk eQTLs or fetal bulk sQTLs, we used nominal p values from fetal bulk eQTLs or sQTLs for corresponding progenitor/neuron primary eSNP-eGene or sSNP-intron junction pairs to compute the π1 value.
We considered an LD-based overlap of e/sQTLs between two datasets when the index e/sSNPs were in LD (r2 > 0.8 where LD was calculated in our sample population) and the eSNP-eGene/sSNP-intron pairs were shared. To determine the total number of eSNP-eGene/sSNP-intron pairs as the universe for enrichment analyses, we pruned all variants associated with each gene per gene for r2 > 0.01 by using PLINK command plink --indep-pairwise 50 5 0.01. To determine whether different proportions of sharing were observed between two cell types, we performed an odds ratio test described here.66
To test for temporal specificity of cell-type-specific e/sQTL data, we downloaded GTEx adult brain e/sQTL data.25 We called loci from the two datasets as colocalized when (1) index adult brain e/sQTLs are found within LD buddies of cell-type-specific e/sQTLs at LD r2 > 0.8 (where LD is calculated using either the European population from 1000 Genomes or our study’s population) and (2) the cell-type-specific e/sQTL data conditioned on index adult brain e/sQTLs, the cell-type-specific index e/sQTL no longer survives the global significance threshold.
LD-thresholded colocalization with brain disorders and traits GWAS
To find eQTLs and sQTLs colocalized with index GWAS loci, we performed LD-thresholded colocalization analysis for each cell type separately.67 We used summary statistics of GWASs for schizophrenia (SCZ)1 (MIM: 181500), major depression disorder (MDD)68 (MIM: 608516), bipolar disorder (BP)2 (MIM: 125480), educational attainment (EA),69 neuroticism,70 IQ,5 cognitive performance (CP),69 attention-deficit/hyperactivity disorder (ADHD)6 (MIM: 143465), Alzheimer disease (AD)71 (MIM: 104300), Parkinson disease (PD)72 (MIM: 168600), insomnia,73 epilepsy74 (MIM: 600669), autism spectrum disorder (ASD)75 (MIM: 209850), and cortical thickness and surface area from the ENIGMA project.4 We used liftover to convert the positions of variants in GWAS summary statistics from hg19 to hg38 with liftOver function from R rtracklayer package.76 Variant rsids were assigned with dbSNP151 based on positions of variants in summary statistics data. To define index GWAS SNPs at genome-wide significant threshold p value (5 × 10−8), we implemented a clumping procedure, where we defined two LD-independent GWAS signals so as to have pairwise LD r2 < 0.5 based on LD matrix computed with European population of 1000 Genomes (1000G European phase 3). Prior to clumping, duplicated rsIDs in 1000G EUR genotype files were assigned with unique names, and BIM files were modified for each chromosome. Following a unique id assignment, BIM files were merged back to BED and FAM files with --bmerge function of PLINK1.9 software (plink --bfile BED file --bmerge modified_BIM file). Since all GWASs we leveraged in our colocalization analysis have been conducted in populations of European ancestry, and our study population is multi-ancestry, we computed LD r2 separately within these two different populations. We considered the index eQTL or sQTL SNP coincident with the index GWAS SNP if the pairwise LD r2 between them was greater than 0.8 based on either the LD matrix computed via either European 1000 Genomes Phase 3 data or our study population. Following that, we performed a conditional eQTL/sQTL analysis by conditioning on the coincident index GWAS SNP. If the association of index QTL and gene expression or intron excision was no longer significant based on p value thresholds defined with eigenMT-FDR method for each dataset, we identified that cell-type-specific and fetal bulk eQTL/sQTL as a colocalized loci with the given GWAS trait. Since GTEx raw data are not available publicly, conditional analysis was not performed to infer colocalization.
Transcription factor motif analysis
We used motifbreakR to detect the disruption of the transcription motif binding site where there was a variant within a chromatin accessibility peak (Figure 4D).77
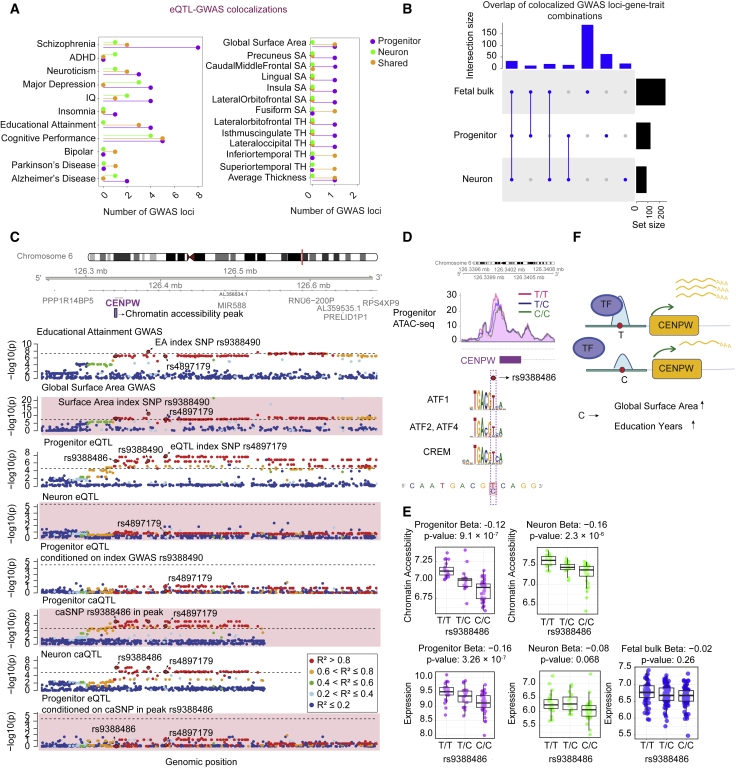
Figure 4.
Colocalization of cell-type-specific eQTLs with GWAS for brain-related traits
(A) Number of GWAS loci colocalized with progenitor (purple)- or neuron (green)-specific eQTLs or both cell types (orange). Each GWAS trait is listed on the y axis (SA, surface area; TH, thickness).
(B) LD-based overlap of colocalized GWAS loci-gene pairs per trait combinations across progenitor, neuron, and fetal bulk eQTL colocalizations for the traits listed in (A).
(C) Genomic track showing regional association of variants with educational attainment (EA), global surface area (GSA), and CENPW expression in progenitors and neurons, −log10 of association p values on the y axis, and genomic location of each variant on the x axis. Progenitor eSNP rs4897179 (3rd row) was coincident with index SNP (rs9388490) for both EA (1st row) and GSA GWAS (2nd row), and conditioning progenitor eSNP rs4897179 on rs9388490 showed colocalization of the two signals (5th row). Also, rs4897179 was colocalized with another variant (rs9388486) located in the chromatin accessibility peak at the promoter of CENPW (6th and 8th rows). Genomic tracks were color-coded based on LD r2 relative to the variant rs9388486. Dashed line indicates significance threshold.
(D) Plot showing the chromatin accessibility peak (chr6:126,339,531–126,340,960) in progenitors across different genotypes of rs938848. The C allele of rs9388486 disrupted binding motifs of transcription factors including CREM, ATF1, ATF2, and ATF4.
(E) Boxplots showing chromatin accessibility across rs9388486 genotypes in progenitors (purple) and neurons (green) (top). Boxplots showing VST normalized CENPW expression across rs9388486 genotypes in progenitors (purple), neurons (green), and fetal bulk (blue) (bottom).
(F) A schematic showing that one or more of the implicated transcription factors (TF) has decreased preference to bind at the C allele, which results in lower CENPW expression, increase in global surface area, and educational attainment.
TWAS analysis
We performed transcriptome-wide association analysis for progenitor and neurons separately with FUSION software.78 First, we obtained a set of variants shared between the genotypes from 1000 Genomes European phase 333 and our study population restricted to variants described for eQTL analysis and removed monomorphic variants within European genotype data. We estimated cis-heritability of genes (including variants within ±1 MB window of the TSS) and intron junctions (including variants within ±200 kb window of two ends of intron junctions) with GCTA software79 by controlling for the same covariates for global gene expression/splicing and 10 PCs of global genotypes used in e/sQTL analysis. VST normalized gene expressions were further subject to quantile normalization for heritability estimation. 1,703/973 genes and 6,552/6,578 intron junctions were significantly cis-heritable in progenitors/neurons for heritability p value < 0.01. To determine the method to be used to estimate the genetic component of gene expression/splicing (weights), we performed leave-one-out cross validation80 for the prediction models including LASSO regression,81 Elastic-net regression82 and EMMAX43 within FUSION software. We used the weights computed from the prediction model with the highest cross validation R2 (the highest performance) per gene/intron junction for downstream analysis for progenitor, neuron, and fetal bulk brain tissue. To evaluate the reproducibility of TWAS analysis, we pseudo-randomly (maintaining similar proportions of donor ancestry) down-sampled the fetal bulk eQTL data to the sample size of progenitor (ndonor = 85) and neuron (ndonor = 74) data twice per cell type, and calculated weights. For adult brain bulk tissue data, we obtained the weights of genes and intron junctions from CommonMind Consortium study.83 Also for the reproducibility of TWAS analysis, we used the weights of genes from GTEx adult frontal cortex (BA9) v.7 model.62
Before running TWAS analysis, we prepared GWAS summary statistics for schizophrenia (SCZ),1 major depression disorder (MDD),68 educational attainment (EA),69 neuroticism,70 IQ,5 Alzheimer disease (AD),71 Parkinson disease (PD),72 and global surface area (GSA) and average thickness from ENIGMA study4 with following adaptations: (1) we obtained common variants found both in genotype files from our study and in GWAS summary statistics; (2) we calculated z-score by dividing the beta coefficient by the standard error if the beta coefficient was available in the summary statistics, or dividing the natural logarithm of odds ratio by the standard error if odds ratio was given in the summary statistics; (3) we matched the sign of the z-score based on the allelic directionality of weights from FUSION software.
To perform TWAS analysis, we tested the association between the predicted gene expression/splicing (w) and brain traits listed above (Z) by implementing the algorithm ZTWAS = w’ Z/sqrt(w’Dw) where D is the LD matrix as the covariance among all cis-variants from the FUSION software.78,83 Since the population structure of our dataset was different from European neuropsychiatric GWASs, we performed TWAS analysis separately with different LD estimates computed based on our study or European population from 1000 Genomes Phase 3 as the covariance. For variants missing in GWAS summary statistics which existed in our study’s genotypes, we implemented IMPG imputation84 allowing imputation of maximum 40% of missing variants within the FUSION algorithm.
To identify genes/intron junctions not driven by co-expression, we defined jointly independent genes/intron junctions through performing summary-statistic-based joint analysis,85 where we replaced SNPs with genes/intron junctions as described in previous work83 within the FUSION software. Implementing genes/intron junctions to the model one at a time in decreasing order of significance, we evaluated whether the conditional TWAS test remained significant. Those with significant conditional TWAS association were defined as jointly independent.
Results
Transcriptomic profiles of primary human progenitors and neurons recapitulates cell-type-specific characteristics of cortical development
We established an in vitro culture of primary human neural progenitor cell (phNPC) lines derived from genotyped human fetal brain tissue (n = 89 unique donors) at 12–19 post conception weeks (PCW) (14–21 gestation weeks), that recapitulates the developing human neocortex26,86, 87, 88 (Figure 1A, Material and methods). Immunofluorescence of the cells showed that undifferentiated progenitors were PAX6 and SOX2 positive (90%–95%), consistent with a homogeneous culture of radial glia89,90 (Figure 1B). At 5 weeks post-differentiation, phNPC cultures were transduced with a virus which expresses EGFP in neurons (AAV2-hSyn1-EGFP), which enabled us to isolate neurons via FACS sorting at 8 weeks post-differentiation (Figures 1A, 1B, S1A, and S1B, Material and methods).
We acquired transcriptomic profiles of progenitors and neurons via RNA sequencing, observing a strong correlation of libraries from the same donor cultured at different times (Figure S1C). After correction for technical confounds (Figure S1D), progenitors and neurons clustered separately by principal component analysis (PCA) of global gene expression, indicating global transcriptomic differences by cell type (Figure 1C). Both cell types showed expected expression of a variety of known cell-type-specific markers (Figure S1E). Next, we identified differentially expressed genes, which were enriched in cell cycle and neurotransmission gene ontology terms, upregulated in progenitors and neurons, respectively (Figures 1D and S1F, Table S1).
We evaluated how well the in vitro progenitors and neurons we generated model in vivo neurodevelopment. We implemented the transition mapping (TMAP) approach for a global assessment of transcriptomic overlap between in vitro cultures and in vivo post-mortem human brain samples, as described in our previous work26 (Material and methods). We compared the transition from progenitor to neurons with laser capture microdissection of cortical laminae from postmortem human fetal brain at 15–21 PCW.41 We observed the strongest overlap in the transition from progenitors to neurons with the transition from outer subventricular zone (oSVZ) to intermediate zone (IZ) or subplate zone (SP) (Figure S1G), supporting the in vivo fidelity of our culture system representing neurogenesis during mid-fetal development.
Cell-type-specific genetically altered gene expression via local expression quantitative loci (eQTL) analysis
To investigate the impact of genetic variation on gene expression, we performed a local eQTL analysis by testing the association of each gene’s expression levels with genetic variants residing within ±1 Mb window of its transcription start site (TSS)62,91 (Figure S2A, see Material and methods). We implemented a linear mixed effects model (LMM) to stringently control for population stratification using a kinship matrix as a random effect with inferred technical confounders as fixed effects, separately for each cell type (λGC for progenitor = 1.028 and λGC for neuron = 1.007; see Material and methods, Figures S2B–S2E). After retaining associations that were lower than 5% false discovery rate with a hierarchical multiple testing correction46,47 (Material and methods), we obtained conditionally independent eQTLs (Figures S3A and S3B, see Material and methods). We identified 1,741 eGenes with 2,079 eSNP-eGene pairs in progenitors and 840 eGenes with 872 eGene-eSNP pairs in neurons (Figure S3C and Table S2). As a complementary analysis, we performed eQTLs using a linear model approach (FastQTL)61 followed by an adaptive permutation. We detected 90%/93% of eGenes and 87%/90% of primary eSNP-eGene pairs discovered via the LMM approach in progenitor/neuron were also identified using the standard linear model, indicating that our LMM approach was highly robust and reproducible (Figure S3D).
To determine whether our detected eQTLs were driven by in vitro artifacts, we tested whether eGenes were enriched in genes with discordant expression between our in vitro culture and the in vivo brain. We selected low-fidelity genes as those with opposing directions of differential expression effect size between in vivo oSVZ versus SP and in vitro progenitor versus neuron (Figure S1G). We did not observe an enrichment of cell-type-specific eGenes within this low fidelity gene list in neurons or progenitors (Figure S4A). This observation suggests that the potential confounding effect of in vitro conditions in our model system was not a major driver of cell-type-specific eGene discovery.
We next evaluated QTL sharing across cell types using multiple different methods to increase confidence in the findings: (1) LD-based overlap, i.e., high LD between significant index SNPs indicates a shared effect, (2) m-values,92 i.e., posterior probability of the shared effect, and (3) π1,63 i.e., the proportion of QTLs selected in one dataset that are true positives in another. We observed that 14.8%/35.5% of progenitor/neuron conditionally independent eSNP-eGene pairs were shared with the other cell type using LD-based overlap (Figure S3C). 53.1%/69.3% of progenitor/neuron primary eSNP-eGene pairs were shared with the other cell type with m-value92 > 0.9 (Figure S4B). Also, the fraction of progenitor/neuron primary eSNP-eGene pairs that are true associations in neuron/progenitor eQTLs (π1) was 76.9%/91.4%, when subset to gene-SNP pairs that were detectable in both datasets (Figure S4C). A higher shared effect for neuron primary eQTLs with progenitor eQTLs than progenitor primary eQTLs with neuron eQTLs suggested similar genetic effects on transcriptomes in immature neurons with their parent cells, whereas parent progenitor cells have unique features, such as proliferation ability, that are not present in neurons.
We determined whether eSNPs were enriched in specific functional chromatin annotations in fetal human brain,51 (Figure 2A). Both progenitor- and neuron-specific eSNPs were enriched in promoters and actively transcribed sites present in the fetal brain, and progenitors were enriched in enhancers regions and depleted in quiescent chromatin regions. Importantly, 40.8%/38.8% of progenitor/neuron-specific significant eQTLs (restricted to variants tested for allele-specific expression [ASE] analysis), respectively, were supported by cell-type-specific ASE, that is less susceptible to cross donor technical confounding, like population stratification55,62,93 (Figures S4D–S4G, Table S2). For the significant eQTLs tested but unsupported by ASE, low power in the ASE analysis where only heterozygous donors were tested may have masked their significant detection in the ASE data. Also, the eQTLs supported by ASE sites were highly concordant in effect size and direction (Figure 2B), providing further confidence in the identified allelic effects on gene expression.
Comparing cell-type-specific eQTLs to bulk eQTLs
We aimed to determine the utility of our cell-type-specific eQTL study by comparison to pre-existing bulk brain eQTL studies. Comparing our results to a bulk fetal cortical wall eQTL dataset from a previous study using a partially overlapping set of donors,19 we observed that 26.2%/45% of progenitor/neuron conditionally independent eSNP-eGene pairs were shared with the fetal bulk eQTL using the LD-based overlap (Figure S5A; odds ratio test between cell type sharing with fetal bulk: p value: 6.5 × 10−25). 45.9%/67.1% of progenitor/neuron primary eSNP-eGene pairs were also detected in the fetal bulk eQTLs (Figure 2C, m-value > 0.9 indicates shared effects; odds ratio test between cell type sharing with fetal bulk: p value: 2.47 × 10−27). Also, the fraction of progenitor/neuron primary eSNP-eGene pairs that were true associations (π1) in fetal bulk eQTLs were 74.9%/92% when subset to gene-SNP pairs that were detectable in both datasets (Figure 2D; see Figure S5B for results with imputed missing eSNP-eGene pairs). Taken together, our observations show that although many genetic effects on gene expression are observed in both bulk and cell-type-specific eQTL data, novel regulatory mechanisms can be identified using cell-type-specific eQTLs, especially in progenitors, which can provide additional information beyond existing prenatal datasets.18, 19, 20
We next explored the temporal specificity of cell-type-specific eQTLs by utilizing adult brain bulk cortical eQTL data from the GTEx project.25 We observed 18.9%/28.3% of conditionally independent eSNP-eGene pairs in progenitors and neurons, respectively, were also found in adult brain eQTL data (LD-based overlap; Figure S5C). That suggests substantial independent genetic mechanisms regulating genes from development to adulthood, as observed previously.20
Cell-type-specific splicing quantitative trait loci (sQTL)
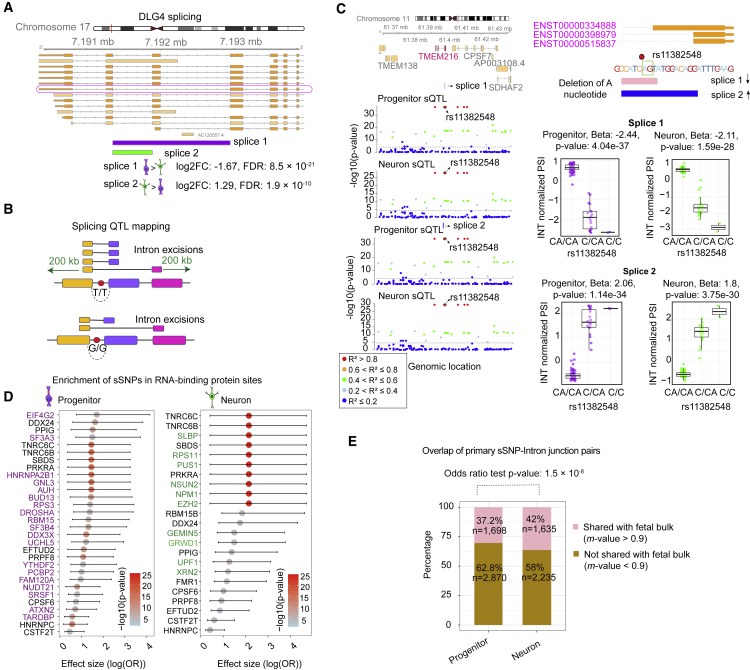
Given the previously known impact of genetic variation on alternative splicing,9,11,19,94 we next performed a splicing quantitative loci (sQTL) analysis separately within progenitors and neurons. We quantified alternative intron excisions as percent spliced in (PSI) by implementing the LeafCutter software, an annotation free approach that allows for discovery of novel isoforms.59 We found 35,238 and 36,070 intron excisions present more often in progenitors and neurons, respectively (|log2FC| > 0.5, see Material and methods, Table S3). As a specific example, we found a differential alternative splicing site within the DLG4 (MIM: 602887) encoding the postsynaptic density protein 95 (PSD-95). An exon skipping splice site supporting nonsense-mediated decay (splice 1, ENST00000491753) was upregulated in progenitors; while another splice site supporting multiple protein coding transcripts (splice 2) was upregulated in neurons (Figure 3A). Post-transcriptional repression of PSD-95 expression in neural progenitors via nonsense mediated decay at splice site 1 has been previously experimentally validated,95,96 giving strong confidence in the cell-type-specific splicing calls.
Figure 3.
Cell-type-specific sQTL analysis
(A) Differential splicing of two intron junctions within DLG4. Splice 1 (chr17:7,191,358–7,192,945) supports a previously validated nonsense-mediated decay transcript (ENST00000491753) with higher expression in progenitors, whereas splice 2 (chr17:7,191,358–7,191,893) has higher expression in neurons.
(B) A schematic illustrating splicing QTL mapping. Association of variants locating within 200 kb distance from each end of intron junctions were tested. The T allele is associated with more frequent splicing of the shorter intron junction.
(C) Two intron junctions supporting an alternative 3′ splicing site for TMEM216 regulated by variant rs11382548 located at the splice site. The regional association of variants to two introns is shown in the genomic tracks on the left colored by pairwise LD r2 relative to variant rs11382548, association p values on the y axis, and genomic location of each variant on the x axis. Dashed line indicates significance threshold. Gene model of TMEM216 is shown in the upper right with the position of the variant rs11382548 (closest variant to the splice site), green box indicates the splice site. Boxplots in the lower right show quantile normalized PSI values for splice 1 (chr11:61,397,975–61,398,261) and splice 2 (chr11:61,397,975–61,398,270) at variant rs11382548.
(D) Enrichment of cell-type-specific sSNPs within RNA-binding protein (RBP) binding sites based on a CLIP-seq dataset. The significantly enriched RBPs based on −log10(enrichment p value) are listed on the y axis, and the x axis shows the effect size from enrichment test with 95% upper and lower confidence interval, where data points colored by −log10(p value) from the enrichment test and cell-type-specific RBPs are colored with purple for progenitors at the left, and as green for neuron at the right.
(E) Overlap percentage of cell-type-specific sSNP-intron junction pairs shared with fetal bulk sQTLs for progenitors and neurons at m-value > 0.9. Odds ratio (OR) test p values are shown.
For the sQTL analysis, we implemented an association test between PSI of each intron excision and genetic variants located within a ±200 kb window from the start and end of the splice junctions (Figures 1A and 3B). We retained significant associations which were lower than 5% false discovery rate by implementing a hierarchical multiple testing correction (see Material and methods) and applied conditional analysis to identify independent sQTLs (Figures S3A and S6A–S6C). We identified 4,568 intron excisions associated with 5,900 conditionally independent sSNPs-intron junction pairs in progenitors and 3,870 intron excisions associated with 4,396 conditionally independent sSNPs-intron junction pairs in neurons (Figure S6D, Table S3). Similar to the eQTL analysis, we additionally performed sQTLs using the standard linear model (FastQTL)61 followed by an adaptive permutation, and we detected 79.8%/78.7% of significant introns and 77.3%76.5% of primary sSNP-intron pairs discovered via the LMM approach in progenitor/neuron were also identified using the standard linear model (Figure S6E).
Regarding the cell-type specificity of sQTLs, we found that 22.4%/30% of progenitor/neuron conditionally independent sSNP-intron junction pairs were shared with other cell types using the LD-based overlap (Figure S6D). 59.4%/57.3% of progenitor/neuron primary sSNP-intron junction pairs were shared with m-value > 0.9 (Figure S7A). The fraction of primary progenitor/neuron sSNP-intron junction pairs that are true associations in neuron/progenitor sQTLs (π1) was 87.3%/85.3% when subsetting to sSNP-intron junction pairs that were detectable in both datasets compared (Figure S7B). However, this analysis may have overestimated sQTL sharing, because 21.5%/30.2% of progenitor/neuron primary sSNP-intron pairs were not detectable in neuron/progenitor sQTL data, which was a higher missing data rate as compared to eQTLs where 6.3%/11% of progenitor/neuron primary eGene-eSNP pairs were not detectable in neuron/progenitor eQTL data. To account for this, we also computed π1 accounting for the missing data (Figure S7B), which suggested substantially more cell-type-specific sQTLs.
As an example, we found that the indel variant rs11382548 creating a canonical splice acceptor sequence impacted two different intron excisions supporting alternative 3′ splice sites for TMEM216 (MIM: 613277) (Figure 3C). Deletion of the A nucleotide at a canonical splice acceptor site of the last exon of TMEM216 leads to disruption of the alternative splicing event for transcript ENST00000334888 and increased usage of transcript ENST00000398979 and ENST00000515837 in both progenitors and neurons. This sQTL may be relevant to neurogenesis because knockdown of the TMEM216 reduces division of both apical and intermediate progenitor cells during corticogenesis.97
Interestingly, many splice sites were previously unannotated in the gene models we used (Ensembl Release 104). We detected 8.2%/10.6% cryptic at the 5′ end, 11.4%/11.5% cryptic 3′ end, and 8.8%/10.8% cryptic at both ends for significant intron excisions within progenitors/neurons.
Leveraging RNA binding sites of 172 RNA-binding proteins in total from CLIP-seq databases,60 we also found that 37 RNA-binding proteins were enriched in progenitor sQTLs and 23 RNA binding proteins were enriched in neuron sQTLs60 (Figure 3D, Table S3). Strikingly, 24 and 10 of these RNA-binding proteins were specifically enriched in progenitor- and neuron-specific sQTLs, respectively. Among RBP binding sites specifically enriched for progenitor sQTLs, we found TARDBP, prominently expressed in neural progenitors98 and known to play a role in neural progenitor proliferation.99 In neurons, we detected enrichment of the EZH2 which regulates neuronal differentiation.100 These observations suggest that sQTLs interfere with the binding sites of RBPs that play cell-type-specific splicing roles during neural development.
To determine whether variants associated with alternative splicing also alter expression of the same genes, we compared cell-type-specific sQTLs with cell-type-specific eQTLs. Only 16.6% and 5.8% of sGenes, the genes that harbor intron excisions, were also eGenes for progenitors and neurons eQTLs, respectively (Figure S7C, upper panel). Furthermore, we also found that only 2.8% and 1.3% of conditionally independent sSNP-sGene pairs overlapped (pairwise LD R2 > 0.8) with conditionally independent eSNP-eGene pairs for progenitors and neurons, respectively (Figure S7C, lower panel). Also, we found that 5.9%/4.4% of progenitor/neuron primary sSNP-sGene pairs were shared with progenitor/neuron eQTLs with the m-value > 0.9, and the fraction of progenitor/neuron primary sQTLs that are true associations in progenitor/neuron eQTLs (π1) was 45%/43% when subsetting to SNP-Gene pairs that were detectable in both datasets. These results indicate that sQTLs generally function through independent mechanisms from eQTLs.
We next examined whether cell type specificity provides additional identification of sQTLs beyond what has previously been detected with bulk RNA-seq. 37.2%/42% of progenitor/neuron sSNP-intron junction pairs were also detected in the fetal bulk sQTLs (Figure 3E, m-value > 0.9 indicates shared effects; odds ratio test between cell type sharing with fetal bulk: p value: 1.5 × 10−8, see Figure S7D for LD-based overlap for conditionally independent sSNP-intron junction pairs and Figure S7E for π1 based overlap). A smaller overlap of progenitor sQTLs with bulk cortical fetal tissue as compared to neuron sQTLs indicated that our cell-type-specific model system allowed for novel discovery of progenitor sQTLs. Also, we found 5.8%/7% of conditionally independent sSNP-intron junction pairs in progenitors and neurons, respectively, were shared with adult brain bulk cortical sQTL data from GTEx25 (LD-based overlap; Figure S7F), showing temporal specificity of cell-type-specific sQTLs.
Using cell-type-specific e/sQTLs to propose regulatory mechanisms of brain-related GWASs
We sought to explain the regulatory mechanism of individual loci associated with neuropsychiatric disorders, brain structure traits, and other brain-relevant traits by leveraging genetic variants regulating cell-type-specific gene expression and splicing. We co-localized GWAS loci of these traits with cell-type-specific eQTLs and sQTLs using a conditional analysis to ensure the loci were shared across traits67 (see Material and methods for the list of GWASs used for this analysis).
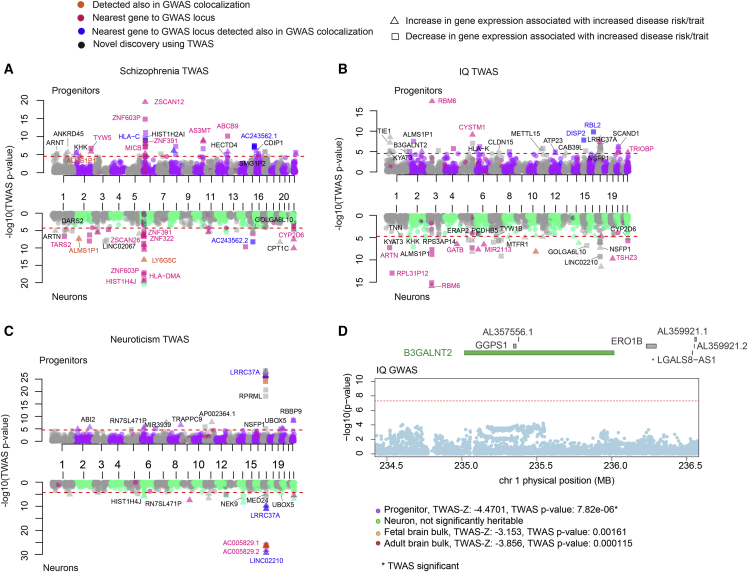
We discovered 41, 13, and 20 GWAS loci that co-localized specifically with progenitor eQTL, specifically with neuron eQTLs, or with both cell types, respectively (Figure 4A, Table S4). These observations show that the same genetic variants impact gene expression, neuropsychiatric traits, and brain structure in a cell-type-specific manner. Importantly, 98 trait associated loci-gene pairs (one locus could be associated with multiple different genes) were not found using fetal bulk cortical tissue eQTLs, where tissue heterogeneity may have masked their detection (Figure 4B).
Next, we leveraged our cell-type-specific chromatin accessibility QTL (caQTL) dataset17 together with eQTLs in order to explain the regulatory mechanism underlying GWAS loci associated with brain relevant traits. As a specific example, we found a colocalization of a locus within the CENPW (MIM: 611264) across caQTLs, eQTLs, and GWASs for global surface area (GSA) and for educational attainment (EA) (Figure 4C). The progenitor index eSNP rs4897179 that was not detected in bulk cortical fetal tissue eQTLs (nominal p value = 3.26 × 10−7 in progenitors, nominal p value = 0.068 in neurons, and nominal p value = 0.26 in fetal cortical bulk tissue), for the CENPW eGene, was colocalized with variant rs9388490, which is the index SNP for both GSA and EA GWAS (nominal p value = 4.95 × 10−12 in GSA GWAS, and nominal p value = 1.43 × 10−8 in EA GWAS). Also, we found that a SNP (rs9388486) located within a chromatin accessible peak region 107 bp upstream of TSS of the CENPW was colocalized with the index eSNP. We therefore consider rs9388486 as the potential causal variant and noted that the C allele disrupts the motifs of the transcription factors CREM, ATF2, ATF4, and ATF1 (Figure 4D). CENPW is required for appropriate kinetochore formation and centriole splitting during mitosis,101 and increased CENPW levels lead to apoptosis in the developing zebrafish central nervous system.102 Overall, these observations propose a cell-type-specific mechanism whereby the C allele at variant rs9388486 disrupts transcription factor binding and diminishes accessibility at the CENPW promoter, resulting in decreased CENPW expression levels in progenitors (Figures 4E and 4F), presumably altering neurogenesis or reducing apoptosis, leading to increased cortical surface area and higher cognitive function.
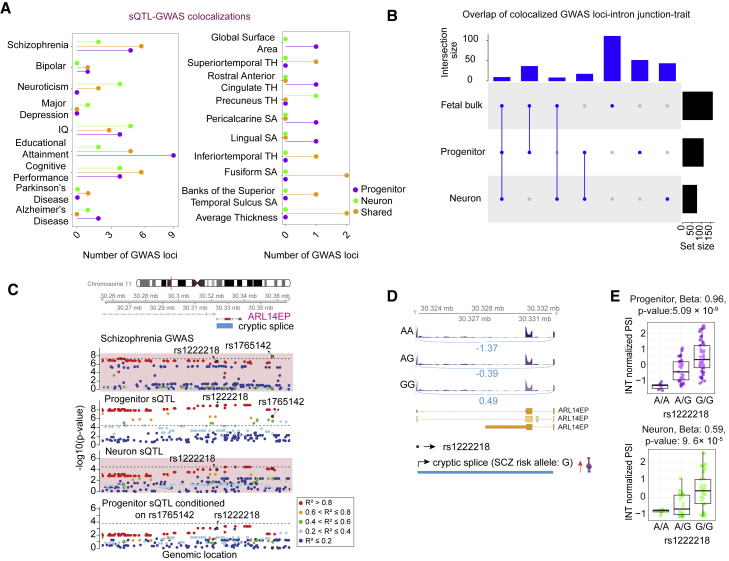
We also aimed to examine cell-type-specific splicing QTLs colocalized with GWAS loci. We observed 29, 20, and 34 GWAS loci in total that co-localized with specifically progenitor/neuron sQTLs and sQTLs present in both cell types (Figure 5A, Table S4). Similar to eQTL colocalizations, we observed that 111 trait-associated loci-intron junction pairs were detected only with cell-type-specific sQTL (one locus could be associated with multiple intron junctions), but not fetal bulk cortical sQTLs (Figure 5B). Interestingly, we detected a progenitor-specific sSNP (rs1222218) regulating a novel alternative exon skipping event for ARL14EP (MIM: 612295) was colocalized with a SCZ index SNP (rs1765142)1 (Figure 5C). The risk allele for SCZ led to more frequent skipping of the exon, supporting expression of a novel isoform (Figures 5D and 5E). The cryptic splice junction has been previously discovered in GTEx within a variety of tissues including adipose and lung, but not in the adult brain.25 ARL14EP has been shown to play a role in axonal development in the mouse neurons.103 Here, we propose a novel transcript of this gene with expression in progenitors as a risk factor for SCZ.
Figure 5.
Colocalization of cell-type-specific sQTLs with GWAS for brain-related traits
(A) Number of GWAS loci colocalized with progenitor (purple)- or neuron (green)-specific sQTLs or both cell types (orange). Each GWAS trait is listed on the y axis (SA, surface area; TH, thickness).
(B) LD-based overlap of colocalized GWAS loci-intron junction pairs per trait across progenitor, neuron, and fetal bulk sQTL colocalizations for the traits listed in (A).
(C) Genomic tracks color-coded based on pairwise LD r2 relative to the variant rs1222218 showing regional association of variants with SCZ and an unannotated alternative splicing event for ARL14EP in progenitors and neurons, association p values on the y axis, and genomic location of each variant on the x axis. A cryptic exon skipping splice site (chr11:30,323,202–30,332,866) was associated with progenitor sSNP (rs1222218) colocalized with SCZ GWAS index SNP (rs1765142). Dashed line indicates significance threshold.
(D) Sashimi plots with the gene model of ARL14EP and the genomic position of the unannotated splice site (blue) overlapping with ARL14EP. Average INT normalized PSI values for the splice site are shown for each genotype group. Schizophrenia risk allele G increases the frequency of the exon skipping event in progenitors.
(E) Boxplots showing INT normalized PSI values for splice across rs1222218 genotypes in progenitors and neurons.
Genetic imputation of cell-type-specific GWAS susceptibility genes and alternative splicing
Next, we imputed genes and alternative splicing associated with brain-related traits by integrating the polygenic impact of cell-type-specific regulatory variants with GWAS risk variants in a transcriptome-wide association study (TWAS) approach.78 We found 1,703/973 genes and 6,552/6,578 intron junctions as significantly cis-heritable in progenitors/neurons (heritability p value < 0.01). We found the cis-heritable impact of 124/102 genes and 372/370 intron junctions in progenitor/neuron significantly correlated with at least one brain-related trait (Table S5). Of those significant TWAS genes/introns, we separated conditionally independent genetic predictors from the co-expressed ones and defined them as jointly independent.83 We performed cell-type-specific TWASs on both gene expression and splicing for schizophrenia (jointly independent genes: 23/26; jointly independent introns: 65/62 in progenitor/neuron), IQ (jointly independent genes: 25/24; jointly independent introns: 42/63 in progenitor/neuron), and neuroticism (jointly independent genes: 13/15 neuron; jointly independent introns: 39/34 in progenitor/neuron) (Figures 6A–6C and S8A–S8C). Also, we found novel loci not discovered in colocalization analysis per trait, demonstrating the additional power of TWASs compared to a single-marker testing approach.
Figure 6.
Prediction of differential gene expression during human brain development via TWAS
(A) Manhattan plots for schizophrenia, IQ, and neuroticism TWAS for progenitors (purple-gray, top) and neurons (green-gray, bottom) where the LD matrix used was based on a European population. Each dot shows −log10 (TWAS p value) for each gene on the y axis, gene names were color coded based on discovery also in colocalization analysis (orange), defined as the nearest gene to GWAS locus (dark pink), being in both these two categories (blue), and discovered only in TWAS analysis (black). Only joint independent genes are labeled (positively and negatively correlated genes represented by triangle and square, respectively, and red line used for TWAS significant threshold).
(B) Manhattan plots for IQ TWAS, as described in (A).
(C) Manhattan plots for neuroticism TWAS, as described in (A).
(D) IQ TWAS results for B3GALNT2, regional association of variants to IQ trait shown at the top, and statistics from each TWAS study shown at the bottom (red line used for genome-wide significant threshold 5 × 10−8).
We evaluated the reproducibility of TWAS results to ensure that the cell type and temporal specificity discovered were not merely due to the sample size of our cell-type-specific QTL study. We observed that SCZ TWAS results using weights from a smaller sample sized adult brain eQTL data from GTEx (n = 118)62 showed a high correlation with TWAS results performed with weights derived from the independent CommonMind Consortium (CMC) adult brain eQTL (n = 452),14 whereas low correlation was observed between SCZ TWAS with weights derived from adult brain as compared to fetal bulk brain or cell-type-specific eQTL data (Figure S9A). Similarly, SCZ TWAS results with weights calculated from two fetal bulk eQTL datasets down-sampled to the size of progenitor (n = 85) and neuron (n = 74) datasets showed a high correlation indicating that reproducible TWAS is achievable in these sample sizes (Figure S9B). These results provide evidence that the limited size of our study was not the major driver for the observed cell-type- and temporal-specific TWAS results. Also, despite the difference in population structure between our dataset and European neuropsychiatric GWASs, we observed that TWAS genes/introns were highly overlapped when different LD estimates were used (Figure S10A).
We next compared our cell-type-specific TWAS approach to TWAS analyses performed using weights calculated from bulk cortical fetal tissue19 and adult brain e/sQTLs from the CMC.14,83 Most TWAS findings were specific to a cell type or temporal e/sQTL dataset, rather than broadly detected, indicating that different developmental or cell type e/sQTL datasets contribute complementary information about genes influencing risk for neuropsychiatric disorders or other brain traits (Figure S10B and Table S5 for comparison). As an example, despite IQ GWASs falling short of the genome-wide significance threshold at B3GALNT2 (MIM: 610194) locus, we detected that genetically imputed B3GALNT2 expression was significantly correlated with IQ in progenitors, but not in neuron, fetal bulk tissue or in CMC adult brain tissue (Figure 6D). Mutations in the B3GALNT2 play a role in glycosylation of α-dystroglycan and were associated with intellectual disability in individuals with congenital muscular dystrophy (MDDGA1 [MIM: 615181]).104 Overall, here we showed that an increase in B3GALNT2 expression in progenitors is associated with lower IQ, suggesting this gene’s early cell-type-specific impact on cognitive function.
Within the cell-type-specific splicing TWAS, we found an intron junction of MRM2 (MIM: 606906) more frequently spliced that was associated with increased risk for schizophrenia specifically in progenitor cells (TWAS-Z: 6.54), but it was not significantly cis-heritable within neuron, fetal bulk, or adult bulk data (Figure S10C). MRM2 is a mitochondrial rRNA methyltransferase,105 and was found to be associated with intellectual disability106 and mitochondrial encephalopathy (MELAS [MIM: 540000]).105 We propose a cell-type-specific developmental basis for alternative splicing of the MRM2 associated with risk for schizophrenia.
Discussion
Here, we investigated the influence of genetic variation on brain-related traits within a cell-type-specific model system recapitulating a critical time period of human brain development, neurogenesis. Our analysis discovered features of gene regulation that will be complementary to previous eQTLs and sQTLs identified in bulk human brain in that: (1) we identified thousands of novel eQTLs, ASEs, and sQTLs during brain development that are enriched in regulatory elements present during neurogenesis; (2) most e/sQTLs in progenitors/neurons were not identified in previous fetal bulk post-mortem tissue datasets using LD-based overlap indicating the importance of cell type specificity for identifying genetic influences on gene regulation; (3) using this resource, we are able to propose cell-type-specific variant-gene/transcript-trait(s) pathways to further explore molecular and developmental causes of neuropsychiatric disorders; (4) by integrating the polygenic effects across traits and gene expression, we are able to impute cell-type-specific gene expression/alternative splicing dysregulation in individuals with neuropsychiatric disorders in time periods prior to disease onset.
As one example of a cell-type-specific variant-gene-trait pathway, we discovered a locus near the CENPW colocalized across cell-type-specific caQTL, eQTL, brain size, and cognitive function. Through the integration of multi-omic gene-to-trait databases, we hypothesize that the C allele at rs9388486 leads to decreased TF binding of up to four transcription factors (ATF1/2/4, CREM) in progenitors, resulting in decreased chromatin accessibility at the promoter peak, decreased expression of CENPW, leading to increased cortical surface area, and increased cognitive function. CENPW has a strong role in proliferation, as it is required for kinetochore formation during mitosis.107 This is consistent with progenitor proliferation influencing surface area, as described in the radial unit hypothesis.108 Increased levels of CENPW may cause death of progenitor cells either by directly being an apoptotic inducer or by triggering apoptosis in response to an imbalance in cell homeostasis with excessive mitotic activity.102 In all, we demonstrate how integration across multi-level biological data can be used to propose functional mechanisms underlying complex traits, and future studies may be able to develop computational models to propose causal pathways across multi-omic QTL data.9,109,110 Such information will be crucial to both design efficient functional validation experiments as well as to leverage GWAS loci to advance treatment targets for neuropsychiatric disorders.
Though the most commonly proposed regulatory mechanism by which non-coding genetic variation influences complex traits is through gene expression levels,91 our data also support mechanisms by which genetic variants associated with cell-type-specific alternative splicing influence complex brain-relevant traits. Importantly, we observed sQTLs impacting previously unannotated cell-type-specific alternative splicing events that are also colocalized with brain-relevant GWASs. For example, we found a progenitor-specific sSNP regulating one unannotated exon skipping splice site for the ARL14EP also colocalized with an index SNP for schizophrenia GWAS, indicating a developmental molecular pathway contributing to schizophrenia risk.
Our cell-type-specific TWAS analysis identified that alteration in expression of multiple genes and transcripts are associated with risk for different neuropsychiatric conditions. We followed a unique TWAS approach allowing us to explore cell type and temporal specificity by leveraging existing fetal brain bulk and adult e/sQTLs together with the cell-type-specific data we generated here. This type of analysis allows the imputation of the genetically regulated component of differential expression within cell types years prior to disease onset. As such, it allows the knowledge of gene expression differences that cannot be gained from post-mortem tissue of affected individuals versus control subjects, which must be acquired after diagnosis. This window into developmental gene expression differences may be particularly important to understand disease risk, as these results are not subject to confounding by medication use or the altered experiences of the environment of individuals living with a neuropsychiatric illness.111 Nevertheless, further support for such data could be gained from iPSC lines modeling early developmental time periods from large populations of affected individuals versus control subjects.
With our cell-type-specific model, we propose how and when genetics influence brain-related traits through gene expression and splicing. The sample size of our study (n = 89 independent donors) is consistent with other previously published cell-based QTLs,21,24,112,113 and cell-type resolution may have led to novel and higher powered eQTL discovery masked in bulk tissue. However, it is also possible that the novel loci identified here contain false positives due to relatively low sample size as compared to post-mortem datasets19,114 or are caused by in vitro cell culture artifacts. eGenes identified in this study are not enriched in genes with low fidelity in our in vitro system, nevertheless the replication of the cell-type-specific study using scRNA-seq from developing fetal brain tissue or cell-type-specific iPSC-derived eQTL datasets24,115 of independent donors derived from a multi-ancestry population will be crucial to mitigate these concerns. This in vitro system has particular utility in that, in the future, it may be used to determine the impact of genetic variation in response to activation of specific pathways or response to environmental stimuli.23 By pursuing cell type, temporal, and environmental specificity of eQTLs, we expect that a greater degree of mechanisms underlying risk for neuropsychiatric disorders and brain-relevant traits can be uncovered.
Acknowledgments
This work was supported by NIH (R00MH102357, U54EB020403, R01MH118349, R01MH120125), Brain Research Foundation, and NC TraCS Pilot funding to J.L.S. D.H.G. was supported by NIH (R37 MH060233, R01 MH094714, UO1MH116489, and R01 MH110927). The following core facilities were utilized for this project: UNC Neuroscience Center Microscopy Core (P30NS045892), UNC Mammalian Genotyping Core, CGIBD Advanced Analytics Core (NIH grant P30 DK034987), UNC Flow Cytometry Core Facility, UNC Vector Core, and UNC Research Computing. Additional core facilities utilized for this project were: UCLA CFAR (5P30 AI028697) and the UCLA Neuroscience Genomics Core. We thank Dr. Karen L. Mohlke and Dr. Yun Li for helpful comments, Dr. Eric Wexler for the idea of the phNPC eQTL, and Dr. Stephen Montgomery for clarifying the eigenMT method.
Declaration of interests
The authors declare no competing interests.
Published: August 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.07.011.
Data and code availability
Data will be available within dbGaP upon publication with study accession number phs002493.v1.p1, and code is available at https://bitbucket.org/steinlabunc/expression_splicing_qtls_public/src/master/.
Web resources
AAV2-hSyn1-eGFP, https://www.addgene.org/50465/
eigenMT, https://github.com/joed3/eigenMT
ENIGMA protocol, http://enigma.ini.usc.edu/wp-content/uploads/2012/07/ENIGMA2_1KGP_cookbook_v3.pdf
FastQC, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
FastQTL, http://fastqtl.sourceforge.net/
GRCh38 release92 reference genome, https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.38/
Homo_sapiens.GRCh38.92 GTF file, http://ftp.ensembl.org/pub/release-92/gtf/homo_sapiens/Homo_sapiens.GRCh38.92.gtf.gz
HapMap3 genotype data, https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html
LeafCutter, https://davidaknowles.github.io/leafcutter/
METASOFT, http://genetics.cs.ucla.edu/meta_jemdoc/index.html
Online Mendelian Inheritance in Man, https://www.omim.org
Supplemental information
Sheet 1: Differential gene expression analysis progenitor vs neurons (FDR < 0.05): gene is the ensemblID, logFC is the expression fold change logFC > 0 indicates a gene more frequently expressed in neurons than progenitors; AveExpr is the average vst normalized expression of all samples. t is the expression fold change divided by its standard error 37. P.Value is the nominal p-value from the testing differential expression; adj.P.Val is the Benjamini-Hochberg FDR adjusted p-value; B is log-odds for the differentially expressed gene in limma.
Sheet 1-3: List of cell-type specific conditionally independent eQTLs for progenitor, neurons and fetal bulk: snp is the variant tested in QTL; beta is the beta coefficient; pvalue is the nominal p-value; gene is the ensemblID of the gene tested; rank is the eQTL order; chr is the chromosome number, BP is the genomic position of the variant; cond.beta is the beta after conditional analysis; cond.pval is the p-value after conditional analysis; A1 is the effect allele. rsid is the rs id of the allele matching in 1000 Genome Phase 3 (NA if rsid is not available for the genomic position of the variant in 1000 Genome data; ∗ if multiple variants exist for the same genomic position). Sheet 4-5: Allele specific expression analysis (FDR < 0.05). SNP is the variant tested for allele specific expression analysis, baseMean is the average of the normalized count values divided by size factors from DESeq236; log2FoldChange is the expression fold change logFC > 0 indicates reads more frequently expressed in donors with reference allele than donors with alternative allele; lfcSE is the standard error estimate for log2FoldChange; stat is the test statistics performed in DESEq2; pvalue is the nominal p-value from the testing differential expression; padj is the Benjamini-Hochberg FDR adjusted p-value; refAllele is the reference allele of the variant.
Sheet 1: Differential splicing analysis progenitor vs neurons (FDR < 0.05): intron is the splice junction, logFC is the expression fold change logFC > 0 indicates a gene more frequently expressed in neurons than progenitors; AveExpr is the average vst normalized expression of all samples. t is the expression fold change divided by its standard error37. P.Value is the nominal p-value from the testing differential expression; adj.P.Val is the Benjamini-Hochberg FDR adjusted p-value; B is log-odds for the differentially expressed intron in limma; gene is the gene symbol of the gene that introns junctions overlap with; ensemblID is the ensemblID of that gene. Sheet 2-4: List of cell-type specific conditionally independent sQTLs for progenitor, neuron and fetal bulk sQTLs: snp is the variant tested; beta is the beta coefficient, pval is the nominal p-value; intron is the intron junction as chromosome:start position:end position format; rank is the order of sQTL after conditional analysis; chr is the chromosome, start is the start position of the junction; end is the end position of the junction; clusterID is the cluster identified from Leafcutter, cluster is the clusterID combined with chromosome number, verdict is the annotation status; gene is the gene symbol of the gene that introns junctions overlap with; ensemblID is the ensemblID of that gene; transcripts is the transcripts where intron junction overlap with; constitutive.score: degree of the junction shown in each transcript; cond.beta is the beta coefficient after conditional analysis (for primary QTLs, it is identical to beta); cond.pval is the p-value after conditional analysis (for primary QTLs, it is identical to pval), A1 is the effect allele; rsid is the rs id of the allele matching in 1000 Genome Phase 3. Sheet 5: Enrichment of RNA binding protein (RBP) sites within cell-type specific sQTLs. PThresh is the p-value threshold used for enrichment; OR is the odd ratio; Pvalue is enrichment p-value; Beta is the beta coefficient after enrichment test via GARFIELD50; SE is the standard error; CI95_lower is the lower bound of 95% confidence interval; CI95 upper is the upper bound of 95% confidence interval; NAnnotThesh is the is the number of annotated variants at the p-value threshold; NAnnot is the total number of variants after pruning; NThresh is the number of variant passing p-value threshold after pruning; N is the number of variants remained after pruning; linkID is the ID in annotation file; Annotation is the RNA-binding protein; Celltype is the cell type used for enrichment test.
Colocalization of GWAS for neuropsychiatric disease and other brain related traits with cell-type specific e/sQTLs and fetal bulk e/sQTLs: e/sQTLsnp is the e/sSNP; inibeta is the beta coefficient before conditioning on GWAS SNP; pval is the nominal p-value prior to conditional analysis, gene/intron is the ensemblID of gene/intron junction associated with the e/sSNP; Condbeta is the beta estimate of e/sQTL after conditional analysis; Condpval is the p-value after conditional analysis; r2 is the linkage disequilibrium (LD) r2; pop is the population used to estimate LD r2 (European population, with “European” or the population used in the QTL study with “Study”); symbol of the symbol of the gene (for eQTLs); biotype is the biotype of the gene for eQTLs; trait is the trait for GWAS; trait is the GWAS study; A1 is the effect allele for e/sQTL index SNP; GWASsnp is the variant e/sSNP colocalized with; rsid is the rs id of the allele matching in 1000 Genome Phase 3.
Sheet 1-8: List of cell-type specific/fetal bulk/adult bulk TWAS gene and introns for neuropsychiatric disease and other brain related traits. Output from FUSION79: ID is the gene ensemblID or intron id; CHR is the chromosome number; HSQ is the heritability; BEST.GWAS.ID is the GWAS SNP in the locus with the most significant association; BEST.GWAS.Z is the z-score of the best GWAS SNP; EQTL.ID is the best e/sQTL in the locus; EQTL.R2 is the cross-validation R2 of the best e/sQTL in the locus; EQTL.Z is the z-score of the best e/sQTL in the locus; EQTL.GWAS.Z is the GWAS Z-score for this e/sQTL; NSNP is the number of SNPs in the locus; NWGT is the number of snps with non-zero weights; MODEL is the best performing model; MODELCV.R2 is the the cross-validation R2 of the best performing model; MODELCV.PV is the p-value from the cross-validation of the best performing model; TWAS.Z is the TWAS z-score; TWAS.P is the TWAS p-value; trait is the GWAS trait; pop is the population used to estimate LD; joint_independent is the status if a gene/intron jointly independent (YES, if it is independent; NO, if it is not independent; NA, if it was not tested for the trait). Sheet 9-10: Summary of heritability (p-value < 0.01) and cross validation r2 from prediction models across cell-type specific/fetal bulk/adult bulk for gene and intron TWAS: hsq is the mean heritability of the genes/introns; hsq.se is the mean standard error of estimated heritability; hsq.pv is the mean p-value of the heritability; emmax.rsq is the mean cross-validation R2 training via EMMAX with p-value as emmax.pval; lasso.rsq is mean the cross-validation R2 via LASSO with p-value as lasso.pval; enet.rsq is mean the cross-validation R2 via elastic net with p-value as enet.pval; blup.rsq is mean the cross-validation R2 via BLUP with p-value as blup.pval; bslmm.rsq is the mean cross-validation R2 via BSLMM with p-value as bslmm.pval; top1.rsq is the mean cross-validation R2 via standard marginal e/sQTL Z-scores computation with p-value as top1.pval. 95 % confidence intervals per parameter are shown their below. Sheet 11-12: SCZ TWAS for GTEx Brain frontal cortex and downsampled fetal bulk data. Output from FUSION78: PANEL: Data type; ID is the gene ensemblID or intron id; CHR is the chromosome number; HSQ is the heritability; BEST.GWAS.ID is the GWAS SNP in the locus with the most significant association; BEST.GWAS.Z is the z-score of the best GWAS SNP; EQTL.ID is the best e/sQTL in the locus; EQTL.R2 is the cross-validation R2 of the best e/sQTL in the locus; EQTL.Z is the z-score of the best e/sQTL in the locus; EQTL.GWAS.Z is the GWAS Z-score for this e/sQTL; NSNP is the number of SNPs in the locus; NWGT is the number of snps with non-zero weights; MODEL is the best performing model; MODELCV.R2 is the the cross-validation R2 of the best performing model; MODELCV.PV is the p-value from the cross-validation of the best performing model; TWAS.Z is the TWAS z-score; TWAS.P is the TWAS p-value; trait is the GWAS trait. Table S1.
References
- 1.Pardiñas A.F., Holmans P., Pocklington A.J., Escott-Price V., Ripke S., Carrera N., Legge S.E., Bishop S., Cameron D., Hamshere M.L., GERAD1 Consortium. CRESTAR Consortium Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stahl E.A., Breen G., Forstner A.J., McQuillin A., Ripke S., Trubetskoy V., Mattheisen M., Wang Y., Coleman J.R.I., Gaspar H.A., eQTLGen Consortium. BIOS Consortium. Bipolar Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howard D.M., Adams M.J., Clarke T.-K., Hafferty J.D., Gibson J., Shirali M., Coleman J.R.I., Hagenaars S.P., Ward J., Wigmore E.M., 23andMe Research Team. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasby K.L., Jahanshad N., Painter J.N., Colodro-Conde L., Bralten J., Hibar D.P., Lind P.A., Pizzagalli F., Ching C.R.K., McMahon M.A.B., Alzheimer’s Disease Neuroimaging Initiative. CHARGE Consortium. EPIGEN Consortium. IMAGEN Consortium. SYS Consortium. Parkinson’s Progression Markers Initiative. Enhancing NeuroImaging Genetics through Meta-Analysis Consortium (ENIGMA)—Genetics working group The genetic architecture of the human cerebral cortex. Science. 2020;367:367. [Google Scholar]
- 5.Savage J.E., Jansen P.R., Stringer S., Watanabe K., Bryois J., de Leeuw C.A., Nagel M., Awasthi S., Barr P.B., Coleman J.R.I. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 2018;50:912–919. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., Baldursson G., Belliveau R., Bybjerg-Grauholm J., Bækvad-Hansen M., ADHD Working Group of the Psychiatric Genomics Consortium (PGC) Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium. 23andMe Research Team Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matoba N., Liang D., Sun H., Aygün N., McAfee J.C., Davis J.E., Raffield L.M., Qian H., Piven J., Li Y. Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl. Psychiatry. 2020;10:265. doi: 10.1038/s41398-020-00953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser H.B., Xie X. Common polymorphic transcript variation in human disease. Genome Res. 2009;19:567–575. doi: 10.1101/gr.083477.108. [DOI] [PubMed] [Google Scholar]
- 9.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandal M.J., Zhang P., Hadjimichael E., Walker R.L., Chen C., Liu S., Won H., van Bakel H., Varghese M., Wang Y., PsychENCODE Consortium Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:362. doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata A., Matsumoto N., Kato T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat. Commun. 2017;8:14519. doi: 10.1038/ncomms14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y., Levy S., Gogos J.A., Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raj B., Blencowe B.J. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M., Santpere G., Imamura Kawasawa Y., Evgrafov O.V., Gulden F.O., Pochareddy S., Sunkin S.M., Li Z., Shin Y., Zhu Y., BrainSpan Consortium. PsychENCODE Consortium. PsychENCODE Developmental Subgroup Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:362. doi: 10.1126/science.aat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Torre-Ubieta L., Stein J.L., Won H., Opland C.K., Liang D., Lu D., Geschwind D.H. The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell. 2018;172:289–304.e18. doi: 10.1016/j.cell.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang D., Elwell A.L., Aygün N., Krupa O., Wolter J.M., Kyere F.A., Lafferty M.J., Cheek K.E., Courtney K.P., Yusupova M. Cell-type-specific effects of genetic variation on chromatin accessibility during human neuronal differentiation. Nat. Neurosci. 2021;24:941–953. doi: 10.1038/s41593-021-00858-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Brien H.E., Hannon E., Hill M.J., Toste C.C., Robertson M.J., Morgan J.E., McLaughlin G., Lewis C.M., Schalkwyk L.C., Hall L.S. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19:194. doi: 10.1186/s13059-018-1567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker R.L., Ramaswami G., Hartl C., Mancuso N., Gandal M.J., de la Torre-Ubieta L., Pasaniuc B., Stein J.L., Geschwind D.H. Genetic Control of Expression and Splicing in Developing Human Brain Informs Disease Mechanisms. Cell. 2020;181:745. doi: 10.1016/j.cell.2020.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Werling D.M., Pochareddy S., Choi J., An J.-Y., Sheppard B., Peng M., Li Z., Dastmalchi C., Santpere G., Sousa A.M.M. Whole-Genome and RNA Sequencing Reveal Variation and Transcriptomic Coordination in the Developing Human Prefrontal Cortex. Cell Rep. 2020;31:107489. doi: 10.1016/j.celrep.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuomo A.S.E., Seaton D.D., McCarthy D.J., Martinez I., Bonder M.J., Garcia-Bernardo J., Amatya S., Madrigal P., Isaacson A., Buettner F., HipSci Consortium Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression. Nat. Commun. 2020;11:810. doi: 10.1038/s41467-020-14457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fairfax B.P., Makino S., Radhakrishnan J., Plant K., Leslie S., Dilthey A., Ellis P., Langford C., Vannberg F.O., Knight J.C. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umans B.D., Battle A., Gilad Y. Where Are the Disease-Associated eQTLs? Trends Genet. 2021;37:109–124. doi: 10.1016/j.tig.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerber J., Seaton D.D., Cuomo A.S.E., Kumasaka N., Haldane J., Steer J., Patel M., Pearce D., Andersson M., Bonder M.J., HipSci Consortium Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation. Nat. Genet. 2021;53:304–312. doi: 10.1038/s41588-021-00801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein J.L., de la Torre-Ubieta L., Tian Y., Parikshak N.N., Hernández I.A., Marchetto M.C., Baker D.K., Lu D., Hinman C.R., Lowe J.K. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–12. [Google Scholar]
- 28.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 30.van de Geijn B., McVicker G., Gilad Y., Pritchard J.K. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaneau O., Marchini J., Zagury J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 33.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jun G., Flickinger M., Hetrick K.N., Romm J.M., Doheny K.F., Abecasis G.R., Boehnke M., Kang H.M. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 39.Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44(W1):W83-9. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plaisier S.B., Taschereau R., Wong J.A., Graeber T.G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller J.A., Ding S.-L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cahill K.M., Huo Z., Tseng G.C., Logan R.W., Seney M.L. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci. Rep. 2018;8:9588. doi: 10.1038/s41598-018-27903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J., Zaitlen N.A., Goddard M.E., Visscher P.M., Price A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014;46:100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price A.L., Zaitlen N.A., Reich D., Patterson N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Q.Q., Ritchie S.C., Brozynska M., Inouye M. Power, false discovery rate and Winner’s Curse in eQTL studies. Nucleic Acids Res. 2018;46:e133. doi: 10.1093/nar/gky780. e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis J.R., Fresard L., Knowles D.A., Pala M., Bustamante C.D., Battle A., Montgomery S.B. An Efficient Multiple-Testing Adjustment for eQTL Studies that Accounts for Linkage Disequilibrium between Variants. Am. J. Hum. Genet. 2016;98:216–224. doi: 10.1016/j.ajhg.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobbyn A., Huckins L.M., Boocock J., Sloofman L.G., Glicksberg B.S., Giambartolomei C., Hoffman G.E., Perumal T.M., Girdhar K., Jiang Y., CommonMind Consortium Landscape of Conditional eQTL in Dorsolateral Prefrontal Cortex and Co-localization with Schizophrenia GWAS. Am. J. Hum. Genet. 2018;102:1169–1184. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen R., Hottenga J.-J., Nivard M.G., Abdellaoui A., Laport B., de Geus E.J., Wright F.A., Penninx B.W.J.H., Boomsma D.I. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum. Mol. Genet. 2017;26:1444–1451. doi: 10.1093/hmg/ddx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iotchkova V., Ritchie G.R.S., Geihs M., Morganella S., Min J.L., Walter K., Timpson N.J., Dunham I., Birney E., Soranzo N., UK10K Consortium GARFIELD classifies disease-relevant genomic features through integration of functional annotations with association signals. Nat. Genet. 2019;51:343–353. doi: 10.1038/s41588-018-0322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ernst J., Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 2015;33:364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021 doi: 10.1101/060012. [DOI] [Google Scholar]
- 54.Castel S.E., Levy-Moonshine A., Mohammadi P., Banks E., Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castel S.E., Levy-Moonshine A., Mohammadi P., Banks E., Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohammadi P., Castel S.E., Brown A.A., Lappalainen T. Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res. 2017;27:1872–1884. doi: 10.1101/gr.216747.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cotto K.C., Feng Y.-Y., Ramu A., Skidmore Z.L., Kunisaki J., Richters M., Freshour S., Lin Y., Chapman W.C., Uppaluri R. RegTools: Integrated analysis of genomic and transcriptomic data for discovery of splicing variants in cancer. bioRxiv. 2021 doi: 10.1101/436634. [DOI] [Google Scholar]
- 59.Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., Pritchard J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y.-C.T., Di C., Hu B., Zhou M., Liu Y., Song N., Li Y., Umetsu J., Lu Z.J. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics. 2015;16:51. doi: 10.1186/s12864-015-1273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ongen H., Buil A., Brown A.A., Dermitzakis E.T., Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 63.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han B., Eskin E. Interpreting meta-analyses of genome-wide association studies. PLoS Genet. 2012;8:e1002555. doi: 10.1371/journal.pgen.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dabney A., Storey J.D., Warnes G.R. R Package Version 1; 2010. qvalue: Q-value estimation for false discovery rate control. [Google Scholar]
- 66.Rosenblatt J.D., Stein J.L. R package version 1.22. 0; 2014. RRHO: test overlap using the rank-rank hypergeometric test. [Google Scholar]
- 67.Civelek M., Wu Y., Pan C., Raulerson C.K., Ko A., He A., Tilford C., Saleem N.K., Stančáková A., Scott L.J. Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits. Am. J. Hum. Genet. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M.F., eQTLGen. 23andMe. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J.J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., Nguyen-Viet T.A., Bowers P., Sidorenko J., Karlsson Linnér R., 23andMe Research Team. COGENT (Cognitive Genomics Consortium) Social Science Genetic Association Consortium Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagel M., Watanabe K., Stringer S., Posthuma D., van der Sluis S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat. Commun. 2018;9:905. doi: 10.1038/s41467-018-03242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jansen I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S., Sealock J., Karlsson I.K., Hägg S., Athanasiu L. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., 23andMe Research Team. System Genomics of Parkinson’s Disease Consortium. International Parkinson’s Disease Genomics Consortium Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jansen P.R., Watanabe K., Stringer S., Skene N., Bryois J., Hammerschlag A.R., de Leeuw C.A., Benjamins J.S., Muñoz-Manchado A.B., Nagel M., 23andMe Research Team Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet. 2019;51:394–403. doi: 10.1038/s41588-018-0333-3. [DOI] [PubMed] [Google Scholar]
- 74.International League Against Epilepsy Consortium on Complex Epilepsies Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun. 2018;9:5269. doi: 10.1038/s41467-018-07524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., Pallesen J., Agerbo E., Andreassen O.A., Anney R., Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium. BUPGEN. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. 23andMe Research Team Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lawrence M., Gentleman R., Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coetzee S.G., Coetzee G.A., Hazelett D.J. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–3849. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meijer R.J., Goeman J.J. Efficient approximate k-fold and leave-one-out cross-validation for ridge regression. Biom. J. 2013;55:141–155. doi: 10.1002/bimj.201200088. [DOI] [PubMed] [Google Scholar]
- 81.Tibshirani R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Series B Stat. Methodol. 1996;58:267–288. [Google Scholar]
- 82.Zou H., Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 2005;67:301–320. [Google Scholar]
- 83.Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., McCarroll S., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pasaniuc B., Zaitlen N., Shi H., Bhatia G., Gusev A., Pickrell J., Hirschhorn J., Strachan D.P., Patterson N., Price A.L. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics. 2014;30:2906–2914. doi: 10.1093/bioinformatics/btu416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375, S1–S3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosen E.Y., Wexler E.M., Versano R., Coppola G., Gao F., Winden K.D., Oldham M.C., Martens L.H., Zhou P., Farese R.V., Jr., Geschwind D.H. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Konopka G., Wexler E., Rosen E., Mukamel Z., Osborn G.E., Chen L., Lu D., Gao F., Gao K., Lowe J.K., Geschwind D.H. Modeling the functional genomics of autism using human neurons. Mol. Psychiatry. 2012;17:202–214. doi: 10.1038/mp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palmer T.D., Schwartz P.H., Taupin P., Kaspar B., Stein S.A., Gage F.H. Cell culture. Progenitor cells from human brain after death. Nature. 2001;411:42–43. doi: 10.1038/35075141. [DOI] [PubMed] [Google Scholar]
- 89.Hansen D.V., Lui J.H., Parker P.R.L., Kriegstein A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 90.Gómez-López S., Wiskow O., Favaro R., Nicolis S.K., Price D.J., Pollard S.M., Smith A. Sox2 and Pax6 maintain the proliferative and developmental potential of gliogenic neural stem cells In vitro. Glia. 2011;59:1588–1599. doi: 10.1002/glia.21201. [DOI] [PubMed] [Google Scholar]
- 91.Albert F.W., Kruglyak L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 92.Sul J.H., Han B., Ye C., Choi T., Eskin E. Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet. 2013;9:e1003491. doi: 10.1371/journal.pgen.1003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat. Rev. Genet. 2010;11:533–538. doi: 10.1038/nrg2815. [DOI] [PubMed] [Google Scholar]
- 94.Monlong J., Calvo M., Ferreira P.G., Guigó R. Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat. Commun. 2014;5:4698. doi: 10.1038/ncomms5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng S., Gray E.E., Chawla G., Porse B.T., O’Dell T.J., Black D.L. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012;15:381–388, S1. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zheng S. Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression. Int. J. Dev. Neurosci. 2016;55:102–108. doi: 10.1016/j.ijdevneu.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo J., Higginbotham H., Li J., Nichols J., Hirt J., Ghukasyan V., Anton E.S. Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun. 2015;6:7857. doi: 10.1038/ncomms8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sephton C.F., Good S.K., Atkin S., Dewey C.M., Mayer P., 3rd, Herz J., Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vogt M.A., Ehsaei Z., Knuckles P., Higginbottom A., Helmbrecht M.S., Kunath T., Eggan K., Williams L.A., Shaw P.J., Wurst W. TDP-43 induces p53-mediated cell death of cortical progenitors and immature neurons. Sci. Rep. 2018;8:8097. doi: 10.1038/s41598-018-26397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pereira J.D., Sansom S.N., Smith J., Dobenecker M.-W., Tarakhovsky A., Livesey F.J. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McKinley K.L., Cheeseman I.M. Large-Scale Analysis of CRISPR/Cas9 Cell-Cycle Knockouts Reveals the Diversity of p53-Dependent Responses to Cell-Cycle Defects. Dev. Cell. 2017;40:405–420.e2. doi: 10.1016/j.devcel.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee S., Koh W., Kim H.-T., Kim C.-H., Lee S. Cancer-upregulated gene 2 (CUG2) overexpression induces apoptosis in SKOV-3 cells. Cell Biochem. Funct. 2010;28:461–468. doi: 10.1002/cbf.1678. [DOI] [PubMed] [Google Scholar]
- 103.Peter C.J., Saito A., Hasegawa Y., Tanaka Y., Nagpal M., Perez G., Alway E., Espeso-Gil S., Fayyad T., Ratner C. In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene. Nat. Commun. 2019;10:4112. doi: 10.1038/s41467-019-12013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maroofian R., Riemersma M., Jae L.T., Zhianabed N., Willemsen M.H., Wissink-Lindhout W.M., Willemsen M.A., de Brouwer A.P.M., Mehrjardi M.Y.V., Ashrafi M.R. B3GALNT2 mutations associated with non-syndromic autosomal recessive intellectual disability reveal a lack of genotype-phenotype associations in the muscular dystrophy-dystroglycanopathies. Genome Med. 2017;9:118. doi: 10.1186/s13073-017-0505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garone C., D’Souza A.R., Dallabona C., Lodi T., Rebelo-Guiomar P., Rorbach J., Donati M.A., Procopio E., Montomoli M., Guerrini R. Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum. Mol. Genet. 2017;26:4257–4266. doi: 10.1093/hmg/ddx314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Freude K., Hoffmann K., Jensen L.-R., Delatycki M.B., des Portes V., Moser B., Hamel B., van Bokhoven H., Moraine C., Fryns J.-P. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 2004;75:305–309. doi: 10.1086/422507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim H., Lee M., Lee S., Park B., Koh W., Lee D.J., Lim D.-S., Lee S. Cancer-upregulated gene 2 (CUG2), a new component of centromere complex, is required for kinetochore function. Mol. Cells. 2009;27:697–701. doi: 10.1007/s10059-009-0083-2. [DOI] [PubMed] [Google Scholar]
- 108.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park Y., Sarkar A., Nguyen K., Kellis M. Causal Mediation Analysis Leveraging Multiple Types of Summary Statistics Data. arXiv. 2019 1901.08540. [Google Scholar]
- 110.Ng B., White C.C., Klein H.-U., Sieberts S.K., McCabe C., Patrick E., Xu J., Yu L., Gaiteri C., Bennett D.A. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Harrison P.J. Using our brains: the findings, flaws, and future of postmortem studies of psychiatric disorders. Biol. Psychiatry. 2011;69:102–103. doi: 10.1016/j.biopsych.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 112.Alasoo K., Rodrigues J., Mukhopadhyay S., Knights A.J., Mann A.L., Kundu K., Hale C., Dougan G., Gaffney D.J., HIPSCI Consortium Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.-B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de Klein N., Tsai E.A., Vochteloo M., Baird D., Huang Y., Chen C.-Y., van Dam S., Deelen P., Bakker O.B., El Garwany O. Brain expression quantitative trait locus and network analysis reveals downstream effects and putative drivers for brain-related diseases. bioRxiv. 2021 doi: 10.1101/2021.03.01.433439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bonder M.J., Smail C., Gloudemans M.J., Frésard L., Jakubosky D., D’Antonio M., Li X., Ferraro N.M., Carcamo-Orive I., Mirauta B., HipSci Consortium. iPSCORE consortium. Undiagnosed Diseases Network. PhLiPS consortium Identification of rare and common regulatory variants in pluripotent cells using population-scale transcriptomics. Nat. Genet. 2021;53:313–321. doi: 10.1038/s41588-021-00800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1: Differential gene expression analysis progenitor vs neurons (FDR < 0.05): gene is the ensemblID, logFC is the expression fold change logFC > 0 indicates a gene more frequently expressed in neurons than progenitors; AveExpr is the average vst normalized expression of all samples. t is the expression fold change divided by its standard error 37. P.Value is the nominal p-value from the testing differential expression; adj.P.Val is the Benjamini-Hochberg FDR adjusted p-value; B is log-odds for the differentially expressed gene in limma.
Sheet 1-3: List of cell-type specific conditionally independent eQTLs for progenitor, neurons and fetal bulk: snp is the variant tested in QTL; beta is the beta coefficient; pvalue is the nominal p-value; gene is the ensemblID of the gene tested; rank is the eQTL order; chr is the chromosome number, BP is the genomic position of the variant; cond.beta is the beta after conditional analysis; cond.pval is the p-value after conditional analysis; A1 is the effect allele. rsid is the rs id of the allele matching in 1000 Genome Phase 3 (NA if rsid is not available for the genomic position of the variant in 1000 Genome data; ∗ if multiple variants exist for the same genomic position). Sheet 4-5: Allele specific expression analysis (FDR < 0.05). SNP is the variant tested for allele specific expression analysis, baseMean is the average of the normalized count values divided by size factors from DESeq236; log2FoldChange is the expression fold change logFC > 0 indicates reads more frequently expressed in donors with reference allele than donors with alternative allele; lfcSE is the standard error estimate for log2FoldChange; stat is the test statistics performed in DESEq2; pvalue is the nominal p-value from the testing differential expression; padj is the Benjamini-Hochberg FDR adjusted p-value; refAllele is the reference allele of the variant.
Sheet 1: Differential splicing analysis progenitor vs neurons (FDR < 0.05): intron is the splice junction, logFC is the expression fold change logFC > 0 indicates a gene more frequently expressed in neurons than progenitors; AveExpr is the average vst normalized expression of all samples. t is the expression fold change divided by its standard error37. P.Value is the nominal p-value from the testing differential expression; adj.P.Val is the Benjamini-Hochberg FDR adjusted p-value; B is log-odds for the differentially expressed intron in limma; gene is the gene symbol of the gene that introns junctions overlap with; ensemblID is the ensemblID of that gene. Sheet 2-4: List of cell-type specific conditionally independent sQTLs for progenitor, neuron and fetal bulk sQTLs: snp is the variant tested; beta is the beta coefficient, pval is the nominal p-value; intron is the intron junction as chromosome:start position:end position format; rank is the order of sQTL after conditional analysis; chr is the chromosome, start is the start position of the junction; end is the end position of the junction; clusterID is the cluster identified from Leafcutter, cluster is the clusterID combined with chromosome number, verdict is the annotation status; gene is the gene symbol of the gene that introns junctions overlap with; ensemblID is the ensemblID of that gene; transcripts is the transcripts where intron junction overlap with; constitutive.score: degree of the junction shown in each transcript; cond.beta is the beta coefficient after conditional analysis (for primary QTLs, it is identical to beta); cond.pval is the p-value after conditional analysis (for primary QTLs, it is identical to pval), A1 is the effect allele; rsid is the rs id of the allele matching in 1000 Genome Phase 3. Sheet 5: Enrichment of RNA binding protein (RBP) sites within cell-type specific sQTLs. PThresh is the p-value threshold used for enrichment; OR is the odd ratio; Pvalue is enrichment p-value; Beta is the beta coefficient after enrichment test via GARFIELD50; SE is the standard error; CI95_lower is the lower bound of 95% confidence interval; CI95 upper is the upper bound of 95% confidence interval; NAnnotThesh is the is the number of annotated variants at the p-value threshold; NAnnot is the total number of variants after pruning; NThresh is the number of variant passing p-value threshold after pruning; N is the number of variants remained after pruning; linkID is the ID in annotation file; Annotation is the RNA-binding protein; Celltype is the cell type used for enrichment test.
Colocalization of GWAS for neuropsychiatric disease and other brain related traits with cell-type specific e/sQTLs and fetal bulk e/sQTLs: e/sQTLsnp is the e/sSNP; inibeta is the beta coefficient before conditioning on GWAS SNP; pval is the nominal p-value prior to conditional analysis, gene/intron is the ensemblID of gene/intron junction associated with the e/sSNP; Condbeta is the beta estimate of e/sQTL after conditional analysis; Condpval is the p-value after conditional analysis; r2 is the linkage disequilibrium (LD) r2; pop is the population used to estimate LD r2 (European population, with “European” or the population used in the QTL study with “Study”); symbol of the symbol of the gene (for eQTLs); biotype is the biotype of the gene for eQTLs; trait is the trait for GWAS; trait is the GWAS study; A1 is the effect allele for e/sQTL index SNP; GWASsnp is the variant e/sSNP colocalized with; rsid is the rs id of the allele matching in 1000 Genome Phase 3.
Sheet 1-8: List of cell-type specific/fetal bulk/adult bulk TWAS gene and introns for neuropsychiatric disease and other brain related traits. Output from FUSION79: ID is the gene ensemblID or intron id; CHR is the chromosome number; HSQ is the heritability; BEST.GWAS.ID is the GWAS SNP in the locus with the most significant association; BEST.GWAS.Z is the z-score of the best GWAS SNP; EQTL.ID is the best e/sQTL in the locus; EQTL.R2 is the cross-validation R2 of the best e/sQTL in the locus; EQTL.Z is the z-score of the best e/sQTL in the locus; EQTL.GWAS.Z is the GWAS Z-score for this e/sQTL; NSNP is the number of SNPs in the locus; NWGT is the number of snps with non-zero weights; MODEL is the best performing model; MODELCV.R2 is the the cross-validation R2 of the best performing model; MODELCV.PV is the p-value from the cross-validation of the best performing model; TWAS.Z is the TWAS z-score; TWAS.P is the TWAS p-value; trait is the GWAS trait; pop is the population used to estimate LD; joint_independent is the status if a gene/intron jointly independent (YES, if it is independent; NO, if it is not independent; NA, if it was not tested for the trait). Sheet 9-10: Summary of heritability (p-value < 0.01) and cross validation r2 from prediction models across cell-type specific/fetal bulk/adult bulk for gene and intron TWAS: hsq is the mean heritability of the genes/introns; hsq.se is the mean standard error of estimated heritability; hsq.pv is the mean p-value of the heritability; emmax.rsq is the mean cross-validation R2 training via EMMAX with p-value as emmax.pval; lasso.rsq is mean the cross-validation R2 via LASSO with p-value as lasso.pval; enet.rsq is mean the cross-validation R2 via elastic net with p-value as enet.pval; blup.rsq is mean the cross-validation R2 via BLUP with p-value as blup.pval; bslmm.rsq is the mean cross-validation R2 via BSLMM with p-value as bslmm.pval; top1.rsq is the mean cross-validation R2 via standard marginal e/sQTL Z-scores computation with p-value as top1.pval. 95 % confidence intervals per parameter are shown their below. Sheet 11-12: SCZ TWAS for GTEx Brain frontal cortex and downsampled fetal bulk data. Output from FUSION78: PANEL: Data type; ID is the gene ensemblID or intron id; CHR is the chromosome number; HSQ is the heritability; BEST.GWAS.ID is the GWAS SNP in the locus with the most significant association; BEST.GWAS.Z is the z-score of the best GWAS SNP; EQTL.ID is the best e/sQTL in the locus; EQTL.R2 is the cross-validation R2 of the best e/sQTL in the locus; EQTL.Z is the z-score of the best e/sQTL in the locus; EQTL.GWAS.Z is the GWAS Z-score for this e/sQTL; NSNP is the number of SNPs in the locus; NWGT is the number of snps with non-zero weights; MODEL is the best performing model; MODELCV.R2 is the the cross-validation R2 of the best performing model; MODELCV.PV is the p-value from the cross-validation of the best performing model; TWAS.Z is the TWAS z-score; TWAS.P is the TWAS p-value; trait is the GWAS trait. Table S1.
Data Availability Statement
Data will be available within dbGaP upon publication with study accession number phs002493.v1.p1, and code is available at https://bitbucket.org/steinlabunc/expression_splicing_qtls_public/src/master/.