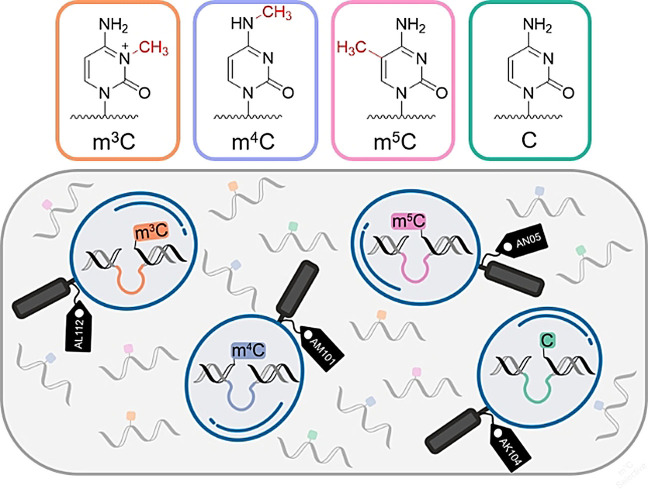
Abstract
Deoxyribozymes are emerging as modification‐specific endonucleases for the analysis of epigenetic RNA modifications. Here, we report RNA‐cleaving deoxyribozymes that differentially respond to the presence of natural methylated cytidines, 3‐methylcytidine (m3C), N 4‐methylcytidine (m4C), and 5‐methylcytidine (m5C), respectively. Using in vitro selection, we found several DNA catalysts, which are selectively activated by only one of the three cytidine isomers, and display 10‐ to 30‐fold accelerated cleavage of their target m3C‐, m4C‐ or m5C‐modified RNA. An additional deoxyribozyme is strongly inhibited by any of the three methylcytidines, but effectively cleaves unmodified RNA. The mXC‐detecting deoxyribozymes are programmable for the interrogation of natural RNAs of interest, as demonstrated for human mitochondrial tRNAs containing known m3C and m5C sites. The results underline the potential of synthetic functional DNA to shape highly selective active sites.
Keywords: deoxyribozymes, epitranscriptomics, in vitro selection, RNA modification, site-specific RNA cleavage
In vitro selected deoxyribozymes respond to the presence of methylcytidines m3C, m4C and m5C in RNA with either enhanced or strongly diminished cleavage rates, and thereby directly reveal the presence of the modified cytidines in target RNA.
Posttranscriptional nucleotide modifications are indispensable for the functional diversity of cellular RNA. More than 170 types of modified nucleotides are known,[1] but detection and quantification remain challenging. Transcriptome‐wide maps are available for some modifications,[2] but the predicted sites and derivatization levels must be further validated by alternative methods.[3] Recently, we reported RNA‐cleaving deoxyribozymes that reliably report the presence of N 6‐alkyladenosines.[4] Deoxyribozymes responsive to N 6‐methyladenosine (m6A) have been used to interrogate m6A levels in lncRNA, snoRNA and mRNA in DGACH sequence motifs,[4a, 5] and the results were in agreement with those obtained by the current benchmark in the field (the SCARLET assay).[6] We found that DNA enzymes activated by m6A reached 5–10‐fold faster cleavage rates for modified vs. unmodified substrate.[4a] The bulkier and more hydrophobic N 6‐isopentenyladenosine (i6A) in the RNA had a much stronger activating effect on deoxyribozyme‐catalyzed RNA cleavage, leading up to 2500‐fold faster cleavage of the i6A‐modified versus unmodified RNA substrate.[4b]
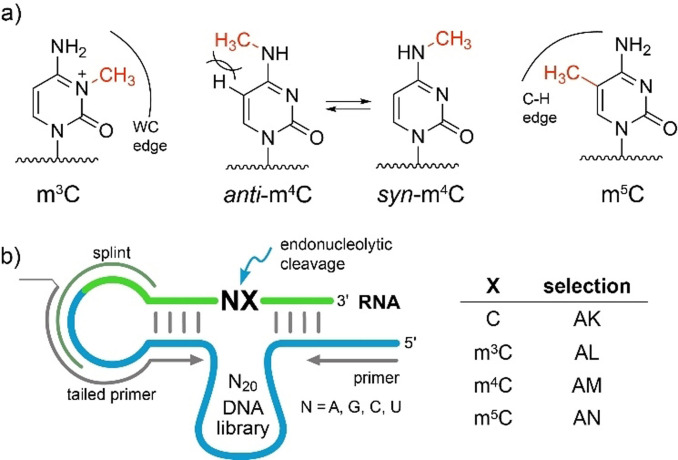
The finding of m6A‐ and i6A‐sensitive deoxyribozymes inspired the search for DNA catalysts that target other RNA modifications. Deoxyribozymes that are able to differentiate the three natural monomethylated cytidine isomers 3‐methylcytidine (m3C), N 4‐methylcytidine (m4C) and 5‐methylcytidine (m5C), and distinguish them from unmodified cytidine, are of particular interest. These methylated cytidines exhibit different structural properties depending on the location of the methyl group (Figure 1 a). The methyl group in m3C disrupts Watson–Crick base pairing and confers a positive charge to the nucleobase.[7] As a result, m3C may promote the formation of alternative secondary structures. Similarly to m6A, the methyl group in m4C is attached to the exocyclic amino group, and depending on its conformation, m4C can either retain (in anti‐conformation) or disrupt Watson–Crick base pairing (in syn‐conformation).[8] The anti‐conformation is enforced in a base‐paired structure but destabilized by steric repulsion (Figure 1 a). In contrast to m4C, the methyl group in m5C does not disturb base pairing directly but may interact with neighboring nucleotides via the C−H‐edge. The m5C modification is known to enhance base stacking.[9] Targeting these methylated cytidine isomers also addresses fundamental questions on scope and plasticity of DNA catalysis.
Figure 1.
a) Structures of three natural methylated cytidines and their structural consequences. m3C blocks the Watson–Crick (WC) edge and confers a positive charge to the nucleobase. m4C retains WC base pairing in anti‐conformation which is destabilized by steric clash between 5‐H and N 4‐Me. In syn‐conformation, m4C hinders WC base pairing. m5C enhances hydrophobicity in the major groove and improves base stacking. b) Design of in vitro selection libraries. Splint for ligation and primer binding sites are indicated. Detailed selection is shown in Figure S1.
To identify modification‐specific catalysts, we performed gel‐based in vitro selection experiments[4] (arbitrarily named AL, AM and AN, Figure 1 b) using an N20 DNA library and three synthetic RNA substrates, each containing one of the three monomethylated cytidines (R2‐R4, Table S1). Negative selection rounds were carried out with unmodified RNA (R1) to enhance the selectivity for the modified RNA. In this way, DNA enzymes that can cleave both unmodified and modified RNA were eliminated (Figure S1). In parallel, we sought DNA enzymes that preferentially cleave unmodified RNA and are inhibited by mxC (AK selection). In this case, the positive selection was performed with R1 and the negative selection used an equimolar mixture of R2‐R4. To evolve potentially universal DNA catalysts that respond to the cytidine modifications within all four possible dinucleotide sequence motifs NC, the RNA substrates contained a degenerate nucleotide upstream of the target cytidine.
After 18 rounds of selection, the enriched DNA pools were cloned, and 10 clones of each selection were sequenced. We identified 36 unique sequences (D6‐D41, Table S2), suggesting a high diversity in the selection libraries. To obtain further insights into the library composition and to potentially identify further DNAzyme candidates, we applied next‐generation sequencing (NGS) for a deeper analysis of AK, AL, AM and AN selection pools. In total, eight libraries were sequenced, including rounds 7 and 18 from each of the four in vitro selections. Deoxyribozymes specific for m3C, m4C or m5C should be enriched in selection libraries AL, AM or AN, respectively, and depleted in selection AK. Conversely, the deoxyribozymes inhibited by a modification would be highly enriched in selection AK and depleted in selections AL, AM or AN. In this way, using log2 fold change in abundance (log2fCA) between selection rounds 7 and 18 we identified nine additional candidate DNA sequences (D42‐D50, Table S2).[10]
Overall, 45 DNA sequences were then tested individually for their ability to cleave sixteen 3′‐fluorescently labeled RNA substrates R5‐R20 in independent cleavage reactions. The degenerate position was substituted with one of the four canonical ribonucleotides. Together R5‐R20 represented every possible combination of four different junctions and four cytidine variants. After removing the 5′‐overhang and 3′‐loop region, 28 deoxyribozymes retained trans‐cleaving activity (Table S3).
Using RNase T1 digestion and alkaline hydrolysis ladders, we determined the cleavage site for every active DNA enzyme. In accordance with the initial design, all tested deoxyribozymes cleaved the RNA substrate near the target cytidine (Table S3). The activity data partially supported our hypothesis that differences in enrichment in the selection libraries correlate with the specificity of deoxyribozymes. The correlation was good for DNAzymes found in the AK, AL and AM selections. However, poor correlation was observed for the selection AN, as the majority of the AN DNA enzyme candidates cleaved both m5C‐modified and unmodified RNA nearly to the same extent despite large differences in fold change abundance values (Table S3).
Interestingly, the inspection of predicted secondary structures revealed that none of the active deoxyribozymes contained the 8–17 motif, which had been repeatedly found in independent in vitro selections of RNA‐cleaving deoxyribozymes.[4a, 11] Its absence is especially surprising for AK selection, which used unmodified RNA as substrate. Probably, the 8–17 variants, that were initially present in the selection pools, were not particularly selective for either modified or unmodified RNA, and therefore were depleted during negative selection rounds. None of previously reported 8–17 variants was particularly efficient at cleaving N|C junctions.[11]
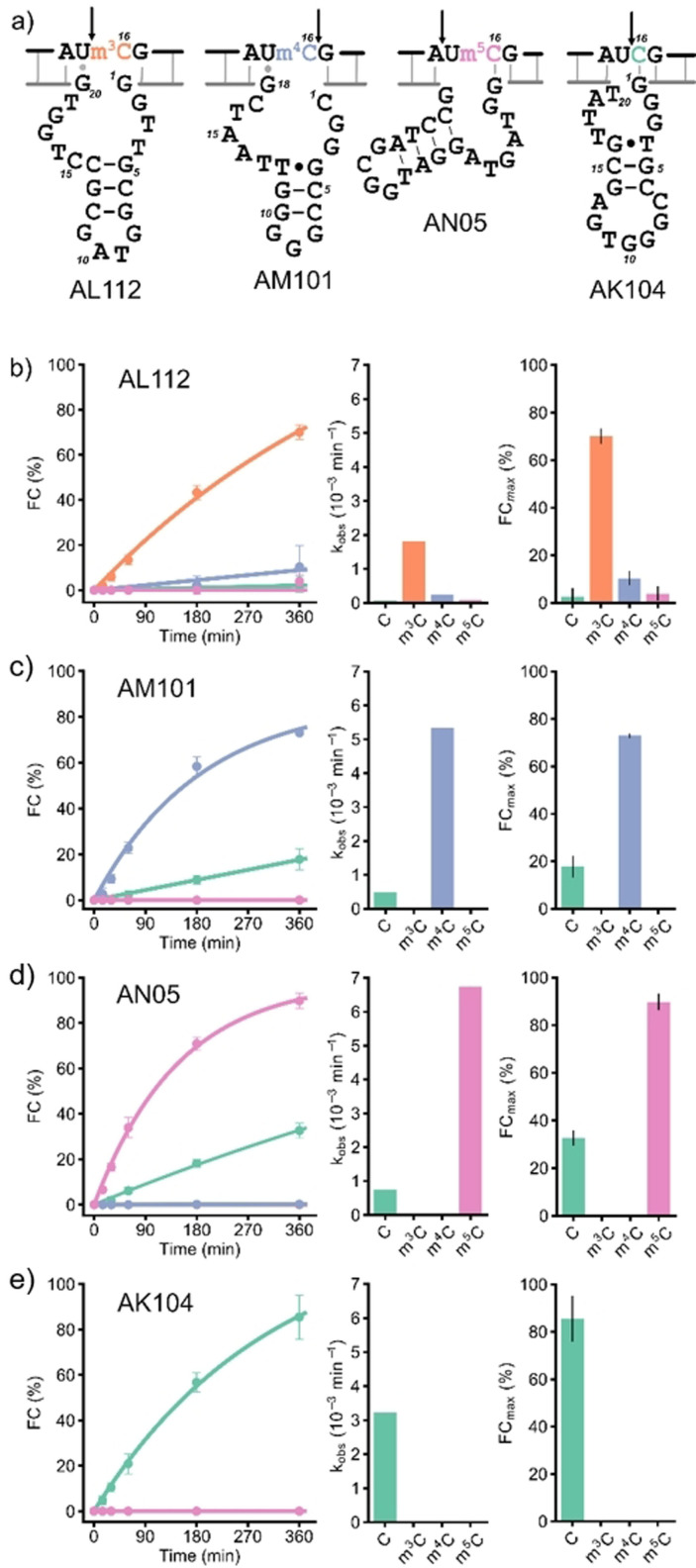
With respect to the degenerate position in the RNA substrate that was introduced to evolve for sequence generality, only deoxyribozyme AL112 distinguished a cytidine modification almost equally well in all four NC sequence contexts. On the other hand, many DNA catalysts were good at differentiating methylated from unmethylated RNA in one or two sequence contexts. In fact, we identified a corresponding deoxyribozyme for almost every combination of modification and junction (Table S3). For the UC junction in particular, we found that AL112, AM101, AN05 and AK104 (Figure 2 a) deoxyribozymes together achieved efficient differentiation for all three monomethylated cytidines. Among these, AL112, AM101 and AN05 preferentially cleaved Um3C‐, Um4C‐ and Um5C‐containing RNAs, respectively (fraction cleaved (FC) for the corresponding modified RNA FCUmXC≥70 %, and unmodified RNA FCUC≤30 % after 6 h, see Figure 2 b and Table S3), while AK104 efficiently cleaved only the UC substrate and was inhibited by all the modified nucleotides (FCUC=85 %, FCUmXC≈0 after 6 h). Therefore, this set of deoxyribozymes was further characterized.
Figure 2.
a) Sequences and predicted secondary structures of selected deoxyribozymes found in this study. The arrow indicates the cleavage site. Kinetic characterization of b) AL112, c) AM101, d) AN05, e) AK104 with four RNA substrates R8 (UC), R12 (Um3C), R16 (Um4C) and R20 (Um5C). b)—e): Single‐turnover kinetics plots (left). Observed rate constants k obs (middle). Cleavage yields FCmax after 6 h incubation (right: mean of three independent experiments with error bars showing SD); representative gel images are in the supporting information). Reaction conditions: 1 μM RNA, 10 μM deoxyribozyme, 20 mM MgCl2, 50 mM Tris‐HCl, 150 mM NaCl, pH 7.5, 37 °C.
First, we confirmed the cleavage sites determined by the gel‐based assay (Figure S3) by mass spectrometry. Consistent with the general mechanism proposed for DNA‐catalyzed RNA cleavage,[12] the cleavage products for all four selected catalysts contained 2′,3′‐cyclic phosphate and 5′‐OH termini (Figure S4).
Deoxyribozyme AL112 preferentially cleaved m3C‐modified RNA in all four NC sequence contexts with the highest cleavage yield for the UC junction (FCUm3C=70 %, Figure S5). In the UC context, AL112 showed a ca. 30‐fold higher k obs value for m3C‐RNA compared to unmodified RNA (Figure 2 b). The cleavage site was situated directly upstream of the modified nucleotide m3C. Analysis of the predicted secondary structure showed that presence of m3C might be required for disruption of the dG1‐rC16 base pair that otherwise inhibited RNA cleavage for the NC‐, Nm4C‐ and Nm5C‐RNA substrates (Figure 2 b, Table S3).
Deoxyribozyme AM101 was specific for m4C and cleaved Um4C‐RNA ca. 10 times faster than UC‐RNA. The cleavage site of AM101 was one nucleotide downstream of the modification site. Analysis of the secondary structure revealed the presence of a dG18‐rU15 wobble base pair, which might be responsible for selectivity of AM101 for the UC junction (Figure 2 a). However, in the CC sequence context, where formation of the stronger Watson–Crick base pair dG18‐rC15 is expected, a significant decrease in cleavage activity was observed (Figure S6, Table S3). Interestingly, AM101 also discriminated against N 4,N 4‐dimethylcytidine (m4,4C), which was cleaved at nearly the same rate as unmodified RNA (Figure S7).
The deoxyribozyme AN05 for cleavage of m5C‐modified RNA was found in Sanger sequencing data of the AN selection. Analysis of NGS data showed that it was also present in the AK selection, where it was strongly depleted from round 7 to round 18 (Table S3). In the UC context, AN05 cleaved only m5C‐modified and unmodified substrates (FC =90 %, FCUC=33 %), and was completely inhibited by m3C and m4C. The kinetic discrimination ability of AN05 was comparable to AM101: AN05 cleaved Um5C‐RNA ca. 10 times faster than the corresponding unmodified substrate (Figure 2 b, S8, and Table S3). The cleavage site of AN05 was located one nucleotide upstream of the target cytosine. The predicted secondary structure contained the dG1‐rC16 base pair that might stabilize an active site which is responsible for preference of AN05 to m5C and C (Figure 2 a). In case of m3C and m4C, this base pair might be destabilized or disrupted and thus lead to inhibition of cleavage.
The recognition of m5C by AN05 raised the question about the oxidized variants 5‐hydroxymethylcytosin (hm5C) and 5‐formylcytosin (f5C), previously reported in RNA as products of TET2 and ALKBH1 enzymes.[13] AN05 cleaved hm5C‐containing RNA slightly slower but to a similar extent as m5C RNA, while f5C was comparable to unmodified RNA (Figure S9). Surprisingly, this result is in line with bisulfite sequencing, which is unable to distinguish m5C from hm5C, and f5C from C.[14] Given the functional importance of hm5C and f5C in natural RNA,[13] future in vitro selection experiments may be directed to evolve deoxyribozymes specific for the individual oxidized m5C variants.
Deoxyribozyme AK104 efficiently cleaved unmodified RNA (FC ≥85 % after 6 h) and was inhibited by all cytosine modifications in AC, GC and UC sequence contexts. In this case, the cleavage site was situated directly upstream of the target cytosine. However, when the RNA substrates with the CC junction were used, AK104 cleaved the m3C‐containing RNA with a moderate yield (FC ca. 30 %) and was inactive with the unmodified substrate. The cleavage site for the Cm3C‐RNA was shifted one nucleotide upstream (Figure S10, Table S3). This odd behavior can be explained by disruption of the dG1‐rC16 base pair by m3C in Am3C‐, Gm3C‐ and Um3C‐RNAs and formation of an alternative GC base pair between dG1 and rC15 in case of Cm3C‐RNA. The new base pair leads to repositioning of the AK104 active site and partial rescue of the cleavage activity. On the other hand, both rC15 and rC16 in the CC‐substrate can form base pairs with dG2 and dG1, respectively, thereby disrupting the functional active site and abolish the catalytic activity of AK104. Thus, AK104 is a RNA‐cleaving DNA enzyme for DC junctions (D=A,G,U), with high selectivity for unmodified RNA even under accelerated cleavage conditions at elevated Mg2+ concentrations. None of the modified RNAs was cleaved, and RNAs with rC16 changed to U, G, or A were also not cleaved (Figures S11, S12, S13).
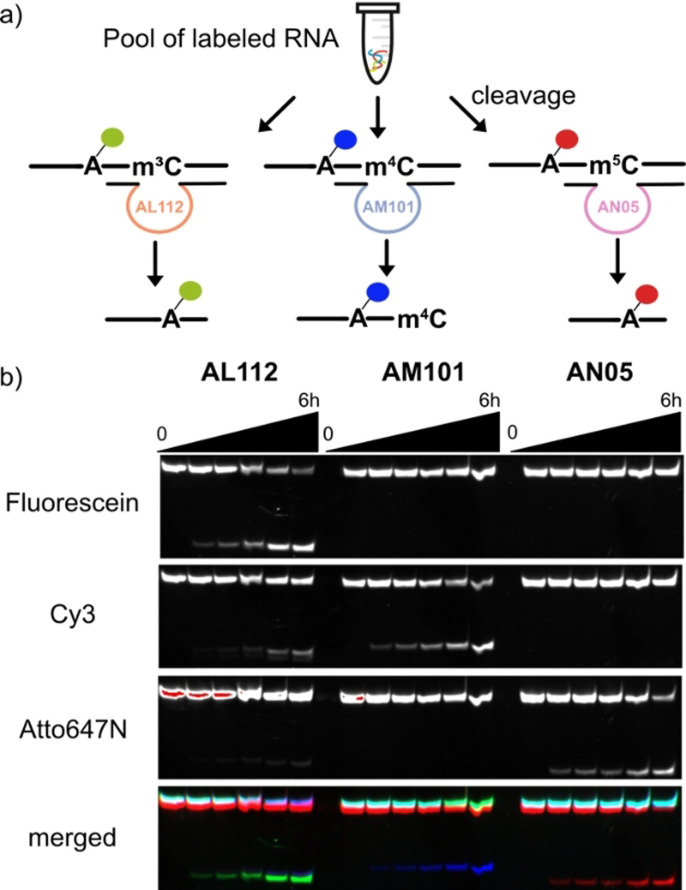
Next, we examined the deoxyribozymes in the more challenging situation when all three RNA isomers (Um3C‐, Um4C‐ and Um5C‐RNAs) are present at the same time. For this experiment, we labeled the three methylated UC‐RNA substrates (R12, R16 and R20) at the 2′‐OH of rA9 with fluorescent N 6‐(6‐aminohexyl)‐ATP derivatives, carrying 6FAM, Cy3 and ATTO‐647N, respectively.[15] An equimolar mixture of the three labeled RNAs was then incubated with 10‐fold excess of the AL112, AM101 or AN05 deoxyribozyme, respectively (Figure 3 a). The cleavage reactions were analyzed by denaturing PAGE and imaged using a dedicated channel for each fluorescent label (Figure 3 b). As expected, AL112 showed preferential cleavage of Um3C‐RNA with negligible signals in the m4C‐ and m5C‐channels. At the same time, AM101 and AN05 cleaved only their cognate substrates with no detectable cleavage products in other channels. The results demonstrated that the specificity of each DNAzyme for its target modification was retained, and this collection of deoxyribozymes can be used to examine the identity of methylated cytidines in UC motifs. In this regard, m4C‐specific RNA cleavage is particularly important, as in contrast to m3C and m5C, there are no specific chemical reactions reported for the detection of m4C in RNA.[16]
Figure 3.
a) Schematic representation of selective RNA cleavage with AL112, AM101 and AN05. RNA substrates R12, R16 and R20 were separately labeled with 6FAM, Cy3 and Atto647N, respectively,[15] pooled and the mixture was subjected to cleavage with each of the three modification‐specific DNAzymes. b) PAGE analysis of the experiment. Individual imaging channels for the three fluorophores are shown in black and white, and the merged image in false‐color representation. Reaction conditions: 1 μM RNA mixture, 10 μM deoxyribozyme, 20 mM MgCl2, 50 mM Tris‐HCl, 150 mM NaCl, 37 °C.
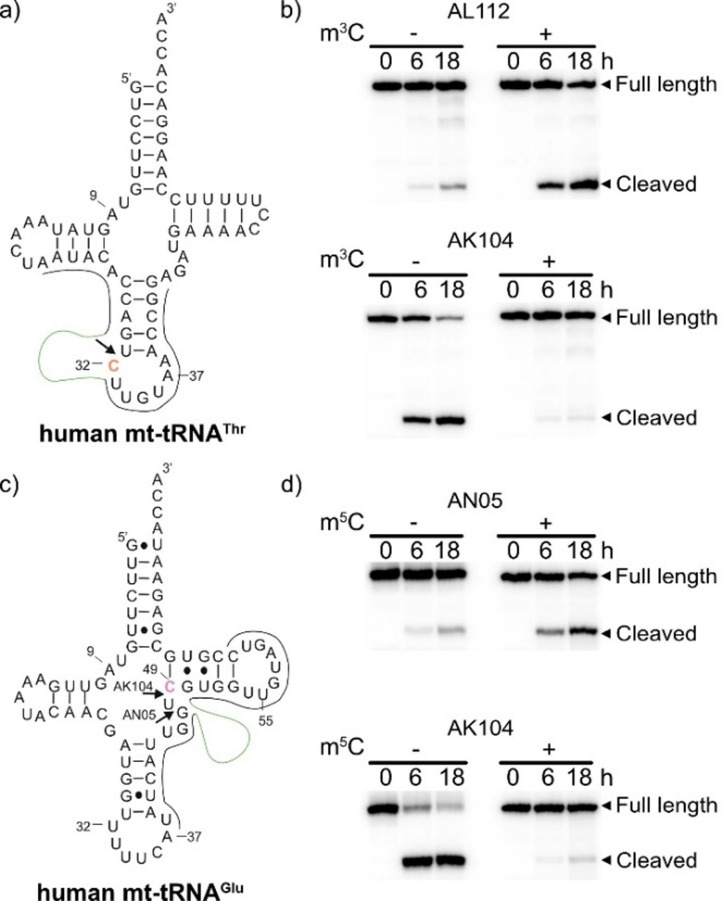
Next, we tested deoxyribozymes AL112, AN05 and AK104 on longer RNA substrates with complex secondary structures. To this end, we chose human mt‐tRNAThr, which contains a known m3C in the anticodon loop (C32, Figure 4 a),[17] and mt‐tRNAGlu with a known m5C at C49 (Figure 4 c).[17] The single‐modified tRNAs were assembled from synthetic RNA fragments by splinted ligation with T4 RNA ligase, and the binding arms of deoxyribozymes were designed to cleave the tRNAs upstream of the corresponding modification sites.[18] The results of the cleavage reactions for unmodified and modified tRNAs were compared (Figure 4 b,d). AK104 cleaved both unmodified tRNAs with a high yield (>90 % after 18 h) and was nearly completely inhibited by m3C and by m5C. AL112 and AN05 cleaved m3C‐modified mt‐tRNAThr and m5C‐modified mt‐tRNAGlu, respectively, both somewhat less efficiently than their cognate substrates R12 and R20 (58 % and 42 % after 18 h), but the unmodified tRNAs did not undergo any significant cleavage. Therefore, AL112 and AN05 can faithfully identify m3C‐ and m5C‐modified tRNA, respectively.
Figure 4.
a) Secondary structure of mt‐tRNAThr with target C32 in orange. b) PAGE analysis of mt‐tRNAThr cleaved with AL112 and AK104. c) Secondary structure of mt‐tRNAGlu with target C49 in violet. d) PAGE analysis of mt‐tRNAGlu cleaved with AN05 and AK104. The cleavage reactions were performed at 37 °C. The arrows indicate the cleavage sites. Binding sites for DNAzymes are shown (binding arms in gray, catalytic core in green).
In summary, we have identified and characterized a series of RNA‐cleaving deoxyribozymes that differentially respond to the presence of methylated cytidines in RNA. On the one hand, the methyl groups in m3C, m4C and m5C can dramatically inhibit deoxyribozyme‐catalyzed RNA cleavage (as in case of AK104). On the other hand, the presence of methylcytidines can lead to accelerated cleavage of modified RNA (as in case of AL112, AM101 and AN05). Owing to these kinetic differences, new deoxyribozymes can be used to examine the identity of methylcytidines and the degree of their methylation at selected target sites. The effect of the methyl group on catalytic activity might be related to steric, electrostatic (m3C) or hydrophobic interaction of the modified nucleotide with its neighbors and Mg2+ in the active site. The detailed structural basis for the selective recognition of methylcytidines remains to be identified.
As the enriched libraries for selection of methylcytidine‐sensitive deoxyribozymes remained highly diverse even after 18 rounds of selection, we expect that many DNA enzymes with potentially interesting properties remained undiscovered. To this end, massively parallel activity assays that are able to screen cleavage reactions of thousands of candidate deoxyribozymes are required. Given the recent success in using deep sequencing for large‐scale characterization of (deoxy)ribozyme kinetics,[19] new catalytic DNA motifs can likely be revealed in the future. The related experiments are currently ongoing in our laboratory and expected to uncover hidden treasures in in vitro selection libraries.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was supported by the DFG (SPP1784) and by the ERC (grant no. 682586). A.L. acknowledges funding by a PhD scholarship from the German Academic Exchange Service (DAAD). M.V.S. thanks the Graduate School of Life Sciences at the University of Würzburg for a Postdoc Plus fellowship. Illumina sequencing was performed at the Core Unit Systems Medicine at the University of Würzburg. Open access funding was enabled and organized by Projekt DEAL.
A. Liaqat, M. V. Sednev, C. Stiller, C. Höbartner, Angew. Chem. Int. Ed. 2021, 60, 19058.
In memory of Professor Siegfried Hünig
References
- 1.
- 1a.Boccaletto P., Machnicka M. A., Purta E., Piątkowski P., Bagiński B., Wirecki T. K., de Crécy-Lagard V., Ross R., Limbach P. A., Kotter A., Helm M., Bujnicki J. M., Nucleic Acids Res. 2018, 46, D303–D307; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b.McCown P. J., Ruszkowska A., Kunkler C. N., Breger K., Hulewicz J. P., Wang M. C., Springer N. A., Brown J. A., Wiley Interdiscip. Rev. RNA 2020, 11, e1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xuan J.-J., Sun W.-J., Lin P.-H., Zhou K.-R., Liu S., Zheng L.-L., Qu L.-H., Yang J.-H., Nucleic Acids Res. 2018, 46, D327–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a.Helm M., Motorin Y., Nat. Rev. Genet. 2017, 18, 275–291; [DOI] [PubMed] [Google Scholar]
- 3b.Linder B., Jaffrey S. R., Cold Spring Harbor Perspect. Biol. 2019, 11, a032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a.Sednev M. V., Mykhailiuk V., Choudhury P., Halang J., Sloan K. E., Bohnsack M. T., Höbartner C., Angew. Chem. Int. Ed. 2018, 57, 15117–15121; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 15337–15341; [Google Scholar]
- 4b.Liaqat A., Stiller C., Michel M., Sednev M. V., Höbartner C., Angew. Chem. Int. Ed. 2020, 59, 18627–18631; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 18786–18790; [Google Scholar]
- 4c.Micura R., Höbartner C., Chem. Soc. Rev. 2020, 49, 7331–7353. [DOI] [PubMed] [Google Scholar]
- 5.Bujnowska M., Zhang J., Dai Q., Heideman E. M., Fei J., J. Biol. Chem. 2020, 295, 6992–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu N., Parisien M., Dai Q., Zheng G., He C., Pan T., RNA 2013, 19, 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a.Flemmich L., Heel S., Moreno S., Breuker K., Micura R., Nat. Commun. 2021, 12, 3877; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b.Mao S., Haruehanroengra P., Ranganathan S. V., Shen F., Begley T. J., Sheng J., ACS Chem. Biol. 2021, 16, 76–85. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a.Micura R., Pils W., Höbartner C., Grubmayr K., Ebert M. O., Jaun B., Nucleic Acids Res. 2001, 29, 3997–4005; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b.Mao S., Sekula B., Ruszkowski M., Ranganathan S. V., Haruehanroengra P., Wu Y., Shen F., Sheng J., Nucleic Acids Res. 2020, 48, 10087–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a.Trixl L., Lusser A., Wiley Interdiscip. Rev. RNA 2019, 10, e1510; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b.Harcourt E. M., Kietrys A. M., Kool E. T., Nature 2017, 541, 339–346; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c.Wang S., Kool E. T., Biochemistry 1995, 34, 4125–4132. [DOI] [PubMed] [Google Scholar]
- 10.We picked the candidates with the largest differences between log2fCA and log2fCA,neg (see Table S3).
- 11.
- 11a.Santoro S. W., Joyce G. F., Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b.Li J., Zheng W., Kwon A. H., Lu Y., Nucleic Acids Res. 2000, 28, 481–488; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11c.Cruz R. P. G., Withers J. B., Li Y., Chem. Biol. 2004, 11, 57–67; [DOI] [PubMed] [Google Scholar]
- 11d.Schlosser K., Li Y., Biochemistry 2004, 43, 9695–9707; [DOI] [PubMed] [Google Scholar]
- 11e.Lam J. C. F., Withers J. B., Li Y., J. Mol. Biol. 2010, 400, 689–701. [DOI] [PubMed] [Google Scholar]
- 12.
- 12a.Breaker R. R., Joyce G. F., Chem. Biol. 1994, 1, 223–229; [DOI] [PubMed] [Google Scholar]
- 12b.Silverman S. K., Nucleic Acids Res. 2005, 33, 6151–6163; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12c.Schlosser K., Li Y., ChemBioChem 2010, 11, 866–879. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a.He C., Bozler J., Janssen K. A., Wilusz J. E., Garcia B. A., Schorn A. J., Bonasio R., Nat. Struct. Mol. Biol. 2021, 28, 62–70; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b.Haag S., Sloan K. E., Ranjan N., Warda A. S., Kretschmer J., Blessing C., Hubner B., Seikowski J., Dennerlein S., Rehling P., Rodnina M. V., Höbartner C., Bohnsack M. T., EMBO J. 2016, 35, 2104–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.
- 14a.Song C.-X., Yi C., He C., Nat. Biotechnol. 2012, 30, 1107–1116; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b.Dai Y., Yuan B.-F., Feng Y.-Q., RSC Chem. Biol. 2021, 10.1039/D1CB00022E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.RNA labeling was performed with FH14 ribozyme: Ghaem Maghami M., Scheitl C. P. M., Höbartner C., J. Am. Chem. Soc. 2019, 141, 19546–19549. [DOI] [PubMed] [Google Scholar]
- 16.
- 16a.Behm-Ansmant I., Helm M., Motorin Y., J. Nucleic Acids 2011, 408053; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Heiss M., Kellner S., RNA Biol. 2017, 14, 1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., Okada S., Mito M., Iwasaki S., Ma D., Zhao X., Asano K., Lin H., Kirino Y., Sakaguchi Y., Suzuki T., Nat. Commun. 2020, 11, 4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.To facilitate access of the deoxyribozymes, disruptor oligonucleotides were included complementary to the 5′ or 3′ end of the acceptor stem, respectively (see D56 and D59 in the Supporting Information, Table S2).
- 19.
- 19a.Kobori S., Yokobayashi Y., Angew. Chem. Int. Ed. 2016, 55, 10354–10357; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 10510–10513; [Google Scholar]
- 19b.Dhamodharan V., Kobori S., Yokobayashi Y., ACS Chem. Biol. 2017, 12, 2940–2945; [DOI] [PubMed] [Google Scholar]
- 19c.Pressman A. D., Liu Z., Janzen E., Blanco C., Müller U. F., Joyce G. F., Pascal R., Chen I. A., J. Am. Chem. Soc. 2019, 141, 6213–6223; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19d.Andreasson J. O. L., Savinov A., Block S. M., Greenleaf W. J., Nat. Commun. 2020, 11, 1663; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19e.Yokobayashi Y., Acc. Chem. Res. 2020, 53, 2903–2912; [DOI] [PubMed] [Google Scholar]
- 19f.Shen Y., Pressman A., Janzen E., Chen I. A., Nucleic Acids Res. 2021, 10.1093/nar/gkab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information