Abstract
States of oral health and disease reflect the compositional and functional capacities of, as well as the interspecies interactions within, the oral microbiota. The oral cavity exists as a highly dynamic microbial environment that harbors many distinct substrata and microenvironments that house diverse microbial communities. Specific to the oral cavity, the nonshedding dental surfaces facilitate the development of highly complex polymicrobial biofilm communities, characterized not only by the distinct microbes comprising them, but cumulatively by their activities. Adding to this complexity, the oral cavity faces near‐constant environmental challenges, including those from host diet, salivary flow, masticatory forces, and introduction of exogenous microbes. The composition of the oral microbiome is shaped throughout life by factors including host genetics, maternal transmission, as well as environmental factors, such as dietary habits, oral hygiene practice, medications, and systemic factors. This dynamic ecosystem presents opportunities for oral microbial dysbiosis and the development of dental and periodontal diseases. The application of both in vitro and culture‐independent approaches has broadened the mechanistic understandings of complex polymicrobial communities within the oral cavity, as well as the environmental, local, and systemic underpinnings that influence the dynamics of the oral microbiome. Here, we review the present knowledge and current understanding of microbial communities within the oral cavity and the influences and challenges upon this system that encourage homeostasis or provoke microbiome perturbation, and thus contribute to states of oral health or disease.
1. INTRODUCTION
The field of human microbiome research has undergone a revolution in its approach toward understanding how microorganisms influence the physiology of their host.1 Development of culture‐independent methods has resulted in increased detection and classification of microbial species within microbial communities.2 technologies, biomarker sequencing, and shotgun metagenomics have become standard tools used to determine the composition and genetic makeup of the human microbiome.3 Other “‐omics” technologies, such as proteomics and metabolomics, support mechanistic hypotheses involved in causal microbial pathways that are related to states of health and disease.4, 5 Since Antonie van Leeuwenhoek first discovered the existence of microbes in the 1700s while analyzing dental plaque under a microscope, the composition of oral microbial communities has been extensively studied.6 Over 250 species from the oral cavity have been isolated in culture and characterized, including several key pathogens, such as Streptococcus mutans, Porphyromonas gingivalis, Tannerella forsythia, and Aggregatibacter actinomycetemcomitans, involved in the etiology of dental caries and periodontal disease.7, 8, 9 An integrated approach toward understanding states of oral disease from the polymicrobial perspective has emerged over time, attributing disease pathology not only to key pathogens but rather to networks of co‐occurring microbes, the collective activities of which contribute to pathogenesis.9, 10, 11, 12 As such, the importance of understanding the divergences, between oral health and disease, in the microbes comprising the system as well as their relative abundance and functional activity, in addition to genetic factors and ecological pressures that drive such changes, is a primary focus of research within the field of oral health research.9, 13, 14, 15, 16, 17 In recent decades, genetic approaches have shed light on the functional capacity of members of oral microbiomes, the mechanistic underpinnings of caries and periodontal disease pathogenesis, and the complex dynamics and fitness factors of key organisms in oral microbiomes.3, 18
The oral microbial ecosystem is constantly exposed to exogenous foreign substances.17 Such circumstances are defining factors for founding microbes and their ability to persist in this environment, and make for distinct relationships between microbe and host that rely on selective pressures. Pioneer microbial colonizers of the oral cavity, such as Streptococcus mitis, Streptococcus sanguinis, Streptococcus gordonii, and Streptococcus salivarius, display core characteristics that make them well suited to this specific niche as they are able to bind selectively to tongue and cheek cells before the teeth emerge and can outcompete other microbial species.17, 19 Emerging teeth acquire a protective glycoprotein coat, which sets in motion successional microbial colonization, resulting in the development of complex polymicrobial biofilm communities, namely dental plaque.20 These complex dental plaque matrices create unique microenvironments that harbor acidic and anaerobic microenvironments, and thus select for organisms distinct from those growing directly on the tooth surface.21
Diet provides nutritional resources for the oral microbiota and also serves as a selective pressure by enriching for organisms best adapted to utilize specific host‐derived dietary resources.22 Major historical dietary shifts throughout evolution are accompanied by significant changes in the oral microbiota.4, 23 Lifestyle changes taking place at the Neolithic Revolution, and the later Industrial Revolution, resulted in development of the Westernized Diet, characterized by dietary staples such as farmed animal meats, dairy products, refined vegetable oils, and processed cereal grains that substantially diverged from pre‐agricultural diets. Today, such dietary constituents are staples of the American diet, as well as in many developed and developing countries. Such changes in diet were paralleled by pathologic changes to the oral microbiota, including greater representation of acid‐producing and acid‐tolerant organisms and periodontal pathogens.17, 22, 24, 25 While diet influences the oral microbiome,22, 24, 26 recent data indicate that the oral microbiome influences the dietary preferences of its host. Certain bacteria, such as some Clostridia and Prevotella species, have been associated with taste thresholds, such as sweet, sour, salty, and bitter, plausibly representing a mechanism by which the oral microbiota influences dietary preferences to sustain its membership and persistence in the oral cavity.27, 28 Oral hygiene habits are another consistent source of influence on the oral microflora.29, 30, 31 Toothbrushing and flossing can be powerful means to disrupt plaque, the microbial inhabitants of which can cause tooth demineralization and gingival inflammation long‐term.32 Toothbrushes themselves, however, can also serve as reservoirs for pathogenic bacteria that can then inoculate the oral cavity, bringing into view the importance of properly sanitizing and storing personal dental hygiene equipment.28, 33 Novel toothpastes have also entered the market to intentionally to shape the oral microbiota via proteins designed to foster species associated with healthy oral bacterial communities, while other products have more general antimicrobial properties.33, 34 Mouthwashes are also designed to have the same effect as toothpaste in that they reduce microbial load via antimicrobial and bactericidal mechanisms.35, 36, 37
The influence of the oral microbiota is not confined to this location.38 Oral cavity‐associated microbes have been detected in many distant organ sites, including the small intestines, lungs, heart, placenta, and brain.39 Many associations between oral microbes, specifically those implicated in periodontal disease, and other common chronic conditions, such as cardiovascular disease and high blood pressure, have been established.40 Data on the mechanistic connections involved in the development of disease at sites distant from the oral cavity remain sparse, but early research demonstrates that oral cavity‐associated microbes can influence immune responses and disease pathogenesis outside the oral cavity, and that their ability to colonize ectopic sites depends on the current state of health of that site.39, 41 These data suggest that the oral microbiome may serve as a reservoir for pathobionts that can either contribute to or exacerbate disease at remote body niches or organ systems. The oral microbiome has become an increasingly important component of recommendations and practices in dental medicine.42 New approaches to modulate oral microbiomes are being presented. For example, some oral probiotics are being designed to increase the alkalinity of the oral cavity and plaque and others are developed to target pathogenic species, such as S mutans.43, 44, 45 Administration of supplements, such as arginine, can also substantially affect the composition and metabolic output of an oral microbial community and represents another modular handle.43
2. RESEARCH TECHNIQUES
2.1. Sequence‐based culture‐independent approaches to assess the microbiome
The field of human microbiome research has revolutionized our view of the role of microbes on mammalian development and health. Traditional approaches revolved around culturing clinical samples in vitro, prior to testing their roles in pathogenesis using in vitro or in vivo assays. A major advancement in this domain has been the development of germ‐free mice—animals bred, fed, and raised under sterile conditions—which offer a useful tool for studying the microbiome.46 First conceptualized by Louis Pasteur in 1885 but with uses only fully appreciated in recent decades, this approach allows experimentation in mammals with either no pre‐existing microbiome or a highly defined microbial background.46 Seminal studies using germ‐free mice confirmed the role of the gut microbiome in a number of diseases, including obesity,47 Kwashiorkor,48 and autism‐spectrum disorder,49 by inducing disease features in recipient animals following transfer of a patient‐associated microbiome. Studies using germ‐free mice have also been employed in the context of the oral microbiome, demonstrating the role of diabetes in disrupting the equilibrium of the oral microbiota50 and confirming the role of the oral microbiota in periodontal disease pathogenesis.51, 52 Studies in germ‐free mice have also recently identified a novel role of masticatory forces in eliciting immune surveillance responses in the gingiva in a microbiota‐independent manner, consistent with the role of gingival tissues as a physiological barrier in the face of ongoing masticatory challenges.53, 54
In vitro biofilm culture systems using human saliva or defined media have served as a useful surrogate for oral biofilm research and have shed light on the mechanisms involved in microbial adherence, species interactions, antibiofilm treatments, and organization of the microbial community.55, 56, 57, 58, 59 Biofilms, with anaerobic centers and aerobic peripheries, exhibit gradients of oxygenation.60, 61 As oral bacteria can be fastidious, slow growing, or require specialized growth media, and many are strict anaerobes, samples should be collected and cultivated appropriately.62, 63 Thus, in vitro models need to be designed to take into account selective pressures in the oral cavity, including salivary flow, species‐mediated biofilm succession, and inflammatory substrates, in the development of subgingival biofilm communities.56, 64, 65 In vivo and in vitro biofilm models have been combined in an ex vivo oral biofilm growth model to facilitate the complex interactions taking place in the oral cavity during microbial biofilm succession.66 While powerful insights into oral biofilms have been obtained from in vitro biofilm studies, the difficulty of recapitulating the complexity of the oral cavity in vitro complicates the use of such approaches to investigate microbial communities and understand their holistic functional qualities in their entirety.
Culture‐independent methods have improved our understanding of microbial diversity in the oral cavity. Advances in sequencing and mass spectrometry technologies have permitted assessment of microbial community membership, their functional activities, and molecular products.9, 14, 67, 68, 69, 70 Sequence‐based approaches to assess microbial composition include biomarker approaches that focus on a single kingdom‐specific ubiquitous microbial gene or region that exhibits sequence hypervariability (eg, the 16S ribosomal RNA gene in bacteria or the interspacer region in fungi).71 Such hypervariable genes or regions permit identification of microbial community members but can be limited in their capacity to resolve phylogenetically related species or strains and do not provide information on functional gene content of the microbial members present.72 As such, they are useful for cataloging differences in microbiota composition, particularly in large studies, because the approach is relatively inexpensive compared with other, more highly resolving, sequence‐based approaches.72, 73 Newer approaches have developed strategies to assess the presence of bacteria in environments with a low‐burden microbial signal.74, 75 One such application is depletion of abundant sequences by hybridization, in which the nonmicrobial DNA burden is depleted via CRISPR‐associated endonuclease Cas9 targeting to enhance bacterial signals in samples with a low bacterial burden.74, 76
Unlike 16S ribosomal RNA sequencing, shotgun metagenomics sequences permit a parallel assessment of all microbial kingdoms (bacterial, fungal, viral) in a given sample.77, 78 This approach employs random fragmentation and adapter ligation sequence in an unbiased manner all extracted DNA, enabling more in‐depth analyses of the pan‐genomic gene content in microbiomes.3 Whole genomes of organisms, in addition to strain tracking, can be extracted from the data, allowing for evolutionary analysis of specific organisms associated with a particular disease or environment.79, 80, 81 The usefulness of these techniques has been greatly accelerated by the advent of next‐generation sequencing technologies, which provide increased read‐depth, improved accuracy, and are higher throughput than older methods, such as Sanger sequencing. Three prominent companies have dominated this field so far, with Illumina MiSeq offering shorter‐read sequencing, and PacBio and Oxford Nanopore technologies providing longer read lengths.77 There are pros and cons of both short‐ and long‐read lengths: short‐read technology provides abundant sequencing data that is less error prone than long‐read technology; however, it can be difficult to assemble complete genomes using short read‐lengths due to limitations in the technology to distinguish repetitive elements. By contrast, long‐read technology provides more read‐length but at the cost of higher error rates.82 The generation of high‐molecular‐weight DNA was a limitation that prevented long‐read technology from demonstrating its full potential in advancing shotgun metagenomic methodologies; however, recent improvements in sample preparation have resulted in increased interest for use of this technology in metagenomic studies.83, 84, 85 Using shotgun metagenomic sequencing, it is now possible to study members of the microbiome other than bacteria, including the oral virome, in the context of oral diseases, together with periodontal disease.86 Results obtained from the small number of studies performed using shotgun metagenomic sequencing suggest that the oral virome may be as significant in disease pathogenesis as the oral bacteriome.87, 88 Metatranscriptomics, or sequencing of mRNA in a sample, provides a snapshot of transcriptionally active microbes.89 RNA has low stability and thus a short half‐life. Hence, the ability of transcriptomic approaches to detect RNA from functionally active and viable bacteria overcomes the limitations of metagenomic approaches. Advances in RNA sequencing, such as random hexamer priming, permit assessment of microbial and host transcriptomes in parallel and are now being explored to assess the interactome.90 Single‐cell sequencing was developed initially for immune‐profiling purposes but has been adapted to permit assessments of single microbial cells.91 Assessing individual cells from environmental samples increases the detection rate of unculturable organisms while providing the opportunity to ask more novel questions related to the functional capacities and significant roles of single organisms within complex microbial communities.92, 93 Application of transcriptomic approaches has proved extremely significant for delineating both microbial and host gene expression in the context of oral health and pathology.14, 68, 94
The field faces some key hurdles, namely the challenge of separating out the highly complex mixtures that are typical of clinical samples while simultaneously visualizing many molecules of a diverse chemical nature. In periodontal disease, there is a characteristic shift in the composition of oral bacteria that is in part mediated by bacterial metabolites.95 Despite its drawbacks, use of metabolomics could provide valuable mechanistic insights into how and why this shift occurs, and may offer clues to critical time points at which therapeutic or lifestyle interventions may be beneficial. Ultimately, longitudinal, integrated multimodal analyses, involving a range of high‐resolution profiling techniques, represent, together with clinical data, the next frontier to understanding microbial host interactions from species level to the molecular level and the implications of these on oral health.
2.2. Metabolomics and proteomics
Recent developments in high‐resolution profiling techniques have additionally focused on the profile of small molecules, such as metabolites and proteins, that are detected via liquid or gas chromatography‐mass spectrometry or nuclear magnetic resonance spectroscopy. Metabolomics represents the study of molecules in biological samples, which, in the case of human samples, may be produced by the host or its microbiome.5 Metabolomics provides insight into the metabolic and functional activities of the host and its microbiome and intricate interspecies interactions encoded within this pangenome.89 Metabolite production is influenced by the availability of energy sources, environmental stressors, and competition among microbes within a system.5 Metabolomics provides important information related to changes in functional and metabolic pathways via analysis of divergent metabolite profiles presented in the context of health or disease.96, 97 In periodontal disease, there is a characteristic shift in the profile of oral bacteria that is in part mediated by bacterial metabolites, which comprise a chemical communication network. Despite its drawbacks, metabolomics could provide valuable mechanistic insights into how and why this shift occurs, and offer clues to potential therapeutic or lifestyle interventions. Longitudinal, integrated multimodal analyses are ideal for investigating the species that are present, active, and that interact with host cells over time. By contrast, proteomics seeks to analyze the proteome, namely, the profile of all proteins within an organism, tissue, cell, or biological fluid, or subcomponent of any of these.98 Such applications provide insight related to expression and modulation of proteins under specific conditions, such as in health or disease.99 These analyses present the opportunity to identify proteins present within a sample, as well as the abundance, post‐translational modifications, isoforms, and molecular interactions of proteins.100 Such technologies use 1‐ or 2‐dimensional gel electrophoresis/mass spectrometry or liquid chromatography/mass spectrometry.100 Proteomic applications have been applied to understand changes in the proteome that diverge states of periodontal health from disease and to further characterize various periodontal disease states, including gingivitis, mild, moderate, chronic, and aggressive periodontitis.70, 101, 102, 103, 104
3. COMMUNITY ASSEMBLY OF THE ORAL MICROBIOME
3.1. Oral colonization in early life
The oral cavity is a site of first encounters. As the gatekeeper of the alimentary canal, the oral cavity is the first organ to encounter ingested food and drink, exogenous microbes, allergens, and antigens before they pass further into the gastrointestinal and/or respiratory tracts.54 These direct environmental exposures in the absence of keratinized epithelium pose the oral cavity as a highly susceptible site for infection.105 Regardless, specialized immune‐cell networks in the oral cavity respond to the challenges of this fluctuating environment via tissue‐specific cues and exclusive immunologic responses that are tailored to the oral cavity.53, 106, 107, 108 In line with the role of oral mucosa as a physiological barrier, the functions of immune networks within this mucosa reflect the site‐specific challenges faced within the oral cavity. For example, they contribute to homeostasis in response to masticatory forces and trigger immune responses to the development of pathologic microbial communities.52, 53
Oral immune ontogeny, as with the gut, develops by 11 weeks of gestation, at which point cellular components related to the prenatal secretary immune system demonstrate organization of tissue into Peyer's patches.109 Current research findings suggest that the prenatal oral cavity is sterile until birth after which colonization with microorganisms occurs upon exposure to the external environment.110 However, oral bacteria have been detected at various sites within the uterus.110, 111 Interestingly, in a study of 12 mother‐neonate pairs, it was found that the microbiota of the neonatal oral cavity displayed clear associations with that of the placenta and was not significantly altered by the birth canal or maternal microbiotas, suggesting that the neonatal microbiota may have a prenatal origin.112 As such, studies pertaining to this question warrant future investigation. Pathogenic bacteria from the oral cavity found at various sites within the uterus are associated with adverse pregnancy outcomes, such as preterm delivery and preeclampsia.113, 114, 115 Studies investigating the placental microbiome to understand its role in preterm pregnancies have identified bacteria associated with periodontal disease, suggesting a relationship between the oral and placental microbiomes.113, 114, 115, 116 Oral microbes from genera including Streptococcus, Fusobacterium, Neisseria, Prevotella, and Porphyromonas have been recovered from placenta.111 Interestingly, the placental microbiota more closely resembles that of the maternal oral microbiome than that of the gut.116 Animal studies have helped to confirm the direct role of the oral microbiota, and more specifically periodontal disease‐related pathogens, in adverse pregnancy outcomes.117 Similarly, bacteria of oral origin, namely Fusobacterium nucleatum, were identified in samples of amniotic fluid and cord blood from women with pregnancy complications, suggesting oral translocation via hematogenous mechanisms.115 Fusobacterium nucleatum has also been associated with stillbirth.118
Following birth, overt colonization of the oral cavity with microbes occurs within 8‐16 hours as a result of transmission of microbes vertically (through exposure to maternal skin and vaginal microbiomes), from the diet via oral fixation by the infant, and horizontally (from human interactions additional to those already mentioned).119, 120 The infant mouth becomes colonized by early oral colonizers associated with the infant's mode of delivery,121 demonstrated by finding distinct differences in the bacterial phyla predominant in the oral cavities of babies delivered vaginally compared with those delivered by Cesarean section.121 Firmicutes, Bacteroides, and Actinobacteria were found to be most abundant, respectively, in babies delivered vaginally, while Bacteroides, Proteobacteria, and Firmicutes were most abundant in babies delivered by Cesarean section. Regarding mode of delivery, vast differences in relative abundance at the genus level have been observed for most phyla, with the most marked increases being observed for Lactobacillus species in children delivered vaginally and for Petrimonas species in children delivered by Cesarean section. Lactobacillus species are common constituents of the vaginal microbiome, with strong consistency found between lactobacilli in the microbiota of the vagina and those in the oral cavity of infants delivered vaginally after a natural labor and birth.
The composition of the oral microbiome is shaped throughout life by factors including host genetics and maternal transmission, as well as by environmental factors, such as dietary habits, oral hygiene practice, medications, stress levels, and systemic factors (Figure 1).24, 129 Rather than being fixed, the composition of the oral microbiota changes throughout life, consistent with the oral cavity being a dynamic microbial environment. Eruption of primary deciduous dentition introduces new substrata for microbial colonization, thus introducing ecological shifts within the oral microbiome.130, 131 Additional changes throughout one's life, including to dietary habits, age, hygiene regimens, and behaviors (such as tobacco and alcohol use), also influence changes to the oral microbiome.132 A 2020 study of crowd‐sourced oral microbial swabs, taken from a large sample representative of the general population, showed that bacterial diversity of oral microbiomes seems to decline with age,133 an observation that has also been made in lower gastrointestinal microbiomes in relation to diet and health status.134 As children age, their oral microbiomes tend to stabilize, a feature attributed to the establishment of independent oral‐hygiene maintenance habits, acquisition of permanent dentition, and consumption of an adult diet with more‐or‐less defined dietary patterns.135
FIGURE 1.
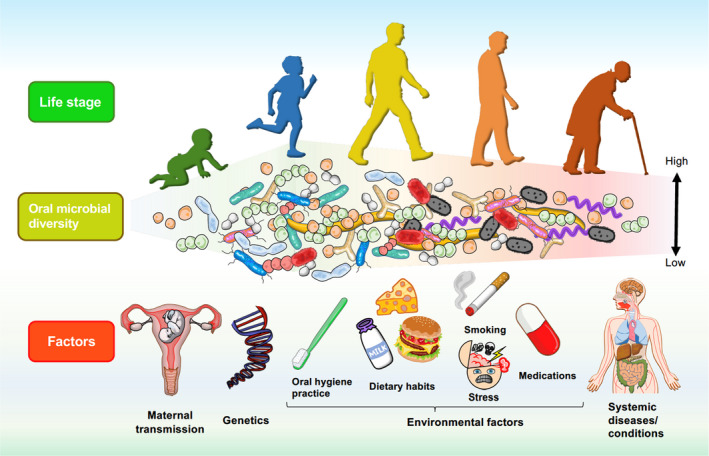
The Oral Microbiome: From First Encounters to Lifelong Encounters. Prenatal. The prenatal oral cavity is thought to be sterile until birth, with colonization occurring soon after delivery. The composition of the oral microbiota in infants has been shown to correlate with mode of delivery. However, infants share an oral microbiota similar to that of their mothers, suggesting that the infant oral microbiota may derive from hematogenous or intrauterine transmission from the mother. Detection of oral microbes of maternal origin among several intrauterine locations, as well as associations with adverse pregnancy outcomes, demonstrate the role of the maternal oral microbiome in prenatal health and suggests in utero colonization. Early life. Microbial colonization begins shortly following birth through vertical transmission from the mother, transmission from the diet, and transmission from infant‐to‐human interactions. Microbial diversity increases upon eruption of primary teeth as this process permits the expansion of microbial niches in the oral cavity. Eruption of primary teeth also results in deviation from the maternal oral microbiota. As children age, their oral microbiotas begin to stabilize. Adult life. The oral microbiota continues to be shaped throughout life by genetic and environmental factors. Environmental factors that influence the composition and function of the oral microbiome include diet, stress, oral hygiene practices, drinking alcohol, and smoking. Genetic factors are linked to conserved phylogenetic and functional microbial signatures related to development of dental caries and heritable predisposition to periodontal disease. Aging and systemic disease. Oral microbiome diversity has been shown to decrease with age. The phylogeny and functional signatures of the oral microbiome are linked to states of dental and periodontal diseases, as well as being implicated in various systemic diseases, including cardiovascular disease, cancers, and Alzheimer's disease. Systemic conditions, such as stress and diabetes, can additionally affect the oral microbiome. Figure courtesy of Dr Ryutaro Kuraji, Assistant Professor, Department of Life Science Dentistry, The Nippon Dental University, Tokyo, Japan; Department of Periodontology, The Nippon Dental University School of Life Dentistry at Tokyo, Tokyo, Japan; Visiting Assistant Professor, Department of Orofacial Sciences, School of Dentistry, University of California San Francisco, San Francisco, CA, USA
The relative importance, on the oral microbiota, of host genetics versus environmental factors has been debated.123, 135, 136, 137, 138, 139 In a large study of many sets of young and middle‐aged Swedish twins, the effect of host genetic factors on the salivary microbiota, predicted metabolic functions, and immune responses to oral bacteria were explored.108 Here, the use of young and middle‐aged twins granted an understanding of genetics versus environment, as the young twins in the study were living together and exposed to similar environmental backgrounds, whereas the middle‐aged twins shared genetic factors but had lived apart for many years. The presence and relative abundance of all species identified were influenced by environmental factors, and thus differed according to the environmental factors to which they had been exposed. Conversely, the influence of host genetic factors on these parameters were variable at the species level, mainly demonstrating strong effects on the presence and relative abundance of 27 bacterial species. The bacteria associated with host genetic factors were caries‐associated species, including S mutans, Scardovia wiggisae, and Stomatobaculum longum. Genetic factors were also associated with predicted metabolic pathways among the salivary microbiota, most specifically in relation to carbohydrate metabolism. In support of this, a different study found that heritability of microbial functions related to acid production was nearly 76%.122 Moreover, heritability of caries is nearly 50% among Swedish twins.140 In terms of immune responses, genetic factors were more strongly associated with serum antibody responses to the putative periodontal pathogen P gingivalis. Notably, strong host genetic factors existed for several taxa and microbial metabolic functions relevant to caries development in the younger cohort, while strong host genetic factors on serum antibody levels against known periodontal pathogens existed in the older age group.108
3.2. Biofilm colonization
The mucosal surfaces serve as the chief substrata available for microbial colonization in the infant oral cavity.141 The most frequently detected early colonizers of the predental oral cavity include Streptococcus, Staphylococcus, and Fusobacterium species.142, 143 Streptococci are capable of adhering to epithelial cells and are a dominant bacterial group in breast milk.144 As such, streptococcal species constitute the majority of the infant oral microbiota. Streptococcus salivarius demonstrates the highest relative abundance among newborns and shows a steady decrease after 3 months of age.119 Additional early colonizers, such as Gemella, Rothia, Granulicatella, and Haemophilus, present at 3‐6 months of age and increase in abundance with time.141, 142 In a cross‐sectional study on acquisition of the oral microbiota by infants, Mason and colleagues found that in 85% of infants, the composition of the oral microbial community was similar to that of their mothers, suggesting a significant role of the mother in introducing oral microbial communities to their children. Moreover, maternal smoking was associated with increased levels of F nucleatum and Campylobacter conscisus among infants. The eruption of deciduous teeth creates additional niches, such as nonshedding enamel surfaces, dentogingival borders, and a subgingival environment, for colonization of microbes.143, 145 The eruption of teeth leads to divergence of the infant oral microbiota from the maternal microbiota, and such changes persist among mixed and permanent dentition states.
The human oral microbiota is markedly diverse among individuals.29 However, despite dissimilar phylogeny, functional signatures among the oral microbiota are often conserved from one individual to another.13, 14 Among infants, phylogenetic divergences across individuals are observed prior to eruption of deciduous teeth143; however, functional signatures remain conserved.143 Marked expansion of oral microbial phylogenetic and functional diversity is observed with the eruption of deciduous teeth, suggesting the significance of related microbial ecosystems and parallel changes in dietary habits (solid foods) to oral microbiome diversity.143 Interestingly, salivary microbial communities from oral cavities of primary teeth‐only cohorts demonstrate greater microbial diversity than do pre‐dentate, mixed dentition, and permanent teeth cohorts. Tooth eruption introduces the relative abundance of Streptococcus, Gemella, Granulicatella, and Veillonella species.143 Expansion of microbial functions at the time of tooth eruption reflect changes in the oral ecosystem, including increased expression of genes related to adhesion, biofilm formation, membrane transport, cell mobility, secretion systems, chemotaxis, flagella assembly, and oxidative phosphorylation.143 Exfoliation of primary dentition, the presence of mixed dentition, and the emergence of permanent teeth continue to alter the oral microbiome in early life and childhood.143
Although the oral microbiome in infants evolves with advancing age, initial colonizers of the oral cavity remain as permanent colonizers that influence this colonization trajectory into adulthood.146 The significance of primary colonizers suggests that such pioneer organisms play a key role in determining development of the oral microbiome and thus the long‐term oral health status of individuals.146 Not only does the composition of one's oral microbiome locally impact oral health, it may also affect systemic health throughout life.38 This is demonstrated by the association of various disease states with the oral microbiome, including not only caries and periodontal disease but also oral, esophageal, pancreatic, and colorectal cancers, cardiovascular disease, and inflammatory bowel disease.147, 148, 149, 150, 151, 152, 153 In line with the associations suggested between oral health and systemic health later in life, disruptions in the oral microbiota early in life have been suggested to play a role in systemic disease states among children. Such conditions include tooth decay and abscess formation among young children, child weight gain, pediatric appendicitis, and pediatric inflammatory bowel diseases.154, 155, 156, 157, 158 Together, such associations between the oral microbiome and systemic health, in line with the role of the maternal oral microbiome in establishing intrauterine and infant microbial communities, encourage future studies that provide insight into the underlying mechanisms of such correlations.
3.3. Composition in adult life
After the colon, the oral microbiome represents the second most diverse microbiota in the human host, with over 700 bacterial species presently identified.35 The NIH Human Microbiome Project assessed the microbial compositions, encompassing both relative abundance and representative microbes, in 9 intraoral sites from 200 participants and found that, compared with other colonized body sites (such as the gut and the skin), the oral microbiome has the largest core set of microbes that are shared among unrelated individuals.130, 159 Moreover, the Human Microbiome Project found that the main genus among intraoral sites, at greater than 10% abundance and present in more than 75% of samples, was Streptococcus, specifically Streptococcus oralis, S mitis, and Streptococcus peroris. Other core members with greater than 1% abundance across at least 80% of samples from 1 or more sites, included those from the family Pasteurellaceae, those from the genera Gemella, Veillonella, Prevotella, Fusobacterium, Porphyromonas, Neisseria, Capnocytophaga, Corynebacterium, and Actinomyces, and those from the orders Lactobacillales and Lachnospiraceae.160 However, specifically identifying the exact composition of the oral microbiota among individuals is difficult because of the nature of the oral cavity: an open system that is regularly exposed to food, drink, exogenous microbes, air, and human contact.9
3.4. The commensal microbiota and oral health
The commensal oral microbiota is important in maintaining oral health. One such mechanism by which commensals promote oral health is via resistance to colonization by pathogens, in which commensals outcompete pathogenic species for colonization substrata, thus allowing few opportunities for integration by exogenous pathogens.161 Some oral commensals, such as S sanguinis, demonstrate direct antagonism against oral pathogens. Streptococcus sanguinis, Streptococcus cristatus, S salivarius, S mitis, Actinomyces naeslundii, and Haemophilus parainfluenzae have been shown to decrease the ability by P gingivalis to adhere to substrate.162, 163 Moreover, clinical isolates from individuals with good oral health status contained strains of Streptococcus, Actinomyces, and Bifidobacterium, all of which were shown to inhibit growth of P gingivalis.164 Lactococcus lactis, a commensal of the oral microbiota, produces nisin, a bacteriocin that has been shown to reduce oral tumorigenesis and extend the life span of mice with tumors.165, 166 In addition, nisin was recently demonstrated in vitro to mitigate pathogen‐mediated oral tumorigenesis, cancer cell migration, and cell invasion of oral squamous cell carcinoma in vitro, thus suggesting an anti‐cancer role for commensal species.167 The oral microbiome is also involved in systemic nutrient cycling in relation to nitrate metabolism.168, 169, 170 Nearly 25% of ingested nitrate is transported, via the enterosalivary circuit, to the oral cavity: here, oral microbes reduce nitrate to nitrite, which is taken into the bloodstream during digestion and converted to nitric oxide. Nitric oxide is important to cardiovascular health as it exerts vasodilation and antihypertensive effects.9, 168, 170 Such findings demonstrate the importance of the commensal oral microbiome in maintaining oral health and promoting systemic health. Dysfunction of nitric oxide signaling is associated with pulmonary hypertension, obesity, and cardiovascular disease.171 Studies have found that the use of antibacterial mouthwash can increase blood pressure as a result of its inhibitory effect on the oral microbiome.172, 173, 174 Some commensal organisms are able to buffer the acid produced by caries‐associated bacteria by raising the pH of saliva through the production of alkaline metabolic by‐products.175 The Stephan Response describes the re‐establishment of pH homeostasis following pulses of carbohydrate in the oral cavity: in this reaction, pathways generating alkali, including those producing arginine, ornithine, citrulline, glutamate, serine, threonine, and urea, are stimulated following glucose intake.176 Organisms involved in re‐establishing pH homeostasis include S gordonii, S sanguinis, and Lactobacillus casei.177
3.5. Microbial dysbiosis and oral disease
Conversely, states of oral disease are induced by dysbiosis of the oral microbiome.178, 179 Such dysbiosis is prompted by various factors, including host diet, inflammatory responses, systemic disruptions, and habits such as alcohol intake, smoking tobacco and consuming alcohol.50, 180, 181, 182, 183, 184, 185, 186 Dental caries is attributed to high dietary intake of carbohydrates, leading to increased production of acid by microbes (which reduces the buffering capabilities of saliva), reductions in salivary pH, increased production of biofilm exopolysaccharide matrix (that entraps and concentrates acids on enamel surfaces), and induction of positive‐feedback loops that encourage outgrowth of aciduric and acidogenic species, including S mutans and Lactobacillus species.8, 9, 22, 181, 187, 188, 189 Carious lesions, if left untreated, may lead to more advanced oral pathologies, such as abscess formation or pulpal involvement.190 Untreated dental caries is the most common reason for hospitalization of youth.155 Fleming and Afful found that untreated carious lesions among adolescents comprise nearly 15% of the total cases of caries among adolescents aged 12‐19 years, with untreated carious lesions primarily occurring in non‐Hispanic black youth (17.1%), followed by Hispanic (13.5%), non‐Hispanic white (11.7%), and non‐Hispanic Asian (10.5%) youth.191 Moreover, the prevalence of untreated caries decreased from 18.6% for youths living below the federal poverty line to 7% among youths in families with incomes 300% above the federal poverty line.191
Periodontal disease is caused by dysbiosis of subgingival microbial communities that adversely affects the host immune system such that it creates and maintains unmitigated inflammation in gingival and periodontal tissues, thus preventing immune subversion and tissue recovery.10 Key species in oral biofilm are recognized as etiological agents in the development of periodontal disease: these species include the red complex bacteria—T forsythia, Treponema denticola, and P gingivalis—that exhibit the capacity to drive the processes involved in periodontal disease pathogenesis by orchestrating restructure of the microbiota and promoting inflammation.178 Inflammatory and immune‐mediated processes, directly instigated by the microbes in the local plaque environment, mediate the development of gingivitis and periodontal disease.21 Early inflammatory processes, such as gingivitis, can be mitigated through oral hygiene and removal of the bacterial biofilm. Stable plaque, which can also acquire opportunistic pathogens, can cause long‐term inflammation that leads to periodontal disease.21 While the inflammation characteristics of gingivitis are mostly reversible, periodontitis results in long‐term, irreversible damage to the periodontal tissues and alveolar bone that manifests as tissue detachment from the tooth, formation of a pocket wherein pathogenic bacteria can proliferate further, and eventual tooth loss in advanced stages of the disease.192, 193, 194, 195
3.6. Oral microbiome in systemic disease
In addition to causing local (periodontal) disease, oral infection has implications in systemic disease. Three primary mechanisms linking oral infection to systemic pathology have been identified: spread of infection from the oral cavity as a result of transient bacteremia; circulation of microbial toxins; and systemic inflammation caused by adverse immunologic responses to oral microbes.196 Saliva contains 109 bacteria per milliliter, with the bacteria present in saliva being shed from the tongue, cheeks, and dental and gingival plaque.197 Between 0.75 and 1.5 L of saliva are produced per day by the oral cavity and the majority is swallowed either alone or with food or drink.198 Oral bacteria are therefore frequently transferred to the gut. Most oral bacteria are not well adapted to survive in a healthy lower gastrointestinal tract; however, increased levels of oral‐associated microbes are found in the gut of patients with inflammatory bowel disease, HIV, liver cirrhosis, and colon cancer.39, 41 These diseases are often associated with a perturbed gut microbiota, suggesting an increased ability of oral microbes to colonize ectopically in the context of immune dysregulation. For instance, Klebsiella species isolated from the oral cavities of patients with Crohn's disease have been shown to be effective colonizers of the gut of germ‐free mice.39 Oral Klebsiella species can also stimulate T‐helper‐cell induction and cause severe colonic inflammation in genetically susceptible mouse models through upregulating interferon‐inducible genes in host dendritic cells and colonic epithelial cells, demonstrating stimulation of the gut‐associated lymphoid tissue. It is hypothesized that disease states, such as chronic bowel inflammation, may enable aerotolerant species from the oral cavity to colonize distant sites more effectively.39
Periodontal disease shares many associative and causative links with systemic disease, and it increases susceptibility to systemic diseases via shared risk factors, such as by harboring pathogenic gram‐negative anaerobes in subgingival biofilms, and by fashioning the periodontium as a reservoir for pro‐inflammatory mediators.199 Subgingival biofilms have a large bacterial load and, because of their close proximity to deeper periodontal tissues, may serve as reservoirs of lipopolysaccharide and gram‐negative pathogens for systemic circulation. When present in systemic circulation, lipopolysaccharide induces pathologic outcomes in the vasculature, including production of inflammatory infiltrate within vessel walls and intravascular coagulation.200, 201 Moreover, lipopolysaccharide induces secretion of inflammatory factors that promote platelet aggregation and adhesion, formation of lipid foam cells, and accumulation of cholesterol deposits.196 Diseased periodontal tissues additionally stores pro‐inflammatory mediators associated with periodontitis.51, 183, 202 Such pro‐inflammatory molecules can directly access the circularity (cardiovascular) system and induce systemic effects, such as coagulation and thrombosis, platelet aggregation and adhesion, and accumulation of cholesterol deposits.196 As such, cardiovascular disease demonstrates a close association with oral infection.203, 204 Periodontal disease predisposes humans to cardiovascular pathologies, such as atherosclerosis and myocardial infarction, as a result of inducing increased levels of systemic pro‐inflammatory cytokines, inflammatory cells and infiltrates, and white blood cell counts.199
Rheumatoid arthritis is a chronic autoimmune disease characterized by synovial inflammation that can result in damage to articular cartilage and bone.205 Studies have revealed a link between rheumatoid arthritis and dysbiosis of the gut and oral cavity microbiomes, and a long‐term correlation between periodontal disease and rheumatoid arthritis exists.206 When rheumatoid arthritis is treated, the oral microbiome is partially restored, and vice versa.207, 208 Antibodies against P gingivalis virulence factors are elevated in patients with rheumatoid arthritis in a way that correlates with disease severity.209, 210 Recently, significant progress has been made in uncovering the mechanistic underpinning of this association. Development of rheumatoid arthritis occurs largely through a loss of tolerance for proteins that have undergone citrullination (ie, posttranslational modification of a positively charged arginine into a neutral citrulline group).205 Citrullination of arginine occurs via the action of peptidyl arginine deiminase, an enzyme found in both humans and P gingivalis.211 Infection with P gingivalis and the subsequent development of microbial dysbiosis stimulates a pro‐inflammatory immune response in the gingiva in response to the lipopolysaccharides and endotoxins produced by increased bacterial pathogens.212 This response is characterized by increased activation of pathologic T cell populations by antigen‐presenting complexes and production of pro‐inflammatory cytokines.52 Porphyromonas gingivalis also expresses peptidyl arginine deaminase, which citrullinates gingival proteins and bacterial peptides, and also undergoes autocitrullination.208 Citrullination of local proteins in the gingiva stimulates production of antibodies to both citrullinated peptidyl arginine deiminase and P gingivalis. Local protein citrullination in the gingiva promotes production of anti‐citrullinated peptidyl arginine deaminase, and antibodies against P gingivalis. Human peptidyl arginine deaminase induces the same action in joints. Porphyromonas gingivalis peptidyl arginine deaminase citrullinated α‐enolase can also generate antibodies that cross react with similar host‐citrullinated α‐enolaseyes, eliciting autoantibody activity through molecular mimicry. Ultimately, long‐standing inflammation evolves into chronic rheumatoid arthritis.
Porphyromonas gingivalis and periodontal disease have also been identified as significant risk factors for the development of amyloid beta plaques, dementia, and Alzheimer's disease.213 Gingipains, another virulence factor of P gingivalis, are implicated in Alzheimer's disease pathology. A diagnosis of Alzheimer's disease correlates with the gingipain load in the brain and also with other P gingivalis‐specific virulence factors, including P gingivalis lipopolyaccharide. Gingipains colocalize with tau protein, a protein in the brain which becomes misfolded and insoluble in patients with Alzheimer's disease. It has now been demonstrated that gingipains cleave tau protein and contribute to tau tangle formation resulting disease. Studies in mice have shown that oral P gingivalis consistently invades the brain in a gingipain‐assisted manner, and that injection of gingipains into the brain increases neuronal degeneration to a level higher than that found in controls.213
Periodontitis is also associated with increased risk of oral squamous cell carcinoma.214, 215 A recent study by Kamarajan and colleagues investigated the mechanisms by which T denticola, P gingivalis, and F nucleatum promote oral carcinogenesis. Their findings demonstrate that T denticola, P gingivalis, and F nucleatum enhance cell migration, invasiveness, stemness, and oral tumorigenesis of oral squamous cell carcinomas but have little effect on cell proliferation and apoptosis.167 Moreover, a mechanistic understanding of this was attributed to crosstalk between oral pathogens and integrin/focal adhesion kinase and toll‐like receptor/Myd88 signaling pathways. Interestingly, these interactions were reversed by treatment with the bacteriocin, nisin.167
The relationship of the oral microbiome with systemic health is bidirectional, meaning that systemic disease may also influence the oral microbiome. Type 2 diabetes mellitus, and the hyperglycemia related to this disease, are associated with increased risk of periodontitis as a result of the vascular complications of diabetes and increased levels of pro‐inflammatory molecules.216, 217, 218 Differences in the composition of subgingival plaque have been found among patients with periodontitis, with or without type 2 diabetes; specifically, patients with diabetes harbor higher proportions of Capnocytophaga species, F nucleatum, Eikenella corrodens, and A actinomycetemcomitans than patients without diabetes.219 Capnocytophaga species are saccharolytic organisms that have been shown to demonstrate increased proteolytic potential in the presence of elevated glucose levels.220 Fusobacterium nucleatum and A actinomycetemcomitans are well recognized in periodontal disease,220, 221 and F nucleatum is noted for its role in the development of anaerobic microenvironments that facilitate the outgrowth of anaerobic periodontal pathogens.222 Moreover, E corrodens is associated with chronic and aggressive periodontitis. Interestingly, increased proportions of saccharolytic bacteria, including Streptococcus, Neisseria, and Veillonella, are speculated to occur among patients with diabetes as a result of the increased glucose contents in serum and gingival crevicular fluid of such patients, which selects for the outgrowth of these organisms.219, 223 Chronic psychological stress is also classified as a risk indicator for periodontal disease and may serve as an early sign of increased risk for development of periodontal disease. Emerging evidence suggests that chronic stress and related diseases, such as depression and anxiety, may be significant factors contributing to the development of oral dysbiosis, progression of periodontal/peri‐implant disease, and inconsistent wound healing following periodontal therapy.125, 224, 225, 226 Moreover, the stress hormone, cortisol, has been shown to directly induce shifts in the composition of the oral microbial community and in its gene expression profiles in vitro, which reproduces results found for the gene expression profiles of the oral microbial community in periodontal disease and during its progression.94
4. PLAQUE FORMATION AND PERIODONTAL DISEASE
4.1. Biogeography and ecology of the oral cavity
Many distinct microenvironments exist in the oral cavity, and these are used as distinct niches for microbial colonization. The nonshedding enamel surfaces, the gingival crevice, the mucosal epithelium of the cheeks, the tongue dorsum, and tonsillar crypts, together present the oral cavity as a complex microbial environment with wide‐ranging topography.227, 228 The hard, nonshedding dental enamel surfaces present a unique opportunity for formation of mature microbial biofilm. The enamel surfaces are coated by an acquired salivary pellicle, composed of proteins, lipids, glycoproteins, and glycolipids that permit initial adherence by primary colonizers.229, 230 The oral cavity as a microbial environment is further complicated by spatial gradients created by salivary flow, nutrient availability, oxygen concentration, saliva, and gingival crevicular fluids,61, 227, 231, 232 in addition to regular environmental disturbances caused by eating and mastication, facial movement related to speaking and expression, and oral hygiene practices.227 Variation in the amount and velocity of salivary flow experienced by these sites, dictated by proximity to salivary glands, affects the composition of microbial communities harbored in distinct niches.231 Within the biofilm, chemical and nutritional spatial gradients are created as a result of microbial colonization patterns, microbial succession, microbial metabolism, and nutrient cycling. For example, colonization by Corynebacterium provides long filaments on which distinct microenvironments are created, and streptococci at the periphery of oral biofilms produce lactate, which serves as a preferred substrate for catabolism by Veillonella, Corynebacterium, and Eubacterium species.61, 233 Moreover, aerobic streptococci create an anaerobic environment via the production of carbon dioxide, lactate, and acetate, facilitating the growth of anaerobic Fusobacterium , Leptotrichia, and Capnocytophaga species.61
Many microbial subtypes are specialized to reside in either dental plaque, on the dorsum of the tongue, or on keratinized gingiva.234 For example, among Streptococcus species in the oral cavity, S salivarius and Streptococcus parasanguinis are specialized for colonization on tongue dorsal surfaces, whereas S sanguinis and S gordonii instead reside in dental plaque communities.234 Strong site preference is additionally observed among several Actinomyces species, as well as in Corynebacterium, Fusobacterium, and Prevotella species. Alternatively, some species, such as S mitis, H parainfluenzae, and Porphyromonas pasteri do not demonstrate site specialization and are found in a variety of habitats. However, most microbes in the mouth demonstrate site specialization. Site specialization is dictated by the ability of a microbe to adhere, colonize, grow, and divide at that site.227, 234
4.2. Oral biofilm colonization
Primary colonizers of dental biofilm with specific adhesion factors form weak, long‐range physicochemical interactions with the pellicle to facilitate receptor‐mediated binding.235, 236 Primary colonizers are primarily gram‐positive cocci and rods, especially Streptococcus species, such as S salivarius.237 Traditional models of microbial biofilm succession follow a sequential process in which the initial attachment of primary colonizers, namely Streptococcus, Actinomyces, Gemella, Veillonella, Rothia, and Neisseria species, bind to dental pellicle and facilitate subsequent colonization by F nucleatum, which functions as a bridging species between early and late colonizers.235 From approximately 18 hours to 4 days following colonization of dental biofilm, primary colonizers remain the predominant species among biofilm communities.238 However the proportions of anaerobes, such as Porphyromonas, Fusbobacterium, Prevotella, and Capnocytophaga, gradually increase.235 Spectral imaging and fluorescence in situ hybridization offer a different perspective of biofilm structure, in which a radially arranged consortium of bacteria forms along the annulus of filamentous Corynebacteria.61 This model shows that microbial localization is organized in such a way that position reflects functional and metabolic capacities, with anaerobic taxa toward the interior and aerobic organisms at the periphery.61 While traditional models recognize Fusobacterium as critical for connecting early and late biofilm successors, findings from this study instead found Cornybacterium as the cornerstone of biofilm development.61, 236 Once established, the new community of bacteria then begins the process of replication, maturation, and formation of a complex biofilm that can contain hundreds of species.20, 239
4.3. Microbial and immunologic involvement in periodontal disease
Biofilms become more pathologic as maturation progresses, with the proportion of pathogenic species increasing over time as the maturing biofilm selects for anaerobic species through metabolic and physical interactions.232, 240 Biofilm communities maintained at the dentogingival border stimulate immunologic responses at the gingiva, further driving dysbiosis and reinforcing inflammation.241 Sustained inflammatory responses induce tissue destruction and deepening of the gingival crevice, leading to formation of an anaerobic subgingival pocket. Homeostatic immune responses in the gingiva are disturbed following the integration of keystone pathogens, such as P gingivalis. The direct interactions of keystone pathogens with members of the microbial community, combined with their ability to elicit, yet also to subvert, host immune responses, result in the creation of a dysbiotic microbial community that thrives under inflamed conditions. The integration of periodontal pathogens, even at low abundance, is able to alter the composition of the microbiota and results in destruction of periodontal tissues by inflammatory processes that they induce.51 Via its interactions with the commensal microbiota, P gingivalis is able to promote disease by exploiting the pro‐inflammatory responses of the host, through which the dysbiotic microbiota induces the pathologic bone loss characteristic of periodontitis. Crosstalk between complement and pattern recognition receptors in response to dysbiosis further reinforces the immunomicrobial nature of periodontitis, in which these factors permit dysbiosis via inflammatory destruction of periodontal tissues and the provision of proteinaceous metabolic by‐products that further drive dysbiosis and tissue destruction.241 Microbial dysbiosis is also necessary for additional inflammatory responses characteristic of periodontal disease, including local expansion of pathologic interleukin‐17+/CD4+ T‐helper (T‐helper 17) cells.52 High T‐helper 17 cell and neutrophil populations are maintained in inflamed gingival tissues via reciprocal reinforcement, mediated primarily via the action of the inflammatory cytokines interleukin‐6, interleukin‐17, interleukin‐23, tumor necrosis factor‐alpha, interferon‐gamma, and granulocyte‐colony stimulating factor, as well as by interleukin‐8, C‐C motif chemokine ligand 2, and C‐C motif chemokine ligand 20 chemokines,241 further driving reciprocal reinforcement of microbial dysbiosis and pathologic inflammation in periodontal tissues.
The development of pathologic subgingival biofilms is largely dependent on the organisms present in homeostatic biofilm communities.240 A study by Thurnheer and colleagues sought to define microbial succession during the transition of from supragingival to subgingival plaque by recapitulating, in vitro, the environmental pressures faced during the transition of biofilm from aerophilic to microaerophilic and then to anaerobic conditions.64 At the end of the microaerophilic phase, at which point primary and secondary colonizers had been introduced, biofilm thickness doubled relative to the biofilm thickness at the end of the aerobic phase in which only primary colonizers were present in the system. Moreover, biofilm viability increased during the transition to the anaerobic phase. Interestingly, a novel role for aerobic exopolysaccharide synthesis in subgingival biofilm formation was identified, in which treatment with dextranase reduced bacterial numbers in subgingival communities,64 thus highlighting the importance of supragingival biofilms in the formation of subgingival biofilm.64 The importance of commensal species to pathogenesis is further demonstrated by the requirement for commensal microbial communities in the instigation of disease by periodontal pathogens.51, 52 Moreover, S gordonii has been shown to increase the virulence of A actinomycetemcomitans by producing lactate via streptococcal carbon metabolism.242 Pathogenic species may also enhance the outgrowth of other pathogens through symbiotic relationships. For example, P gingivalis stimulates the growth of T denticola via the production of isobutyric acid, and T denticola produces succinic acid that enhances the growth of P gingivalis.243 Such interactions demonstrate the role of microbial interactions in promoting disease via multifaceted physical and metabolic interactions that are the cornerstone of the polymicrobial synergy and dysbiosis model.178
5. DIET AND THE ORAL MICROBIOME
5.1. Paleolithic‐Neolithic transition: the introduction of grains
The introduction of agriculture ~ 10,000 years ago at the Mesolithic‐Neolithic transition drastically changed dietary composition, such that grain‐derived carbohydrates emerged as a main staple of the human diet.244 Increased consumption of cereal grains at this transition was accompanied by the emergence of dental and periodontal diseases, which is further reflected by distinct shifts in the oral microbiota.245, 246, 247 Little evidence of dental and periodontal disease exists for pre‐Neolithic hunter‐gatherer societies.245 Changes in the proportions and representation of oral microbial species at the Mesolithic‐Neolithic transition are evident in samples of preserved dental calculus from the Mesolithic that demonstrate a compositionally distinct microbial profile from samples of dental calculus collected from the Neolithic, Bronze Age, and Medieval periods, and modern times.24 Interestingly, the microbial profiles of plaque samples from the Neolithic onward largely cluster together in terms of proportions and species of microbes present, despite additional dietary changes over time: compared with the proportions and species of oral microbes present in pre‐agriculture plaque samples, such samples from farming populations of the Neolithic onward are dominated by caries‐associated species, such as those from the family Veillonellaceae, as well as taxa associated with periodontal disease, including P gingivalis, T forsythia, and T denticola.24 The distinction between the microbial profiles found in Mesolithic and post‐Mesolithic eras suggests that increased carbohydrate consumption introduced profound effects on the composition of the oral microbiota that have been maintained over time.24 Such changes in the pre‐Neolithic oral microbiota have been further exacerbated by the evolution of food‐processing techniques that emerged during the Industrial Revolution, ~ 200 years ago, consistent with the invention of mechanical steel roller mills in the 19th century, in which the nutritive properties of grains were significantly altered by the isolation of starch‐rich components (flour) and removal of the outer bran layer that provides micronutrients and dietary fiber.248, 249, 250 Moreover, processed carbohydrate consumption has been associated with increased incidence of periodontal disease.8, 24, 26, 181, 188, 251 Processing of grains further impacted several additional aspects of diet, including total nutritive content, starch bioavailability, and texture. Current World Health Organization guidelines recommend that starch‐derived carbohydrates constitute > 55% of daily energy intake. This encompasses cereal grains, starchy tubers, whole grains, legumes, etc., in which complex carbohydrate consumption is favored widely over processed carbohydrate consumption in regard to health. In the United States, carbohydrates constitute > 50% of total energy intake, with refined cereal grains representing > 85% of total cereal grains consumed. Moreover, refined sugars constitute approximately 20% of caloric intake.252, 253, 254
5.2. Ancestral diets and oral health
Dietary staples and processing techniques that emerged during the Neolithic and Industrial Revolution periods significantly changed several central components of the human diet, including total macronutrient content, glycemic load, fatty acid composition, sodium and potassium levels, micronutrient levels, dietary pH, and fiber content.253, 255, 256 The pre‐agricultural diet was largely limited to nonprocessed, wild fruits and vegetables, legumes, nuts, and wild animals.253 Staples of the modern diet, including dairy products, refined carbohydrates, vegetable oils, and alcohol, that were not nutritional features of the pre‐agricultural diet, now constitute ~ 70% of the total daily energy intake by people in the US.255 The transition from hunter‐gatherer lifestyles to farming societies is associated with a decline in oral health as well as several additional disease states, including (but not limited to) cardiovascular disease, inflammatory bowel diseases, rheumatic diseases, many cancers, and obesity.24, 245, 253, 257 Modern day hunter‐gatherer societies, although scarce, offer a unique opportunity to observe the impact of ancestral diets on human health in the absence of Westernized dietary influences. An oral health study among the Hadza hunter‐gatherers of Tanzania offered a rare opportunity to study the impact of ancestral diets among “bush dwellers” and increasingly Westernized diets on the oral health status among “villagers.”258 The findings from this study were consistent with the hypothesis that agricultural societies demonstrate poorer dental health than hunter‐gatherer societies: women who lived in village settings and consumed a primarily agricultural diet had significantly greater incidences of caries and periodontal disease than those who lived in a bush setting and consumed a wild‐food diet including legumes, wild game, berries, and uncultivated tubers.24, 258, 259 However, men living in the bush did not uphold the same oral health status as their female counterparts, predictively because of cultural influences, including high consumption of honey and tobacco use.258
5.3. Dietary texture and oral health
The low rates of periodontitis among bush‐dwelling women compared with women who lived in village settings was suggested to result not only from compositional differences in their diets, but also from textural differences.258 It was hypothesized that consumption of fibrous, uncultivated tubers by bush dwellers offered increased masticatory challenge that was effective in helping to interrupt plaque formation by mechanically abrading dentogingival surfaces.258, 260 The benefits of dietary texture on oral health are more commonly recognized among animal studies.23, 261, 262Wild animals have an oral health status superior to that of animals living in captivity because of the absence of masticatory challenge among foods given to domestic and captive animals.23, 261, 263, 264, 265 Interestingly, periodontal disease among captive animals was reversed when their diets were increased in texture to provide masticatory challenge to dentogingival surfaces and alleviate plaque formation.264, 265 A novel role for dietary texture/hardness in periodontal immunity was recently demonstrated by Dutzan et al.53 Mice consuming soft or hard diets demonstrated differential immune responses in gingival tissues: interleukin‐17‐producing T cells accumulated (in an interleukin‐6‐dependent manner) in periodontal tissue of mice fed a hard diet.53 In addition, mechanical damage induced by consumption of foods with increased texture led to elevated expression of epithelial defensins and neutrophil chemoattractants and promoted increased neutrophil counts in gingival tissues.53 Although it was found that increased mechanical damage prompted periodontal bone loss, consistent with destruction of hard tissue in periodontal disease, it was speculated that mechanically induced T‐helper 17 cells may elicit barrier protection at the gingiva.53, 54 These findings were further elucidated by an additional study in which the periodontal T‐helper 17 populations governing health and disease were differentiated: homeostatic T‐helper 17 cells were found to be induced by mechanical stimuli in a microbiome‐independent manner, whereas periodontitis‐associated T‐helper 17 cells were found to accumulate in a microbiome‐dependent manner and to require both interleukin‐6 and interleukin‐23.52, 54 Cumulatively, these findings offer a greater mechanistic understanding of how dietary nutrition and texture alike alter the oral microbiota and immune response at the gingiva.
5.4. Dietary fiber and oral health
Total macronutrient shifts as a result of increased grain consumption at the Paleolithic‐Neolithic transition are well recognized.253 However, the nutritional value of grains is dictated by multiple factors, including grain particle size (ie, degree of processing) and preparation method (ie, cooking).250, 266 Such processing and preparation methods subsequently affect food texture and the bioavailability of inner starch components.248, 250, 267, 268 Processing techniques seek to remove the outer, fibrous bran layer of grains to isolate the inner starch‐rich endosperm components.248, 250, 269 Removal of this outer bran layer not only depletes fiber content but decreases dietary texture due to a loss of grain integrity.249, 268, 269, 270, 271, 272 The role of dietary fiber consumption in oral health status has been recognized predominantly from a correlative perspective.258, 259, 273, 274 In a study of fiber consumption and periodontal disease incidence among US adults, it was determined that consumption of whole grains significantly decreased periodontal disease incidence among adults aged 30 years and older.274 Conversely, low whole grain consumption was associated with increased incidence of periodontitis.274 Consumption of whole grains and fruit has also been shown to reduce periodontal disease progression and improve periodontal disease status among high‐risk subjects.273, 275 Lowered consumption of carbohydrates has additionally been determined to decrease markers of gingival and periodontal inflammation.26, 276 Such correlative studies have offered a clinical perspective regarding the role of dietary carbohydrates and/or dietary fiber on worsened and improved periodontal disease status, respectively. The impact of increased carbohydrate consumption and increased fiber consumption, however, has yet to be understood from mechanistic perspectives. Periodontal disease is a multifaceted disease state involving intricate crosstalk between the oral microbiome and the periodontal and systemic immune systems.10 While the impact of diet on the clinical parameters of periodontal disease has been noted, the impact of diet on microbial and immunologic parameters has yet to be fully delineated. Despite these limitations, a handful of studies have identified changes to the oral microbiota and/or periodontal immunity in response to dietary interventions. For example, a study by Woelber and Tennert26 found that reduced consumption of processed carbohydrate significantly altered the proportions of microbes in supragingival plaque and salivary samples, such that the numbers of S mitis, Granulicatella adiacens, Actinomyces species, and Fusobacterium species were reduced.277 Baumgartner et al251 additionally identified significant shifts in the proportions of microbes in dental plaque in response to “stone‐age” dietary interventions in the absence of oral hygiene, in which counts of the periodontal pathogens T forsythia and A actinomycetemcomitans, as well as Streptococcus species, from tongue samples were reduced following 4 weeks of dietary intervention.251 Although the amount of plaque on teeth was higher at the experiment endpoint than at baseline, decreased bleeding on probing and periodontal pocket depths were noted and, despite abstinence from oral hygiene, gingival inflammation did not increase.251 The effect of dietary texture on oral health status is a relatively unexplored area of research; however, recent studies have offered insight to this emerging field.273, 274, 275, 278 A study by Sedghi and colleagues278 found that the addition of dietary fiber as a mechanical influence in the oral cavity prompted significant changes to the murine dentogingival microbiota.278 Interestingly, this study found that the most significant changes influenced by dietary fiber occurred in the presence of sugar compared with sugar alone.278 When combined with sucrose, fiber significantly increased species richness, compared with a decrease in the Firmicutes to Bacteroides ratio prompted by a sucrose‐only diet.278 Moreover, the addition of fiber led to a decrease in Corynebacterium species, which are recognized as a cornerstone of oral biofilm formation.61, 278 These findings have implications for altering microbial dysbiosis elicited by modern‐day diets and, as such, the role of fiber on the oral microbiome warrants further investigation.
6. FOOD METABOLISM IN THE ORAL CAVITY
6.1. Digestion and macromolecule metabolism in the oral cavity
Digestion of starch is initiated in the oral cavity through the action of salivary alpha‐amylase that catalyzes the hydrolysis of starch by cleaving α‐1,4‐glycosidic linkages to yield smaller saccharide moieties, such as maltose, maltotriose, and alpha‐limit dextrins,279, 280 which can be utilized by early biofilm colonizers as well as by some periodontal pathogens.8, 281, 282 The Agricultural (~10,000 years ago) and Industrial (~200 years ago) Revolutions resulted in historic boosts of sugar intake, and the rates of sugar intake have been rising ever since.17 A 2014 study estimated that added sugar consumption among adults in the US has increased by more than 30% over the last 3 decades alone.283 The incidence of dental caries has increased in parallel with rising sugar consumption.284 Excess carbohydrate, particularly processed simple sugars, promotes the growth of rapidly growing saccharolytic microbes whose growth advantage permits out‐competition of slower‐growing species with alternative nutritional requirements.285 Saccharolytic bacteria, including Streptococcus, Actinomyces, and Veillonella species, degrade carbohydrates via the Embden‐Meyerhof‐Parnas pathways to produce acidic by‐products, including lactate, acetate, ethanol, and formate, which cause demineralization of dental enamel and the development of carious lesions.22, 188, 286 A diet rich in carbohydrate intake promotes bacterial acidogenicity and acidurance by increasing the permeability of the cell membrane to protons, induction of H+‐ATPase activity, and stimulation of metabolic pathways involved in acid neutralization and alkalination. Such processes encourage the outgrowth of acidogenic bacteria, such as S mutans, lactobacilli, and bifidobacteria that enhance the carcinogenicity of dental plaque.8 Pathways in which sugar is metabolized are also shared among some periodontal pathogens, such as Fusobacterium and Prevotella, which can likewise cause acidification.8
Carbohydrate use by S mutans is a well‐recognized feature in the pathogenicity of dental caries.22, 188, 286 Expansion of the genetic repertoire of S mutans to include increased uptake of carbohydrate and stimulation of genes involved in carbohydrate metabolism appears to have been a key evolutionary turning point, as population genomics studies predict that the S mutans population started growing exponentially 10,000 years ago, around the advent of human agriculture.24, 25 Streptococcus mutans uses dietary sucrose to produce exopolysaccharide matrix via glucosyltransferases and fructosyltransferases.181 The exopolysaccharide matrix and related glucan constituents synthesized by glucosyltransferases provide binding sites for further bacterial colonization and contribute to the resilience, bulk, and physical integrity of biofilm communities.188 Furthermore, exopolysaccharides create chemical and metabolic gradients within biofilms that affect chemical and nutrient cycling as well as diffusion, thus altering the microenvironment and pH within biofilms consortium.187, 287, 288 For transporting mono‐ and disaccharides, S mutans primarily uses the phosphoenolpyruvate:sugar phosphotransferase system. While readily metabolizable mono‐ and disaccharides are preferred, S mutans also transports oligosaccharides through ATP‐binding cassette transporters, such as the multiple sugar metabolism and maltose transport complex systems.289 Carbohydrate catabolite repression is another important mechanism by which bacteria utilize carbohydrates, and this may offer increased fitness.290, 291 Carbohydrate catabolite repression enables metabolism of nonpreferred carbohydrates if preferred sources, such as glucose, are not available. Carbohydrate catabolite repression functions through a complex regulatory framework that relies on feedback from the build up of glycolytic intermediates and through regulating expression of glycolytic genes by EII permeases. This adaptation enables S mutans to optimize extraction of energy from carbohydrate types that pass through the oral cavity. The carbohydrate catabolite repression gene, catabolite control protein A (ccpA), is located upstream of the S mutans bacteriocin production regulon and has been shown to regulate the expression of this by controlling the expression of S mutans competence stimulating peptide.292
During the development of periodontal disease, protein‐rich and neutral‐alkaline environmental pressures promote the outgrowth of periodontal pathogens. Inflammatory processes promote the outgrowth of inflammophilic bacteria via the production of proteinaceous substrates resulting from tissue destruction.8, 293 Deepening of the gingival crevice and increased production of gingival crevicular fluid increases the relative abundance of protein‐degrading bacteria.8 Proteins can be metabolized into peptides and amino acids, such as aspartate, serine, and cysteine, by both host proteases and peptidases. Amino acids are fermented to produce short‐chain fatty acids, such as propionate, butyrate, succinate, acetate, and formate.8 Amino acid fermentation neutralizes acidic environments, making the environment more favorable for outgrowth of additional periodontitis‐associated pathogens.8 Some bacteria, such as Prevotella intermedia, exist as both proteolytic and saccharolytic, depending on environmental pressures.294
6.2. Influence of microbiota on taste perception and dietary preference
There has been extensive research on how nutrition and diet shape the microbiota, but comparably little research on how the microbiota influences dietary behaviors and preferences. Recently insights have been made into relationships between the oral microbiota and taste perception. Taste influences eating behaviors and therefore has major implications in overall health. There is well‐known host genetic variability in taste perception that leads to different thresholds of sweet, sour, salty, and bitter taste between individuals. The most commonly studied variable genetic taste element is the taste 2 receptor member 38 (TAS2R38) gene, which encodes a bitter taste receptor. Three major haplotypes of this gene exist: proline‐alanine‐valine/proline‐alanine‐valine (PAV/PAV), or “supertasters”; proline‐alanine‐valine/alanine‐valine‐isoleucine (PAV/AVI), or “medium tasters”; and alanine‐valine‐isoleucine/alanine‐valine‐isoleucine (AVI/AVI), or “nontasters.” The phenotypes are generally determined by a taste test of perceived bitterness of the compound 6‐n‐propylthiouracil. A recent study of healthy subjects defined as nontasters or supertasters (medium tasters were excluded) examined the potential link between taste and the tongue microbiota.27 Lower thresholds for sweet, sour, salty, and bitter tastes were observed for supertasters than for tasters, and the density of fungiform papillae was higher in supertasters. Through 16S ribosomal RNA gene profiling, the study also found higher relative abundances of Actinomyces, Oribacterium, Campylobacter, Solobacterium, and Catonella species in the tongue dorsum microbiotas of supertasters than in that of nontasters. A follow‐up study by the same group showed preliminary associations between some bacterial taxa on the tongue dorsum with gustatory functions. Additional associations were made between the bacterial taxa on the tongue dorsum and vegetable‐rich diets (such as Prevotella) or diets rich in proteins and fats (such as Clostridia).295 These studies present the possibility that microbes may influence dietary preferences. Although conclusive mechanistic evidence in support of this hypothesis remains limited, it is plausible that microbes may modulate the expression levels of taste receptors in the mouth or that they may stimulate or inhibit these types of receptors through metabolite secretion. Future studies using functional microbiome analysis techniques can elucidate how diet‐microbiota‐host genetic interactions influence overeating, unhealthy eating, and the dental and periodontal implications of these behaviors.
6.3. SARS‐CoV‐2 Infection and Dysgeusia
The coronavirus disease 2019 (COVID‐19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is associated with many symptoms including sore throat, dry cough, muscle aches, and chills, as well as enteric symptoms including diarrhea, nausea, and vomiting.296 While many of these symptoms are shared with the common cold and influenza, a symptom more specific to COVID‐19 infection is loss of taste and smell, in which loss of taste, or dysgeusia, occurs in approximately 88.8% of cases.297, 298 Such chemosensory deficits experienced in COVID‐19 are often short‐lived, with most symptoms resolving in 7‐10 days, with sense of smell often returning slightly earlier than taste.299 Angiotensin‐converting enzyme 2, the receptor that mediates entry of SARS‐CoV‐2 into cells, is expressed in the oral mucosa and is enriched in human tongue cells among both gustatory and nongustatory tongue epithelium.300, 301 While the specific underlying mechanism of dysgeusia in COVID‐19 is not completely known, various hypotheses have emerged as new information continues to unfold. A neurologic mechanism has been suggested, in which direct damage to nonneural cells in the olfactory epithelium via SARS‐CoV‐2 may also result in loss of taste.302 Another hypothesis includes the direct disturbance of angiotensin‐converting enzyme 2‐expressing cells in the taste buds and peripheral taste neurosensory chemoreceptors,303 or damage to cranial nerves involved in gustatory sensation, including cranial nerves 7, 9, and 10. Involvement of inflammatory response pathways has also been suggested, in which the angiotensin‐converting enzyme 2 receptors lining the oral mucosa are used by the virus to enter epithelial cells.304, 305 Entry by SARS‐CoV‐2 into the oral mucosa via angiotensin‐converting enzyme 2 receptors may elicit an inflammatory response.306 Tissue hypoxia and tissue injury resulting from anemia and/or impaired oxygen transport exists as another potential mechanism by which COVID‐19 alters taste.307 Hypozincemia is another possible mechanism, in which zinc chelation via immune mechanisms and related molecules may increase with the inflammatory processes implicated in COVID‐19. Such changes may induce localized changes in zinc homeostasis and hypozincemia of oral gustatory cells, thus resulting in taste disturbances that are also common in zinc deficiency.298, 308
The significance of the oral microbiome in coinfection with SARS‐CoV‐2 is a growing area of study. COVID‐19 has also been associated with coinfection by additional viruses, fungi, and bacteria, some of which are derived from the oral cavity. Poor oral hygiene, cough, abnormal inhalation, and in some cases mechanical ventilation are suggested to be mechanisms by which oral microorganisms may gain entry to the lower respiratory tract. For example, metagenomic sequencing of bronchoalveolar lavage fluid among patients with COVID‐19 demonstrated an increased presence of oral and upper‐respiratory‐tract bacteria.309 Opportunistic oral bacteria, including Veillonella parvula, Capnocytophaga gingivalis, Leptotrichia buccalis, and Prevotella melaninogenia were overrepresented in bronchoalveolar lavage fluid of patients with COVID‐19, suggesting that the oral cavity may serve as a reservoir of bacteria implicated in coinfection with SARS‐CoV‐2 in COVID‐19.310 Such findings implicate a possible role for co‐infection by the oral microbiota and SARS‐CoV‐2 in the lungs of COVID‐19 patients,311 and highlight the importance of the oral microbiota in COVID‐19 infection and thus justify future studies in this area of research.
7. ORAL HYGIENE AND THERAPIES: IMPLEMENTING ORAL MICROBIOME LEARNINGS IN DENTAL MEDICINE
7.1. Toothbrushes
Toothbrushes have been used since the 19th century for a variety of oral health‐related purposes.312 Toothbrushing is the cornerstone of most oral hygiene routines in developed countries. The American Dental Association recommends brushing one's teeth twice a day for 2 minutes with a soft‐bristled toothbrush, along with flossing between teeth once per day. Toothbrushing mechanically displaces oral bacteria and plaque from the dentition and gingiva in order to limit long‐term damage from their metabolic by‐products. The American Dental Association recommends replacing one's toothbrush every 3‐4 months, or once bristles become worn out.32 Toothbrushes are consistently exposed to microbes from the mouth and from the environment in which they are stored. A previous review highlights that during normal use, toothbrushes become heavily contaminated from sources including the oral cavity, environment, hands, aerosol, and storage containers.28 Previous studies have demonstrated that toothbrushes can inoculate the oral cavity with microorganisms, but there has been little research on how the microbes harbored on the toothbrush influence the oral microbiota as a whole.55 A 2020 study in which the microbial diversity in toothbrushes and oral cavities of 20 participants was examined using high‐throughput sequencing, found that the oral and toothbrush microbiota shared similar Shannon Indices and that certain species were enriched in toothbrushes, including Acinetobacter baumannii, Staphylococcus aureus, and Candida albicans.33 Such species have been implicated in infectious disease, neurodegenerative disease, cardiovascular disease, and cancer. However, it is still unclear how readily these pathogens and other toothbrush bacteria can seed and survive in the oral cavity; nonetheless, this highlights that toothbrushes may be reservoirs of bacteria, which are reintroduced to the host on a daily basis.
7.2. Toothpaste
The effects of toothpaste on the oral microbiota have been investigated more thoroughly than the effects of toothbrushes. In the same study as previously described,33 the researchers also compared the oral and toothbrush microbiotas of participants who used either traditional Chinese medicine toothpaste or antibacterial toothpaste. While both types of toothpaste effectively reduced the numbers of a selection of pathogenic bacteria, they also suppressed oral S salivarius and L salivarius, both of which are considered beneficial. The toothpastes reduced bacterial numbers to differing degrees: ingredients in the Chinese toothpaste, such as honeysuckle, pseudo‐ginseng, and mint, which replace antibacterial ingredients like sodium fluoride, permitted growth of higher numbers of bacteria. Other toothpaste formulations have been developed formulated to shift oral ecology, rather than to completely decimate the microbiota. ZendiumTM contains proteins designed to promote oral health and limit disease‐associated organisms. Amyloglucosidase, glucose oxidase, and lactoperoxidase are added to promote production of hydrogen peroxide and hypothiocyanite, both of which exert antibacterial activity against plaque‐forming species. Zendium also contains lysozyme, lactoferrin, and IgG. A randomized clinical study comparing Zendium toothpaste with a control toothpaste containing fluoride showed that brushing with Zendium increased the abundance of organisms associated with gum health and decreased the numbers of those associated with periodontal disease. These results prove an important concept, namely that oral ecology can be manipulated via the introduction of biochemical mediators. Future studies in this area will probe the metabolic output of these restructured communities, providing a more complete picture of the potential benefits shown in these preliminary results.34
7.3. Mouthwash: old and new theories
Antimicrobial mouthwashes have long been used as antiseptics because of their ability to kill pathogens, such as methicillin‐resistant S aureus, C albicans, S mutans, P gingivalis, and some viruses.37 These treatments are nonspecific and can wipe out large swathes of the commensal oral microbiota. Mouthwashes often claim to kill “99% of germs” in the mouth, a title that the modern understanding of oral microbes in health no longer condones due to improved understanding of the crucial role played by the oral microbiome in initiating food digestion and maintaining oral and systemic health.35 Most mouthwashes contain alcohol, hydrogen peroxide, or chlorhexidine, or a mixture of these active ingredients. Alcohol, although widely used in high concentrations as an antiseptic agent for surfaces, in most mouthwashes actually has little capacity to kill microbes because the amount of ethanol present is 25% or less. Furthermore, alcohol‐containing mouthwashes can exacerbate bad breath, a common driver of mouthwash use, by reducing production of saliva in the mouth.17, 71 Chlorhexidine digluconate is one of the most frequently used agents to kill bacteria, especially in dental clinics, and has potent broad‐spectrum antiseptic properties against gram‐positive and gram‐negative bacteria. It is also highly adherent to oral surfaces, making it an effective plaque‐disrupting agent.36
The antimicrobial properties of mouthwashing agents have been well studied. Until recently, however, their effects on the function of oral microbial communities have been scarcely investigated. A recent 2020 study assessed the effects of repeated use of chlorhexidine mouthwash on the salivary microbiome and plasma biomarkers in 36 orally healthy patients: shifts in the proportions of oral microbes were found after 7 days of use of chlorhexidine mouthwash, most notably a greater abundance of species from the phyla Firmicutes and Proteobacteria, in conjunction with reductions of species from the genus Bacteroides, the phylum Saccharibacteria (formerly TM7), Candidate Division SR1, and Fusobacteria. These changes were accompanied by significant reductions in salivary pH and saliva buffering capacity and increased salivary lactate. Reduced salivary pH, increased salivary lactate, and reduction in the buffering capacity of saliva are associated with tooth decay and periodontal diseases. Chlorhexidine mouthwash also decreased the concentrations of nitrite in the mouth and plasma.36 Oral nitrite can affect the concentrations of nitric oxide (which is vasodilatory) present in the bloodstream.173 Preliminary research points to a reduction of systolic blood pressure by the production of nitrite from oral microbiota through this pathway.313 The oral nitrate‐reducing capacity of bacteria in the mouth was also significantly reduced in the chlorhexidine mouthwash group compared with the control group. Nitrite forms in the mouth by bacterial reduction of nitrate obtained either from the diet or endogenously (from the synthesis of nitric oxide).314 Species of the genera Veillonella and Actinomyces are thought to be the major reducers of nitrate in the oral cavity.315 In the chlorhexidine mouthwash group, the abundance of Actinomyces decreased significantly, possibly contributing to the lower reductive capacity observed. While more studies are needed to flesh out the contributions of oral bacteria to nitric oxide‐associated clinical parameters, preliminary evidence suggests meaningful influences that should be considered in the use of chlorhexidine‐containing and other antimicrobial mouthwashes.
7.4. Probiotics
Oral probiotics are a method of direct introduction of particular species into the oral microbiome to manipulate the pre‐existing microbiota or elicit a biochemical and/or physiological response. A few different approaches have been employed to generate anti‐caries probiotic formulations. One primary target is to harness the alkalogenicity of certain species to combat acidification and tooth demineralization by pathogenic organisms. A major source of alkalinity in the oral cavity is the breakdown of arginine to ornithine, carbon dioxide, and 2 molecules of basic ammonia. Administration of arginine alone has been shown to be effective at inhibiting the initiation of dental caries, and arginine has recently been included as a supplement in some toothpastes.315 Many common oral microbes maintain this metabolic capacity. Microbial metabolism of arginine is accomplished primarily by the arginine deaminase system, which consists minimally of 3 different enzymes. Researchers have studied the effects of introducing probiotic strains of bacteria with superior arginolytic capacity. Lactobacillus brevis is known to produce high levels of arginine deaminase, and in a study in which L brevis‐containing lozenges were administered to 21 patients with periodontal disease, the treatment led to complete amelioration of all clinical parameters analyzed in all patients.42 It is now recognized that the arginine deaminase system is important in more than just pH modulation—it also influences the inflammatory immune response of the host to periodontal pathogens, which can damage host tissue. In the study above, of L brevis, activity of the inflammatory markers prostaglandin E2, interferon‐gamma, and Matrix metalloproteinase were substantially decreased in treated patients. As arginine is also a substrate for nitric oxide synthase, a mediator of multiple inflammatory pathways, it is hypothesized that increasing the competition for this substrate using the arginine deaminase system decreases nitric oxide production, thus reducing inflammatory activation.42
Another angle that has been taken in designing probiotics is the targeting of specific pathogenic bacteria that are known potentiators of dental caries and periodontal disease. Streptococcus mutans is a key contributor to dental biofilms associated with the formation of dental caries,44 and plays a major role in the development and establishment of extracellular polysaccharide matrices, which increase the virulence of the biofilm and facilitate persistence of an acidic environment in the mouth.316 Certain bacteria obtained from plaque of caries‐free persons have exhibited inhibitory properties toward pathogens, such as S mutans. In addition to strongly expressing the arginine deaminase pathway, the novel strain of Streptococcus, called A12, produced hydrogen peroxide in large quantities along with specific virulence factors that could arrest S mutans growth and interrupt S mutans signaling pathways.45 A12 also demonstrated the capacity to prevent S mutans from killing other arginine deiminase system (ADS) producers by degrading the S mutans bacteriocin‐inducing peptide signal— accomplished through secreting a protease that blocked the competence‐stimulating peptide signaling system. This protease, termed challisin, was first discovered in a certain S gordonii strain and was thought to be specific to this organism.317 These results thus demonstrate the existence of organisms with convincing anti‐caries phenotypes that may be leveraged to establish healthy oral microbiomes. However, these qualities are very specific at the species and strain levels, and tight quality assurance will be needed for incorporation into probiotic formulations.
Throughout the last decade, human microbiome research has focused on the gut and its connection with various diseases. Interest has now shifted to other areas of the human body with an increased focus on the oral microbiome. Hippocrates’ famous quote, “All disease begins in the gut” still holds true today, but we must remember that everything in the gut must pass through the oral cavity. This has led to an increased interest in the oral microbiome and its relationship to health and disease. Advancements in sequencing technologies combined with longitudinal multimodal approaches have increased our understanding of the role of the oral microbiome in health and in systemic diseases. Evidence connecting the relationship between dysbiosis of the oral microbiome and various systemic diseases has raised awareness of the importance of the oral microbiome in regulating health. Full knowledge of the role of the oral microbiome in health, and methods to modulate these ecosystems, can provide an alternative approach to disease prevention and treatment.
ACKNOWLEDGMENTS
This work was supported by the NIH Research Project Grant Program [R01DE025225] to YK, AAP Sunstar Innovation Grant to YK, Berkelhammer Basic Science Fund to YK, and the Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant [5T32DE007306‐24] to LS.
Sedghi L, DiMassa V, Harrington A, Lynch SV, Kapila YL. The oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontol 2000. 2021;87:107–131. 10.1111/prd.12393
Sedghi and DiMassa are co‐first authors with equal contribution
Funding information
This work was supported by funding from NIH R01 DE025225 grant and Berkelhammer Basic Science Funds to YLK.
REFERENCES
- 1.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24(4):392‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57(1):369‐394. [DOI] [PubMed] [Google Scholar]
- 3.Bikel S, Valdez‐Lara A, Cornejo‐Granados F, et al. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems‐level understanding of human microbiome. Comput Struct Biotechnol J. 2015;13:390‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petruschke H, Anders J, Stadler PF, Jehmlich N, von Bergen M. Enrichment and identification of small proteins in a simplified human gut microbiome. J Proteomics. 2020;213:103604. [DOI] [PubMed] [Google Scholar]
- 5.Vernocchi P, Del Chierico F, Putignani L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol. 2016;7:1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gest H. The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the royal society. Notes Rec R Soc Lond. 2004;58(2):187‐201. [DOI] [PubMed] [Google Scholar]
- 7.Holt SC, Ebersole JLPorphyromonas Gingivalis, Treponema Denticola, and Tannerella Forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72‐122. 10.1111/j.1600-0757.2005.00113.x [DOI] [PubMed] [Google Scholar]
- 8.Takahashi N. Oral Microbiome metabolism: from “who are they?” To “what are they doing?”. J Dent Res 2015;94(12):1628‐1637. 10.1177/0022034515606045 [DOI] [PubMed] [Google Scholar]
- 9.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137‐143. 10.1016/j.phrs.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 10.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15(1):30‐44. 10.1038/nri3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajishengallis G, Darveau RP, Curtis MA. The keystone‐pathogen hypothesis. Nat Rev Microbiol. 2012;10(10):717‐725. 10.1038/nrmicro2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21(3):172‐183. 10.1016/j.molmed.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yost S, Duran‐Pinedo AE, Teles R, Krishnan K, Frias‐Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015;7(1):1‐19. 10.1186/s13073-015-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 2014;5(2): 10.1128/mBio.01012-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilian M, Chapple ILC, Hannig M, et al. The oral microbiome – an update for oral healthcare professionals. Br Dent J. 2016;221(10):657‐666. 10.1038/sj.bdj.2016.865 [DOI] [PubMed] [Google Scholar]
- 16.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721‐5732. 10.1128/JCM.43.11.5721-5732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker JL, Edlund A. Exploiting the oral microbiome to prevent tooth decay: has evolution already provided the best tools? Front Microbiol. 2019;9:3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Hemme C, Beleno J, et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 2018;12(5):1210‐1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long SS, Swenson R. Determinants of the developing oral flora in normal newborns. Appl Environ Microbiol. 1976;32(4):494‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2(13):1599‐1607. [DOI] [PubMed] [Google Scholar]
- 21.Kriebel K, Hieke C, Müller‐Hilke B, Nakata M, Kreikemeyer B. Oral biofilms from symbiotic to pathogenic interactions and associated disease‐connection of periodontitis and rheumatic arthritis by peptidylarginine deiminase. Front Microbiol. 2018;9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moye ZD, Zeng L, Burne RA. Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans . J Oral Microbiol. 2014;6(1):24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler CJ, Malik R, Browne GV, Norris JM. Diet may influence the oral microbiome composition in cats. Microbiome. 2016;4: 1‐9. 10.1186/s40168-016-0169-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adler CJ, Dobney K, Weyrich LS, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the neolithic and industrial revolutions. Nat Genet. 2013;45(4):450‐455. 10.1038/ng.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornejo OE, Lefébure T, Pavinski Bitar PD, et al. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans . Mol Biol Evol. 2013;30(4):881‐893. 10.1093/molbev/mss278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woelber JP, Tennert C. Chapter 13: diet and periodontal diseases. Impact Nutr. Diet Oral Health 2020;28:125‐133. 10.1159/000455380 [DOI] [PubMed] [Google Scholar]
- 27.Cattaneo C, Gargari G, Koirala R, et al. New insights into the relationship between taste perception and oral microbiota composition. Sci Rep. 2019;9(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazelle MR, Munro CL. Toothbrush contamination: a review of the literature. Nurs Res Pract. 2012;2012:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall MW, Singh N, Ng KF, et al. Inter‐personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes. 2017;3(1):1‐7. 10.1038/s41522-016-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tribble GD, Angelov N, Weltman R, et al. Frequency of tongue cleaning impacts the human tongue microbiome composition and enterosalivary circulation of nitrate. Front Cell Infect Microbiol. 2019;9: 10.3389/fcimb.2019.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koopman JE, van der Kaaij NCW, Buijs MJ, et al. The effect of fixed orthodontic appliances and fluoride mouthwash on the oral microbiome of adolescents – a randomized controlled clinical trial. PLoS One. 2015;10(9):e0137318. 10.1371/journal.pone.0137318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terezhalmy GT, Bsoul SA, Bartizek RD, Biesbrock AR. Plaque removal efficacy of a prototype manual toothbrush versus an ADA reference manual toothbrush with and without dental floss. J Contemp Dent Pr. 2005;6(3):1‐13. [PubMed] [Google Scholar]
- 33.Shang Q, Gao Y, Qin T, Wang S, Shi Y, Chen T. Interaction of oral and toothbrush microbiota affects oral cavity health. Front Cell Infect Microbiol. 2020;10:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams S, Arnold D, Murphy B, et al. A randomised clinical study to determine the effect of a toothpaste containing enzymes and proteins on plaque oral microbiome ecology. Sci Rep. 2017;7(1):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain A, Bhaskar DJ, Gupta D, et al. Comparative evaluation of honey, chlorhexidine gluconate (0.2%) and combination of xylitol and chlorhexidine mouthwash (0.2%) on the clinical level of dental plaque: a 30 days randomized control trial. Perspect Clin Res. 2015;6(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuda K, Adachi M, Iijima K. The efficacy of antimicrobial mouth rinses in oral health care. Bull Tokyo Dent Coll. 1998;39(1):7‐14. [PubMed] [Google Scholar]
- 38.Sampaio‐Maia B, Caldas I, Pereira M, Perez‐Mongiovi D, Araujo R. The oral microbiome in health and its implication in oral and systemic diseases. Adv Appl Microbiol. 2016;97:171‐210. [DOI] [PubMed] [Google Scholar]
- 39.Atarashi K, Suda W, Luo C, et al. Ectopic colonization of oral bacteria in the intestine drives Th1 cell induction and inflammation. Science. 2017;358(6361):359‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen I. From the Acta Prize Lecture 2014: The Periodontal‐Systemic Connection Seen from a Microbiological Standpoint: Summary of the Acta Odontologica Scandinavia Price Lecture 2014 Presented at the Meeting of the IADR/Pan European Region in Dubrovnik. September 10–13. 2014. Acta Odontol Scand. 2015;73(8):563‐568. [DOI] [PubMed] [Google Scholar]
- 41.d’Ettorre G, Oliva A, DeAngelis M, Vullo VRE. Ectopic colonization of oral bacteria in the intestine drives Th1 cell induction and. Inflammation. Science. 2019;358:359‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riccia DD, Bizzini F, Perilli M, et al. Anti‐inflammatory effects of Lactobacillus brevis (CD2) on periodontal disease. Oral Dis. 2007;13(4):376‐385. 10.1111/j.1601-0825.2006.01291.x [DOI] [PubMed] [Google Scholar]
- 43.Ástvaldsdóttir Á, Naimi‐Akbar A, Davidson T, et al. Arginine and caries prevention: a systematic review. Caries Res. 2016;50(4):383‐393. [DOI] [PubMed] [Google Scholar]
- 44.Beighton D. The complex oral microflora of high‐risk individuals and groups and its role in the caries process. Community Dent Oral Epidemiol. 2005;33(4):248‐255. [DOI] [PubMed] [Google Scholar]
- 45.Huang X, Palmer SR, Ahn S‐J, et al. A Highly arginolytic streptococcus species that potently antagonizes Streptococcus mutans . Appl Environ Microbiol. 2016;82(7):2187‐2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ‐free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. 2018;9:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027‐1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 48.Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548‐554. 10.1126/science.1229000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Goffau MC, Lager S, Sovio U, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572(7769):329‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao E, Mattos M, Vieira GHA, et al. Diabetes enhances IL‐17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe. 2017;22(1):120‐128.e4. 10.1016/j.chom.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajishengallis G, Liang S, Payne MA, et al. Low‐abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10(5):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dutzan N, Kajikawa T, Abusleme L, et al. A dysbiotic microbiome triggers T H 17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med. 2018;10(463):eaat0797. 10.1126/scitranslmed.aat0797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutzan N, Abusleme L, Bridgeman H, et al. On‐going mechanical damage from mastication drives homeostatic Th17 cell responses at the oral barrier. Immunity. 2017;46(1):133‐147. 10.1016/j.immuni.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dutzan N, Konkel JE, Greenwell‐Wild T, Moutsopoulos NM. Characterization of the human immune cell network at the gingival barrier. Mucosal Immunol. 2016;9(5):1163‐1172. 10.1038/mi.2015.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bunetel L, Tricot‐Doleux S, Agnani G, Bonnaure‐Mallet M. In vitro evaluation of the retention of three species of pathogenic microorganisms by three different types of toothbrush. Oral Microbiol Immunol. 2000;15(5):313‐316. [DOI] [PubMed] [Google Scholar]
- 56.Samarian DS, Jakubovics NS, Luo TL, Rickard AH. Use of a high‐throughput in vitro microfluidic system to develop oral multi‐species biofilms. JoVE. 2014;94:e52467. 10.3791/52467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radaic A, Ye C, Parks B, et al. Modulation of pathogenic oral biofilms towards health with nisin probiotic. J Oral Microbiol. 2020;12(1):1809302‐ 10.1080/20002297.2020.1809302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, Zhou X, Zhou X, et al. Effects of different substrates/growth media on microbial community of saliva‐derived biofilm. FEMS Microbiol Lett. 2017;364(13): 10.1093/femsle/fnx123 [DOI] [PubMed] [Google Scholar]
- 59.Tian Y, He X, Torralba M, et al. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol. 2010;25(5):357‐367. 10.1111/j.2041-1014.2010.00585.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marsh PD, Bradshaw DJ. Physiological approaches to the control of oral biofilms. Adv Dent Res. 1997;11(1):176‐185. [DOI] [PubMed] [Google Scholar]
- 61.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113(6):E791‐E800. 10.1073/pnas.1522149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kilian M, Chapple I, Hannig M, et al. The oral microbiome–an update for oral healthcare professionals. Br Dent J. 2016;221(10):657‐666. [DOI] [PubMed] [Google Scholar]
- 63.Edlund A, Yang Y, Hall AP, et al. An in vitrobiofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome. 2013;1(1):25. 10.1186/2049-2618-1-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thurnheer T, Bostanci N, Belibasakis GN. Microbial dynamics during conversion from supragingival to subgingival biofilms in an in vitro model. Mol Oral Microbiol. 2016;31(2):125‐135. 10.1111/omi.12108 [DOI] [PubMed] [Google Scholar]
- 65.Ammann TW, Belibasakis GN, Thurnheer T. Impact of early colonizers on in vitro subgingival biofilm formation. PLoS One. 2013;8(12):e83090. 10.1371/journal.pone.0083090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klug B, Santigli E, Westendorf C, Tangl S, Wimmer G, Grube M. From mouth to model: combining in vivo and in vitro oral biofilm growth. Front Microbiol. 2016;7: 1448. 10.3389/fmicb.2016.01448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Califf KJ, Schwarzberg‐Lipson K, Garg N, et al. Analysis of periodontal pocket microbial communities pre‐ and posttreatment. mSystems. 2017;2(3):e00016‐17. 10.1128/mSystems.00016-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duran‐Pinedo AE, Chen T, Teles R, et al. Community‐wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014;8(8):1659‐1672. 10.1038/ismej.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ngo LH, Veith PD, Chen Y‐Y, Chen D, Darby IB, Reynolds EC. Mass spectrometric analyses of peptides and proteins in human gingival crevicular fluid. J Proteome Res. 2010;9(4):1683‐1693. 10.1021/pr900775s [DOI] [PubMed] [Google Scholar]
- 70.Silva‐Boghossian CM, Colombo APV, Tanaka M, Rayo C, Xiao Y, Siqueira WL. Quantitative proteomic analysis of gingival crevicular fluid in different periodontal conditions. PLoS One. 2013;8(10):e75898. 10.1371/journal.pone.0075898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pérez‐Cobas AE, Gomez‐Valero L, Buchrieser C. Metagenomic approaches in microbial ecology: an update on whole‐genome and marker gene sequencing analyses. Microb Genomics. 2020;6(8): mgen000409. 10.1099/mgen.0.000409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poretsky R, Rodriguez‐R LM, Luo C, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S RRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9(4):e93827. 10.1371/journal.pone.0093827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S RRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl_1):4516‐4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu W, Crawford ED, O’Donovan BD, et al. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high‐abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17(1):41. 10.1186/s13059-016-0904-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Podar M, Abulencia CB, Walcher M, et al. Targeted access to the genomes of low‐abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73(10):3205‐3214. 10.1128/AEM.02985-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rackaityte E, Halkias J, Fukui EM, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. 2020;26:599‐607. 10.1038/s41591-020-0761-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han D, Gao P, Li R, et al. Multicenter assessment of microbial community profiling using 16s rRNA gene sequencing and shotgun metagenomic sequencing. J Adv Res. 2020;26:111‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35(9):833‐844. 10.1038/nbt.3935 [DOI] [PubMed] [Google Scholar]
- 79.Mikheenko A, Saveliev V, Gurevich A. MetaQUAST: Evaluation of Metagenome Assemblies. Bioinformatics. 2016;32(7):1088‐1090. [DOI] [PubMed] [Google Scholar]
- 80.Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP—a flexible pipeline for genome‐resolved metagenomic data analysis. Microbiome. 2018;6(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nayfach S, Rodriguez‐Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26(11):1612‐1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pearman WS, Freed NE, Silander OK. Testing the advantages and disadvantages of short‐and long‐read eukaryotic metagenomics using simulated reads. BMC Bioinformatics. 2020;21:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moss EL, Maghini DG, Bhatt AS. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat Biotechnol. 2020;38(6):701‐707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mostafa HH, Fissel JA, Fanelli B, et al. Metagenomic next‐generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID‐19 patients. MBio. 2020;11(6):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sevim V, Lee J, Egan R, et al. Shotgun metagenome data of a defined mock community using oxford nanopore. PacBio and Illumina Technologies Sci Data. 2019;6(1):1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao L, Kang M, Zhang MJ, et al. Polymicrobial periodontal disease triggers a wide radius of effect and unique virome. NPJ Biofilms Microbiomes. 2020;6(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shkoporov AN, Clooney AG, Sutton TD, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26(4):527‐541. [DOI] [PubMed] [Google Scholar]
- 88.Vemuri R, Shankar EM, Chieppa M, Eri R, Kavanagh K. Beyond just bacteria: functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths. Microorganisms. 2020;8(4):483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nguyen T, Sedghi L, Ganther S, Malone E, Kamarajan P, Kapila YL. Host‐microbe interactions: profiles in the transcriptome, the proteome, and the metabolome. Periodontol 2000. 2020;82(1):115‐128. 10.1111/prd.12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharma PV, Thaiss CA. Host‐microbiome interactions in the era of single‐cell biology. Front Cell Infect Microbiol. 2020;10:569070. 10.3389/fcimb.2020.569070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woyke T, Doud DF, Schulz F. The Trajectory of microbial single‐cell sequencing. Nat Methods. 2017;14(11):1045. [DOI] [PubMed] [Google Scholar]
- 92.McClelland H, Jones C, Chubiz L, Fike D, Bradley A. Direct observation of the dynamics of single‐cell metabolic activity during microbial diauxic growth. MBio. 2020;11(2):e01519‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balachandran M, Cross KL, Podar M. Single‐cell genomics and the oral microbiome. J Dent Res. 2020;99(6):613‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duran‐Pinedo AE, Solbiati J, Frias‐Lopez J. The effect of the stress hormone cortisol on the metatranscriptome of the oral microbiome. NPJ Biofilms Microbiomes. 2018;4(1):1‐4. 10.1038/s41522-018-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aleti G, Baker JL, Tang X, et al. Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. MBio. 2019;10(2):e00321‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang J. Microbial metabolomics. Curr Genomics. 2011;12(6):391‐403. 10.2174/138920211797248619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh M, Saxena M, Saimbi C, Arif J, Roy R. Metabolic profiling by H NMR spectroscopy of saliva shows clear distinction between control and diseased case of periodontitis. Metabolomics. 2017;13(11):1‐9. 10.1007/s11306-017-1245-4 27980501 [DOI] [Google Scholar]
- 98.Husi H, Albalat A. Chapter 9 ‐ Proteomics. In Handbook of Pharmacogenomics and Stratified Medicine; Padmanabhan, S., Ed.; Academic Press: San Diego, 2014; pp 147–179. doi: 10.1016/B978‐0‐12‐386882‐4.00009‐8.
- 99.Proteomics ‐ an overview | ScienceDirect Topic, https://www‐sciencedirect‐com.ucsf.idm.oclc.org/topics/neuroscience/proteomics. Accessed December 7, 2020. [Google Scholar]
- 100.Coorssen JR. Proteomics. In: Maloy S, Hughes K, eds. Brenner’s Encyclopedia of Genetics, 2nd edition. San Diego: Academic Press; 2013:508–510. [Google Scholar]
- 101.Belstrøm D, Jersie‐Christensen RR, Lyon D, et al. Metaproteomics of saliva identifies human protein markers specific for individuals with periodontitis and dental caries compared to orally healthy controls. PeerJ. 2016;4:e2433. 10.7717/peerj.2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bostanci N, Belibasakis GN. Gingival crevicular fluid and its immune mediators in the proteomic era. Periodontol 2000. 2018;76(1):68‐84. 10.1111/prd.12154 [DOI] [PubMed] [Google Scholar]
- 103.Rylev M, Abduljabar AB, Reinholdt J, et al. Proteomic and immunoproteomic analysis of aggregatibacter actinomycetemcomitans JP2 clone strain HK1651. J. Proteomics. 2011;74(12):2972‐2985. 10.1016/j.jprot.2011.07.022 [DOI] [PubMed] [Google Scholar]
- 104.Trindade F, Amado F, Oliveira‐Silva RP, et al. Toward the definition of a peptidome signature and protease profile in chronic periodontitis. PROTEOMICS – Clin Appl. 2015;9(9–10):917‐927. 10.1002/prca.201400191 [DOI] [PubMed] [Google Scholar]
- 105.Winning TA, Townsend GC. Oral mucosal embryology and histology. Clin Dermatol. 2000;18(5):499‐511. 10.1016/S0738-081X(00)00140-1 [DOI] [PubMed] [Google Scholar]
- 106.Oral Mucosal Immunity and Microbiome. In: Belibasakis GN, Hajishengallis G, Bostanci N, Curtis MA, eds. Advances in Experimental Medicine and Biology. Cham: Springer International Publishing; 2019; Vol. 1197. 10.1007/978-3-030-28524-1 [DOI] [PubMed] [Google Scholar]
- 107.Wu R‐Q, Zhang D‐F, Tu E, Chen Q‐M, Chen W. The mucosal immune system in the oral cavity—an orchestra of T cell diversity. Int J Oral Sci. 2014;6(3):125‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Esberg A, Haworth S, Kuja‐Halkola R, Magnusson PKE, Johansson I. Heritability of oral microbiota and immune responses to oral bacteria. Microorganisms. 2020;8(8):1126. 10.3390/microorganisms8081126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smith DJ, Taubman MA. Ontogeny of immunity to oral microbiota in humans. Crit Rev Oral Biol Med Off Publ Am Assoc Oral Biol. 1992;3(1–2):109‐133. 10.1177/10454411920030010201 [DOI] [PubMed] [Google Scholar]
- 110.Perez‐Muñoz ME, Arrieta M‐C, Ramer‐Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The Placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tuominen H, Collado MC, Rautava J, Syrjänen S, Rautava S. Composition and maternal origin of the neonatal oral cavity microbiota. J. Oral Microbiol. 2019;11(1):1663084. 10.1080/20002297.2019.1663084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cobb CM, Kelly PJ, Williams KB, Babbar S, Angolkar M, Derman RJ. The oral microbiome and adverse pregnancy outcomes. Int J Womens Health. 2017;9:551‐559. 10.2147/IJWH.S142730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han YW. Oral health and adverse pregnancy outcomes – what’s next? J Dent Res. 2011;90(3):289‐293. 10.1177/0022034510381905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vander Haar EL, So J, Gyamfi‐Bannerman C, Han YW. Fusobacterium nucleatum and adverse pregnancy outcomes: epidemiological and mechanistic evidence. Anaerobe. 2018;50:55‐59. 10.1016/j.anaerobe.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gomez‐Arango LF, Barrett HL, McIntyre HD, et al. Contributions of the maternal oral and gut microbiome to placental microbial colonization in overweight and obese pregnant women. Sci Rep. 2017;7:2860. 10.1038/s41598-017-03066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fardini Y, Chung P, Dumm R, Joshi N, Han YW. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun. 2010;78(4):1789‐1796. 10.1128/IAI.01395-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Han YW, Fardini Y, Chen C, et al. Term stillbirth caused by oral fusobacterium nucleatum. Obstet Gynecol. 2010;115(2 Pt 2):442‐445. 10.1097/AOG.0b013e3181cb9955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nelson‐Filho P, Borba IG, Mesquita KSF, et al. Dynamics of microbial colonization of the oral cavity in newborns. Braz Dent J. 2013;24(4):415‐419. 10.1590/0103-6440201302266 [DOI] [PubMed] [Google Scholar]
- 120.Könönen E. Development of oral bacterial flora in young children. Ann Med. 2000;32(2):107‐112. 10.3109/07853890009011759 [DOI] [PubMed] [Google Scholar]
- 121.Li H, Wang J, Wu L, et al. The impacts of delivery mode on infant’s oral microflora. Sci Rep. 2018;8(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bretz WA, Corby PMA, Hart TC, et al. Dental caries and microbial acid production in twins. Caries Res. 2005;39(3):168‐172. 10.1159/000084793 [DOI] [PubMed] [Google Scholar]
- 123.Zheng Y, Zhang M, Li J, et al. Comparative analysis of the microbial profiles in supragingival plaque samples obtained from twins with discordant caries phenotypes and their mothers. Front Cell Infect Microbiol. 2018;8:361. 10.3389/fcimb.2018.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lassalle F, Spagnoletti M, Fumagalli M, et al. oral microbiomes from hunter‐gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet. Mol Ecol. 2018;27(1):182‐195. 10.1111/mec.14435 [DOI] [PubMed] [Google Scholar]
- 125.Castro MML, de Ferreira RO, Fagundes NCF, et al. Association between psychological stress and periodontitis: a systematic review. Eur J Dent. 2020;14(1):171‐179. 10.1055/s-0039-1693507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mishiro T, Oka K, Kuroki Y, et al. Oral microbiome alterations of healthy volunteers with proton pump inhibitor. J Gastroenterol Hepatol. 2018;33(5):1059‐1066. 10.1111/jgh.14040 [DOI] [PubMed] [Google Scholar]
- 127.Bui FQ, Almeida‐da‐Silva CLC, Huynh B, et al. Association between periodontal pathogens and systemic disease. Biomed J. 2019;42(1):27‐35. 10.1016/j.bj.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Poole AC, Goodrich JK, Youngblut ND, et al. Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe. 2019;25(4):553‐564.e7. 10.1016/j.chom.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 129.Krishnan K, Chen T, Paster BJ. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017;23(3):276‐286. 10.1111/odi.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694‐1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336(6086):1255‐1262. 10.1126/science.1224203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. 2019;18:153303381986735. 10.1177/1533033819867354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Burcham ZM, Garneau NL, Comstock SS, Tucker RM, Knight R, Metcalf JL. Patterns of oral microbiota diversity in adults and children: a crowdsourced population study. Sci Rep. 2020;10(1):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178‐184. [DOI] [PubMed] [Google Scholar]
- 135.Gomez A, Nelson KE. The oral microbiome of children: development, disease, and implications beyond oral health. Microb Ecol. 2017;73(2):492‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Du Q, Li M, Zhou X, Tian K. A comprehensive profiling of supragingival bacterial composition in chinese twin children and their mothers. Antonie Van Leeuwenhoek. 2017;110(5):615‐627. 10.1007/s10482-017-0828-4 [DOI] [PubMed] [Google Scholar]
- 137.Papapostolou A, Kroffke B, Tatakis DN, Nagaraja HN, Kumar PS. Contribution of host genotype to the composition of health‐associated supragingival and subgingival microbiomes. J Clin Periodontol. 2011;38(6):517‐524. 10.1111/j.1600-051X.2011.01718.x [DOI] [PubMed] [Google Scholar]
- 138.Shaw L, Ribeiro ALR, Levine AP, et al. The human salivary microbiome is shaped by shared environment rather than genetics: evidence from a large family of closely related individuals. MBio. 2017;8(5):e01237‐17. 10.1128/mBio.01237-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Demmitt BA, Corley RP, Huibregtse BM, et al. Genetic influences on the human oral microbiome. BMC Genom. 2017;18(1):1‐15. 10.1186/s12864-017-4008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Haworth S, Esberg A, Lif Holgerson P, et al. Heritability of caries scores, trajectories, and disease subtypes. J Dent Res. 2020;99(3):264‐270. 10.1177/0022034519897910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xiao J, Fiscella KA, Gill SR. Oral microbiome: possible harbinger for children’s health. Int J Oral Sci. 2020;12(1):1‐13. 10.1038/s41368-020-0082-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dzidic M, Collado MC, Abrahamsson T, et al. Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 2018;12(9):2292‐2306. 10.1038/s41396-018-0204-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mason MR, Chambers S, Dabdoub SM, Thikkurissy S, Kumar PS. Characterizing oral microbial communities across dentition states and colonization niches. Microbiome. 2018;6(1):1‐10. 10.1186/s40168-018-0443-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Boix‐Amorós A, Collado MC, Mira A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol. 2016;7:492. 10.3389/fmicb.2016.00492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu X, He J, Xue J, et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol. 2015;17(3):699‐710. 10.1111/1462-2920.12502 [DOI] [PubMed] [Google Scholar]
- 146.Sampaio‐Maia B, Monteiro‐Silva F. Acquisition and maturation of oral microbiome throughout childhood: an update. Dent Res J. 2014;11(3):291. [PMC free article] [PubMed] [Google Scholar]
- 147.Yost S, Stashenko P, Choi Y, et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. Int J Oral Sci. 2018;10(4):1‐10. 10.1038/s41368-018-0037-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Snider EJ, Compres G, Freedberg DE, et al. barrett’s esophagus is associated with a distinct oral microbiome. Clin Transl Gastroenterol. 2018;9(3): 10.1038/s41424-018-0005-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shillitoe EJ. The microbiome of oral cancer. Crit Rev Oncog. 2018;23(3–4): :153‐160. 10.1615/CritRevOncog.2018027422 [DOI] [PubMed] [Google Scholar]
- 150.Klimesova K, Jiraskova Zakostelska Z, Tlaskalova‐Hogenova H. Oral bacterial and fungal microbiome impacts colorectal carcinogenesis. Front. Microbiol. 2018;9:774. 10.3389/fmicb.2018.00774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gaiser RA, Halimi A, Alkharaan H, et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut. 2019;68(12):2186‐2194. 10.1136/gutjnl-2018-317458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brennan CA, Garrett WS. Fusobacterium nucleatum — symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156‐166. 10.1038/s41579-018-0129-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lockhart PB, Bolger AF, Papapanou PN, et al. Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association? Circulation. 2012;125(20):2520‐2544. 10.1161/CIR.0b013e31825719f3 [DOI] [PubMed] [Google Scholar]
- 154.Sheiham A. Dental caries affects body weight, growth and quality of life in pre‐school children. Br Dent J. 2006;201(10):625‐626. 10.1038/sj.bdj.4814259 [DOI] [PubMed] [Google Scholar]
- 155.Acharya A, Khan S, Hoang H, Bettiol S, Goldberg L, Crocombe L. Dental conditions associated with preventable hospital admissions in Australia: a systematic literature review. BMC Health Serv. Res. 2018;18:921. 10.1186/s12913-018-3733-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Craig SJC, Blankenberg D, Parodi ACL, et al. Child weight gain trajectories linked to oral microbiota composition. Sci Rep. 2018;8(1):14030. 10.1038/s41598-018-31866-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Blod C, Schlichting N, Schülin S, et al. the oral microbiome—the relevant reservoir for acute pediatric appendicitis? Int J Colorectal Dis. 2018;33(2):209‐218. 10.1007/s00384-017-2948-8 [DOI] [PubMed] [Google Scholar]
- 158.Docktor MJ, Paster BJ, Abramowicz S, et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2012;18(5):935‐942. 10.1002/ibd.21874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li K, Bihan M, Methé BA. Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS One. 2013;8(5):e63139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J. Bacteriol. 2010;192(19):5002‐5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38(3):409‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Van Hoogmoed CG, Geertsema‐Doornbusch GI, Teughels W, Quirynen M, Busscher HJ, Van der Mei HC. Reduction of periodontal pathogens adhesion by antagonistic strains. Oral Microbiol Immunol. 2008;23(1):43‐48. 10.1111/j.1399-302X.2007.00388.x [DOI] [PubMed] [Google Scholar]
- 163.Wu J, Xie H. Role of arginine deiminase of streptococcus cristatus in porphyromonas gingivalis colonization. Antimicrob Agents Chemother. 2010;54(11):4694‐4698. 10.1128/AAC.00284-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van Essche M, Loozen G, Godts C, et al. Bacterial antagonism against periodontopathogens. J Periodontol. 2013;84(6):801‐811. 10.1902/jop.2012.120261 [DOI] [PubMed] [Google Scholar]
- 165.Shin JM, Gwak JW, Kamarajan P, Fenno JC, Rickard AH, Kapila YL. Biomedical applications of nisin. J Appl Microbiol. 2016;120(6):1449‐1465. 10.1111/jam.13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kamarajan P, Hayami T, Matte B, et al. Nisin ZP, a bacteriocin and food preservative, inhibits head and neck cancer tumorigenesis and prolongs survival. PLoS One. 2015;10(7):e0131008. 10.1371/journal.pone.0131008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kamarajan P, Ateia I, Shin JM, et al. Periodontal pathogens promote cancer aggressivity via tlr/myd88 triggered activation of integrin/FAK signaling that is therapeutically reversible by a probiotic bacteriocin. PLOS Pathog. 2020;16(10):e1008881. 10.1371/journal.ppat.1008881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Govoni M, Jansson EÅ, Weitzberg E, Lundberg JO. The increase in plasma nitrite after a dietary nitrate load is markedly attenuated by an antibacterial mouthwash. Nitric Oxide. 2008;19(4):333‐337. 10.1016/j.niox.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 169.Zhurakivska K, Troiano G, Caponio VCA, et al. Do Changes in oral microbiota correlate with plasma nitrite response? A systematic review. Front Physiol. 2019;10:1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bondonno CP, Liu AH, Croft KD, et al. Antibacterial Mouthwash blunts oral nitrate reduction and increases blood pressure in treated hypertensive men and women. Am J Hypertens. 2015;28(5):572‐575. [DOI] [PubMed] [Google Scholar]
- 171.Koch CD, Gladwin MT, Freeman BA, Lundberg JO, Weitzberg E, Morris A. Enterosalivary nitrate metabolism and the microbiome: intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. Free Radic Biol Med. 2017;105:48‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bondonno CP, Liu AH, Croft KD, et al. Antibacterial mouthwash blunts oral nitrate reduction and increases blood pressure in treated hypertensive men and women. Am J Hypertens. 2015;28(5):572‐575. [DOI] [PubMed] [Google Scholar]
- 173.Kapil V, Haydar SM, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. Physiological role for nitrate‐reducing oral bacteria in blood pressure control. Free Radic Biol Med. 2013;55:93‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cutler C, Kiernan M, Willis J, et al. Post‐exercise hypotension and skeletal muscle oxygenation is regulated by nitrate‐reducing activity of oral bacteria. Free Radic Biol Med. 2019;143:252‐259. [DOI] [PubMed] [Google Scholar]
- 175.Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 2000;193(1):1‐6. 10.1111/j.1574-6968.2000.tb09393.x [DOI] [PubMed] [Google Scholar]
- 176.Edlund A, Yang Y, Yooseph S, et al. Meta‐omics uncover temporal regulation of pathways across oral microbiome genera during in Vitro sugar metabolism. ISME J. 2015;9(12):2605‐2619. 10.1038/ismej.2015.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Senneby A, Davies J, Svensäter G, Neilands J. Acid tolerance properties of dental biofilms in vivo. BMC Microbiol. 2017;17:165. 10.1186/s12866-017-1074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27(6):409‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16(12):745‐759. 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang Y, Wang S, Wu C, et al. Oral microbiome alterations associated with early childhood caries highlight the importance of carbohydrate metabolic activities. MSystems. 2019;4(6):e00450‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Klein MI, DeBaz L, Agidi S, et al. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS One. 2010;5(10):e13478. 10.1371/journal.pone.0013478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chapple ILC. Potential mechanisms underpinning the nutritional modulation of periodontal inflammation. J Am Dent Assoc. 2009;140(2):178‐184. 10.14219/jada.archive.2009.0131 [DOI] [PubMed] [Google Scholar]
- 183.Cekici A, Kantarci A, Hasturk H, Van Dyke TE. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000. 2014;64(1):57‐80. 10.1111/prd.12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.De Pablo P, Dietrich T, McAlindon TE. Association of Periodontal disease and tooth loss with rheumatoid arthritis in the US population. J Rheumatol. 2008;35(1):70‐76. [PubMed] [Google Scholar]
- 185.Wu J, Peters BA, Dominianni C, et al. Cigarette smoking and the oral microbiome in a large study of american adults. ISME J. 2016;10(10):2435‐2446. 10.1038/ismej.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pitiphat W, Merchant AT, Rimm EB, Joshipura KJ. Alcohol consumption increases periodontitis risk. J Dent Res. 2003;82(7):509‐513. 10.1177/154405910308200704 [DOI] [PubMed] [Google Scholar]
- 187.Cugini C, Shanmugam M, Landge N, Ramasubbu N. the role of exopolysaccharides in oral biofilms. J Dent Res. 2019;98(7):739‐745. 10.1177/0022034519845001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192(12):3024‐3032. 10.1128/JB.01649-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bowen WH, Koo H. Biology of Streptococcus mutans ‐derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45(1):69‐86. 10.1159/000324598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Monse B, Heinrich‐Weltzien R, Benzian H, Holmgren C, Helderman WVP. PUFA – An index of clinical consequences of untreated dental caries. Community Dent Oral Epidemiol. 2010;38(1):77‐82. 10.1111/j.1600-0528.2009.00514.x [DOI] [PubMed] [Google Scholar]
- 191.Eleanor F, Joseph A. Prevalence of Total and Untreated Dental Caries Among Youth: United States, 2015–2016, http://pubmed.ncbi.nlm.nih.gov/29717975/. Accessed December 9, 2020 [Google Scholar]
- 192.Koenig JE, Spor A, Scalfone N, et al. succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 2011;108(Supp. 1):4578‐4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li M, Wang M, Donovan SM. Early development of the gut microbiome and immune‐mediated childhood disorders. Sem Reprod Med. 2014;32(01):74‐086. 10.1055/s-0033-1361825 [DOI] [PubMed] [Google Scholar]
- 194.Wu R‐Q, Zhang D‐F, Tu E, Chen Q‐M, Chen W. The mucosal immune system in the oral cavity—an orchestra of t cell diversity. Int J Oral Sci. 2014;6(3):125‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yu JC, Khodadadi H, Malik A, et al. Innate immunity of neonates and infants. Front Immunol. 2018;9:1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic diseases caused by oral infection. Clin Microbiol Rev. 2000;13(4):547‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4(11):430‐435. [DOI] [PubMed] [Google Scholar]
- 198.Dawes C. Salivary flow patterns and the health of hard and soft oral tissues. J. Am Dent Assoc. 2008;139:18S‐24S. [DOI] [PubMed] [Google Scholar]
- 199.Page RC. The pathobiology of periodontal diseases may affect systemic diseases: inversion of a paradigm. Ann Periodontol. 1998;3(1):108‐120. 10.1902/annals.1998.3.1.108 [DOI] [PubMed] [Google Scholar]
- 200.Marcus AJ, Hajjar DP. Vascular transcellular signaling. J Lipid Res. 1993;34(12):2017‐2031. [PubMed] [Google Scholar]
- 201.Mattila KJ. Viral and bacterial infections in patients with acute myocardial infarction. J Intern Med. 1989;225(5):293‐296. 10.1111/j.1365-2796.1989.tb00084.x [DOI] [PubMed] [Google Scholar]
- 202.Zekeridou A, Mombelli A, Cancela J, Courvoisier D, Giannopoulou C. Systemic inflammatory burden and local inflammation in periodontitis: what is the link between inflammatory biomarkers in serum and gingival crevicular fluid? Clin Exp Dent Res. 2019;5(2):128‐135. 10.1002/cre2.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kinane DF. Causation and pathogenesis of periodontal disease. Periodontol 2000. 2001;25(1):8‐20. 10.1034/j.1600-0757.2001.22250102.x [DOI] [PubMed] [Google Scholar]
- 204.Glurich I, Grossi S, Albini B, et al. Systemic inflammation in cardiovascular and periodontal disease: comparative study. Clin Diagn Lab Immunol. 2002;9(2):425‐432. 10.1128/CDLI.9.2.425-432.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365(23):2205‐2219. [DOI] [PubMed] [Google Scholar]
- 206.De Pablo P, Dietrich T, McAlindon TE. Association of periodontal disease and tooth loss with rheumatoid arthritis in the US population. J Rheumatol. 2008;35(1):70‐76. [PubMed] [Google Scholar]
- 207.Kobayashi T, Yokoyama T, Ishida K, Abe A, Yamamoto K, Yoshie H. Serum cytokine and periodontal profiles in relation to disease activity of rheumatoid arthritis in Japanese adults. J Periodontol. 2010;81(5):650‐657. 10.1902/jop.2010.090688 [DOI] [PubMed] [Google Scholar]
- 208.Bingham CO, Moni M. Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr Opin Rheumatol. 2013;25(3):345‐353. 10.1097/BOR.0b013e32835fb8ec [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ortiz P, Bissada NF, Palomo L, et al. Periodontal therapy reduces the severity of active rheumatoid arthritis in patients treated with or without tumor necrosis factor inhibitors. J Periodontol. 2009;80(4):535‐540. 10.1902/jop.2009.080447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Erciyas K, Sezer U, Üstün K, et al. Effects of periodontal therapy on disease activity and systemic inflammation in rheumatoid arthritis patients. Oral Dis. 2013;19(4):394‐400. 10.1111/odi.12017 [DOI] [PubMed] [Google Scholar]
- 211.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis . Peptidylarginine Deiminase Infect Immun. 1999;67(7):3248‐3256. 10.1128/IAI.67.7.3248-3256.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.How KY, Song KP, Chan KGPorphyromonas dingivalis: an overview of periodontopathic pathogen below the gum line. Front Microbiol. 2016;7:53. 10.3389/fmicb.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dominy SS, Lynch C, Ermini F, et al. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small‐molecule inhibitors. Sci Adv. 2019;5(1):eaau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Meyer MS, Applebaum KM, Furniss CS, et al. Human papillomavirus‐16 modifies the association between fruit consumption and head and neck squamous cell carcinoma. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2008;17(12):3419‐3426. 10.1158/1055-9965.EPI-08-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hayashi C, Gudino CV, Gibson FC, Genco CA. Review: pathogen‐induced inflammation at sites distant from oral infection: bacterial persistence and induction of cell‐specific innate immune inflammatory pathways. Mol Oral Microbiol. 2010;25(5):305‐316. 10.1111/j.2041-1014.2010.00582.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mealey BL, Rose LF. Diabetes mellitus and inflammatory periodontal diseases. Curr Opin Endocrinol Diabetes Obes. 2008;15(2):135‐141. 10.1097/MED.0b013e3282f824b7 [DOI] [PubMed] [Google Scholar]
- 217.Vieira Ribeiro F, de Mendonça AC, Santos VR, et al. Cytokines and bone‐related factors in systemically healthy patients with chronic periodontitis and patients with type 2 diabetes and chronic periodontitis. J Periodontol. 2011;82(8):1187‐1196. 10.1902/jop.2011.100643 [DOI] [PubMed] [Google Scholar]
- 218.Duarte PM, Oliveira MCGD, Tambeli CH, Parada CA, Casati MZ, Nociti FH. Overexpression of interleukin‐1β and interleukin‐6 may play an important role in periodontal breakdown in type 2 diabetic patients. J Periodontal Res. 2007;42(4):377‐381. 10.1111/j.1600-0765.2006.00961.x [DOI] [PubMed] [Google Scholar]
- 219.Casarin RCV, Barbagallo A, Meulman T, et al. Subgingival biodiversity in subjects with uncontrolled type‐2 diabetes and chronic periodontitis. J Periodontal Res. 2013;48(1):30‐36. 10.1111/j.1600-0765.2012.01498.x [DOI] [PubMed] [Google Scholar]
- 220.Spratt DA, Greenman J, Schaffer AG. Capnocytophaga gingivalis: effects of glucose concentration on growth and hydrolytic enzyme production. Microbiology. 1996;142(8):2161‐2164. 10.1099/13500872-142-8-2161 [DOI] [PubMed] [Google Scholar]
- 221.Gholizadeh P, Pormohammad A, Eslami H, Shokouhi B, Fakhrzadeh V, Kafil HS. Oral pathogenesis of aggregatibacter actinomycetemcomitans. Microb Pathog. 2017;113:303‐311. 10.1016/j.micpath.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 222.Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of porphyromonas gingivalis in oxygenated and carbon‐dioxide‐depleted environments. Microbiology. 2002;148(2):467‐472. 10.1099/00221287-148-2-467 [DOI] [PubMed] [Google Scholar]
- 223.Ficara AJ, Levin MP, Grower M, Kramer GD. A Comparison of the glucose and protein content of gingival fluid from diabetics and nondiabetics. J Periodontal Res. 1975;10(3):171‐175. 10.1111/j.1600-0765.1975.tb00022.x [DOI] [PubMed] [Google Scholar]
- 224.Decker A, Askar H, Tattan M, Taichman R, Wang H‐L. The assessment of stress, depression, and inflammation as a collective risk factor for periodontal diseases: a systematic review. Clin Oral Investig. 2020;24(1):1‐12. 10.1007/s00784-019-03089-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gomaa N, Glogauer M, Nicolau B, et al. Stressed‐out oral immunity: a gateway from socioeconomic adversity to periodontal disease. Psychosom Med. 2020;82(2):126‐137. 10.1097/PSY.0000000000000774 [DOI] [PubMed] [Google Scholar]
- 226.Spector AM, Postolache TT, Akram F, Scott AJ, Wadhawan A, Reynolds MA. Psychological stress: a predisposing and exacerbating factor in periodontitis. Curr Oral Health Rep. 2020;7(3):208‐215. 10.1007/s40496-020-00282-2 [DOI] [Google Scholar]
- 227.Mark Welch JL, Dewhirst FE, Borisy GG. Biogeography of the oral microbiome: the site‐specialist hypothesis. Annu Rev Microbiol. 2019;73(1):335‐358. 10.1146/annurev-micro-090817-062503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9(5):488‐500. 10.1007/s13238-018-0548-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Chawhuaveang DD, Yu OY, Yin IX, Lam WY‐H, Mei ML, Chu C‐H. Acquired salivary pellicle and oral diseases: a literature review. J Dent Sci. 2020;16:523‐529. 10.1016/j.jds.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Cavalcanti IMG, Cury AADB, Jenkinson HF, Nobbs AH. Interactions between Streptococcus oralis, actinomyces oris, and Candida albicans in the development of multispecies oral microbial biofilms on salivary pellicle. Mol Oral Microbiol. 2017;32(1):60‐73. 10.1111/omi.12154 [DOI] [PubMed] [Google Scholar]
- 231.Proctor DM, Fukuyama JA, Loomer PM, et al. A spatial gradient of bacterial diversity in the human oral cavity shaped by salivary flow. Nat Commun. 2018;9: 10.1038/s41467-018-02900-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26(3):229‐242. 10.1016/j.tim.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lof M, Janus MM, Krom BP. Metabolic Interactions between bacteria and fungi in commensal oral biofilms. J Fungi. 2017;3(3):40. 10.3390/jof3030040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Eren AM, Borisy GG, Huse SM, Welch JLM. Oligotyping Analysis of the human oral microbiome. Proc Natl Acad Sci USA 2014;111(28):E2875‐E2884. 10.1073/pnas.1409644111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jakubovics NS. Intermicrobial interactions as a driver for community composition and stratification of oral biofilms. J Mol Biol. 2015;427(23):3662‐3675. 10.1016/j.jmb.2015.09.022 [DOI] [PubMed] [Google Scholar]
- 236.Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42(1):47‐79. 10.1111/j.1600-0757.2006.00187.x [DOI] [PubMed] [Google Scholar]
- 237.Parashar A, Parashar S, Zingade A, Gupta S, Sanikop S. Interspecies communication in oral biofilm: an ocean of information. Oral Sci Int. 2015;12(2):37‐42. [Google Scholar]
- 238.Takeuchi H, Yamamoto K. Ultrastructural analysis of structural framework in dental plaque developing on synthetic carbonate apatite applied to human tooth surfaces. Eur J Oral Sci. 2001;109(4):249‐259. 10.1034/j.1600-0722.2001.00029.x [DOI] [PubMed] [Google Scholar]
- 239.Marsh PD. Dental plaque as a biofilm and a microbial community‐implications for health and disease. BioMed Central. 2006;6:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hajishengallis G, Lamont RJ. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 2016;24(6):477‐489. 10.1016/j.tim.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014;35(1):3‐11. 10.1016/j.it.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ramsey MM, Rumbaugh KP, Whiteley M. metabolite cross‐feeding enhances virulence in a model polymicrobial infection. PLOS Pathog. 2011;7(3):e1002012. 10.1371/journal.ppat.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Grenier D. Nutritional interactions between two suspected periodontopathogens, treponema denticola and Porphyromonas gingivalis . Infect Immun. 1992;60(12):5298‐5301. 10.1128/IAI.60.12.5298-5301.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Oelze V, Münster A, Nicklisch N, Meller H, Dresely V, Alt K. Early neolithic diet and animal husbandry: stable isotope evidence from three linearbandkeramik (LBK) sites in central Germany. J Archaeol Sci. 2010;38:270‐279. 10.1016/j.jas.2010.08.027 [DOI] [Google Scholar]
- 245.Aufderheide AC, Rodríguez‐Martín C, Rodriguez‐Martin C, Langsjoen O. The Cambridge Encyclopedia of Human Paleopathology. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 246.Grine FE, Gwinnett AJ, Oaks JH. Early hominid dental pathology: interproximal caries in 1.5 million‐year‐old paranthropus robustus from swartkrans. Arch Oral Biol. 1990;35(5):381‐386. 10.1016/0003-9969(90)90185-d [DOI] [PubMed] [Google Scholar]
- 247.Kerr NW. Prevalence and natural history of periodontal disease in prehistoric scots (Pre‐900 AD). J Periodontal Res. 1998;33(3):131‐137. 10.1111/j.1600-0765.1998.tb02303.x [DOI] [PubMed] [Google Scholar]
- 248.Nowakowski D, Sosulski FW, Hoover R. The effect of pin and attrition milling on starch damage in hard wheat flours. Starch ‐ Stärke. 1986;38(8):253‐258. 10.1002/star.19860380802 [DOI] [Google Scholar]
- 249.Stark JR, Yin XS. the effect of physical damage on large and small barley starch granules. Starch ‐ Stärke. 1986;38(11):369‐374. 10.1002/star.19860381103 [DOI] [Google Scholar]
- 250.Li E, Dhital S, Hasjim J. Effects of grain milling on starch structures and flour/starch properties. Starch ‐ Stärke. 2014;66(1–2):15‐27. 10.1002/star.201200224 [DOI] [Google Scholar]
- 251.Baumgartner S, Imfeld T, Schicht O, Rath C, Persson RE, Persson GR. The impact of the stone age diet on gingival conditions in the absence of oral hygiene. J Periodontol. 2009;80(5):759‐768. 10.1902/jop.2009.080376 [DOI] [PubMed] [Google Scholar]
- 252.Martínez Steele E, Popkin BM, Swinburn B, Monteiro CA. The share of ultra‐processed foods and the overall nutritional quality of diets in the us: evidence from a nationally representative cross‐sectional study. Popul Health Metr. 2017;15(1):6. 10.1186/s12963-017-0119-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Cordain L, Eaton SB, Sebastian A, et al. Origins and evolution of the western diet: health implications for the 21st century. Am J Clin Nutr. 2005;81(2):341‐354. 10.1093/ajcn.81.2.341 [DOI] [PubMed] [Google Scholar]
- 254.Halvorsrud K, Lewney J, Craig D, Moynihan PJ. Effects of starch on oral health: systematic review to inform WHO guideline. J Dent Res. 2019;98(1):46‐53. 10.1177/0022034518788283 [DOI] [PubMed] [Google Scholar]
- 255.Cordain L, Miller JB, Eaton SB, Mann N, Holt SH, Speth JD. Plant‐animal subsistence ratios and macronutrient energy estimations in worldwide hunter‐gatherer diets. Am J Clin Nutr. 2000;71(3):682‐692. 10.1093/ajcn/71.3.682 [DOI] [PubMed] [Google Scholar]
- 256.Gerrior S, Bente L. Nutrient Content of the U.S. Food Supply, 1909–99: A Summary Report; Center for Nutrition Policy and Promotion. U.S. Department of Agriculture; 2002. [Google Scholar]
- 257.Carrera‐Bastos P, Fontes‐Villalba M, O’Keefe JH, Lindeberg S, Cordain L. The western diet and lifestyle and diseases of civilization, https://www.dovepress.com/the‐western‐diet‐and‐lifestyle‐and‐diseases‐of‐civilization‐peer‐reviewed‐article‐RRCC. Accessed Jun 15, 2019. 10.2147/RRCC.S16919 [DOI] [Google Scholar]
- 258.Crittenden AN, Sorrentino J, Moonie SA, Peterson M, Mabulla A, Ungar PS. Oral health in transition: the hadza foragers of Tanzania. PLoS One. 2017;12(3):e0172197. 10.1371/journal.pone.0172197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Borges‐Yañez SA, Maupomé G, Martinez‐Gonzalez M, Cervantez‐Turrubiante L, Gutiérrez‐Robledo LM. Dietary fiber intake and dental health status in urban‐marginal, and rural communities in central Mexico. J Nutr. Health Aging. 2004;8(5):333‐339. [PubMed] [Google Scholar]
- 260.Schoeninger MJ, Bunn HT, Murray SS, Marlett JA. Composition of tubers used by hadza foragers of Tanzania. J Food Compos Anal. 2001;14(1):15‐25. 10.1006/jfca.2000.0961 [DOI] [Google Scholar]
- 261.Quest BW. Oral health benefits of a daily dental chew in dogs. J Vet Dent. 2013;30(2):84‐87. 10.1177/089875641303000203 [DOI] [PubMed] [Google Scholar]
- 262.Watson A. Diet and periodontal disease in dogs and cats. Aust Vet J. 1994;71(10):313‐318. 10.1111/j.1751-0813.1994.tb00905.x [DOI] [PubMed] [Google Scholar]
- 263.Clarke D, Cameron A. Relationship between diet, dental calculus and periodontal disease in domestic and feral cats in Australia. Aust Vet J. 1998;76(10):690‐693. 10.1111/j.1751-0813.1998.tb12284.x [DOI] [PubMed] [Google Scholar]
- 264.Kapoor V, Antonelli T, Parkinson JA, Hartstone‐Rose A. oral health correlates of captivity. Res Vet Sci. 2016;107:213‐219. 10.1016/j.rvsc.2016.06.009 [DOI] [PubMed] [Google Scholar]
- 265.Antonelli T, Leischner C, Ososky J, Hartstone‐Rose A. The Effect of captivity on the oral health of the critically endangered black‐footed ferret (Mustela Nigripes). Can J Zool. 2015;94: 10.1139/cjz-2015-0135 [DOI] [Google Scholar]
- 266.Romano A, Mackie A, Farina F, Aponte M, Sarghini F, Masi P. Characterisation, in vitro digestibility and expected glycemic index of commercial starches as uncooked ingredients. J Food Sci Technol. 2016;53(12):4126‐4134. 10.1007/s13197-016-2375-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Edwards CH, Grundy MM, Grassby T, et al. Manipulation of starch bioaccessibility in wheat endosperm to regulate starch digestion, postprandial glycemia, insulinemia, and gut hormone responses: a randomized controlled trial in healthy ileostomy participants. Am J Clin Nutr. 2015;102(4):791‐800. 10.3945/ajcn.114.106203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Grundy MM‐L, Edwards CH, Mackie AR, Gidley MJ, Butterworth PJ, Ellis PR. Re‐evaluation of the mechanisms of dietary fibre and implications for macronutrient bioaccessibility, digestion and postprandial metabolism. Br J Nutr. 2016;116(5):816‐833. 10.1017/S0007114516002610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Dhital S, Shrestha AK, Gidley MJ. Effect of cryo‐milling on starches: functionality and digestibility. Food Hydrocoll. 2010;24(2/3):152‐163. [Google Scholar]
- 270.Tran TTB, Shelat KJ, Tang D, Li E, Gilbert RG, Hasjim J. Milling of rice grains. the degradation on three structural levels of starch in rice flour can be independently controlled during grinding. J Agric Food Chem. 2011;59(8):3964‐3973. 10.1021/jf105021r [DOI] [PubMed] [Google Scholar]
- 271.Singh N, Katyal M, Virdi AS, et al. Effect of grain hardness, fractionation and cultivars on protein, pasting and dough rheological properties of different wheat flours. Int J Food Sci Technol. 2018;53(9):2077‐2087. 10.1111/ijfs.13794 [DOI] [Google Scholar]
- 272.Tamaki S, Hisamatsu M, Teranishi K, Adachi T, Yamada T. Structural change of maize starch granules by ball‐mill treatment. Starch ‐ Stärke. 1998;50(8):342‐348. [DOI] [Google Scholar]
- 273.Schwartz N, Kaye EK, Nunn ME, Spiro A, Garcia RI. High‐fiber foods reduce periodontal disease progression in men aged 65 and older: the veterans affairs normative aging study/dental longitudinal study. J Am Geriatr Soc. 2012;60(4):676‐683. 10.1111/j.1532-5415.2011.03866.x [DOI] [PubMed] [Google Scholar]
- 274.Nielsen SJ, Trak‐Fellermeier MA, Joshipura K, Dye BA. Dietary fiber intake is inversely associated with periodontal disease among US adults. J Nutr. 2016;146(12):2530‐2536. 10.3945/jn.116.237065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kondo K, Ishikado A, Morino K, et al. A high‐fiber, low‐fat diet improves periodontal disease markers in high‐risk subjects: a pilot study. Nutr Res. 2014;34(6):491‐498. 10.1016/j.nutres.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 276.Hujoel P. Dietary carbohydrates and dental‐systemic diseases. J Dent Res. 2009;88(6):490‐502. 10.1177/0022034509337700 [DOI] [PubMed] [Google Scholar]
- 277.Tennert C, Reinmuth A‐C, Bremer K, et al. An oral health optimized diet reduces the load of potential cariogenic and periodontal bacterial species in the supragingival oral plaque: a randomized controlled pilot study. Microbiol Open. 2020;9(8):e1056 10.1002/mbo3.1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Sedghi L, Byron C, Jennings R, Chlipala GE, Green SJ, Silo‐Suh L. Effect of dietary fiber on the composition of the murine dental microbiome. Dent J. 2019;7(2):58. 10.3390/dj7020058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Freitas D, Le Feunteun S. Oro‐gastro‐intestinal digestion of starch in white bread, wheat‐based and gluten‐free pasta: unveiling the contribution of human salivary α‐amylase. Food Chem. 2019;274:566‐573. 10.1016/j.foodchem.2018.09.025 [DOI] [PubMed] [Google Scholar]
- 280.Bornhorst GM, Singh RP. Bolus formation and disintegration during digestion of food carbohydrates. Compr Rev Food Sci Food Saf. 2012;11(2):101‐118. 10.1111/j.1541-4337.2011.00172.x [DOI] [Google Scholar]
- 281.Linke HAB, Birkenfeld LH. Clearance and metabolism of starch foods in the oral cavity. Ann Nutr Metab. 1999;43(3):131‐139. 10.1159/000012778 [DOI] [PubMed] [Google Scholar]
- 282.Jain A, Jain R, Jain S. Determination of salivary amylase activity. In: Jain A, Jain R, Jain S, eds. Basic Techniques in Biochemistry, Microbiology and Molecular Biology: Principles and Techniques. Springer Protocols Handbooks; New York, NY: Springer US; 2020:223‐225. 10.1007/978-1-4939-9861-6_52 [DOI] [Google Scholar]
- 283.Ng M, Fleming T, Robinson M, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2014;384(9945):766‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Natella S, Divan V, Rana M, Mills C. Sugar Consumption at a Crossroads. Credit Suisse Res. Inst. 2013. [Google Scholar]
- 285.Satokari R. High Intake of sugar and the balance between pro‐and anti‐inflammatory gut bacteria. Nutrients. 2020;12(5):1348. 10.3390/nu12051348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Klein MI, Duarte S, Xiao J, Mitra S, Foster TH, Koo H. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Appl Environ Microbiol. 2009;75(3):837‐841. 10.1128/AEM.01299-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Thurnheer T, Gmür R, Shapiro S, Guggenheim B. Mass transport of macromolecules within an in vitro model of supragingival plaque. Appl Environ Microbiol. 2003;69(3):1702‐1709. 10.1128/AEM.69.3.1702-1709.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Vroom JM, Grauw KJD, Gerritsen HC, et al. Depth penetration and detection of ph gradients in biofilms by two‐photon excitation microscopy. Appl Environ Microbiol. 1999;65(8):3502‐3511. 10.1128/AEM.65.8.3502-3511.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Tao L, Sutcliffe I, Russell R, Ferretti J. Transport of sugars, including sucrose, by the msm transport system of Streptococcus mutans . J Dent Res. 1993;72(10):1386‐1390. [DOI] [PubMed] [Google Scholar]
- 290.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11(2):87‐93. [DOI] [PubMed] [Google Scholar]
- 291.Titgemeyer F, Hillen W. Global Control of Sugar Metabolism: A Gram‐Positive Solution. In: Lactic Acid Bacteria: Genetics, Metabolism and Applications. New York, NY: Springer; 2002;59‐71. [PubMed] [Google Scholar]
- 292.Abranches J, Nascimento MM, Zeng L, et al. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans . J Bacteriol. 2008;190(7):2340‐2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Takahashi N, Washio J, Mayanagi G. Metabolomics of supragingival plaque and oral bacteria. J Dent Res. 2010;89(12):1383‐1388. 10.1177/0022034510377792 [DOI] [PubMed] [Google Scholar]
- 294.Saito K, Takahashi N, Horiuchi H, Yamada T. Effects of glucose on formation of cytotoxic end‐products and proteolytic activity of prevotella intermedia, prevotella nigrescens and Porphyromonas gingivalis . J Periodontal Res. 2001;36(6):355‐360. 10.1034/j.1600-0765.2001.360602.x [DOI] [PubMed] [Google Scholar]
- 295.Cattaneo C, Riso P, Laureati M, Gargari G, Pagliarini E. Exploring associations between interindividual differences in taste perception, oral microbiota composition, and reported food intake. Nutrients. 2019;11(5):1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.CDC . Coronavirus Disease 2019 (COVID‐19). https://www.CDC.gov/coronavirus/2019‐ncov/daily‐life‐coping/stress‐coping/index.html. Accessed Sep 27, 2020 [Google Scholar]
- 297.Yan CH, Faraji F, Prajapati DP, Boone CE, DeConde AS. Association of chemosensory dysfunction and COVID‐19 in patients presenting with influenza‐like symptoms. Int Forum Allergy Rhinol. 2020;10(7):806‐813. 10.1002/alr.22579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Lozada‐Nur F, Chainani‐Wu N, Fortuna G, Sroussi H. Dysgeusia in COVID‐19: possible mechanisms and implications. Oral Surg Oral Med Oral Pathol Oral Radiol. 2020;130(3):344‐346. 10.1016/j.oooo.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Butowt R, von Bartheld CS. Anosmia in COVID‐19: underlying mechanisms and assessment of an olfactory route to brain infection. Neuroscientist. 2020. 10.1177/1073858420956905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wang Z, Zhou J, Marshall B, Rekaya R, Ye K, Liu H‐X. SARS‐CoV‐2 receptor ACE2 is enriched in a subpopulation of mouse tongue epithelial cells in nongustatory papillae but not in taste buds or embryonic oral epithelium. ACS Pharmacol Transl Sci. 2020;10:1‐8. 10.1021/acsptsci.0c00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Shigemura N, Takai S, Hirose F, Yoshida R, Sanematsu K, Ninomiya Y. Expression of renin‐angiotensin system components in the taste organ of mice. Nutrients. 2019;11(9):2251. 10.3390/nu11092251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Obiefuna S, Donohoe C. Neuroanatomy, nucleus gustatory. In: StatPearls. Treasure Island, FL: StatPearls Publishing; 2020. [PubMed] [Google Scholar]
- 303.Finsterer J, Stollberger C. Causes of hypogeusia/hyposmia in SARS‐CoV2 infected patients. J Med Virol. 2020;92(10):1793‐1794. 10.1002/jmv.25903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019‐NCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12(1):1‐5. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Liu L, Wei Q, Alvarez X, et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011;85(8):4025‐4030. 10.1128/JVI.02292-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Wang H, Zhou M, Brand J, Huang L. Inflammation and taste disorders: mechanisms in taste buds. Ann NY Acad Sci. 2009;1170:596‐603. 10.1111/j.1749-6632.2009.04480.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Couzin‐FrankelApr. 28, J.; 2020; Pm, 3:40. Why don’t some coronavirus patients sense their alarmingly low oxygen levels?. http://www.sciencemag.org/news/2020/04/why‐don‐t‐some‐coronavirus‐patients‐sense‐their‐alarmingly‐low‐oxygen‐levels. Accessed Dec 13, 2020 [Google Scholar]
- 308.Takeda N, Takaoka T, Ueda C, Toda N, Kalubi B, Yamamoto S. Zinc deficiency in patients with idiopathic taste impairment with regard to angiotensin converting enzyme activity. Auris Nasus Larynx. 2004;31(4):425‐428. 10.1016/j.anl.2004.09.006 [DOI] [PubMed] [Google Scholar]
- 309.Shen Z, Xiao Y, Kang L, et al. Genomic diversity of severe acute respiratory syndrome‐coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. 2020;71(15):713‐720. 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Bao L, Zhang C, Dong J, Zhao L, Li Y, Sun J. Oral microbiome and SARS‐CoV‐2: beware of lung co‐infection. Front. Microbiol. 2020;11:1840. 10.3389/fmicb.2020.01840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Stuart LF. The history of oral hygiene products: how far have we come in 6000 Years? Periodontol 2000. 1997;15:7‐14. [DOI] [PubMed] [Google Scholar]
- 313.Zhurakivska K, Troiano G, Caponio VCA, et al. Do changes in oral microbiota correlate with plasma nitrite response? A systematic review. Front Physiol. 2019;10:1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lundberg JO, Weitzberg E, Gladwin MT. The nitrate–nitrite–nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov. 2008;7(2):156‐167. [DOI] [PubMed] [Google Scholar]
- 315.Hyde ER, Andrade F, Vaksman Z, et al. Metagenomic analysis of nitrate‐reducing bacteria in the oral cavity: implications for nitric oxide homeostasis. PLoS One. 2014;9(3):e88645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Koo H, Xiao J, Klein M, Jeon J. Exopolysaccharides produced by streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192(12):3024‐3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Wang B‐Y, Kuramitsu HK. Interactions between oral bacteria: inhibition of streptococcus mutans bacteriocin production by Streptococcus gordonii . Appl Environ Microbiol. 2005;71(1):354‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]


