Abstract
Since 1971, the Protein Data Bank (PDB) has served as the single global archive for experimentally determined 3D structures of biological macromolecules made freely available to the global community according to the FAIR principles of Findability–Accessibility–Interoperability–Reusability. During the first 50 years of continuous PDB operations, standards for data representation have evolved to better represent rich and complex biological phenomena. Carbohydrate molecules present in more than 14,000 PDB structures have recently been reviewed and remediated to conform to a new standardized format. This machine-readable data representation for carbohydrates occurring in the PDB structures and the corresponding reference data improves the findability, accessibility, interoperability and reusability of structural information pertaining to these molecules. The PDB Exchange MacroMolecular Crystallographic Information File data dictionary now supports (i) standardized atom nomenclature that conforms to International Union of Pure and Applied Chemistry-International Union of Biochemistry and Molecular Biology (IUPAC-IUBMB) recommendations for carbohydrates, (ii) uniform representation of branched entities for oligosaccharides, (iii) commonly used linear descriptors of carbohydrates developed by the glycoscience community and (iv) annotation of glycosylation sites in proteins. For the first time, carbohydrates in PDB structures are consistently represented as collections of standardized monosaccharides, which precisely describe oligosaccharide structures and enable improved carbohydrate visualization, structure validation, robust quantitative and qualitative analyses, search for dendritic structures and classification. The uniform representation of carbohydrate molecules in the PDB described herein will facilitate broader usage of the resource by the glycoscience community and researchers studying glycoproteins.
Keywords: carbohydrate structure, glycan, glycosylation, oligosaccharide, Protein Data Bank
Introduction
The first demonstration that form or 3D molecular structure dictates function in biology came with the Watson and Crick deoxyribonucleic acid (DNA) double helix structure (Watson and Crick 1953). Since their work revealed the structural basis of intergenerational information transfer and DNA replication, ~175,000 experimentally determined 3D structures of proteins and nucleic acids determined by researchers working on all inhabited continents have been archived in the Protein Data Bank (PDB). Founded in 1971 as the first open-access digital data resource in biology (Protein Data Bank 1971), the PDB is the single global archive housing richly annotated 3D structures of proteins, DNA and ribonucleic acid (RNA) (Berman et al. 2003; wwPDB Consortium 2019). The archive has impacted fundamental biology, biomedicine, bioenergy and biotechnology/bioengineering by enabling atomic-level understanding of naturally occurring and engineered biomolecules (Markosian et al. 2018; Burley et al. 2019; Armstrong et al. 2020; Feng et al. 2020; Burley et al. 2021). The Worldwide Protein Data Bank (wwPDB, wwpdb.org) (Berman et al. 2003; wwPDB Consortium 2019) manages the PDB according to the FACT principles of Fairness–Accuracy–Confidentiality–Transparency (van der Aalst et al. 2017) and the FAIR principles of Findability-Accessibility-Interoperability-Reusability (Wilkinson et al. 2016). The vision of the wwPDB is to sustain freely accessible, interoperating Core Archives of structure data and metadata for biological macromolecules as an enduring public good to promote basic and applied research and education across the sciences. wwPDB member organizations include data centers in the United States (RCSB Protein Data Bank [RCSB PDB]) (Berman et al. 2000; Rose et al. 2020; Burley et al. 2021), the United Kingdom (Protein Data Bank in Europe [PDBe]) (Mir et al. 2018; Armstrong et al. 2020) and Japan (Protein Data Bank Japan [PDBj]) (Kinjo et al. 2018) plus the specialist nuclear magnetic resonance (NMR) data resource Biological Magnetic Resonance Bank (BMRB) (Ulrich et al. 2008). The PDB is an open access archive that has been accredited as a trustworthy data science resource by CoreTrustSeal (https://www.coretrustseal.org/).
Each incoming PDB structure determined using macromolecular crystallography (MX), NMR spectroscopy, electron microscopy (3DEM), or microcrystal electron diffraction (microED) experimental methods encompasses 3D atomic coordinates, experimental data and metadata, such as sample polymer sequences, source organism(s), structure determination process and details of data collection. PDB archival data conforms to the PDB Exchange MacroMolecular Crystallographic Information File (PDBx/mmCIF) (Westbrook and Fitzgerald 2009) data dictionary, the Chemical Component Dictionary (CCD, https://www.wwpdb.org/data/ccd) (Westbrook et al. 2015) and the Biologically Interesting Molecule Reference Dictionary (BIRD, https://www.wwpdb.org/data/bird) (Dutta et al. 2014). The PDBx/mmCIF data dictionary defines data content for deposition, validation, biocuration, remediation and secure archiving of PDB structures. The CCD is an external reference dictionary describing all chemical components found in PDB structures, with each component assigned a unique alphanumeric identifier. This dictionary contains detailed chemical descriptions for standard and modified amino acids/nucleotides, small molecule ligands and solvent molecules. The BIRD is a secondary external reference dictionary that contains information about biologically interesting peptide-like or glycopeptide-antibiotic and inhibitor molecules represented in the PDB archive. For such molecules, the BIRD provides information about the chemistry, biology and structure features of the molecule and a dual representation as both a single compound and as an oligomer with component sequence.
Carbohydrates support myriad biological and biochemical functions. They play key roles in energy generation, cell signaling, cell–cell recognition, formation of various structural components of extracellular matrices and many other biologically important processes (Varki 2017b). Within organisms from all three kingdoms of life, carbohydrates are as universal and essential as nucleic acids, proteins, lipids and metabolites (Marth 2008), but their structures and functions were not extensively studied until their biological significance became more apparent (Varki 2017b). The importance of 3D macromolecular structure information (its shape and interaction with small molecules) in generating insight into biological processes and advancing discovery and development of therapeutic agents and is well documented (Westbrook and Burley 2019; Goodsell et al. 2020; Westbrook et al. 2020). Understanding the structure and organization of carbohydrates is critical to discerning their biochemical and biological roles in human health and disease. More than 14,000 PDB structures reveal, confirm or explain the key functions of carbohydrates and serve as foundations for future research.
The first carbohydrate structure in the PDB was Hyaluronan (PDB ID 1HYA) (Winter et al. 1975), which is involved in lubrication, extracellular matrix organization, cell–cell and cell–matrix interactions supporting organogenesis and cell migration and long-range signaling (when released following tissue injury). The earliest protein–carbohydrate cocrystal structures revealed Lysozyme C bound to a substrate-like trisaccharide (PDB ID 9LYZ) (Kelly et al. 1979) and the N-glycosylated Fc portion of a human immunoglobulin G (IgG) (PDB IDs 1FC1 and 1FC2) (Deisenhofer 1981) in which protein glycosylation was first observed in 3D. These landmark structures contributed important insights into the innate and adaptive immune systems.
Since these pioneering studies, more than 14,000 PDB depositions have included carbohydrates, revealing or confirming the biological and biomedical significance of such carbohydrate molecules and their interactions with proteins. During the COVID-19 pandemic, more than 100 SARS-CoV-2 protein–carbohydrate complex structures have been deposited into the PDB. For example, PDB ID 6WPS revealed recognition of glycan-containing epitopes by the immune system in 3D with the structure of a SARS-CoV-2 spike glycoprotein bound to a neutralizing antibody Fab fragment (Pinto et al. 2020). Thousands of PDB structures have glycan structures present at the glycosylation sites. Another group of carbohydrates occurring in hundreds of PDB structures are metabolites, such as maltose, sucrose, lactose and fragments of cellulose. Maltose binding to cyclodextrin glycosyltransferase revealed the presence of a raw starch-binding motif that is conserved among diverse starch-converting enzymes (Lawson et al. 1994). PDB ID 1LES elucidated conformation-dependent sucrose–lectin interactions (Casset et al. 1995). Other commonly studied carbohydrates in the PDB are as follows: (i) glycosaminoglycans, such as heparin fragments complexed with annexin (PDB ID 2HYV) (Shao et al. 2006) and basic fibroblast growth factor (PDB ID 1BFC) (Faham et al. 1996); (ii) blood group epitopes, such as the blood group A Lewis B antigen terminal binding to Helicobacter pylori adhesin (PDB ID 5F7N) (Moonens et al. 2016) and the blood group A type II antigen binding to the VP8* domain of the human rotavirus spike protein VP4 (PDB ID 4DS0) (Hu et al. 2012) and (iii) synthetic carbohydrates, such as the antidiabetic drug pseudotetrasaccharide acarbose bound to human maltase-glucoamylase (PDB ID 2QMJ) (Sim et al. 2008) and the anticoagulant fondaparinux bound to platelet factor 4 (PDB ID 4R9W) (Cai et al. 2015).
The PDB currently houses >14,000 carbohydrate-containing structures. This subset of the archive has grown considerably over the decades (from ~100 in 1999 to >1000 in 2017 to ~14,000 in 2020; Supplementary Figure S1). Most of these carbohydrate-containing PDB structures consist of a carbohydrate bound to a protein. More than half are the result of glycosylation, or glycoconjugate (covalent adduct) formation between glycans and amino acid sidechains. Most early carbohydrate-containing structures in the PDB were determined by MX. In recent years, 3DEM has been come into its own as a structure determination method (Mitra 2019). It has been extensively used to study glycan structures (e.g., SARS-CoV-2 protein-carbohydrate complex PDB ID 6WPS) and the large human oligosaccharyltransferase (OST) complex (PDB IDs 6S7O and 6S7T) (Ramirez et al. 2019).
The PDB data dictionary was originally developed to describe linear protein and nucleic acid polymers. Consequently, the complex dendritic nature of carbohydrate structures found in nature made data representation, archiving and dissemination by the PDB challenging. Initially, carbohydrates occurring within PDB structures were represented at the atomic level as monosaccharides or oligosaccharides, some covalently bound to proteins and others forming noncovalent complexes. Historically, some of the carbohydrates themselves were not correctly represented in the atomic coordinates (e.g., nonstandard atom naming and incorrect stereochemistry), and linkages with one another or with proteins were either missing or wrongly specified in the PDB data files. Lack of consistent representation for carbohydrates within the PDB severely limited integration of 3D structural information with other carbohydrate data resources, such as Glycam (Woods 2005), GlyGen (York et al. 2020), ProCarbDB (Copoiu et al. 2020) and GlyTouCan (Tiemeyer et al. 2017). Regrettably, glycobiology scientists and other experts were unable to fully utilize the rich structural information available in the PDB.
The wwPDB partnership is committed to maintaining consistency and accuracy across the PDB archive. For more than a decade, the wwPDB has addressed data management challenges by regularly reviewing data processing procedures and carrying out structure remediation efforts to improve data representation accuracy and consistency (Henrick et al. 2008; Lawson et al. 2008; Dutta et al. 2014). In the face of growing numbers of carbohydrate structures in the PDB (Supplementary Figure S1) and increasing interest in understanding how carbohydrates and glycoproteins contribute to human health and disease, community feedback and recommendations were collected by the wwPDB via workshops, professional society meetings and email communication with domain experts in the glycoscience and structural biology communities. Based on feedback from these communities, carbohydrate-containing structures in the PDB were remediated. The principal goals of this archive-wide remediation were as follows: (i) standardization of CCD nomenclature following International Union of Pure and Applied Chemistry-International Union of Biochemistry and Molecular Biology (IUPAC-IUBMB) recommendations, introduction of uniform branched entity representations of oligosaccharides; (ii) adoption of descriptors commonly used by the glycoscience community and (iii) biocuration of glycosylation sites in the PDB structures. For the avoidance of doubt, no atomic coordinates (x, y and z values) were changed during the course of the archive-wide remediation.
Carbohydrate representation was addressed as follows:
atoms: standard atom labels in the CCD at the atomic level;
residues: consistent classification and naming of monosaccharides in the CCD at the residue level;
branched polymers: uniform oligosaccharide sequence and linear descriptors in the atomic coordinate files at entity level and
glycosylation sites: proper annotation of glycosylation sites in the atomic coordinate files at the protein structure level.
Standard nomenclatures and consistent data representation compatible with various representations in common use by the glycobiology community now enable for the first time full Findability, Accessibility, Interoperability and Reusability of carbohydrates present in the PDB structures.
Results
Chemical representation of monosaccharides
Monosaccharides, the building blocks of carbohydrates, have been reviewed and their nomenclature has been standardized in the PDB in accord with the 1996 IUPAC recommendations (McNaught 1996). A total of 1040 carbohydrates described in the CCD were reviewed and updated. First, six duplicate monosaccharides were removed. Second, 164 carbohydrates that proved on closer inspection to be oligosaccharides were reclassified as collections of monosaccharides. Third, five ligands of constituent monosaccharides covalently bound to amino acids were separately reclassified as monosaccharides and amino acids. Their original descriptions were obsoleted from the CCD.
A total of 862 unique monosaccharides remained in the CCD following remediation. Among them, 833 are cyclic monosaccharides (see Table I and Supplementary Table SI). Most monosaccharides are D-saccharides and pyranoses. All monosaccharides enumerated in the CCD were updated with standardized chemical names, synonyms, atom labels, modification versus basic sugars tags, chemical types (e.g., isomers), structure feature types (e.g., anomers) and symbol identifiers (e.g., IUPAC condensed symbols). An example of a remediated CCD definition is provided in Supplementary Appendix SI. Chemical names provided in the PDBx/mmCIF data dictionary item _chem_comp.name have been uniformly updated to include stereo- and ring-specific systematic names as recommended by IUPAC (McNaught 1996). Trivial (or common) names were added to the new machine-parsable PDBx/mmCIF synonym category, _pdbx_chem_comp_synonyms. For example, the most abundant monosaccharide present in the PDB, 2-acetamido-2-deoxy-beta-D-glucopyranose, is identified in the CCD by the code of NAG. The commonly used compound name (without stereo specification) N-acetyl-D-glucosamine is provided as a synonym.
Table I.
Summary of the remediated PDB cyclic hemiacetal/hemiketal monosaccharides represented in the CCD as of July 2020
| Features | Types | Number of CCD IDs |
|---|---|---|
| Basic versus modified | Basic | 125 |
| Modified | 708 | |
| Isomer | D-saccharide | 702 |
| L-saccharide | 128 | |
| Ring size | Pyranose | 696 |
| Furanose | 87 | |
| Dihydropyran | 39 | |
| Dihydrofuran | 5 | |
| Thiopyranose | 4 | |
| Anomer | Alpha anomer | 381 |
| Beta anomer | 343 | |
| Acetal versus ketal | Aldose | 695 |
| Ketose | 126 |
IUPAC recommended using symbols for denoting monosaccharides to simplify oligosaccharide descriptions. The IUPAC extended-form symbols, as described in Section 2-Carb-38.3 and the condensed-form symbol specified in 2-Carb-38.4 of the 1996 recommendations (McNaught 1996), are provided in the PDBx/mmCIF data item _pdbx_chem_comp_identifier.identifier. For NAG, the condensed IUPAC extended symbol (β-D-GlcpNAc) and the condensed symbol with addition of the D/L isomer and p/f ring size (DGlcpNAcb) are provided within the remediated CCD definition. Carbohydrate features, including isomer, ring size, anomer and aldose/ketose classification, are described in the PDBx/mmCIF category _pdbx_chem_comp_feature. Hereafter, monosaccharides will be described using the IUPAC extended symbol with the PDB CCD identifier provided in parentheses.
A total of 125 cyclic monosaccharides were classified as basic monosaccharides following IUPAC recommendations (McNaught 1996) and the Symbol Nomenclature for Glycans (SNFG) (Neelamegham et al. 2019). Community-developed SNFG text symbols mapping to 2D and 3D graphics (Varki et al. 2015; Thieker et al. 2016; Neelamegham et al. 2019) are also annotated by using the PDBx/mmCIF item _pdbx_chem_comp_identifier.identifier in the CCD. As illustrated in Figure 1, the SNFG text symbol for α-D-Glcp (GLC) is “Glc” and is displayed as blue circle in 2D and blue sphere in 3D, and the SNFG text symbol for β-D-GlcpNAc (NAG) is “GlcNAc,” that is, blue square in 2D and blue cube in 3D. At present, there are 708 modified monosaccharides that are primarily derived from these basic monosaccharides as deoxy sugars, amino sugars, aldonic and uronic acids and glycosides. If the substructure of a modified sugar overlaps with a basic sugar, the corresponding basic sugar is flagged as its parent in the PDBx/mmCIF item _chem_comp.mon_nstd_parent_comp_id and atom mapping between modified sugar and the basic sugar parent is provided using the PDBx/mmCIF category _pdbx_chem_comp_atom_related in the CCD. Chemical modifications also include substitution of the hydroxyl groups by halogens, chalcogens, nitrogen and phosphorus, plus sulfur and selenium replacements of ring oxygens in pyranoses and furanoses.
Fig. 1.
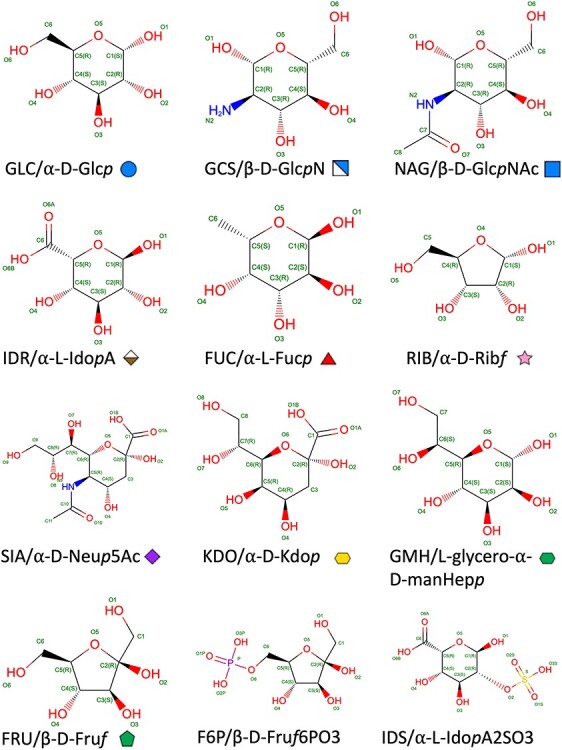
Examples of the current monosaccharide representations for different classes of SNFG symbols, sizes, D/L isomers, ring sizes, anomers, and aldose versus ketose. The 2D chemical diagram displays atom type and number, bond order and stereochemical marker. CCD identifier, the IUPAC extended symbol and the 2D SNFG graphic symbol are provided for each monosaccharide. Following Section 2-Carb-38.3 (McNaught 1996), an extended IUPAC symbol starts with α/β anomer and contains a D/L isomer indicator, the capitalized 3-letter code, and ends with the ring size indicated by the italic letter p for pyranose and f for furanose. Common modifications, if present, are shown with symbols (e.g., “N” for amine derivative and “NAc” for acetal-amine derivatives).
Each monosaccharide is assigned to one of the following carbohydrate types in the PDBx/mmCIF item _chem_comp.type in the CCD definition: “D-saccharide”; “D-saccharide, alpha linking”; “D-saccharide, beta linking”; “L-saccharide”; “L-saccharide, alpha linking”; “L-saccharide, beta linking” and “saccharide.” Alpha/beta linking type is determined based on whether the monosaccharide has hemiacetal or hemiketal reducing end. If the hemiacetal/hemiketal hydroxyl group is replaced by nonchalcogen groups (e.g., halogens), or blocked by forming a glycoside with a nonsugar component, or modified to a double bond (e.g., lactone, dihydropyran/dihydrofuran ring), the alpha/beta linking types are not specified.
Atom naming for monosaccharides
Standardization of monosaccharides in the CCD is essential for accurate oligosaccharide representation across the PDB archive. The hemiacetal/hemiketal annotation in the CCD identifies the reducing end of a monosaccharide in relation to the next monosaccharide. Standardized atom numbering enables proper recognition of the functional group and locants of a glycosidic bond. The anomer annotation specifies the stereo configuration on the glycosidic linkage.
For monosaccharides in the CCD, atom naming was standardized using element type followed by standard atom numbering (Markley et al. 1998). Atom numbering follows Section 2-Carb-2.2 of the IUPAC recommendations (McNaught 1996). Carbon atoms in monosaccharides are numbered using the primary acetal/ketal carbonyl group as reference. For aldose, the carbonyl group is identified as position #1; for ketose, the carbonyl group receives the lowest possible locant (i.e., position #2 for most ketoses occurring in the PDB such as fructose). For Api (YYM), apiose in its cyclic hemiacetal form, carbon atoms on the ring were numbered from #1 to 4, and the branching carbon is numbered as #5. For acidic sugars (e.g., uronic acids), letters A and B of the oxygen atoms are used to distinguish between the double and single bonds in the carbonyl group, respectively, as shown in Figure 1.
For atom labels of the substitution groups and oxygen atoms in hydroxyl/aldehyde/ketone groups, the connected carbon atom number was used (Section 2-Carb-7.1) (McNaught 1996). Sequential numbers were used for additional atoms. For example, β-D-GlcpNAc (NAG), the nitrogen on the amine substituent connected to C2 is labeled N2. Carbons of the acetyl group are labeled C7 and C8 sequentially, following the numbering in the pyranose ring (C1–C6). Sequential numbering in the atom labels for most monosaccharides is used for consistency and simplicity. Figure 1 shows the standardized nomenclature and atom labels for PDB monosaccharides based on IUPAC recommendations. Modified sugars are labeled according to the basic sugars. For thio- or amino sugars, the substitute sulfur or nitrogen atoms follow the same numbering of the replaced oxygen atoms.
Representation and naming of oligosaccharides
An oligosaccharide consists of two or more covalently connected monosaccharides linked by glycosidic bonds (typically between the hemiacetal or hemiketal of one monosaccharide and a hydroxyl group of another). Unlike a protein that can be described as a linear sequence of constituent amino acids, an oligosaccharide cannot be so described because of the branching and differences in connectivity between monosaccharides at different locants (e.g., 1–3, 1–4 and 1–6 glycosidic linkages). Therefore, in consultation of the community experts and following IUPAC recommendations, a new, branched entity representation was introduced into the PDBx/mmCIF dictionary to describe oligosaccharides (Figure 2). Oligosaccharides are classified as a new type “branched” in _entity.type and as an “oligosaccharide” in _pdbx_entity_branch.type. Each monosaccharide constituting an oligosaccharide entity is listed in the _pdbx_entity_branch_list PDBx/mmCIF category. Unlike polypeptides, wherein the peptide bond is implicitly assumed by the linear sequence, the connectivity of the glycosidic bonds for an oligosaccharide entity are explicitly described at the entity level in the PDBx/mmCIF category _pdbx_entity_branch_link. Since an oligosaccharide cannot be described in a linear sequence of constituent monosaccharides in polymer type, its topology and nomenclature are described in the form of a string, linear descriptor that is commonly used by the glycoscience community. wwPDB has adopted community used linear descriptors (_pdbx_entity_branch_descriptor) which provide complete chemical descriptions of a branched entity. In addition, the PDBx/mmCIF category _pdbx_branch_scheme supports monosaccharide residue mapping between the branched entity and the atomic coordinates, including residue alphanumeric identifiers, residue numbers and chain identifiers. Annotation of glycosylation sites is provided at _struct_conn.pdbx_role at a PDB structure level. PDB examples with data representation in these PDBx/mmCIF categories can be found in the Supplementary Appendix SII and in the wwPDB website (https://www.wwpdb.org/documentation/carbohydrate-remediation).
Fig. 2.
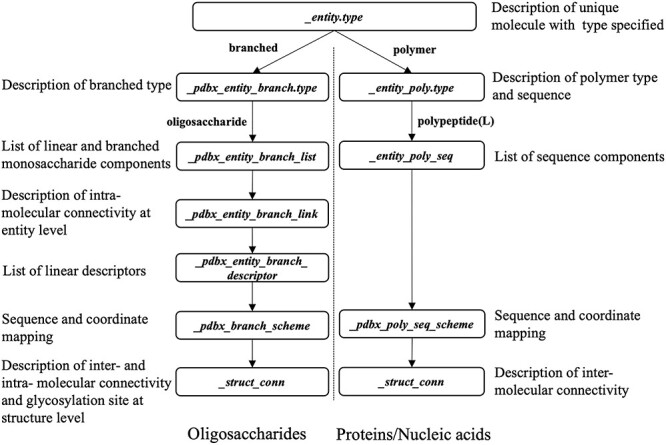
Representation of oligosaccharides compared to proteins/nucleic acids (branched vs. polymer entity representation as specified in the PDBx/mmCIF data dictionary). For carbohydrates, glycosylation sites are annotated using _struct_conn.pdbx_role.
An oligosaccharide with branches can be accurately described using a topological graph. The community-developed 2D SNFG graphical representation (Varki et al. 2015; Neelamegham et al. 2019) was adopted and implemented within the wwPDB structure validation report, with topological connectivity for each oligosaccharide using the wurcs2pic software (Matsubara et al. 2017; Tsuchiya et al. 2017) as illustrated in Figure 3. Reproducible topological description of oligosaccharides depends on consistent indexing (ordering) of monosaccharide components within the PDBx/mmCIF item pdbx_branch_entity_list and standardized linkage descriptions in pdbx_branch_entity_link. Each unique oligosaccharide is given a PDB systematic name in _entity.pdbx_description and linear descriptors in pdbx_entity_branch_descriptor based on its topology. Each unique oligosaccharide (branched entity) is assigned a unique asym identifier in the PDBx/mmCIF item _struct_asym.id, which points to atomic coordinates in _atom_site.label_asym_id. Unique chain IDs (atom_site.auth_asym_id) are assigned to each oligosaccharide in the PDB structure, and the constituent monosaccharides are consistently numbered from 1 to n to generate unambiguous linear descriptors, so users can easily identify an oligosaccharide and its interaction with other components.
Fig. 3.
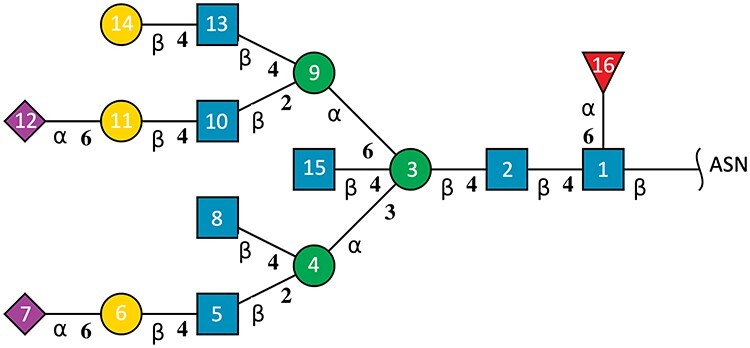
Example depiction of a glycosylated oligosaccharide (branched entity #16) in PDB ID 5T3X. Each monosaccharide is represented by a 2D SNFG symbol generated by the wurcs2pic software (Tsuchiya et al. 2017): blue square for GlcNAc, green circle for Man, yellow circle for Gal, purple diamond for Neu5Ac and red triangle for Fuc. Greek symbols indicate the anomeric form of the hemiacetal/hemiketal. The number at the connection line represents the locant of alcoholic hydroxy where the glycosidic bond is formed. Residue numbers inside SNFG symbols illustrate the ordering of monosaccharides within a branched entity. The GlcNAc at the glycosylation site is numbered as 1, and the main branch with the longest chain at the lowest locant as 1–7. Following depth-first logic, the GlcNAc of the deepest side branch is numbered as 8; 9–12 are then numbered as the longest side branch at branching point 3. The rest of side branches are numbered accordingly.
Consistent atom labeling in atom nomenclature standardization of monosaccharides is used to describe glycosidic linkage (e.g., 1–2, 1–3, 1–4 and 1–6 linkages from an aldose hemiacetal group to 2, 3, 4, 6 hydroxyl group of the connected monosaccharide, respectively, or 2–3, 2–4 and 2–6 linkages from a ketose hemiketal group to 2, 3, 4, 6 hydroxyl group of the connected monosaccharide, respectively). Uniform numbering of carbohydrate residues is essential for branched entity representation and generation of linear descriptors, hence the ordering (residue numbering) starts at the reducing end (#1), where the glycosylation occurs, proceeding to the nonreducing end, following the IUPAC recommended ordering scheme (Section 2-Carb-37.2 for linear oligosaccharides and Section 2-Carb-37.3 for branched oligosaccharides) (McNaught 1996) to define unambiguously oligosaccharide topology. The reducing end is where the hemiacetal/hemiketal group is either free or reacts to noncarbohydrate components, such as asparagine (ASN) glycosylation sites. In addition, the reducing end of a monosaccharide may be chemically converted into “downstream-end” derivatives such as alditol, aldonic acid or glycoside (Section 2-Carb-37.2) (McNaught 1996). When such modifications occur, the “downstream-end” is considered to be the reducing end and is indexed as for #1. Likewise, S- and N-glycosidic bonds in thio- and amino substitutions on the hydroxyl groups or hemiacetals/hemiketals are treated in the same way as the standard O-glycosidic bond. If there exists more than one branch in an oligosaccharide, the longest chain is defined as the main branch and is indexed first. If the two longest chains are of equal length, the chain with the lower locant (the atom number of the connected alcoholic hydroxyl group) at the branch point is defined as the main branch. Once the identity of the main branch is fixed, the remaining monosaccharides are then indexed in a depth-first logic, within which higher priority of side branches is given for deeper branching points away from the starting index #1. Unique residue numbers can then be assigned to monosaccharides of an oligosaccharide as shown in Figure 3, depicting a topology of an oligosaccharide covalently bound to human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 in PDB ID 5T3X (Gristick et al. 2016).
Using standardized residue numbers and glycosidic bonds, linear carbohydrate descriptors are generated to provide full description of the oligosaccharides, including connectivity information. The wwPDB incorporated widely used software tools for generating common linear descriptors of each unique oligosaccharide within a branched entity (_pdbx_entity_branch_descriptor) to promote Reusability and Interoperability. These descriptors are Glycam Condensed Sequence, which are generated based on GMML prototype script (Woods 2005), WURCS (Matsubara et al. 2017) generated using PDB2Glycan software (Tsuchiya et al. 2017) and LINUCS generated using PDB-CARE software (Lutteke and von der Lieth 2004). All three types of linear descriptors are shown in Figure 4A for a fucosylated N-glycan core found on the surface of the SARS-CoV-2 spike glycoprotein in PDB ID 6WPS (Pinto et al. 2020). See Figure 4B for the 3D view of the same N-glycan. The image was generated using the Molstar visualization software (Sehnal et al. 2018). In addition to linear descriptors mentioned herein, the PDB systematic name for this oligosaccharide is also provided in _entity.pdbx_description as alpha-D-mannopyranose-(1–3)-[alpha-D-mannopyranose-(1–6)]beta-D-mannopyranose-(1–4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1–4)-[alpha-L-fucopyranose-(1–6)]2-acetamido-2-deoxy-beta-D-glucopyranose, wherein monosaccharide full names are used, glycosidic linkages are specified and branches are enclosed in square brackets as recommended by IUPAC Section 2-Carb-37.3 (McNaught 1996). Any common name of an oligosaccharide is captured in _entity_name_com.name. The naming convention of cyclic oligosaccharides follows the nomenclature recommended by IUPAC Section 2-Carb-37.4 (McNaught 1996). For example, a cyclic oligosaccharide with six 1–4 linked α-D-Glcp (GLC, alpha-D-glucopyranose) is given a PDB systematic name cyclohexakis-(1–4)-(alpha-D-glucopyranose) and a common name, alpha-cyclodextrin.
Fig. 4.
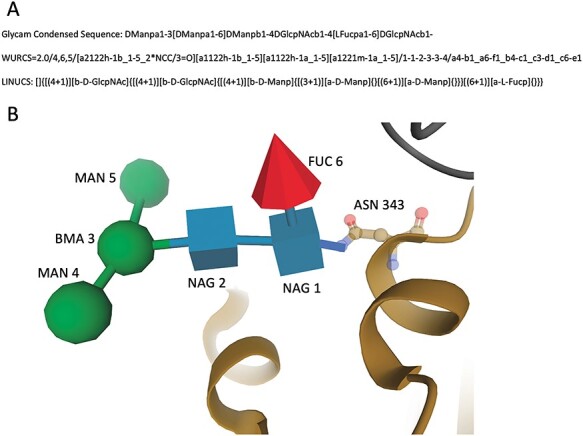
Example depiction of the fucosylated N-glycan core on SARS-CoV-2 spike glycoprotein (PDB ID 6WPS). (A) Linear descriptors are generated for branched entity 5 in three formats: Glycam Condensed Sequence, WURCS and LINUCS. (B) 3D SNFG view of the oligosaccharide chain N for branched entity 5 complexed with protein (brown).
As of February 2021, approximately, 90% of the current PDB holdings have been determined by MX. For these structures, it is not uncommon for certain portions of a biological macromolecule to be unresolvable in experimental electron density maps. Segments of polypeptide chains, nucleic acid strands or carbohydrates may be fully or partially missing from the atomic coordinates (i.e., they could not be modeled from available experimental data). If a particular monosaccharide was not represented within the atomic coordinates of the structure, it is not included in the branch entity. Partially modeled monosaccharides are also included in the branched entity as complete components, using the chemical identity provided by the structure depositor. When two monosaccharides are joined by a glycosidic bond, the connecting oxygen atom is by convention assigned to the monosaccharide having this atom as an alcoholic hydroxyl oxygen. For older PDB structures wherein the glycosidic oxygen was assigned to the residue as hemiacetal/hemiketal oxygen, the connecting oxygen atom was reassigned to the linked molecule during this remediation. For oligosaccharides with any glycosidic bond formed between the hemiacetals/hemiketals of one monosaccharide and the hemiacetals/hemiketals of another, the order of residue numbering is arbitrary because there is no reducing end monosaccharide. There are 443 such cases in the PDB archive, and these were reviewed manually to ensure that the representation is consistent among PDB structures. Finally, for structures with alternate conformations of different monosaccharides at the same position in an oligosaccharide, such as a free hemiacetal in equilibrium of both alpha- and beta-anomer conformation in PDB ID 5ELC (Heggelund et al. 2016), both conformations are identified as microheterogeneity in the branched entity. Systematic names and linear descriptors are generated based on the first conformer listed in the atomic coordinates. A single monosaccharide modeled without connection to another monosaccharide is defined as a nonpolymer ligand and is treated like any other nonpolymer ligand in the PDB.
Annotation of glycosylation sites
Review of the PDB archive revealed that as of July 2020, the PDB archive contained >7000 structures exhibiting N-glycosylation and >400 structures exhibiting O-glycosylation (Table II). Rarer cases of S-glycosylation and C-mannosylation also occur in the PDB (Table II). During remediation, all glycosylation annotations were reviewed and updated as required. Once the glycosylation types were verified and classified, controlled vocabulary in the PDBx/mmCIF dictionary was added for automated annotation in the new structures. Verification of a monosaccharide–amino acid covalent linkage at the glycosylation site was carried out using established glycosylation rules (Spiro 2002), publications referenced by papers reporting individual PDB structures and supporting experimental data.
Table II.
Summary of glycosylation types and sites in the PDB as of July 2020
| Types | Glycosylated amino acids | Number of PDB structures | Bond length range (99% coverage) |
|---|---|---|---|
| N-linked glycosylation | ASN/ARG | 7073 | 1.20–1.60 Å |
| O-linked glycosylation | SER/THR/TYR | 473 | 1.10–1.55 Å |
| S-linked glycosylation | CYS | 6 | 1.55–2.00 Å |
| C-linked mannosylation | TRP | 16 | 1.05–1.65 Å |
N-glycosylation of ASN residues, involving the process of transferring the oligosaccharide precursor en bloc from dolichol-pyrophosphate to nascent polypeptide by OST, forms a stereo-specific β-D-GlcpNAc-ASN N-glycosidic bond (Dempski and Imperiali 2002; Varki 2017a). The biochemical mechanism of glycan transfer is conserved among eukaryotes and bacteria (Mohorko et al. 2011; Napiorkowska et al. 2017; Varki 2017b; Harada et al. 2019). Therefore, the connecting glucosamine monosaccharide should be modeled in the beta anomeric form according to the biological process. Inconsistently modeled alpha form α-D-GlcpNAc (NDG) at N-glycosylation sites were deemed inaccurate and changed to β-D-GlcpNAc (NAG) during remediation. Chirality caveats were highlighted in atomic coordinate files and wwPDB validation reports for 200 carbohydrate-containing structures.
The small number of cases for which the monosaccharide–amino acid covalent linkage cannot be described using established rules (Spiro 2002) were verified by confirming both glycosylation linkage and the stereo specificity with the structure publication and supporting experimental data (i.e., experimental electron density map). For example, glycosylation on arginine by β-D-GlcpNAc (NAG) was not discussed in the Spirio’s influential review (Spiro 2002) but was documented in a publication describing PDB IDs 6AC0 and 6AC5 (Ding et al. 2019). The 1.45 Å resolution experimental electron density map for PDB ID 6AC5 shows clear evidence as to the identity of the monosaccharide and the stereo-specificity of its connection to ARG. Once a monosaccharide–amino acid covalent linkage at the glycosylation site is verified in a new PDB structure, it is then considered valid for all similar structures deposited thereafter, even in cases wherein unambiguous supporting experimental data are lacking. Following this procedure, monosaccharide–amino acid covalent linkages for N-glycosylation, O-glycosylation and C-mannosylation were identified and classified (Supplementary Table SII). C-mannosylation was observed in 16 PDB structures (e.g., PDB ID 6RUR) (Pedersen et al. 2019) and S-glycosylation was detected in six PDB structures (e.g., PDB ID 2MIJ) (Stepper et al. 2011; Garcia De Gonzalo et al. 2014).
Other types of glycosylation through a third or fourth monosaccharide component of an oligosaccharide with amino acids were also reviewed. For the avoidance of doubt, P-glycosylation through phosphate was not detected in any existing PDB structures. It was previously thought that glypiation through glycophosphatidylinositol (GPI) and ethanolamine was present in PDB IDs 5GJV and 5GJW (Wu et al. 2016). Upon further review of the electron density map, the evidence for covalent bonds to ethanolamine was deemed to be equivocal. Neither PDB ID was annotated as an example of glypiation during the remediation.
Detected glycosylation types for N-glycosylation, O-glycosylation, C-mannosylation and S-glycosylation were explicitly annotated in _struct_conn.pdbx_role using a controlled vocabulary. Glycosylation bond lengths have also been reviewed during this remediation. For example, among ~7000 PDB structures, the average length of C–N bonds between C1 of β-D-GlcpNAc (NAG) and ND2 of ASN was ~1.45 Å (SD ~ 0.05 Å). Bond lengths for the four types of amino acid glycosylation are enumerated in Table II. Although most of the observed glycosylation bonds in PDB structures fall within reasonable ranges, uncertainties in atomic positions in lower resolution MX structures made it difficult to confirm glycosylation linkages for some PDB structures, particularly those deposited to the archive before 2000. Annotation of glycosylation sites for lower resolution MX structures was based on the existing monosaccharide–amino acid covalent linkages recorded in struct_conn verified by depositors at the time of deposition and biocuration. For more recent PDB depositions, the RCSB PDB MAcromolecular Exchange and Input Tool (MAXIT) software (Feng 1996) automatically detects glycosylation linkages when the interatomic distance between the designated monosaccharide and an amino acids fall within a generously allowed range that encompasses 99% of observed bond lengths as specified in Table II. We note that most of the more recently deposited carbohydrate structures have chemically reasonable glycosylation bond lengths. We believe that this finding reflects the following: (i) better resolution of MX structures determined from cryogenically preserved samples; (ii) improvements in structure refinement programs that apply geometrically optimized restraints on saccharide conformers and glycosylation bonds (Lebedev et al. 2012; Agirre 2017) and (iii) wwPDB OneDep validation software identification of incorrect stereoisomers or improper glycosidic bonds corrected during OneDep structure deposition, validation and biocuration (Gore et al. 2017; Feng et al. 2021).
Biologically interesting molecules
The BIRD (Dutta et al. 2014) was originally created to remediate peptide inhibitors and antibiotics to provide both polymeric and single ligand information regarding sequence, chemistry, biology and structure of small molecules composed of multiple CCD components. The BIRD enables Findability of oligopeptides, etc., which are present in the PDB archive. As part of the carbohydrate remediation, the BIRD was extended to include oligosaccharides that are frequently referred to by a common name (e.g., maltose, sucrose, lactose and raffinose). Representation of these oligosaccharides was changed from single ligands to branched entities consisting of connected monosaccharides. BIRD definitions include trivial/common name, systematic name, list of sequence components/monosaccharides in the polymer/branched entity, connectivity of these sequence components/monosaccharides, chemical formula, SMILES and InChI chemical descriptors of the oligosaccharide and molecular function.
In aggregate, 134 new BIRD definitions for common oligosaccharides were created during this remediation. Dual representation of oligosaccharides in both oligomeric (branched entity) and single ligand forms present in the BIRD occurs in 130 PDB structures (e.g., see 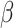 -lactose case illustrated in Figure 5). A branched entity consisting of two constituent monosaccharide components (with chemical formula and molecular weight) was adopted as the remediated BIRD definition (ID PRD_900004) of this disaccharide. Several cyclic oligosaccharides in the PDB are also represented in BIRD. PRD_900015 is the shortest alpha-cyclodextrin in the PDB, consisting of six α-D-Glcp (GLC) residues (Figure 5). Another class of BIRD molecule constituent encompasses the 20 blood group antigen terminal patterned oligosaccharides synthesized for binding studies with specific target proteins (Supplementary Table SIII). Lewis Y antigen (BIRD ID PRD_900015) is depicted in Figure 5 to exemplify how blood group antigens are now managed in the PDB.
-lactose case illustrated in Figure 5). A branched entity consisting of two constituent monosaccharide components (with chemical formula and molecular weight) was adopted as the remediated BIRD definition (ID PRD_900004) of this disaccharide. Several cyclic oligosaccharides in the PDB are also represented in BIRD. PRD_900015 is the shortest alpha-cyclodextrin in the PDB, consisting of six α-D-Glcp (GLC) residues (Figure 5). Another class of BIRD molecule constituent encompasses the 20 blood group antigen terminal patterned oligosaccharides synthesized for binding studies with specific target proteins (Supplementary Table SIII). Lewis Y antigen (BIRD ID PRD_900015) is depicted in Figure 5 to exemplify how blood group antigens are now managed in the PDB.
Fig. 5.
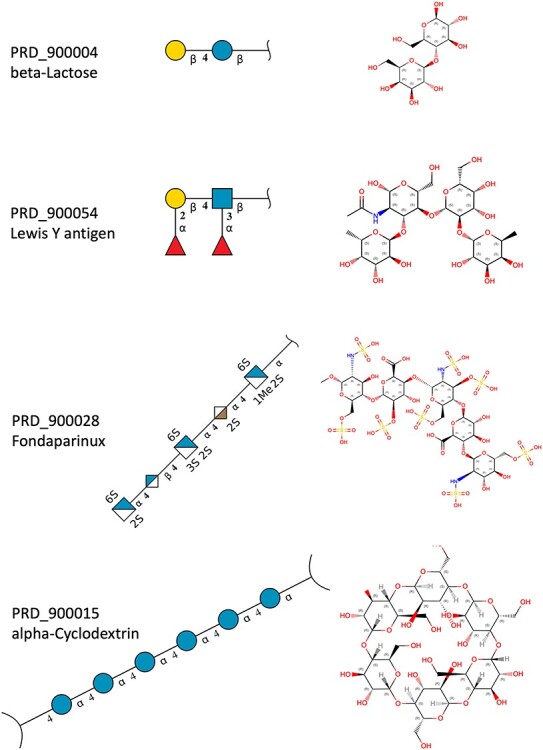
Examples of different classes of BIRD molecules: 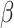 -Lactose, a well-known linear oligosaccharide; Lewis Y antigen, a blood group antigen; Fondaparinux, a US Food and Drug Administration-approved heparin-like anticoagulant drug;
-Lactose, a well-known linear oligosaccharide; Lewis Y antigen, a blood group antigen; Fondaparinux, a US Food and Drug Administration-approved heparin-like anticoagulant drug; 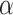 -cyclodextrin, a cyclic oligosaccharide. Two graphical representations are provided for each example, including 2D SNFG diagram of connected monosaccharides (as branched entity) and the chemical structure (as a single ligand).
-cyclodextrin, a cyclic oligosaccharide. Two graphical representations are provided for each example, including 2D SNFG diagram of connected monosaccharides (as branched entity) and the chemical structure (as a single ligand).
wwPDB validation report enhancement for carbohydrates
The growing number of PDB structures containing carbohydrates has increased awareness of the saccharide components (Agirre 2017; Varki 2017b; de Meirelles et al. 2020). wwPDB validation reports (Gore et al. 2017) have been enhanced to enable quality assessment for carbohydrates in PDB structures. Each oligosaccharide present in a branched entity is listed with both systematic name and 2D SNFG image. Residue-property plots for quality assessment at the branched entity chain level are provided for each oligosaccharide instance, with geometrical quality assessments computed at the residue level. Visualization of oligosaccharide validation (2D diagrams highlighting geometric validation criteria) and, for MX structures, 3D views of electron density maps and atomic model fit to experimental data are included (Feng et al. 2021).
Remediation outcomes for oligosaccharide-containing PDB structures
During remediation, 13,584 oligosaccharides were identified and converted to branched entities in 8282 PDB structures. Among these 13,584 oligosaccharide entities were 1706 unique oligosaccharides, including glycan conjugates, carbohydrate metabolites, heparin fragments, analogs of naturally occurring carbohydrates, and cyclodextrins. Tables III and IV provide breakdowns in terms of oligosaccharide size, type and branching. The longest oligosaccharide currently present in the PDB archive is a 36-mer heparin (PDB ID 3IRL) (Khan et al. 2010). The largest cyclic oligosaccharide is a 26-mer cycloamylose (PDB ID 1C58) (Gessler et al. 1999). Supplementary Table SIV lists the oligosaccharides commonly observed in the PDB. The most abundant oligosaccharides in the PDB are fragments of N-glycosylation glycans, such as the disaccharide DGlcpNAcb1-4DGlcpNAcb1 (Glycam Condensed Sequence of 1–4 linked β-D-GlcpNAc [NAG]) found in 3124 PDB structures, the trisaccharide DManpb1-4DGlcpNAcb1-4DGlcpNAcb1 (in 1056 PDB structures) and the pentasaccharide DManpa1–3[DManpa1–6]DManpb1-4DGlcpNAcb1-4DGlcpNAcb1 (in 431 PDB structures). These saccharides are all part of N-glycan cores (Varki 2017a). They were observed at different levels of completeness depending on the quality of experimental data used for structure determination. The reducing end β-D-GlcpNAc (NAG) is typically well visualized in experimental electron density maps (for MX structures) or in electric potential maps (for 3DEM structures) due to covalent linkage with the glycosylated protein. The longer the distance from the reducing end, the more flexible monosaccharides typically become, making them more difficult to discern from the experimental data.
Table III.
Carbohydrates in the PDB grouped by size (monosaccharide counts). These 13,584 oligosaccharides occur in 8,282 PDB entries
| Size of oligosaccharide (# of monosaccharides) | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11+ |
|---|---|---|---|---|---|---|---|---|---|---|
| # of oligosaccharide instances | 5763 | 2973 | 1600 | 1183 | 788 | 566 | 322 | 203 | 125 | 61 |
| # of unique oligosaccharides | 340 | 290 | 255 | 221 | 217 | 150 | 104 | 59 | 31 | 39 |
Table IV.
Carbohydrates in the PDB grouped by type and number of branches as of July 2020. These 13,584 oligosaccharides occur in 8,282 PDB entries
| Types | Cyclic | Noncyclic | ||||||
|---|---|---|---|---|---|---|---|---|
| # of branches | 0 | 1 | 0 | 1 | 2 | 3 | 4 | 5 |
| # of oligosaccharide instances | 85 | 2 | 9880 | 2648 | 877 | 84 | 5 | 3 |
| # of unique oligosaccharides | 12 | 2 | 971 | 454 | 220 | 40 | 5 | 2 |
The longest complex-type N-glycan represented in the PDB (as of July 2020) was a 16-mer oligosaccharide (PDB ID 5T3X, Figure 3) (Gristick et al. 2016). This glycosylated HIV-1 envelope glycoprotein trimer contains three proteins plus 18 oligosaccharide chains with 113 monosaccharide components. Previously, identifying each oligosaccharide and its monosaccharide components from inconsistent representation of carbohydrates in this atomic coordinate file was challenging, to say the least. Following remediation, each of the 18 oligosaccharide branched entities is labeled with a unique chain id (auth_asym_id) and is consistently represented. PDB data consumers can now unambiguously identify and examine each oligosaccharide and its monosaccharide components. Some PDB structures contain large numbers of oligosaccharides attached to many polypeptide chains. For example, PDB ID 5SZS (Walls et al. 2016) contains 62 oligosaccharide chains attached to 3 N-glycosylated protein chains at glycosylation sites; PDB IDs 6CDE and 6CDI (Xu et al. 2018) each have 51 oligosaccharide chains attached to 6 N-glycosylated protein chains and PDB ID 4YD9 (Gai et al. 2015) contains 50 oligosaccharide chains attached to 20 N-glycosylated protein chains. Before introduction of branched entities and proper biocuration of glycosylation types, it was difficult to find a specific glycan and its glycosylation site. Now PDB data consumers can easily identify the glycan-linked proteins and the composition of each glycan. Supplementary Table SV exemplifies the glycan sequence patterns observed at the glycosylation sites in the PDB.
As of December 2020, 163 structures of SARS-Cov-2 proteins with bound carbohydrates were present in the PDB archive; 32 were remediated in July 2020. The remainder were processed with an updated version of the wwPDB global OneDep system for deposition, validation and biocuration (Gore et al. 2017; Young et al. 2017; Young et al. 2018) that generated consistent branched entity representations. Following this remediation, complete chemical descriptions of the oligosaccharides present in SARS-CoV-2 spike glycoprotein structures (e.g., PDB IDs 6WPS) (Pinto et al. 2020) and 6XCM (Ramirez et al. 2019) ensure that users are able to fully appreciate how this target of antibodies is decorated with carbohydrates and how some of the bound sugars shield polypeptide chain epitopes from the host immune system.
Discussion
All carbohydrates (occurring in ~14,000 structures) present in the PDB archive have been remediated to a common biocuration standard using a uniform representation that follows IUPAC recommendations. Once all 1040 monosaccharide components were correctly classified, their atoms correctly named and chemistry consistently defined in the CCD, ~13,500 oligosaccharide instances could be organized into uniformly represented branched entities with explicit mapping between the constituents of the branched entity and the atomic coordinates. In addition, 134 BIRD definitions were created to provide alternative representations of branched oligosaccharides and chemical descriptions of carbohydrates present in 2059 PDB structures. Uniform representation of carbohydrates in the PDB now ensures that every carbohydrate-containing structure is properly validated and biocurated. Introduction of the branched entity biopolymer for carbohydrates now ensures that their biochemical composition and structural organization are fully and accurately described in the structure data files stored in the PDB.
Remediation rendered carbohydrate data in the PDB archive:
(1) Findable: Every unique monosaccharide has a unique chemical name and symbol with its own CCD code and every oligosaccharide has its own systematic name and linear descriptor with explicit enumeration and connectivity of monosaccharide components.
(2) Accessible: Multiple styles for monosaccharide and oligosaccharide identifiers have been adopted to make them both human and machine readable.
(3) Interoperable: This remediation adopted IUPAC recommendations, suggestions from community experts and community-accepted descriptors, thereby strengthening carbohydrate data exchange and interpretation.
(4) Reusable: Reference data have been reviewed for accuracy, and the schema for defining carbohydrates in the PDB is publicly available.
The new PDBx/mmCIF data standards introduced for carbohydrates are fully extensible. For example, should there be a new type of linear descriptor for oligosaccharides that is broadly accepted and its use recommended by community experts, it can be easily added. PDBx/mmCIF is also flexible, permitting facile extension to include new types of glycosylation within the controlled vocabulary of the dictionary and wwPDB biocuration processes. Uniform representation of carbohydrates across the archive enables large-scale structural bioinformatics studies of carbohydrates by PDB data consumers for the first time. Historically, significant postprocessing of PDB data was required to enable identification of carbohydrate-containing structures.
One caveat that PDB data consumers should be aware of is the accuracy of atomic coordinates for some carbohydrates represented in the PDB, particularly in regions of structures that were not well resolved during structure determination. In PDB ID 1B5F (Frazao et al. 1999), for example, the glycan sequence attached to ASN-257 of chain D was previously modeled with α-D-Manp (MAN) connected to the second β-GlcpNAc (NAG) from the glycosylation site. Remediation revealed that the atomic coordinates for the mannose moiety should have been built into the electron density map as β-D-Manp (BMA) (Frazao et al. 1999). During remediation, its chemical identity was updated from α-D-Manp (MAN) to β-D-Manp (BMA), and a caveat record was added to the structure data file warning that the anomeric configuration in the atomic coordinates is incorrect. Although we have attempted to identify and annotate such cases during the remediation, resource constraints made it impossible for us to verify every case at the atomic level due to lack of experimental data and/or publication references or resource limitations. Significantly improved management of carbohydrate containing structures during the wwPDB deposition, validation and biocuration process with the OneDep system will help ensure that few, if any, incorrect structures become part of the PDB archive going forward.
Some carbohydrate-containing compounds represented in the PDB were not addressed during this oligosaccharide-focused remediation. Heavily modified saccharides, for example, are treated as nonsaccharide ligands. Two common examples of such ligands are detergents with saccharide headpieces (e.g., 1,2-distearoyl-monogalactosyl-diglyceride; CCD ID LMG) and saccharide-carrier complexes (e.g., UDP-glucose; CCD ID UPG). Carbohydrate-containing compounds with nonstandard or noncontinuous glycosidic bonds were also excluded from remediation. For example, the commonly used antibiotic erythromycin A (CCD ID ERY) consists of a macrolide connecting two monosaccharides. Due to the complexity of such compounds, branched entity representation was deemed inappropriate. A better representation for such cases may be developed in the future, resources permitting.
Materials and methods
Remediation software tools
Since 2014, the wwPDB OneDep software system has supported web-based one-structure-at-a-time deposition (Young et al. 2017), validation (Gore et al. 2017; Smart et al. 2018a) and biocuration (Young et al. 2018), using an OneDep Workflow system that manages tasks within the data pipeline. The wwPDB supports data depositors (structural biologists) regionally with OneDep server instances at RCSB PDB (for the Americas and Oceania), PDBe (for Europe and Africa) and PDBj (for Asia and the Middle East). OneDep also supports structure data file versioning and ensures quality/completeness of incoming data (Shao et al. 2017; Smart et al. 2018b), benefiting many millions of PDB data consumers around the world.
An important advantage of the OneDep system accrues from inclusion of information regarding provenance. Each structure data file is audited and versioned whenever it is updated, ensuring that changes can be easily tracked and/or reverted. Another critical advantage of OneDep is enforcement of consistent biocuration practices across all wwPDB data centers by underlying backend software known as MAXIT (Feng 1996). The inhouse-developed MAXIT software suite was updated at the start of this remediation to identify carbohydrate components, standardize nomenclature according to the CCD and provide branched entity representation in a batch mode at RCSB PDB initially for data review.
Community engagement
Before carbohydrate data remediation began, requirements were set in consultation with glycoscience community experts. Workshops (e.g., IUPAC Joint Commission on Biochemical Nomenclature task force), professional society meetings and email communication with knowledgeable individuals were all used to solicit input. Consensus recommendations were as follows: standardizing nomenclature following IUPAC-IUBMB recommendations, making representation uniform so that glycosylation sites are findable, treating oligosaccharides as distinct entities and incorporating linear descriptors in common use by the glycoscience community. With the benefit of community feedback, a new PDBx/mmCIF data model for representing oligosaccharides (i.e., the branched entity) was proposed to the wwPDB PDBx/mmCIF Working Group and 3D visualization software developers for feedback in October 2018. In addition, we collaborated with the glycoscience community to incorporate their software for generating linear descriptors for oligosaccharides in Condensed IUPAC, WURCS and LINUCS formats. Once the new carbohydrate data model was finalized and defined in the PDBx/mmCIF dictionary, the MAXIT software tool was modified to provide uniform carbohydrate representation. Various example files of branched entity representations and CCD descriptors for monosaccharides were then provided via a public GitHub site for community review and incorporation into external software tools more than 6 months before the rollout of the remediated data. In parallel, plans for the remediation project were broadly disseminated using community mailing lists (structural biology, glycoscience and community software developers) at professional society meetings (e.g., Carbohydrates Gordon Research Conference, International Carbohydrate Symposium, American Crystallographic Association and American Society for Biochemistry and Molecular Biology) and were advertised on the wwPDB website (https://www.wwpdb.org/documentation/carbohydrate-remediation) and social media.
Remediation procedures
Remediation scripts were developed at RCSB PDB and shared with wwPDB partners. The scripts were then modified to conform to the setup at PDBe and PDBj. Each wwPDB data center ran these scripts for all relevant structures that were previously deposited and processed. The scripts read PDBx/mmCIF atomic coordinate files for each structure from OneDep, standardized nomenclature, generated branched representations, used community software to generate linear descriptors and then copied updated files back to OneDep with a new audit revision history and version number. Remediated files were then shared among wwPDB partners for quality assurance. Once the remediated data files were checked and any errors corrected, they were moved into to the public PDB archive. Announcements of these changes were broadcast to MX, NMR, 3DEM and PDB user communities. A public version of MAXIT software version 11+ was made available for download at the time of the carbohydrate remediation rollout (https://sw-tools.rcsb.org/apps/MAXIT/index.html).
Authors’ contributions
The carbohydrate remediation project is a wwPDB collaborative project that was carried out principally by RCSB PDB at Rutgers, the State University of New Jersey. Data analysis and assessments, chemical component standardization, creation of BIRD definitions, glycosylation rules setting, hands-on data remediation and file checking on remediated data were performed by C.S. Backend software MAXIT for carbohydrate identification, nomenclature standardization, branched representation, integration of community software to generate linear descriptors, OneDep User Interface for carbohydrate representation change and BIRD searching and creation, enhancements of carbohydrates in the validation reports and automated remediation scripts were developed by Z.F. Data extension of PDBx/mmCIF dictionary for branched group was done by J.D.W. and E.P. The modification of automated remediation scripts to conform to local setup at PDBe and PDBj were done by J.B. Generation of remediated files using automated scripts was carried out by Z.F. at RCSB PDB, J.B. at PDBe and Y.I. at PDBj. Project management was provided by J.Y.Y. Overall project direction was provided by J.Y.Y., S.K.B., S.V. and G.K. The article was written by C.S. and J.Y.Y. and edited by S.K.B.
Supplementary Material
Acknowledgements
First and foremost, we thank the tens of thousands of structural biologists who have deposited structures to the PDB since 1971 and the many millions of individuals from around the world who have used and continue to use PDB data. We also thank our collaborators, David Montgomery and Dr. Robert Woods at Complex Carbohydrate Research Center at the University of Georgia, USA, for contribution of GMML script prototype, Dr. Issaku Yamada and his team at The Noguchi Institute, Japan, for contribution of PDB2Glycan software and Thomas Lutteke at Glycosciences.de, Germany, for contribution of PDB-CARE software, for the generation of linear descriptors. Furthermore, we thank the wwPDB biocuration team for feedback on requirements setting and software testing and Li Chen and Vladimir Guranovic for data release to the FTP. Finally, we acknowledge Drs. Kim Henrick, Shuchismita Dutta and Helen M. Berman for earlier carbohydrate analysis efforts.
Contributor Information
Chenghua Shao, Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Zukang Feng, Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
John D Westbrook, Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA; Rutgers Cancer Institute of New Jersey, Robert Wood Johnson Medical School, New Brunswick, NJ 08903, USA.
Ezra Peisach, Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
John Berrisford, Protein Data Bank in Europe (PDBe), European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridgeshire CB10 1SD, UK.
Yasuyo Ikegawa, Protein Data Bank Japan (PDBj), Institute for Protein Research, Osaka University, Osaka 565-0871, Japan.
Genji Kurisu, Protein Data Bank Japan (PDBj), Institute for Protein Research, Osaka University, Osaka 565-0871, Japan.
Sameer Velankar, Protein Data Bank in Europe (PDBe), European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridgeshire CB10 1SD, UK.
Stephen K Burley, Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA; Rutgers Cancer Institute of New Jersey, Robert Wood Johnson Medical School, New Brunswick, NJ 08903, USA; Research Collaboratory for Structural Bioinformatics Protein Data Bank, San Diego Supercomputer Center, University of California, La Jolla, San Diego, CA 92093, USA; Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Jasmine Y Young, Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Funding
National Institutes of Health Common Fund Glycoscience Program through the National Cancer Institute cooperative agreement (U01 CA221216 to Dr. Robert Woods at Complex Carbohydrate Research Center at the University of Georgia in collaboration with Dr. J.Y.Y. as the RCSB PDB subawardee at Rutgers, the State University of New Jersey); National Science Foundation (DBI-1832184), the US Department of Energy (DE-SC0019749) and the National Cancer Institute, National Institute of Allergy and Infectious Diseases and National Institute of General Medical Sciences of the National Institutes of Health (R01GM133198) to RCSB PDB; European Molecular Biology Laboratory-European Bioinformatics Institute and Wellcome Trust (104948 to PDBe); Database Integration Coordination Program from the National Bioscience Database Center (NBDC)-Japan Science and Technology Agency (JST), the Platform Project for Supporting in Drug Discovery and Life Science Research from Japan Agency for Medical Research and Development (AMED) and the joint usage program of Institute for Protein Research, Osaka University (PDBj).
Conflict of interest statement
None declared.
Abbreviations
BIRD, Biologically Interesting Molecules Reference Dictionary; CCD, Chemical Component Dictionary; MAXIT, MAcromolecular Exchange and Input Tool; PDB, Protein Data Bank; PDBe, Protein Data Bank in Europe; PDBj, Protein Data Bank Japan; PDBx/mmCIF, PDB Exchange MacroMolecular Crystallographic Information File; RCSB PDB, Research Collaboratory for Structural Bioinformatics Protein Data Bank; wwPDB, Worldwide Protein Data Bank.
Data availability statement
All remediated data files, including atomic coordinate files, chemical component definitions, BIRD definitions and wwPDB validation reports, are accessible at the PDB archive, https://ftp.wwpdb.org/pub/pdb/ and wwPDB website via PDB DOI link for each structure, e.g., https://www.wwpdb.org/pdb?id=pdb_0000[PDB 4-letter ID] (DOI: 10.2210/pdb[PDB ID]/pdb). In addition, preremediated and remediated atomic coordinate files are versioned at the PDB versioned archive, https://ftp-versioned.wwpdb.org/pdb_versioned/data/.
References
- Agirre J. 2017. Strategies for carbohydrate model building, refinement and validation. Acta Crystallogr D Struct Biol. 73:171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong DR, Berrisford JM, Conroy MJ, Gutmanas A, Anyango S, Choudhary P, Clark AR, Dana JM, Deshpande M, Dunlop R, et al. 2020. Pdbe: Improved findability of macromolecular structure data in the pdb. Nucleic Acids Res. 48:D335–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H. 2003. Announcing the worldwide protein data bank. Nat Struct Biol. 10:980. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, et al. 2019. Rcsb protein data bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 47:D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow G, Christie CH, Dalenberg K, Costanzo LD, Duarte JM, et al. 2021. Rcsb protein data bank: Powerful new tools for exploring 3d structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering, and energy sciences. Nucleic Acid Res. 49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Yarovoi SV, Zhu Z, Rauova L, Hayes V, Lebedeva T, Liu Q, Poncz M, Arepally G, Cines DB, et al. 2015. Atomic description of the immune complex involved in heparin-induced thrombocytopenia. Nat Commun. 6:8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casset F, Hamelryck T, Loris R, Brisson JR, Tellier C, Dao-Thi MH, Wyns L, Poortmans F, Perez S, Imberty A. 1995. Nmr, molecular modeling, and crystallographic studies of lentil lectin-sucrose interaction. J Biol Chem. 270:25619–25628. [DOI] [PubMed] [Google Scholar]
- Copoiu L, Torres PHM, Ascher DB, Blundell TL, Malhotra S. 2020. Procarbdb: A database of carbohydrate-binding proteins. Nucleic Acids Res. 48:D368–D375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meirelles JL, Nepomuceno FC, Pena-Garcia J, Schmidt RR, Perez-Sanchez H, Verli H. 2020. Current status of carbohydrates information in the protein data bank. J Chem Inf Model. 60:684–699. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J. 1981. Crystallographic refinement and atomic models of a human fc fragment and its complex with fragment-b of protein-a from staphylococcus-aureus at 2.9-a and 2.8-a resolution. Biochemistry. 20:2361–2370. [PubMed] [Google Scholar]
- Dempski RE Jr, Imperiali B. 2002. Oligosaccharyl transferase: Gatekeeper to the secretory pathway. Curr Opin Chem Biol. 6:844–850. [DOI] [PubMed] [Google Scholar]
- Ding JJ, Pan X, Du, Yao Q, Xue J, Yao HW, Wang DC, Li S, Shao F. 2019. Structural and functional insights into host death domains inactivation by the bacterial arginine glcnacyltransferase effector. Mol Cell. 74:922–935.e6. [DOI] [PubMed] [Google Scholar]
- Dutta S, Dimitropoulos D, Feng Z, Persikova I, Sen S, Shao C, Westbrook J, Young J, Zhuravleva MA, Kleywegt GJ, et al. 2014. Improving the representation of peptide-like inhibitor and antibiotic molecules in the protein data bank. Biopolymers. 101:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. 1996. Heparin structure and interactions with basic fibroblast growth factor. Science. 271:1116–1120. [DOI] [PubMed] [Google Scholar]
- Feng, Z (1996). Maxit: Macromolecular Exchange and Input Tool, https://sw-tools.rcsb.org/apps/MAXIT/index.html.
- Feng Z, Verdiguel N, Di Costanzo L, Goodsell DS, Westbrook JD, Burley SK, Zardecki C. 2020. Impact of the protein data bank across scientific disciplines. Data Sci J. 19:1–14. [Google Scholar]
- Feng Z, Westbrook JD, Sala R, Smart OS, Bricogne G, Matsubara M, Yamada I, Tsuchiya S, Aoki-Kinoshita KF, Hoch JC, et al. 2021. Enhanced validation of small-molecule ligands and carbohydrates in the protein data bank. Structure. 29:393–400.e1. doi: 10.1016/j.str.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazao C, Bento I, Costa J, Soares CM, Verissimo P, Faro C, Pires E, Cooper J, Carrondo MA. 1999. Crystal structure of cardosin a, a glycosylated and arg-gly-asp-containing aspartic proteinase from the flowers of cynara cardunculus l. J Biol Chem. 274:27694–27701. [DOI] [PubMed] [Google Scholar]
- Gai Z, Matsuno A, Kato K, Kato S, Khan MRI, Shimizu T, Yoshioka T, Kato Y, Kishimura H, Kanno G, et al. 2015. Crystal structure of the 3.8-mda respiratory supermolecule hemocyanin at 3.0 a resolution. Structure. 23:2204–2212. [DOI] [PubMed] [Google Scholar]
- Garcia De Gonzalo CV, Zhu L, Oman TJ, van der Donk WA. 2014. Nmr structure of the s-linked glycopeptide sublancin 168. ACS Chem Biol. 9:796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler K, Uson I, Takaha T, Krauss N, Smith SM, Okada S, Sheldrick GM, Saenger W. 1999. V-amylose at atomic resolution: X-ray structure of a cycloamylose with 26 glucose residues (cyclomaltohexaicosaose). Proc Natl Acad Sci U S A. 96:4246–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, Zardecki C, Di Costanzo L, Duarte JM, Hudson BP, Persikova I, Segura J, Shao C, Voigt M, Westbrook JD, et al. 2020. Rcsb protein data bank: Enabling biomedical research and drug discovery. Protein Sci. 29:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore S, Sanz Garcia E, Hendrickx PMS, Gutmanas A, Westbrook JD, Yang H, Feng Z, Baskaran K, Berrisford JM, Hudson BP, et al. 2017. Validation of structures in the protein data bank. Structure. 25:1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick HB, von Boehmer L, West AP Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, et al. 2016. Natively glycosylated hiv-1 env structure reveals new mode for antibody recognition of the cd4-binding site. Nat Struct Mol Biol. 23:906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Ohkawa Y, Kizuka Y, Taniguchi N. 2019. Oligosaccharyltransferase: A gatekeeper of health and tumor progression. Int J Mol Sci. 20:6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggelund JE, Burschowsky D, Bjornestad VA, Hodnik V, Anderluh G, Krengel U. 2016. High-resolution crystal structures elucidate the molecular basis of cholera blood group dependence. PLoS Pathog. 12:e1005567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, et al. 2008. Remediation of the protein data bank archive. Nucleic Acids Res. 36:D426–D433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. 2012. Cell attachment protein vp8* of a human rotavirus specifically interacts with a-type histo-blood group antigen. Nature. 485:256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JA, Sielecki AR, Sykes BD, James MN, Phillips DC. 1979. X-ray crystallography of the binding of the bacterial cell wall trisaccharide nam-nag-nam to lysozyme. Nature. 282:875–878. [DOI] [PubMed] [Google Scholar]
- Khan S, Gor J, Mulloy B, Perkins SJ. 2010. Semi-rigid solution structures of heparin by constrained x-ray scattering modelling: New insight into heparin-protein complexes. J Mol Biol. 395:504–521. [DOI] [PubMed] [Google Scholar]
- Kinjo AR, Bekker GJ, Wako H, Endo S, Tsuchiya Y, Sato H, Nishi H, Kinoshita K, Suzuki H, Kawabata T, et al. 2018. New tools and functions in data-out activities at Protein Data Bank Japan (PDBj). Protein Sci. 27:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Dutta S, Westbrook JD, Henrick K, Berman HM. 2008. Representation of viruses in the remediated pdb archive. Acta Cryst D. D64:874–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, van Montfort R, Strokopytov B, Rozeboom HJ, Kalk KH, de Vries GE, Penninga D, Dijkhuizen L, Dijkstra BW. 1994. Nucleotide sequence and x-ray structure of cyclodextrin glycosyltransferase from bacillus circulans strain 251 in a maltose-dependent crystal form. J Mol Biol. 236:590–600. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. 2012. Jligand: A graphical tool for the ccp4 template-restraint library. Acta Cryst D. 68:431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutteke T, von der Lieth CW. 2004. Pdb-care (pdb carbohydrate residue check): A program to support annotation of complex carbohydrate structures in pdb files. BMC Bioinform. 5:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. 1998. Recommendations for the presentation of nmr structures of proteins and nucleic acids. Iupac-iubmb-iupab inter-union task group on the standardization of data bases of protein and nucleic acid structures determined by nmr spectroscopy. J Biomol NMR. 12:1–23. [DOI] [PubMed] [Google Scholar]
- Markosian C, Di Costanzo L, Sekharan M, Shao C, Burley SK, Zardecki C. 2018. Analysis of impact metrics for the protein data bank. Sci Data. 5:180212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth JD. 2008. A unified vision of the building blocks of life. Nat Cell Biol. 10:1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M, Aoki-Kinoshita KF, Aoki NP, Yamada I, Narimatsu H. 2017. Wurcs 2.0 update to encapsulate ambiguous carbohydrate structures. J Chem Inf Model. 57:632–637. [DOI] [PubMed] [Google Scholar]
- McNaught AD. 1996. International union of pure and applied chemistry and international union of biochemistry and molecular biology—Joint commission on biochemical nomenclature—Nomenclature of carbohydrates—Recommendations 1996. Pure Appl Chem. 68:1919–2008. [DOI] [PubMed] [Google Scholar]
- Mir S, Alhroub Y, Anyango S, Armstrong DR, Berrisford JM, Clark AR, Conroy MJ, Dana JM, Deshpande M, Gupta D, et al. 2018. PDBe: Towards reusable data delivery infrastructure at protein data bank in Europe. Nucleic Acids Res. 46:D486–D492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra AK. 2019. Visualization of biological macromolecules at near-atomic resolution: Cryo-electron microscopy comes of age. Acta Crystallogr F Struct Biol Commun. 75:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohorko E, Glockshuber R, Aebi M. 2011. Oligosaccharyltransferase: The central enzyme of n-linked protein glycosylation. J Inherit Metab Dis. 34:869–878. [DOI] [PubMed] [Google Scholar]
- Moonens K, Gideonsson P, Subedi S, Bugaytsova J, Romao E, Mendez M, Norden J, Fallah M, Rakhimova L, Shevtsova A, et al. 2016. Structural insights into polymorphic abo glycan binding by Helicobacter pylori. Cell Host Microbe. 19:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napiorkowska M, Boilevin J, Sovdat T, Darbre T, Reymond JL, Aebi M, Locher KP. 2017. Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat Struct Mol Biol. 24:1100–1106. [DOI] [PubMed] [Google Scholar]
- Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lutteke T, O'Boyle N, Packer NH, Stanley P, Toukach P, et al. 2019. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 29:620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen DV, Gadeberg TAF, Thomas C, Wang Y, Joram N, Jensen RK, Mazarakis SMM, Revel M, El Sissy C, Petersen SV, et al. 2019. Structural basis for properdin oligomerization and convertase stimulation in the human complement system. Front Immunol. 10:2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 583:290–295. [DOI] [PubMed] [Google Scholar]
- Protein Data Bank . 1971. Crystallography: Protein data bank. Nature (London), New Biol. 233:223–223.16063295 [Google Scholar]
- Ramirez AS, Kowal J, Locher KP. 2019. Cryo-electron microscopy structures of human oligosaccharyltransferase complexes ost-a and ost-b. Science. 366:1372–1375. [DOI] [PubMed] [Google Scholar]
- Rose Y, Duarte JM, Lowe R, Segura J, Bi C, Bhikadiya C, Chen L, Rose AS, Bittrich S, Burley SK, et al. 2020. RCSB protein data bank: Architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. J Mol Biol. 433:166704–166712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D, Rose A, Koca J, Burley S, Velankar S. 2018. Mol*: Towards a common library and tools for web molecular graphics. In: Proceedings of the Workshop on Molecular Graphics and Visual Analysis of Molecular Data. Brno, Czech Republic: Eurographics Association. p. 29–33. [Google Scholar]
- Shao C, Yang H, Westbrook JD, Young JY, Zardecki C, Burley SK. 2017. Multivariate analyses of quality metrics for crystal structures in the protein data bank archive. Structure. 25:458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Zhang F, Kemp MM, Linhardt RJ, Waisman DM, Head JF, Seaton BA. 2006. Crystallographic analysis of calcium-dependent heparin binding to annexin a2. J Biol Chem. 281:31689–31695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim L, Quezada-Calvillo R, Sterchi EE, Nichols BL, Rose DR. 2008. Human intestinal maltase-glucoamylase: Crystal structure of the n-terminal catalytic subunit and basis of inhibition and substrate specificity. J Mol Biol. 375:782–792. [DOI] [PubMed] [Google Scholar]
- Smart OS, Horsky V, Gore S, Svobodova Varekova R, Bendova V, Kleywegt GJ, Velankar S. 2018a. Validation of ligands in macromolecular structures determined by x-ray crystallography. Acta Crystallogr D Struct Biol. 74:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart OS, Horsky V, Gore S, Svobodova Varekova R, Bendova V, Kleywegt GJ, Velankar S. 2018b. Worldwide protein data bank validation information: Usage and trends. Acta Crystallogr D Struct Biol. 74:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro RG. 2002. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 12:43R–56R. [DOI] [PubMed] [Google Scholar]
- Stepper J, Shastri S, Loo TS, Preston JC, Novak P, Man P, Moore CH, Havlicek V, Patchett ML, Norris GE. 2011. Cysteine s-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 585:645–650. [DOI] [PubMed] [Google Scholar]
- Thieker DF, Hadden JA, Schulten K, Woods RJ. 2016. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 26:786–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeyer M, Aoki K, Paulson J, Cummings RD, York WS, Karlsson NG, Lisacek F, Packer NH, Campbell MP, Aoki NP, et al. 2017. Glytoucan: An accessible glycan structure repository. Glycobiology. 27:915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Aoki NP, Shinmachi D, Matsubara M, Yamada I, Aoki-Kinoshita KF, Narimatsu H. 2017. Implementation of glycanbuilder to draw a wide variety of ambiguous glycans. Carbohydr Res. 445:104–116. [DOI] [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. 2008. Biomagresbank. Nucleic Acids Res. 36:D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Aalst WMP, Bichler M, Heinzl A. 2017. Responsible data science. Bus Inf Syst Eng. 59:311–313. [Google Scholar]
- Varki A. 2017a. Essentials of Glycobiology. 3rd ed ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Varki A. 2017b. Biological roles of glycans. Glycobiology. 27:3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Tortorici MA, Frenz B, Snijder J, Li W, Rey FA, DiMaio F, Bosch BJ, Veesler D. 2016. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 23:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. 1953. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 171:737–738. [DOI] [PubMed] [Google Scholar]
- Westbrook JD, Burley SK. 2019. How structural biologists and the protein data bank contributed to recent FDA new drug approvals. Structure. 27:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook JD, Fitzgerald PMD. 2009. In: Bourne PE, Gu J, editors. editorsStructural Bioinformatics. 2nd ed ed. Hoboken (NJ): John Wiley & Sons, Inc. p. 271–291. [Google Scholar]
- Westbrook JD, Shao C, Feng Z, Zhuravleva M, Velankar S, Young J. 2015. The chemical component dictionary: Complete descriptions of constituent molecules in experimentally determined 3D macromolecules in the protein data bank. Bioinformatics. 31:1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook JD, Soskind R, Hudson BP, Burley SK. 2020. Impact of protein data bank on anti-neoplastic approvals. Drug Discov Today. 25:837–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, et al. 2016. The fair guiding principles for scientific data management and stewardship. Sci Data. 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter WT, Smith PJ, Arnott S. 1975. Hyaluronic acid: Structure of a fully extended 3-fold helical sodium salt and comparison with the less extended 4-fold helical forms. J Mol Biol. 99:219–235. [DOI] [PubMed] [Google Scholar]
- Woods RJ. 2005. (2005–2020) Glycam Web. Athens, Georgia: Complex Carbohydrate Research Center, University of Georgia. (http://www.Glycam.Com). [Google Scholar]
- Wu, Yan Z, Li Z, Qian X, Lu, Dong M, Zhou Q, Yan N. 2016. Structure of the voltage-gated calcium channel ca(v)1.1 at 3.6 a resolution. Nature. 537:191–196. [DOI] [PubMed] [Google Scholar]
- wwPDB Consortium . 2019. Protein data bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O'Dell S, Rawi R, Sastry M, et al. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of hiv-1. Nat Med. 24:857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Mazumder R, Ranzinger R, Edwards N, Kahsay R, Aoki-Kinoshita KF, Campbell MP, Cummings RD, Feizi T, Martin M, et al. 2020. GlyGen: Computational and informatics resources for glycoscience. Glycobiology. 30:72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JY, Westbrook JD, Feng Z, Peisach E, Persikova I, Sala R, Sen S, Berrisford JM, Swaminathan GJ, Oldfield TJ, et al. 2018. Worldwide protein data bank biocuration supporting open access to high-quality 3D structural biology data. Database. 2018:bay002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JY, Westbrook JD, Feng Z, Sala R, Peisach E, Oldfield TJ, Sen S, Gutmanas A, Armstrong DR, Berrisford JM, et al. 2017. Onedep: Unified wwpdb system for deposition, biocuration, and validation of macromolecular structures in the pdb archive. Structure. 25:536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All remediated data files, including atomic coordinate files, chemical component definitions, BIRD definitions and wwPDB validation reports, are accessible at the PDB archive, https://ftp.wwpdb.org/pub/pdb/ and wwPDB website via PDB DOI link for each structure, e.g., https://www.wwpdb.org/pdb?id=pdb_0000[PDB 4-letter ID] (DOI: 10.2210/pdb[PDB ID]/pdb). In addition, preremediated and remediated atomic coordinate files are versioned at the PDB versioned archive, https://ftp-versioned.wwpdb.org/pdb_versioned/data/.


