Abstract
Antimicrobial resistance genes, including multidrug efflux pumps, evolved long before the ubiquitous use of antimicrobials in medicine and infection control. Multidrug efflux pumps often transport metabolites, signals and host-derived molecules in addition to antibiotics or biocides. Understanding their ancestral physiological roles could inform the development of strategies to subvert their activity. In this study, we investigated the response of Acinetobacter baumannii to polyamines, a widespread, abundant class of amino acid-derived metabolites, which led us to identify long-chain polyamines as natural substrates of the disinfectant efflux pump AmvA. Loss of amvA dramatically reduced tolerance to long-chain polyamines, and these molecules induce expression of amvA through binding to its cognate regulator AmvR. A second clinically-important efflux pump, AdeABC, also contributed to polyamine tolerance. Our results suggest that the disinfectant resistance capability that allows A. baumannii to survive in hospitals may have evolutionary origins in the transport of polyamine metabolites.
Subject terms: Antibiotics, Bacteriology, Bacterial genetics
Francesca Short et al. identify natural polyamines as substrates for a drug efflux pump in the clinically-relevant pathogen, Acinetobacter baumannii. Their results suggest that efflux pumps now known to be involved in drug resistance may have originated as polyamine transporters.
Introduction
Antimicrobial resistance (AMR) is a critical public health challenge of the twenty-first century. Infections caused by antibiotic-resistant bacteria are predicted to cause 10 million annual deaths by 2050 if urgent action is not taken1. Though extensive AMR is a clinical phenomenon, the genes responsible have deep evolutionary origins long predating the use of antibiotics in medicine2,3. Naturally occurring antibiotics are typically found at subinhibitory levels, while synthetic drugs and biocides are not present in natural (unpolluted) microbial environments at all. As such, AMR genes are proposed to play additional roles, for example, in bacterial signalling, metabolism and virulence2,4,5. Multidrug efflux pumps (MDEPs) are an important category of AMR determinant, and often have core physiological functions6,7. These functions broadly fall into removal of harmful exogenous molecules (for example, mammalian antimicrobial peptides or plant flavonoids) or secretion of endogenous molecules (such as siderophores, quorum-sensing signals or metabolites)2,6,7. Understanding the origins and ancestral functions of AMR genes, including MDEPs, could help to predict their future evolution or point to new ways to subvert their activity.
Acinetobacter baumannii is a notorious opportunistic pathogen classified within the ‘ESKAPE’ group of bacterial species that are responsible for the majority of antibiotic-resistant infections8,9. Most A. baumannii infections are caused by two dominant multidrug-resistant lineages (designated ‘international clonal lineages’ ICLs 1 and 2), which have spread worldwide10. A. baumannii has a particular ability to survive for prolonged times in hospital environments11, and this is due in part to high disinfectant tolerance conferred by its repertoire of MDEPs11,12. Multiple A. baumannii MDEPs contribute to disinfectant resistance, for example, AdeABC13, AdeIJK13, AmvA14, AceI15 and AbeS16, all confer resistance to one or both of the widely used disinfectants chlorhexidine and benzalkonium chloride. Though disinfectant resistance is an adaptive advantage for the modern clinical era, many A. baumannii efflux pump genes are conserved across the species or genus, suggesting a shared primordial function2,12. Recently it was shown that the physiological substrates of AceI—a chlorhexidine efflux pump encoded in the core genome of A. baumannii15,17—are likely to be short-chain polyamines18.
Polyamines are an ancient class of metabolites comprising two or more amine moieties connected by aliphatic chain(s); the most common biological polyamines being putrescine, cadaverine, spermidine and spermine (Fig. 1a)19. These molecules have central roles in all three kingdoms of life and can be present intracellularly at high (mM) concentrations19,20. In bacteria, polyamines have been implicated in species-specific functions that include biofilm formation, cell growth, oxidative stress resistance and nitrogen storage, among others21. Many pathogenic bacteria depend on polyamine synthesis or import for their pathogenesis22–25. Polyamines can exert their functions in bacteria directly by virtue of their general biochemical properties or they can serve as signals, which act through specific receptors even at low concentrations24,26. Whether produced endogenously or present in the environment, polyamines can be toxic in high amounts. Several efflux systems have been reported to facilitate polyamine transport, such as members of the small multidrug resistance27, major facilitator superfamily (MFS)28, and PACE families18.
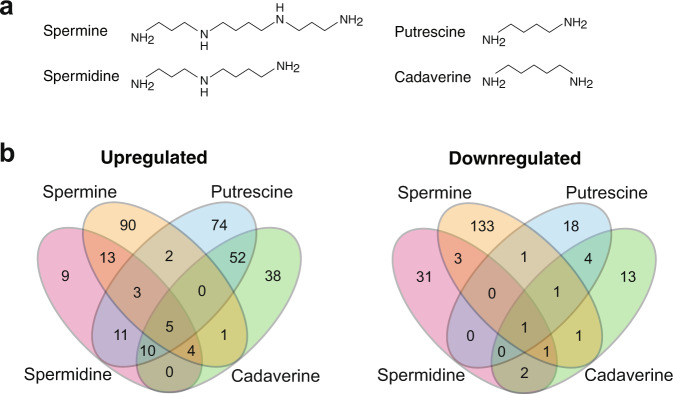
Fig. 1. Distinct regulons of spermine, spermidine, putrescine and cadaverine.
a Structures of the four polyamines used in this study. b Venn diagram of upregulated and downregulated gene sets. Note that some targets showed expression changes in opposite directions following treatment with different polyamines, as shown in Supplementary Fig. 1.
In this study, we have investigated the transcriptomic response of A. baumannii to high levels of the four major biological polyamines (putrescine, cadaverine, spermidine and spermine), with a view to defining their physiological roles and identifying transporters responsible for efflux of these molecules. The efflux pump genes aceI, adeABC and amvA were all strongly induced by different polyamines. We show that AdeABC and AmvA are required for tolerance to the long-chain polyamines spermine and spermidine and demonstrate increased spermidine accumulation in cells lacking the MFS transporter AmvA. Finally, we also show that spermine and spermidine induce amvA expression and bind to its repressor, AmvR. Our results strongly suggest that long-chain polyamines are physiological substrates of the conserved A. baumannii efflux pump AmvA.
Results
RNA sequencing (RNA-Seq) of A. baumannii following polyamine shock
We performed transcriptomics on the widely used, ICL1 A. baumannii strain AB5075-UW following exposure to high levels of exogenous polyamines. The minimum inhibitory concentrations (MICs) of all molecules were very high at 10 mg ml−1 for spermine and 40 mg ml−1 for spermidine, putrescine and cadaverine. RNA was extracted from duplicate log-phase A. baumannii AB5075-UW cultures supplemented with putrescine di-hydrochloride, cadaverine di-hydrochloride, spermidine tri-hydrochloride or spermine tetra-hydrochloride at 1/8 MIC. RNA-Seq reads were mapped, normalised and fold-changes calculated as described (see ‘Methods’). Each biological replicate gave rise to >10 million reads with >98% mapping to the A. baumannii AB5075-UW genome (Supplementary Table 1). Genes showing significantly altered expression in the presence of polyamines were defined as those with log2 fold change >1 or <−1 and a corrected p value <0.05 relative to the control.
Individual polyamines induce distinct transcriptional responses
A total of 499 genes showed altered expression following treatment with one or more polyamines, with individual regulons ranging from 93 (spermidine) to 259 (spermine) genes (Fig. 1b, Supplementary Fig 1A and Supplementary Data 1). The diamines putrescine and cadaverine each caused upregulation of a large number of genes (157 and 110, respectively) and downregulation of relatively few (25 and 23). For the tetraamine spermine and triamine spermidine, the number of genes showing increased and decreased expression was more balanced (spermine: 118 up/141 down; spermidine: 55 up/38 down). Though the majority of gene expression changes were specific to just one polyamine (372 genes), there was substantial overlap in the genes regulated by putrescine and cadaverine, which had 73 common targets (Fig. 1b and Supplementary Fig 1A). Interestingly, putrescine and spermine showed divergent regulation of 21 genes. These genes included ABUW_0068–71 (involved in amino acid metabolism), ABUW_2096–2099 (fatty acid metabolism) and ABUW_2448–2456 (fatty acid metabolism). Nine genes were differentially expressed with all four polyamines: the adeABC efflux pump genes, the transcriptional regulator amvR (ABUW_1678; also called smvR), and ABUW_0233 were induced, the periplasmic OB-fold protein-encoding ABUW_1352 was repressed, and three genes (ABUW_2448, ABUW_2449 and ABUW-2453) were induced by putrescine, cadaverine and spermidine but repressed by spermine.
Functional categories of polyamine-responsive genes
A summary of the clusters of orthologous groups (COG) functional classifications of polyamine-responsive genes is shown in Fig. 2a. Enrichment of specific gene ontology (GO) terms within the polyamine-regulated genes was tested using the TopGO package29. A summary of polyamine-regulated genes of interest is given in Supplementary Table 2.
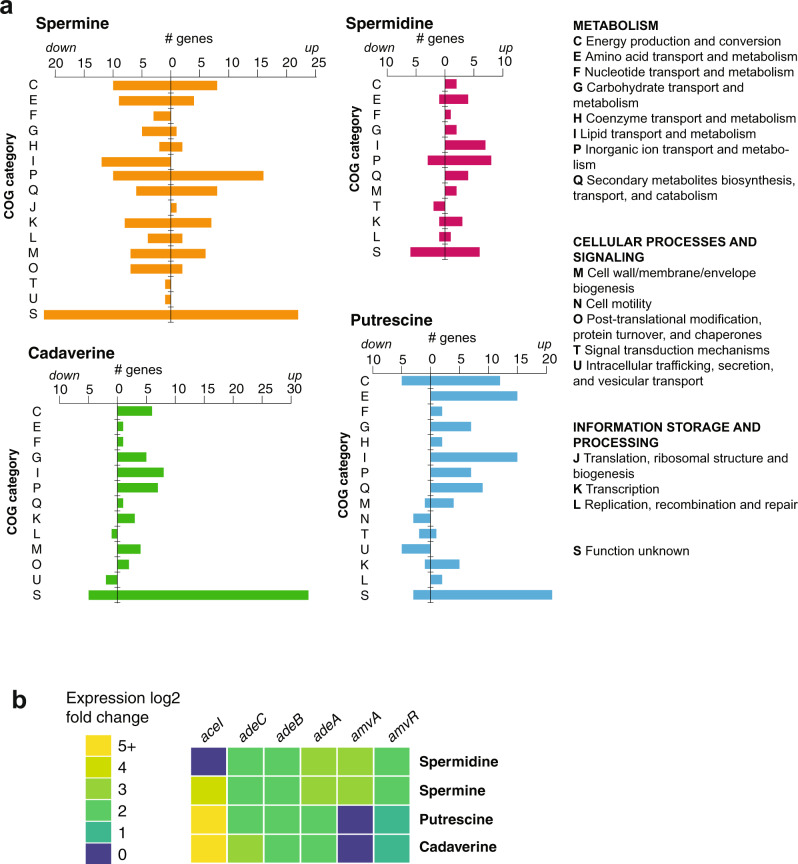
Fig. 2. Polyamine-regulated gene functional categories and phenotypes.
a Functional categories of genes regulated by spermine, spermidine, putrescine and cadaverine. COG categories of the genes showing expression changes in response to each molecule are shown. Regulated genes came from multiple functional classes, and lipid/inorganic ion transport and metabolism genes were highly represented. b Discontinuous heatmap showing induction of the efflux pump genes aceI (PACE family), adeABC (RND family) and amvA (MFS family) and its regulator amvR by different polyamines.
Frequent targets included metabolic genes, particularly those involved in energy production and conversion, lipid transport and metabolism, inorganic ion transport and amino acid metabolism (COGs C, I, P and E, respectively). Transcription genes (COG K) were also common polyamine-responsive targets, particularly with spermine. As expected, given that polyamines are amidated molecules and their metabolism is tightly linked to that of amino acids, several operons involved in utilisation of amino acids were regulated by polyamine shock. This included genes for histidine utilisation (induced by putrescine and spermidine), leucine metabolism genes ABUW_2453–5 (induced by putrescine and cadaverine, repressed by spermine) and many amino acid transporters. Polyamine shock also regulated genes involved in fatty acid utilisation such as genes of the ato butyrate/acetoacetate degradation operon (induced by putrescine and cadaverine), ABUW_1572-4 (induced by spermidine, putrescine and cadaverine) and the ABUW_2447–58 operon for leucine and fatty acid degradation.
Regulation of virulence genes and phenotypes in response to polyamine shock
Virulence of A. baumannii depends on multiple factors, including secretion systems, pili, siderophores, efflux pumps and other defence systems30. Multiple genes from virulence-related pathways were induced or repressed by exogenous polyamines (Supplementary Table 2). Many iron-acquisition genes were polyamine-responsive, such as the siderophore synthesis and uptake genes induced by spermidine and spermine. In addition, polyamines downregulated the expression of several virulence-related cell surface structures including the csu pilus (suppressed by cadaverine), genes of the Type VI secretion system used for interbacterial competition (repressed by cadaverine and spermidine) and Type IV secretion system and competence genes (suppressed by putrescine). Polyamine-regulated stress resistance genes included those for copper resistance (induced by spermine, putrescine and cadaverine), heavy metal resistance (induced by cadaverine and spermine) and hydrogen peroxide resistance. Finally, polyamines appeared to affect the expression of genes involved in horizontal gene transfer, as well as horizontally acquired elements themselves. In addition to the Type IV pili mentioned above, which are required for horizontal gene transfer in A. baumannii30, some enzymes of the CRISPR-Cas phage resistance locus were repressed by spermine, and the expression of genes within the predicted prophage regions31 was variably affected by spermidine and putrescine (Supplementary Table 2).
To determine whether the distinct transcriptional effects of exogenous polyamines were associated with changes in virulence-related phenotypes, serum resistance and biofilm formation assays were performed (Supplementary Fig 1B, C). A. baumannii AB5075-UW showed a delayed susceptibility to serum killing, with a loss of viability between 1 and 3 h (Supplementary Fig 1B). Spermidine and spermine at 1/8 MIC both caused a drastic reduction in serum survival that was apparent after 30 min, while putrescine or cadaverine supplementation increased serum survival to close to 100%. A. baumannii AB5075-UW showed relatively low biofilm formation after 40-h static incubation in rich medium (Supplementary Fig 1B). The presence of putrescine, cadaverine or spermidine at 1/8 MIC did not affect static biofilm formation in rich media, while spermine caused a small decrease. These results do not support a major role for polyamines in biofilm regulation in A. baumannii AB5075-UW despite transcriptional regulation of some relevant genes (e.g. the csu pilus). The opposing effects of short- and long-chain polyamines on serum resistance are consistent with their divergent transcriptional regulation, though we did not notice any specific regulatory targets that would explain their effects on serum resistance.
Multidrug efflux pumps (MDEPs) regulated by polyamine shock
A. baumannii encodes a wide repertoire of MDEPs, which confer the ability to withstand many disinfectants and antibiotics. Drug efflux pumps are often induced by their substrates6. The efflux pump genes aceI, adeABC and amvA all showed increased expression in the presence of polyamines, with different combinations inducing each one (Fig. 2b). The PACE family transporter gene aceI was the most dramatically upregulated, with a log2-fold change of ~8 in the presence of putrescine and cadaverine and ~5 with spermine, while spermidine showed a weak (log2-fold change of 2) induction that was not statistically significant. This fits with the recent finding that short-chain diamines (including putrescine and cadaverine) are physiological substrates of AceI, while the longer-chain polyamines spermine and spermidine are not substrates but do weakly induce aceI expression18. The adeABC genes, encoding an RND family transporter, showed a 2–3 log2-fold increase in expression in the presence of any of the four molecules. This apparent induction by polyamines was surprising given that A. baumannii strain AB5075 carries a mutation in the regulator AdeS, which causes constitutive adeABC expression32,33. The MFS transporter gene amvA was induced by spermidine and spermine but not by putrescine and cadaverine. The amvA repressor gene amvR34, which is encoded 125-bp upstream of amvA on the opposite strand, was also induced by polyamine shock; however, the cognate regulators of aceI and adeABC (aceR and adeRS, respectively, both transcriptional activators) were not.
AmvA is a spermine and spermidine efflux system
As there are no characterised spermine or spermidine efflux pumps in A. baumannii, we sought to determine whether AmvA and/or AdeABC may perform these roles. First, growth in the presence of polyamines was measured to test whether AmvA or AdeABC may be responsible for removing harmful levels of these molecules from the cell (Fig. 3a and Table 1). Mutation of amvA dramatically decreased A. baumannii AB5075-UW resistance to spermine (16-fold reduction in MIC) and spermidine (64-fold reduction). Mutation of amvR, which was previously shown to increase amvA expression by ~6-fold34, increased the MIC of spermine 2-fold from 10 to 20 mg ml−1 but had no effect on growth in the presence of spermidine. Note that the A. baumannii AB5075 spermidine MIC was extremely high (40 mg ml−1)—it is possible that the amvR mutant does have increased AmvA-mediated spermidine efflux, but that this is not sufficient to increase the MIC >40 mg ml−1. Alternatively, amvA expression may be fully derepressed with 40 mg ml−1 spermidine such that mutation of amvR does not further increase expression. An adeB mutant showed an eightfold reduction in spermine MIC and a twofold reduction in spermidine MIC. The MICs of putrescine and cadaverine were not affected by amvR, amvA or adeB mutation, and none of these mutations resulted in a growth defect (Supplementary Fig 2). These observations support the hypothesis that spermine and spermidine may be the natural substrates of the well-characterised drug efflux pumps AmvA and AdeABC. A pangenome analysis of complete A. baumannii genomes was conducted to explore the distribution of these efflux pumps across the species (see ‘Methods’, Supplementary Data 2). While adeABC is variably present (Supplementary Data 2 and ref. 35), amvA is found in all A. baumannii genomes, within a region of very high synteny comprised of other conserved genes (Fig. 3b). Due to its very strong phenotype, and its absolute conservation hinting at a deeply rooted role in A. baumannii biology, AmvA was selected for further characterisation.
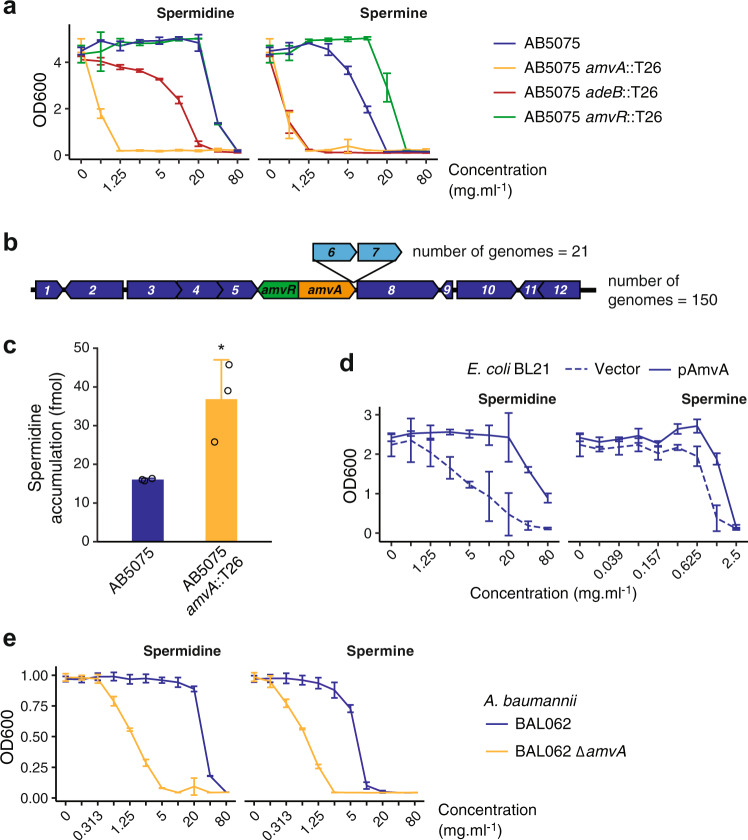
Fig. 3. Induction of efflux pumps by polyamine shock identifies spermine and spermidine as AmvA substrates.
a Growth of A. baumannii AB5075 and mutants in the presence of spermine and spermidine. Results are mean ± SD for two biological and two technical replicates. b Genomic context of amvAR in 172 A. baumannii strains. The genes are present in 100% of complete A. baumannii genomes in a region of very high synteny. Core genes are shown in dark blue, accessory genes in light blue, and the number of genomes possessing each gene path is indicated. Annotated gene functions are: 1—aceI PACE family transporter, 2—acetyl-CoA acyltransferase, 3—short-chain dehydrogenase/reductase, 4—MaoC domain protein dehydratase, 5—beta-lactamase, 6—hypothetical protein, 7—hypothetical protein, 8—dnaX DNA pol III subunit tau, 9—hypothetical protein, 10—phospholipase C, 11—thioesterase family protein, 12—iron-containing alcohol dehydrogenase. c Intracellular accumulation of 3H-spermidine in A. baumannii AB5075 and its AB5075::amvA mutant. Late exponential-phase cells were washed into assay buffer and incubated with 10.8 nM 3H-spermidine (see ‘Methods’), and the amount of accumulated radioactivity measured by liquid scintillation counting. Results shown are from three independent biological replicates. *p < 0.05, one-way ANOVA. d Growth of E. coli BL21 in the presence of spermidine and spermine, with or without AmvA overexpression. Results are mean ± SD for three biological and two technical replicates. e Growth of A. baumannii BAL062 and its ∆amvA mutant in the presence of spermidine and spermine. Results are mean ± SD for two biological and two technical replicates.
Table 1.
Polyamine MIC changes in amvA, amvR and adeB mutants of A. baumannii AB5075.
| MIC (mg ml−1) [fold change] | ||||
|---|---|---|---|---|
| Polyamines | WT | amvA::T26 | amvR::T26 | adeB::T26 |
| Spermine | 10 | 0.625 [16] | 20 [0.5] | 1.25 [8] |
| Spermidine | 40 | 0.625 [64] | 40 | 20 [2] |
| Putrescine | 40 | 40 | 40 | 40 |
| Cadaverine | 40 | 40 | 40 | 40 |
| MIC (µg ml−1) [fold change] | ||||
| Reported AmvA substrates | WT | amvA::T26 | ||
| Acriflavine | 64 | 16 [4] | ||
| Benzalkonium chloride | 32 | 32 | ||
| Chlorhexidine | 15 | 7.5 [2] | ||
| Erythromycin | 250 | 250 | ||
| Ethidium bromide | 32 | 32 | ||
| Methyl viologen | 100 | 25 [4] | ||
An amvA mutant of A. baumannii AB5075 was tested against a selection of its previously reported substrates14 in order to compare its effect on resistance to these molecules with its effect on polyamine resistance (Table 1). Mutation of amvA did not affect the MIC of three of the six substrates tested, while three substrates—acriflavine, chlorhexidine and methyl viologen—showed a modest (twofold or fourfold) MIC reduction in AB5075 amvA::T26. Note that previous investigation of the AmvA substrate range used a drug-sensitive A. baumannii strain AC0037; AB5075-UW has a different complement of efflux pumps that could potentially mask some AmvA activities. Overall, AmvA has a much greater impact on resistance to toxic levels of polyamines than on resistance to its previously reported substrates.
We then tested whether loss of amvA affects accumulation of spermidine in cells (Fig. 3c). A. baumannii AB5075-UW and A. baumannii AB5075-UW tn-amvA were grown to exponential phase, washed, and incubated with 10.8 pmol [3H]-spermidine. Mutation of amvA significantly increased the amount of intracellular [3H]-spermidine, though accumulation of exogenous spermidine was very low in the conditions tested (approximately 15 fmol in 2 × 108 cells), perhaps because of the low concentration of radioactive substrate used, because the cells retained a functional AdeABC efflux system, or because the accumulation of spermidine is likely to rely largely on the activity of uptake systems.
Following the finding that AmvA influences both polyamines tolerance and accumulation in A. baumannii AB5075, we wished to determine whether AmvA could promote spermidine and spermine resistance in a bacterium lacking this MDEP. Escherichia coli BL21 cells overexpressing AmvA showed full growth in the presence of spermidine up to 20 mg ml−1, while growth of the vector-only control strain decreased at ≥2.5 mg ml−1 spermidine (Fig. 3d). AmvA also increased resistance to spermine to a lesser extent (Fig. 3d).
Finally, we examined the effect of amvA mutation on a distinct A. baumannii strain, BAL062, which is a representative of the ICL2 lineage (AB5075-UW is an ICL1 strain). Similarly to our findings in AB5075-UW, the BAL062 ∆amvA mutant showed dramatically reduced tolerance to exogenous spermidine (32-fold MIC reduction) and spermine (8-fold MIC reduction) (Fig. 3e).
AmvR is a spermine and spermidine-responsive repressor of amvA
We next aimed to find out whether expression of amvA is regulated by its polyamine substrates and whether AmvR is responsible for this regulation. Expression of amvA was measured following exposure to spermidine or spermine at 1/16 MIC (~10 and ~2 mM, respectively) for 30 min, in both wild-type and amvR mutant A. baumannii AB5075 (Fig. 4a). In the wild-type strain, amvA expression increased ~6-fold with spermidine and ~2-fold with spermine. The amvR mutant strain showed a ~10-fold increase in the expression of amvA relative to the wild-type strain, and the expression did not increase further on polyamine treatment. Regulation of amvA in AB5075 was also explored using a reporter fusion vector comprising the amvR-amvA intergenic region upstream of green fluorescent protein (GFP; Supplementary Fig 3). Expression of GFP was significantly higher in AB5075 amvR::T26 than the wild-type strain (Supplementary Fig 3A) and did not increase further with spermidine treatment (Supplementary Fig 3B). The amvAprom-GFP reporter was strongly induced by 10 mM spermidine but not by putrescine or cadaverine at 10 mM (Supplementary Fig 3C), while spermine was toxic at this concentration. Note that the polyamine amvAprom-GFP induction experiments were conducted in A. baumannii ATCC 17978 (a widely used antibiotic-sensitive strain) using a gentamicin-resistant vector backbone, due to slight toxicity and high baseline activity of the zeocin-resistant vector used in AB5075 (Supplementary Fig 3A, B). Taken together, these results demonstrate that AmvR is a repressor of amvA, but AmvR repression is lifted in the presence of long-chain polyamines.
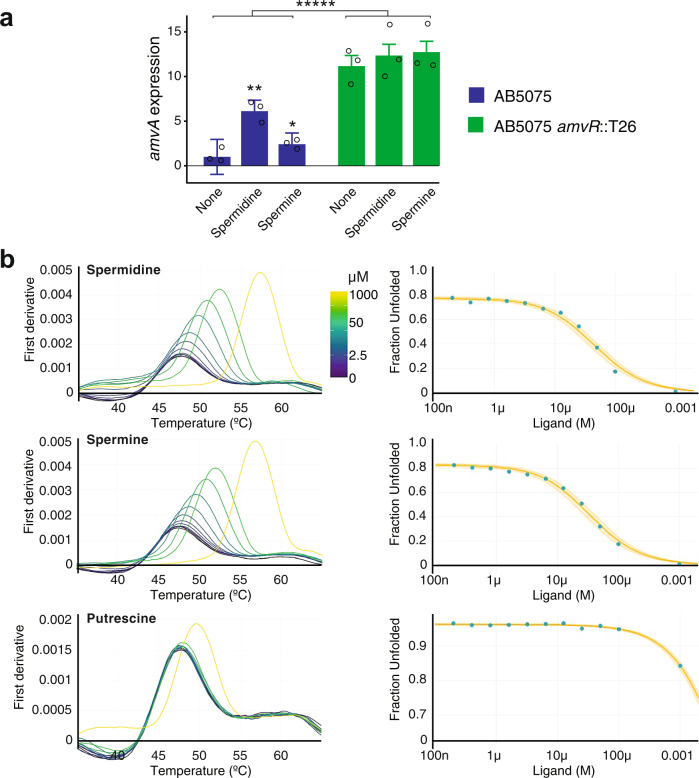
Fig. 4. AmvR and long-chain polyamines control amvA expression.
a AmvR-dependent induction of amvA by long-chain polyamines. Quantitative real-time PCR of amvA transcript in AB5075 and AB5075 amvR::T26 following addition of long-chain polyamines at 1/16 WT MIC (10 mM spermidine, 2 mM spermine). AB5075 amvR::T26 showed dramatically increased amvA transcript levels, which did not increase further on polyamine addition. All fold changes are expressed relative to the untreated AB5075 control. Results shown are the geometric mean ± standard deviation of three biological replicates, which were each comprised of two technical replicates. A two-factor ANOVA showed a significant effect of strain background (*****p < 0.00001) and polyamine treatment (p = 0.0013, comparison not shown) on amvA expression levels. A one-way ANOVA with Dunnett’s post hoc test was used to compare polyamine-treated samples with the untreated sample of the same strain (*p < 0.05, **p < 0.01). b Nano-DSF analysis of purified AmvR shows spermine and spermidine binding at low µM affinity. Melt curves and fitted unfolded protein fraction are shown for AmvR in the presence of increasing polyamine concentrations. Spermidine and spermine prevented protein unfolding at low concentrations (<10 µM), while putrescine only influenced protein unfolding at the highest concentration of 1 mM. The complete experiment was performed in duplicate, and results from the second replicate are provided in Supplementary Fig. 4. Calculated binding affinities are provided in Supplementary Tables 3 and 4.
We then tested whether AmvR has binding capacity for long-chain polyamines, with the aim of finding out if long-chain polyamines may induce amvA expression by direct binding of its repressor. Purified AmvR carrying a C-terminal hexahistidine tag was characterised by analytical size-exclusion chromatography (SEC) to be ~50 kDa in solution, consistent with its native, stable dimeric form (Supplementary Fig 4A). AmvR was first screened for interaction with polyamines using differential scanning fluorimetry (DSF). This method monitors changes to the thermal stability of a protein, observed as a dye response during unfolding, to indicate potential ligand binding36. Spermine and spermidine at 0.2% (5.7 and 7.8 mM, respectively) both elevated the thermal stability of AmvR by +4 °C while putrescine caused little change (+1 °C shift, i.e. below the significance threshold of the DSF experiment).
We then utilised a nano DSF (nDSF)-based isothermal approach to determine the binding constant (Kd) of AmvR for spermine and spermidine. The label-free nDSF assay monitors intrinsic tryptophan and tyrosine fluorescence upon protein unfolding, and using unfolding curves at varying ligand concentrations, the isothermal analysis of nDSF data facilitates Kd determination by fitting the unfolded protein fraction vs. ligand concentrations at a selected temperature37,38. As shown in Fig. 4b, the isothermal analysis of the nDSF data for AmvR/spermine and AmvR/spermidine at 49 °C (heating rate of 2 °C min−1) yields a low micromolar affinity, with Kd of 5.4 and 9.5 µM, respectively (the change of heat capacity, ∆Cp fitted). In contrast, isothermal analysis of the nDSF data for AmvR/putrescine at 51 °C yields Kd of 260 µM. Using the ∆Cp set to zero instead of fitted ∆Cp did not result in any differences in the measured Kd in this experiment (Supplementary Tables 3 and 4). Binding affinities were also measured in a replicate experiment with a slightly faster heating rate (Supplementary Fig 4C and Supplementary Tables 3 and 4) and showed minimal differences.
These specific responses indicate that, in isolation, AmvR can form complexes with spermine or spermidine as ligands with low micromolar affinity, supporting the model of AmvR as a polyamine-responsive repressor of amvA.
Discussion
Bacterial drug efflux pumps can have important physiological roles in addition to their resistance functions6,39. Here we have investigated the effects of four of the predominant biological polyamines on A. baumannii and identified a role for one of its major efflux pumps, AmvA, in the transport of long-chain polyamines. To our knowledge, this is the first spermidine and spermine efflux system to be identified in A. baumannii.
Our findings support the hypothesis that polyamines have broad biological roles in A. baumannii. Four hundred and ninety-nine genes had altered expression (representing 1/6 of the annotated CDS in AB5075), including many virulence-associated loci such as those encoding siderophores, pili and secretion systems (Figs. 1 and 2, Supplementary Fig. 1A, Supplementary Table 2 and Supplementary Data 1). A limitation is that our transcriptomics was performed on cells shocked with high levels of exogenous polyamines, ranging from 18 mM (spermine) to 56 mM (putrescine). These concentrations were selected to show the full range of polyamine-responsive genes and to elicit strong induction of candidate transporters but are higher than reported intracellular concentrations of polyamines in Gram-negative bacteria (e.g. spermine ~6 mM, putrescine ~20 mM40) or in most host environments. The transcriptional responses defined here could arise from metabolic feed, biophysical modulation of DNA or RNA structure, activation of specific regulatory pathways or a combination of all three. Experiments at physiological polyamine levels would help to define the biological roles of these molecules in more detail and determine whether A. baumannii utilises polyamines as specific virulence-regulating signals, as shown in some other bacterial species24,26.
Several lines of evidence indicate that long-chain polyamines are biological substrates of AmvA: (1) loss of this efflux pump dramatically reduced spermine and spermidine tolerance in both ICL-1 and ICL-2 A. baumannii strains, (2) loss of amvA increased intracellular spermidine accumulation, (3) amvA overexpression in E. coli increased long-chain polyamine resistance, (4) amvA expression is increased in response to these molecules, and (4) regulation of amvA depends on its cognate regulator, AmvR, which binds to long-chain polyamines with low µM affinity. Note that amvA expression is not induced by its other substrates, including ciprofloxacin41, deoxycholate42, chlorhexidine15 or benzalkonium chloride43.
AmvA is clinically important in A. baumannii; it confers resistance to widely used disinfectants (e.g. chlorhexidine, benzalkonium), and in hospital-adapted strains, its expression is increased14. AmvA is a member of the DHA-2 (drug H+ antiporter-2) subfamily of MFS transporters and the first member of this family shown to transport long-chain polyamines. DHA-2 proteins comprise 14 transmembrane helices44 and have a large, central hydrophobic cavity with several acidic residues that facilitate both proton and substrate binding7,45,46 and determine substrate charge specificity47. The substrate profile of AmvA includes a range of hydrophobic, cationic compounds with different degrees of protonation at physiological pH14,48. Spermine and spermidine are both long, flexible hydrophobic molecules with multiple protonated sites at physiological pH—it is likely that their transport requires particular active site properties, seen in DHA-2 proteins, that would also permit efflux of other substrates. Note that spermidine efflux has been demonstrated for the more distantly related DHA-1 transporter Blt, of Bacillus subtilis28, which was also originally characterised for its drug-resistance activity49.
We propose that the clinically relevant anti-biocide activity of the well-known A. baumannii efflux pump AmvA stems from an ancestral role in homoeostasis of long-chain polyamines. Interestingly, multiple bacterial pathogens including Staphylococcus aureus, Proteus mirabilis and members of the Enterobacteriaceae possess AmvA homologues (QacA, SmvA), which, like AmvA, confer increased biocide resistance (particularly to chlorhexidine) and are upregulated or have increased prevalence in clinical strains50–53. AmvA shares 50–55% amino acid identity with the enterobacterial SmvA proteins, while S. aureus QacA is more distantly related (30% amino acid identity). It is tempting to speculate that some of these clinically important AmvA homologues may also have physiological roles in polyamine transport.
A further question is why A. baumannii would require specific efflux systems for spermine and spermidine? Such systems may protect from exogenous polyamines encountered in host environments. Polyamine levels can reach the mM range in plants, the gut, and in serum of patients suffering from some diseases54–57. In particular, a recent study showed serum levels of spermidine and spermine at >500 µg ml−1—close to their respective MICs in AB5075-UW amvA or adeB mutants (Fig. 3b)57. Alternatively, efflux of endogenous polyamines by AmvA (and perhaps AdeABC) may provide adaptive advantages unrelated to detoxification. For example, rapid putrescine efflux helps to counter osmotic stress in E. coli58. A. baumannii produces a range of endogenous polyamines, including spermidine59–62; however, their biological role(s) are not yet fully understood.
The ability of A. baumannii to withstand biocides and antibiotics is a major barrier to the effective control of this pathogen. Here we have shown that a key biocide resistance determinant, AmvA, is a spermidine and spermine efflux system and provide preliminary evidence that AdeABC may share this activity. Together with previous findings on AceI18, our work shows that three well-known A. baumannii efflux pumps, of three distinct transporter families, function in polyamine transport. AmvA, AdeABC and AceI are each induced by, and provide tolerance to, different combinations of polyamines (Fig. 5), and we speculate that polyamine tolerance in A. baumannii is determined by the respective expression levels, substrate profiles and transport capacity of these three efflux systems. All three of these polyamine tolerance determinants also provide some resistance to the synthetic biocide chlorhexidine, which may be a common secondary substrate among polyamine efflux systems. Overall, our results suggest a strong link between homoeostasis of the ancient, highly abundant polyamine class of molecules and the broad and flexible efflux pump activity that allows bacterial pathogens—including A. baumannii—to survive treatment with disinfectants.
Fig. 5. Schematic of known links between polyamines and efflux pumps in A. baumannii.
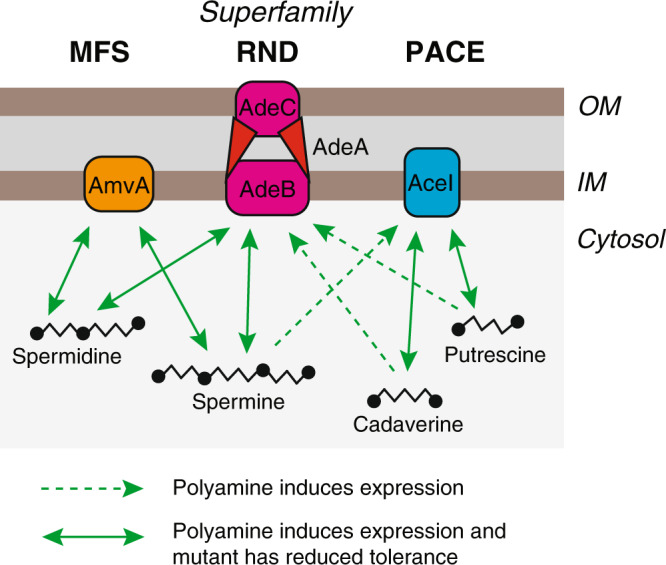
AmvA, AdeABC and AceI of the MFS, RND and PACE efflux pump families are each induced by, and confer tolerance to, distinct subsets of the four major biological polyamines.
Methods
Bacterial growth
Strains, plasmids and oligonucleotides used in this study are given in Supplementary Tables 5–7. A. baumannii was cultivated in Muller–Hinton (MH) broth at 37 °C for all experiments with the exception of reporter assays in A. baumannii AB5075-UW pFLS45, which were conducted in low-salt LB. When necessary, media were supplemented with polyamine salt compounds: spermidine tri-hydrochloride, putrescine di-hydrochloride, cadaverine di-hydrochloride or spermine tetra-hydrochloride. Antibiotics used were zeocin (150 µg ml−1 for A. baumannii; 25 µg ml−1 E. coli), gentamicin (10 µg ml−1), kanamycin (50 µg ml−1) and ampicillin (50 µg ml−1).
Antimicrobial susceptibility testing
MICs were determined by broth dilution63 in 96-well plates with a final volume of 200 µl (AB5075, E. coli) or 100 µl (BAL062). Polyamine salts were added to the MH media (buffered to pH 7.8 using HEPES salt) to final concentrations of 80 mg ml−1. Plates were incubated at 37 °C overnight and end-point growth at OD600 measured. A. baumannii strains were used in MIC experiments directly from overnight cultures, at an inoculum of ~106 cells ml−1, and plates were incubated without shaking. E. coli BL21 cells carrying either pTTQ18R6SH6 or pTTQ18R6SH6-AmvA (previously called AedF)48 were prepared as follows: cells were grown overnight in MHII, subcultured at 1:10 and induced with 0.05 mM IPTG for 1 h at 37 °C 200 rpm, and used to inoculate MIC broth dilution plates at approx. 2.5 × 105 cfu ml−1. Ampicillin and 0.05 mM IPTG were included in media to maintain amvA expression, and plates were sealed with breathable film and incubated with shaking for 24 h prior to OD600 measurement.
RNA-Seq and analysis
A. baumannii AB5075-UW cells were grown overnight and subcultured in MHII broth and grown to mid-exponential phase (5 ml, OD600 = 0.6). Cultures were treated with polyamine salts (1/8 MIC) for 30 min at 37 °C, with shaking. Control cultures contained no polyamine salt compounds. Cell pellets were recovered (5000 g, 15 min at 4 °C) and lysed in QIAzol reagent (Qiagen). Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions and DNA was removed with DNaseI (TURBO DNA-freeTM Kit, Invitrogen). Removal of rRNA and RNA-Seq were conducted by the Ramaciotti Centre for Genomics (Sydney, Australia) as follows: rRNA was depleted using Ribozero (Invitrogen), libraries were prepared using a TruSeq Stranded RNA-seq Prep Kit (Illumina) and sequencing was performed on an Illumina NextSeq machine with a read length of 75 bp. Sequencing statistics are given in Supplementary Table 1.
The raw data from the samples were analysed using EDGE-PRO with Bowtie 264. Reads were normalised and differential expression analysis was conducted with the DESeq R package65. A negative binomial model was used to test the significance of differential expression between control cells and polyamine-treated cells. Genes with adjusted p values <0.05 and presenting at least 2-fold differences in expression were considered to be differentially expressed.
Functional enrichment analyses were conducted by first assigning COG and GO terms to AB5075-UW genes using EggNOG mapper66. Significantly enriched GO terms within the biological process, molecular function and cellular compartment ontologies were identified using the TopGO package with the ‘weight01’ algorithm and Fisher’s exact test.
Serum resistance assays
Bacteria were grown overnight in MHII broth, subcultured 1:100 in fresh medium and grown to late exponential phase (OD600 = 1). Cultures were then washed once in phosphate-buffered saline (PBS) and diluted 1:100 in sterile PBS; 50 µl diluted culture was added to 100 µl pre-warmed human serum (Sigma) and incubated at 37 °C. Samples were taken at set time points, serially diluted and plated for enumeration of viable bacteria.
Biofilm formation assays
In all, 2.5 ml cultures of A. baumannii AB5075-UW with or without polyamine supplementation were grown for 40 h at 37 °C in 15 ml polystyrene tubes. Attached biofilm in the tubes was quantified by crystal violet staining as described67.
Mutagenesis of A. baumannii BAL062
The A. baumannii BAL062 ∆amvA strain was constructed by linear DNA transformation68. Cells were prepared for transformation by washing an overnight culture three times in 10% (w/v) sucrose, then resuspending to a final OD600 of 20. In all, 200 µl of cell suspension was mixed with 3 µg DNA fragment and electroporated (cuvette gap 2 mm, electroporation parameters: 2500 kV, 200 Ω, 25 µF, recovery 2 h 37 °C in 1 ml LB).
Growth curves
For 96-well plate-format growth curves, individual wells were inoculated with 150 µl of MHII medium seeded with the strain of interest at an OD600 of 0.05. The plate was sealed with gas-permeable film (Aeraseal™) and incubated in a Tecan Infinite 200 plate reader at 37 °C with shaking and OD600 measurement every 15 min. For aerobic growth curves, 5 ml of MHII medium in a 50 ml flask was inoculated at OD600 0.025 and cultures were grown at 37 °C with continuous shaking at 200 RPM.
Pangenome analysis
All complete A. baumannii genome assemblies were downloaded from https://www.ncbi.nlm.nih.gov/assembly/ on March 26, 2020. Complete assemblies were used to minimise the effects of contig boundaries on gene neighbourhood analysis. Genome sequences were annotated using prokka69 version 1.14.6 with the following commands passed to the program: ‘prokka --prodigaltf prodigal_training.txt --proteins AB5075_UWversion.gb --genus Acinetobacter --species baumannii’. The training model was generated using default settings, and the pipeline set to preferentially assign genes based on the A. baumannii AB5075-UW genome annotation. Gene content analysis across the population was conducted using Panaroo70 version 1.1.2 with core gene alignment. Gene neighbourhood information was extracted from the final pangenome network using the get_neighbourhood script distributed with Panaroo. The gene presence–absence table, which includes accession IDs for the genomes examined, is provided as Supplementary Data 2.
Spermidine transport assays
A. baumannii AB5075-UW and AB5075-UW tn-amvA were grown overnight in MHII broth, then diluted 1:100 in the same medium and grown to late exponential phase (OD600 = 1.0). Spermidine was added at 1/16 MIC to induce efflux pump expression, and cultures were incubated for a further 30 min. Cells were washed twice in assay buffer (20 mM HEPES pH 8.0, 145 mM NaCl, 5 mM KCl) and resuspended to a final OD600 of 1.0 in assay buffer supplemented with 0.5% (w/v) succinate to energise the cells. Accumulation assays were performed at room temperature with 1 ml resuspended cells. Bacterial samples were incubated with 10.8 pmol 3H-spermidine (46.1 mCi mmol−1, Perkin-Elmer) for 2 min, then 200 µl of each reaction was immediately filtered through a 0.2 µM nitrocellulose membrane and washed with 2 ml assay buffer. Filters were placed in 5 ml PE Emulsifier-Safe scintillation fluid, and the amount of radiation retained on each filter (indicating intracellular 3H) was measured by liquid scintillation.
DNA manipulations
The linear fragment for mutagenesis of A. baumannii BAL062 to remove its amvA gene was constructed by overlap PCR using primers AP2064 and FS175–179 (Supplementary Table 7) to generate a DNA fragment comprising the kanamycin resistance gene from pCR2.1 flanked by the 1000 bp sequences 5′ and 3′ of amvA.
To construct the reporter vector pFLS43 and pFLS45, the GFP marker from plasmid pDiGc was edited by overlap PCR using FS9 + FS10 (outer primers) and FS65 + FS66 (overlap primers) to remove its internal NdeI site and introduced into pCR2.1 by TOPO cloning (Invitrogen). The amvA-amvR intergenic region from A. baumannii AB5075-UW was amplified using primers FS77 + FS78 and cloned upstream of the GFP-terminator fragment from pDiGc using the NdeI site. The promoter-GFP insert was subcloned from pCR2.1 into pVRL1 and pVRL1-Z using restriction enzymes SacI and XhoI. The AmvR expression vector was constructed by amplifying amvR from A. baumannii ATCC17978 genomic DNA using primers AmvR-F and AmvR-R and cloning into the expression vector pTTQ18RGSH6.
Quantitative real-time PCR (qPCR)
Three biological replicate overnight cultures each of AB5075 wild type and AB5075 amvR::T26 were prepared from single colonies on freshly streaked MH agar plates. Subcultures were grown in MHII medium to OD600 0.5–0.6 and treated with either 2 mM spermine, 10 mM spermidine or no additive for 30 min. The cultures were pelleted at 7000 × g 4 °C for 5 min. Total RNA was extracted by the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions and residual genomic DNA was removed by DNase I (TURBO DNA-free Kit, ThermoFisher). KAPA SYBR FAST One-Step Kit (Merck) was used for cDNA synthesis and qPCR in LightCycler 480 II (Roche) with the following cycling parameters: cDNA synthesis and reverse transcriptase (RT) inactivation—42 °C 5 min–95 °C 2 min; qPCR cycling—(95 °C 3 s–60 °C 20 s) × 40; dissociation—60–95 °C at 30 s °C−1. Expression of amvA was measured using the ∆∆Ct method with gadph as the reference gene. Primer sequences are provided in Supplementary Table 7.
Promoter fusion experiments
Reporter plasmid pFLS45, comprising GFP under the control of the amvA promoter, was transformed into A. baumannii AB5075-UW and A. baumannii AB075 tn-amvR and maintained with zeocin selection in low-salt LB. Reporter plasmid pFLS43, which has the same structure but with gentamicin resistance, was transformed into A. baumannii ATCC17978 and maintained in gentamicin-supplemented MHII medium. Expression without induction (Supplementary Fig 3A) was measured following growth in 10 ml cultures to an OD of 2.0. Induction by polyamines was measured by growing cells to late exponential phase in 10 ml cultures, then transferring cultures to a 96-well plate format (100 µl per well), adding polyamine salts, incubating the plate at 37 °C and measuring fluorescence and OD over time. For all experiments, expression was determined by measuring culture fluorescence (ex 485/em 520) and OD600 in a 96-well plate in a Pherastar plate reader. GFP fluorescence/OD was calculated for each well, with cells lacking the reporter vector used as a background control. Results shown represent three biological replicates and six technical replicates.
Expression and purification of AmvR
AmvR protein was expressed by growing BL21 pAmvR cells in ZYP-rich autoinduction medium60 to an OD of 1.2–1.4 at 25 °C (approx. 24 h incubation). Recovered cells were resuspended in 30 ml AmvR buffer (50 mM HEPES pH 8.0, 300 mM NaCl) supplemented with 5 mM imidazole and 5% glycerol and stored at −80 °C. Cell aliquots were thawed in the presence of lysozyme (1 µg ml−1) and DNase I (5 µg ml−1) and lysed by cell disruptor (Constant Systems) at 30 KPsi. Clarified cell lysates (following 40 min centrifugation at 11,000 × g and 0.2 μm filtration) were loaded with a peristaltic pump onto prepacked columns (1 ml) of Ni-Sepharose media (His trap, GE Healthcare) pre-equilibrated in the same buffer. The column was then washed with 50 column volumes of AmvR buffer + 50 mM imidazole, and protein was eluted in AmvR buffer + 500 mM imidazole. Eluate fractions were pooled and further purified using SEC (Superdex HiLoad 200 16/600 column, GE Healthcare) in AmvR buffer (50 mM HEPES pH 8.0, 300 mM NaCl) supplemented with reducing agent tris(2-carboxyethyl)phosphine (TCEP, 1 mM) and glycerol (5% v/v). Protein-containing fractions were pooled, concentrated using centrifugation (Centricon 10 kDa molecular weight (MW) cut-off) and snap-frozen in liquid N2. The purity of the recovered His6-tagged product was verified by using sodium dodecyl sulfate–polyacrylamide gel electrophoresis, showing a single band at ~25 kDa when visualised with Coomassie blue dye. MW of AmvR in solution was estimated using analytical SEC procedures on a Superdex 200 10/300 GL column (GE-Healthcare), by comparing its elution volume to those of a set of MW standards (HMW and LMW calibration sets, GE Healthcare).
Differential scanning fluorimetry
DSF was performed according to standard methods36. SYPRO Orange (100×, Invitrogen) was diluted into AmvR buffer and mixed with purified AmvR (final concentration 1 mg ml−1). Twenty-microlitre aliquots of AmvR-SYPRO orange mixture were transferred to a 96-well plate in triplicate, polyamine salts were added at 0.2% (w/v), which corresponds to 5.7 mM spermine and 7.85 mM spermidine, and the solutions were gently mixed by plate centrifugation (1000 RPM, 1 min). The plate was heated at 1 °C min−1 over 25–95 °C in a real-time qPCR machine (Mx3005P, Stratagene) and change in fluorescence intensity (dR, automatically baseline corrected) at 610 nm was measured. Derivative curves were used to calculate the transition midpoint and assign melting temperature.
nDSF binding study
Ligand interactions for AmvR (15 µM) were monitored in duplicate in HEPES buffer (50 mM, with 300 mM NaCl, pH 8.0) supplemented with TCEP (1 mM) and glycerol (5% v/v). Potential ligands (spermidine, spermine, putrescine) were tested from 0.2 to 100 µM following 2-fold serial dilutions in the same buffer. Additional end-point measurements were made at 1 mM for each compound. Following room temperature incubation (15 min), samples were transferred to standard grade capillaries (Prometheus NT.48 series, Nanotemper). Samples were heated over 20–95 °C at 2 °C min−1 (replicate 1) or 1.5 °C min−1 (replicate 2) with a Prometheus NT.48 fluorimeter (Nanotemper) controlled by PR.ThermControl. Excitation power was pre-adjusted to obtain fluorescence readings >2000 relative fluorescence units for emission at 330 nm (F330) and 350 nm (F350).
The isothermal analysis of nDSF data was performed using the web server FoldAffinity38 utilising the approach of Bai et al.37. Briefly, each fluorescence curve was fitted by six free parameters (melting temperature Tm, unfolding enthalpy ∆H and initial and final intercepts and slopes). Curve fitting was subsequently improved by a global fit of slopes in conjunction with other parameters still fitted individually to curves. From best fits, the fraction of unfolded protein was determined for each ligand concentration for one or several temperatures of interest. Finally, the fraction unfolding vs. ligand concentrations were incorporated into a 1:1 binding model for construction of isothermal plots. All isothermal analysis utilised change of heat capacity (∆Cp) values suggested by the FoldAffinity server38, as well as by setting ∆Cp to zero. The temperature with the lowest numerical fitting error of binding constant (Kd) was selected for the isothermal plots shown in this study.
Statistics and reproducibility
All microbiological experiments reported in the manuscript are from three biological replicates, defined as samples originating from separate overnight cultures each started from a single, separate colony, with technical replicates included as appropriate. Technical replicates were used for qRT-PCR quantification, and for all MIC, growth curve and reporter gene expression experiments were performed in a 96-well plate format and were defined as reactions performed on the same cDNA sample (for qRT-PCR experiments) or wells seeded with the same culture (for plate-format bacteriological experiments). Statistical significance for microbiological experiments was, in most cases, determined by analysis of variance with an appropriate post hoc test. For qRT-PCR experiments, where results are expected to be log-distributed, data were log2-transformed prior to statistical analysis and geometric means are reported. Specific numbers of replicates for each experiment (biological and technical), and details of the specific statistical test(s) used, are provided in the figure legends. In experiments where both biological and technical replicates were used, the mean of the technical replicate values derived from the same biological replicate was used for subsequent analyses.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank members of the Paulsen laboratory for useful discussions. We thank the Ramaciotti Centre, University of New South Wales for performing RNA sequencing. We also gratefully acknowledge the use of equipment in the Structural Biology Facility within the Mark Wainwright Analytical Centre, University of New South Wales, funded in part by the Australian Research Council Linkage Infrastructure, Equipment and Facilities Grant: ARC LIEF 190100165. This work was funded by grant 1120298 from the National Health and Medical Research Council of Australia. KAH is supported by an Australian Research Council Future Fellowship FT180100123. FLS is supported by an Australian Research Council DECRA fellowship DE200101524.
Author contributions
K.A.H. and I.T.P. conceived and designed the study. Q.L. performed RNA sequencing experiments and initial analysis. B.S. performed nanoDSF experiments, and H.E.C. developed the AmvR purification protocol and performed initial DSF experiments, with supervision from B.A.M. V.N. performed exploratory MIC measurements. L.L. and F.T.P. performed qRT-PCR experiments. F.L.S. completed all other experiments and analyses and wrote the manuscript, with input from all other authors.
Data availability
Raw RNA sequencing data associated with this study are available from the European Nucleotide Archive, accession PRJEB40527. Source data for figures is provided as Supplementary Data 3.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review informationCommunications Biology thanks J. Mark Sutton and the other anonymous reviewers for their contribution to the peer review of this work. Primary handling editor: George Inglis. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02629-6.
References
- 1.O’Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. The review on antimicrobial resistance. (2016).
- 2.Henderson PJF, et al. Physiological functions of bacterial “multidrug” efflux pumps. Chem. Rev. 2021;121:5417–5478. doi: 10.1021/acs.chemrev.0c01226. [DOI] [PubMed] [Google Scholar]
- 3.Perry J, Waglechner N, Wright G. The prehistory of antibiotic resistance. Cold Spring Harb. Perspect. Med. 2016;6:1–9. doi: 10.1101/cshperspect.a025197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez JL, et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 2009;33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 6.Blanco P, et al. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms. 2016;4:14. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du D, et al. Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 8.Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 9.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2017;16:91. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE. 2010;5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne S, Courvalin P, Périchon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011;55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajamohan G, Srinivasan VB, Gebreyes WA. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J. Antimicrob. Chemother. 2009;65:228–232. doi: 10.1093/jac/dkp427. [DOI] [PubMed] [Google Scholar]
- 14.Rajamohan G, Srinivasan VB, Gebreyes WA. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2010;65:1919–1925. doi: 10.1093/jac/dkq195. [DOI] [PubMed] [Google Scholar]
- 15.Hassan KA, et al. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA. 2013;110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan VB, Rajamohan G, Gebreyes WA. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:5312–5316. doi: 10.1128/AAC.00748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan KA, Liu Q, Henderson PJF, Paulsen IT. Homologs of the Acinetobacter baumannii aceI transporter represent a new family of bacterial multidrug efflux systems. MBio. 2015;6:1–5. doi: 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan KA, et al. Short-chain diamines are the physiological substrates of PACE family efflux pumps. Proc. Natl. Acad. Sci. USA. 2019;116:18015–18020. doi: 10.1073/pnas.1901591116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michael AJ. Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 2016;291:14896–14903. doi: 10.1074/jbc.R116.734780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 2015;427:3389–3406. doi: 10.1016/j.jmb.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008;68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- 22.Ware D, Jiang Y, Lin W, Swiatlo E. Involvement of potD in Streptococcus pneumoniae polyamine transport and pathogenesis. Infect. Immun. 2006;74:352–361. doi: 10.1128/IAI.74.1.352-361.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armbruster CE, et al. Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection. PLoS Pathog. 2019;15:1–34. doi: 10.1371/journal.ppat.1007653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobe RC, et al. Spermine inhibits Vibrio cholerae biofilm formation through the NspS–MbaA polyamine signaling system. J. Biol. Chem. 2017;292:17025–17036. doi: 10.1074/jbc.M117.801068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang SBin, et al. speG is required for intracellular replication of Salmonella in various human cells and affects its polyamine metabolism and global transcriptomes. Front. Microbiol. 2017;8:1–23. doi: 10.3389/fmicb.2017.02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobley L, et al. Spermidine promotes Bacillus subtilis biofilm formation by activating expression of the matrix regulator slrR. J. Biol. Chem. 2017;292:12041–12053. doi: 10.1074/jbc.M117.789644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higashi K, et al. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 2008;190:872–878. doi: 10.1128/JB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolridge DP, et al. Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilis multidrug transporter Blt. J. Biol. Chem. 1997;272:8864–8866. doi: 10.1074/jbc.272.14.8864. [DOI] [PubMed] [Google Scholar]
- 29.Alexa, A. Gene set enrichment analysis. Encycl. Syst. Biol. 10.1007/978-1-4419-9863-7_100552 (2013).
- 30.Morris FC, Dexter C, Kostoulias X, Uddin MI, Peleg AY. The mechanisms of disease caused by Acinetobacter baumannii. Front. Microbiol. 2019;10:1601. doi: 10.3389/fmicb.2019.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouts DE. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon E-J, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 2013;57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leus IV, et al. Inactivation of AdeABC and AdeIJK efflux pumps elicits specific nonoverlapping transcriptional and phenotypic responses in Acinetobacter baumannii. Mol. Microbiol. 2020;114:1049–1065. doi: 10.1111/mmi.14594. [DOI] [PubMed] [Google Scholar]
- 34.Hassan KA, et al. Fluorescence-based flow sorting in parallel with transposon insertion site sequencing identifies multidrug efflux systems in Acinetobacter baumannii. MBio. 2016;7:3–8. doi: 10.1128/mBio.01200-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C, Bilya SR, Xu W. adeABC efflux gene in Acinetobacter baumannii. N. Microbes N. Infect. 2019;30:100549. doi: 10.1016/j.nmni.2019.100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 37.Bai N, Roder H, Dickson A, Karanicolas J. Isothermal analysis of ThermoFluor data can readily provide quantitative binding affinities. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-018-37072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niebling S, et al. FoldAffinity: binding affinities from nDSF experiments. Sci. Rep. 2021;11:9572. doi: 10.1038/s41598-021-88985-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcalde-Rico M, Hernando-Amado S, Blanco P, Martínez JL. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol. Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geisinger E, et al. The landscape of phenotypic and transcriptional responses to ciprofloxacin in Acinetobacter baumannii: acquired resistance alleles modulate drug-induced SOS response and prophage replication. MBio. 2019;10:1–19. doi: 10.1128/mBio.01127-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López M, et al. Response to bile salts in clinical strains of Acinetobacter baumannii lacking the adeABC efflux pump: virulence associated with quorum sensing. Front. Cell. Infect. Microbiol. 2017;7:1–13. doi: 10.3389/fcimb.2017.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knauf GA, et al. Exploring the antimicrobial action of quaternary amines against Acinetobacter baumannii. MBio. 2018;9:1–13. doi: 10.1128/mBio.02394-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH. The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paulsen IT. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 2003;6:446–451. doi: 10.1016/j.mib.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 46.Majumder P, et al. Dissection of protonation sites for antibacterial recognition and transport in QacA, a multi-drug efflux transporter. J. Mol. Biol. 2019;431:2163–2179. doi: 10.1016/j.jmb.2019.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. USA. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hassan KA, et al. Roles of DHA2 family transporters in drug resistance and iron homeostasis in Acinetobacter spp. J. Mol. Microbiol. Biotechnol. 2011;20:116–124. doi: 10.1159/000325367. [DOI] [PubMed] [Google Scholar]
- 49.Ahmed M, et al. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J. Bacteriol. 1995;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wand ME, Jamshidi S, Bock LJ, Rahman KM, Sutton JM. SmvA is an important efflux pump for cationic biocides in Klebsiella pneumoniae and other Enterobacteriaceae. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-018-37730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelling H, et al. Derepression of the smvA efflux system arises in clinical isolates of Proteus mirabilis and reduces susceptibility to chlorhexidine and other biocides. Antimicrob. Agents Chemother. 2019;63:1–15. doi: 10.1128/AAC.01535-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villagra NA, Hidalgo AA, Santiviago CA, Saavedra CP, Mora GC. SmvA, and not AcrB, is the major efflux pump for acriflavine and related compounds in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2008;62:1273–1276. doi: 10.1093/jac/dkn407. [DOI] [PubMed] [Google Scholar]
- 53.Grkovic S, Brown MH, Roberts NJ, Paulsen IT, Skurray RA. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 1998;273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 54.Bartos F, Bartos D, Grettie DP, Campbell RA. Polyamine levels in normal human serum. Biochem. Biophys. Res. Commun. 1977;75:1689–1699. doi: 10.1016/0006-291X(77)91469-3. [DOI] [PubMed] [Google Scholar]
- 55.Kingsnorth AN, Lumsden AB, Wallace HM. Polyamines in colorectal cancer. Br. J. Surg. 1984;71:791–794. doi: 10.1002/bjs.1800711019. [DOI] [PubMed] [Google Scholar]
- 56.Ramos-Molina B, Queipo-Ortuño MI, Lambertos A, Tinahones FJ, Peñafiel R. Dietary and gut microbiota polyamines in obesity- and age-related diseases. Front. Nutr. 2019;6:1–15. doi: 10.3389/fnut.2019.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ibrahim SA, Zainulabdeen JA, Jasim HM. The significance of spermidine and spermine in association with atherosclerosis in sera of Iraqi patients. Biomed. Pharmacol. J. 2018;11:1389–1396. doi: 10.13005/bpj/1502. [DOI] [Google Scholar]
- 58.Schiller D, Kruse D, Kneifel H, Kramer R, Burkovski A. Polyamine transport and role of potE in response to osmotic stress in Escherichia coli. J. Bacteriol. 2000;182:6247–6249. doi: 10.1128/JB.182.21.6247-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamana K, Matsuzaki S. Diaminopropane occurs ubiquitously in Acinetobacter as the major polyamine. J. Gen. Appl. Microbiol. 1992;38:191–194. doi: 10.2323/jgam.38.191. [DOI] [Google Scholar]
- 60.Skiebe E, et al. Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int. J. Med. Microbiol. 2012;302:117–128. doi: 10.1016/j.ijmm.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Auling G, et al. Analysis of the polyphosphate-accumulating microflora in phosphorus- eliminating, anaerobic-aerobic activated sludge systems by using diaminopropane as a biomarker for rapid estimation of Acinetobacter spp. Appl. Environ. Microbiol. 1991;57:3585–3592. doi: 10.1128/aem.57.12.3585-3592.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kämpfer P, Bark K, Busse HJ, Auling G, Dott W. Numerical and chemotaxonomy of polyphosphate accumulating Acinetobacter strains with high polyphosphate: AMP phosphotransferase (PPAT) activity. Syst. Appl. Microbiol. 1992;15:409–419. doi: 10.1016/S0723-2020(11)80215-8. [DOI] [Google Scholar]
- 63.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 64.Magoc T, Wood D, Salzberg SL. EDGE-pro: estimated degree of gene expression in prokaryotic genomes. Evol. Bioinform. Online. 2013;9:127–136. doi: 10.4137/EBO.S11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huerta-Cepas J, et al. Fast genome-wide functional annotation through orthology assignment by EggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biswas, I. & Mettlach, J. in Acinetobacter baumannii Methods and Protocols (eds Biswas, I. & Rather, P. N.) 159–166 (Humana Press, 2013).
- 68.Aranda J, et al. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol. 2010;10:279. doi: 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 70.Tonkin-Hill G, et al. Producing polished prokaryotic pangenomes with the panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA sequencing data associated with this study are available from the European Nucleotide Archive, accession PRJEB40527. Source data for figures is provided as Supplementary Data 3.