Abstract
Chloroplast biogenesis and development are highly complex processes requiring interactions between plastids and nuclear genomic products. Pentatricopeptide repeat (PPR) proteins play an essential role in the development of chloroplasts; however, it remains unclear how RNA editing factors influence soybean development. In this study, a Glycine max pale green leaf 2 mutant (Gmpgl2) was identified with decreased chlorophyll contents. Genetic mapping revealed that a single-nucleotide deletion at position 1949 bp in the Glyma.05g132700 gene in the Gmpgl2 mutant, resulting in a truncated GmPGL2 protein. The nuclear-encoded GmPGL2 is a PLS-type PPR protein that localizes to the chloroplasts. The C-to-U editing efficiencies of rps16, rps18, ndhB, ndhD, ndhE, and ndhF were reduced in the Gmpgl2 mutant. RNA electrophoresis mobility shift assay (REMSA) analysis further revealed that GmPGL2 binds to the immediate upstream sequences at RNA editing sites of rps16 and ndhB in vitro, respectively. In addition, GmPGL2 was found to interact with GmMORF8, GmMORF9, and GmORRM6. These results suggest that GmPGL2 participates in C-to-U RNA editing via the formation of a complex RNA editosome in soybean chloroplasts.
Keywords: soybean, RNA editing, pentatricopeptide repeat protein, genetic mapping, chloroplast function
Introduction
The chloroplast is a vital photosynthetic organelle for plant growth and development. It is a semi-autonomous organelle with its own DNA genome. The chloroplast proteome contains approximately 3,000 proteins, and only approximately 160 proteins are encoded by the chloroplast genome while the remainder are imported (Martin et al., 2002). The proteins encoded by the chloroplast genome include components of the chloroplast ribosome and NADH dehydrogenase-like complex, which influence plastid protein synthesis and photosystem I cyclic electron transport, respectively (Laughlin et al., 2019). RNA editing plays an important role in the biogenesis and functioning of the mitochondria and chloroplasts. The conversion of cytidines (Cs) to uridines (Us), via a deamination reaction, representing the main RNA editing mechanism in plants (Stern et al., 2004). RNA editing converts hundreds of Cs to Us at specific positions in the plastid and mitochondrial transcripts; moreover, editing often creates start or stop codons (Stern et al., 2010; Small et al., 2020). Meanwhile, a lack of RNA editing may have severe consequences, such as impaired chloroplast biogenesis (Yu et al., 2009; Ma et al., 2017; Jiang et al., 2018; Lv et al., 2020) and embryo lethality (Li et al., 2014, 2019; Sun et al., 2015a).
Several proteins, including pentatricopeptide repeat (PPR) proteins (Barkan et al., 2012; Yin et al., 2013), multiple organellar RNA editing factors (MORF, also known as RIPs, RNA editing factor interacting proteins; Sun et al., 2013; Ma et al., 2017; Jiang et al., 2018), organelle RNA recognition motif (ORRM) proteins (Sun et al., 2013), organelle zinc-finger (OZ) proteins (Sun et al., 2015b), and protoporphyrinogen oxidase 1 (PPO1; Tillich et al., 2009), are involved in RNA editing. The PPR proteins are characterized by 31–36 amino acid (aa) tandem repeats that fold into a pair of anti-parallel alpha helices to facilitate specific binding to target RNA sequences (Fujii and Small, 2011; Barkan et al., 2012; Yin et al., 2013). Most PPR proteins are predicted to be localized to the chloroplast, mitochondrion, or both these organelles (Colcombet et al., 2013). The PPR proteins are further divided into P- and PLS class proteins based on their structure. The P-class proteins contain an array of canonical PPR (P) motifs with 35 aa that participate in RNA-processing activities; by regulating translation initiation, intron splicing, RNA maturation, and RNA stability (Kotera et al., 2005; Haili et al., 2016; Aryamanesh et al., 2017). The PLS-class is composed of not only canonical P motifs (35 aa) but also L (36 aa) and S (31 aa) variants (O’Toole et al., 2008). At their C-terminus, many PLS-class proteins extend to contain a plant-specific conserved E domain; half of the PLS proteins with this E domain are further extended to contain a DYW motif of 100 aa with cytidine deaminase (Schallenberg Ruedinger et al., 2013). The RNA editing reaction requires the C-terminal domains of the PLS-class proteins, as the E domain is essential for editing (Okuda et al., 2007, 2009; Chateigner Boutin et al., 2013; Hayes et al., 2013; Wagoner et al., 2015). Molecular and phylogenetic studies suggest that the terminal DYW domain of PLS-class proteins is also required for the editing activity (Boussardon et al., 2012). PLS-class proteins primarily participate in RNA editing in organelles. In addition, the PPR proteins are essential for the normal activities of the mitochondria and chloroplasts as the majority of the PPR protein mutants display varied physiological phenotypes, such as pigment deficiency (Pyo et al., 2013; Huang et al., 2018), photosynthetic defects (Cai et al., 2009; Johnson et al., 2010), seedling lethality (Sun et al., 2018; Li et al., 2019), and restricted growth (Sung et al., 2010; Hu et al., 2012; Xiao et al., 2018). Recent studies have shown that MORF2, MORF8, and MORF9 are localized to plastids and are required for chloroplast RNA editing (Yan et al., 2017; Huang et al., 2019; Zhang et al., 2019; Zhao et al., 2019); ORRM1 and ORRM6 are also localized to plastids and participate in chloroplast RNA editing (Searing et al., 2020).
Soybean is an important source of edible oil and proteins for human and animal nutrition. The demand for soybean is continuously increasing with the rapid increase in human consumption and industrial use of soybean products (Ainsworth et al., 2012). However, the current rate of increase in soybean yield is insufficient to meet the growing demand. An analysis of historical soybean germplasm revealed that breeders have increased soybean yield by improving the plant harvest index, canopy light interception, and seasonal conversion efficiency, as well as by effectively utilizing of solar energy for the production of plant biomass (Morrison et al., 1999; Koester et al., 2014). The recently released cultivars have a higher daily carbon gain, chlorophyll content, and sink capacity than older cultivars. However, the maximum photosynthetic capacity, mesophyll conductance, and nighttime respiration have remained unchanged (Koester et al., 2016). Recent advances in synthetic biology and molecular biology have enabled the development of technologies for redesigning photosynthesis, thereby meeting the global food and bioenergy demand (Zhu et al., 2020a). Therefore, it is crucial to understand the molecular basis of soybean chloroplast function for yield improvement, particularly with respect to certain gaps in knowledge, such as the role of RNA editing in the regulation of chloroplast genes. Glyma.20G187000 encodes the chloroplast-localized protein ORRM1 that regulates chloroplast RNA editing and photosynthesis (Zhu et al., 2020b). The soybean genome encodes approximately 400 PPR proteins; however, the fundamental molecular functions of most of these proteins remain unknown (Su et al., 2019).
Herein, to identify novel factors involved in chloroplast development, a Glycine max pale green leaf 2 (Gmpgl2) mutant was developed. Map-based cloning revealed that a chloroplast-localized GmPGL2 protein is mutated in Glyma.05g132700. GmPGL2, together with GmMORF8, GmMORF9, and GmORRM6, participates in chloroplast transcript editing. Our study provides evidence that the chloroplast-localized GmPGL2 proteins regulate the normal functioning of organelles, particularly the chloroplast.
Materials and Methods
Plant Materials and Chlorophyll Analysis
The Gmpgl2 mutant was screened in June 2011 from the 60CO γ-radiation mutagenized Hedou12 (HD12) population as described previously (Cheng et al., 2016a). To purify the genetic background, the Gmpgl2 mutant plants were backcrossed for four generations in the Chang-Chun experimental field of Northeast Institute of Geography and Agroecology, CAS.
We collected fresh leaves from 18-day-old plants and determined their chlorophyll content using a spectrophotometer as described previously (Feng et al., 2019). Chlorophyll fluorescence was measured using FluorPen (Czech). Minimal chlorophyll fluorescence (F0) was measured at 650nm following the storage of leaves in the dark for 30min. Maximal chlorophyll fluorescence (Fm) was measured during a 1-s pulse of saturated white light (2,500μmolm−2s−1). The maximum quantum yield of photosystem II electron transport was calculated using the following formula: Fv/Fm =(Fm-F0)/Fm, where Fv indicates the maximum variable fluorescence.
Nuclear Acid Extraction and Analysis
Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Germany). The sequences of anchor markers used for initial mapping were published previously (Song et al., 2015). For fine mapping the GmPGL2 locus, new primers of InDel markers were synthesized for polymerase chain reaction (PCR; Supplementary Table S1). The candidate genes were amplified by PCR, and the PCR products were sequenced by Sangon Biotech (Shanghai, China). The phylogenetic and syntenic analyses were carried out as described previously (Dai et al., 2018).
The total RNA was extracted from tissue samples using TRIzol reagent (Qiagen, Germany) according to the manufacturer’s instructions. RNA samples were reverse transcribed using primer Script I (TaKaRa, Japan). An 18-mer oligo (dT) primer for nuclear-encoded genes or random primers for plastid genes were used for first strand cDNA synthesis. Quantitative real-time PCR (qRT-PCR) was performed using the SYBR® Premix Ex Taq™ Kit (TaKaRa, Japan) on an MX3005P Real-Time PCR System; the primers used are listed in Supplementary Table S1. The PCR program was as follows: 95°C for 15min, followed by 40cycles at 95°C for 10s, 58°C for 20s, and 72°C for 20s. Actin11 was used as the reference gene (Jian et al., 2008). Three biological replicates were used for gene expression analysis.
Plasmid Construction and Transformation
The CDS of GmPGL2 was amplified from HD12 using KOD DNA polymerase (Toyobo, Japan); the PCR products were cloned into the TA cloning vector pMD18-T. The GmPGL2 gene was cloned into the binary vector pCAMBIA3301 (CAMBIA, United States), using HindIII and EcoRI restriction endonuclease enzymes, and the pCAMBIA3301-GmPGL2 plasmid construct was generated. This plasmid was introduced into Agrobacterium tumefaciens (strain EHA 105) for the transformation of cotyledonary explants of soybean (Gao et al., 2020).
Bioinformatics Analysis
The homologs of GmPGL2 protein were characterized by BLAST tool in Phytozome1; the phylogenetic and microsyntynteny analysis were performed as described previously (Tang et al., 2020). The signal peptide of GmPGL2 protein was analyzed by TargetP2.0 program.2 The conserved motifs of GmPGL2 domain were predicted as described by Ian Small group (Cheng et al., 2016b; Gutmann et al., 2020). Target RNA sites prediction of GmPGL2 protein used the “PPR CODE PREDICTION WEB SERVER (Ver. 1.6.11)”3 (Yan et al., 2019). Some PPR codes are based on PPR code dataset from Kobayashi et al. (2019).
Subcellular Localization and Microscopy
The full-length CDS and the 594-bp region (GmPGL21-198) of the GmPGL2 gene were fused to green fluorescent protein (GFP) at the C terminal, and then amplified and cloned into the modified 3301H vector at the XmaI and HindIII sites. We prepared two constructs, namely, 35:GmPGL2-GFP and 35S:GmPGL21-198-GFP, which were introduced into A. tumefaciens (strain EHA105) and subsequently used to infiltrate Nicotiana benthamiana leaves as described previously (Waadt and Kudla, 2008). The GFP fluorescence signals were detected using a LSM510 laser scanning confocal microscope (Carl Zeiss, Germany). Transmission electron microscopy was performed according to a previously described method (Kwon and Cho, 2008).
RNA Editing Analysis Through RNA-Sequencing
The total RNA was isolated from the leaves of 12- and 18-day-old HD12 and Gmpgl2 seedlings. Thereafter, rRNAs were removed to retain mRNAs and non-coding RNAs (ncRNAs). The enriched mRNAs and ncRNAs were cut into short fragments in fragmentation buffer and reverse transcribed into cDNA with random primers. The second-strand cDNAs were synthesized using DNA polymerase I, RNase H, dNTPs, and buffer. Next, the cDNA fragments were purified using the QiaQuick PCR Extraction Kit and end-repaired. Poly(A) was added to the fragments, which were finally ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeq™ 4,000 by Gene Denovo Biotechnology Co. (China). The reads containing adapters and low-quality reads were removed, and RNA sequences were eliminated using the alignment tool Bowtie2 (Langmead and Salzberg, 2012). The remaining reads were considered for assembling contigs (cDNA sequences) for transcriptome analysis. The rRNA-mapped reads of each sample were then mapped to the reference genome using TopHAT2 version 2.0.3.12 (Kim et al., 2013). The transcripts of the chloroplast genes were identified by referring to the soybean chloroplast genome database.4 RNA editing of a gene was considered to occur if the fold-change in the mRNA variants with single-nucleotide polymorphisms (SNPs) at the editing sites was ≥2 for reference reads and ≥3 for variant reads, and the mutation frequency was between 0.1 and 0.9 (Bahn et al., 2012; Ramaswami et al., 2013). Three biological replicate samples were analyzed for each developmental stage.
Recombinant Protein Expression and RNA Electrophoresis Mobility Shift Assay
The cDNA fragments of GmPGL2 were amplified with specific primers OL13194 and OL13195 (Supplementary Table S1) and cloned into the pCold vector (TaKaRa, Japan) to generate recombinant His-GmPGL2. The recombinant protein was purified across columns equipped with Ni2+ affinity resin (Ni-NTA Resin, GenScript; Supplementary Figure S1). RNA probes were synthesized and labeled with 6-FAM at the 3' end by GenScript (Nanjing, China). For REMSAs, the method was similar to a previously described protocol with little modification (Xiao et al., 2018). The recombinant protein was incubated with a labeled RNA probe in a reaction mixture including 2x binding buffer (100mM Na phosphate (pH 7.5), 10units RNasin, 0.1mgml−1 BSA, 10mM dithiothreitol, 2.5mgml−1 heparin, and 300mM NaCl). The mixture was incubated at 25°C for 30min followed by separation through 5% native polyacrylamide gel electrophoresis (PAGE) in 1xMOPS buffer (50mM MOPS, 50mM Tris–HCl, 1mM EDTA, pH7.3). After electrophoresis, the gels were imaged using a fluorescent biological image analysis system, Tanon 4600SF (Tianneng, China). Three concentrations (5, 10 and 50 pmol) of unlabeled probes were added to the reaction mixture as competitive probes.
Yeast Two-Hybrid (Y2H) and Luciferase Complementation Imaging Assay
The Gal4-based Y2H assay was performed using the Matchmaker Gold Yeast Two-Hybrid System (TaKaRa, Japan) according to the manufacturer’s instructions. The cDNA of GmPGL2 and mutated GmPGL2 (GmPGL2m) were cloned into the GAL4-binding domain vector (pGBKT7-BD), and the cDNAs of GmMORF1, GmMORF2, GmMORF8, GmMORF9, GmORRM1, and GmORRM6 were cloned into the GAL4 activation domain vector (pGADT7-AD). Combinations of constructs were co-transformed into the yeast strain Y2H Gold (TaKaRa, Japan). The co-transformants were cultured on SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp (QDO) media for 48h at 28°C to verify protein interactions.
The open reading frames of GmPGL2 and GmMORF8, GmMORF9, and GmORRM6 without stop codon were cloned into pCAMBIA1300-nLUC (NLUC) and pCAMBIA1300-cLUC (CLUC), respectively, yielding the GmPGL2-NLUC and MORF8-CLUC, and MORF9-CLUC and ORRM6-CLUC constructs, respectively. These constructs were introduced into A. tumefaciens (strain EHA105) and then used to infiltrate N. benthamiana leaves for the luciferase complementation imaging (LCI) assay as described previously (Wang et al., 2020). After incubation for 48h under a 16-h light/8-h dark cycle, the leaves were injected with D-luciferin at the final concentration of 1mM. Luciferase signals were imaged using the Tanon 4600SF system (Tianneng, China). GmAPC13a and GmILPA1 (Gao et al., 2017) were used as the positive controls for Y2H and LCI analysis.
Statistical Analyses
All samples had at least three biological replicates. The statistical analyses were performed using R software (version 3.6.2) as described previously (Tang et al., 2020). Asterisks indicate significant differences as determined by p values (*p<0.05; *p<0.01; and **p<0.001).
Results
Isolation and Phenotypic Characterization of the Gmpgl2 Mutant
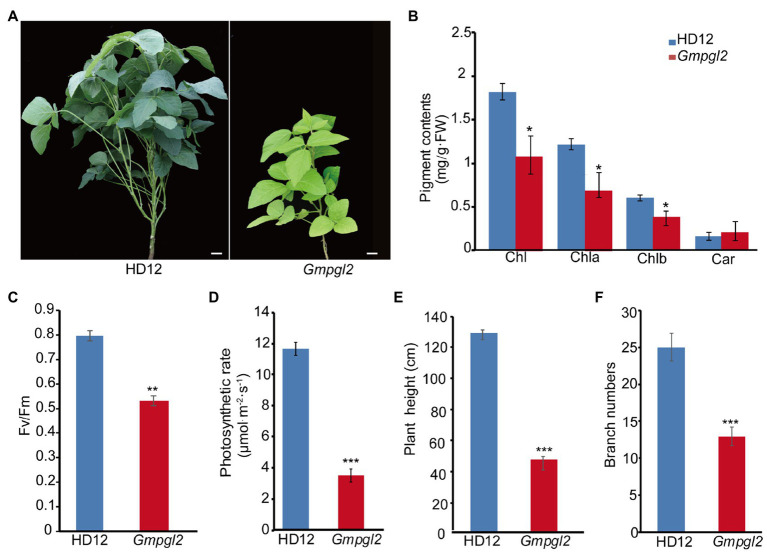
To elucidate the mechanism of chloroplast development in soybean, we isolated the Gmpgl2 mutant by screening nearly 10,000 60Co-γ radiation-induced M2 mutants for pale green leaf from a mutagenesis population of HD12 (Cheng et al., 2016a). The pale green leaves of the Gmpgl2 mutant were clearly identified upon the emergence of the first true leaf and throughout the developmental process (Figure 1A). The components of total chlorophyll (Chl), chlorophyll a (Chla), and chlorophyll b (Chlb) in the leaves of the Gmpgl2 mutants decreased by 40.4, 42.7, and 35.7% of the respective values in the wild-type leaves. The carotenoid (Car) content in the leaves of the mutant plants was 80% that in the leaves of wild-type plants (Figure 1B). Furthermore, the ratio of Chl/Car in the Gmpgl2 mutant was lower than in the wild type, which might be due to a substantial decrease in chlorophyll synthesis compared to carotenoid synthesis.
Figure 1.
Phenotypes of Hedou12 (HD12; wild type) and Gmpgl2 (mutant) plants. (A) Leaves of HD12 and Gmpgl2. Bar=5cm. (B) Pigment content in 18-day-old HD12 and Gmpgl2 seedlings. Chla, chlorophyll a; Chlb, chlorophyll b; and Car, carotenoid. (C) Photosystem II maximum quantum yield (Fv/Fm) photosynthetic rate of 18-day-old HD12 and Gmpgl2 seedlings. (D) Photosynthetic rate of 18-day-old HD12 and Gmpgl2 seedlings. Values are mean±SE of three biological replicates. Asterisks denote significant differences from HD12, as determined using Student’s t-test. (E) Height of HD12 and Gmpgl2 mutant plants. (F) Number of branches in HD12 and Gmpgl2 mutant plants.
The photosystem II maximum quantum yield of Gmpgl2 was only 67.1% of that of HD12, and the photosynthetic rate of Gmpgl2 was 3.41±0.39μmol−1m2·s−1, representing 70.6% of that of HD12 (Figures 1C,D). The Gmpgl2 plants were shorter with fewer branches than HD12 plants (Figures 1E,F). Together, these results showed that the mutation in GmPGL2 leads to defects in chloroplast biogenesis.
Genetic Mapping Reveals That GmPGL2 Encodes a PPR Protein
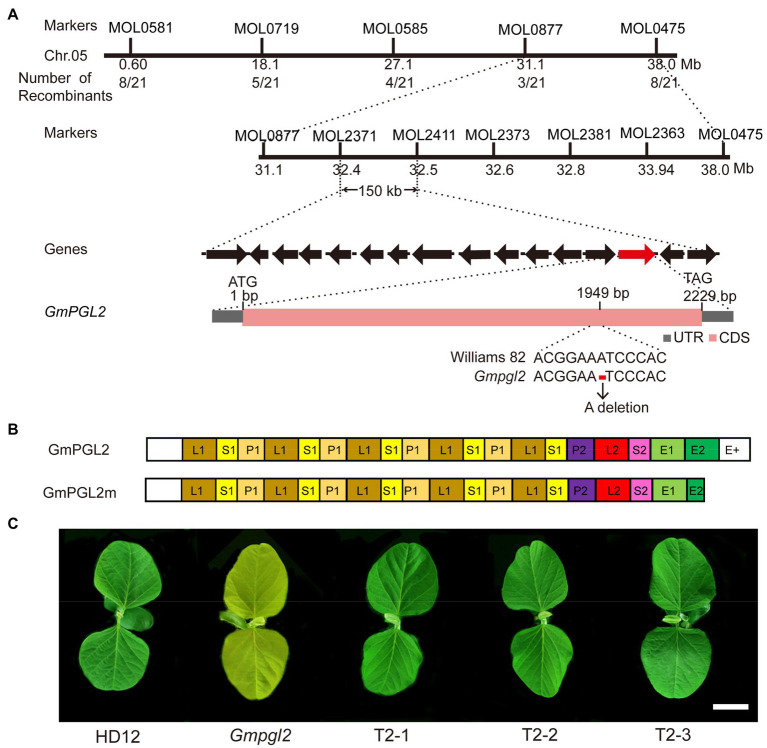
In an attempt to identify the GmPGL2 gene, the Gmpgl2 mutant was crossed with the cultivar Williams 82 to generate a segregation population for mapping. The F1 plants were normal, and the F2 plants segregated in the ration of 3:1 (green:pale green=210:60; χ2=0.53; p>0.05), indicating that the Gmpgl2 phenotype was controlled by a single recessive nuclear gene. Using 165 insertion/deletion (InDel) markers developed earlier (Song et al., 2015), the GmPGL2 locus was initially mapped to a 6.9-Mb region between the MOL0877 and MOL0475 markers of chromosome 5 (Figure 2A). Thereafter, the GmPGL2 gene was further mapped to a 150-kb region between markers MOL2371 and MOL2411 using 702 F3 individuals with the pale green phenotype. Fifteen genes in this region were predicted according to the annotation in the reference genome of Williams 82 (Glycine max Wm82.a2.v1, see footnote 1). Analyses of the coding sequences of these 15 genes in HD12 and Gmpgl2 revealed a single adenine (A) deletion at 1949bp of the Glyma.05g132700 gene resulting in a frame-shift mutation (Figure 2A; Supplementary Table S2).
Figure 2.
Map-based cloning of the GmPGL2 gene. (A) Map-based cloning of the GmPGL2 allele. (i) The GmPGL2 mutation was narrowed to a 150-kb region between the InDel markers MOL2371 and MOL2411 on chromosome 5. (ii) The black arrows represent the 15 putative genes in this 150-kb genomic region; the candidate gene GmPGL2 (Glyma.05g132700) is indicated with the red arrow. (iii) Schematic diagram of the Glyma.05g132700 gene. ATG and TAG, the start and stop codons, respectively, are shown. A deletion point mutation at the 1949bp nucleotide position is shown. (B) Schematic diagram of the GmPGL2 protein. The GmPGL2 gene encodes a PPR protein of the PLS-type carrying a E+ domain. The GmPGL2 protein has 17 PPR motifs; P, L, and S represent the PPR motifs of various repeat lengths; E+ is a domain required for RNA editing. (C) Complementation of the Gmpgl2 mutant. Phenotypes of HD12, GmPGL2, and a 35S:GmPGL2 complemented Gmpgl2 plant. Scale bars=3cm.
To further confirm that the mutation in the Glyma.05g132700 gene was responsible for the pale green leaf phenotype of Gmpgl2, the coding sequence (CDS) of Glyma.05G132700, driven by the cauliflower mosaic virus 35S promoter, was transformed into the Gmpgl2 mutant via A. tumefaciens. Three independent transgenic lines carrying the 35S:GmPGL2 expression cassette were obtained in the Gmpgl2 background. All transgenic lines completely rescued the Gmpgl2 phenotype (Figure 2C), confirming Glyma.05g132700 as GmPGL2.
GmPGL2 Encodes a PPR Protein Localized to the Chloroplast
Analyses of the deduced GmPGL2 amino acid sequence indicated that the GmPGL2 protein contains 17 PPR motifs, an E1/E2 domain, and a E+ domain, and is thus classified as a PPR-E+ subclass protein, similar to that in other PPR proteins reported previously (Rivals et al., 2006). A single-nucleotide deletion at 1949bp of its CDS (Figure 2A) results in a frame-shift mutation and generates a truncated protein that lacks a portion of the of E2 and E+ motif (Figure 2B).
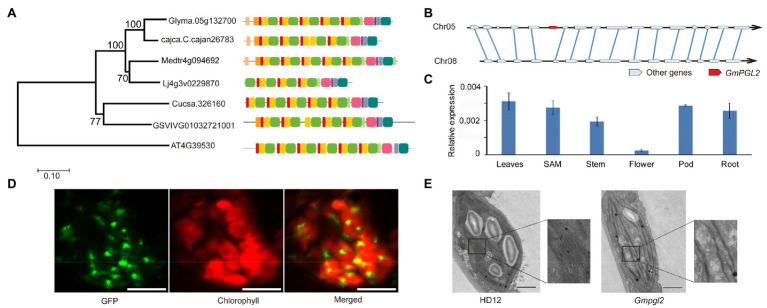
The GmPGL2 protein sequence was used to identify its homologs in Arabidopsis thaliana, Medicago truncatula, Lotus corniculatus, Cajanus cajan, Cucumis sativus, and Vitis vinifera to construct a phylogenetic tree. GmPGL2 showed 74.3, 75.3, and 80.1% identity with homologs cajca.C.cajan26783, Medtr4g094692, and Lj4g3v0229880 of C. cajan, M. truncatula, and L. corniculatus, respectively (Figure 3A). Following whole genome duplication, it is expected that soybean would carry two homologous GmPGL2 genes. However, only one GmPGL2 copy was observed in the phylogenetic analysis. To confirm this finding, we performed synteny analysis of the 143,208-bp region around GmPGL2. The syntenic analysis results revealed that 32,522,450–32,670,694bp of chromosome 5 and 6,566,793–6,686,020bp of chromosome 8 are more likely duplicated blocks in the soybean genome. As expected, no homolog of GmPGL2 was found on chromosome 8 (Figure 3B). These results suggested that GmPGL2 is a single copy gene in the soybean genome. To further understand the function of GmPGL2, the expression levels of GmPGL2 in different tissues were examined using qRT-PCR. GmPGL2 was expressed in all tested tissues and at higher levels in leaf than in stem, flower, pod, and root (Figure 3C), suggesting that GmPGL2 may have a vital role in leaves.
Figure 3.
GmPGL2 is a single copy gene. (A) Phylogenetic tree of GmPGL2 and homologous GmPGL2 proteins from Arabidopsis thaliana, Medicago truncatula, Lotus corniculatus, Cajanus cajan, Cucumis sativus, and Vitis vinifera, constructed using MEGA 5 with the neighbor-joining method. (B) Synteny plot of the homologous GmPGL2 regions. Solid blue lines show the pairs of homologous genes of the two homologous chromosomes. (C) Expression of GmPGL2 in different tissues. (D) Subcellular localization of GmPGL2. GFP signals of the 3301H-GmPGL21-198-GFP fusion protein were localized to the chloroplast of epidermal cells of N. benthamiana leaves. Green fluorescence signals, chlorophyll auto-fluorescence signals, and a merged image are shown. Bar=10μm. (E) Transmission electron micrographs of the chloroplasts of 18-day-old HD12 and Gmpgl2 seedlings. Bar=2μm.
TargetP5 prediction analysis showed a chloroplast-targeting signal at the N terminus (1–198 aa) of the GmPGL2 protein. To localize GmPGL2, the full-length GmPGL2 (without a stop codon) was fused with the GFP and transformed into N. benthamiana; however, no fluorescence signal was detected in the leaf cells of N. benthamiana. We suspected that full-length GmPGL2 with GFP may have been too large to be efficiently expressed, or over-expression of the full-length protein may be detrimental to the cells. Subsequently, a shortened 198-aa N-terminal sequence was fused with GFP (35S,GmPGL21-198-GFP) to detect its localization, and the fluorescent signals were detected as punctuated dots localized to chloroplasts (Figure 3D). No GFP signal was detected in other compartments of the cell, suggesting that the GmPGL2 protein is localized to the chloroplast. The phenomenon of full-length PPR protein fused to GFP without a fluorescence signal has been observed previously (Wang et al., 2019).
Next, we observed the chloroplast ultrastructure of HD12 and Gmpgl2 leaves. In the mesophyll cells of HD12 leaves, chloroplasts showed typical structures with continuous stacking of the grana (Figure 3E). In contrast, the ultrastructural analysis of chloroplasts in the leaves of the mutant showed less stacking of the grana than HD12 (Figure 3E).
GmPGL2 Is Required for the C-to-U Editing During Leaf Development
To understand the role of RNA editing in the development of soybean leaves, we analyzed the variations in chloroplast RNA sequences obtained from high throughput RNA-Seq data between HD12 and Gmpgl2 at each of the following two developmental stages: (i) Stage 1 (S1): first trifoliate leaf of 12-day-old seedlings and (ii) Stage 2 (S2): first trifoliate leaf of 18-day-old seedlings.
Based on SNPs of the observed chloroplast RNA sequences and referenced sequences, 43 candidate RNA editing target sites were identified in the two stages in HD12 (Table 1). Thirty-eight of the 43 RNA editing sites were found to be located in the coding regions of 17 genes; whereas the other were located in the downstream, intergenic, and intron regions of ndhK-12704, ndhJ-14209, rps12-106113, rps12-138416, and rps16-55714, respectively. The 36 editing sites result in amino acid changes, except for ndhC-10779 and petB-74300. These editing sites result in six types of amino acid changes, namely, serine to leucine, proline to leucine, serine to phenylalanine, threonine to methionine, histidine to tyrosine, and threonine to leucine. We then compared the editing efficiency of HD12 and Gmpgl2 during these two developmental stages.
Table 1.
RNA editing efficiency between the HD12 and Gmpgl2 seedlings.
| Gene name | Editing sites | HD12-S1 | HD12-S2 | Gmpgl2-S1 | Gmpgl2-S2 | Amino acid change |
|---|---|---|---|---|---|---|
| ndhC | 10779 | 29.58% | 28.57% | 28.98% | 28.73% | Unchanged |
| ndhC | 11062 | 95.76% | 97.45% | 94.38% | 91.28% | Ser-Leu |
| ndhK | 12704 | 2.89% | 11.38% | 0.41% | 4.91% | Downstream |
| ndhJ | 14209 | 2.88% | 17.12% | 2.62% | 10.17% | Intergenic |
| rps14 | 23651 | 87.54% | 75.43% | 77.29% | 68.19% | Ser-Leu |
| rpoB | 34587 | 63.16% | 52.80% | 66.03% | 52.01% | Ser-Phe |
| rpoB | 34800 | 60.97% | 70.40% | 71.28% | 63.21% | Ser-Leu |
| rpoB | 34815 | 59.00% | 75.71% | 63.50% | 70.22% | Ser-Leu |
| rpoB | 36249 | 89.79% | 86.59% | 80.93% | 80.49% | Ser-Phe |
| rpoC1 | 37529 | 75.08% | 75.36% | 75.79% | 67.91% | Ser-Leu |
| rpoC1 | 38778 | 74.02% | 62.27% | 60.08% | 47.24% | Ser-Leu |
| rpoC2 | 43303 | 3.32% | 15.51% | 3.11% | 2.56% | Ser-Leu |
| rps2 | 45133 | 94.23% | 95.49% | 87.95% | 85.52% | Thr-Ile |
| rps2 | 45247 | 87.55% | 93.77% | 81.33% | 80.77% | Ser-Leu |
| atpF | 48596 | 86.50% | 90.61% | 72.21% | 74.35% | Pro-Leu |
| rps16 | 55714 | 81.84% | 89.00% | 90.59% | 76.07% | intron |
| rps16 | 56313 | 79.02% | 77.48% | 0.00% | 0.00% | Ser-Leu |
| accD | 57518 | 87.08% | 90.13% | 85.64% | 74.74% | Ser-Leu |
| psaI | 58512 | 89.09% | 91.48% | 85.18% | 83.38% | His-Tyr |
| rps18 | 66641 | 84.39% | 72.97% | 57.89% | 50.63% | Ser-Leu |
| petB | 74300 | 26.13% | 26.77% | 22.00% | 24.78% | Unchanged |
| petB | 74899 | 93.59% | 96.18% | 93.81% | 88.49% | Ser-Leu |
| rps12 | 106113 | 36.84% | 0% | 0.00% | 0% | Intergenic |
| ndhA | 115982 | 79.04% | 89.52% | 77.35% | 84.16% | Ser-Leu |
| ndhA | 117982 | 62.86% | 89.87% | 63.66% | 85.68% | Ser-Phe |
| ndhE | 119873 | 64.74% | 80.82% | 42.93% | 52.57% | Pro-Leu |
| ndhD | 120618 | 41.35% | 83.96% | 18.75% | 55.69% | Thr-Met |
| ndhD | 120999 | 73.33% | 84.51% | 57.33% | 71.82% | Thr-Ile |
| ndhD | 121290 | 65.86% | 80.65% | 69.50% | 82.43% | Ser-Leu |
| ndhD | 121494 | 64.94% | 81.79% | 55.67% | 78.83% | Ser-Leu |
| ndhD | 121914 | 44.95% | 46.69% | 28.91% | 37.30% | Ser-Leu |
| ndhF | 124681 | 44.09% | 87.06% | 35.59% | 55.98% | Ser-Leu |
| rps12 | 138416 | 80.00% | 71.91% | 51.23% | 49.59% | Intron |
| ndhB | 139627 | 92.33% | 80.00% | 68.57% | 78.67% | Ser-Leu |
| ndhB | 140020 | 83.63% | 96.65% | 74.06% | 90.27% | Thr-Met |
| ndhB | 140064 | 64.08% | 91.35% | 50.08% | 81.80% | His-Tyr |
| ndhB | 140215 | 73.86% | 92.19% | 47.35% | 71.96% | Pro-Leu |
| ndhB | 140224 | 76.93% | 89.32% | 61.82% | 72.92% | Ser-Phe |
| ndhB | 140999 | 78.49% | 91.12% | 61.73% | 75.21% | Ser-Leu |
| ndhB | 141005 | 82.14% | 90.21% | 60.49% | 66.95% | Ser-Leu |
| ndhB | 141281 | 77.03% | 87.68% | 53.58% | 59.70% | Ser-Leu |
| ndhB | 141424 | 61.89% | 84.78% | 33.91% | 51.22% | His-Tyr |
| ndhB | 141650 | 69.47% | 91.50% | 49.78% | 79.97% | Pro-Leu |
In S1, the editing efficiency of 20 editing sites decreased by more than 10% in Gmpgl2 compared with that in HD12; these sites were atpF-48596; ndhB-139627, -141281, -141424, -141005, -140215, -140224, -140999, -140064, and -141650; ndhD-120618, -121914, and -120999; ndhE-119873; rpoC1-38778; and rps14-23651, rps16-56313, and rps18-66641. Two of which (rps12-106113 and rps12-138416) were located in the intron region, whereas the remaining editing sites caused CDS alterations in 13 genes (Table 1).
In S2, five of the above editing sites (ndhB-140064 and -139627, ndhD-121914, rps14-23651, and rps12-106113) restored to less than 10% of the RNA editing efficiency between the HD12 and mutant seedlings, whereas five new editing sites (ndhF-124681, accD-57518, rps2-45247, rpoC2-43303, and rps16-55714) reduced more than 10% of the editing efficiency of the HD12 seedlings. These newly detected five editing sites led to four amino acids changes, as rps16-55714 was located in the intron region. Another 20 editing sites of 12 genes altered the editing efficiency of the Gmpgl2 seedlings during development stage 2 (Table 1).
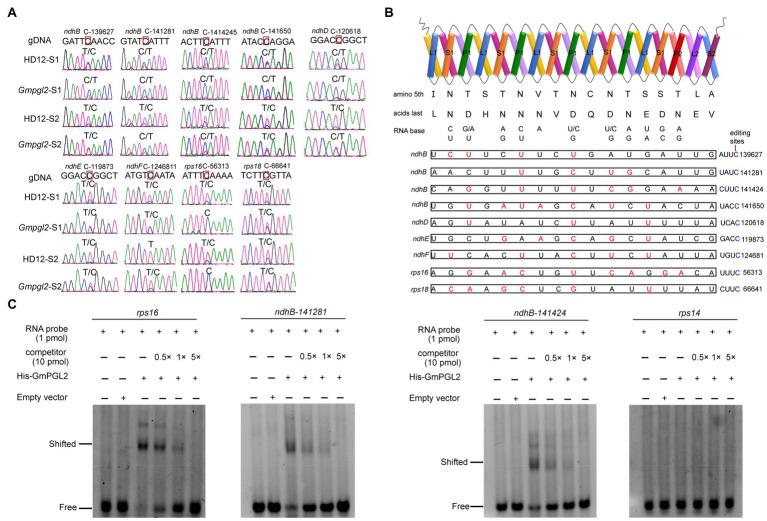
To validate the editing sites detected in chloroplast RNA sequences, we sequenced the cDNA of the above 36 amino acid-changed chloroplast editing sites (Figure 4A and Supplementary Figure S2). Nine putative editing sites were verified to differ between the HD12 and Gmpgl2 seedlings in the two developmental stages: ndhB-139627, -141281, -141424, and -141650; ndhD-120618; ndhE-119873; ndhF-124681; rps16-56313; and rps18-66641. The editing site of rps16-56313 was altered most drastically. In HD12, the transcripts were edited in both developmental stages, whereas no editing was detected in Gmpgl2. The editing sites of ndhB-139627, -141281, -141424, -141650; ndhD-120618; ndhE-119873; ndhF-124681; and rps18-66641 were also reduced in the two developmental stages in the Gmpgl2 mutant compared with those in HD12 (Figure 4A). The C-to-U editing caused serine to leucine substitutions in the ndhB-139627, ndhB-141281, ndhF-124681, rps16-56313, and rps18-66641 transcripts; a change from histidine to tyrosine in the ndhB-141424 transcripts; a change from proline to leucine in ndhB-141650 and ndhE-119873 transcripts, and a change from threonine to methionine in the ndhD-120618 transcripts.
Figure 4.
Analysis of GmPGL2 targeted genes and their interactions. (A) RNA editing sites validated by sequencing of the RT-PCR products. Green, black, red, and blue peaks represent A, G, T, and C, respectively. S1, leaves of 12-day-old seedlings; S2, leaves of 18-day-old seedlings. The names of the RNA editing sites are shown on top of the chromatograms. The edited bases are indicated by red square. (B) Schematic representation of GmPGL2 bound to its target sites. The combinations of positions 5 and last amino acid in each PPR motif of GmPGL2 were aligned to the nucleotide upstream of the editing site. Permissible matched nucleotides are indicated in red, and the edited sites C are indicated in blue. (C) REMSA of GmPGL2 protein with its target probes, rps16, and ndhB-141281, 141424. rps14 probe was served as a negative control.
Above results implied that GmPGL2 participated in nine editing sites (ndhB-139627, -141281, -141424, -141650, ndhD-120618, ndhE-119873, ndhF-124681, rps16-56313, and rps18-66641) during young leaf development of soybean, and the rps16-56313 was completely abolished in the Gmpgl2 mutant.
GmPGL2 Protein Can Directly Bind to Its Targets in vitro
To evaluate the PPR-RNA recognition model, we analyzed the associations between GmPGL2 and chloroplast genes (Figure 4B). We found that the match indices between GmPGL2 and RNA editing efficiency of above nine sites, ndhB-139627, -141281, -141424, -141650, ndhD-120618, ndhE-119873, ndhF-124681, rps16-56313, and rps18-66641, were 4/17, 4/17, 6/17, 7/17, 3/17, 5/17, 5/17, 8/17, and 6/17, respectively (Figure 4B).
To further confirm that GmPGL2 actually binds to above sites, ndhB-141281, ndhB-141424, and rps16-56313 were selected to carry out the REMSA with the FAM-labeled RNA probes. The retarded bands appeared when His-GmPGL2 protein was incubated with these three labeled probes, however only free RNA probe bands were detected with the rps14 probe as the negative control. The binding capacity to the labeled probes gradually decreased following the increased competitor concentration (Figure 4C), indicating that GmPGL2 binds to ndhB-141281, ndhB-141424, and rps16-56313 directly in vitro.
GmPGL2 Influences the Expression of Nuclear- and Plastid-Encoded Genes and Regulates Chloroplast Development
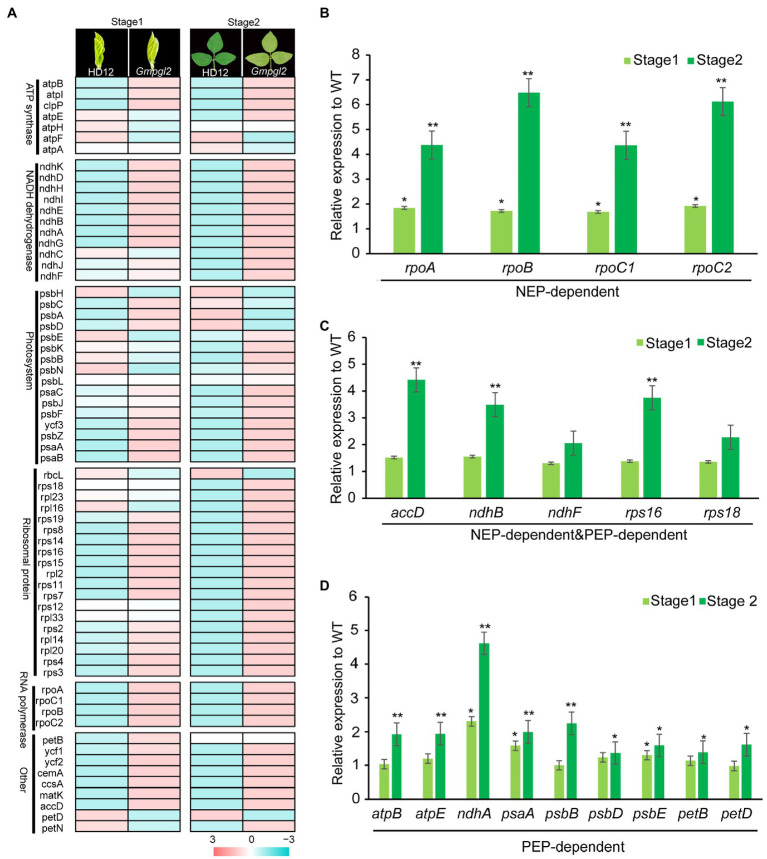
In each of the two developmental stages, 66 chloroplast genes in HD12 and Gmpgl2 seedlings were detected by RNA-Seq (Figure 5A). In the S1 developmental stage, a pairwise comparison of genes between HD12 and Gmpgl2 seedlings revealed 11 differentially expressed genes (DEGs), namely, rps15, rps16, rpl2, rpoA, rpoC2, ndhA, ndhG, ycf2, matK, psbZ, and cemA. Meanwhile, in the S2 developmental stage, 30 DEGs were detected, namely accD, matK, ndhA, ndhB, ndhD, ndhG, ndhH, ndhI, psaC, rpl2, rpl14, rpl16, rpl20, rpl23, rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps15, rps16, rps19, rpoA, rpoB, rpoC1, rpoC2, ycf1, and ycf2 (Figure 5A). Most of DEGs encoded different components of the chloroplast-specific ribosomal proteins, suggesting that the mutation of GmPGL2 influenced the expression of a large number of chloroplast genes.
Figure 5.
(A) Expression of chloroplast genes in the leaves of 12-day-old (Stage1) and 18-day-old (Stgae2) HD12 and Gmpgl2 seedlings. (B-D) Comparison expression levels of chloroplast genes transcribed by NEP (B), both NEP and PEP (C), and PEP (D) between HD12 and Gmpgl2 seedlings. The values are the mean ratio±standard deviation with three biological repeats. The asterisks indicate significant differences between WT and Gmpgl2 (Student’s test; *p<0.05; *p<0.01).
The expression of chloroplast-encoded genes is strongly associated with chloroplast development, and it is coordinately transcribed by nuclear-encoded RNA polymerase (NEP) and plastid-encoded RNA polymerase (PEP; Huang et al., 2018; Xiao et al., 2018). To investigate the role of GmPGL2 in chloroplast development, the expression levels of PEP- and NEP-dependent genes were compared between Gmpgl2 and HD12 seedlings. The transcription levels of the most selected genes in Gmpgl2 were upregulated in the young leaf developmental stage 2 compared with HD12 (Figures 5B–D). The results were consistent with those of RNA-Seq, suggesting that GmPGL2 most likely influences the expression of nuclear- and plastid-encoded genes during chloroplast development.
GmPGL2 Interacts With MORF8, MORF9, and ORRM6
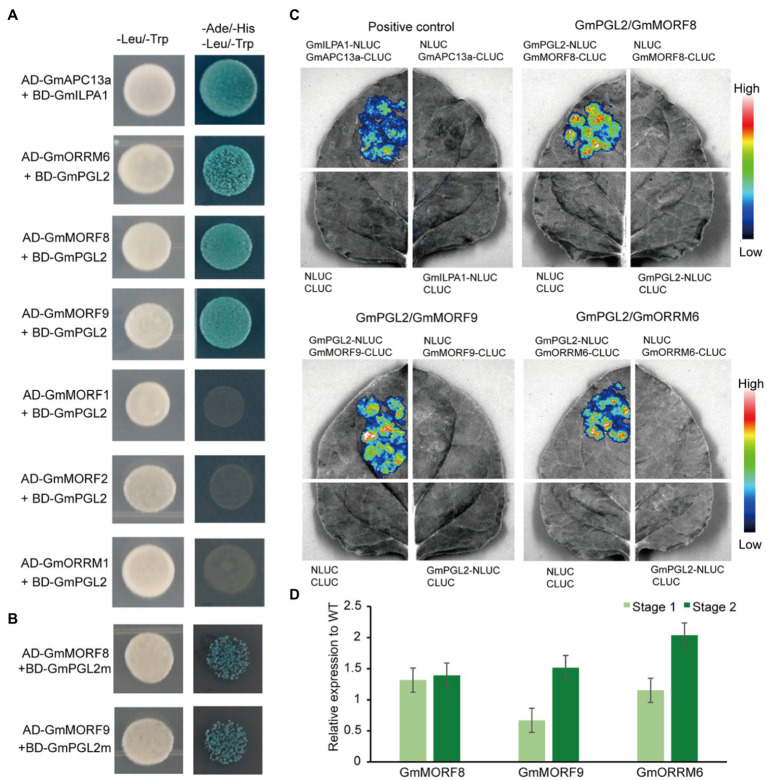
To investigate GmPGL2 interaction proteins, the candidates of MORF and ORRM proteins were identified by searching for homologs from the relevant database (see footnote 1). Four MORF proteins and two ORRM proteins were selected to detect in vitro protein interaction through yeast two-hybrid assay, and they are GmMORF1 (Glyma.08G188700), GmMORF2 (Glyma.04G042700), GmMORF8 (Glyma.13G271400), GmMORF9 (Glyma.15G064300), GmORRM1 (Glyma.20G187000), and GmORRM6 (Glyma.16G217600; Figure 6A). GmMORF8/GmPGL2, GmMORF9/GmPGL2, and GmORRM6/GmPGL2 co-transformants were able to grow in the quadrupole drop-out (QDO) medium, whereas GmMORF1/GmPGL2, GmMORF2/GmPGL2, and GmORRM1/GmPGL2 co-transformants could not. We also found that the interactions of mutated GmPGL2 (GmPGL2m) protein with GmMORF8 and GmMORF9 were decreased (Figure 6B). These results suggested that the mature form of GmPGL2 interacts with GmMORF8, GmMORF9, and GmORRM6, but not with GmMORF1, GmMORF2, and GmORRM1.
Figure 6.
GmPGL2 interacts with GmMORFs and GmORRMs. GmAPC13a and GmILPA1 represent the positive control. (A) Yeast two-hybrid assay between GmPGL2 and some GmMORFs and GmORRMs. (B) Yeast two-hybrid assay between mutated GmPGL2 (GmPGL2m) and GmMORF8, GmMORF9, respectively. (C) Luciferase complementation imaging was performed to confirm the interactions between GmPGL2 and GmMORF8, GmMORF9, and GmORRM6 in N. benthamiana, respectively. The fluorescent signal intensity represents the strength of interaction. (D) The expression levels of GmMORF8, GmMORF9, and GmORRM6 during two developmental stages of young leaves in wild type and Gmpgl2. The values are the mean ratio±standard deviation with three biological repeats. None significant difference was determined by Student’s t-test between wild type and mutant.
Luciferase complementation imaging was performed to further investigate these interactions. High luciferase activity was detected after the co-expression of GmMORF8, GmMORF9, and GmORRM6 fused to C-terminal luciferase (CLUC) and GmPGL2 fused to N-terminal luciferase (NLUC; Figure 6C). These findings indicated that GmPGL2 might affect RNA editing by interacting with GmMORF8, GmMORF9, and GmORRM6. However, the expression levels of GmMORF8, GmMORF9, and GmORRM6 were not significantly different between HD12 and Gmpgl2 mutants during the two developmental stages (Figure 6D). It implied that the loss of E+ motif in Gmpgl2 mutant might serve minor role among their interactions, which was not the binding domain of PPR protein with MORF and ORRM proteins (Hayes et al., 2015; Small et al., 2020).
Discussion
As a post-transcriptional modification process, RNA editing fine-tunes gene expression and functions by altering specific nucleotides of a transcript (Oldenkott et al., 2020). In flowering plants, RNA editing generally changes cytidine to uridine in plastids and mitochondria, playing important roles in organelle biogenesis, adaptation to environmental changes, and signal transduction; PPR, MORF, and ORRM proteins play curial roles in plant RNA editing (Lu, 2018; Zhang et al., 2019; Small et al., 2020). Here, we found that the mutation of PLS-PPR protein, GmPGL2, caused the abnormal chloroplast development. GmPGL2 protein recognized nine RNA editing sites in six chloroplast transcripts in soybean, including ndhB-139627, -141281, -141424, -141650, ndhD-120618, ndhE-119873, ndhF-124681, rps16-56313, and rps18-66641. The editing capacity of GmPGL2 varies in these nine sites during the different stages of young leaf development. These six genes belong to the subunit of NDH complex and ribosomal protein. NdhB, NdhD, NdhE, and NdhF proteins are the subunit of complex of NDH, which is encoded by a combination of genes residing in the plastid and nuclear genomes. Rps16 and Rps18 proteins are the subunit of ribosomal proteins that translate the chloroplast-encoded proteins. The defects of post-transcriptional processing of rps16 and rps18 might decrease the chloroplast translation efficiency. GmPGL2 interacted with GmMORF8, GmMORF9, and GmORRM6 in vitro (Figure 6B). Therefore, we propose that GmPGL2 participates in soybean RNA editing together with GmMORFs and GmORRMs.
Previous studies in Arabidopsis, rice, and maize have demonstrated that a lack of RNA editing is often associated with changes in the expression levels of chloroplast genes (Jiang et al., 2018). In most cases, the expression level of NEP increased that of PEP decreased in mutants, such as chloroplast biogenesis 19 (clb19), pigment-deficient mutant1 (pdm1), and pigment-defective mutant 2 (pdm2; Chateigner Boutin et al., 2008; Du et al., 2017). However, in the Gmpgl2 mutant, the expression of NEP and PEP increased in developmental stages S1 and S2. Recently, this kind of changes has been observed in the knockout of SLC1 gene, which encodes a P subgroup of PPR protein in rice (Lv et al., 2020). In slc1 mutant, the transcript levels of 3 chloroplast ribosomal RNAs and 16 chloroplast development-related and photosynthesis-related genes were also significant increased. This phenomenon was attributed to preclude the intron splicing of rps16 in the slc1 mutant, which blocked the post-transcriptional processing and translation of rps16, and failed to assemble the normal 70S ribosomes (Lv et al., 2020). In our study, the rps16-56313 was the unique editing site, which was completely abolished in the Gmpgl2 mutant. We infer that the lack of an edited functional Rps16 protein might compromise the function of the ribosome complex in chloroplasts. As a result, many of the chloroplast proteins may not be translated to the optimal levels, and this may be compensated in the Gmpgl2 mutant by increased gene transcription.
In summary, the defective development of chloroplasts observed in the Gmpgl2 mutant can be attributed to failure of RNA editing at the subunit of NDH complex and ribosomal protein-related genes, and GmPGL2 plays a crucial role in chloroplast development and normal plant growth.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: NCBI repository, accession number: PRJNA660896 (https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA660896).
Author Contributions
SY and QW conceived the project and designed the research. XF performed most of the experiments. YZ, HY, and JL helped to clone the gene. CZ carried out the REMSA experiment. KT and GL performed bioinformatics analysis. XF and SY prepared the figures and wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by the Chinese Academy of Sciences (ZDRW-ZS-2019-2 and XDA24030303) and the National Natural Science Foundation of China (31971901).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Madan Bhattacharyya (Iowa State University) and Prof. Ning Jiang (Michigan State University) for critically reading the manuscript.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.690973/full#supplementary-material
References
- Ainsworth E. A., Yendrek C. R., Skoneczka J. A., Long S. P. (2012). Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant Cell Environ. 35, 38–52. 10.1111/j.1365-3040.2011.02378.x [DOI] [PubMed] [Google Scholar]
- Aryamanesh N., Ruwe H., Sanglard L. V. P., Eshraghi L., Bussell J. D., Howell K. A., et al. (2017). The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 173, 1164–1176. 10.1104/pp.16.01840, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J. H., Lee J. H., Li G., Greer C., Peng G., Xiao X. (2012). Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 22, 142–150. 10.1101/gr.124107.111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Rojas M., Fujii S., Yap A., Chong Y. S., Bond C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8:e1002910. 10.1371/journal.pgen.1002910, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussardon C., Salone V., Avon A., Berthome R., Hammani K., Okuda K., et al. (2012). Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24, 3684–3694. 10.1105/tpc.112.099507, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Ji D., Peng L., Guo J., Ma J., Zou M., et al. (2009). LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 150, 1260–1271. 10.1104/pp.109.136812, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner Boutin A. L., RamosVega M., Guevara Garcia A., Andres C., Gutierrez Nava M. d. l. L., Cantero A., et al. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602. 10.1111/j.1365-313X.2008.03634.x, PMID: [DOI] [PubMed] [Google Scholar]
- Chateigner Boutin A. L., des Francs Small C. C., Fujii S., Okuda K., Tanz S. K., Small I. (2013). The E domains of pentatricopeptide repeat proteins from different organelles are not functionally equivalent for RNA editing. Plant J. 74, 935–945. 10.1111/tpj.12180 [DOI] [PubMed] [Google Scholar]
- Cheng W., Gao J., Feng X., Shao Q., Yang S., Feng X. (2016a). Characterization of dwarf mutants and molecular mapping of a dwarf locus in soybean. J. Integr. Agric. 15, 2228–2236. 10.1016/S2095-3119(15)61312-0 [DOI] [Google Scholar]
- Cheng S., Gutmann B., Zhong X., Ye Y., Fisher M. F., Bai F., et al. (2016b). Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547. 10.1111/tpj.13121 [DOI] [PubMed] [Google Scholar]
- Colcombet J., Lopez Obando M., Heurtevin L., Bernard C., Martin K., Berthome R., et al. (2013). Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 10, 1557–1575. 10.4161/rna.26128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai A., Yang S., Zhou H., Tang K., Li G., Leng J., et al. (2018). Evolution and expression divergence of the CYP78A subfamily genes in soybean. Genes 9:611. 10.3390/genes9120611, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhang J., Qu S., Zhao Y., Su B., Lv X., et al. (2017). The pentratricopeptide repeat protein pigment-defective mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant and Cell Physiol. 58, 747–759. 10.1093/pcp/pcx004, PMID: [DOI] [PubMed] [Google Scholar]
- Feng X., Yang S., Tang K., Zhang Y., Leng J., Ma J., et al. (2019). GmPGL1, a thiamine thiazole synthase, is required for the biosynthesis of thiamine in soybean. Front. Plant Sci. 10:1546. 10.3389/fpls.2019.01546, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Small I. (2011). The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191, 37–47. 10.1111/j.1469-8137.2011.03746.x [DOI] [PubMed] [Google Scholar]
- Gao J., Yang S., Cheng W., Fu Y., Leng J., Yuan X., et al. (2017). GmILPA1, encoding an APC8-like protein, controls leaf petiole angle in soybean. Plant Physiol. 174, 1167–1176. 10.1104/pp.16.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Yang S., Tang K., Li G., Gao X., Liu B., et al. (2020). GmCCD4 controls carotenoid content in soybeans. Plant Biotechnol. J. 19, 801–813. 10.1111/pbi.13506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann B., Royan S., Schallenberg R. M., Lenz H., Castleden I. R., McDowell R., et al. (2020). The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol. Plant 13, 215–230. 10.1016/j.molp.2019.11.002, PMID: [DOI] [PubMed] [Google Scholar]
- Haili N., Planchard N., Arnal N., Quadrado M., Vrielynck N., Dahan J., et al. (2016). The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH dehydrogenase subunit 7 mRNA in Arabidopsis. Plant Physiol. 170, 354–366. 10.1104/pp.15.01591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M. L., Dang K. N., Diaz M. F., Mulligan R. M. (2015). A conserved glutamate residue in the C-terminal deaminase domain of pentatricopeptide repeat proteins is required for RNA editing activity. J. Biol. Chem. 290, 10136–10142. 10.1074/jbc.M114.631630, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M. L., Giang K., Berhane B., Mulligan R. M. (2013). Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. J. Biol. Chem. 288, 36519–36529. 10.1074/jbc.M113.485755, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang K., Huang W., Liu G., Gao Y., Wang J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in hong-lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24, 109–122. 10.1105/tpc.111.093211, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Li Z. R., Yu Q. B., Ye L. S., Cui Y. L., Molloy D. P., et al. (2019). MORF2 tightly associates with MORF9 to regulate chloroplast RNA editing in Arabidopsis. Plant Sci. 278, 64–69. 10.1016/j.plantsci.2018.10.020, PMID: [DOI] [PubMed] [Google Scholar]
- Huang W., Zhu Y., Wu W., Li X., Zhang D., Yin P., et al. (2018). The pentatricopeptide repeat protein SOT5/EMB2279 is required for plastid rpl2 and trnK intron splicing. Plant Physiol. 177, 684–697. 10.1104/pp.18.00406, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian B., Liu B., Bi Y., Hou W., Wu C., Han T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 9:59. 10.1186/1471-2199-9-59, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Zhang J., Rong L., Feng Y., Wang Q., Song Q., et al. (2018). ECD1 functions as an RNA-editing trans-factor of rps14-149 in plastids and is required for early chloroplast development in seedlings. J. Exp. Bot. 69, 3037–3051. 10.1093/jxb/ery139, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S., et al. (2010). MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22, 234–248. 10.1105/tpc.109.066266, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Yagi Y., Nakamura T. (2019). Comprehensive prediction of target RNA editing sites for PLS-class PPR proteins in Arabidopsis thaliana. Plant Cell Physiol. 60, 862–874. 10.1093/pcp/pcy251, PMID: [DOI] [PubMed] [Google Scholar]
- Koester R. P., Nohl B. M., Diers B. W., Ainsworth E. A. (2016). Has photosynthetic capacity increased with 80 years of soybean breeding? An examination of historical soybean cultivars. Plant Cell Environ. 39, 1058–1067. 10.1111/pce.12675 [DOI] [PubMed] [Google Scholar]
- Koester R. P., Skoneczka J. A., Cary T. R., Diers B. W., Ainsworth E. A. (2014). Historical gains in soybean (Glycine max Merr.) seed yield are driven by linear increases in light interception, energy conversion, and partitioning efficiencies. J. Exp. Bot. 65, 3311–3321. 10.1093/jxb/eru187, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330. 10.1038/nature03229, PMID: [DOI] [PubMed] [Google Scholar]
- Kwon K. C., Cho M. H. (2008). Deletion of the chloroplast-localized AtTerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. Plant J. 55, 428–442. 10.1111/j.1365-313X.2008.03523.x [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin T. G., Bayne A. N., Trempe J. F., Savage D. F., Davies K. M. (2019). Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566, 411–421. 10.1038/s41586-019-0921-0, PMID: [DOI] [PubMed] [Google Scholar]
- Li X. L., Huang W. L., Yang H. H., Jiang R. C., Sun F., Wang H. C., et al. (2019). EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytol. 221, 896–907. 10.1111/nph.15425 [DOI] [PubMed] [Google Scholar]
- Li X. J., Zhang Y. F., Hou M., Sun F., Shen Y., Xiu Z. H., et al. (2014). Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 79, 797–809. 10.1111/tpj.12584 [DOI] [PubMed] [Google Scholar]
- Lu Y. (2018). RNA editing of plastid-encoded genes. Photosynthetica 56, 48–61. 10.1007/s11099-017-0761-9 [DOI] [Google Scholar]
- Lv J., Shang L., Chen Y., Han Y., Yang X., Xie S., et al. (2020). OsSLC1 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling survival. Rice 13:25. 10.1186/s12284-020-00385-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Hu Y., Ju Y., Jiang Q., Cheng Z., Zhang Q., et al. (2017). A novel tetratricopeptide repeat protein, WHITE TO GREEN1, is required for early chloroplast development and affects RNA editing in chloroplasts. J. Exp. Bot. 68, 5829–5843. 10.1093/jxb/erx383, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U. S. A. 99, 12246–12251. 10.1073/pnas.182432999, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M. J., Voldeng H. D., Cober E. R. (1999). Physiological changes from 58 years of genetic improvement of short-season soybean cultivars in Canada. Agron. J. 91, 685–689. 10.2134/agronj1999.914685x [DOI] [Google Scholar]
- O’Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz Linneweber C., et al. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25, 1120–1128. 10.1093/molbev/msn057, PMID: [DOI] [PubMed] [Google Scholar]
- Okuda K., Chateigner Boutin A. L., Nakamura T., Delannoy E., Sugita M., Myouga F., et al. (2009). Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21, 146–156. 10.1105/tpc.108.064667, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2007). Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. U. S. A. 104, 8178–8183. 10.1073/pnas.0700865104, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B., Burger M., Hein A. C., Jorg A., Senkler J., Braun H. P., et al. (2020). One C-to-U RNA editing site and two independently evolved editing factors: testing reciprocal complementation with DYW-type PPR proteins from the Moss Physcomitrium (Physcomitrella) patens and the flowering plants macadamia integrifolia and Arabidopsis. Plant Cell 32, 2997–3018. 10.1105/tpc.20.00311, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo Y. J., Kwon K. C., Kim A., Cho M. H. (2013). Seedling lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 163, 1844–1858. 10.1104/pp.113.227199, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Zhang R., Piskol R., Keegan L. P., Deng P., O’Connell M. A., et al. (2013). Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 10, 128–132. 10.1038/nmeth.2330, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals E., Bruyere C., Toffano Nioche C., Lecharny A. (2006). Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 141, 825–839. 10.1104/pp.106.077826, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallenberg Ruedinger M., Lenz H., Polsakiewicz M., Gott J. M., Knoop V. (2013). A survey of PPR proteins identifies DYW domains like those of land plant RNA editing factors in diverse eukaryotes. RNA Biol. 10, 1549–1556. 10.4161/rna.25755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searing A. M., Satyanarayan M. B., O’Donnell J. P., Lu Y. (2020). Two organelle RNA recognition motif proteins affect distinct sets of RNA editing sites in the Arabidopsis thaliana plastid. Plant Direct 4:e00213. 10.1002/pld3.213, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I. D., Schallenberg Ruedinger M., Takenaka M., Mireau H., Ostersetzer Biran O. (2020). Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 101, 1040–1056. 10.1111/tpj.14578 [DOI] [PubMed] [Google Scholar]
- Song X., Wei H., Cheng W., Yang S., Zhao Y., Li X., et al. (2015). Development of INDEL markers for genetic mapping based on whole genome resequencing in soybean. G3-Genes Genomes Genetics 5, 2793–2799. 10.1534/g3.115.022780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Goldschmidt Clermont M., Hanson M. R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155. 10.1146/annurev-arplant-042809-112242, PMID: [DOI] [PubMed] [Google Scholar]
- Stern D. B., Hanson M. R., Barkan A. (2004). Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 9, 293–301. 10.1016/j.tplants.2004.04.001, PMID: [DOI] [PubMed] [Google Scholar]
- Su H., Li B., Song X., Ma J., Chen J., Zhou Y., et al. (2019). Genome-wide analysis of the DYW subgroup PPR gene family and identification of GmPPR4 responses to drought stress. Int. J. Mol. Sci. 20:5667. 10.3390/ijms20225667, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Germain A., Giloteaux L., Hammani K., Barkan A., Hanson M. R., et al. (2013). An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. U. S. A. 110, E1169–E1178. 10.1073/pnas.1220162110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Huang J., Zhong S., Gu H., He S., Qu L. (2018). Novel DYW-type pentatricopeptide repeat (PPR) protein BLX controls mitochondrial RNA editing and splicing essential for early seed development of Arabidopsis. J. Genet. Genomics 45, 155–168. 10.1016/j.jgg.2018.01.006, PMID: [DOI] [PubMed] [Google Scholar]
- Sun T., Shi X., Friso G., Van Wijk K., Bentolila S., Hanson M. R. (2015b). A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 11:e1005028. 10.1371/journal.pgen.1005028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Wang X., Bonnard G., Shen Y., Xiu Z., Li X., et al. (2015a). Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmF(N) editing, mitochondrial function and seed development in maize. Plant J. 84, 283–295. 10.1111/tpj.12993 [DOI] [PubMed] [Google Scholar]
- Sung T. Y., Tseng C. C., Hsieh M. H. (2010). The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J. 63, 499–511. 10.1111/j.1365-313X.2010.04258.x [DOI] [PubMed] [Google Scholar]
- Tang K., Yang S., Feng X., Wu T., Leng J., Zhou H., et al. (2020). GmNAP1 is essential for trichome and leaf epidermal cell development in soybean. Plant Mol. Biol. 103, 609–621. 10.1007/s11103-020-01013-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Hardel S. L., Kupsch C., Armbruster U., Delannoy E., Gualberto J. M., et al. (2009). Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. U. S. A. 106, 6002–6007. 10.1073/pnas.0808529106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Kudla J. (2008). In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). Cold Spring Harbor Protocols 2008. 10.1101/pdb.prot4995, PMID: [DOI] [PubMed] [Google Scholar]
- Wagoner J. A., Sun T., Lin L., Hanson M. R. (2015). Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 290, 2957–2968. 10.1074/jbc.M114.622084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liang X., Bao Y., Yang S., Zhang X., Yu H., et al. (2020). A malectin-like receptor kinase regulates cell death and pattern-triggered immunity in soybean. EMBO Rep. 21:e50442. 10.15252/embr.202050442, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu X., Yang Y., Huang J., Sun F., Lin J., et al. (2019). Empty pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PLoS Genet. 15:e1008305. 10.1371/journal.pgen.1008305, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Zhang Q., Qin X., Xu Y., Ni C., Huang J., et al. (2018). Rice PPS1 encodes a DYW motif-containing pentatricopeptide repeat protein required for five consecutive RNA-editing sites of nad3 in mitochondria. New Phytol. 220, 878–892. 10.1111/nph.15347 [DOI] [PubMed] [Google Scholar]
- Yan J., Yao Y., Hong S., Yang Y., Shen C., Zhang Q., et al. (2019). Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 47, 3728–3738. 10.1093/nar/gkz075, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang Q., Guan Z., Wang Q., Li L., Ruan F., et al. (2017). MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nat. Plants 3:17037. 10.1038/nplants.2017.37, PMID: [DOI] [PubMed] [Google Scholar]
- Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., et al. (2013). Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. 10.1038/nature12651, PMID: [DOI] [PubMed] [Google Scholar]
- Yu Q. B., Jiang Y., Chong K., Yang Z. N. (2009). AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 59, 1011–1023. 10.1111/j.1365-313X.2009.03930.x [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shen L., Ren D., Hu J., Chen G., Zhu L., et al. (2019). Characterization, expression, and interaction analyses of OsMORF gene family in Rice. Genes 10:694. 10.3390/genes10090694, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Huang J., Chory J. (2019). GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. U. S. A. 116, 10162–10167. 10.1073/pnas.1820426116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. G., Ort D. R., Parry M. A. J., von Caemmerer S. (2020b). A wish list for synthetic biology in photosynthesis research. J. Exp. Bot. 71, 2219–2225. 10.1093/jxb/eraa075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Pan Y., Liu Z., Liu Y., Zhong D., Duan Z., et al. (2020a). Mutation of YL results in a yellow leaf with chloroplast RNA editing defect in soybean. Int. J. Mol. Sci. 21:4275. 10.3390/ijms21124275 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: NCBI repository, accession number: PRJNA660896 (https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA660896).