Abstract
Autologous photoreceptor cell replacement is one of the most promising approaches currently under development for the treatment of inherited retinal degenerative blindness. Unlike endogenous stem cell populations, induced pluripotent stem cells (iPSCs) can be differentiated into both rod and cone photoreceptors in high numbers, making them ideal for this application. That said, in addition to photoreceptor cells, state of the art retinal differentiation protocols give rise to all of the different cell types of the normal retina, the majority of which are not required and may in fact hinder successful photoreceptor cell replacement. As such, following differentiation photoreceptor cell enrichment will likely be required. In addition, to prevent the newly generated photoreceptor cells from suffering the same fate as the patient's original cells, correction of the patient's disease‐causing genetic mutations will be necessary. In this review we discuss literature pertaining to the use of different cell sorting and transfection approaches with a focus on the development and use of novel next generation microfluidic devices. We will discuss how gold standard strategies have been used, the advantages and disadvantages of each, and how novel microfluidic platforms can be incorporated into the clinical manufacturing pipeline to reduce the complexity, cost, and regulatory burden associated with clinical grade production of photoreceptor cells for autologous cell replacement.
Keywords: autologous stem cell transplantation, induced pluripotent stem cells, retina, retinal photoreceptors, stem/progenitor cell
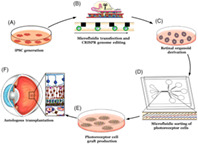
Incorporation of novel microfluidic transfection and cell sorting strategies into an autologous photoreceptor cell replacement pipeline. A‐F, Autologous photoreceptor cell replacement for the treatment of inherited retinal degenerative blindness begins with induced pluripotent stem cell (iPSC) generation (A), followed by CRISPR correction of the patient's disease‐causing genetic mutations (B), generation of 3D retinal organoids (C), dissociation and enrichment of photoreceptor precursor cells (D), seeding of photoreceptor cell grafts (E), and subretinal transplantation (F).
Significance statement.
This article reports on how state of the art microfluidic devices are being used for stem cell transfection and post‐differentiation cell enrichment. The authors discuss how microfluidic approaches avoid the use of specialized reagents and can be used to reduce the regulatory burden and address manufacturing challenges that are associated with autologous cell replacement.
1. INTRODUCTION
Development of patient derived induced pluripotent stem cells (iPSCs), next generation sequencing and genome editing technologies have fueled the field of personalized medicine, which can be defined broadly as use of the patient's own data to inform diagnosis and develop customized treatments. Autologous cell replacement is at the leading edge of this field. The most promising autologous cell replacement strategies currently under development rely on the use of induced pluripotent stem cells, which require tissue specific differentiation prior to transplantation. For instance, we and others have shown that following subretinal transplantation iPSC‐derived photoreceptor precursors cells are able to restore retinal function in animal models of retinal degenerative blindness.1, 2, 3, 4, 5, 6, 7, 8, 9, 10 Following decades of development, the vast majority of the differentiation protocols reported faithfully recapitulate normal retinal development, which means that they give rise to each of the different cell types found in the retina.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40 For therapeutic photoreceptor cell replacement, enrichment of photoreceptor precursor cells away from the unwanted cell types (ie, retinal ganglion cells, bipolar inter neurons, retinal pigmented epithelial [RPE] cells, etc) will be desirable.
In addition to requiring the use of sorting technologies, autologous stem cell therapies for inherited diseases are also likely to require a method for delivering reagents to the interior of cells in order to modify their genome prior to differentiation and transplantation. For instance, when using patient‐derived induced pluripotent stem cells to generate photoreceptors for the treatment of inherited retinal degenerative blindness, the disease‐causing genetic defect that initially resulted in photoreceptor cell dysfunction and death will likely need to be repaired prior to transplantation. As we and others have shown, repair can be accomplished by delivering macromolecules such as Cas9 and homology dependent repair (HDR) sequences to patient derived iPSCs followed by clonal selection and cell line expansion.41, 42, 43
In this review, we will discuss common techniques for both cell sorting and transfection and show how current “gold standard” methods compare with new state of the art microfluidic approaches for stem cell processing. In doing so we will discuss how novel microfluidic strategies have the potential to decrease the cost, complexity and regulatory burden associated with conventional approaches.
2. CELL SORTING TECHNIQUES
2.1. Label based strategies
Currently, fluorescence‐activated cell sorting (FACS) and magnetic‐activated cell sorting (MACS) constitute the gold standard of cell sorting approaches. Both techniques are selective and capable of processing cells with very high throughput. Selectivity is most commonly conferred via the use of antibodies designed to target specific cell surface antigens.
For FACS, cells are first labeled using fluorescent antibodies, dyes, or genetic reporters. They are then passed one cell at a time through a series of lasers, which each excite the cell at a specific wavelength. The light emitted by these fluorophores is then detected by photodetectors, which allow for the identification of the fluorescent signal present. Based on the markers detected, a FACS machine can then deposit different cell populations into separate containers using electrostatic force to deflect single cell liquid droplets. This approach has been used widely for mouse photoreceptor cell isolation and subsequent subretinal transplantation. For instance, several groups have used reporter mouse strains engineered to express green fluorescent protein under control of the NRL promoter to isolate photoreceptor precursor cells at various stages of retinal development to evaluate the role of cellular maturation on functional integration following transplantation.44, 45, 46 Similarly, using genetically modified pluripotent stem cell fluorescent reporter lines several groups have adopted this sorting strategy for evaluating human retinal development in vitro and identify photoreceptor precursor cells following isolation and subretinal transplantation in vivo.47, 48, 49 Unfortunately, this strategy relies on endogenous expression of a cell type specific fluorophore and as such is unlikely to be useful for clinical application. This is especially true for photoreceptor cell replacement where endogenous expression of a fluorescent protein such as GFP would likely interfere with normal visual function.
Unlike FACS, MACS relies exclusively on the use of antibodies to attach paramagnetic beads to the surface of cells, which allows these cells to be pulled out of suspension using a strong magnet. Once isolated, antibody bound beads can be released and cells used for downstream applications. For instance, several groups have used cluster of differentiation antigen 73 (CD73) antibody bound magnetic beads to isolate photoreceptor precursor cells that retain the ability to integrate into the rodent retina following transplantation.50, 51, 52, 53, 54 Like many cell surface markers, CD73 has been reported to be expressed on a variety of different cell types.55, 56, 57 For enhanced specificity, Lakowski and colleagues demonstrated that a panel of five cluster of differentiation antigens (ie, CD73+, CD24+, CD133+, Cd47+, and CD15‐) could be used to isolate photoreceptor precursor cells from embryonic stem cells.55 Unfortunately, the MACS approach is not well suited for multiple marker mediated isolation of specific cell populations as selecting for multiple markers would require sequential isolation, increasing the difficulty of the procedure while also increasing cell loss. As such, the above‐described study was performed using FACS. Interestingly, as shown in Figure 1, in both developing fetal and adult human retina CD73 appears to be predominantly expressed on a subset of Rod photoreceptor cells. While the lack of expression in non‐photoreceptor cell types in the human retina is desirable, the fact that CD73 does not appear to be expressed by cone photoreceptor cells, even during early retinal development, is of concern for cell replacement, as cone photoreceptor cells will be required to restore high acuity vision. Interestingly, Gagliardi and colleagues convincingly demonstrate that human CD73 enriched photoreceptor precursors have the ability to give rise to cells expressing blue and red/green cone opsin following subretinal transplantation.50 As such, additional lineage tracing studies to demonstrate the fate of CD73‐positive progenitor cells are needed if this approach is to be used clinically.
FIGURE 1.
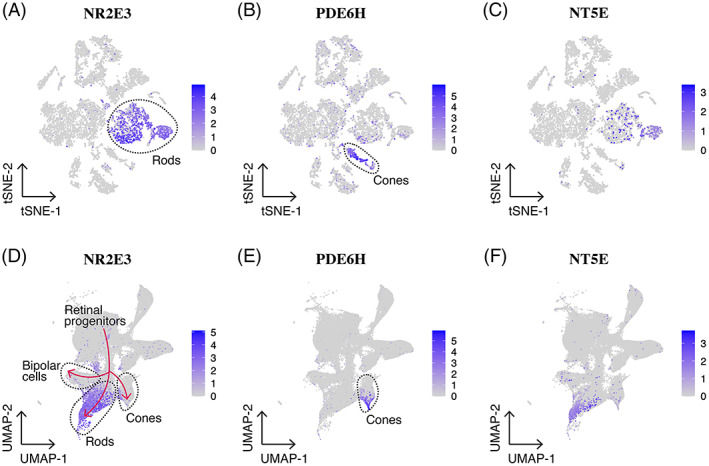
CD73 expression in the adult and developing fetal human retina. A‐C, Single‐cell RNA sequencing of adult human retinal cells58 demonstrates that the NT5E gene that encodes CD73 is expressed by a select population of mature rod photoreceptor cells only. D‐F, Reprocessed single‐cell RNA sequencing of developing fetal human retinal cells59 demonstrates that NT5E is predominantly expressed by committed rod photoreceptor cells, with very infrequent expression in retinal progenitor cells and developing cone photoreceptor cells
While MACS and FACS can be useful for cell sorting, as indicated above, to be clinically relevant cell surface antigens and corresponding antibodies are required. Unfortunately, in many cases targetable antigens either do not exist or are not expressed solely on the cell type of interest, making these approaches challenging to apply successfully. For instance, until recently cell surface antigens useful for isolation of corneal limbal stem cells remained elusive. Although we were able to demonstrate that ABCB5 was expressed on limbal stem cells and could be used for cellular enrichment to successfully treat animal models of limbal stem cell deficiencies, this marker is not Limbal stem cell specific.60, 61, 62 Similarly, markers that are often used for stem cell derived vascular endothelial cell enrichment (eg, CD31) are expressed on a variety of different endothelial cell populations and monocytic cells, making them less than ideal for isolation of tissue specific vascular subtypes.63, 64, 65
2.2. Label free strategies
In regenerative medicine, therapeutic stem cells or their progeny are typically transplanted into patients to rebuild injured tissues. For clinical cell replacement, all reagents applied to the cells must be fully characterized and guaranteed to be safe in order to gain regulatory approval. The use of fluorescent and/or magnetic bead bound antibodies that target specific cell surface antigens for cell sorting, increases the labor and regulatory burden associated with clinical translation. For instance, conjugation of antibodies to cells destined for transplant followed by sorting is both time consuming and has the potential to alter cell function and compromise both safety and the effectiveness of the treatment. In addition, FACS, which requires a complex specialized piece of equipment, presents significant manufacturing challenges. For autologous cell replacement for instance, FACS protocols must reliably prevent cross contamination of patient derived cell lines following sequential sorting. This would likely require a complete sterilization between each sorting run. Sorting strategies that utilize presterilized single use devices that do not require specialized reagents would be much more desirable. To address concerns associated with label‐based cell enrichment, a variety of different label‐free microfluidic cell sorting approaches are being developed. Microfluidic sorting methods are typically designed to sort cells based on physical characteristics such as diameter and stiffness, which avoids reliance on the presence of unique cell surface antigens and reagents. These platforms are generally implemented as cheap, disposable chips driven by an external pump (eg, syringe pump or peristaltic pump). Such label free approaches are ideal for isolation of cells with unreliable or absent cell surface antigens and terminally differentiated stem cell progeny that are destined for clinical cell replacement. Two of the most commonly used microfluidic device designs published to date are (1) deterministic lateral displacement (DLD) and (2) hydrodynamic focusing. There are also many label free cytometry approaches being developed, such as ghost cytometry,66 impedance cytometry67 and deformability cytometry,68 which will likely play an important role in cellular analysis going forward. However, these platforms have not yet been integrated into cell sorters, so we will not be covering them in depth in this review.
As shown in Figure 2A, in deterministic lateral displacement (DLD) a series of microposts are used to move cells perpendicular to the direction of fluid flow in a manner related to their size and stiffness. Several groups have successfully used this approach to separate a variety of different cell types.71, 72, 73 For instance, Xavier and colleagues recently demonstrated how this approach could be used to isolate skeletal progenitor cells from human bone marrow.71 Specifically, they showed that by using DLD they could successfully isolate skeletal progenitor cells, which were stiffer than leukocytes and contained within the larger cell fraction of bone marrow. These isolated progenitor cells retained their ability to form clones in cultures indicating that they retained their stem cell potential. One of the drawbacks of DLD however, is that it is susceptible to clogging. As cell debris and/or aggregates cannot easily escape the active region of the DLD device they become trapped, disrupting the flow pattern required for sorting, which in turn negatively impacts enrichment efficiency.
Hydrodynamic focusing uses hydrodynamic forces to cause cells to come to an equilibrium position a channel. As illustrated in Figure 2B, this equilibrium position is determined by balancing of lift and drag forces, which is a function of cell size and biomechanical properties (ie, stiffness and viscosity). Like DLD, hydrodynamic focusing has been used successfully to sort a variety of different cell types, including endogenous stem cells.74, 75, 76 For instance, Hur and colleagues were able show how hydrodynamic focusing could be used to isolate adrenal cortical progenitor cells from a mixed cell population obtained from digestions of murine adrenal glands.74 Separation was accomplished by exploiting the differing strengths of cell‐cell adhesion between undifferentiated precursors and differentiated somatic cells. Given that the cell‐cell junctions are stronger between differentiated cells than undifferentiated cells, a digestion protocol was developed which resulted in complete dissociation of progenitor cells leaving the differentiated somatic cells in larger clumps. The clumps and single cells could then be hydrodynamically sorted by size to effectively separate progenitor cells from the contaminating somatic cells with little to no reduction in viability (ie, not significantly different from unsorted control samples). As enriched adrenal progenitor cells could subsequently be expanded in culture, it is conceivable that these cells could be given back to the patient from which they were derived in order to restore adrenal function. One drawback of this technique is that it takes relatively large differences in cell size and/or stiffness to result in detectable differences in equilibrium position.
FIGURE 2.
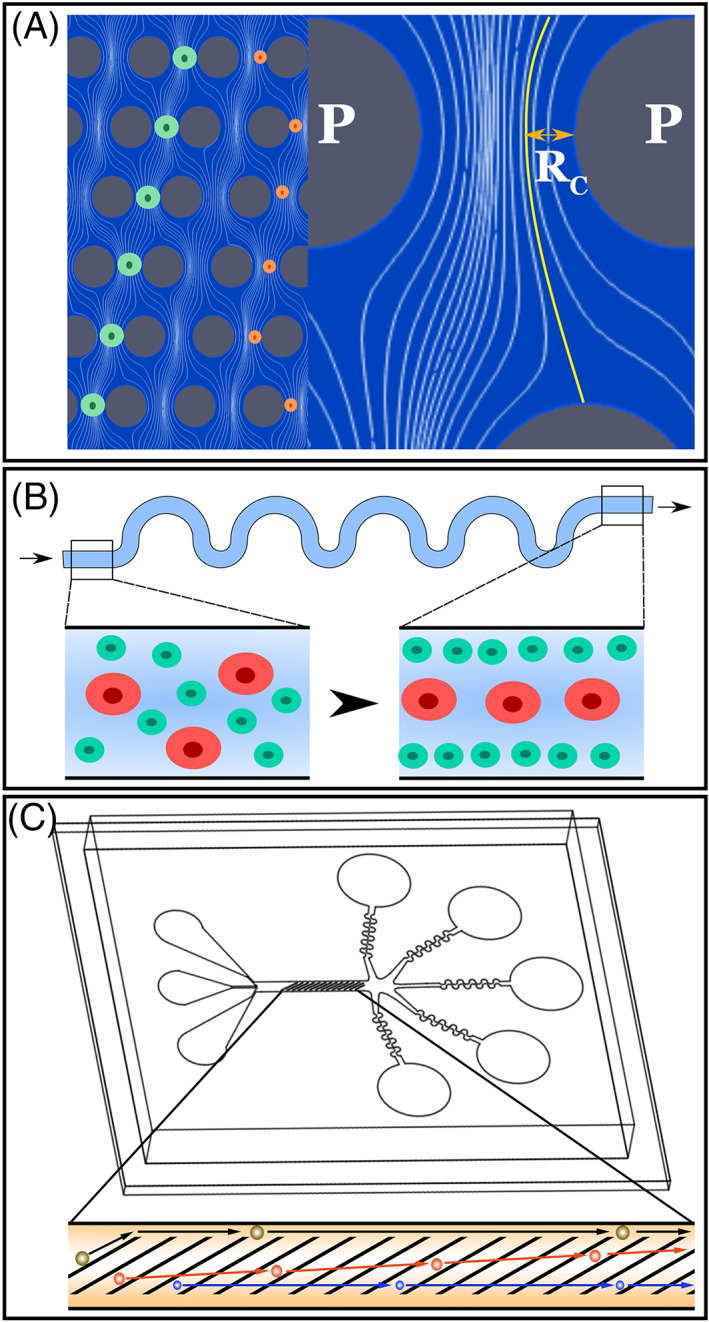
Next‐generation microfluidic cell sorting strategies. A‐C, Microfluidic cell sorting strategies that utilize deterministic lateral displacement (A, adapted from McGrath et al,69 streamlines obtained via CFD simulation), hydrodynamic focusing (B, adapted from Di Carlo et al70) and cellular compression with lateral deflection (C). For DLD cells start in a given streamline. If the cell is larger than a critical size, they are displaced laterally into a new streamline due to interaction with the posts (P). For hydrodynamic focusing the balance between a wall interaction force, a shear gradient lift force and stokes drag dictate the presence of equilibrium positions for cells suspended in a liquid. Over time, cells will tend to occupy equilibrium positions within the channel, resulting in size based focusing. For cellular compression with lateral deflection, the sorting platform consists of a series of diagonal constrictions designed to deflect cells laterally relative to the main direction of flow in a manner related to their size and mechanical properties
Due to the issues mentioned above, neither DLD nor hydrodynamic focusing are ideal for sorting of pluripotent stem cell derived photoreceptor precursor cells from each of the different cell types derived following retinal differentiation.12, 13, 14, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 Specifically, following retinal organoid dissociation clusters of RPE and/or inner retinal cell types is common, as such the potential for DLD device clogging is high. Similarly, the differences in size between retinal cells that are liberated from organoids are less than ideal for efficient hydrodynamic sorting. To address these concerns, we recently developed the microfluidic cell sorting platform illustrated in Figure 2C. Using this device cells are exposed to repeated compressions by a series of thin ridge constrictions oriented diagonally to the direction of flow. In our hands, these repeated compressions do not have a significant impact on cell viability.77 As cells interact with these constrictions, which are designed to be smaller than the cell's diameter, they deflect laterally in the device in a manner related to their size and stiffness. The performance of this platform is dependent on the gap size and as such must be reoptimized for each application. The design of the constrictions allows cell debris and aggregates to move along the constrictions into a gutter and exit the device without causing clogs. At the end of the device there are five independent wells from which sorted cell populations can be collected. To evaluate the utility of this platform, we recently performed a series of experiments using iPSC derived photoreceptor precursor and RPE cell lines as well as human donor retina.78 When photoreceptor precursor and RPE cells were mixed at a ratio of 37% RPE to 63% photoreceptor precursor cell and injected into the device at a concentration of 2x106 cells per mL, we were able to reliably separate the photoreceptor precursor cells away from the RPE cells. Similarly, when human donor retina was dissociated and sorted using the device, we were able to isolate independent samples enriched for RPE (largest fraction in well 1), rod and cone photoreceptor cells (largest fraction in well 4) and inner retinal neurons, including retinal ganglion cells (enriched in wells 1‐3).78 Although we believe that there is still room for further improvement, these findings demonstrate that retinal cells can be separated into discrete populations by exploiting differences in cell size, stiffness and viscosity79, 80, 81 (ie, without the need for antigen specific antibodies or fluorescent markers). Moreover, naturally occurring stem cells may be processed in a similar manner to enrich for desired cell phenotypes.82
3. MICROFLUIDIC MEDIATED CELL TRANSFECTION
Since autologous iPSCs possess the same genome as the patient from whom they are derived, the use of these cells for treatment of inherited disease will likely require correction of the disease‐causing genetic variants that resulted in death of the target cell type prior to differentiation. Genome editing approaches that utilize zinc finger nucleases, Talens or CRISPR/Cas9 to induce double strand DNA breaks and homology directed repair of target genetic loci, have been used for this purpose.83, 84, 85 In addition, next‐generation CRISPR technologies including base editors, primer editors and RNA‐targeting Cas effectors, which can be used to restore gene function in the absence of double strand DNA break induction, are promising new approaches.86 For these strategies to work, delivery of nucleic acid and/or protein to patient derived iPSCs, followed by genetic screening and expansion of genetically corrected clones, is required. Although several strategies for delivering genome editing reagents to iPSCs exist, the most widely used are (1) viral transduction, (2) chemical transfection, and (3) electroporation. Viral transduction takes advantage of the native ability of viruses to insert their DNA into infected cells. By replacing the genetic material required for replication, viruses can be engineered to deliver sequences of interest to a variety of different cell types with high efficiency. For instance, adeno associated viruses (AAV) are used extensively in both preclinical and clinical settings to safely deliver therapeutic DNA to photoreceptor cells and mitigate disease progression.87, 88, 89, 90, 91 Unfortunately, the packaging capacity of AAVs is limited to approximately 4.7 kb, making them less useful for delivery of the genome editing reagents required to induce homology directed repair in patient derived iPSCs in vitro since CRISPR/Cas9 and repair templates exceed this limit. For this reason, lentiviruses, which have a payload of approximately 8 kb, have been wildly used for genome editing of patient derived iPSCs.92, 93, 94, 95 Although successful, the major drawback associated with the use of lentivirus is that they carry with them increased risk of inducing insertional mutagenesis resulting from random insertion of the genetic payload into the host cell's genome.96 In addition, viruses are only useful for delivery of nucleic acids, which precludes their use for CRISPR mediated genome editing via delivery of ribonuclear proteins, which are often significantly more efficient.97, 98 Chemical reagents such as lipofectamine are quite common for delivery of macromolecules given that they are not subject to the packaging size limitation that viruses are subject to. These techniques involve conjugating a positively charged carrier with target nucleic acids. This positively charged complex is then attracted to the cell membrane, whereby it is taken up via endocytosis. One of the greatest advantages of this approach is the ease and speed with which it can be utilized. Specifically, unlike viral transduction, which requires engineering of a viral plasmid and packaging into viral particles by trained technical staff using well characterized safety protocols, lipofection can be safely performed using almost any expression plasmid. As with lipofection, electroporation also avoids concerns associated with cargo size limits. Electroporation works via exposure of cells to large electric fields that cause pores to form transiently in the cell membrane through which macromolecules can enter. Both lipofection and electroporation have been used extensively by us and others for CRISPR mediated correction of patient derived iPSCs with excellent success.83, 99 The major drawback of both techniques is that they use proprietary reagents that would be subject to FDA regulation when used for clinical cell replacement. As these reagents are not typically produced under and compliant with cGMP the regulatory burden associated with their use is high.
To address these concerns, we and others have pioneered the use of novel microfluidic approaches designed to deliver macromolecules to cells without the need for specialized reagents. In general, microfluidic transfection strategies rely on physical deformation of a cell to induce pore formation. Macromolecules (nucleic acids, proteins, etc) are then delivered by either diffusion or convective transport. These transfections are generally very fast (less than an hour of processing time) after which the cells can be replated and expanded for downstream selection, clonal expansion, and validation.
In 2008, Hallow and colleagues demonstrated that fluid shear forces can be used to deliver a macromolecule cargo into cell lines.100 In 2012 Sharei and colleagues reported the development of a microfluidic cellular transfection device that was designed to deliver macromolecules to cells by passing them through a narrow constriction at high flow rates.101 By squeezing the cells into a small channel, the authors demonstrate that they could cause pores to form in the cell membrane through which a payload suspended in the surrounding buffer could be passed into the cell through diffusive transport (Figure 3A). By using mechanical forces to induce pore formation, this approach removes the need for proprietary reagents necessary for common transfection strategies such as lipofection and electroporation. In addition, devices are relatively simple, which allows for mass production and distribution as single use disposable units eliminating the need for complex pieces of equipment and worry about cross contamination of independent patient samples. Collectively these advantages are significant when trying to deploy the technology under current Good Manufacturing Practice (cGMP) for autologous clinical cell replacement. This approach, however, is not without its drawbacks. Most importantly, given that the payload enters the cell through passive, diffusive transport, delivery efficiency when using this technique is inversely related to payload size. For instance, when delivering fluorescent dextran molecules of varying size to murine embryonic stem cells, Sharei et al demonstrated a drop in delivery efficiency from approximately 50% for 3 kDa dextran to approximately 25% when 70 kDa dextran was used.101
FIGURE 3.
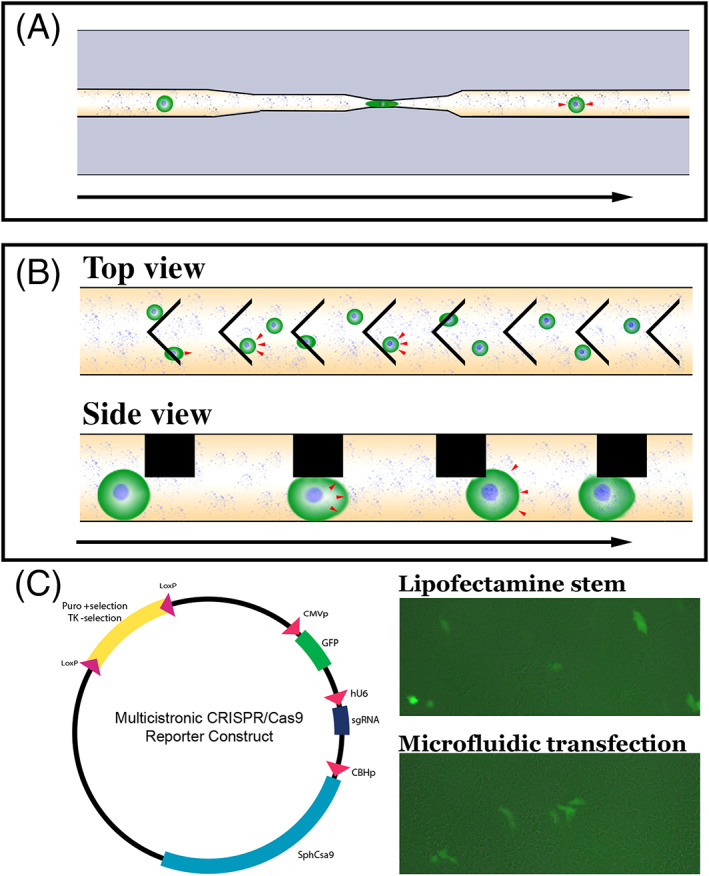
Next‐generation microfluidic cell devices. A,B, Schematics depicting diffusive (A) and convective (B) microfluidic cell transfection device designs. Red arrowheads denote net direction of macromolecular transport across the cell membrane. Black arrow denotes direction of fluid flow. C, Schematic and preliminary data demonstrating microfluidic delivery of a large multicistronic construct containing sgRNAs, Cas9, GFP reporter and associated promoters to patient derived iPSCs. Delivery of the same construct using Lipofectamine Stem, which we have previously used for CRISPR correction of patient iPSCs,85 has been included for comparison
To address issues associated with reduced delivery efficiencies with increased cargo size, our group recently developed another microfluidic delivery platform, which subjects cells to repeated, rapid compressions.77 During these rapid compressions, the volume of the compressed cell decreases, followed by partial volume recovery in between compressions (Figure 3B). As shown in Figure 3C, in preliminary experiments we have been able to deliver a large multicistronic CRISPR construct containing gene specific sgRNA sequences, Cas9 and a GFP report construct with associated promoters, to human patient derived iPSCs at an efficiency similar to that obtained using lipofectamine stem. As CRISPR mediated genomic correction of iPSCs is typically achieved by transient transfection followed by clonal selection and expansion,102, 103 high transfection efficiency is not a requirement for genomic correction. That said, unlike the above‐described approach, the volume changes created using this device cause active, convective transport of the payload suspended in the surround buffer across the cell membrane. The most important consequence of this convective delivery mechanism is that delivery efficiency is not a strong function of payload size. For instance, a relatively constant delivery efficiency of greater than 80% was demonstrated for delivery of dextrans that were 4 kDa and 2000 kDa in size to K562 myelogenous leukemia cells. With some optimization (see Section 4), we believe that efficient transfection of patient derived iPSCs will be possible. As with the diffusive transfection approach described above, use of this convective transfection strategy does not require specialized reagents or proprietary materials. Likewise, devices can be readily fabricated and provided as sterilized single use devices, which is ideal for manufacturing of autologous cell replacement products.
In addition to cGMP compatibility of both the techniques described above are amenable to massive parallelization. Due to the small size of each individual channel, many channels can be fabricated on a single chip with shared inputs and outputs, drastically increasing throughput without increasing device cost or the usability of the platform.
4. FUTURE PERSPECTIVES
While the microfluidic techniques presented in this paper have the potential to enable the manufacture of safe, high potency cell therapies there are some shortfalls, which the community is continuing to address. First, while label‐free microfluidic sorting strategies have advantages over “gold standard” techniques in terms of cost and safety, they are still not as selective as FACS or MACS. While we expect the performance of these new devices to continue to improve, it may be that microfluidic cell sorting will never perform as well as antibody‐based methods, provided that appropriate markers exist. It is the belief of the authors that emerging label‐free microfluidic sorting techniques can complement “gold‐standard” techniques by serving as an alternative when either, (1) appropriate targets for antibody labels do not exist, (2) the conjugation of antibodies to cells presort could interfere with the cell's utility in downstream applications, (3) sterilizing multiuse instrumentation becomes burdensome, or (4) the cost of purchasing/operating large commercial sorters becomes prohibitive.
As discussed above, the performance of microfluidic transfection technologies is highly dependent on proper selection of device geometry to ensure sufficient deformation of cells to facilitate transfection without destroying the cells or clogging the device. While this optimization is more straightforward with a relatively homogeneous cell population, things rapidly become complicated when attempting to transfect heterogeneous samples. One can imagine that for a fixed device geometry, cells of differing size in a mixed population will respond differently. The smallest cells in the population may not be adequately deformed to allow for transfection, while the largest cells may be damaged due to excessive deformation or simply clog the device, resulting in loss of valuable cell product and time. It is possible that this issue can be addressed by “presorting” a heterogeneous cell population into more homogeneous fractions, and then transfecting each fraction containing useful cells using separate devices.
In addition to the technical limitations of emerging technologies, there is also the issue that new microfluidic sorting and transfection platforms currently do not exist as “out of the box” solutions. This is especially problematic when attempting to implement the technology into an existing clinical production pipeline. Unlike existing technologies such as FACS that have been in widespread use and familiar to the FDA, implementation of novel microfluidic devices will likely require extensive validation and pose a significant regulatory burden. Generally, device geometries and process parameters need to be retuned for new applications, and this optimization generally must be done by someone familiar with the platform of interest. While these issues will likely become less pronounced as the technologies mature, we believe that increased communication and collaboration between the stem cell and microfluidics communities is essential in ensuring that these new microfluidic platforms are developed and distributed in a way that is most beneficial to the research needs of the stem cell community and most translatable to the treatment of patients.
5. SUMMARY
Autologous photoreceptor cell replacement for the treatment of inherited retinal degenerative blindness is at the forefront of personalized medicine. Unfortunately, complexities associated with this approach present significant manufacturing challenges. In general, biologics manufacturing is designed for mass production of a single product to treat a large number of individuals (eg, vaccine production). Autologous photoreceptor cell replacement requires that a unique line of iPSCs be generated, genetically corrected, and differentiated for every patient in need, in order to prevent immune rejection. Following differentiation, photoreceptor precursor cells must be isolated and delivered independent of the other retinal cell types that are generated during the differentiation process. Although many of the reagents required for manufacturing of autologous iPSC‐photoreceptor cells are cGMP compliant, gold standard approaches used for genetic correction of patient derived iPSCs and sorting of photoreceptor precursor cells following differentiation require reagents and equipment that are not well suited to clinical manufacturing. As described in this review, novel microfluidic strategies are being developed for both cellular enrichment and transfection that do not require specialized reagents and complex equipment. By incorporating these microfluidic strategies into autologous photoreceptor cell manufacturing pipelines, we believe that it will be possible to greatly reduce the cost, protocol complexity, and regulatory burden associated with production of autologous photoreceptor cells for treatment of retinal degenerative blindness.
CONFLICT OF INTEREST
T.S. declared leadership position, advisory role, and ownership interest with CellFE. The other authors declared no potential conflicts of interest.
AUTHOR CONTRIBUTION
N.E.S., A.P.V., R.F.M.: drafted the manuscript, contributed to the acquisition, analysis, and interpretation of the data; T.S. and B.A.T.: drafted the manuscript, contributed to the acquisition, analysis, and interpretation of the data, provided Final approval of the version to be published and are in agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
ACKNOWLEDGMENTS
This work was performed under a Project Award Agreement from the National Institute for Innovation in Manufacturing Biopharmaceuticals (NIIMBL) and financial assistance award 70NANB17H002 from the U.S. Department of Commerce, National Institute of Standards and Technology.
Stone NE, Voigt AP, Mullins RF, Sulchek T, Tucker BA. Microfluidic processing of stem cells for autologous cell replacement. STEM CELLS Transl Med. 2021;10(10):1384–1393. 10.1002/sctm.21-0080
Funding information National Institute for Innovation in Manufacturing Biopharmaceuticals (NIIMBL), Grant/Award Number: PC2.2‐12; U.S. Department of Commerce, National Institute of Standards and Technology, Grant/Award Number: 70NANB17H002
Contributor Information
Todd Sulchek, Email: todd.sulchek@me.gatech.edu.
Budd A. Tucker, Email: budd-tucker@uiowa.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Tucker BA, Park I‐H, Qi SD, et al. Transplantation of adult mouse iPS cell‐derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS One. 2011;6(4):e18992. 10.1371/journal.pone.0018992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamba DA, McUsic A, Hirata RK, et al. Generation, purification and transplantation of photoreceptors derived from human induced pluripotent stem cells. PLoS One. 2010;5(1):e8763. 10.1371/journal.pone.0008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zerti D, Hilgen G, Dorgau B, et al. Transplanted pluripotent stem cell‐derived photoreceptor precursors elicit conventional and unusual light responses in mice with advanced retinal degeneration. Stem Cells. 2021. 10.1002/stem.3365. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Lin B, McLelland BT, Aramant RB, et al. Retina organoid transplants develop photoreceptors and improve visual function in RCS rats with RPE dysfunction. Invest Ophthalmol Vis Sci. 2020;61(11):34. 10.1167/iovs.61.11.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos‐Ferreira TF, Borsch O, Ader M. Rebuilding the missing part‐a review on photoreceptor transplantation. Front Syst Neurosci. 2016;10:105. 10.3389/fnsys.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goureau O, Orieux G. Photoreceptor cell transplantation for future treatment of retinitis pigmentosa. Med Sci (Paris). 2020;36(6–7):600‐606. 10.1051/medsci/2020097. [DOI] [PubMed] [Google Scholar]
- 7.Gagliardi G, Ben M'Barek K, Goureau O. Photoreceptor cell replacement in macular degeneration and retinitis pigmentosa: a pluripotent stem cell‐based approach. Prog Retin Eye Res. 2019;71:1‐25. 10.1016/j.preteyeres.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Mandai M, Watanabe A, Kurimoto Y, et al. Autologous induced stem‐cell‐derived retinal cells for macular degeneration. N Engl J Med. 2017;376(11):1038‐1046. 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- 9.Sharma R, Khristov V, Rising A, et al. Clinical‐grade stem cell‐derived retinal pigment epithelium patch rescues retinal degeneration in rodents and pigs. Sci Transl Med. 2019;11(475). 10.1126/scitranslmed.aat5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley LA, Burnight ER, DeLuca AP, et al. cGMP production of patient‐specific iPSCs and photoreceptor precursor cells to treat retinal degenerative blindness. Sci Rep. 2016;6. 10.1038/srep30742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiley LA, Anfinson KR, Cranston CM, et al. Generation of xeno‐free, cGMP‐compliant patient‐specific iPSCs from skin biopsy. Curr Protoc Stem Cell Biol. 2017;42(1):4A.12.1‐4A.12.14. 10.1002/cpsc.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tucker BA, Mullins RF, Streb LM, et al. Patient‐specific iPSC‐derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. eLife. 2013;2. 10.7554/eLife.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong X, Gutierrez C, Xue T, et al. Generation of three dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014;5:4047. 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle‐like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29(8):1206‐1218. 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106(39):16698‐16703. 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallman A, Capowski EE, Wang J, et al. Investigating cone photoreceptor development using patient‐derived NRL null retinal organoids. Commun Biol. 2020;3. 10.1038/s42003-020-0808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahlin KJ, Maruotti J, Zack DJ. Modeling retinal dystrophies using patient‐derived induced pluripotent stem cells. Adv Exp Med Biol. 2014;801:157‐164. 10.1007/978-1-4614-3209-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueda K, Onishi A, Ito S‐I, et al. Generation of three‐dimensional retinal organoids expressing rhodopsin and S‐ and M‐cone opsins from mouse stem cells. Biochem Biophys Res Commun. 2018;495(4):2595‐2601. 10.1016/j.bbrc.2017.12.092. [DOI] [PubMed] [Google Scholar]
- 19.Collin J, Zerti D, Queen R, et al. CRX expression in pluripotent stem cell‐derived photoreceptors marks a transplantable subpopulation of early cones. Stem Cells. 2019;37(5):609‐622. 10.1002/stem.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichman S, Terray A, Slembrouck A, et al. From confluent human iPS cells to self‐forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci USA. 2014;111(23):8518‐8523. 10.1073/pnas.1324212111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellough CB, Sernagor E, Moreno‐Gimeno I, et al. Efficient stage‐specific differentiation of human pluripotent stem cells toward retinal photoreceptor cells. Stem Cells. 2012;30(4):673‐686. 10.1002/stem.1037. [DOI] [PubMed] [Google Scholar]
- 22.Rowland TJ, Buchholz DE, Clegg DO. Pluripotent human stem cells for the treatment of retinal disease. J Cell Physiol. 2012;227(2):457‐466. 10.1002/jcp.22814. [DOI] [PubMed] [Google Scholar]
- 23.La Torre A, Lamba DA, Jayabalu A, et al. Production and transplantation of retinal cells from human and mouse embryonic stem cells. Methods Mol Biol. 2012;884:229‐246. 10.1007/978-1-61779-848-1_16. [DOI] [PubMed] [Google Scholar]
- 24.Tucker BA, Scheetz TE, Mullins RF, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia‐related gene male germ cell‐associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci USA. 2011;108(34):E569‐E576. 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z‐B, Okamoto S, Osakada F, et al. Modeling retinal degeneration using patient‐specific induced pluripotent stem cells. PLoS One. 2011;6(2):e17084. 10.1371/journal.pone.0017084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reh TA, Lamba D, Gust J. Directing human embryonic stem cells to a retinal fate. Methods Mol Biol. 2010;636:139‐153. 10.1007/978-1-60761-691-7_9. [DOI] [PubMed] [Google Scholar]
- 27.Osakada F, Jin Z‐B, Hirami Y, et al. In vitro differentiation of retinal cells from human pluripotent stem cells by small‐molecule induction. J Cell Sci. 2009;122(pt 17):3169‐3179. 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- 28.Lamba DA, Gust J, Reh TA. Transplantation of human embryonic stem cell‐derived photoreceptors restores some visual function in Crx‐deficient mice. Cell Stem Cell. 2009;4(1):73‐79. 10.1016/j.stem.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osakada F, Ikeda H, Mandai M, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26(2):215‐224. 10.1038/nbt1384. [DOI] [PubMed] [Google Scholar]
- 30.Boucherie C, Mukherjee S, Henckaerts E, et al. Brief report: self‐organizing neuroepithelium from human pluripotent stem cells facilitates derivation of photoreceptors. Stem Cells. 2013;31(2):408‐414. 10.1002/stem.1268. [DOI] [PubMed] [Google Scholar]
- 31.Nakano T, Ando S, Takata N, et al. Self‐formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771‐785. 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Fligor CM, Huang K‐C, Lavekar SS, et al. Differentiation of retinal organoids from human pluripotent stem cells. Methods Cell Biol. 2020;159:279‐302. 10.1016/bs.mcb.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Zerti D, Dorgau B, Felemban M, et al. Developing a simple method to enhance the generation of cone and rod photoreceptors in pluripotent stem cell‐derived retinal organoids. Stem Cells. 2020;38(1):45‐51. 10.1002/stem.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallam D, Hilgen G, Dorgau B, et al. Human‐induced pluripotent stem cells generate light responsive retinal organoids with variable and nutrient‐dependent efficiency. Stem Cells. 2018;36(10):1535‐1551. 10.1002/stem.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eiraku M, Takata N, Ishibashi H, et al. Self‐organizing optic‐cup morphogenesis in three‐dimensional culture. Nature. 2011;472:51‐56. 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 36.Deng W‐L, Gao M‐L, Lei X‐L, et al. Gene correction reverses ciliopathy and photoreceptor loss in iPSC‐derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 2018;10(4):1267‐1281. 10.1016/j.stemcr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Zhang Y, Zhang Y‐Y, et al. Human embryonic stem cell‐derived organoid retinoblastoma reveals a cancerous origin. PNAS. 2020;117(52):33628‐33638. 10.1073/pnas.2011780117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Lowe A, Dharmat R, et al. Generation, transcriptome profiling, and functional validation of cone‐rich human retinal organoids. PNAS. 2019;116(22):10824‐10833. 10.1073/pnas.1901572116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan D, Xia X‐X, Zhou H, et al. COCO enhances the efficiency of photoreceptor precursor differentiation in early human embryonic stem cell‐derived retinal organoids. Stem Cell Res Ther. 2020;11(1):366. 10.1186/s13287-020-01883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homma K, Usui S, Kaneda M. Knock‐in strategy at 3′‐end of Crx gene by CRISPR/Cas9 system shows the gene expression profiles during human photoreceptor differentiation. Genes Cells. 2017;22(3):250‐264. 10.1111/gtc.12472. [DOI] [PubMed] [Google Scholar]
- 41.Jang Y‐Y, Ye Z. Gene correction in patient‐specific iPSCs for therapy development and disease modeling. Hum Genet. 2016;135(9):1041‐1058. 10.1007/s00439-016-1691-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burnight ER, Giacalone JC, Cooke JA, et al. CRISPR‐Cas9 genome engineering: treating inherited retinal degeneration. Prog Retin Eye Res. 2018;65:28‐49. 10.1016/j.preteyeres.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brookhouser N, Raman S, Potts C, et al. May I cut in? Gene editing approaches in human induced pluripotent stem cells. Cells. 2017;6(1). 10.3390/cells6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacLaren RE, Pearson RA, MacNeil A, et al. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444(7116):203‐207. 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
- 45.Homma K, Okamoto S, Mandai M, et al. Developing rods transplanted into the degenerating retina of Crx‐knockout mice exhibit neural activity similar to native photoreceptors. Stem Cells. 2013;31(6):1149‐1159. 10.1002/stem.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T, Akimoto M, Imai H, et al. Chondroitinase ABC treatment enhances synaptogenesis between transplant and host neurons in model of retinal degeneration. Cell Transplant. 2007;16(5):493‐503. 10.3727/000000007783464966. [DOI] [PubMed] [Google Scholar]
- 47.Aboualizadeh E, Phillips MJ, McGregor JE, et al. Imaging transplanted photoreceptors in living nonhuman primates with single‐cell resolution. Stem Cell Rep. 2020;15(2):482‐497. 10.1016/j.stemcr.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaewkhaw R, Swaroop M, Homma K, et al. Treatment paradigms for retinal and macular diseases using 3‐D retina cultures derived from human reporter pluripotent stem cell lines. Invest Ophthalmol Vis Sci. 2016;57(5):ORSFl1‐ORSFl11. 10.1167/iovs.15-17639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam PT, Gutierrez C, Del Rio‐Tsonis K, Robinson ML. Generation of a retina reporter hiPSC line to label progenitor, ganglion, and photoreceptor cell types. Transl Vis Sci Technol. 2020;9(3):21. 10.1167/tvst.9.3.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gagliardi G, Ben M'Barek K, Chaffiol A, et al. Characterization and transplantation of CD73‐positive photoreceptors isolated from human iPSC‐derived retinal organoids. Stem Cell Rep. 2018;11(3):665‐680. 10.1016/j.stemcr.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reichman S, Slembrouck A, Gagliardi G, et al. Generation of storable retinal organoids and retinal pigmented epithelium from adherent human iPS cells in xeno‐free and feeder‐free conditions. Stem Cells. 2017;35(5):1176‐1188. 10.1002/stem.2586. [DOI] [PubMed] [Google Scholar]
- 52.Santos‐Ferreira T, Völkner M, Borsch O, et al. Stem cell‐derived photoreceptor transplants differentially integrate into mouse models of cone‐rod dystrophy. Invest Ophthalmol Vis Sci. 2016;57(7):3509‐3520. 10.1167/iovs.16-19087. [DOI] [PubMed] [Google Scholar]
- 53.Santos‐Ferreira T, Postel K, Stutzki H, et al. Daylight vision repair by cell transplantation. Stem Cells. 2015;33(1):79‐90. 10.1002/stem.1824. [DOI] [PubMed] [Google Scholar]
- 54.Eberle D, Santos‐Ferreira T, Grahl S, et al. Subretinal transplantation of MACS purified photoreceptor precursor cells into the adult mouse retina. J Vis Exp. 2014;84:e50932. 10.3791/50932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lakowski J, Gonzalez‐Cordero A, West EL, et al. Transplantation of photoreceptor precursors isolated via a cell surface biomarker panel from embryonic stem cell‐derived self‐forming retina. Stem Cells. 2015;33(8):2469‐2482. 10.1002/stem.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishimoto S, Mizuno T, Takahashi K, et al. CD140b and CD73 are markers for human induced pluripotent stem cell‐derived erythropoietin‐producing cells. FEBS Open Bio. 2020;10(3):427‐433. 10.1002/2211-5463.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Estrela C, Carmo Souza PO, Barbosa MG, et al. Mesenchymal stem cell marker expression in periapical abscess. J Endod. 2019;45(6):716‐723. 10.1016/j.joen.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 58.Voigt AP, Whitmore SS, Flamme‐Wiese MJ, et al. Molecular characterization of foveal versus peripheral human retina by single‐cell RNA sequencing. Exp Eye Res. 2019;184:234‐242. 10.1016/j.exer.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Y, Shiau F, Yi W, et al. Single‐cell analysis of human retina identifies evolutionarily conserved and species‐specific mechanisms controlling development. Dev Cell. 2020;53(4):473‐491.e9. 10.1016/j.devcel.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frank NY, Pendse SS, Lapchak PH, et al. Regulation of progenitor cell fusion by ABCB5 P‐glycoprotein, a novel human ATP‐binding cassette transporter. J Biol Chem. 2003;278(47):47156‐47165. 10.1074/jbc.M308700200. [DOI] [PubMed] [Google Scholar]
- 61.Ma J, Lin JY, Alloo A, et al. Isolation of tumorigenic circulating melanoma cells. Biochem Biophys Res Commun. 2010;402(4):711‐717. 10.1016/j.bbrc.2010.10.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson BJ, Schatton T, Zhan Q, et al. ABCB5 identifies a therapy‐refractory tumor cell population in colorectal cancer patients. Cancer Res. 2011;71(15):5307‐5316. 10.1158/0008-5472.CAN-11-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulfaul K, Giacalone JC, Voigt AP, et al. Stepwise differentiation and functional characterization of human induced pluripotent stem cell‐derived choroidal endothelial cells. Stem Cell Res Ther. 2020;11(1):409. 10.1186/s13287-020-01903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang F, Zhu Y, Chen J, et al. Efficient endothelial and smooth muscle cell differentiation from human pluripotent stem cells through a simplified insulin‐free culture system. Biomaterials. 2021;271:120713. 10.1016/j.biomaterials.2021.120713. [DOI] [PubMed] [Google Scholar]
- 65.Pars S, Achberger K, Kleger A, et al. Generation of functional vascular endothelial cells and pericytes from keratinocyte derived human induced pluripotent stem cells. Cells. 2021;10(1). 10.3390/cells10010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ota S, Horisaki R, Kawamura Y, et al. Ghost cytometry. Science. 2018;360(6394):1246‐1251. 10.1126/science.aan0096. [DOI] [PubMed] [Google Scholar]
- 67.Petchakup C, Li KHH, Hou HW. Advances in single cell impedance cytometry for biomedical applications. Micromachines (Basel). 2017;8(3). 10.3390/mi8030087. [DOI] [Google Scholar]
- 68.Mietke A, Otto O, Girardo S, et al. Extracting cell stiffness from real‐time deformability cytometry: theory and experiment. Biophys J. 2015;109(10):2023‐2036. 10.1016/j.bpj.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGrath J, Jimenez M, Bridle H. Deterministic lateral displacement for particle separation: a review. Lab Chip. 2014;14(21):4139‐4158. 10.1039/C4LC00939H. [DOI] [PubMed] [Google Scholar]
- 70.Di Carlo D, Irimia D, Tompkins RG, et al. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Natl Acad Sci USA. 2007;104(48):18892‐18897. 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xavier M, Holm SH, Beech JP, et al. Label‐free enrichment of primary human skeletal progenitor cells using deterministic lateral displacement. Lab Chip. 2019;19(3):513‐523. 10.1039/c8lc01154k. [DOI] [PubMed] [Google Scholar]
- 72.Liu Z, Huang F, Du J, et al. Rapid isolation of cancer cells using microfluidic deterministic lateral displacement structure. Biomicrofluidics. 2013;7(1). 10.1063/1.4774308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Civin CI, Ward T, Skelley AM, et al. Automated leukocyte processing by microfluidic deterministic lateral displacement. Cytometry Part A. 2016;89(12):1073‐1083. 10.1002/cyto.a.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hur SC, Brinckerhoff TZ, Walthers CM, et al. Label‐free enrichment of adrenal cortical progenitor cells using inertial microfluidics. PLoS One. 2012;7(10):e46550. 10.1371/journal.pone.0046550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun J, Li M, Liu C, et al. Double spiral microchannel for label‐free tumor cell separation and enrichment. Lab Chip. 2012;12(20):3952‐3960. 10.1039/C2LC40679A. [DOI] [PubMed] [Google Scholar]
- 76.Geislinger TM, Eggart B, Braunmüller S, et al. Separation of blood cells using hydrodynamic lift. Appl Phys Lett. 2012;100(18):183701. 10.1063/1.4709614. [DOI] [Google Scholar]
- 77.Liu A, Islam M, Stone N, et al. Microfluidic generation of transient cell volume exchange for convectively driven intracellular delivery of large macromolecules. Mater Today (Kidlington). 2018;21(7):703‐712. 10.1016/j.mattod.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stone NE, Voigt AP, Cooke JA, et al. Label‐free microfluidic enrichment of photoreceptor cells. Exp Eye Res. 2020;199:108166. 10.1016/j.exer.2020.108166. [DOI] [PubMed] [Google Scholar]
- 79.Wang G, Mao W, Byler R, et al. Stiffness dependent separation of cells in a microfluidic device. PLoS One. 2013;8(10):e75901. 10.1371/journal.pone.0075901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tasadduq B, Lam W, Alexeev A, et al. Enhancing size based size separation through vertical focus microfluidics using secondary flow in a ridged microchannel. Sci Rep. 2017;7(1):17375. 10.1038/s41598-017-17388-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang G, Crawford K, Turbyfield C, et al. Microfluidic cellular enrichment and separation through differences in viscoelastic deformation. Lab Chip. 2015;15(2):532‐540. 10.1039/c4lc01150c. [DOI] [PubMed] [Google Scholar]
- 82.Bongiorno T, Gura J, Talwar P, et al. Biophysical subsets of embryonic stem cells display distinct phenotypic and morphological signatures. PLoS One. 2018;13(3):e0192631. 10.1371/journal.pone.0192631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burnight ER, Gupta M, Wiley LA, et al. Using CRISPR‐Cas9 to generate gene‐corrected autologous iPSCs for the treatment of inherited retinal degeneration. Mol Ther. 2017;25(9):1999‐2013. 10.1016/j.ymthe.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bohrer LR, Wiley LA, Burnight ER, et al. Correction of NR2E3 associated enhanced S‐cone syndrome patient‐specific iPSCs using CRISPR‐Cas9. Genes (Basel). 2019;10(4). 10.3390/genes10040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giacalone JC, Sharma TP, Burnight ER, et al. CRISPR‐Cas9‐based genome editing of human induced pluripotent stem cells. Curr Protoc Stem Cell Biol. 2018;44:5B.7.1‐5B.7.22. 10.1002/cpsc.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zeballos MA, Gaj T. Next‐generation CRISPR technologies and their applications in gene and cell therapy. Trends Biotechnol. 2020;39:692‐705. 10.1016/j.tibtech.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giacalone JC, Andorf JL, Zhang Q, et al. Development of a molecularly stable gene therapy vector for the treatment of RPGR‐associated X‐linked retinitis pigmentosa. Hum Gene Ther. 2019;30(8):967‐974. 10.1089/hum.2018.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wiley LA, Burnight ER, Kaalberg EE, et al. Assessment of ddeno‐associated virus serotype tropism in human retinal explants. Hum Gene Ther. 2018;29(4):424‐436. 10.1089/hum.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wiley LA, Burnight ER, Drack AV, et al. Using patient‐specific induced pluripotent stem cells and wild‐type mice to develop a gene augmentation‐based strategy to treat CLN3‐associated retinal degeneration. Hum Gene Ther. 2016;27(10):835‐846. 10.1089/hum.2016.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cideciyan AV, Jacobson SG, Drack AV, et al. Effect of an intravitreal antisense oligonucleotide on vision in Leber congenital amaurosis due to a photoreceptor cilium defect. Nat Med. 2019;25(2):225‐228. 10.1038/s41591-018-0295-0. [DOI] [PubMed] [Google Scholar]
- 91.Russell S, Bennett J, Wellman JA, et al. Efficacy and safety of voretigene neparvovec (AAV2‐hRPE65v2) in patients with RPE65‐mediated inherited retinal dystrophy: a randomised, controlled, open‐label, phase 3 trial. Lancet. 2017;390(10097):849‐860. 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jin Y, Shen Y, Su X, et al. Effective restoration of dystrophin expression in iPSC Mdx‐derived muscle progenitor cells using the CRISPR/Cas9 system and homology‐directed repair technology. Comput Struct Biotechnol J. 2020;18:765‐773. 10.1016/j.csbj.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J‐P, Li X‐L, Neises A, et al. Different effects of sgRNA length on CRISPR‐mediated gene knockout efficiency. Sci Rep. 2016;6:28566. 10.1038/srep28566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chu HW, Rios C, Huang C, et al. CRISPR‐Cas9‐mediated gene knockout in primary human airway epithelial cells reveals a proinflammatory role for MUC18. Gene Ther. 2015;22(10):822‐829. 10.1038/gt.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J‐P, Li X‐L, Li G‐H, et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9‐mediated double‐stranded DNA cleavage. Genome Biol. 2017;18(1):35. 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lundstrom K. Viral vectors in gene therapy. Diseases. 2018;6(2):42. 10.3390/diseases6020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim S, Kim D, Cho SW, et al. Highly efficient RNA‐guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012‐1019. 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mout R, Ray M, Yesilbag Tonga G, et al. Direct cytosolic delivery of CRISPR/Cas9‐Ribonucleoprotein for efficient gene editing. ACS Nano. 2017;11(3):2452‐2458. 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sinha D, Steyer B, Shahi PK, et al. Human iPSC modeling reveals mutation‐specific responses to gene therapy in a genotypically diverse dominant maculopathy. Am J Hum Genet. 2020;107(2):278‐292. 10.1016/j.ajhg.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hallow DM, Seeger RA, Kamaev PP, et al. Shear‐induced intracellular loading of cells with molecules by controlled microfluidics. Biotechnol Bioeng. 2008;99(4):846‐854. 10.1002/bit.21651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharei A, Zoldan J, Adamo A, et al. A vector‐free microfluidic platform for intracellular delivery. Proc Natl Acad Sci USA. 2013;110(6):2082‐2087. 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Byrne SM, Church GM. Crispr‐mediated gene targeting of human induced pluripotent stem cells. Curr Protoc Stem Cell Biol. 2015;35:5A.8.1‐5A.8.22. 10.1002/9780470151808.sc05a08s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Byrne SM, Mali P, Church GM. Genome editing in human stem cells. Methods Enzymol. 2014;546:119‐138. 10.1016/B978-0-12-801185-0.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


