Abstract
A recurrent translocation between chromosome 1 (Pbx1) and 19 (E2A) leading to the expression of the E2A-Pbx1 fusion oncoprotein occurs in ∼5 to 10% of acute leukemias in humans. It has been proposed that some of the oncogenic potential of E2A-Pbx1 could be mediated through heterocomplex formation with Hox proteins, which are also involved in human and mouse leukemias. To directly test this possibility, mouse bone marrow cells were engineered by retroviral gene transfer to overexpress E2A-Pbx1a together with Hoxa9. The results obtained demonstrated a strong synergistic interaction between E2A-Pbx1a and Hoxa9 in inducing growth factor-independent proliferation of transduced bone marrow cells in vitro and leukemic growth in vivo in only 39 ± 2 days. The leukemic blasts which coexpress E2A-Pbx1a and Hoxa9 showed little differentiation and produced cytokines such as interleukin-3, granulocyte colony-stimulating factor, and Steel. Together, these studies demonstrate that the Hoxa9 and E2A-Pbx1a gene products collaborate to produce a highly aggressive acute leukemic disease.
Homeodomain (HD)-containing Hox gene products, regulators of pattern formation and tissue identity during embryogenesis (15), have also been identified as potential regulators of hemopoietic cell proliferation and differentiation (17). Several lines of evidence now also directly implicate Hox genes in human and murine leukemias. These include (i) the expression of Hoxa9 as a fusion protein Hoxa9-NUP98, in a subset of human myeloid leukemias (2, 21); (ii) the activation of Hoxa7 and Hoxa9 by retroviral insertional mutagenesis in myeloid leukemias in BXH-2 mice (22), and (iii) the development of leukemias in mice transplanted with bone marrow cells engineered to retrovirally overexpress Hoxb8, Hoxb3, Hoxa10, or Hoxa9 (13, 25, 32, 39).
A number of studies have demonstrated that Hox proteins collaborate in the in vitro DNA binding with a group of HD-containing proteins comprising Pbx and Meis families (34–36). This cooperative interaction between Pbx and Hox proteins is important since genetic and molecular studies in mice and in Drosophila have shown that Pbx1 (or its Drosophila homolog exd) is required for some of the biological functions of Hox proteins (1, 4, 19, 29, 30). The relevance of this Hox-Pbx interaction for malignant transformation has been demonstrated, as we recently showed that Hoxb3- or Hoxb4-induced transformation of Rat-1 fibroblasts is dependent on the presence of endogenous Pbx1 (14). An oncogenic collaboration between Hox and Meis proteins has also been established both by proviral insertion (22) and by retroviral overexpression studies (13). The Hox-Pbx interacting surfaces have been the focus of a number of studies and include, in addition to the HDs of both proteins, a tryptophan-containing motif located N terminal to the HD of Hox (found in several Hox proteins) and a region of 20 to 25 conserved amino acids called HCM (Hox cooperativity motif) located C terminal to the HD of Pbx1 (6, 24, 26, 27). The structure of the Hoxb1-Pbx1 complex bound to DNA was recently solved by crystallographic studies and confirmed the importance of these motives for Hox-Pbx1 interactions (28). Interestingly, these studies also demonstrated that the HCM motif of Pbx1 is part of its HD giving rise to a fourth α helix (28).
One member of the Pbx family, Pbx1, is also involved in human malignancy. In the t(1;19)(q23;p13.3) chromosomal translocation (11, 23), found in 10 to 20% of human pediatric pre-B acute lymphoblastic leukemias (3), most of the Pbx1 coding sequence, including the segment encoding the HD, is fused to the 5′ half of the E2A gene, which encodes two transcription activation domains but lacks both the DNA binding and dimerization domains of E2A (11, 23). In addition to the involvement of the E2A-Pbx1 fusion gene in human acute lymphoblastic leukemia, various transformation assays have clearly demonstrated that the E2A-Pbx1 fusion protein is oncogenic. Transgenic mice expressing the E2A-Pbx1 cDNA in lymphoid cells developed T-cell lymphoblastic lymphomas (8), and mice reconstituted with bone marrow cells engineered by retrovirus-mediated gene transfer to overexpress E2A-Pbx1 developed growth factor-dependent acute myeloid leukemias (AML) (10).
The in vitro cooperative DNA binding properties of Pbx1 and E2A-Pbx1 with Hox proteins are not significantly different (18). This has lead to one current hypothesis, that at least in part, the transforming capacity of E2A-Pbx1 could be mediated by Hox gene products. In agreement with this possibility, deletion of all of the Pbx1 sequence from the E2A-Pbx1 fusion protein completely abrogates transformation of NIH 3T3 cells, indicating that the Pbx1 half of the fusion protein is essential for its transforming abilities (12, 20). Furthermore, the HCM of the Pbx1 half of the E2A-Pbx1 fusion protein is essential for cellular transformation induced by E2A-Pbx1 (5). This finding is very interesting in light of the recent crystallographic studies mentioned above which have redefined the HCM as part of an extended HD in Pbx1 (28). The concept that E2A-Pbx1-induced transformation is Hox dependent was challenged by studies which showed that the helices 1 to 3 of the HD in E2A-Pbx1 are dispensable for the capacity of this fusion protein to transform NIH 3T3 cells and T lymphocytes (12, 20). However, it was recently shown that the HD of E2A-Pbx1 is necessary to block cellular differentiation of myeloid progenitor cells, a process central to leukemic transformation (12). Together, these studies thus suggest that cellular transformation induced by E2A-Pbx1 may involve more than one pathway (mechanisms).
In contrast to E2A-Pbx1, the Pbx proteins lack inherent transforming potential (12–14, 20), and fusion with E2A is essential for the transforming ability of E2A-Pbx1. Structure-function and mutagenesis experiments have demonstrated that the two transcriptional activation domains in E2A are essential for mediating both the malignant transformation of NIH 3T3 cells (20) and blocking the differentiation of myeloid progenitor cells (12). Furthermore, using a Pbx-responsive sequence which allows cooperative DNA binding between Hox and Pbx1 (or E2A-Pbx1), it was shown in a reporter assay that E2A-Pbx1, but not Pbx1, could induce significant transcriptional activity of the reporter gene and that Hox proteins had the capacity to modulate the transactivating activity of E2A-Pbx1 (18). Together, these observations suggest that the oncogenic potential of E2A-Pbx1 is directly linked to the transcriptional activating function of the chimeric protein, and that this activity can be modulated by Hox gene products.
To examine whether Hox proteins and the E2A-Pbx1a fusion protein could collaborate to transform primary hematopoietic cells, we engineered mouse bone marrow cells, by retroviral gene transfer, to cooverexpress E2A-Pbx1a with Hoxa9, and the oncogenic collaboration between these two genes was directly tested in vitro and in vivo following transplantation of retrovirally transduced cells.
MATERIALS AND METHODS
Animals.
All mice were originally bought from The Jackson Laboratory (Bar Harbor, Maine) and then bred and maintained in the specific-pathogen-free animal facility of the Clinical Research Institute of Montreal (IRCM). Donors of primary bone marrow cells were over 12-week-old (C57BL/6Ly-Pep3b × C3H/HeJ)F1 [(PepC3)F1] mice, and recipients were 7- to 12-week-old (C57BL/6J × C3H/HeJ)F1 [(B6C3)F1] mice. All animals were housed in ventilated microisolator cages and provided with sterilized food and acidified water.
Generation of recombinant retroviruses.
The MSCV-Hoxa9-pgk-neo retroviral vector was described before (13). The human E2A-Pbx1a cDNA (a kind gift of Mark Kamps, San Diego, Calif.) was subcloned into the EcoRI site of MSCV-pgk-PAC retroviral vector (which confers puromycin resistance). Both MSCV vectors were kindly provided by R. Hawley (The Toronto Hospital Research Institute, Toronto, Ontario, Canada). High-titer helper-free retroviral producer cells were generated from GP+E-86 and BOSC-23 viral packaging cells and tested as reported previously (13).
Retroviral infection of primary murine bone marrow cells.
Bone marrow cells obtained from (PebC3)F1 mice injected 4 days earlier with 5-fluorouracil (50 mg/kg of body weight; Sigma) were prestimulated in the presence of interleukin-6 (IL-6), IL-3, and Steel factor and cocultivated on GP+E-86 viral producer cells engineered to produce the MSCV-Hoxa9-pgk-neo or the MSCV-E2A-Pbx1a-pgk-puro recombinant retrovirus. The procedure for cell harvesting, prestimulation, cocultivation, and bone marrow transplantation were performed as described elsewhere (13, 31). For double infections of bone marrow cells with Hoxa9 and E2A-Pbx1a, the respective viral producer cells were counted and seeded at equal numbers 24 h prior to the addition of bone marrow cells. Cells were cocultured for 48 h, with a medium change after 24 h. In an effort to enhance the efficiency of retroviral transduction of bone marrow cells, viral supernatants from Hoxa9- and E2A-Pbx1a-transfected BOSC-23 cells were added to the respective cocultures. The gene transfer efficiencies to primitive hemopoietic cells, as assessed by the in vitro colony formation of G418-resistant (containing the neo control or Hoxa9 retrovirus) or puromycin-resistant (containing the puro control or E2A-Pbx1a retrovirus) myeloid clonogenic progenitors, were 50, 40, 72, and 20% for the neo, puro, Hoxa9, and E2A-Pbx1a retroviruses, respectively. The efficiency of double infection of myeloid progenitor cells with the Hoxa9 and E2A-Pbx1a retroviruses was 4% as assessed by resistance of progenitor cells to both G418 and puromycin. All growth factors were used as diluted supernatants from appropriately transfected COS cells, as prepared at the IRCM. All reagents used in this study including media and serum were purchased at GIBCO Life Technologies.
Transplantations.
The primary mice used in this study were generated by injecting intravenously into lethally irradiated (900 cGy, 179 cGy/min, 137Cs gamma rays; J. L. Shepherd, San Fernando, Calif.) 7- to 12-week-old (B6C3)F1 mice 2 × 105 bone marrow cells derived from the (PepC3)F1 mice immediately following their harvesting from the cocultivation with viral producer cells. To determine the transplantability of the diseases that developed in some of the primary mice, lethally irradiated or nonirradiated secondary (B6C3)F1 recipients were injected with 106 bone marrow and/or spleen cells obtained from the primary mice. For some of the leukemias that developed in the Hoxa9 + E2A-Pbx1a mice, the frequency of the leukemia repopulating cells (LRC) in bone marrow of these mice was determined by injecting various numbers of bone marrow cells (5 to 106 cells per mouse) into lethally irradiated (B6C3)F1 mice, along with a life-sparing dose of 105 normal bone marrow cells derived from (B6C3)F1 mice. LRC frequency in primary recipients was estimated by applying Poisson statistics to the proportion of negative recipients at different dilutions as described previously (37).
In vitro cultures.
For myeloid clonogenic progenitor assays, cells were plated on 35-mm-diameter petri dishes (Corning, Fisher) in a 1.1 ml culture mixture containing 0.8% methylcellulose in alpha medium supplemented with 10% fetal calf serum, 5.7% bovine serum albumin, 10−5 M β-mercaptoethanol, 1 U of human urinary erythropoietin (Epo) per ml, 10% WEHI-conditioned medium (tested to contain 50 ng of IL-3 per ml), 2 mM glutamine, and 200 mg of transferrin per ml, in the presence or absence of 1.3 mg of G418 per ml and/or 1.5 μg of puromycin per ml. To test the ability of the retrovirally infected myeloid progenitor cells to form colonies in the absence of added growth factors, cells were plated in the above-described methylcellulose medium lacking both WEHI-conditioned medium and Epo. Bone marrow cells harvested from the cocultivation with virus-producing cells or recovered from reconstituted mice were plated at a concentration of 2 × 103 to 8 × 103 or 3 × 104 cells/ml, respectively. Spleen cells from neo control mice were plated at a concentration of 3 × 106 cells/ml, whereas those of the other reconstituted mice at 105 cells/ml. Bone marrow or spleen cells from the reconstituted mice were plated at these same concentrations in the methylcellulose cultures lacking WEHI-conditioned medium and Epo. Colonies were scored on days 12 to 14 of incubation as derived from CFU–granulocyte-macrophage, burst-forming unit erythroid, or granulocyte-erythrocyte-macrophage-megakaryocyte CFU according to standard criteria (9). For some of the experiments, identification of colony types was confirmed by Wright staining of cytospin preparations of colonies. Bone marrow and/or spleen cells from the transplanted recipients were cultured in liquid cultures of Iscove’s medium containing 10% fetal calf serum, 10−5 M β-mercaptoethanol, 2 mM glutamine, and 200 mg of transferrin per ml, in the presence or absence of 5 ng of IL-3 or 0.5 ng of granulocyte-macrophage colony-stimulating factor (GM-CSF) per ml.
Titration of growth factors.
The presence of bioactive IL-3, G-CSF, and Steel present in conditioned medium obtained from 4-day-old cultures of cells obtained from the spleens of various mice and seeded at 106 cells/ml was tested by [3H]thymidine incorporation assay to measure the proliferation of Ba/F3 (IL-3-responsive), NFS-60 (G-CSF- and IL-3-responsive) and TF-1 (Steel-responsive) cells. Titration of the bioactive substances was performed with COS cell-derived IL-3, G-CSF, and Steel as described above. The specificity of each growth factor to induce [3H]thymidine incorporation into the DNA of the appropriate cell line was verified by using antisera specific to IL-3 (50% neutralizing dose [ND50] = 0.0015 to 0.025 μg/ml), G-CSF (ND50 = 0.4 to 0.8 μg/ml), and GM-CSF (ND50 = 0.05 to 0.15 μg/ml) (all from R&D). Complete and specific neutralization of [3H]thymidine incorporation was obtained for each supernatant tested except for that derived from 4.27 cells, where antibodies to both IL-3 and G-CSF were required for complete inhibition of proliferation of NFS-60 cells.
DNA and RNA analyses.
To assess proviral integration, Southern hybridization analyses were performed as described elsewhere (13). Total cellular RNA was isolated with the TRIzol reagent, and Northern blot analysis was performed as described previously (7). The probes used were an XhoI/SalI fragment of pMC1neo (neo) (38), a HindIII/ClaI fragment of MSCV-pgk-PAC (puro), or the full-length 1.4-kb Hoxa9 and 2.9-kb E2A-Pbx1a cDNAs and were labeled with 32P by random primer extension as described elsewhere (16). To assess the relative amounts of total RNA loaded, membranes were probed for 18S RNA by using end-labeled oligonucleotide 5′- ACG GTA TCT GAT CGT CCT CGA ACC-3′.
Nuclear extract preparation.
A total of 2 × 107 cells were washed twice in phosphate-buffered saline, incubated for 10 min at 0°C in 600 μl of lysis buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA), and then disrupted by passage through a 26-guage needle. Nuclei were collected by centrifugation at 500 rpm for 5 min and resuspended in 60 μl of extraction buffer (20 mM HEPES [pH 7.9], 400 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 5% glycerol) to allow elution of nuclear proteins by vigorous shaking for 1 h at 4°C. Nuclei were spun down by 2 min of centrifugation at 12,000 rpm, and the recovered supernatant was further cleared by 20 min of centrifugation at 40,000 rpm at 4°C. The protease inhibitors phenylmethylsulfonyl fluoride (2 mM), aprotinin (10 μg/ml), leupeptin (1 μg/ml), pepstatin A (10 μg/ml), and antipain (10 μg/ml) were added to both lysis and extraction buffers. Protein concentration was determined by the MicroBCA assay as recommended by manufacturer (Sigma, St. Louis, Mo.).
Western blot analysis.
Thirty-microgram aliquots of proteins separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis were transferred to Immobilon P membranes (Millipore, Bedford, Mass.). Membranes were blocked with 5% nonfat milk in TBST (20 mM Tris-Cl [pH 7.6], 140 mM NaCl, 0.05% Tween 20) and then subjected to Western blot analysis with anti-E2A (E47; Santa Cruz Biotechnology Inc, Santa Cruz, Calif.) and anti-Hoxa9 (generously provided by H. Jeffrey Lawrence, San Francisco, Calif.). Bound antibodies were detected by using horseradish peroxidase-conjugated anti-rabbit antibody (Sigma) followed by enhanced chemiluminescence (Amersham, Buckinghamshire, England).
RESULTS
Altered colony formation and growth factor dependence of myeloid progenitor cells overexpressing Hoxa9 together with E2A-Pbx1a.
The Hoxa9 and E2A-Pbx1a cDNAs were introduced downstream of the long terminal repeat of either the MSCVneoEB (Hoxa9) or MSCVpac (E2A-Pbx1a) retroviral vector (Fig. 1). Immediately following the infection of mouse bone marrow cells with the Hoxa9, E2A-Pbx1a, or Hoxa9 and E2A-Pbx1a retroviruses, the frequency of transduced myeloid colony-forming cells (CFC) was determined in methylcellulose cultures in the presence and absence of hematopoietic growth factors (Fig. 2A and B).
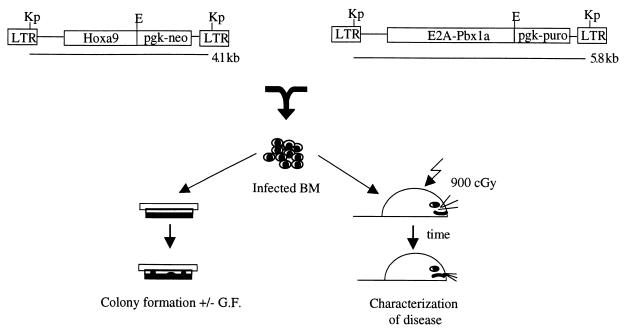
FIG. 1.
Structures of the retroviruses and overview of the experimental strategy used in this study. Diagrammatic representation of the integrated Hoxa9 and E2A-Pbx1a proviruses and the experiments described in this study. The expected sizes of the full-length viral transcripts are shown. Restriction sites indicated are KpnI (Kp) and EcoRI (E). LTR, long terminal repeat. G.F., growth factor.
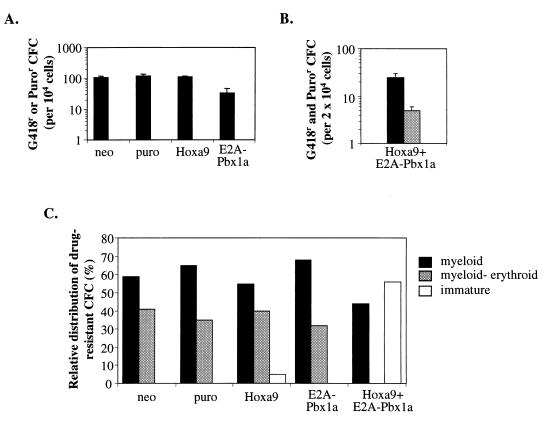
FIG. 2.
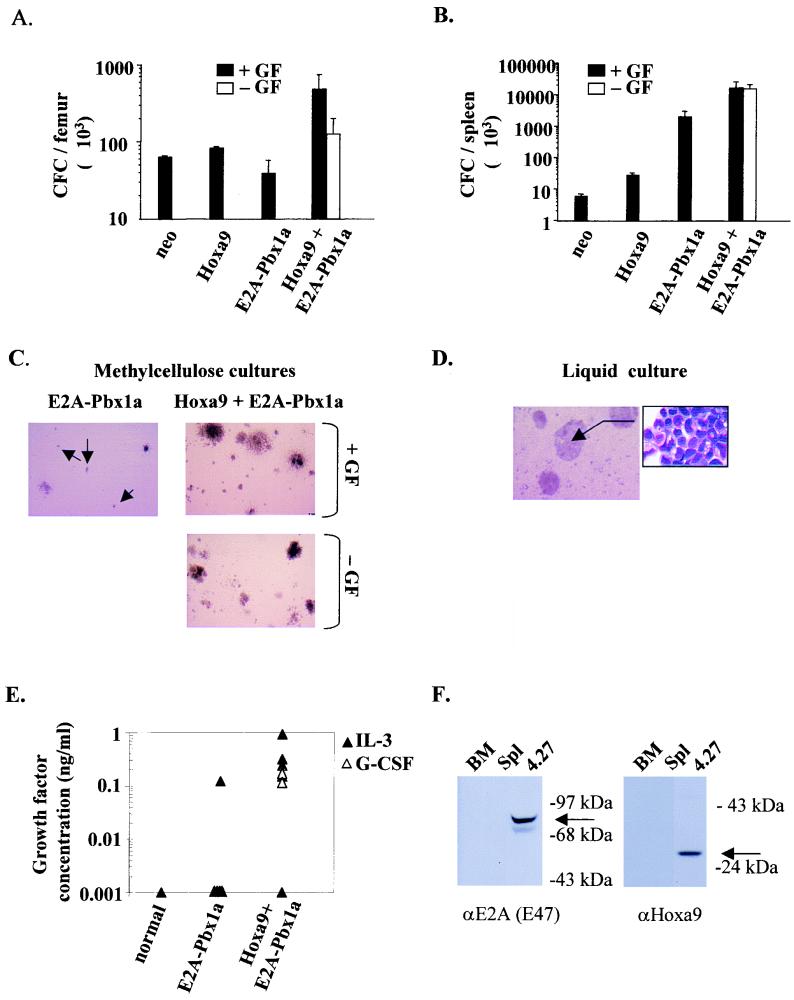
Absence of differentiation and cytokine-independent growth in vitro characterizes myeloid CFC overexpressing both Hoxa9 and E2A-Pbx1a. (A) Total number of G418- or puromycin-resistant colonies generated per 104 bone marrow cells immediately following infection with neo control, puro control, E2A-Pbx1a, and Hoxa9 retroviruses, in the presence of added growth factors. No colonies grew in the absence of added growth factors. (B) Total number of G418- and puromycin-resistant colonies generated from 2 × 104 bone marrow cells immediately following infection with Hoxa9 and E2A-Pbx1a retroviruses, in the presence (black bars) and absence (gray bars) of added growth factors. Results shown in panels A and B represent the mean ± standard deviation of the number of G418 (neo and Hoxa9)- and/or puromycin (puro and E2A-Pbx1a)-resistant colonies detected on four different plates. (C) Well-isolated G418-, puromycin- or G418- and puromycin-resistant colonies were randomly picked (n = 16 to 20 for each infection type, from two different plates) on days 10 to 12 of incubation and examined by Wright staining. Colony types detected were classified as myeloid (mainly containing granulocytes and/or macrophages), myeloid-erythroid (containing granulocytes, macrophages, megakaryocytes, and erythrocytes), and immature (containing undifferentiated blast-like cells).
The in vitro growth of CFC engineered to overexpress Hoxa9 or E2A-Pbx1a alone remained dependent on the presence of added growth factors (Fig. 2A and B), whereas ∼20% of the bone marrow CFC which cooverexpressed both Hoxa9 and E2A-Pbx1a were capable of colony formation in the absence of added growth factors, thus demonstrating the collaboration between these two genes in inducing autonomous growth in vitro (Fig. 2B).
In the presence of added growth factors, transduced (i.e., drug-resistant) CFC overexpressing Hoxa9 or E2A-Pbx1a alone were capable of generating all types of colonies normally present in such cultures, as revealed by cytological examinations of Wright-stained G418- or puromycin-resistant colonies (Fig. 2C). In contrast, about half of the CFC transduced with Hoxa9 and E2A-Pbx1a generated colonies which contained only immature blast cells, a colony type rarely detected in control cultures (Fig. 2C). The generation of these blast colonies was accompanied by the absence of primitive GEMM (myeloid-erythroid) colonies, whereas the proportion GM and M (myeloid) colonies were within normal range (Fig. 2C), indicating that the cooverexpression of Hoxa9 and E2A-Pbx1a can inhibit the in vitro differentiation of primitive (i.e., CFU-GEMM) but not mature (i.e., CFU-GM or CFU-M) CFC. Together, these data suggest that the cooverexpression of Hoxa9 and E2A-Pbx1 could be sufficient to directly transform bone marrow cells by inducing autonomous, cytokine-independent proliferation and simultaneously suppressing differentiation of primitive CFCs.
Cooverexpression of Hoxa9 with E2A-Pbx1a is acutely transforming.
The capacity of Hoxa9- and E2A-Pbx1a-transduced bone marrow cells to induce acute leukemia in vivo was directly tested by transplanting Hoxa9- and E2A-Pbx1a-transduced bone marrow cells into lethally irradiated syngenic mice. In addition to the neo and puro controls, mice were also transplanted with bone marrow cells engineered to overexpress Hoxa9 or E2A-Pbx1a alone. To facilitate comparison of the dose of transduced cells transplanted in each group, the number of transduced myeloid progenitor cells (G418 and/or puromycin resistant) and the estimated number of long-term repopulating cells (LTRC) that were injected per mouse are shown in Table 1.
TABLE 1.
Absolute numbers of myeloid CFC and LTRC resistant to G418 (transduced with neo or Hoxa9) and/or puromycin (puro or E2A-Pbx1a) transplanted per mouse
| Virus | No. of mice transplanted | No. of transduced CFC injected/mousea
|
Estimated no. of transduced LTRC injected/mouseb
|
||||
|---|---|---|---|---|---|---|---|
| G418r | Puror | G418r Puror | G418r | Puror | G418r Puror | ||
| neo | 6 | 2,100 | 21 | ||||
| puro | 4 | 2,340 | 23 | ||||
| Hoxa9 | 7 | 2,232 | 22 | ||||
| E2A-Pbx1a | 6 | 650 | 7 | ||||
| Hoxa9 + E2A-Pbx1a | 6 | 1,764 | 954 | 252 | 18 | 10 | 3 |
Determined as follows: (number of bone marrow cells injected per mouse) × (CFC frequency in injected bone marrow inoculum) × (percentage of CFC resistant to puromycin and/or G418).
All recipients of Hoxa9- and E2A-Pbx1a-transduced bone marrow cells (Hoxa9 + E2A-Pbx1a mice) developed acute leukemia in 39 ± 2 days (Table 2). In contrast, when overexpressed alone, neither Hoxa9 nor E2A-Pbx1a was acutely transforming. In fact, it took between 80 and 167 days for leukemia to developed in these recipients (Table 2; see also below).
TABLE 2.
Characteristics of diseases that developed in primary recipientsa
| Mouse group (n) | Time of death or sacrifice (days post-Tx)b | No. with
|
No. of NC (106)/femur (% BM blast)b | Spleen wt (g)b | Clonality | Disease | |
|---|---|---|---|---|---|---|---|
| HL paralyses | PBL blastosis | ||||||
| neo control (4) | 56c | 0 | 0 | 19.0 ± 4 (<1) | 0.1 ± 0.01 | Oligo/poly | NOD |
| Hoxa9 nonleukemic (3) | 56c | 0 | 0 | 19.0 ± 5 (<1) | 0.13 ± 0.01 | Oligo/poly | NOD |
| Hoxa9 leukemic (4) | 167 ± 32d | 0 | 4 | 22.5 ± 5 (70 ± 20) | 0.6 ± 0.1 | Mono/bi | AML |
| E2A-Pbx1a (6) | 51 ± 3e | 6 | 0 | 7.0 ± 2 (25 ± 8) | 0.7 ± 0.3 | Poly | MPS |
| Hoxa9 + E2A-Pbx1a (6) | 39 ± 2 | 0 | 6 | 17.0 ± 8 (90 ± 5) | 0.8 ± 0.3 | Oligo | AML |
Abbreviations: Tx, transplantation; HL, hind leg; PBL, peripheral blood; NC, nucleated cells; NOD, no observed disease; AML, acute myeloid leukemia; MPS, myeloproliferative disorder.
Results are expressed as mean values ± SD.
The neo control and Hoxa9 nonleukemic mice were healthy at the time of sacrifice. Additional data for the three nonleukemic Hoxa9 mice (mice 1 to 3) sacrificed 56 days after transplantation are presented in Fig. 3, 6A and B, and 7B.
Sacrificed when presenting with acute leukemia (mice 4 to 7 in Fig. 4A and B and 7B).
Sacrificed when presenting with hind leg paralyses.
At the time of sacrifice, the Hoxa9 + E2A-Pbx1a mice were runted, were short of breath, and had very high (>50,000/μl) peripheral leukocyte counts (Fig. 3). In these mice, bone marrow, spleen, and lymph nodes as well as liver, lungs, and brain were highly infiltrated with leukemic blasts (Fig. 3 and data not shown). Morphological (Fig. 3), histochemical, and fluorescence-activated cell sorting (FACS) analyses showed that 30 to 40% of the leukemic blasts were of myeloid origin, with moderate Sudan black staining (myeloid), and coexpressed low levels of Mac-1 and Gr-1 (data not shown). None (<1%) of these leukemic cells expressed B- or T-cell markers such as B220, CD4, or CD8, and these cells were also negative for periodic acid-Schiff reaction characteristic of lymphoblastic leukemia (data not shown). Thus, each of the leukemias in Hoxa9 + E2A-Pbx1a mice had the capacity for partial myeloid differentiation, with a significant proportion of the blasts (60 to 70%) remaining uncommitted to either the myeloid or lymphoid lineage.
FIG. 3.
Cytopathological examination of hematopoietic and nonhematopoietic tissues from neo, Hoxa9, E2A-Pbx1a, and Hoxa9 + E2A-Pbx1a mice after Wright staining of peripheral blood smears (PB), bone marrow (BM) cytospins, and touch preparations of spleen (SPL), lung (LU), liver (LI), and brain (BR) tissues from representative neo control, E2A-Pbx1a, nonleukemic Hoxa9, and Hoxa9 + E2A-Pbx1a mice described in Table 2. Note the infiltration by immature blast cells in all six tissues of the Hoxa9 + E2A-Pbx1a mouse. In contrast, the spleen is the only tissue in the E2A-Pbx1a mouse that is infiltrated by such a cell type in addition to immature granulocytic cells. An increase in immature granulocytic cells is also detected in the spleen of the Hoxa9 mouse. Magnification, ×100 for all except the peripheral blood (×20). a, granulocyte; b, lymphocyte; c, blast; d, immature granulocyte; e, erythroblast; f, hepatocyte; g, erythrocyte.
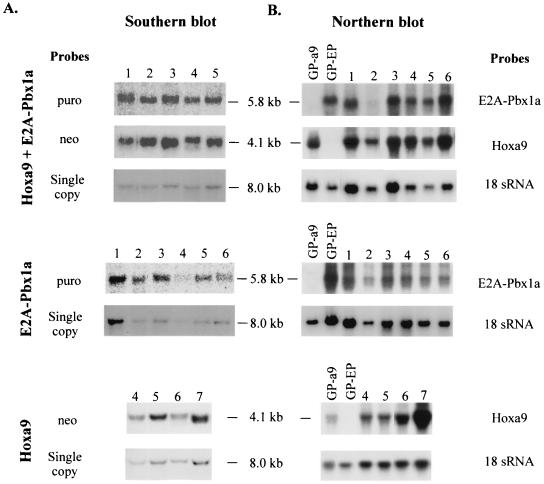
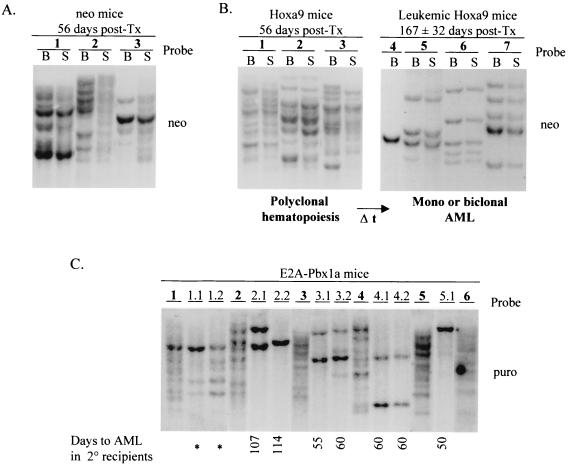
Southern blot analysis of DNA isolated from leukemic cells present in the bone marrow or spleen of the Hoxa9 + E2A-Pbx1a mice verified the presence of the Hoxa9 (4.1-kb) and E2A-Pbx1a (5.8-kb) proviruses in these cells (Fig. 4A, top panel). Furthermore, Northern blot analysis of total RNA from these same tissues detected high levels of the retrovirally derived Hoxa9 and E2A-Pbx1a messages in all of these mice (Fig. 4B, top panel).
FIG. 4.
Demonstration of the presence and expression of the integrated Hoxa9 and E2A-Pbx1a proviruses in primary mice. (A) Southern blot analyses of genomic DNA isolated from the bone marrow or spleen of the primary Hoxa9 + E2A-Pbx1a (top), E2A-Pbx1a (middle), and Hoxa9 (bottom) mice described in Table 2. DNA was digested with KpnI to release the integrated E2A-Pbx1a (5.8-kb) or Hoxa9 (4.1-kb) proviral fragment. The membranes were hybridized with a neo-specific probe to detect the Hoxa9 provirus and a puro-specific probe to detect the E2A-Pbx1a provirus. To provide a single-copy control of loading, the membranes were subsequently probed with full-length Hoxa9 cDNA probe. Note that the signal shown for the 8.0-kb single copy is that of endogenous Hoxa9. (B) Northern blot analysis of total RNA (10 μg) isolated from bone marrow or spleen cells of the primary Hoxa9 + E2A-Pbx1a (top), E2A-Pbx1a (middle), and Hoxa9 (bottom) mice described in Table 2. The membranes were hybridized with full-length E2A-Pbx1a or Hoxa9 cDNA probe. Each number assigned to a lane in panels A and B identifies a specific primary Hoxa9, E2A-Pbx1a, or Hoxa9 + E2A-Pbx1a mouse that is also identified with this same number in Fig. 5 or 7. GP-a9 or -EP, GP+E-86 Hoxa9 or E2A-Pbx1a viral producer cells.
Analysis of secondary recipients of leukemic bone marrow cells derived from Hoxa9 + E2A-Pbx1a mice.
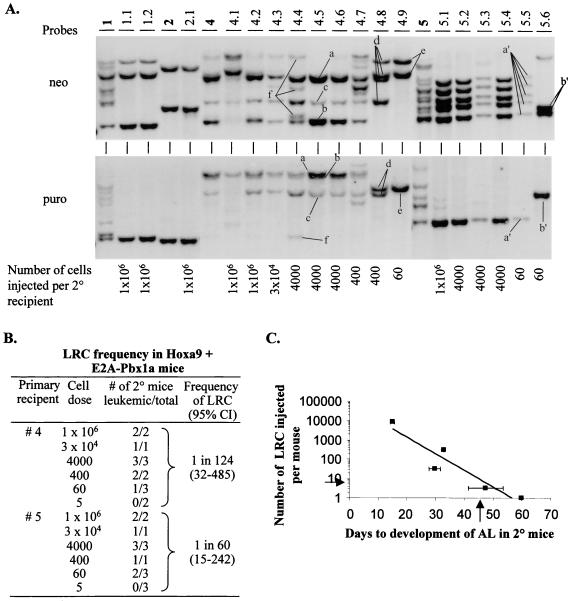
The acute leukemia induced by cooverexpression of Hoxa9 and E2A-Pbx1a was readily transplantable to secondary recipients, and clonal analysis of proviral integration sites demonstrated that the leukemias were derived from leukemic clone(s) present in the original donor leukemia (Fig. 5A). To determine the frequency of the biologically relevant cell in Hoxa9 + E2A-Pbx1a mice that can generate leukemia in secondary recipients (the leukemia repopulating cell [LRC]), bone marrow cells from Hoxa9 + E2A-Pbx1a mice 4 and 5 were injected at limiting dilution into lethally irradiated secondary recipients, along with a radioprotective dose of normal bone marrow cells. The frequency of the LRC was estimated at 1 in 124 and 1 in 60 bone marrow cells for Hoxa9 + E2A-Pbx1a mice 4 and 5, respectively (Fig. 5B). Interestingly, these studies also demonstrated a linear correlation between the number of LRC transplanted and the time required for the development of acute leukemia in the secondary recipients (Fig. 5C).
FIG. 5.
Clonal analysis of the acute leukemia that developed in primary and secondary Hoxa9 + E2A-Pbx1a recipients. (A) Southern blot analysis of DNA isolated from bone marrow of primary and secondary Hoxa9 + E2A-Pbx1a mice. The DNA was digested with EcoRI, which cuts the integrated provirus once, thus generating a unique fragment for each proviral integration site. The membranes were first hybridized with a neo-specific probe to detect Hoxa9 proviral fragments (top panel) and subsequently with a puro-specific probe to detect E2A-Pbx1a proviral fragments (bottom panel). Each primary recipient of Hoxa9- and E2A-Pbx1a-transduced cells is identified with a specific number shown in bold, and its secondary recipients are identified with derivatives thereof (1.1, 1.2, etc.). The number of leukemic cells transplanted per secondary recipients is indicated below each lane. For clarity, different leukemic Hoxa9- and E2A-Pbx1a-transduced clones detected in secondary recipients of mouse 4 are labeled from a to f; in secondary recipients of mouse 5, they are labeled a′ and b′. (B) Evaluation by limiting dilution analysis of the frequency of the LRC in Hoxa9 + E2A-Pbx1a mice 4 and 5. Clonality of the majority of the secondary recipients used in this assay are shown in panel A. (C) Graphic display of the correlation between the number of LRC transplanted per secondary recipient of Hoxa9 + E2A-Pbx1a mouse 4 and the time needed for the development of leukemia in those recipients. The arrow on the x axis indicate the predicted time required for seven LRC (arrow on y axis) to give rise to overt leukemia in secondary mice (see Discussion). AL, acute leukemia; 2°, secondary; Cl, confidence interval.
Southern blot analysis of genomic DNA isolated from the leukemic cells of the secondary recipients of Hoxa9 + E2A-Pbx1a mice 4 and 5 confirmed the presence of Hoxa9 (detected with the neo probe)- and E2A-Pbx1a (detected with the puro probe)-transduced clones in all mice that succumbed to acute leukemia, including those receiving only 60 bone marrow cells, estimated to contain only ∼1 LRC (Fig. 5A, mice 4.9, 5.5, and 5.6). In each secondary recipient, the numbers of leukemic clones detected either with the neo or the puro probe were identical, as shown for clones a, b, and c, which are detected as dominant clones in seven of nine secondary recipients of bone marrow cells from Hoxa9 + E2A-Pbx1a mouse 4 (Fig. 5A). These three clones were, however, not detected in mice 4.8 and 4.9, which were repopulated either by clone d (mouse 4.8), which contains Hoxa9 provirus at four integration sites and E2A-Pbx1a at two, or by clone e (mouse 4.9), which contains Hoxa9 provirus at two integration sites and E2A-Pbx1a at one (Fig. 5A). In addition, other clones could be detected with both probes, e.g., clone f in mouse 4.4, which contains Hoxa9 provirus at three integration sites and E2A-Pbx1a at one. Expression of the transduced genes in leukemic cells of all of these secondary recipients was confirmed by Northern blot analysis that detected high levels of expression of both the Hoxa9 and E2A-Pbx1a retrovirally derived mRNAs (data not shown).
Together, these data confirm that the acute leukemia that developed in the primary Hoxa9 + E2A-Pbx1a mice was caused by leukemic cells overexpressing both Hoxa9 and E2A-Pbx1a. These cells thus outcompeted those overexpressing either Hoxa9 or E2A-Pbx1a alone, which were in ∼6-fold (Hoxa9) and ∼3-fold (E2A-Pbx1a) excess in the inoculum originally transplanted in the primary recipients (Table 1). Furthermore, these studies also demonstrated that between one (mouse 2) and seven (mouse 4) transplantable leukemic clones were present in the primary Hoxa9 + E2A-Pbx1a mice (Fig. 5A).
Leukemic progenitor cells overexpressing Hoxa9 and E2A-Pbx1a do not require hematopoietic growth factors to proliferate in vitro.
The leukemic cells in the bone marrow and spleen of Hoxa9 + E2A-Pbx1a mice formed colonies in methylcellulose cultures supplemented with growth factors with plating efficiencies of 2.4% ± 1.2% and 1.7% ± 0.2% for bone marrow and spleen cells, respectively, or with 8- and 2,800-fold increases compared to neo control mice (Fig. 6A and B). For the five mice tested, nearly 100% of their leukemic progenitor cells were resistant to both G418 and puromycin, demonstrating that these cells were all transduced with both Hoxa9 and E2A-Pbx1a. Interestingly, and in sharp contrast to bone marrow and spleen cells derived from nonleukemic Hoxa9 or E2A-Pbx1a mice sacrificed 56 and 51 ± 3 days after transplantation, the leukemic cells from Hoxa9 + E2A-Pbx1a mice had a similar capacity for colony formation in the presence or absence of added hematopoietic growth factors (Fig. 6A to C).
FIG. 6.
Growth factor-independent proliferation of leukemic blasts isolated from primary recipients of bone marrow cells overexpressing Hoxa9 and E2A-Pbx1a. The number of colonies generated from bone marrow (A) and spleen (B) cells of neo control, Hoxa9, E2A-Pbx1a, and Hoxa9 + E2A-Pbx1a mice, in the presence or absence of added growth factors, are shown. The results shown represent the mean ± standard deviation of the number of myeloid CFC in one femur and in the spleen of neo control (n = 2) and Hoxa9 (n = 3) mice at 56 days following transplantation and that of E2A-Pbx1a (n = 6) and Hoxa9 + E2A-Pbx1a (n = 6) mice when sacrificed as outlined in Table 2. (C) Microscopic view of colonies generated in methylcellulose cultures of spleen cells from E2A-Pbx1a and Hoxa9 + E2A-Pbx1a mice 13 days after initiation of the cultures. The arrows indicate the presence of small macrophage colonies that dominated the cultures from the E2A-Pbx1a mice. GF, growth factor. (D) Liquid culture of Hoxa9- and E2A-Pbx1a-overexpressing cells after 2 months of in vitro growth, demonstrating the growth of these cells in macroscopic clumps (magnification, ×10). The callous shows a Wright-stained cytospin preparation of the cells in these clumps, showing the immature morphology of these cells (magnification, ×100). (E) The presence of bioactive IL-3 and G-CSF in conditioned medium obtained from 4-day-old cultures of cells obtained from the spleens of a normal mouse, or from E2A-Pbx1a (n = 5) and Hoxa9 + E2A-Pbx1a (n = 6) mice and seeded at 106 cells/ml, was tested as outlined in Materials and Methods. (F) Western blot analysis of cell lysates from bone marrow (BM) and spleen (Spl) cells of normal mice and from one culture of Hoxa9 + E2A-Pbx1a leukemic cells. The membrane was probed with anti-E2A monoclonal antibody and then with polyclonal antisera directed against Hoxa9. The position of the 85-kDa E2A-Pbx1a protein and the 36-kDa Hoxa9 protein is indicated (arrows).
To test whether overexpression of Hoxa9 and E2A-Pbx1a in these leukemic cells was inducing their production of hemopoietic growth factors, supernatants harvested from liquid cultures initiated with spleen cells from six Hoxa9 + E2A-Pbx1a mice were tested for the presence of IL-3, G-CSF, and Steel. Five of six leukemic samples tested produced measurable levels (0.1 to 1 ng/ml) of IL-3 and/or G-CSF (Fig. 6E; one sample produced both growth factors), and in the remaining sample secretion of Steel, but not IL-3 or G-CSF, was detected (data not presented). No production of G-CSF or Steel could be detected in the supernatants from the five cultures that were initiated with spleen cells from E2A-Pbx1a primary mice (not leukemic; see below), and only one of these contained measurable amounts of IL-3 (Fig. 6E).
Consistent with their production of growth factors, the leukemic cells from the Hoxa9 + E2A-Pbx1a mice were capable of proliferation in liquid cultures with or without added growth factors for a prolonged period of time. These cells grew in macroscopic clumps (Fig. 6D) containing mostly viable cells (n = 3 of three different leukemic cells tested) that were morphologically identical to the original donor leukemia, i.e., immature blast cells with rare differentiated elements (Fig. 6D). Clonal analysis performed on DNA isolated from these cells demonstrated that each cell line was derived from the same clone(s) identified in the spleen of a corresponding leukemic mouse (data not shown). In addition, it was possible to demonstrate by Western blot analysis that these cell lines expressed high levels of E2A-Pbx1a and Hoxa9 (Fig. 6F).
In contrast to the Hoxa9 + E2A-Pbx1a leukemic cells, the spleen cells from the E2A-Pbx1a mice did not survive (80 to 90% mortality at day 5) or proliferate in the absence of added growth factors. However, in the presence of IL-3 or GM-CSF, these cells proliferated initially, but while forming a feeder layer, the cultures became exhausted in 4 to 5 weeks.
Overexpression of E2A-Pbx1a leads to the development of a highly polyclonal myeloproliferative syndrome (MPS).
The recipients of Hoxa9-transduced bone marrow cells thrived normally for an extended period of time. At 56 days posttransplantation, three of these mice were sacrificed to allow comparison with the leukemic Hoxa9 + E2A-Pbx1a mice. At this time, hemopoietic regeneration by Hoxa9-transduced cells in these mice was polyclonal, similar to that detected in the neo control mice (Table 2 and Fig. 7A and B). The only hematological abnormality detected in these mice was in their spleens, which were slightly enlarged (Table 2), with some increase in cells with immature granulocytic morphology (Fig. 3) and myeloid CFC (Fig. 6A and B). As we previously reported (13), the remaining four Hoxa9 mice eventually developed mono- or biclonal AML with latency of 167 ± 32 days (Table 2 and Fig. 7B).
FIG. 7.
Southern blot analysis of DNA isolated from bone marrow of primary neo (A) and Hoxa9 (B) mice and primary and secondary E2A-Pbx1a mice (C). The DNA was digested with EcoRI, which cuts the integrated provirus once, thus generating unique fragments specific for the proviral integration site(s). The membranes were hybridized with a neo-specific probe to detect the neo control and Hoxa9 proviral fragments and a puro-specific probe to detect those of the E2A-Pbx1a provirus. Each primary mouse is identified with specific number in bold, and its secondary recipients are identified with derivatives thereof. All secondary recipients were transplanted with 1 × 106 to 2 × 106 bone marrow or spleen cells from the donor mouse. Where applicable, the time (in days posttransplantation [post-Tx]) for the development of acute leukemia in the secondary E2A-Pbx1a mice is shown. Asterisks denote secondary (2°) mice that developed MPS.
The recipients of E2A-Pbx1a-transduced bone marrow, however, developed progressive hind limb paralysis and for that reason were sacrificed at 51 ± 3 days following transplantation (Table 2). At the time of their sacrifice, these mice had normal blood cell counts and the cellular constituents of their bone marrow were not overtly altered except for higher than normal proportions of mature neutrophils and monocytes, with some (<30%) immature forms (Fig. 3). Their bone marrow cellularity was, however, decreased threefold compared to the control neo mice (Table 2), and all animals had large spleens (Table 2), characterized by an accumulation of immature granulocytic cells together with few blast-like cells (Fig. 3). These immature cells did, however, not infiltrate their lymph nodes and thymus or nonhematopoietic tissue such as liver, lung, or brain (Fig. 3). The in vitro clonogenic progenitor assays revealed that the numbers of bone marrow-derived CFC in recipients of E2A-Pbx1a-transduced bone marrow cells were not significantly altered (Fig. 6A), whereas the numbers of their spleen-derived CFC were ∼300-fold higher than those determined for the neo control at 56 days following transplantation (Fig. 6B). Of the bone marrow and the spleen CFC, 64% ± 30% and 85% ± 30%, respectively, were puromycin resistant, demonstrating the in vivo expansion of E2A-Pbx1a-transduced myeloid CFC. Although E2A-Pbx1a-transduced CFC were capable of generating all types of colonies normally present in these cultures, the majority (up to 60%) generated small macrophage colonies of 40 to 50 cells (Fig. 6C). Together, these observations suggest that E2A-Pbx1a induced the expansion of a pool of relatively mature CFC, which preferentially homed to and/or proliferated in the spleen and, although not verified, in the lumbosacral region of the spinal cord, leading to hind limb paralysis.
Southern blot analyses of DNA isolated from the bone marrow and spleens of the affected mice verified the presence of the intact E2A-Pbx1a provirus (Fig. 4A, middle panel), and high levels of the E2A-Pbx1a retrovirus-derived mRNA were detected in these tissues by Northern blot analysis (Fig. 4B, middle panel). Interestingly, Southern blot analysis of proviral integration sites indicated that the E2A-Pbx1a mice were repopulated by numerous E2A-Pbx1a-transduced clones (Fig. 7C). This finding, together with the expansion of myeloid CFC with low proliferative potential in these mice, suggests that they suffered from MPS. In support of that view, with the exception of mouse 1, this polyclonal disease that developed in E2A-Pbx1a mice could not be transplanted to secondary recipients, which succumbed to monoclonal AML with a latency of 75 ± 23 days (Fig. 7C). These secondary mice were, in contrast to the primary E2A-Pbx1a recipients, characterized by high leukocyte counts and infiltration by myeloid blasts of the bone marrow, spleen, and lymph nodes as well as liver and lungs (data not shown). The low efficiency in transplanting the polyclonal MPS, as well as the long latency and the monoclonal origin of the leukemic disease in secondary recipients, demonstrates that the development of E2A-Pbx1a-associated leukemia depends on accumulation of additional genetic events.
DISCUSSION
Several studies have suggested that cellular transformation induced by E2A-Pbx1 may result from collaborative interactions with products of the Hox gene complex. The studies described herein provided the first direct evidence demonstrating that E2A-Pbx1 collaborates with a Hox gene product (i.e., Hoxa9) and that this collaboration is sufficient to acutely transform primary bone marrow cells. Furthermore, bone marrow cells which coexpressed these two genes were able to proliferate in the absence of exogenous hematopoietic growth factors that, at least in part, can be explained by their production of IL-3 and/or G-CSF.
Cooverexpression of Hoxa9 and E2A-Pbx1a is sufficient to directly transform primitive hematopoietic cells.
Three main observations in this study strongly suggest that cooverexpression of Hoxa9 and E2A-Pbx1a is sufficient to directly transform primitive bone marrow cells. These are (i) the ability of cooverexpression of Hoxa9 and E2A-Pbx1a to block differentiation and induce cytokine-independent growth of freshly infected primitive CFC (Fig. 2), (ii) the relatively short time required for the development of acute leukemia in the primary Hoxa9 + E2A-Pbx1a mice (39 ± 2 days), and (iii) the evidence that secondary recipients which receive a fixed number of Hoxa9- and E2A-Pbx1a-transduced LRC developed leukemia within a similar time frame (or even longer) as the primary recipients repopulated with comparable numbers of Hoxa9- and E2A-Pbx1a-transduced leukemic clones. For example, primary mouse 4, which had seven detectable leukemic clones (labeled a to f in Fig. 5A), exhibited full-blown leukemia in 40 days, whereas its secondary recipients of seven LRC would develop leukemia in 45 days (Fig. 5C). This finding suggests that the seven leukemic clones active in primary recipient 4 were already transformed when they were originally transplanted into this recipient.
Since the cooverexpression of Hoxa9 and E2A-Pbx1a appears to be sufficient to transform a given subpopulation of bone marrow cells, then the frequency of that target cell population can be estimated by comparing the number of transduced cells in the transplanted bone marrow inoculum to the number of leukemic clones detected in each primary recipient. Table 1 shows that on average, 252 and 3 Hoxa9 and E2A-Pbx1a-transduced CFC and LTRC, respectively, were transplanted per primary recipient. Between one and seven leukemic clones were recovered per primary recipient, which suggests that the target for leukemic transformation is a primitive bone marrow cell which is found in a similar frequency as LTRC.
E2A-Pbx1a enhances proliferation of committed progenitors in vivo.
Recipients of E2A-Pbx1a-transduced bone marrow cells developed a polyclonal MPS characterized by large spleens and hind leg paralysis. With the exception of one mouse, this disease could not be transplanted to secondary recipients, suggesting that the target cell was not a stem cell but rather a more differentiated (and more frequent) short-lived progenitor cell. In support of this conclusion, the number of clones containing the E2A-Pbx1a provirus was much higher than the number of transduced stem cells injected per mouse (Table 1 and Fig. 7B). In addition, this MPS evolved to clonal AML in secondary recipients following a latency period which exceeded that required for disease to occur in primary mice, proving that E2A-Pbx1a alone was incapable of full leukemic transformation of stem cells.
Previous work by Kamps and Baltimore (10) using retroviral expression of E2A-Pbx1a in murine bone marrow cells demonstrated that E2A-Pbx1a causes monoclonal AML with long latencies (155 ± 45 days), indicating that E2A-Pbx1a alone was insufficient to transform hemopoietic cells and that additional somatic mutations were essential for the development of AML. However, none of the primary recipients of E2A-Pbx1a-transduced bone marrow cells described by Kamps and Baltimore developed the MPS observed in our studies (10). Several experimental differences between these two studies may explain this difference. First, different retroviral vectors were used to transduce and express the E2A-Pbx1a gene in mouse bone marrow cells (the pGD retroviral vector versus the MSCV vector used here), suggesting that different expression levels or promoter specificity could contribute to the differences observed. In support of that view, we were unable to induce the expansion of GM-CSF-responsive progenitor cells as described in their study (10). Second, the hemopoietic growth factors and conditions used to infect bone marrow cells differed significantly between the two studies.
One common feature of the disease that developed in the primary recipients of E2A-Pbx1a-transduced cells in these two studies was the high frequency of hind limb paralysis. This, combined with the unusually low number of cells in the bone marrow (three- to fourfold reduction) of the E2A-Pbx1a mice described here, suggests the presence of a unique set of adhesion molecules on the surface of E2A-Pbx1a-transduced bone marrow progenitors allowing them to home preferentially to the spleen and potentially also to nervous tissues (possibly the lumbosacral plexus).
Growth factor independence and production by Hoxa9- and E2A-Pbx1a-transduced bone marrow cells.
Hoxa9- and E2A-Pbx1a-transduced bone marrow cells had the capacity to grow in the absence of growth factors when they were analyzed both immediately following retroviral gene transfer (Fig. 2B) and after being harvested from the bone marrow or spleens of leukemic mice (Fig. 6A to C). Except for bone marrow cells engineered to express either a signal-transducing kinase such as Tel-Jak2 (33) or an activated growth factor receptor (or a growth factor such as IL-3), we believe that these studies are the first to demonstrate the capacity of transcription factors to directly induce growth factor-independent proliferation of primary bone marrow cells (i.e., immediately following gene transfer [Fig. 2B]).
Furthermore, it appears that at least three different growth factors (i.e., IL-3, G-CSF, and Steel) are being produced by the cells obtained from the spleen of Hoxa9 + E2A-Pbx1a mice. Curiously, the majority of leukemic blasts produced detectable levels of only one growth factor (e.g., IL-3 or G-CSF), suggesting that Hoxa9 and E2A-Pbx1a may regulate a master program which controls the production of several hemopoietic growth factors. Alternatively, it cannot be excluded that Hoxa9 and E2A-Pbx1a regulate the production of a growth factor that was not tested here (GM-CSF, IL-6, etc.). Furthermore, the variations in the production of these factors may also reflect the heterogeneity in the population of hemopoietic progenitors that were transformed even if the cells were phenotypically undifferentiated by several criteria (morphology, FACS, etc.).
Together, the studies presented herein prove the existence of a collaboration in leukemic transformation between the fusion oncoprotein E2A-Pbx1a and one of its presumed DNA binding partners, Hoxa9. Future goals will include the determination of Hox gene expression patterns in E2A-Pbx1-expressing human leukemia samples and determining whether other Hox gene products can also collaborate with the E2A-Pbx1 oncoprotein to transform primary hemopoietic cells. Also, it will be important to determine whether this oncogenic collaboration involves cooperative DNA binding between these two proteins.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Cancer Institute of Canada. Unnur Thorsteinsdottir is the recipient of a Leukemia Research Fund of Canada fellowship, and Guy Sauvageau is an MRC Clinician-Scientist Scholar.
We acknowledge Corey Largman and Jeffrey Lawrence for their generous gift of anti-serum to Hoxa9. In addition, we acknowledge Mireille Mathieu and François Letendre, l’Hôpital Hôtel-Dieu de Montréal, for their assistance with peripheral blood counts and histochemistry, and we thank Christiane Lafleur and Stephane Matte for their expertise and help regarding the maintenance and manipulation of the animals kept at the specific-pathogen-free facility of the IRCM. The support of Nathalie Tessier is also acknowledged for FACS analyses.
REFERENCES
- 1.Azpiazu N, Morata G. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev. 1998;12:261–273. doi: 10.1101/gad.12.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrow J, Shearman A M, Stanton V P, Becher R, Collins T, Williams A J, Dubé I D, Katz F, Kwong Y L, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman D E. The t(7;11)(p15;p15) translocation in acute myeloid leukemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 3.Carroll A J, Crist W M, Parmley R T, Roper M, Cooper M D, Finley W M. Pre-B cell leukemia associated with chromosome translocation 1;19. Blood. 1984;63:721–724. [PubMed] [Google Scholar]
- 4.Chan S K, Jaffe L, Capovilla M, Botas J, Mann R S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 5.Chang C P, de Vivo I, Cleary M L. The Hox cooperativity motif of the chimeric oncoprotein E2a-Pbx1 is necessary and sufficient for oncogenesis. Mol Cell Biol. 1997;17:81–88. doi: 10.1128/mcb.17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C P, Shen W F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 7.Davis L, Kuehl M, Battey J. Preparation and analysis of RNA from eukaryotic cells. Norwalk, Conn: Appleton and Lange; 1994. [Google Scholar]
- 8.Dedera D A, Waller E K, LeBrun D P, Sen-Majumdar A, Stevens M E, Barsh G S, Cleary M L. Chimeric homeobox gene E2A-PBX1 induces proliferation, apoptosis, and malignant lymphomas in transgenic mice. Cell. 1993;74:833–843. doi: 10.1016/0092-8674(93)90463-z. . (Erratum, 75:826.) [DOI] [PubMed] [Google Scholar]
- 9.Humphries R K, Eaves A C, Eaves C J. Self-renewal of hemopoietic stem cells during mixed colony formation in vitro. Proc Natl Acad Sci USA. 1981;76:3629–3633. doi: 10.1073/pnas.78.6.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamps M P, Baltimore D. E2A-Pbx1, the t(1;19) translocation protein of human pre-B-cell acute lymphocytic leukemia, causes acute myeloid leukemia in mice. Mol Cell Biol. 1993;13:351–357. doi: 10.1128/mcb.13.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamps M P, Murre C, Sun X H, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in Pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 12.Kamps M P, Wright D D, Lu Q. DNA-binding by oncoprotein E2A-Pbx1 is important for blocking differentiation but dispensable for fibroblast transformation. Oncogene. 1996;12:19–30. [PubMed] [Google Scholar]
- 13.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg A M, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1 but not PBX1. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krosl J, Baban S, Krosl G, Rozenfeld S, Largman C, Sauvageau G. Proliferation and transformation by HOXB4 and HOXB3 genes involves cooperation with PBX. Oncogene. 1998;16:3403–3412. doi: 10.1038/sj.onc.1201883. [DOI] [PubMed] [Google Scholar]
- 15.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence H J, Sauvageau G, Ahmadi A, Lau T, Lopez A R, Le Beau M M, Link M, Largman C. Stage and lineage-specific expression of the HOXA10 homeobox gene in normal and leukemic hematopoietic cells. Exp Hematol. 1995;23:1160–1166. [PubMed] [Google Scholar]
- 17.Lawrence H J, Sauvageau G, Humphries R K, Largman C. The role of Hox Homeobox genes in normal and leukemic hematopoiesis. Stem Cells. 1996;14:281–290. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- 18.Lu Q, Knoepfler P S, Scheele J, Wright D D, Kamps M P. Both Pbx1 and E2A-Pbx1 bind the DNA motif ATCAATCAA cooperatively with the products of multiple murine Hox genes, some of which are themselves oncogenes. Mol Cell Biol. 1995;15:3786–3795. doi: 10.1128/mcb.15.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maconochie M K, Nonchev S, Studr M, Chan S K, Poepperl H, Sham M H, Mann R S, Krumlauf R. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhobdomere 4 is regulated by Hoxb1. Genes Dev. 1997;11:1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- 20.Monica K, LeBrun D P, Dedera D A, Brown R, Cleary M L. Transformation properties of the E2a-Pbx1 chimeric oncoprotein: fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Mol Cell Biol. 1994;14:8304–8314. doi: 10.1128/mcb.14.12.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T, Largaespada D A, Lee M P, Johnson L A, Ohyashiki K, Toyama K, Chen S J, Willman C L, Chen I M, Feinberg A P, Jenkins N A, Copeland N G, Shaughnessy J D. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15; p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura T, Largaespada D A, Shaughnessy J D, Jenkins N A, Copeland N G. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 23.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M C. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–546. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 24.Peltenburg L T, Murre C. Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J. 1996;15:3385–3393. [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins A C, Cory S. Conditional immortalization of mouse myelomonocytic, megakaryocytic and mast cell progenitors by the Hox-2.4 homeobox gene. EMBO J. 1993;12:3835–3846. doi: 10.1002/j.1460-2075.1993.tb06062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelan M L, Featherstone M S. Distinct HOX N-terminal arm residues are responsible for specificity of DNA recognition by HOX monomers and HOX-PBX heterodimers. J Biol Chem. 1997;272:8635–8643. doi: 10.1074/jbc.272.13.8635. [DOI] [PubMed] [Google Scholar]
- 27.Phelan M L, Rambaldi I, Featherstone M S. Cooperative interaction between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piper D E, Batchelor A H, Chang C P, Cleary M L, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 29.Pöpperl H, Bienz M, Studer M, Chan S K, Aparicio S, Brenner S, Mann R S, Krumlauf R. Segmental expansion of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 30.Rocco G D, Mavilio F, Zappavigna V. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 1997;16:3644–3654. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauvageau G, Thorsteinsdottir U, Eaves C J, Lawrence H J, Largman C, Lansdrop P M, Humphries R K. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 32.Sauvageau G, Thorsteinsdottir U, Hough M R, Hugo P, Lawrence H J, Largman C, Humphries R K. Over-expression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997;6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 33.Schwaller J, Frantsve J, Aster J, Williams I R, Tomasson M H, Ross T S, Peeters P, Van Rompaey L, Van Etten R A, Ilaria R, Jr, Marynen P, Gilliland D G. Transformation of hematopoietic cell lines to growth-factor independence and induction of fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion genes. EMBO J. 1998;17:5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen C P, Chang, Rozenfeld S, Sauvageau G, Humphries R K, Lu M, Lawrence H J, Cleary M J, Largman C. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen W F, Montgomery J C, Rozenfeld S, Moskow J J, Lawrence H J, Buchberg A M, Largman C. AbdB-like proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen W F, Rozenfeld S, Lawrence H J, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J Biol Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- 37.Szilvassy S J, Humphries R K, Lansdorp P M, Eaves A C, Eaves C J. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulations strategy. Proc Natl Acad Sci USA. 1990;87:8736–8740. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targetted in mouse embryo-derived stem cell. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 39.Thorsteinsdottir U, Sauvageau G, Hough M R, Lawrence H J, Largman C, Humphries R K. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorsteinsdottir, U., G. Sauvageau, and K. Humphries. Enhanced polyclonal regeneration of hematopoietic stem cells overexpressing HOXB4 following bone marrow transplantation. Blood, in press.