Abstract
Lymphogranuloma venereum (LGV), the invasive infection of the sexually transmissible infection (STI) Chlamydia trachomatis, is caused by strains from the LGV biovar, most commonly represented by ompA-genotypes L2b and L2. We investigated the diversity in LGV samples across an international collection over seven years using typing and genome sequencing. LGV-positive samples (n=321) from eight countries collected between 2011 and 2017 (Spain n=97, Netherlands n=67, Switzerland n=64, Australia n=53, Sweden n=37, Hungary n=31, Czechia n=30, Slovenia n=10) were genotyped for pmpH and ompA variants. All were found to contain the 9 bp insertion in the pmpH gene, previously associated with ompA-genotype L2b. However, analysis of the ompA gene shows ompA-genotype L2b (n=83), ompA-genotype L2 (n=180) and several variants of these (n=52; 12 variant types), as well as other/mixed ompA-genotypes (n=6). To elucidate the genomic diversity, whole genome sequencing (WGS) was performed from selected samples using SureSelect target enrichment, resulting in 42 genomes, covering a diversity of ompA-genotypes and representing most of the countries sampled. A phylogeny of these data clearly shows that these ompA-genotypes derive from an ompA-genotype L2b ancestor, carrying up to eight SNPs per isolate. SNPs within ompA are overrepresented among genomic changes in these samples, each of which results in an amino acid change in the variable domains of OmpA (major outer membrane protein, MOMP). A reversion to ompA-genotype L2 with the L2b genomic backbone is commonly seen. The wide diversity of ompA-genotypes found in these recent LGV samples indicates that this gene is under immunological selection. Our results suggest that the ompA-genotype L2b genomic backbone is the dominant strain circulating and evolving particularly in men who have sex with men (MSM) populations.
Keywords: whole genome sequencing, selective pressure, outer membrane protein, sexually transmitted infections, homosexuality, surveillance, molecular epidemiology, evolution, LGV
Data Summary
All genome data have been submitted to the European Nucleotide Archive (ENA) under project number PRJEB19884 (https://www.ncbi.nlm.nih.gov/bioproject/PRJEB19884). Data for ompA variants have been submitted under project PRJEB37762 with the accession numbers given in Table S1 (available in the online version of this article).
Impact Statement.
Lymphogranoma venereum (LGV) is a destructive and serious sexually transmitted infection (STI) caused by a more aggressive and invasive variant of chlamydia. It affects mainly men who have sex with men. Over the last 20 years, one LGV variant has dominated the diagnosed cases reported globally. We have investigated the recent evolution of this variant, and find that it seems to be continuing to adapt to its hosts and environment. This is particularly apparent in the ompA gene, commonly used to categorize or type strains of chlamydia. Many mutations in ompA, including novel ones which have not been seen before, have now been identified. In particular, a single mutation in ompA has resulted in ompA-genotype L2 in this clade, which generally refers to a strain with a different genomic structure. The variant that we have studied may now be the dominant circulating clone, which presents a challenge for typing. The changes in the ompA gene may aid the bacteria to induce reinfections, by avoiding host immunity. Further surveillance of this disease is warranted.
Introduction
Chlamydia trachomatis is the most common agent of bacterial sexually transmissible infection (STI) globally, with an estimated 127 million new cases of chlamydia infection in 2016 worldwide [1]. Lymphogranuloma venereum (LGV) is an invasive disease causing ulcerative anogenital infection, extending often to regional lymph nodes, and is commonly associated with men who have sex with men (MSM) in many high-income countries. A diagnosis of LGV chlamydia is of clinical importance, as the infection requires prolonged antimicrobial treatment and, if not detected and treated, may be associated with serious clinical complications [2, 3].
OmpA-serotyping was the traditional method for typing C. trachomatis strains, now superseded by ompA-genotyping. The ompA-genotypes A–C are associated with ocular infections (trachoma), D–K with urogenital infections, and L1–L3 with LGV. OmpA is the major outer membrane protein (also known as MOMP), a porin and highly immunogenic surface-exposed antigenic determinant [4, 5]. Within OmpA, four variable domains (VDs) have been described, which are proposed to be surface-exposed and therefore subject to immune selection [4, 6]. No crystal structure of chlamydial OmpA exists as yet.
Among MSM infected with LGV C. trachomatis, L2 was the dominant circulating OmpA-serotype until the early 2000s [7], when ompA-genotype L2b was found to be responsible for an outbreak in the Netherlands associated with severe proctitis [8], which spread globally [9]. Retrospectively, it was found that ompA-genotype L2b isolates were circulating in the USA as early as 1979–1985 [8, 10]. The difference in the ompA gene between L2 and L2b is an SNP causing the N162S amino acid substitution within VD2. On the whole genome level, the reference strains ompA-genotypes L2/434/Bu and L2b/UCH-1 are differentiated by 573 SNPs across their 1.04 Mb genome [11], and have almost identical gene content. The highest variation between the genomes is found within tarp, encoding a translocated actin recruiting phosphoprotein, and pmpH, encoding an immunogenic auto-transporting polymorphic membrane protein, with possible adhesin function. Isolates within ompA-genotype L2b differ from each other by up to 20 SNPs [12], reflecting a relatively recent common ancestor.
Laboratory diagnosis of LGV as opposed to urogenital strains relies on in-house molecular tests [13–16] or can be based on epidemiological and clinical findings [3]. While differences in LGV ompA-genotypes do not affect treatment recommendations, they are of epidemiological importance in continuing to survey the dynamics and evolution of LGV in MSM populations. However, in most laboratories, LGV typing is still not performed, despite IUSTI guidelines [17].
In order to diagnostically distinguish ompA-genotype L2b from other LGVs, a real-time PCR (RT-PCR) was previously developed, based on a characteristic 9 bp insertion found in the pmpH gene of ompA-genotype L2b isolates relative to ompA-genotype L2 [18], referred to here as pmpH-genotype L2b. Recent work has shown that the pmpH-genotype L2b is no longer specifically associated with ompA-genotype L2b, since it is also present in strains carrying ompA-genotype L2 [19]. Several further publications indicate that ompA-genotype L2 is making a resurgence [19–24] and that additional diversity in LGV ompA-genotypes is present in circulating strains (Table S1) [10, 20, 22–27]. It is important to note that strains and their genomes are currently categorized by their ompA-genotype, despite increasing data suggesting that ompA-genotype does not reflect the rest of the genomic backbone [28–30]. Throughout this paper, we try to distinguish between ompA-genotype (defined by ompA sequence), pmpH-genotype (defined by the 9 bp insertion in L2b strains) and genomic backbone (to date generally associated with the ompA-genotype, for example in the first published reference strains).
Recombination is a driver of C. trachomatis evolution [9, 12, 31]; clinical isolates with recombinations between LGV and urogenital strains have been described [28, 30, 32], including a recent L2b-D/Da hybrid strain identified in Portugal, which resulted from a recombination across ompA in an L2b genomic background [30]. Therefore, a plausible hypothesis for this observation of these pmpH–ompA discrepancies is that recombination between the genomes of ompA-genotype L2b and ompA-genotype L2 has occurred. To investigate the pmpH–ompA discrepancy phenomenon and to determine the current genome dynamics of LGV strains at high resolution, we used ompA- and pmpH-genotyping along with whole genome sequencing (WGS) of clinical samples sampled between 2011 and 2017 across Europe and Australia.
Methods
Sample collection
Samples yielding positive amplification for LGV targets, in particular those of pmpH-genotype L2b, were assembled by our consortium, providing a collection as comprehensive and international as possible (Table S2). Our dataset was selected to explore the particular phenomenon of pmpH-genotype L2b linked to other ompA-phenotypes and, as such, is not a random survey of circulating strains. Samples were named after the country of origin (AU, Australia; CZ, Czechia; CH, Switzerland; ES, Spain; FR, France; HU, Hungary; NL, Netherlands; SL, Slovenia; SW, Sweden), sample number and ompA-genotype. Samples positive for pmpH-genotype L2b and ompA-genotype L2 were labelled ‘L2new’. Detailed diagnostic methods in the various source countries are provided in the supplementary data.
pmpH-genotyping was performed as previously described [18]. pmpH-genotype L2b was defined as possessing the 9 bp insertion TCTAGTAGT at position 1884. The ompA gene was amplified using the primer pair P1 : 5′-ATGAAAAAACTCTTGAAATCGG-3′ and OMP2 : 5′-ACTGTAACTGCGTATTTGTCTG-3′ [33], and capillary-sequenced using primer OMP2. Traces were compiled and manually inspected in CLC Genomics Workbench (version 9.5.3, Qiagen).
Whole genome sequencing
SureSelect target capture was used as previously described [12] on a selection of 95 isolates including selected samples from a previous study on LGV diversity [19]. The SureSelect baits were custom designed around known variable regions from all sequenced ompA-genotypes [9]. Sequence capture (48-plex) was performed according to the manufacturer’s protocols (Agilent). After library preparation [34], multiplexed (96-plex) samples were sequenced using Illumina Hiseq V4 (Illumina) with 75 bp paired-end reads.
Sequence analysis
Resulting data were mapped using bwa [35] against reference genomes L2/434/Bu (EMBL accession AM884176) and L2b/UCH-1 (EMBL accession AM884177) using a minimum read identity threshold of 0.9 (minimum 90 % mapping identity of the read to a reference) to avoid mapping of contaminating reads. Bcftools was used for variant calling, with the following options: minimum number of reads matching SNP=8, minimum number of reads matching SNP per strand=3, SNP/mapping quality ratio cutoff=0.8, minimum base call quality to call a SNP=50, minimum mapping quality to call a SNP=20. After extracting mapping parameters and finding that L2b/UCH-1 was the better reference for this study, this was the reference used in all subsequent analyses. Samples meeting coverage criteria (over 15× mean coverage) are detailed in Table S3: 53 provided insufficient coverage, probably due to sample quality and chlamydial load. All called SNPs were checked manually using BAM files visualized in Artemis [36]. The SNP phylogeny was generated using RAxML v8.2.8 AVX and GTRGAMMA model with 100 bootstraps. Read data were similarly mapped against the plasmid sequence from L2b/UCH-2 (EMBL accession NC_020956). Indel investigation was beyond the scope of this phylogenetic study. Alignments of the ompA gene sequences and amino acid sequences were performed within Seaview [37] using Clustal Omega with default settings. The ompA phylogeny was generated in Seaview from a muscle alignment of ompA gene sequences, using PhyML and the GTR model and default settings with 100 bootstraps. Sequences of the pmpH gene were confirmed by extracting the gene sequence from Unicycler v0.4.8 [38] assemblies using Artemis [36] and aligning against the reference L2/434/Bu and L2b/UCH-1 pmpH gene sequences within Seaview using Clustal Omega with default settings.
Recombination and time tree analysis
Read data from the wider LGV clade were analysed as above. Gubbins v2.4.1 [39] was run on 69 genomes from the L2b clade, providing 63 genomes with sufficient data. Linear root-to-tip analysis was performed in TempEst v1.5.1 [40] using the calculated root based on outgroups. BactDating v1.0.11 [41] was run in R Studio v1.2.5033 [42] with R v3.6.2 [43] on the resulting dataset. All models were run, each with 1 million iterations, and compared: arc (additive relaxed clock) was found to be the best model [44].
Antigenicity prediction
The Immune Epitope Database (IEDB.org), B cell epitope prediction using either Kolaskar and Tongaonkar Antigenicity and Bepipred Linear Epitope Prediction) was used to model antigenicity of omopA epitopes. Results are given as an antigenicity score, with higher values indicating higher antigenicity (http://tools.iedb.org/bcell/help/).
Results
Typing shows that pmpH-genotype L2b is associated with several ompA genotypes
A total of 389 LGV-positive samples from eight countries from 2011 to 2017 (Spain n=97, Netherlands n=67, Switzerland n=64, Australia n=53, Sweden n=37, Hungary n=31, Czechia n=30, Slovenia n=10) were obtained. Of these, 321 (83 %) gave positive and interpretable results for pmpH-genotyping and ompA-sequencing (Table S2: Spain n=86, Netherlands n=63, Switzerland n=50, Australia n=38, Sweden n=34, Hungary n=28, Czechia n=20, Slovenia n=2). All the samples were obtained from men and, where sexual orientation was provided (Netherlands, Hungary and Slovenia), all were from MSM. The mean age of the patients, where data were available (n=313), was 37.9 years (range 19–72 years).
All the LGV samples included in this study were found to be pmpH-genotype L2b (n=321). However, ompA-sequencing revealed ompA-genotype L2b (n=83; 26 %) and ompA-genotype L2 (n=180; 56 %) as well as sequences which vary from these by up to 2 bp: L2b (n=45; nine variant types) and L2 (n=7; three variant types) (Table S1). We also identified ompA-genotype L1 variants (n=4), a D variant (n=1) and a single mixed L2/L2b infection (n=1). As our focus is on ompA-genotypes L2/L2b, these latter samples were not analysed further, except to note that the Swiss D variant ompA sequence matches that of the L2b-D/Da hybrid (MN094864) [30] across base pairs 193–1021. The diversity of the ompA-variants sampled is shown in Fig. 1. The phylogeny cannot be fully resolved due to homoplasy (the same mutation in two separate branches of the phylogeny), and one base reversion at position G485A causing the S162N amino acid change. Notably, all the nucleotide substitutions (n=13; as well as three further SNPs from previous studies) lead to predicted amino acid changes in OmpA (Table S1).
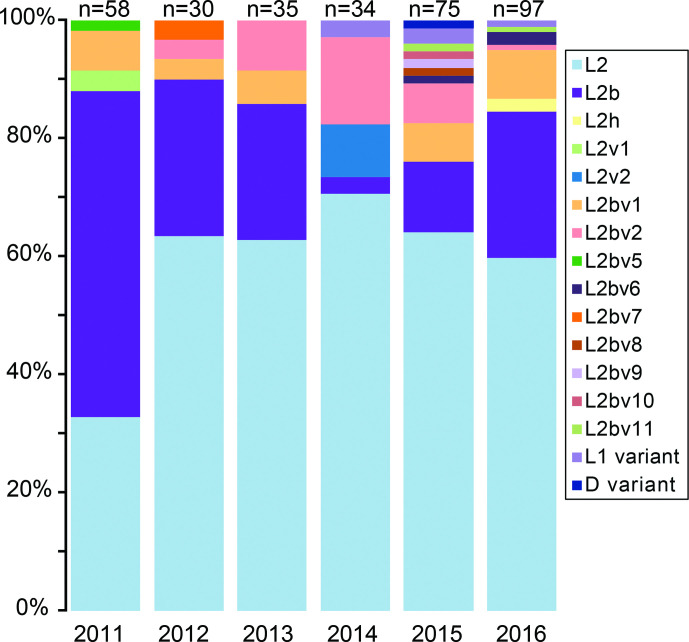
Fig. 1.
Diversity of ompA by sequence clustering. ompA sequences of the cohort tested were compared with sequences from NCBI (accession numbers given). A PhyML tree was created from a muscle alignment of ompA gene sequences in Seaview using the GTR model and default settings, and using ompA-genotype L2 as the root. Full-length ompA sequences were extracted from whole genome sequences and used to complement the PCR data where possible (see further below). The numbers of samples in our study carrying these variants are shown on the right. Distributions are given by country, that with the most samples of this ompA-genotype given first: AU, Australia; CZ, Czechia; CH, Switzerland; ES, Spain; HU, Hungary; NL, Netherlands; SW, Sweden. SNP numbers on branches refer to bases in the reference gene from AM884176, L2/434/Bu; all are non-synonymous. Homoplasic SNPs (those found twice in the tree) are coloured green and blue (bold) respectively. The SNP distinguishing ompA-genotype L2 from L2b is shown in red (bold), as is the ‘revertant’ SNP in sample E91_L2h (according to this phylogeny). L2f_EU676181_128C/07 is identical to the L2/434/Bu reference. Not all sequences are full length: in particular, L2c_EF460796, L2d_EF460797 and L2bv4_KU518892_CV544 do not cover all the assigned SNPs. L2e_EF460798 was excluded from the analysis as it comprises only 150 bp and covers a part of the ompA gene which does not align with many other sequences: it carries a single SNP, C954T. Bootstrap values are low (0–61 %), reflecting the low sequence diversity and presence of homoplasies.
Large ompA diversity in our sample collection
Typing data from our whole collection show a decrease in ompA-genotype L2b samples while rates of ompA-genotype L2 increased from 2010 to 2015 (Fig. 2). The trends vary per country, with data from parts of Europe showing strains carrying ompA-genotype L2 increasing from 2011 (France, Switzerland), being dominant in several countries since 2011 (Spain, Sweden, Hungary), or maintaining a stable proportion (Netherlands) (Fig. S1). Of the further ompA-genotypes (assigning new genotypes with every SNP difference), many (n=10) are specific to single countries (Fig. 1).
Fig. 2.
Distribution of ompA-genotypes temporally from our combined cohort. The SNPs within these ompA-genotypes are shown in Table S1. The data are broken down by country in Fig. S1.
WGS analysis of pmpH-genotype L2b and ompA variants
A selection of 95 samples, chosen from those with lower Ct values (Table S2) to represent the widest geographical, temporal and ompA-genotype diversity, underwent sequence capture and WGS. Genome data for further analysis were obtained from 42 samples (44 %), representing at least one genome from each country, with the exception of Hungary. The phenomenon of ompA-genotype L2 and pmpH-genotype L2b (‘L2new’) is represented by 26 genomes, and ompA-genotype L2b by three. Of the 12 L2/L2b ompA-variants identified above, eight are represented by at least one sample with a complete genome sequence. The ompA-genotypes without genome data are: L2bv6, L2bv8, L2bv10 and L2bv11. All sequenced genomes show much higher identity to the genome of L2b/UCH1 (differing by one to eight SNPs) than that of L2/434/Bu (differing by ≥245 SNPs). The presence of the 9 bp insertion in the pmpH gene was confirmed in 37 of the 42 samples: the assemblies of five samples (ES89_L2bv7, ES96_L2v3, FR1378_L2new, NLB19_L2new and AU39_L2Bv1) were too fragmented over the pmpH locus to analyse, owing to low sequence coverage in these samples.
Each genome shows one to eight SNP differences compared to the genome of L2b/UCH1 from 2006, and all derive from this L2b genomic backbone (Fig. 3). The phylogeny describes the ‘L2new’ samples in this study, with ompA-genotype L2 and pmpH-genotype L2b, having arisen through an initial SNP reversion at genome position 59 342 (G485A in ompA; OmpA S162N), and then undergoing further mutation. Thus, the full genome phylogeny (Fig. 3) does not agree with the ompA gene phylogeny (Fig. 1), as the ‘L2new’ genomes are a further development from the L2b genomic backbone, and do not represent the genome of the reference L2/434/Bu.
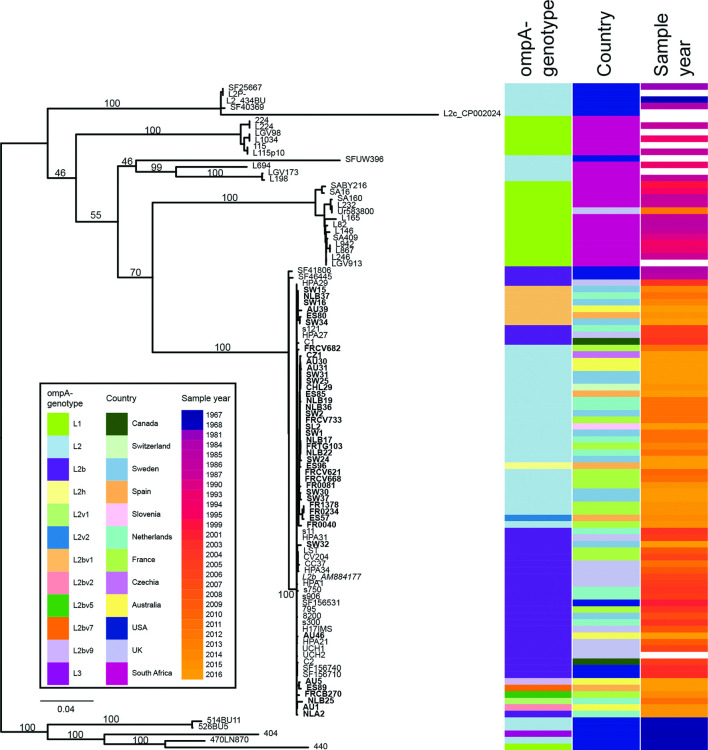
Fig. 3.
Phylogeny of whole genome sequences of 42 sequenced LGV isolates. The reference strain L2b/UCH1_AM884177 was used as the root of the tree, based on additional analysis using outgroups SF41806 and SF46445. Columns to the right of the phylogeny indicate ompA-genotype, country and year of sampling for each sample. The evolution of ompA-genotype L2 from the ompA-genotype L2b backbone is clearly shown, with the star representing the SNP causing this ompA-genotype change. All samples were pmpH-genotype L2b. The same phylogeny with SNP locations on branches is given in Fig. S3. The figure was generated using Phandango [70].
As both recombination and mutation are potential sources of genome variation, we aimed to investigate which mechanism was responsible for these SNPs. The distribution of SNPs across the genome is shown in Fig. S2. With the exception of the ompA gene with nine SNPs, there are five loci with two SNPs within 1 kb, and the rest are spread along the genome. Recombination analysis on a dataset including these and other available genomes from the L2b clade [12] resulted in a collection of 63 LGV genomes, between which no recombinations were identified (data not shown). To investigate whether the SNPs identified above have been seen before in LGV genomes, we analysed in detail these 59 genomes, which altogether possess 1894 SNPs relative to L2b/UCH1. Of the 66 SNPs identified in this study, five have been seen (separately) in previously sequenced LGV strains: four within ompA and one causing a Met-Val change at codon 12 within CTLon_0845/rbsU, encoding a sigma regulatory family protein-PP2C phosphatase, which was seen previously in two UK strains (Table S4). In all cases, no SNPs which fall adjacent to these were identified in the genomes in this study, as would be expected to co-transfer during recombination: this is also the case at the ompA locus, with the closest SNP found over 13 kb away, suggesting that the ompA SNPs in these genomes did not occur as part of a larger recombination. The rate of SNP accumulation, with one to eight SNPs over 10 years (2006–2016), broadly concurs with the mutation rate calculated for the LGV lineage of 2.15×10−7 SNPs per site per year (0.23 SNPs per genome per year) [12]. As such, our data suggest that these SNPs arose by mutation.
A comparison of these data with previously published genome sequences from the L2b clade [12] shows the context of this ongoing evolution (Fig. 4). This phylogenetic tree also highlights the wide presence of ompA-genotype L2 across the phylogeny, showing that this feature is not monophyletic (Fig. 4).
Fig. 4.
Phylogeny of whole genome sequences available for LGV-clade samples. The phylogeny is based on reference strain L2b/UCH1_AM884177 (italicized), including 59 additional LGV genomes for context [12]. Sequences from this study are in bold. Columns to the right of the phylogeny indicate ompA-genotype, country and year of sampling for each sample [12]. The tree is rooted according to the full species phylogeny [12]. This phylogeny agrees largely with that in Fig. 3, with the exception of the location of NLB25, differently located due to the homoplasy at position 59 312 (Fig. S3). Bootstraps of 100 replicates are shown on key branches; the bootstrap to the branch with the ompA L2 SNP is 46. Bar, number of substitutions per site.
The isolates at the root of the L2b clade are from the USA in the 1980s, and the earliest isolate from the branch representing the L2b outbreak is from the USA in 2001 (SF156531), which is genomically identical to the reference ompA-type L2b isolate L2b/UCH1, among others. We attempted to date this clade, including the USA 1980s isolates onwards. A first root-to-tip regression analysis shows that the genomes have a temporal signal, but the mutation rate of 0.44 SNPs per genome per year does not agree with previously published data (Fig. S4). Bayesian analysis of the data using BactDating gives a mutation rate of 0.213 SNPs per genome per year (95 % CI: 0.147–0.283, rate equivalent to 2.1×10−7 SNPs per site per year) and a putative date for the ancestor of ompA-genotype L2b clade of 1935 CE (common era) (95 % CI: 1903–1959) (Fig. S4).
The genomes, particularly ompA, are under selective pressure
Across the phylogeny of the 42 genomes from this study, a total of 66 SNPs were identified (Fig. 3): six (9 %) are intergenic, 15 [23 %; 25 % of SNPs found within coding sequences (CDSs)] are synonymous, and 45 (68 %; 75 % of SNPs found within CDSs) are non-synonymous, one of which causes a premature stop codon in incA (CTLon_0370) (Table S4).
Nine of the 66 SNPs (14 %) are located within a single gene, ompA, all of which are non-synonymous (Table S4). This gene comprises 1185 bp of the 1 038 863 bp genome (0.1 %), showing that SNPs are clearly enriched in this gene. Of the nine SNPs in the ompA gene, six are novel compared to sequenced LGV genomes [12] (Table S4). The three that are seen elsewhere in the phylogeny also often exist in combination with other ompA SNPs, and in some cases these combinations have also been seen in previously sequenced genomes. Two of the ompA SNPs (58 830 and 59 312, causing amino acid changes S333G and K172T) are homoplasic, found in two locations in our phylogeny. Together, these findings suggest either independent mutations in separate lineages, or possible recombination, although no adjacent SNPs were identified, which would be a signature of recombination (see above).
The 48 CDSs affected by mutations identified across the phylogeny include 33 with at least one non-synonymous mutation. A comparison of these CDSs with the 34 identified as having a possible role in pathoadaptation [45] find four in common: CTLon_0243/CTL0247 (ChlaDub1), CTLon_0247/CTL0251 (pmpH), CTLon0370/CTL0374 (incA) and CTLon0845/CTL0851 (rbsU) (Table S4). Note that tarp (translocated actin recruiting phosphoprotein) gene repeats were not considered in this analysis as the mapping and assembly of this region is known to be problematic.
None of these SNPs are located in any of the multilocus sequence typing (MLST)/multilocus variable-number tandem-repeat analysis (MLVA) genes from any of the schemes [46–49], and as such all these 42 samples are from ST44 (Pannekoek scheme [48]), ST1 (Dean scheme [49]) and ST58 (Uppsala scheme [46, 47]). No SNPs were identified within the plasmid.
Antigenic impact of OmpA substitutions
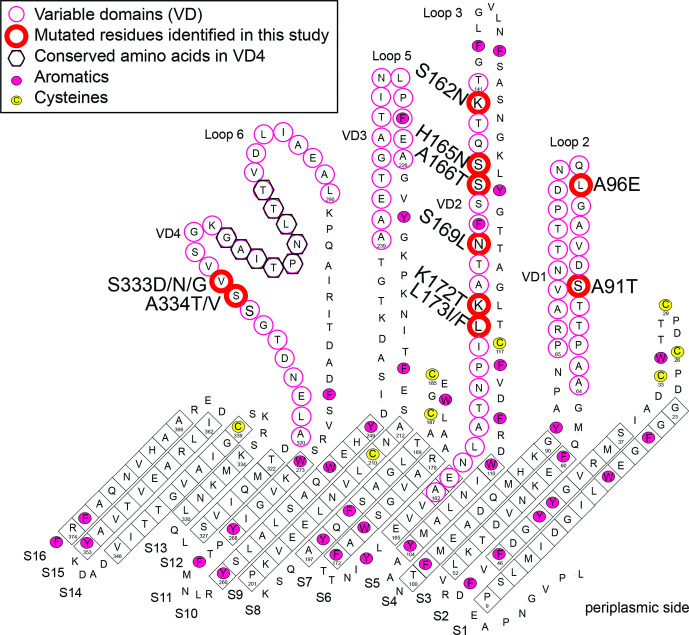
All the 13 SNPs identified in the samples in our study (WGS and genotyping) within ompA cause amino acid changes, each of which is located on the external face in the variable domains (VD1, VD2 and VD4), as previously modelled [6] (Fig. 5). The S162N substitution changes the ompA-genotype from L2b to L2, and vice versa. The majority of substitutions are from polar amino acids to polar or hydrophobic residues, in agreement with previous findings [50, 51]. With the K172T substitution, a charged amino acid is replaced by a polar one.
Fig. 5.
Location of altered amino acids within OmpA. This is from the model developed in [6], based on the C. trachomatis serovar C ompA sequence accession number DQ116399 and associated amino acid numbering. Amino acid substitutions differing from those in ompA-genotype L2b are given next to the position of the amino acid according to L2b/UCH-1 numbering: all are located in variable domains VD1, VD2 and VD4.
The substitutions in OmpA are all located in regions found to be antigenic in C. trachomatis ompA-genotype L2 in rabbits [51]. These regions contain B cell epitopes as defined in the trachoma biovar of C. trachomatis [50, 52]. Modelling of OmpA epitope antigenicity indicates that the L2b version of OmpA with the serine at position 162 has marginally higher antigenicity (scores of 0.939 and 0.589 respectively with B cell epitope prediction using either Kolaskar and Tongaonkar Antigenicity or Bepipred Linear Epitope Predictions) than L2 with the glutamine at position 162 (0.906 and 0.529) (Tables S5 and S6).
Discussion
Investigations into circulating C. trachomatis genotypes are very useful in an epidemiological context. In this study, we show that the evolution of the LGV outbreak L2b-lineage between 2006 and 2016 has resulted particularly in diversity in the immunogenic OmpA protein. Our main finding is the reversion of this L2b genomic backbone to ompA-genotype L2, an event which appears to have occurred recently, between 2006 and 2011, at least once. This confounds traditional typing, as the L2b genomic backbone is now linked to the L2 ompA-genotype. The original hypothesis of recombination between these lineages appears to be unlikely, based on the phylogeny, low SNP numbers, lack of identified recombinations within this clade and absence of co-transferred adjacent SNPs. A more likely scenario is that the ompA gene is under strong selective pressure on specific immunogenic residues, such that chance mutations in these residues are selected for in the circulating strains. It has previously been shown that ompA is under positive selection [53, 54]; this has been hypothesized to be a way to promote immune evasion and enable repeat infections [9]. Among LGV strains, there is high variability of ompA [45], partially as a result of high levels of recombination. A probable recombination event between ompA-genotype D/Da and the L2b genomic backbone has been recently reported as a seemingly successful combination [30]: from ompA evidence, this variant may also be present in Switzerland. This high ompA variability is reflected in our study, where seven novel ompA-genotypes have been identified. It is a key observation that all the 13 mutations in ompA identified in this study cause amino acid changes in the variable domains of OmpA, and thus may affect the epitopes targeted by the hosts’ immune system. Many of the ompA-genotypes are specific to single countries, whereas others are found throughout our international collection.
The ompA-genotype L2b strain was responsible for an outbreak among MSM starting in the early 2000s, thought to have arisen in the USA [2] and rapidly spread globally. We have dated the ancestor of the L2b clade to 1935, in agreement with Hadfield et al. [12], although it is important to bear in mind that genomic data from outbreaks may confound the time tree algorithm [55]. The clonality of the data can also mean that phylogenies are difficult to accurately calculate. We found this L2b genomic backbone present across the many countries we sampled, despite changes in the ompA-genotype, which argues that this is a successful and expanding lineage, diversifying over time. It is possible that this success is due to increased virulence encoded by the ompA-genotype L2b genome compared to other LGV lineages, although there are few functional differences predicted between the genomes of ompA-genotype L2/434/Bu and ompA-genotype L2b/UCH-1 [11]. Another possibility is that this represents a clonal expansion of an epidemiologically present strain among high-risk groups with international sexual networks, fuelled by changes in sexual practice over the past decades. The observed combination of the L2b genomic backbone with the L2 ompA appears to be expanding successfully, perhaps selected to improve infection chances in certain populations with some pre-existing immunity to ompA-genotype L2b.
The L2 variant of OmpA is predicted to be slightly less antigenic than that of L2b, which may be a selective advantage to a naïve host, or perhaps this variant succeeds as it favours reinfection. This may explain the wide presence of this ompA-genotype across the LGV phylogeny. Exploration of this, and whether the immunological impact of human immunodeficiency virus (HIV) co-infection could affect immune recognition and selective pressure would be a fascinating area for further study. Future studies should investigate the fitness, virulence, clinical outcomes and re-infection rates in patients infected with LGV.
Our study found that 75 % of the SNPs in protein coding regions across the whole genome phylogeny are non-synonymous. While this is similar to the expected maximum of 23 % synonymous mutations based on C. trachomatis codon usage [45], it suggests that the mutated sites in these isolates are under positive selection, supporting previous observations in the L2b clade of 90.2 % of the SNPs in CDSs being non-synonymous [45]. Four of the 33 CDSs identified as having non-synonymous mutations in our study were also identified as having an overrepresentation of non-synonymous mutations in a previous study on LGV [45], suggesting that there is indeed a pressure on specific targets in this lineage. Three of these genes are probably involved in interaction with or evasion of the immune system: ChlaDub1 [56], pmpH [57] and incA [58]. Whether this potential patho-adaptation is responsible for increased virulence or increased numbers of asymptomatic cases [59] is not clear, and would be useful to test in future studies.
The nomenclature used for C. trachomatis lineages is ever more confusing. The strain and genome in C. trachomatis are currently referred to by the genotype of a single gene (ompA). It is increasingly clear that ompA-genotype does not reflect the genomic heritage of strains [16, 30, 60]. The ompA sequence of L2b is only a single SNP (A485G) from that of ompA-genotype L2, but in the traditional nomenclature represents a different strain, and genomic backbone. Our data reiterate how unreliable this ompA-genotype distinction is, with isolates sequenced here with the L2b genomic backbone showing reversion of the ompA mutation to L2 (G485A). Further confusion arose with the definition of ‘hypervirulent L2c’ [28], which has an ompA-genotype L2 and a chimeric genome between L2 and D genomic backbones. The ‘L2c’ nomenclature in this case refers to the genomic context, which is unprecedented and has led to problems in the literature, with strains carrying ompA-genotype L2 believed to be the ‘hypervirulent L2c’ [29, 61, 62], whereas they may have represented cases of the ‘L2new’ described here. From a clinical microbiology and phylogenetic point of view, the ompA-genotyping concept is misleading, as it refers to a useful typing target which does not necessarily equate to ancestral lineage. In the cases described here, the ompA-genotype L2b backbone is better described by the pmpH-genotype rather than the ompA-genotype, which confounds attempts at following epidemiology with single or even dual targets. We argue that there is an urgent requirement for a new nomenclature. As most nomenclatures are devised around existing typing tools, a genomovar nomenclature based on the whole genome may be too complex. The Uppsala database http://mlstdb.bmc.uu.se/ [46, 63] already has an internal ompA nomenclature as well as the five-target MLST scheme, and it would be interesting to see how combinations of these would allow discrimination across the whole LGV dataset.
C. trachomatis has provided us with yet another diagnostic conundrum [64–69], elucidated by genomics to show how the ompA-genotype L2b outbreak continues to spread globally, diversifying its ompA gene. The impact and implications of this need to be investigated.
Supplementary Data
Funding information
J.C.G. was supported by the Instituto de Salud Carlos III (Plan Estatal de I+D+ i 2013–2016), Grant PI16-01242.
Acknowledgements
Many thanks to Dr Victoria Feher and Professor Rommie Amaro for the original figure of the modelled OmpA, annotated in Fig. 5. Thanks to Dr Alfredo Mari, University of Basel, for help with Fig. S1. Thanks to Prof. Xavier Didelot for assistance with BactDating. We thank Elisabeth Schultheiss for excellent technical assistance in ompA sequencing. Many thanks also to Dr Simon Harris for help with pipeline clarifications, and Dr Madlen Stange and Dr Fanny Wegner for help with selection analysis.
Author contributions
Conceptualization and methodology: H.M.B.S-S., D.G., A.E. Formal analysis: H.M.B.S-S., A.B. Resources: S.M.B., B.V., B.H., J.K., I.C., O.P., C.B., T.P., D.K., E.B., H.Z., F.R., B.d.B., J.C.G., N.R.T. Supervision: D.G., M.B., S.A.M., D.A.L., J.C.G., H.J.C.d.V., N.R.T. Writing – original draft preparation: H.M.B.S-S. Writing – review and editing: all authors.
Conflicts of interest
The authors declare that there are no conflicts of interest
Footnotes
Abbreviations: CDS, coding sequence; LGV, lymphogranuloma venereum; MLST, mulit locus sequence typing; MLVA, multi locus variable-number tandem-repeat analysis; MOMP, major outer membrane protein; MSM, men who have sex with men; SNP, Single Nucleotide Polymorphism; STI, sexually transmitted infection; VD, variable domain; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary tables and four supplementary figures are available with the online version of this article.
References
- 1.Rowley J, Vander Hoorn S, Korenromp E, Low N, Unemo M. Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull World Health Organ. 2019;97:548–562p. doi: 10.2471/BLT.18.228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Vries HJ, Zingoni A, Kreuter A, Moi H, White J. European Guideline on the Management of Lymphogranuloma. Venereum: IUSTI; 2013. [DOI] [PubMed] [Google Scholar]
- 3.Stoner BP, Cohen SE. Lymphogranuloma venereum 2015: clinical presentation, diagnosis, and treatment. Clin Infec Dis. 2015;61:S865–S873. doi: 10.1093/cid/civ756. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Marañón MJ, Bush RM, Peterson EM, Schirmer T, de la Maza LM. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein science: a publication of the Protein Society. 2002;11:1854–1861. doi: 10.1110/ps.3650102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun G, Pal S, Sarcon AK, Kim S, Sugawara E. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis . J Bacteriol. 2007;189:6222–6235. doi: 10.1128/JB.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feher VA, Randall A, Baldi P, Bush RM, de la Maza LM. A 3-dimensional trimeric beta-barrel model for Chlamydia MOMP contains conserved and novel elements of Gram-negative bacterial porins. PLoS One. 2013;8:e68934. doi: 10.1371/journal.pone.0068934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van de Laar M. The emergence of LGV in Western Europe: what do we know, what can we do? Euro Surveill. 2006;11:1–2. doi: 10.2807/esm.11.09.00641-en. [DOI] [PubMed] [Google Scholar]
- 8.Spaargaren J, Fennema HSA, Morré SA, de Vries HJC, Coutinho RA. New lymphogranuloma venereum Chlamydia trachomatis variant, Amsterdam. Emerg Infect Dis. 2005;11:1090–1092. doi: 10.3201/eid1107.040883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris SR, Clarke IN, Seth-Smith HMB, Solomon AW, Cutcliffe L. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nature Genetics. 2012;44:364–366. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christerson L, de Vries HJ, de Barbeyrac B, Gaydos CA, Henrich B. Typing of lymphogranuloma venereum Chlamydia trachomatis strains. Emerg Infect Dis. 2010;16:1777–1779. doi: 10.3201/eid1611.100379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson NR, Holden MTG, Carder C, Lennard N, Lockey SJ. Chlamydia trachomatis: Genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 2008;18:161–171. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadfield J, Harris S, Seth-Smith H, Parmar S, Andersson P. Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion. Gen Res. 2017;27:1220–1229. doi: 10.1101/gr.212647.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morré SA, Spaargaren J, Fennema JSA, de Vries HJC, Coutinho RA, et al. Real-time polymerase chain reaction to diagnose lymphogranuloma Venereum. Emerg Infect Dis. 2005;11:1311–1312. doi: 10.3201/eid1108.050535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CY, Chi KH, Alexander S, Ison CA, Ballard RC. A real-time quadriplex PCR assay for the diagnosis of rectal lymphogranuloma venereum and non-lymphogranuloma venereum Chlamydia trachomatis infections. Sex Transm Infect. 2008;84:273–276. doi: 10.1136/sti.2007.029058. [DOI] [PubMed] [Google Scholar]
- 15.Quint KD, Bom RJ, Quint WG, Bruisten SM, van der Loeff MF. Anal infections with concomitant Chlamydia trachomatis genotypes among men who have sex with men in Amsterdam, the Netherlands. BMC Infect Dis. 2011;11:63. doi: 10.1186/1471-2334-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Versteeg B, Bruisten SM, Pannekoek Y, Jolley KA, Maiden MCJ. Genomic analyses of the Chlamydia trachomatis core genome show an association between chromosomal genome, plasmid type and disease. BMC genomics. 2018;19:130. doi: 10.1186/s12864-018-4522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries HJC, de Barbeyrac B, de Vrieze NHN, Viset JD, White JA. 2019 European guideline on the management of lymphogranuloma venereum. J Eur Acad Dermatol Venereol. 2019;33:1821–1828. doi: 10.1111/jdv.15729. [DOI] [PubMed] [Google Scholar]
- 18.Verweij SP, Catsburg A, Ouburg S, Lombardi A, Heijmans R. Lymphogranuloma venereum variant L2b-specific polymerase chain reaction: insertion used to close an epidemiological gap. Clin Microbiol Infect. 2011;17:1727–1730. doi: 10.1111/j.1469-0691.2011.03481.x. [DOI] [PubMed] [Google Scholar]
- 19.Touati A, Peuchant O, Henin N, Bebear C, de Barbeyrac B. The L2b real-time PCR targeting the pmpH gene of Chlamydia trachomatis used for the diagnosis of lymphogranuloma venereum is not specific to L2b strains. Clin Microbiol Infect. 2016;22:574. doi: 10.1016/j.cmi.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Peuchant O, Touati A, Sperandio C, Hénin N, Laurier-Nadalié C, et al. Changing pattern of Chlamydia trachomatis strains in lymphogranuloma Venereum outbreak, France, 2010-2015. Emerg Infect Dis. 2016;22:1945–1947. doi: 10.3201/eid2211.160247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaksson J, Carlsson O, Airell Å, Strömdahl S, Bratt G, et al. Lymphogranuloma venereum rates increased and Chlamydia trachomatis genotypes changed among men who have sex with men in Sweden 2004-2016. J Med Microbiol. 2017;66:1684–1687. doi: 10.1099/jmm.0.000597. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Dominguez M, Puerta T, Menendez B, Gonzalez-Alba JM, Rodriguez C. Clinical and epidemiological characterization of a lymphogranuloma venereum outbreak in Madrid, Spain: co-circulation of two variants. Clin Microbiol Infect. 2014;20:219–225. doi: 10.1111/1469-0691.12256. [DOI] [PubMed] [Google Scholar]
- 23.Cole MJ, Field N, Pitt R, Amato-Gauci AJ, Begovac J. Substantial underdiagnosis of lymphogranuloma venereum in men who have sex with men in Europe: preliminary findings from a multicentre surveillance pilot. Sex Transm Infect. 2019;96:137–142. doi: 10.1136/sextrans-2019-053972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marangoni A, Foschi C, Tartari F, Gaspari V, MC R. Lymphogranuloma venereum genovariants in men having sex with men in Italy. Sex Transm Infect. 2020 doi: 10.1136/sextrans-2020-054700. [DOI] [PubMed] [Google Scholar]
- 25.Gomes JP, Nunes A, Florindo C, Ferreira MA, Santo I. Lymphogranuloma venereum in Portugal: unusual events and new variants during 2007. Sex Transm Dis. 2009;36:88–91. doi: 10.1097/OLQ.0b013e31818b1e27. [DOI] [PubMed] [Google Scholar]
- 26.Kendall BA, Tardif KD, Schlaberg R. Chlamydia trachomatis L serovars and dominance of novel L2b ompA variants, U.S.A. Sex Transm Infect. 2014;90:336. doi: 10.1136/sextrans-2013-051478. [DOI] [PubMed] [Google Scholar]
- 27.Stary G, Meyer T, Bangert C, Kohrgruber N, Gmeinhart B. New Chlamydia trachomatis L2 strains identified in a recent outbreak of lymphogranuloma venereum in Vienna, Austria. Sex Transm Dis. 2008;35:377–382. doi: 10.1097/OLQ.0b013e31815d6df8. [DOI] [PubMed] [Google Scholar]
- 28.Somboonna N, Wan R, DM O, Pettengill MA, Joseph SJ. Hypervirulent Chlamydia trachomatis clinical strain is a recombinant between lymphogranuloma venereum (L(2)) and D lineages. Mol Biol Evol. 2011;2:e00045-00011. doi: 10.1128/mBio.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seth-Smith HM, Galan JC, Goldenberger D, Lewis DA, Peuchant O. Concern regarding the alleged spread of hypervirulent lymphogranuloma venereum Chlamydia trachomatis strain in Europe. Euro Surveill. 2017;22:15. doi: 10.2807/1560-7917.ES.2017.22.15.30511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borges V, Cordeiro D, Salas AI, Lodhia Z, Correia C. Chlamydia trachomatis: when the virulence-associated genome backbone imports a prevalence-associated major antigen signature. Microb Genom. 2019;5:11.:e000313. doi: 10.1099/mgen.0.000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph SJ, Didelot X, Rothschild J, de Vries HJC, Morré SA, et al. Population genomics of Chlamydia trachomatis: Insights on drift, selection, recombination, and population structure. Mol Biol Evol. 2012;29:3933–3946. doi: 10.1093/molbev/mss198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Domínguez M, González-Alba JM, Puerta T, Martínez-García L, Menéndez B, et al. Spread of a new Chlamydia trachomatis variant from men who have sex with men to the heterosexual population after replacement and recombination in OMPA and PMPH genes. Clin Microbiol Infect. 2017;23:761–766. doi: 10.1016/j.cmi.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Lysén M, Osterlund A, Rubin CJ, Persson I, Persson I, et al. Characterization of OMPA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J Clin Microbiol. 2004;42:1641–1647. doi: 10.1128/jcm.42.4.1641-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics (Oxford, England: 2009. pp. 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutherford KM, Parkhill J, Crook J, Horsnell T, Rice P, et al. Artemis: Sequence Visualization and Annotation Bioinformatics. Oxford, England: 2000. pp. 944–945. [DOI] [PubMed] [Google Scholar]
- 37.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 38.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen. Virus Evolution. 2016;2 doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 2018;46:e134. doi: 10.1093/nar/gky783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.RStudio Team Rstudio: Integrated development for R version 1.2.5033. 2021 http://www.rstudio.com
- 43.R Core Team R: A language and environment for statistical computing version 3.6.2. 2021 https://www.r-project.org
- 44.Didelot X, Siveroni I, Volz EM. Additive uncorrelated relaxed clock models for the dating of genomic epidemiology phylogenies. Mol Biol Evol. 2020 doi: 10.1093/molbev/msaa193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borges V, Gomes JP. Deep comparative genomics among Chlamydia trachomatis lymphogranuloma venereum isolates highlights genes potentially involved in pathoadaptation. Infect Genet Evol. 2015;32:74–88. doi: 10.1016/j.meegid.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 46.Klint M, Fuxelius HH, Goldkuhl RR, Skarin H, Rutemark C, et al. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J Clin Microbiol. 2007;45:1410–1414. doi: 10.1128/JCM.02301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrmann B, Isaksson J, Ryberg M, Tångrot J, Saleh I, et al. Global multilocus sequence type analysis of Chlamydia trachomatis strains from 16 Countries. J Clin Microbiol. 2015;53:2172–2179. doi: 10.1128/JCM.00249-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pannekoek Y, Morelli G, Kusecek B, Morré SA, Ossewaarde JM, et al. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis . BMC Microbiol. 2008;8:42. doi: 10.1186/1471-2180-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dean D, Bruno WJ, Wan R, Gomes JP, Devignot S, et al. Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg Infect Dis. 2009;15:1385–1394. doi: 10.3201/eid1509.090272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunes A, Nogueira PJ, Borrego MJ, Gomes JP. Adaptive evolution of the Chlamydia trachomatis dominant antigen reveals distinct evolutionary scenarios for B- and T-cell epitopes: worldwide survey. PLoS One. 2010;5:10. doi: 10.1371/journal.pone.0013171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong G, RC B. Antigenic determinants of the chlamydial major outer membrane protein resolved at a single amino acid level. Infect Immun. 1991;59:1141–1147. doi: 10.1128/IAI.59.3.1141-1147.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayes LJ, Pickett MA, Conlan JW, Ferris S, Everson JS, et al. The major outer-membrane proteins of Chlamydia trachomatis serovars A and B: intra-serovar amino acid changes do not alter specificities of serovar- and C subspecies-reactive antibody-binding domains. J Gen Microbiol. 1990;136:1559–1566. doi: 10.1099/00221287-136-8-1559. [DOI] [PubMed] [Google Scholar]
- 53.Joseph SJ, Didelot X, Gandhi K, Dean D, Read TD. Interplay of recombination and selection in the genomes of Chlamydia trachomatis . Biol Direct. 2011;6:28. doi: 10.1186/1745-6150-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunelle BW, Sensabaugh GF. Nucleotide and phylogenetic analyses of the Chlamydia trachomatis ompA gene indicates it is a hotspot for mutation. BMC Res Notes. 2012;5:53. doi: 10.1186/1756-0500-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray GGR, Wang F, Harrison EM, Paterson GK, Mather AE, et al. The effect of genetic structure on molecular dating and tests for temporal signal. Methods Ecol Evol. 2016;7:80–89. doi: 10.1111/2041-210X.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Negrate G, Krieg A, Faustin B, Loeffler M, Godzik A. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10:1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 57.Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, et al. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: Evidence for serovar da recombination and correlation with tissue tropism. J Bacteriol. 2006;188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai PY, Hsu MC, Huang CT, SY L. Human antibody and antigen response to IncA antibody of Chlamydia trachomatis . Int J Immunopathol Pharmacol. 2007;20:156–161. [PubMed] [Google Scholar]
- 59.van Aar F, Kroone MM, de Vries HJ, Gotz HM, van Benthem BH. Increasing trends of lymphogranuloma venereum among HIV-negative and asymptomatic men who have sex with men, the Netherlands, 2011 to 2017. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.14.1900377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris SR, Clarke IN, Seth-Smith HMB, Solomon AW, Cutcliffe LT, et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 2012;44:413–419.:S411. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matičič M, Klavs I, Videčnik Zorman J, Vidmar Vovko D, Kogoj R, et al. Confirmed inguinal lymphogranuloma venereum genovar L2c in a man who had sex with men, Slovenia, 2015. Euro Surveill. 2016;21:2–5. doi: 10.2807/1560-7917.ES.2016.21.5.30129. [DOI] [PubMed] [Google Scholar]
- 62.Petrovay F, Balla E, Erdosi T. Emergence of the lymphogranuloma venereum L2c genovariant, Hungary, 2012 to 2016. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.5.30455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bom RJM, Christerson L, Schim van der Loeff MF, Coutinho RA, Herrmann B, et al. Evaluation of high-resolution typing methods for Chlamydia trachomatis in samples from heterosexual couples. J Clin Microbiol. 2011;49:2844–2853. doi: 10.1128/JCM.00128-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herrmann B, Törner A, Low N, Klint M, Nilsson A, et al. Emergence and spread of Chlamydia trachomatis variant, Sweden. Emerg Infect Dis. 2008;14:1462–1465. doi: 10.3201/eid1409.080153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ripa T, Nilsson P. A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2006;11:E061109. doi: 10.2807/esw.11.45.03076-en. [DOI] [PubMed] [Google Scholar]
- 66.Ripa T, Nilsson PA. A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false-negative nucleic acid amplification tests. Sex Transm Dis. 2007;34:255–256. doi: 10.1097/OLQ.0b013e31805ce2b9. [DOI] [PubMed] [Google Scholar]
- 67.Seth-Smith HMB, Harris SR, Persson K, Marsh P, Barron A, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rantakokko-Jalava K, Hokynar K, Hieta N, Keskitalo A, Jokela P, et al. Chlamydia trachomatis samples testing falsely negative in the aptima Combo 2 test in Finland, 2019. Euro Surveillance: Bulletin Europeen sur les maladies Transmissibles = European Communicable Disease Bulletin. 2019;24:1900298. doi: 10.2807/1560-7917.ES.2019.24.22.1900298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Unemo M, Getman D, Hadad R, Cole M, Thomson N, et al. Letter to the editor: Chlamydia trachomatis samples testing falsely negative in the aptima combo 2 test in Finland, 2019. Euro Surveillance: Bulletin Europeen sur les maladies Transmissibles = European Communicable Disease Bulletin. 2019;24:1900354. doi: 10.2807/1560-7917.ES.2019.24.24.1900354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, et al. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.