Abstract
Ligand-dependent biosensors are valuable tools for coupling the intracellular concentrations of small molecules to easily detectable readouts such as absorbance, fluorescence, or cell growth. While ligand-dependent biosensors are widely used for monitoring the production of small molecules in engineered cells and for controlling or optimizing biosynthetic pathways, their application to directed evolution for biocatalysts remains underexplored. As a consequence, emerging continuous evolution technologies are rarely applied to biocatalyst evolution. Here, we develop a panel of ligand-dependent biosensors that can detect a range of small molecules. We demonstrate that these biosensors can link enzymatic activity to the production of an essential phage protein to enable biocatalyst-dependent phage-assisted continuous evolution (PACE) and phage-assisted continuous selection (PACS). By combining these phage-based evolution and library selection technologies, we demonstrate that we can evolve enzyme variants with improved and expanded catalytic properties. Finally, we show that the genetic diversity resulting from a highly mutated PACS library is enriched for active enzyme variants with altered substrate scope. These results lay the foundation for using phage-based continuous evolution and selection technologies to engineer biocatalysts with novel substrate scope and reactivity.
Short abstract
Ligand-dependent biosensors were engineered to enable enzyme-dependent continuous evolution and selection. These technologies were used to engineer enzyme variants with improved catalytic properties.
Introduction
Enzymes are increasingly valuable tools for chemical synthesis due to their high catalytic proficiency and selectivity under mild, environmentally friendly conditions.1 Natural enzymes can sometimes be repurposed as catalysts for synthesis, but in most cases, enzymes with catalytic properties suitable for transformations of interest cannot be readily obtained from nature.2,3 Instead, directed evolution, which involves iterative cycles of mutagenesis followed by functional screening, can be used to improve enzyme activity, substrate scope, stability, and selectivity.4−9 Due to the slow speed and modest throughput of current enzyme directed evolution pipelines, however, the potential for enzymes to impact chemical synthesis is often limited.10,11
In general, modern enzyme engineering approaches involve some combination of biochemical/bioinformatic analysis and/or genome mining to identify a suitable parent enzyme and directed evolution to improve the activity of interest in the parent enzyme.12−14 These efforts are often facilitated by automated systems that can typically assay enzyme library sizes of ∼104 variants, but such systems are costly to procure and maintain. Moreover, reprogramming enzymes to act on new substrates or with new selectivity is often challenging because the native activity and selectivity that make enzymes so attractive in the first place often limit their scope toward non-native substrates. In some cases, approaches such as substrate walking, in which enzymes are evolved for activity on substrates with increasing structural similarity to the target, can be used to gradually improve substrate scope and activity, even under the constraints of limited library screening throughput.15−17 While substrate walking is effective, the time spent developing activity for the related substrates is substantial and slows the development of the target reaction. New technologies that significantly increase the size of libraries that can be screened and thus improve the speed of directed evolution could facilitate the broader adoption of directed enzyme evolution for organic synthesis.
In vivo continuous evolution has the potential to dramatically accelerate directed evolution by eliminating manual cycles of mutation, translation, selection, and replication required by conventional directed evolution methods.18−20 For example, phage-assisted continuous evolution (PACE)21,22 has been used to evolve novel function in proteases,23,24 DNA-binding proteins,25 T7 RNA polymerase (T7 RNAP),21,26−30 and a range of other enzyme classes31−33 in a time span of only days-to-weeks without costly instrumentation or user intervention. PACE involves linking the activity of a protein of interest (POI), which has been encoded into the bacteriophage genome, to the phage life cycle such that viral replication results in optimization of the POI for the desired function. The link is established by a biosensor system that modulates the production of pIII (encoded by gIII), a native bacteriophage protein essential for phage replication, in response to POI function. Phage encoding the evolving gene are propagated on a constant supply of Escherichia coli cells, which contain a plasmid for the expression of pIII and a mutagenesis plasmid, added to a fixed volume vessel. Because the E. coli cells are continually added, and the system (the “lagoon”)21 is constantly diluted, the E. coli cells do not have time to divide before leaving the vessel and therefore cannot evolve. The rapid replication time of phage allows multiple generations to propagate between lagoon dilutions, ensuring that mutations are allowed to accumulate. With a proper linkage between a carried gene of interest and an inducible expression system for gIII, the phage replication rate is dependent on the desired activity of interest. In other words, phage harboring POI variants with improved function generate more infectious progeny, leading to positive selection, while those encoding inactive proteins cannot and are selected against. A related approach, phage-assisted noncontinuous evolution (PANCE), was recently used to evolve methanol dehydrogenase variants with 3.5-fold improved Vmax for the conversion of methanol to formaldehyde via a ligand-dependent biosensor.22,34 Despite this promising result, the potential utility of biocatalyst evolution using PACE or other in vivo directed evolution methods35−39 is limited by the availability of biosensors, genetic circuits that detect an input signal and drive a genetic output,40 for a reaction product of interest.
In this study, we aimed to develop and validate selection systems for the in vivo directed evolution of biocatalysts using PACE and phage-assisted continuous selection (PACS). First, to maximize the scope of products that can be detected and thus used to drive PACE, we engineered a diverse set of ligand-dependent biosensors capable of detecting a range of small molecules. Using a combination of bacterial transcription factors, ligand-inducible regulators, and chemically induced dimerization systems, we generated biosensing systems for acrylate, IPTG, estrogen, and abscisic acid. We characterized the sensitivity and specificity of each biosensor and then developed each biosensor into a gene circuit that can drive phage replication in response to enzyme catalysis (Figure 1a). Next, we developed a modified PACE apparatus to allow for host cell preincubation with a small molecule substrate, which is critical for biocatalyst evolution under the fast speed of PACE. To validate the new approach, we demonstrate rapid, continuous evolution of esterases. Moreover, we show that the same biosensor-driven phage replication process involved in PACE enables PACS for the high-throughput screening of esterase library variants (Figure 1b). These phage-assisted approaches provide a powerful means to rapidly evolve and identify esterase variants with improved properties for chemical synthesis. More broadly, this study establishes workflows and a methodology needed to adopt PACE21 and PACS41 for a range of other biocatalysts and reaction types.
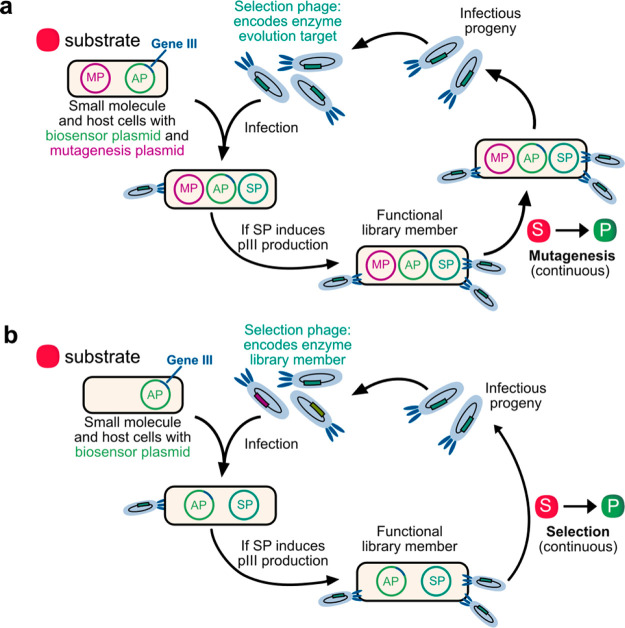
Figure 1.
In vivo continuous evolution approaches can be far faster than conventional directed evolution methods and require less human intervention, unlocking new chemical potential in enzymes. (a) Overview of small-molecule-dependent PACE for the evolution of selective biocatalysts. (b) Overview of small-molecule-dependent PACS for the selection of selective biocatalysts.
Results
Developing Biosensors for Biocatalyst PACE
Representative ligand-dependent biosensors were selected based on their potential applicability for an analysis of diverse substrates and reactions and the availability of information regarding sensor performance (e.g., dynamic range, residual activity in the absence of inducer, sensitivity, and cross-reactivity) (Figure 2a). Transcriptional repressors bind to a promoter sequence and block RNA polymerase (RNAP) from transcribing a target gene. Transcription is triggered when repressor proteins bind a ligand, affording an effective sensor for specific and sensitive ligand detection.42−44 AcuR, which binds acrylate, was selected as a representative member of this family due to its high dynamic range45 and its established utility for metabolic engineering applications.46 The lacUV5 promoter, a mutated promoter from the E. coli lac operon, is one of the most commonly used promoters in molecular biology. It can drive high levels of gene expression in response to isopropyl β-d-1-thiogalactopyranoside (IPTG) and can be regulated by the LacI repressor.47,48 Chemically induced dimerization systems utilize two proteins that bind only in the presence of a ligand and have been extensively used to identify protein–protein interactions and to control protein localization.49−51 The estrogen receptor and abscisic acid systems were selected as representative ligand-inducible dimerization systems. The human estrogen receptor ligand binding domain (LBD) and the SRC coactivator receptor (RID) were identified by yeast two-hybrid data as estradiol-dependent interacting proteins.52−54 These domains have been widely used to study estrogen signaling and have been engineered to respond to different ligands. Similarly, the abscisic acid (ABA) system relies on dimerization domains (ABI and PYL) that bind in the presence of ABA.29,55
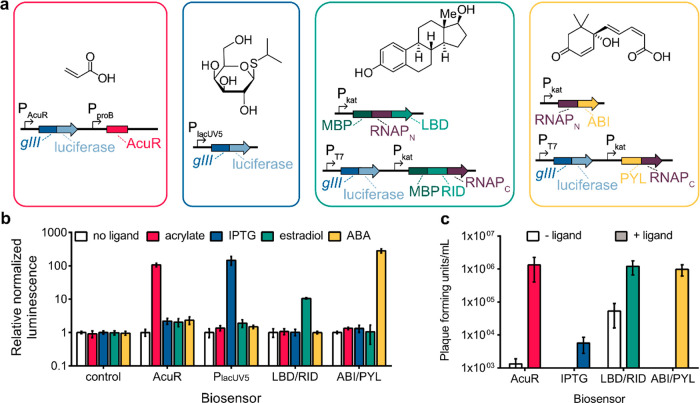
Figure 2.
Development of biosensors for ligand detection. (a) Ligands and vector systems to link gIII expression and the presence of small-molecule substrates. (b) Luciferase output of ligand-dependent biosensors. E. coli expressing plasmids shown in part a were incubated in the absence or presence of a small molecule for 3 h and then analyzed for luminescence. (c) Phage replication of ligand-dependent biosensors. E. coli expressing plasmids shown in part a were incubated in the absence or presence of ligand and phage for 6 h. Phage cultures were then collected and analyzed for phage replication. Error bars are the standard deviation: (b) n = 4 replicates and (c) n = 3 replicates.
The AcuR and lacUV5 biosensors were each constructed as single plasmids with the transcriptional repressor (AcuR) or inducible promoter (lacUV5) and gIII translationally coupled to a bacterial luciferase reporter system. The ligand-inducible dimerization systems were fused to our previously developed split T7 RNAP (RNAP)28 via flexible linkers. A functional RNAP is assembled when the domains interact in the presence of ligand resulting in T7 promoter-driven expression of gIII and translationally coupled luciferase. For each of these biosensors, the ribosomal binding site sequences and plasmid copy number were optimized to provide low background and high dynamic range (Figures S1–S4). The cross-reactivity of each sensor with our panel of probes (acrylate, IPTG, estradiol, and ABA) was then examined using a luciferase reporter assay (Figure 2b and Figure S5). All sensors responded to their cognate ligands by at least 10-fold over the noncognate ligands examined. The greatest orthogonality was observed with the ABA sensor which responded ∼250-fold for ABA over the other probes. Phage growth assays with each biosensor in the presence or absence of ligand were then conducted (Figure 2c and Figure S6), and in each case, phage growth was enhanced with the addition of a small molecule, demonstrating that diverse biosensors can be used to link the presence of different ligands to gIII expression.
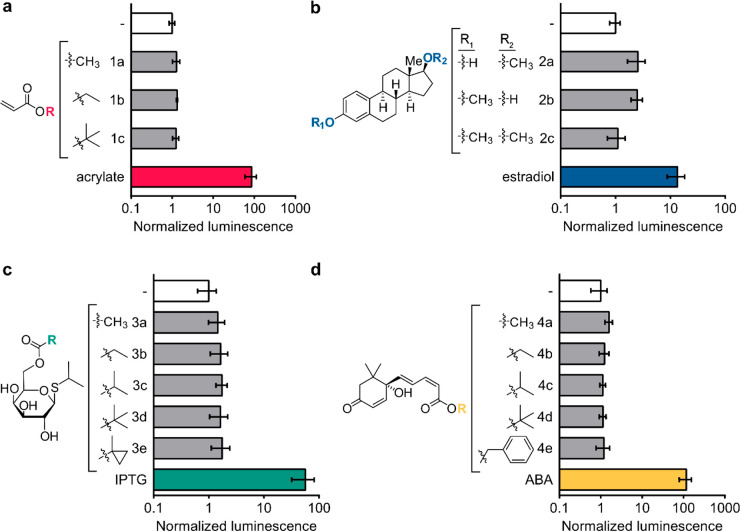
Each biosensor was next evaluated against derivatives of its cognate ligand to establish whether the sensors could be used to quantify enzymatic reactions that reveal the ligand itself. A panel of derivatives for each ligand was prepared, and the biosensor response was determined by luciferase assay (Figure 3 and Figure S7). Luciferase expression was selective for the native ligand relative to the synthetic derivatives in all cases. For example, RNAP assembly in response to ABA yielded a robust luciferase signal (Figure 3d), while no induction was detected with ABA esters 4a–4e, highlighting the specificity of the biosensor for the native ligand even over structurally similar derivatives.
Figure 3.
Small-molecule biosensors can be used to detect selective biocatalysis. (a) E. coli expressing the AcuR biosensor was incubated in the absence or presence of acrylate esters or acrylate for 3 h and then analyzed for luminescence. (b) E. coli expressing the estradiol biosensor was incubated in the absence or presence of methylated estradiol or estradiol for 3 h and then analyzed for luminescence. (c) E. coli expressing the IPTG biosensor was incubated in the absence or presence of IPTG esters or IPTG for 3 h and then analyzed for luminescence. (d) E. coli expressing the ABA biosensor was incubated in the absence or presence of ABA esters or ABA for 3 h and then analyzed for luminescence. Error bars are the standard deviation for n = 4 replicates.
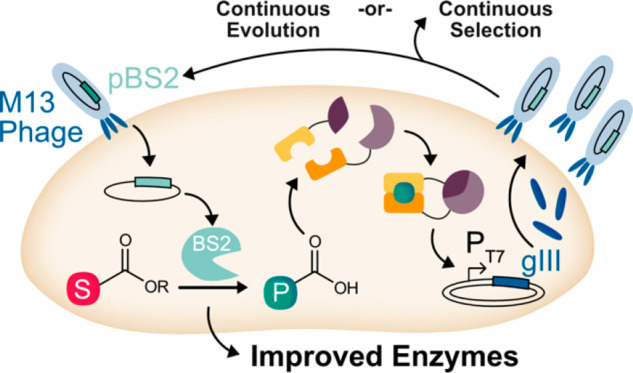
Esterase-Catalyzed Hydrolysis Supports Phage Replication
Given that the biosensors investigated could link phage replication to the presence of a specific ligand and that the ABA biosensor performed well for the detection of ABA and not masked ABA esters, we aimed to activate gIII expression via esterase-catalyzed hydrolysis of substrates 4a–4e. In general, esterases are versatile biocatalysts due to their stability and compatibility with mild reaction conditions.56 The exquisite stereoselectivity that can be achieved for hydrolysis of some substrates has made esterases attractive for organic synthesis.56,57 However, naturally occurring esterases may not possess suitable activity or enantioselectivity for a desired transformation. We therefore aimed to develop a rapid platform for esterase evolution to expand their utility for chemical synthesis.
BS2 esterase from Bacillus subtilis was selected as an initial evolution target since this enzyme and several mutants have been efficiently expressed in E. coli and were shown to act on sterically hindered esters and tertiary alcohols.58−60 As a first step toward linking the phage life cycle to ABA produced through esterase hydrolysis, BS2 activity on substrates 4a–4e was examined by HPLC (Figure 4a). BS2 efficiently catalyzed the hydrolysis of 4a, but little or no activity was observed on 4c or 4d. Based on these results, we envisioned that 4a would act as a positive control (as an efficiently hydrolyzed substrate of BS2) and could also be used for a substrate walking approach in which improved activity on 4a is used as a first step to identify variants with activity on 4c and 4d. To test the feasibility of this approach, we aimed to detect replication of selection phage (SP) expressing BS2 in the presence of the ABA derivatives. M13 phage were engineered to replace gIII with BS2 and then combined with host E. coli cells containing the ABA biosensor in the presence of ABA, 4a–4d, or no ligand. Phage growth was only detected with 4a or 4b and displayed a similar trend to BS2 activity (Figure 4b). These results provide evidence that the ABA biosensor can link phage replication to a ligand that is generated via BS2 hydrolysis of a precursor substrate.
Figure 4.
Esterase-catalyzed hydrolysis of abscisic acid (ABA) can support phage replication. (a) BS2 activity on ABA esters. ABA esters (750 μM) were incubated with 5 μM BS2 over 60 min, and percent conversion to ABA was determined. (b) BS2 phage replication with ABA derivatives. Phage carrying BS2 were incubated with E. coli expressing ABA biosensor in the absence or presence of ABA esters or ABA for 6 h. Phage cultures were then collected and analyzed for phage replication. Error bars are the standard error for n = 3 replicates.
Esterase Evolution Using PACE
To initiate biocatalyst PACE, we modified the conventional PACE setup to include a substrate “holding tank” prior to the lagoon (Figure 5a,b). This addition allows for preincubation of the host cells expressing the biosensor with substrate to increase the likelihood of substrate entering the E. coli before being transferred to the lagoon containing the SP (Figure S8). One holding tank can seed up to four lagoons, which enables replicate evolution experiments and reduces required substrate quantities. To validate this approach, we conducted a mock PACE experiment to determine if active phage could be enriched from an excess of inactive phage in the presence of 4a. A holding tank with 4a was assembled, and a lagoon was seeded with 500-fold excess of active BS2 phage relative to control phage that encode for human rhinovirus-14 (HRV) protease, which cannot process the esterase substrate and should thus not replicate efficiently. Fresh E. coli host cells containing the ABA biosensor were continuously flowed into the holding tank for incubation prior to entering the lagoon. Phage samples were collected over time and assayed by PCR with primers for the phage backbone to determine the relative HRV:BS2 population. The BS2 phage population was detectable by PCR within 24 h and was maintained in the lagoon over the next 24 h. In contrast, the HRV population diluted out over the first 24 h, and complete washout of the inactive phage was observed by 30 h (Figure 5c).
Figure 5.
Evolution of esterase variants with activity on ABA esters. (a) Schematic of PACE for biocatalyst evolution. ABA derivative is preincubated with E. coli expressing ABA biosensor 3 h prior to transfer to a lagoon containing phage carrying BS2. Only BS2 phage that can hydrolyze the derivative to ABA will be able to replicate on the host cells and produce gIII to generate infectious progeny. (b) Biocatalyst PACE setup with a holding tank for host cell preincubation with ABA derivatives. (c) Phage competition in biocatalyst PACE. ABA ester 4a was preincubated with host cells, and phage carrying BS2 or HRV were mixed (1:500) as shown in part b. Phage samples were collected 0–48 h after addition to the lagoon and then analyzed by PCR. Bioconversions of BS2 and PACE BS2 variants on (d) ABA ester 4a or (e) ABA ester 4b.
Based on these results, we initiated PACE of BS2 to improve its activity on substrate 4a. For the first 48 h, ABA was also pumped into the lagoons (turning the sensor on) to determine the efficiency of phage propagation and to provide an evolutionary drift period. ABA was decreased over time and eventually completely removed, such that only 4a was added to select for catalysis-dependent evolution, and PACE was continued for an additional 48 h. BS2 variants at the end of the experiments were subcloned into an arabinose-inducible construct and assayed in E. coli with the ABA biosensor (Figure 1a) for luciferase output, which revealed that variants with improved catalytic performance evolved during PACE (Figure S9). The top four hits were selected and rescreened for activity using the HPLC assay and confirmed up to 2-fold improved hydrolysis of 4a (Figure 5d).
These four variants were then subjected to an additional round of PACE with 4b to evolve enzymes with activity on a more sterically hindered substrate. PACE was conducted using 4a and 4b for 48 h, followed by 4b alone for 72 h. The phage successfully replicated during the 5 days of PACE, but no new mutations were detected in the BS2 population. The same PACE experiment was conducted using phage expressing WT BS2, and complete washout was observed, suggesting that the variants from PACE using 4a had sufficient activity to survive on 4b. To support this idea, we confirmed that all four variants had improved activity on 4b (Figure 5e). Further attempts to tune selection pressure based on concentration of 4b and number of days on substrate resulted in either no new mutations or phage extinction.
While our attempts to evolve BS2 via PACE were successful, the overall improvements in catalytic activity were only 2-fold. We hypothesized that the lack of functional improvement despite increased selection pressure during PACE (e.g., by varying flow rate, substrate concentration, mutagenesis rate, incubation time, etc.) and the low phage titers observed (∼1 × 106 PFU/mL) could result from a low effective library size under conditions that permitted phage replication. We reasoned that phage-assisted continuous selection (PACS)41 could rectify this problem since it allows for the use of in vitro mutagenesis strategies while providing a powerful selection platform based on the same link between POI function and phage replication required for PACE.
Esterase Selection Using PACS
To establish the feasibility of improving BS2 via PACS using the ABA biosensor, a library of BS2 variants was generated from WT BS2 and the four PACE variants noted above via error prone PCR with a high error rate to ensure sampling of as many unique variants as possible61 (Figure S10). Aliquots of library phage were cultured with an ester-caged fluorescein62,63 to confirm that the library contained active variants, and significant fluorescence was observed (Figure S11). Biocatalyst PACS, which is identical to the PACE system except omitting the mutagenesis plasmid, was then initiated with the BS2 library and 4b to enrich for variants with improved activity on 4b. After 24 h, the BS2 phage library was still able to replicate, but a BS2 phage WT control showed phage extinction. Library phage samples were then subcloned into a pET28 vector for expression in BL21(λDE3) and subsequent screening for esterase hydrolysis in lysate.
Rapid qualitative analysis of 900 variants obtained from BS2 PACS was conducted using MISER-MS (multiple injections in a single experimental run).64 90 of the top variants were reanalyzed by UHPLC, and 10 variants with at least a 2-fold improvement over WT for the hydrolysis of 4b in lysate were identified (Figure S12). Variants 10D2 and 10D5 were particularly notable as they fully hydrolyzed ester 4b in lysate. Sequencing revealed that these variants (Table S2) had identical genotypes and contained six new coding mutations, which would be a very high number of mutations to emerge from a single round of traditional directed evolution. These results confirm that PACS can be utilized for the high-throughput screening of esterase library variants to rapidly identify biocatalysts with improved activity.
Substrate Scope of Evolved Esterases
A subset of esterases with improved activity on 4b were purified and screened against 4a–4d (Figure 6a,b and Figures S13–S17). For all enzymes except 10D2, the efficiency of the hydrolysis is directly correlated to the steric bulk of the ester (Me > Et ≫ iPr, tBu). A steady state kinetic analysis of 10D2 showed that the change in preference for bulkier esters is attributed to an increase in KM for the methyl substrate 4a, which increases from 111 μM for WT BS2 to approximately 5615 μM for 10D2. KM for ethyl substrate 4b also increased from 143 to 1265 μM, but the compensatory 50-fold increase in Kcat results in a more efficient reaction (Table S3).
Figure 6.
Evaluation of substrate scope for selected hits from a PACS screen. (a) Reaction scheme for ABA esters evaluated. 750 μM substrate was incubated with 5 μM BS2 variant for 1 h. (b) Conversion of each reaction normalized to the WT reaction. Each bar represents the average of three reactions. Yield of ABA obtained from a calibration curve using 2-acetamidophenol as an internal standard (calibration curve Figure S17). (c) Reaction scheme for the hydrolysis of “truncated” ABA fragment, 3,3-dimethyl acrylate (DMA) esters. 5 mM substrate was incubated with 10 μM BS2 variant for 1 h. (d) Conversion of each reaction normalized to the WT reaction. Each bar represents the average of three reactions. Yield of 3,3-DMA obtained from a calibration curve using 5-bromoindole as the internal standard (calibration curve Figure S18).
We also evaluated the activity of our evolved esterases on several structurally distinct substrates (5–8) protected with a simple 3,3-dimethyl acrylate (DMA) group in place of the full ABA fragment (Figure 6c,d and Figure S18). Interestingly, 10D2 was found to have minimal activity on any of the other substrates evaluated. In contrast, the two mutants found to be broadest in substrate scope, 7A7 and 8F11, contain 4 (P90T, I130 V, N269S, L339P) and 3 (P76R, T220A, D320G) unique coding mutations, respectively. For 8F11, which was over 2-fold improved over WT BS2 for hydrolysis of 6, all the mutations are found on the surface of the protein far from the active site. This finding demonstrates that PACS is not limited to improving activity on the substrate used for selection and can give a population of active enzymes within which improved activity for a variety of reactions can be found, as in another recently reported example of continuous evolution for biocatalyst development.36
Encouraged by the range of activity observed in the substrate screen using enzymes identified based on their activity on ABA esters, we wanted to establish whether the PACS library might contain variants with improved activity on DMA-protected substrates. WT-BS2 provided low conversion of 5, for example, and none of the esterases selected for activity on 4b showed significantly improved activity on this substrate, making it an attractive challenge for the library. The esterase compilation plate was screened against 5, and several enzymes were identified with improved conversion relative to WT (Figure S19). One of these, 1E12 (I21M, K362E, P473M), was found to be 1.8-fold improved for the hydrolysis of 5 over WT after IMAC purification, outperforming any enzyme from the previously analyzed set (Figure S20).
Discussion
While directed evolution has expanded the utility of biocatalysts, relatively long times and significant instrumentation are generally required for the evolution process.10 Most critically, limitations in library sizes often preclude dramatic enzyme reprogramming efforts, thus limiting the potential of biocatalysts to provide solutions to key problems in synthesis. Continuous directed evolution can address these limitations by replacing the manual steps in directed evolution with a continuous process in which enzyme evolution and selection occur concurrently in host cells. PACE requires that the activity of an evolving protein be linked to the production of pIII, an essential protein for phage propagation. To address this requirement for biocatalyst PACE, we established that various transcription regulators would work as ligand-dependent biosensors and that different modes of mutagenesis/selection are possible for enzyme evolution and selection in phage-based systems.
Despite extensive use of ligand-dependent biosensors for detecting small molecules in cells,65−67 their use in assays for directed evolution is rare,68−70 and only one example of their use for continuous evolution systems has been reported.34 In this study, we engineered multiple biosensors to detect diverse small-molecule ligands and yield pIII or a reporter gene as an output. The sensor designs included transcription factors, ligand-inducible promoters, and dimerization systems, all of which responded robustly to their cognate ligands. No response was observed for a panel of derivatives of each ligand, indicating that the sensors can be used to detect selective biocatalysts. Based on these sensors, P450 enzymes could be screened for demethylase activity (methyl ether hydroxylation/acetal hydrolysis) with methylated steroids71 and the estradiol sensor.52 The ligands for the remaining sensors were masked with esters for the detection of esterase-catalyzed hydrolysis.57 Continued development of selective and sensitive ligand-dependent biosensors could further expand continuous evolution of biocatalysts, enabling further diversity of the substrates and reactions that could be accessed via biocatalyst PACE or PACS.
To initiate biocatalyst PACE, we modified the conventional PACE setup to include a holding tank for preincubation of the ligand derivative and the host cells. While active BS2 phage could be enriched over inactive phage, esterase PACE with the ABA biosensor and 4a–4b resulted in only up to a 2-fold improvement in enzyme activity. Attempts to further improve enzyme activity by modulating the selection pressure through ABA ester derivative concentration or length of evolution on the substrate did not afford any new esterase variants. This finding suggests that there is a window of biocatalyst activity necessary for this PACE platform and that this approach may be most useful for gaining functional variants rather than improved variants. In contrast, a high-throughput screening of a BS2 phage library with PACS rapidly selected biocatalysts with improved catalytic activity. Variants 10D2 and 10D5, in particular, highlight the success of PACS for a substrate walking approach, as significant activity on 4c and 4d was detected while no activity was observed on either substrate with WT BS2.
A limitation of evolving enzyme activity using model substrates like 4a–4d is that improved activity on the model substrate is often achieved at the expense of substrate scope—activity on other substrates of interest. This issue is particularly problematic when substrates are designed to release fluorescent or (as in the current study) biologically active fragments since those fragments are not found in typical organic substrates. The low selection pressure in our PACS systems allows for “neutral drift” of the enzyme,72 where variants with diminished activity on the target substrate are still allowed to persist in the lagoon. Although these variants were not always improved for the target substrate relative to WT, the presence of this mild selection enriches the library in active mutants. We found that screening these PACS-derived, functionally enriched libraries for activity using substrate “mimics” that contain truncated versions of the target substrate yielded enzymes that were significantly improved for structurally distinct substrates.
It is noteworthy that, in both this study and the only previous example of PACE applied to biocatalysis, it was necessary to modify the standard PACE format to facilitate biocatalyst evolution. In our study, low phage titers indicate low rates of phage replication in the selection time frame (i.e., while E. coli is circulating in the lagoon). These low phage titers correspond to a small library size, and given the relatively low mutation rate observed for the targeted gene, we suspect that sampling of the sequence space to identify improved mutants was insufficient for the selection pressure applied during PACE. The balance of tuning the “selection window” such that selection pressure is high enough to require beneficial mutation accumulation while ensuring the selection pressure is low enough to allow for phage replication remains an issue in biocatalyst-dependent PACE campaigns.
Conclusions
This study established that gIII expression can be linked to enzyme activity on small-molecule substrates to enable enzyme optimization via PACE and PACS. In addition to expanding the set of biosensor designs that have been linked to gIII expression, we demonstrated the potential of biocatalyst-dependent PACE by evolving an esterase using a designed ABA biosensor. Moreover, we developed a related PACS approach, leveraging the same genetic tools, in conjunction with in vitro mutagenesis to rapidly evaluate the activity of a large library of highly mutated esterases. In addition to obtaining a mutant with 48-fold improvement in kcat for the probe substrate, we showed that the diverse enzymes obtained from the selection process can be further screened to identify promiscuous activity. Many of the variants identified contained mutations outside the esterase active site and would have been missed by targeted mutagenesis approaches, and the large number of mutations identified would have likely required multiple rounds of conventional directed evolution to discover.
We also showed how a broader application of PACE for biocatalyst evolution will require deepening our understanding of the “selection window”. Despite the utility of PACE for evolving proteins directly tied to gIII transcription or translation, true continuous evolution of enzymatic activity on small molecules using this platform remains challenging. This and another recently published study36 demonstrate that (semi)continuous evolution platforms provide a powerful way to develop biocatalysts. Recent improvements to the PACE methodology promise to not only increase throughput but also further our understanding of the fundamental aspects of continuous evolution. When compounding these advances with the improvements to biosensor development and complementary methods of substrate sequestration or substrate “sinks”,73,74 the ability to apply new biosensors in directed evolution campaigns is likely to play a significant role in the future of biocatalysis.
Acknowledgments
This study was supported by a grant from the National Institute of General Medical Sciences (R01 GM115665) to J.C.L., by a CAREER Award from the National Science Foundation (NSF, 1749364) to B.C.D., and by Camille and Henry Dreyfus Foundation Teacher Scholar Awards to both J.C.L. and B.C.D.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00811.
Experimental details on ester synthesis, biosensor engineering, PACE/PACS development, and substrate scope analysis (PDF)
The authors declare the following competing financial interest(s): B.C.D. has filed a patent on the proximity-dependent split RNAP technology and is a founder and holds equity in Tornado Bio, Inc., a company developing RNA-programmable therapies.
Supplementary Material
References
- Hughes G.; Lewis J. C. Introduction: Biocatalysis in Industry. Chem. Rev. 2018, 118 (1), 1–3. 10.1021/acs.chemrev.7b00741. [DOI] [PubMed] [Google Scholar]
- Burton S. G.; Cowan D. A.; Woodley J. M. The Search for the Ideal Biocatalyst. Nat. Biotechnol. 2002, 20 (1), 37–45. 10.1038/nbt0102-37. [DOI] [PubMed] [Google Scholar]
- Madhavan A.; Arun K. B.; Binod P.; Sirohi R.; Tarafdar A.; Reshmy R.; Awasthi M. K.; Sindhu R. Design of Novel Enzyme Biocatalysts for Industrial Bioprocess: Harnessing the Power of Protein Engineering, High Throughput Screening and Synthetic Biology. Bioresour. Technol. 2021, 325, 124617. 10.1016/j.biortech.2020.124617. [DOI] [PubMed] [Google Scholar]
- Andorfer M. C.; Lewis J. C. Understanding and Improving the Activity of Flavin-Dependent Halogenases via Random and Targeted Mutagenesis. Annu. Rev. Biochem. 2018, 87 (87), 159–185. 10.1146/annurev-biochem-062917-012042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F. H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem., Int. Ed. 2018, 57 (16), 4143–4148. 10.1002/anie.201708408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. D.; Labthavikul S. T.; Otey C. R.; Arnold F. H. Protein Stability Promotes Evolvability. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (15), 5869–5874. 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommarius A. S.; Paye M. F. Stabilizing Biocatalysts. Chem. Soc. Rev. 2013, 42 (15), 6534–6565. 10.1039/c3cs60137d. [DOI] [PubMed] [Google Scholar]
- Polizzi K. M.; Bommarius A. S.; Broering J. M.; Chaparro-Riggers J. F. Stability of Biocatalysts. Curr. Opin. Chem. Biol. 2007, 11 (2), 220–225. 10.1016/j.cbpa.2007.01.685. [DOI] [PubMed] [Google Scholar]
- Qu G.; Li A.; Acevedo-Rocha C. G.; Sun Z.; Reetz M. T. The Crucial Role of Methodology Development in Directed Evolution of Selective Enzymes. Angew. Chem., Int. Ed. 2020, 59 (32), 13204–13231. 10.1002/anie.201901491. [DOI] [PubMed] [Google Scholar]
- Truppo M. D. Biocatalysis in the Pharmaceutical Industry: The Need for Speed. ACS Med. Chem. Lett. 2017, 8 (5), 476–480. 10.1021/acsmedchemlett.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard D. J.; Woodley J. M. Biocatalysis for Pharmaceutical Intermediates: the Future is Now. Trends Biotechnol. 2007, 25 (2), 66–73. 10.1016/j.tibtech.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Fisher B. F.; Snodgrass H. M.; Jones K. A.; Andorfer M. C.; Lewis J. C. Site-Selective C-H Halogenation Using Flavin-Dependent Halogenases Identified via Family-Wide Activity Profiling. ACS Cent. Sci. 2019, 5 (11), 1844–1856. 10.1021/acscentsci.9b00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro-Riggers J. F.; Polizzi K. M.; Bommarius A. S. Better library Design: Data-Driven Protein Engineering. Biotechnol. J. 2007, 2 (2), 180–191. 10.1002/biot.200600170. [DOI] [PubMed] [Google Scholar]
- Lorenz P.; Eck J. Metagenomics and Industrial Applications. Nat. Rev. Microbiol. 2005, 3 (6), 510–516. 10.1038/nrmicro1161. [DOI] [PubMed] [Google Scholar]
- Affaticati P. E.; Dai S. B.; Payongsri P.; Hailes H. C.; Tittmann K.; Dalby P. A. Structural Analysis of an Evolved Transketolase Reveals Divergent Binding Modes. Sci. Rep. 2016, 6, 35716. 10.1038/srep35716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savile C. K.; Janey J. M.; Mundorff E. C.; Moore J. C.; Tam S.; Jarvis W. R.; Colbeck J. C.; Krebber A.; Fleitz F. J.; Brands J.; et al. Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science 2010, 329 (5989), 305–309. 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- Payne J. T.; Poor C. B.; Lewis J. C. Directed Evolution of RebH for Site-Selective Halogenation of Large Biologically Active Molecules. Angew. Chem., Int. Ed. 2015, 54 (14), 4226–4230. 10.1002/anie.201411901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran A. H.; Liu D. R. In vivo Continuous Directed Evolution. Curr. Opin. Chem. Biol. 2015, 24, 1–10. 10.1016/j.cbpa.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Oelsnitz S.; Ellington A. Continuous Directed Evolution for Strain and Protein Engineering. Curr. Opin. Biotechnol. 2018, 53, 158–163. 10.1016/j.copbio.2017.12.020. [DOI] [PubMed] [Google Scholar]
- Rix G.; Liu C. C. Systems for in vivo Hypermutation: a Quest for Scale and Depth in Directed Evolution. Curr. Opin. Chem. Biol. 2021, 64, 20–26. 10.1016/j.cbpa.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt K. M.; Carlson J. C.; Liu D. R. A System for the Continuous Directed Evolution of Biomolecules. Nature 2011, 472 (7344), 499–503. 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. M.; Wang T. A.; Liu D. R. Phage-Assisted Continuous and Non-Continuous Evolution. Nat. Protoc. 2020, 15 (12), 4101–4127. 10.1038/s41596-020-00410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B. C.; Packer M. S.; Badran A. H.; Liu D. R. A System for the Continuous Directed Evolution of Proteases Rapidly Reveals Drug-Resistance Mutations. Nat. Commun. 2014, 5, 5352. 10.1038/ncomms6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer M. S.; Rees H. A.; Liu D. R. Phage-Assisted Continuous Evolution of Proteases with Altered Substrate Specificity. Nat. Commun. 2017, 8, 956. 10.1038/s41467-017-01055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard B. P.; Badran A. H.; Zuris J. A.; Guilinger J. P.; Davis K. M.; Chen L. W.; Tsai S. Q.; Sander J. D.; Joung J. K.; Liu D. R. Continuous Directed Evolution of DNA-Binding Proteins to Improve TALEN Specificity. Nat. Methods 2015, 12 (10), 939–942. 10.1038/nmeth.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B. C.; Leconte A. M.; Allen B.; Esvelt K. M.; Liu D. R. Experimental Interrogation of the Path Dependence and Stochasticity of Protein Evolution Using Phage-Assisted Continuous Evolution. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (22), 9007–9012. 10.1073/pnas.1220670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J. C.; Badran A. H.; Guggiana-Nilo D. A.; Liu D. R. Negative Selection and Stringency Modulation in Phage-Assisted Continuous Evolution. Nat. Chem. Biol. 2014, 10 (3), 216–222. 10.1038/nchembio.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J. Y.; Zinkus-Boltz J.; Dickinson B. C. Evolution of a Split RNA Polymerase as a Versatile Biosensor Platform. Nat. Chem. Biol. 2017, 13 (4), 432–438. 10.1038/nchembio.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J. Y.; Kentala K.; Dickinson B. C. Multidimensional Control of Cas9 by Evolved RNA Polymerase-Based Biosensors. ACS Chem. Biol. 2018, 13 (2), 431–437. 10.1021/acschembio.7b00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J.; Disare M.; Dickinson B. C. Evolution of C-Terminal Modification Tolerance in Full-Length and Split T7 RNA Polymerase Biosensors. ChemBioChem 2019, 20 (12), 1547–1553. 10.1002/cbic.201900325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M. F.; Zhao K. V. T.; Eton E.; Lapinaite A.; Newby G. A.; Thuronyi B. W.; Wilson C.; Koblan L. W.; Zeng J.; Bauer D. E.; Doudna J. A.; Liu D. R. Phage-Assisted Evolution of an Adenine Base Editor with Improved Cas Domain Compatibility and Activity. Nat. Biotechnol. 2020, 38 (7), 883–891. 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran A. H.; Guzov V. M.; Huai Q.; Kemp M. M.; Vishwanath P.; Kain W.; Nance A. M.; Evdokimov A.; Moshiri F.; Turner K. H.; Wang P.; Malvar T.; Liu D. R. Continuous Evolution of Bacillus thuringiensis Toxins Overcomes Insect Resistance. Nature 2016, 533 (7601), 58–63. 10.1038/nature17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson D. I.; Fan C. G.; Guo L. T.; Miller C.; Soll D.; Liu D. R. Continuous Directed Evolution of Aminoacyl-tRNA Synthetases. Nat. Chem. Biol. 2017, 13 (12), 1253–1260. 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth T. B.; Woolston B. M.; Stephanopoulos G.; Liu D. R. Phage-Assisted Evolution of Bacillus methanolicus Methanol Dehydrogenase 2. ACS Synth. Biol. 2019, 8 (4), 796–806. 10.1021/acssynbio.8b00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M. S.; Podracky C. J.; Liu D. V. R. The Developing Toolkit of Continuous Directed Evolution. Nat. Chem. Biol. 2020, 16 (6), 610–619. 10.1038/s41589-020-0532-y. [DOI] [PubMed] [Google Scholar]
- Rix G.; Watkins-Dulaney E. J.; Almhjell P. J.; Boville C. E.; Arnold F. H.; Liu C. C. Scalable Continuous Evolution for the Generation of Diverse Enzyme Variants Encompassing Promiscuous Activities. Nat. Commun. 2020, 11 (1), 5644. 10.1038/s41467-020-19539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z. W.; Wong B. G.; Ravikumar A.; Arzumanyan G. A.; Khalil A. S.; Liu C. C. Automated Continuous Evolution of Proteins in vivo. ACS Synth. Biol. 2020, 9 (6), 1270–1276. 10.1021/acssynbio.0c00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman C. M.; Papa L. J.; Hendel S. J.; Moore C. L.; Suen P. H.; Weickhardt A. F.; Doan N. D.; Kumar C. M.; Uil T. G.; Butty V. L.; et al. An Adaptable Platform for Directed Evolution in Human Cells. J. Am. Chem. Soc. 2018, 140 (51), 18093–18103. 10.1021/jacs.8b10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J. G.; Olsen R. H. J.; Lansu K.; Patel M.; White K.; Cockrell A. S.; Singh D.; Strachan R. T.; Wacker D.; Roth B. L. VEGAS as a Platform for Facile Directed Evolution in Mammalian Cells. Cell 2019, 178 (3), 748–761. 10.1016/j.cell.2019.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A.; Zinkus-Boltz J.; Dickinson B. C. Recent Advances in Developing and Applying Biosensors for Synthetic Biology. Nano Futures 2019, 3 (4), 042002. 10.1088/2399-1984/ab4b78. [DOI] [Google Scholar]
- Zinkus-Boltz J.; DeValk C.; Dickinson B. C. A Phage-Assisted Continuous Selection Approach for Deep Mutational Scanning of Protein-Protein Interactions. ACS Chem. Biol. 2019, 14 (12), 2757–2767. 10.1021/acschembio.9b00669. [DOI] [PubMed] [Google Scholar]
- van Rossum T.; Kengen S. W. M.; van der Oost J. Reporter-based Screening and Selection of Enzymes. FEBS J. 2013, 280 (13), 2979–2996. 10.1111/febs.12281. [DOI] [PubMed] [Google Scholar]
- Fernandez-Lopez R.; Ruiz R.; de la Cruz F.; Moncalian G. Transcription Factor-Based Biosensors Enlightened by the Analyte. Front. Microbiol. 2015, 6, 648. 10.3389/fmicb.2015.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr R.; Frunzke J. Transcription Factor-Based Biosensors in Biotechnology: Current State and Future Prospects. Appl. Microbiol. Biotechnol. 2016, 100 (1), 79–90. 10.1007/s00253-015-7090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. K.; Guzman C. D.; Taylor N. D.; Raman S.; Anderson K.; Church G. M. Synthetic Biosensors for Precise Gene Control and Real-Time Monitoring of Metabolites. Nucleic Acids Res. 2015, 43 (15), 7648–7660. 10.1093/nar/gkv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. K.; Church G. M. Genetically Encoded Sensors Enable Real-Time Observation of Metabolite Production. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (9), 2388–2393. 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T. C.; Liu W. L.; Brinck M. S.; Davis J. E.; Shek J.; Bower G.; Einav T.; Insigne K. D.; Phillips R.; Kosuri S.; et al. Multiplexed Characterization of Rationally Designed Promoter Architectures Deconstructs Combinatorial Logic for IPTG-Inducible Systems. Nat. Commun. 2021, 12 (1), 325. 10.1038/s41467-020-20094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. R.; Westerhoff H. V.; Michelsen O. The Use of Lac-Type Promoters in Control Analysis. Eur. J. Biochem. 1993, 211 (1–2), 181–191. 10.1111/j.1432-1033.1993.tb19885.x. [DOI] [PubMed] [Google Scholar]
- Stanton B. Z.; Chory E. J.; Crabtree G. R. Chemically Induced Proximity in Biology and Medicine. Science 2018, 359 (6380), eaao5902. 10.1126/science.aao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fegan A.; White B.; Carlson J. C. T.; Wagner C. R. Chemically Controlled Protein Assembly: Techniques and Applications. Chem. Rev. 2010, 110 (6), 3315–3336. 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- Voss S.; Klewer L.; Wu Y. W. Chemically Induced Dimerization: Reversible and Spatiotemporal Control of Protein Function in Cells. Curr. Opin. Chem. Biol. 2015, 28, 194–201. 10.1016/j.cbpa.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Chen Z. L.; Katzenellenbogen B. S.; Katzenellenbogen J. A.; Zhao H. M. Directed Evolution of Human Estrogen Receptor Variants with Significantly Enhanced Androgen Specificity and Affinity. J. Biol. Chem. 2004, 279 (32), 33855–33864. 10.1074/jbc.M402118200. [DOI] [PubMed] [Google Scholar]
- Chen Z. L.; Zhao H. M. Rapid Creation of a Novel Protein Function by in vitro Coevolution. J. Mol. Biol. 2005, 348 (5), 1273–1282. 10.1016/j.jmb.2005.02.070. [DOI] [PubMed] [Google Scholar]
- Chockalingam K.; Chen Z. L.; Katzenellenbogen J. A.; Zhao H. M. Directed Evolution of Specific Receptor-Ligand Pairs for Use in the Creation of Gene Switches. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (16), 5691–5696. 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F. S.; Ho W. Q.; Crabtree G. R. Engineering the ABA Plant Stress Pathway for Regulation of Induced Proximity. Sci. Signaling 2011, 4 (164), rs2. 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornscheuer U. T. Microbial Carboxyl Esterases: Classification, Properties and Application in Biocatalysis. FEMS Microbiol. Rev. 2002, 26 (1), 73–81. 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- Romano D.; Bonomi F.; de Mattos M. C.; Fonseca T. D.; de Oliveira M. D. F.; Molinari F. Esterases as Stereoselective Biocatalysts. Biotechnol. Adv. 2015, 33 (5), 547–565. 10.1016/j.biotechadv.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Schmidt M.; Henke E.; Heinze B.; Kourist R.; Hidalgo A.; Bornscheuer U. T. A Versatile Esterase from Bacillus subtilis: Cloning, Expression, Characterization, and its Application in Biocatalysis. Biotechnol. J. 2007, 2 (2), 249–253. 10.1002/biot.200600174. [DOI] [PubMed] [Google Scholar]
- Fotakopoulou I.; Barbayianni E.; Constantinou-Kokotou V.; Bornscheuer U. T.; Kokotos G. Enzymatic Removal of Carboxyl Protecting Groups. III. Fast Removal of Allyl and Chloroethyl Esters by Bacillus subtilis Esterase (BS2). J. Org. Chem. 2007, 72 (3), 782–786. 10.1021/jo061871f. [DOI] [PubMed] [Google Scholar]
- Heinze B.; Kourist R.; Fransson L.; Hult K.; Bornscheuer U. T. Highly Enantioselective Kinetic Resolution of Two Tertiary Alcohols Using Mutants of an Esterase from Bacillus subtilis. Protein Eng., Des. Sel. 2007, 20 (3), 125–131. 10.1093/protein/gzm003. [DOI] [PubMed] [Google Scholar]
- Drummond D. A.; Iverson B. L.; Georgiou G.; Arnold F. H. Why High-Error-Rate Random Mutagenesis Libraries are Enriched in Functional and Improved Proteins. J. Mol. Biol. 2005, 350 (4), 806–816. 10.1016/j.jmb.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Tian L.; Yang Y. L.; Wysocki L. M.; Arnold A. C.; Hu A.; Ravichandran B.; Sternson S. M.; Looger L. L.; Lavis L. D. Selective Esterase-Ester Pair for Targeting Small Molecules with Cellular Specificity. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (13), 4756–4761. 10.1073/pnas.1111943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A.; Kentala K.; Beck M. W.; An W. W.; Lippert A. R.; Lewis J. C.; Dickinson B. C. Development of a Split Esterase for Protein-Protein Interaction-Dependent Small-Molecule Activation. ACS Cent. Sci. 2019, 5 (11), 1768–1776. 10.1021/acscentsci.9b00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C. J.; Gong X.; Schafer W.; Pratt E. C.; Brkovic T.; Pirzada Z.; Cuff J. F.; Kosjek B. MISER Chromatography (Multiple Injections in a Single Experimental Run): the Chromatogram is the Graph. Tetrahedron: Asymmetry 2010, 21 (13–14), 1674–1681. 10.1016/j.tetasy.2010.05.029. [DOI] [Google Scholar]
- Liu Y.; Wang M. Design, Optimization and Application of Small Molecule Biosensor in Metabolic Engineering. Front. Microbiol. 2017, 8, 2012. 10.3389/fmicb.2017.02012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Jensen M. K.; Keasling J. D. Development of Biosensors and Their Application in Metabolic Engineering. Curr. Opin. Chem. Biol. 2015, 28, 1–8. 10.1016/j.cbpa.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Liu D.; Evans T.; Zhang F. Z. Applications and Advances of Metabolite Biosensors for Metabolic Engineering. Metab. Eng. 2015, 31, 35–43. 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Kardashliev T.; Pitzler C.; Shehzad A.; Lue H. Q.; Bernhagen J.; Zhu L. L.; Schwaneberg U. A Competitive Flow Cytometry Screening System for Directed Evolution of Therapeutic Enzyme. ACS Synth. Biol. 2015, 4 (7), 768–775. 10.1021/sb500343g. [DOI] [PubMed] [Google Scholar]
- Kwon K. K.; Lee D. H.; Kim S. J.; Choi S. L.; Rha E.; Yeom S. J.; Subhadra B.; Lee J.; Jeong K. J.; Lee S. G. Evolution of Enzymes with New Specificity by High-Throughput Screening Using DmpR-Based Genetic Circuits and Multiple Flow Cytometry Rounds. Sci. Rep. 2018, 8, 2659. 10.1038/s41598-018-20943-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. K.; Ju S. B.; Lee H. W.; Lee J. Y.; Oh S. H.; Kwon K. K.; Sung B. H.; Lee S. G.; Yeom S. J. Biosensor-Based Directed Evolution of Methanol Dehydrogenase from Lysinibacillus xylanilyticus. Int. J. Mol. Sci. 2021, 22 (3), 1471. 10.3390/ijms22031471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. C.; Mantovani S. M.; Fu Y.; Snow C. D.; Komor R. S.; Wong C. H.; Arnold F. H. Combinatorial Alanine Substitution Enables Rapid Optimization of Cytochrome P450(BM3) for Selective Hydroxylation of Large Substrates. ChemBioChem 2010, 11 (18), 2502–2505. 10.1002/cbic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran A. H.; Liu D. R. Development of Potent in vivo Mutagenesis Plasmids with Broad Mutational Spectra. Nat. Commun. 2015, 6, 8425. 10.1038/ncomms9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolston B. M.; Roth T.; Kohale I.; Liu D. R.; Stephanopoulos G. Development of a Formaldehyde Biosensor with Application to Synthetic Methylotrophy. Biotechnol. Bioeng. 2018, 115 (1), 206–215. 10.1002/bit.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A.; Ho J. M.; Parks S. E.; Bennett M. R. Macrolide Biosensor Optimization through Cellular Substrate Sequestration. ACS Synth. Biol. 2021, 10 (2), 258–264. 10.1021/acssynbio.0c00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.