Abstract
t(12;21) is the most frequent translocation found in pediatric B-cell acute lymphoblastic leukemias. This translocation fuses a putative repressor domain from the TEL DNA-binding protein to nearly all of the AML-1B transcription factor. Here, we demonstrate that fusion of the TEL pointed domain to the GAL4 DNA-binding domain resulted in sequence-specific transcriptional repression, indicating that the pointed domain is a portable repression motif. The TEL pointed domain functioned equally well when the GAL4 DNA-binding sites were moved 600 bp from the promoter, suggesting an active mechanism of repression. This lead us to demonstrate that wild-type TEL and the t(12;21) fusion protein bind the mSin3A corepressor. In the fusion protein, both TEL and AML-1B contribute mSin3 interaction domains. Deletion mutagenesis indicated that both the TEL and AML-1B mSin3-binding domains contribute to repression by the fusion protein. While both TEL and AML-1B associate with mSin3A, TEL/AML-1B appears to bind this corepressor much more stably than either wild-type protein, suggesting a mode of action for the t(12;21) fusion protein.
AML1 is one of the most frequently mutated genes in human leukemias (11, 40). It is targeted directly by the t(8;21), t(16;21), and t(3;21) in various forms of acute myelogenous leukemia (AML), as well as by the t(12;21) in B-cell acute lymphoblastic leukemia (ALL) (18, 20, 47, 50, 55). AML-1 is also the indirect target of the inv(16), which alters core binding factor β (CBFβ), the heterodimerizing partner of AML-1 (7, 39, 67). Each of these translocations results in the formation of a dominant negative fusion protein that interferes with normal transcriptional activation by AML-1B (the largest isoform of AML-1 [45]).
The product of the t(12;21)(p13;q22), TEL/AML-1B, is the most frequent translocation found in pediatric B-cell ALL (55, 61). This fusion protein contains the first 336 amino acids (aa) of an ets factor termed TEL (translocation, ets, leukemia; also known as ETV6) linked to residues 21 to 480 of AML-1B (20, 33, 54). The translocation deletes the DNA-binding domain of TEL but retains the AML-1 runt homology domain (RHD; the DNA-binding motif). Like the other RHD-containing translocation proteins, TEL/AML-1B interferes with AML-1B-dependent transcription (17, 27, 66, 70). TEL/AML-1B also inhibits basal transcription from a promoter construct containing the T-cell receptor β (TCRβ) enhancer (27). A second determinant in the majority of cases containing TEL/AML-1B is that the other allele of TEL is deleted (1, 3, 6, 9, 13, 23, 51–53, 63, 65, 71). Thus, TEL is also a putative tumor suppressor, whose loss may cooperate with the t(12;21) to induce leukemia.
The ets transcription factors are characterized by their homologous DNA-binding domain (the ets domain). The founding members of this gene family were identified by virtue of their oncogenic activity in avian retroviruses (58, 69). The oncogenic ets factors appear to transform cells by activating transcription of target genes containing consensus ets factor binding sites (37, 68). Analysis of the transcriptional regulatory functions of the t(12;21) fusion protein led to the suggestion that TEL acts as a transcriptional repressor (27). In addition, wild-type TEL can repress the macrophage colony-stimulating factor receptor promoter (17). Several other ets factors have also been shown to repress transcription. ETS-1 can both activate and repress transcription, depending on the promoter context (19). In addition, the ets-related factor 2 (ERF-2) protein appears to be a constitutive repressor (59). However, the mechanism of repression for these ets factors has not been defined.
TEL was originally identified at the breakpoint of the t(5;12) in patients with chronic myelomonocytic leukemia (21). This translocation fuses the amino terminus of TEL to the tyrosine kinase domain of the platelet-derived growth factor receptor β (12). More recently, the amino terminus of TEL has been found fused to the catalytic domains of the ABL and JAK2 tyrosine kinases in patients with AML and T-cell ALL, respectively (22, 35). TEL is also fused to NTRK3 in congenital fibrosarcomas (31, 56). In each of these cases, the TEL portion of the protein appears to function as a homodimerization motif that is necessary for activation of the kinase catalytic domain.
Although the addition of the N-terminal portion of TEL to AML-1B results in the formation of a transcriptional repressor, the mechanism of this repression has not been determined. Here, we report that the conserved pointed domain of TEL, present in the TEL/AML-1B fusion protein, acts as a portable transcriptional repression domain. Repression by this TEL domain is active, in that it occurs at a distance. Consistent with this result, the pointed domain of TEL is sufficient for association with the mSin3A corepressor. We provide evidence that TEL/AML-1B physically interacts with mSin3A. Moreover, both AML-1B and TEL sequences contribute to this mSin3A association. While both the t(12;21) and t(8;21) fusion proteins interact with mSin3A, TEL/AML-1B-mediated repression is distinct from that of AML-1/ETO. Thus, both TEL and AML-1B contain distinct repression domains that are fused by the t(12;21) to create a constitutive repressor.
MATERIALS AND METHODS
Plasmids.
The reporter plasmids and cytomegalovirus (CMV)-based pCMV5 expression constructs used have been described elsewhere (27, 43, 70). The pCMV-AML-1B(Δ208–237) mutant is described in reference 43. The XbaI fragment from this construct was used to replace those in pCMV-TEL/AML1B and pCMV-TEL/AML(155–792) to make pCMV-TEL/AML(Δ520–549) and pCMV-TEL/AML(155–520/549–792), respectively. The C-terminally deleted pCMV-TEL/AML(1–782) was made by standard PCR methods. The GAL4 DNA-binding domain (aa 1 to 147) cDNA was the generous gift of Ivan Sadowski. The GAL4/TEL(1–333) construct was made by fusing the EcoRI-XbaI fragment from pCMV5-TEL/AML1 in frame to the GAL4 DNA-binding domain sequences and contains the first 336 aa of TEL and 7 aa of AML-1B. The GAL4/TEL(1–127) and the GAL4/TEL(41–127) constructs were made from GAL4/TEL(1–333) by standard PCR methods.
The GAL-thymidine kinase (TK)-luciferase construct was kindly provided by Ming-Jer Tsai (60). The GAL-simian virus 40 (SV40)-chloramphenicol acetyltransferase (CAT) construct was made from the TCRβ-SV40-CAT plasmid by using standard PCR methods to replace the two AML-1B binding sites in the TCRβ enhancer with consensus GAL4 DNA-binding sites (32). The GAL-600-SV40-CAT construct was made by inserting a 1.6-kb ApaI fragment from the 3′ noncoding region of the E2F3 cDNA (64) into the ApaI site between the enhancer and promoter. This plasmid was then cut with SmaI and BglII, blunt ended with Klenow fragment DNA polymerase, and religated. For those mutations made by PCR, isolated fragments were subcloned into pBluescript KS (Stratagene), and the entire fragment was sequenced.
The construct for Flag-tagged TEL was made by adding a C-terminal Asp-Tyr-Asp-Asp-Asp-Asp-Lys peptide by standard PCR methods, using the internal BamHI site and the polylinker Asp718 site for subcloning. The TEL point mutants were created using the PCR and mutagenic oligonucleotides as described previously (28). Plasmid pCMV5-TEL/AML-1B was used as the template for PCR. The sense-strand mutagenic oligonucleotides used were Mut-1 (L94S; 5′ GGC AAA GCT TCC CTG CTG CTG ACC 3′), Mut-2 (F102N, R105G, S106F; 5′ A GAG GAC AAT CGC TAT GGA TTT CC 3′), Mut-3 (I82V, L113S, L117S; 5′ GGT GAT GTG TCC TAT GAA CTC TCT CAG CAT ATT TTG 3′), and Mut-4 (L113D; 5′ GGT GAT GTG GAC TAT GAA CTC CTT) (mutations are underlined). Note that the I82V change in Mut-3 occurred during the PCR. The 5′ and 3′ primers for the second PCR overlapped the unique XhoI and XbaI sites in TEL/AML-1B for subcloning and incorporated an additional E45A change that did not affect TEL dimerization or mSin3A binding. PCR mixtures were incubated for 1 min at 95°C, 2 min at 50°C, and 1 min at 72°C for 25 cycles.
Cell culture and transfection.
C33A cells and Cos-7 cells were maintained in Dulbecco’s modified Eagle medium (DMEM; BioWhittaker) containing 10% fetal calf serum (BioWhittaker), 2 mM L-glutamine (BioWhittaker), and 10 μg of gentamicin (Sigma) per ml. Transfections were performed on 4 × 106 C33A cells per 100-mm-diameter dish by the calcium phosphate coprecipitation method as previously described (24). Coprecipitation was performed with 2 to 4 μg of reporter plasmid, 5 μg of expression plasmid, and 5 μg of the internal control secreted alkaline phosphatase (SEAP) expression plasmid pCMV-SEAP, supplemented with pBluescript KS to 25 μg. For Cos-7 cells, transfections were performed on 2 × 106 cells per 100-mm-diameter dish by the DEAE-dextran method (14), using 3 μg of each of the indicated plasmids. Control transfections were performed with vector only.
NIH 3T3 cells were maintained in DMEM supplemented with 10% calf serum (BioWhittaker), 2 mM l-glutamine, and 10 μg of gentamicin per ml. Transfections were performed on 3.5 × 105 cells per 60-mm-diameter dish with 15 μl of Superfect reagent (Qiagen), 1 μg of reporter plasmid, 2.5 μg of pBluescript KS, 100 ng of repressor plasmid, and 0.5 μg of the internal control plasmid pCMV-SEAP.
Transcription assays.
Cells and supernatants were harvested 44 to 48 h posttransfection. For CAT assays, cells were washed twice with phosphate-buffered saline (PBS; pH 7.4) and lysed in 300 μl of 250 mM Tris (pH 7.4) by sequential freeze-thaw cycles. CAT assays were performed as previously described (24), and percent acetylation was quantitated on a Molecular Dynamics PhosphorImager using ImageQuant software. For luciferase assays, cells were washed twice with PBS and lysed in 200 μl of reporter lysis buffer (Promega). Aliquots of 10 μl were tested for luciferase activity by using the Promega luciferase reagent assay according to the manufacturer’s instructions.
pCMV-SEAP or pRSV-SEAP was included as an internal control for transfection efficiency (27, 70). SEAP activity was quantitated as described elsewhere (8) except that incubations were performed at room temperature. CAT and luciferase activities were then normalized with respect to SEAP activity.
Antibodies.
The anti-RHD AML-1 was generated to a glutathione S-transferase (GST)–AML-1(50–179) fusion protein, which was used to immunize New Zealand White rabbits. This antibody is specific to AML-1 (data not shown) and is available from Calbiochem. The anti-TEL antibodies were directed to a peptide containing the first 29 aa of TEL linked to keyhole limpet hemocyanin, which was used to immunize New Zealand White rabbits. The mSin3A antibody (K-20) was purchased from Santa Cruz Biotechnologies.
Immunoprecipitation.
Labeled or unlabeled cells were lysed 48 h posttransfection in antibody buffer containing either 0.5% NP-40 or 0.1 to 0.5% sodium dodecyl sulfate (SDS)–0.5% sodium deoxycholate–0.5% Triton X-100 in a mixture consisting of 10 mM Tris (pH 7.4), 140 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, and 100 μM sodium orthovanadate as indicated. Cells were labeled 40 h posttransfection with 100 μCi of [35S]methionine and [35S]cysteine (PROMIX; Amersham) for 3 h in methionine- and cysteine-free DMEM (BioWhittaker) that contained 2% dialyzed calf serum (BioWhittaker). Cells were then lysed in antibody buffer. Lysates were sonicated, cleared by centrifugation, and incubated with 100 μl of formalin-fixed Staphylococcus aureus (Pansorbin; Calbiochem) for 30 min to remove nonspecific protein binding. After centrifugation for 5 min at 4°C, the supernatants were collected and incubated for 1 h with primary antibody. A 50% slurry of protein A-Sepharose (15 μl; Pharmacia) was then added for 1 h to collect the immune complexes. The beads were washed three times with antibody buffer, and the immune complexes were eluted by boiling in 1× Laemmli buffer. Samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on SDS–7.5 or 10% polyacrylamide gels prior to immunoblot analysis.
In vitro transcription-translation reactions and glutathione-agarose precipitation assays.
mSin3A was transcribed and translated in vitro in the presence of PROMIX (Amersham), using the Promega T7 coupled reticulocyte lysate system according to the manufacturer’s instructions. The template, pVZ-mSin3A, was generously provided by Robert Eisenman (Fred Hutchinson Cancer Research Center, Seattle, Wash.). GST-TEL fusion proteins were purified with glutathione-Sepharose 4B beads (Pharmacia). After a 4-h binding reaction at 4°C, the beads were washed three times with a high-salt buffer (50 mM HEPES [pH 7.5], 450 mM NaCl, 25% glycerol, 0.5 mM dithiothreitol, 0.1% bovine serum albumin, 1 mM EDTA, 1× Complete protease inhibitor) and washed one time in buffer lacking bovine serum albumin. The amounts of the fusion proteins bound to the beads were determined by SDS-PAGE and staining with Coomassie R250. The amount of mSin3A was determine by autoradiography.
Immunoblot analysis.
Proteins were transferred to nitrocellulose membranes (Hybond C; Amersham), blocked with 5% nonfat dry milk in PBS–0.1% Tween 20, and then probed with primary and secondary antibodies (43) by using an enhanced chemiluminescence analysis system (Amersham).
RESULTS
The pointed domain of TEL is a portable transcriptional repression domain.
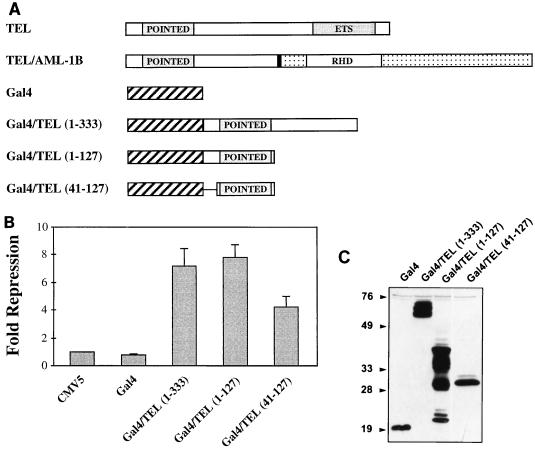
The addition of the N-terminal 336 aa of TEL to AML-1B by the t(12;21) converts this transcriptional regulator into a repressor (27). To determine whether the TEL residues present in the fusion protein could function as a portable repressor domain, we fused the N terminus of TEL/AML-1B to the GAL4 DNA-binding domain (Fig. 1A). GAL4/TEL(1–333) repressed transcription from a plasmid containing four GAL4 binding sites upstream of a minimal TK promoter ∼7-fold (Fig. 1B). Within these TEL sequences lies a motif homologous to one found in the Drosophila pointed protein (the pointed domain). Therefore, we tested whether this domain is sufficient for repression by fusing either the N-terminal 127 aa or the pointed domain alone (residues 47 to 127) to the GAL4 DNA-binding domain (Fig. 1A). The GAL4/TEL(1–127) mutant worked as well as GAL4/TEL(1–333) (Fig. 1B). The isolated pointed domain retained nearly 60% of this activity (Fig. 1B). The chimeric GAL4/TEL proteins were expressed at similar levels (Fig. 1C). These results suggest that the pointed domain is sufficient for transcriptional repression.
FIG. 1.
The pointed domain of TEL is sufficient for transcriptional repression. (A) Illustration of the TEL sequences fused to the GAL4 DNA-binding domain used in these experiments. POINTED, a conserved motif found in a subset of ets factors; RHD, runt homology domain, the DNA-binding domain of AML-1; ETS, ets factor DNA-binding domain. The solid vertical line denotes the t(12;21) fusion point in TEL/AML1B. (B) Transcriptional repression by the pointed domain fused to the GAL4 DNA-binding domain. NIH 3T3 cells were transfected with the indicated plasmids, and luciferase activity was measured 48 h later. Values were normalized to the levels of SEAP expressed from the CMV immediate-early promoter, which was used as an internal control for transfection efficiency. Fold repression represents the promoter activity from cells transfected with expression plasmids compared to those transfected with vector. (C) Immunoblot analysis to confirm that each GAL4/TEL protein is stably expressed. Sizes are indicated in kilodaltons.
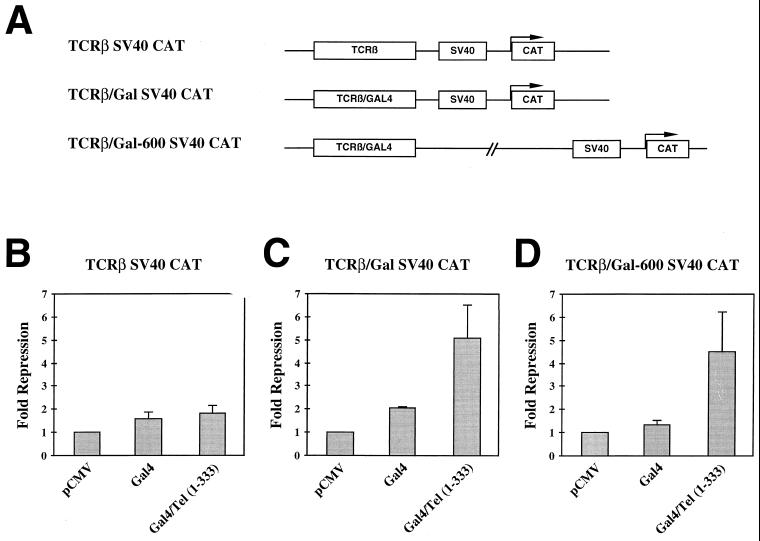
TEL/AML-1B repressed transcription from an SV40 minimal promoter when the promoter was fused to the TCRβ enhancer, which contains binding sites for AML-1B (27). To determine whether the Gal4/TEL(1–333) fusion protein could also inhibit reporter gene expression from the TCRβ enhancer/SV40 promoter, we used a plasmid in which the AML-1B binding sites in the enhancer were replaced with GAL4 binding sites (Fig. 2A). The GAL4 DNA-binding domain alone and GAL4/TEL(1–333) had only small effects on transcription from the original TCRβ plasmid (Fig. 2B). By contrast, GAL4/TEL(1–333) repressed transcription from the SV40 promoter ∼5-fold, whereas GAL4 alone had only a 2-fold effect. This repression was dependent on the presence of the GAL4 DNA-binding sites (Fig. 2C).
FIG. 2.
TEL contains a portable repressor domain. (A) Schematic diagram of the GAL4-SV40-CAT reporter constructs used in these experiments. (B and C) Transcriptional repression by GAL4/TEL. C33A cells were transfected with the TCRβ-SV40 (B) or the TCRβ-GAL-SV40-CAT (C) Reporter plasmid and the expression constructs shown. CAT activity was determined 48 h later. Values were normalized for transfection efficiency by using pCMV-SEAP as an internal control. Fold repression represents the promoter activity from cells transfected with expression plasmids compared to those transfected with vector. (D) GAL4/TEL acts at a distance. C33A cells were transfected with the TCRβ-GAL-600-SV40-CAT plasmid and the expression constructs shown. Fold repression represents the promoter activity from cells transfected with expression plasmids compared to those transfected with the vector.
Given that TEL sequences must be brought to DNA to repress (here via the GAL4 DNA-binding domain), TEL does not appear to function by squelching transcription. Therefore, it appears that TEL is an active repressor. To further test this hypothesis, we asked whether the GAL4/TEL(1–333) fusion protein could act at a distance. A 600-bp DNA fragment was placed between the enhancer and promoter of the GAL-SV40 construct, and this reporter was tested for responsiveness to the GAL4 fusion proteins. The GAL4 DNA-binding domain had no effect on transcription from this reporter (Fig. 2D). However, GAL4/TEL(1–333) still repressed transcription from the SV40 promoter four- to fivefold (Fig. 2D). Thus, the TEL sequences can function from short or long range with equal effectiveness, supporting our hypothesis that TEL-mediated repression is an active process.
The TEL sequences present in TEL/AML-1B stabilize the interaction between AML-1B and mSin3A.
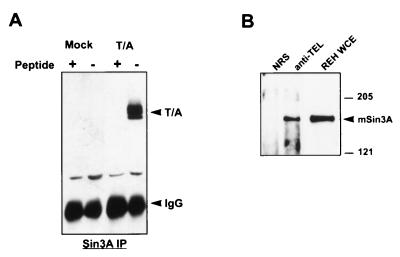
AML-1B physically interacts with mSin3A to repress transcription of the p21WAF1/CIP1 promoter (43). In coimmunoprecipitation assays from cell lysates, sequences adjacent to the RHD are required for associating with mSin3A (43). Because the entire mSin3A interaction domain (SID) is retained in TEL/AML-1B, we tested whether the fusion protein could also associate with the mSin3A corepressor. TEL/AML-1B was transiently expressed in Cos-7 cells, and cell lysates were immunoprecipitated with antibodies that recognize endogenous mSin3A. Coimmunoprecipitating TEL/AML-1B was then detected by immunoblot analysis. TEL/AML-1B specifically associated with mSin3A under high-stringency conditions (Fig. 3A). This association was specific, as addition of the antigenic peptide to the cell lysate effectively blocked coimmunoprecipitation with mSin3A. In addition, we used a t(12;21)-containing cell line, REH, to determine whether the endogenous TEL/AML-1 interacts with endogenous mSin3A. Here, we performed the reciprocal experiment by immunoprecipitating REH cell lysates with anti-TEL or normal rabbit serum and detecting mSin3A by immunoblot analysis (Fig. 3B). mSin3A was found in TEL, but not in control, immune complexes. Thus, TEL/AML-1 and mSin3A associate in a t(12;21)-containing leukemic cell line.
FIG. 3.
TEL/AML-1B coimmunoprecipitates with mSin3A. (A) Overexpressed TEL/AML-1 associates with endogenous mSin3A. Lysates were made from Cos-7 cells that had been transfected with plasmids expressing TEL/AML1B (T/A) or untransfected control cells. These lysates were immunoprecipitated (IP) with anti-mSin3A immunoglobulin G (IgG) in the absence or presence of the mSin3A antigenic peptide. (B) Endogenous TEL/AML-1 is associated with endogenous mSin3A. REH cell lysates were immunoprecipitated with anti-TEL, and associated mSin3A was detected by immunoblot analysis. NRS, normal rabbit serum; WCE, whole-cell extract. Sizes are indicated in kilodaltons.
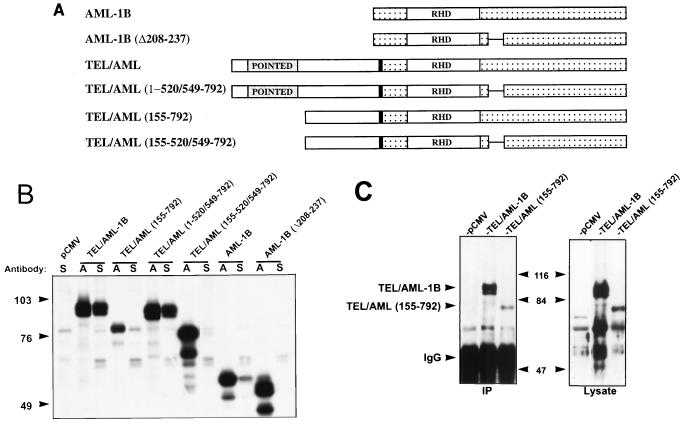
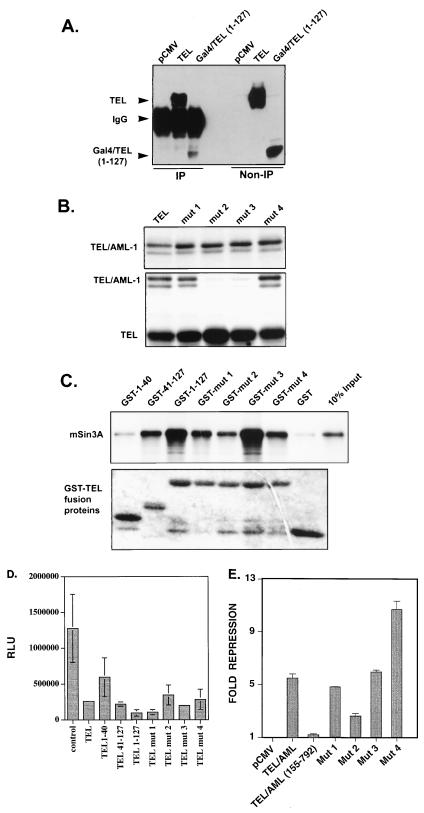
To define the domains of TEL/AML-1B that contact mSin3A, we created a series of TEL/AML-1B deletion mutants (Fig. 4A) and then compared the abilities of the fusion protein and AML-1B to interact with mSin3A. Because AML-1B comigrates with the immunoglobulin G heavy chain, we radiolabeled the transfected Cos-7 cells and tested for mSin3 coimmunoprecipitation (Fig. 4B). Unexpectedly, more TEL/AML-1B was coimmunoprecipitated with anti-mSin3A than AML-1B, suggesting that most of the TEL/AML-1B was bound to mSin3A (Fig. 4B). As previously observed, deletion of residues 208 to 237 in AML-1B was sufficient to abolish the mSin3A interaction (Fig. 4B) (43). Unexpectedly, deletion of this same domain in TEL/AML-1B [TEL/AML-1B(1–520/549–792)] did not affect the mSin3A association, suggesting that TEL sequences contributed to this association (Fig. 4B). Removal of the N terminus of TEL/AML-1B reduced, but did not ablate, mSin3A binding (Fig. 4B, TEL/AML 155–792). Because an endogenous mSin3A-associated protein comigrated with this mutant, we confirmed this association by immunoprecipitation followed by immunoblot analysis (Fig. 4C; note that the nonspecific bands in the lysate panel may be endogenous AML-1). Deletion of both the N terminus of TEL and the AML-1B mSin3A-binding domain [TEL/AML(155–520/549–792)] significantly reduced the amount of TEL/AML-1B that associated with mSin3A (Fig. 4B). However, this double mutant did retain some ability to associate with mSin3A compared to the AML-1B(Δ208–237) mutant (Fig. 4B). Thus, the TEL sequences C terminal to residue 155 may contribute a weak mSin3A-binding domain to the fusion protein, but the N-terminal 155 aa and the AML-1B SID are required for a stronger association with mSin3A.
FIG. 4.
TEL and AML-1B sequences contribute to mSin3A binding by TEL/AML-1B. (A) Schematic diagram of AML-1B, TEL/AML-1B, and deletion mutants of these proteins. (B) Mapping the SID in TEL/AML-1B. Cells were transfected with plasmids expressing the proteins depicted in panel A and radiolabeled with [35S]methionine. Cell lysates were prepared in PBS containing 0.5% Triton X-100, 0.5% sodium deoxycholate, and 0.5% SDS and immunoprecipitated with AML-1 (RHD) (lanes A) and mSin3A (lanes S) antibodies. The immune complexes were separated on an SDS–10% polyacrylamide gel. Several lanes in which the immunoprecipitations were done at low stringency are not shown, but each lane is from the same gel. (C) Immunoblot of unlabeled cell lysates expressing the indicated proteins immunoprecipitated (IP) with mSin3A antibodies. Immunoblots were probed with anti-RHD antibodies. Note that the bands in each of the three lanes in the lysate panel may be endogenous AML-1. Sizes are indicated in kilodaltons. IgG, immunoglobulin G.
Because TEL/AML-1B(1–520/549–792) associated with mSin3A better than wild-type AML-1B (Fig. 4B), we examined whether TEL sequences were sufficient for mSin3A binding. Under the same high-stringency conditions that were used to define the TEL/AML-1B and AML-1B association with mSin3A, no TEL coimmunoprecipited with mSin3A (data not shown). However, under lower-stringency conditions (0.5% NP-40), TEL coimmunoprecipitated with mSin3A (Fig. 5A). To identify the SID(s), we used the available GAL4/TEL chimeric proteins. GAL4/TEL(1–127) associated with mSin3A, albeit at lower levels (Fig. 5A). This finding is consistent with its ability to repress transcription (Fig. 2). Unfortunately, we could not determine whether residues 155 through 333 were sufficient for binding mSin3A [as was indicated by the association with the TEL/AML-1(155–520/549–792) double mutant (Fig. 4B)] because a GAL4/TEL(155–333) protein was not stably expressed (data not shown). Nonetheless, TEL did associate with mSin3A, and residues 1 to 127 contain an SID.
FIG. 5.
The N terminus of TEL mediates an interaction with mSin3A. (A) TEL associates with endogenous mSin3A. TEL and GAL4-TEL(1–127) were transfected into Cos-7 cells, and cell lysates were immunoprecipitated (IP) with anti-mSin3A serum. Coprecipitating TEL and GAL4-TEL(1–127) were detected by immunoblot using anti-TEL antibodies. IgG, immunoglobulin G. (B) Identification of TEL dimerization mutants. Cos cells were transfected with the indicated pCMV5 expression plasmids and pulse-labeled with [35S]methionine and [35S]cysteine for 3 h. Cell lysates were split equally for purification and immunoprecipitation. In the upper panel, TEL/AML-1 fusion proteins were immunoprecipitated with anti-RHD antibodies to confirm expression. In lower panel, lysate proteins bound to Flag-tagged TEL were purified by using anti-Flag antibody-coupled Sepharose beads and separated by SDS-PAGE on a 10% gel. (C) In vitro association of TEL and mSin3A. The indicated GST-TEL fusion proteins were used to pull down in vitro-synthesized mSin3A. (D) Definition of TEL repression domains. The indicated GAL4/TEL fusion proteins were tested for the ability to repress a GAL4-TK-reporter construct. RLU, relative light units. (E) TEL/AML-1-mediated repression is independent of dimerization. The indicated plasmids were assayed in C33A cells by using a TCRβ enhancer linked to the SV40 promoter and CAT as a reporter. SEAP activity expressed from the CMV immediate-early promoter was used to correct for transfection efficiency. Mut, mutant.
The N-terminal 127-aa region of TEL contains the pointed domain, which mediates TEL and TEL/AML-1 homodimerization. To differentiate whether TEL represses transcription through mSin3A binding or homodimerization, we performed PCR-based mutagenesis of the pointed domain to identify mutants that would no longer homodimerize. To test dimerization through the pointed domain, we took advantage of the difference in size between TEL and TEL/AML-1B. These proteins were coexpressed, and metabolically labeled TEL/AML-1B was detected in TEL immune complexes. We were able to identify two TEL mutants that were impaired in the ability to dimerize with TEL/AML-1B (Fig. 5B).
Next, we tested these mutants for the ability to bind to mSin3A. We created GST-TEL fusion proteins containing the first 127 aa of wild-type TEL or the pointed domain mutants. In addition, we subdivided the N-terminal domain to determine if the pointed domain is sufficient for mSin3A binding. The mSin3A corepressor was synthesized in vitro, and the GST-TEL fusion proteins were used to purify mSin3A from reticulocyte lysates. Compared to native GST, the N-terminal 127 aa bound mSin3A, consistent with the coimmunoprecipitation results (Fig. 5C). In addition, the pointed domain, but not residues 1 to 40, was sufficient to mediate a mSin3A interaction (Fig. 5C). These results were confirmed in coimmunoprecipitation assays (data not shown). In addition, the point mutants that failed to dimerize retained the ability to bind mSin3A, although mutant 2 showed a reduced affinity (Fig. 5C). Thus, the pointed domain of TEL interacts with mSin3A in vivo and in vitro.
GAL4-TEL(1–127) and GAL4-TEL(41–127) retained the ability to repress transcription, consistent with their ability to bind mSin3A (Fig. 2 and 5C). Therefore, we subcloned the first 40 aa of TEL and the pointed domain mutations into the GAL4 vector to test these mutants in this assay. Both mutants 2 and 3 retained the ability to repress transcription from the GAL4-TK reporter plasmid, although mutant 2 was consistently a weaker repressor. This repression correlates with these mutants ability to bind mSin3, because mutant 2 was the only mutant that was impaired for mSin3A binding (Fig. 5C and D). Thus, mSin3A binding and not dimerization correlates with TEL’s ability to repress transcription. Although in this assay the first 40 aa of TEL appeared to repress transcription weakly (approximately twofold [Fig. 5D]), these results were not statistically significant and were similar to those observed for GAL4 alone (Fig. 2).
We confirmed the results with the pointed domain mutants by using TEL/AML-1B and a second reporter plasmid. Our previous work had indicated that in C33A cells, TEL/AML-1B-mediated repression is dependent on the N-terminal 155 aa of TEL (27). Therefore, the point mutants were subcloned into TEL/AML-1 and tested for the ability to repress the TCRβ enhancer linked to the SV40 minimal promoter (27). Each mutant repressed transcription in comparison to the N-terminal TEL/AML-1B deletion mutant (Fig. 5E). Once again, mutant 2 was somewhat impaired for repression.
Both TEL and AML-1B sequences can contribute to transcriptional repression.
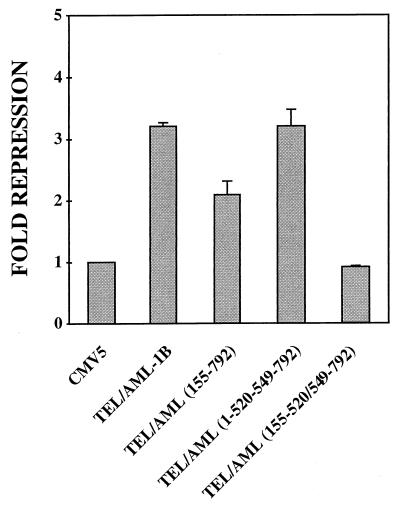
To determine whether mSin3A contributes to TEL/AML-1B-mediated repression, we used TEL/AML-1B mutants lacking the mSin3A-binding domains in transient transfection assays with the p21WAF1/CIP1 promoter (Fig. 6). To assess whether AML-1B sequences contributed to this repression, we used a cell type (NIH 3T3) in which AML-1 represses transcription (43). Deletion of the AML-1B mSin3A-binding domain had little or no effect on the ability of the fusion protein to repress transcription by this promoter [TEL/AML(1–520/549–792) (Fig. 6)]. Deletion of the TEL N-terminal SID [TEL/AML-1(155–792) impaired the ability of the fusion protein to inhibit transcription from the p21WAF1/CIP1 promoter. However, deletion of both the N-terminal TEL SID and the AML-1B mSin3A-binding motif further impaired the fusion protein [TEL/AML(155–520/549–792) (Fig. 6)]. Thus, both TEL and AML-1B sequences contributed to repression of the p21WAF1/CIP1 promoter by TEL/AML-1B.
FIG. 6.
Repression by TEL/AML-1B requires mSin3 binding. Transcriptional repression by TEL/AML-1B and mutants that delete the TEL pointed domain and the AML-1B mSin3A-binding domain (diagram of the mutants is shown in Fig. 4A). Assays were performed as described in the legend to Fig. 1 except that the WWP-luciferase reporter plasmid (16) was used. SEAP activity expressed from the CMV immediate-early promoter was used to correct for transfection efficiency.
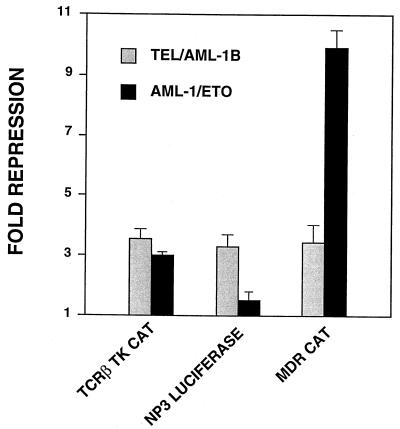
The activity of TEL/AML-1B differs from that of AML-1/ETO.
AML-1/ETO, the product of the t(8;21), also physically interacts with mSin3A and represses transcription (42). However, AML-1/ETO did not affect basal transcription from a TCRβ-SV40-CAT plasmid, whereas TEL/AML-1B did (27). Therefore, we directly compared the abilities of TEL/AML-1B and AML-1/ETO to repress transcription from AML-1B-responsive promoters (Fig. 7). When the TCRβ enhancer was linked to the TK promoter (rather than the SV40 promoter [45]), AML-1/ETO repressed transcription of the TCRβ-TK-CAT construct almost as well as TEL/AML-1B did (∼3-fold). As observed previously, AML-1/ETO had virtually no effect on basal transcription from the NP3 promoter, which was repressed by TEL/AML-1B (Fig. 7). For the multidrug resistance 1 gene (MDR1) promoter, this situation was reversed. While TEL/AML-1B only moderately repressed basal transcription from this promoter (3- to 4-fold), AML-1/ETO repressed its transcription nearly 10-fold, virtually eliminating reporter gene expression (Fig. 7). Considering that TEL/AML-1B and AML-1/ETO are associated with ALL and AML, respectively, our results suggest that differences in activity between TEL/AML-1B and AML-1/ETO could be an important determinant of leukemic phenotype.
FIG. 7.
TEL/AML-1B repression differs from that of AML-1/ETO. C33A cells were transfected with the indicated reporter plasmids and either pCMV5-TEL/AML-1B, pCMV5-AML-1/ETO, or the pCMV5 vector. Data shown are the means of duplicate samples calculated as for Fig. 1.
DISCUSSION
In this study we have begun to dissect the mechanism(s) of transcriptional repression by TEL/AML-1B. By testing GAL4/TEL fusion proteins on two different promoters, we have identified the conserved pointed domain present in TEL and TEL/AML-1B as a repression domain. This motif is sufficient to repress transcription when tethered to a DNA-binding domain. Repression by TEL appears to be active, as repression occurred from within a proximal promoter or a distal enhancer. This repression appears to be mediated by the mSin3 corepressor as both TEL and TEL/AML-1B associated with the mSin3A corepressor. The latter interaction is dependent on both the AML-1B and TEL sequences in TEL/AML1B. Thus, both TEL and AML-1B sequences contribute to repression mediated by the t(12;21) fusion protein.
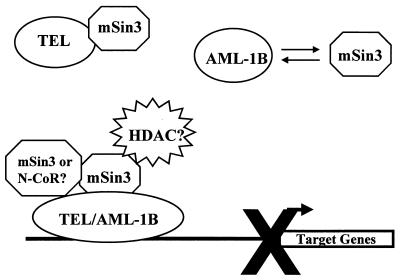
AML-1B acts as a core element for enhancers and promoters by interacting with other DNA-binding proteins and histone acetyltransferases to activate transcription (30). However, AML-1B also associates with the mSin3 and Groucho corepressors to repress transcription (4, 38, 43). The addition of the TEL mSin3-binding domain(s) converts AML-1B to a constitutive repressor (27) (Fig. 6 and 7). Under high-stringency conditions, more TEL/AML-1B associated with mSin3A than did wild-type AML-1B (Fig. 4B) or TEL (which showed no association with mSin3 under these conditions [data not shown]). Based on the weak association of TEL/AML-1B(155–520/549–792) with mSin3A, it is also possible that TEL sequences between residues 155 and 333 bind weakly to mSin3A to contribute to this interaction. Alternatively, this latter association could be indirect. If so, this association could be mediated by a second, yet to be identified corepressor (e.g., N-CoR or SMRT), which then contacts mSin3A. We propose a model in which TEL and AML-1B normally contact the mSin3 corepressors, but where this interaction is regulated (Fig. 8). The addition of TEL sequences to AML-1B in the t(12;21) fusion protein then traps AML-1B in an unregulated repression complex.
FIG. 8.
Model for TEL/AML1B-mediated transcriptional repression. In the schematic diagram depicting TEL and AML-1B interacting with mSin3A, the t(12;21) fuses these two repressors to create a stable, unregulated interaction with the mSin3 corepressor for promoter-specific and active transcriptional repression.
TEL may block the transactivation of positive-acting ets factors through heterodimerization (34) or repress transcription through homodimerization (44). However, our results indicate that TEL can also repress transcription by recruiting the mSin3 proteins through the N-terminal 127 aa and perhaps residues 155 to 333 (Fig. 5 and 6). Several other ets family members have been shown to repress transcription (10, 15, 19, 49, 59, 73). However, the only other constitutive repressor, ERF-2, lacks a pointed domain (59). In addition, ERF-2 does not interact with mSin3 proteins or nuclear hormone corepressors, suggesting that it acts by mechanism distinct from TEL (26). Our identification of the association of TEL and mSin3A provides the first insight into how this putative tumor suppressor might act to counterbalance oncogenic ets factors that activate transcription.
In many cases, the mSin3 corepressors link DNA-binding proteins to histone deacetylases to actively repress transcription (25, 26, 36). To date, we have been unable to coimmunoprecipitate TEL or TEL/AML-1B with histone deacetylases (41). This may be because the putative associations of TEL and TEL/AML-1B with histone deacetylases are indirect and are mediated by mSin3A. If so, the conditions needed to solubilize TEL/AML-1B (0.3% SDS is required for extraction of all of the protein) might disrupt the mSin3A-histone deacetylase interaction. Also, the TEL-mSin3A association was relatively weak, as it was observed only under mild conditions. Thus, TEL’s association with mSin3 may not be stable enough for indirect associations with other components of the corepressor complex to be observed. Alternatively, the mSin3 corepressors can act independently of histone deacetylases (72). Future investigations will be required to elucidate how mSin3 represses transcription in the context of TEL and TEL/AML-1B (Fig. 8).
The t(12;21) is associated with childhood B-cell ALL, whereas the t(8;21) is found in cases of M2 AML (57). Possible explanations for these distinct phenotypes have included the hypothesis that the fusion proteins are expressed from different promoters or that the translocations occur in different hematopoietic progenitor cells. The t(8;21) is present in trilineage progenitor cells but transforms only cells in the granulocytic lineage (46). Therefore, another level of specificity must be intrinsic to AML-1/ETO. Both the t(12;21) fusion protein and the t(8;21) fusion protein retain the AML-1 DNA-binding domain and recruit the mSin3 corepressors to repress transcription. However, these fusion proteins display distinct architectures. TEL is fused to the N terminus of nearly all of AML-1B, whereas ETO replaces the C terminus of AML-1B. We have demonstrated that these differences give the t(12;21) and t(8;21) fusion proteins promoter-specific activities (Fig. 7). We propose this difference as a contributing factor that leads to the distinct leukemic phenotypes generated by these translocations.
The observation that TEL/AML-1B recruits the mSin3A corepressor has implications for our general understanding of the mechanisms of leukemogenesis. The mSin3 proteins repress transcription either by recruiting histone deacetylases or by physically interacting with components of the basal transcriptional machinery (2, 5, 25, 26, 29, 36, 48, 62, 72). The fusion proteins encoded by the t(8;21), t(15;17), t(11;17), and now the t(12;21) all interact with mSin3 proteins and repress transcription, indicating a common mechanism of action (but with different target gene regulation). Given that these are some of the most frequent chromosomal translocations in acute leukemia, pharmaceutical compounds that target the corepressor complex might serve as therapeutic agents for a large number of patients.
ACKNOWLEDGMENTS
We thank Yue Hou for technical assistance, and we thank David Strom, John Nip, and Noel Lenny for plasmids, insightful discussions, and computer assistance.
This work was supported by NIH/NCI grants RO1-CA64140 (S.W.H.), RO1-CA77274 (S.W.H.), and PO1 CA71907-03 (J.R.D.), by American Cancer Society grant JFRA-591 (S.W.H.), by the American Lebanese and Syrian Associated Charities (J.R.D.), by a Center grant from the National Cancer Institute (CA68485), and by the Vanderbilt Cancer Center. R.F. and J.J.W. (F32-CA77167) are recipients of National Research Service Awards from NIH. B.L. is a fellow of the Leukemia Society of America.
REFERENCES
- 1.Agape P, Gerard B, Cave H, Devaux I, Vilmer E, Lecomte M C, Grandchamp B. Analysis of ETV6 and ETV6-AML1 proteins in acute lymphoblastic leukaemia. Br J Haematol. 1997;98:234–239. doi: 10.1046/j.1365-2141.1997.1973014.x. [DOI] [PubMed] [Google Scholar]
- 2.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 3.Andreasson P, Johansson B, Arheden K, Billstrom R, Mitelman F, Hoglund M. Deletions of CDKN1B and ETV6 in acute myeloid leukemia and myelodysplastic syndromes without cytogenetic evidence of 12p abnormalities. Genes Chromosomes Cancer. 1997;19:77–83. doi: 10.1002/(sici)1098-2264(199706)19:2<77::aid-gcc2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 4.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 6.Baccichet A, Sinnett D. Frequent deletion of chromosome 12p12.3 in children with acute lymphoblastic leukaemia. Br J Haematol. 1997;99:107–114. doi: 10.1046/j.1365-2141.1997.3663180.x. [DOI] [PubMed] [Google Scholar]
- 7.Bae S C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. EMBO J. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 9.Bernard O A, Romana S P, Poirel H, Berger R. Molecular cytogenetics of t(12;21) (p13;q22) Leukemia Lymphoma. 1996;23:459–465. doi: 10.3109/10428199609054854. [DOI] [PubMed] [Google Scholar]
- 10.Borras F E, Lloberas J, Maki R A, Celada A. Repression of I-A beta gene expression by the transcription factor PU.1. J Biol Chem. 1995;270:24385–24391. doi: 10.1074/jbc.270.41.24385. [DOI] [PubMed] [Google Scholar]
- 11.Caligiuri M A, Strout M P, Gilliland D G. Molecular biology of acute myeloid leukemia. Semin Oncol. 1997;24:32–44. [PubMed] [Google Scholar]
- 12.Carroll M, Tomasson M H, Barker G F, Golub T R, Gilliland D G. The TEL/platelet-derived growth factor beta receptor (PDGF beta R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF beta R kinase-dependent signaling pathways. Proc Natl Acad Sci USA. 1996;93:14845–14850. doi: 10.1073/pnas.93.25.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cave H, Cacheux V, Raynaud S, Brunie G, Bakkus M, Cochaux P, Preudhomme C, Lai J L, Vilmer E, Grandchamp B. ETV6 is the target of chromosome 12p deletions in t(12;21) childhood acute lymphocytic leukemia. Leukemia. 1997;11:1459–1464. doi: 10.1038/sj.leu.2400798. [DOI] [PubMed] [Google Scholar]
- 14.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–703. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 15.Darby T G, Meissner J D, Ruhlmann A, Mueller W H, Scheibe R J. Functional interference between retinoic acid or steroid hormone receptors and the oncoprotein Fli-1. Oncogene. 1997;15:3067–3082. doi: 10.1038/sj.onc.1201503. [DOI] [PubMed] [Google Scholar]
- 16.el Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Fears S, Gavin M, Zhang D E, Hetherington C, Ben-David Y, Rowley J D, Nucifora G. Functional characterization of ETV6 and ETV6/CBFA2 in the regulation of the MCSFR proximal promoter. Proc Natl Acad Sci USA. 1997;94:1949–1954. doi: 10.1073/pnas.94.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamou T, Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M. The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood. 1998;91:4028–4037. [PubMed] [Google Scholar]
- 19.Goldberg Y, Treier M, Ghysdael J, Bohmann D. Repression of AP-1-stimulated transcription by c-Ets-1. J Biol Chem. 1994;269:16566–16573. [PubMed] [Google Scholar]
- 20.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golub T R, Barker G F, Lovett M, Gilliland D G. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 22.Golub T R, Goga A, Barker G F, Afar D E, McLaughlin J, Bohlander S K, Rowley J D, Witte O N, Gilliland D G. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golub T R, McLean T, Stegmaier K, Carroll M, Tomasson M, Gilliland D G. The TEL gene and human leukemia. Biochim Biophys Acta. 1996;1288:M7–M10. doi: 10.1016/0304-419x(96)00015-7. . (Review.) [DOI] [PubMed] [Google Scholar]
- 24.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 26.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 27.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussell M F, Gilliland D G, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton R M, Ho S N, Pullen J K, Hunt H D, Cai Z, Pease L R. Gene splicing by overlap extension. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 29.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 30.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knezevich S R, Garnett M J, Pysher T J, Beckwith J B, Grundy P E, Sorensen P H. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58:5046–5048. [PubMed] [Google Scholar]
- 32.Kodadek T. How does the GAL4 transcription factor recognize the appropriate DNA binding sites in vivo? Cell Mol Biol Res. 1993;39:355–360. [PubMed] [Google Scholar]
- 33.Kozu T, Miyoshi H, Shimizu K, Maseki N, Kaneko Y, Asou H, Kamada N, Ohki M. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 34.Kwiatkowski B A, Bastian L S, Bauer T R, Jr, Tsai S, Zielinska-Kwiatkowska A G, Hickstein D D. The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. J Biol Chem. 1998;273:17525–17530. doi: 10.1074/jbc.273.28.17525. [DOI] [PubMed] [Google Scholar]
- 35.Lacronique V, Boureux A, Della Valle V, Poirel H, Quang C T, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard O A. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 36.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 37.Lenny N, Westendorf J J, Hiebert S W. Transcriptional regulation during myelopoiesis. Mol Biol Rep. 1997;24:157–168. doi: 10.1023/a:1006859700409. . (Review.) [DOI] [PubMed] [Google Scholar]
- 38.Levanon D, Goldstein R E, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TEL/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 40.Look A T. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 41.Lutterbach, B. 1999. Unpublished data.
- 42.Lutterbach B, Westendorf J J, Linggi B, Patten A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Glass C K, Rosenfeld M G, Seto E, Hiebert S W. ETO, a target of the t(8;21) in acute leukemia, interacts with N-CoR, mSin3, and histone deacetylases. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutterbach, B., J. J. Westendorf, B. Linggi, E. Seto, and S. W. Hiebert. AML-1, a target of multiple chromosomal translocations in acute leukemia, interacts with mSin3 and represses transcription from the p21Waf1/Cip1 promoter. Submitted for publication.
- 44.McLean T W, Ringold S, Neuberg D, Stegmaier K, Tantravahi R, Ritz J, Koeffler H P, Takeuchi S, Janssen J W, Seriu T, Bartram C R, Sallan S E, Gilliland D G, Golub T R. TEL/AML-1 dimerizes and is associated with a favorable outcome in childhood acute lymphoblastic leukemia. Blood. 1996;88:4252–4258. [PubMed] [Google Scholar]
- 45.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyamoto T, Nagafuji K, Akashi K, Harada M, Kyo T, Akashi T, Takenaka K, Mizuno S, Gondo H, Okamura T, Dohy H, Niho Y. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996;87:4789–4796. [PubMed] [Google Scholar]
- 47.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 49.Nozaki M, Onishi Y, Kanno N, Ono Y, Fujimura Y. Molecular cloning of Elk-3, a new member of the Ets family expressed during mouse embryogenesis and analysis of its transcriptional repression activity. DNA Cell Biol. 1996;15:855–862. doi: 10.1089/dna.1996.15.855. [DOI] [PubMed] [Google Scholar]
- 50.Nucifora G, Begy C R, Erickson P, Drabkin H A, Rowley J D. The 3;21 translocation in myelodysplasia results in a fusion transcript between the AML1 gene and the gene for EAP, a highly conserved protein associated with the Epstein-Barr virus small RNA EBER 1. Proc Natl Acad Sci USA. 1993;90:7784–8. doi: 10.1073/pnas.90.16.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poirel H, Lacronique V, Mauchauffe M, Le Coniat M, Raffoux E, Daniel M T, Erickson P, Drabkin H, MacLeod R A, Drexler H G, Ghysdael J, Berger R, Bernard O A. Analysis of TEL proteins in human leukemias. Oncogene. 1998;16:2895–2903. doi: 10.1038/sj.onc.1201817. [DOI] [PubMed] [Google Scholar]
- 52.Raynaud S, Cave H, Baens M, Bastard C, Cacheux V, Grosgeorge J, Guidal-Giroux C, Guo C, Vilmer E, Marynen P, Grandchamp B. The 12;21 translocation involving TEL and deletion of the other TEL allele: two frequently associated alterations found in childhood acute lymphoblastic leukemia. Blood. 1996;87:2891–2899. [PubMed] [Google Scholar]
- 53.Romana S P, Le Coniat M, Poirel H, Marynen P, Bernard O, Berger R. Deletion of the short arm of chromosome 12 is a secondary event in acute lymphoblastic leukemia with t(12;21) Leukemia. 1996;10:167–170. [PubMed] [Google Scholar]
- 54.Romana S P, Mauchauffe M, Le Coniat M, Chumakov I, Le Paslier D, Berger R, Bernard O A. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 55.Romana S P, Poirel H, Leconiat M, Flexor M A, Mauchauffe M, Jonveaux P, Macintyre E A, Berger R, Bernard O A. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 56.Rubin C M, Larson R A, Anastasi J, Winter J N, Thangavelu M, Vardiman J W, Rowley J D, LeBeau M M. t(3;21)(q26;q22) translocation: a recurring chromosomal abnormality in therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood. 1990;76:2594–2598. [PubMed] [Google Scholar]
- 57.Rubnitz J E, Pui C H. Recent advances in the biology and treatment of childhood acute lymphoblastic leukemia. Curr Opin Hematol. 1997;4:233–241. doi: 10.1097/00062752-199704040-00003. [DOI] [PubMed] [Google Scholar]
- 58.Seth A, Papas T S. The c-ets-1 proto-oncogene has oncogenic activity and is positively autoregulated. Oncogene. 1990;5:1761–1767. [PubMed] [Google Scholar]
- 59.Sgouras D N, Athanasiou M A, Beal G J, Jr, Fisher R J, Blair D G, Mavrothalassitis G J. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. Dev Biol. 1985;109:321–335. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shibata H, Nawaz Z, Tsai S Y, O’Malley B W, Tsai M J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 61.Shurtleff S A, Buijs A, Behm F G, Rubnitz J E, Raimondi S C, Hancock M L, Chan G C, Pui C H, Grosveld G, Downing J R. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- 62.Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Seiser C, Loidl P, Luscher B. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- 63.Stegmaier K, Pendse S, Barker G F, Bray-Ward P, Ward D C, Montgomery K T, Krauter K S, Reynolds C, Sklar J, Donnelly M, et al. Frequent loss of heterozygosity at the TEL gene locus in acute lymphoblastic leukemia of childhood. Blood. 1995;86:38–44. [PubMed] [Google Scholar]
- 64.Strom D K, Cleveland J L, Chellappan S, Nip J, Hiebert S W. E2F-1 and E2F-3 are functionally distinct in their ability to promote myeloid cell cycle progression and block granulocyte differentiation. Cell Growth Differ. 1998;9:59–69. [PubMed] [Google Scholar]
- 65.Takeuchi S, Seriu T, Bartram C R, Golub T R, Reiter A, Miyoshi I, Gilliland D G, Koeffler H P. TEL is one of the targets for deletion on 12p in many cases of childhood B-lineage acute lymphoblastic leukemia. Leukemia. 1997;11:1220–1223. doi: 10.1038/sj.leu.2400743. [DOI] [PubMed] [Google Scholar]
- 66.Uchida H, Downing J R, Miyazaki Y, Frank R, Zhang J, Nimer S D. Three distinct domains in TEL-AML1 are required for transcriptional repression of the IL-3 promoter. Oncogene. 1999;18:1015–1022. doi: 10.1038/sj.onc.1202383. [DOI] [PubMed] [Google Scholar]
- 67.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- 69.Watson D K, Ascione R, Papas T S. Molecular analysis of the ets genes and their products. Crit Rev Oncog. 1990;1:409–436. [PubMed] [Google Scholar]
- 70.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-α, inhibits C/EBP-α-dependent transcription, and blocks granulocyte differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wlodarska I, Baens M, Peeters P, Aerssens J, Mecucci C, Brock P, Marynen P, Van den Berghe H. Biallelic alterations of both ETV6 and CDKN1B genes in a t(12;21) childhood acute lymphoblastic leukemia case. Cancer Res. 1996;56:2655–2661. [PubMed] [Google Scholar]
- 72.Wong J, Patterton D, Imhof A, Guschin D, Shi Y B, Wolffe A P. Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J. 1998;17:520–534. doi: 10.1093/emboj/17.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Lichtenheld M G. Non-killer cell-specific transcription factors silence the perforin promoter. J Immunol. 1997;158:1734–1741. [PubMed] [Google Scholar]